- 1Department of Human Genetics, Radboud University Medical Center, Nijmegen, Netherlands
- 2Donders Institute for Brain, Cognition and Behaviour, Radboud University, Nijmegen, Netherlands
- 3Department of Child and Adolescent Psychiatry/Psychology, Erasmus Medical Center, Rotterdam, Netherlands
- 4Department of Psychiatry, Erasmus Medical Center, Rotterdam, Netherlands
- 5Department of Epidemiology, Erasmus Medical Center, Rotterdam, Netherlands
- 6Department of Cognitive Neuroscience, Radboud University Medical Center, Nijmegen, Netherlands
- 7Karakter Child and Adolescent Psychiatry, Radboud University Medical Center, Nijmegen, Netherlands
- 8Department of Psychiatry, Radboud University Medical Center, Nijmegen, Netherlands
Reactive and proactive subtypes of aggression have been recognized to help parse etiological heterogeneity of this complex phenotype. With a heritability of about 50%, genetic factors play a role in the development of aggressive behavior. Imaging studies implicate brain structures related to social behavior in aggression etiology, most notably the amygdala and striatum. This study aimed to gain more insight into the pathways from genetic risk factors for aggression to aggression phenotypes. To this end, we conducted genome-wide gene-based cross-trait meta-analyses of aggression with the volumes of amygdala, nucleus accumbens and caudate nucleus to identify genes influencing both aggression and aggression-related brain volumes. We used data of large-scale genome-wide association studies (GWAS) of: (a) aggressive behavior in children and adolescents (EAGLE, N = 18,988); and (b) Magnetic Resonance Imaging (MRI)-based volume measures of aggression-relevant subcortical brain regions (ENIGMA2, N = 13,171). Second, the identified genes were further investigated in a sample of healthy adults (mean age (SD) = 25.28 (4.62) years; 43% male) who had genome-wide genotyping data and questionnaire data on aggression subtypes available (Brain Imaging Genetics, BIG, N = 501) to study their effect on reactive and proactive subtypes of aggression. Our meta-analysis identified two genes, MECOM and AVPR1A, significantly associated with both aggression risk and nucleus accumbens (MECOM) and amygdala (AVPR1A) brain volume. Subsequent in-depth analysis of these genes in healthy adults (BIG), including sex as an interaction term in the model, revealed no significant subtype-specific gene-wide associations. Using cross-trait meta-analysis of brain measures and psychiatric phenotypes, this study generated new hypotheses about specific links between genes, the brain and behavior. Results indicate that MECOM and AVPR1A may exert an effect on aggression through mechanisms involving nucleus accumbens and amygdala volumes, respectively.
Introduction
Aggression is a common but heterogeneous phenotype often associated with psychiatric disorders that may be harmful to others (Baron and Deborah, 1994; Miczek et al., 2002). The term covers a wide range of human behaviors, varying from verbal aggression and bullying to physical violence. Together, these behaviors have been associated with a large emotional and financial burden on society, while interventions typically still have small effects (McGuire, 2008; Bakker et al., 2017). To address aggression-related negative outcomes more successfully, a better understanding of the genes and neural mechanisms that control this behavior is essential.
Although heritability estimates differ as a function of the population and the type of aggression that is investigated, twin studies show that about 50% of the variance in aggression can be explained by genetic influences, implicating a role for genetics in the development of aggressive behavior (Tuvblad and Baker, 2011; Veroude et al., 2016). Despite this considerable heritability of aggression, the identification of specific genetic risk factors has been difficult. One factor complicating gene-finding is the largely polygenic nature of aggression. While some monogenic disorders leading to aggression phenotypes do exist (the most well-known example perhaps being Brunner syndrome; Brunner et al., 1993), multiple genetic variants, each with a small effect size, contribute to the aggression phenotype in most individuals. Because of the hypothesized polygenic model of multiple common variants with small effects underlying aggression, studies have investigated the role of these common variants by conducting association studies. Next to early candidate genetic approaches relying on a priori biological hypotheses, several genome-wide hypothesis-generating approaches to gene finding have now also been conducted. The main focus of candidate gene studies of aggression has been on genes related to brain neurotransmitter function, in particular to serotonergic and dopaminergic genes, and on genes related to neuroendocrine signaling, like sex-steroid receptors and stress-related circuitry (Iofrida et al., 2014; Fernàndez-Castillo and Cormand, 2016; Waltes et al., 2016). Genome-wide association studies (GWAS) of aggression have investigated a wide range of aggression related phenotypes. Interestingly, two genes showed evidence for association based on more than one GWAS, NFKB1 and A2BP1. NFKB1 encodes the nuclear factor of kappa light polypeptide gene enhancer in B-cells 1, a transcription regulator involved in axonal regeneration and degeneration. A2BP1 (also called RBFOX1) encodes the RNA binding protein, fox-1 homolog (C. elegans) 1, a neuron-specific RNA splicing factor that regulates the expression of large genetic networks during early neuronal development (Fernàndez-Castillo and Cormand, 2016). Recently, a large-scale GWAS meta-analysis was conducted within the framework of the early genetics and lifecourse epidemiology (EAGLE) consortium, including nearly 19,000 subjects. The researchers combined GWAS data on childhood and adolescent aggression from nine population-based cohorts, and found suggestive evidence of association for a region on chromosome 2, near a gene involved in the regulation of excitatory synapse development (Pappa et al., 2015). While most other GWASs of aggression were relatively small-scaled, top-finding of these studies together with bioinformatics approaches have highlighted the importance of neurodevelopmental and synaptic plasticity genes for aggression risk (Fernàndez-Castillo and Cormand, 2016).
Investigation of the neural correlates of aggression has highlighted the involvement of several brain phenotypes in aggression. The main forms of cognitive dysfunction that have been recognized in the context of aggression are decreased empathy, an increased acute threat response and impaired decision-making. Different neural systems are thought to underlie these cognitive impairments. In short, the main neural substrates of empathic processing are thought to be the ventromedial prefrontal cortex (vmPFC) and the amygdala, while the acute threat response is mediated by the amygdala-hypothalamus-periaqueductal gray neural system, and poor decision-making in individuals with aggressive behavior has been related the striatum and vmPFC (Blair et al., 2016). Imaging studies point towards an important role for subcortical brain regions in the neurobiology of aggressive phenotypes (Siever, 2008). Of specific interest in the context of aggression are the amygdala and the striatal subregions nucleus accumbens and caudate nucleus (Blair et al., 2016). The amygdala has been strongly linked to aggression through its role in emotion processing and threat reactivity (Fusar-Poli et al., 2009; Mobbs et al., 2010). A large number of studies have reported differences in the size of the amygdala between aggressive and comparison subjects, predominantly volume reductions (Sterzer et al., 2007; Fairchild et al., 2013; Zhang et al., 2013; Pardini et al., 2014; Wallace et al., 2014; Caldwell et al., 2015; Thijssen et al., 2015; Noordermeer et al., 2016). The striatum has been associated with aggression through its central role in reward sensitivity, processing of punishment and regulation of avoidance behaviors (Finger et al., 2008, 2011; Crowley et al., 2010; White et al., 2013). Impairments in these functions are thought to be the basis of poor decision-making in individuals with aggressive behavior (Fairchild et al., 2009; Blair et al., 2016). Both volume reductions and volume increases of the striatum have been related to aggressive phenotypes, especially for the caudate nucleus and nucleus accumbens (Nosarti et al., 2005; McAlonan et al., 2007; Ducharme et al., 2011; Schiffer et al., 2011; Fairchild et al., 2013; Cha et al., 2015). Brain volume has been shown to be heritable, and the Enhancing Neuro Imaging Genetics through Meta-Analysis (ENIGMA) consortium recently conducted a GWAS meta-analysis on volumes of seven subcortical brain structures and intracranial volume, to identify genetic variants that influence brain structure (Hibar et al., 2015). Identification of such genetic variants may help to uncover mechanisms underlying neuropsychiatric disorders.
Since both aggression risk and brain volumes are heritable, one may hypothesize that part of the genes contributing to aggression neurobiology do so by influencing aggression-related brain volumes. Identification of these genes may highlight specific pathways from gene to aggressive behavior via the brain. However, research into the underlying genetic and neurobiological mechanisms of aggression is complicated by the fact that aggression is a behaviorally and etiologically complex phenomenon. Efforts have been made to recognize different subtypes of aggressive behavior, presumed to differ in their underlying neurobiology. A frequently used system divides aggression into three subtypes; proactive aggression, reactive aggression due to external provocation or threat, and reactive aggression due to internal frustration (Dodge and Coie, 1987; Raine et al., 2006; Brugman et al., 2017; Smeets et al., 2017). Proactive aggression has been related to psychopathic traits and delinquent behavior (Cima et al., 2013). In this subtype, dysfunction in neural circuitry involving (venteromedial) prefrontal and striatal areas is thought to underlie observed difficulties with decision making and reinforcement learning, while a decreased responsiveness of the amygdala to distress cues is thought to reflect deficits in emotional empathy (Blair, 2013). Reactive subtypes of aggression, on the other hand, have been associated with impulsivity, anxiety and hostile interpretation bias (Bubier and Drabick, 2009; Brugman et al., 2015). It has been suggested that reactive forms of aggression are mediated by an overly responsive amygdala-related threat response circuitry, which is dependent on regulation by cortical brain regions (Blair, 2013). Hence, different pathways to the maladaptive behavior are thought to exist. An added complication for the identification of genetic and neurobiological mechanisms underlying aggression are the marked sex-differences in aggressive behaviors. Sex-differences in aggression are pronounced with respect to prevalence and with respect to type of aggression displayed (Hill, 2002; Collett et al., 2003; Stephenson et al., 2014). For example, males are overrepresented among patients with aggression-related disorders such as conduct disorder (Hill, 2002), and are more prone to display physical aggression compared to females (Baillargeon et al., 2007). Identification of subtype- and sex-dependent genetic association may help in elucidating specific links from gene to brain to behavior.
Based on the above, the aim of the current study was two-fold. First, we sought to identify genes influencing both aggression and aggression-related brain volumes. To this end, we conducted genome-wide gene-based cross-trait meta-analyses of aggression and amygdala, nucleus accumbens and caudate nucleus volume, using GWAS meta-analysis data of two large-scale consortia (EAGLE, N = 18,988; ENIGMA2, N = 13,171). Second, we aimed to assess subtype- and sex-specificity of association for identified genes. For this, we conducted gene-wide association analyses with aggression subtypes for these genes in a population sample of healthy adults with available genome-wide genotyping and questionnaire data on aggression subtypes (Brain Imaging Genetics, BIG, N = 501).
Materials and Methods
Samples
EAGLE
GWAS Meta-Analysis (GWAS-MA) data on aggression were obtained from the EAGLE consortium which investigated childhood aggressive behavior using nine population-based studies with a total of 18,988 subjects (mean age = 8.44 years, SD = 4.16; Pappa et al., 2015). Different well-validated parent-report questionnaires were used to assess aggressive behavior. Depending on study sample, aggressive behavior was assessed with the aggression scale of the Childhood Behavioral Checklist (CBCL), the conduct problem scale of the Strengths and Difficulties Questionnaire (SDQ), or comparable items in general questionnaires. Scores derived from SDQ and CBCL questionnaires were shown to be highly correlated and interchangeable for the assessment of children’s behavior problems (Goodman and Scott, 1999). Genomic data were imputed to the HapMap reference panel (release 22) and comprised only samples of European ancestry. GWAS was performed for each cohort, followed by removal of single nucleotide polymorphisms (SNPs) with low minor allele frequency (<0.05) and imputation quality (RSQ < 0.3 or INFO < 0.4). Results were combined using the sample-size weighted z-score method as implemented in METAL (Willer et al., 2010), controlling for genomic inflation. Access to the summary statistics was requested through http://www.tweelingenregister.org/EAGLE. All sites involved in this study obtained approval from local research ethics committees, and written parental consent was obtained for all participants.
ENIGMA2
GWAS-MA data on the aggression-related subcortical volumes of nucleus accumbens, amygdala and caudate nucleus were obtained from the ENIGMA consortium. The ENIGMA consortium conducted GWAS-MA on intracranial volume (ICV) and seven subcortical brain volumes, to identify common genetic variants contributing to volume differences. They used MRI brain scans and genome-wide genotype data of 13,171 subjects of European ancestry from 28 cohorts (discovery sample). Brain scans were examined and processed at each site following a standardized protocol. Subcortical volumes had been adjusted for ICV to identify specific genetic contributions to individual volumes. Genomic data comprised only European samples and were imputed to the 1000 Genomes, v3 phase1 reference panel using MaCH for phasing and minimac for imputation (Fuchsberger et al., 2015). GWAS was performed at each site, and SNPs with an imputation score of RSQ < 0.5 and minor allele count <10 were removed. Results were combined using an inverse-variance-weighted model as implemented in the software package METAL (Willer et al., 2010), controlling for genomic inflation. Further details of the original analysis can be found in Hibar et al. (2015). Access to the summary statistics of ENIGMA was requested through the ENIGMA website1. All sites involved in this study obtained approval from local research ethics committees or Institutional Review Boards, and all participants gave written informed consent.
BIG
To assess subtype-specific association of identified genes and for mediation analysis, data from the BIG study was used. This study was conducted at the Donders Institute for Brain, Cognition and Behavior (Franke et al., 2010), and consists of self-reported healthy adults who participated in smaller-scale imaging studies at the institute. Participants gave consent to use their acquired brain data, donated saliva and performed online testing. In the current study, a sub-sample of 501 subjects with available Reactive Proactive Questionnaire (RPQ) data (Raine et al., 2006), genome-wide genotype data, and structural MRI data was used (age range 18–45 years, 215 male/286 female).
All participants were of Caucasian descent and were screened using a self-report questionnaire for the following exclusion criteria before study participation: a history of somatic disease potentially affecting the brain, current or past psychiatric or neurological disorder, medication (except hormonal contraceptives) or illicit drug use during the past 6 months, history of substance abuse, current or past alcohol dependance, pregnancy, lactation, menopause and magnetic resonance imaging contraindications (Gerritsen et al., 2012). All participants gave written informed consent, and the study was approved by the regional ethics committee (Commissie Mensgebonden Onderzoek/CMO).
Behavioral and Genetic Measures in BIG
Aggression Questionnaire
The RPQ was used to assess subtypes of aggression in the BIG study (Raine et al., 2006). The RPQ is a self-report questionnaire consisting of 23 items. For each item, subjects are asked to indicate, how often they have engaged in a given type of behavior, like “had temper tantrums”. Items are rated on a three-point Likert scale (“never” = 0, “sometimes” = 1, “often” = 2). Responses were summed to yield the three factors that best described the RPQ in earlier exploratory factor analysis (Brugman et al., 2017; Smeets et al., 2017) as well as in the current sample (van Donkelaar et al., 2017): “proactive aggression” (range 0–12), “reactive aggression due to internal frustration” (range 0–9), and “reactive aggression due to external provocation or threat” (range 0–10). RPQ proactive aggression scores were dichotomized into high- and low-scoring (score ≥ 2 and score ≤ 1, respectively), because of a highly positively skewed distribution in both males and females (Supplementary Figure S1). An overview of RPQ items can be found in Supplementary Table S1.
Genotyping and Imputation
Genetic analyses for the BIG study were carried out at the Department of Human Genetics of the Radboud university medical center. Saliva samples were collected using Oragene kits (DNA Genotek, Kanata, Canada), and genomic DNA was extracted as specified by the manufacturer. Genome-wide genotyping was performed on three different genotyping platforms; Affymetrix Genome-Wide Human SNP Array 6.0 (Affymetrix Inc., Santa Clara, CA, USA), Infinium PsychArray-24 v1.1 BeadChip2, and Infinium OmniExpress-24 array2. Quality control steps and imputation were performed using the Ricopili Rapid Imputation Consortium Pipeline3. Pre-imputation quality control included pre-filtering of SNPs with call rate <0.95, filtering of individuals with a genotyping rate <0.98 or inbreeding coefficient >0.02, filtering of SNPs with a call rate <0.98 or Hardy-Weinberg p-value < 1e-06, removal of invariant SNPs, and removal of population ancestry outliers. SHAPEIT4 and IMPUTE2 (Howie et al., 2009) software were used for haplotype phasing and imputation with 1000 Genomes Phase 3.v5a reference data. For the current study, best-guess genotypes were inferred with a minimum probability threshold of 0.8. Post-imputation quality control included a strict SNP imputation quality threshold ≥0.8, removal of duplicated and related individuals (pi hat > 0.25), removal of individuals with a call rate below 95%, and removal of SNPs with a call rate below 95%, a minor allele frequency of less than 1%, or failing the Hardy-Weinberg equilibrium test at a threshold of p ≤ 1e-6.
Analyses
Genome-Wide Gene-Based Cross-Trait Meta-Analyses
We first conducted genome-wide cross-trait meta-analyses of aggression and three different aggression-related brain volume measures (amygdala, nucleus accumbens and caudate nucleus). We used summary statistic data of two large-scale GWAS of: (1) aggressive behavior in children and adolescents (EAGLE, N = 18,988); and (2) MRI-based volume measures of the aggression-relevant brain regions (ENIGMA2, N = 13,171). First, four separate genome-wide gene-based analyses with a 50 kb flanking region around genes were performed for the summary statistic data of aggression and the volumes of amygdala, nucleus accumbens and caudate nucleus using MAGMA v1.06 (de Leeuw et al., 2015). SNPs were mapped onto genes using 1000 Genomes Phase 3 reference data followed by computation of gene p-values by aggregating the effect of common variants within the genes. Next, fixed-effects meta-analyses were performed of aggression with amygdala volume, aggression with caudate nucleus volume and aggression with nucleus accumbens volume, using the weighted Stouffer’s Z method as implemented in MAGMA software. Results were considered significant if they reached the Bonferroni-corrected P-value-threshold for testing 18,310 genes (p < 2.731e-6). Significant genes with stronger association p-values in meta-analysis, compared to the separate analyses of aggression and brain volume, were reported and selected for further investigation.
Gene-Wide Association Analyses for Aggression Subtypes
Gene-wide association of selected genes with three subtypes of aggression, reactive aggression due to internal frustration, reactive aggression due to external provocation or threat and proactive aggression, was assessed. One phenotypic outlier (>4 SD) was removed for all analyses. Gene-wide analyses again included a 50 kb flanking region. Three base gene analysis models are available in MAGMA, each of them sensitive to different genetic architectures: Principal Component Analysis, mean-SNP and top-SNP models. For the current analysis, a multi-model approach was used, combining the results from the base analysis models into an aggregate p-value. Separate analyses were run for subjects genotyped on the three different genotyping arrays. Age and four population components derived from multidimensional scaling analysis were included as covariates. Sex was included as an interaction term in the model, yielding a gene p-value for main and interaction effects combined (P_full). This was followed by meta-analysis of the full model output of the three genotyping arrays, using the weighted Stouffer’s Z method as implemented in MAGMA software (de Leeuw et al., 2015). To protect against type I error, the conventional significance threshold (0.05) was lowered to correct for multiple comparisons, testing two genes and using the effective number of independent tests (Meff, see Li and Ji, 2005) for the aggression subtype outcomes, calculated to be 2.5 (taking into account the correlation matrix of the three aggression measures). Hence, results were considered significant if they reached a significance threshold of 0.01. Given the strong sex effects in aggression research (Collett et al., 2003; Stephenson et al., 2014) explorative analysis of sex are reported for reasons of completeness.
Results
Genome-Wide Gene-Based Cross-Trait (Aggression-Brain) Meta-Analyses
The MDS1 and EVI1 complex locus gene (MECOM) was significantly associated with the cross-trait construct of aggression and nucleus accumbens volume (p = 4.94e-07), and the Vasopressin Receptor 1A gene (AVPR1A) showed significant association with the cross-trait construct of aggression and amygdala volume (p = 1.64e-06). Both associations were more significant compared to the separate analyses of aggression and the respective brain volume (Table 1).
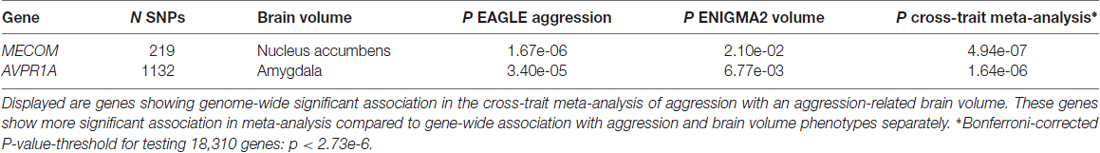
Table 1. Significant results of the genome-wide cross-trait meta-analyses of aggression and aggression-related brain volumes using gene-wide association statistics.
Gene-Wide Association Analyses for Aggression Subtypes
The general characteristics of the 501 participants from the BIG sample included in the aggression subtype analysis are shown in Supplementary Table S2. Gene-wide association analyses with three aggression subtypes were conducted for AVPR1A and MECOM to identify gene-behavior relationships, including sex as an interaction term in the model. Association of the AVPR1A gene with the score for reactive aggression due to external provocation or threat was nominally significant (P_full = 0.016), but did not reach the corrected significance threshold (Table 2). Explorative analyses in males and females separately showed a stronger contribution to this effect for males (P_males = 0.037; P_females = 0.517). The MECOM gene was not associated with any of the aggression subtypes in the population sample.

Table 2. Gene-wide association results for aggression subtypes in healthy adults from the BIG sample (N = 501).
Discussion
Using cross-trait meta-analyses of gene-wide association statistics, this study identified two genes as potentially pleiotropic loci for aggression and aggression-related subcortical brain volumes. We identified MECOM as a gene potentially contributing to both aggression risk and nucleus accumbens volume, and we identified AVPR1A as a gene potentially contributing to both aggression risk and amygdala volume. Subsequently, we investigated subtype-specific and sex-dependent association of these genes in an independent sample. No associations with aggression subtypes could be confirmed in this sample, although sex-dependent association of AVPR1A with reactive aggression due to external provocation/threat reached the nominal significance level.
The MDS1 and EVI1 complex locus gene (MECOM) codes for a protein known as transcriptional regulator and oncoprotein (Yoshimi and Kurokawa, 2011). MECOM plays an important role in early development, with Evi1 homozygous mutant mouse embryos dying approximately 10.5 days post coitum showing disrupted cell proliferation and disrupted development of cardiovascular and neural systems (Hoyt et al., 1997). The association p-value for MECOM in the study of aggression improved in the cross-trait meta-analysis of this behavioral trait with nucleus accumbens volume. According to our hypothesis, this might indicate that it exerts its effect on aggression through mechanisms involving the nucleus accumbens. However, we did not observe the association of MECOM with aggression when investigating specific subtypes of aggression in our own, smaller sample of adults. To our knowledge, little is known about MECOM in relation to psychiatric behavioral phenotypes so far, and future work needs to investigate this association in more detail.
Our study provides further evidence for a role of candidate gene AVPR1A in aggression. The Arginine Vasopressin Receptor 1A gene (AVPR1A) codes for the primary receptor of AVP in the brain. AVP is a neuropeptide strongly implicated in complex social and emotional behaviors, including aggression, through a host of animal studies (Ebstein et al., 2010). Also in humans, AVP was shown to play a role in enhancing aggressive behavior. For example, evidence exists for a positive correlation between aggression and cerebro-spinal fluid AVP in humans (Coccaro et al., 1998). Additional evidence comes from genetic association studies. The original aggression GWAS-MA that we used for cross-trait meta-analysis reported gene-wide association of the AVPR1A gene with childhood aggression (P = 1.61e-03), using VEGAS gene-based analysis and correcting for 21 candidate genes tested (Pappa et al., 2015). Using the MAGMA multi-model approach, which has the advantage of yielding a more even distribution of statistical power and sensitivity for a wider range of different supposed underlying genetic architectures compared to other methods (de Leeuw et al., 2015), an even lower p-value was reported in the current study. The cross-trait meta-analysis of aggression and amygdala volume resulted in gene-wide genome-wide significance. Other human genetic association studies of variants in the AVPR1A gene reported association with anger (Moons et al., 2014), gender-specific nominally significant association with pervasive aggression (Malik et al., 2014), but no association in an early study of antisocial traits (Prichard et al., 2007). While we report nominally significant subtype-specific gene-wide association of the AVPR1A gene with reactive aggression due to external provocation or threat in a sample of healthy adults, this finding is not significant after correcting for multiple testing. We can speculate that association of AVPR1A to this subtype of aggression, which specifically measures social responses to threat and provocation by others or actions of self-defense in response to others, would be in line with existing data highlighting the importance of AVP in social context and social communication. Vasopressin signaling is thought to be an important determinant of the intensity and range of social responses displayed in different social situations (Albers, 2012). For example, AVP can alter the extent to which social stimuli are threatening, by modulating sensory information (Thompson et al., 2006). Sex-dependent effects of AVP have also been found in animal research, finding opposite effects of both vasopressin and V1a receptor blockade on aggressive behavior. For example, AVP injection increases aggression in male hamsters but decreases it in females, while injection of V1a receptor antagonists has the opposite results (Gutzler et al., 2010). This data suggests that there may be a difference between males and females in the effects of vasopressin signaling on aggression. Less is known about sex differences in V1a receptor expression. Nevertheless, research in a number of species indicates that receptor distribution might vary in a sex-dependent manner as well (reviewed in Albers, 2015). Moreover, gonadal hormones can modulate the expression of vasopressin and vasopressin receptors (e.g., Dubois-Dauphin et al., 1994; Young et al., 2000), thus partly explaining sex differences in the vasopressin system. Hence, reducing phenotypic heterogeneity and taking into account sex-related heterogeneity may facilitate the search for genes involved in the etiology of aggression.
Our cross-trait meta-analysis results indicate that amygdala volume might serve as a (proxy for related) mechanisms through which the vasopressin receptor could influence aggressive behavior, and that MECOM may exert its effect on aggression through mechanisms involving the nucleus accumbens. Thus, we provide specific hypotheses about shared genetic risk and generated specific hypotheses about links from gene to brain to behavior for future studies to focus on. It is often assumed in imaging genetics research that genetic risk for a neurodevelopmental disorder passes through the brain phenotype to behavior. However, another possibility is that genetic factors influencing behavior also influence the brain in a way that is independent of the behavioral phenotype of interest (Kendler and Neale, 2010). Different mediation analysis methods have been developed to explore these relationships between genetic variants, brain phenotypes and behavior. Historically, the causal steps approach has been popular in mediation analysis. It uses a set of tests of significance for each path in a causal system, although a more powerful way to determine significance of a mediated effect is bootstrapping (Hayes and Scharkow, 2013). Other methods use causal modeling to describe the direction of association between different variables, using exploratory structure learning algorithms to find conditional independencies, which makes it possible to infer parts of the structure of a structural equation model (SEM) and make predictions about causation (Sokolova et al., 2017). Only a few imaging genetics studies have investigated this issue of causality earlier. Those studies showed that only part of the brain regions showing genotype effects actually do mediate between genetics and the behavior under study (Sokolova et al., 2015; van der Meer et al., 2015), proving the importance of such multilevel investigations to elucidate the biological mechanisms, by which brain alterations may be involved in aggression etiology. Currently, available methods for making causal inferences focus on SNP-level investigations, and future studies would benefit from the development of approaches for aggregating common genetic variant data to gene- or gene-set-level in mediation frameworks.
This study has several strengths and limitations. The current study used the largest data-sets available to investigate pleiotropic genetic factors for aggression and brain volumes at gene-level. Cross-trait meta-analysis of brain measures and psychiatric phenotypes is a useful way of detecting shared genetic risk and generating new hypotheses about specific links between genes, the brain and behavior (Franke et al., 2016). We were also able to use a well-phenotyped population cohort to investigate specific subtypes of aggression. While the limitations of using saliva in this cohort as a source of DNA for genotyping may be particularly relevant for the analysis of copy number variants (Fabre et al., 2011), replication of our work using DNA isolated from other sources would further strengthen the confidence in our results.
The added value of using data from such smaller cohorts over large consortium based data lies in the possibility of in-depth phenotyping and reducing sources of heterogeneity that come with the use of pooled data-sets. Nevertheless, we did not detect significant sex- and subtype-specific associations of MECOM and AVPR1A with aggression in this sample. Possibly, we still lacked power to detect small genetic effects due to sample size, or effects might be larger in non-healthy samples. Hence, future studies are needed to further investigate the specific hypotheses about gene-brain-behavior relationships generated by the current study. As this study only investigated selected subcortical MRI measures, future work should also be extended to include cortical regions as well as connectivity measures that have been shown to play a role in aggression (Meyer-Lindenberg et al., 2006).
In summary, we identified MECOM and AVPR1A as genes contributing to aggression risk in conjunction with nucleus accumbens and amygdala brain volume, respectively. Future studies may elucidate causality of gene-brain-behavior relationships. Comprehension of sex-specific physiological pathways associated with aggression subtypes is needed to enhance our understanding of the determinants of aggression, and only by understanding the mechanisms underlying different forms of aggression will we be able to develop effective treatment approaches and minimize the social costs of aggression.
Author Contributions
MMJD, MH, JKB, BF and JB contributed to the conception and design of the study. MMJD, IP and HT contributed to data analysis and/or interpretation. MMJD wrote the first draft of the manuscript. All authors contributed to manuscript revision, read and approved the submitted version.
Conflict of Interest Statement
BF discloses having received educational speaking fees from Shire and Medice. JKB has been in the past 3 years a consultant to/member of advisory board of/and/or speaker for Janssen Cilag BV, Eli Lilly, Lundbeck, Shire, Roche, Novartis and Servier. He is not an employee of any of these companies, and not a stock shareholder of any of these companies. He has no other financial or material support, including expert testimony, patents, royalties.
The other authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This works was supported by the Netherlands Organization for Scientific Research (Nederlandse Organisatie voor Wetenschappelijk Onderzoek, NWO), i.e., the NWO Brain & Cognition Excellence Program (grant 433-09-229) and the Vici Innovation Program (grant 016-130-669 to BF). Additional support was received from the European Community’s Seventh Framework Programme (FP7/2007–2013) under grant agreements n° 602805 (Aggressotype), n° 602450 (IMAGEMEND), and n° 278948 (TACTICS) as well as from the European Community’s Horizon 2020 Programme (H2020/2014–2020) under grant agreements n° 643051 (MiND) and n° 667302 (CoCA). The work was also supported by grants for the ENIGMA Consortium (Foundation for the National Institutes of Health (NIH); grant number U54 EB020403) from the BD2K Initiative of a cross-NIH partnership.
This work was carried out on the Dutch national e-infrastructure with the support of SURF Cooperative.
The Enhancing NeuroImaging Genetics through Meta-Analysis (ENIGMA) Consortium provided summary statistics of the consortium findings to this project. The original publication of those findings as well as the list of contributing samples and people can be found on the ENIGMA website: http://enigma.ini.usc.edu.
The EArly Genetics and Lifecourse Epidemiology (EAGLE) consortium provided summary statistics of the consortium findings to this project. Contributing samples and people can be found in the original publication of those findings (Pappa et al., 2015).
Footnotes
- ^ http://enigma.ini.usc.edu/download-enigma-gwas-results/
- ^ www.illumina.com
- ^ https://sites.google.com/a/broadinstitute.org/ricopili/home
- ^ https://mathgen.stats.ox.ac.uk/genetics_software/shapeit/shapeit.html
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnbeh.2018.00061/full#supplementary-material
References
Albers, H. E. (2012). The regulation of social recognition, social communication and aggression: vasopressin in the social behavior neural network. Horm. Behav. 61, 283–292. doi: 10.1016/j.yhbeh.2011.10.007
Albers, H. E. (2015). Species, sex and individual differences in the vasotocin/vasopressin system: relationship to neurochemical signaling in the social behavior neural network. Front. Neuroendocrinol. 36, 49–71. doi: 10.1016/j.yfrne.2014.07.001
Baillargeon, R. H., Zoccolillo, M., Keenan, K., Côté, S., Pérusse, D., Wu, H. X., et al. (2007). Gender differences in physical aggression: a prospective population-based survey of children before and after 2 years of age. Dev. Psychol. 43, 13–26. doi: 10.1037/0012-1649.43.1.13
Bakker, M. J., Greven, C. U., Buitelaar, J. K., and Glennon, J. C. (2017). Practitioner review: psychological treatments for children and adolescents with conduct disorder problems—a systematic review and meta-analysis. J. Child Psychol. Psychiatry 58, 4–18. doi: 10.1111/jcpp.12590
Blair, R. J. (2013). The neurobiology of psychopathic traits in youths. Nat. Rev. Neurosci. 14, 786–799. doi: 10.1038/nrn3577
Blair, R. J., Veroude, K., and Buitelaar, J. K. (2016). Neuro-cognitive system dysfunction and symptom sets: a review of fMRI studies in youth with conduct problems. Neurosci. Biobehav. Rev. doi: 10.1016/j.neubiorev.2016.10.022 [Epub ahead of print].
Brugman, S., Cornet, L. J., Smeijers, D., Smeets, K., Oostermeijer, S., Buitelaar, J. K., et al. (2017). Examining the reactive proactive questionnaire in adults in forensic and non-forensic settings: a variable- and person-based approach. Aggress. Behav. 43, 155–162. doi: 10.1002/ab.21671
Brugman, S., Lobbestael, J., Arntz, A., Cima, M., Schuhmann, T., Dambacher, F., et al. (2015). Identifying cognitive predictors of reactive and proactive aggression. Aggress. Behav. 41, 51–64. doi: 10.1002/ab.21573
Brunner, H. G., Nelen, M., Breakefield, X. O., Ropers, H. H., and van Oost, B. A. (1993). Abnormal behavior associated with a point mutation in the structural gene for monoamine oxidase A. Science 262, 578–580. doi: 10.1126/science.8211186
Bubier, J. L., and Drabick, D. A. (2009). Co-occurring anxiety and disruptive behavior disorders: the roles of anxious symptoms, reactive aggression, and shared risk processes. Clin. Psychol. Rev. 29, 658–669. doi: 10.1016/j.cpr.2009.08.005
Caldwell, J. Z., Armstrong, J. M., Hanson, J. L., Sutterer, M. J., Stodola, D. E., Koenigs, M., et al. (2015). Preschool externalizing behavior predicts gender-specific variation in adolescent neural structure. PLoS One 10:e0117453. doi: 10.1371/journal.pone.0117453
Cha, J., Fekete, T., Siciliano, F., Biezonski, D., Greenhill, L., Pliszka, S. R., et al. (2015). Neural correlates of aggression in medication-naive children with ADHD: multivariate analysis of morphometry and tractography. Neuropsychopharmacology 40, 1717–1725. doi: 10.1038/npp.2015.18
Cima, M., Raine, A., Meesters, C., and Popma, A. (2013). Validation of the dutch reactive proactive questionnaire (RPQ): differential correlates of reactive and proactive aggression from childhood to adulthood. Aggress. Behav. 39, 99–113. doi: 10.1002/ab.21458
Coccaro, E. F., Kavoussi, R. J., Hauger, R. L., Cooper, T. B., and Ferris, C. F. (1998). Cerebrospinal fluid vasopressin levels: correlates with aggression and serotonin function in personality-disordered subjects. Arch. Gen. Psychiatry 55, 708–714. doi: 10.1001/archpsyc.55.8.708
Collett, B. R., Ohan, J. L., and Myers, K. M. (2003). Ten-year review of rating scales. VI: scales assessing externalizing behaviors. J. Am. Acad. Child Adolesc. Psychiatry 42, 1143–1170. doi: 10.1097/00004583-200310000-00006
Crowley, T. J., Dalwani, M. S., Mikulich-Gilbertson, S. K., Du, Y. P., Lejuez, C. W., Raymond, K. M., et al. (2010). Risky decisions and their consequences: neural processing by boys with Antisocial Substance Disorder. PLoS One 5:e12835. doi: 10.1371/journal.pone.0012835
de Leeuw, C. A., Mooij, J. M., Heskes, T., and Posthuma, D. (2015). MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput. Biol. 11:e1004219. doi: 10.1371/journal.pcbi.1004219
Dodge, K. A., and Coie, J. D. (1987). Social-information-processing factors in reactive and proactive aggression in children’s peer groups. J. Pers. Soc. Psychol. 53, 1146–1158. doi: 10.1037//0022-3514.53.6.1146
Dubois-Dauphin, M., Theler, J. M., Ouarour, A., Pévet, P., Barberis, C., and Dreifuss, J. J. (1994). Regional differences in testosterone effects on vasopressin receptors and on vasopressin immunoreactivity in intact and castrated Siberian hamsters. Brain Res. 638, 267–276. doi: 10.1016/0006-8993(94)90659-9
Ducharme, S., Hudziak, J. J., Botteron, K. N., Ganjavi, H., Lepage, C., Collins, D. L., et al. (2011). Right anterior cingulate cortical thickness and bilateral striatal volume correlate with child behavior checklist aggressive behavior scores in healthy children. Biol. Psychiatry 70, 283–290. doi: 10.1016/j.biopsych.2011.03.015
Ebstein, R. P., Israel, S., Chew, S. H., Zhong, S., and Knafo, A. (2010). Genetics of human social behavior. Neuron 65, 831–844. doi: 10.1016/j.neuron.2010.02.020
Fabre, A., Thomas, E., Baulande, S., Sohier, E., Hoang, L., Soularue, P., et al. (2011). Is saliva a good alternative to blood for high density genotyping studies: SNP and CNV comparisons? J. Biotechnol. Biomaterial. 1:119. doi: 10.4172/2155-952x.1000119
Fairchild, G., Hagan, C. C., Walsh, N. D., Passamonti, L., Calder, A. J., and Goodyer, I. M. (2013). Brain structure abnormalities in adolescent girls with conduct disorder. J. Child Psychol. Psychiatry 54, 86–95. doi: 10.1111/j.1469-7610.2012.02617.x
Fairchild, G., van Goozen, S. H., Stollery, S. J., Aitken, M. R., Savage, J., Moore, S. C., et al. (2009). Decision making and executive function in male adolescents with early-onset or adolescence-onset conduct disorder and control subjects. Biol. Psychiatry 66, 162–168. doi: 10.1016/j.biopsych.2009.02.024
Fernàndez-Castillo, N., and Cormand, B. (2016). Aggressive behavior in humans: genes and pathways identified through association studies. Am. J. Med. Genet. B Neuropsychiatr. Genet. 171, 676–696. doi: 10.1002/ajmg.b.32419
Finger, E. C., Marsh, A. A., Blair, K. S., Reid, M. E., Sims, C., Ng, P., et al. (2011). Disrupted reinforcement signaling in the orbitofrontal cortex and caudate in youths with conduct disorder or oppositional defiant disorder and a high level of psychopathic traits. Am. J. Psychiatry 168, 152–162. doi: 10.1176/appi.ajp.2010.10010129
Finger, E. C., Marsh, A. A., Mitchell, D. G., Reid, M. E., Sims, C., Budhani, S., et al. (2008). Abnormal ventromedial prefrontal cortex function in children with psychopathic traits during reversal learning. Arch. Gen. Psychiatry 65, 586–594. doi: 10.1001/archpsyc.65.5.586
Franke, B., Stein, J. L., Ripke, S., Anttila, V., Hibar, D. P., van Hulzen, K., et al. (2016). Genetic influences on schizophrenia and subcortical brain volumes: large-scale proof of concept. Nat. Neurosci. 19, 420–431. doi: 10.1038/nn.4228
Franke, B., Vasquez, A. A., Veltman, J. A., Brunner, H. G., Rijpkema, M., and Fernández, G. (2010). Genetic variation in CACNA1C, a gene associated with bipolar disorder, influences brainstem rather than gray matter volume in healthy individuals. Biol. Psychiatry 68, 586–588. doi: 10.1016/j.biopsych.2010.05.037
Fuchsberger, C., Abecasis, G. R., and Hinds, D. A. (2015). minimac2: faster genotype imputation. Bioinformatics 31, 782–784. doi: 10.1093/bioinformatics/btu704
Fusar-Poli, P., Placentino, A., Carletti, F., Landi, P., Allen, P., Surguladze, S., et al. (2009). Functional atlas of emotional faces processing: a voxel-based meta-analysis of 105 functional magnetic resonance imaging studies. J. Psychiatry Neurosci. 34, 418–432.
Gerritsen, L., Tendolkar, I., Franke, B., Vasquez, A. A., Kooijman, S., Buitelaar, J., et al. (2012). BDNF Val66Met genotype modulates the effect of childhood adversity on subgenual anterior cingulate cortex volume in healthy subjects. Mol. Psychiatry 17, 597–603. doi: 10.1038/mp.2011.51
Goodman, R., and Scott, S. (1999). Comparing the strengths and difficulties questionnaire and the child behavior checklist: is small beautiful? J. Abnorm. Child Psychol. 27, 17–24. doi: 10.1023/A:1022658222914
Gutzler, S. J., Karom, M., Erwin, W. D., and Albers, H. E. (2010). Arginine-vasopressin and the regulation of aggression in female Syrian hamsters (Mesocricetus auratus). Eur. J. Neurosci. 31, 1655–1663. doi: 10.1111/j.1460-9568.2010.07190.x
Hayes, A. F., and Scharkow, M. (2013). The relative trustworthiness of inferential tests of the indirect effect in statistical mediation analysis: does method really matter? Psychol. Sci. 24, 1918–1927. doi: 10.1177/0956797613480187
Hibar, D. P., Stein, J. L., Renteria, M. E., Arias-Vasquez, A., Desrivières, S., Jahanshad, N., et al. (2015). Common genetic variants influence human subcortical brain structures. Nature 520, 224–229. doi: 10.1038/nature14101
Hill, J. (2002). Biological, psychological and social processes in the conduct disorders. J. Child Psychol. Psychiatry 43, 133–164. doi: 10.1111/1469-7610.00007
Howie, B. N., Donnelly, P., and Marchini, J. (2009). A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 5:e1000529. doi: 10.1371/journal.pgen.1000529
Hoyt, P. R., Bartholomew, C., Davis, A. J., Yutzey, K., Gamer, L. W., Potter, S. S., et al. (1997). The Evi1 proto-oncogene is required at midgestation for neural, heart, and paraxial mesenchyme development. Mech. Dev. 65, 55–70. doi: 10.1016/s0925-4773(97)00057-9
Iofrida, C., Palumbo, S., and Pellegrini, S. (2014). Molecular genetics and antisocial behavior: where do we stand? Exp. Biol. Med. (Maywood) 239, 1514–1523. doi: 10.1177/1535370214529508
Kendler, K. S., and Neale, M. C. (2010). Endophenotype: a conceptual analysis. Mol. Psychiatry 15, 789–797. doi: 10.1038/mp.2010.8
Li, J., and Ji, L. (2005). Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity (Edinb) 95, 221–227. doi: 10.1038/sj.hdy.6800717
Malik, A. I., Zai, C. C., Berall, L., Abu, Z., Din, F., Nowrouzi, B., et al. (2014). The role of genetic variants in genes regulating the oxytocin-vasopressin neurohumoral system in childhood-onset aggression. Psychiatr. Genet. 24, 201–210. doi: 10.1097/YPG.0000000000000044
McAlonan, G. M., Cheung, V., Cheung, C., Chua, S. E., Murphy, D. G., Suckling, J., et al. (2007). Mapping brain structure in attention deficit-hyperactivity disorder: a voxel-based MRI study of regional grey and white matter volume. Psychiatry Res. 154, 171–180. doi: 10.1016/j.pscychresns.2006.09.006
McGuire, J. (2008). A review of effective interventions for reducing aggression and violence. Philos. Trans. R. Soc. Lond. B Biol. Sci. 363, 2577–2597. doi: 10.1098/rstb.2008.0035
Meyer-Lindenberg, A., Buckholtz, J. W., Kolachana, B., R Hariri, A., Pezawas, L., Blasi, G., et al. (2006). Neural mechanisms of genetic risk for impulsivity and violence in humans. Proc. Natl. Acad. Sci. U S A 103, 6269–6274. doi: 10.1073/pnas.0511311103
Miczek, K. A., Fish, E. W., De Bold, J. F., and de Almeida, R. M. (2002). Social and neural determinants of aggressive behavior: pharmacotherapeutic targets at serotonin, dopamine and γ-aminobutyric acid systems. Psychopharmacology 163, 434–458. doi: 10.1007/s00213-002-1139-6
Mobbs, D., Yu, R., Rowe, J. B., Eich, H., FeldmanHall, O., and Dalgleish, T. (2010). Neural activity associated with monitoring the oscillating threat value of a tarantula. Proc. Natl. Acad. Sci. U S A 107, 20582–20586. doi: 10.1073/pnas.1009076107
Moons, W. G., Way, B. M., and Taylor, S. E. (2014). Oxytocin and vasopressin receptor polymorphisms interact with circulating neuropeptides to predict human emotional reactions to stress. Emotion 14, 562–572. doi: 10.1037/a0035503
Noordermeer, S. D., Luman, M., and Oosterlaan, J. (2016). A systematic review and meta-analysis of neuroimaging in oppositional defiant disorder (ODD) and conduct disorder (CD) taking attention-deficit hyperactivity disorder (ADHD) into account. Neuropsychol. Rev. 26, 44–72. doi: 10.1007/s11065-015-9315-8
Nosarti, C., Allin, M. P., Frangou, S., Rifkin, L., and Murray, R. M. (2005). Hyperactivity in adolescents born very preterm is associated with decreased caudate volume. Biol. Psychiatry 57, 661–666. doi: 10.1016/j.biopsych.2004.12.003
Pappa, I., St Pourcain, B., Benke, K., Cavadino, A., Hakulinen, C., Nivard, M. G., et al. (2015). A genome-wide approach to children’s aggressive behavior: the EAGLE consortium. Am. J. Med. Genet. B Neuropsychiatr. Genet. 171, 562–572. doi: 10.1002/ajmg.b.32333
Pardini, D. A., Raine, A., Erickson, K., and Loeber, R. (2014). Lower amygdala volume in men is associated with childhood aggression, early psychopathic traits, and future violence. Biol. Psychiatry 75, 73–80. doi: 10.1016/j.biopsych.2013.04.003
Prichard, Z. M., Jorm, A. F., Mackinnon, A., and Easteal, S. (2007). Association analysis of 15 polymorphisms within 10 candidate genes for antisocial behavioural traits. Psychiatr. Genet. 17, 299–303. doi: 10.1097/ypg.0b013e32816ebc9e
Raine, A., Dodge, K., Loeber, R., Gatzke-Kopp, L., Lynam, D., Reynolds, C., et al. (2006). The reactive-proactive aggression questionnaire: differential correlates of reactive and proactive aggression in adolescent boys. Aggress. Behav. 32, 159–171. doi: 10.1002/ab.20115
Schiffer, B., Müller, B. W., Scherbaum, N., Hodgins, S., Forsting, M., Wiltfang, J., et al. (2011). Disentangling structural brain alterations associated with violent behavior from those associated with substance use disorders. Arch. Gen. Psychiatry 68, 1039–1049. doi: 10.1001/archgenpsychiatry.2011.61
Siever, L. J. (2008). Neurobiology of aggression and violence. Am. J. Psychiatry 165, 429–442. doi: 10.1176/appi.ajp.2008.07111774
Smeets, K. C., Oostermeijer, S., Lappenschaar, M., Cohn, M., van der Meer, J. M., Popma, A., et al. (2017). Are proactive and reactive aggression meaningful distinctions in adolescents? A variable- and person-based approach. J. Abnorm. Child Psychol. 45, 1–14. doi: 10.1007/s10802-016-0149-5
Sokolova, E., Hoogman, M., Groot, P., Claassen, T., Vasquez, A. A., Buitelaar, J. K., et al. (2015). Causal discovery in an adult ADHD data set suggests indirect link between DAT1 genetic variants and striatal brain activation during reward processing. Am. J. Med. Genet. B Neuropsychiatr. Genet. 168, 508–515. doi: 10.1002/ajmg.b.32310
Sokolova, E., Oerlemans, A. M., Rommelse, N. N., Groot, P., Hartman, C. A., Glennon, J. C., et al. (2017). A causal and mediation analysis of the comorbidity between attention deficit hyperactivity disorder (ADHD) and autism spectrum disorder (ASD). J. Autism. Dev. Disord. 47, 1595–1604. doi: 10.1007/s10803-017-3083-7
Stephenson, Z., Woodhams, J., and Cooke, C. (2014). Sex differences in predictors of violent and non-violent juvenile offending. Aggress. Behav. 40, 165–177. doi: 10.1002/ab.21506
Sterzer, P., Stadler, C., Poustka, F., and Kleinschmidt, A. (2007). A structural neural deficit in adolescents with conduct disorder and its association with lack of empathy. Neuroimage 37, 335–342. doi: 10.1016/j.neuroimage.2007.04.043
Thijssen, S., Ringoot, A. P., Wildeboer, A., Bakermans-Kranenburg, M. J., El Marroun, H., Hofman, A., et al. (2015). Brain morphology of childhood aggressive behavior: a multi-informant study in school-age children. Cogn. Affect. Behav. Neurosci. 15, 564–577. doi: 10.3758/s13415-015-0344-9
Thompson, R. R., George, K., Walton, J. C., Orr, S. P., and Benson, J. (2006). Sex-specific influences of vasopressin on human social communication. Proc. Natl. Acad. Sci. U S A 103, 7889–7894. doi: 10.1073/pnas.0600406103
Tuvblad, C., and Baker, L. A. (2011). Human aggression across the lifespan: genetic propensities and environmental moderators. Adv. Genet. 75, 171–214. doi: 10.1016/B978-0-12-380858-5.00007-1
van der Meer, D., Hoekstra, P. J., Zwiers, M., Mennes, M., Schweren, L. J., Franke, B., et al. (2015). Brain correlates of the interaction between 5-HTTLPR and psychosocial stress mediating attention deficit hyperactivity disorder severity. Am. J. Psychiatry 172, 768–775. doi: 10.1176/appi.ajp.2015.14081035
van Donkelaar, M. M. J., Hoogman, M., Shumskaya, E., Buitelaar, J. K., Bralten, J., and Franke, B. (2017). Monoamine and neuroendocrine gene-sets associate with frustration-based aggression in a gender-specific manner. Eur. Neuropsychopharmacol. doi: 10.1016/j.euroneuro.2017.11.016 [Epub ahead of print].
Veroude, K., Zhang-James, Y., Fernandez-Castillo, N., Bakker, M. J., Cormand, B., and Faraone, S. V. (2016). Genetics of aggressive behavior: an overview. Am. J. Med. Genet. B Neuropsychiatr. Genet. 171, 3–43. doi: 10.1002/ajmg.b.32364
Wallace, G. L., White, S. F., Robustelli, B., Sinclair, S., Hwang, S., Martin, A., et al. (2014). Cortical and subcortical abnormalities in youths with conduct disorder and elevated callous-unemotional traits. J. Am. Acad. Child Adolesc. Psychiatry 53, 456.e1–465.e1. doi: 10.1016/j.jaac.2013.12.008
Waltes, R., Chiocchetti, A. G., and Freitag, C. M. (2016). The neurobiological basis of human aggression: a review on genetic and epigenetic mechanisms. Am. J. Med. Genet. B Neuropsychiatr. Genet. 171, 650–675. doi: 10.1002/ajmg.b.32388
White, S. F., Pope, K., Sinclair, S., Fowler, K. A., Brislin, S. J., Williams, W. C., et al. (2013). Disrupted expected value and prediction error signaling in youths with disruptive behavior disorders during a passive avoidance task. Am. J. Psychiatry 170, 315–323. doi: 10.1176/appi.ajp.2012.12060840
Willer, C. J., Li, Y., and Abecasis, G. R. (2010). METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26, 2190–2191. doi: 10.1093/bioinformatics/btq340
Yoshimi, A., and Kurokawa, M. (2011). Evi1 forms a bridge between the epigenetic machinery and signaling pathways. Oncotarget 2, 575–586. doi: 10.18632/oncotarget.304
Young, L. J., Wang, Z., Cooper, T. T., and Albers, H. E. (2000). Vasopressin (V1a) receptor binding, mRNA expression and transcriptional regulation by androgen in the Syrian hamster brain. J. Neuroendocrinol. 12, 1179–1185. doi: 10.1046/j.1365-2826.2000.00573.x
Zhang, L., Kerich, M., Schwandt, M. L., Rawlings, R. R., McKellar, J. D., Momenan, R., et al. (2013). Smaller right amygdala in Caucasian alcohol-dependent male patients with a history of intimate partner violence: a volumetric imaging study. Addict. Biol. 18, 537–547. doi: 10.1111/j.1369-1600.2011.00381.x
Keywords: aggression, genetics, MRI, brain imaging, neurobiology
Citation: van Donkelaar MMJ, Hoogman M, Pappa I, Tiemeier H, Buitelaar JK, Franke B and Bralten J (2018) Pleiotropic Contribution of MECOM and AVPR1A to Aggression and Subcortical Brain Volumes. Front. Behav. Neurosci. 12:61. doi: 10.3389/fnbeh.2018.00061
Received: 01 December 2017; Accepted: 15 March 2018;
Published: 03 April 2018.
Edited by:
Nelly Alia-Klein, Icahn School of Medicine at Mount Sinai, United StatesReviewed by:
Zhen Yuan, University of Macau, ChinaSara Palumbo, Università degli Studi di Pisa, Italy
Copyright © 2018 van Donkelaar, Hoogman, Pappa, Tiemeier, Buitelaar, Franke and Bralten. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Janita Bralten, amFuaXRhLmJyYWx0ZW5AcmFkYm91ZHVtYy5ubA==
† These authors share last authorship.
 Marjolein M. J. van Donkelaar
Marjolein M. J. van Donkelaar Martine Hoogman
Martine Hoogman Irene Pappa
Irene Pappa Henning Tiemeier3,4,5
Henning Tiemeier3,4,5 Barbara Franke
Barbara Franke Janita Bralten
Janita Bralten