- 1Secció de Física Estadística i Interdisciplinària, Departament de Física de la Matèria Condensada, Universitat de Barcelona, Barcelona, Spain
- 2Institut de Nanociència i Nanotecnologia, Universitat de Barcelona, Barcelona, Spain
- 3Chemistry Department, Royal College of Surgeons in Ireland (RCSI), Dublin, Ireland
Nanoparticles (NPs) in contact with biological fluid adsorb biomolecules into a corona. This corona comprises proteins that strongly bind to the NP (hard corona) and loosely bound proteins (soft corona) that dynamically exchange with the surrounding solution. While the kinetics of hard corona formation is relatively well understood, thanks to experiments and robust simulation models, the experimental characterization and simulation of the soft corona present a more significant challenge. Here, we review the current state of the art in soft corona characterization and introduce a novel open-source computational model to simulate its dynamic behavior, for which we provide the documentation. We focus on the case of transferrin (Tf) interacting with polystyrene NPs as an illustrative example, demonstrating how this model captures the complexities of the soft corona and offers deeper insights into its structure and behavior. We show that the soft corona is dominated by a glassy evolution that we relate to crowding effects. This work advances our understanding of the soft corona, bridging experimental limitations with improved simulation techniques.
1 Introduction
Nanotechnology is a rapidly growing industry with emerging applications across various fields. Although nanostructures have been present in human life for a long time (Freestone et al., 2007), the understanding and development of advanced nanomaterials are relatively new (Qiu et al., 2017). In particular, there is an increasing interest in comprehending the behavior of nanoparticles (NPs) within biological systems (Dawson and Yan, 2021). The unique size, structure, and chemical properties of NPs introduce a wide range of new applications in many areas of research and technology, including therapeutics and diagnostics (Trinh et al., 2022).
When NPs are introduced into a biological environment, they quickly become coated by surrounding biomolecules, such as proteins, unless specifically designed not to do so (Cedervall et al., 2007b; Cedervall et al., 2007a; Lynch and Dawson, 2008). Research has shown that it is not the NP itself, but rather the biomolecules on its surface, that determine its interaction with living organisms. The layer of adsorbed proteins is known as the protein corona and defines the biological identity of the NPs (Lynch et al., 2009; Walczyk et al., 2010; Monopoli et al., 2012).
Due to their size, NPs can distribute throughout organisms, reaching various cellular and organic compartments, and, in specific cases, they can even breach biological barriers, such as the blood-brain barrier (Kreuter et al., 2002; Wagner et al., 2012). Indeed, the size and shape of nanoparticles can influence the formation of the protein corona, and their effects on the overall protein composition remain a subject of debate (Lundqvist et al., 2008; Tenzer et al., 2013; Talamini et al., 2017; Xu et al., 2018; Madathiparambil Visalakshan et al., 2020; Bilardo et al., 2022). In medicine, NPs can serve as therapeutic tools, enhancing previous techniques like drug delivery as they extend overall circulation time and the drug’s efficacy (Kumari et al., 2016), with the corona influencing the therapeutic outcome (Salvati, 2024). However, concerns have also been raised about the safety of prolonged or chronic exposure to NPs and the potential role of the corona in triggering the immune response (Oberdörster et al., 2005; Deng et al., 2011; Savolainen et al., 2013). Therefore, given the potential of nanotechnology, it is crucial to understand whether it poses a threat to organisms and the environment to ensure safe clinical translation of these new biomedical nanotools (Corbo et al., 2016; Boselli et al., 2024).
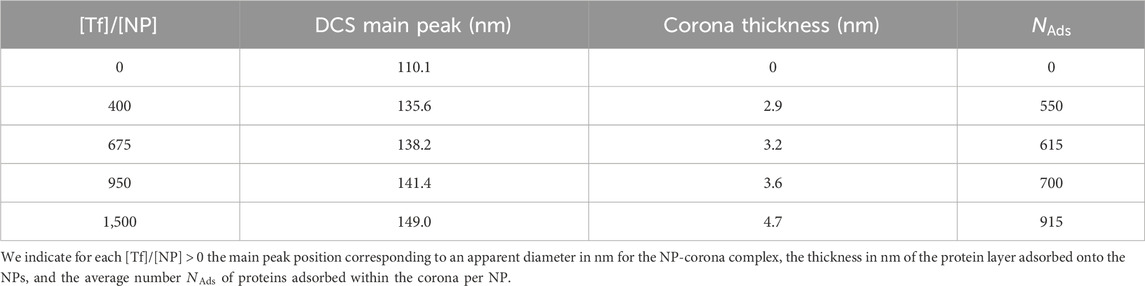
Numerous experiments have been conducted to understand how blood plasma proteins bind to a nanoparticle. The findings indicate that proteins adhere to the surface of the NP, forming a corona around it, consisting of two different components (Milani et al., 2012): a hard corona (HC) comprising tightly bound proteins in direct contact with the surface, and a more dynamic soft corona (SC) that is in constant exchange with the protein solution (Figure 1).
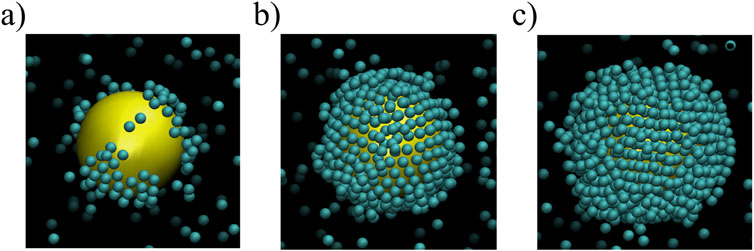
Figure 1. Coarse-grained configurations of Tf in suspension with polystyrene NPs. The relative concentration is
Experiments show that the HC binds almost irreversibly to the NP, while the SC binds reversibly (Lynch et al., 2009; Milani et al., 2012). The HC is considered the most biologically relevant part as it interacts with cells and biological machinery via the receptors. On the other hand, the SC plays a role in mediating transient and dynamic interactions with biological systems (García-Álvarez and Vallet-Regí, 2021; Bai et al., 2021). Characterizing the SC is challenging due to its transient nature and weak binding affinities, but recent advances help to understand its role. For example, using cryo-transmission electron microscopy (cryo-TEM) combined with synchrotron radiation circular dichroism (CD) allows real-time in situ insights into the transient nature and dynamic evolution of the SC. This approach reveals how weakly bound proteins within the SC undergo continuous exchange and reorganization (Sanchez-Guzman et al., 2020).
Also, in situ click-chemistry is used to map and identify the proteins within the SC, enabling precise tracking of protein-NP interactions. This technique reveals that the dynamic and reversible nature of the SC proteins significantly influences NP uptake and interaction with cellular membranes (Mohammad-Beigi et al., 2020).
Furthermore, the analysis of the protein corona on gold NPs by Liquid Chromatography-Tandem Mass Spectrometry (LC-MS/MS) and Enzyme-Linked Immunosorbent Assay (ELISA) has found that only 27% of the adsorbed proteins were functional for binding (Zhang et al., 2020). This indicates that the corona, rather than a simple monolayer, is an assembly of layers comprising a foundation layer (i.e., the HC) and an intermediate corona (IC) plus a binding layer, with the last two usually identified as SC.
Recent studies have combined different experimental methods to characterize
Additionally, a “fishing” method has been developed using bio-layer interferometry and LC-MS/MS to monitor the dynamic formation and evolution of the protein corona on chiral
To understand how different layers form on the surface of NPs, particularly the SC, it is essential to characterize the kinetics of protein adsorption into the corona. The rate at which proteins are adsorbed and the protein corona formation depend greatly on protein concentrations and affinities. Competition and cooperation between different types of proteins are crucial mechanisms in understanding kinetic processes, such as the Vroman effect (Vroman and Adams, 1969; Vroman and Adams, 1986; LeDuc et al., 1995; Vilaseca et al., 2013). Only biomolecules that reside in the protein corona for longer than the characteristic timescale of a biological phenomenon are likely to be relevant to the process. Experiments have shown how tissues, organs, and other biological systems respond depending on how long biomolecules stay at the NP surface (Tran et al., 2017). Therefore, it is reasonable to assume that proteins with a long residence time near the surface are the ones that give the NP its biological identity. On the other hand, the proteins that attach to the NP only temporarily, depending on the current environment, may not be relevant.
These two different timescales suggest a memory effect on the NP. Exposing the NP to a new environment with different biomolecules could result in a partially new corona composition. Biomolecules that have not been replaced in the protein corona serve as a record of previous environments visited by the NP. This was studied in an earlier work, where it was demonstrated, with experiments and simulations, the memory effect on silica NPs suspended in solution with Human Serum Albumin (HSA) and Transferrin (Tf) using simulations and experiments (Vilanova et al., 2016).
Numerical simulations can provide valuable insights into the multilayer adsorption of proteins in the corona. However, they represent a challenge when compared with the experiments. Indeed, experimental techniques like Fluorescence Correlation Spectroscopy (FCS), used to quantify protein adsorption, typically involve very low concentrations of NPs (Rusu et al., 2004; Milani et al., 2012). At the same time, in biological cases of interest, protein concentrations are often very high, as in the case of SDS-PAGE (Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis) experiments (Lundqvist et al., 2011; Pitek et al., 2012).
These conditions correspond to the worst-case scenario in molecular simulation. Indeed, low NP concentrations require extensive systems, eventually made of a large simulation box containing only a single NP. However, the need for high protein concentrations leads to an exponential increase in the number of proteins, making it demanding to simulate the processes. Furthermore, the problem becomes more challenging when simulation time scales need to match actual experiments’ time frames, typically in minutes or even hours.
To achieve significant results within a specific timescale, it is crucial to select an appropriate simulation scheme, which is determined by the integration time step and the level of coarse-graining used in the simulation technique. Indeed, a full-atom description of thousands of interacting proteins is out of the present reach of even the most powerful computer clusters. Therefore, researchers resort to coarse-grained approaches that reduce the degrees of freedom involved in the biological systems at the nanoscale and simplify the description of the relevant interparticle interactions. Meaningful information can still be obtained using a minimal system description focusing on the most appropriate degrees of freedom.
While a general description of the solvent is essential for many biological processes (March et al., 2021; Durà-Faulí et al., 2023), the effects of hydration on protein interactions can be accounted for using effective potentials, especially when the model’s transferability at different thermodynamic conditions is not crucial. Additionally, employing the Langevin integration scheme for the equations of motion allows for the simulation of the thermal energy contribution from water (Vilanova et al., 2016). In this simplified description, proteins are the primary focus, and their number influences the simulation dimensions.
Better computational performances can optimize numerical simulations, and parallelization is a straightforward way to achieve this goal. Over the past decade, Graphical Processing Units (GPUs) have emerged as a cost-effective and computationally efficient choice for implementing molecular simulations (Walters et al., 2008; Anderson et al., 2008; Harvey et al., 2009; Trott et al., 2010). Hence, the codes we describe and use in the following are developed within the
Specifically, we developed BUBBLES (“BUBBLES is a User-friendly Bundle for Bio-nano Large-scale Efficient Simulations”), a suite of tools designed for simulating the interactions and kinetics of NPs in biological environments, modeled as aqueous solutions containing proteins (Vilanova, 2015). Here, as a case study, we use BUBBLE to analyze the SC kinetics of Tf adsorbing onto polystyrene (PSCOOH) NPs.
Polystyrene is a common polymer used in the production of plastic, such as in packaging materials, food containers, and disposable cups. These NPs have a low density and high surface area-to-volume ratio, making them useful in applications like electronics or biomedical research, despite health concerns arising (Kik et al., 2020).
On the other hand, Tf is one of the most abundant proteins in human plasma (Schenk et al., 2008). It is a glycoprotein with a molecular weight of around 80 kDa, composed of two subunits of equal size joined by a disulfide bridge. Each subunit has a single site for binding iron (Gomme et al., 2005). Iron atoms are absorbed in the intestine, bound to Tf in the plasma, and then transported to storage and utilization sites such as the bone marrow and the liver (Gkouvatsos et al., 2012).
We aim to describe the results observed in experiments involving polystyrene NPs in solutions containing Tf (Milani et al., 2012) and to predict by numerical simulations of our coarse-grained model the short-time dynamics of the SC formation. Next, to validate the model’s applicability in protein-rich environments, we use Differential Centrifugal Sedimentation (DCS) to assess NPs size distribution after exposure to Tf.
2 Materials and methods
When simulating biological systems interacting with nanoscale objects using atomistic simulations, handling many proteins for long simulation time scales is prohibitive. Additionally, a significant amount of resources are needed to simulate the solvent. To address this, we developed a coarse-grain approach that provides a simplified system description.
This method retains all the essential molecular details, making it suitable for studying bio-nano interactions at the mesoscale. Our approach is based on an implicit solvent description, which uses phenomenological parameters and significantly reduces the computational cost.
However, the strong approximations in the model make it not transferable to different thermodynamic conditions or protein-NP combinations. To adjust the model’s parameters, preliminary experiments are necessary to measure the adsorption isotherms of each protein onto the NPs of interest. Nevertheless, these experiments are easily manageable and represent no impediment to calibrating the model’s parameters at the desired thermodynamic condition.
2.1 Experimental details
2.1.1 Materials
Polybead® Carboxylate Microspheres 0.10 µm (PSCOOH, nominally 100 nm) were purchased from Polysciences Inc. (Warrington, United States). The colloidal stability of the nanoparticles was ensured by measuring their size distribution in PBS before protein exposure. Phosphate-Buffered Saline (PBS) tablets, D-(+)-sucrose (99.9%) and Human Transferrin (T8158) were purchased from Sigma Aldrich Ireland. We dissolved one PBS tablet in 200 mL of ultrapure water to obtain a 0.01 M phosphate buffer, 0.0027 M potassium chloride and 0.137 M sodium chloride solution with a pH of 7.4 at 25°C.
2.1.2 Preparation of the NP-Corona complexes
The protein corona samples were prepared using protocols previously developed in the lab (Soliman et al., 2024b). Different volumes of PBS were added to low protein binding 1.5 mL microtubes to make the final solution volume 1 mL. Then, various volumes from a 10 mg/mL Tf stock were added to the microtubes to achieve different experimental concentrations (0, 400, 675, 950, and 1,500 [Tf]/[NP]). After that, 7.5 µL from a stock solution of polystyrene microspheres was added to reach a final NP concentration of 1 mg/mL. The solution was then directly injected into the analytical centrifuge.
2.1.3 NP physico-chemical characterisation
Differential Centrifugal Sedimentation (DCS) experiments were conducted using a CPS Disc Centrifuge DC24000 (Analytik Ltd.) with a sucrose gradient ranging from 8% to 24%. We used polystyrene NPs with a diameter of 0.522 µm to calibrate each sample measurement. The travel time of spherical particles with uniform density from the disk’s center to the detector correlates directly with their size. Variations in arrival times allow for the differentiation between populations, which are considered apparent sizes. We calculated the shell thickness using a core-shell model with a protein layer density estimated to be 1.15 g/cm3 (Perez-Potti et al., 2021). Dynamic Light Scattering (DLS) and Zeta Potential measurements were performed using Zetasizer Nano ZS (Malvern). The sample cuvettes were equilibrated at 25°C for 90 s. For each measurement, the number of runs and duration were automatically determined and repeated three times. Measurements and data analysis were performed according to standard procedures (Soliman et al., 2024b).
2.2 Model
For the case of interest here, we simulate the materials and proteins used by Milani et al. (2012). Specifically, we coarse-grain the NPs as spheres with radius
We represent the Tf as globular proteins described by coarse-grain parameters as in Vilanova et al. (2016). Specifically, Tf has a mass
2.2.1 Particle interactions
We adopt an implicit water approach to account for the water’s effects, with effective interaction potentials between the proteins and the proteins and the NP. Our experiments show that Tf does not aggregate under our experimental conditions. Therefore, we model the effective interaction between two proteins in solution as a soft-sphere repulsive potential, as described in Equation 1
where
As demonstrated in Vilanova et al. (2016), the protein-NP interaction can be described within the framework of the Derjaguin, Landau, Verwey, and Overbeek (DLVO) theory for charged solutes in a solvent, whose stability is controlled by the balance between van der Waals attraction and electrostatic interaction, often leading to a short range electric double-layer repulsion, and a strong contact attraction (Derjaguin and Landau, 1993; Verwey et al., 1948; Agmo Hernández, 2023). In the context of adsorption problems, instead, the interaction energy must become positive at very short distances due to the Born repulsion—not considered in the original DLVO theory—preventing the interpenetration of proteins and NP (Adamczyk and Weronski, 1999).
The resulting potential interaction between NP and Tf is described by Equation 2 as a function of the distance
with Equation 3
and Equation 4
where
Figure 2. Schematic definition of the model. (A) The continuous line represents the protein–NP interaction potential (Equation 2) as a function of the distance between the protein center and the NP surface. The protein is represented as a sphere (light gray) with the Tf hydrodynamics radius. Only a small portion of the NP surface (dark gray) is sketched as a reference. (B) Protein-protein 3-body interaction potential (continuous line, Equation 5 for the case in which one protein (dark gray) is adsorbed onto the NP surface (not shown) and another protein (light gray) is approaching the first from the solution, as a function of the center of mass distance between the two proteins. (C) Snapshot of a portion of the simulation box showing the reaction region (white background) with the NP (large yellow sphere) in the center and proteins (small pink spheres) in the solution. The buffer region (red background) encompasses the reaction region.
Once a complete layer of protein forms on the NP surface, other proteins in the solution cannot interact directly with it due to the limited range of the NP-Tf interaction (Equation 2). However, experimental results suggest that proteins near the NP surface can still interact by forming new layers (Milani et al., 2012). While not all the proteins in the first layer are necessarily part of the HC, characterized by almost irreversible protein adsorption, those in the subsequent layers are typically weakly adsorbed and form the SC (Sharma et al., 2024).
We hypothesize that the aggregation of Tf occurs because of conformational changes in the proteins that are adsorbed within the HC. The rationale behind this hypothesis lies in the observation that the Tf is not prone to aggregation when it is suspended in solution under the specific thermodynamic conditions we are considering. This is consistent with the repulsive protein-protein interaction we use in Equation 1. Therefore, the emerging attractive force between free Tf and HC proteins must be due to the effects on the HC proteins caused by the surface of the NPs.
Specifically, when Tf is folded in solution, its hydrophobic amino acids are mainly shielded from water, as occurs for other structured mesophilic proteins (Bianco et al., 2017). Still, Tf can undergo partial unfolding when tightly bound to the NP, as it has been reported for many other proteins forming a corona (Park, 2020), exposing residues with a significant affinity toward other Tf proteins in the solution. As a result, proteins on the surface are highly likely to attract other proteins to minimize the overall free energy of the system. We assume that this process depends on how strongly each Tf binds to the NP via the Eq (ef DLVO), i.e., it depends on the Tf-NP surface distance.
Hence, we describe this attractive interaction between two Tf proteins
where the characteristic interaction energy between proteins,
The second term in Equation 5 is a local attractive Gaussian well potential, centered at
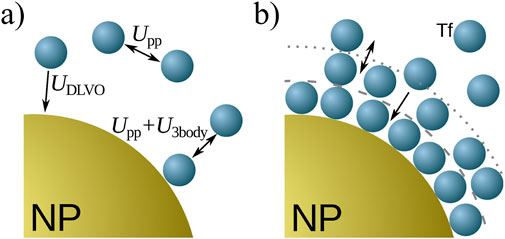
Figure 3. Schematic summary of the model. (A) Interactions among proteins (blue spheres) and the NP (golden sphere). The interaction potentials are given by Equations 1–5. (B) The corona structure. Protein layers form due to the attraction between proteins near the NP, mimicking their structural changes (not shown) once in the corona.
2.3 Computational details
The code we use here is available online as the simulation package BUBBLES (Vilanova, 2015) and was initially introduced by Vilanova et al. in Vilanova et al. (2016). Within this work, we provide a user-friendly tutorial, available on the reference website to use BUBBLES to study HC and SC formation for NPs of different chemistry in contact with solutions including one or more proteins.
We simulate systems at different Tf concentrations,
The NP concentration remains constant at
2.3.1 Langevin dynamics
We use Langevin dynamics to simulate protein diffusion. Langevin dynamics extend the Newtonian equations of motion to account for the effects of a surrounding molecular environment, such as particles in a solvent, with an implicit description (Schneider and Stoll, 1978). The set of equations of motion of a given protein
where
is the result of the interparticle forces described in Equation 8
acting on the protein
Two additional terms appear in the inertial part of the system to account for the proteins’ interaction with the solvent. The first is the viscous force
2.3.2 Buffer implementation
In the experiments, the concentration of NPs,
The main idea is to use a particle reservoir to regulate protein concentration within the area of interest. A similar approach involves conducting simulations in the macrocanonical ensemble, using the chemical potential to control the concentration (Oberholzer et al., 1997). Our method does not require particle insertion/deletion events, which can be inefficient at high concentrations. Additionally, this method allows us to simulate a system with a constant number of particles, which is very convenient, especially for GPU-based numerical simulations, due to device memory management restrictions.
We divide the simulation box into two centered cubic regions: an inner region of volume
During the simulation, we calculate the difference
between the concentration
If
3 Results and discussion
3.1 Fraction bound and model calibration
The experiments by Milani et al. were carried out at the NP concentration of 0.1 mg/mL and a [Tf]/[NP] of up to approximately 1,000 (Milani et al., 2012). Even with our coarse-grained approach, the low NP and high protein concentration combination made numerical simulations impractical. We perform simulations at various higher NP concentrations to address this issue and then extrapolate the results to match the experimental conditions.
The majority of the parameters of our model are given by the experimental setup and discussed in Section. 2.2. Those for Equations 2–4 are set from the adsorption isotherms of a Tf monolayer, as discussed in Vilanova et al. (2016). To evaluate the phenomenological parameters for the interaction relevant to the SC (Equation 5), we set the NP concentration
where
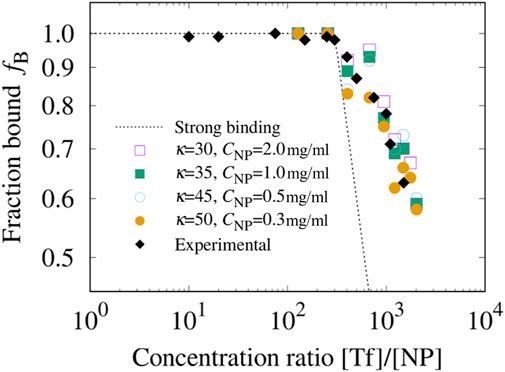
Figure 4. Experimental Tf fraction bound polystyrene NPs compared with the model’s calculations. We use the experimental fraction bound (black diamonds) as a function of the relative concentration [Tf]/[NP] in Milani et al. (2012). to calibrate the model’s parameter
As long as
In the absence of a soft corona, for [Tf]/[NP]
Our model reproduces this behavior associated with the formation of the SC. Once a layer is formed, the next layer can only form at a greater distance from the NP surface. This means that the interaction between proteins, decreases as their average distance from the surface increases. As a result, the new layer will attract fewer proteins, leading to a decrease in the slope of
We find that for
We observe that the estimates for
where
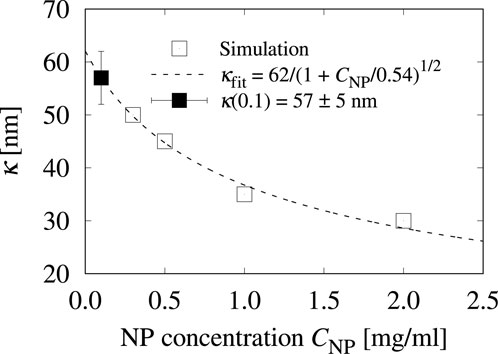
Figure 5. The three-body characteristic distance
It is important to note that
On the other hand, the parameter
3.2 Protein corona density profile
First, we examine the structure of the protein corona in simulated solutions at relative concentrations [Tf]/[NP]
We calculate the local density of proteins within distances
where
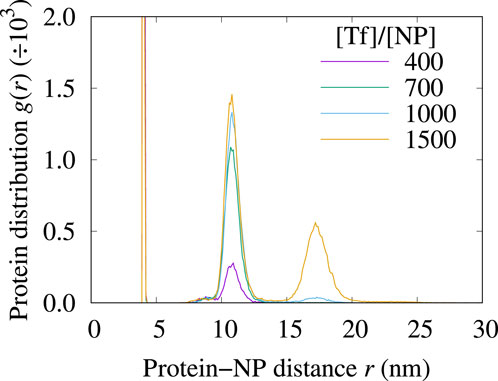
Figure 6. The density profiles
In this range of [Tf]/[NP] values, the RDF displays up to 3 peaks corresponding to the different protein corona layers. The first peak is centered at
The second layer, at approximately
The third peak, centered around
We do not observe a saturation of the
3.3 Protein corona dynamics and glassy behavior
3.3.1 Irreversible and reversible adsorption
Next, we investigate the adsorption kinetics of Tf onto the NP. To validate the interpretation from the RDF analysis of the three layers as hard and soft corona, we track the positions of the proteins within them (Figure 7).
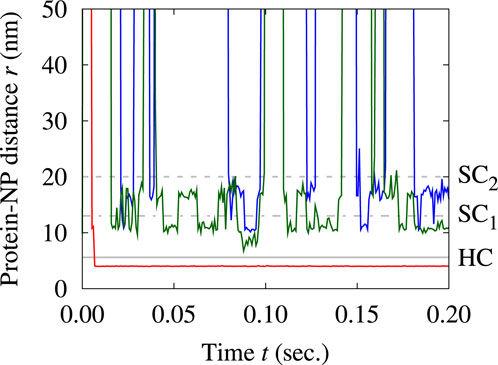
Figure 7. Evolution of the distance from the NP surface of three single proteins over time. We select three proteins among those that spend most of their time within the first (red track), the second (green track), and the third layer (blue track). The gray lines mark the largest distances for each layer, as defined in Figure 6: continuous at 5.6 nm for HC, dotted-dashed at 13 nm for
The tracking confirms the strong adsorption of proteins in the first layer, as they show no signs of displacement once adsorbed. On the other hand, proteins in the other two layers are constantly exchanged with the suspension. They can detach from the protein corona, return to the protein solution, and eventually get reabsorbed, regardless of whether they are in the second or third layer. Our simulations suggest that the proteins in the second layer have longer residence times than those in the third layer, indicating higher stability of the inner layer of the soft corona than the outer layer.
Proteins in the outermost layer sometimes interchange with those in the second layer, but we do not find exchanges of any proteins in the two outer layers with the first layer. All these observations consistently associate the innermost layer with the HC and the two outermost layers with the SC, validating the conclusions drawn from the RDF analysis.
Further understanding can be reached by analyzing the time evolution of the number
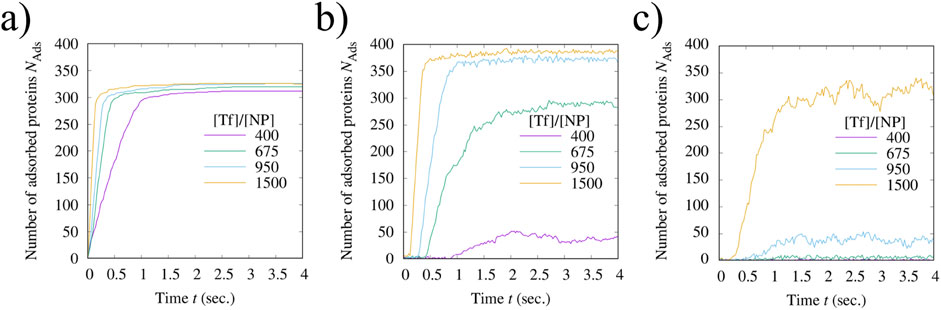
Figure 8. Simulation results for the number of Tf proteins adsorbed into the corona of a polystyrene NP over time at four different relative concentrations [Tf]/[NP]. The time evolution of this number varies among (A) the first, (B) the second, and (C) the third layer, with larger fluctuations for the outer layers, consistent with our interpretation of the first as the HC and the others as the SC. In each panel, the considered values of [Tf]/[NP] are 400 (indigo lines), 675 (green lines), 950 (turquoise lines), and 1,500 (orange lines). The simulation conditions are as in Figure 6.
In the second layer (Figure 8B),
These fluctuations increase, instead, in the third layer (Figure 8C), being consistently higher than in the second layer, indicating that the corona gains stability closer to the NP surface. The
3.3.2 The soft corona glassy behavior
To better characterize the soft corona dynamics, we analyze the autocorrelation function with Equation 13 for the proteins populating the
where
The two SC layers exhibit distinct behaviors, as shown in Figure 9. The
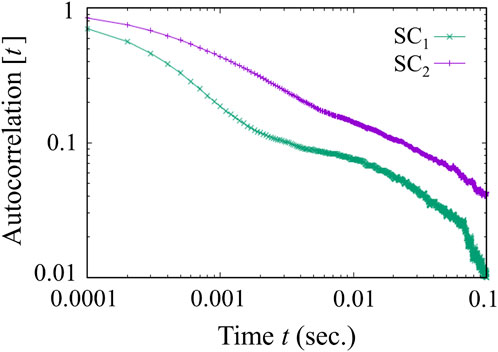
Figure 9. Density autocorrelation function of the soft corona over time. While the outer layer
Conversely, the inner layer of the soft corona,
The relaxation behavior in the SC’s inner layer implies that the corona’s dynamic evolution in this volume is affected by factors such as crowding from other proteins within the corona. These factors can restrict protein mobility, leading to the development of dynamically frozen protein clusters or aggregates, which contribute to the relaxation behavior observed.
3.4 Model validation
To experimentally validate our in silico model for the protein soft corona simulation, we use DCS to evaluate the size distribution of polystyrene NPs before and after exposure to Tf at varying concentrations. Unlike the reference experiment for our modeling, (Milani et al. (2012), our 100 nm PSCOOH polystyrene NPs exhibit an effective radius that closely matches the nominal radius. Specifically, in pristine conditions, our DCS data reveal a sharp, monodisperse distribution of NPs with an average diameter of
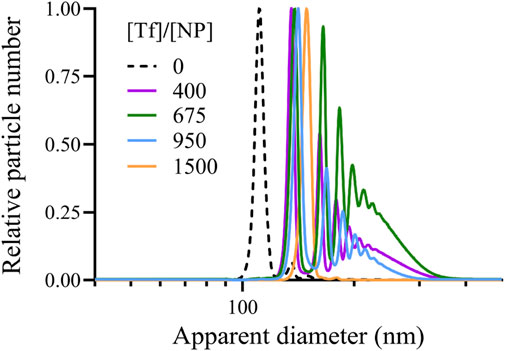
Figure 10. DCS measurements of the (relative) number of polystyrene NPs as a function of their apparent diameter after exposure to Tf proteins at relative concentration [Tf]/[NP]. We find one peak at large [Tf]/[NP] marking the apparent diameter and a main peak—corresponding to a single NP-corona complex diameter—followed by secondary maxima at lower [Tf]/[NP] due to a larger tendency to form clusters under these diluted conditions. [Tf]/[NP] goes from 0 (dashed line) to 400 (indigo line), 675 (green line), to 950 (blue line) to 1,500 (yellow line).
We observe only one peak in the DCS measurements at [Tf]/[NP]
To make a quantitative comparison of the simulation predictions with the experimental measurements, we estimate for each condition the average number
The experiments show a satisfactory quantitative agreement with the predictions based on the simulations of our coarse-grained model (Figure 11). We find that the model slightly underestimates or overestimates
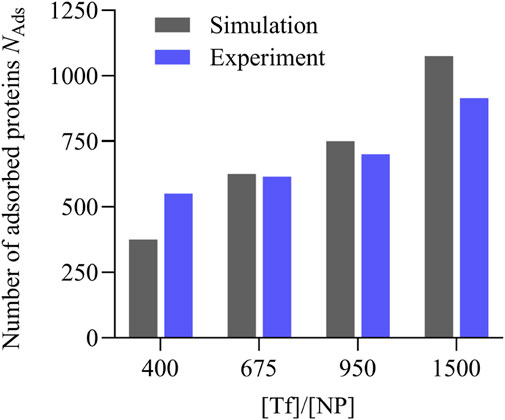
Figure 11. Comparison between the simulation predictions and the experimental results for the number of adsorbed Tf proteins per polystyrene NP within the hard and soft corona under different [Tf]/[NP] conditions. The simulation predictions (gray bars) are estimated from the large-time averages of the sets in Figure 8, and the experimental values (blue bars) are taken from Table 1.
4 Discussion, conclusion, and perspectives
When NPs come into contact with biological fluids, biomolecules adhere to their surfaces, forming a corona composed of multiple layers that influence how these NPs interact with cells and their biological effects. In protein-rich environments, such as blood or serum, strong protein-NP interactions form a hard corona. In contrast, weaker protein-protein and protein-NP interactions give rise to a soft corona. The hard corona is characterized by proteins that adsorb irreversibly onto the NP, whereas the soft corona consists of a fluid layer of loosely bound proteins associated with the NP. Understanding the dynamics of each component of the protein corona is essential for various biological applications of nanotechnology (Sharma et al., 2024).
However, measuring the soft corona poses significant experimental challenges (Guo et al., 2024). In-situ detection methods, which aim to preserve the protein corona in its physiological environment, demand advanced technical skills and involve complex procedures, such as tagging proteins with fluorescent markers and using dynamic light scattering. These modifications risk altering protein binding capacities, thereby complicating accurate measurements. Ex-situ methods, such as ultraviolet-visible spectroscopy and liquid chromatography-mass spectrometry, struggle with the incomplete separation of NP-protein complexes, which can lead to misidentifying the protein corona constituents. Additionally, excessive centrifugal force during separation may result in the loss of protein corona components, further complicating accurate detection.
Therefore, the composition of the protein corona has traditionally been examined, either in vivo or in vitro, through static incubation methods. Nevertheless, this approach limits our ability to explore the dynamics of the corona components over biologically relevant timescales. Here, we present new findings for a computational method that enables us to integrate experimental results and comprehensively describe the dynamics of both the hard and soft corona.
We extend the modeling and computational approach we previously introduced, incorporating the study of the soft corona into analyzing the short- and long-term kinetics of the hard corona formed by different proteins competing for the NP surface (Vilanova et al., 2016). The method employs a coarse-grained description of globular proteins interacting with the NP through colloid-like effective potential energies. This approach allows for molecular simulations over time scales on the order of seconds within the framework of Langevin dynamics once a few phenomenological parameters are extracted from preliminary experiments with monocomponent protein solutions. These simulation predictions can then be extended to any time scale and validated against laboratory results, utilizing a non-Langmuir adsorption theory introduced in Vilanova et al. (2016).
The extension introduced here incorporates a three-body interaction between proteins and the NP, mimicking the denaturation effect that the nanosurface can induce on proteins (Park, 2020). We assume that this effect exponentially decays as the distance between the NP’s surface and any interacting proteins increases. The alignment of our simulation results with experimental data validates our model a posteriori.
As a case study, we consider the experimental conditions described in Milani et al. (2012), which evaluated the formation of the hard and soft corona by Tf proteins in suspension with polystyrene NPs. The experimental setup presents challenges due to the highly diluted concentrations of NPs and proteins, necessitating macroscopically large systems that are computationally prohibitive to simulate. To address this, we develop a scaling strategy to replicate the overall experimental behavior in smaller systems with higher concentrations and effective parameters. Using as an example the parameter describing the exponential decay of the three-body protein-protein-NP interaction, we show how this strategy allows us to extrapolate the quantity of interest in the limit of the experimental conditions.
After establishing the model, we used it to investigate the structure and kinetics of the corona. Our structural analysis reveals that the corona is composed of three distinct layers. The layer closest to the NP’s surface is the sharpest, while the outermost layer is the broadest. The first and second layers become populated at low and moderate protein concentrations, whereas the third layer forms only at the highest concentrations.
The protein dynamics in each layer reveals that the first layer, which is in direct contact with the NP, contains proteins tightly bound to the surface. These proteins adsorb irreversibly over the simulation time scales, forming the hard corona. In contrast, the proteins in the second and third layers are reversibly bound to the corona, constituting the soft corona.
Our study shows that the first (HC) and second (
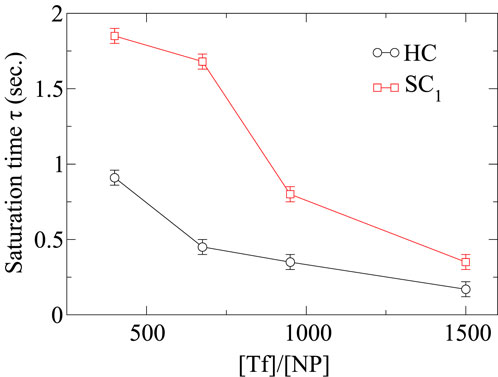
Figure 12. Saturation times
The in-depth analysis of the dynamics of the soft corona reveals that
We validate our model predictions by directly estimating, under similar experimental conditions, the number of Tf proteins adsorbed onto 100 nm PSCOOH polystyrene NPs. Despite the differences, mainly related to the nominal size of the NPs, our experiments show that the computational model can predict within a 10% error the number of adsorbed proteins at the different relative concentrations [Tf]/[NP] considered here. We observe that the model is, overall, predicting a faster-increasing number of proteins adsorbed with the increase of the [Tf]/[NP] ratio relative to the experiments. This difference is possibly due to the agglomeration of the NPs observed in the experiment at increasing [Tf]/[NP]. The clustering of more than one NP-corona complex could 1) decrease the surface area available for protein binding and 2) lead to an experimental underestimation in the number of adsorbed Tf due to the density value used for Tf.
Lastly, the computational model does not consider Tf structural changes upon adsorption. Instead, recent findings from cryo-TEM corona show a uniform corona layer rather than a packing of globular proteins, suggesting structural deformation of the protein structure (Sheibani et al., 2021). This deformation could correspond to the protein flattening out, which would reduce the number of adsorbed Tf.
The overall conclusion is that the close correlation between the experimental and computational models in the present case study demonstrates satisfactory performance in protein-rich environments, indicating the applicability of our approach to biologically relevant conditions. For this reason, to benefit the scientific community, we provide an open-source interactive tutorial with all the steps required to perform the simulations, defining and implementing a buffer of molecules capable of controlling the concentration of proteins in the vicinity of the NP (Vilanova, 2015).
Future research in this field of NP-corona interactions with biological systems should expand beyond the limited scope of studying the competition among just a few types of proteins. Biological media, such as blood plasma, are incredibly complex and contain over 3,700 identified proteins, leading to a highly competitive environment for the formation of the corona. This phenomenon, known as the Vroman effect, highlights the dynamic nature of protein adsorption, where proteins with higher mobility and lower affinity initially occupy the NP surface, only to be replaced by proteins with higher affinity over time.
In addition to proteins, several other molecules in whole blood can contribute to the formation of the NP corona (Lundqvist et al., 2017), including lipids, carbohydrates, nucleic acids, metabolites, complement factors, and antibodies (Soliman et al., 2024a). For example, lipids such as cholesterol and phospholipids, glycans—which are carbohydrate molecules that play a role in cell recognition and signaling—or small metabolites like glucose, hormones, and vitamins can adsorb onto NPs, altering their surface properties and influencing their interactions with cells (Singh et al., 2021).
On the other hand, circulating DNA is present in the blood of all individuals (Van Der Vaart and Pretorius, 2008), and its concentration increases in cancer patients (Jahr et al., 2001). Furthermore, depending on the state of different diseases, blood plasma also contains a variety of RNA types, including mRNAs, noncoding RNAs, and fragments of rRNAs, snoRNAs, and miRNAs (Savelyeva et al., 2017; Semenov et al., 2008), suggesting that the NP-corona composition also depends on the health condition of the host.
Moreover, complement factors that are part of the immune system and immunoglobulins or antibodies, which are glycoproteins produced by plasma cells, can mark the NP for clearance by immune cells (Singh et al., 2021). Therefore, it will be critical in the future to understand how all these components collectively form the biomolecular hard and soft corona, which significantly impacts the biological identity and fate of NPs in the body (Soliman et al., 2024a).
To gain a comprehensive understanding of NP behavior in biological environments, it will be crucial to investigate how these biomolecules operate under healthy or disease conditions. This approach will provide insights into real-world scenarios and how stressful conditions influence the NPs’ biological identity and subsequent interactions with cells and tissues.
Moreover, the study of the dynamics and aging behavior of the biomolecular corona will be essential. As discussed here, the corona’s composition and structure can change over time, exhibiting glassy behavior characterized by slow dynamics and structural rearrangements. Understanding these processes is vital for predicting the fate and stability of NPs in biological environments. The glassy state of the corona may play a significant role in determining the long-term interactions of NPs with biological systems, impacting their efficacy and safety in medical applications.
Considering the corona’s glassy states is particularly important for the development of stable pharmaceuticals. Ensuring that the corona remains properly formulated when NPs are immersed in biological fluids is crucial for maintaining their intended function and avoiding unintended side effects. This knowledge can guide the design of NP-based drug delivery systems, enhancing their stability and performance in the complex biological milieu.
From a computational perspective, the scientific community will need to develop new approaches to account for the interaction of NP-corona complexes with cells, particularly focusing on their interaction with cell membranes. Recent findings indicate that the membrane interface is more complex than traditionally thought (Martelli et al., 2021). This interface extends further than previously suggested, including several water layers up to 2.5 nm (Martelli et al., 2018), and possesses an internal structure composed of both bound and unbound water (Calero and Franzese, 2019). This structure arises from the specific hydrogen-bond network of the hydration water (Bianco et al., 2012).
Current coarse-grain membrane models do not incorporate this hydrogen-bond network (Yesylevskyy et al., 2010). As a result, they fail to replicate the dynamic and thermodynamic anomalies of water (de los Santos and Franzese, 2011), which are crucial to understanding the physics of water (Leoni et al., 2021) and proteins (Bianco et al., 2017). This limitation hinders the study of interactions between the biomolecules forming the corona and the cell membrane.
Progress in this area has been made by developing a quantitatively accurate model of water under life-relevant conditions (Coronas and Franzese, 2024; Coronas et al., 2024). This model is reliable, efficient, scalable, and transferable, meeting the requirements for biological simulations. Such advancements are essential for accurately simulating the complex interactions at the NP-corona and cell membrane interface.
In summary, future studies should adopt a holistic approach that considers the vast array of molecules present in biological media and the competitive interactions that occur on NP surfaces and within the soft corona. Additionally, a deeper understanding of the dynamic and aging behaviors of the biomolecular corona, as well as the water-mediated interaction with the cell membrane, will be instrumental in advancing NP applications in medicine, leading to the development of more effective and safer nanotherapeutics.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Author contributions
OV: Data curation, Formal Analysis, Investigation, Methodology, Software, Validation, Visualization, Writing–original draft. AM-S: Formal Analysis, Investigation, Visualization, Writing–original draft, Writing–review and editing. MM: Conceptualization, Funding acquisition, Methodology, Supervision, Writing–review and editing. GF: Conceptualization, Formal Analysis, Funding acquisition, Methodology, Project administration, Supervision, Writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. OV acknowledges financial support from IN2UB. AM-S and MM acknowledge the support from H2020 grant no. 952924 (SUNSHINE). GF acknowledges the support by a) MCIN/AEI/10.13039/501100011033 and “ERDF A way of making Europe” grant number PID 2021-124297NB-C31, b) the Ministry of Universities 2023-2024 Mobility Subprogram within the Talent and its Employability Promotion State Program (PEICTI 2021-2023), and c) the Visitor Program of the Max Planck Institute for The Physics of Complex Systems for supporting a visit started in November 2022. All authors acknowledge the support of NVIDIA Corporation’s Applied Research Accelerator Program for granting an RTX A5000 GPU used for the calculations presented here. NVIDIA Corporation was not involved in the study design, collection, analysis, interpretation of data, the writing of this article, or the decision to submit it for publication.
Acknowledgments
OV and GF thank Alejandro Rodríguez Ruiz and Delia López Jareño for preliminary tests of the simulation package BUBBLES.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
1We checked that in mono-component protein solutions of Tf our results have no qualitative dependence on small variation of the NP radius by preliminary calculations for
2For
References
Adamczyk, Z., and Weronski, P. (1999). Application of the DLVO theory for particle deposition problems. Adv. Colloid Interface Sci. 83, 137–226. doi:10.1016/S0001-8686(99)00009-3
Agmo Hernández, V. (2023). An overview of surface forces and the dlvo theory. ChemTexts 9, 10. doi:10.1007/s40828-023-00182-9
Anderson, J. a., Lorenz, C. D., and Travesset, A. (2008). General purpose molecular dynamics simulations fully implemented on graphics processing units. J. Comput. Phys. 227, 5342–5359. doi:10.1016/j.jcp.2008.01.047
Bai, X., Wang, J., Mu, Q., and Su, G. (2021). In vivo protein corona formation: characterizations, effects on engineered nanoparticles’ biobehaviors, and applications. Front. Bioeng. Biotechnol. 9, 646708. doi:10.3389/fbioe.2021.646708
Baimanov, D., Wang, J., Zhang, J., Liu, K., Cong, Y., Shi, X., et al. (2022). In situ analysis of nanoparticle soft corona and dynamic evolution. Nat. Commun. 13, 5389. doi:10.1038/s41467-022-33044-y
Bianco, V., Franzese, G., Dellago, C., and Coluzza, I. (2017). Role of water in the selection of stable proteins at ambient and extreme thermodynamic conditions. Phys. Rev. X 7, 021047. doi:10.1103/PhysRevX.7.021047
Bianco, V., Iskrov, S., and Franzese, G. (2012). Understanding the role of hydrogen bonds in water dynamics and protein stability. J. Biol. Phys. 38, 27–48. doi:10.1007/s10867-011-9235-7
Bilardo, R., Traldi, F., Vdovchenko, A., and Resmini, M. (2022). Influence of surface chemistry and morphology of nanoparticles on protein corona formation. WIREs Nanomedicine Nanobiotechnology 14, e1788. doi:10.1002/wnan.1788
Boselli, L., Castagnola, V., Armirotti, A., Benfenati, F., and Pompa, P. P. (2024). Biomolecular corona of gold nanoparticles: the urgent need for strong roots to grow strong branches. Small 20, 2306474. doi:10.1002/smll.202306474
Brooks, C. L., and Karplus, M. (1983). Deformable stochastic boundaries in molecular dynamics. J. Chem. Phys. 79, 6312–6325. doi:10.1063/1.445724
Brooks, C. L., and Karplus, M. (1989). Solvent effects on protein motion and protein effects on solvent motion. J. Mol. Biol. 208, 159–181. doi:10.1016/0022-2836(89)90093-4
Brünger, A., Brooks, C. L., Karplus, M., and Brunger, A. (1984). Stochastic boundary conditions for molecular dynamics simulations of ST2 water. Chem. Phys. Lett. 105, 495–500. doi:10.1016/0009-2614(84)80098-6
Calero, C., and Franzese, G. (2019). Membranes with different hydration levels: the interface between bound and unbound hydration water. J. Mol. Liq. 273, 488–496. doi:10.1016/j.molliq.2018.10.074
Cao, M., Cai, R., Zhao, L., Guo, M., Wang, L., Wang, Y., et al. (2021). Molybdenum derived from nanomaterials incorporates into molybdenum enzymes and affects their activities in vivo. Nat. Nanotechnol. 16, 708–716. doi:10.1038/s41565-021-00856-w
Cedervall, T., Lynch, I., Foy, M., Berggård, T., Donnelly, S. C., Cagney, G., et al. (2007a). Detailed identification of plasma proteins adsorbed on copolymer nanoparticles. Angew. Chem. - Int. Ed. 46, 5754–5756. doi:10.1002/anie.200700465
Cedervall, T., Lynch, I., Lindman, S., Berggård, T., Thulin, E., Nilsson, H., et al. (2007b). Understanding the nanoparticle-protein corona using methods to quantify exchange rates and affinities of proteins for nanoparticles. Proc. Natl. Acad. Sci. U. S. A. 104, 2050–2055. doi:10.1073/pnas.0608582104
Corbo, C., Molinaro, R., Parodi, A., Toledano Furman, N. E., Salvatore, F., and Tasciotti, E. (2016). The impact of nanoparticle protein corona on cytotoxicity, immunotoxicity and target drug delivery. Nanomedicine 11, 81–100. doi:10.2217/nnm.15.188
Coronas, L. E., and Franzese, G. (2024). Phase behavior of metastable water from large-scale simulations of a quantitatively accurate model near ambient conditions: the liquid–liquid critical point. J. Chem. Phys. 161, 164502. doi:10.1063/5.0219313
Coronas, L. E., Vilanova, O., and Franzese, G. (2024). A transferable molecular model for accurate thermodynamic studies of water in large-scale systems. A transferable Mol. Model accurate Thermodyn. Stud. water large-scale Syst. doi:10.21203/rs.3.rs-4243098/v1
Dawson, K. A., and Yan, Y. (2021). Current understanding of biological identity at the nanoscale and future prospects. Nat. Nanotechnol. 16, 229–242. doi:10.1038/s41565-021-00860-0
de los Santos, F., and Franzese, G. (2011). Understanding diffusion and density anomaly in a coarse-grained model for water confined between hydrophobic walls. J. Phys. Chem. B 115, 14311–14320. doi:10.1021/jp206197t
Deng, Z. J., Liang, M., Monteiro, M., Toth, I., and Minchin, R. F. (2011). Nanoparticle-induced unfolding of fibrinogen promotes mac-1 receptor activation and inflammation. Nat. Nanotechnol. 6, 39–44. doi:10.1038/nnano.2010.250
Derjaguin, B., and Landau, L. (1993). Theory of the stability of strongly charged lyophobic sols and of the adhesion of strongly charged particles in solutions of electrolytes. Prog. Surf. Sci. 43, 30–59. doi:10.1016/0079-6816(93)90013-L
Durà-Faulí, B., Bianco, V., and Franzese, G. (2023). Hydrophobic homopolymer’s coil–globule transition and adsorption onto a hydrophobic surface under different conditions. J. Phys. Chem. B 127, 5541–5552. doi:10.1021/acs.jpcb.3c00937
Freestone, I., Meeks, N., Sax, M., and Higgitt, C. (2007). The lycurgus cup—a roman nanotechnology. Gold Bull. 40, 270–277. doi:10.1007/bf03215599
García-Álvarez, R., and Vallet-Regí, M. (2021). Hard and soft protein corona of nanomaterials: analysis and relevance. Nanomaterials 11, 888. doi:10.3390/nano11040888
Gkouvatsos, K., Papanikolaou, G., and Pantopoulos, K. (2012). Regulation of iron transport and the role of transferrin. Biochimica Biophysica Acta (BBA)-General Subj. 1820, 188–202. doi:10.1016/j.bbagen.2011.10.013
Gomme, P. T., McCann, K. B., and Bertolini, J. (2005). Transferrin: structure, function and potential therapeutic actions. Drug Discov. today 10, 267–273. doi:10.1016/s1359-6446(04)03333-1
Gotze, W., and Sjogren, L. (1992). Relaxation processes in supercooled liquids. Rep. Prog. Phys. 55, 241–376. doi:10.1088/0034-4885/55/3/001
Guo, F., Luo, S., Wang, L., Wang, M., Wu, F., Wang, Y., et al. (2024). Protein corona, influence on drug delivery system and its improvement strategy: a review. Int. J. Biol. Macromol. 256, 128513. doi:10.1016/j.ijbiomac.2023.128513
Harvey, M. J., Giupponi, G., and De Fabritiis, G. (2009). ACEMD: accelerating biomolecular dynamics in the microsecond time scale. J. Chem. Theory Comput. 5, 1632–1639. doi:10.1021/ct9000685
Jahr, S., Hentze, H., Englisch, S., Hardt, D., Fackelmayer, F. O., Hesch, R. D., et al. (2001). Dna fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 61 (4), 1659–1665.
Jareño, L., and Delia, F. G. (2015). Numerical and theoretical study of nano-bio interactions. Unpublished.
Kik, K., Bukowska, B., and Sicińska, P. (2020). Polystyrene nanoparticles: sources, occurrence in the environment, distribution in tissues, accumulation and toxicity to various organisms. Environ. Pollut. 262, 114297. doi:10.1016/j.envpol.2020.114297
Kreuter, J., Shamenkov, D., Petrov, V., Ramge, P., Cychutek, K., Koch-Brandt, C., et al. (2002). Apolipoprotein-mediated transport of nanoparticle-bound drugs across the blood-brain barrier. J. Drug Target. 10, 317–325. doi:10.1080/10611860290031877
Kumar, P., Franzese, G., Buldyrev, S. V., and Stanley, H. E. (2006). Molecular dynamics study of orientational cooperativity in water. Phys. Rev. E 73, 041505. doi:10.1103/PhysRevE.73.041505
Kumari, P., Ghosh, B., and Biswas, S. (2016). Nanocarriers for cancer-targeted drug delivery. J. Drug Target. 24, 179–191. doi:10.3109/1061186X.2015.1051049
LeDuc, C. A., Vroman, L., and Leonard, E. F. (1995). A mathematical model for the vroman effect. Ind. Eng. Chem. Res. 34, 3488–3495. doi:10.1021/ie00037a037
Leoni, F., Calero, C., and Franzese, G. (2021). Nanoconfined fluids: uniqueness of water compared to other liquids. ACS Nano 15, 19864–19876. doi:10.1021/acsnano.1c07381
Lundqvist, M., Augustsson, C., Lilja, M., Lundkvist, K., Dahlbäck, B., Linse, S., et al. (2017). The nanoparticle protein corona formed in human blood or human blood fractions. PLOS ONE 12, 01758711–e175915. doi:10.1371/journal.pone.0175871
Lundqvist, M., Stigler, J., Cedervall, T., Berggård, T., Flanagan, M. B., Lynch, I., et al. (2011). The evolution of the protein corona around nanoparticles: a test study. ACS Nano 5, 7503–7509. doi:10.1021/nn202458g
Lundqvist, M., Stigler, J., Elia, G., Lynch, I., Cedervall, T., and Dawson, K. A. (2008). Nanoparticle size and surface properties determine the protein corona with possible implications for biological impacts. Proc. Natl. Acad. Sci. 105, 14265–14270. doi:10.1073/pnas.0805135105
Lynch, I., and Dawson, K. A. (2008). Protein-nanoparticle interactions. Nano Today 3, 40–47. doi:10.1016/S1748-0132(08)70014-8
Lynch, I., Salvati, A., and Dawson, K. A. (2009). Protein-nanoparticle interactions: what does the cell see? Nat. Nanotechnol. 4, 546–547. doi:10.1038/nnano.2009.248
Madathiparambil Visalakshan, R., González García, L. E., Benzigar, M. R., Ghazaryan, A., Simon, J., Mierczynska-Vasilev, A., et al. (2020). The influence of nanoparticle shape on protein corona formation. Small 16, 2000285. doi:10.1002/smll.202000285
March, D., Bianco, V., and Franzese, G. (2021). Protein unfolding and aggregation near a hydrophobic interface. Polymers 13, 156. doi:10.3390/polym13010156
Martelli, F., Calero, C., and Franzese, G. (2021). Redefining the concept of hydration water near soft interfaces. Biointerphases 16, 020801. doi:10.1116/6.0000819
Martelli, F., Ko, H.-Y., Borallo, C. C., and Franzese, G. (2018). Structural properties of water confined by phospholipid membranes. Front. Phys. 13, 136801. doi:10.1007/s11467-017-0704-8
Milani, S., Bombelli, F. B., Pitek, A. S., Dawson, K. A., and Rädler, J. O. (2012). Reversible versus irreversible binding of transferrin to polystyrene nanoparticles: soft and hard corona. ACS Nano 6, 2532–2541. doi:10.1021/nn204951s
Mohammad-Beigi, H., Hayashi, Y., Zeuthen, C. M., Eskandari, H., Scavenius, C., Juul-Madsen, K., et al. (2020). Mapping and identification of soft corona proteins at nanoparticles and their impact on cellular association. Nat. Commun. 11, 4535. doi:10.1038/s41467-020-18237-7
Monopoli, M. P., Åberg, C., Salvati, A., and Dawson, K. A. (2012). Biomolecular coronas provide the biological identity of nanosized materials. Nat. Nanotechnol. 7, 779–786. doi:10.1038/nnano.2012.207
Oberdörster, G., Oberdörster, E., and Oberdörster, J. (2005). Nanotoxicology: an emerging discipline evolving from studies of ultrafine particles. Environ. Health Perspect. 113, 823–839. doi:10.1289/ehp.7339
Oberholzer, M. R., Wagner, N. J., and Lenhoff, A. M. (1997). Grand canonical Brownian dynamics simulation of colloidal adsorption. J. Chem. Phys. 107, 9157–9167. doi:10.1063/1.475207
Park, S. J. (2020). Protein–nanoparticle interaction: corona formation and conformational changes in proteins on nanoparticles. Int. J. Nanomedicine 15, 5783–5802. doi:10.2147/IJN.S254808
Perez-Potti, A., Lopez, H., Pelaz, B., Abdelmonem, A., Soliman, M. G., Schoen, I., et al. (2021). In depth characterisation of the biomolecular coronas of polymer coated inorganic nanoparticles with differential centrifugal sedimentation. Sci. Rep. 11, 6443. doi:10.1038/s41598-021-84029-8
Pitek, A. S., O’Connell, D., Mahon, E., Monopoli, M. P., Bombelli, F. B., and Dawson, K. A. (2012). Transferrin coated nanoparticles: study of the bionano interface in human plasma. PLoS One 7, e40685. doi:10.1371/journal.pone.0040685
Qiu, C., Zhang, Z., Xiao, M., Yang, Y., Zhong, D., and Peng, L.-M. (2017). Scaling carbon nanotube complementary transistors to 5-nm gate lengths. Science 355, 271–276. doi:10.1126/science.aaj1628
Rusu, L., Gambhir, A., McLaughlin, S., and Rädler, J. (2004). Fluorescence correlation spectroscopy studies of Peptide and protein binding to phospholipid vesicles. Biophys. J. 87, 1044–1053. doi:10.1529/biophysj.104.039958
Salvati, A. (2024). The biomolecular corona of nanomedicines: effects on nanomedicine outcomes and emerging opportunities. Curr. Opin. Biotechnol. 87, 103101. doi:10.1016/j.copbio.2024.103101
Sanchez-Guzman, D., Giraudon-Colas, G., Marichal, L., Boulard, Y., Wien, F., Degrouard, J., et al. (2020). In situ analysis of weakly bound proteins reveals molecular basis of soft corona formation. ACS Nano 14, 9073–9088. doi:10.1021/acsnano.0c04165
Savelyeva, A. V., Kuligina, E. V., Bariakin, D. N., Kozlov, V. V., Ryabchikova, E. I., Richter, V. A., et al. (2017). Variety of rnas in peripheral blood cells, plasma, and plasma fractions. BioMed Res. Int. 2017, 1–10. doi:10.1155/2017/7404912
Savolainen, K., Backman, U., Brouwer, D., Fadeel, B., and Fernandes, T. (2013). Nanosafety in Europe 2015-2025: towards safe and sustainable nanomaterials and nanotechnology innovations nanosafety in Europe towards safe and sustainable nanomaterials and nanotechnology innovations (Finnish institue of occupational health). Helsinki: Finnish Institute of Occupational Health. doi:10.13140/2.1.3084.8969
Schenk, S., Schoenhals, G. J., de Souza, G., and Mann, M. (2008). A high confidence, manually validated human blood plasma protein reference set. BMC Med. genomics 1, 41–28. doi:10.1186/1755-8794-1-41
Schneider, T., and Stoll, E. (1978). Molecular-dynamics study of a three-dimensional one-component model for distortive phase transitions. Phys. Rev. B 17, 1302–1322. doi:10.1103/PhysRevB.17.1302
Semenov, D. V., Baryakin, D. N., Kamynina, T. P., Kuligina, E. V., and Richter, V. A. (2008). Fragments of noncoding rna in plasma of human blood. Ann. N. Y. Acad. Sci. 1137, 130–134. doi:10.1196/annals.1448.030
Sharma, N., Yadav, D., Kumari, S., Ghosh, D., Shandilya, M., Hassan, M. I., et al. (2024). Exploring nano-protein corona dynamics: tracing the hard-to-soft corona transition with trypsin and graphene oxide in a silver nanocomposite model. ChemistrySelect 9, e202402816. doi:10.1002/slct.202402816
Sheibani, S., Basu, K., Farnudi, A., Ashkarran, A., Ichikawa, M., Presley, J. F., et al. (2021). Nanoscale characterization of the biomolecular corona by cryo-electron microscopy, cryo-electron tomography, and image simulation. Nat. Commun. 12, 573. doi:10.1038/s41467-020-20884-9
Singh, N., Marets, C., Boudon, J., Millot, N., Saviot, L., and Maurizi, L. (2021). In vivo protein corona on nanoparticles: does the control of all material parameters orient the biological behavior? Nanoscale Adv. 3, 1209–1229. doi:10.1039/D0NA00863J
Soliman, M. G., Martinez-Serra, A., Antonello, G., Dobricic, M., Wilkins, T., Serchi, T., et al. (2024a). Understanding the role of biomolecular coronas in human exposure to nanomaterials. Environ. Sci. Nano 11, 4421–4448. doi:10.1039/D4EN00488D
Soliman, M. G., Martinez-Serra, A., Dobricic, M., Trinh, D. N., Cheeseman, J., Spencer, D. I., et al. (2024b). Protocols for isolation and characterization of nanoparticle biomolecular corona complexes. Front. Toxicol. 6, 1393330. doi:10.3389/ftox.2024.1393330
Talamini, L., Violatto, M. B., Cai, Q., Monopoli, M. P., Kantner, K., Krpetić, Ž., et al. (2017). Influence of size and shape on the anatomical distribution of endotoxin-free gold nanoparticles. ACS Nano 11, 5519–5529. doi:10.1021/acsnano.7b00497
Tenzer, S., Docter, D., Kuharev, J., Musyanovych, A., Fetz, V., Hecht, R., et al. (2013). Rapid formation of plasma protein corona critically affects nanoparticle pathophysiology. Nat. Nano 8, 772–781. doi:10.1038/nnano.2013.181
Tran, L., Bañares, M. A., and Rallo, R. (2017). Modelling the toxicity of nanoparticles, vol. 947 of advances in experimental medicine and biology. Cham: Springer International Publishing. doi:10.1007/978-3-319-47754-1
Trinh, D. N., Gardner, R. A., Franciosi, A. N., McCarthy, C., Keane, M. P., Soliman, M. G., et al. (2022). Nanoparticle biomolecular corona-based enrichment of plasma glycoproteins for n-glycan profiling and application in biomarker discovery. ACS Nano 16, 5463–5475. doi:10.1021/acsnano.1c09564
Trott, C. R., Winterfeld, L., and Crozier, P. S. (2010). General-purpose molecular dynamics simulations on GPU-based clusters. arXiv Prepr. Arxiv1009. 4330, 1–12. doi:10.48550/arXiv.1009.4330
Van Der Vaart, M., and Pretorius, P. J. (2008). Circulating dna. Ann. N. Y. Acad. Sci. 1137, 18–26. doi:10.1196/annals.1448.022
Verwey, E. J. W., Overbeek, J. T. G., and van Nes, K. (1948). Theory of the stability of lyophobic colloids: the interaction of sol particles having an electric double layer. Elsevier Publishing Company. doi:10.1038/162315b0
Vilanova, O. (2015). Github. Available at: https://github.com/bubbles-suite/BUBBLES.
Vilanova, O., Mittag, J. J., Kelly, P. M., Milani, S., Dawson, K. A., Rädler, J. O., et al. (2016). Understanding the kinetics of protein-nanoparticle corona formation. ACS Nano 10, 10842–10850. doi:10.1021/acsnano.6b04858
Vilaseca, P., Dawson, K. A., and Franzese, G. (2013). Understanding and modulating the competitive surface-adsorption of proteins through coarse-grained molecular dynamics simulations. Soft Matter 9, 6978–6985. doi:10.1039/c3sm50220a
Vroman, L., and Adams, A. L. (1969). Findings with the recording ellipsometer suggesting rapid exchange of specific plasma proteins at liquid/solid interfaces. Surf. Sci. 16, 438–446. doi:10.1016/0039-6028(69)90037-5
Vroman, L., and Adams, A. L. (1986). Adsorption of proteins out of plasma and solutions in narrow spaces. J. Colloid Interface Sci. 111, 391–402. doi:10.1016/0021-9797(86)90042-1
Wagner, S., Zensi, A., Wien, S. L., Tschickardt, S. E., Maier, W., Vogel, T., et al. (2012). Uptake mechanism of ApoE-modified nanoparticles on brain capillary endothelial cells as a blood-brain barrier model. PLoS One 7, e32568. doi:10.1371/journal.pone.0032568
Walczyk, D., Bombelli, F. B., Monopoli, M. P., Lynch, I., and Dawson, K. A. (2010). What the cell sees in bionanoscience. J. Am. Chem. Soc. 132, 5761–5768. doi:10.1021/ja910675v
Walters, J. P., Balu, V., Chaudhary, V., Kofke, D., and Schultz, A. (2008). Accelerating molecular dynamics simulations with GPUs. PDCCS, 44–49.
Xu, M., Soliman, M. G., Sun, X., Pelaz, B., Feliu, N., Parak, W. J., et al. (2018). How entanglement of different physicochemical properties complicates the prediction of in vitro and in vivo interactions of gold nanoparticles. ACS Nano 12, 10104–10113. doi:10.1021/acsnano.8b04906
Yesylevskyy, S. O., Schäfer, L. V., Sengupta, D., and Marrink, S. J. (2010). Polarizable water model for the coarse-grained martini force field. PLoS Comput. Biol. 6, e1000810. doi:10.1371/journal.pcbi.1000810
Keywords: biomolecular corona, protein adsorption kinetics, protein-nanoparticle interactions, coarse-grain modeling, molecular simulations, transferrin
Citation: Vilanova O, Martinez-Serra A, Monopoli MP and Franzese G (2025) Characterizing the hard and soft nanoparticle-protein corona with multilayer adsorption. Front. Nanotechnol. 6:1531039. doi: 10.3389/fnano.2024.1531039
Received: 19 November 2024; Accepted: 19 December 2024;
Published: 17 January 2025.
Edited by:
Bingqing Wei, University of Delaware, United StatesReviewed by:
Giorgia Brancolini, National Research Council (CNR), ItalySnehasis Bhakta, Cooch Behar College, India
Copyright © 2025 Vilanova, Martinez-Serra, Monopoli and Franzese. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marco P. Monopoli, bWFyY29tb25vcG9saUByY3NpLmll; Giancarlo Franzese, Z2ZyYW56ZXNlQHViLmVkdQ==
†These authors have contributed equally to this work
 Oriol Vilanova1,2†
Oriol Vilanova1,2† Alberto Martinez-Serra
Alberto Martinez-Serra Marco P. Monopoli
Marco P. Monopoli Giancarlo Franzese
Giancarlo Franzese