- Department of Biomedical Sciences, University of Padova, Padua, Italy
Identifying the function and composition of the protein corona (i.e., the set of host proteins interacting with nanoparticles) is considered a crucial step in the development of nanoparticles for medical and pharmacological applications. Evidence suggests that host proteins can alter NP stability, biocompatibility, and pharmacokinetics features. Therefore, in this review, we provide an updated conceptual, methodological, and experimental guideline for the study of the NP protein corona. We surveyed recent literature (2009–2024) focusing on in vitro and in vivo studies. We show that several methods, including shot-gun proteomics, protein identification after in-gel digestion, and TMT proteomics, must be carefully applied and integrated to shed light on this complex phenomenon. Hence, we discuss in detail the relative protocols, highlighting the importance of the experimental conditions, ranging from the administration route to basic, but determinant, parameters like the kind of biological host fluids, the incubation times and the NP concentrations. Additionally, we propose a series of protocols that involve studying the protein corona using purified serum or plasma proteins, as well as sera depleted of specific complement proteins, to investigate the role of their deposition on the nanoparticle surface. We also explore how the role of the protein corona in inducing uptake by phagocytic cells can be examined; finally, we discuss several methodological approaches to study the effects of different coatings on the composition of the protein corona. Available data indicated that it is possible to characterize and punctually study the differential adsorption of specific proteins onto the nanoparticle surface. This allows designing NP chemical coatings features to actively guide the protein corona formation, thus improving nanotheranostic development.
1 Introduction
A crucial consideration when studying a nanosystem is that once in a biological fluid, nanoparticles interact with various soluble molecules, which can lead to potential adverse effects (e.g., off target effect). Additionally, proteins (but also lipids and carbohydrates) can adhere to the surface of the nanoparticles, forming what is known as the “biomolecular corona,” term coined by Kenneth A. Dawson’s group in 2007 (Cedervall et al., 2007). The biomolecular corona can influence numerous cellular processes in response to the injection: it is universally accepted that the biological fate and biodistribution of nanoparticles depend not on the nanoparticles themselves but on the nanoparticles “dressed” with their protein corona. This protein-coated nanoparticle is what cells actually encounter and respond to.
The frequency of contact between a protein and a nanomaterial is higher the more abundant the protein is. In fact, the higher its concentration, the greater the likelihood that it will interact with the surface of the nanoparticle. Once binding is established, if proteins have a high binding energy they remain attached for hours to nanoparticles; while, in the opposite case (low binding energy), they dissociate within minutes leaving space for the binding of less plentiful proteins (low association rate) with higher affinity: this exchange mechanism is due to the “Vroman effect” (Walkey and Chan, 2012; Foroozandeh and Aziz, 2015; Gupta and Roy, 2020). This creates a multiplayer structure, which is generally divided into two parts: the hard corona and the soft corona (Garcia-Cao et al., 2012; Pederzoli et al., 2017). The hard corona consists of proteins tightly bound to the nanoparticle surface, with interactions that take seconds to minutes to establish and can last for hours. The outer layer of proteins (the soft corona) consists of proteins that take hours to interact with the nanoparticle surface, therefore they have low affinity for the surface and a high dissociation rate (Gupta and Roy, 2020; García-Álvarez and Vallet-Regí, 2021).
The formation of the protein corona depends on many factors, both intrinsic and extrinsic, as reviewed elsewhere (Panico et al., 2022; Zhao et al., 2024). For example, among the intrinsic factors, the larger the size of a nanoparticle, the lower the curvature of its surface, resulting in less protein absorption. Conversely, for particles of equal size, a larger surface area (such as a star-shaped compared to a spherical particle) increases the capacity for protein absorption on the surface. If the nanoparticle has a surface charge, this will alter its affinity for positively or negatively charged proteins. An increased hydrophobicity will result in a greater number of proteins bound to the surface; a nanoparticle with a very rough surface will have fewer proteins bound to it. Extrinsic factors that can alter the formation of the protein corona in vitro include temperature, pH, and the ionic strength of the solutions (Panico et al., 2022). Also variations among species are crucial (Sahneh et al., 2015; Solorio-Rodríguez et al., 2017; Lee et al., 2020). Finally, it is necessary to take into account that when nanoparticles are in the bloodstream, they are subjected to shear stress and it has been shown that the composition of the protein corona depends on the conditions (dynamic or static) of the in vitro tests (Palchetti et al., 2016; Yu et al., 2018; Digiacomo et al., 2019).
Among the proteins that associate with nanoparticles are immune system proteins, especially complement proteins. Once activated, complement proteins act as opsonins, like C3b, and anaphylatoxins such as C3a and C5a. Opsonins are also known to mediate phagocytosis by monocytes and macrophages, phagocytic cells within the reticuloendothelial system (RES) and mononuclear phagocyte system (MPS) (Moghimi et al., 2004; Moghimi et al., 2011; Hamad et al., 2008; Neun et al., 2018). These cells are particularly exposed to nanoparticles following intravenous administration due to their specialized ability to capture particulate matter. Studying how nanoparticles interact with these cells and how the protein corona influences these interactions is crucial for understanding nanoparticle behavior in biological systems. Nanoparticles that are effectively taken up by these phagocytic cells are likely to be swiftly cleared from circulation and accumulate in key filtering organs such as the liver and spleen. Conversely, nanoparticles that evade phagocytic uptake may have extended circulation times and potentially reach various other tissues (Moghimi and Szebeni, 2003).
Therefore, the protein corona influences the biodistribution, cellular uptake, and clearance of nanoparticles (Singh et al., 2021). By studying the interactions between nanoparticles and the proteins that bind to their surfaces, researchers can predict how these particles will behave in a biological environment. This knowledge is vital for optimizing drug delivery systems (Guo et al., 2024), ensuring that therapeutics are effectively targeted to specific tissues or cells. Moreover, the protein corona can modulate the biological response to nanoparticles, affecting their safety and biocompatibility (Mahmoudi, 2022). For instance, the presence of certain proteins may trigger immune responses (inducing the production of immune modulators, such as cytokines), the nanoparticle’ clearance (due to opsonization and subsequent phagocytosis activation), the triggering of the coagulation cascade, the aggregation of the nanomaterials, potentially leading to adverse effects. Moreover, the protein corona can modulate the targeting of the nanoparticle, therefore modifying the therapeutic capabilities of the nanosystem (Sanchez-Moreno et al., 2015). Thus, characterizing the protein corona can help in designing nanoparticles that minimize immunogenicity and enhance therapeutic outcomes; besides, understanding the protein corona can provide insights into the mechanisms of action of nanoparticles in various biomedical applications, including cancer therapy, diagnostics, and regenerative medicine (Docter et al., 2015; Rampado et al., 2020; Xu et al., 2023; Khalilov et al., 2023). The study of the protein corona is indispensable for advancing the field of nanomedicine: by elucidating how nanoparticles interact with biological systems, researchers can enhance the design and functionality of nanocarriers, ultimately leading to improved patient outcomes and the successful translation of nanotechnology into clinical practice.
For many years, we have been studying the protein corona with different types of nanoparticles: amorphous silica NPs (Fedeli et al., 2014; Fedeli et al., 2015), organic silica NPs (Tavano et al., 2018), polymeric NPs made of lipoic acid (Trzciński et al., 2021). Over time, we have refined the methodology and the protocol to study targeted effects of the protein corona on various aspects of biocompatibility. In this review, Chapter 2 will discuss the differences in studying the protein corona in serum or plasma, highlighting the importance of carefully collecting these biological fluids and determining the appropriate concentrations to mimic the physiological conditions nanoparticles will encounter when injected in vivo. In Chapter 3, we will address the significance of choosing the incubation times and nanoparticle concentrations. Chapter 4 will cover the various methods available for isolating the protein corona and the techniques we use in our laboratory. Chapter 5 will focus on the methods we use to study protein content of the corona (through in-gel digestion or shotgun approaches). In Chapter 6, we will discuss some methodological strategies we have used to investigate the function of proteins in the corona, such as using cocktails of purified proteins, sera depleted of specific proteins, or examining the interaction of nanoparticles with phagocytic cells. In Chapter 7, we will address the big issue of how to study the role of nanoparticle coatings in the formation of the protein corona and the methodological approaches we have used to compare different coatings on the surface of the same nanoparticle. Finally, for the future perspectives, in Chapter 8, we will discuss the importance and ongoing research efforts to find the correct methods for studying the protein corona in vivo. Our literature review focused on publications from the last 15 years, specifically addressing the study of the protein corona and the methodologies used to uncover its biological significance. We carefully selected studies that provided insights into how the protein corona forms, its interaction with nanoparticles, and the techniques employed to analyze its composition and function. This approach allowed us to present a comprehensive overview of the evolution of research in this field and highlight key advancements in methods used to understand the biological impact of the protein corona in biomedical applications.
2 The physiological buffers for protein corona assessment
The intravenous route is the most commonly used method for administering nanoparticles, as it allows direct access to the bloodstream. Consequently, most studies on the protein corona have focused on interactions within serum or plasma environments. This focus is critical because the protein corona that forms in the blood can significantly influence nanoparticle behavior, distribution, and efficacy in biological systems.
It has been shown that the protein corona’s composition varies based on whether nanoparticles interact with whole blood, EDTA-treated blood, plasma, or serum (Mirshafiee et al., 2016; Schöttler et al., 2016; Lundqvist et al., 2017). The animal species used, such as rats, mice, bovines, or humans, significantly impacts protein corona formation. In humans, factors such as age, sex, diet, health status, and individual variability also influence the formation of the protein corona (Akhter et al., 2021). Other studies have revealed that minor variations, such as the selection of different media (e.g., RPMI, PBS), could influence the protein corona (PC) formed on nanoparticles (Strojan et al., 2017).
The source of the serum is important, and using bovine-derived proteins to study the NP protein corona for nanosystems that would be used in human beings is questionable. Despite the high homology between bovine and human proteins, the impact of biomolecules in the nanoparticle corona may be more pronounced when using human serum (or plasma) and their effects on human phagocytic cells, given that human proteins have a higher affinity for human receptors compared to bovine proteins. Moreover, there are notable quantitative differences between human and bovine serum, indicating that the type of serum used does indeed have an effect (Kim et al., 2014). For these reasons, initially we decided to compare commercial bovine serum and human plasma, but then we focused just on human serum.
The human serum (HS) we used for our experiments was obtained from venous blood of healthy human donors, collected into BD Vacutainer Z tubes (REF 364917, Becton Dickinson, NJ, United States of America) in the absence of any clot activator and additive. It is in fact well known that clot activators, and in particular silica clot activators, induce the depletion of different complement factors, as reviewed by Moghimi and Simberg (2022). For each donor we collected at least 8 tubes (11 mL each). Clotting was allowed to take place at 37°C, for no more than 1 h. Blood samples are then centrifuged at 1250 g for 5 min to obtain sera. Sera were collected and then aliquoted and stored at −20°C. Before use, the serum was thawed at 37°C for 5 min. In our experiments, we used serum samples from at least two different donors and never pooled serum samples from different donors. Pooled platelet poor human plasma from healthy donors (HP) supplemented with 22% v/v of citrate phosphate dextrose adenine solution (CPDA-1) as an anticoagulant was kindly provided by Centro Trasfusionale of the Hospital of Padua (ULSS 16), frozen in aliquots in liquid nitrogen and stored at −20°C. Before use, plasma was thawed at 37°C for 5 min. Citrate, by chelating Ca2+ and Mg2+ ions, disrupts the complement system, inhibiting all three activation pathways (classical, alternative, and lectin-dependent) (Moghimi et al., 2020).
We also utilized serum from different species other than humans. For mice, the serum was always used fresh and not frozen, in particular for the study of complement activation, it is in fact well-known that repeated freeze and thaw processes induce C1q degradation (Lachmann, 2010). For serum production, blood samples were allowed to clot spontaneously at 37°C; clots were then centrifuged at 4,000 rpm for 10 min; serum was immediately used for experiments.
The choice of the percentage of serum or plasma to use during protein corona studies is a very important parameter (Gräfe et al., 2016; Poulsen and Payne, 2022). In our initial studies with amorphous silica NPs, the incubation medium was RPMI 1640 supplemented with 10% FCS or 10% HP. The protein corona of poly-lipoic NPs (10.1021/acs.biomac.0c01321) was studied at different serum or plasma concentrations (10%, 20%, or 50%). In our subsequent studies with Organic Silica NPs, we performed the incubation in a more physiological buffer containing the highest possible amount of serum (75%), and we also considered other important methodological aspects: the concentration of the nanoparticle preparation, the NP final concentration in the assay and several experimental conditions (e.g., presence of inhibitors, purified proteins, etc., see below).
3 Choosing appropriate conditions for protein corona formation: temperature time and NP concentration
When we studied the corona protein of amorphous silica NPs, we performed the experiments at different incubation times (15 min, 3 h and 6 h) and we used two incubation temperatures, either 37°C or 4°C. In general, 37°C is the temperature used because the study of nanoparticle-protein interaction is conducted with human or mammalian systems in mind. Lower temperatures (4°C) can help preserve the integrity of proteins, but this temperature does not mimic physiological conditions (Böhmert et al., 2020; Rampado et al., 2020; Mahmoudi, 2022). Generally, a 60-min incubation is considered standard for corona protein formation (Böhmert et al., 2020), but in our experience (Fedeli et al., 2015) the main polypeptides of the protein corona remained largely unchanged for both short (15 min) and longer incubation times (up to 6 hours), demonstrating the observed composition is kinetically stable; therefore, we preferred to use a 15–30 min incubation time for all our subsequent research on the protein corona. Another important parameter to consider is the concentration of the nanoparticles. Protein corona was studied at different NP concentrations (up to 160 μg/mL), but in our experience (Fedeli et al., 2015) a low concentration of nanoparticles (∼40 μg/mL) better simulates what happens in vivo when nanoparticles are injected into the bloodstream.
4 Methods for the isolation of the protein corona-coated NPs
Isolation of the protein corona-coated NPs is essential, typically achieved through size exclusion chromatography, magnetic separation, field flow fractionation, and, as we did in our laboratory, centrifugation and ultracentrifugation.
4.1 Size exclusion chromatography
Size exclusion chromatography is a separation technique that sorts molecules based on their size. It employs a porous matrix within a column, where smaller molecules enter the pores and elute later, while larger molecules bypass the pores and elute earlier. This method is particularly advantageous for separating protein-nanoparticle complexes from unbound proteins due to its gentle nature, which maintains the integrity of the complexes. After the incubation of the nanoparticles with a biological fluid, the mixture is loaded onto a size exclusion column. The choice of column material and pore size is crucial, as it determines the separation efficiency. For protein corona isolation, columns with a pore size suitable for separating nanoparticle-protein complexes from free proteins are typically used. The sample is eluted through the column using an appropriate buffer. As the sample moves through the column, nanoparticle-protein complexes, being larger, elute first, followed by smaller unbound proteins; the eluted fractions are collected systematically (Cedervall et al., 2007; Böhmert et al., 2020; Kristensen et al., 2021).
4.2 Magnetic separation
This technique takes advantage of the unique properties of magnetic nanoparticles (Sakulkhu et al., 2015; Bonvin et al., 2017). By incorporating magnetic materials into nanoparticles, an external magnetic field can be applied to quickly and selectively separate the nanoparticles and their associated proteins from a complex biological mixture. Nanoparticles are engineered with a magnetic core, typically composed of iron oxide or other magnetic materials. These nanoparticles are then exposed to biological fluids, such as blood serum or plasma, to allow the formation of the protein corona. Once the protein corona has formed, an external magnetic field is applied to the mixture. The magnetic nanoparticles respond to this field, allowing them to be rapidly separated from the non-magnetic components in the solution. The separated nanoparticles, along with their protein corona, are collected for further analysis. This method has also been used for lipid nanoparticles (Francia et al., 2024): Cullis and co-authors prepared iron-oxide-loaded lipid nanoparticles that allow a fast and precise approach for isolating lipid nanoparticle–corona complexes, preserving the integrity of the particles.
4.3 Field flow fractionation
Field flow fractionation (FFF) operates on the principle of separating particles based on differences in their size, shape, and mass. Unlike traditional chromatographic methods, FFF does not rely on a stationary phase; instead, it uses a field (such as a cross-flow, thermal, or centrifugal field) perpendicular to the flow of the sample to achieve separation (Tadjiki et al., 2024). The external field pushes the particles towards the channel bottom, while diffusion acts in the opposite direction. Equilibrium is reached when these opposing forces balance out. At this point, particles of different sizes form distinct clouds: smaller particles create more diffuse clouds and move faster through the channel, while larger particles form narrower clouds and move slowly.
4.4 Centrifugation and ultracentrifugation
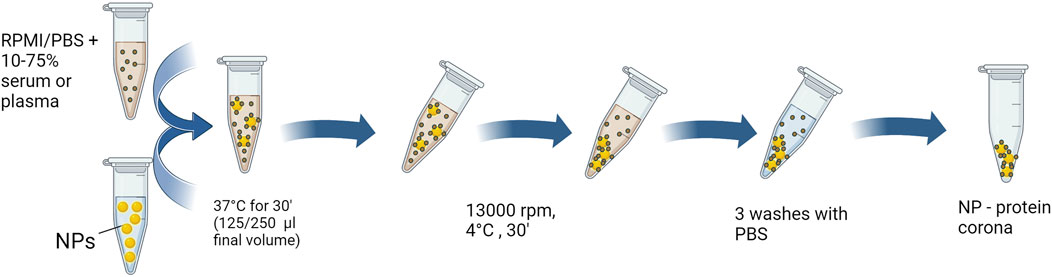
Centrifugation is the most used method for protein corona isolation and it is the method we currently use in our laboratory (see Figure 1). In our laboratory we focused on studying the hard corona that coated the nanoparticles: after incubation with serum or plasma, amorphous silica NPs were diluted with ice-cold PBS in polycarbonate tubes (Beckman Coulter, cat. number 355603), then immediately recovered by ultracentrifugation (45 min, 100,000 g at 4°C, XL-70 Ultracentrifuge Beckman, fixed angle 50 Ti rotor). They were subsequently washed twice with 10.5 mL of ice-cold PBS at pH 7.4. In our work (Fedeli et al., 2015) we have also isolated corona proteins using another method, the centrifugation over a sucrose cushion (Tenzer et al., 2013); even with this procedure, the protein pattern of the nanoparticle corona was similar to that obtained with a simple centrifugation of samples.
For poly lipoic-nanoparticles and organic silica NPs, after incubation at 37°C, a centrifugation was sufficient to recover NPs: samples were therefore centrifuged for 30 min at 13,000 rpm at 4°C. This was followed by three consecutive washes with 1 mL of cold PBS, centrifuging the nanoparticles each time at 13,000 rpm for 30 min at 4°C, in order to obtain the hard corona. For poly lipoic-nanoparticles we observed a very low recovery of proteins, resulting in them being almost corona-free (Trzciński et al., 2021).
In each experiment, mock samples are always included, incubating serum or plasma in the absence of NPs, to estimate the nonspecific protein background after centrifugation and washings.
5 The analysis of protein content on the NP protein corona
5.1 Assessing the overall protein content present on the protein corona of nanoparticles
We usually use a protein assay to quantify the proteins bound to the NPs surface. This measurement is essential to determine if the amount of protein bound to the nanoparticle changes depending on factors such as the coating and to ensure that a sufficient quantity of protein has been obtained for subsequent proteomic analyses (and thus, potentially set the experiment with a final reaction volume that is double or triple). It is also crucial in this case to prepare mock samples, using only the nanoparticles without incubation with serum and plasma proteins. It is also important to quantify the proteins in the supernatants obtained from the first centrifugation of the nanoparticles, after incubation with serum or plasma: this allows to estimate how much protein does not bind to the nanoparticles and how much forms the hard corona after the washes. After the first centrifugation step, supernatant must be collected; after the washing steps, NP pellets are resuspended in 25 μL of PBS 1x. We then quantified the total amount of protein by means of Bradford assay: 10 μL of resuspended pellets and collected supernatants were added in duplicate to a 96-well plate together with standard curve samples (prepared with known concentration of Bovine Serum Albumin). A volume of 200 μL of Bradford reagent (Merck) is subsequently added to each well. The absorbance of the samples and standard curve is analyzed using the spectrophotometer at wavelengths around 595 nm. In other studies, the amount of proteins was measured by micro-BCA (Lebreton et al., 2023) or BCA assay (Mohammad-Beigi et al., 2020; Meng et al., 2022). By adsorbing varying amounts of proteins onto their surface to form a protein corona, NPs can undergo steric stabilization to different degrees, leading to aggregation and resulting in a spectrum of color changes. These changes can be quantitatively characterized using UV-vis absorption spectroscopy. The linear working range of this method was found to be wider than that of the commercial Bradford Assay and comparable to the Micro BCA assay (Ho et al., 2015). Bradford is good to quantify a massive protein corona while for low-binding-NPs it is better BCA.
5.2 Protein corona analysis via in gel digestion
5.2.1 SDS-PAGE and protein staining
To visually compare the composition of surface-adsorbed proteins across different particle chemistries and incubation parameters, SDS page analysis is a very easy-cheap technique. This involves incubating nanoparticle samples under predetermined conditions of temperature and time. After the incubation period, and subsequent centrifugation steps, surface-adsorbed proteins are eluted prior to loading onto SDS-page gels. The composition and relative abundance of proteins eluted from nanoparticles could be analysed from the SDS-gel through Coomassie Staining or Silver Staining (Daramy et al., 2023). For SDS-PAGE, we dissolve the pellet of NPs after washings in non-reducing loading sample buffer (62.5 mM Tris-HCl, pH 6.8, 2% SDS, 25% glycerol, 0.01% bromophenol blue). Samples are heated at 95°C for 5′and equal volumes are loaded on polyacrylamide gels. Electrophoresis is run for 60’ – 90′ at 25 mA/gel and 100 V. At the end, gels are recovered, directly stained by Colloidal Coomassie or Silver Staining procedure, depending on the experiment. Silver staining procedure is carried out at RT as described in Figure 2A. Coomassie staining is carried out at RT, with SimplyBlue SafeStain (Invitrogen) following the manufacturer instructions. After staining, band densitometry is performed using ImageJ software, after background subtraction, to assess the intensity of each band and, consequently, the amount of protein present in the gel.
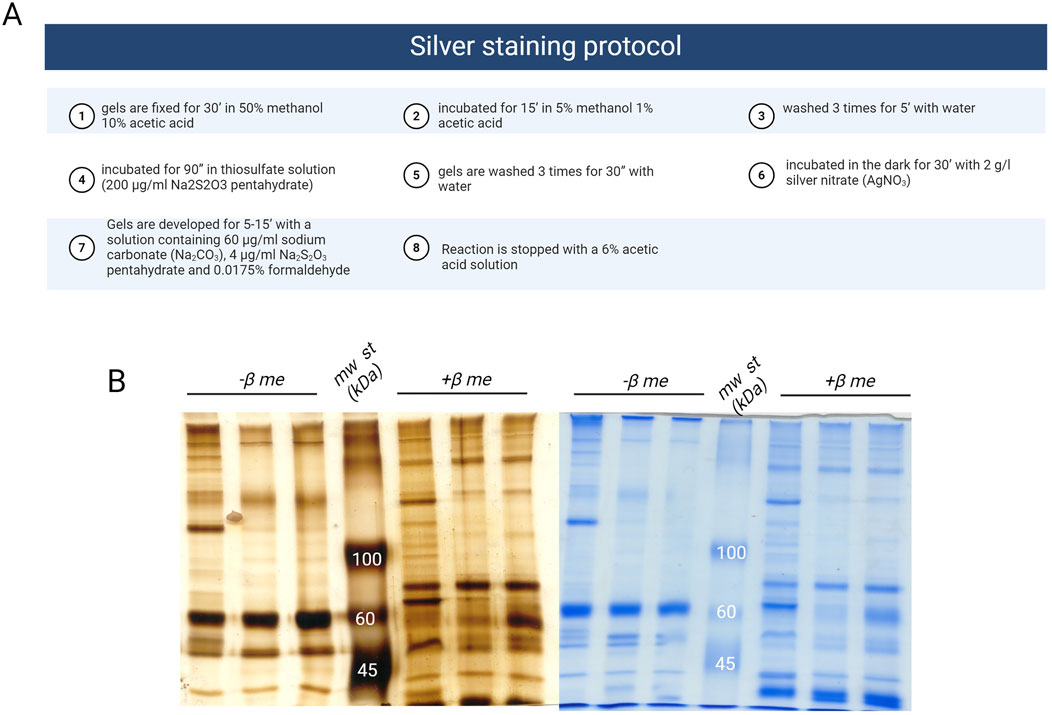
Figure 2. (A) Protocol for silver staining (Created with Biorender); (B) representative images of corona protein samples stained with silver (on the left) or with coomassie (on the right); mw st: molecular weight standard, β me: β mercaptoethanol.
5.2.2 In-gel digestion, MS/MS protein identification and database research
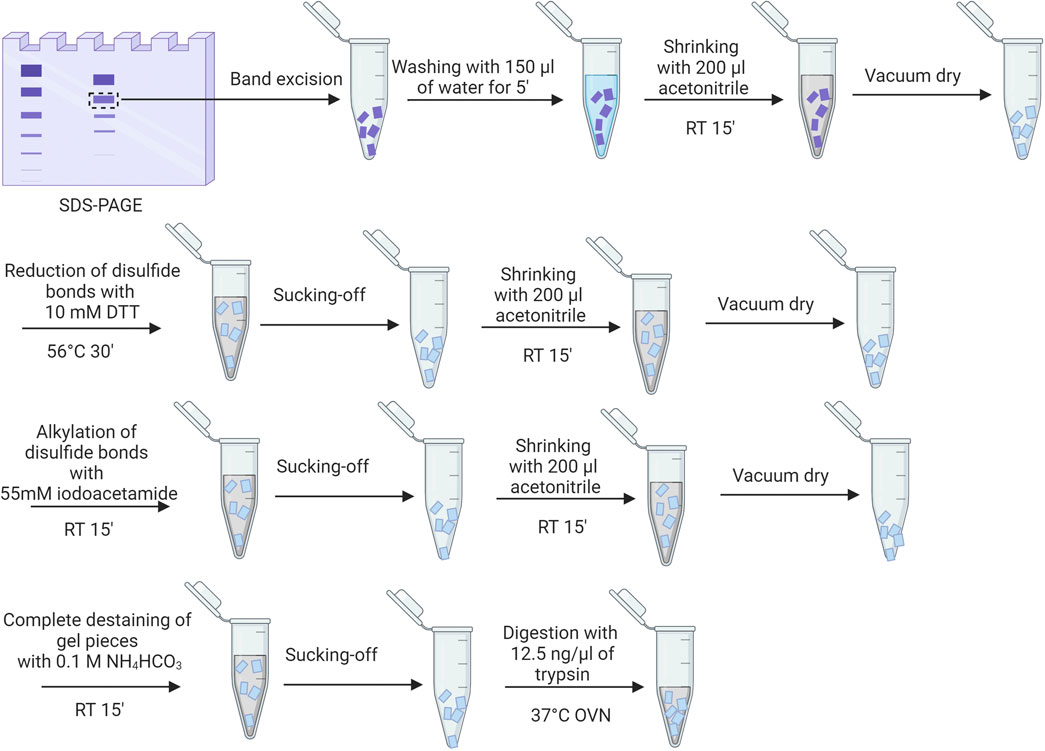
After staining with Coomassie, the bands of interest are processed as visualized in Figure 3, as described also elsewhere (Shevchenko et al., 2006; Lundqvist et al., 2017; Mekseriwattana et al., 2022). After trypsin digestion, the gel fragments are centrifuged, and the supernatant is saved for potential MALDI analysis. To extract peptides, the gel pieces are first incubated with 25 mM NH4HCO3 at 37°C for 15 min while shaking. Then, acetonitrile is added, and the mixture is incubated again at 37°C for an additional 15 min with shaking. The supernatant is collected after centrifugation. Subsequently, the gel pieces are treated with 5% formic acid for 15 min at 37°C with shaking, followed by another 15-min incubation with acetonitrile at 37°C with shaking. The supernatant from this step is collected, combined with the previous supernatant, and concentrated in a vacuum centrifuge. The peptides are reconstituted in 20 µL of 5% acetonitrile with 0.1% formic acid for electrospray MS analysis, and data is collected on a Q-Tof AGILENT 6520 mass spectrometer. Spectra are analyzed with Micromass MassLynx V4.1 software and MASCOT, using specific parameters to search against the Swiss-Prot and NCBI databases.
Using this methodology, we were able to study the differences in the protein corona of amorphous (Fedeli et al., 2015) and organic silica nanoparticles (Tavano et al., 2018) in the presence of fetal bovine serum or human plasma. The differences are analyzed in the following figure (Figure 4). We discovered that the protein hard-corona composition varies significantly based on the protein source used, either FCS or HP. We identified two distinct sets of proteins as major constituents of the coronas derived from FCS and HP, with 7 and 12 proteins respectively for amorphous silica NPs, and with 5 and 11 proteins for organic silica NPs. For the first type of NP, only Apo A-I, Apo A-II, complement factor H, and albumin (HSA or BSA) were common to both sets. For the second type, only albumin (HSA or BSA) and ApoA1 were in common. The predominant proteins for FCS were α2-macroglobulin, ApoA1, and hemoglobin for amorphous silica NPs and just hemoglobin for organic silica NPs, whereas for HP, they were HRG, ApoAI, ApoAII, and fibrinogen for amorphous silica, and ApoAI and clusterin for organic silica. Notably, FCS lacked fibrinogen and kininogen, as these proteins are involved in clot formation during FCS production, and HRG was also absent. Interestingly, HRG, one of the most prevalent proteins in the protein corona of amorphous silica nanoparticles, was completely absent in the protein corona of organic silica nanoparticles. In the case of poly lipoic-nanoparticles, we have found that after three washings nanoparticles were nearly corona free, and in silver-stained gel the only detectable protein was albumin, both with HP and FCS (Biomacromolecules 2021).
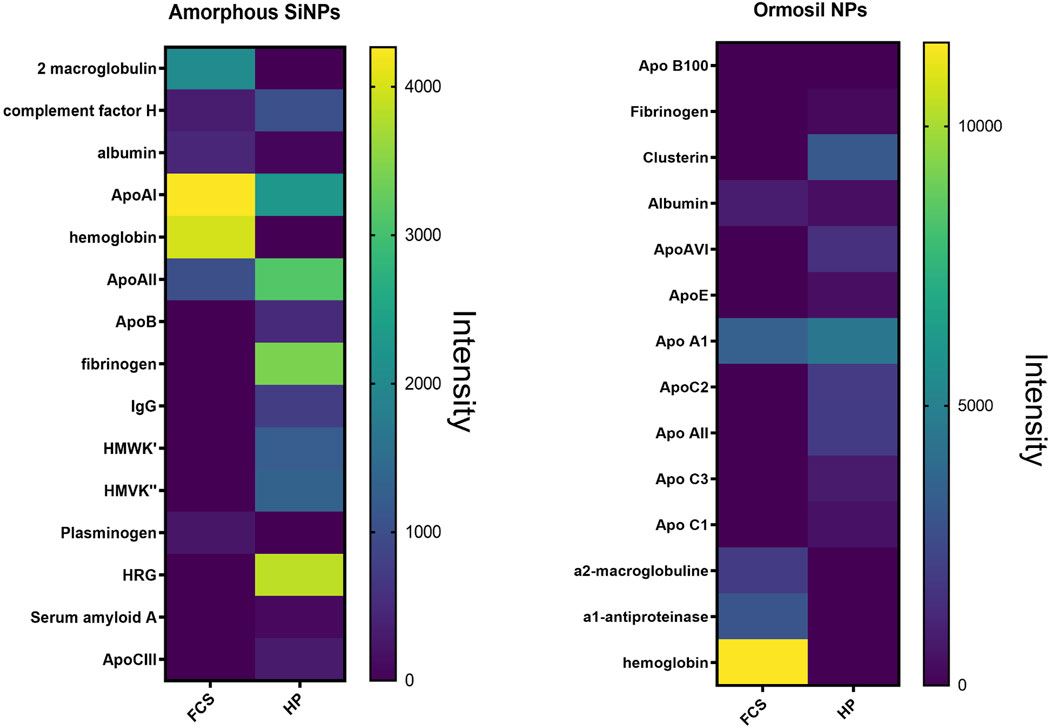
Figure 4. Heat map of the band intensity analysis obtained after protein corona experiments and silver staining with amorphous silica nanoparticles (left panel) and organic silica nanoparticles (right panel).
We also need to point out that an important parameter to consider when studying the protein corona is the final concentration of nanoparticles in the assay. As demonstrated in our study (Fedeli et al., 2015), when a very low concentration of nanoparticles is used, only the proteins with the highest affinity for the nanoparticle surface are likely to bind effectively, simplifying the protein corona composition. At higher nanoparticle concentrations, both the number of proteins and their heterogeneity significantly increase (see the following Figure 5).
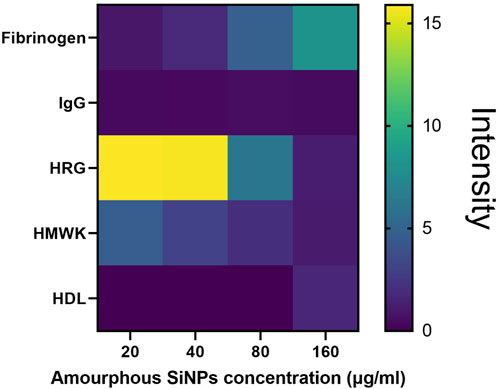
Figure 5. Heat map of the protein quantities present in the corona of amorphous silica nanoparticles (calculated though densitometric analysis of the bands in silver staining) according to the concentration of nanoparticles used.
5.3 Protein corona analysis via label free approach
The gold standard technique to specifically identify the composition of the protein corona is mass spectrometry (Blume et al., 2020).
LC-MS/MS is employed for characterizing the protein corona’s composition, which involves identifying the various proteins adsorbed onto NPs using proteomic techniques. This investigation necessitates enzymatic digestion of proteins, performed in solution directly to the NPs-PC. Subsequently, NPs-PC undergoes digestion and LC-MS analysis. Protein identification is facilitated through common proteomics protocols, often involving protein sequencing. While MS can offer quantitative protein data, conventional LC-SI-MS/MS workflow typically yield semi-quantitative results using label-free methods, estimating protein abundances from peptide counts and ion intensities in a single LC-MS/MS analysis. Achieving absolute protein quantities within the corona necessitated specific standardization, a challenge particularly pronounced when analyzing numerous proteins, as in protein corona studies (Fuentes-Cervantes et al., 2023). After incubation in the physiological buffer, using the selected nanoparticle concentration and incubation time, we treat samples as visualized in Figure 6, as described also elsewhere (Ashkarran et al., 2022).
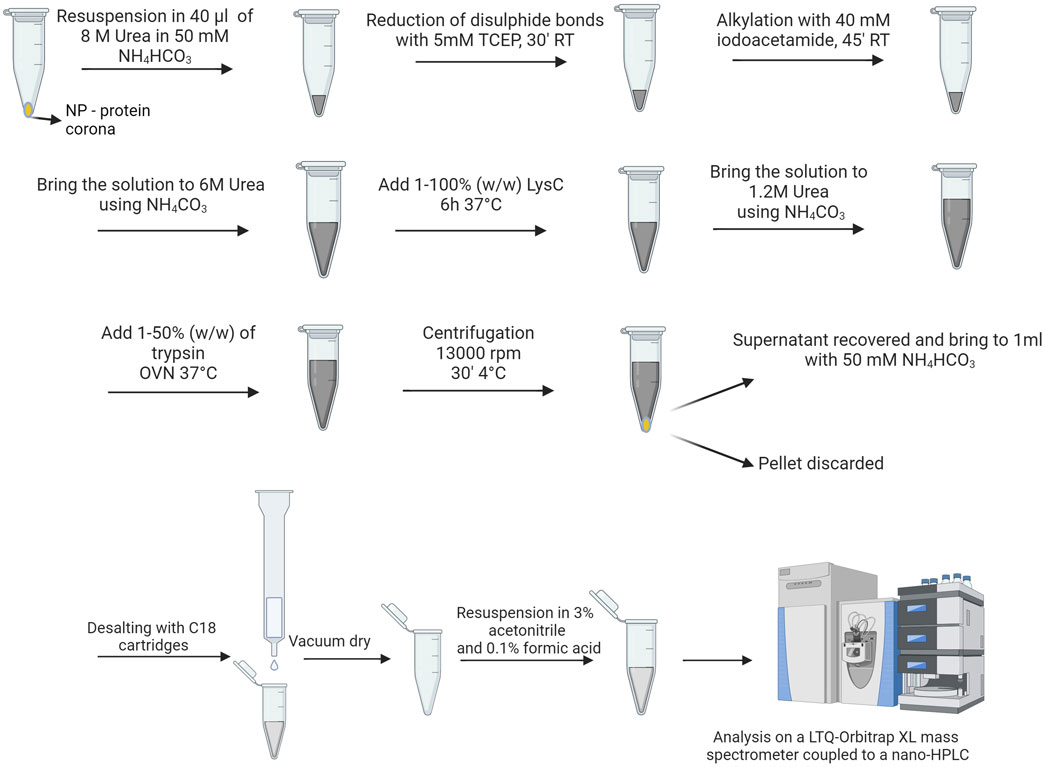
Figure 6. Procedure for preparing protein corona samples for shot-gun analysis (Created with Biorender).
To load peptides we then use pico-frit columns (New Objective, NJ, United States of America), packed with C18 material (Phenomenex, CA, United States of America) followed by separation using an acetonitrile gradient containing 0.1% formic acid. The gradient ranges from 3% to 40% over a period of 45 min, at a flow rate of 250 nL/minute The ion source capillary is maintained at a temperature of 200°C, with a spray voltage between 1.2 and 1.3 kV. By analyzing the mass-to-charge ratio (m/z), individual peptides in the sample can be identified and matched to their respective proteins. For data analysis we use MaxQuant software and peptide identifications are cross-referenced with the UniProt Human and UniProt Mouse databases, depending on the serum source used in the experiment.
Below (Figure 7) is a representative figure of data obtained with this type of analysis (Tavano et al., 2018). Obviously, the shot gun analysis allowed us to more accurately calculate the relative quantities of different proteins in the protein corona of our nanoparticles (see Figure 4, right panel). Most of the proteins present in the protein corona studied via in-gel were comparable to those found through shotgun analysis. However, the shotgun approach enabled us to highlight the presence of complement proteins, various immunoglobulin isotypes, and a broad panel of apolipoproteins, which did not emerge in the in-gel analysis.
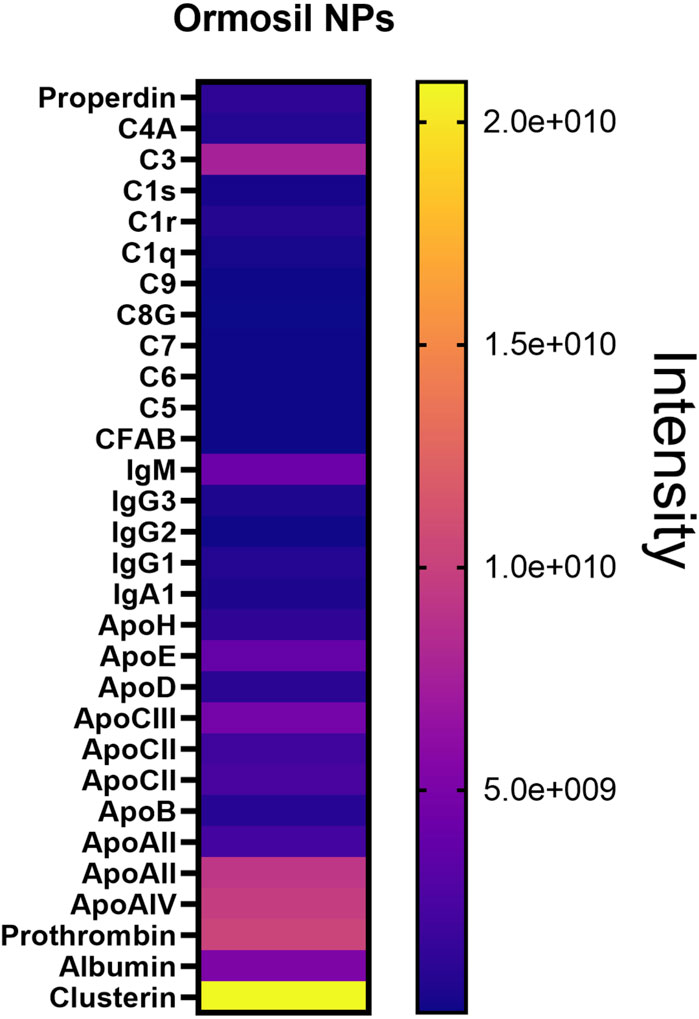
Figure 7. Shotgun analysis of major proteins found in the human serum protein corona of Ormosil NPs.
5.4 Protein corona analysis via tandem mass tags approach
From the wide spectrum of mass spectrometry techniques, TMT-MS shows outstanding characteristics (Zhang and Elias, 2017). Tandem mass tags (TMT) are reliable and efficient systems for multiplexed, relative protein quantitation of up to 18 samples from cells, tissues, or biological fluids. Each mass tagging reagent in a label set has the same nominal mass (i.e., they are isobaric) and chemical structure, consisting of a reactive group, a spacer arm (mass normalizer), and a mass reporter. The isobaric nature of TMT labeling reagents enables the simultaneous identification and quantification of proteins in various samples using tandem mass spectrometry. To have a trustful comparison of PCs between different particles, TMT-MS is the gold standard method (Liessi et al., 2021).
In our laboratory, we found out that TMT is better for comparative approaches but that can detect less proteins (Morbidelli, Papini, Tavano, unpublished data). In the case of this approach, all samples are prepared in duplicate, at the same time and under the same experimental conditions to allow a comparison using equal volumes of each preparation. Samples are reduced, alkylated and trypsin digested as specified for the label-free approach, dried under vacuum and suspended in equal volumes of 100 mM tri-ethyl-ammonium bicarbonate (TEAB). Peptides are then labeled with TMT tags (Thermo Fisher Scientific). Before pooling, the labeling efficiency is evaluated for every sample by performing a LC-MS/MS analysis (see Figure 8). The analysis or raw data is carried out against the relative section of the Uniprot database (as specified above). Trypsin is set as an enzyme with up to 2 missed cleavages allowed. Peptide and fragment tolerance is set at 10 ppm and 0.6 Da, respectively. Carbamidomethyl Cys and TMT labeling (N-term and K) are set as fixed modifications, while Met oxidation is set as variable modifications. The intensities of TMT reporter ions are used by the software to calculate the relative abundance of proteins across the different conditions.
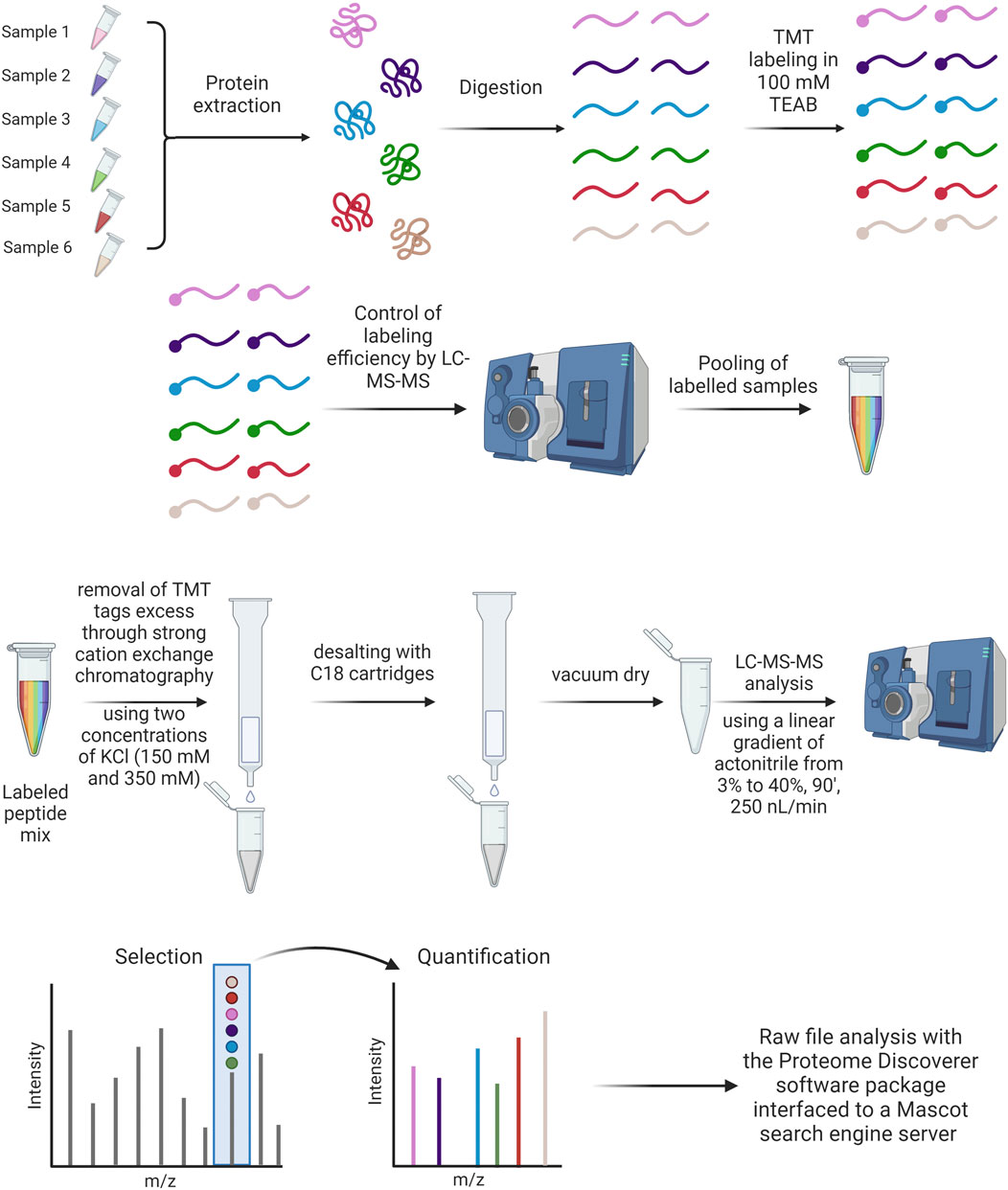
Figure 8. TMT labeling protocol and treatment of samples after TMT labeling analysis (Created with Biorender).
6 Decoding the protein corona: methods to investigate protein corona functions
6.1 Study of the protein corona with purified proteins from human serum
The knowledge of the role of single proteins adsorbed to NPs would allow the design of nanoconstructs that, for example, bind proteins protective against cellular toxicity. To examine the specific roles of key human plasma proteins in forming the corona on amorphous silica nanoparticles, we combined purified fractions of human high-density lipoproteins (HDL), high-density lipoproteins (LDL), and Very Low Density Lipoprotein (VLDL) with purified human plasma IgG fractions (comprising IgG1, IgG2, IgG3, and IgG4 classes), Histidine-rich glycoprotein (HRG), Kininogen-1, Fibrinogen, and serum albumin (Fedeli et al., 2015). These proteins were mixed at concentrations reflecting their average levels in 10% human plasma to create a simplified protein medium (HRG 15 μg/mL; IgG 700 μg/mL; HSA 5 mg/mL; HDL 150 μg/mL; LDL 78 μg/mL; VLDL 12 μg/mL; Kin-1 8 μg/mL; Fibr 300 μg/mL). HRG was purified from human plasma, as explained in Fedeli et al. IgG, serum albumin, HDL, LDL, VLDL, kininogen-1, and fibrinogen were purchased by different companies. We observed (see Figure 9) that the protein corona formed on nanoparticles incubated with purified proteins displayed a polypeptide profile closely resembling that seen with human plasma. Notably, the absence of HRG in the corona mix led to an increased association of Kininogen with the nanoparticles. Conversely, the omission of other proteins did not markedly alter the corona composition, which remained predominantly characterized by a high abundance of HRG. This absence likely allows other proteins with lower affinity for the silica surface to bind to the NPs. Using this methodology allowed us to focus on studying the binding dynamics of various protein to the nanoparticle surface, depending on the presence or absence of these proteins in the biological medium.
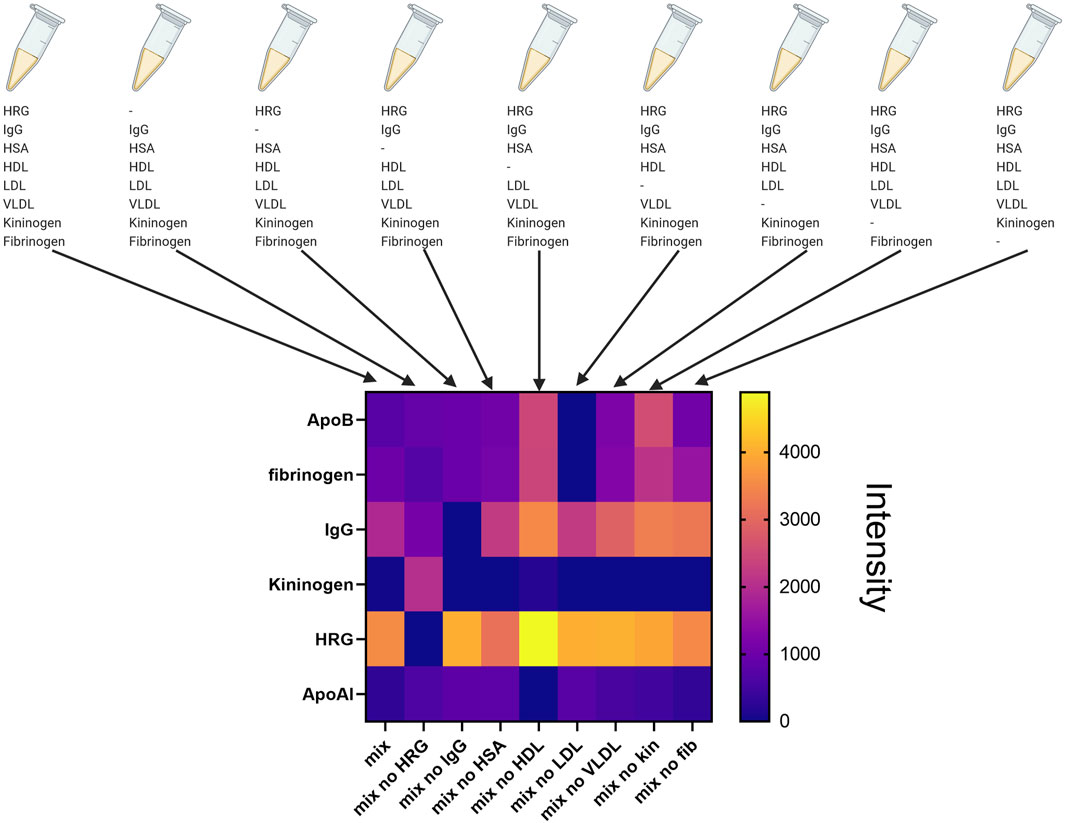
Figure 9. Composition of the protein corona formed after incubation of SiO2-NPs in the presence of the complete mix of purified proteins or in the presence of the mix of purified protein lacking a single protein of the mixture (Created with Biorender).
6.2 Study of the protein corona with serum depleted of specific proteins
The process of selectively eliminating specific factors in order to better understand their roles in biological fluids is more compelling than studying the isolated factors. The selective elimination can be achieved through immune depletion using specific antibodies. In our studies about the protein corona of amorphous silica nanoparticles, human plasma was depleted of HRG, the most abundant protein found in the corona protein. The depletion was achieved by passing human plasma through a phosphocellulose resin (P11, Whatman), followed by collection of the flow-through. The collected plasma was then dialyzed against PBS (for 18 h at 4°C) and quantified for protein concentration. In the corona experiments performed with HRG-depleted plasma, similarly treated and dialyzed normal human plasma was used as control, with adjustments made for differences in protein concentration. To prevent coagulation, both plasmas were supplemented with sodium citrate (0.38% v/v) and were immediately used for corona experiments. We found (Fedeli et al., 2015) that in the absence of HRG, the protein corona of amorphous silica nanoparticles was enriched in Fibrinogen (4 times), albumin (2.5 times) and Apo A-I (2 times).
6.3 Study of complement protein deposition on the surface of nanoparticles
Complement activation induced by nanoparticles is a significant area of research in nanomedicine. The complement system consists of proteins that help to identify and eliminate pathogens. When nanoparticles are introduced into the body, they can trigger this system, leading to various immunological responses. nanoparticles can activate the complement system through multiple mechanisms, primarily dependent on their surface properties (La-Beck et al., 2021). Positively charged nanoparticles are more likely to attracts and bind complement proteins, leading to activation; functional groups on the nanoparticle surface, such as hydroxyl, amine, or carboxyl groups, can bind to complement proteins, triggering activation (Behzadi et al., 2017). The complement system can be activated via three main pathways: classical pathway, lectin pathway and alternative pathway (Moghimi et al., 2011). It is well-known that the activation of the classical and lectin pathways of the complement system is Ca2+-dependent, whereas Mg2+ is essential for the operation of the alternative pathway (Sahu and Lambris, 2001; Merle et al., 2015). In our study Tavano et al. (2018) we selectively chelated calcium by treating the nanoparticles during incubation with human serum with EGTA (10 mM) and Mg2+ (2 mM), inhibiting in this way the classical and lectin pathways; on the other hand, we chelated both calcium and magnesium using EDTA (10 mM), to inhibit the alternative pathway. To study changes in the composition of the corona protein due to complement activation by interaction with the nanoparticle surface, we performed shotgun proteomic experiments by incubating the nanoparticles with control serum or serum in the presence of the specific chelators. The results we obtained with organic silica nanoparticles are depicted in Figure 10: it is evident that the surface of the nanoparticle induces the deposition of C3, which is inhibited by both chelants; simultaneously, the deposition of IgM on the surface increases. The attachment of apolipoproteins to the nanoparticle surface is inhibited when complement activation is blocked, particularly for ApoE, ApoC3, and ApoA1.
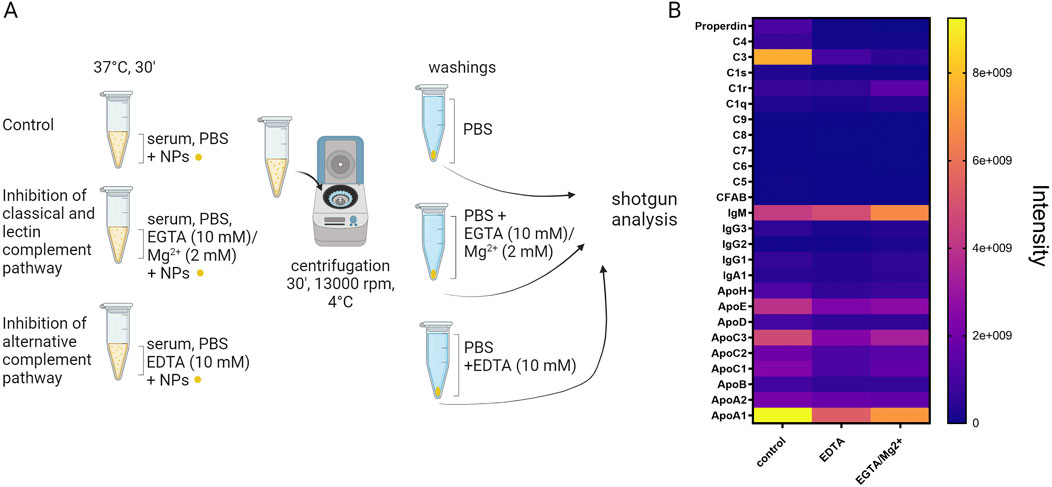
Figure 10. (A) protocol for studying complement activation induced by the surface of nanoparticles (Created with Biorender); (B) heat map showing the abundance intensity of proteins detected by shotgun analysis of the protein corona on Ormosil nanoparticles incubated with human serum and treated with EDTA or EGTA and Magnesium to inhibit complement activation.
6.4 The role of the protein corona in the uptake by phagocytic cells
Monocytes and macrophages, which are phagocytic cells within the reticuloendothelial system (RES) and mononuclear phagocyte system (MPS) are particularly exposed to nanoparticles following intravenous administration due to their specialized ability to capture particulate matter. The presence of a protein corona on the surface of nanoparticles signifies an intricate interplay between biomacromolecules and NPs, dictating their behavior in terms of pharmacokinetics and pharmacodynamics. As proteins interact with NPs, they induce modifications in their surface characteristics, rendering them susceptible to clearance by Mononuclear phagocyte system (MPS) (Khan et al., 2022). Studying how nanoparticles interact with these cells and how the protein corona influences these interactions is crucial for understanding nanoparticle behavior in biological systems (Yan et al., 2013; Cai et al., 2024; Liu et al., 2024). Nanoparticles that are effectively taken up by these phagocytic cells are likely to be swiftly cleared from circulation and accumulate in key filtering organs such as the liver and spleen (Gustafson et al., 2015). Conversely, nanoparticles that evade phagocytic uptake may have extended circulation times and potentially reach various other tissues.
In our laboratory, we investigated how the protein corona can modulate the association and subsequent uptake of both amorphous silica nanoparticles and organic silica nanoparticles. We conducted these experiments with both human phagocytic cells (such as monocytes and macrophages) and human non-phagocytic cells (like lymphocytes and epithelial cells). We believe that working with primary cells is the best choice for studying in vitro mechanisms that closely resemble what occurs in vivo. Therefore, uptake experiments are conducted with phagocytic cells purified from fresh buffy coats from healthy donors, which are provided to us by the transfusion center of the Hospital of Padova. The buffy coats are processed on the same day they are prepared: through two successive centrifugation gradients (the first on Ficoll and the second on Percoll), we obtain monocytes and lymphocytes (see Figure 11), as described also elsewhere (Repnik et al., 2003; Meital et al., 2019; Domínguez-Andrés et al., 2021; Angioni et al., 2023). Neutrophils, on the other hand, are purified through preliminary dextran sedimentation, followed by centrifugation on a Ficoll-Paque gradient and then lysing of red blood cells through a hypotonic shock.
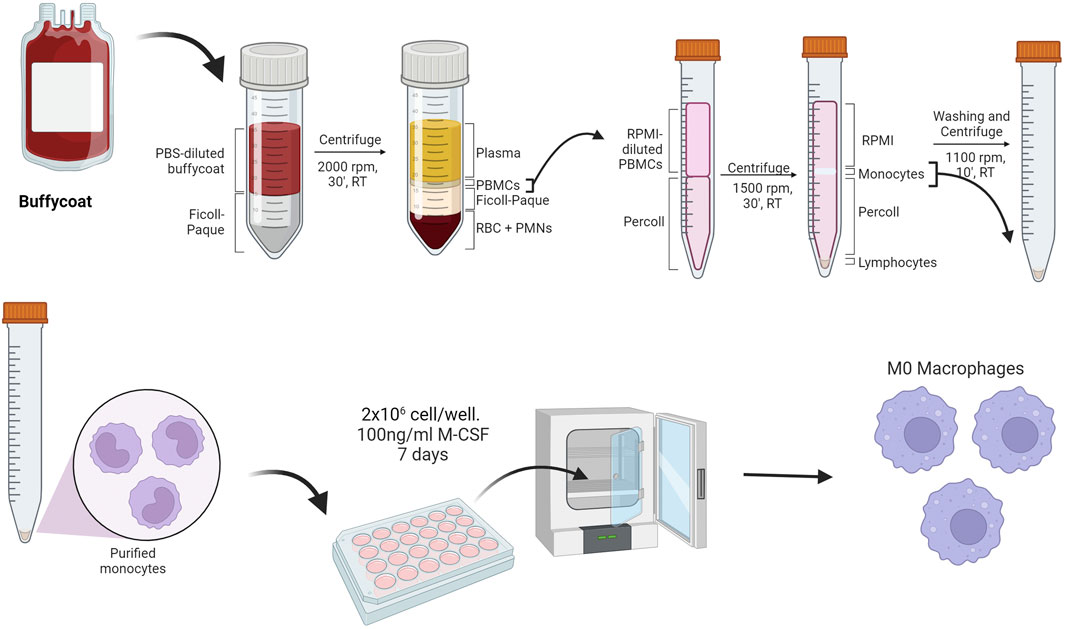
Figure 11. Method for the purification of monocytes from buffy coat and subsequent differentiation into macrophages (Created with Biorender).
To study the interaction of amorphous silica nanoparticles with phagocytic cells, the nanoparticles were incubated with differentiated macrophages, cells were then washed, and the fluorescence associated with the cells was analyzed using flow cytometry (see Figure 12). To investigate the importance of the protein corona in cell-nanoparticle interactions, incubations were conducted in the presence of serum or plasma and also with only the HRG protein (Fedeli et al., 2014), the major component of the protein corona of amorphous silica nanoparticles (as shown in Figure 5). We found that NP uptake by macrophages after 3 h was significantly inhibited by the presence of plasma proteins and HRG alone, highlighting again that the protein corona was fundamental for the interaction of amorphous silica NPs with these cells.
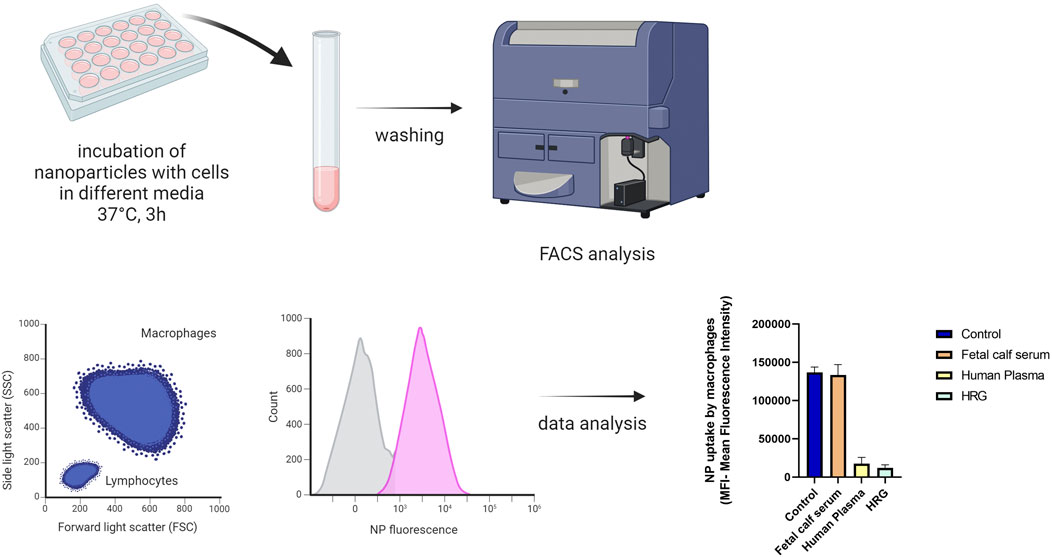
Figure 12. Protocol for measuring the uptake of amorphous silica nanoparticles after incubation with macrophages; the histogram represents the mean fluorescence intensity data of macrophages incubated with amorphous silica NPs in the presence of no proteins (control), FSC, HP or HRG alone (Created with Biorender).
7 Coating matters: how surface design influences protein corona composition, complement activation, and phagocytic cell recognition of nanoparticles
Since now, we have seen how to unveil the protein corona composition. Knowledge acquired in the protein corona field allows scientists to design NPs that can selectively bind or avoid specific proteins (Kopac, 2021). To mitigate and to regulate the formation of a protein corona, NPs are usually coated with polymers designed to avoid nonspecific interactions, forming the so-called coating.
7.1 PEG
Poly (ethylene glycol) (PEG) is a polymer known for its hydrophilic nature and flexibility, which effectively minimizes protein and biomacromolecule adsorption; it has been widely utilized to prolong the circulation duration of protein therapeutics, demonstrating significant success. Additionally, PEG coatings are commonly employed to mitigate nonspecific cellular uptake of nanoparticle drug carriers and other undesired interactions within biological contexts (Yang and Lai, 2017).
Incorporating PEG and PEG-containing copolymers onto nanoparticle surfaces significantly prolongs their blood circulation lifespan by several orders of magnitude. This technique establishes a hydrophilic protective barrier around the nanoparticles, effectively repelling opsonins protein absorption through steric repulsion forces, thereby obstructing and delaying the initial stage of opsonization (Owensiii and Peppas, 2006).
This stealth coating PEG and other polymers prevents adsorption of blood serum proteins, increases circulation time, and enhances the probability of particle permeation into tumor tissue (Mout et al., 2012).
Regardless of the polymer used for the coating, the overall charge of the surface plays a central role in the interaction between nanoparticles and the protein corona. Common terminal groups for polymers are positively charged amines, negatively charged carboxyl groups, a combination of both (zwitterionic), or non-charged methoxy groups. In particular, synthetic zwitterionic materials have been studied as PEG alternatives. In fact, they have a strong hydration rate that confers them stealth properties and low immunogenicity. In particular, NPs coated with amines exhibited quicker and more extensive protein absorption, but lower stability in serum. Carboxyl-PEG and methoxy PEG NPs demonstrated reduced protein corona assembly. Intriguingly, zwitterionic-PEG NPs exhibited the lowest protein absorption. The heightened interactions with methoxy-PEG NPs can be explained by the ability of proteins to interact with non-charged groups through hydrophobic interactions. Additionally, zwitterionic particles demonstrated the highest stability in serum (Rampado et al., 2020).
7.2 Stealth polymers alternative to PEG
In the recent decades, an alternative for PEG has been searched to compensate for critical aspects of this molecule and to allow more chemical versatility, useful to add functionalities and modulate the surface features. Several alternatives have been proposed such as polyvinylpyrrolidone (PVP) (Escamilla-Rivera et al., 2019), zwitterions, peptides (Leal et al., 2020) and carbohydrate moieties.
Joh et al. worked on the modification of linear PEG chains in shorter hyperbranched polymers that could decrease the antigenicity of PEG guarantying stealth properties, presenting PEOGMA as an alternative to linear PEG (Joh et al., 2019). Another alternative is poly-glycerols (PGs) polymers. PG-based drugs present correct pharmacokinetics properties and low immunogenicity. However, it was proven that they tend to accumulate in the liver and kidneys. PGs have been shown to be less susceptible to oxidative or thermal stress than PEG (Hoang Thi et al., 2020).
Poly (acrylamide) and poly (methacrylamide) are well known materials already used for biomedical applications. They are biocompatible and make nanosystem stealth, decreasing the clearance. Although these polymers are biocompatible, their monomers are highly toxic. For this reason, they are not suitable as PEG alternatives (D’souza and Shegokar, 2016).
The poly-N-2-Hydroxypropyl-methacrylamide (PHPMA) has demonstrated an excellent preclinical efficacy as a carrier for chemotherapeutic drugs and has recently entered clinical trials. However, studies into the immune response of PHPMA remain elusive (Hoang Thi et al., 2020).
Among these alternatives, polyalkyloxazolines (PAOXA) and polyalkyloxazines (PAOZI) have been proposed by many studies and attract a wide interest (Adams and Schubert, 2007; Hoogenboom, 2009; Viegas et al., 2011). Oxazolines are hydrophilic, tertiary polyamides with the amide group in the side chain, as well as being structural isomers of polypeptides. Three isomers of oxazolines exist depending on the location of the double bond, 2-, 3-, and 4-oxazoline, where 2-oxazoline is by far the most studied. They can be further substituted on the 2, 4, or 5 position of the 2-oxazoline ring, however substitution on the 4th or 5th position is more difficult to polymerize due to steric crowding. POZs have structural similarities to PEG and polypeptides due to having (C-C-N) backbone repeat units and amide side chains. POZ has higher chemical stability due its N-vicinal C–H bond causing a lower polarization compared to PEG which has an O-vicinal C–H bond. Although PEGylated polymers are highly water soluble, amphipathic, very flexible and hydrated in water, non-toxic, and may be produced with low polydispersity, POZ can be further tuned in size and structure using a living cationic ring opening polymerization (CROP) as well as facile surface functionalization and fluorescent labeling via click chemistry thus overcoming PEGs low drug loading capacity.
7.3 Methodological approaches to compare coatings on the same nanoparticle
In order to compare the biocompatibility of different coatings on the surface of organic silica nanoparticles, NPs were synthesized with no coating, a PEG coating, and a poly (2-methyl-2-oxazoline) (PMOXA) coating (Tavano et al., 2018).
The comparison of coatings was conducted by evaluating several parameters: the composition of the protein corona of the three types of nanoparticles (obtained through shotgun analysis), the assessment of complement activation (via ELISA assays), and the study of the uptake of different types of nanoparticles by macrophages.
Shotgun analysis was performed as described in chapter 5.3 (see Figure 13A). We found that the PMOXA coating induces the formation of a protein corona enriched in complement proteins (such as C3, C5, C6, C9, and complement factor B), with respect to uncoated or PEG-coated nanoparticles.
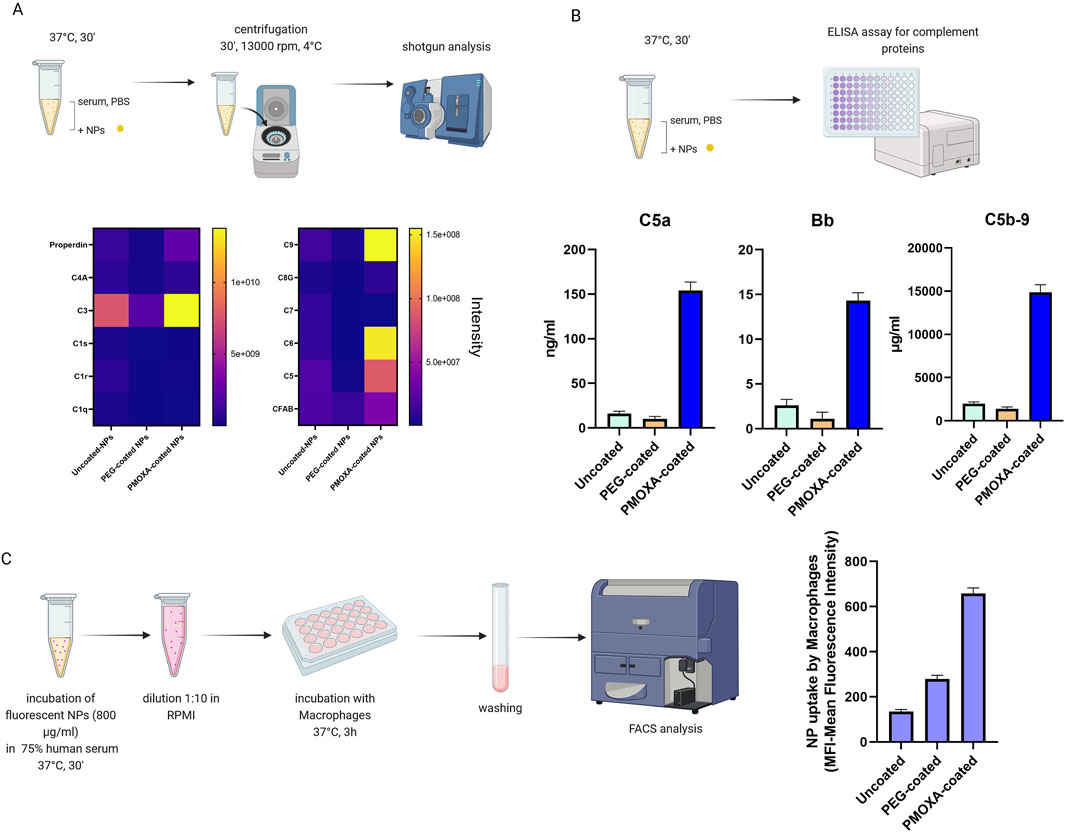
Figure 13. (A) Heat map analysis of the protein corona of organic silica nanoparticles (uncoated, PEG-coated. PMOXA-coated); (B) measurement of the release of C5a, Bb and (s)C5b-9 induced by the nanoparticles through ELISA assay; (C) FACS analysis of the uptake of uncoated- PEG-coated, and PMOXA-coated organic silica nanoparticles by macrophages; nanoparticles were pre-treated in 75% human serum and subsequently diluted in RPMI medium for the incubation with cells (Created with Biorender).
The activation of the complement system in human serum (see Figure 13B) was accessed by measuring the nanoparticle-induced release of complement activation products C5a (a protein fragment released from complement component C5), Bb (the fragment of complement factor B that results from activation of the alternative pathway), and sC5b-9 (the terminal complement complex) by specific ELISA kits, provided by Quidel. In this way we found that the PMOXA coating is responsible for the activation of the complement system and the subsequent release of C5a, Bb, and s-C5b9; instead, the PEG coating did not induce complement activation.
For the uptake experiments (Figure 13C), we preferred to pre-incubate the nanoparticles in 75% serum for 30 min at 37°C, then dilute 1 to 10 in cell medium (RPMI without serum), and finally incubate the nanoparticles with cells, to closely mimic what happens in vivo. We revealed that PMOXA coating results in an uptake of organic silica nanoparticles that is 5 times greater than that of the same uncoated nanoparticles and 2.5 times greater than that of PEG-coated nanoparticles.
To better understand the dependence of PMOXA-coated nanoparticle macrophage uptake on complement activation induced by the nanoparticle surface we used four approaches (Tavano et al., 2018): 1) the role of activation of the classical, lectin, and alternative pathways in NP uptake was studied as described above (chapter 6.3), using EGTA/Mg2+ or EDTA during the pre-incubation of nanoparticles with serum and during the incubation with macrophages; 2) the role of individual components of the complement activation cascade in the uptake of PMOXA-coated NPs was studied using commercially available sera depleted of C3, C1q, C4, and B factors, provided by Comptech (see Figure 14B); these depleted sera were used for pre-incubation with the nanoparticles (including in the experiment also a “super control”, where the depleted serum was reconstituted with the specific purified protein), diluted in RPMI (as described in Figure 13C) and then incubate with macrophages; 3) the role of the Ig deposition on the NP surface in the NP uptake was studied using sera depleted of IgG or IgM: IgM depletion was obtained using an anti-human IgM agarose matrix (Sigma); prechilled and PBS-equilibrated agarose beads were mixed one to one with cold human serum at 4°C for 60 min; the agarose beads were eliminated by centrifugation and the IgM depleted serum was recovered and immediately frozen in liquid nitrogen. To recover the bound IgM, beads were treated with 100 mM glycine (pH2.8) for 5 min and then centrifuged; after collecting the supernatants, 1M Tris-HCl was added to restore the physiological pH; then IgM solution was dialyzed overnight at 4°C against PBS. For IgG antibody depletion, human serum was incubated with an equal amount of protein A sepharose beads (Amersham) for 60 min at 4°C; sepharose beads were removed by centrifugation and IgG were recovered as described for IgM; IgG- or IgM depleted sera were used during the pre-incubation of the nanoparticles, then diluted and used for the incubation with macrophages; also in this case we prepare a “super control”, re-pleting serwith IgM or IgG; 4) the role of lectin pathway in the recognition of nanoparticles by macrophages has been studied using two sugars, N-acetyl glucosamine and mannose, which compete with mannose-binding lectin and ficolins, proteins involved in the lectin pathway, for binding to their substrates. The sugars, at a concentration of 25 nM, were used during the pre-incubation of the nanoparticles with the serum.
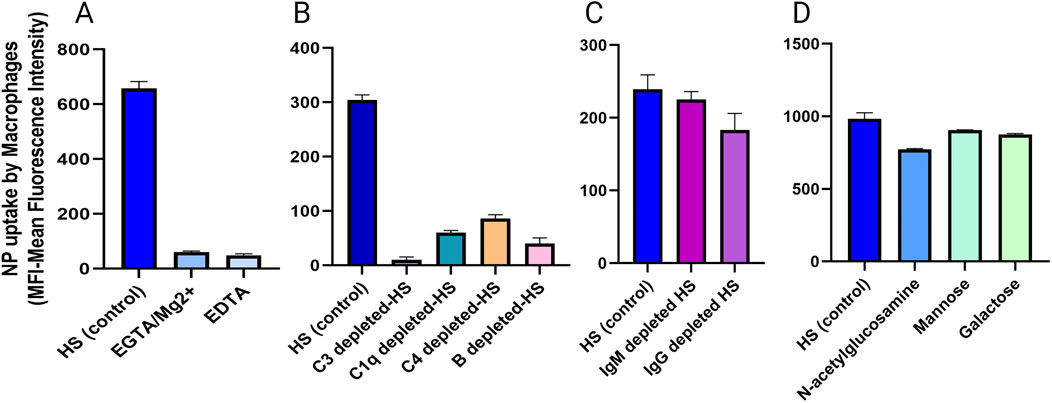
Figure 14. analysis of nanoparticle uptake by macrophages in the presence of (A) inhibitory of classical, lectin and alternative complement pathway, (B) commercial depleted sera, (C) IgM or IgG depleted sera, (D) sugar-treated sera (Created with Biorender).
In this way, we were able to dissect the nanoparticle uptake process. As shown in Figure 14A, PMOXA-coated-NP uptake was completely inhibited in the presence of EGTA/Mg2+ and EDTA; the absence of C3 induces a 95% inhibition of PMOXA-coated NP uptake, whereas the absence of C1q, C4, and factor B results in a lesser inhibition (70%–80% depending on the complement factor, but still significant); neither Ig depletion nor lectin pathway blockade are significant in the uptake of PMOXA-coated NPs.
A crucial point we want to emphasize is the importance of using serum from different individual donors, rather than pooled sera, and to perform separate experiments with each donor’s serum. This approach takes in account of the interindividual variations in complement activity (Gaya Da Costa et al., 2018; Lipsa et al., 2023), which can significantly impact the efficiency of nanoparticle recognition and engulfment by phagocytes. By analyzing sera from multiple donors, it is possible to capture a broader spectrum of opsonic activity and better understand how these differences affect nanoparticle-phagocyte interactions. This allows to gain insights into the variability in immune response among different individuals and provides a more robust understanding of the role of complement proteins in nanoparticle uptake (La-Beck et al., 2021). For instance, PEG-coated nanoparticles were differently uptaken by macrophages, depending on the donor’s serum (Tavano et al., 2018). Another critical point is the choice of the source of the serum: in nanomedicine evaluations, murine models and materials derived from mice are commonly used. However, there are significant species-specific differences between humans and these animals, particularly in the innate immune system’s response to particulate matter (Li et al., 2021). Mice and human immune systems have differences that must be taken into account (Mestas and Hughes, 2004). In our experience, for example, PMOXA-coated nanoparticles, which are strongly phagocytosed by human macrophages, are not taken up at all by murine macrophages (Tavano et al., 2018). This highlights the importance of carefully testing the compatibility of nanomaterials with human cells and sera.
8 Future perspective: studying the protein corona in vivo
Studying the PCs formed in vivo could offer profound insight into their biological impact and accelerate their transition to clinical applications (Singh et al., 2021). Hence, the intricate composition of biological fluids and variations in flow velocity are additional variables that must be considered when assessing the behavior of the nanoparticles in vivo (Simon et al., 2021).
Nanoparticles can enter the animal model body through three main routes: direct injection, inhalation and oral intake (Rizvi and Saleh, 2018). Once they enter systemic circulation, the first phenomenon that occurs is the interaction between the particles and proteins before they are distributed to various organs (Mu et al., 2014). Analytical methods for studying the composition and structure of PCs can be divided into in situ and ex situ characterization. Ex situ techniques involve isolating protein-bound engineered nanoparticles from their physiological environment, followed by cleaving the bound proteins for further analysis. In contrast, in situ techniques directly provide relevant information about PC formation as NPs disperse within the physiological environment (Bai et al., 2021).
One example of the study of the PCs ex situ is the work of Sakulkhu et al. (2015). Polymer-coated superparamagnetic iron oxide nanoparticles (SPIONs) were injected intravenously in rat’s tails. The rats were anesthetized with 5% isoflurane and euthanized 15 min after the injection. Various organs and blood samples were collected. The blood was allowed to clot at room temperature and then centrifuged to separate serum from blood cells. Serum was then loaded into a column in a magnetic reactor to separate the hard corona-SPION complexes. The trapped NPs were then washed and the PCs were analyzed by LC-MS/MS.
Another approach for the ex situ characterization is the immunoprecipitation. Hacene et al. (2021), described an immunoprecipitation assay for nanoparticles coated with PEG using anti-PEG antibodies cross-linked to magnetic beads to extract NPs with their corona from the biological fluids.
On the other hand, the in situ characterization of the protein corona is extremely challenging: Sanchez-Guzman et al. (2020) proposed a new technique utilizing cryo-EM and synchrotron-radiation circular dichroism (CD) to analyze weakly bound proteins and uncover the molecular basis of the PC. Additionally, the hydrodynamic radius of PC-NP complexes in a complex matrix was analyzed through NMR by Carril et al. (2017). Fluorescence correlation spectroscopy (Shang and Nienhaus, 2017) and high-speed dark-field microscopy (Liu et al., 2021) have been proposed to address the issue of the in situ protein corona. One last example is the work of Lo giudice and co-authors in which molecular motifs can be identified through flow cytometry combined with microfluidics in biological milieus (Lo Giudice et al., 2016).
Different animal models can be used to study the interactions between nanoparticles and biological fluids. The choice of the model usually depends on the application of the nanosystem. For instance, zebrafish is used for assessing vital parameters, examining water pollution, and conducting genotoxicity and reproduction toxicity studies. Mouse model is commonly used in nano-toxicological research, biodistribution, pharmacokinetic and pharmacodynamic studies. Rats can be used in nanomedicine research as animal models to study the effect of nanoparticles on various diseases and conditions. Mouse and rats are the most commonly used animal models for nanomedical applications. The pig model is considered the most sensitive among various animal models used in nanomedicine. In particular, the pig model’s sensitivity to lipid NP injection makes it a valuable tool for studying nanoparticle behavior and their effects on the body. Drosophila melanogaster is gradually becoming a valuable model for assessing the toxicity of engineered nanomaterials. Understanding how different animal models react to nanoparticles helps researchers select the most appropriate model for specific studies in the field of nanomedicine (Chrishtop et al., 2021).
9 Conclusion
This review systematically examines the methods available to uncover the significance of proteins binding to nanoparticles when exposed to biological fluids. Throughout various sections, we explored a range of methodologies implemented in our laboratory for studying the protein corona. A key focus of the review is on how different factors, such as nanoparticle surface coatings, incubation times, and biological medium conditions, influence the formation of the protein corona. By comparing these parameters, we can better understand how the biological identity of nanoparticles is shaped, which is crucial for predicting their behavior in biological systems.
In particular, we delved into specific techniques for isolating and analyzing the protein corona, such as mass spectrometry for protein identification and methods to study the functional impact of bound proteins on nanoparticle interactions with immune cells. The review also highlights the importance of employing robust, precise, and reproducible methodologies in nanomedicine research to ensure that findings are reliable and applicable in clinical settings. Ultimately, understanding the interactions between proteins and nanoparticles is fundamental for successful design of nanoparticle-based drugs. Employing accurate characterization techniques during the preparation of the protein corona, combined with rigorous methodological approaches, is essential to advancing nanomedicine and accelerating its translation into effective clinical treatments.
Author contributions
MM: Conceptualization, Data curation, Methodology, Writing–original draft, Writing–review and editing. EP: Conceptualization, Funding acquisition, Writing–review and editing. RT: Conceptualization, Data curation, Methodology, Writing–original draft, Writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was supported by DOR and BIRD grants from the University of Padova and by the European Union’s Horizon 2020 programme, H2020-EU.1.3. – Excellent Science–Marie Skłodowska-Curie Actions, grant agreement ID 956544 (DIRNANO: Directing the Immune Response through Designed Nanomaterials). Open Access funding provided by Universitádegli Studi di Padova University of Padua, Open Science Committee.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Adams, N., and Schubert, U. S. (2007). Poly(2-oxazolines) in biological and biomedical application contexts. Adv. Drug Deliv. Rev. 59, 1504–1520. doi:10.1016/j.addr.2007.08.018
Akhter, M. H., Khalilullah, H., Gupta, M., Alfaleh, M. A., Alhakamy, N. A., Riadi, Y., et al. (2021). Impact of protein corona on the biological identity of nanomedicine: understanding the fate of nanomaterials in the biological milieu. Biomedicines 9, 1496. doi:10.3390/biomedicines9101496
Angioni, R., Bonfanti, M., Caporale, N., Sánchez-Rodríguez, R., Munari, F., Savino, A., et al. (2023). RAGE engagement by SARS-CoV-2 enables monocyte infection and underlies COVID-19 severity. Cell Rep. Med. 4, 101266. doi:10.1016/j.xcrm.2023.101266
Ashkarran, A. A., Gharibi, H., Voke, E., Landry, M. P., Saei, A. A., and Mahmoudi, M. (2022). Measurements of heterogeneity in proteomics analysis of the nanoparticle protein corona across core facilities. Nat. Commun. 13, 6610. doi:10.1038/s41467-022-34438-8
Bai, X., Wang, J., Mu, Q., and Su, G. (2021). In vivo protein corona formation: characterizations, effects on engineered nanoparticles’ biobehaviors, and applications. Front. Bioeng. Biotechnol. 9, 646708. doi:10.3389/fbioe.2021.646708
Behzadi, S., Serpooshan, V., Tao, W., Hamaly, M. A., Alkawareek, M. Y., Dreaden, E. C., et al. (2017). Cellular uptake of nanoparticles: journey inside the cell. Chem. Soc. Rev. 46, 4218–4244. doi:10.1039/C6CS00636A
Blume, J. E., Manning, W. C., Troiano, G., Hornburg, D., Figa, M., Hesterberg, L., et al. (2020). Rapid, deep and precise profiling of the plasma proteome with multi-nanoparticle protein corona. Nat. Commun. 11, 3662. doi:10.1038/s41467-020-17033-7
Böhmert, L., Voß, L., Stock, V., Braeuning, A., Lampen, A., and Sieg, H. (2020). Isolation methods for particle protein corona complexes from protein-rich matrices. Nanoscale Adv. 2, 563–582. doi:10.1039/c9na00537d
Bonvin, D., Chiappe, D., Moniatte, M., Hofmann, H., and Mionić Ebersold, M. (2017). Methods of protein corona isolation for magnetic nanoparticles. Analyst 142, 3805–3815. doi:10.1039/C7AN00646B
Cai, R., Baimanov, D., Yuan, H., Xie, H., Yu, S., Zhang, Z., et al. (2024). Protein corona-directed cellular recognition and uptake of polyethylene nanoplastics by macrophages. Environ. Sci. Technol. 58, 14158–14168. doi:10.1021/acs.est.4c05215
Carril, M., Padro, D., Del Pino, P., Carrillo-Carrion, C., Gallego, M., and Parak, W. J. (2017). In situ detection of the protein corona in complex environments. Nat. Commun. 8, 1542. doi:10.1038/s41467-017-01826-4
Cedervall, T., Lynch, I., Lindman, S., Berggard, T., Thulin, E., Nilsson, H., et al. (2007). Understanding the nanoparticle-protein corona using methods to quantify exchange rates and affinities of proteins for nanoparticles. Proc. Natl. Acad. Sci. 104, 2050–2055. doi:10.1073/pnas.0608582104
Chrishtop, V. V., Prilepskii, A. Y., Nikonorova, V. G., and Mironov, V. A. (2021). Nanosafety vs. nanotoxicology: adequate animal models for testing in vivo toxicity of nanoparticles. Toxicology 462, 152952. doi:10.1016/j.tox.2021.152952
Daramy, K., Punnabhum, P., Hussain, M., Minelli, C., Pei, Y., Rattray, N. J. W., et al. (2023). Nanoparticle isolation from biological media for protein corona analysis: the impact of incubation and recovery protocols on nanoparticle properties. J. Pharm. Sci. S0022-3549 (23), 2826–2836. doi:10.1016/j.xphs.2023.12.021
Digiacomo, L., Palchetti, S., Giulimondi, F., Pozzi, D., Zenezini Chiozzi, R., Capriotti, A. L., et al. (2019). The biomolecular corona of gold nanoparticles in a controlled microfluidic environment. Lab. Chip 19, 2557–2567. doi:10.1039/C9LC00341J
Docter, D., Strieth, S., Westmeier, D., Hayden, O., Gao, M., Knauer, S. K., et al. (2015). No king without A crown – impact of the nanomaterial-protein corona on nanobiomedicine. Nanomedicine (Lond.) 10, 503–519. doi:10.2217/nnm.14.184
Domínguez-Andrés, J., Arts, R. J. W., Bekkering, S., Bahrar, H., Blok, B. A., De Bree, L. C. J., et al. (2021). In vitro induction of trained immunity in adherent human monocytes. Star. Protoc. 2, 100365. doi:10.1016/j.xpro.2021.100365
D’souza, A. A., and Shegokar, R. (2016). Polyethylene glycol (PEG): a versatile polymer for pharmaceutical applications. Expert Opin. Drug Deliv. 13, 1257–1275. doi:10.1080/17425247.2016.1182485
Escamilla-Rivera, V., Solorio-Rodriguez, A., Uribe-Ramirez, M., Lozano, O., Lucas, S., Chagolla-López, A., et al. (2019). Plasma protein adsorption on Fe3O4-PEG nanoparticles activates the complement system and induces an inflammatory response. IJN 14, 2055–2067. doi:10.2147/IJN.S192214
Fedeli, C., Segat, D., Tavano, R., Bubacco, L., De Franceschi, G., De Laureto, P. P., et al. (2015). The functional dissection of the plasma corona of SiO 2 -NPs spots histidine rich glycoprotein as a major player able to hamper nanoparticle capture by macrophages. Nanoscale 7, 17710–17728. doi:10.1039/C5NR05290D
Fedeli, C., Segat, D., Tavano, R., De Franceschi, G., De Laureto, P. P., Lubian, E., et al. (2014). Variations of the corona HDL:albumin ratio determine distinct effects of amorphous SiO 2 nanoparticles on monocytes and macrophages in serum. Nanomedicine (Lond.) 9, 2481–2497. doi:10.2217/nnm.14.22
Foroozandeh, P., and Aziz, A. A. (2015). Merging worlds of nanomaterials and biological environment: factors governing protein corona formation on nanoparticles and its biological consequences. Nanoscale Res. Lett. 10, 221. doi:10.1186/s11671-015-0922-3
Francia, V., Zhang, Y., Cheng, M. H. Y., Schiffelers, R. M., Witzigmann, D., and Cullis, P. R. (2024). A magnetic separation method for isolating and characterizing the biomolecular corona of lipid nanoparticles. Proc. Natl. Acad. Sci. U.S.A. 121, e2307803120. doi:10.1073/pnas.2307803120
Fuentes-Cervantes, A., Ruiz Allica, J., Calderón Celis, F., Costa-Fernández, J. M., and Ruiz Encinar, J. (2023). The potential of ICP-MS as a complementary tool in nanoparticle–protein corona analysis. Nanomaterials 13, 1132. doi:10.3390/nano13061132
García-Álvarez, R., and Vallet-Regí, M. (2021). Hard and soft protein corona of nanomaterials: analysis and relevance. Nanomaterials 11, 888. doi:10.3390/nano11040888
Garcia-Cao, I., Song, M. S., Hobbs, R. M., Laurent, G., Giorgi, C., de Boer, V. C. J., et al. (2012). Systemic elevation of PTEN induces a tumor-suppressive metabolic state. Cell 149, 49–62. doi:10.1016/j.cell.2012.02.030
Gaya Da Costa, M., Poppelaars, F., Van Kooten, C., Mollnes, T. E., Tedesco, F., Würzner, R., et al. (2018). Age and sex-associated changes of complement activity and complement levels in a healthy caucasian population. Front. Immunol. 9, 2664. doi:10.3389/fimmu.2018.02664
Gräfe, C., Weidner, A., Lühe, M. V. D., Bergemann, C., Schacher, F. H., Clement, J. H., et al. (2016). Intentional formation of a protein corona on nanoparticles: serum concentration affects protein corona mass, surface charge, and nanoparticle–cell interaction. Int. J. Biochem. and Cell Biol. 75, 196–202. doi:10.1016/j.biocel.2015.11.005
Guo, F., Luo, S., Wang, L., Wang, M., Wu, F., Wang, Y., et al. (2024). Protein corona, influence on drug delivery system and its improvement strategy: a review. Int. J. Biol. Macromol. 256, 128513. doi:10.1016/j.ijbiomac.2023.128513
Gupta, M. N., and Roy, I. (2020). How corona formation impacts nanomaterials as drug carriers. Mol. Pharm. 17, 725–737. doi:10.1021/acs.molpharmaceut.9b01111
Gustafson, H. H., Holt-Casper, D., Grainger, D. W., and Ghandehari, H. (2015). Nanoparticle uptake: the phagocyte problem. Nano Today 10, 487–510. doi:10.1016/j.nantod.2015.06.006
Hacene, Y. C., Loiseau, A., Maio, V. D. P., Grenier, P., Boisselier, E., and Bertrand, N. (2021). Isolating nanoparticles from complex biological media by immunoprecipitation. Nano Lett. 21, 4530–4538. doi:10.1021/acs.nanolett.0c05056
Hamad, I., Hunter, A. C., Szebeni, J., and Moghimi, S. M. (2008). Poly(ethylene glycol)s generate complement activation products in human serum through increased alternative pathway turnover and a MASP-2-dependent process. Mol. Immunol. 46, 225–232. doi:10.1016/j.molimm.2008.08.276
Ho, Y. T., Poinard, B., Yeo, E. L. L., and Kah, J. C. Y. (2015). An instantaneous colorimetric protein assay based on spontaneous formation of a protein corona on gold nanoparticles. Analyst 140, 1026–1036. doi:10.1039/C4AN01819B
Hoang Thi, T. T., Pilkington, E. H., Nguyen, D. H., Lee, J. S., Park, K. D., and Truong, N. P. (2020). The importance of poly(ethylene glycol) alternatives for overcoming PEG immunogenicity in drug delivery and bioconjugation. Polymers 12, 298. doi:10.3390/polym12020298
Hoogenboom, R. (2009). Poly(2-oxazoline)s: a polymer class with numerous potential applications. Angew. Chem. Int. Ed. 48, 7978–7994. doi:10.1002/anie.200901607
Joh, D. Y., Zimmers, Z., Avlani, M., Heggestad, J. T., Aydin, H. B., Ganson, N., et al. (2019). Architectural modification of conformal PEG-bottlebrush coatings minimizes anti-PEG antigenicity while preserving stealth properties. Adv. Healthc. Mater. 8, 1801177. doi:10.1002/adhm.201801177
Khalilov, R., Bakishzade, A., and Nasibova, A. (2023). Future prospects of biomaterials in nanomedicine. Adv. Biol. and Earth Sci. 8 (Special Issue), 5–10. doi:10.62476/abes.9s5
Khan, S., Sharifi, M., Gleghorn, J. P., Babadaei, M. M. N., Bloukh, S. H., Edis, Z., et al. (2022). Artificial engineering of the protein corona at bio-nano interfaces for improved cancer-targeted nanotherapy. J. Control Release 348, 127–147. doi:10.1016/j.jconrel.2022.05.055
Kim, J. A., Salvati, A., Åberg, C., and Dawson, K. A. (2014). Suppression of nanoparticle cytotoxicity approaching in vivo serum concentrations: limitations of in vitro testing for nanosafety. Nanoscale 6, 14180–14184. doi:10.1039/C4NR04970E
Kopac, T. (2021). Protein corona, understanding the nanoparticle-protein interactions and future perspectives: a critical review. Int. J. Biol. Macromol. 169, 290–301. doi:10.1016/j.ijbiomac.2020.12.108
Kristensen, K., Münter, R., Kempen, P. J., Thomsen, M. E., Stensballe, A., and Andresen, T. L. (2021). Isolation methods commonly used to study the liposomal protein corona suffer from contamination issues. Acta Biomater. 130, 460–472. doi:10.1016/j.actbio.2021.06.008
La-Beck, N. M., Islam, Md. R., and Markiewski, M. M. (2021). Nanoparticle-induced complement activation: implications for cancer nanomedicine. Front. Immunol. 11, 603039. doi:10.3389/fimmu.2020.603039
Lachmann, P. J. (2010). Preparing serum for functional complement assays. J. Immunol. Methods 352, 195–197. doi:10.1016/j.jim.2009.11.003
Leal, J., Peng, X., Liu, X., Arasappan, D., Wylie, D. C., Schwartz, S. H., et al. (2020). Peptides as surface coatings of nanoparticles that penetrate human cystic fibrosis sputum and uniformly distribute in vivo following pulmonary delivery. J. Control. Release 322, 457–469. doi:10.1016/j.jconrel.2020.03.032
Lebreton, V., Legeay, S., Vasylaki, A., Lagarce, F., and Saulnier, P. (2023). Protein corona formation on lipidic nanocapsules: influence of the interfacial PEG repartition. Eur. J. Pharm. Sci. 189, 106537. doi:10.1016/j.ejps.2023.106537
Lee, S. Y., Son, J. G., Moon, J. H., Joh, S., and Lee, T. G. (2020). Comparative study on formation of protein coronas under three different serum origins. Biointerphases 15, 061002. doi:10.1116/6.0000396
Li, Y., Wang, G., Griffin, L., Banda, N. K., Saba, L. M., Groman, E. V., et al. (2021). Complement opsonization of nanoparticles: differences between humans and preclinical species. J. Control. Release 338, 548–556. doi:10.1016/j.jconrel.2021.08.048
Liessi, N., Maragliano, L., Castagnola, V., Bramini, M., Benfenati, F., and Armirotti, A. (2021). Isobaric labeling proteomics allows a high-throughput investigation of protein corona orientation. Anal. Chem. 93, 784–791. doi:10.1021/acs.analchem.0c03134
Lipsa, D., Ruiz Moreno, A., Desmet, C., Bianchi, I., Geiss, O., Colpo, P., et al. (2023). Inter-individual variations: a challenge for the standardisation of complement activation assays. IJN 18, 711–720. doi:10.2147/IJN.S384184
Liu, N., Liang, Y., Wei, T., Huang, X., Zhang, T., and Tang, M. (2024). Protein corona exacerbated inflammatory response in macrophages elicited by CdTe quantum dots. NanoImpact 33, 100494. doi:10.1016/j.impact.2024.100494
Liu, S., Wang, Z., Jiang, X., Gan, J., Tian, X., Xing, Z., et al. (2021). Denatured corona proteins mediate the intracellular bioactivities of nanoparticles via the unfolded protein response. Biomaterials 265, 120452. doi:10.1016/j.biomaterials.2020.120452
Lo Giudice, M. C., Herda, L. M., Polo, E., and Dawson, K. A. (2016). In situ characterization of nanoparticle biomolecular interactions in complex biological media by flow cytometry. Nat. Commun. 7, 13475. doi:10.1038/ncomms13475
Lundqvist, M., Augustsson, C., Lilja, M., Lundkvist, K., Dahlbäck, B., Linse, S., et al. (2017). The nanoparticle protein corona formed in human blood or human blood fractions. PLoS ONE 12, e0175871. doi:10.1371/journal.pone.0175871
Mahmoudi, M. (2022). The need for improved methodology in protein corona analysis. Nat. Commun. 13, 49. doi:10.1038/s41467-021-27643-4
Meital, L. T., Coward, A. S., Windsor, M. T., Bailey, T. G., Kuballa, A., and Russell, F. D. (2019). A simple and effective method for the isolation and culture of human monocytes from small volumes of peripheral blood. J. Immunol. Methods 472, 75–78. doi:10.1016/j.jim.2019.04.005
Mekseriwattana, W., Thiangtrongjit, T., Reamtong, O., Wongtrakoongate, P., and Katewongsa, K. P. (2022). Proteomic analysis reveals distinct protein corona compositions of citrate- and riboflavin-coated SPIONs. ACS Omega 7, 37589–37599. doi:10.1021/acsomega.2c04440
Meng, Y., Chen, J., Liu, Y., Zhu, Y., Wong, Y.-K., Lyu, H., et al. (2022). A highly efficient protein corona-based proteomic analysis strategy for the discovery of pharmacodynamic biomarkers. J. Pharm. Analysis 12, 879–888. doi:10.1016/j.jpha.2022.07.002
Merle, N. S., Church, S. E., Fremeaux-Bacchi, V., and Roumenina, L. T. (2015). Complement system Part I - molecular mechanisms of activation and regulation. Front. Immunol. 6, 262. doi:10.3389/fimmu.2015.00262
Mestas, J., and Hughes, C. C. W. (2004). Of mice and not men: differences between mouse and human immunology. J. Immunol. 172, 2731–2738. doi:10.4049/jimmunol.172.5.2731
Mirshafiee, V., Kim, R., Mahmoudi, M., and Kraft, M. L. (2016). The importance of selecting a proper biological milieu for protein corona analysis in vitro: human plasma versus human serum. Int. J. Biochem. and Cell Biol. 75, 188–195. doi:10.1016/j.biocel.2015.11.019
Moghimi, S. M., Andersen, A. J., Ahmadvand, D., Wibroe, P. P., Andresen, T. L., and Hunter, A. C. (2011). Material properties in complement activation. Adv. Drug Deliv. Rev. 63, 1000–1007. doi:10.1016/j.addr.2011.06.002
Moghimi, S. M., Hunter, A. C., Dadswell, C. M., Savay, S., Alving, C. R., and Szebeni, J. (2004). Causative factors behind poloxamer 188 (Pluronic F68, FlocorTM)-induced complement activation in human sera. Biochimica Biophysica Acta (BBA) - Mol. Basis Dis. 1689, 103–113. doi:10.1016/j.bbadis.2004.02.005
Moghimi, S. M., and Simberg, D. (2022). Critical issues and pitfalls in serum and plasma handling for complement analysis in nanomedicine and bionanotechnology. Nano Today 44, 101479. doi:10.1016/j.nantod.2022.101479
Moghimi, S. M., Simberg, D., Papini, E., and Farhangrazi, Z. S. (2020). Complement activation by drug carriers and particulate pharmaceuticals: principles, challenges and opportunities. Adv. Drug Deliv. Rev. 157, 83–95. doi:10.1016/j.addr.2020.04.012
Moghimi, S. M., and Szebeni, J. (2003). Stealth liposomes and long circulating nanoparticles: critical issues in pharmacokinetics, opsonization and protein-binding properties. Prog. Lipid Res. 42, 463–478. doi:10.1016/S0163-7827(03)00033-X
Mohammad-Beigi, H., Hayashi, Y., Zeuthen, C. M., Eskandari, H., Scavenius, C., Juul-Madsen, K., et al. (2020). Mapping and identification of soft corona proteins at nanoparticles and their impact on cellular association. Nat. Commun. 11, 4535. doi:10.1038/s41467-020-18237-7
Mout, R., Moyano, D. F., Rana, S., and Rotello, V. M. (2012). Surface functionalization of nanoparticles for nanomedicine. Chem. Soc. Rev. 41, 2539. doi:10.1039/c2cs15294k
Mu, Q., Jiang, G., Chen, L., Zhou, H., Fourches, D., Tropsha, A., et al. (2014). Chemical basis of interactions between engineered nanoparticles and biological systems. Chem. Rev. 114, 7740–7781. doi:10.1021/cr400295a
Neun, B. W., Ilinskaya, A. N., and Dobrovolskaia, M. A. (2018). “Analysis of complement activation by nanoparticles,” in Characterization of nanoparticles intended for drug delivery. Editor S. E. McNeil (New York, NY: Springer New York), 149–160. doi:10.1007/978-1-4939-7352-1_13
Owensiii, D., and Peppas, N. (2006). Opsonization, biodistribution, and pharmacokinetics of polymeric nanoparticles. Int. J. Pharm. 307, 93–102. doi:10.1016/j.ijpharm.2005.10.010
Palchetti, S., Colapicchioni, V., Digiacomo, L., Caracciolo, G., Pozzi, D., Capriotti, A. L., et al. (2016). The protein corona of circulating PEGylated liposomes. Biochimica Biophysica Acta (BBA) - Biomembr. 1858, 189–196. doi:10.1016/j.bbamem.2015.11.012
Panico, S., Capolla, S., Bozzer, S., Toffoli, G., Dal Bo, M., and Macor, P. (2022). Biological features of nanoparticles: protein corona formation and interaction with the immune system. Pharmaceutics 14, 2605. doi:10.3390/pharmaceutics14122605
Pederzoli, F., Tosi, G., Vandelli, M. A., Belletti, D., Forni, F., and Ruozi, B. (2017). Protein corona and nanoparticles: how can we investigate on? WIREs Nanomed Nanobiotechnol 9, e1467. doi:10.1002/wnan.1467
Poulsen, K. M., and Payne, C. K. (2022). Concentration and composition of the protein corona as a function of incubation time and serum concentration: an automated approach to the protein corona. Anal. Bioanal. Chem. 414, 7265–7275. doi:10.1007/s00216-022-04278-y
Rampado, R., Crotti, S., Caliceti, P., Pucciarelli, S., and Agostini, M. (2020). Recent advances in understanding the protein corona of nanoparticles and in the formulation of “stealthy” nanomaterials. Front. Bioeng. Biotechnol. 8, 166. doi:10.3389/fbioe.2020.00166
Repnik, U., Knezevic, M., and Jeras, M. (2003). Simple and cost-effective isolation of monocytes from buffy coats. J. Immunol. Methods 278, 283–292. doi:10.1016/S0022-1759(03)00231-X
Rizvi, S. A. A., and Saleh, A. M. (2018). Applications of nanoparticle systems in drug delivery technology. Saudi Pharm. J. 26, 64–70. doi:10.1016/j.jsps.2017.10.012
Sahneh, F. D., Scoglio, C. M., Monteiro-Riviere, N. A., and Riviere, J. E. (2015). Predicting the impact of biocorona formation kinetics on interspecies extrapolations of nanoparticle biodistribution modeling. Nanomedicine (Lond.) 10, 25–33. doi:10.2217/nnm.14.60
Sahu, A., and Lambris, J. D. (2001). Structure and biology of complement protein C3, a connecting link between innate and acquired immunity. Immunol. Rev. 180, 35–48. doi:10.1034/j.1600-065X.2001.1800103.x
Sakulkhu, U., Mahmoudi, M., Maurizi, L., Coullerez, G., Hofmann-Amtenbrink, M., Vries, M., et al. (2015). Significance of surface charge and shell material of superparamagnetic iron oxide nanoparticle (SPION) based core/shell nanoparticles on the composition of the protein corona. Biomater. Sci. 3, 265–278. doi:10.1039/C4BM00264D
Sanchez-Guzman, D., Giraudon--Colas, G., Marichal, L., Boulard, Y., Wien, F., Degrouard, J., et al. (2020). In situ analysis of weakly bound proteins reveals molecular basis of soft corona formation. ACS Nano 14, 9073–9088. doi:10.1021/acsnano.0c04165
Sanchez-Moreno, P., Buzon, P., Boulaiz, H., Peula-Garcia, J. M., Ortega-Vinuesa, J. L., Luque, I., et al. (2015). Balancing the effect of corona on therapeutic efficacy and macrophage uptake of lipid nanocapsules. Biomaterials 61, 266–278. doi:10.1016/j.biomaterials.2015.04.049
Schöttler, S., Klein, K., Landfester, K., and Mailänder, V. (2016). Protein source and choice of anticoagulant decisively affect nanoparticle protein corona and cellular uptake. Nanoscale 8, 5526–5536. doi:10.1039/C5NR08196C
Shang, L., and Nienhaus, G. U. (2017). In situ characterization of protein adsorption onto nanoparticles by fluorescence correlation spectroscopy. Acc. Chem. Res. 50, 387–395. doi:10.1021/acs.accounts.6b00579
Shevchenko, A., Tomas, H., Havlis, J., Olsen, J. V., and Mann, M. (2006). In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 1, 2856–2860. doi:10.1038/nprot.2006.468
Simon, J., Kuhn, G., Fichter, M., Gehring, S., Landfester, K., and Mailänder, V. (2021). Unraveling the in vivo protein corona. Cells 10, 132. doi:10.3390/cells10010132
Singh, N., Marets, C., Boudon, J., Millot, N., Saviot, L., and Maurizi, L. (2021). In vivo protein corona on nanoparticles: does the control of all material parameters orient the biological behavior? Nanoscale Adv. 3, 1209–1229. doi:10.1039/d0na00863j
Solorio-Rodríguez, A., Escamilla-Rivera, V., Uribe-Ramírez, M., Chagolla, A., Winkler, R., García-Cuellar, C. M., et al. (2017). A comparison of the human and mouse protein corona profiles of functionalized SiO 2 nanocarriers. Nanoscale 9, 13651–13660. doi:10.1039/C7NR04685E
Strojan, K., Leonardi, A., Bregar, V. B., Križaj, I., Svete, J., and Pavlin, M. (2017). Dispersion of nanoparticles in different media importantly determines the composition of their protein corona. PLoS ONE 12, e0169552. doi:10.1371/journal.pone.0169552
Tadjiki, S., Sharifi, S., Lavasanifar, A., and Mahmoudi, M. (2024). Advancing in situ analysis of biomolecular corona: opportunities and challenges in utilizing field-flow fractionation. ACS Bio Med. Chem. Au 4, 77–85. doi:10.1021/acsbiomedchemau.4c00001
Tavano, R., Gabrielli, L., Lubian, E., Fedeli, C., Visentin, S., Polverino De Laureto, P., et al. (2018). C1q-Mediated complement activation and C3 opsonization trigger recognition of stealth poly(2-methyl-2-oxazoline)-coated silica nanoparticles by human phagocytes. ACS Nano 12, 5834–5847. doi:10.1021/acsnano.8b01806
Tenzer, S., Docter, D., Kuharev, J., Musyanovych, A., Fetz, V., Hecht, R., et al. (2013). Rapid formation of plasma protein corona critically affects nanoparticle pathophysiology. Nat. Nanotech 8, 772–781. doi:10.1038/nnano.2013.181
Trzciński, J. W., Morillas-Becerril, L., Scarpa, S., Tannorella, M., Muraca, F., Rastrelli, F., et al. (2021). Poly(lipoic acid)-based nanoparticles as self-organized, biocompatible, and corona-free nanovectors. Biomacromolecules 22, 467–480. doi:10.1021/acs.biomac.0c01321
Viegas, T. X., Bentley, M. D., Harris, J. M., Fang, Z., Yoon, K., Dizman, B., et al. (2011). Polyoxazoline: chemistry, properties, and applications in drug delivery. Bioconjugate Chem. 22, 976–986. doi:10.1021/bc200049d
Walkey, C. D., and Chan, W. C. W. (2012). Understanding and controlling the interaction of nanomaterials with proteins in a physiological environment. Chem. Soc. Rev. 41, 2780–2799. doi:10.1039/C1CS15233E
Xu, Q., Rajendrakumar, S. K., and Khirallah, J. (2023). “Role of protein corona on nanoparticle-mediated organ and cell-targeted delivery,” in Encyclopedia of nanomaterials (Elsevier), 506–518. doi:10.1016/B978-0-12-822425-0.00052-X
Yan, Y., Gause, K. T., Kamphuis, M. M. J., Ang, C.-S., O’Brien-Simpson, N. M., Lenzo, J. C., et al. (2013). Differential roles of the protein corona in the cellular uptake of nanoporous polymer particles by monocyte and macrophage cell lines. ACS Nano 7, 10960–10970. doi:10.1021/nn404481f
Yang, Q., and Lai, S. K. (2017). “Engineering well-characterized PEG-coated nanoparticles for elucidating biological barriers to drug delivery,” in Cancer nanotechnology. Editor R. Zeineldin (New York, NY: Springer), 125–137. doi:10.1007/978-1-4939-6646-2_8
Yu, K., Andruschak, P., Yeh, H. H., Grecov, D., and Kizhakkedathu, J. N. (2018). Influence of dynamic flow conditions on adsorbed plasma protein corona and surface-induced thrombus generation on antifouling brushes. Biomaterials 166, 79–95. doi:10.1016/j.biomaterials.2018.03.009
Zhang, L., and Elias, J. E. (2017). “Relative protein quantification using tandem mass tag mass spectrometry,” in Proteomics: methods and protocols. Editors L. Comai, J. E. Katz, and P. Mallick (New York, NY: Springer), 185–198. doi:10.1007/978-1-4939-6747-6_14
Keywords: nanoparticle, protein corona, coating, protocols, shot-gun proteomics, in-gel digestion, TMT proteomics, nanoparticle uptake by phagocytes
Citation: Morbidelli M, Papini E and Tavano R (2024) Essential protocols for decoding the composition and the functional effects of the nanoparticle protein corona. Front. Nanotechnol. 6:1500567. doi: 10.3389/fnano.2024.1500567
Received: 23 September 2024; Accepted: 14 October 2024;
Published: 23 October 2024.
Edited by:
Cristina Satriano, University of Catania, ItalyCopyright © 2024 Morbidelli, Papini and Tavano. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Regina Tavano, cmVnaW5hLnRhdmFub0B1bmlwZC5pdA==
 Maria Morbidelli
Maria Morbidelli Emanuele Papini
Emanuele Papini Regina Tavano
Regina Tavano