
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Nanotechnol. , 16 April 2024
Sec. Environmental Nanotechnology
Volume 6 - 2024 | https://doi.org/10.3389/fnano.2024.1384465
The culture filtrate of Hypocrea lixii GGRK4 played a vital role as a reducing and stabilizing agent in the mycosynthesis of silver nanoparticles (AgNPs) using silver nitrate (AgNO3). The extracellular extract derived from fungi emerged as a noteworthy option for synthesizing AgNPs due to its potential composition of metabolites, including enzymes and other bioactive substances. Hence, the presence of a dark brown color serves as a key indicator for the biosynthesis of AgNPs through the reduction of Ag (I) ions to Ag by the fungal culture filtrate. To facilitate the synthesis of AgNPs, a combination of hybrid technologies, specifically the “one factor at a time” approach and statistical tools such as response surface methodology, was used using a face-centered central composite design (FCCCD). Utilizing a modified CX medium with pH of 5.02 supported the fungi synthesizing AgNPs at a temperature of 30°C. The multi-efficient AgNPs were characterized through various techniques, including UV–visible spectrophotometry, zeta size and potential analysis using a zeta size analyzer, transmission electron microscopy (TEM), X-ray diffraction (XRD), Fourier-transform infrared spectroscopy (FTIR), and fluorescence spectroscopy. The biosynthesized AgNPs have significant associated functional groups, revealed by FTIR analysis. TEM histogram analysis showed that these multi-efficient AgNPs have a size of 17.34 nm. Similarly, they have emission and excitation spectra of 450 nm and 390 nm, respectively, revealed by fluorescence spectrum analysis. Compared to the standard, the biosynthesized AgNPs have significant antibacterial and free radical scavenging properties and dye degradation capability. Additionally, the half-maximal inhibitory concentration (IC50) value was found statistically significant based on t-test analysis. Finally, the biosynthesized AgNPs could be used in potential applications encompassing ecofriendly degradation, antimicrobial activity, and therapeutic applications, such as free radical scavenging properties.
Nanotechnology, due to its structural fidelity for many years, contributed to almost every field of science; nanotechnology is regarded as the collective expertise of nowadays (Dejen et al., 2023). Several physical and chemical methodologies exist currently for nanoparticle synthesis, but due to their cost ineffectiveness and harmfulness to the environment, they are not widely implemented (Singh et al., 2013; Qamar et al., 2023). Nanoparticles are nanoscale particles between 1 and 100 nm in size or even smaller (Khan and Hossain, 2022). Nanoparticles made up of metals have gained significant attention due to their widespread applications in the field of nanochemistry. To synthesize nanoparticles, the use of natural machinery has been approached to obtain environment-friendly nanomaterial for successful remediation (Bhattacharjee et al., 2024; Valle-García et al., 2023). Utilizing any biological element such as bacteria, yeast, plants, fungi, and actinomycetes is a brilliant approach for an ecologically relevant way called green synthesis (Lateef et al., 2016).
Amongst other nanometals, silver nanoparticles (AgNPs) have gained considerable recognition due to their vast nanotechnological implementations, such as optical elements, chemical sensing, pharmaceutical components, chemical reactions, and antibacterial activity (Jorge de Souza et al., 2019). AgNPs gained the attention of scientific communities due to their immense antibacterial activity against drug-resistant pathogens (Huq, 2020; Fatima et al., 2021; More et al., 2023). In terms of microorganisms, filamentous fungi are considered the most ideal since they secrete more enzymes, are simpler to manage, and can thrive under either simple or complicated conditions (Sameena and Thoppil, 2022). It has been observed that the main factor causing the change of Ag+ to Ag0 is the presence of bioactive compounds such as enzymes, amino acids, and other reducing agents in the culture filtrate (Ottoni et al., 2017). Silver is a valuable metal that occurs naturally, most often as a mineral metal that contains different components. Numerous nano-silver products have been developed as a result of the strong antibacterial qualities of silver. Nowadays, the significant application of AgNPs in surgical equipment and other medical devices like bandages is on the rise (Muangman et al., 2006). Recent studies have focused on the amazing biological and physicochemical properties of AgNPs (Ankush et al., 2022; Vidyasagar et al., 2023). This is due to the diverse prospective applications of AgNPs in fields such as optics, environmental engineering, electronics, catalysis, and biomedicine (Govindappa et al., 2018). Several filamentous fungi, such as Aspergillus sp., Candida albicans, and Fusarium oxysporum, are extensively used for the production of AgNPs (Bhat et al., 2015; Rajput et al., 2016; Vijayan et al., 2016).
Previously, traditional methods such as physical and chemical processes were used for the synthesis of nanoparticles (NPs). Most of the chemical methods are expensive and release various toxic agents, which have an impact on the environment and human beings. Nowadays, the greener synthesis of NPs has a novel and significant avenue in the field of nanotechnology (Ahmad et al., 2019; Ramya, 2022). However, the synthesis of NPs using the fungal extract is the method agreed upon as the green and ecofriendly synthesis process. In addition to that, there are various studies available on the optimization processes in AgNP synthesis, mainly to reduce their size and improve their stability (Ajitha et al., 2015; Núñez et al., 2018). However, even if there are well-established methodologies for the synthesis of NPs, it is necessary to investigate significant and efficient synthesis methods that require short reaction times and are inexpensive to obtain NPs with multi-efficient ecofriendly applications. The traditional one-factor-at-a-time (OFAT) method is an overlooked interaction between different variables, and this process is not enough to predict the significant interactions between the variables. Hence, the design of the experiment (DOE) method must be used in order to accurately determine the possible interactions, and the face-centered central composite design (FCCCD) under the response surface methodology (RSM) can be a powerful tool for complex process optimization. RSM can provide an in-depth understanding of the pairwise coupled, curvilinear, and straight effects of the factors on the response (Kong et al., 2014). For experimental designs, the Box–Behnken design (BBD), central composite design (CCD), Doehlert matrix (DM), etc., are available as multivariate approaches. CCD is more efficient than BBD and DM in terms of design efficiency (Jung et al., 2015). Therefore, Javier and Camacho (2022) conducted experiments for the process optimization of AgNP synthesis using the RSM optimization tool.
AgNPs are used in several applications, including diagnostics, antimicrobial treatment, textiles, building materials, electronics, and the remediation of environmental issues. Nanoparticles have applications in many different fields for a variety of purposes. As a result, biologically produced nanoparticles offer a wider range of applications than chemically produced nanoparticles. Among other nanometals, AgNPs receive more attention because of their numerous applications in nanotechnology, including their use as optical components, sensors for chemicals, medicinal components, chemical reactions, and antibacterial activities (Burdușel et al., 2018). Thus, various microorganisms have been used to create functional nanoparticles. It is also widely recognized that the size, shape, and crystal structure of AgNPs have a significant impact on various ecofriendly applications. In addition, fungal cultures are easier to cultivate on an industrial scale and in the laboratory (Velhal et al., 2016). The antibacterial activity of AgNPs has been widely investigated, yet their mode of action is not fully elucidated. In another study, researchers reported the significant potential of AgNPs for dye degradation and antibacterial activity (Rafique et al., 2019). Furthermore, the accumulation of hydrogen peroxide (H2O2) leads to the creation of O− free radicals, which disrupt cell membranes. The scavenging capability of green synthesized AgNPs could be attributed to the functional groups adhered to them. Several studies related to the antioxidant properties of AgNPs have been investigated (Ahmed and Mustafa, 2020). Additionally, AgNPs have also been used as catalytic agents for the degradation of toxic dyes and have gained more attention in recent years. Several studies investigated the catalytic activity of AgNPs against numerous hazardous dyes, such as Rhodamine B dye, Phloxine B, methyl orange, thymol blue, Auramine O, and Congo red (Albeladi et al., 2020; Chandhru et al., 2020). Many researchers reported up to 90%–100% degradation of different dyes using green synthesized AgNPs (Chand et al., 2020; Pandey et al., 2020; Nazir et al., 2021). The presence of phenolic groups in the water system raises a major concern worldwide. Hence, the bioconversion of these compounds is necessary to clean the environment. However, AgNPs undergo catalytic reactions based on oxidation and reduction processes. The conversion of nitroaromatics has led to the production of valuable compounds. Therefore, a study reported the use of AgNPs as a catalyst for the biotransformation of p-nitrophenol into p-aminophenol compounds (Singh et al., 2018; Veisi et al., 2019).
In the present study, the presence of reductant biomolecules and enzymes from newly isolated fungal culture Hypocrea lixii GGRK4 was reported for the mycosynthesis of AgNPs. Therefore, the following aspects were investigated:
a) Process optimization for greener synthesis of AgNPs using statistical optimization tools such as the response surface methodology.
b) Characterization of the mycosynthesized AgNPs using different analytical techniques, i.e., Fourier-transform infrared spectroscopy (FTIR), X-ray diffraction (XRD), zeta size, zeta potential, transmission electron microscopy (TEM), and fluorescence spectroscopy.
c) Evaluation of the antibacterial activity of AgNPs against Gram-positive and Gram-negative bacteria.
d) Estimation of the free radical scavenging property and half-maximal inhibitory concentration (IC50) value of mycosynthesized AgNPs.
e) Determination of the catalytic efficiency of methylene dye degradation and biotransformation of p-nitrophenol.
The potato dextrose agar (PDA), yeast extract, peptone, nutrient agar, nutrient broth, Mueller–Hinton agar (MHA), sucrose, glucose, and lactose were purchased from HiMedia (Thane, Maharashtra, India). Chemicals such as ammonium sulfate, magnesium sulfate heptahydrate, dipotassium phosphate, methylene blue, sodium borohydride, sodium hydroxide, hydrochloric acid, copper sulfate, ferric chloride, sulfuric acid, sodium chloride, and barium chloride were purchased from Merck, India. AgNO3 was purchased from Sigma-Aldrich, Bangalore, India. All the chemicals used in this study are analytical grade.
The fungal culture H. lixii GGRK4 was obtained from the laboratory storage facility. One mycelium disk (8 mm diameter) of the culture was transferred on a PDA medium containing a Petri dish and incubated at 30°C for 3–5 days. The PDA slant was used for sub-culturing and stored at 4°C for further use.
In a 250-mL Erlenmeyer flask, 50 mL of optimized CX broth medium (see Supplementary Table S1 for composition) was inoculated with three mycelium disks (8 mm diameter) and incubated at 150 rpm and 30°C for 3 days. The cell-free culture filtrate (CFS) was separated by filtering fungal biomass using Whatman No. 1 filter paper, which was further clarified by centrifugation at 10,000 rpm and 4°C for 15 min. For the mycosynthesis process, a reaction mixture containing 1.75 mL of cleared CFS and 20 mL of 2 mM AgNO3 solution was incubated at room temperature under the incidence of light for 12 h. A brown color was recorded in the spectrum at a range of 200–800 nm using a UV-vis spectrophotometer (Spectro-UV8 MRC, United Kingdom).
Various process parameters for the mycosynthesis of AgNPs by H. lixii GGRK4 were optimized using a single-factor screening method. The physicochemical parameters such as AgNO3 concentration, CFS volume, reaction time, temperature, and pH were screened. Initially, five different media (Supplementary Table S1) were screened for the most suitable fungal culture for the biosynthesis of AgNPs. The effect of temperature (25°C–35°C) on the growth of the fungal culture filtrate supernatant was assessed for AgNP mycosynthesis. The effect of the concentration ranges of AgNO3 (0.5, 1.0, 1.5, 2.0, 2.5, 3.0, 3.5, 4.0, 4.5, and 5.0 mM) was studied for the mycosynthesis of AgNPs. Furthermore, the effect of different volumes of CFS on AgNP production was assessed. The effect of the reaction time on the AgNP mycosynthesis process was studied by synthesizing the reaction mixture at varying incubation time ranges (2, 4, 6, 8, 10, 14, and 20 h). All sets of tests were examined continuously by a control condition containing silver nitrate solution and the cell-free culture filtrate.
Statistical optimization was performed to obtain the optimum parameters for the AgNP mycosynthesis. For this, the optimization model, i.e., the CCD, was obtained using Design-Expert software (DOE ver. 13.0). Three independent input parameters at five levels are given in Supplementary Table S2. To find the most important factors for the highest yield, a total of 20 experimental matrices (Table 1) were obtained. The substantial correlation between all the variables was estimated using ANOVA and 3D surface response. Finally, the final experiment was conducted using the predicted ideal conditions of the model (silver nitrate concentration of 3.411 mM, volume of 1.931 mL of CFS, and a reaction period of 3 h and 21 min) to validate the model.
The zeta potential and zeta size of the biosynthesized AgNPs were studied using a zetasizer (Malvern Panalytical). The functional groups of AgNPs were determined by scanning the samples using an FTIR spectrophotometer at a range of 375–4,000 cm−1 (Bruker, Germany) (Shankar et al., 2020). The transmission electron microscope analysis was carried out using the JEM-2001 transmission electron microscope (JEOL Ltd., Tokyo, Japan) at 200 kV. In brief, AgNPs were diluted at a ratio of 1:200, and 5 µL of AgNPs were transferred and deposited to prepare a grid/matrix, allowing it to dry for 15 min. The size distribution of mycosynthesized AgNPs was determined using ImageJ 1.53e (NIH, Bethesda, MD).
An X-ray diffractometer with a high beam (Rigaku MiniFlex 600) was additionally used to find the crystal-like nature of AgNPs. The spectra were obtained within 10–70° of 2θ (scattering angle). The X-ray diffraction technique utilizing Cu Kα radiation was used to examine the phase development of the dried AgNP powder sample. The current and voltage of the generator were set to 25 mA and 35 KV, respectively.
The Debye–Scherrer formula was used to evaluate the AgNP crystalline size as follows:
Here,
The inter-planar spacing (d) was estimated using the following formula:
The Lattice constant was estimated using the following formula:
The range of emission and excitation wavelengths of AgNPs can be determined using a fluorescence spectrophotometer. The fluorescence spectroscopy of biosynthesized AgNPs was evaluated by using the method proposed by Qiu et al. (2022).
The antibacterial activity of AgNPs was tested using the agar disk diffusion method against Gram-positive (Staphylococcus aureus MTCC 96 and Bacillus pumilus MTCC 121) and Gram-negative (Escherichia coli MTCC 43 and Pseudomonas aeruginosa MTCC 424) test organisms. A measure of 100 µL of bacterial culture was spread uniformly on MHA media. The sterile disks were immersed in different concentrations (100, 200, 300, 400, and 500 μL/mL) of AgNP solution. Afterward, the immersed disks were placed on Petri plates and incubated for 24 h at 37°C. The zone of inhibition was measured by taking the readings in triplicate.
The dye degradation efficacy of AgNPs against methylene blue (MB) was determined by performing a reaction. First, 20 mL of 5 mg/mL MB was taken in a reaction tube and mixed with 30 µg AgNPs and 200 µL sodium borohydride (NaBH4). A control sample was also prepared by the addition of 20 mL of 5 mg/mL MB dye and 30 µg NaBH4. The tubes were then observed for color change at regular intervals of time and the absorbance at 662 nm using a UV–visible spectrophotometer.
The percentage of degradation was calculated using the following formula:
where Abs0 is the initial concentration and Abst is the final concentration.
The ability of the biosynthesized AgNPs to undergo biotransformation was evaluated through the transformation of p-nitrophenol into p-aminophenol. In brief, 1 mL of freshly prepared NaBH4 solution (30 mM) was mixed with 150 μL of p-nitrophenol (2 mM). Finally, 10 μL of biosynthesized nanoparticles were added, and 3 mL of reaction volume was maintained using distilled water. The biotransformation was performed at room temperature and was monitored using a UV-vis spectrophotometer (Spectro-UV8 MRC, United Kingdom). At varied time intervals (0–180 min), the absorption peak at 400 nm decreased, while the peak at 300 nm increased.
The 2,2-diphenyl-1-picrylhydrazyl (DPPH) test was used to assess the ability of AgNPs to scavenge free radicals. In brief, 200 µM DPPH solution was prepared in ethanol and used as the substrate. The reaction mixture contained 2.5 mL of AgNPs having different concentrations (50, 100, 150, 250, and 500 μg/mL) and 1 mL of DPPH solution. The reaction sets were gently shaken and incubated under dark conditions for up to 30 min at room temperature, and the absorbance was measured at 518 nm using a UV-vis spectrophotometer (Spectro-UV8 MRC, United Kingdom). The percentage scavenging activity was measured by comparing the absorbance to that of the control (which solely contained DPPH). Ascorbic acid solution was used as the positive control. The test was carried out in triplicate. The percentage of inhibition could be determined by using following formula:
The IC50 values were determined using GraphPad Prism software (Aykul and Martinez-Hackert, 2016). First, we evaluated the % scavenging activity data taken at different concentrations of the biosynthesized AgNPs and the standard (ascorbic acid). To obtain the IC50 values, we fitted the % scavenging activity as a function for fit spline analysis using the non-linear regression algorithm for log (concentration) versus normalized response implemented in GraphPad.
All investigations were performed in triplicate, and data were interpreted as the mean ± SD calculated using GraphPad Prism (ver 8) software. Statistical analysis was performed using one-way ANOVA, followed by Tukey’s test, and p < 0.05 was regarded as significant. The XRD analysis was performed using OriginPro 2021 software.
In the present study, biological approaches are preferred for reasons such as their convenient accessibility, inexpensiveness, and ecological properties (Ahmed et al., 2022). The color shift from pale milky to dark brown verified the reduction of silver nitrate into AgNPs in the fungal culture filtrate supernatant. The conversion of Ag+ to Ag0 is mostly caused by the presence of biomolecules in the culture filtrate supernatant of the fungus, including amino acids, polysaccharides, enzymes/proteins, and vitamins. Mukherjee et al. (2001) demonstrated and reported that the surface of fungi played a significant role in trapping Ag+ and successively reducing it into nanoparticles by extracellular enzymes secreted by the fungal system. Several other studies reported that the nitrate reductase activity of the isolates had a potent ability to synthesize NPs, which supports the hypothesis of the enzymatic reduction of AgNO3 into AgNPs (Saifuddin et al., 2009; Hamedi et al., 2014; Ottoni et al., 2017). The significant participation of these bioactive molecules has been further assessed by FTIR analysis (see Section 3.3.2). The FTIR investigation revealed the connections between silver and biologically active molecules that play an important role in the formation and constancy of NPs as capping agents. The carboxylic acid group obtained from the amino acid of proteins has strong connections among the NPs due to the negative charge (Neethu et al., 2018; Ibrahim et al., 2021). A measure of 2 mM silver nitrate solution was introduced to the fungal culture filtrate of H. lixii GGRK4 at 30°C in the presence of white light under static conditions to synthesize silver nanoparticles. The initial confirmation of the synthesis of AgNPs was the color change of the reaction mixture from colorless to dark brown (Figure 1A). The presence of a band at a surface plasmon resonance (SPR) of 464 nm of AgNPs in the UV–visible spectrum indicated their synthesis (Figure 1B). The SPR concept describes the excitation of electrons in the conductive band surrounding silver particles. The absorption band observed at 464 nm closely matches the value reported by previous studies (Elgorban et al., 2016; Ottoni et al., 2017; Neethu et al., 2018). However, spectroscopic analysis of the silver nitrate solution in the absence of the culture filtrate supernatant revealed two bands at 222 and 240 nm. Additionally, it was shown that no synthesis of fungal biomass occurred, and that the mycosynthesis of AgNPs exclusively occurred extracellularly.
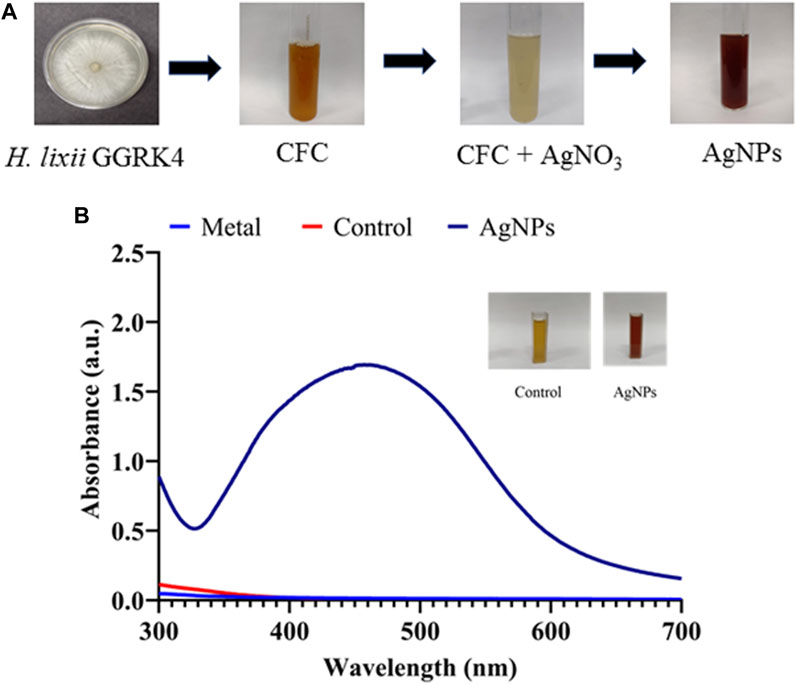
Figure 1. Mycosynthesis of silver nanoparticles (AgNPs): (A) step-wise biosynthesis process and (B) UV-vis spectrum of biosynthesized AgNPs.
The medium composition has an impact on metabolite production and microbial growth, which are crucial processes in the biological synthesis of AgNPs (Shankar et al., 2020). In the present study, the CX medium containing lactose and peptone maintained healthy fungal growth and encouraged the production of extracellularly synthesized AgNPs (Figure 2A). In a related study, researchers produced AgNPs by the cultivation of F. oxysporum in 10 distinct medium sources (Birla et al., 2013). A study reported that glucose was used as a carbon source for the production of AgNPs using Aspergillus species (Zomorodian et al., 2016). Another study investigated various media, such as Czapek Dox (CZAPEK), Richard medium (RM), glucose yeast extract peptone (GYP), potato dextrose broth (PDB), protease production (PP) media, and Sabouraud’s dextrose (SB), for the enhanced synthesis of AgNPs (Saxena et al., 2016). Various concentrations of AgNO3 ranging from 0.5 to 5 mM were used to determine to best suitable concentration. It was found that 2 mM of AgNO3 reported the optimum concentration for the maximum synthesis of AgNPs (Figure 2B). In the best possible reaction circumstances, the amount of culture filtrate has a considerable and important impact on the yield of AgNPs. The results show that as the volume of the culture filtrate supernatant increases, the synthesis of AgNPs increases (Figure 2C). This is because it now requires more protein or an enzyme to convert AgNO3 into AgNPs. Higher amounts of “reducing agents” (enzymes) in the broth medium result in the greater production of AgNPs (Singh et al., 2013). Several concentrations of AgNO3 were used for the production of AgNPs using a fungal culture filtrate of Aspergillus oryzae (Phanjom and Ahmed, 2017). A similar type of experiment was conducted by Omran et al. (2018). Similarly, many researchers conducted experiments with different concentrations of dry biomass of culture with optimum concentrations of AgNO3 and reported that increasing the amount of dry biomass of culture improved the synthesis of AgNPs (Saxena et al., 2016; Omran et al., 2018). pH of the media plays a crucial role in the maintenance and production of microbial metabolites, and in our study, the highest biosynthesis of AgNPs was found at pH 5.02. Omran et al. (2018) reported that the highest amount of AgNPs were synthesized at pH 7.0. It was also observed that a lower synthesis of AgNPs was obtained when increasing pH due to the denaturation of the enzymes, which can lead to a further decrease in Ag+ reduction. Additionally, temperature is another crucial factor that affects the proliferation and production of metabolites by fungi. Our study reports that the maximum amount of AgNPs were synthesized at 30°C, followed by 25°C (Figure 2D). Similarly, thermophilic mold Humicola sp. produced AgNPs at 50°C (Syed et al., 2013). Furthermore, similar experiments were performed using Humicola sp. to synthesize gadolinium oxide nanoparticles at 50°C (Khan et al., 2014). The present study demonstrated that the maximum AgNP synthesis was at 14 h of reaction time (Figure 2E). The yield of biosynthesized AgNPs was the maximum under light conditions (Figure 2F). Similar observations were obtained by researchers, and they concluded that the presence of light encouraged Klebsiella pneumoniae to produce more AgNPs (Mokhtari et al., 2009). For the first time, it has been documented that Penicillium polonicum ARA10, an endophytic fungus, produces AgNPs in the presence of light (Neethu et al., 2018). In addition, a similar investigation was performed using laser light during the AgNP synthesis (Zomorodian et al., 2016). In contrast, another study observed the formation of gadolinium oxide nanoparticles in the dark (Khan et al., 2014).
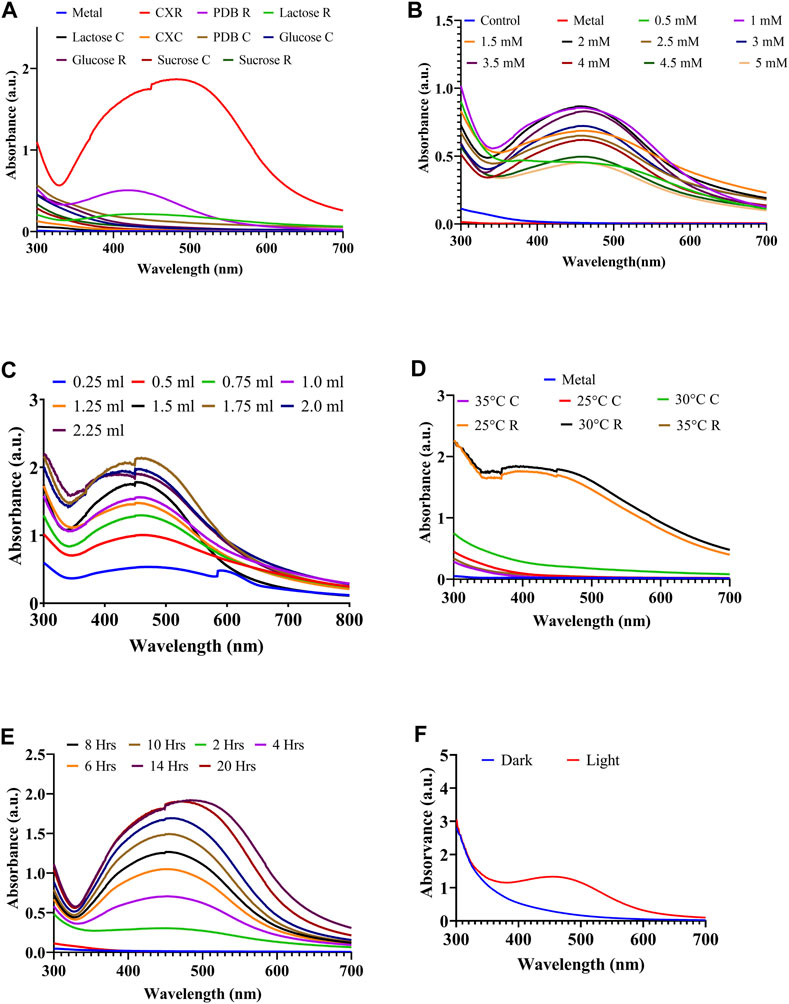
Figure 2. Process Optimization of AgNPs biosynthesis through one factor at a time approach: (A) effect of different media, (B) effect of AgNO3 concentration, (C) effect of different CFS volume, (D) effect of temperature, (E) effect of reaction time, and (F) effect of light condition.
By using the FCCCD to examine the interaction effects of all three parameters (Table 2), the AgNP production medium was further optimized. With 3 independent variables at 5 levels (+, +1, 0, −1, and –), a total of 20 tests were run. The quadratic model was obtained as the best match compared to other models based on the model summary (Supplementary Table S3). Table 1 displays the observed and predicted responses. The range of responses to the experiment was found as 36.68%–98.89% (Table 1). ANOVA was used to analyze the model fitness and suitability, as shown in Table 2. With a calculated “F-” value of 28.73 and a “p-” value of < 0.0001, ANOVA demonstrates the model suitability (Table 2). In the intended space, statistical significance for the interactions between the four variables was established. The maximum AgNP production was indicated by the linearly negative effects of the variables A and D. The accuracy of the model was determined by the coefficient of determination R2, which was 96.28%, and adjusted R2, which was 94.92%, and the predicted R2 indicated a significant correlation between the input variables and response. The lack of fit was deemed unimportant to the validation of the model for the current investigation. Consequently, it is possible to navigate the design space using the projected model. The following quadratic equation represents the combined influence of the model process parameters based on the response:
where A = silver nitrate, B = CFS, and C = reaction time.
The interactive effect of the variables in the form of a 3D surface was plotted for the AgNP yield responses obtained in the CCD, as shown in Figure 3. A high-intensity and narrow SPR peak at 350–420 nm was observed when the color changed from clear transparent to dark brown using UV–visible spectroscopy. The experimental value was obtained quite near the predicted response; hence, this model was validated. Finally, it was concluded that the biosynthesis of AgNPs was significantly enhanced by the statistical tool (RSM), and it can be used in various fields.
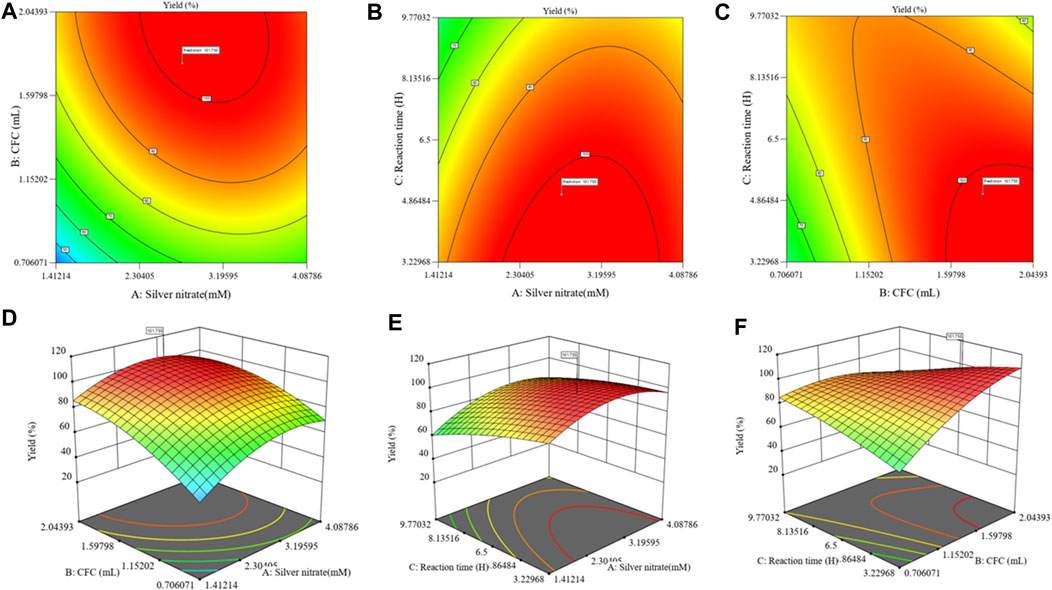
Figure 3. Contour and 3D response surface plots showing the effects of variables and their interaction during the biosynthesis of AgNP activity: (A, D) effect of silver nitrate vs. CFC volume; (B, E) silver nitrate vs. reaction time; (C, F) reaction time vs. CFS volume.
The bioreduction of silver ions in the filtrate is a step in the extracellular manufacture of AgNPs using the fungus H. lixii GGRK4. After 12 h of incubation, the synthesized AgNPs from H. lixii GGRK4 displayed the maximal absorbance peak at 462 nm, suggesting that the bioreduction of AgNO3 occurred with the cell-free filtrate. To determine the stability, periodic samples of the reaction mixture were taken from the reaction mixture at regular intervals. After a complete reduction for 89 h, the AgNPs produced were extremely stable. A spherical morphology of NPs was shown by the TEM analysis, with a diameter of 10–45 nm and an average diameter of 17.34 ± 0.410 nm (Figure 4). The analysis of the TEM histogram was performed using Origin software. The present study was supported by various reports using Aspergillus spp. (Rudrappa et al., 2022; Wang et al., 2021). Prior to diluting the AgNPs, the hydrodynamic size distribution and zeta potential of biosynthesized AgNPs were measured using a zeta analyzer. The zeta potential of AgNP was found to be −14.4 mV, which indicated that the AgNPs were relatively stable (Figure 5A). However, the amino residues were completely protonated under the acidic condition, indicating the mutual electrostatic repulsion, and the narrow size distribution curve of biosynthesized AgNPs at pH 5.02 (average Dh = 116.4 nm) with a polydispersity index (PDI) of 0.249 was observed (Figure 5B). The DLS approach analyzes the varying scattered intensity from the suspended nanoparticles in the solution to determine the diffusion coefficient of the nanoparticles undergoing Brownian motion. The particle diffusion coefficient was inversely proportional to the scattered light swings on a particular time scale from the suspension of nanoparticles. Since kinetic and hydrodynamic conditions affect how particles diffuse in a solution, DLS has become a popular tool for sizing particles in diluted solutions. However, the repeatability and uncertainty of DLS measurements are frequently insufficient to determine the true size of nanoparticles (Takahashi et al., 2008). In the case of TEM analysis, a study reported that the difference may occur due to the multiple-domain diffraction caused by crystal twinning (Ma et al., 2012). Based on our findings, we can conclude that the biosynthesized AgNPs have a mixed population consisting of crystal structures and single-crystalline nanoparticles. Wongpreecha et al. (2018) investigated the size of AgNPs using a zetasizer, which showed that the size of their nanoparticles was 74.7 nm, which was greater than the TEM size. Similarly, Golpour et al. reported a DLS particle size of 91.78 nm (Golpour et al., 2024). Various researchers have documented closely similar size ranges of 30–150 nm (Sarkar et al., 2011; Verma and Maheshwari, 2018). When using biofunctionalized nanoparticles in drug delivery systems, colloidal solutions are essential for preserving the monodispersity and stability of the particles. The zeta potential is one such method that verifies the applicability of nanoparticles based on those mentioned criteria. Singh et al. reported that there was a difference in XRD and SEM analysis of AgNP size, which was supported by our study (Singh et al., 2020).
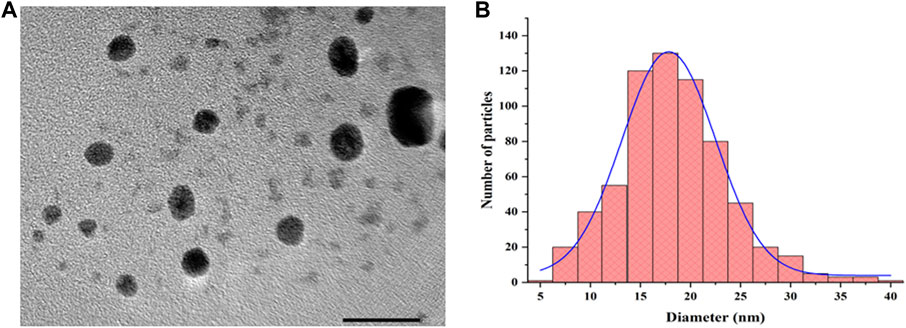
Figure 4. Transmission electron microscopy characterization: (A) image of AgNPs at 200 kV and (B) size distribution histogram.
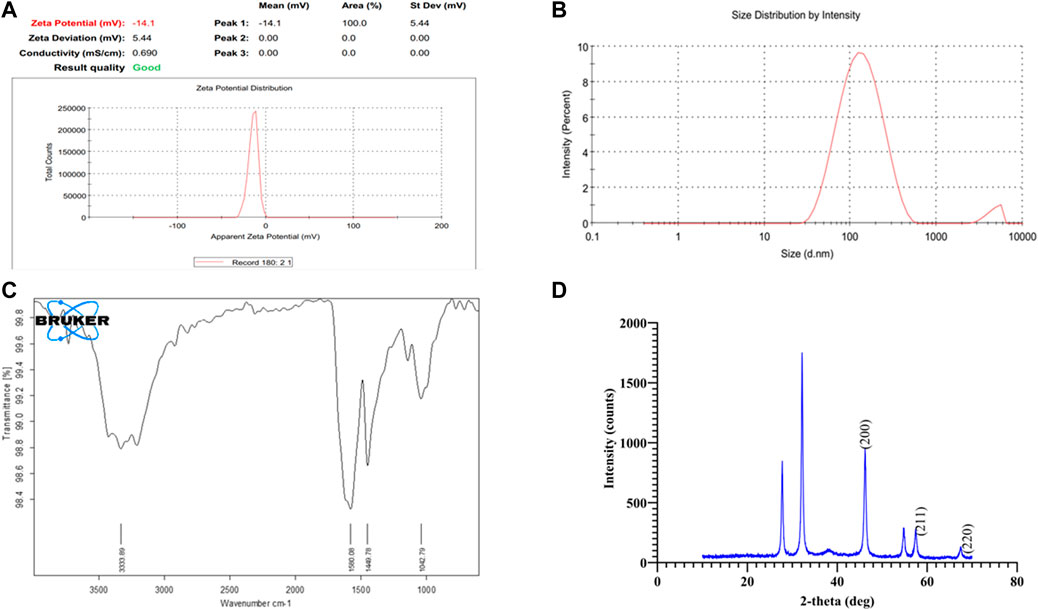
Figure 5. Characterization of biosynthesized AgNPs: (A) zeta potential, (B) zeta size, (C) Fourier-transform infrared spectroscopy analysis, and (D) X-ray diffraction analysis.
The FTIR analysis was performed to evaluate the AgNP-associated functional group. The obtained spectra of FTIR revealed that the bands at 3,333.89, 1,580.08, 1,449.78, and 1,042.79 cm−1 showed the amino acid functional groups (Figure 5C). The presence of primary and secondary amines with amides was suggested by N–H stretching in the IR spectrum between 3,250 and 3,400 cm−1. Additionally, the N–H stretching between 1,580 and 1,650 cm−1 suggests the presence of primary amines. The intensity of 1,500–1,400-cm−1 bands revealed the C–C stretching of an aromatic ring. Similarly, the C–N stretching at bands between 1,250 and 1,020 cm−1 represents the aliphatic amines. These findings supported the presence of bioactive molecules, which help in the biosynthesis of AgNPs (Shankar et al., 2020). The protein is present around AgNO3, which is responsible for the stabilizing, capping, and reducing agents (Shah et al., 2015; Ibrahim et al., 2021). The present findings were supported by several reports (Schröfel et al., 2014; Abdel-Hafez et al., 2016; Prabakaran et al., 2016; Omran et al., 2018; Abdelmoneim et al., 2022).
The XRD data analysis revealed a total of six intense bands (Figure 5D). The AgNPs had an average size of 20.37 nm. The crystalline nature of silver nanoparticles was visible in the XRD pattern, as recorded by Venkatesan et al. (2014). The three different intensities (46.20, 57.45, and 67.45°) at 2θ in the diffractogram have been marked and correspond to (hkl) values such as (200), (211), and (220) planes of silver and are labeled in Figure 5D. The extensive study regarding these three peaks is given in Table 3 and confirms the face-centered cubic crystal structure of the AgNPs. This study is supported by the study conducted by Mehta et al. (2017). The crystalline nature of silver nanoparticles was visible in the XRD pattern, as recorded by Venkatesan et al. (2014). The value of the inter-planar spacing obtained is nearly the same. Furthermore, a lattice constant of 3.93 Å was found in these bands. These findings are well supported and hold good agreement with the standard value of silver. Furthermore, the findings were consistent with those of previous investigations that reported similar diffraction peaks for silver nanoparticles (Trivedi, 2014; Venkatesan et al., 2014).
Fluorescence emission spectra of freshly prepared AgNPs were checked by changing the emission wavelength from 450 nm to 490 nm, from which the maximum emission was observed at 450 nm (Figure 6A). This might be due to an increase in the excitation wavelength, which results in the relaxation of electrons to a lower energy level, causing emissions at a longer wavelength. The excitation spectra of biosynthesized AgNPs were obtained at 390 nm (Figure 6B).
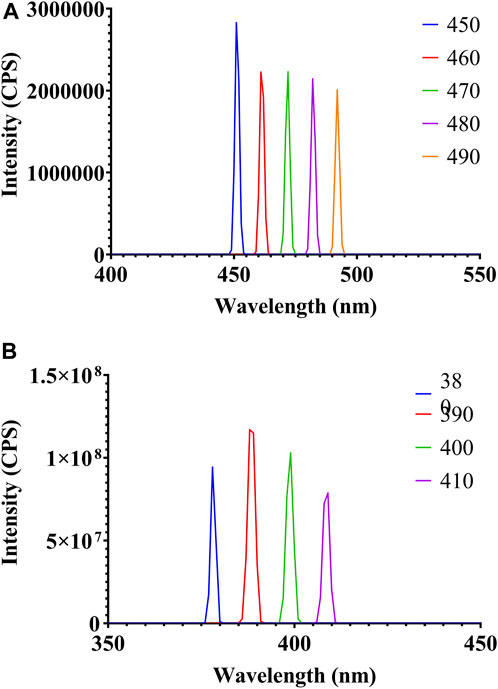
Figure 6. Fluorescence spectroscopy analysis of biosynthesized AgNPs: (A) emission spectra and (B) excitation spectra.
The biosynthesized AgNPs showed significant antibacterial activity against all tested organisms, i.e., Gram-positive (S. aureus MTCC 96 and B. pumilus MTCC 121) and Gram-negative (E. coli MTCC 43 and P. aeruginosa MTCC 424) bacteria. Table 4 shows that the highest antibacterial activity was obtained against S. aureus MTCC 96, followed by E. coli MTCC 43. Among the different AgNP concentrations, 300 μL/mL showed the maximum zone of inhibition (17 mm) against S. aureus MTCC 96 (Figure 7). On the other hand, the highest zone of inhibition with 11.1 mm and 9 mm was observed against P. aeruginosa MTCC 424 and B. pumilus MTCC 121, respectively (Table 4). The possible reason may be that Ag+ ions attract and associate together with the bacterial cell-membrane, resulting in the modifications in the bacterial structure with major cell membrane damage. Moreover, it could be due to the inactivation or loss of bacterial enzymes due to the significant involvement of Ag+ ions with the sulfhydryl group (-SH) of the bacterial enzyme. Hence, a possible mechanism of antibacterial activity is shown in Figure 8. Several recent studies reported and supported the antibacterial activity of synthesized AgNPs (Przemieniecki et al., 2022; Tripathi and Goshisht, 2022; Ivankovic et al., 2023).
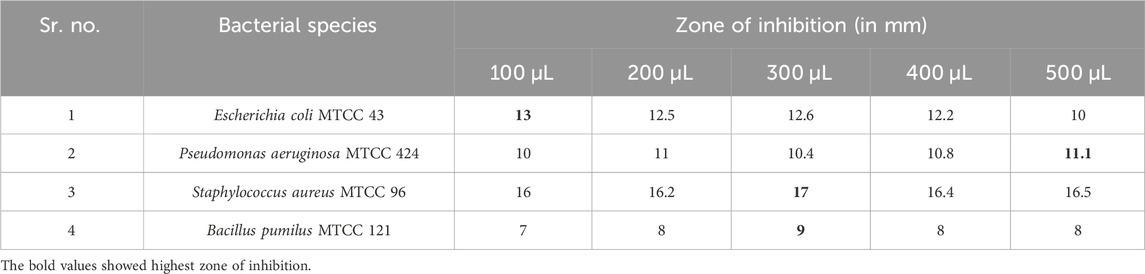
Table 4. Zone of inhibition of mycosynthesized AgNPs against Gram-positive and Gram-negative bacteria.
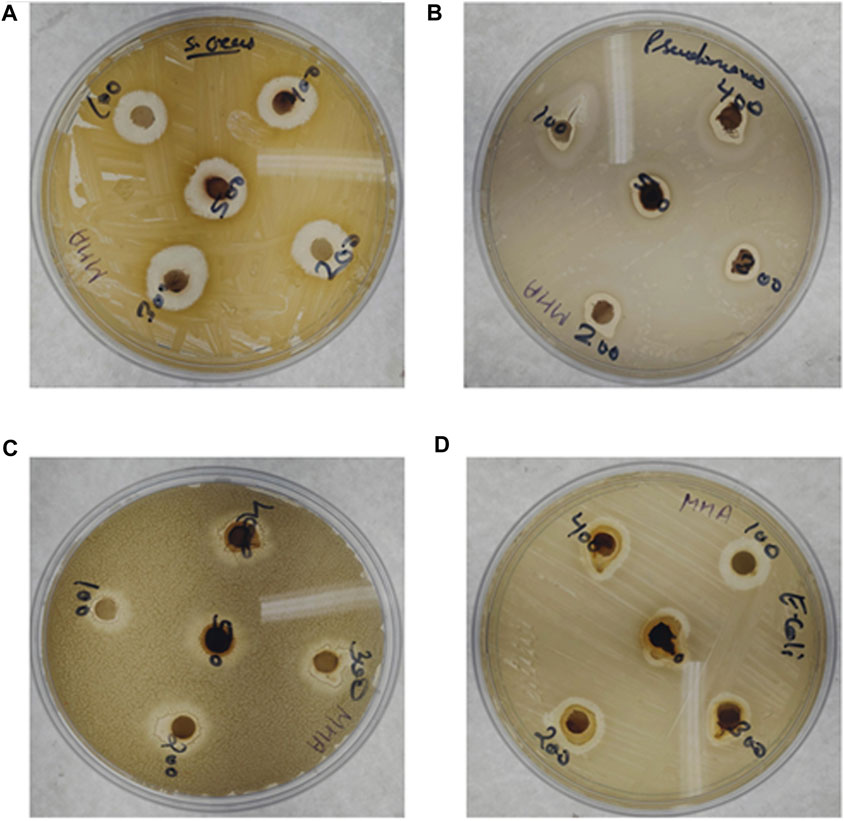
Figure 7. Antibacterial efficacy of biosynthesized AgNPs: (A) Staphylococcus aureus MTCC 96, (B) Pseudomonas aeruginosa MTCC 424, (C) Bacillus pumilus MTCC 121, and (D) Escherichia coli MTCC 43.
The DPPH method was performed for evaluating the free radical scavenging activity of mycosynthesized AgNPs using CFS of H. lixii GGRK4. DPPH is a well-known stable free radical and can receive hydrogen and electrons from the donors, followed by change in color from purple to yellow (Rajkumar and Sundar, 2022). These investigations were performed in triplicate, and the results are reported as the mean value. In the present study, 50–500 μg/mL of AgNPs revealed a scavenging percentage of 32.34–78.30% in response to DPPH. As shown in Figure 9A, the AgNPs showed the highest inhibition efficiency of 78.30%, and ascorbic acid exhibited 69.74% of free radical scavenging activity at 500 μg/mL. The outputs make it evident that the DPPH radical scavenging capability of mycosynthesized AgNPs was concentration-dependent and improved with an increase in AgNP concentration from 50 to 500 μg/mL (Figure 9A). This antioxidant action might be attributed to capping components found in the culture filtrate and on metal surfaces (Ahmed et al., 2023).
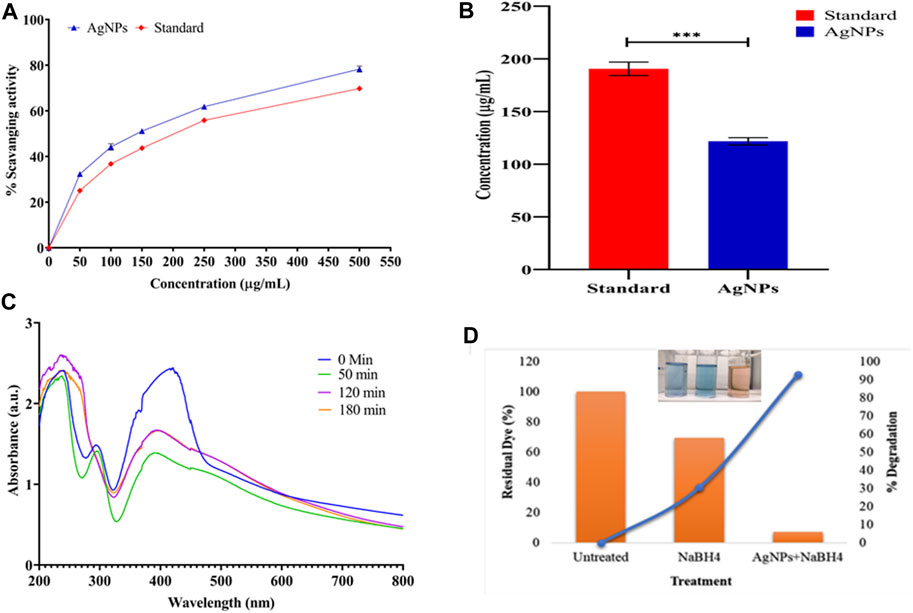
Figure 9. Ecofriendly application of biosynthesized AgNPs: (A) antioxidant property of AgNPs, (B) IC50 of biosynthesized AgNPs, (C) catalytic efficacy on the bioconversion of p-nitro to the p-amino group, and (D) residual dye and % degradation of dye (clear degradation shown in the inset image).
The results of the IC50 value of AgNPs and ascorbic acid against DPPH free radicals are shown in Figure 9B. Compared to the standard, AgNPs had the lowest IC50 value (121.74 ± 3.44 μg/mL) with the highest % scavenging activity. The standard has the lowest % scavenging activity with the highest IC50 value of 190.55 ± 6.39 μg/mL. The ANOVA study revealed significant p-values with 0.0001. The lower IC50 value indicates the greater potency for the antioxidant activity of the biosynthesized AgNPs.
The biotransformation activity of AgNPs was significantly observed. The decrease in the intensity at 400 nm and increase in the intensity at 300 nm indicate the bioconversion of p-nitro to p-amino (Figure 9C). Similar findings have been reported while using gold and platinum nanoparticles in combination with NaBH4 (Wunder et al., 2010; Das et al., 2012). Halligudra et al. (2022) reported that Fe3O4–MoS2 nanoflowers cause a significant reduction in the p-nitrophenol compound (Halligudra et al., 2022). Similarly, Zhang et al. experimented with the cellulose/silver-based composite material, demonstrating efficient degradability potential against p-nitrophenol (Zhang et al., 2023).
Methylene blue was significantly degraded in the presence of AgNPs (serving as a catalyst). First, the absorption spectrum of MB dye was documented at 662 nm. After 20 min of treatment, the absorption almost completely disappeared, signifying the complete degradation of the MB chromophore due to the dissolution of its aromatic ring. In a reaction mixture, 20 mL MB dye (5 mg/L) reacted with 200 µL NaBH4 (reducing agent), along with 30 µg AgNPs, resulting in 92.75% (residue 7.25%) degradation of the dye, which was extremely high compared to the reaction without the AgNP catalyst, which was 30.6% (residue 69.4%) in 20 min (Figure 9D). The biosynthesized AgNPs exhibited superior catalytic activity to others, showing efficient activity in converting methylene blue at 0.131 min−1 at a dose of 30 μg/mL. Similar results were observed by several researchers (Kumar and Bharadvaja, 2022; Mechouche et al., 2022; Nguyen et al., 2022; Gokul Eswaran et al., 2023; Shaban et al., 2024). The following steps describe the mechanism of the reduction process: (i) adsorption of the dye–
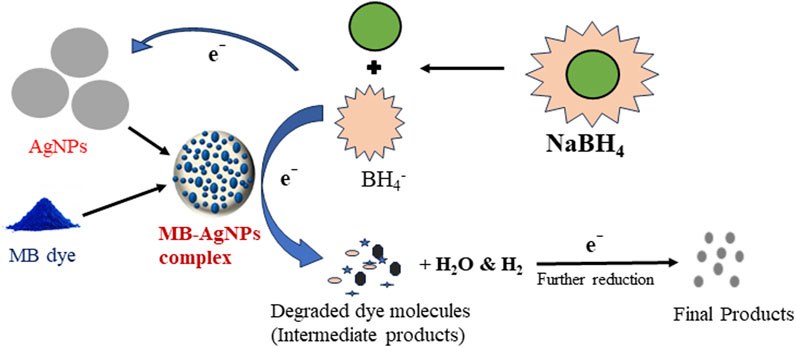
Figure 10. Possible mechanism of the methylene blue (MB) dye degradation process using AgNPs by the NaBH4 nucleophile approach.
In the OFAT approach, various physio–chemical parameters were optimized. These parameters were then further optimized for the maximum biosynthesis of AgNPs using the statistical tool (RSM) with a CCD approach. ANOVA analysis confirmed the significant fitness and adequacy of the model. All variables were found to fit well within the design approach. The coefficient of determination (R2) with a value of 0.9628 indicated a strong correlation between the actual and predicted responses with the polynomial quadratic model obtained. The validation of the responses authenticated the predicted outcomes from the RSM model. Ultimately, it was concluded that the statistical tool (RSM) greatly enhanced the biosynthesis of AgNPs and shows potential for various applications across different fields. In addition, the biosynthesized AgNPs showed a crystalline nature and inter-planar spacing, along with significant agreement compared to the standard data. Furthermore, it has efficient antibacterial activity against pathogenic bacteria. Similarly, the biosynthesized AgNPs have a strong and significant biotransformation and hazardous dye degradation ability. The AgNPs showed excellent free radical scavenging properties. Therefore, based on the above applicability, the mycosynthesized AgNPs have multi-efficient functionality and will be used in upcoming biotechnological avenues.
The raw data supporting the conclusion of this article will be made available by the authors, without undue reservation.
GG: conceptualization, formal analysis, writing–original draft, and writing–review and editing. DK: methodology and writing–original draft. RK: funding acquisition, project administration, resources, supervision, validation, visualization, and writing–review and editing.
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
The authors acknowledge the Department of Science and Technology, New Delhi, Government of India, for providing infrastructure assistance under the FIST grant (Grant No. 1196 SR/FST/LS-I/ 2017/4). GKG acknowledges the University research scholarship. The authors acknowledge M.D. University, Rohtak, for providing research facilities that allowed them to complete this study.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnano.2024.1384465/full#supplementary-material
Abdel-Hafez, S. I. I., Nafady, N. A., Abdel-Rahim, I. R., Shaltout, A. M., Daròs, J. A., and Mohamed, M. A. (2016). Assessment of protein silver nanoparticles toxicity against pathogenic Alternaria solani. 3 Biotech. 6 (2), 199. doi:10.1007/s13205-016-0515-6
Abdelmoneim, H. M., Taha, T. H., Elnouby, M. S., and AbuShady, H. M. (2022). Extracellular biosynthesis, OVAT/statistical optimization, and characterization of silver nanoparticles (AgNPs) using Leclercia adecarboxylata THHM and its antimicrobial activity. Microb. Cell Factories 21 (1), 277. doi:10.1186/s12934-022-01998-9
Ahmad, S., Munir, S., Zeb, N., Ullah, A., Khan, B., Ali, J., et al. (2019). Green nanotechnology: a review on green synthesis of silver nanoparticles — an ecofriendly approach. Int. J. Nanomedicine 14, 5087–5107. doi:10.2147/IJN.S200254
Ahmed, F., Kalia, A., and Ahmad, A. (2023). Marine fungi and yeast: a green approach for production of bionanoparticles. Elsevier.
Ahmed, R. H., and Mustafa, D. E. (2020). Green synthesis of silver nanoparticles mediated by traditionally used medicinal plants in Sudan. Int. Nano Lett. 10 (1), 1–14. doi:10.1007/s40089-019-00291-9
Ahmed, S. F., Mofijur, M., Rafa, N., Chowdhury, A. T., Chowdhury, S., Nahrin, M., et al. (2022). Green approaches in synthesising nanomaterials for environmental nanobioremediation: technological advancements, applications, benefits and challenges. Environ. Res. 204, 111967. doi:10.1016/j.envres.2021.111967
Ajitha, B., Ashok Kumar Reddy, Y., and Sreedhara Reddy, P. (2015). Enhanced antimicrobial activity of silver nanoparticles with controlled particle size by pH variation. Powder Technol. 269, 110–117. doi:10.1016/j.powtec.2014.08.049
Albeladi, S. S. R., Malik, M. A., and Al-Thabaiti, S. A. (2020). Facile biofabrication of silver nanoparticles using Salvia officinalis leaf extract and its catalytic activity towards Congo red dye degradation. J. Mater. Res. Technol. 9 (5), 10031–10044. doi:10.1016/j.jmrt.2020.06.074
Ankush, K., Vishakha, V., Anjana, D., Kamal, J., Sanjay, K., and Rohit, B. (2022). A review on silver nanoparticles focusing on applications in biomedical sector. Int. J. Pharm. Sci. Dev. Res. 8 (1), 057–063. doi:10.17352/ijpsdr.000043
Aykul, S., and Martinez-Hackert, E. (2016). Determination of half-maximal inhibitory concentration using biosensor-based protein interaction analysis. Anal. Biochem. 508, 97–103. doi:10.1016/j.ab.2016.06.025
Bhat, M. A., Nayak, B. K., and Nanda, A. (2015). Evaluation of bactericidal activity of biologically synthesised silver nanoparticles from Candida albicans in combination with ciprofloxacin. Mater. Today Proc. 2 (9), 4395–4401. doi:10.1016/j.matpr.2015.10.036
Bhattacharjee, N., Som, I., Saha, R., and Mondal, S. (2024). “A critical review on novel eco-friendly green approach to synthesize zinc oxide nanoparticles for photocatalytic degradation of water pollutants. Int. J. Environ. Anal. Chem. 104 (3), 489–516. doi:10.1080/03067319.2021.2022130
Birla, S. S., Gaikwad, S. C., Gade, A. K., and Rai, M. K. (2013). Rapid synthesis of silver nanoparticles from Fusarium oxysporum by optimizing physicocultural conditions. Sci. World J. 2013, 1–12. doi:10.1155/2013/796018
Burdusel, A. C., Gherasim, O., Grumezescu, A. M., Mogoantă, L., Ficai, A., and Andronescu, E. (2018). Biomedical applications of silver nanoparticles: an up-to-date overview. Nanomaterials 8 (9), 681. doi:10.3390/nano8090681
Chand, K., Cao, D., Eldin Fouad, D., Hussain Shah, A., Qadeer Dayo, A., Zhu, K., et al. (2020). Green synthesis, characterization and photocatalytic application of silver nanoparticles synthesized by various plant extracts. Arabian J. Chem. 13 (11), 8248–8261. doi:10.1016/j.arabjc.2020.01.009
Chandhru, M., Rani, S. K., and Vasimalai, N. (2020). Reductive degradation of toxic six dyes in industrial wastewater using diaminobenzoic acid capped silver nanoparticles. J. Environ. Chem. Eng. 8 (5), 104225. doi:10.1016/j.jece.2020.104225
Cherian, T., Ragavendran, C., Remesh, R. K. V., Jacob, J., Jamal, W., Kamaraj, C., et al. (2023). Bio-functionalization, stabilization and potential functionalities of hyaluronate macromolecules capped copper oxide nanoparticles. J. Environ. Chem. Eng. 11 (6), 111051. doi:10.1016/j.jece.2023.111051
Das, S. K., Dickinson, C., Lafir, F., Brougham, D. F., and Marsili, E. (2012). Synthesis, characterization and catalytic activity of gold nanoparticles biosynthesized with Rhizopus oryzae protein extract. Green Chem. 14 (5), 1322–1334. doi:10.1039/c2gc16676c
Dejen, K. D., Sabir, F. K., Ananda Murthy, H. C., Ayanie, G. T., Shume, M. S., and Bekele, E. T. (2023). “Green synthesis of nanomaterials for environmental remediation,” in Green nanoremediation (Springer International Publishing), 27–65. doi:10.1007/978-3-031-30558-0_2
Elgorban, A. M., Al-Rahmah, A. N., Sayed, S. R., Hirad, A., Mostafa, A. A. F., and Bahkali, A. H. (2016). Antimicrobial activity and green synthesis of silver nanoparticles using Trichoderma viride. Biotechnol. Biotechnol. Equip. 30 (2), 299–304. doi:10.1080/13102818.2015.1133255
Fatima, F., Siddiqui, S., and Khan, W. A. (2021). Nanoparticles as novel emerging therapeutic antibacterial agents in the antibiotics resistant era. Biol. Trace Elem. Res. 199 (7), 2552–2564. doi:10.1007/s12011-020-02394-3
Gokul Eswaran, S., Shahid Afridi, P., and Vasimalai, N. (2023). Effective multi toxic dyes degradation using bio-fabricated silver nanoparticles as a green catalyst. Appl. Biochem. Biotechnol. 195 (6), 3872–3887. doi:10.1007/s12010-022-03902-y
Golpour, M., Ebrahimnejad, P., Gatabi, Z. R., Najafi, A., Davoodi, A., Khajavi, R., et al. (2024). Green tea-mediated synthesis of silver nanoparticles: enhanced anti-cancer activity and reduced cytotoxicity melanoma and normal murine cell lines. Inorg. Chem. Commun. 161, 111989. doi:10.1016/j.inoche.2023.111989
Govindappa, M., Hemashekhar, B., Arthikala, M. K., Ravishankar Rai, V., and Ramachandra, Y. L. (2018). Characterization, antibacterial, antioxidant, antidiabetic, anti-inflammatory and antityrosinase activity of green synthesized silver nanoparticles using Calophyllum tomentosum leaves extract. Results Phys. 9, 400–408. doi:10.1016/j.rinp.2018.02.049
Halligudra, G., Paramesh, C. C., Gururaj, R., Giridasappa, A., Sabbanahalli, C., Channapillekoppalu Siddegowda, A. K., et al. (2022). Magnetic Fe3O4 supported MoS2 nanoflowers as catalyst for the reduction of p-nitrophenol and organic dyes and as an electrochemical sensor for the detection of pharmaceutical samples. Ceram. Int. 48 (23), 35698–35707. doi:10.1016/j.ceramint.2022.06.188
Hamedi, S., Shojaosadati, S. A., Shokrollahzadeh, S., and Hashemi-Najafabadi, S. (2014). Extracellular biosynthesis of silver nanoparticles using a novel and non-pathogenic fungus, Neurospora intermedia: controlled synthesis and antibacterial activity. World J. Microbiol. Biotechnol. 30 (2), 693–704. doi:10.1007/s11274-013-1417-y
Huq, M. A. (2020). Green synthesis of silver nanoparticles using pseudoduganella eburnea MAHUQ-39 and their antimicrobial mechanisms investigation against drug resistant human pathogens. Int. J. Mol. Sci. 21 (4), 1510. doi:10.3390/ijms21041510
Ibrahim, S., Ahmad, Z., Manzoor, M. Z., Mujahid, M., Faheem, Z., and Adnan, A. (2021). Optimization for biogenic microbial synthesis of silver nanoparticles through response surface methodology, characterization, their antimicrobial, antioxidant, and catalytic potential. Sci. Rep. 11 (1), 770. doi:10.1038/s41598-020-80805-0
Ivankovic, T., Turk, H., Hrenovic, J., Schauperl, Z., Ivankovic, M., and Ressler, A. (2023). Antibacterial activity of silver doped hydroxyapatite toward multidrug-resistant clinical isolates of Acinetobacter baumannii. J. Hazard. Mater. 458, 131867. doi:10.1016/j.jhazmat.2023.131867
Javier, K. R. A., and Camacho, D. H. (2022). Dataset on the optimization by response surface methodology for the synthesis of silver nanoparticles using Laxitextum bicolor mushroom. Data Brief 45, 108631. doi:10.1016/j.dib.2022.108631
Jorge de Souza, T. A., Rosa Souza, L. R., and Franchi, L. P. (2019). Silver nanoparticles: an integrated view of green synthesis methods, transformation in the environment, and toxicity. Ecotoxicol. Environ. Saf. 171, 691–700. doi:10.1016/j.ecoenv.2018.12.095
Jung, J. P., Hu, D., Domian, I. J., and Ogle, B. M. (2015). An integrated statistical model for enhanced murine cardiomyocyte differentiation via optimized engagement of 3D extracellular matrices. Sci. Rep. 5, 18705. doi:10.1038/srep18705
Khan, S., and Hossain, M. K. (2022). Classification and properties of nanoparticles. Nanoparticle-Based Polym. Compos., 15–54. doi:10.1016/B978-0-12-824272-8.00009-9
Khan, S. A., Gambhir, S., and Ahmad, A. (2014). Extracellular biosynthesis of gadolinium oxide (Gd2O3) nanoparticles, their biodistribution and bioconjugation with the chemically modified anticancer drug taxol. Beilstein J. Nanotechnol. 5 (1), 249–257. doi:10.3762/bjnano.5.27
Kong, Y., Zou, P., Miao, L., Qi, J., Song, L., Zhu, L., et al. (2014). Medium optimization for the production of anti-cyanobacterial substances by Streptomyces sp. HJC-D1 using response surface methodology. Environ. Sci. Pollut. Res. 21 (9), 5983–5990. doi:10.1007/s11356-014-2532-5
Kumar, L., and Bharadvaja, N. (2022). Biosynthesis, characterization, and evaluation of antibacterial and photocatalytic dye degradation activities of silver nanoparticles biosynthesized by Chlorella sorokiniana. Biomass Convers. Biorefinery. doi:10.1007/s13399-022-03433-w
Lateef, A., Akande, M. A., Ojo, S. A., Folarin, B. I., Gueguim-Kana, E. B., and Beukes, L. S. (2016). Paper wasp nest-mediated biosynthesis of silver nanoparticles for antimicrobial, catalytic, anticoagulant, and thrombolytic applications. 3 Biotech. 6 (2), 140. doi:10.1007/S13205-016-0459-X
Ma, R., Levard, C., Marinakos, S. M., Cheng, Y., Liu, J., Michel, F. M., et al. (2012). Size-controlled dissolution of organic-coated silver nanoparticles. Environ. Sci. Technol. 46 (2), 752–759. doi:10.1021/es201686j
Mechouche, M. S., Merouane, F., Messaad, C. E. H., Golzadeh, N., Vasseghian, Y., and Berkani, M. (2022). Biosynthesis, characterization, and evaluation of antibacterial and photocatalytic methylene blue dye degradation activities of silver nanoparticles from Streptomyces tuirus strain. Environ. Res. 204, 112360. doi:10.1016/j.envres.2021.112360
Mehta, B. K., Chhajlani, M., and Shrivastava, B. D. (2017). Green synthesis of silver nanoparticles and their characterization by XRD. J. Phys. Conf. Ser. 836 (1), 012050. doi:10.1088/1742-6596/836/1/012050
Mokhtari, N., Daneshpajouh, S., Seyedbagheri, S., Atashdehghan, R., Abdi, K., Sarkar, S., et al. (2009). Biological synthesis of very small silver nanoparticles by culture supernatant of Klebsiella pneumonia: the effects of visible-light irradiation and the liquid mixing process. Mater. Res. Bull. 44 (6), 1415–1421. doi:10.1016/j.materresbull.2008.11.021
More, P. R., Pandit, S., Filippis, A. D., Franci, G., Mijakovic, I., and Galdiero, M. (2023). Silver nanoparticles: bactericidal and mechanistic approach against drug resistant pathogens. Microorganisms 11 (2), 369. doi:10.3390/microorganisms11020369
Muangman, P., Chuntrasakul, C., Silthram, S., Suvanchote Rn, S., Benjathanung Hn, R., Kittidacha Rn, S., et al. (2006). Comparison of efficacy of 1% silver sulfadiazine and ActicoatTM for treatment of partial-thickness burn wounds. J. Med. Assoc. Thai 89 (7), 953–961.
Mukherjee, P., Ahmad, A., Mandal, D., Senapati, S., Sainkar, S. R., Khan, M. I., et al. (2001). Fungus-Mediated synthesis of silver nanoparticles and their immobilization in the mycelial matrix: a novel biological approach to nanoparticle synthesis. Nano Lett. 1 (10), 515–519. doi:10.1021/nl0155274
Nazir, A., Farooq, S., Abbas, M., Alabbad, E. A., Albalawi, H., Alwadai, N., et al. (2021). Synthesis, characterization and photocatalytic application of Sophora mollis leaf extract mediated silver nanoparticles. Z. Fur Phys. Chem. 235 (12), 1589–1607. doi:10.1515/zpch-2020-1803
Neethu, S., Midhun, S. J., Sunil, M. A., Soumya, S., Radhakrishnan, E. K., and Jyothis, M. (2018). Efficient visible light induced synthesis of silver nanoparticles by Penicillium polonicum ARA 10 isolated from Chetomorpha antennina and its antibacterial efficacy against Salmonella enterica serovar Typhimurium. J. Photochem. Photobiol. B Biol. 180, 175–185. doi:10.1016/j.jphotobiol.2018.02.005
Nguyen, T. A. N., Raghavendra, V. B., Sindhu, R., Alshiekheid, M., Sabour, A., Krishnan, R., et al. (2022). Green fabrication of silver nanoparticles using Chloroxylon swietenia leaves and their application towards dye degradation and food borne pathogens. Food Chem. Toxicol. 165, 113192. doi:10.1016/j.fct.2022.113192
Núñez, R. N., Veglia, A. V., and Pacioni, N. L. (2018). Improving reproducibility between batches of silver nanoparticles using an experimental design approach. Microchem. J. 141, 110–117. doi:10.1016/j.microc.2018.05.017
Omran, B. A., Nassar, H. N., Fatthallah, N. A., Hamdy, A., El-Shatoury, E. H., and El-Gendy, N. S. (2018). Characterization and antimicrobial activity of silver nanoparticles mycosynthesized by Aspergillus brasiliensis. J. Appl. Microbiol. 125 (2), 370–382. doi:10.1111/jam.13776
Ottoni, C. A., Simões, M. F., Fernandes, S., dos Santos, J. G., da Silva, E. S., de Souza, R. F. B., et al. (2017). Screening of filamentous fungi for antimicrobial silver nanoparticles synthesis. Amb. Express 7 (1), 31. doi:10.1186/S13568-017-0332-2
Pandey, S., Do, J. Y., Kim, J., and Kang, M. (2020). Fast and highly efficient catalytic degradation of dyes using κ-carrageenan stabilized silver nanoparticles nanocatalyst. Carbohydr. Polym. 230, 115597. doi:10.1016/j.carbpol.2019.115597
Phanjom, P., and Ahmed, G. (2017). Effect of different physicochemical conditions on the synthesis of silver nanoparticles using fungal cell filtrate of Aspergillus oryzae (MTCC No. 1846) and their antibacterial effect. Adv. Nat. Sci. Nanosci. Nanotechnol. 8 (4), 045016. doi:10.1088/2043-6254/aa92bc
Prabakaran, K., Ragavendran, C., and Natarajan, D. (2016). Mycosynthesis of silver nanoparticles from: beauveria bassiana and its larvicidal, antibacterial, and cytotoxic effect on human cervical cancer (HeLa) cells. RSC Adv. 6 (51), 44972–44986. doi:10.1039/c6ra08593h
Przemieniecki, S. W., Oćwieja, M., Ciesielski, S., Halecki, W., Matras, E., and Gorczyca, A. (2022). Chemical structure of stabilizing layers of negatively charged silver nanoparticles as an effector of shifts in soil bacterial microbiome under short-term exposure. Int. J. Environ. Res. Public Health 19 (21), 14438. doi:10.3390/ijerph192114438
Qamar, O. A., Jamil, F., Hussain, M., Bae, S., Inayat, A., Shah, N. S., et al. (2023). Advances in synthesis of TiO2 nanoparticles and their application to biodiesel production: a review. Chem. Eng. J. 460, 141734. doi:10.1016/j.cej.2023.141734
Qiu, X., Gu, J., Yang, T., Ma, C., Li, L., Wu, Y., et al. (2022). Sensitive determination of Norfloxacin in milk based on β-cyclodextrin functionalized silver nanoparticles SERS substrate. Spectrochimica Acta - Part A Mol. Biomol. Spectrosc. 276, 121212. doi:10.1016/j.saa.2022.121212
Rafique, M., Sadaf, I., Tahir, M. B., Rafique, M. S., Nabi, G., Iqbal, T., et al. (2019). Novel and facile synthesis of silver nanoparticles using Albizia procera leaf extract for dye degradation and antibacterial applications. Mater. Sci. Eng. C 99, 1313–1324. doi:10.1016/j.msec.2019.02.059
Rajkumar, G., and Sundar, R. (2022). Biogenic one-step synthesis of silver nanoparticles (AgNPs) using an aqueous extract of Persea americana seed: characterization, phytochemical screening, antibacterial, antifungal and antioxidant activities. Inorg. Chem. Commun. 143, 109817. doi:10.1016/j.inoche.2022.109817
Rajput, S., Werezuk, R., Lange, R. M., and Mcdermott, M. T. (2016). Fungal isolate optimized for biogenesis of silver nanoparticles with enhanced colloidal stability. Langmuir 32 (34), 8688–8697. doi:10.1021/acs.langmuir.6b01813
Ramya, E. (2022). “Green synthesis of metal nanostructures and its nonlinear optical properties,” in Nonlinear optics - nonlinear nanophotonics and novel materials for nonlinear optics (Intechopen). doi:10.5772/intechopen.99449
Rudrappa, M., Rudayni, H. A., Assiri, R. A., Bepari, A., Basavarajappa, D. S., Nagaraja, S. K., et al. (2022). Plumeria alba-mediated green synthesis of silver nanoparticles exhibits antimicrobial effect and anti-oncogenic activity against glioblastoma U118 MG cancer cell line. Nanomaterials 12 (3), 493. doi:10.3390/nano12030493
Saha, J., Begum, A., Mukherjee, A., and Kumar, S. (2017). A novel green synthesis of silver nanoparticles and their catalytic action in reduction of Methylene Blue dye. Sustain. Environ. Res. 27 (5), 245–250. doi:10.1016/j.serj.2017.04.003
Saifuddin, N., Wong, C. W., and Yasumira, A. A. N. (2009). Rapid biosynthesis of silver nanoparticles using culture supernatant of bacteria with microwave irradiation. E-Journal Chem. 6 (1), 61–70. doi:10.1155/2009/734264
Sameena, V. P., and Thoppil, J. E. (2022). Green synthesis of silver nanoparticles from Euphorbia and its biological activities. Nanotechnol. Environ. Eng. 7 (2), 377–392. doi:10.1007/s41204-022-00232-6
Sarkar, J., Dey, P., Saha, S., and Acharya, K. (2011). Mycosynthesis of selenium nanoparticles. Micro Nano Lett. 6 (8), 599–602. doi:10.1049/mnl.2011.0227
Saxena, J., Sharma, P. K., Sharma, M. M., and Singh, A. (2016). Process optimization for green synthesis of silver nanoparticles by Sclerotinia sclerotiorum MTCC 8785 and evaluation of its antibacterial properties. SpringerPlus 5 (1), 861. doi:10.1186/S40064-016-2558-X
Schröfel, A., Kratošová, G., Šafařík, I., Šafaříková, M., Raška, I., and Shor, L. M. (2014). Applications of biosynthesized metallic nanoparticles - a review. Acta Biomater. 10 (10), 4023–4042. doi:10.1016/j.actbio.2014.05.022
Shaban, S. M., Taha, A. A., Elged, A. H., Taha, S. T., Sabet, V. M., Kim, D. H., et al. (2024). Insights on Gemini cationic surfactants influence AgNPs synthesis: controlling catalytic and antimicrobial activity. J. Mol. Liq. 397, 124071. doi:10.1016/j.molliq.2024.124071
Shah, M., Fawcett, D., Sharma, S., Tripathy, S. K., and Poinern, G. E. J. (2015). Green synthesis of metallic nanoparticles via biological entities. Materials 8 (11), 7278–7308. doi:10.3390/ma8115377
Shankar, A., Kumar, V., Kaushik, N. K., Kumar, A., Malik, V., Singh, D., et al. (2020). Sporotrichum thermophile culture extract-mediated greener synthesis of silver nanoparticles: eco-friendly functional group transformation and anti-bacterial study. Curr. Res. Green Sustain. Chem. 3, 100029. doi:10.1016/j.crgsc.2020.100029
Singh, A., Gaud, B., and Jaybhaye, S. (2020). Optimization of synthesis parameters of silver nanoparticles and its antimicrobial activity. Mater. Sci. Energy Technol. 3, 232–236. doi:10.1016/j.mset.2019.08.004
Singh, J., Mehta, A., Rawat, M., and Basu, S. (2018). Green synthesis of silver nanoparticles using sun dried tulsi leaves and its catalytic application for 4-Nitrophenol reduction. J. Environ. Chem. Eng. 6 (1), 1468–1474. doi:10.1016/j.jece.2018.01.054
Singh, R., Wagh, P., Wadhwani, S., Gaidhani, S., Kumbhar, A., Bellare, J., et al. (2013). Synthesis, optimization, and characterization of silver nanoparticles from Acinetobacter calcoaceticus and their enhanced antibacterial activity when combined with antibiotics. Int. J. Nanomedicine 8, 4277–4290. doi:10.2147/IJN.S48913
Syed, A., Saraswati, S., Kundu, G. C., and Ahmad, A. (2013). Biological synthesis of silver nanoparticles using the fungus Humicola sp. and evaluation of their cytoxicity using normal and cancer cell lines. Spectrochimica Acta - Part A Mol. Biomol. Spectrosc. 114, 144–147. doi:10.1016/j.saa.2013.05.030
Takahashi, K., Kato, H., Saito, T., Matsuyama, S., and Kinugasa, S. (2008). Precise measurement of the size of nanoparticles by dynamic light scattering with uncertainty analysis. Part. Part. Syst. Charact. 25 (1), 31–38. doi:10.1002/ppsc.200700015
Tripathi, N., and Goshisht, M. K. (2022). Recent advances and mechanistic insights into antibacterial activity, antibiofilm activity, and cytotoxicity of silver nanoparticles. ACS Appl. Bio Mater. 5 (4), 1391–1463. doi:10.1021/acsabm.2c00014
Trivedi, P. (2014). Statistically optimized synthesis of silver nanocubes from peel extracts of citrus limetta and potential application in waste water treatment. J. Microb. Biochem. Technol. s1 (01), 4. doi:10.4172/1948-5948.s4-004
Valle-García, J. D., Ali, A., Patra, J. K., Kerry, R. G., Das, G., and Fernández-Luqueño, F. (2023). “Integration of eco-friendly biological and nanotechnological strategies for better agriculture: a sustainable approach. Agric. Environ. Nanotechnol: Novel Technol. and Their Ecol. Imp., 647–674. doi:10.1007/978-981-19-5454-2_24
Veisi, H., Kazemi, S., Mohammadi, P., Safarimehr, P., and Hemmati, S. (2019). Catalytic reduction of 4-nitrophenol over Ag nanoparticles immobilized on Stachys lavandulifolia extract-modified multi walled carbon nanotubes. Polyhedron 157, 232–240. doi:10.1016/j.poly.2018.10.014
Velhal, S. G., Kulkarni, S. D., and Latpate, R. V. (2016). Fungal mediated silver nanoparticle synthesis using robust experimental design and its application in cotton fabric. Int. Nano Lett. 6 (4), 257–264. doi:10.1007/S40089-016-0192-9
Venkatesan, B., Subramanian, V., Tumala, A., and Vellaichamy, E. (2014). Rapid synthesis of biocompatible silver nanoparticles using aqueous extract of Rosa damascena petals and evaluation of their anticancer activity. Asian Pac. J. Trop. Med. 7 (S1), S294–S300. doi:10.1016/S1995-7645(14)60249-2
Verma, P., and Maheshwari, S. K. (2018). Preparation of sliver and selenium nanoparticles and its characterization by dynamic light scattering and scanning electron microscopy. J. Microsc. Ultrastruct. 6 (4), 182–187. doi:10.4103/JMAU.JMAU_3_18
Vidyasagar, N., Patel, R. R., Singh, S. K., and Singh, M. (2023). Green synthesis of silver nanoparticles: methods, biological applications, delivery and toxicity. Mater. Adv. 4 (8), 1831–1849. doi:10.1039/d2ma01105k
Vijayan, S., Divya, K., George, T. K., and Jisha, M. S. (2016). Biogenic synthesis of silver nanoparticles using endophytic fungi fusarium oxysporum isolated from withania somnifera (L.), its antibacterial and cytotoxic activity. J. Bionanoscience 10 (5), 369–376. doi:10.1166/jbns.2016.1390
Wan, H., Li, C., Mahmud, S., and Liu, H. (2021). Kappa carrageenan reduced-stabilized colloidal silver nanoparticles for the degradation of toxic azo compounds. Colloids Surfaces A Physicochem. Eng. Aspects 616, 126325. doi:10.1016/j.colsurfa.2021.126325
Wang, D., Xue, B., Wang, L., Zhang, Y., Liu, L., and Zhou, Y. (2021). Fungus-mediated green synthesis of nano-silver using Aspergillus sydowii and its antifungal/antiproliferative activities. Sci. Rep. 11 (1), 10356. doi:10.1038/s41598-021-89854-5
Wang, H., Zhang, G., Mia, R., Wang, W., Xie, L., Lü, S., et al. (2022). Bioreduction (Ag+ to Ag0) and stabilization of silver nanocatalyst using hyaluronate biopolymer for azo-contaminated wastewater treatment. J. Alloys Compd. 894, 162502. doi:10.1016/j.jallcom.2021.162502
Wongpreecha, J., Polpanich, D., Suteewong, T., Kaewsaneha, C., and Tangboriboonrat, P. (2018). One-pot, large-scale green synthesis of silver nanoparticles-chitosan with enhanced antibacterial activity and low cytotoxicity. Carbohydr. Polym. 199, 641–648. doi:10.1016/j.carbpol.2018.07.039
Wunder, S., Polzer, F., Lu, Y., Mei, Y., and Ballauff, M. (2010). Kinetic analysis of catalytic reduction of 4-nitrophenol by metallic nanoparticles immobilized in spherical polyelectrolyte brushes. J. Phys. Chem. C 114 (19), 8814–8820. doi:10.1021/JP101125J
Zhang, S., Xu, X., Ye, Z., Liu, Y., Wang, Q., Chen, Q., et al. (2023). A large-nanosphere/small-nanosphere (cellulose/silver) antibacterial composite with prominent catalytic properties for the degradation of p-nitrophenol. Appl. Surf. Sci. 608, 155192. doi:10.1016/j.apsusc.2022.155192
Keywords: silver nanoparticles, extracellular biosynthesis, antimicrobial efficacy, antioxidant property, response surface methodology
Citation: Gupta GK, Koli D and Kapoor RK (2024) Statistical optimization for greener synthesis of multi-efficient silver nanoparticles from the Hypocrea lixii GGRK4 culture filtrate and their ecofriendly applications. Front. Nanotechnol. 6:1384465. doi: 10.3389/fnano.2024.1384465
Received: 09 February 2024; Accepted: 27 March 2024;
Published: 16 April 2024.
Edited by:
Francis Boluwaji Elehinafe, Covenant University, NigeriaReviewed by:
Tijo Cherian, Mar Athanasios College for Advanced Studies Tiruvalla (MACFAST), IndiaCopyright © 2024 Gupta, Koli and Kapoor. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guddu Kumar Gupta, Z3VkZHVndXB0YTM2N0BnbWFpbC5jb20=; Rajeev Kumar Kapoor, cGF0ZW50LmFnZW50LmJpb3RlY2hAZ21haWwuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.