- 1Micro-Nano-Bio Integration Center, Optical-Bio Microsystems Laboratory, Concordia University, Montreal, Canada
- 2Department of Sensors and Biomedical Technology, VIT, Vellore, India
In 2006, Whitesides, writing about microfluidics, said that microfluidics is in early adolescence and it is not yet clear how it will develop. Today, almost 20 years later, microfluidics became a fully developed, highly sophisticated, multidisciplinary field that had entirely honoured its early promise. Its strength stems from the knowledge and know-how, coming from multiple disciplines such as physics of fluids, engineering, and microfabrication in the beginning, followed, more recently, by cell biological research, in full bloom nowadays. In microfluidic devices, the environment of cells such as chemical and mechanical gradients can be reproduced, making biological studies even more compelling. The red thread of this review paper follows the new insights and discoveries in both traditional macro- and microfluidic cell culture brought into the cell biology field, especially in the culture of stem cells, filled with promise in the field of regenerative medicine. Microfluidic devices provide an environment that is much closer to that of in vivo cell culture than the conventional culture platforms, where large amounts of cells are cultured and the environment of individual cells cannot be distinguished. The convenience of live cell imaging, portability, and the integration of sensors to precisely, control various parameters, has expanded cell biologists’ arsenal In addition, microfluidic devices, integrated with different functionalities, that is, the automated cell culture systems, will be discussed as well.
1 Introduction
Stem cells are a special category of cells that have potential for tissue engineering and other cell-based therapies, because they can transform themselves into other types of functional cells. Until recently, scientists, mainly, worked with embryonic stem cells which are pluripotent, and non-embryonic, or with mesenchymal stem cells that are multipotent, that is, they only can differentiate into specific stem cells (Zhang and Austin, 2012; McKee and Chaudhry, 2017; Zhang et al., 2017; Aghlmandi et al., 2021).
Embryonic stem cells as well as induced pluripotent stem cells, are, especially, interesting for cell replacement therapy (Konagaya et al., 2015). It has been suggested that the dysregulation of stem cell properties may induce certain types of cancer (Reya et al., 2001; Dalerba and Clarke, 2007; Ramalho-Santos and Willenbring, 2007). The large amounts of potent cells for cell-therapy applications are supplied mostly by traditional large-scale culture and, more recently, by automated culture systems. The choice to focus the discussion of cell culture on stem cells was dictated by the importance of regenerative medicine. Fuchs and Segre in their excellent review paper (Fuchs and Segre, 2000) called stem cells “A New Lease on Life”. The large-scale culture methods were significantly improved recently (Liu et al., 2010; Konagaya et al., 2015). To maintain induced pluripotent stem cells in an undifferentiated state for a long time, automated culture methods were developed (see also (Terstegge et al., 2007; Thomas et al., 2009; Hussain et al., 2013; Soares et al., 2014)). Even more recently, autonomous robotic AI systems were developed that allow the optimization of cell culture protocols (Sebastian, 2022). All these changes have transformed traditional macroscale culture into a most interesting technique, with a wide perspective. In the traditional cell culture of the past, the microenvironment of stem cells that determines their fate, could not be controlled but in the emerging robotic AI systems and in microfluidic cell culture, this becomes possible.
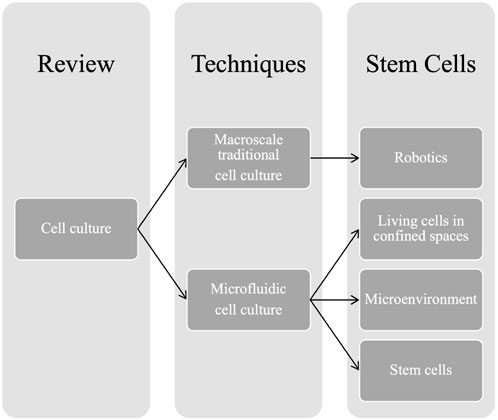
In this review, the recent applications of automated macro- and microfluidic cultures in cell biology are discussed, and their future perspectives are evaluated (Scheme 1). The comparison is not confrontational, and no conclusion on the superiority of the different cell culture methods is imposed onto the readers.
2 Macroscale cell culture traditional culture and robotics
Cell culturing is a basic experimental technique, a versatile tool in cell biology research. It is essential to cell biology, and tissue engineering, as cells can be cultured in large amounts. Mammalian cells are almost all, adherent, needing a surface, appropriate for attachment and proliferation. In vitro experiments with adherent human cells, currently, are performed, by using a two-dimensional (2D) monolayer platform, where the cells are plated onto a polystyrene plate that is surface-treated, to stimulate cell binding. This model is called a two-dimensional (2D) monolayer platform.
Conventional (traditional) cell culture methods rely on Petri dishes, cell-culture shake flasks, microtiter plates, and bioreactors. Reagents, and cell suspensions have to be individually pipetted, a laborious process. The Petri dish was invented over a century ago by Petri, a technician in the laboratory of Koch, the well-known German bacteriologist who discovered the TB bacterium. Since then, the Petri dish became a staple in cell culturing (Zhang, 2004).
Conventional in vitro methods allow the growth of most cells in humidity, temperature, and CO2-controlled incubators, and precise protocols have been established for different cell lines. However, these methods are quite expensive, because of the amounts of cells and reagents and the time they take up.
The macroscale environment is very different from that of the real biological systems, while three-dimensional platforms (3D) represent much better the natural context of cells in tissues. The 3D architecture (Zhang, 2004) represents much better a cell in its in situ environment. In the 3D platform, cells are in contact with each other through a soft extracellular material and not a rigid flat surface as in the case of the 2D culture.
Even now, the most used culture platform, is the 2D macroscopic cell culture, by using Petri dishes or flasks. Although these platforms are less expensive than animal models and result in reproducible quantitative studies, the physiological relevance of the information retrieved from these in vitro studies is often debatable. The structure and physiology of living tissues, as well as the dynamic 3D environments in vivo (Huang et al., 2022) are not reproduced correctly in the 2D platforms but, in spite of all these disadvantages, 2D remains still the method of choice for research laboratories and the pharmaceutical industry while 3D culture, gradually, is gaining popularity.
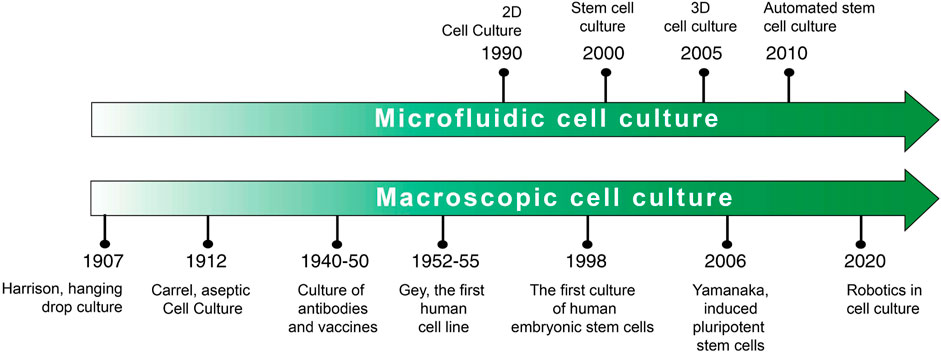
Scheme 2 a timeline of the two most important cell culture techniques is shown below. It has to be emphasized that, presently, the so-called traditional cell culture is quite different from the way it was a century ago. Many recent improvements and refinements along the way have relieved some of the hurdles and opened new ways to culture cells.
Generally, 2D cell culture requires passaging of cells, until a confluence level of 70%–80% is reached. Then, they are split between flasks to provide them with enough space to grow effectively. The use of robots that can automate this process, eliminates, not only subjective judgment, but also automates processes such as harvesting and passaging. Interestingly, studies that compared the automated to the manual culture, have shown no statistical differences in cell count and viability of cells when harvested, demonstrating the significant advantage of automation to save time, without compromising efficacy. Additionally, these systems allow unattended operation over a long time period, as well as allowing for evaluation of variables, including, pH, nutrient or waste concentration, cell concentration, and viability (Chapman, 2003). The use of robotics in automated cell culture can reduce human intervention and error and increase the efficacy (Sebastian, 2022).
Kanda et al. (Kanda et al., 2022), recently, has demonstrated how to optimize the process by using robotics and an artificial intelligence system, called LabDroid Maholo has been developed with an integrated imaging system and computer algorithms (Sasamata et al., 2021; Kanda et al., 2022) that have improved, significantly, the cell culture protocols. For example, the system was programmed to differentiate stem cells into retinal pigment epithelial cells and the level of cell differentiation was analyzed by using computer algorithms, and statistical analyses.
It has been found that combining robotics and artificial intelligence to optimize complex cell culture protocols is a viable way to use macroscale cell culture to differentiate stem cells for therapeutic purposes (Figure 1).
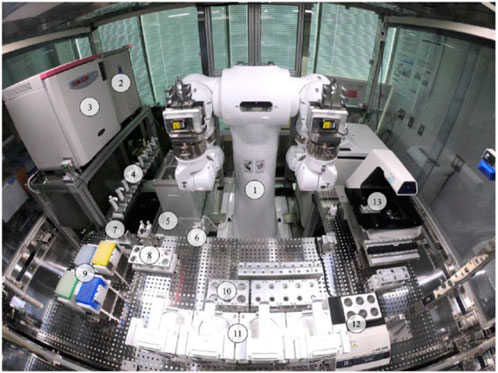
FIGURE 1. LabDroid Maholo including peripheral equipment. A bird's eye view of the LabDroid booth: (1) dual-arm robot, (2) refrigerator, (3) CO2 incubator, (4) micropipettes, (5) dust bin, (6) aspirator, (7) tip sensor, (8) 50 mL tube subrack, (9) pipette tips, (10) 50 mL tube main rack, (11) six-well plate rack, (12) dry bath, and (13) microscope. Reproduced under CC BY-NC-ND 4.0, from Ochiai et al. (2021).
As mentioned previously, stem cells, having the ability to self-renew and differentiate in specialized cell types, are extremely important in regenerative medicine. They may be especially useful to study early human development (Imreh et al., 2006; van Noort et al., 2009). The use of stem cells for clinical applications requires high quality and quantity of cells.
Although robotic platforms such as LabDroid Maholo will need further improvements, it is an important step toward the reproducible manufacturing of cells, especially, for regenerative treatments. Other automated platforms are currently available, and they may incorporate biosafety cabinets and incubators as well (Crombie et al., 2017). Less expensive pipetting robots, for academic settings, have also been developed (Dettinger et al., 2022) and they can be used, without prior knowledge, to automate many experimental procedures in standard cell culture vessels. These studies are extremely important as stem cells hold enormous promise for fundamental biological studies and as well as for cell-based therapies.
These innovative approaches to macroscale cell culture show that this culture became again a viable technique, creating niches for applications in special fields such as, for example, stem cell studies. Most probably, in the future, new developments will emerge, and macroscale cell culture will continue to be used for different purposes, in parallel with new technologies such as microfluidic cell culture, microcarriers, and microencapsulation (McKee and Chaudhry, 2017).
3 Microfluidic cell culture
3.1 Introduction
“Learning to think on an entirely different scale is a new and exciting challenge in microfluidics” (Beebe et al., 2002).
Microfluidics is the science and technology of the controlled manipulation of low volumes of liquids (nanoliters or less), constrained to microchannels with, usually, rectangular cross-sections, and diameters around hundreds of micrometres. In such small channels, the behaviour of a liquid is significantly different from that at the macro scale as the flow gains completely new features at this scale. The effect that become dominant in microfluidics includes laminar flow, and the mixing, mainly occurs by diffusion, at the interface between laminar layers, compared to the macrosystems, where turbulent flow determines fast convective processes. When proper mixing configurations are integrated with microfluidics, due to the small distances in the channels, mixing is completed rapidly.
Microfluidics involves the design of these devices and the study of the behaviour of fluids in the narrow microchannels. In addition to the channels, microfluidic devices contain also miniaturized components for fluid’s manipulation, such as microvalves, micropumps, micromixers, and micro separators. The small length scales, associated with microfluidics, enable faster analysis, requiring lower amounts of, possibly, expensive reagents.
It has to be mentioned that microfluidics is often considered a new field but, actually, it was born in the 1990s when the seminal paper by Andreas Manz (Manz et al., 1990) was published. The paper envisioned, for the first time, an integrated platform for performing a multitude of analysis steps. Microfluidics, developed by the confluence of a number of concepts, techniques, and materials lends from previously existing fields, especially, analytical chemistry (microanalysis) and later on, material science, physics of fluids, and microelectronics.
As stressed by George Whitesides, an eminent personality in this field (Whitesides, 2006), “microfluidics can be considered as having four “parents” that contributed to building it up: molecular biology, molecular analysis, national security, and microelectronics’. Gas-phase chromatography and capillary electrophoresis, allowing the separation of chemical compounds, by flowing very small amounts of a sample in capillaries, was developed already in the 1950s. These methods inspired researchers to improve the separation, by reducing even more the size of capillaries. However, the most important precursor of microfluidics is the microchip (the word “chip” used for “microfluidic chip” is borrowed from microelectronics). Although at the beginning of microfluidics, photolithography was used with mainly standard semiconductor materials, silicon, and glass, soon, due to lower cost and more convenient properties, they were displaced by polymers. The main drawback of Silicon microfluidic chips is their opacity, making it impossible to use optical detection. For this reason, microfluidics split from microelectronics and semiconductors technology and developed techniques that facilitated significantly the further development of the field. Research for military purposes stimulated the effort in the development of microfluidics and molecular biology strongly contributed to microfluidics’ birth and evolution. In the 1980s, the interest of scientists in sequencing nucleic acids as well as developing precision equipment for the PCR (polymerase chain reaction) technique, led to the development of machines capable of working with very small samples.
Today, microfluidic technology represents a distinct, well-consolidated research field, with well-defined boundaries and extensive applications in various domains such as synthesis of nanoparticles, point-of-care devices, biosensors, drug screening and delivery systems, wearable biosensors, devices for cell analysis, and cell sorting. Also, microfluidic systems can be used for point-of-care clinical applications, or even deployed in remote locations. The development of inkjet printheads, and DNA sequencing chips are only some of the multiple applications of microfluidics. In addition, Micro Total Analysis Systems (μTAS) or lab-on-a-chip, may integrate complex analytical processes that commonly solicit a whole laboratory, in a small device, without off-chip sample preparation. They are also able to perform separations and detection with high resolution and sensitivity. The convergence of microfluidics and nanoparticle synthesis allowed the preparation of nanoparticles with a narrow size distribution, important for developing new functional materials (Badilescu and Packirisamy, 2012).
3.2 Culturing cells on the microscale
As Young and Beebe stressed in their excellent tutorial review paper, ‘culturing cells in microscale environments requires knowledge of multiple disciplines, including physics, biochemistry, and engineering’ (Paguirigan and Beebe, 2008; Young EW and Beebe D, 2010). They have shown that at the beginning of microfluidics, research was focused more on understanding the chemical and physical phenomena at the microscale, and only later on, started the study of complex structures such as cells.
Over the past decades, the potential of microfluidics to change cell biology research had become gradually a reality (Scheme 3). The similitude of the dimensions of microfluidic channels and the size of cells, and the ability to manipulate small volumes of fluid in micrometre-scale channels, together with many other important advantages of microfluidics such as flexibility, rapid energy dissipation, and a fast-prototyping fabrication for customized experimental design, make microfluidics suitable for cell culture and for cells’ subsequent analysis (Tehranirokh et al., 2013; Mehling and Tay, 2014; Kerk et al., 2021). Microfluidics reduced cell populations to a few hundred cells, compared to macroscopic cell cultures that contain around 104–107 cells (Halldorsson et al., 2015). Microfluidics allows precise control over experimental conditions via custom-designed chip architectures, automation, and coupling to downstream analysis platforms (Yeon and Park, 2007). One of the key benefits of microfluidics is the ability to closely mimic the cell’s natural microenvironment, by creating chemical gradients. The composition of the media can be optimized rapidly because of the small volumes of liquids involved.
Also, microfluidics allows the relatively easy patterning of molecules and cells and high levels of experimental automation. In microfluidics, the consumption of nutrients is faster than in traditional culture, because of the higher surface-to-volume ratio (Torino et al., 2018). For this reason, microfluidic culture often needs an integrated perfusion system.
In the course of time, two major microfluidic technologies have been developed: continuous fluid flow in enclosed microchannels and droplet-based cell culture systems, involving, either immiscible droplets in enclosed microchannels, or digital microfluidic manipulation of droplets on planar surfaces (Ng et al., 2015). In digital microfluidics, discrete droplets are manipulated on a hydrophobic surface. The first lab-on-a-chip platform for complete mammalian cell culture was introduced by Wheeler (Barbulovic-Nad et al., 2010) and it was, at the time, the first microfluidic platform able to implement all the steps, required for mammalian cell culture, including passaging. The results obtained by using digital microfluidics were found to be similar to those obtained by macroscale cell culture experiments. An electrical potential is applied to an array of insulated electrodes to actuate the droplets Cells that grow while suspended in a liquid culture medium may be further manipulated by the use of electrical, optical, or magnetic forces. To enhance suspension culture performance, mamallian and microbial cells are encapsulated in hydrogel beads as a scaffold to create a 3D environment (Mulas et al., 2020; Kerk et al., 2021).
Separating cells, positioning them in confined regions can be done by using cell patterning methods (Takayama et al., 1999). To perform patterned cell deposition, the authors used parallel liquid streams in capillary networks to pattern the substrate with adhesion promoters and inhibitors (cell non-adhesive molecules, for example, polyethylene glycol), followed by depositing cells in patterns of parallel stripes. This method generates patterns inside the channels of microfluidic networks. Other techniques, such as microcontact printing of collagen patterns, were also used to permit cell attachment on PDMS substrates (de Silva et al., 2004; Huang et al., 2022).
Three-dimensional culture in digital microfluidics was first demonstrated by Fiddes (Fiddes et al., 2012). He cultured NIH-3T3 cells in hydrogel discs for 7 days, by pipetting the cell suspension and agarose onto the bottom plate. After assembling the top plate, the mixture was cooled to help the gelation of the agarose.
Cells can also be encapsulated, by using hydrogel materials such as collagen, gelatin, fibrin, alginate, and agarose (Wan, 2012). Droplet microfluidics technologies are able to encapsulate the cells into picoliter-sized droplets for growing and subsequent analysis.
A plethora of microfluidic devices, based on two-dimensional and three-dimensional cultures have been developed. As mentioned previously, the most important advantages of microscale technology are better spatial and temporal environmental control and the ease of patterning molecules and cells. At the microscale, complex 3D environments can be created, for example, mimicking microvasculature (Mannino et al., 2018), by incorporating endothelial cell monolayers into microchannels to generate blood vessels-on-a-chip. Endotheliazed microfluidic technology has allowed the study of microvascular phenomena, impossible to investigate previously.
The first applications of microfluidics to cell culture and subsequent analysis were using devices made of silicon and glass because polystyrene, frequently used in macroscopic cell cultures, has limited use in microfluidics, due to the difficult mold fabrication and bonding. A number of challenges of PDMS-based microfluidic cell culture are summarized in reference (Tanyeri et al., 2018). One of the most important requirements for PDMS-based devices is a special surface treatment to promote cell adhesion and proliferation on the hydrophobic surface of PDMS. Typically, the surface of PDMS is treated with oxygen plasma and then coated with fibronectin or collagen which are adhesion-promoting. It has been shown that surfaces need to be only moderately hydrophilic, in order to avoid the antifouling properties of highly hydrophilic surfaces that may preclude cell culture. Selective treatment of PDMS has been recommended with only the culture chambers made more hydrophilic.
Small molecules absorbed by the porous PDMS may have a negative effect on cell culture applications. To avoid this, PDMS can be coated with low-permeability materials such as parylene and wax (Tanyeri et al., 2018).
Progress in the area of biology-related microfluidic systems has been mostly in proof of-principle demonstrations and it has been delayed because the behavior of living cells in confined spaces was difficult to understand. Cell culture techniques cannot be directly transferred from macroscale to microfluidic environments, without a deep understanding of the physics of the microscale. Some recent reports have also begun to reveal challenges with existing microfluidic methods, and have, in some cases, provided possible solutions that may lead to new directions in the field (Velve-Casquillas et al., 2010).
3.3 The cellular microenvironment gradient microgenerators
The most important benefit of using microfluidics for biology is the ability to tailor the cellular microenvironment. Unlike the traditional macroscale culture, where the parameters can be controlled only for a population of cells, at the microscale, the spatial and temporal gradients in the microenvironment of cells can be created and controlled.
The microenvironment of a cell is defined by chemical and mechanical parameters. The chemical environment is composed of soluble molecules around the cell, associated with the composition of the culture medium, and the mechanical environment is composed of the extracellular matrix (ECM), associated with the substrate composition.
Cells reside in a milieu composed of soluble factors, cell-matrix interactions, and cell-cell contacts, living within an environment with specific physicochemical properties (pH, oxygen tension, temperature, and osmolality) (Figure 2).
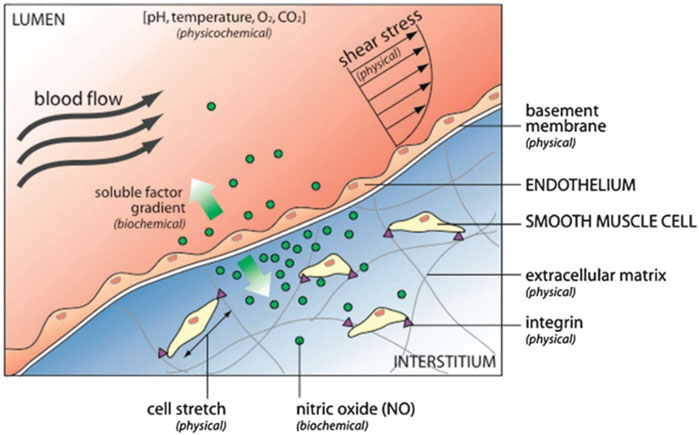
FIGURE 2. The cell microenvironment consists of physical, biochemical, and physicochemical factors. For example, the endothelium that lines blood vessels is exposed to hemodynamic shear stress (external physical force) that stimulates biochemical response releasing nitric oxide. Nitric oxide diffuses to neighboring smooth muscle cells, where it regulates cell contraction and relaxation. Reproduced from Young EW and Beebe D. (2010) with permission from the Royal Society of Chemistry.
Microfluidics has the ability, not only to locally address parameters of the cell environment but also to change them automatically. Natural stimuli that happen in biological processes can act by producing a chemical gradient. Concentration gradients using hydrogel from collagen or agars to create concentration gradients were found unstable (Velve-Casquillas et al., 2010) and other macroscopic methods, based on chambers, separated by membranes, generated unstable gradients as well (Zigmond, 1977). Precisely defined chemical gradients in microfluidic devices have been successfully generated by microscale gradient generators based on single-layer PDMS.
The simplest microgenerator is based on the laminar flow in a T-sensor and the shape of the gradient depends on the flow rate. However, in this device, cells may be subjected to mechanical stress that may damage them. In addition, long experiments require large volumes of often expensive reagents. The gradients are created by diffusion at the interface of two streams, then, flowed over cells to expose them to the gradients (Paguirigan and Beebe, 2008). In addition to the flow-based gradient generators, static gradient devices can be integrated more easily, and even for long experiments, minimal reagents are required (Young EW and Beebe D, 2010).
The cell microenvironment results from biochemical and physical factors and has an important effect on the growth and development of neighbouring tissues. For example, the local microenvironment of stem cells, called the stem cell niche, regulates their survival, self-renewal, and differentiation (Discher et al., 2009). Most cells in the body are attached to the surrounding extracellular matrix (ECM) via cell surface integrins, responsible for transducing mechanical signals from the adhesion sites to the cytoskeletal machinery. It has been shown that the mechanical properties of cells are essential to the mechanisms by which cells sense forces and transduce them into chemical signals (Janmey and McCulloch, 2007). Migration, proliferation, and differentiation of cells are all driven by chemical signals. Since gradients have an important role in many processes, recent studies are incorporating them into their assays to understand better the effect of gradients on cells. Microfluidics is expected to play a significant role in the implementation of such gradient assays, because it can establish stable gradients (Keenan T and Folch, 2008). Some recent studies examine the combinatorial effects of soluble factor signaling and cell-matrix interactions on cell behavior.
3.4 Stem cells in microfluidics
Stem cells are unspecialized pluripotent cells able to produce any type of cell in the human body. They can migrate to the disease-affected area, and play a pivotal role in wound healing, repair, and growth. They are highly self-regulating, but the fate of each stem cell is governed by the chemical and mechanical stimuli a stem cell receives during the period of growth and development. They are sensitive to shear stress caused by the flow of the medium, and morphology of the substrate/extracellular matrix.
As mentioned previously, until recently, scientists, primarily, worked with two kinds of stem cells from animals and humans: embryonic stem cells, which are pluripotent, and very attractive for therapeutical applications, and non-embryonic or “adult” stem cells (mesenchymal) that are multipotent, that is, they only differentiate into a limited number of stem cells (Burdick and Vunjak-Novakovic, 2009; Velve-Casquillas et al., 2010; Zhang and Austin, 2012; Tehranirokh et al., 2013; Sugiura et al., 2016; Luni et al., 2022) In 1981, scientists derived embryonic stem cells from early mouse embryos, and later, they discovered a method to derive stem cells from human embryos and grow them in the laboratory. These cells are called human embryonic stem cells. Stem cells have the capacity to both self-renew and give rise to descendants which can commit themselves in a number of distinct directions. Stem cell behavior is extremely sensitive to environmental cues that can be manipulated in microfluidic cell culture (Scheme 4).
Analyses of stem cells can be done in a much expanded way in a microfluidic device than in a conventional tissue culture dish. Conventional tissue culture dishes have been used to test different culture conditions of stem cell growth and differentiation. However, this approach requires significant amounts of stem cells, which reports the average response of the population and it is very difficult to precisely control the cell number. It is also expensive and labour-intensive to maintain hundreds of dishes over a long time.
Microfluidics offers a new way to perform high-throughput screening, using a much lower amounts of starting cells, and the dynamic adjustment of culture conditions (James et al., 2012; Jensen and Teng, 2020).
Figure 3 shows other approaches used for the three-dimensional culture of stem cells and Figure 4 shows the stem cell niche.
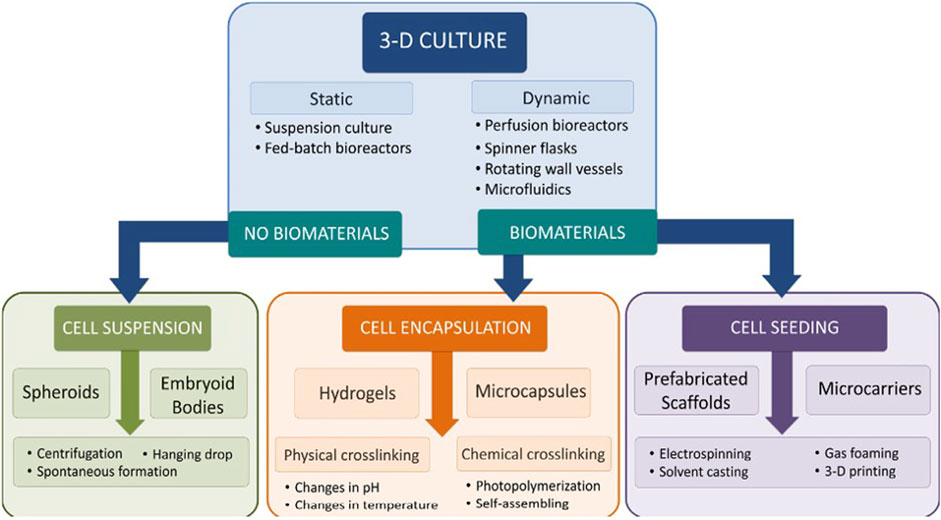
FIGURE 3. Schematic of approaches used for three-dimensional culture of stem cells. Reproduced from McKee and Chaudry (2017) under CC BY-NC-ND 4.0.
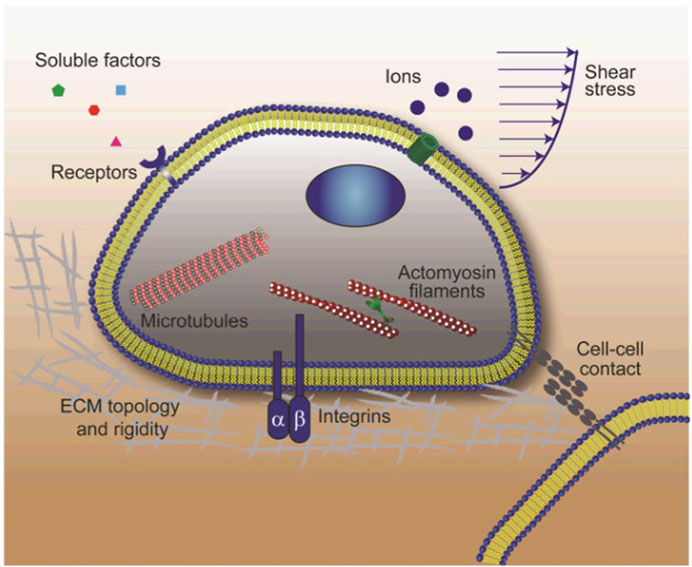
FIGURE 4. Schematic showing the stem cell niche where interactions between stem cells and their local microenvironment regulate stem cell fate. The stem cell niche is a complex dynamically regulated 3-D microenvironment comprising soluble biochemical and insoluble biomechanical cues, adhesive signals as well as well as signals arising from direct cell-cell contacts. Soluble biochemical cues include small ions, growth factors, cytokines, etc. Insoluble biophysical signals consist of matrix rigidity and topology, fluid shear-stress, and other mechanical forces exerted by adjacent cells or owing to tissue growth and loading. Stem cells sense and respond to these biophysical stimuli through different mechanosensory components including heterodimeric integrins, mechanosensitive ion channels, cycloskeleton structures (actin microfilaments, microtubules, and intermediate filaments) and cell-cell contacts. Reproduced from Sun Y et al. (2012), with permission from Wiley Periodicals, Inc.
3D stem cells culture systems are, either scaffold-based, or scaffold-free systems. Among the scaffold-free systems, the most used is the hanging drop culture where individual cells aggregate under the effect of their own gravity to form 3D spheres. This method is simple and low cost and allows the co-culturing of different types of cells but scale-up is difficult.
Another quite simple method, easier to scale-up than the hanging drop culture is the rotating bioreactor culture, a dynamic culture, where the cell suspension is continuously stirred, distributing uniformly the nutrients and oxygen. This method maximizes the contact between the cells and the spheroids can be formed easily. However, stem cells are sensitive and may be easily damaged by the shear stress generated by the continuous stirring. . Scaffold-based culture systems may use natural, or artificial (synthetic) materials as scaffolds. In this regard, it has to be mentioned that both natural and synthetic hydrogels are most widely used scaffold materials. Other natural materials are collagen, fibrin, gelatin, and composite hydrogels with good biocompatibility and mechanical properties. Synthetic hydrogels scaffold materials have a better-defined composition but poor biocompatibility (Scheme 5).
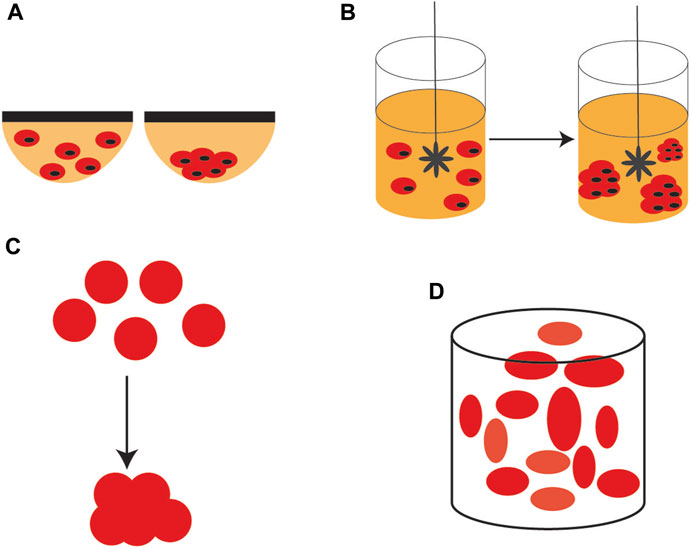
SCHEME 5. 3D stem cell culture systems. (A) hanging drop culture (B) rotating bioreactor culture (C) formation of scaffolds, and (D) self-assembling scaffolds.
Because of the need for stem cells in cell-based therapies, automated cell culture techniques became extremely important. One of the main requirements in culturing stem cells is a controlled proliferation and differentiation (Ertl et al., 2014; Zhang et al., 2017; Aghlmandi et al., 2021). There are now important research efforts focused on microfluidic studies of stem cells, not only on culture, but also on tissue engineering and organ models.
Microenvironmental control for stem cells is extremely important as they are highly sensitive. Microfluidic perfusion culture allows the production of small-volume cultures, with precisely controlled microenvironments and is applicable to high-throughput cellular environment screening. Microfluidic technology has great potential to improve stem cell (SC) cultures, whose promise in cell–based therapies is limited by the inability to precisely control their behavior in culture. Compared to traditional culture tools, microfluidic platforms provide much greater control over the cell microenvironment and rapid optimization of media composition, using relatively small numbers of cells. By controlling fluidic properties like convection, diffusion, and reaction, microfluidics can tune the microenvironment around stem cells in a variety of ways. Microfluidics can probe important biological processes like differentiation and evolution in more biologically relevant conditions than conventional tissue culture dishes.
3.5 Microfluidic culture of stem cells. Examples
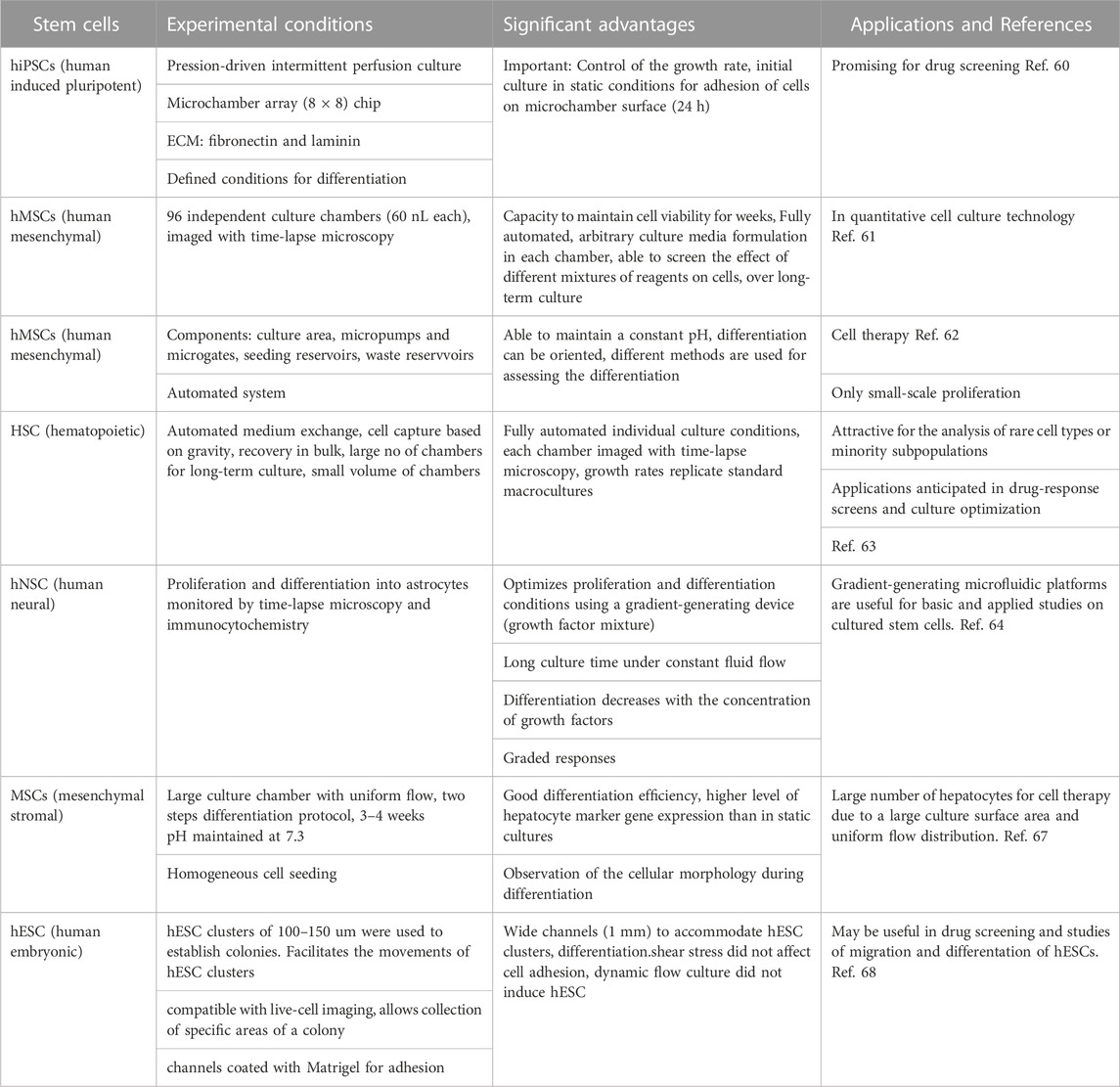
Recent work on the microfluidic culture of different stem cells is summarized below (see also Table 1).
- It has been shown that microfluidics can create dynamic microenvironments that can accelerate the growth rate of some stem cells such as hiPSCs. To investigate the growth under perfusion, a microfluidic perfusion culture system, using a microchamber array chip, was developed (Yoshimitsu et al., 2014). The authors found that both fibronectin and laminin were beneficial for culturing the cells in PDMS microfluidic devices. Interestingly, the authors found that, under pressure-driven perfusion culture conditions, the cells growth faster than under static culture conditions. The system was used to study the self-renewal and differentiation cultures of hiPSCs, and it was found that their state can be controlled by using the perfusion system.
- A fully automated, highly integrated cell culture system, with the capacity to screen 96 independent culture chambers and maintain cell viability for a long time was developed by Gomez-Sjoberg et al. The device was able to establish different parameters in each chamber and change them separately. The authors worked with human mesenchymal stem cells in each chamber and studied the effect of inoculation number on their proliferation, differentiation, and motility, over several weeks (Gómez-Sjöberg et al., 2007).
- A prototype microfluidic system for the long-term culture and differentiation of Human mesenchymal stem cells (MSCs), composed mainly of seeding and waste reservoirs and stem cell culture areas, was developed by Wu et al. (Wu et al., 2009). The main advantage of this complex system is the capacity to maintain a constant pH, an important parameter that may determine the direction of the differentiation process. Depending of the need of tissue lineages for cell therapy, in this microfluidic system, the differentiation process can be oriented and studied by different methods. In this system, the cells can be cultured for several weeks.
A long-time culture and proliferation of hematopoietic stem cells (HSC) have been carried out by Lecault et al. in a programmable microfluidic platform, containing thousands of small-scale (nanoliter scale) chambers (Lecault et al., 2011). The control of proliferation of HSC on this device can be done at the single cell level. It is interesting to mention that in this device, cells are not trapped mechanically but gravity is used to immobilize them, without any perturbation. Cells are not disturbed either by the programmable medium exchanges and no nutrient limitation has been observed in long-time experiments. After culture, cells are recovered in bulk, by inverting and flushing the device. The authors found growth rates similar to those in standard macrocultures. They anticipate that this device will be especially useful for culture optimization and cell characterization.
- Chemical gradients have been established in microfluidics devices by either laminar flow or controlled diffusion. For example, the proliferation and differentiation of human neural stem cells (hNSCs) from the developing cerebral cortex were optimized by culturing them in a gradient-generating microfluidics platform (Chung et al., 2005). The cells were cultured more than a week, exposed constantly to a gradient of a growth factor mixture. Time-lapse microscopy and immunocytochemistry were used to monitor the proliferation and differentiation of neural stem cells into astrocytes. The authors found that both proliferation and differentiation varied with the concentration of the growth factor. The device is made up of a gradient chamber and two control chambers. The microchannel network that produces the gradient (“Christmas tree”) generates a combinatorial mixture of soluble factors, utilizing laminar flow and diffusive mixing. Interesingly, the authors found in this work that, while proliferation is proportional to the concentration of growth factors, astrocyte differentiation was found inversely proportional to concentration of growth factors but this result has not be discussed. This microfluidic platform should be useful in the future for studies of a variety of cultured stem cells.
- In addition to growth factors, oxygen and temperature gradients have also been established in microfluidics devices. By using diffusion to localize oxygen delivery the authors could modulate the intracellular reactive oxygen species. It was found that embryos can adapt to the effects of temperature steps by increasing their development rate to compensate for the fluctuating environment (Lucchetta et al., 2005; Lo et al., 2010). These two papers focus on creation of different gradients in microfluidic devices but are not specifically stem cell studies, therefore, will not be discussed in this section.
- An innovative microfluidic cell culture device for efficient hepatic differentiation of mesenchimal stromal cells (MSCs) has been designed and fabricated by Yen et all. (Ju et al., 2008).
The proliferation and hepatic differentiation of mouse MSCs have been studied in a device with an unusually large culture chamber (culture area 400 mm2), allowing a uniform flow distribution, creating an optimal environment for the hepatic differentiation, and accelarating it, compared with a conventional culture dish. A point of interest of this work is the presence of a continuous flow, supplying nutrients for the differentiation and removing the unhealthy cells. At the same time, the effect of fluid shear stress on the hepatic differentiation has been investigated. Also, the authors used a two-step hepatic differentiation protocol with growth factors that support hepatogenesis. Microscopic observations allowed real-time observation of cell morphology. The authors are confident that the cells differentiated in their microfluidic device will be suitable for cell therapy.
Individual human embryonic stem cell (hESC) colonies in dynamic or static conditions have been studied in a microfluidic culture system, allowing to treat in a different way desired parts of a colony (Villa-Diaz et al., 2009). Figure 5 shows a microfluidic device adapted for hESC culture. The authors found that both static and dynamic cultures for 96 h results in the same properties of hESC. They have demonstrated that, neither adhesion, nor proliferation and self-renewal of hESCs are affected by a dynamical culture at flow rates up to 1.5 mL/h. Portions of hESC colonies can be targeted, This may allow the study of propagation signals from one end to the other end of a colony. This experimental platform is useful in drug screening and studies of spatially regulated signaling, and differentiation of hESCs. The human embryonic stem cells may be differentiated into different lineages.
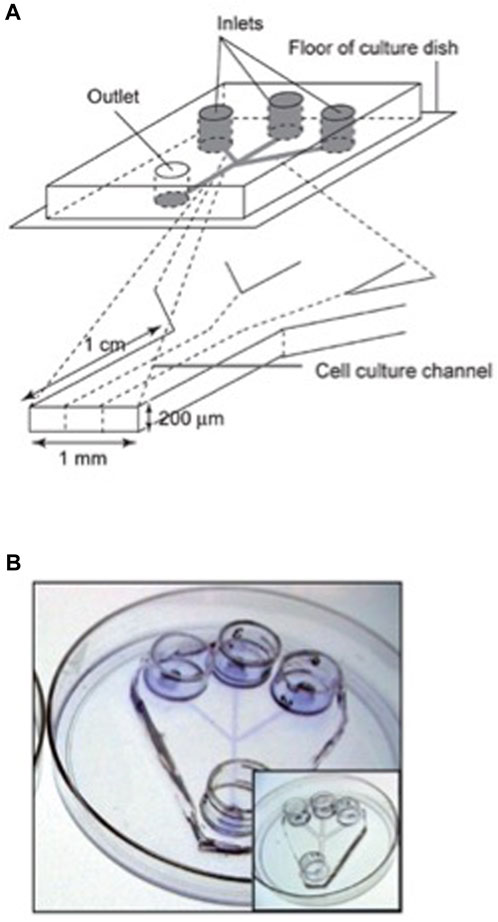
FIGURE 5. Design of a microfluidic device adapted for hESC culture. (A) Schematic representation of the microfluidic device, with three inlets channels that converge into a single main channel. The channels size was customized to accommodate and facilitate the introduction and movement of hESC clusters and posterior colony formation. Each channel feature is 200 μm in height, 1 mm in width and 1 cm in length, and the entire device fits in a 100 mm tissue culture dish. This microfluidic device is able to generate laminar flow in the cell-culture channel. (B) Channels were coated with Matrigel to provide extracellular matrix (ECM) components that allow hESC attachment and colony formation. The photograph shows a microfluidic device stained with Commassie blue to demonstrate the ECM coating. The insert shows a microfluidic device without ECM coating. Reproduced from Villa-Diaz IG et al. (2009) with permission from the Royal Society of Chemistry.
Table 1 summarizes the above microfluidic stem cell experiments, including a critical appraisal.
It is difficult to compare the experiments described in the text and summarized in the table. They are all different, with various designs and proliferation and differentiation conditions. The general trend seems to be automation, allowing any change, simultaneously, in the experimental conditions in thousands of culture chambers, during the long-time experiments. It has to be mentioned that microfluidic culture is convenient for selecting the best scaffold materials for differentiation of stem cells. The results of the experiments described in this paper show that microfluidic culture of stem cells is still in the stage of data collection on the way differentiation of stem cells happens in confined spaces. In addition, culture scale up in microfluidic devices, necessary for cell therapy, will need further advances in design of devices and changes in the methods. However, each work, discussed here, brings important novelties that do contribute to the progress of one of the most important research fields today, microfluidic stem cell culture.
Apart from differentiation and proliferation, stem cell migration behavior gained high importance due to their application in regenerative medicines. Stem cell migration is a fundamental process that occurs during embryogenesis, organogenesis, immune response, and most importantly, cancer progression (Pijuan Marquilles et al., 2019). Various microfluidic models have been reported to model the cell migration that occurs during the above processes, neural embryogenesis models (Kurosaka and Kashina, 2008; Lee et al., 2014; Bae et al., 2016), and immune response (Vesperini et al., 2021). In the traditional migration assay or wound healing assay, the cells are cultured on a plate, and once confluence is achieved, a scratch is made, removing a section on the cells, creating a cell-free zone, then, the migration of the cells is measured. However, in a microfluidic platform, in addition to the various advantages already mentioned, the device allows single-cell level tracking (Yazdanpanah et al., 2023), the creation of drug gradients similar to in vivo situations, and the compartmentalization of cells and molecules. In a microfluidic device, the entire channel is cultured with cells and later the cells from the desired area are removed by flushing liquids via specific channels. Here, the cells remain intact and are not injured as in the case of the conventional scratch assays.
Stem cell migration studies have gained further importance with the development of regenerative therapy, where stem cells are directly introduced into the body and they must traverse considerable distances to reach the desired location. By installing diffusion barriers, in vivo-like environments were recaptured on a multichannel microfluidic device to find out drug gradients and evaluate the effect of Substance P, a stem cell mobilizer, on the migration of human bone marrow-derived mesenchymal stem cells (BM-MSCs) (Kim et al., 2023).
4 Conclusion and outlook
This paper reviews recent advancements in both macro-scale and microfluidic culture. It shows that microfluidics is attractive in many aspects, especially reconstructing the physiological environment, and isolating rare stem cells. As in microfluidics, many factors, physiologically relevant, can be controlled precisely, microfluidic devices are well-suited to biological experiments and provide an ideal platform for the study of stem cells, especially, their behavior in cultures. Compared to traditional culture tools, microfluidic platforms provide much better control over the cell microenvironment and the composition of the media can be optimized rapidly. The macroscopic culture platforms, in spite of the recent improvements, do not have yet the capability to create concentration and other type of gradients. However, in our opinion, there are issues that need to be solved before microfluidics can be widely applied to the resolution of fundamental biological problems, rather than simply to be a proof of concepts. It is important to make microfluidics devices easy to use by biologists in general and, particularly, by stem cell biologists. To help to solve this issue it would be important to standardize and commercialize the microfluidics technologies developed in academic labs. It will be necessary to be able to scale up microfluidic devices in order to meet the requirements, regarding large-scale applications. Also, it would be useful to train biologists who do not have experience in fluid physics or microfabrication as well as engineers who may not have a deep knowledge in the biology of stem cells.
As Paguirigan and Beebe (Paguirigan and Beebe, 2008) have shown, there is a “disconnect” between the engineers, who design and fabricate the devices, and the biologists who will use them. It seems to the authors of this review paper that, this disconnect still exists, regarding language (terminology), mindset, etc., and will have to be attenuated for a better collaboration.
Regarding the materials to make microfluidics devices for biology experiments, especially, stem cell research, PDMS which is a common choice today, will have to be, at least partially replaced, by more biocompatible, cell-friendly materials.
Absorption of molecules in PDMS can change the local chemical concentration and could reduce the reproducibility of experiments. Therefore, it is important to look in the future for more biocompatible materials for stem cell research, especially, if cells are exposed to PDMS for longer duration. Also, highly integrated systems are needed to address a specific question in stem cell biology from different aspects. Nevertheless, we believe microfluidics will become in the near future a powerful tool for both fundamental understanding and medical applications of stem cells.
The effect of microgravity on differentiation and cell growth in stem cells and cancer stem cells has been studied recently by (Grimm et al., 2020). The results of this study are extremely important for stem cell research.
A number of authors advanced the idea that microfluidic culture systems have the potential to become a substitute for macroscale techniques but, there is still much work to be done (Halldorsson et al., 2015; Tanyeri et al., 2018; Coluccio et al., 2019). Most importantly, high reproducibility requirements and precise culture protocols have to be developed for the microscale. Due to all the recent improvements, we believe that macroscopic cell cultures will continue to be useful for culturing large amounts of cells required by biomedical applications.
Author contributions
SB: Writing–original draft, Writing–review and editing, Supervision, Validation. SP: Conceptualization, Investigation, Methodology, Writing–original draft, Formal Analysis. MP: Writing–review and editing, Conceptualization, Supervision.
Funding
The authors declare that no financial support was received for the research, authorship, and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Aghlmandi, A., Nikshad, A., Safaralizadeh, R., Warkiani, M. E., Aghebati-Maleki, L., and Yousefi, M. (2021). Microfluidics as efficient technology for the isolation and characterization of stem cells. EXCLI J. 20, 426–443. doi:10.17179/excli2020-3028
Badilescu, S., and Packirisamy, M. (2012). Microfluidics-nano-integration for synthesis and sensing. Polymers 4 (2), 1278–1310. doi:10.3390/polym4021278
Bae, J., Lee, N., Choi, W., Lee, S., Ko, J. J., Han, B. S., et al. (2016). “Use of microfluidic technology to monitor the differentiation and migration of human ESC-derived neural cells,” in Bioreactors in stem cell biology: Methods and protocols. Methods in molecular biology. Editor K. Turksen (New York, NY: Springer), 223–235. doi:10.1007/7651_2016_337
Barbulovic-Nad, I., Au, H., and Wheeler, R. (2010). A microfluidic platform for complete mammalian cell culture. Lab. Chip 10 (12), 1536–1542. doi:10.1039/c002147d
Beebe, D. J., Mensing, G. A., and Walker, G. M. (2002). Physics and applications of microfluidics in biology. Annu. Rev. Biomed. Eng. 4 (1), 261–286. doi:10.1146/annurev.bioeng.4.112601.125916
Burdick, J. A., and Vunjak-Novakovic, G. (2009). Engineered microenvironments for controlled stem cell differentiation. Tissue Eng. Part A 15 (2), 205–219. doi:10.1089/ten.tea.2008.0131
Chapman, T. (2003). Lab automation and robotics: automation on the move. Nature 421 (6923), 661–663. doi:10.1038/421661a
Chung, B. G., Flanagan, L. A., Rhee, S. W., Schwartz, P. H., Lee, A. P., Monuki, E. S., et al. (2005). Human neural stem cell growth and differentiation in a gradient-generating microfluidic device. Lab. Chip 5 (4), 401–406. doi:10.1039/b417651k
Coluccio, M. L., Perozziello, G., Malara, N., Parrotta, E., Zhang, P., Gentile, F., et al. (2019). Microfluidic platforms for cell cultures and investigations. Microelectron. Eng. 208, 14–28. doi:10.1016/j.mee.2019.01.004
Crombie, D. E., Daniszewski, M., Liang, H. H., Kulkarni, T., Li, F., Lidgerwood, G. E., et al. (2017). Development of a modular automated system for maintenance and differentiation of adherent human pluripotent stem cells. SLAS Discov. Adv. Sci. Drug Discov. 22 (8), 1016–1025. doi:10.1177/2472555217696797
Dalerba, P., and Clarke, M. F. (2007). Cancer stem cells and tumor metastasis: first steps into uncharted territory. Cell Stem Cell 1 (3), 241–242. doi:10.1016/j.stem.2007.08.012
de Silva, M. N., desai, R., and Odde, D. J. (2004). Micro-patterning of animal cells on PDMS substrates in the presence of serum without use of adhesion inhibitors. Biomed. Microdevices 6 (3), 219–222. doi:10.1023/b:bmmd.0000042051.09807.8c
Dettinger, P., Kull, T., Arekatla, G., Ahmed, N., Zhang, Y., Schneiter, F., et al. (2022). Open-source personal pipetting robots with live-cell incubation and microscopy compatibility. Nat. Commun. 13 (1), 2999. doi:10.1038/s41467-022-30643-7
Discher, D. E., Mooney, D. J., and Zandstra, P. W. (2009). Growth factors, matrices, and forces combine and control stem cells. Science 324 (5935), 1673–1677. doi:10.1126/science.1171643
Ertl, P., Sticker, D., Charwat, V., Kasper, C., and Lepperdinger, G. (2014). Lab-on-a-chip technologies for stem cell analysis. Trends Biotechnol. 32 (5), 245–253. doi:10.1016/j.tibtech.2014.03.004
Fiddes, L. K., Luk, V. N., Au, S. H., Ng, A. H. C., Luk, V., Kumacheva, E., et al. (2012). Hydrogel discs for digital microfluidics. Biomicrofluidics 6 (1), 014112–1411211. doi:10.1063/1.3687381
Fuchs, E., and Segre, J. A. (2000). Stem cells: A new Lease on life. Cell 100 (1), 143–155. doi:10.1016/s0092-8674(00)81691-8
Gómez-Sjöberg, R., Leyrat, A. A., Pirone, D. M., Chen, C. S., and Quake, S. R. (2007). Versatile, fully automated, microfluidic cell culture system. Anal. Chem. 79 (22), 8557–8563. doi:10.1021/ac071311w
Grimm, D., Wehland, M., Corydon, T. J., Richter, P., Prasad, B., Bauer, J., et al. (2020). The effects of microgravity on differentiation and cell growth in stem cells and cancer stem cells. Stem Cells Transl. Med. 9 (8), 882–894. doi:10.1002/sctm.20-0084
Halldorsson, S., Lucumi, E., Gómez-Sjöberg, R., and Fleming, R. M. T. (2015). Advantages and challenges of microfluidic cell culture in polydimethylsiloxane devices. Biosens. Bioelectron. 63, 218–231. doi:10.1016/j.bios.2014.07.029
Huang, X., Huang, Z., Gao, W., Gao, W., He, R., Li, Y., et al. (2022). Current advances in 3D dynamic cell culture systems. Gels 8 (12), 829. doi:10.3390/gels8120829
Hussain, W., Moens, N., Veraitch, F. S., Hernandez, D., Mason, C., and Lye, G. J. (2013). Reproducible culture and differentiation of mouse embryonic stem cells using an automated microwell platform. Biochem. Eng. J. 77, 246–257. doi:10.1016/j.bej.2013.05.008
Imreh, M. p., Gertow, K., Cedervall, J., Unger, C., Holmberg, K., Szöke, K., et al. (2006). In vitro culture conditions favoring selection of chromosomal abnormalities in human ES cells. J. Cell Biochem. 99 (2), 508–516. doi:10.1002/jcb.20897
James, L. X., Valadez, A. V., Zuo, P., and Nie, Z. (2012). Microfluidic 3D cell culture: potential application for tissue-based bioassays. Bioanalysis 4 (12), 1509–1525. doi:10.4155/bio.12.133
Janmey, P. A., and McCulloch, C. A. (2007). Cell mechanics: integrating cell responses to mechanical stimuli. Annu. Rev. Biomed. Eng. 9 (1), 1–34. doi:10.1146/annurev.bioeng.9.060906.151927
Jensen, C., and Teng, Y. (2020). Is it time to start transitioning from 2D to 3D cell culture? Front. Mol. Biosci. 7, 33. doi:10.3389/fmolb.2020.00033
Ju, X., Li, D., Gao, N., Shi, Q., and Hou, H. (2008). Hepatogenic differentiation of mesenchymal stem cells using microfluidic chips. Biotechnol. J. 3 (3), 383–391. doi:10.1002/biot.200700152
Kanda, G. N., Tsuzuki, T., Terada, M., Sakai, N., Motozawa, N., Masuda, T., et al. (2022). Robotic search for optimal cell culture in regenerative medicine. eLife 11, e77007. doi:10.7554/elife.77007
Keenan T, M., and Folch, A. (2008). Biomolecular gradients in cell culture systems. Lab. Chip 8 (1), 34–57. doi:10.1039/b711887b
Kerk, Y. J., Jameel, A., Xing, X. H., and Zhang, C. (2021). Recent advances of integrated microfluidic suspension cell culture system. Eng. Biol. 5 (4), 81–97. doi:10.1049/enb2.12015
Kim, J., Kim, J., Park, H. J., Jeon, E. J., and Cho, S. W. (2023). A microfluidic platform for simulating stem cell migration using in vivo-like gradients of stem cell mobilizer. Korean J. Chem. Eng. 40 (4), 903–909. doi:10.1007/s11814-023-1390-6
Konagaya, S., Ando, T., Yamauchi, T., Suemori, H., and Iwata, H. (2015). Long-term maintenance of human induced pluripotent stem cells by automated cell culture system. Sci. Rep. 5 (1), 16647. doi:10.1038/srep16647
Kurosaka, S., and Kashina, A. (2008). Cell biology of embryonic migration. Birth Defects Res. Part C Embryo Today Rev. 84 (2), 102–122. doi:10.1002/bdrc.20125
Lecault, V., VanInsberghe, M., Sekulovic, S., Knapp, D. J. H. F., Wohrer, S., Bowden, W., et al. (2011). High-throughput analysis of single hematopoietic stem cell proliferation in microfluidic cell culture arrays. Nat. Methods 8 (7), 581–586. doi:10.1038/nmeth.1614
Lee, N., Park, J. W., Kim, H. J., Yeon, J. H., Kwon, J., Ko, J. J., et al. (2014). Monitoring the differentiation and migration patterns of neural cells derived from human embryonic stem cells using a microfluidic culture system. Mol. Cells 37 (6), 497–502. doi:10.14348/molcells.2014.0137
Liu, Y., Hourd, P., Chandra, A., and Williams, D. J. (2010). Human cell culture process capability: A comparison of manual and automated production. J. Tissue Eng. Regen. Med. 4 (1), 45–54. doi:10.1002/term.217
Lo, F., Sinkala, E., and Eddington, T. D. (2010). Oxygen gradients for open well cellular cultures via microfluidic substrates. Lab. Chip 10 (18), 2394–2401. doi:10.1039/c004660d
Lucchetta, E. M., Lee, J. H., Fu, L. A., Patel, N. H., and Ismagilov, R. F. (2005). Dynamics of Drosophila embryonic patterning network perturbed in space and time using microfluidics. Nature 434 (7037), 1134–1138. doi:10.1038/nature03509
Luni, C., Gagliano, O., and Elvassore, N. (2022). Derivation and differentiation of human pluripotent stem cells in microfluidic devices. Annu. Rev. Biomed. Eng. 24 (1), 231–248. doi:10.1146/annurev-bioeng-092021-042744
Mannino, R. G., Qiu, Y., and Lam, W. A. (2018). Endothelial cell culture in microfluidic devices for investigating microvascular processes. Biomicrofluidics 12 (4), 042203. doi:10.1063/1.5024901
Manz, A., Graber, N., and Widmer, H. M. (1990). Miniaturized total chemical analysis systems: A novel concept for chemical sensing. Sens. Actuators B Chem. 1 (1), 244–248. doi:10.1016/0925-4005(90)80209-i
McKee, C., and Chaudhry, G. R. (2017). Advances and challenges in stem cell culture. Colloids Surf. B Biointerfaces 159, 62–77. doi:10.1016/j.colsurfb.2017.07.051
Mehling, M., and Tay, S. (2014). Microfluidic cell culture. Curr. Opin. Biotechnol. 25, 95–102. doi:10.1016/j.copbio.2013.10.005
Mulas, C., Hodgson A, C., Kohler T, N., Agley C, C., Humphreys, P., Kleine-Brüggeney, H., et al. (2020). Microfluidic platform for 3D cell culture with live imaging and clone retrieval. Lab. Chip 20 (14), 2580–2591. doi:10.1039/d0lc00165a
Ng, A. H. C., Chamberlain, M. D., Situ, H., Lee, V., and Wheeler, A. R. (2015). Digital microfluidic immunocytochemistry in single cells. Nat. Commun. 6 (1), 7513. doi:10.1038/ncomms8513
Ochiai, K., Motozawa, N., Terada, M., Horinouchi, T., Masuda, T., Kudo, T., et al. (2021). A variable scheduling maintenance culture platform for mammalian cells. SLAS Technol. Transl. Life Sci. Innov. 26 (2), 209–217. doi:10.1177/2472630320972109
Paguirigan, A. L., and Beebe, D. J. (2008). Microfluidics meet cell biology: bridging the gap by validation and application of microscale techniques for cell biological assays. BioEssays 30 (9), 811–821. doi:10.1002/bies.20804
Pijuan Marquilles, J., Barceló Gómez, C., Moreno, D. F., Maiques Carlos, O., Sisó, P., Martí Laborda, R. M., et al. (2019). In vitro cell migration, invasion, and adhesion assays: from cell imaging to data analysis. Front. Cell Dev. Biol. 7, 107. doi:10.3389/fcell.2019.00107
Ramalho-Santos, M., and Willenbring, H. (2007). On the origin of the term stem cell. Cell Stem Cell 1 (1), 35–38. doi:10.1016/j.stem.2007.05.013
Reya, T., Morrison, S. J., Clarke, M. F., and Weissman, I. L. (2001). Stem cells, cancer, and cancer stem cells. Nature 414 (6859), 105–111. doi:10.1038/35102167
Sasamata, M., Shimojo, D., Fuse, H., Nishi, Y., Sakurai, H., Nakahata, T., et al. (2021). Establishment of a robust platform for induced pluripotent stem cell research using Maholo LabDroid. SLAS Technol. Transl. Life Sci. Innov. 26 (5), 441–453. doi:10.1177/24726303211000690
Sebastian, S. (2022). Implementing robotics and artificial intelligence. eLife 11, e80609. doi:10.7554/elife.80609
Soares, F. A. C., Sheldon, M., Rao, M., Mummery, C., and Vallier, L. (2014). International coordination of large-scale human induced pluripotent stem cell initiatives: wellcome trust and ISSCR workshops white paper. Stem Cell Rep. 3 (6), 931–939. doi:10.1016/j.stemcr.2014.11.006
Sugiura, T., Hibino, N., Breuer, C. K., and Shinoka, T. (2016). Tissue-engineered cardiac patch seeded with human induced pluripotent stem cell derived cardiomyocytes promoted the regeneration of host cardiomyocytes in a rat model. J. Cardiothorac. Surg. 11 (1), 163. doi:10.1186/s13019-016-0559-z
Sun, Y., Weng, S., and Fu, J. (2012). Microengineered synthetic cellular microenvironment for stem cells. WIREs Nanomedicine Nanobiotechnology 4 (4), 414–427. doi:10.1002/wnan.1175
Takayama, S., McDonald, J. C., Ostuni, E., Liang, M. N., Kenis, P. J. A., Ismagilov, R. F., et al. (1999). Patterning cells and their environments using multiple laminar fluid flows in capillary networks. Proc. Natl. Acad. Sci. 96 (10), 5545–5548. doi:10.1073/pnas.96.10.5545
Tanyeri, M., and Tay, S. (2018). “Chapter 1 - viable cell culture in PDMS-based microfluidic devices,” in Methods in cell biology. Microfluidics in cell biology Part C: Microfluidics for cellular and subcellular analysis. Editors D. A. Fletcher, J. Doh, and M. Piel (Cambridge, Massachusetts: Academic Press). Available from: https://www.sciencedirect.com/science/article/pii/S0091679X18301304.
Tehranirokh, M., Kouzani, A. Z., Francis, P. S., and Kanwar, J. R. (2013). Microfluidic devices for cell cultivation and proliferation. Biomicrofluidics 7 (5), 051502. doi:10.1063/1.4826935
Terstegge, S., Laufenberg, I., Pochert, J., Schenk, S., Itskovitz-Eldor, J., Endl, E., et al. (2007). Automated maintenance of embryonic stem cell cultures. Biotechnol. Bioeng. 96 (1), 195–201. doi:10.1002/bit.21061
Thomas, R. J., Anderson, D., Chandra, A., Smith, N. M., Young, L. E., Williams, D., et al. (2009). Automated, scalable culture of human embryonic stem cells in feeder-free conditions. Biotechnol. Bioeng. 102 (6), 1636–1644. doi:10.1002/bit.22187
Torino, S., Corrado, B., Iodice, M., and Coppola, G. (2018). PDMS-based microfluidic devices for cell culture. Inventions 3 (3), 65. doi:10.3390/inventions3030065
Underhill, G. H., and Bhatia, S. N. (2007). High-throughput analysis of signals regulating stem cell fate and function. Curr. Opin. Chem. Biol. 11 (4), 357–366. doi:10.1016/j.cbpa.2007.05.036
van Noort, D., Ong, S. M., Zhang, C., Zhang, S., Arooz, T., and Yu, H. (2009). Stem cells in microfluidics. Biotechnol. Prog. 25 (1), 52–60. doi:10.1002/btpr.171
Velve-Casquillas, G., Le Berre, M., Piel, M., and Tran, P. T. (2010). Microfluidic tools for cell biological research. Nano Today 5 (1), 28–47. doi:10.1016/j.nantod.2009.12.001
Vesperini, D., Montalvo, G., Qu, B., and Lautenschläger, F. (2021). Characterization of immune cell migration using microfabrication. Biophys. Rev. 13 (2), 185–202. doi:10.1007/s12551-021-00787-9
Villa-Diaz, L. G., Torisawa, Y., Uchida, T., Ding, J., Nogueira-de-Souza, N. C., O’Shea, K. S., et al. (2009). Microfluidic culture of single human embryonic stem cell colonies. Lab. Chip 9 (12), 1749–1755. doi:10.1039/b820380f
Wan, J. (2012). Microfluidic-based synthesis of hydrogel particles for cell microencapsulation and cell-based drug delivery. Polymers 4 (2), 1084–1108. doi:10.3390/polym4021084
Whitesides, G. M. (2006). The origins and the future of microfluidics. Nature 442 (7101), 368–373. doi:10.1038/nature05058
Wu, H. W., Lin, X. Z., Hwang, S. M., and Lee, G. B. (2009). The culture and differentiation of amniotic stem cells using a microfluidic system. Biomed. Microdevices 11 (4), 869–881. doi:10.1007/s10544-009-9304-x
Yazdanpanah Moghadam, E., Sonenberg, N., and Packirisamy, M. (2023). Microfluidic wound-healing assay for ECM and microenvironment properties on microglia BV2 cells migration. Biosensors 13 (2), 290. doi:10.3390/bios13020290
Yeon, J., and Park, J. K. (2007). Microfluidic cell culture systems for cellular analysis. BioChip J. 1, 17–27.
Yoshimitsu, R., Hattori, K., Sugiura, S., Kondo, Y., Yamada, R., Tachikawa, S., et al. (2014). Microfluidic perfusion culture of human induced pluripotent stem cells under fully defined culture conditions. Biotechnol. Bioeng. 111 (5), 937–947. doi:10.1002/bit.25150
Young Ew, K., and Beebe D, J. (2010). Fundamentals of microfluidic cell culture in controlled microenvironments. Chem. Soc. Rev. 39 (3), 1036–1048. doi:10.1039/b909900j
Zhang, J., Wei, X., Zeng, R., Xu, F., and Li, X. (2017). Stem cell culture and differentiation in microfluidic devices toward organ-on-a-chip. Future Sci. OA 3 (2), FSO187. doi:10.4155/fsoa-2016-0091
Zhang, Q., and Austin, R. H. (2012). Applications of microfluidics in stem cell biology. BioNanoScience 2 (4), 277–286. doi:10.1007/s12668-012-0051-8
Keywords: macroscopic culture, robotics, AI, microfluidic culture, stem cell culture, stem cell differentiation
Citation: Badilescu S, Parimalam SS and Packirisamy M (2023) Culturing cells for life: innovative approaches in macroscopic and microfluidic cultures, with an emphasis on stem cells. Front. Nanotechnol. 5:1264498. doi: 10.3389/fnano.2023.1264498
Received: 20 July 2023; Accepted: 01 September 2023;
Published: 29 September 2023.
Edited by:
Kevin M. Koo, The University of Queensland, AustraliaReviewed by:
Wei Shao, Westlake University, ChinaShuvashis Dey, The University of Queensland, Australia
Copyright © 2023 Badilescu, Parimalam and Packirisamy. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Simona Badilescu, c2ltb25hYmFkaWxlc2N1MEBnbWFpbC5jb20=
 Simona Badilescu
Simona Badilescu Subhathirai Subramaniyan Parimalam2
Subhathirai Subramaniyan Parimalam2