
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Nanotechnol. , 15 September 2023
Sec. Biomedical Nanotechnology
Volume 5 - 2023 | https://doi.org/10.3389/fnano.2023.1230407
 Elham Ghazizadeh1,2*†
Elham Ghazizadeh1,2*† Zahra Nasery1†
Zahra Nasery1†Biosensors are of significant importance today in the rapid, sensitive, and reliable detection of biological molecules in many fields, including medicine; owing to this fact, the development of a strong and reliable diagnostic agent is a very interesting topic. Because of their unique features, among other nanomaterials, lipid-based vesicles such as liposomes, exosomes, and microvesicles represent a type of biocompatible and versatile biosensing membrane surface for rapid biomarker detection and diagnosis of diseases, enhancing the assay sensitivity and decreasing the detection limit. In this review, we have reviewed the recent diagnostic application of lipid-based vesicles as biosensing substances in both conventional and novel techniques for identifying targets, especially in medicine and biotechnology sciences. Eventually, we have highlighted several recent promising developments in a new generation of biosensors based on liposome–nanomaterial hybrids and exosomes for analyzing targets and possible further advances in the future.
Lipid-based vesicles are biophospholipid bilayer vesicles such as liposomes and exosomes, which can be used in diagnosis and therapy. The term “vesicles” is employed particularly for liposomes, which are structures formed spontaneously by polar lipid particles or amphiphilic particles, each having a polar head bunch and a long hydrophobic tail. The interesting focal points of these lipid vesicles are to extend the effectiveness and helpfulness of drugs, increase the soundness of drugs through typifying, decrease the harmfulness of the drugs typified, make a difference to diminish the harmfulness of touchy tissues, and have the capacity to pair with committed ligands to realize focused treatments; liposomes are, moreover, non-toxic, adaptable, biocompatible, biodegradable, and non-immunogenic for systemic or non-systemic organizations (Wagner and Vorauer-Uhl, 2011). These interesting properties have driven liposome applications in different areas of nanobiotechnology, chemical and biochemical analytics, nourishment and horticulture businesses, makeup, and quality or medicinal conveyance. In addition to clinical applications, liposomes can also be utilized for explanatory purposes (Bozzuto and Molinari, 2015). The ease of functionalization of the inalienable utilitarian groups of lipids and their capacity to typify receptive atoms in their hydrophilic compartment make it possible to create a detecting substrate for effective bioaffinity detection. So the foremost critical necessity for biosensing is the arrangement of steady vesicles with a tall embodiment capacity and with dynamic destinations for atomic intuition (Antimisiaris et al., 2018). Liposomes, combined with successful advances such as fluorescence, calorimetry, and optical spectroscopy, can display expansive flag speakers and accomplish ultrasensitive tests. Biosensors are one of the achievements of the 1960s that have been developed with the advancement of life sciences, chemistry, and engineering over the past few years. Lipid-based vesicle biosensors are analytical tools that can exploit biological intelligence to detect compounds and interact with them. The product of this reaction can be a chemical, optical, or electrical output. On the other hand, their widespread use in diagnostic processes has led to the development of a new generation of these vesicles along with other nanomaterials such as quantum dots, nanotubes, nanodiamonds, and graphene (Nsairat et al., 2022; Pattni et al., 2015). In this article, we will specifically discuss the application of lipid-based vesicles, such as liposomes, exosomes, and microvesicles, in a variety of biosensors used in medicine (Figure 1).
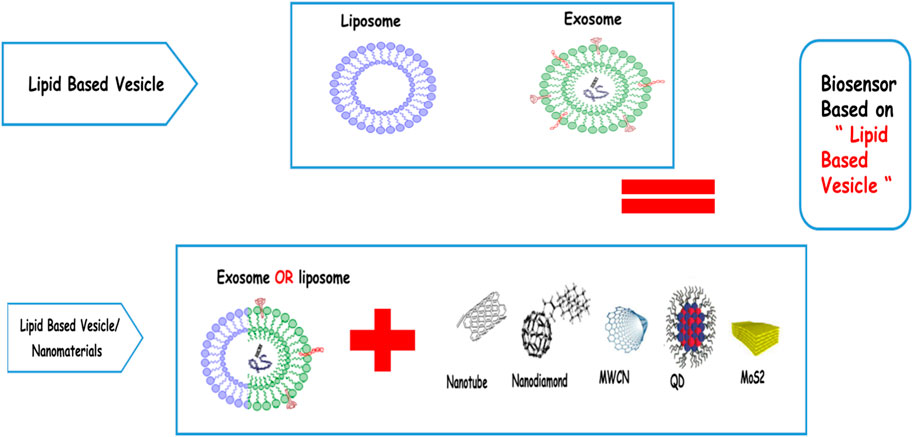
FIGURE 1. Schematic design about using different types of lipid-based vesicles with different types of nanomaterials.
In concept, optical biosensors serve as an elective alternative to customary explanatory measures; they are nano-biomolecular devices containing a bio-recognition-detecting component that coordinates with an optical transducer system to deliver a signal relative to the concentration of a measured analyte. Optical biosensing methodologies are basically preferred because of their high sensitivity, robustness, and potential to be coordinated on a single chip without complexity in their pretreatment and probable impact on target atoms (Andra et al., 2022). For a long time, many analysts have connected the optical or fiber-optical tests within the inside fluid environment of liposomes or in screens of conjugates with the liposome layers to make strides in the concentration and sensitivity of the biosensors (Filipczak et al., 2020). In an interesting study, Zhang et al. illustrated a liposome-based chemosensory by embedding the fluorescent sensors (calcein, spiropyran, and monochlorobimane) into the layer of the liposomes for the specific detection of extracellular analytes such as Zn2+, Ca2+, and GSH for biomedical applications, especially in the IVF setting (Kirby et al., 1980). In order to screen particle channel capacities, Szekacs et al. proposed a novel sensor stage for optical waveguide lightmode spectroscopy (OWLS) assays by depositing liposomes containing the particle channels onto the hydrophilic polytetrafluoroethylene (PTFE) layer. The pharmacological performance of the sensor was assessed by observing the movement of Na+ and Cl- particles through gramicidin and GABAA receptor channels individually (Patil and Jadhav, 2014). Huang et al. proposed a test chemotherapy agent delivery stage based on the fiber-optic activated topical release of photo-thermally responsive liposomes embedded with Au nanoparticles, employing a 200-µm optical fiber and a 65-mW continuous wave diode laser. The measured pattern of the activated fluorescein discharge from the liposomes illustrated that the AuNP-embedded liposomes appeared to have a more effective activated release than conventional liposomes without AuNPs (Meure et al., 2008). Clear et al. planned a liposome-based nanosensor containing lipid-conjugated optical tests for observing pH in biomedical tests (Figure 2A). In order to fine-tune the sensor pKa, they applied two lipid-anchored pH-sensitive optical tests conjugated within the liposome layer, each with enhanced deep-red cyanine dyes containing indoline nitrogen atoms. They also accomplished the next sensing dynamic extension by changing the liposome layer composition from zwitterionic POPC to anionic POPC: POPS (1:1), which raised the pKa value of the two optical probes by 0.6 and 0.9, respectively (Chatin et al., 2015). By taking advantage of PDA properties, Park et al. created coordinate PDA liposome-based multiplex biosensor arrays employing a novel immobilization method for the simultaneous and quantitative detection of six species of pathogens. They chose ethylenediamine as an ideal interlinker between individual PDA liposomes, with a final concentration of 1 mM. Their novel technique improved the signal intensity and sensitivity of PDA chips for bioanalysis (Xia et al., 2019). Liposomes have been connected to SPR biosensors for the characterization of cell membrane-related natural components and drug-induced layer impacts. The interaction between a few drugs and the immobilized liposomes has been checked in real time using the liposomes directly attached to the SPR biosensor surface (Silindir et al., 2012; Beltrán-Gracia et al., 2019). The ProteOn GLC Lipid Kit and the Biacore L1 sensor chip are two examples of sensor chips on the market that use covalently connecting alkyl groups to the chip surface to capture liposomes through lipophilic intelligence (Larsson et al., 2020). The ProteOn liposome capturing kit, including the LCP sensor chip and the LCP capturing reagent kit, advertised a novel strategy for liposome capture. The ProteOn XPR36 framework may be a label-free SPR biosensor, with the array 6 × 6 arrangement providing an ideal platform to directly analyze liposomes and small-molecule drugs intelligently. In order to show the effective performance of liposomes on localized surface plasmon resonance (LSPR) detection, Kawasaki et al. examined the biosensing capabilities using the immobilization of liposome or phospholipid single-layer structures on Au nanostructures of LSPR sensor chips. They concluded that the DMPC single layer was more effective in enhancing the sensitivity than the liposomes, probably since its measure was closer to the range of the near-field effect. Their proposed biosensor clearly recognized the 100-nM target protein of carbonic anhydrase from bovine (CAB) with an evaluated detection range of 10 nM from wavelength determination by interaction with the phospholipid single layer in Figure 2B(a, b) (Kawasaki et al., 2018). Liposome-based fluorescence biosensors have become a popular instrument for studying lipid–protein interactions by exploiting the signal amplification method (Jara-Quijada et al., 2023). Scaglione and Rintoul created a sensitive liposome-based fluorometric biosensor to assess the function of crevice intersections using the liposome containing the fluorescent probe NBD-PE. Quenching of the probe with cobalt chloride permitted the measurement of the permeability of the liposomes in the presence and absence of major intrinsic protein 26 (MIP26) (Imamura et al., 2017). Imamura and colleagues created a label-free fluorescently displayed biosensor manufactured with different phospholipid liposomes, encapsulating calcein molecules to separate different target proteins and their concentrations. They used three methodologies in their work: 1) considering a proper concentration of a fluorescent particle for encapsulation within the liposome; 2) incorporating cholesterol into the liposome membrane to improve the fluidity of the membrane and the resultant fluorescence; and 3) assessing the interaction qualities of different phospholipid liposome species with target molecules to recognize the most effective phospholipids. They also found that the protein discrimination by the exhibited sensor with a single shot was way better than that with many shots of a microwell (Weitsman et al., 2017). For identifying the intramural heterogeneity of EGFR activity, Weitsman and his colleagues designed a genetically encoded Crkll-based biosensor focused on lipopolyplexes bearing EGFR-binding peptides that experience conformational changes in phosphorylation by EGFR. They found a critical correlation between high EGFR activity in tumor cells and macrophage-tumor cell proximity, which partially accounts for the intramural heterogeneity in EGFR activity. Motamarry et al. (2019) displayed a real-time in vivo fluorescence imaging strategy for observing and evaluating tumor drug uptake during warming by exploiting the inherent fluorescence of Dox and in vivo microscopy probes. Their result illustrated that time lapse in vivo fluorescence imaging can screen medication uptake in real time during hyperthermia-interceded medication delivery, and in vivo fluorescence intensity can predict tumor Dox accumulation (Wang et al., 2020). Nanosensor detection systems have provided an attractive alternative to focused imaging of cancer cells. Wang et al. have also developed a fluorescence “turn-on” nanosensor for the location of MUC1 in living cells by designing a polydiacetylene (PDA) liposome-based sensing framework functionalized with a Cy3-labeled MUC1 binding aptamer with a limit of discovery around 0.8 nM. In the presence of MUC1, the highly specific interaction with the Cy3-aptamer leads to the dye dissociation from the liposome interface, recovering the red fluorescence (Petaccia et al., 2017).
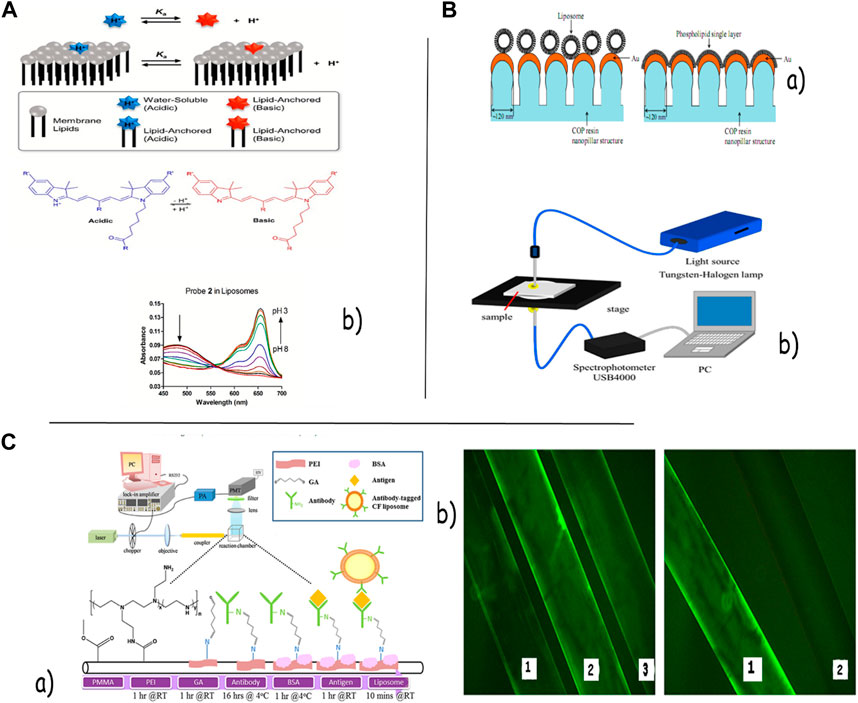
FIGURE 2. (A) Structure of acidic and basic forms of pH-responsive dyes. Representative absorbance spectra for probes 1 and 2 in liposomes composed of 10:90 POPS: POPC. (b) LSPR measurement system (B).(a) Schematic diagram of chemical modification to the optical fibers and the optical setup of our TIRF-FOB system. (b) Image taken using a fluorescence microscope representing PEI-modified optical fibers coated with the FITC-label antibody via GA (C). (a) Schematic diagram of chemical modification of this TIRF-FOB system. (b) Image taken using a fluorescence microscope, representing PEI-modified and non-PEI-modified optical fibers coated with the FITC-labeled antibody. No observable amount of the FITC-labeled antibody was coated at fiber (b). A liposome-based fluorescence nanosensor detection framework has been proposed as an attractive alternative to the time-consuming, refined strategies for the design of more effective and rapid detectors for pathogens. Petaccia et al. proposed a liposome-based approach for the discovery of bacteria in potable water by misusing cationic liposomes functionalized with a delicate surface potential fluorophore, 4-heptadecylumbelliferone (C17-HC). A fluorescent signal was created upon an interaction between the liposome-based nonsense and three bacterial strains that included an alteration in the surface potential experienced by C17-HC. Furthermore, Gao et al. (2017) set up a facile and efficient turn-on detection procedure for LPS by misusing its basic design and primitive work with a restraint of 0.65 nM. In this system, the insertion of LPS into the phospholipid bilayers of the liposomes seems to anticipate the fluorescent quenching of the fluorophore-labeled liposomes caused by the binding of amphiphilic Au nanoparticles. Vamvakaki et al. (2005) designed an optical liposome-based biosensor to detect the activity of the unstable protein acetylcholinesterase. This biosensor consisted of porins embedded into the lipid bilayer of the liposome as an analyte transporter and pyranine as a pH-sensitive fluorescence test for observing protein action inside the liposome. The performance of the liposome biosensor was quick and reproducible, and the framework was also stable enough to immobilize into a sol–gel network. Chang et al. (2016) planned an ultrasensitive liposome-based biosensor for protein location with a limit of detection of 2.0 attomoles by combining the total inside reflection fluorescence (TIRF) and fiber-optic (Fob) procedures, which coordinated a liposomal fluorescent amplifier and a sandwich immunoassay arranged with TIRF-FOB. Figure 2 shows the schematic chart of the chemical modification of this TIRF-FOB framework, where fluorescence is excited through evanescent waves and amplified through antibody-tagged, fluorescent molecule-entrapped liposomes [Figure 2C(a, b) (Kim and Lee, 2019)]. Chen et al. proposed a Lipo@HRP&ABTS optical nanoprobe by loading HRP and its substrate, ABTS, into liposomal nanoparticles for the detection of inflammation forms using an H2O2-responsive chromogenic assay (Wang et al., 2016). In order to overcome the limitations to intensity-based location frameworks and the cost of externally incorporating fluorescent colors into integrated detection frameworks. SEO and colleagues planned a convenient detection instrument for the ratiometric fluorescent detection of the anthrax biomarker by creating a red-fluorescent blended polydiacetylene acid (PDA)-determined liposome with a built-in reference dye, in which Eu-EDTA complexes are contained (Lu et al., 2012).
Several liposome-based colorimetric biosensors have been outlined to screen for very low levels of pesticides by joining a dynamic protein inside the nanoenvironment of the liposomes. A plasmonic colorimetric sensor for highly sensitive visual detection of telomerase activity in cancer cells has been created by integrating an etching substrate, Au nanobipyramids, with a liposome-based signal enhancement procedure. In reaction to high expression of telomerase, HRP was discharged from the liposomes, following the expansion of the TMB substrate, and then, the formation of TMB2+ led to the morphological advancement of Au NBPs, which gave rise to tremendous LSPR responses with location limits comparable to one HeLa cell for localized surface plasmon resonance spectrometer estimation and a visual detection limit of 20 cells (Gao et al., 2017). Jung et al. created a novel detecting procedure for nucleic acids increased by the common PCR response using a polydiacetylene (PDA) liposome-based colorimetric sensor. Upon the addition of nucleic acids with a concentration of 100 nM and, in this way, ionic interactions between PDA liposomes and nucleic acids, a dramatic color alteration from blue to red was observed by the exposed eye (Chen et al., 2017). In another study, they coordinated bidentate aptamer sets with the PDA liposome sensor to attain coordinated colorimetric detection of target proteins at sub-micromolar concentrations with high specificity. They proposed that the liposome aggregation activated by particular, multi-site binding to the target protein was dependable for the improved colorimetric reaction (Seo et al., 2017). Jannah and Kim outlined a pH-responsive carboxyl-terminated PDA liposome sensor for colorimetric detection of urease activity in pathogenic microbes and cancer based on the blue–red transition of the PDA with a discovery limit of 70 pU/mL (Wang et al., 2019). A liposome-based colorimetric biosensor has also been created for convenient visual detection of pathogenic bacteria. For illustration, Wu et al. assessed an aptamer-based biosensor for particular colorimetric detection of E. coli O157:H7 by conjugating the truncated lipopolysaccharide authoritative aptamer on the surface of the PDA vesicle using peptide holding between the carboxyl group of the vesicle and the amine group of the aptamer. Then, 104–108 CFU/mL of E. coli O157:H7 cells was readily discernable by the aptasensor within 2 h in clinical fecal examples (Jung et al., 2010).
The liposomes have been integrated into two forms of electrochemical biosensor stages: strip-organized immunoassays and microfluidic chips. Within the final decade, many analysts created an arrangement of strip-organized based immunoassay based lateral stream guideline (Jannah and Kim, 2019). In a modern technique, a novel electrochemical immunosensor based on liposome signal enhancement and immunochromatographic methods for the detection of theophylline was displayed. It was composed of a disposable screen-printed electrode and a nitrocellulose strip containing a signal-era zone with immobilized ferrocyanide-loaded liposomes. The theophylline concentration is amperometrically detected by the screen-printed cathode with a location constraint of 5 μg/mL upon liposome disruption and release of the electroactive ferrocyanides (Qu et al., 2010). Ho et al. described a competitive-type immunoanalytical stage technology based on the competition between the analyte biotin and biotin-tagged potassium ferrocyanide-encapsulated liposomes for binding to a few anti-biotin antibody-binding sites. This biosensor system uses PAH-change, nano-sized Au particles assembled on a screen-printed cathode (nanoAu-SPE) for electrochemical detection (Siebert et al., 1993). An interesting electrochemical biosensor has been used by Seo et al. for neomycin discovery with a high sensitivity down to 2.3 nM, in which the fluorescent emission of rhodamine 6G (R6G) dyes was efficiently quenched on the surfaces of phopspholipid liposome by the formation of H-aggregates. Particular interactions between neomycin and phospholipids lead to the displacement of R6G colors in favor of the neomycin molecules, inducing the sensory signal (Lee et al., 1997). DOTAP-AuNP, DOPE-AuNP, and double DOTAP-DOPE-AuNP are three common liposome-based electrochemical biosensors that have been used for ultrasensitive DNA or protein detection, each of which expresses unique characteristics. Narayanan et al. reported, for the first time, a spherical liposome-AuNP nanocomposite by sandwiching round DOTAP in a gold nanoparticle bed and directly immobilizing on the gold transducer surface to separate between three complementary, non-complementary, and single mismatch targets within the presence of [Fe (CN) 6] −3 and double-stranded particular interlocutor methylene blue. Then, it is followed by the potential use of the sDOTAP-AuNP-DNA composite for the electrochemical DNA transfection of E. coli or other cells (Ho et al., 2009). The effect of the brief and long-chain thiol monolayers on the tethering of binary liposome mixtures of DOTAP and DOPE in the absence and presence of gold nanoparticles to upgrade the sensor stability and sensitivity for label-free DNA and protein sensing was assayed by Divya and Dharuman. In this organization, higher detection happens because of the increased number of captured probes within the presence of [Fe (CN) 6] −3 on the DOPE-DOTAP-AuNP on the MPA layer compared to MUDA, AUT, and cysteamine hydrochloride. Results showed a large increase in the charge transfer resistance and hybridization discrimination impact with a wide linearity from 1 × 10-14 to 1 × 10-9 with the least detection limit of 1 × 10–14 M for DNA sensing and a 1 ng/mL detection limit with a linear range from 100 ng to 1 µg for protein sensing (Seo et al., 2015).
Bhuvana et al. outlined a liposome-based nanocomposite by covalently immobilizing a 1,2-dioleoyl-sn-glycerol-3-phosphoethanolamine (DOPE) liposome on the MPA layer on the gold surface for electrochemical sensing of name-free DNA. The single-stranded DNA (ssDNA) probes were immobilized on the DOPE-AuNP nano-composite through a basic gold–thiol linkage to separate between the hybridized and single base mismatch targets in the presence of [Fe (CN) 6] 3-/4- and [Ru (NH3) 6] 3+ redox probes without signal amplification with a detection limit of 0.1 × 10–12 M. Their results collectively affirmed the non-fused round nature of the DOPE on the MPA layer and the high steadiness of the DOPE-AuNP nanocomposite on the gold surface (Figure 3A) (Narayanan et al., 2014). Yazdi et al. outlined a DOPC-MoS2/AuNP crossover as a natural bed for the discovery of microRNAs. This is the primary report of a combination of a cationic DOPC liposome (dioleoylphosphatidylcholine) with MoS2 to enhance the sensitivity of liposomal sensors. Since AuNP can attach to MoS2 and DOPC, there are more places to connect and recognize miRNAs. So they moved the sensitivity of the sensor forward (LOD = 10 a.m.) (Divya and Dharuman, 2017). In another report by Yazdi et al., diverse amperometric electrochemical designs of liposomes for the revelation of intense lymphoblastic leukemia cells were examined. They used lectin and boronic acid-conjugated liposomes for the disclosure of Molt-4 cells, focusing on the characteristics of glycosylation (Figure 3C) (Divya and Dharuman, 2017). Yu et al. created an immobilized liposome electrode (ILE)-based sensor integrating the pattern recognition (PARC) technique to quantify conformational changes of the proteins by measuring the current deviations amperometrically created based on the status of the proteins under different stress conditions. A self-assembled monolayer (SAM) manufactures the ILE using 16-mercaptohexadecanoic acid formed on a gold electrode. The liposomes are then immobilized on the SAM layer onto the gold anode surface by amino coupling to create the electric signal because of the electrolyte release from the liposome membrane, according to protein structures such as naïve, liquid globule, and denature states (Gao et al., 2017). Damhorst et al. displayed an ELISA-like lab-on-a-chip platform for electrical biosensing of surface-immobilized antigens based on functionalized liposome ion-releasing impedance spectroscopy to overcome the limitations to expensive components and inadequate portability. They outlined the microfluidic cathode impedance detecting device, which comprises a gold contact pad with micromanipulator tests, a fluidic chamber secured with PDMS, and a fluidic outlet and channel ports. The evaluation of the anti-gp120 IgG functionalized liposomes by genuine time impedance observing of particle discharge upon heating illustrated that the lower limit of detection is around 1,000 vesicles/µL. They also used the device for qualitative sensing of HIV because of the need for optimizing the device geometry, antibody immobilization strategies, and conventions for virus capture (Yazdi et al., 2020). In recent years, the substitution of biological receptors with entirely synthetic molecularly imprinted polymers (MIPs) in point-of-care diagnostic devices has advertised many benefits related to the excellent chemical and thermal stability of MIPs coupled with their reproducible and cost-effective manufacture. Johari-Ahar et al. represented a novel MIP-based electrochemical sensor using a liposomal amplification strategy by altering a gold screen-printed electrode (Au-SPE)-based biosensor with molecularly imprinted polymers as the capture element. Subsequently, they amplified the signal using antibody-conjugated nano-liposomes for concurrent detection of EGFR and VEGF as cancer biomarkers in real tests. In this sensing device, the nano-liposomes, which were loaded with Cd (II) and Cu (II) electroactive agents and enhanced with particular antibodies, were used to create electrochemical signals for the detection of EGFR with a discovery limit of 0.01 pg/mL and VEGF with a detection limit of 0.005 pg/mL. They highlighted that the SPE-based biosensing device has the capability of being coordinated with lab-on-chip, microfluidics, or micro total analysis systems (Figure 3B) (Ghazizadeh and Neshastehriz, 2020). Recently, research into the direct electron transfer of redox enzymes has also attracted significant attention for creating third-generation enzymatic electrochemical biosensors. Polymeric liposomes (polysomes), as a type of biocompatible and conductive material, have been connected to self-assembled redox enzymes to overcome the unfavorable enzyme orientation at routine terminals and realize coordinated electrochemistry (Yu et al., 2018). Garcia et al. established a novel ionic liquid-based polymerized liposome by polymerizing self-assembled ionic liquid-altered lipids by exploiting the improved physical stability of liposomes as a supporting matrix for transforming the existing state of ionic liquid from fluid to solidified nanomaterial (Damhorst et al., 2013). Tain et al. detailed the primary study of such multifunctionalized liposomes in the fields of biosensors and bioelectronics. They related the ionic liquid polysomes (IL-polysomes) with a redox enzyme, horseradish peroxidase, to construct a coordinate-changed HRP/IL-polysomes/polyvinyl alcohol/glassy carbon electrode to assess the enzymatic activity. Their results showed that the IL-polysomes, compared to unpolymerized ionic liquid-based liposomes (IL-liposomes), showed significant stability and biocompatibility while protecting their unique morphology and bilayer membrane structure on the GC electrode because of the cross-linking of polymerized lipids. The coordinate IL-polysomes with HRP/PVA/GC electrodes showed significant electrocatalytic execution toward H2O2 reduction with a wide straight range, good reproducibility, and long-term stability (Johari-Ahar et al., 2018). Despite progress in the advancement of liposome-nanomaterial crossbreeds and exosomes in recent years, there are still uncertain issues, such as low stability and leakage of signaling markers, because of a combination of liposomes and less effective strategies for disrupting liposomes in bioassays.
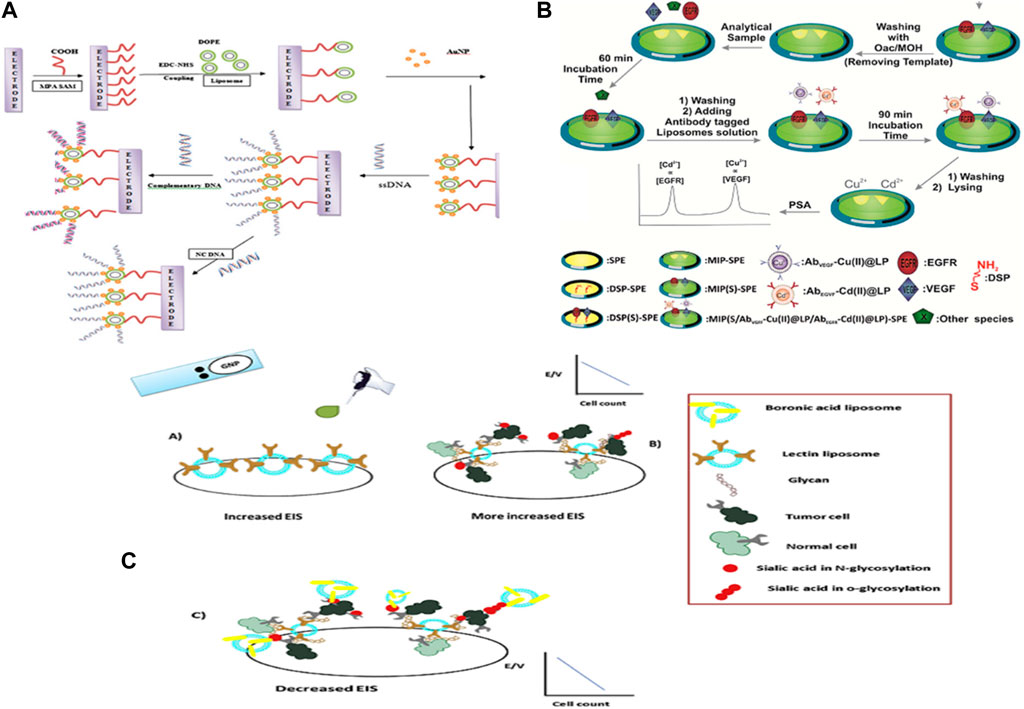
FIGURE 3. (A) Formation of a spherical DOPE–AuNP nano-composite and DNA detection. (B) Dropping the Molt-4 cells also caused an increase in EIS. High sensitivity of Molt-4 cells was observed with lectin liposome. (C) Dropping the boronic acid liposome caused a decrease in EIS. In this stage, higher detection occurred due to the target screening of sialic acid in Molt-4 cells.
In order to improve the diagnostic efficiency of liposomes, studies have been conducted on the use of other nanomaterials along with liposomes to form hybrids or supermolecules. A new generation of natural lipid vesicles called exosomes has been developed to take a new step.
Two-dimensional and three-dimensional nanomaterials have a wide capacity to hybridize with liposomes. Imran et al. used self-assembled DOTAP liposomes on graphene oxide/reduced graphene oxide (GO/rGO) composites supported on a gold electrode for the ultra-sensitive and selective label-free detection of target DNA in the presence of gold nanoparticles, with the lowest detection limit of 1.29 × 10−17 M and a broad linear range of 10–17-10–8 M. The liposomes can be introduced as tethered, intact vesicles for improved drug and DNA delivery applications in therapeutics and diagnosis (Figure 4A). The bilayered liposome/AuNP structures on AuE, GO, and rGO do not appear to have any selectivity for DNA detection in graphene materials on a gold surface by physisorption without utilizing thiol self-assembled monolayers. They used this vesicle structure for label-free sensing of DNA in the energetic range from 10 to 15 to 10–9 nM [Figure 4B(a, b)] (Zhang and Sun, 2021). Combining liposomes with semiconductor quantum dots (QDs) shows incredible potential for diagnosis systems. Ye et al. also labeled MCF7 cells with CdTe or CdHgTe QDs typified in liposomes. L-QDs were efficiently internalized by MCF7 cells in a time-dependent way without cytotoxic impacts. In in vivo imaging, strong fluorescence in the imaging process and a solid fluorescence signal throughout the entire body were observed instantly after L-QD injection (Andres Aranguren et al., 2020). Voura et al. infused the tail veins of C57BL/6 mice with L-QD-labeled B16F10 cells. This research illustrated that L-QD has no recognizable toxicity to the labeled cells or the host animal. It appeared that metastatic tumor cell extravasation could be tracked using an L-QD signal (Mahmoudi-Badiki et al., 2016). In Fahmida’s research, quantum dots (QDs) embedded within the lipid bilayer of liposomes and methylene blue (MB) encapsulated in the internal center of liposomes played a part in the dual-signaling modulator of CHIK-VP within the femtogram to nanogram per milliliter level with a LOD of 32 fg mL−1. After CHIK-VP addition, the nanocomposites and APTES-coated Fe3O4 nanoparticles (Fe3O4 NPs) were conjugated with antibodies to make a sandwich structure and isolated from the medium attractively. The chloroforms of nanoconjugates, QDs, and MB are discharged from the liposomes and give fluorescence and electrochemical signals, respectively (Imran et al., 2019). In a later investigation, Douda et al. used amine-functionalized nanodiamonds (NDs) typified in liposomes to increase the optical response in theranostic liposomes and all-optical sensors. In the advancement of theranostic liposomes and all-optical sensors, the amine-functionalized NDs were successfully typified in lecithin liposomes arranged by the green and conventional strategies (Ye et al., 2013). In Yazdi et al.’s investigation, it appeared that the DOPC-MoS2/AuNP crossover can act as an amplifier bed to identify miR-21 with high sensitivity. It appeared that MoS2 improved the sensitivity of liposomal sensors clearly in point-of-care tests (Ghazizadeh and Neshastehriz, 2020).
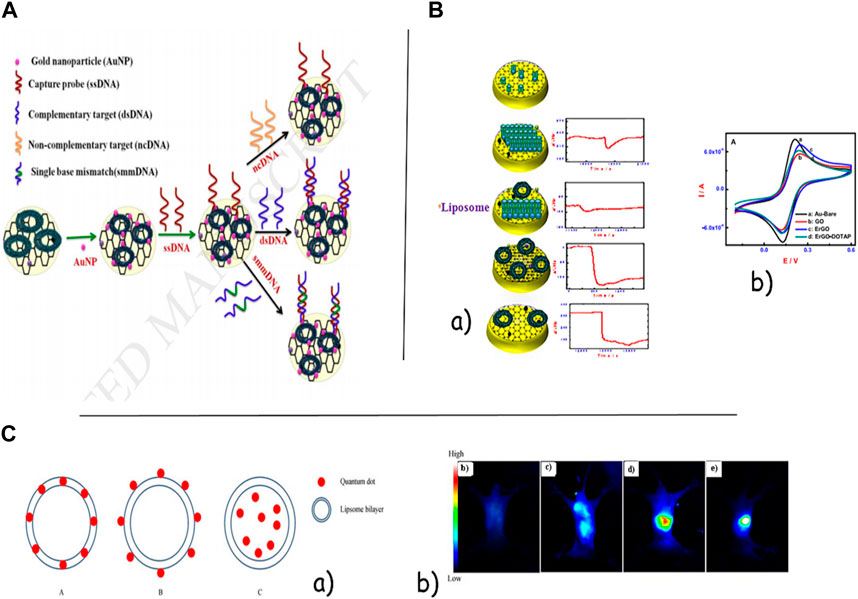
FIGURE 4. (A). Schematic image of graphene oxide-supported liposomes for liposomal sensors clearly in point-of-care test label-free electrochemical DNA biosensing. (B). Ultra-sensitive and selective label-free electrochemical DNA detection at layer-by-layer self-assembled graphene oxide and vesicle liposome nanoarchitecture. (C).(a) Quantum dot incorporation models of hybrid delivery systems. A, hydrophobic quantum dot-encapsulated shell model; B, hydrophilic quantum dot-linked shell model; C, hydrophilic quantum dot-encapsulated core model. (b) Temporal internalization of L-QD by confocal microscopy and representative NIR images. These images represented the denuded mouse after lipid-CdHgTe was injected via the tail vein for 10 seconds, and 5, 10, and 20 min.
Exosomes are liposome-like characteristic vesicles with many parts in typical physiological work (Voura et al., 2004). It is challenging to picture a few tumor tissues because of the lack of cancer cells. It can use exosomes as a sensitive marker for the detection of small numbers of cancer cells. Their application in cancerous imaging, particularly for lung cancers, was considered in many later investigations (Nasrin et al., 2020). Zong et al. and Jiang et al. showed exceptional determination images of live cells and the metastatic action of breast tumor cells by stacking silicon quantum bits (30 nm) and gold–carbon quantum specks onto the outside layer of the exosomes (50–100 nm) separately (Douda et al., 2020; Ghazizadeh and Neshastehriz, 2020). Srivastava et al. used press oxide nanoparticles (5–10 nm) and doxorubicin in exosomes determined from lung fibroblast cells in a dual action that could capture a high-resolution tumor of lung fibroblast cells (Andaloussi et al., 2013). By combining the benefits of echogenic conventional micro-bubbles with nano-sized exosomes, Osborn et al. showed the echogenic exosomes as an unused ultrasound differentiation agent. To begin with every report, the exosomes with a concentration of 40 µg mL-1 exhibited vital scattered responses and resulted in basically higher brightness without harmful quality or immunogenic response that can be used as a viable ultrasound-responsive medication delivery system [Figure 5A(b)] (Beach et al., 2014). Shi et al., by abusing exosomes radiolabeled with fluorescence and PEG in mice, captured the images with higher resolution from 4T1 breast cancer cells at the side, providing a visual uptake (Soung et al., 2017). Ghazizadeh et al. showed that exosomes can act as amplifiers in electrochemical sensors. They affirmed this approach with miRNAs in the MCF-7 exosome-p19 composite. They appeared to be exosomes derived from different stages of breast cancer with diverse electrochemical behaviors because of diverse biomarker expressions. In Ghazizadeh et al., the electrochemical behavior of exosomes as a natural bed was confirmed. They used exosomes and p19 proteins for the detection of 21 miRNAs, serving as a common demonstration. Eventually, it was observed that the MCF-7 exosome-p19 composite could identify the 21 miRNAs with high sensitivity (LOD = 1 a.m.) (Figure 5B) (Jiang et al., 2018).
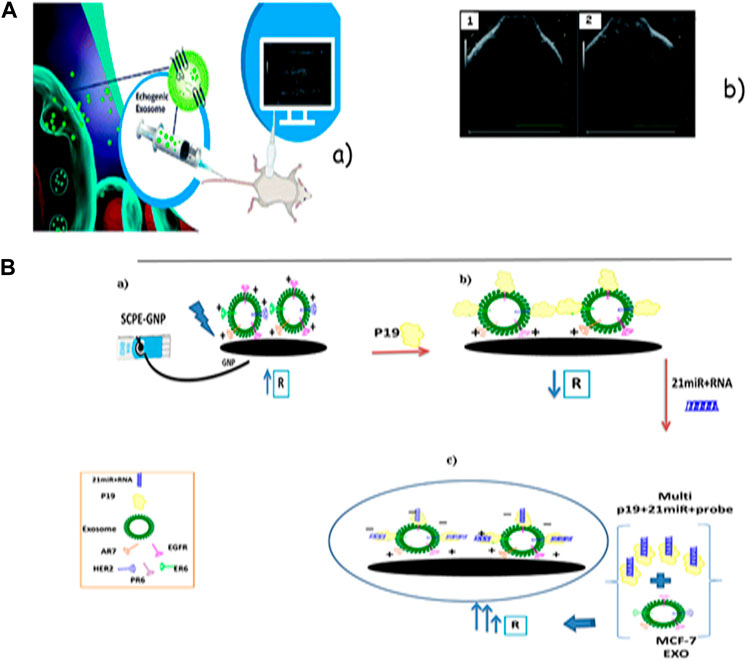
FIGURE 5. (A) Schematic image of echogenic exosomes as ultrasound contrast agents. (B) Ultrasound images before and after the injection of echogenic exosomes into the synovial space of a Sprague–Dawley rat. Images were taken with a Vevo 3,100 Imaging System and a 40 MHz transducer. (B) Schematic schema for the stages of the exosomal electrochemical sensor model for the detection of 21 miRNAs: (a) immobilization of the MCF-10A exosome with different biomarkers on the SCPE-GNP; (b) adding p19 to the MCF-10A exosome/SCPE-GNP; (c) adding a hybrid of the 21miR-probe to the p19/MCF-10A exosome/SCPE-GNP.
In another curiously novel study, Ghazizizedeh et al. suggested a double stage to identify any spiked infections and specific SARS-CoV-2 antigens consecutively. They presented a common bed-receptor surface as microparticle vesicle-galactins1 (MV-gal1) with the capacity of glycan binding to screen spiked viruses. The sensor has the capacity to screen for any spiked viruses and detect COVID-19 (LOD: 4.57 102 copies/mL) by utilizing the natural bed receptor and a specific antibody in the point-of-care test (Jiang et al., 2018).
In order to diagnose, treat, and examine diseases, it is necessary to have an image with high quality and resolution (Srivastava et al., 2016; Zong et al., 2018; Gao et al., 2017; Ghazizadeh et al., 2021). Today, lipid-based vesicles (such as liposomes, exosomes, and microvesicles) are one of the most important biocompatible nanomaterials, which show high power in imaging because of their high power in diagnosis and imaging, especially in living cells. However, there is an abundance of created concepts, and there are many curious challenges ahead before the complete potential of lipid-based vesicle sensors can be realized into a biochip that is appropriate for the detection of analyses in complex physiological media (human serum or human whole blood). These incorporations progress and are also about the stability of lipid-based vesicles, minimizing the leakage of typified markers and avoiding liposomes against fusion for the best ability of sensing. To date, changing with another material (sugar, protein, polymer, nanomaterials, etc.) has been used to protect liposome keenness and to ensure films from solute leakage amid drying. Using liposomal hybridization with other nanomaterials in order to increase diagnostic efficiency and develop optimal synthetic methods for liposome encapsulation will provide an important step in the application of broader studies in this field. The introduction of multiplex innovation complements a continuous move toward personalized medicine. It is exceedingly likely that multiplex assays will optimize their popularity, and overcoming the challenges of the aforementioned viewpoints will enable the development of liposome- and lipid bilayer-based biosensors for fast, precise, and convenient point-of-care clinical conclusions.
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Andaloussi, S. E., Mäger, I., Breakefield, X. O., and Wood, M. J. (2013). Extracellular vesicles: Biology and emerging therapeutic opportunities. Nat. Rev. Drug Discov. 12 (5), 347–357. doi:10.1038/nrd3978
Andra, V. V., Pammi, S. V. N., Bhatraju, L. V. K. P., and Ruddaraju, L. K. (2022). A comprehensive review on novel liposomal methodologies, commercial formulations, clinical trials and patents. BioNanoScience 12, 274–291. doi:10.1007/s12668-022-00941-x
Andres Aranguren, B., Torres, E., Johann, F., Osma, J. F., and Cruz, J. C. (2020). Synthesis of nanoscale liposomes via low-cost microfluidic systems. Micromachines 11 (12), 1050. doi:10.3390/mi11121050
Antimisiaris, S. G., Mourtas, S., and Marazioti, A. (2018). Exosomes and exosome-inspired vesicles for targeted drug delivery. Pharmaceutics 10 (4), 218. doi:10.3390/pharmaceutics10040218
Beach, A., Zhang, H. G., Ratajczak, M. Z., and Kakar, S. S. (2014). Exosomes: An overview of biogenesis, composition and role in ovarian cancer. J. ovarian Res. 7 (1), 14–11. doi:10.1186/1757-2215-7-14
Beltrán-Gracia, E., López-Camacho, A., Higuera-Ciapara, I., Velázquez-Fernández, J. B., and Vallejo-Cardona, A. A. (2019). Nanomedicine review: Clinical developments in liposomal applications. Cancer Nanotechnol. 10 (1), 11–40. doi:10.1186/s12645-019-0055-y
Bozzuto, G., and Molinari, A. (2015). Liposomes as nanomedical devices. Int. J. nanomedicine 10, 975–999. doi:10.2147/ijn.s68861
Chang, Y. F., Fu, C., Chen, Y. T., Jou, A. F. J., Chen, C. C., Chou, C., et al. (2016). Use of liposomal amplifiers in total internal reflection fluorescence fiber-optic biosensors for protein detection. Biosens. Bioelectron. 77, 1201–1207. doi:10.1016/j.bios.2015.10.012
Chatin, B., Mével, M., Devallière, J., Dallet, L., Haudebourg, T., Peuziat, P., et al. (2015). Liposome-based formulation for intracellular delivery of functional proteins. Mol. Therapy-Nucleic Acids 4, e244. doi:10.1038/mtna.2015.17
Chen, Q., Liang, C., Sun, X., Chen, J., Yang, Z., Zhao, H., et al. (2017). H2O2-responsive liposomal nanoprobe for photoacoustic inflammation imaging and tumor theranostics via in vivo chromogenic assay. Proc. Natl. Acad. Sci. 114 (21), 5343–5348. doi:10.1073/pnas.1701976114
Damhorst, G. L., Smith, C. E., Salm, E. M., Sobieraj, M. M., Ni, H., Kong, H., et al. (2013). A liposome-based ion release impedance sensor for biological detection. Biomed. microdevices 15 (5), 895–905. doi:10.1007/s10544-013-9778-4
Divya, K. P., and Dharuman, V. (2017). Supported binary liposome vesicle-gold nanoparticle for enhanced label free DNA and protein sensing. Biosens. Bioelectron. 95, 168–173. doi:10.1016/j.bios.2017.04.022
Douda, J., González-Vargas, C., Mota-Díaz, I., Basiuk, E., Hernández-Contreras, X., Fuentes-García, J., et al. (2020). Photoluminescent properties of liposome-encapsulated amine-functionalized nanodiamonds. Nano Express 1 (3), 030009. doi:10.1088/2632-959x/abc1c5
Filipczak, N., Pan, J., Torchilin, L. P., and Torchilin, V. P. (2020). Recent advancements in liposome technology. Adv. Drug Deliv. Rev. 156, 4–22. doi:10.1016/j.addr.2020.06.022
Gao, J., Li, Z., Zhang, O., Wu, C., and Zhao, Y. (2017). Tunable accessibility of dye-doped liposomes towards gold nanoparticles for fluorescence sensing of lipopolysaccharide. Analyst 142 (7), 1084–1090. doi:10.1039/c7an00019g
Ghazizadeh, E., and Neshastehriz, A. (2020). Different liposome patterns to detection of acute leukemia based on electrochemical cell sensor. Anal. Chim. acta 1109, 122–129. doi:10.1016/j.aca.2020.02.060
Ghazizadeh, E., Ali, N., Kaji Yazdi, M., Saievar-Iranizad, E., and Einali, S. (2021). Dual electrochemical sensing of spiked virus and SARS-CoV-2 using natural bed-receptor (MV-gal1). Sci. Rep. 11, 22969. doi:10.1038/s41598-021-02029-0
Ho, J. A., Chiu, J. K., Hong, J. C., Lin, C. C., Hwang, K. C., and Hwu, J. R. R. (2009). Gold-nanostructured immunosensor for the electrochemical sensing of biotin based on liposomal competitive assay. J. Nanosci. Nanotechnol. 9 (4), 2324–2329. doi:10.1166/jnn.2009.se40
Imamura, R., Murata, N., Shimanouchi, T., Yamashita, K., Fukuzawa, M., and Noda, M. (2017). A label-free fluorescent array sensor utilizing liposome encapsulating calcein for discriminating target proteins by principal component analysis. Sensors 17 (7), 1630. doi:10.3390/s17071630
Imran, H., Manikandan, P. N., and Dharuman, V. (2019). Ultra-sensitive and selective label free electrochemical DNA detection at layer-by-layer self-assembled graphene oxide and vesicle liposome nano-architecture. J. Electroanal. Chem. 835, 10–21. doi:10.1016/j.jelechem.2019.01.011
Jannah, F., and Kim, J-M. (2019). pH-sensitive colorimetric polydiacetylene vesicles for urease sensing. Dyes Pigments 169, 15–21. doi:10.1016/j.dyepig.2019.04.072
Jara-Quijada, E., Pérez-Won, M., Tabilo-Munizaga, G., Lemus-Mondaca, R., González-Cavieres, L., Palma-Acevedo, A., et al. (2023). Liposomes loaded with green tea polyphenols—optimization, characterization, and release kinetics under conventional heating and pulsed electric fields. Food Bioprocess Technol. 2023, 1–13. doi:10.1007/s11947-023-03136-8
Jiang, X., Zong, S., Chen, C., Zhang, Y., Wang, Z., and Cui, Y. (2018). Gold–carbon dots for the intracellular imaging of cancer-derived exosomes. Nanotechnology 29 (17), 175701. doi:10.1088/1361-6528/aaaf14
Johari-Ahar, M., Karami, P., Ghanei, M., Afkhami, A., and Bagheri, H. (2018). Development of a molecularly imprinted polymer tailored on disposable screen-printed electrodes for dual detection of EGFR and VEGF using nano-liposomal amplification strategy. Biosens. Bioelectron. 107, 26–33. doi:10.1016/j.bios.2018.02.005
Jung, Y. K., Kim, T. W., Park, H. G., and Soh, H. T. (2010). Specific colorimetric detection of proteins using bidentate aptamer-conjugated polydiacetylene (PDA) liposomes. Adv. Funct. Mater. 20 (18), 3092–3097. doi:10.1002/adfm.201001008
Kawasaki, M., Yoshikawa, H., Saito, M., Jiang, S., Akeyama, T., Tamiya, E., et al. (2018). A new type of LSPR sensor featuring immobilized liposome or phospholipid single layer. Proceedings 2, 791. doi:10.3390/proceedings2130791
Kim, C., and Lee, K. (2019). Polydiacetylene (PDA) liposome-based immunosensor for the detection of exosomes. Biomacromolecules 20 (9), 3392–3398. doi:10.1021/acs.biomac.9b00641
Kirby, C., Clarke, J., and Gregoriadis, G. (1980). Effect of the cholesterol content of small unilamellar liposomes on their stability in vivo and in vitro. Biochem. J. 186 (2), 591–598. doi:10.1042/bj1860591
Larsson, E., Hubert, M., and Lundmark, R. (2020). Analysis of protein and lipid interactions using liposome co-sedimentation assays Methods. Mol. Biol. 2169, 119–127. doi:10.1007/978-1-0716-0732-9_11
Lee, K. S., Lee, W. Y., and Park, J. K. (1997). “Disposable thick-film liposome immunosensor for theophylline using immunochromatographic matrix,” in Proceedings of International Solid State Sensors and Actuators Conference (Transducers' 97), Chicago, IL, USA, 19-19 June 1997 (IEEE).
Lu, K. Y., Tao, S. C., Yang, T. C., Ho, Y. H., Lee, C. H., Lin, C. C., et al. (2012). Profiling lipid–protein interactions using nonquenched fluorescent liposomal nanovesicles and proteome microarrays. Mol. Cell. Proteomics 11 (11), 1177–1190. doi:10.1074/mcp.m112.017426
Mahmoudi-Badiki, T., Alipour, E., Hamishehkar, H., and Golabi, S. M. (2016). Dopamine-loaded liposome and its application in electrochemical DNA biosensor. J. biomaterials Appl. 31 (2), 273–282. doi:10.1177/0885328216650378
Meure, L. A., Foster, N. R., and Dehghani, F. (2008). Conventional and dense gas techniques for the production of liposomes: A review. Aaps Pharmscitech 9 (3), 798–809. doi:10.1208/s12249-008-9097-x
Motamarry, A., Negussie, A. H., Rossmann, C., Small, J., Wolfe, A. M., Wood, B. J., et al. (2019). Real-time fluorescence imaging for visualization and drug uptake prediction during drug delivery by thermosensitive liposomes. Int. J. Hyperth. 36 (1), 816–825. doi:10.1080/02656736.2019.1642521
Narayanan, J. S., Bhuvana, M., and Dharuman, V. (2014). Sandwiching spherical 1, 2-dioleoyltrimethylammoniumpropane liposome in gold nanoparticle on solid transducer for electrochemical ultrasensitive DNA detection and transfection. Biosens. Bioelectron. 58, 326–332. doi:10.1016/j.bios.2014.02.033
Nasrin, F., Chowdhury, A. D., Ganganboina, A. B., Achadu, O. J., Hossain, F., Yamazaki, M., et al. (2020). Fluorescent and electrochemical dual-mode detection of Chikungunya virus E1 protein using fluorophore-embedded and redox probe-encapsulated liposomes. Microchim. Acta 187 (12), 674–711. doi:10.1007/s00604-020-04656-2
Nsairat, H., Khater, D., Sayed, U., Odeh, F., Al Bawa, A., and Alshaer, W. (2022). Liposomes: Structure, composition, types, and clinical applications. Heliyon 8 (5), e09394. doi:10.1016/j.heliyon.2022.e09394
Patil, Y. P., and Jadhav, S. (2014). Novel methods for liposome preparation. Chem. Phys. lipids 177, 8–18. doi:10.1016/j.chemphyslip.2013.10.011
Pattni, B. S., Chupin, V. V., and Torchilin, V. P. (2015). New developments in liposomal drug delivery. Chem. Rev. 115 (19), 10938–10966. doi:10.1021/acs.chemrev.5b00046
Petaccia, M., Bombelli, C., Sterbini, F. P., Papi, M., Giansanti, L., Bugli, F., et al. (2017). Liposome-based sensor for the detection of bacteria. Sensors Actuators B Chem. 248, 247–256. doi:10.1016/j.snb.2017.03.124
Qu, B., Guo, L., Chu, X., Wu, D. H., Shen, G. L., and Yu, R. Q. (2010). An electrochemical immunosensor based on enzyme-encapsulated liposomes and biocatalytic metal deposition. Anal. Chim. acta 663 (2), 147–152. doi:10.1016/j.aca.2010.01.050
Seo, H., Singha, S., and Ahn, K. H. (2017). Ratiometric fluorescence detection of anthrax biomarker with EuIII-edta functionalized mixed poly (diacetylene) liposomes. Asian J. Org. Chem. 6 (9), 1257–1263. doi:10.1002/ajoc.201700158
Seo, S., Kwon, M. S., Phillips, A. W., Seo, D., and Kim, J. (2015). Highly sensitive turn-on biosensors by regulating fluorescent dye assembly on liposome surfaces. Chem. Commun. 51 (50), 10229–10232. doi:10.1039/c5cc01621e
Siebert, S. T. A., Reeves, S. G., and Durst, R. A. (1993). Liposome immunomigration field assay device for Alachlor determination. Anal. Chim. acta 282 (2), 297–305. doi:10.1016/0003-2670(93)80214-6
Silindir, M., Erdoğan, S., Özer, A. Y., and Maia, S. (2012). Liposomes and their applications in molecular imaging. J. drug Target. 20 (5), 401–415. doi:10.3109/1061186x.2012.685477
Soung, Y. H., Ford, S., Zhang, V., and Chung, J. (2017). Exosomes in cancer diagnostics. Cancers 9 (1), 8. doi:10.3390/cancers9010008
Srivastava, A., Amreddy, N., Babu, A., Panneerselvam, J., Mehta, M., Muralidharan, R., et al. (2016). Nanosomes carrying doxorubicin exhibit potent anticancer activity against human lung cancer cells. Sci. Rep. 6 (1), 38541–38615. doi:10.1038/srep38541
Vamvakaki, V., Fournier, D., and Chaniotakis, N. A. (2005). Fluorescence detection of enzymatic activity within a liposome based nano-biosensor. Biosens. Bioelectron. 21 (2), 384–388. doi:10.1016/j.bios.2004.10.028
Voura, E. B., Jaiswal, J. K., Mattoussi, H., and Simon, S. M. (2004). Tracking metastatic tumor cell extravasation with quantum dot nanocrystals and fluorescence emission-scanning microscopy. Nat. Med. 10 (9), 993–998. doi:10.1038/nm1096
Wagner, A., and Vorauer-Uhl, K. (2011). Liposome technology for industrial purposes. J. drug Deliv. 2011, 1–9. doi:10.1155/2011/591325
Wang, D., Zhang, Y., Zhao, X., and Xu, Z. (2019). Plasmonic colorimetric biosensor for visual detection of telomerase activity based on horseradish peroxidase-encapsulated liposomes and etching of Au nanobipyramids. Sensors Actuators B Chem. 296, 126646. doi:10.1016/j.snb.2019.126646
Wang, D-E., Gao, X., You, S., Chen, M., Ren, L., Sun, W., et al. (2020). Aptamer-functionalized polydiacetylene liposomes act as a fluorescent sensor for sensitive detection of MUC1 and targeted imaging of cancer cells. Sensors Actuators B Chem. 309, 127778. doi:10.1016/j.snb.2020.127778
Wang, D. E., Zhao, L., Yuan, M. S., Chen, S. W., Li, T., and Wang, J. (2016). Fabrication of polydiacetylene liposome chemosensor with enhanced fluorescent self-amplification and its application for selective detection of cationic surfactants. ACS Appl. Mater. interfaces 8 (41), 28231–28240. doi:10.1021/acsami.6b10794
Weitsman, G., Mitchell, N. J., Evans, R., Cheung, A., Kalber, T. L., Bofinger, R., et al. (2017). Detecting intratumoral heterogeneity of EGFR activity by liposome-based in vivo transfection of a fluorescent biosensor. Oncogene 36 (25), 3618–3628. doi:10.1038/onc.2016.522
Xia, Y., Xu, C., Zhang, X., Ning, P., Wang, Z., Tian, J., et al. (2019). Liposome-based probes for molecular imaging: From basic research to the bedside. Nanoscale 11 (13), 5822–5838. doi:10.1039/c9nr00207c
Yazdi, M. K., Ghazizadeh, E., Noroozi, M., and Neshastehriz, A. (2020). Design of a DOPC-MoS 2/AuNP hybrid as an organic bed with higher amplification for miR detection in electrochemical biosensors. Anal. Bioanal. Chem. 412 (13), 3209–3219. doi:10.1007/s00216-020-02579-8
Ye, C., Wang, Y., Li, C., Yu, J., and Hu, Y. (2013). Preparation of liposomes loaded with quantum dots, fluorescence resonance energy transfer studies, and near-infrared in-vivo imaging of mouse tissue. Microchim. Acta 180 (1-2), 117–125. doi:10.1007/s00604-012-0907-1
Yu, H., Son, Y. H., Kim, H. J., Kim, K., Chang, P. S., and Jung, H. S. (2018). Amperometric detection of conformational change of proteins using immobilized-liposome sensor system. Sensors 18 (1), 136. doi:10.3390/s18010136
Zhang, G., and Sun, X. (2021). Lipid in chips: A brief review of liposomes formation by microfluidics. Int. J. Nanomedicine 16, 7391–7416. doi:10.2147/ijn.s331639
Keywords: biosensor, lipid-based vesicle, exosome, liposome, microvesicle
Citation: Ghazizadeh E and Nasery Z (2023) The viewpoint of nanolipid vesicles (liposomes, exosomes, and microvesicles) as biosensors in medical health advances. Front. Nanotechnol. 5:1230407. doi: 10.3389/fnano.2023.1230407
Received: 08 June 2023; Accepted: 25 July 2023;
Published: 15 September 2023.
Edited by:
Arindam Pramanik, Amity University, IndiaReviewed by:
Ahmad Mobed, Tabriz University of Medical Sciences, IranCopyright © 2023 Ghazizadeh and Nasery. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Elham Ghazizadeh, ZWxnaEB0Zi51bmkta2llbC5kZQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.