
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Nanotechnol. , 04 August 2022
Sec. Nanomaterials
Volume 4 - 2022 | https://doi.org/10.3389/fnano.2022.939197
This article is part of the Research Topic New Generation Nano-Biomaterials and their Potential Application in Drug Delivery and Bio-sensing View all 4 articles
In the modern era of rapid development and advancement in cancer therapeutics and management, there is a growing awareness in the application of exosomes as a potential tool to target cancer cells. Exosomes are cell-derived nano-vesicles that modulate intercellular communications and transport. Due to their ideal native structure and characteristics, exosomes have emerged as a promising nanocarrier for clinical use. Nevertheless, their medical application is coupled with some intrinsic restrictions which hinder their widespread use. In order to make exosomes more effective, they are engineered at the cellular level to develop designer exosomes. The focus of this review is to summarize the various exosome bio-engineering approaches aimed at the development of designer exosomes and their application in cancer treatment.
Exosomes are nano-vesicles released by almost all types of cells into the extracellular microenvironment. It comprises a small fraction of extracellular vesicles (EVs) along with microvesicles (MVs), and apoptotic bodies (Liu et al., 2019). These three fractions of EVs differ in their source of origin and size range. Exosomes arises from multivesicular bodies (MVBs), which are derived from the protrusion of the cell’s plasma membrane. They are typically 30–150 nm in diameter (Dutta, 2021). Apoptotic bodies are generated through cellular fragmentation via apoptosis. Their size ranges may vary from 50 to 5,000 nm in diameter, with a tendency to be on the larger side (Jafari et al., 2020). MV formation occurs by direct outward budding, or pinching, of the cell’s plasma membrane. Their size range typically varies from 100 nm up to 1 μm in diameter (Doyle and Wang, 2019). The cargo, or the composition of EVs consists of lipids, nucleic acids (DNA, RNA, non-coding RNA), and proteins (plasma membrane associated proteins, cytosolic proteins, and those participating in lipid metabolism) which are resistant to protease and nuclease attack in the extracellular space due to inclusion within the exosomal membrane (Abels and Breakefield, 2016). There is a growing interest in exploring emerging roles of exosomes as intercellular messengers and their potential in disease diagnosis and treatment. Exosomal cargo closely mirrors the content of their parent cell and could be delivered to both the neighbouring and distant cells. As a result, critical inspection of exosomal cargo contents shall offer new opportunities for detection and treatment of a range of diseases (yujie et al., 2021). Thus, exosomes evolved as natural drug delivery vehicles for their ability to travel long distances in extracellular fluids and transport cellular cargo to the target cells with high specificity and efficiency (Butreddy et al., 2021). Various anti-cancer drugs and products including paclitaxel, doxorubicin, curcumin, celastrol, and ß-Elemene have been successfully packaged within exosomes and many are currently undergoing clinical trials (Kim et al., 2016; Song et al., 2021). Despite the immense potential of exosomes, growth of this potential carrier in clinical setting has been below satisfactory levels due to challenges imposed by incompetent separation and refinement methods from the heterogeneous EV population, and difficulties in characterization due to lack of specific biomarkers (Li et al., 2017). In this review, we summarize the limitation of natural exosomes along with the current knowledge and opportunities in exosome bioengineering to produce targeted and drug-loaded exosomes.
One of the major gridlock in the medical application of exosomes as drug vehicles is the lack of a standardized criterion for the separation of exosomes from EVs (Li et al., 2017; Merchant et al., 2017). The various methods of exosomes isolation includes 1) ultracentrifugation-orientated methods; 2) immunoaffinity capture-dependent methods; 3) size-based isolation techniques; 4) precipitation; 5) microfluidics-based isolation techniques (Li et al., 2017). Some other approaches involve immunoisolation, density gradients, precipitation, filtration, and size exclusion chromatography (SEC). These techniques have various specificity and recovery potential. While methods like precipitation kits/polymer and high speed ultracentrifugation devoid of gradient has high retrival and reduced precision, approaches like size-exclusion chromatography (Stranska et al., 2018) has intermediary retrieval rate and intermediary precision and techniques like filtration coupled with SEC (Thery et al., 2018), immunoaffinity capture-based protocols, and microfluidics-based isolation protocols have reduced recovery rate but highly precise. Still, emerging purification and detection protocols, such as magnetic adhesion (Qi et al., 2016) and flow cytometry (Morales-Kastresana et al., 2017) holds promise to improve exosome isolation from EVs. But till date no method has been developed that can yield clinical-grade exosomes economically and reproducibly.
Exosomal cargo (mRNA, protein, DNA, lipids) significantly varies from 1 cell type to another (Kalluri and LeBleu, 2020). Acute myeloid leukemia cells derived exosomes contains significant levels of TGFβ1, which undergo a receptor-ligand interaction with the recipient cells facilitating tumor progression (Raimondo et al., 2015). Glioma cells derived exosomes contains significant levels of oncogenic receptor EGFRvIII which transforms the glioma cells which are devoid of the same provoking oncogenic signals linked to the AKT pathway in the recipient cells. Oncogenic fibroblast derived exosomes contains a substantial amount of ADAM10 which triggers the cancer cell migration rate through GTPase-mediated RhoA and NOTCH pathway (Harishkumar et al., 2021). Additionally, reports confirmed that endothelial cells which are subjected to hypoxic stress or TNF-α (model for inflammation and endothelial activation) modulated both the protein and the mRNA content of exosomes derived from these cells, whereas when subjected to high glucose load (model for hyperglycemia) or mannose concentrations (osmotic control for glucose) did not influence the exosomal protein or mRNA profiles. (de Jong et al., 2012). Harmati et al., established that oxidative stress elicited Ki-67 expression in melanoma-derived exosomes and cytostatic stress-exposed exosomes promoted melanoma cell migration (Harmati et al., 2019). Accumulating evidences suggested that 1) molecular profiling of exosomes, are controlled by the micro-environmental conditions, 2) this alteration of the exosomal-cargo might influence therapy as well as treatment efficacy. Therefore, the condition is which exosomes are isolated from the parental cells should be critically maintained for each batch to ensure minimal variability in the content of exosomal-cargo.
Proficient packaging of therapeutic content within exosomes to employ them in the field of targeted therapeutics possess another challenge to establish them for clinical applications (Luan et al., 2017). Although various therapeutic cargoes can be packaged in exosomes, like liposomes, the loading efficiency for liposomes is relatively higher than that for exosomes (Vader et al., 2016). Thus, the accurate packaging of exogenous drugs within the exosomes is a massive obstacle as exosomes themselves contain cargo derived from the parent cell, resulting in limited space for additional cargo. (Mund and Pelham, 2009; Lai et al., 2013; Li S. P. et al., 2018). Therefore engineering exosomes with reduced parental content can increase the loading efficiency of drugs within exosomes.
There are quite a few drug loading approaches, which can be grouped into two major categories: 1) endogeneous approaches (i.e., during exosome synthesis); and 2) exogeneous approaches (i.e., after exosome extraction). Endogeneous approaches include, transfection (Akao et al., 2011; Ohno et al., 2013; Batrakova and Kim, 2015) and co-incubation (Pascucci et al., 2014; Lee et al., 2015), of the parental cells with the drug to be packaged within exosomes. This process leads to encapsulation of the drug within exosomes during their formation process. Post-loading protocols, includes electroporation (Tian et al., 2014), incubation (Sun et al., 2010), sonication methods (Kim et al., 2016), extrusion methods (Batrakova et al., 2015), and freeze/thaw cycle (Haney et al., 2015), where drugs are directly loaded into the exosomes. However, loading of exosomes with drugs or nanoparticles by keeping the structural integrity intact is crucial for attaining an efficient loading and encapsulation. To resolve this concern, some authors proposed employment of a passive method employing in situ generation of the nanostructures directly inside the vesicles. Furthermore, exosome functionalization is critical to house the exogenous hydrophilic macromolecules either through pre-loading or post-loading approaches (Stremersch et al., 2016).
The conversion of this drug-delivery platform into therapeutics experiences a major obstacle concerning a clinical-grade synthesis approach that assures best quality and bulk quantity (Ohno et al., 2013; Batrakova et al., 2015). Various research groups have made efforts with different methods, researchers to obtain GMP grade exosomes (Momen-Heravi et al., 2013). Lamparski et al., explained a stepwise generation, characterization and purification method for GMP-grade antigen presenting cells derived exosomes as a possible vaccine to boost the immune system against cancer (Lamparski et al., 2022). Recently, Mendt et al., created a protocol for large-scale, bioreactor-based production of clinical-grade exosomes employing GMP standards. To maintain production of GMP-grade exosomes incorporated with therapeutic payloads one has to ensure sterile generation of exosomes in adequate quantities, without significant batch-to-batch variation for clinical testing (Mendt et al., 2018). Apparently, till date there are no state-of-the-art protocols that fulfill the ideal methodology for the generation of GMP-grade exosomes at an industrial level, where size distribution, reproducibility, scalability, potency, surface charge, and purity of the final product are a decisive issue (Pachler et al., 2017). Likewise, the origin of cell source still remains unresolved. Exosomes mimics parent cell attributes, where the origin of the cell type may affect targeting and biological attributes of exosomes. Consequently, the perfect exosome donor cell along with exosomal payload has to be monitored carefully to reach a consensus.
In the past 2 decades, a variety of artificially synthesized nanoparticles like liposomes (containing a synthetic lipid bilayer used as a vehicle for therapeutic cargo and genetic information like mRNAs and DNAs), polymer-based NPs (such as, polylactic-co-glycolic acid-coated nanoparticles), dendrimers (branched polymers used for carrying therapeutic drugs and genetic material), aptamers (that are small single-stranded DNAs or RNAs that binds to particular ligands) and various other biomolecules have built in a novel platform for precise release of therapeutic cargo and genetic materials to target cells and tissues (Raposo and Stoorvogel, 2013; Hood, 2016). Although despite the advancement, multiple obstacles still persist in using these nanovesicles which includes, bioavailability, biocompatibility, bioaccumulation, cost, short half-life, and undesirable side-effects such as cytotoxic effects on some cells in vivo, preventing wide-spread clinical applications (Hood and Wickline, 2012; Raposo and Stoorvogel, 2013).
Whereas exosomes are naturally produced from mostly all cell types under physiological conditions and their cargo closely resembles that of the parental cell (Dutta, 2021). Exosomes isolated from different cellular origin have distinct features, cargo, and hence different consequences on their target cells (Sancho-Albero et al., 2019). These two core characteristics of exosomes have immense biomedical relevance. Also exosomes are small therefore, they can escape immune reactions, such as fusion with membranes and phagocytosis, thus evading by lysosomal engulfment. These key properties of exosomes have fueled intensive research on their biomedical uses, including regeneration of tissues (Jing et al., 2018), cancer treatment and diagnosis (Lan et al., 2018), vaccine development (Aline et al., 2004), drug delivery (Gomari et al., 2018), and gene therapy (Mathiyalagan and Sahoo, 2017). However, introducing natural exosomes in the clinical field to deliver therapeutic cargo is quite challencing and cumbersome. The therapeutic significance of using exosomes as a drug delivery system requires the creation of precisely controlled and targeted exosomes. Furthermore, various loading technique is employed to package additional therapeutic molecules within exosomes.
Incidentally, designer exosomes or targeted designer exosomes loaded with appropriate therapeutic molecule could probably tackle these limitations to a greater extent. In the subsequent segments, various approaches executed for the advancement of designer exosomes are explained (see Figure 1).
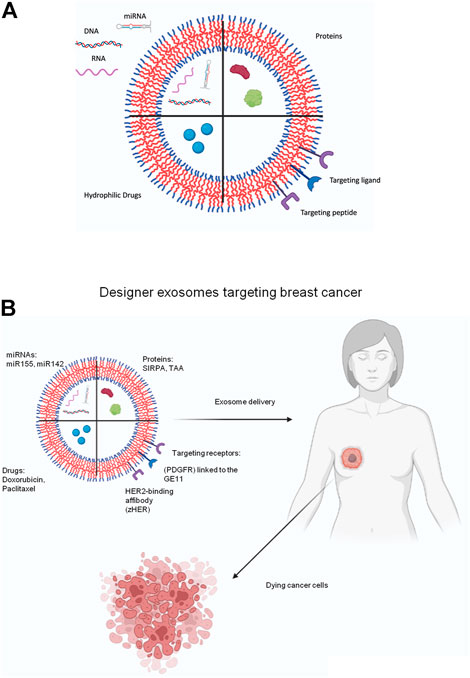
FIGURE 1. (A) Schematic representation of the different types exosome engineering approaches. Different therapeutic molecules can be either loaded into lumen or displayed on the exosome surface for clinical purposes. Various DNA, RNA, miRNA molecules can be packaged or various protein molecules. Targeting ligands or targeting peptides can be displayed on the surface. Various anti-cancer drugs can loaded into the exosomes. (B) Schematic diagram showing treatment of breast cancer using designer exosomes. Exosomal cargo such as miRNA, protein, drugs can be specifically targeted to breast cancer cells using specific targeting receptors that binds to cognate ligands on breast cancer cells.
Exosome engineering methods aims to achieve 1) precision targeting of exosomes to a particular tissue type or cells; 2) exosomal loading with exogenous molecules like nucleic acids, drugs, proteins, or fortification of an endogenous molecule into the exosomal lumen or on their surface. The multiple techniques employed in the two key exosome engineering approaches are discussed below:
Nowadays, the receptor-ligand based targeting approach has been believed as a highly precise method for targeted delivery by showcasing ligands that identify their cognate receptors on certain cell types. One straightforward method is transfection-based ectopic expression of the desired ligand in the parent cell which leads to ligand accumulation within exosomes. Ohno et al., manipulated donor cells to synthesize platelet-derived growth factor receptor (PDGFR) linked to the GE11 peptide (Ohno et al., 2013) which binds specifically to EGFR. This system was used to target let-7a to EGFR-producing xenograft breast tumor tissue in recombination activating gene 2 (RAG2) knockout mice. Furthermore, Tian et al. generated immature dendritic cells (imDCs) expressing Lamp2b-αv integrin-specific iRGD peptide fusion proteins, which was used to precisely recognize and release Doxorubicin to αv breast cancer cells which are integrin-positive in nature and significantly inhibit tumor development (Van der meel et al., 2014). Lamp2B has also been used to express IL3 on exosomes which targets interleukin-3 receptor α (IL-3Rα), highly expressed on CML (Bellavia et al., 2017). Additionally HER2-binding affibody (zHER) fused to the N-terminus of Lamp2 efficiently targeted HCT-116 colon cancer cells and specifically delivered the cargoes (5-FU and anti-miRNA-21) to HER2-expressing tumors in vivo (Liang et al., 2020). tLyP-1 peptide (CGNKRTR), binds neuropilin-1 (NRP1) and neuropilin-2 (NRP2) receptors expressed on NSCLC cells are used to deliver siRNAs effectively to these cells (Bai et al., 2020). Despite the efficacy of this genetic method, its utility is limited as the targeting peptide can be degraded in some situations. Although a glycosylation motif (GNSTM) can be added to the N-terminus of the Lamp2B fusion peptide to enhance the stability (Hung and Leonard, 2015).
Another method is direct chemical assembling of ligands on exosomal surface. Chemical modification directly accumulated ligands on the parental cell membrane or exosomal membrane to fabricate targeted exosomes. Wang et al. produced biotin and avidin labeled human umbilical vein endothelial cells which can target the biotin receptor and lectin enriched tumor cells respectively, in the phospholipid membrane (Wang J. et al., 2017). These exosomes precisely unloaded the drug into tumor cells which causes apoptosis. Similarly, exosomes labelled with folate was used to deliver erastin to treat triple negative breast cancer. The choice between these methods depends on the varied size ranges, constitutions, and composition of homing-molecules. Furthermore, membrane-labeled lipidomimetic chains-grafted hyaluronic acid or A33 antibody to target exosomes to colon cancer cells expressing A33 more interestingly to cancer stem cell-like drug resistant cancer cells expressing CD44, resulting in inhibition of tumor intensification (Li Y. et al., 2018; Liu et al., 20,119).
Aptamers are tiny, RNA molecules, single-stranded DNA, or xeno nucleic acid (XNA). These can be fashioned with elevated recognition potential and affinity towards PCR-based desired targets, this procedure is defined as systematic evolution of ligands by exponential enrichment (SELEX) (Zhang et al., 2019). Aptamers are widely utilized in exosomal surface modification for targeted delivery. Wan et al., synthesized a nucleolin-targeting aptamer AS1411 grafted paclitaxel loaded EVs (via sonication method) for targeting breast cancer cells in vivo (Wan et al., 2018). Pi et al. modified EVs to display aptamer targeted to prostate-specific membrane antigen and packaged the engineered EVs with survivin siRNA which restricted the intensification of a prostate cancer xenograft. Furthermore, EV-dependent delivery method coupled EGFR aptamer could restrict orthotropic breast cancer (Pi et al., 2018). Since inception nearly 30 years ago, only pegaptanib, an RNA aptamer against vascular endothelial growth factor (VEGF), has been approved by the FDA for the treatment of macular degeneration (Ng et al., 2006). Therefore development to aptamer technology for effective preclinical and clinical efficacy study is crucial. One potential barrier for their translation is the loss of targeting competence under in vivo conditions. Several factors can contribute to the loss of in vivo targeting efficacy of aptamers including, clearance due to the immune response, aggregation, or enzymatic cleavage of the aptamer.
Exosomes with anchoring specific peptide on their surface is an emerging technique that has been employed for successful targeted delivery of therapeutic cargo via engineered exosomes. Alvarez-Erviti et al. generated Lamp2b expressing DCs (an exosomal membrane protein) coupled with neuron-specific RVG peptide. The isolated exosomes were loaded with siRNA using electroporation. These bio-engineered RVG-specific exosomes successfully unloaded siRNAs precisely to brain cells including neurons, microglia, and oligodendrocytes (Alvarez-Erviti et al., 2011a). Similarly, Zhan et al. bio-engineered blood derived exosomes via coupling of magnetic particles to L17E (an endosomolytic peptides) on the exosomal surface, followed by packaging of doxorubicin and cholesterol-modified miRNA21 inhibitor (miR-21i). It enhanced the tumor accumulation of the exosomes and augmented potential to escape endosomes, leading to proficient delivery of cargoes within cancer cells in vitro and in vivo (Zhan et al., 2020). Arginine-rich CPPs is known to induce macropinocytosis, which can lead to efficient cellular uptake of the exosomes-tagged with octaarginine peptide R8. The N-terminus of the peptides was steary-lated and thereafter was inserted into the exosomal membrane. But the concentration of peptide inserted plays a crucial role in cellular uptake of modified exosomes. Also the functionality of the exosomal membrane can be compromised due to insertion of the hydrophobic moiety into the exosomal membrane (Nakase et al., 2004).
The exosomal surface can be modified using chemicals, although less explored. Interestingly the amine groups of exosomal surface proteins can be labelled with alkyne groups which can then be bio-orthogonally conjugated to azide-containing reagents via a copper-catalyzed azide-alkyne cycloaddition reactions (click reaction) (Smyth et al., 2014). This method can be used to modify the exosomal surface with both a small molecular dye as well as larger azide-containing model protein. Click reaction has been used to conjugate glioma-targeting peptide RGE peptide (RGERPPR) and c (RGDyK) (Arg-Gly-Asp-D-Tyr-Lys) to exosomal surface. These conjugated exosomes successfully penetrated the BBB and either targeted tumor regions or suppressed inflammatory responses and cellular apoptosis in a transient middle cerebral artery occlusion mouse model respectively (Tian et al., 2018). Introduction of amphipathic molecules into the exosomal membrane is another chemical modification strategy. Polyethylene glycol (PEG)-grafted 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DSPE-PEG) is known to accumulate in the exosome membrane and it was used to label exosomes with DSPE-PEG-RGD. These RGD exosomes were combined with tumor-specific targeting ligand folate which ensured specific targeting to tumor cells. This method can also useful for miRNA and siRNA delivery (Kim et al., 2018). The modification or exosomal surface structures may influence both exosome trafficking and function in vivo. The number of surface macromolecules modified might also influence the efficacy and immunogenicity of exosomes or even clearance by the liver and spleen. Chemical modification also results in increased aggregation of exosomes which will make them more readily phagocytosed by antigen presenting cells. This could potentially abrogate the intended function of engineered exosomes in therapeutic application.
Other targeting methods include pH gradient/surface charge-driven targeted delivery as due to the increased rate of lactate production and intracellular glycolysis, tumor microenvironment pH is lowered compared to normal tissues, thereby making pH-responsive drug delivery platform a convincing tool to treat tumors and minimize side effects of drugs (Yu et al., 2014). In another, method magnetism-guided targeted delivery of exosome-based superparamagnetic nanovesicles clusterings are used as a targeted drug delivery vehicle to treat cancer (Qi et al., 2016) these vehicles also crossed the blood brain barrier smoothly and targeted glioma cells by specific transfer of chemotherapeutic drugs (Jia et al., 20,118). These methods have been briefly discussed in Table 1.
There are two main approaches for loading and displaying functional molecules on exosomes, 1) parental cell-based, 2) direct exosome engineering. The next two subsections.
In the parental cell-based method, the cells that are the origin of the exosomes are manipulated and engineered prior to exosome isolation from parental cells. Functional entities can be targeted to either exosomal surface or lumen using genetic editing of the parental cells.
The most widely used technique for targeting a protein to the exterior of exosomal membrane is using an exosomal signal peptide tagged to the protein to be loaded. Lysosome-associated membrane protein 2b (Lamp2b) contains an exosomal signal peptide is an exosomal membrane protein. The fusion of a protein of interest with Lamp2b is widespread for showcasing the protein on the exosomal surface as a targeting entity, ligand, or receptor. Furthermore, coupling of a glycosylation motif with Lamp2b-fusion proteins could further boost exosome delivery by shielding the surface-accessible fusion protein from enzymatic degradation to the target cells (Barile and Vassalli, 2017). Other commonly used molecules for exosomal surface display are tetraspanins (CD63, CD9, CD81) (Stickney et al., 2016), glycosylphosphatidylinositol (GPI) (Kooijmans et al., 2016), lactadherin (C1C2 domain) (Rountree et al., 2011), and platelet-derived growth factor receptors (PDGFRs) (Ohno et al., 2013).
The methods of molecular packaging into the exosomal lumen have started developing recently. Transfection of the gene of interest into donor cells results in loading of the desired protein within exosomes through natural encapsulation processes. Post isolation and purification steps, engineered exosomes can be collected (Jafari et al., 2020). Additionally, it was observed ubiquitin-fused protein accumulated in exosomes. The target protein enrichment was increased by ∼10-fold within exosomes following fusion to ubiquitin (Cheng and Schorey, 2016). However, shielding of the modified protein from degradation is still a major concern. The WW-tag mediated ubiquitination is another strategy for the loading proteins into exosomes (Mund and Pelham, 2009). In another strategy Di Bonito et al. have synthesized fusion protein combining Nef exosome-anchoring protein and HPV-E7 expressing DNA vector (Di Bonito et al., 2017). Recently, mutant HIV-I Nef, which lacks the enzymatic activity and function of Nef was engineered for categorization of proteins within exosomes. Mutant Nef-GFP fusion protein useful for monitoring transfection and loading efficiency when successfully packaged into exosomes (McNamara et al., 2018). In a recent advancement mutant Nef was fused to several pathogens including influenza virus NP, Ebola virus VP24, VP40, Crimean-Congo hemorrhagic fever NP, and hepatitis C virus NS3 etc. Which facilitated expression of stable fusion proteins and their optimal loading into exosomes (Anticoli et al., 2018). In a recent approach researchers used archaeal ribosomal protein L7Ae that directly binds to the C/D box RNA structure. Attachment of L7Ae to the C-terminus of CD63 and insertion of C/D box into the reporter gene’s 3′-UTR can efficiently package the mRNA of the reporter gene into the exosomes (Rozhdestvensky et al., 2003).
Yim et al., 2016, developed EXPLORs (exosomes for protein loading via optically reversible PPIs) employing optically reversible protein–protein interaction (PPI) and using photoreceptor cryptochrome 2 (CRY2) and CIBN PPI modules. In this approach intracellular delivery of protein cargo into exosomes were ensured by exposure to blue light. A reporter protein was coupled to CRY2 and CIBN protein was fused to the CD9 (a exosomal membrane protein) to acquire two fusion proteins (CIBN-EGFP-CD9 and mCherry-CRY2). The blue light exposure induced a reversible PPI between CIBN and CRY2 that leads to the reporter fusion protein (mCherry-CRY2) direction to the exosomal lumen through interaction with CIBN in the CIBN-EGFP-CD9 complex (Yim et al., 2016). A novel method was also developed for loading of mRNA into exosomes called the EXOtic (Exosomal Transfer Into Cells) device. Using EXOtic device, the catalase enzyme mRNA containing a C/Dbox sequence on its 3′ UTR was packaged within exosomes (Kojima et al., 2018). Li et al., 2019a created an unique RNA delivery EV by engineering a fusion protein CD9-HUR which has extremely high affinity for miRNA (Li Z. et al., 2019). Although these gene-editing technologies are efficient it is highly time-consuming and capital intensive thereby reducing the translational vaue.
Various small nucleic acid molecules such as miRNAs, siRNAs and therapeutic molecules such as chemotherapeutics drugs (doxorubicin, paclitaxel etc.) can be loaded within exosomes by direct exosome editing. These methods are less complex than compared to parental cell-based methods. A simple method is direct incubation of the drugs with exosomes, where the drugs slowly disseminate into the exosomes alongside the concentration gradient and the loading efficacy depends on the hydrophobic quotient of the drug molecules (Pascucci et al., 2014). Sonication is another method where donor cell derived exosomes are mixed with drugs and then sonicated with homogenizer probe. The mechanical shear force produced from the sonicator probe interferes with the integrity of the exosomal membrane and permits the drug to disperse through the exosomal lumen during this membrane shearing (Kim et al., 2016). Electroporation uses electric field to create small perforations within the exosomal membrane which allows diffusion of the molecules into the interior of the exosomes via the pores (Johnsen et al., 2016). Another method for direct loading of molecules into exosomes is extrusion, where exosomes (from the donor cells) are mixed with the molecule to be packaged and loaded into a syringe-based liquid extruder with a ∼100–400 nm porous membrane. The exosomal membrane is disturbed during extrusion and vigorously mixed with the drug which leads to packaging of the drug with the exosomes (Fuhrmann et al., 2015). Other methods for direct engineering includes freeze-thaw, bio-conjugation, click chemistry, and cloaking (Luan et al., 2017), although these methods does not provide a constant source of bio-engineered exosomes the technical feasibility of these methods has lead to their widespread acceptance (see Figure 2).
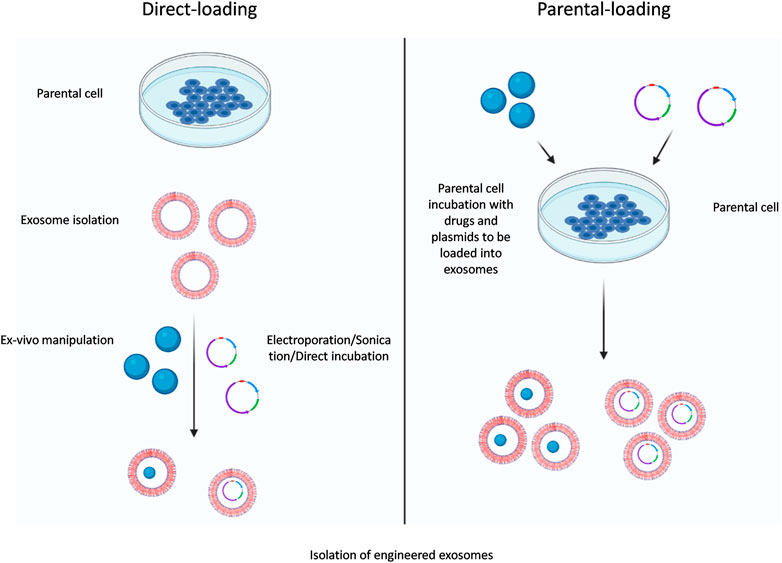
FIGURE 2. Exosomal-loading approaches. Two main approaches of exosome loading are parental cell-based and direct-loading engineering methods.
These parental cell-based or direct-loading methods can be useful to package chemotherapeutic drugs into exosomes which can targeted to cancer cells using specific targeting modules. As GE11-based targeted designer exosome was used to target EGFR + breast cancer cells (Ohno et al., 2013) and RGE targeting peptide was used to target NRP-1 on glioma cells (Jia et al., 2018). Additional research needs to be carried out to further elucidate additional exosome-based targeting strategies. The exosomes-loading method has been briefly listed in Table 2.
The therapeutic potential of exosomes are now being clinically investigated. As of June 2022, a key word search in the US-NIH clinical trial database with “exosomes” and disease “cancer” gives 117 clinical studies search result. A number of pre-clinical studies have evaluated the efficacy of exosomes as a therapeutic efficacy of exosomes, while the clinical evaluation is still ongoing. Considering the limitations of natural exosomes trials have mostly involved engineered exosomes. A Phase II is underway evaluate autologous dendritic cell-derived exosomes loaded with MAGE (Melanoma Associated Antigen) peptides in targeting non-small cell lung cancer trial (clinicaltrials.gov/NCT01159288). Similarly, a Phase I study is recruiting participants to evaluate the efficacy of plant-derived exosomes to deliver curcumin to normal and malignant colon tissue (clinicaltrials.gov/NCT01294072). Clinical trials which uses exosomes as a therapeutic tool has been listed in Table 3.
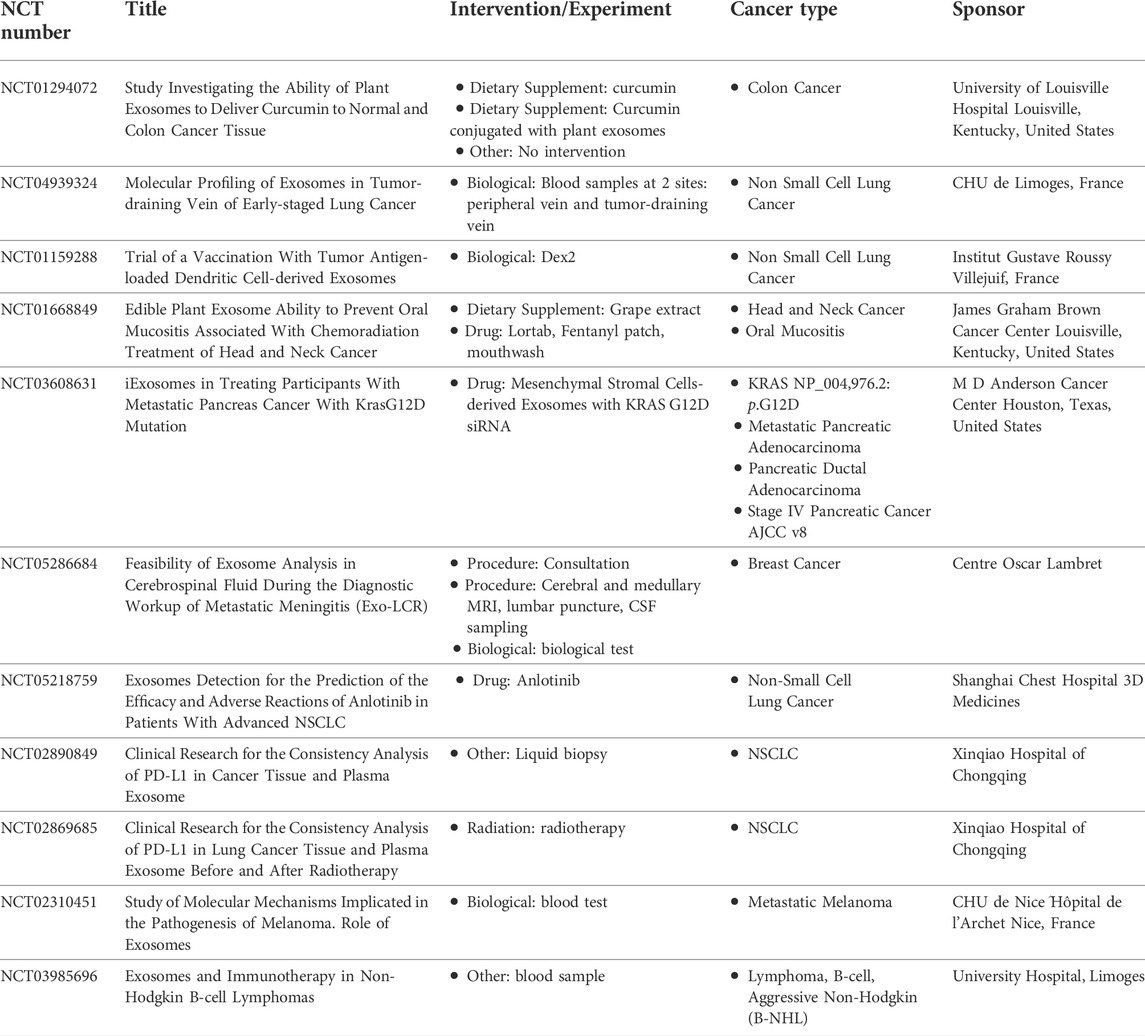
TABLE 3. Exosomes-based clinical trials registered in ClinicalTrials.gov (https://www.clinicaltrials.gov/).
The development of therapeutic exosomes will require advanced approaches for exosome isolation, engineering of exosomes to target them and load them with therapeutic cargo or the synthesis of semi-synthetic, more highly defined exosome-like therapeutic nano-vehicles. For successful exosome therapeutics development, the Chemistry, Manufacturing, and Control (CMC) and development for good manufacturing practice (GMP) grade therapeutic exosome production needs to be established. It covers multiple area like establishing master cell bank (MCB), process for isolating large quantity of exosome and quality control (QC) development for therapeutic exosome production.
The dawn of nanotechnology heralded a new chapter in drug delivery. Exosomes have also evolved as a promising vehicle for drug delivery owing to their strong biocompatibility and low immunogenicity (Chen et al., 2021). However there are various limitations connected with the usage of natural exosomes as a drug delivery vehicle. Therefore, extensive research was performed to find a suitable alternative. This has lead to modification of exosomes to circumvent their intrinsic limitations. Despite the advancement outlined in this review, there exists many limitations associated with the clinical use of engineered exosomes. The methods and the utility of exosome engineering are expanding rapidly. For example, common chemotherapeutic drugs target all rapidly proliferating cells in the body leading to significant adverse effects due to therapy-induced cytotoxicity. In this regard, engineering exosomes to carry chemotherapeutic drug and displaying targeting molecules specifically targeted to tumor cells are is a unique platform to eradicate tumor cells and sparing the healthy cells (Gilligan and Dwyer, 2017). With the advancements on bioavailability, biocompatibility, numerous cargo loading potential, deep tissue penetration, and surface modification tolerability, exosomes are emerging as the new era of natural carriers (Bunggulawa et al., 2018). However, certain caveats remain in the large-scale production of high quality-controllable engineered exosomes. Another concern in the development of exosome-based delivery systems is the origin of exosomes which needs to be carefully considered as because the source of exosomes influences the content of exosomes. This variation in the composition of exosomes can greatly alter the therapeutic outcome (Butreddy et al., 2021). Furthermore, exosomes also carry a multitude of factors which can potentially lead to tumor progression. Exosomes carry caspase-3 may also inhibit cell death by apoptosis or enhance tumor cell survival by preventing accumulation of chemotherapeutics drugs. An effective solution will be to produce empty exosomes which can be loaded with the drug of interest. This approach will not only remove potential tumor promoting factors but also increase the space within exosomes to carry therapeutic cargoes. Generally with the advancement of large-scale production approaches for natural exosomes and engineered exosomes, the settlement of therapeutic and diagnostic platforms related with exosomes may be promising in the upcoming years. Nevertheless in recent years, a few companies have been established to manufacture bio-engineered exosome-based platforms. This review will encourage and inspire scientists to create new strategies for efficacious, stable and safe targeted delivery platforms in the near future.
AD conceived the idea of the paper. The manuscript was jointly drafted by AD and SP. All authors gave final approval for publication. All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
The authors are grateful to KIIT-TBI and BIG-BIRAC team for their constant support. All figures were created with BioRender.com.
AD and SP is employed by EXSURE PVT LTD.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abels, E. R., and Breakefield, X. O. (2016). Introduction to extracellular vesicles: Biogenesis, RNA cargo selection, content, release, and uptake. Cell. Mol. Neurobiol. 36, 301–312. doi:10.1007/s10571-016-0366-z
Akao, Y., Iio, A., Itoh, T., Noguchi, S., Itoh, Y., Ohtsuki, Y., et al. (2011). Microvesicle-mediated RNA molecule delivery system using monocytes/macrophages. Mol. Ther. 19, 395–399. doi:10.1038/mt.2010.254
Aline, F., Bout, D., Amigorena, S., Roingeard, P., and Dimier-Poisson, I. (2004). Toxoplasma gondii antigen-pulsed-dendritic cell-derived exosomes induce a protective immune response against T. gondii infection. Infect. Immun. 72, 4127–4137. doi:10.1128/iai.72.7.4127-4137.2004
Alvarez-Erviti, L., Seow, Y., Yin, H., Betts, C., Lakhal, S., Wood, M. J., et al. (2011a). Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat. Biotechnol. 29, 341–345. doi:10.1038/nbt.1807
Alvarez-Erviti, L., Seow, Y., Yin, H., Betts, C., Lakhal, S., Wood, M. J., et al. (2011b). Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat. Biotechnol. 29, 341–345. doi:10.1038/nbt.1807
Anticoli, S., Manfredi, F., Chiozzini, C., Arenaccio, C., Olivetta, E., Ferrantelli, F., et al. (2018). An exosome-based vaccine platform imparts cytotoxic T lymphocyte immunity against viral antigens. Biotechnol. J. 13, e1700443. doi:10.1002/biot.201700443
Bai, J., Duan, J., Liu, R., Du, Y., Luo, Q., Cui, Y., et al. (2020). Engineered targeting tLyp-1 exosomes as gene therapy vectors for efficient delivery of siRNA into lung cancer cells. Asian J. Pharm. Sci. 15, 461–471. doi:10.1016/j.ajps.2019.04.002
Barile, L., and Vassalli, G. (2017). Exosomes: Therapy delivery tools and biomarkers of diseases. Pharmacol. Ther. 174, 63–78. doi:10.1016/j.pharmthera.2017.02.020
Batrakova, E. V., and Kim, M. S. (2015). Using exosomes, naturally-equipped nanocarriers, for drug delivery. J. Control. Release 219, 396–405. doi:10.1016/j.jconrel.2015.07.030
Bellavia, D., Raimondo, S., Calabrese, G., Forte, S., Cristaldi, M., Patinella, A., et al. (2017). Interleukin 3- receptor targeted exosomes inhibit in vitro and in vivo Chronic Myelogenous Leukemia cell growth. Theranostics 7, 1333–1345. doi:10.7150/thno.17092
Bunggulawa, E. J., Wang, W., Yin, T., Wang, N., Durkan, C., Wang, Y., et al. (2018). Recent advancements in the use of exosomes as drug delivery systems. J. Nanobiotechnology 16, 81. doi:10.1186/s12951-018-0403-9
Butreddy, A., Kommineni, N., and Dudhipala, N. (2021). Exosomes as naturally occurring vehicles for delivery of biopharmaceuticals: Insights from drug delivery to clinical perspectives. Nanomater. (Basel, Switz. 11, 1481. doi:10.3390/nano11061481
Chen, H., Wang, L., Zeng, X., Schwarz, H., Nanda, H. S., Peng, X., et al. (2021). Exosomes, a new star for targeted delivery. Front. Cell Dev. Biol. 9, 751079. doi:10.3389/fcell.2021.751079
Cheng, Q., Shi, X., Han, M., Smbatyan, G., Lenz, H. J., Zhang, Y., et al. (2018). Reprogramming exosomes as nanoscale controllers of cellular immunity. J. Am. Chem. Soc. 140, 16413–16417. doi:10.1021/jacs.8b10047
Cheng, Y., and Schorey, J. S. (2016). Targeting soluble proteins to exosomes using a ubiquitin tag. Biotechnol. Bioeng. 113, 1315–1324. doi:10.1002/bit.25884
de Jong, O. G., Verhaar, M. C., Chen, Y., Vader, P., Gremmels, H., Posthuma, G., et al. (2012). Cellular stress conditions are reflected in the protein and RNA content of endothelial cell-derived exosomes. J. Extracell. vesicles 1, 18396. doi:10.3402/jev.v1i0.18396
Di Bonito, P., Chiozzini, C., Arenaccio, C., Anticoli, S., Manfredi, F., Olivetta, E., et al. (2017). Antitumor HPV E7-specific CTL activity elicited by in vivo engineered exosomes produced through DNA inoculation. Int. J. Nanomedicine 12, 4579–4591. doi:10.2147/ijn.s131309
Didiot, M. C., Hall, L. M., Coles, A. H., Haraszti, R. A., Godinho, B. M., Chase, K., et al. (2016). Exosome-mediated delivery of hydrophobically modified siRNA for huntingtin mRNA silencing. Mol. Ther. 24, 1836–1847. doi:10.1038/mt.2016.126
Doyle, L. M., and Wang, M. Z. (2019). Overview of extracellular vesicles, their origin, composition, purpose, and methods for exosome isolation and analysis. Cells 8, 727. doi:10.3390/cells8070727
Dutta, A. (2021). Exosomes-based cell-free cancer therapy: A novel strategy for targeted therapy. Immunol. Med. 44, 116–123. doi:10.1080/25785826.2020.1818482
Ferrantelli, F., Arenaccio, C., Manfredi, F., Olivetta, E., Chiozzini, C., Leone, P., et al. (2019). The intracellular delivery of anti-HPV16 E7 scFvs through engineered extracellular vesicles inhibits the proliferation of HPV-infected cells. Int. J. Nanomedicine 14, 8755–8768. doi:10.2147/ijn.s209366
Fuhrmann, G., Serio, A., Mazo, M., Nair, R., and Stevens, M. M. (2015). Active loading into extracellular vesicles significantly improves the cellular uptake and photodynamic effect of porphyrins. J. Control. Release 205, 35–44. doi:10.1016/j.jconrel.2014.11.029
Gilligan, K. E., and Dwyer, R. M. (2017). Engineering exosomes for cancer therapy. Int. J. Mol. Sci. 18, 1122. doi:10.3390/ijms18061122
Goh, W. J., Lee, C. K., Zou, S., Woon, E. C., Czarny, B., Pastorin, G., et al. (2017). Doxorubicin-loaded cell-derived nanovesicles: An alternative targeted approach for anti-tumor therapy. Int. J. Nanomedicine 12, 2759–2767. doi:10.2147/ijn.s131786
Gomari, H., Forouzandeh Moghadam, M., and Soleimani, M. (2018). Targeted cancer therapy using engineered exosome as a natural drug delivery vehicle. Onco. Targets. Ther. 11, 5753–5762. doi:10.2147/ott.s173110
Haney, M. J., Klyachko, N. L., Zhao, Y., Gupta, R., Plotnikova, E. G., He, Z., et al. (2015). Exosomes as drug delivery vehicles for Parkinson's disease therapy. J. Control. Release 207, 18–30. doi:10.1016/j.jconrel.2015.03.033
Harishkumar, M., Radha, M., Yuichi, N., Muthukalianan, G. K., Kaoru, O., Shiomori, K., et al. (2021). Designer exosomes: Smart nano-communication tools for translational medicine. Bioeng. (Basel, Switz. 8, 158. doi:10.3390/bioengineering8110158
Harmati, M., Gyukity-Sebestyen, E., Dobra, G., Janovak, L., Dekany, I., Saydam, O., et al. (2019). Small extracellular vesicles convey the stress-induced adaptive responses of melanoma cells. Sci. Rep. 9, 15329. doi:10.1038/s41598-019-51778-6
Hood, J. L. (2016). Post isolation modification of exosomes for nanomedicine applications. Nanomedicine Lond. Engl. 11, 1745–1756. doi:10.2217/nnm-2016-0102
Hood, J. L., and Wickline, S. A. (2012). A systematic approach to exosome-based translational nanomedicine. WIREs. Nanomed. Nanobiotechnol. 4, 458–467. doi:10.1002/wnan.1174
Hung, M. E., and Leonard, J. N. (2015). Stabilization of exosome-targeting peptides via engineered glycosylation. J. Biol. Chem. 290, 8166–8172. doi:10.1074/jbc.m114.621383
Jafari, D., Shajari, S., Jafari, R., Mardi, N., Gomari, H., Ganji, F., et al. (2020). Designer exosomes: A new platform for biotechnology therapeutics. BioDrugs 34, 567–586. doi:10.1007/s40259-020-00434-x
Jeyaram, A., Lamichhane, T. N., Wang, S., Zou, L., Dahal, E., Kronstadt, S. M., et al. (2020). Enhanced loading of functional miRNA cargo via pH gradient modification of extracellular vesicles. Mol. Ther. 28, 975–985. doi:10.1016/j.ymthe.2019.12.007
Jia, G., Han, Y., An, Y., Ding, Y., He, C., Wang, X., et al. (2018). NRP-1 targeted and cargo-loaded exosomes facilitate simultaneous imaging and therapy of glioma in vitro and in vivo. Biomaterials 178, 302–316. doi:10.1016/j.biomaterials.2018.06.029
Jing, H., He, X., and Zheng, J. (2018). Exosomes and regenerative medicine: State of the art and perspectives. Transl. Res. 196, 1–16. doi:10.1016/j.trsl.2018.01.005
Johnsen, K. B., Gudbergsson, J. M., Skov, M. N., Christiansen, G., Gurevich, L., Moos, T., et al. (2016). Evaluation of electroporation-induced adverse effects on adipose-derived stem cell exosomes. Cytotechnology 68, 2125–2138. doi:10.1007/s10616-016-9952-7
Kalluri, R., and LeBleu, V. S. (2020). The biology, function, and biomedical applications of exosomes. Sci. (New York, N.Y.) 367, eaau6977. doi:10.1126/science.aau6977
Kanuma, T., Yamamoto, T., Kobiyama, K., Moriishi, E., Masuta, Y., Kusakabe, T., et al. (2017). CD63-Mediated antigen delivery into extracellular vesicles via DNA vaccination results in robust CD8+ T cell responses. J. Immunol. 198, 4707–4715. doi:10.4049/jimmunol.1600731
Kim, M. S., Haney, M. J., Zhao, Y., Mahajan, V., Deygen, I., Klyachko, N. L., et al. (2016). Development of exosome-encapsulated paclitaxel to overcome MDR in cancer cells. Nanomedicine Nanotechnol. Biol. Med. 12, 655–664. doi:10.1016/j.nano.2015.10.012
Kim, M. S., Haney, M. J., Zhao, Y., Yuan, D., Deygen, I., Klyachko, N. L., et al. (2018). Engineering macrophage-derived exosomes for targeted paclitaxel delivery to pulmonary metastases: In vitro and in vivo evaluations. Nanomedicine Nanotechnol. Biol. Med. 14, 195–204. doi:10.1016/j.nano.2017.09.011
Kojima, R., Bojar, D., Rizzi, G., Hamri, G. C., El-Baba, M. D., Saxena, P., et al. (2018). Designer exosomes produced by implanted cells intracerebrally deliver therapeutic cargo for Parkinson's disease treatment. Nat. Commun. 9, 1305. doi:10.1038/s41467-018-03733-8
Kooijmans, S. A., Aleza, C. G., Roffler, S. R., van Solinge, W. W., Vader, P., Schiffelers, R. M., et al. (2016). Display of GPI-anchored anti-EGFR nanobodies on extracellular vesicles promotes tumour cell targeting. J. Extracell. vesicles 5, 31053. doi:10.3402/jev.v5.31053
Lai, R. C., Yeo, R. W., Tan, K. H., and Lim, S. K. (2013). Exosomes for drug delivery - a novel application for the mesenchymal stem cell. Biotechnol. Adv. 31, 543–551. doi:10.1016/j.biotechadv.2012.08.008
Lamparski, H. G., Metha-Damani, A., Yao, J. Y., Patel, S., Hsu, D. H., Ruegg, C., et al. (2002). Production and characterization of clinical grade exosomes derived from dendritic cells. J. Immunol. methods 270, 211–226. doi:10.1016/s0022-1759(02)00330-7
Lan, F., Qing, Q., Pan, Q., Hu, M., Yu, H., Yue, X., et al. (2018). Serum exosomal miR-301a as a potential diagnostic and prognostic biomarker for human glioma. Cell. Oncol. 41, 25–33. doi:10.1007/s13402-017-0355-3
Lee, J., Kim, J., Jeong, M., Lee, H., Goh, U., Kim, H., et al. (2015). Liposome-based engineering of cells to package hydrophobic compounds in membrane vesicles for tumor penetration. Nano Lett. 15, 2938–2944. doi:10.1021/nl5047494
Lee, J., Lee, H., Goh, U., Kim, J., Jeong, M., Lee, J., et al. (2016). Cellular engineering with membrane fusogenic liposomes to produce functionalized extracellular vesicles. ACS Appl. Mat. Interfaces 8, 6790–6795. doi:10.1021/acsami.6b01315
Li, P., Kaslan, M., Lee, S. H., Yao, J., and Gao, Z. (2017). Progress in exosome isolation techniques. Theranostics 7, 789–804. doi:10.7150/thno.18133
Li, S. P., Lin, Z. X., Jiang, X. Y., and Yu, X. Y. (2018a). Exosomal cargo-loading and synthetic exosome-mimics as potential therapeutic tools. Acta Pharmacol. Sin. 39, 542–551. doi:10.1038/aps.2017.178
Li, S., Wu, Y., Ding, F., Yang, J., Li, J., Gao, X., et al. (2020). Engineering macrophage-derived exosomes for targeted chemotherapy of triple-negative breast cancer. Nanoscale 12, 10854–10862. doi:10.1039/d0nr00523a
Li, X., Corbett, A. L., Taatizadeh, E., Tasnim, N., Little, J. P., Garnis, C., et al. (2019a). Challenges and opportunities in exosome research-Perspectives from biology, engineering, and cancer therapy. Apl. Bioeng. 3, 011503. doi:10.1063/1.5087122
Li, Y., Gao, Y., Gong, C., Wang, Z., Xia, Q., Gu, F., et al. (2018b). A33 antibody-functionalized exosomes for targeted delivery of doxorubicin against colorectal cancer. Nanomedicine Nanotechnol. Biol. Med. 14, 1973–1985. doi:10.1016/j.nano.2018.05.020
Li, Z., Zhou, X., Wei, M., Gao, X., Zhao, L., Shi, R., et al. (2019b). In vitro and in vivo RNA inhibition by CD9-HuR functionalized exosomes encapsulated with miRNA or CRISPR/dCas9. Nano Lett. 19, 19–28. doi:10.1021/acs.nanolett.8b02689
Liang, G., Kan, S., Zhu, Y., Feng, S., Feng, W., Gao, S., et al. (2018). Engineered exosome-mediated delivery of functionally active miR-26a and its enhanced suppression effect in HepG2 cells. Int. J. Nanomedicine 13, 585–599. doi:10.2147/ijn.s154458
Liang, G., Zhu, Y., Ali, D. J., Tian, T., Xu, H., Si, K., et al. (2020). Engineered exosomes for targeted co-delivery of miR-21 inhibitor and chemotherapeutics to reverse drug resistance in colon cancer. J. Nanobiotechnology 18, 10. doi:10.1186/s12951-019-0563-2
Limoni, S. K., Moghadam, M. F., Moazzeni, S. M., Gomari, H., and Salimi, F. (2019). Engineered exosomes for targeted transfer of siRNA to HER2 positive breast cancer cells. Appl. Biochem. Biotechnol. 187, 352–364. doi:10.1007/s12010-018-2813-4
Liu, J., Ye, Z., Xiang, M., Chang, B., Cui, J., Ji, T., et al. (2019). Functional extracellular vesicles engineered with lipid-grafted hyaluronic acid effectively reverse cancer drug resistance. Biomaterials 223, 119475. doi:10.1016/j.biomaterials.2019.119475
Luan, X., Sansanaphongpricha, K., Myers, I., Chen, H., Yuan, H., Sun, D., et al. (2017). Engineering exosomes as refined biological nanoplatforms for drug delivery. Acta Pharmacol. Sin. 38, 754–763. doi:10.1038/aps.2017.12
Mathiyalagan, P., and Sahoo, S. (2017). Exosomes-based gene therapy for MicroRNA delivery. Methods Mol. Biol. 1521, 139–152. doi:10.1007/978-1-4939-6588-5_9
McNamara, R. P., Costantini, L. M., Myers, T. A., Schouest, B., Maness, N. J., Griffith, J. D., et al. (2018). Nef secretion into extracellular vesicles or exosomes is conserved across human and simian immunodeficiency viruses. mBio 9, e02344–17. doi:10.1128/mbio.02344-17
Mendt, M., Kamerkar, S., Sugimoto, H., McAndrews, K. M., Wu, C. C., Gagea, M., et al. (2018). Generation and testing of clinical-grade exosomes for pancreatic cancer. JCI insight 3, e99263. doi:10.1172/jci.insight.99263
Merchant, M. L., Rood, I. M., Deegens, J., and Klein, J. B. (2017). Isolation and characterization of urinary extracellular vesicles: Implications for biomarker discovery. Nat. Rev. Nephrol. 13, 731–749. doi:10.1038/nrneph.2017.148
Momen-Heravi, F., Balaj, L., Alian, S., Mantel, P. Y., Halleck, A. E., Trachtenberg, A. J., et al. (2013). Current methods for the isolation of extracellular vesicles. Biol. Chem. 394, 1253–1262. doi:10.1515/hsz-2013-0141
Morales-Kastresana, A., Telford, B., Musich, T. A., McKinnon, K., Clayborne, C., Braig, Z., et al. (2017). Labeling extracellular vesicles for nanoscale flow cytometry. Sci. Rep. 7, 1878. doi:10.1038/s41598-017-01731-2
Mund, T., and Pelham, H. R. (2009). Control of the activity of WW-HECT domain E3 ubiquitin ligases by NDFIP proteins. EMBO Rep. 10, 501–507. doi:10.1038/embor.2009.30
Nakase, I., Niwa, M., Takeuchi, T., Sonomura, K., Kawabata, N., Koike, Y., et al. (2004). Cellular uptake of arginine-rich peptides: Roles for macropinocytosis and actin rearrangement. Mol. Ther. 10, 1011–1022. doi:10.1016/j.ymthe.2004.08.010
Ng, E. W., Shima, D. T., Calias, P., Cunningham, E. T., Guyer, D. R., Adamis, A. P., et al. (2006). Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nat. Rev. Drug Discov. 5, 123–132. doi:10.1038/nrd1955
Ohno, S., Takanashi, M., Sudo, K., Ueda, S., Ishikawa, A., Matsuyama, N., et al. (2013). Systemically injected exosomes targeted to EGFR deliver antitumor microRNA to breast cancer cells. Mol. Ther. 21, 185–191. doi:10.1038/mt.2012.180
Pachler, K., Lener, T., Streif, D., Dunai, Z. A., Desgeorges, A., Feichtner, M., et al. (2017). A Good Manufacturing Practice-grade standard protocol for exclusively human mesenchymal stromal cell-derived extracellular vesicles. Cytotherapy 19, 458–472. doi:10.1016/j.jcyt.2017.01.001
Pascucci, L., Coccè, V., Bonomi, A., Ami, D., Ceccarelli, P., Ciusani, E., et al. (2014). Paclitaxel is incorporated by mesenchymal stromal cells and released in exosomes that inhibit in vitro tumor growth: A new approach for drug delivery. J. Control. Release 192, 262–270. doi:10.1016/j.jconrel.2014.07.042
Pi, F., Binzel, D. W., Lee, T. J., Li, Z., Sun, M., Rychahou, P., et al. (2018). Nanoparticle orientation to control RNA loading and ligand display on extracellular vesicles for cancer regression. Nat. Nanotechnol. 13, 82–89. doi:10.1038/s41565-017-0012-z
Qi, H., Liu, C., Long, L., Ren, Y., Zhang, S., Chang, X., et al. (2016). Blood exosomes endowed with magnetic and targeting properties for cancer therapy. ACS Nano 10, 3323–3333. doi:10.1021/acsnano.5b06939
Raposo, G., and Stoorvogel, W. (2013). Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 200, 373–383. doi:10.1083/jcb.201211138
Raimondo, S., Saieva, L., Corrado, C., Fontana, S., Flugy, A., Rizzo, A., et al. (2015). Chronic myeloid leukemia-derived exosomes promote tumor growth through an autocrine mechanism. Cell Commun. Signal. 13, 8. doi:10.1186/s12964-015-0086-x
Rountree, R. B., Mandl, S. J., Nachtwey, J. M., Dalpozzo, K., Do, L., Lombardo, J. R., et al. (2011). Exosome targeting of tumor antigens expressed by cancer vaccines can improve antigen immunogenicity and therapeutic efficacy. Cancer Res. 71, 5235–5244. doi:10.1158/0008-5472.can-10-4076
Rozhdestvensky, T. S., Tang, T. H., Tchirkova, I. V., Brosius, J., Bachellerie, J. P., and Hüttenhofer, A. (2003). Binding of L7Ae protein to the K-turn of archaeal snoRNAs: A shared RNA binding motif for C/D and H/ACA box snoRNAs in archaea. Nucleic acids Res. 31, 869–877. doi:10.1093/nar/gkg175
Stranska, R., Gysbrechts, L., Wouters, J., Vermeersch, P., Bloch, K., Dierickx, D., et al. (2018). Comparison of membrane affinity-based method with size-exclusion chromatography for isolation of exosome-like vesicles from human plasma. J. Transl. Med. 16, 1. doi:10.1186/s12967-017-1374-6
Sancho-Albero, M., Navascués, N., Mendoza, G., Sebastián, V., Arruebo, M., Martín-Duque, P., et al. (2019). Exosome origin determines cell targeting and the transfer of therapeutic nanoparticles towards target cells. J. Nanobiotechnology 17, 16. doi:10.1186/s12951-018-0437-z
Smyth, T., Petrova, K., Payton, N. M., Persaud, I., Redzic, J. S., Graner, M. W., et al. (2014). Surface functionalization of exosomes using click chemistry. Bioconjug. Chem. 25, 1777–1784. doi:10.1021/bc500291r
Song, H., Liu, B., Dong, B., Xu, J., Zhou, H., Na, S., et al. (2021). Exosome-based delivery of natural products in cancer therapy. Front. Cell Dev. Biol. 9, 650426. doi:10.3389/fcell.2021.650426
Stickney, Z., Losacco, J., McDevitt, S., Zhang, Z., and Lu, B. (2016). Development of exosome surface display technology in living human cells. Biochem. biophysical Res. Commun. 472, 53–59. doi:10.1016/j.bbrc.2016.02.058
Stremersch, S., Vandenbroucke, R. E., Van Wonterghem, E., Hendrix, A., De Smedt, S. C., Raemdonck, K., et al. (2016). Comparing exosome-like vesicles with liposomes for the functional cellular delivery of small RNAs. J. Control. Release 232, 51–61. doi:10.1016/j.jconrel.2016.04.005
Sun, D., Zhuang, X., Xiang, X., Liu, Y., Zhang, S., Liu, C., et al. (2010). A novel nanoparticle drug delivery system: The anti-inflammatory activity of curcumin is enhanced when encapsulated in exosomes. Mol. Ther. 18, 1606–1614. doi:10.1038/mt.2010.105
Théry, C., Witwer, K. W., Aikawa, E., Alcaraz, M. J., Anderson, J. D., Andriantsitohaina, R., et al. (2018). Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the international society for extracellular vesicles and update of the MISEV2014 guidelines. J. Extracell. vesicles 7, 1461450. doi:10.1080/20013078.2018.1461450
Tian, T., Zhang, H. X., He, C. P., Fan, S., Zhu, Y. L., Qi, C., et al. (2018). Surface functionalized exosomes as targeted drug delivery vehicles for cerebral ischemia therapy. Biomaterials 150, 137–149. doi:10.1016/j.biomaterials.2017.10.012
Tian, Y., Li, S., Song, J., Ji, T., Zhu, M., Anderson, G. J., et al. (2014). A doxorubicin delivery platform using engineered natural membrane vesicle exosomes for targeted tumor therapy. Biomaterials 35, 2383–2390. doi:10.1016/j.biomaterials.2013.11.083
Vader, P., Mol, E. A., Pasterkamp, G., and Schiffelers, R. M. (2016). Extracellular vesicles for drug delivery. Adv. drug Deliv. Rev. 106, 148–156. doi:10.1016/j.addr.2016.02.006
van der Meel, R., Fens, M. H., Vader, P., van Solinge, W. W., Eniola-Adefeso, O., Schiffelers, R. M., et al. (2014). Extracellular vesicles as drug delivery systems: Lessons from the liposome field. J. Control. Release 195, 72–85. doi:10.1016/j.jconrel.2014.07.049
Wan, Y., Wang, L., Zhu, C., Zheng, Q., Wang, G., Tong, J., et al. (2018). Aptamer-conjugated extracellular nanovesicles for targeted drug delivery. Cancer Res. 78, 798–808. doi:10.1158/0008-5472.can-17-2880
Wang, J., Li, W., Zhang, L., Ban, L., Chen, P., Du, W., et al. (2017a). Chemically edited exosomes with dual ligand purified by microfluidic device for active targeted drug delivery to tumor cells. ACS Appl. Mat. Interfaces 9, 27441–27452. doi:10.1021/acsami.7b06464
Wang, Y., Chen, X., Tian, B., Liu, J., Yang, L., Zeng, L., et al. (2017b). Nucleolin-targeted extracellular vesicles as a versatile platform for biologics delivery to breast cancer. Theranostics 7, 1360–1372. doi:10.7150/thno.16532
Wei, H., Chen, J., Wang, S., Fu, F., Zhu, X., Wu, C., et al. (2019). A nanodrug consisting of doxorubicin and exosome derived from mesenchymal stem cells for osteosarcoma treatment in vitro. Int. J. Nanomedicine 14, 8603–8610. doi:10.2147/ijn.s218988
Xi, X. M., Xia, S. J., and Lu, R. (2021). Drug loading techniques for exosome-based drug delivery systems. Pharmazie 76, 61–67. doi:10.1691/ph.2021.0128
Xiao, Z., Shangguan, D., Cao, Z., Fang, X., and Tan, W. (2008). Cell-specific internalization study of an aptamer from whole cell selection. Chem. Eur. J. 14, 1769–1775. doi:10.1002/chem.200701330
Ye, Z., Zhang, T., He, W., Jin, H., Liu, C., Yang, Z., et al. (2018). Methotrexate-loaded extracellular vesicles functionalized with therapeutic and targeted peptides for the treatment of glioblastoma multiforme. ACS Appl. Mat. Interfaces 10, 12341–12350. doi:10.1021/acsami.7b18135
Yim, N., Ryu, S. W., Choi, K., Lee, K. R., Lee, S., Choi, H., et al. (2016). Exosome engineering for efficient intracellular delivery of soluble proteins using optically reversible protein-protein interaction module. Nat. Commun. 7, 12277. doi:10.1038/ncomms12277
Yu, X., Yang, X., Horte, S., Kizhakkedathu, J. N., and Brooks, D. E. (2014). A pH and thermosensitive choline phosphate-based delivery platform targeted to the acidic tumor microenvironment. Biomaterials 35, 278–286. doi:10.1016/j.biomaterials.2013.09.052
Yujie, L., Duan, L., Lu, J., and Xia, J. (2021). Engineering exosomes for targeted drug delivery. Theranostics 11, 3183–3195. doi:10.7150/thno.52570
Zhan, Q., Yi, K., Qi, H., Li, S., Li, X., Wang, Q., et al. (2020). Engineering blood exosomes for tumor-targeting efficient gene/chemo combination therapy. Theranostics 10, 7889–7905. doi:10.7150/thno.45028
Zhang, Y., Lai, B. S., and Juhas, M. (2019). Recent advances in aptamer discovery and applications. Mol. (Basel, Switz. 24, 941. doi:10.3390/molecules24050941
Keywords: exosomes, cancer therapeutics, designer exosomes, exosome engineering, parental-cell loading, direct exosome loading
Citation: Dutta A and Paul S (2022) Advancement in exosome-based cancer therapeutics: A new era in cancer treatment. Front. Nanotechnol. 4:939197. doi: 10.3389/fnano.2022.939197
Received: 08 May 2022; Accepted: 08 July 2022;
Published: 04 August 2022.
Edited by:
Abhishek Kumar, Washington University in St. Louis, United StatesReviewed by:
Suchi Gupta, All India Institute of Medical Sciences, IndiaCopyright © 2022 Dutta and Paul. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Abhishek Dutta, YWJoaXNoZWsuZHV0dGFAZXhzdXJlLmlu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.