
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Nanotechnol., 21 January 2022
Sec. Biomedical Nanotechnology
Volume 3 - 2021 | https://doi.org/10.3389/fnano.2021.798928
This article is part of the Research TopicAdvanced Nanomaterials and Nanostructure-Based Sensors for Biomedical ApplicationsView all 5 articles
Since its invention, atomic force microscopy (AFM) has come forth as a powerful member of the “scanning probe microscopy” (SPM) family and an unparallel platform for high-resolution imaging and characterization for inorganic and organic samples, especially biomolecules, biosensors, proteins, DNA, and live cells. AFM characterizes any sample by measuring interaction force between the AFM cantilever tip (the probe) and the sample surface, and it is advantageous over other SPM and electron micron microscopy techniques as it can visualize and characterize samples in liquid, ambient air, and vacuum. Therefore, it permits visualization of three-dimensional surface profiles of biological specimens in the near-physiological environment without sacrificing their native structures and functions and without using laborious sample preparation protocols such as freeze-drying, staining, metal coating, staining, or labeling. Biosensors are devices comprising a biological or biologically extracted material (assimilated in a physicochemical transducer) that are utilized to yield electronic signal proportional to the specific analyte concentration. These devices utilize particular biochemical reactions moderated by isolated tissues, enzymes, organelles, and immune system for detecting chemical compounds via thermal, optical, or electrical signals. Other than performing high-resolution imaging and nanomechanical characterization (e.g., determining Young’s modulus, adhesion, and deformation) of biosensors, AFM cantilever (with a ligand functionalized tip) can be transformed into a biosensor (microcantilever-based biosensors) to probe interactions with a particular receptors of choice on live cells at a single-molecule level (using AFM-based single-molecule force spectroscopy techniques) and determine interaction forces and binding kinetics of ligand receptor interactions. Targeted drug delivery systems or vehicles composed of nanoparticles are crucial in novel therapeutics. These systems leverage the idea of targeted delivery of the drug to the desired locations to reduce side effects. AFM is becoming an extremely useful tool in figuring out the topographical and nanomechanical properties of these nanoparticles and other drug delivery carriers. AFM also helps determine binding probabilities and interaction forces of these drug delivery carriers with the targeted receptors and choose the better agent for drug delivery vehicle by introducing competitive binding. In this review, we summarize contributions made by us and other researchers so far that showcase AFM as biosensors, to characterize other sensors, to improve drug delivery approaches, and to discuss future possibilities.
Understanding the process of biosensing, characterizing biosensors (Pontinha et al., 2011; Kim et al., 2015; Chiorcea-Paquim et al., 2018; Chiorcea-Paquim et al., 2018), measuring interaction forces between ligand–receptor pairs (Sarkar et al., 2019), and utilizing self-assembled nanoparticles and other drug delivery carriers to improve drug delivery approaches are of utmost importance in the field of biomedicine, nanotechnology (Goldsbury and Scheuring 2002; Wu et al., 2002; Kreplak 2016), and biophysics. Atomic force microscopy (AFM) (Lal and John 1994; Parot et al., 2007), with its unique capability to perform high-resolution imaging and characterize organic and inorganic samples in ambient air, liquid, and vacuum, is advantageous over other scanning probe microscopy (SPM) techniques to investigate all the scenarios mentioned above. Other than topographical imaging, AFM (Cuellar et al., 2013; Senapati et al., 2013; Vahabi et al., 2013) offers the opportunity to measure nanomechanical properties like elastic modulus, adhesion, deformation, and dissipation of any substate, and AFM cantilever tip acts a nanoindenter in that case. In addition, AFM probe might be transformed into a specific biosensor to measure piconewton (pN) range interaction forces with particular cellular receptor on live cells with high specificity and high signal-to-noise ratio. AFM-based force spectroscopy (AFM-FS) (Guo et al., 2016) is an outstanding way of understanding the interaction forces and binding kinetics of protein–protein and ligand–receptor interactions on live cells at single-molecule level using protein or ligand functionalized tip. In this review, we summarize how AFM has been utilized to understand the topographical and nanomechanical properties of multiple drug delivery carriers/vehicles as well as to learn about the binding dynamics of the targeted receptor/tissue/organ with these carriers. AFM can also help in selecting a better candidate for drug delivery vehicle by introducing competitive binding. We also shed some light on how AFM has not only been used as a biosensor but also to characterize the biosensors.
Atomic force microscope (AFM) as shown in Figure 1A is a member of SPM family, and it was invented by Binning et al. in 1986 (Binnig et al., 1986). AFM has the capability to yield three-dimensional topographical images of conducting and nonconducting samples in any environment without using costly and time-consuming sample preparation techniques including freezing, drying, tagging the sample with dyes, or using metal coating on the sample like electron microscopy (EM). AFM consists of a probe (flexible cantilever with a sharp tip at the end), piezoelectric scanner, cantilever deflection detecting system or a spilt photodiode, AFM electronics and feedback loop, and cantilever and sample holders (shown in Figure 1B). The characterization of any substrate by AFM (Koehler et al., 2017; Li et al., 2017) is done by measuring the force of interaction between the cantilever tip and sample surface (obtained via measurement of cantilever deflection). The probe is first approached toward the substrate manually, and this is followed by a fine motion of the cantilever tip, controlled by the piezoelectric scanner, and it depends on the force set point chosen by the user. A laser beam is focused at the reflective coating on the back of the cantilever and the beam reflects off to a position sensitive detector (PSD) or split photodiode. When the cantilever tip moves over the hills and valleys of the sample topography, a constant force (chosen by the user) of interaction is maintained between the tip and substrate in AFM contact mode by keeping the cantilever deflection constant. This process is performed by the feedback loop that negotiates the piezoelectric scanner height, recorded by a computer. This information is used to create the topographical image of the sample. As the cantilever tip comes very close to the sample surface, the cantilever deflects due to interaction forces between the cantilever tip and the sample. As a result, the laser spot deviates to a different position on the photodiode, creating voltage difference between four sections of the split photodiode. The voltage difference is used to find out the interaction force acting on the cantilever tip. Obtaining topography data via measuring cantilever deflection is only specific to AFM contact mode. Tapping mode of AFM performs sample topography mapping by gently tapping the substrate with an oscillating AFM cantilever tip, and the oscillation amplitude varies with substrate topography. The topographical information is achieved by measuring these amplitude variations and terminating the z feedback loop to reduce these changes. Because of the several advantages over other contemporary techniques, AFM is an ideal characterization tool to promote a better understanding of the characterization of drug delivery vehicles.
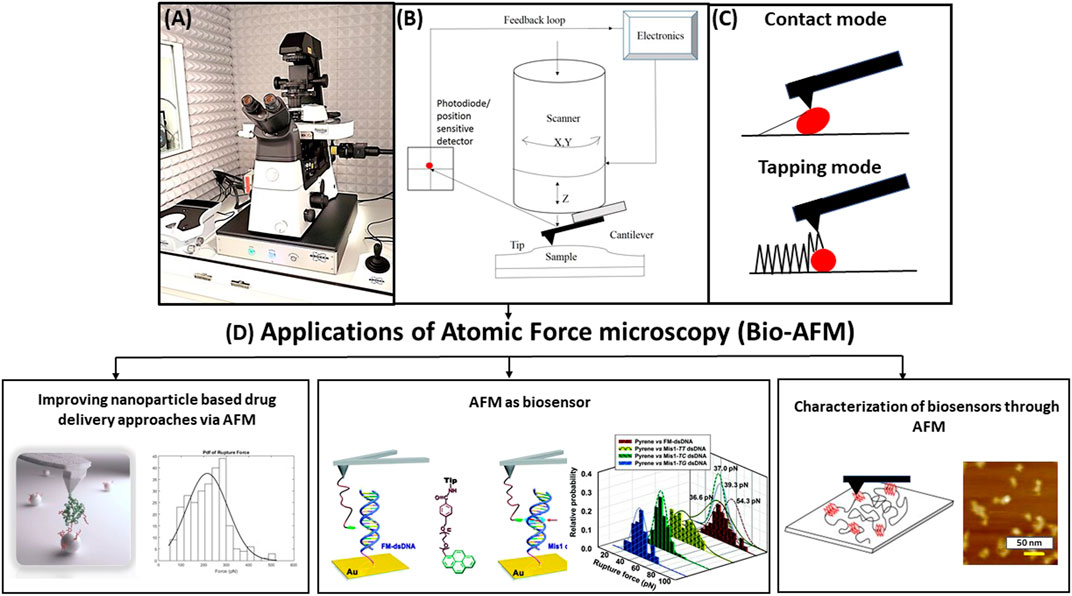
FIGURE 1. (A) Schematic diagram of bio–atomic force microscopy (AFM) setup sitting inside a vibration isolation hood. (B) Basic working principle of AFM showing the major components like flexible cantilever with a sharp tip at the end, piezoelectric scanner, and cantilever deflection detection system, i.e., split photodiode and position sensitive detector, AFM electronics, feedback loop, cantilever, and sample holder. (C) Two basic modes of operation: contact mode (continuous contact of tip and sample) and tapping mode (intermittent tapping by the tip on sample). (D) Applications of bio-AFM to (1) improve nanoparticle-based drug delivery approaches (i.e., measuring the size, shape, and morphology of the drug delivery carriers and finding a suitable carrier by measuring the most probable unbinding force and binding probability (Jones et al., 2017); (2) serve as a biosensor (for example, biosensing of DNA via AFM by measuring most probable unbinding forces between matched and mismatched pairs of dsDNA) (Wei et al., 2013); and (3) characterize the morphology, roughness, and nanomechanical properties of the biosensors (for example, characterization of G-quadruplex electrochemical biosensor was performed using AFM as shown here) (Chiorcea-Paquim et al., 2015).
AFM measurements can be performed in different modes (Dufrene et al., 2017) such as contact mode and tapping/non-contact mode (shown in Figure 1C). The cantilever tip scans across the substrate and the probe is in continuous contact with the sample in contact mode. Changes in the cantilever deflection are tracked via the photodiode detector and a constant deflection is preserved between the probe and the substrate by employing a feedback loop and shifting the piezo scanner vertically. As a result, tip–sample interaction force is also maintained at a constant value in contact mode. Spring constant of the AFM probe can be as low as 0.01 N/m. The force acting on the probe is calculated from Hooke’s law as
where x = cantilever deflection, F = force, and k = cantilever spring constant. AFM has the capacity to measure the deflection of cantilever as low as 0.1 nm, leading to the lowest force measured by AFM as 1 pN. AFM probe is oscillated marginally below its resonance frequency with oscillation amplitudes of approximately 20–100 nm in the tapping mode. The cantilever only intermittently taps the sample at the very bottom of each oscillation swing and the feedback loop maintains a constant amplitude of oscillation in tapping mode. As tapping mode reduces the tip–sample contact, it also reduces lateral forces, preventing both tip and sample from getting damaged. As described above, there are some differences between the two major modes of AFM operation. AFM cantilever tip is in constant contact with the sample in contact mode, whereas the tip is brought close to the sample so that it lightly taps the sample at the bottom end of cantilever oscillation in tapping mode. A constant tip–sample interaction force is maintained in contact mode, whereas a constant oscillation amplitude is maintained in tapping mode. Contact mode is harsher on soft biological samples like live cells and biomolecules due to the presence of high lateral and frictional forces (causing damage to both tip and sample) compared to tapping mode. The difference between tapping mode and PeakForce tapping mode is that the latter is an improved version that provides the user control over the maximum force exerted on the tip and an opportunity to have a turnkey solution for improved imaging and characterization independent of the experience level of the user (by using ScanAsyst auto control of the scanning parameters) (Liu et al., 2018) As PeakForce tapping mode lowers the chances of sample and tip wear significantly, it is an appropriate mode for extracting the topographical information of biosensors, biomolecules, and drug delivery carriers such as self-assembled nanoparticles. Further development and improvement of peak force tapping mode (Li et al., 2021) led to the invention of peak force QNM mode (Sweers et al., 2011; Adamcik et al., 2012; Dokukin and Sokolov 2012) that has the ability of capturing and analyzing individual force curves at each contact point and performing the necessary calculations “on the fly” to produce high-resolution maps of nanomechanical properties of the substrate. For obvious reasons, peak force QNM mode has revolutionized the process of AFM characterization of biological specimens and made it considerably high throughput. Figure 1D demonstrates various applications of bio-AFM (Hoh and Hansma 1992; Shao and Yang 1995; Czajkowsky et al., 2000; Hansma 2001; Goldsbury and Scheuring 2002; Malkin et al., 2002; Alonso and Goldmann 2003; Besch et al., 2003; Gadegaard 2006; Shahin and Barrera 2008; Goldsbury et al., 2009; Ramachandran et al., 2011; Kreplak 2016; Dufrene et al., 2017; Braet and Taatjes 2018; Gao et al., 2018; Cheong et al., 2019; Nandi and Ainavarapu 2021), and we are about to summarize these in this review. Unlike the AFM imaging modes where the probe is scanned over the surface of the substrate, the cantilever tip is first approached toward the substrate until tip–sample contact happens and then retracted in AFM dynamic force spectroscopy (AFM-DFS) (Sulchek et al., 2005; Neuert et al., 2006; Thormann et al., 2006; Diezemann and Janshoff 2008; Alessandrini et al., 2012; Sengupta et al., 2014; Sluysmans et al., 2018; Ju 2019; Reiter-Scherer et al., 2019; Alhalhooly et al., 2021) and single-molecule force spectroscopy (AFM-SMFS) experiments. These two are similar methods with slight differences. AFM-DFS is a procedure utilized for the measurement of binding properties and the force required for manipulating biomolecular complex or biomolecules, whereas AFM-SMFS is a method that provides an unparallel control and sensitivity to manipulate and investigate the nanomechanical properties of single molecules. The difference between the two AFM force spectroscopy modes mentioned above and chemical force microscopy (CFM) is that CFM employs a chemically modified AFM cantilever tip to gauge the spatial distribution of various chemical groups on the substrate. Recognition AFM is quite different from the modes mentioned so far as it utilizes a magnetic cantilever that is regulated by an external magnetic field and oscillated in liquid. All these modes will be described in more details in the following sections of this review paper.
“Recognition AFM” or recognition imaging has facilitated usage of AFM (Stroh et al., 2004; Ebner et al., 2005; Hinterdorfer and Dufrene 2006; Tang et al., 2007; Dufrene and Hinterdorfer 2008; Takahashi et al., 2009; Chtcheglova and Hinterdorfer 2018; Koehler et al., 2019) as biosensor and enhanced its performance as a “characterization and detection tool” of biomolecules. A magnetic cantilever, stimulated by an external magnetic field, is oscillated in liquid in topography and recognition imaging (TREC) as shown in Figures 2A–B (Ebner et al., 2005). Loss of energy from the downswing to the upswing and back and hydrodynamic drag on the AFM probe make the quality factor of the oscillation quite low. As a result, the force due to magnetic field on the cantilever becomes the major force. Generally, an antibody or aptamer attached to the cantilever tip via a bifunctional PEG linker is used as the recognition element. The PEG linkers with long length are chosen for this purpose to reduce non-specific adhesion and increase degree of accessibility to the ligand. The amplitude of oscillation on the downswing decreases when the antibody or aptamer functionalized tip interacts with the sample features. This reduced amplitude is not carried over to the upswing. On the other hand, recognition events cause a reduction of the upswing, and the downswing remains unaffected. Therefore, the resulting signal vs. time plot can be divided in half and converted in an image with the upper part disclosing locations of recognition events and the lower part yielding the topographical image as demonstrated clearly in Figures 2C–D (Ebner et al., 2005).
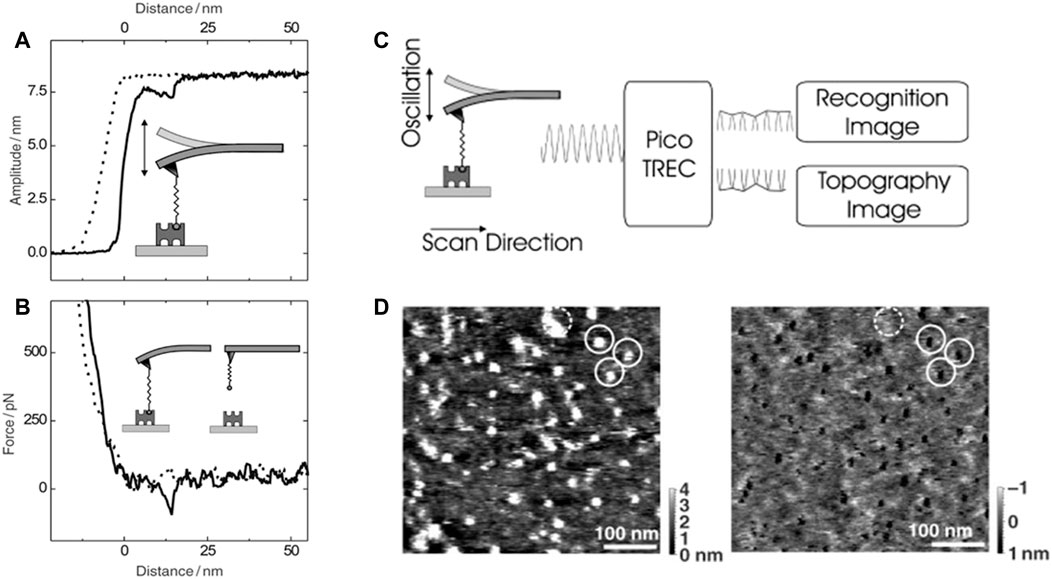
FIGURE 2. (A–B) Amplitude–distance and force–distance cycle demonstrating a single rupture of avidin–biotin bond. (A) A single rupture event was observed at 14 nm in the amplitude-distance cycle causing amplitude to decrease by ≈ 1 nm. (B) The same rupture event was simultaneously observed in the force–distance cycle with rupture force of ≈80 pN. (C) Principle of TREC imaging: The cantilever oscillation comprises of lower and upper parts, yielding topography and recognition images simultaneously. (D) Mica substrate was functionalized with Avidin and imaged with a biotin functionalized AFM cantilever tip. Bright dots in the left topography image is well correlated with dark spots in right recognition image; scan size, 500 nm; spots within the dashed circle in the topographical image represent structural components and devoid of specific interaction (Ebner et al., 2005).
This review comprises of five major sections including this one (Section 1: introduction). Section 2 summarizes how AFM can be utilized to improve drug delivery approaches. Section 3 focuses on how AFM itself can act as a biosensor for sensing DNA, RNA, drug molecules, proteins, and so on. Section 4 explains the working principle of peak force QNM mode of AFM that has improved the nanomechanical characterization of biomolecules. This section also summarizes the characterization of biosensors, explored using AFM. In Section 5, we discuss the possible improvements and future steps that can make bio-AFM more effective and high throughput.
Any novel therapeutic strategy to treat and cure diseases faces a significant challenge due to lack of appropriate drug delivery systems (DDSs) and improper delivery of the drug/therapeutics (Garg and Kokkoli, 2005; Parot et al., 2007; Lamprecht et al., 2014) to the organ or tissue of interest. An ideal DDS should be able to maintain desired drug levels for long time, and protect biologically active DDSs’ constituents (proteins and peptides) from degrading. Using a smaller amount of drugs with a reduced number of dosages is also another quality, an ideal DDS should possess. The type of DDS is usually chosen depending on the target cell surface receptor or organ or tissue and the structure of the therapeutics being delivered. The categories of carriers for these systems include cellular carriers (live cell and virus), particle carriers (microspheres, polymeric micelles, nanoparticles, liposomes, solid lipid particles, and polymeric systems), and soluble carriers like peptides, proteins, and polysaccharides (Smith et al., 2018). Among these abovementioned carriers, particle-based carriers have great potential. These are more efficient than the other ones due to the perfect incorporation of the drug within these carriers maintaining the drug’s efficiency and stability. A wide variety of nanocarriers like polymeric system, micelleplexes, carbon nanospheres, porous silica, and graphene are crucial in delivering drugs to designated places even with poor solubility and permeability. The release mechanism of these carriers varies from rapid, slow, and local to targeted delivery. Targeted drug delivery approaches are in the spotlight when it comes to novel therapeutic strategies. These targeted strategies have leverage over other medical strategies as these can prevent side effects and toxicity of chemotherapeutics by avoiding drug release to unaffected or healthy cells. Targeted drug delivery can significantly enhance the drug concentration reaching the target tissue or cell and increasing therapeutic response and therapeutic index. We summarize the research studies that employ AFM to properly characterize drug delivery carriers for optimal usage of carriers in specific scenarios.
Measurement of the particle size involved in drug delivery can be performed by AFM more accurately as it enables high-speed and high-resolution (in nanometer range) imaging of the drug delivery vehicles (Garg and Kokkoli 2005) in its native liquid environment. A lot of improvement has been done to make AFM high speed. High-speed AFM (HS-AFM) (Ando 2019) can visualize dynamic events of soft biological samples occurring in liquid microenvironments and identify morphological variations occurring inside live cells. In HS-AFM, all components are optimized for their prompt response and vibrations caused by the fast scanning of the AFM sample stage are efficiently decreased. Among HS-AFM components, z piezo scanner and the cantilever have a slower response as these are mechanical devices. Commercially available short cantilevers are chosen for shortening the response time of the cantilevers to make AFM high speed. AFM does not require the usage of fitting functions or complicated prolonged sample preparation techniques. AFM modes such as tapping mode, and Peak Force QNM modes are nondestructive to the sample as they reduce the lateral forces significantly by intermittently tapping the samples. Liposomes (Storrs et al., 1995), one of particle type carriers of DDSs appeared as flat ellipsoids in AFM imaging performed by Storrs et al. and size of the liposomes (Spyratou et al., 2009), determined using AFM aligned with the results obtained via transmission EM (TEM). The size and diameter of the liposomes (Kanno et al., 2002) conjugated with a transmembrane protein (vesicles) were accurately determined by Kanno et al. using AFM (diameters: 550–1,500 nm and height: 1.8–2.5 nm) as demonstrated in Figures 3A–D (Kanno et al., 2002). AFM topographical imaging also revealed flattened shape of these vesicles upon their physical adsorption on the freshly cleaved mica substrate. Solid lipid nanoparticles (SLNs), another particle type carrier of DDSs, were successfully imaged using AFM showing flattened nanoparticles (Shahgaldian et al., 2003a). Dubes et al. showed via AFM imaging that SLNs (Dubes et al., 2003) derived from amphiphilic cyclodextrins were circular in shape and found in clusters of 15–30 particles (diameter: 359 ± 50 nm, height: 140 ± 27 nm) as demonstrated in Figure 3E–G (Dubes et al., 2003). AFM measurements of size and diameter of these nanoparticles were reported by Montasser et al. and proved to be more accurate than DLS and TEM measurements, revealing bimodal distribution of the nanoparticle population (Montasser et al., 2002) in terms of size and diameter. Limitations of AFM characterization include cantilever tip induced activation of cytoskeleton proteins such as talin, vinculin, and a resolution limit of 50 nm of the nanoparticles inside live cells.
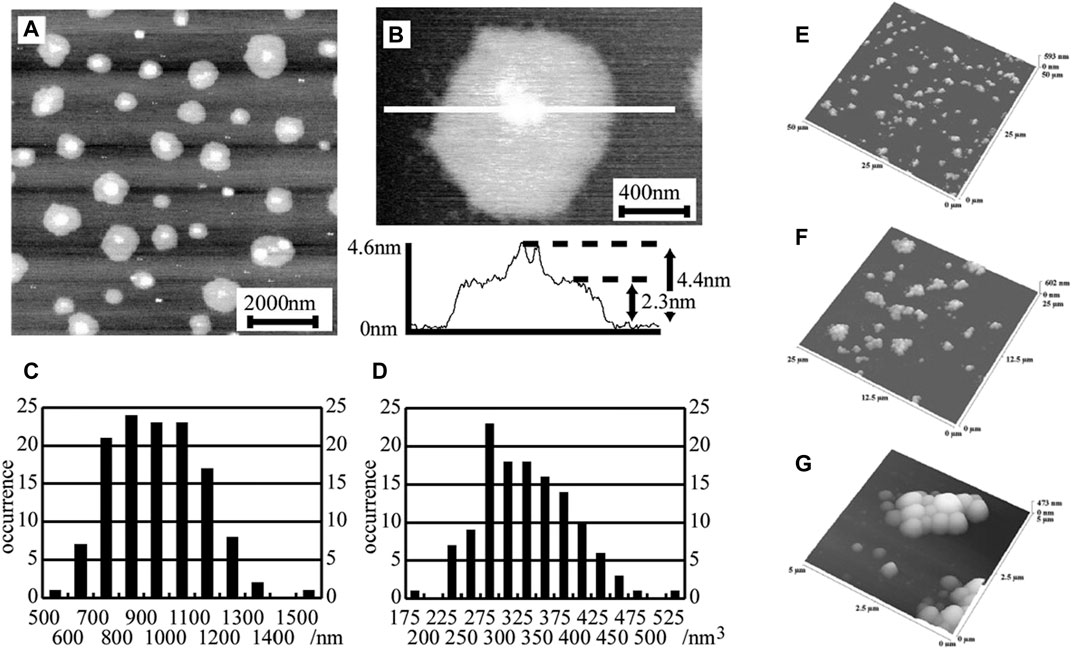
FIGURE 3. AFM images for the of HBsAg L particles plated on mica surfaces. (A) With scale of 10,000 × 10,000 × 5 nm and (B) with scale of 2,000 × 2,000 × 4 nm. The section analysis of AFM demonstrates a flattened disc shaped vesicle comprising of a lipid monolayer with height of 1.8–2.5 nm. The central white dot represents HBsAg L protein with height of 4–18 nm. (C) Histogram of the diameters of the flattened structures measured from AFM images. (D) Histogram of the size of the vesicles calculated from the diameters of the flattened disc structures (Kanno et al., 2002). (E–G) Non-contact mode AFM images of the SLNs (β-CD21C6 derived) at scan ranges of (E) 50 μm, (F) 25 μm, and (G) 5 μm (Dubes et al., 2003).
AFM is not only helpful for accurate size measurement but also important to reveal the structure and shape of these nanoparticles. Characterizing particle shape is crucial as it can affect carrier degradation and transport pathways to deliver drugs. Zhang et al., Gupta et al., and Dong et al. demonstrated that SLNs had spherical shape as individual particles and existed as clusters (Dong and Feng 2004; Gupta and Wells 2004; Zhang et al., 2004) when they formed complexes. The effect of shell cross-linking on the structure and shape of the nanoparticles was demonstrated by Qi et al., and the nanoparticles demonstrated less deformation upon cross-linking (Qi et al., 2004). Cationic liposomes or SLNs (promising gene delivery vehicle) formed bonds with DNA and Almofti et al. investigated the morphology of these complexes by varying liposome: DNA charge ratio to observe the proper encapsulation of DNA (Almofti et al., 2003). Efficiency of DDSs employing nanoparticles as carriers was investigated using both experimental (via AFM) and computational approaches by Ramezanpour et al. (Ramezanpour et al., 2016). Viscosity-independent diameter measurement of an antimycotic nanomedicine (AmBisome) was performed by Watanabe et al. using AFM (Watanabe et al., 2018). AFM tapping mode imaging by Zhang et al. revealed that intestinal mucous-penetrating core–shell nanocomplexes were spherical in shape and diameter varied in the range of 100–300 nm (Zhang et al., 2018). Masarudin et al. performed AFM imaging of the chitosan nanoparticles (CNPs) to yield the morphological information of shape, size, and diameter. Diameter of these particles was measured to be 45 nm (Masarudin et al., 2015). Loaded SLNs with anticancer drug and empty SLNs were imaged using AFM, and their sizes (Akanda et al., 2015) were determined as 169.8 ± 21.0 and 117.8 ± 11.0 nm, respectively, by Akanda et al. AFM measurements (Cai et al., 2016) of size, shape, and diameter of these particles turned out to be more accurate than results from the other contemporary techniques as AFM sample preparation was less time consuming, samples were not dehydrated, and the results did not suffer from inaccurate or higher values due to metal coating on samples.
Nanoparticles such as nanocapsules, nanosized emulsions, and nanospheres, used for drug delivery are generally less than 200–500 nm in size. Their multiple traits, i.e., porosity, surface roughness, and shape, make them ideal candidates for precise delivery. Eaton et al. performed a direct comparison of AFM, TEM, and SEM results regarding nanoparticle shape, size, and diameter (Eaton et al., 2017). Tapping mode AFM imaging was used by Bazylińska et al. to pursue characterization of PEG-ylated nanocarriers, encapsulating both hydrophobic thiazole dye and hydrophilic DNA (Bazylinska et al., 2017). Successful encapsulation of a P13K inhibitor, buparlisib, in nanoparticles for treating leukomina was observed using AFM (Bousmail et al., 2017). Through these studies, nanoparticles in size range of 100–200 nm were found to be efficient in encapsulating hydrophilic drugs to hinder tumor growth (Arora et al., 2017). The size, shape, and structures of black phosphorus nanoparticles were explored with AFM revealing the fact that these nanoparticles possessed a platelet-like structure with sizes varying in the range of 100–500 nm. For drug delivery in ovarian cancer cells, these nanoparticles were functionalized with oxaliplatin and cisplatin (Caporali et al., 2017).
Roughness of the substrate is an important aspect of drug delivery carriers’ characterization as it can shed light on the drug release mechanism. AFM provides a trustworthy measurement (Smith et al., 2003) of the surface roughness as the measurement is usually performed in tapping mode or PeakForce QNM mode where the probe is not in continuous contact with the sample, preventing and lowering any chance of damage, modification, and degradation of the sample due to lateral shear forces. Roughness expressed as arithmetic roughness average (Ra) is quantified via calculating the deviation of height from a mean height by drawing a line across the AFM images. Masking of any significant traits of the sample surface can also be avoided in AFM by using simpler sample preparation steps devoid of coating with metal, freezing, and tagging with dyes used in other microscopy techniques. For example, polymeric nanoparticles have been imaged by SEM, showing a smooth surface of these particles, whereas AFM imaging with smaller length scale showed cracks and caves on the surface (Kumar et al., 2015; Crucho and Barros 2017). These features are clear indications of diffusive drug release mechanism. The roughness of both silver nanoparticles coated with anticoagulant and stabilizing agent (chitosan) and uncoated silver nanoparticles were simultaneously investigated with AFM yielding particle diameters of 160 and 30 nm, respectively (Kim et al., 2015). Reduction in roughness values was observed with higher chitosan concentration, possibly due to smooth coating formation. Silver nanoparticles used as drug delivery carriers to target A549 lung cancer cells were successfully investigated using both AFM and TEM (Shimpi and Jha 2017). Roughness measurement of silk nanoparticles, used for drug delivery was performed by Kumar and Singh et al. with AFM (Kumar and Singh 2017). Shahgaldian et al. performed surface roughness measurements of gel dispersed SLNs (used for topical medicine) and discovered the existence of SLNs as non-aggregated structures (Shahgaldian et al., 2003b). AFM has also been utilized to study the effects of ionization radiation on surface roughness of microsphere (particle type drug delivery carrier) and non-irradiated microspheres appeared to be smoother in AFM images (Montanari et al., 2003).
PeakForce QNM mode is based on peak force tapping mode of AFM and has unprecedented power to determine quantitative nanomechanical properties like adhesion, stiffness or elastic modulus, deformation, and dissipation of conducting as well nonconducting samples specially biomolecules, live cells, and so on. PeakForce QNM mode can help with the characterization of biological samples like tracking particles of interest via adhesion mapping, diagnosing cancer using stiffness values of live cells, and fathoming cellular responses to drugs and quantifying it. Reggente et al. successfully examined and identified stiffer magnetic nanoparticles embedded inside soft tissue, facilitating improvement in drug delivery approaches, and nanotoxicology (Reggente et al., 2017). AFM has also been used to perform nanomechanical property characterization of platelets on carbon nanocoating to demonstrate the variation of nanomechanical properties of platelets on nanomaterials (Karagkiozaki et al., 2010).
Other than high-resolution imaging, roughness measurement, and nanomechanical property characterization, AFM is also extremely advantageous for the measurement of interaction forces, and important parameters of binding kinetics (binding probability, most probable rupture force, association and dissociation rates, and affinity) for protein–protein or ligand–receptor interactions on live cells at single-molecule level. For force measurements in protein–protein interactions, AFM cantilever tip (functionalized with one of the proteins) is initially brought close to the substrate (functionalized with the other protein) so that the proteins have the opportunity to bind to each other. Afterward, the cantilever is retracted with constant speed to observe the minimum force (unbinding or rupture force) that is required to break the complex bond. Keeping in mind that unbinding force is a stochastic variable (due to thermally activated process of dissociation of the complex bond), experimentalists have to perform multiple sets of a large number (e.g., 1,000) of force curves on varying locations to obtain statistically sound results. Any set of these (e.g., 1,000) force curves performed with functionalized cantilever tip on live cells/functionalized substrate will contain single rupture events, double rupture events, multiple rupture events, and no rupture events. The most probable rupture force is determined from the peak of the distribution or histogram of the specific single rupture events. By dividing the number of force curves showing at least one bond rupture event by the total number of force measurement trials in a set, binding probability is obtained.
AFM-FS (Stroh et al., 2004; Hinterdorfer and Dufrene 2006; Tang et al., 2007; Dufrene and Hinterdorfer 2008; Takahashi et al., 2009) is crucial due to its wide range of applications (Mayyas et al., 2010). Other than determining the binding kinetics and interaction forces, AFM-FS has also been utilized to understand the unfolding and folding of nucleic acids with multiple repeated units and single proteins, as well as to study molecule–material interface and molecule–molecule interactions. AFM-FS has also been helpful to understand the cellular mechanics. Quantitative analysis and detection have also been possible using AFM-SMFS techniques (Mayyas et al., 2010). Polyproteins were first investigated by the Dougan group (Hughes and Dougan 2016) using AFM-SMFS techniques (employing both force-clamp and force-extension modes). Schönherr et al. (Schönherr et al., 2000) investigated supramolecular host–guest interactions using AFM-SMFS technique in liquid and measured unbinding forces between ferrocene moiety functionalized cantilever tip and β-cyclodextrin receptor functionalized self-assembled monolayers coated on gold substrate. Recent research studies to characterize G protein–coupled receptors were also executed using AFM-SMFS by the Muller group (Sapra et al., 2019). The function and structure of cellular membranes, explored with AFM-SMFS techniques was reported by the Wang group (Shi et al., 2018). Furthermore, AFM-SMFS has also been utilized to imagine proteins, live cells, DNA, antibodies, bacteria, and multiple drug delivery carriers.
Self-assembled nanoparticles or polymeric nanomaterials have been heavily employed to transport drug molecules or agents securely to specific locations. Apprehending the binding dynamics of these nanoparticle-based carriers with targeted receptors on live cells at single-molecule level is significantly challenging. AFM, with its capacity to image with sub-nanometer resolution and probe these nanoparticles and cellular receptors with pN resolution, can be a great tool in drug design and drug delivery. AFM cantilever tip can be coated with the particular targeted cellular receptor, and the substrate can be plated with polymeric nanoparticles (used as drug delivery carrier) or vice versa. AFM is a convenient tool to monitor the response of live cells to drugs molecules and to find out a better drug delivery agent by implementing competitive binding. Jones et al. (Jones et al., 2017) measured interaction forces between folate receptor alpha or FRα and polymeric nanoparticles linked to its ligand, folic acid (FA) as demonstrated in the schematic diagram in Figure 4A. FRα is an overexpressed receptor in most diseases like fibrosis and cancer. Thus, it appears as a promising target for the targeted drug delivery approach. In this study (Jones et al., 2017), the nanoparticles consisted of micelles linked to FA and attached with triblock copolymer: PEI-g-PCL-b-PEG-FA, and these nanaoparticles were imaged using tapping mode (Figures 4B–C). The interaction forces and binding kinetics between these small interfering RNA–tagged nanoparticles and FRα were explored using bio-AFM in comparison with free FA. The binding probability was 0.462 and 0.573 for FA functionalized substrate and the micelleplexes functionalized substrates, respectively, whereas the cantilever was coated with folate receptors. The most probable rupture force was 78.6 and 215.8 pN for those two cases, respectively, as evident from Figures 4D–E. The significant difference in most probable rupture forces was caused by the formation of multiple bonds on the folate-decorated micelleplexes. By combining the results of AFM and flow cytometry, this study concluded that multivalent micelleplexes formed bonds with FRα with a greater binding force and binding probability than monovalent FA. To observe if free FA addition can outcompete the binding of FRα to micelleplexes, free FA was injected repeatedly in the flow cell. Figure 4F demonstrates the binding probability vs. the concentration of injected FA. The results of this study strongly supported the notion that multivalent nanoparticles had a greater receptor binding and high concentration of monovalent ligand could not outcompete that. This study concluded that injection of high concentration of competing ligand (FA) disrupted the stability of micelleplexes.
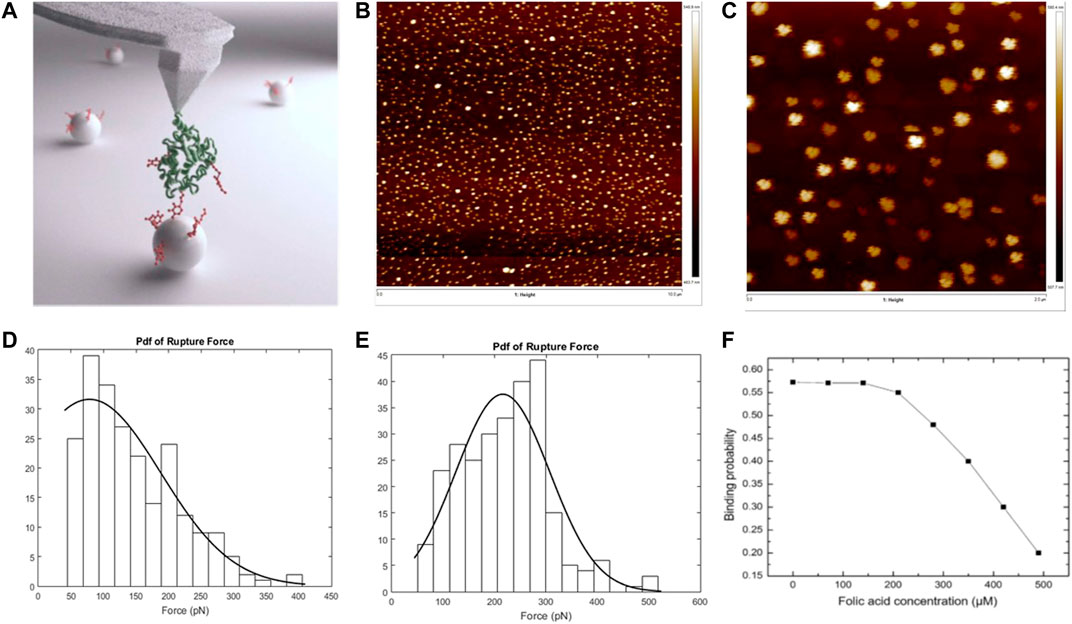
FIGURE 4. Atomic force microscopy in understanding the binding dynamics of the folate receptor alpha (FRα) with self-assembled nanoparticles (micelleplexes) vs. free folic acid. (A) Graphical abstract representing interaction between AFM cantilever tip functionalized with FRα and substrate functionalized with folic acid conjugated nanoparticles. (B) Tapping mode AFM images of micelleplexes: topographical image of micelleplexes on a glass coverslip of 25 × 25 mm; scan size, 10 μm. (C) Zoomed in version of the topographical image shown in (B); scan size, 2 μm. (D) Rupture force histogram plotted for substrate functionalized with free folic acid and tip functionalized with folate receptor. (E) Rupture force histogram for substrate functionalized with folate-decorated nanoparticles and tip functionalized with folate receptor. (F) Binding probability versus concentration of folic acid after injecting folic acid in the flow cell with the substrate, functionalized with folate-decorated nanoparticles and cantilever tip functionalized with folate receptor (Jones et al., 2017).
Biosensors (Caballero et al., 2003) have three constituents: a delivery system for sample, a sensor device implementing various optical and physical principles, and a substrate for the sensor (for immobilization of one of the interaction partners). AFM-SMFS has been utilized in biosensors (based on molecular recognition) meant for RNA, protein, DNA, drug molecules, enzyme, and antibody–antigen. Implementation of HS-AFM scanning, high-resolution imaging, and force spectroscopy techniques provide opportunities to image biomolecules and bionanostructures (Parot et al., 2007) like antibody, protein, DNA, RNA, virus, and live cell, as well as to study molecular interactions and develop label-free biosensors (Li et al., 2016). Although AFM is advantageous as a label-free technique, the combination of AFM and fluorescence microscopy is sometimes employed to investigate specific interactions like protein–DNA interactions. While AFM is excellent in shedding light on structural details of biomolecules and similar substrates, it faces challenges to differentiate among particles of similar sizes in a complex system. Fluorescence imaging techniques can readily access this information using site-specific fluorescent tags, enabling detection of multiple constituents simultaneously. A combination of these two techniques may be useful in these cases. Fluorescence microscopy has been successfully combined with AFM-SMFS (Steffens et al., 2012) to detect molecules of interest on the cantilever tip and substrate, study biomolecular interaction on the cellular surface, and to detect drug and measure the density of receptors in ligand–receptor interactions at single-molecule level on live cells.
Because of its high-resolution, specificity, and non-destructive nature, Recognition AFM has been successfully applied as a biosensor to investigate and detect proteins, DNA, RNA, and live cell membrane proteins. Detection of the toxin and ricin attached to a gold substrate at single-molecule level was performed by Chen et al. using an anti-ricin antibody–coated cantilever tip (Chen et al., 2009) and implementing the Recognition AFM approach. The binding strength of antibody-ricin interaction was measured as 64.89 ± 1.67 pN using DFS. On the basis of the binding strength and koff values measured by AFM, DNA aptamer was proved to be a more efficient sensing molecule than the antibody in a follow-up study (Wang et al., 2012) conducted by Wang et al. AFM-based biosensing is advantageous for many reasons such as obtaining data with single-molecule specificity. For AFM-based biosensors, reducing non-specific interactions is critical as these interactions can result in false positives. Prior to modification, coating the substrate with an anti-fouling layer (Sharma et al., 2004) has also been performed by Sharma et al. to reduce non-specific adhesions.
CFM is an advanced variant in the AFM family that has the objective to study multiple aspects of chemical and structural details of molecules and polymers and their effect on the chemical reactions between the substrate and AFM probe. In CFM (Clear and Nealey 1999; Takano et al., 1999; Okabe et al., 2000; Fiorini et al., 2001; Kreller et al., 2002; Brewer and Leggett 2004; Gourianova et al., 2005; Cameron et al., 2006; Dague et al., 2007; Dufrene 2008; Foster et al., 2009; Sirghi et al., 2009; Barattin and Voyer 2011; Teobaldi et al., 2011; Alsteens et al., 2012; Picas et al., 2012; Beaussart et al., 2014; Hibino and Nakano-Nishida 2014), cantilever tips are chemically functionalized with particular functional groups for performing specific functions in a system. CFM relies on two approaches: microcantilever-based biosensors (MC-B) (Subramanian and Catchmark 2007; Lang and Gerber 2008; Gruber et al., 2011; Johnson and Mutharasan 2012; Peiner and Wasisto 2019; Bennett et al., 2020; Lee and Lee 2020; Mamou et al., 2021) and nanomechanical cantilever sensors (NCS) to achieve these goals of characterizing and understanding the system at single-molecule level with high reproducibility and repeatability. The microcantilever-based biosensing utilizes the specific binding of biomolecules for analytical sensing. These biosensors are advantageous due to their capability of label-free detection and small sizes (dimensions are roughly 100 × 30 × 0.6 μm). MC-Bs (Zhang et al., 2018; Zhao et al., 2019) are physical sensors made of silicon that react to the variation in surface stress as a result of biological or chemical processes. If manufactured with small force constants, then these sensors can yield high sensitivity while measuring surface stress and forces. Assimilation of molecules on any surface of these MC-Bs causes a differential surface stress as the adsorption-induced forces change cantilever deflection. The resonant frequency of the cantilever also varies due to mass loading. Adsorption-induced change in cantilever bending and variation in resonant frequency can both be monitored simultaneously through these sensors. As explained in the introduction of this review, the cantilever deflection is usually measured by laser spot movement in PSD and voltage difference among sections of the diode caused by that. As MC-Bs require accurate measurement of the surface deflections, the cantilever bending may be measured via a deflection sensor embedded into the cantilever. MC-Bs have been applied in titration-AFM to obtain adhesive forces and apparent elastic modulus, to detect different chemical groups, and to study specific interactions.
The working principle of a NCS relies on the adsorption of analytes on the cantilever surface (coated with a sensing layer). This absorption results in induced surface stress that causes an increase in the apparent mass of the cantilever. Upon functionalization of the cantilever with sensitive materials like polymers, metal, and enzymes, the specific interactions (reversible or irreversible) between the sensitive coating and analyte molecules of interest transform the cantilever into a sensitive biosensor. For reversible interactions, the interaction of the analyte with the sensor surface yields a response and sensor retains its original state when the analyte molecules are withdrawn. In case of irreversible interactions, the sensor materials catalyze the chemical reaction between the analyte and sensor surface, and the analyte gets internalized. Major controllers of sensitivity (produced by irreversible interactions) are the degree of uniformity of the sensor surface coating and external stimuli affecting the molecular reorganization. Few studies where NCS (Fagan et al., 2000; Mertens et al., 2019; Peiner and Wasisto 2019) was used efficiently to achieve this goal are mentioned here. For example, adsorption of gas can cause bending and deflection of functionalized cantilever. Thundat et al. (Thundat et al., 1994a; Thundat et al., 1995) investigated the absorption of water vapor in an inorganic sensing layer by functionalizing one side of silicon cantilever with a various hygroscopic thin films. This study demonstrated that cantilevers coated with phosphoric acid (H3PO4) caused a reduction of resonant frequency with reduction of relative humidity, but usage of cantilevers functionalized with gelatin film increased the resonant frequency. Detection of several gases and volatile organic compounds can be achieved by functionalizing cantilevers with polymer coatings. The study by Then et al. (Then et al., 2006) showed that these polymeric coatings on the probe surface absorbed the analyte molecules causing the polymer matrix to swell up and yielding varied cantilever stress. Electrochemical AFM (EC-AFM) (Dos Ramos et al., 2015; Zhang et al., 2015; Ma et al., 2020) is another crucial mode that enables AFM measurement using a non-conducting cantilever tip during electrochemical reactions, whereas AFM does not take part in electrochemical reactions. The oxidation reactions occurring at the anode and the reduction reactions occurring at the cathode can both be studied using a special electrochemical cell with EC-AFM. Real-time effects of electric fields on the chemical performance and interaction forces of the target biochemical system have been effectively explored using EC-AFM–based DFS (Zhang et al., 2015) in multiple studies.
It is crucial to implement efficient methods for RNA detection as RNA is critically important in decoding, expression, and regulation of genes. Hepatitis C virus (HCV) RNA was successfully detected by Jung et al. (Jung et al., 2013) using AFM-SMFS techniques. In this study, the detection DNA was covalently conjugated to the top of the dendron (already immobilized on the AFM cantilever tip), and the capture probe DNA was covalently conjugated to the top of the dendron, immobilized to the glass slide. Following proper hybridization of HCV RNAs to the capture DNA, the interaction forces were measured between the detection DNA connected to the AFM tip and captured RNAs using AFM. By utilizing AFM-SMFS–based force mapping and analyzing force–distance curves, RNA sample was detected with a concentration of even 5 fM.
As DNA (Thundat et al., 1992a; Thundat et al., 1992b; Thundat et al., 1994b) matching and mismatching will result in different unbinding/rupture forces, AFM-SMFS may be adopted as a DNA sensor. Albrecht et al. (Albrecht et al., 2003) established a force-based programmable biosensor for the first time using this approach by substituting the cantilever spring with a polymeric anchor and a known fluorescently labeled molecular bond. Once the cantilever was retracted from the substrate, the two surfaces were separated causing extension of the polymeric anchor, rupture of the complex bond, and release of the fluorescent label. This technique identified single-stranded DNA (ssDNA) match and mismatch with enhanced sensitivity. Interaction forces between pyrene functionalized cantilever tip and matched or mismatched double-stranded DNA (dsDNA) (attached to gold substrate) were measured by Jiang et al. (Jiang et al., 2010). This study clearly demonstrated that rupture/unbinding forces were reduced due to the occurrence of mismatched sites during pyridine–dsDNA interaction, leading to new pathways to identify dsDNA mismatch. In another study conducted by Wei et al. (Wei et al., 2013), a particular ssDNA oligomer sequence was successfully detected using the label-free, force-based sensing mentioned above and by comparing rupture force between ssDNA with complementary ssDNA vs. the rupture force between ssDNA and graphite substrate as evident from Figures 5A–G (Wei et al., 2013). As a control experiment, AFM cantilever tip was functionalized with a particular sequence of ssDNA (D1) and the rupture/unbinding force was measured between the modified tip and graphite substrate in DI water. D1-decorated cantilever tip was incubated in 1 nM complementary sequences of target ssDNA (cD1) and a mismatched ssDNA (mD1), leading to hybridization and mis-hybridization, respectively. The results of this study showed that average values of plateau force for the hybridized DNA (D1 + cD1) reduce to approximately 35.5% with no significant variation of plateau force of the mis-hybridized DNA (D1 + mD1) compared to that of D1.
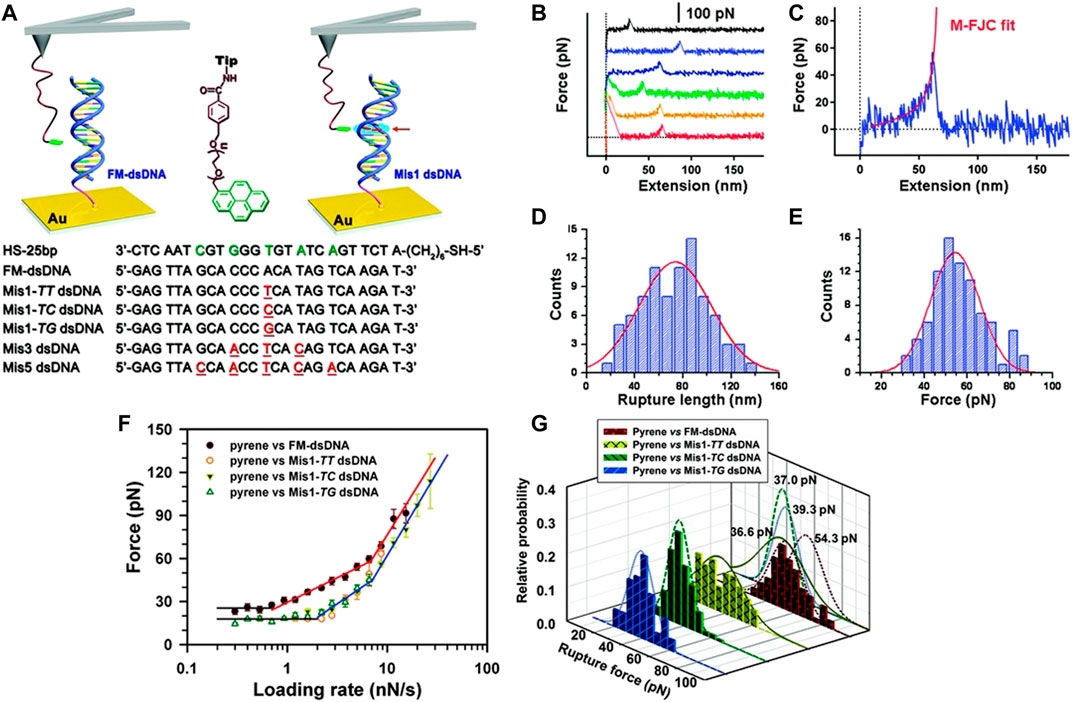
FIGURE 5. (A) Schematic diagrams of the process of interaction force measurement between dsDNA and pyrene using AFM single-molecule force spectroscopy (SMFS). The red arrow represents the mismatched site. All the oligonucleotide sequences utilized in the assay are mentioned at the bottom the red underlining represents the mismatched bases. (B) Force curves produced with pyrene functionalized cantilever tip and FM-dsDNA functionalized Au substrate at room temperature. (C) M-FJC model fitting was performed with the force curve. (D) All the rupture lengths of force curves were accumulated to generate a histogram and yield the most probable rupture length as 73 (±30) nm. (E) The most probable rupture force was calculated from the peak of rupture force histogram as 54.3 (±12) pN. (F) Logarithm of the most probable rupture forces for interactions between pyrene–FM dsDNA and pyrene–Mis1 dsDNA vs. loading rates are plotted here. (G) Rupture force histograms of the interaction forces and corresponding Gaussian fits between pyrene and the four different types of dsDNA: Mis1–TT dsDNA (solid), FM dsDNA (short dashed), Mis1–TC dsDNA (dashed), and Mis1–TG dsDNA (dotted) (Jiang et al., 2010).
Detection of proteins is an important function of AFM-based biosensors. A force-based protein biochip was invented by Blank and co-workers (Blank et al., 2003). Single-molecule binding forces were quantified using an assay. The assay was based on the measurement of differential unbinding forces by comparing interaction forces of DNA hybrids with ligand–receptor interactions. DNA zippers allowed to differentiate between non-specific and specific interactions and detect antibodies and proteins with high sensitivity. AFM-SMFS was also efficiently used by Wei et al. (Wei et al., 2013) for the identification of lysozyme with high sensitivity. In this study, anti-lysozyme aptamer (D2) was fixed to the cantilever tip, and the unbinding forces between a SiO2 substrate and lysozyme modified AFM probe (500 pN) were much higher compared to the forces between SiO2 substrate and D2-conjugated AFM probe (70 pN). This significant difference in unbinding forces served as the identification markers of lysozymes. Sarkar et al. (Sarkar et al., 2019) isolated DDR1–collagen interactions on live cells and quantified dissociation and association rates of the protein complex. DDR is an overexpressed cell surface receptor in many types of cancers. This study estimated different binding probabilities for collagen I–DDR binding and collagen I–integrin binding and determined the density of receptors on live cells using AFM.
Identification and recognition of antigen–antibody (Willemsen et al., 1998) and their interactions are of primary importance for biosensing platforms. High-resolution AFM-FS and topographical imaging were combined in the force–volume mode by Kienberger et al. (Kienberger et al., 2006) to measure interaction forces between lysozyme functionalized mica and anti-lysozyme antibody functionalized tip. This study displayed entirely different images when the tip was functionalized with antibody versus when the experiment was performed with nonfunctionalized tip and the binding sites on the lysozyme were identified. The binding probability decreased noticeably as the antibody was blocked in the solution. Casalini and co-workers (Casalini et al., 2015) functionalized cantilever tip with antigen IL-4 (interleukin-4) and the substrate with anti–IL-4 for the measurement of antigen–antibody interaction forces and binding kinetics like unbinding length, binding probability, and adhesion energy, as demonstrated in Figures 6A–J (Casalini et al., 2015).
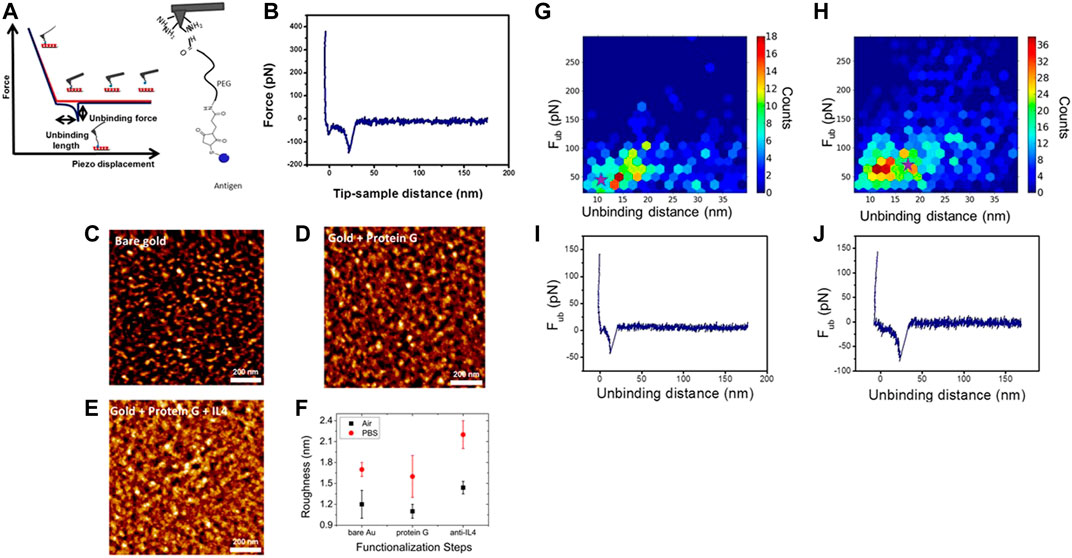
FIGURE 6. (A) (Left) Schematic diagram of a force–distance curve: With the cantilever tip approaching toward the substrate, tip–sample contact occurs. The cantilever tip bends upward due to repulsive forces and the probe is retracted. When the recognition event occurs, the cantilever tip is bent downward due to adhesion. The cantilever tip jumps out of contact when force gradient becomes greater than the cantilever spring constant. The maximum adhesion force, calculated as the difference between the baseline and the minimum during retraction, is the rupture/unbinding force. (Right) L4–PEG linker complex is adhered to an amino-decorated AFM cantilever tip. (B) Force–distance curve demonstrates a specific rupture event. (C) AFM image of just gold. (D) AFM image of protein G (PG)–decorated gold. (E) AFM image of anti-IL4 + PG–plated gold. (F) Roughness measurements of the samples shown in (C–E) were performed in air and in PBS solution with AFM. Roughness decreases upon PG deposition but increases following the antibody coating in both air and PBS. (G–H) Histograms of the unbinding distance and unbinding force in two dimensions using the HSC6NH2-based protocol PG-based protocol, respectively. (I–J) Force vs. tip sample separating distance plots corresponding to the hexagons marked with stars in (G) and (H), respectively (Casalini et al., 2015)
Various biochemical reactions are catalyzed by enzymes, and AFM-SMFS can be instrumental to understand the mechanism of enzymes. The catalytic activity of the enzyme was explored by Ditzler et al. by creating stable active E. coli dihydrofolate reductase (ecDHFR) monolayer on gold coated substrate (Ditzler et al., 2011). The interaction forces were measured in this study at single-molecule level between methotrexate (binding inhibitor) functionalized cantilever tip and enzyme functionalized gold substrate. Binding kinetics of enzymatic dextran elongations were studied by Mori et al. using AFM-SMFS in another study using DSase functionalized mica substrate and dextran-immobilized cantilever tip (Mori et al., 2011). The reaction dynamics and the real-time enzymatic activity were monitored by obtaining continuous force distance curves on a particular DSase every second. The results of this study conveyed the shift in the rupture force peak by nanometer range due to tip–sample contact time of few seconds and the catalytic elongation rate constant was calculated as 2.7 s−1.
AFM-SMFS is also in the spotlight for detection of numerous drug molecules like adenosine using molecular recognition events between drug molecules and ssDNA aptamers. Detection of adenosine monophosphate (AMP) was performed by Nguyen et al. by measuring single small-molecule unbinding forces of a split aptamer using AFM-FS (Nguyen et al., 2011). The sample preparation for this study was performed by using a split bipartite aptamer: The cantilever tip was functionalized with one portion of it, and the sample surface was functionalized with the other portion. This study showed an enhancement in rupture forces between two oligos in the presence of AMP in the solution vs. control experiments when AMP was absent. The detection method demonstrated in this study showed immense potential to develop more drug sensors. In another recent study, AFM force–based sensor was developed (Li J et al., 2015) for the detection of adenosine with higher sensitivity and probe the molecular recognition events between DNA aptamer and adenosine. This label-free detection procedure provides an easier and less time-consuming way of identification of other similar chemical molecules.
AFM-SMFS can also be used for the detection of metallic ions that are dangerous to human health. An AFM-SMFS–based aptasensor was recently developed (Li Q et al., 2015) for detecting mercury ions with high sensitivity. Unbinding forces were measured between graphite substrate and ssDNA (specific to Hg2+) functionalized cantilever tip for this study and an increase in unbinding forces was noticed due to addition of Hg2+ in the flow cell with water. This detection technique can be similarly used for other metal ions that are harmful to the environment and the human body. Crown–metal–crown complexations were studied by Kuo et al. In this study (Kuo et al., 2015), the cantilever tip and the substrate were functionalized with two crowns creating crown–metal–crown assembly upon addition of metallic ions during AFM force spectroscopy measurements. Through the measurement of the unbinding force of this complex during AFM-SMFS peeling experiments, crucial parameters of binding kinetics like binding probability, energy barrier, and dissociation rate were also determined using the Bell–Evans model. In Bell–Evans model (Evans 2001), survival probability or S(t) of a complex bond means the fraction of complex molecular bonds that has not experienced rupture/dissociation, and it is represented as
According to Bell–Evans model, the probability distribution of rupture forces is represented as
where Koff is the dissociation rate constant, f is the force, and df/dt =
AFM was initially developed for generating three-dimensional surface profile of organic and inorganic samples. AFM has been used for monitoring and reporting the topographical changes and the variation in substrate roughness of multiple biosensors. Development of PeakForce QNM mode (Sweers et al., 2011; Adamcik et al., 2012; Dokukin and Sokolov 2012; Nakajima et al., 2014) of AFM has revolutionized the process of characterization by allowing users to obtain maps of nanomechanical properties such as adhesion, Young’s modulus, and deformation with high resolution and acquisition speed. This mode was developed on the basis of tapping mode that regulates the maximum or peak force on the sample and lowers friction and shear forces significantly by intermittent contact of AFM probe and the sample surface. Peak force QNM mode analyzes the force–distance curve for each tapping of the probe on the fly, and all the nanomechanical properties are mapped with the same resolution as AFM height image. Figure 7A demonstrates force applied on the cantilever and Z-position versus time for one full cycle of probe motion, and the dashed line shows Z-position of the modulation. The force applied on the probe is represented by solid blue and solid read while approaching toward the substrate and retracting from the substrate, respectively. The cantilever tip is away from the substrate, resulting in the absence of any significant force on the cantilever at point A. With the approach of the cantilever tip, attractive forces like capillary forces, van der Waals force, and electrostatic force pull the cantilever tip toward the substrate, as shown by the negative force at point B. The tip–sample contact occurs following that, and the force increases until peak force occurs at point C (peak force set point was chosen by the user). The probe is then retracted from the substrate, and the force decreases until it reaches a minimum at point D, representing the adhesion force. Point E represents the pull-off point where long-range forces acting on the probe reduce to a minimum value. Adhesion force is the minimum force mentioned in Figure 7B. The probable source of adhesion force can be the attractive forces (van der Waals, capillary, and electrostatic forces) acting between the sample and tip surface. Adhesion force enhances with higher values of cantilever tip radius. The area above the withdrawing curve and beneath the zero-force reference is mentioned as work of adhesion. Integration of (force * velocity) represented by the yellow area between “retract” and “approach” curve in Figure 7B over one period of vibration is considered as dissipation.
where
where (d − d0) is the substrate deformation, R is the end radius of the cantilever tip, and (F − Fadh) is the force acting on the probe w.r.t. adhesion. E* is the reduced modulus that depends on the passion ratio (γ).
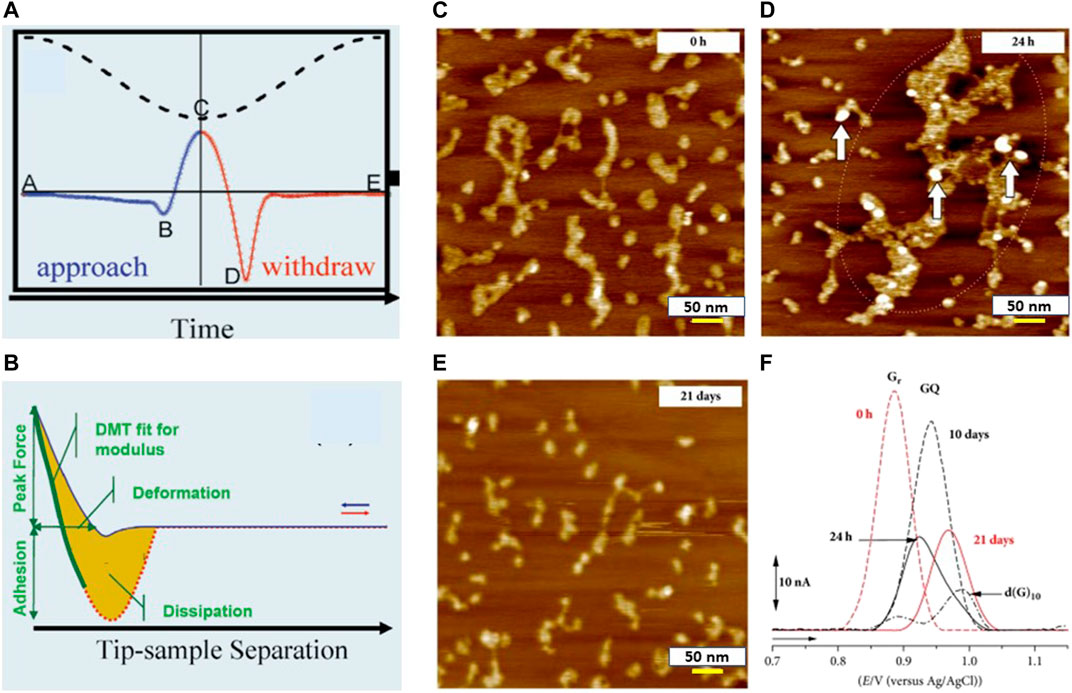
FIGURE 7. (A) Plot of applied force on the AFM probe and Z position of the piezoelectric scanner vs. time: point B represents the beginning of tip–sample contact, point C represents the peak force, and point D represents adhesion (courtesy: Application note # 128, Bruker). (B) Applied force vs. tip–sample separation plot where the sum of cantilever deflection and z piezo position represents the tip–sample separation (courtesy: Application note # 128, Bruker). (C–E) AFM images of poly(G) in pH 7.0 (5 µg/ml) when K+ ions were present in the solution with incubation time of (C) 0 h, (D) 24 h, and (E) 21 days. (F) DP voltammograms baseline of 100 µg ml−1 poly (G) with different durations of incubation (Chiorcea-Paquim et al., 2018).
Chiorcea et al. demonstrated successful characterization of electrochemical DNA-biosensor that was used for rapid DNA detection and evaluation of nucleic acid damage (Chiorcea-Paquim et al., 2015; Chiorcea-Paquim, et al., 2018). Various protocols for immobilizing dsDNA at the HOPG electrode surface were validated via AFM imaging and nanomechanical property measurements. This study showed that a thin dsDNA film (thickness measured via section analysis of AFM as 0.94 ± 0.2 nm and roughness value as 0.36 nm) appeared as a network with holes, uncovering many areas of the HOPG electrode surface, whereas the thick dsDNA film produced a uniform layer (height: 3–50 nm) of increased roughness (RMS value of roughness was measured as 3.41 nm) on electrode surface without gaps. Therefore, the thick layer of the dsDNA film turned out to be more effective through AFM measurements as it blocks nonspecific binding of the HOPG substrate and drug molecules by forming a uniform layer. High-resolution AFM images were utilized by Oliveira et al. for characterizing various protocols to immobilize DNA films on the surface of carbon electrodes before and after exposure to hazardous compounds (Oliveira and Oliveira-Brett 2010). Synthetic 10-mer homo- and hetero-oligodeoxynucleotides (10-mer ODNs) of familiar base sequences were absorbed on the HOPG surface. Their morphology and mechanical properties were explored using AFM to obtain a better understanding of the interaction mechanism of the HOPG surface with a specific nucleic acid. AFM imaging performed in air demonstrated various degrees of coverage and adsorption patterns on the HOPG substrate for the ODNs, correlated with different base sequences and structures of the specific ODNs. Chiorcea-Paquim et al. explored the characterization of G-Quadruplex electrochemical biosensors using AFM. G-quadruplex electrochemical biosensors (Chiorcea-Paquim, et al., 2018) are becoming increasingly important because of their highly sensitive electrochemical response to the structural changes of DNA from ssDNA, dsDNA, and hairpin DNA. Enhanced specific binding and of G-quadruplex with the aptamer provides these biosensors the leverage of higher sensitivity to detect and evaluate live cells, proteins, metal ions, ligands, and small organic molecules. Existence of long-chain polynucleotides: poly (dG) and poly(G) in human and other genomes at both RNA and DNA levels, are obvious, and these polynucleotides are used as constituents of DNA-electrochemical biosensors to detect interaction between of G-rich sections of DNA with drug molecules. This study showed that the poly(G) single strands formed self-assembly into small GQ regions for short incubation times when monovalent Na+ or K+ ions were present, whereas longer incubation periods led to formation of large poly(G) GQ aggregates with low adsorption (shown in Figure 7C–E (Chiorcea-Paquim et al., 2015). Voltammograms of freshly prepared poly(G) solutions in Figure 7F (Braet and Taatjes 2018) showed only oxidation peak of the G residues that reduced with longer incubation times. The maximum value of GQ oxidation peak current was observed following incubation of 10 days and plateaued after 17 days. The combination of AFM-DFS (Strunz et al., 1999), theory of stochastic system, and steered molecular dynamics (Zhang et al., 2010; Wong 2018; Spiriti and Wong 2021) have created a new research area that explores mechanical aspects of ligand–receptor interactions (unbinding/binding) and muscle proteins as they stretch. This hybrid method has been utilized in generating information about the energy landscape that regulates mechanical functions and the multiple components of proteins that carry forces.
Bio-AFM continues to be successfully and heavily utilized for high-resolution imaging, measuring shapes, sizes, roughness, and nanomechanical characterization of drug delivery vehicles. AFM results of these measurements have also been compared to similar techniques (DLS and TEM/SEM) for validation in multiple research studies conducted till date, and AFM seemed to produce more accurate measurements than these techniques. The applications of AFM-FS and AFM-SMFS techniques presented in this review paper clearly demonstrate their advantage over other characterization techniques as AFM can image biomolecules, bio-nanostructures, biosensors, and live cells in their native physiological environment with resolution in the sub-nanometer range. AFM-SMFS technique is a powerful approach to monitor molecular recognition and molecular interactions and to build biosensor architectures for dye-free and label-free biosensing of protein, DNA, enzyme, drug molecules, and other analytes. It still remains a promising approach and will attract more interest in the coming years for biosensing and characterization of biosensors, encouraging and facilitating new developments in the area of biomedical engineering and nanotechnology. However, the following scopes of improvement should be taken into consideration to make this technique more efficient and high throughput. Only experimentalists with extensive training in bio-AFM and particular skill sets can operate it. It also requires continuous human supervision. Breaking of cantilever tip after long period of functionalization and damaging live cell samples due to lack of optimization of the loading forces are major problems that make this method low throughput (Xie and Ren 2019a; Xie and Ren 2019b). Over the past couple of years, researchers in the material science community have begun to combine artificial intelligence (AI) and machine learning approaches (Huang et al., 2018; Müller et al., 2019; Alldritt et al., 2020; Gordon and Moriarty 2020; Krull et al., 2020) with AFM for various pattern recognition and data post-processing tasks. Also, some initial research happened to select appropriate AFM scanning areas and data modeling using deep learning. However, the potential of AI-AFM approaches in characterization of live cells is mostly unexplored. For example, AI-driven automation opportunity can help with sample specific loading forces to protect both tip and sample and auto-select appropriate live cell samples to improve the quality of data for nanomechanical property measurements. Other than this possible improvement, AFM probes should be developed that are already functionalized with specific biomolecules to exhibit increased activity in case of molecular recognition events. Other than extracting roughness and topographical information of different biosensors, nanomechanical properties (adhesion and Young’s modulus) of these biosensors need to be characterized too using AFM to increase their efficacy. Also, bio-AFM setup may be combined with fluorescence microscopy more to obtain a better understanding of the locations of the molecule of interest.
The author confirms being the sole contributor of this work and has approved it for publication.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Adamcik, J., Lara, C., Usov, I., Jeong, J. S., Ruggeri, F. S., Dietler, G., et al. (2012). Measurement of Intrinsic Properties of Amyloid Fibrils by the Peak Force QNM Method. Nanoscale 4 (15), 4426–4429. doi:10.1039/c2nr30768e
Akanda, M. H., Rai, R., Slipper, I. J., Chowdhry, B. Z., Lamprou, D., Getti, G., et al. (2015). Delivery of Retinoic Acid to LNCap Human Prostate Cancer Cells Using Solid Lipid Nanoparticles. Int. J. Pharm. 493 (1-2), 161–171. doi:10.1016/j.ijpharm.2015.07.042
Albrecht, C., Blank, K., Lalic-Mülthaler, M., Hirler, S., Mai, T., Gilbert, I., et al. (2003). DNA: a Programmable Force Sensor. Science 301 (5631), 367–370. doi:10.1126/science.1084713
Alessandrini, A., Seeger, H. M., Caramaschi, T., and Facci, P. (2012). Dynamic Force Spectroscopy on Supported Lipid Bilayers: Effect of Temperature and Sample Preparation. Biophysical J. 103 (1), 38–47. doi:10.1016/j.bpj.2012.05.039
Alhalhooly, L., Mamnoon, B., Kim, J., Mallik, S., and Choi, Y. (2021). Dynamic Cellular Biomechanics in Responses to Chemotherapeutic Drug in Hypoxia Probed by Atomic Force Spectroscopy. Oncotarget 12 (12), 1165–1177. doi:10.18632/oncotarget.27974
Alldritt, B., Hapala, P., Oinonen, N., Urtev, F., Krejci, O., Federici Canova, F., et al. (2020). Automated Structure Discovery in Atomic Force Microscopy. Sci. Adv. 6 (9), eaay6913. doi:10.1126/sciadv.aay6913
Alonso, J. L., and Goldmann, W. H. (2003). Feeling the Forces: Atomic Force Microscopy in Cell Biology. Life Sci. 72 (23), 2553–2560. doi:10.1016/s0024-3205(03)00165-6
Alsteens, D., Dupres, V., Yunus, S., Latgé, J.-P., Heinisch, J. J., and Dufrêne, Y. F. (2012). High-resolution Imaging of Chemical and Biological Sites on Living Cells Using Peak Force Tapping Atomic Force Microscopy. Langmuir 28 (49), 16738–16744. doi:10.1021/la303891j
Ando, T. (2019). High-speed Atomic Force Microscopy. Curr. Opin. Chem. Biol. 51, 105–112. doi:10.1016/j.cbpa.2019.05.010
Arora, D., Kumar, A., Gupta, P., Chashoo, G., and Jaglan, S. (2017). Preparation, Characterization and Cytotoxic Evaluation of Bovine Serum Albumin Nanoparticles Encapsulating 5-methylmellein: A Secondary Metabolite Isolated from Xylaria Psidii. Bioorg. Med. Chem. Lett. 27 (23), 5126–5130. doi:10.1016/j.bmcl.2017.10.064
Barattin, R., and Voyer, N. (2011). Chemical Modifications of Atomic Force Microscopy Tips. Methods Mol. Biol. 736, 457–483. doi:10.1007/978-1-61779-105-5_28
Bazylinska, U., Pietkiewicz, J., Rossowska, J., Chodaczek, G., Gamian, A., and Wilk, K. A. (2017). Polyelectrolyte Oil-Core Nanocarriers for Localized and Sustained Delivery of Daunorubicin to Colon Carcinoma MC38 Cells: The Case of Polysaccharide Multilayer Film in Relation to PEG-Ylated Shell. Macromol Biosci. 17 (5). doi:10.1002/mabi.201600347
Beaussart, A., Ngo, T. C., Derclaye, S., Kalinova, R., Mincheva, R., Dubois, P., et al. (2014). Chemical Force Microscopy of Stimuli-Responsive Adhesive Copolymers. Nanoscale 6 (1), 565–571. doi:10.1039/c3nr05256g
Bennett, I., Pyne, A. L. B., and McKendry, R. A. (2020). Cantilever Sensors for Rapid Optical Antimicrobial Sensitivity Testing. ACS Sens. 5 (10), 3133–3139. doi:10.1021/acssensors.0c01216
Besch, S., Snyder, K. V., Zhang, P. C., and Sachs, F. (2003). Adapting the Quesant Nomad Atomic Force Microscope for Biology and Patch-Clamp Atomic Force Microscopy. Cbb 39 (3), 195–210. doi:10.1385/cbb:39:3:195
Binnig, G., Quate, C. F., and Gerber, C. (1986). Atomic Force Microscope. Phys. Rev. Lett. 56 (9), 930–933. doi:10.1103/physrevlett.56.930
Blank, K., Mai, T., Gilbert, I., Schiffmann, S., Rankl, J., Zivin, R., et al. (2003). A Force-Based Protein Biochip. Proc. Natl. Acad. Sci. 100 (20), 11356–11360. doi:10.1073/pnas.1934928100
Bousmail, D., Amrein, L., Fakhoury, J. J., Fakih, H. H., Hsu, J. C. C., Panasci, L., et al. (2017). Precision Spherical Nucleic Acids for Delivery of Anticancer Drugs. Chem. Sci. 8 (9), 6218–6229. doi:10.1039/c7sc01619k
Braet, F., and Taatjes, D. J. (2018). Foreword to the Special Issue on Applications of Atomic Force Microscopy in Cell Biology. Semin. Cel Dev. Biol. 73, 1–3. doi:10.1016/j.semcdb.2017.06.025
Brewer, N. J., and Leggett, G. J. (2004). Chemical Force Microscopy of Mixed Self-Assembled Monolayers of Alkanethiols on Gold: Evidence for Phase Separation. Langmuir 20 (10), 4109–4115. doi:10.1021/la036301e
Caballero, B., Trugo, L. C., and Finglas, P. M. (2003). Encyclopedia of Food Sciences and Nutrition. Academic.
Cai, X. J., Woods, A., Mesquida, P., and Jones, S. A. (2016). Assessing the Potential for Drug-Nanoparticle Surface Interactions to Improve Drug Penetration into the Skin. Mol. Pharmaceutics 13 (4), 1375–1384. doi:10.1021/acs.molpharmaceut.6b00032
Cameron, N. S., Ott, A., Roberge, H. l. n., and Veres, T. (2006). Chemical Force Microscopy for Hot-Embossing Lithography Release Layer Characterization. Soft Matter 2 (7), 553–557. doi:10.1039/b600936k
Caporali, M., Serrano-Ruiz, M., Telesio, F., Heun, S., Nicotra, G., Spinella, C., et al. (2017). Decoration of Exfoliated Black Phosphorus with Nickel Nanoparticles and its Application in Catalysis. Chem. Commun. 53 (79), 10946–10949. doi:10.1039/c7cc05906j
Carlos B. Oliveira, S., and Maria Oliveira-Brett, A. (2010). DNA-electrochemical Biosensors: AFM Surface Characterisation and Application to Detection of In Situ Oxidative Damage to DNA. Cchts 13 (7), 628–640. doi:10.2174/1386207311004070628
Casalini, S., Dumitru, A. C., Leonardi, F., Bortolotti, C. A., Herruzo, E. T., Campana, A., et al. (2015). Multiscale Sensing of Antibody-Antigen Interactions by Organic Transistors and Single-Molecule Force Spectroscopy. ACS Nano 9 (5), 5051–5062. doi:10.1021/acsnano.5b00136
Chen, G., Ning, X., Park, B., Boons, G.-J., and Xu, B. (2009). Simple, Clickable Protocol for Atomic Force Microscopy Tip Modification and its Application for Trace Ricin Detection by Recognition Imaging. Langmuir 25 (5), 2860–2864. doi:10.1021/la803523b
Cheong, L.-Z., Zhao, W., Song, S., and Shen, C. (2019). Lab on a Tip: Applications of Functional Atomic Force Microscopy for the Study of Electrical Properties in Biology. Acta Biomater. 99, 33–52. doi:10.1016/j.actbio.2019.08.023
Chiorcea-Paquim, A.-M., Pontinha, A. D. R., Eritja, R., Lucarelli, G., Sparapani, S., Neidle, S., et al. (2015). Atomic Force Microscopy and Voltammetric Investigation of Quadruplex Formation between a Triazole-Acridine Conjugate and Guanine-Containing Repeat DNA Sequences. Anal. Chem. 87 (12), 6141–6149. doi:10.1021/acs.analchem.5b00743
Chiorcea-Paquim, A. M., Eritja, R., and Oliveira-Brett, A. M. (2018). Electrochemical and AFM Characterization of G-Quadruplex Electrochemical Biosensors and Applications. J. Nucleic Acids 2018, 5307106. doi:10.1155/2018/5307106
Chtcheglova, L. A., and Hinterdorfer, P. (2018). Simultaneous AFM Topography and Recognition Imaging at the Plasma Membrane of Mammalian Cells. Semin. Cel Dev. Biol. 73, 45–56. doi:10.1016/j.semcdb.2017.08.025
Clear, S. C., and Nealey, P. F. (1999). Chemical Force Microscopy Study of Adhesion and Friction between Surfaces Functionalized with Self-Assembled Monolayers and Immersed in Solvents. J. Colloid Interf. Sci. 213 (1), 238–250. doi:10.1006/jcis.1999.6139
Crucho, C. I. C., and Barros, M. T. (2017). Polymeric Nanoparticles: A Study on the Preparation Variables and Characterization Methods. Mater. Sci. Eng. C 80, 771–784. doi:10.1016/j.msec.2017.06.004
Cuellar, J. L., Llarena, I., Iturri, J. J., Donath, E., and Moya, S. E. (2013). A Novel Approach for Measuring the Intrinsic Nanoscale Thickness of Polymer Brushes by Means of Atomic Force Microscopy: Application of a Compressible Fluid Model. Nanoscale 5 (23), 11679–11685. doi:10.1039/c3nr02929h
Czajkowsky, D. M., Iwamoto, H., and Shao, Z. (2000). Atomic Force Microscopy in Structural Biology: from the Subcellular to the Submolecular. J. Electron Microsc. 49 (3), 395–406. doi:10.1093/oxfordjournals.jmicro.a023821
Dague, E., Alsteens, D., Latgé, J.-P., Verbelen, C., Raze, D., Baulard, A. R., et al. (2007). Chemical Force Microscopy of Single Live Cells. Nano Lett. 7 (10), 3026–3030. doi:10.1021/nl071476k
Diezemann, G., and Janshoff, A. (2008). Dynamic Force Spectroscopy: Analysis of Reversible Bond-Breaking Dynamics. J. Chem. Phys. 129 (8), 084904. doi:10.1063/1.2968543
Dinesh Kumar, V., Verma, P. R. P., and Singh, S. K. (2015). Development and Evaluation of Biodegradable Polymeric Nanoparticles for the Effective Delivery of Quercetin Using a Quality by Design Approach. LWT - Food Sci. Tech. 61 (2), 330–338. doi:10.1016/j.lwt.2014.12.020
Ditzler, L. R., Sen, A., Gannon, M. J., Kohen, A., and Tivanski, A. V. (2011). Self-assembled Enzymatic Monolayer Directly Bound to a Gold Surface: Activity and Molecular Recognition Force Spectroscopy Studies. J. Am. Chem. Soc. 133 (34), 13284–13287. doi:10.1021/ja205409v
Dokukin, M. E., and Sokolov, I. (2012). Quantitative Mapping of the Elastic Modulus of Soft Materials with HarmoniX and PeakForce QNM AFM Modes. Langmuir 28 (46), 16060–16071. doi:10.1021/la302706b
Dong, Y., and Feng, S.-S. (2004). Methoxy Poly(ethylene Glycol)-Poly(lactide) (MPEG-PLA) Nanoparticles for Controlled Delivery of Anticancer Drugs. Biomaterials 25 (14), 2843–2849. doi:10.1016/j.biomaterials.2003.09.055
Dos Ramos, L., de Beer, S., Hempenius, M. A., and Vancso, G. J. (2015). Redox-Induced Backbiting of Surface-Tethered Alkylsulfonate Amphiphiles: Reversible Switching of Surface Wettability and Adherence. Langmuir 31 (23), 6343–6350. doi:10.1021/acs.langmuir.5b01105
Dubes, A., Parrot-Lopez, H., Abdelwahed, W., Degobert, G., Fessi, H., Shahgaldian, P., et al. (2003). Scanning Electron Microscopy and Atomic Force Microscopy Imaging of Solid Lipid Nanoparticles Derived from Amphiphilic Cyclodextrins. Eur. J. Pharmaceutics Biopharmaceutics 55 (3), 279–282. doi:10.1016/s0939-6411(03)00020-1
Dufrêne, Y. F., Ando, T., Garcia, R., Alsteens, D., Martinez-Martin, D., Engel, A., et al. (2017). Imaging Modes of Atomic Force Microscopy for Application in Molecular and Cell Biology. Nat. Nanotech 12 (4), 295–307. doi:10.1038/nnano.2017.45
Dufrêne, Y. F. (2008). Atomic Force Microscopy and Chemical Force Microscopy of Microbial Cells. Nat. Protoc. 3 (7), 1132–1138. doi:10.1038/nprot.2008.101
Dufrêne, Y. F., and Hinterdorfer, P. (2008). Recent Progress in AFM Molecular Recognition Studies. Pflugers Arch. - Eur. J. Physiol. 456 (1), 237–245. doi:10.1007/s00424-007-0413-1
Eaton, P., Quaresma, P., Soares, C., Neves, C., de Almeida, M. P., Pereira, E., et al. (2017). A Direct Comparison of Experimental Methods to Measure Dimensions of Synthetic Nanoparticles. Ultramicroscopy 182, 179–190. doi:10.1016/j.ultramic.2017.07.001
Ebner, A., Kienberger, F., Kada, G., Stroh, C. M., Geretschläger, M., Kamruzzahan, A. S. M., et al. (2005). Localization of Single Avidin-Biotin Interactions Using Simultaneous Topography and Molecular Recognition Imaging. Chemphyschem 6 (5), 897–900. doi:10.1002/cphc.200400545
Enache, T. A., Chiorcea-Paquim, A.-M., and Oliveira-Brett, A. M. (2018). Amyloid Beta Peptide VHHQ, KLVFF, and IIGLMVGGVV Domains Involved in Fibrilization: AFM and Electrochemical Characterization. Anal. Chem. 90 (3), 2285–2292. doi:10.1021/acs.analchem.7b04686
Evans, E. (2001). Probing the Relation between Force-Lifetime-And Chemistry in Single Molecular Bonds. Annu. Rev. Biophys. Biomol. Struct. 30, 105–128. doi:10.1146/annurev.biophys.30.1.105
Fagan, B., Tipple, C. A., Xue, Z., Sepaniak, M. J., and Datskos, P. G. (2000). Modification of Micro-cantilever Sensors with Sol-Gels to Enhance Performance and Immobilize Chemically Selective Phases. Talanta 53 (3), 599–608. doi:10.1016/s0039-9140(00)00533-6
Fiorini, M., McKendry, R., Cooper, M. A., Rayment, T., and Abell, C. (2001). Chemical Force Microscopy with Active Enzymes. Biophysical J. 80 (5), 2471–2476. doi:10.1016/s0006-3495(01)76215-7
Foster, A. S., Barth, C., and Henry, C. R. (2009). Chemical Identification of Ions in Doped NaCl by Scanning Force Microscopy. Phys. Rev. Lett. 102 (25), 256103. doi:10.1103/physrevlett.102.256103
Gadegaard, N. (2006). Atomic Force Microscopy in Biology: Technology and Techniques. Biotech. Histochem. 81 (2-3), 87–97. doi:10.1080/10520290600783143
Gao, L., Zhao, H., Li, T., Huo, P., Chen, D., and Liu, B. (2018). Atomic Force Microscopy Based Tip-Enhanced Raman Spectroscopy in Biology. Int. J. Mol. Sci. 19 (4). doi:10.3390/ijms19041193
Garg, A., and Kokkoli, E. (2005). Characterizing Particulate Drug-Delivery Carriers with Atomic Force Microscopy. IEEE Eng. Med. Biol. Mag. 24 (1), 87–95. doi:10.1109/memb.2005.1384106
Goldsbury, C., and Scheuring, S. (2002). Introduction to Atomic Force Microscopy (AFM) in Biology. Curr. Protoc. Protein Sci. Chapter 17, Unit–7. Unit 17 17. doi:10.1002/0471140864.ps1707s29
Goldsbury, C. S., Scheuring, S., and Kreplak, L. (2009). Introduction to Atomic Force Microscopy (AFM) in Biology. Curr. Protoc. Protein Sci. Chapter 17, Unit–19. Unit 17 17 11. doi:10.1002/0471140864.ps1707s58
Gordon, O. M., and Moriarty, P. J. (2020). Machine Learning at the (Sub)atomic Scale: Next Generation Scanning Probe Microscopy. Mach. Learn. Sci. Technol. 1 (2), 023001. doi:10.1088/2632-2153/ab7d2f
Gourianova, S., Willenbacher, N., and Kutschera, M. (2005). Chemical Force Microscopy Study of Adhesive Properties of Polypropylene Films: Influence of Surface Polarity and Medium. Langmuir 21 (12), 5429–5438. doi:10.1021/la0501379
Gruber, K., Horlacher, T., Castelli, R., Mader, A., Seeberger, P. H., and Hermann, B. A. (2011). Cantilever Array Sensors Detect Specific Carbohydrate−Protein Interactions with Picomolar Sensitivity. ACS Nano 5 (5), 3670–3678. doi:10.1021/nn103626q
Guo, S., Zhu, X., Jańczewski, D., Lee, S. S. C., He, T., Teo, S. L. M., et al. (2016). Measuring Protein Isoelectric Points by AFM-Based Force Spectroscopy Using Trace Amounts of Sample. Nat. Nanotech 11 (9), 817–823. doi:10.1038/nnano.2016.118
Gupta, A. K., and Wells, S. (2004). Surface-modified Superparamagnetic Nanoparticles for Drug Delivery: Preparation, Characterization, and Cytotoxicity Studies. IEEE Trans.on Nanobioscience 3 (1), 66–73. doi:10.1109/tnb.2003.820277
Hansma, H. G. (2001). Surface Biology of DNA by Atomic Force Microscopy. Annu. Rev. Phys. Chem. 52, 71–92. doi:10.1146/annurev.physchem.52.1.71
Hibino, M., and Nakano-Nishida, T. (2014). Chemical Force Microscopy Using Functionalized ZnO Whisker Probe Tips. J. Nanosci Nanotechnol 14 (4), 3080–3086. doi:10.1166/jnn.2014.8592
Hinterdorfer, P., and Dufrêne, Y. F. (2006). Detection and Localization of Single Molecular Recognition Events Using Atomic Force Microscopy. Nat. Methods 3 (5), 347–355. doi:10.1038/nmeth871
Hoh, J. H., and Hansma, P. K. (1992). Atomic Force Microscopy for High-Resolution Imaging in Cell Biology. Trends Cel Biol. 2 (7), 208–213. doi:10.1016/0962-8924(92)90248-l
Huang, B., Li, Z., and Li, J. (2018). An Artificial Intelligence Atomic Force Microscope Enabled by Machine Learning. Nanoscale 10 (45), 21320–21326. doi:10.1039/c8nr06734a
Hughes, M. L., and Dougan, L. (2016). The Physics of Pulling Polyproteins: a Review of Single Molecule Force Spectroscopy Using the AFM to Study Protein Unfolding. Rep. Prog. Phys. 79 (7), 076601. doi:10.1088/0034-4885/79/7/076601
Jiang, Z., Zhang, Y., Yu, Y., Wang, Z., Zhang, X., Duan, X., et al. (2010). Study on Intercalations between Double-Stranded DNA and Pyrene by Single-Molecule Force Spectroscopy: toward the Detection of Mismatch in DNA. Langmuir 26 (17), 13773–13777. doi:10.1021/la102647p
Johnson, B. N., and Mutharasan, R. (2012). Biosensing Using Dynamic-Mode Cantilever Sensors: a Review. Biosens. Bioelectron. 32 (1), 1–18. doi:10.1016/j.bios.2011.10.054
Jones, S. K., Sarkar, A., Feldmann, D. P., Hoffmann, P., and Merkel, O. M. (2017). Revisiting the Value of Competition Assays in Folate Receptor-Mediated Drug Delivery. Biomaterials 138, 35–45. doi:10.1016/j.biomaterials.2017.05.034
Ju, L. (2019). Dynamic Force Spectroscopy Analysis on the Redox States of Protein Disulphide Bonds. Methods Mol. Biol. 1967, 115–131. doi:10.1007/978-1-4939-9187-7_7
Jung, J., Lifland, A. W., Zurla, C., Alonas, E. J., and Santangelo, P. J. (2013). Quantifying RNA-Protein Interactions In Situ Using Modified-MTRIPs and Proximity Ligation. Nucleic Acids Res. 41 (1), e12. doi:10.1093/nar/gks837
Kanno, T., Yamada, T., Iwabuki, H., Tanaka, H., Kuroda, S. i., Tanizawa, K., et al. (2002). Size Distribution Measurement of Vesicles by Atomic Force Microscopy. Anal. Biochem. 309 (2), 196–199. doi:10.1016/s0003-2697(02)00291-9
Karagkiozaki, V. C., Logothetidis, S. D., Kassavetis, S. N., and Giannoglou, G. D. (2010). Nanomedicine for the Reduction of the Thrombogenicity of Stent Coatings. Int. J. Nanomedicine 5, 239–248. doi:10.2147/ijn.s7596
Kienberger, F., Ebner, A., Gruber, H. J., and Hinterdorfer, P. (2006). Molecular Recognition Imaging and Force Spectroscopy of Single Biomolecules. Acc. Chem. Res. 39 (1), 29–36. doi:10.1021/ar050084m
Kim, S. Y., Naskar, D., Kundu, S. C., Bishop, D. P., Doble, P. A., Boddy, A. V., et al. (2015). Formulation of Biologically-Inspired Silk-Based Drug Carriers for Pulmonary Delivery Targeted for Lung Cancer. Sci. Rep. 5, 11878. doi:10.1038/srep11878
Koehler, M., Fis, A., Gruber, H. J., and Hinterdorfer, P. (2019). AFM-based Force Spectroscopy Guided by Recognition Imaging: A New Mode for Mapping and Studying Interaction Sites at Low Lateral Density. Methods Protoc. 2 (1). doi:10.3390/mps2010006
Koehler, M., Macher, G., Rupprecht, A., Zhu, R., Gruber, H., Pohl, E., et al. (2017). Combined Recognition Imaging and Force Spectroscopy: A New Mode for Mapping and Studying Interaction Sites at Low Lateral Density. Sci. Adv. Mater. 9 (1), 128–134. doi:10.1166/sam.2017.3066
Kreller, D. I., Gibson, G., vanLoon, G. W., and Horton, J. H. (2002). Chemical Force Microscopy Investigation of Phosphate Adsorption on the Surfaces of Iron(III) Oxyhydroxide Particles. J. Colloid Interf. Sci. 254 (2), 205–213. doi:10.1006/jcis.2002.8575
Kreplak, L. (2016). Introduction to Atomic Force Microscopy (AFM) in Biology. Curr. Protoc. Protein Sci. 85 (17 17 11), 17–21. doi:10.1002/cpps.14
Krull, A., Hirsch, P., Rother, C., Schiffrin, A., and Krull, C. (2020). Artificial-intelligence-driven Scanning Probe Microscopy. Commun. Phys. 3 (1), 1–8. doi:10.1038/s42005-020-0317-3
Kumar, S., and Singh, S. K. (2017). Fabrication and Characterization of Fibroin Solution and Nanoparticle from Silk Fibers ofBombyx Mori. Particulate Sci. Tech. 35 (3), 304–313. doi:10.1080/02726351.2016.1154908
Kuo, T.-Y., Tseng, W.-H., and Chen, C.-h. (2015). Force Spectroscopy of Metal-Crown Ether Multivalency: Effect of Local Environments on Energy Landscape and Sensing Kinetics. Angew. Chem. Int. Ed. 54 (32), 9213–9217. doi:10.1002/anie.201503948
Lal, R., and John, S. A. (1994). Biological Applications of Atomic Force Microscopy. Am. J. Physiol. 266 (1 Pt 1), C1–C21. doi:10.1152/ajpcell.1994.266.1.C1
Lamprecht, C., Hinterdorfer, P., and Ebner, A. (2014). Applications of Biosensing Atomic Force Microscopy in Monitoring Drug and Nanoparticle Delivery. Expert Opin. Drug Deliv. 11 (8), 1237–1253. doi:10.1517/17425247.2014.917078
Lang, H. P., and Gerber, C. (2008). Microcantilever Sensors. Top. Curr. Chem. 285, 1–27. doi:10.1007/128_2007_28
Lee, S. H., and Lee, S. (2020). Fabrication and Characterization of Roll-To-Roll-Coated Cantilever-Structured Touch Sensors. ACS Appl. Mater. Inter. 12 (41), 46797–46803. doi:10.1021/acsami.0c14889
Li, J., Li, Q., Ciacchi, L., and Wei, G. (2015). Label-free Sensing of Adenosine Based on Force Variations Induced by Molecular Recognition. Biosensors 5 (1), 85–97. doi:10.3390/bios5010085
Li, M., Dang, D., Liu, L., Xi, N., and Wang, Y. (2017). Imaging and Force Recognition of Single Molecular Behaviors Using Atomic Force Microscopy. Sensors (Basel) 17 (1). doi:10.3390/s17010200
Li, M., Xi, N., and Liu, L. (2021). Peak Force Tapping Atomic Force Microscopy for Advancing Cell and Molecular Biology. Nanoscale 13 (18), 8358–8375. doi:10.1039/d1nr01303c
Li, Q., Michaelis, M., Wei, G., and Colombi Ciacchi, L. (2015). A Novel Aptasensor Based on Single-Molecule Force Spectroscopy for Highly Sensitive Detection of Mercury Ions. Analyst 140 (15), 5243–5250. doi:10.1039/c5an00708a
Li, Q., Zhang, T., Pan, Y., Ciacchi, L. C., Xu, B., and Wei, G. (2016). AFM-based Force Spectroscopy for Bioimaging and Biosensing. RSC Adv. 6 (16), 12893–12912. doi:10.1039/c5ra22841g
Liu, L., Wei, Y., Liu, W., Sun, T., Wang, K., Wang, Y., et al. (2018). Progress in the Applications of High-Speed Atomic Force Microscopy in Cell Biology. Nan Fang Yi Ke Da Xue Xue Bao 38 (8), 931–937. doi:10.3969/j.issn.1673-4254.2018.08.05
Ma, X., Gosai, A., and Shrotriya, P. (2020). Resolving Electrical Stimulus Triggered Molecular Binding and Force Modulation upon Thrombin-Aptamer Biointerface. J. Colloid Interf. Sci. 559, 1–12. doi:10.1016/j.jcis.2019.09.080
Malkin, A. J., Plomp, M., and McPherson, A. (2002). Application of Atomic Force Microscopy to Studies of Surface Processes in Virus Crystallization and Structural Biology. Acta Crystallogr. D Biol. Crystallogr. 58 (Pt 10 Pt 1), 1617–1621. doi:10.1107/s090744490201274x
Mamou, D., Nsubuga, L., Lisboa Marcondes, T., Høegh, S. O., Hvam, J., Niekiel, F., et al. (2021). Surface Modification Enabling Reproducible Cantilever Functionalization for Industrial Gas Sensors. Sensors (Basel) 21 (18). doi:10.3390/s21186041
Masarudin, M. J., Cutts, S. M., Evison, B. J., Phillips, D. R., and Pigram, P. J. (2015). Factors Determining the Stability, Size Distribution, and Cellular Accumulation of Small, Monodisperse Chitosan Nanoparticles as Candidate Vectors for Anticancer Drug Delivery: Application to the Passive Encapsulation of [14C]-Doxorubicin. Nsa 8, 67–80. doi:10.2147/nsa.s91785
Mayyas, E., Bernardo, M., Runyan, L., Sohail, A., Subba-Rao, V., Pantea, M., et al. (2010). Dissociation Kinetics of an Enzyme−Inhibitor System Using Single-Molecule Force Measurements. Biomacromolecules 11 (12), 3352–3358. doi:10.1021/bm100844x
Mertens, J., Cuervo, A., and Carrascosa, J. L. (2019). Nanomechanical Detection ofEscherichia Coliinfection by Bacteriophage T7 Using Cantilever Sensors. Nanoscale 11 (38), 17689–17698. doi:10.1039/c9nr05240b
Montanari, L., Cilurzo, F., Selmin, F., Conti, B., Genta, I., Poletti, G., et al. (2003). Poly(lactide-co-glycolide) Microspheres Containing Bupivacaine: Comparison between Gamma and Beta Irradiation Effects. J. Controlled Release 90 (3), 281–290. doi:10.1016/s0168-3659(03)00153-6
Montasser, I., Fessi, H., and Coleman, A. W. (2002). Atomic Force Microscopy Imaging of Novel Type of Polymeric Colloidal Nanostructures. Eur. J. Pharmaceutics Biopharmaceutics 54 (3), 281–284. doi:10.1016/s0939-6411(02)00087-5
Mori, T., Asakura, M., and Okahata, Y. (2011). Single-molecule Force Spectroscopy for Studying Kinetics of Enzymatic Dextran Elongations. J. Am. Chem. Soc. 133 (15), 5701–5703. doi:10.1021/ja200094f
Müller, P., Abuhattum, S., Möllmert, S., Ulbricht, E., Taubenberger, A. V., and Guck, J. (2019). Nanite: Using Machine Learning to Assess the Quality of Atomic Force Microscopy-Enabled Nano-Indentation Data. BMC bioinformatics 20 (1), 1–9.
Nakajima, K., Ito, M., Wang, D., Liu, H., Nguyen, H. K., Liang, X., et al. (2014). Nano-palpation AFM and its Quantitative Mechanical Property Mapping. Microscopy (Tokyo) 63 (3), 193–208. doi:10.1093/jmicro/dfu009
Nandi, T., and Ainavarapu, S. R. K. (2021). Applications of Atomic Force Microscopy in Modern Biology. Emerg. Top. Life Sci. 5 (1), 103–111. doi:10.1042/etls20200255
Neuert, G., Albrecht, C., Pamir, E., and Gaub, H. E. (2006). Dynamic Force Spectroscopy of the Digoxigenin-Antibody Complex. FEBS Lett. 580 (2), 505–509. doi:10.1016/j.febslet.2005.12.052
Nguyen, T.-H., Steinbock, L. J., Butt, H.-J., Helm, M., and Berger, R. (2011). Measuring Single Small Molecule Binding via Rupture Forces of a Split Aptamer. J. Am. Chem. Soc. 133 (7), 2025–2027. doi:10.1021/ja1092002
Okabe, Y., Furugori, M., Tani, Y., Akiba, U., and Fujihira, M. (2000). Chemical Force Microscopy of Microcontact-Printed Self-Assembled Monolayers by Pulsed-Force-Mode Atomic Force Microscopy. Ultramicroscopy 82 (1-4), 203–212. doi:10.1016/s0304-3991(99)00143-6
Parot, P., Dufrêne, Y. F., Hinterdorfer, P., Le Grimellec, C., Navajas, D., Pellequer, J.-L., et al. (2007). Past, Present and Future of Atomic Force Microscopy in Life Sciences and Medicine. J. Mol. Recognit. 20 (6), 418–431. doi:10.1002/jmr.857
Peiner, E., and Wasisto, H. S. (2019). Cantilever Sensors. Sensors (Basel) 19 (9). doi:10.3390/s19092043
Picas, L., Milhiet, P.-E., and Hernández-Borrell, J. (2012). Atomic Force Microscopy: a Versatile Tool to Probe the Physical and Chemical Properties of Supported Membranes at the Nanoscale. Chem. Phys. Lipids 165 (8), 845–860. doi:10.1016/j.chemphyslip.2012.10.005
Pontinha, A. D. R., Jorge, S. M. A., Chiorcea Paquim, A.-M., Diculescu, V. C., and Oliveira-Brett, A. M. (2011). In Situ evaluation of Anticancer Drug Methotrexate-DNA Interaction Using a DNA-Electrochemical Biosensor and AFM Characterization. Phys. Chem. Chem. Phys. 13 (12), 5227–5234. doi:10.1039/c0cp02377a
Qi, K., Ma, Q., Remsen, E. E., Clark, C. G., and Wooley, K. L. (2004). Determination of the Bioavailability of Biotin Conjugated onto Shell Cross-Linked (SCK) Nanoparticles. J. Am. Chem. Soc. 126 (21), 6599–6607. doi:10.1021/ja039647k
Radwan Almofti, M., Harashima, H., Shinohara, Y., Almofti, A., Baba, Y., and Kiwada, H. (2003). Cationic Liposome-Mediated Gene Delivery: Biophysical Study and Mechanism of Internalization. Arch. Biochem. Biophys. 410 (2), 246–253. doi:10.1016/s0003-9861(02)00725-7
Ramachandran, S., Teran Arce, F., and Lal, R. (2011). Potential Role of Atomic Force Microscopy in Systems Biology. Wires Syst. Biol. Med. 3 (6), 702–716. doi:10.1002/wsbm.154
Ramezanpour, M., Leung, S. S., Delgado-Magnero, K. H., Bashe, B. Y., Thewalt, J., and Tieleman, D. P. (2016). Computational and Experimental Approaches for Investigating Nanoparticle-Based Drug Delivery Systems. Biochim. Biophys. Acta 1858 (7 Pt B), 1688–1709. doi:10.1016/j.bbamem.2016.02.028
Reggente, M., Passeri, D., Angeloni, L., Scaramuzzo, F. A., Barteri, M., De Angelis, F., et al. (2017). Detection of Stiff Nanoparticles within Cellular Structures by Contact Resonance Atomic Force Microscopy Subsurface Nanomechanical Imaging. Nanoscale 9 (17), 5671–5676. doi:10.1039/c7nr01111c
Reiter-Scherer, V., Cuellar-Camacho, J. L., Bhatia, S., Haag, R., Herrmann, A., Lauster, D., et al. (2019). Force Spectroscopy Shows Dynamic Binding of Influenza Hemagglutinin and Neuraminidase to Sialic Acid. Biophysical J. 116 (8), 1577. doi:10.1016/j.bpj.2019.03.032
Sapra, K. T., Spoerri, P. M., Engel, A., Alsteens, D., and Müller, D. J. (2019). Seeing and Sensing Single G Protein-Coupled Receptors by Atomic Force Microscopy. Curr. Opin. Cel Biol. 57, 25–32. doi:10.1016/j.ceb.2018.10.006
Sarkar, A., Sohail, A., Dong, J., Prunotto, M., Shinki, K., Fridman, R., et al. (2019). Live Cell Measurements of Interaction Forces and Binding Kinetics between Discoidin Domain Receptor 1 (DDR1) and Collagen I with Atomic Force Microscopy. Biochim. Biophys. Acta (Bba) - Gen. Subjects 1863 (11), 129402. doi:10.1016/j.bbagen.2019.07.011
Schönherr, H., Beulen, M. W., Bügler, J., Huskens, J., van Veggel, F. C., Reinhoudt, D. N., et al. (2000). Individual Supramolecular Host− Guest Interactions Studied by Dynamic Single Molecule Force Spectroscopy. J. Am. Chem. Soc. 122 (20), 4963–4967.
Senapati, S., Manna, S., Lindsay, S., and Zhang, P. (2013). Application of Catalyst-free Click Reactions in Attaching Affinity Molecules to Tips of Atomic Force Microscopy for Detection of Protein Biomarkers. Langmuir 29 (47), 14622–14630. doi:10.1021/la4039667
Sengupta, E., Yan, Y., Wang, X., Munechika, K., and Ginger, D. S. (2014). Dynamic Force Spectroscopy of Photoswitch-Modified DNA. ACS Nano 8 (3), 2625–2631. doi:10.1021/nn406334b
Shahgaldian, P., Da Silva, E., Coleman, A. W., Rather, B., and Zaworotko, M. J. (2003a). Para-acyl-calix-arene Based Solid Lipid Nanoparticles (SLNs): a Detailed Study of Preparation and Stability Parameters. Int. J. Pharm. 253 (1-2), 23–38. doi:10.1016/s0378-5173(02)00639-7
Shahgaldian, P., Quattrocchi, L., Gualbert, J., Coleman, A. W., and Goreloff, P. (2003b). AFM Imaging of Calixarene Based Solid Lipid Nanoparticles in Gel Matrices. Eur. J. Pharmaceutics Biopharmaceutics 55 (1), 107–113. doi:10.1016/s0939-6411(02)00123-6
Shahin, V., and Barrera, N. P. (2008). Providing Unique Insight into Cell Biology via Atomic Force Microscopy. Int. Rev. Cytol. 265, 227–252. doi:10.1016/s0074-7696(07)65006-2
Shao, Z., and Yang, J. (1995). Progress in High Resolution Atomic Force Microscopy in Biology. Quart. Rev. Biophys. 28 (2), 195–251. doi:10.1017/s0033583500003061
Sharma, S., Johnson, R. W., and Desai, T. A. (2004). XPS and AFM Analysis of Antifouling PEG Interfaces for Microfabricated Silicon Biosensors. Biosens. Bioelectron. 20 (2), 227–239. doi:10.1016/j.bios.2004.01.034
Shi, Y., Cai, M., Zhou, L., and Wang, H. (2018). The Structure and Function of Cell Membranes Studied by Atomic Force Microscopy. Semin. Cel Dev. Biol. 73, 31–44. doi:10.1016/j.semcdb.2017.07.012
Shimpi, N. G., and Jha, M. (2017). Green Synthesis of Silver Nanoparticles Using tabernaemontana Divaricata and In-Vitro Cytotoxicity Investigation against Human Lung Adenocarcinoma. INTERNATIONAL JOURNAL Pharm. Sci. Res. 8 (12), 5100–5110.
Sirghi, L., Bretagnol, F., Mornet, S., Sasaki, T., Gilliland, D., Colpo, P., et al. (2009). Atomic Force Microscopy Characterization of the Chemical Contrast of Nanoscale Patterns Fabricated by Electron Beam Lithography on Polyethylene Glycol Oxide Thin Films. Ultramicroscopy 109 (3), 222–229. doi:10.1016/j.ultramic.2008.10.022
Sluysmans, D., Devaux, F., Bruns, C. J., Stoddart, J. F., and Duwez, A.-S. (2018). Dynamic Force Spectroscopy of Synthetic Oligorotaxane Foldamers. Proc. Natl. Acad. Sci. USA 115 (38), 9362–9366. doi:10.1073/pnas.1712790115
Smith, J. R., Breakspear, S., and Campbell, S. A. (2003). AFM in Surface Finishing: Part II. Surface Roughness. Trans. IMF 81 (3), B55–B58. doi:10.1080/00202967.2003.11871499
Smith, J. R., Olusanya, T. O. B., and Lamprou, D. A. (2018). Characterization of Drug Delivery Vehicles Using Atomic Force Microscopy: Current Status. Expert Opin. Drug Deliv. 15 (12), 1211–1221. doi:10.1080/17425247.2018.1546693
Spiriti, J., and Wong, C. F. (2021). Qualitative Prediction of Ligand Dissociation Kinetics from Focal Adhesion Kinase Using Steered Molecular Dynamics. Life (Basel) 11 (2). doi:10.3390/life11020074
Spyratou, E., Mourelatou, E. A., Makropoulou, M., and Demetzos, C. (2009). Atomic Force Microscopy: a Tool to Study the Structure, Dynamics and Stability of Liposomal Drug Delivery Systems. Expert Opin. Drug Deliv. 6 (3), 305–317. doi:10.1517/17425240902828312
Steffens, C., Leite, F. L., Bueno, C. C., Manzoli, A., and Herrmann, P. S. D. P. (2012). Atomic Force Microscopy as a Tool Applied to Nano/biosensors. Sensors 12 (6), 8278–8300. doi:10.3390/s120608278
Storrs, R. W., Tropper, F. D., Li, H. Y., Song, C. K., Sipkins, D. A., Kuniyoshi, J. K., et al. (1995). Paramagnetic Polymerized Liposomes as New Recirculating MR Contrast Agents. J. Magn. Reson. Imaging 5 (6), 719–724. doi:10.1002/jmri.1880050617
Stroh, C., Wang, H., Bash, R., Ashcroft, B., Nelson, J., Gruber, H., et al. (2004). Single-molecule Recognition Imaging Microscopy. Proc. Natl. Acad. Sci. 101 (34), 12503–12507. doi:10.1073/pnas.0403538101
Strunz, T., Oroszlan, K., Schafer, R., and Guntherodt, H.-J. (1999). Dynamic Force Spectroscopy of Single DNA Molecules. Proc. Natl. Acad. Sci. 96 (20), 11277–11282. doi:10.1073/pnas.96.20.11277
Subramanian, S., and Catchmark, J. M. (2007). Cantilever-based Chemical Sensors for Detecting Catalytically Produced Reactions and Motility Forces Generated via Electrokinetic Phenomena. Small 3 (11), 1934–1940. doi:10.1002/smll.200700325
Sulchek, T. A., Friddle, R. W., Langry, K., Lau, E. Y., Albrecht, H., Ratto, T. V., et al. (2005). Dynamic Force Spectroscopy of Parallel Individual Mucin1-Antibody Bonds. Proc. Natl. Acad. Sci. 102 (46), 16638–16643. doi:10.1073/pnas.0505208102
Sweers, K., van der Werf, K., Bennink, M., and Subramaniam, V. (2011). Nanomechanical Properties of α-synuclein Amyloid Fibrils: a Comparative Study by Nanoindentation, Harmonic Force Microscopy, and Peakforce QNM. Nanoscale Res. Lett. 6 (1), 270. doi:10.1186/1556-276x-6-270
Takahashi, H., Hizume, K., Kumeta, M., H Yoshimura, S. K., and Takeyasu, K. (2009). Single-molecule Anatomy by Atomic Force Microscopy and Recognition Imaging. Arch. Histol. Cytol. 72 (4-5), 217–225. doi:10.1679/aohc.72.217
Takano, H., Kenseth, J. R., Wong, S.-S., O'Brie, J. C., and Porter, M. D. (1999). Chemical and Biochemical Analysis Using Scanning Force Microscopy. Chem. Rev. 99 (10), 2845–2890. doi:10.1021/cr9801317
Tang, J., Krajcikova, D., Zhu, R., Ebner, A., Cutting, S., Gruber, H. J., et al. (2007). Atomic Force Microscopy Imaging and Single Molecule Recognition Force Spectroscopy of Coat Proteins on the Surface ofBacillus Subtilis Spore. J. Mol. Recognit. 20 (6), 483–489. doi:10.1002/jmr.828
Teobaldi, G., Lämmle, K., Trevethan, T., Watkins, M., Schwarz, A., Wiesendanger, R., et al. (2011). Chemical Resolution at Ionic crystal Surfaces Using Dynamic Atomic Force Microscopy with Metallic Tips. Phys. Rev. Lett. 106 (21), 216102. doi:10.1103/physrevlett.106.216102
Then, D., Vidic, A., and Ziegler, C. (2006). A Highly Sensitive Self-Oscillating Cantilever Array for the Quantitative and Qualitative Analysis of Organic Vapor Mixtures. Sensors Actuators B: Chem. 117 (1), 1–9. doi:10.1016/j.snb.2005.07.069
Thormann, E., Hansen, P. L., Simonsen, A. C., and Mouritsen, O. G. (2006). Dynamic Force Spectroscopy on Soft Molecular Systems: Improved Analysis of Unbinding Spectra with Varying Linker Compliance. Colloids Surf. B: Biointerfaces 53 (2), 149–156. doi:10.1016/j.colsurfb.2006.08.015
Thundat, T., Allison, D. P., Warmack, R. J., Brown, G. M., Jacobson, K. B., Schrick, J. J., et al. (1992a). Atomic Force Microscopy of DNA on Mica and Chemically Modified Mica. Scanning Microsc. 6 (4), 911–918.
Thundat, T., Allison, D. P., Warmack, R. J., and Ferrell, T. L. (1992b). Imaging Isolated Strands of DNA Molecules by Atomic Force Microscopy. Ultramicroscopy 42-44 (Pt B), 1101–1106. doi:10.1016/0304-3991(92)90409-d
Thundat, T., Allison, D. P., and Warmack, R. J. (1994a). Stretched DNA Structures Observed with Atomic Force Microscopy. Nucl. Acids Res. 22 (20), 4224–4228. doi:10.1093/nar/22.20.4224
Thundat, T., Chen, G. Y., Warmack, R. J., Allison, D. P., and Wachter, E. A. (1995). Vapor Detection Using Resonating Microcantilevers. Anal. Chem. 67 (3), 519–521. doi:10.1021/ac00099a006
Thundat, T., Warmack, R. J., Chen, G. Y., and Allison, D. P. (1994b). Thermal and Ambient‐induced Deflections of Scanning Force Microscope Cantilevers. Appl. Phys. Lett. 64 (21), 2894–2896. doi:10.1063/1.111407
Vahabi, S., Nazemi Salman, B., and Javanmard, A. (2013). Atomic Force Microscopy Application in Biological Research: a Review Study. Iran J. Med. Sci. 38 (2), 76–83.
Wang, B., Guo, C., Zhang, M., Park, B., and Xu, B. (2012). High-resolution Single-Molecule Recognition Imaging of the Molecular Details of Ricin-Aptamer Interaction. J. Phys. Chem. B 116 (17), 5316–5322. doi:10.1021/jp301765n
Watanabe, A., Takagi, M., Murata, S., and Kato, M. (2018). Stability and Drug Release Studies of an Antimycotic Nanomedicine Using HPLC, Dynamic Light Scattering and Atomic Force Microscopy. J. Pharm. Biomed. Anal. 148, 149–155. doi:10.1016/j.jpba.2017.09.030
Wei, G., Steckbeck, S., Köppen, S., and Colombi Ciacchi, L. (2013). Label-free Biosensing with Single-Molecule Force Spectroscopy. Chem. Commun. 49 (31), 3239–3241. doi:10.1039/c3cc40506k
Willemsen, O. H., Snel, M. M. E., van der Werf, K. O., de Grooth, B. G., Greve, J., Hinterdorfer, P., et al. (1998). Simultaneous Height and Adhesion Imaging of Antibody-Antigen Interactions by Atomic Force Microscopy. Biophysical J. 75 (5), 2220–2228. doi:10.1016/s0006-3495(98)77666-0
Wong, C. F. (2018). Steered Molecular Dynamics Simulations for Uncovering the Molecular Mechanisms of Drug Dissociation and for Drug Screening: A Test on the Focal Adhesion Kinase. J. Comput. Chem. 39 (19), 1307–1318. doi:10.1002/jcc.25201
Wu, A., Li, Z., Yu, L., Wang, H., and Wang, E. (2002). A Relocated Technique of Atomic Force Microscopy (AFM) Samples and its Application in Molecular Biology. Ultramicroscopy 92 (3-4), 201–207. doi:10.1016/s0304-3991(02)00133-x
Xie, S., and Ren, J. (2019a). High-speed AFM Imaging via Iterative Learning-Based Model Predictive Control. Mechatronics 57, 86–94. doi:10.1016/j.mechatronics.2018.11.008
Xie, S., and Ren, J. (2019b). Recurrent-neural-network-based Predictive Control of Piezo Actuators for Trajectory Tracking. Ieee/asme Trans. Mechatron. 24 (6), 2885–2896. doi:10.1109/tmech.2019.2946344
Zhang, B., Lim, T. S., Vedula, S. R. K., Li, A., Lim, C. T., and Tan, V. B. C. (2010). Investigation of the Binding Preference of Reovirus σ1 for Junctional Adhesion Molecule A by Classical and Steered Molecular Dynamics. Biochemistry 49 (8), 1776–1786. doi:10.1021/bi901942m
Zhang, H., Molino, P. J., Wallace, G. G., and Higgins, M. J. (2015). Quantifying Molecular-Level Cell Adhesion on Electroactive Conducting Polymers Using Electrochemical-Single Cell Force Spectroscopy. Sci. Rep. 5, 13334. doi:10.1038/srep13334
Zhang, L., Hu, Y., Jiang, X., Yang, C., Lu, W., and Yang, Y. H. (2004). Camptothecin Derivative-Loaded Poly(caprolactone-Co-Lactide)-B-PEG-B-Poly(caprolactone-Co-Lactide) Nanoparticles and Their Biodistribution in Mice. J. Controlled Release 96 (1), 135–148. doi:10.1016/j.jconrel.2004.01.010
Zhang, S., Ma, L., Ma, K., Xu, B., Liu, L., and Tian, W. (2018). Label-Free Aptamer-Based Biosensor for Specific Detection of Chloramphenicol Using AIE Probe and Graphene Oxide. ACS Omega 3 (10), 12886–12892. doi:10.1021/acsomega.8b01812
Zhang, X., Cheng, H., Dong, W., Zhang, M., Liu, Q., Wang, X., et al. (2018). Design and Intestinal Mucus Penetration Mechanism of Core-Shell Nanocomplex. J. Controlled Release 272, 29–38. doi:10.1016/j.jconrel.2017.12.034
Keywords: atomic force microscopy, biosensing, drug delivery, nanomechanical characterization, biosensors
Citation: Sarkar A (2022) Biosensing, Characterization of Biosensors, and Improved Drug Delivery Approaches Using Atomic Force Microscopy: A Review. Front. Nanotechnol. 3:798928. doi: 10.3389/fnano.2021.798928
Received: 20 October 2021; Accepted: 13 December 2021;
Published: 21 January 2022.
Edited by:
Deqiang Wang, Chongqing Institute of Green and Intelligent Technology (CAS), ChinaReviewed by:
Jayne Carol Garno, Louisiana State University, United StatesCopyright © 2022 Sarkar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anwesha Sarkar, YW53ZXNoYXNAaWFzdGF0ZS5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.