- 1Department of Biological Engineering, Massachusetts Institute of Technology, Cambridge, MA, United States
- 2The Koch Institute for Integrative Cancer Research, Massachusetts Institute of Technology, Cambridge, MA, United States
- 3Department of Materials Science and Engineering, Massachusetts Institute of Technology, Cambridge, MA, United States
With the emergence of global pandemics such as the Black Death (Plague), 1918 influenza, smallpox, tuberculosis, HIV/AIDS, and currently the COVID-19 outbreak caused by the SARS-CoV-2 virus, there is an urgent, pressing medical need to devise methods of rapid testing and diagnostics to screen a large population of the planet. The important considerations for any such diagnostic test include: 1) high sensitivity (to maximize true positive rate of detection); 2) high specificity (to minimize false positives); 3) low cost of testing (to enable widespread adoption, even in resource-constrained settings); 4) rapid turnaround time from sample collection to test result; and 5) test assay without the need for specialized equipment. While existing testing methods for COVID-19 such as RT-PCR (real-time reverse transcriptase polymerase chain reaction) offer high sensitivity and specificity, they are quite expensive – in terms of the reagents and equipment required, the laboratory expertise needed to run and interpret the test data, and the turnaround time. In this review, we summarize the recent advances made using carbon nanotubes for sensors; as a nanotechnology-based approach for diagnostic testing of viral pathogens; to improve the performance of the detection assays with respect to sensitivity, specificity and cost. Carbon nanomaterials are an attractive platform for designing biosensors due to their scalability, tunable functionality, photostability, and unique opto-electronic properties. Two possible approaches for pathogen detection using carbon nanomaterials are discussed here: 1) optical sensing, and 2) electrochemical sensing. We explore the chemical modifications performed to add functionality to the carbon nanotubes, and the physical, optical and/or electronic considerations used for testing devices or sensors fabricated using these carbon nanomaterials. Given this progress, it is reason to be cautiously optimistic that nanosensors based on carbon nanotubes, graphene technology and plasmonic resonance effects can play an important role towards the development of accurate, cost-effective, widespread testing capacity for the world’s population, to help detect, monitor and mitigate the spread of disease outbreaks.
Introduction
From its emergence in December 2019, the world has been battling a fierce outbreak of the coronavirus disease 2019 (COVID-19), caused by a viral infection of the human severe acute respiratory syndrome (SARS-CoV-2) virus. Since the declaration of the COVID-19 outbreak to be a “global pandemic” by the World Health Organization (WHO) on March 11, 2020, the infection has continued to rage on, with record numbers of infection cases and high mortality rates throughout many regions of the world. As of the end of June 2021, there have been over 180 million confirmed cases of COVID-19 worldwide, including over 3.9 million deaths (WHO Coronavirus (COVID-19) Dashboard. Geneva: World Health Organization, 2020).
The mechanism of infection of the SARS-CoV-2 coronavirus in human subjects has been well studied (V’kovski et al., 2021). Briefly, the first step of infection is to bind to entry receptors on the cell surface, such as angiotensin-converting enzyme 2 (ACE2). Therefore, the expression and tissue distribution of ACE2 consequently influences the viral pathogenicity in the host organism. For instance, it has been shown that ACE2 is abundantly present in humans in the lung epithelia and small intestine (Hamming et al., 2004), especially in type I and type II pneumocytes of the lower respiratory tract. This explains the possible routes of entry and infection of coronaviruses such as SARS-CoV and SARS-CoV-2, and their pathogenesis particularly in the lung. Post receptor-mediated endocytosis, the coronaviruses replicate their genomic RNA inside the host cell to produce full length copies, which are then incorporated into newly produced viral particles. It is well known that viruses constantly evolve through genetic mutation, and new variants occur in different regions or populations of the world. The same is true for the SARS-CoV-2 virus. Four of the named variants of concern which are especially prevalent in mid 2021, which have emerged over the past year are B.1.1.7 (Alpha variant, first detected in the UK), B.1.351 (Beta variant, first detected in South Africa in Dec. 2020), P.1 (Gamma variant, first identified in travelers from Brazil), and B.1.617.2 (Delta variant, originally identified in India in Dec. 2020). Of these variants, the Alpha and Delta variants have been labeled as “SARS-CoV-2 Variants of Concern” by the U.S. Centers for Disease Control and Prevention (CDC) because of the evolved virus’ increased transmission capability, and reduced neutralization by antibodies in the serum post COVID vaccination by currently available vaccines. Recent studies have also reported that vaccine effectiveness against the Delta variant is slightly lower, compared to the other variants. For example, in a comparison study (Lopez Bernal et al., 2021) of the BNT162b2 (Pfizer-BioNTech developed mRNA vaccine) and the ChAdOx1 nCoV-19 (AstraZeneca developed replication-deficient chimpanzee adenoviral vector, containing the SARS-CoV-2 structural surface glycoprotein antigen (spike protein)), it was found that one dose of either vaccine showed low efficacy in preventing infection by the Delta variant. However, with two doses, the BNT162b2 vaccine showed 93.7 and 88.0% effectiveness, while the ChAdOx1 nCoV-19 vaccine showed 74.5 and 67.0% effectiveness against the Alpha and Delta variants, respectively. The other concerning aspect about these newly-emerged variants is the reduced sensitivity of these variants to antibody neutralization from antibodies present in the serum. Particularly, the Delta variant was found to be resistant to anti-NTD antibodies and anti-RBD antibodies, including the monoclonal antibody Bamlanivimab which lost the emergency use authorization (EUA) issued by the United States FDA. Furthermore, antibodies isolated from convalescent patients’ serum (at up to 12 mo post COVID-19 infection) were found (Planas et al., 2021) to be 4-fold less potent against the Delta variant, relative to the Alpha variant. Similar to the previous study, it was found that one dose of either the Pfizer or AstraZeneca vaccine barely inhibited the Delta variant. The good news is that with two doses of the vaccine, a neutralizing response was generated in up to 95% of the vaccinated population, albeit with lower antibody titers which are three to five-fold less effective against the Delta variant compared to the Alpha variant.
Given the emergence of these more potent, highly transmissible strains of SARS-CoV-2, there is a great, unmet clinical need for the availability of large-scale, low-cost testing platforms; both laboratory-based and point-of-care testing (Tymm et al., 2020) of the virus responsible for the coronavirus disease 2019 (COVID-19). By intelligently designing testing systems based on a combination of ideas borrowed from molecular biology, nanomaterials, analytical chemistry, signal processing and instrumentation, these types of tests offer significant promise for providing fast, accurate testing for the diagnosis of viral and other pathogenic diseases in a highly efficient, cost-effective and scalable manner to be deployed globally on a massive scale (Choi, 2020). Nanobiosensors, which are used to detect the presence of various biomolecules or microorganisms such as bacteria and viruses, work by amplifying small amounts of signal, based on the method of signal transduction (Sharma et al., 2021). Numerous forms of biosensors have been explored based on the principle of detection, such as resonant biosensors (coupled acoustic wave), optical biosensors (light), thermal biosensors (heat), ion-sensitive biosensors (change in surface potential), electrochemical biosensors (change in electrical properties of the medium), and amperometric or potentiometric biosensors (electron activity, or redox potential). Along with the role of nanotechnology in diagnostics and sensing, it is anticipated that nanomaterials as carriers for “next-generation” vaccines against diseases such as COVID-19 is an extremely important and promising area of research (Varahachalam et al., 2021); given the highly sophisticated and precious cargo (such as DNA, mRNA, engineered antigen-presenting cells, proteins, etc.) needed to be delivered to activate the immune system’s defenses against pathogenic infection.
With the recent advances (Hong et al., 2015; Bardhan, 2017) in synthesis, sorting, purification, characterization and functionalization techniques, these carbon nanomaterials are emerging as promising candidates for a number of scalable technologies; especially in the field of biosensors for rapid diagnostics. These low-dimensional nanomaterials, such as carbon nanotubes and graphene are exceptionally advantageous for biomedical applications–because of their unique physical, chemical, mechanical and opto-electronic properties and large surface area, coupled with dimensions that are comparable with those of biomolecules of interest such as DNA, proteins or viruses. As a point of reference, each SARS-CoV-2 virion is estimated (Chen N. et al., 2020) to be ∼50–200 nm in diameter; which is well-suited for analyte capture along the longitudinal dimension of an single-walled carbon nanotube (typical dimensions: ∼ 500–1000 nm in length, ∼ 1 nm in diameter), or on a 2D graphene sheet (which can range from 100 nm - a few microns in lateral dimensions). In this report, we offer a perspective of the recent advances in the field of rapid testing of SARS-CoV-2 using carbon-based nanomaterials, including graphene and carbon nanotubes, and plasmonic nanosensors, as depicted in Figure 1. Graphene and its derivatives are particularly well-suited to be used for designing nanobiosensors (Novoselov et al., 2012), as summarized in Table 1. Since the focus of this article is to discuss the application of these nanosensors for COVID-19 detection, we do not go into great detail about the numerous, wide-ranging applications of carbon-based nanosensors, and the reader is referred to three excellent reviews on this topic for further exploration (Tîlmaciu and Morris, 2015; Banerjee, 2018; Shahdeo et al., 2020).
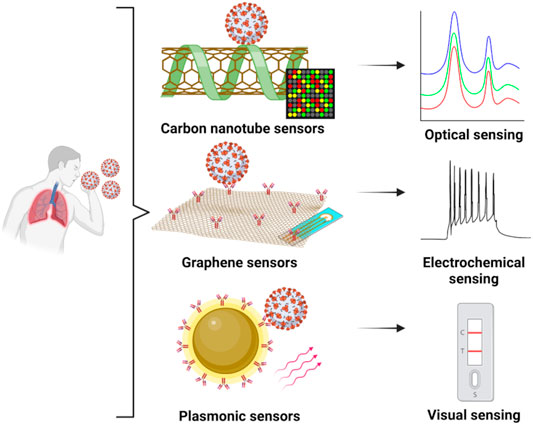
FIGURE 1. Nanosensors based on graphene, carbon nanotubes or plasmonic nanomaterials and their application towards the detection of COVID-19 in sample specimens from human subjects: concepts based on optical, electrochemical or visual readout assays. Created with BioRender.com.
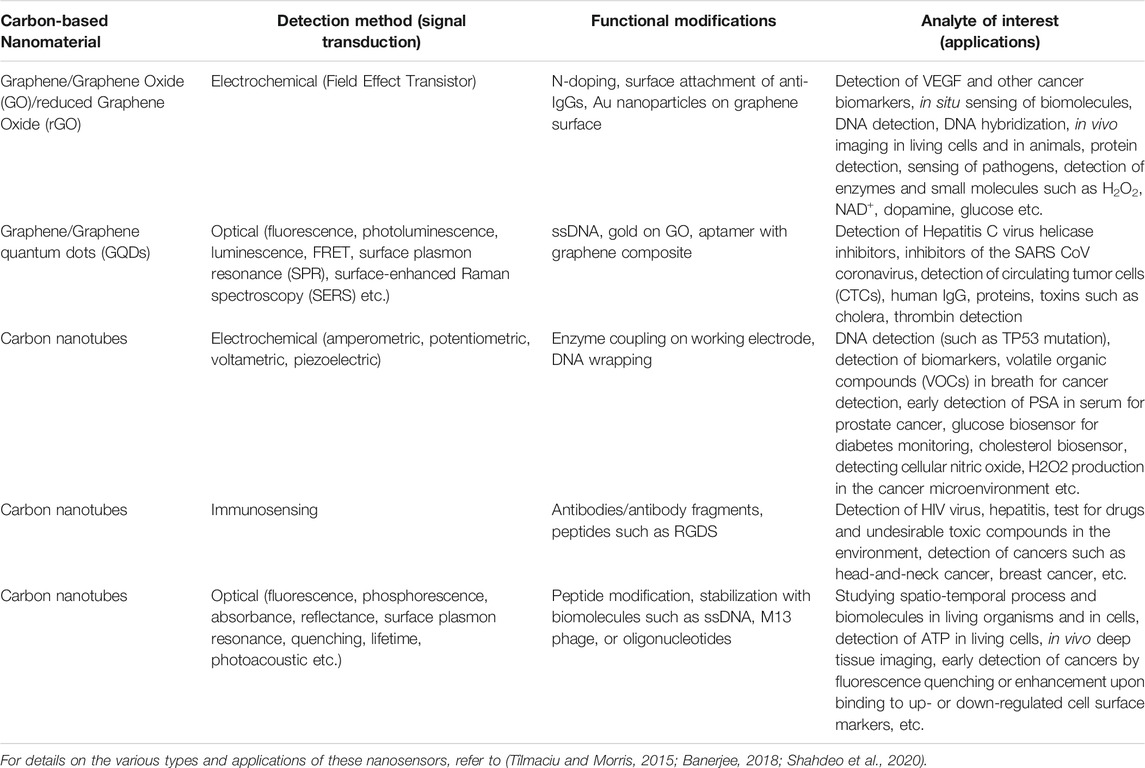
TABLE 1. Graphene and its derivative carbon-based nanomaterials: representative applications in nanobiosensing.
Before we jump into the discussion of some of the COVID-19 nanosensors using carbon-based nanomaterials reported in the literature, it is important to get a brief overview of the surface functionalization techniques used for these materials. Broadly speaking, there are two classes of functionalization methods: 1) non-covalent surface interactions (through methods such as supramolecular π-π stacking and hydrophobic interactions), and 2) covalent surface modification with other functional groups. Examples of the former include methods used to wrap SWNTs and graphene sheets using surfactants (O’Connell et al., 2002), polymers (Liu et al., 2009; Welsher et al., 2009; Bardhan and Belcher, 2020), DNA (Zheng et al., 2003), proteins and other biological templates such as M13 viruses (Dang et al., 2011; Bardhan et al., 2014; Ghosh et al., 2014; Ceppi et al., 2019) to achieve good dispersion and additional functionality such as targeting moieties to biomolecules of interest. In recent times, viruses have been exploited by engineering the surface of the viral capsid through genetic and other chemical methods, with the resulting virus-like particles (VLPs) and viral nanoparticles having important implications in drug delivery, chemotherapy, vaccine production, immunotherapy, and molecular imaging, to name a few (Aljabali et al., 2021). On the contrary, covalent functionalization techniques also present their unique set of advantages due to the high stability of covalent bonding mechanisms. For example, the high surface area and large number of oxygen-rich functional groups (Kumar et al., 2014, 2016) in graphene oxide (GO) and reduced graphene oxide (rGO) make them ideally suited as substrates for immobilizing biomolecules of interest (Reina et al., 2021), as illustrated in Figure 2A, which can then be used to capture cells (Chen et al., 2015; Bardhan et al., 2017) or analytes of interest (Pumera, 2011) for pathogen detection. These 2D materials can also be further developed with special properties, such as an antibacterial surface coating designed by incorporating cobalt ferrite into GO as a “magnetic paint” (Arun et al., 2019), with induced generation of reactive oxygen species used as the anti-bacterial mechanism. In contrast to two-dimensional (2D) materials such as graphene and its derivatives, one-dimensional (1D) materials such as carbon nanotubes come with their own set of unique challenges and opportunities. For example, single-walled carbon nanotubes (SWNTs) are an attractive candidate for biosensing, due to their photoluminescence in the 900–1,400 nm range, large Stokes’ shift (separation between excitation and emission wavelengths), low-autofluorescence background, relative insensitivity to photobleaching compared with organic dyes, the ability to be functionalized with targeting/drug delivery agents for virucidal action. CNTs can also be tailored to have binding affinity towards different receptors or molecules on the surface of the target of interest - in this case, the SARS-CoV-2 virus as depicted in Figure 2B, and the binding strength can be tuned by adjusting parameters such as the tube diameter, chirality, and surface functionalization, as predicted through molecular docking simulation studies (Patel et al., 2021). Given these promising attributes, the logical extension of the use of carbon-based nanomaterials for the purpose of designing rapid, low-cost, point-of-care diagnostic solutions for COVID-19 detection has been explored in the past year, and we review the recent advances in Section 2 and Section 3 below, with a special focus on optical and electrochemical sensors.
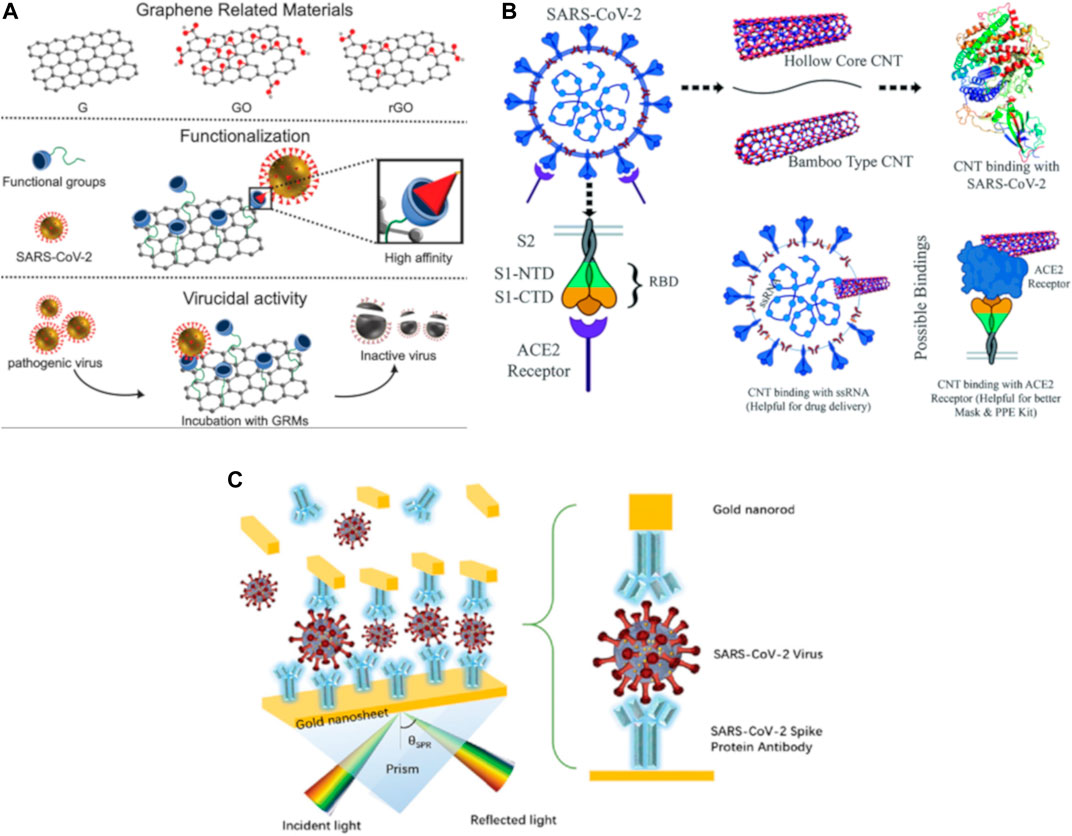
FIGURE 2. An overview of the numerous possibilities for the use of graphene, carbon nanotubes and plasmonic nanosensors for COVID-19 sensing, diagnostics and treatment. (A) Graphene and its derivative materials (G, graphene; GO, graphene oxide; and rGO, reduced graphene oxide) can be used to selectively bind to the SARS-CoV-2 virus through chemical functionalization with specific functional groups, and this can be harnessed to detect, capture and even neutralize the viral pathogen. Reprinted (adapted) with permission from (Reina et al., 2021). (B) Molecular docking simulations showing the binding interaction between multi-walled carbon nanotubes (CNTs) and targets of interest: the receptor-binding domain, ssRNA, or the ACE2 receptor corresponding to the SARS-CoV-2 virus. The binding affinity can be tuned depending on CNT tube structure, diameter, chirality, and other factors. Reprinted (adapted) from (Patel et al., 2021), CC BY 3.0 license. (C) Representative conceptual structure of a plasmonic immuno-assay based on a “sandwich-type” structure, demonstrating the concept of using gold nanorods to enhance the electric field, and tuning the angle of reflection of the surface plasmon resonance. Reprinted (adapted) from (Das et al., 2020). Copyright Wiley-VCH GmbH. Reproduced with permission.
While sensors based on carbon-based nanomaterials such as graphene and CNTs are relatively new, appearing mainly in the last 10–20 yr, plasmonic nanosensors for biosensing applications have been under development for nearly 40 yr. In a very simplistic explanation, the phenomenon of plasmon resonance refers to the changes in electric field brought about by the oscillation of the free electrons of a metallic nanoparticle (typically Au or Ag), which is influenced by the wavelength of incident light, the dielectric medium, as well as the proximity to other metal particles (plasmonic coupling), resulting in strong amplification of the electric field. Plasmonic nanoparticles are strong absorbers and scatterers of light, and the optical response can be tuned by change the nanoparticle shape, size and composition, the wavelength and angle of the excitation light source, as well as the configuration of the nanoparticle and its interaction with the target biomolecule of interest, as demonstrated in the simulation studies (Das et al., 2020) in Figure 2C. Compared to other sensing modalities for virus detection such as gene sequencing, polymerase chain reaction, enzyme-linked immunosorbent assay (ELISA) which all require expensive sophisticated instrumentation, expert sample handling and long time for detection, sensors based on plasmonic phenomena such as surface plasmon resonance, localized surface plasmon resonance, surface-enhanced Raman scattering, surface-enhanced fluorescence or IR absorption spectroscopy can offer significant advantages (Anker et al., 2008): high sensitivity, low cost, minimal sample pre-treatment, rapid turnaround time from sample collection to assay result, and simple instrumentation, to name a few. There have been numerous comprehensive reviews covering the use of plasmonic biosensors for detection of viral (Dengue virus, Zika, Ebola, HIV, Hepatitis B, Avian influenza, Norovirus, etc.) pathogens, and the reader is referred to two such recent manuscripts for further learning (Hassan et al., 2021; Shrivastav et al., 2021). Given the focus of the present review, however, we choose to highlight the recent reports on the use of plasmonic nanosensors for detection of the SARS-CoV-2 virus. Numerous innovative techniques have been tried in the wake of the COVID-19 pandemic to enhance the sensitivity of these plasmonic nanosensors, such as in situ hybridization of the target gene of the virus, dual-mode sensing, colorimetric readout with RNA amplification, and others, as discussed in Section 2 and Section 3 below.
Optical Sensing of COVID-19
One important class of biosensors being developed for rapid, accurate detection of the SARS-CoV-2 virus is optical nanosensors (Maddali et al., 2021). Out of the various types of optical sensors, those based on the phenomenon of localized surface plasmon resonance (LSPR) are suitable for detecting various kinds of analytes. Typically, metal nanoparticles are optimally placed near fluorophores to enhance the fluorophore excitation rate via local enhancement of the electric field (Halas et al., 2011). This can also impact the radiative decay rate of the excited state of the fluorophore, and open up new non-radiative channels of decay, resulting in changes in the quantum yield and the lifetime of the fluorophore. For example, using an M13 virus-based framework, a 24-fold enhancement in the fluorescence of Cy3 dye was reported (Huang et al., 2019), by controlling the distance between the dye and silver nanoparticles (AgNPs) and the PEG spacer layer. Building on this approach, surface plasmon-enhanced fluorescence imaging in the short-wave infrared (SWIR) wavelength region has been successfully deployed for other areas of interest in bioimaging, such as in the imaging of cancers. For instance, high brightness fluorescence probes based on a combination of gold nanorods decorated with an SWIR dye, IR-E1050, was shown (Huang et al., 2021) to enable in vivo imaging of sub-millimeter sized tumors, in a mouse model of ovarian cancer. In addition to the numerous advances made in the field of nanosensors for molecular diagnostics, a suite of technological innovations in optics hardware and image processing algorithms, such as hyperspectral imaging (Dang et al., 2019) and plasmonic metamaterials (Lee et al., 2019) to name a few, has played an instrumental role in pushing the limits of opto-electronic detection capabilities. Therefore, when faced with the challenging circumstances in the COVID-19 pandemic, it has been naturally useful to come up with ways to adapt these known techniques of optical nanosensors used in other fields (such as cancer imaging) and make them suitable for the purposes of rapid detection (Shrivastav et al., 2021) of the SARS-CoV-2 viral pathogen. A summary of these methods is reported in Table 2.
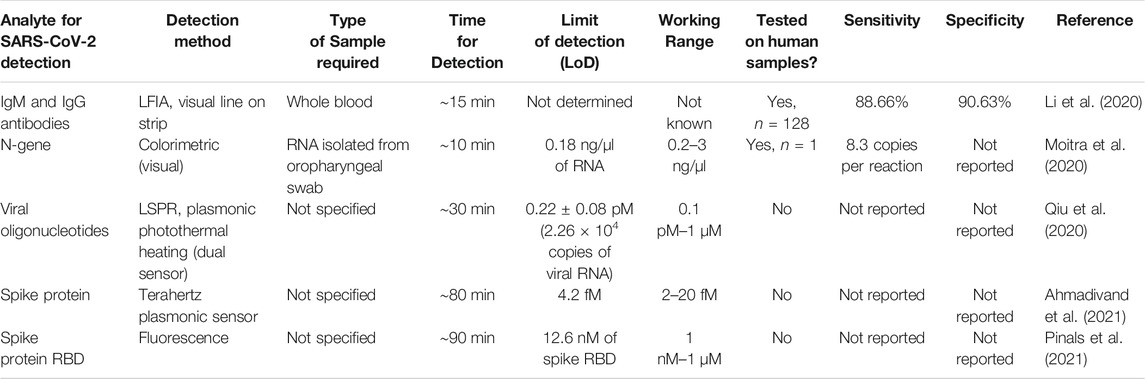
TABLE 2. Summary of reported methods for detecting SARS-CoV-2 using optical biosensors based on carbon and plasmonic nanomaterials.
More recently, in the context of the COVID-19 pandemic, various research groups have worked on the idea of adapting plasmonic resonance as a method of detection for the SARS-CoV-2 viral pathogen. In one of the first reported applications of the use of this technology, a lateral flow immunoassay (LFIA) chip was developed (Li et al., 2020) as fast, rapid (<15 min) point-of-care diagnostic chip for the detection of the presence of IgM and IgG antibodies against the SARS-CoV-2 virus in blood serum, shown in Figure 3A. As the patient sample flows through the device, the antibodies, if present (indicating COVID-19 infection) undergo a binding event with the Au-COVID-19 antigen conjugates placed on the conjugate pad. This binding results in a change in color of the bound complex due to localized surface plasmon resonance with the Au nanoparticles, and can be visually detected as the appearance of a line on the diagnostic strip. This assay requires only about 20 µl of whole blood sample, or 10 µl of serum/plasma from the patient, and can return a test result within 15 min, with a reported overall sensitivity of ∼88.66% and a specificity ∼90.63%. Similar LFIA platforms have been subsequently reported (Chen Z. et al., 2020), with slight variations in the readout signal by modifying the optical agent (using lanthanide-doped nanoparticles instead of Au nanoparticles, for example).
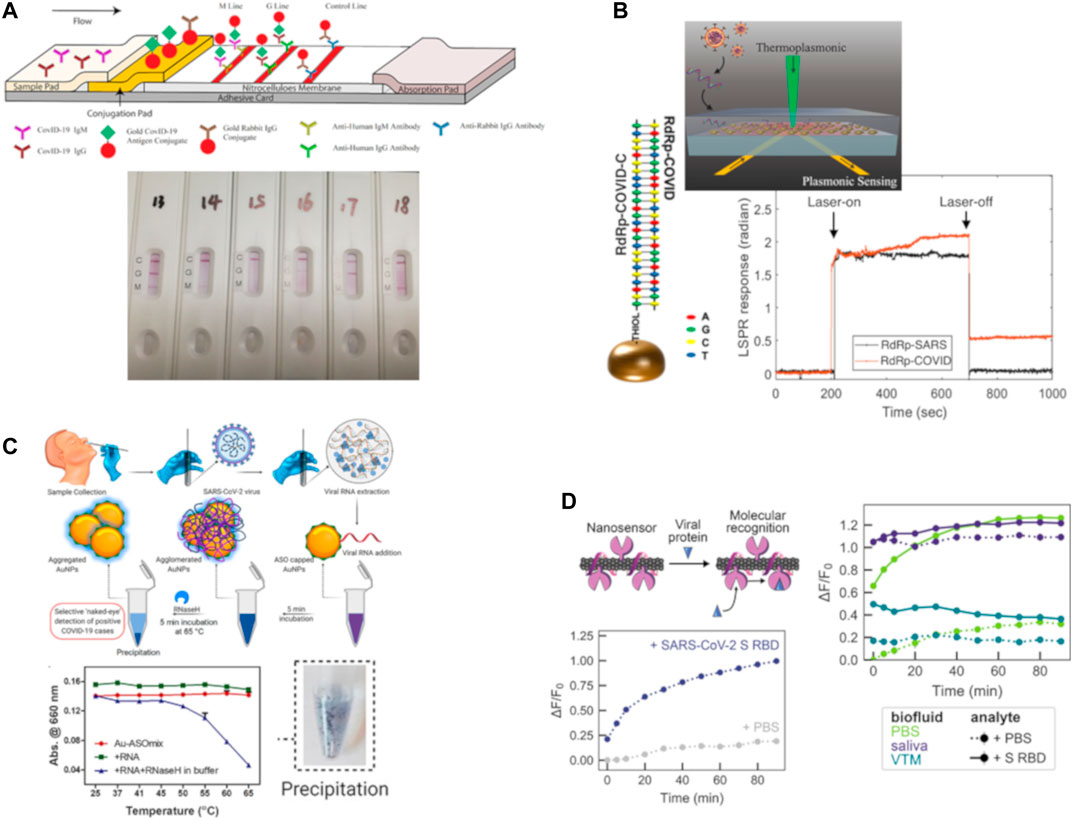
FIGURE 3. Representative examples of optical nanosensors for COVID-19 detection. (A) A lateral flow immuno-assay (LFIA) test strip, combined testing for both IgM and IgG antibodies against SARS-CoV-2. The bottom photo shows sample results from human patients. Credit: Reprinted (adapted) from (Li et al., 2020), CC BY 4.0 license. (B) Schematic of the dual-functional (plasmonic photothermal + localized surface plasmon resonance) biosensor, with the hybridization of the two complementary strands of the RdRp gene of SARS-CoV-2. Laser-activated plasmonic photothermal enhancement enables distinguishing between two similar viral sequences, SARS-CoV (responsible for the SARS epidemic in 2003) and SARS-CoV-2 (responsible for COVID-19). Reprinted (adapted) with permission from (Qiu et al., 2020). Request for further permissions to reuse should be directed to American Chemical Society. (C) Protocol for selective naked-eye detection of SARS-CoV-2 RNA, mediated by antisense oligonucleotides-capped Au nanoparticles. Plot shows relative change in absorbance at 660 nm, upon incubation with RNase H at 65°C for 5 min. Photo shows precipitation of the AuNPs, indicating a positive test visual indication. Credit: Reprinted (adapted) from (Moitra et al., 2020), part of the ACS COVID-19 subset. Copyright © 2020, American Chemical Society. (D) Conceptual visualization of the SARS-CoV-2 spike protein binding to single-walled carbon nanotubes (SWNTs) functionalized with the ACE2 protein. Plot showing the change in fluorescence of SWNTs upon binding of the SARS-CoV-2 spike protein, and improved nanosensor performance achieved by mitigating biofouling by passivation with a phospholipid-PEG polymer. Reprinted (adapted) with permission from (Pinals et al., 2021). Copyright © 2021, American Chemical Society.
In another instance, a dual-functional plasmonic biosensor was designed (Qiu et al., 2020), combining the detection capability of LSPR resonance signal with the heat generation capability of plasmon-induced photothermal effect for highly specific detection, as shown in Figure 3B. In this approach, the authors took advantage of the large optical cross-section of the plasmonic nanoparticles, namely two-dimensional Au “nano islands,” which results in partial non-radiative decay of the excited light, causing large photothermal heating. This local photothermal heating was employed to induce in situ hybridization of the RNA-dependent RNA polymerase sequence, and the complementary DNA, for highly sensitive, accurate and label-free detection of the SARS-CoV-2 virus. Importantly, given the similarity between the genetic targets of the SARS-CoV-2 and SARS-CoV viruses, this method was able to successfully distinguish between the two by improving the specificity of hybridization at the elevated temperature of 41°C, achieved using the plasmonic photothermal heating. This dual-functional biosensor was shown to have a range from 0.1 pM to 1 µM for detecting oligonucleotides, with the lower limit of detection reported to be ∼0.22 ± 0.08 pM. In terms of the applicability of this biosensor for clinical detection, it was projected that the sensor could be capable of detecting as few as 2.26 × 104 copies; which is significantly smaller than the viral load after the onset of COVID infection, estimated to be >1 × 106 copies per ml from respiratory samples of infected individuals.
Another unique approach to the detection of SARS-Cov-2 using a “naked-eye” diagnostic test was reported (Moitra et al., 2020) using plasmonic nanoparticles. Gold nanoparticles, or AuNPs, which were thiolated and capped with suitable anti-sense oligonucleotides which are specific to the N-gene (the nucleocapsid phosphoprotein) of the SARS-CoV-2 virus. These AuNPs agglomerate only in the presence of the target RNA sequence of the SARS-CoV-2 RNA, and this causes a change in its surface plasmon resonance, with an ∼40 nm red shift in the absorption spectrum. Upon addition of RNaseH, this is further amplified, resulting in the precipitation of AuNPs which is visually detectable as a colorimetric assay, as illustrated in Figure 3C. For this approach, the dynamic range is 0.2–3 ng/μl, and the lower limit of detection is 0.18 ng/μl of RNA. This method enables a visual naked-eye detection of the COVID-19 virus in <10 min from viral RNA, and according to the researchers, this diagnostic nanosensor passes the World Health Organization’s (WHO’s) ASSURED criteria; having the characteristics of being Affordable, Sensitive, Specific, User-friendly, Rapid and robust, sophisticated Equipment-free, and Delivered to the end-users. It should be noted here, however, that diagnostic assays based on the N-protein of the SARS-CoV-2 virus are generally less reliable compared to those relying on the detection of the spike or S-protein of the virus. Furthermore, it is interesting to consider how the choice of the diagnostic assay makes a remarkable difference in the sensitivity, specificity and accuracy. As discussed in the following section, the same group of researchers had previously reported (Alafeef et al., 2020) an electrochemical sensing using almost identical chemistry of anti-sense oligonucleotides, but arranged in the form of an electrochemical biosensor on a graphene platform. While the latter has been reported to have nearly 100% sensitivity, accuracy and specificity, the colorimetric assay described here does suffer from relatively lower sensitivity.
More recently, there has been tremendous progress in femtomolar-level detection capability (Ahmadivand et al., 2021) of the SARS-CoV-2 virus spike protein, using terahertz plasmonic metasensors. For this technique, the Spike S1 antibody reacting to the SARS-CoV-2 virus spike protein was conjugated with gold nanoparticles, and then dispersed onto the platform surface of a toroidal-shaped Terahertz metamolecule. The presence of the analyte of interest (in this case, the captured spike proteins) is then measured by the shift in the position of the toroidal dipole moment. Based on a signal-to-noise ratio (SNR) cutoff of 4, the authors report a lower limit of detection of ∼4.2 fM of the spike protein; about two orders-of-magnitude better than the LoD of the aforementioned dual-functional biosensor. Another advantage of this technique is the relatively rapid analysis; requiring only about 80 min from sample to result. This kind of an immunobiosensor has the potential to be deployed on a wide scale as a low-cost, selective, rapid and highly sensitive point-of-care diagnostic to meet the challenges of a global pandemic.
For a more direct approach to optical sensing, it is appealing to consider the use of the intrinsic near-infrared fluorescence properties of single-walled carbon nanotubes (SWNTs). In a recent report (Pinals et al., 2021), an innovative approach was used comprising of a protein-SWNT nanosensor construct, which can bind to the spike protein’s receptor binding domain on the SARS-CoV-2 virus particle. By using a non-covalent functionalization strategy to immobilize the ACE2 proteins on the surface of the SWNT, the researchers were able to retain the fluorescence properties of the SWNT, as shown in Figure 3D. These nanosensors were shown to have impressive binding capability to the SARS-CoV-2 spike protein, with a lower limit of detection of 12.6 nM of spike protein. While this nanosensor is less sensitive than what is required for the detection of realistic viral loads in clinical samples of COVID-19 (typically in the range of ∼10–104 viral copies per µl, corresponding to ∼5 fM–5 pM of the spike protein), there are ways in which the performance of the nanosensor could be improved further to reduce the bio-fouling problem which affects the sensor’s fluorescence response. This type of optical diagnostic nanosensor could lead the way for rapid, point-of-care testing for SARS-CoV-2 and other pathogens, which could be helpful in management of pandemics such as COVID-19 and other emerging diseases.
Electrochemical Sensing of COVID-19
Graphene and its derivatives (such as graphene oxide, GO, or reduced graphene oxide, rGO) have been explored in versatile ways (Jiang et al., 2020) for the detection of viral pathogens, including the Zika, Ebola, MERS and HIV viruses, to name a few. More recently, there has been significant progress made towards the use of techniques such as CRISPR-associated nuclease (Cas) proteins, in the form of a CRISPR-enhanced graphene-based field-effect transistor fabricated as a “CRISPR-Chip” (Hajian et al., 2019), to enable rapid and selective detection of a target sequence contained within genomic DNA. Using the selective binding event of the target DNA to the deactivated Cas9 CRISPR complex, it is possible to obtain a readout signal from the graphene FET in as little as 15 min without amplification, with a limit of detection of ∼1.7 fM of the target gene. A summary of these methods is reported in Table 3.
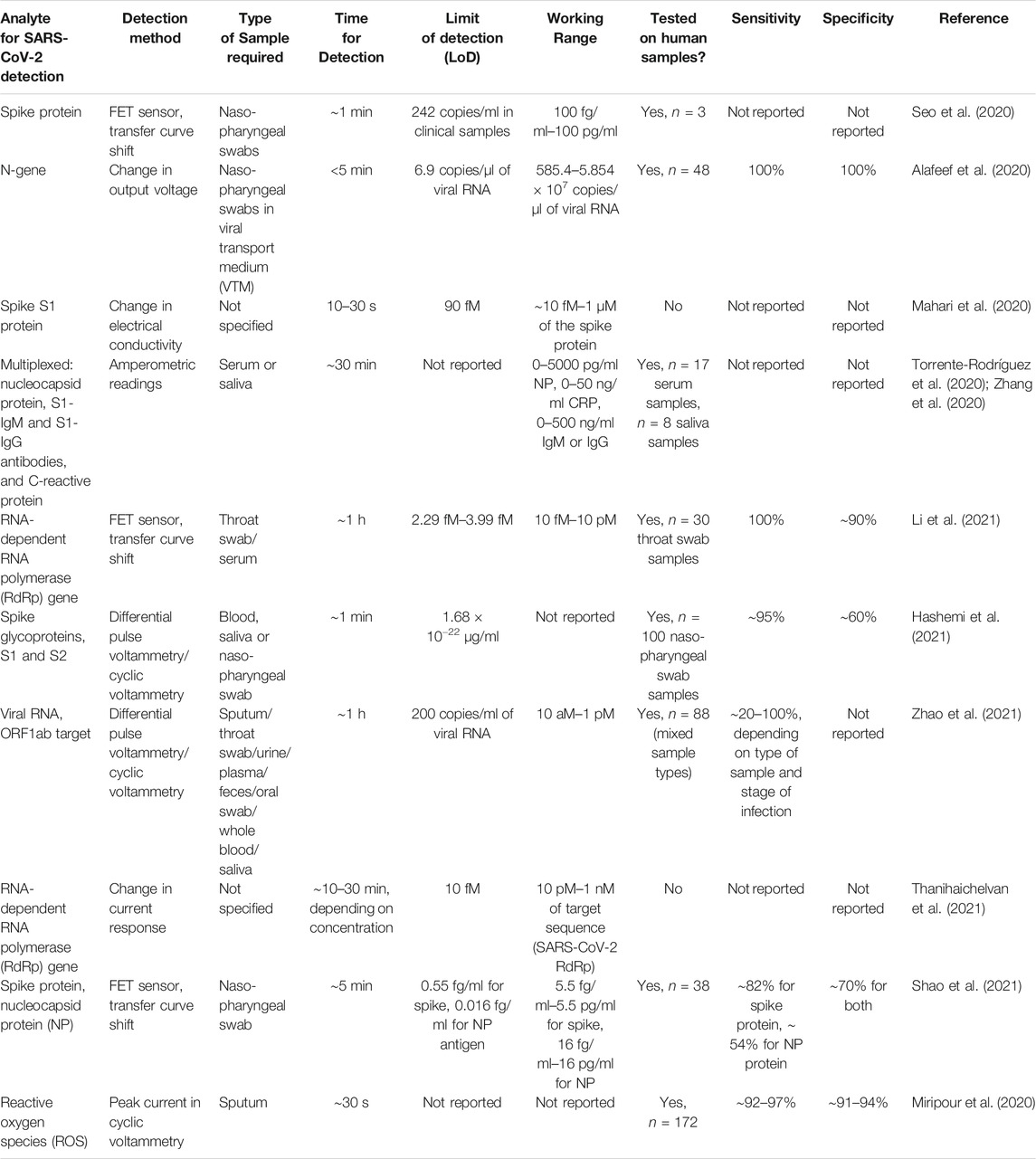
TABLE 3. Summary of reported methods for detecting SARS-CoV-2 using electrochemical biosensors based on carbon nanomaterials.
In one of the earliest reports (Seo et al., 2020) of the use of electrochemical sensors for the detection of the SARS-CoV-2 virus, a graphene-based field effect transistor (FET) was used, coated with an antibody against the spike protein of the virus, as illustrated in Figure 4A. Graphene, which is a two-dimensional material of hexagonal sp2 carbon, offers unique advantages for biosensing due to its properties such as high electronic conductivity, high carrier mobility, and large exposed surface area. Through a 1-step coupling process using 1-pyrenebutyric acid N-hydroxysuccinimide ester (PBASE) which is an efficient interface coupling agent, the SARS-CoV-2 spike antibody was conjugated on the surface of the graphene sheet. This study reported a lower limit of detection of 1 fg/ml of the spike protein. This FET sensor was demonstrated to be able to detect the antigen from cultured virus samples, as well as from nasopharyngeal swabs from n = 3 human patients, and clinically-relevant samples diluted down by a factor of 105 (as few as 242 copies/ml). Importantly, the FET sensor was able to distinguish the spike protein of the SARS-CoV-2 virus from the protein of the MERS-CoV virus.
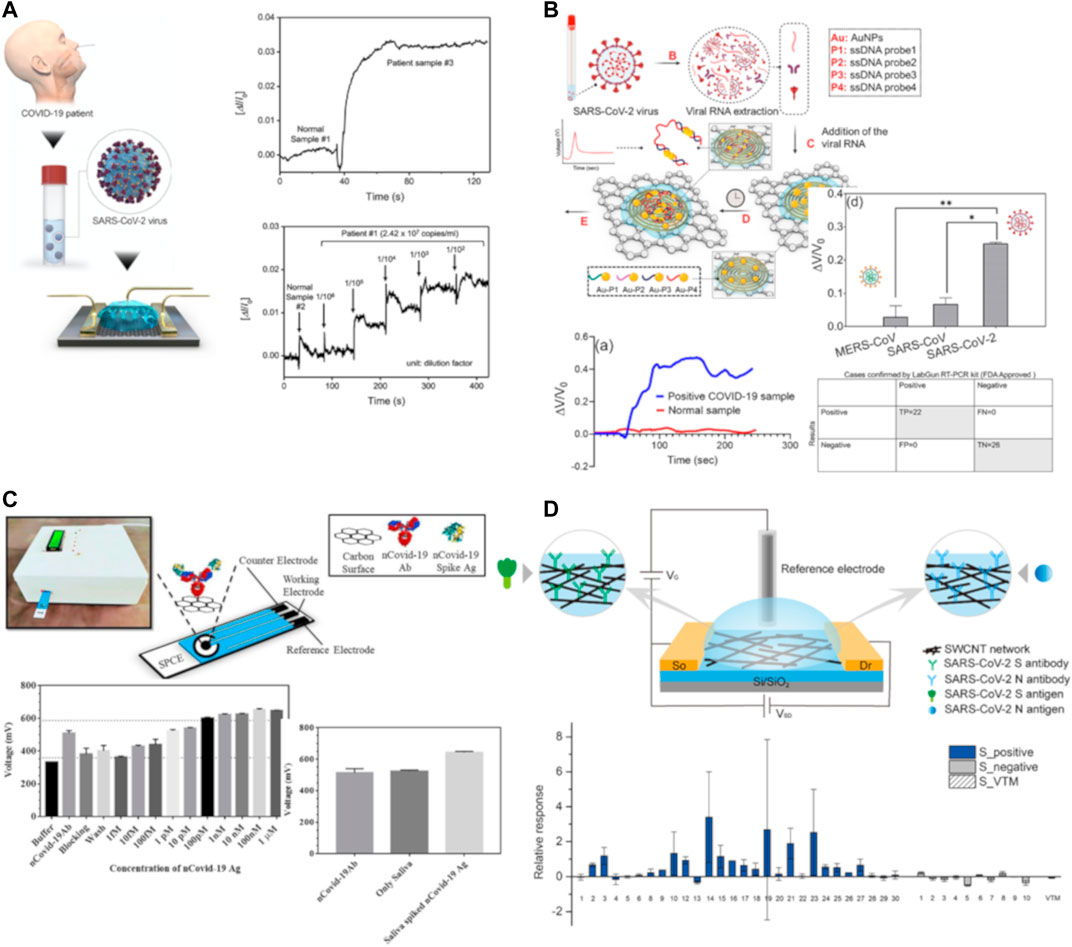
FIGURE 4. Representative examples of electrochemical nanosensors for COVID-19 detection. (A) Graphene-based field effect transistor (FET), conjugated with an antibody against the spike protein of SARS-CoV-2. Plots show response signal from normal v. COVID-19 positive patient, and dose-dependent response curve. Credit: Reprinted (adapted) from (Seo et al., 2020), part of the ACS COVID-19 subset. Copyright © 2020, American Chemical Society. (B) Paper-based electrochemical sensor based on Au nanoparticles capped with antisense oligonucleotides targeting the N-gene of SARS-CoV-2, on a graphene substrate. Protocol shows workflow from human sample to RNA extraction, 5 min incubation and sensor readout. Plots show sensor response difference between COVID-19 positive and healthy samples, the ability to distinguish between SARS-CoV-2 and related coronaviruses (MERS-CoV or SARS-CoV), and confusion matrix of the classification results of the sensor on n = 48 clinical samples, compared to the standard RT-PCR assay results. Credit: Reprinted (adapted) from (Alafeef et al., 2020), part of the ACS COVID-19 subset. Copyright © 2020, American Chemical Society. (C) eCovSens device, based on a screen printed carbon electrode with an nCovid-19 antibody immobilized on a graphene surface. Plots show sensor response to the nCovid-19 antigen at different concentrations (1 fM–1 µM), and saliva-spiked samples. Reprinted (adapted) with permission from (Mahari et al., 2020). (D) Liquid-gated single-walled carbon-nanotube FET for detection of the spike protein or nucleocapsid protein of SARS-CoV-2 virus. Response of spike antibody-functionalized FET sensor to clinical samples, containing both COVID-19 positive and negative cases, and blank viral transport medium (VTM). Sensitivity of detection was 82% (23 out of 28 COVID-19 positive detected), compared to the standard nucleic acid amplification test results. Credit: Reprinted (adapted) from (Shao et al., 2021), part of the ACS COVID-19 subset. Copyright © 2021, American Chemical Society.
Yet another interesting implementation of a rapid (<5 min), low-cost electrochemical sensor was reported (Alafeef et al., 2020) based on a graphene sensor platform decorated with Au nanoparticles, resulting in surface-enhanced Raman scattering. By using antisense oligonucleotides specific to the N-gene of the SARS-CoV-2 virus, the authors designed a combination nanosensor to simultaneously target 2 separate regions within the N-gene, which increased the sensor performance compared to other electrochemical assays, as shown in Figure 4B. The authors reported that this test was successful in distinguishing SARS-CoV-2 from related viral infections, caused by the SARS-CoV and MERS-CoV pathogens, and the output signal could be obtained in as little as 5 min of incubation, with a sensitivity of 231 copies/µl and a limit of detection ∼6.9 copies/µl, with a linear response in the range of 585.4–5.854 × 107 copies of viral RNA per µl. In a clinical test sample of n = 48, the sensor was able to identify COVID-19 positive and negative samples with a sensitivity, specificity and accuracy of 100% (as verified with an FDA EUA-approved RT-PCR diagnostic kit). Finally, it is worth highlighting that the authors estimated that these electrochemical sensors could be deployed very inexpensively, at a cost of ∼ $10 per sensor, which makes them relatively affordable for use on a wide scale to combat the global pandemic of COVID-19.
Rapid, portable, low-cost diagnostics of COVID-19 are strongly needed in order to manage and mitigate the spread of the coronavirus pandemic in large population areas; especially under resource-constrained settings. Researchers in India have devised a novel biosensor, which is a printed circuit board-based electrochemical device, dubbed “eCovSens” (Mahari et al., 2020) for ultra-sensitive detection of the spike protein domain I of the SARS-CoV-2 virus, as depicted in Figure 4C. This device works on the principle of detecting changes in the electrical conductivity upon binding of the analyte of interest (the SARS-CoV-2 virion) to its antibody, immobilized on to the surface of a screen printed carbon electrode. The working range of this device was reported to be ∼10 fM–1 μM, under optimal conditions; which is similar to the performance of a more complex, expensive potentiostat-based immunosensor. In spiked saliva samples, the eCovSens sensor had a limit of detection ∼90 fM, and can be used as a rapid diagnostic tool for detection of COVID-19 within 10–30 s. This low-cost, portable instrument is very suitable for point-of-care diagnostics, as it can be battery operated, and is stable up to 4 wk with no change in voltage, compared to the need for expensive equipment such as a potentiostat which can only be run in a laboratory setting.
Similar to the previous approach, another case of a flex printed circuit board (FPCB) DNA sensor for the detection of SARS-CoV-2 was reported (Damiati et al., 2021) using graphene as a working electrode, based on the hybridization of the captured DNA molecule using a simple streptavidin-biotin interaction platform. The functionalized electrode was then used to test the detection capability against a synthetic strand of ssDNA, which is similar to the ORF1ab sequence of the SARS-CoV-2 virus. After capture, the hybridization reaction was performed at 37°C for 30 min; followed by differential pulse voltammetry measurements to analyze the captured ssDNA sequence versus the target sequence. This sensor enabled label-free, rapid and sensitive detection of the target DNA sequence in a working range of 0.1–106 pg/ml. The calculated limit of detection of the sensor was estimated to be ∼32.88 fg/ml, corresponding to about 5 × 105 copies/µl, and in a similar range of sensitivity comparable to that achieved by nucleic acid tests such as loop-mediated isothermal amplification (LAMP). Further investigation of this highly sensitive sensor needs to be done, to determine its performance with viral RNA from human clinical samples.
Given the need for large-scale, low-cost deployment of Point-of-care diagnostic solutions for COVID-19, the next innovation is particularly intriguing. Dubbed as RapidPlex, it is a graphene-based, multiplexed wireless telemedicine platform (Zhang et al., 2020), designed by researchers at CalTech. Using a mass-produced laser-engraved graphene platform with four working electrodes (Torrente-Rodríguez et al., 2020), the method can quantitatively detect biomarkers in blood and saliva of human subjects, including those of the SARS-CoV-2 nucleocapsid protein, antibodies against the spike protein (S1-IgM and S1-IgG), and C-reactive protein; which can then be used to diagnose aspects of the disease, viz. viral infection (NP), immune response (IgG and IgM), and disease severity (CRP). Upon testing in human serum samples from n = 17 subjects, (10 COVID-19 positive and seven negative by RT-PCR test results), and human saliva samples from n = 8 subjects, (5 COVID-19 positive and three negative by RT-PCR test results), the authors observed that the biosensors showed much higher signals in COVID-19 positive patients’ serum and saliva; with target capture in as little as 1 min, which shows the potential of this platform to be used for monitoring population-level spread of the viral infection.
Another detection technique is based on reduced graphene oxide-based FET nanosensors designed by using Au nanoparticles, functionalized with a peptide nucleic acid, phosphorodiamidate morpholino oligos (PMO) (Li et al., 2021). By hybridization of the PMO with the SARS-CoV-2 RdRp gene, the sensor shift can be detected rapidly within 2 min, without the need for amplification. The lower limit of detection of this sensor was in the femtomolar range, with LoD ∼0.37 fM in PBS, ∼ 2.29 fM in throat swab samples, and ∼3.99 fM in serum. In tests with n = 30 clinical throat swab samples, the sensor demonstrated outstanding accuracy, with the area under the ROC curve found to be ∼0.995. A Kappa test comparison of the sensor’s performance with the RT-PCR results yielded a Kappa index ∼0.92, showing near perfect agreement with the gold standard detection test. One of the unique aspects of this nanosensor is the re-usability, which offers the potential for cost savings: after denaturing the PMO-RNA hybrid duplex, it is possible to reintroduce the RdRp sequence to the nanosensor’s surface, and the signal output was very similar after three such cycles of hybridization and denaturing. This PCR-free, direct detection technique can be used for a quick, rapid screening tool in a sensitive, reliable and accurate manner for large-scale testing in COVID-19 hotspots.
While the next electrochemical detection system is based on using the same classes of materials, viz. graphene oxide (GO) with gold nanostars (Au NS), this one differs from the previous ones by detecting traces of viral glycoproteins in biological media; instead of relying on the detection of biomarkers or other molecules such as RNA obtained through extraction from viral particles. The active part of the “NanoSystem” biosensor (Hashemi et al., 2021) consists of GO coated with 8-hydroxyquinoline, 1-ethyl-3-(3-dimethylaminopropyl carbodiimide) (EDC) and N-hydroxysuccinimide (NHS) coupled with Au NS. The main mechanism of detection is hypothesized to be the adsorption of viral glycoprotein-based structures, which is enhanced by the Au NS hybridization with the GO-8H-EDC-NHS surface, which provides an increase in the active surface area and a larger number of reactive hydroxyl functional groups for protein binding. The sensor was shown to be able to distinguish between various strains of human or animal viruses, including the SARS-CoV-2 virus, infectious bronchitis virus (IBV), avian influenza and Newcastle Disease Virus (LaSota and V4 strains). The limit of detection of this nanosensor was reported to be ∼1.68 × 10−22 μg/ml, for the detection of the SARS-CoV-2 virus in spiked biological media. Upon further testing for SARS-CoV-2 detection with n = 100 clinical samples of nasopharyngeal swab specimens, the performance of this sensor was compared with test results from the standard RT-PCR assay. The sensor was reported to have a sensitivity of ∼95%, with a somewhat lower specificity of ∼60% due to a large number of false positives. However, this nanosensor-based platform shows promising potential for the rapid detection (<1 min) of a number of different viral diseases; even in trace amounts.
Smartphone-based diagnostics are certainly worth exploring further, for the purposes of making wide-scale, low-cost, point-of-care diagnostics more accessible to the population. Towards this end, it is worth discussing the electrochemical sensor based on a supersandwich-type recognition strategy (Zhao et al., 2021), based on sulfocalixarene (SCX8)-functionalized reduced graphene oxide, and decorated with the capture probe CP/Au@Fe3O4 nanocomposites for targeting the RNA of SARS-CoV-2. In spiked samples with the target for detection of the ORF1ab gene of SARS-CoV-2, the sensor demonstrated outstanding performance, with a limit of detection of 3 aM. Further, in n = 88 RNA samples taken from 25 clinically COVID-positive and eight recovered patients, the sensor performance was tested, with the samples ranging in different forms such as sputum, throat swabs, urine, feces, plasma, oral swabs, serum, whole blood and saliva. Comparing with the ground truth through RT-PCR assay results, this nanosensor performed superior in detecting upper respiratory samples with 100% accuracy, and performed better than or similar to RT-PCR for the other types of samples. Moreover, the minimum limit of detection of viral RNA for this nanosensor was reported to be ∼200 copies/ml, which is superior to most of the other reported assays in the literature, including the commercially available RT-PCR tests which have a limit of detection of the order of ∼500–100 copies/ml. Thus, through the use of electrochemical detection using a smartphone readout, this point-of-care diagnostic can be used without the need for expensive laboratory equipment; such as those needed for amplification-based tests.
Although the bulk of research has focused on the development of FET nanosensors based on 2D materials such as graphene and its derivatives (graphene oxide or reduced graphene oxide), there have been some efforts towards the use of carbon nanotube (CNT)-based field effect transistors (FETs). In one such instance, CNT FETs were fabricated on a polymer substrate, and the sensor is made by immobilizing the complementary sequence of the RdRp gene of the SARS-CoV-2 virus on the CNT channel (Thanihaichelvan et al., 2021). Using synthetic target sequences, the authors showed a limit of detection of ∼10 fM. More work needs to be done to develop this sensor, and tests on real human clinical samples need to validate the detection performance relative to the gold standard RT-PCR assay. In another instance, researchers used high-purity semiconducting single-walled carbon nanotubes-based field effect transistors (FETs), decorated with binding antibodies against the SARS-CoV-2 spike protein and against the nucleocapsid protein (Shao et al., 2021). Based on calibration test results, these nanosensors were shown to have a dynamic range of 5.5 fg/ml–5.5 ng/ml for the spike protein antigen, and 16 fg/ml–16 pg/ml for the nucleocapsid protein antigen. The limit of detection was reported to be ∼0.55 fg/ml for the spike antigen detection, and ∼0.016 fg/ml for the nucleocapsid antigen. However, in clinical testing with n = 38 subjects (nasopharyngeal swab specimens in viral transport medium), the FET biosensors did not perform as well as some of the other FET nanosensors described above in this review, as shown in Figure 4D. For the spike antibody-based test, there was a 17.8% false negative rate compared to the standard nucleic acid amplification test, and was even worse still for the nucleocapsid antibody-based test; suggesting potential issues with fouling of the biosensor or cross-reactivity with other proteins in the sample. These issues highlight some of the challenges involved in designing nanosensors which can offer a combination of high sensitivity, specificity and accuracy at low cost, without the need for amplification of viral genetic material, and with a rapid readout time. Interestingly, multi-walled carbon nanotubes have also been used to design nanosensors for indirect detection of COVID-19. In a novel approach, researchers built a COVID-19-induced reactive oxygen species (ROS) detector system, comprised of multi-walled carbon nanotubes (MWCNTs) grown on the tips of steel needles to form three electrodes in a specific geometric configuration (Miripour et al., 2020). Upon connecting the probe sensor and incubating the working electrode in fresh human sputum samples, the researchers generated a calibration curve of sputum ROS levels, correlated with the diagnostic ranges of COVID detection. The ranges for negative, suspicious and positive COVID-19 detection were determined to be <190 μA, between 190–230 μA, and >230 µA peak current, respectively. In clinical testing on n = 172 patients, the test showed 97% sensitivity, 91% specificity, and 97% accuracy on 142 known subjects (COVID-19 test status already determined by clinical testing), and 92% sensitivity, 94% specificity and 94% accuracy on 30 candidates who were recommended for CT scan. The main benefit of this diagnostic sensor-based technique is the ability to offer a rapid screening tool for assessing the health of suspected infectious people, with test results in under 30 s.
Outlook and Future Research Directions
Upon surveying the landscape for the development of optical and electrochemical-based nanosensors for the detection of COVID-19, it becomes immediately apparent that the amount of innovation in the last 18 mo driven by the urgency of the pandemic situation has been truly remarkable, and mind-boggling. Where does the field go from here? Some areas of research are worth investigating for future improvements, as suggested below:
• Human specimen testing: Given the global nature of the pandemic, which continues to wreak havoc in many parts of the world in mid-2021 despite the emergency use authorization (EUA) of multiple, effective and safe vaccines (albeit with a slow global rollout, with only ∼2.7 billion total vaccine doses administered as of the end of June 2021 (WHO Coronavirus (COVID-19) Dashboard. Geneva: World Health Organization, 2020), out of a population of ∼7.7 billion) – there remains a strong need for the availability of low-cost, rapid, point-of-care diagnostic solutions to better manage population-level spread and mitigate the impacts of COVID-19. Therefore, it is of utmost importance that researchers coming up with new designs of biosensors and assays for SARS-CoV-2 detection should actually test and report the performance of these sensors with clinical samples (such as sputum, saliva, naso-pharyngeal swab, blood, serum or plasma) from both COVID-19 infected and healthy human subjects, and cross-validate their results with the gold standard RT-PCR assay. Many of the novel sensors reported here have already reported clinical data with remarkable levels of Sensitivity and Specificity of detection (seeTable 2 and Table 3), however, merely testing the nanosensors on artificially spiked samples with the target molecule of interest does not guarantee faithful reproduction in clinical samples which may have other confounding factors or biofouling issues to negatively impact sensor performance. It is imperative for researchers to test their nanosensors on clinical samples and report the relevant statistical metrics (True Positive, True Negative, False Positive, False Negative, Sensitivity or Recall, Specificity, Accuracy, Precision, Area under the Receiver Operating Characteristic, or ROC curve) benchmarked against the ground truth (RT-PCR test results), for other researchers to compare and improve their own designs. Furthermore, with the worldwide deployment of vaccines against SARS-CoV-2 well underway, it becomes extremely important to ensure that the nanobiosensors being designed for COVID-19 detection do not produce false positive results of infection, or confound results generated due to immunity provided by COVID-19 vaccination. This is particularly of concern for those diagnostic tests which rely on the detection of the spike protein, or IgM/IgG antibodies, and the U.S. FDA currently does not recommend antibody testing either for diagnostic purposes or for assessing one’s immunity post COVID-19 vaccination. In the future, it is worth channeling the research community’s efforts to design new diagnostic nanosensors based on the detection of viral RNA, or the RdRp gene, or some multiplexed assay (presence of multiple analytes such as antibodies against the SARS-CoV-2 nucleocapsid and spike proteins) to conclusively differentiate viral infection from an immune response to vaccination.
• Machine Learning (ML) in nanosensor design: While many of the nanosensors discussed in the above review show great performance when designed with the right affinity ligand to capture and bind the target of interest, often the sensors are faced with detection challenges owing to low amounts of target molecules, or noisy background. This is where applying ML to “intelligent” biosensors can provide a boost to the sensor performance, in various ways (Cui et al., 2020): categorization of the signal, anomaly detection, noise reduction, and object/pattern recognition, to name a few. As an example, researchers have recently reported (Yan et al., 2020) a high performance plasmonic nanosensor by combining a particle swarm optimization model with a multi-layer perceptron artificial neural network to design the optimized nanostructure with the highest sensitivity, ∼142,500 nm/RIU (refractive index unit); which is about three orders-of-magnitude higher than the sensitivity of recently reported optical nanosensors. However, as mentioned by the researchers, ML results cannot be standalone solutions, and need to be reconciled with the challenges of actually being able to fabricate and synthesize these materials in the real world. Towards this goal, another advantage of these ML models is the ability to rapidly tweak the design of the nanosensor (to make them somewhat easier to fabricate), and use the model to predict sensor performance, which is about four orders-of-magnitude faster than using traditional methods based on calculation of electromagnetic fields. In another example of an immunoinformatics approach (Panda et al., 2020), virtual drug screening of existing antiviral compounds with the goal of “repurposing” current pharmaceutical drugs enabled the rapid identification of an polymerase inhibitor, PC786, with the mechanism of binding illustrated by the molecular docking simulations performed in silico, and could then be used to guide the design of T-cell and B-cell epitopes against SARS-CoV-2 for vaccine production. Recently, efforts have been made to compile databases (Sahoo et al., 2021) of the structural glycoproteins of beta coronaviruses (such as SARS-CoV-2) and the T-cell and B-cell epitopes for each protein, with built-in tools to help researchers perform similarity search, cross genome comparison, phylogenetic analysis and multiple sequence alignment to help speed up vaccine research and development. Using such techniques, it is now possible to rapidly design a multi-epitope construct (Mahapatra et al., 2020) for the purpose of designing peptide-based vaccines against pathogens such as SARS-CoV-2, without having to rely on expensive, time-consuming traditional methods such as mouse models of infection.
• Point-of-care diagnostics at low cost: Researchers working on developing new nanobiosensors for COVID-19 need to take into account the World Health Organization’s ASSURED criteria (Affordable, Sensitive, Specific, User-friendly, Rapid, Equipment-free, Delivered). While these criteria were originally laid out by the WHO for the sexually transmitted disease diagnostics initiative, these guidelines are broadly applicable to a wide array of disease conditions (Drain et al., 2014), and can be adopted for evaluating new point-of-care nanosensors being designed for COVID-19. It is not a coincidence that “affordability” is listed at the top of the ASSURED criteria, as a large fraction of the world’s population lives in resource-constrained settings with limited access to healthcare. What is considered affordable? Typically (Land et al., 2019), a lateral flow immuno-assay (LFIA) type diagnostic is considered “low-cost” if it is ∼US $1, while a molecular test is considered “low-cost” if it costs < US $10. Besides cost, it is also important to try and meet the other objectives to the extent possible: high sensitivity and specificity compared to a laboratory-based reference standard assay (in the case of COVID-19: the RT-PCR nucleic acid test), user-friendly (very few steps required, minimal or no sample preparation before assay results), rapid turnaround from sample collection to result (ideally < 1–2 h), equipment-free (without the need for specialized analysis equipment), and deliverable, i.e. to the point-of-care. Out of the many nanosensors discussed in this review, only one claims (Moitra et al., 2020) to meet the ASSURED criteria; most of the others fall short in some aspect. To their credit, some of the nanosensors in this review are low-cost (Alafeef et al., 2020; Moitra et al., 2020), simple equipment-free paper-based diagnostics (Li et al., 2020) or portable smartphone-based detection systems (Mahari et al., 2020; Zhao et al., 2021); all of which offer promising potential to expand the coverage of these diagnostic tests in resource-constrained settings. In this direction, researchers are encouraged to think about iterating and improving upon their nanosensor designs to objectively satisfy the ASSURED criteria, towards making a real positive impact on society by helping mitigate the scourge of COVID-19.
Author Contributions
NB, PJ, and AB jointly outlined the concepts, discussed the sections, and researched the literature for this review article. NB and AB wrote the text of the article. PJ helped with designing the artwork and illustrations. All authors reviewed and edited the manuscript.
Funding
NB is grateful for the support by a Mazumdar-Shaw International Oncology Fellowship. AB would like to gratefully acknowledge funding provided by the Massachusetts Consortium on Pathogen Readiness (MassCPR) and the China Evergrande Group, the Marble Center for Cancer Nanomedicine, the Koch Institute Frontier Research Program.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ahmadivand, A., Gerislioglu, B., Ramezani, Z., Kaushik, A., Manickam, P., and Ghoreishi, S. A. (2021). Functionalized Terahertz Plasmonic Metasensors: Femtomolar-Level Detection of SARS-CoV-2 Spike Proteins. Biosens. Bioelectron. 177, 112971. doi:10.1016/j.bios.2021.112971
Alafeef, M., Dighe, K., Moitra, P., and Pan, D. (2020). Rapid, Ultrasensitive, and Quantitative Detection of SARS-CoV-2 Using Antisense Oligonucleotides Directed Electrochemical Biosensor Chip. ACS Nano 14, 17028–17045. doi:10.1021/acsnano.0c06392
Aljabali, A. A., Hassan, S. S., Pabari, R. M., Shahcheraghi, S. H., Mishra, V., Charbe, N. B., et al. (2021). The Viral Capsid as Novel Nanomaterials for Drug Delivery. Future Sci. OA FSO744, 31. doi:10.2144/fsoa-2021-0031
Anker, J. N., Hall, W. P., Lyandres, O., Shah, N. C., Zhao, J., and Van Duyne, R. P. (2008). Biosensing with Plasmonic Nanosensors. Nat. Mater. 7, 442–453. doi:10.1038/nmat2162
Arun, T., Verma, S. K., Panda, P. K., Joseyphus, R. J., Jha, E., Akbari-Fakhrabadi, A., et al. (2019). Facile Synthesized Novel Hybrid Graphene Oxide/cobalt Ferrite Magnetic Nanoparticles Based Surface Coating Material Inhibit Bacterial Secretion Pathway for Antibacterial Effect. Mater. Sci. Eng. C 104, 109932. doi:10.1016/j.msec.2019.109932
Banerjee, A. N. (2018). Graphene and its Derivatives as Biomedical Materials: Future Prospects and Challenges. Interf. Focus 8, 20170056. doi:10.1098/rsfs.2017.0056
Bardhan, N. M. (2017). 30 Years of Advances in Functionalization of Carbon Nanomaterials for Biomedical Applications: A Practical Review. J. Mater. Res. 32, 107–127. doi:10.1557/jmr.2016.449
Bardhan, N. M., and Belcher, A. M. (2020). “Polymer-Functionalized NIR-Emitting Nanoparticles: Applications in Cancer Theranostics and Treatment of Bacterial Infections,” in Near Infrared-Emitting Nanoparticles for Biomedical Applications. Editors A. Benayas, E. Hemmer, G. Hong, and D. Jaque (Cham, Switzerland: Springer International Publishing), 231–277. doi:10.1007/978-3-030-32036-2_10
Bardhan, N. M., Ghosh, D., and Belcher, A. M. (2014). Carbon Nanotubes as In Vivo Bacterial Probes. Nat. Commun. 5, 4918. doi:10.1038/ncomms5918
Bardhan, N. M., Kumar, P. V., Li, Z., Ploegh, H. L., Grossman, J. C., Belcher, A. M., et al. (2017). Enhanced Cell Capture on Functionalized Graphene Oxide Nanosheets Through Oxygen Clustering. ACS Nano 11, 1548–1558. doi:10.1021/acsnano.6b06979
Ceppi, L., Bardhan, N. M., Na, Y., Siegel, A., Rajan, N., Fruscio, R., et al. (2019). Real-Time Single-Walled Carbon Nanotube-Based Fluorescence Imaging Improves Survival After Debulking Surgery in an Ovarian Cancer Model. ACS Nano 13, 5356–5365. doi:10.1021/acsnano.8b09829
Chen, G.-Y., Li, Z., Theile, C. S., Bardhan, N. M., Kumar, P. V., Duarte, J. N., et al. (2015). Graphene Oxide Nanosheets Modified with Single-Domain Antibodies for Rapid and Efficient Capture of Cells. Chem. – Eur. J. 21, 17178–17183. doi:10.1002/chem.201503057
Chen, N., Zhou, M., Dong, X., Qu, J., Gong, F., Han, Y., et al. (2020a). Epidemiological and Clinical Characteristics of 99 Cases of 2019 Novel Coronavirus Pneumonia in Wuhan, China: A Descriptive Study. The Lancet 395, 507–513. doi:10.1016/S0140-6736(20)30211-7
Chen, Z., Zhang, Z., Zhai, X., Li, Y., Lin, L., Zhao, H., et al. (2020b). Rapid and Sensitive Detection of Anti-SARS-CoV-2 IgG, Using Lanthanide-Doped Nanoparticles-Based Lateral Flow Immunoassay. Anal. Chem. 92, 7226–7231. doi:10.1021/acs.analchem.0c00784
Choi, J. R. (2020). Development of Point-of-Care Biosensors for COVID-19. Front. Chem. 8, 517. doi:10.3389/fchem.2020.00517
Cui, F., Yue, Y., Zhang, Y., Zhang, Z., and Zhou, H. S. (2020). Advancing Biosensors with Machine Learning. ACS Sens 5, 3346–3364. doi:10.1021/acssensors.0c01424
Damiati, S., Søpstad, S., Peacock, M., Akhtar, A. S., Pinto, I., Soares, R., et al. (2021). Flex Printed Circuit Board Implemented Graphene-Based DNA Sensor for Detection of SARS-CoV-2. IEEE Sens. J. 21, 13060–13067. doi:10.1109/JSEN.2021.3068922
Dang, X., Bardhan, N. M., Qi, J., Gu, L., Eze, N. A., Lin, C.-W., et al. (2019). Deep-tissue Optical Imaging of Near Cellular-Sized Features. Sci. Rep. 9, 3873. doi:10.1038/s41598-019-39502-w
Dang, X., Yi, H., Ham, M.-H., Qi, J., Yun, D. S., Ladewski, R., et al. (2011). Virus-templated Self-Assembled Single-Walled Carbon Nanotubes for Highly Efficient Electron Collection in Photovoltaic Devices. Nat. Nanotechnol. 6, 377–384. doi:10.1038/nnano.2011.50
Das, C. M., Guo, Y., Yang, G., Kang, L., Xu, G., Ho, H.-P., et al. (2020). Gold Nanorod Assisted Enhanced Plasmonic Detection Scheme of COVID-19 SARS-CoV-2 Spike Protein. Adv. Theor. Simul. 3, 2000185. doi:10.1002/adts.202000185
Drain, P. K., Hyle, E. P., Noubary, F., Freedberg, K. A., Wilson, D., Bishai, W., et al. (2014). Evaluating Diagnostic Point-of-Care Tests in Resource-Limited Settings. Lancet Infect. Dis. 14, 239–249. doi:10.1016/S1473-3099(13)70250-0
Ghosh, D., Bagley, A. F., Na, Y. J., Birrer, M. J., Bhatia, S. N., and Belcher, A. M. (2014). Deep, Noninvasive Imaging and Surgical Guidance of Submillimeter Tumors Using Targeted M13-Stabilized Single-Walled Carbon Nanotubes. Proc. Natl. Acad. Sci. 111, 13948–13953. doi:10.1073/pnas.1400821111
Hajian, R., Balderston, S., Tran, T., deBoer, T., Etienne, J., Sandhu, M., et al. (2019). Detection of Unamplified Target Genes via CRISPR–Cas9 Immobilized on a Graphene Field-Effect Transistor. Nat. Biomed. Eng. 3, 427–437. doi:10.1038/s41551-019-0371-x
Halas, N. J., Lal, S., Chang, W.-S., Link, S., and Nordlander, P. (2011). Plasmons in Strongly Coupled Metallic Nanostructures. Chem. Rev. 111, 3913–3961. doi:10.1021/cr200061k
Hamming, I., Timens, W., Bulthuis, M. L. C., Lely, A. T., Navis, G. J., and Goor, H. van. (2004). Tissue Distribution of ACE2 Protein, the Functional Receptor for SARS Coronavirus. A First Step in Understanding SARS Pathogenesis. J. Pathol. 203, 631–637. doi:10.1002/path.1570
Hashemi, S. A., Golab Behbahan, N. G., Bahrani, S., Mousavi, S. M., Gholami, A., Ramakrishna, S., et al. (2021). Ultra-sensitive Viral Glycoprotein Detection NanoSystem Toward Accurate Tracing SARS-CoV-2 in Biological/non-Biological Media. Biosens. Bioelectron. 171, 112731. doi:10.1016/j.bios.2020.112731
Hassan, M. M., Sium, F. S., Islam, F., and Choudhury, S. M. (2021). A Review on Plasmonic and Metamaterial Based Biosensing Platforms for Virus Detection. Sens. Bio-sens. Res. 33, 100429. doi:10.1016/j.sbsr.2021.100429
Hong, G., Diao, S., Antaris, A. L., and Dai, H. (2015). Carbon Nanomaterials for Biological Imaging and Nanomedicinal Therapy. Chem. Rev. 19, 10816–10906. doi:10.1021/acs.chemrev.5b00008
Huang, S., Lin, C.-W., Qi, J., Iyer, A. M., He, Y., Li, Y., et al. (2021). Surface Plasmon-Enhanced Short-Wave Infrared Fluorescence for Detecting Sub-millimeter-sized Tumors. Adv. Mater. 33, 2006057. doi:10.1002/adma.202006057
Huang, S., Qi, J., deQuilettes, D. W., Huang, M., Lin, C.-W., Bardhan, N. M., et al. (2019). M13 Virus-Based Framework for High Fluorescence Enhancement. Small 15, 1901233. doi:10.1002/smll.201901233
Jiang, Z., Feng, B., Xu, J., Qing, T., Zhang, P., and Qing, Z. (2020). Graphene Biosensors for Bacterial and Viral Pathogens. Biosens. Bioelectron. 166, 112471. doi:10.1016/j.bios.2020.112471
Kumar, P. V., Bardhan, N. M., Chen, G.-Y., Li, Z., Belcher, A. M., and Grossman, J. C. (2016). New Insights into the thermal Reduction of Graphene Oxide: Impact of Oxygen Clustering. Carbon 100, 90–98. doi:10.1016/j.carbon.2015.12.087
Kumar, P. V., Bardhan, N. M., Tongay, S., Wu, J., Belcher, A. M., and Grossman, J. C. (2014). Scalable Enhancement of Graphene Oxide Properties by Thermally Driven Phase Transformation. Nat. Chem. 6, 151–158. doi:10.1038/nchem.1820
Land, K. J., Boeras, D. I., Chen, X.-S., Ramsay, A. R., and Peeling, R. W. (2019). REASSURED Diagnostics to Inform Disease Control Strategies, Strengthen Health Systems and Improve Patient Outcomes. Nat. Microbiol. 4, 46–54. doi:10.1038/s41564-018-0295-3
Lee, Y. Y., Kim, R. M., Im, S. W., Balamurugan, M., and Nam, K. T. (2019). Plasmonic Metamaterials for Chiral Sensing Applications. Nanoscale 12, 58–66. doi:10.1039/C9NR08433A
Li, J., Wu, D., Yu, Y., Li, T., Li, K., Xiao, M.-M., et al. (2021). Rapid and Unamplified Identification of COVID-19 with Morpholino-Modified Graphene Field-Effect Transistor Nanosensor. Biosens. Bioelectron. 183, 113206. doi:10.1016/j.bios.2021.113206
Li, Z., Yi, Y., Luo, X., Xiong, N., Liu, Y., Li, S., et al. (2020). Development and Clinical Application of a Rapid IgM-IgG Combined Antibody Test for SARS-CoV-2 Infection Diagnosis. J. Med. Virol. 92, 1518–1524. doi:10.1002/jmv.25727
Liu, Z., Tabakman, S. M., Chen, Z., and Dai, H. (2009). Preparation of Carbon Nanotube Bioconjugates for Biomedical Applications. Nat. Protoc. 4, 1372–1381. doi:10.1038/nprot.2009.146
Lopez Bernal, J., Andrews, N., Gower, C., Gallagher, E., Simmons, R., Thelwall, S., et al. (2021). Effectiveness of Covid-19 Vaccines Against the B.1.617.2 (Delta) Variant. N. Engl. J. Med. doi:10.1056/NEJMoa2108891
Maddali, H., Miles, C. E., Kohn, J., and O’Carroll, D. M. (2021). Optical Biosensors for Virus Detection: Prospects for SARS-CoV-2/COVID-19. ChemBioChem 22, 1176–1189. doi:10.1002/cbic.202000744
Mahapatra, S. R., Sahoo, S., Dehury, B., Raina, V., Patro, S., Misra, N., et al. (2020). Designing an Efficient Multi-Epitope Vaccine Displaying Interactions with Diverse HLA Molecules for an Efficient Humoral and Cellular Immune Response to Prevent COVID-19 Infection. Expert Rev. Vaccin. 19, 871–885. doi:10.1080/14760584.2020.1811091
Mahari, S., Roberts, A., Shahdeo, D., and Gandhi, S. (2020). eCovSens-Ultrasensitive Novel In-House Built Printed Circuit Board Based Electrochemical Device for Rapid Detection of nCovid-19 Antigen, A Spike Protein Domain 1 of SARS-CoV-2. bioRxiv [Preprint]. Available at: https://www.biorxiv.org/content/10.1101/2020.04.24.059204v3. (Accessed May 11, 2020). doi:10.1101/2020.04.24.059204
Miripour, Z. S., Sarrami-Forooshani, R., Sanati, H., Makarem, J., Taheri, M. S., Shojaeian, F., et al. (2020). Real-time Diagnosis of Reactive Oxygen Species (ROS) in Fresh Sputum by Electrochemical Tracing; Correlation between COVID-19 and Viral-Induced ROS in Lung/respiratory Epithelium during This Pandemic. Biosens. Bioelectron. 165, 112435. doi:10.1016/j.bios.2020.112435
Moitra, P., Alafeef, M., Dighe, K., Frieman, M. B., and Pan, D. (2020). Selective Naked-Eye Detection of SARS-CoV-2 Mediated by N Gene Targeted Antisense Oligonucleotide Capped Plasmonic Nanoparticles. ACS Nano 14, 7617–7627. doi:10.1021/acsnano.0c03822
Novoselov, K. S., Fal′ko, V. I., Colombo, L., Gellert, P. R., Schwab, M. G., and Kim, K. (2012). A Roadmap for Graphene. Nature 490, 192–200. doi:10.1038/nature11458
O’Connell, M. J., Bachilo, S. M., Huffman, C. B., Moore, V. C., Strano, M. S., Haroz, E. H., et al. (2002). Band Gap Fluorescence from Individual Single-Walled Carbon Nanotubes. Science 297, 593–596. doi:10.1126/science.1072631
Panda, P. K., Arul, M. N., Patel, P., Verma, S. K., Luo, W., Rubahn, H.-G., et al. (2020). Structure-based Drug Designing and Immunoinformatics Approach for SARS-CoV-2. Sci. Adv. 6, eabb8097. doi:10.1126/sciadv.abb8097
Patel, S., Srivastav, A. K., Gupta, S. K., Kumar, U., Mahapatra, S. K., Gajjar, P. N., et al. (2021). Carbon Nanotubes for Rapid Capturing of SARS-COV-2 Virus: Revealing A Mechanistic Aspect of Binding Based on Computational Studies. RSC Adv. 11, 5785–5800. doi:10.1039/D0RA08888A
Pinals, R. L., Ledesma, F., Yang, D., Navarro, N., Jeong, S., Pak, J. E., et al. (2021). Rapid SARS-CoV-2 Spike Protein Detection by Carbon Nanotube-Based Near-Infrared Nanosensors. Nano Lett. 21, 2272–2280. doi:10.1021/acs.nanolett.1c00118
Planas, D., Veyer, D., Baidaliuk, A., Staropoli, I., Guivel-Benhassine, F., Rajah, M. M., et al. (2021). Reduced Sensitivity of SARS-CoV-2 Variant Delta to Antibody Neutralization. Nature 9, 1–7. doi:10.1038/s41586-021-03777-9
Pumera, M. (2011). Graphene in Biosensing. Mater. Today 14, 308–315. doi:10.1016/S1369-7021(11)70160-2
Qiu, G., Gai, Z., Tao, Y., Schmitt, J., Kullak-Ublick, G. A., and Wang, J. (2020). Dual-Functional Plasmonic Photothermal Biosensors for Highly Accurate Severe Acute Respiratory Syndrome Coronavirus 2 Detection. ACS Nano 14, 5268–5277. doi:10.1021/acsnano.0c02439
Reina, G., Iglesias, D., Samorì, P., and Bianco, A. (2021). Graphene: A Disruptive Opportunity for COVID-19 and Future Pandemics? Adv. Mater. 33, 2007847. doi:10.1002/adma.202007847
Sahoo, S., Mahapatra, S. R., Parida, B. K., Rath, S., Dehury, B., Raina, V., et al. (2021). DBCOVP: A Database of Coronavirus Virulent Glycoproteins. Comput. Biol. Med. 129, 104131. doi:10.1016/j.compbiomed.2020.104131
Seo, G., Lee, G., Kim, M. J., Baek, S.-H., Choi, M., Ku, K. B., et al. (2020). Rapid Detection of COVID-19 Causative Virus (SARS-CoV-2) in Human Nasopharyngeal Swab Specimens Using Field-Effect Transistor-Based Biosensor. ACS Nano 14, 5135–5142. doi:10.1021/acsnano.0c02823
Shahdeo, D., Roberts, A., Abbineni, N., and Gandhi, S. (2020). Graphene Based Sensors. Comprehensive Analytical Chemistry 91, 175–199. doi:10.1016/bs.coac.2020.08.007
Shao, W., Shurin, M. R., Wheeler, S. E., He, X., and Star, A. (2021). Rapid Detection of SARS-CoV-2 Antigens Using High-Purity Semiconducting Single-Walled Carbon Nanotube-Based Field-Effect Transistors. ACS Appl. Mater. Inter. 13, 10321–10327. doi:10.1021/acsami.0c22589
Sharma, P. K., Dorlikar, S., Rawat, P., Malik, V., Vats, N., Sharma, M., et al. (2021). “Nanotechnology and its Application: A Review,” in Nanotechnology in Cancer Management. Editors K. R. Khondakar, and A. Kaushik (Elsevier), 1–33. doi:10.1016/B978-0-12-818154-6.00010-X
Shrivastav, A. M., Cvelbar, U., and Abdulhalim, I. (2021). A Comprehensive Review on Plasmonic-Based Biosensors Used in Viral Diagnostics. Commun. Biol. 4, 1–12. doi:10.1038/s42003-020-01615-8
Thanihaichelvan, M., Surendran, S. N., Kumanan, T., Sutharsini, U., Ravirajan, P., Valluvan, R., et al. (2021). Selective and Electronic Detection of COVID-19 (Coronavirus) Using Carbon Nanotube Field Effect Transistor-Based Biosensor: A Proof-Of-Concept Study. Mater. Today Proc. 2021, 1. doi:10.1016/j.matpr.2021.05.011
Tîlmaciu, C.-M., and Morris, M. C. (2015). Carbon Nanotube Biosensors. Front. Chem. 3, 1. doi:10.3389/fchem.2015.00059
Torrente-Rodríguez, R. M., Lukas, H., Tu, J., Min, J., Yang, Y., Xu, C., et al. (2020). SARS-CoV-2 RapidPlex: A Graphene-Based Multiplexed Telemedicine Platform for Rapid and Low-Cost COVID-19 Diagnosis and Monitoring. Matter 3, 1981–1998. doi:10.1016/j.matt.2020.09.027
Tymm, C., Zhou, J., Tadimety, A., Burklund, A., and Zhang, J. X. J. (2020). Scalable COVID-19 Detection Enabled by Lab-On-Chip Biosensors. Cell. Mol. Bioeng. 13, 313–329. doi:10.1007/s12195-020-00642-z
Varahachalam, S. P., Lahooti, B., Chamaneh, M., Bagchi, S., Chhibber, T., Morris, K., et al. (2021). Nanomedicine for the SARS-CoV-2: State-Of-The-Art and Future Prospects. Int. J. Nanomedicine 16, 539–560. doi:10.2147/IJN.S283686
V’kovski, P., Kratzel, A., Steiner, S., Stalder, H., and Thiel, V. (2021). Coronavirus Biology and Replication: Implications for SARS-CoV-2. Nat. Rev. Microbiol. 19, 155–170. doi:10.1038/s41579-020-00468-6
Welsher, K., Liu, Z., Sherlock, S. P., Robinson, J. T., Chen, Z., Daranciang, D., et al. (2009). A Route to Brightly Fluorescent Carbon Nanotubes for Near-Infrared Imaging in Mice. Nat. Nanotechnol. 4, 773–780. doi:10.1038/nnano.2009.294
World Health Organization (2020). WHO Coronavirus (COVID-19) Dashboard. Geneva, Switzerland: World Health Organization. Available at: https://covid19.who.int. (Accessed June 28, 2021).
Yan, R., Wang, T., Jiang, X., Zhong, Q., Huang, X., Wang, L., et al. (2020). Design of High-Performance Plasmonic Nanosensors by Particle Swarm Optimization Algorithm Combined with Machine Learning. Nanotechnology 31, 375202. doi:10.1088/1361-6528/ab95b8
Zhang, Z., Tang, Z., Farokhzad, N., Chen, T., and Tao, W. (2020). Sensitive, Rapid, Low-Cost, and Multiplexed COVID-19 Monitoring by the Wireless Telemedicine Platform. Matter 3, 1818–1820. doi:10.1016/j.matt.2020.11.001
Zhao, H., Liu, F., Xie, W., Zhou, T.-C., OuYang, J., Jin, L., et al. (2021). Ultrasensitive Supersandwich-type Electrochemical Sensor for SARS-CoV-2 from the Infected COVID-19 Patients Using a Smartphone. Sens. Actuators B Chem. 327, 128899. doi:10.1016/j.snb.2020.128899
Keywords: COVID-19, SARS-CoV-2, detection, point-of-care diagnostics, graphene, carbon nanotubes, plasmonic, nanosensors
Citation: Bardhan NM, Jansen P and Belcher AM (2021) Graphene, Carbon Nanotube and Plasmonic Nanosensors for Detection of Viral Pathogens: Opportunities for Rapid Testing in Pandemics like COVID-19. Front. Nanotechnol. 3:733126. doi: 10.3389/fnano.2021.733126
Received: 29 June 2021; Accepted: 05 August 2021;
Published: 25 August 2021.
Edited by:
Ajeet Kaushik, Florida Polytechnic University, United StatesReviewed by:
Suresh K. Verma, KIIT University, IndiaAparajita Singh, Florida International University, United States
Copyright © 2021 Bardhan, Jansen and Belcher. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Angela M. Belcher, YmVsY2hlckBtaXQuZWR1
 Neelkanth M. Bardhan
Neelkanth M. Bardhan Peter Jansen
Peter Jansen Angela M. Belcher
Angela M. Belcher