
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Nanotechnol. , 09 June 2021
Sec. Biomedical Nanotechnology
Volume 3 - 2021 | https://doi.org/10.3389/fnano.2021.694838
This article is part of the Research Topic Nanotechnology for Precision Cancer Therapy: Advances in gene therapy, immunotherapy, and 3D bioprinting View all 11 articles
 Guruprasadh Swaminathan1†
Guruprasadh Swaminathan1† Aisha Shigna1†
Aisha Shigna1† Aviral Kumar1‡
Aviral Kumar1‡ Vishnu Vardhan Byroju2‡
Vishnu Vardhan Byroju2‡ Varsha Reddy Durgempudi1
Varsha Reddy Durgempudi1 Lekha Dinesh Kumar1*
Lekha Dinesh Kumar1*Cancer is a significant health hazard of the 21st century, and GLOBOCAN predicts increasing cancer incidence in the coming decades. Though several conventional treatment modalities exist, most of them end up causing off-target and debilitating effects, and drug resistance acquisition. Advances in our understanding of tumor molecular biology offer alternative strategies for precise, robust, and potentially less toxic treatment paradigms for circumventing the disease at the cellular and molecular level. Several deregulated molecules associated with tumorigenesis have been developed as targets in RNA interference (RNAi) based cancer therapeutics. RNAi, a post-transcriptional gene regulation mechanism, has significantly gained attention because of its precise multi-targeted gene silencing. Although the RNAi approach is favorable, the direct administration of small oligonucleotides has not been fruitful because of their inherent lower half-lives and instability in the biological systems. Moreover, the lack of an appropriate delivery system to the primary site of the tumor that helps determine the potency of the drug and its reach, has limited the effective medical utilization of these bio-drugs. Nanotechnology, with its unique characteristics of enhanced permeation and better tumor-targeting efficiency, offers promising solutions owing to the various possibilities and amenability for modifications of the nanoparticles to augment cancer therapeutics. Nanoparticles could be made multimodal, by designing and synthesizing multiple desired functionalities, often resulting in unique and potentially applicable biological structures. A small number of Phase I clinical trials with systemically administered siRNA molecules conjugated with nanoparticles have been completed and the results are promising, indicating that, these new combinatorial therapies can successfully and safely be used to inhibit target genes in cancer patients to alleviate some of the disease burden. In this review, we highlight different types of nano-based delivery strategies for engineering Nano-RNAi-based bio drugs. Furthermore, we have highlighted the insights gained from current research that are entering the preclinical evaluation and information about initial clinical developments, shaping the future for next generation cancer therapeutics.
With the growing global burden and economic impact, cancer has become the most significant public health challenge of this century. Cancer, an exemplar of deregulated genetic function, arises from a complex interplay of genes and their products. Conventional therapeutic regimens include surgical resection, chemotherapy, radiotherapy, and anticancer drugs or a combination of all these. Despite the fact that these treatment approaches have increased the overall patient survival for many cancers in their early stages, the lack of specificity, off-target effects, and acquisition of drug resistance stand as a hindrance in the path of considering these treatment methods as effective and patient-friendly. The significant distress caused by these side effects in patients is often a leading impediment in the course of adopting these as mainstream treatments. With the identification of several novel molecules, their mechanisms and pathways involved in cancer, it has become even more imperative to further distinguish and isolate the molecules which could help in the management of the disease.
Recently, much attention has been paid to the development of cancer therapeutics based on RNA molecules. RNA interference (RNAi), a process that involves sequence-specific gene silencing, is an endogenous post-transcriptional regulation process that consists of non-coding RNAs like microRNAs (miRNAs), small interfering RNAs (siRNAs), long non-coding RNAs (lncRNAs), and circular RNAs (circRNAs), which target and silence messenger RNAs (mRNAs) in a sequence-specific manner (Chalbatani et al., 2017). With the added benefit of simultaneously targeting multiple genes that share homology, RNAi is fast, economical, and has the potential for site-specific targeting. The primary role of RNAi in cells would be to downregulate the expression of their target gene and thereby the encoded proteins thus bringing about the desired results (Mahmoodi Chalbatani et al., 2019). Through careful sequence selection and synthesis of tailored non-coding RNAs (ncRNAs), this powerful approach could be used to circumvent conventional cancer therapy limitations, thus paving the way for next generation therapeutics (Lorenzer et al., 2015).
The in vitro experimental data obtained using RNA interference has shown promising results, demonstrating its feasibility to be taken from bench to bedside. Though advantageous in in vitro scenario, several impediments lie on its path in achieving gene silencing in vivo and making gene therapy a reality. The major hurdles being instability of RNA molecules, its low transfection efficiency and half-life, lack of site-specific targeting and distribution in the target tissue, heterogeneity of the tumor, etc. add up to the challenges of onsite delivery of drugs using RNAi. The unique characteristics of nanoparticles have enabled researchers to successfully demonstrate them as efficient chaperones for the delivery of RNAi molecules to the primary site of the tumor. Nanoparticles usually exist as particulate dispersions or solid particles with sizes ranging between 10–1,000 nm (Nagal and Singlab, 2013). Their enhanced permeability and retention (EPR) effect, capacity to prevent RNAi molecules from undergoing enzymatic degradation, and transportation efficiency across the cell membrane make them a perfect carrier of RNAi molecules for targeted therapy (Xin et al., 2017). The conjugation of nanocarriers with the RNAi molecules widely opens the door to targeting mediators in cancer progression, identifying molecular targets, and engineering delivery vehicles conjugated with DNA/RNA as therapeutic devices, thus representing an ideal approach. Currently, a small number of Phase I clinical trials with systemically administered ncRNA molecules conjugated with different nanoparticles as delivery vehicles are already complete, indicating that these new therapeutics can safely inhibit targeted gene products in patients with cancer. In spite of elucidation of the immense potential of RNAi in cancer therapy in many explorative research trials, there is still a long way to go for successfully translating this exciting result from bench to bedside.
In this review, we discuss the importance of harnessing ncRNAs as next generation cancer therapeutics. We also discuss critical delivery strategies that are based on nanoparticles and the parameters to be considered for RNAi-based drug development. Furthermore, we have highlighted the insights gained from current research that are entering the preclinical evaluation and information about initial clinical developments, shaping the future for RNAi-based therapeutics in cancer.
The practice of using chemicals to reduce the burden of cancer saw its beginning in the early decades of 20th century and has continued to treat this disease over the years. While surgery and radiation therapy were leading the cancer treatment, their failure to target inoperable metastatic lesions placed chemotherapy at an advantageous position. Therefore, most of the times an evidence driven choice was made to use a combination of these three modalities to induce remission of cancer (DeVita and Chu, 2008). While chemotherapy had become the standard care in numerous cancers, the ill effects of the chemicals on multiple organ systems of the body were also well known. The debilitating after effects and overall decreased quality of life that patients endure, directed research interest towards innovating novel strategies that are equally, if not more, effective as chemotherapy while being better tolerable by the human body (Gegechkori et al., 2017; Pearce et al., 2017).
Chemotherapy is harmful due to the lack of specificity and patients placed under any chemotherapeutic regimen report dissatisfaction due to undesired symptoms generated. It is also often an added factor for depression and anxiety in patients diagnosed with cancer (Niedzwiedz et al., 2019). Subjects placed on chemotherapy experience side effects that can range from mild (nausea, vomiting and diarrhea) to life threatening (cardiomyopathy with decreased ejection fraction, development of secondary malignancies such as leukemias) after effects. These can arise during the treatment phase (long term effects) or years later (latent effects) (Stein et al., 2010). Chemotherapy regimen places patients in a loop wherein for minimizing adverse effects of one drug, they will be forced to take additional drugs and the cycle continues, often affecting their mental health and emotional state (Pearce et al., 2017). Pregnancy is another physiological state where chemotherapy is deemed unsafe and the detrimental effects include intrauterine growth restriction, limb anomalies and stillbirth etc. (Koren et al., 2013).
Chemotherapy affects rapidly dividing cells without differentiating the neoplastic and non-neoplastic cell. Gastrointestinal tract, germ cells, hematopoietic cells and hair follicles are prime targets, considering their replicative potential (Agarwal, 2016). Nonspecific targeting of gastric mucosal cells presents as diarrhea (immediate onset and late onset) is common with drugs like topoisomerase II inhibitors (irinotecan, topotecan), 5-Fluorouracil, taxanes (docetaxel/paclitaxel), antibodies against epidermal growth factor receptors and calcineurin inhibitors. Though there are treatment options like opioid based formulae and somatostatin analogs that are used to alleviate the pain and discomfort caused due to chemotherapy, these drugs too present with their very own set of side effects (Rock and Kono, 2008).
Meiotic cells are also affected by chemotherapy and this raises concerns about fecundability in female (often resulting in premature ovarian failure) and male subjects alike. While ovarian follicles and the endocrine axis are disrupted in females, males have diminished semen quality with spermatozoa that are low in number and/or of poor quality. Secondary sexual characteristics are affected in pubertal and prepubescent children undergoing chemotherapy which has an impact on their self-confidence (Vakalopoulos et al., 2015; Waimey et al., 2015; Poorvu et al., 2019). Chemotherapy induced alopecia is reported in numerous clinical and standard of practice trials which are considered a distressing effect as it affects the emotional state and body image of subjects (Saraswat et al., 2019). Hair loss is observed on the scalp, axillae, groin and all over non-glabrous skin surfaces which indicates the effects chemotherapy has on vellus and terminal hairs in a non-specific manner (Muth, 2017). Chemotherapy has an impact on childhood cancer subjects to an extent of impacting self-esteem in over two thirds of them and inducing significant stress among their parents. Chemotherapy in children in certain cases invalidates their germ cells and gonads resulting in decreased fertility and the psychological impact this creates often needs anticipatory counseling in their adolescence. Understanding the pathophysiology of chemotherapy induced alopecia to a certain extent has led to the use of techniques and medications such as hypothermia, YHO618 to minimize hair loss and improve hair growth but substantial research still needs to be done as no prevention and treatment guidelines are currently in place (You et al., 2019; Rossi et al., 2020).
Nervous system impact in subjects undergoing chemotherapy is well studied and includes impact on the central and peripheral nervous system manifesting as cognitive dysfunction, neuropathy and mental health concerns (Stone and DeAngelis, 2016). Chemotherapy results in a variety of neurological complications that include mood disorders, seizures, memory impairment ocular toxicity, ototoxicity and uncertain effects when there is unspecified neuro-degeneration (Yang and Moon, 2013). Peripheral neuropathy is well noted with a variety of chemotherapeutic drugs like paclitaxel, vincristine, bortezomib and cisplatin. As the peripheral nervous system includes nerves constituting the autonomic and enteric nervous systems, effects such as gastroparesis and orthostatic hypotension can be observed post chemotherapy. These neurological effects have been understudied and the mechanisms and their prevention or treatment is limited (Zajaczkowską et al., 2019; Loprinzi et al., 2020).
Secondary malignancies are encountered years after chemotherapeutic regimens and drugs that are known to have such effects include alkylating agents (cyclophosphamide, ifosphamide, nitrogen mustards, melphalan), daunorubicin and doxorubicin. A life-long follow up is recommended to screen for the development of leukemias and lymphomas which are the most common secondary malignancies (Vega-Stromberg, 2003). The risk of secondary malignancies is higher in subjects placed on chemotherapy independent of irradiation status indicating the impact chemotherapy has on cells and their replicative potential (Nutalapati and Jain, 2018). Chemotherapy impacts essential organ systems, overall quality of life, causes functionally impairing fatigue, distorts body image and self-esteem, compromises fecundability and with the risk of unpredictable secondary malignancies.
Therefore, there is an imminent need to replace chemotherapy with biological therapies. With the advent of various next generation technologies in genomics, transcriptomics, proteomics and high throughput data analysis, the molecular path to next generation drugs has been set in. One of the promising technologies is in the area of non-coding RNA based drugs that are considered to be next generation molecules in the field of molecular medicine. These molecules are classified as miRNAs, lncRNAs and circRNAs etc. They can act independently or in co-ordination with circular endogenous RNAs (ceRNAs) to bring about the regulation of genes. MicroRNAs (miRNAs) are short 22–25 nt non-coding RNAs that regulate a wide array of biological process including carcinogenesis. These molecules have revolutionized the field of cancer ever since their discovery as they have the capability of silencing multiple targets involved in one or different pathways thus bringing about a co-ordination among large oncogene regulatory networks. These miRNAs (miRs) are known to be deregulated in different stages of cancer development as they are known to regulate the expression of genes involved in onset and development of tumors, and such miRNAs are called oncomiRs. Thus these molecules along with numerous agents that can efficiently be part of molecular targeted therapy are being developed that can target cellular processes, such as angiogenesis, signal transduction, cell cycle regulation, apoptosis induction, protein translation, and metastases (Rishabh et al., 2021). To enhance the likelihood of achieving a positive response to this targeted therapy, further studies have to be done to understand the precise molecular mechanisms of cancer in each patient, then carefully designing bio-drugs to deal with the situation paving the way for personalized cancer therapeutics.
RNAi is an evolutionary, endogenous, conserved, post-transcriptional, gene silencing pathway that involves double-stranded RNA-mediated degradation of mRNA. Usually mediated by small RNA molecules like siRNAs, lncRNAs, miRNAs, ceRNAs; RNAi in eukaryotes initiated by the RNase III enzyme called DICER that cleaves long double-stranded RNAs (dsRNAs) into mature small interfering RNAs (siRNAs) with an overhang at the 3’ end. Constituting a passenger strand and a guide strand, siRNAs are incorporated into the RNA Induced Silencing Complex (RISC), where the guide strand complement pairs with the target mRNA sequences and initiates endonucleolytic cleavage through the action of induced Argonaute protein (Borges and Martienssen, 2015). Among these, miRNAs are well studied and known regulators of gene expression in various cellular processes like cell development, differentiation, apoptosis, and proliferation (Hutvágner and Zamore, 2002). The disruption of miRNAs and their cellular functions has been reported to be responsible for the initiation and progression of various cancers and other fatal human ailments. Though their functions are elucidated as oncogenes and tumour suppressors, the complete function of miRNAs, especially in humans, is not fully understood. Thus the function of RNAi and its biological significance within the organism’s body in which it is present is also a mystery to the scientific world. The accurate and expeditious nature of RNA interference technology makes it a popular strategy to learn the gene expression of various organisms. Loss of function studies and systemic RNAi-based genetic screens performed on organisms like plants, C. elegans, and drosophila highlighted the role of this technique in functional genomics. The fact that transfecting mammalian cells with long dsRNAs induced interferon response leading to global gene silencing and, consequently apoptosis, hindered the use of RNAi in mammalian cells in the earlier stages. This was at a time when the structures of these molecules were not elucidated. However, chemical synthesis of siRNA molecules specific to mRNA targets in later years has enabled efficient targeting and silencing of a particular gene, leading to the widespread use of the same in mammalian cells (Karagiannis and El-Osta, 2005).
With a complex etiology, cancer cells with their characteristic lesion often resemble their normal counterparts except for the difference in a series of genetic alterations (Wilda et al., 2002). It is well known that the mutations in the gate keeping genes are the ones that give rise to tumorigenesis in the first place. Subsequent mutations in different genes that belong to cellular oncogenes (c-onc), viral oncogenes (v-onc), tumor suppressor genes (TSG) classes also cause tumor initiation and progression. Different experimental approaches have been used to compare tumor cells with normal cells and have shown that deregulation of a set of non-coding RNAs (ncRNAs) that might be acting as a master switch, might be vital in their differences. The earliest example of a non-coding RNA involved in cancer was the H19 gene (Hao et al., 1993). Cellular oncogenes usually encode various proteins like growth factors and their receptors, signal transducers, and transcription factors associated with tumor initiation and progression. The downregulation of the K-Ras gene through the RNAi pathway is among the first RNAi-mediated deregulations discovered. The silencing of the K-Ras gene, was found to trigger anchorage-independent growth and thus tumorigenesis (Brummelkamp et al., 2002). Various other cellular oncogenes that code for Bcl-2, CDK-2, Mdm-2, PKC-α, TGF-β1, H-Ras, VEGF, and GFP proteins are cognate targets for siRNAs, effectively leading to suppression of cancer cell proliferation. Viral oncogenes, integrated into the host genome through DNA and RNA viruses, are also found to be regulated by RNAi. In cervical cancers and hepatocellular carcinomas (HCC) that are caused by HPV and HBV viruses, respectively, these viral oncogenes turn out to be the target for cancer treatment (McCaffrey et al., 2003; Yoshinouchi et al., 2003). Belonging to the tumor suppressor gene prototype, Retinoblastoma protein (RbP) is found to undergo somatic inactivation in various cancers. In order to study RbP in cancer, components of the dE2F/dDP/RBF pathway were silenced by RNAi, and a remarkable division of labor between family members of this pathway was analyzed through examination of gene expression changes (Agami, 2002). DNA damage-based biological response is usually governed by the effectiveness of DNA repair processes called the checkpoint cell cycle responses and induction of processes that favor or inhibit pathways leading to apoptosis of the cell. The viability of the cell may not be affected by the defects in DNA repair enzymes, but it can lead to genomic instability, and thereby enhancing the rate of genetic changes and thus the rate of tumor formation. Suppression of Rad51 gene, a DNA damage repair gene, by RNAi method, elucidated the role of the same after both endogenous and exogenous double-strand break formation. This was complemented by mutating SPO11 and MRE11 proteins (Takanami et al., 2003). RNAi-based functional genomic approaches have also enabled the identification of chromosomal DNA degradation mediating nucleases like CPS6 and NUC1. siRNAs developed against the ATR gene have also been in use to analyze its role in checkpoint responses, along with p53 binding protein. These have turned out to be a significant target in clinical application since then (Cortez et al., 2001; Morales et al., 2003).
During invasion and metastasis, tumor cells can undertake either single-cell locomotive or cohort migration strategies. The membrane-anchored glycoprotein RECK inhibits tumor metastasis and angiogenesis by negatively regulating matrix metalloproteinases (MMPs). RNAi-mediated inhibition of RECK in CL-1 human lung cancer cells exterminated the inhibitory effect of trichostatin A (TSA), and HDAC inhibitor, on MMP-2 activation (Liu et al., 2003). RNAi studies done on MDA-MB-231 human breast cancer cells showed the overexpression of CXC chemokine receptor-4 (CXCR4) could be controlled by knockdown of the gene and thereby metastasis. Matrix metalloproteases MMP-9 and cathepsin B promote invasion and metastasis of gliomas through matrix degradation. Studies showed that the suppression of gene expression of these proteins is caused due to short hairpin RNAs in SNB19 cells, leading to a significant reduction in cell-cell interaction of human microvascular endothelial cells (Lakka et al., 2004).
Angiogenesis is a major contributor to the development and growth of cancer. Various proteins involved in angiogenesis have been targeted by RNAi technology. The angiogenic factor VEGF is regulated by the interaction of its GC-rich motif with that of Sp protein. RNAi-based studies have been done on Sp proteins to analyze their role in the regulation of VEGF. Sequential knockdown of the Sp1, Sp3, or Sp4 gene with the help of siRNA showed that all three proteins regulated transactivation in pancreatic cancer cells transfected with the pVEGF1, pVEGF2, and pVEGF3 constructs containing VEGF promoter inserts. This indicates that Sp1, Sp3, and Sp4 cooperatively regulate VEGF expression in tumor cells (Abdelrahim et al., 2004). Another molecule that regulates the function of VEGF is HIF. RNAi approach has been used successfully to determine functional differences between HIF-1α and HIF-2α in different human cell lines (Warnecke et al., 2004). HIF-1α knockdown in cervical cancer cell lines reduced hypoxia-induced mRNA stimulation of glucose transporter 1 GLUT1, lactate dehydrogenase A (LDH-A), VEGF, carbonic anhydrase IX (CA IX), and HIF prolyl hydroxylase 2 (PHD2) by 40–60%. In contrast, HIF-2α knockdown had no effect on these HIF target genes. Tie-2 is a small molecule inhibitor of endothelial cell-specific tyrosine kinases, which, when interrupted by RNAi, make endothelial cells lose their viability. This was further investigated and found to be due to AKT signaling inhibition, leading to increased thrombospondin expression (Niu et al., 2004). The tumor growth occurs due to the imbalance between cell proliferation and apoptosis. The livin gene expressed in numerous cancers is found to have an anti-apoptotic function. A study based on vector-based livin-siRNAs plasmid found that it silenced endogenous livin gene expression through association with caspase-3 activation, thus, in turn, enhancing apoptotic rate (Crnkovic-Mertens et al., 2003). Bcl-2 and xIAP are other anti-apoptotic factors. Studies showed that the sensitivity of MCF-7 breast cancer cells to treatment with the drugs etoposide and doxorubicin is increased after silencing of Bcl-2 or xIAP by siRNA (Lima et al., 2004). RNAi is developing from a powerful tool utilized to elucidate the function of novel genes to a potential therapeutic modality in cancer therapy. Further research is needed to understand and unravel the precise mechanism, by which RNA interference modulates gene expression inside the body, especially during cancer, and to exploit this powerful tool and use it to its full potential.
With the development of novel therapeutic strategies like gene silencing and genome editing, diverse RNA molecules have come to the forefront as potential therapeutic molecules (Fire et al., 1998; Cong et al., 2013; Mali et al., 2013). The fact that they can interact with all the major biological molecules like DNA, RNA, and protein led to the idea of RNA therapeutics, which is now the primary reason for the expansion in the range of druggable targets, including conventional proteins and previously undruggable transcripts and genes. Basically, RNA can take up three different roles in therapy. Firstly, RNAs can form oligonucleotide molecules called aptamers that can bind to the extracellular, cell surface, or intracellular proteins targeted by small molecule drugs thus bringing about silencing (Gragoudas, 2004). Secondly, siRNAs, miRNAs, and their mimics/knockdown molecules can target specific mRNAs leading to gene silencing or control of gene expression for treatment of diseases. Thirdly, mRNA molecules can be transfected in the cells where they can get translated into protein and thus be a part of vaccination or protein replacement therapy (Sahin et al., 2014; Lieberman, 2018). Though scientists knew that the transcription of many genes in eukaryotic cells are repressed, genes are transcribed into mRNA that never gets translated, post-transcriptional mechanisms are in place to add another level of control over this already existing complex system, identification of the function of these endogenous and exogenous molecules revolutionized the way gene expression can be manipulated (Christopher et al., 2016).
MicroRNA plays a significant role in gene regulation transcriptionally and translationally. The fact that one miRNA can compromise the expression of several different target genes simultaneously indicates that it has a very different role in pharmaceutical targeting when compared to siRNAs. Ambros and colleagues discovered the first miRNA lin-4 in 1994 (Lee et al., 1993). As an extension to this discovery, various other miRNA molecules like let-7 were identified and were observed to perform sequence-specific RNA-RNA interaction with mRNA, leading to inhibition of gene expression (Rawoof et al., 2020). These non-coding RNA molecules were found to possess highly conserved nature suggesting that these molecules are essential gene regulatory factors. Hsa-miR-15a and miR-16–1 were the first cancer-related miRNAs discovered. These were tumor-suppressing miRNAs found in chronic lymphocytic leukemia that induced apoptosis through repression of the Bcl-2 gene (Calin et al., 2002). Further studies on the deletion of miR-15 and miR-16–1 cluster in mice recapitulated chronic lymphocytic leukemia-associated phenotypes observed in humans. This convincingly demonstrated that these two miRNAs are crucial for tumor suppression (Klein et al., 2010). Hsa-miR-143 and miR-145 are often found deregulated in lung cancer, resulting in decreased expression of both miRNAs. The abnormal expression of certain miRNAs can also be due to the deregulation of various transcription factors like c-Myc and p53. c-Myc is found to repress the transcriptional activity of tumor-suppressive miRNAs like miR-15a, miR-26, miR-29, miR-30, and let-7, thus setting in proliferation (Chang et al., 2008). Tumor suppressor gene p53, on the other hand, positively regulates the expression of various miRNAs like miR-34, miR-605, miR-1246, and miR-107.
The failure of chemotherapy in cancer has paved the way for development of miRNA molecules as therapeutic targets, especially for patients with drug resistance issues. The fact that there exists an alteration in the expression of these miRNAs in cancer makes it possible to manipulate miRNA expression in such tumor tissues. These manipulations are often facilitated by injecting miRNAs similar to the use of antisense mRNAs. By identifying signature miRNAs, their mechanism of action in cancer, and strategies to attain this miRNA molecule delivered at the target, miRNA therapeutics can be applied more efficiently in cancer therapy and thus would emerge as a novel therapeutic tool (Kumar et al., 2020).
After the discovery of the first long non-coding RNA (H19) associated with a tumor, several ncRNAs have been identified, implicated to have functions in different cancers. These ncRNAs usually constitute small and long ncRNA, and some of these long ncRNAs can even give rise to miRNAs (Metzler et al., 2004; Eis et al., 2005). Recent studies have revealed the role of lncRNA in cancer development and in maintaining the hallmarks of cancer. This is mainly by virtue of their ability to interact with the major biological molecules like DNA, RNA, and protein, or even a combination of them. LINE-1, HERV, Satellites, and TERRA are specific lncRNAs that can contribute to disease progression through immune response activation. Other than this, LINE-1 is also found to support chromatin formation. TERRA, on the other hand, supports alternative lengthening of telomeres (Parasramka et al., 2016). Highly upregulated lncRNA in liver cancer (HULC) is a molecule that has a prominent role in the early stages of cancer progression. The binding of the promoter region of HULC to that of CREB protein is found to activate an autoregulatory loop involving HULC and miR-372, which further leads to phosphorylation of CREB and thus the chain of similar events (Wang et al., 2010). HULC also promotes HCC proliferation by targeting and suppressing p18 and IGF2BP1 (Hämmerle et al., 2013). Metastasis associated lung adenocarcinoma transcript 1 (MALAT1) is another lncRNA upregulated in HCC and non small cell lung carcinoma (NSCLC) cells that is associated with the alternative splicing of various precursor mRNAs (Ji et al., 2003; Schalken et al., 2003; Lee et al., 2011; Lai et al., 2012). Detection of certain lncRNAs in body fluids and serums has propounded the idea of using these molecules as cancer biomarkers. Plasma levels of HULC are higher in patients with HCC when compared to healthy humans. PCA3 is a prostate-specific lncRNA found usually upregulated in patients and thus is used as a biomarker (Lee et al., 2011). Prostate-Specific Gene 1 (PCGEM1), small dendritic non-translatable RNA BC1, and DD3 ncRNA genes are also over expressed in prostate tumors compared to normal and primary tumor specimens. DD3 is an ideal molecule for cancer therapy as it has not been detected in any of the healthy tissues like breast, bladder, testis, gastrointestinal organ, and musculoskeletal tissue (Schalken et al., 2003). Similarly, BC200 ncRNA, a brain-specific small cytoplasmic RNA, is found to be expressed in cancers like breast, cervix, esophagus, lung, ovary, parotid, and tongue though absent in healthy individuals (Chen et al., 1997).
The circRNAs are functional non-coding RNAs that lack 3’ and 5’ ends, existing as circular transcripts. Expressed tissue specifically, these transcripts, identified to be coded by thousands of genes, were till recently considered aberrant spliced by-products. Nigro and colleagues in 1991 identified the first circRNA in mammalian cells (Nigro et al., 1991). They observed the transcript of the DCC tumor suppressor gene to lack 3’ and 5’ end and existed as circular molecules. They termed it as scrambled exons. With the development of high-throughput sequencing and bioinformatics pipelines, the identification of more and more circRNAs became accessible in the past decade (Santer et al., 2019). The most frequently described function of circRNAs is that of sponging. With one circRNA having the ability to target more than one miRNA, these non-coding RNAs bind to miRNA molecules and prevent them from binding onto its canonical targets. The circRNA CDR1as have more than 70 conserved binding sites for miRNA-7 (miR-7), inhibiting its activity (Memczak et al., 2013). Studies in liver cancer showed that circHIPK3 was upregulated and could sponge at least nine different tumor suppressor miRNAs with a total of 18 potential miRNA-binding sites (Zheng et al., 2016). Some of the circRNAs were found to have the protein-interacting ability. Circular RNA, circ-FOXO3 is one of them, with expression levels negatively correlating with cell proliferation, inhibition of circ-FOXO3 function promoted cell proliferation, whereas overexpression repressed cell cycle progression (Du et al., 2016). Though numerous challenges are there on the path of knocking down this circular transcript, various gain and loss of function experiments are being conducted that can elucidate the function of circRNAs. The success of such research will decide the potential of using circRNAs as diagnostic and therapeutic tool for clinical application.
Eventhough we celebrate the potentiality of RNA interference, there are immense challenges to be overcome to take this from bench to bedside. The RNAi molecules being negatively charged, their capability to get infused through a cell membrane and to stay stable in the cellular matrix is questionable. Though studies that anchor on increasing the half-life of these molecules in the serum through chemical modification exist, the chance of them becoming immunotolerant inside an organism's body is meager. Thus, the development of carrier molecules that is in nanoscale, which can stay undetected or are pH-sensitive, is required for the development of RNAi to a potent therapeutic molecule. With the advancement in nanotechnology and its application in medicine and pharmaceutical companies, more and more molecules are being experimented for stable and safe delivery into the target tissue.
The advent of gene therapy in clinical realm has facilitated the use of nucleic acids or oligonucleotides like plasmid DNAs, small interfering RNAs, short hairpin RNA etc. in therapeutic approaches, opening a new paradigm for treating wide range of genetic, innate and acquired diseases including cancer. A comprehensive approach in the oligonucleotide delivery deals with two major aspects. Firstly, to assimilate the oligos into a type of carrier that helps determine their precise morphology, tissue distribution, half-lives and other biological interactions, and secondly, to attempt the possible modifications in the nucleotide itself, say with a targeting ligand without altering the nature of the conjugate (Juliano, 2016). In order to address the complications associated with therapeutic intervention, efforts are being made to comply homeostasis within the therapeutic findings and shell out the prospects of desired outcomes. Drug delivery is a crucial part of treatment having its own attributes and advantages. However, regardless of the significant potential of bio-drugs in gene silencing, a key limitation in transition to bedside is the lack of appropriate delivery systems that do not reduce the drug efficacy. Even though the phenomenon of RNAi is promising, direct administration of small oligos has not been fruitful because of the lower half-lives of the oligos and presence of nucleases in biological fluids. Furthermore, its low molecular weight hinders oligos to effectively penetrate the endothelial lining and diffuse into the extra-cellular matrix. In addition to these biological perturbations, nucleic acid itself possesses negative charge which lowers its penetration efficiency through the negatively charged cellular membrane. Also, the endosomal pathway offers harsh environment, resulting in degradation of these bio-molecules. The pursuit to make a viable novel antisense drug has given birth to various strategies, delivery approaches and modifications that address limitations associated with direct assimilation of bio-molecules. These strategies facilitate the targeted carriage of bio-molecules, evading biological barriers of the body and they confer better in vivo stability, tissue bioavailability, and targeted cellular delivery to the bio-drugs (Figure 1). Both viral and non-viral vectors have been developed for improving target cell penetration as well as minimizing toxicity by off-target hybridization.
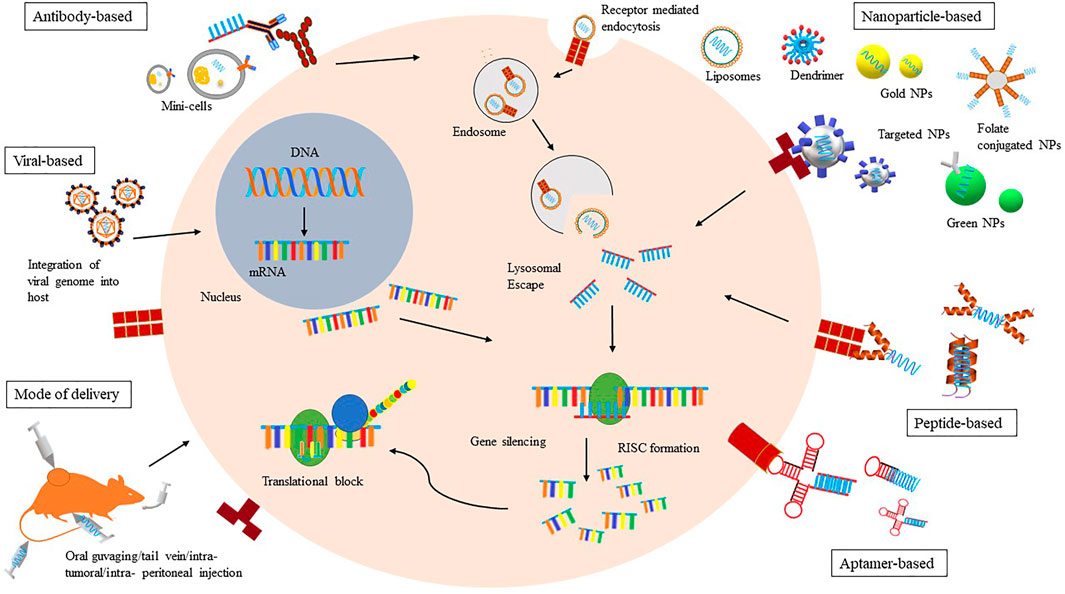
FIGURE 1. A schematic representation of various modes of Bio-drug delivery. Various modes of in vivo drug delivery like oral guvaging, tail vein (hydrodynamic), intratumoral, intraperitoneal etc have been shown here. Drug delivery methods such as viral based and non-viral based like antibody, mini cell and various nanoparticles like liposomal, dendrimer, gold, peptide and aptamer based is also been demonstrated. The action of the RNAi drug by gene silencing within the cell by post transcriptional and translational block after receptor based endocytosis has been displayed.
The multifaceted nature of science has paved the way for novel combinatorial and synergistic approaches in clinical terms from bench to bedside. Nanoparticles are versatile vehicles that can be used as drug carriers because they can be tweaked to address the limitations of delivery systems by permitting concurrent transport of numerous therapeutic agents for active therapy. Nanotechnology enables the design and assemblage of intended drugs with unprecedented control over their size and shape. Nanoscale methods have focused on developing adaptable bio-composites by conjugating therapeutic agents of interest into them, offering prominent advantages in drug delivery and imaging. Intracellular targeting describes the site where the drug has to be delivered; whether a tissue, an organelle or a compartment. Nanoparticles being sub-cellular and sub-micron in size, have the ability to penetrate deep into tissues or any other desired region by crossing the epithelial lining, ultimately getting permeated into the cell for achieving their therapeutic effects (Vinogradov et al., 2002). The enhanced permeation and retention effect (EPR effect) in cancerous tissues is essentially exploited by adopting various drug delivery methods, particularly with nano-based techniques. The defective tissue morphology and leaky vasculature of cancers increase the EPR effect, making them accurate targets for effective drug delivery (Brannon-peppas and Blanchette, 2004). The nanoparticle-based delivery systems can further be segregated into solid, lipid, and polymer based and in the next section we deal with each of these three systems taking appropriate examples.
Recently biocompatible magnetic nanoparticles have been used to deliver small oligos with high gradient external magnets attracting the drug complex, to the target site within the body providing great transfection efficiency (Nagal and Singlab, 2013), (Xin et al., 2017). Halloysite nanotube (HNT) based carrier was recently developed to deliver antisense oligonucleotides to target anti-apoptotic genes in cancer. In order to study the functionality of the carrier as well as the target gene, HNT carriers were surface modified with γ-aminopropyltriethoxysilane and the oligonucleotides were labelled with fluorescein. Confocal and transmission electron microscopic images recorded efficient cellular uptake and the variable gene expression (Shi et al., 2011). Conducive to construct efficient delivery systems, efforts were directed to preserve the bioactivity of oligonucleotides throughout the administration, from start to target site. Two different strategies have been devised which could be used to bind siRNA on gold nanoparticles. A covalent approach, based on thiolated siRNA bound to nanoparticle surface, and an ionic approach based on the electrostatic interaction between the negatively charged siRNA backbones. Both methodologies proved to be efficient for siRNA delivery, attaining precise gene silencing in both in vitro and in vivo biological systems (Tortiglione and de la Fuente, 2019). In leukemia mouse models, administration of cancer cell-specific mRNA drug conjugated with gold nanoparticles resulted in efficient drug delivery, appropriate release, and competent cellular uptake in both in vitro and in vivo experiments (Gossai et al., 2019). Also, gold nanoparticles have shown to act as a functional scaffold for various medicinal applications such as drug delivery, imaging etc. A gold nanoparticle (AuNP) based non-viral delivery system was created to direct siRNA into prostate cancer cells. Consequently, polyethylenimine (PEI) capped AuNPs were synthesized and combined with folic acid, a targeting ligand for folate receptors (AuNPs-PEI-FA) which could effectively integrate siRNA through electrostatic interactions. Flow cytometric analysis showed that, AuNPs-PEI-FA could exactly deliver siRNA into LNCaP cells, a prostate cancer cell line overexpressing prostate specific membrane antigen (PSMA). To inspect and counter-validate the complex, internalization of siRNA into PC-3 cells, a prostate cancer cell line not expressing PSMA or folate receptors, remained unattained via AuNPs-PEI-FA, signifying the potential of AuNPs-PEI-FA for targeted delivery in treatment of prostate cancer (Rahme et al., 2019).
Lipid nanoparticles (LNPs) are the most progressive nanocarriers for siRNA delivery due to their high encapsulation efficacy and potent gene knockdown ability. For precisely targeting the subsets of lymphocytes and to offer RNAi payloads in a cell-specific fashion, targeted lipid nanoparticles (tLNPs) were used as a unique favorable approach holding abundant potential for the manipulation of gene expression. This method exploits particular antibodies that adorn the lipid nanoparticle that is aimed at targeting lymphocyte subsets (Hazan-Halevy et al., 2019). Antisense oligonucleotides were designed and conjugated with Liposome-Polycation-DNA (LPD) nanoparticles linked with animoside, a targeting ligand to study the down-regulation of an anti-apoptotic protein called survivin in lung cancer (Hill and Carolina, 2006). 1,2-dioleoyl-sn-glycero-3-phosphatidylcholine (DOPC) based nanoliposomes were used to deliver siRNA directly into the tumor cells in vivo, with greater efficiency. Xenograft and orthotopic tumor models treated with DOPC incorporated siRNA, resulted in substantial regression in the tumor size and target genes causing no distress or toxicity (Ozpolat et al., 2010). Multi-stage delivery systems demonstrated the advancement of nanovectors. Nanovectors with spherical, quasi-hemispherical and discoidal silicon microparticles have unique roles in cellular adhesion and internalization, and the fabrication of such nanovectors have considered size, shape, and design. In vivo studies report increased therapeutic efficiency of liposome coated siRNA when administered through multi-stage systems as compared to free nanoparticles (Serda et al., 2011). Cervical cancer being one of the most life threatening types of cancer among the women, is generally known to be resistant to chemotherapy. Formulations of different solid lipid nanoparticles (SLNs) having chemotherapeutic agent and genetic material (Paclitaxel and siRNA against Bcl-2 oncogene) were tailored respectively. The intent of the study was to understand efficacy with respect to release amount of siRNA, when administered through vaginal suppositories (Büyükköroğlu et al., 2019). Multifunctional envelope typed nano device (MEND) was designed to address the novelty of non-viral gene delivery system. It comprises a complex nucleic acid core, a ligand for specific targeting, fusogenic lipids to enhance endosomal escape and a cell-penetrating peptide to increase the intracellular availability of novel drugs (Hatakeyama et al., 2011). Specific antisense oligonucleotides were incorporated into Cationic amphiphilic bolaamphiphiles (CABs), which were created and tested for their potential to form nano-sized vesicles based on various properties. Results demonstrated significant effect in treating C. difficile infection (CDI) by effective targeting and controlling the disease. For treatment options, antisense therapies promise to be a feasible alternative to the conventional antibiotic therapies (Sharma et al., 2018).
Numerous carriers have exhibited promising results in terms of delivering antitumoral oligonucleotides like siRNAs to xenograft tumor models, where the formulation was administered through intravenous mode. Chitosan-coated poly (isobutylcyanoacrylate) nanoparticles were used as transporter molecules to deliver siRNAs in xenograft tumor mice models. siRNAs coupled with the nanoparticles by adsorption made the polyelectrolyte complexes formed between the chitosan and nucleic acids very stable and non-toxic (Vauthier, 2019). Treatment of mammary tumors with intratumoral delivery of c-Myc shRNA complexed to rhodamine labelled nanoparticles in Brca2/p53 conditional knockout mouse models, resulted in regression of tumors, prolonging the survival, indicating the suppression of tumors treated with c-Myc shRNA linked-nanoparticles compared to control (Tangudu et al., 2015). Some malignancies like pancreatic cancers, being refractory for regular treatment options (chemotherapy), have the potential to play an important role in precision medicine for cancers by silencing the expression of genes when small-interfering RNAs (siRNAs) are used. For targeted carriage, star-shaped polymeric nanoparticles were developed which self-assembles siRNA into them, for delivery into pancreatic cancer cells in orthotopic mouse pancreatic tumor models. Results indicated substantial reduction in the tumor size and slow recovery in health of the mice after successful administration (McCarroll et al., 2019). Spherical Nucleic Acids (SNAs) are nanostructures that typically consist of a core where functional repositories such as siRNA, miRNA mimics or antagonists could be housed and used as gene regulators. It is known to confer heightened resistance towards degradation, facilitate ease of cell entry in the absence of transfection vehicles. In murine models of glioblastoma, precisely administered siRNA or miRNA conjugated with SNAs, penetrate blood-brain and blood-tumor barriers, regressing tumor progression (Tommasini-Ghelfi et al., 2019).
For tumor-targeted drug delivery, currently aptamers are being used when addressing cancer treatment with a therapeutic approach. In a recent study, a 64Cu-labeled modified A10 aptamer was designed to target prostate cancer by conjugating with the drug (p-SCN-Bn-NOTA), labeled with 64 Cu radioisotope. This extends the hope for diagnostic potential, non-invasive imaging, and to trace their bio-distribution in vivo paving the way for clinical applications (Kang et al., 2019). Tumor-specific carrier systems for siRNA delivery have extended the scope for reaching targets in treatment of cancer. For targeted combinatorial approach, Folate-conjugated siRNA polyplexes were developed based on sequence-defined oligomer platforms. In vivo and in vitro experiments validated the efficiency of these polyplexes in gene silencing, receptor-directed killing of cancer cells (Lee and Wagner, 2019). Synergistic combinations of oligonucleotides and drugs are the new edges in finding effective therapeutic approaches. To expound active combinations in triple negative breast cancer cell models, resourceful protocols to screen siRNA libraries were developed. For validation of effective transfection reagent for intracellular delivery of siRNA, lipid-grafted low molecular weight (1200Da) Polyethylenimines (PEIs) were used owing to its advantages (Thapa et al., 2019). Optimized DNA nanosuitcases was constructed as tractable DNA prisms that can encapsulate and selectively release siRNA upon recognizing an oligonucleotide trigger. It was designed with much adaptability of oligonucleotides, where it could be made to respond to an oligonucleotide trigger of interest like shRNA or miRNA, thus aiding dual therapeutic approaches (Bujold et al., 2016). Dendrimers being nano sized and radially symmetric molecules have been used as a carrier in gene delivery systems. For specific gene delivery in head and neck squamous cell carcinomas (HNSCC), a fluorescently labeled folic acid-decorated polyamidoamine (PAMAM) dendrimer was conjugated with folic acid (FA) as targeting moiety. When linked with a plasmid or siRNA, this complex is known to significantly enhance the gene transfection or knockdown efficiency in mouse xenograft models (Xu and Yang, 2019). Trastuzumab (TZ) is a monoclonal antibody that binds explicitly to human epidermal growth factor 2 (HER2) receptors and is presently being used for the treatment of HER2-positive breast cancer. A targeted carrier system made up of trastuzumab-conjugated poly (amido) amine dendrimers demonstrated a strategy for site-specific carriage of siRNA to HER2-positive breast cancer cells, subsequently resulting in definite targeting without off-target effects (Kulhari et al., 2019). Higher levels of miRNAs in cancer cells are frequently related with oncogenic effects. Targeted delivery of synthetically modified antagomiR molecules to malignant myeloid cells and B cells by linking with single stranded phophorothioated oligodeoxynucleotides (PSO), reported to persuasively reduce the levels of target miRNA. Therefore, the PSO-antagomiR has demonstrated its role, by virtue of regulating the expression of downstream protein targets, both in vitro and in vivo (Su et al., 2019). Synergistic approach by delivery of small interfering RNA (siRNA) and chemotherapeutic agent (cisplatin) for Human antigen R (HuR) mRNA in cancer cells by means of a polyamidoamine (PAMAM) dendrimer, was developed. In vitro results established that this strategy was effective in lung cancer cell lines H1299 and A549, along with reduced toxicity in normal lung fibroblast MRC9 cells (Shivdasani, 2006). During invasion and metastasis of cancer from a primary organ, patients have to undergo treatment with anti-tumor drugs by means of systemic administration, instead of local injection. Unfortunately, when oligonucleotides are administered, they tend to be degraded by ribonucleases in the blood leading to poor accumulation of the effector molecules in target sites. To overcome this, double-stranded RNA/DNA chimera (dsRDC) was used as a strategy instead of small interfering RNAs in in vivo breast cancer models. This kind of chimera sidesteps off-target properties leading to the formation of RISC complex by the sense strand, thereby conferring more durability and stability in bloodstream (Taniguchi and Imai, 2019). DNA nanostructures offer a simple yet powerful technique for the self-assembly of designed oligonucleotides into them. Multivalent DNA nanostructures consisting of unmethylated CpG motifs were prepared and tested for their ability to withstand nuclease degradation, stability, and transfection efficiency. Studies demonstrated that CpG motifs are recognized by toll-like receptors of the cell, proving their specificity and high potential in drug targeting (Li et al., 2011). Engineered gold nano-shell based oligonucleotides were therapeutically designed to discharge its cargo on mandate upon illumination with near-infrared laser irradiation at 800 nm. A fluorescently labeled Green fluorescent protein (GFP) expressing human lung cancer H1299 cell line in vitro, was used to determine the cellular uptake, controlled release, and gene silencing induced by the desired siRNA (Huschka et al., 2012).
Peptide-based nanoparticles form a stable complex of proteins and nucleic acids. These nanoparticles facilitate efficient delivery of cargo into cells highlighting the importance of cellular uptake, independent of the endosomal pathway. Cell-penetrating peptides initiate electrostatic interactions with proteoglycans, allowing modification of actin network in the extracellular matrix, thereby facilitating the uptake of nanoparticles (Crombez et al., 2008). In one study, a morpholino based oligonucleotide was constructed and linked to a tumor-targeting RGD peptide to form oligoconjugates. These were further bound to a single molecule of human serum albumin. The resultant nanoconjugate showed a greater enhancement in receptor-specific intracellular delivery of oligonucleotides and markedly boosting the functional activity at low nanomolar concentrations, overcoming the limitations of cytotoxicity (Ming et al., 2013). The delivery of siRNA through modular L1, a peptide carrier bearing CXCR4 targeting ligand was evaluated for its ability to condense siRNA, mediating endosomal escape. This resulted in forming complexes with siRNA to provide efficient VEGFA gene knockdown by substantial reduction of VEGFA gene expression in EA.hy 926 endothelial cells and in A172 glioblastoma cells (Egorova et al., 2019). Biodegradable nanoparticles developed from poly D, L-lactide-co-glycolide (PLGA) were studied for their localization, release, and delivery of therapeutic proteins, plasmid DNA, and small molecules suggesting that they are efficient in overcoming the endo-lysosomal compartment, thus moving into the cytosol after administration (Panyam and Labhasetwar, 2012). An innovative biomolecule compound Atelocollagen was developed to deliver tumor-specific siRNAs in nude mice. Atelocollagen being positively charged forms a complex with siRNA by electrostatic interactions. In orthotopic tumor models of prostate cancer in nude mice, this complex overcame degradation by nucleases and was proficiently transported in the bloodstream to the target tumor tissues, where the siRNA/atelocollagen complex integrated through endocytosis, promoting anti-metastasis and tumor regression (Takei, 2019). The usage of cationic polymer Polyethylenimine-Polycaprolactone-Polyethylene glycol (PEI-PCL-PEG) showed increased efficiency as it formed a superior complex with nucleic acids and thus proved better for gene delivery. Moreover, this polymer permits targeting moieties to be coupled to the micelleplex, the overexpressed receptors found within tumors. With this approach, indium labeled siRNA encapsulated with PEI-PCL-PEG nanoparticles and folic acid targeting ligand was developed for their evaluation in in vivo tumor targeting in orthotopic ovarian cancer models (Feldmann et al., 2019). Bacterial minicells, nanosized enucleated cells, have been exploited as a productive delivery method for nucleotide based therapeutics. Minicells retain all of the molecular components of the parent cell, except chromosome, lacking the ability to divide. Due to numerous advantages like convenience, efficient packaging of si/shRNA, stability, and easy surface modification with antibodies/ligand for active targeting, they can be manifested selectively to tumor cell-surface receptors with any targeting moieties conjugated on minicells. The functionality of minicells was observed in vivo in xenograft mice tumor models and in in vitro selected cancer cell lines (Jivrajani and Nivsarkar, 2019).
The detailed understanding of molecular targets underlying cancer has paved way for personalized therapy, overcoming the intrinsic complexity and asymptomatic nature of various cancers. With an ability to interfere with the function of specific molecular targets, targeted therapy controverts the conventional and empirical approach of developing cytotoxic chemotherapeutics. The past decades have witnessed the manipulation of diverse molecules like tyrosine inhibitors, serine/threonine inhibitors, small molecule drug conjugates, and monoclonal antibodies into developing targeted therapeutics with anti-cancer activity. Although these molecules were effective compared to the conventional methods, researchers found it challenging to circumvent the challenges related to protein durability. This is where RNA interference interceded and revolutionized targeted therapies. With an uncanny ability to target cancer-related genes, the RNAi pathway employs non-coding RNAs to bind messenger RNAs and silence gene expression in eukaryotic cells. Although profuse alternatives for gene silencing exist like CRISPR/Cas and TALENs, RNA interference remains the most favored option due to its precise functional mechanism, high potentiality, high specificity of gene silencing, and scanty side effects (Mansoori et al., 2014). The emergence of RNAi-based cancer therapies has materialized the usage of nanoscale particles as delivery molecules. With an enhanced ability to accumulate in tumor cells compared to normal cells due to the EPR effect, nanoparticles protect the RNAi molecules from undergoing enzymatic degradation and prevent them from being recognized by the immune system, thereby showing higher transportation efficiency compared to other carriers (Xin et al., 2017). However, a detailed preclinical characterization of the nano molecule and studies on the mode of offloading the loaded non-coding RNAs by these carrier particles have to be analyzed both in vitro and in vivo before taking these molecules to clinical trials. Evaluation of rate, extent, and perpetuation of silencing and the time-frame of delivery of the nanocarriers to the cancer niche is crucial in preclinical studies and the information generated enhances the chance of success in clinical trials (Wu et al., 2014). Almost every cancer tissue with a prominent biomarker and a potential to regress after gene silencing has been targeted for RNAi-based anti-cancer therapy. We are summarizing the results of pre-clinical obtained with nanoparticle-RNAi system in each type of cancer.
Advancement in gene therapy has accentuated the potentiality of RNAi molecules in breast cancer therapy over recent years. Lu Han et al. targeted HOX transcript antisense intergenic RNA (HOTAIR) by using specific siRNAs and by transfecting it into three human breast cancer cell lines (MCF-7, MDA-MB-231, and SKBR-3) resulting in repressed proliferation, invasion, and migration of cancer cells (Han et al., 2018). Studies have analyzed the interaction between miR-145 and Hepatitis B virus X-Interacting Protein (HBXIP), an oncoprotein promoting breast cancer progression and metastasis. Anti-miR-145 and si-HBXIP upon transfection revealed that miR-145 directly targets HBXIP and is a useful therapeutic target (Jiang et al., 2019). In order to knock down the expression of Ubiquitin-associated protein 2-like (UBAP2L), an oncogene associated with various cancers like prostate cancer, colorectal cancer, breast cancer, and glioma, shRNAs targeting the same has been transfected into breast cancer cell lines like ZR-75–30 and T-47D, successfully regressing the tumor (He et al., 2018). Other than these, Ca+2-dependent phospholipid and membrane-binding proteins, Annexin A3 (ANXA3), underwent gene knockdown in MDA-MB-231 cells with the help of sh-ANXA3 containing lentiviral vectors leading to inhibition of cell proliferation, migration, and invasion (Zhou et al., 2017).
A large intergenic non-coding RNA-regulator of reprogramming (lincRNA-ROR) is a newly identified lncRNA associated with initiation, development, and metastasis of multiple tumors, including breast cancer. This lincRNA-ROR has been shown to enhance the resistance of radio- and chemo-therapy of cancer cells along with the ability to inhibit the gemcitabine (Gem)-induced apoptosis and autophagy of breast cancer cells (Takahashi et al., 2014; Chen et al., 2016a; Yang et al., 2017a). Even after tamoxifen (TAM) has been clinically proven to reduce the risk of development of breast cancer, its resistance towards therapy is still a drawback. Thus, shRNA targeting lincRNA-ROR has been transfected into MDA-MB-321 BC cells. This showed decreased cell proliferation and increased TAM sensitivity by reducing its resistance (Lu et al., 2019). TAM is also found to promote apoptosis by suppressing the activation of the P13K/Akt/mTOR signaling pathway. The PRDI-BF1 and RIZ (PR) domain zinc finger protein 14 (PRDM14) is found to be overexpressed mostly in three out of five breast cancer patients, out of which some exhibit gene amplification. siRNA targeted against PRDM14 in nude mice successfully reduced size of breast tumors and lung metastases (Taniguchi and Imai, 2019).
Previous studies have revealed the hindrance of colon cancer proliferation and invasion in vitro via the down-regulation of TSPAN1 expression (Chen et al., 2010). Enacting the role of a potential prognostic factor in colorectal carcinoma, TSPAN1, through its inhibition by miRNA mediated RNAi, paved the way to the deduction of miR-638 as a tumor suppressor miRNA (Zhang et al., 2014). Similarly, a knock-down study on s100p gene in colon cancer cells using lentivirus-mediated RNAi, indicated significant reduction of tumor growth and liver metastasis in vivo (Jiang et al., 2011). Via in-silico screening, forced overexpression of miR-124 in colon cancer cells reduced Collagen prolyl-4-hydroxylase α subunit 1 (P4HA1) expression and subsided its malignant phenotype. The malignant phenotype was also diminished by treating the cancer cells with diethyl-pythiDC, a small molecule inhibitor of P4HA1 (Agarwal et al., 2019). Colo-320 cells were transfected with two complementary pairs of short-hairpin siRNA oligomers specific to human BRCP/ABCG2 cDNA sequence which led to a significant reduction in protein expression and chemoresistance (Hu et al., 2017).
Colon cancer-associated transcript 2 (CCAT2) is an lncRNA which is found to be highly overexpressed in microsatellite-stable colorectal cancer. A meta-analysis on the same revealed that the overexpression of CCAT2 gene can be used as a novel prognostic factor in several cancers (Fan et al., 2017). Thus, shRNAi plasmids targeting CCAT2 have been transfected into human gastric cancer BGC-823 cell lines. The corresponding silencing of the CCAT2 gene reduced the tumor cell proliferation and promoted apoptosis and autophagy in BGC-823 cells (Yu et al., 2018). The regulatory interlink between HOTAIR and insulin like growth factor 2 mRNA‐binding protein 2 (IGF2BP2) has been shown to influence the invasion and migration of colon cancer LoVo cells (Wu et al., 2019). The lncRNA, upon targeting and silencing HOTAIR, could significantly lower the proliferation of tumor cells as well as promote apoptosis through suppression of IGF2BP2 and Epithelial to Mesenchymal Transition (EMT). LINC01234, a highly abundant mammalian noncoding RNA has been proven to be a novel molecule in the field of tumor biology with associations with almost all types of cancers like breast, gastric, ovarian and colon cancers (Guo et al., 2016; Gu et al., 2017; Guo et al., 2017). Serine hydroxymethyltransferase 2 (SHMT2) is an enzyme regulating the serine/glycine metabolism pathway, a pathway identified to be critical in tumor metabolism since they have high energy and anabolic consumption capability. shRNA interference vectors targeting LINC01234 and SHMT2 were injected into BALB/c nude mice which had already been transplanted with tumor through subcutaneous injections of LoVo and HCT119 cell lines. Knockdown of LINC01234 arrested serine/glycine metabolism pathway thereby suppressing the cell proliferation and invasion (Lin et al., 2019a). Furthermore, LINC01234 was identified to be functioning as a competitive endogenous RNA (ceRNA) for miR-642a-5p, thus implying that LINC01234-miR642a-5p-SHMT2 axis plays a critical role in colon cancer proliferation.
Nuclear paraspeckle assembly transcript 1 (NEAT1), one of the nuclear lncRNAs is found to be associated with the deteriorated prognosis of colorectal cancer (CRC). siRNA and antisense oligonucleotides targeting NEAT1 as well as knockdown of miR-193a-3p modulated KRAS proteins, thus providing a therapeutic and diagnostic marker for CRC (Zhu et al., 2019). Similarly, the function of Tripartite Motif Containing 25 (TRIM29) was explored in colorectal cancer cells by transfecting HT-29 and SW1116 cells with si-TRIM29. This knockdown reduced tumor cell proliferation, migration and invasion followed by reducing the phosphorylation levels of JAK2 and STAT3, which are the regulators of JAK/STAT signaling pathway (Xu et al., 2016).
A novel oncogenic lncRNA, Sprouty4-Intron 1 (SPRY4-IT1) is found to be upregulated in multiple cancers regulating cell growth, invasion and apoptosis (Li et al., 2017a). To knockdown its expression, CRC (LoVo and SW480) cell lines were transfected with siRNA sequences specific to SPRY4-IT1 (Shen et al., 2017). Thus, cell proliferation, migration and invasion of CRC cells were suppressed via EMT-related gene modulation by negatively regulating the expression of miR-101-3p (target of SPRY4-IT1).
Cervical cancer is one among the most common types of cancer found in women worldwide. Several non-coding RNAs have been associated with cervical cancer and a few of them were taken forward for preclinical analysis. In 2012, Gibb et al found the abnormal expression of growth arrest specific 5 (GAS5) lncRNA by performing serial analysis of gene expression (SAGE) in cervical cancer (Gibb et al., 2012). In order to downregulate GAS5, RNAi sequence targeting this lncRNA was transfected into cervical cancer HeLA cell lines using liposome INTERFERin and its regulatory role was unraveled (Li et al., 2018a). Mitogen-activated protein kinase 1 (MAPK1), identified as a direct target of miR-329-3p, is upregulated in cervical cancer tissues and found to depict an inverse correlation with miR-329-3p expression. The knockdown of MAPK1 by transfecting human cervical cancer cell lines with si-MAPK1 significantly inhibited tumor cell growth, migration and invasion (Li et al., 2017b). These results were similar to the results obtained by overexpression of miR-329-3p inferring its therapeutic ability in cervical cancer.
Diverse cell-penetrating peptides (CPPs) have been in use to deliver anti-VEGF siRNA into cancer cells (Choi et al., 2010; Kanazawa et al., 2012; Egorova et al., 2016; Chung et al., 2017). With inadequate cell-type specificity being a major drawback, CPPs like BR2 specific to cancer cells have been reported (Lim et al., 2013). An amalgamation of siVEGF and BR2, when delivered intracellularly in HeLa cell lines, down regulated VEGF levels notably reducing toxicity, increasing antitumor efficacy indicated a specific and efficient delivery of siRNA by BR2 CPP (Lee et al., 2018).
Prostate cancer (PCa) is the most prevalent visceral cancer in men worldwide (Chen et al., 2015). Special AT-rich sequence-binding protein 1 (SATB1), a transcriptional factor is found to be overexpressed in various types of malignant cancers like nasopharyngeal carcinoma, cutaneous malignant melanoma, osteosarcoma and small cell lung cancer (Chen et al., 2014a; Hou et al., 2016; Lee and Pelletier, 2016; Xiong et al., 2017a). Three siRNAs targeting SATB1 region containing nucleotides 2,147–2,185 of complementary DNA were synthesized and were transfected into prostate cancer cell line DU145 using siPORT lipid transfection reagent (Mi et al., 2016). Tumor cell growth, invasion and migration capabilities were significantly inhibited in vitro implying that SATB1 siRNA could be a potential agent for human PCa therapy. Playing a major role in Ca+2 homeostasis, transient receptor potential cation channel subfamily M member 8 (TRPM8) has also emerged as a potential therapeutic target in PCa (Fidaleo et al., 2015), (Cao et al., 2017). The specific siRNA complementary to TRPM8 complexed with Lipofectamine 2000 was transfected into prostate cancer (LNCaP) cells thus, inhibiting cell proliferation and enhancing epirubic in chemosensitivity of cancer cells via promoting apoptosis (Chen et al., 2012). Prostate stem cell antigen (PSCA) is a glycosylphosphatidylinositol (GPI)-anchored cell membrane glycoprotein which is highly expressed in prostate cancer (Gu et al., 2000). Prior study by Zhigang Zhao and team produced in vitro validation that 21-nt long siRNA targeting PSCA could significantly inhibit cell proliferation and invasion of human PCa (PC-3M) cells (Zhao et al., 2016). The same group of researchers in the pursuit of validating in vivo, injected si-PSCA subcutaneously into male SCID mice bearing PCa xenografts (Leconet et al., 2018). The expression of PSCA was completely shut down as shown by immunohistochemical analysis as well as the reduced tumor growth volumes and metastasis onset. As ZEB1 was overexpressed in malignant cancers like prostate cancer (Heinrich et al., 2018), when the antisense lncRNA ZEB1-AS1 was expressed it positively regulated ZEB1 expression promoting tumor growth and metastasis via functioning as an oncogene in primary liver cancer (Huang et al., 2020). Based on these observations when two different siRNA vectors targeting ZEB1-AS1 were transfected into prostate cancer cell lines they inhibited ZEB1 overexpression and thereby the cell proliferation (Su et al., 2017).
In 2019, Fei cao et al., reported that targeting epidermal growth factor receptor (EGFR) mutations by RNAi can possibly treat residual lung cancer. PEG-PEI-EGFR siRNA nanocomposite was created by conjugating PEG-PEI polymer with siRNA targeting EGFR. This when injected intratumorally significantly reduced cell proliferation and invasion of tumor cells in BALB/c nude mice which had been subcutaneously injected with HCC827 lung cancer cells, subsequently rejecting the human lung xenograft model in mice (Cao et al., 2019). Growth arrest-specific 5 (GAS5) is known to be a vital tumor suppressor lncRNA in some types of cancers. NSCLC cells were transfected with plasmid vectors of GAS5 siRNA sequences. This study determined that miR-205 was a direct target of GAS5 in lung cancer and the Phosphatase and Tensin homologue (PTEN) was a direct target of miR-205. Upregulation of GAS5 aided in the suppression of tumor growth and invasion via miR-205/PTEN axis (Dong et al., 2019). On the contrary to this, X inactive-specific transcript (XIST) is known to function as an oncogenic lncRNA in bladder cancer, CRC and NSCLC (Fang et al., 2016; Xiong et al., 2017b; Chen et al., 2017). In order to unzip the correlation between XIST and TGF-β induced EMT, siRNA sequences targeting XIST and zinc finger E-box-binding protein (ZEB2) were transfected into human NSCLC (A549) cells using Lipofectamine 2000 transfection reagent. The knockdown of XIST repressed ZEB2 expression thereby inhibiting TGF-β induced EMT, cell proliferation and invasion. In addition to in vitro validation, the sh-XIST sequences were intravenously injected into female nude mice along with stable A549 cells. The decreased pulmonary metastasis observed from metastatic nodules on the lung surface revealed the role of XIST and ZEB2 in promoting EMT (Li et al., 2018b).
Programmed death receptor 1 (PD-1), being a specific programmed death receptor ligand 1 (PD-L1), plays a major role in immune escape by malignant tumor cells and is found to be overexpressed in most of the cancer types. The antitumor effects of dendritic cell (DC) immunization have been observed to be inhibited by the signaling functions of PD-1/PD-L1. The shutdown of the PD-L1 expression by lentivirus-mediated RNAi in SCID-hu mice injected with human pancreatic cancer PaTu8988 cells (Wang et al., 2019), reduced the tumor growth and lung metastasis thereby increasing the survival time of SCID-hu mice. Staphylococcal nuclease domain-containing protein 1 (SND1), also termed as P100 is a major component of RISC complex which regulates gene expression at both transcriptional and translational levels. Various studies have revealed the interaction of P100 with c-Myc, STAT5 and STAT6 (Dash et al., 1996; Yang et al., 2002; Välineva et al., 2005). Anna et al (year) reported the upregulation and chemoresistant characteristics of P100 in lung cancer (Zagryazhskaya et al., 2015). So, when miR-320a targeting P100 was transfected into human lung cancer cell lines, it inhibited cell migration, thus potentiating the biomarker effects in prognosis of lung cancer (Xing et al., 2018). Cancer cells require enhanced lipid biosynthesis since the fatty acids and phospholipids supplied by the gut and the liver alone are insufficient when compared to the consequent increase in the expression and activity of crucial enzymes like fatty acid synthase (FASN). Ning Zhan and his team constructed a lentiviral vector containing shRNA complementary to FASN and then transfected it into human non-small cell lung cancer (A549) cells making use of Lipofectamine 2000 as transfection reagent (Zhan et al., 2018). They observed that the FASN knockdown increased the radiosensitivity of NSCLC cells by effectively promoting apoptosis.
Aquaporins (AQP) which are integral membrane proteins facilitating water transport between cells are known to be prominently abundant in diverse malignant tumors, promoting cell proliferation and angiogenesis (Hu et al., 2006), (Verkman et al., 2008). In particular, AQP3 has been reported to be regulating cancer cell growth, invasion and migration (Lieberman, 2018), (Christopher et al., 2016) and hence inhibition of it can decrease the proliferation of NSCLC cells (Chen et al., 2015). So, two siRNA oligonucleotides targeting AQP3 were transfected into lung cancer (XWLC-05) cells and nude mice were subcutaneously inoculated with lung cancer cells transfected with si-AQP3. Cellular experiments revealed the inhibition of cell proliferation and invasion promoting apoptosis whereas reduced tumor growth was observed in in vivo studies (Xiong et al., 2017a). Lung cancer (A549) cell lines were transfected with four siRNA oligonucleotides cloned into lentiviral expression vectors to silence RNA helicase (DHX9), a protein overexpressed in a wide variety of cancer (Fidaleo et al., 2015; Lee and Pelletier, 2016; Mi et al., 2016). Subsequently using enoxacin to inhibit cell proliferation, this particular knockdown of DHX9 completely suppressed lung cancer cell growth (Cao et al., 2017). The combined effect of RNAi and tyrosine kinase inhibitors (TKIs) by targeting EGFR in NSCLC cells and subsequently silencing the gene in the cancer has also been reported (Chen et al., 2012).
Being rarely diagnosed at early stages, gastric cancer (GC), though could be treated using surgery, radiation, and chemotherapy at its earlier stages is very unresponsive to the same at its advanced and metastatic stage. The use of RNA interference has been thus emphasized in gastric cancer treatment to enhance both diagnosis and prognosis. The MYCL1 being a proto-oncogene, has been found to be upregulated in most of the gastric cancer cells. The usage of lentiviral MYCL1 shRNA expression vectors to knockdown the overexpression of this gene reduced migration and invasion of MGC-803 cell line in vitro (Qin et al., 2019). This effect of MYCL1 gene on migration phenotype can be exploited to enhance the prognosis rate of gastric cancer patients. Plasmacytoma variant translocation 1 (PVT1), has been reported to be a potential lncRNA biomarker for Gastric cancer patients. The shRNA targeting lncRNA PVT1 when transfected into BGC823 and AGS cells using lipofectamine , the expression of c-Myc was reduced which in turn decreased migration and invasion ability of tumor cells (Ren et al., 2019).
Methyltransferase-like 3 (METTL3) is known not only for N6-methyladenosine (m6A) modification of mRNA but also for regulating the translation of oncogenes. Human gastric cancer cell lines, AGS and MKN45 when transfected by shRNA targeting METTL3, it significantly decreased Bcl2 and increased Bax and active Caspase-3 implying activation on the apoptotic pathway. Thus, downregulation of METTL3 suppressed the proliferation and invasion of human gastric cancer cells (Lin et al., 2019b). Many researchers have reported the role of lncRNA LINC00978 in lung cancer, breast cancer and gastric cancer (Yang et al., 2015; Ke et al., 2017; Fu et al., 2018). In order to understand the biological function of this lncRNA in gastric cancer, Min Fu and his colleagues targeted the lncRNA using its specific shRNA in human gastric cancer (MGC-803 and SGC-7901) cell lines using LipoFiter as transfection reagent. The knockdown of this overexpressed lncRNA induced apoptosis by suppressing cell cycle progression. Also, since the migration and invasion of GC cells were found to be inhibited, LINC00978 could serve as a prognostic biomarker for gastric cancers (Fu et al., 2018).
Another lncRNA involved in cell proliferation, apoptosis and tumor development in liver and gastric cancer is small nucleolar RNA host gene 15 (SNHG15) (Zhang et al., 2016). The knockdown of this gene, in human NSCLC (A549) cells using Lipofectamine 2000, using specific siRNA oligonucleotides significantly suppressed cell proliferation, invasion and metastatic abilities of cancer cells thereby initiating apoptosis (Chen et al., 2016b). In order to investigate the role of TRIM25 in gastric cancer, two different siRNA oligonucleotides specific to TRIM25 were transfected into GC cell lines (MGC-803 and AGS). This silencing of TRIM25 expression decreased cell migration and invasion by regulating TGF-β signaling pathway (Dong et al., 2018). Being a transmembrane protein, the IQ motif containing GTPase-activating protein three gene (IQGAP3) is known to be upregulated in gastric cancer cells and could serve as therapeutic target (Yasui et al., 2011). So, the siRNA sequences targeting IQGAP3 were conjugated with Lipofectamine RNAi-MAX and then transfected into human GC (MKN-1) cells. Performing spheroid colony formation assay elucidated that both the number and size of the spheres formed by GC cells were remarkably reduced (Oue et al., 2018). Another transmembrane glycoprotein shown to be overexpressed in GC is liver-intestine cadherin (CDH17). Lentiviral-mediated RNAi performed by transfecting human GC (MKN28) cells with shRNA oligonucleotides specific to CDH17, promoted apoptosis by notably inhibiting cell proliferation of GC cells. Furthermore, subcutaneously injected GC mice xenografts showed significant lessening of tumor growth and volume thereby restricting tumorigenicity (Su et al., 2008).
First ever trials of RNAi in humans were focused on assessing the potentiality and activity of lipid nanoparticles (LNPs) formulated with siRNAs targeting VEGF and kinesin spindle protein (KSP) in liver (Tabernero et al., 2013). With this, not only intravenous administration of ALN-VSP liver was shown to be effective and safe but also complete regression of liver metastases was observed in endometrial cancer. Endometrial cancer (EC) being the most common malignant female genital cancer in the world, one of the candidate biomarkers of EC, chitinase-3-like protein 1 (YKL-40) is overexpressed in patients diagnosed (Fan et al., 2013) has a pivotal role in cancer cell proliferation (Brøchner et al., 2012), angiogenesis (Francescone et al., 2011) and anti-apoptosis (Lee et al., 2009). In order to silence the expression of YKL-40, siRNA oligomers targeting it were first cloned into retroviral vectors and then transfected into EC (HEC-1A) cells using Lipofectamine 2000 followed by lentiviral-mediated transduction (Li et al., 2018c). Tumor cell attributes like cell proliferation, migration and invasion were found to be inhibited consequently increasing the mean cellular apoptotic rate. One of the oncogenic lncRNAs, HOTAIR, has been reported to be mediating chemoresistance to cancer cells (Liu et al., 2013). In order to ascertain the role of this lncRNA in endometrial cancer, the Ishikawa human EC cell line along with cisplatin-resistant clones were transfected with three si-HOTAIRs with the help of Lipofectamine 2000 as transfection reagent (Sun et al., 2017). This knockdown of HOTAIR inhibited the cell proliferation of EC cells thereby decreasing their cisplatin-induced chemoresistance.
The oncogenic transmembrane glycoprotein MUC1, being overexpressed in several tumors, upon silencing via RNAi inhibited the growth of Hepatocellular carcinoma cells indicating its crucial role in tumorigenicity (Li et al., 2014). The following study performed by the same group of researchers indicated that MUC1 initiates the autocrine TGF-β signaling in HCC cells by activating JNK/AP-1 pathway (Li et al., 2015). JNK, being a member of the MAPK family, has been displaying its role in HCC development and progression (Ozpolat et al., 2010). In order to collectively target both MUC1 and JNK, this team (Wang et al., 2017) firstly transfected HCC (SMMC-7721) cells with si-MUC1 and si-JNK1/2 or SP600125 (inhibitor of JNK) separately complexed with X-fect transfection reagent. Cell viability of MUC1-transfected cells was decreased and showed similar results with si-JNK and its inhibitor. Since unmodified siRNA is not stable in bloodstream and does not get transported through cell membranes (Whitehead et al., 2009), both the si-MUC1 and si-JNK were chemically modified by conjugating with 5’-cholesterol and 2’-O-methyl groups and then intratumorally injected into tumor bearing mice which in fact did not show variable results compared with negative control. Thus, a more effective delivery method has to be innovated or developed to inhibit JNK/AP-1 pathway.
Most of the human malignancies which have epithelial origin of cells are associated with overexpressed EGFR. So, shRNA targeted against EPS8 (EGFR pathway substrate 8) conjugated with polyamidoamine dendrimers through receptor-mediated delivery has been utilized to assess the phenotype of head and neck squamous carcinoma cells (Yuan et al., 2019). This showed that it inhibited cell growth, proliferation, migration and malignancy of tumor cells. Signal peptidase complex 18 (SPC18), encoded by SEC11A gene has been reported to induce the secretion of transforming growth factor (TGF-∝), which phosphorylates EGFR and stimulate pathways involved in cell proliferation and anti-apoptosis. So, siRNA targeting SEC11A was transfected into human bladder cancer (T24 and KMBC-2) cell lines using Lipofectamine RNAi-MAX. The knockdown of SEC11A prominently inhibited cell growth and invasiveness contrary to the forced expression of SEC11A (Shigematsu et al., 2019). Small Nucleolar RNA Host gene 7 (SNHG7), an lncRNA has been found to be overexpressed in bladder cancer tissues and cell lines. Thus, such overexpression has been downregulated by siRNA-mediated RNA interference. This downregulation not only had regulatory effect on Epithelial-Mesenchymal Transition (EMT) related markers but also inhibited cell proliferation and invasion thereby promoting apoptosis (Zhong et al., 2018; Katoch et al., 2021).
Bone cancer pain (BCP) has both the components: nociceptive and neuropathic. The nociceptive component is associated with the release of algogenic substances by tumor and thus plays a pivotal role in inducing cancer-related deaths. The neuromedin U receptor 2 (NMUR2) has been shown to be upregulated in rat models which were injected with walker 256 cells (Peng et al., 2019). Thus, siRNA targeted to silence NMUR2 successfully relieved pain by assessing paw withdrawal threshold (PWT) value.
Recent studies have shown that the hypoxia pathway is dysregulated in pancreatic cancer. Also, glucose transporter-1 being a transmembrane transporter is upregulated in many types of cancers and is responsible for transformation into malignant tumor. Folic acid receptor is overexpressed on almost all the malignant tumor cell surfaces since it is the essential vitamin for proliferative cells. The siRNA complementary to HIF-1α which has been transfected with graphene oxide nanoparticles targeting folate receptor was injected intravenously into nude mouse tumor models with pancreatic cancer PaTu8988 cells (Wan et al., 2019). This shutdown of the expression of HIF-1α and GLUT1 under hypoxic conditions significantly inhibited the proliferation and metastasis of tumor cells and thereby speeding up the apoptosis of tumor cells. In addition, hypoxic state of tumor tissues was reduced, which is usually higher in malignant tumor cells.
Tumor-derived exosomes (TEXs) are not only widely distributed in body fluids of cancer models but also have significant molecular level effects on immune cells like NK cells (Andre et al., 2002; Kim et al., 2007; Dai et al., 2008; Reiners et al., 2014). In order to determine the mechanistic role of oral cancer-derived exosomes (OCEXs), Yang and his colleagues performed in vitro knockdown of NF-κβ-activating kinase-associated protein 1 (NAP1) using siRNA in human oral cancer (OC) cell lines using Lipofectamine RNAi-MAX transfection reagent, and observed that OCEX derived NAP1 induces activation of interferon regulatory factor 3 (IRF-3). Thus, exosomal NAP1 enhances the tumor-suppressing functions of NK cells specifically cytotoxicity towards tumor cells and could be used as immunotherapy for treating oral cancer (Wang et al., 2018).
Research has shown that, Human antigen R (HuR) being the most ubiquitously expressed RNA-binding protein in eukaryotes is one of the key regulators in post-transcriptional gene expression mechanisms (Schwanhüusser et al., 2011) consequently causing several types of cancers like breast cancer, ovarian cancer and colon cancer (Govindaraju and Lee, 2013; Wang et al., 2013). Balden et al. demonstrated that HuR is overexpressed also in thyroid cancer (Baldan et al., 2016) and to determine its biological activity, the siRNA targeting HuR were transfected into anaplastic thyroid cancer cell lines (SW1736 and 8505C) using Dharma-FECT 1 as transfection reagent and this, reduced colony forming ability (Allegri et al., 2018). Apart from using the RNAi mechanism to silence HuR, SAHA an FDA approved drug was also used in mouse epidermal JB6 C141 cells.
Urinary bladder cancer (UBC) is another most common genitourinary malignancy worldwide. ASAP1-Intron 1 (ASAP1-IT1), is an intronic transcript of ASAP1 whose overexpression has been implied in cancer development (Guo et al., 2018). Human UBC cell lines were transfected with si-ASAP1-IT1 cloned into a lentiviral vector using Lipofectamine 3,000 transfection reagent thereby reducing stemness of UBC (Yang et al., 2017b). Also, Snail-1 is one of the crucial transcription factors in the process of regulation of EMT. Musavi Shenas et al. transfected EJ-138 bladder cancer cells with three different siRNA sequences targeting Snail-1 (Musavi Shenas et al., 2017). The expression of a few miRNAs involved in EMT such as miR-29b, -21 and -203 was shown to be reduced significantly by the knockdown of Snail-1. In addition, Snail-1 siRNA aided in induction of apoptosis as measured by the TUNEL test.
The human trophoblast cell surface antigen 2 (TROP2), is a cell surface glycoprotein and its overexpression in several cancers has been linked to tumorigenicity, poor survival and metastasis (Cubas et al., 2009). Though Chen et al. reported the association of TROP2 overexpression with poor prognosis in gall bladder carcinoma (GBC), mechanisms underlying proliferation and metastasis are still unknown (Chen et al., 2014b). To determine this, human GBC cell lines were transfected with shRNA targeting TROP2 conjugated with Lipofectamine 2000 reagent. In addition to this, BALB/c mice were subcutaneously injected with GBC cells transfected with sh-TROP2. The in vitro results confirmed the inhibition of cell proliferation, invasion and migration of GBC cells whereas, in vivo studies showed the postponed EMT of cancer cells analyzed by immunohistochemistry (Li et al., 2017c).
In a world where the development of new drugs is painfully slow and the pipeline of new therapeutics frightfully thin, the discovery of RNA interference has paved way for the development of relatively rapid approaches towards cancer drug design. The transition from preclinical work to clinical trials requires a novel therapeutic that shows sufficient promise to be effective and safe in treating human diseases. The fact that the number of nanocarriers that have reached the phase of clinical approval is meager and the high diversity of the nano platform makes concluding a particular nanoparticle to be “promising” is complicated (Mirkin et al., 2015). However, with the success and validation in in vivo models, various RNAi and nanoparticle-based molecules were taken for clinical trials in the past decade.
Approximately 25 different siRNA/shRNA molecules have been approved for clinical trials till now for anti-cancer therapy (Ozcan et al., 2015). Unlike other antisense therapies that have a stoichiometric effect on mRNA, siRNA’s ability to reduce off-target toxicity along with its ability to act with increased potency, makes it a better contender than any other therapeutic molecules. These RNAi molecules can be transported to the patient’s body via either natural carriers like bacteria or virus and using synthetic nanocarriers, the strategic use of these depends on the application, the required duration of the therapeutic, and the targeted tissue (Burnett et al., 2011; Chakraborty et al., 2019). Usually in clinical trials, the natural carriers are used to deliver to target cells ex vivo, which are then reinfused back inside the patient's body. These bacteria and viruses often possess shRNAs that will undergo intracellular modifications to become functional siRNAs. Synthetic carriers like nanoparticles are often delivered systemically through IV injections (Whitehead et al., 2009).
Being that most predominant molecules coming under the category of ‘FDA approved small RNA drugs’ to enter the paradigm of clinical application, nano-conjugated miRNA molecules have not still entered the United States government’s website recording clinical trials. Several pharmaceutical and biotech companies have launched miRNA projects based on miRNA mimics and antagomiRs. Several companies like Dharmacon, BioSyn and GenScript tried to produce natural and chemically modified RNA, to stabilize and reduce the high reactive nature of RNA molecules. While some companies have shown some success, most of them are struggling to impact patient outcomes. For example, FDA halted the Phase1 clinical trial by MiRNA Therapeutics on miRNA mimic, MRX34, as a consequence of adverse side effects in patients caused due to lack of immune tolerance (Ling et al., 2017). MiRagen Therapeutics is actively developing MRG-106, an LNA antagomiR that targets miR-155 and is now in phase1 and phase2 clinical trials. Several companies work in preclinical and large screening studies to identify potential biologic miRNA such as CURNA program by Opko, Alnylam Pharmaceuticals, Interna Technologies and Mello Biotech (Bonneau et al., 2019). The first siRNA-based delivery system that was approved for clinical trials was that of the cyclodextrin polymer (CDP). Denoted as CALAA-01, this CDP polymer attached to polyethylene glycol (PEG), steric stabilization agent, and human transferrin targeted transferrin receptors that are usually found upregulated in cancer cells (Davis, 2009). Calando Pharmaceuticals (Pasadena, CA, United States) in 2010 conjugated siRNA targeting M2 subunit of ribonucleotide reductase to CALAA-01 regressing the tumor growth (Davis et al., 2010). Though there was an intratumoral downregulation of ribonucleotide reductase, the fact that the trial took place only in a few patients made these results preliminary.
Liposomes, being a potential agent in transferring siRNA to the target tissue, many liposome conjugated siRNAs have been sanctioned by the FDA for clinical trials. With an ability to improve siRNA entrapment efficiency, 1,2-dioleoyl sn-glycero-3- phosphatidylcholine (DOPC) when used to encapsulate EphA2 siRNA showed promising results in orthotopic ovarian carcinoma. This conjugate was taken for Phase I clinical trial and is currently in the recruiting phase (Landen et al., 2005). Atu027 is a lipoplexed siRNA that can target the protein kinase N3. Constituting a positively charged AtuFect01, a neutral, fusogenicDPhyPE helper lipid and the PEGylated lipid MPEG-2000-DSPE (Schultheis et al., 2014), Atu027, due to its success in treating lymph node metastases in mouse models of the prostate, pancreatic cancer, and lung metastasis, was approved for clinical trials (Aleku et al., 2008). Silence Therapeutics (London, United Kingdom) conducted Phase I clinical trials and found that it is tolerable and does not show dose-dependent toxicity (Strumberg et al., 2012). The efficacy of Atu027 in clinical trials is also being tested with gemcitabine in metastatic and advanced pancreatic cancer patients.
Stable Nucleic Acid Lipid Particles (SNALP), due to their high bioavailability and enhanced permeability and retention, has become a notable carrier of siRNA in clinical trials. Found to be well tolerated at high dose, SNALP encapsulated version of siRNA targeting PLK1 was taken for Phase I trials by Tekmira Pharmaceuticals Corporation (Burnaby, BC, Canada). Moreover, SNALP was used to create the first dual-targeted siRNA drug. ALN-VSP02, developed by Alnylam Pharmaceuticals (Cambridge, MA, United States), targeted both VEGF and KSP and in Phase I trial was well tolerated at high doses in hepatic and extra hepatic tumor patients (Morrissey et al., 2005; Shen et al., 2012). The first dual-target nanoparticle-based RNAi showed promising results as one of the patients exhibited complete response, and the rest were stable for eight to twelve months of treatment (Mahmoodi Chalbatani et al., 2019). DCR-PHXC-101 is another lipid formulation that has entered the Phase I trial. It targets the transcription factor Myc and is showing metabolic response in patients with solid tumors, multiple myeloma, and lymphoma (Tolcher et al., 2015). Besides these, miniature biodegradable polymeric matrices were loaded with KRASG12D-targeting siRNA to target the K-Ras gene in pancreatic cancer patients. Phase I and II clinical trials on the same showed no evidence of cancer progression (Strand et al., 2019). A summary of the nano-conjugated siRNAs in clinical trials has been presented in the table below (Table 1).
Despite RNA interference being a promising therapeutic strategy in cancer therapy, it faces a lot of hurdles during the process of being recruited from bench to bedside. Potent and effective gene silencing in the tumor cells is mandatory for determining the efficiency of siRNA-based drugs in combating cancer. To achieve such effective gene knockdown in vivo and in clinical trials, efficient delivery of RNAi molecules to the target tissues is required (Petrocca and Lieberman, 2011). The efficient uptake of siRNA molecules by the cell is usually intercepted due to its high negative charge. These molecules need to survive in the extracellular matrices that constitute the RNA degrading endonucleases. Although endocytosis of these molecules is an elegant strategy to enter the cells, the hassle of releasing the RNA efficiently from the endosome and preventing it from undergoing endosomal degradation is unavoidable. This, along with the fact that siRNAs undergo rapid excretion via the renal system after intravenous injection, is what reduces the potency of naked siRNA-based therapy (Bora et al., 2012).
Another major obstacle to RNAi-based therapeutic strategy is intracellular delivery. This is primarily in the case of treating disseminating cancer cells that cannot be removed through surgery. To combat this, many biotechnological and pharmaceutical companies have come up with multiple delivery strategies. Chemical conjugation of siRNAs to specific molecules, being one of them, has, to an extent, increased the half-life of these molecules in vivo and has thus enhanced the potency of the same in the serum (Braasch et al., 2003; Chiu and Rana, 2003; Layzer et al., 2004). Modification of these nucleic acid molecules so as to have extended cholesterol, glycan, and folate branches that bind to cell receptors and incorporating these within nanocarriers, liposomes, and aptamers to do cell-specific targeting have shown promising results when induced in mice and non-human primates (Elmén et al., 2008). Nevertheless, the fact that only a few siRNAs among these have entered clinical trials and various companies are still working to resolve this RNAi-based molecule delivery problem shows that none of these current solutions are ideal, and the payoff for the same could be enormous.
Besides accomplishing target specificity, developing therapeutic strategies that give minor off-target effects is another major challenge (Jackson et al., 2003). The ultimate answer to such a problem would be to develop an efficient delivery system. Site specific delivery of siRNA, if appropriate, would reduce the number of normal cells exposed, favoring the uptake by cancerous cells and thus reducing the potential off-target effects. This would also minimize the dose to be administered, lowering potential dose-related detrimental side effects. Some of the siRNA molecules can also induce immunostimulatory effects. RNAi-based molecules can induce the innate immune system to release inflammatory cytokines, such as interferon-α, tumor necrosis factor-α, and interleukin-6, which may lead to considerable toxicity (Elbashir et al., 2002; Karikó et al., 2004; Hornung et al., 2005).
However, despite all these drawbacks, RNAi-based cancer therapeutics is still the best option available in bringing about apoptosis of highly malignant cells in a specific manner especially for the tumor cells resistant to conventional therapy. The development of personalized cancer therapies that combine monoclonal antibodies with RNAi has come up due to their ability to complement each other's vulnerabilities (Schultheis et al., 2016). This is also considered more effective than using either alone. RNAi being a revolutionary tool to treat several diseases, through rational strategies that take into consideration all the cautionary drawbacks, could be developed into a powerful weapon that can combat cancer.
Arising from a complex interplay of oncogenes and tumor suppressor genes, cancer is a collective manifestation of deregulated gene functions. In spite of the innovation of various approaches of therapeutic modalities like chemotherapy, radiotherapy, and immunotherapy to combat cancer, the fact that these strategies have encountered numerous limitations compelled the scientific world to go on a quest for a better treatment paradigm. This is where RNA interference entered basic and clinical research, and revolutionized it. Termed as ‘next generation therapeutic molecules’, alongside recombinant protein and monoclonal antibody, RNAi has shown propitious results in vitro due to its high efficacy, and specificity. Despite its potential, several hurdles arise on the path of using RNAi in vivo due to its instability and short half-life in the serum. To overcome these problems, use of non-viral vector delivery systems that protect the RNAi molecules from degradation have been extensively explored. Drug delivery systems at the nanoscale have a promising future with the demonstrated superiority (by virtue of targeted approach, minimal collateral damage and accumulation at pharmacological levels at the desired site) compared to the traditional chemotherapy modalities. The addition of active targeting molecules to nanoparticles is found to enhance the tumor-targeting and transfection efficiency towards tumor cells. However, the use of nanoparticles too has its own limitations due to cytotoxicity, hurdle of delivery, and immunogenicity that prevents the reach of RNAi from bench to bedside. As constant progress is being made to achieve improved tumor targeting accuracy, interaction of these nanoparticles with the immune system of the host remains an interesting current and future prospect (Zolnik et al., 2010; Bolkestein et al., 2016). The increased uptake of liposomal preparations in therapeutics by tumor cells has been attributed to certain defining features of malignancy such as increased vascular permeability, the microenvironment that is created by the neighboring stroma, the presence or absence of sustentacular cells, the lymphatic composition and mononuclear cell presence etc (Bolkestein et al., 2016). The interaction of chemical mediators such as bradykinin, nitrous oxide and physical factors (such as the smooth muscle in the blood vessel wall) in controlling nanoparticle movement and reach to the tumor site have been demonstrated in certain studies as the real hindrances in large scale adoption of nanoparticles as therapeutic vehicles (Stylianopoulos et al., 2012). These factors are fundamental to the enhanced permeability and retention effect (EPR effect) which holds great importance in nanoparticle and tumor biology (Stylianopoulos et al., 2012; Bolkestein et al., 2016; Golombek et al., 2018).
The immune status of the preclinical model appears to play a role in dictating tumor targeting efficacy of RNAi-nanoparticle therapeutics. Some factors which have been demonstrated to have an effect are opsonization (which is the process of making a target ‘ready’ for an immune system interaction), structural and electrochemical features of the nanoparticle and the duration of nanoparticle presence in the host (retention effect) (Zahednezhad et al., 2019). Mice models with deficient adaptive immune system were demonstrated to have low levels of neoplastic growth inhibition when the kinase inhibitor sorafenib was used, indicating the probable prerequisite of an intact adaptive immune status for the nanoparticles to function (Zhao et al., 2018). The interaction of nanoparticles with immune system components like the complement system and mononuclear phagocyte system has led to clinically observed effects such as CARPA (complement activation-related pseudoallergy) and increased clearance of therapeutic nanoparticles. The variability in antitumor efficacy with the use of CTLA-4 and PD-1 inhibitors (implying the immune status) has led to the question of validity of preclinical models being used (La-Beck and Gabizon, 2017).
The extent of interaction between the immune system and nanoparticles drives the need to test these drug delivery systems in immuno-competent models and humanized mice models (which are often achieved by reconstituting SCID mice with human bone marrow introduction). As the immune system is complex, interaction with other subgroups such as the Th1 and Th2 systems needs to be studied further to have better outcomes (La-Beck and Gabizon, 2017). The gap between the pre-clinical and clinical stages of nanoparticle therapeutics needs to be bridged and the progress might be seen when hurdles such as clearance and toxicity of nanoparticles are circumvented and the use of humanized models is adopted for large scale studies (Rosenblum et al., 2018). With backing of the advantages they offer, this combinatorial therapeutics have the potential to be game changers in attacking cancer cells and tissues and are expected to yield revolutionizing clinical outcomes. Such a future is possible through constant improvement in the materials used in the nanoparticle drug delivery system and by moving towards an individualized and patient centric approach (Golombek et al., 2018), (Navya et al., 2019).
In spite of the remarkable progress made till now in terms of increasing the efficiency of nanobased RNAi delivery system with novel complexation or conjugation approaches, there are still specific challenges posed by cell types in different target organs to be resolved. Therefore, as an important strategy, RNAi therapeutics must be validated for varied targets to obviate this challenge. It would then be conceivable to promptly advance RNAi therapeutics innovatively and empirically to target different tissues and organs by fine tuning nano-based delivery system through experimental studies. Furthermore, the detailed molecular mechanism behind the RNA interference and its off-target effects are to be deciphered as the multi-targeting network of their pathways are often very complicated and challenging. Considering the immense influence our understanding of RNAi regulatory network in neoplasia has in the development of RNAi based anti-cancer therapeutics, there is a compelling need to decipher such networks to place RNAi based therapeutics on a firm footing. Even though novel clinical studies on the same have ushered hope, there is a necessity for the development of more multi-centered, randomized, and placebo-controlled treatment methods that would be effective irrespective of the racial or ethnic variations among the patients. With the development of safe, biocompatible, and biodegradable targeted delivery systems for the clinical application and with proper knowledge on the RNAi networks, this promising alliance of nanotechnology and RNA interference is sure to transform anti-cancer therapeutic methods and interventions in future.
LDK has conceptualized the topic, critically reviewed, edited and rewritten many parts of the MS. GS and AS have written the major aspects of the manuscript. AK contributed the figure, introduction, conclusion and editing of the manuscript. VB has written the clinical aspects of the review, and VD contributed to the preclinical section of the manscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors are extremely grateful to V. Dinesh Kumar for his critical evaluation, suggestions, and help in editing the manuscript. AS was supported by INSPIRE scholarship for higher education from Department of Science and Technology (DST), Government of India. AK was supported by the fellowship from the Ministry of Education (MOE), Government of India.
Abdelrahim, M., Smith, R., Burghardt, R., and Safe, S. (2004). Role of Sp Proteins in Regulation of Vascular Endothelial Growth Factor Expression and Proliferation of Pancreatic Cancer Cells. Cancer Res. 64, 6740–6749. doi:10.1158/0008-5472.CAN-04-0713
Agami, R. (2002). RNAi and Related Mechanisms and Their Potential Use for Therapy. Curr. Opin. Chem. Biol. 6, 829–834. doi:10.1016/S1367-5931(02)00378-2
Agarwal, M. (2016). Is Cancer Chemotherapy Dying?. Asian J. Transfus. Sci. 10, 1. doi:10.4103/0973-6247.182735
Aleku, M., Schulz, P., Keil, O., Santel, A., Schaeper, U., Dieckhoff, B., et al. (2008). Atu027, a Liposomal Small Interfering RNA Formulation Targeting Protein Kinase N3, Inhibits Cancer Progression. Cancer Res. 68, 9788–9798. doi:10.1158/0008-5472.CAN-08-2428
Allegri, L., Mio, C., Russo, D., Filetti, S., and Baldan, F. (2018). Effects of HuR Downregulation on Anaplastic Thyroid Cancer Cells. Oncol. Lett. 15, 575–579. doi:10.3892/ol.2017.7289
Andre, F., Schartz, N. E. C., Movassagh, M., Flament, C., Pautier, P., Morice, P., et al. (2002). Malignant Effusions and Immunogenic Tumour-Derived Exosomes. Lancet 360, 295–305. doi:10.1016/S0140-6736(02)09552-1
Baldan, F., Mio, C., Allegri, L., Conzatti, K., Toffoletto, B., Puppin, C., et al. (2016). Identification of Tumorigenesis-Related mRNAs Associated with RNA-Binding Protein HuR in Thyroid Cancer Cells. Oncotarget 7, 63388–63407. doi:10.18632/oncotarget.11255
Bolkestein, M., De Blois, E., Koelewijn, S. J., Eggermont, A. M. M., Grosveld, F., De Jong, M., et al. (2016). Investigation of Factors Determining the Enhanced Permeability and Retention Effect in Subcutaneous Xenografts. J. Nucl. Med. 57, 601–607. doi:10.2967/jnumed.115.166173
Bonneau, E., Neveu, B., Kostantin, E., Tsongalis, G. J., and De Guire, V. (2019). How Close Are miRNAs from Clinical Practice? A Perspective on the Diagnostic and Therapeutic Market. EJIFCC 30, 114–127.
Bora, R., Gupta, D., Mukkur, T. K. S., and Saini, K. S. (2012). RNA Interference Therapeutics for Cancer: Challenges and Opportunities (Review). Mol. Med. Rep. 6, 9–15. doi:10.3892/mmr.2012.871
Borges, F., and Martienssen, R. A. (2015). The Expanding World of Small RNAs in Plants. Nat. Rev. Mol. Cel Biol. 16, 727–741. doi:10.1038/nrm4085
Braasch, D. A., Jensen, S., Liu, Y., Kaur, K., Arar, K., White, M. A., et al. (2003). RNA Interference in Mammalian Cells by Chemically-Modified RNA. Biochemistry 42, 7967–7975. doi:10.1021/bi0343774
Brannon-peppas, L., and Blanchette, J. O. (2004). Nanoparticle and Targeted Systems for Cancer Therapy. Adv. Drug Deliv. Rev. 56, 1649–1659. doi:10.1016/j.addr.2004.02.014
Brøchner, C. B., Johansen, J. S., Larsen, L. A., Bak, M., Mikkelsen, H. B., Byskov, A. G., et al. (2012). YKL-40 Is Differentially Expressed in Human Embryonic Stem Cells and in Cell Progeny of the Three Germ Layers. J. Histochem. Cytochem. 60, 188–204. doi:10.1369/0022155411433331
Brummelkamp, T. R., Bernards, R., and Agami, R. (2002). Stable Suppression of Tumorigenicity by Virus-Mediated RNA Interference. Cancer Cell 2, 243–247. doi:10.1016/S1535-6108(02)00122-8
Bujold, K. E., Hsu, J. C. C., and Sleiman, H. F. (2016). Optimized DNA “Nanosuitcases” for Encapsulation and Conditional Release of siRNA. J. Am. Chem. Soc. 138, 14030–14038. doi:10.1021/jacs.6b08369
Burnett, J. C., Rossi, J. J., and Tiemann, K. (2011). Current Progress of siRNA/shRNA Therapeutics in Clinical Trials. Biotechnol. J. 6, 1130–1146. doi:10.1002/biot.201100054
Büyükköroğlu, G., Şenel, B., and Yenilmez, E. (2019). Vaginal Suppositories with siRNA and Paclitaxel-Incorporated Solid Lipid Nanoparticles for Cervical Cancer: Preparation and In Vitro Evaluation. Methods Mol. Biol. 1974, 303–328. doi:10.1007/978-1-4939-9220-1_22
Calin, G. A., Dumitru, C. D., Shimizu, M., Bichi, R., Zupo, S., Noch, E., et al. (2002). Nonlinear Partial Differential Equations and Applications: Frequent Deletions and Down-Regulation of Micro- RNA Genes miR15 and miR16 at 13q14 in Chronic Lymphocytic Leukemia. Proc. Natl. Acad. Sci. 99, 15524–15529. doi:10.1073/pnas.242606799
Cao, F., Wan, C., Xie, L., Qi, H., Shen, L., Chen, S., et al. (2019). Localized RNA Interference Therapy to Eliminate Residual Lung Cancer after Incomplete Microwave Ablation. Thorac. Cancer 10, 1369–1377. doi:10.1111/1759-7714.13079
Cao, S., Sun, R., Wang, W., Meng, X., Zhang, Y., Zhang, N., et al. (2017). RNA Helicase DHX9 May Be a Therapeutic Target in Lung Cancer and Inhibited by Enoxacin. Am. J. Transl. Res. 9, 674–682.
Chakraborty, S., Kumar, A., Faheem, M. M., Katoch, A., Kumar, A., Jamwal, V. L., et al. (2019). Vimentin Activation in Early Apoptotic Cancer Cells Errands Survival Pathways during DNA Damage Inducer CPT Treatment in colon Carcinoma Model. Cell Death Dis 10, 467. doi:10.1038/s41419-019-1690-2
Chalbatani, G. M., Dana, H., Gharagozlo, E., Mahmoodzad, H., Zeinalinia, E., Rezaeian, O., et al. (2017). Microrna a New Gate in Cancer and Human Disease: A Review. J. Biol. Sci. 17, 247–254. doi:10.3923/jbs.2017.247.254
Chang, T.-C., Yu, D., Lee, Y.-S., Wentzel, E. A., Arking, D. E., West, K. M., et al. (2008). Widespread microRNA Repression by Myc Contributes to Tumorigenesis. Nat. Genet. 40, 43–50. doi:10.1038/ng.2007.30
Chen, D.-l., Chen, L.-z., Lu, Y.-x., Zhang, D.-s., Zeng, Z.-l., Pan, Z.-z., et al. (2017). Long Noncoding RNA XIST Expedites Metastasis and Modulates Epithelial-Mesenchymal Transition in Colorectal Cancer. Cel Death Dis 8, e3011. doi:10.1038/cddis.2017.421
Chen, G., Kronenberger, P., Teugels, E., Umelo, I. A., and De Grève, J. (2012). Targeting the Epidermal Growth Factor Receptor in Non-small Cell Lung Cancer Cells: The Effect of Combining RNA Interference with Tyrosine Kinase Inhibitors or Cetuximab. BMC Med. 10, 28. doi:10.1186/1741-7015-10-28
Chen, J., Wang, T., Zhou, Y.-C., Gao, F., Zhang, Z.-H., Xu, H., et al. (2014). Aquaporin 3 Promotes Epithelial-Mesenchymal Transition in Gastric Cancer. J. Exp. Clin. Cancer Res. 33, 38. doi:10.1186/1756-9966-33-38
Chen, J., Wang, Z., Xu, D., Liu, Y., and Gao, Y. (2015). Aquaporin 3 Promotes Prostate Cancer Cell Motility and Invasion via Extracellular Signal-Regulated Kinase 1/2-mediated Matrix Metalloproteinase-3 Secretion. Mol. Med. Rep. 11, 2882–2888. doi:10.3892/mmr.2014.3097
Chen, L., Yuan, D., Zhao, R., Li, H., and Zhu, J. (2010). Suppression of TSPAN1 by RNA Interference Inhibits Proliferation and Invasion of colon Cancer Cells In Vitro. Tumori J. 96, 744–750. doi:10.1177/030089161009600517
Chen, M.-B., Wu, H.-F., Zhan, Y., Fu, X.-L., Wang, A.-K., Wang, L.-S., et al. (2014). Prognostic Value of TROP2 Expression in Patients with Gallbladder Cancer. Tumor Biol. 35, 11565–11569. doi:10.1007/s13277-014-2469-9
Chen, S.-x., Yin, J.-f., Lin, B.-c., Su, H.-f., Zheng, Z., Xie, C.-y., et al. (2016). Upregulated Expression of Long Noncoding RNA SNHG15 Promotes Cell Proliferation and Invasion through Regulates MMP2/MMP9 in Patients with GC. Tumor Biol. 37, 6801–6812. doi:10.1007/s13277-015-4404-0
Chen, W., Böcker, W., Brosius, J., and Tiedge, H. (1997). Expression of Neural BC200 RNA in Human Tumours. J. Pathol. 183, 345–351. doi:10.1002/(sici)1096-9896(199711)183:3<345::aid-path930>3.0.co;2-8
Chen, Y.-M., Liu, Y., Wei, H.-Y., Lv, K.-Z., and Fu, P.-F. (2016). Large Intergenic Non-coding RNA-ROR Reverses Gemcitabine-Induced Autophagy and Apoptosis in Breast Cancer Cells. Oncotarget 7, 59604–59617. doi:10.18632/oncotarget.10730
Chiu, Y.-L., and Rana, T. M. (2003). siRNA Function in RNAi: A Chemical Modification Analysis. RNA 9, 1034–1048. doi:10.1261/rna.5103703
Choi, Y.-S., Lee, J. Y., Suh, J. S., Kwon, Y.-M., Lee, S.-J., Chung, J.-K., et al. (2010). The Systemic Delivery of siRNAs by a Cell Penetrating Peptide, Low Molecular Weight Protamine, Biomaterials, 31, 1429–1443. doi:10.1016/j.biomaterials.2009.11.001
Christopher, A. F., Kaur, R. P., Kaur, G., Kaur, A., Gupta, V., and Bansal, P. (2016). MicroRNA Therapeutics: Discovering Novel Targets and Developing Specific Therapy. Perspect. Clin. Res. 7, 68–74. doi:10.4103/2229-3485.179431
Chung, J. Y., Ul Ain, Q., Lee, H. L., Kim, S.-M., and Kim, Y.-H. (2017). Enhanced Systemic Anti-angiogenic siVEGF Delivery Using PEGylated Oligo-D-Arginine. Mol. Pharmaceutics 14, 3059–3068. doi:10.1021/acs.molpharmaceut.7b00282
Cong, L., Ran, F. A., Cox, D., Lin, S., Barretto, R., Habib, N., et al. (2013). Multiplex Genome Engineering Using CRISPR/Cas Systems, Science, 339, 819–823. doi:10.1126/science.1231143
Cortez, D., Guntuku, S., Qin, J., and Elledge, S. J. (2001). ATR and ATRIP: Partners in Checkpoint Signaling. Science 294, 1713–1716. doi:10.1126/science.1065521
Crnkovic-Mertens, I., Hoppe-Seyler, F., and Butz, K. (2003). Induction of Apoptosis in Tumor Cells by siRNA-Mediated Silencing of the livin/ML-IAP/KIAP Gene. Oncogene 22, 8330–8336. doi:10.1038/sj.onc.1206973
Crombez, L., Morris, M., Deshayes, S., Heitz, F., and Divita, G. (2008). Peptide-Based Nanoparticle for Ex Vivo and In Vivo Dug Delivery. Cpd 14, 3656–3665. doi:10.2174/138161208786898842
Cubas, R., Li, M., Chen, C., and Yao, Q. (2009). Trop2: A Possible Therapeutic Target for Late Stage Epithelial Carcinomas. Biochim. Biophys. Acta (Bba) - Rev. Cancer 1796, 309–314. doi:10.1016/j.bbcan.2009.08.001
Dai, S., Wei, D., Wu, Z., Zhou, X., Wei, X., Huang, H., et al. (2008). Phase I Clinical Trial of Autologous Ascites-Derived Exosomes Combined with GM-CSF for Colorectal Cancer. Mol. Ther. 16, 782–790. doi:10.1038/mt.2008.1
Dash, A. B., Orrico, F. C., and Ness, S. A. (1996). The EVES Motif Mediates Both Intermolecular and Intramolecular Regulation of C-Myb. Genes Dev. 10, 1858–1869. doi:10.1101/gad.10.15.1858
Davis, M. E. (2009). The First Targeted Delivery of siRNA in Humans via a Self-Assembling, Cyclodextrin Polymer-Based Nanoparticle: From Concept to Clinic. Mol. Pharmaceutics 6, 659–668. doi:10.1021/mp900015y
Davis, M. E., Zuckerman, J. E., Choi, C. H. J., Seligson, D., Tolcher, A., Alabi, C. A., et al. (2010). Evidence of RNAi in Humans from Systemically Administered siRNA via Targeted Nanoparticles. Nature 464, 1067–1070. doi:10.1038/nature08956
DeVita, V. T., and Chu, E. (2008). A History of Cancer Chemotherapy. Cancer Res. 68, 8643–8653. doi:10.1158/0008-5472.CAN-07-6611
Dong, L., Li, G., Li, Y., and Zhu, Z. (2019). Upregulation of Long Noncoding RNA GAS5 Inhibits Lung Cancer Cell Proliferation and Metastasis via miR-205/PTEN axis. Med. Sci. Monit. 25, 2311–2319. doi:10.12659/MSM.912581
Dong, Y. Z., Meng, X. M., and Li, G. S. (2018). Long Non-coding RNA SNHG15 Indicates Poor Prognosis of Non-small Cell Lung Cancer and Promotes Cell Proliferation and Invasion. Eur. Rev. Med. Pharmacol. Sci. 22, 2671–2679. doi:10.26355/eurrev_201805_14963
Du, W. W., Yang, W., Liu, E., Yang, Z., Dhaliwal, P., and Yang, B. B. (2016). Foxo3 Circular RNA Retards Cell Cycle Progression via Forming Ternary Complexes with P21 and CDK2. Nucleic Acids Res. 44, 2846–2858. doi:10.1093/nar/gkw027
Egorova, A. A., Maretina, M. A., and Kiselev, A. V. (2019). VEGFA Gene Silencing in CXCR4-Expressing Cells via siRNA Delivery by Means of Targeted Peptide Carrier. Methods Mol. Biol. 1974, 57–68. doi:10.1007/978-1-4939-9220-1_5
Egorova, A., Shubina, A., Sokolov, D., Selkov, S., Baranov, V., and Kiselev, A. (2016). CXCR4-targeted Modular Peptide Carriers for Efficient Anti-VEGF siRNA Delivery. Int. J. Pharmaceutics 515, 431–440. doi:10.1016/j.ijpharm.2016.10.049
Eis, P. S., Tam, W., Sun, L., Chadburn, A., Li, Z., Gomez, M. F., et al. (2005). Accumulation of miR-155 and BIC RNA in Human B Cell Lymphomas. Proc. Natl. Acad. Sci. 102, 3627–3632. doi:10.1073/pnas.0500613102
Elbashir, S. M., Harborth, J., Weber, K., and Tuschl, T. (2002). Analysis of Gene Function in Somatic Mammalian Cells Using Small Interfering RNAs. Methods 26, 199–213. doi:10.1016/S1046-2023(02)00023-3
Elmén, J., Lindow, M., Schütz, S., Lawrence, M., Petri, A., Obad, S., et al. (2008). LNA-mediated microRNA Silencing in Non-human Primates. Nature 452, 896–899. doi:10.1038/nature06783
Fan, J.-t., Si, X.-h., Liao, Y., and Shen, P. (2013). The Diagnostic and Prognostic Value of Serum YKL-40 in Endometrial Cancer. Arch. Gynecol. Obstet. 287, 111–115. doi:10.1007/s00404-012-2546-5
Fan, Y.-H., Fang, H., Ji, C.-x., Xie, H., Xiao, B., and Zhu, X.-G. (2017). Long Noncoding RNA CCAT2 Can Predict Metastasis and Poor Prognosis: A Meta-Analysis. Clinica Chim. Acta 466, 120–126. doi:10.1016/j.cca.2017.01.016
Fang, J., Sun, C.-C., and Gong, C. (2016). Long Noncoding RNA XIST Acts as an Oncogene in Non-small Cell Lung Cancer by Epigenetically Repressing KLF2 Expression. Biochem. Biophysical Res. Commun. 478, 811–817. doi:10.1016/j.bbrc.2016.08.030
Feldmann, D. P., Jones, S., Douglas, K., Shields, A. F., and Merkel, O. M. (2019). Microfluidic Assembly of siRNA-Loaded Micelleplexes for Tumor Targeting in an Orthotopic Model of Ovarian Cancer. Methods Mol. Biol. 1974, 355–369. doi:10.1007/978-1-4939-9220-1_24
Fidaleo, M., Svetoni, F., Volpe, E., Miñana, B., Caporossi, D., and Paronetto, M. P. (2015). Genotoxic Stress Inhibits ewing Sarcoma Cell Growth by Modulating Alternative Pre-mRNA Processing of the RNA Helicase DHX9. Oncotarget 6, 31740–31757. doi:10.18632/oncotarget.5033
Fire, A., Xu, S., Montgomery, M. K., Kostas, S. A., Driver, S. E., and Mello, C. C. (1998). Potent and Specific Genetic Interference by Double-Stranded RNA in caenorhabditis Elegans. Nature 391, 806–811. doi:10.1038/35888
Francescone, R. A., Scully, S., Faibish, M., Taylor, S. L., Oh, D., Moral, L., et al. (2011). Role of YKL-40 in the Angiogenesis, Radioresistance, and Progression of Glioblastoma. J. Biol. Chem. 286, 15332–15343. doi:10.1074/jbc.M110.212514
Fu, M., Huang, Z., Zang, X., Pan, L., Liang, W., Chen, J., et al. (2018). Long Noncoding RNA LINC 00978 Promotes Cancer Growth and Acts as a Diagnostic Biomarker in Gastric Cancer. Cell Prolif 51, e12425. doi:10.1111/cpr.12425
Gegechkori, N., Haines, L., and Lin, J. J. (2017). Long-Term and Latent Side Effects of Specific Cancer Types. Med. Clin. North America 101, 1053–1073. doi:10.1016/j.mcna.2017.06.003
Gibb, E. A., Becker-Santos, D. D., Enfield, K. S. S., Guillaud, M., Niekerk, D. v., Matisic, J. P., et al. (2012). Aberrant Expression of Long Noncoding RNAs in Cervical Intraepithelial Neoplasia. Int. J. Gynecol. Cancer 22, 1557–1563. doi:10.1097/IGC.0b013e318272f2c9
Golombek, S. K., May, J.-N., Theek, B., Appold, L., Drude, N., Kiessling, F., et al. (2018). Tumor Targeting via EPR: Strategies to Enhance Patient Responses. Adv. Drug Deliv. Rev. 130, 17–38. doi:10.1016/j.addr.2018.07.007
Gossai, N. P., Naumann, J. A., Li, N., Zamora, E. A., Gordon, D. J., Piccirilli, J. A., et al. (2019). Drug Conjugated Nanoparticles Activated by Cancer Cell Specific mRNA. Oncotarget 7, 38243–38256. doi:10.18632/oncotarget.9430
Govindaraju, S., and Lee, B. S. (2013). Adaptive and Maladaptive Expression of the Mrna Regulatory Protein HuR. Wjbc 4, 111. doi:10.4331/wjbc.v4.i4.111
Gragoudas, E. S. (2004). VEGF Inhibition Study in Ocular Neovascularization-1 (VISION=1): Efficacy Results from Phase II/III MacugenTM (Pegaptanib Sodium) Clinical Trials. Iovs 45, 2364.
Gu, J., Li, Y., Fan, L., Zhao, Q., Tan, B., Hua, K., et al. (2017). Identification of Aberrantly Expressed Long Non-coding RNAs in Stomach Adenocarcinoma. Oncotarget 8, 49201–49216. doi:10.18632/oncotarget.17329
Gu, Z., Thomas, G., Yamashiro, J., Shintaku, I. P., Dorey, F., Raitano, A., et al. (2000). Prostate Stem Cell Antigen (PSCA) Expression Increases with High gleason Score, Advanced Stage and Bone Metastasis in Prostate Cancer. Oncogene 19, 1288–1296. doi:10.1038/sj.onc.1203426
Guo, L., Peng, Y., Meng, Y., Liu, Y., Yang, S., Jin, H., et al. (2017). Expression Profiles Analysis Reveals an Integrated miRNA-lncRNA Signature to Predict Survival in Ovarian Cancer Patients with Wild-type BRCA1/2. Oncotarget 8, 68483–68492. doi:10.18632/oncotarget.19590
Guo, L., Zhou, Y., Chen, Y., Sun, H., Wang, Y., and Qu, Y. (2018). LncRNA ASAP1-IT1 Positively Modulates the Development of Cholangiocarcinoma via Hedgehog Signaling Pathway. Biomed. Pharmacother. 103, 167–173. doi:10.1016/j.biopha.2018.04.015
Guo, W., Wang, Q., Zhan, Y., Chen, X., Yu, Q., Zhang, J., et al. (2016). Transcriptome Sequencing Uncovers a Three-Long Noncoding RNA Signature in Predicting Breast Cancer Survival. Sci. Rep. 6, 27931. doi:10.1038/srep27931
Hämmerle, M., Gutschner, T., Uckelmann, H., Ozgur, S., Fiskin, E., Gross, M., et al. (2013). Posttranscriptional Destabilization of the Liver-specific Long Noncoding RNA HULC by the IGF2 mRNA-Binding Protein 1 (IGF2BP1). Hepatology 58, 1703–1712. doi:10.1002/hep.26537
Han, L., Zhang, H.-C., Li, L., Li, C.-X., Di, X., and Qu, X. (2018). Downregulation of Long Noncoding RNA HOTAIR andEZH2Induces Apoptosis and Inhibits Proliferation, Invasion, and Migration of Human Breast Cancer Cells. Cancer Biother. Radiopharm. 33, 241–251. doi:10.1089/cbr.2017.2432
Hao, Y., Crenshaw, T., Moulton, T., Newcomb, E., and Tycko, B. (1993). Tumour-suppressor Activity of H19 RNA. Nature 365, 764–767. doi:10.1038/365764a0
Hatakeyama, H., Akita, H., and Harashima, H. (2011). A Multifunctional Envelope Type Nano Device (MEND) for Gene Delivery to Tumours Based on the EPR Effect: A Strategy for Overcoming the PEG Dilemma. Adv. Drug Deliv. Rev. 63, 152–160. doi:10.1016/j.addr.2010.09.001
Hazan-Halevy, I., Rosenblum, D., Ramishetti, S., and Peer, D. (2019). Systemic Modulation of Lymphocyte Subsets Using siRNAs Delivered via Targeted Lipid Nanoparticles BT - RNA Interference and Cancer Therapy, Methods Mol. Biol., 1974, 151–159. doi:10.1007/978-1-4939-9220-1_11
He, J., Chen, Y., Cai, L., Li, Z., and Guo, X. (2018). UBAP2L Silencing Inhibits Cell Proliferation and G2/M Phase Transition in Breast Cancer. Breast Cancer 25, 224–232. doi:10.1007/s12282-017-0820-x
Heinrich, M.-C., Göbel, C., Kluth, M., Bernreuther, C., Sauer, C., Schroeder, C., et al. (2018). PSCA Expression Is Associated with Favorable Tumor Features and Reduced PSA Recurrence in Operated Prostate Cancer. BMC Cancer 18, 612. doi:10.1186/s12885-018-4547-7
Hill, C., and Carolina, N. (2006). Targeted Delivery of Antisense Oligodeoxynucleotide and Small Interference RNA into Lung Cancer Cells. Mol. Pharm. 3, 579–588. doi:10.1021/mp060039w
Hornung, V., Guenthner-Biller, M., Bourquin, C., Ablasser, A., Schlee, M., Uematsu, S., et al. (2005). Sequence-specific Potent Induction of IFN-α by Short Interfering RNA in Plasmacytoid Dendritic Cells through TLR7. Nat. Med. 11, 263–270. doi:10.1038/nm1191
Hou, S.-Y., Li, Y.-P., Wang, J.-H., Yang, S.-L., Wang, Y., Wang, Y., et al. (2016). Aquaporin-3 Inhibition Reduces the Growth of NSCLC Cells Induced by Hypoxia. Cell. Physiol. Biochem. 38, 129–140. doi:10.1159/000438615
Hu, J., Li, J., Yue, X., Wang, J.-c., Wang, J.-f., Liu, J.-z., et al. (2017). Targeting BCRP/ABCG2 by RNA Interference Enhances the Chemotherapy Sensitivity of Human colon Cancer Side Population Cells. J. Huazhong Univ. Sci. Technol. [Med. Sci. 37, 231–236. doi:10.1007/s11596-017-1720-1
Hu, J., Verkman, A. S., Hu, J., and Verkman, A. S. (2006). Increased Migration and Metastatic Potential of Tumor Cells Expressing Aquaporin Water Channels. FASEB j. 20, 1892–1894. doi:10.1096/fj.06-5930fje
Huang, Z., Zhou, J.-K., Peng, Y., He, W., and Huang, C. (2020). The Role of Long Noncoding RNAs in Hepatocellular Carcinoma. Mol. Cancer 19, 77. doi:10.1186/s12943-020-01188-4
Huschka, R., Barhoumi, A., Liu, Q., Roth, J. A., Ji, L., and Halas, N. J. (2012). Gene Silencing by Gold Nanoshell-Mediated Delivery and Laser-Triggered Release of Antisense Oligonucleotide and siRNA. ACS Nano 6, 7681–7691. doi:10.1021/nn301135w
Hutvágner, G., and Zamore, P. D. (2002). A microRNA in a Multiple-Turnover RNAi Enzyme Complex. Science 297, 2056–2060. doi:10.1126/science.1073827
Jackson, A. L., Bartz, S. R., Schelter, J., Kobayashi, S. V., Burchard, J., Mao, M., et al. (2003). Expression Profiling Reveals Off-Target Gene Regulation by RNAi. Nat. Biotechnol. 21, 635–637. doi:10.1038/nbt831
Ji, P., Diederichs, S., Wang, W., Böing, S., Metzger, R., Schneider, P. M., et al. (2003). MALAT-1, a Novel Noncoding RNA, and Thymosin β4 Predict Metastasis and Survival in Early-Stage Non-small Cell Lung Cancer. Oncogene 22 (39), 8031–8041. doi:10.1038/sj.onc.1206928
Jiang, L., Lai, Y.-K., Zhang, J., Wang, H., Lin, M. C. M., He, M.-l., et al. (2011). Targeting S100p Inhibits colon Cancer Growth and Metastasis by Lentivirus-Mediated RNA Interference and Proteomic Analysis. Mol. Med. 17, 709–716. doi:10.2119/molmed.2011.00008
Jiang, Y., Wang, D., Ren, H., Shi, Y., and Gao, Y. (2019). MiR-145-targeted HBXIP Modulates Human Breast Cancer Cell Proliferation. Thorac. Cancer 10, 71–77. doi:10.1111/1759-7714.12903
Jivrajani, M., and Nivsarkar, M. (2019). Minicell-based Targeted Delivery of shRNA to Cancer Cells: An Experimental Protocol. Methods Mol. Biol. 1974, 111–139. doi:10.1007/978-1-4939-9220-1_9
Juliano, R. L. (2016). The Delivery of Therapeutic Oligonucleotides. Nucleic Acids Res. 44, 6518–6548. doi:10.1093/nar/gkw236
Kanazawa, T., Sugawara, K., Tanaka, K., Horiuchi, S., Takashima, Y., and Okada, H. (2012). Suppression of Tumor Growth by Systemic Delivery of Anti-VEGF siRNA with Cell-Penetrating Peptide-Modified MPEG-PCL Nanomicelles. Eur. J. Pharmaceutics Biopharmaceutics 81, 470–477. doi:10.1016/j.ejpb.2012.04.021
Kang, L., Rosenkrans, Z. T., and Cai, W. (2019). 64Cu-Labeled Aptamers for Tumor-Targeted Radionuclide Delivery BT - RNA Interference and Cancer Therapy, Methods Mol. Biol., 1974, 223–231. doi:10.1007/978-1-4939-9220-1_17
Karagiannis, T. C., and El-Osta, A. (2005). RNA Interference and Potential Therapeutic Applications of Short Interfering RNAs. Cancer Gene Ther. 12, 787–795. doi:10.1038/sj.cgt.7700857
Karikó, K., Bhuyan, P., Capodici, J., Ni, H., Lubinski, J., Friedman, H., et al. (2004). Exogenous siRNA Mediates Sequence-independent Gene Suppression by Signaling through Toll-like Receptor 3. Cells Tissues Organs 177, 132–138. doi:10.1159/000079987
Katoch, A., Nayak, D., Faheem, M. M., Kumar, A., Sahu, P. K., Gupta, A. P., et al. (2021). Natural Podophyllotoxin Analog 4DPG Attenuates EMT and Colorectal Cancer Progression via Activation of Checkpoint Kinase 2. Cell Death Discov. 7, 25. doi:10.1038/s41420-021-00405-3
Ke, D., Li, H., Zhang, Y., An, Y., Fu, H., Fang, X., et al. (2017). The Combination of Circulating Long Noncoding RNAs AK001058, INHBA-AS1, MIR4435-2HG, and CEBPA-AS1 Fragments in Plasma Serve as Diagnostic Markers for Gastric Cancer. Oncotarget 8, 21516–21525. doi:10.18632/oncotarget.15628
Kim, S. H., Bianco, N. R., Shufesky, W. J., Morelli, A. E., and Robbins, P. D. (2007). MHC Class II+ Exosomes in Plasma Suppress Inflammation in an Antigen-specific and Fas Ligand/Fas-dependent Manner. J. Immunol. 179, 2235–2241. doi:10.4049/jimmunol.179.4.2235
Klein, U., Lia, M., Crespo, M., Siegel, R., Shen, Q., Mo, T., et al. (2010). The DLEU2/miR-15a/16-1 Cluster Controls B Cell Proliferation and its Deletion Leads to Chronic Lymphocytic Leukemia. Cancer Cell 17, 28–40. doi:10.1016/j.ccr.2009.11.019
Koren, G., Carey, N., Maxwell, C., Nulman, I., Gagnon, R., and Senikas, V. (2013). Cancer Chemotherapy and Pregnancy. J. Obstet. Gynaecol. Can. 35 (3), 263–278. doi:10.1016/S1701-2163(15)30999-3
Kulhari, H., Jangid, A. K., and Adams, D. J. (2019). Monoclonal Antibody-Conjugated Dendritic Nanostructures for siRNA Delivery. Methods Mol. Biol. 1974, 195–201. doi:10.1007/978-1-4939-9220-1_14
Kumar, L. D., Golani, A., and Kumar, L. D. (2020). EMT in Breast Cancer Metastasis an Interplay of microRNAs Signaling Pathways and Circulating Tumor Cells. Front. Biosci. 25, 979–1010. doi:10.2741/4844
La-Beck, N. M., and Gabizon, A. A. (2017). Nanoparticle Interactions with the Immune System: Clinical Implications for Liposome-Based Cancer Chemotherapy. Front. Immunol. 8, 416. doi:10.3389/fimmu.2017.00416
Lai, M.-c., Yang, Z., Zhou, L., Zhu, Q.-q., Xie, H.-y., Zhang, F., et al. (2012). Long Non-coding RNA MALAT-1 Overexpression Predicts Tumor Recurrence of Hepatocellular Carcinoma after Liver Transplantation. Med. Oncol. 29, 1810–1816. doi:10.1007/s12032-011-0004-z
Lakka, S. S., Gondi, C. S., Yanamandra, N., Olivero, W. C., Dinh, D. H., Gujrati, M., et al. (2004). Inhibition of Cathepsin B and MMP-9 Gene Expression in Glioblastoma Cell Line via RNA Interference Reduces Tumor Cell Invasion, Tumor Growth and Angiogenesis. Oncogene 23, 4681–4689. doi:10.1038/sj.onc.1207616
Landen, C. N., Chavez-Reyes, A., Bucana, C., Schmandt, R., Deavers, M. T., Lopez-Berestein, G., et al. (2005). Therapeutic EphA2 Gene Targeting In Vivo Using Neutral Liposomal Small Interfering RNA Delivery. Cancer Res. 65, 6910–6918. doi:10.1158/0008-5472.CAN-05-0530
Layzer, J. M., McCaffrey, A. P., Tanner, A. K., Huang, Z., Kay, M. A., and Sullenger, B. A. (2004). In Vivo activity of Nuclease-Resistant siRNAs. RNA 10, 766–771. doi:10.1261/rna.5239604
Leconet, W., Liu, H., Guo, M., Le Lamer-Déchamps, S., Molinier, C., Kim, S., et al. (2018). Anti-PSMA/CD3 Bispecific Antibody Delivery and Antitumor Activity Using a Polymeric Depot Formulation. Mol. Cancer Ther. 17, 1927–1940. doi:10.1158/1535-7163.MCT-17-1138
Lee, C. G., Hartl, D., Lee, G. R., Koller, B., Matsuura, H., Da Silva, C. A., et al. (2009). Role of Breast Regression Protein 39 (BRP-39)/chitinase 3-like-1 in Th2 and IL-13-induced Tissue Responses and Apoptosis. J. Exp. Med. 206, 1149–1166. doi:10.1084/jem.20081271
Lee, D.-J., and Wagner, E. (2019). Combinatorial siRNA Polyplexes for Receptor Targeting BT - RNA Interference and Cancer Therapy, Methods Mol. Biol., 1974, 83–98. doi:10.1007/978-1-4939-9220-1_7
Lee, G. L., Dobi, A., and Srivastava, S. (2011). Diagnostic Performance of the PCA3 Urine Test. Nat. Rev. Urol. 8, 123–124. doi:10.1038/nrurol.2011.10
Lee, R. C., Feinbaum, R. L., and Ambros, V. (1993). The C. elegans Heterochronic Gene Lin-4 Encodes Small RNAs with Antisense Complementarity to Lin-14. Cell 75, 843–854. doi:10.1016/0092-8674(93)90529-Y
Lee, T., and Pelletier, J. (2016). The Biology of DHX9 and its Potential as a Therapeutic Target. Oncotarget 7, 42716–42739. doi:10.18632/oncotarget.8446
Lee, Y. W., Hwang, Y. E., Lee, J. Y., Sohn, J.-H., Sung, B. H., and Kim, S. C. (2018). VEGF siRNA Delivery by a Cancer-specific Cell-Penetrating Peptide. J. Microbiol. Biotechnol. 28, 367–374. doi:10.4014/jmb.1711.11025
Li, C., Wan, L., Liu, Z., Xu, G., Wang, S., Su, Z., et al. (2018). Long Non-coding RNA XIST Promotes TGF-β-Induced Epithelial-Mesenchymal Transition by Regulating miR-367/141-ZEB2 axis in Non-small-cell Lung Cancer. Cancer Lett. 418, 185–195. doi:10.1016/j.canlet.2018.01.036
Li, J., Chen, Y., Chen, Z., He, A., Xie, H., Zhang, Q., et al. (2017). SPRY4-IT1: A Novel Oncogenic Long Non-coding Rna in Human Cancers. Tumour Biol. 39, 101042831771140. doi:10.1177/1010428317711406
Li, J., Pei, H., Zhu, B., Liang, L., Wei, M., He, Y., et al. (2011). Self-Assembled Multivalent DNA Nanostructures for Noninvasive Intracellular Delivery of Immunostimulatory CpG Oligonucleotides. ACS Nano 5, 8783–8789. doi:10.1021/nn202774x
Li, L., Fan, J., Li, D., Liu, Y., Shrestha, P., Zhong, C., et al. (2018). Influence of YKL-40 G-ene RNA I-nterference on the B-iological B-ehaviors of E-ndometrial C-ancer HEC-1A C-ells. Oncol. Lett. 16, 1777–1784. doi:10.3892/ol.2018.8814
Li, Q., Liu, G., Shao, D., Wang, J., Yuan, H., Chen, T., et al. (2015). Mucin1 Mediates Autocrine Transforming Growth Factor Beta Signaling through Activating the C-Jun N-Terminal Kinase/activator Protein 1 Pathway in Human Hepatocellular Carcinoma Cells. Int. J. Biochem. Cel Biol. 59, 116–125. doi:10.1016/j.biocel.2014.11.012
Li, Q., Wang, F., Liu, G., Yuan, H., Chen, T., Wang, J., et al. (2014). Impact of Mucin1 Knockdown on the Phenotypic Characteristics of the Human Hepatocellular Carcinoma Cell Line SMMC-7721. Oncol. Rep. 31, 2811–2819. doi:10.3892/or.2014.3136
Li, W., Liang, J., Zhang, Z., Lou, H., Zhao, L., Xu, Y., et al. (2017). MicroRNA-329-3p Targets MAPK1 to Suppress Cell Proliferation, Migration and Invasion in Cervical Cancer. Oncol. Rep. 37, 2743–2750. doi:10.3892/or.2017.5555
Li, X., Teng, S., Zhang, Y., Zhang, W., Zhang, X., Xu, K., et al. (2017). TROP2 Promotes Proliferation, Migration and Metastasis of Gallbladder Cancer Cells by Regulating PI3K/AKT Pathway and Inducing EMT. Oncotarget 8, 47052–47063. doi:10.18632/oncotarget.16789
Li, Y., Wan, Y. P., and Bai, Y. (2018). Correlation between Long Strand Non-coding RNA GASS Expression and Prognosis of Cervical Cancer Patients. Eur. Rev. Med. Pharmacol. Sci. 22, 943–949. doi:10.26355/eurrev_201802_14375
Lieberman, J. (2018). Tapping the RNA World for Therapeutics. Nat. Struct. Mol. Biol. 25, 357–364. doi:10.1038/s41594-018-0054-4
Lim, K. J., Sung, B. H., Shin, J. R., Lee, Y. W., Kim, D. J., Yang, K. S., et al. (2013). A Cancer Specific Cell-Penetrating Peptide, BR2, for the Efficient Delivery of an scFv into Cancer Cells. PLoS One 8, e66084. doi:10.1371/journal.pone.0066084
Lima, R. T., Martins, L. M., Guimarães, J. E., Sambade, C., and Vasconcelos, M. H. (2004). Specific Downregulation of Bcl-2 and xIAP by RNAi Enhances the Effects of Chemotherapeutic Agents in MCF-7 Human Breast Cancer Cells. Cancer Gene Ther. 11, 309–316. doi:10.1038/sj.cgt.7700706
Lin, C., Zhang, Y., Chen, Y., Bai, Y., and Zhang, Y. (2019). Long Noncoding RNA LINC01234 Promotes Serine Hydroxymethyltransferase 2 Expression and Proliferation by Competitively Binding miR-642a-5p in colon Cancer. Cel Death Dis 10, 137. doi:10.1038/s41419-019-1352-4
Lin, S., Liu, J., Jiang, W., Wang, P., Sun, C., Wang, X., et al. (2019). METTL3 Promotes the Proliferation and Mobility of Gastric Cancer Cells. Open Med. 14, 25–31. doi:10.1515/med-2019-0005
Ling, H., Girnita, L., Buda, O., and Calin, G. A. (2017). Non-coding RNAs: The Cancer Genome Dark Matter that Matters!. Clin. Chem. Lab. Med. 55, 705–714. doi:10.1515/cclm-2016-0740
Liu, L. T., Chang, H. C., Chiang, L. C., and Hung, W. C. (2003). Histone Deacetylase Inhibitor Up-Regulates RECK to Inhibit MMP-2 Activation and Cancer Cell Invasion. Cancer Res. 63, 3069–3072.
Liu, Z., Sun, M., Lu, K., Liu, J., Zhang, M., Wu, W., et al. (2013). The Long Noncoding RNA HOTAIR Contributes to Cisplatin Resistance of Human Lung Adenocarcinoma Cells via Downregualtion of p21WAF1/CIP1 Expression. PLoS One 8, e77293. doi:10.1371/journal.pone.0077293
Loprinzi, C. L., Lacchetti, C., Bleeker, J., Cavaletti, G., Chauhan, C., Hertz, D. L., et al. (2020). Prevention and Management of Chemotherapy-Induced Peripheral Neuropathy in Survivors of Adult Cancers: ASCO Guideline Update. Jco 38, 3325–3348. doi:10.1200/JCO.20.01399
Lorenzer, C., Dirin, M., Winkler, A.-M., Baumann, V., and Winkler, J. (2015). Going beyond the Liver: Progress and Challenges of Targeted Delivery of siRNA Therapeutics. J. Controlled Release 203, 1–15. doi:10.1016/j.jconrel.2015.02.003
Lu, P.-W., Li, L., Wang, F., and Gu, Y.-T. (2019). Inhibitory Role of Large Intergenic Noncoding RNA-ROR on Tamoxifen Resistance in the Endocrine Therapy of Breast Cancer by Regulating the PI3K/Akt/mTOR Signaling Pathway. J. Cel. Physiol. 234, 1904–1912. doi:10.1002/jcp.27066
Mahmoodi Chalbatani, G., Dana, H., Gharagouzloo, E., Grijalvo, S., Eritja, R., Logsdon, C. D., et al. (2019). Small Interfering RNAs (siRNAs) in Cancer Therapy: a Nano-Based Approach. Ijn 14, 3111–3128. doi:10.2147/IJN.S200253
Mali, P., Yang, L., Esvelt, K. M., Aach, J., Guell, M., DiCarlo, J. E., et al. (2013). RNA-guided Human Genome Engineering via Cas9. Science 339, 823–826. doi:10.1126/science.1232033
Mansoori, B., Sandoghchian Shotorbani, S., and Baradaran, B. (2014). RNA Interference and its Role in Cancer Therapy. Adv. Pharm. Bull. 4, 313–321. doi:10.5681/apb.2014.046
McCaffrey, A. P., Nakai, H., Pandey, K., Huang, Z., Salazar, F. H., Xu, H., et al. (2003). Inhibition of Hepatitis B Virus in Mice by RNA Interference. Nat. Biotechnol. 21, 639–644. doi:10.1038/nbt824
McCarroll, J. A., Sharbeen, G., Kavallaris, M., and Phillips, P. A. (2019). The Use of star Polymer Nanoparticles for the Delivery of siRNA to Mouse Orthotopic Pancreatic Tumor Models. Methods Mol. Biol. 1974, 329–353. doi:10.1007/978-1-4939-9220-1_23
Memczak, S., Jens, M., Elefsinioti, A., Torti, F., Krueger, J., Rybak, A., et al. (2013). Circular RNAs Are a Large Class of Animal RNAs with Regulatory Potency. Nature 495, 333–338. doi:10.1038/nature11928
Metzler, M., Wilda, M., Busch, K., Viehmann, S., and Borkhardt, A. (2004). High Expression of Precursor MicroRNA-155/BIC RNA in Children with Burkitt Lymphoma. Genes Chromosom. Cancer 39, 167–169. doi:10.1002/gcc.10316
Mi, J., Ray, P., Liu, J., Kuan, C.-T., Xu, J., Hsu, D., et al. (2016). In Vivo Selection against Human Colorectal Cancer Xenografts Identifies an Aptamer that Targets RNA Helicase Protein DHX9. Mol. Ther. - Nucleic Acids 5, e315. doi:10.1038/mtna.2016.27
Ming, X., Carver, K., and Wu, L. (2013). Albumin-based Nanoconjugates for Targeted Delivery of Therapeutic Oligonucleotides. Biomaterials 34, 7939–7949. doi:10.1016/j.biomaterials.2013.06.066
Mirkin, C. A., Meade, T. J., Petrosko, S. H., and Stegh, A. H. (2015). Nanotechnology-based Precision Tools for the Detection and Treatment of Cancer. Springer, 166.
Morales, J. C., Xia, Z., Lu, T., Aldrich, M. B., Wang, B., Rosales, C., et al. (2003). Role for the BRCA1 C-Terminal Repeats (BRCT) Protein 53BP1 in Maintaining Genomic Stability. J. Biol. Chem. 278, 14971–14977. doi:10.1074/jbc.M212484200
Morrissey, D. V., Lockridge, J. A., Shaw, L., Blanchard, K., Jensen, K., Breen, W., et al. (2005). Potent and Persistent In Vivo Anti-HBV Activity of Chemically Modified siRNAs. Nat. Biotechnol. 23, 1002–1007. doi:10.1038/nbt1122
Musavi Shenas, S. M. H., Mansoori, B., Mohammadi, A., Salehi, S., Kaffash, B., Talebi, B., et al. (2017). SiRNA-mediated Silencing of Snail-1 Induces Apoptosis and Alters Micro RNA Expression in Human Urinary Bladder Cancer Cell Line. Artif. Cell Nanomedicine, Biotechnol. 45, 969–974. doi:10.1080/21691401.2016.1198361
Nagal, A., and Singlab, R. (2013). Nanoparticles in Different Delivery Systems: A Brief Review. Indo Glob. J. Pharm. Sci. 3 (2), 96–106.
Navya, P. N., Kaphle, A., Srinivas, S. P., Bhargava, S. K., Rotello, V. M., and Daima, H. K. (2019). Current Trends and Challenges in Cancer Management and Therapy Using Designer Nanomaterials. Nano Convergence 6, 23. doi:10.1186/s40580-019-0193-2
Niedzwiedz, C. L., Knifton, L., Robb, K. A., Katikireddi, S. V., and Smith, D. J. (2019). Depression and Anxiety Among People Living with and beyond Cancer: A Growing Clinical and Research Priority. BMC Cancer 19 (1), 943. doi:10.1186/s12885-019-6181-4
Nigro, J. M., Cho, K. R., Fearon, E. R., Kern, S. E., Ruppert, J. M., Oliner, J. D., et al. (1991). Scrambled Exons. Cell 64, 607–613. doi:10.1016/0092-8674(91)90244-S
Niu, Q., Perruzi, C., Voskas, D., Lawler, J., Dumont, D., and Benjamin, L. E. (2004). Inhibition of Tie-2 Signaling Induces Endothelial Cell Apoptosis, Decreases Akt Signaling and Induces Endothelial Cell Expression of the Endogenous Anti-angiogenic Molecule, Thrombospondin-1. Cancer Biol. Ther. 3, 402–405. doi:10.4161/cbt.3.4.735
Nutalapati, S., and Jain, S. R. (2018). Risk of Second Malignancies in Breast Cancer Patients Who Received Chemotherapy: A SEER Analysis. Jco 36, 1565. doi:10.1200/jco.2018.36.15_suppl.1565
Oue, N., Yamamoto, Y., Oshima, T., Asai, R., Ishikawa, A., Uraoka, N., et al. (2018). Overexpression of the Transmembrane Protein IQGAP3 Is Associated with Poor Survival of Patients with Gastric Cancer. Pathobiology 85, 192–200. doi:10.1159/000481890
Ozcan, G., Ozpolat, B., Coleman, R. L., Sood, A. K., and Lopez-Berestein, G. (2015). Preclinical and Clinical Development of siRNA-Based Therapeutics. Adv. Drug Deliv. Rev. 87, 108–119. doi:10.1016/j.addr.2015.01.007
Ozpolat, B., Sood, A. K., and Lopez-Berestein, G. (2010). Nanomedicine Based Approaches for the Delivery of siRNA in Cancer. J. Intern. Med. 267, 44–53. doi:10.1111/j.1365-2796.2009.02191.x
Panyam, J., and Labhasetwar, V. (2012). Biodegradable Nanoparticles for Drug and Gene Delivery to Cells and Tissue. Adv. Drug Deliv. Rev. 64, 61–71. doi:10.1016/j.addr.2012.09.023
Parasramka, M. A., Maji, S., Matsuda, A., Yan, I. K., and Patel, T. (2016). Long Non-coding RNAs as Novel Targets for Therapy in Hepatocellular Carcinoma. Pharmacol. Ther. 161, 67–78. doi:10.1016/j.pharmthera.2016.03.004
Pearce, A., Haas, M., Viney, R., Pearson, S.-A., Haywood, P., Brown, C., et al. (2017). Incidence and Severity of Self-Reported Chemotherapy Side Effects in Routine Care: A Prospective Cohort Study. PLoS One 12, e0184360. doi:10.1371/journal.pone.0184360
Peng, S., Lu, Y., Li, P., Liu, P., Shi, X., Liu, C., et al. (2019). The Short Interference RNA (siRNA) Targeting NMUR2 Relieves Nociception in a Bone Cancer Pain Model of Rat through PKC-ERK and PI3K-AKT Pathways. Biochem. Biophysical Res. Commun. 512, 616–622. doi:10.1016/j.bbrc.2019.03.067
Petrocca, F., and Lieberman, J. (2011). Promise and challenge of RNA Interference-Based Therapy for Cancer. Jco 29, 747–754. doi:10.1200/JCO.2009.27.6287
Poorvu, P. D., Frazier, A. L., Feraco, A. M., Manley, P. E., Ginsburg, E. S., Laufer, M. R., et al. (2019). Cancer Treatment-Related Infertility: A Critical Review of the Evidence. JNCI Cancer Spectr. 3 (1), pkz008. doi:10.1093/jncics/pkz008
Qin, P., Wang, H., Zhang, F., Huang, Y., and Chen, S. (2019). Targeted Silencing of MYCL1 by RNA Interference Inhibits Migration and Invasion of MGC‐803 Gastric Cancer Cells. Cell Biochem. Funct. 37, 266–272. doi:10.1002/cbf.3395
Rahme, K., Guo, J., and Holmes, J. D. (2019). Bioconjugated Gold Nanoparticles Enhance siRNA Delivery in Prostate Cancer Cells BT - RNA Interference and Cancer Therapy. Methods Protoc. 1974, 291–301. doi:10.1007/978-1-4939-9220-1_21
Rawoof, A., Swaminathan, G., Tiwari, S., Nair, R. A., and Dinesh Kumar, L. (2020). LeukmiR: A Database for miRNAs and Their Targets in Acute Lymphoblastic Leukemia. Database 2020, 2020. doi:10.1093/database/baz151
Reiners, K. S., Dassler, J., Coch, C., and Pogge von Strandmann, E. (2014). Role of Exosomes Released by Dendritic Cells And/or by Tumor Targets: Regulation of NK Cell Plasticity. Front. Immunol. 5, 91. doi:10.3389/fimmu.2014.00091
Ren, X., Cao, D., Yang, L., Li, X., Zhang, W., Xiao, Y., et al. (2019). High Expression of Long Non-coding RNA PVT1 Predicts Metastasis in Han and Uygur Patients with Gastric Cancer in Xinjiang, China. Sci. Rep. 9, 548. doi:10.1038/s41598-018-36985-x
Rishabh, K., Khadilkar, S., Kumar, A., Kalra, I., Kumar, A. P., and Kunnumakkara, A. B. (2021). MicroRNAs as Modulators of Oral Tumorigenesis-A Focused Review. Ijms 22, 2561. doi:10.3390/ijms22052561
Rock, K. L., and Kono, H. (2008). The Inflammatory Response to Cell Death. Annu. Rev. Pathol. Mech. Dis. 3, 99–126. doi:10.1146/annurev.pathmechdis.3.121806.151456
Rosenblum, D., Joshi, N., Tao, W., Karp, J. M., and Peer, D. (2018). Progress and Challenges towards Targeted Delivery of Cancer Therapeutics. Nat. Commun. 9, 1410. doi:10.1038/s41467-018-03705-y
Rossi, A., Caro, G., Fortuna, M. C., Pigliacelli, F., D'Arino, A., and Carlesimo, M. (2020). Prevention and Treatment of Chemotherapy-Induced Alopecia. Dermatol. Pract. Concept 10 (3), e2020074. doi:10.5826/dpc.1003a74
Agarwal, S., Chakravarthi, B. V., Behring, M., Kim, H.-G., Hale, K., Alsubaie, A. M., et al. Abstract 851: MicroRNA-124 Modulated Collagen-Prolyl Hydroxylase P4HA1 Expression Regulates colon Cancer Progression, Mol. Cell Biol., 79. (2019). doi:10.1158/1538-7445.am2019-851
Sahin, U., Karikó, K., and Türeci, Ö. (2014). mRNA-based Therapeutics - Developing a New Class of Drugs. Nat. Rev. Drug Discov. 13, 759–780. doi:10.1038/nrd4278
Santer, L., Bär, C., and Thum, T. (2019). Circular RNAs: A Novel Class of Functional RNA Molecules with a Therapeutic Perspective. Mol. Ther. 27, 1350–1363. doi:10.1016/j.ymthe.2019.07.001
Saraswat, N., Chopra, A., Sood, A., Kamboj, P., and Kumar, S. (2019). A Descriptive Study to Analyze Chemotherapy-Induced Hair Loss and its Psychosocial Impact in Adults: Our Experience from a Tertiary Care Hospital. Indian Dermatol. Online J. 10, 426–430. doi:10.4103/idoj.idoj_471_18
Schalken, J. A., Hessels, D., and Verhaegh, G. (2003). New Targets for Therapy in Prostate Cancer: Differential Display Code 3 (DD3PCA3), a Highly Prostate Cancer-specific Gene. Urology 62, 34–43. doi:10.1016/S0090-4295(03)00759-3
Schultheis, B., Strumberg, D., Kuhlmann, J., Wolf, M., Link, K., Seufferlein, T., et al. (2016). A Phase Ib/IIa Study of Combination Therapy with Gemcitabine and Atu027 in Patients with Locally Advanced or Metastatic Pancreatic Adenocarcinoma. Jco 34, 385. doi:10.1200/jco.2016.34.4_suppl.385
Schultheis, B., Strumberg, D., Santel, A., Vank, C., Gebhardt, F., Keil, O., et al. (2014). First-in-human Phase I Study of the Liposomal RNA Interference Therapeutic Atu027 in Patients with Advanced Solid Tumors. Jco 32, 4141–4148. doi:10.1200/JCO.2013.55.0376
Schwanhüusser, B., Busse, D., Li, N., Dittmar, G., Schuchhardt, J., Wolf, J., et al. (2011). Global Quantification of Mammalian Gene Expression Control. Nature 473, 337–342. doi:10.1038/nature10098
Serda, R. E., Godin, B., Blanco, E., Chiappini, C., and Ferrari, M. (2011). Multi-stage Delivery Nano-Particle Systems for Therapeutic Applications. Biochim. Biophys. Acta (Bba) - Gen. Subjects 1810, 317–329. doi:10.1016/j.bbagen.2010.05.004
Sharma, A. K., Krzeminski, J., Weissig, V., Hegarty, J. P., Stewart, D. B., and Stewart, D. B. (2018). Cationic Amphiphilic Bolaamphiphile-Based Delivery of Antisense Oligonucleotides Provides a Potentially Microbiome Sparing Treatment for C. difficile. J. Antibiot. 71, 713–721. doi:10.1038/s41429-018-0056-9
Shen, F., Cai, W.-S., Feng, Z., Chen, J.-w., Feng, J.-h., Liu, Q.-c., et al. (2017). Long Non-coding RNA SPRY4-IT1 Pormotes Colorectal Cancer Metastasis by Regulate Epithelial-Mesenchymal Transition. Oncotarget 8, 14479–14486. doi:10.18632/oncotarget.10407
Shen, H., Sun, T., and Ferrari, M. (2012). Nanovector Delivery of siRNA for Cancer Therapy. Cancer Gene Ther. 19, 367–373. doi:10.1038/cgt.2012.22
Shi, Y. F., Tian, Z., Zhang, Y., Shen, H. B., and Jia, N. Q. (2011). Functionalized Halloysite Nanotube-Based Carrier for Intracellular Delivery of Antisense Oligonucleotides. Nanoscale Res. Lett. 6 (1), 1–7. doi:10.1186/1556-276X-6-608
Shigematsu, Y., Oue, N., Sekino, Y., Sakamoto, N., Sentani, K., Uraoka, N., et al. (2019). SEC11A Expression Is Associated with Basal-like Bladder Cancer and Predicts Patient Survival. Pathobiology 86, 208–216. doi:10.1159/000497206
Shivdasani, R. A. (2006). MicroRNAs: Regulators of Gene Expression and Cell Differentiation. Blood 108, 3646–3653. doi:10.1182/blood-2006-01-030015
Stein, A., Voigt, W., and Jordan, K. (2010). Review: Chemotherapy-Induced Diarrhea: Pathophysiology, Frequency and Guideline-Based Management. Ther. Adv. Med. Oncol. 2, 51–63. doi:10.1177/1758834009355164
Stone, J. B., and DeAngelis, L. M. (2016). Cancer-treatment-induced Neurotoxicity-Focus on Newer Treatments. Nat. Rev. Clin. Oncol. 13, 92–105. doi:10.1038/nrclinonc.2015.152
Strand, M. S., Krasnick, B. A., Pan, H., Zhang, X., Bi, Y., Brooks, C., et al. (2019). Precision Delivery of RAS-Inhibiting siRNA to KRAS Driven Cancer via Peptide-Based Nanoparticles. Oncotarget 10, 4761–4775. doi:10.18632/oncotarget.27109
Strumberg, D., Schultheis, B., Traugott, U., Vank, C., Santel, A., Keil, O., et al. (2012). Phase I Clinical Development of Atu027, a siRNA Formulation Targeting PKN3 in Patients with Advanced Solid Tumors. Cp 50, 76–78. doi:10.5414/CPP50076
Stylianopoulos, T., Wong, C., Bawendi, M. G., Jain, R. K., and Fukumura, D. (2012). Multistage Nanoparticles for Improved Delivery into Tumor Tissue. Methods Enzymol. 508, 109–130. doi:10.1016/B978-0-12-391860-4.00006-9
Su, M.-C., Yuan, R.-H., Lin, C.-Y., and Jeng, Y.-M. (2008). Cadherin-17 Is a Useful Diagnostic Marker for Adenocarcinomas of the Digestive System. Mod. Pathol. 21, 1379–1386. doi:10.1038/modpathol.2008.107
Su, W., Xu, M., Chen, X., Chen, N., Gong, J., Nie, L., et al. (2017). Long Noncoding RNA ZEB1-AS1 Epigenetically Regulates the Expressions of ZEB1 and Downstream Molecules in Prostate Cancer. Mol. Cancer 16, 142. doi:10.1186/s12943-017-0711-y
Su, Y.-L., Swiderski, P., Marcucci, G., and Kortylewski, M. (2019). Targeted Delivery of miRNA Antagonists to Myeloid Cells In Vitro and In Vivo. Methods Mol. Biol. 1974, 141–150. doi:10.1007/978-1-4939-9220-1_10
Sun, M.-Y., Zhu, J.-Y., Zhang, C.-Y., Zhang, M., Song, Y.-N., Rahman, K., et al. (2017). Autophagy Regulated by lncRNA HOTAIR Contributes to the Cisplatin-Induced Resistance in Endometrial Cancer Cells. Biotechnol. Lett. 39, 1477–1484. doi:10.1007/s10529-017-2392-4
Tabernero, J., Shapiro, G. I., LoRusso, P. M., Cervantes, A., Schwartz, G. K., Weiss, G. J., et al. (2013). First-in-humans Trial of an RNA Interference Therapeutic Targeting VEGF and KSP in Cancer Patients with Liver Involvement. Cancer Discov. 3, 406–417. doi:10.1158/2159-8290.CD-12-0429
Takahashi, K., Yan, I. K., Kogure, T., Haga, H., and Patel, T. (2014). Extracellular Vesicle-Mediated Transfer of Long Non-coding RNA ROR Modulates Chemosensitivity in Human Hepatocellular Cancer. FEBS Open Bio 4, 458–467. doi:10.1016/j.fob.2014.04.007
Takanami, T., Takanami, T., Mori, A., Takahashi, H., Horiuchi, S., and Higashitani, A. (2003). Caenorhabditis elegans Ce-Rdh-1/rad-51 Functions after Double-Strand Break Formation of Meiotic Recombination. Chromosom. Res. 11, 125–135. doi:10.1023/A:1022863814686
Takei, Y. (2019). siRNA-Based Drug Targeting Human Bcl-xL against Cancers BT - RNA Interference and Cancer Therapy. Methods Mol. Biol. 1974, 31–40. doi:10.1007/978-1-4939-9220-1_3
Tangudu, N. K., Verma, V. K., Clemons, T. D., Beevi, S. S., Hay, T., Mahidhara, G., et al. (2015). RNA Interference Using c-Myc–Conjugated Nanoparticles Suppresses Breast and Colorectal Cancer Models. Mole. Cancer Ther. 14 (5), 1259–1269.
Taniguchi, H., and Imai, K. (2019). Silencing PRDM14 via Oligonucleotide Therapeutics Suppresses Tumorigenicity and Metastasis of Breast Cancer. Methods Mol. Biol. 1974, 233–243. doi:10.1007/978-1-4939-9220-1_18
Thapa, B., Remant, K., and Uludağ, H. (2019). siRNA Library Screening to Identify Complementary Therapeutic Pairs in Triple-Negative Breast Cancer Cells BT - RNA Interference and Cancer Therapy, Methods Mol. Biol., 1974, 1–19. doi:10.1007/978-1-4939-9220-1_1
Tolcher, A. W., Papadopoulos, K. P., Patnaik, A., Rasco, D. W., Martinez, D., Wood, D. L., et al. (2015). Safety and Activity of DCR-MYC, a First-In-Class Dicer-Substrate Small Interfering RNA (DsiRNA) Targeting MYC, in a Phase I Study in Patients with Advanced Solid Tumors. Jco 33, 11006. doi:10.1200/jco.2015.33.15_suppl.11006
Tommasini-Ghelfi, S., Lee, A., Mirkin, C. A., and Stegh, A. H. (2019). Synthesis, Physicochemical, and Biological Evaluation of Spherical Nucleic Acids for RNAi-Based Therapy in Glioblastoma. Methods Mol. Biol. 1974, 371–391. doi:10.1007/978-1-4939-9220-1_25
Tortiglione, C., and de la Fuente, J. M. (2019). Synthesis of Gold Nanoparticles for Gene Silencing BT - RNA Interference and Cancer Therapy. Methods Protoc. 1974, 203–214. doi:10.1007/978-1-4939-9220-1_15
Vakalopoulos, I., Dimou, P., Anagnostou, I., and Zeginiadou, T. (2015). Impact of Cancer and Cancer Treatment on Male Fertility. Hj 14 (4), 579–589. doi:10.14310/horm.2002.1620
Välineva, T., Yang, J., Palovuori, R., and Silvennoinen, O. (2005). The Transcriptional Co-activator Protein P100 Recruits Histone Acetyltransferase Activity to STAT6 and Mediates Interaction between the CREB-Binding Protein and STAT6. J. Biol. Chem. 280, 14989–14996. doi:10.1074/jbc.M410465200
Vauthier, C. (2019). Preparation of a Carrier to Achieve In Vivo Delivery of siRNA: The Example of Chitosan-Coated Poly(isobutylcyanoacrylate) Nanoparticles. Methods Mol. Biol. 1974, 181–194. doi:10.1007/978-1-4939-9220-1_13
Vega-Stromberg, T. (2003). Chemotherapy-induced Secondary Malignancies. J. Infusion Nurs. 26, 353–361. doi:10.1097/00129804-200311000-00004
Verkman, A. S., Hara-Chikuma, M., and Papadopoulos, M. C. (2008). Aquaporins-new Players in Cancer Biology. J. Mol. Med. 86, 523–529. doi:10.1007/s00109-008-0303-9
Vinogradov, S. V., Bronich, T. K., and Kabanov, A. V. (2002). Nanosized Cationic Hydrogels for Drug Delivery: Preparation, Properties and Interactions with Cells. Adv. Drug Deliv. Rev. 54, 135–147. doi:10.1016/s0169-409x(01)00245-9
Waimey, K. E., Smith, B. M., Confino, R., Jeruss, J. S., and Pavone, M. E. (2015). Understanding Fertility in Young Female Cancer Patients. J. Women's Health 24, 812–818. doi:10.1089/jwh.2015.5194
Wan, R., Luan, Y., Wu, Z., Deng, S., Peng, M., Xu, L., et al. (2019). The Target Therapeutic Effect of Functionalized Graphene Oxide Nanoparticles Graphene Oxide-Polyethylene Glycol-Folic Acid-1-Pyrenemethylamine Hydrochloride-Mediated RNA Interference of HIF-1α Gene in Human Pancreatic Cancer Cells. J. Biomater. Appl. 34, 155–177. doi:10.1177/0885328219847019
Wang, J., Guo, Y., Chu, H., Guan, Y., Bi, J., and Wang, B. (2013). Multiple Functions of the RNA-Binding Protein HuR in Cancer Progression, Treatment Responses and Prognosis. Ijms 14, 10015–10041. doi:10.3390/ijms140510015
Wang, J., Liu, X., Wu, H., Ni, P., Gu, Z., Qiao, Y., et al. (2010). CREB Up-Regulates Long Non-coding RNA, HULC Expression through Interaction with microRNA-372 in Liver Cancer. Nucleic Acids Res. 38, 5366–5383. doi:10.1093/nar/gkq285
Wang, J., Ni, W.-h., Hu, K.-b., Zhai, X.-y., Xie, F., Jie, J., et al. (2017). Targeting MUC1 and JNK by RNA Interference and Inhibitor Inhibit the Development of Hepatocellular Carcinoma. Cancer Sci. 108, 504–511. doi:10.1111/cas.13144
Wang, J., Sun, M., Zhu, X., Zhao, H., Mao, D., Zhang, Z., et al. (2019). Lentivirus-mediated RNA I-nterference T-argeting P-rogrammed D-eath R-eceptor L-igand�1 Increases the Immunologic Anti-tumor E-ffect of D-endritic C-ell V-accination against P-ancreatic C-ancer in SCID-hu M-ice. Oncol. Lett. 18, 1539–1547. doi:10.3892/ol.2019.10426
Wang, Y., Qin, X., Zhu, X., Chen, W., Zhang, J., and Chen, W. (2018). Oral Cancer-Derived Exosomal NAP1 Enhances Cytotoxicity of Natural Killer Cells via the IRF-3 Pathway. Oral Oncol. 76, 34–41. doi:10.1016/j.oraloncology.2017.11.024
Warnecke, C., Zaborowska, Z., Kurreck, J., Erdmann, V. A., Frei, U., Wiesener, M., et al. (2004). Differentiating the Functional Role of Hypoxia‐inducible Factor (HIF)‐1α and HIF‐2α (EPAS‐1) by the Use of RNA Interference: Erythropoietin Is a HIF‐2α Target Gene in Hep3B and Kelly Cells. FASEB j. 18, 1462–1464. doi:10.1096/fj.04-1640fje
Whitehead, K. A., Langer, R., and Anderson, D. G. (2009). Knocking Down Barriers: Advances in siRNA Delivery. Nat. Rev. Drug Discov. 8, 129–138. doi:10.1038/nrd2742
Wilda, M., Fuchs, U., Wössmann, W., and Borkhardt, A. (2002). Killing of Leukemic Cells with a BCR/ABL Fusion Gene by RNA Interference (RNAi). Oncogene 21, 5716–5724. doi:10.1038/sj.onc.1205653
Wu, S. Y., Lopez-Berestein, G., Calin, G. A., and Sood, A. K. (2014). RNAi Therapies: Drugging the Undruggable. Sci. Translational Med. 6, 240ps7. doi:10.1126/scitranslmed.3008362
Wu, X. L., Lu, R. Y., Wang, L. K., Wang, Y. Y., Dai, Y. J., Wang, C. Y., et al. (2019). Long Noncoding RNA HOTAIR Silencing Inhibits Invasion and Proliferation of Human colon Cancer LoVo Cells via Regulating IGF2BP2. J. Cel. Biochem. 120, 1221–1231. doi:10.1002/jcb.27079
Xin, Y., Huang, M., Guo, W. W., Huang, Q., Zhang, L. z., and Jiang, G. (2017). Nano-based Delivery of RNAi in Cancer Therapy. Mol. Cancer 16, 134. doi:10.1186/s12943-017-0683-y
Xing, A., Pan, L., and Gao, J. (2018). p100 Functions as a Metastasis Activator and Is Targeted by Tumor Suppressing microRNA-320a in Lung Cancer. Thorac. Cancer 9, 152–158. doi:10.1111/1759-7714.12564
Xiong, G., Chen, X., Zhang, Q., Fang, Y., Chen, W., Li, C., et al. (2017). RNA Interference Influenced the Proliferation and Invasion of XWLC-05 Lung Cancer Cells through Inhibiting Aquaporin 3. Biochem. Biophysical Res. Commun. 485, 627–634. doi:10.1016/j.bbrc.2017.02.013
Xiong, Y., Wang, L., Li, Y., Chen, M., He, W., and Qi, L. (2017). The Long Non-coding RNA XIST Interacted with MiR-124 to Modulate Bladder Cancer Growth, Invasion and Migration by Targeting Androgen Receptor (AR). Cel. Physiol. Biochem. 43, 405–418. doi:10.1159/000480419
Xu, L., and Yang, H. (2019). Folate-decorated Polyamidoamine Dendrimer Nanoparticles for Head and Neck Cancer Gene Therapy. Methods Mol. Biol. 1974, 393–408. doi:10.1007/978-1-4939-9220-1_26
Xu, W., Xu, B., Yao, Y., Yu, X., Cao, H., Zhang, J., et al. (2016). RNA Interference against TRIM29 Inhibits Migration and Invasion of Colorectal Cancer Cells. Oncol. Rep. 36, 1411–1418. doi:10.3892/or.2016.4941
Yang, J., Aittomäki, S., Pesu, M., Carter, K., Saarinen, J., Kalkkinen, N., et al. (2002). Identification of P100 as a Coactivator for STAT6 that Bridges STAT6 with RNA Polymerase II. EMBO J. 21, 4950–4958. doi:10.1093/emboj/cdf463
Yang, L., Xue, Y., Liu, J., Zhuang, J., Shen, L., Shen, B., et al. (2017). Long Noncoding RNA ASAP1-IT1 Promotes Cancer Stemness and Predicts a Poor Prognosis in Patients with Bladder Cancer. neo 64, 847–855. doi:10.4149/neo_2017_606
Yang, M., and Moon, C. (2013). Neurotoxicity of Cancer Chemotherapy. Neural Regen. Res. 8, 1606–1614. doi:10.3969/j.issn.1673-5374.2013.17.009
Yang, P., Yang, Y., An, W., Xu, J., Zhang, G., Jie, J., et al. (2017). The Long Noncoding RNA-ROR Promotes the Resistance of Radiotherapy for Human Colorectal Cancer Cells by Targeting the p53/miR-145 Pathway. J. Gastroenterol. Hepatol. 32, 837–845. doi:10.1111/jgh.13606
Yang, Q., Xu, E., Dai, J., Liu, B., Han, Z., Wu, J., et al. (2015). A Novel Long Noncoding RNA AK001796 Acts as an Oncogene and Is Involved in Cell Growth Inhibition by Resveratrol in Lung Cancer. Toxicol. Appl. Pharmacol. 285, 79–88. doi:10.1016/j.taap.2015.04.003
Yasui, W., Sentani, K., Sakamoto, N., Anami, K., Naito, Y., and Oue, N. (2011). Molecular Pathology of Gastric Cancer: Research and Practice. Pathol. - Res. Pract. 207, 608–612. doi:10.1016/j.prp.2011.09.006
Yoshinouchi, M., Yamada, T., Kizaki, M., Fen, J., Koseki, T., Ikeda, Y., et al. (2003). In Vitro and In Vivo Growth Suppression of Human Papillomavirus 16-positive Cervical Cancer Cells by E6 siRNA. Mol. Ther. 8, 762–768. doi:10.1016/j.ymthe.2003.08.004
You, J.-s., Guo, L., Huang, M., Shi, X.-l., Lin, M.-d., Guo, Z., et al. (2019). The Effect and Mechanism of YH0618 Granule on Chemotherapy- Induced Hair Loss in Patients with Breast Cancer: Study Protocol for a Randomized, Double-Blind, Multi-center Clinical Trial. Trials 20 (1), 719. doi:10.1186/s13063-019-3893-3
Yu, Z. Y., Wang, Z., Lee, K. Y., Yuan, P., and Ding, J. (2018). Effect of Silencing colon Cancer-Associated Transcript 2 on the Proliferation, Apoptosis and Autophagy of Gastric Cancer BGC-823 Cells. Oncol. Lett. 15, 3127–3132. doi:10.3892/ol.2017.7677
Yuan, Q., Yeudall, W. A., Lee, E., and Yang, H. (2019). Targeted Inactivation of EPS8 Using Dendrimer-Mediated Delivery of RNA Interference. Int. J. Pharmaceutics 557, 178–181. doi:10.1016/j.ijpharm.2018.12.060
Zagryazhskaya, A., Surova, O., Akbar, N. S., Allavena, G., Gyuraszova, K., Zborovskaya, I. B., et al. (2015). Tudor Staphylococcal Nuclease Drives Chemoresistance of Non-small Cell Lung Carcinoma Cells by Regulating S100A11. Oncotarget 6, 12156–12173. doi:10.18632/oncotarget.3495
Zahednezhad, F., Saadat, M., Valizadeh, H., Zakeri-Milani, P., and Baradaran, B. (2019). 305. Release, 194–209. doi:10.1016/j.jconrel.2019.05.030Liposome and Immune System Interplay: Challenges and PotentialsJ. Controlled Release
Zajaczkowską, R., Kocot-Kępska, M., Leppert, W., Wrzosek, A., Mika, J., and Wordliczek, J. (2019). Mechanisms of Chemotherapy-Induced Peripheral Neuropathy. Int. J. Mol. Sci. 20 (6), 1451. doi:10.3390/ijms20061451
Zhan, N., Li, B., Xu, X., Xu, J., and Hu, S. (2018). Inhibition of FASN Expression Enhances Radiosensitivity in Human Non-small Cell Lung Cancer. Oncol. Lett. 15, 4578–4584. doi:10.3892/ol.2018.7896
Zhang, J., Fei, B., Wang, Q., Song, M., Yin, Y., Zhang, B., et al. (2014). MicroRNA-638 Inhibits Cell Proliferation, Invasion and Regulates Cell Cycle by Targeting Tetraspanin 1 in Human Colorectal Carcinoma. Oncotarget 5, 12083–12096. doi:10.18632/oncotarget.2499
Zhang, J. H., Wei, H. W., and Yang, H. G. (2016). Long Noncoding RNA SNHG15, a Potential Prognostic Biomarker for Hepatocellular Carcinoma. Eur. Rev. Med. Pharmacol. Sci. 20, 1720–1724.
Zhao, Z.-B., Long, J., Zhao, Y.-Y., Yang, J.-B., Jiang, W., Liu, Q.-Z., et al. (2018). Adaptive Immune Cells Are Necessary for the Enhanced Therapeutic Effect of Sorafenib-Loaded Nanoparticles. Biomater. Sci. 6, 893–900. doi:10.1039/c8bm00106e
Zhao, Z., He, J., Kang, R., Zhao, S., Liu, L., and Li, F. (2016). RNA Interference Targeting PSCA Suppresses Primary Tumor Growth and Metastasis Formation of Human Prostate Cancer Xenografts in SCID Mice. Prostate 76, 184–198. doi:10.1002/pros.23110
Zheng, Q., Bao, C., Guo, W., Li, S., Chen, J., Chen, B., et al. (2016). Circular RNA Profiling Reveals an Abundant circHIPK3 that Regulates Cell Growth by Sponging Multiple miRNAs. Nat. Commun. 7, 11215. doi:10.1038/ncomms11215
Zhong, X., Long, Z., Wu, S., Xiao, M., and Hu, W. (2018). LncRNA-SNHG7 Regulates Proliferation, Apoptosis and Invasion of Bladder Cancer Cells Assurance Guidelines. J. B.U.ON. 23, 776–781.
Zhou, T., Li, Y., Yang, L., Liu, L., Ju, Y., and Li, C. (2017). Silencing of ANXA3 Expression by RNA Interference Inhibits the Proliferation and Invasion of Breast Cancer Cells. Oncol. Rep. 37, 388–398. doi:10.3892/or.2016.5251
Zhu, Z., Du, S., Yin, K., Ai, S., Yu, M., Liu, Y., et al. (2019). Knockdown Long Noncoding RNA Nuclear Paraspeckle Assembly Transcript 1 Suppresses Colorectal Cancer through Modulating miR-193a-3p/KRAS. Cancer Med. 8, 261–275. doi:10.1002/cam4.1798
Keywords: cancer, combinatorial therapy, nanotechnology, non-coding RNAs, RNA interference, targeted drug delivery
Citation: Swaminathan G, Shigna A, Kumar A, Byroju VV, Durgempudi VR and Dinesh Kumar L (2021) RNA Interference and Nanotechnology: A Promising Alliance for Next Generation Cancer Therapeutics. Front. Nanotechnol. 3:694838. doi: 10.3389/fnano.2021.694838
Received: 13 April 2021; Accepted: 21 May 2021;
Published: 09 June 2021.
Edited by:
Christiane Pienna Soares, Sao Paulo State Universty, BrazilReviewed by:
Anirudh Sharma, University of Minnesota, United StatesCopyright © 2021 Swaminathan, Shigna, Kumar, Byroju, Durgempudi and Dinesh Kumar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lekha Dinesh Kumar, bGVraGFAY2NtYi5yZXMuaW4=
†These authors have contributed equally to this work and share first authorship
‡These authors have contributed equally to this work and share second authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.