
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Nanotechnol. , 25 June 2021
Sec. Biomedical Nanotechnology
Volume 3 - 2021 | https://doi.org/10.3389/fnano.2021.674360
This article is part of the Research Topic Advances in Self-Assembled Nanocarriers for Drug Delivery View all 5 articles
Here, we have demonstrated that on modification of linear polyethylenimine (lPEI, LP) with amphiphilic 3-bromopropyltetramethylguanidinium (PTMG) linker, the transfection efficiency exhibited by the modified polymers decreased while cell viability improved. A series of LP-PTMG polymers was synthesized by the reaction of varying amounts of 3-bromopropyl tetramethylguanidinium linker with lPEI (25 kDa). These modified polymers interacted efficiently with pDNA and formed nanosized complexes as shown by dynamic light scattering analysis. The size of the complexes in the series LP-PTMG/pDNA was observed in the range of ∼178–205 nm. The interaction of modified polymers with plasmid DNA was stronger than linear PEI as evidenced by heparin release assay which showed ∼83% pDNA release from LP-PTMG-3/pDNA complexes in comparison to ∼95% in lPEI/pDNA complexes on treatment with same amount of heparin suggesting the formation of self-assembled structures in modified polymers. The transfection studies in HeLa and Chinese hamster ovary cells showed a decrease in transfection efficiency of LP-PTMG polymers, the reason for this may be strong binding of modified polymers with pDNA due to accumulation of charge on the surface. This finding showed the significance of optimum binding of polymer and DNA to form polyplexes as well as release of DNA from the polyplexes.
Gene therapy, considered to treat acquired or genetic diseases by precisely transferring genetic material as therapeutics into particular cells of a patient, has become a fascinating field since the use of 1st human gene therapy trial in 1990s (Xiao et al., 2018). The clinical application of gene therapy depends primarily on the safety and efficiency of gene delivery vectors. Nucleic acid delivery vectors are generally categorized as viral and non-viral ones. The concerns associated with the use of viral vectors is their biosafety issue and immunogenic nature which hamper their use in clinical trials (Mali, 2013). On the other hand, the non-viral vectors for gene therapy are attracting attention of the researchers to develop them as therapeutic carriers mainly due to safety, biocompatibility and easy synthesis/manipulation. Moreover, the non-viral vectors in comparison to viral vectors can carry large sizes of cargo molecules i.e., therapeutic nucleic acids (Durymanov and Reineke, 2018). Non-viral or chemically synthesized vector systems such as polymers, peptides, cationic lipids, and dendrimers, etc. offer prospective routes for compacting DNA for systemic delivery (Araújo et al., 2018; Zakeri et al., 2018). As viral vectors have evolved mechanisms to conquer cellular barrier and integrate DNA into host cells by escaping the host immune defence system but limitations associated with non-viral vectors are their inadequate transfection efficiency due to a variety of extra- and intracellular barriers.
Among various cationic polymers, polyethylenimines have been used enormously as vector for gene delivery, due to intrinsic attribute of compacting nucleic acids as well as possession of proton sponge feature (Boussif et al., 1995; Behr, 1997; Guillem and Aliño, 2004). PEI possesses a high density of different amine groups which can be protonated. Linear PEI (lPEI) possesses mainly secondary amines. lPEI has been assumed as an alternative to bPEI due to its reduced toxicity in comparison to bPEI. But it exhibits low transfection efficiency in comparison to bPEI due to its poor solubility as well as it forms loose polyplexes with nucleic acids which leads to decomplexation prior to reaching the target site resulting in damage to unprotected DNA by nucleases and other enzymes. Various modifications have been incorporated to tackle these issues, leading to enhancement in transfection efficiency of linear PEI (Zakeri et al., 2018). Recently, tetramethylguanidinium modified branched polyethylenimine polymers have been reported to exhibit high transfection efficiency with low toxicity (Mahato et al., 2012; Mahato et al., 2013; Yadav et al., 2014b; Mahato et al., 2014). Tetramethylguanidine (TMG), being a superbase, provides higher charge density with an ability to condense nucleic acids more strongly. Keeping this point in mind, it was anticipated that substitution of TMG moieties on linear polyethylenimine would improve pDNA binding as well as cellular uptake that would result in improved transfection efficiency without compromising on toxicity. The enhanced uptake would be due to efficient interactions between linear polyethylenimine-propyl TMG (LP-PTMG) polymers and cell membrane.
Therefore, in the present study, TMG bearing amphiphilic linker molecule, 3-bromopropyl tetramethylguanidinium, was synthesized and used for the tethering onto the backbone of lPEI. lPEI was grafted with amphiphilic cationic linker, propyl tetramethylguanidinium (PTMG), having hydrophobic alkyl chain and positively charged N, N, N, N-tetrametylguanidinium group. A series of amphiphilic LP-PTMG polymers with varying amounts of PTMG linker was obtained. Further, the series of LP-PTMG polymers was physicochemically characterized and assessed for their cellular toxicity and gene delivery (transfection) efficiency in HeLa and CHO cells. The toxicity and transfection efficiency of series of LP-PTMG polymers was evaluated in comparison to native lPEI (25 kDa) as well as widely used branched PEI (bPEI).
HeLa (Human cervical cancer) cells were maintained in high glucose Dulbeccơs modified eagle’s media (DMEM) and CHO (Chinese hamster ovary) cells in HAM’S F-12 media, supplemented with 10% heat inactivated FBS along with antibiotic cocktail of streptomycin and penicillin (1%) with all the media components dissolved in autoclaved MiliQ water. The cells were grown at 37°C in an incubator containing 5% CO2 and 95% relative humidity.
For transfection experiments, the pDNA for enhanced green fluorescent protein (EGFP) gene was transformed in E. coli DH5α bacterial strain. The plasmid was isolated from transformed bacterial culture using Endofree Maxi-Prep kit from Qiagen. The concentration and quality of the extracted pDNA was determined by recording the absorbance at 260 nm and taking ratio of absorbance at 260/280.
The synthesis of LP-PTMG polymers was accomplished as described in our previously published article (Yadav et al., 2018). Briefly lPEI (25 kDa) was conjugated to 3-bromopropyl tetramethylguanidium linker by mixing both the components in appropriate amounts in methanol at 60°C for 24 h. Concentration followed by dialysis in water yielded LP-PTMG polymers. The series of LP-PTMG polymers with 4, 6, 8, and 10% attempted substitutions was named as LP-PTMG-1, LP-PTMG-2, LP-PTMG-3, and LP-PTMG-4, respectively.
pDNA complexes of LP-PTMG polymers were fabricated in aqueous environment at different w/w ratios of 1.66, 3.33, 5.0, 6.66, 8.33, and 10.0. Briefly, pDNA (1 μl, 300 ng/μl) was mixed with different amounts of LP-PTMG polymer solutions (1 mg/ml) followed by addition of 20% dextrose (5 µl). The final reaction mixture was prepared upto 20 µl with H2O followed by incubation at an ambient temperature for 30 min before using them for physico-chemical and biological studies.
The size and zeta potential of series of LP-PTMG/pDNA polyplexes were measured using Zetasizer Nano ZS instrument. The hydrodynamic diameter of the modified polyplexes at their best working w/w ratio of 8.33, in terms of transfection, in aqueous environment and 10% FBS were calculated in triplicates in automatic mode by Zetasizer using dynamic light scattering analysis. Each measurement presented an average value of twenty runs and the results representing the average value of three experiments carried out independently.
Similarly, the surface charge on the polyplexes were measured in terms of zeta potential values in aqueous environment as well as 10% FBS. The zeta potential of polyplexes at w/w ratio of 8.33 were calculated on a Zetasizer Nano-ZS by carrying out thirty runs in triplicates. The average values of zeta potential are presented automatically using Smoluchowski approximation from electrophoretic mobility which is calculated by performing laser Doppler electrophoresis on sample. Morphology as well as size of LP-PTMG-3/pDNA polyplex were further examined transmission electron microscopically (TEM) at the same w/w ratio. For TEM imaging, 1% w/v solution of uranyl acetate was used as negative stain to enhance the contrast in TEM image of the LP-PTMG-3/pDNA polyplex.
lPEI and LP-PTMG polymers’ ability to resist protonation was demonstrated experimentally employing acid-base titration assay (Benns et al., 2000). A suspension of LP-PTMG polymer (3 mg/ml) in 30 ml 0.1 N NaCl was taken in 50 ml falcon tube and pH of this solution was increased upto pH 10 with addition of 0.1 N NaOH solution. Then 20 µL aliquots of 0.1 N HCl were added to this solution and measured the pH values after each addition. The addition of 0.1N HCl was continued till the pH value reached pH 3.0. A graph was sketched between amounts of HCl consumed vs. pH of the solution which demonstrated the buffering capacity of the system. It is considered that a polymer having good buffering capacity either resists a change in pH or exhibits a very minor variation in pH when a small quantity of strong acid or base is added.
EMSA was performed to determine the quantity of LP-PTMG polymer necessary for complete offsetting of negative charge of pDNA taken in the experiment. The lPEI and LP-PTMG polyplexes with 0.3 μg pDNA were formed at w/w ratios of polymer/pDNA at 0.16, 0.26, 0.40, 0.50, 0.66, 0.83, and 1.0. The polyplexes were kept at 25°C for 0.5 h followed by addition of 2 µl of Orange G loading dye (10x). Agarose gel (0.8%) with ethidium bromide staining dye was casted on a casting tray. The polyplexes were loaded on wells of agarose gel dipped in 1x TAE (Tris-acetic acid-EDTA) buffer and electrophoresis was carried out at 100 V for 1 h in 1x TAE buffer. After 1 h, electrophoresis was stopped and the bands were visualized under ultraviolet transilluminator using G:BOX gel documentation system.
In the present work, in vitro transfection studies were carried out on HeLa and CHO cells. For cell seeding, ∼10,000 cells were added to each well in 96-well plates and placed in CO2 incubator for 24–36 h for adhering to surface of wells. The media was removed from the wells and washed with 1x PBS once followed by addition of 60 μL serum free DMEM. pDNA complexes of LP-PTMG and lPEI polymers were prepared at w/w ratios of 1.66, 3.33, 5.0, 6.66, 8.33, and 10.0. The polyplexes were kept at RT for 0.5 h. The pDNA complex of bPEI was also formed at w/w ratio of 2.66. The pDNA complexes of LP-PTMG, lPEI, and bPEI were added gently onto the cells in 96-well plates and kept in an incubator maintaining temperature of 37 °C, 95% relative humidity and 5% CO2. On completion of incubation time of 3 h, the media was removed gently from the wells and added 100 μL DMEM containing 10% FBS to each well followed by incubation in CO2 incubator upto 45 h. After 45 h, GFP gene expression was visualized under inverted fluorescence microscope (Nikon Eclipse TE 2000-S, Japan) having C-F1 epifluorescence filter block (B-2A) comprising of an excitation filter (Ex 450–490 nm), Dichroic mirror (DM 505) and barrier filter (BA 520).
pDNA release on heparin treatment was performed to evaluate the strength of the so formed pDNA complexes with PTMG modified PEI polymers. Complexes of lPEI and LP-PTMG-3 polymers were prepared with pDNA at w/w ratio of 8.33 and kept for incubation at room temperature for 0.5 h. Subsequently, varying amount of heparin (0.2–3.0U) was added followed by incubation for 20 min at room temperature. The samples were loaded onto 0.8% agarose gel pre-stained with EtBr and electrophoresis was carried out at 100 V for 60 min in 1x TAE buffer. Subsequently, the bands were observed under UV transilluminator and amount of pDNA released on competitive binding of heparin with polymers was quantified by densitometry using gene tool software (Syngene) (Goyal et al., 2011).
FACS analysis was carried out to determine the number of cells transfected in a population of cells. CHO and HeLa cells were cultured in 24-well plates and transfection experiments were performed as mentioned above in vitro transfection studies. After 45 h of incubation, the media was gently removed followed by washing with 1x PBS. The 100 µl 1x-trypsin solution was added to cells for trypsinization and subsequently, 500 µl DMEM was added to wells. The trypsinized cells were collected in 1.5 ml centrifuge tubes and centrifuged at 5,000 rpm for 5 min at 4°C. The cell pellets were washed twice with 500 μl of 1x PBS. Subsequently, the cell suspension was made in 200 μl of 1x PBS. Percent EGFP gene transfected in the cells was quantified by Guava Easycyte Plus Flow cytometry system consisting of Cytosoft Software. The statistics of cells fluorescence above the control level were counted as transfected cells while the cells from non-transfected wells were used as control.
The cell viability of HeLa and CHO cells on treatment with LP-PTMG/pDNA, lPEI/pDNA, bPEI/pDNA and the commercially available transfection reagent, Lipofectamine/pDNA, polyplexes was estimated by MTT assay (Tripathi et al., 2012). The mitochondria of live cells contain enzyme succinate dehydrogenase which reduces MTT dye to dark purple colored formazan crystals. These formazan crystals were dissolved in organic solvents. The intensity of solubilized product was measured on a spectrophotometer at 540 nm. For cell viability studies, the transfection was performed at same w/w ratios as done in vitro transfection studies. After 45 h, the media was aspirated from wells and 200 μl of MTT solution (0.5 mg/ml dissolved in media) was introduced in each well. The plate was placed inside a CO2 incubator for 2–3 h at 37°C. After 2–3 h of incubation, the media was removed and the formazan crystals formed at the bottom of the wells were dissolved in 100 µl MTT lysis buffer followed by measuring the intensity of color at 540 nm on multiwell plate reader (MRX, Dynatech Laboratories). For calculating the cell viability percentage, the cells from untreated wells were considered as having 100% cell viability. The relative cell viability (%) in comparison to untreated cells were examined using formulae: (Abssample/Abscontrol) x 100 (Yadav et al., 2014a).
Propyl-tetramethylguanidinium-grafted lPEI polymers i.e. LP-PTMG polymer series (4%, 6%, 8%, and 10%), were prepared and characterized as reported in our earlier publication (Yadav et al., 2018).
The charge on the surface of the particles is an important criteria for gene delivery vectors to assist in binding to nucleic acids and entry into the cells. It was anticipated that conjugation of amphiphilic PTMG to lPEI would incorporate amphiphilic character which would facilitate LP-PTMG polymers to form self-assembled structures bringing the charged guanidinium moieties on the surface of the structures. This was evidenced by the zeta potential values of these polymers in water where surface charge on the modified polymer series i.e. LP-PTMG was found to be in the range of 46.5–54.8 mV, while the zeta potential of native lPEI polymer was 18.1 mV. This drastic increase in zeta potential value of LP-PTMG polymer series might be due to rearrangement of LP-PTMG polymers into self-assembled structures with cationic charged guanidinium groups were more exposed on to the surface of these structures. These modified amphiphilic LP-PTMG polymers interacted electrostatically with a negatively charged pDNA producing nano-sized particles which were assessed for their size as well as zeta potential values using DLS. The average size of the LP-PTMG/pDNA complexes in water at w/w 8.33 was found to be ∼178–205 nm (Table 1), whereas in 10% FBS, the size decreased to ∼30–40 nm (Table 1). The reduction in size of the particles in the presence of serum might be due to stabilization of particles by serum proteins which might have resulted in reduction in aggregation tendency as well as reduction in hydration tendency around the serum stabilized individual particles (Ogris et al., 1998; Pichon et al., 2001; Lungwitz et al., 2005; Konopka et al., 2006; Yadav et al., 2014a). Further, we checked the stability of LP-PTMG/pDNA particles by DLS up to 8 h and found no significant variation in the size of the particles.
The average zeta potential of LP-PTMG/pDNA polyplexes at w/w ratio 8.33 in water analyzed by zetasizer was found to be ∼ + 20 to +30 mV (Table 1) while in 10% FBS, the zeta potential decreased to–13.15 mV to–15.36 mV. The zeta potential on the grafted LP-PTMG/pDNA polyplexes was more in contrast to unmodified lPEI polymer/pDNA polyplexes. Furthermore, the zeta potential values of particles in 10% FBS was found to be reversed in polarity. Similar results have been observed in earlier studies also (Ogris et al., 1998; Pichon et al., 2001; Lungwitz et al., 2005; Konopka et al., 2006; Yadav et al., 2014a).
The morphology of the particles as well as size was further observed by TEM and found that LP-PTMG-3/pDNA complexes at w/w 8.33 formed spherical shaped particles with average size ∼40–50 nm (Figure 1). The reason for small size of particles observed in TEM in comparison to that observed by DLS is due to measurement of particles in dry state in TEM and solution state in DLS which calculates hydrodynamic diameter of the particles (Mahato et al., 2012; Priyam et al., 2018).
Each of the PTMG-grafted lPEI samples were analyzed for their optimal binding concentration with 0.3 µg of pDNA using EMSA. Amount of the samples were taken from 0.05–0.3 μg. pDNA without any polymer served as control. Complexes of LP-PTMG series of polymers retarded the mobility of pDNA at w/w ratio of 0.4 each, whereas native lPEI showed complete neutralization with pDNA at w/w ratio of 0.5. PTMG-modified lPEI i.e., LP-PTMG polymers retarded pDNA at lower w/w in comparison to lPEI (Figure 2). This might be due to higher charge on the surface of LP-PTMG polymers due to PTMG group. These results revealed that modified polymers carried slightly higher charge than the native lPEI.
After endocytosis of PEI/pDNA complex, the release of complex from endosome occur due to intrinsic buffering capacity of lPEI polymer (Tseng et al., 2005; Singh et al., 2015). The influence on buffering capacity of lPEI on grafting with propyl tetramethylguanidinium was evaluated by gradually adding 0.1 N HCl to the polymer solution and recording the change in pH. As shown in Figure 3, the native lPEI polymer exhibited a good buffering capacity in the pH range 3–10. The decrease or increase in buffering capacity of PEI after modification depends upon the type and extent of modification. Here, the strong basicity of guanidinium group of PTMG might have influenced the buffering capacity of the lPEI which could be responsible for endosomal escape of the polyplexes. Among the series, LP-PTMG-1 polymer showed a slight increase in buffering capacity, while LP-PTMG-2 to LP-PTMG-4 showed slightly decreased buffering capacity in comparison to native lPEI.
pDNA release assay was performed to check the binding ability and stability of the LP-PTMG/pDNA complexes at w/w ratio of 8.33 using an anionic heparin polysaccharide in increasing units (0.2–3.0 U). It was found that ∼95% of pDNA was released by native lPEI on addition of 1.2 U of heparin while LP-PTMG-3 complex released ∼83% of pDNA on adding the same amount of heparin (Figure 4). This suggested that the presence of TMG moieties in place of secondary amines of PEI strengthened the interaction with pDNA and these interactions provided excessive stability to the complex which subdued the release of pDNA from the polyplexes.
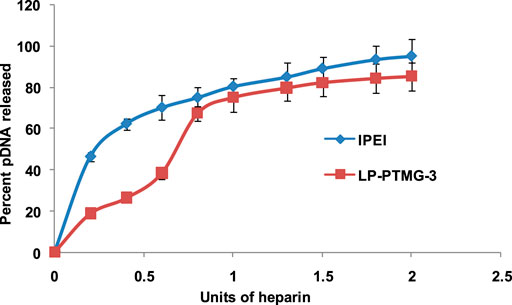
FIGURE 4. pDNA release assay of lPEI and LP-PTMG-3 polymers. The experiment was performed thrice and error bars represent the standard deviation.
The cytotoxicity of lPEI, bPEI, Lipofectamine and LP-PTMG series of polyplexes was performed on HeLa and CHO cell lines by MTT assay at the same w/w ratios as used in the transfection studies (Tripathi et al., 2012). The cell viability increased on an increase in percent grafting of PTMG on lPEI (Figure 5). Compared to native lPEI/pDNA complex, PTMG-grafted lPEI series i.e., LP-PTMG/pDNA complexes showed marginally better cell viability (∼89–91%). bPEI/pDNA and Lipofectamine/pDNA complexes exhibited higher cytotoxicity.
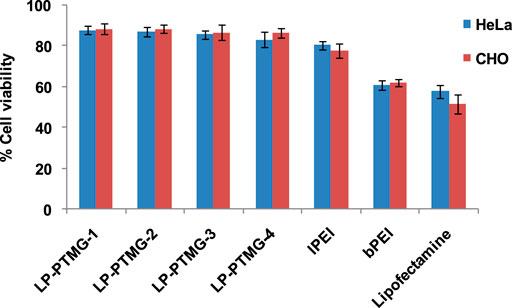
FIGURE 5. Cell viability profile of LP-PTMG/pDNA, lPEI/pDNA, bPEI/pDNA and Lipofectamine/pDNA complexes on CHO and HeLa cells.
The transfection efficiency of LP-PTMG/pDNA complexes was evaluated by FACS analysis in CHO and HeLa cells. After preliminary examination of transfection at different w/w ratios, the final transfection experiments were done at w/w ratio of 5.0, 6.66, and 8.33 for LP-PTMG/pDNA and lPEI/pDNA polyplexes. The transfection efficiency was compared with the standard transfection reagent, bPEI/pDNA complex (w/w 2.66). Transfection efficiency of pDNA complexes of the series of PTMG-modified lPEI polymers i.e. LP-PTMG/pDNA complexes decreased in comparison to native lPEI/pDNA complex in CHO as well as HeLa cells (Figures 6A,B). The best transfection efficiency exhibited among the series was of LP-PTMG-3/pDNA polyplex at w/w ratio of 8.33. The LP-PTMG-3/pDNA complex transfected ∼11.55 and 11.83% cells at w/w ratio of 6.66 and 8.33 in CHO cells while at the same w/w ratios, lPEI/pDNA complex transfected ∼13.86 and 16.87% cells, respectively (Figure 6A). Similarly, in HeLa cells, LP-PTMG-3/pDNA complex transfected ∼7.6 and 8.44% cells at w/w ratios of 6.66 and 8.33, while, at the similar w/w ratios, lPEI/pDNA complex transfected ∼8.56 and 11.36% cells, respectively (Figure 6B). The decrease in transfection efficiency of LPTG polymers in comparison to native lPEI might be because of tight binding of pDNA with LPTG polymers which might have hampered the release of pDNA from the polyplexes inside the cells. The results of the projected study are in agreement with the results reported in previously investigated reports. Similar results have been observed by Sainlos et al. (2005) where the guanidination of amine groups of an aminoglycoside, kanamycin, conjugated with cholesterol, resulted in the reduced transfection efficiency in comparison to non-guanidinylated counterpart. In another study, Bono et al. (2019) also observed that on guanidinylation of neomycin conjugated to PAMAM dendrimers, transfection efficacy decreased.
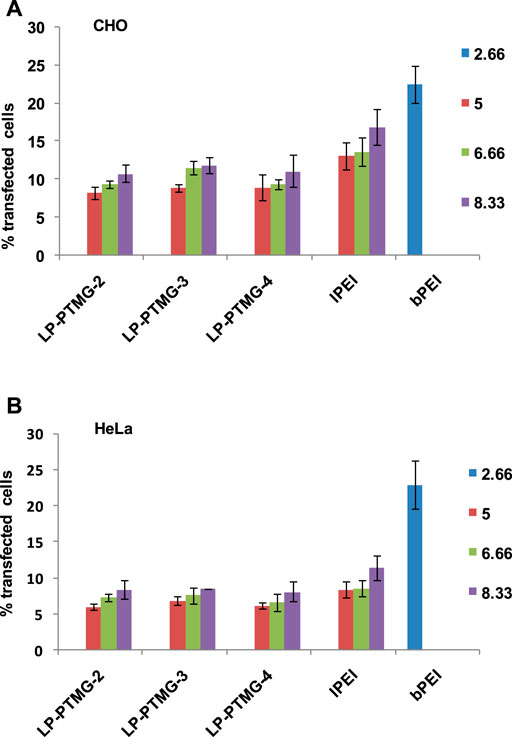
FIGURE 6. Transfection efficiency of LP-PTMG/pDNA, lPEI/pDNA and bPEI/pDNA complexes in (A) CHO, and (B) HeLa cells as determined by FACS analysis.
The fluorescence images of HeLa cells on transfection with lPEI/pDNA and LP-PTMG-3/pDNA complexes at their best working w/w ratio of 8.33 are depicted in Figure 7. Visually, there was not much difference observed in the transfection efficiency exhibited by the pDNA complexes of lPEI and LP-PTMG polymers.
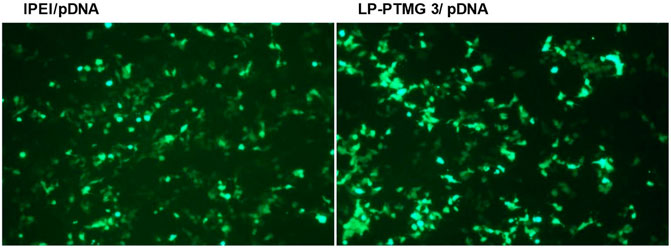
FIGURE 7. Fluorescence images observed on transfection of Hela Cells with lPEI/pDNA and LP-PTMG/pDNA complexes at w/w ratio of 8.33.
The functionalization or modification of the polymers for gene delivery does not always result in the generation of more efficient vectors as compared to the native ones. As it is well accepted that modification of the polymers with guanidinium groups favours their cellular uptake but the polyplex decomplexation has to take place in an efficient way to enable the release of pDNA and its transcription to go on. Here, in the projected study, we found that modification of secondary amines of linear PEI with highly basic tetramethylguanidinium groups resulted in a decrease in buffering capacity as well as strong binding with pDNA, resulting in the higher stability of the polyplexes, thus finally affecting the transfection efficiency of the polymers. Grafting of PTMG on lPEI decreased the pDNA release efficiency from LP-PTMG/pDNA complexes. The conversion of secondary amines of lPEI into substituted guanidinium moieties caused a decrease in the transfection efficiency. However, this study demonstrated that modification of linear PEI with tetramethylguanidinium groups further improved the cell viability of linear PEI. These findings established the fact that the physicochemical properties of these polyplexes could further be helpful in understanding the structure-activity relationship of these polymers in a better way.
The raw data supporting the conclusion of this article will be made available by the authors, without undue reservation.
SY: Conceptualization, methodology, and compilation AS: Reviewing and editing PK: Editing and supervising.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
SY acknowledges financial support from ICMR for providing Research Associateship vide reference no. 45/39/2020-Nan/BMS.
Araújo, R., Santos, S., Igne Ferreira, E., and Giarolla, J. (2018). New Advances in General Biomedical Applications of PAMAM Dendrimers. Molecules 23, 2849. doi:10.3390/molecules23112849
Behr, J.-P. (1997). The Proton Sponge: a Trick to Enter Cells the Viruses Did Not Exploit. CHIMIA Int. J. Chem. 51, 34–36.
Benns, J. M., Choi, J.-S., Mahato, R. I., Park, J.-S., and Kim, S. W. (2000). pH-Sensitive Cationic Polymer Gene Delivery Vehicle: N-Ac-Poly(l-Histidine)-Graft-Poly(l-Lysine) Comb Shaped Polymer. Bioconjug. Chem. 11, 637–645. doi:10.1021/bc0000177
Bono, N., Pennetta, C., Bellucci, M. C., Sganappa, A., Malloggi, C., Tedeschi, G., et al. (2019). Role of Generation on Successful DNA Delivery of PAMAM-(Guanidino)Neomycin Conjugates. ACS Omega 4, 6796–6807. doi:10.1021/acsomega.8b02757
Boussif, O., Lezoualc'h, F., Zanta, M. A., Mergny, M. D., Scherman, D., Demeneix, B., et al. (1995). A Versatile Vector for Gene and Oligonucleotide Transfer into Cells in Culture and In Vivo: Polyethylenimine. Proc. Natl. Acad. Sci. 92, 7297–7301. doi:10.1073/pnas.92.16.7297
Durymanov, M., and Reineke, J. (2018). Non-viral Delivery of Nucleic Acids: Insight into Mechanisms of Overcoming Intracellular Barriers. Front. Pharmacol. 9, 971. doi:10.3389/fphar.2018.00971
Goyal, R., Tripathi, S. K., Tyagi, S., Ravi Ram, K., Ansari, K. M., Shukla, Y., et al. (2011). Gellan Gum Blended PEI Nanocomposites as Gene Delivery Agents: Evidences from In Vitro and In Vivo Studies. Eur. J. Pharm. Biopharm. 79, 3–14. doi:10.1016/j.ejpb.2011.01.009
Guillem, V. M., and Aliño, S. F. (2004). Transfection Pathways of Nonspecific and Targeted PEI-Polyplexes. Gene Ther. Mol. Biol. 8, 369–384.
Konopka, K., Overlid, N., Nagaraj, A. C., and Düzgüneş, N. (2006). Serum Decreases the Size of Metafectene-And Genejammer-DNA Complexes but Does Not Affect Significantly Their Transfection Activity in SCCVII Murine Squamous Cell Carcinoma Cells. Cell Mol. Biol. Lett. 11, 171. doi:10.2478/s11658-006-0015-5
Lungwitz, U., Breunig, M., Blunk, T., and Göpferich, A. (2005). Polyethylenimine-based Non-viral Gene Delivery Systems. Eur. J. Pharm. Biopharm. 60, 247–266. doi:10.1016/j.ejpb.2004.11.011
Mahato, M., Kumar, P., and Sharma, A. K. (2013). Amphiphilic Polyethylenimine Polymers Mediate Efficient Delivery of DNA and siRNA in Mammalian Cells. Mol. Biosyst. 9, 780–791. doi:10.1039/c3mb25444e
Mahato, M., Rana, G., Kumar, P., and Sharma, A. K. (2012). Tetramethylguanidinium-polyallylamine (Tmg-PA): A New Class of Nonviral Vector for Efficient Gene Transfection. J. Polym. Sci. A. Polym. Chem. 50, 2344–2355. doi:10.1002/pola.26017
Mahato, M., Yadav, S., Kumar, P., and Sharma, A. K. (2014). Synthesis and Evaluation of Tetramethylguanidinium-Polyethylenimine Polymers as Efficient Gene Delivery Vectors. Biomed. Res. Int. 2014, 1–11. doi:10.1155/2014/459736
Mali, S. (2013). Delivery Systems for Gene Therapy. Indian J. Hum. Genet. 19, 3. doi:10.4103/0971-6866.112870
Ogris, M., Steinlein, P., Kursa, M., Mechtler, K., Kircheis, R., and Wagner, E. (1998). The Size of DNA/transferrin-PEI Complexes Is an Important Factor for Gene Expression in Cultured Cells. Gene Ther. 5, 1425–1433. doi:10.1038/sj.gt.3300745
Pichon, C., Gonçalves, C., and Midoux, P. (2001). Histidine-rich Peptides and Polymers for Nucleic Acids Delivery. Adv. Drug Deliv. Rev. 53, 75–94. doi:10.1016/s0169-409x(01)00221-6
Priyam, A., Nagar, P., Sharma, A. K., and Kumar, P. (2018). Mussel-inspired Polydopamine-Polyethylenimine Conjugated Nanoparticles as Efficient Gene Delivery Vectors for Mammalian Cells. Colloids Surf. B: Biointerfaces 161, 403–412. doi:10.1016/j.colsurfb.2017.10.063
Sainlos, M., Hauchecorne, M., Oudrhiri, N., Zertal-Zidani, S., Aissaoui, A., Vigneron, J.-P., et al. (2005). Kanamycin A-Derived Cationic Lipids as Vectors for Gene Transfection. ChemBioChem 6, 1023–1033. doi:10.1002/cbic.200400344
Singh, B., Maharjan, S., Park, T.-E., Jiang, T., Kang, S.-K., Choi, Y.-J., et al. (2015). Tuning the Buffering Capacity of Polyethylenimine with Glycerol Molecules for Efficient Gene Delivery: Staying in or Out of the Endosomes. Macromol. Biosci. 15, 622–635. doi:10.1002/mabi.201400463
Tripathi, S. K., Yadav, S., Gupta, K. C., and Kumar, P. (2012). Synthesis and Evaluation of N-(2,3-dihydroxypropyl)-PEIs as Efficient Vectors for Nucleic Acids. Mol. Biosyst. 8, 1426–1434. doi:10.1039/c2mb05516c
Tseng, W.-C., Fang, T.-Y., Su, L.-Y., and Tang, C.-H. (2005). Dependence of Transgene Expression and the Relative Buffering Capacity of Dextran-Grafted Polyethylenimine. Mol. Pharmaceutics 2, 224–232. doi:10.1021/mp050007t
Xiao, Y., Shi, K., Qu, Y., Chu, B., and Qian, Z. (2018). Engineering Nanoparticles for Targeted Delivery of Nucleic Acid Therapeutics in Tumor. Mol. Ther. Methods Clin. Dev. 12, 1–18. doi:10.1016/j.omtm.2018.09.002
Yadav, S., Mahato, M., Jha, D., Ahmadi, Z., Gautam, H. K., and Sharma, A. K. (2018). Enhanced Antibacterial Activity of Tetramethylguanidinium-Conjugated Linear Polyethylenimine Polymers. Int. J. Polymeric Mater. Polymeric Biomater. 67, 889–895. doi:10.1080/00914037.2017.1393679
Yadav, S., Mahato, M., Pathak, R., Jha, D., Kumar, B., Deka, S. R., et al. (2014a). Multifunctional Self-Assembled Cationic Peptide Nanostructures Efficiently Carry Plasmid DNA In Vitro and Exhibit Antimicrobial Activity with Minimal Toxicity. J. Mater. Chem. B 2, 4848–4861. doi:10.1039/c4tb00657g
Yadav, S., Priyam, A., Mahato, M., R Deka, S., and K Sharma, A. (2014b). Ligands with Delocalized Charge Density and Hydrophobicity Significantly Affect the Transfection Efficacy of the PAMAM Dendrimer. Pharm. Nanotechnol. 2, 196–207. doi:10.2174/2211738503666150421234612
Keywords: Gene delivery, cytotoxicity, Nanoparticles, polyethylenimine, Self-assembly
Citation: Yadav S, Sharma AK and Kumar P (2021) Tight Binding of Plasmid DNA With Self-Assembled Tetramethylguanidinium Conjugated Polyethylenimine Suppresses Transfection Efficiency. Front. Nanotechnol. 3:674360. doi: 10.3389/fnano.2021.674360
Received: 01 March 2021; Accepted: 14 June 2021;
Published: 25 June 2021.
Edited by:
Aliasgar Shahiwala, Dubai Pharmacy College, United Arab EmiratesReviewed by:
Bapu Surnar, University of Miami Hospital, United StatesCopyright © 2021 Yadav, Sharma and Kumar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Santosh Yadav, c2FudG9zaC55ODdAZ21haWwuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.