
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Nanotechnol. , 22 February 2021
Sec. Nanodevices
Volume 3 - 2021 | https://doi.org/10.3389/fnano.2021.628861
This article is part of the Research Topic The DNA Molecule as an object of Nanotechnology and the Creation of Helical-Structured Metamaterials and Metasurfaces View all 5 articles
 Wenxing Huo
Wenxing Huo Wei Ling
Wei Ling Zilun Wang
Zilun Wang Ya Li
Ya Li Mingxing Zhou
Mingxing Zhou Miaoning Ren
Miaoning Ren Xueting Li
Xueting Li Jiameng Li
Jiameng Li Zhiqiang Xia
Zhiqiang Xia Xinyu Liu
Xinyu Liu Xian Huang*
Xian Huang*The appearance of next generation sequencing technology that features short read length with high measurement throughput and low cost has revolutionized the field of life science, medicine, and even computer science. The subsequent development of the third-generation sequencing technologies represented by nanopore and zero-mode waveguide techniques offers even higher speed and long read length with promising applications in portable and rapid genomic tests in field. Especially under the current circumstances, issues such as public health emergencies and global pandemics impose soaring demand on quick identification of origins and species of analytes through DNA sequences. In addition, future development of disease diagnosis, treatment, and tracking techniques may also require frequent DNA testing. As a result, DNA sequencers with miniaturized size and highly integrated components for personal and portable use to tackle increasing needs for disease prevention, personal medicine, and biohazard protection may become future trends. Just like many other biological and medical analytical systems that were originally bulky in sizes, collaborative work from various subjects in engineering and science eventually leads to the miniaturization of these systems. DNA sequencers that involve nanoprobes, detectors, microfluidics, microelectronics, and circuits as well as complex functional materials and structures are extremely complicated but may be miniaturized with technical advancement. This paper reviews the state-of-the-art technology in developing essential components in DNA sequencers and analyzes the feasibility to achieve miniaturized DNA sequencers for personal use. Future perspectives on the opportunities and associated challenges for compact DNA sequencers are also identified.
The sequence of nucleic acids in DNA contains the secret of life. Therefore, determining the sequence of the four bases, namely adenine, guanine, cytosine, and thymine, is crucial for biological research as well as numerous applied fields such as medicine, virology, anthropology, and forensics (Heather and Chain, 2016; Shendure et al., 2017). The first-generation DNA sequencing technology named “Sanger method” contributes to the birth of commercial DNA sequencers in 1986 (Hunkapiller et al., 1991), followed by the second-generation sequencing technologies (also known as the next-generation sequencing, NGS) with high-throughput, low measurement cost, and short read length (Bramhachari and Naidu, 2019; Kumar et al., 2019). Emerging third-generation sequencing technologies represented by two single-molecule sequencing approaches based on zero mode waveguides (ZMW) and nanopores demand no template amplification while offering real-time, long-read sequencing with potential capability to achieve portable and readily DNA sequencing.
The third generation of sequencing technologies represented by the MinION developed by Oxford Nanopore Technologies (ONT) and the Sequel II by PacBio have demonstrated vital use in surveillance such as Zika (Faria et al., 2016), African swine fever (Jia et al., 2020), Ebola (Quick et al., 2016), and the very recent COVID-19 (Lopez-Alvarez et al., 2020), whose genomes can be sequenced within 14 h after receiving the sample (Fauver et al., 2020). Sequencing platforms such as MinION have also been introduced to educate medical students (Cervantes et al., 2020) through a brief hands-on workshop, indicating the possibility to rapidly cultivate large numbers of skillful operators in a short period of time. These commercial systems reveal new possibilities to achieve rapid point-of-care or in-field testing for personal applications. In addition, easy access to DNA sequencers for quickly determining personal health conditions and identify species of analytes may prevent the outbreak of public health crises and global pandemics.
Despite great technical achievement in the third generation of sequencing technology, both the nanopore-based and ZMW based technologies possess individual shortages. For example, the systematic error rates of the individual reads (Jain et al., 2018; Logsdon et al., 2020) limit the application of the MinION, while the PacBio systems are not portable in size. The operation of these systems is still not optimized for personal use as other commercially available consumer electronics. The future development path of DNA sequencers may follow the same route as many other biological and medical analytical systems that were originally bulky in sizes and eventually became portable, wearable, and implantable with high measurement precision. The development of relevant technologies such as nanotechnology, microelectronics, and microfluidics may eventually lead to more miniaturized DNA sequencers. Table 1 summaries the features of ONT MinION, the PacBio Sequel II and the future miniaturized sequencers. This paper reviews the state-of-the-art technology in developing essential components in DNA sequencers and analyzes the feasibility to achieve miniaturized DNA sequencers for personal use. Future perspectives on the opportunities and associated challenges for compact DNA sequencers are also identified.
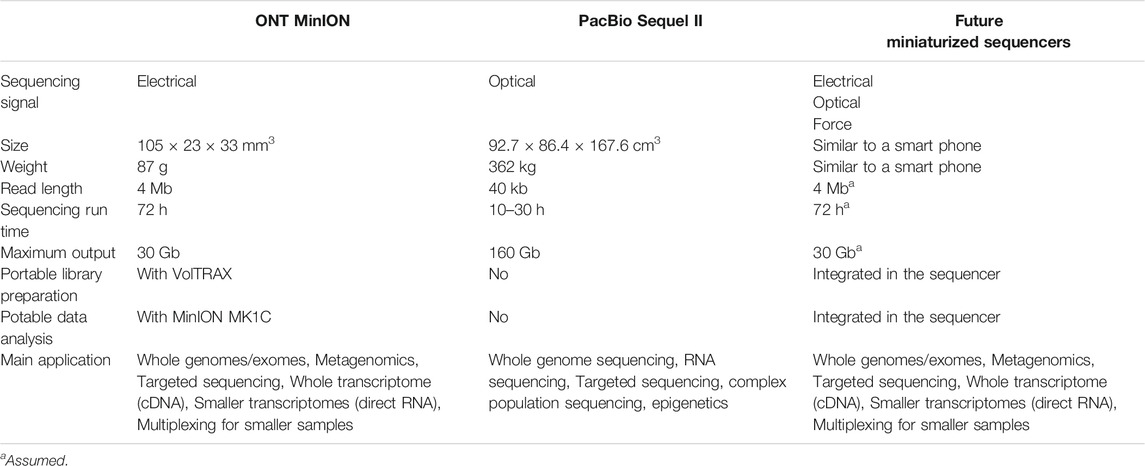
TABLE 1. The comparation of ONT MinION, the PacBio Sequel II and the future miniaturized sequencers.
Before discussing the possibilities of miniaturized DNA sequencers for personal use, it is necessary to dissect the sequencing systems to analyze the opportunities and challenges for each component. Despite different principles for DNA sequencers, a typical DNA sequencing process may generally involve steps such as library preparation, DNA sequencing, and data processing. Correspondingly, a miniaturized DNA sequencer may contain essential components such as microfluidics, nanoprobes, acquisition circuits/systems, data analysis, and display (Figure 1). Library preparation is a vital process to ensure high-quality results. The protocols for library preparation of different sequencing platforms contain major steps such as quantification, mechanical fragmentation, enzymatic reactions, purification, and amplification (Hess et al., 2020). Microfluidics that can parallelly and automatically handle minimum volumes of samples is favorable for portable library preparation. The nanoprobes that refer to sensors with similar scales as DNA output specific signals in response to different nucleobases. Large numbers of nanoprobe arrays result in high-throughput sequencing. Different types of nanoprobes may generate varied electrical, chemical, force, and optical signals, demanding corresponding signal acquisition systems that can be as simple as a USB disk. Data analysis of sequencing results can either be processed by a computer or a dedicated signal processor. Despite that a fully miniaturized system has not yet been achieved, some existing microfluidics and nanoprobes have already demonstrated the tendency toward a fully portable system for personal use, while many critical techniques such as signal acquisition and data analysis circuits remain to be developed.
As for library preparation in miniaturized DNA sequencing, the best solution should be the miniaturized, integratable, and high-throughput microfluidic devices, which have been widely used in chemical synthesis, biopharmaceutical, medical diagnosis, and tissue culture (Zhong et al., 2019). Microfluidic devices have been applied in NGS with the major research effort to reduce the cost and reagent consumption, where the chamber used for mixing, cleaning, and reaction can be up to the micro-nano liter level. The strategies and technologies may also be applied to single-molecule sequencing. Tan et al., (2013) demonstrated a microfluidic device based on multilayer soft lithography. The device allows parallelly introducing 16 samples, and enable enzymatic reactions as well as purification steps. Kim et al., (2017) implemented Illumina's complete Nextera protocol on a similar automatic microfluidic system that included enzymatic reaction, purification, and amplification steps for minimized sample input (Figure 2A). Murphy et al., (2020) presented a microfluidic droplet-based system for NGS library preparation, which reduced reagent consumption by 10× and allowed an extremely low DNA input requirement of 10−11 g/library. Ruan et al., (2020) also developed an automatic sample preparation platform, Digital-WGS, based on digital microfluidics for high-performance single-cell whole-genome sequencing (WGS). Digital microfluidics has also been adopted by several companies for automatic library preparation, such as the Illumina NeoPrep and the NuGEN Mondrian system that based on electrowetting (Tan et al., 2013; Coelho et al., 2017). The commercially available automated library preparation system VolTRAX by ONT (Figure 2B) is also based on electrowetting (Li and Kim, 2020). The compact system that can be powered by USB has a volume of 58 × 64 × 134 mm3 and a weight of 301 g. The VolTRAX allows users to develop their own protocols for fluorescence detection or polymerase chain reaction (PCR). Agrawal et al., (2020) presented a 3D printed poly(dimethylsiloxane) (PDMS) device for DNA extraction, which was specifically developed for long-read genomics (Figure 2C). All examples above indicate that the microfluidics technology has been well-developed with a variety of microfluidic components, which can be readily used for library preparation for both the second and the third generations of DNA sequencing technology. More systematic reviews about microfluidic components suitable for DNA sequencers have been provided elsewhere (Hess et al., 2020). Although the microfluidics is well developed, it still needs further research on more sophisticated functions and technologies to fully integrate with the miniaturized sequencers.
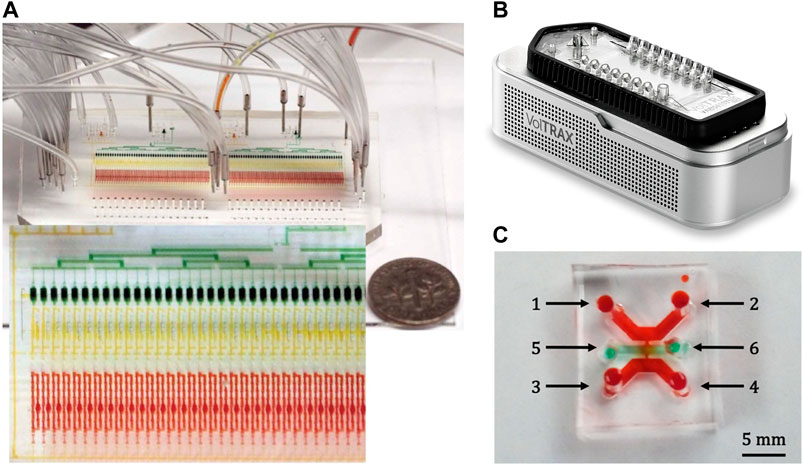
FIGURE 2. Microfluidics systems for DNA sequencing. (A) The 96× 36 nl microfluidic sample preparation device filled with food coloring to highlight features: the reactor (red), filter (yellow), and reservoir (green) units. Reproduced from (Kim et al., 2017) with permission from Nature Publishing Group. (B) The photo of the VolTRAX system from the website of ONT. (C) DNA extraction device for long-read genomics. Reproduced from (Agrawal et al., 2020) with permission from American Chemical Society.
After the library preparation process, the integrated microfluidic device can transport the DNA samples to the nanoprobes, where the sequencing data is measured from the interaction between them. The nanoprobes have been extensively explored in varied contexts. They possess similar scales as DNA chains and measure specific DNA signals through optical, electrical, and mechanical approaches. Most nanoprobe devices have already possessed miniaturized formats, and thus can be readily integrated into miniaturized DNA sequencers. However, different sensing principles require varied peripheral components, leading to different complexity for miniaturizing these components and feasibility to achieve high-throughput personal DNA sequencing. Table 2 summaries the possible nanoprobes for miniaturized DNA sequencers, including ZMWs, nanopores, nanopipettes, and field-effect transistors (FET). Among them, the nanopore might be the most promising one to achieve the goal of a miniaturized DNA sequencer for personal use. The following section reviews the state-of-the-art techniques for nanoprobes and analyses the possibility to achieve miniaturized DNA sequencers based on different techniques.
The ZMW devices contain nanophotonic confinement structures that enhance the fluorescent intensity on the bottom of the structures while minimizing background light interference from free-flowing molecules labeled with fluorescent dyes. The ZMW effect becomes significant in nanowells with a diameter of 100 nm and a height of ∼100 nm. When affixing each of the nanowells with polymerase on the bottom (Figure 3A), the polymerase can synthesize double strand DNA using templates from measured single strand DNA and materials from four DNA bases labeled with different fluorescent dyes in the surrounding solution. The ZMW effect allows observation of fluorescent color from the DNA base in use while minimizing the influence of background fluorescent. The basecalling is then conducted to identify the nucleotides according to the corresponding fluorescence of the dye. A typical single molecule real-time (SMRT) sequencing procedure is shown in Figure 3B (Rhoads and Au, 2015). A fluorescently labeled nucleotide is incorporated into the DNA template by the polymerase, then the corresponding fluorescence (yellow for base C as an example here) signal arises. When the fluorescent tag is cleaved off and diffuses out of the ZMW, the fluorescence pulse ends and the polymerase translocates to the next position. The next nucleotide is then incorporated into the DNA template by the polymerase to repeat the same steps.
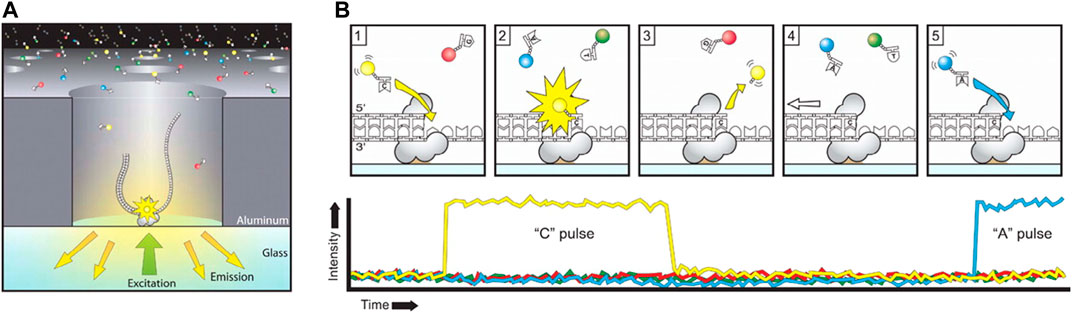
FIGURE 3. The principle of PacBio sequencing. (A) A schematic diagram of the SMRT sequencing. The adaptor binds to a polymerase immobilized at the bottom of a ZMW, where the light excitation and emission occurs. (B) The SMRT sequencing procedure. Each of the four nucleotides is labeled with a different fluorescent dye, which produces a light pulse when the corresponding nucleotide is held by the polymerase. Reproduced from (Rhoads and Au, 2015) with permission from Elsevier.
The ZMW technique has been developed for almost 20 years. The pioneering work of ZMW can be dated back to 2003, when Levene et al., (2003) demonstrated that ZMW can be used to optically detect individual molecules that diffuse into nanowells, which limited the numbers of analytes to only a few Zeptoliters (Figure 4A). Zambrana-Puyalto et al., (2019) fabricated hybrid metal-dielectric nanoslots on a bilayer film of silicon and gold on a transparent glass substrate (Figure 4B). The optical response has been measured at three different wavelengths, including 587, 633, and 676 nm. Enhanced ZWM effect was observed in the near-infrared spectral region. Wu et al., (2019) found that undercutting can significantly enhance the fluorescence signal of Au ZMWs at the cost of increasing the excitation volume. On the other hand, reducing the radial size of the ZMW can both reduce the excitation volume and enhance the fluorescence. Klimov (2019) proved that selection of the permittivity of the metal film and the dielectric substrate could cause the excitation of "leakage" surface plasmon wave and "forbidden light", which greatly changed the relationship between the radiative and nonradiative decay channels (Figure 4C). As a result, the "leakage" wave significantly increased the emissivity into the dielectric substrate. The discovered effect can be used to design smart ZMW devices with high-efficiency single-molecule detection. Martin et al., (2016) studied the effect of gold and aluminum ZMW on the fluorescence of two fluorophores, Atto647N and Atto565. This work showed that 200 nm gold ZMW was more suitable for single-molecule fluorescence studies using red light, while aluminum is more suitable for green light.
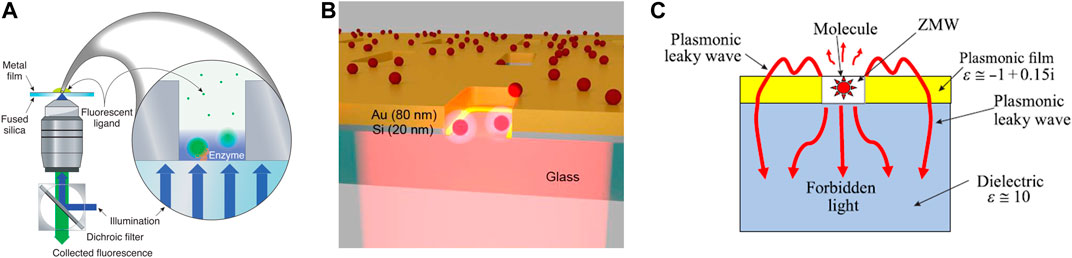
FIGURE 4. Representative zero-mode waveguide devices for DNA sequencing. (A) Experimental setup for detecting translocation of labeled DNA molecules. The illuminating light is shown in blue. Emitted light (fluorescence) is shown in green. Reproduced from (Levene et al., 2003) with permission from AAAS. (B) The multilayer structure of a hybrid metal-dielectric plasmonic ZMW for enhanced single-molecule detection. Reproduced from (Zambrana-Puyalto et al., 2019) with permission from Royal Society of Chemistry. (C) ZMW for effective single-molecule detection by excitation of leaky plasmonic waves and forbidden light. Reproduced from (Klimov, 2019) with permission from American Physical Society.
The sequencers based on the ZMW effect have been successfully commercialized by PacBio. Each sequencer contains an SMRT cell with up to eight millions of ZMW nanowells, each of which is 100 nm in diameter and 70 nm in depth (Foquet et al., 2008). Despite the small footprint of individual SMRT cells, the peripheral components including the light source and photodetectors are still very bulky. As a result, the latest Sequel II System from PacBio has an overall dimension of 92.7 × 86.4 × 167.6 cm3 and a weight of 362 kg (PacBio, 2020). The major obstacle for achieving a small ZMW sequencer may come from the optical system that includes light sources and detectors. Potential solutions have been presented later in this review.
Nanopore sequencing is a label-free and amplification-free approach for single-molecule recognition and detection of DNA bases. The specific signal from the interaction of nanopore with nucleotides provides base-specific information of DNA and even RNA (Shen et al., 2020). Nanopore sequencing can be achieved through different sensing principles with varied nanopore materials.
A typical nanopore device measures the ion blockade current during the translocation movement of DNA through biological nanopores formed by proteins on lipid bilayers. The fundamental concept has been presented in Figure 5A. The translocation of the DNA chain through the nanopore leads to blockage of the path of ions, resulting in a significant decrease in current measured by electrodes between the two sides of the nanopore. The translocation events that exhibit as spikes and pulses in the measured ionic current are specific in frequency, dwell time, and magnitudes among different DNA bases. To achieve high-resolution sequencing, the diameter of the nanopore should be comparable to that of the DNA molecule (1–2 nm), and the thickness should be smaller than the distance between two adjacent bases (∼0.5 nm) (Gasparyan et al., 2019). The MinION sequencer developed by ONT may be the first device that is close to the format of a fully miniaturized system. It is based on protein nanopores and ionic current sensing. The device features a small dimension of 10× 3.2 × 2 cm3 and a weight of 90 g. It is powered by a USB and is connected to computers for data processing. This device does not contain an integrated microfluidic system for automatic sample handling. Another portable system SmidgION, which is also developed by ONT, can work together with smartphones for more portable applications.
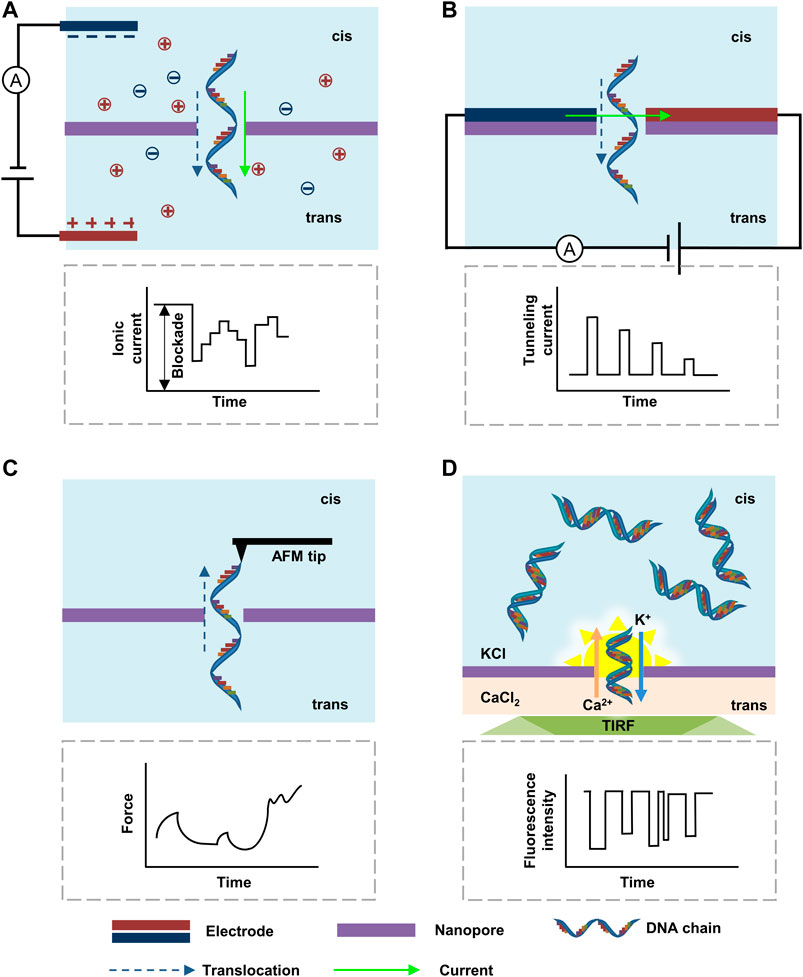
FIGURE 5. Nanopore sequencing based on (A) ionic current blockade; (B) tunneling current; (C) force; (D) electrode-free DiffusiOptoPhysiology.
Besides the measurement of the changes in the ionic current, the transversal tunneling current between two electrodes located on the same side of a nanopore with a distance similar to the size of the nanopore provides higher resolution for DNA sequencing (Figure 5B). As the resistance of nucleotide is much larger than that of the paired metallic electrodes, an analogical metal-“dielectric”-metal junction is formed around the nanopore during the translocation, causing varied tunneling current whose amplitude is corresponding to the nucleotide types. The transverse tunneling current is at the scale of nano-ampere that is approximately three orders higher than the ionic blockade current. Therefore, it can be measured at high frequencies without concern about the problem of translocation speed, leading to a much higher sequencing speed than the ionic current method (Gasparyan et al., 2019). However, as the mass-production of paired electrodes with nanogap remains challenging, the application of high throughput transversal tunneling current monitoring is still unpractical.
Apart from the collection of electrical signals, optical and force signals measured at the atom level have also been achieved (Zwolak and Di Ventra, 2008). The force signal that arises from the steric effect during the translocation of DNA through a small nanopore (Figure 5C) was first performed by Keyser et al., (2006) with the combing of optical tweezers and ionic-current detection. The resolution could be further improved by using atomic force microscopy (AFM). Nelson et al., (2014) simultaneously measured the force and the blockade current with a customized AFM. Moreover, Si et al., (2020) demonstrated that the DNA transport through nanopores could be manipulated by AFM, which slowed down the translocation velocity of DNA to ∼100 nm s−1, allowing the signal acquisition circuit to measure with higher precision and longer response time. The AFM with antibodies attached to its tip can also be used to detect methylation in individual DNA strands (Marszalek, 2010). The high resolution and tunable speeds make AFM-based force signal detection a promising method for DNA sequencing. However, the AFM systems are normally complicated and cumbersome, demanding further miniaturization and integration. In addition, the dependence of tips for the measurement of force signals brings huge challenges to the realization of massively parallel sequencing.
An optical sensing method defined as DiffusiOptoPhysiology (DOP) was created (Wang et al., 2019). The configuration of DOP included asymmetric electrolyte buffers, where the cis side was filled with KCl, Fluo-8, and ethylene diamine tetraacetic acid (EDTA), and the trans side was filled with CaCl2 (Figure 5D). Driven by the chemical gradient, Ca2+ and Fluo-8 diffused through the nanopore and subsequently bound together to form FluoCa, which emitted fluorescence around the nanopore for sensing of small molecules, macromolecules, and biomacromolecules. The image was recorded by an inverted microscope equipped with a total internal reflection fluorescence (TIRF). By further optimizing electrolytes and channel sizes, DOP may be able to make parallel measurement of thousands of nanopores with a low cost of < $1 in consumables for single use.
The nanopores can be constructed based on biological materials and solid-state materials. Biological nanopores offer smaller pore size and adjustable DNA translation velocity, however, these nanopores may suffer from instability and short longevity due to the degradation of proteins. As a result, alternative solutions based on solid-state nanopores (SSNs) have been proposed with high robustness and reliability as well as long lifespan. These nanopores also offer better compatibility to the fabrication and integration processes with other on-chip electronic devices. SSNs can be made through silicon-based membranes, two-dimensional membranes, and polymer membranes. SSNs can be commonly achieved by drilling through electron or ion beam (Figure 6A). The Si3N4 SSNs showed the excellent signal-to-noise ratio (SNR) due to relatively low noise at high frequencies when compared with biological nanopores and MoS2 SSNs (Fragasso et al., 2020). The noise can be further reduced by using sapphire as the supporting substrate of SSNs (Xia et al., 2020).
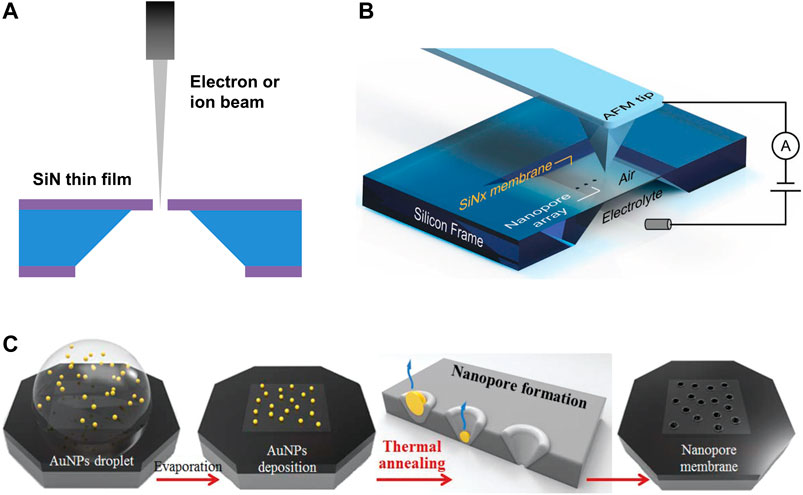
FIGURE 6. Silicon-based nanopores. (A) Schematic diagram of the nanopore fabrication process by electron/ion beam drilling. (B) Schematic diagram of nanopore fabrication via tip-controlled local breakdown. Reproduced from (Zhang et al., 2019) with permission from John Wiley and Sons. (C) Schematic diagram of the scalable fabrication process of Si3N4 nanopores via thermal annealing of Au nanoparticles. Reproduced from (Park et al., 2018) with permission from Royal Society of Chemistry.
In 2001, the first solid-state nanopores were fabricated on a free-standing Si3N4 film by a focused ion beam (FIB) (Li et al., 2001). Storm et al., (2003) further used the transmission electron microscope (TEM) to drill nanopores with a diameter of a few nanometers. However, the processing speed and cost are not acceptable for large-scale nanopore arrays. Another promising alternative method to achieve SSNs is based on controlled dielectric breakdown (CBD). Yanagi et al., (2017) used the CBD approach to create nanopores on sub-10-nm-thick Si3N4 membranes. Smaller nanopores were created in thinner membranes due to the smaller increment of breakdown current. Carlsen et al., (2017) demonstrated SSNs localization by CBD with selective membrane thinning. The results proved that the helium ion beam could reduce the thickness of the Si3N4 film to about 1 nm with sub-10 nm accuracy, indicating great promise for DNA sequencing that required an enhanced signal-to-noise ratio. Yamazaki et al., (2018) presented an electrokinetic process that combined laser-controlled etching and dielectric breakdown for fabricating Si3N4 nanopores of arbitrary dimensions as small as 1–2 nm in diameter and thickness. Zhang et al., (2019) proposed a tip-controlled local breakdown method with a conductive AFM tip for fast nanopore formation with diameter close to or under 5 nm on Si3N4 nanomembranes (10–20 nm in thickness) (Figure 6B). The nanopore formation was 100× faster than classic breakdown. However, the abrasion of AFM tips may induce large variations in the nanopores, resulting in difficulties in the scalable fabrication. Another scalable fabrication method for nanopores via thermal annealing of Au nanoparticles (AuNPs) was demonstrated by (Park et al., 2018). When the annealing temperature was close to the melting point of Au, the surface melting of the AuNPs resulted in a quasi-liquid (or quasi-solid) state of Au. Then the evaporation of Au on the interface of the substrate and the melted AuNPs led to direct formation of nanopores. This thermal annealing approach enabled one-step fabrication of Si3N4 nanopores with adjustable diameters of 8–63 nm (Figure 6C). Moreover, massive production of uniform SSNs is still a big challenge for above mentioned methods due to low throughput, nonuniformity, and high cost.
Two-dimensional (2D) materials with thickness comparable to the distance between two neighboring DNA bases have also been used to construct nanopores (Su et al., 2021). The fabrication processes of 2D nanopores are compatible with other silicon-based SSNs. Graphene has been considered to be an attractive nanopore material due to its superior physical and chemical properties. Garaj et al., (2010) and Schneider et al., (2010) used graphene sheets 2–25 nm in thickness for double-stranded DNA detection. Garaj et al., (2013) produced a graphene nanopore that has an inherent resolution of ≤0.6 nm (Figure 7A). To further improve detection sensitivity, a hydrophilic substance has been applied to the graphene pores, resulting in noncovalent interaction between the nanopores and the DNA. As a result, the speed of DNA translocation has been reduced to allow more time for the sensing circuit to conduct precise measurement (Schneider et al., 2013). In addition to graphene, other 2D materials such as molybdenum disulfide (MoS2) have also been used for DNA sequencing. Graf et al., (2019) implemented a single layer MoS2-based FET device with a nanopore for simultaneous detection of the transverse current and the ionic current (Figure 7B). To address the issues of the small size of MoS2-based nanopore devices, a large-area fabrication protocol including a wafer-scale monolayer MoS2 synthesis, Si/SiNx substrate fabrication, and wafer-scale transfer printing was demonstrated by Thakur et al., (2020) (Figure 7C). The MoS2 nanopores were created by electrochemical reaction, which has the potential for scalable fabrication of nanopores. The 2D nanopores is also most likely to achieve the tunneling current detection due to the advanced nanofabrication technologies. However, the repeatability and stability of the 2D nanopores should be further improved.
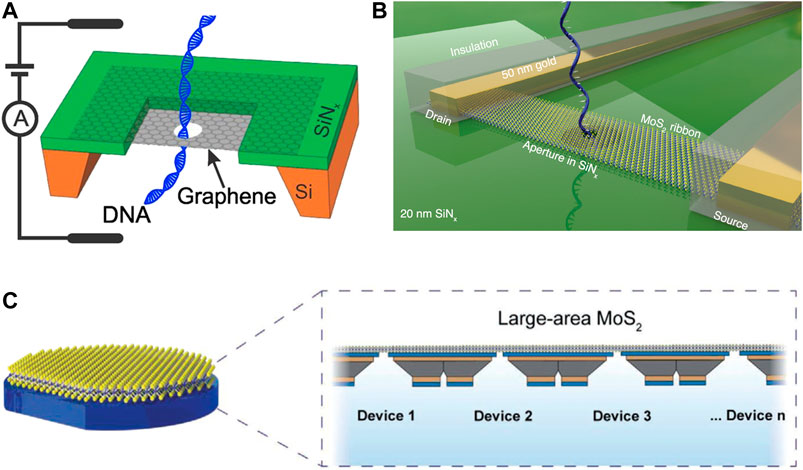
FIGURE 7. 2D nanopores for DNA sequencing. (A) DNA translocating through a nanopore in graphene mounted on a SiNx frame. Reproduced from (Garaj et al., 2013) with permission from PNAS. (B) MoS2 FET-nanopore device for transverse current detection. Reproduced from (Graf et al., 2019) with permission from American Chemical Society. (C) Wafer-scale (7.62 cm) devices of MoS2 nanopores. Reproduced from (Thakur et al., 2020) with permission from John Wiley and Sons.
Polymer membranes show unique advantages due to their flexibility and mass production efficiency. Various fabrication techniques have been developed to generate nanoporous polymer membranes, including several top-down approaches such as lithography, pattern transfer, and track-etching and bottom-up approaches represented by immersion precipitation, phase separation, reactive pore formation, and polyelectrolyte multilayer (Bernards and Desai, 2010). Development in track-etched nanopores (Figure 8A) was summarized by Kaya and Keçeci (2020) and Ma et al., (2020). Choi et al., (2019) demonstrated sub-10 nm nanopores in freestanding SU-8 membranes via a simple, cost-effective, and scalable fabrication method, in which the initial nanopores were prepared by nanoimprint lithography (NIL), and then the pore size was finely controlled by a polymer reflow process (Figure 8B). The fabricated SU-8 nanopore membrane has been successfully used for the ion blockade current measurement. Kim et al., (2019) presented nanopores with 4–10 nm diameter on sub-10 nm-thick polyurea membrane fabricated by molecular layer deposition (Figure 8C). The highly negative surface charge density (−51 mC m−2 at pH 8) improved the detection efficiency of positively charged proteins due to the electrostatic interaction. Therefore, the polymer based nanopores can provide promising candidates for inexpensive DNA sequencers that are crucial to the large-scale application for personal use. However, the difficulty in integration with other platforms presents a significant challenge to the implementation of miniaturized sequencers.
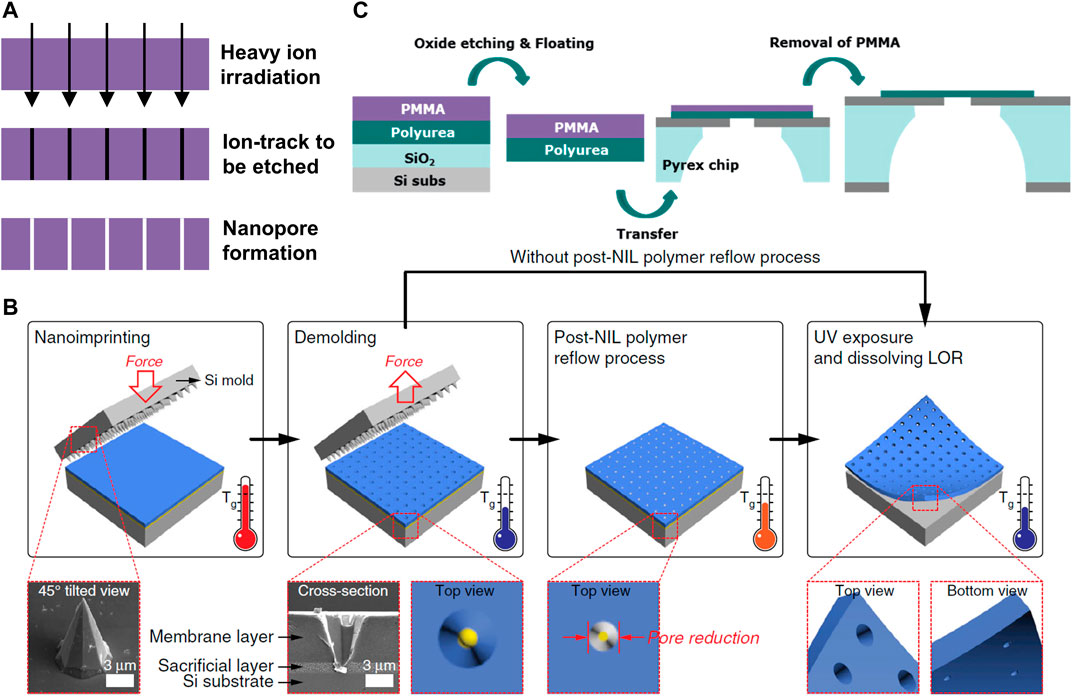
FIGURE 8. Polymer-based nanopores. (A) Fabrication process of track-etching technology. (B) The scalable process of nanopores in a freestanding polymer membrane via NIL and polymer reflowing. Reproduced from (Choi et al., 2019) with permission from Nature Publishing Group. (C) Fabrication process of the polyurea nanopore. Reproduced from (Kim et al., 2019) with permission from Royal Society of Chemistry.
Compared with biological and solid-state nanopores, nanopipettes have great potential for DNA detection due to its stability, ease of manufacturing, and good compatibility with other technologies (Figure 9A). Steinbock et al., (2010) reported the detection of λ-DNA with a 50 nm nanopipette. The translocation duration of unfolded λ-DNA was 1.56 ms, giving only 0.03 μs per base pair. The folded state of single λ-DNA molecules was identified, indicating that this simple and cheap method is suitable for single molecule characterization. Xu et al., (2017) proposed a low-cost and convenient approach to shrink the pore size of nanopipettes through sodium silicate hydrolysis. They successfully reduced the pore size of the nanopipette from 42 nm to 6.4 nm. The magnitude of the blocking current measured in the 6.4 nm pore was two times higher than that of the 42 nm pore. Crick et al., (2015) coated a nanopipette with graphene nanopores to detect DNA. The nanopore size could be tuned from fully closed to fully open (25 nm) by the electroetching method. The translocations of DNA through varied devices were studied, where pore with smaller sizes showed more subfeatures of DNA and the opened pore was able to observe the trends. Youn et al., (2016) used nanopipettes functionalized with DNA as probes to selectively detect single-stranded DNA. The translocation duration of poly(dA)20 through non-functionalized nanopipettes was 0.14 ms, while that in nanopipettes functionalized with complementary sequences was 0.45 ms. The longer duration time was due to the interaction between probe and target single-stranded DNA, which is dependent on the base pair binding strength. The nanopipette can also be used to fingerprint the ribosomes and polysomes from the peak amplitude and the dwell time (Raveendran et al., 2020). Despite the advantages of long read length, fast reading speed, and low cost, the achievement of miniaturized nanopipette arrays with uniform pore sizes is still quite challenging. Massively parallel sequencing techniques based on arrays of nanopipette is difficult to achieve.
FET is a novel alternative method for DNA sequencing with advantages of huge potential for miniaturization and seamless integration with complementary metal oxide semiconductor (CMOS) processes (Figure 9B). Wasfi et al., (2020) investigated z-shaped FET with nanopores for rapid DNA detection by first-principle transport calculations. The electronic signature in transmission spectrum and current can be identified for each DNA base. Three kinds of CMOS-based label-free sensors for future personalized DNA sequencing were presented by Jiang et al., (2016), including a high-sensitivity ion-sensitive FET sensor with pH-to-time-to-voltage conversion, a dual-mode sensor that combined imaging and chemical sensing for high accuracy, and a THz metamaterial sensor for electrical resonance detection. Islam et al., (2020) summarized VLSI biosensors for nucleotide detection and DNA sequencing, and proposed a new approach using nanowire FET to realize rapid measurement of long-strand DNA with high sensitivity and accuracy. The proposed approach requires a motor protein to process the nucleotide and act as a biological read head that may suffer from the stability issue. Moreover, such device has only been proved through simulation rather than making into a real device. Thus, it is still impractical to only use FET to sequence DNA. However, it may still be feasible to combine the strengths of FETs and nanopores for miniaturized DNA sequencers.
When the DNA chains translocate through the nanopores, the electrical signals generated from the interaction between DNA and nanopores need to be collected by circuits and processed by processors. The acquisition of electrical signals has already been accomplished by miniaturized circuits. Using sophisticate integrated chips, the signal acquisition circuit within the MinION device has been reduced to the size as a USB disk. Meilitech corporation has developed a hand-held pA-level microcurrent detection system, which has a dimension of 120 × 63 × 20 mm3 and a weight of 147 g. The signal acquisition circuit can be further miniaturized using dedicated microchips or more compact components, such as ingestible devices (Van Helleputte et al., 2020) that integrated signal sensing, wireless communication, power supply, and power management in small footprint. Comparing with the ingestible devices, the wireless communication and power modules are not the primary concerns for the DNA sequencing circuit. Thus the signal acquisition circuit in miniaturized DNA sequencer can be allowed to occupy more device space for more precise and more rapid signal sensing.
The development of “NGS processors” for NGS sequencing may also offer an opportunity to miniaturize DNA sensing circuits. Wu et al., (2017) presented an integrated system-on-a-chip (SoC) data processor fabricated using a 40-nm CMOS technique. The processor that was only 2.8 × 2.8 mm2 in dimension consumed 135 mW at 200 MHz operation frequency under a supply voltage of 0.9 V. The analysis time decreased from hours to 10 min with 1 GB external memory, achieving 43065× and 8971× (3208× and 402×) higher energy efficiency (throughput-to-area ratio) than the high-end CPU and GPU solutions, respectively. With such a high processing capability and small size, it has great potential to be used in portable sequencing devices. Researchers have also proposed the field-programmable gate array (FPGA) accelerator for nanopore sequencing. Comparing with the conventional analysis based on the CPU alone, Wu et al., (2020) designed and implemented a low-power, real-time FPGA to accelerate the basecalling. By introducing the FPGA accelerator, which worked in series with the CPU-accelerator through a PCIe link, new measurement events were first sent to FPGA for computation and aggregation, and then returned to the CPU, allowing continuous flow of measurement while avoiding multiple parallel assignment solely to the CPU. With this improvement, one human genome can be analyzed in less than 5 min, a 172× improvement over the CPU-only basecaller. Moreover, this FPGA-accelerated system achieved a measured energy efficiency of about 1000× better than the CPU alone.
Besides the electrical signals, the future miniaturized sequencers may be also based on optical and force signals. Therefore, other miniaturized components for the acquisition of these signals, imaging systems for optical and AFM for force, need to be investigated for greater chances to realize the miniaturized DNA sequencers.
For sequencers that rely on optical detection, the imaging systems are the limiting factors for the miniaturization of the sequencers. The light sources of imaging systems are generally supplied by lasers that generate high intensity and narrow bandwidth monochromatic light. Di Sieno et al., (2017) presented a miniaturized pulsed laser source based on the combination of a CMOS application specific integrated circuit (ASIC) driver coupled to a heterostructure laser diode. The laser source could provide short and high energy (∼100 ps, ∼0.5 nJ) pulses at up to 1 MHz with a laser transmitter as small as 12 × 6 mm2 (Figure 10A) under a power consumption of 50–160 mW. Another compact miniaturized laser module emitting 1.6 W of laser light with wavelength around 576 nm was demonstrated by Sahm et al., (2018). The entire laser module can be packaged into a small dimension of 47 × 76 × 15 mm3. In addition to the design of the laser module, the utilization of advanced materials would also promote the development of miniaturized DNA sequencers. Liao et al., (2019) summarized the advances of halide perovskite-based miniaturized solid-state laser. The advantages of high photoluminescence quantum yield, low non-radiative recombination rates, large and balanced charge-carrier mobilities, and high gain coefficients make the halide perovskite materials suitable for the application of lasers.
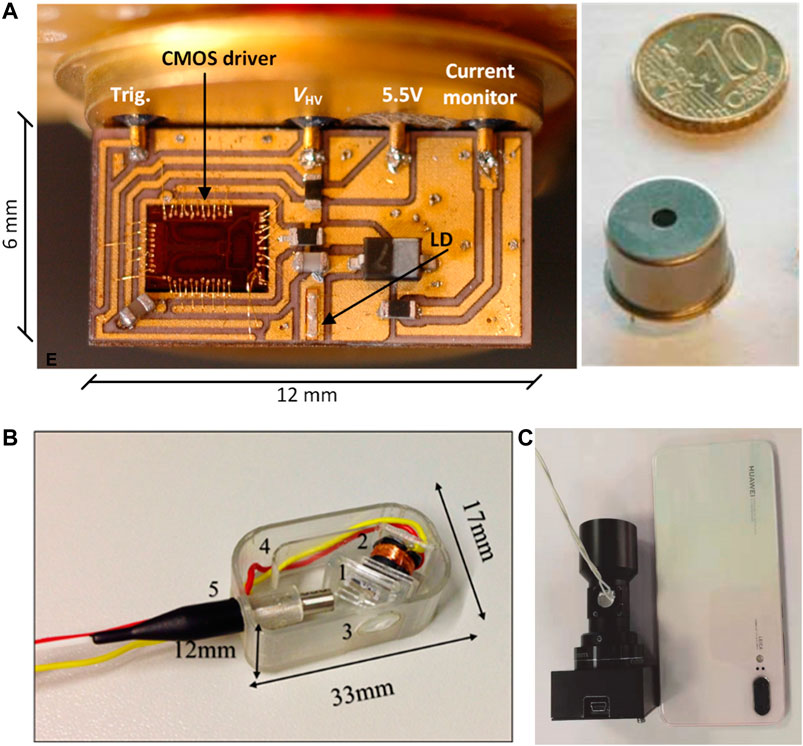
FIGURE 10. (A) Images of the pulsed laser transmitter and a packaged laser transmitter in comparison with the size of a coin. Reproduced from (Di Sieno et al., 2017) with permission from SPIE. (B) Three-dimensional-printed miniaturized confocal imager. The shown components are (1) scanner, (2) coil, (3) lens, (4) electrical interconnect channel, and (5) input fiber. Reproduced from (Savas et al., 2018) with permission from SPIE. (C) A miniaturized fluorescence imaging device compared with a cell phone. Reproduced from (Pan et al., 2020) with permission from World Scientific Publishing Co. Pte Ltd.
Complex imaging systems have also been developed with small footprints. A three-dimensional-printed miniaturized confocal imaging system was demonstrated with a footprint of 17 × 33 mm2 (Figure 10B), The system contained a laser scanner with 10 × 10 mm2 frame size, lens with 6 mm aperture and 10 mm focal length, and a housing to pack all components (Savas et al., 2018). The low-cost and comparative performance of the 3D-printed imager to conventional glass-lens and micro-electromechanical systems (MEMS) scanners makes this method promising for miniaturized DNA sequencers. In addition, the scanning micromirrors based on MEMS used for optical coherence tomography (Gorecki and Bargiel, 2020) may also applied to the imaging systems for the miniaturized DNA sequencers. A portable fluorescence imaging device was introduced for skin cancer detection (Pan et al., 2020), which was smaller than 3.5 × 3.5 × 9.5 mm3 with a weight of 150 g and power of 1.5 W (Figure 10C). The lateral resolution was 14 μm with an object field of 1.2 × 1.0 cm2. The miniaturized fluorescence microscope with a small dimension of 8.4 × 13 × 22 mm3and a weight of 1.9 g could be made from commercially available parts such as a semiconductor light source and a CMOS camera (Ghosh et al., 2011). Another encouraging development is a miniaturized, low-cost Raman spectrometer with CMOS based hyperspectral (HS) filter technology (Vunckx et al., 2020). By selecting the set of HS filters, it was able to achieve a Raman spectrometer with specific bands for target applications. More advances in the miniaturization of fluorescence sensing were summarized by Măriuţa et al., (2020). All above development can facilitate the miniaturized optical system to achieve the goal of fully miniaturized DNA sequencers based on optical signals.
Besides electrical or optical detection, force measurement using compact on-chip AFM may be a promising approach for miniaturized DNA sequencers. Ruppert et al., (2017) presented an on-chip AFM based on a silicon-on-insulator MEMS. The system contained electrostatic actuators for in-plane actuation, a piezoelectric transducer for out-of-plane actuation, and a microcantilever for tapping-mode atomic force microscopy. A much smaller single-chip AFM (sc-AFM), which integrated triaxial scanners, sensors, and a sharp tip was designed and fabricated by a standard 0.35 μm CMOS process with a dimension of 1.2 × 0.8 × 0.3 mm3(Strathearn et al., 2017). Furthermore, a commercially available sc-AFM named ‘nGauge’ was developed by ISCPI Corp. The device was 1,000-time smaller and 100-time cheaper than the conventional AFM systems. The nGauge AFM was able to scan an area up to 20 × 20 μm2 with root mean square noise of less than 2 nm. Moreover, an optical pickup unit (OPU) could be used as a vertical detection system in AFM for high-speed scanning (Russell-Pavier et al., 2018). The high-speed AFM with OPU could perform high-fidelity measurements at visible length scales in short timescales with a pixel rate of 2 megapixels s−1 and a tip velocity of 10 mm s−1. Despite compact AFMs for force monitoring have been developed, these systems are still based on single probe detection that may be suitable for single DNA nanoprobe, unable to satisfy the demands for high-throughput and parallel DNA sequencing. In addition, small force sensing has strict requirements to the surrounding environments, interference such as vibration, liquid flow, temperature variation may all impose significant influence on the results. Thus, comparing with optical and electrical sensing, the mechanical sensing approaches still demand further improvement to accommodate daily use environment.
The raw data from the acquisition systems need to be analyzed to provide accessible information to people. Many efforts have been made in software and algorithms for data analysis, where some might be directly applied to the miniaturized DNA sequencers. However, the bioinformatics tools need to be developed by considering the limited computing power of the miniaturized DNA sequencers. Dozens of bioinformatics tools for nanopore sequencing, including base calling, variant detection, and genome annotation and assembly, have been developed and summarized in various literatures (Krehenwinkel et al., 2019; Amarasinghe et al., 2020; Makałowski and Shabardina, 2020). Sahlin et al., (2021) presented a computational method named isONcorrect for correcting errors in the DNA sequencing data by the ONT sequencers with a median accuracy of 98.9–99.6%. Chen et al., (2021) developed an error correction and de novo assembly tool, NECAT, to overcome complex error characteristics in nanopore reads. The tool integrated methods such as progressive error correction, adaptive supporting reads and alignment selection, and two-stage assembly were included, resulting in high-quality assembly of nanopore reads. The genome assembly ability of HiFi reads in PacBio Sequel II and ultralong reads in ONT PromethION systems was compared. The results indicated that ONT ultralong reads showed higher contiguity while PacBio HiFi reads generated assemblies with considerably fewer errors (Lang et al., 2020). Wei et al., (2019) proposed a novel shapelet-based machine learning approach to process seriously overlapped blockade current signals. The learning time-series shapelet algorithm is also suitable for processing noisy nanopore data. Ni et al., (2019) developed a deep learning method to detect DNA methylation states from nanopore sequencing data and achieved over 90% accuracy for detecting DNA methylation on N6-adenine (6mA) and C5-cytosine (5mC) using only 2× coverage of reads. Phelan et al., (2019) presented online and stand-alone versions of the TBProfiler web server for rapid and accurate detection of resistance to anti-tuberculous drugs with whole-genome sequencing data obtained from the MinION. Boža et al., (2020) developed a new base caller Deepnano-biltz that could keep up with two MinION sequencers on a common laptop CPU (i7-7700HQ) without heavy GPU usage. Oliva et al., (2020) successfully transported 11 out of 23 programs to Android smartphone. It was demonstrated that 1 GB data was processed in 5–10 min by DSK/BCALM2, and in less than a minute by Kraken on the smartphone. These three tools on the smartphone were able to process files up to 16 GB. Despite many algorithms and software have been successfully migrated to the mobile platform, issues such as occupation of hardware resources and excessive heat during operation remain to be resolved. In addition, some mainstream assembly pipeline, such as Canu, cannot be compiled and tested on mobile platforms. Thus, optimization of the data analysis algorithms while improving the capability of the hardware is still needed for achieving miniaturized sequencers for personal use.
The development of sequencing technologies and related miniaturized systems above indicates the possibility to achieve miniaturized DNA sequencers for personal use. Each component of such a DNA sequencer has already been or are being miniaturized. A large number of related technologies can help to facilitate the development of miniaturized DNA sequencers. The existing portable MinION sequencer in combination with supporting systems such as VolTRAX and MK1C has demonstrated the possibility to achieve miniaturized DNA sequencers for personal use. By considering the huge technical advances that lead to miniaturization of the ENIAC that weighed 27 tons and occupied 167 m2 to small and handheld cellphones and tablets, one may foresee that the miniaturized DNA sequencers for personal use will not be an unreachable dream, but an achievable goal with the rapid development of science and technology. The future miniaturized DNA sequencers may include the following parts. First, automatic digital microfluidic devices with minimal volumes are suitable for the library preparation. The manipulation of small droplets that contains magnetic or other functional nanoparticles may be worth to be studied. Secondly, the best candidate for the nanoprobe may be large arrays of SSNs modified with proteins or enzyme to improve selectivity and switched by FET arrays. Thirdly, the heterogeneous integrated circuit with SoC and FPGA chips and optical system for signal acquisition can take full advantage of advanced technologies of additive manufacturing and flexible electronics to achieve miniaturization and high performance. Lastly, versatile bioinformatics tools for data analysis can be based on highly developed artificial intelligence algorithms.
However, there are still many technical challenges to be addressed. First, the sequencing accuracy needs to be further improved without increasing the costs, time-consumption, and operation complexity. Some applications have low requirements for nucleotide-level accuracy, for example, prenatal testing for Down’s syndrome only requires counting the number of the chromosome. However, the error rates should be reduced to 0.1% for some specific applications such as biodiversity assessments (Loit et al., 2019). A great amount of research and development has been studied to continuously improve the structure and the function of nanopores for higher sequencing accuracy (Kumar et al., 2019; Shen et al., 2020), but some methods may be at the expense of time or complexity (Lu et al., 2016). Using electron tunneling current instead of ionic current is expected to improve the resolution of DNA sequence since the paired electrodes in the transverse direction are more sensitive (Graf et al., 2019). A simple way is to introduce macromolecular crowding to increase the detection sensitivity of DNA and proteins using an SSN (Chau et al., 2020). A remarkable 1000-fold increase in the molecule count for the globular protein β-galactosidase was observed. Another simple method is to use time-varying cross membrane voltage to overcome the primary error modes (Noakes et al., 2019). The driving voltage across a Mycobacterium smegmatis porin A (MspA) nanopore varied from 100 to 200 mV with a frequency of 200 Hz to change the translocation behavior of the DNA strand. The single-passage de novo basecalling accuracy was improved from 62.7 ± 0.5% with a constant driving voltage to 79.3 ± 0.3% with a variable driving voltage. The combination of highly developed biological and solid-state nanopores can improve the of quality of the signals. The improvement may also be realized by the direct integration of nanopores with CMOS chips to decrease the physical distance and lower the noise.
In addition, signal process algorithms suitable for portable analysis should be further developed. Currently, most sequencing data were analyzed by powerful computers or the cloud, which may not be available for personal miniaturized DNA sequencers in some cases. Although ONT MinION Mk1C is small (140 × 30 × 114 mm3, 450 g), it costs more than $4,500, which is as high as other high-performance laptops. Moreover, it is not open source so that it may be difficult to develop pipelines within the system. On the other hand, Oliva et al., (2020) made effort to transport nanopore analysis tools to Android smartphones and benchmarked their performance. Although some of them have been successfully operated, issues such as high operation temperature and limited computation resources hinder the realization of portable analysis. Rapid development of computing technologies and micro/nanoelectronics may eventually lead to high performance portable computer with low power consumption for portable DNA analysis. Unlike conventional von Neumann-based computers, the human brain exhibits excellent performance on solving certain problems such as face identification, speech recognition, and pattern recognition at a low power consumption (Boybat et al., 2018). The neuromorphic computing that mimics the human brain has been regarded as a promising candidate for the next-generation computing technique (Kendall and Kumar, 2020). Its features such as parallel computing, low-power consumption, high-density integration, and biocompatibility are also very suitable for DNA sequencing analysis. Intriguingly, transistors can serve as nanoprobes for DNA sequencing (Islam et al., 2020) as well as neuron nodes for neuromorphic computing (Dai et al., 2019; Ni et al., 2020; Zhu et al., 2020), offering great promise to combine the sensing and data processing together.
Lastly, the whole miniaturized sequencing platform should be easy to operate and rapidly deployable for large-scale applications. The portable DNA sequencers have been used for teaching medical students, but handling such equipment still requires basic training in biology. Users of personal DNA sequencers may prefer simple, user-friendly, and automatic procedures instead of complex manual operations and complicated technical instructions. Standard kits and protocols are also intensively needed for typical users that have only received minimal training. Furthermore, improving the durability of nanoprobes and reducing the usage of reagents is also necessary for future personal sequencers.
The development of miniaturized DNA sequencers is a multidiscipline work that involves contribution from microfluidic devices, nanoprobes, signal acquisition, and data analysis. These essential components have been miniaturized in different levels, and hold the promise to achieve complete miniaturized systems. Some existing portable systems such as MinION and SmidgION have revealed the promising possibility of device miniaturization and large-scale applications. Comparing with optical and mechanical sensing approaches, electrical detection based on ionic current measurement can readily achieve high-density arrays and suitable for multichannel high-throughput sensing, making this approach favorable for more compact DNA sensing platforms. The optical and mechanical sensing mechanisms are still relatively bulky in size but have the capability to be further miniaturized. With the development of micro/nano electronics and the introduction of new ASICs designed specifically for DNA sequencing, the dimensions of both the signal acquisition and data analysis circuits can be further reduced, while the performance can be further enhanced, suggesting high feasibility of DNA sequencers for personal use. After addressing current limitations such as sequencing accuracy, portable algorithms, and easy operation, the miniaturized DNA sequencers for personal use will come true. The successful implementation of miniaturized sequencer may generate a significant impact on human society through changing people’s lifestyles and reshaping healthcare systems. It will eventually lead to a revolution in precision medicine, healthcare, and public health, and enable better capabilities of human beings in preventing diseases and extending lifespan.
WH and XH both organized the literature research and contributed to the final version of the manuscript. All authors contributed to the draft of the manuscript.
This work was supported by the Independent Innovation Fund in Tianjin University.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Agrawal, P., Reifenberger, J. G., and Dorfman, K. D. (2020). 3D printing-enabled DNA extraction for long-read genomics. ACS Omega 5 (33), 20817–20824. doi:10.1021/acsomega.0c01912
Amarasinghe, S. L., Su, S., Dong, X., Zappia, L., Ritchie, M. E., and Gouil, Q. (2020). Opportunities and challenges in long-read sequencing data analysis. Genome Biol. 21 (1), 30. doi:10.1186/s13059-020-1935-5
Bernards, D. A., and Desai, T. A. (2010). Nanoscale porosity in polymer films: fabrication and therapeutic applications. Soft Matter 6 (8), 1621–1631. doi:10.1039/B922303G
Boža, V., Perešíni, P., Brejová, B., and Vinař, T. (2020). DeepNano-blitz: a fast base caller for MinION nanopore sequencers. Bioinformatics 36(14), 4191–4192. doi:10.1093/bioinformatics/btaa297
Boybat, I., Le Gallo, M., Nandakumar, S. R., Moraitis, T., Parnell, T., Tuma, T., et al. (2018). Neuromorphic computing with multi-memristive synapses. Nat. Commun. 9 (1), 2514. doi:10.1038/s41467-018-04933-y
Bramhachari, P. V., and Naidu, B. P. (2019). Next-generation sequencing of nucleic acids: technology and applications. Polymorphism 2, 66–81.
Carlsen, A. T., Briggs, K., Hall, A. R., and Tabard-Cossa, V. (2017). Solid-state nanopore localization by controlled breakdown of selectively thinned membranes. Nanotechnology 28 (8), 085304. doi:10.1088/1361-6528/aa564d
Cervantes, J., Perry, C., and Wang, M. C. (2020). Teaching next-generation sequencing to medical students with a portable sequencing device. Perspect. Med. Education doi:10.1007/s40037-020-00568-2
Chau, C. C., Radford, S. E., Hewitt, E. W., and Actis, P. (2020). Macromolecular crowding enhances the detection of DNA and proteins by a solid-state nanopore. Nano Lett. 20 (7), 5553–5561. doi:10.1021/acs.nanolett.0c02246
Chen, Y., Nie, F., Xie, S. Q., Zheng, Y. F., Dai, Q., Bray, T., et al. (2021). Efficient assembly of nanopore reads via highly accurate and intact error correction. Nat. Commun. 12 (1), 60. doi:10.1038/s41467-020-20236-7
Choi, J., Lee, C. C., and Park, S. (2019). Scalable fabrication of sub-10 nm polymer nanopores for DNA analysis. Microsystems Nanoengineering 5 (1), 1–10. doi:10.1038/s41378-019-0050-9
Coelho, B., Veigas, B., Fortunato, E., Martins, R., Águas, H., Igreja, R., et al. (2017). Digital microfluidics for nucleic acid amplification. Sensors (Basel) 17 (7), 17071495. doi:10.3390/s17071495
Crick, C. R., Sze, J. Y., Rosillo-Lopez, M., Salzmann, C. G., and Edel, J. B. (2015). Selectively sized graphene-based nanopores for in Situ single molecule sensing. ACS Appl. Mater. Inter. 7 (32), 18188–18194. doi:10.1021/acsami.5b06212
Dai, S., Zhao, Y., Wang, Y., Zhang, J., Fang, L., Jin, S., et al. (2019). Recent advances in transistor-based artificial synapses. Adv. Funct. Mater. 29 (42), 1903700. doi:10.1002/adfm.201903700
Di Sieno, L., Nissinen, J., Hallman, L., Martinenghi, E., Contini, D., Pifferi, A., et al. (2017). Miniaturized pulsed laser source for time-domain diffuse optics routes to wearable devices. J. Biomed. Opt. 22 (8), 085004. doi:10.1117/1.JBO.22.8.085004
Faria, N. R., Sabino, E. C., Nunes, M. R., Alcantara, L. C., Loman, N. J., and Pybus, O. G. (2016). Mobile real-time surveillance of Zika virus in Brazil. Genome Med. 8 (1), 97. doi:10.1186/s13073-016-0356-2
Fauver, J. R., Petrone, M. E., Hodcroft, E. B., Shioda, K., Ehrlich, H. Y., Watts, A. G., et al. (2020). Coast-to-Coast spread of SARS-CoV-2 during the early epidemic in the United States. Cell 181 (5), 990–996.e5. doi:10.1016/j.cell.2020.04.021
Foquet, M., Samiee, K. T., Kong, X., Chauduri, B. P., Lundquist, P. M., Turner, S. W., et al. (2008). Improved fabrication of zero-mode waveguides for single-molecule detection. J. Appl. Phys. 103 (3), 034301. doi:10.1063/1.2831366
Fragasso, A., Schmid, S., and Dekker, C. (2020). Comparing current noise in biological and solid-state nanopores. ACS Nano 14 (2), 1338–1349. doi:10.1021/acsnano.9b09353
Garaj, S., Hubbard, W., Reina, A., Kong, J., Branton, D., and Golovchenko, J. A. (2010). Graphene as a subnanometre trans-electrode membrane. Nature 467 (7312), 190–193. doi:10.1038/nature09379
Garaj, S., Liu, S., Golovchenko, J. A., and Branton, D. (2013). Molecule-hugging graphene nanopores. Proc. Natl. Acad. Sci. USA 110 (30), 12192–12196. doi:10.1073/pnas.1220012110
Gasparyan, L., Mazo, I., Simonyan, V., and Gasparyan, F. (2019). DNA sequencing: current state and prospects of development. Open J. Biophys. 09 (3), 169. doi:10.4236/ojbiphy.2019.93013
Ghosh, K. K., Burns, L. D., Cocker, E. D., Nimmerjahn, A., Ziv, Y., Gamal, A. E., et al. (2011). Miniaturized integration of a fluorescence microscope. Nat. Methods 8 (10), 871–878. doi:10.1038/nmeth.1694
Gorecki, C., and Bargiel, S. (2020). MEMS scanning mirrors for optical coherence tomography. Photonics 8 (1), 8010006. doi:10.3390/photonics8010006
Graf, M., Lihter, M., Altus, D., Marion, S., and Radenovic, A. (2019). Transverse detection of DNA using a MoS2 nanopore. Nano Lett. 19 (12), 9075–9083. doi:10.1021/acs.nanolett.9b04180
Heather, J. M., and Chain, B. (2016). The sequence of sequencers: the history of sequencing DNA. Genomics 107 (1), 1–8. doi:10.1016/j.ygeno.2015.11.003
Hess, J. F., Kohl, T. A., Kotrová, M., Rönsch, K., Paprotka, T., Mohr, V., et al. (2020). Library preparation for next generation sequencing: a review of automation strategies. Biotechnol. Adv. 41, 107537. doi:10.1016/j.biotechadv.2020.107537
Hunkapiller, T., Kaiser, R. J., Koop, B. F., and Hood, L. (1991). Large-scale and automated DNA sequence determination. Science 254 (5028), 59–67. doi:10.1126/science.1925562
Islam, M. A., Datta, P. K., and Myler, H. (2020). VLSI structures for DNA sequencing—a survey. Bioengineering 7 (2), 49. doi:10.3390/bioengineering7020049
Jain, M., Koren, S., Miga, K. H., Quick, J., Rand, A. C., Sasani, T. A., et al. (2018). Nanopore sequencing and assembly of a human genome with ultra-long reads. Nat. Biotechnol. 36 (4), 338–345. doi:10.1038/nbt.4060
Jia, L., Jiang, M., Wu, K., Hu, J., Wang, Y., Quan, W., et al. (2020). Nanopore sequencing of African swine fever virus. Sci. China Life Sci. 63 (1), 160–164. doi:10.1007/s11427-019-9828-1
Jiang, Y., Liu, X., Huang, X., Shang, Y., Yan, M., and Yu, H. (2016). “Lab-on-CMOS: a multi-modal CMOS sensor platform towards personalized DNA sequencing,” in IEEE International Symposium on Circuits and Systems, May 22–25, 2016, Montreal, QC, Canada, 2266–2269.
Kaya, D., and Keçeci, K. (2020). Review—track-etched nanoporous polymer membranes as sensors: a review. J. Electrochem. Soc. 167 (3), 037543. doi:10.1149/1945-7111/ab67a7
Kendall, J. D., and Kumar, S. (2020). The building blocks of a brain-inspired computer. Appl. Phys. Rev. 7 (1), 011305. doi:10.1063/1.5129306
Keyser, U. F., Koeleman, B. N., van Dorp, S., Krapf, D., Smeets, R. M. M., Lemay, S. G., et al. (2006). Direct force measurements on DNA in a solid-state nanopore. Nat. Phys. 2 (7), 473–477. doi:10.1038/nphys344
Kim, H. J., Choi, U. J., Kim, H., Lee, K., Park, K. B., Kim, H. M., et al. (2019). Translocation of DNA and protein through a sequentially polymerized polyurea nanopore. Nanoscale 11 (2), 444–453. doi:10.1039/C8NR06229C
Kim, S., De Jonghe, J., Kulesa, A. B., Feldman, D., Vatanen, T., Bhattacharyya, R. P., et al. (2017). High-throughput automated microfluidic sample preparation for accurate microbial genomics. Nat. Commun. 8, 13919. doi:10.1038/ncomms13919
Klimov, V. V. (2019). Smart design of zero-mode waveguide Nanodevices for effective detection of single-molecule fluorescence. Phys. Rev. Appl. 12 (1), 014049. doi:10.1103/PhysRevApplied.12.014049
Krehenwinkel, H., Pomerantz, A., and Prost, S. (2019). Genetic biomonitoring and biodiversity assessment using portable sequencing technologies: current uses and future directions. Genes 10 (11), 858. doi:10.3390/genes10110858
Kumar, K. R., Cowley, M. J., and Davis, R. L. (2019). Next-generation sequencing and emerging technologies. Semin. Thromb. Hemost. 45 (7), 661–673. doi:10.1055/s-0039-1688446
Lang, D., Zhang, S., Ren, P., Liang, F., Sun, Z., Meng, G., et al. (2020). Comparison of the two up-to-date sequencing technologies for genome assembly: HiFi reads of Pacific Biosciences Sequel II system and ultralong reads of Oxford Nanopore. GigaScience 9, giaa123. doi:10.1093/gigascience/giaa123
Levene, M. J., Korlach, J., Turner, S. W., Foquet, M., Craighead, H. G., and Webb, W. W. (2003). Zero-mode waveguides for single-molecule analysis at high concentrations. Science 299 (5607), 682–686. doi:10.1126/science.1079700
Li, J., and Kim, C. C. (2020). Current commercialization status of electrowetting-on-dielectric (EWOD) digital microfluidics. Lab Chip 20 (10), 1705–1712. doi:10.1039/D0LC00144A
Li, J., Stein, D., McMullan, C., Branton, D., Aziz, M. J., and Golovchenko, J. A. (2001). Ion-beam sculpting at nanometre length scales. Nature 412 (6843), 166–169. doi:10.1038/35084037
Liao, Q., Jin, X., and Fu, H. (2019). Tunable halide perovskites for miniaturized solid-state laser applications. Adv. Opt. Mater. 7 (17), 1900099. doi:10.1002/adom.201900099
Logsdon, G. A., Vollger, M. R., and Eichler, E. E. (2020). Long-read human genome sequencing and its applications. Nat. Rev. Genet. 21 (10), 597–614. doi:10.1038/s41576-020-0236-x
Loit, K., Adamson, K., Bahram, M., Puusepp, R., Anslan, S., Kiiker, R., et al. (2019). Relative performance of MinION (Oxford nanopore technologies) versus Sequel (pacific biosciences) third-generation sequencing instruments in identification of agricultural and forest fungal pathogens. Appl. Environ. Microbiol. 85 (21), e01368–e01319. doi:10.1128/AEM.01368-19
Lopez-Alvarez, D., Parra, B., and Cuellar, W. J. (2020). Genome sequence of SARS-CoV-2 isolate cali-01, from Colombia, obtained using Oxford nanopore MinION sequencing. Microbiol. Resour. Announc 9 (26), e00573-20. doi:10.1128/MRA.00573-20
Lu, H., Giordano, F., and Ning, Z. (2016). Oxford nanopore MinION sequencing and genome assembly. Genomics Proteomics Bioinformatics 14 (5), 265–279. doi:10.1016/j.gpb.2016.05.004
Măriuţa, D., Colin, S., Barrot-Lattes, C., Le Calvé, S., Korvink, J. G., Baldas, L., et al. (2020). Miniaturization of fluorescence sensing in optofluidic devices. Microfluidics and Nanofluidics 24 (9), 65. doi:10.1007/s10404-020-02371-1
Ma, T., Janot, J. M., and Balme, S. (2020). Track‐etched nanopore/membrane: from fundamental to applications. Small Methods 4 (9), 366. doi:10.1002/smtd.202000366
Makałowski, W., and Shabardina, V. (2020). Bioinformatics of nanopore sequencing. J. Hum. Genet. 65 (1), 61–67. doi:10.1038/s10038-019-0659-4
Marszalek, P. E. (2010). DNA sequencing: detecting methylation with force. Nat. Nanotechnol 5 (11), 765–766. doi:10.1038/nnano.2010.223
Martin, W. E., Srijanto, B. R., Collier, C. P., Vosch, T., and Richards, C. I. (2016). A comparison of single-molecule emission in aluminum and gold zero-mode waveguides. J. Phys. Chem. A. 120 (34), 6719–6727. doi:10.1021/acs.jpca.6b03309
Murphy, T. W., Hsieh, Y. P., Zhu, B., Naler, L. B., and Lu, C. (2020). Microfluidic platform for next-generation sequencing library preparation with low-input samples. Anal Chem. 92 (3), 2519–2526. doi:10.1021/acs.analchem.9b04086
Nelson, E. M., Li, H., and Timp, G. (2014). Direct, concurrent measurements of the forces and currents affecting DNA in a nanopore with comparable topography. ACS Nano 8 (6), 5484–5493. doi:10.1021/nn405331t
Ni, P., Huang, N., Zhang, Z., Wang, D. P., Liang, F., Miao, Y., et al. (2019). DeepSignal: detecting DNA methylation state from Nanopore sequencing reads using deep-learning. Bioinformatics 35 (22), 4586–4595. doi:10.1093/bioinformatics/btz276
Ni, Y., Wang, Y., and Xu, W. (2020). Recent process of flexible Transistor-Structured memory. Small 1905332. doi:10.1002/smll.201905332
Noakes, M. T., Brinkerhoff, H., Laszlo, A. H., Derrington, I. M., Langford, K. W., Mount, J. W., et al. (2019). Increasing the accuracy of nanopore DNA sequencing using a time-varying cross membrane voltage. Nat. Biotechnol. 37 (6), 651. doi:10.1038/s41587-019-0096-0
Oliva, M., Milicchio, F., King, K., Benson, G., Boucher, C., and Prosperi, M. (2020). Portable nanopore analytics: are we there yet? Bioinformatics 36 (16), 4399–4405. doi:10.1093/bioinformatics/btaa237
PacBio (2020). Sequel II System Brochure: delivering highly accurate long reads. Available at: https://www.pacb.com/literature/sequel-ii-system-brochure-delivering-highly-accurate-long-reads/.
Pan, J., Liu, Q., Sun, H., Zheng, W., Wang, P., Wen, L., et al. (2020). A miniaturized fluorescence imaging device for rapid early skin cancer detection. J. Innovative Opt. Health Sci. 2050026. doi:10.1142/s1793545820500261
Park, T., Lee, S. J., Cha, J. H., and Choi, W. (2018). Scalable fabrication of nanopores in membranes via thermal annealing of Au nanoparticles. Nanoscale 10 (47), 22623–22634. doi:10.1039/C8NR06441E
Phelan, J. E., O'Sullivan, D. M., Machado, D., Ramos, J., Oppong, Y. E. A., Campino, S., et al. (2019). Integrating informatics tools and portable sequencing technology for rapid detection of resistance to anti-tuberculous drugs. Genome Med. 11 (1), 41. doi:10.1186/s13073-019-0650-x
Quick, J., Loman, N. J., Duraffour, S., Simpson, J. T., Severi, E., Cowley, L., et al. (2016). Real-time, portable genome sequencing for Ebola surveillance. Nature 530 (7589), 228–232. doi:10.1038/nature16996
Raveendran, M., Leach, A. R., Hopes, T., Aspden, J. L., and Actis, P. (2020). Ribosome fingerprinting with a solid-state nanopore. ACS Sens 5 (11), 3533–3539. doi:10.1021/acssensors.0c01642
Rhoads, A., and Au, K. F. (2015). PacBio sequencing and its applications. Genomics Proteomics Bioinformatics 13 (5), 278–289. doi:10.1016/j.gpb.2015.08.002
Ruan, Q., Ruan, W., Lin, X., Wang, Y., Zou, F., Zhou, L., et al. (2020). Digital-WGS: automated, highly efficient whole-genome sequencing of single cells by digital microfluidics. Sci. Adv. 6 (50), eabd6454. doi:10.1126/sciadv.abd6454
Ruppert, M. G., Fowler, A. G., Maroufi, M., and Moheimani, S. O. R. (2017). On-chip dynamic mode atomic force microscopy: a silicon-on-insulator MEMS approach. J. Microelectromechanical Syst. 26 (1), 215–225. doi:10.1109/jmems.2016.2628890
Russell-Pavier, F. S., Picco, L., Day, J. C. C., Shatil, N. R., Yacoot, A., and Payton, O. D. (2018). ‘Hi-Fi AFM’: high-speed contact mode atomic force microscopy with optical pickups. Meas. Sci. Technology 29 (10), 105902. doi:10.1088/1361-6501/aad771
Sahlin, K., Sipos, B., James, P. L., and Medvedev, P. (2021). Error correction enables use of Oxford Nanopore technolog for reference-free transcriptome analysis. Nat. Commun. 12 (1), 2. doi:10.1038/s41467-020-20340-8
Sahm, A., Werner, N., Hofmann, J., Bege, R., and Paschke, K. (2018). Compact miniaturized laser module emitting more than 1.6 W of yellow light at 576 nm. IEEE Photon. Technology Lett. 30 (21), 1878–1881. doi:10.1109/lpt.2018.2870524
Savas, J., Khayatzadeh, R., Civitci, F., Gokdel, Y. D., and Ferhanoglu, O. (2018). Toward fully three-dimensional-printed miniaturized confocal imager. Opt. Eng. 57 (4), 041402. doi:10.1117/1.OE.57.4.041402
Schneider, G. F., Kowalczyk, S. W., Calado, V. E., Pandraud, G., Zandbergen, H. W., Vandersypen, L. M., et al. (2010). DNA translocation through graphene nanopores. Nano Lett. 10 (8), 3163–3167. doi:10.1021/nl102069z
Schneider, G. F., Xu, Q., Hage, S., Luik, S., Spoor, J. N., Malladi, S., et al. (2013). Tailoring the hydrophobicity of graphene for its use as nanopores for DNA translocation. Nat. Commun. 4, 2619. doi:10.1038/ncomms3619
Shen, B., Piskunen, P., Nummelin, S., Liu, Q., Kostiainen, M. A., and Linko, V. (2020). Advanced DNA nanopore technologies. ACS Appl. Bio Mater. 3 (9), 5606–5619. doi:10.1021/acsabm.0c00879
Shendure, J., Balasubramanian, S., Church, G. M., Gilbert, W., Rogers, J., Schloss, J. A., et al. (2017). DNA sequencing at 40: past, present and future. Nature 550 (7676), 345–353. doi:10.1038/nature24286
Si, W., Yang, H., Wu, G., Chen, C., and Yu, M. (2020). Manipulation of DNA transport through solid-state nanopores by atomic force microscopy. Mater. Res. Express 7 (9), 095404. doi:10.1088/2053-1591/abb856
Steinbock, L. J., Otto, O., Chimerel, C., Gornall, J., and Keyser, U. F. (2010). Detecting DNA folding with nanocapillaries. Nano Lett. 10 (7), 2493–2497. doi:10.1021/nl100997s
Storm, A. J., Chen, J. H., Ling, X. S., Zandbergen, H. W., and Dekker, C. (2003). Fabrication of solid-state nanopores with single-nanometre precision. Nat. Mater. 2 (8), 537–540. doi:10.1038/nmat941
Strathearn, D., Sarkar, N., Lee, G., Olfat, M., and Mansour, R. R. (2017). The benefits of miniaturization of an atomic force microscope”, in IEEE 30th International Conference on micro Electro mechanical Systems (MEMS), Las Vegas, NV, United States, January 22–26, 2017, 1363–1366.
Su, S., Wang, X., and Xue, J. (2021). Nanopores in two-dimensional materials: accurate fabrication. Mater. Horizons. doi:10.1039/D0MH01412E
Tan, S. J., Phan, H., Gerry, B. M., Kuhn, A., Hong, L. Z., Min Ong, Y., et al. (2013). A microfluidic device for preparing next generation DNA sequencing libraries and for automating other laboratory protocols that require one or more column chromatography steps. PLoS One 8 (7), e64084. doi:10.1371/journal.pone.0064084
Thakur, M., Macha, M., Chernev, A., Graf, M., Lihter, M., Deen, J., et al. (2020). Wafer-scale fabrication of nanopore devices for single-molecule DNA biosensing using MoS2. Small Met. 4 (11), 2000072. doi:10.1002/smtd.202000072
Van Helleputte, N., Even, A. J. G., Leonardi, F., Stanzione, S., Song, M., Garripoli, C., et al. (2020). Miniaturized electronic circuit design challenges for ingestible devices. J. Microelectromechanical Syst. 29 (5), 1–8. doi:10.1109/JMEMS.2020.3006580
Vunckx, K., Geelen, B., Garcia Munoz, V., Lee, W., Chang, H., Van Dorpe, P., et al. (2020). “Towards a miniaturized application-specific Raman spectrometer,” in Sensing for Agriculture and food quality and Safety XII. Bellingham, Washington, United States: International Society for Optics and Photonics. doi:10.1117/12.2557790
Wang, Y., Wang, Y., Du, X., Yan, S., Zhang, P., Chen, H. Y., et al. (2019). Electrode-free nanopore sensing by DiffusiOptoPhysiology. Sci. Adv. 5 (9), eaar3309. doi:10.1126/sciadv.aar3309
Wasfi, A., Awwad, F., and Ayesh, A. I. (2020). DNA sequencing via Z-shaped graphene nanoribbon field effect transistor decorated with nanoparticles using first-principle transport simulations. New J. Phys. 22 (6), e87. doi:10.1088/1367-2630/ab87ef
Wei, Z. X., Ying, Y. L., Li, M. Y., Yang, J., Zhou, J. L., Wang, H. F., et al. (2019). Learning shapelets for improving single-molecule nanopore sensing. Anal Chem. 91 (15), 10033–10039. doi:10.1021/acs.analchem.9b01896
Wu, M., Liu, W., Hu, J., Zhong, Z., Rujiralai, T., Zhou, L., et al. (2019). Fluorescence enhancement in an over-etched gold zero-mode waveguide. Opt. Express 27 (13), 19002–19018. doi:10.1364/OE.27.019002
Wu, Y. C., Chang, C. H., Hung, J. H., and Yang, C. H. (2017). A 135-mW fully integrated data processor for next-generation sequencing. IEEE Trans. Biomed. Circuits Syst. 11 (6), 1216–1225. doi:10.1109/TBCAS.2017.2760109
Wu, Z., Hammad, K., Ghafar-Zadeh, E., and Magierowski, S. (2020). FPGA-accelerated 3rd generation DNA sequencing. IEEE Trans. Biomed. Circuits Syst. 14 (1), 65–74. doi:10.1109/TBCAS.2019.2958049
Xia, P., Zuo, J., Paudel, P., Choi, S., Chen, X., Rahman Laskar, M. A., et al. (2021). Sapphire-supported nanopores for low-noise DNA sensing. Biosens. Bioelectron. 174, 112829. doi:10.1016/j.bios.2020.112829
Xu, X., Li, C., Zhou, Y., and Jin, Y. (2017). Controllable shrinking of glass capillary nanopores down to sub-10 nm by wet-chemical silanization for signal-enhanced DNA translocation. ACS Sens 2 (10), 1452–1457. doi:10.1021/acssensors.7b00385
Yamazaki, H., Hu, R., Zhao, Q., and Wanunu, M. (2018). Photothermally assisted thinning of silicon nitride membranes for ultrathin asymmetric nanopores. ACS Nano 12 (12), 12472–12481. doi:10.1021/acsnano.8b06805
Yanagi, I., Fujisaki, K., Hamamura, H., and Takeda, K.-i. (2017). Thickness-dependent dielectric breakdown and nanopore creation on sub-10-nm-thick SiN membranes in solution. J. Appl. Phys. 121 (4), 4974286. doi:10.1063/1.4974286
Youn, Y., Lee, C., Kim, J. H., Chang, Y. W., Kim, D. Y., and Yoo, K. H. (2016). Selective detection of single-stranded DNA molecules using a glass nanocapillary functionalized with DNA. Anal Chem. 88 (1), 688–694. doi:10.1021/acs.analchem.5b02540
Zambrana-Puyalto, X., Ponzellini, P., Maccaferri, N., Tessarolo, E., Pelizzo, M. G., Zhang, W., et al. (2019). A hybrid metal-dielectric zero mode waveguide for enhanced single molecule detection. Chem. Commun. (Camb) 55 (65), 9725–9728. doi:10.1039/C9CC04118D
Zhang, Y., Miyahara, Y., Derriche, N., Yang, W., Yazda, K., Capaldi, X., et al. (2019). nanopore formation via tip-controlled local breakdown using an atomic force microscope. Small Methods 3 (7), 1900147. doi:10.1002/smtd.201900147
Zhong, Q., Ding, H., Gao, B., He, Z., and Gu, Z. (2019). Advances of microfluidics in biomedical engineering. Adv. Mater. Tech. 4 (6), 1800663. doi:10.1002/admt.201800663
Zhu, J., Zhang, T., Yang, Y., and Huang, R. (2020). A comprehensive review on emerging artificial neuromorphic devices. Appl. Phys. Rev. 7 (1), 011312. doi:10.1063/1.5118217
Keywords: nanopore, DNA, sequencing, miniaturized, nanotechnology
Citation: Huo W, Ling W, Wang Z, Li Y, Zhou M, Ren M, Li X, Li J, Xia Z, Liu X and Huang X (2021) Miniaturized DNA Sequencers for Personal Use: Unreachable Dreams or Achievable Goals. Front. Nanotechnol. 3:628861. doi: 10.3389/fnano.2021.628861
Received: 13 November 2020; Accepted: 22 January 2021;
Published: 22 February 2021.
Edited by:
P. Ravi Selvaganapathy, McMaster University, CanadaReviewed by:
Rajesh Patel, Veer Narmad South Gujarat University, IndiaCopyright © 2021 Huo, Ling, Wang, Li, Zhou, Ren, Li, Li, Xia, Liu and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xian Huang, aHVhbmd4aWFuQHRqdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.