
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Neuroanat. , 29 July 2021
Volume 15 - 2021 | https://doi.org/10.3389/fnana.2021.711955
This article is part of the Research Topic Brain Imaging for Glycobiology View all 7 articles
 Md. Mahmudul Hasan1
Md. Mahmudul Hasan1 Mst. Afsana Mimi1
Mst. Afsana Mimi1 Md. Al Mamun1
Md. Al Mamun1 Ariful Islam1
Ariful Islam1 A. S. M. Waliullah1
A. S. M. Waliullah1 Md. Mahamodun Nabi1
Md. Mahamodun Nabi1 Zinat Tamannaa1
Zinat Tamannaa1 Tomoaki Kahyo1,2*
Tomoaki Kahyo1,2* Mitsutoshi Setou1,2,3
Mitsutoshi Setou1,2,3Glycans are diverse structured biomolecules that play crucial roles in various biological processes. Glycosylation, an enzymatic system through which various glycans are bound to proteins and lipids, is the most common and functionally crucial post-translational modification process. It is known to be associated with brain development, signal transduction, molecular trafficking, neurodegenerative disorders, psychopathologies, and brain cancers. Glycans in glycoproteins and glycolipids expressed in brain cells are involved in neuronal development, biological processes, and central nervous system maintenance. The composition and expression of glycans are known to change during those physiological processes. Therefore, imaging of glycans and the glycoconjugates in the brain regions has become a “hot” topic nowadays. Imaging techniques using lectins, antibodies, and chemical reporters are traditionally used for glycan detection. However, those techniques offer limited glycome detection. Mass spectrometry imaging (MSI) is an evolving field that combines mass spectrometry with histology allowing spatial and label-free visualization of molecules in the brain. In the last decades, several studies have employed MSI for glycome imaging in brain tissues. The current state of MSI uses on-tissue enzymatic digestion or chemical reaction to facilitate successful glycome imaging. Here, we reviewed the available literature that applied MSI techniques for glycome visualization and characterization in the brain. We also described the general methodologies for glycome MSI and discussed its potential use in the three-dimensional MSI in the brain.
Glycans are diverse and complex structured sugar chains and crucial to many biological processes in all living organisms. Organisms polymerize simple sugars to process oligo- and polysaccharides, commonly referred to as glycans, when freely or covalently linked to proteins and lipids. Glycans are made up of several sugar residues with distinct glycosidic bonds, forming a complex, branched structure. As a result, various glycan structures have been generated in organisms with different glycosylation forms (Cipolla et al., 2011; Coff et al., 2020). Glycans are covalently linked to proteins and modulate their functions through direct interactions that control protein conformation, stabilization, and turnover. During protein biosynthesis, amino acids may be modified by the covalent binding of several functional groups. This protein modification type is known as post-translational modification (PTM) (Veillon et al., 2018). The glycosylation of proteins and lipids occurs in the endoplasmic reticulum (ER) and the Golgi apparatus of the cells.
In contrast to the genome and proteome, glycans are generated in non-templated processes and are carefully regulated at different levels in ERs and Golgi apparatuses (Reily et al., 2019). There are also no “completed” glycan structures. During the glycosylation, proteins may be modified, resulting in various glycans that differ uniformly by interactions, length, number of antennas, and composition (Ruhaak et al., 2018).
In glycosylation, carbohydrates are linked to a protein (as N-linked or O-linked glycans), lipid, or glycan substrate (Iqbal et al., 2018; Mueller et al., 2018). In the protein glycosylation processes, asparagine, serine, threonine, tryptophan (Ihara, 2015.), hydroxyproline, hydroxylysine (Song and Mechref, 2013), and rarely tyrosine residues are connected by the binding with carbohydrate moieties, resulting in a mass increase of the substrate protein. The glycosylation of proteins and lipids is known to be associated with brain development, cell adhesion, molecular trafficking, signal transduction, and differentiation (Ohtsubo and Marth, 2006). Aberrant glycosylation including, chemical modification, cutting or elongation of glycan structures on the cellular surface, critical receptors on the cell surface, and neurotransmitter transporters, are associated with various neurological abnormalities, including immune responses, neurodegenerative disorders, mental disorders, and brain tumors (Ohtsubo and Marth, 2006; Hwang et al., 2010; Iqbal et al., 2018). It is indispensable to understand the intrinsic molecular mechanisms, cellular profiling, and glycome imaging to determine the brain function and behavior in neurological disorders associated with glycosylation. Chemical reporters and glycan-binding proteins such as lectins and antibodies techniques have been typically used for specific glycan determination and imaging. However, those traditional approaches for glycan imaging are inadequate to cover the wide range of mammalian glycans (Eshghi et al., 2014). Therefore, it is essential to use alternate imaging methods to complement the detailed information acquired from histostaining methods.
Mass spectrometry imaging (MSI) has emerged as a method of choice for reliable brain imaging to study the roles of glycosylation in the brain. MSI has overcome some of the difficulties of conventional histostaining methods due to its high ionization efficiency. Unlike traditional affinity-based detection methods such as immunohistochemistry, MSI techniques do not require prior information from the analytes of interest. This feature, which is a unique characteristic of MSI, is especially useful for discovery study (Eshghi et al., 2014). MSI is a unique method that incorporates histology and mass spectrometry (MS). It allows label-free detection of hundreds to thousands of compounds in a single tissue section without extraction, purification, or separation (Eriksson et al., 2013; Buck et al., 2015). In addition, MSI can combine quantitative techniques that may allow for direct quantitative imaging of various analytes from a tissue section. Recent developments in MS instrumentations, methods, and data analysis have propelled quantitative glycomics studies to greater levels (Zaia, 2008). Matrix-assisted laser desorption ionization-mass spectrometry imaging (MALDI-MSI), desorption electrospray ionization mass spectrometry imaging (DESI-MSI), and secondary ion mass spectrometry (SIMS) imaging are three of the most popular ionization technologies used in MSI (McDonnell and Heeren, 2007). So far, MALDI-MSI has been applied to visualize glycome due to its higher sensitivity, ionization efficiency and high mass accuracy over a broad mass-to-charge (m/z) range.
The capability of MSI to capture and visualize spatial distributions of biomolecules such as proteins (Heijs et al., 2015; Piehowski et al., 2020), lipids (Wildburger, 2017; Islam et al., 2020), glycans (Mori et al., 2019; Heijs et al., 2020), and metabolites (Cornett et al., 2008; Sugiura and Setou, 2010) in brain tissues with high mass accuracy and resolution enables the identification of compounds through accurate mass matching. Therefore, it is essential to further MSI application knowledge and accelerate medical diagnostics development (Neubert and Walch, 2013; Schubert et al., 2016). MSI platforms have witnessed rapid growth, and they now can provide comprehensive chemical information at subcellular spatial resolution (Zavalin et al., 2012). Current technological developments in instrumentation and software have made MSI an analytical tool capable of identifying and characterizing a wide range of molecular species while simultaneously imaging their spatial distributions with accurate mass measurements. This ability of simultaneous molecular detection, spatial imaging, and relative abundance of biomolecules helps MSI applications in the research field of brain glycobiology. Currently, it has been shown that glycans released from glycoconjugates that have been immobilized in a solid phase could be examined directly by MSI techniques.
Here we have focused mainly on the methodologies of glycome imaging in animal and human brains using MSI techniques. In addition, we have discussed the limitations of the current methodology and the future development for the study of glycome by MSI.
Glycosylation is a common enzymatic mechanism in which a carbohydrate donor is glycosidically linked to a functional group of another molecule (proteins or lipids) that produces glycoconjugates. In the protein and lipid glycosylation process, oligosaccharides are attached to proteins or lipids such as N-linked or O-linked glycans. In the consensus sequence of Asn-X-Ser/Thr, N-glycans are linked to the nitrogen of the amide-side chain in the asparagine residue, where X can be any amino acid other than proline. On the other hand, O-glycans are covalently attached to the oxygen of the hydroxyl group of serine, threonine, or tyrosine (Van Den Steen et al., 1998; Imperiali and O’Connor, 1999). Glycosylation is a ubiquitous PTM of proteins in all three domains of life, which plays a vital role in determining protein structure, function, and stability (Moremen et al., 2012). Glycolipids are also synthesized through this process in eukaryotes and prokaryotes and play diverse biological roles (Kopitz, 2017). Approximately 700 proteins are associated with glycosylation in mammals. Among them, about 200 proteins are glycosyltransferases (Moremen et al., 2012). These proteins play a critical role in producing glycoconjugates possessing diverse functions in several organs in mammals, including the brain. About 70% of brain proteins are present as glycoproteins. They mainly contain a higher number of high-mannose, hexose, and N-acetyl hexosamine (HexNAc) with fucose and sialic acid (Lee et al., 2020; Tena and Lebrilla, 2021). Many brain lipids also present as glycoconjugates that play various biological functions in the brain (Tena and Lebrilla, 2021).
Glycosylation has pivotal roles in brain development, physiology, and functions, including regulating synaptic processes and neural excitability by controlling neural transmission (Scott and Panin, 2014; Klarić et al., 2020). In the brain, neurons, oligodendrocytes, and astrocytes are developed from neural stem cells, which depend on cell surface molecules present on the neural stem cells and their interactions with other molecules and cells (Yale et al., 2018). Moreover, cells derived from neural stem cells also depend on their interaction with other cells and molecules through glycosylation (Yale et al., 2018). Therefore, dysregulated glycosylation can develop several neurological disorders. For example, N-glycosylation is involved in developing the human nervous system, which controls the folding, trafficking, localization, adhesion, cell-cell interaction, and enzymatic activity of proteins (Freeze et al., 2012). O-glycosylation also regulates the functions of thousands of proteins in the brain and significantly affects neuroprotection, memory function, and neuronal signaling. Impaired biosynthesis of glycoconjugates due to altered N- or O-glycosylation can cause several neurological diseases, including cerebellar atrophy, stroke, epilepsy, neuropathy, delayed development, paralysis, and tremors (Freeze et al., 2012; Reily et al., 2019). The analysis of post-mortem brain samples shows that people with schizophrenia have aberrant glycosylation (Williams et al., 2020). Dysregulated fucosylation and sialylation of both N- and O-glycans also play a critical role in brain cancer development and progression (Veillon et al., 2018). Aberrant glycosylation of proteins associated with inhibitory and excitatory neurotransmission, including glutamate transporter, gamma-aminobutyric acid receptors, and quisqualate receptors, was recently reported in individuals with schizophrenia.
Additionally, differential expression of glycosyltransferases was observed in a study using the post-mortem brain of schizophrenia patients (Williams et al., 2020). Zhang et al. have reported the N-glycoproteome profile of Alzheimer’s disease (AD) and the unaffected brains using mass spectrometry-based proteomics analysis. This study revealed dysregulated N-glycosylation mediated altered pathways in human AD brains, including synaptic dysfunction, altered lysosomal function, dysregulated cell adhesion, and cell signaling (Zhang et al., 2020). As well as proteins, lipids glycosylation also has pivotal roles in the development of the central nervous system (CNS), synaptic plasticity, and regeneration, and its dysregulation causes several neurological disorders. Glycolipids are the cell membrane’s structural components and act as ligands for signaling molecules, control cell-to-cell communication, and form lipid rafts. The most common glycolipids are glycoglycerolipids, cerebrosides, and gangliosides (Brandenburg and Holst, 2015). They are abundant in eukaryotic cells, including, neuronal and glial cells of the CNS, and play a crucial role in controlling these cell’s functions (Brekk et al., 2020). Altered glycolipids in the brain due to dysregulated glycosylation can cause several neurodegenerative diseases, including Parkinson’s disease, AD, Huntington’s disease, and amyotrophic lateral sclerosis in humans (Brekk et al., 2020; Moll et al., 2020).
Although glycobiology has opened a new window for glycan-based therapeutics for several diseases such as breast cancer and human immunodeficiency virus (HIV-1), glycan-based therapy for neurological diseases is yet to be developed (Doran et al., 2018; Reily et al., 2019). Substantial benefits must be achieved for neurological disorders of glycosylation by early detection and management. Hopefully, further development in glycobiology can offer better therapeutic approaches for treating neurological disorders in the future.
Glycan-binding proteins, including lectins and antibodies, have traditionally been used to recognize and imaging glycans or glycoconjugate complexes in brain tissues. Lectins are naturally occurring proteins identified in most organisms (Lis and Sharon, 1998) and have been widely used for the detection (Pilobello and Mahal, 2007) and enrichment (Hirabayashi, 2004) of glycoconjugates. These naturally occurring lectins are also used to research brain disorders based on glycan expression (Gabius, 2009). The significant biological glycan residues such as sialic acids, mannose, fucose, and galactose can be identified from wheat germ agglutinin (WGA), Concanavalin A; Lens culinaris agglutinin, Aleuria aurantia lectin, and peanut agglutinin (PNA); Ricinus communis Agglutinin; Griffonia simplificola lectin-I lectins, respectively. WGA and cholera toxin B (CTB) is a neuronal tracer based on lectin that is endocytosed readily by brain cells after binding to glycolipids of the surface in the cells (Jobling et al., 2012), making them helpful in understanding the complex and functional neuronal network connections in both anterograde and retrograde directions (Parker et al., 2013, 2015; Lee et al., 2017). WGA tracer has been used in the mouse brain tissue to study neuronal synapses (Yoshihara, 2002) and for transneuronal tracing purposes in several animal species, including Drosophila (Tabuchi et al., 2000) and rodent brains (Libbrecht et al., 2017). Glycans were visualized using lectins on tissue sections during mouse secondary neurulation (Griffith and Wiley, 1989) and human thymus (Paessens et al., 2007). Although lectins have contributed significantly to glycosylation profiles and neural networks in the brain, they appear to have a low affinity for their glycan epitope and require multi-valence for high-avidity binding (Kiessling and Pohl, 1996). In addition, lectins are usually tissue-impermeable, and sometimes they are toxic (Schwarz et al., 1999; Ohba and Bakalova, 2003).
Previously, antibodies against protein antigens have been developed and characterized against several glycan structures for particular neuronal cells (Jacobsen et al., 2016). These functional monoclonal anti-glycan antibodies remain elusive due to the difficulties experienced throughout their choice and development (Sterner et al., 2016). The most commercially available anti-glycan antibodies are IgM and IgG subtypes, used for imaging of O-glycans, N-glycans, and so on (Cummings and Etzler, 2009). Generally, they have high glycan epitopes specificity and affinity. Like lectins, antibodies have limited permeability in tissues and have been used for specific glycans (Laughlin and Bertozzi, 2009).
Biorthogonal chemical reporter techniques have been used to target and imaging sialoglycans in living organisms (Laughlin et al., 2008; Cheng et al., 2016). This method uses endogenous metabolic tracers that consist of sialic acid analogs or N-acetylmannosamine (ManNAc) linked with chemical reporters such as azide. In vivo brain imaging of these glycans manipulation techniques has been reported recently (Gagiannis et al., 2007; Sampathkumar et al., 2008; Xie et al., 2016; Shajahan et al., 2017). However, it is difficult to monitor the sialoglycans in the brain tissues because of the labeled glycan’s impermeability through the blood–brain barrier (BBB). In order to address this problem, carrier-mediated transport systems have been developed. For example, modified ManNAc conjugated with neuroactive carriers including choline present in BBB mediated access into mice brain by intravenous injection (Shajahan et al., 2017). Although the techniques above are advantageous for brain glycan imaging, there are some drawbacks: they are only used for a limited number of glycan targets, they are not labeled free, and spontaneous imaging methods are often toxic.
MSI is an emerging tool that offers label-free imaging of the tissue glycome allowing the detection of several glycans at a time, with the spatial distribution and regional heterogeneity with accurate mass matching. Thus, some of the difficulties of histological staining with lectins, antibodies, and chemical reporters can be resolved (Iqbal et al., 2018).
The MSI technique consists of an ionization source, the mass analyzer, and the detector. A specimen is examined/scanned under ambient conditions or vacuum using a light beam, lasers, or ions; as a result, analytes are ionized and desorbed simultaneously. The most commonly used ionization methods for MSI are MALDI, DESI, secondary ion beam, and laser ablation post-ionization. The selection of an appropriate ionization approach is crucial to ionize the specific analyte of interest efficiently. After the ionization process, the next step is to transfer them into the mass analyzer for separation and characterization. Various mass analyzers have been coupled to MSI, including time-of-flight (TOF), TOF/TOF, quadrupole TOF mass analyzers (QTOF), linear ion trap, Orbitrap, and Fourier-transform ion cyclotron resonance (FT-ICR), and they offer a varying degree of mass resolution, accuracy, and speed. Typically, MALDI is integrated into TOF systems, offering a mass error of <10 ppm and mass resolution up to 50,000. The TOF/TOF system offers MS/MS facilities with a mass resolving power of >60,000 and a mass error of <1 ppm. The TOF/TOF system currently offers rapid and highly reproducible fragmentation with structural characterizations of glycome formulated to simplify the linkage-specific substituent information on the terminal units (Mechref et al., 2003; Yu et al., 2006). These benefits are often missing in traditional TOF or QTOF systems. The QTOF mass analyzer provides the MS/MS capability (Loboda et al., 2000), while the Orbitrap combined with MALDI-MSI provides up to 100,000–200,000 mass resolution, <5 ppm mass error, and MS/MS capability (Makarov, 2000; Zubarev and Makarov, 2013). FT-ICR combined with MALDI offers the ultrahigh mass-resolving power (m/upDeltam50% > 27,00,000 at m/z 400) and mass accuracy (80 ppb RMS) that enable confident identification of tens of thousands of unique elemental compositions (Marshall, 2000; Smith et al., 2018).
Spatial resolution in MALDI-MSI depends on four major factors: (1) analyte delocalization during sample preparation, (2) matrix homogeneity, (3) laser spot size, and (4) sensitivity (Dueñas et al., 2016). Sensitivity has become a critical factor that has improved by advanced sample preparation and instrumentation. Advanced technologies have been utilized to improve spatial resolution, usually ranging from 50 to 200 μm for tissue MSI analysis (Römpp and Spengler, 2013). The high-quality protein images have been achieved at 5 μm spatial resolution using a Gaussian laser beam and an aspheric lens in MALDI-FT-ICR MSI (Zavalin et al., 2014). Using transmission geometry MALDI-TOF-MSI technology, up to 1 μm spatial resolution images have been achieved in mouse brain tissue analyses (Zavalin et al., 2012). Similarly, by modifying the beam-delivery optics, up to 5 μm spatial resolution has been achieved by the Orbitrap in biological tissues (Korte et al., 2015).
Of the most frequently used MSI techniques, TOF and Orbitrap systems are attractive for high throughput analysis, whereas TOF/TOF and FT-ICR systems are more suitable for discovery and research platforms. After the ions are separated based on their m/z ratio in the analyzer, ions need to be detected. The detector converts charged ions/current flow into electric signals digitally. The commonly used detectors are the Faraday cup, photographic plate, an electron multiplier (Potts, 1987). Nowadays, an image current detector is used in modern mass spectrometers with FT-ICR and orbitrap (Wu et al., 2020).
Molecular detection also depends on the sensitivity of the MSI instruments. While DESI and SIMS offer better sensitivity for the low mass molecules (<500 Da), MALDI has greater sensitivity for the high mass molecules (>500 Da) (Nabi et al., 2021). The recent addition of MALDI-2 MSI and 21 tesla MALDI-FT-ICR techniques offers the highest sensitivity, mass resolution, accuracy, spatial resolution, molecular details, dynamic range, and MS/MS capabilities (Smith et al., 2018; Niehaus et al., 2019; Bowman et al., 2020; Heijs et al., 2020). On the other hand, on-tissue treatment with buffer-free phospholipase C reduced the ion suppression effects, thereby ion signal intensities of mono-, di-, and oligohexosylceramides were enhanced by up to 10-fold in flash-frozen (FF) tissue sections without decreasing the high lateral resolution of MSI analyses (Vens-Cappell et al., 2016). These latest MSI instrumentations and methodologies may improve the glycome detection, characterization, and visualization in biological tissues.
Careful sample preparation is crucial for successful MSI analysis of glycomics from a thin tissue section. Therefore, a series of laborious and delicate sample pre-treatment is required as protein denaturation, deglycosylation, stabilization for efficient glycome MSI in the tissue section.
Formalin-fixed paraffin-embedded (FFPE) tissue should be cryosectioned (∼3–7 μm) and mounted on a conductive glass slide such as indium tin oxides-coated slides (ITO-coated) (Figure 1, method-I). On the other hand, fresh-frozen or FF tissue should be cryosectioned (∼5–10 or ∼5–18 μm) and mounted on a conductive slide or MALDI target plate to achieve high-resolution ion images (Figure 1, method-II and III). Then mounted FFPE tissue proteins should be denatured by dry heating (1 h at 60°C) (Drake et al., 2018). FFPE tissue must be dewaxed properly to remove paraffin, which may obscure the N-glycan signal. Deparaffinization of FFPE tissue includes washing in xylene solution and rehydrating it in a series of ethanol rinses at room temperature. In addition, FF tissue must be rinsed in organic solvents (e.g., CHCl3) to remove salts, lipids, and other metabolites to improve N-glycans detection significantly (Yang and Caprioli, 2011). MSI of gangliosides can be achieved from FF tissues without enzyme pre-treatments and derivatization (Whitehead et al., 2011; Jackson et al., 2018). After dewaxing FFPE tissue, heat-induced antigen retrieval should be performed using citraconic anhydride buffer into a pre-warmed device to break protein crosslinks (Drake et al., 2018). Antigen retrieval breaks chemical crosslinking formed by formalin, allowing for efficient digestion. It is crucial for unmasking hidden or latent epitopes in preparation and allows for enzymatic access for releasing glycans from the proteins in the tissue. For FF tissue sections, antigen retrieval is not necessary (Raghunathan et al., 2019).
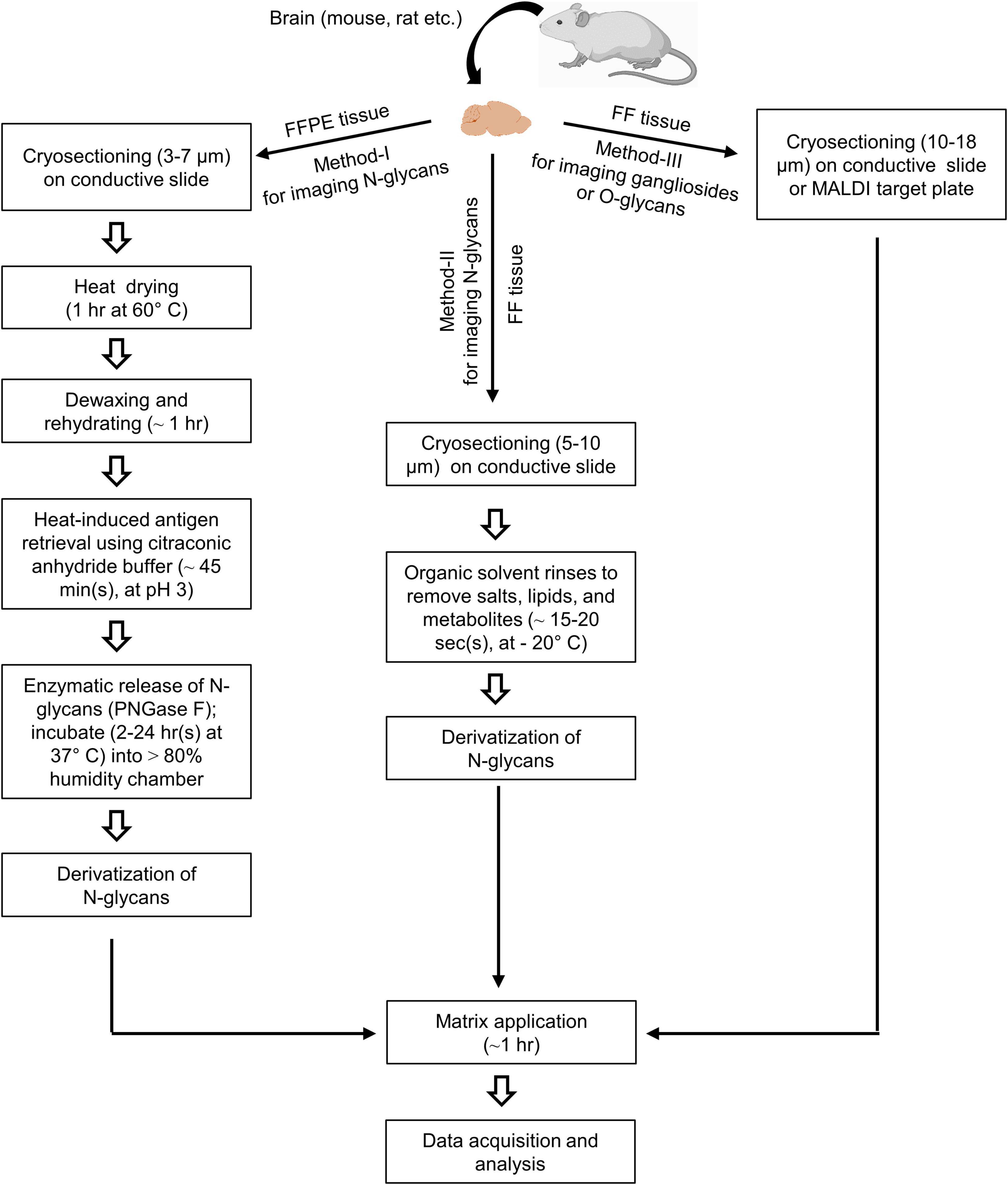
Figure 1. Schematic workflow of mass spectrometry imaging (MSI) for glycome in the brain. An FFPE tissue section is mounted on an indium tin oxide-coated (ITO-coated) glass slide and dried by heating. Then section is deparaffinized and rehydrated by xylene and a series of ethanol rinsing, respectively. The epitope activation and tissue proteins are denatured by the treatment with a basic heat-induced antigen retrieval buffer (citraconic anhydride buffer, at pH 3) (Method-I). On the other hand, FF tissue (Method-II) rinses in an organic solvent for removing salts, lipids, and metabolites. Another FF tissue is directly mounted on the conductive ITO-coated glass slide or MALDI target plate for matrix spraying and MSI analysis (Method-III). Enzymatic release of N-glycans is performed using PNGase F under incubation at 37°C (Method-I). On-tissue derivatization is required (Method-I and II) before matrix deposition to stabilize glycans and enhance the ionization efficiency. After automated matrix spraying, the sections are subjected to a laser source for data acquisition and analysis.
The release of glycans from glycoproteins is crucial and facilitates its analysis by increasing sensitivity and simplicity. The methods for releasing glycans vary between N-linked and O-linked glycans and can be divided into chemical and enzymatic reactions (Table 1). The chemical methods can release both N-linked and O-linked glycans, but those methods may degrade the released glycans and proteins, forming other by-products (Koutsioulis et al., 2008). Hence, the enzymatic release of glycans is widely applied for MSI analysis. Peptide-N-glycosidase F (PNGase F) is a conventionally used enzyme to release N-glycans (Figure 2A-i). It is an amidase enzyme that dissociates N-glycans and asparagine residues from the innermost N-acetylglucosamine (GlcNAc). The PNGase F is used to release high mannose, complex, and hybrid N-glycans due to its broad specificity. The only limitation for such enzymes is that one cannot cleave if an α(1→3)-fucose is linked to the core GlcNAc residue (Veillon et al., 2017). Glycopeptidase A hydrolyzes oligosaccharides containing a fucose residue α(1→3)-linked to the core GlcNAc residue (Figure 2A-ii). Endoglycosidase H cleaves the β-1,4 glycosidic linkage of high-mannose N-linked glycans between GlcNAc1 and GlcNAc2 residues (Figure 2A-iii). They are also known as chitobiose. While most N-linked oligosaccharides can be removed using PNGase F, there are no universal and efficient enzymes for releasing O-linked glycans. For example, endo-α-N-acetylgalactosaminidase only cleaves O-glycans with a core-1 such as β(1→3)-linkage to the GalNAc structure. Another novel O-glycosidase has a slightly broader specificity, cleaves both the core-1 and then core-3 structures (Koutsioulis et al., 2008; Guthrie and Magnelli, 2016). In another way, monosaccharides must be sequentially hydrolyzed by a series of exoglycosidases until only the Gal-β(1→3)-GalNAc core remains. α(2→3,6,8,9)-neuraminidase enzyme is capable of efficient cleavage of the NeuAc-α(2→8)-NeuAc bond (Figure 2B-i). Disialylated (NeuNAc) O-linked core-2 hexasaccharide is sequentially degraded by using α(2→3,6,8,9) neuraminidase (Figure 2B-i). β(1→4)-galactose (Gal) residues are removed by β(1→4)-galactosidase, and N-acetylglucosamine (GlcNAc) residues are cleaved by N-acetylglucosaminidase (Figure 2B-ii). Finally, O-glycosidase (endoglycosidase) hydrolyzes the serine or threonine-linked unsubstituted O-glycan core [Gal-β(1→3)-GalNAc] (Figure 2B-iii). Any type of modification in the core structure can stop the action of O-glycosidase. These enzymes can be applied on the tissue of interest by an automated sprayer.
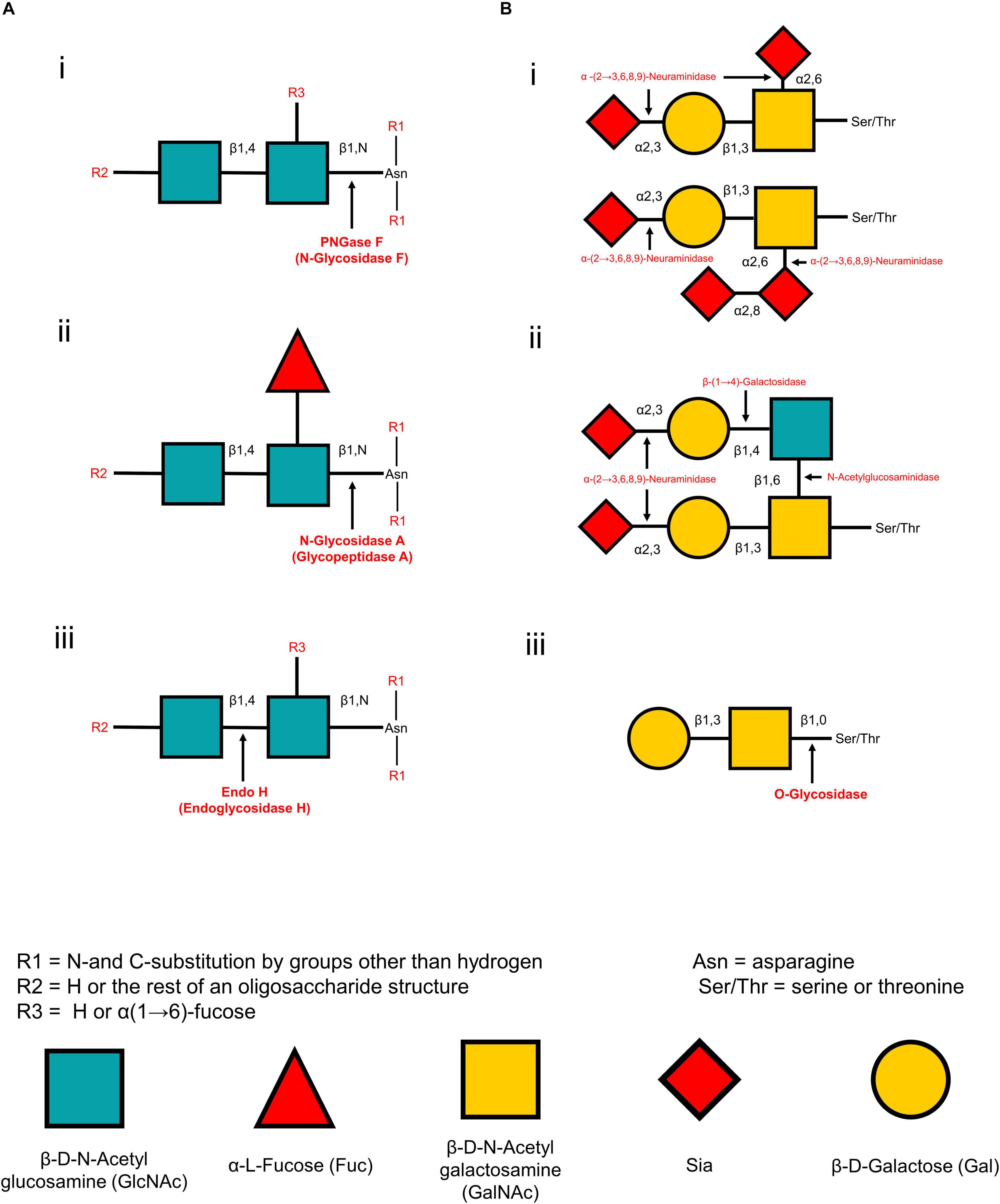
Figure 2. Cleaving sites for enzymatic deglycosylation of glycans. N-glycan strategies (A) and O-glycan strategies (B). PNGase F cleaves all asparagine-linked complex, hybrid, or high mannose oligosaccharides unless the core contains an α(1→3)-fucose. Cleaving site for PNGase F (A-i). N-glycosidase A hydrolyzes oligosaccharides containing a fucose residue α(1→3)-linked to the asparagine-linked N-acetylglucosamine (A-ii). Endo H cleaves the β-1,4 glycosidic linkage of high-mannose N-linked glycans between GlcNAc1 and GlcNAc2 (A-iii). In a sequential glycolytic cleavage, disialylated and trisialylated O-linked glycans have the sialic acid residues (NeuNAc) removed by α(2→3,6,8,9)-neuraminidase to release core-1 type O-glycans (B-i). Disialylated O-linked Core-2 hexasaccharide or sialic acid residues are sequentially degraded by α(2→3,6,8,9)-neuraminidase, β(1→4)-galactosidase and N-acetylglucosaminidase to release core-1 type O-glycans (B-ii). Cleavage site for O-glycosidase on the core-1 type O-glycans after sequential glycolytic degradation (B-iii).
After being released, glycans are subjected to derivatization before matrix application and MSI analysis (Figure 1, method-I and II). Derivatization of released glycans is necessary to stabilize glycan structures and improve ionization efficiency (Holst et al., 2016). Among different approaches, the permethylation and amidation approaches are widely used in both released N-linked and O-linked glycans (Morelle et al., 2009).
The choice of a matrix is crucial in MSI analysis for tissue glycome. Homogeneous coating of the matrix allows the formation of co-crystals between matrix molecules and analytes. In addition, the matrix molecules must have the absorbing property for laser energy to aid in the soft ionization of analytes. Various matrices have been developed for glycome MSI and MS analysis listed in Table 2. The most common matrices for the MSI analysis of glycome are 2,5-dihydroxybenzoic acid (DHB) and α-cyano-4-hydroxycinnamic acid (CHCA). Usually, the matrices are sprayed on the tissue using an automated robotic sprayer such as HTX M5 SprayerTM (Stanback et al., 2021), HTX TM-SprayerTM (Shi et al., 2019), ImagePrep sprayer (Powers et al., 2014). After matrix coating, the tissue sections are sent to an MSI instruments for data acquisition and analysis. Assignments or annotation of glycans can be performed by online databases such as GlycoWorkbench and GlycanBuilder (Ceroni et al., 2007, 2008) are the tools used for the drawing of glycan structures and automatically matching these models and their theoretical fragments with the experimental mass spectra. Consortium for functional glycomics1 for permethylated glycans, PeakFinder tool2, GlycanMass3, GlycomeDB4, Glycosciences5, PRIDE Archive search for glycoproteins6, GlycoMod tool7 is used for N-glycan-, O-glycan-, and fragments of glycosaminoglycan annotations, METLIN8, and Human Metabolome Database9 (Kunzke et al., 2017; Yang et al., 2017; Saldova and Wilkinson, 2020).
The MSI study for glycome has recently become a rapidly growing field of research. The tissue-specific or disease-associated glycome, which may act as molecular signatures for diagnosis, can be detected by direct imaging analyses and profiling of the glycome in the tissue sections. Diverse glycans are expressed in different brain tissues reported previously (Chen et al., 1998; Kleene and Schachner, 2004; Fang et al., 2016).
While the MSI study of lipidomics in brain tissues has been conducted extensively in the last decades, glycomics was insufficient. It may be caused by challenges for laborious and delicate sample preparation using tissue sections. Diverse glycome structures, characterization, glycosylation patterns were reported by several MS methods using brain tissue in normal and disease conditions (Chen et al., 1998; Geyer et al., 2001; Liedtke et al., 2001; Ohl et al., 2003; Bleckmann et al., 2009; Breloy et al., 2012; Gizaw et al., 2016; Samal et al., 2020). In addition, the spatial information of glycome in the tissue section is lost through this glycomics procedure due to the sample homogenization (Eshghi et al., 2014).
For the first time, a MALDI-MSI method has been applied to spatially profile the location and distribution of multiple N-linked glycans species in mouse brain tissues in 2013, followed by PNGase F digestion (Powers et al., 2013). This method has been developed to profile the multiple glycan species simultaneously released from intracellular organelle and cell surface glycoproteins. Thus extracted glycans were structurally analyzed by the MS method. In another study, the MSI technique was primarily used to directly analyze 42 N-glycans in FFPE mouse brain tissue sections (Eshghi et al., 2014). This procedure includes sectioning FFPE tissues, deparaffinization, rehydration, denaturing tissue proteins, releasing N-linked glycans followed by PNGase F digestion, matrix coating, and analyzing N-glycans by MALDI-MSI. The novel sub-atmospheric pressure (SubAP)/MALDI-MS system coupled with a Q Exactive HF hybrid quadrupole-orbitrap mass spectrometer was used for characterization and spatial visualization of 55 N-glycans in FFPE mouse brain tissue sections (Shi et al., 2019). PNGase F was used to promote the release of N-glycan from the brain tissues in these studies. A comprehensive MSI protocol was developed to visualize in situ N-glycans (at least 40 or more individual N-glycans were mapped) using FFPE and FF tissues (Drake et al., 2018). PNGase F-based N-glycan identification in situ multimodal MSI technique could then be applied to visualize N-linked glycans and proteins from the identical FFPE tissue section (Heijs et al., 2016). The novel combination of PNGase F with glycosidase (sialidase) was found to recognize N-glycan with greater sensitivity (Powers et al., 2013). MALDI-MSI and its advanced MALDI-2-MSI have recently been reported to show the distribution of several N-glycans (Heijs et al., 2020) in the human cerebellum from post-mortem brain tissue. In their analysis, MALDI-2-MSI has demonstrated that the sensitivity for detecting molecular [M–H]– species of N-glycans increased by about three orders of magnitude, and sensitivity has increased by about a factor of 10 in positive ion mode analysis compared to the current gold standard. They also reported that enormously high structural information of complex N-glycans was obtained directly from thin tissue sections in the human cerebellum and upon low-energy collision-induced dissociation tandem MS. However, MALDI-MSI techniques are capable of visualizing hundreds of glycome with spatial information in a single brain tissue followed by PNGase F treated or non-treated sample (Figure 3A; Stanback et al., 2021), which is limited by the typical antibody staining methods. Figure 3A showed the representative ion images of Figure 3A-iv Hex5HexNAc4Fuc1 at m/z 1809, (Figure 3A-v) Hex7HexNAc2 at m/z 1581, (Figure 3A-vi) Hex9HexNAc2 at m/z 1905, and (Figure 3A-vii) the overlay ion image of those N-glycans in FFPE and FF mouse brain tissues by the standard MALDI-MSI protocols described by Stanback et al., 2021.
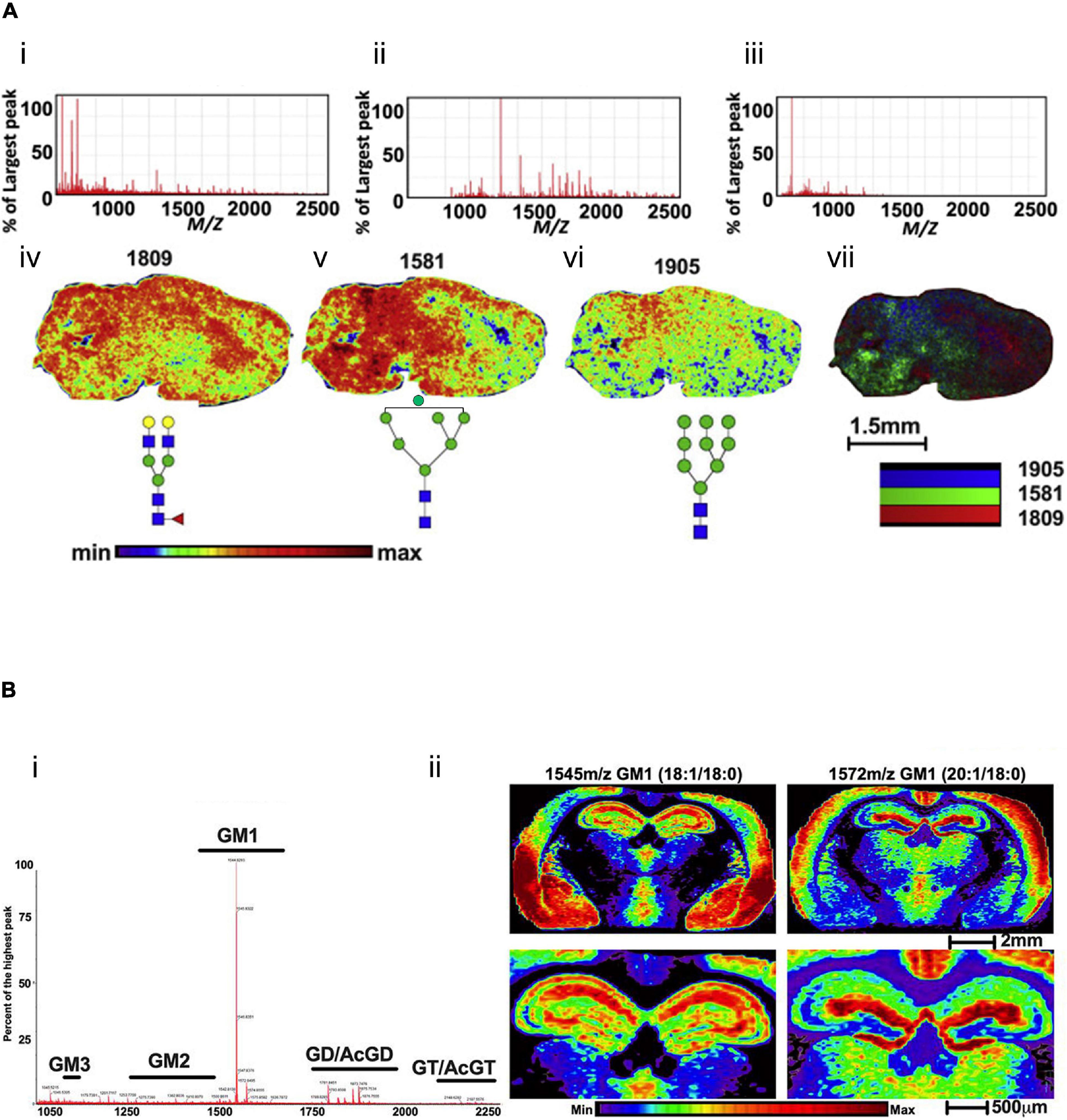
Figure 3. Mass spectra and ion images for glycome in mouse brain tissues. (A) PNGase F digestion and MALDI-MSI analysis of N-glycans in mouse brain tissue. Extracted mass spectra of released N-glycans and matrix ions for (A-i) both, (A-ii) N-glycans and (A-iii) matrix. Representative ion images of (A-iv) Hex5HexNAc4Fuc1 at m/z 1809, (A-v) Hex7HexNAc2 at m/z 1581, (A-vi) Hex9HexNAc2 at m/z 1905, and (A-vii) shows the overlay image of m/z 1809 (red), m/z 1581 (green), and m/z 1905 (blue). Scale bar, 1.5 mm. The mass spectra (B-i) and ion images (B-ii) of gangliosides in the mouse brain coronal section without pre-treatment of the enzyme. Gangliosides are mostly localized in the cortex and hippocampus of the mouse brain tissue section using the CHCA matrix. Scale bar, 2 mm (upper panel), 500 μm (lower panel). The figures of panels (A,B) are reprinted and modified from Stanback et al. (2021) and Andres et al. (2020), respectively, with free-reuse permission.
PNGase F-based MSI procedures have widely been used for tissue localization of N-glycans. Special care should be taken in sample preparation steps to get efficient results. This broad specificity enzyme is applicable for a wide range of N-linked glycans MSI analysis and cannot release other glycans such as O-linked glycans.
Kunzke et al. (2017) published a report on O-glycans MSI analysis. They performed MSI of native and O-linked glycans fragmentation from tissue microarray and FFPE cancer tissues without prior digestion or chemical reactions using MALDI-FT-ICR-MSI (Kunzke et al., 2017). By employing their protocol, four O-linked glycans have been detected and visualized. In addition, On-tissue spatially resolved glycoproteomics strategies combined with MALDI-MSI demonstrated the global dysregulation of N-linked, O-linked glycans in canine glioma biopsies tissue (Malaker et al., 2020). This preliminary report directly links glycan imaging with intact glycopeptide identification and characterization using canine glioma FFPE tissues.
N-linked glycans and their diverse heterogeneity in different biological tissues have been analyzed in detail by the different MSI techniques. However, MSI analysis for O-linked glycome has not been widely conducted in brain tissues and other organs. This is primarily attributed to the absence of a standardized enzyme with broad specificity, enabling to release of a diverse range of O-glycans, and the lack of knowledge about their O-linked counterparts. Intact O-glycans can be released chemically using a β-elimination reaction. The release of O-glycans by β-elimination or other chemical reactions is widely used for MS analysis. In this method, released glycans can be degraded, and existing knock-on effects must be overcome (Saldova and Wilkinson, 2020). The chemical method is not suitable for tissue release of glycans as it contains chemicals itself and is challenging to remove before MSI analysis (Veillon et al., 2017). O-glycans can be sequentially trimmed using a combination of exoglycosidases and an endoglycosidase (Figure 2B). Although this method can preserve a structure of proteins and activity, it may degrade the glycans (Guthrie and Magnelli, 2016, 14(2)). Thus, unlike N-linked glycans, MSI analysis of O-glycans from a thin biological tissue has not been widely investigated. Currently, no known broad specificity enzymes can release O-glycans from the protein in tissue samples. As a result, MSI analysis of O-glycans is far more complicated than for N-glycans. Universal O-glycosidases must be developed for the MSI analyses of O-glycans. Although several recombinant enzymes and chemical deglycosylation strategies are commercially available. However, more sophisticated enzymatic, chemical, and trimming methods should be developed for MSI of O-linked glycans. On the other hand, O-linked glycans can be detected and visualized as conjugated or fragmented directly from FF or FFPE brain tissues without prior digestion or chemical reactions (Kunzke et al., 2017).
In addition to glycans, glycoconjugates (glycolipids) are also essential biomolecules in the brain that result from glycosylation. Apart from glycan identification, MSI techniques have been utilized to detect several glycoconjugates in tissue sections. Ganglioside-monosialic acid (GM1) is a subtype of a glycolipid known as an important area of research for studying AD. An altered GM1 to GM2/GM3 ganglioside metabolism was observed using multimodal TOF-SIMS and MALDI-based MSI strategy in the transgenic mouse model and the human brain of AD (Hirano-Sakamaki et al., 2015; Michno et al., 2019; Scholarship et al., 2020). The gangliosides (i.e., C18- or C20-sphingosine) were detected using MALDI-MSI in the frontal brain or dentate gyrus in mouse brain tissue (Sugiura et al., 2008). A prototype MSI of the iMScope (Shimadzu) has been applied to monitor the accumulation of GM2 in the hypothalamus, hippocampus, and cerebellum of the Sandhoff disease model mice (Kitakaze et al., 2016). This cutting-edge technology is also known as a mass microscope. Aside from GM1, GM2, and GM3, another glycolipid GD1 was detected and visualized spatially by MALDI-MSI in the mouse and rat brain tissue (Sugiura et al., 2008; Caughlin et al., 2017). Recently, transmission-mode geometry MALDI-MSI (t-MALDI–MSI) and MALDI-2-MSI are used to visualize several glycolipids in subcellular resolution with higher sensitivity in the cerebellum of the mouse brain tissues (Niehaus et al., 2019). N-linked, O-linked glycoconjugates, and S-linked glycopeptides have been characterized and visualized in the FFPE tissue of glioma biopsies (Malaker et al., 2020). GM1, a most abundant ganglioside family in the brain, has been visualized with higher spatial resolution in FF mouse brain tissue without enzymatic pre-treatments using SYNAPT G2-Si high definition mass spectrometry (Waters, Milford, MA, United States) (Andres et al., 2020). MALDI-FT-ICR MSI has been applied to visualize tissue localization of glycosphingolipids accumulation in Gaucher disease of mouse brain tissues (Jones et al., 2017). The unique distribution of specific glycosphingolipids family has been visualized by MALDI-MSI, DESI-MSI, and SIMS imaging techniques in different tissues, cells, and model membranes (Luberto et al., 2019). A combination of laser microdissection and liquid chromatography-electrospray ionization mass spectrometry (LC-ESI-MS) was employed for highly sensitive tissue localization of most glycolipid structural isomers in the mouse brain tissues (Ikeda and Taguchi, 2010).
Using MALDI-MSI, several types of gangliosides were spatially visualized in FF mice brain tissue without PNGase F digestion (Figure 3B; Andres et al., 2020). These analyses demonstrated that gangliosides GM1 are most abundant in the mouse brain (Figure 3B-i). In addition, a coronal image of the whole brain reveals differential cortex localization for GM1 18:1/18:0 and GM1 20:1/18:0, two of the most abundant ganglioside subtypes (Figure 3B-ii). GM1 18:1/18:0 is primarily localized in the piriform, amygdala nucleus, and striatum, where the 20:1/18:0 is localized in the anterior region of layers 1 and 2 of primary and supplemental somatosensory and the dorsal auditory areas in the brain.
Currently, there is no glycans-cleaving enzyme from glycolipids. MSI of glycolipids in brain tissues as conjugated forms is the only way for imaging analysis. The dedicated MSI technologies (i.e., sensitive for conjugate molecules, MS/MS capability) are required for glycolipids detection and visualization.
Several label-free and isotopic labeling strategies have been described extensively as the most frequently used quantitative methods based on separation techniques such as LC-MS for glycome in biological samples (Zaia, 2008, 2010; Hu et al., 2013). However, MSI is a qualitative technique and considered a semi-quantitative method for biomolecules in tissues. The quantification of tissue biomolecules (lipids, proteins, and carbohydrates) by the current MSI methods is challenging due to several limitations. These include (a) the high dependence of the detected signals on the matrix deposition, (b) the MALDI ionization yield of specific target molecules, and finally, (c) the ion suppression effect on the tissue section (Hamm et al., 2012). Several attempts have been reported for quantitative analysis by MSI in tissue biomolecules. For example, semi-quantitative analysis of gangliosides in mouse brain tissues has been reported using the combination of ESI and MALDI-MSI (Zhang et al., 2016). The small tissue biomolecules such as drugs, metabolites are quantitively analyzed by the MSI method using tissue extinction calculation as a normalization factor without a labeled standard (Hamm et al., 2012). This technique is limited to small tissue biomolecules and not suited for larger biomolecules such as glycans. The quantification in tissue biomolecules by the MSI methods is under the developmental stage. Hopefully, the quantitive analysis of glycome in brain tissues by MSI methods will be achieved in the near future.
Matrix-free ionization processes such as surface-assisted laser desorption ionization (SALDI) and laser desorption ionization (LDI) are emerging as alternative MSI techniques that can provide complementary insight into molecular distributions with high ionization efficiency in the biological tissue sections. In contrast to MALDI-MSI, the matrix-free LD-MSI processes are freed from interference induced by matrix-derived ion peaks in the lower mass range. In addition, desorption ionization using through-hole alumina membrane, one of the SALDI methods, drastically lowers the sample pre-treatment time and does not require a skilled technician or dedicated instruments for matrix application, providing higher reproducibility in mass accuracy and intensity (Hasan et al., 2021).
Previously, the matrix-free LDI methods have been reported, such as gold nanoparticles (Su and Tseng, 2007; Nayak and Knapp, 2010), desorption ionization on porous silicon (Cha, 2008), nanostructured weathering steel (Etxebarria et al., 2014), silver nanostructures (Sekuła et al., 2015), chemical vapor deposited graphene surfaces (Merino et al., 2020), carbon nanoparticles and graphene nanosheets (Banazadeh et al., 2018) for glycome MS and MSI analysis. Matrix-free LDI methods have made data interpretation easier for glycome MSI analysis compare to MALDI-MSI. In addition, most LDI techniques are designed for the highly sensitive, enhanced ionization efficiency for low-mass molecules. Therefore, these LDI methods could be applicable for MSI analysis of low-mass glycolipids.
Most MSI studies are conducted in a 2D fashion where only a single section of the entire sample volume is tested. Biological processes happen within a tissue volume and can be efficiently investigated as a whole organ to achieve comprehensive information regarding spatial and molecular complexity. By registering and stacking serial tissue sections, MSI techniques can produce 3D volumes imaging demonstrating the molecular distributions in the whole tissue and organ/animal (Seeley and Caprioli, 2012). The benefit of analyzing volumetric data has led to a quick rise in the application of 3D-MSI analysis using a single sample (Vos et al., 2021). In this process, multiple 2D mass spectrometric images can be reconstructed for 3D mapping of the molecules in the entire tissue section or organ through specialized image processing software. The exploration of 3D MSI has always been challenging due to complex MSI procedures and dedicated image processing software. Some investigations regarding 3D MSI have already been done using DESI (Eberlin et al., 2010), laser ablation electrospray ionization (LAESI) (Nemes et al., 2009), MALDI (Giordano et al., 2016), and SIMS (Seeley and Caprioli, 2012). The most prominent points are the MSI analyzer and image processing software. Consider those, the TOF, Orbitrap, and FT-ICR analyzers are most suitable for 3D MSI (Makarov et al., 2006). The BioMap (Novartis, Basel, Switzerland), MATLAB (Mathworks, Natick, MA, United States), Maya software (Autodesk, Inc.), FastRBF Interpolation Toolbox (FarField Technology, Inc.), Image J are most frequently used for image data processing (Seeley and Caprioli, 2012). The potentiality of MSI techniques to create 3D representations of biomolecules in a whole organ or tissue with the image reconstruction process and high throughput processing has been reviewed (Mamun et al., 2021). The 3D volume of brain MSI for glycome has not been investigated yet. It can be an effective technique for the 3D volume of glycome imaging in entire brain tissues. The advent of 3D IMS methods has allowed for more detailed ion images of connections between signaling pathways and disease processes in various complex organs like the brain. The combination of this technology with other methods of imaging has shown the capacity to bridge the distance between non-invasive functional imaging and fundamental biology. We expect that developments of 3D MSI will continue to advance and become a significant technology in brain glycobiology in the near future.
Glycan expression changes are thought to have a detrimental effect on brain function, leading to pathological brain diseases, including neurodegenerative disorders. Visualization of glycome and glycosylation patterns in complex brain tissues has been revolutionized by employing simultaneous label-free MSI techniques. The current state of MSI has generated a global snapshot of N-linked glycans in FFPE and FF brain tissue sections, showing the tissue localization, distribution, and relative abundance of glycan subtypes. MSI analysis of the counterpart, O-linked glycans, is rarely investigated due to its complicated procedures. Discovery of broad specificity O-glycosidases and development of highly sensitive MSI instruments and on-tissue quantification strategy will extend glycome research in brain tissues to analyze brain function and behavior.
MH, TK, and MS conceptualized this manuscript. All authors drafted the manuscript and have reviewed and agreed with the publication of this manuscript.
This study was supported by the Japan Agency for Medical Research and Development (AMED) under the grant number of JP20gm0910004, MEXT project for promoting public utilization of advanced research infrastructure (Imaging Platform) under the grant number of JPMXS0410300220, and the Japan Society for the Promotion of Science (JSPS) KAKENHI under the grant number of JP18H05268.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Andres, D. A., Young, L. E. A., Gentry, M. S., and Sun, R. C. (2020). Spatial profiling of gangliosides in mouse brain by mass spectrometry imaging. J. Lipid Res. 61:1537. doi: 10.1194/jlr.ILR120000870
Banazadeh, A., Peng, W., Veillon, L., and Mechref, Y. (2018). Carbon nanoparticles and graphene nanosheets as MALDI matrices in glycomics: a new approach to improve glycan profiling in biological samples. J. Am. Soc. Mass Spectrom. 29, 1892–1900. doi: 10.1007/s13361-018-1985-z
Bleckmann, C., Geyer, H., Lieberoth, A., Splittstoesser, F., Liu, Y., Feizi, T., et al. (2009). O-glycosylation pattern of CD24 from mouse brain. Biol. Chem. 390, 627–645. doi: 10.1515/BC.2009.044
Bowman, A. P., Blakney, G. T., Hendrickson, C. L., Ellis, S. R., Heeren, R. M. A., and Smith, D. F. (2020). Ultra-high mass resolving power, mass accuracy, and dynamic range MALDI mass spectrometry imaging by 21-T FT-ICR MS. Anal. Chem. 92, 3133–3142. doi: 10.1021/acs.analchem.9b04768
Brandenburg, K., and Holst, O. (2015). Glycolipids: Distribution and Biological Function. Chichester, UK: John Wiley & Sons, Ltd, 1–10. doi: 10.1002/9780470015902.a0001427.pub3
Brekk, O. R., Honey, J. R., Lee, S., Hallett, P. J., and Isacson, O. (2020). Cell type-specific lipid storage changes in Parkinson’s disease patient brains are recapitulated by experimental glycolipid disturbance. Proc. Natl. Acad. Sci. U.S.A. 117, 27646–27654. doi: 10.1073/pnas.2003021117
Breloy, I., Pacharra, S., Aust, C., and Hanisch, F. G. (2012). A sensitive gel-based global O-glycomics approach reveals high levels of mannosyl glycans in the high mass region of the mouse brain proteome. Biol. Chem. 393, 709–717. doi: 10.1515/hsz-2012-0214
Buck, A., Ly, A., Balluff, B., Sun, N., Gorzolka, K., Feuchtinger, A., et al. (2015). High-resolution MALDI-FT-ICR MS imaging for the analysis of metabolites from formalin-fixed, paraffin-embedded clinical tissue samples. J. Pathol. 237, 123–132. doi: 10.1002/path.4560
Buckeridge, M. S., and Grant Reid, J. S. (1994). Purification and properties of a novel β-galactosidase or exo-(1 → 4)-β-d-galactanase from the cotyledons of germinated Lupinus angustifolius L. seeds. Planta 192, 502–511. doi: 10.1007/BF00203588
Cabezas, J. A. (1991). Some questions and suggestions on the type references of the official nomenclature (IUB) for sialidase(s) and endosialidase [3]. Biochem. J. 278, 311–312. doi: 10.1042/bj2780311
Caughlin, S., Maheshwari, S., Weishaupt, N., Yeung, K. K. C., Cechetto, D. F., and Whitehead, S. N. (2017). Age-dependent and regional heterogeneity in the long-chain base of a-series gangliosides observed in the rat brain using MALDI imaging. Sci. Rep. 7, 1–12. doi: 10.1038/s41598-017-16389-z
Ceroni, A., Dell, A., and Haslam, S. M. (2007). The glycanbuilder: a fast, intuitive and flexible software tool for building and displaying glycan structures. Source Code Biol. Med. 2:3. doi: 10.1186/1751-0473-2-3
Ceroni, A., Maass, K., Geyer, H., Geyer, R., Dell, A., and Haslam, S. M. (2008). GlycoWorkbench: a tool for the computer-assisted annotation of mass spectra of glycans. J. Proteome Res. 7, 1650–1659. doi: 10.1021/pr7008252
Cha, S. (2008). Laser Desorption/Ionization Mass Spectrometry for Direct Profiling and Imaging of Small Molecules From Raw Biological Materials. Available online at: https://lib.dr.iastate.edu/rtd/15643 (accessed April 17, 2021).
Chai, W., Feizi, T., Yuen, C.-T., and Lawson, A. M. (1997). Nonreductive release of O-linked oligosaccharides from mucin glycoproteins for structure/function assignments as neoglycolipids: application in the detection of novel ligands for E-selectin. Glycobiology 7, 861–872. doi: 10.1093/glycob/7.6.861
Chan, K., Lanthier, P., Liu, X., Sandhu, J. K., Stanimirovic, D., and Li, J. (2009). MALDI mass spectrometry imaging of gangliosides in mouse brain using ionic liquid matrix. Anal. Chim. Acta 639, 57–61. doi: 10.1016/j.aca.2009.02.051
Chen, Y. J., Wing, D. R., Guile, G. R., Dwek, R. A., Harvey, D. J., and Zamze, S. (1998). Neutral N-glycans in adult rat brain tissue - Complete characterisation reveals fucosylated hybrid and complex structures. Eur. J. Biochem. 251, 691–703. doi: 10.1046/j.1432-1327.1998.2510691.x
Cheng, B., Xie, R., Dong, L., and Chen, X. (2016). Metabolic remodeling of cell-surface sialic acids: principles, applications, and recent advances. ChemBioChem 17, 11–27. doi: 10.1002/cbic.201500344
Cipolla, L., Gregori, M., and So, P.-W. (2011). Glycans in magnetic resonance imaging: determinants of relaxivity to smart agents, and potential applications in biomedicine. Curr. Med. Chem. 18, 1002–1018. doi: 10.2174/092986711794940851
Coff, L., Chan, J., Ramsland, P. A., and Guy, A. J. (2020). Identifying glycan motifs using a novel subtree mining approach. BMC Bioinform. 21:42. doi: 10.1186/s12859-020-3374-4
Collin, M., and Olsén, A. (2001). EndoS, a novel secreted protein from Streptococcus pyogenes with endoglycosidase activity on human IgG. EMBO J. 20, 3046–3055. doi: 10.1093/emboj/20.12.3046
Colsch, B., and Woods, A. S. (2010). Localization and imaging of sialylated glycosphingolipids in brain tissue sections by MALDI mass spectrometry. Glycobiology 20, 661–667. doi: 10.1093/glycob/cwq031
Cornett, D. S., Frappier, S. L., and Caprioli, R. M. (2008). MALDI-FTICR imaging mass spectrometry of drugs and metabolites in tissue. Anal. Chem. 80, 5648–5653. doi: 10.1021/ac800617s
Cummings, R. D., and Etzler, M. E. (2009). Antibodies and Lectins in Glycan Analysis. Cold Spring Harbor Laboratory Press. Available online at: http://www.ncbi.nlm.nih.gov/pubmed/20301245 (accessed February 9, 2021)
Dai, Y., Whittal, R. M., Bridges, C. A., Isogai, Y., Hindsgaul, O., and Li, L. (1997). Matrix-assisted laser desorption ionization mass spectrometry for the analysis of monosulfated oligosaccharides. Carbohydr. Res. 304, 1–9. doi: 10.1016/S0008-6215(97)00195-X
Doran, R. C., Tatsuno, G. P., O’Rourke, S. M., Yu, B., Alexander, D. L., Mesa, K. A., et al. (2018). Glycan modifications to the gp120 immunogens used in the RV144 vaccine trial improve binding to broadly neutralizing antibodies. PLoS One 13:e0196370. doi: 10.1371/journal.pone.0196370
Drake, R. R., Powers, T. W., Norris-Caneda, K., Mehta, A. S., and Angel, P. M. (2018). In situ imaging of N-glycans by MALDI imaging mass spectrometry of fresh or formalin-fixed paraffin-embedded tissue. Curr. Protoc. Protein Sci. 94:e68. doi: 10.1002/cpps.68
Dreisewerd, K., Kölbl, S., Peter-Katalinić, J., Berkenkamp, S., and Pohlentz, G. (2006). Analysis of native milk oligosaccharides directly from thin-layer chromatography plates by matrix-assisted laser desorption/ionization orthogonal-time-of-flight mass spectrometry with a glycerol matrix. J. Am. Soc. Mass Spectrom. 17, 139–150. doi: 10.1016/j.jasms.2005.10.003
Dueñas, M. E., Carlucci, L., and Lee, Y. J. (2016). Matrix recrystallization for MALDI-MS imaging of maize lipids at high-spatial resolution. J. Am. Soc. Mass Spectrom. 27, 1575–1578. doi: 10.1007/s13361-016-1422-0
Eberlin, L. S., Ifa, D. R., Wu, C., and Graham Cooks, R. (2010). Three-dimensional vizualization of mouse brain by lipid analysis using ambient ionization mass spectrometry. Angew. Chemie - Int. Ed. 49, 873–876. doi: 10.1002/anie.200906283
Eriksson, C., Masaki, N., Yao, I., Hayasaka, T., and Setou, M. (2013). MALDI imaging mass spectrometry—a mini review of methods and recent developments. Mass Spectrom. 2, S0022–S0022. doi: 10.5702/massspectrometry.s0022
Eshghi, S. T., Yang, S., Wang, X., Shah, P., Li, X., and Zhang, H. (2014). Imaging of N-linked glycans from formalin-fixed paraffin-embedded tissue sections using MALDI mass spectrometry. ACS Chem. Biol. 9, 2149–2156. doi: 10.1021/cb500405h
Etxebarria, J., Calvo, J., and Reichardt, N. C. (2014). Nanostructured weathering steel for matrix-free laser desorption ionisation mass spectrometry and imaging of metabolites, drugs and complex glycans. Analyst 139, 2873–2883. doi: 10.1039/c4an00216d
Fang, P., Wang, X. J., Xue, Y., Liu, M. Q., Zeng, W. F., Zhang, Y., et al. (2016). In-depth mapping of the mouse brain N-glycoproteome reveals widespread N-glycosylation of diverse brain proteins. Oncotarget 7, 38796–38809. doi: 10.18632/oncotarget.9737
Freeze, H. H., and Kranz, C. (2010). Endoglycosidase and glycoamidase release of N-linked glycans. Curr. Protoc. Mol. Biol. 2010, Chater8:Unit8.15. doi: 10.1002/0471142727.mb1713as89
Freeze, H. H., Eklund, E. A., Ng, B. G., and Patterson, M. C. (2012). Neurology of inherited glycosylation disorders. Lancet Neurol. 11, 453–466. doi: 10.1016/S1474-4422(12)70040-6
Fujita, K., Kobayashi, K., Iwamatsu, A., Takeuchi, M., Kumagai, H., and Yamamoto, K. (2004). Molecular cloning of mucor hiemalis endo-β-N-acetylglucosaminidase and some properties of the recombinant enzyme. Arch. Biochem. Biophys. 432, 41–49. doi: 10.1016/j.abb.2004.09.013
Fukuyama, Y., Funakoshi, N., Takeyama, K., Hioki, Y., Nishikaze, T., Kaneshiro, K., et al. (2014). 3-aminoquinoline/p-coumaric acid as a MALDI matrix for glycopeptides, carbohydrates, and phosphopeptides. Anal. Chem. 86, 1937–1942. doi: 10.1021/ac4037087
Furuki, K., Toyo’oka, T., and Ban, K. (2017). Highly sensitive glycosylamine labelling of O-glycans using non-reductive β-elimination. Anal. Bioanal. Chem. 409, 2269–2283. doi: 10.1007/s00216-016-0171-z
Gabius, H.-J. (2009). The Sugar Code: Fundamentals of Glycosciences, Wiley Blackwell, Weinheim. ∗city bub.
Gagiannis, D., Gossrau, R., Reutter, W., Zimmermann-Kordmann, M., and Horstkorte, R. (2007). Engineering the sialic acid in organs of mice using N-propanoylmannosamine. Biochim. Biophys. Acta Gen. Subj. 1770, 297–306. doi: 10.1016/j.bbagen.2006.09.023
Geyer, H., Bahr, U., Liedtke, S., Schachner, M., and Geyer, R. (2001). Core structures of polysialylated glycans present in neural cell adhesion molecule from newborn mouse brain. Eur. J. Biochem. 268, 6587–6599. doi: 10.1046/j.0014-2956.2001.02613.x
Giordano, S., Morosi, L., Veglianese, P., Licandro, S. A., Frapolli, R., Zucchetti, M., et al. (2016). 3D mass spectrometry imaging reveals a very heterogeneous drug distribution in tumors. Sci. Rep. 6, 1–8. doi: 10.1038/srep37027
Gizaw, S. T., Ohashi, T., Tanaka, M., Hinou, H., and Nishimura, S. I. (2016). Glycoblotting method allows for rapid and efficient glycome profiling of human Alzheimer’s disease brain, serum and cerebrospinal fluid towards potential biomarker discovery. Biochim. Biophys. Acta Gen. Subj. 1860, 1716–1727. doi: 10.1016/j.bbagen.2016.03.009
Goetz, J. A., Novotny, M. V., and Mechref, Y. (2009). Enzymatic/chemical release of O-glycans allowing MS analysis at high sensitivity. Anal. Chem. 81, 9546–9552. doi: 10.1021/ac901363h
Griffith, C. M., and Wiley, M. J. (1989). The distribution of cell surface glycoconjugates during mouse secondary neurulation. Anat. Embryol. (Berl). 180, 567–575. doi: 10.1007/BF00300554
Guthrie, E., and Magnelli, P. E. (2016). Using glycosidases to remove, trim, or modify glycans on therapeutic proteins. Bioprocess Int. 14. ∗page,Google Scholar
Hamaguchi, T., Ito, T., Inoue, Y., Limpaseni, T., Pongsawasdi, P., and Ito, K. (2009). Purification, characterization and molecular cloning of a novel endo-β-N-acetylglucosaminidase from the basidiomycete. Flammulina Velutipes. Glycobiology 20, 420–432. doi: 10.1093/glycob/cwp188
Hamm, G., Bonnel, D., Legouffe, R., Pamelard, F., Delbos, J. M., Bouzom, F., et al. (2012). Quantitative mass spectrometry imaging of propranolol and olanzapine using tissue extinction calculation as normalization factor. J. Proteomics 75, 4952–4961. doi: 10.1016/j.jprot.2012.07.035
Harvey, D. J. (2011). Analysis of carbohydrates and glycoconjugates by matrix-assisted laser desorption/ionization mass spectrometry: an update for the period 2005-2006. Mass Spectrom. Rev. 30, 1–100. doi: 10.1002/mas.20265
Hasan, M. M., Eto, F., Al Mamun, M., Sato, S., Islam, A., Waliullah, A. S. M., et al. (2021). Desorption ionization using through-hole alumina membrane offers higher reproducibility than 2,5-dihydroxybenzoic acid, a widely used matrix in Fourier transform ion cyclotron resonance mass spectrometry imaging analysis. Rapid Commun. Mass Spectrom. 35:35. doi: 10.1002/rcm.9076
Heijs, B., Carreira, R. J., Tolner, E. A., De Ru, A. H., Van Den Maagdenberg, A. M. J. M., Van Veelen, P. A., et al. (2015). Comprehensive analysis of the mouse brain proteome sampled in mass spectrometry imaging. Anal. Chem. 87, 1867–1875. doi: 10.1021/ac503952q
Heijs, B., Holst, S., Briaire-De Bruijn, I. H., Van Pelt, G. W., De Ru, A. H., Van Veelen, P. A., et al. (2016). Multimodal mass spectrometry imaging of N-glycans and proteins from the same tissue section. Anal. Chem. 88, 7745–7753. doi: 10.1021/acs.analchem.6b01739
Heijs, B., Potthoff, A., Soltwisch, J., and Dreisewerd, K. (2020). MALDI-2 for the enhanced analysis of N-linked glycans by mass spectrometry imaging. Anal. Chem. 92, 13904–13911. doi: 10.1021/acs.analchem.0c02732
Hirabayashi, J. (2004). Lectin-based structural glycomics: Glycoproteomics and glycan profiling. Glycocon. J. 21, 35–40. doi: 10.1023/B:GLYC.0000043745.18988.a1
Hirano-Sakamaki, W., Sugiyama, E., Hayasaka, T., Ravid, R., Setou, M., and Taki, T. (2015). Alzheimer’s disease is associated with disordered localization of ganglioside GM1 molecular species in the human dentate gyrus. FEBS Lett. 589, 3611–3616. doi: 10.1016/j.febslet.2015.09.033
Holst, S., Heijs, B., De Haan, N., Van Zeijl, R. J. M., Briaire-De Bruijn, I. H., Van Pelt, G. W., et al. (2016). Linkage-specific in situ sialic acid derivatization for N-glycan mass spectrometry imaging of formalin-fixed paraffin-embedded tissues. Anal. Chem. 88, 5904–5913. doi: 10.1021/acs.analchem.6b00819
Hu, Y., Zhou, S., Khalil, S. I., Renteria, C. L., and Mechref, Y. (2013). Glycomic profiling of tissue sections by LC-MS. Anal. Chem. 85, 4074–4079. doi: 10.1021/ac400106x
Huang, C. C., and Aminoff, D. (1972). Enzymes that destroy blood group specificity. V. the oligosaccharase of Clostridium perfringens. J. Biol. Chem. 247, 6737–6742. doi: 10.1016/S0021-9258(19)44648-6
Hwang, H., Zhaang, J., Chung, K. A., Leverenz, J. B., Zabetian, C. P., Peskind, E. R., et al. (2010). Glycoproteomics in neurodegenerative diseases. Mass Spectrom. Rev. 29, 79–125. doi: 10.1002/mas.20221
Ikeda, K., and Taguchi, R. (2010). Highly sensitive localization analysis of gangliosides and sulfatides including structural isomers in mouse cerebellum sections by combination of laser microdissection and hydrophilic interaction liquid chromatography/electrospray ionization mass spectrometry with theoretically expanded multiple reaction monitoring. Rapid Commun. Mass Spectrom. 24, 2957–2965. doi: 10.1002/rcm.4716
Imperiali, B., and O’Connor, S. E. (1999). Effect of N-linked glycosylatian on glycopeptide and glycoprotein structure. Curr. Opin. Chem. Biol. 3, 643–649. doi: 10.1016/S1367-5931(99)00021-6
Iqbal, S., Fard, M. G., Everest-Dass, A., Packer, N. H., and Parker, L. M. (2018). Understanding cellular glycan surfaces in the central nervous system. Biochem. Soc. Trans. 47, 89–100. doi: 10.1042/BST20180330
Islam, A., Takeyama, E., Al Mamun, M., Sato, T., Horikawa, M., Takahashi, Y., et al. (2020). Green nut oil or DHA supplementation restored decreased distribution levels of DHA containing phosphatidylcholines in the brain of a mouse model of dementia. Metabolites 10:153. doi: 10.3390/metabo10040153
Iwase, H., and Hotta, K. (1993). Release of O-linked glycoprotein glycans by endo-alpha-N-acetylgalactosaminidase. Methods Mol. Biol. 14, 151–159. doi: 10.1385/0-89603-226-4:151
Jackson, S. N., Muller, L., Roux, A., Oktem, B., Moskovets, E., Doroshenko, V. M., et al. (2018). AP-MALDI mass spectrometry imaging of gangliosides using 2,6-dihydroxyacetophenone. J. Am. Soc. Mass Spectrom. 29, 1463–1472. doi: 10.1007/s13361-018-1928-8
Jacobsen, J. H. W., Parker, L. M., Everest-Dass, A. V., Schartner, E. P., Tsiminis, G., Staikopoulos, V., et al. (2016). Novel imaging tools for investigating the role of immune signalling in the brain. Brain. Behav. Immun. 58, 40–47. doi: 10.1016/j.bbi.2016.04.014
Jobling, M. G., Yang, Z., Kam, W. R., Lencer, W. I., and Holmes, R. K. (2012). A single native ganglioside GM1-binding site is sufficient for cholera toxin to bind to cells and complete the intoxication pathway. MBio 3, e401–e412. doi: 10.1128/mBio.00401-12
Jones, E. E., Zhang, W., Zhao, X., Quiason, C., Dale, S., Shahidi-Latham, S., et al. (2017). Tissue localization of glycosphingolipid accumulation in a gaucher disease mouse brain by LC-ESI-MS/MS and high-resolution MALDI imaging mass spectrometry. SLAS Discov. 22, 1218–1228. doi: 10.1177/2472555217719372
Kaneshiro, K., Fukuyama, Y., Iwamoto, S., Sekiya, S., and Tanaka, K. (2011). Highly sensitive MALDI analyses of glycans by a new aminoquinoline-labeling method using 3-aminoquinoline/α-cyano-4-hydroxycinnamic acid liquid matrix. Anal. Chem. 83, 3663–3667. doi: 10.1021/ac103203v
Kiessling, L. L., and Pohl, N. L. (1996). Strength in numbers: non-natural polyvalent carbohydrate derivatives. Chem. Biol. 3, 71–77. doi: 10.1016/S1074-5521(96)90280-X
Kitakaze, K., Mizutani, Y., Sugiyama, E., Tasaki, C., Tsuji, D., Maita, N., et al. (2016). Protease-resistant modified human β-hexosaminidase B ameliorates symptoms in GM2 gangliosidosis model. J. Clin. Invest. 126, 1691–1703. doi: 10.1172/JCI85300
Klarić, T. S., Salopek, M., Micek, V., Gornik Kljaić, O., and Lauc, G. (2020). Post-natal developmental changes in the composition of the rat neocortical N-glycome. Glycobiology 31, 636–648. doi: 10.1093/glycob/cwaa108
Kleene, R., and Schachner, M. (2004). Glycans and neural cell interactions. Nat. Rev. Neurosci. 5, 195–208. doi: 10.1038/nrn1349
Kopitz, J. (2017). Lipid glycosylation: a primer for histochemists and cell biologists. Histochem. Cell Biol. 147, 175–198. doi: 10.1007/s00418-016-1518-4
Korte, A. R., Yandeau-Nelson, M. D., Nikolau, B. J., and Lee, Y. J. (2015). Subcellular-level resolution MALDI-MS imaging of maize leaf metabolites by MALDI-linear ion trap-Orbitrap mass spectrometer. Anal. Bioanal. Chem. 407, 2301–2309. doi: 10.1007/s00216-015-8460-5
Koutsioulis, D., Landry, D., and Guthrie, E. P. (2008). Novel endo-α- N -acetylgalactosaminidases with broader substrate specificity. Glycobiology 18, 799–805. doi: 10.1093/glycob/cwn069
Kunzke, T., Balluff, B., Feuchtinger, A., Buck, A., Langer, R., Luber, B., et al. (2017). Native glycan fragments detected by MALDI-FT-ICR mass spectrometry imaging impact gastric cancer biology and patient outcome. Oncotarget 8, 68012–68025. doi: 10.18632/oncotarget.19137
Laštovièková, M., Chmelik, J., and Bobalova, J. (2009). The combination of simple MALDI matrices for the improvement of intact glycoproteins and glycans analysis. Int. J. Mass Spectrom. 281, 82–88. doi: 10.1016/j.ijms.2008.12.017
Laughlin, S. T., and Bertozzi, C. R. (2009). Imaging the glycome. Proc. Natl. Acad. Sci. U.S.A. 106, 12–17. doi: 10.1073/pnas.0811481106
Laughlin, S. T., Baskin, J. M., Amacher, S. L., and Bertozzi, C. R. (2008). In vivo imaging of membrane-associated glycans in developing zebrafish. Science 320, 664–667. doi: 10.1126/science.1155106
Lecchi, P., Le, H. M. T., and Pannell, L. K. (1995). 6-Aza-2-thiothymine: a matrix for MALDI spectra of oligonucleotides. Nucleic Acids Res. 23, 1276–1277. doi: 10.1093/nar/23.7.1276
Lee, D., Huang, T. H., De La Cruz, A., Callejas, A., and Lois, C. (2017). Methods to investigate the structure and connectivity of the nervous system. Fly (Austin). 11, 224–238. doi: 10.1080/19336934.2017.1295189
Lee, J., Ha, S., Kim, M., Kim, S. W., Yun, J., Ozcan, S., et al. (2020). Spatial and temporal diversity of glycome expression in mammalian brain. Proc. Natl. Acad. Sci. U.S.A. 117, 28743–28753. doi: 10.1073/pnas.2014207117
Lesur, D., Duhirwe, G., and Kovensky, J. (2019). High resolution MALDI-TOF-MS and MS/MS: application for the structural characterization of sulfated oligosaccharides. Eur. J. Mass Spectrom. 25, 428–436. doi: 10.1177/1469066719851438
Libbrecht, S., Van den Haute, C., Malinouskaya, L., Gijsbers, R., and Baekelandt, V. (2017). Evaluation of WGA–Cre-dependent topological transgene expression in the rodent brain. Brain Struct. Funct. 222, 717–733. doi: 10.1007/s00429-016-1241-x
Liedtke, S., Geyer, H., Wuhrer, M., Geyer, R., Frank, G., Gerardy-Schahn, R., et al. (2001). Characterization of N-glycans from mouse brain neural cell adhesion molecule. Glycobiology 11, 373–384. doi: 10.1093/glycob/11.5.373
Lis, H., and Sharon, N. (1998). Lectins: carbohydrate-specific proteins that mediate cellular recognition. Chem. Rev. 98, 637–674. doi: 10.1021/cr940413g
Loboda, A. V., Krutchinsky, A. N., Bromirski, M., Ens, W., and Standing, K. G. (2000). A tandem quadrupole/time-of-flight mass spectrometer with a matrix-assisted laser desorption/ionization source: design and performance. Rapid Commun. Mass Spectrom. 14, 1047–1057. doi: 10.1002/1097-0231(20000630)14:12<1047::AID-RCM990<3.0.CO;2-E
Luberto, C., Haley, J. D., and Del Poeta, M. (2019). Imaging with mass spectrometry, the next frontier in sphingolipid research? A discussion on where we stand and the possibilities ahead. Chem. Phys. Lipids 219, 1–14. doi: 10.1016/j.chemphyslip.2019.01.001
Makarov, A. (2000). Electrostatic axially harmonic orbital trapping: a high-performance technique of mass analysis. Anal. Chem. 72, 1156–1162. doi: 10.1021/ac991131p
Makarov, A., Denisov, E., Kholomeev, A., Balschun, W., Lange, O., Strupat, K., et al. (2006). Performance evaluation of a hybrid linear ion trap/orbitrap mass spectrometer. Anal. Chem. 78, 2113–2120. doi: 10.1021/ac0518811
Malaker, S. A., Quanico, J., Romero, A. R., Kobeissy, F., Aboulouard, S., Tierny, D., et al. (2020). On-tissue spatially-resolved glycoproteomics guided by N-glycan imaging reveal global dysregulation of canine glioma glycoproteomic landscape. bioRxiv[preprint]. doi: 10.1101/2020.10.02.324434
Maley, F., Trimble, R. B., Tarentino, A. L., and Plummer, T. H. (1989). Characterization of glycoproteins and their associated oligosaccharides through the use of endoglycosidases. Anal. Biochem. 180, 195–204. doi: 10.1016/0003-2697(89)90115-2
Mamun, A., Islam, A., Eto, F., Sato, T., Kahyo, T., and Setou, M. (2021). Mass spectrometry-based phospholipid imaging: methods and findings. Expert Rev. Proteomics 17, 843–854. doi: 10.1080/14789450.2020.1880897
Marshall, A. G. (2000). Milestones in fourier transform ion cyclotron resonance mass spectrometry technique development. Int. J. Mass Spectrom. 200, 331–356. doi: 10.1016/S1387-3806(00)00324-9
McDonnell, L. A., and Heeren, R. M. A. (2007). Imaging mass spectrometry. Mass Spectrom. Rev. 26, 606–643. doi: 10.1002/mas.20124
Mechref, Y., and Novotny, M. V. (1998). Matrix-assisted laser desorption/ionization mass spectrometry of acidic glycoconjugates facilitated by the use of spermine as a co-matrix. J. Am. Soc. Mass Spectrom. 9, 1293–1302. doi: 10.1016/S1044-0305(98)00106-8
Mechref, Y., and Novotny, M. V. (2010). High-sensitivity analytical approaches to the analysis of N-glycans. Handbook Glycomics 1, 3–43. doi: 10.1016/B978-0-12-373600-0.00001-9
Mechref, Y., Novotny, M. V., and Krishnan, C. (2003). Structural characterization of oligosaccharides using MALDI-TOF/TOF tandem mass spectrometry. Anal. Chem. 75, 4895–4903. doi: 10.1021/ac0341968
Merino, J. P., Serna, S., Criado, A., Centeno, A., Napal, I., Calvo, J., et al. (2020). Mass spectrometry of carbohydrate-protein interactions on a glycan array conjugated to CVD graphene surfaces. 2D Mater. 7, 024003. doi: 10.1088/2053-1583/ab6030
Michno, W., Wehrli, P. M., Zetterberg, H., Blennow, K., and Hanrieder, J. (2019). GM1 locates to mature amyloid structures implicating a prominent role for glycolipid-protein interactions in Alzheimer pathology. Biochim. Biophys. Acta Proteins Proteomics 1867, 458–467. doi: 10.1016/j.bbapap.2018.09.010
Mohr, M. D., OlafBörnsen, K., and Widmer, H. M. (1995). Matrix-assisted laser desorption/ionization mass spectrometry: improved matrix for oligosaccharides. Rapid Commun. Mass Spectrom. 9, 809–814. doi: 10.1002/rcm.1290090919
Moll, T., Shaw, P. J., and Cooper-Knock, J. (2020). Disrupted glycosylation of lipids and proteins is a cause of neurodegeneration. Brain 143, 1332–1340. doi: 10.1093/brain/awz358
Morelle, W., Faid, V., Chirat, F., and Michalski, J. C. (2009). Analysis of N-and O-linked glycans from glycoproteins using MALDI-TOF mass spectrometry. Methods Mol. Biol. 534, 5–21. doi: 10.1007/978-1-59745-022-5_1
Moremen, K. W., Tiemeyer, M., and Nairn, A. V. (2012). Vertebrate protein glycosylation: diversity, synthesis and function. Nat. Rev. Mol. Cell Biol. 13, 448–462. doi: 10.1038/nrm3383
Mori, N., Mochizuki, T., Yamazaki, F., Takei, S., Mano, H., Matsugi, T., et al. (2019). MALDI imaging mass spectrometry revealed atropine distribution in the ocular tissues and its transit from anterior to posterior regions in the whole-eye of rabbit after topical administration. PLoS One 14:e0211376. doi: 10.1371/journal.pone.0211376
Mueller, T. M., Simmons, M. S., Helix, A. T., Haroutunian, V., and Meador-Woodruff, J. H. (2018). Glycosylation enzyme mRNA expression in dorsolateral prefrontal cortex of elderly patients with schizophrenia: Evidence for dysregulation of multiple glycosylation pathways. bioRxiv[preprint]. doi: 10.1101/369314
Nabi, M. M., Al Mamun, M., Islam, A., Hasan, M. M., Waliullah, A. S. M., Tamannaa, Z., et al. (2021). Mass spectrometry in the lipid study of cancer. Expert Rev. Proteomics 18, 201–219. doi: 10.1080/14789450.2021.1912602
Nayak, R., and Knapp, D. R. (2010). Matrix-free LDI mass spectrometry platform using patterned nanostructured gold thin film. Anal. Chem. 82, 7772–7778. doi: 10.1021/ac1017277
Nemes, P., Barton, A. A., and Vertes, A. (2009). Three-dimensional imaging of metabolites in tissues under ambient conditions by laser ablation electrospray ionization mass spectrometry. Anal. Chem. 81, 6668–6675. doi: 10.1021/ac900745e
Neubert, P., and Walch, A. (2013). Current frontiers in clinical research application of MALDI imaging mass spectrometry. Expert Rev. Proteomics 10, 259–273. doi: 10.1586/epr.13.19
Niehaus, M., Soltwisch, J., Belov, M. E., and Dreisewerd, K. (2019). Transmission-mode MALDI-2 mass spectrometry imaging of cells and tissues at subcellular resolution. Nat. Methods 16, 925–931. doi: 10.1038/s41592-019-0536-2
Niehaus, M., Soltwisch, J., Belov, M. E., and Dreisewerd, K. (2019). Transmission-mode MALDI-2 mass spectrometry imaging of cells and tissues at subcellular resolution. Nat. Methods 16, 925–931. doi: 10.1038/s41592-019-0536-2
Nonami, H., Tanaka, K., Fukuyama, Y., and Erra-Balsells, R. (1998). β-Carboline alkaloids as matrices for UV-matrix-assisted laser desorption/ionization time-of-flight mass spectrometry in positive and negative ion modes. Analysis of proteins of high molecular mass, and of cyclic and acyclic oligosaccharides. Rapid Commun. Mass Spectrom. 12, 285–296. doi: 10.1002/(SICI)1097-0231(19980331)12:6<285::AID-RCM158<3.0.CO;2-4
O’Neill, R. A. (1996). Enzymatic release of oligosaccharides from glycoproteins for chromatographic and electrophoretic analysis. J. Chromatogr. A 720, 201–215. doi: 10.1016/0021-9673(95)00502-1
Ohba, H., and Bakalova, R. (2003). Relationships between degree of binding, cytotoxicity and cytoagglutinating activity of plant-derived agglutinins in normal lymphocytes and cultured leukemic cell lines. Cancer Chemother. Pharmacol. 51, 451–458. doi: 10.1007/s00280-003-0607-y
Ohl, C., Albach, C., Altevogt, P., and Schmitz, B. (2003). N-glycosylation patterns of HSA/CD24 from differnet cell lines and brain homogenates: a comparison. Biochimie 85, 565–573. doi: 10.1016/S0300-9084(03)00107-X
Ohtsubo, K., and Marth, J. D. (2006). Glycosylation in cellular mechanisms of health and disease. Cell 126, 855–867. doi: 10.1016/j.cell.2006.08.019
Paessens, L. C., García-Vallejo, J. J., Fernandes, R. J., and van Kooyk, Y. (2007). The glycosylation of thymic microenvironments. a microscopic study using plant lectins. Immunol. Lett. 110, 65–73. doi: 10.1016/j.imlet.2007.03.005
Papac, D. I., Wong, A., and Jones, A. J. S. (1996). Analysis of acidic oligosaccharides and glycopeptides by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Anal. Chem. 68, 3215–3223. doi: 10.1021/ac960324z
Parker, L. M., Kumar, N. N., Lonergan, T., and Goodchild, A. K. (2013). Neurochemical codes of sympathetic preganglionic neurons activated by glucoprivation. J. Comp. Neurol. 521, 2703–2718. doi: 10.1002/cne.23310
Parker, L. M., Kumar, N. N., Lonergan, T., McMullan, S., and Goodchild, A. K. (2015). Distribution and neurochemical characterization of neurons in the rat ventrolateral medulla activated by glucoprivation. Brain Struct. Funct. 220, 117–134. doi: 10.1007/s00429-013-0642-3
Patel, T., Bruce, J., Merry, A., Bigge, C., Parekh, R., Wormald, M., et al. (1993). Use of hydrazine to release in intact and unreduced form both N- and O-linked oligosaccharides from glycoproteins. Biochemistry 32, 679–693. doi: 10.1021/bi00053a037
Piehowski, P. D., Zhu, Y., Bramer, L. M., Stratton, K. G., Zhao, R., Orton, D. J., et al. (2020). Automated mass spectrometry imaging of over 2000 proteins from tissue sections at 100-μm spatial resolution. Nat. Commun. 11, 1–12. doi: 10.1038/s41467-019-13858-z
Pilobello, K. T., and Mahal, L. K. (2007). Lectin microarrays for glycoprotein analysis. Methods Mol. Biol. 385, 193–203. doi: 10.1007/978-1-59745-426-1_14
Pitt, J. J., and Gorman, J. J. (1997). Oligosaccharide characterization and quantitation using 1-phenyl-3- methyl-5-pyrazolone derivatization and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Anal. Biochem. 248, 63–75. doi: 10.1006/abio.1997.2080
Potts, P. J. (1987). Mass Spectrometry: Principles and Instrumentation,” A Handbook of Silicate Rock Analysis. Boston, MA: Springer, 497–522. doi: 10.1007/978-1-4615-3270-5_16
Powers, T. W., Jones, E. E., Betesh, L. R., Romano, P. R., Gao, P., Copland, J. A., et al. (2013). Matrix assisted laser desorption ionization imaging mass spectrometry workflow for spatial profiling analysis of N-linked Glycan expression in tissues. Anal. Chem. 85, 9799–9806. doi: 10.1021/ac402108x
Powers, T. W., Neely, B. A., Shao, Y., Tang, H., Troyer, D. A., Mehta, A. S., et al. (2014). MALDI imaging mass spectrometry profiling of N-glycans in formalin-fixed paraffin embedded clinical tissue blocks and tissue microarrays. PLoS One 9:e106255. doi: 10.1371/journal.pone.0106255
Raghunathan, R., Sethi, M. K., and Zaia, J. (2019). On-slide tissue digestion for mass spectrometry based glycomic and proteomic profiling. MethodsX 6, 2329–2347. doi: 10.1016/j.mex.2019.09.029
Reily, C., Stewart, T. J., Renfrow, M. B., and Novak, J. (2019). Glycosylation in health and disease. Nat. Rev. Nephrol. 15, 346–366. doi: 10.1038/s41581-019-0129-4
Römpp, A., and Spengler, B. (2013). Mass spectrometry imaging with high resolution in mass and space. Histochem. Cell Biol. 139, 759–783. doi: 10.1007/s00418-013-1097-6
Ruhaak, L. R., Xu, G., Li, Q., Goonatilleke, E., and Lebrilla, C. B. (2018). Mass spectrometry approaches to glycomic and glycoproteomic analyses. Chem. Rev. 118, 7886–7930. doi: 10.1021/acs.chemrev.7b00732
Saldova, R., and Wilkinson, H. (2020). Current methods for the characterization of o-glycans. J. Proteome Res. 19, 3890–3905. doi: 10.1021/acs.jproteome.0c00435
Samal, J., Saldova, R., Rudd, P. M., Pandit, A., and O’Flaherty, R. (2020). Region-specific characterization of N-glycans in the striatum and substantia nigra of an adult rodent brain. Anal. Chem. 92, 12842–12851. doi: 10.1021/acs.analchem.0c01206
Sampathkumar, S.-G., Li, A., and Yarema, K. (2008). Sialic acid and the central nervous system: perspectives on biological functions, detection, imaging methods and manipulation. CNS Neurol. Disord. Drug Targets 5, 425–440. doi: 10.2174/187152706777950729
Scholarship, W., Harris, A., and Yeung, S. (2020). Ganglioside Detection from Formalin Fixed Human Brain Tissue Ganglioside Detection from Formalin Fixed Human Brain Tissue Utilizing MALDI Imaging Mass Spectrometry Utilizing MALDI Imaging Mass Spectrometry. Available online at: https://ir.lib.uwo.ca/etdhttps://ir.lib.uwo.ca/etd/6898 (accessed May 18, 2021).
Schubert, K. O., Weiland, F., Baune, B. T., and Hoffmann, P. (2016). The use of MALDI-MSI in the investigation of psychiatric and neurodegenerative disorders: a review. Proteomics 16, 1747–1758. doi: 10.1002/pmic.201500460
Schwarz, R. E., Wojciechowicz, D. C., Picon, A. I., Schwarz, M. A., and Paty, P. B. (1999). Wheatgerm agglutinin-mediated toxicity in pancreatic cancer cells. Br. J. Cancer 80, 1754–1762. doi: 10.1038/sj.bjc.6690593
Scott, H., and Panin, V. M. (2014). N-Glycosylation in regulation of the nervous system. Adv. Neurobiol. 9, 367–394. doi: 10.1007/978-1-4939-1154-7_17
Seeley, E. H., and Caprioli, R. M. (2012). 3D imaging by mass spectrometry: a new frontier. Anal. Chem. 84, 2105–2110. doi: 10.1021/ac2032707
Sekuła, J., Nizioł, J., Rode, W., and Ruman, T. (2015). Silver nanostructures in laser desorption/ionization mass spectrometry and mass spectrometry imaging. Analyst 140, 6195–6209. doi: 10.1039/c5an00943j
Shajahan, A., Parashar, S., Goswami, S., Ahmed, S. M., Nagarajan, P., and Sampathkumar, S. G. (2017). Carbohydrate-neuroactive hybrid strategy for metabolic glycan engineering of the central nervous system in vivo. J. Am. Chem. Soc. 139, 693–700. doi: 10.1021/jacs.6b08894
Shi, Y., Li, Z., Felder, M. A., Yu, Q., Shi, X., Peng, Y., et al. (2019). Mass spectrometry imaging of N-glycans from formalin-fixed paraffin-embedded tissue sections using a novel subatmospheric pressure ionization source. Anal. Chem. 91, 12942–12947. doi: 10.1021/acs.analchem.9b02995
Shimma, S., Kubo, A., Satoh, T., and Toyoda, M. (2012). Detailed structural analysis of lipids directly on tissue specimens using a MALDI-spiraltof-Reflectron TOF mass spectrometer. PLoS One 7:e37107. doi: 10.1371/journal.pone.0037107
Smith, D. F., Podgorski, D. C., Rodgers, R. P., Blakney, G. T., and Hendrickson, C. L. (2018). 21 tesla FT-ICR mass spectrometer for ultrahigh-resolution analysis of complex organic mixtures. Anal. Chem. 90, 2041–2047. doi: 10.1021/acs.analchem.7b04159
Snovida, S. I., Rak-Banville, J. M., and Perreault, H. (2008). On the use of DHB/aniline and DHB/N,N-dimethylaniline matrices for improved detection of carbohydrates: automated identification of oligosaccharides and quantitative analysis of sialylated glycans by MALDI-TOF mass spectrometry. J. Am. Soc. Mass Spectrom. 19, 1138–1146. doi: 10.1016/j.jasms.2008.04.033
Song, E., and Mechref, Y. (2013). LC-MS/MS identification of the o-glycosylation and hydroxylation of amino acid residues of collagen α-1 (II) chain from bovine cartilage. J. Proteome Res. 12, 3599–3609. doi: 10.1021/pr400101t
Song, X., Ju, H., Lasanajak, Y., Kudelka, M. R., Smith, D. F., and Cummings, R. D. (2016). Oxidative release of natural glycans for functional glycomics. Nat. Methods 13, 528–534. doi: 10.1038/nmeth.3861
Stanback, A. E., Conroy, L. R., Young, L. E. A., Hawkinson, T. R., Markussen, K. H., Clarke, H. A., et al. (2021). Regional N-glycan and lipid analysis from tissues using MALDI-mass spectrometry imaging. STAR Protoc. 2:100304. doi: 10.1016/j.xpro.2021.100304
Sterner, E., Flanagan, N., and Gildersleeve, J. C. (2016). Perspectives on anti-glycan antibodies gleaned from development of a community resource database. ACS Chem. Biol. 11, 1773–1783. doi: 10.1021/acschembio.6b00244
Su, C. L., and Tseng, W. L. (2007). Gold nanoparticles as assisted matrix for determining neutral small carbohydrates through laser desorption/ionization time-of-flight mass spectrometry. Anal. Chem. 79, 1626–1633. doi: 10.1021/ac061747w
Sugiura, Y., and Setou, M. (2010). Imaging mass spectrometry for visualization of drug and endogenous metabolite distribution: toward in situ pharmacometabolomes. J. Neuroimmune Pharmacol. 5, 31–43. doi: 10.1007/s11481-009-9162-6
Sugiura, Y., Shimma, S., Konishi, Y., Yamada, M. K., and Setou, M. (2008). Imaging mass spectrometry technology and application on ganglioside study; visualization of age-dependent accumulation of C20-ganglioside molecular species in the mouse hippocampus. PLoS One 3:e3232. doi: 10.1371/journal.pone.0003232
Suzuki, S., Kakehi, K., and Honda, S. (1996). Comparison of the sensitivities of various derivatives of oligosaccharides in LC/MS with fast atom bombardment and electrospray ionization interfaces. Anal. Chem. 68, 2073–2083. doi: 10.1021/ac951144c
Tabuchi, K., Sawamoto, K., Suzuki, E., Ozaki, K., Sone, M., Hama, C., et al. (2000). GAL4/UAS-WGA system as a powerful tool for tracingDrosophila transsynaptic neural pathways. J. Neurosci. Res. 59, 94–99. doi: 10.1002/(SICI)1097-4547(20000101)59:1<94::AID-JNR11<3.0.CO;2-Q
Takasaki, S., Mizuochi, T., and Kobata, A. (1982). Hydrazinolysis of asparagine-linked sugar chains to produce free oligosaccharides. Methods Enzymol. 83, 263–268. doi: 10.1016/0076-6879(82)83019-X
Tarentino, A. L., and Plummer, T. H. (1994). Enzymatic deglycosylation of asparagine-linked glycans: purification, properties, and specificity of oligosaccharide-cleaving enzymes from Flavobacterium meningosepticum. Methods Enzymol. 230, 44–57. doi: 10.1016/0076-6879(94)30006-2
Tarentino, A. L., Gomez, C. M., and Plummer, T. H. (1985). Deglycosylation of asparagine-linked glycans by peptide: N-glycosidase F. Biochemistry 24, 4665–4671. doi: 10.1021/bi00338a028
Tarentino, A. L., Plummer, T. H., and Maley, F. (1974). The release of intact oligosaccharides from specific glycoproteins by endo β N acetylglucosaminidase H. J. Biol. Chem. 249, 818–824. doi: 10.1016/s0021-9258(19)43002-0
Tena, J., and Lebrilla, C. B. (2021). Glycomic profiling and the mammalian brain. Proc. Natl. Acad. Sci. U.S.A. 118, e2022238118. doi: 10.1073/PNAS.2022238118
Trastoy, B., Klontz, E., Orwenyo, J., Marina, A., Wang, L. X., Sundberg, E. J., et al. (2018). Structural basis for the recognition of complex-type N-glycans by endoglycosidase S. Nat. Commun. 9, 1–11. doi: 10.1038/s41467-018-04300-x
Tretter, V., Altmann, F., and Marz, L. (1991). Peptide-N4-(N-acetyl-beta-glucosaminyl)asparagine amidase F cannot release glycans with fucose attached alpha1 3 to the asparagine-linked N-acetylglucosamine residue. Eur. J. Biochem. 199, 647–652. doi: 10.1111/j.1432-1033.1991.tb16166.x
Trimble, R. B., and Tarentino, A. L. (1991). Identification of distinct endoglycosidase (endo) activities in flavobacterium meningosepticum: Endo F1, endo F2, and endo F3. Endo F1 and endo H hydrolyze only high mannose and hybrid glycans. J. Biol. Chem. 266, 1646–1651. doi: 10.1016/s0021-9258(18)52343-7
Van Den Steen, P., Rudd, P. M., Dwek, R. A., and Opdenakker, G. (1998). Concepts and principles of O-linked glycosylation. Crit. Rev. Biochem. Mol. Biol. 33, 151–208. doi: 10.1080/10409239891204198
Veillon, L., Fakih, C., Abou-El-Hassan, H., Kobeissy, F., and Mechref, Y. (2018). Glycosylation changes in brain cancer. ACS Chem. Neurosci. 9, 51–72. doi: 10.1021/acschemneuro.7b00271
Veillon, L., Zhou, S., and Mechref, Y. (2017). Quantitative glycomics: a combined analytical and bioinformatics approach. Methods Enzymol. 585, 431–477. doi: 10.1016/bs.mie.2016.11.006
Vens-Cappell, S., Kouzel, I. U., Kettling, H., Soltwisch, J., Bauwens, A., Porubsky, S., et al. (2016). On-tissue phospholipase C digestion for enhanced MALDI-MS imaging of neutral glycosphingolipids. Anal. Chem. 88, 5595–5599. doi: 10.1021/acs.analchem.6b01084
Vos, D. R. N., Ellis, S. R., Balluff, B., and Heeren, R. M. A. (2021). Experimental and data analysis considerations for three-dimensional mass spectrometry imaging in biomedical research. Mol. Imaging Biol. 23, 149–159. doi: 10.1007/s11307-020-01541-5
Walker, J. M., Hounsell, E. F., Davies, M. J., and Smith, K. D. (2003). Chemical release of O-linked oligosaccharide chains. Protein Proto. Handbook 8, 817–818. doi: 10.1385/1-59259-169-8:817
Whitehead, S. N., Chan, K. H. N., Gangaraju, S., Slinn, J., Li, J., and Hou, S. T. (2011). Imaging mass spectrometry detection of gangliosides species in the mouse brain following transient focal cerebral ischemia and long-term recovery. PLoS One 6:808. doi: 10.1371/journal.pone.0020808
Wildburger, N. C. (2017). MALDI-imaging mass spectrometry of brain lipids. Neuromethods 1203, 45–59. doi: 10.1007/978-1-4939-6946-3_4
Williams, S. E., Mealer, R. G., Scolnick, E. M., Smoller, J. W., and Cummings, R. D. (2020). Aberrant glycosylation in schizophrenia: a review of 25 years of post-mortem brain studies. Mol. Psychiatry 25, 3198–3207. doi: 10.1038/s41380-020-0761-1
Wu, Z., Bagarolo, G. I., Thoröe-Boveleth, S., and Jankowski, J. (2020). “Lipidomics”: mass spectrometric and chemometric analyses of lipids. Adv. Drug Deliv. Rev. 159, 294–307. doi: 10.1016/j.addr.2020.06.009
Xie, R., Dong, L., Du, Y., Zhu, Y., Hua, R., Zhang, C., et al. (2016). In vivo metabolic labeling of sialoglycans in the mouse brain by using a liposome-assisted bioorthogonal reporter strategy. Proc. Natl. Acad. Sci. U.S.A. 113, 5173–5178. doi: 10.1073/pnas.1516524113
Yale, A. R., Nourse, J. L., Lee, K. R., Ahmed, S. N., Arulmoli, J., Jiang, A. Y. L., et al. (2018). Cell surface N-glycans influence electrophysiological properties and fate potential of neural stem cells. Stem Cell Reports 11, 869–882. doi: 10.1016/j.stemcr.2018.08.011
Yamamoto, S., Muramatsu, H., and Muramatsu, T. (2005). Mutational studies on endo-β-N-acetylglucosaminidase D which hydrolyzes core portion of asparagine-linked complex type oligosaccharides. Glycoconj. J. 22, 35–42. doi: 10.1007/s10719-005-0847-7
Yang, J., and Caprioli, R. M. (2011). Matrix sublimation/recrystallization for imaging proteins by mass spectrometry at high spatial resolution. Anal. Chem. 83, 5728–5734. doi: 10.1021/ac200998a
Yang, S., Höti, N., Yang, W., Liu, Y., Chen, L., Li, S., et al. (2017). Simultaneous analyses of N-linked and O-linked glycans of ovarian cancer cells using solid-phase chemoenzymatic method. Clin. Proteomics 14:3. doi: 10.1186/s12014-017-9137-1
Yoshihara, Y. (2002). Visualizing selective neural pathways with WGA transgene: combination of neuroanatomy with gene technology. Neurosci. Res. 44, 133–140. doi: 10.1016/S0168-0102(02)00130-X
Yu, S. Y., Wu, S. W., and Khoo, K. H. (2006). Distinctive characteristics of MALDI-Q/TOF and TOF/TOF tandem mass spectrometry for sequencing of permethylated complex type N-glycans. Glycoconj. J. 23, 355–369. doi: 10.1007/s10719-006-8492-3
Zaia, J. (2008). Mass spectrometry and the emerging field of glycomics. Chem. Biol. 15, 881–892. doi: 10.1016/j.chembiol.2008.07.016
Zaia, J. (2010). Mass spectrometry and glycomics. Omi. J. Integr. Biol. 14, 401–418. doi: 10.1089/omi.2009.0146
Zavalin, A., Todd, E. M., Rawhouser, P. D., Yang, J., Norris, J. L., and Caprioli, R. M. (2012). Direct imaging of single cells and tissue at sub-cellular spatial resolution using transmission geometry MALDI MS. J. Mass Spectrom. 47:i. doi: 10.1002/jms.3132
Zavalin, A., Yang, J., Haase, A., Holle, A., and Caprioli, R. (2014). Implementation of a gaussian beam laser and aspheric optics for high spatial resolution MALDI imaging MS. J. Am. Soc. Mass Spectrom. 25, 1079–1082. doi: 10.1007/s13361-014-0872-5
Zhang, Q., Ma, C., Chin, L. S., and Li, L. (2020). Integrative glycoproteomics reveals protein n-glycosylation aberrations and glycoproteomic network alterations in Alzheimer’s disease. Sci. Adv. 6:eabc5802. doi: 10.1126/sciadv.abc5802
Zhang, Y., Wang, J., Liu, J., Han, J., Xiong, S., Yong, W., et al. (2016). Combination of ESI and MALDI mass spectrometry for qualitative, semi-quantitative and in situ analysis of gangliosides in brain. Sci. Rep. 6, 1–11. doi: 10.1038/srep25289
Keywords: glycans, glycosylation, mass spectrometry imaging, brain tissue, three-dimensional MSI
Citation: Hasan MM, Mimi MA, Mamun MA, Islam A, Waliullah ASM, Nabi MM, Tamannaa Z, Kahyo T and Setou M (2021) Mass Spectrometry Imaging for Glycome in the Brain. Front. Neuroanat. 15:711955. doi: 10.3389/fnana.2021.711955
Received: 19 May 2021; Accepted: 07 July 2021;
Published: 29 July 2021.
Edited by:
Takeshi Yoshimura, Osaka University, JapanReviewed by:
Yoshinao Wada, Osaka Women’s and Children’s Hospital, JapanCopyright © 2021 Hasan, Mimi, Mamun, Islam, Waliullah, Nabi, Tamannaa, Kahyo and Setou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tomoaki Kahyo, a2FoeW9AaGFtYS1tZWQuYWMuanA=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.