- 1Addgene, Watertown, MA, United States
- 2Stanley Center for Psychiatric Research, Broad Institute of MIT and Harvard, Cambridge, MA, United States
- 3Cold Spring Harbor Laboratory, Cold Spring Harbor, NY, United States
- 4Allen Institute for Brain Science, Seattle, WA, United States
- 5Penn Vector Core, Gene Therapy Program, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, United States
- 6Department of Genetics, Harvard Medical School, Howard Hughes Medical Institute, Boston, MA, United States
- 7Department of Ophthalmology, Harvard Medical School, Howard Hughes Medical Institute, Boston, MA, United States
Cell-type-specific expression of molecular tools and sensors is critical to construct circuit diagrams and to investigate the activity and function of neurons within the nervous system. Strategies for targeted manipulation include combinations of classical genetic tools such as Cre/loxP and Flp/FRT, use of cis-regulatory elements, targeted knock-in transgenic mice, and gene delivery by AAV and other viral vectors. The combination of these complex technologies with the goal of precise neuronal targeting is a challenge in the lab. This report will discuss the theoretical and practical aspects of combining current technologies and establish best practices for achieving targeted manipulation of specific cell types. Novel applications and tools, as well as areas for development, will be envisioned and discussed.
Introduction
Understanding neural networks as they relate to development, behavior, and learning is a critical objective of neuroscience. These questions can be addressed, in part, by understanding the role of specific neural cells and brain regions, as well as the impact of individual molecules in these circuits. The successful execution of these neurobiology studies requires methods that are highly targetable, efficient, and precise. In this regard, recombinant adeno-associated viral vectors (herein referred to as AAV) are powerful tools that can be used both to target and manipulate specific neuronal subtypes (defined based on gene expression, location, and connectivity) and non-neuronal cell types within the nervous system.
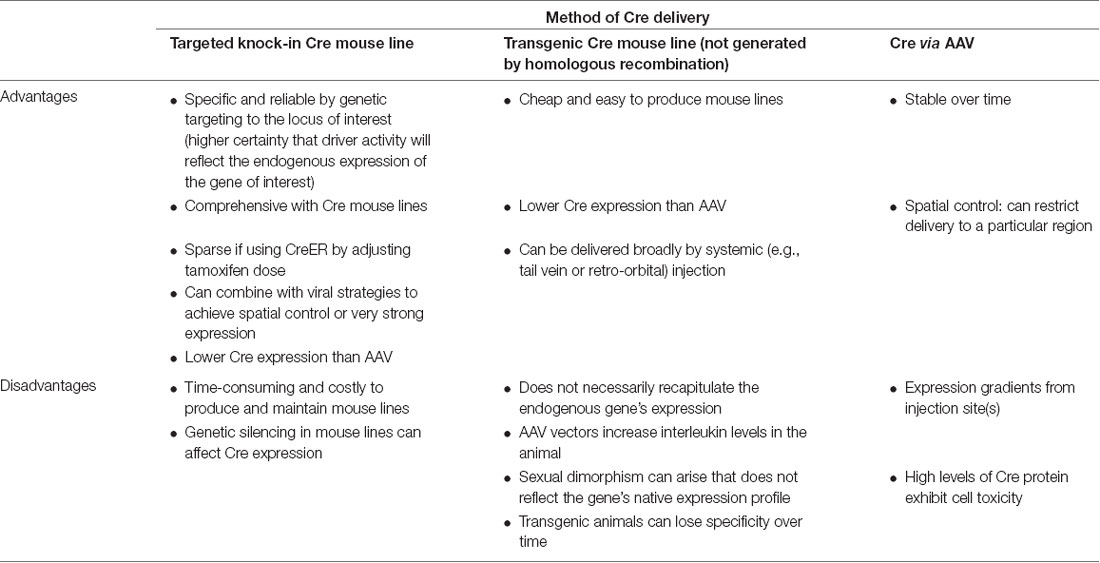
Scientists using AAV for gene transfer and/or neuronal targeting must consider various questions about experimental design, including: (1) how to best deliver/administer AAV (Figure 1A); (2) which AAV serotype to use (Figure 1B); and (3) how to drive gene expression with gene regulatory elements (both within the AAV genome and the host animal or cell line; Figures 1C–F). These and many other factors can affect how efficiently cells of interest are targeted by AAV. Further, experimental parameters such as AAV titer and dosage can impact AAV efficiency, and these details are often omitted from experimental methods in the literature and can be expensive and timely to determine empirically for each study. Overall, designing an experiment with AAV is multifaceted and suboptimal experimental design can drastically reduce the quality of results. In this report, we will discuss practical aspects of using AAV and considerations for designing experiments.
Figure 1. Several aspects of experimental design affect neuronal targeting and manipulation including (A) viral delivery method, (B) composition of viral capsid proteins, (C) promoters and/or enhancers driving transgene expression, (D) IRES or 2A elements for multicistronic expression coupled with fluorescent proteins (FP) or protein epitopes, (E) post-translational regulatory elements such as WPRE or 3′-UTR, and (F) Recombinase (Cre, CreER, Flp, or FlpER) expression from transgenic driver lines (inserted genomically via targeted or random integration) and ligand-dependent or recombinase-dependent expression elements such as TRE or lox sites, respectively. Abbreviations: TRE, tetracycline-response element; lox, LoxP sequence; IRES, internal ribosomal entry site; 2A, 2A sequence for self-cleavage; FP, fluorescent protein; WPRE, woodchuck hepatitis virus posttranscriptional regulatory element; 3′-UTR, 3′-untranslated sequence.
Selecting The Route of Administration and Capsid
AAV tropism, as dictated by AAV capsid proteins, is an important factor affecting transduction efficiency and specificity across cell types. Since the mechanism of AAV transduction is through the interaction of the AAV capsid with cell surface proteins and glycans, protein composition of the capsid (i.e., the AAV serotype) and the cell surface (i.e., based on cell type) determine transduction efficiency. Additionally, as the landscape of cell surface molecules varies across species, the efficiency of AAV may subsequently vary considerably across species and strains (Watakabe et al., 2015; El-Shamayleh et al., 2016; Hordeaux et al., 2018; Huang et al., 2019). Consequently, serotype and route of delivery should be carefully considered when designing experiments (Figures 1A,B). For an overview of the primary receptors for AAV serotypes, see Schultz and Chamberlain (2008).
Direct Intraparenchymal Delivery
When injected directly into the brain, many of the naturally-occurring AAV capsids, which share homology ranging from 65% to 99% (Drouin and Agbandje-McKenna, 2013), have distinct but significantly overlapping tropisms and distribution characteristics. AAV1, AAV2, AAV5, AAV8, AAV9 and the engineered variant AAV-DJ are commonly used to target local populations of neurons after direct injections (Table 1).
In addition to exhibiting local transduction, several serotypes exhibit transduction distal to the injection site (Burger et al., 2004; Cearley and Wolfe, 2006, 2007; Klein et al., 2006, 2008; Li et al., 2006; Reimsnider et al., 2007; Sondhi et al., 2007; Taymans et al., 2007; Cearley et al., 2008; Hollis et al., 2008; Hadaczek et al., 2009; Masamizu et al., 2011; Bu et al., 2012) and the mechanisms of these phenotypes are active areas of investigation (Castle et al., 2014). The AAV vector purification method has also been shown to impact transduction patterns (Klein et al., 2008).
Furthermore, there are important differences in how far different capsid variants spread from the injection site—AAV2 and AAV-DJ diffusion are more confined and, therefore, these capsids are often chosen for applications that require precise targeting. While expression from AAV2 is mostly neuronal, several serotypes, including AAV1, AAV5, AAV8 and AAV9, also transduce astrocytes and oligodendrocytes.
Despite the tremendous volume of work on serotype-dependent expression patterns and the complexity of the mechanisms both hypothesized and shown to drive these phenotypes, the ability to predict confidently the expression pattern in a particular experimental setup still requires empirical evidence. A non-exhaustive list of reported characteristics for several serotypes is outlined (Table 2) and can be used to narrow down suitable serotypes, though the importance of empirical validation at the onset of each study cannot be understated. Importantly, results reported in this table may vary based on anatomical region, though the results have not been summarized in this report to that degree.
Systemic Delivery
Several natural AAV capsids cross the blood-brain barrier (BBB). In contrast to direct injections, intravenous injections of AAV can provide a central nervous system (CNS)-wide gene delivery. This activity is present across several species and is most pronounced when AAV is administered to the neonate. Neonatal injections of AAV9 and rh.10 have been used to transduce neurons broadly across the CNS. However, when delivered at later developmental stages including in the adult, transduction is more limited and primarily restricted to endothelial cells and astrocytes, with transduction occurring in 1–2% of neurons in the forebrain (Foust et al., 2009; Dufour et al., 2014; Deverman et al., 2016). In this context, engineered AAV capsids have provided new and dramatically more efficient options for widespread gene delivery to the CNS. The first of these vectors, AAV-PHP.B, enabled researchers to deliver genes to more than 50% of neurons and astrocytes across numerous brain regions with a single non-invasive injection (Deverman et al., 2016). Achieving this efficiency requires relatively high viral doses (~1 × 1014 vector genomes/kg), thus requiring large volumes of high titer virus. A further-evolved AAV-PHP.B variant, AAV-PHP.eB, addresses this issue and can achieve >50% transduction of most neuron and astrocyte populations even with a 20-fold reduction in dose (Chan et al., 2017). While the activity of the PHP capsids is not universally observed across all species, or even strains of a given species, the receptor engaged by the PHP capsids during AAV transduction has been identified and can be used to predict permissivities of cell or tissue types to these engineered capsids (Hordeaux et al., 2019; Huang et al., 2019). In addition, the same group has developed an additional AAV variant (the PHP.S variant) that can efficiently transduce dorsal root ganglia and other peripheral neuron populations following systemic administration, which should enable the study of these otherwise difficult to target peripheral neuron populations (Chan et al., 2017).
CSF Delivery
The third option for gene delivery to the CNS is to inject vectors into the cerebral spinal fluid (CSF). Several access points can be used: the lateral ventricle (intracerebroventricular, ICV), the cisterna magna (CM), subpial (Miyanohara et al., 2016) or the intrathecal (IT) space along the spinal cord. When performed in neonates, ICV AAV administration can provide widespread gene delivery. In the adult, ICV and CM injections result in gene delivery in multiple brain regions, however, the expression is not uniform across all brain regions and superficial structures are preferentially targeted. Beyond neurons, ICV injections also provide access to periventricular cell populations. For example, after ICV injection, AAV4 can be used to transduce the ependymal cells (Liu et al., 2005), and two engineered AAV capsids, SCH9 and AAV4.18, enable transduction of subventricular zone neural progenitors (Murlidharan et al., 2015; Ojala et al., 2018). IT injection can be used to deliver genes to spinal cord motor neurons and dorsal root ganglions (Foust et al., 2010; Federici et al., 2012; Schuster et al., 2014).
Retrograde and Anterograde Transport for Circuit Studies
AAV vectors are commonly used as part of circuit studies. Numerous natural AAV serotypes exhibit retrograde trafficking activity from their uptake at axon terminals (see Table 2). However, retrograde transduction with natural serotypes such as AAV1, AAV2, AAV6, and AAV9 requires high vector doses due to the relative inefficiency of this transduction mechanism. More recently Tervo et al. (2016) and Davidsson et al. (2018) have created modified capsids AAV2-Retro and AAV MNM008, respectively, that provide efficient transduction of neurons that send axon projections into the injection site. Transduction efficiencies of both capsids are shown to be circuit-dependent, and thus capsids should be validated for circuits of interest when planning experiments. Zingg et al. (2017) report that AAV1 and AAV9 exhibit transsynaptic anterograde transport at high titers, specifically showing that AAV1-directed Cre can activate Cre-dependent transgene expression in a post-synaptic neuron. Importantly, they note that retrograde transmission also can occur, making interpretation clear only for circuits where there is a unidirectional pattern of connectivity.
AAV Can Cause Toxicity at High Doses
Although AAV is less inflammatory than some other viruses, it is not inert with respect to the innate (Rogers et al., 2011) or adaptive (Mingozzi and High, 2011) immune system, and may also perturb other cellular activities. Several studies have found neurotoxicity when the virus was delivered systemically or via direct injections into the CNS, or into the sub-retinal space of the retina. For rabies monosynaptic tracing studies, AAV is often delivered as a helper virus to supply TVA and rabies G protein. A recent study found that high doses of helper did not enable rabies infection or tracing, while diluted preparations did (Lavin et al., 2019). Although the mechanism was not determined, the authors suggest that the high doses were toxic. Similarly, neurotoxicity was seen in piglets injected systemically (Hinderer et al., 2014). In the retina, toxicity was associated with dose, if the viral promoter was expressed in a support cell type, the retinal pigmented epithelium, but not if the viral promoter activity was restricted to photoreceptor cells (Xiong et al., 2019). Innate immunity has been shown to result from stimulation of TLR9, a sensor of unmethylated CpG’s, in studies of AAV infection of muscle (Zhu et al., 2009) and liver (Martino et al., 2011). Inclusion of a short TLR9 blocking oligonucleotide within the AAV genome has been shown to alleviate this problem in some instances (Chan et al., 2019). It is thus worth carefully considering this aspect of virus dose when setting up an experiment.
In summary, a consensus of opinion has not been reached regarding the best serotype for each cell type, brain region or application. Choosing the optimal serotype requires reviewing the literature most relevant to the planned experiment and performing pilot testing for new or at least for challenging applications. As new engineered capsids with unique features continue to be developed, the available options will become more numerous and more powerful (Deverman et al., 2016; Tervo et al., 2016; Chan et al., 2017; Davidsson et al., 2018; Ojala et al., 2018).
Controlling Gene Expression With Regulatory Elements
Cre and Flp recombinase-dependent expression elements within AAV vectors remain the go-to system for restricting transgene expression to genetically defined cell types in model organisms. However, few Cre or Flp transgenic lines have been developed in other mammalian species. Furthermore, breeding multiple transgenic lines to generate the desired offspring can be time consuming and expensive. Therefore, there is significant interest in developing the means to achieve similar expression specificity in nontransgenic animals using flexible vector-based approaches that will translate across species.
Cis-regulatory elements can be used to control transgene expression from AAV genomes. These elements include promoters and enhancers (Figure 1C), as well as introns, micro-RNA recognition sequences, and internal ribosome entry sites (IRES; Figure 1D) that can be used to tailor RNA processing, stability, and translation to the experimental needs. Here we will discuss how these regulatory elements can be used to restrict AAV-mediated gene expression.
Enhancers and Promoters
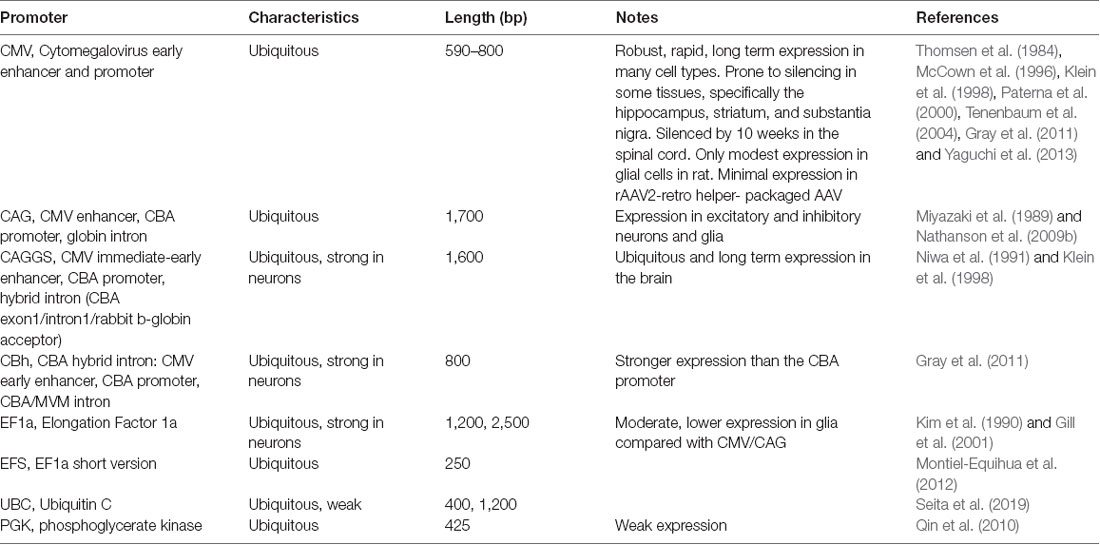
Enhancer and promoters (hereafter referred to as promoters for simplicity) can generally be divided into two classes: general/ubiquitous and cell type-specific. Typically, ubiquitous promoters provide high-level, long-term expression in most cell types, though some, such as the viral CMV promoter, have been shown to exhibit silencing in specific tissues over time (McCown et al., 1996; Klein et al., 1998; Paterna et al., 2000; Tenenbaum et al., 2004; Table 3).
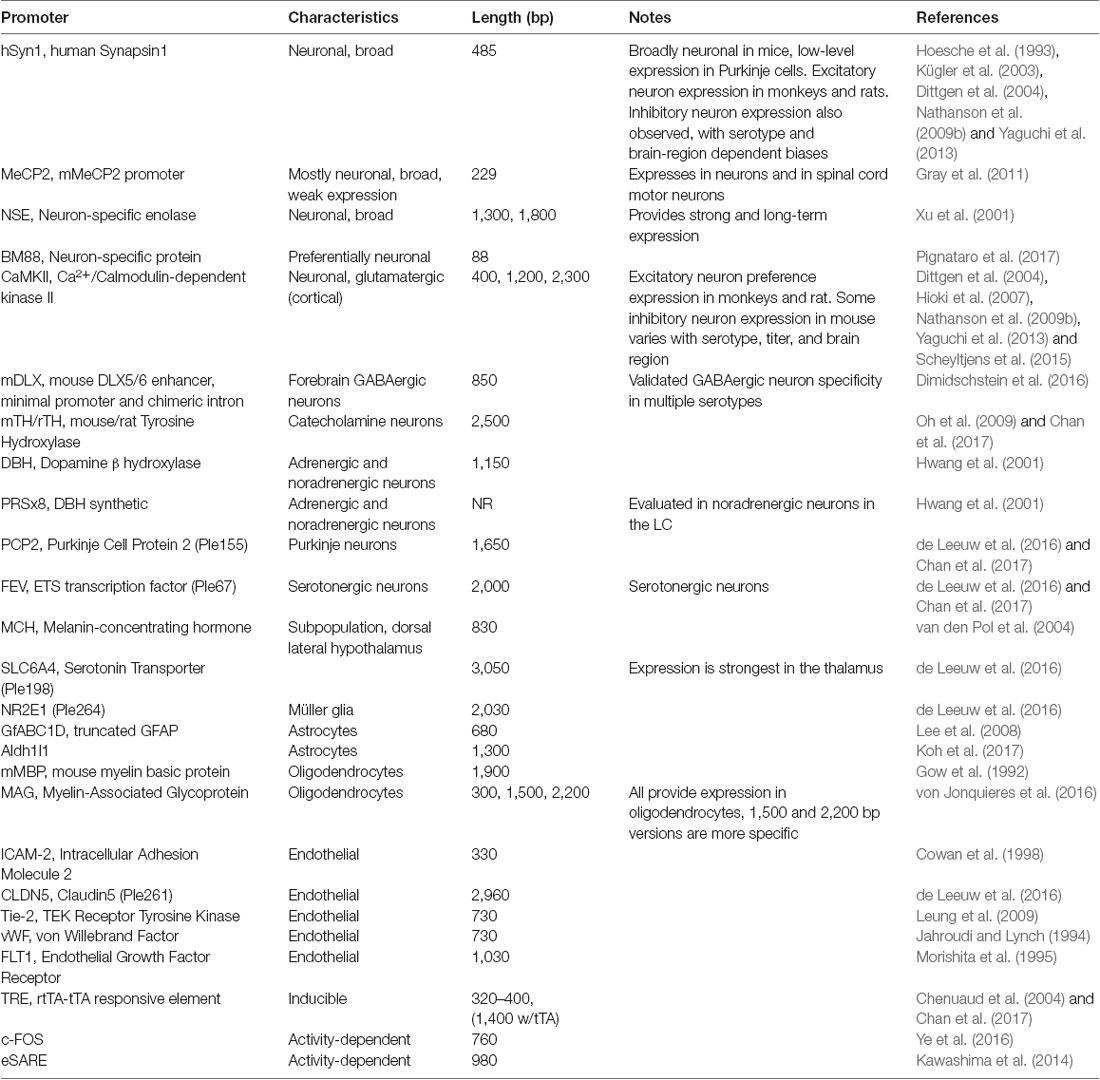
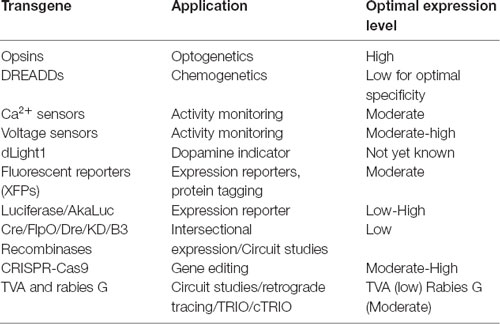
AAV and other single-stranded DNA viruses evolutionarily exhibit some of the smallest viral genomes (Campillo-Balderas et al., 2015), which has generally provided a selective pressure toward shorter promoter sequences. In contrast, the regulatory elements that control mammalian gene transcription are often distributed over thousands to hundreds of thousands of bases. Due to the limited packaging capacity of the AAV genome, identifying AAV-compatible promoters has been challenging and the development of shortened promoters is an area of active study (Nathanson et al., 2009a; de Leeuw et al., 2016). A list of cell type-specific promoters compatible with AAV vectors is provided in Table 4. High expression levels are not optimal for every application and alternative regulatory elements, such as those from the mammalian MeCP2 or PGK genes (Table 3) may be suitable for experiments where high-level viral enhancer driven expression is not desired. General recommendations for expression levels of various types of transgenes are summarized in Table 5.
Multicistronic Vectors
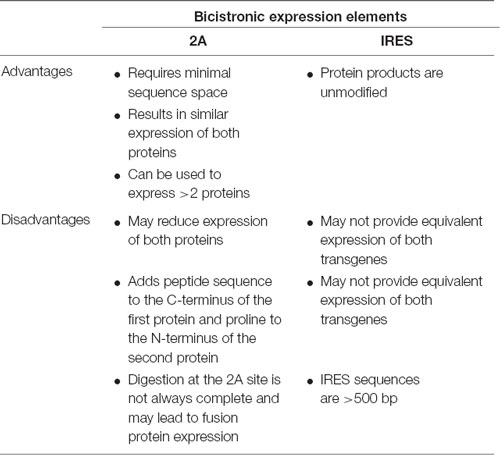
Although AAV vectors have a limited packaging capacity, it is possible to express multiple short transgenes from a single vector using one of several approaches: (1) separate translational units where each cDNA is controlled by separate 5′ and 3′ regulatory elements; (2) using IRES sequences to insert two separate translational units into a single mRNA; or (3) the use of viral 2A sequences to generate separate proteins from the same translational unit (Figure 1D). Table 6 highlights considerations for choosing between the use of IRES sequences and 2A “self-cleaving” peptides.
One important consideration when evaluating expression strategies and determining specificity is that the expression levels required for reporter detection may not match what is necessary for the activity of an opsin, DREADD or recombinase. For example, fluorescent proteins are commonly used to evaluate gene regulatory elements and vector design. However, fluorescent reporter assays may give the false impression of specificity if high levels of expression are seen in target cell types and low-level expression goes undetected in off-target populations. If these same regulatory elements are then used to drive DREADDs or Cre, which can mediate their effects at low expression levels, then the specificity may appear reduced. If this goes unexamined, then the interpretation of experimental results could be compromised. Moreover, though they are commonly used, fluorescent proteins are not necessarily inert and can lead to immune responses in larger animals (Samaranch et al., 2014), and over-expression related toxicities in mice.
Post-transcriptional Regulatory Elements
Transgene expression can also be controlled post-transcriptionally through the use of elements impacting RNA splicing, nuclear export, stability, and translation into proteins (Figure 1E). Inclusion of an intron can have positive impacts on expression levels. Introns have also been combined creatively with recombinase sites and partially inverted transgenes to achieve tight intersectional control of transgene expression (Fenno et al., 2014, 2017). Many recombinant AAV genomes also include a woodchuck hepatitis virus posttranscriptional regulatory element (WPRE), which can dramatically enhance expression. For several examples of how the inclusion of a WPRE affects expression from AAV vectors please see de Leeuw et al. (2016).
Complementary miRNA target sites (TS) are frequently engineered into the 3′-untranslated region (3′-UTR) of recombinant AAV vector genomes to mitigate off-target transgene expression. These sequences are complementary to miRNAs expressed within off-target cell types but not within the target population. miRNA binding to the perfectly complementary miRNA TS results in degradation of the RNA. Inclusion of multiple copies of the short miRNA TS sequences can dramatically lower off-target transgene expression. For example, by incorporating three copies each of miR-1 and miR-122, which are specifically expressed in muscle and liver, respectively, Xie et al. (2011) reduced transgene expression from intravenously administered AAV9 in muscle, heart, and liver, while maintaining brain expression. miRNA TS that enhance the restriction of lentiviral mediated gene expression to GABAergic neurons have also been identified (Xie et al., 2011; Keaveney et al., 2018), as have miRNA TS that result in more selective GCaMP6f expression from a strong ubiquitous promoter (Challis et al., 2019). Given their short lengths, miRNA TS can be multiplexed within the same genome, making them attractive elements for reducing expression outside of the cell type of interest.
Conditional Expression
The mammalian CNS contains extremely diverse types of neurons. These neuronal cell types can be distinguished by their intrinsic gene expression profiles, which are differentially regulated throughout development. Binary expression systems of drivers and reporters can be used to drive transgene expression in genetically-defined cells (reviewed in Huang et al., 2014).
There are two general strategies for using binary expression systems to access genetically-defined cell types: transactivation-based systems (based on tetracycline (TET)-response elements, TRE) and recombination-based systems (based on lox-site recombination, lox) described in (Taniguchi, 2014; Figure 1F). Gene targeting techniques can be used to insert Cre or Flp site-specific recombinases in the mouse genome. These knock-in driver mouse lines express Cre or Flp under the activity of a target gene’s endogenous promoter. Thus, Cre and Flp driver mouse lines constitute a genetic switch to turn on a recombinase-dependent reporter or effector. Since the recombinase is only expressed in cells defined by the target gene’s endogenous promoter activity, this system allows labeling and manipulation of neurons defined by the targeted gene’s expression pattern. A recently developed strategy can direct Cre or Flp activity to cells expressing GFP (Tang et al., 2017). Nanobodies directed against GFP have been engineered as fusion proteins with Cre or Flp. The recombinases are active only in GFP-expressing cells. This method can be used for the activity of only one recombinase, or can be used intersectionally, e.g., mating GFP and Cre mouse lines, infecting with a GFP-dependent Flp-nanobody fusion, and then using a Cre+ and Flp-dependent readout (see further below for intersectional methods). For temporal control, one can use TET-inducible systems to provide rapid and detectable expression at a given time point (Hioki et al., 2009; Sadakane et al., 2015). The TET system also has the advantage of amplified gene expression (Watakabe et al., 2012; Chan et al., 2017). This may be particularly important in studies on non-human primates since transduced cells could be maintained long-term without affecting cell integrity (Sadakane et al., 2015).
The reporter or effector whose expression is dependent on the driver’s activity is introduced in vivo by either crossing the driver to a transgenic reporter mouse line, using a viral vector, or electroporating the DNA construct into the cells. With the widely used Cre or Flp drivers, the conditional reporter expression depends on the recombination of specific lox or frt sites, respectively. In the case of the TET system, a tet-on, reverse tetracycline transactivator (rtTA) or tet-off (tTA) driver is paired with an AAV vector with a promoter harboring tet-responsive elements (TRE).
Targeting at Random vs. Targeting to a Specific Gene’s Locus
Knock-in Cre and Flp recombinases or GFP can be inserted into the genome either randomly or at a particular gene locus (Figure 1F). Conventional transgenic and BAC transgenic approaches result in relatively random insertions into the genome. However, knock-in mouse lines targeted to a specific locus by homologous recombination have the advantage that the expression of the inserted gene will recapitulate the expression pattern within cells of the endogenous gene of interest. There are several advantages to using targeted knock-in driver mouse lines (Table 7). Targeted knock-in of Cre or Flp or GFP into a specific gene’s transcription/translation initiation site can result in the recapitulation of expression of the endogenous locus. When using this strategy, it should be noted that individuals can exhibit variable levels of silencing. Typically, an optimal “non-silencing” male should be identified and used for genetic crosses. However, the offspring of this male may exhibit silencing and must be revalidated.
Temporal Control With Tamoxifen
To control the expression of a reporter in a subset of neurons that uniquely and transiently express a certain marker at a particular time point, tamoxifen-inducible recombinases (Feil et al., 2009), so-called CreER or FlpER, can be used as drivers. In CreER driver mice, activation of the expressed Cre recombinase requires administration of the estrogen receptor modulator drug tamoxifen to the animal, allowing for temporal control of recombination.
The typical tamoxifen dose varies from 10 to 200 mg/kg, depending on the desired degree of recombination at the reporter allele. In effect, this controls the sparseness/density of the labeling of the targeted neuronal population. The timing for tamoxifen administration depends on the temporal characteristics of the promoter driving CreER in the specific population to be targeted. Importantly, the half-life of tamoxifen (approximately 48 h) must be taken into consideration. Tamoxifen preparation is detailed in the published protocol in (Vaughan et al., 2015).
Tamoxifen can be administered in one of three ways, depending on the desired developmental time point for activating the CreER driver: oral gavage to the mother for embryonic induction; subcutaneous injection to offspring for early postnatal induction, or intraperitoneal injection in offspring for late postnatal and adult induction.
Best Practices for Induction of CreER or FlpER Mouse Lines
To generally improve the reliability of results obtained with CreER or FlpER mouse lines, several tamoxifen doses should be evaluated. If the level of recombination of the reporter achieved with a high dose of tamoxifen is low, administration of 4-hydroxytamoxifen (the native form of tamoxifen) can improve activation of CreER (Jahn et al., 2018). Tamoxifen needs to be metabolized in the liver to reach its active 4-hydroxytamoxifen form. Tamoxifen is often preferred over 4-hydroxytamoxifen for routine applications in highly active CreER driver lines due to lower cost and improved solubility. Following tamoxifen induction, CreER or FlpER will be active for ~48 h, which may impact induction during developmentally active time periods. 4-hydroxytamoxifen is a better choice for tighter control of the window of activation. Administration of tamoxifen or 4-hydroxytamoxifen by gavage to the pregnant mother to induce the pups at embryonic timepoints can lead to problems of miscarriage or poor mothering. When administering tamoxifen at embryonic time-points, use of Swiss or CD1 compared to C57Bl6 mice can improve outcomes in two ways: they produce larger litters and females are better mothers, which overall can improve pup survival.
Validating a Knock-in Driver Mouse Line
Knock-in driver mouse lines are designed to control the expression of reporter probes, sensors, and effectors in genetically-defined cell-types. It is important to validate that the driver mouse line expressing a site-specific recombinase (e.g., Cre, CreER, Flp, FlpER) reflects the endogenous expression pattern of the gene targeted by the knock-in driver. Various approaches can be used separately or jointly to validate a knock-in driver mouse line: (1) Crossing the mice with a suitable reporter like Rosa26-CAG-LSL-td-tomato (Ai14) or Rosa26-CAG-LSL-h2B-GFP and assessing brain-wide expression; (2) Immunostaining of the target regions; and (3) dual fluorescent in situ hybridization (dual fISH) with probe for reporter (e.g., RFP or GFP for Rosa26-CAG-LSL-td-tomato (Ai14) or Rosa26-CAG-LSL-h2B-GFP, respectively). Note that assessing Cre lines by crossing to a reporter line gives an integrated view of Cre activity over the lifetime of the animal. To assess Cre activity in the target cell population at the particular age of interest, a viral vector with a Cre-dependent reporter in a Cre line can be used. Similarly, for CreER lines tamoxifen induction should be performed at different developmental time-points to assess temporal specificity.
In addition, leakiness of both mouse lines and viruses must be assessed prior to the interpretation of experimental results that rely on the complete restriction of transgene expression. Leakiness of a mouse line can be assessed when validating the mouse line, ensuring that expression of the site-specific recombinase or activator (i.e., Cre, CreER, Flp, FlpER or tTA) is consistent with the gene of interest in its native form. Crossing the driver to a reporter (e.g., fluorescent reporter) and performing dual fluorescent in situ hybridization (dual fISH), with probes for the fluorescent reporter and for the gene of interest allows one to check that endogenous expression of the gene and activity of the driver are present in the same cells. Using knock-in driver mouse lines instead of randomly inserted BAC or plasmid driver mouse lines allows for targeted expression, with a higher likelihood of expression within cells expressing the endogenous gene of interest. Leakiness of the tamoxifen-inducible driver mouse line can be assessed by crossing the inducible driver CreER or FlpER to a Cre or Flp dependent fluorescent reporter and checking for expression of the fluorescent protein without administering tamoxifen. Finally, leakiness of the Cre- or Flp-dependent or tTA-activated AAV can be checked by injecting the virus in a mouse that has been crossed to a fluorescent reporter mouse line for the driver/activator. If expression from the virus and from the mouse reporter line match, this indicates that the AAV is specific to the driver/activator. One further evaluation of a given virus prep for background recombination (or leak) is to inject it into a mouse line that does not encode the cognate recombinase. Recombination can occur during the growth of the AAV plasmid in bacteria, and/or during virus preparation in mammalian cells, and should be assessed using this test.
Viral Strategies for Targeting Defined Populations
Combining various experimental techniques can enable the precise targeting of specific neuronal populations of interest. The general advantage of using combinatorial approaches is that the specificity of cell targeting can be improved with each additional technique. Applying known characteristics of AAV and the genetically encoded transgenes they carry, neuronal populations can be specifically targeted and manipulated based on their locations and connectivity. Here we will review several targeting strategies.
Axon Terminals
A viral encoded transgene can be targeted to axon terminals. Such terminals can then be targeted optogenetically in specific areas by only delivering light to the region harboring the terminals of interest (Figure 2A). This technique can be further restricted such that only axons of a particular cell type are targeted by using cell-type-specific promoters to drive the AAV expression (Figure 2B). Additional specificity can be achieved by choosing an injection site, AAV serotype, and titer so that only desired cell types are infected (Nassi et al., 2015).
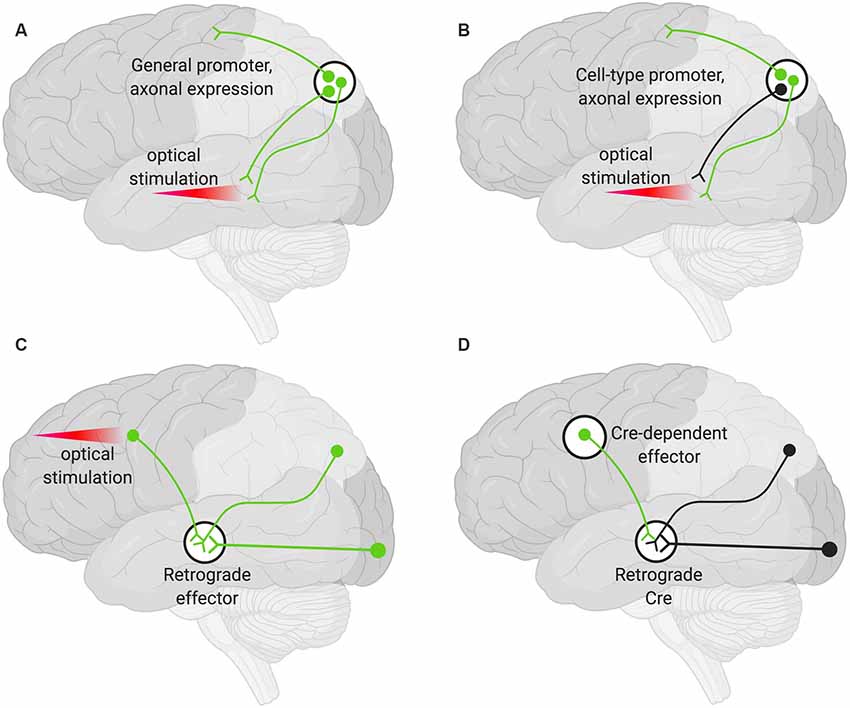
Figure 2. Various strategies for neuronal targeting using AAV. Delivery of neuronal effectors via AAV labels (green) axons and terminals with cell bodies at the injection site. (A) Effectors under a general promoter express in all transduced neurons with cell bodies at the injection site. Specific regions can be optically stimulated (red beam). (B) Effectors under cell-type promoters express only within a cell type. (C) Effectors delivered via a retrograde AAV express in all transduced neurons with axons that project into the injection site. Cell bodies in regions of interest can be optogenetically stimulated (red beam). (D) Delivery of a retrograde AAV expressing Cre recombinase (Retrograde Cre) to the projection site coupled with local delivery of a Cre-dependent effector limits expression to neurons within specific circuits.
Projection Neuron Targeting
Targeting the cell bodies of projection neurons can be important for manipulating only those neurons that terminate at a particular site. To target these neurons, an AAV capable of retrograde transduction (e.g., AAV2-Retro) can be delivered to projection sites and the cell bodies of those neurons terminating at that region will be targeted (Tervo et al., 2016). This method alone will not give rise to pathway specificity. However, if used to deliver optogenetic tools, light can be delivered only to the region harboring the cell bodies of interest (Figure 2C; Nassi et al., 2015).
Finally, specific populations of projection neurons can be targeted by coupling local delivery of a Cre-dependent, inducible neuromodulator (e.g., a DREADD or opsin) with retrograde delivery of Cre to the site where the targeted projection neurons originate (Figure 2D). Using this approach, a subset of projection neurons (Figure 2D), rather than all the projection neurons (Figure 2C), then express the Cre-dependent neuromodulator. This has the advantage that effectors (e.g., the DREADD ligand) can then be delivered systemically rather than locally and still only manipulate the subset of projection neurons that have been targeted (Nassi et al., 2015).
Areas for Development
The arsenal of new sensors, actuators, recombinases, genes, RNA and base editing enzymes, and other genetically encoded tools for studying the nervous system is rapidly growing. AAV vectors remain the most versatile and powerful approach for delivering these tools to the CNS. Nevertheless, delivery challenges remain and efforts are ongoing to develop new vectors that address several key needs including: (1) improved widespread CNS gene transfer via IV and ICV routes; (2) AAV vectors capable of more efficient trans-synaptic anterograde transport; (3) vector solutions for delivering transgenes too large to fit in a single AAV virus; (4) capsids that specifically target defined neural cell types and neuronal subtypes; and (5) viral vectors that enable transduction of microglia.
Overall, consistency and repeatability of both existing and newly developed AAV tools can be improved by following best practices and guidelines. While powerful technologies are being developed, each has technical limits that need to be considered both when designing experiments and when interpreting results. To this end, improved platforms for sharing information, including technical guidelines and best practices, will serve the research community by enabling technologies to be used to their fullest capacities consistently across labs.
Author Contributions
LH, KG, MR, IE, SB, JK, and MF planned and hosted the workshop where this material was discussed. BD, KM, AC, KW, and CC discussed and established the best practices for the field based on their expertise. LH, BD, and KM wrote the manuscript, which was then edited by AC, KW, CC, KG, MR, IE, SB, JK, and MF.
Conflict of Interest
LH, KG, MR, IE, SB, JK, and MF were employed by Addgene, a nonprofit repository that distributes research materials including plasmids and AAV.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Gowan Tervo, Sarada Viswanathan, and John Dingus for helpful discussions. We are grateful for the generous support by The Kavli Foundation to hold the meeting that gave rise to this report. We also thank the Research Team and all other members of Addgene for advice and support during the preparation of this manuscript. Figures in this manuscript were created with biorender.com.
References
Aschauer, D. F., Kreuz, S., and Rumpel, S. (2013). Analysis of transduction efficiency, tropism and axonal transport of AAV serotypes 1, 2, 5, 6, 8 and 9 in the mouse brain. PLoS One 8:e76310. doi: 10.1371/journal.pone.0076310
Borel, F., Gernoux, G., Cardozo, B., Metterville, J. P., Toro Cabreja, G. C., Song, L., et al. (2016). Therapeutic RAAVrh10 mediated sod1 silencing in adult SOD1G93A mice and nonhuman primates. Hum. Gene Ther. 27, 19–31. doi: 10.1089/hum.2015.122
Bu, J., Ashe, K. M., Bringas, J., Marshall, J., Dodge, J. C., Cabrera-Salazar, M. A., et al. (2012). Merits of combination cortical, subcortical, and cerebellar injections for the treatment of niemann-pick disease type A. Mol. Ther. 20, 1893–1901. doi: 10.1038/mt.2012.118
Burger, C., Gorbatyuk, O. S., Velardo, M. J., Peden, C. S., Williams, P., Zolotukhin, S., et al. (2004). Recombinant AAV viral vectors pseudotyped with viral capsids from serotypes 1, 2, and 5 display differential efficiency and cell tropism after delivery to different regions of the central nervous system. Mol. Ther. 10, 302–317. doi: 10.1016/j.ymthe.2004.05.024
Campillo-Balderas, J. A., Lazcano, A., and Becerra, A. (2015). Viral genome size distribution does not correlate with the antiquity of the host lineages. Front. Ecol. Evol. 3:143. doi: 10.3389/fevo.2015.00143
Carty, N., Lee, D., Dickey, C., Ceballos-Diaz, C., Jansen-West, K., Golde, T. E., et al. (2010). Convection-enhanced delivery and systemic mannitol increase gene product distribution of AAV vectors 5, 8, and 9 and increase gene product in the adult mouse brain. J. Neurosci. Methods 194, 144–153. doi: 10.1016/j.jneumeth.2010.10.010
Castle, M. J., Gershenson, Z. T., Giles, A. R., Holzbaur, E. L. F., and Wolfe, J. H. (2014). Adeno-associated virus serotypes 1, 8, and 9 share conserved mechanisms for anterograde and retrograde axonal transport. Hum. Gene Ther. 25, 705–720. doi: 10.1089/hum.2013.189
Cearley, C. N., Vandenberghe, L. H., Parente, M. K., Carnish, E. R., Wilson, J. M., and Wolfe, J. H. (2008). Expanded repertoire of AAV vector serotypes mediate unique patterns of transduction in mouse brain. Mol. Ther. 16, 1710–1718. doi: 10.1038/mt.2008.166
Cearley, C. N., and Wolfe, J. H. (2006). Transduction characteristics of adeno-associated virus vectors expressing cap serotypes 7, 8, 9, and Rh10 in the mouse brain. Mol. Ther. 13, 528–537. doi: 10.1016/j.ymthe.2005.11.015
Cearley, C. N., and Wolfe, J. H. (2007). A single injection of an adeno-associated virus vector into nuclei with divergent connections results in widespread vector distribution in the brain and global correction of a neurogenetic disease. J. Neurosci. 27, 9928–9940. doi: 10.1523/JNEUROSCI.2185-07.2007
Challis, R. C., Ravindra Kumar, S., Chan, K. Y., Challis, C., Beadle, K., Jang, M. J., et al. (2019). Systemic AAV vectors for widespread and targeted gene delivery in rodents. Nat. Protoc. 14, 379–414. doi: 10.1038/s41596-018-0097-3
Chan, K. Y., Jang, M. J., Yoo, B. B., Greenbaum, A., Ravi, N., Wu, W. L., et al. (2017). Engineered AAVs for efficient noninvasive gene delivery to the central and peripheral nervous systems. Nat. Neurosci. 20, 1172–1179. doi: 10.1038/nn.4593
Chan, Y. K., Wang, S. K., Letizia, A. J., Chan, Y., Lim, E. T., Graveline, A. R., et al. (2019). Engineering adeno-associated 450 viral vectors to evade innate immune and inflammatory responses. In review.
Chenuaud, P., Larcher, T., Rabinowitz, J. E., Provost, N., Joussemet, B., Bujard, H., et al. (2004). Optimal design of a single recombinant adeno-associated virus derived from serotypes 1 and 2 to achieve more tightly regulated transgene expression from nonhuman primate muscle. Mol. Ther. 9, 410–418. doi: 10.1016/j.ymthe.2003.12.015
Colella, P., Ronzitti, G., and Mingozzi, F. (2018). Emerging issues in AAV-mediated in vivo gene therapy. Mol. Ther. Methods Clin. Dev. 8, 87–104. doi: 10.1016/j.omtm.2017.11.007
Cowan, P. J., Tsang, D., Pedic, C. M., Abbott, L. R., Shinkel, T. A., D’Apice, A. J. F., et al. (1998). The human ICAM-2 promoter is endothelial cell-specific in vitro and in vivo and contains critical Sp1 and GATA binding sites. J. Biol. Chem. 273, 11737–11744. doi: 10.1074/jbc.273.19.11737
Cucchiarini, M., Ren, X. L., Perides, G., and Terwilliger, E. F. (2003). Selective gene expression in brain microglia mediated via adeno-associated virus type 2 and type 5 vectors. Gene Ther. 10, 657–667. doi: 10.1038/sj.gt.3301925
Davidson, B. L., Stein, C. S., Heth, J. A., Martins, I., Kotin, R. M., Derksen, T. A., et al. (2000). Recombinant adeno-associated virus type 2, 4, and 5 vectors: transduction of variant cell types and regions in the mammalian central nervous system. Proc. Natl. Acad. Sci. U S A 97, 3428–3432. doi: 10.1073/pnas.97.7.3428
Davidsson, M., Wang, G., Aldrin-Kirk, P., Cardoso, T., Nolbrant, S., Hartnor, M., et al. (2018). Barcoded rational AAV vector evolution enables systematic in vivo mapping of peptide binding motifs. SSRN Electron. J. doi: 10.2139/ssrn.3245528
Deverman, B. E., Pravdo, P. L., Simpson, B. P., Kumar Ravindra, S., Chan, K. Y., Banerjee, A., et al. (2016). Cre-dependent selection yields AAV variants for widespread gene transfer to the adult brain. Nat. Biotechnol. 34, 204–209. doi: 10.1038/nbt.3440
de Leeuw, C. N. De., Korecki, A. J., Berry, G. E., Hickmott, J. W., Lam, S. L., Lengyell, T. C., et al. (2016). RAAV-compatible minipromoters for restricted expression in the brain and eye. Mol. Brain 9:52. doi: 10.1186/s13041-016-0232-4
Dimidschstein, J., Chen, Q., Tremblay, R., Rogers, S. L., Saldi, G. A., Guo, L., et al. (2016). A viral strategy for targeting and manipulating interneurons across vertebrate species. Nat. Neurosci. 19, 1743–1749. doi: 10.1038/nn.4430
Dittgen, T., Nimmerjahn, A., Komai, S., Licznerski, P., Waters, J., Margrie, T. W., et al. (2004). Lentivirus-based genetic manipulations of cortical neurons and their optical and electrophysiological monitoring in vivo. Proc. Natl. Acad. Sci. U S A 101, 18206–18211. doi: 10.1073/pnas.0407976101
Dodiya, H. B., Bjorklund, T., Stansell, J. III., Mandel, R. J., Kirik, D., and Kordower, J. H. (2010). Differential transduction following basal ganglia administration of distinct pseudotyped AAV capsid serotypes in nonhuman primates. Mol. Ther. 18, 579–587. doi: 10.1038/mt.2009.216
Drouin, L. M., and Agbandje-McKenna, M. (2013). Adeno-associated virus structural biology as a tool in vector development. Future Virol. 8, 1183–1199. doi: 10.2217/fvl.13.112
Dufour, B. D., Smith, C. A., Clark, R. L., Walker, T. R., and McBride, J. L. (2014). Intrajugular vein delivery of AAV9-RNAi prevents neuropathological changes and weight loss in Huntington’s disease mice. Mol. Ther. 22, 797–810. doi: 10.1038/mt.2013.289
During, M. J., Samulski, R. J., Elsworth, J. D., Kaplitt, M. G., Leone, P., Xiao, X., et al. (1998). in vivo expression of therapeutic human genes for dopamine production in the caudales of MPTP-treated monkeys using an AAV vector. Gene Ther. 5, 820–827. doi: 10.1038/sj.gt.3300650
El-Shamayleh, Y., Ni, A. M., and Horwitz, G. D. (2016). Strategies for targeting primate neural circuits with viral vectors. J. Neurophysiol. 116, 122–134. doi: 10.1152/jn.00087.2016
Federici, T., Taub, J. S., Baum, G. R., Gray, S. J., Grieger, J. C., Matthews, K. A., et al. (2012). Robust spinal motor neuron transduction following intrathecal delivery of AAV9 in pigs. Gene Ther. 19, 852–859. doi: 10.1038/gt.2011.130
Feil, S., Valtcheva, N., and Feil, R. (2009). Inducible Cre mice. Methods Mol. Biol. 530, 343–363. doi: 10.1007/978-1-59745-471-1_18
Fenno, L. E., Mattis, J., Ramakrishnan, C., and Deisseroth, K. (2017). A guide to creating and testing new INTRSECT constructs. Curr. Protoc. Neurosci. 80, 4.39.1–4.39.24. doi: 10.1002/cpns.30
Fenno, L. E., Mattis, J., Ramakrishnan, C., Hyun, M., Lee, S. Y., He, M., et al. (2014). Targeting cells with single vectors using multiple-feature boolean logic. Nat. Methods 11, 763–772. doi: 10.1038/nmeth.2996
Foust, K. D., Nurre, E., Montgomery, L. C., Hernandez, A., Chan, C. M., and Kaspar, B. K. (2009). Intravascular AAV9 preferentially targets neonatal neurons and adult astrocytes. Nat. Biotechnol. 27, 59–65. doi: 10.1038/nbt.1515
Foust, K. D., Wang, X., McGovern, V. L., Braun, L., Bevan, A. K., Haidet, A. M., et al. (2010). Rescue of the spinal muscular atrophy phenotype in a mouse model by early postnatal delivery of SMN. Nat. Biotechnol. 28, 271–274. doi: 10.1038/nbt.1610
Gill, D. R., Smyth, S. E., Goddard, C. A., Pringle, I. A., Higgins, C. F., Colledge, W. H., et al. (2001). Increased persistence of lung gene expression using plasmids containing the ubiquitin C or elongation factor 1α promoter. Gene Ther. 8, 1539–1546. doi: 10.1038/sj.gt.3301561
Gow, A., Friedrich, V. L., and Lazzarini, R. A. (1992). Myelin basic protein gene contains separate enhancers for oligodendrocyte and schwann cell expression. J. Cell Biol. 119, 605–616. doi: 10.1083/jcb.119.3.605
Gray, S. J., Foti, S. B., Schwartz, J. W., Bachaboina, L., Taylor-Blake, B., Coleman, J., et al. (2011). Optimizing promoters for recombinant adeno-associated virus-mediated gene expression in the peripheral and central nervous system using self-complementary vectors. Hum. Gene Ther. 22, 1143–1153. doi: 10.1089/hum.2010.245
Gray, S. J., Nagabhushan Kalburgi, S., McCown, T. J., and Jude Samulski, R. (2013). Global CNS gene delivery and evasion of anti-AAV-neutralizing antibodies by intrathecal AAV administration in non-human primates. Gene Ther. 20, 450–459. doi: 10.1038/gt.2012.101
Grimm, D., Lee, J. S., Wang, L., Desai, T., Akache, B., Storm, T. A., et al. (2008). in vitro and in vivo gene therapy vector evolution via multispecies interbreeding and retargeting of adeno-associated Viruses. J. Virol. 82, 5887–5911. doi: 10.1128/JVI.00254-08
Gurda, B. L., De Guilhem De Lataillade, A., Bell, P., Zhu, Y., Yu, H., Wang, P., et al. (2016). Evaluation of AAV-mediated gene therapy for central nervous system disease in canine mucopolysaccharidosis VII. Mol. Ther. 24, 206–216. doi: 10.1038/mt.2015.189
Hadaczek, P., Forsayeth, J., Mirek, H., Munson, K., Bringas, J., Pivirotto, P., et al. (2009). Transduction of nonhuman primate brain with adeno-associated virus serotype 1: vector trafficking and immune response. Hum. Gene Ther. 20, 225–237. doi: 10.1089/hum.2008.151
Halbert, C. L., Miller, A. D., McNamara, S., Emerson, J., Gibson, R. L., Ramsey, B., et al. (2006). Prevalence of neutralizing antibodies against adeno-associated virus (AAV) types 2, 5, and 6 in cystic fibrosis and normal populations: implications for gene therapy using AAV Vectors. Hum. Gene Ther. 17, 440–447. doi: 10.1089/hum.2006.17.ft-194
Hammond, S. L., Leek, A. N., Richman, E. H., and Tjalkens, R. B. (2017). Cellular selectivity of AAV serotypes for gene delivery in neurons and astrocytes by neonatal intracerebroventricular injection. PLoS One 12:e0188830. doi: 10.1371/journal.pone.0188830
Hillier, D., Fiscella, M., Drinnenberg, A., Trenholm, S., Rompani, S. B., Raics, Z., et al. (2017). Causal evidence for retina-dependent and independent visual motion computations in mouse cortex. Nat. Neurosci. 20, 960–968. doi: 10.1038/nn.4566
Hinderer, C., Bell, P., Vite, C. H., Louboutin Pierre, J., Grant, R., Bote, E., et al. (2014). Widespread gene transfer in the central nervous system of cynomolgus macaques following delivery of AAV9 into the cisterna magna. Mol. Ther. Methods Clin. Dev. 1:14051. doi: 10.1038/mtm.2014.51
Hioki, H., Kameda, H., Nakamura, H., Okunomiya, T., Ohira, K., Nakamura, K., et al. (2007). Efficient gene transduction of neurons by lentivirus with enhanced neuron-specific promoters. Gene Ther. 14, 872–882. doi: 10.1038/sj.gt.3302924
Hioki, H., Kuramoto, E., Konno, M., Kameda, H., Takahashi, Y., Nakano, T., et al. (2009). High-level transgene expression in neurons by lentivirus with tet-off system. Neurosci. Res. 63, 149–154. doi: 10.1016/j.neures.2008.10.010
Hoesche, C., Sauerwald, A., Veh, R. W., Krippl, B., and Kilimann, M. W. (1993). The 5′-flanking region of the rat synapsin I gene directs neuron-specific and developmentally regulated reporter gene expression in transgenic mice. J. Biol. Chem. 268, 26494–26502.
Hollis, E. R. II., Kadoya, K., Hirsch, M., Samulski, R. J., and Tuszynski, M. H. (2008). Efficient retrograde neuronal transduction utilizing self-complementary AAV1. Mol. Ther. 16, 296–301. doi: 10.1038/sj.mt.6300367
Hordeaux, J., Wang, Q., Katz, N., Buza, E. L., Bell, P., and Wilson, J. M. (2018). The neurotropic properties of AAV-PHP.B are limited to C57BL/6J mice. Mol. Ther. 26, 664–668. doi: 10.1016/j.ymthe.2018.01.018
Hordeaux, J., Yuan, Y., Clark, P. M., Wang, Q., Martino, R. A., Sims, J. J., et al. (2019). The GPI-linked protein LY6A drives AAV-PHP.B transport across the blood-brain barrier. Mol. Ther. 27, 912–921. doi: 10.1016/j.ymthe.2019.02.013
Huang, Q., Chan, K. Y., Tobey, I. G., Chan, Y. A., Poterba, T., Boutros, C. L., et al. (2019). Delivering genes across the blood-brain barrier: LY6A, a novel cellular receptor for AAV-PHP.B capsids. BioRxiv [Preprint]. doi: 10.1101/538421
Huang, Z. J., Taniguchi, H., He, M., and Kuhlman, S. (2014). Cre-dependent adeno-associated virus preparation and delivery for labeling neurons in the mouse brain. Cold Spring Harb. Protoc. 2014, 190–194. doi: 10.1101/pdb.prot080382
Hwang, D. Y., Carlezon, W. A. Jr., Isacson, O., and Kim, K. S. (2001). A high-efficiency synthetic promoter that drives transgene expression selectively in noradrenergic neurons. Hum. Gene Ther. 12, 1731–1740. doi: 10.1089/104303401750476230
Jahn, H. M., Kasakow, C. V., Helfer, A., Michely, J., Verkhratsky, A., Maurer, H. H., et al. (2018). Refined protocols of tamoxifen injection for inducible DNA recombination in mouse astroglia. Sci. Rep. 8:5931. doi: 10.1038/s41598-018-24085-9
Jahroudi, N., and Lynch, D. C. (1994). Endothelial-cell-specific regulation of von willebrand factor gene expression. Mol. Cell. Biol. 14, 999–1008. doi: 10.1128/mcb.14.2.999
Kaplitt, M. G., Leone, P., Samulski, R. J., Xiao, X., Pfaff, D. W., O’Malley, K. L., et al. (1994). Long-term gene expression and phenotypic correction using adeno-associated virus vectors in the mammalian brain. Nat. Genet. 8, 148–154. doi: 10.1038/ng1094-148
Kaspar, B. K., Erickson, D., Schaffer, D., Hinh, L., Gage, F. H., and Peterson, D. A. (2002). Targeted retrograde gene delivery for neuronal protection. Mol. Ther. 5, 50–56. doi: 10.1006/mthe.2001.0520
Kawashima, T., Okuno, H., and Bito, H. (2014). A new era for functional labeling of neurons: activity-dependent promoters have come of age. Front. Neural Circuits 8:37. doi: 10.3389/fncir.2014.00037
Keaveney, M. K., Tseng, H. A., Ta, T. L., Gritton, H. J., Man, H. Y., and Han, X. (2018). A MicroRNA-based gene-targeting tool for virally labeling interneurons in the rodent cortex. Cell Rep. 24, 294–303. doi: 10.1016/j.celrep.2018.06.049
Kim, D. W., Uetsuki, T., Kaziro, Y., Yamaguchi, N., and Sugano, S. (1990). Use of the human elongation factor 1α promoter as a versatile and efficient expression system. Gene 91, 217–223. doi: 10.1016/0378-1119(90)90091-5
Klein, R. L., Dayton, R. D., Leidenheimer, N. J., Jansen, K., Golde, T. E., and Zweig, R. M. (2006). Efficient neuronal gene transfer with AAV8 leads to neurotoxic levels of tau or green fluorescent proteins. Front. Cell Dev. Biol. 13, 517–527. doi: 10.1016/j.ymthe.2005.10.008
Klein, R. L., Dayton, R. D., Tatom, J. B., Henderson, K. M., and Henning, P. P. (2008). AAV8, 9, Rh10, Rh43 vector gene transfer in the rat brain: effects of serotype, promoter and purification method. Mol. Ther. 16, 89–96. doi: 10.1038/sj.mt.6300331
Klein, R. L., Meyer, E. M., Peel, A. L., Zolotukhin, S., Meyers, C., Muzyczka, N., et al. (1998). Neuron-specific transduction in the rat septohippocampal or nigrostriatal pathway by recombinant adeno-associated virus vectors. Gene Ther. 150, 183–194. doi: 10.1006/exnr.1997.6736
Koh, W., Park, Y. M., Lee, S. E., and Lee, C. J. (2017). AAV-mediated astrocyte-specific gene expression under human ALDH1L1 promoter in mouse thalamus. Exp. Neurobiol. 26, 350–361. doi: 10.5607/en.2017.26.6.350
Kügler, S., Kilic, E., and Bähr, M. (2003). Human synapsin 1 gene promoter confers highly neuron-specific long-term transgene expression from an adenoviral vector in the adult rat brain depending on the transduced area. Gene Ther. 10, 337–347. doi: 10.1038/sj.gt.3301905
Lavin, T. K., Jin, L., Lea, N. E., and Wickersham, I. (2019). Monosynaptic tracing success depends critically on helper virus concentrations. BioRxiv [Preprint]. doi: 10.1101/736017
Lee, Y., Messing, A., Su, M., and Brenner, M. (2008). GFAP promoter elements required for region-specific and astrocyte-specific expression. GLIA 56, 481–493. doi: 10.1002/glia.20622
Leung, J. W. C., Chung, S. S. M., and Chung, S. K. (2009). Endothelial endothelin-1 over-expression using receptor tyrosine kinase tie-1 promoter leads to more severe vascular permeability and blood brain barrier breakdown after transient middle cerebral artery occlusion. Brain Res. 1266, 121–129. doi: 10.1016/j.brainres.2009.01.070
Li, S. F., Wang, R. Z., Meng, Q. H., Li, G. L., Hu, G. J., Dou, W. C., et al. (2006). Intra-ventricular infusion of RAAV1-EGFP resulted in transduction in multiple regions of adult rat brain: a comparative study with RAAV2 and RAAV5 vectors. Brain Res. 1122, 1–9. doi: 10.1016/j.brainres.2006.09.042
Liu, G., Martins, I. H., Chiorini, J. A., and Davidson, B. L. (2005). Adeno-associated virus type 4 (AAV4) targets ependyma and astrocytes in the subventricular zone and RMS. Gene Ther. 12, 1503–1508. doi: 10.1038/sj.gt.3302554
Lo, W. D., Qu, G., Sferra, T. J., Clark, R., Chen, R., and Johnson, P. R. (1999). Adeno-associated virus-mediated gene transfer to the brain: duration and modulation of expression. Hum. Gene Ther. 10, 201–213. doi: 10.1089/10430349950018995
Lukashchuk, V., Lewis, K. E., Coldicott, I., Grierson, A. J., and Azzouz, M. (2016). AAV9-mediated central nervous system-targeted gene delivery via cisterna magna route in mice. Mol. Ther. Methods Clin. Dev. 3:15055. doi: 10.1038/mtm.2015.55
Mandel, R. J., Rendahl, K. G., Spratt, S. K., Snyder, R. O., Cohen, L. K., and Leff, S. E. (1998). Characterization of intrastriatal recombinant adeno-associated virus- mediated gene transfer of human tyrosine hydroxylase and human GTP- cyclohydrolase I in a rat model of Parkinson’s disease. J. Neurosci. 18, 4271–4284. doi: 10.1523/jneurosci.18-11-04271.1998
Marchiò, S., Sidman, R. L., Arap, W., and Pasqualini, R. (2016). Brain endothelial cell-targeted gene therapy of neurovascular disorders. EMBO Mol. Med. 8, 592–594. doi: 10.15252/emmm.201606407
Martino, A. T., Suzuki, M., Markusic, D. M., Zolotukhin, I., Ryals, R. C., Moghimi, B., et al. (2011). The genome of self-complementary adeno-associated viral vectors increases toll-like receptor 9-dependent innate immune responses in the liver. Blood 117, 6459–6468. doi: 10.1182/blood-2010-10-314518
Masamizu, Y., Okada, T., Ishibashi, H., Takeda, S., Yuasa, S., and Nakahara, K. (2010). Efficient gene transfer into neurons in monkey brain by adeno-associated virus 8. Neuroreport 21, 447–451. doi: 10.1097/wnr.0b013e328338ba00
Masamizu, Y., Okada, T., Kawasaki, K., Ishibashi, H., Yuasa, S., Takeda, S., et al. (2011). Local and retrograde gene transfer into primate neuronal pathways via adeno-associated virus serotype 8 and 9. Neuroscience 193, 249–258. doi: 10.1016/j.neuroscience.2011.06.080
Mastakov, M. Y., Baer, K., Xu, R., Fitzsimons, H., and During, M. J. (2001). Combined injection of RAAV with mannitol enhances gene expression in the rat brain. Mol. Ther. 3, 225–232. doi: 10.1006/mthe.2001.0246
McCown, T. J., Xiao, X., Li, J., Breese, G. R., and Samulski, R. J. (1996). Differential and persistent expression patterns of CNS gene transfer by an adeno-associated virus (AAV) vector. Brain Res. 713, 99–107. doi: 10.1016/0006-8993(95)01488-8
Mingozzi, F., and High, K. A. (2011). Immune responses to AAV in clinical trials. Curr. Gene Ther. 11, 321–330. doi: 10.2174/156652311796150354
Miyanohara, A., Kamizato, K., Juhas, S., Juhasova, J., Navarro, M., Marsala, S., et al. (2016). Potent spinal parenchymal AAV9-mediated gene delivery by subpial injection in adult rats and pigs. Mol. Ther. Methods Clin. Dev. 3:16046. doi: 10.1038/mtm.2016.46
Miyazaki, J., Takaki, S., Araki, K., Tashiro, F., Tominaga, A., Takatsu, K., et al. (1989). Expression vector system based on the chicken β-actin promoter directs efficient production of interleukin-5. Gene 79, 269–277. doi: 10.1016/0378-1119(89)90209-6
Montiel-Equihua, C. A., Zhang, L., Knight, S., Saadeh, H., Scholz, S., Carmo, M., et al. (2012). The β-globin locus control region in combination with the EF1α short promoter allows enhanced lentiviral vector-mediated erythroid gene expression with conserved multilineage activity. Mol. Ther. 20, 1400–1409. doi: 10.1038/mt.2012.50
Morishita, K., Johnson, D. E., and Williams, L. T. (1995). A novel promoter for vascular endothelial growth factor receptor (Flt-1) that confers endothelial-specific gene expression. J. Biol. Chem. 270, 27948–27953. doi: 10.1074/jbc.270.46.27948
Murlidharan, G., Corriher, T., Ghashghaei, H. T., and Asokan, A. (2015). Unique glycan signatures regulate adeno-associated virus tropism in the developing brain. J. Virol. 89, 3976–3987. doi: 10.1128/jvi.02951-14
Nassi, J. J., Cepko, C. L., Born, R. T., and Beier, K. T. (2015). Neuroanatomy goes viral!. Front. Neuroanat. 9:80. doi: 10.3389/fnana.2015.00080
Nathanson, J. L., Jappelli, R., Scheeff, E. D., Manning, G., Obata, K., Brenner, S., et al. (2009a). Short promoters in viral vectors drive selective expression in mammalian inhibitory neurons, but do not restrict activity to specific inhibitory cell-types. Front. Neural Circuits 3:19. doi: 10.3389/neuro.04.019.2009
Nathanson, J. L., Yanagawa, Y., Obata, K., and Callaway, E. M. (2009b). Preferential labeling of inhibitory and excitatory cortical neurons by endogenous tropism of adeno-associated virus and lentivirus vectors. Neurosci. 161, 441–450. doi: 10.1016/j.neuroscience.2009.03.032
Niwa, H., Yamamura, K., and Miyazaki, J. (1991). Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108, 193–199. doi: 10.1016/0378-1119(91)90434-d
Oh, M. S., Hong, S. J., Huh, Y., and Kim, K. S. (2009). Expression of transgenes in midbrain dopamine neurons using the tyrosine hydroxylase promoter. Gene Ther. 16, 437–440. doi: 10.1038/gt.2008.148
Ojala, D. S., Sun, S., Santiago-Ortiz, J. L., Shapiro, M. G., Romero, P. A., and Schaffer, D. V. (2018). in vivo selection of a computationally designed SCHEMA AAV library yields a novel variant for infection of adult neural stem cells in the SVZ. Mol. Ther. 26, 304–319. doi: 10.1016/j.ymthe.2017.09.006
Paterna, J. C., Moccetti, T., Mura, A., Feldon, J., and Büeler, H. (2000). Influence of promoter and WHV post-transcriptional regulatory element on AAV-mediated transgene expression in the rat brain. Gene Ther. 7, 1304–1311. doi: 10.1038/sj.gt.3301221
Peel, A. L., Zolotukhin, S., Schrimsher, G. W., Muzyczka, N., and Reier, P. J. (1997). Efficient transduction of green fluorescent protein in spinal cord neurons using adeno-associated virus vectors containing cell type-specific promoters. Gene Ther. 4, 16–24. doi: 10.1038/sj.gt.3300358
Pignataro, D., Sucunza, D., Vanrell, L., Lopez-Franco, E., Dopeso-Reyes, I. G., Vales, A., et al. (2017). Adeno-associated viral vectors serotype 8 for cell-specific delivery of therapeutic genes in the central nervous system. Front. Neuroanat. 11:2. doi: 10.3389/fnana.2017.00002
Qin, J. Y., Zhang, L., Clift, K. L., Hulur, I., Xiang, A. P., Ren, B. Z., et al. (2010). Systematic comparison of constitutive promoters and the doxycycline-inducible promoter. PLoS One 5:e10611. doi: 10.1371/journal.pone.0010611
Reimsnider, S., Manfredsson, F. P., Muzyczka, N., and Mandel, R. J. (2007). Time course of transgene expression after intrastriatal pseudotyped RAAV2/1, RAAV2/2, RAAV2/5, and RAAV2/8 transduction in the rat. Mol. Ther. 15, 1504–1511. doi: 10.1038/sj.mt.6300227
Rogers, G. L., Martino, A. T., Aslanidi, G. V., Jayandharan, G. R., Srivastava, A., and Herzog, R. W. (2011). Innate immune responses to AAV vectors. Front. Microbiol. 2:194. doi: 10.3389/fmicb.2011.00194
Ruzo, A., Marcó, S., García, M., Villacampa, P., Ribera, A., Ayuso, E., et al. (2012). Correction of pathological accumulation of glycosaminoglycans in central nervous system and peripheral tissues of MPSIIIA mice through systemic AAV9 gene transfer. Hum. Gene Ther. 23, 1237–1246. doi: 10.1089/hum.2012.029
Sadakane, O., Masamizu, Y., Watakabe, A., Terada, S. I., Ohtsuka, M., Takaji, M., et al. (2015). Long-term two-photon calcium imaging of neuronal populations with subcellular resolution in adult non-human primates. Cell Rep. 13, 1989–1999. doi: 10.1016/j.celrep.2015.10.050
Salegio, E. A., Samaranch, L., Kells, A. P., Mittermeyer, G., San Sebastian, W., Zhou, S., et al. (2013). Axonal transport of adeno-associated viral vectors is serotype-dependent. Gene Ther. 20, 348–352. doi: 10.1038/gt.2012.27
Samaranch, L., Salegio, E. A., Sebastian, W. S., Kells, A. P., Bringas, J. R., Forsayeth, J., et al. (2013). Strong cortical and spinal cord transduction after AAV7 and AAV9 delivery into the cerebrospinal fluid of nonhuman primates. Hum. Gene Ther. 24, 526–532. doi: 10.1089/hum.2013.005
Samaranch, L., Salegio, E. A., Sebastian, W. S., Kells, A. P., Foust, K. D., Bringas, J. R., et al. (2011). Adeno-associated virus serotype 9 transduction in the central nervous system of nonhuman primates. Hum. Gene Ther. 23, 382–389. doi: 10.1089/hum.2011.200
Samaranch, L., Sebastian, W. S., Kells, A. P., Salegio, E. A., Heller, G., Bringas, J. R., et al. (2014). AAV9-mediated expression of a non-self protein in nonhuman primate central nervous system triggers widespread neuroinflammation driven by antigen-presenting cell transduction. Mol. Ther. 22, 329–337. doi: 10.1038/mt.2013.266
Scheyltjens, I., Laramée, M. E., Van den Haute, C., Gijsbers, R., Debyser, Z., Baekelandt, V., et al. (2015). Evaluation of the expression pattern of RAAV2/1, 2/5, 2/7, 2/8, and 2/9 serotypes with different promoters in the mouse visual cortex. J. Comp. Neurol. 14, 2019–2042. doi: 10.1002/cne.23819
Schultz, B. R., and Chamberlain, J. S. (2008). Recombinant adeno-associated virus transduction and integration. Mol. Ther. 16, 1189–1199. doi: 10.1038/mt.2008.103
Schuster, D. J., Dykstra, J. A., Riedl, M. S., Kitto, K. F., Belur, L. R., McIvor, R. S., et al. (2014). Biodistribution of adeno-associated virus serotype 9 (AAV9) vector after intrathecal and intravenous delivery in mouse. Front. Neuroanat. 8:42. doi: 10.3389/fnana.2014.00042
Seita, Y., Tsukiyama, T., Azami, T., Kobayashi, K., Iwatani, C., Tsuchiya, H., et al. (2019). Comprehensive evaluation of ubiquitous promoters suitable for the generation of transgenic cynomolgus monkeys. Biol. Reprod. 100, 1440–1452. doi: 10.1093/biolre/ioz040
Sondhi, D., Hackett, N. R., Peterson, D. A., Stratton, J., Baad, M., Travis, K. M., et al. (2007). Enhanced survival of the LINCL mouse following CLN2 gene transfer using the Rh.10 Rhesus macaque-derived adeno-associated virus vector. Mol. Ther. 15, 481–491. doi: 10.1038/sj.mt.6300049
Stoica, L., Ahmed, S. S., Gao, G., and Sena-Esteves, M. (2013). Gene transfer to the CNS using recombinant adeno-associated virus. Curr. Protoc. Microbiol. 14:14D.5. doi: 10.1002/9780471729259.mc14d05s29
Sullivan, J. A., Stanek, L. M., Lukason, M. J., Bu, J., Osmond, S. R., Barry, E. A., et al. (2018). Rationally designed AAV2 and AAVrh8R capsids provide improved transduction in the retina and brain. Gene Ther. 25, 205–219. doi: 10.1038/s41434-018-0017-8
Tang, J. C. Y., Rudolph, S., and Cepko, C. L. (2017). Viral delivery of GFP-dependent recombinases to the mouse brain. Methods Mol. Biol. 1642, 109–126. doi: 10.1007/978-1-4939-7169-5_8
Taniguchi, H. (2014). Genetic dissection of gabaergic neural circuits in mouse neocortex. Front. Cell. Neurosci. 8:8. doi: 10.3389/fncel.2014.00008
Taymans, J. M., Vandenberghe, L. H., Van Den Haute, C., Thiry, I., Deroose, C. M., Mortelmans, L., et al. (2007). Comparative analysis of adeno-associated viral vector serotypes 1, 2, 5, 7 and 8 in mouse brain. Hum. Gene Ther. 18, 195–206. doi: 10.1089/hum.2006.178
Tenenbaum, L., Chtarto, A., Lehtonen, E., Velu, T., Brotchi, J., and Levivier, M. (2004). Recombinant AAV-mediated gene delivery to the central nervous system. J. Gene Med. 6, S212–S222. doi: 10.1002/jgm.506
Tervo, D. G. R., Hwang, B. Y., Viswanathan, S., Gaj, T., Lavzin, M., Ritola, K. D., et al. (2016). A designer AAV variant permits efficient retrograde access to projection neurons. Neuron 92, 372–382. doi: 10.1016/j.neuron.2016.09.021
Thomsen, D. R., Stenberg, R. M., Goins, W. F., and Stinski, M. F. (1984). Promoter-regulatory region of the major immediate early gene of human cytomegalovirus. Proc. Natl. Acad. Sci. U S A 81, 659–663. doi: 10.1073/pnas.81.3.659
Tordo, J., O’Leary, C., Antunes, A. S. L. M., Palomar, N., Aldrin-Kirk, P., Basche, M., et al. (2018). A novel adeno-associated virus capsid with enhanced neurotropism corrects a lysosomal transmembrane enzyme deficiency. Brain 141, 2014–2031. doi: 10.1093/brain/awy126
Vaughan, A., Vaughan, A., Brumwell, A., and Chapman, H. (2015). Tamoxifen administration for lineage tracing using CreERT2 mice. Protoc. Exchange.doi: 10.1038/protex.2015.018
van den Pol, A. N., Acuna-Goycolea, C., Clark, K. R., and Ghosh, P. K. (2004). Physiological properties of hypothalamic mch neurons identified with selective expression of reporter gene after recombinant virus infection. Neuron 42, 635–652. doi: 10.1016/s0896-6273(04)00251-x
von Jonquieres, G., Fröhlich, D., Klugmann, C. B., Wen, X., Harasta, A. E., Ramkumar, R., et al. (2016). Recombinant human myelin-associated glycoprotein promoter drives selective AAV-mediated transgene expression in oligodendrocytes. Front. Mol. Neurosci. 9:13. doi: 10.3389/fnmol.2016.00013
von Jonquieres, G., Mersmann, N., Klugmann, C. B., Harasta Editha, A., Lutz, B., Teahan, O., et al. (2013). Glial promoter selectivity following AAV-delivery to the immature brain. PLoS One 8:e65646. doi: 10.1371/journal.pone.0065646
Watakabe, A., Kato, S., Kobayashi, K., Takaji, M., Nakagami, Y., Sadakane, O., et al. (2012). Visualization of cortical projection neurons with retrograde TET-Off lentiviral vector. PLoS One 7:e46157. doi: 10.1371/journal.pone.0046157
Watakabe, A., Ohtsuka, M., Kinoshita, M., Takaji, M., Isa, K., Mizukami, H., et al. (2015). Comparative analyses of adeno-associated viral vector serotypes 1, 2, 5, 8 and 9 in marmoset, mouse and macaque cerebral cortex. Neurosci. Res. 93, 144–157. doi: 10.1016/j.neures.2014.09.002
Xie, J., Xie, Q., Zhang, H., Ameres, S. L., Hung, J. H., Su, Q., et al. (2011). MicroRNA-regulated, systemically delivered RAAV9: a step closer to cns-restricted transgene expression. Mol. Ther. 19, 526–535. doi: 10.1038/mt.2010.279
Xiong, W., Wu, D. M., Xue, Y., Wang, S. K., Chung, M. J., Ji, X., et al. (2019). AAV cis-regulatory sequences are correlated with ocular toxicity. Proc. Natl. Acad. Sci. U S A 116, 5785–5794. doi: 10.1073/pnas.1821000116
Xu, R., Janson, C. G., Mastakov, M., Lawlor, P., Young, D., Mouravlev, A., et al. (2001). Quantitative comparison of expression with adeno-associated virus (AAV-2) brain-specific gene cassettes. Gene Ther. 8, 1323–1332. doi: 10.1038/sj.gt.3301529
Yaguchi, M., Ohashi, Y., Tsubota, T., Sato, A., Koyano, K. W., Wang, N., et al. (2013). Characterization of the properties of seven promoters in the motor cortex of rats and monkeys after lentiviral vector-mediated gene transfer. Hum. Gene Ther. Methods 24, 333–344. doi: 10.1089/hgtb.2012.238
Yazdan-Shahmorad, A., Tian, N., Kharazia, V., Samaranch, L., Kells, A., Bringas, J., et al. (2018). Widespread optogenetic expression in macaque cortex obtained with MR-guided, convection enhanced delivery (CED) of AAV vector to the thalamus. J. Neurosci. Methods 293, 347–358. doi: 10.1016/j.jneumeth.2017.10.009
Ye, L., Allen, W. E., Thompson, K. R., Tian, Q., Hsueh, B., Ramakrishnan, C., et al. (2016). Wiring and molecular features of prefrontal ensembles representing distinct experiences. Cell 165, 1776–1788. doi: 10.1016/j.cell.2016.05.010
Zhang, H., Yang, B., Mu, X., Ahmed, S. S., Su, Q., He, R., et al. (2011). Several RAAV vectors efficiently cross the blood-brain barrier and transduce neurons and astrocytes in the neonatal mouse central nervous system. Mol. Ther. 19, 1440–1448. doi: 10.1038/mt.2011.98
Zhang, Y., Zheng, Y., Zhang, Y. P., Shields, L. B. E., Hu, X., Yu, P., et al. (2010). Enhanced adenoviral gene delivery to motor and dorsal root ganglion neurons following injection into demyelinated peripheral nerves. J. Neurosci. Res. 88, 2374–2384. doi: 10.1002/jnr.22394
Zhu, J., Huang, X., and Yang, Y. (2009). The TLR9-MyD88 pathway is critical for adaptive immune responses to adeno-associated virus gene therapy vectors in mice. J. Clin. Invest. 119, 2388–2398. doi: 10.1016/s1525-0016(16)39131-6
Zincarelli, C., Soltys, S., Rengo, G., and Rabinowitz, J. E. (2008). Analysis of AAV serotypes 1-9 mediated gene expression and tropism in mice after systemic injection. Mol. Ther. 16, 1073–1080. doi: 10.1038/mt.2008.76
Keywords: AAV, neuroscience, viral vectors, cell-type specificity, gene delivery, intersectional methods, targeted neuronal manipulation, virus technologies
Citation: Haery L, Deverman BE, Matho KS, Cetin A, Woodard K, Cepko C, Guerin KI, Rego MA, Ersing I, Bachle SM, Kamens J and Fan M (2019) Adeno-Associated Virus Technologies and Methods for Targeted Neuronal Manipulation. Front. Neuroanat. 13:93. doi: 10.3389/fnana.2019.00093
Received: 22 August 2019; Accepted: 30 October 2019;
Published: 26 November 2019.
Edited by:
Jose L. Lanciego, University of Navarra, SpainReviewed by:
Els Henckaerts, KU Leuven, BelgiumAkiya Watakabe, RIKEN Center for Brain Science (CBS), Japan
Copyright © 2019 Haery, Deverman, Matho, Cetin, Woodard, Cepko, Guerin, Rego, Ersing, Bachle, Kamens and Fan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Melina Fan, bWZhbkBhZGRnZW5lLm9yZw==
 Leila Haery
Leila Haery Benjamin E. Deverman2
Benjamin E. Deverman2 Connie Cepko
Connie Cepko Meghan A. Rego
Meghan A. Rego Joanne Kamens
Joanne Kamens