- 1Instituto de Neurociencias de Alicante, Universidad Miguel Hernandez, Consejo Superior de Investigaciones Científicas (UMH-CSIC), Alicante, Spain
- 2Instituto Murciano de Investigacion Biomedica IMIB-Arrixaca (CIBERSAM), Murcia, Spain
In the study of central nervous system morphogenesis, the identification of new molecular markers allows us to identify domains along the antero-posterior and dorso-ventral (DV) axes. In the past years, the alar and basal plates of the midbrain have been divided into different domains. The precise location of the alar-basal boundary is still under discussion. We have identified Barhl1, Nhlh1 and Six3 as appropriate molecular markers to the adjacent domains of this transition. The description of their expression patterns and the contribution to the different mesencephalic populations corroborated their role in the specification of these domains. We studied the influence of Sonic Hedgehog on these markers and therefore on the specification of these territories. The lack of this morphogen produced severe alterations in the expression pattern of Barhl1 and Nhlh1 with consequent misspecification of the basolateral (BL) domain. Six3 expression was apparently unaffected, however its distribution changed leading to altered basal domains. In this study we confirmed the localization of the alar-basal boundary dorsal to the BL domain and demonstrated that the development of the BL domain highly depends on Shh.
Introduction
The midbrain, located between the forebrain and the hindbrain, is subdivided, along the antero-posterior axis, into two mesomeres (m1–m2) (Hidalgo-Sánchez et al., 2005; Moreno-Bravo et al., 2012; Puelles et al., 2012). The rostral mesomere (m1) includes the tectal gray and the superior and inferior colliculi among other populations. The caudal mesomere is rather small and corresponds to the pre-isthmic region (Puelles et al., 2012). Along the dorso-ventral axis (DV) the midbrain is subdivided into roof plate, alar plate, basal plate and floor plate. This DV patterning is due to the effect of two opposing organizing regions, the roof plate and the floor plate. These regions secrete signaling molecules establishing gradients, e.g., Wnt1 and BMP from the roof plate and Shh from the floor plate. These gradients are translated by neuroblasts into positional information. This triggers differentiation programs that specify different neuronal populations along the DV axis (Basler et al., 1993; Dickinson et al., 1995; Lee and Jessell, 1999; Patten and Placzek, 2000; Chizhikov and Millen, 2005; Placzek and Briscoe, 2005; Ingham and Placzek, 2006; Szabó et al., 2009a,b). In the midbrain this DV organization is referred to as tectum (dorsal) and tegmentum (ventral), which acquire their alar and basal characteristics under the influence of roof and floor plate organizers respectively. These alar and basal regions are subsequently subdivided into smaller domains based on different gene expression patterns (Nakatani et al., 2007; Kala et al., 2009; Moreno-Bravo et al., 2012; Puelles et al., 2012). The alar plate is compartmentalized in alar dorsal (AD), alar lateral (AL) and alar ventrolateral (AVL) domains, whereas the basal plate is divided into basolateral (BL), basal intermediate (BI) and basal medial (BM) domains (Nakatani et al., 2007; Kala et al., 2009; Moreno-Bravo et al., 2012; Puelles et al., 2012; see Figure 1A). The BL domain is an intricate transition region extending ventrally form the alar-basal boundary into the basal plate, which displays a major morphological complexity. The whole BL domain expressing Nkx2.2 can be further subdivided into dorsal (dBL) and ventral (vBL) subdomains. The dBL is Gata2 positive and gives rise to GABAergic neurons and the vBL is Pax6 positive and gives rise to glutamatergic neurons (Kala et al., 2009). The fact that this domain generates different neuronal subtypes highlights the complexity of this area.
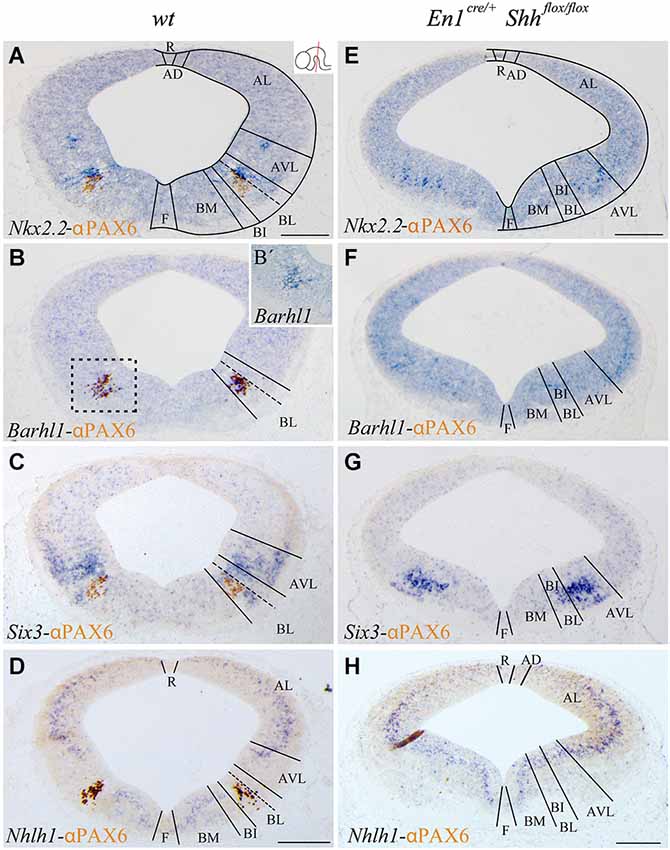
Figure 1. Barhl1, Six3 and Nhlh1 expression pattern along the midbrain DV axis. Coronal mesencephalic sections of wild type (A–D) and En1cre/+; Shhflox/flox (E–H) embryos at E12.5 processed by immunochemistry and in situ hybridization (A–H). Schematic subdivision from dorsal to ventral using Nkx2.2 and PAX6 as reference (A). The Barhl1 expression was restricted to the ventral region of the BL domain (B). Square in B shows magnification of this area before immunochemistry (B’). The Six3 expression included the entire AVL and BL domains (C). The Nhlh1 expression was exclusively negative for the BL domain (D). In the conditional mutant, the whole basal plate showed a reduction in size. Nkx2.2 was detected only in the AVL and it was completely lost, as PAX6, in the BL/BI domains (E). Barhl1 expression was also lost in the BL/BI domains (F). Six3 expression was more compacted in the BL/BI and AVL domains (G). The expression of Nhlh1 was continuous along the DV axis, being also positive for the altered BL/BI domains (H). Inset in A shows the level of the sections. Abbreviations: AD, alar dorsal; AL, alar lateral; AVL, alar ventrolateral; BI, basal intermediate; BL, basolateral; BM, basal medial; F, floor; R, roof. Scale bar = 200 μm.
It has been reported that the basal midbrain exhibits distinct phenotypes depending on the Shh signaling alteration level. Loss of function mutations generates a complete ablation of floor and basal plate neural structures (Chiang et al., 1996; Fogel et al., 2008). A specific conditional inactivation in the En1 expression domain (midbrain and rhombomere 1) leads to a reduction in dopaminergic neurons, a disorganization of the red nucleus (RN) and a complete loss of the oculomotor complex (III) among other abnormalities (Perez-Balaguer et al., 2009).
The aim of this work is to describe new markers that could serve as tools to analyze and describe the basal and alar domains surrounding the alar-basal boundary. We chose Barhl1, Nhlh1 and Six3 for this detailed characterization and also studied possible roles in Shh signaling using a Shh conditional inactivation driven by En1. We selected Barhl1, Nhlh1 and Six3, based on gene expression databases as Eurexpress1 and Allen Brain Atlas,2 because they are specifically expressed in basal mesencephalic compartments among other territories. These transcription factors have proven useful tools for the characterization of basal midbrain organization and have helped to understand the phenotypic alterations of the Shh conditional mutant. We described their expression pattern in both E12.5 and E15.5. At E12.5 the DV domains are being generated, the neuronal precursors are still proliferating in the ventricular area and much of them are migrating to their final position. At E15.5, the basal midbrain is completely stablished and the nuclei are already visible.
Materials and Methods
Animals
The conditional Shh mutant mouse contains the Shhtm2AMC allele where the exon 2 is flanked by LoxP sequences as previously reported (Lewis et al., 2001). The En1cre/+ transgenic line was previously described (Kimmel et al., 2000). The conditional mutant embryos were generated crossing double heterozygous males (En1cre/+; Shhflox/+) with homozygous females (Shhflox/flox) as previously described (Perez-Balaguer et al., 2009). The day when the vaginal plug was detected was considered as embryonic day 0.5 (E0.5). Embryos were fixed in phosphate buffer solution (PBS 1x, NaCl 13 mM, Sigma S3014; KCl 0.3 mM, Sigma P9541; Na2HPO4 1 mM, Sigma S3264 and KH2PO4 0.2 mM, Sigma P9791) with 4% paraformaldehyde (PFA, Panreac 141451.1211) overnight at 4°C and cryoprotected for storage at −20°C. Embryos were washed twice in 100% butanol (Panreac 14.682.1211) previously to be wax embedded (Gemcut emerald paraffin, Catalog no. 24364-1) and then sectioned in parallel series (7 μm thick). A total of 20 pregnant females were used. Each experiment was triplicated to confirm the result. All mouse experiments were performed according to protocols approved by the Universidad Miguel Hernandez CEIE committee.
In Situ Hybridization
The sections were dewaxed at 65°C and completely rehydrated. To facilitate probe penetration, tissue was incubated during 4 min with proteinase K (0.1 mg/ml) in PBS-T (PBS 1x with 0.1% Tween 20, Sigma P1379) and post fixed in 4% PFA. The sections were washed in PBS-T and prehybridized for 1 h in hybridization buffer comprised of 50% deionized formamide (Amresco, 0606), SALT 1X (NaCl 0.2 M, Sigma S3014, tris-HCl 9 mM Sigma T3253, Tris-Base 1 mM, Sigma T6066, NaH2PO4·2H2O 5 mM, Scharlau SO0334, Na2HPO4 5 mM, Sigma S3264 and EDTA 5 mM, Sigma E5134) Denharts 2X (Bio Basic Canada D0062), Dextran sulfate 0.2 mM (Amresco, 0198) and 0.1% tRNA (Sigma R6625). The RNA probes were obtained from Source Bioscience/ ImaGenes (Barhl1, IRAVp968G10116D; Nhlh1, IRAVp968D02101D) or constructions kindly provided by Dr. J. Rubenstein (Nkx2.2) and Dr. L. Puelles (Six3). These digoxigenin-labeled RNA probes (DIG-11-UTP, Roche Diagnostics, 11209256910) were denaturalized at 80°C and incubated with the tissue in hybridization buffer overnight at 62°C. The next day sections were washed in wash solution with 50% SSC 1x pH 7.0 (NaCl 0.15 M, Sigma S3014 and Na3C6H5O7·2H2O 15 mM, Sigma C8532), 25% formamide (Sigma, F7503) and 0.1% tween 20 at 65°C and incubated with MABT 1x pH 7.4 (NaCl 40 mM, maleic acid 20 mM, NaOH 40 mM and 0.1% tween 20) with 10% sheep serum (Sigma, S2263) and 20% blocking reagent (Roche, 10057177103). After blocking, tissue was incubated overnight at 4°C in the same solution with an alkaline phosphatase-coupled anti-digoxigenin antibody (1:3500, Roche, 11093274910). Excess of non-specific anti-digoxigenin antibody was extensively washed in MABT. Finally, the sections were washed with NTMT (NaCl 0.1 M, Sigma S3014, Tris-HCl 0.1 M pH 9.5, Sigma T3253, MgCl2·6H2O 0.05 M, VWR 1.05833 and 0.1% tween-20) and incubated overnight at room temperature in NTMT with 0.45 μl/ml of 4-Nitro blue tetrazolium chloride (NBT, 75 mg/ml, Roche, 70227721) and 3.5 μl/ml of 5-Bromo-4-Chloro-3-indolyl-phosphate, (BCIP, 50 mg/ml, Roche 11585002001). The NBT/BCIP was used for the colorimetric reaction to detect the presence of the hybridized probes. The alkaline phosphatase reacts with these substrates and produces a solid blue precipitate.
Immunohistochemistry
The sections were dewaxed, completely rehydrated and for antigen retrieval boiled in sodium citrate 0.1 M pH 6. The sections were washed in PBS-T and incubated in PBS-T with 1.5% H2O2 for 30 min to inactivate endogenous peroxidase. After inactivation, tissue was washed in PBT and blocked 1 h in PBS-T with 0.1% albumin bovine serum (BSA, A2153, Sigma) and 10% lysine 1 M (L5626, Sigma). Next, sections were incubated overnight at room temperature in PBS-T with 0.1% BSA and 0.01% sodium azide (S2002, Sigma) with αNKX2.2 (1:5, raised in mouse, Hybridoma Bank, 74.5A5) or αPAX6 (1:200, raised in rabbit, Covance, PRB-278P). The day after, the tissue was rinsed in PBS-T and incubated 1 h with αMouse (1:200, raised in goat, Vector Labs, BA-2020) or αRabbit (1:200, raised in goat, Vector Labs, BA-1000) biotinylated secondary antibody. Afterwards, the sections were washed in PBS-T and incubated in PBS-T with Avidin–Biotin Complex (1:500, Vectastain PK-4000) for 1 h. Finally, tissue was washed in PBS-T and Tris 0.1 M pH 7 and the immunolabeling was revealed in Tris 0.1 M with 1% 3-3′ diaminobenzidine tetrahydroc (DAB, Acros Organics W0572M) and 0.003% H2O2 leading to a brown precipitate (0.025% Ammonium Nickel Sulphate was added to obtain black precipitate).
Results
Role of Shh in the Development of DV Mesencephalic Domains
With the aim to better characterize the different domains of the basal midbrain, we studied the expression pattern of Barhl1, Nhlh1 and Six3. These markers have been proven to be useful for a better understanding of the complexity of the basal midbrain, specifically in the BL domain. We used Nkx2.2 and PAX6 as a well-known reference for a precise localization of these markers. At E12.5 Nkx2.2 delimited the BL domain and a small area in the AVL whereas PAX6 was only restricted to the vBL domain (Figure 1A). In situ hybridization for the new markers showed a discrete expression of Barhl1 in the vBL, overlapping with the PAX6 positive domain (Figures 1B,B’). Six3 displayed a complex expression pattern in the alar-basal transition including the AVL and BL domain (Figure 1C). Nhlh1 was widely expressed along the DV axis of the midbrain, including all the domains of the alar region, the BM and BI domains in the basal plate and the floor plate. Only the BL domain was negative for this marker (Figure 1D).
After the characterization of the precise localization of our markers, our aim was to investigate the influence of the basal organizer in the specification of the BL domain. We used the conditional mutant En1cre/+; Shhflox/flox to abolish the expression of Shh in the midbrain. This strain allows us to investigate the effect of the loss of Shh on the expression pattern of Barhl1, Nhlh1 and Six3. The entire basal plate showed a complete disorganization and the expression of our markers presented the following alterations. At E12.5, Nkx2.2 expression was only detected in the AVL domain. Both Nkx2.2 and PAX6 were completely lost in the BL domain (Figure 1E). Barhl1, that colocalized with PAX6, was also absent at E12.5 (Figure 1F). Six3 expression was maintained in the alar-basal transition, but the expression domain was more compact (Figure 1G), probably due to the incorrect specification of the basal territory that receives the Six3 positive neurons. Nhlh1 expression also displayed clear alterations. The discontinuity of expression found in the BL domain disappeared in the conditional mutant (Figure 1H).
We confirmed the morphological interpretation of the expression patterns by comparison with Gad2 and vGluT2 distribution and basic Nissl staining. The distribution of GABAergic (Figures 2A,D) and glutamatergic neurons (Figures 2B,E) showed an altered distribution in the conditional mutant when compared with control. In the basic Nissl staining, we verified the abnormal development of the DV domains described above (Figures 2C,F).
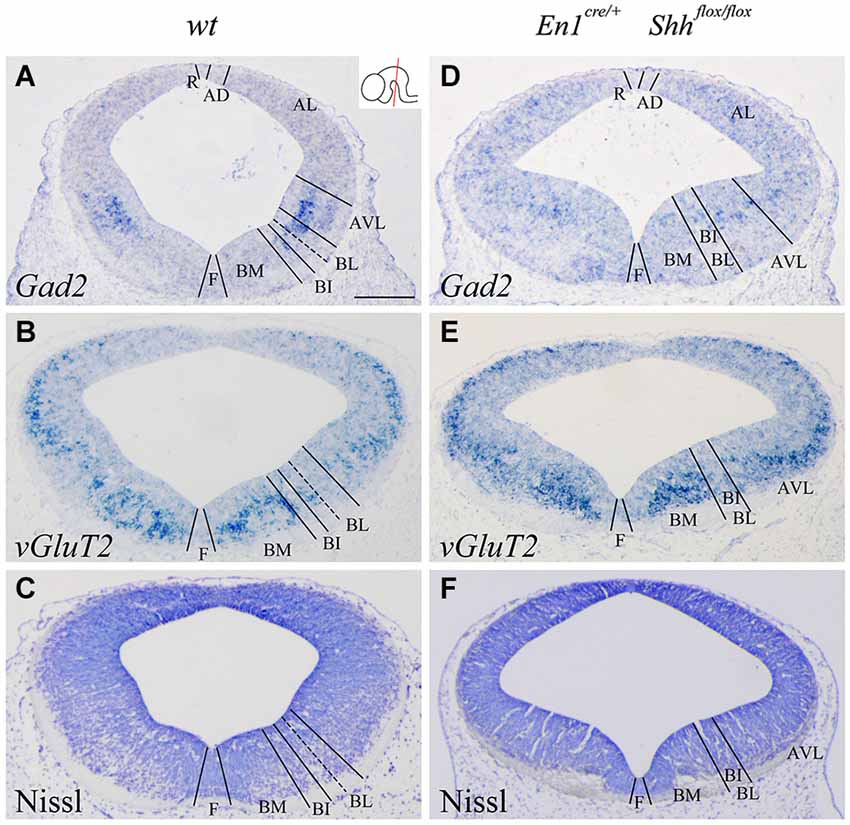
Figure 2. Alterations of the neuronal precursors in the absence of Shh. Coronal mesencephalic sections of wild type (A–C) and En1cre/+; Shhflox/flox (D–F) embryos at E12.5 processed by in situ hybridization (A,B,D,E) and basic Nissl staining (C,F). Neuronal markers for GABAergic (A) and glutamatergic (B) neurons showed their contribution to the DV domains. The absence of Shh showed the altered development of both GABAergic (D) and glutamatercig (E) precursors. Basic Nissl staining showed the abnormal developing of the basal plate when compared with control (C,F). Inset in A shows the level of the sections. Abbreviations: AD, alar dorsal; AL, alar lateral; AVL, alar ventrolateral; BI, basal intermediate; BL, basolateral; BM, basal medial; F, floor; R, roof. Scale bar = 200 μm.
We conclude that Barhl1 is a useful marker for the vBL; whereas Six3 covers the AVL and the BL domains and Nhlh1 could serve as a negative marker for the BL as it labels all the other domains. The changes in expression pattern in the conditional mutant indicate a clear defect in the specification of the BL domain in the absence of the morphogen Shh.
Differentiation of Midbrain Neuronal Populations in the Altered Basal Domains
We analyzed the expression pattern of these markers at E15.5 to associate the domains observed at E12.5 with differentiated neuronal populations. Nkx2.2 and PAX6 expression was used again as a reference. Nkx2.2 was still detected in the AVL and BL domains, but migrated cells were also located in the mantle layer of the BI and BM domains. PAX6 labeled the vBL domain and a few migrated scattered cells were detected in the BI and BM domains (Figure 3A). Barhl1 extended its expression to the alar plate, including the superior colliculus (SC) and the bed nucleus of the brachium of the inferior colliculus (BIC) in the AVL (Figure 3B). In the vBL, coincident with the location of PAX6 positive neurons (compare Figures 3B,D), Barhl1 defined a discrete population lateral to the oculomotor complex that is part of the pararubral formation (mPaRF, Figure 3B). Expression of Six3 was observed in the mantle layer of the AVL, BL, BI and BM. It corresponded to the mesencephalic reticular formation (mRF) of the AVL, BL and BI. Six3 positive cells located in the BM correspond with tangentially migrating neurons forming the Substantia nigra pars reticulata (SNR; Figure 3C), the population located lateral to the dopaminergic Substantia nigra pars compacta (Figure 3C). At this stage, Nhlh1 expression was completely downregulated (Figure 3D).
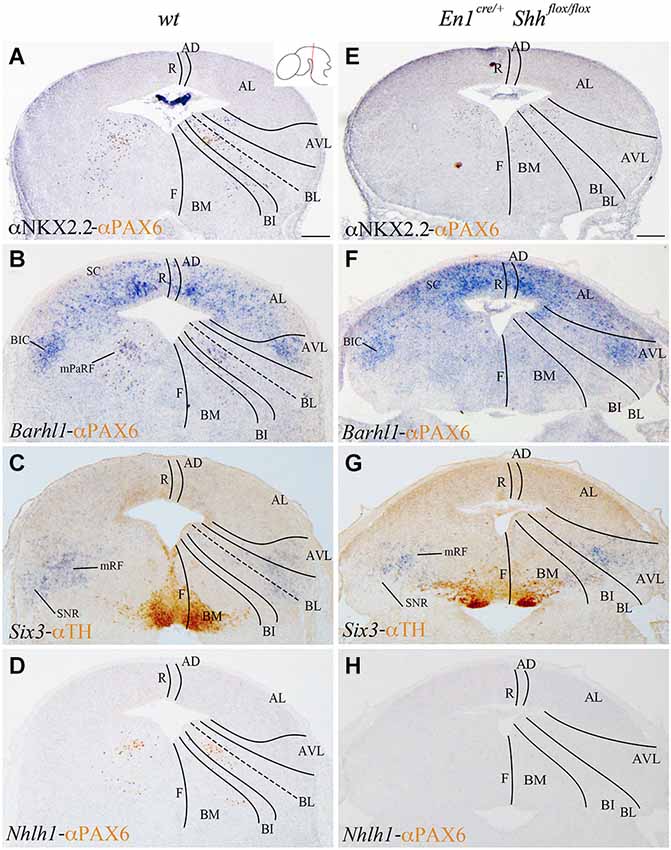
Figure 3. Contribution of Barhl1, Six3 and Nhlh1 to the basal populations. Coronal mesencephalic sections of wild type (A–D) and En1cre/+; Shhflox/flox (E–F) embryos at E15.5 processed by immunochemistry (A,E) or immunochemistry and in situ hybridization (B–D,F–H). Schematic subdivision from dorsal to ventral using Nkx2.2 and PAX6 as reference (A). The expression pattern of Barhl1 showed its contribution to the whole alar plate and the mPaRF in the basal plate (B). The expression pattern of Six3 showed its contribution to the mRF and SNR (limited by TH positive neurons; (C). Nhlh1 was downregulated at this stage (D). The absence of Shh caused a reduction of Nkx2.2 (E) and the lost of PAX6 and Barhl1 (F) in the basal midbrain. Six3 expression presented a scattered and wider distribution in the mRF (G). Nhlh1 was not expressed as in the wild type (H). Inset in A shows the level of the sections. Abbreviations: AD, alar dorsal; AL, alar lateral; AVL, alar ventrolateral; BI, basal intermediate; BIC, bed nucleus of the brachium of the inferior colliculus; BL, basolateral; BM, basal medial; F, floor; mPaRF, mesencephalic pararubral formation; mRF, mesencephalic reticular formation; R, roof; SC, superior colliculus; SNC, Substantia nigra pars compacta; SNR, Substantia nigra pars reticulata. Scale bar = 200 μm.
In order to further investigate the effect of loss of Shh on the differentiation of the neuronal populations we also studied the expression of these markers at E15.5. Nkx2.2 expression was almost absent in the conditional mutant; we detected a small number of positive neurons in the mantle layer of the AVL, BI/BL and BM domains (Figure 3E). PAX6 positive neurons were absent (Figures 3E,F,H). Barhl1 expression in the alar plate was apparently normal. It was present in the SC and BIC (Figure 3F). In contrast, its expression in the basal plate was completely lost (Figure 3F). The expression of Six3 in the mantle layer barely differed from the wild type expression. In the conditional mutant the expression pattern was similar but more compact (Figure 3G). Nhlh1 was not expressed as it was described previously in the control (Figure 3H). The misspecification observed in the BI and BL domains were corroborated by the study of Gad2 and vGluT2 distribution and basic Nissl staining. The distribution of GABAergic neurons was clearly altered. The three populations described, mRF, SNR and ventral tegmental area (VTA) presented a strong reduction in the number of neurons (Figures 4A,D). The glutamatergic neurons did not display obvious changes. Nevertheless, the RN displayed a clear alteration in its neuronal distribution, occupying a wider territory (Figures 4B,E). This phenotype was confirmed by the analysis of basic Nissl staining (Figures 4C,F).
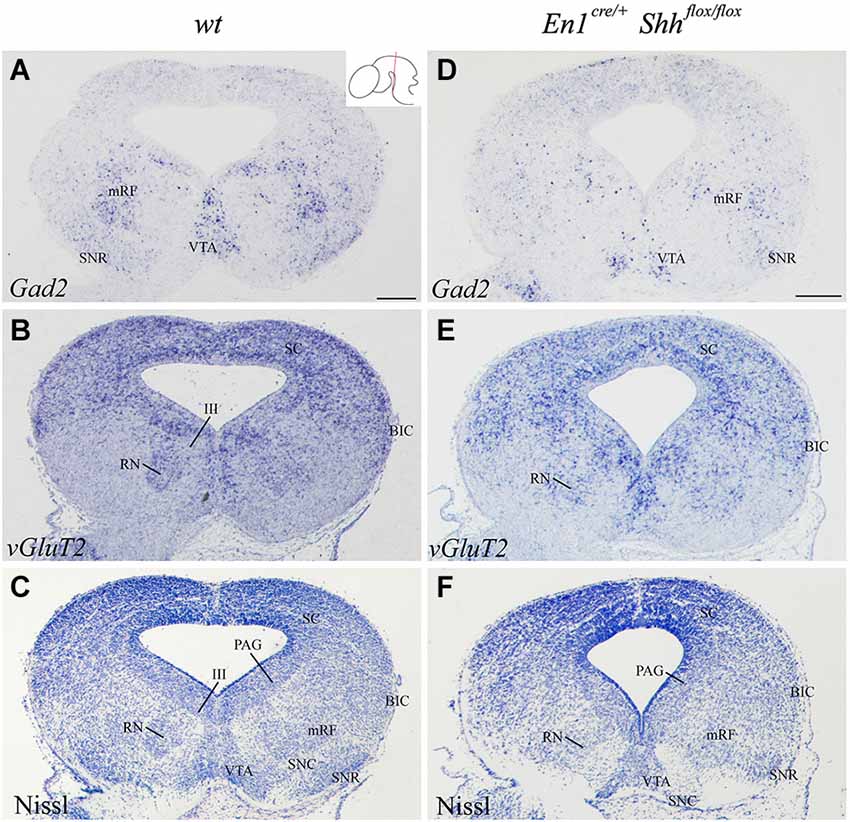
Figure 4. Alteration of the basal population in the absence of Shh. Coronal mesencephalic sections of wild type (A–C) and En1cre/+; Shhflox/flox (D–F) embryos at E15.5 processed by in situ hybridization (A,B,D,E) and basic Nissl staining (C,F). The absence of Shh affected the GABAergic (A,D) and glutamatergic (B,E) populations, here verified by Nissl staining (C,F). Inset in A shows the level of the sections. Abbreviations: III, oculomotor complex; BIC, bed nucleus of the brachium of the inferior colliculus; mRF, mesencephalic reticular formation; PAG, periaqueductal gray; RN, red nucleus; SC, superior colliculus; SNC, Substantia nigra pars compacta; SNR, Substantia nigra pars reticulate; VTA, ventral tegmental area. Scale bar = 200 μm.
In summary, our markers allowed us to follow the development of all the DV domains at late embryonic stages and we were able to identify neuronal subpopulations that were generated in each of these domains. Additionally, we demonstrated that Shh is required for the proper specification of the mesencephalic BL domain. Its absence results in alterations of this domain causing the loss or miss-expression of Nkx2.2, Pax6, Barhl1, Nhlh1 and Six3 leading to an incorrect differentiation of neuronal populations located in this territory.
Discussion
New Markers for the DV Axis and Their Contribution to the Mesencephalic Neuronal Population
The study of gene expression patterns along the DV axis of the midbrain helped us to further characterize the alar and basal plate subdivisions (Nakatani et al., 2007; Kala et al., 2009; Puelles et al., 2012). In the present work we have identified Barhl1, Nhlh1 and Six3 as useful markers for a detailed characterization of these subdivisions (Figure 5). The Barhl1 expression pattern, at E12.5, specifically defined the vBL. It mimics the distribution of the PAX6 protein. Both markers allowed us to describe two distinct subdomains within the BL midbrain region (Figure 5). One subdomain located in the dBL is negative for these markers and gives rise to GABAergic neurons. The second subdomain located in the vBL is Barhl1 and Pax6 positive and generates glutamatergic neurons (Kala et al., 2009; Figure 5). At E15.5, Barhl1 also presented an expression domain in the alar plate defining the SC and the BIC, while in the basal plate it defined a discrete population close to the oculomotor complex. Barhl1 and Pax6 have been implicated in the specification of other glutamatergic populations. In the cerebellum Pax6, expressed in migrating granular cells, is implicated in the proper migration of these neurons (Engelkamp et al., 1999). In the same region, the expression of Barhl1 coincides with Pax6. However, there is no implication for a functional interaction. The current opinion is that they work independently (Landsberg et al., 2005). In the cerebellum, Barhl1 is also involved in the generation of the glutamatergic granular cells, but is involved in a pathway that controls the migration and survival of their progenitors (Bulfone et al., 2000; Li et al., 2004; Lopes et al., 2006; Rachidi and Lopes, 2006). At E12.5, the expression of Barhl1 as occurs in the cerebellum, should be related to the proper migration and survival of the glutamatergic neurons generated in the vBL. At E15.5, Barhl1 in the alar plate is involved in the maintenance and survival, but not in the differentiation of the SC neurons (Li and Xiang, 2006).
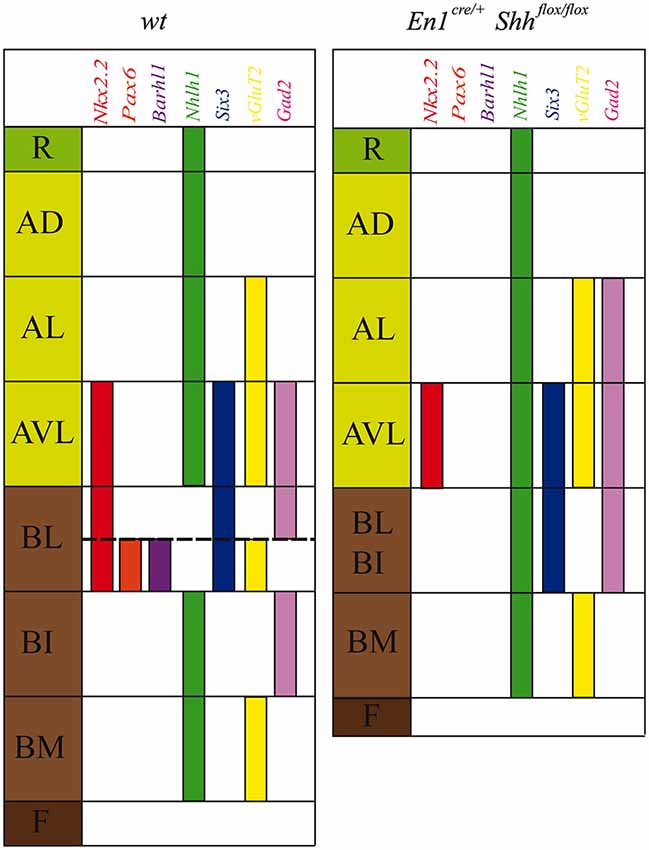
Figure 5. Schematic representation of DV mesencephalic domains. Model of the DV patterning in the midbrain domains at E12.5 in the wild type and the conditional mutant. The lack of Shh generated an altered development of the basal plate and a miss-expression of Nkx2.2, Pax6, Barhl1, Nhlh1, Six3, vGluT2 and Gad2.
At E12.5, Six3 labeled the domains adjacent to the alar-basal boundary, defining both the AVL and the BL domains (Figure 5). In the BL, there was a clear difference between the vBL and the dBL, showing a stronger expression in the latter. This transcription factor is a well-known marker for the GABAergic lineage, expressed in neuronal precursors after the onset of Gata2 expression (Virolainen et al., 2012). The GABAergic nature of the AVL and dBL (Kala et al., 2009) indicates a role of Six3 in the development of these neuronal populations. At E15.5 its expression extended into the mantle layer covering almost all the GABAergic mRF, the pararubral nucleus and the SNR (Paxinos and Watson, 1998; Paxinos et al., 2009; Moreno-Bravo et al., 2012; Madrigal et al., 2015).
Finally, Nhlh1 expression at E12.5 served as a negative marker for the BL domain, as it is the only negative domain along the DV axis (Figure 5). Unfortunately, at E15.5, its expression was completely down regulated. This early Nhlh1 expression has been related to a possible role in promoting migration or in preventing postmitotic cells from re-entering the cell cycle erroneously (Li et al., 1999; Schmid et al., 2007).
In summary, we have identified a new set of genetic markers for the characterization of mesencephalic DV domains at both sides of the alar-basal boundary. They additionally allowed us to identify neuronal populations generated in these domains.
The Basolateral Domain of the Midbrain Depends on Shh for Its Specification
Shh is secreted from the ventricular layer of the floor and basal plates and has an important role in the development of mesencephalic basal populations (Hynes et al., 1995; Litingtung and Chiang, 2000; Agarwala and Ragsdale, 2002; Abeliovich and Hammond, 2007; Joksimovic et al., 2009; Perez-Balaguer et al., 2009). Little is known about possible roles in the specification of the domains adjacent to the alar-basal boundary. In order to unveil if Barhl1, Nhlh1 and Six3 are regulated by Shh we used the conditional mutant En1cre/+; Shhflox/flox. The basal plate of this mutant is strongly affected in the BM domain, the III is lost, the SNC and the VTA are strongly reduced and the RN is altered (Hynes et al., 1995; Abeliovich and Hammond, 2007; Joksimovic et al., 2009; Perez-Balaguer et al., 2009). The analysis of Barhl1, Nhlh1 and Six3 finally allows us to better describe these defects in the basal midbrain. This was complex until today due to the reduced number of specific markers for the BI and BL. One of the most evident defects was the loss of BL domain identity where all of the markers were affected (Figure 5). At E12.5, Barhl1 expression in the vBL domain was absent. This coincides with the loss of Pax6 expression in the same domain (Perez-Balaguer et al., 2009). At E15.5, Barhl1 was expressed normally in the alar plate, since the absence of Shh does not affect the specification of the alar plate (Echevarría et al., 2003; Chizhikov and Millen, 2005; Vieira et al., 2010; Martinez-Ferre and Martinez, 2012; Martinez-Ferre et al., 2013). However, a small Barhl1 positive population located close to the oculomotor complex was completely lost. It is plausible to hypothesize that this population originated from the Barhl1 positive vBL domain described at E12.5. Six3 expression at E12.5, in the absence of Shh, was similar to the expression pattern observed in controls. Nevertheless, its pattern appeared more compact probably due to a general reduction in the size of this domain. At E15.5, Six3 expression continued without apparent alterations. Nevertheless, Six3 positive neurons located in the mantle layer were disorganized. This anomalous distribution could be due to a general alteration of the mRF in the mesencephalic basal plate. However, the absence of Shh does not affect Six3 expression itself. A possible explanation for that could be that the fate of GABAergic neurons has already been determined by Gata2, which is expressed before the onset of Six3 expression in the basal midbrain (Virolainen et al., 2012). The fact, that GABAergic nuclei that populate the basal midbrain arise from Shh negative ventricular domains, further supports this hypothesis (Joksimovic et al., 2009; Madrigal et al., 2015). Finally, Nhlh1 functions as a negative marker for the BL domain at E12.5. In the absence of Shh, it is expressed along the entire DV axis including the altered BL domain, suggesting again a loss of identity of this area.
In conclusion, our data corroborate the strong influence of Shh on the specification of the most dorsal domain of the mesencephalic basal plate, the BL domain. This strong Shh dependence allowed us to validate its basal identity. Therefore, the dorsal limit of the BL domain help us to precisely locate the alar-basal boundary; all the domains ventral to this boundary are Shh dependents.
Author Contributions
All authors had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. Conceived and designed the experiments: Jesus E. Martinez-Lopez, Salvador Martinez and Eduardo Puelles; Performed the experiments: Jesus E. Martinez-Lopez, Juan A. Moreno-Bravo and M. Pilar Madrigal; Analyzed the data: Jesus E. Martinez-Lopez and Eduardo Puelles; Wrote the article: Jesus E. Martinez-Lopez and Eduardo Puelles; Obtained funding: Salvador Martinez and Eduardo Puelles.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Work supported by “Ministerio de Economía y Competitividad” BFU2010-16548 and BFU2013-48230-P (FEDER Fonds), Generalitat Valenciana: PROMETEO (PROMETEOII/2014/014) to E. Puelles; European commission (EUCOMMTOOLS, contract 261492) and Instituto de Salud Carlos III. RD12/0019/0024 to S.M.; J.A. Moreno-Bravo was supported by the Predoctoral Program of the “Consejo Superior de Investigaciones Científicas-Junta de Ampliación de Estudios”, co-financed by the European Social Fund. The Instituto de Neurociencias is a “Centre of Excellence Severo Ochoa”. We thank N. Mecklenburg for critical reading of the manuscript.
Footnotes
References
Abeliovich, A., and Hammond, R. (2007). Midbrain dopamine neuron differentiation: factors and fates. Dev. Biol. 304, 447–454. doi: 10.1016/j.ydbio.2007.01.032
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Agarwala, S., and Ragsdale, C. W. (2002). A role for midbrain arcs in nucleogenesis. Development 129, 5779–5788. doi: 10.1242/dev.00179
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Basler, K., Edlund, T., Jessell, T. M., and Yamada, T. (1993). Control of cell pattern in the neural tube: regulation of cell differentiation by dorsalin-1, a novel TGF beta family member. Cell 21, 687–702. doi: 10.1016/0092-8674(93)90249-p
Bulfone, A., Menguzzato, E., Broccoli, V., Marchitiello, A., Gattuso, C., Mariani, M., et al. (2000). Barhl1, a gene belonging to a new subfamily of mammalian homeobox genes, is expressed in migrating neurons of the CNS. Hum. Mol. Genet. 9, 1443–1452. doi: 10.1093/hmg/9.9.1443
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chiang, C., Litingtung, Y., Lee, E., Young, K. E., Corden, J. L., Westphal, H., et al. (1996). Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function. Nature 383, 407–413. doi: 10.1038/383407a0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chizhikov, V. V., and Millen, K. J. (2005). Roof plate-dependent patterning of the vertebrate dorsal central nervous system. Dev. Biol. 277, 287–295. doi: 10.1016/j.ydbio.2004.10.011
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Dickinson, M. E., Selleck, M. A., McMahon, A. P., and Bronner-Fraser, M. (1995). Dorsalization of the neural tube by the non-neural ectoderm. Development 121, 2099–2106.
Echevarría, D., Vieira, C., Gimeno, L., and Martínez, S. (2003). Neuroepithelial secondary organizers and cell fate specification in the developing brain. Brain Res. Rev. 43, 179–191. doi: 10.1016/j.brainresrev.2003.08.002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Engelkamp, D., Rashbass, P., Seawright, A., and van Heyningen, V. (1999). Role of Pax6 in development of the cerebellar system. Development 126, 3585–3596.
Fogel, J. L., Chiang, C., Huang, X., and Agarwala, S. (2008). Ventral specification and perturbed boundary formation in the mouse midbrain in the absence of Hedgehog signaling. Dev. Dyn. 237, 1359–1372. doi: 10.1002/dvdy.21536
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hidalgo-Sánchez, M., Martínez-de-la-Torre, M., Alvarado-Mallart, R.-M., and Puelles, L. (2005). A distinct preisthmic histogenetic domain is defined by overlap of Otx2 and Pax2 gene expression in the avian caudal midbrain. J. Comp. Neurol. 483, 17–29. doi: 10.1002/cne.20402
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hynes, M., Porter, J. A., Chiang, C., Chang, D., Tessier-Lavigne, M., Beachy, P. A., et al. (1995). Induction of midbrain dopaminergic neurons by Sonic hedgehog. Neuron 15, 35–44. doi: 10.1016/0896-6273(95)90062-4
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ingham, P. W., and Placzek, M. (2006). Orchestrating ontogenesis: variations on a theme by sonic hedgehog. Nat. Rev. Genet. 7, 841–850. doi: 10.1038/nrg1969
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Joksimovic, M., Anderegg, A., Roy, A., Campochiaro, L., Yun, B., Kittappa, R., et al. (2009). Spatiotemporally separable Shh domains in the midbrain define distinct dopaminergic progenitor pools. Proc. Natl. Acad. Sci. U S A 106, 19185–19190. doi: 10.1073/pnas.0904285106
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kala, K., Haugas, M., Lilleväli, K., Guimera, J., Wurst, W., Salminen, M., et al. (2009). Gata2 is a tissue-specific post-mitotic selector gene for midbrain GABAergic neurons. Development 136, 253–262. doi: 10.1242/dev.029900
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kimmel, R. A., Turnbull, D. H., Blanquet, V., Wurst, W., Loomis, C. A., and Joyner, A. L. (2000). Two lineage boundaries coordinate vertebrate apical ectodermal ridge formation. Genes Dev. 14, 1377–1389. doi: 10.1101/gad.14.11.1377
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Landsberg, R. L., Awatramani, R. B., Hunter, N. L., Farago, A. F., DiPietrantonio, H. J., Rodriguez, C. I., et al. (2005). Hindbrain rhombic lip is comprised of discrete progenitor cell populations allocated by Pax6. Neuron 48, 933–947. doi: 10.1016/j.neuron.2005.11.031
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lee, K. J., and Jessell, T. M. (1999). The specification of dorsal cell fates in the vertebrate central nervous system. Annu. Rev. Neurosci. 22, 261–294. doi: 10.1146/annurev.neuro.22.1.261
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lewis, P. M., Dunn, M. P., McMahon, J. A., Logan, M., Martin, J. F., St-Jacques, B., et al. (2001). Cholesterol modification of sonic hedgehog is required for long-range signaling activity and effective modulation of signaling by Ptc1. Cell 105, 599–612. doi: 10.1016/s0092-8674(01)00369-5
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Li, S., Qiu, F., Xu, A., Price, S. M., and Xiang, M. (2004). Barhl1 regulates migration and survival of cerebellar granule cells by controlling expression of the neurotrophin-3 gene. J. Neurosci. 24, 3104–3114. doi: 10.1523/jneurosci.4444-03.2004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Li, S., and Xiang, M. (2006). Barhl1 is required for maintenance of a large population of neurons in the zonal layer of the superior colliculus. Dev. Dyn. 235, 2260–2265. doi: 10.1002/dvdy.20858
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Li, C. M., Yan, R. T., and Wang, S. Z. (1999). Misexpression of a bHLH gene, cNSCL1, results in abnormal brain development. Dev. Dyn. 215, 238–247. doi: 10.1002/(sici)1097-0177(199907)215:3<238::aid-aja6>3.3.co;2-6
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Litingtung, Y., and Chiang, C. (2000). Control of Shh activity and signaling in the neural tube. Dev. Dyn. 219, 143–154. doi: 10.1002/1097-0177(2000)9999:9999<::aid-dvdy1050>3.3.co;2-h
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lopes, C., Delezoide, A. L., Delabar, J. M., and Rachidi, M. (2006). BARHL1 homeogene, the human ortholog of the mouse Barhl1 involved in cerebellum development, shows regional and cellular specificities in restricted domains of developing human central nervous system. Biochem. Biophys. Res. Commun. 339, 296–304. doi: 10.1016/j.bbrc.2005.11.021
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Madrigal, M. P., Moreno-Bravo, J. A., Martínez-López, J. E., Martínez, S., and Puelles, E. (2015). Mesencephalic origin of the rostral substantia nigra pars reticulata. Brain Struct. Funct. doi: 10.1007/s00429-014-0980-9. [Epub ahead of print].
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Martinez-Ferre, A., and Martinez, S. (2012). Molecular regionalization of the diencephalon. Front. Neurosci. 6:73. doi: 10.3389/fnins.2012.00073
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Martinez-Ferre, A., Navarro-Garberi, M., Bueno, C., and Martinez, S. (2013). Wnt signal specifies the intrathalamic limit and its organizer properties by regulating Shh induction in the alar plate. J. Neurosci. 33, 3967–3980. doi: 10.1523/JNEUROSCI.0726-12.2013
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Moreno-Bravo, J. A., Martinez-Lopez, J. E., and Puelles, E. (2012). Mesencephalic neuronal populations: new insights on the ventral differentiation programs. Histol. Histopathol. 27, 1529–1538.
Nakatani, T., Minaki, Y., Kumai, M., and Ono, Y. (2007). Helt determines GABAergic over glutamatergic neuronal fate by repressing Ngn genes in the developing mesencephalon. Development 134, 2783–2793. doi: 10.1242/dev.02870
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Patten, I., and Placzek, M. (2000). The role of Sonic hedgehog in neural tube patterning. Cell. Mol. Life Sci. 57, 1695–1708. doi: 10.1007/pl00000652
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Paxinos, G., and Watson, C. (1998). The Rat Brain in Stereotaxic Coordinates. 4th Edn. San Diego: Academic Press.
Paxinos, G., Watson, C., Carrive, P., Kirkcaldie, M., and Ashwell, K. (2009). Chemoarchitectonic Atlas of the Rat Brain. 2nd Edn. San Diego: Elsevier Academic Press.
Perez-Balaguer, A., Puelles, E., Wurst, W., and Martinez, S. (2009). Shh dependent and independent maintenance of basal midbrain. Mech. Dev. 126, 301–313. doi: 10.1016/j.mod.2009.03.001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Placzek, M., and Briscoe, J. (2005). The floor plate: multiple cells, multiple signals. Nat. Rev. Neurosci. 6, 230–240. doi: 10.1038/nrn1628
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Puelles, E., Martinez-de-la-Torre, M., Watson, C., and Puelles, L. (2012). “Midbrain”, in the Mouse Nervous System. San Diego: Elsevier Academic Press.
Rachidi, M., and Lopes, C. (2006). Differential transcription of Barhl1 homeobox gene in restricted functional domains of the central nervous system suggests a role in brain patterning. J. Neurosci. 24, 35–44. doi: 10.1016/j.ijdevneu.2005.11.015
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Schmid, T., Krüger, M., and Braun, T. (2007). NSCL-1 and -2 control the formation of precerebellar nuclei by orchestrating the migration of neuronal precursor cells. J. Neurochem. 102, 2061–2072. doi: 10.1111/j.1471-4159.2007.04694.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Szabó, N. E., Zhao, T., Cankaya, M., Theil, T., Zhou, X., and Alvarez-Bolado, G. (2009a). Role of neuroepithelial Sonic hedgehog in hypothalamic patterning. J. Neurosci. 29, 6989–7002. doi: 10.1523/JNEUROSCI.1089-09.2009
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Szabó, N. E., Zhao, T., Zhou, X., and Alvarez-Bolado, G. (2009b). The role of Sonic hedgehog of neural origin in thalamic differentiation in the mouse. J. Neurosci. 29, 2453–2466. doi: 10.1523/JNEUROSCI.4524-08.2009
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Vieira, C., Pombero, A., García-Lopez, R., Gimeno, L., Echevarria, D., and Martínez, S. (2010). Molecular mechanisms controlling brain development: an overview of neuroepithelial secondary organizers. Int. J. Dev. Biol. 54, 7–20. doi: 10.1387/ijdb.092853cv
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Virolainen, S. M., Achim, K., Peltopuro, P., Salminen, M., and Partanen, J. (2012). Transcriptional regulatory mechanisms underlying the GABAergic neuron fate in different diencephalic prosomeres. Development 139, 3795–3805. doi: 10.1242/dev.075192
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: midbrain, Shh, Barhl1, Nhlh1, Six3, basolateral domain, alar-basal boundary
Citation: Martinez-Lopez JE, Moreno-Bravo JA, Madrigal MP, Martinez S and Puelles E (2015) Mesencephalic basolateral domain specification is dependent on Sonic Hedgehog. Front. Neuroanat. 9:12. doi: 10.3389/fnana.2015.00012
Received: 21 November 2014; Accepted: 27 January 2015;
Published online: 17 February 2015.
Edited by:
James A. Bourne, Australian Regenerative Medicine Institute, AustraliaReviewed by:
Yu-Qiang Ding, Tongji Unversity, ChinaJihane Homman-Ludiye, Monash University, Australia
Copyright © 2015 Martinez-Lopez, Moreno-Bravo, Madrigal, Martinez and Puelles. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution and reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Eduardo Puelles, Instituto de Neurociencias de Alicante, Universidad Miguel Hernandez, Consejo Superior de Investigaciones Científicas (UMH-CSIC), Avda Ramon y Cajal s/n, E-03550 Alicante, Spain e-mail: epuelles@umh.es
 Jesus E. Martinez-Lopez
Jesus E. Martinez-Lopez Juan A. Moreno-Bravo
Juan A. Moreno-Bravo M. Pilar Madrigal
M. Pilar Madrigal Salvador Martinez
Salvador Martinez Eduardo Puelles
Eduardo Puelles