
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Neuroanat. , 02 March 2011
volume 5 - 2011 | https://doi.org/10.3389/fnana.2011.00011
This article is part of the Research Topic Adaptive Function and Brain Evolution View all 20 articles
The expression of the Nkx2.2 gene is involved in the organization of the alar-basal boundary in the forebrain of vertebrates. Its expression in different diencephalic and telencephalic regions, helped to define distinct progenitor domains in mouse and chick. Here we investigated the pattern of Nkx2.2 protein distribution throughout the development of the forebrain of the anuran amphibian, Xenopus laevis. We used immunohistochemical and in situ hybridization techniques for its detection in combination with other essential territorial markers in the forebrain. No expression was observed in the telencephalon. In the alar hypothalamus, Nkx2.2 positive cells were scattered in the suprachiasmatic territory, but also in the supraopto-paraventricular area, as defined by the expression of the transcription factor Orthopedia (Otp) and the lack of xDll4. In the basal hypothalamus Nkx2.2 expressing cells were localized in the tuberal region, with the exception of the arcuate nucleus, rich in Otp expressing cells. In the diencephalon it was expressed in all three prosomeres (P1–P3) and not in the zona limitans intrathalamica. The presence of Nkx2.2 expressing cells in P3 was restricted to the alar portion, as well as in prosomere P2, whereas in P1 the Nkx2.2 expressing cells were located in the basal plate and identified the alar/basal boundary. These results showed that Nkx2.2 and Sonic hedgehog are expressed in parallel adjacent stripes along the anterior–posterior axis. The results of this study showed a conserved distribution pattern of Nkx2.2 among vertebrates, crucial to recognize subdivisions that are otherwise indistinct, and supported the relevance of this transcription factor in the organization of the forebrain, particularly in the delineation of the alar/basal boundary of the forebrain.
During the last 10 years our understanding of the organization of the developing forebrain has dramatically changed, in part as a consequence of the impressive number of morphological, chemoarchitectonic, embryological, and, primarily, genoarchitectonic data. In most of the recent studies, the columnar conception of the brain organization (Herrick, 1910) has been frequently challenged by the interpretation of the data in a current prosomeric model (Puelles and Rubenstein, 1993, 2003), which is emerging as the most useful tool in the evolutionary genoarchitectonic analysis. In this scheme, a prosomere, as all neural segments, is composed of four longitudinal zones: roof, alar plate, basal plate, and floor. Their boundaries are defined by molecular patterns and cellular fates commonly established by dorso-ventral patterning of the forebrain wall (Puelles and Rubenstein, 1993, 2003) that show an impressive grade of conservation across vertebrates in morphological and gene expression terms (Bardet et al. 2008, 2010; Puelles et al., 2000; Bachy et al., 2001, 2002; Brox et al., 2003, 2004; Moreno et al., 2003, 2004, 2005, 2008a,b, 2010; Osorio et al., 2005, 2006; Flames et al., 2007; García-Lopez et al., 2008; Abellán and Medina, 2009; Ferran et al., 2009; Domínguez et al., 2010; Morona et al., 2011).
In this scenario, the expressions of key patterning genes involved both in regional and cellular specification processes are currently being analyzed in detail in the main vertebrate models, in order to establish precise traits of the forebrain evolution. Among these developmental regulators is Nkx2.2, a member of the vertebrate homeodomain transcription factor gene family homologous to the Drosophila NK2/ventral nervous system defective (vnd) gene (Kim and Nirenberg, 1989; Price et al., 1992; Jiménez et al., 1995). It was originally identified as a gene that is expressed in ventral regions of the developing vertebrate central nervous system (Price et al., 1992), and is closely related to Sonic hedgehog (Shh), a powerful morphogen that controls progenitor proliferation, regional patterning, and cell fate in the developing brain (for review see Fuccillo et al., 2006). It is involved in a wide range of regionalization mechanisms including the early specification of progenitor cell identity and cell fate processes in the ventral neural tube, in response to graded Shh signaling (Briscoe and Ericson, 1999; Briscoe et al., 2000). It is also implicated in the establishment of the alar-basal boundary (Puelles and Rubenstein, 1993; Vieira et al., 2005) and in the specification of the diencephalic patterning, helping to define distinct progenitor domains (Ericson et al., 1997; Vue et al., 2007; Kataoka and Shimogori, 2008; Ferran et al., 2009).
Most of the data about Nkx2.2 expression in the forebrain were obtained in amniotes, primarily mouse, and chick, and only fragmentary data on Nkx2.2 expression have been reported in anamniotes, especially fishes and amphioxus (Holland et al., 1998; Schäfer et al., 2005). Surprisingly, the expression of Nkx2.2 has not been analyzed in the forebrain of amphibians, which constitute the only anamniote group of tetrapods. Of interest, in numerous recent studies it is shown that the forebrain organization in amphibians shares many key features with amniotes, mainly in terms of genetic specification, as revealed when the prosomeric paradigm is used in the interpretation of many territorial gene expression patterns (Bachy et al., 2001, 2002; Brox et al., 2003, 2004; Moreno et al., 2004, 2008a,b; van den Akker et al., 2008; Domínguez et al., 2010; Morona et al., 2011). In particular, we have recently reported the distribution of xShh in the forebrain of the anuran amphibian Xenopus laevis during development (Domínguez et al., 2010) and, following this line of research, herein we have analyzed the pattern of distribution of Nkx2.2, a functionally and anatomically related transcription factor in vertebrates. The comparative analysis, following the prosomeric model, serves to assess evolutionary trends. We have characterized phenotypically the developing Nkx2.2 expressing forebrain subdivisions and neurons by means of the combination of Nkx2.2 expression with forebrain essential regulators and markers, such as Nkx2.1, Tbr1, Pax6, GABA, Pax7, Orthopedia (Otp), xDll4, xShh, tyrosine hydroxylase (TH), mesotocin (MST), and somatostatin (SOM). The results of this study showed an extremely conserved distribution pattern of Nkx2.2 among vertebrates, crucial to delineate subdivisions that were otherwise indistinct, and supported the relevance of this transcription factor in the establishment and organization of the forebrain.
For the present study, adults, juveniles, and tadpoles of X. laevis were used. Embryos and larvae were classified according to Nieuwkoop and Faber (1967). Embryonic (42–45), premetamorphic (46–52), prometamorphic (53–58), and metamorphic (59–65) stages were used, minimizing as much as possible the number of animals used. All animals were treated according to the regulations and laws of the European Union (86/609/EEC) and Spain (Royal Decrees 1201/2005) for care and handling of animals in research, after approval from the University to conduct the experiments described.
Adult Xenopus were purchased from commercial suppliers (Xenopus Express; France), and the different developing specimens were obtained by in vitro fertilization and maintained in tap water at 20°C throughout their development. At appropriate times, embryos and larvae were deeply anesthetized in a 0.4-mg/ml solution of tricaine methanesulfonate (MS222, Sigma-Aldrich, Steinheim, Germany). The adults, juveniles, and late larvae were perfused transcardially with 0.9% sodium chloride, followed by cold 4% paraformaldehyde in a 0.1-M phosphate buffer (PB, pH 7.4). The brains were removed and kept in the same fixative for 2–3 h. Subsequently, they were immersed in a solution of 30% sucrose in PB for 4–6 h at 4°C until they sank, embedded in a solution of 20% gelatin with 30% sucrose in PB, and stored for 6 h in a 3.7% formaldehyde solution at 4°C. The brains were cut on a freezing microtome at 40 μm (adults) or 20–30 μm (juveniles and late larvae) in the transverse or sagittal plane and sections were collected and rinsed in cold PB. The embryos and premetamorphic larvae were fixed by immersion overnight at 4°C in MEMFA [0.1 M MOPS (4-morpholinopropanesulfonic acid) 2 mM ethyleneglycoltetraacetic acid, 1 mM MgSO4, 3.7% formaldehyde], then they were processed in toto and finally sectioned at 14–16 μm thickness in the transverse, horizontal, or sagittal plane on a freezing microtome.
A immunohistofluorescence procedure was conducted with the primary antibody on the free-floating sections that, in all cases, was diluted in 5–10% normal serum of the species in which the secondary antibody was raised in PB with 0.1% Triton X-100 (Sigma) and 2% bovine serum albumin (BSA, Sigma).The protocol included two steps, as follows: (1) Incubation for 72 h at 4°C in the dilution of the primary antibody mouse anti-Nkx2.2 (1:500; Developmental Studies Hybridoma Bank, DSHB, Iowa City, USA. Clone: 74.5A5) and (2) the second incubation was conducted for 90 min at room temperature with the labeled secondary antibody Alexa 488- conjugated goat anti-mouse (1:500; Molecular Probes; catalog reference: A21042). After being rinsed, the sections were mounted on glass slides and coverslipped with Vectashield mounting medium (Vector Laboratories, Burlingame, CA, USA; catalog number: H1000).
The cocktails of primary antibodies were diluted in PB with 0.1% Triton X-100 and used for 60 h at 4°C. They always included mouse anti-Nkx2.2 (1:500; DSHB) in combination with: rabbit anti-Otp (1:1000; produced by “PickCell laboratories” Amsterdam, The Netherlands; according to the protocol described in Lin et al., 1999), rabbit anti-MST (diluted 1:2000; donated by Dr. J. M. Guerné Université de Strasbourg, France), rabbit anti-SOM (1:1000; Incstar, Wisconsin, USA, Code number: 20067), rabbit anti-Nkx2.1 (1:500; Biopat Immunotechnologies, Italy, Code number: PA0100), rabbit anti-Tbr1 (1:250; Santa Cruz Biotechnology, Inc., USA, Code number: sc-48816), rabbit anti-TH (diluted 1:1000; Chemicon International, Inc., USA, Code number: P22941), rabbit anti-Pax6 (1:200; Covance, California, USA, Code number: PBR-278P), and rabbit anti-GABA (1:3000; Sigma-Aldrich, Steinheim, Germany, Code number: A2052). The secondary antibodies were used in appropriated combinations and were: Alexa 488-conjugated goat anti-mouse (1:500, Molecular Probes) and Alexa 594-conjugated goat anti-rabbit (1:500, Molecular Probes; catalog number: A11012). In all cases, secondary antibodies were diluted in PB with 0.1% Triton X-100 for 90 min at room temperature. After rinsing, the sections were mounted on glass slides and coverslipped with Vectashield.
For double histofluorescence labeling experiments, we combined the immunohistochemistry for Nkx2.2 with in situ hybridization for the following markers: xShh (provided by Dr. Randal Moon. University of Washington; Ekker et al., 1995) and xDll4 (provided by Dr. Nancy Papalopulu. University of Manchester; Papalopulu and Kintner, 1993).
For in situ hybridization, which was performed first, antisense digoxigenin (DIG)-labeled riboprobes for these markers were synthesized according to the protocol described in Bachy et al. (2001), linearizing the clones in Bluescript KS with Bam H1 (Promega, Madison, USA) and transcribing with T3 (Promega) for xShh, with Not1 (Promega, Madison, USA) and T3 (Promega) for xDll4. The embryos and premetamorphic larvae were processed in toto after progressive re-hydration and pretreatments (see Bachy et al., 2001), and the late larvae were processed in floating sections (see Moreno et al., 2004).
Hybridization step was done with 3 μl/ml of a DIG-labeled RNA probe, in a 50% formamide containing medium overnight at 55°C. The solution used for prehybridization (at 60°C for 1 h) and hybridization contained 50% deionized formamide (Fluka, Steinheim, Germany), 5× standard saline citrate (Sigma-Aldrich, Steinheim, Germany), 2% blocking reagent (Roche Diagnostics, Mannheim, Germany), 0.1% Tween-20, 0.5% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS; Sigma-Aldrich), 1 mg/ml of yeast tRNA (Sigma-Aldrich), 5 mM of ethylenediaminetetraacetic acid (Sigma-Aldrich), and 50 g/ml of heparin (Sigma-Aldrich) in water.
Hybridization was detected using an alkaline phosphatase coupled anti-DIG antibody (Roche Diagnostics, dilution 1:1500). Alkaline phosphatase staining was developed with Fast red tablets (Roche Diagnostics). The in situ hybridization was followed by the immunohistochemistry for mouse anti-Nkx2.2 (1:500; DSHB) revealed with Alexa 488-conjugated goat anti-mouse (diluted 1:500, Molecular Probes). Subsequently, embryos and early larvae were immersed in a solution of 30% sucrose in PB until they sank, embedded in a solution of 20% gelatin and 30% sucrose in PB, and stored overnight at 4°C in a solution of 4% formaldehyde and 30% sucrose in PB. Sections were cut at 14–25 μm thickness in the frontal, sagittal, and horizontal plane on a freezing microtome.
The sections were analyzed with an Olympus BX51 microscope that was equipped for fluorescence with appropriate filter combinations. Selected sections were photographed by using a digital camera (Olympus DP72). Contrast and brightness of the photomicrographs were adjusted in Adobe PhotoShop CS3 (Adobe Systems, San Jose, CA) and figures were mounted in Canvas 11 (ACD Systems, Canada).
The distribution of the Nkx2.2 protein has been analyzed in the prosencephalon of X. laevis from embryonic stages through the adult. In the following sections we analyze, using both immunohistochemistry and in situ hybridization procedures, the spatio-temporal sequence of Nkx2.2 expression during forebrain development in single (Figure 1) and double (Figures 2–5) stained material. All of the markers used in combination with Nkx2.2 have been previously used in the analysis of the Xenopus forebrain development, and the nomenclature used in the present study largely follows that used in our preceding mapping studies of the anuran forebrain (Moreno et al., 2004, 2008a,b; Morona and González, 2008; Domínguez et al., 2010; Morona et al., 2011). A schematic representation of the Nkx2.2 distribution in the case of the early (premetamorphic) forebrain is shown in Figure 6.
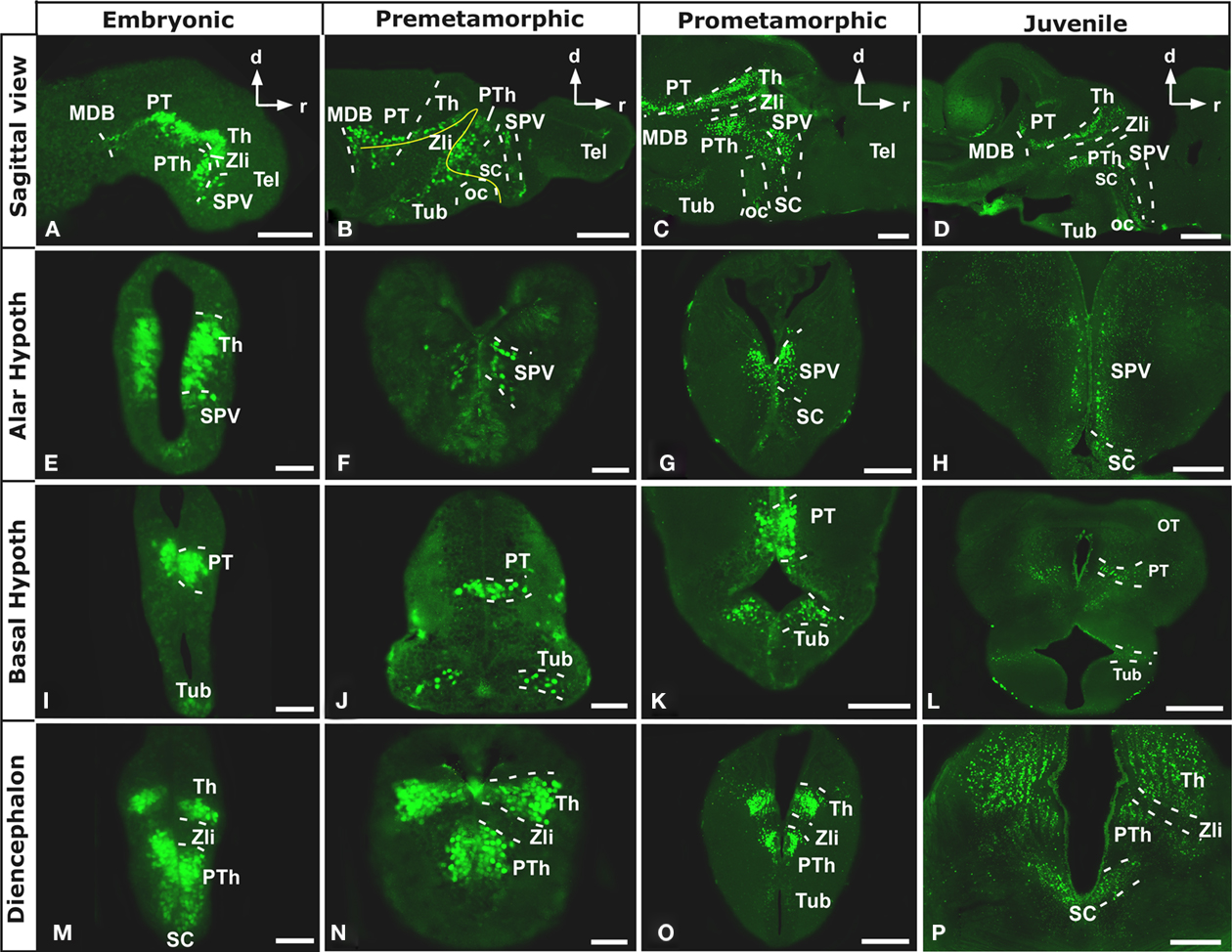
Figure 1. Photomicrographs of sagittal (A–D) and transverse (E–P) sections through the Xenopus forebrain subdivisions along the different representative developmental stages. The sagittal sections show the almost continuous Nkx2.2 expression from anterior hypothalamic areas to the most caudal regions of the forebrain (A–D). Nkx2.2 is not expressed in telencephalic areas, from embryonic stages through the adult. Nkx2.2 expression starts in the SPV territory of the alar hypothalamus (E–H). In the basal hypothalamus Nkx2.2 expression is restricted to the tuberal hypothalamus (I–L). In the diencephalon, Nkx2.2 is observed in the three prosomeres (P1–P3) and the Zli lacks Nkx2.2 expressing cells (I–P). Abbreviations: Arc, nucleus arcuatus; BH, basal hypothalamus; MB, mammillary band; MDB, midbrain-diencephalic boundary; OB, olfactory bulb; oc, optic chiasm; OT, optic tectum; P1-P3, diencephalic prosomeres 1–3; Pa, pallium; POC, preoptic commissural area/commissural septo-preoptic area; PO, preoptic area; PT, pretectum; PTh, prethalamus; PThE, prethalamic eminence; PV, paraventricular nucleus; SC, suprachiasmatic nucleus; SPa, subpallium; SPV, supraopto-paraventricular area; Tel, telencephalon; Th, thalamus; TP, posterior tubercle; Tub, tuberal area; ZI, zona incerta; Zli, zona limitans intrathalamica. Scale bars: 500 μm (D,L), 200 μm (C,G,H,K,O,P), 100 μm (A,B), and 50 μm (E,F,I,J,M,N).
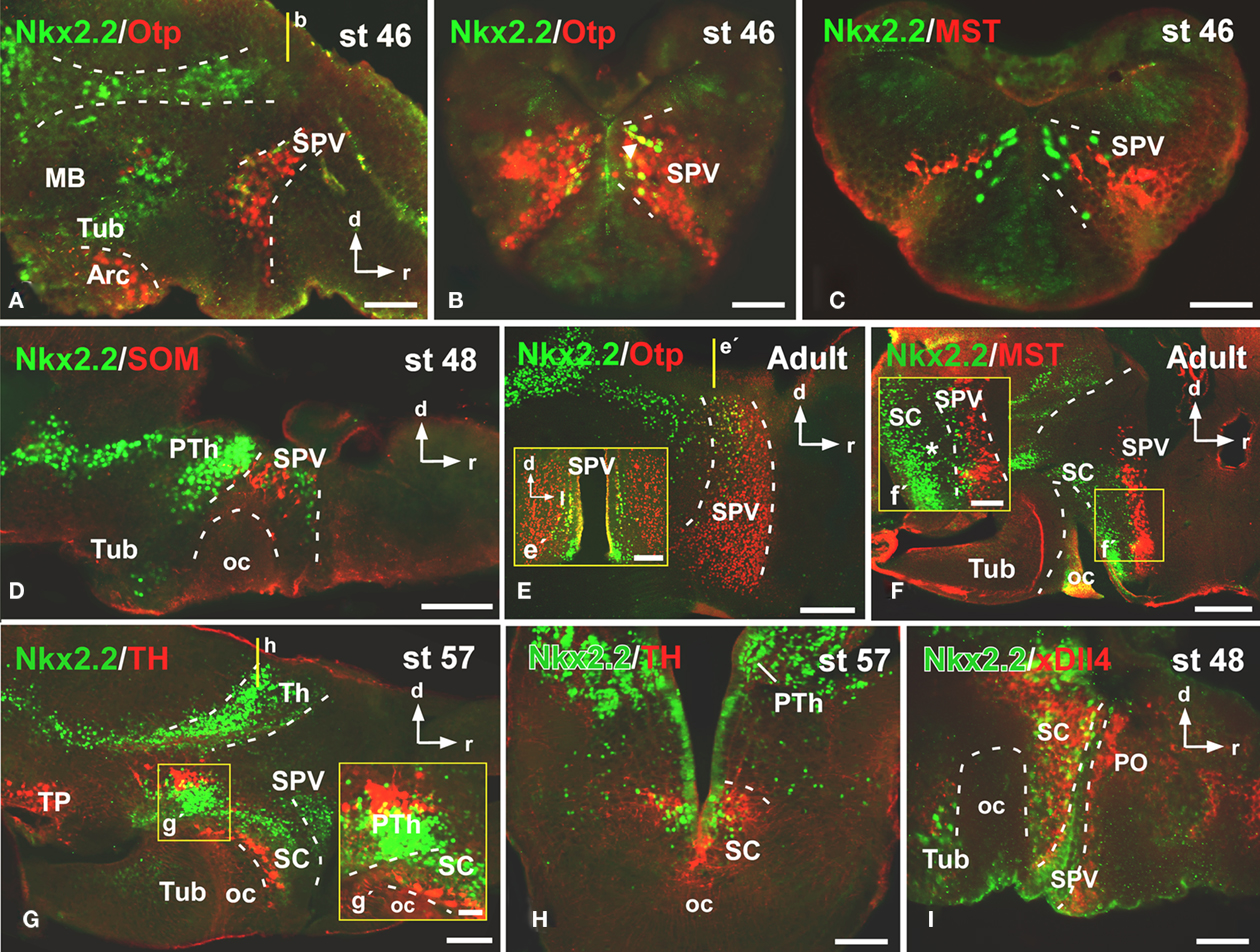
Figure 2. Photomicrographs of sagittal (A,D–G,I) and transverse (B,C,E′,H) sections through the alar hypothalamus illustrating the Nkx2.2 expression in combination with several hypothalamic markers. The most anterior Nkx2.2 expressing cells are observed in the SPV (the Otp expressing territory) during development (A,B,E,E′), in the region where it colocalizes with mesotocin (C,F,F′) and somatostatin (D), but only a subpopulation of the SPV cells coexpress Nkx2.2 and Otp [see arrow in (B,E,E′)]. At suprachiasmatic levels, the Nkx2.2 positive cells are located in the catecholaminergic (G,G′,H) and xDll4 (I) expressing region, constituting an independent territory within this domain (G–I). The yellow lines in (A,E,G) indicate the level of the sections of (B,E′,H), respectively. Yellow boxes in (F,G) indicate the higher magnifications shown in (F′,G′), respectively. Abbreviations as in Figure 1. Scale bars: 500 μm (F), 200 μm (E,E′,G), 100 μm (D,H) and 50 μm (A,B,C,F′,G′,I).
In Xenopus, Nkx2.2 immunoreactive (Nkx2.2-ir) cells were not observed in telencephalic areas, neither evaginated nor non-evaginated territories, from early developmental stages through the juvenile, when the brain morphology is close to that observed in adults (Figure 1).
The most anterior expression detected was observed in the hypothalamic territory, helping to the identification of the alar and basal domains (Figures 1A–D). Form early (Figures 1A,B) to late (Figures 1C,D) developmental stages, Nkx2.2-ir cells were observed in the supraopto-paraventricular (SPV) region of the alar hypothalamus (Figures 1E–H). At embryonic developmental stages (Figure 1E), virtually all the cells observed in the ventricular (vz) and subventricular layers (svz) of this zone were Nkx2.2-ir, whereas from early larval (Figures 1F,G) through juvenile stages the cells decreased notably occupying a band in the svz (Figure 1H). In addition, in the alar hypothalamus scattered Nkx2.2 cells were observed in the suprachiasmatic (SC) territory that became more numerous from late developmental stages and through the adulthood (Figures 1A–D,G,H,P). These cells formed a continuous band above the optic chiasm (oc) that was especially evident in sagittal sections (Figures 1C,D). In the basal hypothalamus (BH), scarce Nkx2.2-ir cells were found in the svz of the medial part of the tuberal region, whereas the most posterior portion of this basal region was devoid of Nkx2.2 positive cells (Figures 1I–L). Caudally, Nkx2.2 expressing cells were detected in the diencephalon, defining the midbrain-diencephalic boundary (MDB; Figures 1A–D) with the exception of the zona limitans intrathalamica (Zli), which was devoid of Nkx2.2 expression (Figures 1A–D,M–P). In prosomeres 2 (P2) and 3 (P3) Nkx2.2 positive cells were restricted to the alar portion (Figures 1M–P). At early stages of development, these cells were observed in the vz and svz of P2 and P3, respectively (Figures 1M,N), whereas from prometamorphic stages Nkx2.2-ir cells were only detected in the svz and mz (Figures 1O,P). In P1, the Nkx2.2-ir cells were located in the alar/basal boundary, representing the ventral limit of the pretectum (Figures 1I–L).
In order to further characterize the localization of Nkx2.2-ir cells within the forebrain, we carried out double labeling experiments throughout development, using different prosencephalic markers (Figures 2–5).
To analyze the distribution of the Nkx2.2-ir cells in the alar hypothalamus we combined Nkx2.2 with the transcription factor Orthopedia (Otp; Figures 2A,B,E), the neuropeptides mesotocin (MST; Figures 2C,F) and somatostatin (SOM; Figure 2D), the dopaminergic marker tyrosine hydroxylase (TH; Figures 2G,H), and the transcription factor xDll4 (Figure 2I). From early premetamorphic stages, the double staining for Nkx2.2/Otp (Figure 2A) defined the position of a restricted population of Nkx2.2 expressing cells in the svz of the SPV. Whereas almost all the SPV cells expressed Otp (Figure 2B), only a subpopulation of the SPV cells coexpressed Nkx2.2 and Otp (see arrowhead in Figure 2B), just in the most posterior portion of the SPV, defining the limit between the SPV and the SC (Figures 2E,E′). This was also confirmed by the double labeling Nkx2.2/MST (Figure 2C) and Nkx2.2/SOM (Figure 2D).
At SC levels, the double staining for Nkx2.2/MST (Figure 2F), Nkx2.2/TH (Figures 2G,H), and Nkx2.2/xDll4 (Figure 2I) confirmed the position of Nkx2.2-ir cells in this territory, anterior to the oc and posterior to the SPV (Figure 2F′). In this area, the Nkx2.2-ir cells were found in the catecholaminergic (Figures 2G,H) and xDll4-expressing region (Figure 2I). During the development the Nkx2.2 expressing cells were numerous and from the early larvae stages these cells showed a clear patterning, however it is from late developmental stages when these cells clearly formed an independent and exclusive subpopulation in the SC, situated in the ventral portion avoiding the anterior region (see asterisk in Figure 2F′).
From early premetamorphic stages, the double staining for Nkx2.2/Nkx2.1 (Figure 3A) confirmed the Nkx2.2 expression in the svz of the BH. The double staining Nkx2.2/Otp confirmed the position of Nkx2.2 positive cells in the tuberal area of the BH, but avoiding the nucleus arcuatus (Arc), and the mammillary band (MB), both rich in Otp expressing cells (Figures 3B,C). In addition, the lack of Nkx2.2 expression in the mammillary region was confirmed by the Nkx2.2/TH (Figure 3D) and Nkx2.2/xDll4 double staining (Figure 3E).
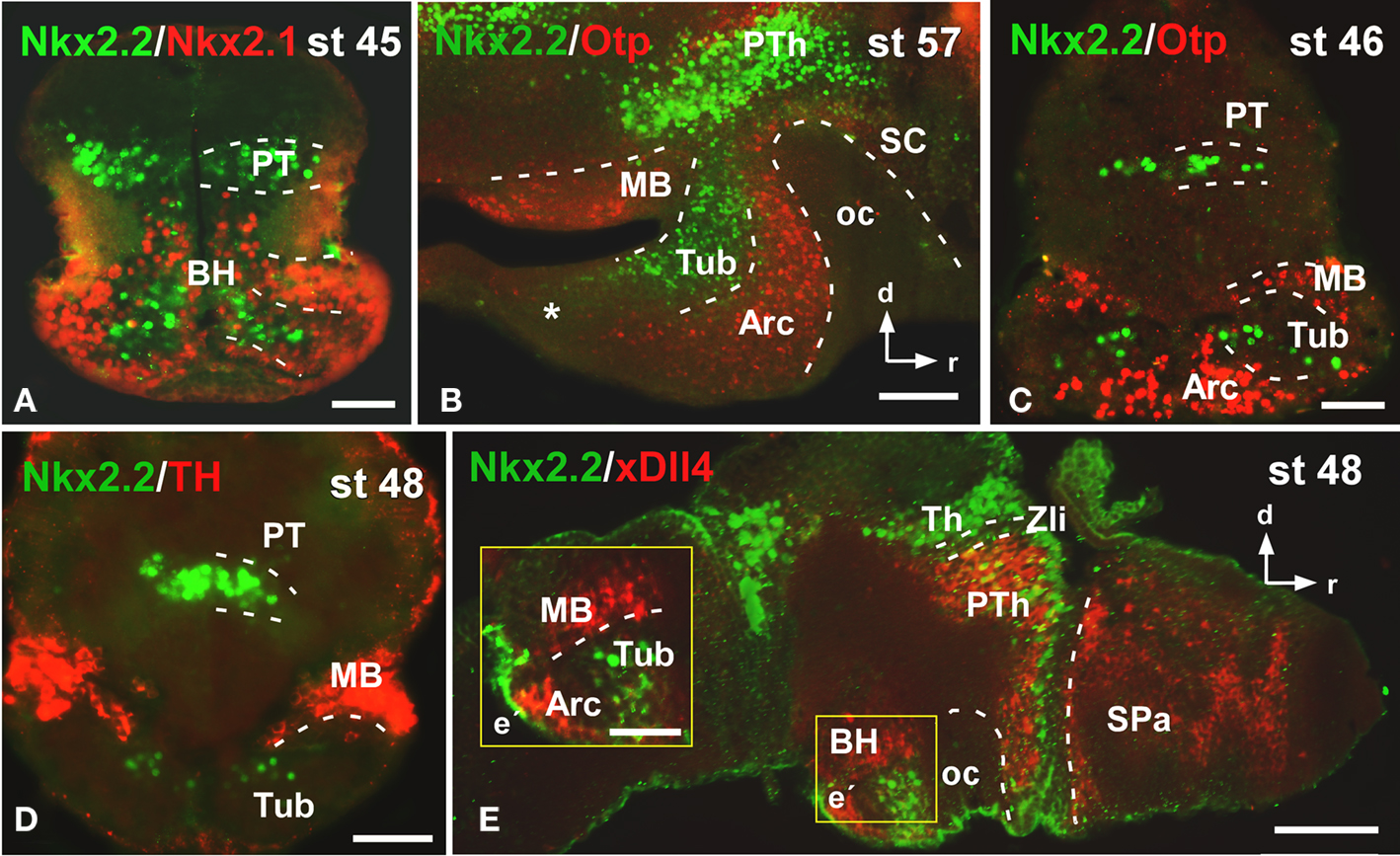
Figure 3. Photomicrographs of transverse (A,C,D) and sagittal (B,E) sections through the basal hypothalamus illustrating the Nkx2.2 expression in combination with several hypothalamic markers. The Nkx2.1 expression confirms that the limited Nkx2.2 expression found in the basal hypothalamic territory is situated in the tuberal region (A), restricted to the anterior part [asterisk in (B)] and avoiding the nucleus arcuatus, labeled for Otp (B,C), and the mammillary band, defined by TH (D) and xDll4 (E,E′). Yellow box in e indicates the area shown in higher magnification in e´. Abbreviations as in Figure 1. Scale bars: 100 μm (B,E), 50 μm (A,C,D,E′).
Caudally in the diencephalon, the banded staining pattern obtained for the pair Nkx2.2/Tbr1 confirmed the lack of Nkx2.2 expressing cells in the prethalamic eminence (PThE), rich in Tbr1 expression (Figure 4A), but allowed the identification of Nkx2.2-ir cells in the thalamus and prethalamus. The expression observed in the alar portion of P3 and P2 was confirmed by the double staining Nkx2.2/Pax6 (Figure 4B), Nkx2.2/xDll4 (Figure 4C), and Nkx2.2/GABA (Figure 4D). In addition, specifically the Nkx2.2-ir cells in P3 were noted in the zona incerta (ZI; Figures 4E,F), where Nkx2.1 expression is present (Moreno et al., 2008a) and a conserved catecholaminergic group is located (Smeets and González, 2000). The combination of Nkx2.2/xShh confirmed the lack of Nkx2.2 expressing cells in the zona limitans intrathalamica (Zli; Figure 4G) defined by the xShh-expression (Domínguez et al., 2010).
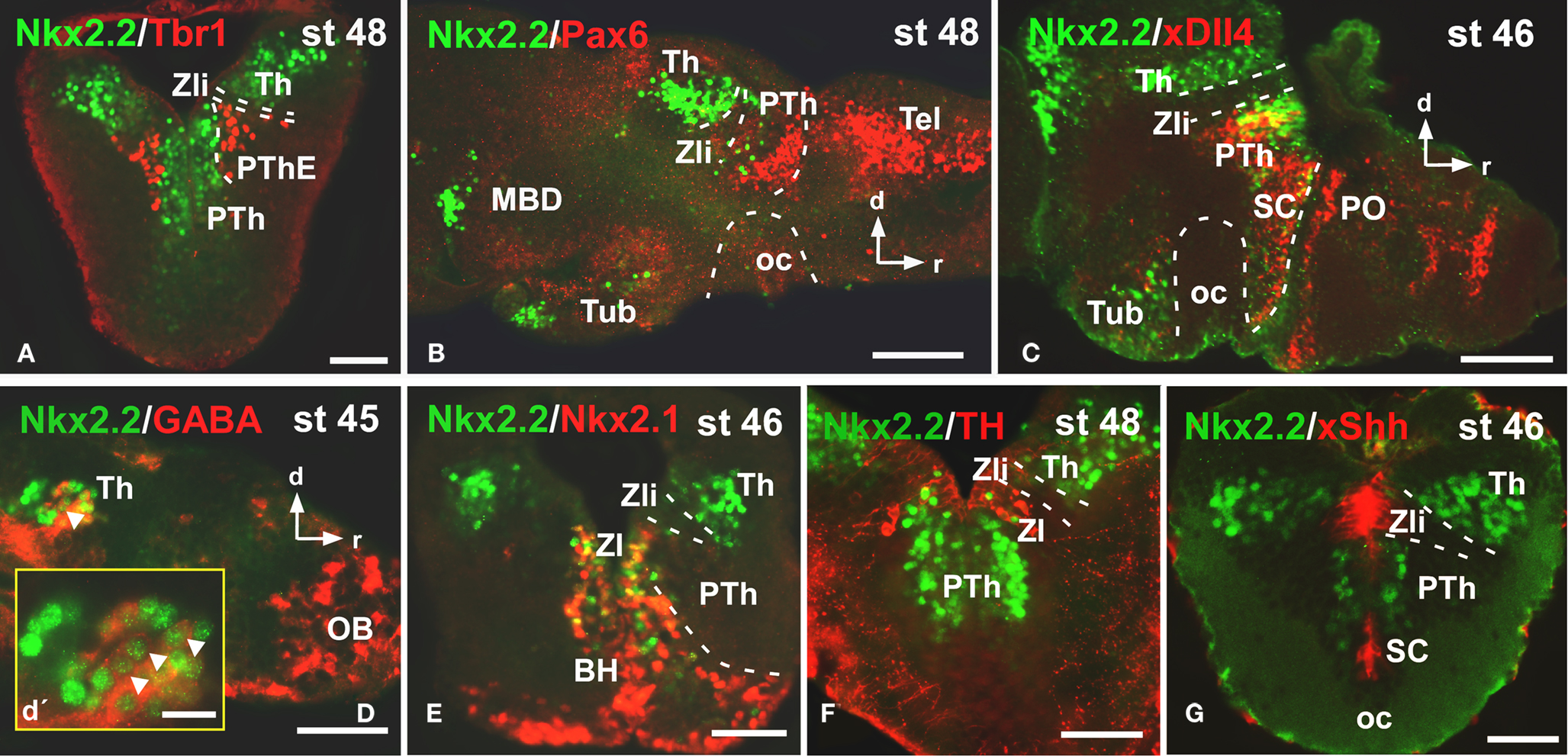
Figure 4. Photomicrographs of transverse (A,E,F,G) and sagittal (B,C,D) sections through the diencephalon illustrating the Nkx2.2 expression in combination with diverse diencephalic markers. The prethalamic eminence, rich in Tbr1 (A), was devoid of Nkx2.2 expression, in contrast to the prethalamus, defined by the Pax6 (B) and xDll4 expressions (C). The yellow box in d shows a higher magnification of the Nkx2.2/GABA double labeled cells found in the thalamus [see arrowheads in (D,D′)]. The double labeling Nkx2.2/Nkx2.1 (E) and Nkx2.2/TH (F) define the position of the Nkx2.2 cells in the zona incerta of the prethalamus. The xShh diencephalic expression defines the lack of Nkx2.2 expression in the Zli (zona limitans intrathalamica) (G). Abbreviations as in Figure 1. Scale bars: 100 μm (B,C,D), 50 μm (A,E,F,G), 25 μm (D′).
Finally, Nkx2.2 has been functionally related to the morphogen Shh implied in the alar-basal boundary establishment (for review see Fuccillo et al., 2006). We have recently reported the precise distribution of xShh in the Xenopus developing forebrain (Domínguez et al., 2010) and in the present study we have analyzed the extent of the Nkx2.2 expressing cells along the entire forebrain in combination with xShh, in transverse (Figures 5A,B) and horizontal sections (Figure 5C) to analyze their relation with the Xenopus forebrain axis. Of note, this double labeling shows that the pattern of distribution of Nkx2.2-ir cells formed a series of bands that extended along the anterior–posterior axis in parallel to adjacent stripes labeled for xShh (Figure 5).
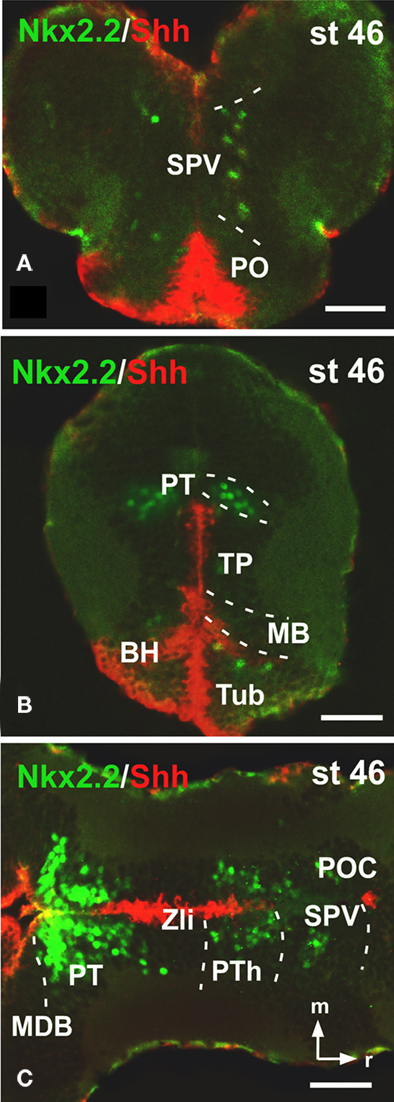
Figure 5. Photomicrographs of transverse (A,B) and horizontal (C) sections through the forebrain illustrating Nkx2. 2 expression in combination with xShh. The expressions of Nkx2.2 and xShh extend along the anterior–posterior axis in parallel adjacent stripes, forming longitudinal columns along the forebrain. Abbreviations as in Figure 1. Scale bars: 50 μm.
All the results obtained from the double labeling analysis confirmed the localization of the Nkx2.2-ir cells in the regions described above and summarized in Figure 6.
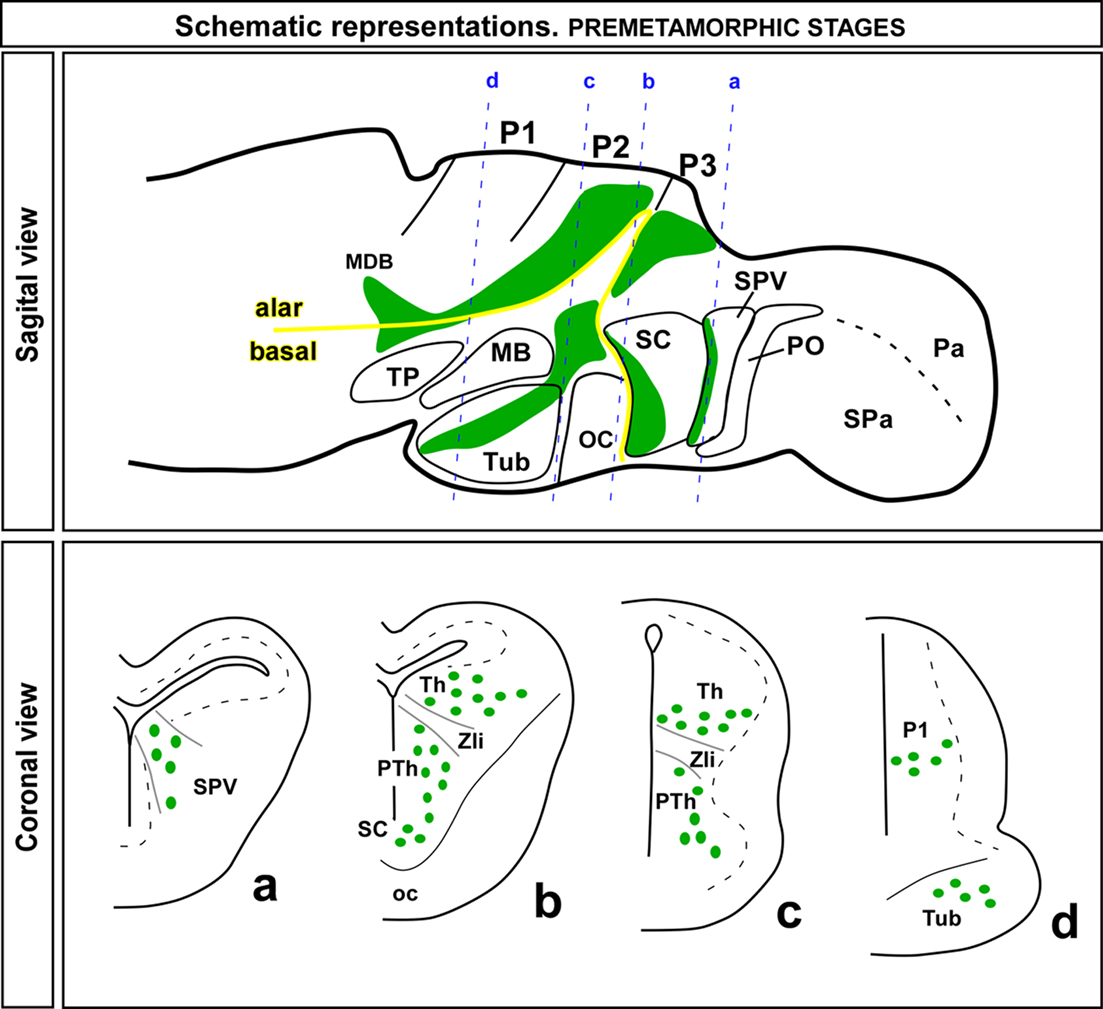
Figure 6. Schematic drawings of sagittal and coronal sections through a premetamorphic brain of Xenopus laevis illustrating the distribution of Nkx2.2 expressing zones (green regions in the sagittal view) and cells (green dots in the coronal view) along the forebrain. The appropriated levels of the coronal sections are indicated in the sagittal view. Abbreviations as in Figure 1.
Nkx2.2 was the first gene of the NK2 homeobox class to be demonstrated in all the deuterostomes, showing homology in all the models analyzed. Even possible homologies were suggested in the nervous system between invertebrates and vertebrates given the high conservations in the expression and function of this gene (Holland et al., 1998). The presence of two paralogs, Nkx2.2a and Nkx2.2b, was demonstrated in zebrafish and phylogenetic and expression analysis suggests that these genes arose by a fish-specific gene duplication and acquired differential transcriptional control. Subsequently one paralog was lost and only the Nkx2.2a is considered ortholog of the mammal gene (Schäfer et al., 2005) and, in addition, its expression domain is very conserved. However, both paralogs are regulated by Shh (Barth and Wilson, 1995; Schäfer et al., 2005).
In general terms, in the vertebrate central nervous system Nkx2.2 has been implicated in the ventral neuronal patterning, at early developmental stages (Briscoe and Ericson, 1999), defining the alar/basal boundary along to Shh-expression (Puelles and Rubenstein, 1993; Vieira et al., 2005). This is found even in amphioxus, in which there is not obvious anatomical boundary separating alar and basal regions along the dorso-ventral axis of the cerebral vesicle (Holland et al., 1998).
In all vertebrates analyzed Nkx2.2 is expressed in the prosencephalon where it is known to be involved in the specification of progenitor domains and in establishing regionalization patterns. Moreover, it contributes to the differentiation of distinct areas and their compartmentalization, as well as to the acquisition of cellular identity and the regulation of the distribution of the earliest neurons in the brain (Wilson et al., 1993; Barth and Wilson, 1994; Vieira and Martínez, 2006; Vue et al., 2007). Thus, the analysis in anamniote and amniote vertebrates of the spatio-temporal distribution of this transcription factor is considered of relevance in the understanding of the establishment of the different subdivisions within the forebrain and in evaluating the degree of conservation across vertebrates (Price et al., 1992; Barth and Wilson, 1995; Holland et al., 1998; Schäfer et al., 2005; Vieira and Martínez, 2006; Vue et al., 2007; Ferran et al., 2009). In the present study we have provided for the first time in amphibians a detailed analysis of its distribution in X. laevis, a tetrapod anamniote with a forebrain organization that in genoarchitectonic terms shares many features with its counterpart in amniotes (Bachy et al., 2001, 2002; Brox et al., 2003, 2004; Moreno et al., 2004, 2008a,b; van den Akker et al., 2008; Domínguez et al., 2010; Morona et al., 2011). To fully understand the precise topological distribution of Nkx2.2 expression, its combination with the respective expression of different forebrain markers has been shown to be extremely useful. Thus, we have analyzed the distribution of Nkx2.2 in combination with the localization of Nkx2.1, Otp, Pax6, GABA, Tbr1, TH, MST, and SOM, and in situ hybridization for the detection of xShh and xDll4.
The expression of Nkx2.1 has been deeply analyzed in the prosencephalon of many vertebrates and it has served to identify the preoptic (PO), SC, and tuberal territories (Tub) during development (Rohr et al., 2001; González et al., 2002a,b; Moreno et al., 2008a; van den Akker et al., 2008), like the developmental regulatory gene xDll4 (Papalopulu and Kintner, 1993; Puelles and Rubenstein, 1993; Shimamura and Rubenstein, 1997; Puelles et al., 2000; Bachy et al., 2002; Marín et al., 2002; Brox et al., 2003). In addition, the localization of TH highlighted the regionalization of the SC and the ZI in P3 (González et al., 1993, 1994; Milán and Puelles, 2000; Smeets and González, 2000). Specifically in the hypothalamus, Otp expression served in the identification of the SPV region and it also defines the Arc and the MB within the BH (Bardet et al., 2008; Domínguez et al., 2010). The presence of MST (or its homolog oxytocin) and SOM in the magnocellular neurons of the SPV is also a shared feature between amniotes and anamniotes (Bläsher and Heinrichs, 1982; González et al., 1995, 2003; Petkó and Orosz, 1996; López et al., 2007). In the diencephalon, the morphogen xShh was crucial to delimit the Zli, as well as to define the alar-basal boundary and the longitudinal columns that establish the dorso-ventral patterning (Puelles and Rubenstein, 2003; Domínguez et al., 2010). The homeobox gene Tbr1 was extremely useful in the recognition of the PThE (Puelles et al., 2000; Brox et al., 2004) and the relative expression of xDll4, Pax6, and GABA served to identify the boundaries of the three diencephalic prosomeres (Bachy et al., 2002; Brox et al., 2003; Moreno et al., 2008b; Morona et al., 2011).
In mammals, the most anterior Nkx2.2 expression was observed in the medial ganglionic eminence (MGE), also called pallidal domain, which consists of, at least, five progenitor domains globally defined by the strong expression of Nkx2.1, weak expression of Nkx2.2, and lack of Pax6 and Shh-expressions (Flames et al., 2007). In the case of the chicken, three subdivisions were recently identified in the pallidal domain, comparable to those described for mammals (Abellán and Medina, 2009). In chicken, the Nkx2.2 expression was not described in detail but it seems to be weakly expressed in the subpallium (see Figures 4F,G in Vieira et al., 2005 and see Figures 1K,Q in Bardet et al., 2010) in contrast to the strong expression found in mammals (Flames et al., 2007). Differently, in Xenopus (present results) and in the turtle (Moreno et al., 2010) Nkx2.2 expression has not been observed in the svz of the MGE. This interesting result might be in correlation with the presence of Shh-expression in the subventricular zone of the mouse MGE (García-Lopez et al., 2008) and chicken (Abellán and Medina, 2009) and its absence in the pallidal region of amphibians (Domínguez et al., 2010). However, Shh and Nkx2.2 are not always expressed in the same regions.
Also in the telencephalon, the PO region in mammals contains at least two distinct progenitor domains (Flames et al., 2007). The ventricular zone of the PO is uniquely defined by the simultaneous expression of Nkx2.1, Nkx2.2, and Shh, and most prominently by the lack of detectable levels of Gsh2, Lhx6, Lhx7, or Olig2 expression (Flames et al., 2007). Like in the case of the MGE, in Xenopus (present results) but also in turtle (Moreno et al., 2010), there is not Nkx2.2 expression in the POA. However, in this case it is not correlated with the lack of the Shh-expression, because Shh-expression is observed in the preoptocommissural area (Domínguez et al., 2010).
The boundary between the telencephalon and the hypothalamus has been recently defined, first in chicken (Bardet et al., 2006) and later in mouse (Flames et al., 2007), as the preoptohypothalamic zone (POH). It constitutes the border of the subpallium and was described by its characteristic and specific expression of Nkx2.2 as a narrow territory separating longitudinally the PO from the magnocellular hypothalamus (Bardet et al., 2006). This region contains progenitor cells that express Dlx2, Dlx5, Pax6, Olig2, and Gsh2, lacks expression of Nkx2.1, Shh, Nkx6.2, and Dbx1 and can be further subdivided into two areas because the POH1, but not the POH2, expresses high levels of Nkx2.2 in mammals (Flames et al., 2007; Bardet et al., 2010). In Xenopus, Nkx2.2 expression is not detected in a comparable region, given that the most anterior expression found coincides with the Otp SPV expressing zone within the hypothalamus (see below).
The hypothalamus constitutes a very complex structure, as regards its functionality, cell complexity, hodology, development, and morphology. Therefore, it is not surprising that the elucidation of its genoarchitecture is particularly important to understand how is generated and maintained the diversity in the hypothalamus. That is one of the reasons that makes fascinating the hypothalamus and, thereby, the focus of analysis in many comparative studies. This has as its counterparts the numerous studies in which the nomenclature and interpretation of the nuclei and boundaries are constantly under debate (Szabó et al., 2009b; Bardet et al., 2010; Domínguez et al., 2010; Shimogori et al., 2010).
On the basis of combined expression analysis, at least two different longitudinal alar domains have been proposed in the chick hypothalamus: the Dlx- and Shh-negative SPV area, which lies under the border of the FoxG1-positive telencephalic field, and the subparaventricular area, which lies under it and is adjacent to the Shh-positive basal plate, and expresses Dlx5 and Nkx2.2 (Bardet et al., 2010). In Xenopus, the most anterior Nkx2.2 positive cells were localized in a Shh−/Nkx2.1−/xDll4−/Otp+ territory forming a thin strip of cells that delimit the region just anterior to the SC (present results) and their localization within this distinct region is highlighted because some of these cells contain both Otp and Nkx2.2.
The SC region is complex in terms of nuclei organization that most likely reflects a complex genetic specification. In general, the SC is defined as a Dlx-expressing zone in all vertebrates analyzed (present results)Bachy et al., 2002; Brox et al., 2003; Puelles and Rubenstein, 2003; Flames et al., 2007; . In mammals, this region does not express Nkx2.1 and Shh, in contrast to the situation in other amniotes(Medina, 2008) and anamniotes (Medina, 2008; Moreno et al., 2008a; Domínguez et al., 2010). In particular, although these two markers were not specifically described in the SC region of the chick, their presence can be inferred from the published mapping studies (compare Figures 6I,J from Puelles et al., 2000 and Figures 1D,P,F,J from Bardet et al., 2010) suggesting possible evolutionary differences between birds and mammals in this area and/or the existence of different progenitor domains.
The SC of X. laevis has been carefully studied in the last years, mapping the distribution of numerous markers that identified different regions within this territory. In general terms, mainly on the basis of calcium binding proteins expression, rostral, and caudal SC zones were defined (Milán and Puelles, 2000; Morona and González, 2008). In addition, based on the expression of neuropeptide Y and TH (Kramer et al., 2001) three different nuclei were identified, the ventrolateral, dorsomedial, and caudal nuclei. All of them differ from each other in location, shape, number of cells, and function (Kramer et al., 2001) and, therefore, it is logic to think that also in genetic specification. In terms of genoarchitecture, the expression of several genes of the LIM-homeodomain family appears to define distinct territories within this SC area (Moreno et al., 2004, 2008a). Among others, Islet1 is expressed in a cell population that forms a curved band surrounding externally the Nkx2.1/Shh vz expression, i.e., the only area where Nkx2.2, TH, and xGAD67/xDll4 populations seem to be intermingled (present results; Brox et al., 2003; Moreno et al., 2008a; Domínguez et al., 2010). Noteworthy, it is known that Nkx2.2 and Shh are involved in the control of the development of midbrain dopaminergic neurons (Prakash and Wurst, 2006) and, given the close spatial relationship between Shh, Nkx2.2, and TH expressing neurons in the SC of Xenopus, a possible implication of Shh/Nkx2.2 may exist for the acquisition of the dopaminergic phenotype in this region.
Finally in the BH, like in the rest of the forebrain regions, the elucidation of genetic combinatorial expression patterns has served to characterize and define regions, and their comparative analysis in different vertebrates has been used as a useful tool to establish their grade of conservation. Thus, it was defined that Nkx2.1 expression in the BH is required to maintain molecular characteristics of the developing hypothalamus (Kimura et al., 1996; Takuma et al., 1998; Sussel et al., 1999; Marín et al., 2002; van den Akker et al., 2008), but other members of this gene family also seem to have roles in hypothalamic formation because, for example, Nkx2.2 or Nkx6.1 mutant mice have ventral to dorsal transformations (Briscoe and Ericson, 1999; Sander et al., 2000). In addition, in all vertebrates studied the transcription factor Otp is expressed in the arcuate nucleus and the oblique perimammillary band of the BH (Simeone et al., 1994; Puelles and Rubenstein, 2003; Del Giacco et al., 2006; Bardet et al., 2008), where contributes to progenitor cell proliferation, survival, and migration (Goshu et al., 2004), and operates in the proper differentiation of several neurohormone-secreting nuclei (Acampora et al., 1999, 2000; Wang and Lufkin, 2000; Michaud, 2001; Blechman et al., 2007; Eaton and Glasgow, 2007; Ryu et al., 2007; Del Giacco et al., 2008; Eaton et al., 2008). Additionally, the activity of Otp is essential for the induction of the dopaminergic phenotype in the hypothalamus of vertebrates (Del Giacco et al., 2008). In Xenopus, the mammillary area has been defined by TH and dopamine or histamine immunohistochemistry (Airaksinen and Panula, 1990; González et al., 1993, 1994; Milán and Puelles, 2000) and further identified by the expression of Otp (Domínguez et al., 2010) and the lack of Nkx2.2 (present results). In contrast, the Nkx2.2 combinatorial expression patterns identified different zones within the Nkx2.1 expressing tuberal hypothalamus in Xenopus: an intermediate Nkx2.2+/Otp− zone and the Nkx2.2−/Otp+ zone that constitutes the arcuate nucleus (Bardet et al., 2008; Domínguez et al., 2010; present results). Of note, the lack of Nkx2.2 expressing cells in the most posterior region of the tuberal area, coincides with a total absence of xShh-expression in the same territory (Domínguez et al., 2010).
In the last 10 years an impressive number of morphological, chemo- and genoarchitectural data has helped to the interpretation in the diencephalon of the boundaries, extent of the areas and identification of distinct nuclei and subnuclei. These data have often been gathered not only for mammals but also in a number of non-mammalian species and the evolutionary perspective shows the impressive degree of conservation of this forebrain region, not only regarding the expression of different genes but also their functions. In all vertebrates analyzed, the alar diencephalon early develops into three major neuroepithelial domains along the anterior– posterior (A/P) axis, known as the prethalamus (P3), the thalamus (P2), and the pretectum (P1), and it is limited by two boundaries, the most anterior one that lies along the supraopto-mammillary region and the posterior one that lies between the pretectum and the mesencephalon (Puelles and Rubenstein, 2003; Moreno et al., 2004, 2008b; Vieira et al., 2005; Ferran et al., 2009; Morona et al., 2011). Also in all the vertebrates exists the Zli as a secondary morphogenetic organizer in diencephalic histogenesis that appears as a transverse ventricular ridge between the prethalamus and the thalamus from neural tube stages (Echevarría et al., 2003; Puelles and Rubenstein, 2003). The final position, boundaries, organizations, and functions are in the end defined by signaling molecules such as Shh or Fgf8 (Echevarría et al., 2003; Vieira et al., 2005, 2010; Kataoka and Shimogori, 2008), which act regulating the expression of developmental genes that will specify compartmentalization and cell fate in the diencephalon (Echevarría et al., 2003; Hashimoto-Torii et al., 2003; Kiecker and Lumsden, 2004).
In mammals, from early embryonic stages Nkx2.2 is expressed in a distinct pattern in all three diencephalic prosomeres (Price et al., 1992; Puelles and Rubenstein, 2003; Vue et al., 2007; Kataoka and Shimogori, 2008) and is induced by Shh, which diffuses from the ventral territory and from the Zli, and is directly or indirectly repressed by Pax6 (Pratt et al., 2000; Kiecker and Lumsden, 2004). However, the interaction Pax6–Nkx2.2 it is not simple and reciprocal given that the downregulation of Pax6 in the posterior thalamus is not followed by the upregulation of Nkx2.2 (Pratt et al., 2000). In many studies, the influence of Shh over Nkx2.2 has been demonstrated (Barth and Wilson, 1995; Kiecker and Lumsden, 2004; Vieira and Martínez, 2006), however there are also evidences about their independence in different models. This is the case of the mammalian Nkx2.2 expression in the posterior region of the Zli, which is under the control of Fgf8 and not Shh (Kataoka and Shimogori, 2008), but also in the cave-living form of the teleost fish Astyanax mexicanus, which shows a clear expansion of Shh-expression at the ventral midline but is not correlated with a larger Nkx2.2 expression domain; in contrast to other transcription factors expressed in the forebrain such us Nkx2.1 (Menuet et al., 2007).
In the diencephalon of mouse and chicken, the gene Nkx2.2 is expressed during development in a rostroventral band of the thalamus (Puelles and Rubenstein, 1993; Martínez-de-la-Torre et al., 2002). On the basis of the Nkx2.2 expression, a distinct progenitor domain has been characterized in the developing thalamus (also marked by Mash1; Vue et al., 2007; Kataoka and Shimogori, 2008), which is controlled by FGF signaling, with independence of the Shh activity (Kataoka and Shimogori, 2008). Later in the thalamic development, the expressions of Nkx2.2 and Gad67 were detected in different nuclei (Vue et al., 2007; Kataoka and Shimogori, 2008), specifically in the posterior ventral lateral geniculate nucleus (vLGN), concluding that at least a portion of the vLGN, classically a considered a prethalamic nuclei (Kitamura et al., 1997), belongs to the set of retinorecipient thalamic nuclei that do not project to the cortex (Paxinos, 1994) and arises from the thalamic domain (Vue et al., 2007; Kataoka and Shimogori, 2008).
In the chick, the combined expression of Gbx2, Nkx2.2, and Pax6 (Martínez-de-la-Torre et al., 2002), the cadherin expression (Redies, 2000), and fate maps analysis (García-Lopez et al., 2004) allowed the identification in the thalamus of four subdivisions, the anteroventral, dorsal, intermediate, and ventral regions. Interestingly, Nkx2.2 expression was only detected into the anteroventral region (adjacent to the Zli) and its derivatives, among which are the set retinorecipient thalamic nuclei (Martínez-de-la-Torre et al., 2002) that express Nkx2.2 and are proposed as thalamic derivates (Vue et al., 2007).
In Xenopus, we have found an extended Nkx2.2 positive region along the developing thalamus that might define a distinct progenitor domain whose cells would contribute to the formation of other diencephalic nuclei. The Nkx2.2 thalamic expression is observed in different nuclei expressing diverse transcription factors such as x-Lhx2/9 (Moreno et al., 2004) and scattered GAD67 (Brox et al., 2003). In addition, it is suggested that in Xenopus the Nkx2.2 gene is a good candidate involved in the acquisition of the GABAergic phenotype in the thalamus, the pretectum, and the basal plate of the caudal diencephalon, where there is not Dll expression (Brox et al., 2003; present results). In mammals and chick it has been proposed that the area adjacent to the Shh-expression of the Zli is induced by Shh to express Nkx2.2, and this area has been proposed to be the source of the subpopulation of GABAergic interneurons observed in the thalamus (Fode et al., 2000; Martínez-de-la-Torre et al., 2002; Puelles et al., 2004). In this context, we have observed Nkx2.2/GABA double labeled cells in the thalamus (present results), a region devoid of Dll4 expression (Brox et al., 2003; present results), suggesting that Nkx2.2 could be implicated in the acquisition of the GABAergic phenotype, like in amniotes.
Interestingly, also in Xenopus, Nkx2.2 expression is observed in the prethalamus, and its precise localization is corroborated by the colocalization of the Nkx2.2 expressing cells in the territory of P3 that is xDll4+/Nkx2.1+/Pax6+/TH+/xShh+ (present results; Bachy et al., 2002; Brox et al., 2003; Moreno et al., 2008a,b; Domínguez et al., 2010).
As regards the Zli, it expresses Shh in all vertebrates analyzed and is involved in the correct acquisition of the P2 and P3 gene expression and regionalization pattern (Braun et al., 2003; Hashimoto-Torii et al., 2003; Kiecker and Lumsden, 2004; Scholpp et al., 2006; Szabó et al., 2009a). In this context, Nkx2.2 expression in the thalamus and prethalamus is induced by Shh secreted by Zli cells (Kiecker and Lumsden, 2004; Vieira et al., 2005; Vieira and Martínez, 2006). Thus, the xShh-expression in the Zli found in Xenopus suggests that also in amphibians Shh is likely involved in the specification of the diencephalic territory (Domínguez et al., 2010), and that xShh-expression in the Zli could lead to establish the Nkx2.2 expression pattern in the P2/P3 territory of amphibians, like in amniotes.
Finally, the Nkx2.2 expression observed in the thalamus of the mouse and chicken continues into P1 (Puelles and Rubenstein, 1993; Martínez-de-la-Torre et al., 2002). Studies about the pretectal molecular regionalization in chick and Xenopus have described that the precise Nkx2.2 expression within P1 marks the alar/basal boundary, representing a tentative ventral limit of the pretectum (Ferran et al., 2009; Morona et al., 2011; present results).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This work supported by grants from Spanish MICINN and the UCM (Grant numbers: BFU2009-12315 and BSCH-UCM GR58/08). We are grateful to Dr. Ruth Morona for the fruitful discussions about the diencephalic regionalization and to Dr. Jesús M. López for the critical reading of the manuscript.
Abellán, A., and Medina, L. (2009). Subdivisions and derivatives of the chicken subpallium based on expression of LIM and other regulatory genes and markers of neuron subpopulations during development. J. Comp. Neurol. 515, 465–501.
Acampora, D., Postiglione, M. P., Avantaggiato, V., Di Bonito, M., and Simeone, A. (2000). The role of Otx and Otp genes in brain development. Int. J. Dev. Biol. 44, 669–677.
Acampora, D., Postiglione, M. P., Avantaggiato, V., Di Bonito, M., Vaccarino, F. M., Michaud, J., and Simeone, A. (1999). Progressive impairment of developing neuroendocrine cell lineages in the hypothalamus of mice lacking the orthopedia gene. Genes Dev. 13, 2787–2800.
Airaksinen, M. S., and Panula, P. (1990). Comparative neuroanatomy of the histaminergic system in the brain of the frog Xenopus laevis. J. Comp. Neurol. 292, 412–423.
Bachy, I., Berthon, J., and Rétaux, S. (2002). Defining pallial and subpallial divisions in the developing Xenopus forebrain. Mech. Dev. 117, 163–172.
Bachy, I., Vernier, P., and Rétaux, S. (2001). The LIM-homeodomain gene family in the developing Xenopus brain: conservation and divergences with the mouse related to the evolution of the forebrain. J. Neurosci. 21, 7620–7629.
Bardet, S. M., Cobos, I., Puelles, E., Martínez-De-La-Torre, M., and Puelles, L. (2006). Chicken lateral septal organ and other circumventricular organs form in a striatal subdomain abutting the molecular striatopallidal border. J. Comp. Neurol. 499, 745–767.
Bardet, S. M., Ferran, J. L., Sánchez-Arrones, L., and Puelles, L. (2010). Ontogenetic expression of sonic hedgehog in the chicken subpallium. Front. Neuroanat. 4:28. doi: 10.3389/fnana.2010.00028
Bardet, S. M., Martínez-de-la-Torre, M., Northcutt, R. G., Rubenstein, J. L., and Puelles, L. (2008). Conserved pattern of OTP-positive cells in the paraventricular nucleus and other hypothalamic sites of tetrapods. Brain Res. Bull. 75, 231–235.
Barth, K. A., and Wilson, S. W. (1994). Specification of neuronal identity in the embryonic CNS. Semin. Dev. Biol. 5, 349–358.
Barth, K. A., and Wilson, S. W. (1995). Expression of zebrafish nk2.2 is influenced by sonic hedgehog/vertebrate hedgehog-1 and demarcates a zone of neuronal differentiation in the embryonic forebrain. Development 121, 1755–1768.
Bläsher, S., and Heinrichs, M. (1982). Immunoreactive neuropeptide systems in avian embryos (domestic mallard, domestic fowl, Japanese quail). Cell Tissue Res. 223, 287–303.
Blechman, J., Borodovsky, N., Eisenberg, M., Nabel-Rosen, H., Grimm, J., and Levkowitz, G. (2007). Specification of hypothalamic neurons by dual regulation of the homeodomain protein orthopedia. Development 134, 4417–4426.
Braun, M. M., Etheridge, A., Bernard, A., Robertson, C. P., and Roelink, H. (2003). Wnt signaling is required at distinct stages of development for the induction of the posterior forebrain. Development 130, 5579–5587.
Briscoe, J., and Ericson, J. (1999). The specification of neuronal identity by graded sonic hedgehog signalling. Semin. Cell Dev. Biol. 10, 353–362.
Briscoe, J., Pierani, A., Jessell, T. M., and Ericson, J. (2000). A homeodomain protein code specifies progenitor cell identity and neuronal fate in the ventral neural tube. Cell 101, 435–445.
Brox, A., Puelles, L., Ferreiro, B., and Medina, L. (2003). Expression of the genes GAD67 and distal-less-4 in the forebrain of Xenopus laevis confirms a common pattern in tetrapods. J. Comp. Neurol. 461, 370–393.
Brox, A., Puelles, L., Ferreiro, B., and Medina, L. (2004). Expression of the genes Emx1, Tbr1, and Eomes (Tbr2) in the telencephalon of Xenopus laevis confirms the existence of a ventral pallial division in all tetrapods. J. Comp. Neurol. 474, 562–577.
Del Giacco, L., Pistocchi, A., Cotelli, F., Fortunato, A. E., and Sordino, P. (2008). A peek inside the neurosecretory brain through orthopedia lenses. Dev. Dyn. 237, 2295–2303.
Del Giacco, L., Sordino, P., Pistocchi, A., Andreakis, N., Tarallo, R., Di Benedetto, B., and Cotelli, F. (2006). Differential regulation of the zebrafish orthopedia 1 gene during fate determination of diencephalic neurons. BMC Dev. Biol. 6, 50. doi: 10.1186/1471-213X-6-50
Domínguez, L., González, A., and Moreno, N. (2010). Sonic hedgehog expression during Xenopus laevis forebrain development. Brain Res. 1347, 19–32.
Eaton, J. L., and Glasgow, E. (2007). Zebrafish orthopedia (otp) is required for isotocin cell development. Dev. Genes Evol. 217, 149–158.
Eaton, J. L., Holmqvist, B., and Glasgow, E. (2008). Ontogeny of vasotocin-expressing cells in zebrafish: selective requirement for the transcriptional regulators orthopedia and single-minded 1 in the preoptic area. Dev. Dyn. 237, 995–1005.
Echevarría, D., Vieira, C., Gimeno, L., and Martínez, S. (2003). Neuroepithelial secondary organizers and cell fate specification in the developing brain. Brain Res. Brain Res. Rev. 43, 179–191.
Ekker, S. C., McGrew, L. L., Lai, C. J., Lee, J. J., von Kessler, D. P., Moon, R. T., and Beachy, P. A. (1995). Distinct expression and shared activities of members of the hedgehog gene family of Xenopus laevis. Development 121, 2337–2347.
Ericson, J., Rashbass, P., Schedl, A., Brenner-Morton, S., Kawakami, A., van Heyningen, V., Jessell, T. M., and Briscoe, J. (1997). Pax6 controls progenitor cell identity and neuronal fate in response to graded Shh signaling. Cell 90, 169–180.
Ferran, J. L., de Oliveira, E. D., Merchán, P., Sandoval, J. E., Sánchez-Arrones, L., Martínez-De-La-Torre, M., and Puelles, L. (2009). Genoarchitectonic profile of developing nuclear groups in the chicken pretectum. J. Comp. Neurol. 517, 405–451.
Flames, N., Pla, R., Gelman, D. M., Rubenstein, J. L., Puelles, L., and Marín, O. (2007). Delineation of multiple subpallial progenitor domains by the combinatorial expression of transcriptional codes. J. Neurosci. 27, 9682–9695.
Fode, C., Ma, Q., Casarosa, S., Ang, S. L., Anderson, D. J., and Guillemot, F. (2000). A role for neural determination genes in specifying the dorsoventral identity of telencephalic neurons. Genes Dev. 14, 67–80.
Fuccillo, M., Rutlin, M., and Fishell, G. (2006). Removal of Pax6 partially rescues the loss of ventral structures in Shh null mice. Cereb. Cortex 16(Suppl. 1), 96–102.
García-Lopez, M., Abellán, A., Legaz, I., Rubenstein, J. L., Puelles, L., and Medina, L. (2008). Histogenetic compartments of the mouse centromedial and extended amygdala based on gene expression patterns during development. J. Comp. Neurol. 506, 46–74.
García-Lopez, R., Vieira, C., Echevarría, D., and Martínez, S. (2004). Fate map of the diencephalon and the zona limitans at the 10-somites stage in chick embryos. Dev. Biol. 268, 514–530.
González, A., López, J. M., and Marín, O. (2002a). Expression pattern of the homeobox protein NKX2-1 in the developing Xenopus forebrain. Brain Res. Gene Expr. Patterns 1, 181–185.
González, A., López, J. M., Sánchez-Camacho, C., and Marín, O. (2002b). Regional expression of the homeobox gene NKX2-1 defines pallidal and interneuronal populations in the basal ganglia of amphibians. Neuroscience 114, 567–575.
González, A., Marín, O., Tuinhof, R., and Smeets, W. J. (1994). Ontogeny of catecholamine systems in the central nervous system of anuran amphibians: an immunohistochemical study with antibodies against tyrosine hydroxylase and dopamine. J. Comp. Neurol. 346, 63–79.
González, A., Moreno, N., Morona, R., and López, J. M. (2003). Somatostatin-like immunoreactivity in the brain of the urodele amphibian Pleurodeles waltl. Colocalization with catecholamines and nitric oxide. Brain Res. 965, 246–258.
González, A., Muñóz, A., Muñóz, M., Marín, O., and Smeets, W. J. (1995). Ontogeny of vasotocinergic and mesotocinergic systems in the brain of the South African clawed frog Xenopus laevis. J. Chem. Neuroanat. 9, 27–40.
González, A., Tuinhof, R., and Smeets, W. J. (1993). Distribution of tyrosine hydroxylase and dopamine immunoreactivities in the brain of the South African clawed frog Xenopus laevis. Anat. Embryol. (Berl.) 187, 193–201.
Goshu, E., Jin, H., Lovejoy, J., Marion, J. F., Michaud, J. L., and Fan, C. M. (2004). Sim2 contributes to neuroendocrine hormone gene expression in the anterior hypothalamus. Mol. Endocrinol. 18, 1251–1262.
Hashimoto-Torii, K., Motoyama, J., Hui, C. C., Kuroiwa, A., Nakafuku, M., and Shimamura, K. (2003). Differential activities of sonic hedgehog mediated by Gli transcription factors define distinct neuronal subtypes in the dorsal thalamus. Mech. Dev. 120, 1097–1111.
Herrick, C. J. (1910). The Morphology of the forebrain in amphibia and reptilia. J. Comp. Neurol. 20, 413–547.
Holland, L. Z., Venkatesh, T. V., Gorlin, A., Bodmer, R., and Holland, N. D. (1998). Characterization and developmental expression of AmphiNk2-2, an NK2 class homeobox gene from Amphioxus. (phylum chordata; subphylum cephalochordata). Dev. Genes Evol. 208, 100–105.
Jiménez, F., Martin-Morris, L. E., Velasco, L., Chu, H., Sierra, J., Rosen, D. R., and White, K. (1995). vnd, a gene required for early neurogenesis of Drosophila, encodes a homeodomain protein. EMBO J. 14, 3487–3495.
Kataoka, A., and Shimogori, T. (2008). Fgf8 controls regional identity in the developing thalamus. Development 135, 2873–2881.
Kiecker, C., and Lumsden, A. (2004). Hedgehog signaling from the ZLI regulates diencephalic regional identity. Nat. Neurosci. 7, 1242–1249.
Kim, Y., and Nirenberg, M. (1989). Drosophila NK-homeobox genes. Proc. Natl. Acad. Sci. U.S.A. 86, 7716–7720.
Kimura, S., Hara, Y., Pineau, T., Fernández-Salguero, P., Fox, C. H., Ward, J. M., and González, F. J. (1996). The T/ebp null mouse: thyroid-specific enhancer-binding protein is essential for the organogenesis of the thyroid, lung, ventral forebrain, and pituitary. Genes Dev. 10, 60–69.
Kitamura, K., Miura, H., Yanazawa, M., Miyashita, T., and Kato, K. (1997). Expression patterns of Brx1 (Rieg gene), sonic hedgehog, Nkx2.2, Dlx1 and Arx during zona limitans intrathalamica and embryonic ventral lateral geniculate nuclear formation. Mech. Dev. 67, 83–96.
Kramer, B. M., Welting, J., Berghs, C. A., Jenks, B. G., and Roubos, E. W. (2001). Functional organization of the suprachiasmatic nucleus of Xenopus laevis in relation to background adaptation. J. Comp. Neurol. 432, 346–355.
Lin, X., State, M. W., Vaccarino, F. M., Greally, J., Hass, M., and Leckman, J. F. (1999). Identification, chromosomal assignment, and expression analysis of the human homeodomain containing gene Orthopedia (OTP). Genomics 60, 96–104.
López, J. M., Moreno, N., Morona, R., Muñóz, M., Domínguez, L., and González, A. (2007). Distribution of somatostatin-like immunoreactivity in the brain of the caecilian Dermophis mexicanus (amphibia: gymnophiona): comparative aspects in amphibians. J. Comp. Neurol. 501, 413–340.
Marín, O., Baker, J., Puelles, L., and Rubenstein, J. L. (2002). Patterning of the basal telencephalon and hypothalamus is essential for guidance of cortical projections. Development 129, 761–773.
Martínez-de-la-Torre, M., Garda, A. L., Puelles, E., and Puelles, L. (2002). Gbx2 expression in the late embryonic chick dorsal thalamus. Brain Res. Bull. 57, 435–438.
Medina, L. (2008). “Evolution and embryological development of forebrain,” in Enciclopedic Reference of Neuroscience, eds M. D. Binder and N. Hirokawa (Springer-Verlag), 1172–1192.
Menuet, A., Alunni, A., Joly, J. S., Jeffery, W. R., and Rétaux, S. (2007). Expanded expression of sonic hedgehog in Astyanax cavefish: multiple consequences on forebrain development and evolution. Development 134, 845–855.
Michaud, J. L. (2001). The developmental program of the hypothalamus and its disorders. Clin. Genet. 60, 255–263.
Milán, F. J., and Puelles, L. (2000). Patterns of calretinin, calbindin, and tyrosine-hydroxylase expression are consistent with the prosomeric map of the frog diencephalon. J. Comp. Neurol. 419, 96–121.
Moreno, N., Bachy, I., Rétaux, S., and González, A. (2003). Pallial origin of mitral cells in the olfactory bulbs of Xenopus. Neuroreport 14, 2355–2358.
Moreno, N., Bachy, I., Rétaux, S., and González, A. (2004). LIM-homeodomain genes as developmental and adult genetic markers of Xenopus forebrain functional subdivisions. J. Comp. Neurol. 472, 52–72.
Moreno, N., Bachy, I., Rétaux, S., and González, A. (2005). LIM-homeodomain genes as territory markers in the brainstem of adult and developing Xenopus laevis. J. Comp. Neurol. 485, 240–254.
Moreno, N., Domínguez, L., Rétaux, S., and González, A. (2008a). Islet1 as a marker of subdivisions and cell types in the developing forebrain of Xenopus. Neuroscience 154, 1423–1439.
Moreno, N., Rétaux, S., and González, A. (2008b). Spatio-temporal expression of Pax6 in Xenopus forebrain. Brain Res. 1239, 92–99.
Moreno, N., Morona, R., López, J. M., and González, A. (2010). Subdivisions of the turtle Pseudemys scripta subpallium based on the expression of regulatory genes and neuronal markers. J. Comp. Neurol. 518, 4877–4902.
Morona, R., Ferran, J. L., Puelles, L., and González, A. (2011). Embryonic genoarchitecture of pretectum in Xenopus laevis: a conserved pattern in tetrapods. J. Comp. Neurol. 519, 1024–1050.
Morona, R., and González, A. (2008). Calbindin-D28k and calretinin expression in the forebrain of anuran and urodele amphibians: further support for newly identified subdivisions. J. Comp. Neurol. 511, 187–220.
Nieuwkoop, P. D., and Farber, J. (1967). Normal Table of Xenopus laevis (Daudin). Amsterdam: North-Holland.
Osorio, J., Mazan, S., and Rétaux, S. (2005). Organization of the lamprey (Lampetra fluviatilis) embryonic brain: insights from LIM-homeodomain, Pax and hedgehog genes. Dev. Biol. 288, 100–112.
Osorio, J., Megías, M., Pombal, M. A., and Rétaux, S. (2006). Dynamic expression of the LIM-homeodomain gene Lhx15 through larval brain development of the sea lamprey (Petromyzon marinus). Gene Expr. Patterns 6, 873–878.
Papalopulu, N., and Kintner, C. (1993). Xenopus distal-less related homeobox genes are expressed in the developing forebrain and are induced by planar signals. Development 117, 961–975.
Petkó, M., and Orosz, V. (1996). Distribution of somatostatin-immunoreactive structures in the central nervous system of the frog, Rana esculenta. J. Hirnforsch. 37, 109–120.
Prakash, N., and Wurst, W. (2006). Genetic networks controlling the development of midbrain dopaminergic neurons. J. Physiol. 575, 403–410.
Pratt, T., Vitalis, T., Warren, N., Edgar, J. M., Mason, J. O., and Price, D. J. (2000). A role for Pax6 in the normal development of dorsal thalamus and its cortical connections. Development 127, 5167–5178.
Price, M., Lazzaro, D., Pohl, T., Mattei, M. G., Rüther, U., Olivo, J. C., Duboule, D., and Di Lauro, R. (1992). Regional expression of the homeobox gene Nkx-2.2 in the developing mammalian forebrain. Neuron 8, 241–255.
Puelles, L., Kuwana, E., Puelles, E., Bulfone, A., Shimamura, K., Keleher, J., Smiga, S., and Rubenstein, J. L. (2000). Pallial and subpallial derivatives in the embryonic chick and mouse telencephalon, traced by the expression of the genes Dlx-2, Emx-1, Nkx-2.1, Pax-6, and Tbr-1. J. Comp. Neurol. 424, 409–438.
Puelles, L., Martínez, S., Martínez-de-la-Torre, M., and Rubenstein, J. L. (2004). “Gene maps and related histogenetic domains in the forebrain and midbrain,” in The Rat Nervous System, 3rd Edn, ed. G. Paxinos (San Diego: Elsevier), 3–25.
Puelles, L., and Rubenstein, J. L. (1993). Expression patterns of homeobox and other putative regulatory genes in the embryonic mouse forebrain suggest a neuromeric organization. Trends Neurosci. 16, 472–479.
Puelles, L., and Rubenstein, J. L. (2003). Forebrain gene expression domains and the evolving prosomeric model. Trends Neurosci. 26, 469–476.
Rohr, K. B., Barth, K. A., Varga, Z. M., and Wilson, S. W. (2001). The nodal pathway acts upstream of hedgehog signaling to specify ventral telencephalic identity. Neuron 29, 341–351.
Ryu, S., Mahler, J., Acampora, D., Holzschuh, J., Erhardt, S., Omodei, D., Simeone, A., and Driever, W. (2007). Orthopedia homeodomain protein is essential for diencephalic dopaminergic neuron development. Curr. Biol. 17, 873–880.
Sander, M., Paydar, S., Ericson, J., Briscoe, J., Berber, E., German, M., Jessell, T. M., and Rubenstein, J. L. (2000). Ventral neural patterning by Nkx homeobox genes: Nkx6.1 controls somatic motor neuron and ventral interneuron fates. Genes Dev. 14, 2134–2139.
Schäfer, M., Kinzel, D., Neuner, C., Schartl, M., Volff, J. N., and Winkler, C. (2005). Hedgehog and retinoid signalling confines nkx2.2b expression to the lateral floor plate of the zebrafish trunk. Mech. Dev. 122, 43–56.
Scholpp, S., Wolf, O., Brand, M., and Lumsden, A. (2006). Hedgehog signalling from the zona limitans intrathalamica orchestrates patterning of the zebrafish diencephalon. Development 133, 855–864.
Shimamura, K., and Rubenstein, J. L. (1997). Inductive interactions direct early regionalization of the mouse forebrain. Development 124, 2709–2718.
Shimogori, T., Lee, D. A., Miranda-Angulo, A., Yang, Y., Wang, H., Jiang, L., Yoshida, A. C., Kataoka, A., Mashiko, H., Avetisyan, M., Qi, L., Qian, J., and Blackshaw, S. (2010). A genomic atlas of mouse hypothalamic development. Nat. Neurosci. 13, 767–775.
Simeone, A., D’Apice, M. R., Nigro, V., Casanova, J., Graziani, F., Acampora, D., and Avantaggiato, V. (1994). Orthopedia, a novel homeobox-containing gene expressed in the developing CNS of both mouse and Drosophila. Neuron 13, 83–101.
Smeets, W. J., and González, A. (2000). Catecholamine systems in the brain of vertebrates: new perspectives through a comparative approach. Brain Res. Brain Res. Rev. 33, 308–379.
Sussel, L., Marín, O., Kimura, S., and Rubenstein, J. L. (1999). Loss of Nkx2.1 homeobox gene function results in a ventral to dorsal molecular respecification within the basal telencephalon: evidence for a transformation of the pallidum into the striatum. Development 126, 3359–3370.
Szabó, N. E., Zhao, T., Zhou, X., and Alvarez-Bolado, G. (2009a). The role of sonic hedgehog of neural origin in thalamic differentiation in the mouse. J. Neurosci. 29, 2453–2466.
Szabó, N. E., Zhao, T., Cankaya, M., Theil, T., Zhou, X., and Alvarez-Bolado, G. (2009b). Role of neuroepithelial sonic hedgehog in hypothalamic patterning. J. Neurosci. 29, 6989–7002.
Takuma, N., Sheng, H. Z., Furuta, Y., Ward, J. M., Sharma, K., Hogan, B. L., Pfaff, S. L., Westphal, H., Kimura, S., and Mahon, K. A. (1998). Formation of Rathke’s pouch requires dual induction from the diencephalon. Development 125, 4835–4840.
van den Akker, W. M., Brox, A., Puelles, L., Durston, A. J., and Medina, L. (2008). Comparative functional analysis provides evidence for a crucial role for the homeobox gene Nkx2.1/Titf-1 in forebrain evolution. J. Comp. Neurol. 506, 211–223.
Vieira, C., Garda, A. L., Shimamura, K., and Martínez, S. (2005). Thalamic development induced by Shh in the chick embryo. Dev. Biol. 284, 351–363.
Vieira, C., and Martínez, S. (2006). Sonic hedgehog from the basal plate and the zona limitans intrathalamica exhibits differential activity on diencephalic molecular regionalization and nuclear structure. Neuroscience 143, 129–140.
Vieira, C., Pombero, A., García-Lopez, R., Gimeno, L., Echevarría, D., and Martínez, S. (2010). Molecular mechanisms controlling brain development: an overview of neuroepithelial secondary organizers. Int. J. Dev. Biol. 54, 7–20.
Vue, T. Y., Aaker, J., Taniguchi, A., Kazemzadeh, C., Skidmore, J. M., Martin, D. M., Martin, J. F., Treier, M., and Nakagawa, Y. (2007). Characterization of progenitor domains in the developing mouse thalamus. J. Comp. Neurol. 505, 73–91.
Wang, W., and Lufkin, T. (2000). The murine Otp homeobox gene plays an essential role in the specification of neuronal cell lineages in the developing hypothalamus. Dev. Biol. 227, 432–449.
Keywords: prosencephalon, hypothalamus, in situ hybridization, evolution, forebrain patterning, thalamus
Citation: Domínguez L, González A and Moreno N (2011) Ontogenetic distribution of the transcription factor Nkx2.2 in the developing forebrain of Xenopus laevis. Front. Neuroanat. 5:11. doi: 10.3389/fnana.2011.00011
Received: 05 November 2010;
Paper pending published: 29 December 2010;
Accepted: 16 February 2011;
Published online: 02 March 2011.
Edited by:
Fernando Martinez-Garcia, Universidad de Valencia, SpainReviewed by:
Loreta Medina, Universidad de Lleida, SpainCopyright: © 2011 Domínguez, González and Moreno. This is an open-access article subject to an exclusive license agreement between the authors and Frontiers Media SA, which permits unrestricted use, distribution, and reproduction in any medium, provided the original authors and source are credited.
*Correspondence: Nerea Moreno, Faculty of Biology, Department of Cell Biology, University Complutense of Madrid, C/José Antonio Novais 2, Madrid E-28040, Spain. e-mail:bmVyZWFAYmlvLnVjbS5lcw==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.