- 1Department of Cell Biology and Physiology, University of Kansas Medical Center, Kansas City, MO, United States
- 2University of Kansas Alzheimer's Disease Center, University of Kansas Medical Center, Fairway, KS, United States
- 3Department of Neurology, University of Kansas Medical Center, Kansas City, MO, United States
- 4University of Kansas Diabetes Institute, University of Kansas Medical Center, Kansas City, MO, United States
Introduction: Apolipoprotein E4 (APOE4) is the strongest genetic risk factor for Alzheimer's disease (AD), yet it's unclear how this allele mediates risk. APOE4 carriers experience reduced mobility and faster decline in muscle strength, suggesting skeletal muscle involvement. Mitochondria are critical for muscle function and although we have reported defects in muscle mitochondrial respiration during early cognitive decline, APOE4-mediated effects on muscle mitochondria are unknown.
Methods: Here, we sought to determine the impact of APOE4 on skeletal muscle bioenergetics using young, male and female APOE3 (control) and APOE4 targeted replacement mice (n = 8 per genotype/sex combination). We examined the proteome, mitochondrial respiration, fiber size, and fiber-type distribution in skeletal muscle.
Results: We found that APOE4 alters mitochondrial pathway expression in young mouse muscle in a sex-dependent manner without affecting respiration and fiber size or composition relative to APOE3. In both sexes, the expression of mitochondrial pathways involved in electron transport, ATP synthesis, and heat production by uncoupling proteins and mitochondrial dysfunction significantly differed between APOE4 and APOE3 muscle. For pathways with predicted direction of activation, electron transport and oxidative phosphorylation were upregulated while mitochondrial dysfunction and sirtuin signaling were downregulated in female APOE4 vs. APOE3 muscle. In males, sulfur amino acid metabolism was upregulated in APOE4 vs. APOE3 muscle.
Discussion: This work highlights early involvement of skeletal muscle in a mouse model of APOE4-linked AD, which may contribute to AD pathogenesis or serve as a biomarker for brain health.
1 Introduction
Approximately 6.9 million people over the age of 65 are living with Alzheimer's disease (AD; 2024), yet no drugs significantly improve prognosis. Three variations of the APOE gene in humans differentially influence AD risk. APOE4 increases risk, accounting for ~65% of AD cases (Mayeux et al., 1998). In contrast, APOE2 protects against AD and APOE3 does not affect risk (Anonymous, 2024). Risk may further be modified by sex and age as female APOE4 carriers are more likely than males to develop mild cognitive impairment between 55 and 70 years old (Neu et al., 2017).
Despite the established relationship between APOE genotype and AD, it is not fully understood how APOE4 mediates risk. Metabolic function is a prime area of interest because APOE encodes a lipid transport protein, and the APOE4 variant is associated with alterations in energy metabolism in the brain and other organ systems (Lumsden et al., 2020; Farmer et al., 2021; Wilkins et al., 2017; Reiman et al., 1996; Arnold et al., 2020). APOE4 carriers display earlier reductions in brain glucose metabolism (Reiman et al., 1996) and studies have linked APOE4 to changes in brain mitochondrial dynamics (Schmukler et al., 2020; Simonovitch et al., 2019), respiration (Area-Gomez et al., 2020), and energy homeostasis (Qi et al., 2021). Although APOE4 may have systemic effects, fewer studies have focused on the role of APOE4 in peripheral tissues. Individuals carrying an APOE4 allele have increased cholesterol levels and are more likely to develop cardiovascular disease (Lumsden et al., 2020), which is an AD risk factor (Livingston et al., 2024). There is also evidence that APOE4 reduces weight and fat mass (Ando et al., 2022), alters the relationship between plasma metabolites and AD biomarkers (Arnold et al., 2020), and reduces resting energy expenditure (Farmer et al., 2021) in a sex-dependent manner, supporting a role for APOE in modulating whole-body metabolism and sex as a modifying factor.
Comprising ~40% of body weight and accounting for 26% of basal metabolic rate (Mathias, 2022), skeletal muscle plays a major role in regulating whole-body metabolism and may influence brain health as outlined in recent reviews by Brisendine and Drake (2023) and Liu et al. (2023). Of note, Matthews et al. has shown that skeletal muscle-specific overexpression of transcription factor B (TFEB) improves AD-associated pathology in mice (Matthews et al., 2023). Given the role of TFEB in regulating protein turnover and mitochondrial function, this suggests that muscle metabolism may be an important and understudied factor in modulating brain health. Further, we have shown that lipid-supported oxygen consumption is reduced in human muscle in the early stages of cognitive decline (Morris et al., 2021). Few studies have examined the effects of APOE4 specifically on skeletal muscle, despite clinical evidence that skeletal muscle function is impacted by this allele. Melzer et al. found that APOE4 carriers perform worse on mobility tests in older age (Melzer et al., 2005) and Buchman et al. reported faster motor decline in APOE4 carriers that was largely attributed to changes in muscle strength (Buchman et al., 2009). Further, Huebbe et al. found that the expression of a few proteins involved in fatty acid oxidation are increased in APOE4 compared to APOE3 muscle in mice fed a high-fat diet (Huebbe et al., 2015). However, no one has determined the effects of APOE4 on the whole-muscle proteome and mitochondrial function.
Here, we used APOE4 and APOE3 targeted replacement mice to assess the impact of APOE genotype on skeletal muscle mitochondrial bioenergetics at 4 months old, which is roughly equivalent to a young adult human. We chose this age because AD pathology begins decades before the development of clinical symptoms (Villemagne et al., 2013). Thus, leveraging mice at younger ages provides an opportunity to study the role of skeletal muscle in early disease. APOE3 mice served as our controls because this allele is the most prevalent APOE isoform in humans and does not affect AD risk (Anonymous, 2024). We hypothesized that APOE4 would drive muscle bioenergetic dysfunction and be reflected by reduced mitochondrial protein expression, impaired mitochondrial respiratory function, and a shift in fiber-type distribution in APOE4 compared to APOE3 mice. We found that APOE4 affects the skeletal muscle proteome of 4-month-old mice in a sex-dependent manner, without affecting fiber size, fiber-type composition, or oxygen consumption. Metabolic pathways that differed between APOE4 and APOE3 mice in our study are affected by AD or APOE genotype in other tissues, including those involved in amino acid metabolism, oxidative phosphorylation, and sirtuin signaling (Yin et al., 2018; Haytural et al., 2021; Toledo et al., 2017; Valla et al., 2010). This work highlights the need to understand the involvement of skeletal muscle in individuals at genetic risk for AD and the potential modulatory role of sex.
2 Materials and methods
2.1 Animals
Four-month-old male and female APOE3 and APOE4 targeted replacement mice (n = 8 per genotype/sex combination) from Taconic Biosciences (Germantown, NY) were used for all analyses. These mice are genetically modified to express human APOE isoforms through gene knock-in and replacement of mouse Apoe (Knouff et al., 1999; Sullivan et al., 1997). Mice were fed a standard chow diet (14% kcal fat, Teklad Global Rodent Diets, 8604). All procedures for the use of animals in this study were approved by the Institutional Animal Care and Use Committee at the University of Kansas Medical Center.
2.2 Body mass and composition
Mice were weighed and body composition was analyzed the week of sacrifice using an EchoMRI TM 1100 Analyzer (EchoMRI, Houston, TX).
2.3 Tissue preparation
Mice were anesthetized with 90 mg/kg ketamine and 10 mg/kg xylazine and then immediately euthanized prior to muscle collection. The left gastrocnemius was dissected free of fat and connective tissue before respirometry analysis, and the right gastrocnemius was flash-frozen in liquid nitrogen and stored at −80°C for mass spectrometry. The left quadriceps was embedded in OCT and stored at −80°C for immunofluorescent staining.
2.4 Muscle proteome
Frozen gastrocnemius tissue from the right hindlimb was crushed with a pestle and placed in homogenization buffer (1% Triton X-100, 50 mM HEPES, 12 mM Na pyrophosphate, 100 mM NaF, 10 mM EDTA, protease inhibitor, phosphatase inhibitor) with a stainless-steel bead. Samples were then homogenized on the TissueLyser II (Qiagen, Germantown, MD) programmed to beat samples at 20 Hz for 2 min. After beating, samples were immediately placed on ice for 10 min and beating was repeated two more times prior to centrifuging at 15,000 g for 25 min at 4°C. The supernatant containing the lysed proteins was removed and protein content was measured using a bicinchoninic acid assay (Thermo Fisher, Waltham, MA). Protein lysates (50 ug) were sent to the IDEA National Resource for Proteomics at the University of Arkansas for Medical Sciences (UMAS). Samples were processed as previously described (Mccoin et al., 2022). Briefly, proteins were extracted, digested, separated, eluted, and ionized prior to running on an Orbitrap Exploris 480 mass spectrometer (Thermo Fisher).
2.5 Immunofluorescent staining
OCT-embedded quadriceps muscle was used to cut 10 μm transverse sections on a Cryostar NX50 Cryostat (Thermo Fisher) at −22°C and stored at −20°C until fiber-typing. Prior to staining, sections were hydrated in PBST for 5 min, blocked for 1 h in blocking solution (10% FBS, 6% w/w BSA, and 0.10% Triton X-100 in PBS) and washed three times with PBST. Sections were then incubated at 4°C overnight in primary antibodies (1:100) against type 1 (BA-F8 mIgG2b), type IIa (SC-71 mIgG1), and type IIb (BF-F3 mIgM) fibers all purchased from Developmental Studies Hybridoma Bank (Iowa City, IA). Visualization of the basement membrane was detected with a laminin antibody (1:200, Abcam ab11575, Cambridge, MA). After overnight incubation in primary antibodies, sections were washed three times with PBST and stained for 1 h with secondary antibodies conjugated to fluorophores for visualization (mIgG2b-AF350 A21140, mIgG1-AF488 A21121, mIgM-AF555 A21426, and rbIgG-AF647 A21244). Sections were then washed three times with PBST. Images were acquired on a Zeiss Observer 7 microscope (Carl Zeiss Microscopy, Jena, Germany) and analyzed with MuscleJ2 (Danckaert et al., 2023) for fiber-type, cross-sectional area, and minimum Feret diameter. Minimum Feret diameter is the smallest diameter that can be measured across a muscle fiber and may be superior to cross-sectional area as a marker of muscle fiber size because it is not as sensitive to changes in fiber size due to sectioning (Briguet et al., 2004). Unstained fibers were defined as type IIx fibers. Approximately 3,570 muscle fibers on average were assessed from one muscle section per mouse.
2.6 Respirometry analyses
Gastrocnemius muscle was placed in ice cold buffer X immediately following dissection, cleared of fat and connective tissue, then carefully separated and teased into muscle bundles (~0.4–0.8 mg). Buffer X (pH 7.1) included the following constituents: 50 mM K-MES, 7.23 mM K2EGTA, 2.77 mM CaK2EGTA, 20 mM Imidazole, 20 mM Taurine, 5.7 mM Na-ATP, 14.3 mM Na-PCr, and 6.56 mM MgCl2-6H2O. Teased bundles were permeabilized with saponin (30 μg/ml) for 30 min at 4°C then rinsed with buffer Z (105 mM K-MES, 30 mM KCl, 10 mM K2HPO4, 5 mM MgCl2-6H2O, 0.5M EGTA and 0.5% w/w fatty acid-free BSA; pH 7.1). Oxygen consumption of permeabilized fibers was then measured on the Oroboros Oxygraph-2k (Oroboros Instruments, Innsbruck, Austria) at 37°C in buffer Z with 20 mM creatine monohydrate. For carbohydrate-supported respiration, basal oxygen consumption was measured at steady state in the presence of 0.01 mM blebbistatin, 0.5 mM malate, and 5 mM potassium pyruvate. State 3 respiration was then measured by adding 2.5 mM ADP followed by 5 mM glutamate to assess complex I-linked state 3 respiration, 10 mM succinate to assess complex II-linked state 3S respiration, and 0.00005 mM FCCP in sequential steps until maximal respiration was reached to assess uncoupled respiration. For lipid-supported respiration, oxygen consumption was measured at steady state in the presence of 0.01 mM blebbistatin, 0.5 mM malate, 0.5 mM CoA, 5 mM carnitine, and 0.02 mM palmitoyl-CoA. State 3 respiration was then measured by adding 4 mM ADP followed by 0.02 mM palmitoyl-carnitine (PC) to assess state 3+PC respiration then 10 mM succinate to assess complex II-linked state 3S respiration. 0.0015 mM FCCP was then added until maximal, uncoupled respiration was reached. After completing respirometry analyses, muscle bundles were dried in a lyophilizer (FreeZone 2.5 L, Labconco, Kansas City, MO) and weighed with a microbalance (MX5, Mettler Toledo, Columbus, OH). Average steady-state oxygen consumption values for each respiratory state were normalized to dried muscle weights. Coupling efficiency was determined by calculating the ratio of basal leak respiration to state 3 respiration and subtracting this value from 1.
2.7 Statistical analyses
Spectronaut software (Biognosys version 18.3) was used to analyze mass spectrometry data. The UnitProt Mus musculus database (September 2023) was used to search and identify mouse proteins using directDIA with a false discovery threshold of 1%. ProteiNorm (Graw et al., 2020) was used to evaluate the quality of MS2 intensities and variance stabilizing normalization was applied (Huber et al., 2002). Linear models for Microrray Data (limma) with empirical Bayes (eBayes) smoothing to the standard errors (Ritchie et al., 2015; Timothy J Thurman et al., 2023) was performed with proteoDA. Proteins with a p-value ≤ 0.05 for between genotype differences within each sex were considered significant. Mitocarta3.0 was used to identify and filter differentially expressed mitochondrial proteins (Rath et al., 2021) and Ingenuity Pathway Analysis (IPA, Quiagen) was used for pathway analysis of the whole-muscle proteome and the MitoCarta3.0-filtered dataset. The p-value of overlap was used for pathway enrichment analysis in IPA to determine if the expression of a pathway significantly differed between genotypes (≤ 0.05). IPA activation z-scores were used to predict the direction of pathway or upstream regulator activity. Upstream regulators flagged for bias were excluded. Gene names are used in place of protein names as this is the standard format used by IPA.
All other statistical analyses were performed in SPSS 29.0. Boxplot analysis was used to detect, inspect and remove outliers >3 box lengths from the edge of the box. After verifying ANOVA assumptions, two-way ANOVA was then used to assess for a sex by genotype interaction and main effects. Significant interaction effects were followed up with the Fisher's LSD test. Two sample t-tests were used to assess genotype effects within sex where indicated. Statistical significance for all tests was determined based on p ≤ 0.05.
3 Results
3.1 Body composition
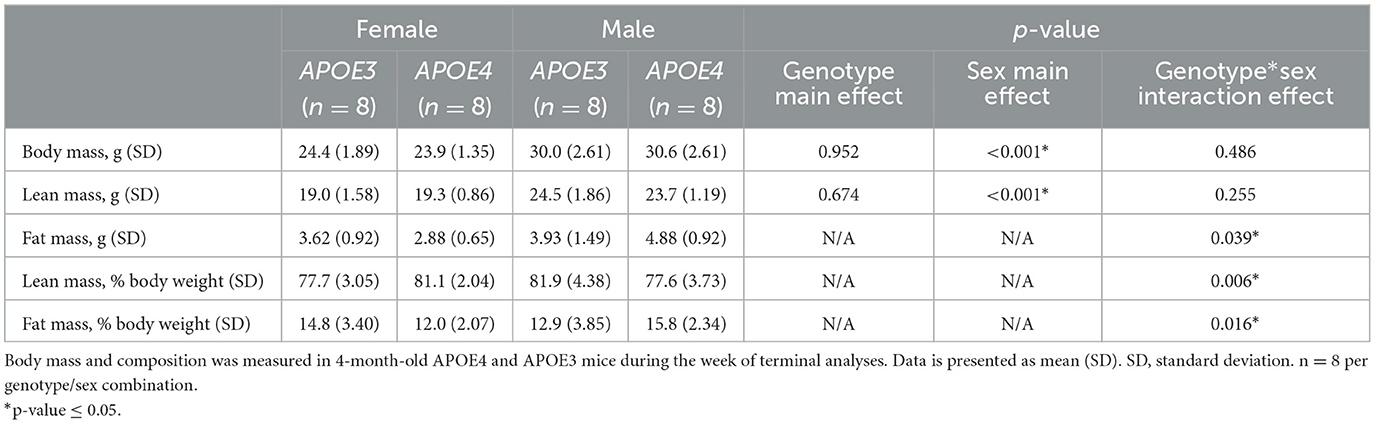
Male mice weighed more than female mice while APOE genotype did not affect body mass at 4-months-old (Table 1). Given findings that APOE4 may alter fat mass in women particularly in the early stages of cognitive decline (Ando et al., 2022), we were also interested in body composition. Male mice had greater lean mass than female mice regardless of APOE genotype and there was a genotype by sex interaction effect on fat mass, percent fat mass, and percent lean mass. Post-hoc analyses revealed that fat mass and percent fat mass were greater in APOE4 males compared to APOE4 females, while there was no sex effect in APOE3 mice. Percent lean mass was lower in APOE4 versus APOE3 males and greater in male versus female APOE3 mice.
3.2 Muscle proteome
3.2.1 APOE4-driven shifts in whole-muscle proteome of male and female mice
Proteomic characterization of gastrocnemius muscle identified 3,062 unique proteins across all samples. One hundred and ninety proteins in females and 268 proteins in males were differentially expressed in APOE4 vs. APOE3 muscle in each sex, respectively (Figures 1A, B). Only 17 proteins were affected by APOE4 in both males and females (Figure 1B) and of these, 9 proteins were upregulated or downregulated in the same direction while 8 proteins were regulated in the opposite direction (Table 2). Pathway analysis of significantly altered proteins in muscle revealed that APOE4 affected the expression of pathways involved in metabolism, including insulin secretion in females, sulfur amino acid metabolism in males, and mitochondrial dysfunction in both sexes (Supplementary Table 1).
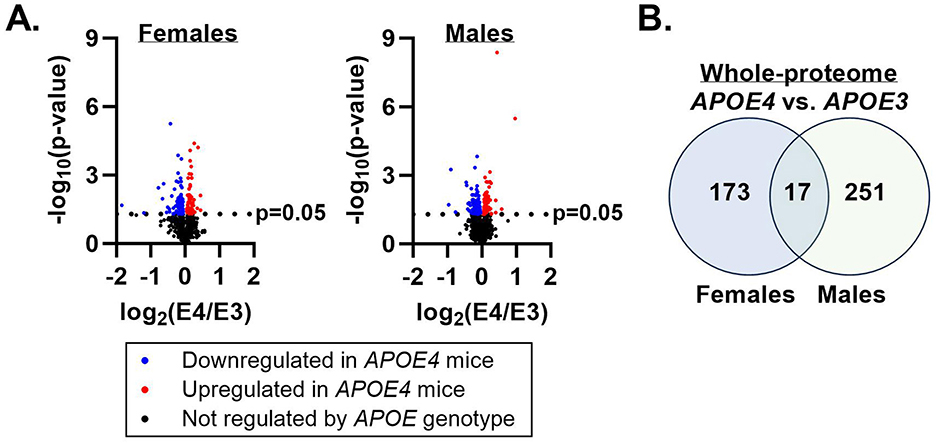
Figure 1. Apolipoprotein E4 (APOE4)-driven shifts in whole-muscle proteome. Mass spectrometry was performed to assess the effect of APOE genotype on the muscle proteome in male and female mice. Proteins differentially expressed in APOE4 vs. APOE3 mice (p-value ≤ 0.05) are represented by the colored dots in the volcano plots (A). The number of proteins uniquely impacted by APOE genotype in each sex and the number of proteins altered by APOE genotype in both sexes are depicted in a Venn diagram (B). n = 8 per genotype/sex combination.
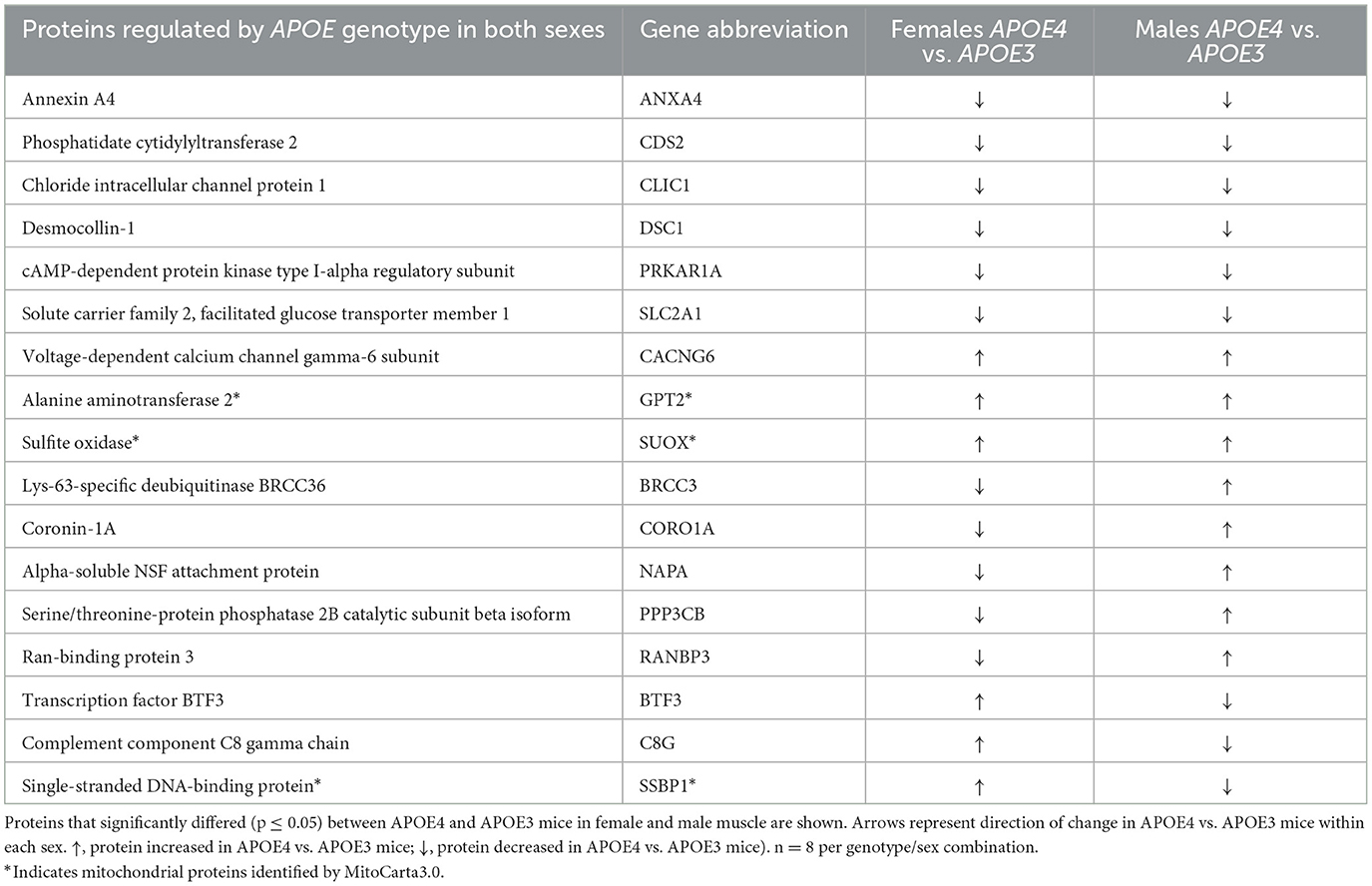
Table 2. Proteins regulated by apolipoprotein E (APOE) genotype in both female and male skeletal muscle.
3.2.2 APOE4-driven shifts in muscle mitochondrial pathways of male and female mice
Since our primary interest is the effect of APOE genotype and sex on muscle mitochondria, we referenced MitoCarta3.0 (Rath et al., 2021) to filter our dataset to only analyze mitochondrial proteins. We found that 35 proteins in females and 42 proteins in males that are differentially expressed between APOE4 and APOE3 muscle are part of the mitochondrial proteome and only 3 of these proteins, mitochondrial alanine transaminase (GPT2), single-stranded DNA binding protein 1 (SSBP1), and sulfite oxidase (SUOX), are affected by APOE genotype in both sexes (Figure 2A, Table 2). GPT2 and SUOX are both involved in amino acid metabolism (Sakagishi, 1995; Isabelle Papet et al., 2018) and are upregulated in APOE4 vs. APOE3 muscle in both sexes. SSPB1 is implicated in mitochondrial DNA (mtDNA) maintenance and replication (Tiranti et al., 1995) and is upregulated by APOE4 in female muscle and downregulated by APOE4 in male muscle.
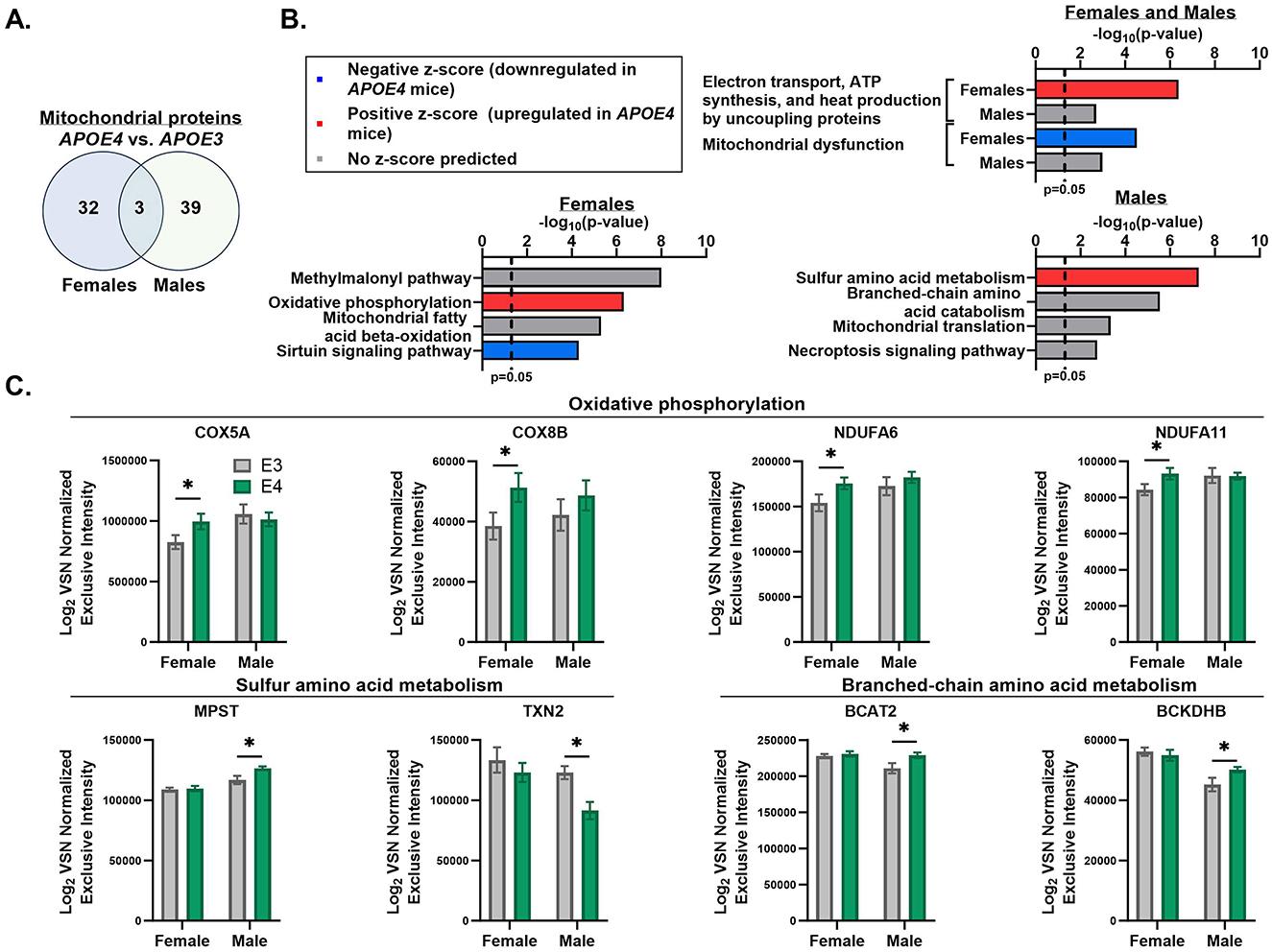
Figure 2. Apolipoprotein E4 (APOE4)-driven shifts in muscle mitochondrial pathways. MitoCarta3.0 was used to assess mitochondrial proteins that were differentially expressed between APOE4 and APOE3 muscle in male and female mice (p-value ≤ 0.05). The number of mitochondrial proteins uniquely impacted by APOE genotype in each sex and the number of mitochondrial proteins altered by APOE genotype in both sexes are depicted in a Venn diagram (A). Ingenuity pathway analysis was used to assess the top six pathways that differed between APOE4 and APOE3 muscle in males and females using the MitoCarta3.0-filtered dataset. Highly redundant pathways were excluded. Two of these pathways were affected by APOE genotype in both sexes and are shown in the top graph. The other four pathways for each sex are shown in the bottom graphs (B). Normalized intensities for selected proteins from APOE genotype-regulated pathways are plotted for each sex (C). n = 8 per genotype/sex combination. COX5A, Cytochrome c oxidase subunit 5A; COX8B, cytochrome c oxidase subunit 8B; NDUFA6, NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 6; NDUFA11, NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 11; MPST, 3-mercaptopyruvate sulfurtransferase; TXN2, thioredoxin; BCAT2, branched-chain-amino-acid aminotransferase; BCKDHB, 2-oxoisovalerate dehydrogenase subunit beta; VSN, variance stabilization normalization. *p ≤ 0.05.
We then used IPA to determine the top mitochondrial pathways regulated by APOE genotype using the MitoCarta3.0-filtered dataset (Figure 2B). In both sexes, mitochondrial dysfunction and electron transport, ATP synthesis, and heat production by uncoupling proteins were among the top pathways that significantly differed between APOE4 and APOE3 muscle. In females, the pathway involving electron transport, ATP synthesis, and heat production was upregulated while mitochondrial dysfunction was downregulated in APOE4 vs. APOE3 muscle. While these pathways also varied between APOE4 and APOE3 muscle in males, there was no prediction on the direction of pathway expression. This can occur when enough proteins belonging to a specific pathway differ between groups, resulting in a significant enrichment score, but the pattern of protein expression is inconsistent with one direction of pathway activity or there is not enough data to predict an activation z-score. In females, upregulated mitochondrial dysfunction was driven by reduced expression of proteins involved in oxidative phosphorylation and mitochondrial fission. In males, differences in mitochondrial dysfunction between APOE4 and APOE3 muscle was driven by altered expression of proteins involved in mitochondrial DNA replication and repair, energy metabolism, and redox balance. In female muscle, oxidative phosphorylation was also upregulated by APOE4 while sirtuin signaling was downregulated by APOE4. In male muscle, sulfur amino acid metabolism was upregulated by APOE4. Other mitochondrial pathways that significantly differed between APOE4 and APOE3 muscle without a predication on the direction of pathway change include the methylmalonyl pathway and fatty acid β-oxidation in females, and branched-chain amino acid metabolism, mitochondrial translation, and necroptosis signaling in males. Normalized intensities for select proteins from genotype-enriched pathways are graphed in Figure 2C.
In addition to proteins localized to mitochondria, we identified extra-mitochondrial proteins impacted by APOE4 that may indirectly influence mitochondrial energy metabolism. This includes calcium voltage-gated channel auxiliary subunit gamma 6 (CACNG6), which limits calcium influx (Burgess et al., 2001) and was upregulated in APOE4 vs. APOE3 muscle in both sexes (Table 2). Further, solute carrier family 2 facilitated glucose transporter member 1 (SLC2A1) was downregulated in APOE4 vs. APOE3 muscle in both sexes (Table 2).
3.2.3 Potential upstream regulators
Next, we used IPA to predict upstream regulators that may drive differences in mitochondrial pathway expression in APOE4 vs. APOE3 muscle using the MitoCarta3.0-filtered dataset (Figure 3, Table 3). Using an activation z-score cut-off of ±2, six upstream regulators were identified in females compared to only one in males. This difference is likely explained by IPA's algorithm, which relies on a pattern of protein expression that is consistent with a specific direction of upstream regulator activity to calculate a z-score. All seven of these molecules have been implicated in the pathogenesis or treatment of AD and other metabolic diseases and include rapamycin-insensitive companion of mammalian target of rapamycin (RICTOR), TNF receptor associated protein 1 (TRAP1), PPARG coactivator 1 alpha (PPARGC1A), and insulin receptor (INSR; Koopman and Rüdiger, 2020; Katsouri et al., 2011; Morris and Burns, 2012; Lee et al., 2017).
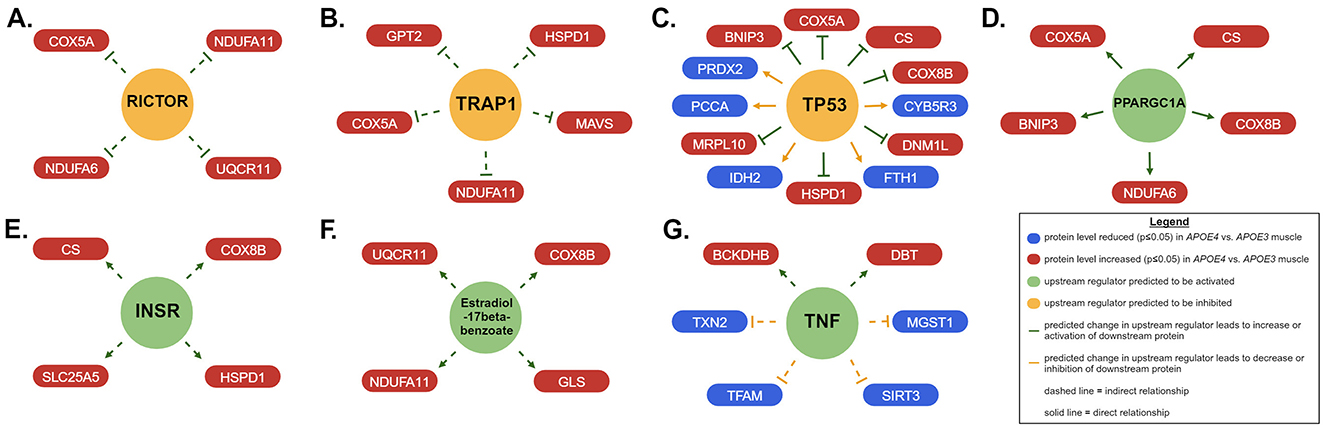
Figure 3. Upstream regulators of mitochondrial proteins impacted by apolipoprotein E4 (APOE4) in muscle. Ingenuity pathway analysis (IPA) was used to predict upstream regulators that drive differences in expression of mitochondrial proteins in skeletal muscle between APOE4 and APOE3 mice. Upstream regulators with an activation z-score of >2 or <-2 are shown at the center of each panel for females (A–F) and males (G). Proteins that drove these results are represented in the red and blue boxes that are connected to each upstream regulator by an arrowhead or inhibitor symbol. n = 8 per genotype/sex combination. The IPA-generated version of this figure was recreated with Biorender.com. BCKDHB, Branched chain keto acid dehydrogenase E1 subunit beta; BNIP3, BCL2 interacting protein 3; COX5A, cytochrome c oxidase subunit 5A; COX8B, cytochrome c oxidase subunit 8B; CS, citrate synthase; CYB5R3, cytochrome b5 reductase 3; DBT, dihydrolipoamide branched chain transacylase E2; DNM1L, dynamin 1 like; FTH1, ferritin heavy chain 1; GLS, glutaminase; GPT2, glutamic-pyruvic transaminase 2; Hsp60, heat shock protein family D; HSPD1, member 1; NADP(+) 2 (IDH2), isocitrate dehydrogenase; INSR, insulin receptor; MAVS, mitochondrial antiviral signaling protein; MGST1, microsomal glutathione S-transferase 1; MRPL10, mitochondrial ribosomal protein L10; NDUFA11, NADH:ubiquinone oxidoreductase subunit A11; NDUFA6, NADH:ubiquinone oxidoreductase subunit A6; PCCA, propionyl-CoA carboxylase subunit alpha; PPARGC1A, PPARG coactivator 1 alpha; PRDX2, peroxiredoxin 2; RICTOR, RPTOR independent companion of MTOR complex 2; SIRT3, sirtuin 3; SLC25A5, solute carrier family 25 member 5; TFAM, transcription factor A, mitochondrial; TNF, tumor necrosis factor; TP53, tumor protein p53; TRAP1, TNF receptor associated protein 1; TXN2, thioredoxin 2; UQCR11, ubiquinol-cytochrome c reductase, complex III subunit XI.
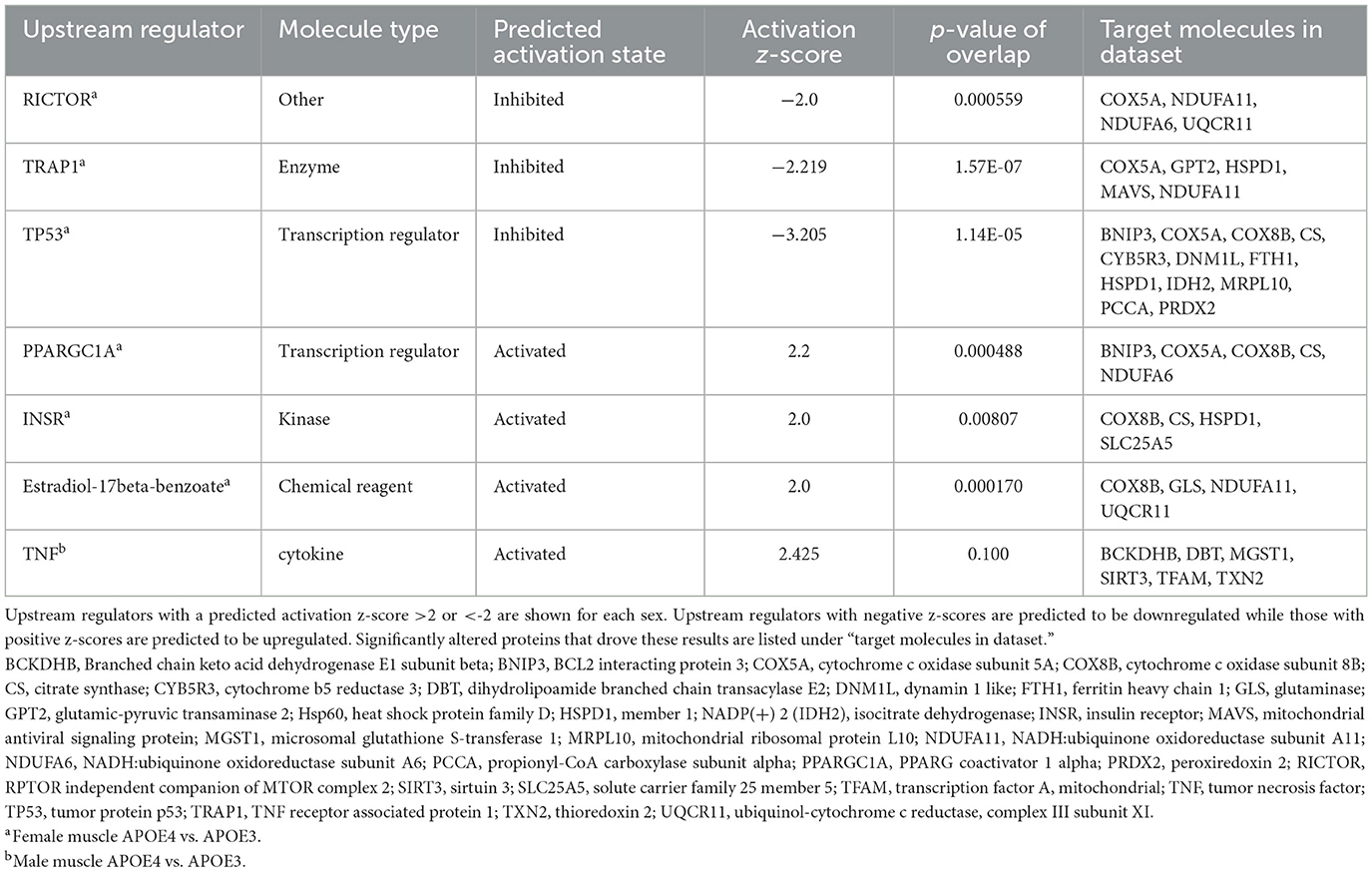
Table 3. Upstream regulators of mitochondrial proteins impacted by apolipoprotein E4 (APOE4) in muscle.
3.3 Muscle fiber type and size
Mitochondrial protein expression and oxidative capacity varies between different isoforms of skeletal muscle fibers. Given our finding that APOE genotype influences the expression of mitochondrial oxidative pathways in skeletal muscle, we were interested to determine if fiber composition is affected. We assessed this in the quadriceps, which is normally composed of ~90% fast-twitch fibers in mice (Zancanaro et al., 2007). We also examined cross-sectional area and minimum Feret diameter to evaluate muscle fiber size. There was no effect of APOE genotype or sex on the relative amounts of oxidative (type I and IIa) or glycolytic (type IIb and IIx) muscle fibers (Supplementary Table 2). There was also no significant effect of APOE genotype or sex on fiber size, although APOE4 mice trended toward reduced cross-sectional area (p = 0.211 for main effect of genotype) and minimum Feret diameter (p = 0.251 for main effect of genotype) compared to APOE3 mice (Figure 4).
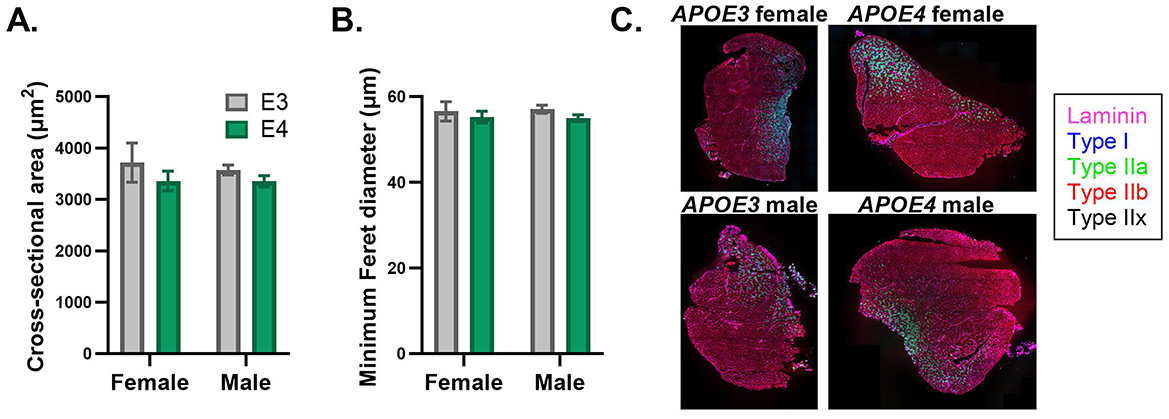
Figure 4. Muscle fiber size is similar between apolipoprotein E4 (APOE4) and APOE3 mice. Cross-sectional area (A) and minimum Feret diameter (B) in the quadriceps was determined by MuscleJ2 using whole-muscle sections stained with immunofluorescent antibodies. Representative images of muscle sections stained with antibodies against laminin, type I fibers, type IIa fibers, and type IIb fibers (C). Pure type IIx fibers are not stained and identified as black fibers. n = 8 per genotype/sex combination.
3.4 Mitochondrial respiration in permeabilized muscle fibers
Although APOE genotype did not significantly impact muscle fiber size or fiber-type distribution, we wanted to know if APOE4-dependent enrichment in mitochondrial oxidative pathways accompanied functional differences in mitochondrial respiration. We did this by assessing oxygen consumption in permeabilized muscle bundles supported by a carbohydrate or lipid source, the primary fuels used by muscle. APOE genotype did not have a significant effect on muscle oxygen consumption with either fuel based on the two-way ANOVA results (Figure 5). Since sex is a significant biological variable in our study, we wanted to examine the effects of genotype within males and females individually. This exploratory analysis revealed significantly greater lipid-supported state 3S (p = 0.028) and uncoupled (p = 0.024) respiration in APOE4 vs. APOE3 male mice while there was no genotype effect in female mice. Future studies with a larger sample size are needed to confirm this finding. There was no difference in coupling efficiency, an indicator of linkage between the generation of a proton gradient through electron transport and ATP production.
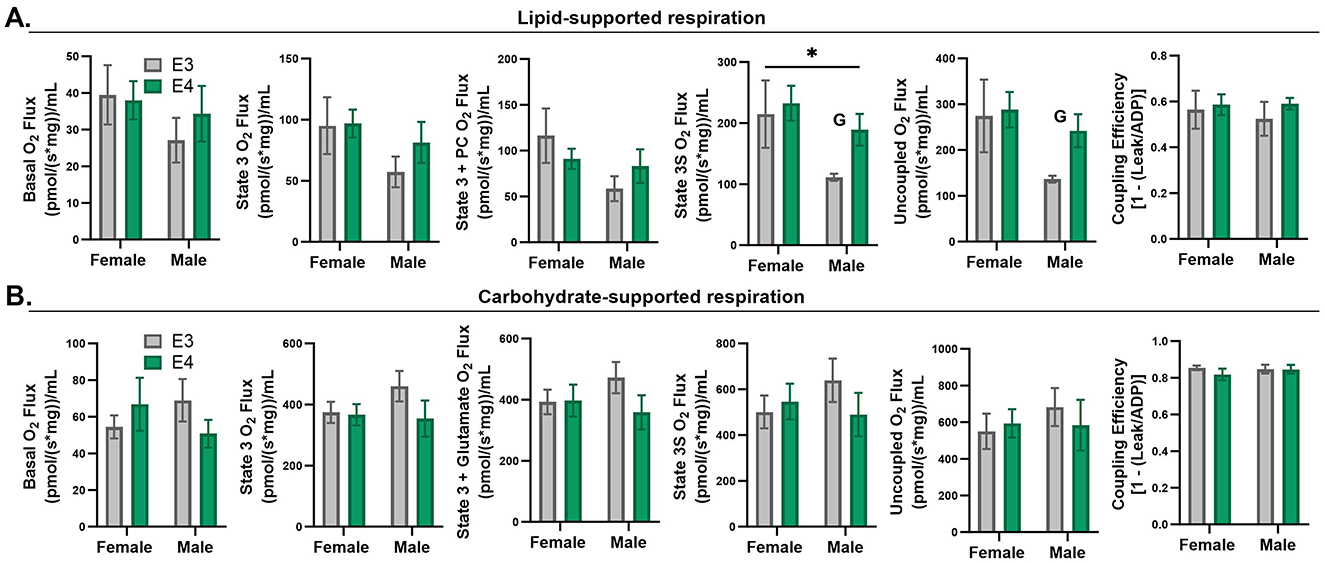
Figure 5. Respiration in permeabilized muscle fibers of apolipoprotein E4 (APOE4) and APOE3 mice. Lipid-supported oxygen consumption measured at basal, state 3 (ADP), state 3+palmitoyl-carnitine (PC), complex II-linked state 3S respiration (succinate), and maximal, uncoupled respiration (FCCP) (A). Carbohydrate-supported oxygen consumption measured at basal, state 3 (ADP), complex-I linked state 3 respiration (glutamate), complex II-linked state 3S respiration (succinate), and maximal, uncoupled respiration (FCCP) (B). Coupling efficiency is shown in the last panel for lipid- and carbohydrate-supported respiration (A, B). *p-value ≤ 0.05 for main effect of sex. G = p-value ≤ 0.05 for exploratory two sample t-test of genotype effect within sex. n = 7–8 per genotype/sex combination.
4 Discussion
Here, we show for the first time that APOE4 impacts the murine skeletal muscle proteome at 4 months old, equivalent to a young adult human. We identified mitochondrial pathways in muscle affected by APOE4 that have been implicated in AD pathogenesis based on cognitive status or APOE genotype (Yin et al., 2018; Haytural et al., 2021; Toledo et al., 2017; Valla et al., 2010; Kish et al., 1992). These pathways largely involve mitochondrial fuel oxidation, supporting findings that APOE4 drives metabolic alterations in systemic tissues (Lumsden et al., 2020; Farmer et al., 2021; Wilkins et al., 2017; Arnold et al., 2020).
Our finding that the effects of APOE4 are largely sex-specific at this age as summarized in Table 4 is consistent with several studies demonstrating sex- and APOE genotype-dependent differences in molecular and physiological measures across various tissues (Farmer et al., 2021; Arnold et al., 2020; Altmann et al., 2014). In both sexes in our study, APOE4 drove changes in the muscle proteome that would support mitochondrial energy metabolism. In females, this was reflected by an upregulation of pathways involved in oxidative phosphorylation and electron transport, ATP synthesis, and heat production by uncoupling proteins. In males, this was noted by upregulated sulfur amino acid metabolism which contributes to mitochondrial oxidative metabolism through the synthesis of coenzyme A (Isabelle Papet et al., 2018). Although most proteins that differed in expression between APOE genotypes were sex dependent, GPT2, SUOX, and SSBP1 were affected in both sexes. GPT2 and SUOX, which were expressed at greater levels in APOE4 vs. APOE3 muscle in males and females, contribute to energy metabolism by providing α-ketoglutarate to the citric acid cycle or by participating in electron transfer to cytochrome c, respectively (Isabelle Papet et al., 2018; Sakagishi, 1995). Our finding that APOE4 upregulates protein expression in young mice in a pattern that would likely support energy metabolism is consistent with a study from Perkins et al., which demonstrated increased levels of proteins involved in glucose transport and oxidative phosphorylation in the posterior cingulate brain region of young adult APOE4 carriers compared to non-carriers (Perkins et al., 2016). We extend this knowledge to skeletal muscle in a mouse model. Interestingly, SSBP1, a protein that is essential for mtDNA stability and replication during mitochondrial biogenesis, was increased by APOE4 in females and decreased by APOE4 in males, reflecting a sex-divergent response to APOE4 in the regulation of the same mitochondrial protein.
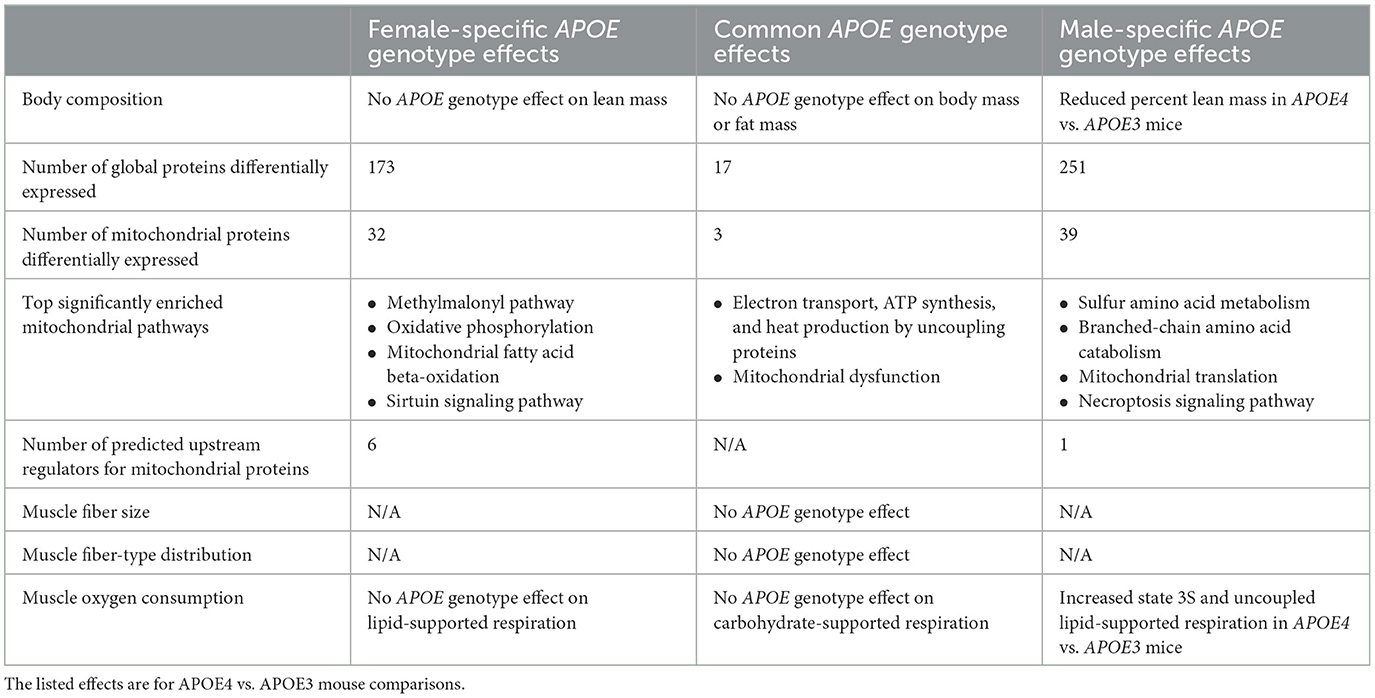
Table 4. Summary of apolipoprotein E (APOE) genotype effects that are unique to females, unique to males, or common to both sexes.
Other pathways impacted by APOE4 in our study, including methylmalonyl metabolism, fatty acid oxidation, and sirtuin signaling in females and branched-chain amino acid catabolism and mitochondrial translation in males, also play important roles in energy metabolism through their direct involvement in fuel oxidation, metabolic signaling, or synthesis of electron transport chain proteins. Given the important role of sirtuin signaling in promoting fatty acid oxidation and mitochondrial function when metabolic demand is high (Chang and Guarente, 2014), the downregulation of sirtuin signaling in female APOE4 muscle could reflect a reduced ability to adapt to metabolic challenge. Additionally, skeletal muscle is a major site for the metabolism of branched-chain amino acids which influence muscle mass and whole-body metabolism, and differed between APOE4 and APOE3 male mice. Reduced levels of branched-chain amino acids have been associated with worse performance on cognitive tests (Toledo et al., 2017). However, the contribution of muscle to this observation has not been explored and warrants further investigation based on our findings.
We also identified potential upstream regulators that could explain APOE genotype- and sex-dependent patterns in mitochondrial protein expression in muscle. Predicted inhibition of RICTOR in APOE4 vs. APOE3 female muscle is consistent with a study from Lee et al. demonstrating reduced activity of mammalian target of rapamycin complex 2 (mTORC2) in brain tissue from individuals with AD. RICTOR is an important component of mTORC2 (Lee et al., 2017). TRAP1, a mitochondrial-specific chaperone protein that regulates oxidative phosphorylation among other roles, was also expected to be downregulated in female APOE4 muscle. This is in agreement with findings that TRAP1 levels are reduced in the AD brain (Koopman and Rüdiger, 2020). Our results also suggested that PPARGC1A was activated in APOE4 female muscle. PPARGC1A encodes peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC1α), which promotes mitochondrial biogenesis, fatty acid oxidation, oxidative phosphorylation, and branched-chain amino acid metabolism in muscle (Kamei et al., 2020). PGC1α protein levels are reduced in brain tissue from individuals with AD (Katsouri et al., 2011) and 12-month old APOE4 mice (Yin et al., 2019). Estradiol-17beta-benzoate was also identified as an upstream regulator, corroborating findings that estrogen signaling is an important component of sex-dependent differences in AD pathophysiology and may be an important treatment target in APOE4 carriers (Saleh et al., 2023). These regulators, among others identified in our study should be evaluated for their roles in driving skeletal muscle and whole-body metabolism in APOE4 carriers and as potential treatment targets or biomarkers. Our finding that the activation status of more upstream regulators could be predicted in females compared to males coincides with our observation that more pathways in females had associated z-scores. This was not due to a difference in the number of mitochondrial proteins altered by APOE genotype between sexes because this was similar in males and females. Instead, this can be explained by which proteins were affected and the direction of their change. Females displayed a pattern of protein expression that aligned better with a particular direction of pathway or upstream regulator activity. This difference in protein regulation reflects an interaction between APOE genotype and sex and the role of hormone and chromosome differences to this effect remains to be explored.
The trend toward reduced muscle fiber size in APOE4 mice based on CSA and minimum Feret diameter, the latter which is less sensitive to changes in fiber-shape during sectioning (Briguet et al., 2004) is consistent with evidence that lean mass is lower in individuals in the early stages of AD relative to cognitively healthy individuals (Burns et al., 2010). This is also in line with our findings that percent lean mass was reduced in APOE4 versus APOE3 male mice. Although we did not detect a difference in lean mass between APOE genotypes in females, it is possible that 4 months is too early to observe changes. Additionally, we did not examine bone mass which is tightly linked to mitochondrial health in skeletal muscle (Tian et al., 2022) and could be impacted in our study. Similar distribution of fiber-types between APOE4 and APOE3 mice regardless of sex suggests that genotype-dependent effects on muscle protein expression are likely not explained by differences in the relative amounts of each fiber type, which display unique proteomic expression profiles (Murgia et al., 2021). Quadriceps was used to determine fiber-type distribution, while gastrocnemius was used for proteomics. Due to limited tissue availability, we could not make both assessments in the same muscles, however both are mixed-fiber type, lower-limb muscles.
Defects in mitochondrial respiratory chain function in skeletal muscle of a mouse model of autosomal dominant AD are not evident until 12 months of age, despite early molecular changes detected in skeletal muscle (Monteiro-Cardoso et al., 2015). Similar to this study, increased expression of mitochondrial respiratory pathways in APOE4 female mice despite normal respiratory function may reflect an underlying respiratory chain defect that is compensated for by increased production of proteins involved in oxidative metabolism. If true, a natural reduction in protein maintenance with age may eventually result in reduced expression or activity of the proteins found to be upregulated in this study. This would be consistent with findings that many proteins impacted by APOE genotype in the brain are upregulated in young APOE4 carriers yet downregulated in older individuals with AD (Roberts et al., 2021). This is also in line with the mitochondrial cascade hypothesis of AD which suggests that compensatory mitochondrial pathways cannot be maintained indefinitely, ultimately leading to mitochondrial dysfunction (Swerdlow et al., 2014). Although mitochondrial respiration was not affected by APOE4 in female muscle, we found in an exploratory analysis that state 3S and uncoupled lipid-supported respiration were increased in APOE4 vs. APOE3 male muscle. This occurred with an upregulation of sulfur amino metabolism, which in addition to supporting metabolism, is also involved in responding to oxidative stress (Isabelle Papet et al., 2018). Glutathione is an important antioxidant that is produced from cysteine, a sulfur containing amino acid. The upregulation of sulfur amino acid metabolism in male APOE4 muscle could therefore reflect a compensatory response to protect against oxidative stress associated with increased muscle mitochondrial activity. More experiments are required to characterize the effects of APOE4 on other outcomes that influence mitochondrial function including membrane potential, calcium homeostasis, and oxidative balance. Future work should also consider how these outcomes are impacted by aging as protein maintenance declines and oxidative balance is impaired.
Our study has several strengths. Our experimental design included molecular and physiological outcomes, and use of male and female mice allowed characterization of sex-related effects. Furthermore, our analysis of whole-tissue sections for fiber-typing, and use of intact muscle bundles to assess mitochondrial oxygen consumption is novel and provides important information about muscle in this model. Muscle bundles should more closely mimic physiologic conditions compared to isolated mitochondria because the mitochondria in bundles retain their subcellular localization. Our study is limited by the lack of gene expression data to determine the correspondence between RNA and protein expression. We acknowledge that gene expression data would enrich our understanding of these mechanisms. However, we chose to focus on protein expression because proteins are the drivers of physiologic function and RNA does not always correlate with protein levels (Upadhya and Ryan, 2022). We also used homozygous APOE3 and APOE4 mice, which do not reflect the full spectrum of APOE allele combinations in humans (Hana Saddiki et al., 2020). However, abnormal levels of AD biomarkers in the majority of older APOE4 homozygotes supports studying homozygotes as a separate, genetically-determined form of AD (Fortea et al., 2024). We also did not include a wild-type mouse group to differentiate effects driven by the introduction of human APOE isoforms into a mouse versus APOE isoform-specific effects. However, extensive characterization of these mice has shown that the introduction of human APOE variants results in functional APOE and APOE protein levels comparable to wild-type levels (Knouff et al., 1999; Sullivan et al., 1997). Importantly, the humanized mice recapitulate many of the phenotypic and biochemical outcomes related to lipid metabolism observed in humans. However, it is important to note that there are some differences in lipid metabolism between these mice and humans that could influence our bioenergetic outcomes, including the distribution of non-HDL cholesterol among lipoprotein particles. Additionally, the interaction between human APOE variants and mouse proteins is not fully understood.
5 Conclusion
In conclusion, APOE4 alters the expression of mitochondrial pathways in skeletal muscle in a sex-dependent manner without affecting oxygen consumption or fiber-type size and distribution. This could represent a compensatory response to maintain mitochondrial function. This information may ultimately be used to deepen our understanding of the relationship between whole-body metabolism and brain health. The potential for proteomic changes in muscle for use to identify novel AD biomarkers and treatment targets should be further explored given consistency in pathways altered in muscle and other tissues in AD and the ease of access to muscle tissue in humans. Follow-up studies should determine the extent to which our findings are reflected in human muscle.
Data availability statement
The dataset used in this study has been uploaded to the Harvard Dataverse repository (https://doi.org/10.7910/DVN/C7BZTX).
Ethics statement
The animal study was approved by the University of Kansas Medical Center Institutional Animal Care and Use Committee. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
CJ: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Validation, Visualization, Writing – original draft. CL: Data curation, Investigation, Writing – review & editing. CM: Investigation, Methodology, Validation, Writing – review & editing. ME: Investigation, Writing – review & editing. JT: Investigation, Methodology, Project administration, Writing – review & editing. HW: Project administration, Resources, Writing – review & editing. JM: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Writing – review & editing. PG: Conceptualization, Project administration, Resources, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the KU ADRC Brain Health Training Program P30 AG072973. Additional author support was provided through R01 AG056062 (JM), T32 AG078114 (CJ), and the University of Kansas Medical Center Biomedical Research Training Program (CJ). Proteomics at UAMS was supported by IDeA National Resource for Quantitative Proteomics and NIH/NIGMS Grant: R24GM137786. The IPA software used in this publication was supported by the Biostatistics and Informatics Shared Resource, funded by the National Cancer Institute Cancer Center Support Grant: P30 CA168524, and the Kansas IDeA Network of Biomedical Research Excellence Bioinformatics Core, supported in part by the National Institute of General Medical Science award: P20GM103418. In addition, respiration studies were supported by the Metabolism Core within the Kansas Center for Metabolism and Obesity Research, supported by a National Institute of General Medicine COBRE award: (P20GM144269). Equipment within the Metabolism Core was purchased with NIH S10OD028598.
Acknowledgments
We would like to thank Elaine Gast and Maryjane Wolf for work related to this project.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnagi.2024.1486762/full#supplementary-material
References
Altmann, A., Tian, L., Henderson, V. W., and Greicius, M. D. (2014). Sex modifies the APOE-related risk of developing Alzheimer disease. Ann. Neurol. 75, 563–573. doi: 10.1002/ana.24135
Ando, T., Uchida, K., Sugimoto, T., Kimura, A., Saji, N., Niida, S., et al. (2022). ApoE4 is associated with lower body mass, particularly fat mass, in older women with cognitive impairment. Nutrients 14:30539. doi: 10.3390/nu14030539
Anonymous (2024). 2024 Alzheimer's disease facts and figures. Alzheimer's Dement. 20, 3708–3821. doi: 10.1002/alz.13809
Area-Gomez, E., Larrea, D., Pera, M., Agrawal, R. R., Guilfoyle, D. N., Pirhaji, L., et al. (2020). APOE4 is associated with differential regional vulnerability to bioenergetic deficits in aged APOE mice. Sci. Rep. 10:4277. doi: 10.1038/s41598-020-61142-8
Arnold, M., Nho, K., Kueider-Paisley, A., Massaro, T., Huynh, K., Brauner, B., et al. (2020). Sex and APOE ε4 genotype modify the Alzheimer's disease serum metabolome. Nat. Commun. 11:1148. doi: 10.1038/s41467-020-14959-w
Briguet, A., Courdier-Fruh, I., Foster, M., Meier, T., and Magyar, J. P. (2004). Histological parameters for the quantitative assessment of muscular dystrophy in the mdx-mouse. Neuromuscul. Disord. 14, 675–682. doi: 10.1016/j.nmd.2004.06.008
Brisendine, M. H., and Drake, J. C. (2023). Early-stage Alzheimer's disease: are skeletal muscle and exercise the key? J. Appl. Physiol. 134, 515–520. doi: 10.1152/japplphysiol.00659.2022
Buchman, A. S., Boyle, P. A., Wilson, R. S., Beck, T. L., Kelly, J. F., and Bennett, D. A. (2009). Apolipoprotein E e4 allele is associated with more rapid motor decline in older persons. Alzheimer's Dis. Assoc. Disord. 23, 63–69. doi: 10.1097/WAD.0b013e31818877b5
Burgess, D. L., Gefrides, L. A., Foreman, P. J., and Noebels, J. L. (2001). A cluster of three novel Ca2+ channel gamma subunit genes on chromosome 19q13.4: evolution and expression profile of the gamma subunit gene family. Genomics 71, 339–350. doi: 10.1006/geno.2000.6440
Burns, J. M., Johnson, D. K., Watts, A., Swerdlow, R. H., and Brooks, W. M. (2010). Reduced lean mass in early Alzheimer disease and its association with brain atrophy. Arch. Neurol. 67, 428–433. doi: 10.1001/archneurol.2010.38
Chang, H. C., and Guarente, L. (2014). SIRT1 and other sirtuins in metabolism. Trends Endocrinol. Metab. 25, 138–145. doi: 10.1016/j.tem.2013.12.001
Danckaert, A., Trignol, A., Le Loher, G., Loubens, S., Staels, B., Duez, H., et al. (2023). MuscleJ2: a rebuilding of MuscleJ with new features for high-content analysis of skeletal muscle immunofluorescence slides. Skelet. Muscle 13:14. doi: 10.1186/s13395-023-00323-1
Farmer, B. C., Williams, H. C., Devanney, N. A., Piron, M. A., Nation, G. K., Carter, D. J., et al. (2021). APO?4 lowers energy expenditure in females and impairs glucose oxidation by increasing flux through aerobic glycolysis. Mol. Neurodegener. 16:62. doi: 10.1186/s13024-021-00483-y
Fortea, J., Pegueroles, J., Alcolea, D., Belbin, O., Dols-Icardo, O., Vaqué-Alcázar, L., et al. (2024). APOE4 homozygozity represents a distinct genetic form of Alzheimer's disease. Nat. Med. 30, 1284–1291. doi: 10.1038/s41591-024-02931-w
Graw, S., Tang, J., Zafar, M. K., Byrd, A. K., Bolden, C., Peterson, E. C., et al. (2020). proteiNorm—a user-friendly tool for normalization and analysis of TMT and label-free protein quantification. ACS Omega 5, 25625–25633. doi: 10.1021/acsomega.0c02564
Hana Saddiki, A. F., Cognat, E., Sabia, S., Engelborghs, S., Wallon, D., Alexopoulos, P., et al. (2020). Age and the association between apolipoprotein E genotype and Alzheimer disease: a cerebrospinal fluid biomarker-based case-control study. PLoS Med. 2020:1003289. doi: 10.1371/journal.pmed.1003289
Haytural, H., Benfeitas, R., Schedin-Weiss, S., Bereczki, E., Rezeli, M., Unwin, R. D., et al. (2021). Insights into the changes in the proteome of Alzheimer disease elucidated by a meta-analysis. Sci. Data 8:312. doi: 10.1038/s41597-021-01090-8
Huber, W., Von Heydebreck, A., Sültmann, H., Poustka, A., and Vingron, M. (2002). Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics 18(Suppl.1), S96–S104. doi: 10.1093/bioinformatics/18.suppl_1.S96
Huebbe, P., Dose, J., Schloesser, A., Campbell, G., Glüer, C. C., Gupta, Y., et al. (2015). Apolipoprotein E (APOE) genotype regulates body weight and fatty acid utilization-Studies in gene-targeted replacement mice. Mol. Nutr. Food Res. 59, 334–343. doi: 10.1002/mnfr.201400636
Isabelle Papet, D. R., Dardevet, D., Mosoni, L., Polakof, S., Peyron, M. A., and Savary-Auzeloux, I. (2018). Chapter 21—sulfur amino acids and skeletal muscle. Nutr. Skelet. Muscle 8, 335–363. doi: 10.1016/B978-0-12-810422-4.00020-8
Kamei, Y., Hatazawa, Y., Uchitomi, R., Yoshimura, R., and Miura, S. (2020). Regulation of skeletal muscle function by amino acids. Nutrients 12:10261. doi: 10.3390/nu12010261
Katsouri, L., Parr, C., Bogdanovic, N., Willem, M., and Sastre, M. (2011). PPARγ co-activator-1α (PGC-1α) reduces amyloid-β generation through a PPARγ-dependent mechanism. J. Alzheimer's Dis. 25, 151–162. doi: 10.3233/JAD-2011-101356
Kish, S. J., Bergeron, C., Rajput, A., Dozic, S., Mastrogiacomo, F., Chang, L. J., et al. (1992). Brain cytochrome oxidase in Alzheimer's disease. J. Neurochem. 59, 776–779. doi: 10.1111/j.1471-4159.1992.tb09439.x
Knouff, C., Hinsdale, M. E., Mezdour, H., Altenburg, M. K., Watanabe, M., Quarfordt, S. H., et al. (1999). Apo E structure determines VLDL clearance and atherosclerosis risk in mice. J. Clin. Invest. 103, 1579–1586. doi: 10.1172/JCI6172
Koopman, M. B., and Rüdiger, S. G. D. (2020). Alzheimer cells on their way to derailment show selective changes in protein quality control network. Front. Mol. Biosci. 7:214. doi: 10.3389/fmolb.2020.00214
Lee, H. K., Kwon, B., Lemere, C. A., De La Monte, S., Itamura, K., Ha, A. Y., et al. (2017). mTORC2 (Rictor) in Alzheimer's disease and reversal of amyloid-β expression-induced insulin resistance and toxicity in rat primary cortical neurons. J. Alzheimer's Dis. 56, 1015–1036. doi: 10.3233/JAD-161029
Liu, C., Wong, P. Y., Chow, S. K. H., Cheung, W. H., and Wong, R. M. Y. (2023). Does the regulation of skeletal muscle influence cognitive function? a scoping review of pre-clinical evidence. J. Orthop. Translat. 38, 76–83. doi: 10.1016/j.jot.2022.10.001
Livingston, G., Huntley, J., Liu, K. Y., Costafreda, S. G., Selbæk, G., Alladi, S., et al. (2024). Dementia prevention, intervention, and care: 2024 report of the Lancet standing Commission. Lancet 2024:1296. doi: 10.1016/S0140-6736(24)01296-0
Lumsden, A. L., Mulugeta, A., Zhou, A., and Hyppönen, E. (2020). Apolipoprotein E (APOE) genotype-associated disease risks: a phenome-wide, registry-based, case-control study utilising the UK Biobank. EBioMedicine 59:102954. doi: 10.1016/j.ebiom.2020.102954
Mathias, D. (2022). Energy Consumption I-Basal Metabolic Rate. Fit and Healthy From 1 to 100 With Nutrition and Exercise (Berlin: Springer Nature Link), 17–18.
Matthews, I., Birnbaum, A., Gromova, A., Huang, A. W., Liu, K., Liu, E. A., et al. (2023). Skeletal muscle TFEB signaling promotes central nervous system function and reduces neuroinflammation during aging and neurodegenerative disease. Cell Rep. 42:113436. doi: 10.1016/j.celrep.2023.113436
Mayeux, R., Saunders, A. M., Shea, S., Mirra, S., Evans, D., Roses, A. D., et al. (1998). Utility of the apolipoprotein E genotype in the diagnosis of Alzheimer's disease. Alzheimer's disease centers consortium on apolipoprotein E and Alzheimer's disease. N. Engl. J. Med. 338, 506–511. doi: 10.1056/NEJM199802193380804
Mccoin, C. S., Franczak, E., Deng, F., Pei, D., Ding, W. X., and Thyfault, J. P. (2022). Acute exercise rapidly activates hepatic mitophagic flux. J. Appl. Physiol. 132, 862–873. doi: 10.1152/japplphysiol.00704.2021
Melzer, D., Dik, M. G., Van Kamp, G. J., Jonker, C., and Deeg, D. J. (2005). The apolipoprotein E E4 polymorphism is strongly associated with poor mobility performance test results but not self-reported limitation in older people. J. Gerontol. A Biol. Sci. Med. Sci. 60, 1319–1323. doi: 10.1093/gerona/60.10.1319
Monteiro-Cardoso, V. F., Castro, M., Oliveira, M. M., Moreira, P. I., Peixoto, F., and Videira, R. A. (2015). Age-dependent biochemical dysfunction in skeletal muscle of triple-transgenic mouse model of Alzheimer's disease. Curr. Alzheimer's Res. 12, 100–115. doi: 10.2174/1567205012666150204124852
Morris, J. K., and Burns, J. M. (2012). Insulin: an emerging treatment for Alzheimer's disease dementia? Curr. Neurol. Neurosci. Rep. 12, 520–527. doi: 10.1007/s11910-012-0297-0
Morris, J. K., Mccoin, C. S., Fuller, K. N., John, C. S., Wilkins, H. M., Green, Z. D., et al. (2021). Mild cognitive impairment and donepezil impact mitochondrial respiratory capacity in skeletal muscle. Function 2:zqab045. doi: 10.1093/function/zqab045
Murgia, M., Nogara, L., Baraldo, M., Reggiani, C., Mann, M., and Schiaffino, S. (2021). Protein profile of fiber types in human skeletal muscle: a single-fiber proteomics study. Skelet. Muscle 11:24. doi: 10.1186/s13395-021-00279-0
Neu, S. C., Pa, J., Kukull, W., Beekly, D., Kuzma, A., Gangadharan, P., et al. (2017). Apolipoprotein E genotype and sex risk factors for Alzheimer disease: a meta-analysis. J. Am. Med. Assoc. Neurol. 74, 1178–1189. doi: 10.1001/jamaneurol.2017.2188
Perkins, M., Wolf, A. B., Chavira, B., Shonebarger, D., Meckel, J. P., Leung, L., et al. (2016). Altered energy metabolism pathways in the posterior cingulate in young adult apolipoprotein E ε4 carriers. J. Alzheimer's Dis. 53, 95–106. doi: 10.3233/JAD-151205
Qi, G., Mi, Y., Shi, X., Gu, H., Brinton, R. D., and Yin, F. (2021). ApoE4 impairs neuron-astrocyte coupling of fatty acid metabolism. Cell Rep. 34:108572. doi: 10.1016/j.celrep.2020.108572
Rath, S., Sharma, R., Gupta, R., Ast, T., Chan, C., Durham, T. J., et al. (2021). MitoCarta3.0: an updated mitochondrial proteome now with sub-organelle localization and pathway annotations. Nucl. Acids Res. 49, D1541–D1547. doi: 10.1093/nar/gkaa1011
Reiman, E. M., Caselli, R. J., Yun, L. S., Chen, K., Bandy, D., Minoshima, S., et al. (1996). Preclinical evidence of Alzheimer's disease in persons homozygous for the ε4 allele for apolipoprotein E. N. Engl. J. Med. 334, 752–758. doi: 10.1056/NEJM199603213341202
Ritchie, M. E., Phipson, B., Wu, D., Hu, Y., Law, C. W., Shi, W., et al. (2015). Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucl. Acids Res. 43:e47. doi: 10.1093/nar/gkv007
Roberts, J. A., Varma, V. R., An, Y., Varma, S., Candia, J., Fantoni, G., et al. (2021). A brain proteomic signature of incipient Alzheimer's disease in young APOE ε4 carriers identifies novel drug targets. Sci. Adv. 7:eabi8178. doi: 10.1126/sciadv.abi8178
Saleh, R. N. M., Hornberger, M., Ritchie, C. W., and Minihane, A. M. (2023). Hormone replacement therapy is associated with improved cognition and larger brain volumes in at-risk APOE4 women: results from the European Prevention of Alzheimer's Disease (EPAD) cohort. Alzheimer's Res. Ther. 15:10. doi: 10.1186/s13195-022-01121-5
Schmukler, E., Solomon, S., Simonovitch, S., Goldshmit, Y., Wolfson, E., Michaelson, D. M., et al. (2020). Altered mitochondrial dynamics and function in APOE4-expressing astrocytes. Cell Death Dis. 11:578. doi: 10.1038/s41419-020-02776-4
Simonovitch, S., Schmukler, E., Masliah, E., Pinkas-Kramarski, R., and Michaelson, D. M. (2019). The effects of APOE4 on mitochondrial dynamics and proteins in vivo. J. Alzheimer's Dis. 70, 861–875. doi: 10.3233/JAD-190074
Sullivan, P. M., Mezdour, H., Aratani, Y., Knouff, C., Najib, J., Reddick, R. L., et al. (1997). Targeted replacement of the mouse apolipoprotein E gene with the common human APOE3 allele enhances diet-induced hypercholesterolemia and atherosclerosis. J. Biol. Chem. 272, 17972–17980. doi: 10.1074/jbc.272.29.17972
Swerdlow, R. H., Burns, J. M., and Khan, S. M. (2014). The Alzheimer's disease mitochondrial cascade hypothesis: progress and perspectives. Biochim. Biophys. Acta 1842, 1219–1231. doi: 10.1016/j.bbadis.2013.09.010
Tian, J., Chung, H. K., Moon, J. S., Nga, H. T., Lee, H. Y., Kim, J. T., et al. (2022). Skeletal muscle mitoribosomal defects are linked to low bone mass caused by bone marrow inflammation in male mice. J. Cachexia Sarcopenia Muscle 13, 1785–1799. doi: 10.1002/jcsm.12975
Timothy J Thurman, C. L. W., Alkam, D., Bird, J. T., Gies, A., Dhusia, K., Robeson, M. S., et al. (2023). proteoDA: a package for quantitative proteomics. J. Open Sour. Softw. 2023:5184. doi: 10.21105/joss.05184
Tiranti, V., Rossi, E., Ruiz-Carrillo, A., Rossi, G., Rocchi, M., Didonato, S., et al. (1995). Chromosomal localization of mitochondrial transcription factor A (TCF6), single-stranded DNA-binding protein (SSBP), and endonuclease G (ENDOG), three human housekeeping genes involved in mitochondrial biogenesis. Genomics 25, 559–564. doi: 10.1016/0888-7543(95)80058-T
Toledo, J. B., Arnold, M., Kastenmüller, G., Chang, R., Baillie, R. A., Han, X., et al. (2017). Metabolic network failures in Alzheimer's disease: a biochemical road map. Alzheimer's Dement. 13, 965–984. doi: 10.1016/j.jalz.2017.01.020
Upadhya, S. R., and Ryan, C. J. (2022). Experimental reproducibility limits the correlation between mRNA and protein abundances in tumor proteomic profiles. Cell Rep. Methods 2:100288. doi: 10.1016/j.crmeth.2022.100288
Valla, J., Yaari, R., Wolf, A. B., Kusne, Y., Beach, T. G., Roher, A. E., et al. (2010). Reduced posterior cingulate mitochondrial activity in expired young adult carriers of the APOE ε4 allele, the major late-onset Alzheimer's susceptibility gene. J. Alzheimer's Dis. 22, 307–313. doi: 10.3233/JAD-2010-100129
Villemagne, V. L., Burnham, S., Bourgeat, P., Brown, B., Ellis, K. A., Salvado, O., et al. (2013). Amyloid β deposition, neurodegeneration, and cognitive decline in sporadic Alzheimer's disease: a prospective cohort study. Lancet Neurol. 12, 357–367. doi: 10.1016/S1474-4422(13)70044-9
Wilkins, H. M., Koppel, S. J., Bothwell, R., Mahnken, J., Burns, J. M., and Swerdlow, R. H. (2017). Platelet cytochrome oxidase and citrate synthase activities in APOE ε4 carrier and non-carrier Alzheimer's disease patients. Redox Biol. 12, 828–832. doi: 10.1016/j.redox.2017.04.010
Yin, J., Han, P., Song, M., Nielsen, M., Beach, T. G., Serrano, G. E., et al. (2018). Amyloid-β increases tau by mediating sirtuin 3 in Alzheimer's disease. Mol. Neurobiol. 55, 8592–8601. doi: 10.1007/s12035-018-0977-0
Yin, J., Nielsen, M., Carcione, T., Li, S., and Shi, J. (2019). Apolipoprotein E regulates mitochondrial function through the PGC-1α-sirtuin 3 pathway. Aging 11, 11148–11156. doi: 10.18632/aging.102516
Zancanaro, C., Mariotti, R., Perdoni, F., Nicolato, E., and Malatesta, M. (2007). Physical training is associated with changes in nuclear magnetic resonance and morphometrical parameters of the skeletal muscle in senescent mice. Eur. J. Histochem. 51, 305–310. doi: 10.4081/1156
Glossary
BCKDHB, 2-oxoisovalerate dehydrogenase subunit beta; MPST, 3-mercaptopyruvate sulfurtransferase; GPT2, alanine aminotransferase 2; NAPA, alpha-soluble NSF attachment protein; AD, Alzheimer's disease; ANXA4, annexin A4; APOE, apolipoprotein E; BNIP3, BCL2 interacting protein 3; BCKDHB, branched chain keto acid dehydrogenase E1 subunit beta; BCAT2, branched-chain-amino-acid aminotransferase; PRKAR1A, cAMP-dependent protein kinase type I-alpha regulatory subunit; CLIC1, chloride intracellular channel protein 1; CS, citrate synthase; C8G, complement component C8 gamma chain; CORO1A, coronin-1A; CYB5R3, cytochrome b5 reductase 3; COX5A, cytochrome c oxidase subunit 5A; COX5A, cytochrome c oxidase subunit 5A; COX8B, cytochrome c oxidase subunit 8B; COX8B, cytochrome c oxidase subunit 8B; DSC1, desmocollin-1; DBT, dihydrolipoamide branched chain transacylase E2; DNM1L, dynamin 1 like; FTH1, ferritin heavy chain 1; GPT2, glutamic-pyruvic transaminase 2; GLS, glutaminase; Hsp60, heat shock protein family D; HSPD1, member 1; IPA, INSR, Ingenuity Pathway Analysis; insulin receptor; NADP(+)2 (IDH2), isocitrate dehydrogenase; BRCC3, lys-63-specific deubiquitinase BRCC36; MGST1, microsomal glutathione S-transferase 1; MAVS, mitochondrial antiviral signaling protein; mtDNA, mitochondrial DNA; MRPL10, mitochondrial ribosomal protein L10; NDUFA11, NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 11; NDUFA6, NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 6; NDUFA11, NADH:ubiquinone oxidoreductase subunit A11; NDUFA6, NADH:ubiquinone oxidoreductase subunit A6; PC, palmitoyl-carnitine; PRDX2, peroxiredoxin 2; CDS2, Phosphatidate cytidylyltransferase 2; PPARGC1A, PPARG coactivator 1 alpha; PCCA, propionyl-CoA carboxylase subunit alpha; RANBP3, ran-binding protein 3; RICTOR, RPTOR independent companion of MTOR complex 2; PPP3CB, serine/threonine-protein phosphatase 2B catalytic subunit beta isoform; SSBP1, single-stranded DNA-binding protein, mitochondrial; SIRT3, sirtuin 3; SLC2A1, solute carrier family 2, facilitated glucose transporter member 1; SLC25A5, solute carrier family 25 member 5; SUOX, sulfite oxidase, mitochondrial; TXN2, thioredoxin; TXN2, thioredoxin 2; TRAP1, TNF receptor associated protein 1; TFAM, transcription factor A, mitochondrial; TFEB, transcription factor B; BTF3, transcription factor BTF3; TNF, tumor necrosis factor; TP53, tumor protein p53; UQCR11, ubiquinol-cytochrome c reductase, complex III subunit XI; VSN, variance stabilization normalization; CACNG6, voltage-dependent calcium channel gamma-6 subunit.
Keywords: APOE4, Alzheimer's disease, skeletal muscle, mitochondria, proteomics, mice
Citation: Johnson CN, Lysaker CR, McCoin CS, Evans MR, Thyfault JP, Wilkins HM, Morris JK and Geiger PC (2024) Skeletal muscle proteome differs between young APOE3 and APOE4 targeted replacement mice in a sex-dependent manner. Front. Aging Neurosci. 16:1486762. doi: 10.3389/fnagi.2024.1486762
Received: 26 August 2024; Accepted: 24 October 2024;
Published: 20 November 2024.
Edited by:
Anamitra Ghosh, Wave Life Sciences Ltd., United StatesReviewed by:
Manasee Gedam, Baylor College of Medicine, United StatesSimone Mastrogiacomo, Aix-Marseille Université, France
Copyright © 2024 Johnson, Lysaker, McCoin, Evans, Thyfault, Wilkins, Morris and Geiger. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Paige C. Geiger, cGdlaWdlckBrdW1jLmVkdQ==
†These authors have contributed equally to this work and share senior authorship
 Chelsea N. Johnson
Chelsea N. Johnson Colton R. Lysaker
Colton R. Lysaker Colin S. McCoin
Colin S. McCoin Mara R. Evans
Mara R. Evans John P. Thyfault
John P. Thyfault Heather M. Wilkins
Heather M. Wilkins Jill K. Morris
Jill K. Morris Paige C. Geiger
Paige C. Geiger