
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Aging Neurosci., 28 June 2024
Sec. Alzheimer's Disease and Related Dementias
Volume 16 - 2024 | https://doi.org/10.3389/fnagi.2024.1400447
This article is part of the Research TopicSynaptic Health as a Prognostic Tool in Alzheimer's and Related DementiasView all 6 articles
 Allen L. Pan1
Allen L. Pan1 Mickael Audrain2
Mickael Audrain2 Emmy Sakakibara1
Emmy Sakakibara1 Rajeev Joshi1
Rajeev Joshi1 Xiaodong Zhu3
Xiaodong Zhu3 Qian Wang4,5
Qian Wang4,5 Minghui Wang4,5
Minghui Wang4,5 Noam D. Beckmann4
Noam D. Beckmann4 Eric E. Schadt4
Eric E. Schadt4 Sam Gandy2,6
Sam Gandy2,6 Bin Zhang4,5
Bin Zhang4,5 Michelle E. Ehrlich2,4,7*
Michelle E. Ehrlich2,4,7* Stephen R. Salton1,8*
Stephen R. Salton1,8*Introduction: Dual specificity protein phosphatase 6 (DUSP6) was recently identified as a key hub gene in a causal VGF gene network that regulates late-onset Alzheimer’s disease (AD). Importantly, decreased DUSP6 levels are correlated with an increased clinical dementia rating (CDR) in human subjects, and DUSP6 levels are additionally decreased in the 5xFAD amyloidopathy mouse model.
Methods: To investigate the role of DUSP6 in AD, we stereotactically injected AAV5-DUSP6 or AAV5-GFP (control) into the dorsal hippocampus (dHc) of both female and male 5xFAD or wild type mice, to induce overexpression of DUSP6 or GFP.
Results: Barnes maze testing indicated that DUSP6 overexpression in the dHc of 5xFAD mice improved memory deficits and was associated with reduced amyloid plaque load, Aß1–40 and Aß1–42 levels, and amyloid precursor protein processing enzyme BACE1, in male but not in female mice. Microglial activation, which was increased in 5xFAD mice, was significantly reduced by dHc DUSP6 overexpression in both males and females, as was the number of “microglial clusters,” which correlated with reduced amyloid plaque size. Transcriptomic profiling of female 5xFAD hippocampus revealed upregulation of inflammatory and extracellular signal-regulated kinase pathways, while dHc DUSP6 overexpression in female 5xFAD mice downregulated a subset of genes in these pathways. Gene ontology analysis of DEGs (p < 0.05) identified a greater number of synaptic pathways that were regulated by DUSP6 overexpression in male compared to female 5xFAD.
Discussion; In summary, DUSP6 overexpression in dHc reduced amyloid deposition and memory deficits in male but not female 5xFAD mice, whereas reduced neuroinflammation and microglial activation were observed in both males and females, suggesting that DUSP6-induced reduction of microglial activation did not contribute to sex-dependent improvement in memory deficits. The sex-dependent regulation of synaptic pathways by DUSP6 overexpression, however, correlated with the improvement of spatial memory deficits in male but not female 5xFAD.
Late-onset Alzheimer’s disease (LOAD) is a progressive neurological disorder that affects more than 5 million elderly in the United States (Matthews et al., 2019). Currently, there are no effective drugs that can permanently prevent the progression of Alzheimer’s disease (AD)-associated cognitive decline. Previously, members of our team at Mount Sinai used multiscale causal network-based approaches to identify the dual specificity protein phosphatase 6 (DUSP6), also known as mitogen-activated protein kinase (MAPK) phosphatase 3 (MKP3), as a key hub in the VGF gene network that regulates AD (Beckmann et al., 2020). DUSP6 is a member of the dual specificity protein phosphatase (DUSP) family that regulates MAPK activity (Chen et al., 2019). MAPK signaling pathways are involved in many cellular processes, including inflammation (Manzoor and Koh, 2012), and AD pathogenesis and progression in brain is associated with and potentially driven by immunological mechanisms, including the upregulation of disease-associated microglial (DAM) genes (Sobue et al., 2021).
Neuroinflammation is a double-edged sword, as it is hypothesized to be either protective or detrimental, perhaps depending on disease stage (Castranio et al., 2023). Microglia are the innate immune cells of the brain, responsible for maintenance of brain homeostasis through the detection and elimination of harmful stimuli. However, activation of microglia in a disease state can exert both injurious and favorable effects in context-dependent manners. MAPKs, including extracellular signal-regulated kinase (ERK), c-Jun N-terminal kinase (JNK), and p38, are involved in the regulation of early immune responses through serial phosphorylation events (Cargnello and Roux, 2011), which are involved in cellular proliferation and inflammation and are usually transient, while prolonged activation can lead to chronic inflammatory microglial responses (Kim et al., 2004) that can be damaging. Phosphoproteomic analysis of microglia, isolated from 5xFAD mice, show a significant increase of phosphorylated ERK (Chen et al., 2021), suggesting an imbalance in MAPK signaling. DUSPs, including DUSP6, are involved in maintaining the homeostatic level of phosphorylated ERK. DUSP6 is a cytoplasmic MAPK phosphatase that has high selectivity for extracellular signal-regulated kinases 1 and 2 (ERK1/2), which is associated with the high affinity binding of its N-terminal kinase-interacting motif (KIM) to ERK1/2 (Arkell et al., 2008). DUSP6 is downregulated in several neurological and neuropsychiatric diseases including AD (Banzhaf-Strathmann et al., 2014), schizophrenia (Kondo et al., 2022), and major depressive disorder (Labonte et al., 2017), the latter frequently co-morbid with AD (Martín-Sánchez et al., 2021; Arnaud et al., 2022).
Amyloid plaques and neurofibrillary tangles (NFTs) are the pathological hallmarks of AD. Microglia are involved in clearance of aggregated proteins, including amyloid beta (Aβ), through phagocytosis. The exact mechanisms involved in the phagocytosis of amyloid plaques by microglia are not fully understood. Microglia recognize Aβ through triggering receptor expressed on myeloid cells 2 (TREM2). Binding of Aβ to TREM2 enhances the interaction of TREM2 with its adaptor TYROBP, which induces downstream signaling and promotes microglial clearance of amyloid plaques (Zhao et al., 2018). TREM2 and phosphorylated ERK (pERK) levels are increased in microglia acutely isolated from the 5xFAD AD mouse model, and pERK is an upstream regulator of DAM gene expression including Trem2 and Tyrobp (Chen et al., 2021). Interestingly, DUSP6 overexpression has been demonstrated to protect against Aβ-induced neural stem cell injury, including against oxidative and ER stress and mitochondrial dysfunction, and to reverse Aβ-induced ERK1/2 activation (Liao et al., 2018).
We investigated the role that DUSP6 plays in AD pathogenesis and progression, determining the effects of adeno-associated virus (AAV)-mediated hippocampal DUSP6 overexpression in 5xFAD mice on AD-related behavioral, neuropathological, and transcriptomic phenotypes. DUSP6 overexpression improved spatial memory, decreased plaque load, and decreased BACE1 expression only in male 5xFAD mice. Reduced hippocampal microglial activation and msh homeobox 3 (Msx3) gene expression were observed in male and female 5xFAD overexpressing DUSP6 (5xFAD-DUSP6), while transcriptomic profiling of female 5xFAD further demonstrated that DUSP6 overexpression downregulated neuroinflammatory and ERK/MAPK signaling pathways. Gene ontology (GO) analysis of differentially expressed genes showed that DUSP6 regulated more pathways associated with synaptic structure and function in male 5xFAD-DUSP6 than in female 5xFAD-DUSP6. Our findings suggest that DUSP6 may function in sex-specific and shared pathways to regulate neurodegeneration, neuroinflammation, and synaptic function in the 5xFAD mouse model.
5xFAD transgenic mice that overexpress human APP (695) with Swedish (K670N, M671L), Florida (I716V) and London (V717I) familial AD (FAD) mutations and human Presenilin1 (PS1) with the M146L and L286V FAD mutations (Oakley et al., 2006) were purchased from Jackson Labs (Bar Harbor, ME; JAX#34840) and were maintained on a mixed B6/SJL genetic background as described (Beckmann et al., 2020). Female and male wild-type (WT) and 5xFAD mice at 4 months of age were stereotactically infused using a twenty-five gauge needle (Hamilton, Reno, NV) with 1.0 μL of AAV5-GFP or AAV5-DUSP6 (4 × 1012vg/ml) into dorsal hippocampus (dHc) (AP = −2.0 mm, ML = ±1.5 mm, and DV = −2.0 mm relative to Bregma) at a rate of 0.2 μL per minute. Data obtained from female and male WT and 5xFAD mice overexpressing GFP, used in this study, were previously published in an analysis of DUSP4 overexpression, and all were co-sequenced with those from DUSP6 overexpressing mice to avoid batch effects (Pan et al., 2022). AAV5-injected mice were allowed to recover for a month before behavioral testing. AAV5-GFP (control), and AAV5-DUSP6 (VectorBuilder Inc., Chicago, IL; AAV-5′ITR-CAG-mDUSP6-WPRE-BGHpA-3′ITR) (AAV5 serotype/AAV2 genotype) were prepared by the Vector Core at the University of North Carolina at Chapel Hill. All mice were housed under standard conditions (12 h light-dark cycle with ad libitum access to food and water). All experimental procedures were conducted in accordance with the NIH guidelines for animal research and were approved by the Institutional Animal Care and Use Committee (IACUC) at the Icahn School of Medicine at Mount Sinai (ISMMS).
The Barnes maze test was performed using a standard apparatus (Barnes, 1979), as described (Audrain et al., 2019). Briefly, 5-month-old 5xFAD or WT mice were habituated in the testing room for 30 min prior to the test. Then the mice were transferred to the center of the platform using a closed chamber where they remained for 10 s prior to exploring the maze for 3 min. Any mice that failed to enter the escape box within 3 min were directed to the escape box by the experimenter, and the latency was recorded as 180 s. Mice were allowed to remain in the escape box for 1 min before being transferred back to their cages. After each test, the platform and the escape box were cleaned with 70% ethanol to eliminate the use of olfactory cues to locate the target hole. Two trials were conducted, and all trials were recorded by video camera and analyzed with ANY-maze video tracking software (Stoelting Co, Wood Dale, USA).
The Barnes maze data for 5xFAD and WT mice overexpressing GFP used in this study were previously published in our DUSP4 overexpression paper (Pan et al., 2022). These mice were analyzed in the same Barnes maze test with those overexpressing DUSP6 that are reported here.
Two days after the final behavioral test, mice were transcardially perfused with 20 mL ice-cold phosphate buffered saline (PBS). The right hemisphere was fixed in 4% PFA for 24 hr followed by incubation in 30% sucrose until the brains sunk to the bottom. Then the brains were cut into 30 μm coronal sections by a cryostat (Leica). The contralateral hemisphere was dissected to isolate dHc, which was cut symmetrically in half. Half of the dHc was homogenized in RIPA buffer (Millipore Sigma) containing phosphatase (Roche) and protease (Roche) inhibitors, centrifuged for 20 min at 15,000 × g and the supernatant was collected, while the other half was used for RNA extraction employing the RNeasy Mini Kit (Qiagen).
The QIAzol® Lysis Reagent (Qiagen) and the miRNeasy® Mini Kit (Qiagen) were used to extract RNAs from hippocampi following the manufacturer’s instructions. The purities and the concentration of the RNA extracts were determined by NanoDrop 2000c (ThermoFisher Scientific). A high-capacity RNA-to-cDNA™ kit (Applied Biosystems, 4387406) was used to reverse transcribe the total RNA into cDNA in a 20 μl reaction volume using the thermal cycler (T100, Bio-Rad). The QuantStudio 7 Flex Real-Time PCR System (Applied Biosystems) was used for Real-time PCR reactions with the Maxima SYBR Green/ROX qPCR Master Mix (ThermoFisher Scientific, K0221). Reactions were carried out in duplicate in 384-well plates (Applied Biosystems) according to the manufacturer’s three-step cycling protocol. The relative gene expression of each transcript was normalized to the reference gene Gapdh with the ΔCt method. The sequences of oligonucleotides used are:
Gapdh 5′-AACGACCCCTTCATTGACCT-3′ and
5′-TGGAAGATGGTGATGGGCTT-3′,
Dusp4 5′-CCTGCTTAAAGGTGGCTATGAGA-3′ and
5′-GGTGCTGGGAGGTACAGGG-3′,
Dusp6 5′-CTCGGATCACTGGAGCCAAAAC-3′ and
5′-TCTGCATGAGGTACGCCACTGT-3′,
Vgf 5′-CGAAGAAGCAGCAGAAGCTC-3′ and
5′-TCGAAGTTCTTGGAGCAAGG-3′,
Sst 5′-CCGTCAGTTTCTGCAGAAGT-3′ and
5′-CAGGGTCAAGTTGAGCATCG-3′,
Bdnf 5′-GAAGAGCTGCTGGATGAGGAC-3′ and
5′-CGAGTTCCAGTGCCTTTTGTC-3′,
Scg2 5′- AGGGTTGACGAGGAACAAA-3′ and
5′-CTGGACTGGGCACTCTCTTC-3′,
Mouse Ps1 5′-CAAAAACAGAGAGCAAGCCC-3′ and
5′-TCTCTCAAGTCACTGAGGGACA-3′,
Human PS1 5′-GCAGTATCCTCGCTGGTGAAGA-3′ and
5′-CAGGCTATGGTTGTGTTCCAGTC-3′,
Ps2 5′-CTGGTGTTCATCAAGTACCTGCC-3′ and
5′-TTCTCTCCTGGGCAGTTTCCAC-3′,
Adam10 5′-TAAGGAATTATGCCATGTTTGCTGC-3′ and
5′-ACTGAACTGCTTGCTCCACTGCA-3′,
Adam17 5′-TTGGAGCAGAACATGACCCTGATGG-3′ and
5′-TGCAGCAGGTGTCGTTGTTCAGGTA-3′,
Bace1 5′-TCTTTTCCCTGCAGCTCTGT-3′ and
5′-ACTGCCCGTGTATAGCGAGT-3′
Nct 5′-CCAAGCTTCCCATTGTGTGC-3′ and
5′-TGCTGAAGGTGCTCTGGATG-3′
Aph1a 5′- GTGCTGCTGTCTCTGTCCTT-3′ and
5′-TCTGTCGGATGGAGATGGGT-3′
Aph1b 5′- CTGGGGCGTTGTGTTCTTTG-3′ and
5′-AAATGCCCAGATGCCCATGA-3′
Aph1c 5′-TTCCTCATCGCTGGTGCTTT-3′ and
5′-CGCTCCGAAGATGAGCAGAT-3′
Cd68 5′-TCCAAGCCCAAATTCAAATC-3′ and
5′-ATGGGTACCGTCACAACCTC-3′
Aif1 5′-GTCGCACTCAGCAACAGG-3′ and
5′-ACTTCTGGTCACAGAGGAACTC-3′
Human APP 5′-GTGGCATTCTTTTGGGGCTG-3′ and
5′-GAACCTGGTCGAGTGGTCAG-3′
Mouse APP 5′-TCCGTGTGATCTACGAGCGCAT-3′ and
5′-GCCAAGACATCGTCGGAGTAGT–3′
RT-PCR data for the following VGF network genes (Dusp4, Dusp6, Sst, Bdnf, mouse Ps1, human Ps1, and Ps2) and the following APP processing enzyme genes (Adam10, Bace1, Nct, Aph1a, Aph1b, and Aph1c), analyzing RNA levels in 5xFAD and WT overexpressing GFP, were previously published with our DUSP4 overexpression RT-PCR data (Pan et al., 2022), all of which were analyzed in the same RT-PCR assay with the DUSP6 data reported here.
Coronal sections (30 μm thickness) from dHc were first washed with PBS, and then incubated with the following primary antibodies in 0.1% Triton X-100 in PBS overnight at 4°C: anti-DUSP6 (1:1,000, Abcam; ab76310); anti-Aβ (1:1,000, 6E10, Biolegend, 803001); anti-GFAP (1:1000, Abcam, ab53554); anti-NeuN (1:1000, Invitrogen, MA5-33103), or anti-IBA1 (1:1,000, Fijifilm, 019-19741). On the second day, sections were rinsed with PBS and incubated for 1 h at room temperature with appropriate secondary antibodies: anti-rabbit Alexa Fluor IgG 488 or 568 (1:1,000, Invitrogen) and anti-mouse IgG Alexa 488 (1:1000, Invitrogen). Then sections were then washed with PBS, and were allowed to dry before mounting with Hardset Vectashield plus DAPI mounting medium (Vector Laboratories, H1500) and sealed with coverslips. Thioflavin S (Sigma-Aldrich, T1892, 1% w/v stock solution) was used for labeling amyloid deposits. Images were acquired by Nikon Eclipse TE 200 and Zeiss LSM 780 microscopes. The images were captured with constant parameters, and the quantification of images was conducted by an operator blinded to the treatment groups. Staining was analyzed by Fiji software (ImageJ, v2.1.0) at the same threshold setting for each immunostained marker.
For microglial number, IBA1-immunolabeled sections were thresholded and particles were analyzed by Fiji (ImageJ, v2.1.0). A “microglial cluster” was counted when three or more microglial somas occurred within, or touching the margins of, a 20 μm2 virtual graticule subregion in the hippocampal region (Paasila et al., 2020). IBA1-immunolabeled sections and Aβ-immunolabeled sections were prepared from separate slices obtained from the same 5xFAD mice and hippocampal region with Aβ presence. Microglial clusters were counted only in the 568 nm (IBA1-positive) channel to minimize the false-positive counts in the presence of Aβ.
NeuN (neuron), IBA1 (microglia), or GFAP (astrocyte) was co-stained with DUSP6 in hippocampal brain sections, as described above. Colocalization was analyzed by JACoP plugin in Fiji (ImageJ, v2.1.0). The output of colocalization was calculated using thresholded Manders’ correlation coefficient of global statistical analysis, considering pixel intensity distributions. At least 4 brain sections from each animal were analyzed, and the percentage of the fraction of DUSP6 in the neurons, microglia, or astrocytes was expressed as the mean ± standard error of the mean (SEM).
Hippocampal amyloid plaques were quantified based on mean gray value of percentage thresholded area. The total numbers of amyloid plaque clusters from each brain section were manually counted using Fiji software (ImageJ, v2.1.0). The results are represented as 6E10 intensity or number of plaques in dHc. The image quantification of 6E10 was performed by an operator blinded to the treatment groups. Thioflavin S-labeled plaque number and size in hippocampi were analyzed by Fiji software (ImageJ, v2.1.0) as described (Christensen and Pike, 2020).
Hippocampal Aβ1–40 and Aβ1–42 from RIPA-extracted supernatants were quantified by human/rat Aβ1–40/1–42 ELISA kits (Wako, #294-64701, #290-6260) following the manufacturer’s instructions. Absolute concentrations of Aβ were normalized to the initial tissue weight.
Protein samples were resolved by electrophoresis on 4–12% Bis-Tris gels (Bio-Rad) and were transferred to polyvinylidene difluoride membranes using the iBlot system (Invitrogen). Membranes were then blocked in Odyssey blocking buffer for 1 h at room temperature before incubation with the following primary antibodies in blocking buffer (Odyssey) and 0.1% Tween-20 at 4°C overnight: anti-DUSP6 (1:1,000, Abcam; ab76310); anti-BACE1 (1:1,000, Abcam; ab2077); or anti-Aβ (1:1,000, Biolegend, 803001). Membranes were washed the next day with 0.1% Tween-20 in PBS followed by incubation in a mixture of secondary antibodies: goat anti-rabbit 800CW (1:15,000, LI-COR, Lincoln, NE) and goat anti-mouse 680LT (1:20,000, LI-COR, Lincoln, NE) in Odyssey blocking buffer with 0.1% Tween-20 and 0.01% SDS at room temperature for 1 hr. After the incubation, the membranes were washed with 0.1% Tween-20 in PBS and then were washed with PBS. After the final wash with PBS, the membranes were analyzed using an Odyssey infrared imager (LI-COR, Lincoln, NE). Bands were quantified using Odyssey Imager analysis software and were normalized using β-actin as an internal loading control.
Mouse hippocampal RNA samples were sequenced by Novogene for transcriptomic profiling using Illumina Novaseq 6000 S4 flow cells. RNA quality of each sample was assessed and only the samples with RNA integrity number (RIN) > 9 were included. Non-directional libraries were constructed with an NEB kit using the manufacturer’s protocol. RNA sequencing assays were performed after ribosomal RNA depletion by Ribo-Zero. RNA reads were aligned to the mm10 reference genome using STAR (Dobin et al., 2013) (version 2.7.5b) to obtain the raw counts for each gene. Differential expression analysis was performed in R (version 3.6.3) using edgeR (Robinson et al., 2010). Genes with counts per million reads (cpm) > 1 in at least 5 samples were included for further analysis. Differentially expressed genes with FDR < 0.05 were considered statistically significant in each comparison.
Transcriptomic data for WT and 5xFAD overexpressing GFP were previously published with our DUSP4 overexpression dataset (Pan et al., 2022), all of which were sequenced in the same batch with the DUSP6 transcriptomics reported here.
The expression patterns of DUSP6 genes in human AD brains, with a sample size of 82-133 per sex, were analyzed using our recently published RNA-seq data in the hippocampal gyrus (PHG) region of postmortem brains of AD and controls from the Mount Sinai Brain Bank (MSBB) (Wang M. et al., 2018; Wang et al., 2021). Here we used the preprocessed data which had been normalized and corrected for known covariates, except sex which was excluded from the covariate correction. The gender-specific gene expression distribution stratified by clinical dementia rating (CDR) scale were presented using boxplots. In addition, we calculated the Spearman correlation coefficients between gene expression values and CDR scale in males and females, separately.
The raw RNA-seq data from hippocampus of 4, 12 and 18 month old 5xFAD mice were obtained from the AMP-AD Portal (2021). Details about the sample processing, library construction and sequencing are available at SYNAPSE (AMP-AD Portal, 2021).
Paired-end 43bp sequencing reads were aligned to mouse reference genome mm10 using STAR aligner v2.5.3a (Dobin et al., 2013) guided by a customized mouse GENCODE gene model release v15. Mapped reads were summarized to gene levels using the featureCounts program v1.6.3 (Liao et al., 2014). Raw count data were normalized as counts per million (CPM) at the log2 scale by the voom function in the R limma package (Ritchie et al., 2015). Expression patterns of Dusp6 were visualized per age-genotype-gender combinations by boxplot. The expression difference between 5xFAD and age-gender-matched control was calculated by one-sided t-test.
hiPSC-derived microglial cells were generated using defined conditions with several modifications to the previously published protocol (Mancuso et al., 2019; Fattorelli et al., 2021). All hiPSC lines with a normal karyotype were regularly checked and confirmed negative for mycoplasma. hiPSCs, maintained in complete mTeSR1 medium (StemCell Technologies) according to WiCell protocols, were dissociated by Accutase (Thermo Fisher Scientific) to obtain a single-cell suspension. Approximately 10,000 cells were plated in each well of an ultra-low-attachment 96-well plate (Corning) in complete mTeSR1 medium supplemented with human BMP4 (50 ng/mL), human VEGF (50 ng/mL), human SCF (20 ng/mL) and 10 μM Rho-associated protein kinase inhibitor (ROCKi, Selleck Chemicals). Embryoid bodies were fed every day from day 1 to day 3, then transferred to 6-well plates (Corning) in the differentiation media containing X-VIVO 15 media (LONZA), 2 mM GlutaMAX, 50 U/mL of penicillin-streptomycin, 0.055 mM 2-mercaptoethanol, and supplemented with human SCF (50 ng/mL), human M-CSF (50 ng/mL), human IL3 (50 ng/mL), human FLT3 (50 ng/mL) and human TPO (5 ng/mL). After 4 days, embryoid bodies were fed with the same differentiation media. On differentiation day 11, a full medium change was performed and embryoid bodies were maintained in differentiation media plus human FLT3 (50 ng/mL), human M-CSF (50 ng/mL) and human GM-CSF (25 ng/mL). On day 18, floating microglial progenitors in the medium were collected and cultured in RPMI 1640 medium (Thermo Fisher Scientific) containing 2 mM GlutaMAX, 50 U/mL of penicillin-streptomycin, 10 ng/mL GM-CSF and 100 ng/mL IL-34 for 2 weeks in order to generate mature microglial cells. All cytokines were purchased from R&D Systems.
hiPSC-derived cortical neurons were generated as described (Chambers et al., 2009; Qi et al., 2017). hiPSCs were dissociated with Accutase and plated at 200,000 cells per cm2 onto Matrigel-coated plates in complete mTeSR1 medium with ROCKi (10 μM). After 1–2 days when cells were 100% confluent, the medium was replaced with differentiation media (DMEM/F12:Neurobasal (1:1), 2 mM GlutaMAX, 1% N2 supplement, 2% B27 minus Vitamin A supplement) containing LDN193189 (100 nM, Stemgent), SB431542 (10 μM, Selleck Chemicals) and XAV939 (1 μM, Tocris) for 10 days of differentiation. Cultures were fed with differentiation media with XAV939 (1 μM) for an additional week to allow the expansion of neural progenitor cells. Neural progenitor cells were then dissociated and replated on poly-l-ornithine/fibronectin/laminin-coated plates and maintained in BrainPhys Basal medium (StemCell Technologies) containing B-27 supplement, BDNF (40 ng/mL, R&D Systems), GDNF (40 ng/mL, R&D Systems), Laminin (1 μg/mL, Life Technologies), L-Ascorbic acid (200 μM, Sigma), dbcAMP (250 μM, Sigma), for neuronal differentiation and maturation, with the addition of SU5402 (10 μM, Selleck Chemicals), DAPT (10 μM, Tocris) and PD0325901 (10 μM, Selleck Chemicals) for the first week of differentiation.
Brain sections (30 μm) were used in RNA in situ hybridization (RNAscope®). RNAscope® fluorescent in situ hybridization (FISH) was performed according to the manufacturer’s instructions (Advanced Cell Diagnostics, Inc.). Briefly, Dusp6 and NeuN mRNAs of the mounted sections were probed with Mouse Dusp6 (Advanced Cell Diagnostics, 429321-C2-R) and Rbfox3-C4 (Advanced Cell Diagnostics, 313311-C4) from ACD at 1:50 dilution. Then the microglial cells were detected by rabbit anti-IBA1 antibody (Fujifilm Wako Chemicals, 01919741) and secondary Alexa Fluor 488 anti-rabbit antibody. Finally, the slides were sealed with Hardset Vectashield plus DAPI mounting medium (Vector Laboratories, H1500). Images were obtained by Zeiss LSM 780 microscopy.
Graphs represent the mean of all samples in each group ± SEM. Sample sizes (n values) and statistical tests are indicated in the figure legends. One-way or two-way ANOVA was used for multiple comparisons. A Student’s t-test was used for simple comparisons. Significance is reported at *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
To determine DUSP6 gene expression patterns in human AD brains, we used our recently published RNA-seq data from the hippocampal gyrus (PHG) region of postmortem brains of AD subjects and controls from the Mount Sinai Brain Bank (MSBB) (Wang M. et al., 2018; Wang et al., 2021). The Spearman correlation coefficients showed that the downregulation of DUSP6 gene expression is correlated with increased clinical dementia rating (CDR) scores in both sexes (Figure 1A). Although our current and published studies with manipulation of DUSP expression (El Gaamouch et al., 2020) utilize 5xFAD mice on a mixed B6/SJL genetic background, the availability of raw RNA-seq data (AMP-AD portal) from 5xFAD mice on a congenic C57BL/6J genetic background allowed us to determine hippocampal Dusp6 gene expression at 4, 12, and 18 months of age (Figure 1B). Both female and male 5xFAD mice showed a significant decrease in hippocampal Dusp6 expression at 4 and 12 months of age, which normalized by 18 months of age (Figure 1B).
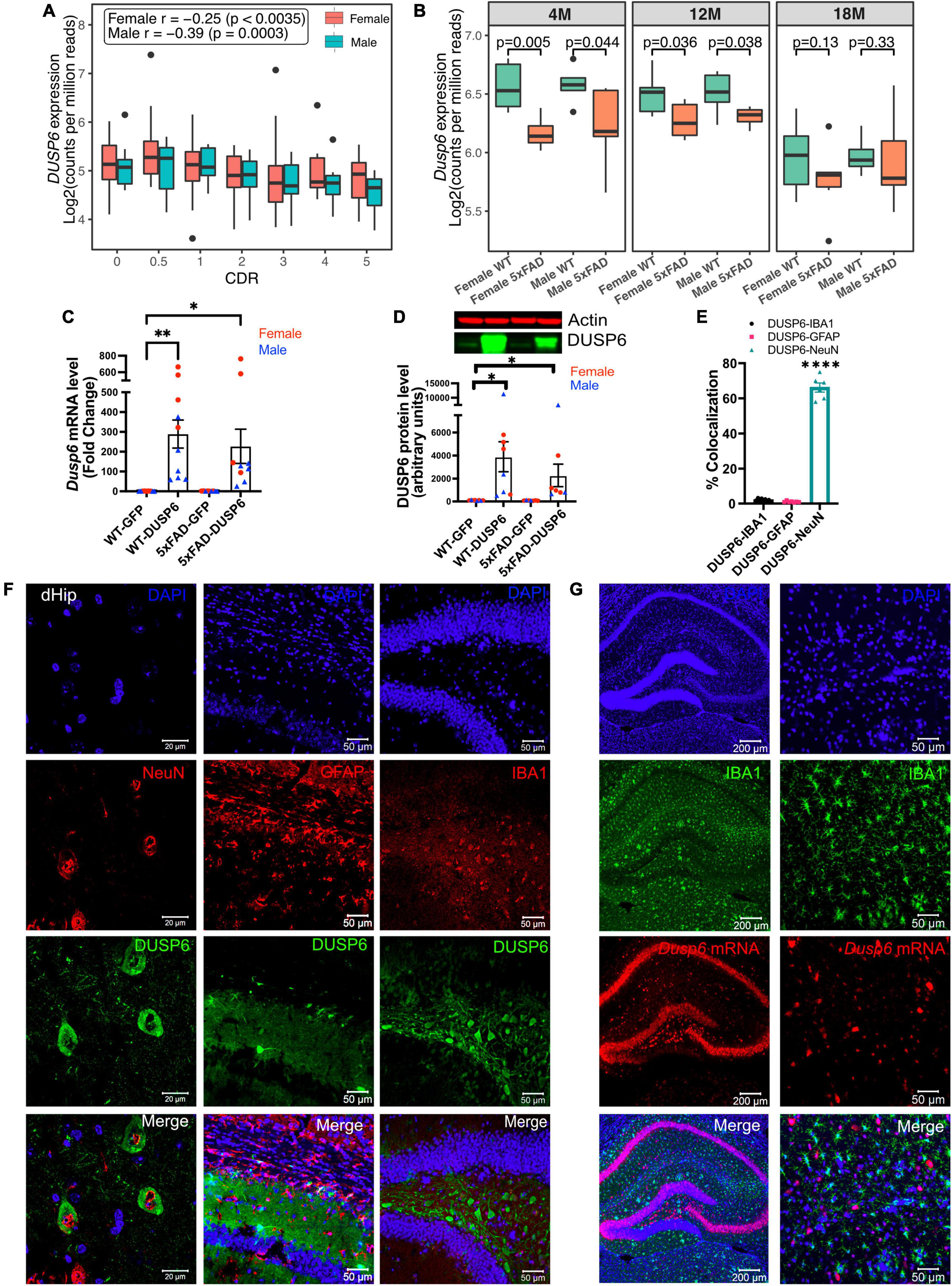
Figure 1. AAV5-mediated overexpression of DUSP6 in dorsal hippocampus (dHc) of 5-month-old 5xFAD and WT mice. (A) Boxplot shows expression of hippocampal DUSP6 mRNA [Log2(counts per million reads)] in AD postmortem brain samples from the Mount Sinai Brain Bank (MSBB) stratified by clinical dementia rating CDR; r, Spearman’s correlation coefficient. n = 82–133/sex. (B) Boxplots compare the expression of hippocampal Dusp6 mRNA among female and male 5xFAD and WT mice at different ages using RNAseq data obtained from the AMP-AD portal (see 2 Materials and methods). n = 4–16/sex/age. (C) RT-PCR and (D) western blot analyses of DUSP6 overexpression in 5xFAD and WT, n = 9–10 mice/group. (E) Graph shows percentage of colocalization using Mander’s correlation coefficient, and the thresholded Mander’s M-values corresponding to the fraction of DUSP6 in NeuN (neurons), IBA1 (microglia), or GFAP (astrocytes) analyzed by the JACoP plugin from ImageJ, n = 6–9 mice/group. (F) Co-staining NeuN (left), GFAP (middle), or IBA1 (right) with DAPI and DUSP6 in hippocampi of WT-DUSP6 mice. (G) RNA scope images of IBA1 protein and Dusp6 mRNA in hippocampi of WT-DUSP6. Scale bars = 20, 50, or 200 μm. Error bars represent means ± SEM. Statistical analyses were performed using a one-way ANOVA followed by a Tukey’s post-hoc test, *p < 0.05, **p < 0.01, ****p < 0.0001.
Utilizing proteinatlas.org and brainrnaseq.org to determine cell-type specific expression of DUSP6 mRNA in human and mouse brain, we found that DUSP6 mRNA was most abundant in endothelial cells, and was additionally expressed in neurons, astroglia, and microglia (Supplementary Figure 1A). Consistent with these data, we detected DUSP6 mRNA in cultured human iPSCs differentiated into either microglia or neurons (Supplementary Figure 1B). To determine whether overexpression of DUSP6 in the 5xFAD mouse model rescued AD-related phenotypes and neuropathology, we stereotactically infused AAV5-DUSP6 or AAV5-GFP (control) into dHc at 4 months of age. Western blot and RT-qPCR analyses confirmed the overexpression of DUSP6 protein and mRNA in WT or 5xFAD mice administered AAV-DUSP6 (Figures 1C, D). To determine in which cell type(s) DUSP6 was overexpressed, we quantified the colocalization of cell-type specific markers IBA1 (microglia), NeuN (neurons), or GFAP (astrocytes), with DUSP6, which showed that overexpressed DUSP6 co-localized with NeuN, but not with GFAP or IBA1 (Figures 1E, F). RNA scope additionally confirmed that AAV-mediated DUSP6 mRNA was not expressed in microglia (Figure 1G) or astrocytes (data not shown). Overexpression of DUSP6 detected predominantly in neurons is consistent with the previously reported neurotropism of AAV5 (Haery et al., 2019).
We assessed spatial learning in male and female mice overexpressing DUSP6 or GFP in the dHc at 5 months of age using the Barnes maze test. During the 5-day training session, male 5xFAD-GFP took longer to enter the hidden tunnel (escape box) (Figure 2A) and traveled less distance in the target quadrant (location of escape box) (Figure 2B), compared to male WT-GFP, consistent with a learning behavior deficit. Male 5xFAD mice overexpressing DUSP6 in dHc (abbreviated 5xFAD-DUSP6) took significantly less time to enter the hidden tunnel compared to those overexpressing GFP (5xFAD-GFP), but spent significantly more time than WT-GFP (Figure 2A), suggesting that DUSP6 overexpression may partially rescue learning behavior deficits in male 5xFAD. No significant differences in distance traveled in the target quadrant were found comparing male 5xFAD-DUSP6 and 5xFAD-GFP during the 5-day training period, although on days 3 and 5, male 5xFAD-DUSP6 did show a trend toward an increase in the percentage of distance traveled in the target quadrant compared to 5xFAD-GFP (Figure 2B). On the other hand, female 5xFAD-DUSP6 showed no significant changes in Barnes maze performance compared to 5xFAD-GFP (Figures 2C, D). These results indicated that DUSP6 overexpression in dHc partially rescued learning behavior deficits in male but not female 5xFAD mice, and contrasted with previously reported dHc DUSP4 overexpression that resulted in rescued learning behavior deficits in female but not male 5xFAD mice (Pan et al., 2022), with all mice analyzed in the same Barnes maze test (see Materials and Methods).
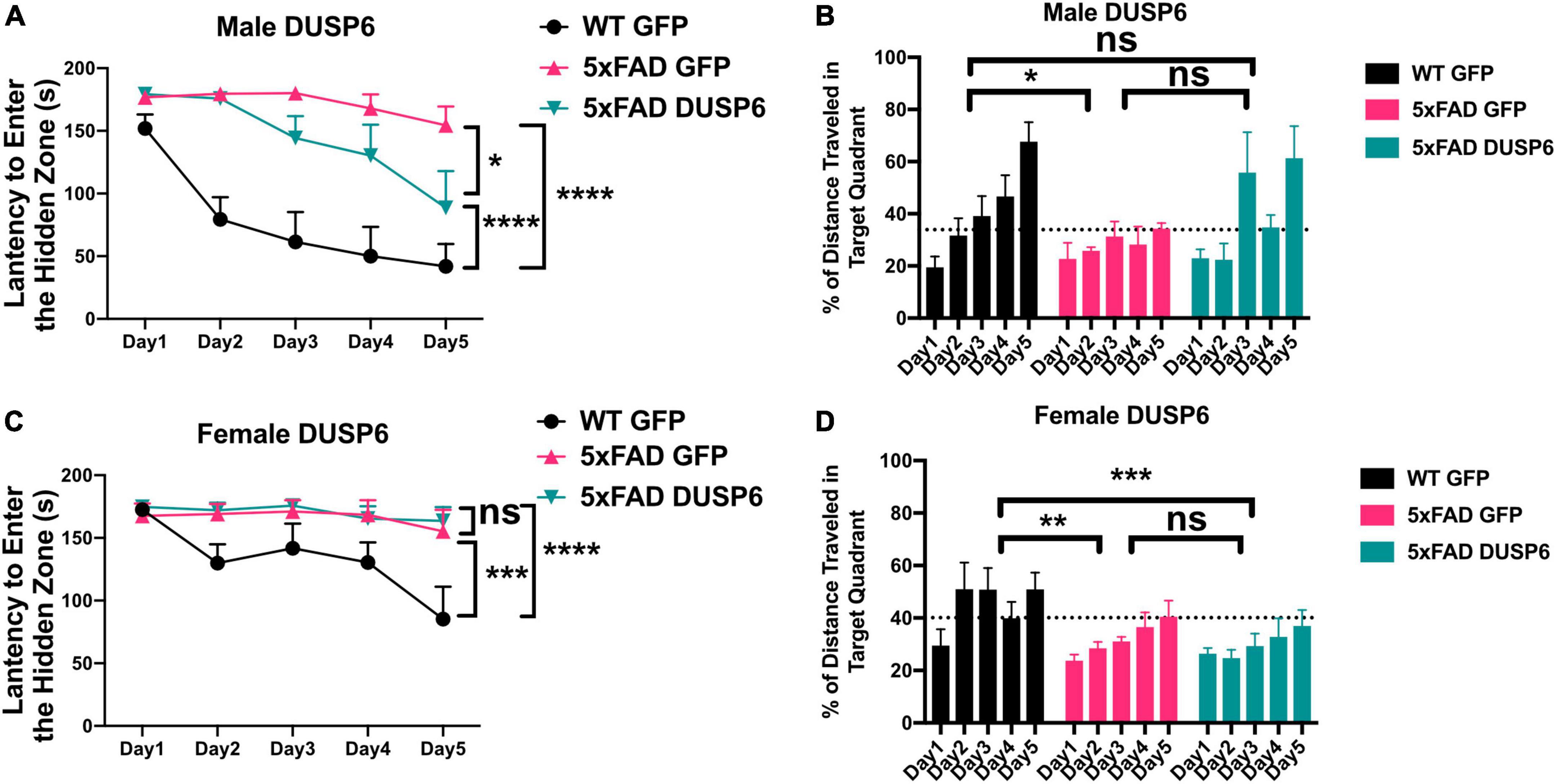
Figure 2. Barnes Maze testing of 5xFAD mice overexpressing DUSP6. (A,B) Male and (C,D) female 5xFAD and WT overexpressing DUSP6 or GFP were tested in the Barnes Maze at 5 months of age, n = 6–7 mice/group. Training was performed in a 5-day session with two trials per day, and the time (A,C) spent to enter the hidden tunnel and the percentage of distance traveled in the target quadrant (B,D) were recorded. Error bars represent means ± SEM. Statistical analyses were performed using a Two-Way ANOVA followed by a Tukey’s post-hoc test, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001; ns nonsignificant.
We sought to determine whether DUSP6 overexpression reduced amyloid burden in 5xFAD mice which could contribute to the partial rescue of learning deficits. DUSP6 overexpression significantly reduced 6E10-immunolabeled amyloid plaque density in male 5xFAD but not female 5xFAD mice (Figures 3A, B). Western blot analysis revealed that levels of human APP-related proteins containing the 6E10 epitope were reduced in male 5xFAD-DUSP6 but not in female 5xFAD-DUSP6, compared to 5xFAD-GFP mice (Figure 3C). However, RT-qPCR analysis showed that human APP and mouse App mRNA levels were not altered by DUSP6 overexpression (Supplementary Figure 4), indicating that DUSP6 did not affect APP at the transcriptional level. Aβ1–40 and Aβ1–42 peptide levels were reduced in male 5xFAD-DUSP6 but not female 5xFAD-DUSP6 mice, compared to sex- and age-matched 5xFAD-GFP mice (Figure 3D). Taken together, these data suggest that DUSP6 is potentially involved in the regulation of hAPP translation, Aβ peptide production, degradation, and/or amyloid plaque clearance.
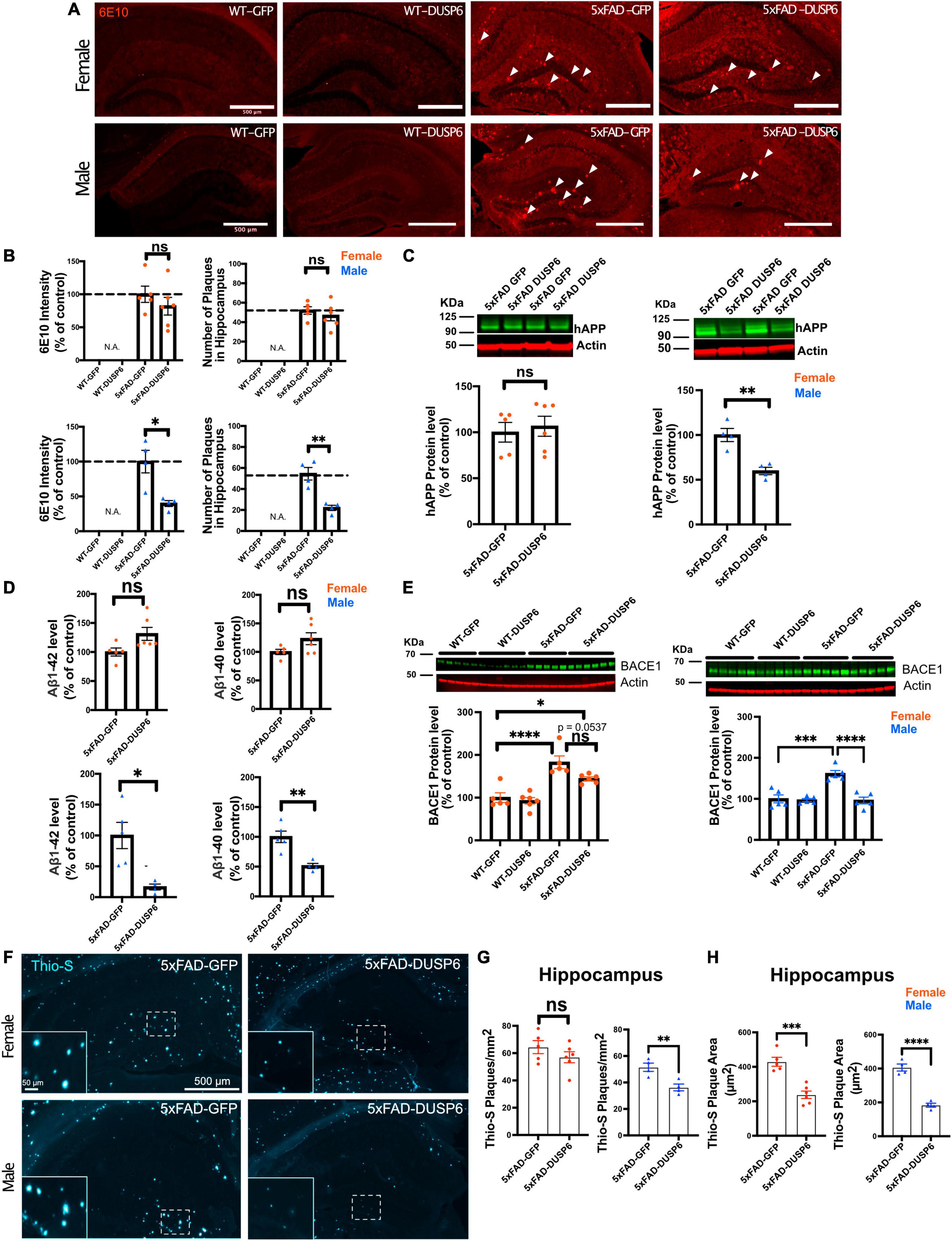
Figure 3. Immunohistochemistry and ELISA analyses of amyloid plaque load in hippocampi of 5xFAD mice overexpressing DUSP6. (A) Representative images of 6E10 staining in male and female 5xFAD and WT mice overexpressing DUSP6 or GFP at 5 months of age. Arrows indicate amyloid plaques in the hippocampus. Scale bar = 500 μm. (B) Quantification of intensity and number of 6E10-positive plaques in the hippocampi of male and female 5xFAD and WT mice overexpressing DUSP6 or GFP at 5 months of age. n = 4–6 mice per group and per sex with 3 coronal sections per animal. (C) Western blot analysis of human APP proteins (clone 6E10) in hippocampus of female and male 5xFAD and WT overexpressing DUSP6 or GFP. n = 4–6 mice per group. (D) Human Aβ1–40 and Aβ1–42 levels were quantified by ELISA in hippocampi of female and male 5xFAD and WT overexpressing DUSP6 or GFP. (E) BACE1 levels in hippocampi of female and male 5xFAD and WT mice overexpressing DUSP6 or GFP were quantified by western blot. n = 5–6 mice per group. (F) Representative images of thioflavin S (Thio-S) staining in male and female 5xFAD mice overexpressing DUSP6 or GFP at 5 months of age. (G) Quantification of the number of Thio-S positive plaques in the hippocampus. n = 4–6 mice/sex/group (H). Quantification of the average plaque area in the hippocampus. n = 4–6 mice/sex/group. Error bars represent means ± SEM. Statistical analyses were performed using a one-way ANOVA followed by a Tukey’s post-hoc test for BACE1 western blots and a Student’s t-test for all other graphs, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001; ns = nonsignificant.
Three key enzymes, α-, β- and γ-secretases, regulate APP processing, and Aβ peptide is produced by the proteolytic action of β- and γ-secretases on APP in the amyloidogenic pathway. We used RT-qPCR to quantify the transcripts encoded by the mouse γ-secretase subunit genes Ps1 and Ps2, the mouse β-secretase APP cleaving enzyme 1 (Bace1), and by transgenic human PS1, in 5xFAD mouse hippocampus. As we have shown previously through the analysis of gene expression in these samples from 5xFAD-GFP and WT-GFP hippocampus, when we investigated the efficacy of DUSP4 overexpression to slow progression of neuropathology in 5xFAD mice (Pan et al., 2022), most of these processing enzyme genes were upregulated in both female and male 5xFAD-GFP compared to WT-GFP, except for Ps2 mRNA levels in male and Bace1 mRNA levels in female, where no changes were observed when compared to WT-GFP mice (Supplementary Figure 2). DUSP6 overexpression in 5xFAD significantly reduced hippocampal Bace1 mRNA levels in males only (Supplementary Figure 2C, right panel), and did not significantly affect expression of any other APP-processing enzymes. Western blotting indicated that BACE1 protein levels were significantly decreased in male 5xFAD mice overexpressing DUSP6, while female 5xFAD mice overexpressing DUSP6 showed a trend of reduction in BACE1 protein levels (Figure 3E).
BACE1 catalyzes the rate liming step in the production of β-amyloid, and the inhibition of BACE1 has been reported to significantly reduce the rate of plaque formation (Peters et al., 2018). To determine whether DUSP6 overexpression altered amyloid plaque formation, we analyzed the number and size of thioflavin S-labeled plaques. The total number of thioflavin S-positive plaques was significantly reduced only in male 5xFAD mice overexpressing DUSP6 (Figures 3F, G), while the average size of thioflavin S-positive plaques was reduced in both female and male 5xFAD mice overexpressing DUSP6 (Figure 3H), suggesting that DUSP6 overexpression reduces the rate of amyloid plaque formation.
To investigate whether DUSP6 overexpression regulated the non-amyloidogenic APP processing pathway, we quantified mRNA levels of α-secretases Adam10 and Adam17 by RT-qPCR. Levels of Adam10 and Adam17 mRNAs in 5xFAD were previously reported as increased at one month of age, and by 9 months of age, Adam10 mRNA was decreased and Adam17 also showed a trend of reduction at the same age (Reinhardt et al., 2014). We found that Adam10 and Adam17 mRNAs were upregulated in female and male 5xFAD-GFP at 5 months of age compared to WT-GFP (Supplementary Figure 2D). Overexpression of DUSP6 in either female or male 5xFAD mice did not affect the levels of Adam10 and Adam17 mRNAs compared to 5xFAD-GFP. Lastly, transcripts encoded by γ-secretase subunit genes including Nct, Aph1a, Aph1b, and Aph1c were quantified, and Nct mRNA levels were upregulated in female and male 5xFAD-GFP compared to WT-GFP, while no changes were observed in 5xFAD mice overexpressing DUSP6 compared to 5xFAD-GFP (Supplementary Figures 2E, F). Taken together, these results indicate that the amelioration of amyloid burden by DUSP6 overexpression is not obviously caused by widespread changes in the expression of the major secretase-type APP-processing enzymes.
To determine whether DUSP6 overexpression affects AD-associated microglial activation in a sex-dependent manner, we assayed microglia-associated markers. We first assessed hippocampal Aif1 (IBA1) and Cd68 mRNA levels in female and male 5xFAD overexpressing GFP by real-time quantitative polymerase chain reaction (RT-qPCR), and found a ∼10-fold increase in Cd68 and a ∼6-fold increase in Aif1 mRNA levels in female 5xFAD-GFP and a ∼13-fold increase in Cd68 and ∼3-fold increase in Aif1 mRNA levels in male 5xFAD-GFP, compared to sex-matched WT-GFP (Figures 4A, B), consistent with AD-associated microglial activation. There were no significant changes in Cd68 and Aif1 mRNA levels in female or male WT-DUSP6 mice, compared to WT-GFP (Figures 4A, B). Both female and male 5xFAD overexpressing DUSP6 had significantly decreased Cd68 and Aif1 mRNA levels compared to 5xFAD-GFP mice (Figures 4A, B). However, only Aif1 mRNA levels showed a consistent reduction in female 5xFAD-DUSP6 vs 5xFAD-GFP in both RNAseq (p = 0.022) and RT-qPCR (p < 0.001) data, while male 5xFAD-DUSP6 vs 5xFAD-GFP showed a reduction by RT-qPCR (p < 0.001) but no significant change in RNAseq (p = 0.52) analysis. The discrepancy between these results may be due to the masking of microglial gene expression as a result of bulk RNA sequencing analysis, which takes average gene expression across all cells in the sample. Therefore, we assayed IBA1 by immunohistochemistry (IHC), finding that the intensity of IBA1-positive staining increased in both female and male 5xFAD-GFP hippocampal sections compared to WT-GFP, while IBA1 staining intensity in the hippocampus of female and male 5xFAD mice overexpressing DUSP6 was significantly reduced compared to 5xFAD-GFP (Figures 4C–F).
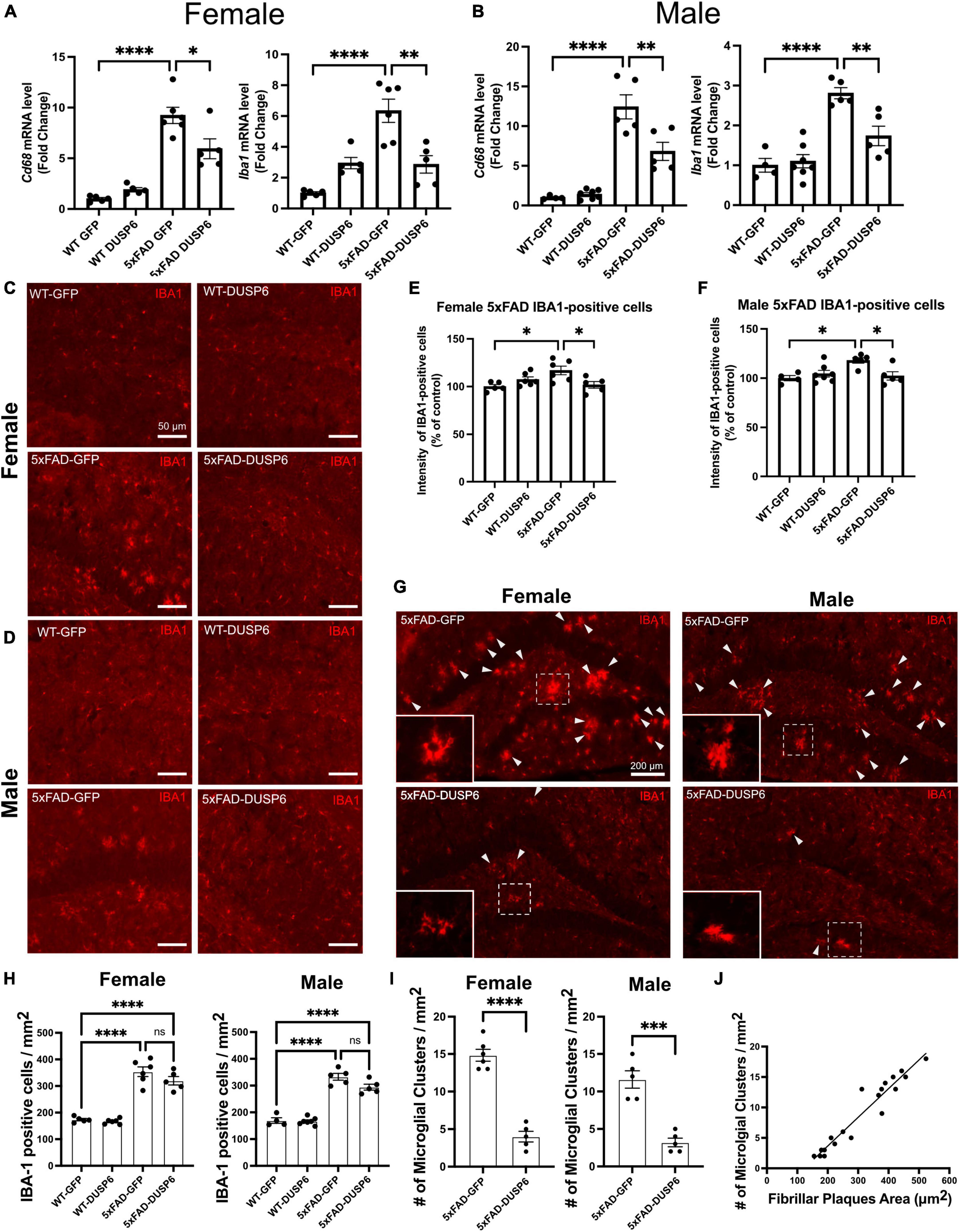
Figure 4. DUSP6 overexpression ameliorates microglial activation in female and male 5xFAD mice. (A,B) RT-qPCR results showed a significant decrease of Aif1 and Cd68 mRNAs in DUSP6-overexpressing female (A) and male (B) 5xFAD mice compared to GFP-overexpressing 5xFAD mice, n = 4–7 mice/group. (C,D) Representative images of microglial cells from female (C) and male (D) dorsal hippocampi labeled with anti-IBA1 (red). Scale bar = 50 μm. (E) Quantification of IBA1 fluorescence intensity from images in C showed that DUSP6 overexpression decreased IBA1 levels in female 5xFAD, n = 5–6 mice/group. (F) Quantification of IBA1 fluorescence intensity from images in D showed that DUSP6 overexpression reduced IBA1 levels in male 5xFAD mice, n = 4–7 mice/group. (G) Representative images of “microglial clusters” (a cluster of microglia is counted when three or more microglial soma occurred within 20 μm of graticule subregion) in the hippocampus. (H) Quantification of IBA1 positive cells in the hippocampus, n = 4–7 mice/group. (I) Quantification of the number of microglial clusters in the hippocampus, n = 5–6 mice/group. (J) The increased number of microglial clusters is correlated with the increased number of fibrillar plaques in a combined group analysis, R2 = 0.92, p < 0.001. Statistical analyses were performed using a Student’s t-test for microglial clusters and a One-way ANOVA followed by a Tukey’s post-hoc test for all other graphs, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001; ns nonsignificant.
Clustering of activated microglia and the interaction between microglia and amyloid plaques have been noted in post-mortem human brain tissue (Serrano-Pozo et al., 2011; Walker et al., 2020). We therefore assessed whether DUSP6 overexpression affected “microglial clustering” in 5xFAD mice by distinguishing clusters of IBA1-positive cells [defined as three or more microglial somas occurring within, or touching the margins of, a 20 μm2 virtual graticule subregion of hippocampus (Paasila et al., 2020)], from individual IBA1-positive cells. There were no significant changes in the total number of IBA1-positive microglia in either female or male 5xFAD overexpressing DUSP6 compared to sex-matched 5xFAD overexpressing GFP (Figure 4H), while the number of “microglial clusters” was reduced in both female and male 5xFAD overexpressing DUSP6 compared to the sex-matched 5xFAD overexpressing GFP (Figures 4G–I). This reduction in “microglia clusters” correlated with a decrease in amyloid plaque size in a combined group analysis, comparing 5xFAD overexpressing DUSP6 and 5xFAD overexpressing GFP (Figure 4J). These results suggest that DUSP6 overexpression may regulate microglia-associated neuroinflammation in 5xFAD mice, but it is not clear whether this is a direct effect of DUSP6 on microglial activation or an indirect outcome of reduced amyloid burden.
To determine the molecular pathways in the dHc of 5-month-old 5xFAD mice that are affected by DUSP6 overexpression, we compared the dHc transcriptomic profiles of female and male WT or 5xFAD overexpressing DUSP6 to WT or 5xFAD overexpressing GFP (transcriptomic data for WT and 5xFAD overexpressing GFP were previously published with our DUSP4 overexpression dataset (Pan et al., 2022), all of which were sequenced in the same batch with the DUSP6 transcriptomics reported here). There were 1469 differentially expressed genes (DEGs) in female WT-DUSP6 relative to female WT-GFP (FDR < 0.05) (Supplementary Figure 3A). Several DEGs including alpha-2-macroglobulin (A2m) (Blacker et al., 1998), syndecan 1 (Sdc1) (Letoha et al., 2019), and interferon-induced transmembrane protein 2 (Ifitm2) (Yao and Yan, 2020), were upregulated by DUSP6 overexpression in female WT mice, and these genes have either been directly or indirectly associated with metabolism of Aß. Another highly upregulated gene in female WT overexpressing DUSP6 is Msx3. Overexpression of MSX3 in microglia protects neurons from injury, and promotes the maturation of oligodendrocyte precursors and remyelination, whereas the deletion of Msx3 in microglia induces apoptosis of oligodendrocytes and prevents neuroprotection (Yu et al., 2015). Inflammatory response pathways were highlighted when pathway enrichment analysis of the DEGs identified in female WT overexpressing DUSP6 was performed (Supplementary Figure 3B). By comparison, there were only 7 DEGs in male WT mice overexpressing DUSP6 compared to those expressing GFP (FDR < 0.05) (Supplementary Figure 3C), but Msx3 is again one of the most upregulated genes.
We then assessed transcriptomics in 5xFAD mice overexpressing DUSP6 or GFP in dHc, compared to WT-GFP. We identified 1828 DEGs in female 5xFAD-GFP compared to WT-GFP, most of which were upregulated (FDR < 0.05) (Figure 5A). Overexpression of DUSP6 in female 5xFAD dHc downregulated 119 DEGs compared to 5xFAD-GFP (FDR < 0.05) (Figure 5B), and 116 of these DEGs overlapped with the upregulated DEGs from female 5xFAD-GFP vs WT-GFP (Supplementary Figure 3D). Enrichr pathway analysis showed, as previously described (Landel et al., 2014; Boza-Serrano et al., 2018; Manji et al., 2019), that many inflammatory pathways in the hippocampi of female 5xFAD were upregulated compared to WT (Figure 5E). Overexpression of DUSP6 (Figure 5F) in female 5xFAD downregulated some of these inflammatory pathways. Ingenuity Pathway Analysis (IPA) predicted regulation of similar pathways, including downregulation of ERK/MAPK (p-value = 0.02, z-score = −1.34), interferon (p-value = 1.26x10-13, z-score = −2.53) and neuroinflammatory (p-value = 5.01x10-14, z-score = −4.69) pathways, and PD-1/PD-L1 (p-value = 3.16x10-15, z-score = 3.64) pathway by DUSP6 overexpression in female 5xFAD (Figure 5G). By comparison, there were only three DEGs observed in male 5xFAD-GFP compared to WT-GFP (Figure 5C), consistent with previous reports that female 5xFAD develop more severe neuropathology than age-matched males (Sadleir et al., 2015, 2018), while 5 DEGs were found in male 5xFAD-DUSP6 when compared to male 5xFAD-GFP (Figure 5D). The murine homeobox gene, Msx3, was notably upregulated in male 5xFAD-DUSP6, male WT-DUSP6, female 5xFAD-DUSP6, and female WT-DUSP6 hippocampus (Figures 5B, D and Supplementary Figures 3A, C), while the Baiap3 gene encoding a Munc13-related protein involved in large dense core vesicle exocytosis was also upregulated by DUSP6 overexpression in male 5xFAD (Figure 5D).
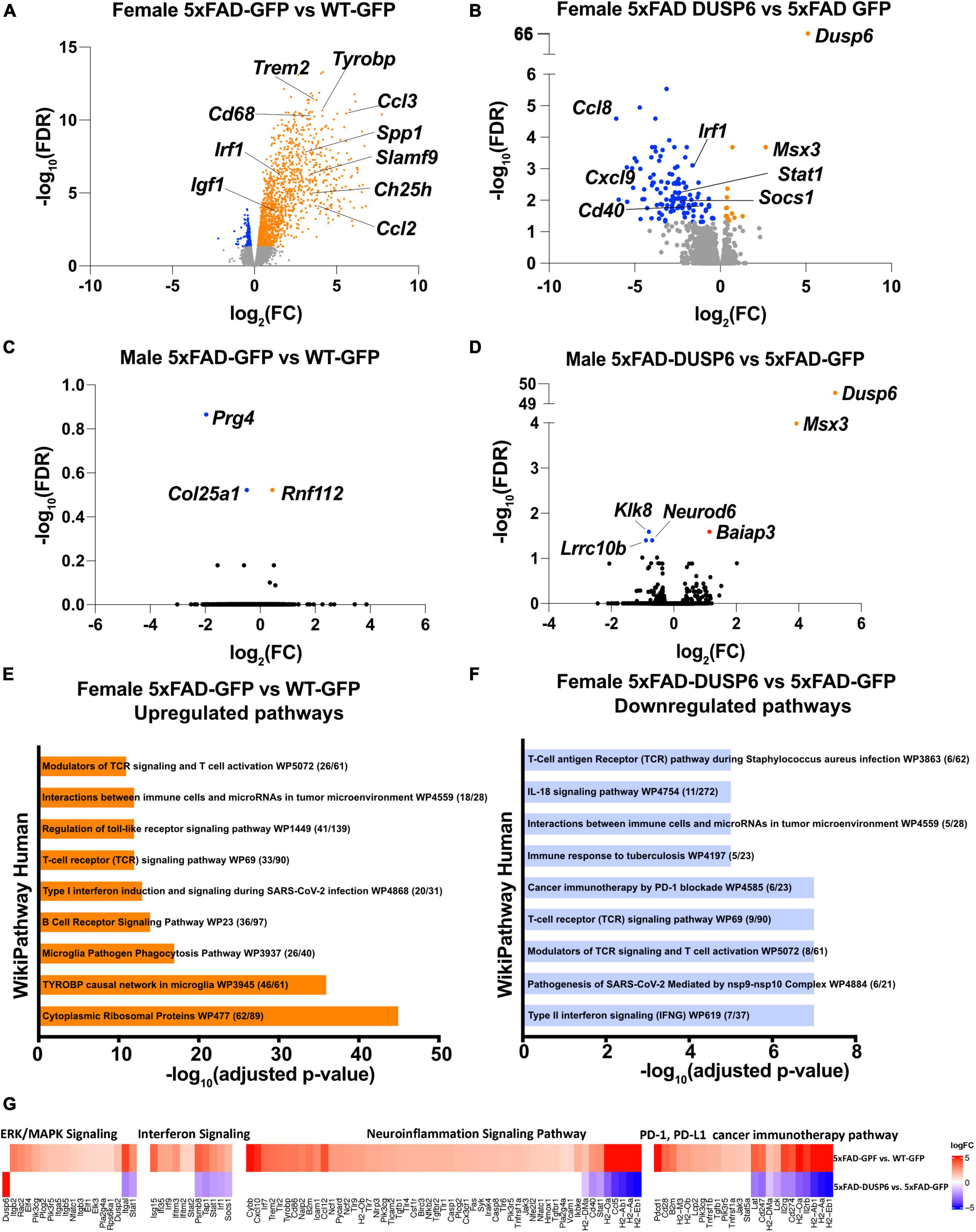
Figure 5. DUSP6 overexpression downregulates differentially expressed genes (DEGs) in female 5xFAD. (A,B) Volcano plot representation of female 5xFAD-GFP vs WT-GFP (A) and female 5xFAD-DUSP6 vs 5xFAD-GFP (B), n = 5 mice per group, threshold for DEGs represented is FDR < 0.05 (orange and blue dots). (C) Volcano plot representation of DEGs from male 5xFAD-GFP vs WT-GFP showed three DEGs. (D) Volcano plot representation of DEGs from male 5xFAD-DUSP6 vs 5xFAD-GFP showed five DEGs. (E,F) Enrichment analysis of DEGs from female 5xFAD GFP vs WT-GFP (E) and female 5xFAD-DUSP6 vs 5xFAD-GFP (F). (G) DEGs from panels (A,B) indicate a downregulation of the PD-1, PD-L1 cancer immunotherapy pathway (p-value = 7.94x10-15, z-score = −3.27), and an upregulation of the neuroinflammation signaling pathway (p-value = 2.51x10-28, z-score = 6.80), interferon signaling (p-value = 6.31x10-11, z-score = 2.89), and ERK/MAPK signaling (p-value = 3.55x10-5, z-score = 3.05) in female 5xFAD-GFP vs WT-GFP. Conversely, in 5xFAD-DUSP6 vs 5xFAD-GFP, the PD-1, PD-L1 cancer immunotherapy pathway (p-value = 3.16x10-15, z-score = 3.64) was upregulated, while the neuroinflammation signaling pathway (p-value = 5.01x10-14, z-score = −4.69), interferon signaling (p-value = 1.26x10-13, z-score = −2.53), and ERK/MAPK signaling (p-value = 0.02, z-score = −1.34) were downregulated.
Because our bulk RNAseq yielded fewer DEGs (FDR < 0.05) in male compared to female 5xFAD vs WT, potentially a result of variability in gene expression resulting from the mixed B6/SJL genetic background compared to previous studies where higher numbers of DEGs were detected in 4 month old male 5xFAD on a C57BL/6J background (537 DEGs) (Bundy et al., 2019), we used less stringent parameters to filter DEGs (p < 0.05). DUSP6 overexpression rescues cognitive deficits in male 5xFAD mice but not in females (Figure 2), in contrast to DUSP4 overexpression, which rescues cognitive deficits in female 5xFAD but not in males (Pan et al., 2022). We therefore compared the transcriptomic analyses of these groups to determine whether alterations in gene expression induced by DUSP overexpression could explain these sex differences in cognitive behavior. Enrichr was used to perform Synapse Gene Ontologies (SynGO) on the sex differences in DEGs (p < 0.05) between female and male 5xFAD overexpressing either DUSP4 or DUSP6. DUSP6 overexpression regulated more synaptic pathways in male than in female 5xFAD, while DUSP4 overexpression regulated more synaptic pathways in female than in male 5xFAD, which is consistent with cognitive rescue in male 5xFAD-DUSP6 (Figure 2) and that previously reported in female 5xFAD-DUSP4 (Pan et al., 2022). The top nine significant pathways (p < 0.05) in male 5xFAD-DUSP6 were involved in synaptic activity including regulation of postsynaptic neurotransmitter receptors, while female 5xFAD-DUSP6 showed seven significant synaptic pathways (p < 0.05) including regulation of synaptic assembly (Figure 6A). Several DEGs, which include Baiap3, Oprk1, Hap1, Fxyd6, Adra2a, Efnb2, Actn2, Nos1, Grin3a, Mapk3, Slc6a7, and Cadm1, were upregulated in male 5xFAD-DUSP6 but not in female 5xFAD-DUSP6, and these DEGs are either directly or indirectly associated with synaptic regulation. The top ten significant pathways in female 5xFAD-DUSP4 were involved in both postsynaptic and presynaptic activity, while male 5xFAD-DUSP4 showed two significant pathways (p < 0.05), one of which was involved in postsynaptic neurotransmitter receptor activity (Figure 6A). Comparison of the synapse-associated DEGs identified by SynGO indicated that there were no overlapping DEGs (p < 0.05) between male and female 5xFAD overexpressing DUSP4, and no overlapping DEGs (p < 0.05) between male and female 5xFAD overexpressing DUSP6, while three DEGs were shared between male 5xFAD-DUSP6 and male 5xFAD-DUSP4, and four DEGs between female 5xFAD-DUSP6 and female 5xFAD-DUSP4 (Figure 6B). DUSP6 regulated more synapse-associated DEGs in male (33 DEGs) than in female (16 DEGs) 5xFAD (p < 0.05), while DUSP4 overexpression regulated more synapse-associated DEGs in female (48 DEGs) than in male (4 DEGs) 5xFAD (Figure 6B). Moreover, analysis using the Mouse Genome Informatics (MGI) database revealed that DUSP6 overexpression regulated DEGs associated with spatial learning, long term potentiation, and cued conditioning behaviors in male 5xFAD mice, while DUSP6 overexpression regulated mainly pro-inflammatory pathways in female 5xFAD (Figure 6C).
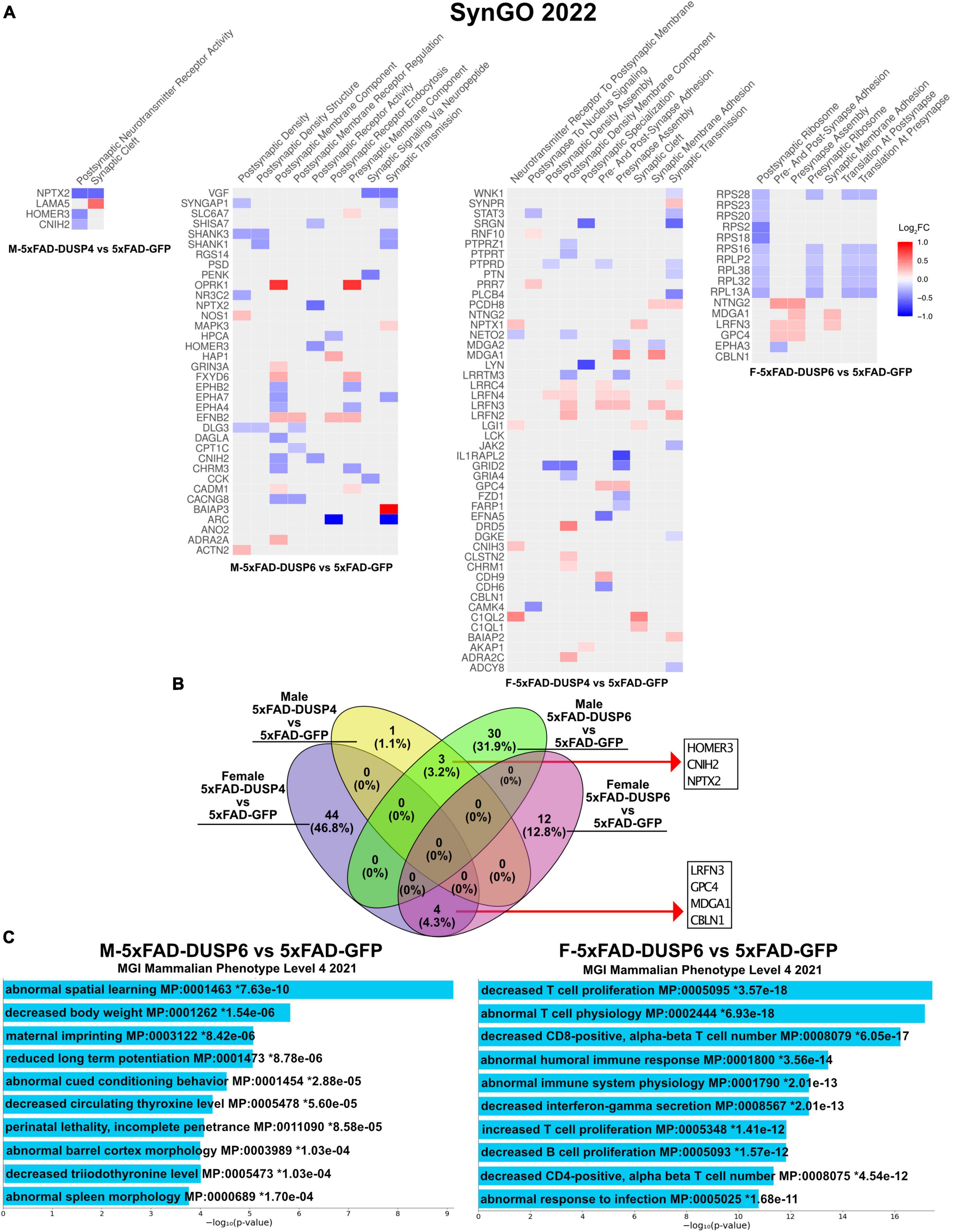
Figure 6. Gene ontology (GO) enrichment analysis of DEGs (p < 0.05) of female and male 5xFAD mice overexpressing DUSP4 or DUSP6. (A) Enrichr was used to perform Synaptic Gene Ontologies (SynGO) analysis of DEGs (p < 0.05) from female and male 5xFAD mice overexpressing DUSP4 or DUSP6. (B) Venn diagram showed the common DEGs between female and male 5xFAD overexpressing DUSP4 or DUSP6. (C) Enrichr was utilized to perform GO enrichment analysis of DEGs from female and male 5xFAD-DUSP6 vs 5xFAD-GFP compared to the Mouse Genome Informatics (MGI).
Using reduced stringency (p < 0.05), comparison of male 5xFAD-DUSP6 vs 5xFAD-GFP RNAseq resulted in 351 DEGs, and female 5xFAD-DUSP6 vs 5xFAD-GFP resulted in 875 DEGs, while comparison of male 5xFAD-DUSP4 vs 5xFAD-GFP resulted in 268 DEGs, and male 5xFAD-DUSP4 vs 5xFAD-GFP resulted in 1438 DEGs. As indicated in the Venn diagram, female 5xFAD-DUSP6 and 5xFAD-DUSP4 share 575 DEGs, while male 5xFAD-DUSP6 and 5xFAD-DUSP4 share 59 DEGs (p < 0.05) (Figure 7A). In addition, the comparison between female and male overexpressing DUSP6 or DUSP4 shows that male and female 5xFAD-DUSP6 share 47 DEGs, while male and female 5xFAD-DUSP4 share 32 DEGs. Enrichr pathway analysis indicated that these four groups share a common pathway: tumor necrosis factor-alpha (TNF-α) signaling via nuclear factor-kappaB (NF-kB) (Figure 7B). The majority of the ten most significant pathways regulated in female 5xFAD-DUSP6 and female 5xFAD-DUSP4 are pro-inflammatory, while only one pathway in male 5xFAD-DUSP6 and male 5xFAD-DUSP4 is pro-inflammatory (Figure 7B), indicating that DUSP4 and DUSP6 may contribute via shared pathways to the reduction of inflammation, primarily in female 5xFAD mice.
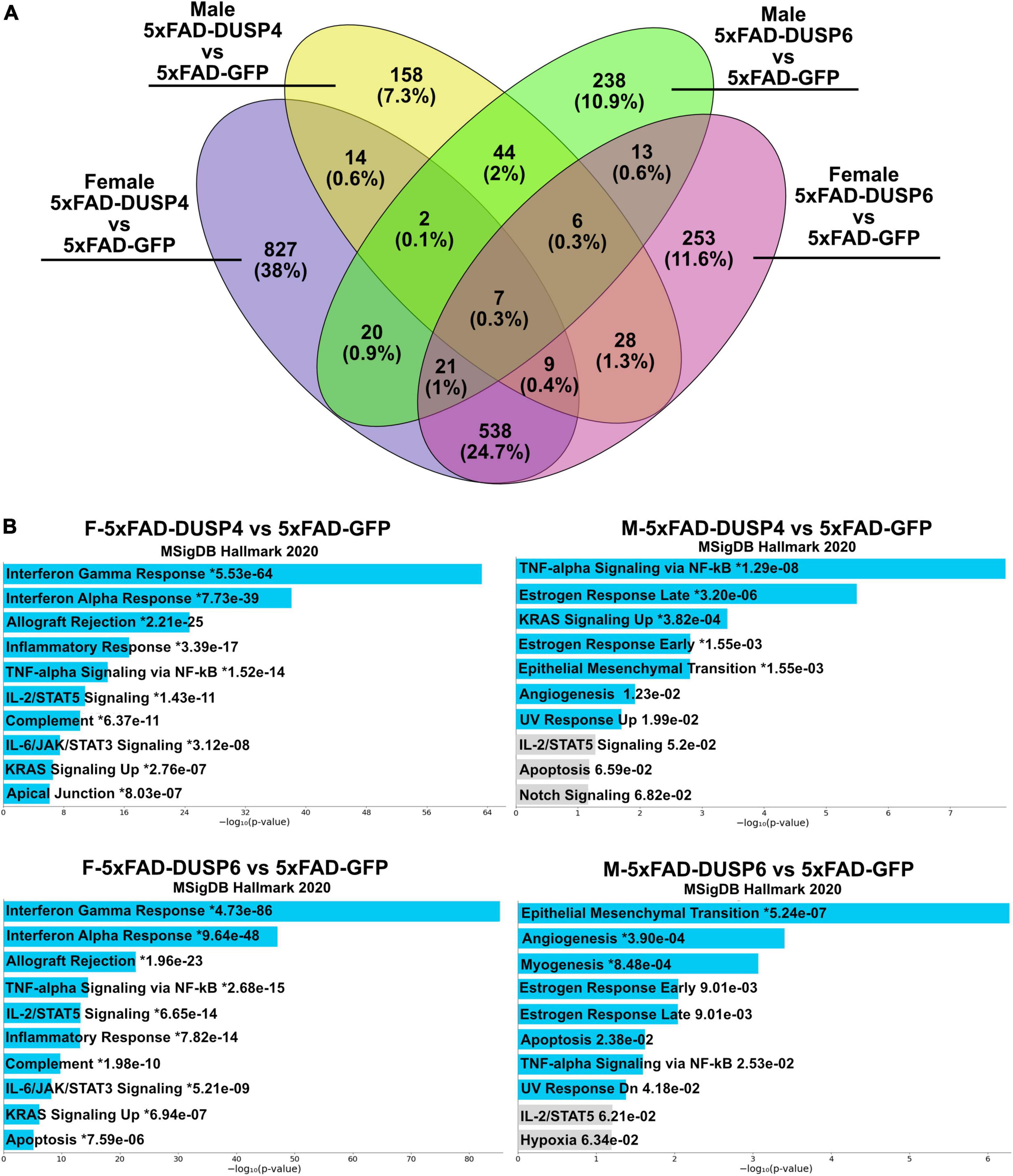
Figure 7. Pathway analysis comparison of female and male 5xFAD mice overexpressing DUSP4 or DUSP6 using DEGs (p < 0.05). (A) Venn diagram showed the shared common DEGs (p < 0.05) in female and male 5xFAD overexpressing DUSP4 or DUSP6. (B) Pathway enrichment analysis of the same DEGs (p < 0.05) by Enrichr, blue bars indicate p < 0.05 and gray bars indicate p > 0.05.
To investigate if DUSP6 overexpression had an effect on the VGF network (Beckmann et al., 2020), we first determined whether Vgf-associated genes are dysregulated in 5-month-old 5xFAD mice by comparing hippocampal Vgf, Bdnf, Sst, and Scg2 mRNA levels in 5xFAD-GFP, WT-GFP and AAV-DUSP6 injected mice (Figures 8A–D). We again observed a reduction of Vgf, Sst, and Scg2 mRNA levels in female 5xFAD-GFP mice compared to WT-GFP (Pan et al., 2022), but no change in male 5xFAD-GFP mice (Figures 8A, C, D). Female 5xFAD-GFP also showed a trend toward reduction in Bdnf mRNA levels, but no change was observed in male 5xFAD-GFP mice (Figure 8B). Following DUSP6 overexpression, hippocampal Vgf and Sst mRNA levels were downregulated in both female and male 5xFAD mice or WT mice overexpressing DUSP6 compared to controls overexpressing GFP (Figures 8A, C). DUSP6 overexpression reduced Bdnf mRNA levels only in male mice (Figure 8B). DUSP6 overexpression did not alter Scg2 mRNA levels in female or male mice (Figure 8D). DUSP6 overexpression therefore alters the expression levels of additional key nodes in the VGF-centered multiscale network.
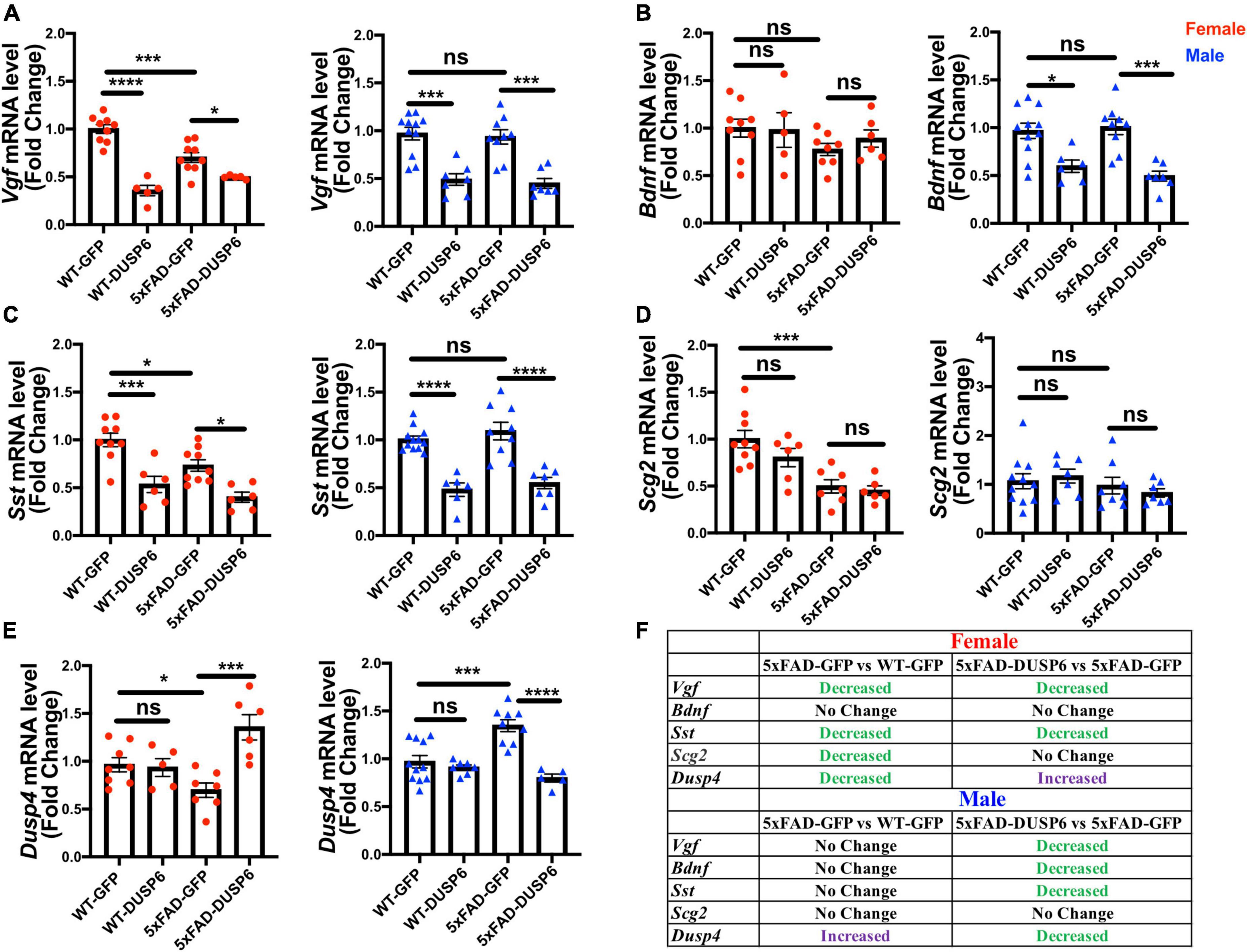
Figure 8. The effects of DUSP6 overexpression on VGF network-associated genes in 5xFAD and WT mice. (A) Hippocampal Vgf mRNA levels in 5xFAD or WT mice overexpressing DUSP6 compared to control (WT-GFP), n = 5–11 mice per group. (B) Hippocampal Bdnf mRNA levels in 5xFAD or WT mice overexpressing DUSP6 compared to the control, n = 5–11 mice per group. (C) Hippocampal Sst mRNA levels in 5xFAD or WT mice overexpressing DUSP6 compared to the control, n = 6–11 mice per group. (D) Hippocampal Scg2 mRNA levels in 5xFAD or WT mice overexpressing DUSP6 compared to control, n = 6–11 mice per group. (E) Hippocampal Dusp4 mRNA levels in 5xFAD or WT overexpressing DUSP6 or GFP were assayed by RT-PCR, n = 5–11 mice per group. (F) Summary table of changes in VGF network-associated gene expression as a result of DUSP6 overexpression. Statistical analyses were performed using a One-Way ANOVA followed by a Tukey’s post-hoc test, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001; ns, nonsignificant.
DUSP6 is located downstream of DUSP4 in the VGF-gene network model (Beckmann et al., 2020). Previously, we determined that DUSP4 overexpression downregulated DUSP6 expression in hippocampus of both female and male 5xFAD (Pan et al., 2022). In 5xFAD overexpressing DUSP6, Dusp4 mRNA levels were upregulated in female 5xFAD-DUSP6 compared to female 5xFAD-GFP (Figure 8E, left), whereas the overexpression of DUSP6 in male 5xFAD restored DUSP4 mRNA to its basal level compared to male 5xFAD-GFP (Figure 8E, right). Overexpression of DUSP6 in WT mice did not affect DUSP4 mRNA levels compared to WT-GFP mice. These data represent another example of a sexually dimorphic effect of DUSP6 overexpression in 5xFAD mice.
DUSP6 is a key hub in the VGF multiscale causal gene network that regulates AD (Beckmann et al., 2020). DUSP6 is involved in the regulation of many signaling pathways, including ERK/MAPK and JNK signaling, but its potential role in AD has not been investigated. Here we provide evidence that DUSP6 gene expression is decreased in 4 and 12 month old 5xFAD male and female mice compared to age- and sex-matched wild types, and that overexpression of DUSP6 in hippocampus reduces memory deficits and amyloid load in male but not female 5xFAD mice. Sex-associated differences have been reported between female and male 5xFAD, with female 5xFAD mice developing more severe AD-associated neuropathology than age-matched males (Sadleir et al., 2015, 2018). GO analysis of DEGs showed that DUSP6 regulated several more synaptic pathways and DEGs in male 5xFAD than female 5xFAD mice, indicating that DUSP6 regulated synaptic activity in a sex-dependent manner. Spatial memory deficits in the Barnes maze were partially rescued in male 5xFAD-DUSP6 but not female 5xFAD-DUSP6 mice. These results suggest that DUSP6 regulated AD-associated pathogenesis in 5xFAD mice in a sex-dependent fashion.
In our studies, DUSP6 overexpression was found to reduce amyloid plaque load in male 5xFAD mice, which was associated with decreased expression of the APP-processing enzyme BACE1, human APP, and Aβ1–40 and Aβ1–42 peptides, in males but not females, while human and mouse APP mRNA levels were not changed. These data suggest that DUSP6 regulates hAPP translation, Aβ peptide production, and/or degradation, rather than APP gene transcription. APP mRNA translation is known to be regulated by several overlapping sites in the 5′ untranslated region (UTR), which interact with iron response protein 1 (IRP1), microRNA miR-346, and interleukin-1 (Rogers G. et al., 1999; Cho et al., 2010; Long et al., 2018), and by miRNA binding sites in the 3′ UTR (Vilardo et al., 2010; Delay et al., 2011). Select miRNAs are regulated in a sex-dependent manner (Kodama et al., 2020), which could provide an underlying mechanism for the gender-specific differences in hAPP protein levels.
Our transcriptomics did reveal significantly higher interleukin-1 alpha (IL1α) mRNA levels in female but not male WT-DUSP6 compared to WT-GFP mice. Increased binding of IL1α to the APP 5′UTR would be predicted to increase APP translation 1 (Rogers G. et al., 1999; Rogers J. et al., 1999; Cho et al., 2010; Long et al., 2018) in DUSP6 overexpressing females compared to DUSP6 overexpressing males. However, we saw no significant effects of DUSP6 overexpression on Il1α mRNA levels in male or female 5xFAD (Supplementary Material 1). Another possible mechanism underlying the gender-specific regulation by DUSP6 of APP protein levels could be mediated by miRNAs that modulate translation of APP processing enzymes and/or other targets. Our transcriptomics analysis identified that miR-124a levels were increased in male 5xFAD compared to WT (p = 0.056), and that DUSP6 overexpression decreased miR-124a levels in male 5xFAD compared to GFP (p = 0.048). MiR-124 plays a pivotal role in the development and function of the nervous system (Åkerblom et al., 2012; Franke et al., 2012; Wang et al., 2022; Zhang W. et al., 2023) through its binding to a number of target RNAs, including the APP processing enzyme BACE1 (An et al., 2017). Our findings did not support miR-124 as a primary regulator of hippocampal BACE1 levels in male 5xFAD-DUSP6 mice, as BACE1 protein and miR-124a levels both decreased. However, levels of miR-124 have been found to be either reduced or elevated in AD brain (Wang et al., 2022; Zhang W. et al., 2023), but in agreement with our findings of increased miR-124a in male 5xFAD, miR124 levels were found to be increased in hippocampus of AD subjects and the Tg2576 AD mouse model (Wang X. et al., 2018), in which miR-124 was shown to target the tyrosine-protein phosphatase non-receptor type 1 (PTPN1). Moreover, overexpression of miR-124 or knockdown of PTPN1 recapitulated AD-like phenotypes in mice, including deficits in synaptic transmission, plasticity, and memory (Wang X. et al., 2018). Similarly, knockdown of miR-124 was shown to improve severe deficits in spatial learning and social interactions, and LTP deficits, in EPAC null mutant mice, via the miR-124-regulated target Zif268 (Yang et al., 2012). The gender-specific mechanism(s) by which DUSP6 overexpression regulates miR-124 and potentially other miRNAs remain to be elucidated.
Our results indicate that DUSP6 overexpression reduced hippocampal Bace1 gene expression and BACE1 protein levels in male but not female 5xFAD mice. BACE1 enzymatic activity was not assessed. Previous studies have shown that BACE1 is phosphorylated, as is its substrate APP, and that phosphorylation of either regulates catalysis (Song et al., 2015; Zhang et al., 2020) as well as intracellular sorting of BACE1 (Toh et al., 2018). The cyclin-dependent kinase 5 (CDK5) has been implicated in AD pathogenesis and plays a critical role in BACE1 regulation, at the transcriptional level via STAT3 (Wen et al., 2008) and at the post-translational level via BACE1 phosphorylation (Song et al., 2015; Allnutt et al., 2020; Requejo-Aguilar, 2023). Crosstalk between ERK and CDK5 pathways has been reported (Zhong et al., 2019), so it remains possible that DUSP6 inactivation of ERK could impact the CDK5/BACE1 pathways that regulate Aβ peptide generation and amyloid plaque deposition.
Emerging evidence indicates that chronic inflammation prior to symptomatic AD onset may be a contributing factor to disease progression, which includes systemic inflammation that damages blood-brain barrier integrity, allowing entry of peripheral immune cells and proinflammatory factors into the brain (Kinney et al., 2018). We noted downregulation of several microglia-associated markers in both female and male 5xFAD mice overexpressing DUSP6, and also increased expression of the Msx3 gene, which was previously shown to regulate microglial M1/M2 polarization and to reduce neuroinflammation (Yu et al., 2015). Because AAV-5 is a neurotropic virus (Haery et al., 2019), and we noted DUSP6 overexpression predominantly in neurons (Figure 1), DUSP6 could indirectly modulate microglial or astroglial gene expression via intercellular crosstalk by regulating the synthesis and/or secretion of proteins from neurons, including neuroinflammatory cytokines and/or chemokines, that bind cognate receptors on nonneuronal cells.
Female 5xFAD-DUSP6 mice demonstrated downregulation of neuroinflammatory, type II interferon, T-cell receptor, and ERK/MAPK signaling pathways. Coactivation of Interferon γ (IFNγ) and Toll-like receptor (TLR)-4 receptors in microglia has been shown to result in severe neuronal dysfunction and neurodegeneration (Papageorgiou et al., 2016), while transcriptomic profiling of primary mouse microglia provides evidence that MAPK signaling pathways, including ERK, regulate pro-inflammatory microglial activation in response to IFNγ (Chen et al., 2021), which would potentially be downregulated by DUSP6 overexpression. In addition to eliciting pro-inflammatory microglial activation, IFNγ regulates AD-associated Aβ plaque deposition and β-secretase expression. For example, IFNγ receptor knockout in Swedish APP transgenic mouse reduces amyloid plaque load at 14 months of age (Yamamoto et al., 2007). A recent study, moreover, showed that IFN-induced transmembrane 3 (IFTM3) protein is involved in the regulation of γ-secretase activity and amyloid plaque deposition in AD (Hur et al., 2020), supporting the notion that IFNγ signaling is involved in the regulation of Aβ production. Building on published studies which identify the role that the spleen tyrosine kinase (SYK) plays in modulating microglial responses to AD-associated Aβ deposition (Ennerfelt et al., 2022) and neuroinflammation (Chu et al., 2021), through the PI3K/AKT/GSK3β signaling pathway, our studies identify a potential role for DUSP6 modulation of the ERK signaling pathway in the mitigation of AD-associated Aβ deposition, neuroinflammation, and AD-associated microglial responses.
Synaptic dysfunction is recognized relatively early in the pathological progression of AD (Pelucchi et al., 2022), and sex-dependent reduction of AD-associated synaptic proteins has been demonstrated in AD animal models (Djordjevic et al., 2021). Using Enrichr analysis of the DEGs (relaxed stringency p < 0.05) from female and male 5xFAD-DUSP6 and compared to the SynGO database, we observed that DUSP6 regulated more than twice the number of DEGs associated with synaptic pathways in male compared to female 5xFAD-DUSP6 mice (Figures 6A, B). In male 5xFAD mice overexpressing DUSP6, nine synaptic pathways were involved, while in female 5xFAD mice, seven synaptic pathways were involved. Among these pathways, differences were observed between sexes in the regulation of synaptic components by DUSP6. Specifically, in males, DUSP6 regulated genes associated with both postsynaptic and presynaptic structures and components [e.g., HOMER, SHANK1/3, OPRK1 (kappa-opioid receptor 1), and BAIBP3 (BAI1-associated protein 3)] while in females, DUSP6 influenced genes related to both presynaptic and postsynaptic adhesion proteins [e.g., NTNG2 (netrin-2), MDGA1 (receptor protein-tyrosine phosphatase mu MAM domain-containing glycosylphosphatidylinositol anchor protein 1), and LRFN3 (Leucine Rich Repeat And Fibronectin Type III Domain Containing 3)]. Several of the DEGs, upregulated by DUSP6 overexpression in male but not female 5xFAD-DUSP6 mouse hippocampus, include Baiap3, Oprk1, Hap1, Fxyd6, Adra2a, Efnb2, Actn2, Nos1, Grin3a, Mapk3, Slc6a7, and Cadm1. All of these have been associated with either AD or other neurodegenerative diseases (Reif et al., 2011; Chen et al., 2014; Ji et al., 2015; Jin et al., 2019; Ting et al., 2019; Aykac and Sehirli, 2020; Lee and Lee, 2020; Georgakopoulos et al., 2021; Muraoka et al., 2021; Kim H. et al., 2022; Zhong et al., 2022; Zhang J. et al., 2023). BAIAP3 is a member of the mammalian uncoordinated 13 (MUNC13) protein family that controls synaptic activity by regulating neurotransmitter and neuropeptide vesicle exocytosis (Wojcik et al., 2013). Baiap3 dysregulation has been associated with both major depressive disorder (Kim H. et al., 2022) and AD (Lalli et al., 2012), and it is regulated in a sex-dependent manner in mouse models and human subjects with anxiety (Wojcik et al., 2013), and also plays a role in the recovery from CNS injury (Lauridsen et al., 2011). Homer and Shank1/3 are scaffolding proteins that play dynamic roles in regulating synaptic development, structure, transmission, and plasticity, and cognitive and neuropsychiatric disease (Harris et al., 2016; Shi et al., 2017; Clifton et al., 2019), and their interaction with PSD95 and the NMDA-receptor can be dysregulated by the binding of phosphorylated tau (Shen et al., 2023). DUSP6 overexpression also increased mRNA levels for the kappa opioid G-protein coupled receptor, which has been implicated in memory and depression (Bilkei-Gorzo et al., 2012; Bodnar, 2023; Wang et al., 2023; Zhu et al., 2024), and additionally, OPRK1 promoter methylation that leads to reduced gene expression has been found to increase AD risk (Ji et al., 2015). Hap1 encodes Huntingtin-associated protein-1 (HAP1), which negatively regulates Aß production in the amyloidogenic pathway (Ting et al., 2019). Interaction of HAP1 and Abelson helper integration site-1 (AHI1) enhances neurotrophic signaling through ERK activation, which leads to cell survival and differentiation (Ting et al., 2019). In female 5xFAD overexpressing DUSP6, LRFN3 expression was increased, and this cell surface protein has been found to regulate memory via interaction with PSD95 and the NMDA receptor (Morimura et al., 2006; Wang et al., 2006; Lie et al., 2021). Finally, also regulated in females, NTNG2 and MDGA1 encode adhesion proteins that play critical roles in axon guidance and synapse formation (Dias et al., 2019), with the latter interestingly regulating APP-mediated GABAergic inhibitory synaptic transmission and novel object recognition memory (Kim J. et al., 2022).
As already noted, GO enrichment analysis indicated that DUSP6 overexpression regulated spatial learning, LTP, and cued conditioning behavior pathways in male but not female 5xFAD-DUSP6 mice. These results parallel improved memory in male but not female 5xFAD-DUSP6, and although synaptic dysfunction is also thought to underlie Major Depressive Disorder (Duman et al., 2016), hyperactivity of 5xFAD precluded our assessment of depression-like behavior in these mice, as previously reported (Yamazaki et al., 2015; Oblak et al., 2021).
Based on these results with DUSP6 overexpression, we returned to the transcriptomic changes that occur following DUSP4 overexpression. GO analysis further identified 48 DEGs that were associated with synaptic development and function in female 5xFAD-DUSP4, including C1ql2, Adra2c, Ntng2, Cdh9, lrfn3, and Nptx1 (Williams et al., 2011; Figueiro-Silva et al., 2015; Lie et al., 2018; Abu-Libdeh et al., 2019; Brocos-Mosquera et al., 2021; Yang et al., 2023), while only 4 synapse-associated DEGs were identified in male 5xFAD-DUSP4. C1ql2 encodes a synaptic cleft protein between mossy fibers and CA3 neurons, which may regulate excitatory synapse formation on hippocampus neurons (Yang et al., 2023). Similarly, Cdh9 encodes Cadherin-9 (CDH9), which regulates hippocampal dentate gyrus-CA3 neuronal synapses (Williams et al., 2011). The sex-dependent regulation of synaptic pathways by DUSP4 is consistent with sex-dependent improvement of memory deficits in female but not male 5xFAD-DUSP4 mice, identified in our previous study (Pan et al., 2022). These results suggest the potential involvement of DUSP4 and DUSP6 in regulating synaptic pathways. However, the current study lacked sufficient hippocampal tissues to explore the regulation of synaptic pathways and proteins by DUSP4 or DUSP6, but one of our ongoing projects indicates that the overexpression of DUSP4 reversed the decline of PSD95 protein only in female APP/PS1 mice (unpublished data), suggesting a sex-dependent regulation of synaptic pathways by DUSP4 that correlated with improved cognition. In addition, previous studies found that hippocampal VGF overexpression rescued reduced PSD95 levels in 5xFAD that correlated with a reduction in their memory deficits (Beckmann et al., 2020).
DUSP6 and DUSP4 were identified as critical hub genes in a VGF multiscale causal network that regulates AD pathogenesis and progression (Beckmann et al., 2020). Hippocampal overexpression of VGF in male and female 5xFAD reduced Aβ accumulation and memory deficits (Beckmann et al., 2020), indicating a protective effect of VGF. DUSP6 was shown to be downregulated in AD (Banzhaf-Strathmann et al., 2014), so we hypothesized that DUSP6 overexpression would mitigate AD-associated neuropathology by upregulating expression of genes in the VGF gene network, including VGF. Similar to our previous studies of hippocampal DUSP4 overexpression in 5xFAD (Pan et al., 2022), DUSP6 overexpression downregulated the expression of several genes in the VGF network, including Vgf, Sst, and Bdnf (males only), indicating a regulatory effect of DUSP6 overexpression on genes in the VGF network, but in an unanticipated direction. One plausible mechanism underlying reduced Vgf, Sst, and Bdnf mRNA levels is that these VGF network genes are transcriptionally regulated by the cAMP response element-binding (CREB) protein (Montminy and Bilezikjian, 1987; Montminy et al., 1990). CREB is activated via phosphorylation of Ser133 by the ribosomal protein S6 kinase 2 (RSK2), that is itself activated by ERK (Xing et al., 1996, 1998; Benito et al., 2011), and ERK inactivation is catalyzed by DUSPs (Chen et al., 2019). This would suggest that chronic DUSP6 or DUSP4 overexpression results in gender-specific rescue of AD-related phenotypes in 5xFAD mice via pathways that may be independent of these genes in the VGF network.
It is well known that sex is a major risk factor for LOAD, and females are at a greater risk of developing AD (Rahman et al., 2019). In fact, sex-dependent differences in AD-associated progression, including the expression of synaptic proteins (Djordjevic et al., 2021) and microglial numbers during AD development (Harry, 2013), are supported by recent studies. Both DUSP4 overexpression in AD animals from our previous study (Pan et al., 2022) and DUSP6 overexpression in AD animals from the current study showed a sex-dependent improvement in spatial learning behavior. In addition, transcriptomic profiling of the hippocampal tissues overexpressing DUSP4 or DUSP6 identified sex-dependent changes in synaptic gene expression. Therefore, DUSP4 and DUSP6 may possess the potential to target AD-associated neuropathology in a sex-specific manner.
The datasets presented in this study can be found in the article/Supplementary material.
The animal study was approved by Institutional Animal Care and Use Committee (IACUC) at the Icahn School of Medicine at Mount Sinai (ISMMS). The study was conducted in accordance with the local legislation and institutional requirements.
AP: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Software, Validation, Writing – original draft, Writing – review and editing. MA: Conceptualization, Writing – review and editing. ES: Formal analysis, Validation, Writing – review and editing. RJ: Data curation, Formal analysis, Validation, Writing – review and editing. XZ: Data curation, Formal analysis, Validation, Writing – review and editing. QW: Data curation, Formal analysis, Software, Validation, Writing – review and editing. MW: Data curation, Formal analysis, Software, Validation, Writing – review and editing. NB: Funding acquisition, Resources, Writing – review and editing. EES: Funding acquisition, Resources, Writing – review and editing. SG: Funding acquisition, Resources, Writing – review and editing. BZ: Funding acquisition, Resources, Writing – review and editing. ME: Funding acquisition, Resources, Writing – review and editing, Conceptualization. SS: Conceptualization, Funding acquisition, Resources, Writing – review and editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was supported by the National Institute of Health U01AG046170 (EES, SG, BZ, MEE), R01AG062355 (SS, BZ, MEE), RF1AG062661 (SS, MEE), R01DK117504 (SS), RF1AG057440 (BZ), RF1AG054014 (BZ), and R01AG057907 (BZ, MEE). This study was also supported by the Cure Alzheimer’s Fund (MEE, SS) and an Alzheimer’s Disease Research Center (ADRC) Research Education Component (REC) Scholar Award to ALP (P30AG066514; Mary Sano, PI).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnagi.2024.1400447/full#supplementary-material
Abu-Libdeh, B., Ashhab, M., Shahrour, M., Daana, M., Dudin, A., Elpeleg, O., et al. (2019). Homozygous frameshift variant in NTNG2, encoding a synaptic cell adhesion molecule, in individuals with developmental delay, hypotonia, and autistic features. Neurogenetics 20, 209–213. doi: 10.1007/s10048-019-00583-4
Åkerblom, M., Sachdeva, R., Barde, I., Verp, S., Gentner, B., Trono, D., et al. (2012). MicroRNA-124 is a subventricular zone neuronal fate determinant. J. Neurosci. 32, 8879–8889.
Allnutt, A., Waters, A., Kesari, S., and Yenugonda, V. (2020). Physiological and pathological roles of Cdk5: Potential directions for therapeutic targeting in neurodegenerative disease. ACS Chem. Neurosci. 11, 1218–1230. doi: 10.1021/acschemneuro.0c00096
AMP-AD Portal (2021). https://www.synapse.org/#WSynapse:syn18637070 (accessed November 01, 2021).
An, F., Gong, G., Wang, Y., Bian, M., Yu, L., and Wei, C. (2017). MiR-124 acts as a target for Alzheimer’s disease by regulating BACE1. Oncotarget 8, 114065–114071.
Arkell, R., Dickinson, R., Squires, M., Hayat, S., Keyse, S., and Cook, S. (2008). DUSP6/MKP-3 inactivates ERK1/2 but fails to bind and inactivate ERK5. Cell Signal. 20, 836–843. doi: 10.1016/j.cellsig.2007.12.014
Arnaud, A., Brister, T., Duckworth, K., Foxworth, P., Fulwider, T., Suthoff, E. D., et al. (2022). Impact of major depressive disorder on comorbidities. J. Clin. Psychiatry 83:21r14328.
Audrain, M., Haure-Mirande, J., Wang, M., Kim, S., Fanutza, T., Chakrabarty, P., et al. (2019). Integrative approach to sporadic Alzheimer’s disease: Deficiency of TYROBP in a tauopathy mouse model reduces C1q and normalizes clinical phenotype while increasing spread and state of phosphorylation of tau. Mol. Psychiatry 24, 1383–1397. doi: 10.1038/s41380-018-0258-3
Aykac, A., and Sehirli, A. (2020). The role of the SLC transporters protein in the neurodegenerative disorders. Clin. Psychopharmacol. Neurosci. 18, 174–187.
Banzhaf-Strathmann, J., Benito, E., May, S., Arzberger, T., Tahirovic, S., Kretzschmar, H., et al. (2014). MicroRNA-125b induces tau hyperphosphorylation and cognitive deficits in Alzheimer’s disease. EMBO J. 33, 1667–1680. doi: 10.15252/embj.201387576
Barnes, C. (1979). Memory deficits associated with senescence: A neurophysiological and behavioral study in the rat. J. Comp. Physiol. Psychol. 93, 74–104.
Beckmann, N., Lin, W., Wang, M., Cohain, A., Charney, A., Wang, P., et al. (2020). Multiscale causal networks identify VGF as a key regulator of Alzheimer’s disease. Nat. Commun. 11:3942.
Benito, E., Valor, L., Jimenez-Minchan, M., Huber, W., and Barco, A. (2011). cAMP response element-binding protein is a primary hub of activity-driven neuronal gene expression. J. Neurosci. 31, 18237–18250.
Bilkei-Gorzo, A., Erk, S., Schürmann, B., Mauer, D., Michel, K., Boecker, H., et al. (2012). Dynorphins regulate fear memory: From mice to men. J. Neurosci. 32, 9335–9343. doi: 10.1523/JNEUROSCI.1034-12.2012
Blacker, D., Wilcox, M., Laird, N., Rodes, L., Horvath, S., Go, R., et al. (1998). Alpha-2 macroglobulin is genetically associated with Alzheimer disease. Nat. Genet. 19, 357–360.
Boza-Serrano, A., Yang, Y., Paulus, A., and Deierborg, T. (2018). Innate immune alterations are elicited in microglial cells before plaque deposition in the Alzheimer’s disease mouse model 5xFAD. Sci. Rep. 8:1550. doi: 10.1038/s41598-018-19699-y
Brocos-Mosquera, I., Miranda-Azpiazu, P., Muguruza, C., Corzo-Monje, V., Morentin, B., Meana, J., et al. (2021). Differential brain ADRA2A and ADRA2C gene expression and epigenetic regulation in schizophrenia. Effect of antipsychotic drug treatment. Transl. Psychiatry 11:643. doi: 10.1038/s41398-021-01762-4
Bundy, J., Vied, C., Badger, C., and Nowakowski, R. (2019). Sex-biased hippocampal pathology in the 5XFAD mouse model of Alzheimer’s disease: A multi-omic analysis. J. Comp. Neurol. 527, 462–475. doi: 10.1002/cne.24551
Cargnello, M., and Roux, P. (2011). Activation and function of the MAPKs and their substrates, the MAPK-activated protein kinases. Microbiol. Mol. Biol. Rev. 75, 50–83.
Castranio, E., Hasel, P., Haure-Mirande, J., Ramirez Jimenez, A., Hamilton, B., Kim, R., et al. (2023). Microglial INPP5D limits plaque formation and glial reactivity in the PSAPP mouse model of Alzheimer’s disease. Alzheimers Dement. 19, 2239–2252. doi: 10.1002/alz.12821
Chambers, S., Fasano, C., Papapetrou, E., Tomishima, M., Sadelain, M., and Studer, L. (2009). Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol. 27, 275–280. doi: 10.1038/nbt.1529
Chen, H., Chuang, H., and Tan, T. (2019). Regulation of dual-specificity phosphatase (DUSP) ubiquitination and protein stability. Int. J. Mol. Sci. 20:2668. doi: 10.3390/ijms20112668
Chen, M., Ramesha, S., Weinstock, L., Gao, T., Ping, L., Xiao, H., et al. (2021). Extracellular signal-regulated kinase regulates microglial immune responses in Alzheimer’s disease. J. Neurosci. Res. 99, 1704–1721.
Chen, Y., Peng, Y., Che, P., Gannon, M., Liu, Y., Li, L., et al. (2014). α(2A) adrenergic receptor promotes amyloidogenesis through disrupting APP-SorLA interaction. Proc. Natl. Acad. Sci. U.S.A. 111, 17296–17301. doi: 10.1073/pnas.1409513111
Cho, H., Cahill, C., Vanderburg, C., Scherzer, C., Wang, B., Huang, X., et al. (2010). Selective translational control of the Alzheimer amyloid precursor protein transcript by iron regulatory protein-1. J. Biol. Chem. 285, 31217–31232. doi: 10.1074/jbc.M110.149161
Christensen, A., and Pike, C. (2020). Staining and quantification of beta-amyloid pathology in transgenic mouse models of Alzheimer’s disease. Methods Mol. Biol. 2144, 211–221.
Chu, E., Mychasiuk, R., Hibbs, M., and Semple, B. (2021). Dysregulated phosphoinositide 3-kinase signaling in microglia: Shaping chronic neuroinflammation. J. Neuroinflamm. 18:276. doi: 10.1186/s12974-021-02325-6
Clifton, N., Trent, S., Thomas, K., and Hall, J. (2019). Regulation and function of activity-dependent homer in synaptic plasticity. Mol. Neuropsychiatry 5, 147–161.
Delay, C., Calon, F., Mathews, P., and Hebert, S. (2011). Alzheimer-specific variants in the 3’UTR of Amyloid precursor protein affect microRNA function. Mol. Neurodegener. 6:70. doi: 10.1186/1750-1326-6-70
Dias, C., Punetha, J., Zheng, C., Mazaheri, N., Rad, A., Efthymiou, S., et al. (2019). Homozygous missense variants in NTNG2, encoding a presynaptic netrin-G2 adhesion protein, lead to a distinct neurodevelopmental disorder. Am. J. Hum. Genet. 105, 1048–1056. doi: 10.1016/j.ajhg.2019.09.025
Djordjevic, J., Albensi, S., Slike, A., Perez, C., and Albensi, B. (2021). Sex differences in the expression of synaptic proteins in the brain of the TgCRND8 mouse model of Alzheimer’s disease. Alzheimers Dement. 17:e056574. doi: 10.1007/s12031-016-0865-x
Dobin, A., Davis, C., Schlesinger, F., Drenkow, J., Zaleski, C., Jha, S., et al. (2013). STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. doi: 10.1093/bioinformatics/bts635
Duman, R., Aghajanian, G., Sanacora, G., and Krystal, J. (2016). Synaptic plasticity and depression: New insights from stress and rapid-acting antidepressants. Nat. Med. 22, 238–249.
El Gaamouch, F., Audrain, M., Lin, W., Beckmann, N., Jiang, C., Hariharan, S., et al. (2020). VGF-derived peptide TLQP-21 modulates microglial function through C3aR1 signaling pathways and reduces neuropathology in 5xFAD mice. Mol. Neurodegener. 15:4. doi: 10.1186/s13024-020-0357-x
Ennerfelt, H., Frost, E., Shapiro, D., Holliday, C., Zengeler, K., Voithofer, G., et al. (2022). SYK coordinates neuroprotective microglial responses in neurodegenerative disease. Cell 185:4135–52.e22. doi: 10.1016/j.cell.2022.09.030
Fattorelli, N., Martinez-Muriana, A., Wolfs, L., Geric, I., De Strooper, B., and Mancuso, R. (2021). Stem-cell-derived human microglia transplanted into mouse brain to study human disease. Nat. Protoc. 16, 1013–1033.
Figueiro-Silva, J., Gruart, A., Clayton, K., Podlesniy, P., Abad, M., Gasull, X., et al. (2015). Neuronal pentraxin 1 negatively regulates excitatory synapse density and synaptic plasticity. J. Neurosci. 35, 5504–5521. doi: 10.1523/JNEUROSCI.2548-14.2015
Franke, K., Otto, W., Johannes, S., Baumgart, J., Nitsch, R., and Schumacher, S. (2012). miR-124-regulated RhoG reduces neuronal process complexity via ELMO/Dock180/Rac1 and Cdc42 signalling. Embo J. 31, 2908–2921. doi: 10.1038/emboj.2012.130
Georgakopoulos, A., Yoon, Y., Voloudakis, G., Dimovasili, C., Chen, L., Tang, C., et al. (2021). Presenilin1 FAD mutants impair ischemia-induced brain neovascularization and neuronal survival decreasing γ-secretase processing of ephrinB2 and ephrinB2-mediated angiogenic functions. Alzheimers Dement. 17:e053186.
Haery, L., Deverman, B., Matho, K., Cetin, A., Woodard, K., Cepko, C., et al. (2019). Adeno-associated virus technologies and methods for targeted neuronal manipulation. Front. Neuroanat. 13:93. doi: 10.3389/fnana.2019.00093
Harris, K., Akbergenova, Y., Cho, R., Baas-Thomas, M., and Littleton, J. (2016). Shank modulates postsynaptic wnt signaling to regulate synaptic development. J. Neurosci. 36, 5820–5832. doi: 10.1523/JNEUROSCI.4279-15.2016
Hur, J., Frost, G., Wu, X., Crump, C., Pan, S., Wong, E., et al. (2020). The innate immunity protein IFITM3 modulates gamma-secretase in Alzheimer’s disease. Nature 586, 735–740.
Ji, H., Wang, Y., Liu, G., Xu, X., Dai, D., Chen, Z., et al. (2015). OPRK1 promoter hypermethylation increases the risk of Alzheimer’s disease. Neurosci. Lett. 606, 24–29. doi: 10.1016/j.neulet.2015.08.027
Jin, J., Liu, L., Chen, W., Gao, Q., Li, H., Wang, Y., et al. (2019). The implicated roles of cell adhesion molecule 1 (CADM1) gene and altered prefrontal neuronal activity in attention-deficit/hyperactivity disorder: A “gene-brain-behavior relationship”? Front. Genet. 10:882. doi: 10.3389/fgene.2019.00882
Kim, H., Kim, J., Lee, H., Shin, E., Kang, H., Jeon, J., et al. (2022). Baiap3 regulates depressive behaviors in mice via attenuating dense core vesicle trafficking in subsets of prefrontal cortex neurons. Neurobiol. Stress 16, 2352–2895. doi: 10.1016/j.ynstr.2021.100423
Kim, J., Kim, S., Kim, H., Hwang, I., Bae, S., Karki, S., et al. (2022). MDGA1 negatively regulates amyloid precursor protein-mediated synapse inhibition in the hippocampus. Proc. Natl. Acad. Sci. U.S.A. 119:e2115326119. doi: 10.1073/pnas.2115326119
Kim, S., Smith, C., and Van Eldik, L. (2004). Importance of MAPK pathways for microglial pro-inflammatory cytokine IL-1 beta production. Neurobiol. Aging 25, 431–439.
Kinney, J., Bemiller, S., Murtishaw, A., Leisgang, A., Salazar, A., and Lamb, B. (2018). Inflammation as a central mechanism in Alzheimer’s disease. Alzheimers Dement. 4, 575–590.
Kodama, L., Guzman, E., Etchegaray, J., Li, Y., Sayed, F., Zhou, L., et al. (2020). Microglial microRNAs mediate sex-specific responses to tau pathology. Nat. Neurosci. 23, 167–171. doi: 10.1038/s41593-019-0560-7
Kondo, M., Norris, A., Yang, K., Cheshire, M., Newkirk, I., Chen, X., et al. (2022). Dysfunction of mitochondria and GABAergic interneurons in the anterior cingulate cortex of individuals with schizophrenia. Neurosci. Res. 185, 67–72. doi: 10.1016/j.neures.2022.09.011
Labonte, B., Engmann, O., Purushothaman, I., Menard, C., Wang, J., Tan, C., et al. (2017). Sex-specific transcriptional signatures in human depression. Nat. Med. 23, 1102–1111.
Lalli, M., Garcia, G., Madrigal, L., Arcos-Burgos, M., Arcila, M., Kosik, K., et al. (2012). Exploratory data from complete genomes of familial alzheimer disease age-at-onset outliers. Hum. Mutat. 33, 1630–1634. doi: 10.1002/humu.22167
Landel, V., Baranger, K., Virard, I., Loriod, B., Khrestchatisky, M., Rivera, S., et al. (2014). Temporal gene profiling of the 5XFAD transgenic mouse model highlights the importance of microglial activation in Alzheimer’s disease. Mol. Neurodegener. 9:33. doi: 10.1186/1750-1326-9-33
Lauridsen, J., Johansen, J., Rekling, J., Thirstrup, K., Moerk, A., and Sager, T. (2011). Regulation of the Bcas1 and Baiap3 transcripts in the subthalamic nucleus in mice recovering from MPTP toxicity. Neurosci. Res. 70, 269–276. doi: 10.1016/j.neures.2011.03.011
Lee, T., and Lee, H. (2020). Prediction of Alzheimer’s disease using blood gene expression data. Sci. Rep. 10:3485.
Letoha, T., Hudak, A., Kusz, E., Pettko-Szandtner, A., Domonkos, I., Josvay, K., et al. (2019). Contribution of syndecans to cellular internalization and fibrillation of amyloid-beta(1-42). Sci. Rep. 9:1393. doi: 10.1038/s41598-018-37476-9
Liao, W., Zheng, Y., Fang, W., Liao, S., Xiong, Y., Li, Y., et al. (2018). dual specificity phosphatase 6 protects neural stem cells from beta-amyloid-induced cytotoxicity through ERK1/2 inactivation. Biomolecules 8:181. doi: 10.3390/biom8040181
Liao, Y., Smyth, G., and Shi, W. (2014). featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. doi: 10.1093/bioinformatics/btt656
Lie, E., Li, Y., Kim, R., and Kim, E. (2018). SALM/Lrfn family synaptic adhesion molecules. Front. Mol. Neurosci. 11:105. doi: 10.3389/fnmol.2018.00105
Lie, E., Yeo, Y., Lee, E., Shin, W., Kim, K., Han, K., et al. (2021). SALM4 negatively regulates NMDA receptor function and fear memory consolidation. Commun. Biol. 4:1138. doi: 10.1038/s42003-021-02656-3
Long, J., Maloney, B., Rogers, J., and Lahiri, D. (2018). Novel upregulation of amyloid-β precursor protein (APP) by microRNA-346 via targeting of APP mRNA 5’-untranslated region: Implications in Alzheimer’s disease. Mol. Psychiatry 24, 345–363.
Mancuso, R., Van Den Daele, J., Fattorelli, N., Wolfs, L., Balusu, S., Burton, O., et al. (2019). Stem-cell-derived human microglia transplanted in mouse brain to study human disease. Nat. Neurosci. 22, 2111–2116.
Manji, Z., Rojas, A., Wang, W., Dingledine, R., Varvel, N., and Ganesh, T. (2019). 5xFAD mice display sex-dependent inflammatory gene induction during the prodromal stage of Alzheimer’s disease. J. Alzheimers Dis. 70, 1259–1274. doi: 10.3233/JAD-180678
Manzoor, Z., and Koh, Y. (2012). Mitogen-activated protein kinases in inflammation. J. Bacteriol. Virol. 42, 169–171.
Martín-Sánchez, A., Piñero, J., Nonell, L., Arnal, M., Ribe, E., Nevado-Holgado, A., et al. (2021). Comorbidity between Alzheimer’s disease and major depression: A behavioural and transcriptomic characterization study in mice. Alzheimers Res. Ther. 13:73. doi: 10.1186/s13195-021-00810-x
Matthews, K., Xu, W., Gaglioti, A., Holt, J., Croft, J., Mack, D., et al. (2019). Racial and ethnic estimates of Alzheimer’s disease and related dementias in the United States (2015–2060) in adults aged =65 years. Alzheimers Dement. 15, 17–24. doi: 10.1016/j.jalz.2018.06.3063
Montminy, M., and Bilezikjian, L. (1987). Binding of a nuclear protein to the cyclic-AMP response element of the somatostatin gene. Nature 328, 175–178. doi: 10.1038/328175a0
Montminy, M., Gonzalez, G., and Yamamoto, K. (1990). Regulation of camp-inducible genes by creb. Trends Neurosci. 13, 184–188.
Morimura, N., Inoue, T., Katayama, K., and Aruga, J. (2006). Comparative analysis of structure, expression and PSD95-binding capacity of Lrfn, a novel family of neuronal transmembrane proteins. Gene 380, 72–83. doi: 10.1016/j.gene.2006.05.014
Muraoka, S., Jedrychowski, M., Iwahara, N., Abdullah, M., Onos, K., Keezer, K., et al. (2021). Enrichment of neurodegenerative microglia signature in brain-derived extracellular vesicles isolated from Alzheimer’s disease mouse models. J. Proteome Res. 20, 1733–1743. doi: 10.1021/acs.jproteome.0c00934
Oakley, H., Cole, S., Logan, S., Maus, E., Shao, P., Craft, J., et al. (2006). Intraneuronal beta-amyloid aggregates, neurodegeneration, and neuron loss in transgenic mice with five familial Alzheimer’s disease mutations: Potential factors in amyloid plaque formation. J. Neurosci. 26, 10129–10140. doi: 10.1523/JNEUROSCI.1202-06.2006
Oblak, A., Lin, P., Kotredes, K., Pandey, R., Garceau, D., Williams, H., et al. (2021). Comprehensive evaluation of the 5XFAD mouse model for preclinical testing applications: A MODEL-AD study. Front. Aging Neurosci. 13:713726. doi: 10.3389/fnagi.2021.713726
Paasila, P., Davies, D., Sutherland, G., and Goldsbury, C. (2020). Clustering of activated microglia occurs before the formation of dystrophic neurites in the evolution of Abeta plaques in Alzheimer’s disease. Free Neuropathol. 1, 1–20. doi: 10.17879/freeneuropathology-2020-2845
Pan, A., Audrain, M., Sakakibara, E., Joshi, R., Zhu, X., Wang, Q., et al. (2022). Dual-specificity protein phosphatase 4 (DUSP4) overexpression improves learning behavior selectively in female 5xFAD mice, and reduces β-amyloid load in males and females. Cells 11:3880. doi: 10.3390/cells11233880
Papageorgiou, I., Lewen, A., Galow, L., Cesetti, T., Scheffel, J., Regen, T., et al. (2016). TLR4-activated microglia require IFN-gamma to induce severe neuronal dysfunction and death in situ. Proc. Natl. Acad. Sci. U.S.A. 113, 212–217. doi: 10.1073/pnas.1513853113
Pelucchi, S., Gardoni, F., Di Luca, M., and Marcello, E. (2022). Synaptic dysfunction in early phases of Alzheimer’s disease. Handb. Clin. Neurol. 184, 417–438.
Peters, F., Salihoglu, H., Rodrigues, E., Herzog, E., Blume, T., Filser, S., et al. (2018). BACE1 inhibition more effectively suppresses initiation than progression of β-amyloid pathology. Acta Neuropathol. 135, 695–710.
Qi, Y., Zhang, X., Renier, N., Wu, Z., Atkin, T., Sun, Z., et al. (2017). Combined small-molecule inhibition accelerates the derivation of functional cortical neurons from human pluripotent stem cells. Nat. Biotechnol. 35, 154–163. doi: 10.1038/nbt.3777
Rahman, A., Jackson, H., Hristov, H., Isaacson, R., Saif, N., Shetty, T., et al. (2019). Sex and gender driven modifiers of Alzheimer’s: The role for estrogenic control across age, race, medical, and lifestyle risks. Front. Aging Neurosci. 11:315. doi: 10.3389/fnagi.2019.00315
Reif, A., Grünblatt, E., Herterich, S., Wichart, I., Rainer, M. K., Jungwirth, S., et al. (2011). Association of a functional NOS1 promoter repeat with Alzheimer’s disease in the VITA cohort. J. Alzheimers Dis. 23, 327–333. doi: 10.3233/JAD-2010-101491
Reinhardt, S., Schuck, F., Grosgen, S., Riemenschneider, M., Hartmann, T., Postina, R., et al. (2014). Unfolded protein response signaling by transcription factor XBP-1 regulates ADAM10 and is affected in Alzheimer’s disease. FASEB J. 28, 978–997. doi: 10.1096/fj.13-234864
Requejo-Aguilar, R. (2023). Cdk5 and aberrant cell cycle activation at the core of neurodegeneration. Neural Regener. Res. 18, 1186–1190. doi: 10.4103/1673-5374.360165
Ritchie, M., Phipson, B., Wu, D., Hu, Y., Law, C., Shi, W., et al. (2015). Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43:e47.
Robinson, M., McCarthy, D. J., and Smyth, G. K. (2010). edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140.
Rogers, G., Richter, N., and Merrick, W. (1999). Biochemical and kinetic characterization of the RNA helicase activity of eukaryotic initiation factor 4A. J. Biol. Chem. 274, 12236–12244.
Rogers, J., Leiter, L., McPhee, J., Cahill, C., Zhan, S., Potter, H., et al. (1999). Translation of the alzheimer amyloid precursor protein mRNA is up-regulated by interleukin-1 through 5’-untranslated region sequences. J. Biol. Chem. 274, 6421–6431.
Sadleir, K., Eimer, W., Cole, S., and Vassar, R. (2015). Aβ reduction in BACE1 heterozygous null 5XFAD mice is associated with transgenic APP level. Mol. Neurodegener. 10:1.
Sadleir, K., Popovic, J., and Vassar, R. (2018). ER stress is not elevated in the 5XFAD mouse model of Alzheimer’s disease. J Biol Chem. 293, 18434–18443.
Serrano-Pozo, A., Mielke, M., Gomez-Isla, T., Betensky, R., Growdon, J., Frosch, M., et al. (2011). Reactive glia not only associates with plaques but also parallels tangles in Alzheimer’s disease. Am. J. Pathol. 179, 1373–1384.
Shen, Z., Sun, D., Savastano, A., Varga, S., Cima-Omori, M., Becker, S., et al. (2023). Multivalent Tau/PSD-95 interactions arrest in vitro condensates and clusters mimicking the postsynaptic density. Nat. Commun. 14:6839. doi: 10.1038/s41467-023-42295-2
Shi, R., Redman, P., Ghose, D., Hwang, H., Liu, Y., Ren, X., et al. (2017). Shank proteins differentially regulate synaptic transmission. eNeuro 4:0163-15.2017 1–12.
Sobue, A., Komine, O., Hara, Y., Endo, F., Mizoguchi, H., Watanabe, S., et al. (2021). Microglial gene signature reveals loss of homeostatic microglia associated with neurodegeneration of Alzheimer’s disease. Acta Neuropathol. Commun. 9:1.
Song, W., Son, M., Lee, H., Seo, H., Kim, J., and Chung, S. (2015). Enhancement of BACE1 Activity by p25/Cdk5-Mediated Phosphorylation in Alzheimer’s disease. PLoS One 10:e0136950. doi: 10.1371/journal.pone.0136950
Ting, L., Lu, H., Yen, S., Ngo, T., Tu, F., Tsai, I., et al. (2019). Expression of AHI1 rescues amyloidogenic pathology in alzheimer’s disease model cells. Mol. Neurobiol. 56, 7572–7582. doi: 10.1007/s12035-019-1587-1
Toh, W., Chia, P., Hossain, M., and Gleeson, P. (2018). GGA1 regulates signal-dependent sorting of BACE1 to recycling endosomes, which moderates Aβ production. Mol. Biol. Cell 29, 191–208.
Vilardo, E., Barbato, C., Ciotti, M., Cogoni, C., and Ruberti, F. (2010). MicroRNA-101 regulates amyloid precursor protein expression in hippocampal neurons. J. Biol. Chem. 285, 18344–18351.
Walker, D., Tang, T., Mendsaikhan, A., Tooyama, I., Serrano, G., Sue, L., et al. (2020). Patterns of expression of purinergic receptor P2RY12, a putative marker for non-activated microglia, in aged and Alzheimer’s disease brains. Int. J. Mol. Sci. 21:678. doi: 10.3390/ijms21020678
Wang, C., Chang, K., Petralia, R., Wang, Y., Seabold, G., and Wenthold, R. J. (2006). A novel family of adhesion-like molecules that interacts with the NMDA receptor. J. Neurosci. 26, 2174–2183. doi: 10.1523/JNEUROSCI.3799-05.2006
Wang, I., Ho, P., and Tsai, K. (2022). MicroRNAs in learning and memory and their impact on Alzheimer’s disease. Biomedicines 10:1856.
Wang, M., Beckmann, N., Roussos, P., Wang, E., Zhou, X., Wang, Q., et al. (2018). The Mount Sinai cohort of large-scale genomic, transcriptomic and proteomic data in Alzheimer’s disease. Sci. Data 5:180185. doi: 10.1038/sdata.2018.185
Wang, X., Liu, D., Huang, H., Wang, Z., Hou, T., Yang, X., et al. (2018). A Novel MicroRNA-124/PTPN1 signal pathway mediates synaptic and memory deficits in Alzheimer’s disease. Biol. Psychiatry 83, 395–405. doi: 10.1016/j.biopsych.2017.07.023
Wang, M., Li, A., Sekiya, M., Beckmann, N., Quan, X., Schrode, N., et al. (2021). Transformative network modeling of multi-omics data reveals detailed circuits, key regulators, and potential therapeutics for Alzheimer’s Disease. Neuron 109:257–72 e14. doi: 10.1016/j.neuron.2020.11.002
Wang, Y., Zan, G., Xu, C., Li, X., Shu, X., Yao, S., et al. (2023). The claustrum-prelimbic cortex circuit through dynorphin/κ-opioid receptor signaling underlies depression-like behaviors associated with social stress etiology. Nat. Commun. 14:7903. doi: 10.1038/s41467-023-43636-x
Wen, Y., Yu, W., Maloney, B., Bailey, J., Ma, J., Marié, I., et al. (2008). Transcriptional regulation of beta-secretase by p25/cdk5 leads to enhanced amyloidogenic processing. Neuron 57, 680–690. doi: 10.1016/j.neuron.2008.02.024
Williams, M., Wilke, S., Daggett, A., Davis, E., Otto, S., Ravi, D., et al. (2011). Cadherin-9 regulates synapse-specific differentiation in the developing hippocampus. Neuron 71, 640–655. doi: 10.1016/j.neuron.2011.06.019
Wojcik, S., Tantra, M., Stepniak, B., Man, K., Müller-Ribbe, K., Begemann, M., et al. (2013). Genetic markers of a Munc13 protein family member, BAIAP3, are gender specifically associated with anxiety and benzodiazepine abuse in 03mice and humans. Mol. Med. 19, 135–148. doi: 10.2119/molmed.2013.00033
Xing, J., Ginty, D., and Greenberg, M. (1996). Coupling of the RAS-MAPK pathway to gene activation by RSK2, a growth factor-regulated CREB kinase. Science 273, 959–963. doi: 10.1126/science.273.5277.959
Xing, J., Kornhauser, J., Xia, Z., Thiele, E., and Greenberg, M. (1998). Nerve growth factor activates extracellular signal-regulated kinase and p38 mitogen-activated protein kinase pathways to stimulate CREB serine 133 phosphorylation. Mol. Cell Biol. 18, 1946–1955. doi: 10.1128/MCB.18.4.1946
Yamamoto, M., Kiyota, T., Horiba, M., Buescher, J., Walsh, S., Gendelman, H., et al. (2007). Interferon-gamma and tumor necrosis factor-alpha regulate amyloid-beta plaque deposition and beta-secretase expression in Swedish mutant APP transgenic mice. Am. J. Pathol. 170, 680–692. doi: 10.2353/ajpath.2007.060378
Yamazaki, H., Jin, Y., Tsuchiya, A., Kanno, T., and Nishizaki, T. (2015). Adipose-derived stem cell-conditioned medium ameliorates antidepression-related behaviors in the mouse model of Alzheimer’s disease. Neurosci. Lett. 609, 53–57. doi: 10.1016/j.neulet.2015.10.023
Yang, L., Wei, M., Xing, B., and Zhang, C. (2023). Extracellular matrix and synapse formation. Biosci. Rep. 43:BSR20212411.
Yang, Y., Shu, X., Liu, D., Shang, Y., Wu, Y., Pei, L., et al. (2012). EPAC null mutation impairs learning and social interactions via aberrant regulation of miR-124 and Zif268 translation. Neuron 73, 774–788. doi: 10.1016/j.neuron.2012.02.003
Yao, A., and Yan, R. (2020). Activity of Alzheimer’s gamma-secretase is linked to changes of interferon-induced transmembrane proteins (IFITM) in innate immunity. Mol. Neurodegener. 15:69. doi: 10.1186/s13024-020-00417-0
Yu, Z., Sun, D., Feng, J., Tan, W., Fang, X., Zhao, M., et al. (2015). MSX3 switches microglia polarization and protects from inflammation-induced demyelination. J. Neurosci. 35, 6350–6365. doi: 10.1523/JNEUROSCI.2468-14.2015
Zhang, W., Jiang, L., Li, M., and Liu, J. (2023). MicroRNA-124: An emerging therapeutic target in central nervous system disorders. Exp. Brain Res. 241, 1215–1226.
Zhang, J., Li, X., Xiao, J., Xiang, Y., and Ye, F. (2023). Analysis of gene expression profiles in Alzheimer’s disease patients with different lifespan: A bioinformatics study focusing on the disease heterogeneity. Front. Aging Neurosci. 15:1072184. doi: 10.3389/fnagi.2023.1072184
Zhang, T., Chen, D., and Lee, T. (2020). Phosphorylation signaling in APP processing in Alzheimer’s disease. Int. J. Mol. Sci. 21:209.
Zhao, Y., Wu, X., Li, X., Jiang, L., Gui, X., Liu, Y., et al. (2018). TREM2 Is a Receptor for beta-Amyloid that Mediates Microglial Function. Neuron 97: 1023–31.e7.
Zhong, W., Wu, A., Berglund, K., Gu, X., Jiang, M., Talati, J., et al. (2022). Pathogenesis of sporadic Alzheimer’s disease by deficiency of NMDA receptor subunit GluN3A. Alzheimers Dement. 18, 222–239.
Zhong, Y., Chen, J., Chen, J., Chen, Y., Li, L., and Xie, Y. (2019). Crosstalk between Cdk5/p35 and ERK1/2 signalling mediates spinal astrocyte activity via the PPARγ pathway in a rat model of chronic constriction injury. J. Neurochem. 151, 166–184.
Keywords: neuroinflammation, Alzheimer’s disease, dual-specificity protein phosphatase 6, mitogen-activated protein kinase, microglial activation
Citation: Pan AL, Audrain M, Sakakibara E, Joshi R, Zhu X, Wang Q, Wang M, Beckmann ND, Schadt EE, Gandy S, Zhang B, Ehrlich ME and Salton SR (2024) Dual-specificity protein phosphatase 6 (DUSP6) overexpression reduces amyloid load and improves memory deficits in male 5xFAD mice. Front. Aging Neurosci. 16:1400447. doi: 10.3389/fnagi.2024.1400447
Received: 13 March 2024; Accepted: 14 June 2024;
Published: 28 June 2024.
Edited by:
Erin Rutherford Hascup, Southern Illinois University School of Medicine, United StatesReviewed by:
Seokjo Kang, Regenerative Medicine Institute, United StatesCopyright © 2024 Pan, Audrain, Sakakibara, Joshi, Zhu, Wang, Wang, Beckmann, Schadt, Gandy, Zhang, Ehrlich and Salton. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michelle E. Ehrlich, bWljaGVsbGUuZWhybGljaEBtc3NtLmVkdQ==; Stephen Salton, c3RlcGhlbi5zYWx0b25AbXNzbS5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.