- 1Department of Neurology, Dongguk University Ilsan Hospital, Dongguk University College of Medicine, Goyang, Republic of Korea
- 2Department of Neurology, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Republic of Korea
- 3Alzheimer’s Disease Convergence Research Center, Samsung Medical Center, Seoul, Republic of Korea
- 4Department of Biostatistics, Epidemiology and Informatics, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, United States
- 5Department of Digital Health, Samsung Advanced Institute for Health Sciences & Technology, Samsung Medical Center, Sungkyunkwan University, Seoul, Republic of Korea
- 6Department of Neurology, Dongtan Sacred Heart Hospital, Hallym University College of Medicine, Hwaseong, Republic of Korea
- 7Department of Radiology and Imaging Sciences, Center for Neuroimaging, Indiana University School of Medicine, Indianapolis, IN, United States
- 8Samsung Genome Institute, Samsung Medical Center, Seoul, Republic of Korea
- 9Department of Artificial Intelligence, Ajou University, Suwon, Republic of Korea
- 10Department of Software and Computer Engineering, Ajou University, Suwon, Republic of Korea
- 11Cell and Gene Therapy Institute, Research Institute for Future Medicine, Samsung Medical Center, Seoul, Republic of Korea
- 12Department of Health Sciences and Technology, Samsung Advanced Institute for Health Sciences & Technology, Samsung Medical Center, Sungkyunkwan University, Seoul, Republic of Korea
- 13Department of Intelligent Precision Healthcare Convergence, Seoul, Republic of Korea
Background: The genetic basis of amyloid β (Aβ) deposition in subcortical vascular cognitive impairment (SVCI) is still unknown. Here, we investigated genetic variants involved in Aβ deposition in patients with SVCI.
Methods: We recruited a total of 110 patients with SVCI and 424 patients with Alzheimer’s disease-related cognitive impairment (ADCI), who underwent Aβ positron emission tomography and genetic testing. Using candidate AD-associated single nucleotide polymorphisms (SNPs) that were previously identified, we investigated Aβ-associated SNPs that were shared or distinct between patients with SVCI and those with ADCI. Replication analyses were performed using the Alzheimer’s Disease Neuroimaging Initiative (ADNI) and Religious Orders Study and Rush Memory and Aging Project cohorts (ROS/MAP).
Results: We identified a novel SNP, rs4732728, which showed distinct associations with Aβ positivity in patients with SVCI (Pinteraction = 1.49 × 10–5); rs4732728 was associated with increased Aβ positivity in SVCI but decreased Aβ positivity in ADCI. This pattern was also observed in ADNI and ROS/MAP cohorts. Prediction performance for Aβ positivity in patients with SVCI increased (area under the receiver operating characteristic curve = 0.780; 95% confidence interval = 0.757–0.803) when rs4732728 was included. Cis-expression quantitative trait loci analysis demonstrated that rs4732728 was associated with EPHX2 expression in the brain (normalized effect size = −0.182, P = 0.005).
Conclusion: The novel genetic variants associated with EPHX2 showed a distinct effect on Aβ deposition between SVCI and ADCI. This finding may provide a potential pre-screening marker for Aβ positivity and a candidate therapeutic target for SVCI.
Introduction
Subcortical vascular cognitive impairment (SVCI), the second most prevalent cause of dementia in East Asia, is characterized by extensive cerebral small vessel disease (CSVD) burdens, which include white matter hyperintensities (WMHs) and multiple lacunes (Román et al., 2002). Although amyloid beta (Aβ) deposition is a pathological hallmark of Alzheimer’s disease-related cognitive impairment (ADCI), it frequently co-exists with SVCI, with approximately 30–40% of patients with SVCI having significant brain Aβ depositions, as measured by positron emission tomography (PET) (Lee et al., 2011, 2014; Kang et al., 2021). Previous studies have also demonstrated that Aβ deposition is associated with poor clinical outcomes in patients with SVCI (Kim et al., 2013a; Lee et al., 2014; Ye et al., 2015).
The aberrant deposition of Aβ in ADCI is related to the decreased Aβ clearance; specifically, decreased Aβ clearance can result from impaired microglial function, enzymatic degradation, perivascular Aβ drainage, and the blood–brain barrier (BBB) function (Grimmer et al., 2012; Tarasoff-Conway et al., 2015). We previously revealed that patients with SVCI showed predominant Aβ deposition in the occipital lobe (Jang et al., 2018) and WMHs were associated with Aβ deposition, particularly in posterior brain regions (Noh et al., 2014). Considering that the posterior regions are vulnerable to ischemic injury, the CSVD burden may impaired Aβ clearance by creating a deficiency in perivascular Aβ drainage and in the BBB (Grinberg and Thal, 2010; Zlokovic, 2011). Therefore, the pathobiology of Aβ deposition in patients with SVCI may differ from that of patients with ADCI (Kim et al., 2013b; Lee et al., 2020).
Regarding Aβ deposition in ADCI, genetic factors play an important role; for example, a number of genetic variants, including APOE ∈4, have been strongly associated with Aβ deposition in the brain (Morris et al., 2010; Yan et al., 2021). However, to the best of our knowledge, no previous study evaluated the genetic basis of Aβ deposition in SVCI.
In the present study, we aimed to identify genetic variants involved with Aβ deposition using single nucleotide polymorphism (SNP) data from patients with SVCI and ADCI. We hypothesized that there may be SNPs associated with Aβ deposition that are shared and distinct between patients with SVCI and ADCI.
Materials and methods
Study participants (discovery data)
We prospectively recruited 110 patients with SVCI and 424 patients with ADCI [284 with amnestic mild cognitive impairment (aMCI) and 140 with AD dementia (ADD)] who underwent Aβ PET at Samsung Medical Center (Seoul, South Korea) between September 2015 and December 2018 and were genotyped using peripheral blood samples in 2019.
Patients with SVCI satisfied the following criteria for SVCI diagnosis: (i) subjective cognitive complaints from either the patient or a caregiver; (ii) objective cognitive impairment below the 16th percentile in any domain, including attention, language, visuospatial, memory, and frontal/executive functions, on the basis of detailed neuropsychological tests (Kang et al., 2003, 2019; Ahn et al., 2010); (iii) significant ischemia on brain magnetic resonance imaging (MRI), defined as periventricular WMH ≥10 mm and deep WMH ≥25 mm, modified from Fazekas’ ischemia criteria, as described in previous studies (Fazekas et al., 1993; Seo et al., 2009), which met the imaging criteria for SVCI proposed by Erkinjuntti et al. (2000); and (iv) focal neurological symptoms or signs.
Patients with aMCI met the following criteria, modified from Peterson’s criteria (Petersen, 2011): (i) normal activities of daily living; (ii) objective memory impairment according to verbal or visual memory tests, which was below the 16th percentile of that in age- and education-matched norms; and (iii) no dementia. Patients with ADD satisfied the core clinical criteria for probable ADD according to the National Institute of Neurological and Communicative Disorders and Stroke and Alzheimer’s Disease and Related Disorders Association (McKhann et al., 2011).
All patients were evaluated through clinical interviews and neurological and neuropsychological examinations. Patients also underwent laboratory tests, including a complete blood count, blood chemistry assessment, vitamin B12, folate evaluation, syphilis serological assessment, and thyroid function test. Brain MRI confirmed the absence of structural lesions, including territorial cerebral infarction, brain tumors, hippocampal sclerosis, and vascular malformations.
All participants provided written informed consent, and the study was approved by the Institutional Review Board of the Samsung Medical Center.
Genotype data
Peripheral blood samples were genotyped using the Illumina Asian Screening Array BeadChip (Illumina, CA, USA), and SNP markers were analyzed. Quality control (QC) was conducted using PLINK software (version 1.9) (Purcell et al., 2007). Patients were excluded according to the following criteria: (i) call rate <95%, (ii) mismatch between reported and genetically inferred sex, (iii) deviation from each population parameter [5 SD from the sample mean based on the first or second genomic principal components (PCs) of genetic ancestry], and (iv) excess heterozygosity rate (5 SD from the mean). If two patients were related to the second or closer degree, as assessed using KING (Manichaikul et al., 2010), one of the two was excluded. SNPs were excluded based on the following criteria: (i) call rate <98%, (ii) minor allele frequency (MAF) <1%, and (iii) a P-value < 10–6 in the Hardy-Weinberg equilibrium test. After QC, genome-wide imputation was performed using Minimac4 software and all available reference haplotypes from HRC-r1.1 at the University of Michigan Imputation Server (Howie et al., 2012; Fuchsberger et al., 2015). For post-imputation QC, we excluded SNPs according to the following criteria: (i) poor imputation quality (r2 ≤ 0.8) and (ii) MAF ≤1%. Among the filtered SNPs, we restricted our analysis to AD-associated SNPs using summary statistics published by the International Genomics of Alzheimer’s Project (IGAP) (Kunkle et al., 2019). IGAP is one of the largest studies (composed of 41,944 AD patients and 21,982 controls), results from which have been validated in a number of subsequent studies. We selected SNPs with genome-wide suggestive associations with AD diagnosis (P < 1 × 10–6) based on summary statistics from IGAP (Kunkle et al., 2019). Finally, 2,548 SNPs were analyzed in this study.
Aβ PET acquisition and visual assessment
Amyloid β PET images were obtained using a Discovery STE PET/CT scanner (GE Medical Systems, WI, USA). The PET images were acquired 90 min after an intravenous injection with 18F-florbetaben or 18F-flutemetamol. The acquisition time was 20 min. Aβ positivity or negativity was determined by well-trained nuclear physicians using visual assessments of florbetaben (Barthel et al., 2011) and flutemetamol (Curtis et al., 2015) PET images. Briefly, positivity for tracer uptake was assessed in four cortical regions (lateral temporal, frontal, parietal, and posterior cingulate cortices) for florbetaben PET and five cortical regions (lateral temporal, frontal, parietal, posterior cingulate cortices, and striatum) for flutemetamol PET. Amyloid PET positivity was defined as having at least one cortical region with evidence of positive uptake.
Replication data
For the first replication analysis, we used data from individuals enrolled in the Alzheimer’s Disease Neuroimaging Initiative (ADNI)-GO/2 dataset, with available genetic, Aβ PET, and WMH volume data. For the second replication analysis, we used data from Religious Orders Study and Rush Memory and Aging Project (ROS/MAP) cohorts (Bennett et al., 2018). The details of the two cohorts are described in Supplementary File.
Statistical methods
Discovery analysis
We performed two analyses to identify genetic variants associated with Aβ positivity that were shared (the same effect) or distinct (the opposite effect) between patients with SVCI and those with ADCI.
First, to identify shared SNPs, we used a logistic regression model with covariates (including age, sex, education, diagnosis, and the first four PCs of genetic ancestry) expressed as: Aβ positivity = β0 + β1 age + β2 sex + β3 education + β4 diagnosis (SVCI or ADCI) + β5 PC1 + β6 PC2 + β7 PC3 + β8 PC4 + β9 SNP (additive model, 0, 1, and 2 as the number of minor alleles).
Second, to identify distinct SNPs between SVCI and ADCI, we included the interaction term in the logistic regression model, expressed as: Aβ positivity = β0 + β1 age + β2 sex + β3 education + β4 diagnosis + β5 PC1 + β6 PC2 + β7 PC3 + β8 PC4 + β9 SNP + β10 SNP × diagnosis. The term of interest in this model was the SNP × diagnosis interaction, which identified SNPs with distinct associations with Aβ between SVCI and ADCI. Considering the number of tested SNPs (n = 2,548), we defined a P-value < 1.96 × 10–5 as statistically significant based on the Bonferroni correction (0.05/2,548).
Replication analysis
Because the ADNI database only recruited patients with ADCI but not with SVCI, we used the WMH volume data, which is a hallmark of SVCI. We used a multivariable logistic regression model, including the WMH volume. To replicate the association of distinct SNPs, we included the interaction term in the logistic regression model, expressed as: Aβ positivity = β0 + β1 age + β2 sex + β3 education + β4 intracranial volume + β5 WMH + β6 SNP + β7 SNP × WMH. This model evaluates whether the association of SNPs with Aβ positivity differs according to the level of WMH.
Regarding the replication in the ROS/MAP cohorts, we leveraged both amyloid and cerebral vessel pathology data. Aβ positivity was determined using the binarized score of the Consortium to Establish a Registry for Alzheimer’s Disease (negative for none to sparse, positive for moderate to frequent) (Bennett et al., 2006). Cerebral vessel pathology was scored based on the severity of arteriosclerosis, as follows: negative for none to mild and positive for moderate to severe (Nag et al., 2015). To replicate the association of distinct SNPs, we included the interaction term in the logistic regression model, expressed as: Aβ positivity = β0 + β1 age at death + β2 sex + β3 education + β4 post-mortem interval + β5 study (ROS or MAP) + β6 cerebral arteriosclerosis + β7 SNP + β8 SNP × cerebral arteriosclerosis. This model evaluates whether the association of SNPs with Aβ positivity differs according to the presence of cerebral arteriosclerosis. In addition, we evaluated whether SNP interacts with the degree of cerebral amyloid angiopathy on Aβ positivity using the following model: Aβ positivity = β0 + β1 age at death + β2 sex + β3 education + β4 post-mortem interval + β5 study (ROS or MAP) + β6 cerebral amyloid angiopathy + β7 SNP + β8 SNP × cerebral amyloid angiopathy. For the replication analyses, we defined a significance level of P < 0.05.
Functional analysis
We characterized the function of the identified SNPs by leveraging bioinformatics tools. First, we checked whether the MAF of SNPs in our data was similar to that in East Asian populations using the 1000 Genome Project dataset (Sherry et al., 2001). Next, we performed enrichment analysis using HaploReg (version 4.1) and cis-expression quantitative trait loci (cis-eQTL) analysis through the Genotype-Tissue Expression portal (Carithers and Moore, 2015)1. Detailed description of the functional analysis is provided in Supplementary File.
Prediction of Aβ positivity using the newly identified SNPs
To test the clinical utility of the newly identified SNPs, we developed multivariable logistic models to predict Aβ positivity in each individual. We performed receiver operating characteristic curve analysis and measured the area under the receiver operating characteristic curve (AUC). As an internal validation, we conducted a 10-fold cross-validation with 100 repeats. Data are reported as the mean AUC and 95% confidence interval (CI).
Results
Clinical characteristics of the study participants
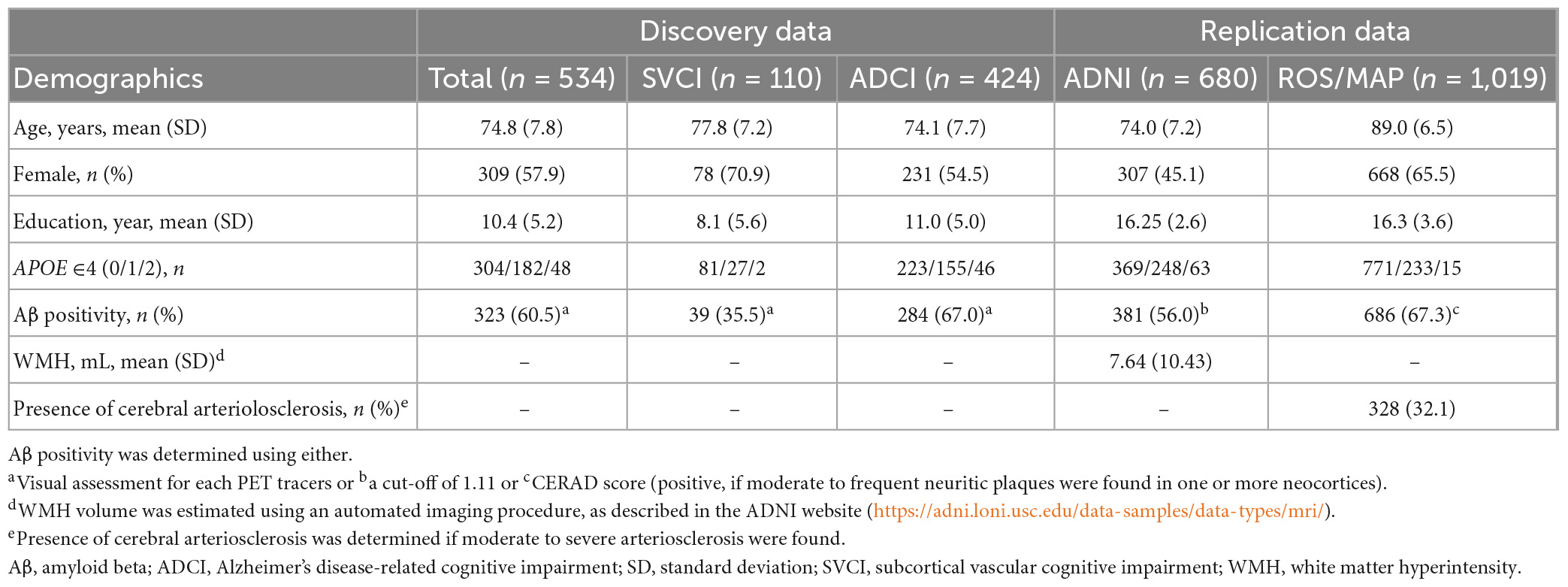
Table 1 shows the baseline demographics of the discovery and replication datasets. In the discovery dataset, 67.0% of the patients with ADCI and 35.5% of those with SVCI showed positive results for Aβ deposition in the brain.
Discovery analysis
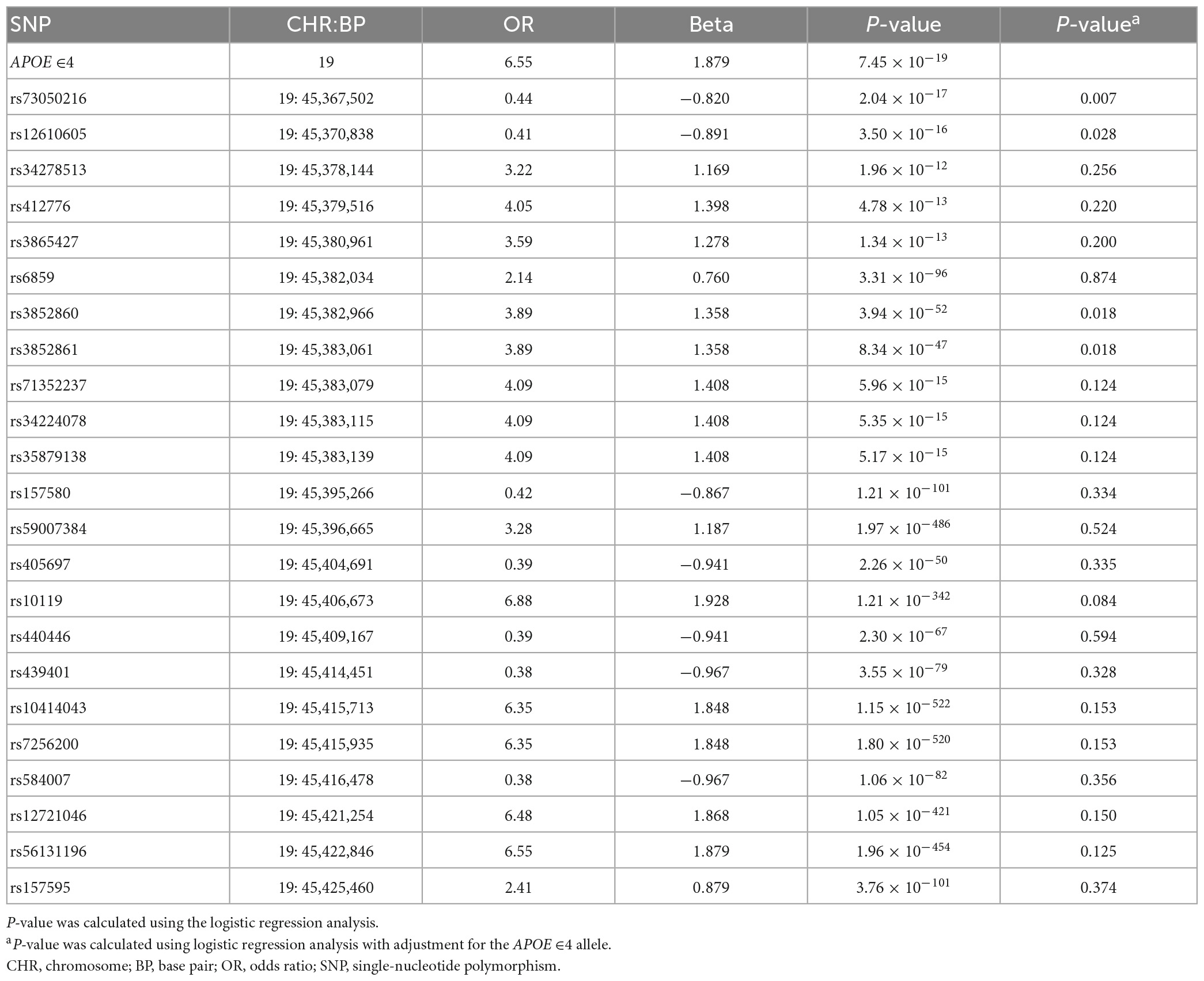
Analysis of Aβ-associated SNPs that were shared between patients with SVCI and those with ADCI revealed 23 SNPs on chromosome 19 (P < 1.961 × 10–5) (Table 2). These significant SNPs were located within a 500-kb region surrounding the APOE gene and they lost genome-wide significance when adjusted for the APOE ∈4 allele.
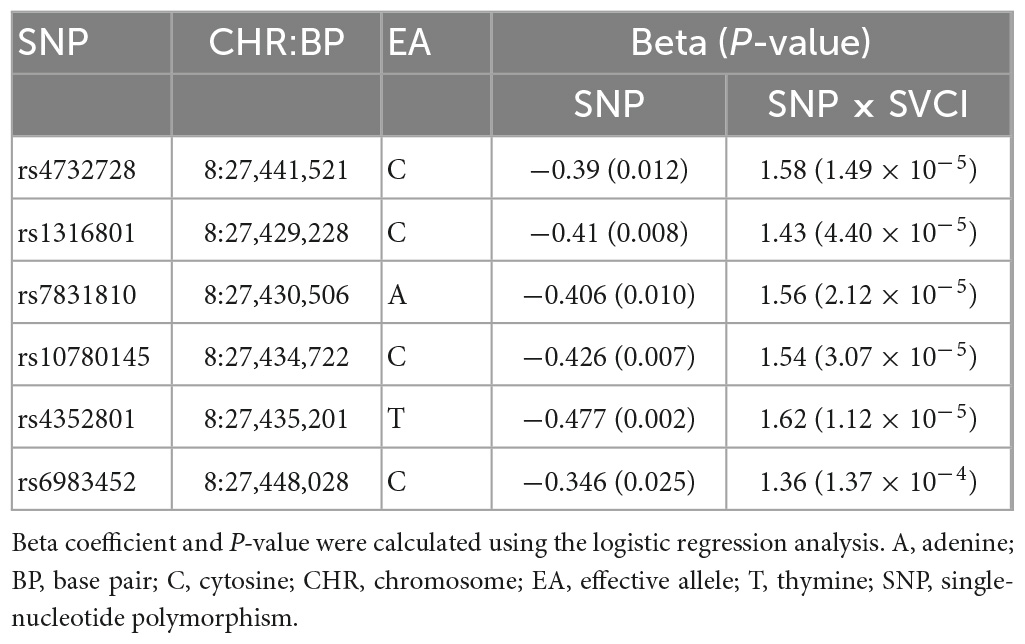
The analysis of Aβ-associated SNPs that were distinct between patients with SVCI and those with ADCI revealed one significant SNP on chromosome 8, rs4732728 (β = 1.58, P = 1.49 × 10–5; Table 3). A similar result was observed after adjusting for the APOE ∈4 allele (β = 1.60, P = 7.19 × 10–5). Subgroup analyses based on the diagnosis (SVCI or ADCI) showed that rs4732728 was associated with a 4.58-fold higher risk of Aβ deposition in SVCI [odds ratio (OR) = 4.58, P = 8.04 × 10–5] and a 1.32-fold lower risk of Aβ deposition in ADCI (OR = 0.76, P = 0.01) (Figure 1). In the regional association plot of rs4732728 (Figure 2), SNPs in high linkage disequilibrium (LD; r2 > 0.8) also had a significant interaction with SVCI on Aβ deposition (Table 3).
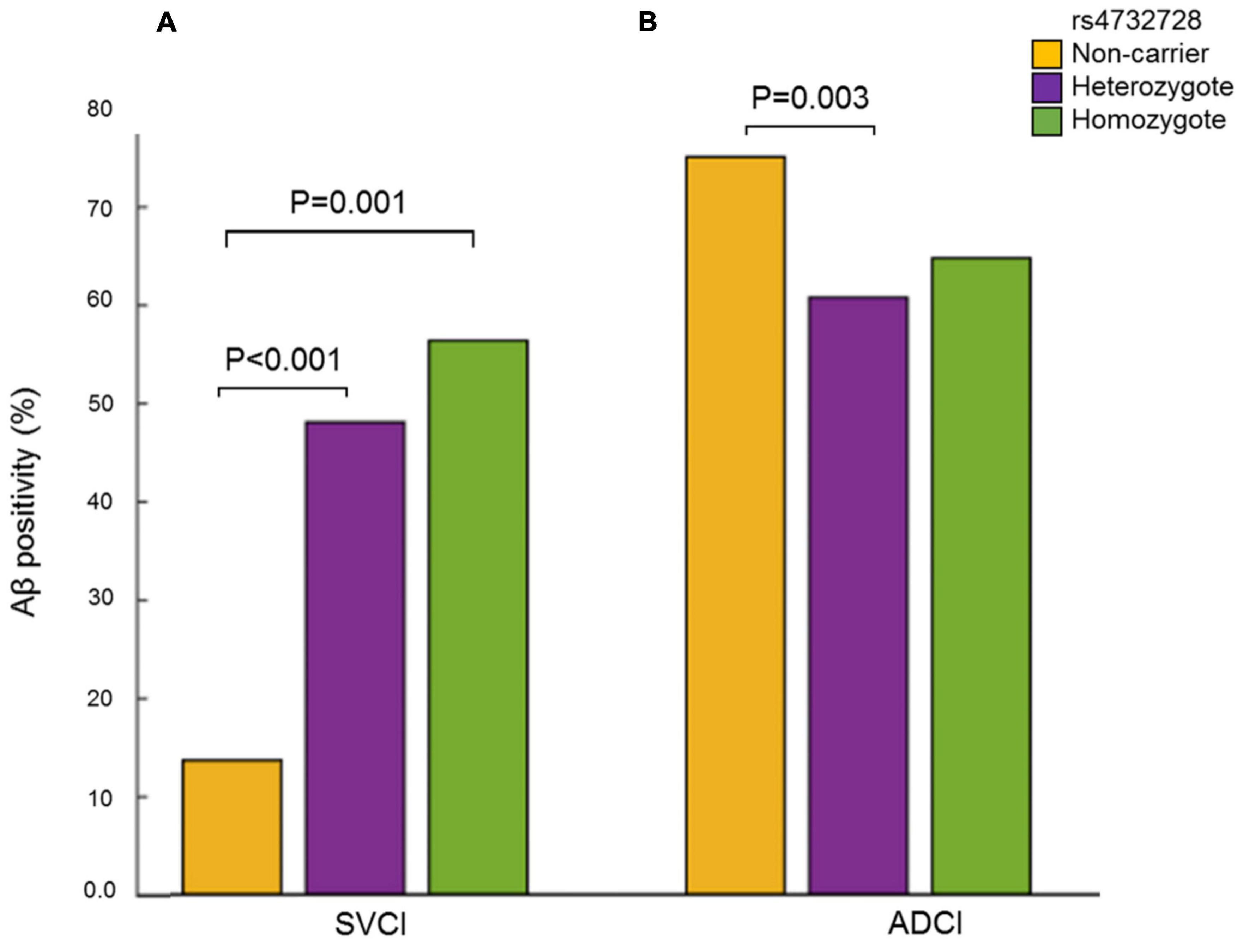
Figure 1. Frequency of Aβ positivity according to carrier status of the rs4732728. (A) SVCI. (B) ADCI. P-values were calculated using the Chi-square test. Aβ, amyloid beta; ADCI, Alzheimer’s disease-related cognitive impairment; SVCI, subcortical vascular cognitive impairment.
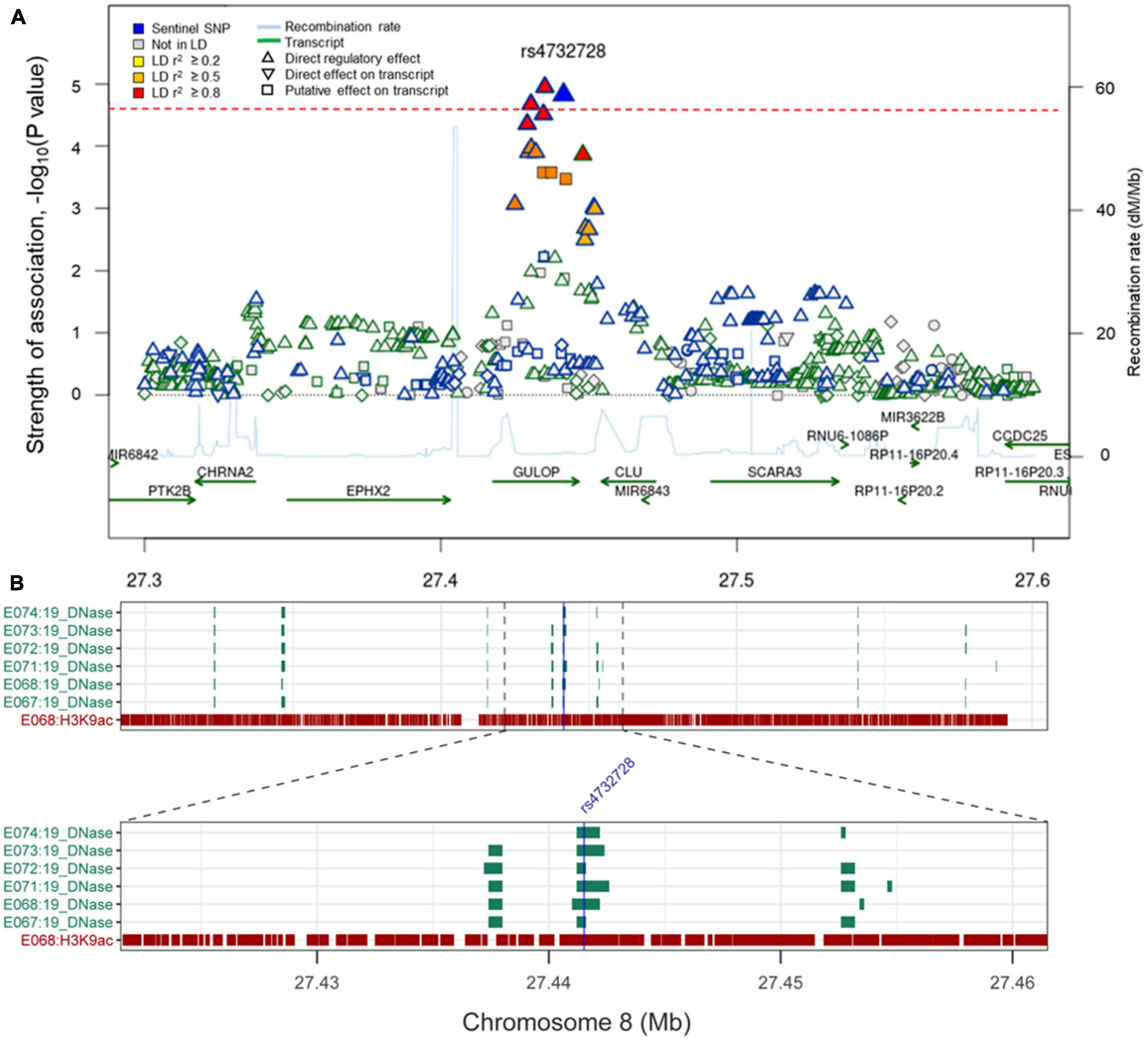
Figure 2. (A) Regional association plot of rs4732728. The red dotted line indicates P-value threshold (1.96 × 10– 5). P-values were calculated using the logistic regression with the interaction term (SNP × diagnosis). The figure was modified from the SNiPA (single-nucleotide polymorphism annotator) (https://snipa.helmholtz-muenchen.de/snipa3). (B) Chromatin state of rs4732728 in brain tissues. Brain angular gyrus (E067), brain anterior caudate (E068), brain hippocampus middle (E071), brain inferior temporal lobe (E072), brain dorsolateral prefrontal cortex (E073), and brain substantia nigra (E074). The figure was based on the Roadmap Epigenomics (https://egg2.wustl.edu/roadmap/web_portal).
Replication analyses
In the ADNI cohort, there was a significant interaction between rs4732728 and the level of WMH on Aβ positivity (β = 0.531, P = 0.02), with the effect being in the same direction as that in the discovery analysis. The positive association between rs4732728 and Aβ positivity increased as the WMH volume increased.
In the ROS/MAP cohorts, there was a significant interaction between rs4732728 and the presence of cerebral arteriosclerosis on Aβ positivity (β = 0.44, P = 0.03), with the effect being in the same direction as that in the discovery analysis. The positive association between rs4732728 and Aβ positivity increased in the presence of cerebral arteriosclerosis. In addition, there was a significant interaction between rs4732728 and the degree of cerebral amyloid angiopathy on Aβ positivity (β = 0.28, P = 0.02).
Functional characterization of rs4732728
The frequency of the effective allele (cytosine) of rs4732728 in the discovery dataset was 0.333, and that of the two replication datasets was 0.593 [cognitively unimpaired (CU) subjects of ADNI (n = 203)] and 0.577 [CU subjects of ROS/MAP (n = 359)], respectively. This was in accordance with the previously reported frequencies of 0.382 and 0.580 for East Asian and European populations (The 1000 Genomes Project Consortium et al., 2015), indicating that the samples used in this study represent each ancestry populations.
We characterized the function of the novel SNP rs4732728 using bioinformatics tools. rs4732728 is located in the intron of gulonolactone (L-) oxidase (GULOP). HaploReg based on ChromHMM annotated rs4732728 as a DNase I hypersensitive site in brain tissues (hippocampus middle, substantia nigra, anterior caudate, inferior temporal lobe, angular gyrus, and dorsolateral prefrontal cortex), indicating that this SNP is in an accessible chromatin region. We also found positive results for the presence of the histone modification mark H3K9ac (active promoter state) of rs6983452 (SNP of high LD with rs4732728), indicating acetylation of the 9th lysine residue of the histone H3 protein, in the anterior caudate and angular gyrus (Figure 2).
In the cis-eQTL analysis using the GTEx database, the rs4732728 and additional five high LD SNPs (rs1316802, rs7831810, rs10780145, rs4352801, and rs6983452) showed significant cis-eQTL effects on epoxide hydrolase 2 (EPHX2) in the brain, and a greater dosage of SNPs decreased the expression of EPHX2 [rs4732728: normalized effect size (NES) = −0.182, P = 0.005; rs1316801: NES = −0.181, P = 0.006; rs7831810: NES = −0.191, P = 3.8 × 10–3; rs10780145: NES = −0.192, P = 3.5 × 10–3; rs4352801: NES = −0.181, P = 0.008; rs6983452: NES = −0.182, P = 4.9 × 10–3; Figure 3). No SNP showed significant cis-eQTL effects on GULOP in the brain.
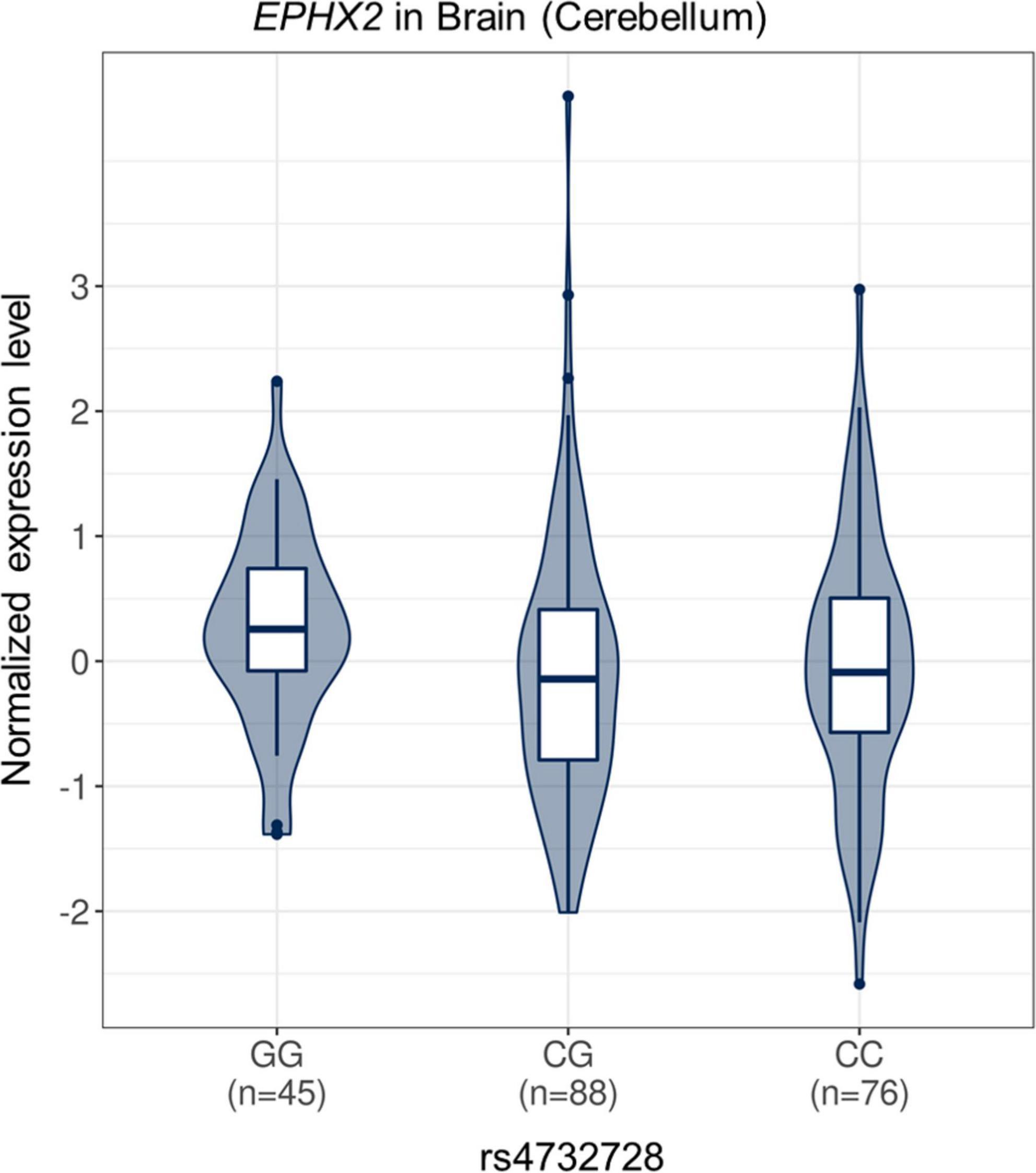
Figure 3. Violin plot of EPHX2 expression according to rs4732728 genotype. The allelic effect of rs4732728 on normalized EPHX2 gene expression levels are shown. The figure was based on the Genotype-Tissue Expression database (http://gtexportal.org). C, cytosine; G, guanine.
Prediction of Aβ positivity in SVCI and ADCI
To test the clinical utility of rs4732728, we developed logistic models to predict Aβ positivity in SVCI and ADCI. In the cross validation, the model including clinical factors (age, sex, and education) and the APOE ∈4 allele showed an AUC of 0.676 (95% CI = 0.659–0.693) and 0.776 (95% CI = 0.767–0.785) in SVCI and ADCI, respectively, (Model 2 of Figure 4). When the model included rs4732728 (Model 3 of Figure 4), a significant increase in the prediction performance was observed in SVCI (AUC = 0.780, 95% CI = 0.757–0.803) but not in ADCI (AUC = 0.777, 95% CI = 0.764–0.790; Figure 4).

Figure 4. ROC curves for the prediction of Aβ positivity. (A) SVCI. (B) ADCI. Solid lines indicate the mean AUC and dotted lines indicate the 95% CIs of the AUC. Each model was developed by multivariable logistic regression. Model 1: Aβ positivity ∼ clinical factors (sex, age, and education). Model 2: Aβ positivity ∼ clinical factor + APOE ∈4. Model 3: Aβ positivity ∼ clinical factor + APOE ∈4 + rs4732728. Aβ, amyloid beta; ADCI, Alzheimer’s disease-related cognitive impairment; AUC, area under the receiver operating characteristic curve; CI, confidence interval; ROC, receiver operating characteristic; SVCI, subcortical vascular cognitive impairment.
Discussion
In the present study, we identified a novel SNP showing a distinct effect on Aβ deposition between SVCI and ADCI. Our major findings are as follows: First, rs4732728 was associated with increased Aβ positivity in SVCI but decreased Aβ positivity in ADCI. The interaction between rs4732728 and CSVD markers on Aβ deposition was replicated in independent ADNI and ROS/MAP cohorts. Second, the functional analysis revealed that rs4732728 was associated with decreased expression levels of EPHX2 in the brain. Finally, rs4732728 contributed to increased accuracy in the prediction of Aβ positivity in patients with SVCI.
We observed that variants in the APOE locus were associated with increased Aβ positivity not only in patients with ADCI but also in those with SVCI. This is accordance with previous study where APOE ∈4 allele increases the risk of Aβ deposition in patients with SVCI (Kim et al., 2013b). Notably, we identified a novel locus showing distinct associations with Aβ positivity in the patient groups. Specifically, patients with SVCI who carried a minor allele (cytosine) of rs4732728 showed an increased risk of Aβ positivity, whereas those with ADCI showed a decreased risk of Aβ positivity. Similar findings were observed in two other independent cohorts comprising participants of European ancestry. This indicates that the identified SNPs may be functional in populations of various ancestries. In addition, different CSVD markers were used among the three datasets; we used WMH volumes in the ADNI dataset, arteriosclerosis severity in the ROSMAP dataset, and the diagnosis of SVCI in the discovery dataset. Nonetheless, the findings were consistent in various measures of vascular pathologies.
The eQTL analysis revealed that the minor allele (cytosine) of rs4732728 was associated with decreased expression levels of EPHX2 in the brain, suggesting that this gene may be a link between rs4732728 and Aβ deposition. EPHX2 encodes an enzyme, epoxide hydrolase, which binds to specific epoxides and converts them to the corresponding diols (Morisseau and Hammock, 2013). In a previous study of AD, the expression of microsomal epoxide hydrolase was increased in the hippocampal tissues of patients with AD (Liu et al., 2006). Furthermore, genetic deletion of soluble epoxide hydrolase was found to reduce Aβ deposition and delay progression of AD in transgenic mice (Lee et al., 2019; Chen et al., 2020; Ghosh et al., 2020). In a previous study of cerebrovascular disease, decreased levels of epoxide hydrolase were associated with increased neuronal survival after ischemic injury via changes in the levels of epoxyeicosatrienoic acids (Koerner et al., 2007). A recent study also demonstrated significant association of EPHX2 genetic variation with cerebrovascular disease (Zhu et al., 2022). The findings of these previous studies suggest that patients with the minor allele (cytosine) of rs4732728 and low levels of EPHX2 would be more resistant to Aβ deposition and ischemic injury than those with the major allele (guanine). These results may explain the distinct associations of rs4732728 with Aβ positivity in patients with SVCI. In SVCI patient with the minor allele (cytosine) of rs473278, Aβ deposition may contribute to cognitive impairment because white matter changes may be less pathogenic to these patients. In contrast, in SVCI patients with the major allele (guanine) of rs4732728, white matter changes are sufficient to cause cognitive impairment since these patients were more susceptible to ischemic injury. Further genomic studies are necessary to elucidate the biological mechanism underlying the distinct actions of rs4732728 on Aβ deposition in patients with SVCI and ADCI.
Identifying patients with SVCI with brain Aβ deposition is important in predicting the prognosis and successful intervention, with the expectation that future treatments may target Aβ. However, currently available diagnostic tools for measuring Aβ are either invasive (cerebrospinal fluid examination) or expensive (PET) (Fargo et al., 2016). In the present study, we demonstrated that genetic data (APOE ∈4 and rs4732728) from blood with clinical information could predict Aβ positivity in patients with SVCI. Considering that the rate of Aβ positivity in our SVCI cohort was 35.5%, 275 patients would be required to perform Aβ PET in order to obtain 100 patients with Aβ deposition. In contrast, when we applied the prediction model including rs4732728, the number of individuals that would need to undergo Aβ PET was reduced by 58%. This result suggests that rs4732728 may play a role as a potential pre-screening marker for Aβ positivity in patients with SVCI. However, this result needs to be validated using independent datasets.
In addition to rs473728, as a pre-screening marker for Aβ positivity in SVCI, our results support the possible therapeutic target of EPHX2 for cerebrovascular disease (Zuloaga et al., 2015). Because drugs that control epoxide hydrolase level are available, the clinical trial can be conducted for SVCI in the future.
The strength of our study is that we performed a genetic study in thoroughly phenotyped patients with ADCI and SVCI using Aβ PET and structural MRI. However, this study has several limitations. First, the sample size was relatively small compared to that of recent genome-wide association studies. Second, the statistical significance level in the replication dataset was small compared to that in the discovery dataset. The difference might result from the heterogeneities between the dataset in terms of pathology measures (Aβ and CSVD), clinical demographics, and genetic backgrounds. Nevertheless, the similar observations in the two independent datasets and the biological relevance of the identified SNPs both strengthen the validity of our findings. Third, we used candidate SNPs that have previously been identified in genome-wide association studies for AD diagnosis. Future whole-genome analyses using larger datasets may identify additional genetic variants that were not tested in this study. Fourth, we could not investigate the biological mechanism underlying the distinct effects of the identified SNPs on Aβ between patients with SVCI and ADCI. Future functional studies using gene editing are necessary to elucidate the underlying mechanisms. Fifth, Aβ PET could not discriminate between different Aβ isoforms. As Aβ shows parenchymal or vascular deposition depending on dominance of Aβ42 or Aβ40 (Yamada, 2012), measuring different Aβ isoforms might be helpful in this study. Finally, as alternative pathomechanisms such as tau, neuroinflammation, and oxidative stress also contribute to both ADCI and SVCI (Román et al., 2002; Gong et al., 2018), mechanisms other than Aβ should be evaluated in the future.
Conclusion
In summary, we identified novel SNPs that showed a distinct effect on Aβ deposition between SVCI and ADCI. The identified SNP showed an additive predictive value for Aβ positivity in patients with SVCI and showed an association with expression of the EPHX2 gene. This finding may provide a potential pre-screening marker for Aβ positivity and a candidate therapeutic target for SVCI.
Data availability statement
The original contributions presented in this study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
Ethics statement
This studies was involving human participants were reviewed and approved by the Samsung Medical Center. The patients/participants provided their written informed consent to participate in this study.
Author contributions
H-RK, H-HW, and SS contributed to the study conception and design. H-RK, JK, HJ, DN, HK, H-HW, and SS performed the material preparation and data collection. H-RK, S-HJ, BK, JPK, SK, KN, H-HW, and SS performed the data analysis. H-RK wrote the first draft of the manuscript. All authors contributed to manuscript revision, read, and approved the final manuscript.
Alzheimer’s Disease Neuroimaging Initiative
Data used in preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (YWRuaS5sb25pLnVzYy5lZHU=). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in the analysis or writing of this manuscript. A complete listing of ADNI investigators can be found at http://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf.
Funding
This research was supported by Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare and Ministry of Science and ICT, Republic of Korea (HU20C0111, HU22C0170, HI19C1132, HI19C1088, and HU22C0042), the National Research Foundation of Korea (NRF) Grant funded by the Korea government (MSIT) (NRF-2019R1A5A2027340), Institute of Information & Communications Technology Planning & Evaluation (IITP) grant funded by the Korea Government (MSIT) (No. 2021-0-02068, Artificial Intelligence Innovation Hub), Future Medicine 20*30 Project of the Samsung Medical Center (#SMX1230081), and the “Korea National Institute of Health” Research Project (2021-ER1006-02). Data collection and sharing for this project were funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number: W81XWH-12-2-0012). ADNI was funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer’s Association, Alzheimer’s Drug Discovery Foundation, Araclon Biotech, BioClinica, Inc., Biogen, Bristol-Myers Squibb Company, CereSpir, Inc., Cogstate, Eisai Inc., Elan Pharmaceuticals, Inc., Eli Lilly and Company, EuroImmun, and F. Hoffmann-La Roche Ltd., and its affiliated company Genentech, Inc., Fujirebio, GE HealthCare, IXICO Ltd., Janssen Alzheimer Immunotherapy Research & Development, LLC., Johnson & Johnson Pharmaceutical Research & Development LLC., Lumosity, Lundbeck, Merck & Co., Inc., Meso Scale Diagnostics, LLC., NeuroRx Research, Neurotrack Technologies, Novartis Pharmaceuticals Corporation, Pfizer Inc., Piramal Imaging, Servier, Takeda Pharmaceutical Company, and Transition Therapeutics. The Canadian Institutes of Health Research were providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization was the Northern California Institute for Research and Education, and the study was coordinated by the Alzheimer’s Therapeutic Research Institute at the University of Southern California. ADNI data were disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
Acknowledgments
We are sincerely grateful to the participants enrolled in the Rush Memory and Aging Project, Rush Minority Aging Research Study, and Religious Orders Study Core.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnagi.2023.1160536/full#supplementary-material
Footnotes
References
Ahn, H., Chin, J., Park, A., Lee, B. H., Suh, M. K., Seo, S. W., et al. (2010). Seoul neuropsychological screening battery-dementia version (SNSB-D): A useful tool for assessing and monitoring cognitive impairments in dementia patients. J. Korean Med. Sci. 25, 1071–1076. doi: 10.3346/jkms.2010.25.7.1071
Barthel, H., Gertz, H., Dresel, S., Peters, O., Bartenstein, P., Buerger, K., et al. (2011). Cerebral amyloid-β PET with florbetaben (18F) in patients with Alzheimer’s disease and healthy controls: A multicentre phase 2 diagnostic study. Lancet Neurol. 10, 424–435. doi: 10.1016/S1474-4422(11)70077-1
Bennett, D. A., Schneider, J. A., Arvanitakis, Z., Kelly, J. F., Aggarwal, N. T., Shah, R. C., et al. (2006). Neuropathology of older persons without cognitive impairment from two community-based studies. Neurology 66, 1837–1844. doi: 10.1212/01.wnl.0000219668.47116.e6
Bennett, D., Buchman, A., Boyle, P., Barnes, L., Wilson, R., and Schneider, J. (2018). Religious orders study and rush memory and aging project. J. Alzheimers Dis. 64, S161–S189. doi: 10.3233/JAD-179939
Carithers, L., and Moore, H. (2015). The genotype-tissue expression (GTEx) project. Biopreserv. Biobank. 13, 307–308. doi: 10.1089/bio.2015.29031.hmm
Chen, W., Wang, M., Zhu, M., Xiong, W., Qin, X., and Zhu, X. (2020). 14, 15-epoxyeicosatrienoic acid alleviates pathology in a mouse model of Alzheimer’s disease. J. Neurosci. 40, 8188–8203. doi: 10.1523/JNEUROSCI.1246-20.2020
Curtis, C., Gamez, J. E., Singh, U., Sadowsky, C. H., Villena, T., Sabbagh, M. N., et al. (2015). Phase 3 trial of flutemetamol labeled with radioactive fluorine 18 imaging and neuritic plaque density. JAMA Neuro. 72, 287–294. doi: 10.1001/jamaneurol.2014.4144
Erkinjuntti, T., Inzitari, D., Pantoni, L., Wallin, A., Scheltens, P., Rockwood, K., et al. (2000). Research criteria for subcortical vascular dementia in clinical trials. J. Neurol. Transm. Suppl. 59, 23–30. doi: 10.1007/978-3-7091-6781-6_4
Fargo, K., Carrillo, M., Weiner, M., Potter, W., and Khachaturian, Z. (2016). The crisis in recruitment for clinical trials in Alzheimer’s and dementia: An action plan for solutions. Alzheimers Dement. 12, 1113–1115. doi: 10.1016/j.jalz.2016.10.001
Fazekas, F., Kleinert, R., Offenbacher, H., Schmidt, R., Kleinert, G., Payer, F., et al. (1993). Pathologic correlates of incidental MRI white matter signal hyperintensities. Neurology 43, 1683–1689. doi: 10.1212/WNL.43.9.1683
Fuchsberger, C., Abecasis, G., and Hinds, D. (2015). minimac2: Faster genotype imputation. Bioinformatics 31, 782–784. doi: 10.1093/bioinformatics/btu704
Ghosh, A., Comerota, M. M., Wan, D., Chen, F., Propson, N. E., Hwang, S. H., et al. (2020). An epoxide hydrolase inhibitor reduces neuroinflammation in a mouse model of Alzheimer’s disease. Sci. Transl. Med. 12:eabb1206. doi: 10.1126/scitranslmed.abb1206
Gong, C., Liu, F., and Iqbal, K. (2018). Multifactorial hypothesis and multi-targets for Alzheimer’s disease. J. Alzheimers Dis. 64, S107–S117. doi: 10.3233/JAD-179921
Grimmer, T., Faust, M., Auer, F., Alexopoulos, P., Förstl, H., Henriksen, G., et al. (2012). White matter hyperintensities predict amyloid increase in Alzheimer’s disease. Neurobiol. Aging 33, 2766–2773. doi: 10.1016/j.neurobiolaging.2012.01.016
Grinberg, L., and Thal, D. (2010). Vascular pathology in the aged human brain. Acta Neuropathol. 119, 277–290. doi: 10.1007/s00401-010-0652-7
Howie, B., Fuchsberger, C., Stephens, M., Marchini, J., and Abecasis, G. (2012). Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat. Genet. 44, 955–959. doi: 10.1038/ng.2354
Jang, H., Park, J., Jang, Y., Kim, H., Lee, J., and Na, D. (2018). Distinct amyloid distribution patterns in amyloid positive subcortical vascular cognitive impairment. Sci. Rep. 8:16178. doi: 10.1038/s41598-018-34032-3
Kang, S. H., Kim, M. E., Jang, H., Kwon, H., Lee, H., Kim, H. J., et al. (2021). Amyloid positivity in the Alzheimer/subcortical-vascular spectrum. Neurology 96, e2201–e2211. doi: 10.1212/WNL.0000000000011833
Kang, S. H., Park, Y. H., Lee, D., Kim, J. P., Chin, J., Ahn, Y., et al. (2019). The cortical neuroanatomy related to specific neuropsychological deficits in Alzheimer’s continuum. Dement. Neurocogn. Disord. 18, 77–95. doi: 10.12779/dnd.2019.18.3.77
Kang, Y., Na, D., and Hahn, S. (2003). Seoul neuropsychological screening battery. Incheon: Human brain research & consulting co.
Kim, H. J., Kang, S. J., Kim, C., Kim, G. H., Jeon, S., Lee, J. M., et al. (2013a). The effects of small vessel disease and amyloid burden on neuropsychiatric symptoms: A study among patients with subcortical vascular cognitive impairments. Neurobiol. Aging 34, 1913–1920. doi: 10.1016/j.neurobiolaging.2013.01.002
Kim, H. J., Ye, B. S., Yoon, C. W., Cho, H., Noh, Y., Kim, G. H., et al. (2013b). Effects of APOE ∈4 on brain amyloid, lacunar infarcts, and white matter lesions: A study among patients with subcortical vascular cognitive impairment. Neurobiol. Aging 34, 2482–2487. doi: 10.1016/j.neurobiolaging.2013.05.009
Koerner, I., Jacks, R., Debarber, A., Koop, D., Mao, P., Grant, D., et al. (2007). Polymorphisms in the human soluble epoxide hydrolase gene EPHX2 linked to neuronal survival after ischemic injury. J. Neurosci. 27, 4642–4649. doi: 10.1523/JNEUROSCI.0056-07.2007
Kunkle, B., Grenier-Boley, B., Sims, R., Bis, J. C., Damotte, V., Naj, A. C., et al. (2019). Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. Nat. Genet. 51, 414–430. doi: 10.1038/s41588-019-0358-2
Lee, H., Lee, K., Chen, C., and Lee, T. (2019). Genetic deletion of soluble epoxide hydrolase delays the progression of Alzheimer’s disease. J. Neuroinflammation 16:267. doi: 10.1186/s12974-019-1635-9
Lee, J. S., Lee, H., Park, S., Choe, Y., Park, Y. H., Cheon, B. K., et al. (2020). Association between APOE ε2 and Aβ burden in patients with Alzheimer-and vascular-type cognitive impairment. Neurology 95, e2354–e2365. doi: 10.1212/WNL.0000000000010811
Lee, J., Kim, S., Kim, G., Seo, S., Park, H., Oh, S., et al. (2011). Identification of pure subcortical vascular dementia using 11C-Pittsburgh compound B. Neurology 77, 18–25. doi: 10.1212/WNL.0b013e318221acee
Lee, M. J., Seo, S. W., Na, D. L., Kim, C., Park, J. H., Kim, G. H., et al. (2014). Synergistic effects of ischemia and β-amyloid burden on cognitive decline in patients with subcortical vascular mild cognitive impairment. JAMA Psychiatry 71, 412–422. doi: 10.1001/jamapsychiatry.2013.4506
Liu, M., Sun, A., Shin, E., Liu, X., Kim, S., Runyons, C. R., et al. (2006). Expression of microsomal epoxide hydrolase is elevated in Alzheimer’s hippocampus and induced by exogenous β−amyloid and trimethyl-tin. Eur. J. Neurosci. 23, 2027–2034. doi: 10.1111/j.1460-9568.2006.04724.x
Manichaikul, A., Mychaleckyj, J., Rich, S., Daly, K., Sale, M., and Chen, W. (2010). Robust relationship inference in genome-wide association studies. Bioinformatics 26, 2867–2873. doi: 10.1093/bioinformatics/btq559
McKhann, G. M., Knopman, D. S., Chertkow, H., Hyman, B. T., Jack, C. R., Kawas, C. H., et al. (2011). The diagnosis of dementia due to Alzheimer’s disease: Recommendations from the national institute on aging-Alzheimer’s association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 7, 263–269. doi: 10.1016/j.jalz.2011.03.005
Morisseau, C., and Hammock, B. (2013). Impact of soluble epoxide hydrolase and epoxyeicosanoids on human health. Annu. Rev. Pharmacol. Toxicol. 53, 37–58. doi: 10.1146/annurev-pharmtox-011112-140244
Morris, J., Roe, C., Xiong, C., Fagan, A. M., Goate, A. M., Holtzman, D. M., et al. (2010). APOE predicts amyloid-beta but not tau Alzheimer pathology in cognitively normal aging. Ann. Neurol. 67, 122–131. doi: 10.1002/ana.21843
Nag, S., Yu, L., Capuano, A. W., Wilson, R. S., Leurgans, S. E., Bennett, D. A., et al. (2015). Hippocampal sclerosis and TDP-43 pathology in aging and Alzheimer disease. Ann. Neurol. 77, 942–952. doi: 10.1002/ana.24388
Noh, Y., Seo, S. W., Jeon, S., Lee, J. M., Kim, J., Kim, G. H., et al. (2014). White matter hyperintensities are associated with amyloid burden in APOE4 non-carriers. J. Alzheimers Dis. 40, 877–886. doi: 10.3233/JAD-130461
Petersen, R. (2011). Clinical practice. Mild cognitive impairment. N. Engl. J. Med. 364, 2227–2234. doi: 10.1056/NEJMcp0910237
Purcell, S., Neale, B., Todd-Brown, K., Thomas, L., Ferreira, M. A., Bender, D., et al. (2007). PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575. doi: 10.1086/519795
Román, G., Erkinjuntti, T., Wallin, A., Pantoni, L., and Chui, H. (2002). Subcortical ischaemic vascular dementia. Lancet Neurol. 1, 426–436. doi: 10.1016/S1474-4422(02)00190-4
Seo, S., Cho, S., Park, A., Chin, J., and Na, D. (2009). Subcortical vascular versus amnestic mild cognitive impairment: Comparison of cerebral glucose metabolism. J. Neuroimaging 19, 213–219. doi: 10.1111/j.1552-6569.2008.00292.x
Sherry, S. T., Ward, M. H., Kholodov, M., Baker, J., Phan, L., Smigielski, E. M., et al. (2001). dbSNP: The NCBI database of genetic variation. Nucleic Acids Res. 29, 308–311. doi: 10.1093/nar/29.1.308
Tarasoff-Conway, J. M., Carare, R. O., Osorio, R. S., Glodzik, L., Butler, T., Fieremans, E., et al. (2015). Clearance systems in the brain-implications for Alzheimer disease. Nat. Rev. Neurol. 11, 457–470. doi: 10.1038/nrneurol.2015.119
The 1000 Genomes Project Consortium, A., Brooks, L. D., Durbin, R. M., Garrison, E. P., Kang, H. M., et al. (2015). A global reference for human genetic variation. Nature 526, 68–74. doi: 10.1038/nature15393
Yamada, M. (2012). Predicting cerebral amyloid angiopathy-related intracerebral hemorrhages and other cerebrovascular disorders in Alzheimer’s disease. Front. Neurol. 3:64. doi: 10.3389/fneur.2012.00064
Yan, Q., Nho, K., Del-Aguila, J. L., Wang, X., Risacher, S. L., Fan, K., et al. (2021). Genome-wide association study of brain amyloid deposition as measured by Pittsburgh Compound-B (PiB)-PET imaging. Mol. Psychiatry 26, 309–321. doi: 10.1038/s41380-018-0246-7
Ye, B. S., Seo, S. W., Kim, J., Kim, G. H., Cho, H., Noh, Y., et al. (2015). Effects of amyloid and vascular markers on cognitive decline in subcortical vascular dementia. Neurology 85, 1687–1693. doi: 10.1212/WNL.0000000000002097
Zhu, X., Li, Y., Yu, T., Li, S., and Chen, M. (2022). A hypothesis-driven study to comprehensively investigate the association between genetic polymorphisms in EPHX2 gene and cardiovascular diseases: Findings from the UK Biobank. Gene 822:146340. doi: 10.1016/j.gene.2022.146340
Zlokovic, B. (2011). Neurovascular pathways to neurodegeneration in Alzheimer’s disease and other disorders. Nat. Rev. Neurosci. 12, 723–738. doi: 10.1038/nrn3114
Keywords: Alzheimer’s disease, amyloid beta, positron emission tomography, subcortical vascular cognitive impairment (SVCI), single nucleotide polymorphism (SNP)
Citation: Kim H-R, Jung S-H, Kim B, Kim J, Jang H, Kim JP, Kim SY, Na DL, Kim HJ, Nho K, Won H-H and Seo SW (2023) Identifying genetic variants for amyloid β in subcortical vascular cognitive impairment. Front. Aging Neurosci. 15:1160536. doi: 10.3389/fnagi.2023.1160536
Received: 07 February 2023; Accepted: 31 March 2023;
Published: 18 April 2023.
Edited by:
Stephen D. Ginsberg, Nathan Kline Institute for Psychiatric Research, United StatesReviewed by:
Ezekiel Gonzalez-Fernandez, University of Mississippi Medical Center, United StatesMichael Malek-Ahmadi, Banner Alzheimer’s Institute, United States
Copyright © 2023 Kim, Jung, Kim, Kim, Jang, Kim, Kim, Na, Kim, Nho, Won and Seo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sang Won Seo, c3c3Mi5zZW9AZ21haWwuY29t; Hong-Hee Won, d29uaGhAc2trdS5lZHU=
†These authors have contributed equally to this work
 Hang-Rai Kim
Hang-Rai Kim Sang-Hyuk Jung
Sang-Hyuk Jung Beomsu Kim5
Beomsu Kim5 Jaeho Kim
Jaeho Kim Hyemin Jang
Hyemin Jang Jun Pyo Kim
Jun Pyo Kim So Yeon Kim
So Yeon Kim Hee Jin Kim
Hee Jin Kim Kwangsik Nho
Kwangsik Nho Hong-Hee Won
Hong-Hee Won