
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Aging Neurosci. , 11 April 2023
Sec. Alzheimer's Disease and Related Dementias
Volume 15 - 2023 | https://doi.org/10.3389/fnagi.2023.1139418
Introduction: The misfolding and aggregation of β-amyloid (Aβ) easily form Aβ fibers, which are continuously deposited in the brain, leading to the massive generation of amyloid plaques, severely destroying neuronal connections, and promoting Alzheimer’s disease (AD) The occurrence and development of AD is one of the pathogenesis of AD. There is an urgent need to develop inhibitors against Aβ aggregation, which is hopefully a potential way to treat AD.
Methods: In this study, we first found the crystal structure of the Aβ1–42 receptor protein from the RCSB PDB protein structure database and used the SYBYL X2.0 software for molecular docking, and then used the Peptide Ranker, Innovagen, DPL, and ToxinPred online websites to perform peptides. Predict the activity score, toxicity and water solubility, and then calculate the affinity constant KD value of polypeptide and Aβ through Surface Plasmon Resonance (SPR) experiment. Subsequently, the CCK-8 kit method was used to determine the toxicity of different concentrations of peptides (3.125, 6.25, 12.5, 25, 50, 100, 200 μM) to PC12 cells, and then the peptides and Aβ according to different concentration ratios (1:4, 1:2, 1:1, 1:0.5, 1:0.25, 0:4), this method is also used to detect the effect of peptides on Aβ-induced neurotoxicity. The thioflavin T (ThT) fluorescence method was used to detect the effects of peptides (50 μM) on Aβ (25 μM) aggregation inhibitory effect.
Results: The results showed that the CScore of YVRHLKYVRHLK peptide molecule docking was 10.0608, the predicted activity score was 0.20, and the KD value was 5.385 × 10−5. The ThT and CCK-8 kit method found that the peptide itself is less toxic to PC12 cells at a concentration of 50 μM, and it has a significant inhibitory effect on the formation of Aβ1–42 aggregates when incubated with Aβ1–42 at a ratio of 1:1 (p < 0.05) and can significantly reduce the PC12 cytotoxicity induced by Aβ1–42 (p < 0.05).
Conclusion: In conclusion, the polypeptide YVRHLKYVRHLK designed in this study has a neuroprotective effect on PC12 cytotoxicity induced by Aβ1–42.
Alzheimer’s disease (AD) is a progressive neurodegenerative disease of the central nervous system that occurs in the elderly. The main pathological feature is the formation of amyloid senile plaques, the main component of which is β-amyloid protein (Aβ), its misfolding and excessive aggregation trigger the disease process of AD, and it is one of the core factors in the occurrence and development of AD (Lin et al., 2014). Under normal circumstances, amyloid precursor protein (APP) is produced under the cleavage of α-secretase (ADAM10) and does not produce Aβ. Instead, it produces neurotrophic factor (sAPPα), which is beneficial to human nerves, which is the non-starch of APP. The amyloidogenic pathway of APP is the enzymatic reaction of β-secretase (BACE1) and γ-secretase (PS1) to produce Aβ. Studies have found that potentiating BACE1 cleavage of APP at both the Asp1 and Glu11 sites, or shifting the cleavage from the Glu11 site to the Asp1 site, can greatly increase the level of C99 and the ratio of C99/C89, increased Aβ production and facilitate neuritic plaque formation (Deng et al., 2013; Zhang et al., 2017). The production and elimination of Aβ in the human body is in a balanced state, but any mutation of presenilin-1 (PSEN), presenilin-2 (PSEN 2) and amyloid precursor protein (APP) genes will cause People suffer from sporadic AD (SAD) or late-onset AD (LOAD) (Uddin and Kabir, 2020). Aβ is composed of 42 amino acids, and α-helix, β-turn and β-sheet form its secondary structure. The β-sheet constitutes the hydrophobic carboxyl terminus, while the α-helix and β-turn constitute the hydrophilic amino terminus. Under physiological conditions, the hydrophobic carboxyl end is hidden and the hydrophilic amino end is exposed, and Aβ is soluble (Wang et al., 2004). Multiple evidence indicated that a large part of Aβ aggregation is driven by hydrophobic sequences (Hilbich et al., 1992; Jarrett et al., 1993; Soto et al., 1995).
Nowadays, the drugs that slow the progression of AD on the market mainly include acetylcholinesterase inhibitors (rivastigmine, galantamine and donepezil), N-methyl-D-aspartic acid (NMDA) antagonists (memantine), intestinal flora regulator (sodium oligomannate, GV-971), Aβ inhibitors (Aducanumab, Gantenerumab, Ban2401 and Alz-801), among which drugs such as rivastigmine, galantamine, donepezil and memantine also have potential side effects, such as Diarrhea, vomiting, nausea, and abdominal cramps (Wang et al., 2019; Behl et al., 2020; Tolar et al., 2020). Aducanumab is a recombinant antibody that uses the epitope mapping and binding kinetics of synthetic peptides to study the crystal structure of the antigen-binding region (Fab) of the aducanumab fragment that binds to Aβ1–11. Computer analysis showed that aducanumab has a weak binding effect to the N-terminal of Aβ, and may adapt to a variety of peptide conformations, which further supports its selectivity for Aβ aggregates (Arndt et al., 2018). In current research, inhibitors for Aβ1–42 protein aggregation have a certain inhibitory effect in previous studies, but the results in subsequent studies are not ideal. Therefore, there is an urgent need to develop a peptide inhibitor against Aβ aggregation. This research creatively uses computer molecular docking technology to simulate and design peptide inhibitors targeting the structural sites of Aβ1–42. First, predict the activity score, electrostatic charge, toxicity, and water solubility of the peptide, and then use the CCK-8 reagent. The box method was used to determine the toxicity of the polypeptide itself to PC12 cells and the effect of the polypeptide on the neurotoxicity induced by Aβ1–42. Then, the inhibitory effect of the peptide inhibitor on the aggregation of Aβ1–42 was detected by the thioflavin T (ThT) staining method. One step is to develop functional foods that can prevent AD and drugs to treat AD to lay a theoretical foundation.
Hexafluoroisopropanol (HFIP) and ThT were purchased from Solarbio. The CM5 chip was purchased from GE in the United States. Human-derived Aβ1–42 standard (95%), P9(Ac-YVRHHHYVRHHH-NH2), P11(Ac-YVRHSVYVRHSV-NH2), P12(Ac-YVRHDLYVRHDL-NH2), P21(Ac-YVRHLKYVRHLK- NH2) was purchased from GL Biochem Ltd. (Shanghai). Cell Counting Kit-8 was purchased from Biosharp Biotechnology (Beijing). The remaining reagents can be purchased from suppliers and can be used without purification.
Rat adrenal pheochromocytoma (PC12) cells were donated by Zhengzhou University School of Basic Medicine (#CRL-1721; RRID: CVCL-0481) in DMEM containing 10% fetal bovine serum, 1% penicillin and 1% streptomycin, culture in a cell incubator with 5% CO2 and 37°C, and passage once every 3 days.
Take the Aβ1–42 monomer out of the refrigerator at −20°C and place it on ice. Add 0.11 mL of HFIP pre-cooled on ice in advance for every 0.5 mg of Aβ1–42 monomer and operate on ice throughout the process. Let stand at room temperature for 1 h to fully dissolve Aβ1–42 and dispense 100 μL per tube. Make HFIP volatilize completely in a fume hood, and freeze at −80°C. Then add DMSO to the centrifuge tube to fully dissolve it to obtain a 2 mmol/LAβ1–42-DMSO solution. Place the centrifuge tube in an ultrasonic bath for 10 min to fully dissolve the peptide membrane. Add phenol red-free DMEM medium to each tube to dilute to the final concentration required for the experiment (Messa et al., 2014).
First dissolve the peptides in ultrapure water to make a 10 mg/ml stock solution and dispense them into 1.5 mL EP tubes. Dilute the stock solution to the corresponding concentration as needed during use to avoid repeated freezing and thawing.
Search the known crystal structure of Aβ1–42 active ingredient from Protein Data Bank, PDB code is 6SH3, as a molecular pair acceptor model. Using FlexX, a semi-flexible, fast and accurate ligand docking method based on incremental construction, the interaction between peptides and Aβ1–42 protein is reflected through FlexX/SYBYL X2.0 software, and Surflex-Dock (SFXC) is selected as the docking mode to generate protomol Related documents, the docking results are expressed relative to the best docking mode. In order to confirm the correctness and reliability of the docking model, before the target small molecule docking, according to the experimental working parameters, the original ligand molecule in the crystal structure was docked to the processed receptor structure active pocket for docking. The total score (total score) is the evaluation criterion. The total score of docking scoring is also called total surflexdock score in English. It divides the binding energy into multiple energy items, such as van der waals force, hydrogen bond, ionic bond, and hydrophobic interaction. Each energy item is multiplied by a coefficient, and the sum is finally calculated to give the final Scoring. The computer virtually screens out the peptides that bind to the Aβ1–42 protein.
Use the online tool Peptide Ranker1 program to predict the biological activity of the peptide inhibitors obtained in the previous stage. Use the online tool DPL2 Evaluate the water solubility of the designed peptide. When the net charge value is ≥2 or ≤−2, the designed peptide has good water solubility. Use the online website ToxinPred3 to predict the toxicity of peptides. ToxinPred can be used to predict the toxicity of peptides, in addition to designing the least toxic peptides and discovering toxic regions in proteins.
Surface Plasmon Resonance (SPR) is a sensitive surface analysis technique that detects changes in dielectric constant caused by molecules adsorbed on heavy metal films. In this study, the 1-(3-Dimethylaminopropyl)-3-ethylcarbodiimide hydrochloride (EDC) and N-Hydroxy succinimide (NHS) in the coupling kit were, respectively, dissolved in 1 mL of activation buffer, divided into 10 parts, 100 μL/part, and stored at −20°C for later use. Before the experiment, first install the CM5 chip in the machine and wash it with HBS-EP buffer 3 times. After screening the appropriate buffer pH, couple the Aβ1–42 protein to the chip, and then dissolve the peptide solution in HBS-EP, dilute to different concentrations, and set a zero concentration and a minimum concentration repeated sample for affinity analysis Perform system maintenance until all samples are measured. After the experiment is over, use the Evaluation software to analyze the results, and use Flow Cell2 to subtract the background signal of channel 1 as the experimental result. The 1:1 binding method is used for fitting calculation and analysis of affinity parameters: binding rate constant ka (1/Ms), dissociation rate constant kd (1/s) and equilibrium dissociation constant KD (M), KD = kd/ ka, the smaller the KD value, the stronger the affinity.
ThT is a chemical fluorescent dye. In recent years, studies have found that ThT can specifically bind to Aβ to enhance its fluorescence. Based on this fluorescence phenomenon, ThT is used as a kind of fluorescent indicator to detect the degree of aggregation of Aβ. In this experiment, the ThT method was used to determine the inhibitory effect of the polypeptide on the aggregation of Aβ1–42.
In this experiment, Aβ1–42 was dissolved in a small amount of DMSO and then diluted with PBS to a mother liquor of 100 μM. The peptide inhibitor was added and diluted with PBS so that the final concentration of Aβ1–42 was 25 μM and the final concentration of peptide inhibitor was 50 μM. The samples containing only Aβ1–42 and Aβ1–42 solution containing peptide inhibitors were incubated separately and together at 37°C for 24 h. Take about 10 μL of the sample, dilute it 20 times with ThT solution, and read the fluorescence intensity value with a microplate reader, 440 nm is the excitation wavelength, and 480 nm is the emission wavelength, and the bandwidth is 5 nm. The fluorescence intensity of the sample containing only Aβ1–42 was set to 100%, and the fluorescence intensity of the PBS without Aβ1–42 and inhibitor was set to background subtraction for normalization. The sample was repeated five times and the average value was taken.
Pipette the cultured cells in a cell culture flask repeatedly to make them evenly distributed. Use a pipete to pipete 10 μL of cell suspension and transfer to a cell counting plate for counting. Count under a microscope to calculate the number of living cells distributed in a large square composed of 16 squares on the cytometer. Cell density (pcs / mL) = (sum of cell count/4) × 104 Calculate the cell concentration, repeat counting 2–3 times for each sample (the value should not be too different each time, otherwise the operation should be repeated). If there are less than 5 cells in each large square, you need to re-centrifuge to adjust the cell concentration.
Use CCK-8 kit to detect cell viability, reflecting the cytotoxicity of peptide inhibitors themselves. The counted PC12 cells were plated at 100 μL/well (8 × 104 cells/well). After put the 96-well plate at 37°C for 12 h, discard the medium, add 100 μL of medium containing different concentrations of peptide inhibitors to each well, and continue to culture 24 h. Add 10 μL of CCK-8 solution to each well (be careful not to generate bubbles in the well, they will affect the reading of the OD (optical density) value), and continue to incubate for 2 h. The wells with DMEM medium solution containing no peptide inhibitors but containing cells were used as the control group. The wells with 100 μL DMEM medium and 10 μL CCK-8 solution but without cells were used as the blank control, and the absorbance was measured at 450 nm.
The cell viability of each group was detected by the CCK-8 kit method, and the effect of peptide inhibitors on the PC12 cytotoxicity caused by the accumulation of Aβ1–42. After incubating the sample containing only Aβ1–42 and the peptide inhibitor co-incubated with Aβ1–42 for 72 h (aging), spread it in a 96-well plate, and after incubating in a 96-well plate for 12 h, discard the medium. Add 10 μL of CCK-8 solution to each well and continue to incubate for 2 h. Add 10 μL of CCK-8 solution to each well and continue to incubate for 2 h. The wells with normal DMEM medium and cell solution were used as the control group. The wells with 100 μL DMEM medium and 10 μL CCK-8 solution but no cells were used as the blank control, and the absorbance was measured at 450 nm.
All experimental data are expressed as Mean ± SEM, and SPSS16.0 software is used for statistical analysis. p < 0.05 indicates that the difference is statistically significant.
The molecular docking scoring results include Cscore, Crash and Polar results. The Cscore value indicates the degree of binding between the peptide ligand and the receptor protein. The better the spatial complementarity and energy matching between the two, the higher the Cscore value. The results of molecular docking in this study are shown in Table 1.
The biological activity, water solubility and toxicity of the four peptides obtained through virtual screening of molecular docking were predicted. The prediction results are shown in Table 2. The prediction scores of the four peptides (P9, P11, P12, and P21) in this study are 0.15, 0.07, 0.20, and 0.20. The activity scores of P12 and P12 are higher. In terms of water solubility, P9 and P11 have poor water solubility, while P12 and P21 have better water solubility. The toxicity of the polypeptide is predicted by ToxinPred, and the results show that none of the four polypeptides is toxic.
Figure 1A shows the pH value of the sodium acetate solution coupled with the chip (for pre-enrichment of the chip). The optimal pH value was pH 4.5 in the SPR experiment. Figure 1B is the coupling result of Aβ1–42 and CM-5 chip. Figure 2 shows the SPR results of the interaction between peptides (P9, P11, P12, P21) and Aβ1–42. It can be seen from the kinetic curve in Figure 2 that the binding characteristics of different peptides to Aβ1–42 protein are also different. In the binding phase of polypeptide and Aβ1–42 protein, the faster the curve rises, the stronger the binding ability. During the dissociation phase, the faster the kinetic curve drops, the faster they dissociate. The kinetic curve of P12 polypeptide rises rapidly at the beginning of binding and reaches saturation in a short time. During the dissociation period, the curve drops rapidly and the dissociation is completed. P21 polypeptide dissociates relatively quickly, but the binding phase also rises rapidly at the beginning, but slowly reaches saturation in the following time, and the binding rate is faster. During the binding phase of P11 and P9 polypeptides, the curve rises steadily, and it takes a long time to reach saturation, and it is relatively slow in the dissociation phase. The calculation results after data fitting show that the KD values of P9, P11, P12, P21 and Aβ1–42 are 1.401 × 10−5, 2.051 × 10−5, 3.266 × 10−7, 5.385 × 10−5 (Table 3). Since the smaller the KD value, the stronger the binding ability, so the binding ability of P12 and Aβ1–42 is the strongest, followed by P21 and P11, and the binding ability of P9 and Aβ1–42 is the weakest.
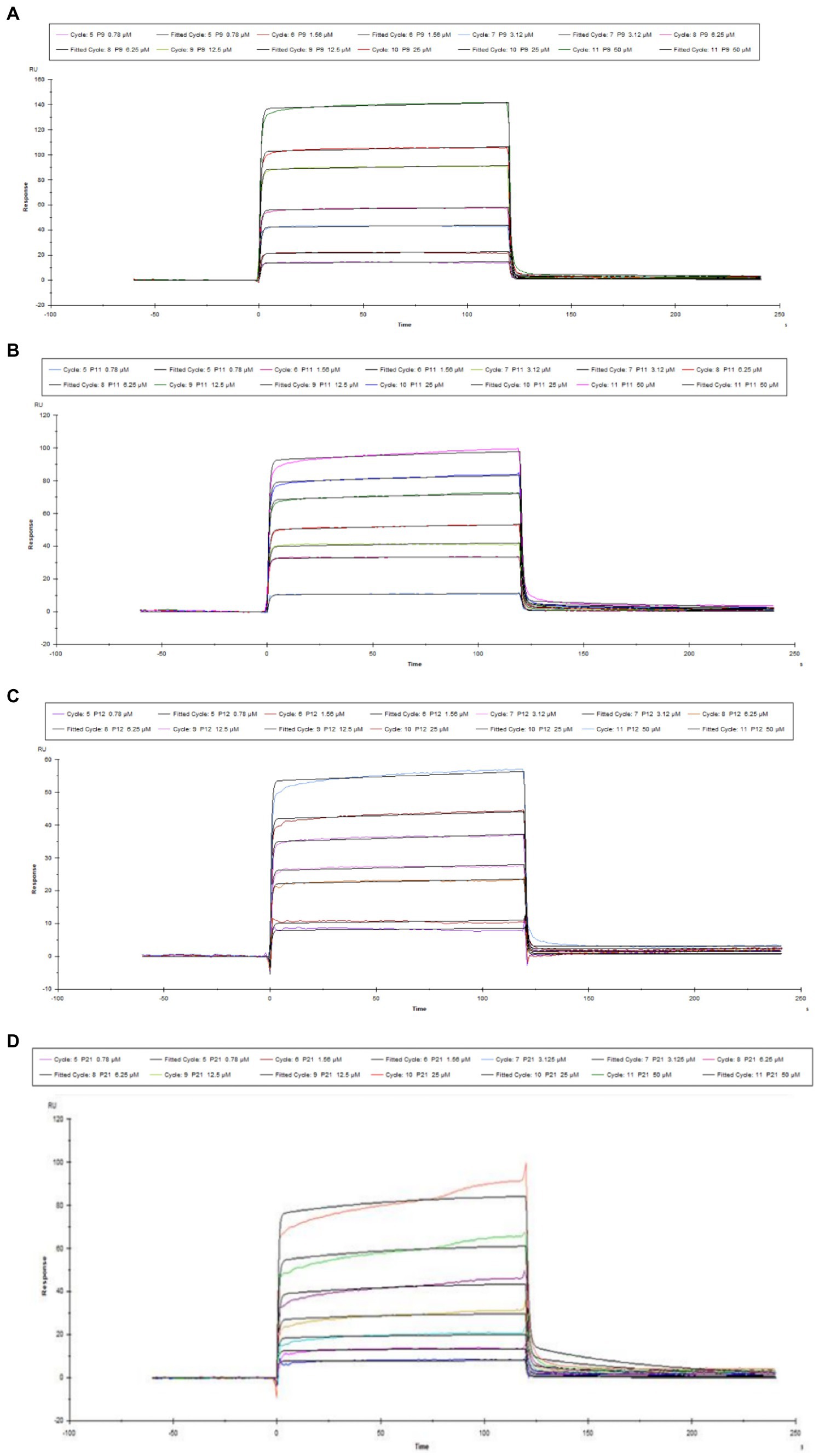
Figure 2. Kinetic curve diagram of the interaction between polypeptide and Aβ1–42. (A) P9 (B) P11 (C) P12 (D) P21.
Although online tools predict the toxicity of peptide inhibitors, in order to avoid the inaccuracy of online tools prediction, further experimental verification of the results is needed. Figure 3 is a graph showing the results of using the CCK-8 method to determine the viability of the polypeptide on PC12 cells, which reflects the toxicity of the polypeptide itself.
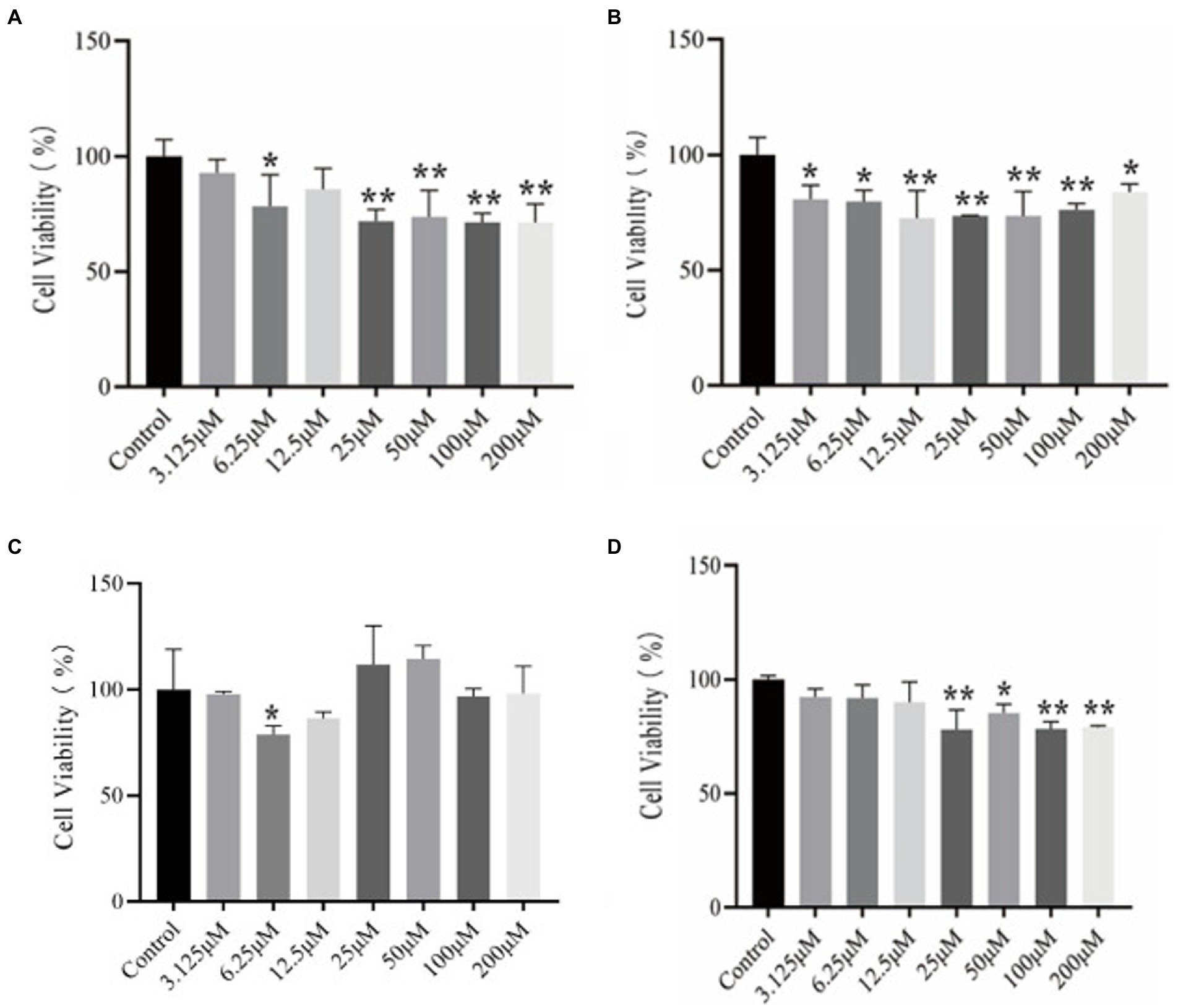
Figure 3. CCK-8 kit method to determine the results of different concentrations of peptides on the viability of PC12 cells. (A) P9 (B) P11 (C) P12 (D) P21.
It can be seen from Figure 3A that the cell viability of P9 at a lower concentration (6.25 μM) began to decrease significantly (p < 0.05), and the cell viability of 25, 50, 100, and 200 μM also decreased significantly, with extremely significant differences (p < 0.01). In Figure 3B, cell viability began to decrease significantly at low concentration (3.125 μM) of P11 (p < 0.05), showing that this concentration can make cells significantly toxic (p < 0.05). Other concentrations also have significant effects on cell viability. Influence (p < 0.05). It can be seen from Figure 3C that P12 has significant effect on the viability of PC12 cells at 6.25 μM (p < 0.05). But there was no significant difference at other concentrations. In Figure 3D, the cell viability of P21 was significantly reduced at 25 μM (p < 0.05), and the cytotoxic peptide concentration produced at this time was higher than that of other peptides, and the cytotoxicity of P21 itself was lower.
The previous research results showed that when the peptide concentration is 50 μM, the peptides in this experiment are almost non-toxic, and the optimal induction concentration of Aβ1–42 through preliminary experiments is 25 μM, so the ThT experiment is at the peptide concentration: Aβ1–42 concentration = 2:1. After co-culturing the peptide with Aβ1–42 for 24 h, the fluorescence intensity was read. The fluorescence intensity of the Aβ positive control group containing only 25 μM of Aβ1–42 without peptides was set to 100% for normalization. The results are shown in Figure 4.
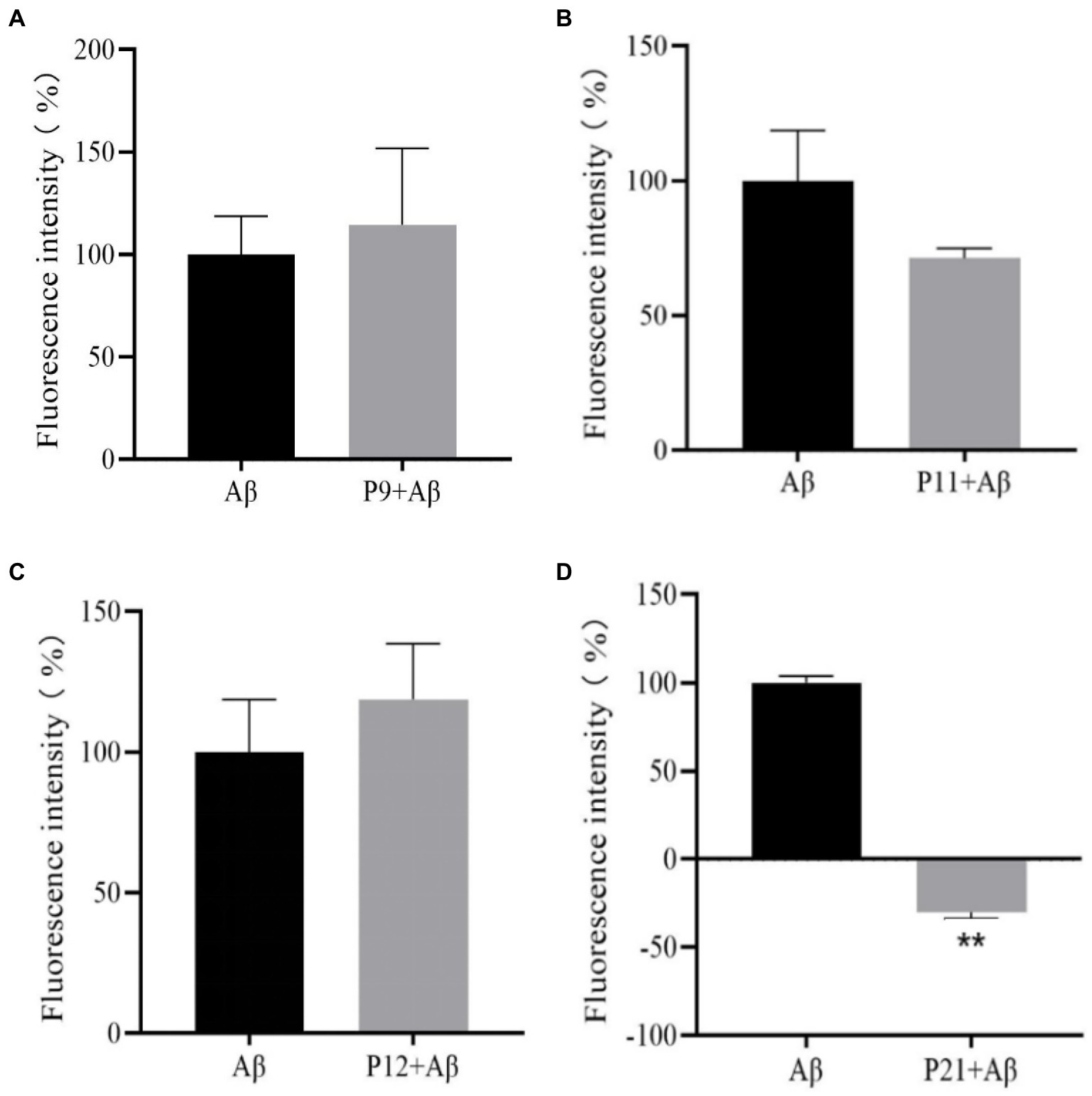
Figure 4. The result graph of the aggregation fluorescence intensity of peptides P9, P11, P12, P21 on Aβ1–42. (A) P9 (B) P11 (C) P12 (D) P21.
It can be seen from Figure 4 that compared with the Aβ positive control group, the fluorescence intensity of P9 and P12 polypeptides has increased, but the difference is not statistically significant (p > 0.05), indicating that P9 and P12 have no inhibitory effect on Aβ aggregation. The fluorescence intensity of P11 polypeptide decreased, but the difference was not statistically significant (p > 0.05), indicating that P11 has a certain inhibitory ability on Aβ aggregation. The co-incubation of P21 and Aβ1–42 protein has a strong inhibitory ability on Aβ aggregation. P21 makes the fluorescence intensity of Aβ1–42 aggregation significantly lower than that of the Aβ group (p < 0.05), which can achieve 100% inhibition.
We comprehensively evaluated the results of the previous peptide self-toxicity and ThT fluorescence intensity, and finally selected the CCK-8 method to evaluate the effect of the P21 peptide on the PC12 cytotoxicity induced by Aβ1–42, as shown in Figure 5. It can be seen from the figure that the inducer containing only Aβ1–42 can cause about 20% of cell death. Incubate the P21 polypeptide with Aβ1–42. The P21 polypeptide has a good inhibitory effect on the PC12 cytotoxicity caused by the aggregation of Aβ1–42 at a low concentration ratio (1:0.25 and 1:0.5). After the concentration ratio of P21 reached 1:1, with the increase of P21 concentration (1:2, 1:4), cell viability continued to increase, reaching the maximum at 1:4, which was close to the normal level, indicating that P21 can inhibit the aggregation process of Aβ1–42 Toxicity to PC12 cells.
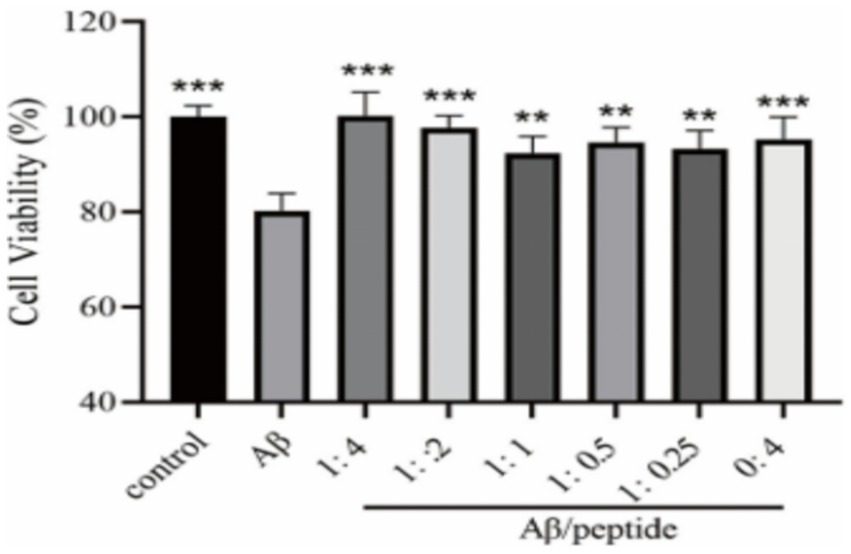
Figure 5. The evaluation result of P21 on the cytotoxicity caused by Aβ1–42 aggregation. **p < 0.01 ***p < 0.001 compared with the Aβ group.
Figure 6 is a diagram showing the relative position of P21 and Aβ1–42 protein after docking. P21 is represented in green, and Aβ1–42 is represented in red. The three-dimensional structure diagram of the combination of P21 and Aβ1–42 is shown in Figure 7. It can be seen from the figure that P21 is located in the center of the groove-like active pocket on the Aβ1–42 protein, which is in line with the Asp-1, Ala-21, Glu-22, Asp-23, Val-24 of the Aβ1–42 protein. The amino acids at Gly-25, Ser-26, Asn-27, and Lys-28 are connected by hydrogen bonds. The core hydrophobic region is composed of Ala-21 and Glu-22, which play a key role in the aggregation of Aβ1–42. It indicates the possible mechanism of action of the peptide we designed and verified the specific binding of the peptide and Aβ1–42 protein.
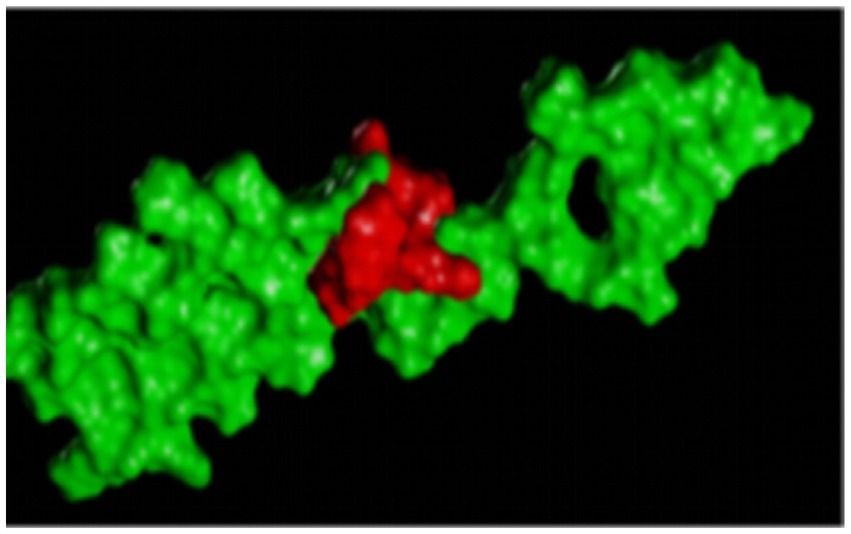
Figure 6. The three-dimensional structure diagram of the docking of the polypeptide and Aβ1–42, the red is the P21 in red, and the Aβ1–42 is in green.
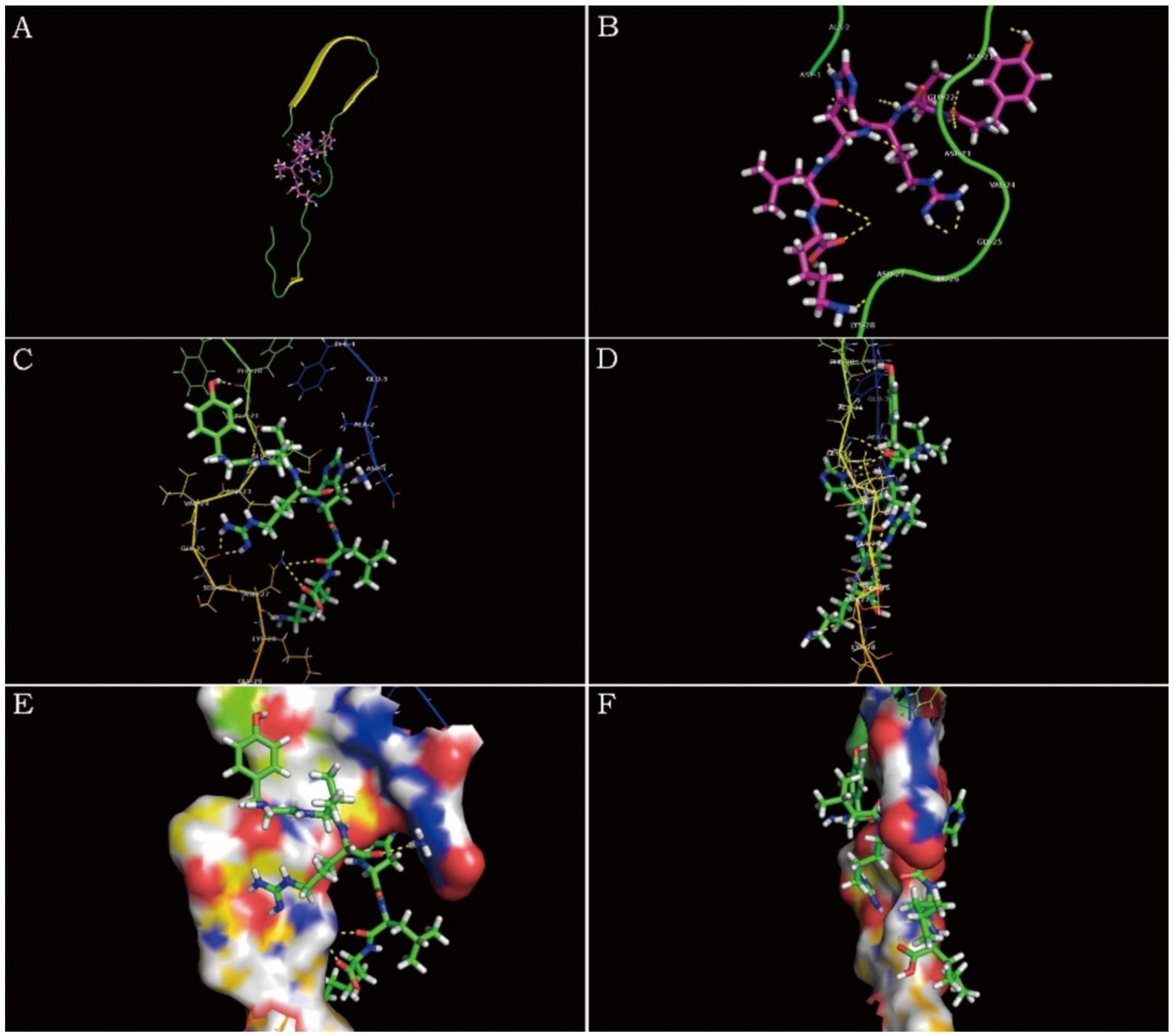
Figure 7. Schematic diagram of P21 and Aβ1–42 protein binding. (A) The three-dimensional structure diagram of the combination of P21 and Aβ1–42 is displayed as a whole. (B–F) Show the combination of P21 and Aβ1–42 from different perspectives.
In this study, the virtual peptide P21 (YVRHLKYVRHLK) designed by molecular docking has a good inhibitory effect on the protein aggregation of Aβ1–42 and reduces the neurotoxicity induced by Aβ1–42, which is consistent with the results of previous studies (Mondal et al., 2019; Kapadia et al., 2021). The mechanism of action may be that it has specific affinity for the hydrophobic amino acid residues in Aβ1–42 (Ala-21 and Glu-22, Aβ21–22), and inhibits the formation of β-sheets within Aβ1–42 and the relationship between β-sheets. The binding between them reduces the formation of soluble oligomers.
Abnormally precipitated Aβ in amyloid plaques becomes one of the hallmark proteins of brain nerve damage in AD patients, and promotes the deterioration of AD. The accumulation of Aβ protein is caused by the interaction between the hydrophobic center and the C-terminal molecule (Kapadia et al., 2021). At present, the development of anti-Aβ aggregation compounds is a promising approach in the treatment of AD, and many clinical trials are ongoing. A molecular dynamics study by Hong Zhou et al. using the core fragment of Aβ (KLVFFA) found that lanosterol binds to the Aβ residues Phe-19 and Phe-20 to form a hydrophobic site, which further induces the decomposition of Aβ protein and the β-sheet layer. Separate. The ThT fluorescence intensity in the cell experiment also showed that lanosterol can reduce the Aβ-induced cytotoxicity in PC12 cells and showed a good inhibitory effect (Zhou et al., 2019). In addition, a study showed that after vitamin K2 intervention, the number of apoptosis caused by Aβ accumulation was significantly reduced, and the ratio of Bax/Bcl-2 decreased, which had a protective effect on the cytotoxicity caused by Aβ deposition. Which has anti-apoptotic and antioxidant effects, may inactivate p38 The MAP kinase pathway is a valuable candidate protective substance against the progress of AD (Hadipour et al., 2020). Pradeepkiran et al. constructed a BACE1 pharmacophore with pepstatin, and screened it by molecular docking studies. It was found that there was an interaction between ligand 1 and BACE1, which reduced the activity of BACE1 and the levels of Aβ40 and 42. In addition, in mutant APP cells treated with ligand 1, mitochondrial biogenesis, mitochondrial fusion and synaptic activity increased, while mitochondrial fission decreased (Pradeepkiran et al., 2020).
In recent years, through the emergence of peptides that inhibit the aggregation of Aβ, extensive research has been conducted on peptides for the treatment of Alzheimer’s disease. However, these peptides have high specificity and low toxicity, which will make them candidate drugs for peptide inhibitors against Aβ aggregation (Fosgerau and Hoffmann, 2015). Peptides designed by computer simulation and applied to Alzheimer’s disease are becoming more and more popular and welcomed by researchers. At present, the peptide inhibitors of Aβ aggregation include LVFF, KLVFF, KLVFF modified peptide (QKLVFF), halogenated KLVFF peptide, FVFLM, and LF (Takahashi and Mihara, 2008; Kouza and Banerji, 2017; Khalili Samani and Mofid, 2020). The amyloid β-sheet mimics designed by Pin-Nan Cheng et al., KLVFFAE (Aβ16–22) and LFFFAED (Aβ17–23) peptides, have the effect of antagonizing Aβ protein aggregation and reducing Aβ toxicity (Cheng et al., 2012). Another study showed that KLVFF analog (QKLVFF, Aβ15–20) is a modified peptide inhibitor of Aβ protein aggregation, which can specifically bind to Aβ1–40 peptide, thereby inhibiting the formation of Aβ fibers (Findeis et al., 1999). Studies have shown that 10 μM iodinated KLVFF peptides (H2N-KLVFF (4-I) -CONH2) and 10 μM Aβ at a concentration ratio of 1:1 show the best resistance compared to other halogenated peptides. Aggregation activity (Khalili Samani and Mofid, 2020). Aggregation activity. The LF peptide (sequence Ac-KQKLLLFLEE-NH2) constructed using Aβ binding elements can form amyloid fibrils, which can effectively co-assemble with mature Aβ1–42 fibrils and effectively inhibit Aβ1–42 oligomerization (Takahashi and Mihara, 2008).In addition, the pituitary adenylate cyclase activating polypeptide (PACAP) attenuates Aβ-induced cell death in PC12 cells by increasing cAMP, and the inactivation of apoptotic factor (caspase-3) indicates that PACAP has a neuroprotective effect (Onoue et al., 2002).
The peptides synthesized by targeting Aβ aggregation have not only been extensively studied in in vitro experiments, but also in many in vivo experiments. For example, the tetrapeptide Ser-Leu-Lys-Pro (SLKP) studied by Pradhan K et al. showed significant neuroprotection against Aβ-mediated toxicity, promoted significant neurite growth, and maintained the growth of rat primary cortical neurons. Healthy form and cross the blood–brain barrier (BBB) (Pradhan et al., 2019). In this study, there is no in vivo study, which will be our follow-up work.
In summary, the aggregation of Aβ1–42 can cause the neurotoxicity of PC12 cells. The CScore of YVRHLKYVRHLK peptide molecule docking in the four peptides involved in this study (YVRHHHYVRHHH, YVRHSVYVRHSV, YVRHDLYVRHDL, YVRHLKYVRHLK) is 10.0608, the predicted activity score is 0.20 and the KD value is 5.385 × 10−5. The ThT and CCK-8 kit method found that the peptide itself is less toxic to PC12 cells at a concentration of 50 μM, and it has a significant inhibitory effect on the formation of Aβ1–42 aggregates when incubated with Aβ1–42 at a ratio of 1:1. It can significantly reduce the PC12 cytotoxicity induced by Aβ1–42, which further shows that the polypeptide YVRHLKYVRHLK designed in this study has neuroprotective effects, which may be due to the high degree of binding between the polypeptide and the amino acid residues 21–22 in Aβ1–42. Caused by the hydrophobicity. YVRHLKYVRHLK can be considered as a potential drug to prevent the progression of Alzheimer’s disease or as a pretreatment to slow down the progression of the disease.
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
The animal experimental protocol was approved by Scientific Ethics Committee of Henan University of Animal Husbandry and Economy (Zhengzhou, China).
YW, SG, and JK designed the whole experiment, provided technical support and technical assistance. YW and SG conducted the experiments. SG and KW wrote the manuscript and analyzed data. KW and JK revised the manuscript. All the authors read and approved the manuscript.
This research was supported by the National Natural Science Foundation of China (NNSFC), by grant NNSFC -31201878, U1204804, and U1604183. It has also been funded by the Excellent Youth Project of the Natural Science Foundation of Henan Province 202300410193.
The authors thanks all co-authors in the team for participating in this study. Thanks to Ning Li and Fangyu Wang. Thanks to the School of Food Science and Technology of Henan Agricultural University and the Key Laboratory of Animal Immunology of Henan Academy of Agricultural Sciences for their great assistance.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Arndt, J. W., Qian, F., Smith, B. A., Quan, C., Kilambi, K. P., Bush, M. W., et al. (2018). Structural and kinetic basis for the selectivity of aducanumab for aggregated forms of amyloid-β. Sci. Rep. 8:6412. doi: 10.1038/s41598-018-24501-0
Behl, T., Kaur, I., Fratila, O., Brata, R., and Bungau, S. (2020). Exploring the potential of therapeutic agents targeted towards mitigating the events associated with amyloid-β cascade in Alzheimer’s disease. Int. J. Mol. Sci. 21:7443. doi: 10.3390/ijms21207443
Cheng, P. N., Liu, C., Zhao, M., Eisenberg, D., and Nowick, J. S. (2012). Amyloid β-sheet mimics that antagonize protein aggregation and reduce amyloid toxicity. Nat. Chem. 4, 927–933. doi: 10.1038/nchem.1433
Deng, Y., Wang, Z., Wang, R., Zhang, X., Zhang, S., Wu, Y., et al. (2013). Amyloid-β protein (Aβ) Glu11 is the major β-secretase site of β-site amyloid-β precursor protein-cleaving enzyme 1(BACE1), and shifting the cleavage site to Aβ Asp1 contributes to Alzheimer pathogenesis. Eur. J. Neurosci. 37, 1962–1969. doi: 10.1111/ejn.12235
Findeis, M. A., Musso, G. M., Arico-Muendel, C. C., Benjamin, H. W., Hundal, A. M., Lee, J. J., et al. (1999). Modified-peptide inhibitors of amyloid β-peptide polymerization. Biochemistry 38, 6791–6800. doi: 10.1021/bi982824n
Fosgerau, K., and Hoffmann, T. (2015). Peptide therapeutics: current status and future directions. Drug Discov. Today 20, 122–128. doi: 10.1016/j.drudis.2014.10.003
Hadipour, E., Tayarani-Najaran, Z., and Fereidoni, M. (2020). Vitamin K2 protects PC12 cells against Aβ(1-42)and H2O2-induced apoptosis via p38 MAP kinase pathway. Nutr. Neurosci. 23, 343–352. doi: 10.1080/1028415X.2018.1504428
Hilbich, C., Kisters-Woike, B., Reed, J., Masters, C. L., and Beyreuther, K. (1992). Substitutions of hydrophobic amino acids reduce the amyloidogenicity of Alzheimer's disease βA4 peptides. J. Mol. Biol. 228, 460–473. doi: 10.1016/0022-2836(92)90835-8
Jarrett, J. T., Berger, E. P., and Lansbury, P. T. Jr. (1993). The carboxy terminus of the beta amyloid protein is critical for the seeding of amyloid formation: implications for the pathogenesis of Alzheimer's disease. Biochemistry 32, 4693–4697. doi: 10.1021/bi00069a001
Kapadia, A., Sharma, K. K., Maurya, I. K., Singh, V., Khullar, M., and Jain, R. (2021). Structural and mechanistic insights into the inhibition of amyloid-β aggregation by Aβ39-42 fragment derived synthetic peptides. Eur. J. Med. Chem. 212:113126. doi: 10.1016/j.ejmech.2020.113126
Khalili Samani, E., and Mofid, M. R. (2020). The effect of terminal groups and halogenation of KLVFF peptide on its activity as an inhibitor of beta-amyloid aggregation. J. Pept. Sci. 26:e3227. doi: 10.1002/psc.3227
Kouza, M., and Banerji, A. (2017). Oligomerization of FVFLM peptides and their ability to inhibit beta amyloid peptides aggregation: consideration as a possible model. Phys. Chem. Chem. Phys. 19, 2990–2999. doi: 10.1039/C6CP07145G
Lin, L. X., Bo, X. Y., Tan, Y. Z., Sun, F. X., Song, M., Zhao, J., et al. (2014). Feasibility of β-sheet breaker peptide-H102 treatment for Alzheimer's disease based on β-amyloid hypothesis. PLoS One 9:e112052. doi: 10.1371/journal.pone.0112052
Messa, M., Colombo, L., del Favero, E., Cantù, L., Stoilova, T., Cagnotto, A., et al. (2014). The peculiar role of the A2V mutation in amyloid-β (Aβ) 1-42 molecular assembly. J. Biol. Chem. 289, 24143–24152. doi: 10.1074/jbc.M114.576256
Mondal, P., das, G., Khan, J., Pradhan, K., Mallesh, R., Saha, A., et al. (2019). Potential neuroprotective peptide emerged from dual neurotherapeutic targets: a fusion approach for the development of anti-Alzheimer's lead. ACS Chem. Neurosci. 10, 2609–2620. doi: 10.1021/acschemneuro.9b00115
Onoue, S., Endo, K., Ohshima, K., Yajima, T., and Kashimoto, K. (2002). The neuropeptide PACAP attenuates β-amyloid (1–42)-induced toxicity in PC12 cells. Peptides 23, 1471–1478. doi: 10.1016/S0196-9781(02)00085-2
Pradeepkiran, J. A., Reddy, A. P., Yin, X., Manczak, M., and Reddy, P. H. (2020). Protective effects of BACE1 inhibitory ligand molecules against amyloid beta-induced synaptic and mitochondrial toxicities in Alzheimer's disease. Hum. Mol. Genet. 29, 49–69. doi: 10.1093/hmg/ddz227
Pradhan, K., das, G., Gupta, V., Mondal, P., Barman, S., Khan, J., et al. (2019). Discovery of neuroregenerative peptoid from amphibian neuropeptide that inhibits amyloid-β toxicity and crosses blood–brain barrier. ACS Chem. Neurosci. 10, 1355–1368. doi: 10.1021/acschemneuro.8b00427
Soto, C., Castaño, E. M., Frangione, B., and Inestrosa, N. C. (1995). The α-helical to β-strand transition in the amino-terminal fragment of the amyloid β-peptide modulates amyloid formation. J. Biol. Chem. 270, 3063–3067. doi: 10.1074/jbc.270.7.3063
Takahashi, T., and Mihara, H. (2008). Peptide and protein mimetics inhibiting amyloid beta-peptide aggregation. Acc. Chem. Res. 41, 1309–1318. doi: 10.1021/ar8000475
Tolar, M., Abushakra, S., Hey, J. A., Porsteinsson, A., and Sabbagh, M. (2020). Aducanumab, gantenerumab, BAN2401, and ALZ-801-the first wave of amyloid-targeting drugs for Alzheimer's disease with potential for near term approval. Alzheimers Res. Ther. 12:95. doi: 10.1186/s13195-020-00663-w
Uddin, M. S., and Kabir, M. T. (2020). Novel anti-Alzheimer's therapeutic molecules targeting amyloid precursor protein processing. Oxidative Med. Cell. Longev. 2020:7039138. doi: 10.1155/2020/7039138
Wang, Z., Chang, L., Klein, W. L., Thatcher, G. R. J., and Venton, D. L. (2004). Per-6-substituted-per-6-deoxy β-cyclodextrins inhibit the formation of β-amyloid peptide derived soluble oligomers. J. Med. Chem. 47, 3329–3333. doi: 10.1021/jm034224e
Wang, X., Sun, G., Feng, T., Zhang, J., Huang, X., Wang, T., et al. (2019). Sodium oligomannate therapeutically remodels gut microbiota and suppresses gut bacterial amino acids-shaped neuroinflammation to inhibit Alzheimer's disease progression. Cell Res. 29, 787–803. doi: 10.1038/s41422-019-0216-x
Zhang, S., Wang, Z., Cai, F., Zhang, M., Wu, Y., Zhang, J., et al. (2017). BACE1 cleavage site selection critical for Amyloidogenesis and Alzheimer's pathogenesis. J. Neurosci. 37, 6915–6925. doi: 10.1523/JNEUROSCI.0340-17.2017
Keywords: Aβ, molecular docking, neuroprotection, neuroscience, PC12 cells +
Citation: Wu Y, Guo S, Wang K and Kang J (2023) The interaction of peptide inhibitors and Aβ protein: Binding mode analysis, inhibition of the formation of Aβ aggregates, and then exert neuroprotective effects. Front. Aging Neurosci. 15:1139418. doi: 10.3389/fnagi.2023.1139418
Received: 07 January 2023; Accepted: 20 March 2023;
Published: 11 April 2023.
Edited by:
Zhiyou Yang, Guangdong Ocean University, ChinaReviewed by:
Hossein Mohammad-Beigi, Technical University of Denmark, DenmarkCopyright © 2023 Wu, Guo, Wang and Kang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jingjing Kang, a2FuZ2pqQGhudWFoZS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.