
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Aging Neurosci. , 03 May 2022
Sec. Neurocognitive Aging and Behavior
Volume 14 - 2022 | https://doi.org/10.3389/fnagi.2022.859313
This article is part of the Research Topic Physical Exercise for Age-Related Neuromusculoskeletal Disorders View all 36 articles
Objective: Regular exercise is a powerful tool that enhances skeletal muscle mass and strength. Lysine acetylation is an important post-translational modification (PTM) involved in a broad array of cellular functions. Skeletal muscle protein contains a considerable number of lysine-acetylated (Kac) sites, so we aimed to investigate the effects of exercise-induced lysine acetylation on skeletal muscle proteins.
Methods: We randomly divided 20 male C57BL/6 mice into exercise and control groups. After 6 weeks of treadmill exercise, a lysine acetylation proteomics analysis of the gastrocnemius muscles of mice was performed.
Results: A total of 2,254 lysine acetylation sites in 693 protein groups were identified, among which 1,916 sites in 528 proteins were quantified. The enrichment analysis suggested that protein acetylation could influence both structural and functional muscle protein properties. Moreover, molecular docking revealed that mimicking protein deacetylation primarily influenced the interaction between substrates and enzymes.
Conclusion: Exercise-induced lysine acetylation appears to be a crucial contributor to the alteration of skeletal muscle protein binding free energy, suggesting that its modulation is a potential approach for improving exercise performance.
Protein lysine acetylation is an indispensable and conserved post-translational modification (PTM) that occurs in prokaryotic and eukaryotic cells (Xing and Poirier, 2012). Acetylation is involved in energy metabolism, gene expression, and cell signaling in both animals and bacteria (Xing and Poirier, 2012; Fukushima and Lopaschuk, 2016; Narita et al., 2019). In recent years, many studies have reported that lysine acetylation regulates a wide range of cellular functions in skeletal muscle (Lundby et al., 2012; Moresi et al., 2015; Tsuda et al., 2018). For example, histone acetylation in the nucleus controls MEF2- and MyoD-dependent transcription, which regulates myoblast differentiation and muscle homeostasis (Moresi et al., 2015). Tsuda found that protein acetylation in the skeletal muscle mitochondria of mice was linked with intermediary metabolism (Tsuda et al., 2018). In addition, Lundby discovered a mass of lysine acetylation sites in 16 different tissues of rats, where the degree of lysine acetylation of muscle was highest (Lundby et al., 2012). Furthermore, they found that proteins involved in muscle contraction and enzymes generating ATP were all acetylated (Lundby et al., 2012), which provided a new and important clue that acetylation modification regulates the energy and metabolism of muscle.
The skeletal muscle is an organ for the maintenance of mobility, and exercise training in turn is known to positively affect the skeletal muscle system (Widegren et al., 2001; Russell et al., 2014; Nelson et al., 2020). Cellular stress triggered by exercise can alter the coordinated activity of acetyltransferases and deacetylases to regulate lysine acetylation (Marton et al., 2010; Menzies and Auwerx, 2013). Previous studies have found dynamic changes in histone acetylation and mitochondrial protein acetylation in the skeletal muscle during exercise (McGee et al., 2009; Overmyer et al., 2015). In our previous study, aerobic training or Scriptaid dramatically improved the exercise performance of mice, including endurance, grip strength, and citrate synthase activity (Huang et al., 2020). These changes were also accompanied by an increase in titin lysine acetylation in gastrocnemius tissue, which was postulated to function as a mediator to regulate muscle protein function at the molecular level (Huang et al., 2020). Given the important role of skeletal muscle in exercise or physical activities (Gan et al., 2018), we speculate that exercise-induced lysine acetylation in muscle may be an important factor mediating exercise performance.
The purpose of this study was to profile protein lysine acetylation in the skeletal muscle of exercise and control groups using different proteomic technologies and bioinformatics tools. Our preliminary data demonstrated widespread protein lysine acetylation in the skeletal muscle of mice after exercise. Subsequent proteomic studies confirmed that the different acetylation patterns of muscle proteins were closely related to their structure and function of muscle. Then, molecular docking data suggested that lysine acetylation modification could change the binding of substrates and enzymes, ultimately influencing enzyme activity and energy transformation. Taken together, exercise-induced lysine acetylation might play an important role in muscle structural and functional proteins properties, indicating the crucial role of protein acetylation in skeletal muscle physiology.
Twenty 5-week-old male C57BL/6 mice were purchased from Beijing HFK Bioscience Co., (Beijing, China) in this study. The animal study was approved by the Tianjin Medical University Animal Care and Use Committee (approval number: SYXK-2019-0004) under the guidelines of the Chinese Academy of Sciences. The mice were housed in a temperature-controlled room (22 ± 2°C) with a 12-h light/dark cycle (light on from 08:00 to 20:00) and free access to food and water. All animals were allowed to acclimatize to the laboratory environment for 1 week.
At the beginning of the experiment, the mice were randomly divided into two groups: a sedentary control group (n = 10) and an exercise group (n = 10). Mice in the control group were kept sedentary for 6 weeks. Mice in the exercise group were familiarized with the treadmill 5 days before the experiment by exercising them for 1 h at 8 m/min; each day thereafter, the speed was increased by 2 m/min until the speed has reached 12 m/min. Then, mice in the exercise group underwent 6 weeks of treadmill exercise for 5 days per week (1 h/day) on a 0% grade at the intensity of 75% VO2max (12 m/min) as previously described (Fernando et al., 1993). During the 1-h session, the mice ran on the treadmill continuously without rest intervals throughout the whole 60-min time. By the end of the 6-week exercise, the body weight of the mice was determined with an ImpediVET analyzer (ImpediMed). Forty-eight hours after the last bout of exercise, mice were fasted for 14 h and then anesthetized with isoflurane. The gastrocnemius was dissected and weighed, frozen in liquid nitrogen, and then stored at −80°C for later analysis.
Each skeletal muscle sample was ground with liquid nitrogen and transferred to a 5-mL centrifuge tube. Then, the samples were sonicated three times on ice using a high-intensity ultrasonic processor (Scientz, Ningbo, China) in lysis buffer [8 M urea, 10 mM dithiothreitol (DTT), 3 μM trichostatin (TSA), 50 mM nicotinamide (NAM), and 1% protease inhibitor cocktail], followed by centrifugation at 12,000 × g at 4°C for 10 min. After that, we discarded the supernatant and washed the remaining precipitate three times. The protein was redissolved in 8 M urea, and its concentration was determined with a BCA kit.
For trypsin digestion, 5 mM DTT was added to implement the reduction of proteins for 30 min at 56°C, and 11 mM iodoacetamide was added to perform alkylation of proteins for 15 min at room temperature in darkness. Then, the protein sample was diluted by adding 100 mM TEAB to a urea concentration of less than 2 M. In the first digestion, trypsin was added at a 1:50 trypsin-to-protein mass ratio, and this digestion was performed at 37°C overnight. For the second digestion, trypsin was added at a mass ratio of 1:100 trypsin/protein and digested for 4 h at 37°C.
After trypsin digestion, the peptide was desalted by a Strata X C18 SPE column (Phenomenex) and vacuum-dried. The peptide was reconstituted in 0.5 M TEAB and processed according to the manufacturer’s protocol for the TMT kit/iTRAQ kit. In brief, one unit of TMT/iTRAQ reagent was thawed and reconstituted in acetonitrile. The peptide mixtures were then incubated for 2 h at room temperature and pooled, desalted, and dried by vacuum centrifugation.
The TMT-labeled samples were fractionated by high-pH reverse-phase HPLC using a Thermo Betasil C18 column (5 μm particles, 10 mm ID, and 250 mm length). In brief, peptides were first separated with a gradient of 8–32% acetonitrile (pH 9.0) over 60 min into 60 fractions. Finally, the peptides were combined and dried by vacuum centrifugation.
Immunoprecipitation was performed to enrich modified peptides. IP buffer solution (100 mM NaCl, 1 mM EDTA, 50 mM Tris–HCl, 0.5% NP-40, pH 8.0) was added to dissolve tryptic peptides. Then, the liquid supernatant was incubated with prewashed antibody beads (acetyl-lysine antibody beads lot number PTM1001, PTM Biolabs Inc., China) at 4°C overnight with gentle shaking. The beads were washed four times with IP buffer solution and two times with double-distilled H2O. Then, we eluted bound peptides with 0.1% trifluoroacetic acid (TFA) and vacuum-dried them. For LC-MS/MS analysis, the obtained peptides were desalted with C18 ZipTips (Millipore) in accordance with the manufacturer’s instructions.
Peptides were dissolved in solvent A (0.1% formic acid) and directly loaded onto a homemade reversed-phase analytical column (15 cm length, 75 μm i.d.). The gradient comprised an increase from 6 to 23% solvent B (0.1% formic acid in 98% acetonitrile) over 26 min, from 23 to 35% for 8 min, increasing to 80% in 3 min and then holding at 80% for the last 3 min, all at a constant flow rate of 400 nL/min on an EASY-nLC 1000 UPLC system. The peptides were subjected to an NSI source followed by tandem mass spectrometry (MS/MS) in a Q Exactive™ Plus (Thermo) coupled online to the UPLC. The applied electrospray voltage was 2.0 kV. The m/z scan range was 350–1,800 for the full scan, and intact peptides were detected in the Orbitrap at a resolution of 70,000. The peptides were then selected for MS/MS using an NCE setting of 28. Ion fragments were detected in the Orbitrap at a resolution of 17,500 (m/z 200). A data-dependent procedure alternated between one MS scan followed by 20 MS/MS scans with 15.0 s dynamic exclusion. Automatic gain control was used to prevent overfilling of the ion trap, and 5e4 ions were accumulated for the generation of MS/MS spectra. The fixed first mass was set as 100 m/z.
Then, we processed the LC-MS/MS data with the MaxQuant search engine (v.1.5.2.8). Tandem mass spectra were searched against the UniProt database concatenated with the reverse decoy database. Trypsin/P was specified as the cleavage enzyme allowing up to four missing cleavages. In the first search and the main search, the mass tolerances for precursor ions were set as 20 and 5 ppm, respectively. Meanwhile, the mass tolerance for fragment ions was set as 0.02 Da. Carbamidomethylation on Cys was specified as a fixed modification, while acetylation modification and oxidation on Met were variable modifications. The false discovery rate (FDR) and the minimum score for modified peptides were adjusted to <1% and >40, respectively. The retrieval parameter settings and quality control of mass spectrometry are shown in Supplementary Material.
For subsequent bioinformatics analysis, normalization with protein quantification to remove the effect of protein expression on modification abundance was performed. Protein acetylation was identified on the basis of the following criteria: t-test with p-value < 0.05, fold change > 1.2 or < 0.83, which were consistent with previous studies (Ren et al., 2013; Wu et al., 2016; Huang et al., 2019; Yu et al., 2021). For hierarchical clustering based on Kac sites of proteins associated with muscle function, we collated all the categories obtained after enrichment along with their P-values. The Gene Ontology (GO) annotation proteome was derived from the UniProt-GOA database,1 and the proteins were classified by GO annotation based on three categories: biological process, cellular component, and molecular function. For each category, a two-tailed Fisher’s exact test was employed to test the enrichment of the identified modified protein against all proteins from the species database. The GO terms with a corrected p-value < 0.05 were considered significant. All differentially expressed modified proteins were searched against the STRING database2 for protein–protein interactions (PPIs). Only interactions between the proteins showing acetylation variance were selected, thereby excluding external candidates. The PPI networks for the identified acetylated proteins were analyzed by Cytoscape 3.9.0 software using interaction data.
AutoDock 4.2 software, an excellent tool to recognize docking affinity between individual ligands and specific proteins (Seeliger and de Groot, 2010; Rizvi et al., 2013), was utilized in all the docking experiments. Protein receptors were taken from UniProt3 in PDB format, and then, specific-site lysine-to-arginine (K-to-R) changes were made in PyMOL 2.5.1, mimicking deacetylated lysine residues. Ligands were obtained from PubChem4 in SDF format and converted to PDB format by Open Babel 3.1.1. After preparing proteins and ligands, we ran Autogrid to calculate the affinity maps, and the size of the grid box was kept large enough to at least allow the ligand to rotate freely. Then, the docking of the protein and ligand was performed. Grid and docking parameter files were given as GPF files and DPF files in AutoDock. Finally, we used Chimaera 1.15 and Notepad++ 7.9.5 to read the results.
Total RNA was extracted from gastrocnemius muscle and reverse-transcribed to cDNA using TransScript One-Step gDNA Removal and cDNA Synthesis SuperMix (TransGen Biotech). The synthesized cDNA was used for PCR with specific cycles. The primer sequences were as follows: MyHC I, 5′-GCCTGGGCTTA CCTCTCTATCAC-3′ (forward), 5′-CTTCTCAGACTTCCGC AGGAA-3′ (reverse); MyHC IIa, 5′-CAGCTGCACCTTCTCG TTTG-3′ (forward), 5′-CCCGAAAACGGCCATCT-3′ (reverse); MyHC IIb, 5′-CAATCAGGAACCTTCGGAACAC-3′ (forward), 5′-GTCCTGGCCTCTGAGAGCAT-3′ (reverse); MyHC IIx, 5′-G GACCCACGGTCGAAGTTG-3′ (forward), 5′-CCCGAAAACG GCCATCT-3′ (reverse); and GAPDH, 5′-GCACAGTCAAGGC CGAGAAT-3′ (forward), 5′-GCCTTCTCCATGGTGGTGAA-3′ (reverse).
PCR amplification was performed using the Roche LightCycler 480 real-time PCR system (Roche Life Science, United States) and the PerfectStart Uni RT-qPCR Kit (TransGen Biotech) according to the manufacturers’ instructions. The relative expression levels of the target genes were determined using the 2–ΔΔCT method.
Gastrocnemius samples were dissected and fixed for 24 h in 10% neutral buffered formalin. Then, we embedded tissues with paraffin and sliced them into 7-μm serial horizontal sections. The gastrocnemius muscle sections were stained with Oil Red O, and the cross-sectional area (CSA) of the muscle fiber was assessed under light microscopy (AF 6000, Leica). Five images per animal (n = 3) and at least 60 muscle cells per image were used in the statistical analysis (ImageJ software). The total average area was used to determine the CSA per square micrometer.
The data are presented as the means ± SEM, and the differences were analyzed using Student’s t-test or two-way ANOVA. All data were considered significantly different when P < 0.05. We used SPSS to make statistical comparisons.
In this study, gastrocnemius muscle samples from the two groups were mapped to identify the lysine acetylation sites of proteins, and the workflow of the experimental procedures is shown in Figure 1A. To verify the initial mass spectrometry data, we detected the lengths of all the obtained peptides and the quality errors. The lengths of most peptides, ranging between 7 and 20, matched the lengths of the tryptic peptides (Supplementary Figure 1), indicating that the sample preparation met the standard for proteomics analysis. The average mass error for all the identified peptides was nearly zero, and first-order mass errors were less than 10 ppm, meaning that the mass accuracy of the MS data met the requirement (Supplementary Figure 2). During the identification, a total of 73,353 secondary spectra were obtained by mass spectrometry. Then, we used mass spectrometry secondary spectra to search against protein theory data. The available efficiency was 5,846, the spectrum utilization rate was 8.0%, and the peptides were resolved into 2,598 peptides and 2,219 acetylated peptides. In addition, 2,254 acetylation sites were identified in 693 proteins, including 1,916 quantified sites in 528 quantified proteins (Figure 1B). Among these proteins, 372 proteins (53.70%) had one Kac site and 108 proteins (15.58%) contained five or more Kac sites (Figure 1C). There were 15 proteins (2.16%) that contained more than 15 Kac sites (Figure 1C). Interestingly, those proteins that contained more than 15 Kac sites were mainly distributed in sarcomeres, especially titin (333 sites) (Huang et al., 2020) and myosin-4 (74 sites). On average, there were 3.25 Kac modification sites per protein (Figure 1D) in the gastrocnemius of mice. In another independent experiment, Lundby et al. (2012) found 3,416 sites from 917 proteins in the rat muscle and 2,811 sites from 941 proteins in the human muscle, where the average numbers of acetylation sites per protein were 3.73 and 2.99, respectively (Figure 1D). Comparison analysis of these data of three species indicated that protein acetylation widely exists in the skeletal muscle of mice, rats, and humans.
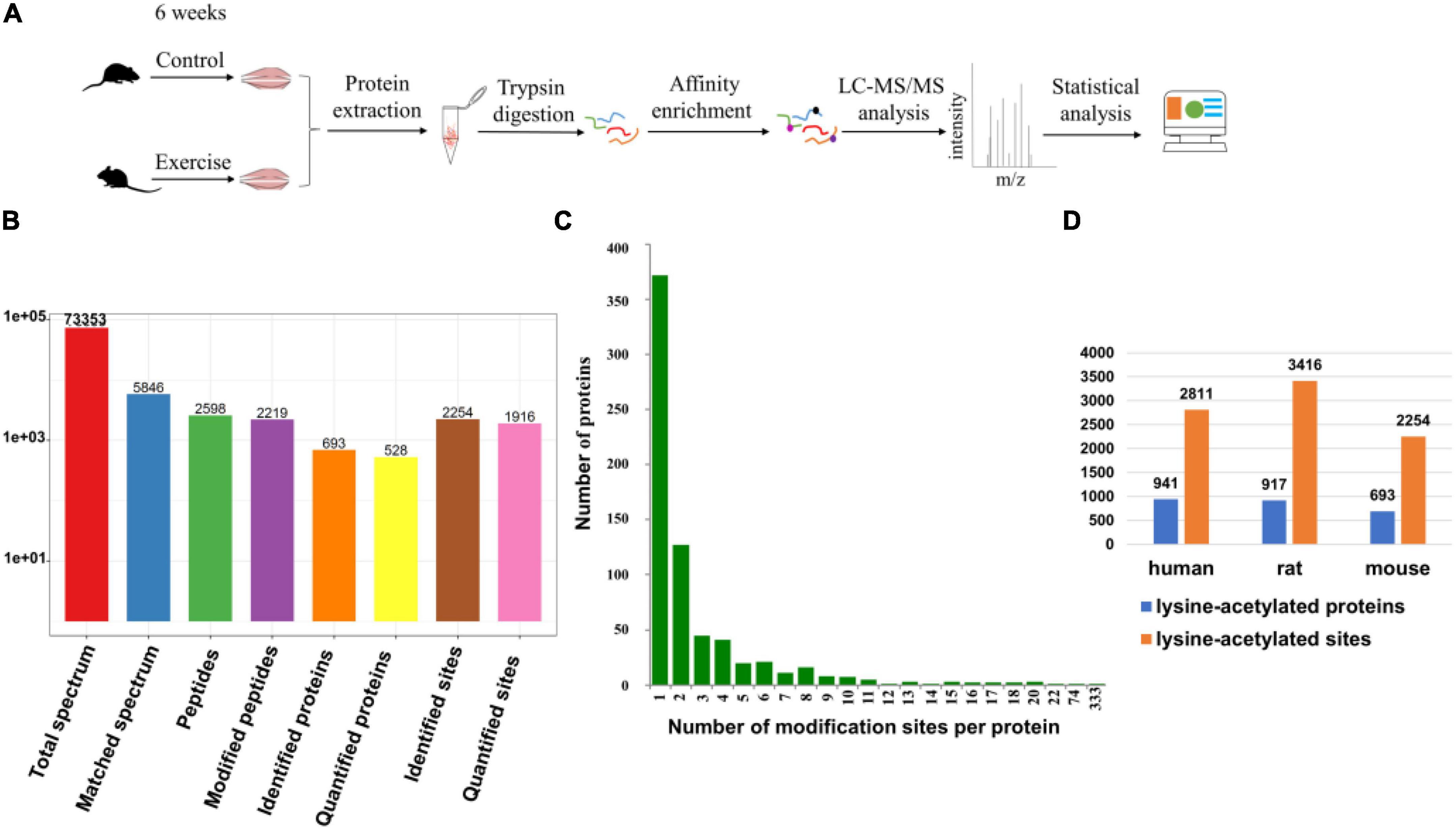
Figure 1. Identification of lysine-acetylated proteins and sites in the gastrocnemius muscle. (A) Schematic of the LC/MS-based quantitative acetylproteomic strategy. (B) The basic statistical figure for MS results. (C) Distribution of acetylated proteins based on the number of acetylated peptides. (D) Comparison of our proteomic study with Lundby’s (Lundby et al., 2012). The figure shows the number of lysine-acetylated proteins and sites identified in muscle from three different species.
Previous studies have shown that physical activity significantly influences muscle features such as muscle mass, muscle force, and muscle fiber type (Kanzleiter et al., 2015; Bacurau et al., 2016; Liu et al., 2021). Different from endurance exercise, which can boost muscle strength, aerobic exercise plays a vital role in increasing the aerobic ability of the muscle. Consistent with these findings, aerobic exercise significantly restrained the body weight increase in the exercise group compared with the control group (Figure 2A). Although gastrocnemius mass and the CSA of gastrocnemius fiber in the exercise group did not change when compared with the control group (Figures 2B,D), we observed that the percentage of MyHC I was increased (Figure 2C), suggesting a potential change of “fast” to “slow” muscle fiber. To investigate the molecular mechanism of exercise on muscle functional properties, we focused on lysine acetylation of muscle proteins. Muscle contraction relies on the correct assembly of myofibrils (Wei et al., 2018), which shows alternating light and dark bands under a microscope corresponding to I-bands and A-bands, respectively (Figure 2E). Myofibrils are composed of tandem arrays of basic functional contractile units called the sarcomeres (Hwang and Sykes, 2015; Wei et al., 2018), which are highly ordered structures composed of thin filaments (actin, tropomyosin, and troponin), thick filaments (myosin), and their associated proteins (such as titin, myomesin, and desmin) (Figure 2E). The sarcomeric function is determined by sarcomeric protein isoform expression and PTMs (Hamdani et al., 2008; Russell and Solis, 2021). In our results, sarcomeric protein levels in gastrocnemius muscle showed no change after exercise (Table 1), but their acetylation levels did change (Table 2). Sarcomeric proteins containing Kac sites were mapped to the protein network database, and the global network graph of these interactions is shown in Figure 2F. Exercise can induce acetylation changes at different sites in these proteins, especially in titin and myosin-4. During muscle contraction, the acetylation levels of 36 Kac sites in titin were upregulated or downregulated, including 10 Kac sites in myosin-4 (Figure 2F). Detailed information on these statistically significant acetylated lysine sites is shown in Table 2 and in Huang’s study (Huang et al., 2020). Although the number of sarcomeric proteins identified in the muscle was much less than that in non-sarcomeric proteins, Kac sites of sarcomeric proteins obviously are more abundant (Figure 2F), especially in titin, myh4, and mybpc2, 333, 74, and 17 Kac sites, respectively (Figure 1C). Thus, lysine acetylation is a crucial PTM of muscle sarcomeric proteins during the exercise process.
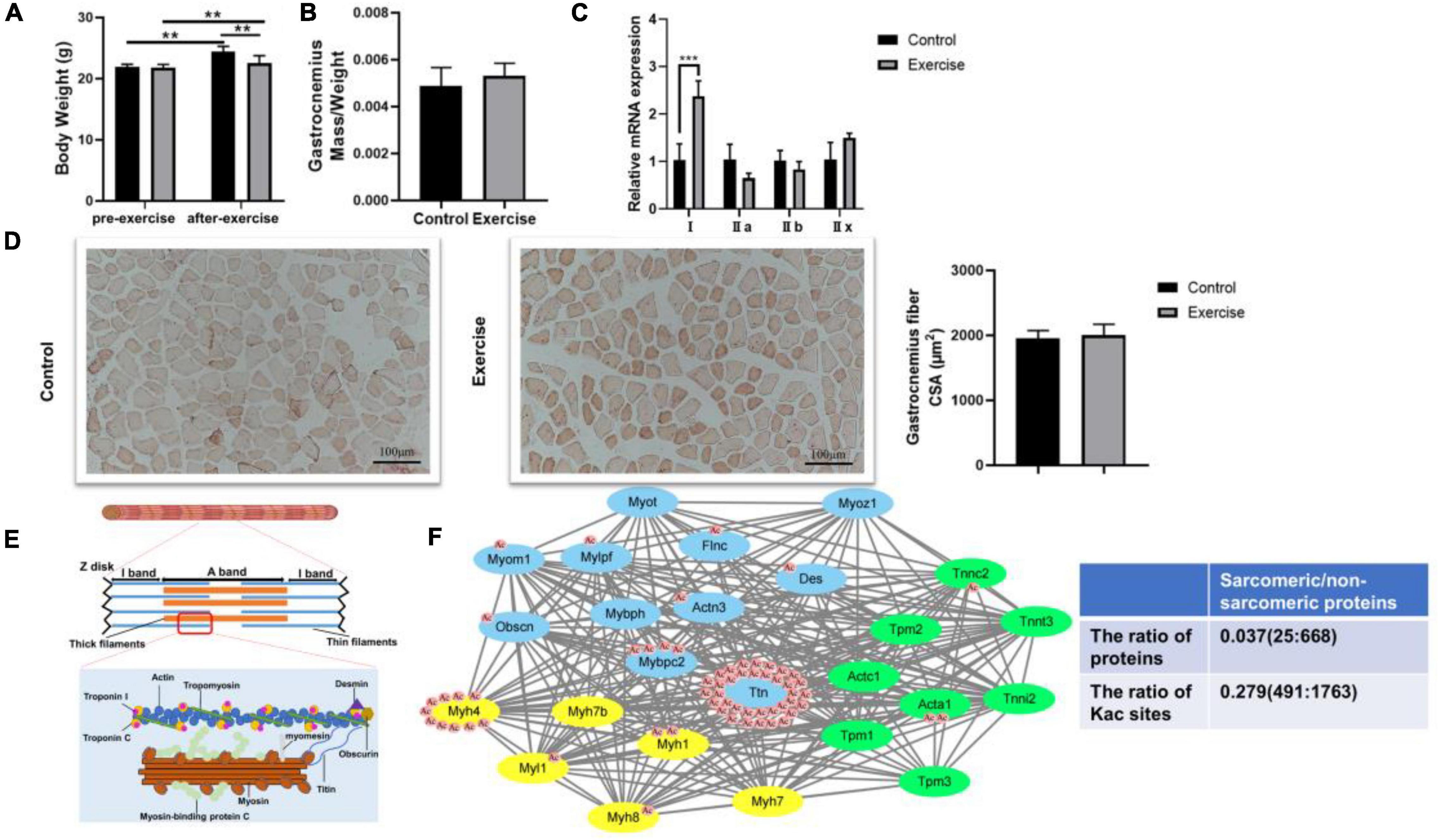
Figure 2. Effect of aerobic exercise on the muscle of mice. (A) Body weight of mice was examined 48 h after treadmill exercise. **P < 0.01. (B) Gastrocnemius muscle mass was measured and normalized to body weight. (C) The gastrocnemius muscle is mainly composed of MyHC IIb and MyHC IIx fiber. Compared with the control group, MyHC I was upregulated in the gastrocnemius of trained mice. Values are means ± SEM; n = 3 in each group; ***P < 0.001. (D) Representative Oil Red O staining of muscle sections and analysis of gastrocnemius fiber CSA in the control and exercise groups. (E) Schematic representation of muscle sarcomeres. Only the main acetylated/deacetylated proteins in the sarcomere and the basic contractile unit of skeletal muscle are shown. (F) Lysine sites of sarcomeric proteins were identified in the mouse skeletal muscle, and a protein–protein interaction network was visualized with STRING. Each green node represents proteins expressed in thin filaments, each yellow node represents proteins expressed in thick filaments, and each blue node represents sarcomere-associated proteins. Some nodes are surrounded by red circles, which represent lysine acetylation induced by exercise. The table shows the comparison of sarcomeric proteins and non-sarcomeric proteins.
To further test the role of lysine acetylation in regulating muscle biological functions, we performed an enrichment analysis. Differential lysine acetylation levels of proteins related to muscle contraction are shown in Figure 3. The site-specific heatmap analysis revealed a clear boundary between the control group and the exercise group, indicating that exercise induced significant changes in the acetylation levels of these proteins, which could be upregulated or downregulated (Figure 3). GO enrichment was performed to better understand the specific role of lysine-acetylated proteins in muscle. A bubble plot of GO enrichment of proteins corresponding to lysine acetylation sites is shown in Figures 4A–C. GO enrichment of biological processes revealed significant protein enrichment in terms related to skeletal muscle adaptation, striated muscle contraction, regulation of muscle contraction, muscle cell development, and others (p < 0.05; Figure 4A). The results of molecular function analysis also showed that proteins related to titin binding, muscle alpha-actinin binding, and calcium ion binding were significantly enriched (p < 0.05; Figure 4B). Cellular component analysis indicated that acetylated proteins were enriched in the muscle myosin complex, I-band, sarcomere, myofibril, and others (p < 0.05; Figure 4C). In addition, a PPI network containing 26 nodes and 67 edges was constructed (Figure 4D). In this study, the PPI network was constructed of acetylated proteins identified as nodes and connected with each other. Many proteins in the PPI network contained numerous Kac sites. In summary, these data suggest that exercise-induced lysine acetylation is critical in muscle protein contraction and structure.
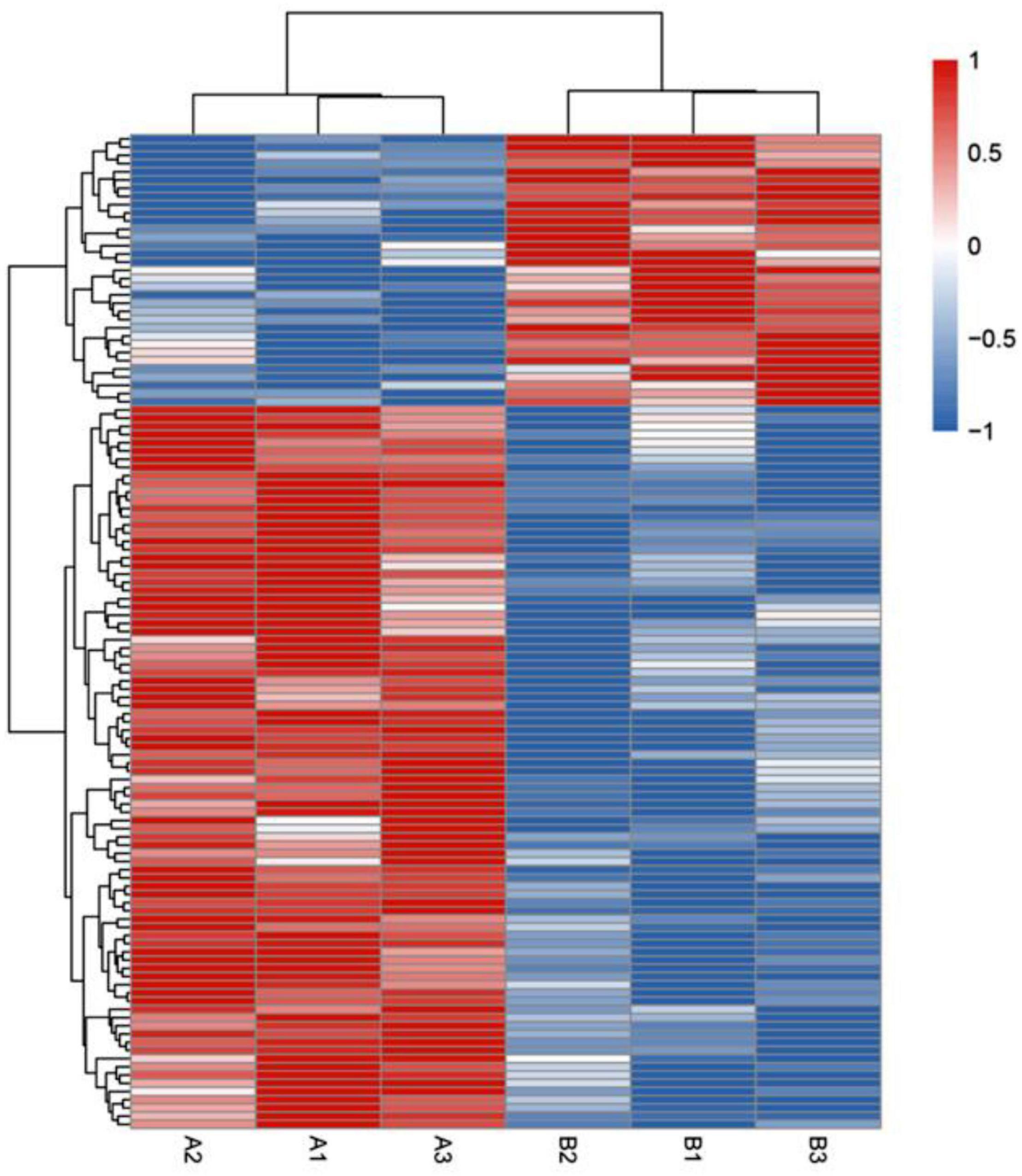
Figure 3. Heatmap showing hierarchical cluster analysis of lysine-acetylated proteins in control groups (A1–A3) and exercise groups (B1–B3). The Euclidean distance was used as the distance metric, and the clustering criteria were complete. The heatmap includes an accompanying supplemental table with the list of the proteins used and their fold change (supplemental “DEP heatmap”).
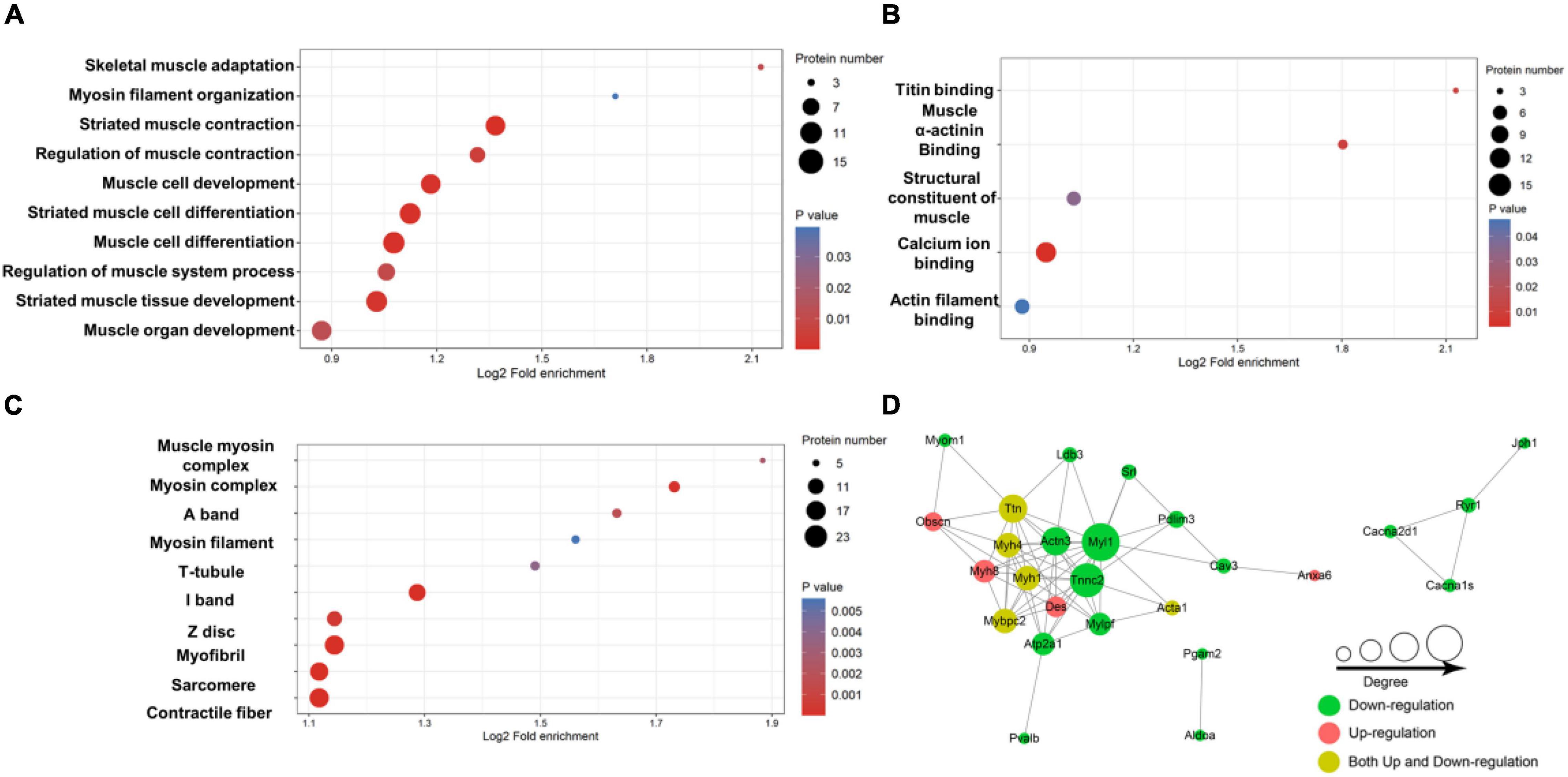
Figure 4. Enrichment analysis of the lysine-acetylated proteins. Enrichment based on GO annotation in terms of biological process (A), molecular function (B), and cellular component (C). (D) Protein–protein interaction network for acetylated proteins. The nodes represent proteins, and their colors represent the upregulation or downregulation of these proteins; green means downregulation (change ratio < 0.83), red means upregulation (change ratio > 1.20), and yellow means both upregulation and downregulation. The size of each node represents the number of directed edges.
Extensive reversible lysine acetylation of metabolic enzymes occurs in both eukaryotes and prokaryotes (Zhao et al., 2010; Nakayasu et al., 2017), which could influence energy balance. Muscle is a high-energy-consuming tissue, so lysine acetylation will control its energy homeostasis (Menzies and Auwerx, 2013). In our study, we identified multiple Kac sites on three enzymes involved in muscle contraction, namely, creatine kinase M-type (Ckm), creatine kinase S-type (Ckmt2), and sarcoplasmic/endoplasmic reticulum calcium ATPase 1 (Atp2a1). There were 22, 8, and 20 Kac sites in Ckm, Ckmt2, and Atp2a1, respectively. There was no alteration in the protein abundance levels of the three enzymes (Table 3), whereas their acetylation levels did change. Interestingly, all Kac sites of the three proteins were deacetylated rather than acetylated. After exercise, the numbers of deacetylated sites of Ckm, Ckmt2, and Atp2a1 were 5, 1, and 10, respectively (Figure 5A and Table 4). Molecular docking is a well-established approach for predicting binding conformations of ligands in active or binding sites of relevant protein targets (Bafna et al., 2020). The catalytic consequences of K9, K11, K25, K247, and K365 deacetylation in Ckm, as well as Ckmt2 and Atp2a1, were evaluated using lysine-to-arginine (K-to-R) mutants in vitro, which mimic deacetylated lysine residues. The docking results showed that deacetylation significantly changed the substrate binding energy and binding regions (Figures 5B,C). For instance, the binding energy between Ckm and creatine was −5.57 kcal/mol, whereas the binding energy between CkmK9R and creatine was −5.21 kcal/mol, and the binding active site changed as well. These data suggest that deacetylation impeded two-molecule interactions and then influenced enzyme activity.
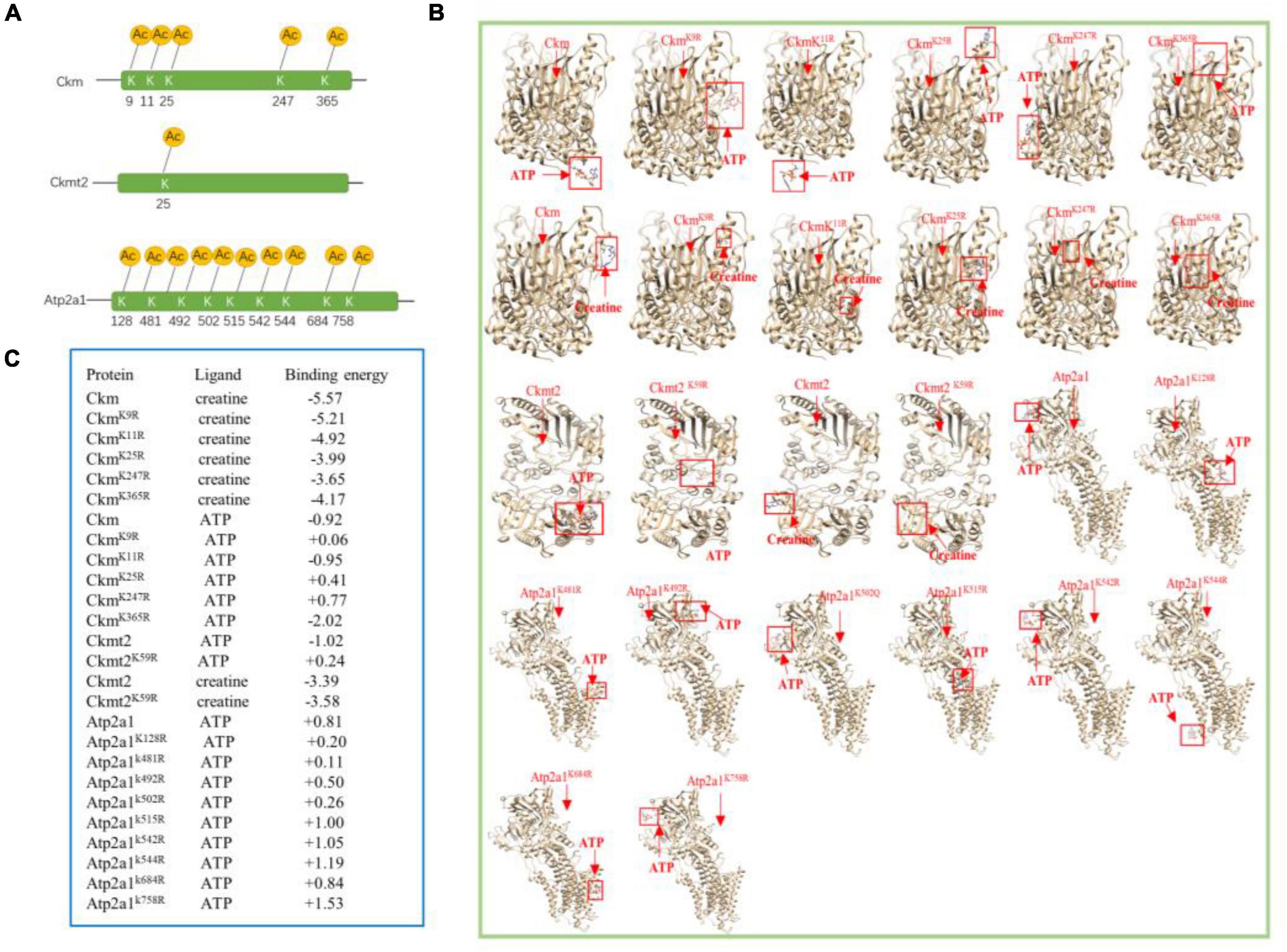
Figure 5. Molecular docking analysis of enzymes. (A) Localizations of the multiacetylated sites in three enzymes that were identified with 5, 1, and 9 acetylated lysine residues. (B) AutoDock 4.2 was used to predict docking interactions of proteins and ligands. The binding positions are highlighted by red boxes. Ckm and Ckmt2 both have two ligands, and Atp2a1 has one ligand. (C) Docking results of the three enzymes.
In short, a particular map of lysine acetylation sites from the skeletal muscle of mice was produced by high-resolution tandem mass spectrometry. Our results revealed that exercise-induced acetylation may play a crucial role in skeletal muscle protein binding free energy, at least to some extent.
In our study, the body weights of the mice in these two groups all increased significantly, and exercise slowed the rate of weight gain (Figure 2A). We did not find a significant difference in the gastrocnemius mass or the CSA of the gastrocnemius fiber between the two groups (Figures 2B,D), but the expression levels of MyHC genes were changed, suggesting that the aerobic training program may be able to switch the fiber from the fast- to the slow-twitch type (Figure 2C). While the results of Oil Red O staining revealed that there was little effect of aerobic exercise on the CSA of muscle, exercise obviously boosted lipid deposition in muscle compared with the sedentary group (Figure 2D), suggesting that physical training increases the energy reserves of muscle. Exercise remains beneficial for the musculoskeletal system, which is a very complex physiological process that requires an appropriate regulatory network (Egan and Zierath, 2013; Hoffman et al., 2015). Protein lysine acetylation/deacetylation in skeletal muscle could be a mediator of this network in mice during exercise. We used the acetylproteomics approach to quantify and map acetyl sites after aerobic exercise in mice. Lundby found that the muscle was the most acetylation-accessible tissue in 16 rat tissues (Lundby et al., 2012), and we also identified large numbers of Kac sites in mouse muscle proteins (Figure 1B). By comparing these two studies, we found that protein acetylation is widespread regardless of species, and the abundance levels of protein lysine acetylation in human, rat, and mouse muscle tissue are similar (Figure 1D). Interestingly, dramatic changes in the acetylation status of numerous cytoplasmic proteins were observed, but the expression levels of KATs and HDACs detected in our proteomic analysis were similar in the two groups (Supplementary Table 1). It is possible that other KATs and HDACs we did not identify in the mass spectrometry are involved in the progress.
In our results, more than half of the proteins (53.70%) had one Kac site and only 15 proteins (2.16%) contained >15 Kac sites (Figure 1C). The importance of acetylation modification is well appreciated in gene regulation, but its regulatory implications extend beyond nuclear histone modifications. We found that sarcomeric proteins accounted for a huge percentage of numerous-site proteins, especially titin (333 sites) and myosin-4 (74 sites). Protein acetylation occurs widely in sarcomeres (Gupta et al., 2008; Samant et al., 2015; Lin et al., 2016) and significantly influences sarcomere function. Gupta provided evidence that the reversible acetylation of sarcomeres plays a role in the regulation of myofilament contractile activity (Gupta et al., 2008). In our study, there were 25 sarcomeric proteins containing Kac sites, which were distributed on thin filaments, thick filaments, or Z-disks. After exercise, the Kac sites of 14 proteins were acetylated or deacetylated (Figure 2F). Thus, exercise triggered protein acetylation or deacetylation in sarcomeres, especially in titin and myosin-4. More importantly, the mean lysine acetylation level of sarcomeric proteins was higher than that of their non-sarcomeric counterparts (Figure 2F). These findings suggested that acetylation modification plays an important role in sarcomeric proteins, which might contribute to muscle contraction. However, the effects of sarcomeric protein acetylation on myofilament contractile activity during exercise remain unclear.
To further understand the potential effect of protein acetylation on muscle structure and function, we performed a bioinformatics analysis. The site-specific heatmap showed that exercise-regulated lysine-acetylated proteins are involved in muscle contraction (Figure 3). According to GO enrichment analysis, the results classification for biological process, molecular function, and cellular component highlighted that these acetylated/deacetylated proteins correspond to different muscle structures or functions (Figures 4A–C). In addition, the protein network analysis indicated that the physiological interactions of these proteins identified in GO enrichment were likely to contribute to their cooperation in muscle (Figure 4D).
Since lysine residues of enzymes are always present around their active sites, protein acetylation influences the catalytic activity of enzymes involved in cellular metabolism (Hernandez-Saavedra et al., 2019; Wang et al., 2019; Min et al., 2020). Lundby reported that multiple enzymes involved in ATP generation are hyperacetylated in skeletal muscle mitochondria (Lundby et al., 2012), and protein acetylation of some enzymes was thought to be involved in impaired fatty acid oxidation and exercise intolerance (Tsuda et al., 2018). In addition, the enzymatic activity of many mitochondrial protein bacterial orthologs is regulated by lysine acetylation (Wang et al., 2010). Interestingly, we found that there were three enzymes directly linked with muscle energy regulation, namely, Ckm, Ckmt2, and Atp2a1. Then, we used AutoDock software to simulate the effects of lysine acetylation on these metabolic enzymes. Compared with wild-type enzymes, mimetic mutants appear to have different substrate-binding energies and binding positions (Figures 5B,C). Consistent with a previous study (Walker et al., 2021), acetylation of Ckm disrupted salt bridge formations between Ckm monomers, including K9, K11, and K25, in this study. In addition, Gorski found that acetylation at K492 played a critical role in regulating SERCA2a (sarcoplasmic/endoplasmic reticulum Ca2+ ATPase 2a) activity in mice (Gorski et al., 2019). Therefore, the deacetylation of these lysine sites was likely to affect the catalytic reaction of Ckm, Ckmt2, and Atp2a1, which may be associated with the impact of acetylation on the dimer formation of Ckm and Ckmt2 (Walker et al., 2021), although site-specific contributions were indistinct.
It is widely accepted that aerobic exercise can increase exercise capacity, such as muscle oxidative capacity, muscle contraction, and muscle endurance (Overmyer et al., 2015; de Oliveira et al., 2017; Huang et al., 2020). Furthermore, muscle protein acetylation levels also significantly changed after 6 weeks of aerobic exercise. Previously, Gupta reported that reversible acetylation of sarcomeric proteins played a role in regulating myofilament contractile activity (Gupta et al., 2008). Similarly, lysine acetylation of cardiac myosin heavy chain isoforms modulates their enzymatic and motor activity (Samant et al., 2015). Some researchers have also demonstrated that altering the muscle structure and function by acetylation of existing sarcomeric proteins can optimize energy usage (Russell and Solis, 2021). With these observations, it is reasonable to speculate that lysine acetylation is associated with alterations in muscle protein binding free energy during exercise.
We showed that exercise changes muscle protein acetylation in mice, which serves as a reference for further Kac studies in human. In these future clinical and mechanistic studies, verification of the correlation between protein acetylation and skeletal muscle physiology or exercise performance should be taken into account, which will improve our understanding of the molecular response to regular exercise. Ongoing acetylproteomics studies will provide a theoretical basis for exercise prescription. In addition, novel approaches targeting the acetylation of skeletal muscle proteins, especially sarcomeric proteins, would provide important clues for the development of an effective intervention for muscle atrophy with aging. In any case, these results from our study still need to be confirmed in clinical trials.
However, there are some limitations to this study. First, the effects of acetylation on the mechanical parameters of sarcomere proteins remain unknown. Then, we did not perform the same experiments on female mice or old mice, which may not be replicated because sex and age are also important biological variables. In addition, other exercise parameters (such as session length, frequency, and intensity) were not tested in our study. Finally, the impact of deacetylation on enzyme activity requires further experimental verification.
Our data strongly suggested that the regulatory scope of exercise-induced lysine acetylation is widespread in skeletal muscle proteins. Exploring the mechanisms of exercise-induced acetylation changes in skeletal muscle can help develop scientific exercise prescriptions. Overall, skeletal muscle exhibits extensive metabolic and energetic remodeling during exercise, and exercise-induced lysine acetylation appears to be a crucial mediator of skeletal muscle proteins.
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the iProX partner repository (Ma et al., 2019) with the dataset identifier PXD032660.
The animal study was reviewed and approved by the SYXK-2019-0004.
DL prepared the figures and drafted the manuscript. LF and YN edited and revised the manuscript. CC, SH, SL, LF, and YN approved the final version of the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by grants from the National Natural Science Foundation of China (NSFC) 31671237 (to YN), 32171135, and 31871206 (to LF).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We are grateful to Xinyu Zhang for her technical support during the experiments.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnagi.2022.859313/full#supplementary-material
Atp2a1, sarcoplasmic/endoplasmic reticulum calcium ATPase 1; Ckm, creatine kinase M-type; Ckmt2, creatine kinase S-type; DTT, dithiothreitol; FDR, false discovery rate; GO, Gene Ontology; Kac, lysine-acetylated; NAM, nicotinamide; PTM, post-translational modification; TFA, trifluoroacetic acid; TSA, trichostatin.
Bacurau, A. V., Jannig, P. R., de Moraes, W. M., Cunha, T. F., Medeiros, A., Barberi, L., et al. (2016). Akt/mTOR pathway contributes to skeletal muscle anti-atrophic effect of aerobic exercise training in heart failure mice. Int. J. Cardiol. 214, 137–147. doi: 10.1016/j.ijcard.2016.03.071
Bafna, K., Krug, R. M., and Montelione, G. T. (2020). Structural similarity of SARS-CoV2 M(pro) and HCV NS3/4A Proteases suggests new approaches for identifying existing drugs useful as COVID-19 therapeutics. ChemRxiv doi: 10.26434/chemrxiv.12153615
de Oliveira, N. C., Portes, L. A., Pettersson, H., Alexanderson, H., and Bostrom, C. (2017). Aerobic and resistance exercise in systemic sclerosis: state of the art. Musculoskeletal Care 15, 316–323. doi: 10.1002/msc.1185
Egan, B., and Zierath, J. R. (2013). Exercise metabolism and the molecular regulation of skeletal muscle adaptation. Cell Metab. 17, 162–184. doi: 10.1016/j.cmet.2012.12.012
Fernando, P., Bonen, A., and Hoffman-Goetz, L. (1993). Predicting submaximal oxygen consumption during treadmill running in mice. Can. J. Physiol. Pharmacol. 71, 854–857. doi: 10.1139/y93-128
Fukushima, A., and Lopaschuk, G. D. (2016). Acetylation control of cardiac fatty acid beta-oxidation and energy metabolism in obesity, diabetes, and heart failure. Biochim. Biophys. Acta 1862, 2211–2220. doi: 10.1016/j.bbadis.2016.07.020
Gan, Z., Fu, T., Kelly, D. P., and Vega, R. B. (2018). Skeletal muscle mitochondrial remodeling in exercise and diseases. Cell Res. 28, 969–980. doi: 10.1038/s41422-018-0078-7
Gorski, P. A., Jang, S. P., Jeong, D., Lee, A., Lee, P., Oh, J. G., et al. (2019). Role of SIRT1 in modulating acetylation of the sarco-endoplasmic reticulum Ca(2+)-ATPase in heart failure. Circ. Res. 124, e63–e80. doi: 10.1161/CIRCRESAHA.118.313865
Gupta, M. P., Samant, S. A., Smith, S. H., and Shroff, S. G. (2008). HDAC4 and PCAF bind to cardiac sarcomeres and play a role in regulating myofilament contractile activity. J. Biol. Chem. 283, 10135–10146. doi: 10.1074/jbc.M710277200
Hamdani, N., Kooij, V., van Dijk, S., Merkus, D., Paulus, W. J., Remedios, C. D., et al. (2008). Sarcomeric dysfunction in heart failure. Cardiovasc. Res. 77, 649–658. doi: 10.1093/cvr/cvm079
Hernandez-Saavedra, D., Moody, L., Xu, G. B., Chen, H., and Pan, Y. X. (2019). Epigenetic regulation of metabolism and inflammation by calorie restriction. Adv. Nutr. 10, 520–536. doi: 10.1093/advances/nmy129
Hoffman, N. J., Parker, B. L., Chaudhuri, R., Fisher-Wellman, K. H., Kleinert, M., Humphrey, S. J., et al. (2015). Global phosphoproteomic analysis of human skeletal muscle reveals a network of exercise-regulated kinases and AMPK substrates. Cell Metab. 22, 922–935. doi: 10.1016/j.cmet.2015.09.001
Huang, C., Yang, X., Zeng, B., Zeng, L., Gong, X., Zhou, C., et al. (2019). Proteomic analysis of olfactory bulb suggests CACNA1E as a promoter of CREB signaling in microbiota-induced depression. J. Proteomics 194, 132–147. doi: 10.1016/j.jprot.2018.11.023
Huang, S., Liu, S., Niu, Y., and Fu, L. (2020). Scriptaid/exercise-induced lysine acetylation is another type of posttranslational modification occurring in titin. J. Appl. Physiol. 128, 276–285. doi: 10.1152/japplphysiol.00617.2019
Hwang, P. M., and Sykes, B. D. (2015). Targeting the sarcomere to correct muscle function. Nat. Rev. Drug. Discov. 14, 313–328. doi: 10.1038/nrd4554
Kanzleiter, T., Jahnert, M., Schulze, G., Selbig, J., Hallahan, N., Schwenk, R. W., et al. (2015). Exercise training alters DNA methylation patterns in genes related to muscle growth and differentiation in mice. Am. J. Physiol. Endocrinol. Metab. 308, E912–E920. doi: 10.1152/ajpendo.00289.2014
Lin, Y. H., Warren, C. M., Li, J., McKinsey, T. A., and Russell, B. (2016). Myofibril growth during cardiac hypertrophy is regulated through dual phosphorylation and acetylation of the actin capping protein CapZ. Cell Signal. 28, 1015–1024. doi: 10.1016/j.cellsig.2016.05.011
Liu, S., Yu, C., Xie, L., Niu, Y., and Fu, L. (2021). Aerobic Exercise improves mitochondrial function in sarcopenia mice through Sestrin2 in an AMPKalpha2-dependent manner. J. Gerontol. A Biol. Sci. Med. Sci. 76, 1161–1168. doi: 10.1093/gerona/glab029
Lundby, A., Lage, K., Weinert, B. T., Bekker-Jensen, D. B., Secher, A., Skovgaard, T., et al. (2012). Proteomic analysis of lysine acetylation sites in rat tissues reveals organ specificity and subcellular patterns. Cell Rep. 2, 419–431. doi: 10.1016/j.celrep.2012.07.006
Ma, J., Chen, T., Wu, S., Yang, C., Bai, M., Shu, K., et al. (2019). iProX: an integrated proteome resource. Nucleic Acids Res. 47, D1211–D1217. doi: 10.1093/nar/gky869
Marton, O., Koltai, E., Nyakas, C., Bakonyi, T., Zenteno-Savin, T., Kumagai, S., et al. (2010). Aging and exercise affect the level of protein acetylation and SIRT1 activity in cerebellum of male rats. Biogerontology 11, 679–686. doi: 10.1007/s10522-010-9279-2
McGee, S. L., Fairlie, E., Garnham, A. P., and Hargreaves, M. (2009). Exercise-induced histone modifications in human skeletal muscle. J. Physiol. 587(Pt 24), 5951–5958. doi: 10.1113/jphysiol.2009.181065
Menzies, K., and Auwerx, J. (2013). An acetylation rheostat for the control of muscle energy homeostasis. J. Mol. Endocrinol. 51, T101–T113. doi: 10.1530/JME-13-0140
Min, Z., Long, X., Zhao, H., Zhen, X., Li, R., Li, M., et al. (2020). Protein lysine acetylation in ovarian granulosa cells affects metabolic homeostasis and clinical presentations of women with polycystic ovary syndrome. Front. Cell Dev. Biol. 8:567028. doi: 10.3389/fcell.2020.567028
Moresi, V., Marroncelli, N., Coletti, D., and Adamo, S. (2015). Regulation of skeletal muscle development and homeostasis by gene imprinting, histone acetylation and microRNA. Biochim. Biophys. Acta 1849, 309–316. doi: 10.1016/j.bbagrm.2015.01.002
Nakayasu, E. S., Burnet, M. C., Walukiewicz, H. E., Wilkins, C. S., Shukla, A. K., Brooks, S., et al. (2017). Ancient regulatory role of lysine acetylation in central metabolism. mBio 8:e1894-17. doi: 10.1128/mBio.01894-17
Narita, T., Weinert, B. T., and Choudhary, C. (2019). Author correction: functions and mechanisms of non-histone protein acetylation. Nat. Rev. Mol. Cell Biol. 20:508. doi: 10.1038/s41580-019-0156-9
Nelson, M. E., Parker, B. L., Burchfield, J. G., Hoffman, N. J., Needham, E. J., Cooke, K. C., et al. (2020). Phosphoproteomics reveals conserved exercise-stimulated signaling and AMPK regulation of store-operated calcium entry. EMBO J. 39:e104246. doi: 10.15252/embj.2019104246
Overmyer, K. A., Evans, C. R., Qi, N. R., Minogue, C. E., Carson, J. J., Chermside-Scabbo, C. J., et al. (2015). Maximal oxidative capacity during exercise is associated with skeletal muscle fuel selection and dynamic changes in mitochondrial protein acetylation. Cell Metab. 21, 468–478. doi: 10.1016/j.cmet.2015.02.007
Ren, Y., Hao, P., Dutta, B., Cheow, E. S., Sim, K. H., Gan, C. S., et al. (2013). Hypoxia modulates A431 cellular pathways association to tumor radioresistance and enhanced migration revealed by comprehensive proteomic and functional studies. Mol. Cell Proteomics 12, 485–498. doi: 10.1074/mcp.M112.018325
Rizvi, S. M., Shakil, S., and Haneef, M. (2013). A simple click by click protocol to perform docking: AutoDock 4.2 made easy for non-bioinformaticians. EXCLI J. 12, 831–857.
Russell, A. P., Foletta, V. C., Snow, R. J., and Wadley, G. D. (2014). Skeletal muscle mitochondria: a major player in exercise, health and disease. Biochim. Biophys. Acta 1840, 1276–1284. doi: 10.1016/j.bbagen.2013.11.016
Russell, B., and Solis, C. (2021). Mechanosignaling pathways alter muscle structure and function by post-translational modification of existing sarcomeric proteins to optimize energy usage. J. Muscle Res. Cell Motil. 42, 367–380. doi: 10.1007/s10974-021-09596-9
Samant, S. A., Pillai, V. B., Sundaresan, N. R., Shroff, S. G., and Gupta, M. P. (2015). Histone deacetylase 3 (HDAC3)-dependent reversible lysine acetylation of cardiac myosin heavy chain isoforms modulates their enzymatic and motor activity. J. Biol. Chem. 290, 15559–15569. doi: 10.1074/jbc.M115.653048
Seeliger, D., and de Groot, B. L. (2010). Ligand docking and binding site analysis with PyMOL and Autodock/Vina. J. Comput. Aided Mol. Des. 24, 417–422. doi: 10.1007/s10822-010-9352-6
Tsuda, M., Fukushima, A., Matsumoto, J., Takada, S., Kakutani, N., Nambu, H., et al. (2018). Protein acetylation in skeletal muscle mitochondria is involved in impaired fatty acid oxidation and exercise intolerance in heart failure. J. Cachexia Sarcopenia Muscle 9, 844–859. doi: 10.1002/jcsm.12322
Walker, M. A., Chavez, J., Villet, O., Tang, X., Keller, A., Bruce, J. E., et al. (2021). Acetylation of muscle creatine kinase negatively impacts high-energy phosphotransfer in heart failure. JCI Insight 6:e144301. doi: 10.1172/jci.insight.144301
Wang, Q., Zhang, Y., Yang, C., Xiong, H., Lin, Y., Yao, J., et al. (2010). Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science 327, 1004–1007. doi: 10.1126/science.1179687
Wang, Y., Wang, F., Bao, X., and Fu, L. (2019). Systematic analysis of lysine acetylome reveals potential functions of lysine acetylation in Shewanella baltica, the specific spoilage organism of aquatic products. J. Proteomics 205:103419. doi: 10.1016/j.jprot.2019.103419
Wei, L., Gregorich, Z. R., Lin, Z., Cai, W., Jin, Y., McKiernan, S. H., et al. (2018). Novel sarcopenia-related alterations in sarcomeric protein post-translational modifications (PTMs) in skeletal muscles identified by top-down proteomics. Mol. Cell Proteomics 17, 134–145. doi: 10.1074/mcp.RA117.000124
Widegren, U., Ryder, J. W., and Zierath, J. R. (2001). Mitogen-activated protein kinase signal transduction in skeletal muscle: effects of exercise and muscle contraction. Acta Physiol. Scand. 172, 227–238. doi: 10.1046/j.1365-201x.2001.00855.x
Wu, Y., Tang, J., Zhou, C., Zhao, L., Chen, J., Zeng, L., et al. (2016). Quantitative proteomics analysis of the liver reveals immune regulation and lipid metabolism dysregulation in a mouse model of depression. Behav. Brain Res. 311, 330–339. doi: 10.1016/j.bbr.2016.05.057
Xing, S., and Poirier, Y. (2012). The protein acetylome and the regulation of metabolism. Trends Plant Sci. 17, 423–430. doi: 10.1016/j.tplants.2012.03.008
Yu, Y., Wang, H., Rao, X., Liu, L., Zheng, P., Li, W., et al. (2021). Proteomic profiling of lysine acetylation indicates mitochondrial dysfunction in the hippocampus of gut microbiota-absent mice. Front. Mol. Neurosci. 14:594332. doi: 10.3389/fnmol.2021.594332
Keywords: aerobic exercise, bioinformatics analysis, lysine acetylation, muscle, proteins
Citation: Liang D, Chen C, Huang S, Liu S, Fu L and Niu Y (2022) Alterations of Lysine Acetylation Profile in Murine Skeletal Muscles Upon Exercise. Front. Aging Neurosci. 14:859313. doi: 10.3389/fnagi.2022.859313
Received: 21 January 2022; Accepted: 04 April 2022;
Published: 03 May 2022.
Edited by:
Howe Liu, University of North Texas Health Science Center, United StatesReviewed by:
Eirini Lionaki, Foundation for Research and Technology Hellas (FORTH), GreeceCopyright © 2022 Liang, Chen, Huang, Liu, Fu and Niu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Li Fu, bGlmdUB0bXUuZWR1LmNu; Yanmei Niu, eWFubWVpLm5pdUB0bXUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.