- 1School of Information Engineering, Tianjin University of Commerce, Tianjin, China
- 2School of Sciences, Tianjin University of Commerce, Tianjin, China
- 3Department of Molecular Imaging, Qingdao Central Hospital, Qingdao University, Qingdao, China
- 4Department of Radiology and Tianjin Key Laboratory of Functional Imaging, Tianjin Medical University General Hospital, Tianjin, China
Late-onset Alzheimer's disease (LOAD) is a common irreversible neurodegenerative disease with heterogeneous genetic characteristics. Identifying the biological biomarkers with the potential to predict the conversion from normal controls to LOAD is clinically important for early interventions of LOAD and clinical treatment. The polygenic risk score for LOAD (AD-PRS) has been reported the potential possibility for reliably identifying individuals with risk of developing LOAD recently. To investigate the external phenotype changes resulting from LOAD and the underlying etiology, we summarize the comprehensive associations of AD-PRS with multiple biomarkers, including neuroimaging, cerebrospinal fluid and plasma biomarkers, cardiovascular risk factors, cognitive behavior, and mental health. This systematic review helps improve the understanding of the biomarkers with potential predictive value for LOAD and further optimizing the prediction and accurate treatment of LOAD.
Introduction
Alzheimer's disease (AD) which accounts for about 70% of dementia is an irreversible progressive polygenic neurodegenerative disease with insidious onset (Kametani and Hasegawa, 2018; Breijyeh and Karaman, 2020; Tank et al., 2022). By age at onset, AD can be classified into early-onset AD (EOAD) and late-onset AD (LOAD). EOAD is an autosomal dominant disease with heritability of more than 70% (Gatz et al., 2006; Wingo et al., 2012) and three responsible mutated genes, the amyloid protein precursor gene (APP), presenilin-1 gene (PSEN1), and presenilin-2 gene (PSEN2), were found to mainly dominate the production, aggregation, and clearance of amyloid β-protein (Aβ) (Cacace et al., 2016). Unlike the EOAD, LOAD occurs in more than 95% of the AD patients with a relatively complex polygenetic mechanism (Zhu et al., 2015; Xiao et al., 2017), and the related external phenotype changes in the very early stage. Although aducanumab can reduce the amyloid deposition in the brain and has been approved by Food and Drug Administration to treat Alzheimer's disease lately, however, controversy about it still exists (Selkoe, 2021; Servick, 2021). Therefore, identifying the biomarkers with the potential to predict the conversion from normal controls to LOAD and the progression of LOAD is clinically very important for early interventions.
In recent years, genome-wide association studies (GWAS) have been widely applied to study complex neuropsychiatric disorders (Ripke et al., 2014; Lello et al., 2019; van der Merwe et al., 2019; Levey et al., 2021; Peyrot and Price, 2021) and more than 200 susceptibility genetic variants have been identified to characterize the polygenetic architecture of LOAD (Chen et al., 2021). To overcome the small effect size of a single genetic variant, some polygenic methods have been developed to quantify the cumulative effects of multiple genetic variants related to complex diseases (Tan et al., 2018; Altmann et al., 2020; Choi et al., 2020), of which the polygenic risk score (PRS) is the most representative and widely used method (Wray et al., 2021). With the release of large-sample GWAS summary statistics for LOAD (Lambert et al., 2013; Weiner et al., 2015; Kunkle et al., 2019), AD-PRS, which measures the cumulative genome-wide-weighted effects of LOAD-risk genetic variants, is being increasingly used with multiple biomarkers to identify the underlying neurobiological mechanisms of LOAD.
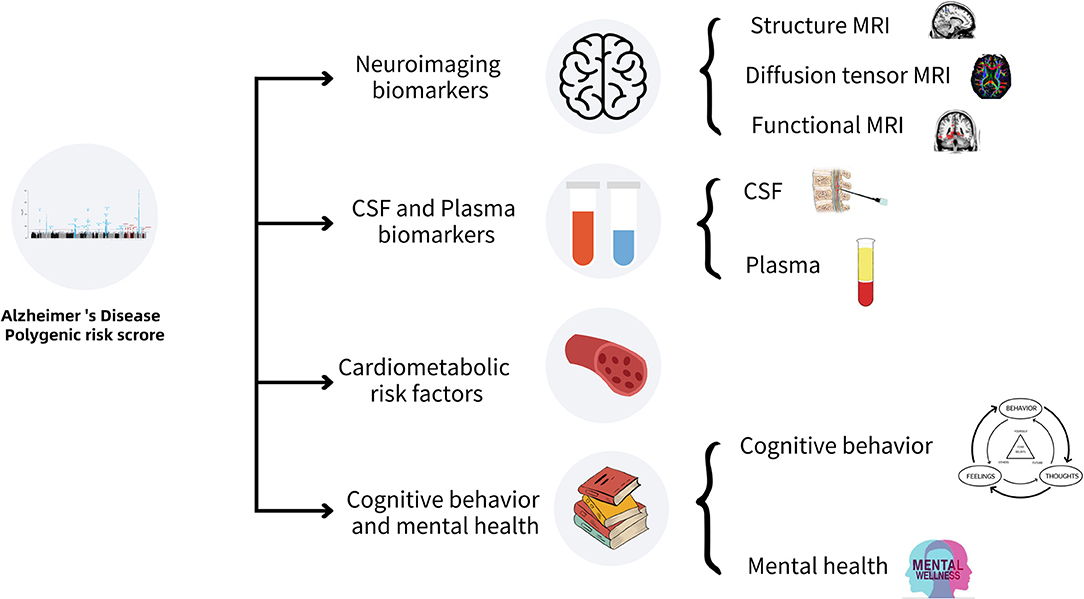
In this review, we summarized the research progress of the associations of AD-PRS with multiple biomarkers, including neuroimaging, cerebrospinal fluid, and plasma, cardiovascular risk factors, cognitive behaviors, and mental health. This review is helpful to identify the biomarkers with the potential to predict the occurrence and development of LOAD, which is clinically important for the early diagnosis and interventions of this complex disease. A schematic summary of the related work in this review is shown in Figure 1 and Table 1.
Associations of AD-PRS With Neuroimaging Biomarkers
Exploring the structural and functional changes through medical imaging techniques is crucial for understanding LOAD development. Because of the advantages of safety and information abundance, magnetic resonance imaging (MRI) has become prominent among various medical imaging techniques. Of the various modalities of MRI, structure MRI (sMRI), diffusion tensor MRI (dMRI), and functional MRI (fMRI) have been mostly applied to study the underlying neural mechanism of LOAD and its clinical diagnosis and treatment by exploring the correlation between AD-PRS and brain phenotypes.
sMRI is one of the most important avenues to illustrate the brain morphological measures, for example, gray matter volume, cortical surface area, and cortical thickness. Studies have found that AD-PRS was associated with reduced gray matter volume (GMV) in the hippocampus (Axelrud et al., 2018) and its subregions (Heidi et al., 2021), left precuneus and right cingulate gyrus cortex (Li et al., 2018), whereas with increased GMV in the right superior frontal gyrus and caudate (Li et al., 2018). Meanwhile, AD-PRS was found to be associated with decreased surface area in the frontal pole (Xiao et al., 2017), decreased cortical thickness in the bilateral medial temporal cortices (Lee et al., 2021), posterior cingulate cortices (Sabuncu et al., 2012), and bilateral entorhinal cortices (Harrison et al., 2016). The changes of these brain regions are some of the most prominent early pathological features of LOAD and can be used as reliable predictive measures for the conversion from normal controls or mild-cognitive impairment to LOAD (Yang et al., 2012).
dMRI is mainly used to measure the microstructural integrity of the white matter through modeling-water diffusivity in the tissue microstructure (Kilimann et al., 2013), with fractional anisotropy (FA) and mean diffusivity as the two most used indices. AD-PRS is associated with decreased FA in the right cingulum bundle in healthy adults (Foley et al., 2017). AD-PRS was also found to be associated with reduced FA and increased mean diffusivity across the whole brain white matter tracts, notably in the inferior occipitofrontal fascicle, superior longitudinal fascicle, cingulum and corpus callosum in the AD patients (Harrison et al., 2020a). Recently, Mirza-Davies et al. (2021) found the visual subnetwork constructed based on dMRI was also correlated with AD-PRS.
fMRI was used to evaluate brain activity by detecting changes associated with blood flow (Smitha et al., 2017), referred to as the blood-oxygen-level-dependent (BOLD) signal in the brain-resting or task-based state. AD-PRS was found to be associated with increased functional connectivity between the right precuneus and the right superior temporal gyrus in the youths, which might impact memory performance and inhibitory control in early life (Axelrud et al., 2019). AD-PRS was also found to be associated with decreased functional connectivity within the temporal cortex in mild-cognitive impairment patients (Su et al., 2017). The hippocampal activation, mostly responsible for episodic memory processing, was severely impaired in the LOAD patients (Zhan et al., 2016; Xiao et al., 2017). However, contrary research findings have been reported between the AD-PRS and hippocampal activation. Chandler et al. (2020) found a significantly positive correlation and Xiao et al. (2017) found a significantly negative correlation during the episodic memory. This divergence may be due to the different task codings and sample size of the studies.
Arterial spin labeling was a functional MRI technology for measuring tissue perfusion to quantify the cerebral blood flow (CBF) in a given period with high time resolution (Rostami et al., 2014). There is a hypothesis proposing that insufficient CBF increases the risk of developing LOAD, leads to the decline of consciousness and dysfunction of LOAD, and even can be treated as an early antecedent of LOAD (Chandler et al., 2021). AD-PRS was found to be negatively correlated with CBF on many brain regions across the younger and older participants, including the frontal pole, middle frontal gyrus, inferior frontal gyrus, insular, frontal medial cortex, and orbitofrontal cortex (Chandler et al., 2019, 2021). These studies may shed light on exploring the key molecular processes that underpin LOAD.
All of the above findings together revealed the close relationship between the cumulative genetic risk of LOAD and the changes in the brain structure and function, providing new perspectives to explain the pathophysiology of LOAD. The combination of the neuroimaging biomarkers with AD-PRS to predict the LOAD development is attracting attention (Harrison et al., 2016, 2020b; Williams et al., 2021) and this is thought to be a promising step toward improving the very early identification of LOAD (Williams et al., 2021).
Associations of AD-PRS With Cerebrospinal Fluid and Plasma Biomarkers
The concentration determination of Aβ, total tau (T-tau), and phosphorylated tau (P-tau) in the cerebrospinal fluid (CSF) are three classical biomarkers for the clinical diagnosis of LOAD (Lee et al., 2019; Shen et al., 2021). The changes of these measures in the brain occur more than 15 years before the onset of symptoms in LOAD patients (Bateman et al., 2012; Dementia, 2021). More studies devoted to the association analysis of AD-PRS and these biomarkers found that AD-PRS was not only correlated with the CSF levels of Aβ42, Aβ42/Aβ40, T-tau, and P-tau in the older adults (Porter et al., 2018a; Li et al., 2020), but could also predict the incidence rate of LOAD and the age at onset (Li et al., 2020). In addition, there was an interaction between AD-PRS and the Aβ42 pathology status to the neurofilament light (NfL) (Skoog et al., 2021). Moreover, the A/T/N criteria including a combined accumulation of amyloid plaques (A), neurofibrillary tangles composed of tau (T), and neurodegeneration (N) can predict the cognitive decline and clinical progression of LOAD (Soldan et al., 2019; Ebenau et al., 2020) and are recommended to be included in the diagnostic categories of LOAD (Foley et al., 2017). AD-PRS also showed a significant correlation with the A/T/N profiles (Ebenau et al., 2021). A study found that the integration of genetic risk across the AD biomarkers like A/T/N may improve the prediction of the disease progression (Moore et al., 2019).
Various inflammations occur in pathologically vulnerable brain regions in LOAD patients (Akiyama, 2000) and many plasma biomarkers of inflammation are useful for early diagnosis and monitoring the progression of LOAD (Kinney et al., 2018; Naveed et al., 2019). AD-PRS was found to be associated with various increased inflammatory biomarkers in the plasma, such as clusterin, complement receptor 1 inhibitor and C-reactive protein (Morgan et al., 2017), osteopontin and neurocan core protein (Zhou et al., 2020), and P-tau 181 (Zettergren et al., 2021). Similar to other biomarkers, the integration of AD-PRS and inflammatory biomarkers can also greatly improve the sensitivity and specificity of predicting LOAD. These findings not only facilitate the development of genetic tools for assessing the individual risk of LOAD but could also improve our understanding of the underlying mechanisms of this disease.
Associations of AD-PRS With Cardiometabolic Risk Factors
Many cardiometabolic risk factors are implicated in the etiology of LOAD and are thought to lie on the pathways linking the genetic variants of LOAD (Korologou-Linden et al., 2019b). Of these factors, cardiovascular risk factors are found to increase the incidence of LOAD (Lin et al., 2019), which may be due to the high genetic association between LOAD and many cardiovascular diseases, such as hypertension (Baumgart et al., 2015), coronary heart disease (Elman et al., 2019), diabetes, and diastolic blood pressure (Richardson et al., 2019). AD-PRS was also found positively associated with other cardiometabolic risk factors such as traumatic brain injury, obesity, and hypertension in adults (Baumgart et al., 2015). However, these associations are not consistent throughout the whole life trajectory. For example, Korologou-linden et al. did not detect evidence to suggest that AD-PRS acts through childhood and adolescent cardiometabolic risk factors (Korologou-Linden et al., 2019b). More studies should be conducted in other large-birth cohorts to examine whether the genetic risk for Alzheimer's disease can be captured in early childhood. If not, further studies should examine whether and why these associations emerge only later, in adulthood, when the variation in the cardiometabolic risk factors is likely to be greater.
The combination of the genetic accumulation risk of LOAD and some vascular risk factors increased the predictive potential of LOAD for the shared genetic heritage (Li et al., 2016). The coronary artery disease (CAD) interacting with the LOAD pathology is highly heritable and CAD-PRS has been widely used to improve cardiovascular risk prediction (Wehby et al., 2018; Elliott et al., 2020; Levin and Rader, 2020). A healthy adult group with higher CAD-PRS and AD-PRS showed a significantly increased risk of developing amnestic mild-cognitive impairment (aMCI) (Elman et al., 2019), which is a state of cognitive deficit that is not severe enough to fulfill the criteria of dementia (Bennett et al., 2002) and showed a much higher probability of developing into LOAD (Chaudhury et al., 2019). In summary, AD-PRS, combined with the PRS of cardiovascular risk factors, has shown a superior predictive value of onset of aMCI and LOAD compared to the independent application of AD-PRS, indicating the importance of infusing multiple PRSs and their interactions.
Association of AD-PRS With Cognitive Behaviors and Mental Health
The impairment of episodic memory and decline in advanced cognitive functions are the earliest and most characteristically clinical manifestations of LOAD (Bäckman et al.,2004). In the early stage, cognitive behaviors and mental health of the LOAD patients are partially impaired, which complicate and intertwine with the occurrence and progression of LOAD. Exploring the association between AD-PRS and cognitive functions has aroused many important findings. For example, AD-PRS was reported to be associated with lower total, verbal, and performance intelligence quotients in childhood and adolescence (Korologou-Linden et al., 2019a), whereas no significant associations were identified in the cognitively normal adult individuals (Li et al., 2018). Moreover, increasing studies showed that AD-PRS had a significant negative correlation with immediate memory and verbal episodic memory, which increases the predictive efficiency of conversion from healthy controls to LOAD (Marden et al., 2016; Porter et al., 2018b). It is worth noting that, in a study of Chinese samples, a significant correlation between AD-PRS and episodic memory ability was not found (Li et al., 2018). The inconsistency may be caused by ethnic differences or the evaluation efficiency of different memory scales.
AD-PRS was found to be closely associated with economic behaviors. Individuals with different levels of AD-PRS showed different saving behaviors and wealth composition (Shin et al., 2019), for instance, individuals with higher AD-PRS are more likely to hold less wealth in the Individual Retirement Accounts and to have simpler managed assets, such as fixed deposits, whereas individuals with lower AD-PRS have more complex managed assets, such as stocks (Shin et al., 2019). In addition, it was suggested that the interaction between higher AD-PRS and lower wealth levels would lead to the early-onset age of LOAD and accelerate its development (Ajnakina et al., 2020).
Mental health is also a vital risk factor affecting the onset and progression of LOAD, and up to 50% of LOAD patients have psychosis symptoms, such as hallucinations and delusions (Creese et al., 2019). Studies have shown that AD-PRS is positively correlated with neuroticism (Duberstein et al., 2011; Terracciano and Sutin, 2019) and hallucinations (Kusters et al., 2020). The association between AD-PRS and cognition was also mediated by these two personality traits (Stephan et al., 2018). Further, a combination between AD-PRS and major depression disorder-PRS has been used to study LOAD and their integration would significantly increase the ability to predict conversion from aMCI to LOAD (Xu et al., 2018). The above results indicated that LOAD shared a highly genetic association with mental health disorders.
Opportunities and Challenges for AD-PRS Applications
AD-PRS has been widely used in many different research fields and has exhibited a huge ability in the prediction of LOAD. However, there was large heterogeneity in AD-PRS considering the huge variations in the calculation pipeline (Choi et al., 2020).
First, the selection of a certain PT threshold from the GWAS summary statistics of the discovery sample was quite important for building PRS in the target sample, because it determined how many SNPs were included for calculation. In the classic AD-PRS calculation method, only those SNPs less than a predefined PT threshold were included (Axelrud et al., 2018). Recently, the optimal PT threshold method was applied widely, in which a series of AD-PRS were typically calculated over a range of thresholds, and the associations between the target trait and each AD-PRS were calculated to find out the best prediction model with the underlying PT threshold accordingly was set as the optimized PT threshold in the calculation of PRS (Choi et al., 2020). Second, after identifying the PT threshold, the calculation strategies of PRS in the target sample also varied. The simple AD-PRS only calculates the number of risk alleles assuming that all SNPs have the same effect on the disease. More commonly, an odds-ratio-weighted PRS was calculated for each individual as the sum of the count of risk alleles multiplied by the corresponding effect sizes across these SNPs. Third, the quality of the base sample and target sample including ethnicity, sample size, and the number of genetic variants used has a great impact on the AD-PRS and will exert the findings. To date, no consensus has been reached about these points and various strategies have been adopted by researchers, which of course will hamper the utility of the AD-PRS for a clinical diagnosis.
Besides the above points, another important question is whether the APOE-ε4 should be included for calculating AD-PRS, which is the largest risk factor for LOAD (Kim et al., 2009). At present, the accuracy of predicting the risk of LOAD by using the PRS method is 84% (Escott-Price et al., 2015, 2017). However, by far, the APOE-ε4 allele (risk) and the APOE-ε2 allele (protective) contributed the largest to this risk, where the predictive accuracy could reach 0.68 (APOE-ε4) and 0.69 (APOE-ε4+APOE-ε2) in the clinical samples (Escott-Price et al., 2015). An important practical and theoretical consideration is to understand how good AD-PRS is when excluding the APOE-ε4 gene risk and no consensus has been reached so far. Thus, associations of the AD-PRS with multiple biomarkers adjusting for APOE locus or not need to be tested.
It should be noted that, although some limitations about AD-PRS still need to be addressed, the advanced development of large-GWAS studies and data-sharing policies are driving the AD-PRS to be constantly optimized and updated for drawing unambiguous conclusions about LOAD. For example, many researchers have identified that AD-PRS was associated with lower hippocampal volume in different target samples using different PTwhen using the publicly available International Genomics of Alzheimer's Project (IGAP) as the base sample (Mormino et al., 2016; Axelrud et al., 2018; Heidi et al., 2021; Tank et al., 2022). The underlying reason may be that the base sample from IGAP or UK Biobank is very large which can reduce the deviation caused by a small sample, and also offer the same risk alleles for the AD-PRS calculation which makes the most important risk alleles always included.
In the future, more studies considering the causal inference between AD-PRS, biomarkers, and LOAD occurrence are needed to infer the underlying mechanism of LOAD. Moreover, the application of AD-PRS would also be critical for drug discovery, as drugs targeting proteins encoded in genetic risk loci would be more likely to be successful in phases II and III clinical trials (King et al., 2019). Thus, AD-PRS have a greater utility in biomedical research and personalized precision medicine in the future.
Author Contributions
QL, XL, and JX contributed to conception and design of the study. QL and XL wrote the first draft of the manuscript. JX wrote sections of the manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (grant no. 81801687), Science & Technology Development Fund of Tianjin Education Commission for Higher Education (grant no. 2019KJ195), and Open Research Project of The Beijing Key Laboratory of High Dynamic Navigation Technology under grant no. HDN2020102.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ajnakina, O., Cadar, D., and Steptoe, A. (2020). Interplay between socioeconomic markers and polygenic predisposition on timing of dementia diagnosis. J. Am. Geriatr. Soc. 68, 1529–1536. doi: 10.1111/jgs.16406
Akiyama, H (2000). Inflammation and Alzheimer's disease. Neurobiol. Aging 21, 383–421. doi: 10.1016/S0197-4580(00)00124-X
Altmann, A., Scelsi, M. A., Shoai, M., de Silva, E., Aksman, L. M., Cash, D. M., et al. (2020). A comprehensive analysis of methods for assessing polygenic burden on Alzheimer's disease pathology and risk beyond APOE. Brain Commun. 2, fcz047. doi: 10.1093/BRAINCOMMS/FCZ047
Axelrud, L. K., Santoro, M. L., Pine, D. S., Talarico, F., Gadelha, A., Manfro, G. G., et al. (2018). Polygenic risk score for Alzheimer's disease: implications for memory performance and hippocampal volumes in early life. Am. J. Psychiatry 175, 555–563. doi: 10.1176/appi.ajp.2017.17050529
Axelrud, L. K., Sato, J. R., Santoro, M. L., Talarico, F., Pine, D. S., Rohde, L. A., et al. (2019). Genetic risk for Alzheimer's disease and functional brain connectivity in children and adolescents. Neurobiol. Aging 82, 10–17. doi: 10.1016/j.neurobiolaging.2019.06.011
Bäckman, L., Jones, S., Berger, A. K., Laukka, E. J., and Small, B. J. (2004). Multiple cognitive deficits during the transition to Alzheimer's disease. J. Intern. Med. 256, 195–204. doi: 10.1111/j.1365-2796.2004.01386.x
Bateman, R. J., Xiong, C., Benzinger, T. L. S., Fagan, A. M., Goate, A., Fox, N. C., et al. (2012). Clinical and biomarker changes in dominantly inherited Alzheimer's disease. N. Engl. J. Med. 367, 795–804. doi: 10.1056/NEJMoa1202753
Baumgart, M., Snyder, H. M., Carrillo, M. C., Fazio, S., Kim, H., and Johns, H. (2015). Summary of the evidence on modifiable risk factors for cognitive decline and dementia: a population-based perspective. Alzheimers. Dement. 11, 718–726. doi: 10.1016/j.jalz.2015.05.016
Bennett, D. A., Wilson, R. S., Schneider, J. A., Evans, D. A., Beckett, L. A., Aggarwal, N. T., et al. (2002). Natural history of mild cognitive impairment in older persons. Neurology 59, 198–205. doi: 10.1212/WNL.59.2.198
Breijyeh, Z., and Karaman, R. (2020). Comprehensive review on Alzheimer's disease: causes and treatment. Molecules 25, 5789. doi: 10.3390/molecules25245789
Cacace, R., Sleegers, K., and Van Broeckhoven, C. (2016). Molecular genetics of early-onset Alzheimer's disease revisited. Alzheimers Dement. 12, 733–748. doi: 10.1016/J.JALZ.2016.01.012
Chandler, H. L., Hodgetts, C. J., Caseras, X., Murphy, K., and Lancaster, T. M. (2020). Polygenic risk for Alzheimer's disease shapes hippocampal scene-selectivity. Neuropsychopharmacology 45, 1171–1178. doi: 10.1038/s41386-019-0595-1
Chandler, H. L., Wise, R. G., Linden, D. E., Williams, J., Murphy, K., Lancaster, T. M., et al. (2021). Alzheimer's genetic risk effects on cerebral blood flow are spatially consistent and proximal to gene expression across the lifespan. bioRxiv. doi: 10.1101/2020.12.31.424949
Chandler, H. L., Wise, R. G., Murphy, K., Tansey, K. E., Linden, D. E. J., and Lancaster, T. M. (2019). Polygenic impact of common genetic risk loci for Alzheimer's disease on cerebral blood flow in young individuals. Sci. Rep. 9, 467. doi: 10.1038/s41598-018-36820-3
Chaudhury, S., Brookes, K. J., Patel, T., Fallows, A., Guetta-Baranes, T., Turton, J. C., et al. (2019). Alzheimer's disease polygenic risk score as a predictor of conversion from mild-cognitive impairment. Transl. Psychiatry 9, 154. doi: 10.1038/s41398-019-0485-7
Chen, H.-H., Petty, L. E., Sha, J., Zhao, Y., Kuzma, A., Valladares, O., et al. (2021). Genetically regulated expression in late-onset Alzheimer's disease implicates risk genes within known and novel loci. Transl. Psychiatry 11, 618. doi: 10.1038/s41398-021-01677-0
Choi, S. W., Mak, T. S.-H., and O'Reilly, P. F. (2020). Tutorial: a guide to performing polygenic risk score analyses. Nat. Protoc. 15, 2759–2772. doi: 10.1038/s41596-020-0353-1
Creese, B., Vassos, E., Bergh, S., Athanasiu, L., Johar, I., Rongve, A., et al. (2019). Examining the association between genetic liability for schizophrenia and psychotic symptoms in Alzheimer's disease. Transl. Psychiatry 9, 273. doi: 10.1038/s41398-019-0592-5
Dementia, A. (2021) Alzheimer's disease facts figures. Alzheimers Dement. 17 327–406. doi: 10.1002/alz.12328
Duberstein, P. R., Chapman, B. P., Tindle, H. A., Sink, K. M., Bamonti, P., Robbins, J., et al. (2011). Personality and risk for Alzheimer's disease in adults 72 years of age and older: a 6-year follow-up. Psychol. Aging 26, 351–362. doi: 10.1037/a0021377
Ebenau, J. L., Lee, S. J., Hulsman, M., Tesi, N., Jansen, I. E., Verberk, I. M. W., et al. (2021). Risk of dementia in APOE ε4 carriers is mitigated by a polygenic risk score. Alzheimers Dement. 13, 1–9. doi: 10.1002/dad2.12229
Ebenau, J. L., Timmers, T., Wesselman, L. M. P., Verberk, I. M. W., Verfaillie, S. C. J., Slot, R. E. R., et al. (2020). ATN classification and clinical progression in subjective cognitive decline. Neurology 95, e46–e58. doi: 10.1212/WNL.0000000000009724
Elliott, J., Bodinier, B., Bond, T. A., Chadeau-Hyam, M., Evangelou, E., Moons, K. G. M., et al. (2020). Predictive accuracy of a polygenic risk score–enhanced prediction model vs a clinical risk score for coronary artery disease. JAMA 323, 636. doi: 10.1001/jama.2019.22241
Elman, J. A., Panizzon, M. S., Logue, M. W., Gillespie, N. A., Neale, M. C., Reynolds, C. A., et al. (2019). Genetic risk for coronary heart disease alters the influence of Alzheimer's genetic risk on mild cognitive impairment. Neurobiol. Aging 84, 237.e5–237.e12. doi: 10.1016/j.neurobiolaging.2019.06.001
Escott-Price, V., Myers, A. J., Huentelman, M., and Hardy, J. (2017). Polygenic risk score analysis of pathologically confirmed Alzheimer disease. Ann. Neurol. 82, 311–314. doi: 10.1002/ana.24999
Escott-Price, V., Sims, R., Bannister, C., Harold, D., Vronskaya, M., Majounie, E., et al. (2015). Common polygenic variation enhances risk prediction for Alzheimer's disease. Brain 138, 3673–3684. doi: 10.1093/brain/awv268
Foley, S. F., Tansey, K. E., Caseras, X., Lancaster, T., Bracht, T., Parker, G., et al. (2017). Multimodal brain imaging reveals structural differences in Alzheimer's disease polygenic risk carriers: a study in healthy young adults. Biol. Psychiatry 81, 154–161. doi: 10.1016/j.biopsych.2016.02.033
Gatz, M., Reynolds, C. A., Fratiglioni, L., Johansson, B., Mortimer, J. A., Berg, S., et al. (2006). Role of genes and environments for explaining Alzheimer disease. Arch. Gen. Psychiatry 63, 168. doi: 10.1001/archpsyc.63.2.168
Harrison, J. R., Bhatia, S., Tan, Z. X., Mirza-Davies, A., Benkert, H., Tax, C. M. W., et al. (2020a). Imaging Alzheimer's genetic risk using diffusion MRI: a systematic review. Neuroimage Clin. 27, 102359. doi: 10.1016/j.nicl.2020.102359
Harrison, J. R., Mistry, S., Muskett, N., and Escott-Price, V. (2020b). From polygenic scores to precision medicine in Alzheimer's disease: a systematic review. J. Alzheimer's Dis. 74, 1271–1283. doi: 10.3233/JAD-191233
Harrison, T. M., Mahmood, Z., Lau, E. P., Karacozoff, A. M., Burggren, A. C., Small, G. W., et al. (2016). An Alzheimer's disease genetic risk score predicts longitudinal thinning of hippocampal complex subregions in healthy older adults. eNeuro 3, ENEURO.0098-16. doi: 10.1523/ENEURO.0098-16.2016
Heidi, T. A., Jiang, J., Koch, F., Mather, K. A., Wen, W., et al. (2021). Associations between Alzheimer's disease polygenic risk scores and hippocampal subfield volumes in 17,161 UK Biobank participants. Neurobiol. Aging 98, 108–115. doi: 10.1016/j.neurobiolaging.2020.11.002
Kametani, F., and Hasegawa, M. (2018). Reconsideration of amyloid hypothesis and tau hypothesis in Alzheimer's disease. Front. Neurosci. 12, 25. doi: 10.3389/FNINS.2018.00025
Kilimann, I., Likitjaroen, Y., Hampel, H., and Teipel, S. (2013). Diffusion tensor imaging to determine effects of antidementive treatment on cerebral structural connectivity in Alzheimer's disease. Curr. Pharm. Des. 19, 6416–6425. doi: 10.2174/1381612811319360003
Kim, J., Basak, J. M., and Holtzman, D. M. (2009). The role of apolipoprotein E in Alzheimer's disease. Neuron 63, 287–303. doi: 10.1016/j.neuron.2009.06.026
King, E. A., Davis, J. W., and Degner, J. F. (2019). Are drug targets with genetic support twice as likely to be approved? Revised estimates of the impact of genetic support for drug mechanisms on the probability of drug approval. PLoS Genet. 15, e1008489. doi: 10.1371/journal.pgen.1008489
Kinney, J. W., Bemiller, S. M., Murtishaw, A. S., Leisgang, A. M., Salazar, A. M., and Lamb, B. T. (2018). Inflammation as a central mechanism in Alzheimer's disease. Alzheimer's Dement. Transl. Res. Clin. Interv. 4, 575–590. doi: 10.1016/j.trci.2018.06.014
Korologou-Linden, R., Anderson, E. L., Jones, H. J., Davey Smith, G., Howe, L. D., and Stergiakouli, E. (2019a). Polygenic risk scores for Alzheimer's disease, and academic achievement, cognitive and behavioural measures in children from the general population. Int. J. Epidemiol. 48, 1972–1980. doi: 10.1093/ije/dyz080
Korologou-Linden, R., O'Keeffe, L., Howe, L. D., Davey-Smith, G., Jones, H. J., Anderson, E. L., et al. (2019b). Polygenic risk score for Alzheimer's disease and trajectories of cardiometabolic risk factors in children. Wellcome Open Res. 4, 125. doi: 10.12688/wellcomeopenres.15359.1
Kunkle, B. W., Grenier-Boley, B., Sims, R., Bis, J. C., Damotte, V., Naj, A. C., et al. (2019). Genetic meta-analysis of diagnosed Alzheimer's disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. Nat. Genet. 51, 414–430. doi: 10.1038/s41588-019-0358-2
Kusters, C. D. J., Paul, K. C., Folle, A. D., Keener, A. M., Bronstein, J. M., Dobricic, V., et al. (2020). Genetic risk scores and hallucinations in patients with Parkinson disease. Neurol. Genet. 6, e492. doi: 10.1212/NXG.0000000000000492
Lambert, J.-C., Ibrahim-Verbaas, C. A., Harold, D., Naj, A. C., Sims, R., Bellenguez, C., et al. (2013). Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nat. Genet. 45, 1452–1458. doi: 10.1038/ng.2802
Lee, J. C., Kim, S. J., Hong, S., and Kim, Y. S. (2019). Diagnosis of Alzheimer's disease utilizing amyloid and tau as fluid biomarkers. Exp. Mol. Med. 51, 1–10. doi: 10.1038/s12276-019-0250-2
Lee, Y., Jeon, S., Kang, S. W., Park, M., Baik, K., Yoo, H. S., et al. (2021). Interaction of CSF α-synuclein and amyloid beta in cognition and cortical atrophy. Alzheimer's Dement. 13, 1–12. doi: 10.1002/dad2.12177
Lello, L., Raben, T. G., Yong, S. Y., Tellier, L. C. A. M., and Hsu, S. D. H. (2019). Genomic prediction of 16 complex disease risks including heart attack, diabetes, breast and prostate cancer. Sci. Rep. 9, 15286. doi: 10.1038/s41598-019-51258-x
Levey, D. F., Stein, M. B., Wendt, F. R., Pathak, G. A., Zhou, H., Aslan, M., et al. (2021). Bi-ancestral depression GWAS in the Million Veteran Program and meta-analysis in andgt;1.2 million individuals highlight new therapeutic directions. Nat. Neurosci. 24, 954–963. doi: 10.1038/s41593-021-00860-2
Levin, M. G., and Rader, D. J. (2020). Polygenic risk scores and coronary artery disease. Circulation 141, 637–640. doi: 10.1161/CIRCULATIONAHA.119.044770
Li, J., Zhang, X., Li, A., Liu, S., Qin, W., Yu, C., et al. (2018). Polygenic risk for Alzheimer's disease influences precuneal volume in two independent general populations. Neurobiol. Aging 64, 116–122. doi: 10.1016/j.neurobiolaging.2017.12.022
Li, J.-Q. Q., Tan, L. L., Wang, H.-F. F., Tan, M.-S. S., Tan, L. L., Xu, W., et al. (2016). Risk factors for predicting progression from mild cognitive impairment to Alzheimer's disease: a systematic review and meta-analysis of cohort studies. J. Neurol. Neurosurg. Psychiatry 87, 476–484. doi: 10.1136/jnnp-2014-310095
Li, W.-W., Wang, Z., Fan, D.-Y., Shen, Y.-Y., Chen, D.-W., Li, H.-Y., et al. (2020). Association of polygenic risk score with age at onset and cerebrospinal fluid biomarkers of Alzheimer's disease in a Chinese cohort. Neurosci. Bull. 36, 696–704. doi: 10.1007/s12264-020-00469-8
Lin, Y., Smith, A. V., Aspelund, T., Betensky, R. A., Smoller, J. W., Gudnason, V., et al. (2019). Genetic overlap between vascular pathologies and Alzheimer's dementia and potential causal mechanisms. Alzheimers Dement. 15, 65–75. doi: 10.1016/j.jalz.2018.08.002
Marden, J. R., Mayeda, E. R., Walter, S., Vivot, A., Tchetgen Tchetgen, E. J., Kawachi, I., et al. (2016). Using an Alzheimer disease polygenic risk score to predict memory decline in black and white americans over 14 years of follow-up. Alzheimer Dis. Assoc. Disord. 30, 195–202. doi: 10.1097/WAD.0000000000000137
Mirza-Davies, A., Foley, S., Caseras, X., Baker, E., Holmans, P., Escott-Price, V., et al. (2021). The impact of genetic risk for Alzheimer's disease on the structural brain networks of young adults. bioRxiv. doi: 10.1101/2021.09.22.461338
Moore, A. M., Filshtein, T. J., Dumitrescu, L., Harrati, A., Elahi, F., Mormino, E. C., et al. (2019). A/T/N polygenic risk score for cognitive decline in old age Annah. bioRxiv. doi: 10.1101/838847
Morgan, A. R., Touchard, S., O'Hagan, C., Sims, R., Majounie, E., Escott-Price, V., et al. (2017). The correlation between inflammatory biomarkers and polygenic risk score in Alzheimer's disease. J. Alzheimers Dis. 56, 25–36. doi: 10.3233/JAD-160889
Mormino, E. C., Sperling, R. A., Holmes, A. J., Buckner, R. L., De Jager, P. L., Smoller, J. W., et al. (2016). Polygenic risk of Alzheimer disease is associated with early- and late-life processes. Neurology 87, 481–488. doi: 10.1212/WNL.0000000000002922
Naveed, M., Mubeen, S., Khan, A., Ibrahim, S., and Meer, B. (2019). Plasma biomarkers: potent screeners of Alzheimer's disease. Am. J. Alzheimers Dis. 34, 290–301. doi: 10.1177/1533317519848239
Peyrot, W. J., and Price, A. L. (2021). Identifying loci with different allele frequencies among cases of eight psychiatric disorders using CC-GWAS. Nat. Genet. 53, 445–454. doi: 10.1038/s41588-021-00787-1
Porter, T., Burnham, S. C., Milicic, L., Savage, G., Maruff, P., Lim, Y. Y., et al. (2018a). Utility of an Alzheimer's disease risk-weighted polygenic risk score for predicting rates of cognitive decline in preclinical Alzheimer's disease: a prospective longitudinal study. J. Alzheimer's Dis. 66, 1193–1211. doi: 10.3233/JAD-180713
Porter, T., Burnham, S. C., Savage, G., Lim, Y. Y., Maruff, P., Milicic, L., et al. (2018b). A polygenic risk score derived from episodic memory weighted genetic variants is associated with cognitive decline in preclinical Alzheimer's disease. Front. Aging Neurosci. 10, 423. doi: 10.3389/fnagi.2018.00423
Richardson, T. G., Harrison, S., Hemani, G., and Smith, G. D. (2019). An atlas of polygenic risk score associations to highlight putative causal relationships across the human phenome. eLife 8, 1–24. doi: 10.7554/eLife.43657
Ripke, S., Neale, B. M., Corvin, A., Walters, J. T. R., Farh, K. H., Holmans, P. A., et al. (2014). Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–427. doi: 10.1038/nature13595
Rostami, E., Engquist, H., and Enblad, P. (2014). Imaging of cerebral blood flow in patients with severe traumatic brain injury in the neurointensive care. Front. Neurol. 5, 114. doi: 10.3389/fneur.2014.00114
Sabuncu, M. R., Buckner, R. L., Smoller, J. W., Lee, P. H., Fischl, B., Sperling, R. A., et al. (2012). The association between a polygenic Alzheimer score and cortical thickness in clinically normal subjects. Cereb. Cortex 22, 2653–2661. doi: 10.1093/cercor/bhr348
Selkoe, D. J (2021). Treatments for Alzheimer's disease emerge. Science 373, 624–626. doi: 10.1126/science.abi6401
Servick, K (2021). Alzheimer's drug approval spotlights blood tests. Science 373, 373–374. doi: 10.1126/science.373.6553.373
Shen, X. N., Huang, Y. Y., Chen, S. D., Guo, Y., Tan, L., Dong, Q., et al. (2021). Plasma phosphorylated-tau181 as a predictive biomarker for Alzheimer's amyloid, tau and FDG PET status. Transl. Psychiatry 11, 1–10. doi: 10.1038/s41398-021-01709-9
Shin, S. H., Lillard, D. R., and Bhattacharya, J. (2019). Understanding the correlation between Alzheimer's disease polygenic risk, wealth, and the composition of wealth holdings. Biodemography Soc. Biol. 65, 323–350. doi: 10.1080/19485565.2020.1769466
Skoog, I., Kern, S., Najar, J., Guerreiro, R., Bras, J., Waern, M., et al. (2021). A non-APOEPolygenic risk score foralzheimer's disease is associated with cerebrospinal fluid neurofilament light in a representative sample of cognitively unimpaired 70-year olds. J. Gerontol. Ser. A Biol. Sci. Med. Sci. 76, 983–990. doi: 10.1093/gerona/glab030
Smitha, K., Akhil Raja, K., Arun, K., Rajesh, P., Thomas, B., Kapilamoorthy, T., et al. (2017). Resting state fMRI: a review on methods in resting state connectivity analysis and resting state networks. Neuroradiol. J. 30, 305–317. doi: 10.1177/1971400917697342
Soldan, A., Pettigrew, C., Fagan, A. M., Schindler, S. E., Moghekar, A., Fowler, C., et al. (2019). ATN profiles among cognitively normal individuals and longitudinal cognitive outcomes. Neurology 92, e1567–e1579. doi: 10.1212/WNL.0000000000007248
Stephan, Y., Sutin, A. R., Luchetti, M., Caille, P., and Terracciano, A. (2018). Polygenic score for Alzheimer disease and cognition: the mediating role of personality. J. Psychiatr. Res. 107, 110. doi: 10.1016/J.JPSYCHIRES.2018.10.015
Su, F., Shu, H., Ye, Q., Xie, C., Yuan, B., Zhang, Z., et al. (2017). Integration of multilocus genetic risk into the default mode network longitudinal trajectory during the Alzheimer's disease process. J. Alzheimers. Dis. 56, 491–507. doi: 10.3233/JAD-160787
Tan, C. H., Fan, C. C., Mormino, E. C., Sugrue, L. P., Broce, I. J., Hess, C. P., et al. (2018). Polygenic hazard score: an enrichment marker for Alzheimer's associated amyloid and tau deposition. Acta Neuropathol. 135, 85–93. doi: 10.1007/s00401-017-1789-4
Tank, R., Ward, J., Flegal, K. E., Smith, D. J., Bailey, M. E. S., Cavanagh, J., et al. (2022). Association between polygenic risk for Alzheimer's disease, brain structure and cognitive abilities in UK Biobank. Neuropsychopharmacology 47, 564–569. doi: 10.1038/s41386-021-01190-4
Terracciano, A., and Sutin, A. R. (2019). Personality and Alzheimer's disease: an integrative review. Personal. Disord. Theory, Res. Treat. 10, 4–12. doi: 10.1037/per0000268
van der Merwe, C., Passchier, R., Mufford, M., Ramesar, R., Dalvie, S., and Stein, D. J. (2019). Polygenic risk for schizophrenia and associated brain structural changes: a systematic review. Compr. Psychiatry 88, 77–82. doi: 10.1016/j.comppsych.2018.11.014
Wehby, G. L., Domingue, B. W., and Wolinsky, F. D. (2018). Genetic risks for chronic conditions: implications for long-term wellbeing. J. Gerontol. Ser. A Biol. Sci. Med. Sci. 73, 477–483. doi: 10.1093/gerona/glx154
Weiner, M. W., Veitch, D. P., Aisen, P. S., Beckett, L. A., Cairns, N. J., Cedarbaum, J., et al. (2015). 2014 Update of the Alzheimer's Disease Neuroimaging Initiative: a review of papers published since its inception. Alzheimer's Dement. 11, e1–e120. doi: 10.1016/j.jalz.2014.11.001
Williams, M. E., Elman, J. A., McEvoy, L. K., Andreassen, O. A., Dale, A. M., Eglit, G. M. L., et al. (2021). 12-year prediction of mild cognitive impairment aided by Alzheimer's brain signatures at mean age 56. Brain Commun. 3, fcab167. doi: 10.1093/braincomms/fcab167
Wingo, T. S., Lah, J. J., Levey, A. I., and Cutler, D. J. (2012). Autosomal recessive causes likely in early-onset Alzheimer disease. Arch. Neurol. 69, 59–64. doi: 10.1001/archneurol.2011.221
Wray, N. R., Lin, T., Austin, J., McGrath, J. J., Hickie, I. B., Murray, G. K., et al. (2021). From basic science to clinical application of polygenic risk scores: a primer. JAMA Psychiatry 78, 101–109. doi: 10.1001/jamapsychiatry.2020.3049
Xiao, E., Chen, Q., Goldman, A. L., Tan, H. Y., Healy, K., Zoltick, B., et al. (2017). Late-Onset Alzheimer's disease polygenic risk profile score predicts hippocampal function. Biol. Psychiatry Cogn. Neurosci. Neuroimaging 2, 673–679. doi: 10.1016/j.bpsc.2017.08.004
Xu, J., Li, Q., Qin, W., Li, M. J., Zhuo, C., Liu, H., et al. (2018). Neurobiological substrates underlying the effect of genomic risk for depression on the conversion of amnestic mild cognitive impairment. Brain 141, 3457–3471. doi: 10.1093/brain/awy277
Yang, J., Pan, P., Song, W., Huang, R., Li, J., Chen, K., et al. (2012). Voxelwise meta-analysis of gray matter anomalies in Alzheimer's disease and mild cognitive impairment using anatomic likelihood estimation. J. Neurol. Sci. 316, 21–29. doi: 10.1016/j.jns.2012.02.010
Zettergren, A., Lord, J., Ashton, N. J., Benedet, A. L., Karikari, T. K., Lantero Rodriguez, J., et al. (2021). Association between polygenic risk score of Alzheimer's disease and plasma phosphorylated tau in individuals from the Alzheimer's Disease Neuroimaging Initiative. Alzheimers. Res. Ther. 13, 17. doi: 10.1186/s13195-020-00754-8
Zhan, Y., Ma, J., Xu, K., Ding, Y., Cui, Y., Yang, Z., et al. (2016). “Impaired episodic memory network in subjects at high risk for Alzheimer's disease,” in 38th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC) (Orlando, FL: IEEE), 4017–4020. doi: 10.1109/EMBC.2016.7591608
Zhou, X., Chen, Y., Ip, F. C. F., Lai, N. C. H., Li, Y. Y. T., Jiang, Y., et al. (2020). Genetic and polygenic risk score analysis for Alzheimer's disease in the Chinese population. Alzheimers Dement. 12, 1–15. doi: 10.1002/dad2.12074
Keywords: late onset Alzheimer's disease, polygenic risk score, biomarker, prediction, brain
Citation: Li Q, Lv X, Jin F, Liao K, Gao L and Xu J (2022) Associations of Polygenic Risk Score for Late-Onset Alzheimer's Disease With Biomarkers. Front. Aging Neurosci. 14:849443. doi: 10.3389/fnagi.2022.849443
Received: 06 January 2022; Accepted: 14 March 2022;
Published: 14 April 2022.
Edited by:
Panteleimon Giannakopoulos, Université de Genève, SwitzerlandReviewed by:
Keeley Brookes, Nottingham Trent University, United KingdomAnbupalam Thalamuthu, University of New South Wales, Australia
Copyright © 2022 Li, Lv, Jin, Liao, Gao and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qiaojun Li, bGlxaWFvanVuQHRqY3UuZWR1LmNu
 Qiaojun Li
Qiaojun Li Xingping Lv
Xingping Lv Fei Jin3
Fei Jin3