- 1Key Laboratory of Optoelectronic Science and Technology for Medicine of Ministry of Education, Fujian Provincial Key Laboratory of Photonics Technology, Fujian Normal University, Fuzhou, China
- 2School of Electronic & Electrical Engineering and Physics, Fujian University of Technology, Fuzhou, China
- 3Fujian Provincial Engineering Technology Research Center of Photoelectric Sensing Application, Fujian Normal University, Fuzhou, China
Medical image segmentation is of important support for clinical medical applications. As most of the current medical image segmentation models are limited in the U-shaped structure, to some extent the deep convolutional neural network (CNN) structure design is hard to be accomplished. The design in this study mimics the way the wave is elastomeric propagating, extending the structure from both the horizontal and spatial dimensions for realizing the Elastomeric UNet (EUNet) structure. The EUNet can be divided into two types: horizontal EUNet and spatial EUNet, based on the propagation direction. The advantages of this design are threefold. First, the training structure can be deepened effectively. Second, the independence brought by each branch (a U-shaped design) makes the flexible design redundancy available. Finally, a horizontal and vertical series-parallel structure helps on feature accumulation and recursion. Researchers can adjust the design according to the requirements to achieve better segmentation performance for the independent structural design. The proposed networks were evaluated on two datasets: a self-built dataset (multi-photon microscopy, MPM) and publicly benchmark retinal datasets (DRIVE). The results of experiments demonstrated that the performance of EUNet outperformed the UNet and its variants.
Introduction
With the popularization of medical imaging technology, such as magnetic resonance imaging (MRI), computed tomography (CT), positron emission tomography (PET), ultrasonic imaging, digital pathology, and microscopy, medical imaging technology plays an important role in clinical medicine to emerge the structure of human organs, tissues, and cells (Taruttis and Ntziachristos, 2015; Papadopoulos et al., 2017; Pang et al., 2020). The morphology of in vivo cells is an intuitive reflection of cell structure and function, and the analysis of it is a significant part of histopathology and clinical medicine to carry out clinical medical diagnosis and genetic research on aging, cell, and tissue expression of Alzheimer’s and Parkinson’s diseases (Li et al., 2010; Haft-Javaherian et al., 2019; Ye et al., 2020). Research on aging morphology and molecular mechanism of skin cells (especially in vivo skin cells) is an effective means for clinical medical research, such as skin aging and refractory skin wounds, and can provide the basis for aging cell research of a variety of aging-related diseases (e.g., cardiovascular disease, osteoporosis, and Parkinson’s disease) (Loeser, 2009; Aspray and Hill, 2019; Sun, 2020). Cortical angiography was used to explore the influences of Alzheimer’s disease and cellular senescence on capillaries (Haft-Javaherian et al., 2019). The incidence of melanoma in patients with Parkinson’s disease was studied by multiphoton imaging tracking, the dynamic process of melanocytic lesions (Balu et al., 2017; Lentsch et al., 2019; Ye et al., 2020). Image segmentation is an active research topic within medical image technology, which accounts for 70% of the international image processing competitions. As a basilic step in the medical image process, medical image segmentation separates the medical image into different regions with analogous characteristics to segment the regions of interest (ROI) and extract interrelated features. The role of medical image segmentation is to improve the intuitiveness of medical images and reduce man-made mistakes. Accurate medical image segmentation is difficult to achieve, owing to the data scarcity and image complexity, such as noisy background and intensity in-homogeneity.
The broad classification of medical image segmentation technology is traditional segmentation, such as thresholds, morphological methods (Chen et al., 2015) and clustering techniques, and deep-learning segmentation rest on CNN. Although various traditional algorithms have been designed to segment the medical images, the accuracy of segmentation is still difficult to improve. The fully convolutional network (FCN) designed by Long et al., 2015 is an end-to-end pixel-wise segmentation with a 20% amelioration of mean IOU than 2012 (Long et al., 2015). On account of significant improvement in the field of medical image segmentation by deep CNN, diverse variants of CNN are employed to segment medical images. As the development of FCN, UNet was applied to biomedical image segmentation by symmetric U-shaped architecture and skip connections yielding more accurate properties (Ronneberge et al., 2015). The improvement of UNet performance is apt to stagnate by the increase of depth. As a consequence, the diverse variants of UNet had been explored in the past 5 years for medical image segmentation (Li H. et al., 2018; Zhang et al., 2018; Liu et al., 2020). Inspired by the ResNet framework, a residual neural network with U-shape named U-ResNet was explored by Ibtehaz and Rahman (2020) and Drozdzal et al. (2016). In the same way, H-DenseUnet had been proposed to segment liver tumor mixed DenseNet to UNet by Li X. et al. (2018) and Cai et al. (2020). Another recent relevant work was Mask-RCNN, a novel UNet structure introduced by Vuola et al. (2019) through combining UNet and Mask-RCNN for nucleus segmentation. A U-shaped structure with dilated convolution path named Modified UNet was implemented by Zhao et al. (2019). UNet + + was employed to alleviate the a priori unknown of depth by Zhou et al. (2020), and the restrictive fusion architecture of UNet skip connections was also optimized in it. A UNet-like network entitled DSC-Net was performed to segment thymoma from CT images by Li et al. (2021). Another variant of UNet called X-Net was designed to segment liver and tumor by Chi et al. (2021). So far, however, the improved models still adopt a U-shaped structure, namely, the symmetrical single branch down-up sampling U-shaped structure. The performance of UNet is difficult to ameliorate, and the technical challenges of expanding the network size of UNet and extracting detailed information have prevented this architecture from being more accurate in medical imaging segmentation. Breaking the U-shaped architecture is an inevitable choice to overcome the technical bottleneck.
The purpose of this study was to design a flexible elastic expansion convolution operation to enhance the boundary detail extraction ability of the model and extend it to the segmentation of public medical image datasets by adjusting parameters through experiments. An elastomeric UNet (EUNet) has been proposed to address the limitations of U-shaped architecture, and the experimental validation demonstrates its effectiveness of it.
Methodology
UNet Model and Its Variants
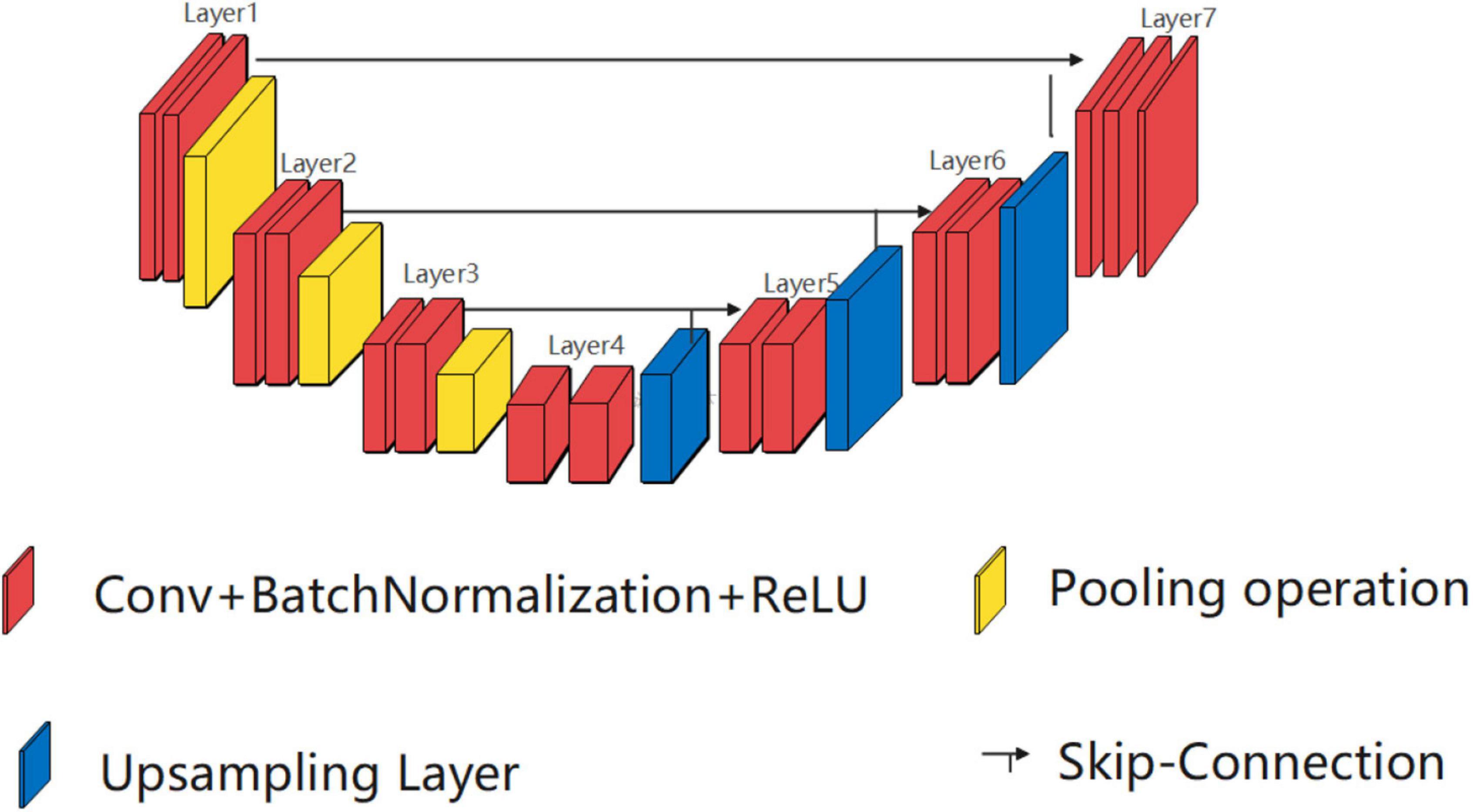
UNet was first proposed stemming from FCN. As shown in Figure 1, the U-shaped architecture of UNet is illustrated using three elements, namely, a contracting path, an expansive path, and skip connections. The key idea is to connect symmetric contracting path and expansive path by skip connections, where the convolutional operations are employed to extract features and upsampled operations are utilized to locate the ROI.
UNet + + is designed to overcome the two defects of UNet, a priori unknown of depth and the restrictive fusion architecture of skip connections. U-shapes with varying depths are integrated to relieve the a priori unknown of depth, and a pruning scheme is devised to accelerate convergence in UNet + +. Aggregation feature scheme instead of skip connections is executed in the up-sampling path to extract multi-scale features (Li X. et al., 2018).
Modified UNet extract varying scale features by introducing dilated convolution into U-shaped architecture (Cai et al., 2020). Each feature extraction operation consists of two steps. The first step is feature extraction operation through two convolution paths: one path is composed of ordinary convolution operation, and the other one uses dilated convolution for feature extraction. The second step is to complete feature fusion through a “concatenate” operation. A bigger receptive field is acquired in this way. Dilated convolution and conventional convolution coverage of receptive fields are the same when the dilate rate equals 1, the coverage goes to 5 × 5 when the dilate rate to 2 (Xia et al., 2020; Amer et al., 2021). Compared with the conventional convolution performed regular range feature extraction on fixed regions, the dilated convolution obtains larger receptive fields by the dilation rate.
Proposed Model: Elastomeric UNet
The proposed model is built on the coding-decoding mechanisms, while novel structural breakthroughs are proposed over the original structure by adding a spatially extended network layer to improve the accuracy. Two types of EUNet, namely, horizontal EUNet and spatial EUNet, are introduced in this section, which include three extension structures of horizontal EUNet and two structures of spatial EUNet.
Horizontal Elastomeric UNet
Owing to the elastomeric wave horizontal transmission, the structure of horizontal EUNet was stretched horizontally by multiple feature extraction and spatial location algorithm to improve the segmentation accuracy. Horizontal EUNet expands the network by means of continuous U-shaped structure cascading and provides three structures according to the amounts of U-shaped structures that are cascaded: single-peak, double-peak, and three-peak horizontal EUNet. In the light of the position of wave peak, each structure is further characterized into two forms, i.e., low peak network and high peak network. There is less difference in performance between low peak and high peak networks, so this study introduces high peak networks.
Single High Peak Horizontal Elastomeric UNet
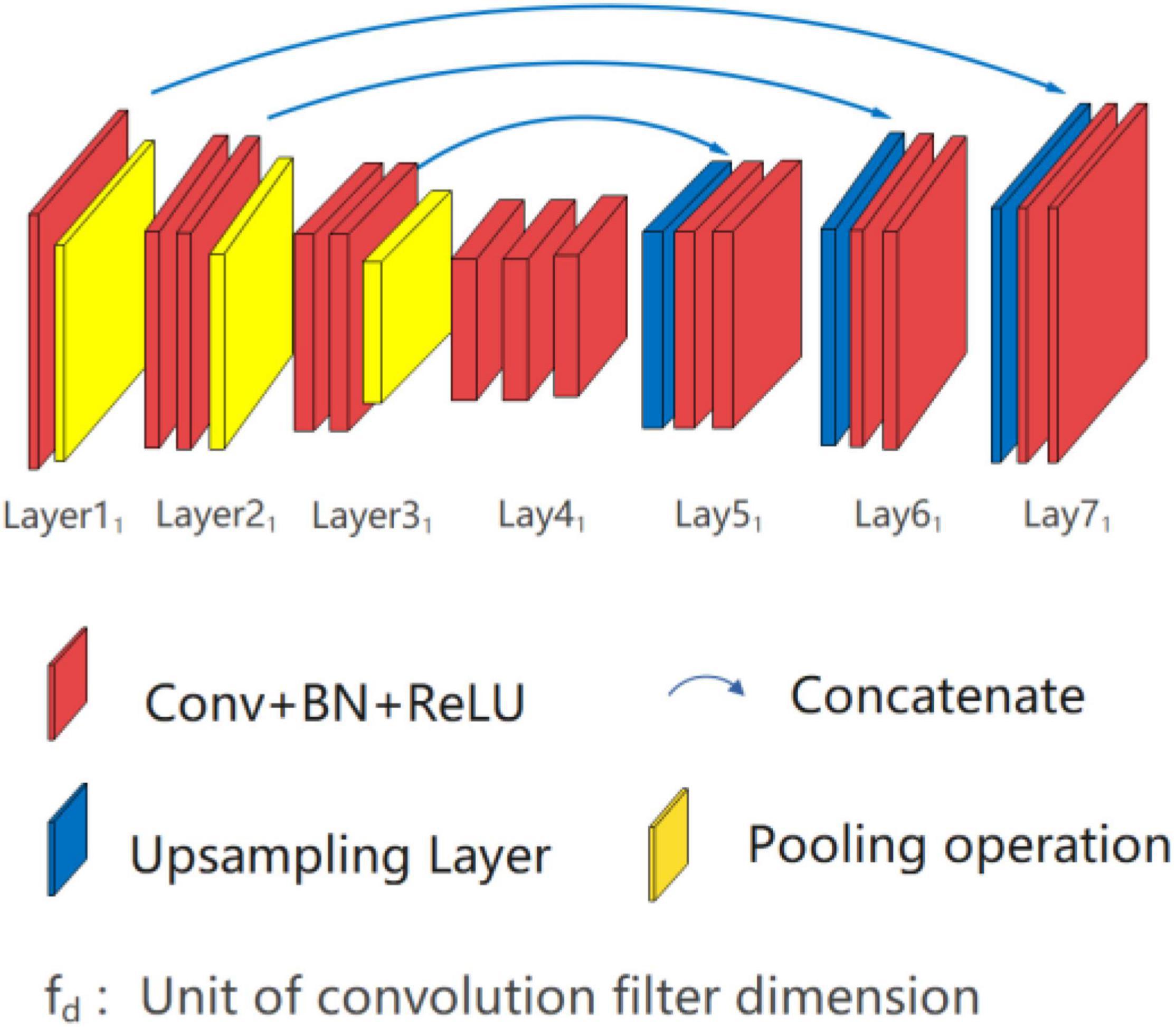
Single high peak horizontal EUNet (SHP-horizontal EUNet) contains a wave peak yielded W-shaped architecture. Due to twice consecutive down-up sampling, the feature extraction and space location operation were performed twice in the network to achieve more accurate pixel-to-pixel segmentation results. Figure 2 shows the structure of SHP-horizontal EUNet U1(4-4)-U2(4-4), which includes two symmetrical down-up paths forming a W-like structure. The input layer Lay1 and the output layer Lay13 of SHP-horizontal EUNet are located at both ends of the “W” shape on the same height and symmetrical to each other. The peak layer is also at the same height as the input/output layer. The outcome of the output layer is the cascaded output of the input/output layer and peak layer. There are two four-layered capture paths for feature extraction, indicating Down1 path (Lay1–Lay4) and Down2 path (Lay7–Lay10), and two extended paths for spatial information location, Up1 path (Lay4–Lay7) and Up2 path (Lay10–Lay13). Each down-layer is comprised of two convolutions to extract information and a pooling operation to reduce dimension for data compression, and each up-layer consists of a concatenate, an up-sampling, and two convolution operations. Batch normalization (BN) layers normalize the input value to improve the convergence of the gradient. Down1-Up1 form U1(4-4) structure, i.e., four layers of down-sampling and four layers of up-sampling, connected through the skip connections, and the structure of corresponding layers is completely symmetric. For instance, the resolution and dimension of Lay2 and Lay6 are the same, which is due to the skip connections. The network configuration is shown in Table 1.
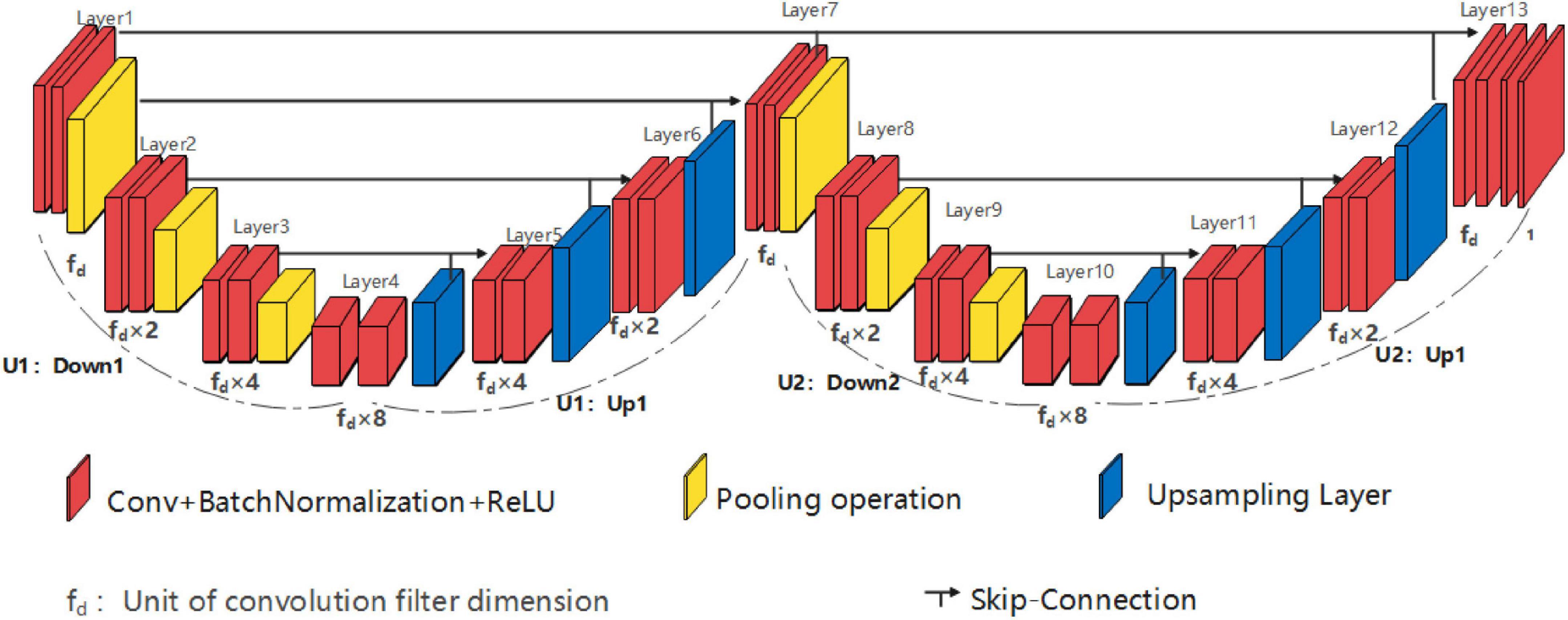
Figure 2. Structure diagram of single high peak horizontal EUNet (SHP-horizontal EUNet) U1(4-4)-U2 (4-4).
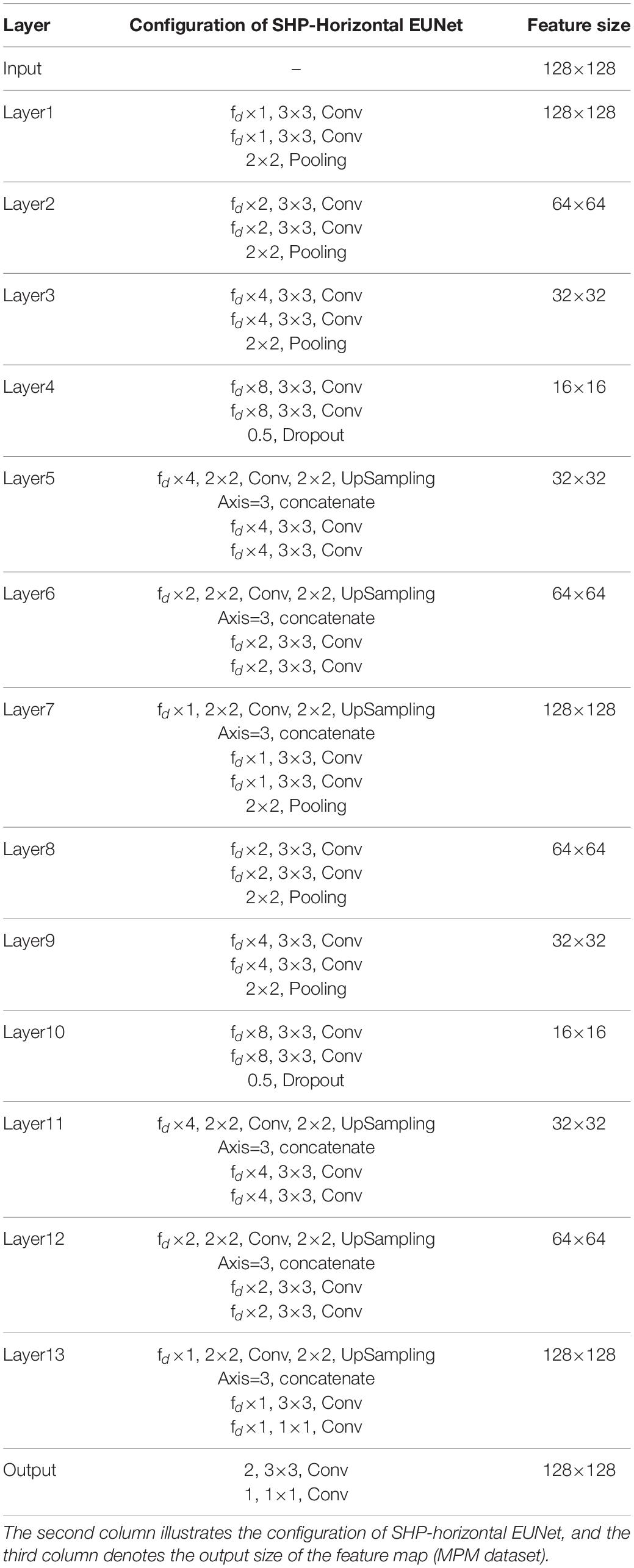
Table 1. Architecture and parameters of single high peak horizontal elastomeric U-Net (SHP-horizontal EUNet).
Researchers can flexibly adjust the structure of the network according to the characteristics of input pictures, such as using the SHP-horizontal EUNet U1(4-4)-U2(3-3). That is to say, the number of the sampling layer for different U-shaped structures is independent which means that researchers can determine how many layers are to be hired accordingly. For the dataset with image resolution between 128 and 600, it is recommended to adopt a small sampling structure with 4–5 layers. It is recommended to adopt a 4–5 layer sampling structure with a convolution filter equaled to (3 × 3). Generally, the below rules need to be followed:
➀ SHP-horizontal EUNet consists of two U-shaped structures. The input layer, wave peak layer, and output layer are at the same height.
➁ The down-sampling and up-sampling paths are completely symmetrical in each U-shaped structure, and different U-shaped structures are independent of each other, for example, there is no connection relationship between Lay2 and Lay8.
➂ The wave peak layer should have three groups of convolutional operations, namely, the cascaded input layer of the output layer, the output layer itself, and the output of the wave peak layer.
Single low peak horizontal EUNet (SLP-horizontal EUNet) can also be designed, and the design principle is similar to SHP-horizontal EUNet. The input layer and the output layer of SLP-horizontal EUNet are also located at both ends of the “W” shape on the same height and symmetrical to each other. The difference between SHP- and SLP-horizontal EUNet is that the peak position is one layer lower than the input/output layer, as shown in Figure 3.
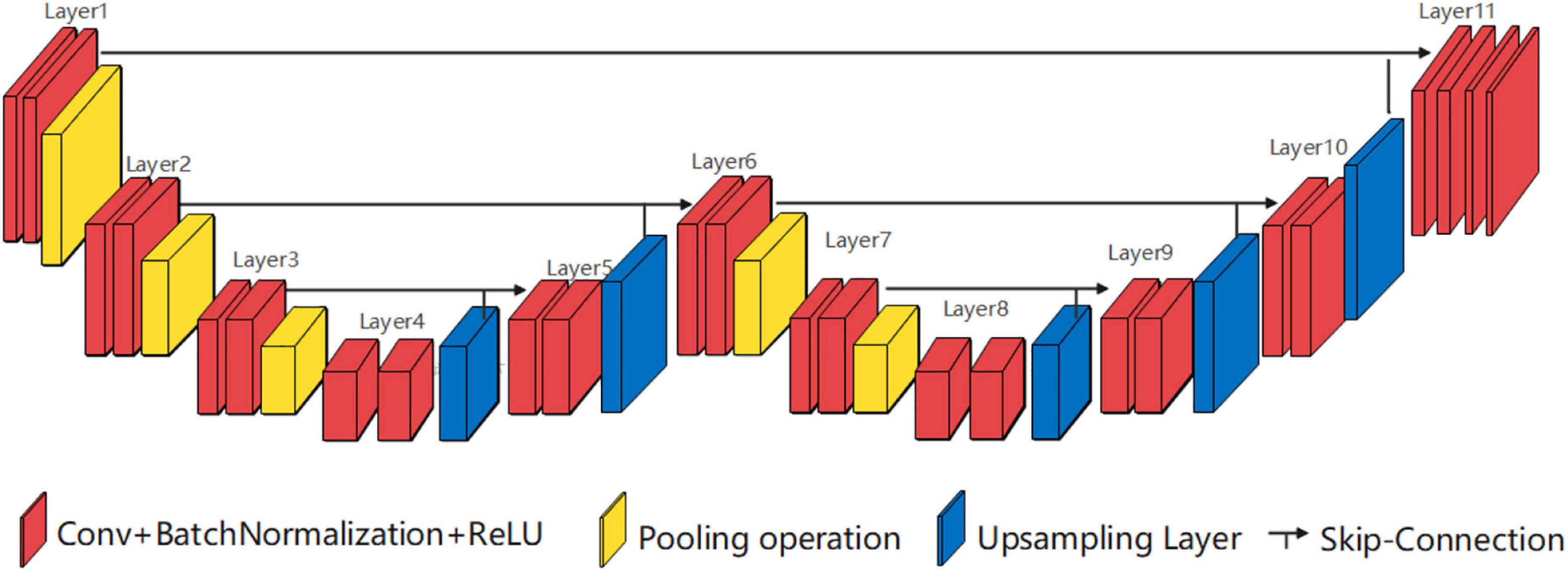
Figure 3. Structure diagram of single low peak horizontal EUNet (SLP-horizontal EUNet) U1(4-3)-U2(3-4).
Double High Peak Horizontal Elastomeric UNet
Similar to the SHP-horizontal EUNet structure, the double high peak horizontal EUNet (DHP-horizontal EUNet) is composed of three independent and fully symmetrical U-shaped structures by in-series connection. As shown in Figure 4, DHP-horizontal EUNet U1(4-4)-U2(4-4)-U3(4-4) is adopted in this section, which can be adjusted by the designer according to their needs, such as DHP-horizontal EUNet U1(5-5)-U2(3-3)- U3(4-4).
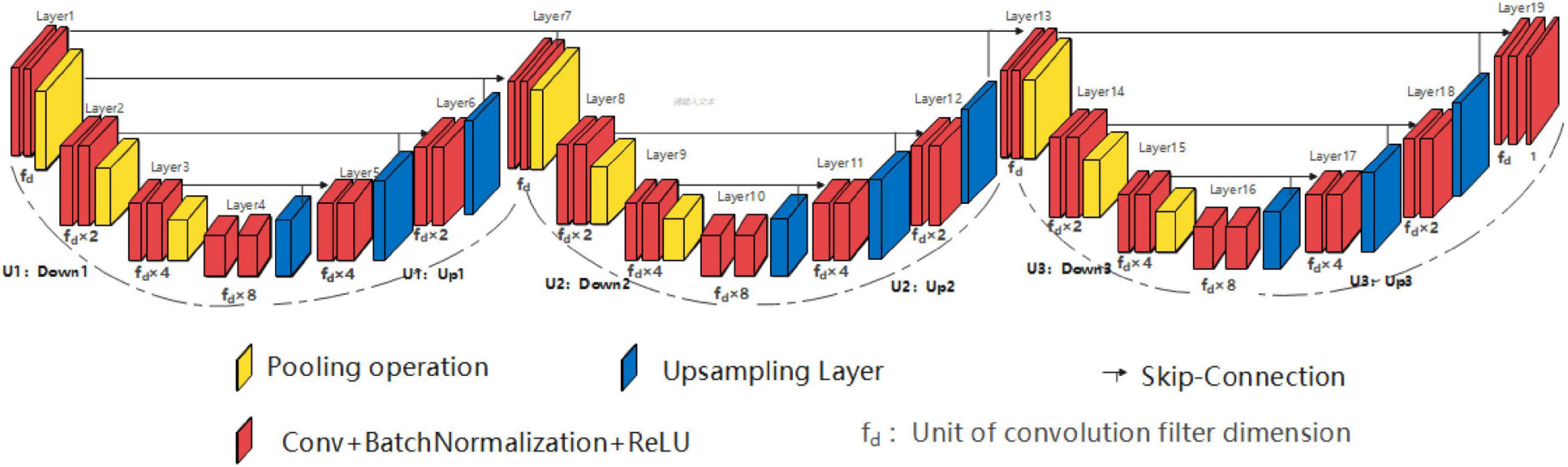
Figure 4. Structure diagram of double high peak horizontal EUNet (DHP-horizontal EUNet) U1(4-4)-U2 (4-4)-U3(4-4).
Three High Peak Horizontal Elastomeric UNet
Consisting of four consecutive independent U-shaped structures, the three high peak horizontal EUNet (THP-horizontal EUNet) resembles SHP-horizontal EUNet structure which has four consecutive symmetrical U-shaped structures. As shown in Figure 5, different U-shaped structure is independent of each other. THP-horizontal EUNet U1(4-4)-U2(4-4)-U3(4-4)-U4(4-4) is exhibited in this section.
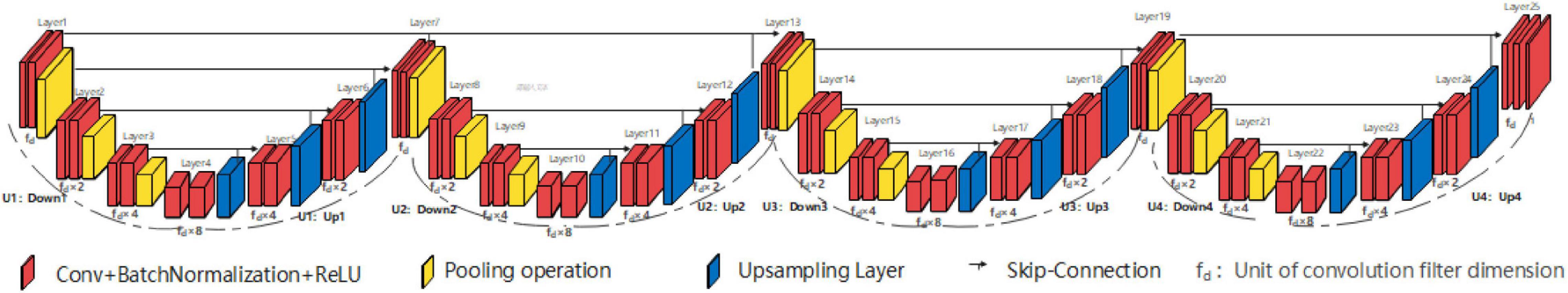
Figure 5. Structure diagram of three high peak horizontal EUNet (THP-horizontal EUNet) U1(4-4)-U2(4-4)-U3(4-4)-U4(4-4).
Spatial Elastomeric UNet
The spatial EUNet extends the network structure from both horizontal and vertical dimensions. The continuous cascade mode of horizontal EUNet is used on the horizontal dimension of spatial EUNet, and random seeds are used to generate multiple independent parallel branches on the vertical dimension. This design provides two spatial EUNet structures, which are double branches and three branches, denoting as U*UNet and U*U*UNet.
Double Branch Network: U*UNet
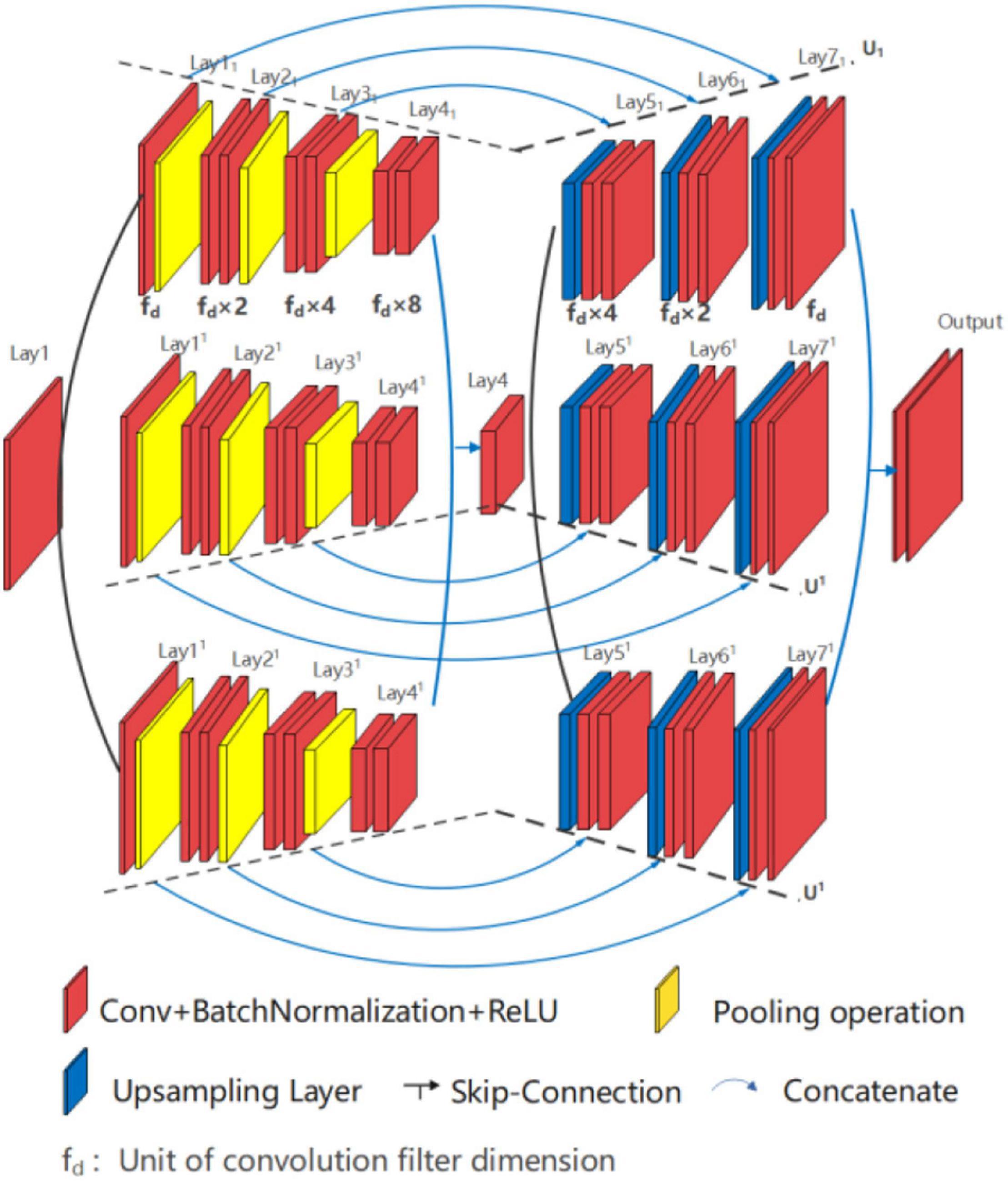
U*UNet, as shown in Figure 6, is composed of two U-shaped parallel U1 and U1, within Lay11-Lay4-Lay17 forms U1 path and Lay11-Lay4-Lay17 forms U1 path. Each path of U*UNet adopting full symmetric down-up sampling mode for feature extraction and location information recovery, such as the path U1, is shown in Figure 7. The separation operation is implemented in the input layer (Lay1) and peak layer (Lay4) to form two paths, and therefore, the convergence operation is executed at the valley (Lay41 and Lay41) and output layer (Lay71 and Lay71). Separation operation utilizes different random seeds for convolution operation, while merge operation is carried out for convergence. In addition, the two U-shaped paths are independent of each other, so multiform nesting, such as Densenet and Resnet, can be performed independently in each path on the basis of design requirements. This study does not do any nesting, but only provides the basic form of the running results.
The architecture design of U*UNet is as follows:
➀ U*UNet is composed of two in-parallel connected U-shaped structures.
➁ Each path contains an independent symmetrical U-shaped structure.
➂ The fuse output is realized through skip connecting via the wave valley and the output layer.
Three Branch Network: U*U*UNet
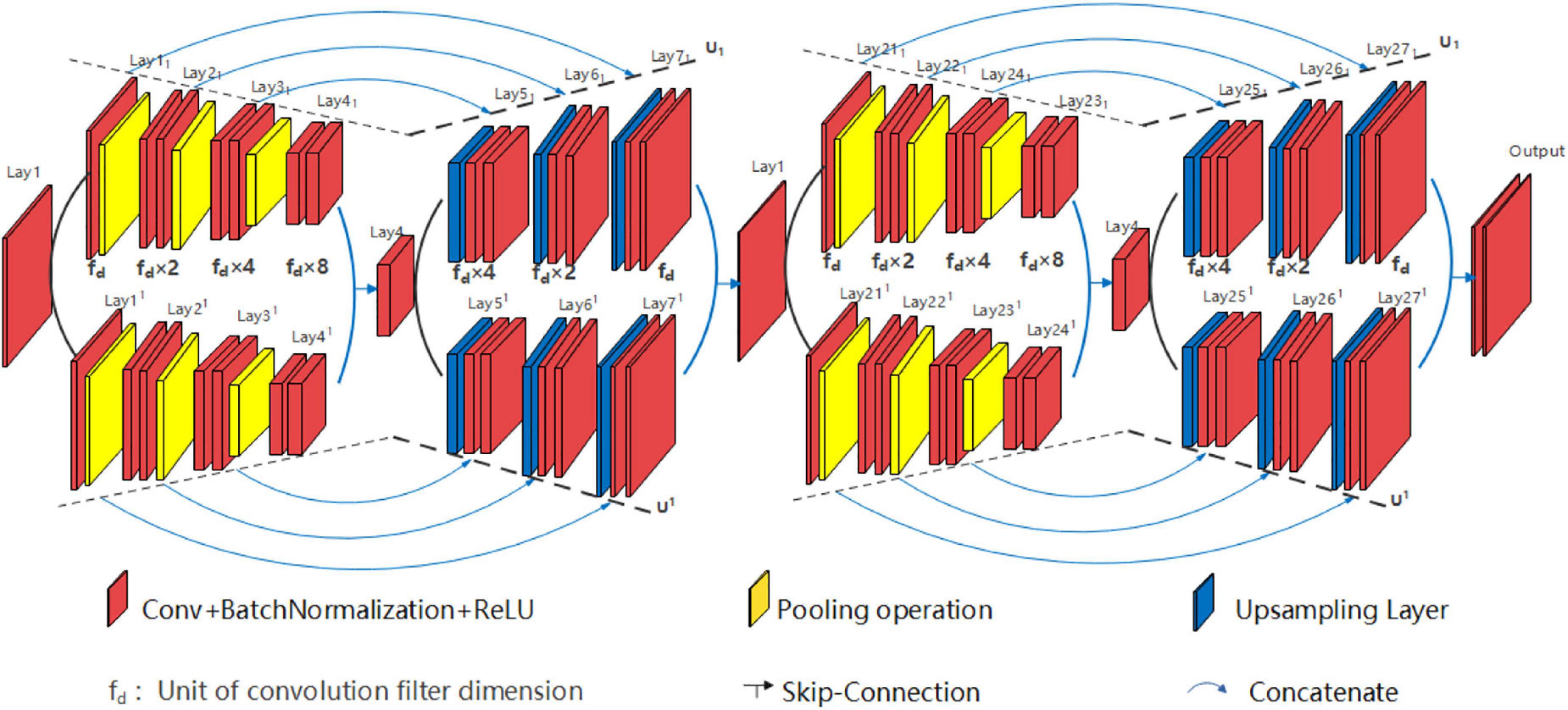
The structure of U*U*UNet is similar to U*UNet, with three U-shaped branches connected in parallel, as shown in Figure 8. Three branches are formed by convolutional division operation through the input layer (Lay1) and the wave valley (Lay4), on which the jump-connection aggregation is further taken place through the wave valley (Lay41, Lay41, and Lay411) and the output layer (Lay71, Lay71, and Lay711). Researchers can take other forms of nested superposition in different branches regarding different research targets.
Various structures of EUNet can be employed, i.e., signal peak U*UNet U1(5)//U1(5)-U2(5)//U2(5) can be devised and signal peak U*UNet U1(3)//U1(3)-U2(5) can also be planned. For example, signal peak U*UNet U1(5)//U1(5)-U2 (5)//U2 (5) is shown in Figure 9. The signal peak U*UNet follows the design principle of horizontal EUNet in the horizontal direction with two consecutive down-up sampling structures. Its space expansion mode is the same as that of U*U Net. To sum up, the designer can design flexibly according to the dataset features, and design requirements for each path of EUNet are independent.
Experiment and Results
Dataset
All architectures proposed in the study were evaluated on two datasets, namely, the multi-photon microscopy (MPM) publicly benchmark retinal datasets (DRIVE). The self-built MPM imaging datasets were obtained by femtosecond home-testing Ti:Sa laser with a field view of 200 × 200 μm. An imaging system was carried out on Inc., Zeiss LSM 510 META with Plan-Neofluar objective. The testing data were taken from the dorsal forearm of the volunteer with a resolution of 128 × 128 pixels. Informed consent from each volunteer was acquired. The MPM dataset contains 90 images in.png format, of which 60 images are used for training, and the remaining 30 images are disposed of for testing. The DRIVE dataset contains 40 color retina images in.tif format, of which 20 images are used for training, and the remaining 20 images are disposed of for testing. The resolution of each image is 584 × 565 pixels at 45° field of view (Taruttis and Ntziachristos, 2015). The common medical image segmentation evaluation metrics suggested in the ISBI 2016 challenge (Ye et al., 2020) were used for benchmarking, including accuracy, precision, specificity, and mIOU. All networks were executed in Keras 2.24 with TensoFlow-GPU backend using Python 3.5.2. The training was implemented on a Microsoft Windows machine configured with GeForce GTX 1080Ti. All segmentation operations for two datasets were performed using the Dice coefficient loss function.
Results
A series of experiments on MPM and DRIVE datasets was conducted to evaluate the performance between EUNet and U-shaped network (UNet, UNet + +, and Dilated UNet). The UNet and its variants were chosen because other deep network structures do not work well for the low resolution of MPM.
The Segmentation Results of the Multi-Photon Microscopy Dataset
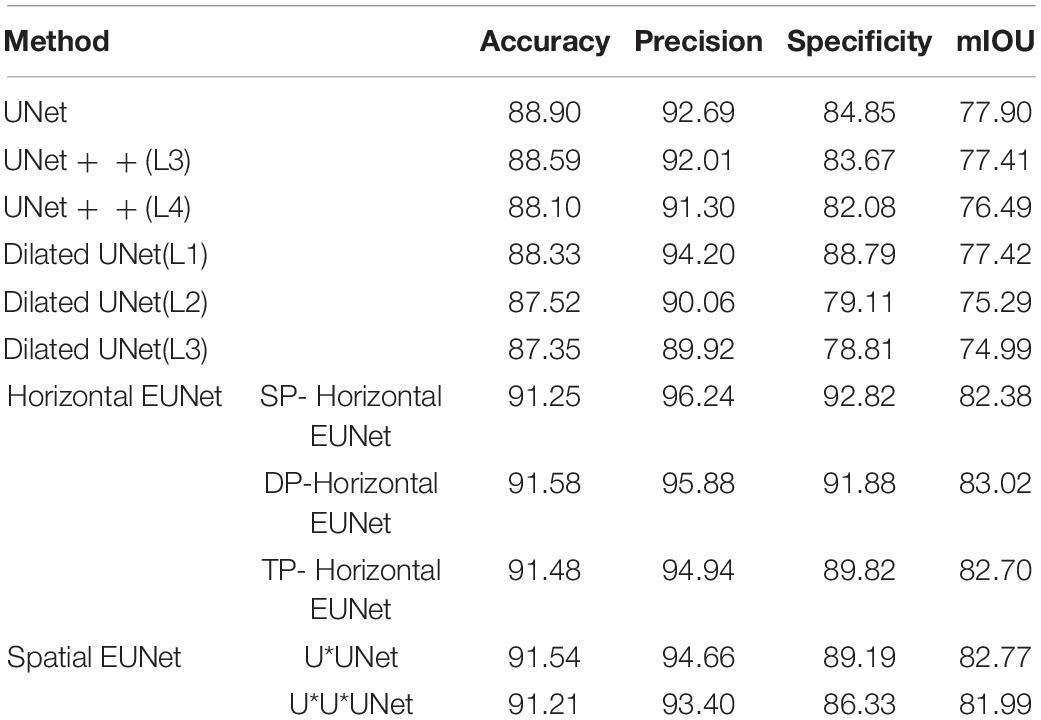
In this section, the proposed EUNet was evaluated based on MPM. EUNet model adopts a 4-layered structure, for example, SHP-horizontal EUNet represents SHP-horizontal EUNet U1 (4-4)-U2 (4-4). The segmentation performance comparison between the proposed EUNet and the U-shaped network is summarized in Table 2, in which the segmentation results are also based on the accuracy, precision, specificity, and mIOU.
UNet + + (L3) and UNet + + (L4) represent three-layered and four-layered nested structures, respectively, while dilated UNet(L1) denotes that the top layer using dilated convolution instead of convolution and (L2) means that the top two layers employ dilated convolution in down-sampling, and so on. UNET + + and dilated UNet perform comparably of MPM dataset, which segmentation accuracy is lower than that of UNet. Dilation convolution is not effective in small resolution datasets, especially with the deepening of dilated convolution. The performance of EUNet proposed in this article all show improvements based upon the U-shape network, where the average accuracy, precision, Specificity, and mIOU achieve 91.41%, 95.02%, 90%, and 82.57% respectively, yielding average improvement of 2.51 and 4.67 points in accuracy and mIOU than UNet respectively. EUNet shows good convergence, as shown in Figure 10.
For a better understanding of performance for each model intuitively, segmentation results of UNet, DHP-horizontal EUNet, and U*UNet are illustrated in Figure 11. It can be seen from the figure that EUNet embodies better boundary segmentation by comparing the outline detail in the second, third, and fourth images. In addition, it is noteworthy that more accurate edge feature extraction is achieved by U*UNet.
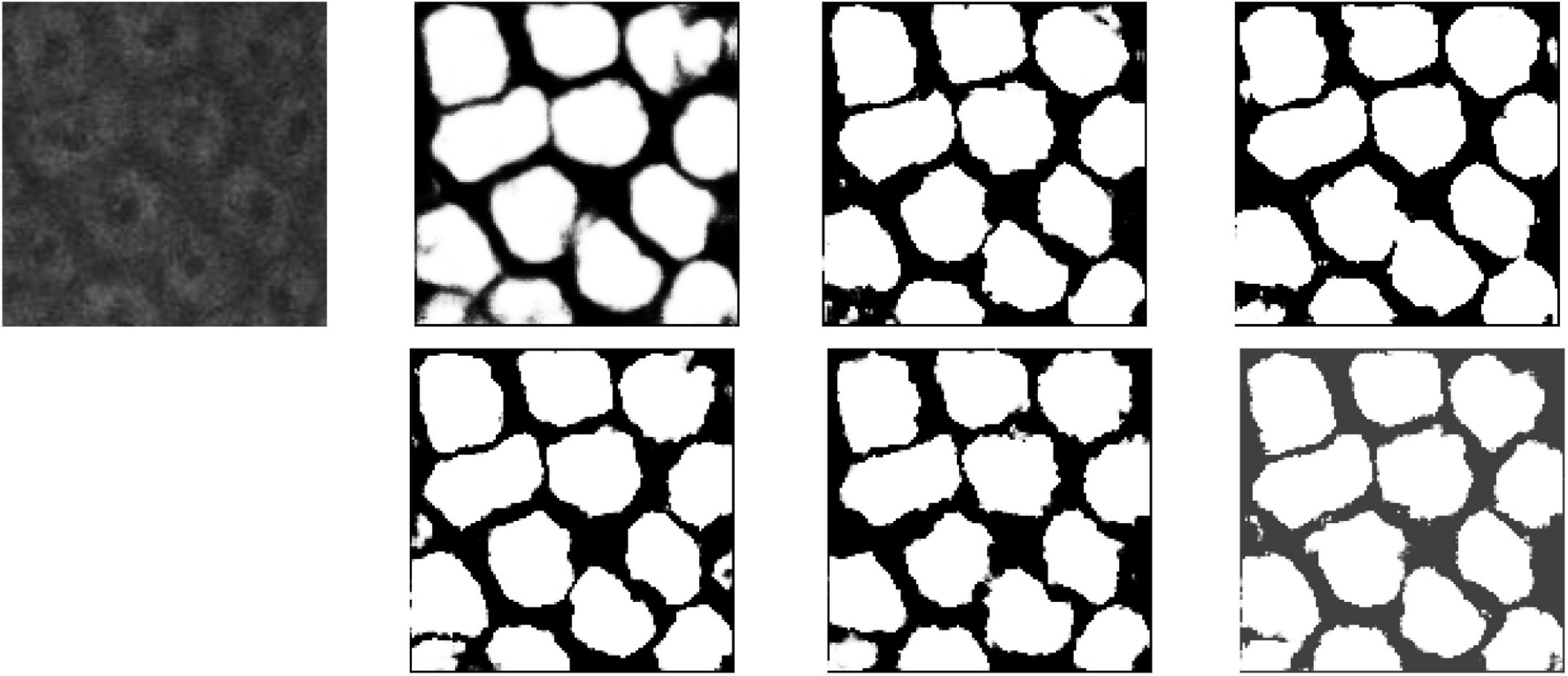
Figure 11. Segmentation results for MPM dataset. Row 1, column 2: UNet; row 1, column 3: UNet + + (L4); row 1, column 4: dilated UNet(L2); row 2, column 2: DHP-horizontal EUNet; row 2, column 3: U*UNet; row 1, column 4: U*U*UNet.
The Segmentation Results of the DRIVE Dataset
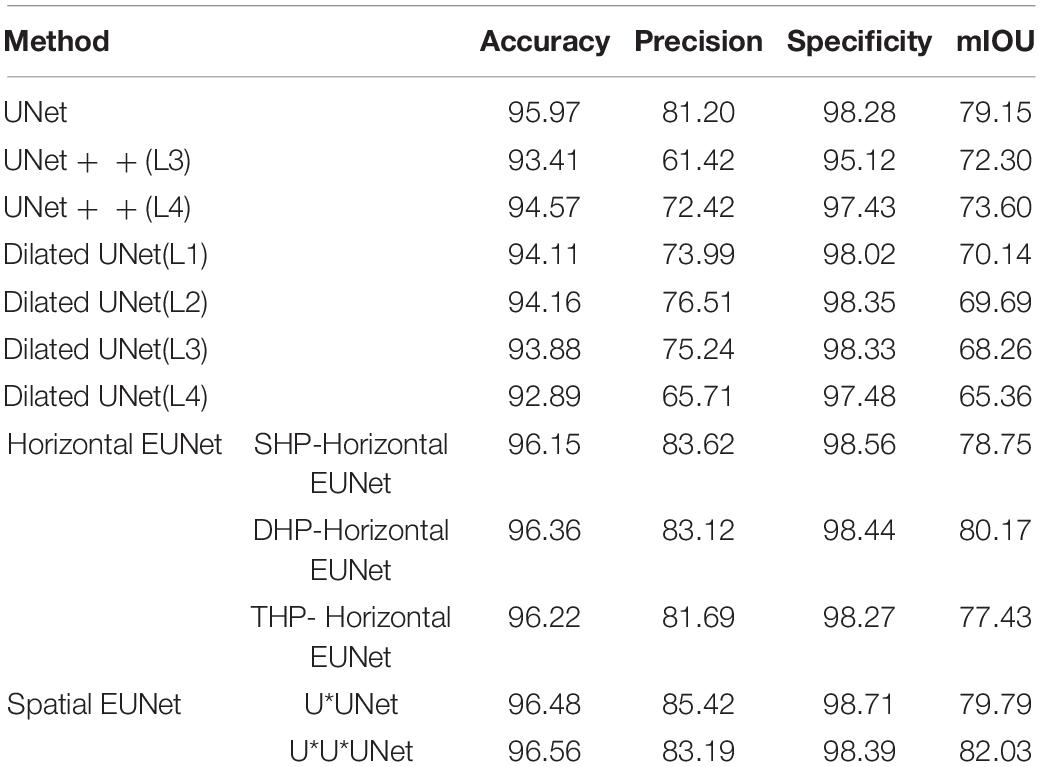
In this section, EUNet and U-shaped networks were tested and verified based on the DRIVE dataset. Each EUNet model adopts a 5-layered structure, for example, SHP-horizontal EUNet represents SHP-horizontal EUNet U1 (5-5)–U2 (5-5). Table 3 shows the segmentation results that were assessed in the light of accuracy, precision, specificity, and mIOU.
As seen, expansion networks bring as a result to significant improvement for DRIVE dateset, which with average improvement of 0.38 points over UNet. Among the space expansion networks, the performance of U*U*UNet is the best, the accuracy of U*U*UNet is up to 96.56%, which is higher than that of UNet 0.59%. The precision of U*UNet is 85.42%, which is higher than that of UNet 4.22%. Experimental results showed that the proposed EUNet provided good performance than UNet. Compared with the UNET in the DRIVE dataset, the Dilated UNet has reduced performance, and the average accuracy is reduced by 2.21 points, which is lower than that of EUNet by 2.59 points. The results show that expansion convolution performs better inmedium resolution datasets, such as DRIVE dataset.
Partial segmentation results are illustrated in Figure 12. As shown in the figure, more details can be described by elastomeric networks. The best background noise processing ability is shown by U*UNet.
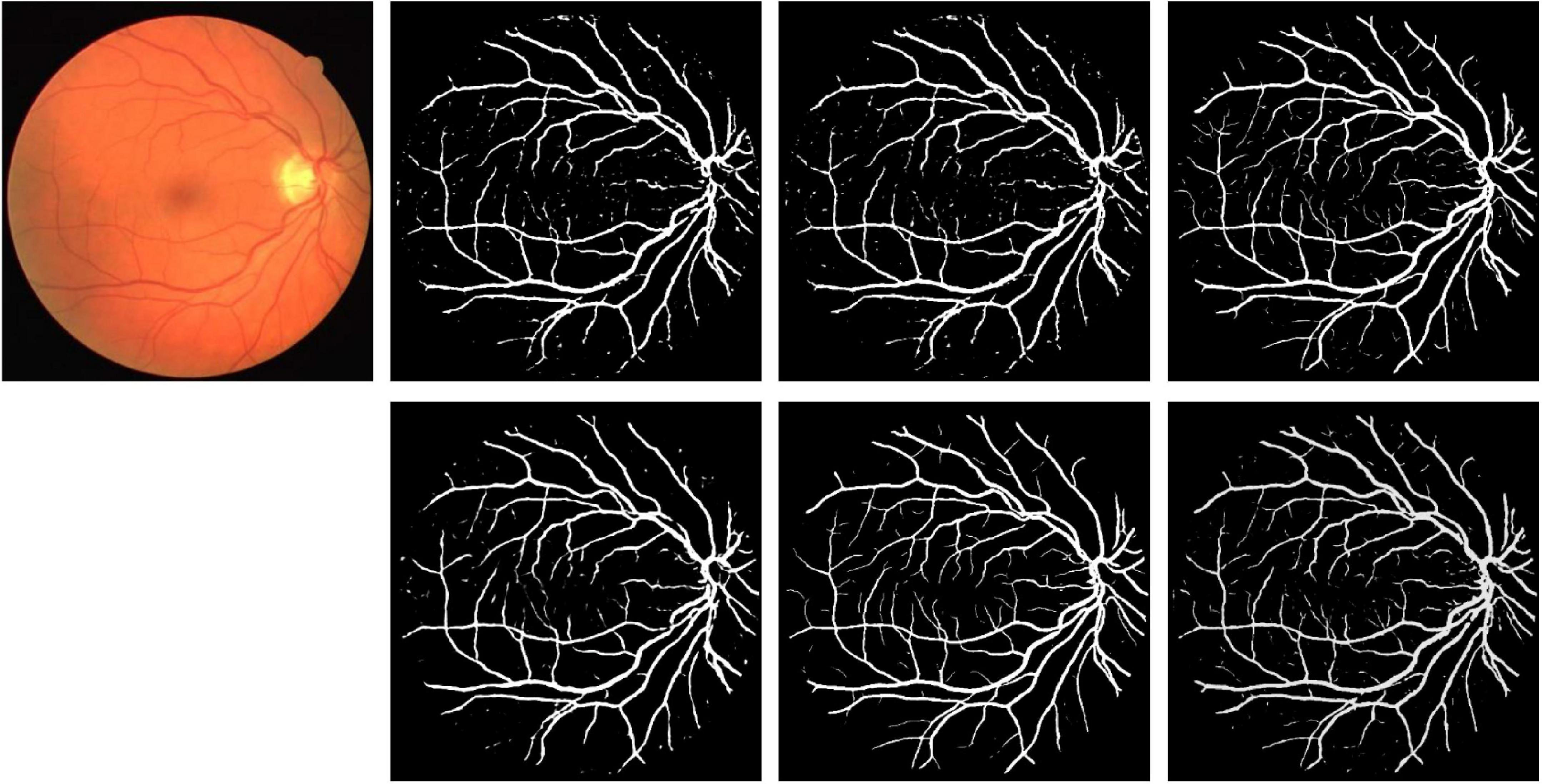
Figure 12. Segmentation results for DRIVE dataset. Row 1, column 2: UNet; row 1, column 3: UNet + + (L4); row 1, column 4: dilated UNet(L2); row 2, column 2: DHP-horizontal EUNet; row 2, column 3: U*Unet; row 1, column 4: U*U*UNet.
Discussion
In general, the overall performance of MPM cell segmentation is limited by the amount of dataset and the resolution size. Based on preliminary work, this study provides EUNet. According to their different structural characteristics, each network embodies different segmentation characteristics, as follows:
In summary, the main contributions of this study include:
(1) We proposed EUNet for medical image segmentation. This network provides the possibility for the malleable development of networks through the horizontal and vertical extension of the elastomeric wave, which greatly breaks through the shackles of traditional structures.
(2) Each corresponding peak layer and valley layer of the branch are operated by fusion convolution in spatial EUNet to realize the multipath information complement and to overcome the position information loss caused by down-sampling.
(3) Every path in this study maintains relative independence, which provides flexible redundancy for researchers. Designers can flexibly design diverse structural variants based on image characteristics and the consideration of Res-Net and Dense-Net. For example, the researcher can carry out multi-form nesting in any U-shaped path regarding the design demand or choose different segmentation model parameters to adjust in different paths (e.g., Conv (3)-Conv (3) can be employed in one path, Conv (3)-Conv (1)-Conv (3) can be utilized in the other path in U*UNet, and so forth).
Conclusion
A novel EUNet is presented in this study to overcome the fixed U-shaped structure of medical image segmentation, which is desirable for the research of Parkinson’s disease and gene aging. Multiple expansion-contraction paths are provided to extract edge detailed features. Convergence is executed to strengthen the fusion of local and global information in peak and valley layers of different branches. The datasets employed in this design were MPM and DRIVE datasets to verify the effectiveness of EUNet. The experimental results demonstrate that the EUNet all show good accuracy and robustness in two datasets. The detailed description is strengthened, and the multi-scale features are fully described by adding feature channels. The consecutive expand-contract path fully explores the dependency of context information and enhances the association feature representation of each semantic class to make the detailed expression of the image edge clearer. Moreover, the model provides a very flexible and extensible structure for future medical image segmentation owing to the relative independence of each U-shaped structure. Researchers can design each U-shaped structural level, functional structure, and parameter model independently according to the characteristics of the segmented images, which will further offer unprecedented room for segmentation resolution improvement.
Data Availability Statement
The data analyzed in this study is subject to the following licenses/restrictions: this is the in vivo skin cell dataset. Requests to access these datasets should be directed to Y2Fpc2lqaW5nQDE2My5jb20=.
Ethics Statement
The studies involving human participants were reviewed and approved by Fujian Normal University. The patients/participants provided their written informed consent to participate in this study. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Author Contributions
YW: responsible for the overall planning of the project. SC: responsible for CNN model design, improvement and implementation, experimental test and data analysis. GC: responsible for providing experimental data and model design guidance. All authors contributed to the article and approved the submitted version.
Funding
This work was supported in part by the National Natural Science Foundation of China (Nos. U1805262 and 61901117), Fujian provincial Research Project, China (Nos. 2021G02006, 2021L3010, and 2020J01879), by the Program for Changjiang Scholars and Innovative Research Team in University (IRT_15R10), by the Special Funds of the Central Government Guiding Local Science and Technology Development (2020L3008 and 2021L3010), by Key provincial scientific and technological innovation projects (2021G02006), the United Fujian Provincial Health and Education Project for Tackling the Key Research of China (2019-WJ-03), and by Fujian Photoelectric Sensing Application Engineering Technology Research Center (2021002).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Amer, A., Ye, X., and Janan, F. (2021). “Residual dilated U-net for the segmentation Of COVID-19 infection from CT images,” in Proceedings of the IEEE/CVF International Conference on Computer Vision (ICCV) Workshops, Seoul, 462–470. doi: 10.1109/ICCVW54120.2021.00056
Aspray, T. J., and Hill, T. R. (2019). Osteoporosis and the ageing skeleton. Subcell. Biochem. 91, 453–476. doi: 10.1007/978-981-13-3681-2_16
Balu, M., Lentsch, G., Korta, D. Z., König, K., Kelly, K. M., Tromberg, B. J., et al. (2017). In vivo multiphoton-microscopy of picosecond-laser induced optical breakdown in human skin, lasers in surgery and medicine. Lasers Surg. Med. 49, 555–562. doi: 10.1002/lsm.22655
Cai, S., Tian, Y., Lui, H., Zeng, H., Wu, Y., and Chen, G. (2020). Dense-UNet: a novel multiphoton in vivo cellular image segmentation model based on a convolutional neural network. Quant. Imaging Med. Surg. 10, 1275–1285. doi: 10.21037/qims-19-1090
Chen, G., Lui, H., and Zeng, H. (2015). Image segmentation for integrated multiphoton microscopy and reflectance confocal microscopy imaging of human skin in vivo. Quant. Imaging Med. Surg. 5, 17–22. doi: 10.3978/j.issn.2223-4292.2014.11.02
Chi, J., Han, X., Wu, C., Wang, H., and Ji, P. (2021). X-Net: multi-branch UNet-like network for liver and tumor segmentation from 3D abdominal CT scans. Neurocomputing 459, 81–96. doi: 10.1016/j.neucom.2021.06.021
Drozdzal, M., Vorontsov, E., Chartrand, G., Kadoury, S., and Pal, C. (2016). “The importance of skip connections in biomedical image segmentation,” in Deep Learning and Data Labeling for Medical Applications. DLMIA 2016, LABELS 2016. Lecture Notes in Computer Science, Vol. 10008, ed. G. Carneiro, D. Mateus, L. Peter, A. Bradley, J. M. R. S. Tavares, V. Belagiannis, et al. (Cham: Springer). doi: 10.1016/j.media.2019.02.006
Haft-Javaherian, M., Fang, L., Muse, V., Schaffer, C. B., Nishimura, N., and Sabuncu, M. R. (2019). Deep convolutional neural networks for segmenting 3D in vivo multiphoton images of vasculature in Alzheimer disease mouse models. PLoS One 14:e0213539. doi: 10.1371/journal.pone.0213539
Ibtehaz, N., and Rahman, M. S. (2020). MultiResUNet : rethinking the U-Net architecture for multimodal biomedical image segmentation. Neural Netw. 121, 74–87. doi: 10.1016/j.neunet.2019.08.025
Lentsch, G., Valdebran, M., Harris, R. M., Barr, R. J., Smith, J., Linden, K., et al. (2019). “Recent advances in multiphoton microscopy for clinical skin imaging,” in Proceedings of the Biophotonics Congress: Optics in the Life Sciences Congress, Vancouver. doi: 10.1364/NTM.2019.NM2C.1
Li, H., Zhygallo, A., and Menze, B. (2018). “Automatic brain structures segmentation 985 using deep residual dilated U-Net,” in Proceedings of the International MICCAI Brainlesion Workshop, Vol. 26, (Berlin: Springer), 385–393. doi: 10.1007/978-3-030-11723-8_39
Li, J., Sun, W., Feng, X., Xing, G., von Deneen, K. M., Wang, W., et al. (2021). A dense connection encoding–decoding convolutional neural network structure for semantic segmentation of thymoma. Neurocomputing 451, 1–11. doi: 10.1016/j.neucom.2021.04.023
Li, X., Chen, H., Qi, X., Dou, Q., Fu, C.-W., and Heng, P.-A. (2018). H-DenseUNet: hybrid densely connected UNet for liver and tumor segmentation from CT volumes. IEEE Trans. Med. Imaging 12, 2663–2674. doi: 10.1109/TMI.2018.2845918
Li, Z., Krishnaswamy, V., Jiang, S., Davis, S. C., Srinivasan, S., Paulsen, K. D., et al. (2010). Rapid magnetic resonance-guided near-infrared mapping to image pulsatile hemoglobin in the breast. Opt. Lett. 35, 3964–3966. doi: 10.1364/OL.35.003964
Liu, L., Cheng, J., Quan, Q., Wu, F.-X., Wang, Y.-P., and Wang, J. (2020). A survey on U-shaped networks in medical image segmentations. Neurocomputing 409, 244–258. doi: 10.1016/j.neucom.2020.05.070
Loeser, R. F. (2009). Aging and osteoarthritis: the role of chondrocyte senescence and aging changes in the cartilage matrix. Osteoarthr. Cartil. 17, 971–979. doi: 10.1016/j.joca.2009.03.002
Long, J., Shelhamer, E., and Darrell, T. (2015). “Fully convolutional networks for semantic segmentation,” in Proceedings of the 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) (Boston, MA: IEEE).
Pang, S., Pang, C., Zhao, L., Chen, Y., Su, Z., Zhou, Y., et al. (2020). SpineParseNet: spine parsing for volumetric MR image by a two-stage segmentation framework with semantic image representation. IEEE Trans. Med. Imaging 40, 262–273. doi: 10.1109/TMI.2020.3025087
Papadopoulos, I. N., Jouhanneau, J.-S., Poulet, J. F. A., and Judkewitz, B. (2017). Scattering compensation by focus scanning holographic aberration probing (F-SHARP). Nat. Photonics 11, 116–123. doi: 10.1038/nphoton.2016.252
Ronneberge, O., Fischer, P., and Brox, T. (2015). “U-Net: convolutional networks for biomedical image segmentation,” in Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015. MICCAI 2015. Lecture Notes in Computer Science, Vol. 9351, eds N. Navab, J. Hornegger, W. Wells, and A. Frangi (Cham: Springer).
Sun, Y. (2020). A Dissertation Submitted to Shanghai Jiao Tong University for the Degree of Philosophy Doctor[D]. Shanghai: Shanghai Jiao Tong University.
Taruttis, A., and Ntziachristos, V. (2015). Advances in real-time multispectral optoacoustic imaging and its applications. Nat. Photonics 9, 219–227. doi: 10.1038/nphoton.2015.29
Vuola, A. O., Akram, S. U., and Kannala, J. (2019). “Mask-RCNN and U-Net ensembled for nuclei segmentation,” in Proceedings of the 2019 IEEE 16th International Symposium on Biomedical Imaging (Venice: IEEE), 8–11.
Xia, H., Sun, W., Song, S., and Mou, X. (2020). Md-Net: multi-scale dilated convolution network for CT images segmentation. Neural Process. Lett. 51, 2915–2927. doi: 10.1007/s11063-020-10230-x
Ye, Q., Wen, Y., Al-Kuwari, N., and Chen, X. (2020). Association between Parkinson’s disease and melanoma: putting the pieces together. Front. Aging Neurosci. 12:60. doi: 10.3389/fnagi.2020.00060
Zhang, J., Jin, Y., Xu, J., Xu, X., and Zhang, Y. (2018). Mdu-net: multi-scale densely connected u-net for biomedical image segmentation. arXiv preprint. arXiv:1812.00352.
Zhao, B., Soraghan, J., Di Caterina, G., and Grose, D. (2019). “Segmentation of head and neck tumours using modified U-net,” in Proceedings of the 27th European Signal Processing Conference (EUSIPCO) (A Coruna: IEEE), 2–6.
Keywords: Parkinson’s disease, medical image segmentation, convolutional neural network (CNN), elastomeric UNet, UNet
Citation: Cai S, Wu Y and Chen G (2022) A Novel Elastomeric UNet for Medical Image Segmentation. Front. Aging Neurosci. 14:841297. doi: 10.3389/fnagi.2022.841297
Received: 22 December 2021; Accepted: 10 January 2022;
Published: 10 March 2022.
Edited by:
Woon-Man Kung, Chinese Culture University, TaiwanCopyright © 2022 Cai, Wu and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yi Wu, d3V5aUBmam51LmVkdS5jbg==; Guannan Chen, ZWRhZG9AZmpudS5lZHUuY24=
 Sijing Cai1,2
Sijing Cai1,2 Yi Wu
Yi Wu