- 1Department of Psychiatry, Faculty of Medicine, Shimane University, Izumo, Japan
- 2Medical Sciences Cluster, Health Service Center, Research and Education Faculty, Kochi University, Kochi, Japan
- 3The Center for Peace, Hiroshima University, Hiroshima, Japan
- 4Department of Legal Medicine, Faculty of Medicine, Shimane University, Izumo, Japan
Heterogeneity of Myeloid Cells in the Brain
In the late 1980's, McGeer et al. observed that major histocompatibility complex (MHC) class II-immunopositive cells with an amoeboid shape were concentrated in the vicinity or center of amyloid plaques in postmortem brains of patients with Alzheimer's disease (AD) (McGeer et al., 1987; Itagaki et al., 1988). This historical neuropathological finding was interpreted to mean that microglia were activated in lesioned areas, and led to the neuroinflammation hypothesis suggesting that activated microglia significantly contribute to AD pathogenesis. Recently, genome-wide association studies have identified variants of the myeloid cell genes triggering receptor expressed on myeloid cell 2 (TREM2) (Jonsson et al., 2013), complement receptor 1 (Lambert et al., 2009), and CD33 (Hollingworth et al., 2011) as novel AD risk genes, sparking renewed interest in microglia from the aspect of genetics (Hashioka et al., 2020).
However, microglia are not the only myeloid cells expressing MHC class II in the brain. Besides parenchymal microglia, the intact brain hosts non-parenchymal specialized myeloid cells such as perivascular, meningeal, and choroid-plexus macrophages, which are referred to as CNS-associated macrophages (CAMs) (Kierdorf et al., 2019). In addition, circulating monocytes are believed to infiltrate the brain and differentiate into macrophages under pathological conditions (Martin et al., 2017). Human microglia, CAMs, and infiltrating monocytes/macrophages express MHC class II as well as certain pan-macrophage markers, such as Iba1 (ionized calcium-binding adapter molecule 1), CD11b, and the fractalkine receptor CX3CR1 (Prinz et al., 2017; Bottcher et al., 2019; Kierdorf et al., 2019). Identification of these brain mononuclear phagocytes was based on their location, morphology, and a small set of surface markers. Such mononuclear cells, therefore, used to be mingled in conventional bulk analyses.
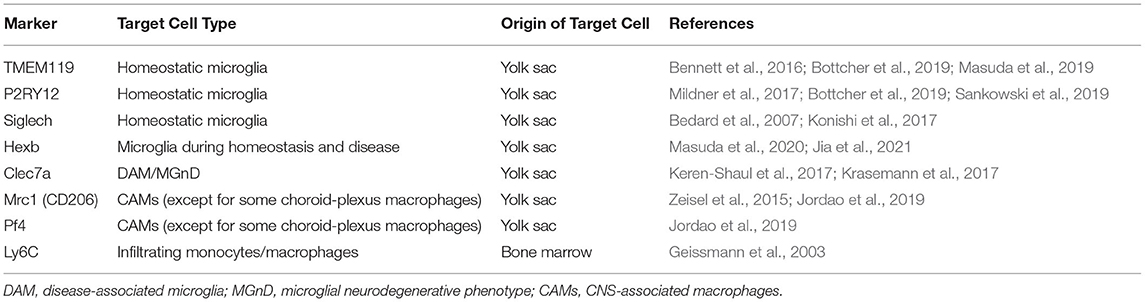
Accumulating evidence indicates that ontogeny and longevity are prominent properties shared by microglia and CAMs, but not by infiltrating monocytes/macrophages (Prinz et al., 2017; Kierdorf et al., 2019). Microglia and CAMs, excluding choroid-plexus macrophages, arise solely from erythromyeloid progenitor cells in the extraembryonic yolk sac and possess extreme longevity and self-renewal potential, without replacement by circulating monocytes (Ginhoux et al., 2010; Goldmann et al., 2016). As an exception for CAMs, choroid-plexus macrophages show mixed ontogeny and a substantial contribution from circulating monocytes (Goldmann et al., 2016). Immigrating monocytes/macrophages, which express a unique monocytic marker Ly6C in mice (Geissmann et al., 2003), originate from the myeloid progenitor lineage in the bone marrow and exhibit a short life with high turnover (Prinz et al., 2017).
Specific Markers Segregating Myeloid Cells in the Brain
Although microglia and most CAM populations share the same prenatal origin (i.e., yolk sac), recent studies with single-cell RNA sequencing have clearly segregated the transcriptome signature specific for microglia from that for CAMs. Specifically, TMEM119 (transmembrane protein 119) and P2RY12 (P2Y purinergic receptor 12) have been identified as core genes specific to microglia in humans (Masuda et al., 2019; Sankowski et al., 2019), while Mrc1 (mannose receptor 1, also called CD206) and Pf4 (platelet factor 4) have been considered as core genes specific to CAMs in mice (Zeisel et al., 2015; Jordao et al., 2019) (Table 1). Indeed, selective microglial expression of TMEM119 and P2RY12 has been confirmed at the protein level in humans. Immunohistochemical analysis of postmortem human brains showed that parenchymal Iba1-immunopositive cells expressed TMEM119 (Bennett et al., 2016; Satoh et al., 2016). On the other hand, the Iba1+ or CD68+ cells, which were presumed to be infiltrating monocytes, did not express TMEM119 in active demyelinating lesions of multiple sclerosis (MS) or necrotic lesions of cerebral infarction (Satoh et al., 2016). P2RY12 immunoreactivity was also observed in parenchymal Iba1+ ramified cells that were supposed to be microglia, but not in CD14+ and CD16+ cells in blood vessels and in the meninges. These cells most likely correspond to peripherally derived monocytes and meningeal macrophages, respectively (Mildner et al., 2017). Postmortem brain study demonstrated that such microglial expression of P2RY12 was decreased in the brains of AD patients and those of MS patients (Mildner et al., 2017). More critically, another advanced single-cell technology, namely cytometry by time of flight (CyTOF), showed that TMEM119 and P2RY12 were expressed on microglia isolated from postmortem human brains and were absent from myeloid cells in human blood and cerebrospinal fluid (Bottcher et al., 2019). Accordingly, in humans, it is tempting to regard TMEM119 and P2RY12 as the most reliable markers that can identify “genuine microglia” in humans, while Siglech (sialic-acid-binding immunoglobulin-like lectin-h) (Bedard et al., 2007; Konishi et al., 2017) and Hexb (beta-hexosaminidase subunit beta) (Masuda et al., 2020; Jia et al., 2021) are also considered as microglia-enriched genes (Table 1).
Pathological Roles of Heterogenous Brain Myeloid Cells in Alzheimer's Disease
The heterogeneous nature of brain myeloid cells raises the question as to whether or not there are differences in pathological roles between microglia, CAMs, and infiltering monocytes in AD. Do “genuine microglia” play specific roles in AD pathogenesis? The answer seems to be yes, since it was demonstrated that macrophages transplanted from the bone marrow in donors could adopt some features of endogenous microglia, but such macrophages were not able to fully recapitulate all microglial properties, such as increased expression of microglial identity genes, even after prolonged residence in the recipient brain (Bennett et al., 2018).
In addition, several studies have reported conflicting results concerning the contribution of microglia, CAMs, and recruited monocytes to AD pathology. For instance, infusion of wildtype monocytes derived from the bone marrow to the peripheral blood of AD transgenic mice led to spontaneous migration of monocytes to amyloid lesions in the absence of irradiation, genetic manipulation, or chemotherapy. Such treated mice showed a decrease in cerebral Aβ levels, which seemed to be associated with monocytic phagocytosis, and ameliorated cognitive deficits (Koronyo et al., 2015). On the other hand, a study using AD transgenic mice demonstrated that peripheral monocytes distinguished from microglia by parabiosis were not significantly recruited to Aβ plaques, whereas resident microglia gathered to surround Aβ plaques (Wang et al., 2016). This controversy may stem from limitations of conventional analytical methods, such as immunohistochemistry and flow cytometry, employed in the aforementioned studies to characterize myeloid cells. These methods can only probe a few preselected proteins as cell surface markers.
But now, can “genuine microglia” reliably be typified by the microglia-specific markers TMEM119 and P2RY12, which were established by the latest single-cell profiling technologies? There seems to be no clear answer, since expression levels of TMEM119 and P2RY12 depend on the microglial activation status. Microglia highly express microglia core genes TMEM119 and P2RY12 in the homeostatic state. After loss of their homeostatic phenotype, however, microglia suppress the expression of TMEM119 and P2RY12 in an activated state referred to as disease-associated microglia (DAM) (Keren-Shaul et al., 2017) or microglial neurodegenerative phenotype (MGnD) (Krasemann et al., 2017). Such DAM/MGnD microglia are closely associated with Aβ plaques and possess pro-inflammatory signatures (Keren-Shaul et al., 2017; Krasemann et al., 2017). Based on these findings, TMEM119 and P2RY12 should be regarded as homeostatic microglia molecules. Therefore, immunohistochemical analysis appears to be difficult to distinguish between the absence of microglia themselves and the presence of microglia in the DAM/MGnD activation state in lesions showing poverty of sole TMEM119 or P2RY12 immunoreactivity.
A recent study using immunocytochemical electron microscopy has uncovered a new microglial phenotype called dark microglia, which are associated with amyloid plaques in APP/PS1 mice (Bisht et al., 2016). Dark microglia display condensed, electron-dense cytoplasm and nucleoplasm, a characteristic giving them a “dark” appearance. They also show a downregulated expression of the homeostatic marker P2RY12 (Bisht et al., 2016). Therefore, gene expression profiles and biological characteristics of dark microglia may be similar to those of the DAM/MGnD microglia.
Novel Genetic Approaches to Define Pathological Roles of Microglia
Advances in genetic manipulation have established certain transgenic or knock-in mouse lines that genetically target microglia. Such mouse lines have been shown to monitor precisely and manipulate microglia regardless of their activation state. In addition, CyTOF combined with fate mapping on APP/PS1 mice has detected a subset of microglia associated with AD-prone neurodegeneration (Mrdjen et al., 2018).
The mice with the recombinase CreERT2 inserted into the locus TMEM119 and the TMEM119-tdTomato knock-in mice have shown clear discrimination of microglia from CAMs, even though non-myeloid brain cells, such as endothelial cells and fibroblasts, and some choroid-plexus macrophages can also be targeted (Kaiser and Feng, 2019; Ruan et al., 2020). In the brains of Tmem119-tdTomato reporter mice that were treated by laser ablation, tdTomato-positive microglia, which were presumably activated, entered the site of injury and dramatically changed their process length without losing the TMEM119-tdTomato signal (Ruan et al., 2020). It is yet to be clarified why TMEM119 expression was preserved even in microglia activated in lesioned areas. Also, P2RY12-CreERT2 knock-in mice were able to specifically label microglia, as shown by P2ry12-CreERT2; Rosa26Rosa26Ai14 reporter mice that expressed TdTomato upon Cre-dependent recombination with minor effects on CAMs (McKinsey et al., 2020). When P2RY12-CreERT2; Rosa26Rosa26Ai14 mice underwent middle cerebral artery occlusion, tdTomato+ microglia exhibited reduced immunoreactivity for P2RY12 and TMEM119 in the ischemic core and penumbra (McKinsey et al., 2020).
To define the pathological roles of “genuine microglia” in AD, it is tempting to apply these mouse lines targeting microglia genetically to experimental AD models. For instance, intrahippocampal injection of Aβ into such mice seems to be technically feasible and to facilitate the transcriptional and functional analysis of microglia in response to Aβ. It should be noted that there is no AD animal model sufficient to reflect all aspects of AD pathology (Drummond and Wisniewski, 2017). In fact, intrahippocampal Aβ injection appears to exaggerate inflammatory responses and to represent an acute brain insult (McLarnon and Ryu, 2008), even though chronic inflammation is considered a critical aspect of AD pathology. Nevertheless, further studies along this line are warranted to elucidate the potential of microglia as key therapeutic targets in AD.
Tracing DAM/MGnD microglia in the brain of AD animal models could also help to reveal the pathological roles of microglia. DAM/MGnD microglia have been identified by high expression of the DAM/MGnD marker Clec7a combined with low expression of the homeostatic microglia marker P2RY12 (Keren-Shaul et al., 2017; Krasemann et al., 2017). While clarifying microglial ontogeny in humans could help to clarify the pathological roles of human microglia, there is no approach to address this issue directly for obvious reasons. However, using single-cell transcriptional profiling approaches, a recent study on macrophages from aborted fetuses implies that human microglia are derived largely from yolk sac progenitors (Bian et al., 2020).
Author Contributions
SH wrote the manuscript. All authors discussed, edited the manuscript, read, and approved the final manuscript.
Funding
This study was supported by JSPS KAKENHI Grant Numbers 19K08018 (SH) and 19H04355 (SH and NK).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
Sincere appreciation is extended to Dr. Edith McGeer for her invaluable support.
References
Bedard, A., Tremblay, P., Chernomoretz, A., and Vallieres, L. (2007). Identification of genes preferentially expressed by microglia and upregulated during cuprizone-induced inflammation. Glia 55, 777–789. doi: 10.1002/glia.20477
Bennett, F. C., Bennett, M. L., Yaqoob, F., Mulinyawe, S. B., Grant, G. A., Hayden Gephart, M., et al. (2018). A Combination of Ontogeny and CNS Environment Establishes Microglial Identity. Neuron 98, 1170–1183 e1178. doi: 10.1016/j.neuron.2018.05.014
Bennett, M. L., Bennett, F. C., Liddelow, S. A., Ajami, B., Zamanian, J. L., Fernhoff, N. B., et al. (2016). New tools for studying microglia in the mouse and human CNS. Proc. Natl. Acad. Sci. U. S. A. 113, E1738–1746. doi: 10.1073/pnas.1525528113
Bian, Z., Gong, Y., Huang, T., Lee, C. Z. W., Bian, L., Bai, Z., et al. (2020). Deciphering human macrophage development at single-cell resolution. Nature 582, 571–576. doi: 10.1038/s41586-020-2316-7
Bisht, K., Sharma, K. P., Lecours, C., Sanchez, M. G., El Hajj, H., Milior, G., et al. (2016). Dark microglia: a new phenotype predominantly associated with pathological states. Glia 64, 826–839. doi: 10.1002/glia.22966
Bottcher, C., Schlickeiser, S., Sneeboer, M. A. M., Kunkel, D., Knop, A., Paza, E., et al. (2019). Human microglia regional heterogeneity and phenotypes determined by multiplexed single-cell mass cytometry. Nat. Neurosci. 22, 78–90. doi: 10.1038/s41593-018-0290-2
Drummond, E., and Wisniewski, T. (2017). Alzheimer's disease: experimental models and reality. Acta Neuropathol. 133, 155–175. doi: 10.1007/s00401-016-1662-x
Geissmann, F., Jung, S., and Littman, D. R. (2003). blood monocytes consist of two principal subsets with distinct migratory properties. Immunity 19, 71–82. doi: 10.1016/S1074-7613(03)00174-2
Ginhoux, F., Greter, M., Leboeuf, M., Nandi, S., See, P., Gokhan, S., et al. (2010). Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science 330, 841–845. doi: 10.1126/science.1194637
Goldmann, T., Wieghofer, P., Jordao, M. J., Prutek, F., Hagemeyer, N., Frenzel, K., et al. (2016). Origin, fate and dynamics of macrophages at central nervous system interfaces. Nat. Immunol. 17, 797–805. doi: 10.1038/ni.3423
Hashioka, S., Inoue, K., Takeshita, H., and Inagaki, M. (2020). Do Alzheimer's disease risk gene products actually act in microglia? Front. Aging Neurosci. 12, 589196. doi: 10.3389/fnagi.2020.589196
Hollingworth, P., Harold, D., Sims, R., Gerrish, A., Lambert, J. C., Carrasquillo, M. M., et al. (2011). Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer's disease. Nat Genet 43, 429–435. doi: 10.1038/ng.803
Itagaki, S., Mcgeer, P. L., and Akiyama, H. (1988). Presence of T-cytotoxic suppressor and leucocyte common antigen positive cells in Alzheimer's disease brain tissue. Neurosci. Lett. 91, 259–264. doi: 10.1016/0304-3940(88)90690-8
Jia, M., Zhang, W., Zhu, J., Huang, C., Zhou, J., Lian, J., et al. (2021). Microglia-specific expression of HEXA and HEXB leads to poor prognosis in glioblastoma patients. Front. Oncol. 11, 685893. doi: 10.3389/fonc.2021.685893
Jonsson, T., Stefansson, H., Steinberg, S., Jonsdottir, I., Jonsson, P. V., Snaedal, J., et al. (2013). Variant of TREM2 associated with the risk of Alzheimer's disease. N. Engl. J. Med. 368, 107–116. doi: 10.1056/NEJMoa1211103
Jordao, M. J. C., Sankowski, R., Brendecke, S. M., Sagar, Locatelli, G., Tai, Y. H., et al. (2019). Single-cell profiling identifies myeloid cell subsets with distinct fates during neuroinflammation. Science 363, eaat7554. doi: 10.1126/science.aat7554
Kaiser, T., and Feng, G. (2019). Tmem119-EGFP and Tmem119-CreERT2 transgenic mice for labeling and manipulating microglia. eNeuro 6. doi: 10.1523/ENEURO.0448-18.2019
Keren-Shaul, H., Spinrad, A., Weiner, A., Matcovitch-Natan, O., Dvir-Szternfeld, R., Ulland, T. K., et al. (2017). A Unique Microglia Type Associated with Restricting Development of Alzheimer's Disease. Cell 169, 1276–1290.e1217. doi: 10.1016/j.cell.2017.05.018
Kierdorf, K., Masuda, T., Jordao, M. J. C., and Prinz, M. (2019). Macrophages at CNS interfaces: ontogeny and function in health and disease. Nat. Rev. Neurosci. 20, 547–562. doi: 10.1038/s41583-019-0201-x
Konishi, H., Kobayashi, M., Kunisawa, T., Imai, K., Sayo, A., Malissen, B., et al. (2017). Siglec-H is a microglia-specific marker that discriminates microglia from CNS-associated macrophages and CNS-infiltrating monocytes. Glia 65, 1927–1943. doi: 10.1002/glia.23204
Koronyo, Y., Salumbides, B. C., Sheyn, J., Pelissier, L., Li, S., Ljubimov, V., et al. (2015). Therapeutic effects of glatiramer acetate and grafted CD115(+) monocytes in a mouse model of Alzheimer's disease. Brain 138, 2399–2422. doi: 10.1093/brain/awv150
Krasemann, S., Madore, C., Cialic, R., Baufeld, C., Calcagno, N., El Fatimy, R., et al. (2017). The TREM2-APOE pathway drives the transcriptional phenotype of dysfunctional microglia in neurodegenerative diseases. Immunity 47, 566–581.e569. doi: 10.1016/j.immuni.2017.08.008
Lambert, J. C., Heath, S., Even, G., Campion, D., Sleegers, K., Hiltunen, M., et al. (2009). Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer's disease. Nat. Genet. 41, 1094–1099. doi: 10.1038/ng.439
Martin, E., Boucher, C., Fontaine, B., and Delarasse, C. (2017). Distinct inflammatory phenotypes of microglia and monocyte-derived macrophages in Alzheimer's disease models: effects of aging and amyloid pathology. Aging Cell 16, 27–38. doi: 10.1111/acel.12522
Masuda, T., Amann, L., Sankowski, R., Staszewski, O., Lenz, M., Ericco, P. D., et al. (2020). Novel Hexb-based tools for studying microglia in the CNS. Nat. Immunol. 21, 802–815. doi: 10.1038/s41590-020-0707-4
Masuda, T., Sankowski, R., Staszewski, O., Bottcher, C., Amann, L., Sagar, et al. (2019). Spatial and temporal heterogeneity of mouse and human microglia at single-cell resolution. Nature 566, 388–392. doi: 10.1038/s41586-019-0924-x
McGeer, P. L., Itagaki, S., Tago, H., and Mcgeer, E. G. (1987). Reactive microglia in patients with senile dementia of the Alzheimer type are positive for the histocompatibility glycoprotein HLA-DR. Neurosci. Lett. 79, 195–200. doi: 10.1016/0304-3940(87)90696-3
McKinsey, G. L., Lizama, C. O., Keown-Lang, A. E., Niu, A., Santander, N., Larpthaveesarp, A., et al. (2020). A new genetic strategy for targeting microglia in development and disease. Elife 9, e54590. doi: 10.7554/eLife.54590.sa2
McLarnon, J. G., and Ryu, J. K. (2008). Relevance of abeta1-42 intrahippocampal injection as an animal model of inflamed Alzheimer's disease brain. Curr. Alzheimer Res. 5, 475–480. doi: 10.2174/156720508785908874
Mildner, A., Huang, H., Radke, J., Stenzel, W., and Priller, J. (2017). P2Y12 receptor is expressed on human microglia under physiological conditions throughout development and is sensitive to neuroinflammatory diseases. Glia 65, 375–387. doi: 10.1002/glia.23097
Mrdjen, D., Pavlovic, A., Hartmann, F. J., Schreiner, B., Utz, S. G., Leung, B. P., et al. (2018). High-dimensional single-cell mapping of central nervous system immune cells reveals distinct myeloid subsets in health, aging, and disease. Immunity 48, 380–395.e386. doi: 10.1016/j.immuni.2018.01.011
Prinz, M., Erny, D., and Hagemeyer, N. (2017). Ontogeny and homeostasis of CNS myeloid cells. Nat. Immunol. 18, 385–392. doi: 10.1038/ni.3703
Ruan, C., Sun, L., Kroshilina, A., Beckers, L., De Jager, P., Bradshaw, E. M., et al. (2020). A novel Tmem119-tdTomato reporter mouse model for studying microglia in the central nervous system. Brain Behav. Immun. 83, 180–191. doi: 10.1016/j.bbi.2019.10.009
Sankowski, R., Bottcher, C., Masuda, T., Geirsdottir, L., Sagar, S., Indram, E., Seredenina, T., et al. (2019). Mapping microglia states in the human brain through the integration of high-dimensional techniques. Nat. Neurosci. 22, 2098–2110. doi: 10.1038/s41593-019-0532-y
Satoh, J., Kino, Y., Asahina, N., Takitani, M., Miyoshi, J., Ishida, T., et al. (2016). TMEM119 marks a subset of microglia in the human brain. Neuropathology 36, 39–49. doi: 10.1111/neup.12235
Wang, Y., Ulland, T. K., Ulrich, J. D., Song, W., Tzaferis, J. A., Hole, J. T., et al. (2016). TREM2-mediated early microglial response limits diffusion and toxicity of amyloid plaques. J. Exp. Med. 213, 667–675. doi: 10.1084/jem.20151948
Keywords: microglia, monocytes, CNS-associated macrophages (CAMs), transmembrane protein 119 (TMEM119), P2Y purinergic receptor 12 (P2RY12), single-cell profiling, Alzheimer's disease
Citation: Hashioka S, Inoue K, Otsuki K, Hayashida M, Wake R, Kawano N, Takeshita H and Inagaki M (2022) Contribution of “Genuine Microglia” to Alzheimer's Disease Pathology. Front. Aging Neurosci. 14:815307. doi: 10.3389/fnagi.2022.815307
Received: 15 November 2021; Accepted: 03 March 2022;
Published: 24 March 2022.
Edited by:
Guanghui Wang, Soochow University, ChinaReviewed by:
Marie-Ève Tremblay, University of Victoria, CanadaCopyright © 2022 Hashioka, Inoue, Otsuki, Hayashida, Wake, Kawano, Takeshita and Inagaki. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sadayuki Hashioka, aGFzaGlva2FAZjIuZGlvbi5uZS5qcA==
 Sadayuki Hashioka
Sadayuki Hashioka Ken Inoue
Ken Inoue Koji Otsuki
Koji Otsuki Maiko Hayashida1
Maiko Hayashida1 Haruo Takeshita
Haruo Takeshita