- 1Yurov's Laboratory of Molecular Genetics and Cytogenomics of the Brain, Mental Health Research Center, Moscow, Russia
- 2Laboratory of Molecular Cytogenetics of Neuropsychiatric Diseases, Veltischev Research and Clinical Institute for Pediatrics of the Pirogov Russian National Research Medical University, Moscow, Russia
- 3Department of Medical Biological Disciplines, Belgorod State University, Belgorod, Russia
It is hard to estimate the profound impact of COVID-19 (SARS-CoV-2 infection) on our life. SARS-CoV-2 infection is the cause of a pandemic and is associated with a severe disease threatening life during and after the manifestation (Tu et al., 2020; Wu et al., 2020; Hu et al., 2021). It is repeatedly noted that the central nervous system is seriously affected by COVID-19 infection. However, the intrinsic effects of COVID-19 on the human brain remain a matter of future research (Pennisi et al., 2020). Pathological brain aging and natural brain deterioration are associated with accumulation and propagation of genome (chromosome) instability (Yurov et al., 2010, 2019; Andriani et al., 2017; Zhang and Vijg, 2018; Iourov et al., 2021). Since genome (chromosome) instability may result from viral infections (Heng, 2019), SARS-CoV-2 interactions with cells of the central nervous system are able to increase the risk for early manifestations of aging-related brain disorders and/or premature brain deterioration mediated by genome and chromosome instability. Recently, a number of studies dedicated to host-coronavirus protein interaction networks have highlighted numerous molecular and cellular processes, which are likely to be altered by SARS-CoV-2 (Gordon et al., 2020a; Lee et al., 2021; Schmidt et al., 2021; Terracciano et al., 2021). Here, we have addressed data on SARS-CoV-2–host protein–protein interactomes for assessing potential COVID-19 effects on aging-related genome (chromosome) instability in the brain.
Data on SARS-CoV-2–host protein–protein interactomes or networks were taken from following articles: Díaz (2020); Gordon et al. (2020a,b); Guzzi et al. (2020); Perrin-Cocon et al. (2020); Schmidt et al. (2021), and Terracciano et al. (2021). Candidate pathways (networks) were grouped according to the association with processes involved in genome stability maintenance, cell cycle regulation, chromatin regulation, DNA metabolism, and cell death. These clusters of pathways are generally associated with brain-specific chromosome/genome instability.
Looking through the SARS-CoV-2–host interactomes, one may find a wide spectrum of different pathways affected by the coronavirus. For more details, see Díaz (2020); Gordon et al. (2020b), and Terracciano et al. (2021). However, each article reported on a small but significant proportion of pathways implicated in genome stability maintenance, DNA regulation, and chromatin organization. Taking into account previous evaluations of processes involved in brain-specific chromosome/genome instability (Jeppesen et al., 2011; Yurov et al., 2011; Bajic et al., 2015; Caneus et al., 2018; Martínez-Cué and Rueda, 2020; Heng et al., 2021), following candidate pathways (pathway clusters) were selected: cell cycle, cell death, centrosome, chromatin organization, DNA damage response, DNA regulation, DNA replication, DNA repair, ER stress, and nucleotide metabolism. The rates of chromosome and genome instability may increase with age being highly dependent on environmental factors (Iourov et al., 2020; Vorsanova et al., 2020). A viral infection may be such a factor (Heng et al., 2021). Therefore, according to interactomic data, SARS-CoV-2 interactions with proteins involved in the aforementioned pathways are able to initiate/stimulate genome and chromosome instability in neuronal cells. Figure 1 schematically depicts possible effects of SARS-CoV-2 infection on the brain in the context of aging-related genome (chromosome) instability.
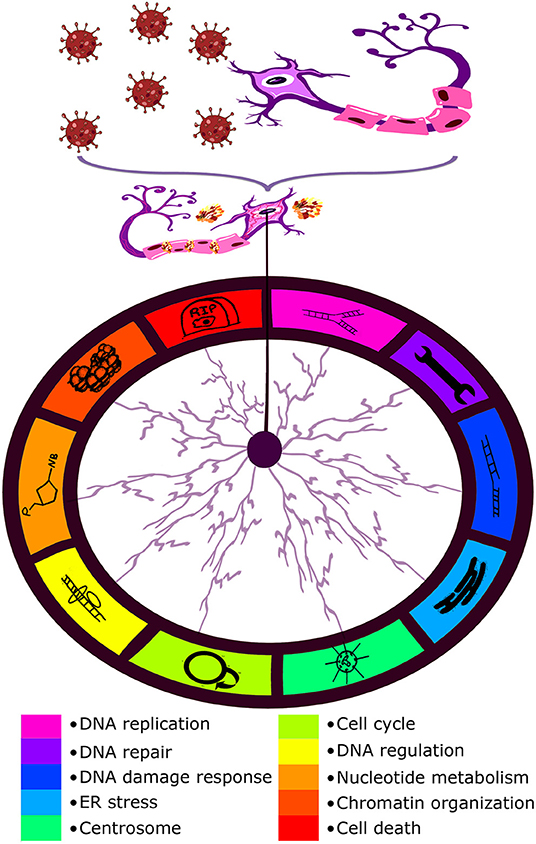
Figure 1. Schematic representation of the possible effect of SARS-CoV-2 infection (COVID-19) on the brain in the context of aging-related genome (chromosome) instability. Interactome analysis of SARS-CoV-2 infection has highlighted a number of pathways to be potentially altered by the virus which are listed at the bottom of the figure.
Brain-specific genomic variations [including aneuploidy (loss/gain of whole chromosomes) and single gene mutations] are associated with a wide spectrum of late-onset brain diseases (Yurov et al., 2010; Rohrback et al., 2018; Iourov et al., 2021). More importantly, chromosome instability mediates neurodegeneration (Iourov et al., 2009; Rohrback et al., 2018; Yurov et al., 2019). Several molecular pathways have been associated with chromosome instability in the neurodegenerating brain including neuronal cell cycle errors, chromosome missegregation, and cellular senescence (Bajic et al., 2015; Caneus et al., 2018; Martínez-Cué and Rueda, 2020). These processes are intimately linked to aging at molecular, cellular, and tissular levels. For instance, premature aging is associated with increased rates of chromosome and genome instability. Natural aging is associated with accumulation and propagation of somatic genome variations (e.g., aneuploidy) and genome instability. Alterations to genome stability maintenance may cause aging-related brain diseases or early manifestations of late-onset neurodegenerative diseases (Yurov et al., 2010; Andriani et al., 2017; Zhang and Vijg, 2018; Iourov et al., 2021). Additionally, DNA regulation and chromatin organization are able to affect genome stability by altering the expression of genes implicated in the pathways demonstrated in Figure 1. Since SARS-CoV-2 interactions with proteins involved in genome stability maintenance pathways are able to contribute to chromosome/genome instability propagation, the coronavirus infection has the potential to cause neurobehavioral abnormalities, neurodegeneration (e.g., Alzheimer's disease) and premature brain deterioration.
SARS-CoV-2 infection possesses an appreciable effect on the organism (Tu et al., 2020; Wu et al., 2020; Hu et al., 2021). Alterations to the central nervous system are observed in individuals with COVID-19 (Pennisi et al., 2020). Here, we express our opinion that SARS-CoV-2 may increase the risk for neurobehavioral alterations and neurodegeneration mediated by aging-related genome (chromosome) instability. Thus, SARS-CoV-2 is a potential risk factor for premature brain deterioration, Alzheimer's disease and other late-onset neurodegenerative diseases. The opinion is supported by addressing SARS-CoV-2-host interactomes. It is to note that chromosome instability mediating complex diseases and aging is not specific for brain diseases (Yurov et al., 2010; Iourov et al., 2019, 2020; Vorsanova et al., 2020). In other words, similar processes may occur in any tissue of an individual with SARS-CoV-2 infection. Additionally, other viruses are able to produce chromosome instability (Heng, 2019). Thus, one should be aware of the complications caused by chromosome and genome instability (e.g., cancer or tissue degeneration) in individuals affected by COVID-19 infection. Moreover, these studies seem to be especially important for those who went through a cytokine storm, since the latter may trigger tissue degeneration. Accordingly, molecular cytogenetic monitoring of chromosome and genome instability is warranted in individuals with SARS-CoV-2 infection to prevent genome instability-mediated and aging-dependent pathologies on time.
Author Contributions
II wrote the manuscript. Both authors conceived the idea and made theoretical contributions.
Funding
Authors were partially supported by RFBR and CITMA according to the Research Project No. 18-515-34005. The work was partially supported by the Government Assignment of the Russian Ministry of Science and Higher Education, Assignment No. AAAA-A19–119040490101-6 (Mental Health Research Center) and by the Government Assignment of the Russian Ministry of Health, Assignment No. 121031000238-1 (Veltischev Research and Clinical Institute for Pediatrics).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
Our communication was dedicated to Yuri Yurov and Ilia Soloviev. We express our sincere gratitude to Vasilisa Iurova for Figure 1.
References
Andriani, G. A., Vijg, J., and Montagna, C. (2017). Mechanisms and consequences of aneuploidy and chromosome instability in the aging brain. Mech. Ageing Dev. 161, 19–36. doi: 10.1016/j.mad.2016.03.007
Bajic, V., Spremo-Potparevic, B., Zivkovic, L., Isenovic, E. R., and Arendt, T. (2015). Cohesion and the aneuploid phenotype in Alzheimer's disease: a tale of genome instability. Neurosci. Biobehav. Rev. 55, 365–374. doi: 10.1016/j.neubiorev.2015.05.010
Caneus, J., Granic, A., Rademakers, R., Dickson, D. W., Coughlan, C. M., Chial, H. J., et al. (2018). Mitotic defects lead to neuronal aneuploidy and apoptosis in frontotemporal lobar degeneration caused by MAPT mutations. Mol. Biol. Cell. 29, 575–586. doi: 10.1091/mbc.E17-01-0031
Díaz, J. (2020). SARS-CoV-2 molecular network structure. Front. Physiol. 11 :870. doi: 10.3389/fphys.2020.00870
Gordon, D. E., Hiatt, J., Bouhaddou, M., Rezelj, V. V., Ulferts, S., Braberg, H., et al. (2020a). Comparative host-coronavirus protein interaction networks reveal pan-viral disease mechanisms. Science 370:eabe9403. doi: 10.1126/science.abe9403
Gordon, D. E., Jang, G. M., Bouhaddou, M., Xu, J., Obernier, K., White, K. M., et al. (2020b). A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 583, 459–468. doi: 10.1038/s41586-020-2286-9
Guzzi, P. H., Mercatelli, D., Ceraolo, C., and Giorgi, F. M. (2020). Master regulator analysis of the SARS-CoV-2/human interactome. J. Clin. Med. 9:982. doi: 10.3390/jcm9040982
Heng, E., Moy, A., Liu, G., Heng, H. H., and Zhang, K. (2021). ER stress and micronuclei cluster: stress response contributes to genome chaos in cancer. Front. Cell Dev. Biol. 9:673188. doi: 10.3389/fcell.2021.673188
Hu, B., Guo, H., Zhou, P., and Shi, Z. L. (2021). Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 19, 141–154. doi: 10.1038/s41579-020-00459-7
Iourov, I. Y., Vorsanova, S. G., Liehr, T., Kolotii, A. D., and Yurov, Y. B. (2009). Increased chromosome instability dramatically disrupts neural genome integrity and mediates cerebellar degeneration in the ataxia-telangiectasia brain. Hum. Mol. Genet. 18, 2656–2569. doi: 10.1093/hmg/ddp207
Iourov, I. Y., Vorsanova, S. G., Yurov, Y. B., and Kutsev, S. I. (2019). Ontogenetic and pathogenetic views on somatic chromosomal mosaicism. Genes. 10:379. doi: 10.3390/genes10050379
Iourov, I. Y., Vorsanova, S. G., Yurov, Y. B., Zelenova, M. A., Kurinnaia, O. S., Vasin, K. S., et al. (2020). The cytogenomic “theory of everything”: chromohelkosis may underlie chromosomal instability and mosaicism in disease and aging. Int. J. Mol. Sci. 21:8328. doi: 10.3390/ijms21218328
Iourov, I. Y., Yurov, Y. B., Vorsanova, S. G., and Kutsev, S. I. (2021). Chromosome instability, aging and brain diseases. Cells. 10:1256. doi: 10.3390/cells10051256
Jeppesen, D. K., Bohr, V. A., and Stevnsner, T. (2011). DNA repair deficiency in neurodegeneration. Prog. Neurobiol. 94, 166–200. doi: 10.1016/j.pneurobio.2011.04.013
Lee, S., Lee, Y. S., Choi, Y., Son, A., Park, Y., Lee, K. M., et al. (2021). The SARS-CoV-2 RNA interactome. Mol. Cell. 81, 2838–2850.e6. doi: 10.1016/j.molcel.2021.04.022
Martínez-Cué, C., and Rueda, N. (2020). Cellular senescence in neurodegenerative diseases. Front. Cell. Neurosci. 14:16. doi: 10.3389/fncel.2020.00016
Pennisi, M., Lanza, G., Falzone, L., Fisicaro, F., Ferri, R., and Bella, R. (2020). SARS-CoV-2 and the nervous system: from clinical features to molecular mechanisms. Int. J. Mol. Sci. 21:5475. doi: 10.3390/ijms21155475
Perrin-Cocon, L., Diaz, O., Jacquemin, C., Barthel, V., Ogire, E., Ramière, C., et al. (2020). The current landscape of coronavirus-host protein-protein interactions. J. Transl. Med. 18:319. doi: 10.1186/s12967-020-02480-z
Rohrback, S., Siddoway, B., Liu, C. S., and Chun, J. (2018). Genomic mosaicism in the developing and adult brain. Dev. Neurobiol. 78, 1026–1048. doi: 10.1002/dneu.22626
Schmidt, N., Lareau, C. A., Keshishian, H., Ganskih, S., Schneider, C., Hennig, T., et al. (2021). The SARS-CoV-2 RNA-protein interactome in infected human cells. Nat. Microbiol. 6, 339–353. doi: 10.1038/s41564-020-00846-z
Terracciano, R., Preianò, M., Fregola, A., Pelaia, C., Montalcini, T., and Savino, R. (2021). Mapping the SARS-CoV-2-host protein-protein interactome by affinity purification mass spectrometry and proximity-dependent biotin labeling: a rational and straightforward route to discover host-directed anti-SARS-CoV-2 therapeutics. Int. J. Mol. Sci. 22:532. doi: 10.3390/ijms22020532
Tu, Y. F., Chien, C. S., Yarmishyn, A. A., Lin, Y. Y., Luo, Y. H., Lin, Y. T., et al. (2020). A review of SARS-CoV-2 and the ongoing clinical trials. Int. J. Mol. Sci. 21:2657. doi: 10.3390/ijms21072657
Vorsanova, S. G., Yurov, Y. B., and Iourov, I. Y. (2020). Dynamic nature of somatic chromosomal mosaicism, genetic-environmental interactions and therapeutic opportunities in disease and aging. Mol. Cytogenet. 13:16. doi: 10.1186/s13039-020-00488-0
Wu, J. T., Leung, K., and Leung, G. M. (2020). Nowcasting and forecasting the potential domestic and international spread of the 2019-nCoV outbreak originating in Wuhan, China: a modelling study. Lancet. 395, 689–697. doi: 10.1016/S0140-6736(20)30260-9
Yurov, Y. B., Vorsanova, S. G., and Iourov, I. Y. (2010). Ontogenetic variation of the human genome. Curr. Genomics. 11, 420–425. doi: 10.2174/138920210793175958
Yurov, Y. B., Vorsanova, S. G., and Iourov, I. Y. (2011). The DNA replication stress hypothesis of Alzheimer's disease. ScientificWorldJournal. 11, 2602–2612. doi: 10.1100/2011/625690
Yurov, Y. B., Vorsanova, S. G., and Iourov, I. Y. (2019). Chromosome instability in the neurodegenerating brain. Front. Genet. 10:892. doi: 10.3389/fgene.2019.00892
Keywords: aging, brain, chromosome instability, COVID-19, neurodegeneration, genome instability, SARS-CoV-2
Citation: Iourov IY and Vorsanova SG (2022) COVID-19 and Aging-Related Genome (Chromosome) Instability in the Brain: Another Possible Time-Bomb of SARS-CoV-2 Infection. Front. Aging Neurosci. 14:786264. doi: 10.3389/fnagi.2022.786264
Received: 30 September 2021; Accepted: 31 January 2022;
Published: 03 March 2022.
Edited by:
Julia Fuchs, Institut National de la Santé et de la Recherche Médicale (INSERM), FranceReviewed by:
Henry H. Q. Heng, Wayne State University, United StatesCopyright © 2022 Iourov and Vorsanova. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ivan Y. Iourov, aXZhbi5pb3Vyb3ZAZ21haWwuY29t
 Ivan Y. Iourov
Ivan Y. Iourov Svetlana G. Vorsanova
Svetlana G. Vorsanova