
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Aging Neurosci., 16 February 2022
Sec. Cellular and Molecular Mechanisms of Brain-aging
Volume 14 - 2022 | https://doi.org/10.3389/fnagi.2022.785741
This article is part of the Research TopicExtracellular Vesicles in Age-Related Neurodegenerative Disease: Biological Mechanisms, Diagnostics, and TherapeuticsView all 6 articles
 Daisy Sproviero1
Daisy Sproviero1 Stella Gagliardi1*
Stella Gagliardi1* Susanna Zucca1,2
Susanna Zucca1,2 Maddalena Arigoni3
Maddalena Arigoni3 Marta Giannini1,4
Marta Giannini1,4 Maria Garofalo1,5
Maria Garofalo1,5 Valentina Fantini4,6
Valentina Fantini4,6 Orietta Pansarasa1
Orietta Pansarasa1 Micol Avenali4,7
Micol Avenali4,7 Matteo Cotta Ramusino4,8
Matteo Cotta Ramusino4,8 Luca Diamanti9
Luca Diamanti9 Brigida Minafra10
Brigida Minafra10 Giulia Perini4,8
Giulia Perini4,8 Roberta Zangaglia10
Roberta Zangaglia10 Alfredo Costa4,8
Alfredo Costa4,8 Mauro Ceroni4,8
Mauro Ceroni4,8 Raffaele A. Calogero3
Raffaele A. Calogero3 Cristina Cereda1
Cristina Cereda1Objectives: There is a lack of effective biomarkers for neurodegenerative diseases (NDs) such as Alzheimer's disease (AD), Parkinson's disease (PD), amyotrophic lateral sclerosis (ALS), and frontotemporal dementia. Extracellular vesicle (EV) RNA cargo can have an interesting potential as a non-invasive biomarker for NDs. However, the knowledge about the abundance of EV-mRNAs and their contribution to neurodegeneration is not clear.
Methods: Large and small EVs (LEVs and SEVs) were isolated from plasma of patients and healthy volunteers (control, CTR) by differential centrifugation and filtration, and RNA was extracted. Whole transcriptome was carried out using next generation sequencing (NGS).
Results: Coding RNA (i.e., mRNA) but not long non-coding RNAs (lncRNAs) in SEVs and LEVs of patients with ALS could be distinguished from healthy CTRs and from other NDs using the principal component analysis (PCA). Some mRNAs were found in commonly deregulated between SEVs of patients with ALS and frontotemporal dementia (FTD), and they were classified in mRNA processing and splicing pathways. In LEVs, instead, one mRNA and one antisense RNA (i.e., MAP3K7CL and AP003068.3) were found to be in common among ALS, FTD, and PD. No deregulated mRNAs were found in EVs of patients with AD.
Conclusion: Different RNA regulation occurs in LEVs and SEVs of NDs. mRNAs and lncRNAs are present in plasma-derived EVs of NDs, and there are common and specific transcripts that characterize LEVs and SEVs from the NDs considered in this study.
Disease-specific mechanisms for neurodegenerative diseases (NDs) like Alzheimer's disease (AD), Parkinson's disease (PD), amyotrophic lateral sclerosis (ALS), and frontotemporal dementia (FTD) are more commonly investigated for specific therapeutic and diagnostic targets (Habib, 2018). NDs have a complex multifactorial nature, and preventive interventions that simultaneously target multiple risk factors and disease mechanisms at an early stage of the diseases might be effective. Therefore, studying the shared mechanisms and common biomarkers can facilitate the development of a better understanding in the prevention of these diseases. In this context, extracellular vesicles [EVs; large EVs (LEVs) and small EVs (SEVs)] are considered intriguing biomarkers. LEVs are vesicles of 100–1,000 nm, shed by budding of the plasma membrane of cells in homeostasis but particularly during pathological conditions, whereas SEVs are vesicles of 30–150 nm, which are formed intracellularly and released by exocytosis of multivesicular bodies (Cocucci and Meldolesi, 2015; Théry et al., 2018). EVs are produced by all types of cells, and they can help the shuttling of proteins, microRNAs (miRNAs), and mRNA in the intercellular space and in different body fluids. There are several studies on the role of EV cargo in cerebrospinal fluid (CSF) samples of patients with NDs (Burgos et al., 2014; Gui et al., 2015; Thompson et al., 2020). However, due to the invasiveness and risk on CSF collection, plasma-derived EVs are being studied. For instance, it has been proved that EVs are potential carriers of misfolded toxic proteins, such as amyloid-beta (Aβ) peptide and tau in AD (Saman et al., 2012; Rajendran et al., 2014; Ghidoni et al., 2018), α-synuclein in PD (Stuendl et al., 2021), and TDP-43 in ALS and FTD (Iguchi et al., 2016; Sproviero et al., 2018, 2019). Increased levels of t-tau, p-tau, and Aβ42 in plasma/serum neurally derived blood exosomes were demonstrated to be an early biomarker for AD and cognitive decline progression (Fiandaca et al., 2015). Moreover, several synaptic proteins were found reduced in blood-derived exosomes from patients with AD and FTD (Goetzl et al., 2015, 2018). Large EVs of patients with ALS carry more SOD1, TDP-43, and FUS compared with those of healthy control (CTR) (Sproviero et al., 2018). It has been reported that EVs are also loaded with different kinds of RNAs involved in intercellular communication. In general, EVs can transport protein coding RNA (i.e., mRNA) and non-coding species of RNA like miRNAs, t-RNA, rRNA, long non-coding RNAs (lncRNA), and piwi-interacting RNA (piRNA) (O'Brien et al., 2020). It is known that RNA metabolism is a common factor in the pathogenesis of NDs, and numerous new studies have been investigating the role of miRNAs in EVs of NDs (Liu et al., 2017; Jiang et al., 2019; Soares Martins et al., 2021). On this subject, we have recently demonstrated that plasma-derived SEVs and LEVs from AD, PD, ALS, and FTD carry different miRNAs and have a common signature among the four NDs (Sproviero et al., 2021). However, there is no evidence in the literature of the transcriptomic analysis of mRNAs and lncRNAs in EVs from NDs. The aim of this study was to fill this gap and describe mRNA and lncRNA involvement in LEVs and SEVs from plasma of patients with NDs.
Patients were all recruited at the IRCCS Mondino Foundation, Pavia (Italy), and they signed an informed consent form approved by the Ethical Committee (for patients with ALS, Protocol n-20180034329; for patients with PD, Protocol n-20170001758; for patients with AD, Protocol n-20170016071; and for patients with FTD, Protocol n-20180049077). Six patients with AD, 9 patients with PD, 6 patients with sporadic ALS (SALS), and 9 patients with FTD were involved (Table 1). All patients were screened for mutations using next generation sequencing (Sure Select QXT Target Enrichment, Agilent Technology), and they did not carry genetic mutations. Diagnosis of AD was based on criteria expressed by Aging-Alzheimer's Association workgroups (McKhann et al., 2011). Extensive neuropsychological evaluation, 3T-MRI, and a 3-year follow-up were performed. Clinical diagnostic criteria of the Movement Disorder Society (MDS) were used for patients with PD; bradykinesia plus rest tremor or rigidity were taken in account. Rascovsky criteria were followed for patients with FTD (Rascovsky et al., 2011; Postuma et al., 2015). Only patients fulfilling clinical criteria for behavioral variant FTD (bv-FTD) or motor neuron disease (MND) were selected. ALS diagnosis was made according to the revised El Escorial Criteria (Brooks et al., 2000). Further clinical characteristics are listed in Tables 3–7 of the previous publication (Sproviero et al., 2021).
Six age-matched healthy volunteers, free from any pharmacological treatment, were recruited and used as healthy CTRs. CTRs were recruited at the Immunohematological and Transfusional Service, IRCCS Foundation “San Matteo,” Pavia (Italy).
Venous blood (15 ml) was collected in sodium citrate tubes from all patients and CTRs and processed as previously described (Sproviero et al., 2018, 2019, 2021; Morasso et al., 2020). In brief, platelet-free plasma was centrifuged at 20,000 × g for 1 h. The pellet was washed in 0.2 μm filter and filtered with 1 × PBS (Sigma-Aldrich, Italy). The supernatant of LEVs was filtered through a 0.2 μm filter and spun in an Optima MAX-TL Ultracentrifuge at 100,000 × g for 1 h at 4°C, and the SEV pellet was washed with 1 ml of filtered 1 × PBS. The purity of LEVs and SEVs was confirmed by the Western blot (WB) analysis (using anti-Annexin V antibody-Santa Cruz Biotechnology, USA, for LEVs and anti-Alix antibody-Abcam, Cambridge, UK, for SEVs), nanotracking particle analysis (NTA), and transmission electron microscopy (TEM) as we described previously (Sproviero et al., 2021).
RNA was extracted from LEV and SEV fractions using the Qiagen miRNeasy Mini kit (Qiagen, Germany), according to the instructions of the manufacturer. RNA was quantified using a Nanodrop ND-100 Spectrophotometer (Nanodrop Technologies, Wilmington, USA) and a 2100 Bioanalyzer (Agilent RNA 6000 Nano Kit, Waldbronn, Germany). The RNA quantity used was ~200ng.
Long RNA libraries (i.e., mRNAs and lncRNAs) were prepared using the Illumina TruSeq Stranded RNA Library Prep (Illumina, USA). Sequencing (i.e., nts paired-end) was performed on the Illumina NextSeq500 system (Illumina, USA).
The raw bcl files were converted into demultiplexed fastq files with bcl2fastq (Illumina, USA) implemented in the docker4seq package (Kulkarni et al., 2018). For the row count analysis, only transcripts with counts above five were considered (see Supplementary Figure 1). No relevant difference among counts in SEVs and LEVs in the four diseases emerged. The gene and isoform quantification was performed as previously described (Gagliardi et al., 2018; Zucca et al., 2019). The differential expression analysis for mRNAs was performed using the R package EBSeq (Langfelder and Horvath, 2008). Coding and non-coding genes were considered differentially expressed and retained for further analysis with |log2(disease sample/healthy control)| ≥ 1 and an FDR ≤ 0.1. We imposed minimum |Log2FC| of 1 and an FDR lower than 0.1 as thresholds to differentially expressed genes (DEGs). To understand common RNAs in the four diseases, we calculated the intersection of deregulated mRNAs and lncRNAs compared with CTRs with http://bioinformatics.psb.ugent.be/webtools/Venn/. The datasets generated and analyzed during this study are available in the NCBI GEO repository [GSE155700].
The gene enrichment analysis was performed on coding genes with the KEGG pathway analysis (Kyoto Encyclopedia of Genes and Genomes, http://www.genome.ad.jp/kegg) and enrichR web tool (Chen et al., 2013; Kuleshov et al., 2016). Only pathways with p < 0.05 are considered statistically significant.
The statistical analysis was carried out using the R Studio software. The Shapiro-Wilk test was used to test variables for normality distribution, and the Levene test was used to test the assumption of homogeneity. Categorical variables were tested using the non-parametric Kruskal-Wallis with Dunn's multiple comparison test, and a value of p < 0.05 was considered significant.
Large extracellular vesicles (LEVs) and SEVs were separated by differential centrifugation. NTA, WB analysis, and TEM were carried out (Supplementary Figure 2) to establish the purity of EVs. The concentration and diameter size of LEVs and SEVs of CTRs and patients are shown in Supplementary Figure 3. The diameter of LEVs in patients with ALS is enhanced compared with that of CTRs (Dunn's test, p < 0.001; Supplementary Figure 3A). The diameter of SEVs among the four NDs had a slight difference (Supplementary Figure 3C). Only patients with PD showed more LEVs compared with CTRs and other groups (*p < 0.05; Supplementary Figure 3B). The number of SEVs of ALS and FTD was enhanced compared with that in CTRs (i.e., Dunn's test, **p < 0.01, and ****p < 0.001; Supplementary Figure 3D). These data have to be confirmed with more patients and CTRs.
Differentially expressed mRNA (DE mRNA) and DE lncRNAs between LEVs and SEVs of the 4 NDs (i.e., AD, PD, ALS, and FTD) and the healthy CTRs were calculated, and the results are shown in Table 2 and Supplementary Table 1. SEVs from patients with ALS and FTD were more enriched in DE mRNAs compared with those in LEVs. As shown in Table 2, LEVs and SEVs from patients with ALS and FTD were equally enriched in lncRNAs. However, no DE mRNAs and lncRNAs were found in plasma-derived EVs from patients with AD, and only 12 DE mRNAs and 1 lncRNA were found in LEVs from plasma of patients with PD.
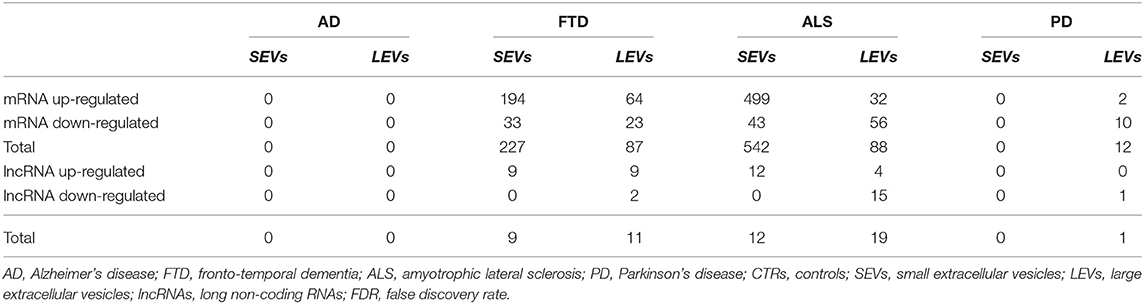
Table 2. Statistically significant differentially expressed RNAs number in small extracellular vesicles (SEVs) and large extracellular vesicles (LEVs) from patients with Alzheimer's disease (AD), frontotemporal dementia (FTD), amyotrophic lateral sclerosis (ALS), and Parkinson's disease (PD) in terms of upregulated transcripts, downregulated transcripts, and total compared with CTRs. Transcripts were considered as differentially expressed when |log2(disease sample/healthy control)|≥1 and an FDR ≤ 0.1.
We have largely demonstrated significant differences between LEVs and SEVs derived from plasma for dimension, markers, protein, and miRNA loading (Sproviero et al., 2018, 2019, 2021). As reported in our previous work (Sproviero et al., 2021), miRNAs can sort differently in LEVs and SEVs of the same disease. So, we moved to investigate the number of different and common deregulated mRNAs and lncRNAs that sort into SEVs and LEVs in the same disease. We could only calculate the intersection between SEVs and LEVs for ALS and FTD, since there were no deregulated mRNAs (with a Log2FC| of 1) in EVs of patients with AD and in SEVs of patients with PD compared with those in CTRs. For FTD, of the 228 mRNAs in SEVs and 114 in LEVs, 39 mRNAs were in common, 36 were upregulated, and 3 were downregulated (Figure 1A). For ALS, of the 522 mRNA in SEVs and 124 in LEVs, 44 were in common (i.e., 33 upregulated and 11 downregulated; Figure 1B). The percentage of common RNAs between SEVs and LEVs are shown in Table 3. For FTD, of the 9 lncRNA in SEVs and 11 in LEVs, 6 were in common (Figure 2A). For ALS, of the 12 lncRNA in SEVs and 19 in LEVs, 4 were in common (Figure 2B).
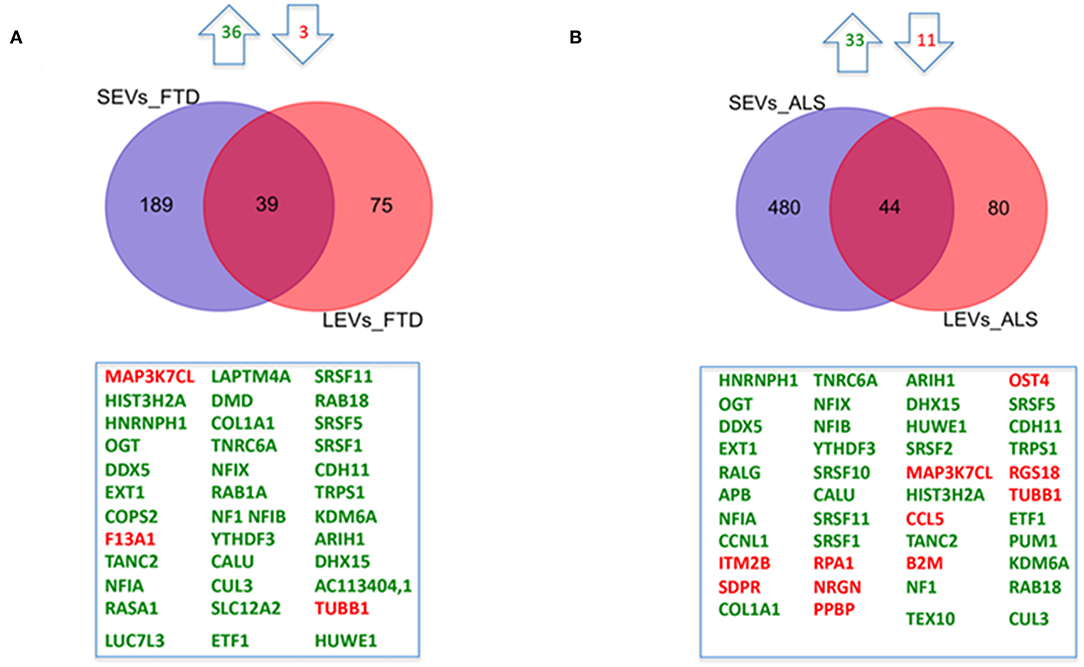
Figure 1. Common packaging of deregulated mRNAs into small extracellular vesicles (SEVs) and large extracellular vesicles (LEVs) from patients with frontotemporal dementia (FTD) and amyotrophic lateral sclerosis (ALS). (A) For FTD, of the 228 mRNA in SEVs and 114 in LEVs, 39 were in common (i.e., 36 upregulated, green, and 3 downregulated, red). (B) For ALS, of the 522 mRNA in SEVs and 124 in LEVs, 44 were in common (i.e., 33 upregulated and 11 downregulated). Differential mRNA expression analysis was carried out using DESeq2 (log2FC > 1, p < 0.05).
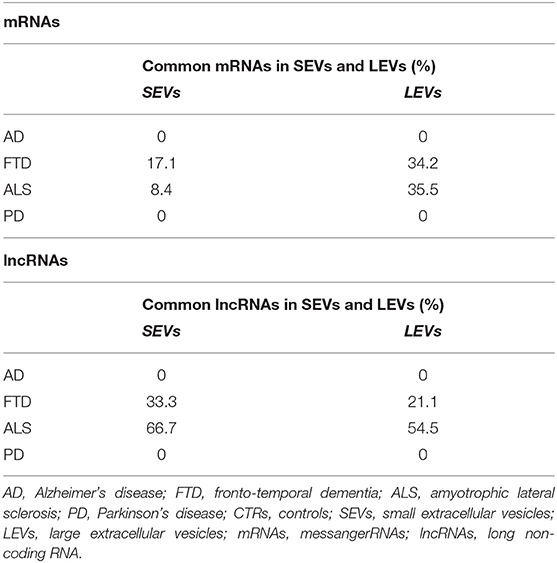
Table 3. Percentage of common mRNAs and long non-coding RNAs (lncRNAs) in SEVs and LEVs in AD, FTD, ALS, and PD.
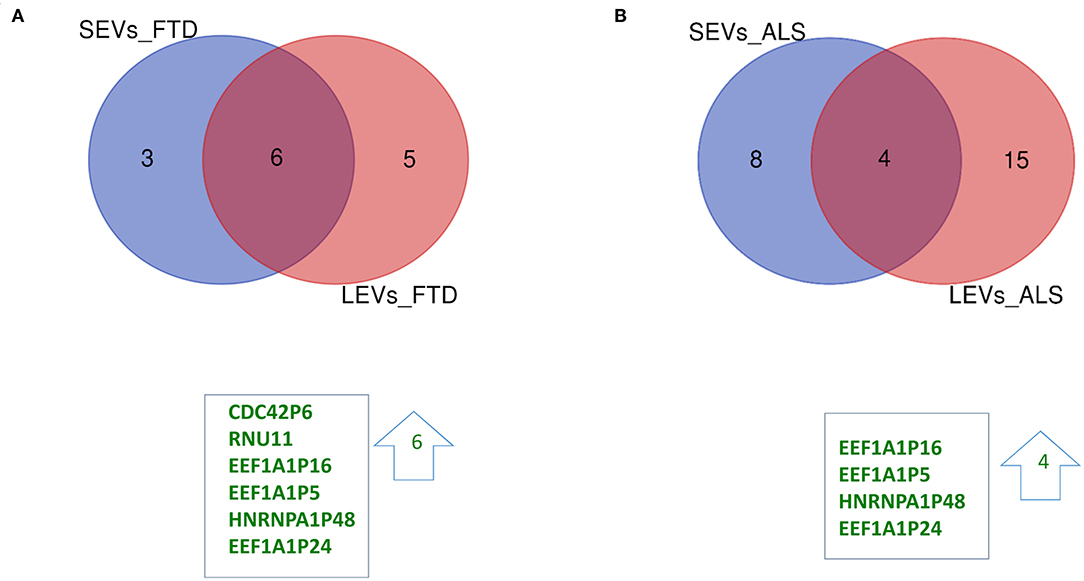
Figure 2. Common packaging of deregulated long non-coding RNAs (lncRNAs) and pseudogenes into SEVs and LEVs from FTD and ALS patients. (A) For FTD, of the 9 lncRNAs in SEVs and 11 in LEVs, 6 were in common (all upregulated, green). (B) For ALS, of the 12 lncRNA in SEVs and 19 in LEVs, 4 were in common (all upregulated, green). Differential lncRNA expression analysis was carried out using DESeq2 (log2FC> 1, p < 0.05).
Differentially expressed mRNAs and lncRNAs in SEVs and LEVs of the four groups (Supplementary Table 1) were analyzed, and the principal component analysis (PCA) was carried out. DE mRNAs and lncRNAs in SEVs and LEVs showed that patients with ALS are well-divided from CTRs (Figures 3, 4), showing a partial overlap with FTD. Common DE mRNAs and lncRNAs between LEVs or SEVs of the four diseases were investigated. In SEVs from patients with FTD and ALS, there were 209 mRNA and 9 lncRNAs in common, and the common pathways of gene ontology biological processes (GO-BPs) were mRNA processing (p = 6.133e-15), RNA splicing, via transesterification reactions, and via spliceosome (Figures 5A, 6A). In LEVs, there was only 1 mRNA MAP3K7CL, a kinase gene, and one antisense, AP003068.3, which are in common among PD, FTD, and ALS (Figures 5B, 6B).
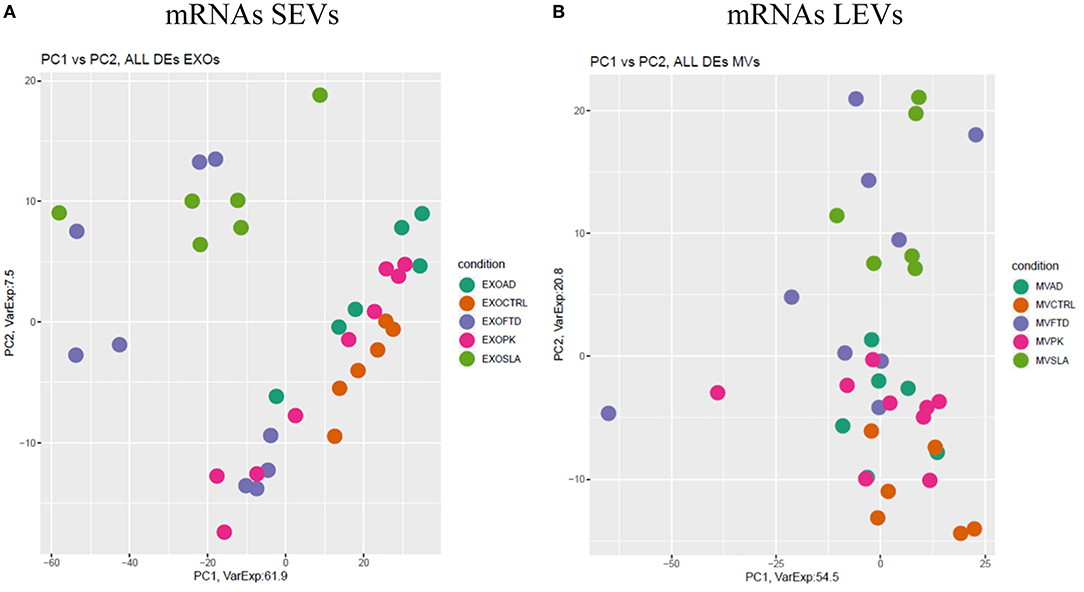
Figure 3. Principal component analysis (PCA) of coding genes differentially expressed in SEVs (A) and LEVs (B) of patients with AD, FTD, ALS, and PD and healthy controls (CTRs). PCA is performed using all the differentially expressed coding genes in at least one disease as predictors in the comparison of each disease to the control state. Each dot represents a sample, and each color represents a disease.
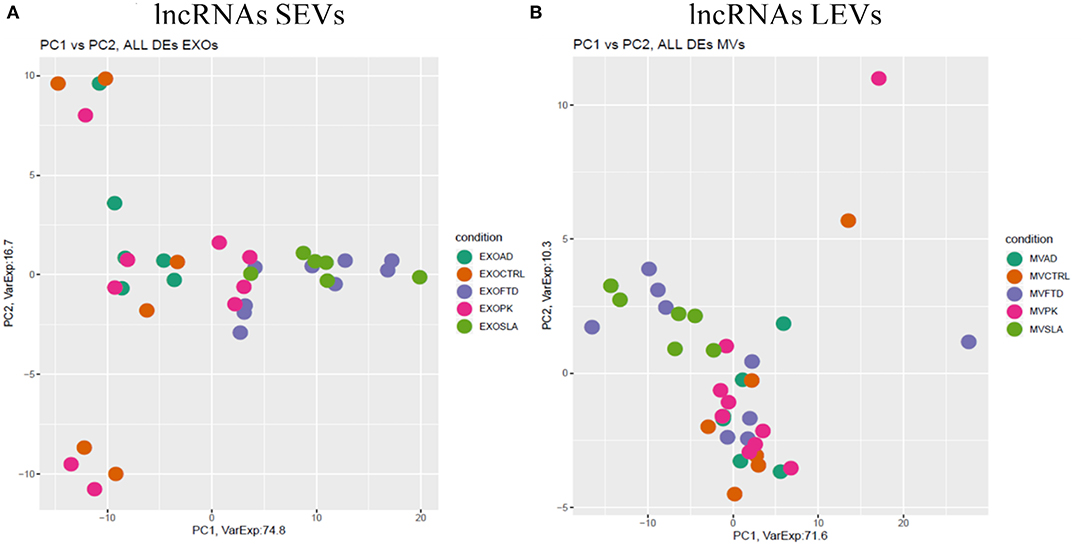
Figure 4. PCA of long non-coding genes differentially expressed in SEVs (A) and LEVs (B) of patients with AD, FTD, ALS, and PD and healthy CTRs. PCA is performed using all DE long non-coding genes in at least one disease as predictors in the comparison of each disease to the control state. Each dot represents a sample, and each color represents a disease. Considering only lncRNAs, both SEVs and LEVs showed a mixed scenario between patients and controls, without any specific characterization of AD, FTD, ALS, or PD.
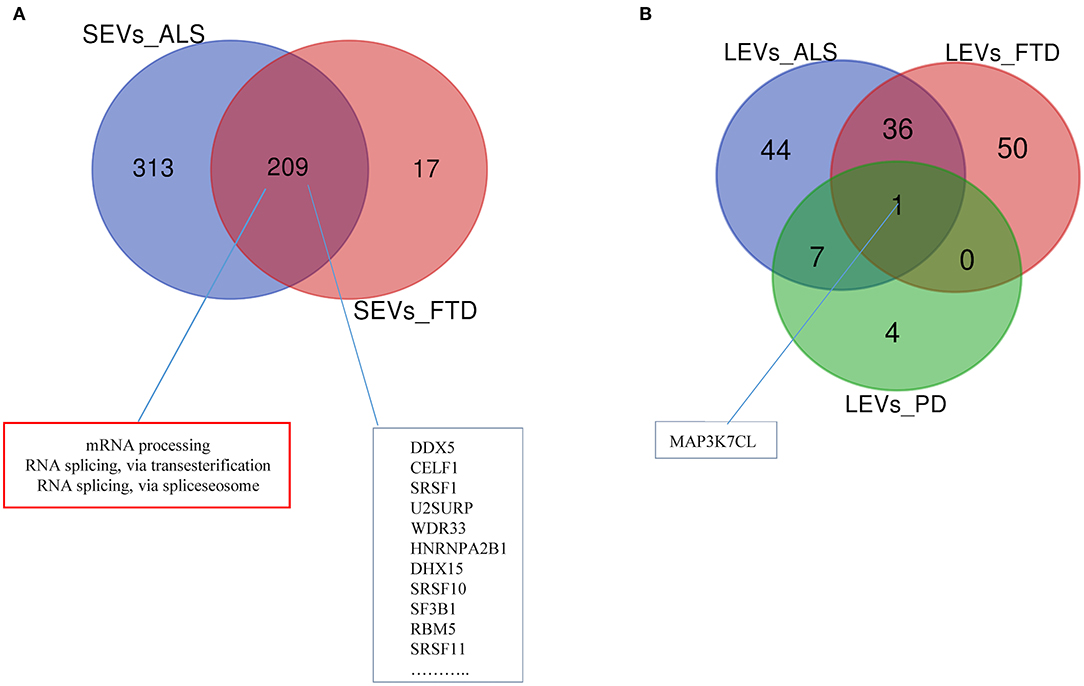
Figure 5. Venn diagram showing numbers of common and unique RNA in SEVs from plasma of patients with ALS and FTD (A) and in LEVs from plasma of ALS, FTD, and PD (B). Common mRNAs and pathways are listed. Differential mRNA expression analysis was carried out using DESeq2 (log2FC> 1, p < 0.05).
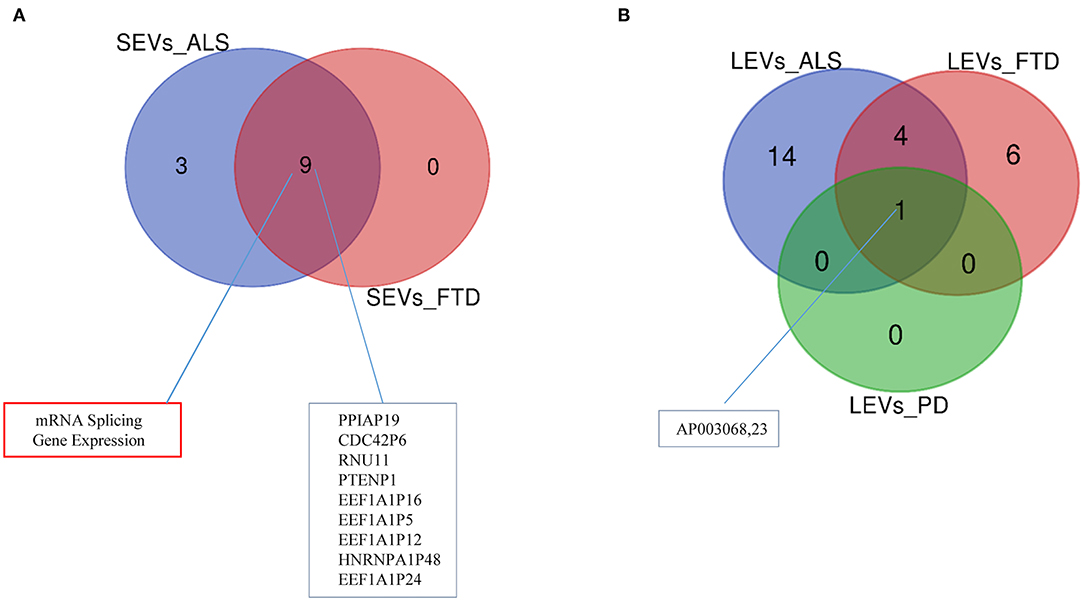
Figure 6. Venn diagram showing numbers of common and unique lncRNAs and pseudogenes in SEVs from plasma of patients with ALS and FTD (A) and in LEVs (B) from plasma of ALS, FTD, and PD. Common lncRNAs and pathways are listed. Differential mRNA expression analysis was carried out using DESeq2 (log2FC > 1, p < 0.05).
The EnrichR analysis (i.e., KEGG pathway and Gene Ontology analysis) for DE mRNA in patients with FTD and ALS compared with that in healthy CTRs has been performed (Supplementary Table 2). GO-BPs identified (1) positive regulation of transmembrane transport and of leukocyte chemotaxis in LEVs from patients with ALS; (2) cellular response to molecule of bacterial origin in LEVs from patients with FTD; (3) positive regulation of mitochondrial membrane permeability involved in apoptotic process and mitochondrial outer membrane permeabilization in LEVs from patients with PD; (4) regulation of transcription from an RNA polymerase II promoter, regulation of transcription, DNA-templated, and negative regulation of gene expression in SEVs from ALS; (5) regulation of cellular catabolic process, phosphatidylinositol metabolic process, and transcription initiation from RNA polymerase III promoted in SEVs from patients with FTD (Supplementary Table 2).
As widely reported in the literature, EVs can transport full-length and fragmented RNAs (Skog et al., 2008). Protein-coding RNAs are synthesized in the nucleus; they undergo splicing, are modified at the ends, and are exported to the cytosol, where they can be released by EVs from the cell. The transcriptomic analysis supported the view that mRNA from the parent cell can be translated in the recipient cell to generate a functional protein. For example, it was found that 17% of the total mRNAs found in EVs were not detected in parental cells, and more than 11% of mRNAs were preferentially included in EVs compared with mRNAs found in cells (Skog et al., 2008).
Long non-coding RNAs, defined as >200 nt transcripts that are not translated into proteins, are involved in different biological processes such as chromatin organization, gene transcription, mRNA turnover, protein translation, and assembly of macromolecular complexes. Numerous lncRNAs have been found in EVs. For example, MALAT1, HOTAIR, lincRNAp21, GAS5, TUG1, and ncRNA-CCND1 were identified in EVs derived from human cervical and breast carcinomas (Gezer et al., 2014).
In this study, we have analyzed SEV and LEV cargo to detect RNAs acting as novel, easily accessible biomarkers for AD, PD, ALS, and FTD. We first compared DE mRNAs and lncRNAs between SEVs and LEVs in the same disease. We only found a partial overlap between SEVs and LEVs for mRNAs (Table 3). We could not evaluate the overlap between DE mRNAs of SEVs and LEVs of AD and PD, since we did not find any DE mRNAs in patients with AD and only few in LEVs of patients with PD. Although there is some overlap between the two types of EVs, there is a significant difference. This could be justified by the different functions of LEVs and SEVs in plasma of patients with ND, as we previously described for dimension, protein, miRNAs, and lipid loading (Sproviero et al., 2018, 2019, 2021). In diseases like FTD and ALS, SEVs were enriched with a greater number of DE mRNAs compared with LEVs. FTD and ALS are linked clinically, pathologically, and mechanistically. Mutations in TDP-43 and FUS/TLS were discovered to be common in both diseases. Moreover, TDP-43 was found to be the major ubiquitinated protein found in both patients with ALS and FTD (Arai et al., 2006; Neumann et al., 2006). These findings highlighted the dysfunctions in RNA metabolism and multiple RNA processing steps as central pathogenic pathways in ALS and FTD (Kwiatkowski et al., 2009; Lagier-Tourenne et al., 2010). However, many ALS/FTD-linked proteins are involved in the autophagy pathways (like ubiquilin-2-UBQLN2-, p62/SQSTM1, and optineurin-OPTN-) and in the endosomal sorting complex (like CHMP2B, a core component of endosomal sorting complex required for transport (ESCRT) complexes) (Ling et al., 2013). Being the exosomal pathway linked to autophagy and to the endolysosomal pathway (Buratta et al., 2020), this might justify the enhanced deregulation of RNA observed in SEVs.
We then analyzed DE mRNAs among the four NDs by the PCA. We found that DE mRNA cargo from ALS and FTD was different from CTRs, particularly, in SEVs, while in LEVs, the only group that did not overlap with CTRs was ALS. PCA related to lncRNAs in LEVs and SEVs could only distinguish ALS from CTRs.
Common genes in SEVs of ALS and FTD were mainly classified as belonging to splicing, a mechanism largely described in the literature for these two NDs (Ito et al., 2017). For example, we found upregulation of serine arginine-rich protein family (SRs), including SRSF7, SRSF3, SRSF1, SRSF2, SRSF11, and SRSF10 in SEVs from patients with ALS and SRSF1, SRSF11, SRSF2, SRSF5, and SRSF10 in SEVs from patients with FTD. SRs are serine/arginine-rich splicing factors that are formed by an N-terminal 1 or 2 RNA recognition domains (RRM domain) and a C-terminal domain enriched with arginine (R) and serine (S) amino acid sequences (RS domain). SR proteins are involved in RNA metabolism and constitutive or alternative splicing, since they bind splicing enhancers [i.e., exonic splicing enhancer (ESE)] and recruit small nuclear RNP (snRNP) (Zheng et al., 2020).
Moreover, upregulation of HNRNPA2B1, Matrin3, and HNRNPA1P48 in SEVs of both patients with ALS and FTD, as well as TARDBP, identified ALS-FTD-related RNA-binding proteins, characteristically deposited in the affected regions of the brain of ALS/FTD (Ling et al., 2013).
One common gene, MAP3K7CL (a kinase gene), and one antisense, AP003068.3, were identified among LEVs of PD, ALS, and FTD. Not much is known about their role in neurodegeneration. Merienne and collaborators identified highly selective transcriptomic signatures in adult mouse striatal direct and indirect spiny projection neurons (SPNs), astrocytes, and microglia (Merienne et al., 2019). MAP3K7CL was one of the genes identified in SPNs of the striatonigral. The loss of striatal efferent neurons has been described in Huntington disease and X-linked recessive dystonia parkinsonism (Reiner et al., 1988; Goto et al., 2005); however, striatal efferent projections can be involved in the TDP-43-related FTLD/ALS disease (Riku et al., 2016).
We found 36 mRNAs and 4 non-coding RNAs in common between LEVs from ALS and FTD. mRNAs are involved in splicing and gene transcription. Even if common RNAs in SEVs and LEVs are both involved in splicing, only 15 mRNAs (i.e., CUL3, HIST3H2A, SRSF11, RAB18, SRSF1, DDX5, TRPS1, NFIA, ARIH1, DHX15, NFIX TUBB1, NF1, NFIB, and HUWE1) are shared, meaning that a specific common signature can be described for the two EV subpopulations.
One striking result from our RNA-Seq analysis is the small number of DE genes found in PD LEVs and the lack of DE genes and lncRNAs/pseudogenes in patients with AD and in SEVs from patients with PD compared with CTRLs. There are existing studies on plasma-derived EVs of patients with AD and their miRNA cargo analyzed with the high-throughput sequencing technology. Three studies confirmed the deregulation of miRNA-342-3p in plasma-derived exosomes from patients with AD (Tan et al., 2014; Lugli et al., 2015; Rani et al., 2017) compared with that in healthy donors. Downregulation of miRNA-21-5p, miRNA-451a (Gámez-Valero et al., 2019), and miRNA-132 and miRNA-212 (Cha et al., 2019) was found in exosomes of patients with AD compared with that in patients with DLB and healthy donors. Other works reported the presence of specific miRNAs in EVs from periphery of AD (Cheng et al., 2020; Thomas et al., 2020). We previously demonstrated a moderate presence of deregulated miRNAs in EVs from patients with PD and AD (Sproviero et al., 2021). Kim et al. (2020) reported a significant enrichment in mitochondrial RNAs in EVs isolated from blood of AD groups; however, there is no mention of DE coding and other non-coding RNAs (Kim et al., 2020).
Specific pathways are also described for each disease, and different DE genes are recognized in LEVs and SEVs of the same disease. Further work is needed to unravel specific signature for each disease.
Our previous work showed a different grade of RNA metabolism involvement in AD, ALS, and PD by investigating mRNAs and lncRNA in peripheral blood mononuclear cells (PBMC). Specifically, we found a great difference in the amount of DE mRNAs in the three diseases (Garofalo et al., 2020). These latter data are in line with the results described in this article, taking in account that patients with FTD were not included in the PBMC study. Further studies are needed on extended cohorts of patients with different severity of the disease to understand if the deregulation of these RNAs may be associated to a specific clinical window (i.e., disease onset and outcome) and progression of the disease.
This study may improve our understanding of mechanisms underlying neurodegeneration overlap, and it might help to develop diagnostics based on detection of RNAs in blood of patients.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
The studies involving human participants were reviewed and approved by the Ethical Committee (for ALS patients Protocol n°-20180034329; for PD patients Protocol n°20170001758; for AD patients Protocol n°20170016071; and for FTD patients Protocol n°20180049077). The patients/participants provided their written informed consent to participate in this study.
DS, SG, and SZ wrote the manuscript. DS, MAr, MGi, MGa, and VF performed the experiments. RC and SZ performed bioinformatic analysis. AC, BM, GP, MAv, LD, RZ, MC, and MR participated in patients and controls recruitment. MAr, DS, OP, and CC set up the experimental plan. OP, RC, and CC reviewed the manuscript. RC and CC supervised this work. All authors reviewed and accepted the final version of this manuscript.
This research was funded by Italian Ministry of Health (Grant N°5*1000 anno 2016, Ricerca Corrente 2018–2020, Young research project GR-2016-02361552, AIFA-co-ALS); EuroNanoMed III JTC 2018; Fondazione Regionale per la Ricerca Biomedica for TRANS–ALS (FRRB 2015-0023); and Fondazione Cariplo 2017 (Extracellular vesicles in the pathogenesis of Frontotemporal Dementia 2017-0747 and Association between frailty trajectories and biological markers of aging 2017-0557).
SZ was employed by EnGenome SRL.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank all patients and their families. We also thank Prof. Fabio Corsi and Mr. Raffaele Allevi (Dipartimento di Scienze Biomediche e Cliniche L. Sacco) for TEM images.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnagi.2022.785741/full#supplementary-material
Supplementary Figure 1. Row counts for small and large extracellular vesicles. (A) Only transcripts with counts above five were considered. (B) No relevant difference between count in SEVs and LEVs in four diseases emerged.
Supplementary Figure 2. Large extracellular vesicles (LEVs) and small extracellular vesicles (SEVs) characterization. (A) Nanosight profile of LEVs and SEVs from plasma of a control (CTR); (B) Representative images obtained by transmission electron microscopy (TEM) of LEVs and SEVs from plasma (Scale bar: 100 nm). (C) Western blot of LEVs and SEVs markers in LEVs and SEVs samples from two CTRs showed the presence of Annexin V only in the LEV pellet and more Alix presence in SEV fraction.
Supplementary Figure 3. Mean diameter and particle count concentration for LEVs and SEVs of the four NDs and CTRs. NTA was performed on LEVs and SEVs extracted from plasma of healthy controls and neurodegenerative diseases (NDs). The mean diameter size and count of LEVs (A,B) and SEVs (C,D) of CTRs and patients are reported in the boxplots (Dunn's test, *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001) (D). These data have to be confirmed with more patients and controls.
Supplementary Table 1. Differentially expressed RNAs in ALS, frontotemporal dementia (FTD), Parkinson's disease (PD), and Alzheimer's disease (AD) groups respect to healthy controls. Transcript ID, gene name, gene type, gene status, measured log2FC, and false discovery rate (FDR) are reported for each transcript. Only transcripts with | log2 (disease sample/healthy control)|≥1 and an FDR ≤ 0.1 are shown.
Supplementary Table 2. Specific mRNAs pathways in ALS, FTD, PD and AD groups. Deregulated mRNAs specific of each disease were analyzed using the EnrichR web tool. KEGG and Gene Ontology Biological Processes with a p < 0.05 are listed.
Arai, T., Hasegawa, M., Akiyama, H., Ikeda, K., Nonaka, T., Mori, H., et al. (2006). TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem. Biophys. Res. Commun. 351, 602–611. doi: 10.1016/j.bbrc.2006.10.093
Brooks, B. R., Miller, R. G., Swash, M., and Munsat, T. L. (2000). El Escorial revisited: revised criteria for the diagnosis of amyotrophic lateral sclerosis. Amyotroph. Lateral Scler. 1, 293–299. doi: 10.1080/146608200300079536
Buratta, S., Tancini, B., Sagini, K., Delo, F., Chiaradia, E., Urbanelli, L., et al. (2020). Lysosomal exocytosis, exosome release and secretory autophagy: The autophagic- and endo-lysosomal systems go extracellular. Int. J. Mol. Sci. 21, 2576. doi: 10.3390/ijms21072576
Burgos, K., Malenica, I., Metpally, R., Courtright, A., Rakela, B., Beach, T., et al. (2014). Profiles of extracellular miRNA in cerebrospinal fluid and serum from patients with Alzheimer's and Parkinson's diseases correlate with disease status and features of pathology. PLoS ONE 9, e94839. doi: 10.1371/journal.pone.0094839
Cha, D. J., Mengel, D., Mustapic, M., Liu, W., Selkoe, D. J., Kapogiannis, D., et al. (2019). miR-212 and miR-132 are downregulated in neurally derived plasma exosomes of Alzheimer's patients. Front. Neurosci. 13, 1208. doi: 10.3389/fnins.2019.01208
Chen, E. Y., Tan, C. M., Kou, Y., Duan, Q., Wang, Z., Meirelles, G. V., et al. (2013). Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 128, 14. doi: 10.1186/1471-2105-14-128
Cheng, L., Vella, L. J., Barnham, K. J., McLean, C., Masters, C. L., and Hill, A. F. (2020). Small RNA fingerprinting of Alzheimer's disease frontal cortex extracellular vesicles and their comparison with peripheral extracellular vesicles. J. Extracell. Vesicles. 9, 1766822. doi: 10.1080/20013078.2020.1766822
Cocucci, E., and Meldolesi, J. (2015). Ectosomes and exosomes: shedding the confusion between extracellular vesicles. Trends Cell Biol. 25, 364–372. doi: 10.1016/j.tcb.2015.01.004
Fiandaca, M. S., Kapogiannis, D., Mapstone, M., Boxer, A., Eitan, E., Schwartz, J. B., et al. (2015). Identification of preclinical Alzheimer's disease by a profile of pathogenic proteins in neurally derived blood exosomes: a case-control study. Alzheimers Dement. 11, 600.e1–607.e1. doi: 10.1016/j.jalz.2014.06.008
Gagliardi, S., Zucca, S., Pandini, C., Diamanti, L., Bordoni, M., Sproviero, D., et al. (2018). Long non-coding and coding RNAs characterization in peripheral blood mononuclear cells and spinal cord from amyotrophic lateral sclerosis patients. Sci. Rep. 8, 2378. doi: 10.1038/s41598-018-20679-5
Gámez-Valero, A., Campdelacreu, J., Vilas, D., Ispierto, L., Reñé, R, Alvarez, R., et al. (2019). Exploratory study on microRNA profiles from plasma-derived extracellular vesicles in Alzheimer's disease and dementia with Lewy bodies. Transl. Neurodegener. 8, 31. doi: 10.1186/s40035-019-0169-5
Garofalo, M., Pandini, C., Bordoni, M., Pansarasa, O., Rey, F., Costa, A., et al. (2020). Alzheimer's, Parkinson's disease and amyotrophic lateral sclerosis gene expression patterns divergence reveals different grade of RNA metabolism involvement. Int. J. Mol. Sci. 21, 9500. doi: 10.3390/ijms21249500
Gezer, U., Ozgur, E., Cetinkaya, M., Isin, M., and Dalay, N. (2014). Long non-coding RNAs with low expression levels in cells are enriched in secreted exosomes. Cell Biol. Int. 38,1076–1079. doi: 10.1002/cbin.10301
Ghidoni, R., Squitti, R., Siotto, M., and Benussi, L. (2018). Innovative biomarkers for Alzheimer's disease: focus on the hidden disease biomarkers. J. Alzheimers Dis. 62, 1507–1518. doi: 10.3233/JAD-170953
Goetzl, E. J., Abner, E. L., Jicha, G. A., Kapogiannis, D., and Schwartz, J. B. (2018). Declining levels of functionally specialized synaptic proteins in plasma neuronal exosomes with progression of Alzheimer's disease. FASEB J. 32, 888–893. doi: 10.1096/fj.201700731R
Goetzl, E. J., Boxer, A., Schwartz, J. B., Abner, E. L., Petersen, R. C., Miller, B. L., et al. (2015). Low neural exosomal levels of cellular survival factors in Alzheimer's disease. Ann. Clin. Transl. Neurol. 2, 769–773. doi: 10.1002/acn3.211
Goto, S., Lee, L. V., Munoz, E. L., Tooyama, I., Tamiya, G., Makino, S., et al. (2005). Functional anatomy of the basal ganglia in X-linked recessive dystonia-Parkinsonism. Ann. Neurol. 58, 7–17. doi: 10.1002/ana.20513
Gui, Y., Liu, H., Zhang, L., Lv, W., and Hu, X. (2015). Altered microRNA profiles in cerebrospinal fluid exosome in Parkinson disease and Alzheimer disease. Oncotarget 6, 37043–37053. doi: 10.18632/oncotarget.6158
Habib, R, Noureen, N, and Nadeem, N (2018). Decoding common features of neurodegenerative disorders: from differentially expressed genes to pathways. Curr. Genom. 19, 300–312. doi: 10.2174/1389202918666171005100549
Iguchi, Y., Eid, L., Parent, M., Soucy, G., Bareil, C., Riku, Y., et al. (2016). Exosome secretion is a key pathway for clearance of pathological TDP-43. Brain 139(Pt 12), 3187–3201. doi: 10.1093/brain/aww237
Ito, D., Hatano, M., and Suzuki, N. (2017). RNA binding proteins and the pathological cascade in ALS/FTD neurodegeneration. Sci. Transl. Med. 9 eaah5436. doi: 10.1126/scitranslmed.aah5436
Jiang, Y., Liu, J., Chen, L., Jin, Y., Zhang, G., Lin, Z., et al. (2019). Serum secreted miR-137-containing exosomes affects oxidative stress of neurons by regulating OXR1 in Parkinson's disease. Brain Res. 1722 146331. doi: 10.1016/j.brainres.2019.146331
Kim, K. M., Meng, Q., Perez de Acha, O., Mustapic, M., Cheng, A., Eren, E., et al. (2020). Mitochondrial RNA in Alzheimer's disease circulating extracellular vesicles. Front. Cell Dev. Biol. 8, 581882. doi: 10.3389/fcell.2020.581882
Kuleshov, M. V., Jones, M. R., Rouillard, A. D., Fernandez, N. F., Duan, Q., Wang, Z., et al. (2016). Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic acids research 44, W90–W97. doi: 10.1093/nar/gkw377
Kulkarni, N., Alessandrì, L., Panero, R., Arigoni, M., Olivero, M., Ferrero, G., et al. (2018). Reproducible bioinformatics project: a community for reproducible bioinformatics analysis pipelines. BMC Bioinform. 19, 349. doi: 10.1186/s12859-018-2296-x
Kwiatkowski, T. J. Jr., Bosco, D. A., Leclerc, A. L., Tamrazian, E., Vanderburg, C. R., Russ, C., et al. (2009). Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science 323, 1205–1208. doi: 10.1126/science.1166066
Lagier-Tourenne, C., Polymenidou, M., and Cleveland, D. W. (2010). TDP-43 and FUS/TLS: emerging roles in RNA processing and neurodegeneration. Hum. Mol. Genet. 19, R46–R64. doi: 10.1093/hmg/ddq137
Langfelder, P., and Horvath, S. (2008). WGCNA: an R package for weighted correlation network analysis. BMC Bioinform. 9, 559. doi: 10.1186/1471-2105-9-559
Ling, S. C., Polymenidou, M., and Cleveland, D. W. (2013). Converging mechanisms in ALS and FTD: disrupted RNA and protein homeostasis. Neuron 79, 416–438. doi: 10.1016/j.neuron.2013.07.033
Liu, E. Y., Cali, C. P., and Lee, E. B. (2017). RNA metabolism in neurodegenerative disease. Dis. Models Mech. 10, 509–518. doi: 10.1242/dmm.028613
Lugli, G., Cohen, A. M., Bennett, D. A., Shah, R. C., Fields, C. J., Hernandez, A. G., et al. (2015). Plasma exosomal miRNAs in persons with and without Alzheimer disease: altered expression and prospects for biomarkers. PLoS ONE 10, e0139233. doi: 10.1371/journal.pone.0139233
McKhann, G. M., Knopman, D. S., Chertkow, H., Hyman, B. T., Jack, C. R. Jr., Kawas, C. H., Klunk, W. E., et al. (2011). The diagnosis of dementia due to Alzheimer's disease: recommendations from the National Institute on Aging-Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimers Dement. 7, 263–269. doi: 10.1016/j.jalz.2011.03.005
Merienne, N., Meunier, C., Schneider, A., Seguin, J., Nair, S. S., Rocher, A. B., et al. (2019). Cell-type-specific gene expression profiling in adult mouse brain reveals normal and disease-state signatures. Cell Rep. 26, 2477–2493.e9. doi: 10.1016/j.celrep.2019.02.003
Morasso, C. F., Sproviero, D., Mimmi, M. C., Giannini, M., Gagliardi, S., Vanna, R., et al. (2020). Raman spectroscopy reveals biochemical differences in plasma derived extracellular vesicles from sporadic Amyotrophic Lateral Sclerosis patients. Nanomed. Nanotechnol. Biol. Med. 29, 102249. doi: 10.1016/j.nano.2020.102249
Neumann, M., Sampathu, D. M., Kwong, L. K., Truax, A. C., Micsenyi, M. C., Chou, T. T., et al. (2006). Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 314, 130–133. doi: 10.1126/science.1134108
O'Brien, K., Breyne, K., Ughetto, S., Laurent, L. C., and Breakefield, X. O. (2020). RNA delivery by extracellular vesicles in mammalian cells and its applications. Nat. Rev. Mol. Cell Biol. 21, 585–606. doi: 10.1038/s41580-020-0251-y
Postuma, R. B., Berg, D., Stern, M., Poewe, W., Olanow, C. W., Oertel, W., et al. (2015). MDS clinical diagnostic criteria for Parkinson's disease. Movement Disord. 30, 1591–1601. doi: 10.1002/mds.26424
Rajendran, L., Bali, J., Barr, M. M., Court, F. A., Krämer-Albers, E. M., Picou, F., et al. (2014). Emerging roles of extracellular vesicles in the nervous system. J. Neurosci. 34, 15482–15489. doi: 10.1523/JNEUROSCI.3258-14.2014
Rani, A., O'shea, A., Ianov, L., Cohen, R. A., Woods, A. J., and Foster, T. C. (2017). miRNA in circulating microvesicles as biomarkers for age-related cognitive decline. Front. Aging. Neurosci. 9, 323. doi: 10.3389/fnagi.2017.00323
Rascovsky, K., Hodges, J. R., Knopman, D., Mendez, M. F., Kramer, J. H., Neuhaus, J., et al. (2011). Sensitivity of revised diagnostic criteria for the behavioural variant of frontotemporal dementia. Brain J. Neurol. 134(Pt 9), 2456–2477. doi: 10.1093/brain/awr179
Reiner, A., Albin, R. L., Anderson, K. D., D'Amato, C. J., Penney, J. B., and Young, A. B. (1988). Differential loss of striatal projection neurons in Huntington disease. Proc. Natl. Acad. Sci. U.S.A. 85, 5733–5737. doi: 10.1073/pnas.85.15.5733
Riku, Y., Watanabe, H., Yoshida, M., Mimuro, M., Iwasaki, Y., Masuda, M., et al. (2016). Marked involvement of the striatal efferent system in TAR DNA-binding protein 43 kDa-related frontotemporal lobar degeneration and amyotrophic lateral sclerosis. J. Neuropathol. Exp. Neurol. 75, 801–811. doi: 10.1093/jnen/nlw053
Saman, S., Kim, W., Raya, M., Visnick, Y., Miro, S., Saman, S., et al. (2012). Exosome-associated tau is secreted in tauopathy models and is selectively phosphorylated in cerebrospinal fluid in early Alzheimer disease. J. Biol. Chem. 287, 3842–3849. doi: 10.1074/jbc.M111.277061
Skog, J., Würdinger, T., van Rijn, S., Meijer, D. H., Gainche, L., Sena-Esteves, M., et al. (2008). Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 10, 1470–1476. doi: 10.1038/ncb1800
Soares Martins, T., Trindade, D., Vaz, M., Campelo, I., Almeida, M., Trigo, G., et al. (2021). Diagnostic and therapeutic potential of exosomes in Alzheimer's disease. J. Neurochem. 156, 162–181. doi: 10.1111/jnc.15112
Sproviero, D., Gagliardi, S., Zucca, S., Arigoni, M., Giannini, M., Garofalo, M., et al. (2021). Different miRNA profiles in plasma derived small and large extracellular vesicles from patients with neurodegenerative diseases. Int. J. Mol. Sci. 22, 2737. doi: 10.3390/ijms22052737
Sproviero, D., La Salvia, S., Colombo, F., Zucca, S., Pansarasa, O., Diamanti, L., et al. (2019). Leukocyte derived microvesicles as disease progression biomarkers in slow progressing amyotrophic lateral sclerosis patients. Front. Neurosci. 13, 344. doi: 10.3389/fnins.2019.00344
Sproviero, D., La Salvia, S., Giannini, M., Crippa, V., Gagliardi, S., Bernuzzi, S., et al. (2018). Pathological proteins are transported by extracellular vesicles of sporadic amyotrophic lateral sclerosis patients. Front. Neurosci. 12, 487. doi: 10.3389/fnins.2018.00487
Stuendl, A., Kraus, T., Chatterjee, M., Zapke, B., Sadowski, B., Moebius, W., et al. (2021). α-synuclein in plasma-derived extracellular vesicles is a potential biomarker of Parkinson's disease. Movement Disord. 36, 2508–2518. doi: 10.1002/mds.28639
Tan, L., Yu, J. T., Tan, M. S., Liu, Q. Y., Wang, H. F., Zhang, W., et al. (2014). Genome-wide serum microRNA expression profiling identifies serum biomarkers for Alzheimer's disease. J. Alzheimers Dis. 40, 1017–1027. doi: 10.3233/JAD-132144
Théry, C., Witwer, K. W., Aikawa, E., Alcaraz, M. J., Anderson, J. D., Andriantsitohaina, R., et al. (2018). Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 7, 1535750. doi: 10.1080/20013078.2018.1461450
Thomas, L., Florio, T., and Perez-Castro, C. (2020). Extracellular vesicles loaded miRNAs as potential modulators shared between glioblastoma, and Parkinson's and Alzheimer's diseases. Front. Cell. Neurosci. 14, 590034. doi: 10.3389/fncel.2020.590034
Thompson, A. G., Gray, E., Mäger, I., Thézénas, M. L., Charles, P. D., Talbot, K., et al. (2020). CSF extracellular vesicle proteomics demonstrates altered protein homeostasis in amyotrophic lateral sclerosis. Clin. Proteomics 17, 31. doi: 10.1186/s12014-020-09294-7
Zheng, X., Peng, Q., Wang, L., Zhang, X., Huang, L., Wang, J., et al. (2020). Serine/arginine-rich splicing factors: the bridge linking alternative splicing and cancer. Int. J. Biol. Sci. 16, 2442–2453. doi: 10.7150/ijbs.46751
Keywords: neurodegenerative diseases, extracellular vesicles, RNA-Seq, mRNA, lncRNA
Citation: Sproviero D, Gagliardi S, Zucca S, Arigoni M, Giannini M, Garofalo M, Fantini V, Pansarasa O, Avenali M, Ramusino MC, Diamanti L, Minafra B, Perini G, Zangaglia R, Costa A, Ceroni M, Calogero RA and Cereda C (2022) Extracellular Vesicles Derived From Plasma of Patients With Neurodegenerative Disease Have Common Transcriptomic Profiling. Front. Aging Neurosci. 14:785741. doi: 10.3389/fnagi.2022.785741
Received: 29 September 2021; Accepted: 13 January 2022;
Published: 16 February 2022.
Edited by:
Tao Sun, Nankai University, ChinaReviewed by:
Satoru Morimoto, Keio University, JapanCopyright © 2022 Sproviero, Gagliardi, Zucca, Arigoni, Giannini, Garofalo, Fantini, Pansarasa, Avenali, Ramusino, Diamanti, Minafra, Perini, Zangaglia, Costa, Ceroni, Calogero and Cereda. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Stella Gagliardi, c3RlbGxhLmdhZ2xpYXJkaUBtb25kaW5vLml0
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.