- 1Proteomics Programme, Paracelsus Medical University, Salzburg, Austria
- 2Department of Dermatology, Medical University of Vienna, Vienna, Austria
- 3Center for Brain Research, Department of Molecular Neurosciences, Medical University of Vienna, Vienna, Austria
- 4Institut für Labortierkunde, Medizinische Universität Wien, Vienna, Austria
Cognitive processes require striatal activity. The underlying molecular mechanisms are widely unknown. For this reason the striatal transcriptome of young (YM), aged cognitively impaired (OMB), and unimpaired (OMG) male rats was analyzed. The global comparison of transcripts reveal a higher number of differences between OMG and YM as compared to OMB and YM. Hierarchical clustering detects differences in up- and down-regulated gene clusters in OMG and OMB when compared to YM. In OMG we found more single genes to be specifically regulated in this group than in OMB when compared to young. These genes were considered as cognition specific, whereas genes shared in OMG and OMB were considered as age specific. OMB specific up-regulated genes are related to negative control of cell differentiation and transcription (Hopx), to phagocytosis (Cd202) and cell adhesion (Pcdhb21), whereas down-regulated genes are related to associative learning, behavioral fear response and synaptic transmission (Gabra5). OMG specific up-regulated genes are in the context of maintenance of transcription and estrogen receptor signaling (Padi2, Anxa3), signal transduction [Rassf4, Dock8)], sterol regulation (Srebf1), and complement activity (C4a, C4b). Down-regulated genes are related to lipid oxidation reduction processes (Far2) and positive regulation of axon extension (Islr2). These relations were supported by pathway analysis, which reveals cholesterol metabolism processes in both aged group and cholesterol biosynthesis specifically in OMG; adipogenesis and focal adhesion in OMB. In OMG glucuronidation, estrogen metabolism, inflammatory responses and TGF beta signaling where detected as specific for this group. Signal transduction of the sphingosine-1-phospate-receptor (S1P) receptor was the main pathway difference in the comparison of OMB and OMG with downregulated genes in the first group. This difference could also be observed in the OMB vs. YM comparison but not in the OMG vs. YM analysis. Thus, an up-regulation of cognition related genes could be observed in OMG compared to OMB rats. The S1P pathway discriminated between OMB and OMG as well as between OMB and OMG. Since this pathway has been described as essential for cognitive processes in the striatum of mice, it may, among steroid hormone signaling, significantly contribute to the maintenance of cognitive processes in OMG.
Introduction
Striatal involvement in spatial learning is well-documented (Luine and Hearns, 1990; Lavoie and Mizumori, 1994; McDonald and White, 1994; Packard and Teather, 1997; Gengler et al., 2005). Torromino et al. (2019) found the cross-regional communication between hippocampus and the ventral striatum to play a critical role in spatial memory formation in mice after water maze training. Simultaneous activation of the dorsal striatum and the hippocampus is required for correct navigation in a double H maze in a cued environment (Gasser et al., 2020). Despite this long standing knowledge the underlying molecular mechanisms are largely unknown. Most transcriptomic and proteomic analyses were performed in the context of disease, drug addiction or pharmacology. Xu et al. (2007) found the striatum less responsive than other brain regions in terms of transcriptome changes related to age in mice. Genes were mostly regulated in the context of transcription regulation, protein synthesis and degradation and signal transduction. Similar gene expression profiles in the striatum and hippocampus have been found in NOS-1 (neural isofrom of nitric oxide synthase) knockdown animals which show impairments of spatial learning and memory including the holeboard test. Highly upregulated genes were related to gamma-Aminobutyric acid (GABA) -ergic signaling and the glucocorticoid receptor (GR). Martín-García et al. (2015) found genes related to extracellular signal-regulated kinase 5 (ERK5) signaling, cytokine signaling and protein ubiquitination in the ventral striatum of mice during extinction learning of operant conditioning.
Hamezah et al. (2018) profiled the proteome of the striatum in male rats at different ages, detecting only 5 proteins differently expressed with age. The affected pathway was the glutathione metabolism with increased expression of the proteins Glutathione S-transferase A3 (GSTA3) and Superoxide Dismutase 1 (SOD1) in 23 months old rats. However, rats were not tested in a cognitive task. Auditory learning in a shuttle box induced regulations of some synaptic proteins like Titin and Casitas B lineage lymphoma (CBL) in mice, the latter involved in protein degrading processes (Kähne et al., 2012). The same training induced also protein rearrangement in the striatum involving GABA and dopamine receptor signaling (Kähne et al., 2016).
Because of the small body of literature regarding age and cognition related effects upon gene expression in the striatum, we combined both aspects in the present study. Therefore, we compared young males with aged ones and divided the latter into bad (OMB) and good (OMG) performers, similar as in the study of De Risi et al. (2020) in mice. This approach provides information about age effects and should reveal mechanisms of cognitive maintenance during aging. The study was designed as a screening test involving a large number of genes. This should provide a basis for own follow up studies on a reduced number of here identified critical genes and pathways involving real time PCR and proteomic studies. The here presented results may also be of interest for other researchers in the field.
Methods
Animals
Animals were bred and maintained in the Core Unit of Biomedical Research, Division of Laboratory Animal Science and Genetics, Medical University of Vienna. One week before and throughout the experiment the animals lived in a separate experimental room and were housed individually in standard Makrolon cages filled with autoclaved woodchips at a temperature of 22 ± 2°C with a humidity of 55 ± 5% and 12 h artificial light/12 h dark cycle (light on at 7:00 a.m.). Individual housing should avoid social rank specific impacts on behavioral performance in the test. Tap water and food (R/M-H Ered II) diets from ssniff®, Soest, Germany) was provided at libitum. The study was carried out according to the guidelines of the Ethics Committee, Medical University of Vienna, and was approved by the Federal Ministry of Education, Science and Culture, Austria.
Three groups of male Sprague–Dawley male rats (8 animals per group) were tested for their transcriptome profile in the striatum: young males (YM), old males with bad (OMB) and good (OMG) memory. YM were 3 months old with an average weight of 440 g, aged rats were 20 months old with an average weight of 650 g. The animal samples represent a subgroup of animals from a previous study. In this study the effects of different diets on memory were studied. The here used animals were fed with the low energy diet as described in Maliković et al. (2018). In order to distinct between old male rats with impaired and unimpaired memory, a hole-board memory test was performed according to a previously described protocol (Maliković et al., 2018; Daba Feyissa et al., 2019). The spatial memory performance was evaluated based on Reference Memory Index (RMI) calculated as: (first + revisits of baited holes) / total visits of all holes). In order to compare individuals with similar levels of motivation, animals with <40 hole-visits in total over the 10 trials were excluded from the analysis. The individuals were considered as good performers (OMG) when RMI >the mean RMI value + standard deviation, and as bad performers (OMB) when RMI <the mean RMI value – standard deviation, with the mean value obtained from all animals included in the analysis over the evaluated training/testing period. To determine statistical significance of the differences between the two groups of old rats, the data was analyzed Graphpad-Prism 5 using a two way repeated measures ANOVA with age as one and training trial as the second factor, followed by a post-hoc comparison (Bonferroni) between groups. The significance was set at p < 0.05.
Four to six weeks after the behavioral test the animals were decapitated, the brains were rapidly removed and dissected on a Para Cooler (RWW Medizintechnik, Hallerndorf, Germany) at 4°C to obtain the striatum. The tissue was immediately stored at −80°C until transcriptomic analysis.
RNA Purification
Total RNA was extracted from rat striatum tissue according to the “Purification of total RNA from animal and human tissue” protocol of the RNeasy Micro Kit (QIAGEN, Hilden, Germany).
In brief, the sample was frozen and shipped on dry ice. After adding 800 μl buffer RLT containing 1% beta-mercaptoethanol the tissue was disrupted and homogenized with Precellys CK14 ceramic beads (1 cycle of 20 s at 6,000 rpm) using a Precellys 24 Homogenisator (Bertin Corp., Rockville, MD, USA). Next the sample was centrifuged for 3 min at full speed and 350 μl of the cleared supernatant was transferred to a new tube. One volume of 70% ethanol was added and the sample was applied to a RNeasy MinElute spin column followed by an on-column DNase digestion and several wash steps. Finally total RNA was eluted in 14 μl of nuclease free water. Purity and integrity of the RNA was assessed on the Agilent 2100 Bioanalyzer with the RNA 6000 Nano LabChip reagent set (Agilent, Palo Alto, CA, USA).
GeneChipTM Microarray Assay
Sample preparation for microarray hybridization was carried out as described in the Applied BiosystemsTM GeneChipTM Whole Transcript (WT) PLUS Reagent Kit User Guide (Thermo Fisher Scientific, Waltham, MA, USA). In brief, 200 ng of total RNA was used to generate double-stranded cDNA. 12 μg of subsequently synthesized cRNA were purified and reverse transcribed into single-stranded (ss) cDNA, whereas unnatural dUTP residues were incorporated. Purified ss cDNA was fragmented using a combination of uracil DNA glycosylase (UDG) and apurinic/apyrimidinic endonuclease 1 (APE 1) followed by a terminal labeling with biotin. 3,8 μg of fragmented and labeled ss cDNA were hybridized to Applied BiosystemsTM GeneChipTM Clariom S rat arrays for 16 h at 45°C and 60 rpm in an Applied BiosystemsTM GeneChipTM hybridization oven 640. Hybridized arrays were washed and stained in an Applied BiosystemsTM GeneChipTM Fluidics Station FS450, and the fluorescent signals were measured with an Applied BiosystemsTM GeneChipTM GeneChip Scanner 3000 7G System. Fluidics and scan functions were controlled by the Applied BiosystemsTM GeneChipTM Command Console v4.3.3 software.
RNA extraction and sample processing were performed at a Genomics Core Facility, “KFB - Center of Excellence for Fluorescent Bioanalytics” (Regensburg, Germany; www.kfb-regensburg.de).
Microarray Data Analysis
Summarized probe set signals in log2 scale were calculated by using the GCCN-SST-RMA algorithm with the Applied BiosystemsTM GeneChipTM Expression Console v1.4 Software. After exporting into Microsoft Excel average signal values, comparison fold changes, significance P-values, hierarchical clustering, Venn-diagram and pathway-analysis were done with the transcriptome analysis console (V. 4.0.2.15, applied biosystems, Thermo Fisher Scientific). Probe sets with a student's t-test P-value lower than 0.05 were considered as significantly regulated.
Results
Behavior
The behavioral results are shown in Figure 1. The two-way ANOVA revealed a significant effect between groups (F2,21 = 54.42, p < 0.0001), a significant trial effect (F9,189 = 11.13, p < 0.0001) and a significant trial x group interaction (F18,189 = 6.34, p = 0.0005). Post-hoc tests revealed no significant difference between trial performance as compared between YM and OMG (p > 0.05, each) except for trial 8 with better performance in OMG (t = 2.90, p < 0.05) whereas OMB had significantly lower RMI compared to YM (p < 0.05) except for trial 4 with no difference (t = 1.29, p > 0.05). OMG rats show significantly higher RMI in all trials when compared to OMB (p < 0.05, each).
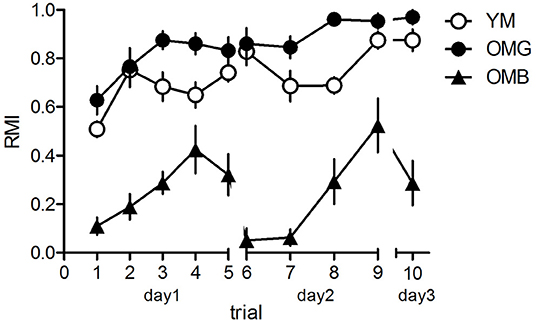
Figure 1. Behavioral results for the hole-board test. Young (YM) and old, cognitively unimpaired males (OMG) showed no differences in reference memory indices (RMI) over trials with the exeption of trial 8 where OMG performed better than YM. Both, YM and OMG exhibited higher RMI compared to cognitively impaired aged males (OMB) over trials with the exception of trial 4 where YM and OMB showed no difference. For statistics see the Results section. Given are the mean values and standard errors of the mean, n = 8 for each group).
Transcriptome
We found 367 significant differences in transcript amounts between OMB and YM and 610 between OMG and YM, whereas between OMG and OMB we found only 59 with a fold change of ≤-1.5 or ≥1.5 and a p-value ≤0.05. In the OMG vs. YM comparison most of the genes that differed between groups were coding (Figure 2). This was also true for the OMB vs. YM in upregulated genes whereas downregulated transcripts show a higher proportion of non-coding and unassigned genes. In the OMB vs. OMG comaparison the percentage of non-coding downregulated genes was even higher with 18.75%.
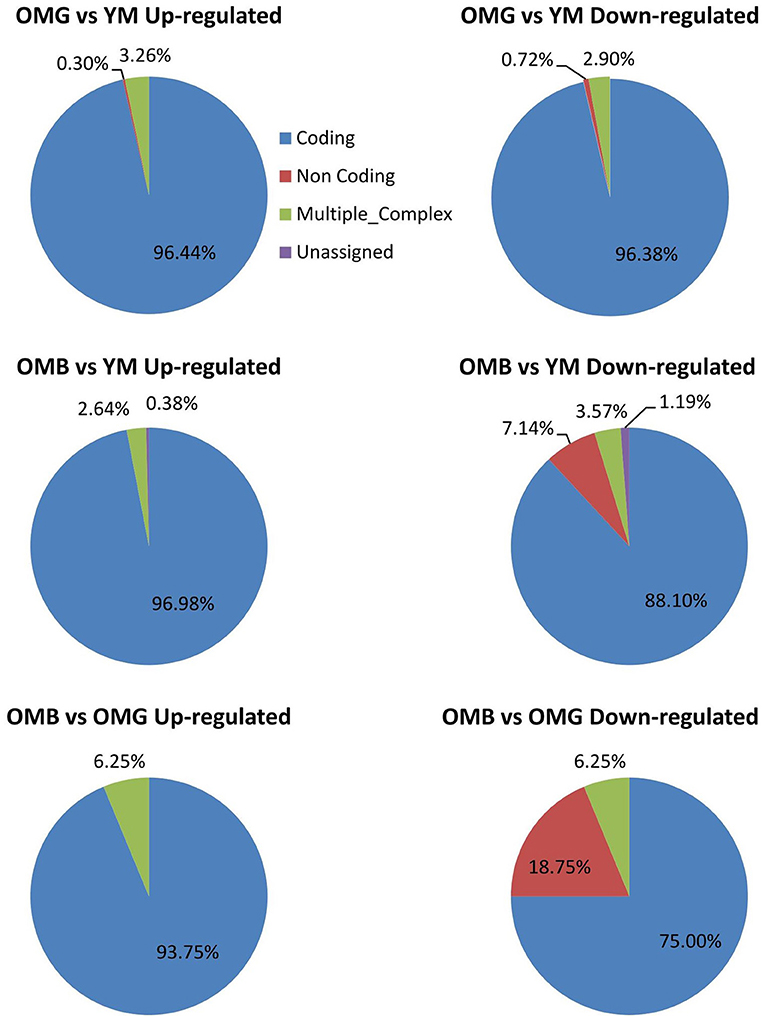
Figure 2. Summary of the transcriptome analysis. Characterization of regulated genes in the OMH vs. YM (upper panel), the OMB vs. YM (middle panel) and the OMB vs. OMG (lower panel) comparisons.
The hierarchical clusters (Figure 3) revealed differences in certain gene clusters. In the upper part of Figure 2 OMG together with YM showed down-regulated clusters (genes Mgat4e, protein N-linked glycosylation, and Olr1555, sensory perception of smell:); (RGD1561318, histone lysin methylation, and Sult1c3, sulfur compound metabolic process); (Stox1, regulation of gene expression and, cellular response to nitrosative stress and Itgb3bp, integrin subunit beta 3 binding protein). Most remarkably one OMG animal did not cluster with OMG and not with YM.
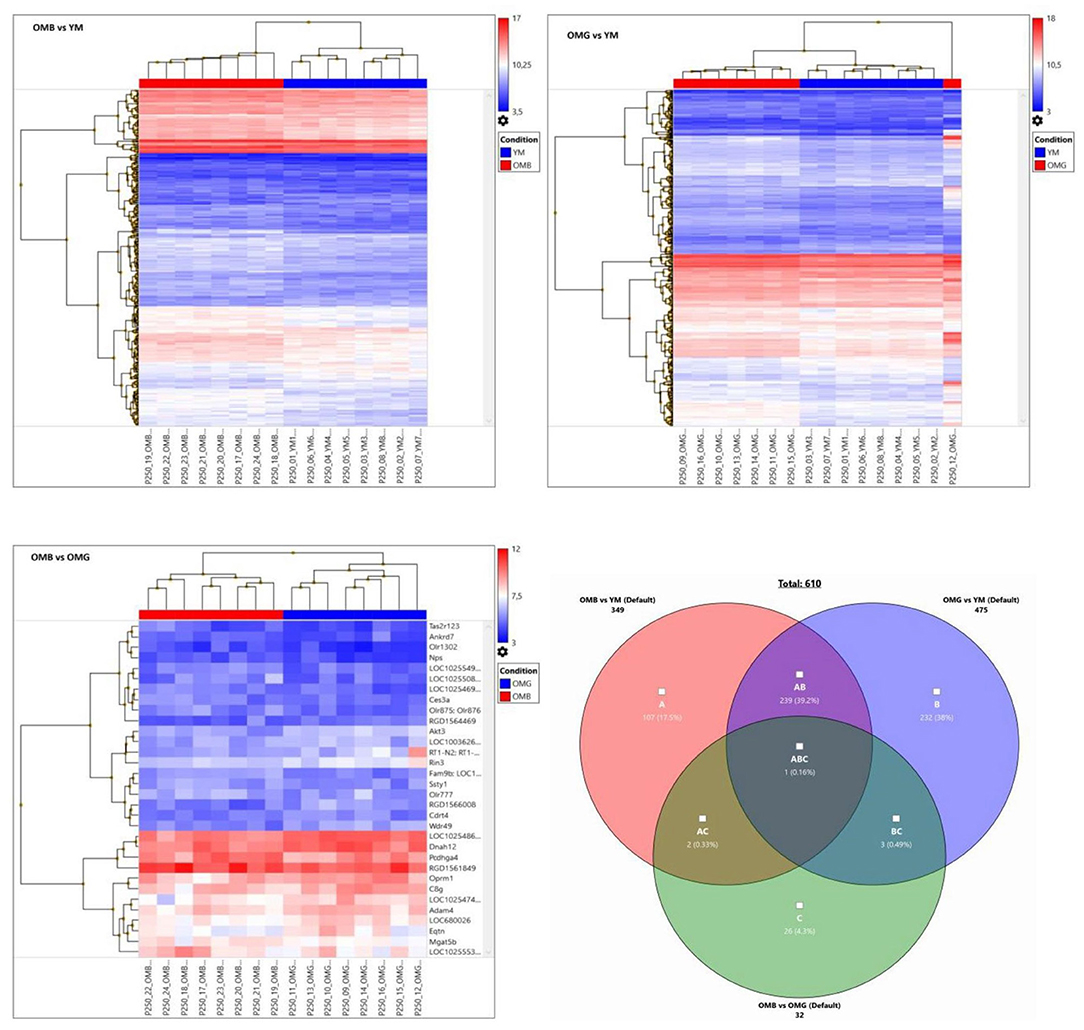
Figure 3. Hierarchical clusters of OMB vs. YM (upper left), OMG vs. (YM upper right) and OMB vs. OMG (lower left) comparisons for genes with a fold change of ≤-1.5 and ≥1.5 and p ≤ 0.05. Autoscale gives log2 signals beyond the middle in red and below in blue. Venn- diagram lower right, showing relations between groups.
For demonstration of individual heterogeneity a random subset of differently expressed genes between aged and young males were extracted and given in Figure 4. In OMB the variation between individual rats is lower than in OMG. In the OMG vs. YM analysis differences were found in clusters (Pde1c, phosphodiesterase 1c involved in cellular signaling, and LOC100362345, hypothetical protein LOC100362345), (RGD1305645, neuropeptide signaling pathway, positive regulation of corticosterone secretion, regulation of cell proliferation, and Stoml3, signal transduction), (Srebf1, sterol regulatory element binding transcription factor 1, and Nqo2, memory, oxidation-reduction process) with an up-regulation in OMG as compared to YM.
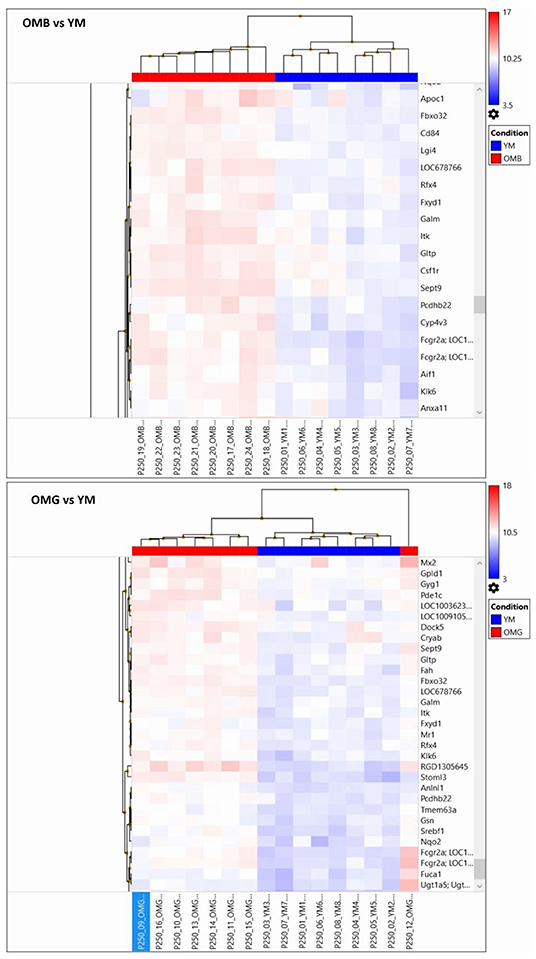
Figure 4. Random extraction of clusters with opposing log2 signals between compared groups in order to demonstrate within group variabilities for the OMB vs. YM (upper panel) and the OMG vs. YM (lower panel) comparisons.
In the OMB vs. YM clusters (Itih3, inter-alpha trypsin inhibitor, and Cml3, histone acetylation); Zcchc24, zinc ion binding, and Tmem192, protein homodimerization activity), (S100b, calcium binding protein B, and Spock1, sparc/osteonectin, cwcv and kazal like domains proteoglycan 1) are upregulated. Specific cluster (Apoc1, cholesterol metabolic process, negative regulation of lipid metabolic process, and Fbxo32, F-box protein 32), (Cd84, Cd84 molecule and Lgi 4, leucine-rich repeat LGI family, member 4), (LOC678766, actin cytosceleton organization, and Rfx4, regulatory factor X4) are upregulated in OMB compared to YM.
In order to reduce the amount of genes and to figure out the most meaningful differences, the criterion was set to a fold cutoff at ≤-2 and ≥2 and p ≤ 0.05. The genes regulated in both comparisons of aged and young males are given in Table 1. Genes that are specifically regulated within one group are given in bold. These genes are considered to be cognition related, whereas shared genes are considered to be age related. In OMB vs. YM the Hopx, homeodomain-only; Slc9a, solute carrier family 9 member A9; Rt1-Ce13, RT1 class I, locus CE13 are upregulated in the first with the highest fold changes. In Table 2 the description and biological processes in which the OMB specific genes are involved are given. For most of the up-regulated genes biological processes are unknown, however the Hopx gene with the highest fold change is related to negative regulation of cell differentiation and transcription. Down-regulated genes are involved in cell adhesion and migration (Itgb6, Integrin alpha-6), associative learning and synaptic transmission gamma-aminobutyric acid type A receptor subunit alpha5 (Gabra5) and G protein coupled signal transduction.

Table 1. Comparison of old vs. young males, cognition-related genes (not shared between OMG and OMB) are given in bold.
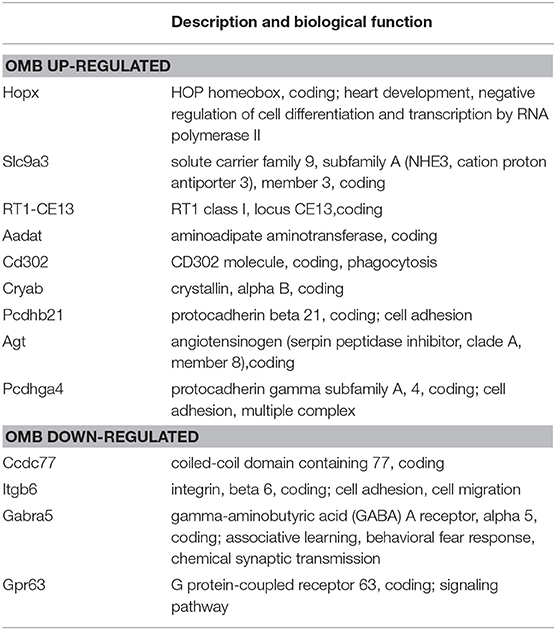
Table 2. OMB specific regulated genes compared to YM, description and biological process (NCBI, gene catalog).
The amount of specifically related genes in OMG rats is higher as compared to OMB (Table 3). Genes related to signal transduction (Rassf4), steroid membrane lipids regulation (Srebf1, Dock8, dedicator of cytokinesis 8; Cyb5r2, cytochrome B5 reductase 2), steroid (estrogenic) signaling and transcription maintenance (Padi2, Peptidyl Arginine Deiminase 2; Anxa3, Annexin A3), glucocorticoid response (Anxa3), and the activation and maintenance of the complement system (C4a, complement C4-A; C4b, complement C4-B) are upregulated. Down-regulated are gene transcripts involved in positive regulation of axon extension (Islr2, immunoglobulin superfamily containing leucine rich repeat 2) and lipid oxidation reduction process (Far2, fatty acyl-CoA reductase 2).

Table 3. OMG specific regulated genes compared to YM, description and biological process (NCBI, gene catalog).
The next step was the identification of pathway differences between groups (Table 4). Here again the criterion was set at ≤-1.5 and ≥1.5 and p ≤ 0.05 in order to detect also weaker differences in order not to miss pathways because of false negative results. Between OMB and OMG rats we found only one pathway to be different, that is the signal transduction of S1P receptor. This difference could also be observed between OMB and YM, but not between OMG and YM. Complement activation and activity appeared in bot aged groups compared to YM. Adiposegenesis, focal adhesion, B cell receptor signaling and Eicosanoid synthesis were specific for OMB, whereas glucoronidation, estrogen metabolism, cholesterol synthesis, Transforming growth factor beta (TGF beta) signaling, monoamine G protein-coupled receptors (GPCRs) and metapathway biotransformation were specific for OMG when compared to YM.
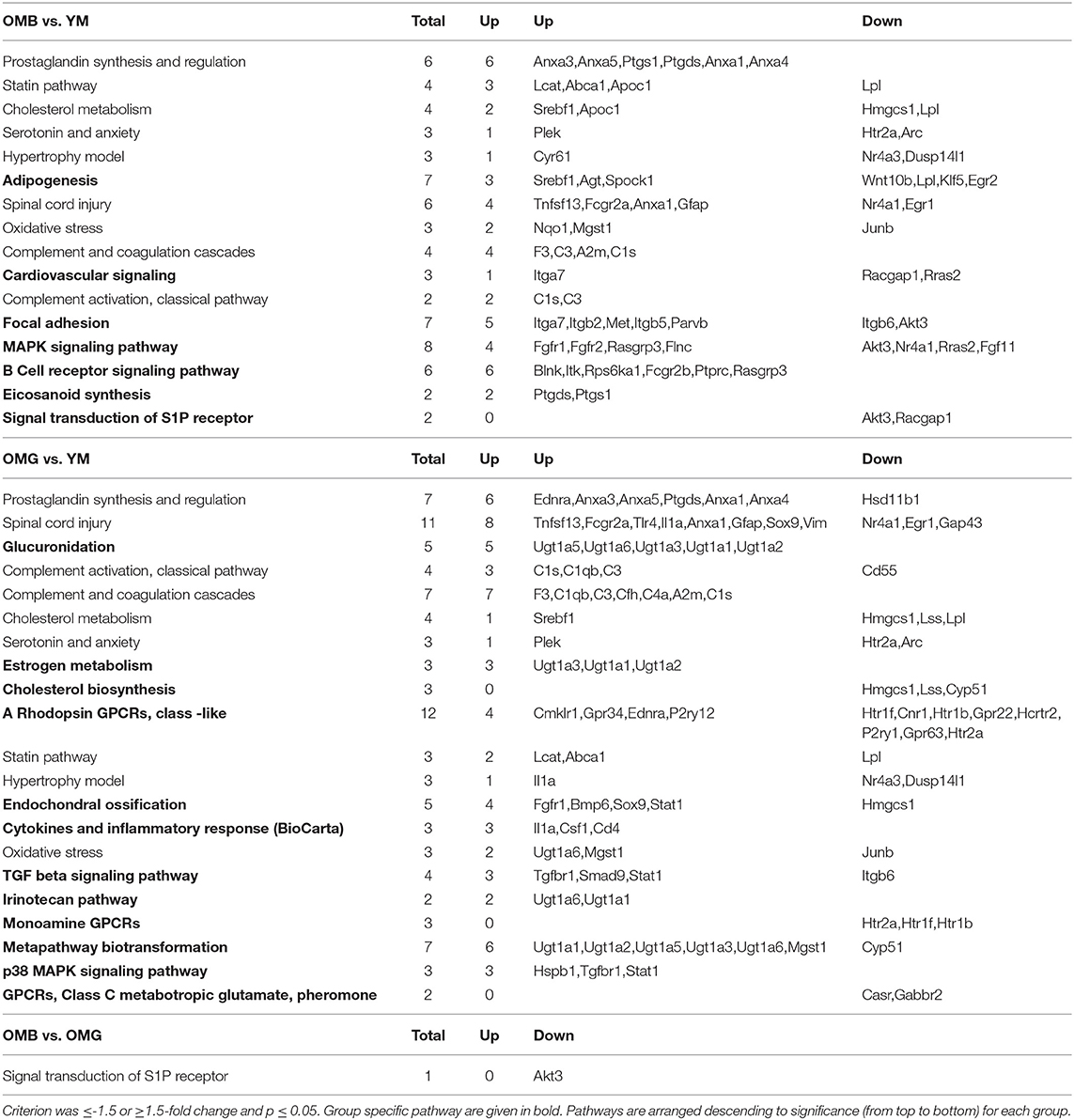
Table 4. Pathway analysis (www.wikipathways.org) for aged vs. young and between aged groups.
Discussion
Aging is not necessarily accompanied with cognitive decline. In a cohort of 125 aged male rats, fed with a low energy diet, we found 43.2% males not to be impaired in the holeboard test (RMI ≥0.8). The comparison of the whole cohort with young males show an aging effect in line with other studies (van der Staay et al., 1990). These authors found a age related decline in spatial reference memory in the holeboard with the most profound effect in 19 and 25 month old rats, the age class we used in the present study. However, in spite of the small samples (8–10 rats) also the aged rats in the van der Staay study show an linear improvement in spatial reference memory over blocks of trials, suggesting intact learning capacity in at least some of the rats (unfortunately no standard deviation is given).
Cognitively unimpaired and impaired aged male rats differed in the expression of genes when compared with YM and between the two groups. These genes were related to different pathways or were differently regulated in similar pathways. Genes related to the prostaglandin synthesis and regulation pathway, were regulated with the highest significance in both aged groups compared to YM. Prostaglandine A1 has been shown to protect striatal cells against excitotoxic injuries (Qin et al., 2001). In both aged groups related gene transcripts were mostly up-regulated, however hydroxysteroid 11-beta-dehydrogenase 1-like protein (Hsd11b1) transcript is downregulated in OMG and not regulated in OMB. This gene is involved in the cellular response to estradiol stimulus (Tagawa et al., 2009), glucocorticoid biosynthesis and catabolic processes (Nobel et al., 2002) by 17β-estradiol induced inhibition of the 11 beta-hydroxysteroid hydrogenase, which catalyzes the production of corticosterone from 11-dehydrocorticosterone in rats. The synthesis of Prostaglandines and lipid metabolism are highly interconnected, the latter also with the statin pathway, adipogenesis and eicosanoid synthesis. Lipid synthesis and regulatory pathways are changed in both groups as compared to young rats.
Lipid composition changes in the hippocampus, a brain structure being strongly involved in spatial memory processes, can also lead to specific differences in spatial learning. In a previous study we found nutrition induced lower sphingomyelin amounts in the hippocampal dentate gyrus in rats with impairments of spatial cognitive flexibility (Maliković et al., 2018). The striatum and the hippocampus play different roles in spatial learning and memory (Torromino et al., 2019) and are highly interconnected. The hippocampus projects to the ventral striatum with the nucleus accumbens that has been identified to be critically involved in spatial long-term memory processes (Ferretti et al., 2010; Torromino et al., 2019) with different functions of the core and shell substructures (Meredith and Totterdell, 1999; Ito et al., 2008; Ito and Hayen, 2011). The dorsal striatum in turn contributes to different aspects of spatial memory like egocentric vs. allocentric spatial learning strategies (De Leonibus et al., 2005). These differences cannot be addressed in the present study. However, the extraction of genes and pathways, discriminating between the aged groups, out of thousands of genes, allows the identification of candidates for further, more specific studies.
Previously we showed that also hypothalamic lipid profiles differed between the aged male groups in our paradigm (Wackerlig et al., 2020). The hypothalamus is involved in the regulation of thyroid and steroid hormone release and homeostasis. Interestingly, OMG and OMB mainly differed in the expression of the S1P receptor and in regulatory and functional pathways of steroid hormones. That is glucuronidation, estrogen metabolism and cholesterol biosynthesis. Glucuronidation means the transformation of small lipophylic molecules, like thyroid and steroid hormones, into more water-soluble metabolites. In a previous study we were able to show that blood levels of thyroid hormones, namely triiodothyronine (T3) and thyroid-stimulating hormone (TSH) discriminate aged cognitively impaired from unimpaired male rats (Maliković et al., 2019). It has been shown that treatment with the thyroid hormone triiodothyronine (T3) upregulates the expression of different isoforms of the UDP –glucuronosyltransferase coded by the gene UGT1a6, but decreased Ugt1a1and Ugt1a5, the first was up-regulated by dexamethasone in primary cultures of rats hepatocytes (Li et al., 1999). The up-regulation of all these isoforms here suggests effects of different hormones. Further some isoforms up-regulated here are involved in estrogen metabolism. The synthesis of all steroid hormones is based on cholesterol mainly in the adrenal glands and gonads. Thus, here the downregulation of cholesterol biosynthesis related genes may be a mechanism to reduce tissue cholesterol amount. Estrogen in male rats can locally be synthesized from testosterone by the enzyme aromatase. It has been shown that application of testosterone but not the non-aromatizable dihydrotestosterone improves spatial working memory in aged male rats (Bimonte-Nelson et al., 2003). Hippocampal local activation of estrogen receptors enhances spatial reference memory, in male rats (Meyer and Korz, 2013a,b; Sase et al., 2014). Given the close cooperation of the hippocampus and the striatum this may also have beneficial effects of spatial memory in the striatum. However, while high external estrogen exposure improves memory in hippocampus-dependent tasks, it can impair it in striatum-dependent tasks (Korol and Pisani, 2015). The effects of estrogens upon learning and memory are complex, can be bidirectional (Korol and Pisani, 2015), suggesting the coordinated involvement of different pathways. Post-training corticosterone administration into the dorsomedial striatum has been shown to enhance spatial place memory consolidation in male rats (Lozano et al., 2013).
The TGF Beta signaling pathway specifically changed in OMG vs. YM is involved in multiple cellular signaling processes including cellular homeostasis. TGF-β1restores hippocampal synaptic plasticity and memory in a mouse model of Alzheimer's disease (Hu et al., 2019). The differences in genes related to the MAPK –pathway in OMB and p38 MAPK –pathway in OMG point to different signaling pathways within the MAPK system with cytokine related activation in OMG.
The downregulation of serotonin receptor coding transcripts in both aged groups are probably related to aging, since other studies report similar effects with aging in rats (Wang et al., 1995; Paulose and Balakrishnan, 2008). Functional alterations of the serotonergic system by pharmacological receptor blockade in the striatum of aged male rats has been found to induce spatial learning and memory impairments (Stemmelin et al., 2000; Rutz et al., 2009). Reduced 5-HT2A receptor binding in the striatum of human Alzheimer's disease patients correlated negatively with anxiety and depression scores (Hasselbalch et al., 2008), pointing to functional consequences of striatal serotonin signaling not only for cognition but also mood. Similarily binding of GABAB receptors decrease with age (Milbrandt et al., 1994; Turgeon and Albin, 1994), whereas receptor density remains the same (Turgeon and Albin, 1994). Why this and the calcium sensing receptor coded by Casr are down-regulated only in OMG is unclear, the revelation of functional consequences requires further studies. In OMG these negative effects upon cognition may be compensated by mechanisms mentioned above, whereas in OMB no compensatory processes rescue cognitive impairments.
Such a response is obviously the regulation of the signal transduction of the sphingosine-1-phospate-receptor. This clearly discriminates OMB from OMG as well as from YM. It is a G protein-coupled receptor with the ligand sphingosine-1-phosphate. Striatal S1P is expressed in the striatopallidal neurons in the striatum of mice and essential for instrumental conditiong (Lobo et al., 2007). S1P is involved in a variety of cellular signaling processes including the increase of intracellular calcium from thapsigargin-sensitive stores (Pébay et al., 2001). Pharmacological activation of the S1P receptor by fingolimod increased the levels of brain derived neurotrophic factor (BDNF) and increased the size of the striatum in treated Mecp2 knockout mice as a model of Rett syndrome. BDNF is essentially involved in learning and memory processes and synaptic plasticity (Black, 1999; Yamada et al., 2002; Bramham and Messaoudi, 2005).
Thus, this pathway, among steroid hormone signaling, may significantly contribute to the maintenance of cognitive processes in OMG.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below:
https://gigamove.rz.rwth-aachen.de/d/id/ip2rGzhL4YCFTh, KFB.
Ethics Statement
The animal study was reviewed and approved by Federal Ministry of Education, Science and Culture, Austria.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Bimonte-Nelson, H. A., Singleton, R. S., Nelson, M. E., Eckman, C. B., Barber, J., Scott, T. Y., et al. (2003). Testosterone, but not nonaromatizable dihydrotestosterone, improves working memory and alters nerve growth factor levels in aged male rats. Exp. Neurol. 181, 301–312. doi: 10.1016/S0014-4886(03)00061-X
Black, I. B. (1999). Trophic regulation of synaptic plasticity. J. Neurobiol. 41, 108–118. doi: 10.1002/(SICI)1097-4695(199910)41:1andlt;108::AID-NEU14andgt;3.0.CO;2-U
Bramham, C. R., and Messaoudi, E. (2005). BDNF function in adult synaptic plasticity: the synaptic consolidation hypothesis. Prog. Neurobiol. 76, 99–125. doi: 10.1016/j.pneurobio.2005.06.003
Daba Feyissa, D., Sialana, F. J., Keimpema, E., Kalaba, P., Paunkov, A., Engidawork, E., et al. (2019). Dopamine type 1- and 2-like signaling in the modulation of spatial reference learning and memory. Behav. Brain Res. 362, 173–180. doi: 10.1016/j.bbr.2019.01.028
De Leonibus, E., Oliverio, A., and Mele, A. (2005). A study on the role of the dorsal striatum and the nucleus accumbens in allocentric and egocentric spatial memory consolidation. Learn. Memory 12, 491–503. doi: 10.1101/lm.94805
De Risi, M., Torromino, G., Tufano, M., Moriceau, S., Pignataro, A., Rivagorda, M., et al. (2020). Mechanisms by which autophagy regulates memory capacity in ageing. Aging Cell 19:e13189. doi: 10.1111/acel.13189
Ferretti, V., Roullet, P., Sargolini, F., Rinaldi, A., Perri, V., Del Fabbro, M., et al. (2010). Ventral striatal plasticity and spatial memory. Proc. Natl. Acad. Sci. U. S. A. 107, 7945–7950. doi: 10.1073/pnas.0911757107
Gasser, J., Pereira de Vasconcelos, A., Cosquer, B., Boutillier, A. L., and Cassel, J. C. (2020). Shifting between response and place strategies in maze navigation: effects of training, cue availability and functional inactivation of striatum or hippocampus in rats. Neurobiol. Learn. Mem. 167:107131. doi: 10.1016/j.nlm.2019.107131
Gengler, S., Mallot, H. A., and Hölscher, C. (2005). Inactivation of the rat dorsal striatum impairs performance in spatial tasks and alters hippocampal theta in the freely moving rat. Behav. Brain Res. 164, 73–82. doi: 10.1016/j.bbr.2005.06.009
Hamezah, H. S., Durani, L. W., Yanagisawa, D., Ibrahim, N. F., Aizat, W. M., Bellier, J. P., et al. (2018). Proteome profiling in the hippocampus, medial prefrontal cortex, and striatum of aging rat. Exp. Gerontol. 111, 53–64. doi: 10.1016/j.exger.2018.07.002
Hasselbalch, S. G., Madsen, K., Svarer, C., Pinborg, L. H., Holm, S., Paulson, O. B., et al. (2008). Reduced 5-HT2A receptor binding in patients with mild cognitive impairment. Neurobiol. Aging 29, 1830–1838. doi: 10.1016/j.neurobiolaging.2007.04.011
Hu, Y., Chen, W., Wu, L., Jiang, L., Liang, N., Tan, L., et al. (2019). TGF-β1 restores hippocampal synaptic plasticity and memory in Alzheimer model via the PI3K/Akt/Wnt/β-catenin signaling pathway. J. Mol. Neurosci. 67, 142–149. doi: 10.1007/s12031-018-1219-7
Ito, R., and Hayen, A. (2011). Opposing roles of nucleus accumbens core and shell dopamine in the modulation of limbic information processing. J. Neurosci. 31, 6001–6007. doi: 10.1523/JNEUROSCI.6588-10.2011
Ito, R., Robbins, T. W., Pennartz, C. M., and Everitt, B. J. (2008). Functional interaction between the hippocampus and nucleus accumbens shell is necessary for the acquisition of appetitive spatial context conditioning. J. Neurosci. 28, 6950–6959. doi: 10.1523/JNEUROSCI.1615-08.2008
Kähne, T., Kolodziej, A., Smalla, K. H., Eisenschmidt, E., Haus, U. U., Weismantel, R., et al. (2012). Synaptic proteome changes in mouse brain regions upon auditory discrimination learning. Proteomics 12, 2433–2444. doi: 10.1002/pmic.201100669
Kähne, T., Richter, S., Kolodziej, A., Smalla, K. H., Pielot, R., Engler, A., et al. (2016). Proteome rearrangements after auditory learning: high-resolution profiling of synapse-enriched protein fractions from mouse brain. J. Neurochem. 138, 124–138. doi: 10.1111/jnc.13636
Korol, D. L., and Pisani, S. L. (2015). Estrogens and cognition: friends or foes? An evaluation of the opposing effects of estrogens on learning and memory. Horm. Behav. 74, 105–115. doi: 10.1016/j.yhbeh.2015.06.017
Lavoie, A. M., and Mizumori, S. J. (1994). Spatial, movement- and reward-sensitive discharge by medial ventral striatum neurons of rats. Brain Res. 638, 157–168. doi: 10.1016/0006-8993(94)90645-9
Li, Y. Q., Prentice, D. A., Howard, M. L., Mashford, M. L., and Desmond, P. V. (1999). The effect of hormones on the expression of five isoforms of UDP-glucuronosyltransferase in primary cultures of rat hepatocytes. Pharm. Res. 16, 191–197.
Lobo, M. K., Cui, Y., Ostlund, S. B., Balleine, B. W., and Yang, X. W. (2007). Genetic control of instrumental conditioning by striatopallidal neuron-specific S1P receptor Gpr6. Nat. Neurosci. 10, 1395–1397. doi: 10.1038/nn1987
Lozano, Y. R., Serafín, N., Prado-Alcalá, R. A., Roozendaal, B., and Quirarte, G. L. (2013). Glucocorticoids in the dorsomedial striatum modulate the consolidation of spatial but not procedural memory. Neurobiol. Learn. Mem. 101, 55–64. doi: 10.1016/j.nlm.2013.01.001
Luine, V., and Hearns, M. (1990). Spatial memory deficits in aged rats: contributions of the cholinergic system assessed by ChAT. Brain Res. 523, 321–324. doi: 10.1016/0006-8993(90)91507-D
Maliković, J., Feyissa, D. D., Hussein, A. M., Höger, H., Lubec, G., and Korz, V. (2018). Moderate differences in feeding diets largely affect motivation and spatial cognition in adult and aged but less in young male rats. Front. Aging Neurosci. 10:249. doi: 10.3389/fnagi.2018.00249
Maliković, J., Feyissa, D. D., Kalaba, P., Marouf, B. S., Höger, H., Hartmann, M. F., et al. (2019). Age and cognitive status dependent differences in blood steroid and thyroid hormone concentrations in intact male rats. Behav. Brain Funct. 15:10. doi: 10.1186/s12993-019-0161-3
Martín-García, E., Fernández-Castillo, N., Burokas, A., Gutiérrez-Cuesta, J., Sánchez-Mora, C., Casas, M., et al. (2015). Frustrated expected reward induces differential transcriptional changes in the mouse brain. Addict. Biol. 20, 22–37. doi: 10.1111/adb.12188
McDonald, R. J., and White, N. M. (1994). Parallel information processing in the water maze: evidence for independent memory systems involving dorsal striatum and hippocampus. Behav. Neural Biol. 61, 260–270. doi: 10.1016/S0163-1047(05)80009-3
Meredith, G. E., and Totterdell, S. (1999). Microcircuits in nucleus accumbens' shell and core involved in cognition and reward. Psychobiology 27, 165–186.
Meyer, K., and Korz, V. (2013a). Age dependent differences in the regulation of hippocampal steroid hormones and receptor genes: relations to motivation and cognition in male rats. Horm. Behav. 63, 376–384. doi: 10.1016/j.yhbeh.2012.12.002
Meyer, K., and Korz, V. (2013b). Estrogen receptor α functions in the regulation of motivation and spatial cognition in young male rats. PLoS ONE 8:e79303. doi: 10.1371/journal.pone.0079303
Milbrandt, J. C., Albin, R. L., and Caspary, D. M. (1994). Age-related decrease in GABAB receptor binding in the Fischer 344 rat inferior colliculus. Neurobiol. Aging 15, 699–703. doi: 10.1016/0197-4580(94)90051-5
Nobel, C. S., Dunås, F., and Abrahmsén, L. B. (2002). Purification of full-length recombinant human and rat type 1 11beta-hydroxysteroid dehydrogenases with retained oxidoreductase activities. Protein Expr. Purif. 26, 349–356. doi: 10.1016/S1046-5928(02)00547-8
Packard, M. G., and Teather, L. A. (1997). Double dissociation of hippocampal and dorsal-striatal memory systems by posttraining intracerebral injections of 2-amino-5-phosphonopentanoic acid. Behav. Neurosci. 111, 543–551. doi: 10.1037/0735-7044.111.3.543
Paulose, C. S., and Balakrishnan, S. (2008). Adrenergic, dopaminergic and serotonergic gene expression in low dose, long time insulin and somatotropin treatment to ageing rats: rejuvenation of brain function. Biogerontology 9, 429–439. doi: 10.1007/s10522-008-9183-1
Pébay, A., Toutant, M., Prémont, J., Calvo, C. F., Venance, L., Cordier, J., et al. (2001). Sphingosine-1-phosphate induces proliferation of astrocytes: regulation by intracellular signalling cascades. Eur. J. Neurosci. 13, 2067–2076. doi: 10.1046/j.0953-816x.2001.01585.x
Qin, Z. H., Wang, Y., Chen, R. W., Wang, X., Ren, M., Chuang, D. M., et al. (2001). Prostaglandin A(1) protects striatal neurons against excitotoxic injury in rat striatum. J. Pharmacol. Exp. Ther. 297, 78–87.
Rutz, S., Majchrzak, M., Siedschlag, V., Barbelivien, A., Harati, H., Rothmaier, A. K., et al. (2009). The modulation of striatal dopamine release correlates with water-maze performance in aged rats. Neurobiol. Aging 30, 957–972. doi: 10.1016/j.neurobiolaging.2007.09.011
Sase, S., Meyer, K., Lubec, G., and Korz, V. (2014). Different expression of membrane-bound and cytosolic hippocampal steroid receptor complexes during spatial training in young male rats. Eur. Neuropsychopharmacol. 24, 1819–1827. doi: 10.1016/j.euroneuro.2014.09.003
Stemmelin, J., Lazarus, C., Cassel, S., Kelche, C., and Cassel, J. C. (2000). Immunohistochemical and neurochemical correlates of learning deficits in aged rats. Neuroscience 96, 275–289. doi: 10.1016/S0306-4522(99)00561-8
Tagawa, N., Yuda, R., Kubota, S., Wakabayashi, M., Yamaguchi, Y., Kiyonaga, D., et al. (2009). 17Beta-estradiol inhibits 11beta-hydroxysteroid dehydrogenase type 1 activity in rodent adipocytes. J. Endocrinol. 202, 131–139. doi: 10.1677/JOE-09-0021
Torromino, G., Autore, L., Khalil, V., Mastrorilli, V., Griguoli, M., Pignataro, A., et al. (2019). Offline ventral subiculum-ventral striatum serial communication is required for spatial memory consolidation. Nat. Commun. 10:5721. doi: 10.1038/s41467-019-13703-3
Turgeon, S. M., and Albin, R. L. (1994). GABAB binding sites in early adult and aging rat brain. Neurobiol. Aging 15, 705–711. doi: 10.1016/0197-4580(94)90052-3
van der Staay, F. J., van Nies, J., and Raaijmakers, W. (1990). The effects of aging in rats on working and reference memory performance in a spatial holeboard discrimination task. Behav. Neural Biol. 53, 356–370. doi: 10.1016/0163-1047(90)90226-V
Wackerlig, J., Köfeler, H. C., Korz, V., Hussein, A. M., Feyissa, D. D., Höger, H., et al. (2020). Differences in hypothalamic lipid profiles of young and aged male rats with impaired and unimpaired spatial cognitive abilities and memory. Front. Aging Neurosci. 12:204. doi: 10.3389/fnagi.2020.00204
Wang, G. J., Volkow, N. D., Logan, J., Fowler, J. S., Schlyer, D., MacGregor, R. R., et al. (1995). Evaluation of age-related changes in serotonin 5-HT2 and dopamine D2 receptor availability in healthy human subjects. Life Sci. 56, 249–253. doi: 10.1016/0024-3205(95)00066-F
Xu, X., Zhan, M., Duan, W., Prabhu, V., Brenneman, R., Wood, W., et al. (2007). Gene expression atlas of the mouse central nervous system: impact and interactions of age, energy intake and gender. Genome Biol. 8:R234. doi: 10.1186/gb-2007-8-11-r234
Keywords: learning, memory, cognition, striatum, aging, GeneChipTM microarray assay
Citation: Korz V, Kremslehner C, Maliković J, Hussein A, Feyissa DD, Nagelreiter I-M, Smidak R, Plasenzotti R, Gruber F and Lubec G (2021) Striatal Transcriptome Reveals Differences Between Cognitively Impaired and Unimpaired Aged Male Rats. Front. Aging Neurosci. 12:611572. doi: 10.3389/fnagi.2020.611572
Received: 29 September 2020; Accepted: 09 December 2020;
Published: 08 January 2021.
Edited by:
Brian C. Clark, Ohio University, United StatesReviewed by:
Giulia Torromino, National Research Council (CNR), ItalyXavier Xifró, University of Girona, Spain
Copyright © 2021 Korz, Kremslehner, Maliković, Hussein, Feyissa, Nagelreiter, Smidak, Plasenzotti, Gruber and Lubec. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gert Lubec, Z2VydC5sdWJlY0BsdWJlY2xhYi5jb20=
 Volker Korz
Volker Korz Christopher Kremslehner2
Christopher Kremslehner2 Jovana Maliković
Jovana Maliković Ahmed Hussein
Ahmed Hussein Daniel Daba Feyissa
Daniel Daba Feyissa Ionela-Mariana Nagelreiter
Ionela-Mariana Nagelreiter Roman Smidak
Roman Smidak Florian Gruber
Florian Gruber Gert Lubec
Gert Lubec