
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Aging Neurosci. , 23 October 2020
Sec. Alzheimer's Disease and Related Dementias
Volume 12 - 2020 | https://doi.org/10.3389/fnagi.2020.589588
 Pingfang Qi1
Pingfang Qi1 Jing Li2
Jing Li2 Shichao Gao3,4
Shichao Gao3,4 Yirong Yuan5
Yirong Yuan5 Yindi Sun6
Yindi Sun6 Na Liu6
Na Liu6 Yuanyuan Li7
Yuanyuan Li7 Gang Wang8
Gang Wang8 Ling Chen8*†
Ling Chen8*† Jing Shi7*†
Jing Shi7*†Alzheimer’s disease (AD) is one of the neurodegenerative brain disorders inducing nearly half of dementia cases, and the diagnosis and treatment of AD are the primary issues clinically. However, there is a lack of effective biomarkers and drugs for AD diagnosis and therapeutics so far. In this study, bioinformatics analysis combined with an experimental verification strategy was used to identify the biomarkers and the quercetin targets for AD diagnosis and treatment. First, differentially expressed genes in the AD brain were identified by microarray data analysis. Second, quercetin, a predominant flavonoid, was used to screen the target genes. Third, the drug–disease network was determined, and the target genes of quercetin treatment were obtained in AD-related HT-22 cell-based assay. Six genes, including MAPT, PIK3R1, CASP8, DAPK1, MAPK1, and CYCS, were validated by the system pharmacology analysis in the hippocampus samples of AD patients. The results suggested that MAPT, PIK3R1, CASP8, and DAPK1 were significantly increased, but MAPK1 and CYCS were significantly decreased in HT-22 cells after Aβ1-42 treatment. Moreover, MAPK1 and CYCS were markedly increased, but MAPT, PIK3R1, CASP8, and DAPK1 were markedly decreased after quercetin treatment in these HT-22 cells. Altogether, MAPT, PIK3R1, CASP8, DAPK1, MAPK1, and CYCS are all the biomarkers for AD diagnosis and the targets of quercetin treatment, and our findings may provide valuable biomarkers for AD diagnosis and treatment.
Alzheimer’s disease (AD) is a neurodegenerative brain disease that is characterized by progressive cognitive impairment and memory loss, and psychiatric symptoms, which mostly induces over half of the dementia cases (Najafi et al., 2016). An estimated 115 million patients will be diagnosed with AD by 2050. These AD patients will progress to dementia within 5 years of diagnosis and will account for approximately half of all dementia cases (Luo et al., 2016). The typical pathogenesis of AD is the accumulation of amyloid-β (Aβ) aggregates, and the hyperphosphorylation of the tau proteins, which together lead to neurofibrillary tangles (NFTs) and synaptic dysfunction (Wallace and Dalton, 2011; Asaad and Lee, 2018). Current AD therapy is only for a single target, the effects are also limited, and always accompanied by many adverse effects (Aliev et al., 2008; Rountree et al., 2009). Therefore, an active compound against one or more of these triggering factors may be a promising strategy for the treatment of AD.
Phytochemical flavonoids are powerful eliminators of reactive oxygen and nitrogen species, which are associated with oxidative stress (Russo et al., 2012). Flavonoids have been proven beneficial in the prevention of neurodegenerative disorders and also may delay the progress of neurodegeneration (Solanki et al., 2015). Previous studies showed that the flavonoids are attributed to the reduction of Aβ toxicity and decreasing oxidative stress, which is important in ameliorating the pathogenesis of AD (Baptista et al., 2014; Deng et al., 2017).
Quercetin (3,5,7,30,40-penta hydroxyflavone), a predominant flavonoid, is one of the most effective antioxidants of plant origin and commonly in edible plants (Brüll et al., 2015). Quercetin has various beneficial effects on human health, such as anticarcinogenic, anti-inflammatory, and anti-infective effects. It also inhibits lipid peroxidation and platelet aggregation and stimulates mitochondrial biogenesis (Li et al., 2016). The quercetin can cross the blood–brain barrier (BBB) and exhibits antioxidant and anti-inflammatory properties in the brain (Youdim et al., 2004), which exerts the neuroprotective effects in neurodegenerative disorders, such as AD (Rishitha and Muthuraman, 2018) and Parkinson’s disease (PD) (El-Horany et al., 2016). Increasing evidence suggested that quercetin has shown therapeutic efficacy in multiple AD animal models, such as mouse and Drosophila models of AD (Richetti et al., 2011; Wang et al., 2014; Sabogal-Guáqueta et al., 2015). Despite a large number of studies on the biological activity of quercetin, the mechanisms of its action on the treatment of AD are still not well understood.
In this study, bioinformatics analyses, including microarray data analysis and an integrated system pharmacology approach, were used to examine the mechanisms of quercetin treatment. First, the quercetin was used to screen the potential target genes, and then, the differentially expressed genes of AD were identified using microarray datasets. Second, the quercetin target genes would be confirmed as the AD-related genes. Finally, the predicted target genes were verified by quantitative polymerase chain reaction (qPCR) with different doses of quercetin treatment in the HT-22 cells. Our study provides a more specific and effective way to offer new insight into the mechanisms of quercetin in the treatment of AD.
The chemical structure of quercetin was obtained from the PubChem database1. Afterward, the quercetin target genes were screened from the PharmMapper database2, the Similarity ensemble approach (SEA) database3, and the function of the target genes were analyzed by the Swiss Target Prediction database4.
Microarray dataset GSE5281 was downloaded from the Gene Expression Omnibus (GEO) database and collected using the GPL570 platform (Affymetrix Human Genome U133 Plus 2.0 Array). This microarray dataset formed the study of “Alzheimer’s disease and the normal aged brain.” This study aims to find the differentially expressed genes (DEGs) of the AD brain (Liang et al., 2008), which is consistent with the design of our study. Difference analysis was performed by R script using limma (Linear models for microarray analysis) R package, p < 0.05 and |logFC| > 1 as cutoff values for screening DEGs. The DEGs of the comparison group were shown as the volcano plot. The heatmap of the expression data was generated using ClustVis online tools5. The expression value of the DEGs was obtained from the GEO2R online tools.
The interaction of quercetin target genes and DEGs of AD patients were obtained by the Venn diagram. Cytoscape 3.7.2 was used to determine the drug–target–disease network. Furthermore, the STRING database6 was used to analyze the protein–protein interactions (PPIs) of the quercetin potential target genes, and the hub genes were counted by R script.
The GO and KEGG enrichment analysis was using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) database7, which is an integrated online biological knowledge base and analytical tool. In this study, the target genes were mapped into DAVID and to identify the biological processes, cellular components, molecular function, and KEGG pathways of the predicted target genes involved. The map of the KEGG signaling pathway was obtained from the KEGG database8.
The peptide Aβ1-42 (Invitrogen, Carlsbad, CA, United States) was completely dissolved in DMSO (Sigma, St. Louis, MO, United States), then cold Ham’s F12 was added (Caisson Labs, Smithfield, UT, United States) and incubated at 4°C overnight. The solution was then centrifuged at 14,000 × g × 10 min at 4°C. The supernatant was carefully transferred to a sterile tube. This Aβ1-42 stock solution was stored at -80°C and equilibrated for 1 h at room temperature before use. Quercetin (Acmec biochemical, Shanghai, China) was diluted to a final concentration of 100 μM in DMSO before use.
The HT-22 cells were cultured in DMEM medium with 10% fetal bovine serum (FBS) and 1% penicillin–streptomycin (PS) and incubated at 37°C in 5% CO2 cell incubator. Cells were divided into four groups, normal control (NC), NC group treatment with quercetin, Aβ1-42 treatment, and Aβ1-42 group treatment with quercetin. When the cells were confluent (>90%), the fresh media containing various concentrations of drugs (10 μM Aβ1-42 for 12 h, and 100 μM quercetin for 48 h) were added to the carefully aspirated wells. Cells were then harvested and prepared for the following tests.
The cells were collected and washed by 4°C PBS. Then, the RNeasy Mini kit (Qiagen, Germany) was used to extract the total RNA and reversed by ThermoScript RT-PCR System (Invitrogen, Carlsbad, CA, United States). The mRNA expression was detected by Talent qPCR kit (TIANGEN, Beijing, China). The primers used in this study are shown in Table 1.
Statistical analysis was calculated by GraphPad Prism software version 7.0 (CA, United States). All data were displayed as the mean ± SEM. The comparison of the two groups was analyzed using the two-tailed Student’s t-test. A value of p < 0.05 was considered significant.
The chemical structure of quercetin was obtained from the PubChem database as shown in Figure 1A. Based on its structure, PharmMapper database, SEA database, and Swiss Target Prediction database were used to predict the potential target genes of quercetin. A total of 277 potential target genes were obtained from those databases, and then, the function of the target genes was classified according to their biochemical criteria, as shown in Figure 1B. These 277 targets mainly include kinase, enzyme, lyase, protease, family A/G protein-coupled receptors, etc.
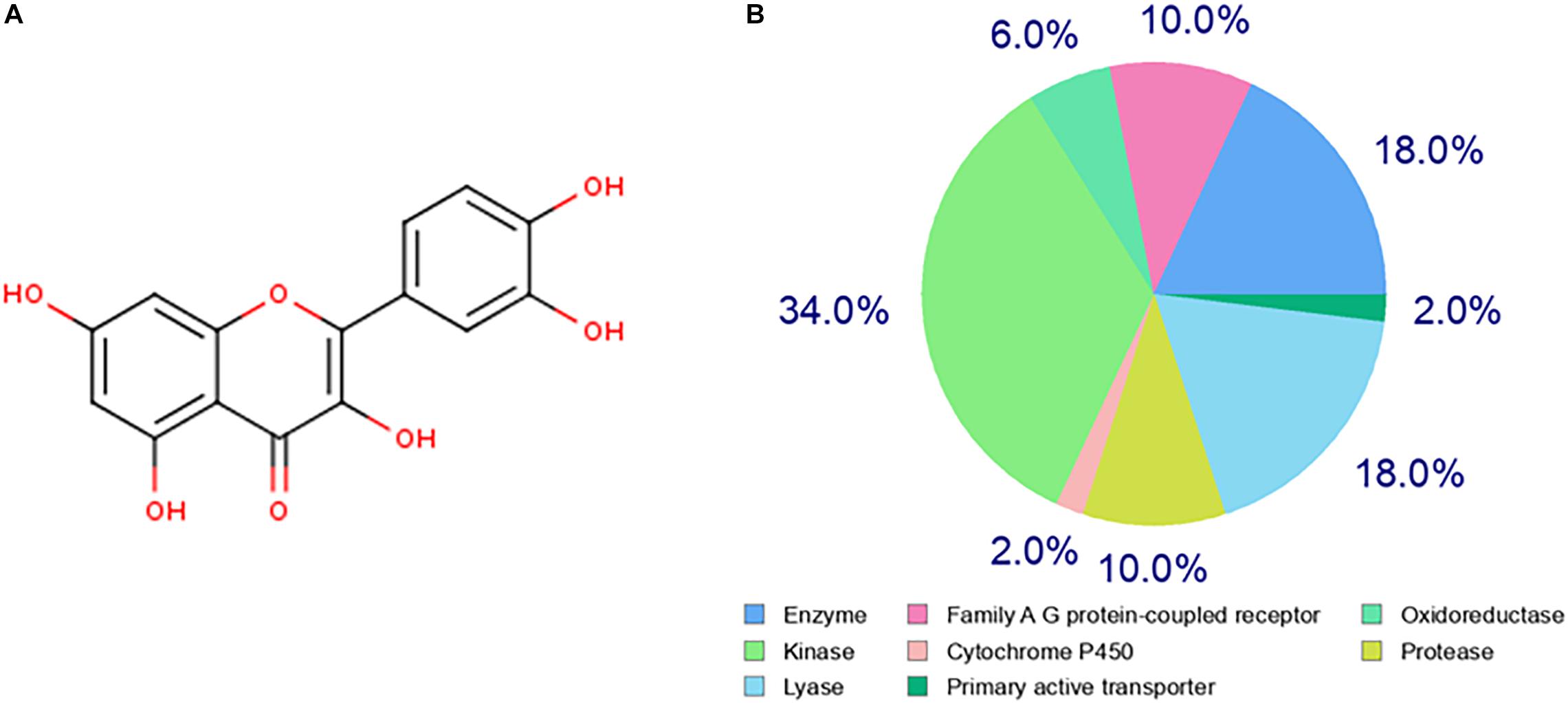
Figure 1. Prediction of the quercetin target genes. (A) The chemical structure of the quercetin. (B) The classification of drug target genes according to their biochemical criteria. Different colors indicate different items.
The hippocampus gene expression datasets GSE5821 was downloaded from the GEO database for analyzing the AD-related gene changes. The gene expression analysis was made on health control and AD groups. As shown in Figures 2A,B, there were a total of 3,256 differentially expressed genes with 1,227 upregulated and 2029 downregulated in AD patients, which may be closely related to the progression of AD.
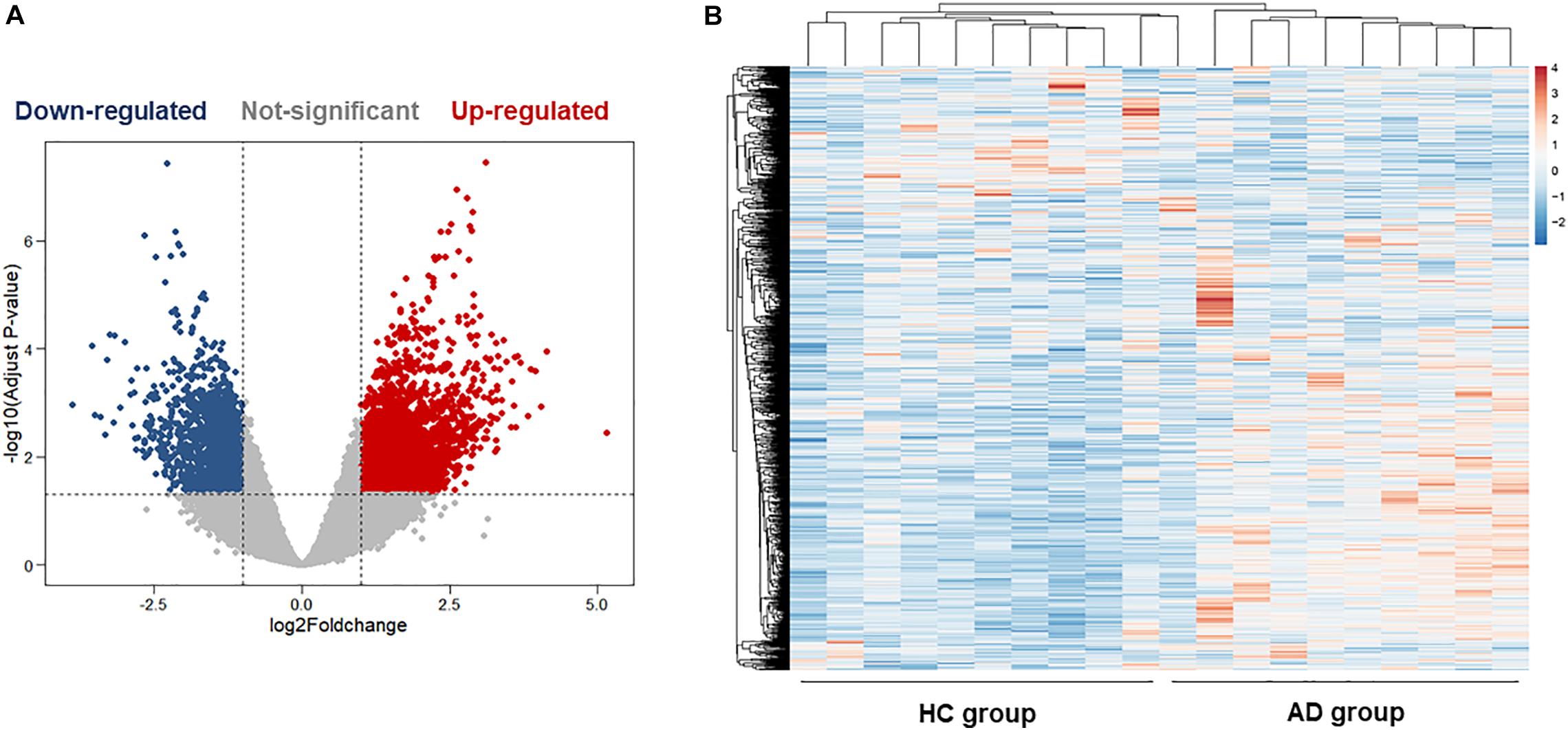
Figure 2. Bioinformatics analysis of differentially expressed genes (DEGs) in the hippocampus of health control (HC) and Alzheimer’s disease (AD) patients. (A) The volcano plot of DEGs in the hippocampus between the HC group and the AD group. (B) Heatmaps of DEGs in the hippocampus of the HC group and the AD group. Colors in the heatmaps indicate the row Z-score among the different datasets. High expression is shown by the red color, and low expression is shown by the blue color.
As shown in Figure 3, we found 277 potential target genes of quercetin treatment, and 3,256 differentially expressed genes of AD. To further analyze the quercetin’s effects on AD, the interaction of drug target genes and AD-related genes was used as a Venn diagram. As shown in Figure 3A, there were 46 genes in both groups, and these target genes were not only AD-related genes but also the drug targets. At the same time, an interactive Quercetin-target gene–AD network was constructed (Figure 3B), which indicated that quercetin might have effect on AD by stimulating or inhibiting these target genes.
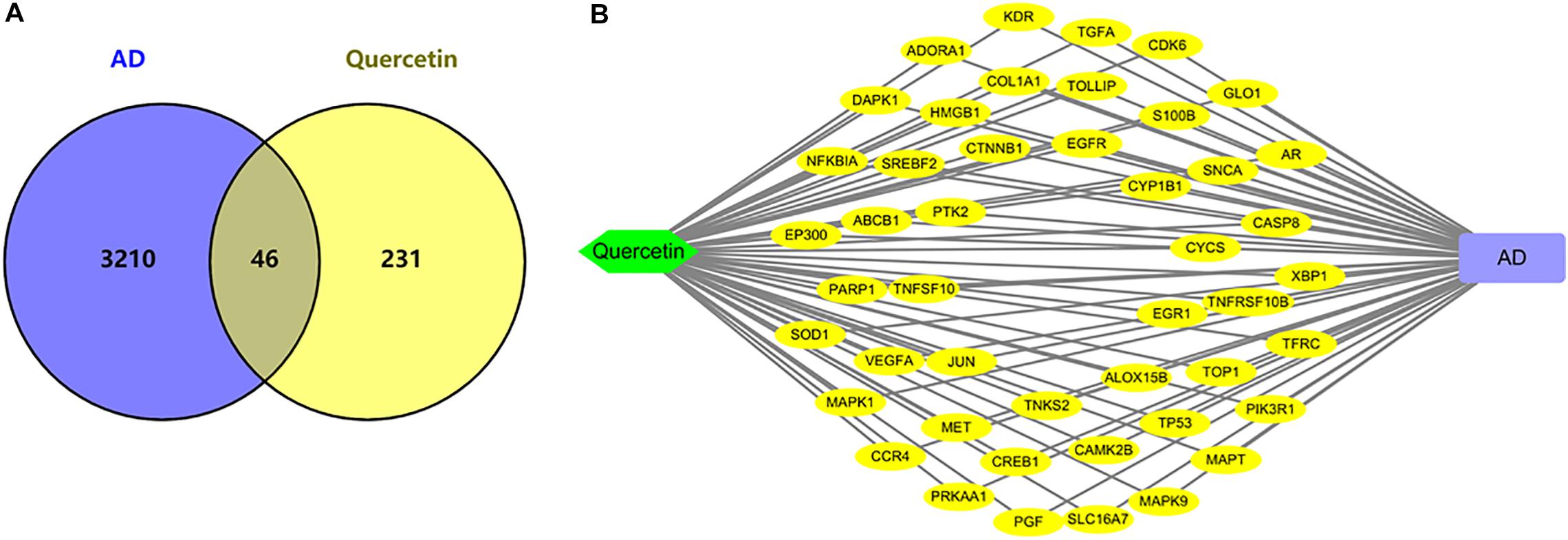
Figure 3. Network analysis of quercetin target genes and AD differentially expressed genes. (A) Venn diagram of quercetin target genes and AD differentially expressed genes. (B) The network of Quercetin, AD, and all the potential target genes. Yellow nodes represented the target genes.
In order to explore the relationship between these 46 potential target genes, the protein–protein interaction (PPI) analysis was done by the STRING database. A network was generated, the hub genes were analyzed, the top 20 hub genes in the network were marked in red, and other genes connected with hub genes were presented by purple nodes (Figure 4A). A bar plot of the number of hub gene links is shown in Figure 4B, and we found that the hub genes, such as TP53, MAPK1, CYCS, CASP8, PIK3R1, and MAPT, may play a critical role in the biological activity in the quercetin treatment process.
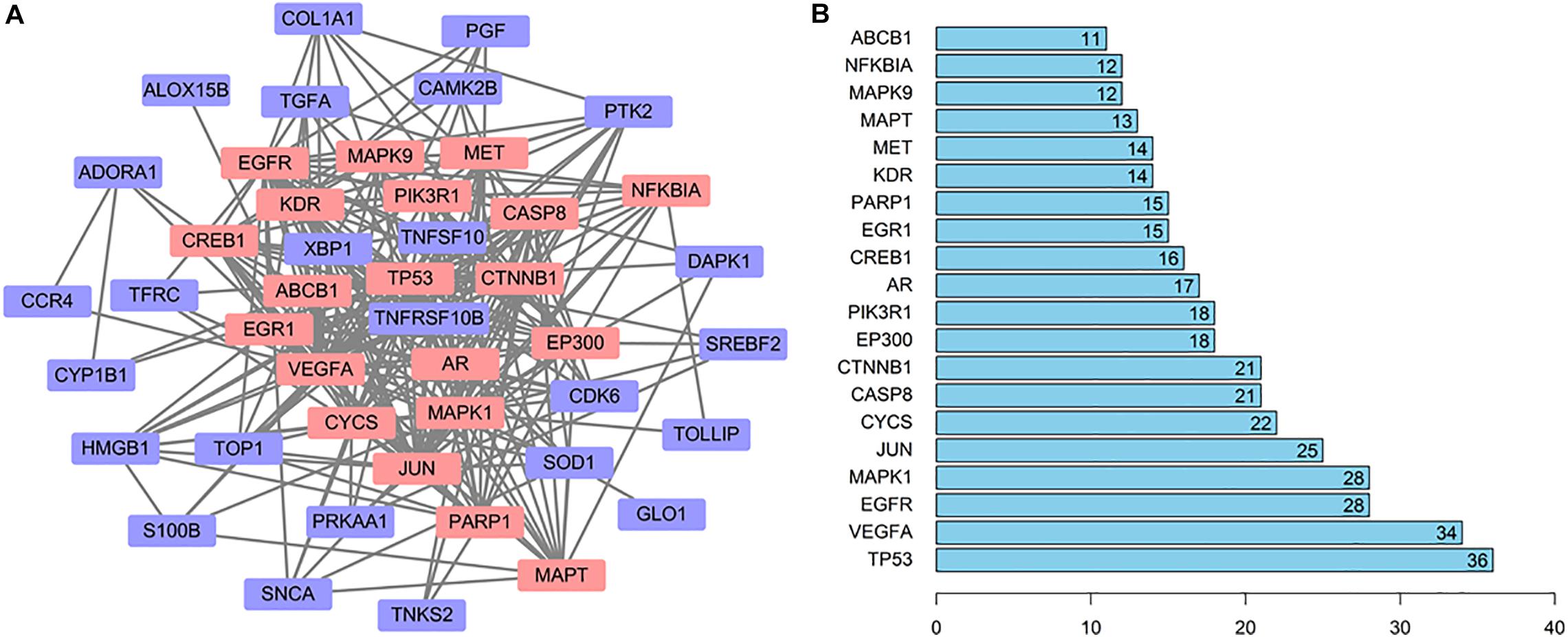
Figure 4. The interaction network of the quercetin target genes. (A) The protein—protein interaction (PPI) network of the quercetin target genes. The orange nodes are the hub genes in the network. (B) Bar plot of the number of hub gene links.
To gain a comprehensive understanding of these target genes, the DAVID database was used to do the GO and KEGG enrichment analysis. A variety of GO enrichment terms were enriched, including 76 biological processes, 13 cellular components, and 12 molecular functions. The top 10 GO terms are shown in Figure 5. We found that the biological processes such as negative regulation of apoptotic process and neuronal migration (Figure 5A), cellular components like nucleus and mitochondrion (Figure 5B), and molecular function like chromatin binding and ATP binding (Figure 5C), were enriched, which may be involved in the biological activity in the quercetin treatment process. In addition, 75 KEGG pathways were enriched (Figure 6A), and the important genes were mainly distributed in the PI3K–AKT signaling pathway (Figure 6B), which indicate that quercetin might ameliorate AD-related symptoms through the PI3K–AKT signaling pathway.
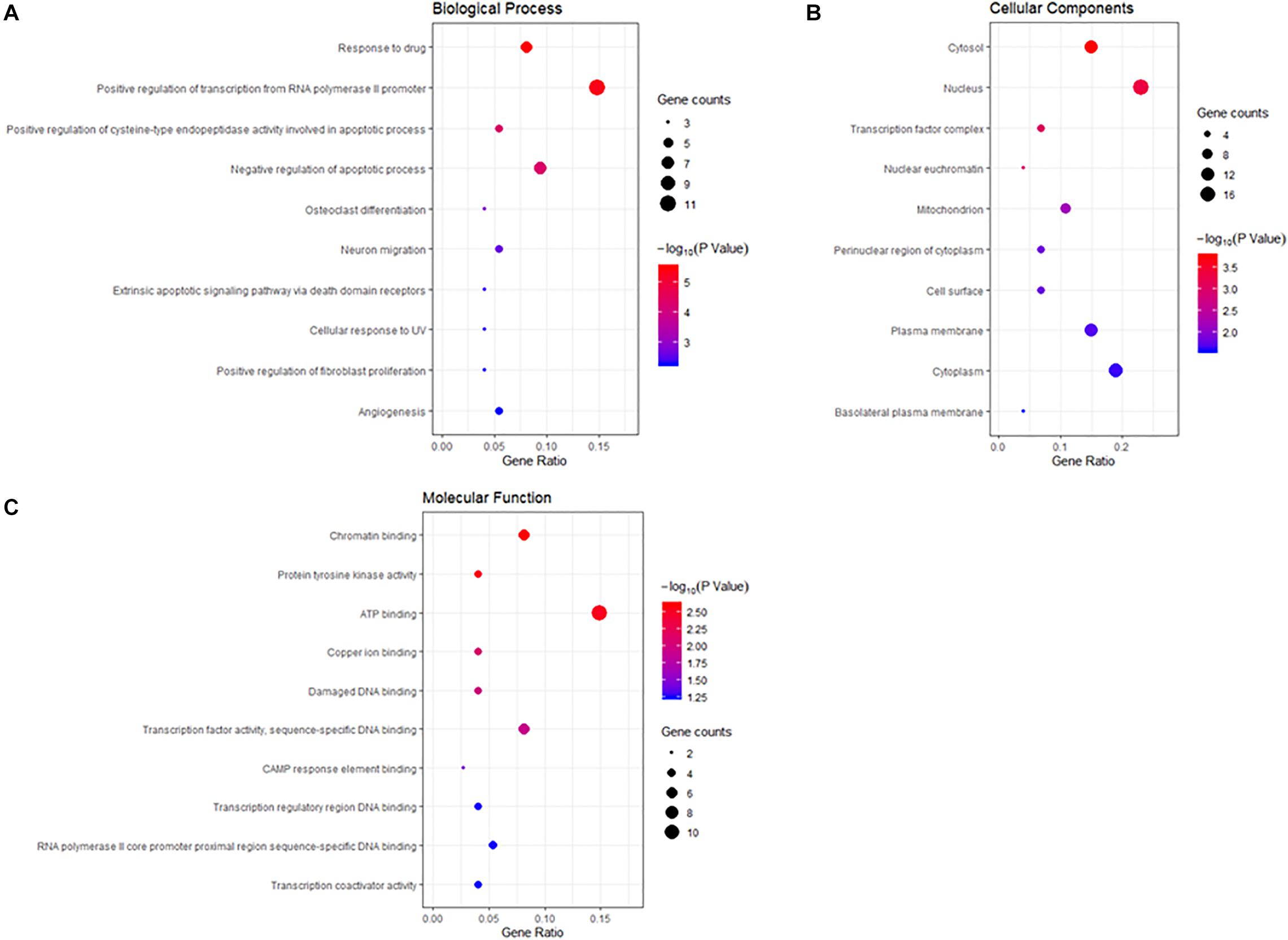
Figure 5. Gene ontology (GO) enrichment analysis of the quercetin target genes. (A) The plot of enriched biological processes. (B) The plot of enriched cellular components. (C) The plot of enriched molecular functions. The number of genes enriched in each GO term is shown as the circle size, the p-value shown as different colors.
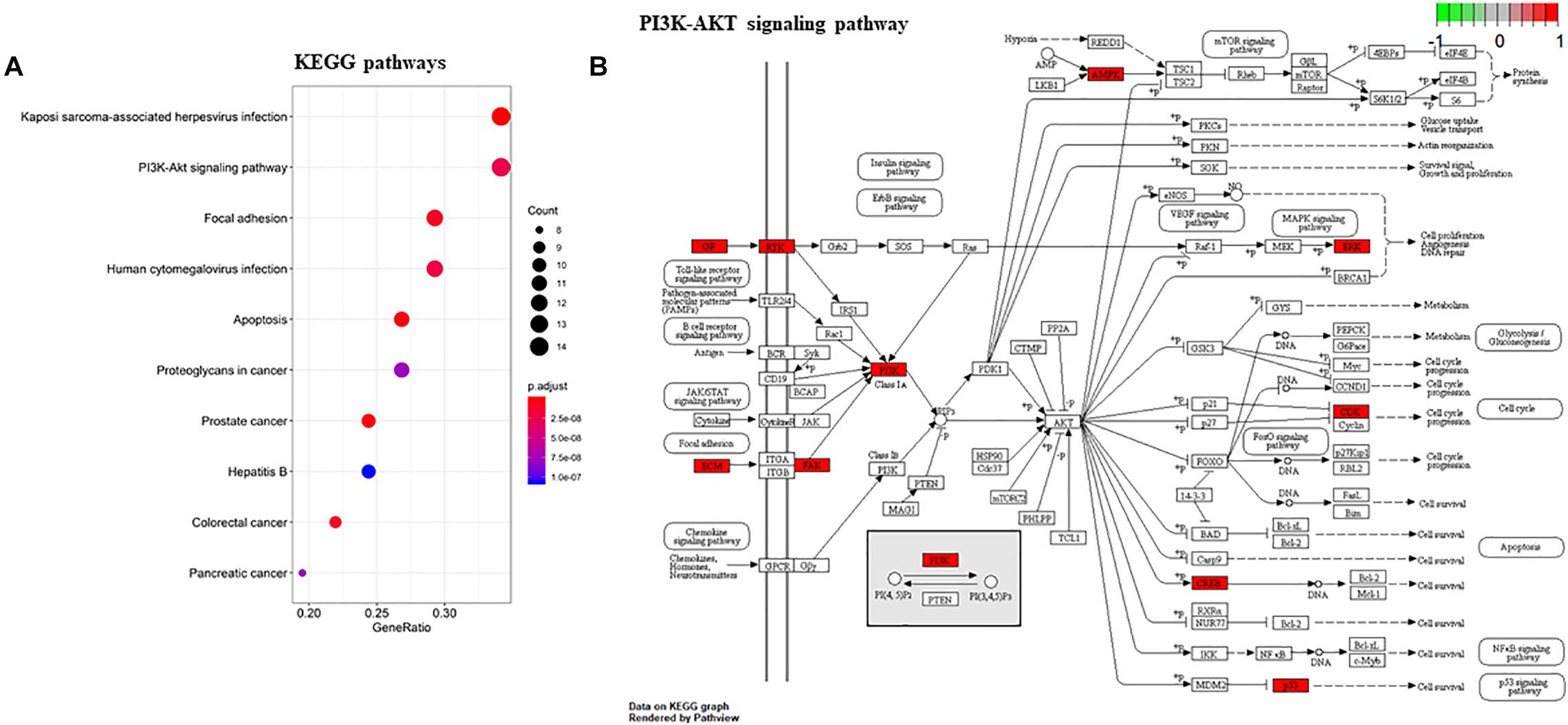
Figure 6. KEGG enrichment analysis of the quercetin target genes. (A) KEGG annotation of target genes. The number of genes enriched in each KEGG term is shown as the circle size, the p-value shown as different colors. (B) The important genes were mainly distributed in the PI3K–AKT signaling pathway. Arrows represent activation effect, T-arrows represent inhibition effect and segments show activation effect or inhibition effect. The red nodes are the intersection genes.
Based on the system pharmacology analysis of quercetin target genes, and the results of the network and functional enrichment analysis, six genes, including MAPT, PIK3R1, CASP8, DAPK1, MAPK1, and CYCS, were selected to validate the AD treatment effects of the quercetin. As shown in Figure 7, MAPT, PIK3R1, CASP8, and DAPK1 were significantly increased (p < 0.05, Figures 7A–D), but MAPK1 and CYCS were significantly decreased (p < 0.05, Figures 7E,F) in the AD group of the GSE5281 dataset.
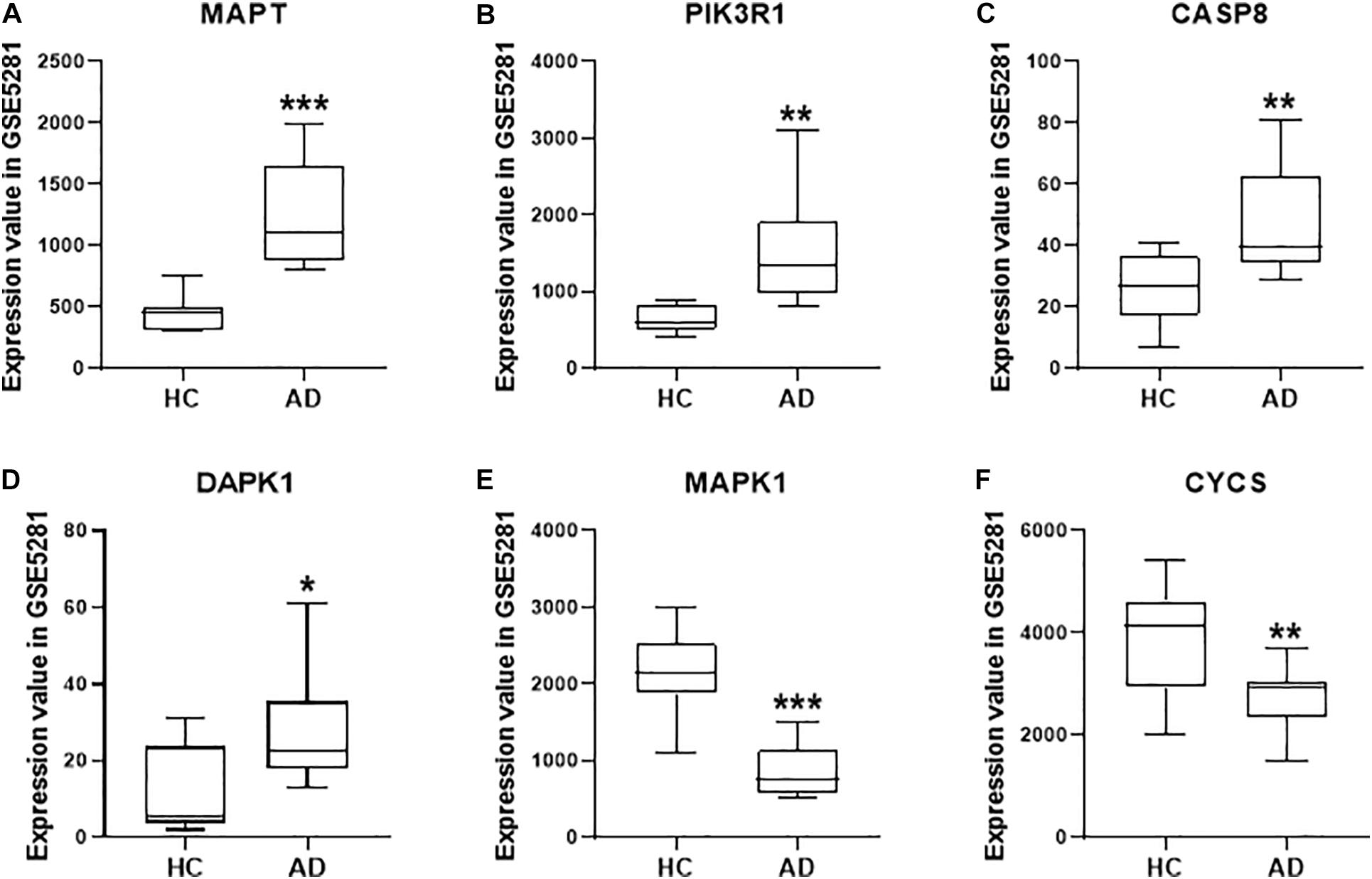
Figure 7. The quercetin target gene expression in HC and AD groups of GSE5281 dataset. The expression of MAPT (A), PIK3R1 (B), CASP8 (C), DAPK1 (D), MAPK1 (E), and CYCS (F) in different groups. Values are presented mean ± SEM (n = 10 in each group). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 vs. HC group.
To further validate whether these six genes are related to AD pathology, we performed a serial of qPCR quantification in the HT-22 cells in the presence of Aβ1-42. As shown in Figure 8, the mRNA levels of MAPT, PIK3R1, CASP8, and DAPK1 were significantly increased (Figures 8A–D, p < 0.05), and MAPK1 and CYCS were significantly decreased (Figures 8E,F, p < 0.05), after treatment of the HT22 cells with Aβ1-42, which were consistent with the GSE5281 dataset results. Moreover, the mRNA levels of MAPT, PIK3R1, CASP8, and DAPK1 were significantly decreased (Figures 8A–D, p < 0.05), and MAPK1 and CYCS were significantly increased (Figures 8E,F, p < 0.05), after treatment of these HT22 cells with quercetin for 48 h. These results support that all six genes are closely related to the AD process as the quercetin treatment targets.
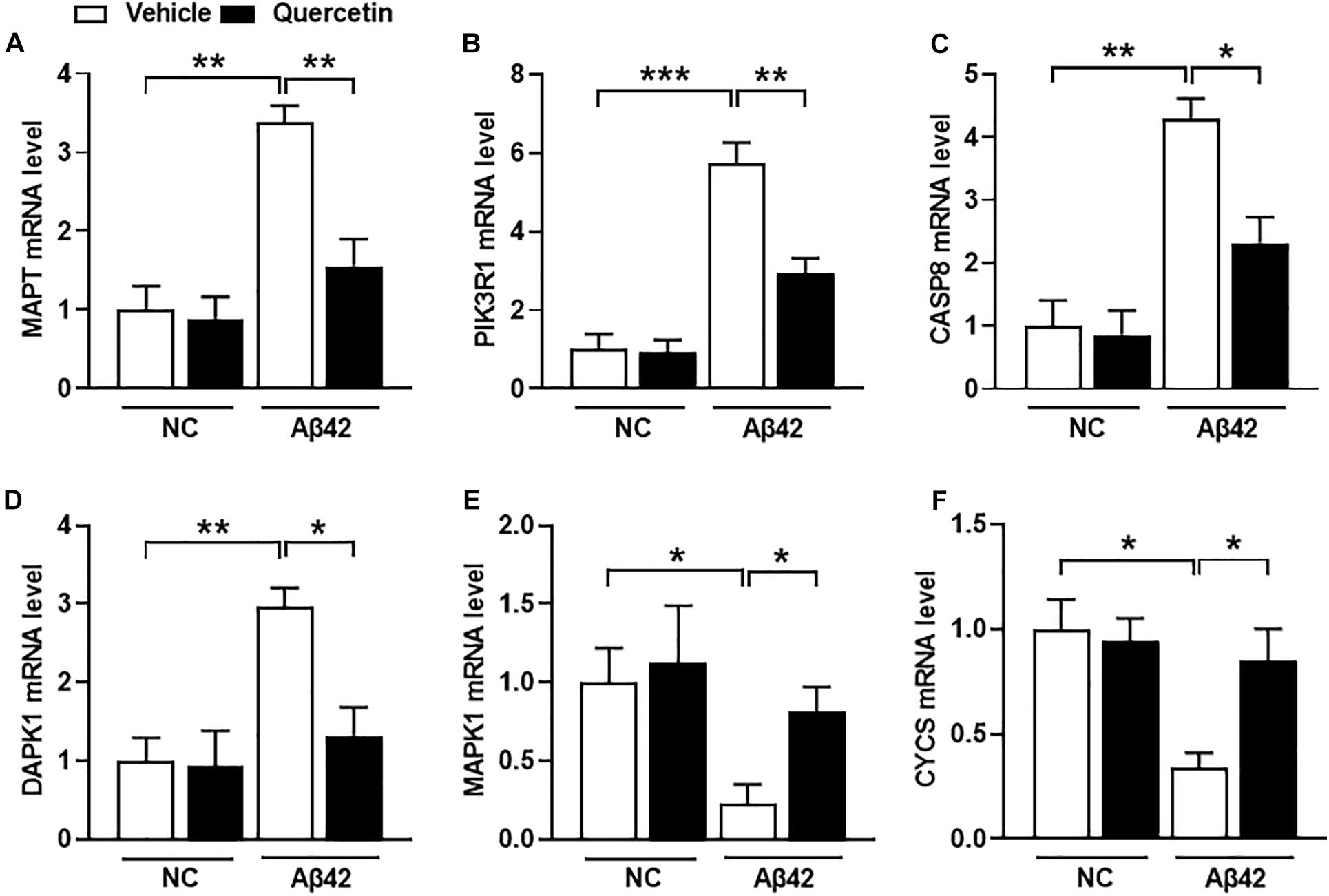
Figure 8. The quercetin potential target genes among different groups of HT22 cells by quantitative polymerase chain reaction (qPCR) analysis. The mRNA level of MAPT (A), PIK3R1 (B), CASP8 (C), DAPK1 (D), MAPK1 (E), and CYCS (F) in different treatment groups. Data were normalized to β-actin, and values are presented as mean ± SEM (n = 5 in each group). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Alzheimer’s disease is closely related to neuroinflammation, impairment of cerebral circulation, altered synaptic function, and cerebral amyloid angiopathy, and these typical pathological changes are considered to be important drug targets for AD treatment (Ghanemi, 2015). There is a lot of evidence that suggests that some dietary supplements could minimize the risk of AD (Hartman et al., 2006). The flavonol quercetin, which can be found in various fruits and vegetables, has received extensive attention due to its potential biological activities (Cooper and Ma, 2017). Some studies suggested that quercetin can protect neurons against the oxidative stress by regulating the process of apoptosis in the neuronal cells (Ansari et al., 2009). Other studies found that quercetin improves cholinergic function and acts as a neuroprotective agent in neurodegenerative diseases (Gupta et al., 2016). Recent studies have been suggested that quercetin may inhibit the NFT formation by attenuating the Aβ aggregation (Zhang et al., 2016). However, the molecular mechanisms of quercetin to ameliorate AD are still unknown.
In our study, we examined the AD-related quercetin target genes through a series of bioinformatics analysis combined with the subsequent experimental verification by real-time RT-PCR assay. Based on these strategies, we obtained 46 genes, which were related to AD pathology and quercetin treatment. To further understand the function of these genes, the PPI analysis and functional enrichment analysis was conducted afterward. We found that the hub genes, such as TP53, MAPK1, CYCS, CASP8, PIK3R1, and MAPT, may play a critical role in the biological pathway in the quercetin treatment process. The negative regulation of the apoptotic process, ATP binding, mitochondrion, and PI3K–AKT signaling pathway may also be involved in the biological activity in the quercetin treatment process. After analysis of these 46 genes by network and functional enrichment analysis, six genes, including MAPT, PIK3R1, CASP8, DAPK1, MAPK1, and CYCS, were chosen for experimental validation. We found that all the six genes were verified to be differentially expressed in the GSE5281 dataset and after the Aβ1-42 treatment. Meanwhile, all the six genes were markedly changed after quercetin treatment, suggesting that they might serve as potential targets in the progression of AD and quercetin treatment.
The microtubule-associated protein tau (MAPT) is an intracellular tau protein mainly restricted to axons in mature neurons (Binder et al., 1985; Kosik et al., 1986), which plays an important role in assembling and stabilizing of microtubules (Mandelkow and Mandelkow, 2012). The aberrant aggregation of MAPT in neurons induced many irreversible, progressive neurodegeneration diseases, such as AD, corticobasal degeneration (CBD), and frontotemporal dementia with parkinsonism linked to chromosome 17 (FTDP-17) (Guo et al., 2017). The MAPT, mislocated to the somatodendritic compartments, is one of the early signs of AD neurodegeneration (Braak et al., 1994; Balaji et al., 2018). In addition, the accumulation of MAPT in dendrites is a critical step in Aβ-induced neurotoxicity (Ittner et al., 2010; Zempel et al., 2010). In this study, we predicted and verified that the level of MAPT was significantly increased in the brain sample of AD patients and after Aβ1-42 treatment in HT-22 cells. Moreover, quercetin treatment can inhibit MAPT expression, which indicates that MAPT is the target in the treatment of quercetin in AD patients. Therefore, it is important to explore the pathological changes of MAPT in the progression of AD, which also has an important implication for tau-targeting therapeutics (Strang et al., 2019).
Death-associated protein kinase 1 (DAPK1) is a death domain-containing and stress-responsive serine/threonine protein kinase (Bialik and Kimchi, 2006). Via its death domain, DAPK1 is a positive mediator of apoptosis-like cell death and can be induced by a variety of death receptor activations (Stevens and Hupp, 2008). The previous study showed that DAPK1 deletion mice can enhance the learning and spatial memory (Yukawa et al., 2006), which may be through the regulation of DAPK1 in tau toxicity by modulating microtubule (MT) assembly and neuronal differentiation (Wu et al., 2011). However, the association between DAPK1 and AD has not been sufficiently explored, and DAPK1 regulation in AD progression is poorly understood. In the present study, our results demonstrated that DAPK1 expression is increased in AD patients and after the Aβ1-42 treatment in HT-22 cells, which can be inhibited by quercetin treatment. This suggests that DAPK1 may be a novel therapeutic target for treating human AD and other tau-related pathologies as a valuable biomarker of the progression of AD.
Caspase-8 (CASP8) belongs to the cysteine–aspartic acid protease (caspase) family and plays a pivotal initiator role in the death receptor pathway of apoptosis (Ashkenazi and Dixit, 1999). CASP8 is thought to activate downstream caspases, particularly caspase-3, which is an early event in the apoptotic cascade (Takahashi, 1999). Caspase-3 is involved in the APP proteolysis (Gervais et al., 1999) and increased in the AD brains (Su et al., 2001), which indicates that CASP8 might be activated in the AD brain (Stadelmann et al., 1999). CASP8 was reported to mediate Aβ-induced neuronal apoptosis in an animal study (Ivins et al., 1999), and Aβ may activate CASP8 through the cross-linking and activation of receptors of the death receptor family, thereby inducing neuronal apoptosis of AD brain (Rohn et al., 2001). In the present study, CASP8 was significantly increased in the AD brain and Aβ1-42-treated HT-22 cells, which can be inhibited after quercetin treatment. This indicates that CASP8 has the function to promote the AD process as the target for quercetin treatment. Although the function of caspases in AD has been proposed, the role of CASP8 in AD still requires further studies.
Mitogen-activated protein kinase 1 (MAPK1), a serine/threonine kinase, is one of the components of the MAPK signaling pathway, which can activate many cellular effects, including the regulation of cell growth, survival, and differentiation (Fang and Richardson, 2005). MAPK1 can induce the overactivation of Dynamin-related protein 1 (Drp1), the mitochondrial fission protein, leading to mitochondrial fragmentation, neuronal apoptosis (Pyakurel et al., 2015), and neurodegeneration (Roe and Qi, 2018). The cytochrome c, somatic (CYCS) is the isoform of cytochrome c (Cyt c), as an integral membrane protein located in the mitochondrial intermembrane, and participant in ATP synthesis of the mitochondria (Ow et al., 2008), and also play an important role in apoptosis (Zhivotovsky et al., 1998). Based on the function in mitochondria energy metabolism as well as in apoptosis, MAPK1 and CYCS were chosen as the candidate genes to investigate the AD progress and quercetin treatment. In the present study, MAPK1 and CYCS were significantly decreased in AD patients and the Aβ1-42-treated HT-22 cells, which can be reversed by quercetin treatment. This indicates that MAPK1 and CYCS were involved in the progression of AD and the quercetin treatment.
Phosphoinositide-3-kinase, regulatory subunit 1 (PIK3R1) is one of the isoforms of phosphoinositide 3-kinase (PI3K). PI3K is a heterodimer that consists of the subunit of p85 and p110 (Ueki et al., 2000). The PIK3R1 gene encodes the PI3K catalytic subunit p85α in most eukaryotic cells (Taniguchi et al., 2006). PI3K pathway plays an essential role in proliferation, apoptosis, and metabolism of both normal and malignant cells (Vanhaesebroeck et al., 2012). It has been reported that PIK3R1 is differentially expressed in many human cancers and is closely related to tumor progression and metastasis (Lin et al., 2015), which make it an important therapeutic target through inhibiting the PI3K/AKT/mTOR pathway (Lahusen et al., 2009). However, the role of PIK3R1 in the AD progress and quercetin treatment remains blank. In this study, we first demonstrated that PIK3R1 was significantly increased after Aβ1-42 treatment, and it was also the target of quercetin treatment. Further studies that focus on the roles of PIK3R1 in the diagnosis and treatment of AD patients are essential and of great interest.
This bioinformatics analysis combined with the experimental verification strategy provides six potential quercetin target genes for the AD treatment, which is the first study to use such an approach for predicting quercetin treatment targets. The limitation of our study is that only HT-22 cells treated with Aβ1-42 monomer were used to simulate the AD status. Alternative approaches, using human neuronal cells, such as iPS-derived neurons, and Aβ1-42 oligomer/aggregate rather than monomer, as well as AD animal models, would further support our findings in the future.
A total of 277 target genes of quercetin and 3,256 differentially expressed genes in AD were obtained in our study, and then the functional analysis suggested that six genes (MAPT, PIK3R1, CASP8, DAPK1, MAPK1, and CYCS) were involved in the progression of AD and the treatment target of AD patients. Our study provides valuable information to investigate the pathogenesis of AD and the potential mechanisms of quercetin in the treatment of AD.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
PQ and JL wrote and prepared the original draft. SG and YY were responsible for the software and data curation. YS and NL were in charge of the resources and validation. YL and GW wrote, reviewed and edited the manuscript. LC and JS were responsible for the conceptualization, funding acquisition, and supervision. All authors approved the manuscript for publication.
This study was supported by the Science and Technology Plan Project of Zhejiang Province (No. 2018C37082) to LC, the Science and Technology Plan Project of Zhejiang Province (No. LGF20H300002) and the “13th Five-Year” Chinese Medicine Key Discipline in Zhejiang Province – Chinese Medicine Quality and Functional Evaluation (2017-XK-A43) to JS, and the Scientific Research Project of Xi’an Municipal Health Commission (No. 2020yb32) to NL.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Aliev, G., Obrenovich, M. E., Reddy, V. P., Shenk, J. C., Moreira, P. I., Nunomura, A., et al. (2008). Antioxidant therapy in Alzheimer’s disease: theory and practice. Mini Rev. Med. Chem. 8, 1395–1406. doi: 10.2174/138955708786369582
Ansari, M. A., Abdul, H. M., Joshi, G., Opii, W. O., and Butterfield, D. A. (2009). Protective effect of quercetin in primary neurons against Aβ(1–42): relevance to Alzheimer’s disease. J. Nutr. Biochem. 20, 269–275. doi: 10.1016/j.jnutbio.2008.03.002
Asaad, M., and Lee, J. H. (2018). A guide to using functional magnetic resonance imaging to study Alzheimer’s disease in animal models. Dis. Models Mech. 11:dmm031724. doi: 10.1242/dmm.031724
Ashkenazi, A., and Dixit, V. M. (1999). Apoptosis control by death and decoy receptors. Curr. Opin. Cell Biol. 11, 255–260. doi: 10.1016/s0955-0674(99)80034-9
Balaji, V., Kaniyappan, S., Mandelkow, E., Wang, Y., and Mandelkow, E. M. (2018). Pathological missorting of endogenous MAPT/Tau in neurons caused by failure of protein degradation systems. Autophagy 14, 2139–2154. doi: 10.1080/15548627.2018.1509607
Baptista, F. I., Henriques, A. G., Silva, A. M. S., Wiltfang, J., and Da Cruz e Silva, O. A. B. (2014). Flavonoids as therapeutic compounds targeting key proteins involved in Alzheimer’s disease. ACS Chem. Neurosci. 5, 83–92. doi: 10.1021/cn400213r
Bialik, S., and Kimchi, A. (2006). The death-associated protein kinases: structure, function, and beyond. Annu. Rev. Biochem. 75, 189–210. doi: 10.1146/annurev.biochem.75.103004.142615
Binder, L. I., Frankfurter, A., and Rebhun, L. I. (1985). The distribution of tau in the mammalian central nervous system. J. Cell Biol. 101, 1371–1378.
Braak, E., Braak, H., and Mandelkow, E. M. (1994). A sequence of cytoskeleton changes related to the formation of neurofibrillary tangles and neuropil threads. Acta Neuropathol. 87, 554–567. doi: 10.1007/bf00293315
Brüll, V., Burak, C., Stoffel-Wagner, B., Wolffram, S., Nickenig, G., Müller, C., et al. (2015). Effects of a quercetin-rich onion skin extract on 24 h ambulatory blood pressure and endothelial function in overweight-to-obese patients with (pre-)hypertension: a randomised double-blinded placebo-controlled cross-over trial. Br. J. Nutr. 114, 1263–1277. doi: 10.1017/S0007114515002950
Cooper, E. L., and Ma, M. J. (2017). Alzheimer Disease: clues from traditional and complementary medicine. J. Tradition. Comp. Med. 7, 380–385. doi: 10.1016/j.jtcme.2016.12.003
Deng, Y. H., Wang, N. N., Zou, Z. X., Zhang, L., Xu, K. P., Chen, A. F., et al. (2017). Multi-target screening and experimental validation of natural products from selaginella plants against Alzheimer’s Disease. Front. Pharmacol. 8:539. doi: 10.3389/fphar.2017.00539
El-Horany, H. E., El-Latif, R. N., Elbatsh, M. M., and Emam, M. N. (2016). Ameliorative effect of quercetin on neurochemical and behavioral deficits in rotenone rat model of Parkinson’s disease: modulating autophagy (Quercetin on Experimental Parkinson’s Disease). J. Biochem. Mol. Toxicol. 30, 360–369. doi: 10.1002/jbt.21821
Fang, J. Y., and Richardson, B. C. (2005). The MAPK signalling pathways and colorectal cancer. Lancet Oncol. 6, 322–327. doi: 10.1016/s1470-2045(05)70168-6
Gervais, F. G., Xu, D., Robertson, G. S., Vaillancourt, J. P., Zhu, Y., Huang, J., et al. (1999). Involvement of caspases in proteolytic cleavage of Alzheimer’s amyloid-beta precursor protein and amyloidogenic A beta peptide formation. Cell 97, 395–406. doi: 10.1016/s0092-8674(00)80748-5
Ghanemi, A. (2015). Alzheimer’s disease therapies: selected advances and future perspectives. Alexandr.J. Med. 51, 1–3. doi: 10.1016/j.ajme.2014.09.006
Guo, T., Noble, W., and Hanger, D. P. (2017). Roles of tau protein in health and disease. Acta Neuropathol. 133, 665–704. doi: 10.1007/s00401-017-1707-9
Gupta, A., Birhman, K., Raheja, I., Sharma, S. K., and Kar, H. K. (2016). Quercetin: a wonder bioflavonoid with therapeutic potential in disease management. Asian Pacific J. Trop. Dis. 6, 248–252. doi: 10.1016/S2222-1808(15)61024-6
Hartman, R. E., Shah, A., Fagan, A. M., Schwetye, K. E., Parsadanian, M., Schulman, R. N., et al. (2006). Pomegranate juice decreases amyloid load and improves behavior in a mouse model of Alzheimer’s disease. Neurobiol. Dis. 24, 506–515. doi: 10.1016/j.nbd.2006.08.006
Ittner, L. M., Ke, Y. D., Delerue, F., Bi, M., Gladbach, A., Van Eersel, J., et al. (2010). Dendritic function of tau mediates amyloid-beta toxicity in Alzheimer’s disease mouse models. Cell 142, 387–397. doi: 10.1016/j.cell.2010.06.036
Ivins, K. J., Thornton, P. L., Rohn, T. T., and Cotman, C. W. (1999). Neuronal apoptosis induced by beta-amyloid is mediated by caspase-8. Neurobiol. Dis. 6, 440–449. doi: 10.1006/nbdi.1999.0268
Kosik, K. S., Joachim, C. L., and Selkoe, D. J. (1986). Microtubule-associated protein tau (tau) is a major antigenic component of paired helical filaments in Alzheimer disease. Proc. Nat. Acad.Sci.U.S.A. 83, 4044–4048. doi: 10.1073/pnas.83.11.4044
Lahusen, T., Henke, R. T., Kagan, B. L., Wellstein, A., and Riegel, A. T. (2009). The role and regulation of the nuclear receptor co-activator AIB1 in breast cancer. Breast Cancer Res. Treat. 116, 225–237. doi: 10.1007/s10549-009-0405-2
Li, Y., Yao, J., Han, C., Yang, J., Chaudhry, M. T., Wang, S., et al. (2016). Quercetin, Inflammation and Immunity. Nutrients 8:167. doi: 10.3390/nu8030167
Liang, W. S., Dunckley, T., Beach, T. G., Grover, A., Mastroeni, D., Ramsey, K., et al. (2008). Altered neuronal gene expression in brain regions differentially affected by Alzheimer’s disease: a reference data set. Physiol. Genom. 33, 240–256. doi: 10.1152/physiolgenomics
Lin, Y., Yang, Z., Xu, A., Dong, P., Huang, Y., Liu, H., et al. (2015). PIK3R1 negatively regulates the epithelial-mesenchymal transition and stem-like phenotype of renal cancer cells through the AKT/GSK3β/CTNNB1 signaling pathway. Sci. Rep. 5:8997. doi: 10.1038/srep08997
Luo, W., Wang, T., Hong, C., Yang, Y. C., Chen, Y., Cen, J., et al. (2016). Design, synthesis and evaluation of 4-dimethylamine flavonoid derivatives as potential multifunctional anti-Alzheimer agents. Eur. J. Med. Chem. 122, 17–26. doi: 10.1016/j.ejmech.2016.06.022
Mandelkow, E. M., and Mandelkow, E. (2012). Biochemistry and cell biology of tau protein in neurofibrillary degeneration. Cold Spring Harb. Perspect. Med. 2:a006247. doi: 10.1101/cshperspect.a006247
Najafi, Z., Saeedi, M., Mahdavi, M., Sabourian, R., Khanavi, M., Tehrani, M. B., et al. (2016). Design and synthesis of novel anti-Alzheimer’s agents: acridine-chromenone and quinoline-chromenone hybrids. Bioorg. Chem. 67, 84–94. doi: 10.1016/j.bioorg.2016.06.001
Ow, Y. P., Green, D. R., Hao, Z., and Mak, T. W. (2008). Cytochrome c: functions beyond respiration. Nat. Rev. Mol. Cell Biol. 9, 532–542. doi: 10.1038/nrm2434
Pyakurel, A., Savoia, C., Hess, D., and Scorrano, L. (2015). Extracellular regulated kinase phosphorylates mitofusin 1 to control mitochondrial morphology and apoptosis. Mol. Cell 58, 244–254. doi: 10.1016/j.molcel.2015.02.021
Richetti, S. K., Blank, M., Capiotti, K. M., Piato, A. L., Bogo, M. R., Vianna, M. R., et al. (2011). Quercetin and rutin prevent scopolamine-induced memory impairment in zebrafish. Behav. Brain Res. 217, 10–15. doi: 10.1016/j.bbr.2010.09.027
Rishitha, N., and Muthuraman, A. (2018). Therapeutic evaluation of solid lipid nanoparticle of quercetin in pentylenetetrazole induced cognitive impairment of zebrafish. Life Sci. 199, 80–87. doi: 10.1016/j.lfs.2018.03.010
Roe, A. J., and Qi, X. (2018). Drp1 phosphorylation by MAPK1 causes mitochondrial dysfunction in cell culture model of Huntington’s disease. Biochem. Biophys. Res. Commun. 496, 706–711. doi: 10.1016/j.bbrc.2018.01.114
Rohn, T. T., Head, E., Nesse, W. H., Cotman, C. W., and Cribbs, D. H. (2001). Activation of caspase-8 in the Alzheimer’s disease brain. Neurobiol. Dis. 8, 1006–1016. doi: 10.1006/nbdi.2001.0449
Rountree, S. D., Chan, W., Pavlik, V. N., Darby, E. J., Siddiqui, S., and Doody, R. S. (2009). Persistent treatment with cholinesterase inhibitors and/or memantine slows clinical progression of Alzheimer disease. Alzheimers Res. Ther. 1:7. doi: 10.1186/alzrt7
Russo, M., Spagnuolo, C., Tedesco, I., Bilotto, S., and Russo, G. L. (2012). The flavonoid quercetin in disease prevention and therapy: facts and fancies. Biochem. Pharmacol. 83, 6–15. doi: 10.1016/j.bcp.2011.08.010
Sabogal-Guáqueta, A. M., Muñoz-Manco, J. I., Ramírez-Pineda, J. R., Lamprea-Rodriguez, M., Osorio, E., and Cardona-Gómez, G. P. (2015). The flavonoid quercetin ameliorates Alzheimer’s disease pathology and protects cognitive and emotional function in aged triple transgenic Alzheimer’s disease model mice. Neuropharmacology 93, 134–145. doi: 10.1016/j.neuropharm.2015.01.027
Solanki, I., Parihar, P., Mansuri, M. L., and Parihar, M. S. (2015). Flavonoid-based therapies in the early management of neurodegenerative diseases. Adv. Nutr. 6, 64–72. doi: 10.3945/an.114.007500
Stadelmann, C., Deckwerth, T. L., Srinivasan, A., Bancher, C., Brück, W., Jellinger, K., et al. (1999). Activation of caspase-3 in single neurons and autophagic granules of granulovacuolar degeneration in Alzheimer’s disease. Evidence for apoptotic cell death. Am. J. Pathol. 155, 1459–1466. doi: 10.1016/s0002-9440(10)65460-0
Stevens, C., and Hupp, T. R. (2008). Novel insights into DAPK autophagic signalling using peptide aptamer combinatorial protein-interaction screens. Autophagy 4, 531–533. doi: 10.4161/auto.5940
Strang, K. H., Golde, T. E., and Giasson, B. I. (2019). MAPT mutations, tauopathy, and mechanisms of neurodegeneration. Lab. Invest. 99, 912–928. doi: 10.1038/s41374-019-0197-x
Su, J. H., Zhao, M., Anderson, A. J., Srinivasan, A., and Cotman, C. W. (2001). Activated caspase-3 expression in Alzheimer’s and aged control brain: correlation with Alzheimer pathology. Brain Res. 898, 350–357. doi: 10.1016/s0006-8993(01)02018-2
Takahashi, A. (1999). Caspase: executioner and undertaker of apoptosis. Int. J. Hematol. 70, 226–232.
Taniguchi, C. M., Emanuelli, B., and Kahn, C. R. (2006). Critical nodes in signalling pathways: insights into insulin action. Nat. Rev. Mol. Cell Biol. 7, 85–96. doi: 10.1038/nrm1837
Ueki, K., Algenstaedt, P., Mauvais-Jarvis, F., and Kahn, C. R. (2000). Positive and negative regulation of phosphoinositide 3-kinase-dependent signaling pathways by three different gene products of the p85alpha regulatory subunit. Mol. Cell Biol. 20, 8035–8046. doi: 10.1128/mcb.20.21.8035-8046.2000
Vanhaesebroeck, B., Stephens, L., and Hawkins, P. (2012). PI3K signalling: the path to discovery and understanding. Nat. Rev. Mol. Cell Biol. 13, 195–203. doi: 10.1038/nrm3290
Wallace, R. A., and Dalton, A. J. (2011). What can we learn from study of Alzheimer’s disease in patients with Down syndrome for early-onset Alzheimer’s disease in the general population? Alzheimers Res. Ther.g 3:13. doi: 10.1186/alzrt72
Wang, D. -M., Li, S. -Q., Wu, W. -L., Zhu, X. -Y., Wang, Y., and Yuan, H. -Y. (2014). Effects of long-term treatment with quercetin on cognition and mitochondrial function in a mouse model of Alzheimer’s disease. Neurochem. Res. 39, 1533–1543. doi: 10.1007/s11064-014-1343-x
Wu, P. R., Tsai, P. I., Chen, G. C., Chou, H. J., Huang, Y. P., Chen, Y. H., et al. (2011). DAPK activates MARK1/2 to regulate microtubule assembly, neuronal differentiation, and tau toxicity. Cell Death Diff. 18, 1507–1520. doi: 10.1038/cdd.2011.2
Youdim, K. A., Shukitt-Hale, B., and Joseph, J. A. (2004). Flavonoids and the brain: interactions at the blood-brain barrier and their physiological effects on the central nervous system. Free Radic. Biol. Med. 37, 1683–1693. doi: 10.1016/j.freeradbiomed
Yukawa, K., Tanaka, T., Bai, T., Li, L., Tsubota, Y., Owada-Makabe, K., et al. (2006). Deletion of the kinase domain from death-associated protein kinase enhances spatial memory in mice. Int. J. Mol. Med. 17, 869–873. doi: 10.3892/ijmm.17.5.869
Zempel, H., Thies, E., Mandelkow, E., and Mandelkow, E. M. (2010). Abeta oligomers cause localized Ca(2+) elevation, missorting of endogenous Tau into dendrites, Tau phosphorylation, and destruction of microtubules and spines. J. Neurosci. 30, 11938–11950. doi: 10.1523/jneurosci
Zhang, X., Hu, J., Zhong, L., Wang, N., Yang, L., Liu, C. C., et al. (2016). Quercetin stabilizes apolipoprotein E and reduces brain Aβ levels in amyloid model mice. Neuropharmacology 108, 179–192. doi: 10.1016/j.neuropharm.2016.04.032
Keywords: bioinformatics analysis, experimental verification, AD biomarkers, AD diagnosis, treatment targets
Citation: Qi P, Li J, Gao S, Yuan Y, Sun Y, Liu N, Li Y, Wang G, Chen L and Shi J (2020) Network Pharmacology-Based and Experimental Identification of the Effects of Quercetin on Alzheimer’s Disease. Front. Aging Neurosci. 12:589588. doi: 10.3389/fnagi.2020.589588
Received: 31 July 2020; Accepted: 18 September 2020;
Published: 23 October 2020.
Edited by:
Ines Moreno-Gonzalez, University of Málaga, SpainReviewed by:
Joana Angélica Loureiro, University of Porto, PortugalCopyright © 2020 Qi, Li, Gao, Yuan, Sun, Liu, Li, Wang, Chen and Shi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ling Chen, Y2hlbmxpbmdoZXp6eEAxNjMuY29t; Jing Shi, c2hpajEzNkBobWMuZWR1LmNu
†These authors co-directed this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.