
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Aging Neurosci. , 05 July 2016
Sec. Cellular and Molecular Mechanisms of Brain-aging
Volume 8 - 2016 | https://doi.org/10.3389/fnagi.2016.00163
 Isidro Ferrer1,2,3*
Isidro Ferrer1,2,3* Paula Garcia-Esparcia1,2,3
Paula Garcia-Esparcia1,2,3 Margarita Carmona1,2,3
Margarita Carmona1,2,3 Eva Carro2,4
Eva Carro2,4 Eleonora Aronica5
Eleonora Aronica5 Gabor G. Kovacs6
Gabor G. Kovacs6 Alice Grison7
Alice Grison7 Stefano Gustincich7
Stefano Gustincich7Olfactory receptors (ORs) and down-stream functional signaling molecules adenylyl cyclase 3 (AC3), olfactory G protein α subunit (Gαolf), OR transporters receptor transporter proteins 1 and 2 (RTP1 and RTP2), receptor expression enhancing protein 1 (REEP1), and UDP-glucuronosyltransferases (UGTs) are expressed in neurons of the human and murine central nervous system (CNS). In vitro studies have shown that these receptors react to external stimuli and therefore are equipped to be functional. However, ORs are not directly related to the detection of odors. Several molecules delivered from the blood, cerebrospinal fluid, neighboring local neurons and glial cells, distant cells through the extracellular space, and the cells’ own self-regulating internal homeostasis can be postulated as possible ligands. Moreover, a single neuron outside the olfactory epithelium expresses more than one receptor, and the mechanism of transcriptional regulation may be different in olfactory epithelia and brain neurons. OR gene expression is altered in several neurodegenerative diseases including Parkinson’s disease (PD), Alzheimer’s disease (AD), progressive supranuclear palsy (PSP) and sporadic Creutzfeldt-Jakob disease (sCJD) subtypes MM1 and VV2 with disease-, region- and subtype-specific patterns. Altered gene expression is also observed in the prefrontal cortex in schizophrenia with a major but not total influence of chlorpromazine treatment. Preliminary parallel observations have also shown the presence of taste receptors (TASRs), mainly of the bitter taste family, in the mammalian brain, whose function is not related to taste. TASRs in brain are also abnormally regulated in neurodegenerative diseases. These seminal observations point to the need for further studies on ORs and TASRs chemoreceptors in the mammalian brain.
Chemical signals acting on appropriate receptors are the most common and precise form of communication. This applies both unicellular organisms with their milieu and to complex individuals in distinct cell types and systems within the same individual. Odorant and pheromone receptors are specialized chemical receptors designed to recognize and respond to volatile molecules by activating down-stream pathways to modulate specific behaviors. This type of receptor includes most of genome in the insects and mammals, and the number of putative ligands is not known but is estimated to be in the thousands. Odorant and pheromone signaling mediates chemical communication among species and among individuals in the same species. This information is crucial in natural settings, enabling individuals to obtain appropriate data about accessibility of food, and to reject toxic and damaging substances; in determining the sexual status of females, permitting mating and reproduction, breeding and nursing; in imprinting; in delimitation of territories and homing; and in attraction, repulsion, perception of danger and navigation. (Hanson, 1999; Ryan, 2002; Blomquist and Vogt, 2003; Doty, 2003; Wyatt, 2003; Brennan and Zufall, 2006; Brewer et al., 2006; Hallem et al., 2006; Wilson and Stevenson, 2006; Menini, 2009; Ihara et al., 2013; Liberles, 2014; Barish and Volkan, 2015; Li and Liberles, 2015; Wicher, 2015).
Odorant or olfactory receptors (ORs) are localized in sensory organs such as the olfactory epithelium in the nasal cavity in mammals, but overwhelming evidence in recent years has shown that the same type of receptors are distributed in many different organs and systems in mammals. These have been called “ectopic olfactory receptors” with distinct but organ- and tissue-specific characteristics. ORs and down-stream and related signaling molecules are also present in non-olfactory regions of the nervous system. Moreover, they are vulnerable to several neurodegenerative and mental diseases, and to drugs.
Olfactory or odorant receptors (ORs) have been identified as members of the G-protein-coupled receptors (GPCRs) family which share characteristic conserved trans-membrane motifs (Buck and Axel, 1991; Kato and Touhara, 2009; Malnic et al., 2010). OR genes constitute the largest family of GPCRs with about 1000 genes in the mouse and about 370 in humans (Buck and Axel, 1991; Glusman et al., 2000a,b; Zozulya et al., 2001; Young et al., 2002; Zhang and Firestein, 2002; Zhang et al., 2004, 2007; Niimura and Nei, 2003, 2005; Malnic et al., 2004; Olender et al., 2004; Kanageswaran et al., 2015). Amphioxi express ORs similar to those found in advanced vertebrates (Niimura, 2012). In later natural history, ORs diversified and two main classes of ORs appeared during the evolution of Chordata: teleost fish have class I which permits the detection of water-soluble odorants, whereas amphibians, reptiles, birds and mammals developed class II specialized in the perception of aerial odorants (Niimura, 2012; Hoover, 2013). Coelacanth, which is in transition between fish and tetrapods has class I ORs but also seven class II OR genes (Picone et al., 2014). The majority of ORs in mammals are classified as class II but between 10% and 20% are class I (Glusman et al., 2001; Zhang and Firestein, 2002; Niimura and Nei, 2003, 2005).
The repertoire of OR expression shows differences among species, not only between clearly separated species such as mice and humans but also among primates including Homo sapiens (Rouquier et al., 1998, 2000; Gimelbrant et al., 2001, 2004; Glusman et al., 2001; Godfrey et al., 2004; Gilad et al., 2005; Quignon et al., 2005; Go and Niimura, 2008). African elephants express the largest number of ORs, approximately 2000 OR genes and 2000 OR pseudogenes, about double the numbers for cows and horses, and a large number when compared with 1000–1200 OR genes in common rodents and around 400 OR genes in most primates including humans (Niimura et al., 2014). Although no similar data are available for Asian elephants, these animals have an extraordinary capacity to perceive odor differences (Rizvanovic et al., 2013). The reasons for OR expression changes during mammalian evolution are not known but they are most probably related to environmental and practical factors. For example, dolphins have reduced numbers of ORs but frugivorous bats among Chiropterans have developed particular OR profiles adapted to their diet (Hayden et al., 2014). Among primates, not only ORs but also the expression of V1Rs has been modified during evolution when compared with other mammals (Yoder and Larsen, 2014). The study of the cranium in Neanderthals has led to the suggestion that those hominids had better olfactory capacity than present humans (Hughes et al., 2014). Finally, to add more complexity, the expression of some genes is subjected to individual variations in a given species (Young et al., 2003).
Activation of ORs leads to the dissociation of the Olfactory G-protein Golf into three subunits: α, β and γ (Li et al., 2013a). olfactory G protein α subunit (Gαolf), Gβ1 and Gγ13 form the functional heterotrimeric G protein in olfactory sensory neurons (Li et al., 2013a). Ric-8B co-localizes and interacts with Gαolf and γ13 in the cilia of olfactory sensory neurons and mediates signaling of ORs (Von Dannecker et al., 2005, 2006; Kerr et al., 2008).
Activation of Gαolf and adenylyl cyclase 3 (AC3) induces an increase in the intracellular concentration of cyclic adenosine monophosphate (cAMP), activation of cAMP-gated channel, external Ca++ influx, and membrane depolarization (Pace et al., 1985; Sklar et al., 1986; Nakamura and Gold, 1987; Jones and Reed, 1989; Lowe et al., 1989; Bakalyar and Reed, 1990; Dhallan et al., 1990; Firestein et al., 1991; Liman and Buck, 1994; Bönigk et al., 1999; Mombaerts, 1999, 2004; Firestein, 2001; Stephan et al., 2009; Billig et al., 2011). General anosmia is produced by a targeted disruption of AC3 in mice (Brunet et al., 1996; Wong et al., 2000). Mice null for the expression of Gαolf do not perceive odors (Belluscio et al., 1998; Luo et al., 2002); conditional Gγ13 knockout mice exhibit altered recognition of several odorants (Li et al., 2013b). More subtle regulation occurs following modulation of βγ subunits of Golf complex which suppress the expression of several odor-related genes, probably through the methylation of specific histones (Ferreira et al., 2014).
Repolarization occurs by extruding intracellular Ca++ through the plasma membrane with the coordinated action of a non-potassium-dependent sodium/calcium exchanger which is modulated by the olfactory marker protein (OMP), a potassium-dependent sodium/calcium exchanger, and a plasma membrane Ca++-ATPase (Pyrski et al., 2007; Kwon et al., 2009; Saidu et al., 2009; Stephan et al., 2011).
ORs and other chemoreceptors require specific cofactors to improve the membrane localization of the receptors (Jungnickel et al., 2001; Saito et al., 2004; Behrens et al., 2006). Co-factors of GPCRs upon binding to specific domains of receptors, facilitate or inhibit cell surface expression (Duvernay et al., 2004, 2009; Hague et al., 2004; Syme et al., 2006). Receptor transporter proteins 1 and 2 (RTP1 and RTP2) and receptor expression enhancing protein 1 (REEP1) enhance OR function by the recruitment of ORs to lipid rafts (Saito et al., 2004). Recently a complex interaction between transient receptor potential 2 channel (TRPC2), which establishes the first electrical signal in the pheromone transduction pathway has been discovered in the peripheral olfactory organs of rats (Mast et al., 2010). Homer protein isoforms 1 b/c and 3 bind to proline-rich sequences on proteins associated with calcium signaling (Worley et al., 2007) and RTP1.
UDP-glucuronosyltransferases (UGTs) are localized in the nuclear membrane and endoplasmic reticulum where glucuronidate xenobiotic and endobiotic compounds are present, thus facilitating their excretion and contributing to detoxification of cells (Tukey and Strassburg, 2000; Mackenzie et al., 2005a,b). Various UGTs have been identified in the olfactory epithelium and olfactory bulb (Leclerc et al., 2002; Heydel et al., 2010) where they probably participate in the removal of compounds produced during the process of OR activation and signaling.
OR expression is not fixed; rather, OR presentation is modulated by noradrenergic inputs (Hague et al., 2004; Eckmeier and Shea, 2014) and odorant stimulation (Kim et al., 2015b), among other factors (Malnic et al., 2010). Interestingly, variations of temperature in the olfactory epithelium of Xenopus tadpoles result in variable activation in the olfactory glomeruli (Kludt et al., 2015). Regulation of olfactory organs and receptors has also been observed with high environmental temperatures in Drosophila (Martin et al., 2011; Riveron et al., 2013). Similar complementary effects of temperature and odors probably occur in the olfactory system in rodents. Curiously, olfaction thresholds increase in hyperbaric situations (Ay et al., 2014). These examples illustrate the effects of physical environments on smell perception. Finally, behavior and odor memories can modify the expression of chemoreceptors in butterflies and honeybees (Briscoe et al., 2013; Claudianos et al., 2014).
Growing evidence indicates that the olfactory epithelium is only one of the multiple regions bearing ORs and probably the only organ in which ORs are charged with reacting to diverse small aerial molecules which are processed into odors. Yet the perception of odors (and their recognition in humans) is not the only function of ORs. More readily, activation of certain ORs switches on complex cellular responses mediated by neural networks, neurotransmitters and hormones which in turn determine sophisticated behaviors. ORs located in non-olfactory tissue and organs are chemoreceptors able to trigger complex response receptors but not precisely linked to any sensorial perception.
ORs located in non-chemosensory organs were categorized as “ectopic” ORs because they were expressed in unexpected regions. However, this term is misleading as it implies that “ectopic” ORs might have functions related to olfaction. Yet mounting evidence indicates that the function of “ectopic” ORs has no relation to the capture of odors. Ectopic ORs were first identified in the testes and sperm and their function seems to be related to discriminatory chemotaxis (Parmentier et al., 1992; Walensky et al., 1995, 1998; Asai et al., 1996; Vanderhaeghen et al., 1997; Branscomb et al., 2000; Spehr et al., 2003; Fukuda et al., 2004; Fukuda and Touhara, 2006; Veitinger et al., 2011).
ORs are also expressed in germinal cells (Goto et al., 2001), embryos (Nef and Nef, 1997; Dreyer, 1998), developing heart (Drutel et al., 1995), developing muscle (Griffin et al., 2009) and axons of olfactory neurons. These observations show the participation of ORs in a wide range of processes at several stages of development (Baker et al., 2015) including angiogenesis (Kim et al., 2015a) and direction of nerve fibers in the olfactory bulb (Vassalli et al., 2002; Feinstein and Mombaerts, 2004; Feinstein et al., 2004).
Certain ORs and functional mediators are also expressed in lung (Zhang et al., 2004; Li et al., 2013b; Gu et al., 2014), spleen (Blache et al., 1998), liver (Yuan et al., 2001), gastrointestinal tract (Yuan et al., 2001; Braun et al., 2007), placenta (Itakura et al., 2006), prostate (Xu et al., 2000; Xia et al., 2001; Neuhaus et al., 2009), erythroid cells and peripheral blood cells (Feingold et al., 1999; Zhao et al., 2013) and eye (Pronin et al., 2014), among other tissues and organs (Feldmesser et al., 2006; De la Cruz et al., 2009; Kang and Koo, 2012; Flegel et al., 2013).
ORs and functional signaling molecules are abundant in the kidney (Pluznick et al., 2009; Rajkumar et al., 2014) where they regulate the glomerular filtration rate (Pluznick et al., 2009; Pluznick, 2014).
Recent transcriptome studies using cDNA arrays in the rat have shown expression of ORs and taste receptors (TASRs) in choroid plexus (Quintela et al., 2013). Interestingly, TASR and OR transduction pathways are regulated by hormones, with the expression of transcripts being different in males and females (Quintela et al., 2013). As in other non-sensory regions, ORs in the choroid plexus do not participate in the identification of odors but rather in the detection of soluble molecules in the CSF (Quintela et al., 2013).
Several ORs are expressed in a number of tissues, whereas certain ORs are only expressed in testis (Flegel et al., 2013). OR-specific functions have been identified in a very small number of cases. Olfr13 (MOR23) participates in the journey of sperm, growth of axonal growth cones and migration of myocytes (Fukuda et al., 2004; Griffin et al., 2009). OR1A and OR2A4 regulate actin cytoskeleton and cytokinesis (Zhang et al., 2012).
ORs responding to short chain fatty acids produced by gut microbiota modulate renin secretion and regulation of blood pressure (Pluznick et al., 2013), while activation of ORs by spicy species in enterochromaffin cells liberates serotonin (Braun et al., 2007; Breer et al., 2012). OR activation in mouse pulmonary macrophages promotes monocyte chemotactic protein-1 production (Li et al., 2013b). ORs in α-cells of pancreatic islets regulate glucagon secretion (Kang et al., 2015). Renal and cardiovascular ORs regulate blood pressure (Pluznick, 2013). Prostate-specific ORs are regulated by a derivative of androstenone (Neuhaus et al., 2009). Complex interactions like those of microbiota-derived products in gut, which activate ORs in gut and kidney, have an effect in the control of blood pressure (Natarajan and Pluznick, 2015). In summary, ORs in non-sensory tissues and organs are not involved in olfaction but rather carry out diverse and unrelated functions. Their effects are local and there is no connection between these receptors and the brain.
A number of ORs have been found in the cerebral cortex of mice (Otaki et al., 2004). Olfr110, Adcy3 and Gnal (Gαolf) mRNAs are also expressed in the cerebral cortex at the ages of new born, and at 3, 6 and 12 months. mRNA expression levels do not vary over the first year of life (Ansoleaga et al., 2013).
Gene array studies have revealed expression of ORs in the human brain and mouse dopaminergic neurons of the substantia nigra (Garcia-Esparcia et al., 2013; Grison et al., 2014). This has been further validated by qRT-PCR analysis (Ansoleaga et al., 2013, 2015; Garcia-Esparcia et al., 2013; Grison et al., 2014). Immunohistochemistry in the adult human brain has shown the presence of OR2H2, OR2A4, and OR6K3 in neurons of the neocortex, hippocampus, dentate gyrus, striatum, thalamus, nuclei of the basal forebrain, hypothalamus, nuclei of the brainstem, cerebellar cortex, dentate nucleus and neurons of the spinal cord.
OR immunostaining predominates in the cytoplasm as a granular precipitate and the intensity is to a certain extent dependent on the size of neurons, as the staining of neurons of the motor nuclei of the cranial nerves, Purkinje cells and CA1 neurons is more intense in comparison with that of cerebellar granule cells and neurons of the dentate gyrus. A mainly cytosolic localization of ORs is supported by sub-cellular fractionation analysis (Garcia-Esparcia et al., 2013). In contrast to the olfactory epithelium, several ORs seem to be expressed in a single neuron in the central nervous system (CNS).
Gαolf is expressed in the striatum, where it may couple dopamine D1 receptors and adenosine A2A receptors to the activation of adenylyl cyclase type 5.1 (Corvol et al., 2001; Hervé, 2011). Nevertheless, Gαolf is not restricted to the striatum; rather it is widely distributed, at least, in the human brain, as well as AC3 (Garcia-Esparcia et al., 2013).
RTP1, RTP2, and REEP1 are expressed in the cytoplasm of neurons, as revealed by immunohistochemistry, paralleling the expression of ORs and down-stream signaling molecules in all brain regions (Garcia-Esparcia et al., 2013).
Finally, various UGTs, including UGT1A6, have been localized in the olfactory mucosa and olfactory bulb (Leclerc et al., 2002). Immunohistochemistry reveals UGT1A6 localization in neurons of the human CNS (Garcia-Esparcia et al., 2013).
Olfr110, Adyc3, and Gnal (Gαolf) mRNAs are also expressed in the cerebral cortex of mice (Ansoleaga et al., 2013; Grison et al., 2014).
Although co-localization studies are needed to determine whether all the components of the receptor signaling pathway are present in the same cell, in vitro studies have identified functional responses of neuronal ORs in the face of external stimuli which cannot be produced in the absence of the signaling pathway. Thus, taking advantage of transgenic labeling, laser capture microdissection coupled to nano cap-analysis of gene expression (nanoCAGE) technology on isolated A9 and A10 cells of the substantia nigra in mice, has revealed that a subset of ORs is expressed in mesencephalic dopaminergic neurons (Grison et al., 2014). OR expression in the mesencephalon has been validated by RT-PCR and in situ hybridization.
Heterologous HEK 293 cells expressing Olfr287 following transfection respond to selected molecules, particularly S- and R-carvones, and decanoic acid. In the same line, cultured isolated dopaminergic neurons are activated by odor-like agonists and carvones as revealed by neuronal Ca++ entry upon stimulation. Ca++ levels return to normal after washout (Grison et al., 2014).
ORs have also been reported in the autonomic nervous system and murine sensory ganglia (Weber et al., 2002; Manteniotis et al., 2013).
Like ORs in other non-sensory organs and regions, the function of ORs in brain differs from the specific detection of molecules by ORs in the nasal cavity, and it is not related to the perception of odors.
OBP are small soluble extracellular proteins related to pheromone and odor transduction, mainly but not exclusively found in olfactory organs such as pheromone-binding and odorant-binding proteins. These proteins trap and convey odorants and pheromones to specific receptors (Vogt and Riddiford, 1981; Vogt et al., 1985, 1991; Pevsner et al., 1986, 1988, 1990; Pelosi and Maida, 1990). In mammals, OBPs belong to the super family of carrier protein lipocalins (Flower, 1996) present in the lateral nasal gland and olfactory epithelium (Pevsner et al., 1988; Dal Monte et al., 1991; Paolini et al., 1998). OBPs are also present in several fluids including vaginal discharge, urine, saliva, tears and amniotic fluid (Singer et al., 1986; Garibotti et al., 1995; Redl, 2000; Briand et al., 2004; Guiraudie-Capraz et al., 2005).
Certain lipocalins, such as lipocalin 2, are produced in brain where they have important functions in regulating inflammatory responses, angiogenesis, and astrocyte and microglial activation (Lee et al., 2011; Hamzic et al., 2013; Jin et al., 2014; Nam et al., 2014; Xing et al., 2014; Wu et al., 2015). Apolipoprotein D (APOD), a member of the lipocalin family and transporter of small hydrophobic ligands, is also expressed in brain (Rassart et al., 2000). The close relationship between OBPs and other lipocalins has prompted the hypothesis, that this type of transporter links chemical communication and immunity (Stopková et al., 2014). Therefore, OBPs in brain and other organs do not necessarily participate in olfaction, but rather are used in other metabolic functions which have nothing in common with smell.
Various natural ligands of OBPs have been recognized in mammals (Robertson et al., 1993; Glasgow et al., 1995; Marchese et al., 1998; Briand et al., 2004; Sanz et al., 2005; Le Danvic et al., 2009; Scaloni et al., 2011). These are small molecules of about 200–400 mW related to derived fatty acids, steroids, and terpenoids (Wetzel et al., 1999; Spehr et al., 2003; Mombaerts, 2004; Sanz et al., 2005; Shirokova et al., 2005; Jacquier et al., 2006; Saito et al., 2006; Keller et al., 2007; Khafizov et al., 2007; Malnic, 2007; Schmiedeberg et al., 2007; Hsu et al., 2008; Nodari et al., 2008; Le Danvic et al., 2009).
In vitro studies have shown that certain peripheral odorants activate ORs expressed in cultured dopaminergic neurons (Grison et al., 2014). However, these observations do not indicate that the same ligands are used in brain under physiological conditions. Rather, distinct molecules delivered from the blood, cerebrospinal fluid, neighboring local neurons and glial cells, distant cells through the extracellular space and the cells’ own self-regulating internal homeostasis may be postulated as putative ligands in brain.
Taste perception is mediated by taste receptors located in the taste buds located mainly on the tongue. Each taste bud contains between 50 and 100 sensory cells, each with the capacity to react and transduce one of the five categories of taste: bitter, sour, umami, sweet, and salty (Iwatsuki and Uneyama, 2012). Bitter receptors bind to α-gustducin which upon activation opens transient channels producing influx of Na+, resulting in cell depolarization (Ishimaru and Matsunami, 2009; Iwatsuki and Uneyama, 2012). α-gustducin is necessary to mediate sweet and bitter taste perception (Wong et al., 1996; Iwatsuki and Uneyama, 2012).
In human, 25 genes encoding bitter taste receptors have been identified (Behrens et al., 2007). As for ORs, ectopic TASRs are localized in many organs and systems (Behrens and Meyerhof, 2010). Recent studies have shown the expression of TASRs and α-gustducin in gastrointestinal tract in both specialized neural cells and selected non-endocrine cells in the mucosa (Höfer et al., 1996; Wu et al., 2002; Dyer et al., 2005; Rozengurt, 2006; Jang et al., 2007; Mace et al., 2007; Margolskee et al., 2007; Egan and Margolskee, 2008; Sternini et al., 2008; Depoortere, 2014; Kokrashvili et al., 2014).
Many TASRs have been related to glucose transport metabolism in which they regulate the expression of specific transporters (Au et al., 2002; Margolskee et al., 2007). Bitter taste receptors are also expressed in the stomach and intestine and their activation produces an increase in the expression of cholecystokinin and peptide YY (Halatchev and Cone, 2005; Chen et al., 2006; Rozengurt et al., 2006). Interestingly, the expression of bitter receptors seems to be partially modulated by diet (Vegezzi et al., 2014).
The role of these receptors in the control of food intake has been elegantly demonstrated by infusion through a nasoduodenal catheter of combinations of tastants; unami alone and the combination of tastants reduced appetite (van Avesaat et al., 2015). Non-caloric sweeteners seem to act on the same sweet receptors and this may explain, at least in part, why diabetes, obesity and metabolic syndrome do not decrease but rather quite the reverse in heavy non-caloric sweetener consumers (Dhingra et al., 2007; Pierce et al., 2007; Lutsey et al., 2008; Davidson et al., 2011; Burke and Small, 2015; Fowler et al., 2015; Pepino, 2015).
Several bitter taste receptors are found in the intestine (Reimann et al., 2012; Kendig et al., 2014; Shirazi-Beechey et al., 2014; Gu et al., 2015). Activation of selected bitter receptors triggers colonic peristaltic movements (Kendig et al., 2014). Also, a complex relationship exists among gut taste receptors, transporters and colonic microbiota (Swartz et al., 2012; Alcock et al., 2014; Kaji et al., 2014).
In contrast to TASRs in the mouth, which transmit information to the brain, TASRs in the digestive system are probably not connected to the brain but rather detect local soluble substances and respond by instructing cells to release hormones and other molecules.
TASR genes, their products and down-stream signaling molecules have also been reported in the rat and mouse choroid plexus (Ren et al., 2009; Quintela et al., 2013), where they have been considered as glucose sensors.
Regarding the nervous system, bitter TASRs are expressed in the brain of mice and rats (Singh et al., 2011; Dehkordi et al., 2012; Voigt et al., 2015). TASR gene expression has also been identified in the human brain (Ansoleaga et al., 2013; Garcia-Esparcia et al., 2013; Grison et al., 2014). Their function in brain is not known. As for ORs, the term TASRs is misleading when referring to these receptors in the nervous system as they are not related to taste.
The concept of Parkinson’s disease has changed with time. First clinically described by J. Parkinson in 1817 in the “Essay of shaking palsy”, it was not until a century later that characteristic neuronal inclusions named Lewy bodies were identified and the substantia nigra was recognized as the main area affected. In the late 60’s of the last century, about 15 years after the discovery of dopamine as the deficient neurotransmitter, the first clinical trials with levodopa (L-DOPA) began to change dramatically the course of the disease. This success, together with the search for pre-clinical symptoms that might help to recognize early stages of the disease and therefore make treatment possible, has had important consequences. Pre-clinical PD should be considered as pre-motor or pre-parkinsonian PD, as several clinical symptoms may precede motor symptoms by many years, including smell disorders, abnormal sleep, variegated neuro-vegetative symptoms, and particular modifications of mood such as apathy and depression. Further, pharmacological and in some cases specific brain stimulation has dramatically increased not only quality of life but also survival. It is not unusual for individuals with PD to live more than 15 years after the clinical diagnosis. Yet this would have other consequences. Since PD is a progressive disease, other brain areas are affected and particular forms of cognitive deterioration, mood disorders, and dementia appear in patients with a long disease course (Ferrer, 2009; Ferrer et al., 2011, 2012; Schneider and Obeso, 2015).
Neuropathological stages of PD have also been established in typical cases. Deposition of aggregated α-synuclein, which is the main component of Lewy bodies and abnormal neurites, first occurs in PD in the olfactory bulb, autonomic nervous system, and selected nuclei of the medulla oblongata at stage 1. Stage 2 involves, in addition, selected nuclei of the pons such as the locus ceruleus. Stage 3 sees spreading to the midbrain and particularly, to the substantia nigra pars compacta, thereby producing dopaminergic denervation in the striatum and other brain regions. Stage 4 involves the limbic system, while stages 5 and 6 progressively compromise the neocortex (Jellinger, 2011; Revesz et al., 2015).
Neuropathological staging apparently supports a structural underpinning to clinical symptoms in PD. Although this is true for pre-parkinsonian and parkinsonian stages of the disease, Lewy bodies that do not properly match with the degree of cognitive impairment and dementia in PD. Alzheimer’s disease (AD) and vascular co-morbidities have been suggested as putative factors (Jellinger, 2011).
About 5–10% of PD cases are familial and inherited with dominant or recessive traits. Autosomal dominant PD has been linked to mutations (but also duplications and triplications) in SNCA (the gene encoding α-synuclein), LRRK2 (encoding leucine-rich repeat kinase 2), and VPS35 (encoding vacuolar protein sorting 35-homolog). Autosomal recessive PD has been associated with mutations in PARK2, encoding parkin, PINK1 (gene product: PTEN-induced kinase-1), PARK7 (gene product: DJ1), ATP13A2 (encoding ATPase type 13A2, PLAG2G6 (gene product: phospholipase A2 group 6, FBX07 (gene product F-box protein 7, and PANK2 (encoding pantothenate kinase 2). Sporadic PD is modulated by several gene variants and undetermined numbers of environmental factors including exogenous pathogens (Houlden and Singleton, 2012; Revesz et al., 2015).
Gene expression analysis followed by qRT-PCR validation in PD has identified a cluster of OR genes whose expression is altered in the frontal cortex; these modifications seem to occur at relatively early stages of the disease as they appear at premotor stages and remain in parkinsonian stages of the process (Garcia-Esparcia et al., 2013). OR2L13, OR1E1, OR2J3, OR52L1 and OR11H1 are down-regulated. Interestingly, OR4F4 mRNA shows a significant augmentation only in PD females.
Taste receptors (TASRs) have been analyzed in parallel; TAS2R5 is down-regulated, and TAS2R10 and TAS2R13 up-regulated, at premotor and parkinsonian stages in the same samples. Up-regulation affects mainly males and down-regulation occurs in one gene in females (Garcia-Esparcia et al., 2013). All deregulated TASRs belong to the bitter taste family.
These observations indicate that: (i) cortical ORs and TASRs are vulnerable to PD; (ii) deregulation of selective OR and TASR genes occurs at relative early stages of the disease; (iii) altered OR and TASR gene expression is not dependent on treatment because changes are observed in incidental cases discovered on post-mortem neuropathological examination (pre-clinical stages not detected during life) in individuals who did not receive drug therapy; (iv) OR gene down-regulation predominates but the expression of some OR genes is preserved, and only a few of them are up-regulated; (v) deregulation of TASR genes at the same stages of the disease and in the same region is more variable; and (vi) deregulation of certain ORs and TASRs in PD is gender-dependent.
Importantly, functional genomics in the same series has shown deregulation of UGT2A3, UGT2B7 and UGT2B10 in PD samples, reinforcing the idea of cooperative function with ORs. Interestingly, all these UGTs cluster on chromosome 4q13.
Regarding the substantia nigra, OR2L13, OR2T33, OR2J3, OR52L1, OR10G8, OR11H1 and OR4F4 are down-regulated in PD (Grison et al., 2014). Decreased expression of all these receptors might be interpreted as the consequence of neuron loss in this region. However, TAS2R4, TAS2R5 and TAS2R50 mRNA expression is increased in the same samples (Grison et al., 2014). Therefore, altered expression of ORs and TASRs in the substantia nigra appears to be highly regulated in PD.
The concept of AD has been modified over the years. The seminal definition first advanced by A. Alzheimer was of a form of pre-senile dementia (appearance of the clinical symptoms before the age of 65 years) characterized by the accumulation of extracellular structures named senile plaques and intra-neuronal tangles of fibrils visualized with specific silver stains. These changes were similar to those seen in a number of cases with dementia appearing before the age of 65 years and identified as senile dementia (of Alzheimer’s type). Senile plaques composed of a core of amyloid together with neurofibrillary tangles composed of hyper-phosphorylated and aggregated tau have been the hallmarks of AD regardless of the age at onset of clinical symptoms. We now know that early onset AD (EOAD) is often inherited and linked to mutations in the three genes APP, PSEN1, and PSEN2, encoding β-amyloid precursor protein, presenilin 1, and presenilin 2, respectively (familial EOAD: EOFAD), whereas late onset AD (LOAD) is currently sporadic and modulated by increased numbers of genetic factors with low penetrance (genetic modulators or variants) particularly APOEε4, and environmental factors (Bertram and Tanzi, 2011; Duyckaerts and Dickson, 2011; Serrano-Pozo et al., 2011; Lowe and Kalaria, 2015).
Like other neurodegenerative diseases with abnormal protein aggregates, AD is a long-lasting biological process in which only the advanced stages of the disease manifest with cognitive impairment and dementia. Therefore, AD is now classified clinically as prodromic or preclinical (without symptoms), mild cognitive impairment of AD origin, and AD (dementia of AD). Tremendous efforts are now underway to identify early biomarkers of AD in living individuals (Knopman, 2011).
Additionally, neuropathological studies of consecutive autopsy cases in young, middle-aged, and elderly individuals with and without cognitive impairment have shown a progression in the localization, distribution, and extent of β-amyloid plaques and neurofibrillary tangles in the non-selected population. Based on these neuropathological findings, the most widely accepted neuropathological classifications divide AD into stages I, II, III, IV, V, and VI depending on the neurofibrillary tangle pathology (Braak and Braak, 1991, 1999), and into stages 0, A, B, and C (Braak and Braak, 1999), or 0, 1, 2, 3, and 4 (Thal et al., 2002) depending on the distribution of senile plaques or β-amyloid deposits.
Following the Braak and Braak nomenclature, stages I–II refer to neurofibrillary tangle involvement of the trans-entorhinal and entorhinal cortices; stages III–IV include additional entanglement of the hippocampus, deep temporal cortex, and limbic regions; and stages V–VI manifest with large numbers of neurofibrillary tangles in the neocortex eventually including primary sensory areas. First stages are subclinical and they can last for decades whereas mild cognitive impairment and dementia occur at stages IV and VI (for review see Ferrer, 2012).
To be more precise, certain individuals in their thirties already show the early changes of AD. About 85% of individuals aged 65 years have AD at stages I–II at least, but only 5% have extended lesions clinically manifested as dementia. Cortical involvement increases with age and about 25% of the population has Alzheimer’s dementia at the age of 85 years (Braak et al., 2011; Ferrer, 2012).
A region on 14q11.2, between 19.2 and 19.49 MB, is linked to earlier age of the appearance of clinical symptoms, as revealed using array comparative genome hybridization and replicated evaluation with Affymetrix array and multiplex ligation-dependent probe amplification (Shaw et al., 2011). This association is independent of and in addition to the association observed for APOE4. The association signal comes from olfactory receptor gene cluster in the region where OR4M1, OR4N2, OR4K2, OR4K5 and OR4K1 are located. The 2nd copy and 3rd copy states have similar age at onset and the 4th copy state has a trend toward earlier age at onset on all APOE4 backgrounds. The 5 + copy subjects were 2 times more likely to be diagnosed with AD by any given age compared to the 2, 3, and 4 copy carriers. The risk of onset before age 72 years for persons who had 5 copies or more was 6-fold higher compared to persons having lower copy numbers (Shaw et al., 2011).
OR gene expression in AD has been studied in the entorhinal cortex and frontal cortex area 8 covering all the stages of disease progression. OR11H1 mRNA is increased and OR4F4 mRNA decreased in the AD entorhinal cortex at stages III–IV and V–VI compared with controls. OR10G8 mRNA expression levels decrease in the entorhinal cortex at late stages of AD (Ansoleaga et al., 2013). OR4F4 mRNA expression levels are increased in the frontal cortex area 8 at stages III–IV, and OR52L1 mRNA at stages III–IV and V–VI (Ansoleaga et al., 2013).
Whether changes in AD can be replicated in transgenic models of AD has been tested in APP/PS1 transgenic mice which bear the Swedish APP (K594M/N595L) mutation and the dE9 deletion in PSEN1 (Borchelt et al., 1996; Aso et al., 2012). APP/PS1 mice were developed by co-injection of the two transgene constructs [mouse/human (Mo/Hu) chimeric APP695 harboring the Swedish (mutation and exon-9-deleted PS1] into pronuclei with a single genomic insertion site resulting in the two transgenes being transmitted as a single mendelian locus (Borchelt et al., 1996).
APP/PS1 transgenic mice exhibit alterations in the central OR signaling pathways. Olfr110 mRNA, the only OR assessed, significantly increases from the age of 3 months onwards, whereas Adcy3 mRNA expression levels are reduced in transgenic mice aged 12 months (Ansoleaga et al., 2013).
Regarding TASRs, increased TAS2R13 mRNA expression is observed at stages III–IV, and TAS2R5 and TAS2R10 mRNA expression at stages V–VI in the entorhinal cortex. Yet no modification in the mRNA expression levels of these TASRs is observed in frontal cortex area 8 at any stage of AD (Ansoleaga et al., 2013).
PSP is a neurodegenerative disease characterized clinically by pseudobulbar palsy, vertical ocular palsy, nuchal dystonia, and dementia. The term was first coined as PSP following description by J.C. Steele, J.C. Richardson, and J. Olszewski in the mid-60’s of the last century. The main pathological finding is the accumulation of abnormally hyper-phosphorylated 4 R tau in neurons in the form of pre-tangles and tangles, astrocytes forming characteristic tufts, and oligodendrocytes in the form of coiled bodies in several brain regions including selected nuclei of the brain stem, diencephalic nuclei, cerebellum, and several areas of the telencephalon. PSP has rarely been reported as a familial disease, presenting most commonly as a sporadic form (Dickson et al., 2011).
The same OR genes assessed in PD and AD have been assessed in the frontal cortex (area 8) of PSP at the terminal stage of the disease. OR11H1, OR2D2, OR2L13, OR2D2, OR2T33, OR4F4, OR10G8 and OR52L1 mRNAs are up-regulated whereas OR51E1 mRNA levels are similar to those of controls (Ansoleaga et al., 2013). Interestingly, all mRNAs of TASRs analyzed, TAS2R4, TAS2R5, TAS2R10, TAS2R13, TAS2R14 and TAS2R50, are up-regulated in the same cortical region in PSP (Ansoleaga et al., 2013).
CJD was first described at the beginning of the second decade of the last century by H. G. Creutzfeldt and A.M. Jakob but only two seminal cases are now considered real CJD. The disease is manifested clinically as rapid dementia plus ataxia, myoclonus, and other neurological symptoms, and neuropathologically as spongiform encephalopathy with neuron loss, spongiform degeneration (small and often confluent brain vacuoles), and microglia and astroglia activation. CJD is the paradigm of human prion diseases rarely caused by mutations in PRNP, the gene encoding the prion protein (fCJF; about 10% of cases), transmission of abnormal prion from one individual to another (iCJD; very rare but most commonly represented by the variant Crreutzfeldt-Jakob disease (vCJD) resulting from the transmission of bovine spongiform encephalopathy-contaminated food), or apparently occurring de novo as sporadic CJD (sCJD, representing the most common form albeit having an incidence of about 1–2 cases per 1,000,000 inhabitants per year). In all these circumstances, the normal prion protein is transformed into an abnormal, predominantly but not exclusively proteinase-resistant form called a prion, which is pathogenic. PrP presents in sporadic cases as type I and type II depending on the molecular weight of the non-glycosylated isoform and the amount of the mono- and di-glycosylated isoforms. Interestingly, CJD is also conditioned by the characteristics of codon 129 in PRNP (methionine/methionine, valine/valine, and methionine/valine) which marks the phenotypes of fCJD, iCJD, and sCJD. Focusing on sCJD, several types may be categorized according to the codon 129 in PRNP and prion type; the most frequent are MM1 and VV2 (Head and Ironside, 2009; Parchi et al., 2009, 2011; Zerr et al., 2009; Budka et al., 2011).
Frontal cortex area 8 has been assessed in sCJD subtypes MM1 and VV2 to investigate regional commonalities and differences between the two main subtypes of sCJD. Involvement of the cerebral cortex predominates in subtype MM1 whereas the cerebellum is severely affected in subtype VV2. PrPRes deposition differs in the two subtypes: a synaptic pattern occurs in sCJD MM1 and perineuronal deposits characterize sCJD VV2 in the cerebral cortex, while PrPRes-plaques are common in the cerebellum in sCJD VV2 (Budka et al., 2011).
OR2L13, OR4F4, OR51E1 and OR52L1 mRNA expression is reduced in the frontal cortex in sCJD MM1; OR11H1 mRNA is down-regulated in sCJD MM1 and VV2; and OR10G8 mRNA expression levels are significantly increased in sCJD VV2 (Ansoleaga et al., 2013). Regarding the cerebellum, OR2D2 is up-regulated in sCJD MM1, while OR4F4 and OR51E1 mRNA decrease in sCJD MM1 and VV2, respectively (Ansoleaga et al., 2013).
TAS2R4, TAS2R10 and TAS2R14 mRNA expression is increased in the frontal cortex area 8 in sCJD MM1 and VV2, but increased TAS2R5 mRNA is only in MM1, and TASR13 mRNA only in VV2 (Ansoleaga et al., 2013).
The study of gene expression performed in the same cortical region (frontal cortex area 8) using the same probes, the same method and the same hands in the same laboratory permits a comparative evaluation of the results obtained in PD, AD, PSP and sCJD subtypes MM1 and VV2. Curiously, AD shows little modification when compared with the other diseases. In contrast, OR gene up-regulation occurs in 8 of 11 analyzed OR genes. Predominant OR mRNA down-regulation is found in sCJD mainly in subtype MM1. Finally, reduced mRNA expression of several ORs occurs in the frontal cortex area 8 in PD in males but up-regulation of one OR occurs in females (Table 1).
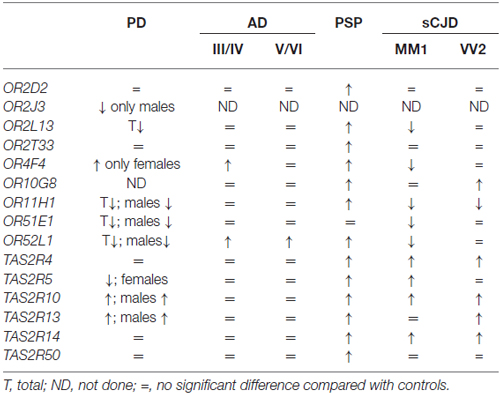
Table 1. Comparative aspects of olfactory (odorant) receptors (ORs) and taste receptors (TASR) gene expression in frontal cortex area 8 in Parkinson’s disease (PD), Alzheimer’s disease (AD) II–IV and V–VI corresponding stages of Braak and Braak, Progressive Supranuclear Palsy (PSP) and sporadic Creutzfeldt-Jakob disease (sCJD) subtypes MM1 and VV2, from observations described in reference Ansoleaga et al. (2013) and Garcia-Esparcia et al. (2013).
Parallel studies of TASR mRNA expression reveal no significant differences in the expression of analyzed genes in AD, up-regulation of six of the six analyzed genes in PSP, predominant up-regulation in sCJD, and increased or depressed mRNA expression of TASR genes depending on gender in PD (Table 1).
These observations show differential regulation of OR and TASR genes in frontal cortex area 8 in PD, AD, PSP and sCJD. These changes cannot be attributed merely to cell death, as some genes are up-regulated, others are down-regulated and still others show no modifications when compared with gene expression in control individuals.
Olfactory signaling in olfactory organs and smell perception are subject to gender differences (Stowers and Logan, 2010). This may be related to distinct and complementary factors such as defined gender responses to sex-specific initial signals (Holy et al., 2000; Luo et al., 2003; Ben-Shaul et al., 2010; Haga et al., 2010) probably secondary to modulation by steroid hormones (Leypold et al., 2002; Stowers et al., 2002; Kimchi et al., 2007; Raskin et al., 2009; Wu et al., 2009; Juntti et al., 2010). The observations in PD indicate that at least a sub-population of cortical cerebral ORs and TASRs in PD is modulated by sex-specific initial signals, sexually-dimorphic specific ligands or steroid hormones and their derivatives (Garcia-Esparcia et al., 2013). No similar data are available for AD, PSP or CJD.
mRNA levels of OR2T1, OR2T33, OR52H1, OR2D2, OR10G8, OR51E1 and OR52L1 are significantly reduced in the prefrontal cortex of patients with schizophrenia. The main changes cluster in 11p15.4, 1q44 and 11q24.2 (Ansoleaga et al., 2015). TAS2R4, TAS2R13, TAS2R5 and TAS2R50 mRNAs are decreased in the prefrontal cortex in the same group of patients (Ansoleaga et al., 2015).
Gene deregulation does not correlate with negative symptoms or with the duration of the disease, but mRNA expression levels of olfactory receptors OR2T1, OR252L1, OR2D2, OR10G8, and taste receptors TAS2R5, TAS2R13, and TAS2R50 show an association with treatment with antipsychotics, particularly chlorpromazine (Ansoleaga et al., 2015).
These findings show that brain deregulation of ORs and TASRs is not restricted to neurodegenerative diseases with abnormal protein aggregates but rather deregulation can also occur in neuropsychiatric disease. Moreover, modulation of certain ORs and TASRs largely depends on antipsychotics in schizophrenia, thus widening the scenario of OR and TASR regulation to variegated pharmacological agents (Ansoleaga et al., 2015).
ORs in regions other than those located in specific olfactory sensory organs have been categorized as “ectopic” ORs. These receptors are widely distributed in many organs in mammals where they play distinct functions not directed to the perception of odors as they are not connected to olfactory centers in brain. In most regions, ectopic ORs await identification of putative ligands and physiological functions. However, the impressively large number of genes and the wide distribution in their expression support a cardinal role for ORs (and TASRs) in homeostasis, possibly with the complementary action of different receptors.
Most work on Gαolf in human brain has been related to its expression in the striatum, its relation with dopamine D1 and adenosine A2A receptors (Corvol et al., 2001; Hervé, 2011) and its possible implications in movement disorders and mental disorders and in animal models in which dopamine is implicated. Gαolf is regulated by dopamine in the striatum, which may have implications in PD (Ruiz-DeDiego et al., 2015). In this line, certain polymorphisms in GNAL have been linked to bipolar and attention deficit/hyperactivity disorders (Berrettini et al., 1998; Laurin et al., 2008). Gαolf protein levels in rat striatum are increased by chronic antidepressant treatment (Taoka et al., 2006). Mutations in GNAL, which encodes Gαolf, are causative of craniocervical dystonia (Fuchs et al., 2013; Vemula et al., 2013; Kumar et al., 2014).
Gαolf and AC3 are not specific to the OR down-stream signaling pathway as they are used in multiple intracellular signaling processes.
Even considering that almost half of the human OR repertoire is probably non-functional (Rouquier and Giorgi, 2007; Go and Niimura, 2008), alterations in gene expression which are differentially regulated in a number of neurodegenerative and mental human diseases make the scenario a subject of attention. This is further driven by the fact that observations in other organs and systems have shown that ORs located in non-olfactory organs are chemoreceptors with unexpected functions.
The presence of a variable number of ORs in a single neuron, the variety of putative odorant ligands and the different feasible responses dependent on the region where the neuron is located and operates make elucidating the function of ORs in the nervous system an apparently tremendous task. The study of single receptors expressed in neural cell lines and combined receptors in particular primary neuron cultures in combination with screening of putative agonists of ORs and suspected OR ligands, most of them probable intrinsic metabolites, will facilitate the task. Design and isolation of small molecules to act on selected ORs functioning as cell-type specific agonists or antagonists seems a promising tool. Reconstruction of the tri-dimensional structure of receptors will help in the analysis of mechanisms of activation and in the recognition and design of functional ligands, and specific agonists and antagonists (Persuy et al., 2015; Wolf and Grünewald, 2015).
The activation of ORs and immune cellular responses by common molecules in several systems (Jang et al., 2013; Li et al., 2013b; Perricone et al., 2013; Stopková et al., 2014; Ward et al., 2015), and the cooperation of certain MHC genes in the expression of ORs in the vomeronasal organ of mice (Leinders-Zufall et al., 2014), lend support to the working hypothesis that the same scenario may occur in brain, where the central olfactory system expressed in neurons and the brain immune system involving microglia, astrocytes and neurons work in cooperation. The fact that the CNS is not an isolated immune organ but rather interacts with the systemic immune system, together with the expression of ORs and related OR factors in brain and choroid plexus directly interacting with blood, reinforces the notion that ORs in the CNS play novel functions unrelated to olfaction.
IF: designed and wrote the article, PG-E and MC: carried out immunohistochemistry; EC: studied the choroid plexus; EA and GGK: studied the developing brain; AG and SG: participate in the discussion and criticism of the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This study was funded by the Seventh Framework Programme of the European Commission, grant agreement 278486: DEVELAGE and the Ministerio de Ciencia e Innovación, Instituto de Salud Carlos III—Fondos FEDER, a way to build Europe FIS PIE14/00034 and PI14/00757. We wish to thank T. Yohannan for editorial assistance.
AC3/AC3, adenylyl cyclase 3 gene and protein, human; AD, Alzheimer’s disease; Adcy3, adenylate cyclase 3 gene, mouse; APOE4, apolipoprotein 4; APP, amyloid precursor protein; cAMP, cyclic adenosine monophosphate; Golf, olfactory G protein composed of three subunits: α, β and γ; Gαolf, olfactory G protein α subunit; GNAL/gnal, guanine nucleotide binding protein (G protein), alpha activating activity polypeptide olfactory type gene, human and mouse respectively, which encodes Gαolf; GPCRs, G-protein-coupled receptors; OBP, olfactory-binding protein; Olfr13 (MOR23), Olfr110, Olfr287, names of olfactory receptor genes, mouse; OMP, olfactory marker protein; ORs, olfactory (odorant) receptors; OR2D2, OR2H2, OR2J3, OR2L13, OR2T33, OR4F4, OR10G8, OR11H1, OR51E1, OR52L1, names of different human olfactory receptor genes; OR2H2, OR2A4, OR6K3, names of olfactory receptor proteins; PD, Parkinson’s disease; PrPRes, Prion protein resistant to proteinase K, Prion; PSEN1, Presenilin 1; PSP, Progressive supranuclear palsy; RTP1 and RTP2, Receptor Transporter Proteins 1 and 2; REEP1, Receptor Expression Enhancing Protein 1; sCJD, sporadic Creutzfeldt-Jakob disease; subtypes MM1, Methionine/methionine at codon 129 of PRNP; VV2, valine/valine at codon 129 of PRNP; PRNP, prion protein gene; TASR, taste receptors; TAS2R4, TAS2R5, TAS2R10, TAS2R13, TAS2R14, TAS2R50, names of different human taste receptor genes; UGTs, UDP-glucuronosyltransferases; UGT1A6, UGT2A3, UGT2B7, UGT2B10, names of different human UGT genes.
Alcock, J., Maley, C. C., and Aktipis, C. A. (2014). Is eating behavior manipulated by the gastrointestinal microbiota? Evolutionary pressures and potential mechanisms. Bioessays 36, 940–949. doi: 10.1002/bies.201400071
Ansoleaga, B., Garcia-Esparcia, P., Llorens, F., Moreno, J., Aso, E., and Ferrer, I. (2013). Dysregulation of brain olfactory and taste receptors in AD, PSP, CJD and AD-related model. Neuroscience 248, 369–382. doi: 10.1016/j.neuroscience.2013.06.034
Ansoleaga, B., Garcia-Esparcia, P., Pinacho, R., Haro, J. M., Ramos, B., and Ferrer, I. (2015). Decrease in olfactory and taste receptor expression in the dorsolateral prefrontal cortex in chronic schizophrenia. J. Psychiatr. Res. 60, 109–116. doi: 10.1016/j.jpsychires.2014.09.012
Asai, H., Kasai, H., Matsuda, Y., Yamazaki, N., Nagawa, F., Sakano, H., et al. (1996). Genomic structure and transcription of a murine odorant receptor gene: differential initiation of transcription in the olfactory and testicular cells. Biochem. Biophys. Res. Commun. 221, 240–247. doi: 10.1006/bbrc.1996.0580
Aso, E., Lomoio, S., López-González, I., Joda, L., Carmona, M., Fernández-Yagüe, N., et al. (2012). Amyloid generation and dysfunctional immunoproteasome activation with disease progression in animal model of familial Alzheimer’s disease. Brain Pathol. 22, 636–653. doi: 10.1111/j.1750-3639.2011.00560.x
Au, A., Gupta, A., Schembri, P., and Cheeseman, C. I. (2002). Rapid insertion of GLUT2 into the rat jejunal brush-border membrane promoted by glucagon-like peptide 2. Biochem. J. 367, 247–254. doi: 10.1042/bj20020393
Ay, H., Salihoglu, M., Altundag, A., Tekeli, H., Memis, A., and Cayonu, M. (2014). The effect of hyperbaric conditions on olfactory functions. Undersea Hyperb. Med. 41, 203–207.
Bakalyar, H. A., and Reed, R. R. (1990). Identification of a specialized adenylyl cyclase that may mediate odorant detection. Science 250, 1403–1406. doi: 10.1126/science.2255909
Baker, N. L., Miller, K. A., Newgreen, D. F., and Farlie, P. G. (2015). Olfr603, an orphan olfactory receptor, is expressed in multiple specific embryonic tissues. Gene Expr. Patterns 19, 30–35. doi: 10.1016/j.gep.2015.06.002
Barish, S., and Volkan, P. C. (2015). Mechanisms of olfactory receptor neuron specification in Drosophila. Wiley Interdiscip Rev. Dev. Biol 4, 609–621. doi: 10.1002/wdev.197
Behrens, M., Bartelt, J., Reichling, C., Winnig, M., Kuhn, C., and Meyerhof, W. (2006). Members of RTP and REEP gene families influence functional bitter taste receptor expression. J. Biol. Chem. 281, 20650–20659. doi: 10.1074/jbc.m513637200
Behrens, M., Foerster, S., Staehler, F., Raguse, J. D., and Meyerhof, W. (2007). Gustatory expression pattern of the human TAS2R bitter receptor gene family reveals a heterogenous population of bitter responsive taste receptor cells. J. Neurosci. 27, 12630–12640. doi: 10.1523/jneurosci.1168-07.2007
Behrens, M., and Meyerhof, W. (2010). Oral and extraoral bitter taste receptors. Results Probl. Cell. Differ. 52, 87–99. doi: 10.1007/978-3-642-14426-4_8
Belluscio, L., Gold, G. H., Nemes, A., and Axel, R. (1998). Mice deficient in G(olf) are anosmic. Neuron 20, 69–81. doi: 10.1016/s0896-6273(00)80435-3
Ben-Shaul, Y., Katz, L. C., Mooney, R., and Dulac, C. (2010). In vivo vomeronasal stimulation reveals sensory encoding of conspecific and allospecific cues by the mouse accessory olfactory bulb. Proc. Natl. Acad. Sci. U S A 107, 5172–5177. doi: 10.1073/pnas.0915147107
Berrettini, W. H., Vuoristo, J., Ferraro, T. N., Buono, R. J., Wildenauer, D., and Ala-Kokko, L. (1998). Human G (olf) gene polymorphisms and vulnerability to bipolar disorder. Psychiatr. Genet. 8, 235–238. doi: 10.1097/00041444-199808040-00006
Bertram, L., and Tanzi, R. E. (2011). “Genetics of Alzheimer’s disease,” in Neurodegeneration: The Molecular Pathology of Dementia and Movement Disorders, eds D. W. Dickson and R. O. Weller (Oxford, NY: Wiley-Blackwell), 51–61.
Billig, G. M., Pál, B., Fidzinski, P., and Jentsch, T. J. (2011). Ca2+-activated Cl- currents are dispensable for olfaction. Nat. Neurosci. 14, 763–769. doi: 10.1038/nn.2821
Blache, P., Gros, L., Salazar, G., and Bataille, D. (1998). Cloning and tissue distribution of a new rat olfactory receptor-like (OL2). Biochem. Biophys. Res. Commun. 242, 669–672. doi: 10.1006/bbrc.1997.8041
Blomquist, G. J., and Vogt, R. G. (2003). Insect Pheromone Biochemistry and Molecular Biology. London: Elsevier, Academic Press.
Bönigk, W., Bradley, J., Müller, F., Sesti, F., Boekhoff, I., Ronnett, G. V., et al. (1999). The native rat olfactory cyclic nucleotide-gated channel is composed of three distinct subunits. J. Neurosci. 19, 5332–5347.
Borchelt, D. R., Thinakaran, G., Eckman, C. B., Lee, M. K., Davenport, F., Ratovitsky, T., et al. (1996). Familial Alzheimer’s disease-linked presenilin 1 variants elevate Aβ1–42/1–40 ratio in vitro and in vivo. Neuron 17, 1005–1013. doi: 10.1016/s0896-6273(00)80230-5
Braak, H., and Braak, E. (1991). Neuropathological staging of Alzheimer-related changes. Acta Neuropathol. 82, 239–259. doi: 10.1007/bf00308809
Braak, H., and Braak, E. (1999). “Temporal sequence of Alzheimer’s disease-related pathology,” in Cerebral Cortex Vol. 14, Neurodegenerative and Age-Related Changes in Structure and Function of Cerebral Cortex, eds A. Peters and J. H. Morrison (New York, NY, Boston, Dordrecht, London, Moscow: Kluwer Academic/Plenum Publishers), 475–512.
Braak, H., Thal, D. R., Ghebremedhin, E., and Del Tredici, K. (2011). Stages of the pathologic process in Alzheimer disease: age categories from 1 to 100 years. J. Neuropathol. Exp. Neurol. 70, 960–969. doi: 10.1097/NEN.0b013e318232a379
Branscomb, A., Seger, J., and White, R. L. (2000). Evolution of odorant receptors expressed in human testes. Genetics 156, 785–797. doi: 10.1002/1521-1878(200009)22:9<803::aid-bies5>3.3.co;2-b
Braun, T., Voland, P., Kunz, L., Prinz, C., and Gratzl, M. (2007). Enterochromaffin cells of the human gut: sensors for spices and odorants. Gastroenterology 132, 1890–1901. doi: 10.1053/j.gastro.2007.02.036
Breer, H., Eberle, J., Frick, C., Haid, D., and Widmayer, P. (2012). Gastrointestinal chemosensation: chemosensory cells in the alimentary tract. Histochem. Cell Biol. 138, 13–24. doi: 10.1007/s00418-012-0954-z
Brennan, P. A., and Zufall, F. (2006). Pheromonal communication in vertebrates. Nature 444, 308–315. doi: 10.1038/nature05404
Brewer, W. J., Castle, D., and Pantelis, C. (eds.) (2006). Olfaction and the Brain. Cambridge: Cambridge University Press.
Briand, L., Blon, F., Trotier, D., and Pernollet, J. C. (2004). Natural ligands of hamster aphrodisin. Chem. Senses 29, 425–430. doi: 10.1093/chemse/bjh044
Briscoe, A. D., Macias-Muñoz, A., Kozak, K. M., Walters, J. R., Yuan, F., Jamie, G. A., et al. (2013). Female behaviour drives expression and evolution of gustatory receptors in butterflies. PLoS Genet. 9:e1003620. doi: 10.1371/journal.pgen.1003620
Brunet, L. J., Gold, G. H., and Ngai, J. (1996). General anosmia caused by a targeted disruption of the mouse olfactory cyclic nucleotide-gated cation channel. Neuron 17, 681–693. doi: 10.1016/s0896-6273(00)80200-7
Buck, L., and Axel, R. (1991). A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell 65, 175–187. doi: 10.1016/0092-8674(91)90418-x
Budka, H., Head, M. W., Ironside, J. W., Gambetti, P., Parchi, P., and Tagliavini, F. (2011). “Sporadic Creutzdfeldt-Jakob disease,” in Neurodegeneration: The Molecular Pathology of Dementia and Movement Disorders, eds D. W. Dickson and R. O. Weller (Oxford, NY: Wiley-Blackwell), 322–335.
Burke, M. V., and Small, D. M. (2015). Physiological mechanisms by which non-nutritive sweeteners may impact body weight and metabolism. Physiol. Behav. 152, 381–388. doi: 10.1016/j.physbeh.2015.05.036
Chen, M. C., Wu, S. V., Reeve, J. R. Jr., and Rozengurt, E. (2006). Bitter stimuli induce Ca2+ signaling and CCK release in enteroendocrine STC-1 cells: role of L-type voltage-sensitive Ca2+ channels. Am. J. Physiol. Cell Physiol. 291, C726–C739. doi: 10.1152/ajpcell.00003.2006
Claudianos, C., Lim, J., Young, M., Yan, S., Cristino, A. S., Newcomb, R. D., et al. (2014). Odor memories regulate olfactory receptor expression in the sensory periphery. Eur. J. Neurosci. 39, 1642–1654. doi: 10.1111/ejn.12539
Corvol, J. C., Studler, J. M., Schonn, J. S., Girault, J. A., and Hervé, D. (2001). Gαolf is necessary for coupling D1 and A2a receptors to adenylyl cyclase in the striatum. J. Neurochem. 76, 1585–1588. doi: 10.1046/j.1471-4159.2001.00201.x
Dal Monte, M., Andreini, I., Revoltella, R., and Pelosi, P. (1991). Purification and characterization of two odorant-binding proteins from nasal tissue of rabbit and pig. Comp. Biochem. Physiol. B 99, 445–451. doi: 10.1016/0305-0491(91)90068-o
Davidson, T. L., Martin, A. A., Clark, K., and Swithers, S. E. (2011). Intake of high-intensity sweeteners alters the ability of sweet taste to signal caloric consequences: implications for the learned control of energy and body weight regulation. Q. J. Exp. Psychol. (Hove) 64, 1430–1441. doi: 10.1080/17470218.2011.552729
Dehkordi, O., Rose, J. E., Fatemi, M., Allard, J. S., Balan, K. V., Young, J. K., et al. (2012). Neuronal expression of bitter taste receptors and downstream signalling molecules in rat brainstem. Brain Res. 1475, 1–10. doi: 10.1016/j.brainres.2012.07.038
De la Cruz, O., Blekham, R., Zhang, X., Nicolae, D., Firestein, S., and Gilad, Y. (2009). A signature of evolutionary constraint on a subset of ectopically expressed olfactory receptor genes. Mol. Biol. Evol. 26, 491–494. doi: 10.1093/molbev/msn294
Depoortere, I. (2014). Taste receptors of the gut: emerging roles in health and disease. Gut 63, 179–190. doi: 10.1136/gutjnl-2013-305112
Dhallan, R. S., Yau, K. W., Schrader, K. A., and Reed, R. R. (1990). Primary structure and functional expression of a cyclic nucleotide-activated channel from olfactory neurons. Nature 347, 184–187. doi: 10.1038/347184a0
Dhingra, R., Sullivan, L., Jacques, P. F., Wang, T. J., Fox, C. S., Meigs, J. B., et al. (2007). Soft drink consumption and risk of developing cardiometabolic risk factors and the metabolic syndrome in middle-aged adults in the community. Circulation 116, 480–488. doi: 10.1161/circulationaha.107.689935
Dickson, D. W., Hauw, J. J., Agid, Y., and Litvan, I. (2011). “Progressive supranuclear palsy and corticobasal degeneration,” in Neurodegeneration: The Molecular Pathology of Dementia and Movement Disorders, eds D. W. Dickson and R. O. Weller (Oxford, NY: Wiley-Blackwell), 135–155.
Dreyer, W. J. (1998). The area code hypothesis revisited: olfactory receptors and other related transmembrane receptors may function as the last digits in a cell surface code for assembling embryos. Proc. Natl. Acad. Sci. U S A 95, 9072–9077. doi: 10.1073/pnas.95.16.9072
Drutel, G., Arrang, J. M., Diaz, J., Wisnewsky, C., Schwartz, K., and Schwartz, J. C. (1995). Cloning of OL1, a putative olfactory receptor and its expression in the developing rat heart. Receptors Channels 3, 33–40.
Duvernay, M. T., Dong, C., Zhang, X., Zhou, F., Nichols, C. D., and Wu, G. (2009). Anterograde trafficking of G protein-coupled receptors: function of the C-terminal F(X)6LL motif in export from the endoplasmic reticulum. Mol. Pharmacol. 75, 751–761. doi: 10.1124/mol.108.051623
Duvernay, M. T., Zhou, F., and Wu, G. (2004). A conserved motif for the transport of G protein-coupled receptors from the endoplasmic reticulum to the cell surface. J. Biol. Chem. 279, 30741–30750. doi: 10.1074/jbc.M313881200
Duyckaerts, C. H., and Dickson, D. (2011). “Neuropathology of Alzheimer’s disease and variants,” in Neurodegeneration: The Molecular Pathology of Dementia and Movement Disorders, eds D. W. Dickson and R. O. Weller (Oxford, NY: Wiley-Blackwell), 62–91.
Dyer, J., Salmon, K. S., Zibrik, L., and Shirazi-Beechey, S. P. (2005). Expression of sweet taste receptors of the T1R family in the intestinal tract and enteroendocrine cells. Biochem. Soc. Trans. 33, 302–305. doi: 10.1042/bst0330302
Eckmeier, D., and Shea, S. D. (2014). Noradrenergic plasticity of olfactory sensory neuron inputs to the main olfactory bulb. J. Neurosci. 34, 15234–15243. doi: 10.1523/JNEUROSCI.0551-14.2014
Egan, J. M., and Margolskee, R. F. (2008). Taste cells of the gut and gastrointestinal chemosensation. Mol. Interv. 8, 78–81. doi: 10.1124/mi.8.2.5
Feingold, E. A., Penny, L. A., Nienhuis, A. W., and Forget, B. G. (1999). An olfactory receptor gene is located in the extended human β-globin gene cluster and is expressed in erythroid cells. Genomics 61, 15–23. doi: 10.1006/geno.1999.5935
Feinstein, P., Bozza, T., Rodriguez, I., Vassalli, A., and Mombaerts, P. (2004). Axon guidance of mouse olfactory sensory neurons by odorant receptors and the β2 adrenergic receptor. Cell 117, 833–846. doi: 10.1016/j.cell.2004.05.013
Feinstein, P., and Mombaerts, P. (2004). A contextual model for axonal sorting into glomeruli in the mouse olfactory system. Cell 117, 817–831. doi: 10.1016/j.cell.2004.05.011
Feldmesser, E., Olender, T., Khen, M., Yanai, I., Ophir, R., and Lancet, D. (2006). Widespread ectopic expression of olfactory receptor genes. BMC Genomics 7:121. doi: 10.1186/1471-2164-7-121
Ferreira, T., Wilson, S. R., Choi, Y. G., Risso, D., Dudoit, S., Speed, T. P., et al. (2014). Silencing of odorant receptor genes by G protein βγ signaling ensures the expression of one odorant receptor per olfactory sensory neuron. Neuron 81, 847–859. doi: 10.1016/j.neuron.2014.01.001
Ferrer, I. (2012). Defining Alzheimer as a common age-related neurodegenerative process not inevitably leading to dementia. Prog. Neurobiol. 97, 38–51. doi: 10.1016/j.pneurobio.2012.03.005
Ferrer, I. (2009). Early involvement of the cerebral cortex in Parkinson’s disease: convergence of multiple metabolic defects. Prog. Neurobiol. 88, 89–103. doi: 10.1016/j.pneurobio.2009.02.004
Ferrer, I., López-Gonzalez, I., Carmona, M., Dalfó, E., Pujol, A., and Martínez, A. (2012). Neurochemistry and the non-motor aspects of PD. Neurobiol. Dis. 46, 508–526. doi: 10.1016/j.nbd.2011.10.019
Ferrer, I., Martinez, A., Blanco, R., Dalfó, E., and Carmona, M. (2011). Neuropathology of sporadic Parkinson disease before the appearance of parkinsonism: preclinical Parkinson disease. J. Neural Transm. (Vienna) 118, 821–839. doi: 10.1007/s00702-010-0482-8
Firestein, S. (2001). How the olfactory system makes sense of scents. Nature 413, 211–218. doi: 10.1038/35093026
Firestein, S., Darrow, B., and Shepherd, G. M. (1991). Activation of the sensory current in salamander olfactory receptor neurons depends on a G protein-mediated cAMP second messenger system. Neuron 6, 825–835. doi: 10.1016/0896-6273(91)90178-3
Flegel, C., Manteniotis, S., Osthold, S., Hatt, H., and Gisselmann, G. (2013). Expression profile of ectopic olfactory receptors determined by deep sequencing. PLoS One 8:e55368. doi: 10.1371/journal.pone.0055368
Flower, D. R. (1996). The lipocalin protein family: structure and function. Biochem. J. 318, 1–14. doi: 10.1042/bj3180001
Fowler, S. P., Williams, K., and Hazuda, H. P. (2015). Diet soda intake is associated with long-term increases in waist circumference in a biethnic cohort of older adults: the San Antonio Longitudinal Study of Aging. J. Am. Geriatr. Soc. 63, 708–715. doi: 10.1111/jgs.13376
Fuchs, T., Saunders-Pullman, R., Masuho, I., Luciano, M. S., Raymond, D., Factor, S., et al. (2013). Mutations in GNAL cause primary torsion dystonia. Nat. Genet. 45, 88–92. doi: 10.1038/ng.2496
Fukuda, N., and Touhara, K. (2006). Developmental expression patterns of testicular olfactory receptor genes during mouse spermatogenesis. Genes Cells 11, 71–81. doi: 10.1111/j.1365-2443.2005.00915.x
Fukuda, N., Yomogida, K., Okabe, M., and Touhara, K. (2004). Functional characterization of a mouse testicular olfactory receptor and its role in chemosensing and in regulation of sperm motility. J. Cell Sci. 117, 5835–5845. doi: 10.1242/jcs.01507
Garcia-Esparcia, P., Schlüter, A., Carmona, M., Moreno, J., Ansoleaga, B., Torrejón-Escribano, B., et al. (2013). Functional genomics reveals dysregulation of cortical olfactory receptors in Parkinson’s disease: novel putative chemoreceptors in the human brain. J. Neuropathol. Exp. Neurol. 72, 524–539. doi: 10.1097/NEN.0b013e318294fd76
Garibotti, M., Christiansen, H., Schmale, H., and Pelosi, P. (1995). Porcine VEG proteins and tear prealbumin. Chem. Senses 20, 69–76. doi: 10.1093/chemse/20.1.69
Gilad, Y., Man, O., and Glusman, G. (2005). A comparison of the human and chimpanzee olfactory receptor gene repertoires. Genome Res. 15, 224–230. doi: 10.1101/gr.2846405
Gimelbrant, A., Haley, S., and McClintock, T. (2001). Olfactory receptor trafficking involves conserved regulatory steps. J. Biol. Chem. 276, 7285–7290. doi: 10.1074/jbc.m005433200
Gimelbrant, A., Skaletski, H., and Chess, A. (2004). Selective pressures on the olfactory receptor repertoire since the human-chimpanzee divergence. Proc. Natl. Acad. Sci. U S A 101, 9019–9022. doi: 10.1073/pnas.0401566101
Glasgow, B. J., Abduragimov, A. R., Farahbakhsh, Z. T., Faull, K. F., and Hubbell, W. L. (1995). Tear lipocalins bind a broad array of lipid ligands. Curr. Eye Res. 14, 363–372. doi: 10.3109/02713689508999934
Glusman, G., Bahar, A., Sharon, D., Pilpel, Y., White, J., and Lancet, D. (2000a). The olfactory receptor gene superfamily: data mining, classification and nomenclature. Mamm. Genome 11, 1016–1023. doi: 10.1007/s003350010196
Glusman, G., Sosinsky, A., Ben-Asher, E., Avidan, N., Sonkin, D., Bahar, A., et al. (2000b). Sequence, structure and evolution of a complete human olfactory receptor gene cluster. Genomics 63, 227–245. doi: 10.1006/geno.1999.6030
Glusman, G., Yanai, I., Rubin, I., and Lancet, D. (2001). The complete human olfactory subgenome. Genome Res. 11, 685–702. doi: 10.1101/gr.171001
Go, Y., and Niimura, Y. (2008). Similar numbers but different repertoires of olfactory receptor genes in humans and chimpanzees. Mol. Biol. Evol. 25, 1897–1907. doi: 10.1093/molbev/msn135
Godfrey, P. A., Malnic, B., and Buck, L. B. (2004). The mouse olfactory receptor gene family. Proc. Natl. Acad. Sci. U S A 101, 2156–2161. doi: 10.1073/pnas.0308051100
Goto, T., Salpekar, A., and Monk, M. (2001). Expression of a testis-specific member of the olfactory receptor gene family in human primordial germ cells. Mol. Hum. Reprod. 7, 553–558. doi: 10.1093/molehr/7.6.553
Griffin, C. A., Kafadar, K. A., and Pavlath, G. K. (2009). MOR23 promotes muscle regeneration and regulates cell adhesion and migration. Dev. Cell 17, 649–661. doi: 10.1016/j.devcel.2009.09.004
Grison, A., Zucchelli, S., Urzì, A., Zamparo, I., Lazarevic, D., Pascarella, G., et al. (2014). Mesencephalic dopaminergic neurons express a repertoire of olfactory receptors and respond to odorant-like molecules. BMC Genomics 15:729. doi: 10.1186/1471-2164-15-729
Gu, X., Karp, P. H., Brody, S. L., Pierce, R. A., Welsh, M. J., Holtzman, M. J., et al. (2014). Chemosensory functions for pulmonary neuroendocrine cells. Am. J. Respir. Cell Mol. Biol. 50, 637–646. doi: 10.1165/rcmb.2013-0199OC
Gu, F., Liu, X., Liang, J., Chen, J., Chen, F., and Li, F. (2015). Bitter taste receptor mTas2r105 is expressed in small intestinal villus and crypts. Biochem. Biophys. Res. Commun. 463, 934–941. doi: 10.1016/j.bbrc.2015.06.038
Guiraudie-Capraz, G., Slomianny, M. C., Pageat, P., Malosse, C., Cain, A. H., Orgueur, P., et al. (2005). Biochemical and chemical supports for a transnasal olfactory continuity through sow maternal fluids. Chem. Senses 30, 241–251. doi: 10.1093/chemse/bji020
Haga, S., Hattori, T., Sato, T., Sato, K., Matsuda, S., Kobayakawa, R., et al. (2010). The male mouse pheromone ESP1 enhances female sexual receptive behaviour through a specific vomeronasal receptor. Nature 466, 118–122. doi: 10.1038/nature09142
Hague, C., Chen, Z., Pupo, A. S., Schulte, N. A., Toews, M. L., and Minneman, K. P. (2004). The N terminus of the human α1D-adrenergic receptor prevents cell surface expression. J. Pharmacol. Exp. Ther. 309, 388–397. doi: 10.1124/jpet.103.060509
Halatchev, I. G., and Cone, R. D. (2005). Peripheral administration of PYY(3–36) produces conditioned taste aversion in mice. Cell Metab. 1, 159–168. doi: 10.1016/j.cmet.2005.02.003
Hallem, E. A., Dahanukar, A., and Carlson, J. R. (2006). Insect odor and taste receptors. Annu. Rev. Entomol. 51, 113–135. doi: 10.1146/annurev.ento.51.051705.113646
Hamzic, N., Blomqvist, A., and Nilsberth, C. (2013). Immune-induced expression of lipocain-2 in brain endothelial cells: relationship with interleukin-6, cyclooxygenassse-2 and the febrile response. J. Neuroendocrinol. 25, 271–280. doi: 10.1111/jne.12000
Hayden, S., Bekaert, M., Goodbla, A., Murphy, W. J., Dávalos, L. M., and Teeling, E. C. (2014). A cluster of olfactory receptor genes linked to frugivory in bats. Mol. Biol. Evol. 31, 917–927. doi: 10.1093/molbev/msu043
Head, M. W., and Ironside, J. W. (2009). Sporadic Creutzfeldt-Jakob disease: discrete subtypes or a spectrum of the disease. Brain 132, 2627–2629. doi: 10.1093/brain/awp225
Hervé, D. (2011). Identification of a specific assembly of the g protein golf as a critical and regulated module of dopamine and adenosine-activated cAMP pathways in the striatum. Front. Neuroanat. 5:48. doi: 10.3389/fnana.2011.00048
Heydel, J. M., Holsztynska, E. J., Legendre, A., Thiebaud, N., Artur, Y., and Le Bon, A. M. (2010). UDP-glucuronosyltransferases (UGTs) in neuro-olfactory tissues: expression, regulation and function. Drug Metab. Rev. 42, 74–97. doi: 10.3109/03602530903208363
Höfer, D., Püschel, B., and Drenckhahn, D. (1996). Taste receptor-like cells in the rat gut identified by expression of alpha-gustducin. Proc. Natl. Acad. Sci. U S A 93, 6631–6634. doi: 10.1073/pnas.93.13.6631
Holy, T. E., Dulac, C., and Meister, M. (2000). Responses of vomeronasal neurons to natural stimuli. Science 289, 1569–1572. doi: 10.1126/science.289.5484.1569
Hoover, K. C. (2013). Evolution of olfactory receptors. Methods Mol. Biol. 1003, 241–249. doi: 10.1007/978-1-62703-377-0_18
Houlden, H., and Singleton, A. B. (2012). The genetics and neuropathology of Parkinson’s disease. Acta Neuropathol. 124, 325–338. doi: 10.1007/s00401-012-1013-5
Hsu, F. F., Nodari, F., Kao, L. F., Fu, X., Holekamp, T. F., Turk, J., et al. (2008). Structural characterization of sulfated steroids that activate mouse pheromone-sensing neurons. Biochemistry 47, 14009–14019. doi: 10.1021/bi801392j
Hughes, G. M., Teeling, E. C., and Higgins, D. G. (2014). Loss of olfactory receptor function in hominin evolution. PLoS One 9:e84714. doi: 10.1371/journal.pone.0084714
Ihara, S., Yoshikawa, K., and Touhara, K. (2013). Chemosensory signals and their receptors in the olfactory neural system. Neuroscience 254, 45–60. doi: 10.1016/j.neuroscience.2013.08.063
Ishimaru, Y., and Matsunami, H. (2009). Transient receptor potential (TRP) channels and taste sensation. J. Dent. Res. 88, 212–218. doi: 10.1177/0022034508330212
Itakura, S., Ohno, K., Ueki, T., Sato, K., and Kanayama, N. (2006). Expression of Golf in the rat placenta: possible implication in olfactory receptor transduction. Placenta 27, 103–108. doi: 10.1016/j.placenta.2004.12.006
Iwatsuki, K., and Uneyama, H. (2012). Sense of taste in the gastrointestinal tract. J. Pharmacol. Sci. 118, 123–128. doi: 10.1254/jphs.11r08cp
Jacquier, V., Pick, H., and Vogel, H. (2006). Characterization of an extended receptive ligand repertoire of the human olfactory receptor OR17–40 comprising structurally related compounds. J. Neurochem. 97, 537–544. doi: 10.1111/j.1471-4159.2006.03771.x
Jang, H. J., Kokrashvili, Z., Theodorakis, M. J., Carlson, O. D., Kim, B. J., Zhou, J., et al. (2007). Gut-expressed gustducin and taste receptors regulate secretion of glucagon-like peptide-1. Proc. Natl. Acad. Sci. U S A 104, 15069–15074. doi: 10.1073/pnas.0706890104
Jang, E., Lee, S., Kim, J. H., Kim, J. H., Seo, J. W., Lee, W. H., et al. (2013). Secreted protein lipocalin-2 promotes microglial M1 polarization. FASEB J. 27, 1176–1190. doi: 10.1096/fj.12-222257
Jellinger, K. A. (2011). “Parkinson’s disease,” in Neurodegeneration: The Molecular Pathology of Dementia and Movement Disorders, eds D. W. Dickson and R. O. Weller (Oxford, NY: Wiley-Blackwell), 194–223.
Jin, M., Kim, J. H., Jang, E., Lee, Y. M., Soo Han, H., Woo, D. K., et al. (2014). Lipocalin-2 deficiency attenuates neuroinflammation and brain injury after transient middle cerebral artery occlusion in mice. J. Cereb. Blood Flow Metab. 34, 1306–1314. doi: 10.1038/jcbfm.2014.83
Jones, D. T., and Reed, R. R. (1989). Golf: an olfactory neuron specific-G protein involved in odorant signal transduction. Science 244, 790–795. doi: 10.1126/science.2499043
Jungnickel, M. K., Marrero, H., Birnbaumer, L., Lémos, J. R., and Florman, H. M. (2001). Trp2 regulates entry of Ca2+ into mouse sperm triggered by egg ZP3. Nat. Cell Biol. 3, 499–502. doi: 10.1038/35074570
Juntti, S. A., Tollkuhn, J., Wu, M. V., Fraser, E. J., Soderborg, T., Tan, S., et al. (2010). The androgen receptor governs the execution, but not programming, of male sexual and territorial behaviours. Neuron 66, 260–272. doi: 10.1016/j.neuron.2010.03.024
Kaji, I., Karaki, S., and Kuwahara, A. (2014). Taste sensing in the colon. Curr. Pharm. Des. 20, 2766–2774. doi: 10.2174/13816128113199990573
Kanageswaran, N., Demond, M., Nagel, M., Schreiner, B. S., Baumgart, S., Scholz, P., et al. (2015). Deep sequencing of the murine olfactory receptor neuron transcriptome. PLoS One 10:e0113170. doi: 10.1371/journal.pone.0113170
Kang, N., Bahk, Y. Y., Lee, N., Jae, Y., Cho, Y. H., Ku, C. R., et al. (2015). Olfactory receptor Olfr544 responding to azelaic acid regulates glucagon secretion in α-cells of mouse pancreatic islets. Biochem. Biophys. Res. Commun. 460, 616–621. doi: 10.1016/j.bbrc.2015.03.078
Kang, N., and Koo, J. (2012). Olfactory receptors in non-chemosensory tissues. BMB Rep. 45, 612–622. doi: 10.5483/bmbrep.2012.45.11.232
Kato, A., and Touhara, K. (2009). Mammalian olfactory receptors: pharmacology, G protein coupling and desensitization. Cell Mol. Life Sci. 66, 3743–3753. doi: 10.1007/s00018-009-0111-6
Keller, A., Zhuang, H. Y., Chi, Q. Y., Vosshall, L. B., and Matsunami, H. (2007). Genetic variation in a human odorant receptor alters odour perception. Nature 449, 468–472. doi: 10.1038/nature06162
Kendig, D. M., Hurst, N. R., Bradley, Z. L., Mahavadi, S., Kuemmerle, J. F., Lyall, V., et al. (2014). Activation of the umami taste receptor (T1R1/T1R3) initiates the peristaltic reflex and pellet propulsion in the distal colon. Am. J. Physiol. Gastrointest. Liver Physiol. 307, G1100–G1107. doi: 10.1152/ajpgi.00251.2014
Kerr, D. S., Von Dannecker, L. E., Davalos, M., Michaloski, J. S., and Malnic, B. (2008). Ric-8B interacts with Gαolf and Gγ13 and co-localizes with Gαolf, Gβ1 and Gγ13 in the cilia of olfactory sensory neurons. Mol. Cell Neurosci. 38, 341–348. doi: 10.1016/j.mcn.2008.03.006
Khafizov, K., Anselmi, C., Menini, A., and Carloni, P. (2007). Ligand specificity of odorant receptors. J. Mol. Model. 13, 401–409. doi: 10.1007/s00894-006-0160-9
Kim, S. H., Yoon, Y. C., Lee, A. S., Kang, N., Koo, J., Rhyu, M. R., et al. (2015a). Expression of human olfactory receptor 10J5 in heart aorta, coronary artery and endothelial cells and its functional role in angiogenesis. Biochem. Biophys. Res. Commun. 460, 404–448. doi: 10.1016/j.bbrc.2015.03.046
Kim, S. Y., Yoo, S. J., Ronnett, G. V., Kim, E. K., and Moon, C. (2015b). Odorant stimulation promotes survival of rodent olfactory receptor neurons via PI3K/Akt activation and Bcl-2 expression. Mol. Cells 38, 535–539. doi: 10.14348/molcells.2015.0038
Kimchi, T., Xu, J., and Dulac, C. (2007). A functional circuit underlying male sexual behaviour in the female mouse brain. Nature 448, 1009–1014. doi: 10.1038/nature06089
Kludt, E., Okom, C., Brinkmann, A., and Schild, D. (2015). Integrating temperature with odor processing in the olfactory bulb. J. Neurosci. 35, 7892–7902. doi: 10.1523/JNEUROSCI.0571-15.2015
Knopman, D. (2011). “Clinical aspects of Alzheimer’s disease,” in Neurodegeneration: The Molecular Pathology of Dementia and Movement Disorders, eds D. W. Dickson and R. O. Weller (Oxford, NY: Wiley-Blackwell), 39–50.
Kokrashvili, Z., Yee, K. K., Ilegems, E., Iwatsuki, K., Li, Y., Mosinger, B., et al. (2014). Endocrine taste cells. Br. J. Nutr. 111, S23–S29. doi: 10.1017/S0007114513002262
Kumar, K. R., Lohmann, K., Masuho, I., Miyamoto, R., Ferbert, A., Lohnau, T., et al. (2014). Mutations in GNAL: a novel cause of craniocervical dystonia. JAMA Neurol. 71, 490–494. doi: 10.1001/jamaneurol.2013.4677
Kwon, H. J., Koo, J. H., Zufall, F., Leinders-Zufall, T., and Margolis, F. L. (2009). Ca extrusion by NCX is compromised in olfactory sensory neurons of OMP mice. PLoS One 4:e4260. doi: 10.1371/journal.pone.0004260
Laurin, N., Ickowicz, A., Pathare, T., Malone, M., Tannock, R., Schachar, R., et al. (2008). Investigation of the G protein subunit Gαolf gene (GNAL) in attention deficit/hyperactivity disorder. J. Psychiatr. Res. 42, 117–124. doi: 10.1016/j.jpsychires.2006.10.010
Leclerc, S., Heydel, J. M., Amossé, V., Gradinaru, D., Cattarelli, M., Artur, Y., et al. (2002). Glucuronidation of odorant molecules in the rat olfactory system: activity, expression and age-linked modifications of UDP-glucuronosyltransferase isoforms, UGT1A6 and UGT2A1 and relation to mitral cell activity. Brain Res. Mol. Brain Res. 107, 201–213. doi: 10.1016/s0169-328x(02)00455-2
Le Danvic, C., Guiraudie-Capraz, G., Abderrahmani, D., Zanetta, J. P., and Nagnan-Le Meillour, P. (2009). Natural ligands of porcine olfactory binding proteins. J. Chem. Ecol. 35, 741–751. doi: 10.1007/s10886-009-9645-1
Lee, S., Kim, J. H., Kim, J. H., Seo, J. W., Han, H. S., Lee, W. H., et al. (2011). Lipocalin-2 Is a chemokine inducer in the central nervous system: role of chemokine ligand 10 (CXCL10) in lipocalin-2-induced cell migration. J. Biol. Chem. 286, 43855–43870. doi: 10.1074/jbc.M111.299248
Leinders-Zufall, T., Ishii, T., Chamero, P., Hendrix, P., Oboti, L., Schmid, A., et al. (2014). A family of nonclassical class I MHC genes contributes to ultrasensitive chemodetection by mouse vomeronasal sensory neurons. J. Neurosci. 34, 5121–5533. doi: 10.1523/JNEUROSCI.0186-14.2014
Leypold, B. G., Yu, C. R., Leinders-Zufall, T., Kim, M. M., Zufall, F., and Axel, R. (2002). Altered sexual and social behaviors in trp2 mutant mice. Proc. Natl. Acad. Sci. U S A 99, 6376–6381. doi: 10.1073/pnas.082127599
Li, Q., and Liberles, S. D. (2015). Aversion and attraction through olfaction. Curr. Biol. 25, R120–R129. doi: 10.1016/j.cub.2014.11.044
Li, F., Ponissery-Saidu, S., Yee, K. K., Wang, H., Chen, M. L., Iguchi, N., et al. (2013a). Heterotrimeric G protein subunit Gγ13 is critical to olfaction. J. Neurosci. 33, 7975–7984. doi: 10.1523/JNEUROSCI.5563-12.2013
Li, J. J., Tay, H. L., Plank, M., Essilfie, A. T., Hansbro, P. M., Foster, P. S., et al. (2013b). Activation of olfactory receptors on mouse pulmonary macrophages promotes monocyte chemotactic protein-1 production. PLoS One 8:e80148. doi: 10.1371/journal.pone.0080148
Liberles, S. D. (2014). Mammalian pheromones. Annu. Rev. Physiol. 76, 151–175. doi: 10.1146/annurev-physiol-021113-170334
Liman, E. R., and Buck, L. B. (1994). A second subunit of the olfactory cyclic nucleotide-gated channel confers high sensitivity to cAMP. Neuron 13, 611–621. doi: 10.1016/0896-6273(94)90029-9
Lowe, J., and Kalaria, R. (2015). “Dementia,” in Greenfield’s Neuropathology, (Vol. 1) eds S. Love, H. Budka, J. W. Ironside and A. Perry (Boca Raton, London, New York, NY: CRC Press, Taylor and Francis group), 858–973.
Lowe, G., Nakamura, T., and Gold, G. H. (1989). Adenylate cyclase mediates olfactory transduction for a wide variety of odorants. Proc. Natl. Acad. Sci. U S A 86, 5641–5645. doi: 10.1073/pnas.86.14.5641
Luo, A. H., Cannon, E. H., Wekesa, K. S., Lyman, R. F., Vandenbergh, J. G., and Anholt, R. R. (2002). Impaired olfactory behavior in mice deficient in the α subunit of G(o). Brain Res. 941, 62–71. doi: 10.1016/s0006-8993(02)02566-0
Luo, M., Fee, M. S., and Katz, L. C. (2003). Encoding pheromonal signals in the accessory olfactory bulb of behaving mice. Science 299, 1196–1201. doi: 10.1126/science.1082133
Lutsey, P. L., Steffen, L. M., and Stevens, J. (2008). Dietary intake and the development of the metabolic syndrome: the atherosclerosis risk in communities study. Circulation 117, 754–761. doi: 10.1161/CIRCULATIONAHA.107.716159
Mace, O. J., Affleck, J., Patel, N., and Kellett, G. L. (2007). Sweet taste receptors in rat small intestine stimulate glucose absorption through apical GLUT2. J. Physiol. 582, 379–392. doi: 10.1113/jphysiol.2007.130906
Mackenzie, P. I., Bock, K. W., Burchell, B., Guillemette, C., Ikushiro, S., Iyanagi, T., et al. (2005a). Nomenclature up-date for the mammalian UDP glycosyltransferase (UGT) gene superfamily. Pharmacogenet. Genomics 15, 677–685. doi: 10.1097/01.fpc.0000173483.13689.56
Mackenzie, P. I., Gregory, P. A., Lewinsky, R. H., Yasmin, S. N., Height, T., McKinnon, R. A., et al. (2005b). Polymorphic variations in the expression of the chemical detoxifying UDP glucuronosyltransferases. Toxicol. Appl. Pharmacol. 207, 77–83. doi: 10.1016/j.taap.2004.12.026
Malnic, B. (2007). Searching for the ligands of odorant receptors. Mol. Neurobiol. 35, 175–181. doi: 10.1007/s12035-007-0013-2
Malnic, B., Godfrey, P. A., and Buck, L. B. (2004). The human olfactory receptor gene family. Proc. Natl. Acad. Sci. U S A 101, 2584–2589. doi: 10.1073/pnas.0307882100
Malnic, B., Gonzalez-Kristeller, D. C., and Gutiyama, L. M. (2010). “Odorant receptors,” in The Neurobiology of Olfaction, ed. A. Menini (Boca Raton: CRC Press).
Manteniotis, S., Lehmann, R., Flegel, C., Vogel, F., Hofreuter, A., Schreiner, B. S., et al. (2013). Comprehensive RNA-Seq expression analysis of sensory ganglia with a focus on ion channels and GPCRs in trigeminal ganglia. PLoS One 8:e79523. doi: 10.1371/journal.pone.0079523
Marchese, S., Pes, D., Scaloni, A., Carbone, V., and Pelosi, P. (1998). Lipocalins of boar salivary glands binding odours and pheromones. Eur. J. Biochem. 252, 563–568. doi: 10.1046/j.1432-1327.1998.2520563.x
Margolskee, R. F., Dyer, J., Kokrashvili, Z., Salmon, K. S., Ilegems, E., Daly, K., et al. (2007). T1R3 and gustducin in gut sense sugars to regulate expression of Na+-glucose cotransporter 1. Proc. Natl. Acad. Sci. U S A 104, 15075–15080. doi: 10.1073/pnas.0706678104
Martin, F., Riveron, J., and Alcorta, E. (2011). Environmental temperature modulates olfactory reception in Drosophila melanogaster. J. Insect Physiol. 57, 1631–1642. doi: 10.1016/j.jinsphys.2011.08.016
Mast, T. G., Brann, J. H., and Fadool, D. A. (2010). The TRPC2 channel forms protein-protein interactions with homer and RTP in the rat vomeronasal organ. BMC Neurosci. 11:61. doi: 10.1186/1471-2202-11-61
Mombaerts, P. (1999). Molecular biology of odorant receptors in vertebrates. Annu. Rev. Neurosci. 22, 487–509. doi: 10.1146/annurev.neuro.22.1.487
Mombaerts, P. (2004). Genes and ligands for odorant, vomeronasal and taste receptors. Nat. Rev. Neurosci. 5, 263–278. doi: 10.1038/nrn1365
Nakamura, T., and Gold, G. H. (1987). A cyclic nucleotide-gated conductance in olfactory receptor cilia. Nature 325, 442–444. doi: 10.1038/325442a0
Nam, Y., Kim, J. H., Seo, M., Kim, J. H., Jin, M., Jeon, S., et al. (2014). Lipocalin-2 protein deficiency ameliorates experimental autoimmune encephalomyelitis: the pathogenic role of lipocalin-2 in the central nervous system and peripheral lymphoid tissues. J. Biol. Chem. 289, 16773–16789. doi: 10.1074/jbc.M113.542282
Natarajan, N., and Pluznick, J. L. (2015). Olfaction in the kidney: ‘smelling’ gut microbial metabolites. Exp. Physiol. 101, 478–481. doi: 10.1113/ep085285
Nef, S., and Nef, P. (1997). Olfaction: transient expression of a putative odorant receptor in the avian notochord. Proc. Natl. Acad. Sci. U S A 94, 4766–4771. doi: 10.1073/pnas.94.9.4766
Neuhaus, E. M., Zhang, W., Gelis, L., Deng, Y., Noldus, J., and Hatt, H. (2009). Activation of an olfactory receptor inhibits proliferation of prostate cancer cells. J. Biol. Chem. 284, 16218–16225. doi: 10.1074/jbc.M109.012096
Niimura, Y. (2012). Olfactory receptor multigene family in vertebrates: from the viewpoint of evolutionary genomics. Curr. Genomics 13, 103–114. doi: 10.2174/138920212799860706
Niimura, Y., Matsui, A., and Touhara, K. (2014). Extreme expansion of the olfactory receptor gene repertoire in African elephants and evolutionary dynamics of orthologous gene groups in 13 placental mammals. Genome Res. 24, 1485–1496. doi: 10.1101/gr.169532.113
Niimura, Y., and Nei, M. (2003). Evolution of olfactory receptor genes in the human genome. Proc. Natl. Acad. Sci. U S A 100, 12235–12240. doi: 10.1073/pnas.1635157100
Niimura, Y., and Nei, M. (2005). Comparative evolutionary analysis of olfactory receptor gene clusters between humans and mice. Gene 346, 13–21. doi: 10.1016/j.gene.2004.09.025
Nodari, F., Hsu, F. F., Fu, X., Holekamp, T. F., Kao, L. F., Turk, J., et al. (2008). Sulfated steroids as natural ligands of mouse pheromone-sensing neurons. J. Neurosci. 28, 6407–6418. doi: 10.1523/jneurosci.1425-08.2008
Olender, T., Feldmesser, E., Atarot, T., Eisenstein, M., and Lancet, D. (2004). The olfactory receptor universe–from whole genome analysis to structure and evolution. Genet. Mol. Res. 3, 545–553.
Otaki, J. M., Yamamoto, H., and Firestein, S. (2004). Odorant receptor expression in the mouse cerebral cortex. J. Neurobiol. 58, 315–327. doi: 10.1002/neu.10272
Pace, U., Hanski, E., Salomon, Y., and Lancet, D. (1985). Odorant-sensitive adenylate cyclase may mediate olfactory reception. Nature 316, 255–258. doi: 10.1038/316255a0
Paolini, S., Scaloni, A., Amoresano, A., Marchese, S., Napolitano, E., and Pelosi, P. (1998). Amino acid sequence, posttranslational modifications, binding and labelling of porcine odorant-binding protein. Chem. Senses 23, 689–698. doi: 10.1093/chemse/23.6.689
Parchi, P., Gambetti, P., and Capellari, S. (2011). “Genetic Creutzfeldt-Jakob disease,” in Neurodegeneration: The Molecular Pathology of Dementia and Movement Disorders, eds D. W. Dickson and R. O. Weller (Oxford, NY: Wiley-Blackwell), 322–333.
Parchi, P., Strammiello, R., Notari, S., Giese, A., Langeveld, J. P., Ladogana, A., et al. (2009). Incidence and spectrum of sporadic Creutzfledt-Jakob disease variants with mixed phenotypes and co-occurrence of PrPsc types: an updated classification. Acta Neuropathol. 118, 659–671. doi: 10.1007/s00401-009-0585-1
Parmentier, M., Libert, F., Schumans, S., Schiffmann, S., Lefort, A., Eggerickx, D., et al. (1992). Expression of members of the putative olfactory receptor gene family in mammalian germ cells. Nature 355, 453–455. doi: 10.1038/355453a0
Pelosi, P., and Maida, R. (1990). Odorant binding proteins in vertebrates and insects: similarities and possible common functions. Chem. Senses 15, 205–215. doi: 10.1093/chemse/15.2.205
Pepino, M. Y. (2015). Metabolic effects of non-nutritive sweeteners. Physiol. Behav. 152, 450–455. doi: 10.1016/j.physbeh.2015.06.024
Perricone, C., Shoenfeld, N., Agmon-Levin, N., de Carolis, C., Perricone, R., and Shoenfeld, Y. (2013). Smell and autoimmunity: a comprehensive review. Clin. Rev. Allergy Immunol. 45, 87–96. doi: 10.1007/s12016-012-8343-x
Persuy, M. A., Sanz, G., Tromelin, A., Thomas-Danguin, T., Gibrat, J. F., and Pajot-Augy, E. (2015). Mammalian olfactory receptors: molecular mechanisms of odorant detection, 3D-modeling and structure-activity relationships. Prog. Mol. Biol. Transl. Sci. 130, 1–36. doi: 10.1016/bs.pmbts.2014.11.001
Pevsner, J., Hou, V., Snowman, A. M., and Snyder, S. H. (1990). Odorant-binding protein. J. Biol. Chem. 265, 6118–6125.
Pevsner, J., Reed, R. R., Feinstein, P. G., and Snyder, S. H. (1988). Molecular cloning of odorant-binding protein: member of a ligand carrier family. Science 241, 336–339. doi: 10.1126/science.3388043
Pevsner, J., Sklar, P. B., and Snuyder, S. H. (1986). Odorant-binding protein: localization to nasal glands and secretions. Proc. Natl. Acad. Sci. U S A 83, 4942–4946. doi: 10.1073/pnas.83.13.4942
Picone, B., Hesse, U., Panji, S., Van Heusden, P., Jonas, M., and Christoffels, A. (2014). Taste and odorant receptors of the coelacanth–a gene repertoire in transition. J. Exp. Zool. B Mol. Dev. Evol. 322, 403–414. doi: 10.1002/jez.b.22531
Pierce, W. D., Heth, C. D., Owczarczyk, J. C., Russell, J. C., and Proctor, S. D. (2007). Overeating by young obesity-prone and lean rats caused by tastes associated with low energy foods. Obesity (Silver Spring) 15, 1969–1979. doi: 10.1038/oby.2007.235
Pluznick, J. L. (2013). Renal and cardiovascular sensory receptors and blood pressure regulation. Am. J. Physiol. Renal. Physiol. 305, F439–444. doi: 10.1152/ajprenal.00252.2013
Pluznick, J. L. (2014). Extra sensory perception: the role of Gpr receptors in the kidney. Curr. Opin. Nephrol. Hypertens. 23, 507–512. doi: 10.1097/MNH.0000000000000048
Pluznick, J. L., Protzko, R. J., Gevorgyan, H., Peterlin, Z., Sipos, A., Han, J., et al. (2013). Olfactory receptor responding to gut microbiota-derived signals plays a role in renin secretion and blood pressure regulation. Proc. Natl. Acad. Sci. U S A 110, 4410–4415. doi: 10.1073/pnas.1215927110
Pluznick, J. L., Zou, D. J., Zhang, X., Yan, Q., Rodriguez-Gil, D. J., Eisner, C., et al. (2009). Functional expression of the olfactory signaling system in the kidney. Proc. Natl. Acad. Sci. U S A 106, 2059–2064. doi: 10.1073/pnas.0812859106
Pronin, A., Levay, K., Velmeshev, D., Faghihi, M., Shestopalov, V. I., and Slepak, V. Z. (2014). Expression of olfactory signaling genes in the eye. PLoS One 9:e96435. doi: 10.1371/journal.pone.0096435
Pyrski, M., Koo, J. H., Polumuri, S. K., Ruknudin, A. M., Margolis, J. W., Schulze, D. H., et al. (2007). Sodium/calcium exchanger expression in the mouse and rat olfactory systems. J. Comp. Neurol. 501, 944–958. doi: 10.1002/cne.21290
Quignon, P., Giraud, M., Rimbault, M., Lavigne, P., Tacher, S., Morin, E., et al. (2005). The dog and rat olfactory receptor repertoires. Genome Biol. 6:R83. doi: 10.1186/gb-2005-6-10-r83
Quintela, T., Gonçalves, I., Carreto, L. C., Santos, M. A., Marcelino, H., Patriarca, F. M., et al. (2013). Analysis of the effects of sex hormone background on the rat choroid plexus transcriptome by cDNA microarrays. PLoS One 8:e60199. doi: 10.1371/journal.pone.0060199
Rajkumar, P., Aisenberg, W. H., Acres, O. W., Protzko, R. J., and Pluznick, J. L. (2014). Identification and characterization of novel renal sensory receptors. PLoS One 9:e111053. doi: 10.1371/journal.pone.0111053
Raskin, K., de Gendt, K., Duittoz, A., Liere, P., Verhoeven, G., Tronche, F., et al. (2009). Conditional inactivation of androgen receptor gene in the nervous system: effects on male behavioral and neuroendocrine responses. J. Neurosci. 29, 4461–4470. doi: 10.1523/JNEUROSCI.0296-09.2009
Rassart, E., Bedirian, A., Do Carmo, S., Guinard, O., Sirois, J., Terrisse, L., et al. (2000). Apolipoprotein D. Biochim. Biophys. Acta 1482, 185–198. doi: 10.1016/S0167-4838(00)00162-X
Redl, B. (2000). Human tear lipocalin. Biochim. Biophys. Acta 1482, 241–248. doi: 10.1016/S0167-4838(00)00142-4
Reimann, F., Tolhurst, G., and Gribble, F. M. (2012). G-protein-coupled receptors in intestinal chemosensation. Cell Metab. 15, 421–431. doi: 10.1016/j.cmet.2011.12.019
Ren, X., Zhou, L., Terwilliger, R., Newton, S. S., and de Araujo, I. E. (2009). Sweet taste signalling functions as a hypothalamic glucose sensor. Front. Integr. Neurosci. 3:12. doi: 10.3389/neuro.07.012.2009
Revesz, T., Ckark, H. B., Holton, J. L., Houlden, H. H., Ince, P., and Halliday, G. M. (2015). “Extrapyramidal diseases of movement,” in Greenfield’s Neuropathology, (Vol. 1) eds S. Love, H. Budka, J. W. Ironside and A. Perry (Boca Raton, London, New York: CRC Press, Taylor and Francis Group), 740–798.
Riveron, J., Boto, T., and Alcorta, E. (2013). Transcriptional basis of the acclimation to high environmental temperature at the olfactory receptor organs of Drosophila melanogaster. BMC Genomics 14:259. doi: 10.1186/1471-2164-14-259
Rizvanovic, A., Amundin, M., and Laska, M. (2013). Olfactory discrimination ability of Asian elephants (Elephas maximus) for structurally-related odorants. Chem. Senses 38, 107–118. doi: 10.1093/chemse/bjs097
Robertson, D. H. L., Beynon, R. J., and Evershed, R. P. (1993). Extraction, characterization and binding analysis of two pheromonally active ligands associated with major urinary protein of the house mouse (Mus musculus). J. Chem. Ecol. 19, 1405–1416. doi: 10.1007/BF00984885
Rouquier, S., Blancher, A., and Giorgi, D. (2000). The olfactory receptor gene repertoire in primates and mouse: evidence for reduction of the functional fraction in primates. Proc. Natl. Acad. Sci. U S A 97, 2870–2874. doi: 10.1073/pnas.040580197
Rouquier, S., and Giorgi, D. (2007). Olfactory receptor gene repertoires in mammals. Mutat. Res. 616, 95–102. doi: 10.1016/j.mrfmmm.2006.11.012
Rouquier, S., Taviaux, S., Trask, B. J., Brand-Arpon, V., van den Engh, G., Demaille, J., et al. (1998). Distribution of olfactory receptor genes in the human genome. Nat. Genet. 18, 243–250. doi: 10.1038/ng0398-243
Rozengurt, E. (2006). Taste receptors in the gastrointestinal tract. I. Bitter taste receptors and α-gustducin in the mammalian gut. Am. J. Physiol. Gastrointest. Liver Physiol. 291, G171–G177. doi: 10.1152/ajpgi.00073.2006
Rozengurt, N., Wu, S. V., Chen, M. C., Huang, C., Sternini, C., and Rozengurt, E. (2006). Colocalization of the α-subunit of gustducin with PYY and GLP-1 in L cells of human colon. Am. J. Physiol. Gastrointest. Liver Physiol. 291, G792–G802. doi: 10.1152/ajpgi.00074.2006
Ruiz-DeDiego, I., Naranjo, J. R., Hervé, D., and Moratalla, R. (2015). Dopaminergic regulation of olfactory G-protein subunit expression in the striatum. Mov. Disord. 30, 1039–1049. doi: 10.1002/mds.26197
Ryan, M. F. (2002). Insect Chemoreception: Fundamental and Applied. Boston, MA: Kluver Academic Publishers.
Saidu, S. P., Weeraratne, S. D., Valentine, M., Delay, R., and Van Houten, J. L. (2009). Role of plasma membrane calcium ATPases in calcium clearance from olfactory sensory neurons. Chem. Senses 34, 349–358. doi: 10.1093/chemse/bjp008
Saito, H., Chi, Q., Zhuang, H., and Matsunami, H. (2006). The functional properties of mammalian odorant receptors. Chem. Senses 31:A65.
Saito, H., Kubota, M., Roberts, R. W., Chi, Q., and Matsunami, H. (2004). RTP family members induce functional expression of mammalian odorant receptors. Cell 119, 679–691. doi: 10.1016/j.cell.2004.11.021
Sanz, G., Schlegel, C., Pernollet, J. C., and Briand, L. (2005). Comparison of odorant specificity of two human olfactory receptors from different phylogenetic classes and evidence for antagonism. Chem. Senses 30, 69–80. doi: 10.1093/chemse/bji002
Scaloni, A., Paolini, S., Brandazza, A., Fantacci, M., Bottiglieri, C., Marchese, S., et al. (2011). Purification, cloning and characterisation of odorant- and pheromone-binding proteins from pig nasal epithelium. Cell Mol. Life Sci. 58, 823–834. doi: 10.1007/pl00000903
Schmiedeberg, K., Shirokova, E., Weber, H. P., Schilling, B., Meyerhof, W., and Krautwurst, D. (2007). Structural determinants of odorant recognition by the human olfactory receptors OR1A1 and OR1A2. J. Struct. Biol. 159, 400–412. doi: 10.1016/j.jsb.2007.04.013
Schneider, S. A., and Obeso, J. A. (2015). Clinical and pathological features of Parkinson’s disease. Curr. Top. Behav. Neurosci. 22, 205–220. doi: 10.1007/7854_2014_317
Serrano-Pozo, A., Frosch, M. P., Masliah, E., and Hyman, B. T. (2011). Neuropathological alterations in Alzheimer’s disease. Cold Spring Harb. Perspect. Med. 1:a006189. doi: 10.1101/cshperspect.a006189
Shaw, C. A., Li, Y., Wisniewska, J., Chasse, S., Zaidi, S. N. Y., Jin, W., et al. (2011). Olfactory copy number association with age at onset of Alzheimer disease. Neurology 76, 1302–1309. doi: 10.1212/WNL.0b013e318221c187
Shirazi-Beechey, S. P., Daly, K., Al-Rammahi, M., Moran, A. W., and Bravo, D. (2014). Role of nutrient-sensing taste 1 receptor (T1R) family members in gastrointestinal chemosensing. Br. J. Nutr. 111, S8–S15. doi: 10.1017/s0007114513002286
Shirokova, E., Schmiedeberg, K., Bedner, P., Niessen, H., Willecke, K., Raguse, J. D., et al. (2005). Identification of specific ligands for orphan olfactory receptors. J. Biol. Chem. 280, 11807–11815. doi: 10.1074/jbc.m411508200
Singer, A. G., Macrides, F., Clancy, A. N., and Agosta, W. C. (1986). Purification and analysis of a proteinaceous aphrodisiac pheromone from hamster vaginal discharge. J. Biol. Chem. 28, 13323–13326.
Singh, N., Vrontakis, M., Parkinson, F., and Chelikani, P. (2011). Functional bitter taste receptors are expressed in brain cells. Biochem. Biophys. Res. Commun. 406, 146–151. doi: 10.1016/j.bbrc.2011.02.016
Sklar, P. B., Anholt, R. R., and Snyder, S. H. (1986). The odorant-sensitive adenylate cyclase of olfactory receptor cells. J. Biol. Chem. 261, 15538–15543.
Spehr, M., Gisselmann, G., Poplawski, A., Riffell, J. A., Wetzel, C. H., Zimmer, R. K., et al. (2003). Identification of testicular odorant receptor mediating human sperm chemotaxis. Science 299, 2054–2058. doi: 10.1126/science.1080376
Stephan, A. B., Shum, E. Y., Hirsh, S., Cygnar, K. D., Reisert, J., and Zhao, H. (2009). ANO2 is the cilial calcium-activated chloride channel that may mediate olfactory amplification. Proc. Natl. Acad. Sci. U S A 106, 11776–11781. doi: 10.1073/pnas.0903304106
Stephan, A. B., Tobochnik, S., Dibattista, M., Wall, C. M., Reisert, J., and Zhao, H. (2011). The Na+/Ca2+ exchanger NCKX4 governs termination and adaptation of the mammalian olfactory response. Nat. Neurosci. 15, 131–137. doi: 10.1038/nn.2943
Sternini, C., Anselmi, L., and Rozengurt, E. (2008). Enteroendocrine cells: a site of ‘taste’ in gastrointestinal chemosensing. Curr. Opin. Endocrinol. Diabetes Obes. 15, 73–78. doi: 10.1097/MED.0b013e3282f43a73
Stopková, R., Dudková, B., Hájková, P., and Stopka, P. (2014). Complementary roles of mouse lipocalins in chemical communication and immunity. Biochem. Soc. Trans. 42, 893–898. doi: 10.1042/BST20140053
Stowers, L., Holy, T. E., Meister, M., Dulac, C., and Koentges, G. (2002). Loss of sex discrimination and male-male aggression in mice deficient for TRP2. Science 295, 1493–1500. doi: 10.1126/science.1069259
Stowers, L., and Logan, D. W. (2010). Sexual dimorphism in olfactory signalling. Curr. Opin. Neurobiol. 20, 770–775. doi: 10.1016/j.conb.2010.08.015
Swartz, T. D., Duca, F. A., de Wouters, T., Sakar, Y., and Covasa, M. (2012). Up-regulation of intestinal type 1 taste receptor 3 and sodium glucose luminal transporter-1 expression and increased sucrose intake in mice lacking gut microbiota. Br. J. Nutr. 107, 621–630. doi: 10.1017/S0007114511003412
Syme, C. A., Zhang, L., and Bisello, A. (2006). Caveolin-1 regulates cellular trafficking and function of the glucagon-like peptide 1 receptor. Mol. Endocrinol. 20, 3400–3411. doi: 10.1210/me.2006-0178
Taoka, H., Hamamura, T., Endo, S., Miki, M., Lee, Y., Miyata, S., et al. (2006). Golf protein levels in rat striatum are increased by chronic antidepressant administration and decreased by olfactory bulbectomy. Life Sci. 79, 462–468. doi: 10.1016/j.lfs.2006.01.033
Thal, D. R., Rüb, U., Orantes, M., and Braak, H. (2002). Phases of β-amyloid deposition in the human brain and its relevance for the development of AD. Neurology 58, 1791–1800. doi: 10.1212/wnl.58.12.1791
Tukey, R. H., and Strassburg, C. P. (2000). Human UDP-glucuronosyltransferases: metabolism, expression and disease. Annu. Rev. Pharmacol. Toxicol. 40, 581–616. doi: 10.1146/annurev.pharmtox.40.1.581
van Avesaat, M., Troost, F. J., Ripken, D., Peters, J., Hendriks, H. F., and Masclee, A. A. (2015). Intraduodenal infusion of a combination of tastants decreases food intake in humans. Am. J. Clin. Nutr. 102, 729–735. doi: 10.3945/ajcn.115.113266
Vanderhaeghen, P., Schurmans, S., Vassart, G., and Parmentier, M. (1997). Molecular cloning and chromosomal mapping of olfactory receptor genes expressed in the male germ line: evidence for their wide distribution in the human genome. Biochem. Biophys. Res. Commun. 237, 283–287. doi: 10.1006/bbrc.1997.7043
Vassalli, A., Rothman, A., Feinstein, P., Zapotocky, M., and Mombaerts, P. (2002). Minigenes impart odorant receptor-specific axon guidance in the olfactory bulb. Neuron 35, 681–696. doi: 10.1016/s0896-6273(02)00793-6
Vegezzi, G., Anselmi, L., Huynh, J., Barocelli, E., Rozengurt, E., Raybould, H., et al. (2014). Diet-induced regulation of bitter taste receptor subtypes in the mouse gastrointestinal tract. PLoS One 9:e107732. doi: 10.1371/journal.pone.0107732
Veitinger, T., Riffell, J. R., Veitinger, S., Nascimento, J. M., Triller, A., Chandsawangbhuwana, C., et al. (2011). Chemosensory Ca2+ dynamics correlate with diverse behavioral phenotypes in human sperm. J. Biol. Chem. 286, 17311–17325. doi: 10.1074/jbc.m110.211524
Vemula, S. R., Puschmann, A., Xiao, J., Zhao, Y., Rudzińska, M., Frei, K. P., et al. (2013). Role of Gα(olf) in familial and sporadic adult-onset primary dystonia. Hum. Mol. Genet. 22, 2510–2519. doi: 10.1093/hmg/ddt102
Vogt, R. G., and Riddiford, L. M. (1981). Pheromone binding and inactivation by moth antennae. Nature 293, 161–163. doi: 10.1038/293161a0
Vogt, R. G., Riddiford, L. M., and Pretswich, G. D. (1985). Kinetic properties of a sex pherpomone-degrading enzyme: the sensillar esterase of Anthereacea polyphemus. Proc. Natl. Acad. Sci. U S A 82, 8827–8831. doi: 10.1073/pnas.82.24.8827
Vogt, R. G., Rybczynski, R., and Lerner, M. R. (1991). Molecular cloning and sequencing of general odorant-binding proteins GOBP1 and GOBP2 from the tobacco hawk moth Manduca sexta: comparison with other insect OBPs and their signal peptides. J. Neurosci. 11, 2972–2984.
Voigt, A., Bojahr, J., Narukawa, M., Hübner, S., Boehm, U., and Meyerhof, W. (2015). Trans-synaptic tracing from taste receptor cells reveals local taste receptor gene expression in gustatory ganglia and brain. J. Neurosci. 35, 9717–9729. doi: 10.1523/JNEUROSCI.0381-15.2015
Von Dannecker, L., Mercadante, A., and Malnic, B. (2005). Ric-8B, an olfactory putative GTP exchange factor, amplifies signal transduction through the olfactory-specific G-protein Gαolf. J. Neurosci. 25, 3793–3800. doi: 10.1523/jneurosci.4595-04.2005
Von Dannecker, L. E., Mercadante, A. F., and Malnic, B. (2006). Ric-8B promotes functional expression of odorant receptors. Proc. Natl. Acad. Sci. U S A 103, 9310–9314. doi: 10.1073/pnas.0600697103
Walensky, L. D., Roskams, A. J., Lefkowitz, R. J., Snyder, S. H., and Ronnett, G. V. (1995). Odorant receptors and desensitization proteins co-localize in mammalian sperm. Mol. Med. 1, 130–141.
Walensky, L. D., Ruat, M., Bakin, R. E., Blackshaw, S., Ronnett, G. V., and Snyder, S. H. (1998). Two novel odorant receptor families expressed in spermatids undergo 5’-splicing. J. Biol. Chem. 273, 9378–9387. doi: 10.1074/jbc.273.16.9378
Ward, A., Hong, W., Favaloro, V., and Luo, L. (2015). Toll receptors instruct axon and dendrite targeting and participate in synaptic partner matching in a Drosophila olfactory circuit. Neuron 85, 1013–1028. doi: 10.1016/j.neuron.2015.02.003
Weber, M., Pehl, U., Breer, H., and Strotmann, J. (2002). Olfactory receptor expressed in ganglia of the autonomic nervous system. J. Neurosci. Res. 68, 176–184. doi: 10.1002/jnr.10164
Wetzel, C. H., Oles, M., Wellerdieck, C., Kurczkowiak, M., Gisselmann, G., and Hatt, H. (1999). Specificity and sensitivity of a human olfactory receptor functionally expressed in human embryonic kidney 293 cells and Xenopus laevis oocytes. J. Neurosci. 19, 7426–7433.
Wicher, D. (2015). Olfactory signaling in insects. Prog. Mol. Biol. Transl. Sci. 130, 37–54. doi: 10.1016/bs.pmbts.2014.11.002
Wilson, D. A., and Stevenson, R. S. (2006). Learning to Smell: Olfactory Perception from Neurobiology to Behaviour. Baltimore, MD: Johns Hopkins University Press.
Wolf, S., and Grünewald, S. (2015). Sequence, structure and ligand binding evolution of rhodopsin-like G protein-coupled receptors: a crystal structure-based phylogenetic analysis. PLoS One 10:e0123533. doi: 10.1371/journal.pone.0123533
Wong, G. T., Gannon, K. S., and Margolskee, R. F. (1996). Transduction of bitter and sweet taste by gustducin. Nature 381, 796–800. doi: 10.1038/381796a0
Wong, S. T., Trinh, K., Hacker, B., Chan, G. C., Lowe, G., Gaggar, A., et al. (2000). Disruption of the type III adenylyl cyclase gene leads to peripheral and behavioral anosmia in transgenic mice. Neuron 27, 487–497. doi: 10.1016/s0896-6273(00)00060-x
Worley, P. F., Zeng, W., Huang, G., Kim, J. Y., Shin, D. M., Kim, M. S., et al. (2007). Homer proteins in Ca2+ signaling by excitable and non-excitable cells. Cell Calcium 42, 363–371. doi: 10.1016/j.ceca.2007.05.007
Wu, L., Du, Y., Lok, J., Lo, E. H., and Xing, C. (2015). Lipocalin-2 enhances angiogenesis in rat brain endothelial cells via reactive oxygen species and iron-dependent mechanisms. J. Neurochem. 132, 622–628. doi: 10.1111/jnc.13023
Wu, M. V., Manoli, D. S., Fraser, E. J., Coats, J. K., Tollkuhn, J., Honda, S., et al. (2009). Estrogen masculinizes neural pathways and sex-specific behaviors. Cell 139, 61–72. doi: 10.1016/j.cell.2009.07.036
Wu, S. V., Rozengurt, N., Yang, M., Young, S. H., Sinnett-Smith, J., and Rozengurt, E. (2002). Expression of bitter taste receptors of the T2R family in the gastrointestinal tract and enteroendocrine STC-1 cells. Proc. Natl. Acad. Sci. U S A 99, 2392–2397. doi: 10.1073/pnas.042617699
Xia, C., Ma, W., Wang, F., Hua, S. B., and Liu, M. (2001). Identification of a prostate-specific G-protein coupled receptor in prostate cancer. Oncogene 20, 5903–5907. doi: 10.1038/sj.onc.1204803
Xing, C., Wang, X., Cheng, C., Montaner, J., Mandeville, E., Leung, W., et al. (2014). Neuronal production of lipocalin-2 as a help-me signal for glial activation. Stroke 45, 2085–2092. doi: 10.1161/STROKEAHA.114.005733
Xu, L. L., Stackhouse, B. G., Florence, K., Zhang, W., Shanmugam, N., Sesterhenn, I. A., et al. (2000). PSGR, a novel prostate-specific gene with homology to a G protein-coupled receptor, is overexpressed in prostate cancer. Cancer Res. 60, 6568–6572.
Yoder, A. D., and Larsen, P. A. (2014). The molecular evolutionary dynamics of the vomeronasal receptor (class 1) genes in primates: a gene family on the verge of a functional breakdown. Front. Neuroanat. 8:153. doi: 10.3389/fnana.2014.00153
Young, J. M., Friedman, C., Williams, E. M., Ross, J. A., Tonnes-Priddy, L., and Trask, B. J. (2002). Different evolutionary processes shaped the mouse and human olfactory receptor gene families. Hum. Mol. Genet. 11, 535–546. doi: 10.1093/hmg/11.5.535
Young, J. M., Shykind, B. M., Lane, R. P., Tonnes-Priddy, L., Ross, E. M., Walker, M., et al. (2003). Odorant receptor expressed sequence tags demonstrate olfactory expression of over 400 genes, extensive alternate splicing and unequal expression levels. Genome Biol. 4:R71. doi: 10.1186/gb-2003-4-11-r71
Yuan, T. T., Toy, P., McClary, J. A., Lin, R. J., Miyamoto, N. G., and Kretschmer, P. J. (2001). Cloning and genetic characterization of an evolutionarily conserved human olfactory receptor that is differentially expressed across species. Gene 278, 41–51. doi: 10.1016/s0378-1119(01)00709-0
Zerr, I., Kallenberg, K., Summers, D. M., Romero, C., Taratuto, A., Heinemann, U., et al. (2009). Updated clinical diagnostic criteria for sporadic Creutzfeldt-Jakob disease. Brain 132, 2659–2668. doi: 10.1093/brain/awp191
Zhang, X., Bedigian, A. V., Wang, W., and Eggert, U. S. (2012). G protein-coupled receptors participate in cytokinesis. Cytoskeleton (Hoboken) 69, 810–818. doi: 10.1002/cm.21055
Zhang, X., De la Cruz, O., Pinto, J. M., Nicolae, D., Firestein, S., and Gilad, Y. (2007). Characterizing the expression of the human olfactory receptor gene family using a novel DNA microarray. Genome Biol. 8:R86. doi: 10.1186/gb-2007-8-5-r86
Zhang, X., and Firestein, S. (2002). The olfactory receptor gene superfamily of the mouse. Nat. Neurosci. 5, 124–133.
Zhang, X., Rogers, M., Tian, H., Zhang, X., Zou, D. J., Liu, J., et al. (2004). High-throughput microarray detection of olfactory receptor gene expression in the mouse. Proc. Natl. Acad. Sci. U S A 101, 14168–14173. doi: 10.1073/pnas.0405350101
Zhao, W., Ho, L., Varghese, M., Yemul, S., Dams-O’Connor, K., Gordon, W., et al. (2013). Decreased level of olfactory receptors in blood cells following traumatic brain injury and potential association with tauopathy. J. Alzheimers Dis. 34, 417–429. doi: 10.3233/JAD-121894
Keywords: olfactory receptors, taste receptors, brain, Alzheimer, Parkinson, progressive supranuclear palsy, Creutzfeldt-Jakob disease, schizophrenia
Citation: Ferrer I, Garcia-Esparcia P, Carmona M, Carro E, Aronica E, Kovacs GG, Grison A and Gustincich S (2016) Olfactory Receptors in Non-Chemosensory Organs: The Nervous System in Health and Disease. Front. Aging Neurosci. 8:163. doi: 10.3389/fnagi.2016.00163
Received: 23 April 2016; Accepted: 21 June 2016;
Published: 05 July 2016.
Edited by:
Filippo Tempia, University of Turin, ItalyReviewed by:
Michael Lardelli, University of Adelaide, AustraliaCopyright © 2016 Ferrer, Garcia-Esparcia, Carmona, Carro, Aronica, Kovacs, Grison and Gustincich. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution and reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Isidro Ferrer, ODA4MmlmYUBnbWFpaWwuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.