- 1Julius Center for Health Sciences and Primary Care, University Medical Center, Utrecht, Netherlands
- 2Department of Neurology, Brain Center Rudolf Magnus, University Medical Center, Utrecht, Netherlands
In the last decade, the in vivo assessment of hippocampal subfields has received increasing attention because of the differential role of hippocampal subfields in several neuropsychiatric diseases (Geuze et al., 2005). Several manual segmentation protocols have been developed for 3–7 T MRI (Mueller et al., 2007; Van Leemput et al., 2008; La Joie et al., 2010; Wisse et al., 2012), some of which are automated (Van Leemput et al., 2008; Yushkevich et al., 2009). One of these automated protocols (Van Leemput et al., 2008, 2009) has recently been implemented in FreeSurfer (Fischl, 2012), a freely available easy-to-use set of automated brain MRI analysis tools. This has made hippocampal subfield segmentation available to everyone with 1.5–3 T MRI data and the method is being used in an increasing number of studies (Teicher et al., 2012; Li et al., 2013; Pereira et al., 2014).
In this commentary, we express our concern with the hippocampal subfield segmentation package in FreeSurfer. In particular, we address issues concerning (1) image acquisition, (2) the parcelation scheme, and (3) validation of this automated segmentation.
The first concern with the hippocampal subfield segmentation package in FreeSurfer is that it requires low resolution (1 mm3) T1 images (whole-brain). Most other manual or automated segmentation methods are developed for high-resolution T2 images (in-plane: 0.20–0.70 mm2, often with partial-brain coverage) (Mueller et al., 2007; Kerchner et al., 2010; La Joie et al., 2010; Wisse et al., 2012). On high-resolution T2 images, contrast between white and gray matter is sufficient to visualize the white matter bands between the dentate gyrus and the cornu ammonis (CA) that are generally used as a boundary between these subfields. The low resolution T1 images on which the FreeSurfer segmentation is applied do not contain this amount of detail. See Figure 1 for a comparison of low resolution T1 and high-resolution T2 images.
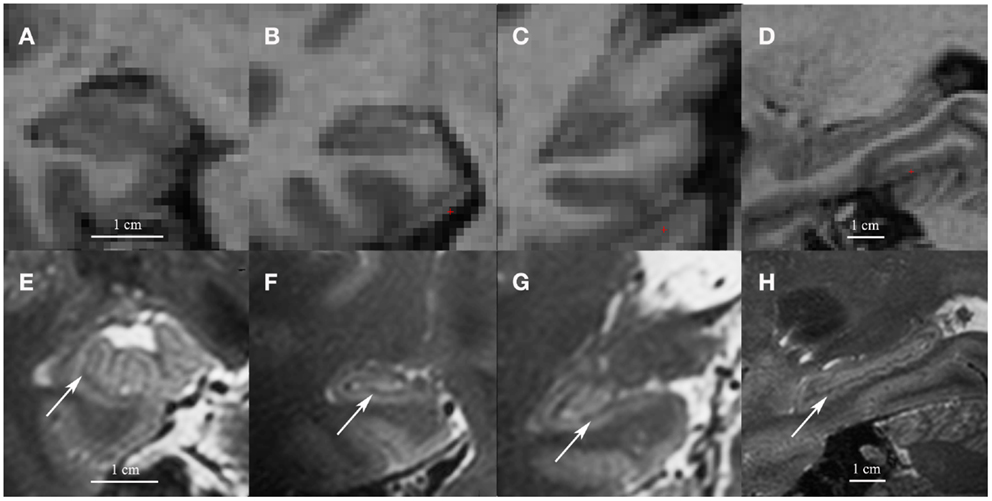
Figure 1. Coronal images of the head (A), body (B) and tail (C) and a sagittal cross-section of the hippocampus (D) on low resolution 1mm3 T1 1.5 T images (A–D) and on high resolution 0.7 mm3 T2 7 T images (E–H). Note the white matter bands between the dentate gyrus and cornu ammonis on the high resolution T2 images (indicated by arrows). Although we show high resolution 7 T T2 images here, the white matter bands between the dentate gyrus and the cornu ammonis can also be visualized on high resolution T2 3–4 T images (Mueller et al., 2007; La Joie et al., 2010; Winterburn et al., 2013).
The second concern is the parcelation scheme used for the FreeSurfer segmentation, which is based on the subfield distribution in one coronal section in the body of the hippocampus (Van Leemput et al., 2008, 2009) and then used to segment subfields along the complete long axis of the hippocampus. However, the presence and position of the subfields differ along the long axis (Duvernoy et al., 2005; Mai et al., 2008; Insausti and Amaral, 2012). Consequently, the locations of the boundaries between subfields in this segmentation protocol are in mismatch with the anatomical atlases in a large part of the long axis. For example, in FreeSurfer, the dentate gyrus is segmented from the anterior pole of the hippocampus, while it only becomes visible 6 mm after the anterior pole of the hippocampus (Insausti and Amaral, 2012). Several segmentation methods exist also for T2 images, manual (La Joie et al., 2010; Wisse et al., 2012) as well as automated (Yushkevich et al., 2009). Because of the complex anatomy of the hippocampal head and tail, these methods either limit the segmentation of subfields to the hippocampal body (Mueller et al., 2007; Yushkevich et al., 2009) or developed a separate segmentation scheme for the head and/or tail (La Joie et al., 2010; Wisse et al., 2012; Winterburn et al., 2013).
As a consequence of the placement of the subfield boundaries in FreeSurfer, large parts of subfields are assigned to neighboring subfields. For example, large parts of CA1 are included in the subiculum and CA2&3. This generates volume estimates that are in contrast with anatomical studies. In studies using the FreeSurfer segmentation package (e.g., Teicher et al., 2012; Boen et al., 2014), CA2&3 is the largest subfield, while CA1 is the smallest. According to anatomical studies, CA1 is the largest and CA2&3 is the smallest subfield (Simic et al., 1997; Rossler et al., 2002). In general, subfield boundaries are difficult to discern in vivo and part of subfields are counted toward neighboring subfields in all segmentation protocols. However, other manual or automated methods generate subfield estimates that are more in line with those of anatomical studies (e.g., Wisse et al., 2012; Winterburn et al., 2013). See Table S1 in Supplementary Material for a comparison of subfield volumes and their percentage distribution within the hippocampus according to several segmentation protocols.
Studies using this FreeSurfer segmentation package to investigate hippocampal subfield volumes in mild cognitive impairment (MCI) and Alzheimer disease (AD) reported results that differ from anatomical studies. Several studies using the FreeSurfer package reported that MCI and AD were mainly related to CA2&3 atrophy (Hanseeuw et al., 2011; Lim et al., 2012). These latter results stand in contrast to the anatomical studies that reported the greatest atrophy in CA1 (Simic et al., 1997; Rossler et al., 2002). Perhaps, CA2&3 atrophy in MCI or AD in studies using FreeSurfer actually represents CA1 atrophy, as a large part of CA1 is counted toward CA2&3 in FreeSurfer. Studies using other manual or automated segmentation methods reported subfield atrophy in AD that more closely matched the results of anatomical studies (Mueller and Weiner, 2009; Pluta et al., 2012; La Joie et al., 2013).
A third concern is that the automated segmentation in FreeSurfer was developed on high-resolution (0.19 mm × 0.19 mm × 0.80 mm) 3 T images and is now applied on low resolution (1 mm3) images. To the best of our knowledge, the protocol was not validated against a manual segmentation on these lower resolutions 1.5–3 T MR images (see also Lim et al., 2012; Pluta et al., 2012). Moreover, it should be noted that the intra-rater reliability of the manual segmentation used for the FreeSurfer package was based on repeated segmentation of two coronal slices rather than on segmentation of the complete long axis of the hippocampus (Van Leemput et al., 2009).
In conclusion, though FreeSurfer provides a useful, broad set of automated brain MRI analysis tools, we have concerns about the current package for automated hippocampal subfield segmentation. The boundaries of the parcelation scheme are in mismatch with known anatomical boundaries. This will impact the reliability of studies using FreeSurfer to investigate subfield atrophy in neuropsychiatric diseases.
Author Contributions
Laura E. M. Wisse: study concept and design, wrote the manuscript, final approval and agreement to be accountable for all aspects of the work. Mirjam I. Geerlings: study concept and design, critical revision of the manuscript for important intellectual content, final approval and agreement to be accountable for all aspects of the work. Geert Jan Biessels: study concept and design, critical revision of the manuscript for important intellectual content, final approval and agreement to be accountable for all aspects of the work.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The research of MIG was supported by a VIDI grant from the Netherlands Organization for Scientific Research (NWO) (grant number 917-66-311). The research of GJB was supported by a VIDI grant (grant number 91711384) from ZonMw, The Netherlands Organization for Health Research and Development and by the Netherlands Heart Foundation (grant number 2010T073).
Supplementary Material
The Supplementary Material for this article can be found online at http://www.frontiersin.org/Journal/10.3389/fnagi.2014.00261/full
Abbreviations
AD, Alzheimer disease; CA, cornu ammonis; MCI, mild cognitive impairment.
References
Boen, E., Westlye, L. T., Elvsashagen, T., Hummelen, B., Hol, P. K., Boye, B., et al. (2014). Smaller stress-sensitive hippocampal subfields in women with borderline personality disorder without posttraumatic stress disorder. J. Psychiatry Neurosci. 39, 127–134. doi: 10.1503/jpn.130070
Duvernoy, H. M., Cattin, E., Naidich, T., Fatterpekar, G. M., Raybaud, C., Risold, P. Y., et al. (2005). The Human Hippocampus. Berlin: Springer Verlag.
Geuze, E., Vermetten, E., and Bremner, J. D. (2005). MR-based in vivo hippocampal volumetrics: 2 findings in neuropsychiatric disorders. Mol. Psychiatry 10, 160–184. doi:10.1038/sj.mp.4001580
Hanseeuw, B. J., Van, L. K., Kavec, M., Grandin, C., Seron, X., and Ivanoiu, A. (2011). Mild cognitive impairment: differential atrophy in the hippocampal subfields. AJNR Am. J. Neuroradiol. 32, 1658–1661. doi:10.3174/ajnr.A2589
Insausti, R., and Amaral, D. G. (2012). “Hippocampal formation,” in The Human Nervous System, eds J. K. Mai and G. Paxinos (San Diego: Elsevier Academic Press), 896–942.
Kerchner, G. A., Hess, C. P., Hammond-Rosenbluth, K. E., Xu, D., Rabinovici, G. D., Kelley, D. A., et al. (2010). Hippocampal CA1 apical neuropil atrophy in mild Alzheimer disease visualized with 7-T MRI. Neurology 75, 1381–1387. doi:10.1212/WNL.0b013e3181f736a1
La Joie, R., Fouquet, M., Mezenge, F., Landeau, B., Villain, N., Mevel, K., et al. (2010). Differential effect of age on hippocampal subfields assessed using a new high-resolution 3T MR sequence. Neuroimage 53, 506–514. doi:10.1016/j.neuroimage.2010.06.024
La Joie, R., Perrotin, A., Sayette, D. L., Egret, S., Doeuvre, L., Belliard, S., et al. (2013). Hippocampal subfield volumetry in mild cognitive impairment, Alzheimer’s disease and semantic dementia. Neuroimage Clin. 3, 155–162. doi:10.1016/j.nicl.2013.08.007
Li, Y. D., Dong, H. B., Xie, G. M., and Zhang, L. J. (2013). Discriminative analysis of mild Alzheimer’s disease and normal aging using volume of hippocampal subfields and hippocampal mean diffusivity: an in vivo magnetic resonance imaging study. Am. J. Alzheimers Dis. Other Demen. 28, 627–633. doi:10.1177/1533317513494452
Lim, H. K., Hong, S. C., Jung, W. S., Ahn, K. J., Won, W. Y., Hahn, C., et al. (2012). Automated segmentation of hippocampal subfields in drug-naive patients with Alzheimer disease. AJNR Am. J. Neuroradiol 34, 747–751. doi:10.3174/ajnr.A3293
Mai, J. K., Paxinos, G., and Voss, T. (2008). Atlas of the Human Brain. New York, NY: Elsevier Academic Press.
Mueller, S. G., Stables, L., Du, A. T., Schuff, N., Truran, D., Cashdollar, N., et al. (2007). Measurement of hippocampal subfields and age-related changes with high resolution MRI at 4T. Neurobiol. Aging 28, 719–726. doi:10.1016/j.neurobiolaging.2006.03.007
Mueller, S. G., and Weiner, M. W. (2009). Selective effect of age, Apo e4, and Alzheimer’s disease on hippocampal subfields. Hippocampus 19, 558–564. doi:10.1002/hipo.20614
Pereira, J. B., Valls-Pedret, C., Ros, E., Palacios, E., Falcon, C., Bargallo, N., et al. (2014). Regional vulnerability of hippocampal subfields to aging measured by structural and diffusion MRI. Hippocampus 24, 403–414. doi:10.1002/hipo.22234
Pluta, J., Yushkevich, P., Das, S., and Wolk, D. (2012). In vivo analysis of hippocampal subfield atrophy in mild cognitive impairment via semi-automatic segmentation of T2-weighted MRI. J. Alzheimers Dis. 31, 85–99. doi:10.3233/JAD-2012-111931
Rossler, M., Zarski, R., Bohl, J., and Ohm, T. G. (2002). Stage-dependent and sector-specific neuronal loss in hippocampus during Alzheimer’s disease. Acta. Neuropathol. 103, 363–369. doi:10.1007/s00401-001-0475-7
Simic, G., Kostovic, I., Winblad, B., and Bogdanovic, N. (1997). Volume and number of neurons of the human hippocampal formation in normal aging and Alzheimer’s disease. J. Comp. Neurol. 379, 482–494. doi:10.1002/(SICI)1096-9861(19970324)379:4<482::AID-CNE2>3.0.CO;2-Z
Teicher, M. H., Anderson, C. M., and Polcari, A. (2012). Childhood maltreatment is associated with reduced volume in the hippocampal subfields CA3, dentate gyrus, and subiculum. Proc. Natl. Acad. Sci. U. S. A. 109, E563–E572. doi:10.1073/pnas.1115396109
Van Leemput, K., Bakkour, A., Benner, T., Wiggins, G., Wald, L. L., Augustinack, J., et al. (2008). Model-based segmentation of hippocampal subfields in ultra-high resolution in vivo MRI. Med. Image Comput. Comput. Assist. Interv. 11, 235–243.
Van Leemput, K., Bakkour, A., Benner, T., Wiggins, G., Wald, L. L., Augustinack, J., et al. (2009). Automated segmentation of hippocampal subfields from ultra-high resolution in vivo MRI. Hippocampus 19, 549–557. doi:10.1002/hipo.20615
Winterburn, J. L., Pruessner, J. C., Chavez, S., Schira, M. M., Lobaugh, N. J., Voineskos, A. N., et al. (2013). A novel in vivo atlas of human hippocampal subfields using high-resolution 3 T magnetic resonance imaging. Neuroimage 74, 254–265. doi:10.1016/j.neuroimage.2013.02.003
Wisse, L. E. M., Gerritsen, L., Zwanenburg, J. J., Kuijf, H. J., Luijten, P. R., Biessels, G. J., et al. (2012). Subfields of the hippocampal formation at 7 T MRI: in vivo volumetric assessment. Neuroimage 61, 1043–1049. doi:10.1016/j.neuroimage.2012.03.023
Keywords: hippocampal subfields, FreeSurfer, automated segmentation, hippocampus, Alzheimer
Citation: Wisse LEM, Biessels GJ and Geerlings MI (2014) A critical appraisal of the hippocampal subfield segmentation package in FreeSurfer. Front. Aging Neurosci. 6:261. doi: 10.3389/fnagi.2014.00261
Received: 30 July 2014; Accepted: 10 September 2014;
Published online: 25 September 2014.
Edited by:
Manuel Menéndez-González, Hospital Álvarez-Buylla, SpainReviewed by:
Nikolai Malykhin, University of Alberta, CanadaCopyright: © 2014 Wisse, Biessels and Geerlings. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence:bS5nZWVybGluZ3NAdW1jdXRyZWNodC5ubA==
 Laura E. M. Wisse
Laura E. M. Wisse Geert Jan Biessels
Geert Jan Biessels Mirjam I. Geerlings
Mirjam I. Geerlings