- 1Biochemistry Department, Faculty of Medicine (for Girls), Al-Azhar University, Cairo, Egypt
- 2Clinical Pathology Department, Faculty of Medicine (for Girls), Al-Azhar University, Cairo, Egypt
- 3Pediatrics Department, Faculty of Medicine (for Girls), Al-Azhar University, Cairo, Egypt
- 4Medical Microbiology and Immunology Department, Faculty of Medicine (for Girls), Al-Azhar University, Cairo, Egypt
- 5Community and Occupational Medicine Department, Faculty of Medicine (for Girls), Al-Azhar University, Cairo, Egypt
- 6Neurology Department, Faculty of Medicine (for Girls), Al-Azhar University, Cairo, Egypt
- 7Radiodiagnosis Department, Faculty of Medicine, Al-Azhar University, Cairo, Egypt
- 8Radiodiagnosis and Intervention Radiology Department, Faculty of Medicine (for Girls), Al-Azhar University, Cairo, Egypt
- 9Department of Internal Medicine, Faculty of Medicine, Assiut University, Asyut, Egypt
- 10Internal Medicine Department, College of Medicine, Princess Nourah Bint Abdulrahman University, Riyadh, Saudi Arabia
- 11Department of Basic Medical Sciences, College of Medicine, Princess Nourah Bint Abdulrahman University, Riyadh, Saudi Arabia
- 12Department of Microbiology and Immunology, Faculty of Pharmacy, Sphinx University, New Assiut, Egypt
- 13Department of Pharmacology, Faculty of Medicine, Assiut University, Asyut, Egypt
- 14Clinical Pathology Department, Faculty of Medicine, Ain Shams University, Cairo, Egypt
- 15Medical Biochemistry and Molecular Biology Department, Faculty of Medicine, Assiut University, Asyut, Egypt
- 16Department of Medical Biochemistry and Molecular Biology, Armed Forces College of Medicine (AFCM), Cairo, Egypt
Background: Globally, approximately 70 million people suffer from epilepsy. Infants constitute a significant percentage of these cases. Hence, there is a significant need for better understanding of the pathophysiology of epilepsy through laboratory and radiological methods for early detection and optimized management. Interleukin enhancer binding factor 3 antisense RNA l (ILF3AS1) is a long non-coding RNA (lncRNA) that enhances the expressions of matrix metalloproteinase 3 (MMP3) and matrix metalloproteinase 9 (MMP9), which are considered to be epileptogenic.
Aim: We aimed to assess the serum expressions of the lncRNAs ILF3AS1, MMP3, and MMP9 along with microRNA-212 (miRNA-212) as predictive biomarkers in children with epilepsy; we also assessed their correlations with magnetic resonance imaging (MRI) findings.
Subjects and Methods: Fifty children with epilepsy and fifty healthy controls were considered in this study. Serum expressions of the lncRNA ILF3AS1 and miRNA-212 were estimated by quantitative real-time polymerase chain reaction (qPCR). Serum concentrations of MMP3 and MMP9 were estimated by enzyme-linked immunosorbent assay (ELISA) in parallel with MRI findings and different baseline biochemical parameters of all the subjects.
Results: The results showed significantly higher levels of lncRNAs ILF3AS1, MMP3, and MMP9 as well as lower levels of miRNA-212 in children with epilepsy compared to the controls. The fold-change of miRNA-212 was a significant negative predictor (odds ratio = 0.153, p = 0.000). The receiver operating characteristic curves (Roc) showed that the areas under the curves for MMP3, MMP9, and lncRNA ILF3AS1 as well as the fold-change for miRNA-212 were 0.659, 0.738, 0.656, and 0.965, respectively. Brain lesions were detected in 15 patients (30%) with epilepsy, whereas the remaining 35 patients (70%) had normal results.
Conclusion: Serum levels of the lncRNA ILF3AS1 among children with epilepsy were higher than those in the control group and were associated with upregulation of both MMP3 and MMP9 as well as downregulation of miRNA-212 expressions, suggesting their predictive utility in monitoring the development of epilepsy; this also means that a treatment plan focusing on the ILF3AS1/miRNA-212/MMP3/MMP9 axis could be an effective strategy for treating epilepsy.
Introduction
Epilepsy accounts for a sizable percentage of the neurological disorders reported around the world (Zeng et al., 2022). Every year, between 50 and 70 million people are affected globally, and 2.4 million new people are diagnosed with epilepsy. Approximately 80% of all epilepsy cases are reported from developing countries, yet only three-quarters of those affected receive the necessary treatment (Cava et al., 2018). The risk of epilepsy is greater in younger and older age groups (Thijs et al., 2019), with recurrent seizures being a hallmark of pediatric epilepsy (Banawalikar et al., 2021).
The clinical techniques used to diagnose epilepsy at present include clinical manifestations, imaging examinations, and electroencephalography (EEG) (Fernandez-Baca Vaca and Park, 2020). Neuroimaging is important for establishing the etiology, providing prognosis, and planning appropriate care. The most effective imaging technique for identifying underlying epileptogenic foci and various causes is magnetic resonance imaging (MRI) (Vattipally and Bronen, 2004; Bernasconi et al., 2011; Wilmshurst et al., 2015).
There are multiple theories explaining the etiology of epilepsy, and one of the primary processes in epilepsy is neuroinflammation, which is an inflammatory response to epileptic convulsions (Zeng et al., 2022). Inflammation is both a cause and a result of temporal lobe epilepsy (TLE) (Wu et al., 2017). However, genetic background also plays a significant role in addition to inflammation (Liu et al., 2022).
Non-coding RNAs (ncRNAs) are not translated into functional proteins; they alter gene translation and interfere with the signaling pathways (Zeng et al., 2022). In addition, they serve as epigenetic factors that both participate in and are dysregulated during epileptogenesis. Abnormal ncRNA expressions have been observed in patients with and animal models of epilepsy. Thus, they may be used as biomarkers for the diagnosis of epilepsy and treatment response prognosis (Manna et al., 2022).
Non-coding RNAs exceeding 200 nucleotides in length are referred to as long non-coding RNAs (lncRNAs) that are thought to contribute to inflammation. LncRNAs serve as competing endogenous RNAs (ceRNAs) that negatively regulate microRNA (miRNA) expressions (Lekka and Hall, 2018) in the form of small ncRNAs that are 20–23 nucleotides in length and regulate gene expression (Manna et al., 2022). A newly discovered lncRNA, ILF3AS1, has been reported to be associated with dysregulated expressions in many types of cancers and epilepsy (Cai et al., 2020).
Several miRNAs are specifically expressed in the brain. Therefore, the circulating brain-enriched miRNAs are valuable sources for diagnosing pathological brain development. Many brain pathologies associated with epileptogenesis are thought to be caused by dysregulated miRNAs, including synaptic plasticity and neuroinflammation (Heiskanen et al., 2023).
Alterations in miRNA expressions may be involved in the pathogenesis of epilepsy by regulating the expressions of inflammatory factors, such as IL-1, INF-α, and tumor necrosis factor alpha (TNF-α). Of note, increased expressions of the reactive astrocytes were observed in the hippocampal sclerosis specimens of TLE patients (Wang and Zhao, 2021).
In the central nervous system (CNS), miRNA-212 has a potent modulatory effect (Brennan and Henshall, 2020); it has been linked to neurological disorders (Herrera-Espejo et al., 2019), and epilepsy-inducing circumstances, such as status epilepticus, rapidly stimulate its expression (Brennan and Henshall, 2020).
As TLE is caused by inflammation, several inflammatory agents have been detected in the brain tissues of patients suffering from refractory epilepsy. Accordingly, data suggest that increased production or activity of matrix metalloproteinase (MMP), an inflammatory mediator, after an insult may influence epileptogenesis (Cai et al., 2020).
Matrix metalloproteinase 9 (MMP9) acts on the inflammatory processes at various levels; it facilitates leukocyte influx via the blood–brain barrier (BBB) and releases chemokines from the extracellular matrix (ECM) to produce a chemotactic signal and stimulate TNF-α (Van Lint and Libert, 2007; Rivera et al., 2010; De Bock et al., 2015).
MMP3 and MMP9 expressions are known to be promoted by ILF3AS1 overexpression, leading to the hypothesis that MMP inhibitors could be reliable treatments for epilepsy. However, the currently used MMP inhibitors are non-specific, with many negative side effects. MMPs themselves may not be suitable for focused treatment as molecules upstream of the MMPs, such as ILF3AS1 (Cai et al., 2020).
Multiple studies have explored the associations between miRNA-212 and epilepsy, but very few studies have explored the association between the lncRNA ILF3AS1 and epilepsy, considering that miRNA-212 is targeted by lncRNA ILF3AS1, causing boomback effects on MMP3 and MMP9. Therefore, the current study was aimed at assessing the interactions between the lncRNA ILF3AS1, MMP3, MMP9, and miRNA-212 in children with epilepsy and healthy children to explore their roles as potential biomarkers and independent risk factors of epilepsy; these results were further correlated with the MRI features among children with epilepsy to better understand the pathogenesis of epilepsy and predict potential targeted therapies.
Patients and methods
Study design and setting
Fifty children with epilepsy and fifty healthy controls of both sexes, aged 6 months to 18 years, were included in this case–control study. The subjects were recruited from the Al Zahraa University Hospital in Cairo, Egypt, between December 2022 and May 2023.
Ethical statement
The study protocols were reviewed and approved by the Research Ethics Committee of the Faculty of Medicine (for Girls) at Al-Azhar University (Approval No. 2022101560). Before starting the study, the parents of all the participants signed an informed consent form after clarifying the purpose of the study and ensuring its safety and confidentiality.
Sampling technique
The sample size was calculated using the online epi info program by considering the confidence level (95%), power of the test (80%), and ratio of controls to cases (1:1), with an estimated odds ratio (OR) of 3 for detection; therefore, the sample size was calculated as 50 cases along with 50 controls. Samples were collected from the hospital departments on randomly selected days (Dean et al., 2024).
Inclusion criteria
Children with epilepsy were recruited from the Pediatric and Neurology departments. These patients were clinically diagnosed in accordance with the International League Against Epilepsy (ILAE) criteria (Dean et al., 2024), and only patients with no family history of epilepsy, no concurrent systemic disorders, no psychiatric or neurological diseases, and normal neurological examination results were included in the study. A comparable number of age- and sex-matched apparently healthy children were recruited from the Pediatric and Neurology outpatient clinics based on selection after a routine checkup. The healthy controls had no history of epilepsy or any other neurological diseases.
Exclusion criteria
Patients were excluded if they had had clinical seizures during the 3 days prior to blood drawing, severe neurological or neuroimmunological illnesses (such as meningitis or encephalitis), a concurrent inflammatory disease, cancer, received immunomodulatory or immunosuppressive drugs during the prior 6 months, surgery or serious trauma within the previous 2 weeks, a severe mental disease, or renal or hepatic insufficiency. In addition, patients with absolute contraindications for MRI examination, such as cardiac resynchronization therapy (CRT) devices, cardiac implantable electronic devices (CIEDs), and implantable cardioverter defibrillators (ICDs), were not included. Lastly, patients with contraindications to iodinated intravenous contrast agent administration were also excluded from this study.
Procedures and assessments
Full history
Information was gathered from each subject, including their age, sex, duration of epilepsy, frequency of attacks, type of seizures, time of last attack, and types of antiepileptic drugs used, to exclude pseudointractability.
Radiological examination
Using the non-focal and temporal lobe epilepsy protocol, a 1.5 T superconducting MR imager (Achieva, Philips Medical System) was used for the scanning process.
MRI scan: Before entering the scanning room, the patient or their parents signed an approved written consent form after being appraised of the procedures.
Positioning for epilepsy MRI: Headfirst supine, with pillows under the legs for added comfort; the head was placed in a head coil for immobilization.
MRI scan localizer: A three-plane localizer was initially used to locate and design the sequences. These localizers were T1-weighted, low-resolution scans generally lasting less than 25 s (Wang et al., 2020).
T2 turbo spin echo (TSE) axial, T2 fluid-attenuated inversion recovery (FLAIR) axial, and diffusion-weighted imaging (DWI)/apparent diffusion coefficient (ADC) axial: The block was positioned parallel to the genu and splenium of the corpus callosum, and axial slices were planned along the sagittal plane. T2 TSE axial and T2 FLAIR parameters: repetition time (TR) of 4000–5500 ms, echo time (TE) of 100–120 ms, slice thickness of 1 mm, flip angle of 130°–150°, phase R > L, matrix of 320 × 320, field of view (FOV) of 210°–230°, gap between slices of 10%, average number of excitations (NEXs) of 2. DWI parameters: TR of 7000–9000 ms, TE of 70–115 ms, flip angle of 130°, average NEXs of 1–2, slice thickness of 1 mm, matrix of 192 × 192, FOV of 210°–230°, phase R > L, gap between slices of 10%, and B value of 0–1000.
T1 spin echo (SE) coronal: The positioning block was tilted perpendicular to the line extending along the genu and splenium of the corpus callosum, and coronal slices were planned along the sagittal plane. T1 parameters: TR of 500–700 ms, TE of 15–25 ms, slice thickness of 3 mm, flip angle of 90°, phase R > L, matrix of 304 × 304, FOV of 210°–230°, gap between slices of 10%, and NEXs (average) of 2.
T2 TSE sagittal: The block was positioned parallel to the midline of the brain, and sagittal slices were planned along the axial plane. T2 TSE sagittal parameters: TR of 4500–6000 ms, TE of 100–120 ms, slice thickness of 2 mm, flip angle of 130°–150°, phase A > P, matrix of 320 × 304, FOV of 210°–230°, gap between slices of 10%, and NEXs (average) of 2.
T1 and T2 TSE coronal oblique 1 mm (epilepsy protocol): The block was tilted perpendicular to the long axis of the hippocampal region, and high-resolution coronal slices were planned along the sagittal plane. T1 parameters: TR of 400–600 ms, TE of 15–25 ms, slice thickness of 1 mm, flip angle of 140°, phase R > L, matrix of 320 × 320, FOV of 170°–190°, gap between slices of 10%, NEXs (average) of 4. T2 parameters: TR of 3000–4000 ms, TE of 100–120 ms, slice thickness of 1 mm, flip angle of 130°–150°, phase A > P, matrix of 170 × 190, FOV of 170°–190°, gap between slices of 10%, and NEXs (average) of 4 (Wang et al., 2020).
T1-weighted contrast + axial: The block was positioned parallel to the genu and splenium of the corpus callosum, with axial slices planned along the sagittal plane. A contrast agent of 0.2 mL/kg (0.1 mmol/kg) was administered intravenously at 2 mL/s, for a maximum dose of 20 mL (Nardone et al., 2014).
Susceptibility-weighted imaging (SWI) or T2* axial: The block was positioned parallel to the genu and splenium of the corpus callosum, with axial slices planned along the sagittal plane.
Laboratory investigations
Serum levels of MMP3 and MMP9 were determined using enzyme-linked immunosorbent assay (ELISA) (Cat nos. E-EL-H1446 and E0936Hu, respectively, Shanghai Korain Biotech Co.) according to the manufacturer instructions. Routine laboratory investigation include a complete blood picture, ALT, AST, serum creatinine, blood urea, serum Calcium (Ca), Pottassium (K), Sodium (Na), and Phosphorus (P) were done for all patients and controls.
Expression levels of lncRNA ILF3AS1 and miRNA-212 using quantitative real-time polymerase chain reaction (q-PCR)
The total RNA was isolated from the whole blood samples of all groups using Direct-zol RNA Miniprep Plus (Cat no. R2072, Zymo Research Corp., United States), after which both the quantity and quality were evaluated using a Beckman dual spectrophotometer (United States). The Super Script IV One-Step real-time PCR kit (Cat no. 12594100, Thermo Fisher Scientific, Waltham, MA, United States) was used to reverse transcribe the extracted RNA, followed by q-PCR using a 96-well-plate step one instrument (Applied Biosystems, United States). The primers used for lncRNA ILF3AS1, miRNA-212, GAPDH, and U6 were as follows.
LncRNA ILF3AS1: forward 5′-TAAACCCCACTGTCTTCC-3′ and reverse 5′-TTCCTTGCTCTTCTTGCTC-3′; GAPDH: forward 5′-AATGGACAACTGGTCGTGGAC-3′ and reverse 5′-CCCTCCAGGGGATCTGTTTG-3′; miRNA-212: forward 5′- CCTAAGGTGGCGAGAAAATG-3′ and reverse 5′- TGCTGGCTGATACCTTCAGA-3′; U6: forward 5′-AGTAAGCCCTTGCTGTCAGTG-3′ and reverse 5′-CCTGGGTCTGATAATGCTGGG-3′. Using the 2−∆∆Cq approach, the miRNA expression levels were measured and normalized to the internal reference genes, namely U6 and GAPDH.
The cyclic conditions used in the q-PCR system were as follows: initial denaturation for 40 cycles each of 10 s at 98°C, 10 s at 55°C, and 30 s at 72°C for the amplification step.
The data were expressed as cycle thresholds (Ct) for the target and housekeeping genes, with normalization to account for the variations in the expressions of each of the target genes; lncRNA ILF3AS1 and miRNA-212 were investigated for the mean critical threshold expressions of the GAPDH and U6 housekeeping genes, respectively, using the ΔΔCt method (Gene Copoeia, Inc. United States). Furthermore, the relative quantification of each target gene was achieved using the 2−ΔΔCT method.
Data management and statistical analyses
Statistical analyses were performed using the Statistical Package for Social Sciences (SPSS) version 22 (IBM Corp., release 2013, IBM SPSS Statistics for Windows, Version 22.0, Armonk, NY, United States). The mean ± standard deviation (SD) values were used to present the numerical data, whereas frequencies (N) and percentages (%) were used to express the categorical variables. The mean rank variations of the two independent groups were compared using the Mann–Whitney U test.
We assured the significance of our results to control the family-wise error rate across the estimated parameters tested by applying Benforoni correction that was carried out by dividing the significance level by the number of tests performed for pairwise comparisons, and the adjusted ɑ were assessed, then we compared each p-value by the ɑ values.
The area under the curve (AUC) values of the receiver operating characteristic (ROC) curves were utilized to assess the diagnostic potentials of the molecular biomarkers and metalloproteinases in the identification of epilepsy. A p-value of less than 0.05 was deemed significant for probability.
Results
In this study, the mean ages of the case and control subjects were 9.4 ± 3.7 and 8 ± 4.5 years, respectively, with no statistically significant difference. The demographics and biochemical parameters related to the enrolled children are shown in Table 1.
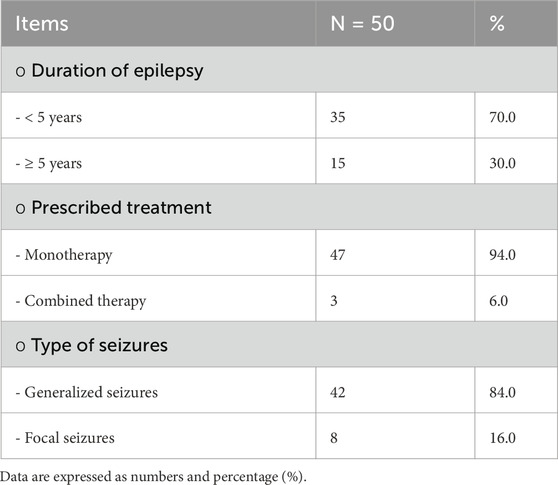
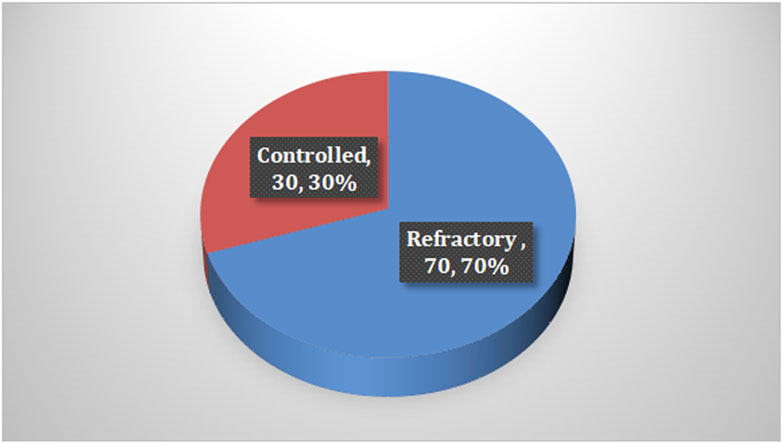
Among the cases, it was found that 70% of children with epilepsy had been suffering for less than 5 years, and the majority of them received levetiracetam and valproic acid as monotherapy. In terms of seizures, 84% of the cases reported having generalized seizures and 70% experienced refractory seizures, with the last attack occurring less than 6 months prior to the study, as shown in Table 2 and Figure 1.
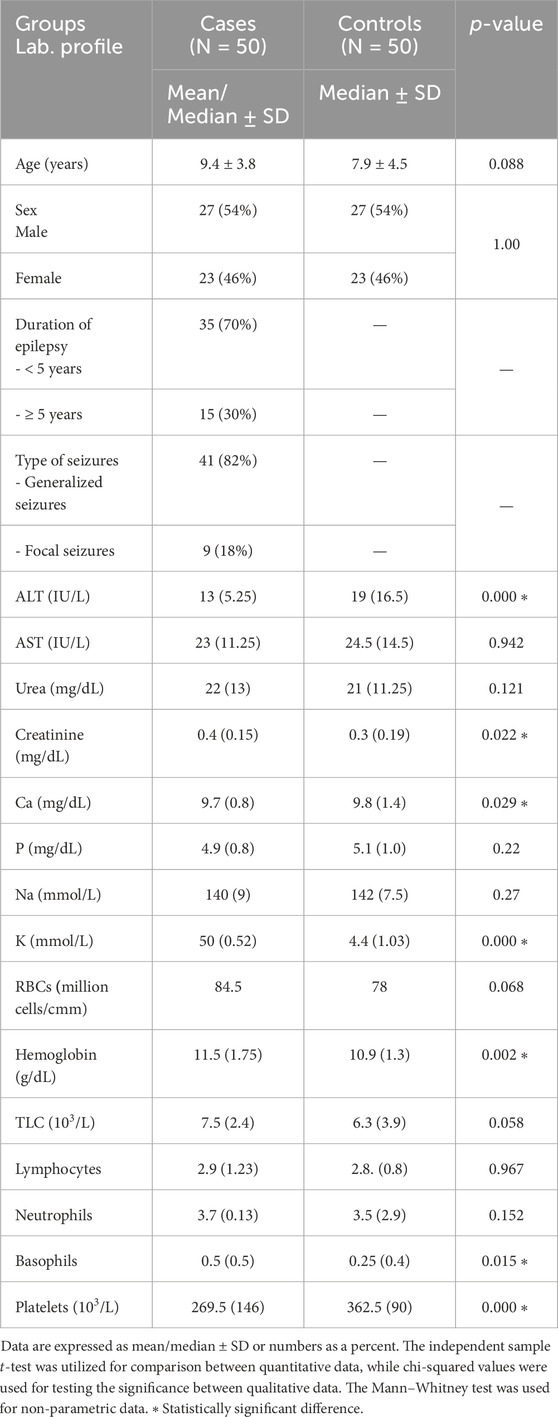
Table 1. Demographics and biochemical parameters between children with epilepsy and healthy children.
Using MRI as the primitive diagnostic tool for epilepsy, it was found that 30% of the cases had lesioned epilepsy, out of which mesial temporal sclerosis (MTS) represented approximately half of the cases, as shown in Table 3 and Figures 2–4.
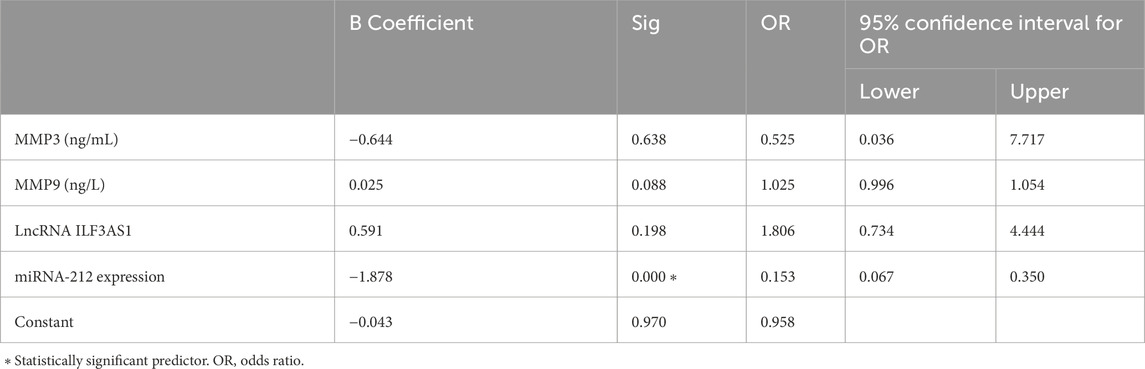
Higher levels of MMP3 and MMP9 were found in the cases compared to controls. Furthermore, higher serum expression levels of lncRNA ILF3AS1 and lower levels of miRNA-212 were found among the cases compared to controls, with statistically significant differences, as shown in Table 4. Among the children with epilepsy, higher levels of MMP9, lncRNA ILF3AS1, and fold-change miRNA-212 were noted among those with lesions detected by MRI than those with normal findings, with no statistically significant differences, as shown in Table 5. Using logistic regression analysis, the serum expression of miRNA-212 alone was detected as a negative predictor of epilepsy with statistical significance, as shown in Table 6.
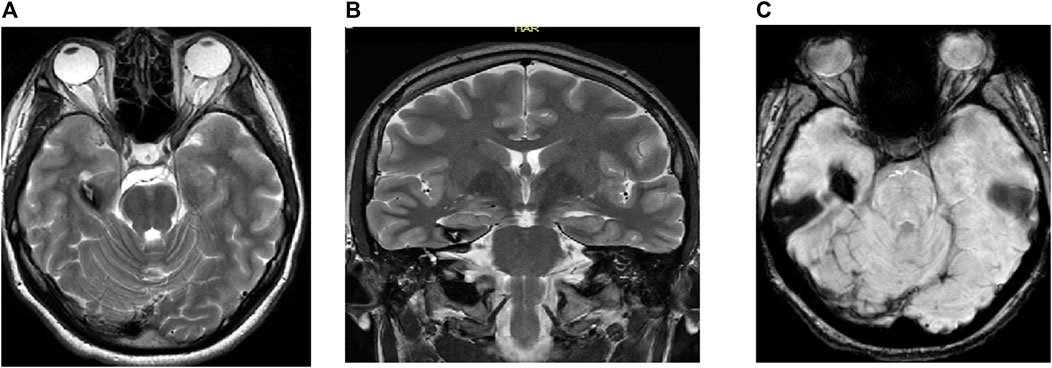
Figure 2. MRI of the brain of a 17-year-old man showing (A) axial T2 weighted image (WI), (B) coronal T2 WI, and (C) gradient echo (GRE) T2*/SWI: Figure show RT hippocampal popcorn lesion with peripheral signal loss due to hemosiderin deposition and prominent blooming in the GRE T2*/SWI image, characteristic of hippocampal cavernoma.
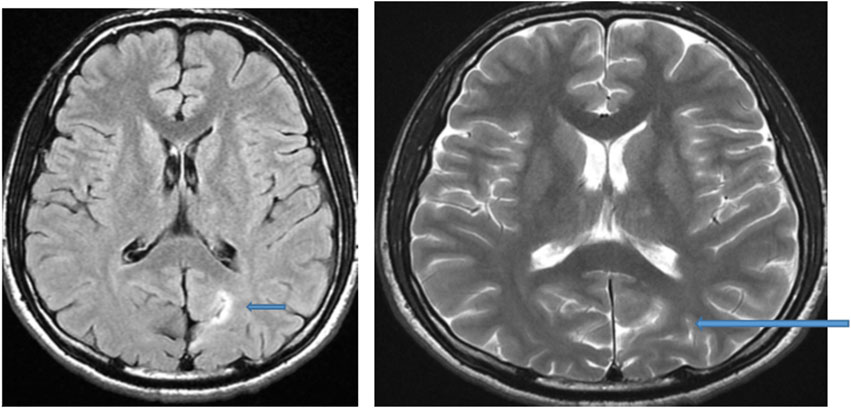
Figure 3. MRI of the brain of a 13-year-old male child: Axial FLAIR and T2-weighted image showing hyperintense focal thickening at the left occipitoparietal sulcus (arrow) compatible with focal cortical dysplasia.
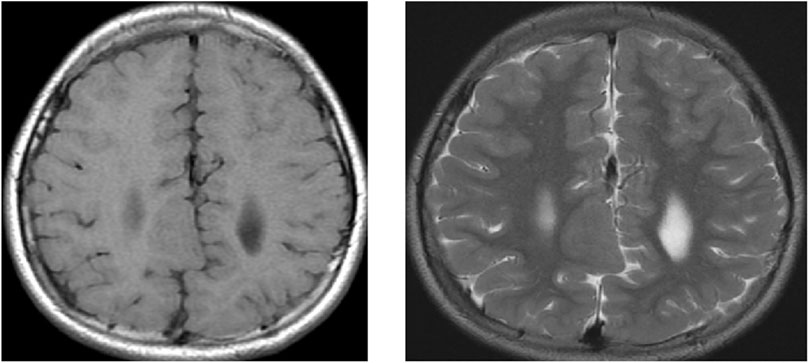
Figure 4. MRI of the brain of a 9-year-old child showing axial T1 weighted image (WI) and axial T2 WI for a large well-defined nodule at the RT subcortical parafalcine region abutting the falx another similar lesion seen at the left frontal region, with both lesions having isointense signals to the gray matter suggestive of gray matter heterotopia.
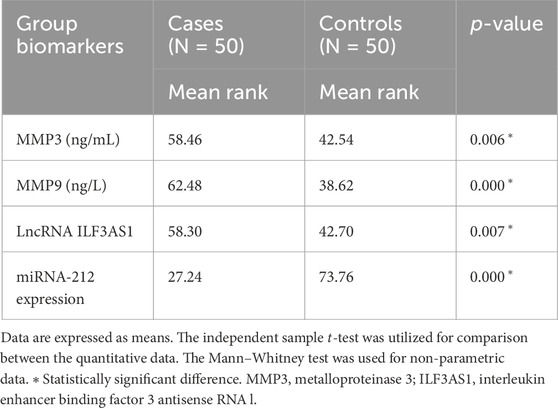
Table 4. Comparison between children with epilepsy and healthy controls regarding serum metalloproteinases and molecular biomarkers.
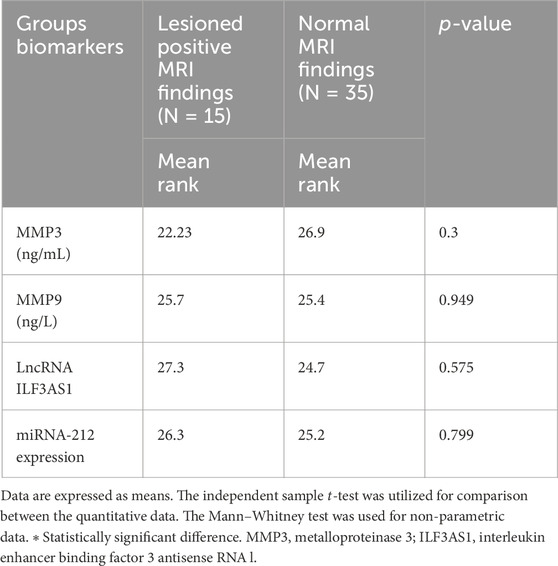
Table 5. Assessment of metalloproteinases and molecular biomarkers among children with epilepsy based on MRI findings.
Table 7 and Figure 5 present the evaluations of the metalloproteinases and molecular biomarkers as diagnostic tools for epilepsy based on the ROC curve. The estimated AUC values were 0.659, 0.738, 0.656, and 0.965 for MMP3, MMP9, lncRNA ILF3AS1, and fold-change miRNA-212, respectively, which are good results for MMP9 and excellent for miRNA-212.
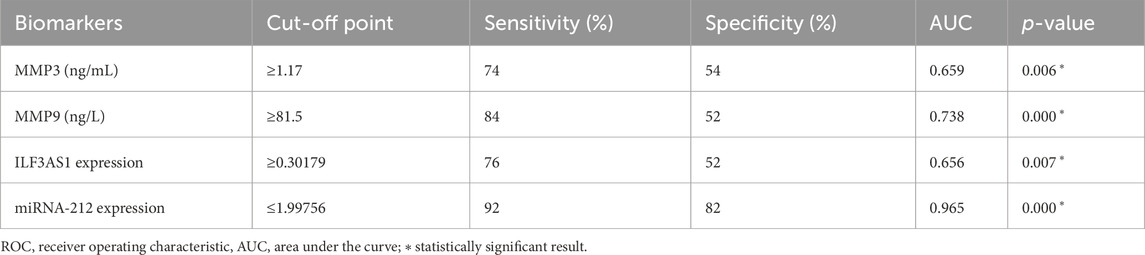
Table 7. ROC curve data for assessment of diagnostic power of metalloproteinases and molecular biomarkers in detecting epilepsy.
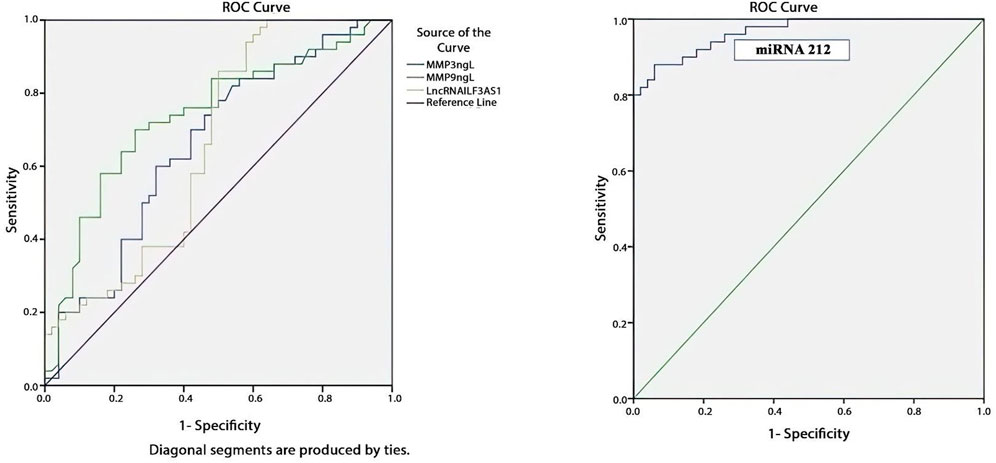
Figure 5. ROC curve showing that fold-change miRNA-212 has the most diagnostic power for detecting epilepsy based on the highest AUC of 0.965, as shown in Table 6.
Discussion
Epilepsy is one of the most prevalent neurological disorders worldwide, affecting almost 50 million individuals of various ages. However, when epilepsy is properly diagnosed and treated, up to 70% of the affected population may be able to live seizure-free (World Health Organization, 2024).
In the present study, 30% of the studied children had suffered from epilepsy for more than 5 years; of these, 84% had suffered from generalized seizures. Furthermore, 76% of the children with epilepsy had refractory-type seizures, with the last episode of seizure being registered within the last 6 months before the study; this is fairly consistent with the findings reported by El-Tallawy et al. (2013) and Khedr et al. (2013), who revealed that generalized tonic–clonic seizures were the most common type of epilepsy. In other studies, focal onset seizures were the most prevalent seizure type (Alshahawy et al., 2018; Ullah et al., 2018).
In this study, MRI was performed to correlate the radiological findings with the cause of epilepsy, where 30% of the patient group had lesioned epilepsy; however, normal findings were obtained in 70% of the cases. These findings differ only slightly from those reported by Kreilkamp et al. (2019), who noted lesioned epilepsy in 33% and normal findings in 67% of the cases.
MTS was the most common type of lesions (46.7%) in our study. However, this is not compatible with the findings of Apolot et al. (2022), who reported acquired structural brain abnormalities (61.22%) as the most common type of lesions in their study.
Apolot et al. (2022) reported hippocampal sclerosis (HS) as the most common structural abnormality, which is consistent with our finding, as noted in 46.7% of the cases.
Numerous genome-wide studies have provided evidence that ncRNA molecules play significant roles in large-scale epigenetic alterations that occur as epilepsy progresses; these are highly expressed in the brain and contribute to the control of physiological and pathophysiological processes, such as immune system activation, oxidative stress, synaptic plasticity, neuronal development, apoptosis, and neurogenesis, all of which are linked to epilepsy (Cai and Wan, 2018; Yao et al., 2019; Manna et al., 2022).
A growing number of studies have shown that epigenetic regulation involving histone modification, DNA methylation, and ncRNAs are implicated in epilepsy (Butler-Ryan and Wood, 2021).
The potential involvement of lncRNAs in epileptogenesis has been suggested based on the altered numbers and diversity of lncRNAs detected in patients with epilepsy, making them appropriate targets for treatment (Liu et al., 2022). Additionally, lncRNAs can control these processes by competitively suppressing miRNAs that are directly implicated in these functions (Ratti et al., 2020).
The aberrant production of miRNAs, which regulate protein levels by binding to their target mRNAs, is assumed to be associated with inflammatory pathways, cell death, neuronal excitability, and synaptic reorganization, which are also closely related to epileptogenesis (Li et al., 2014).
Several studies have examined various miRNAs and lncRNAs as well as their roles in epileptogenesis (Manna et al., 2022), suggesting that these lncRNAs can be used as diagnostic markers or prospective therapeutic targets for epilepsy. However, the findings of these studies have not been verified in independent cohorts (Ghafouri-Fard et al., 2022).
Regarding the lab profiles in the present study, higher levels of the lncRNA ILF3AS1 were found in patients with epilepsy than the matched controls, which is consistent with the results reported by Cai et al. (2020).
Neuroinflammation, BBB dysfunction, and altered synaptic remodeling functions are the potential contributors to seizures (Wang et al., 2024).
Moreover, the serum levels of the metalloproteinases (MMP3 and MMP9) were elevated in the epilepsy group compared to the control group with significant differences.
The seizure activity triggers activation of various pathways within the CNS, including matrix metalloproteases, which result in distinct responses in the brain with the progression of necroptosis as a type of regulated cell death. These responses involve the activation of neurons and astrocytes, consequently leading to increased expressions of proteins and genes (Mohseni-Moghaddam et al., 2024).
This finding is consistent with that of the studies conducted by Broekaart et al. (2020) and Cai et al. (2020), who reported that serum levels of MMP3 and MMP9 were overexpressed in children with epilepsy, suggesting that ILF3AS1 promoted their expression. The present results are also consistent with the findings of Korotkov et al. (2018), who reported elevated MMP3 expressions in the hippocampus of TLE patients, and Rempe et al. (2018), who reported increased MMP9 activity in a TLE rat model (Broekaart et al., 2020).
Kaczmarek et al. (2023) also discussed and enforced the effects of MMP9; they stated that MMP9 is frequently generated and released in excess in reaction to stressors that have the potential to cause epileptogenesis. Moreover, the pathophysiology of epilepsy has been linked to abnormal synaptic plasticity. Increased MMP9 levels are also a result of neuroinflammation, which is another proepileptic brain response (Kaczmarek et al., 2023).
In this study, miRNA-212 was found to be downregulated, which is similar to the findings of Cai et al. (2020), who showed that the serum and hippocampal miRNA-212 levels of TLE patients decreased by 50%; this is also consistent with the findings of Haenisch et al. (2015), who screened for miRNA-212-3p in the hippocampal focal and non-focal samples of brain tissues in temporal neocortex TLE patients.
Several studies have demonstrated that lncRNAs inhibit miRNA expression (Geng et al., 2018; Han et al., 2018; Zhao et al., 2019). In our study, it was found that increased expression of lncRNA ILF3AS1 was associated with lower miRNA-212 expression in the epilepsy patients than the controls with a statistically significant difference, suggesting that lncRNA ILF3AS1 induced downregulation of miRNA-212 expression. This finding is compatible with that of Hu et al. (2019), who demonstrated that ILF3AS1 knockdown suppressed miRNA-212 expression, which in turn lowered osteosarcoma cell proliferation, invasion, and migration while provoking apoptosis.
In summary, we found that lncRNA ILF3AS1, miRNA-212, MMP3, and MMP9 have predictive and diagnostic powers for epilepsy, with AUC values of 0.656, 0.965, 0.659, and 0.738, respectively.
Conclusion
According to our study, peripheral blood gene expression profiling provided the preliminary clues that lncRNAs ILF3AS1 and miRNA-212 are dysregulated in human epilepsy, so they may act as independent predictors of epilepsy.
The serum lncRNA ILF3AS1 expression levels were higher in children with epilepsy and were associated with higher levels of MMP3 and MMP9 as well as lower levels of miRNA-212 than those in the control group. This provides easy, sensitive, and accessible diagnostic biomarker tools for epilepsy.
Therefore, it is plausible that targeting these pathways of the ILF3AS1/miRNA-212/MMP3/MMP9 axis may potentially be a promising approach to improve the prognosis as well as manage epileptic seizures.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors without undue reservation.
Ethics statement
The studies involving humans were approved by the Ethics Committee of the Faculty of Medicine (for Girls) of Al-Azhar University (Approval No. 2022101560). The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation in this study was provided by the legal guardians/next of kin of the participants.
Author contributions
AmM: conceptualization, data curation, formal analysis, funding acquisition, investigation, methodology, project administration, resources, software, supervision, validation, visualization, writing–original draft, and writing–review and editing. WE: conceptualization, data curation, formal analysis, funding acquisition, investigation, methodology, project administration, resources, software, supervision, validation, visualization, writing–original draft, and writing–review and editing. SM: investigation, methodology, writing–original draft, and writing–review and editing. AA: investigation, methodology, writing–original draft, and writing–review and editing. EM: data curation, investigation, writing–original draft, and writing–review and editing. HN: data curation, investigation, writing–original draft, and writing–review and editing. AG: methodology, validation, writing–original draft, and writing–review and editing. SA: investigation, validation, writing–original draft, and writing–review and editing. DF: formal analysis, software, writing–original draft, and writing–review and editing. RE: conceptualization, data curation, writing–original draft, and writing–review and editing. YA: conceptualization, data curation, investigation, writing–original draft, and writing–review and editing. AH: data curation, investigation, writing–original draft, and writing–review and editing. BZ: data curation, investigation, writing–original draft, and writing–review and editing. MK: conceptualization, data curation, writing–original draft, and writing–review and editing. NA: conceptualization, data curation, funding acquisition, validation, writing–original draft, and writing–review and editing. SS: conceptualization, data curation, investigation, writing–original draft, and writing–review and editing. AD: conceptualization, data curation, investigation, writing–original draft, and writing–review and editing. MS: investigation, writing–original draft, and writing–review and editing. MG: conceptualization, investigation, writing–original draft, and writing–review and editing. SE: conceptualization, methodology, writing–original draft, and writing–review and editing. AlM: conceptualization, investigation, methodology, writing–original draft, and writing–review and editing. MD: conceptualization, data curation, formal analysis, funding acquisition, investigation, methodology, project administration, resources, software, supervision, validation, visualization, writing–original draft, and writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. The authors thank Princess Nourah Bint Abdulrahman University, Riyadh, Saudi Arabia, for the Researcher Support (project number: PNURSP2024R339) and publication of this article.
Acknowledgments
The authors thank Princess Nourah Bint Abdulrahman University, Riyadh, Saudi Arabia, for the Researcher Support. The authors also thank Prof. Dr. Manal H. Maabady for her assistance.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations or those of the publisher, editors, and reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Abbreviations
ADC, apparent diffusion coefficient; AUC, area under the curve; BBB, blood–brain barrier; ceRNAs, competing endogenous RNAs; CIED, cardiac implantable electronic device; CNS, central nervous system; CRT, cardiac resynchronization therapy; Ct, cycle threshold; DWI, diffusion-weighted imaging; ECM, extracellular matrix; EEG, electroencephalography; ELISA, enzyme-linked immunosorbent assay; FCD, focal cortical dysplasia; FLAIR, fluid-attenuated inversion recovery; FOV, field of view; GRE, gradient echo; HS, hippocampal sclerosis; ICD, implantable cardioverter defibrillator; ILF3AS1, interleukin enhancer binding factor 3 antisense RNA l; lncRNA, long non-coding RNA; miRNA-212, microRNA 212; MMP, matrix metalloproteinase; MRI, magnetic resonance imaging; MTS, mesial temporal sclerosis; ncRNA, non-coding RNA; NEX, number of excitations; OR, odds ratio; p-value, probability value; q-RTPCR, quantitative real-time polymerase chain reaction; ROC, receiver operator characteristic; SD, standard deviation; SE, spin echo; SWI, susceptibility-weighted imaging; TE, echo time; TLE, temporal lobe epilepsy; TNF-α, tumor necrosis factor alpha; TR, repetition time; TSE, turbo spin echo.
References
Alshahawy, A. K., Darwish, A. H., Elsaid Shalaby, S., and Mawlana, W. (2018). Prevalence of idiopathic epilepsy among school children in Gharbia Governorate, Egypt. Brain Dev. 40 (4), 278–286. doi:10.1016/j.braindev.2017.12.009
Apolot, D., Erem, G., Nassanga, R., Kiggundu, D., Tumusiime, C. M., Teu, A., et al. (2022). Brain magnetic resonance imaging findings among children with epilepsy in two urban hospital settings, Kampala-Uganda: a descriptive study. BMC Med. Imaging 22 (1), 175. doi:10.1186/s12880-022-00901-7
Banawalikar, N., Adiga, S., Adiga, U., Shenoy, V., Kumari, S., Shetty, P., et al. (2021). Association of UGT1A6 gene polymorphism with clinical outcome in pediatric epileptic patients on sodium valproate monotherapy. Braz J. Med. Biol. Res. 54 (9), e11097. doi:10.1590/1414-431X2021e11097
Bernasconi, A., Bernasconi, N., Bernhardt, B. C., and Schrader, D. (2011). Advances in MRI for ‘cryptogenic’ epilepsies. Nat. Rev. Neurol. 7 (2), 99–108. doi:10.1038/nrneurol.2010.199
Brennan, G. P., and Henshall, D. C. (2020). MicroRNAs as regulators of brain function and targets for treatment of epilepsy. Nat. Rev. Neurol. 16, 506–519. doi:10.1038/s41582-020-0369-8
Broekaart, D. W. M., van Scheppingen, J., Anink, J. J., Wierts, L., van Het Hof, B., Jansen, F. E., et al. (2020). Increased matrix metalloproteinases expression in tuberous sclerosis complex: modulation by microRNA 146a and 147b in vitro. Neuropathol. Appl. Neurobiol. 46 (2), 142–159. doi:10.1111/nan.12572
Butler-Ryan, R., and Wood, I. C. (2021). The functions of repressor element 1-silencing transcription factor in models of epileptogenesis and post-ischemia Metab. Brain Dis.
Cai, X., Long, L., Zeng, C., Ni, G., Meng, Y., Guo, Q., et al. (2020). LncRNA ILF3-AS1 mediated the occurrence of epilepsy through suppressing hippocampal miR-212 expression. Aging (Albany NY) 12 (9), 8413–8422. doi:10.18632/aging.103148
Cai, Y., and Wan, J. (2018). Competing endogenous RNA regulations in neurodegenerative disorders: current challenges and emerging insights. Front. Mol. Neurosci. 11, 370. doi:10.3389/fnmol.2018.00370
Cava, C., Manna, I., Gambardella, A., Bertoli, G., and Castiglioni, I. (2018). Potential role of miRNAs as theranostic biomarkers of epilepsy. Mol. Ther. Nucleic Acids 13, 275–290. doi:10.1016/j.omtn.2018.09.008
Dean, A. G., Sullivan, K. M., and Soe, M. M. (2024). OpenEpi: open-source epidemiologic Statistics for public Health, version. Available at: www.OpenEpi.com.
De Bock, M., Wang, N., Decrock, E., Bultynck, G., and Leybaert, L. (2015). Intracellular cleavage of the cx43 C-terminal domain by matrix-metalloproteases: a novel contributor to inflammation? Mediat. Inflamm. 2015, 257471. doi:10.1155/2015/257471
El-Tallawy, H. N., Farghaly, W. M., Shehata, G. A., Abdel-Hakeem, N. M., Rageh, T. A., Abo-Elftoh, N. A., et al. (2013). Epidemiology of epilepsy in new valley governorate, Al kharga district, Egypt. Epilepsy Res. 104 (1-2), 167–174. doi:10.1016/j.eplepsyres.2012.08.010
Fernandez-Baca Vaca, G., and Park, J. T. (2020). Focal EEG abnormalities and focal ictal semiology in generalized epilepsy. Seizure 77, 7–14. doi:10.1016/j.seizure.2019.12.013
Geng, J. F., Liu, X., Zhao, H. B., Fan, W. F., Geng, J. J., and Liu, X. Z. (2018). LncRNA UCA1 inhibits epilepsy and seizure-induced brain injury by regulating miR-495/Nrf2-ARE signal pathway. Int. J. Biochem. Cell Biol. 99, 133–139. doi:10.1016/j.biocel.2018.03.021
Ghafouri-Fard, S., Hussen, B. M., Jamali, E., Branicki, W., Taheri, M., and Akbari Dilmaghani, N. (2022). Role of lncRNAs and circRNAs in epilepsy. Ageing Res. Rev. 82, 101749. doi:10.1016/j.arr.2022.101749
Haenisch, S., Zhao, Y., Chhibber, A., Kaiboriboon, K., Do, L. V., Vogelgesang, S., et al. (2015). SOX11 identified by target gene evaluation of miRNAs differentially expressed in focal and non-focal brain tissue of therapy-resistant epilepsy patients. Neurobiol. Dis. 77, 127–140. doi:10.1016/j.nbd.2015.02.025
Han, C. L., Ge, M., Liu, Y. P., Zhao, X. M., Wang, K. L., Chen, N., et al. (2018). Long non-coding RNA H19 contributes to apoptosis of hippocampal neurons by inhibiting let-7b in a rat model of temporal lobe epilepsy. Cell Death Dis. 9 (6), 617. doi:10.1038/s41419-018-0496-y
Heiskanen, M., Das Gupta, S., Mills, J. D., van Vliet, E. A., Manninen, E., Ciszek, R., et al. (2023). Discovery and validation of circulating microRNAs as biomarkers for epileptogenesis after experimental traumatic brain injury-the EPITARGET cohort. Int. J. Mol. Sci. 24 (3), 2823. doi:10.3390/ijms24032823
Herrera-Espejo, S., Santos-Zorrozua, B., Álvarez-González, P., Lopez-Lopez, E., and Garcia-Orad, Á. (2019). A systematic review of MicroRNA expression as biomarker of late-onset alzheimer's disease. Mol. Neurobiol. 56 (12), 8376–8391. doi:10.1007/s12035-019-01676-9
Hu, X. H., Dai, J., Shang, H. L., Zhao, Z. X., and Hao, Y. D. (2019). SP1-mediated upregulation of lncRNA ILF3-AS1 functions a ceRNA for miR-212 to contribute to osteosarcoma progression via modulation of SOX5. Biochem. Biophys. Res. Commun. 511, 510–517. doi:10.1016/j.bbrc.2019.02.110
Kaczmarek, K. T., Protokowicz, K., and &Kaczmarek, L. (2023). Matrix metalloproteinase-9: a magicdrug target in neuropsychiatry? J. Neurochem. 00, 1–12. doi:10.1111/jnc.15976
Khedr, E. M., Shawky, O. A., Ahmed, M. A., Elfetoh, N. A., Al Attar, G., Ali, A. M., et al. (2013). A community based epidemiological study of epilepsy in Assiut Governorate/Egypt. Epilepsy Res. 103 (2-3), 294–302. doi:10.1016/j.eplepsyres.2012.08.006
Korotkov, A., Broekaart, D. W. M., van Scheppingen, J., Anink, J. J., Baayen, J. C., Idema, S., et al. (2018). Increased expression of matrix metalloproteinase 3 can be attenuated by inhibition of microRNA-155 in cultured human astrocytes. J. Neuroinflammation 15 (1), 211. doi:10.1186/s12974-018-1245-y
Kreilkamp, B. A. K., Das, K., Wieshmann, U. C., Biswas, S., Marson, A. G., and Keller, S. S. (2019). Neuroradiological findings in patients with “non-lesional” focal epilepsy revealed by research protocol. Clin. Radiol. 74 (1), 78.e1–78. doi:10.1016/j.crad.2018.08.013
Lekka, E., and Hall, J. (2018). Noncoding RNAs in disease. FEBS Lett. 592 (17), 2884–2900. doi:10.1002/1873-3468.13182
Li, M. M., Li, X. M., Zheng, X. P., Yu, J. T., and Tan, L. (2014). MicroRNAs dysregulation in epilepsy. Brain Res. 1584, 94–104. doi:10.1016/j.brainres.2013.09.049
Liu, S., Fan, M., Ma, M. D., Ge, J. F., and Chen, F. H. (2022). Long non-coding RNAs: potential therapeutic targets for epilepsy. Front. Neurosci. 16, 986874. doi:10.3389/fnins.2022.986874
Manna, I., Fortunato, F., De Benedittis, S., Sammarra, I., Bertoli, G., Labate, A., et al. (2022). Non-coding RNAs: new biomarkers and therapeutic targets for temporal lobe epilepsy. Int. J. Mol. Sci. 23 (6), 3063. doi:10.3390/ijms23063063
Mohseni-Moghaddam, P., Khaleghzadeh-Ahangar, H., and Atabaki, R. (2024). Role of necroptosis, a regulated cell death, in seizure and epilepsy. Neurochem. Res. 49, 1–13. doi:10.1007/s11064-023-04010-x
Nardone, B., Saddleton, E., Laumann, A. E., Edwards, B. J., Raisch, D. W., McKoy, J. M., et al. (2014). Pediatric nephrogenic systemic fibrosis is rarely reported: a RADAR report. Pediatr. Radiol. 44 (2), 173–180. doi:10.1007/s00247-013-2795-x
Ratti, M., Lampis, A., Ghidini, M., Salati, M., Mirchev, M. B., Valeri, N., et al. (2020). MicroRNAs (miRNAs) and long non-coding RNAs (lncRNAs) as new tools for cancer therapy: first steps from bench to bedside. Target Oncol. 15 (3), 261–278. doi:10.1007/s11523-020-00717-x
Rempe, R. G., Hartz, A. M. S., Soldner, E. L. B., Sokola, B. S., Alluri, S. R., Abner, E. L., et al. (2018). Matrix metalloproteinase-mediated blood-brain barrier dysfunction in epilepsy. J. Neurosci. 38 (18), 4301–4315. doi:10.1523/JNEUROSCI.2751-17.2018
Rivera, S., Khrestchatisky, M., Kaczmarek, L., Rosenberg, G. A., and Jaworski, D. M. (2010). Metzincin proteases and their inhibitors: foes or friends in nervous system physiology? J. Neurosci. 30 (46), 15337–15357. doi:10.1523/JNEUROSCI.3467-10.2010
Thijs, R. D., Surges, R., O’Brien, T. J., and Sander, J. W. (2019). Epilepsy in adults. lancet 393, 689–701. doi:10.1016/S0140-6736(18)32596-0
Ullah, S., Ali, N., Khan, A., Ali, S., and Nazish, H. R. (2018). The epidemiological characteristics of epilepsy in the province of khyber pakhtunkhwa, Pakistan. Front. Neurol. 9, 845. doi:10.3389/fneur.2018.00845
Van Lint, P., and Libert, C. (2007). Chemokine and cytokine processing by matrix metalloproteinases and its effect on leukocyte migration and inflammation. J. Leukoc. Biol. 82 (6), 1375–1381. doi:10.1189/jlb.0607338
Vattipally, V. R., and Bronen, R. A. (2004). MR imaging of epilepsy: strategies for successful interpretation. Neuroimaging Clin. N. Am. 14 (3), 349–372. doi:10.1016/j.nic.2004.04.002
Wang, I., Bernasconi, A., Bernhardt, B., Blumenfeld, H., Cendes, F., Chinvarun, Y., et al. (2020). MRI essentials in epileptology: a review from the ILAE Imaging Taskforce. Epileptic Disord. 22 (4), 421–437. doi:10.1684/epd.2020.1174
Wang, J., and Zhao, J. (2021). MicroRNA dysregulation in epilepsy: from pathogenetic involvement to diagnostic biomarker and therapeutic agent development. Front. Mol. Neurosci. 14, 650372. doi:10.3389/fnmol.2021.650372
Wang, Q., Lin, Z., Yao, C., Liu, J., Chen, J., and Diao, L. (2024). Meta-analysis of MMP-9 levels in the serum of patients with epilepsy. Front. Neurosci. 18, 1296876. doi:10.3389/fnins.2024.1296876
Wilmshurst, J. M., Gaillard, W. D., Vinayan, K. P., Tsuchida, T. N., Plouin, P., Van Bogaert, P., et al. (2015). Summary of recommendations for the management of infantile seizures: task force report for the ILAE commission of pediatrics. Epilepsia 56 (8), 1185–1197. doi:10.1111/epi.13057
World Health Organization (2024). Epilepsy. Available at: https://www.who.int/en/news-room/fact-sheets/detail/epilepsy.
Wu, Z., Zhou, L., Ding, G., and Cao, L. (2017). Overexpressions of miR-212 are associated with poor prognosis of patients with pancreatic ductal adenocarcinoma. Cancer Biomark. 18 (1), 35–39. doi:10.3233/CBM-160671
Yao, R. W., Wang, Y., and Chen, L. L. (2019). Cellular functions of long noncoding RNAs. Nat. Cell Biol. 21 (5), 542–551. doi:10.1038/s41556-019-0311-8
Zeng, C., Hu, J., Chen, F., Huang, T., and Zhang, L. (2022). The coordination of mTOR signaling and non-coding RNA in regulating epileptic neuroinflammation. Front. Immunol. 13, 924642. doi:10.3389/fimmu.2022.924642
Keywords: epilepsy, lncRNA ILF3AS1, miRNA-212, MMP3, MMP9, magnetic resonance imaging
Citation: Mohammed AR, Emam WA, Mohammed SA, Abd Elalim AA, Mansour ENA, Nasr HM, Ghamry AA, Alkhawagah SM, Fathy DSA, Elattar RS, Abish YGI, Hussein A, Zaghloul BA, Khairallah MK, Alharbi N, Seif Eldin S, Dawood AF, Sabet MA, Gamea MG, Elshishtawy Ibrahim SE, Mosa AA and Dahpy MA (2024) LncRNA ILF3AS1, MMP3, and MMP9 as well as miRNA-212 as emerging novel biomarkers for childhood epilepsy. Front. Mol. Biosci. 11:1434023. doi: 10.3389/fmolb.2024.1434023
Received: 17 May 2024; Accepted: 17 July 2024;
Published: 23 August 2024.
Edited by:
Mario Mastrangelo, Sapienza University of Rome, ItalyReviewed by:
Chitra Rawat, Cleveland Clinic, United StatesLaura Duran-Lozano, Vall d’Hebron Institute of Oncology (VHIO), Spain
Copyright © 2024 Mohammed, Emam, Mohammed, Abd Elalim, Mansour, Nasr, Ghamry, Alkhawagah, Fathy, Elattar, Abish, Hussein, Zaghloul, Khairallah, Alharbi, Seif Eldin, Dawood, Sabet, Gamea, Elshishtawy Ibrahim, Mosa and Dahpy. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marwa A. Dahpy, bWFyd2FkYWhweUB5YWhvby5jb20=, bWFyd2FkYWhweUBhdW4uZWR1LmVn; Amena Rezk Mohammed, YW1lbmFyZXprLm1lZGdAYXpoYXIuZWR1LmVn
†ORCID: Marwa A. Dahpy, orcid.org/0000-0002-6007-5691
 Amena Rezk Mohammed1*
Amena Rezk Mohammed1* Wafaa Abdelaziz Emam
Wafaa Abdelaziz Emam Salwa Seif Eldin
Salwa Seif Eldin Suzan Eid Elshishtawy Ibrahim
Suzan Eid Elshishtawy Ibrahim Marwa A. Dahpy
Marwa A. Dahpy