
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Mol. Biosci. , 10 September 2024
Sec. RNA Networks and Biology
Volume 11 - 2024 | https://doi.org/10.3389/fmolb.2024.1388476
This article is part of the Research Topic RNA Processing in Chronic Diseases View all 6 articles
Myasthenia Gravis (MG) is a chronic autoimmune disease that primarily affects the neuromuscular junction, leading to muscle weakness in patients with this condition. Previous studies have identified several dysfunctions in thymus and peripheral blood mononuclear cells (PBMCs), such as the formation of ectopic germinal centers in the thymus and an imbalance of peripheral T helper cells and regulatory T cells, that contribute to the initiation and development of MG. Recent evidences suggest that noncoding RNA, including miRNA, lncRNA and circRNA may play a significant role in MG progression. Additionally, the network between these noncoding RNAs, such as the competing endogenous RNA regulatory network, has been found to be involved in MG progression. In this review, we summarized the roles of miRNA, lncRNA, and circRNA, highlighted their potential application as biomarkers in diagnosing MG, and discussed their potential regulatory networks in the abnormal thymus and PBMCs during MG development.
Myasthenia Gravis (MG) is a chronic autoimmune disease that mainly affects the neuromuscular junction (NMJ), leading to weakness of patients’ muscles (Gilhus, 2016; El-Salem et al., 2014). MG can be classified into several subtypes according to the age of disease onset, types of autoantibodies and clinical manifestation (El-Salem et al., 2014). Based on the age of disease onset, the MG is classified into early onset MG (EOMG, ≦ 50-year-old) and late onset MG (LOMG, >50-year-old) (Szczudlik et al., 2014; Huang et al., 2023). The EOMG is more frequent in young women with thymic hyperplasia and LOMG usually occurs in middle-aged and elderly men with thymoma (Berrih-Aknin and Le Panse, 2014). Depending on the type of serum autoantibodies, MG can generally be classified into acetylcholine receptor antibody positive MG (AChR+ MG), muscle-specific tyrosine kinase antibody positive MG (MuSK+ MG), or lipoprotein receptor-related protein 4 antibody positive MG (LRP4+ MG) (Huang et al., 2023). Among them, AChR+ MG is the most frequent subgroup of MG. In some cases, other autoantibodies such as Titin antibody, or ryanodine receptor antibody (RyR-Ab) can also be detected in patients’ serum (Skeie et al., 2006). Based on the clinical manifestation of MG, in other words, depending on the type of muscle involved, MG can be classified into ocular MG (OMG) and generalized MG (GMG) (Monsul et al., 2004). The ocular muscle is typically the initial affected muscle, resulting in ptosis and diplopia, which are usually the first symptoms of MG. Gradually, as the muscles of the limbs and trunk are affected, patients demonstrate limb weakness and may also experience dysarthria and dysphagia (bulbar symptoms), as well as respiratory difficulties. This condition is known as generalized MG. Due to the different characteristic of MG subtypes, the mechanism behind the different types of MG is variable (El-Salem et al., 2014).
Although muscle is the primary target organ of MG, the effector organ is generally considered to be the thymus. Thymic lesions, including thymic hyperplasia and thymoma, play crucial roles in the pathological process of AChR+ MG (Cron et al., 2020). The thymus serves as the organ for T cell maturation. Physiologically, naive T cells can only survive and mature after positive and negative selection in cortical and medullary thymus, respectively (Sprent and Kishimoto, 2002; Ashby and Hogquist, 2024). Positive selection secures T cells with ability to bind MHC complexes and negative selection secures T cells without autoimmune potential (Ashby and Hogquist, 2024). However, for MG patients with thymoma, the negative selection of thymus is largely compromised (Berrih-Aknin and Le Panse, 2014). Tumor cells of thymoma, derived from the cortical thymic epithelial cells (cTECs), lack medullary thymic epithelial cells (mTECs). Moreover, the decreased expression of autoimmune regulator (AIRE) and forkhead box protein P3 (FOPX3), which play a crucial role in preventing autoimmune responses, can also be observed in thymoma (Berrih-Aknin and Le Panse, 2014). As a result, T cells with autoimmune potential may not be deleted effectively in the thymoma of MG patients due to the lack of negative selection. The pathologic mechanism of thymic hyperplasia is distinct from thymoma in MG. The ectopic germinal centers (GCs) formed by numerous B cells could secrete AChR antibodies (Berrih-Aknin and Le Panse, 2014). Moreover, the ectopic high endothelial venules (HEVs) and chemokines such as CXCL13 and CCL21 could recruit the pro-inflammatory cells in thymus, activate inflammation, and compromise the normal immune regulatory process (Berrih-Aknin and Le Panse, 2014).
Although the mechanisms underlying thymoma and hyperplastic thymus-induced MG differ, noncoding RNAs including micro RNA (miRNA), long noncoding RNA (lncRNA) and circular RNA (circRNA), play crucial roles in both processes (Cron et al., 2020). Generally, these noncoding RNAs cannot encode protein, but it can regulate the expression of mRNA through posttranscriptional modification (Cai et al., 2009). MiRNA is short (approximately 20–25 nucleotides) single-stranded noncoding RNA, lncRNA is noncoding RNAs longer than 200 nucleotides, and circRNA is circular non-coding RNA. As important part of epigenetics, increasing evidence shows that noncoding RNA is important in the pathological process of MG (Berrih-Aknin and Le Panse, 2014). Moreover, benefiting from the easy availability and circulating stability of ncRNA, researchers screen out several ncRNAs for diagnosing MG and predicting the effectiveness of treatment for MG (Punga and Punga, 2018; Yoganathan et al., 2022). Newly identified non-coding RNAs, such as rRNA-derived small RNAs and tRNA-derived small RNAs, have also been found to play regulatory roles in various diseases (Zhang et al., 2023; You et al., 2019). However, these non-coding RNAs have not been extensively studied in MG. In this paper, we reviewed the roles of miRNA, lncRNA, and circRNA and their networks that participating in the pathomechanism of MG and summarized the ncRNA which may act as potential biomarkers for MG.
MiRNAs are short (approximately 20–25 nucleotides) single-stranded RNA molecules (Blow et al., 2006). Mature miRNAs can base pair with target mRNAs, post-transcriptionally inhibiting mRNA expression through the RNA-induced silencing complex (Gregory et al., 2005). These miRNAs play significant roles in both physiological and pathological processes within the human body, including immunity, inflammation, and oncogenesis (Singh et al., 2013; Zhang et al., 2007). Notably, miRNAs exhibit stable expression in peripheral blood, facilitated by vesicles and protein complexes (Kai et al., 2018; Cheng et al., 2014). Consequently, miRNAs hold promise as potential diagnostic biomarkers for assessing and predicting treatment responses in MG (Huang et al., 2023; Punga and Punga, 2018; Sabre et al., 2020). In this review, we summarize existing research on miRNA profiles and their diagnostic potential in MG.
The MG related autoantibody is currently the specific tool for diagnosing MG. However, the titer of autoantibody cannot be used to predict therapy response of MG. In recent years, due to their ease of detection and stability in serum, miRNAs have increasingly been recognized as novel biomarkers of diseases diagnosis (Kai et al., 2018). In MG patients, dysregulated miRNAs can be released into peripheral blood from thymic tissue or peripheral blood mononuclear cells (PBMCs) (Cron et al., 2019; Witwer, 2015). Several studies have analyzed the miRNA profiles of different types of MG and found some miRNA could be potentially used for diagnosing MG, predicting the therapeutic response and even distinguishing the various MG subtypes (Punga and Punga, 2018; Sabre et al., 2020). In this section, we comprehensively review the miRNA profiles specific to different MG subtypes (Table 1).
Several miRNAs are dysregulated in MG patients (Huang et al., 2023; Wang and Zhang, 2020). In this section, we primarily review miRNA dysregulated generally in MG. The aberrant miRNAs in each MG subtype will be discussed later.
Among these miRNA, miR-150 and miR-21 are most researched in MG. Compared with both healthy individuals and patients with other autoimmune diseases such as Addison’s disease and Crohn’s disease, these two miRNAs are significantly upregulated in serum of MG patients (Punga et al., 2015). In addition, miR-21 is much higher in serum of females. This may, to some extent, explain why MG is more prevalent in women (Fiorillo et al., 2020). Besides that, miR-30e is also found to be unregulated in serum of MG patients, and high level of miR-30e may indicate the progression of MG (Sabre et al., 2019). Meanwhile, another study also showed that MG patient with elevated miR-30e-5p exhibited a higher rate of MG progression (Beretta et al., 2022).
Furthermore, miRNA can also be affected under the influence of treatment and some miRNAs can even predict the treatment response in MG patients. Serum miR-150 and miR-21 decreases after immunosuppressive treatment (Sabre et al., 2018). Low dose rituximab can also decrease serum miR-150-5p, along with improved symptoms and reduced CD19+ and CD27+ B cells (Zhong et al., 2020). Moreover, international research conducted in Italy and Israel indicated that miR-323b-3p and miR-485-3p in whole blood could predict the response to immunosuppressive agents in MG (Cavalcante et al., 2019). The miR-323b-3p and miR-485-3p decreased in MG patients with poor responses to immunosuppressive treatment. Recently, a researcher explored the dysregulated small extracellular vesicle microRNAs in the plasma of pediatric MG patients. Through RNA sequencing technology, researches identified 24 dysregulated small extracellular vesicle microRNAs (16 downregulated and 9 upregulated). Subsequent validation revealed that plasma miR-143-3p is downregulated in seronegative pediatric MG and can be used for diagnosing pediatric MG (Zhu et al., 2024).
EOMG is one of the most common types of MG. EMOG frequently occurs in younger females and is more likely to accompany thymic hyperplasia (Cron et al., 2018; Fang et al., 2015). It has been indicated that ectopic GC formed in hyperplastic thymus plays crucial role in EOMG (Cron et al., 2018).
Many studies identified several dysregulated miRNAs in EOMG patient, including the upregulation of miR-612, miR-3654, pre-miR-3651, and miR-3651 in PBMCs, miR-150 and miR-21 in serum, and the downregulation of miR-122, miR-140-3p, miR-185, miR-192, miR-20, miR-15b and miR-27a in serum (Nogales-Gadea et al., 2014; Barzago et al., 2016; Punga et al., 2014). Some of these dysregulated miRNAs could be affected by the treatment of MG. In peripheral blood, the abnormally upregulated serum miR-150 could dropped quickly after thymectomy and the abnormally decreased serum miR-20 increased after administration of immunosuppressive agents (Punga et al., 2015). Notably, the level of serum miR-20 negatively correlates with the severity of MG (Chunjie et al., 2015). However, despite the upregulation of thymic miR-150 in MG patients, there is no decline in thymic miR-150 after immunosuppressive therapy (Cron et al., 2019).
It should be noted that the changes of dysregulated miRNA could differ considerably in peripheral blood and thymus. Research explored the role of miR-146a in peripheral blood and hyperplastic thymus in MG patients (Bortone et al., 2020). The results indicated that thymic miR-146a decreased in MG patients without receiving corticosteroids and its level increased to normal levels after treated with corticosteroids. Because corticosteroids have been shown to reduce the number and size of ectopic GC in the hyperplastic thymus of MG patients (Berrih-Aknin, 2016), researchers further explored the relationship between miR-146a and GC in thymus. They found that miR-146a in GC-surrounding tissue rather than in GC was remarkably downregulated in MG patients without receiving corticosteroids. However, the miR-146a in both GC and GC-surrounding tissue is upregulated after treated with corticosteroids. These findings may indicate that corticosteroids could induce the upregulation of miR-146a in MG patients. However, in peripheral blood of corticosteroids-naïve patients, miR-146a was downregulated in serum but upregulated in PBMC. The miR-146a in PBMC then decreased to normal after the administration of corticosteroids. These findings indicated that, in peripheral blood, decreased serum miR-146a might be due to a defect in PBMC secretion of miR-146a and corticosteroids could improve this secretion defect.
LOMG is more commonly seen in elderly patients and more likely to accompany thymoma (Fan et al., 2019). Due to its lower incidence compared with EOMG, there is less research focusing on LOMG.
It has been shown that several miRNAs are dysregulated in the serum of LOMG patients, including the upregulation of miR-106, miR-30e, miR-223, miR-140, and miR-19b and downregulation of miR-122, miR-140, miR-185, miR-192, miR-20b, and miR-15b (Sabre et al., 2018; Nogales-Gadea et al., 2014). Some of these could be affected by immunosuppressive therapy and correlate with the severity of MG. In a study comparing the changes in miRNA over 2 years in LOMG patients, the level of serum miR-150, miR-21 and miR-30e decreased along with an improvement in muscle strength after 1 year follow-up (Sabre et al., 2018). Among a subgroup of LOMG patients, serum miR-150 and miR-21 were found to be lower in the ocular group than in the generalized group. In the generalized group, serum miR-30e, miR-21 and miR-19b was decreased after immunosuppressive therapy. In ocular LOMG, only serum miR-140 was decreased after 1 year follow-up. Moreover, serum miR-150, miR-21, and miR-30e were positively correlated with the Myasthenia Gravis Composite (MGC) score in the subgroup of generalized MG. However, these three miRNAs might not have a potential as biomarkers indicating the severity of LOMG, as all the correlation coefficients are less than 0.3.
Despite being a major subtype of MG, MuSK+ MG only occurs in <5% of MG patients (Gilhus and Verschuuren, 2015). It is generally assumed that pathogenesis of MuSK+ MG is largely unrelated to the thymus. Therefore, clinical guideline recommends using immunosuppressants and rituximab, rather than thymectomy, for treating MuSK+ MG without thymic tumors (El-Salem et al., 2014). Consequently, investigators are more interested in the changes of circulating miRNAs in peripheral blood of MuSK+ MG patients.
It has been shown that serum miR-151a, let-7a, let-7f, and miR-423, plasma miR-210 and miR-324, as well as PBMC miR-340, miR-106b, and miR-27a are upregulated in MuSK+ MG patients (Punga et al., 2016; Tan et al., 2021). Among these, exosomal miR-151a in peripheral blood levels reduced quickly after being treated with immunosuppressants (Zhou et al., 2021). However, due to the limited studies on MuSK+ MG, no miRNAs have been found to be associated with the severity of MuSK+ MG currently. Therefore, identifying miRNAs that can predict the therapeutic response of MuSK+ MG still requires more extensive and in-depth research in the future.
MG can be classified based on the extent of involved muscle, including OMG, in which symptoms are limited to the eye muscles, and GMG, which involves muscles throughout the body, including the limbs and trunk. In the progression of OMG, about 80% of patients with OMG progress to secondary GMG (sGMG) (Wong et al., 2014). During this process, the miRNA profile also changes.
It has been revealed that exosomal miR-106a, miR-4712, miR-320d, and miR-3614-3p is decreased in OMG in contrast to healthy controls (Mu et al., 2024). These four miRNAs are further reduced in GMG compared with the OMG and shows promise as a potential biomarker for diagnosing OMG and GMG (AUC > 0.75), despite limited sample size. Another study with a larger sample size demonstrated the serum miR-30e is higher in sGMG compared with OMG, showing promising potential for distinguishing between OMG and sGMG (AUC > 0.9) (Sabre et al., 2019). While researches on OMG and GMG remains limited, these studies have already identified several miRNAs that can be used for diagnosing OMG, as well as for distinguishing between GMG and OMG. The diagnostic efficacy of these miRNAs still requires validation through prospective studies.
Dysregulated miRNAs play an important role in the pathogenesis of MG, influencing various pathological processes (Berrih-Aknin and Le Panse, 2014). These miRNAs impact immune cell development within the thymus and PBMCs. Additionally, they modulate cytokines and transcription factors, potentially contributing to the thymic alterations observed in MG. These alterations encompass impaired negative selection, ectopic GCs, and angiogenesis (Berrih-Aknin and Le Panse, 2014). In this section, we delve into the specific effects of miRNAs on both the thymus and PBMCs (Figure 1A, by Figdraw).
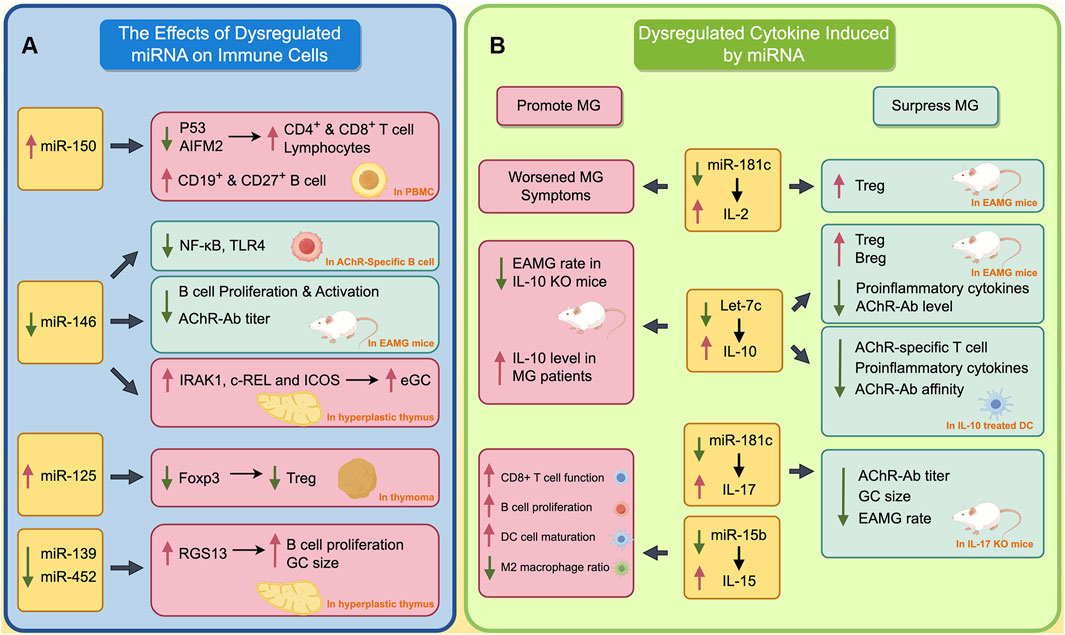
Figure 1. The Impact of Dysregulated miRNAs on Immune Cells and Cytokines in MG. (A) The effects of dysregulated miRNA on immune cells; (B) Dysregulated cytokines induced by miRNA. The red boxes indicate the processes that promote the progression of MG, while green boxes indicate processes that inhibit the progression of MG. A red upward arrow indicates that this process is enhanced, while a green downward arrow indicates that this process is diminished.
miR-150 is the most studied miRNA associated with MG. Normally, miRNA could regulate the expression of multiple transcript factors, such as Batch2 and EGR2, thereby participating in the differentiation of immune cells (Huang et al., 2023; Cron et al., 2020; Cron et al., 2019). It has been found that miR-150 is involved in the differentiation and maturation of T cells, B cells and NK cells (Huang et al., 2015). Due to its significant role in the immune system, the dysregulated regulation of miR-150 is implicated in the development of autoimmune disorders such as MG and multiple sclerosis.
In hyperplastic thymus of MG patients, miR-150 is significantly upregulated and primarily located in the mantle zone of ectopic germinal centers (Cron et al., 2019). In addition, the level of miR-150 was positively correlated with CD19, which is a marker of B cells. In peripheral blood, the level of miR-150 in the PBMCs of MG patients was lower than in healthy controls, while serum miR-150 level was significantly higher than in healthy individual. This is thought to occur due to the release of miR-150 from PBMC into serum under abnormal pathological conditions in peripheral blood of MG patients. Further studies found the inhibition of miR-150 could upregulate the expression of p53 and AIFM2, and thus induce the apoptosis of lymphocytes, CD4+ and CD8+ T cells, which complies with the previously mentioned function of miR-150 in inducing immune cell maturation. Another study found the level of exosomal miR-150 is positively correlated with the level of CD19+B and CD27+B cells (Zhong et al., 2020). This may be attributed to the ability of miR-150 to regulate its target gene c-Myb, thereby influencing the differentiation of B cells into CD19 and CD27 B cells (Xiao et al., 2016).
miR-146 has been identified as a negative regulator of the NF-κB signaling pathway in human immunity (Liu et al., 2015). The activation of miR-146a reduces pro-inflammatory cytokines by inhibiting IRAK1 and TRAF6. In miR-146a deficient mice, NF-κB mediated transcription is abnormally activated (Zhao et al., 2011).
In MG, miR-146a levels are significantly upregulated in PBMC, but downregulated in the hyperplastic thymus (Bortone et al., 2020). This different changing pattern of miR-146a in peripheral and central collectively promotes the progression of MG through different mechanisms. In the mice mode of experimental autoimmune myasthenia gravis (EAMG), inhibition of miR-146 remarkably reduces the expression of TLR4 and NF-κB on AchR specific B cells (Lu et al., 2013). This finding contrasts with previous studies showing the inhibitory effect of miR-146 on the NF-κB signaling pathway, and is assumed to result from a functional deficiency of miR-146a in AchR-specific B cells in MG (Zhao et al., 2011). Another study showed that the inhibition of miR-146 could relieve the myasthenic symptoms of EAMG mice, reduce the production of AChR-Ab, and suppress the proliferation and activation of B cells (Zhang et al., 2015). These two studies demonstrate that, in peripheral blood, miR-146 promotes MG by inducing activation and proliferation of B cell. However, in hyperplastic thymus, downregulated miR-146 may promote the formation of ectopic GC by inducing the expression of IRAK1, c-REL, and ICOS, thereby contributing to the development of MG (Bortone et al., 2020).
As mentioned earlier, dysfunction of immune cells in both thymus and peripheral blood is a characteristic of MG. Apart from miR-150 and miR-146, other miRNAs are also involved in these complicated processes. In TAMG, the abnormally increased miR-125a could inhibit the expression of Foxp3, and thus reduce the Treg cell in thymoma (Li et al., 2016). The defective Treg results in the inability to suppress excessive inflammation in MG, therefore promoting progression of the disease. Similarly, in the hyperplastic thymus of MG patients, the aberrant downregulation of miR-139-5p and miR-452-5p can increase the expression of RGS13, thereby promoting B cell differentiation and increasing the size of ectopic GC (Sengupta et al., 2018).
The abnormally activated inflammatory responses in autoimmune reactions is a crucial pathological process of MG (Huda, 2023). During this process, several miRNAs lead to the aberrant secretion and dysfunction of various cytokines, thus resulting in a hyperactive and inflammatory environment (Berrih-Aknin and Le Panse, 2014). In this section, we will summarize the abnormal cytokines in this process, as well as the miRNAs regulating these cytokines (Figure 1B, by Figdraw).
As IL-2 plays an important role in negative selection in the thymus during T cell maturation and maintaining the proper function of Treg in peripheral, dysregulation of IL-2 can be found in several autoimmune diseases such as SLE, RA, and of course, MG (Li et al., 2022; Kolios et al., 2021). IL-2 plays a complex and diverse regulatory role in MG. It has been found that the serum IL-2 level is significantly upregulated in MG patients, but this increase is not positively correlated with the severity of the disease (Huan et al., 2022). In an EAMG mice model, IL-2 secreted by NKT could expand the Treg cells, and thus relieve the myasthenia symptom of mice (Liu et al., 2005). Furthermore, IL-2/IL-2 mAb complex is found to expand the Treg cells in EAMG mice, and therefore suppress the autoreactive T and B cells, thereby relieving the myasthenia symptom of EAMG mice (Liu et al., 2010). However, another study also found that MG patients whose PBMC have strong capacity to secrete more IL-2 after stimulating, often exhibit more severe generalized myasthenia symptoms (Utsugisawa et al., 2003). This result indicates that IL-2 might promote the development of MG. Previous study showed miR-181c could repress the expression of IL-2 by targeting its 3′UTR. Thus, the downregulation of miR-181c in PBMC of MG patients could induce the expression of IL-2 (Utsugisawa et al., 2003; Zhang et al., 2016).
IL-10 plays a complicated role in progression of MG. Early studies found that serum IL-10 levels are elevated in MG patients, and IL-10 knockout mice have a lower incidence of MG when inducing EAMG models (Poussin et al., 2000; Uzawa et al., 2014). Therefore, IL-10 is thought to promote MG. However, in recent years, studies have found that IL-10 may suppress the development of MG. B cell-derived IL-10 is significantly downregulated in MG patients (Yilmaz et al., 2015). A previous study shows that IL-10-competent B cells increase the proportion of immunoregulatory Treg and Breg cells, reduce the levels of proinflammatory cytokines and AChR-Ab, and therefore alleviate the myasthenic symptom of EAMG mice (Sheng et al., 2015). Furthermore, IL-10-treated DC could reduce the AChR-specific T cell proliferation, level of pro-inflammatory cytokines, and affinity of AChR-Ab (Duan et al., 2004). IL-10 can be regulated by let-7c. The let-7c could bind the 3′UTR of IL-10 and inhibit the expression of IL-10 (Jiang et al., 2012). In MG patients, the let-7c is significantly downregulated, which may contribute to the upregulation of serum IL-10 in MG (Jiang et al., 2012).
IL-17 is thought to be a proinflammatory cytokines. A previous study found that serum IL-17 is unregulated in both AChR+ and MuSK+ MG (Roche et al., 2011; Li et al., 2020). Normally, Treg cells could suppress abnormally overactivated inflammation. However, in the thymus of MG patients, the IL-17 gene family is found to be upregulated, indicating that Treg cells are functionally deficient in MG (Gradolatto et al., 2014; Gradolatto et al., 2012; Villegas et al., 2018). Moreover, it is more difficult to induce EAMG in IL-17 knockout mice compared with wild-type mice (Schaffert et al., 2015). Even in EAMG mode, IL-17 knockout mice exhibit lower levels of myasthenia symptoms and AChR-Ab. The germinal center area in the spleen of IL-17 knockout mice is also smaller (Schaffert et al., 2015; Aguilo-Seara et al., 2017). The aforementioned studies all demonstrate that IL-17 plays a promotive role in the progression of MG. Previous studies have found that miR-15a, miR-320a, and miR-181c are negatively correlated with IL-17 (Zhang et al., 2016; Liu et al., 2016; Cheng et al., 2013). Among these miRNAs, miR-181c could inhibit the expression of IL-17 by targeting its 3′UTR (Zhang et al., 2016). This may indicate that the decrease of miR-181c contributes to the increase in IL-17 to a certain extent in MG patients.
Despite the promotive role that IL-17 plays in MG, it is reported that inhibition of IL-17 by monoclonal antibody could induce MuSK+ MG (Papi et al., 2023). This finding suggests that the role of IL-17 in MG may be complex, and thus the use of IL-17 monoclonal antibodies for MG therapy warrants careful investigation.
The complex pathogenesis of MG is also accompanied by abnormalities in various cytokines, including TNF-α, IL-15, and IL-23. As one of the most well-known pro-inflammatory cytokines, TNF-α is found to be upregulated in both thymus and serum of MG patients (Berrih-Aknin and Le Panse, 2014). Previous studies have indicated that miR-146a may be involved in regulating TNF-α (Nahid et al., 2009). TNF-α is significantly upregulated in miR-146a knockout mice, whereas the mimic of miR-146a was found to inhibit the expression of TNF-α (Yan et al., 2021).
IL-15 is another proinflammatory cytokine that is involved in the regulation of multiple immune cells including enhancing the cytotoxic function of CD8+ T cells, inducing B cell proliferation and immunoglobulin secretion, promoting dendritic cell maturation, and reducing the population of M2 macrophages (Zhang et al., 2021). In MG patients, the serum IL-15 level is significantly upregulated and might correlate with the severity of MG (Li et al., 2019). MiR-15b could inhibit the expression of IL-15 by targeting its 3′UTR (Shi et al., 2015). Thus, the reduction of miR-15b could promote the level of IL-15 in MG.
The pro-inflammatory effect of IL-23 is largely associated with IL-17. In MG, the abnormal thymic epithelial cells (TECs) could secrete IL-23, leading to the activation of Th17 cells and subsequently promoting the release of another pro-inflammatory cytokine IL-17 (Villegas et al., 2019). Though this IL23/Th17 axis, the ectopic GCs can be maintained and lead to persistent inflammatory activation in MG (Villegas et al., 2019). Based on the promotive role that IL-23 plays in MG progression, researchers found that blocking IL-23 can alleviate myasthenia symptoms effectively in EAMG mice (Villegas et al., 2023).
Moreover, miRNA is also involved in the metabolism of drug for MG treatment. CYP3A5 encodes a member of the cytochrome P450 superfamily, playing a crucial role in the metabolism of tacrolimus. A study showed that miR-500a could directly inhibit the expression of CYP3A5, and further experiments demonstrated that this miRNA has the potential to maintain the contraction of tacrolimus in vitro (Meng et al., 2020).
LncRNAs are non-coding RNAs that are more than 200 nucleotides long. circRNAs are circular non-coding RNAs formed by back-splicing of coding and non-coding transcripts (Mattick et al., 2023). Although there is less research on lncRNA and circRNA compared with miRNA in MG, they still play roles in the pathological processes of MG. By high-throughput sequencing, multiple studies have found that lncRNA and circRNA from different samples may play various roles in MG patients. In PBMC, the differentially expressed lncRNAs (DElncRNA) are mainly related to phosphoric ester hydrolase activity and phosphatase activity (Ke et al., 2022). In myotubes treated with AChR-Ab, the DElncRNA is mainly enriched in cellular homeostasis (Hong et al., 2020). In thymoma of TAMG patients, the DElncRNAs were mainly enriched in herpes simplex virus 1 infection, transcription coregulator activity and actin binding (Zhuang et al., 2021). Among these dysregulating lncRNAs and circRNAs, some of them such as IFNG-AS1 in PBMC and circRNA5333-4 in whole blood, are correlated with the severity and AChR-Ab (Lv et al., 2021; Luo et al., 2017).
Within MG pathogenesis, lncRNAs have been found to be primarily involved in the dysregulation of immune cells and the abnormal secretion of cytokines. lncRNA XLOC_003810 is highly expressed in the thymus of MG patients. The upregulated XLOC_003810 can increase the proportion of CD4+T cells, and elevate the levels of pro-inflammatory cytokines, including IFN-γ, TNF-α and IL-1β (Hu et al., 2020). Moreover, XLOC_003810 was also found to disrupt the Treg/Th17 balance by reducing Treg cells and increasing Th17 cells (Niu et al., 2020). Through the aforementioned mechanisms, XLOC_003810 may contribute to abnormal immune activation in MG. Meanwhile, lncRNA GAS5 could upregulate the IL-10 in GMG patients and suppress the Th17 differentiation by targeting miR-23a. Consequently, the abnormal reduced GAS5 in MG promotes the Th17 differentiation, thereby advancing the progression of MG (Peng and Huang, 2022; Xu and Ouyang, 2022). In addition, lncRNA IFNG-AS1 is downregulated in PBMC of MG and IFNG-AS1 level is negatively correlated with MG severity. Further study has found the IFNG-AS1 could regulate the Treg/Th17 balance by increasing the Treg and decreasing the Th17. Furthermore, IFNG-AS1 can upregulate the expression of CD40L and IFN in CD4+ T cells and activate CD4+ T cells in an HLA-DOB- and HLA-DRB1-dependent manner (Luo et al., 2017). The roles of dysregulated lncRNAs in MG are summarized in Figure 2A (by Figdraw) and Table 2.
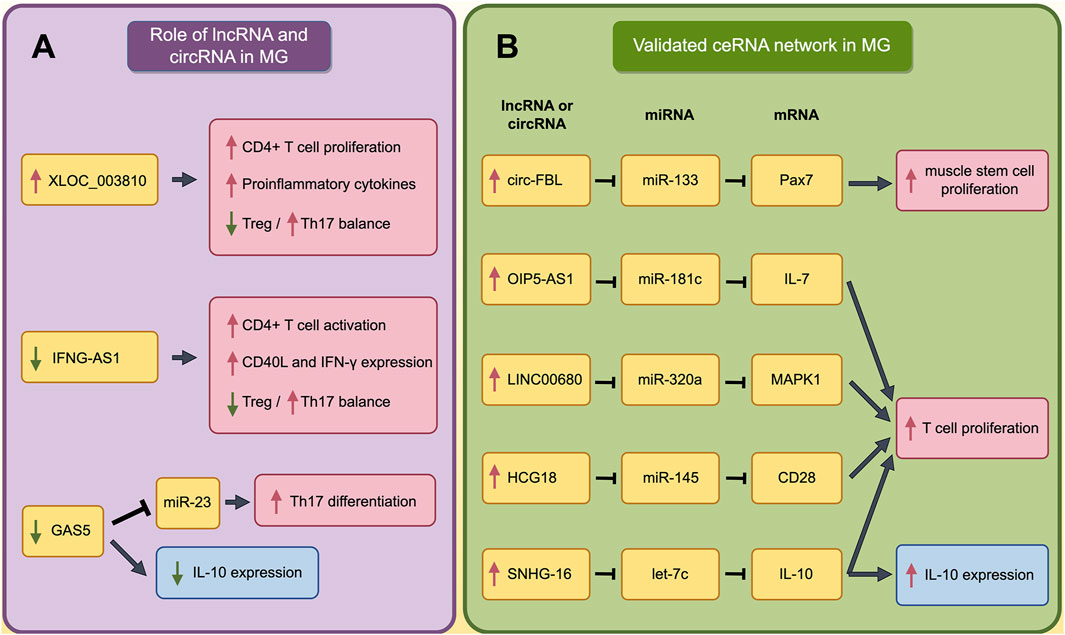
Figure 2. The Impact of Dysregulated lncRNA and validated ceRNA network on Immune Cells and Cytokines in MG. (A) The role of dysregulated lncRNAs and circRNAs in pathogenesis of MG; (B) The validated ceRNA network in pathogenesis of MG. The red boxes indicate the processes that promote the progression of MG, while the blue boxes indicate that these processes play complex roles in the pathogenesis of MG and cannot be simply categorized as promoting or inhibiting MG. A red upward arrow indicates that this process is enhanced, while a green downward arrow indicates that this process is diminished.
By competitively binding to miRNAs, lncRNAs and circRNAs can positively regulate mRNA expression through the competing endogenous RNA (ceRNA) network (Thomson and Dinger, 2016). The ceRNA hypothesis has been shown to be involved in many physiological and pathological processes, such as immune response, inflammation response, tumor development and, of course, MG (Zhang et al., 2020; Wang et al., 2020). Moreover, researchers also introduced several networks such as TF-lncRNA-target gene network and the lncRNA-SNP-mRNA-pathway in MG (Lu et al., 2021; Wang et al., 2021). We would then review the research, focusing on the noncoding RNA-based network to help fully understand the pathological process of MG.
In autoimmune diseases, dysregulated long non-coding RNAs regulate mRNA expression through ceRNA mechanisms, thereby contributing to the progression of autoimmune diseases such as rheumatoid arthritis (RA), dermatomyositis, and systemic lupus erythematosus (SLE) (Wang et al., 2019; Huang et al., 2022; Li et al., 2017). ceRNA networks can be predicted through bioinformatics methods and validated through further experiments (Wang et al., 2020; Wang et al., 2022). In studies involving ceRNA in MG, we found that validation studies mainly focus on the dysregulated ceRNA axis in peripheral tissue of MG patients. In contrast, bioinformatics-predicted ceRNA networks also highlight potential dysregulated ceRNA networks in the thymus and those involving methylated genes.
It has been found the muscle stem cells (as known as satellite cells, SC) proliferated actively in MG patient (Attia et al., 2017). As a marker for SCs, Pax7 expression in MG patients is regulated by ceRNA networks. Researchers have found that the upregulated circ-FBL in MG patients can increase Pax7 levels by competitively binding to miR-133 (Lai et al., 2021). Moreover, other studies have found that several lncRNAs could promote the proliferation of T cells through ceRNA network. LINC00680 is highly expressed in the PBMCs of MG patients, and this lncRNA can upregulate MAPK1 levels by targeting miR-320a (Liu et al., 2022). Similarly, HCG18, which is upregulated in MG patients, can increase CD28 expression by targeting miR-145, and OIP5-AS1 can enhance IL-7 expression by targeting miR-181c. Besides, upregulated SNHG16 promote the expression of IL-10 by sponging let-7c (Wang et al., 2020; Wang et al., 2022; Li et al., 2021). Through these mechanisms, all these four lncRNAs could promote T cell proliferation and contribute to the progression of MG. These processes are summarized in Figure 2B (by Figdraw) and Table 3.
We have previously predicted a ceRNA in thymoma of TAMG by using the RNA-seq data from the TCGA-THYM project (Wang et al., 2022). In the predicted ceRNA network, we further demonstrated that the changes of each component in the LINC00452/miR-204/CHST4 axis comply with the principles of the ceRNA mechanism, and speculate that Treg cells may be regulated by this axis. Another study established two different ceRNA networks based on the methylation level of mRNA and suggested that LINC00173 plays a significant role in the ceRNA network composed of hypomethylated genes (Xu et al., 2021). Additionally, a study predicted a ceRNA network based on dysregulated exosomal lncRNAs (Lu et al., 2021). Researchers found that molecules previously considered to play important roles in MG, such as miR-181c, miR-125, CD28, and CD40, were also arrested in this predicted ceRNA network. This finding provides further evidence that these molecules may be key players in the development of MG.
ceRNA is the most studied and recognized noncoding RNA regulatory mechanism, but some researchers have also explored other modes of networks involving noncoding RNAs in MG. Luo and colleagues constructed the transcription factor-lncRNA-target gene networks in TAMG (Luo et al., 2015). In this network, the transcription factor was thought to regulate gene expression by regulating the lncRNA. This three-element network has mechanisms that are somewhat similar to the ceRNA network. Other research constructed a miRNA-regulated drug-pathway network to predict how the new drug can be for to MG treatment (Cao et al., 2017). Researchers find the alemtuzumab, bevacizumab and efalizumab might have potential to treat MG. However, further validation studies are lacking to support the researchers’ hypotheses.
Currently, detecting noncoding RNA is still not a routine test for MG diagnosing or for treatment response predicting. Although some miRNA showed encouraging potential as a biomarker, the noncoding RNA, especially miRNA, varies from different MG subtypes and different samples (Huang et al., 2023; Sabre et al., 2020). Some miRNA such as miR-146a, showed completely different variation in serum and PBMC before and after treatment (Bortone et al., 2020). Therefore, the specific characteristics of MG such as thymus lesion, autoantibody type and treatment should be carefully concerned when choosing the miRNA as a biomarker in future prospective studies and MG diagnosing.
In addition, although numerous studies have explored the impact of non-coding RNAs and their regulatory networks on the development and progression of MG, RNA processes such as RNA editing and RNA methylation—known to play significant roles in other autoimmune diseases like RA and SLE—are scarcely studied in MG. Previous research has found that adenosine-to-inosine (A-to-I) editing levels are significantly elevated in patients with RA, SLE, and Sjögren’s syndrome (Vlachogiannis et al., 2020; Roth et al., 2018; Wang et al., 2023). Moreover, A-to-I levels significantly decreased in responding RA patients after treatment, indicating that A-to-I editing may play an important role in these autoimmune diseases (Vlachogiannis et al., 2020). However, research on the role of A-to-I editing in MG is currently lacking. In the field of RNA methylation, researchers have found that the levels of m6A RNA methylation in PBMCs and m5C RNA methylation in CD4+ T cells are significantly reduced in SLE patients (Guo et al., 2020; Wu et al., 2022). In MG, researchers have found that patients with different m6A RNA methylation patterns might be associated with varying immune cell infiltration characteristics (Li et al., 2023). But it is regrettable that this study did not measure the m6A RNA methylation levels in MG patients. Therefore, the role of RNA processes in the progression of MG remains to be further explored.
The disturbance of immune homeostasis and abnormally activated inflammation are very important pathological processes in MG (Gilhus, 2016; Berrih-Aknin and Le Panse, 2014; El-Salem et al., 2014). The evidence reviewed above, showing that noncoding RNA can widely influence the both processes by regulating the activation of immune cell, impairing the immunoregulatory functions of Treg and inducing the release of proinflammatory factors (Lu et al., 2013; Li et al., 2016; Zhang et al., 2016). However, the role of noncoding RNA in mechanism behind the TAMG as well as MuSK+ MG is less explored. Further studies are expected to uncover such mechanism for fully understanding the pathologic process of MG.
FW: Writing–original draft, Writing–review and editing. XM: Writing–original draft, Writing–review and editing. YY: Writing–original draft, Writing–review and editing. HZ: Writing–original draft. ZL: Writing–original draft. LZ: Supervision, Writing–review and editing. SD: Supervision, Writing–review and editing. YW: Supervision, Writing–review and editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Science and Technology Department of Sichuan Province Funding Project (Nos. 2023YFS0199 and 2022YFS0091), Chengdu Science and Technology Project (No. 2022-YF05-01833-SN), and Sichuan Cadre Healthcare Research Project (GBKT23020).
We utilized ChatGPT-4o (from OpenAI) to refine some of the sentences in this article. The refined sentences were thoroughly reviewed to ensure that their meaning aligns with our original intent.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Aguilo-Seara, G., Xie, Y., Sheehan, J., Kusner, L. L., and Kaminski, H. J. (2017). Ablation of IL-17 expression moderates experimental autoimmune myasthenia gravis disease severity. Cytokine 96, 279–285. doi:10.1016/j.cyto.2017.05.008
Ashby, K. M., and Hogquist, K. A. (2024). A guide to thymic selection of T cells. Nat. Rev. Immunol. 24 (2), 103–117. doi:10.1038/s41577-023-00911-8
Attia, M., Maurer, M., Robinet, M., Le Grand, F., Fadel, E., Le Panse, R., et al. (2017). Muscle satellite cells are functionally impaired in myasthenia gravis: consequences on muscle regeneration. Acta Neuropathol. 134 (6), 869–888. doi:10.1007/s00401-017-1754-2
Barzago, C., Lum, J., Cavalcante, P., Srinivasan, K. G., Faggiani, E., Camera, G., et al. (2016). A novel infection- and inflammation-associated molecular signature in peripheral blood of myasthenia gravis patients. Immunobiology 221 (11), 1227–1236. doi:10.1016/j.imbio.2016.06.012
Beretta, F., Huang, Y. F., and Punga, A. R. (2022). Towards personalized medicine in myasthenia gravis: role of circulating microRNAs miR-30e-5p, miR-150-5p and miR-21-5p. Cells 11 (4), 740. doi:10.3390/cells11040740
Berrih-Aknin, S. (2016). Role of the thymus in autoimmune myasthenia gravis. Clin. Exp. Neuroimmunol. 7 (3), 226–237. doi:10.1111/cen3.12319
Berrih-Aknin, S., and Le Panse, R. (2014). Myasthenia gravis: a comprehensive review of immune dysregulation and etiological mechanisms. J. Autoimmun. 52, 90–100. doi:10.1016/j.jaut.2013.12.011
Blow, M. J., Grocock, R. J., van Dongen, S., Enright, A. J., Dicks, E., Futreal, P. A., et al. (2006). RNA editing of human microRNAs. Genome Biol. 7 (4), R27. doi:10.1186/gb-2006-7-4-r27
Bortone, F., Scandiffio, L., Marcuzzo, S., Bonanno, S., Frangiamore, R., Motta, T., et al. (2020). miR-146a in myasthenia gravis thymus bridges innate immunity with autoimmunity and is linked to therapeutic effects of corticosteroids. Front. Immunol. 11, 142. doi:10.3389/fimmu.2020.00142
Cai, Y., Yu, X., Hu, S., and Yu, J. (2009). A brief review on the mechanisms of miRNA regulation. Genomics Proteomics Bioinforma. 7 (4), 147–154. doi:10.1016/S1672-0229(08)60044-3
Cao, Y., Lu, X., Wang, J., Zhang, H., Liu, Z., Xu, S., et al. (2017). Construction of an miRNA-regulated drug-pathway network reveals drug repurposing candidates for myasthenia gravis. Int. J. Mol. Med. 39 (2), 268–278. doi:10.3892/ijmm.2017.2853
Cavalcante, P., Mizrachi, T., Barzago, C., Scandiffio, L., Bortone, F., Bonanno, S., et al. (2019). MicroRNA signature associated with treatment response in myasthenia gravis: a further step towards precision medicine. Pharmacol. Res. 148, 104388. doi:10.1016/j.phrs.2019.104388
Cheng, L., Sharples, R. A., Scicluna, B. J., and Hill, A. F. (2014). Exosomes provide a protective and enriched source of miRNA for biomarker profiling compared to intracellular and cell-free blood. J. Extracell. Vesicles 3. doi:10.3402/jev.v3.23743
Cheng, Z., Qiu, S., Jiang, L., Zhang, A., Bao, W., Liu, P., et al. (2013). MiR-320a is downregulated in patients with myasthenia gravis and modulates inflammatory cytokines production by targeting mitogen-activated protein kinase 1. J. Clin. Immunol. 33 (3), 567–576. doi:10.1007/s10875-012-9834-5
Chunjie, N., Huijuan, N., Zhao, Y., Jianzhao, W., and Xiaojian, Z. (2015). Disease-specific signature of serum miR-20b and its targets IL-8 and IL-25, in myasthenia gravis patients. Eur. Cytokine Netw. 26 (3), 61–66. doi:10.1684/ecn.2015.0367
Cron, M. A., Guillochon, É., Kusner, L., and Le Panse, R. (2020). Role of miRNAs in normal and myasthenia gravis thymus. Front. Immunol. 11, 1074. doi:10.3389/fimmu.2020.01074
Cron, M. A., Maillard, S., Truffault, F., Gualeni, A. V., Gloghini, A., Fadel, E., et al. (2019). Causes and consequences of miR-150-5p dysregulation in myasthenia gravis. Front. Immunol. 10, 539. doi:10.3389/fimmu.2019.00539
Cron, M. A., Maillard, S., Villegas, J., Truffault, F., Sudres, M., Dragin, N., et al. (2018). Thymus involvement in early-onset myasthenia gravis. Ann. N. Y. Acad. Sci. 1412 (1), 137–145. doi:10.1111/nyas.13519
Duan, R. S., Adikari, S. B., Huang, Y. M., Link, H., and Xiao, B. G. (2004). Protective potential of experimental autoimmune myasthenia gravis in Lewis rats by IL-10-modified dendritic cells. Neurobiol. Dis. 16 (2), 461–467. doi:10.1016/j.nbd.2004.03.017
El-Salem, K., Yassin, A., Al-Hayk, K., Yahya, S., Al-Shorafat, D., and Dahbour, S. S. (2014). Treatment of MuSK-associated myasthenia gravis. Curr. Treat. Options Neurol. 16 (4), 283. doi:10.1007/s11940-014-0283-8
Fan, L., Ma, S., Yang, Y., Yan, Z., Li, J., and Li, Z. (2019). Clinical differences of early and late-onset myasthenia gravis in 985 patients. Neurol. Res. 41 (1), 45–51. doi:10.1080/01616412.2018.1525121
Fang, F., Sveinsson, O., Thormar, G., Granqvist, M., Askling, J., Lundberg, I. E., et al. (2015). The autoimmune spectrum of myasthenia gravis: a Swedish population-based study. J. Intern Med. 277 (5), 594–604. doi:10.1111/joim.12310
Fiorillo, A. A., Heier, C. R., Huang, Y. F., Tully, C. B., Punga, T., and Punga, A. R. (2020). Estrogen receptor, inflammatory, and FOXO transcription factors regulate expression of myasthenia gravis-associated circulating microRNAs. Front. Immunol. 11, 151. doi:10.3389/fimmu.2020.00151
Gilhus, N. E. (2016). Myasthenia gravis. N. Engl. J. Med. 375 (26), 2570–2581. doi:10.1056/NEJMra1602678
Gilhus, N. E., and Verschuuren, J. J. (2015). Myasthenia gravis: subgroup classification and therapeutic strategies. Lancet Neurol. 14 (10), 1023–1036. doi:10.1016/S1474-4422(15)00145-3
Gradolatto, A., Nazzal, D., Foti, M., Bismuth, J., Truffault, F., Le Panse, R., et al. (2012). Defects of immunoregulatory mechanisms in myasthenia gravis: role of IL-17. Ann. N. Y. Acad. Sci. 1274, 40–47. doi:10.1111/j.1749-6632.2012.06791.x
Gradolatto, A., Nazzal, D., Truffault, F., Bismuth, J., Fadel, E., Foti, M., et al. (2014). Both Treg cells and Tconv cells are defective in the Myasthenia gravis thymus: roles of IL-17 and TNF-α. J. Autoimmun. 52, 53–63. doi:10.1016/j.jaut.2013.12.015
Gregory, R. I., Chendrimada, T. P., Cooch, N., and Shiekhattar, R. (2005). Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell 123 (4), 631–640. doi:10.1016/j.cell.2005.10.022
Guo, G., Wang, H., Shi, X., Ye, L., Yan, K., Chen, Z., et al. (2020). Disease activity-associated alteration of mRNA m(5) C methylation in CD4(+) T cells of systemic lupus erythematosus. Front. Cell Dev. Biol. 8, 430. doi:10.3389/fcell.2020.00430
Hong, Y., Liang, X., and Gilhus, N. E. (2020). AChR antibodies show a complex interaction with human skeletal muscle cells in a transcriptomic study. Sci. Rep. 10 (1), 11230. doi:10.1038/s41598-020-68185-x
Hu, B., Niu, L., Jiang, Z., Xu, S., Hu, Y., and Cao, K. (2020). LncRNA XLOC_003810 promotes T cell activation and inhibits PD-1/PD-L1 expression in patients with myasthenia gravis-related thymoma. Scand. J. Immunol. 92 (1), e12886. doi:10.1111/sji.12886
Huan, X., Zhao, R., Song, J., Zhong, H., Su, M., Yan, C., et al. (2022). Increased serum IL-2, IL-4, IL-5 and IL-12p70 levels in AChR subtype generalized myasthenia gravis. BMC Immunol. 23 (1), 26. doi:10.1186/s12865-022-00501-8
Huang, P., Tang, L., Zhang, L., Ren, Y., Peng, H., Xiao, Y., et al. (2022). Identification of biomarkers associated with CD4(+) T-cell infiltration with gene coexpression network in dermatomyositis. Front. Immunol. 13, 854848. doi:10.3389/fimmu.2022.854848
Huang, X., Zhang, Z., Wang, Y., Xu, M., Du, X., and Zhang, Y. (2023). Circulating miRNAs drive personalized medicine based on subgroup classification in myasthenia gravis patients. Neurol. Sci. 44 (11), 3877–3884. doi:10.1007/s10072-023-06933-3
Huang, X. L., Zhang, L., Li, J. P., Wang, Y. J., Duan, Y., and Wang, J. (2015). MicroRNA-150: a potential regulator in pathogens infection and autoimmune diseases. Autoimmunity 48 (8), 503–510. doi:10.3109/08916934.2015.1072518
Huda, R. (2023). Inflammation and autoimmune myasthenia gravis. Front. Immunol. 14, 1110499. doi:10.3389/fimmu.2023.1110499
Jiang, L., Cheng, Z., Qiu, S., Que, Z., Bao, W., Jiang, C., et al. (2012). Altered let-7 expression in Myasthenia gravis and let-7c mediated regulation of IL-10 by directly targeting IL-10 in Jurkat cells. Int. Immunopharmacol. 14 (2), 217–223. doi:10.1016/j.intimp.2012.07.003
Kai, K., Dittmar, R. L., and Sen, S. (2018). Secretory microRNAs as biomarkers of cancer. Semin. Cell Dev. Biol. 78, 22–36. doi:10.1016/j.semcdb.2017.12.011
Ke, J., Du, X., Cui, J., Yu, L., and Li, H. (2022). LncRNA and mRNA expression associated with myasthenia gravis in patients with thymoma. Thorac. Cancer 13 (1), 15–23. doi:10.1111/1759-7714.14201
Kolios, A. G. A., Tsokos, G. C., and Klatzmann, D. (2021). Interleukin-2 and regulatory T cells in rheumatic diseases. Nat. Rev. Rheumatol. 17 (12), 749–766. doi:10.1038/s41584-021-00707-x
Lai, X., Bi, Z., Yang, X., Hu, R., Wang, L., Jin, M., et al. (2021). Upregulation of circ-FBL promotes myogenic proliferation in myasthenia gravis by regulation of miR-133/PAX7. Cell Biol. Int. 45 (11), 2287–2293. doi:10.1002/cbin.11676
Li, H., Boulougoura, A., Endo, Y., and Tsokos, G. C. (2022). Abnormalities of T cells in systemic lupus erythematosus: new insights in pathogenesis and therapeutic strategies. J. Autoimmun. 132, 102870. doi:10.1016/j.jaut.2022.102870
Li, J., Qiu, D., Chen, Z., Du, W., Liu, J., and Mo, X. (2016). Altered expression of miR-125a-5p in thymoma-associated myasthenia gravis and its down-regulation of foxp3 expression in Jurkat cells. Immunol. Lett. 172, 47–55. doi:10.1016/j.imlet.2016.02.005
Li, L. J., Zhao, W., Tao, S. S., Leng, R. X., Fan, Y. G., Pan, H. F., et al. (2017). Competitive endogenous RNA network: potential implication for systemic lupus erythematosus. Expert Opin. Ther. Targets 21 (6), 639–648. doi:10.1080/14728222.2017.1319938
Li, S., Liu, H., Ruan, Z., Guo, R., Sun, C., Tang, Y., et al. (2023). Landscape analysis of m6A modification regulators related biological functions and immune characteristics in myasthenia gravis. J. Transl. Med. 21 (1), 166. doi:10.1186/s12967-023-03947-5
Li, S., Wang, X., Wang, T., Zhang, H., Lu, X., Liu, L., et al. (2021). Identification of the regulatory role of lncRNA HCG18 in myasthenia gravis by integrated bioinformatics and experimental analyses. J. Transl. Med. 19 (1), 468. doi:10.1186/s12967-021-03138-0
Li, Y., Guptill, J. T., Russo, M. A., Howard, J. F., Massey, J. M., Juel, V. C., et al. (2020). Imbalance in T follicular helper cells producing IL-17 promotes pro-inflammatory responses in MuSK antibody positive myasthenia gravis. J. Neuroimmunol. 345, 577279. doi:10.1016/j.jneuroim.2020.577279
Liu, L., Zhang, H., Lu, X., Li, L., Wang, T., Li, S., et al. (2022). LncRNA LINC00680 acts as a competing endogenous RNA and is associated with the severity of myasthennia gravis. Front. Neurol. 13, 833062. doi:10.3389/fneur.2022.833062
Liu, R., La Cava, A., Bai, X. F., Jee, Y., Price, M., Campagnolo, D. I., et al. (2005). Cooperation of invariant NKT cells and CD4+CD25+ T regulatory cells in the prevention of autoimmune myasthenia. J. Immunol. 175 (12), 7898–7904. doi:10.4049/jimmunol.175.12.7898
Liu, R., Liu, C., Chen, D., Yang, W. H., Liu, X., Liu, C. G., et al. (2015). FOXP3 controls an miR-146/NF-κB negative feedback loop that inhibits apoptosis in breast cancer cells. Cancer Res. 75 (8), 1703–1713. doi:10.1158/0008-5472.CAN-14-2108
Liu, R., Zhou, Q., La Cava, A., Campagnolo, D. I., Van Kaer, L., and Shi, F. D. (2010). Expansion of regulatory T cells via IL-2/anti-IL-2 mAb complexes suppresses experimental myasthenia. Eur. J. Immunol. 40 (6), 1577–1589. doi:10.1002/eji.200939792
Liu, X. F., Wang, R. Q., Hu, B., Luo, M. C., Zeng, Q. M., Zhou, H., et al. (2016). MiR-15a contributes abnormal immune response in myasthenia gravis by targeting CXCL10. Clin. Immunol. 164, 106–113. doi:10.1016/j.clim.2015.12.009
Li, X., Lu, F., Li, W., Zhao, D., Li, X., Zhang, J., et al. (2019). The correlation of the levels of interleukin-15 with late onset myasthenia gravis. Chin. J. Neurology, 446–451. doi:10.3760/cma.j.issn.1006-7876.2019.06.002
Lu, J., Yan, M., Wang, Y., Zhang, J., Yang, H., Tian, F. F., et al. (2013). Altered expression of miR-146a in myasthenia gravis. Neurosci. Lett. 555, 85–90. doi:10.1016/j.neulet.2013.09.014
Lu, W., Lu, Y., Wang, C. F., and Chen, T. T. (2021). Expression profiling and bioinformatics analysis of exosomal long noncoding RNAs in patients with myasthenia gravis by RNA sequencing. J. Clin. Lab. Anal. 35 (4), e23722. doi:10.1002/jcla.23722
Luo, M., Liu, X., Meng, H., Xu, L., Li, Y., Li, Z., et al. (2017). IFNA-AS1 regulates CD4(+) T cell activation in myasthenia gravis though HLA-DRB1. Clin. Immunol. 183, 121–131. doi:10.1016/j.clim.2017.08.008
Luo, Z., Li, Y., Liu, X., Luo, M., Xu, L., Luo, Y., et al. (2015). Systems biology of myasthenia gravis, integration of aberrant lncRNA and mRNA expression changes. BMC Med. Genomics 8, 13. doi:10.1186/s12920-015-0087-z
Lv, J., Ren, L., Han, S., Zhang, J., Zhao, X., Zhang, Y., et al. (2021). Peripheral blood hsa-circRNA5333-4: a novel biomarker for myasthenia gravis. Clin. Immunol. 224, 108676. doi:10.1016/j.clim.2021.108676
Mattick, J. S., Amaral, P. P., Carninci, P., Carpenter, S., Chang, H. Y., Chen, L. L., et al. (2023). Long non-coding RNAs: definitions, functions, challenges and recommendations. Nat. Rev. Mol. Cell Biol. 24 (6), 430–447. doi:10.1038/s41580-022-00566-8
Meng, H. Y., Li, X., Jin, W. L., Yan, C. K., Dong, X. H., Xu, Q., et al. (2020). Multiple genetic factors affecting the pharmacokinetic and pharmacodynamic processes of tacrolimus in Chinese myasthenia gravis patients. Eur. J. Clin. Pharmacol. 76 (5), 659–671. doi:10.1007/s00228-019-02803-0
Monsul, N. T., Patwa, H. S., Knorr, A. M., Lesser, R. L., and Goldstein, J. M. (2004). The effect of prednisone on the progression from ocular to generalized myasthenia gravis. J. Neurol. Sci. 217 (2), 131–133. doi:10.1016/j.jns.2003.08.017
Mu, Y., Huang, X., Yang, Y., Huang, Z., Chen, J., Li, S., et al. (2024). Study of serum exosome miRNA as a biomarker for early onset adult ouclar myastthenia gravis. Gene 896, 148034. doi:10.1016/j.gene.2023.148034
Nahid, M. A., Pauley, K. M., Satoh, M., and Chan, E. K. L. (2009). miR-146a is critical for endotoxin-induced tolerance: implication in innate immunity. J. Biol. Chem. 284 (50), 34590–34599. doi:10.1074/jbc.M109.056317
Niu, L., Jiang, J., Yin, Y., and Hu, B. (2020). LncRNA XLOC_003810 modulates thymic Th17/Treg balance in myasthenia gravis with thymoma. Clin. Exp. Pharmacol. Physiol. 47 (6), 989–996. doi:10.1111/1440-1681.13280
Nogales-Gadea, G., Ramos-Fransi, A., Suárez-Calvet, X., Navas, M., Rojas-García, R., Mosquera, J. L., et al. (2014). Analysis of serum miRNA profiles of myasthenia gravis patients. PLoS One 9 (3), e91927. doi:10.1371/journal.pone.0091927
Papi, C., Granata, G., Galluzzo, M., and Iorio, R. (2023). Myasthenia gravis associated with muscle-specific kinase antibodies in a patient treated with interleukin-17 inhibitor. Neurol. Sci. 44 (3), 1105–1107. doi:10.1007/s10072-022-06498-7
Peng, S., and Huang, Y. (2022). LncRNA GAS5 positively regulates IL-10 expression in patients with generalized myasthenia gravis. Brain Behav. 12 (1), e2457. doi:10.1002/brb3.2457
Poussin, M. A., Goluszko, E., Hughes, T. K., Duchicella, S. I., and Christadoss, P. (2000). Suppression of experimental autoimmune myasthenia gravis in IL-10 gene-disrupted mice is associated with reduced B cells and serum cytotoxicity on mouse cell line expressing AChR. J. Neuroimmunol. 111 (1-2), 152–160. doi:10.1016/s0165-5728(00)00385-4
Punga, A. R., Andersson, M., Alimohammadi, M., and Punga, T. (2015). Disease specific signature of circulating miR-150-5p and miR-21-5p in myasthenia gravis patients. J. Neurol. Sci. 356 (1-2), 90–96. doi:10.1016/j.jns.2015.06.019
Punga, A. R., and Punga, T. (2018). Circulating microRNAs as potential biomarkers in myasthenia gravis patients. Ann. N. Y. Acad. Sci. 1412 (1), 33–40. doi:10.1111/nyas.13510
Punga, T., Bartoccioni, E., Lewandowska, M., Damato, V., Evoli, A., and Punga, A. R. (2016). Disease specific enrichment of circulating let-7 family microRNA in MuSK+ myasthenia gravis. J. Neuroimmunol. 292, 21–26. doi:10.1016/j.jneuroim.2016.01.003
Punga, T., Le Panse, R., Andersson, M., Truffault, F., Berrih-Aknin, S., and Punga, A. R. (2014). Circulating miRNAs in myasthenia gravis: miR-150-5p as a new potential biomarker. Ann. Clin. Transl. Neurol. 1 (1), 49–58. doi:10.1002/acn3.24
Roche, J. C., Capablo, J. L., Larrad, L., Gervas-Arruga, J., Ara, J. R., Sánchez, A., et al. (2011). Increased serum interleukin-17 levels in patients with myasthenia gravis. Muscle Nerve 44 (2), 278–280. doi:10.1002/mus.22070
Roth, S. H., Danan-Gotthold, M., Ben-Izhak, M., Rechavi, G., Cohen, C. J., Louzoun, Y., et al. (2018). Increased RNA editing may provide a source for autoantigens in systemic lupus erythematosus. Cell Rep. 23 (1), 50–57. doi:10.1016/j.celrep.2018.03.036
Sabre, L., Maddison, P., Sadalage, G., Ambrose, P. A., and Punga, A. R. (2018). Circulating microRNA miR-21-5p, miR-150-5p and miR-30e-5p correlate with clinical status in late onset myasthenia gravis. J. Neuroimmunol. 321, 164–170. doi:10.1016/j.jneuroim.2018.05.003
Sabre, L., Maddison, P., Wong, S. H., Sadalage, G., Ambrose, P. A., Plant, G. T., et al. (2019). miR-30e-5p as predictor of generalization in ocular myasthenia gravis. Ann. Clin. Transl. Neurol. 6 (2), 243–251. doi:10.1002/acn3.692
Sabre, L., Punga, T., and Punga, A. R. (2020). Circulating miRNAs as potential biomarkers in myasthenia gravis: tools for personalized medicine. Front. Immunol. 11, 213. doi:10.3389/fimmu.2020.00213
Schaffert, H., Pelz, A., Saxena, A., Losen, M., Meisel, A., Thiel, A., et al. (2015). IL-17-producing CD4(+) T cells contribute to the loss of B-cell tolerance in experimental autoimmune myasthenia gravis. Eur. J. Immunol. 45 (5), 1339–1347. doi:10.1002/eji.201445064
Sengupta, M., Wang, B. D., Lee, N. H., Marx, A., Kusner, L. L., and Kaminski, H. J. (2018). MicroRNA and mRNA expression associated with ectopic germinal centers in thymus of myasthenia gravis. PLoS One 13 (10), e0205464. doi:10.1371/journal.pone.0205464
Sheng, J. R., Quan, S., and Soliven, B. (2015). IL-10 derived from CD1dhiCD5⁺ B cells regulates experimental autoimmune myasthenia gravis. J. Neuroimmunol. 289, 130–138. doi:10.1016/j.jneuroim.2015.10.023
Shi, L., Liu, T., Zhang, M., Guo, Y., Song, C., Song, D., et al. (2015). miR-15b is downregulated in myasthenia gravis patients and directly regulates the expression of interleukin-15 (IL-15) in experimental myasthenia gravis mice. Med. Sci. Monit. 21, 1774–1780. doi:10.12659/MSM.893458
Singh, R. P., Massachi, I., Manickavel, S., Singh, S., Rao, N. P., Hasan, S., et al. (2013). The role of miRNA in inflammation and autoimmunity. Autoimmun. Rev. 12 (12), 1160–1165. doi:10.1016/j.autrev.2013.07.003
Skeie, G. O., Aarli, J. A., and Gilhus, N. E. (2006). Titin and ryanodine receptor antibodies in myasthenia gravis. Acta Neurol. Scand. Suppl. 183, 19–23. doi:10.1111/j.1600-0404.2006.00608.x
Sprent, J., and Kishimoto, H. (2002). The thymus and negative selection. Immunol. Rev. 185, 126–135. doi:10.1034/j.1600-065x.2002.18512.x
Szczudlik, P., Szyluk, B., Lipowska, M., Ryniewicz, B., Kubiszewska, J., Dutkiewicz, M., et al. (2014). Antititin antibody in early- and late-onset myasthenia gravis. Acta Neurol. Scand. 130 (4), 229–233. doi:10.1111/ane.12271
Tan, Y., Zhu, L., Cui, L., and Guan, Y. (2021). Differential expression of miRNA in the peripheral blood mononuclear cells in myasthenia gravis with muscle-specific receptor tyrosine kinase antibodies. Crit. Rev. Eukaryot. Gene Expr. 31 (2), 1–15. doi:10.1615/CritRevEukaryotGeneExpr.2021037369
Thomson, D. W., and Dinger, M. E. (2016). Endogenous microRNA sponges: evidence and controversy. Nat. Rev. Genet. 17 (5), 272–283. doi:10.1038/nrg.2016.20
Utsugisawa, K., Nagane, Y., Obara, D., Kondoh, R., Yonezawa, H., and Tohgi, H. (2003). Interleukin-2 production by peripheral blood mononuclear cells from patients with myasthenia gravis. Eur. Neurol. 49 (3), 160–163. doi:10.1159/000069079
Uzawa, A., Kawaguchi, N., Himuro, K., Kanai, T., and Kuwabara, S. (2014). Serum cytokine and chemokine profiles in patients with myasthenia gravis. Clin. Exp. Immunol. 176 (2), 232–237. doi:10.1111/cei.12272
Villegas, J. A., Bayer, A. C., Ider, K., Bismuth, J., Truffault, F., Roussin, R., et al. (2019). Il-23/Th17 cell pathway: a promising target to alleviate thymic inflammation maintenance in myasthenia gravis. J. Autoimmun. 98, 59–73. doi:10.1016/j.jaut.2018.11.005
Villegas, J. A., Van Wassenhove, J., Le Panse, R., Berrih-Aknin, S., and Dragin, N. (2018). An imbalance between regulatory T cells and T helper 17 cells in acetylcholine receptor-positive myasthenia gravis patients. Ann. N. Y. Acad. Sci. 1413 (1), 154–162. doi:10.1111/nyas.13591
Villegas, J. A., Van Wassenhove, J., Merrheim, J., Matta, K., Hamadache, S., Flaugère, C., et al. (2023). Blocking interleukin-23 ameliorates neuromuscular and thymic defects in myasthenia gravis. J. Neuroinflammation 20 (1), 9. doi:10.1186/s12974-023-02691-3
Vlachogiannis, N. I., Gatsiou, A., Silvestris, D. A., Stamatelopoulos, K., Tektonidou, M. G., Gallo, A., et al. (2020). Increased adenosine-to-inosine RNA editing in rheumatoid arthritis. J. Autoimmun. 106, 102329. doi:10.1016/j.jaut.2019.102329
Wang, F., Zhang, H., Qiu, G., Li, Z., and Wang, Y. (2022a). The linc00452/miR-204/CHST4 Axis regulating thymic tregs might Be involved in the progression of thymoma-associated myasthenia gravis. Front. Neurol. 13, 828970. doi:10.3389/fneur.2022.828970
Wang, J., Cao, Y., Lu, X., Wang, X., Kong, X., Bo, C., et al. (2020). Identification of the regulatory role of lncRNA SNHG16 in myasthenia gravis by constructing a competing endogenous RNA network. Mol. Ther. Nucleic Acids 19, 1123–1133. doi:10.1016/j.omtn.2020.01.005
Wang, J., Yan, S., Yang, J., Lu, H., Xu, D., and Wang, Z. (2019). Non-coding RNAs in rheumatoid arthritis: from bench to bedside. Front. Immunol. 10, 3129. doi:10.3389/fimmu.2019.03129
Wang, L., and Zhang, L. (2020). Emerging roles of dysregulated MicroRNAs in myasthenia gravis. Front. Neurosci. 14, 507. doi:10.3389/fnins.2020.00507
Wang, T., Xu, S., Zhang, H., Lu, X., Li, S., Liu, L., et al. (2021). Competitive endogenous RNA network and pathway-based analysis of LncRNA single-nucleotide polymorphism in myasthenia gravis. Sci. Rep. 11 (1), 23920. doi:10.1038/s41598-021-03357-x
Wang, X., Zhang, H., Lu, X., Li, S., Kong, X., Liu, L., et al. (2022b). LncRNA OIP5-AS1 modulates the proliferation and apoptosis of Jurkat cells by sponging miR-181c-5p to regulate IL-7 expression in myasthenia gravis. PeerJ 10, e13454. doi:10.7717/peerj.13454
Wang, X., Zhu, L., Ying, S., Liao, X., Zheng, J., Liu, Z., et al. (2023). Increased RNA editing sites revealed as potential novel biomarkers for diagnosis in primary Sjögren’s syndrome. J. Autoimmun. 138, 103035. doi:10.1016/j.jaut.2023.103035
Witwer, K. W. (2015). Circulating microRNA biomarker studies: pitfalls and potential solutions. Clin. Chem. 61 (1), 56–63. doi:10.1373/clinchem.2014.221341
Wong, S. H., Huda, S., Vincent, A., and Plant, G. T. (2014). Ocular myasthenia gravis: controversies and updates. Curr. Neurol. Neurosci. Rep. 14 (1), 421. doi:10.1007/s11910-013-0421-9
Wu, J., Deng, L. J., Xia, Y. R., Leng, R. X., Fan, Y. G., Pan, H. F., et al. (2022). Involvement of N6-methyladenosine modifications of long noncoding RNAs in systemic lupus erythematosus. Mol. Immunol. 143, 77–84. doi:10.1016/j.molimm.2022.01.006
Xiao, C., Calado, D. P., Galler, G., Thai, T. H., Patterson, H. C., Wang, J., et al. (2016). MiR-150 controls B cell differentiation by targeting the transcription factor c-myb. Cell 165 (4), 1027. doi:10.1016/j.cell.2016.04.056
Xu, S., Wang, T., Lu, X., Zhang, H., Liu, L., Kong, X., et al. (2021). Identification of LINC00173 in myasthenia gravis by integration analysis of aberrantly methylated- differentially expressed genes and ceRNA networks. Front. Genet. 12, 726751. doi:10.3389/fgene.2021.726751
Xu, Y., and Ouyang, Y. (2022). Long non-coding RNA growth arrest specific 5 regulates the T helper 17/regulatory T balance by targeting miR-23a in myasthenia gravis. J. Int. Med. Res. 50 (6), 3000605211053703. doi:10.1177/03000605211053703
Yan, F., Wufuer, D., Ding, J., and Wang, J. (2021). MicroRNA miR-146a-5p inhibits the inflammatory response and injury of airway epithelial cells via targeting TNF receptor-associated factor 6. Bioengineered 12 (1), 1916–1926. doi:10.1080/21655979.2021.1927545
Yilmaz, V., Oflazer, P., Aysal, F., Parman, Y. G., Direskeneli, H., Deymeer, F., et al. (2015). B cells produce less IL-10, IL-6 and TNF-α in myasthenia gravis. Autoimmunity 48 (4), 201–207. doi:10.3109/08916934.2014.992517
Yoganathan, K., Stevenson, A., Tahir, A., Sadler, R., Radunovic, A., and Malek, N. (2022). Bedside and laboratory diagnostic testing in myasthenia. J. Neurol. 269 (6), 3372–3384. doi:10.1007/s00415-022-10986-3
You, C., He, W., Hang, R., Zhang, C., Cao, X., Guo, H., et al. (2019). FIERY1 promotes microRNA accumulation by suppressing rRNA-derived small interfering RNAs in Arabidopsis. Nat. Commun. 10 (1), 4424. doi:10.1038/s41467-019-12379-z
Zhang, B., Pan, X., Cobb, G. P., and Anderson, T. A. (2007). microRNAs as oncogenes and tumor suppressors. Dev. Biol. 302 (1), 1–12. doi:10.1016/j.ydbio.2006.08.028
Zhang, J., Jia, G., Liu, Q., Hu, J., Yan, M., Yang, B., et al. (2015). Silencing miR-146a influences B cells and ameliorates experimental autoimmune myasthenia gravis. Immunology 144 (1), 56–67. doi:10.1111/imm.12347
Zhang, K., Zhang, L., Mi, Y., Tang, Y., Ren, F., Liu, B., et al. (2020). A ceRNA network and a potential regulatory axis in gastric cancer with different degrees of immune cell infiltration. Cancer Sci. 111 (11), 4041–4050. doi:10.1111/cas.14634
Zhang, L., Liu, J., and Hou, Y. (2023). Classification, function, and advances in tsRNA in non-neoplastic diseases. Cell Death Dis. 14 (11), 748. doi:10.1038/s41419-023-06250-9
Zhang, S., Zhao, J., Bai, X., Handley, M., and Shan, F. (2021). Biological effects of IL-15 on immune cells and its potential for the treatment of cancer. Int. Immunopharmacol. 91, 107318. doi:10.1016/j.intimp.2020.107318
Zhang, Y., Guo, M., Xin, N., Shao, Z., Zhang, X., Zhang, Y., et al. (2016). Decreased microRNA miR-181c expression in peripheral blood mononuclear cells correlates with elevated serum levels of IL-7 and IL-17 in patients with myasthenia gravis. Clin. Exp. Med. 16 (3), 413–421. doi:10.1007/s10238-015-0358-1
Zhao, J. L., Rao, D. S., Boldin, M. P., Taganov, K. D., O'Connell, R. M., and Baltimore, D. (2011). NF-kappaB dysregulation in microRNA-146a-deficient mice drives the development of myeloid malignancies. Proc. Natl. Acad. Sci. U. S. A. 108 (22), 9184–9189. doi:10.1073/pnas.1105398108
Zhong, H., Lu, J., Jing, S., Xi, J., Yan, C., Song, J., et al. (2020). Low-dose rituximab lowers serum Exosomal miR-150-5p in AChR-positive refractory myasthenia gravis patients. J. Neuroimmunol. 348, 577383. doi:10.1016/j.jneuroim.2020.577383
Zhou, Y., Yan, C., Gu, X., Zhou, L., Lu, J., Zhu, W., et al. (2021). Short-term effect of low-dose rituximab on myasthenia gravis with muscle-specific tyrosine kinase antibody. Muscle Nerve 63 (6), 824–830. doi:10.1002/mus.27233
Zhu, M., Wang, Y., Xu, X., Guo, X., Mao, Y., and Gao, F. (2024). Small extracellular vesicle microRNAs in pediatric myasthenia gravis plasma and skeletal muscle. Postgrad. Med. J. 100 (1185), 488–495. doi:10.1093/postmj/qgae015
Keywords: autoimmune disease, Myasthenia Gravis, non-coding RNA, ceRNA, regulatory network
Citation: Wang F, Mei X, Yang Y, Zhang H, Li Z, Zhu L, Deng S and Wang Y (2024) Non-coding RNA and its network in the pathogenesis of Myasthenia Gravis. Front. Mol. Biosci. 11:1388476. doi: 10.3389/fmolb.2024.1388476
Received: 19 February 2024; Accepted: 21 August 2024;
Published: 10 September 2024.
Edited by:
Jiankang Zhou, Chengdu Medical College, ChinaReviewed by:
Liping Ma, Chengdu Medical College, ChinaCopyright © 2024 Wang, Mei, Yang, Zhang, Li, Zhu, Deng and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lei Zhu, lei_zhu@scu.edu.cn; Senyi Deng, senyi_deng@scu.edu.cn; Yun Wang, yunwwang@yeah.net
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.