- 1Faculty of Biology and Biotechnology, HSE University, Moscow, Russia
- 2Shemyakin-Ovchinnikov Institute of Bioorganic Chemistry, Russian Academy of Sciences, Moscow, Russia
Alternative splicing is often deregulated in cancer, and cancer-specific isoform switches are part of the oncogenic transformation of cells. Accumulating evidence indicates that isoforms of the multifunctional cell-surface glycoprotein CD44 play different roles in cancer cells as compared to normal cells. In particular, the shift of CD44 isoforms is required for epithelial to mesenchymal transition (EMT) and is crucial for the maintenance of pluripotency in normal human cells and the acquisition of cancer stem cells phenotype for malignant cells. The growing and seemingly promising use of splicing inhibitors for treating cancer and other pathologies gives hope for the prospect of using such an approach to regulate CD44 alternative splicing. This review integrates current knowledge about regulating CD44 alternative splicing by RNA-binding proteins.
1 Introduction
CD44 is a multifunctional transmembrane glycoprotein that is widely expressed and plays an essential role in physiological activities in normal cells throughout the body. CD44 was first discovered to be present on the cell membrane of haematopoietic cells (Jalkanen et al., 1986) and after that, its expression was noted in different non-haematopoietic cells (Fox et al., 1994). The first studies of the physiological role of CD44 showed that CD44-deficient mice are viable without obvious developmental defects and show no overt abnormalities as adults. However, during development, they had impaired lymphocyte trafficking into the thymus (Protin et al., 1999). During further decades CD44 emerged as a regulator of malignant progression and metastasis formation due to its involvement in cell proliferation, adhesion, cytoskeleton rearrangement, migration, angiogenesis, inflammation, metabolism (regulating glucose and lipid homeostasis) (Zöller, 2011; Senbanjo and Chellaiah, 2017; Chaffer and Goetz, 2018; Chen et al., 2020; Guo Q. et al., 2022; Weng et al., 2022). Notably, CD44 is a commonly accepted marker of cancer stem cells (CSC) of different cancer entities including breast, colon, gastric, pancreas, glioma, ovarian (Zöller, 2011; Hu and Fu, 2012; Yan et al., 2015; Skandalis et al., 2019), and of epithelial to mesenchymal transition (EMT) a process vital for distant metastasis formation (Cho et al., 2012; Zhang et al., 2012; Jiang et al., 2015).
The marked multifunctionality and variability of the CD44 protein are ensured by the existence of its multiple forms, which mainly originate in alternative splicing and are further amplified by extensive and often isoform-specific posttranslational modifications including N- and O-glycosylation, phosphorylation, and glycosaminoglycan attachment (Fox et al., 1994; Ponta et al., 2003; Zöller, 2011; Wang et al., 2018). Thus, CD44 is a family of transmembrane glycoproteins with a high heterogeneity in molecular weight (85–250 kDa). Alternative splicing (AS) is often deregulated in cancer, and cancer-specific isoform switches are part of the oncogenic transformation of cells (Di et al., 2018; Zhang et al., 2021; Shaw et al., 2022; Bradley and Anczuków, 2023). Indeed, accumulating evidence supports the concept that CD44 isoforms play different roles in cancer cells as compared to their normal counterparts (Zöller, 2011; Bhattacharya et al., 2018; Xu et al., 2020). In particular CD44 isoform switches have been shown during EMT and acquisition of CSC properties (Bhattacharya et al., 2018; Zhang et al., 2019). Moreover, the shift of CD44 isoforms is required for EMT (Reinke et al., 2012). The understanding of the mechanisms of alternative splicing and the occurrence of variant isoforms of CD44 is essential not only to a deeper insight into malignant progression but may also provide a new generation of splicing inhibitors as therapies for cancer (Bonnal et al., 2020; Rogalska et al., 2022). The major experimentally tested regulators of alternative splicing of CD44 in cancer have been described earlier by Prochazka and co-authors (Prochazka et al., 2014). Our review will focus on new data concerning RNA binding proteins, which were recently shown as an essential regulator in CD44 isoform switching.
2 Overview of CD44 isoforms
CD44 proteins have a common structure consisting of three major domains: an extracellular or ectodomain (ECD), a transmembrane domain (TMD) and a cytoplasmic or intracellular domain (ICD) (Figure 1) (Ponta et al., 2003; Zöller, 2011; Wang et al., 2018). The ECD comprises an N-terminal globular domain and a membrane-proximal region, which may include variant exons (variable region). All CD44 proteins are encoded by one single gene present on chromosome 11 in humans, which includes 19 exons so that alternative splicing gives rise to plentiful isoforms (Figure 2) (Screaton et al., 1992; Azevedo et al., 2018). According to NCBI database, eight CD44 isoforms are commonly accepted as biologically expressed (Figure 2), and the existence of 27 other isoforms was predicted. Nonetheless, it should be mentioned that some data exist that other CD44 isoforms except these eight do exist (Bánky et al., 2012; Marzese et al., 2015; Kim et al., 2018). In a recent study, full-length mRNA transcripts from diverse normal and cancerous human tissues have been profiled using long-read sequencing techniques (Shi et al., 2023). The RNA sequencing data were collected in the FLIBase repository. Based on the FLIBase data, more than two hundred CD44 isoforms were detected in human cells. The shortest or standard CD44 isoform (CD44s, isoform 4) contains only constant (invariant) exons (the first one to five and the last four 15–17 and 19). Exon 18 is mostly spliced out in humans. CD44s is ubiquitously expressed in most tissues. The ECD of this isoform is composed of only an N-terminal globular domain (Figure 1). Including variant exons v2-v10 (variant exon v1 is not present in humans) into a membrane-proximal region of the ECD gives larger isoforms which are expressed in only a few epithelial tissues, mainly in proliferating cells, and in cancer cells of several cancer entities as well. CD44 variant isoforms are often numbered depending on the inclusion of corresponding exons, e.g., CD44 isoform 1 contains CD44v2-v10, isoform 2 contains CD44v3-v10 and isoform 3 contains CD44v8-v10.
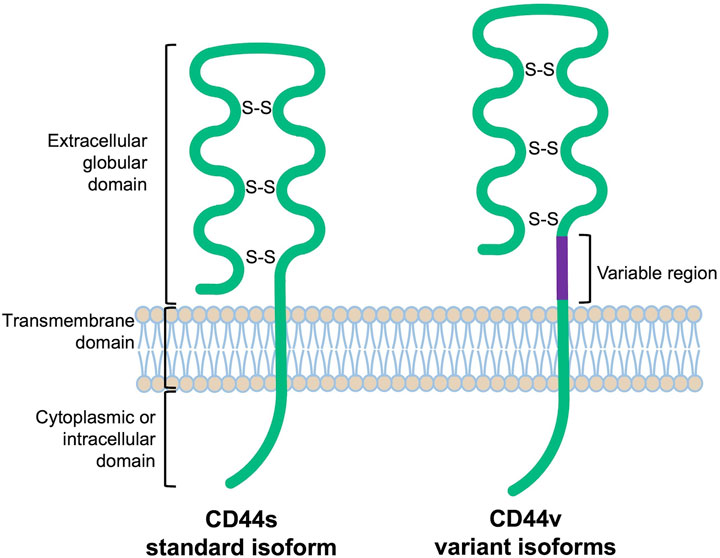
FIGURE 1. Schematic protein structure of CD44 molecules [extrapolated from (Naor et al., 1997)].
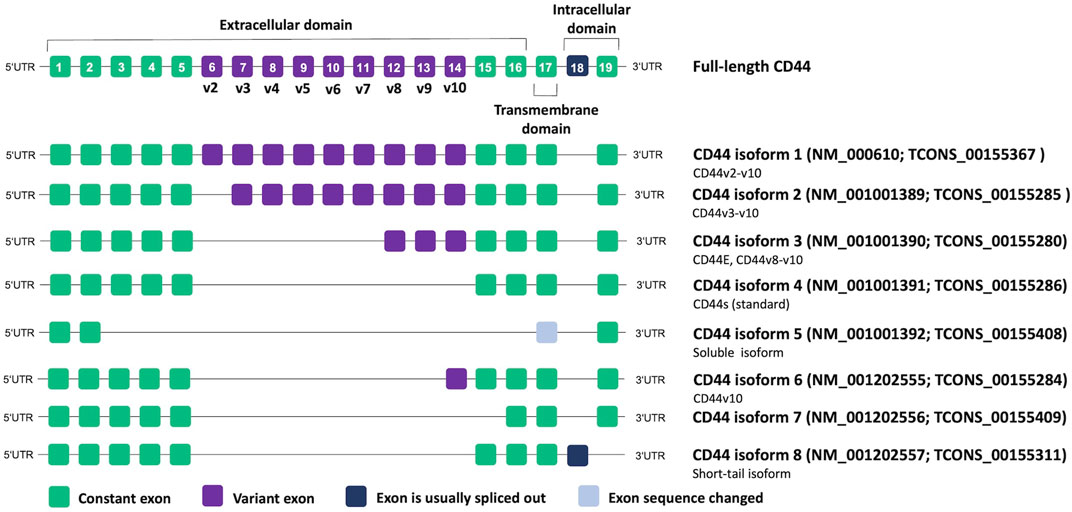
FIGURE 2. CD44 genomic organization and alternative splicing according to NCBI database. Transcript IDs are also provided according to the FLIBase database (Shi et al., 2023).
Importantly, authors of studies often adopt a nomenclature based on commercial names of the used monoclonal antibodies, highlighting the targeted variant exon, and disregard that the analysis of a specific variant exon can result in the detection of all isoforms containing it instead of only one particular protein. Thus, such isoforms as CD44v3, CD44v6, and CD44v9 started being the most associated with cancer (Azevedo et al., 2018). However, most of these studies dealt only with antibodies specific to the corresponding individual exons. Thus, the lack of nomenclature standardization makes it difficult to interpret the results presented in the articles and requires careful conclusions about which isoform/isoforms are actually in question. In our review, we prefer to use CD44 isoform designations according to NCBI nomenclature if possible.
CD44 proteins are primarily considered as cell adhesion molecules as they contain binding sites for hyaluronan (Underhill, 1992), collagen (Ishii et al., 1993), laminins (Ishii et al., 1993; Hibino et al., 2004), fibronectin (Jalkanen and Jalkanen, 1992), E-/P-/L-selectins (Hanley et al., 2006). However, due to their signaling functions, CD44 proteins play essential roles in intercellular communication and numerous other cellular functions associated with it (Ponta et al., 2003; Wang et al., 2018). CD44 variant isoforms encode additional peptides in the membrane-proximal region, which provide binding sites for other molecules including cytokines and growth factors. This configuration allows CD44 transmembrane glycoproteins to emerge as a multidomain platform, which integrates various extracellular information. We will not dwell on the mechanisms of signaling in details, information for a deeper understanding can be found in these excellent reviews (Orian-Rousseau and Sleeman, 2014; Ouhtit et al., 2018; Wang et al., 2018; Mesrati et al., 2021; Guo Q. et al., 2022).
CD44 is a well-known normal intestinal stem cell marker (ISC) (Habowski et al., 2020), and its alternative splicing should be tightly controlled in the crypt-villus axis (Orian-Rousseau and Sleeman, 2014). Thus, ISCs residing at the crypt base in mice express mRNA encoding CD44 isoforms v4–v10, v6-v10, v7-v10 and isoform 3 (v8-v10), but do not express the standard isoform 4 (CD44s) (Zeilstra et al., 2013). Progenitor cells from a transit-amplifying compartment (daughter cells of ISCs) express mRNAs encoding CD44 isoforms containing v6-v10, v7-v10, isoform 3 (v8-10), as well as standard isoform 4 (CD44s). Interestingly, human ISCs display a somewhat different repertoire of CD44 isoforms than mice presenting CD44v6-v10, v7-v10, isoform 3 (v8-10), and standard isoform 4 (Zeilstra et al., 2013). However, neoplastic epithelial cells from microadenomas of familial adenomatous polyposis patients demonstrate an expression profile of CD44 mRNAs more similar to mice ISCs, suggesting involvement of variant isoforms at early stages of human CRC (Zeilstra et al., 2013). An earlier study reported that the lower part of the crypts express CD44 isoform containing exon v9 but not exons v4 and v6 in humans (Mackay et al., 1994).
3 CD44 isoforms in cancer
A huge amount of data indicates that different CD44 isoforms play a role in many types of cancer [reviewed in (Chen et al., 2018; Mesrati et al., 2021; Yaghobi et al., 2021)], and their cellular functions can both overlap and be distinct. Sometimes, the information about the functions of CD44 isoforms is controversial, which complicates our current understanding of their roles in malignancy and cancer progression (Table 1). Thus, elevated expression of CD44 isoform 3 occurs in breast (Yae et al., 2012), gastric (Ishimoto et al., 2011; Lau et al., 2014), bladder (Miyake et al., 2002), esophageal (Kagami et al., 2018), gallbladder cancer (Yamaguchi et al., 2000), thyroid (Kawai et al., 2019), ovarian (Sosulski et al., 2016), colorectal cancer (Boman et al., 2023; Everest-Dass et al., 2023), melanoma (Zhang et al., 2016) and leukemia as well (Holm et al., 2015). While in breast, esophageal, and gallbladder cancer CD44 isoform 3 is associated with a more metastatic phenotype and poor prognosis [e.g., (Yamaguchi et al., 2000; Yae et al., 2012; Kagami et al., 2018)], it does not do so in ovarian cancer (Sosulski et al., 2016). Sometimes, e.g., in the case of colorectal cancer, CD44 isoform 3 has been shown associated with both poor prognosis/higher recurrence rate (Yamaguchi et al., 2016) and good prognosis/lower recurrence rate (Mashita et al., 2014; Everest-Dass et al., 2023), pointing to the possible greater significance of the ratio of CD44 isoforms. Indeed, the high ratio of CD44 isoform 4/CD44 variant exon v9 in patients with colorectal cancer shows a significantly poorer prognosis than the low CD44 isoform 4/CD44 variant exon v9 ratio (Mashita et al., 2014). In prostate cancer, CD44 isoform 3 is associated with CSC features (Zeng et al., 2013). However, in breast (Zhang et al., 2019), pancreatic (Li et al., 2014) and ovarian (Bhattacharya et al., 2018) cancers CSC features are determined by CD44 isoform 4. CD44 isoform 4 has been showing to play a critical role in the mesenchymal phenotype of many cancers (Primeaux et al., 2022), including hepatocellular carcinoma cells (Mima et al., 2012), breast cancer cells (Brown et al., 2011; Preca et al., 2015), colorectal cancer cells (Mashita et al., 2014), ovarian cancer cells (Bhattacharya et al., 2018), and in EMT of CSCs of cutaneous squamous cell carcinoma (Biddle et al., 2013). Several studies showed that a switch in CD44 isoform expression from CD44v to CD44 standard isoform 3 is essential for EMT (Brown et al., 2011; Preca et al., 2015). All of the above-mentioned findings suggest that both CD44 isoform 3 and isoform 4 are expressed in cancer cells but play distinct roles in the different steps of cancer development. Thus, as it has been partially shown in colon cancer, CD44 isoform 4 can play an anti-tumor role during the initial malignant transformation but may later benefit metastasis formation (Everest-Dass et al., 2023). In gallbladder cancer CD44 isoform 4 is associated with a mesenchymal phenotype, increased chemotaxis, increased invasiveness, but lower tumorigenicity (Miwa et al., 2017). At the same time, the CD44 variant exon v9 expression is associated with an epithelial phenotype, decreased chemotaxis, decreased invasiveness, and unexpectedly increased tumorigenicity. In the review (Wang et al., 2018), one can find detailed information about the engagement of CD44 exons v6 and v3 in the maintenance of CSCs and tumor progression. It is plausible that regulation of CD44 splicing allows CSCs to maintain the hybrid E/M state correlated with higher stemness and tumorigenicity (Pradella et al., 2017). Thus, CD44 undergoes isoform switching in cancer cells (Primeaux et al., 2022) and understanding its regulation mechanisms is incredibly important for a deeper insight into malignant progression. Our review will focus on how tumor cells implement CD44 isoform switching. The main players here are certainly RNA-binding proteins as far as AS relies on them to recognize and bind target sequences in pre-mRNAs, which allows for the inclusion or skipping of alternative exons (Table 2).
4 RNA-binding proteins regulating CD44 alternative splicing
RNA binding proteins (RBPs) recognize and bind target sequences in pre-mRNAs, which allows for the inclusion or skipping of alternative exons. Such target sequences could be intronic or exonic splicing enhancers (ISEs or ESEs) or intronic or exonic splicing silencers (ISSs or ESSs), which nucleate the assembly of complexes of regulatory factors that promote or inhibit splice site recognition by the core splicing machinery (Ule and Blencowe, 2019; Rogalska et al., 2022) (Figure 3).
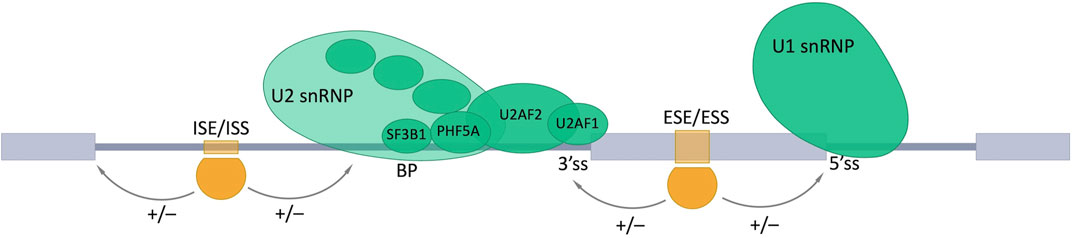
FIGURE 3. Involvement of RNA-binding proteins in the alternative splicing (AS). AS is regulated by combined action of trans- and cis-acting elements. Trans-acting elements are represented by different RBPs (shown as orange shapes). Cis-acting elements are specific nucleotide motifs in pre-mRNA: intronic and exonic splicing enhancers (ISE and ESE), which promote the inclusion (+) of the AS exon by providing the binding sites for activators (shown in orange); intronic and exonic splicing silencers (ISS and ESS) are bound by repressors (shown in orange) and promote exon skipping (−). Exons are represented as gray boxes, introns as gray lines. BP, a branch point; 3’ss, 3′ splice site; 5’ss, 5′ splice site.
4.1 ESRP1 and ESRP2
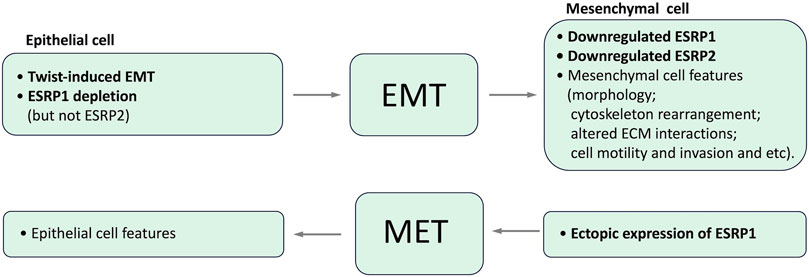
Epithelial splicing regulatory proteins (ESRPs), including ESRP1 and ESRP2 (also known as RBM35A and RBM35B, respectively), are specifically expressed in epithelial cells and identified as core modulators of EMT-related splicing events (Mashita et al., 2014; Katsuno and Derynck, 2021; Liu et al., 2022). In particular ESRP1 and ESRP2 regulate the alternative splicing of a number of proteins important for maintaining an epithelial phenotype. It has been shown that ESRP1 transcription is regulated by Snail and ZEB1, EMT-related transcription factors, through its direct binding to ESRP1 promoter (Reinke et al., 2012; Preca et al., 2015; Chen et al., 2017). Induction of EMT results in the downregulation of ESRP1 and ESRP2, whereas the depletion of ESRP1, but not ESRP2 (Warzecha et al., 2009b), is sufficient to induce mesenchymal splicing patterns (Warzecha et al., 2009b; Jeong et al., 2017). Ectopic expression of ESRP1/mouse Esrp1 in human mesenchymal cells induces epithelial-specific changes in the splicing of ESRP1 target transcripts (Warzecha et al., 2009a; Warzecha et al., 2009b). Interdependence between expression of ESRPs and EMT is summarized in Figure 4.
Numerous studies implicate ESRP1 as a regulator of CD44 isoform switching (see below, Table 2). It has been reported that ESRP1 recognizes GGU/UGG–rich sequences (Dittmar et al., 2023). In particular it has been shown that ESRP1 promotes the inclusion of CD44 variant exon v8 by directly binding to the GGU/UGG–rich motifs located in the intron downstream of this exon (Reinke et al., 2012). Another study indicated the dependence of ESRP1 and ESRP2 upon the presence of ISE/ISS-3 and/or the UGCAUG motif to promote splicing (Warzecha et al., 2009a).
Simultaneous depletion of ESRP1 and ESRP2 in the normal human prostate epithelium cell line PNT2 causes a significant decrease in the inclusion of CD44 variant exons (mainly exons v8–v10 corresponding to CD44 isoform 3) and an increase in the standard CD44 isoform 4 in which all the variable exons are skipped (Warzecha et al., 2009a; Warzecha et al., 2009b). This isoform skipping was also accompanied by increased expression of the mesenchymal isoforms of p120-catenin and FGFR2 and silencing epithelial-specific isoform of ENAH.
The loss of ESRPs expression in the human mammary epithelial cell line HMLE due to the induction of an EMT by the transcription factor Twist resulted in the elevated level of CD44 standard isoform 4 expression and the decreased CD44 variant isoforms expression, indicating codependence of these events (Warzecha et al., 2009a). ShRNA-mediated depletion of ESRP1 in HMLE cells resulted in a shift of expression from CD44 variant isoforms to isoform 4, with only a slight effect on the overall CD44 expression level and accelerated EMT (Brown et al., 2011), while ESRP1 overexpression regulated CD44 alternative splicing in the opposite direction and inhibited Snail-induced EMT in these cells (Reinke et al., 2012). Notably, this isoform switch from CD44 variant isoforms to isoform 4 was essential for forming breast tumors in mice (Brown et al., 2011).
To test whether ectopic expression of the ESRP1 in mesenchymal cells would restore an epithelial splicing pattern, the mesenchymal human breast cancer cell line MDA-MB-231 was transduced with virus encoding FLAG-tagged ESRP1 (Warzecha et al., 2009a). The expression of ESRP1 caused a switch from primarily CD44 isoform 4 expression to variant isoforms (including CD44 isoform 3 and CD44v6-v10). Interestingly, the repeat of the experiment with mesenchymal melanoma cell line MDA-MB-435 (Prasad and Gopalan, 2015) with predominant expression of the standard CD44 isoform 4 did not lead to the same results, overall expression of CD44 was significantly downregulated (Warzecha et al., 2009a). The same authors ectopically expressed mouse Esrp1 in human MDA-MB-231 cells and also showed the increased inclusion of human CD44 variant exons and the decreased expression of human CD44 isoform 4 (Warzecha et al., 2009b). ESRP1 downregulation in 4T1 mouse breast cancer cells also resulted in an isoform switch from CD44 containing variant exons to CD44 isoform 4 (Yae et al., 2012). However, this downregulation led to suppression of lung colonization, presumably due to reduced cell surface expression of the cystine transporter xCT, the stability of which is controlled by CD44 variant isoforms. A more recent study demonstrated that ectopic expression of ESRP1 inhibits the production of standard CD44 isoform 4 and thus inhibits CSC properties of TGFβ-treated HMLE cells, where endogenous ESRP1 expression was low (Zhang et al., 2019). In contrast, ESRP1-depletion possessed enhanced mammosphere-forming ability of TGFβ-treated HMLE cells, and silencing CD44 in these ESRP1-depleted cells abrogated mammosphere formation. In addition, Zhang et al. expressed ESRP1 in a mesenchymal triple-negative breast cancer cell line SUM159 and revealed reduced potential for mammosphere formation, which was rescued by coexpression of CD44 isoform 4 but not CD44v3-v10 (Zhang et al., 2019). A transient knockdown of ESRP1 in human breast cancer MCF7 and human pancreatic adenocarcinoma BxPC-3 cells resulted in a shift of expression from CD44v (containing exon v6) to CD44 isoform 4 without affecting total CD44 level (Preca et al., 2015). Vice versa, overexpression of ESRP1 in undifferentiated, mesenchymal breast cancer MDA-MB231 cells and pancreatic ductal adenocarcinoma Panc-1 cells resulted in a reverse isoform switch accompanied by decreased ZEB1 levels (Preca et al., 2015).
Silencing of ESRP1 significantly decreased the expression of CD44 isoforms containing exon v6 in human MB and LH cells derived from melanoma lymph node metastases (Marzese et al., 2015). These siESRP1-transfected melanoma cells also demonstrated lower migratory potential under hepatocyte growth factor (HGF) treatment (HGF, a factor released during inflammation or tissue disruption, should increase the migration of CD44v6-positive melanoma cells). Further in this study, it has been shown that ESRP1 is epigenetically silenced in human melanoma brain metastasis, and high expression of CD44 containing exon v6 in early stages is a significant predictor of melanoma brain metastasis development (Marzese et al., 2015).
Switching from CD44 variant isoforms containing exon v7 to isoform 4 without change in a total amount of CD44 was observed in human epithelial ovarian cancer cell line HO8910 with a stable suppression of ESRP1 expression (Chen et al., 2017). This switch in expression was accompanied by increasing migratory and invasive capabilities of the ESPRP1 suppressed cells. Moreover, the siRNA-mediated downregulation of CD44 expression, in turn, suppressed migration and invasion of the ESRP1-depleted HO8910 cells, indicating that ESRP1 suppresses HO8910 cell motility mainly by repressing CD44 isoform switching (Chen et al., 2017). An enforced ESRP1 expression in the ovarian cancer cell line SKOV3, significantly reduced the level of the mesenchymal cell-specific CD44 isoform 4 and increased levels of CD44 variant isoforms as well as caused overall switching from mesenchymal to epithelial phenotype of cells (Jeong et al., 2017).
Based on the qPCR measurement in 14 colorectal cancer (CRC) cell lines, it has been demonstrated a higher ESRP1 expression was noted in epithelial phenotype cells than those of mesenchymal phenotype (Mashita et al., 2014). The higher ESRP1 expression strongly correlated with higher expression of CD44 variant exon v9 and lower expression of CD44 isoform 4, respectively. An inverse correlation between the expression of ESPRs and CD44 alternative splicing was observed for CRC cell line LS1034 on mRNA level (Dinger et al., 2020). In particular LS1034 xenografted cancer cells demonstrated an elevated expression of CD44 mRNA isoform 3 compared to cultured cells, whereas ESPR1 expression was reduced and ESRP2 expression was essentially enhanced in the LS1034 xenografts. However, ESRP1 silencing in HCT-116 cell line has shown suppressed CD44 mRNA variant isoforms and enhanced standard isoform 4 expressions, which were accompanied by inducing caspase-independent cell death (Vadlamudi and Kang, 2022).
Higher ESRP1 expression was associated with CD44 variant isoforms, including CD44 isoform 1 (CD44v2-v10), isoform 3 (CD44v8-v10), and CD44v6-v10, in human head and neck squamous cell carcinoma cell lines (Ishii et al., 2014). Si-RNA-mediated silencing of ESRP1 in SAS and HSC4 cells resulted in switching from the CD44 variant isoforms to the CD44 standard isoform 4 (Ishii et al., 2014). In contrast, silencing of ESRP2 did not affect CD44 isoform switching. Although knockdown of both ESRPs enhanced cell motility without effect on cell proliferation. Further, it has been shown that ESRPs suppress cell motility in HNSCC through distinct mechanisms: ESRP1 regulates the dynamics of the actin cytoskeleton through repressing expression of the Rac1b isoform, whereas ESRP2 is involved in the regulation of cell-cell adhesion by suppressing EMТ-associated transcription factors (Ishii et al., 2014).
Several splice variants of ESRP1 were tested for their ability to regulate CD44 alternative splicing (Kim et al., 2018). Overexpression of ESRP1 v1, v4, or v5 in fully differentiated human foreskin fibroblasts resulted in converting CD44v9-v10 to CD44v7-v10. ESRP1 knockdown in undifferentiated H9 human embryonic stem cell induced downregulation of CD44v7-v10 expression as well as the loss of pluripotency of the cells. Thus, regulating the ESRP1-CD44v7-v10 axis is crucial for human pluripotency maintenance and reprogramming of human somatic cells to pluripotent stem cells (iPSCs) (Kim et al., 2018).
Overall, the presented information indicates that ESRP1 is mainly a positive regulator of including variant exons (many of them) in CD44 transcript, whereas ESRP2 is apparently not so much involved in CD44 alternative splicing.
4.2 RBFOX2 and QKI
RBFOX2 is a member of the RNA-binding Fox (RBFOX) protein family (RBFOX1, RBFOX2 and RBFOX3) regulating alternative splicing (Kuroyanagi, 2009). RBFOX1 is expressed in heart, skeletal muscle and neuronal tissues, whereas RBFOX2 is ubiquitously expressed in many tissues from the embryonic-stem-cell stage through adulthood (Jin et al., 2003; Yeo et al., 2009; Kim K. K. et al., 2011; Underwood et al., 2023). RBFOX3 is expressed exclusively in the brain (Kim et al., 2009). All three proteins contain a single conserved RNA-recognition motif (RRM) and recognize the consensus sequence (U)GCAUG in the introns flanking target exons. RBFOX2 prevents the binding of U2AF2 to the 3′-splice site (Ivanova et al., 2023). A general rule for RBFOX2-regulated exon inclusion or skipping in a position-dependent manner has been revealed. In particular, RBFOX2 promotes exon skipping when it binds upstream of the alternative exon but inclusion occurs when it binds downstream of this exon (Yeo et al., 2009). Of note, RBFOX2 mRNA undergoes extensive alternative splicing itself, thus generating many isoforms with a common RRM. The RBFOX2 splice variants show differences in intracellular localization and splicing activity (Nakahata and Kawamoto, 2005). Within the nucleus RBFOX2 can operate their targets using three binding modes: single, multiple or secondary (Damianov et al., 2016; Zhou et al., 2021). In the single binding mode, RBFOX2 is recruited to its target splice sites through a single canonical binding motif, while in the multiple binding mode, RBFOX2 binding sites include the adjacent binding of at least one other RBP partner. In the secondary binding mode, RBFOX2 is recruited to splice sites lacking its canonical binding motif by binding one of its protein partners and likely without direct binding to mRNA. Interestingly, many targets of RBFOX2 are themselves splicing regulators (Yeo et al., 2009). In addition, RBFOX2 is implicated in the biogenesis of some miRNAs (e.g. miR-20b and miR-107) and, thus, in the expression of their downstream targets (Chen et al., 2016). All of these different modes of operation may explicate an ambiguous role of RBFOX2 in cancer progression. Thus, several studies have reported that RBFOX2 is important to specify a mesenchymal splicing signature in breast (Braeutigam et al., 2013), colon and ovarian tissues (Venables et al., 2013). RBFOX2 promotes oncogenic splice-switching and the resulting mesenchymal signature and drives an invasive phenotype in breast cancer (Braeutigam et al., 2013; Ahuja et al., 2020). However, other studies have reported the anti-metastatic role of RBFOX2 in pancreatic cancer (Jbara et al., 2023) and its decreased expression in breast, colon, and prostate adenocarcinomas (Danan-Gotthold et al., 2015), as well as ovarian cancer (Venables et al., 2009). In general, RBFOX2 is considered a mesenchymal marker (Pradella et al., 2017; Lambert and Weinberg, 2021), whereas it also promotes epithelial-specific splicing in some cases (Braeutigam et al., 2013; Baraniak et al., 2023). A possible clue of these contradictions may be that the epithelial state of cells is determined by the ratio of the expression levels of RBFOX2 and ESRP1 (Barriere et al., 2014; Meng et al., 2019).
Based on the analysis of patient TCGA RNA-Seq data of colon adenocarcinoma and corresponding normal colon, RBFOX2 has been predicted to act as a negative regulator of variant exon inclusion in CD44 mRNA (Danan-Gotthold et al., 2015). Interestingly, it was not the case in seven other analyzed cancer types (breast invasive carcinoma, kidney clear cell carcinoma, liver hepatocellular carcinoma, lung adenocarcinoma, prostate adenocarcinoma, head and neck squamous cell carcinoma, and thyroid carcinoma). Our bioinformatic analysis of mRNA-Seq data of 56 colorectal cancer cell lines downloaded from the CCLE database revealed the association of higher expression of RBFOX2 with a higher level of CD44 isoform 4 and a lower level of CD44 isoform 3 (Novosad and Maltseva, 2023).
Si-RNA mediated knockdown of RBFOX2 did not alter the level of inclusion of exons v8-v10 into CD44 mRNA in both non-transformed mammary epithelial cell line NMuMG and epithelial murine breast cancer cell line PY2T under normal conditions and TGF-β-treatment (Braeutigam et al., 2013). Upregulation of the long transcript variant of RBFOX2 and downregulation of short variant RBFOX2 and ESPR1 in mouse breast cancer 4T1 cell line in response to ectopic expression of WNT5A (a non-canonical Wnt signaling) was accompanied by downregulated inclusion of exons v4-v6 (but not v9) in CD44 mRNA (Jiang et al., 2013). These events also were associated with reduced cell migration and fewer spontaneous lung metastasis.
It has been observed that RBFOX2 co-operates with Quaking (QKI) in the splicing regulation of common pre-mRNA targets (Brosseau et al., 2014; Danan-Gotthold et al., 2015; Yang et al., 2023). QKI is an RBP belonging to the signal transduction and activation of RNA (STAR) protein family, which binds specifically to RNA containing ACUAA motifs (Hall et al., 2013). QKI regulates several posttranscriptional processes, including AS, mRNA localization, mRNA stability, and protein translation (Saccomanno et al., 1999; Li et al., 2000; Larocque et al., 2002; Zhao et al., 2010; Hall et al., 2013). Among the three major isoforms of QKI (QKI-5, QKI-6, and QKI-7), only QKI-5 is predominantly localized in the nucleus and is involved in the regulation of AS (Ebersole et al., 1996; Bockbrader and Feng, 2008). QKI binding in the downstream intron promotes exon inclusion while binding in the upstream intron promotes exon skipping (Hall et al., 2013). Of note, QKI strongly induces the mesenchymal and stem-like phenotypes (Li et al., 2018; Mukohyama et al., 2019) and promotes mesenchymal splicing patterns (Lambert and Weinberg, 2021; Yang et al., 2023).
To our knowledge, the involvement of QKI in CD44 splicing regulation has yet to be experimentally confirmed. However, such a possibility was predicted for colorectal cancer based on bioinformatic analysis of mRNA-Seq data of patient tumor samples (Novosad, 2023). In particular QKI has been shown as a potential negative regulator of CD44 isoform 3 formation, what is consistent with the EMT-promoting role of QKI.
4.3 NONO and its protein partners
Non-POU domain-containing octamer-binding protein (NONO, also known as p54nrb) belongs to the Drosophila behavior/human splicing (DBHS) family (Knott et al., 2016). In humans, the DBHS protein family also includes two other members: splicing factor proline/glutamine-rich (SFPQ, also known as PSF) and paraspeckle protein component 1 (PSPC1, also known as PSP1). All three DBHS proteins contain two highly conserved RNA-recognition motifs and a nuclear localization signal and are regarded mainly as nuclear factors. However, they may additionally function intra-cytoplasmically and on the cell surface (Knott et al., 2016). Structural and biological data suggest that DBHS proteins rarely function alone. They are found in the nucleoplasm within the subnuclear domain termed paraspeckles (Knott et al., 2016), which are known to regulate RNA metabolism, including splicing, stabilization and export, as well as DNA repair (Wang and Chen, 2020). NONO and SFPQ were found to be associated with both the hypophosphorylated and hyperphosphorylated forms of RNA polymerase II in HeLa cell extracts, indicating that these two proteins could provide a direct physical link between RNA polymerase and other pre-mRNA processing components (Emili et al., 2002).
NONO is a multipurpose protein engaging in almost every step of gene regulation, including transcriptional activation and inhibition, RNA processing, and DNA repair (Knott et al., 2016). Dysregulation of NONO has been found in many types of cancer entities (Feng et al., 2020). In some of them, such as bladder cancer, lung cancer, prostate cancer, and oesophageal squamous cell carcinoma, glioblastoma multiforme NONO exhibits tumor promoting role, as it induces cell proliferation and inhibits apoptosis. In contrast, in estrogen receptor-negative breast cancer, it demonstrates tumor suppressive functions (Feng et al., 2020; Wang et al., 2022). Recently, it has been shown that NONO induces expression of ZEB1 and CD44 in LN229 glioblastoma cells and patient-derived P3glioblastoma stem-like cells and promoted cells migration and invasion indicating an association between NONO and EMT (Wang et al., 2022). Si-RNA-mediated loss of NONO in U251 and P3 glioblastoma cells reduced levels of proteins involved in the EMT but increased those involved in apoptosis (Wang et al., 2022). Silencing of NONO inhibits EMT and stemness of breast cancer cells, as well as its growth, survival, migration and invasion (Lone et al., 2023).
Using the CD44 minigene reporter system [the variant exons v4 and v5 of the human CD44 gene, along with their surrounding intron sequences, inserted into an intron of the β-globin gene driven by the HSV promoter (Auboeuf et al., 2002)] it has been demonstrated that NONO and SFPQ regulate alternative splicing of CD44 variant exons in HEK293T cells transfected with either NONO vector or SFPQ vector (Liu et al., 2011). NONO decreased the ratio of skipping to inclusion of CD44 exons v4-v5, whereas SFPQ increased this skipping-inclusion ratio (Liu et al., 2011). Interestingly, dephosphorylation of NONO and SFPQ by protein phosphatase 1 (PP1) reduced their alternative splicing activity on CD44 minigene.
Knockdown of SFPQ in lung cancer mesenchymal stem cells resulted in the reduced expression of CD44 isoforms containing exon v6 with concomitant inhibition of cell stemness, proliferation in vitro, and metastasis in vivo (Yang et al., 2022).
Applying the CD44 minigene reporter system by Zhao et al. showed that overexpression of both NONO and SFPQ in HeLa cells significantly increases variant exon inclusion and decreases a level of CD44 isoform 4 (Zhao et al., 2022). Of note, a simple overexpression of NONO or SFPQ in HeLa cells did not significantly affect the transcription level of neither CD44 isoform 4 nor CD44 variant isoforms. In the same work, the authors revealed the interaction of NONO and SFPQ with an angiogenic factor AGGF1 in paraspeckles, which forms an outside rim around the NONO/SFPQ/PSP1 core (Zhao et al., 2022). Interestingly, the overexpression of AGGF1 in HeLa cells in turn resulted in enhanced inclusion of exons v4 and v5 (but not v8-v10 or v10) in CD44 mRNA and decreased level of CD44 isoform 4. The enhanced inclusion of exons v4 and v5 was also detected in HeLa cells co-transfected with CD44 minigene reporter and AGGF1 vector. Also, Zhao et al. detected the decreased ratio of skipping to inclusion of exons v4-v5 in the CD44 minigene in response to overexpression of DHX15 (DEAH-Box Helicase 15), interacting with AGGF1 in HeLa cells (Zhao et al., 2022). Co-overexpression of AGGF1 with NONO, SFPQ, or DHX15 also enhanced the inclusion of exons v4 and v5 in the CD44 minigene.
Thus, the available evidence suggests that NONO, AGGF1, and DHX15 function primarily as an enhancer of the formation of variant CD44 isoforms. At the same time, SFPQ can contribute to both the skipping and inclusion of variant exons in the CD44 transcript.
There is evidence for the interactions of NONO and SFPQ with the ubiquitously expressed heterogeneous nuclear ribonucleoprotein M (hnRNPM) and for the presence of the last one within a subpopulation of paraspeckles (Marko et al., 2010). hnRNPM is a component of the spliceosome machinery and can influence both constitutive and alternative splicing. It typically binds to ESS motives, thus antagonizing the recognition of splice sites and suppressing pre-mRNA splicing (Wahl et al., 2009). hnRNPM is associated with aggressive breast cancer and correlates with increased CD44s in patient specimens (Xu et al., 2014; Sun et al., 2017). Moreover, it has been demonstrated that hnRNPM precisely controls CD44 splice isoform switching during EMT and acts in a mesenchymal-specific manner in breast cancer cells (Xu et al., 2014; Harvey et al., 2018). Silencing hnRNPM completely abolished TGFβ-induced CD44 isoform switching from CD44 variant isoforms to isoform 4 in HMLE cells (Xu et al., 2014). The hnRNPM depletion was also accompanied by a general inhibition of TGFβ-induced EMT in HMLE cells which resulted in the reduction of spontaneous lung metastasis numbers in mice with into the mammary fat pad implanted murine T4 breast cancer cells. Reduced dissemination potential of murine T4 breast cancer cells and human LM2 breast cancer cells (MDA-MB-231-derived lung metastatic cells) with hnRNPM knockdown was also shown after intravenous injection into murine tail vein (Xu et al., 2014). Interestingly, the enforced expression of CD44 isoform 4 overrode the loss of hnRNPM and permits EMT and metastasis formation to occur. In a combination of experiments with several cell lines, Xu et al. also demonstrated that hnRNPM is necessary and sufficient to stimulate CD44 variant exon skipping via its interaction with GU-rich motifs located in introns downstream from variable exons (Xu et al., 2014). In addition, a cell-type restricted activity of hnRNPM has been revealed as it does not promote CD44 exon skipping in HCT116 human colon cancer cells. A possible reason for this observation is that a competition of hnRNPM with ESRP1 for the binding to CD44 pre-mRNA exists (Xu et al., 2014). In their subsequent study, the authors showed that coregulation of alternative splicing by hnRNPM and ESRP1 is widespread and primarily antagonistic in breast cancer cells, although a subset of events is regulated concordantly (Harvey et al., 2018). An overexpression of hnRNPM in MCF-7 human breast cancer cells resulted in a decreased expression in CD44 isoforms containing exon v6 and an increased expression in CD44 isoform 4 with a slight change in the total level of CD44 transcripts (Sun et al., 2017). These changes in expression levels resulted in an increased invasion capacity of MCF-7 cells.
In triple-negative breast cancer cells, hnRNPM has been shown as a binding partner of a mutated form of chromatin regulatory protein MORC2 (microorchidia family CW-type zinc finger 2) (Zhang et al., 2018). The mutation of MORC2 protein consists in substitution of methionine to isoleucine at residue 276 (M276I), a cancer-associated mutation, which enhances the interaction of MORC2 with hnRNPM. This interaction promotes the hnRNPM-mediated splicing switch from the epithelial CD44 variant isoform containing exons v5/v6 to the mesenchymal CD44 isoform 4, ultimately driving EMT. ShRNA-mediated knockdown of hnRNPM reduced the binding of mutant MORC2 to CD44 pre-mRNA. It also reversed the mutant MORC2-induced CD44 splicing switch and EMT, consequently impairing the migration, invasion, and lung metastasis potential of mutant MORC2-expressing cells in mice.
Based on experiments with HMLE and HEK293FT cells, it has been demonstrated that hnRNPM’s splicing activity on CD44 variant exon skipping could be inhibited by the interaction of hnRNPM with AKAP8 (the A-kinase anchoring protein 8), a recently identified RNA-binding protein (Hu et al., 2020). Several observations let the authors speculate that AKAP8 binding to hnRNPM blocks hnRNPM from binding to its RNA targets. Firstly, the AKAP8-hnRNPM interaction became stronger upon RNase treatment. Secondly, depletion of AKAP8 promoted hnRNPM’s ability to bind its consensus RNA sequences and to stimulate exon skipping. Significantly, AKAP8 can bind its own RNA consensus sequences and prevent CD44 variant exon skipping, as well as the other EMT-associated alternative splicing. AKAP8 itself inhibits EMT and breast cancer metastasis to the lung. In the same study, 28 other hnRNPM-interacting splicing factors have been found (Hu et al., 2020). Among them, PTBP1 and hnRNPF promoted exon v8 inclusion in CD44 exon v8 splicing minigene reporter assay, whereas RBM10, RBMX, and hnRNPR promoted exon skipping.
Summarizing the current studies, we can conclude that hnRNPM promotes the exclusion of variant exons from CD44 pre-mRNA in breast cancer. However, the role of hnRNPM in regulating alternative splicing is more complex and may vary in different cell types.
4.4 SR proteins
The serine/arginine (SR)-rich protein family of RNA-binding proteins includes 12 members (SRSF1-12) in humans (Busch and Hertel, 2012; Wagner and Frye, 2021). The alternative nomenclature for SR proteins is presented in (Manley and Krainer, 2010). SR proteins play important roles in both alternative and constitutive splicing. As the regulator of constitutive splicing, they promote the binding of U1 snRNP to a 5′ splice site and the binding of U2 snRNP to a branch point in spliceosome assembly. In general, SR proteins are shown to antagonize hnRNP functions in alternative splicing. Of note, not all SR proteins promote splicing. Thus, depending on their phosphorylation state, SRSF10 and SRSF12 also act as global splicing repressors (Wagner and Frye, 2021). SRSF1 has been described as a mesenchymal splicing factor (Lambert and Weinberg, 2021).
Two screening studies of SR proteins for CD44 splicing were performed by Loh et al. (2014); Loh et al. (2016). Overexpression of SR proteins in MCF7 cells stably expressing the pFlare-V6 plasmid (a kind of CD44 minigene reporter system containing CD44 variant exon v6) showed that SRSF3 and SRSF4 do not affect exon v6 splicing of CD44 pre-mRNA, whereas SRSF1, SRSF6, SRSF9, and SRSF2 induced the exon v6 skipping. However, lentivirus-mediated shRNA treatment of MCF7 cells revealed that reduced expression of SRSF3 and SRSF1 caused a decrease of CD44v6-v10 and CD44v6,v8-v10 isoforms. Reduced expression of SRSF4 and SRSF9 did not induce a significant change in CD44 isoforms. Depletion of SRSF2 (also known as SC35) led to decreased expression of CD44v6 isoform but increased expression of both CD44v6-v10 and CD44v6,v8-v10 isoforms (Loh et al., 2016). These results indicate that CD44 minigene reporter systems could be used for the identification of RBPs’ responsive elements in exons or their flanking introns, but the endogenous regulation mechanisms of CD44 alternative splicing are more complicated in cells, and other events may play a role, e.g., the presence of other exons in CD44 pre-mRNA. This conclusion is confirmed by the results obtained for HEK293 and HCT-116 cells (Oh et al., 2020). Thus, using a minigene-based approach, Oh et al. demonstrated the opposite roles of SRSF9 and Tra2β on CD44 variant exon v10 splicing. While SRSF9 inhibited exon v10 inclusion, Tra2β promoted exon v10 inclusion. They also showed that both proteins functionally bind to exon v10, in which SRSF9 recognizes the AAGAC sequence and Tra2β recognizes the GAAGAAG sequence. However, the knockdown of neither SRSF9 nor Tra2β did not affect endogenous CD44 exon v10 splicing in HEK293T and HCT116 cells.
In the triple-negative breast cancer cell lines HCC1806 and MDA-MB-231, SRSF3 has been identified as a positive regulator of variant exon inclusion in CD44 pre-mRNA, especially exons v8, v9, and v10 (Guo L. et al., 2022). The loss of SRSF3 reduced the abundance of CD44 variant isoforms expression but increased the expression of CD44 standard isoform 4. Accordingly, exogenous expression of SRSF3 induced a significant increase in CD44 variant exon inclusion in the MDA-MB-231 and HCC1806 cells, while the total abundance of CD44 did not change. Interestingly, the reduction of CD44 variant isoform expression due to SRSF3 silencing could be partially rescued through the elevation of another splicing regulator TDP43 (TAR DNA-binding protein-43). Based on overexpression and knockdown experiments, it has been shown that TDP43 promotes variant exons inclusion in CD44 mRNA, especially exons v8, v9, and v10, in triple-negative breast cancer cell lines MDA-MB-231 and HCC 1806 (Guo L. et al., 2022). The AS regulation occurs through the direct interaction of TDP43 with CD44 pre-mRNA. The knockdown of TDP43 reduced stemness features of breast cancer stem cells. SRSF3, in turn, stabilized the TDP43 mRNA by inhibiting non-sense-mediated decay and thereafter provides enough TDP43 proteins for the cooperative network to regulate the splicing of its target genes (Guo L. et al., 2022).
In MGC-803gastric cancer cells, the splicing of CD44 was controlled by SRSF1 (Peng et al., 2019). The depletion of SRSF1 led to a significant decrease in CD44 isoform 3 level (but not in isoforms containing exon v6 or exons v6-v10) and an increase in CD44 isoform 4 level. An overexpression of SRSF1, in turn, induced switching from CD44 isoform 4 to isoform 3.
The splicing activity of SRSF1 and SRSF2 could be counteracted by another SR protein family member namely by NSrp70 (Kim Y. D. et al., 2011; Kim et al., 2016). Based on the CD44 exon v5 minigene assay, it has been shown that NSrp70 and SRSF1/2 have opposite functions in HEK293T cells. The interaction of NSrp70 with SRSF1 and SRSF2 prevented the SRSF1- and SRSF2-induced CD44 exon v5 exclusion.
4.5 Other RNA-binding proteins
Several other RBPs have also been implicated in the splicing regulation of CD44. Based on siRNA-mediated knockdown, it has been shown that PTBP1, RBP recognizing CUCUCU-rich sequences (Oberstrass et al., 2005), induced a significant decrease in expression of CD44 containing exon v6 at mRNA and protein level in two melanoma brain metastases’ cell lines BD and M16 (Marzese et al., 2015). Interestingly, the reduction of PTBP1 affected the migration of BD and M16 cells treated with HGF in opposite directions: M16 showed a significant decrease, while BD showed a significant increase in cell migration. Also, PTBP1 promoted exon v8 inclusion in CD44 exon v8 minigene system in HEK293FT cells (Hu et al., 2020).
CELF1 and ELAVL1 proteins, in addition to their cytoplasmic roles, have been found directly interacting in the nucleus, where they cooperatively control the splicing of CD44 in HeLa cells (David et al., 2022). Namely, they promote the inclusion of exons v7-v10. Correlation analysis of the alternative splicing events of CD44 with expression levels of CELF1 and ELAVL1 based on RNA-Seq data from TCGA revealed that high expression of CELF1 and/or ELAVL1 is correlated with the inclusion of CD44 variable exons in eight tumor types.
In experiment combination, it has been shown that SRm160 (encoded by the SRRM1 gene) is important for the inclusion of most of the endogenous CD44 variable exons in HeLa cells (Cheng and Sharp, 2023). The regulation of CD44 splicing by SRm160 occurs in a Ras-dependent manner. Reduction of SRm160 by siRNA transfection downregulated the endogenous levels of CD44 variant isoforms and correlated with a decrease in HeLa cell invasiveness. In immunoprecipitation assay an association of SRm160 with Sam68 has been revealed (Cheng and Sharp, 2023), which in turn also stimulated the formation of CD44 variant isoforms in a Ras-dependent manner (Matter et al., 2002; Cheng and Sharp, 2023). The patterns of CD44 variant exons’ inclusion in HeLa cells treated with Sam68 siRNA were like those treated with SRm160 siRNA. These results suggest that SRm160 with Sam68 may interact to regulate CD44 splicing.
The splicing activity of Sam68 is dependent on the type of a complex it is a part of (Huot et al., 2009). A large Sam68 complex (>1 MDa) is a ribonucleoprotein complex composed of ∼40 proteins. The treatment of HeLa cells by phorbol 12-myristate 13-acetate or epidermal growth factor induced the disassociation of Sam68 from this large complex and the appearance of Sam68 within the smaller complex. In human MCF-7 and BT-20 breast cancer cells Sam68 exists in equilibrium between a large and a small complex, whereas MDA-MB-231 cells harbors only the smaller Sam68 complex. The appearance of the small Sam68 complex in the cells correlated with the ability of Sam68 to promote the inclusion of exon v5 in the CD44 minigene system and cell migration (Huot et al., 2009). The existence of Sam68 in the form of a protein complex provides multiple opportunities for cell-type-specific regulation of its splicing activity. Thus, the interaction of Sam68 with SND1 in prostate cancer cells leads to a synergic effect with Sam68 on variant exon inclusion in CD44 mRNA (Cappellari et al., 2013). It has been demonstrated that SND1 affected the recruitment of Sam68 and snRNPs on CD44 pre-mRNA. These results, in combination with others provided by (Cappellari et al., 2013), suggest that SND1 acts as a bridge between RNA polymerase II (RNAPII) and Sam68 and has a crucial role in CD44 AS by favoring the recruitment of the spliceosome and the efficient splicing of the variant exons. Knockdown of SND1, or Sam68, reduced proliferation and migration of prostate cancer cells.
In prostate cancer PC3 cells, overexpression of RBM3 protein resulted in decreased expression of CD44 isoform 3 (CD44v8-v10) and an increased expression of CD44 isoform 4 (CD44s) (Zeng et al., 2013). Vice versa, decreasing the expression of RBM3 promoted the expression of CD44 isoform 3 and suppressed the expression of isoform 4. These results suggested that RBM3 promoted switching from CD44 isoform 3 to isoform 4. Such switching, in turn, attenuated CSC-like features of prostate cancer cells. This finding is confirmed by the fact that RBM3 overexpression in PC3 cells showed a significant reduction in tumor formation when cells were inoculated in nude mice.
The MBNL3 protein is a splicing regulator promoting embryonic stem cell differentiation (Han et al., 2013). Knockdown of MBNL3 in acute myeloid leukemia stem cells enhanced the expression of the CD44 isoform 3, which promoted stem cell maintenance (Holm et al., 2015).
Interestingly, in studies with MCF7 and HEK293T cells, it has been found that binding of the acetyltransferase p300 to the CD44 promoter region stimulated the inclusion of variant exons v5-v6 in CD44 mRNA independently of RNAPII transcriptional elongation rate (Siam et al., 2019). The mechanism of AS regulation by p300 included an acetylation of splicing factors, leading to the exclusion of hnRNPM from CD44 pre-mRNA and activation of Sam68.
U2AF2 knockdown and overexpression experiments revealed its positive regulatory role in the inclusion of variant exons and CD44 isoform 3 expression in melanoma cells (Zhang et al., 2016). It has been demonstrated that U2AF2 can bind to weak polypyrimidine tract in the 3′-splicing site to facilitate CD44 isoform 3 splicing. The U2AF2 activity could be inhibited by the CD82 tetraspanin protein by inducing U2AF2 ubiquitination. Knockdown of U2AF2 or CD44 isoform 3 significantly diminished the adhesion to E-selectin of Lu1205M melanoma cells and reduced the number of metastatic lesions. Of note, silencing of a set of other splicing factors in this study, Tra2β, SRp20, ESRP1, YB-1, SRm160, and Sam68, did not show any changes in the expression level of CD44 isoform 3 in melanoma cells (Zhang et al., 2016).
PCBP1 (alpha CP1 or hnRNPE1) has been characterized as a negative regulator of CD44 variants splicing in the human hepatoma cell line HepG2 (Zhang et al., 2010). An enforced expression of PCBP1 inhibited CD44 variant isoforms expression, including v3, v5, v6, v8, and v10 exons, while knockdown of endogenous PCBP1 induced CD44 variant isoforms splicing. The PCBP1 overexpression was accompanied by a decrease in invasive features of tumor cells; the knockdown accordingly promoted invasion.
Reduced expression of hnRNPL (the heterogeneous nuclear ribonucleoprotein L) promoted inclusion of only exon v10 in endogenous CD44 mRNA in MDA-MB-231 cells (Loh et al., 2015). A similar result has been shown using the CD44 exon v10 minigene reporter system in MDA-MB-231 and HCT-116 cells. In addition, it has been revealed that hnRNPL directly interacts with the CA-rich sequence in the intron upstream of CD44 exon v10. This interaction inhibited the recruitment of U2AF2 on intron upstream exon v10 and prevented its splicing.
HNRNPLL, a paralog of HNRNPL, has been demonstrated as a negative regulator of invasion and metastasis of mouse colorectal cancer CMT93 cells, which may be caused in part by its negative regulation of splicing CD44 isoforms containing exon v6 (Sakuma et al., 2018). In human colon cancer SW480 cells, reduced level of HNRNPLL enhanced expression of the CD44 isoforms containing exons v3-v10 and cell invasion activity. Induction of EMT in SW480 cells led to transcriptional downregulation of HNRNPLL and upregulation of exon v6 inclusion in CD44 mRNA.
In the study described by Muys et al. CD44 was the strongest alternatively spliced target of ZMAT3 in HCT116 cells (Muys et al., 2021). Silencing of ZMAT3 resulted in a higher abundance of CD44 variant isoform 1 and isoform 2, a concomitant reduction of the short standard CD44 isoform 4 and an increase in clonogenicity of HCT116 cells. ZMAT3 regulation of CD44 splicing may be related to its binding at pyrimidine-rich sequences of pre-mRNA introns, a crucial sequence element required for 3′ splice site definition. Most commonly, ZMAT3 binding sites consist of Us, with additional significant contribution of A/U-rich elements (AREs). Thus, ZMAT3 might compete with the other ARE-binding RBPs and splicing machinery for binding and interfere with properly recognizing 3′ splice sites (Muys et al., 2021).
In experiments with HeLa cells, A/C-rich elements (ACE) in CD44 exon v4 were recognized by the human YB-1 protein, encoding the YBX1 gene and initially identified as a transcription factor (Stickeler et al., 2001). The YB-1 binding to the exonic ACE stimulated CD44 exons v4 and v5 inclusion in the final transcript.
5 Conclusion and prospects
Alternative splicing of CD44 pre-mRNA and the essential role of CD44 isoforms in cancers are highlighted in this review. Many RBPs have been identified as regulators of CD44 isoform splicing, of which the most studied regulator is ESRP1 (Table 2, the extended version of the table see in Supplementary Table S1). RBPs typically exist as an important part of larger protein complexes that provide multiple opportunities for cell-type specific regulation of their splicing activity. Moreover, RBPs may counteract each other [e.g., as was shown for SRSF1/2 and NSrp70 (Kim Y. D. et al., 2011; Kim et al., 2016) or hnRNPM and AKAP8 (Hu et al., 2020)] and their expression ratio could be important [e.g., as it was shown for RBFOX2 and ESRP1 (Barriere et al., 2014; Meng et al., 2019)]. All of these necessitate further research into the role of RBPs mentioned in the review in each type of cancer and the identification of other possible regulators of CD44 alternative splicing. The importance of studying endogenous AS in cells is worth noting, since the model systems such as the CD44 minigene splicing reporter systems provide only limited information and do not fully reflect endogenous processes in cells.
It is also important to note that, according to sequencing data of full-length RNA transcripts from the FLIBase repository (Shi et al., 2023), CD44 splicing may be more complex than the inclusion or exclusion of cassette exons and may involve changes in the sequence of the exons themselves. How these types of CD44 isoforms are realized and what functional role they play remains to be studied. The fact that six isoforms of eight, confirmed per the NCBI database, are very low expressed and are not even included in the top 20 high-expressed ones (Supplementary Figures S1, S2), according to FLIBase, also deserves particular discussion.
Several studies demonstrated that CD44 is a potential therapeutic target among various malignant entities, e.g., triple-negative basal-like breast cancer, squamous cell carcinomas, and acute myelogenous leukemia (Yan et al., 2015; Xu et al., 2020; Elakad et al., 2022). However, the results of preclinical and clinical trials showed not only the safety and efficacy of existing anti-CD44 therapies but their limited success also [the detailed information were nicely summarized in (Xu et al., 2020; Primeaux et al., 2022; Weng et al., 2022)].
Many studies have been devoted to exploring the possibility of splicing regulation through splicing-switch oligonucleotides (SSOs) which can specifically bind to splicing sites in the pre-mRNA in a complementary pairing manner, preventing RBPs binding and the normal assembly of spliceosome (Figure 5) (Hong, 2017; Du et al., 2021; Roy Burman et al., 2021; Zhang et al., 2021). Such oligonucleotides are analogs of the antisense oligonucleotides (ASOs), which the FDA has approved for the treatment of Duchenne muscular dystrophy (Lim et al., 2017) and spinal muscular atrophy (Corey, 2017). This approach provides a hope for perspective using SSOs as regulators of AS in cancer treatment. Interestingly, the endogenous prototypes of SSOs are miRNAs. It raises the important question: Could natural miRNAs be the regulators of AS in cells? The answer of this question definitively defines direction for future studies.
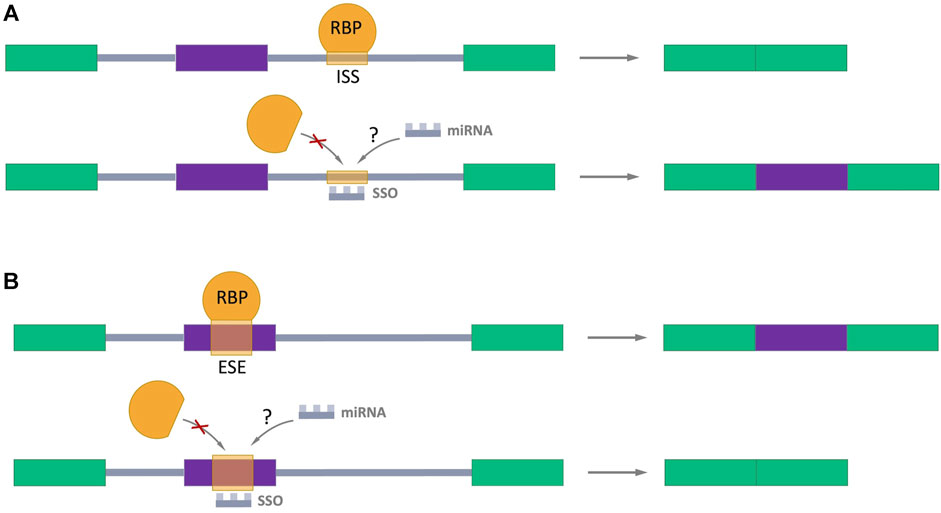
FIGURE 5. Splicing-switch oligonucleotides (SSOs) in AS regulation. (A) An SSO that binds to an intronic splicing silencer (ISS) and prevents binding of RNA-binding protein (RBP) which negatively regulates splicing (shown in orange), leading to exon inclusion. (B) An SSO that binds to an exonic splicing enhancer (ESE) and blocks the binding of RBP which promotes splicing (shown in orange), resulting in exon skipping. Exons are represented as color boxes, introns as gray lines.
Author contributions
DM: Writing–original draft and review. AT: Supervision, Writing–review and editing.
Funding
The authors declare financial support was received for the research, authorship, and/or publication of this article. This study was performed within the framework of the Basic Research Program at HSE University.
Acknowledgments
The authors thank Prof. Udo Schumacher for critically reading the manuscript; Danilia Gabudinova for her help in finding literature on RNA-binding proteins that regulate splicing of CD44.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2023.1326148/full#supplementary-material
SUPPLEMENTARY FIGURE S1 | Mean expression of CD44 isoforms across different cancers according to the FLYBase database.
SUPPLEMENTARY FIGURE S2 | The mainly expressed CD44 splicing isoforms according to the FLIBase database.
Abbreviations
AS, Alternative splicing; ASOs, Antisense Oligos; AREs, A/U-rich elements; ACE, A/C-rich elements; CRC, Colorectal cancer; CSC, Cancer stem cells; ESE, Exonic splicing enhancer; ISS, Intronic splicing silencer; iPSCs, Pluripotent stem cells; RBPs, RNA binding proteins; RNAPII, RNA polymerase II; RRM, RNA-recognition motif; siRNA, Small interfering RNA; shRNA, Small hairpin RNA; SSOs, Splicing-switch oligonucleotides.
References
Ahuja, N., Ashok, C., Natua, S., Pant, D., Cherian, A., Pandkar, M. R., et al. (2020). Hypoxia-induced TGF-β–RBFOX2–ESRP1 axis regulates human MENA alternative splicing and promotes EMT in breast cancer. Nar. Cancer 2, zcaa021. doi:10.1093/NARCAN/ZCAA021
Auboeuf, D., Hönig, A., Berget, S. M., and O’Malley, B. W. (2002). Coordinate regulation of transcription and splicing by steroid receptor coregulators. Science 298, 416–419. doi:10.1126/SCIENCE.1073734
Azevedo, R., Gaiteiro, C., Peixoto, A., Relvas-Santos, M., Lima, L., Santos, L. L., et al. (2018). CD44 glycoprotein in cancer: a molecular conundrum hampering clinical applications. Clin. Proteomics 15, 22. doi:10.1186/s12014-018-9198-9
Bánky, B., Rásó-Barnett, L., Barbai, T., Tímár, J., Becságh, P., and Rásó, E. (2012). Characteristics of CD44 alternative splice pattern in the course of human colorectal adenocarcinoma progression. Mol. Cancer 11, 83–15. doi:10.1186/1476-4598-11-83
Baraniak, A. P., Chen, J. R., and Garcia-Blanco, M. A. (2023). Fox-2 mediates epithelial cell-specific fibroblast growth factor receptor 2 exon choice. Mol. Cell Biol. 26, 1209–1222. doi:10.1128/MCB.26.4.1209-1222.2006
Barriere, G., Fici, P., Gallerani, G., Fabbri, F., Zoli, W., and Rigaud, M. (2014). Circulating tumor cells and epithelial, mesenchymal and stemness markers: characterization of cell subpopulations. Ann. Transl. Med. 2, 109. doi:10.3978/J.ISSN.2305-5839.2014.10.04
Bhattacharya, R., Mitra, T., Ray Chaudhuri, S., and Roy, S. S. (2018). Mesenchymal splice isoform of CD44 (CD44s) promotes EMT/invasion and imparts stem-like properties to ovarian cancer cells. J. Cell Biochem. 119, 3373–3383. doi:10.1002/JCB.26504
Biddle, A., Gammon, L., Fazil, B., and Mackenzie, I. C. (2013). CD44 staining of cancer stem-like cells is influenced by down-regulation of CD44 variant isoforms and up-regulation of the standard CD44 isoform in the population of cells that have undergone epithelial-to-mesenchymal Transition. PLoS One 8, e57314. doi:10.1371/JOURNAL.PONE.0057314
Bockbrader, K., and Feng, Y. (2008). Essential function, sophisticated regulation and pathological impact of the selective RNA-binding protein QKI in CNS myelin development. Future Neurol. 3, 655–668. doi:10.2217/14796708.3.6.655
Boman, B. M., Viswanathan, V., Facey, C. O. B., Fields, J. Z., and Stave, J. W. (2023). The v8-10 variant isoform of CD44 is selectively expressed in the normal human colonic stem cell niche and frequently is overexpressed in colon carcinomas during tumor development. Cancer Biol. Ther. 24, 2195363. doi:10.1080/15384047.2023.2195363
Bonnal, S. C., López-Oreja, I., and Valcárcel, J. (2020). Roles and mechanisms of alternative splicing in cancer — implications for care. Nat. Rev. Clin. Oncol. 17 (8), 457–474. doi:10.1038/s41571-020-0350-x
Bradley, R. K., and Anczuków, O. (2023). RNA splicing dysregulation and the hallmarks of cancer. Nat. Rev. Cancer 23 (3), 135–155. doi:10.1038/s41568-022-00541-7
Braeutigam, C., Rago, L., Rolke, A., Waldmeier, L., Christofori, G., and Winter, J. (2013). The RNA-binding protein Rbfox2: an essential regulator of EMT-driven alternative splicing and a mediator of cellular invasion. Oncogene 33 (9), 1082–1092. doi:10.1038/onc.2013.50
Brosseau, J. P., Lucier, J. F., Nwilati, H., Thibault, P., Garneau, D., Gendron, D., et al. (2014). Tumor microenvironment–associated modifications of alternative splicing. RNA 20, 189–201. doi:10.1261/RNA.042168.113
Brown, R. L., Reinke, L. M., Damerow, M. S., Perez, D., Chodosh, L. A., Yang, J., et al. (2011). CD44 splice isoform switching in human and mouse epithelium is essential for epithelial-mesenchymal transition and breast cancer progression. J. Clin. Invest. 121, 1064–1074. doi:10.1172/JCI44540
Busch, A., and Hertel, K. J. (2012). Evolution of SR protein and hnRNP splicing regulatory factors. Wiley Interdiscip. Rev. RNA 3, 1–12. doi:10.1002/WRNA.100
Cappellari, M., Bielli, P., Paronetto, M. P., Ciccosanti, F., Fimia, G. M., Saarikettu, J., et al. (2013). The transcriptional co-activator SND1 is a novel regulator of alternative splicing in prostate cancer cells. Oncogene 33 (29), 3794–3802. doi:10.1038/onc.2013.360
Chaffer, C. L., and Goetz, J. G. (2018). CD44 orchestrates metastatic teamwork. Dev. Cell 47, 691–693. doi:10.1016/J.DEVCEL.2018.11.038
Chen, C., Zhao, S., Karnad, A., and Freeman, J. W. (2018). The biology and role of CD44 in cancer progression: therapeutic implications. J. Hematol. Oncol. 11 (1), 64–23. doi:10.1186/S13045-018-0605-5
Chen, L., Fu, C., Zhang, Q., He, C., Zhang, F., and Wei, Q. (2020). The role of CD44 in pathological angiogenesis. FASEB J. 34, 13125–13139. doi:10.1096/FJ.202000380RR
Chen, L., Yao, Y., Sun, L., Zhou, J., Miao, M., Luo, S., et al. (2017). Snail driving alternative splicing of CD44 by ESRP1 enhances invasion and migration in epithelial ovarian cancer. Cell. Physiology Biochem. 43, 2489–2504. doi:10.1159/000484458
Chen, Y., Zubovic, L., Yang, F., Godin, K., Pavelitz, T., Castellanos, J., et al. (2016). Rbfox proteins regulate microRNA biogenesis by sequence-specific binding to their precursors and target downstream Dicer. Nucleic Acids Res. 44, 4381–4395. doi:10.1093/NAR/GKW177
Cheng, C., and Sharp, P. A. (2023). Regulation of CD44 alternative splicing by SRm160 and its potential role in tumor cell invasion. Mol. Cell Biol. 26, 362–370. doi:10.1128/MCB.26.1.362-370.2006
Cho, S. H., Park, Y. S., Kim, H. J., Kim, C. H., Lim, S. W., Huh, J. W., et al. (2012). CD44 enhances the epithelial-mesenchymal transition in association with colon cancer invasion. Int. J. Oncol. 41, 211–218. doi:10.3892/ijo.2012.1453
Corey, D. R. (2017). Nusinersen, an antisense oligonucleotide drug for spinal muscular atrophy. Nat. Neurosci. 20 (4), 497–499. doi:10.1038/nn.4508
Damianov, A., Ying, Y., Lin, C. H., Lee, J. A., Tran, D., Vashisht, A. A., et al. (2016). Rbfox proteins regulate splicing as part of a large multiprotein complex LASR. Cell 165, 606–619. doi:10.1016/J.CELL.2016.03.040
Danan-Gotthold, M., Golan-Gerstl, R., Eisenberg, E., Meir, K., Karni, R., and Levanon, E. Y. (2015). Identification of recurrent regulated alternative splicing events across human solid tumors. Nucleic Acids Res. 43, 5130–5144. doi:10.1093/NAR/GKV210
David, G., Reboutier, D., Deschamps, S., Méreau, A., Taylor, W., Padilla-Parra, S., et al. (2022). The RNA-binding proteins CELF1 and ELAVL1 cooperatively control the alternative splicing of CD44. Biochem. Biophys. Res. Commun. 626, 79–84. doi:10.1016/J.BBRC.2022.07.073
Di, C., Syafrizayanti, , Zhang, Q., Chen, Y., Wang, Y., Zhang, X., et al. (2018). Function, clinical application, and strategies of Pre-mRNA splicing in cancer. Cell Death Differ. 26 (7), 1181–1194. doi:10.1038/s41418-018-0231-3
Dinger, T. F., Chen, O., Dittfeld, C., Hetze, L., Hüther, M., Wondrak, M., et al. (2020). Microenvironmentally-driven plasticity of CD44 isoform expression determines engraftment and stem-like phenotype in CRC cell lines. Theranostics 10, 7599–7621. doi:10.7150/THNO.39893
Dittmar, K. A., Jiang, P., Park, J. W., Amirikian, K., Wan, J., Shen, S., et al. (2023). Genome-wide determination of a broad ESRP-regulated posttranscriptional network by high-throughput sequencing. Mol. Cell Biol. 32, 1468–1482. doi:10.1128/MCB.06536-11
Du, J. X., Zhu, G. Q., Cai, J. L., Wang, B., Luo, Y. H., Chen, C., et al. (2021). Splicing factors: insights into their regulatory network in alternative splicing in cancer. Cancer Lett. 501, 83–104. doi:10.1016/J.CANLET.2020.11.043
Ebersole, T. A., Chen, Q., Justice, M. J., and Artzt, K. (1996). The quaking gene product necessary in embryogenesis and myelination combines features of RNA binding and signal transduction proteins. Nat. Genet. 12 (3), 260–265. doi:10.1038/ng0396-260
Elakad, O., Häupl, B., Labitzky, V., Yao, S., Küffer, S., von Hammerstein-Equord, A., et al. (2022). Activation of CD44/PAK1/AKT signaling promotes resistance to FGFR1 inhibition in squamous-cell lung cancer. npj Precis. Oncol. 6 (1), 52–14. doi:10.1038/s41698-022-00296-2
Emili, A., Shales, M., McCracken, S., Xie, W., Tucker, P. W., Kobayashi, R., et al. (2002). Splicing and transcription-associated proteins PSF and p54nrb/NonO bind to the RNA polymerase II CTD. RNA 8, 1102–1111. doi:10.1017/S1355838202025037
Everest-Dass, A., Nersisyan, S., Maar, H., Novosad, V., Schröder-Schwarz, J., Freytag, V., et al. (2023). Spontaneous metastasis xenograft models link CD44 isoform 4 to angiogenesis, hypoxia, EMT and mitochondria-related pathways in colorectal cancer. Mol. Oncol. doi:10.1002/1878-0261.13535
Feng, P., Li, L., Deng, T., Liu, Y., Ling, N., Qiu, S., et al. (2020). NONO and tumorigenesis: more than splicing. J. Cell Mol. Med. 24, 4368–4376. doi:10.1111/JCMM.15141
Fox, S. B., Fawcett, J., Jackson, D. G., Collins, I., Gatter, K. C., Harris, A. L., et al. (1994). Normal human tissues, in addition to some tumors, express multiple different CD44 isoforms. Cancer Res. 54, 4539–4546. Available at: https://aacrjournals.org/cancerres/article/54/16/4539/500485/Normal-Human-Tissues-in-Addition-to-Some-Tumors (Accessed July 11, 2023).
Guo, L., Ke, H., Zhang, H., Zou, L., Yang, Q., Lu, X., et al. (2022a). TDP43 promotes stemness of breast cancer stem cells through CD44 variant splicing isoforms. Cell Death Dis. 13 (5), 428. doi:10.1038/s41419-022-04867-w
Guo, Q., Yang, C., and Gao, F. (2022b). The state of CD44 activation in cancer progression and therapeutic targeting. FEBS J. 289, 7970–7986. doi:10.1111/FEBS.16179
Habowski, A. N., Flesher, J. L., Bates, J. M., Tsai, C. F., Martin, K., Zhao, R., et al. (2020). Transcriptomic and proteomic signatures of stemness and differentiation in the colon crypt. Commun. Biol. 3 (1), 453–517. doi:10.1038/s42003-020-01181-z
Hall, M. P., Nagel, R. J., Fagg, W. S., Shiue, L., Cline, M. S., Perriman, R. J., et al. (2013). Quaking and PTB control overlapping splicing regulatory networks during muscle cell differentiation. RNA 19, 627–638. doi:10.1261/RNA.038422.113
Han, H., Irimia, M., Ross, P. J., Sung, H. K., Alipanahi, B., David, L., et al. (2013). MBNL proteins repress ES-cell-specific alternative splicing and reprogramming. Nature 498 (7453), 241–245. doi:10.1038/nature12270
Hanley, W. D., Napier, S. L., Burdick, M. M., Schnaar, R. L., Sackstein, R., and Konstantopoulos, K. (2006). Variant isoforms of CD44 are P- and L-selectin ligands on colon carcinoma cells. FASEB J. 20, 337–339. doi:10.1096/FJ.05-4574FJE
Harvey, S. E., Xu, Y., Lin, X., Gao, X. D., Qiu, Y., Ahn, J., et al. (2018). Coregulation of alternative splicing by hnRNPM and ESRP1 during EMT. RNA 24, 1326–1338. doi:10.1261/RNA.066712.118
Hibino, S., Shibuya, M., Engbring, J. A., Mochizuki, M., Nomizu, M., and Kleinman, H. K. (2004). Identification of an active site on the laminin alpha5 chain globular domain that binds to CD44 and inhibits malignancy. Cancer Res. 64, 4810–4816. doi:10.1158/0008-5472.CAN-04-0129
Holm, F., Hellqvist, E., Mason, C. N., Ali, S. A., Delos-Santos, N., Barrett, C. L., et al. (2015). Reversion to an embryonic alternative splicing program enhances leukemia stem cell self-renewal. Proc. Natl. Acad. Sci. U. S. A. 112, 15444–15449. doi:10.1073/PNAS.1506943112
Hong, S. (2017). RNA binding protein as an emerging therapeutic target for cancer prevention and treatment. J. Cancer Prev. 22, 203–210. doi:10.15430/JCP.2017.22.4.203
Hu, X., Harvey, S. E., Zheng, R., Lyu, J., Grzeskowiak, C. L., Powell, E., et al. (2020). The RNA-binding protein AKAP8 suppresses tumor metastasis by antagonizing EMT-associated alternative splicing. Nat. Commun. 11 (1), 486. doi:10.1038/s41467-020-14304-1
Hu, Y., and Fu, L. (2012). Targeting cancer stem cells: a new therapy to cure cancer patients. Am. J. Cancer Res. 2, 340–356. Available at: http://www.ncbi.nlm.nih.gov/pubmed/22679565 (Accessed August 24, 2021).
Huot, M. É., Vogel, G., and Richard, S. (2009). Identification of a Sam68 ribonucleoprotein complex regulated by epidermal growth factor. J. Biol. Chem. 284, 31903–31913. doi:10.1074/JBC.M109.018465
Ishii, H., Saitoh, M., Sakamoto, K., Kondo, T., Katoh, R., Tanaka, S., et al. (2014). Epithelial splicing regulatory proteins 1 (ESRP1) and 2 (ESRP2) suppress cancer cell motility via different mechanisms. J. Biol. Chem. 289, 27386–27399. doi:10.1074/JBC.M114.589432
Ishii, S., Ford, R., Thomas, P., Nachman, A., Steele, G., and Jessup, J. M. (1993). CD44 participates in the adhesion of human colorectal carcinoma cells to laminin and type IV collagen. Surg. Oncol. 2, 255–264. doi:10.1016/0960-7404(93)90015-q
Ishimoto, T., Nagano, O., Yae, T., Tamada, M., Motohara, T., Oshima, H., et al. (2011). CD44 variant regulates redox status in cancer cells by stabilizing the xCT subunit of system xc- and thereby promotes tumor growth. Cancer Cell 19, 387–400. doi:10.1016/J.CCR.2011.01.038
Ivanova, O. M., Anufrieva, K. S., Kazakova, A. N., Malyants, I. K., Shnaider, P. V., Lukina, M. M., et al. (2023). Non-canonical functions of spliceosome components in cancer progression. Cell Death Dis. 14 (2), 77–17. doi:10.1038/s41419-022-05470-9
Jalkanen, S., and Jalkanen, M. (1992). Lymphocyte CD44 binds the COOH-terminal heparin-binding domain of fibronectin. J. Cell Biol. 116, 817–825. doi:10.1083/JCB.116.3.817
Jalkanen, S. T., Bargatze, R. F., Herron, L. R., and Butcher, E. C. (1986). A lymphoid cell surface glycoprotein involved in endothelial cell recognition and lymphocyte homing in man. Eur. J. Immunol. 16, 1195–1202. doi:10.1002/EJI.1830161003
Jbara, A., Lin, K. T., Stossel, C., Siegfried, Z., Shqerat, H., Amar-Schwartz, A., et al. (2023). RBFOX2 modulates a metastatic signature of alternative splicing in pancreatic cancer. Nature 617 (7959), 147–153. doi:10.1038/s41586-023-05820-3
Jeong, H. M., Han, J., Lee, S. H., Park, H. J., Lee, H. J., Choi, J. S., et al. (2017). ESRP1 is overexpressed in ovarian cancer and promotes switching from mesenchymal to epithelial phenotype in ovarian cancer cells. Oncogenesis 6 (10), e389. doi:10.1038/oncsis.2017.87
Jiang, W., Crossman, D. K., Mitchell, E. H., Sohn, P., Crowley, M. R., and Serra, R. (2013). WNT5A inhibits metastasis and alters splicing of Cd44 in breast cancer cells. PLoS One 8, e58329. doi:10.1371/JOURNAL.PONE.0058329
Jiang, W., Zhang, Y., Kane, K. T., Collins, M. A., Simeone, D. M., Di Magliano, M. P., et al. (2015). CD44 regulates pancreatic cancer invasion through MT1-MMP. Mol. Cancer Res. 13, 9–15. doi:10.1158/1541-7786.MCR-14-0076
Jin, Y., Suzuki, H., Maegawa, S., Endo, H., Sugano, S., Hashimoto, K., et al. (2003). A vertebrate RNA-binding protein Fox-1 regulates tissue-specific splicing via the pentanucleotide GCAUG. EMBO J. 22, 905–912. doi:10.1093/EMBOJ/CDG089
Kagami, T., Yamade, M., Suzuki, T., Uotani, T., Tani, S., Hamaya, Y., et al. (2018). High expression level of CD44v8-10 in cancer stem-like cells is associated with poor prognosis in esophageal squamous cell carcinoma patients treated with chemoradiotherapy. Oncotarget 9, 34876–34888. doi:10.18632/ONCOTARGET.26172
Katsuno, Y., and Derynck, R. (2021). Epithelial plasticity, epithelial-mesenchymal transition, and the TGF-β family. Dev. Cell 56, 726–746. doi:10.1016/J.DEVCEL.2021.02.028
Kawai, T., Iwata, K., Shinotsuka, Y., Kubo, S., Masuoka, H., Yabuta, T., et al. (2019). CD44v8-10 and CD44s are age-dependently expressed in primary cultured papillary thyroid carcinoma cells and are associated with cell proliferation. Kobe J. Med. Sci. 65, E1–E9. Available at: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6668591/.(Accessed July 17, 2023).
Kim, C. H., Kim, Y. D., Choi, E. K., Kim, H. R., Na, B. R., Im, S. H., et al. (2016). Nuclear speckle-related protein 70 binds to serine/argininerich splicing factors 1 and 2 via an arginine/serine-like region and counteracts their alternative splicing activity. J. Biol. Chem. 291, 6169–6181. doi:10.1074/JBC.M115.689414
Kim, K. K., Adelstein, R. S., and Kawamoto, S. (2009). Identification of neuronal nuclei (NeuN) as Fox-3, a new member of the Fox-1 gene family of splicing factors. J. Biol. Chem. 284, 31052–31061. doi:10.1074/JBC.M109.052969
Kim, K. K., Kim, Y. C., Adelstein, R. S., and Kawamoto, S. (2011a). Fox-3 and PSF interact to activate neural cell-specific alternative splicing. Nucleic Acids Res. 39, 3064–3078. doi:10.1093/NAR/GKQ1221
Kim, Y. D., Kim, H. S., Lee, J., Choi, J. K., Han, E., Jeong, J. E., et al. (2018). ESRP1-induced CD44 v3 is important for controlling pluripotency in human pluripotent stem cells. Stem Cells 36, 1525–1534. doi:10.1002/STEM.2864
Kim, Y. D., Lee, J. Y., Oh, K. M., Araki, M., Araki, K., Yamamura, K. I., et al. (2011b). NSrp70 is a novel nuclear speckle-related protein that modulates alternative pre-mRNA splicing in vivo. Nucleic Acids Res. 39, 4300–4314. doi:10.1093/NAR/GKQ1267
Knott, G. J., Bond, C. S., and Fox, A. H. (2016). The DBHS proteins SFPQ, NONO and PSPC1: a multipurpose molecular scaffold. Nucleic Acids Res. 44, 3989–4004. doi:10.1093/NAR/GKW271
Kuroyanagi, H. (2009). Fox-1 family of RNA-binding proteins. Cell. Mol. Life Sci. 66, 3895–3907. doi:10.1007/S00018-009-0120-5
Lambert, A. W., and Weinberg, R. A. (2021). Linking EMT programmes to normal and neoplastic epithelial stem cells. Nat. Rev. Cancer 21 (5), 325–338. doi:10.1038/s41568-021-00332-6
Larocque, D., Pilotte, J., Chen, T., Cloutier, F., Massie, B., Pedraza, L., et al. (2002). Nuclear retention of MBP mRNAs in the Quaking viable Mice. Neuron 36, 815–829. doi:10.1016/S0896-6273(02)01055-3
Lau, W. M., Teng, E., Chong, H. S., Lopez, K. A. P., Tay, A. Y. L., Salto-Tellez, M., et al. (2014). CD44v8-10 is a cancer-specific marker for gastric cancer stem cells. Cancer Res. 74, 2630–2641. doi:10.1158/0008-5472.CAN-13-2309
Li, J., Choi, P. S., Chaffer, C. L., Labella, K., Hwang, J. H., Giacomelli, A. O., et al. (2018). An alternative splicing switch in FLNB promotes the mesenchymal cell state in human breast cancer. Elife 7, e37184. doi:10.7554/ELIFE.37184
Li, L., Hao, X., Qin, J., Tang, W., He, F., Smith, A., et al. (2014). Antibody against CD44s inhibits pancreatic tumor initiation and postradiation recurrence in mice. Gastroenterology 146, 1108–1118. doi:10.1053/J.GASTRO.2013.12.035
Li, Z., Zhang, Y., Li, D., and Feng, Y. (2000). Destabilization and mislocalization of myelin basic protein mRNAs in quaking dysmyelination lacking the QKI RNA-binding proteins. J. Neurosci. 20, 4944–4953. doi:10.1523/JNEUROSCI.20-13-04944.2000
Lim, K. R. Q., Maruyama, R., and Yokota, T. (2017). Eteplirsen in the treatment of Duchenne muscular dystrophy. Drug Des. Devel Ther. 11, 533–545. doi:10.2147/DDDT.S97635
Liu, L., Xie, N., Rennie, P., Challis, J. R. G., Gleave, M., Lye, S. J., et al. (2011). Consensus PP1 binding motifs regulate transcriptional corepression and alternative RNA splicing activities of the steroid receptor coregulators, p54nrb and PSF. Mol. Endocrinol. 25, 1197–1210. doi:10.1210/ME.2010-0517
Liu, Y., Li, Y., Du, C., Kuang, S., Zhou, X., Zhang, J., et al. (2022). Underlying mechanisms of epithelial splicing regulatory proteins in cancer progression. J. Mol. Med. 100 (11), 1539–1556. doi:10.1007/S00109-022-02257-5
Loh, T. J., Cho, S., Moon, H., Jang, H. N., Williams, D. R., Jung, D. W., et al. (2015). hnRNP L inhibits CD44 V10 exon splicing through interacting with its upstream intron. Biochimica Biophysica Acta (BBA) - Gene Regul. Mech. 1849, 743–750. doi:10.1016/J.BBAGRM.2015.01.004
Loh, T. J., Moon, H., Cho, S., Jung, D. W., Hong, S. E., Kim, D. H., et al. (2014). SC35 promotes splicing of the C5-V6-C6 isoform of CD44 pre-mRNA. Oncol. Rep. 31, 273–279. doi:10.3892/or.2013.2812
Loh, T. J., Moon, H., Jang, H. N., Liu, Y., Choi, N., Shen, S., et al. (2016). SR proteins regulate V6 exon splicing of CD44 pre-mRNA. BMB Rep. 49, 612–616. doi:10.5483/BMBREP.2016.49.11.118
Lone, B. A., Siraj, F., Sharma, I., Verma, S., Karna, S. K. L., Ahmad, F., et al. (2023). Non-POU Domain-Containing Octomer-Binding (NONO) protein expression and stability promotes the tumorigenicity and activation of Akt/MAPK/β-catenin pathways in human breast cancer cells. Cell Commun. Signal. 21, 157. doi:10.1186/S12964-023-01179-0
Mackay, C. R., Terpe, H. J., Stauder, R., Marston, W. L., Stark, H., and Günthert, U. (1994). Expression and modulation of CD44 variant isoforms in humans. J. Cell Biol. 124, 71–82. doi:10.1083/JCB.124.1.71
Manley, J. L., and Krainer, A. R. (2010). A rational nomenclature for serine/arginine-rich protein splicing factors (SR proteins). Genes Dev. 24, 1073–1074. doi:10.1101/GAD.1934910
Marko, M., Leichter, M., Patrinou-Georgoula, M., and Guialis, A. (2010). hnRNP M interacts with PSF and p54nrb and co-localizes within defined nuclear structures. Exp. Cell Res. 316, 390–400. doi:10.1016/J.YEXCR.2009.10.021
Marzese, D. M., Liu, M., Huynh, J. L., Hirose, H., Donovan, N. C., Huynh, K. T., et al. (2015). Brain metastasis is predetermined in early stages of cutaneous melanoma by CD44v6 expression through epigenetic regulation of the spliceosome. Pigment. Cell Melanoma Res. 28, 82–93. doi:10.1111/PCMR.12307
Mashita, N., Yamada, S., Nakayama, G., Tanaka, C., Iwata, N., Kanda, M., et al. (2014). Epithelial to mesenchymal transition might be induced via CD44 isoform switching in colorectal cancer. J. Surg. Oncol. 110, 745–751. doi:10.1002/jso.23705
Matter, N., Herrlich, P., and König, H. (2002). Signal-dependent regulation of splicing via phosphorylation of Sam68. Nature 420, 691–695. doi:10.1038/nature01153
Meng, X., Yang, S., Zhang, J., and Yu, H. (2019). Contribution of alternative splicing to breast cancer metastasis. J. Cancer Metastasis Treat. 5, 21. doi:10.20517/2394-4722.2018.96
Mesrati, M. H., Syafruddin, S. E., Mohtar, M. A., and Syahir, A. (2021). CD44: a multifunctional mediator of cancer progression. Biomolecules 11, 1850. doi:10.3390/BIOM11121850
Mima, K., Okabe, H., Ishimoto, T., Hayashi, H., Nakagawa, S., Kuroki, H., et al. (2012). CD44s regulates the TGF-β-mediated mesenchymal phenotype and is associated with poor prognosis in patients with hepatocellular carcinoma. Cancer Res. 72, 3414–3423. doi:10.1158/0008-5472.CAN-12-0299
Miwa, T., Nagata, T., Kojima, H., Sekine, S., and Okumura, T. (2017). Isoform switch of CD44 induces different chemotactic and tumorigenic ability in gallbladder cancer. Int. J. Oncol. 51, 771–780. doi:10.3892/IJO.2017.4063
Miyake, H., Eto, H., Arakawa, S., Kamidono, S., and Hara, I. (2002). Over expression of CD44v8-10 in urinary exfoliated cells as an independent prognostic predictor in patients with urothelial cancer. J. Urol. 167, 1282–1287. doi:10.1016/S0022-5347(05)65282-2
Mukohyama, J., Isobe, T., Hu, Q., Hayashi, T., Watanabe, T., Maeda, M., et al. (2019). MiR-221 targets QKI to enhance the tumorigenic capacity of human colorectal cancer stem cells. Cancer Res. 79, 5151–5158. doi:10.1158/0008-5472.CAN-18-3544
Muys, B. R., Anastasakis, D. G., Claypool, D., Pongor, L., Li, X. L., Grammatikakis, I., et al. (2021). The p53-induced RNA-binding protein ZMAT3 is a splicing regulator that inhibits the splicing of oncogenic CD44 variants in colorectal carcinoma. Genes Dev. 35, 102–116. doi:10.1101/GAD.342634.120
Nakahata, S., and Kawamoto, S. (2005). Tissue-dependent isoforms of mammalian Fox-1 homologs are associated with tissue-specific splicing activities. Nucleic Acids Res. 33, 2078–2089. doi:10.1093/NAR/GKI338
Naor, D., Sionov, R. V., and Ish-Shalom, D. (1997). CD44: structure, function and association with the malignant process. Adv. Cancer Res. 71, 241–319. doi:10.1016/S0065-230X(08)60101-3
Novosad, V. O. (2023). Identification of significant RNA-binding proteins in the process of CD44 splicing using the boosted beta regression algorithm. Dokl. Biochem. Biophys. 510, 99–103. doi:10.1134/S1607672923700199
Novosad, V. O., and Maltseva, D. V. (2023). The RNA-binding proteins OAS1, ZFP36L2, and DHX58 are involved in the regulation of CD44 mRNA splicing in colorectal cancer cells. Bull. Exp. Biol. Med. 175, 144–149. doi:10.1007/S10517-023-05826-X
Oberstrass, F. C., Auwetor, S. D., Erat, M., Hargous, Y., Henning, A., Wenter, P., et al. (2005). Structure of PTB bound to RNA: specific binding and implications for splicing regulation. Science 309, 2054–2057. doi:10.1126/SCIENCE.1114066
Oh, J., Liu, Y., Choi, N., Ha, J., Pradella, D., Ghigna, C., et al. (2020). Opposite roles of Tra2β and SRSF9 in the v10 exon splicing of CD44. Cancers 12, 3195. doi:10.3390/CANCERS12113195
Orian-Rousseau, V., and Sleeman, J. (2014). CD44 is a multidomain signaling platform that integrates extracellular matrix cues with growth factor and cytokine signals. Adv. Cancer Res. 123, 231–254. doi:10.1016/B978-0-12-800092-2.00009-5
Ouhtit, A., Rizeq, B., Saleh, H. A., Rahman, M. D. M., and Zayed, H. (2018). Novel CD44-downstream signaling pathways mediating breast tumor invasion. Int. J. Biol. Sci. 14, 1782–1790. doi:10.7150/IJBS.23586
Peng, W., Liu, J., Li, C., Ma, R., and Jie, J. (2019). Hnrnpk promotes gastric tumorigenesis through regulating cd44e alternative splicing. Cancer Cell Int. 19, 335. doi:10.1186/s12935-019-1020-x
Ponta, H., Sherman, L., and Herrlich, P. A. (2003). CD44: from adhesion molecules to signalling regulators. Nat. Rev. Mol. Cell Biol. 4, 33–45. doi:10.1038/nrm1004
Pradella, D., Naro, C., Sette, C., and Ghigna, C. (2017). EMT and stemness: flexible processes tuned by alternative splicing in development and cancer progression. Mol. Cancer 16 (1), 8–19. doi:10.1186/S12943-016-0579-2
Prasad, V. V. T. S., and Gopalan, R. O. G. (2015). Continued use of MDA-MB-435, a melanoma cell line, as a model for human breast cancer, even in year, 2014. npj Breast Cancer 1 (1), 15002. doi:10.1038/npjbcancer.2015.2
Preca, B. T., Bajdak, K., Mock, K., Sundararajan, V., Pfannstiel, J., Maurer, J., et al. (2015). A self-enforcing CD44s/ZEB1 feedback loop maintains EMT and stemness properties in cancer cells. Int. J. Cancer 137, 2566–2577. doi:10.1002/IJC.29642
Primeaux, M., Gowrikumar, S., and Dhawan, P. (2022). Role of CD44 isoforms in epithelial-mesenchymal plasticity and metastasis. Clin. Exp. Metastasis 39 (3), 391–406. doi:10.1007/S10585-022-10146-X
Prochazka, L., Tesarik, R., and Turanek, J. (2014). Regulation of alternative splicing of CD44 in cancer. Cell Signal 26, 2234–2239. doi:10.1016/J.CELLSIG.2014.07.011
Protin, U., Schweighoffer, T., Jochum, W., and Hilberg, F. (1999). CD44-Deficient mice develop normally with changes in subpopulations and recirculation of lymphocyte subsets. J. Immunol. 163, 4917–4923. doi:10.4049/JIMMUNOL.163.9.4917
Reinke, L. M., Xu, Y., and Cheng, C. (2012). Snail represses the splicing regulator epithelial splicing regulatory protein 1 to promote epithelial-mesenchymal transition. J. Biol. Chem. 287, 36435–36442. doi:10.1074/JBC.M112.397125
Rogalska, M. E., Vivori, C., and Valcárcel, J. (2022). Regulation of pre-mRNA splicing: roles in physiology and disease, and therapeutic prospects. Nat. Rev. Genet. 24 (4), 251–269. doi:10.1038/s41576-022-00556-8
Roy Burman, D., Das, S., Das, C., and Bhattacharya, R. (2021). Alternative splicing modulates cancer aggressiveness: role in EMT/metastasis and chemoresistance. Mol. Biol. Rep. 48 (1), 897–914. doi:10.1007/S11033-020-06094-Y
Saccomanno, L., Loushin, C., Jan, E., Punkay, E., Artzt, K., and Goodwin, E. B. (1999). The STAR protein QKI-6 is a translational repressor. Proc. Natl. Acad. Sci. U. S. A. 96, 12605–12610. doi:10.1073/PNAS.96.22.12605
Sakuma, K., Sasaki, E., Kimura, K., Komori, K., Shimizu, Y., Yatabe, Y., et al. (2018). HNRNPLL, a newly identified colorectal cancer metastasis suppressor, modulates alternative splicing of CD44 during epithelial-mesenchymal transition. Gut 67, 1103–1111. doi:10.1136/GUTJNL-2016-312927
Screaton, G. R., Bell, M. V., Jackson, D. G., Cornelis, F. B., Gerth, U., and Bell, J. I. (1992). Genomic structure of DNA encoding the lymphocyte homing receptor CD44 reveals at least 12 alternatively spliced exons. Proc. Natl. Acad. Sci. 89, 12160–12164. doi:10.1073/PNAS.89.24.12160
Senbanjo, L. T., and Chellaiah, M. A. (2017). CD44: a multifunctional cell surface adhesion receptor is a regulator of progression and metastasis of cancer cells. Front. Cell Dev. Biol. 5, 18. doi:10.3389/FCELL.2017.00018
Shaw, T. I., Zhao, B., Li, Y., Wang, H., Wang, L., Manley, B., et al. (2022). Multi-omics approach to identifying isoform variants as therapeutic targets in cancer patients. Front. Oncol. 12, 1051487. doi:10.3389/FONC.2022.1051487
Shi, Q., Li, X., Liu, Y., Chen, Z., and He, X. (2023). FLIBase: a comprehensive repository of full-length isoforms across human cancers and tissues. Nucleic Acids Res., gkad745. doi:10.1093/NAR/GKAD745
Siam, A., Baker, M., Amit, L., Regev, G., Rabner, A., Najar, R. A., et al. (2019). Regulation of alternative splicing by p300-mediated acetylation of splicing factors. RNA 25, 813–824. doi:10.1261/RNA.069856.118
Skandalis, S. S., Karalis, T. T., Chatzopoulos, A., and Karamanos, N. K. (2019). Hyaluronan-CD44 axis orchestrates cancer stem cell functions. Cell Signal 63, 109377. doi:10.1016/J.CELLSIG.2019.109377
Sosulski, A., Horn, H., Zhang, L., Coletti, C., Vathipadiekal, V., Castro, C. M., et al. (2016). CD44 splice variant v8-10 as a marker of serous ovarian cancer prognosis. PLoS One 11, e0156595. doi:10.1371/JOURNAL.PONE.0156595
Stickeler, E., Fraser, S. D., Honig, A., Chen, A. L., Berget, S. M., and Cooper, T. A. (2001). The RNA binding protein YB-1 binds A/C-rich exon enhancers and stimulates splicing of the CD44 alternative exon v4. EMBO J. 20, 3821–3830. doi:10.1093/EMBOJ/20.14.3821
Sun, H., Liu, T., Zhu, D., Dong, X., Liu, F., Liang, X., et al. (2017). HnRNPM and CD44s expression affects tumor aggressiveness and predicts poor prognosis in breast cancer with axillary lymph node metastases. Genes Chromosom. Cancer 56, 598–607. doi:10.1002/GCC.22463
Ule, J., and Blencowe, B. J. (2019). Alternative splicing regulatory networks: functions, mechanisms, and evolution. Mol. Cell 76, 329–345. doi:10.1016/J.MOLCEL.2019.09.017
Underhill, C. (1992). CD44: the hyaluronan receptor. J. Cell Sci. 103, 293–298. doi:10.1242/JCS.103.2.293
Underwood, J. G., Boutz, P. L., Dougherty, J. D., Stoilov, P., and Black, D. L. (2023). Homologues of the Caenorhabditis elegans fox-1 protein are neuronal splicing regulators in mammals. Mol. Cell Biol. 25, 10005–10016. doi:10.1128/MCB.25.22.10005-10016.2005
Vadlamudi, Y., and Kang, S. C. (2022). Silencing ESRP1 expression promotes caspase-independent cell death via nuclear translocation of AIF in colon cancer cells. Cell Signal 91, 110237. doi:10.1016/J.CELLSIG.2021.110237
Venables, J. P., Brosseau, J.-P., Gadea, G., Klinck, R., Prinos, P., Beaulieu, J.-F., et al. (2013). RBFOX2 is an important regulator of mesenchymal tissue-specific splicing in both normal and cancer tissues. Mol. Cell Biol. 33, 396–405. doi:10.1128/MCB.01174-12
Venables, J. P., Klinck, R., Koh, C., Gervais-Bird, J., Bramard, A., Inkel, L., et al. (2009). Cancer-associated regulation of alternative splicing. Nat. Struct. Mol. Biol. 16 (6), 670–676. doi:10.1038/nsmb.1608
Wagner, R. E., and Frye, M. (2021). Noncanonical functions of the serine-arginine-rich splicing factor (SR) family of proteins in development and disease. BioEssays 43, 2000242. doi:10.1002/BIES.202000242
Wahl, M. C., Will, C. L., and Lührmann, R. (2009). The spliceosome: design principles of a dynamic RNP machine. Cell 136, 701–718. doi:10.1016/J.CELL.2009.02.009
Wang, X., Han, M., Wang, S., Sun, Y., Zhao, W., Xue, Z., et al. (2022). Targeting the splicing factor NONO inhibits GBM progression through GPX1 intron retention. Theranostics 12, 5451–5469. doi:10.7150/THNO.72248
Wang, Y., and Chen, L. L. (2020). Organization and function of paraspeckles. Essays Biochem. 64, 875–882. doi:10.1042/EBC20200010
Wang, Z., Zhao, K., Hackert, T., and Zöller, M. (2018). CD44/CD44v6 a reliable companion in cancer-initiating cell maintenance and tumor progression. Front. Cell Dev. Biol. 6, 97. doi:10.3389/fcell.2018.00097
Warzecha, C. C., Sato, T. K., Nabet, B., Hogenesch, J. B., and Carstens, R. P. (2009a). ESRP1 and ESRP2 are epithelial cell-type-specific regulators of FGFR2 splicing. Mol. Cell 33, 591–601. doi:10.1016/J.MOLCEL.2009.01.025
Warzecha, C. C., Shen, S., Xing, Y., and Carstens, R. P. (2009b). The epithelial splicing factors ESRP1 and ESRP2 positively and negatively regulate diverse types of alternative splicing events. RNA Biol. 6, 546–562. doi:10.4161/RNA.6.5.9606
Weng, X., Maxwell-Warburton, S., Hasib, A., Ma, L., and Kang, L. (2022). The membrane receptor CD44: novel insights into metabolism. Trends Endocrinol. Metabolism 33, 318–332. doi:10.1016/J.TEM.2022.02.002
Xu, H., Niu, M., Yuan, X., Wu, K., and Liu, A. (2020). CD44 as a tumor biomarker and therapeutic target. Exp. Hematol. Oncol. 9, 36. doi:10.1186/s40164-020-00192-0
Xu, Y., Gao, X. D., Lee, J. H., Huang, H., Tan, H., Ahn, J., et al. (2014). Cell type-restricted activity of hnRNPM promotes breast cancer metastasis via regulating alternative splicing. Genes Dev. 28, 1191–1203. doi:10.1101/GAD.241968.114
Yae, T., Tsuchihashi, K., Ishimoto, T., Motohara, T., Yoshikawa, M., Yoshida, G. J., et al. (2012). Alternative splicing of CD44 mRNA by ESRP1 enhances lung colonization of metastatic cancer cell. Nat. Commun. 3 (1), 883–889. doi:10.1038/ncomms1892
Yaghobi, Z., Movassaghpour, A., Talebi, M., Abdoli Shadbad, M., Hajiasgharzadeh, K., Pourvahdani, S., et al. (2021). The role of CD44 in cancer chemoresistance: a concise review. Eur. J. Pharmacol. 903, 174147. doi:10.1016/J.EJPHAR.2021.174147
Yamaguchi, A., Urano, T., Goi, T., Saito, M., Takeuchi, K., Hirose, K., et al. (2016). Expression of a CD44 variant containing exons 8 to 10 is a useful independent factor for the prediction of prognosis in colorectal cancer patients. J. Clin. Oncol. 14, 1122–1127. doi:10.1200/JCO.1996.14.4.1122
Yamaguchi, A., Zhang, M., Goi, T., Fujita, T., Niimoto, S., Katayama, K., et al. (2000). Expression of variant CD44 containing variant exon v8-10 in gallbladder cancer. Oncol. Rep. 7, 541–544. doi:10.3892/or.7.3.541
Yan, Y., Zuo, X., and Wei, D. (2015). Concise review: emerging role of CD44 in cancer stem cells: a Promising Biomarker and Therapeutic Target. Stem Cells Transl. Med. 4, 1033–1043. doi:10.5966/sctm.2015-0048
Yang, L., Yang, J., Jacobson, B., Gilbertsen, A., Smith, K., Higgins, L. A., et al. (2022). SFPQ promotes lung cancer malignancy via regulation of CD44 v6 expression. Front. Oncol. 12, 862250. doi:10.3389/FONC.2022.862250
Yang, Y., Park, J. W., Bebee, T. W., Warzecha, C. C., Guo, Y., Shang, X., et al. (2023). Determination of a comprehensive alternative splicing regulatory network and combinatorial regulation by key factors during the epithelial-to-mesenchymal transition. Mol. Cell Biol. 36, 1704–1719. doi:10.1128/MCB.00019-16
Yeo, G. W., Coufal, N. G., Liang, T. Y., Peng, G. E., Fu, X. D., and Gage, F. H. (2009). An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nat. Struct. Mol. Biol. 16 (2), 130–137. doi:10.1038/nsmb.1545
Zeilstra, J., Joosten, S. P. J., Van Andel, H., Tolg, C., Berns, A., Snoek, M., et al. (2013). Stem cell CD44v isoforms promote intestinal cancer formation in Apc(min) mice downstream of Wnt signaling. Oncogene 33 (5), 665–670. doi:10.1038/onc.2012.611
Zeng, Y., Wodzenski, D., Gao, D., Shiraishi, T., Terada, N., Li, Y., et al. (2013). Stress-response protein RBM3 attenuates the stem-like properties of prostate cancer cells by interfering with CD44 variant splicing. Cancer Res. 73, 4123–4133. doi:10.1158/0008-5472.CAN-12-1343
Zhang, F. L., Cao, J. L., Xie, H. Y., Sun, R., Yang, L. F., Shao, Z. M., et al. (2018). Cancer-associated MORC2-mutant M276I regulates an hnRNPM-mediated CD44 splicing switch to promote invasion and metastasis in triple-negative breast cancer. Cancer Res. 78, 5780–5792. doi:10.1158/0008-5472.CAN-17-1394
Zhang, H., Brown, R. L., Wei, Y., Zhao, P., Liu, S., Liu, X., et al. (2019). CD44 splice isoform switching determines breast cancer stem cell state. Genes Dev. 33, 166–179. doi:10.1101/gad.319889.118
Zhang, P., Feng, S., Liu, G., Wang, H., Fu, A., Zhu, H., et al. (2016). CD82 suppresses CD44 alternative splicing-dependent melanoma metastasis by mediating U2AF2 ubiquitination and degradation. Oncogene 35 (38), 5056–5069. doi:10.1038/onc.2016.67
Zhang, T., Huang, X. H., Dong, L., Hu, D., Ge, C., Zhan, Y. Q., et al. (2010). PCBP-1 regulates alternative splicing of the CD44 gene and inhibits invasion in human hepatoma cell line HepG2 cells. Mol. Cancer 9, 72–10. doi:10.1186/1476-4598-9-72
Zhang, Y., Qian, J., Gu, C., and Yang, Y. (2021). Alternative splicing and cancer: a systematic review. Signal Transduct. Target. Ther. 6 (1), 78–14. doi:10.1038/s41392-021-00486-7
Zhang, Y. E., Wei, J., Wang, H., Xue, X., An, Y., Tang, D., et al. (2012). Epithelial mesenchymal transition correlates with CD24+CD44+ and CD133+ cells in pancreatic cancer. Oncol. Rep. 27, 1599–1605. doi:10.3892/or.2012.1681
Zhao, J., Xie, W., Yang, Z., Zhao, M., Ke, T., Xu, C., et al. (2022). Identification and characterization of a special type of subnuclear structure: AGGF1-coated paraspeckles. FASEB J. 36, e22366. doi:10.1096/FJ.202101690RR
Zhao, L., Mandler, M. D., Yi, H., and Feng, Y. (2010). Quaking I controls a unique cytoplasmic pathway that regulates alternative splicing of myelin-associated glycoprotein. Proc. Natl. Acad. Sci. U. S. A. 107, 19061–19066. doi:10.1073/PNAS.1007487107
Zhou, D., Couture, S., Scott, M. S., and Abou Elela, S. (2021). RBFOX2 alters splicing outcome in distinct binding modes with multiple protein partners. Nucleic Acids Res. 49, 8370–8383. doi:10.1093/NAR/GKAB595
Keywords: CD44 alternative splicing regulation, CD44 isoform, RNA-binding protein, cancer, ESRP1
Citation: Maltseva D and Tonevitsky A (2023) RNA-binding proteins regulating the CD44 alternative splicing. Front. Mol. Biosci. 10:1326148. doi: 10.3389/fmolb.2023.1326148
Received: 22 October 2023; Accepted: 15 November 2023;
Published: 01 December 2023.
Edited by:
Jiao Li, Sichuan University, ChinaCopyright © 2023 Maltseva and Tonevitsky. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Diana Maltseva, ZC5tYWx0c2V2YUBoc2UucnU=
 Diana Maltseva
Diana Maltseva Alexander Tonevitsky
Alexander Tonevitsky