- 1Orthopaedic Center, Affiliated Hospital of Guangdong Medical University, Zhanjiang, China
- 2Guangdong Engineering Research Center for Translation of Medical 3D Printing Application, Guangdong Provincial Key Laboratory of Medical Biomechanics, National Key Discipline of Human Anatomy, School of Basic Medical Sciences, Southern Medical University, Guangzhou, China
- 3Guangdong Medical Innovation Platform for Translation of 3D Printing Application, The Third Affiliated Hospital of Southern Medical University, Guangzhou, China
Low back pain (LBP) is a major public health problem worldwide and a significant health and economic burden. Intervertebral disc degeneration (IDD) is the reason for LBP. However, we have not identified effective therapeutic strategies to address this challenge. With accumulating knowledge on the role of circular RNAs in the pathogenesis of IDD, we realised that circular RNAs (circRNAs) may have tremendous therapeutic potential and clinical application prospects in this field. This review presents an overview of the current understanding of characteristics, classification, biogenesis, and function of circRNAs and summarises the protective and detrimental circRNAs involved in the intervertebral disc that have been studied thus far. This review is aimed to help researchers better understand the regulatory role of circRNAs in the progression of IDD, reveal their clinical therapeutic potential, and provide a theoretical basis for the prevention and targeted treatment of IDD.
1 Introduction
Low back pain (LBP) is a major public health concern worldwide, and up to 84% of people experiences LBP during their lifetime. Approximately 23% of people experiences chronic LBP, and 11%–12% of people experiences LBP-related disability (Hoy et al., 2012). LBP affects people of all ages and is a major contributor to the global disease burden. For academics and physicians, managing LBP remains difficult despite advancements in its evaluation and treatment (Vlaeyen et al., 2018). A strong correlation exists between IDD and LBP (Risbud and Shapiro, 2014). According to modern evidence-based medicine, many factors cause or accelerate IDD development, including mechanical, genetic, traumatic, inflammatory, and biological factors (Molinos et al., 2015b; Oichi et al., 2020). There is a strong possibility that among these factors, genetic factors may cause the degeneration of the intervertebral disc (IVD) (Zhang et al., 2008).
The IVD is a complex fibrocartilaginous structure comprising the nucleus pulposus (NP), annulus fibrosus (AF), and cartilaginous vertebral endplates (CEP) (Liuke et al., 2005). The NP is a gelatinous structure located at the centre of the disc that produces the extracellular matrix (ECM), including aggrecan and type II collagen, which play crucial roles in maintaining the integrity of the disc (Roughley, 2004; Waxenbaum et al., 2021). The AF is a fibrous, avascular external region of the IVD consisting of type I and type II collagen fibrils that surround the NP (Ashinsky et al., 2019). As the main nutrient supply of the IVD is diffused by the endplates, a thin horizontal layer is necessary for the appropriate functioning of the IVD (Urban et al., 2004; Raj, 2008). Therefore, they play a crucial role in maintaining the proper function of the IVD. The development of IDD is associated with disorders or abnormalities in these three components.
The IVD is unique in that it does not have a rich blood and nutrient supply structure and is the largest non-vascular part of the body (Urban et al., 2004). The IVD cells depend on the capillary supply from the vertebral body (Roberts et al., 2006), and this unique structure of the IVD makes it highly vulnerable to injury, particularly in terms of the supply of nutrients to the disc. The IVD is very sensitive to glucose and oxygen, and a lack of nutrient supply reduces glucose concentration and increases lactate synthesis owing to a decrease in oxygen concentration, resulting in an acidic pH environment (Johnson et al., 2008; Hiyama et al., 2012). Cells have trouble surviving extended exposure to low pH or glucose concentrations because the rate of matrix formation substantially declines at an acidic pH and low oxygen concentrations, leading to disc degeneration (Horner and Urban, 2001; Urban and Roberts, 2003).
The main factors involved in IDD are apoptosis, ECM degradation, and inflammatory factors (Clouet et al., 2009). Metabolic disorders of NP cells under various factors reduce their ability to synthesise ECM components, such as aggrecan and type II collagen, and cause the secretion of a large number of ECM-degrading molecules, such as matrix metalloproteinases (MMP), deintegrins, and a disintegrin and metalloproteinase with thrombospondin-like motifs (ADAMTS), resulting in progressive degradation of the ECM, which is responsible for the mechanical functioning of the disc (Vo et al., 2013; Wang et al., 2015). The association between disc degeneration and inflammation is widely recognised. Inflammation also plays a significant role in disc degeneration, and inflammatory variables may even play a role in the pain induced by IDD (Richardson et al., 2009; Molinos et al., 2015a; Mosley et al., 2017).
NP cells secrete proinflammatory cytokines in response to multiple triggers, including tumour necrosis factor (TNF), interleukin-1β (IL-1β), and interleukin-6 (IL-6) (Le Maitre et al., 2005; Wang et al., 2011). Proinflammatory cytokines lead to the dysregulation of ECM metabolism by upregulating ECM degradation enzymes (e.g., MMP) and downregulating ECM structural components (e.g., type II collagen and aggrecan) (Séguin et al., 2005; Studer et al., 2011; Risbud and Shapiro, 2014). ECM degradation further stimulates NP cells to produce proinflammatory factors, creating a vicious cycle (Quero et al., 2013). Simultaneously, the senescence and death of NP cells are facilitated by metabolic abnormalities in the ECM and the generation of inflammatory substances (Wang et al., 2016). Apoptosis, the degradation of the ECM, and the production of inflammatory factors are interrelated and mutually reinforcing and together contribute to the degeneration of IVDs (Figure 1).
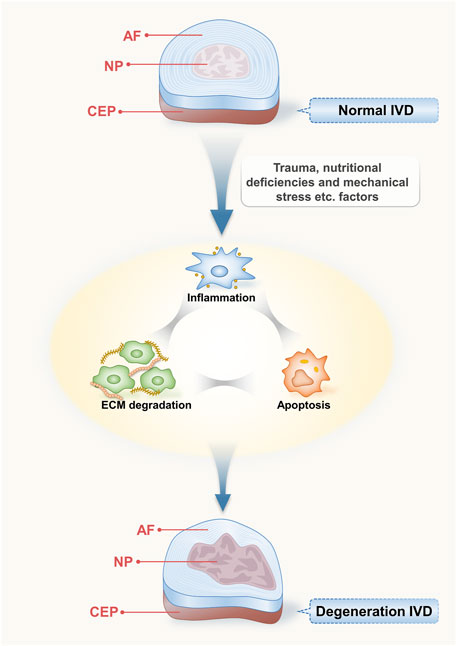
FIGURE 1. Structure of intervertebral disc and its degenerative process. The IVD is an intricate fibrocartilaginous formation comprising the nucleus pulposus (NP), annulus fibrosus (AF), and cartilaginous endplates (CEP); they play a crucial role in maintaining the proper function of the IVD. Various induction factors that affect the disc, such as trauma, malnutrition, and mechanical stress, can lead to inflammation, apoptosis, and ECM degradation, resulting in IDD.
Currently, the main treatment options for disc degeneration include physiotherapy, medication, and surgery, depending on the symptoms and degree of disc degeneration (Knezevic et al., 2017). However, treatment methods and their effects are unsatisfactory. According to some statistics, 10%–40% of patients experiences ‘failed back surgery syndrome’ following surgery, a condition in which patients experience persistent chronic back pain after surgery (Thomson and Jacques, 2009). Patients frequently experience back pain, preventing them from carrying out their daily work and activities or living normally (French et al., 2018). There is an increased risk of developing an addiction to narcotics owing to the necessity for pain relief (Chen et al., 2019). Based on these findings, there is a pressing need to explore new, practical, and improved prognostic approaches to address disc degeneration. In this context, gene therapy based on a class of non-coding RNA biological factors, such as circRNAs, has attracted attention.
The role of non-coding RNAs in various diseases has attracted increasing attention, and the differential expression of circRNAs between patients with IDD and healthy controls has been increasingly investigated (Wang et al., 2019b; Guo et al., 2020a). CircRNAs also promote or inhibit disc degeneration by modulating several pathological processes, including proliferation, apoptosis, phagocytosis, and senescence in the NP and chondrocytes and the expression of key components of the ECM, anabolic regulators of the ECM such as MMP and ADAMTS, and inflammatory cytokines such as IL-1. However, the mechanism of action of circRNAs in IDD is not well understood, and further research is necessary.
2 Properties, classification, and functions of circRNAs
CircRNAs are produced by back-splicing linear precursor mRNAs (pre-mRNAs), a class of non-coding RNAs that are cyclised and covalently bonded (Ashwal-Fluss et al., 2014; Zimmerman et al., 2020). CircRNA molecules were first discovered in RNA viruses in 1976 and in eukaryotic cells in 1991 (Wang et al., 2019a). At that time, circRNAs were considered to be formed by mis-splicing (Cocquerelle et al., 1993). However, in subsequent in-depth studies, numerous circRNAs exhibited tissue- and disease-specific expression patterns with important biological functions (Rybak-Wolf et al., 2015; Kristensen et al., 2018; Nicolet et al., 2018), indicating the need for more research on circRNAs.
Currently, circRNAs are classified into three categories: exonic circular RNAs (ecRNAs), which account for approximately 85% of all circRNAs; exonic, intronic circular RNAs (eicRNAs); and circular intronic RNAs (ciRNAs), all of which are classified based on various exon–intron combinations in the final sequence (Di Agostino et al., 2020; Chen et al., 2021). RNAs include cyclic introns from pre-mRNAs and tRNA intron cyclic RNA (Schmidt et al., 2019). EcRNAs are abundant in the cytoplasm, whereas EIcRNAs are abundant in the nucleus (Li et al., 2015a). Exon-containing circRNAs are the final splicing products and are currently the main products of genome discovery and study (Hsiao et al., 2017). Currently, it is believed that the circular conformation of most circRNAs is formed through back-splicing of exons in precursor mRNA. During this process, the 5′-splicing site joins the upstream 3′- splice site to form a circRNA that is covalently linked at the end by a 3′–5′ phosphodiester bond. This is in contrast to canonical splicing, where the upstream 5′splice site is sequentially jointed to the downstream 3′splice site to produce linear RNA (Zhang et al., 2016; Pagliarini et al., 2020).
A small proportion of the circRNAs formed from tRNA, known as tricRNA, is a unique circular RNA formed by the tRNA splice endonuclease (TSEN) complex (consisting of two structural and two catalytic components), which cleaves the tRNA precursor at the canonical bulge-helix-bulge (BHB) motif to produce introns that are then joined together by RtcB ligase at the ends of the resulting introns (Paushkin et al., 2004; Schmidt et al., 2019). Back-splicing is a critical step in the creation of circRNAs and is controlled by trans-acting factors and cis-acting components (Chuang et al., 2018). Similar to that of other RNA transcripts, the abundance of circRNAs is controlled by their balanced relative rates of synthesis and degradation (Venkatesh et al., 2016). However, circRNAs are more stable than linear RNAs. Their half-life is at least 2-fold longer than that of linear RNAs, exceeding 48 h (Jeck et al., 2013; Enuka et al., 2016), and for some genes, they are 10-fold more abundant than the associated linear mRNAs (Borchardt et al., 2017). Moreover, circRNA expression is tissue- or cell-specific, suggesting the potential to be used as biomarkers for certain human diseases (Rybak-Wolf et al., 2015; Yang et al., 2017a). To date, the following circRNAs have been identified: sponging miRNAs, those interacting with proteins, those regulating parental gene expression, and those competing with linear RNAs (Li et al., 2020). Figure 2 shows the biogenesis and functions of circRNAs.
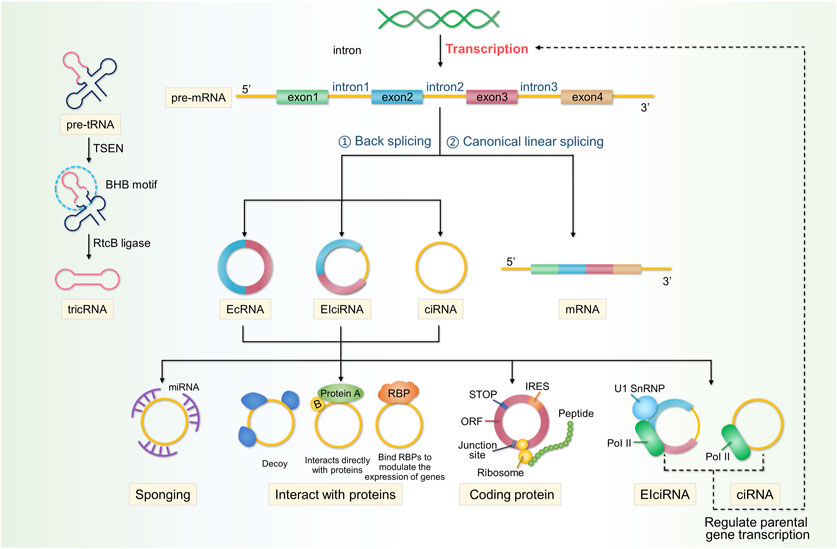
FIGURE 2. Properties, classification, and functions of circRNAs. CircRNAs are produced by the reverse splicing of linear precursor mRNAs (pre-mRNAs), followed by cyclisation. circRNAs are classified into three categories: EcRNAs, EIcRNAs, and ciRNAs. A small proportion of the ciRNAs formed from tRNA, known as tricRNA, which is formed by the TSEN complex cleaving the tRNA precursor at the canonical BHB motif to produce introns, is then joined together by RtcB ligase at the ends of the resulting introns. CircRNAs perform a multitude of functions within cells and can act as sponges for miRNAs. circRNAs interact with proteins in various ways and can affect the expression of parental genes. circRNAs derived from exonic RNA can be translated into encoded proteins. Additionally, circRNAs can compete with linear RNA to affect gene expression.
2.1 microRNA (miRNA) sponges
The role of circRNAs as a sponge has been the most extensively studied and is their main function. Most circRNAs are primarily located in the cytoplasm of the cell, which may be related to post-transcriptional regulation (Salzman et al., 2012; Yang et al., 2017c). MiRNAs are short RNA molecules, measuring of 19–25 nucleotides in length, that regulate the post-transcriptional silencing of target genes (Lu and Rothenberg, 2018). They regulate RNA stability and translation rates by pairing with complementary sites in target RNAs, called miRNA response elements (MREs) (Guo et al., 2010). According to the competitive endogenous RNA (ceRNA) theory, all RNA transcripts with miRNA-binding sites might interact and regulate one another by competing for the same miRNAs (Liu et al., 2019). Therefore, as long as circRNAs possess MREs that bind miRNAs, they can also act as endogenous RNAs that bind to miRNAs and act as sponges, alleviating miRNA-mediated gene repression inhibition (Tay et al., 2014). Well-known examples include circRNA CDR1as, a highly stable circRNA rich in conserved binding sites for miR-7 (up to 60) and a potent sponge for miR-7 target genes (Hansen et al., 2013). Many circRNAs are considerably less abundant than miRNAs and may not always act as sponges (Guo et al., 2014); therefore, they may have additional functions.
2.2 CircRNAs interact with proteins, which is another important function
EicRNAs interact with the host U1 snRNP and RNA Pol II to inhibit the transcription of their parent genes, competing for mRNA production and affecting protein translation (Ashwal-Fluss et al., 2014). CircRNAs can also act as decoys or scaffolds for proteins. Circ-amotl1 binds and stabilises c-Myc in the nucleus and upregulates its target genes (Yang et al., 2017a). In the cytoplasm, circPABPN1 competes with PABPN1 mRNA to bind to HuR and acts as a HuR decoy, inhibiting PABPN1 translation (Abdelmohsen et al., 2017). Circ-amotl1 acts as a scaffold that bind pdk1 and AKT1 to form ternary complexes, exerting a cardioprotective effect in DOX-induced cardiomyopathy (Zeng et al., 2017). Notably, proteins such as RBPs also regulate circRNA biogenesis (Okholm et al., 2020). FUS binds to the introns flanking back-splicing junctions during back-splicing, altering circRNA expression in mouse embryonic stem cell-derived motor neurones (Errichelli et al., 2017). The interactions between proteins and circRNAs are also worth studying.
2.3 Effects on parental gene expression
Some intron-containing circRNAs are immobilised in the nucleus, while ecRNAs are transported to the cytoplasm (Zhang et al., 2013; Li et al., 2015b). Therefore, circRNAs remaining in the nucleus are likely to be involved in transcriptional regulation. As they interact with U1 snRNA and the PoI II complex, EIciRNAs can enhance the transcription of their parental genes (Li et al., 2015b). ciRNA regulates the transcription of its parent genes by promoting the elongation of Pol II (Zhang et al., 2013).
2.4 CircRNAs can encode proteins
The source of exonic RNA in the cytoplasm is the protein-coding exon, which is the basis for the translation of circRNAs into proteins. CircRNAs can be translated, directly encoding functional proteins (Yang et al., 2017b; Legnini et al., 2017; Pamudurti et al., 2017). However, further research is required to elucidate the mechanisms underlying circRNA translation.
2.5 Competition with linear RNA
As most circRNAs originate from coding gene exons and use the same splicing site and spliceosome as linear RNA, the hypothesis that circRNAs compete with pre-mRNA splicing has emerged (Salzman et al., 2012; Ashwal-Fluss et al., 2014; Starke et al., 2015). CircMBL and its surrounding introns contain consistent MBL-binding sites that are recognised by specific MBL isoforms, contribute to the cyclisation of circMBL, and compete with mRNA splicing (Ashwal-Fluss et al., 2014). Therefore, the competition between circRNAs and linear RNAs may affect gene expression levels, leading to the occurrence of some diseases.
3 Mechanisms of the influence of circRNA on IDD
Current research on the specific mechanisms of circRNAs in the development of IDD has focused on five major aspects: cell proliferation and apoptosis, cellular senescence, ECM metabolism, inflammation, and autophagy in IVD cells (Lin et al., 2021; Li et al., 2022a). Pulposus cells are located at the centre of the disc and produce ECM components, whose proliferation and apoptosis play important roles in the pathogenesis of IDD. With age and the development of IDD, the proportion of cells expressing senescence-associated beta-galactosidase (SA-beta-gal) increases, the telomere length shortens, and telomerase activity decreases, indicating varying degrees of senescence in NP cells (Kim et al., 2009; Jeong et al., 2014). The senescence of NP cells affects their ability to proliferate and secrete ECM, promoting the development of IDD. The ECM maintains the structural and functional integrity of the IVDs. Type II collagens and proteoglycans promote ECM synthesis, whereas MMPs and ADAMTS promote ECM catabolism (Vo et al., 2013; Chen et al., 2020a). Imbalances in the proteins related to ECM metabolism affect ECM synthesis and have a direct and profound effect on IDD progression.
Many inflammatory cells and related factors, such as IL-1, TNF-α, and IL-6, have also been found in degenerated IVDs (Lyu et al., 2021). These inflammatory factors affect the development of IDD by activating inflammation-related pathways such as NF-κB, inducing an increase in matrix metalloproteinase levels, and promoting the expression of inflammatory mediators and chemokines, thereby leading to a cascade reaction and exacerbating tissue inflammatory damage (Zhang et al., 2021b). Despite the significance of these mechanisms in IDD, we cannot ignore the role of autophagy in the pathogenesis of IDD (Choi et al., 2016; Xu et al., 2020). Autophagy protects NP cells by scavenging oxidatively damaged mitochondria to prevent cell damage and maintain the energy supply (Ashrafi and Schwarz, 2013; Wang et al., 2018b). Moderately limited autophagy can protect the IVD from degeneration, whereas excessive autophagy or dysfunction caused by various conditions can accelerate the onset of IDD; however, its role in IDD requires further in-depth study (Chen et al., 2017b; Zhang et al., 2020b).
The preceding discussion outlined several contemporary mechanistic approaches concerning the role of circRNAs in IDD. However, a single circRNA can influence IDD through more than one mechanistic pathway. Many circRNAs affect the IDD process via two or more of the aforementioned mechanisms. Based on these findings, we propose the classification of circRNAs into two distinct categories: protective and detrimental circRNAs. This classification depends on whether the circRNAs serve as protective factors for the IVD. Protective circRNAs protect the component structure of the IVD and actively inhibit NP cell senescence and apoptosis, reducing ECM degradation factor production, promoting ECM component expression, inhibiting inflammatory cytokine expression, and regulating the autophagy of IVD cells. In contrast, dermal circRNAs have the opposite effects. Analysing circRNAs from this perspective not only provides a more succinct classification but also supports the practical clinical application of circRNAs in the future. This perspective will help prospective researchers select specific genes for more comprehensive investigations.
The expression of the following circRNAs is significantly reduced or increased in degenerating disc tissues and cells, thereby inhibiting or promoting disc degeneration through the mechanisms described above. Table 1 and Figure 3 show the roles and specific mechanisms of IDD-related circRNAs in detail. Aberrantly expressed circRNAs may serve as diagnostic or therapeutic targets for the clinical treatment of degenerative disc diseases.
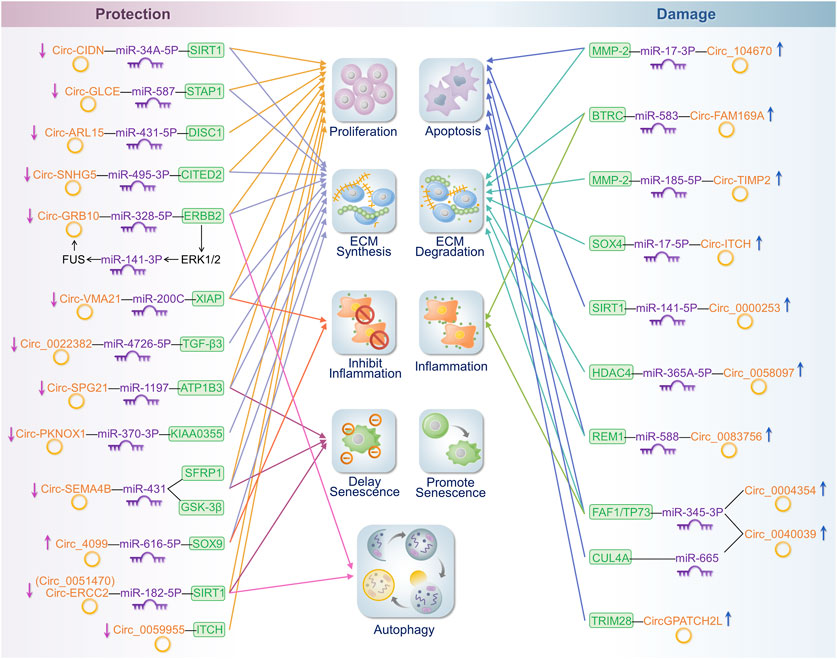
FIGURE 3. Mechanisms of influence of cyclic RNA in IDD. On the left side of the figure are the circRNAs that protect the intervertebral disc. On the right side of the figure are the circRNAs that play a detrimental role in the intervertebral disc. These circRNAs regulate disease progression in IDD by affecting cell proliferation or the apoptosis, synthesis, or degradation of the ECM; promotion or inhibition of inflammatory responses; and the extent of senescence and autophagy.
4 Protective circRNAs of IVD
1) The IVD plays an important role as a load-absorbing and cushioning unit in the spine, which endures mechanical stress daily. Excessive mechanical load can lead to apoptosis and ECM degradation in NP cells, promoting IDD (Adams and Roughley, 2006; Yan et al., 2017; Li et al., 2018). Non-coding RNAs influence the IDD process by acting on CEP, which is a new topic for current research (Zhao et al., 2023). Xiang et al. (Xiang et al., 2020) first verified the apoptosis- and catabolism-promoting effects of compression treatments on human NP cells. They found that circRNA-CIDN is significantly downregulated in compression-treated human NP cells and that the overexpression of circRNA-CIDN inhibits NP cell apoptosis and ECM degradation. The overexpression of Circ-CIDN delays IDD progression in an in vitro rat IVD organ culture model. This study demonstrates that circRNA-CIDN attenuates compressive load damage in human NP cells via the miR-34a-5p-SIRT1 axis. This study provides the first evidence that circRNAs in IDD can affect human NP cells in compression load-induced injury, providing a new perspective on compression-induced disc degeneration and establishing a library of differentially expressed circRNAs that will facilitate further studies.
2) Chen et al. showed that Circ-GLCE is stably present in the cytoplasm of myeloid cells and is significantly downregulated in IDD, as evidenced by microarray and further analysis. The knockdown of Circ-GLCE leads to NP apoptosis and the degradation of the NP ECM. In contrast, the overexpression of Circ-GLCE partially reverses the apoptosis induced via IL-1β treatment of NP cells. In a mouse model of IDD, the overexpression of Circ-GLCE reduces NP cell apoptosis and ECM degradation and delays the progression of IDD in vivo via AAV circ-GLCE treatment. These results indicate that Circ-GLCE attenuates IDD by targeting the miR-587-STAP1 axis to inhibit apoptosis and ECM degradation in NP cells. This represents a promising therapeutic target for the treatment of IDD.
3) Wang et al. (Wang et al., 2021a) used gene microarray technology and found that circARL15 is the most significantly differentially expressed in the ceRNA network and is downregulated in IDD. Functionally, circARL15 overexpression inhibits apoptosis, and miR-431-5p mimics counteract this effect. The direct binding of DISC1 to miR-431-5p was verified via luciferase reporter, co-transfection, and EGFP reporter assays. In a rat model of IDD established by acupuncture, infusion of the circARL15 overexpression plasmid reduced the loss of NP tissue and structural degeneration of the IVD, validating its function in vivo. These findings suggest that circARL15 is involved in IDD through the targeting of the miR-431-5p/DISC1 signalling pathway and regulating NP cell proliferation and apoptosis.
4) CEP is a component of the IVD consisting of hyaline cartilage and extracellular mesenchyme and is responsible for the main blood supply to the disc (Chen et al., 2017b). The disruption of CEP can lead to nutrient deprivation in the ECM and contribute to the induction and development of IDD (Grunhagen et al., 2006). Zhang et al. (Zhang et al., 2021c) investigated the function of circ-SNHG5 in cartilage endplates in IDD. Initially, a diminished expression of circ-SNHG5 was observed in degenerative CEP tissues. Through rigorous iterations of knockdown and rescue experiments, we established that circ-SNHG5 serves as a sponge, inhibiting miR-495-3p. Subsequent experiments involving the co-transfection of HEK293T cells with luciferase reporter plasmids and miR-495-3p mimics revealed a direct interaction between miR-495-3p and CITED2, with the former acting as an upstream suppressor. The overexpression of CITED2 decreased MMP13 levels and increased the expression of collagen II and aggrecan, indicating a healthy ECM. Thus, this study revealed a novel circ-SNHG5/miR-495-3p/CITED2 axis in IDD, offering promising avenues for future targeted therapies aimed at halting disease progression. Although NP cells have garnered significant attention in IDD research, the relatively unexplored chondrocytes of the CEP may provide valuable insights for future IDD studies. Notably, this study was confined to in vitro studies of chondrocytes in the CEP and did not include in vivo animal studies. Further investigations are warranted to examine the expression profiles and potential functions of circRNAs in the CEP.
5) Guo et al. (Guo et al., 2018) found that circ-GRB10 is significantly downregulated in IDD samples; however, it controls the expression of ERBB2 by sponging miR-328-5p. ERBB2 induces apoptosis and autophagy by forming a complex with Beclin 1. However, as the reasons behind the downregulation of circ-GRB10 have not been identified, the authors have conducted a more in-depth study. In further experiments, Guo et al. (Guo et al., 2020c) not only examined the ERBB2/ERK pathway but also investigated the effect of circ-GRB10 on ECM anabolism. By establishing a rat model of IDD via acupuncture, they found that the overexpression of circ-GRB10 significantly elevates ERBB2 levels in NP cells and effectively inhibits the process of IDD in rats. They also elaborated on the upstream mechanisms.
FUS is an RNA-binding protein that regulates the biosynthesis of circ-GRB10-forming exons in NP cells by binding to recognition sites (Errichelli et al., 2017). FUS expression is inhibited by targeting MiR-141-3p. Finally, miR-141-3p is regulated by ERBB2 through the phosphorylation of Erk1/2, as demonstrated via knockdown studies, immunoblotting, and quantitative RT-PCR experiments. Through a series of studies, the authors established a circular signalling pathway formed by circ-GBR10-miR-328-5p phosphorylation of the ERBB2-Erk1/2-miR-141-3p-Fus loop. Notably, circ-GRB10 has multiple RBP-binding sites, and the knockdown of FUS does not completely inhibit circ-GRB10 synthesis. This suggests the existence of regulatory pathways beyond FUS involving circ-GRB10, and further studies are needed to verify this hypothesis.
These studies highlight the role and pathway of circ-GRB10 in IDD; however, research on its clinical aspects is lacking. Wei et al. (Wei et al., 2023) explored the potential of circ-GRB10 as a biomarker in clinical applications. By comparing the IDD group with the other groups, including sacroiliac joint pain, lumbar disc herniation, piriformis syndrome, entrapment neuropathy, and healthy controls, it was found that plasma circ-GRB10 levels were significantly downregulated. Moreover, after standard treatment in each group, the plasma expression of circ-GRB10 was elevated only in patients with IDD. These results suggest that circ-GRB10 is specific to lumbar degenerative disc disease. Measuring plasma GRB10 levels before and after treatment not only improves the diagnostic accuracy of IDD but also suggests its therapeutic efficacy.
6) Cheng et al. (Cheng et al., 2018) found that IDD samples contained considerably lower amounts of circ-VMA21 than control tissues. Circ-VMA21can directly bind to miR-200c in NP cells. Elevated miR-200c levels reduce the expression of X-linked inhibitor of apoptosis protein (XIAP), an inhibitor of the apoptotic family of proteins. It binds to cysteoaspartic enzymes that mediate apoptotic cell death, including caspases 3, 7, and 9, and regulates cysteoaspartic enzyme activation, inhibiting apoptosis (Jost and Vucic, 2020). It also regulates several signalling pathways that regulate cell necrosis, autophagy, and differentiation (Hanifeh and Ataei, 2022). In terms of function, the absence of XIAP significantly counteracts the effect of miR-200c antagonists on TNF-α- and IL-1β-treated NP cells, inducing apoptosis, exacerbating catabolic responses, and reducing the expression of ECM components. Furthermore, the injection of human adenovirus circ-VMA21 into punctured IVDs attenuates degenerative changes in the NP and reduces disc degeneration in a rat model of IVDD. These results confirmed that IDD development can be regulated in NP cells through the circ-VMA21-mir-200C-XIAP axis.
7) Hu et al. (Hu et al., 2022) used human endplate chondrocytes to study IDD because chondrocytes in the endplate can degenerate owing to periodic mechanical tension, and endplate degeneration can lead to IVDD (Holm et al., 2004; Adams and Roughley, 2006; Xiao et al., 2020). The results showed that intermittent cyclic mechanical tension leads to endplate chondrocyte degeneration and circRNA_0022382 downregulation. CircRNA_0022382 regulates the anabolic and catabolic activities of the ECM of endplate chondrocytes by sponging miR-4726-5p and downregulating the expression of the target gene encoding transforming growth factor 3 (TGF-β3). In vivo models of acupuncture in rats show that IDD is alleviated by the intravertebral disc injection of circ_0022382. Mechanistically, the development of IDD generated by intermittent mechanical tension is effectively blocked by the circ_0022382-miR-4726-5p-TGF-β3 axis by inhibiting the development of cartilage endplate cell degeneration.
8) Huang et al. (Huang et al., 2021b) analysed clinically normal IVD and degenerated disc tissues and found that water loss and cellular senescence are present in degenerated disc tissues. Moreover, the expression of has_circ_0035875 (circSPG21) decreases with increasing lesion severity. Transfection experiments showed that the knockdown of circSPG21 leads to NP cell senescence, inhibits ECM anabolism, and promotes catabolism; however, the overexpression had the opposite effect. To establish a mouse model of IDD, researchers induced disc degeneration using sustained pressure on the mouse IVD. They also found that injecting circSPG21 reverses the increase in ECM breakdown in IDD caused by continuous pressure and slows the development of IDD. Additionally, researchers explored the relationship between circSPG21 and miR-1197 and their target gene ATP1B3 and confirmed that circSPG21 reduces inflammatory cytokine-induced senescence, apoptosis, and ECM catabolism in NP cells via the miR-1197-ATP1B3 axis. ATP1B3 is involved in establishing an electrochemical gradient across the plasma membrane for Na+ and K+ ions, controlling osmotic pressure and sustaining neuromuscular excitement (Mahmmoud, 2005; Ogawa et al., 2015). These results suggest that circSPG21 may serve as a biomarker and therapeutic target for the diagnosis of IDD. Furthermore, the downregulation of ATP1B3 may regulate the development of IDD by modulating the exchange of Na+/K+ ions, leading to cellular water loss. The relationship between IDD development and intracellular ion concentrations requires further investigation.
9) Huang et al. (Huang et al., 2021a) found that circPKNOX1 expression is the most downregulated. Immunofluorescence assays and RT-qPCR results indicated that circPKNOX1 promotes matrix synthesis and inhibits the breakdown of the corresponding protease. Mechanistically, circPKNOX1 regulates KIAA0355 (GARRE1) through a pathway mediated by miR-370-3p. KIAA0355, a dual effector of Rac and Rab, exhibits low GTPase activity and is intricately linked to a range of cellular activities (Serres et al., 2023). KIAA0355 knockdown markedly decreases the expression levels of genes associated with ECM formation and increases the expression levels of genes related to ECM degradation in NP cells. This sequence of events accelerates extracellular mesenchymal degradation and promotes IDD progression. The mouse animal model confirmed that increased circPKNOX1 expression reverses the trend of decreased anabolism and increased catabolism due to the compression of NP tissue, providing an effective direction for treating IVDD.
10) Wang et al. (Wang et al., 2018c) found that the expression of circSEMA4B is significantly lower in IDD specimens with severe degeneration than in those with mild degeneration, according to the Pfirrmann classification. Inhibiting circSEMA4B enhances the inhibition of proliferation, senescence, and the degradation of the ECM in NP cells via IL-1β; however, its overexpression partially reverses its effects. Mechanistically, circSEMA4B inhibits Wnt pathway activation by competing with the Wnt pathway inhibitors, SFRP1 and GSK-3β, for binding to mir-431, thereby regulating proliferation, senescence, and ECM degradation in NP cells and tissues. An miRNA can have multiple target genes acting on the cell, while a circRNA can mediate the development of IDD by regulating these genes. Thus, to comprehensively study the function of circRNAs, attention should be paid to the influence of circRNAs and other non-coding RNA networks.
11) Through circRNA gene microarray and qPCR, Wang et al. (Wang et al., 2018a) showed that circ_4099 is the most upregulated circRNA in degenerated NP tissues, with a >2-fold change compared to that of normal tissues. TNF-α is upregulated via circ_4099 through GRP78 in a dose- and time-dependent manner. As the MAPK and NF-κB signalling pathways primarily produce TNF-α, transducing cells with pathway inhibitors, such as sh-p65 and sh-IKKβ, significantly reduces the effect of TNF-α on the induction of expression. Gain- and loss-of-function studies have shown that the overexpression of circ_4099 increases the expression of type II collagen and aggrecan and significantly inhibits the expression of inflammatory factors but does not regulate MMP-3 and ADAMTS-5. Mechanistically, the expression of circ_4099 is upregulated by TNF-α via the MAPK and NF-κB pathways and competitively binds miR-616-5p to reverse the inhibition of SOX9, increasing ECM synthesis in NP cells and playing an important role in ECM metabolism.
Until the present study, circ_4099 was the only circRNA found to be significantly upregulated in degenerating disc tissues and cells and provided protection to the disc. The upregulation of circ_4099 is influenced by the action of TNF-α on GRP78. GRP78, a member of the heat shock protection protein 70 superfamily, is predominantly present in the endoplasmic reticulum (ER) (Kang et al., 2016). As one of the most important ER chaperones, GRP78 plays an important role in many cellular processes, including protein assembly, folding, and translocation and regulates calcium homeostasis (Wang et al., 2009). GRP78 expression is upregulated under many stress conditions, including hypoxia and glucose deficiency, regulates cell proliferation, and prevents death (Farshbaf et al., 2020). However, the exact mechanism of GRP78 and whether it regulates more cirRNAs are unclear, which directs us to further examine the mechanism of cirRNA regulation and explore approaches to significantly upregulate protective circRNAs.
12) Xie et al. (Xie et al., 2019) showed that circERCC2 is the most downregulated gene and functions through the miR-182-5p/SIRT1 axis. Functionally, the overexpression of circERCC2 significantly reduces apoptosis and increases the phagocytosis of TBHP-treated NP cells (Xia et al., 2015; Guo et al., 2017). revealed that SIRT1 acts on phagocytosis and apoptosis via the SIRT1-parkin-mitophagy pathway and plays a therapeutic role in IVDD. SIRT1-si reduces apoptosis in circERCC2-inhibited neuronal cells and blocks the inhibitory effects of circERCC2 on NPcells senescence. Rat puncture IDD experiments showed that injecting circERCC2 inhibits NP cell apoptosis and reduces histological grading by enhancing the autophagic response. In conclusion, the circERCC2-miR-182-5P-SIRT1 axis can promote the phagocytosis of cells, reducing apoptosis, delaying senescence, and effectively regulating IDD development.
13) Kong et al. (Kong et al., 2020) studied the effects of circRNA expression on the actions of their host genes. Itchy E3 ubiquitin protein ligase (ITCH), the host gene for hsa_circ_0059955, promotes the ubiquitination and degradation of p73, a homologue of p53 that induces apoptosis and cell cycle arrest. hsa_circ_0059955 expression is significantly reduced in IVDD tissues. Downregulating hsa_circ_0059955 inhibits proliferation, induces apoptosis, and induces G0/G1 phase arrest in NP cells. Moreover, overexpressing hsa_circ_0059955 significantly attenuates or reverses the progression of IDD in rats. This study suggests that circ_0059955 may affect the transcription and expression of its host gene ITCH, leading to apoptosis and cell cycle arrest in NP cells by inhibiting ITCH. However, they could not completely confirm the action of 0059955 on IDD via ITCH because they did not perform experiments to verify the direct binding between hsa_circ_0059955 and ITCH. Therefore, the mechanism through which circ_0059955 affects the protein levels of its host genes requires further investigation.
5 Detrimental genes in IVD
1) Song et al. (Song et al., 2018) found that circRNA_104670 is one of the most upregulated circRNAs, with nearly 4.5-fold upregulation in patients with IDD. Using Cytoscape to construct circRNA–miRNA–mRNA networks and microarray data, they predicted the presence of the circRNA_104670-miR-17-3p-MMP2 axis, which was experimentally verified. CircRNA_104670 can induce apoptosis, inhibit the growth of NP cells, and accelerate ECM degradation by inhibiting type II collagen expression and promoting MMP-2 expression. Through the injection suppression of circRNA_104670 and the dual gene suppression of circRNA_104670 and miR-17-3p, they found that circRNA_104670 gene-suppressed mice had lower IDD gradings, while the dual gene suppression group exhibited an accelerated IDD process. By analysing and comparing normal and degenerated human NP tissues, the ROC curves showed that circRNA_104670 and miR-17-3p were of good diagnostic significance for IDD, and both correlated with the IDD grading. This suggests that circRNA_104670 and miR-17-3p may serve as biomarkers for IDD diagnostic grades and as therapeutic targets.
2) Wei et al. (Guo et al., 2020b) analysed circRNAs with specificity using a microarray and found that circ-FAM169A is significantly upregulated in degenerative NP tissues and positively correlated with IDD classification (r = 0.919). Functionally, circ-FAM169A overexpression in NP cells increases the expression of MMP-13 and ADAMTS-5 and decreases the production of type II collagen and aggrecan. The authors injected rats with a circ-FAM169A inhibitor after developing a needle-pricked rat model of IDD and showed that inhibiting circ-FAM169A significantly protected the structure of the IVD and delayed the process of IDD. In vitro and in vivo experiments showed that circRNA-FAM169A is upregulated in IDD-degenerating tissues and targets it by sponging miR-583. BTRC is an E3 ubiquitin ligase that forms the SCF complex through its F-box structural domain. It specifically binds to phosphorylated IκBα and mediates its ubiquitination, facilitating the translocation of NF-κB to the nucleus and activating the transcription of related genes (Spencer et al., 1999; Zhang et al., 2018). NF-κB induces the expression of catabolic genes and exacerbates inflammatory factor responses, shifting the equilibrium toward ECM degradation and facilitating the IDD process (Zhang et al., 2021b).
3) Guo et al. (Guo et al., 2020d) showed that the expression levels of circ-TIMP2 in NP tissues from patients with IDD are significantly increased, and circ-TIMP2 directly binds to miR-185-5p in these tissues. MiR-185-5p is significantly downregulated in TNF-α- and IL-1β-treated cells and degenerated NP tissues. Moreover, MMP2 is targeted by miR-185-5p to regulate NP cell functions. As a metalloprotease, MMP-2 degrades collagen II and the polymers found in the ECM (Rutges et al., 2008). These results suggest that circ-TIMP2 acts on NP cells by regulating the expression of miR-185-5p and MMP-2, promoting ECM degradation. In conclusion, MMP-2 may act as a target gene for both circ_104670 and circ-TIMP2 in IVD. Further comprehensive studies are required to determine whether MMP-2 has additional regulatory mechanisms.
4) Zhang et al. (Zhang et al., 2021a) revealed that NP tissues from patients with IDD show elevated levels of circ-ITCH expression compared to normal tissues. Transfection experiments showed that the knockdown of circ-ITCH promotes NP cell proliferation and inhibits ECM degradation; however, circ-ITCH overexpression has the opposite effect. The authors then confirmed that circ-ITCH targets and regulates SOX4 (SRY-box transcription factor 4) gene expression by sponging miR-17-5p. The CCK-8 assay showed that circ-ITCH overexpression inhibits NP cell proliferation and leads to the degradation of ECM components, whereas circ-ITCH knockdown reduces NP cell apoptosis. Moreover, SO4 regulates the progression of IDD by activating the Wnt/β-catenin pathway. The knockdown of circ-ITCH inhibits the expression of Wnt1, β-catenin, c-Myc, and Cyclin d1, and the overexpression of SOX4, using the signalling activator LiCl, reverses this response. This finding indicates that circ-ITCH is involved in ECM degradation in degenerating NP cells via the miR-17-5p/SOX4/Wnt/β-catenin axis.
5) Liao et al. (Liao et al., 2019) found that the in vivo delivery of bone marrow-derived mesenchymal stem cell exosomes (BMSC-exos) modulates ER stress-induced apoptosis and delays IDD in a rat caudal model. Therefore, the role of exosomes in IDD needs to be comprehensively explored. Song et al. (Song et al., 2020) collected NP cells media and isolated exosomes and observed that degenerative NP cells secrete more exosomes than normal NP cells. Degenerating NP cells secrete exosomes that contain additional apoptosis-degrading factors which can promote apoptosis in NP cells. CircRNA_0000253 is the most upregulated circRNA in exosomes generated by degenerating NP cells. Mechanistically, circ_000253 promotes apoptosis and inhibits cell proliferation by competing for the adsorption of miR-141-5p to downregulate SIRT1, which promotes IDD both in vitro and in vivo.
In a rat IDD model established by needling the IVD, the injection of exosomes from the IDD group resulted in IDD in rats, and the severity increased with the grading of IDD from the exosome source. In addition, by injecting AAV-2-circRNA_0000253-siRNA into rats, it was found that inhibiting circRNA_0000253 significantly reduces NP cellsapoptosis and protects the disc structure. Exosomes are vesicles that undergo cellular exocytosis and are composed of lipid-bound nanoparticles (Willms et al., 2018). Exosomes can carry nucleic acids, proteins, mRNAs, and non-coding RNAs and are key mediators of intercellular communication (De Jong et al., 2014). Exosomes are likely to play an important role in the cellular therapy for IDD through various cellular exosome sources as an emerging therapeutic modality (DiStefano et al., 2022; Xia et al., 2022). Notably, Song et al. studied the negative effects of exosomes on the IVD. Their results showed that exosomes variably express different levels of circRNAs in NP cells and act at different levels of disc degeneration, which could be a biological marker for potential therapeutic directions for IDD.
6) Biomechanical stimulation is crucial for the growth and function of endplate cartilage. Excessive mechanical load changes the structure of the endplate cartilage and the components of the ECM, disrupting the nutrient supply, leading to IDD (O'Conor et al., 2013; Vasiliadis et al., 2014). Endplate chondrocytes from primary human endplates were cyclically tensioned at 0.5 Hz with a 10% elongation by Xiao et al. (Xiao et al., 2020). They found altered cytoplasmic morphology and significant downregulation of the AGN, COLII, and SOX9 expression levels in the loaded group compared with those in the control group. The expression of circRNA_0058097 significantly differed and increased with Thompson grading. The stress resilience of endplate chondrocytes was enhanced by inhibiting circRNA_0058097 expression. These studies suggest that the tension-sensitive circRNA_0058097 is present in human endplate chondrocytes and promotes morphological changes in endplate chondrocytes through the circRNA_0058097-miR-365a-5p-HDAC4 axis, leading to an increase in the extracellular interstitial degradation and degeneration of the endplate cartilage.
7. Du et al. (Du et al., 2022) discovered that hsa_circ_0083756 expression is increased in tissues and cells with NP degeneration. In vitro functional studies showed that circ_0083756 knockdown promotes NP cell proliferation and ECM synthesis and inhibits the release of inflammatory factors, while gene overexpression had the opposite effect. In a rat IDD model established by acupuncture, inhibiting circ_0083756 gene expression attenuated the loss of NP tissues and the destruction of IVD structure and reduced the protein expression of ECM degradation and proinflammatory factors. Experiments conducted in vitro and in vivo indicated that hsa_circ_0083756 is upregulated in degenerative NP tissues and cells and promotes the IVDD process by sponging miR-558 to promote TREM1 expression. The Triggering receptor expressed on myeloid cells-1 (TREM-1) gene, located on chromosome 6p21, encodes the TREM-1 glycoprotein receptor, which enhances the expression of proinflammatory cytokines and matrix-degrading enzymes (Panagopoulos et al., 2022). It is likely to promote ECM degradation and apoptosis in NP cells through the TLR4/NF-κB signalling pathway, exacerbating the development of IDD (Zhang et al., 2022).
8) In pathological situations, extensive crosstalk occurs, and cell death commonly occurs in mixed forms rather than in isolation (Karki et al., 2021). Li et al. (Li et al., 2022b) proposed a new proinflammatory cell death modality called PAoptosis, which includes apoptosis and pyroptosis, and is more suitable for studying the mechanisms of inflammatory cell death in IVDD. The apoptotic pathway is mediated by the cleavage of caspase3, which can drive Gasdermin E, leading to pyroptosis with the concomitant secretion of IL-1β (Wang et al., 2017). Current research focuses mostly on the control of miRNAs by a single circRNA; however, the influence of multiple circRNAs belonging to the same parental gene on miRNAs has not been investigated. Syntrophin Beta 2 (SNTB2) is a cytoskeletal protein associated with dystrophin and dystrophin-related proteins that are involved in insulin secretion (Ort et al., 2001; Krautbauer et al., 2019). The authors found that circ_0040039 and circ_0004354 are derived from different exon positions of the same gene, SNTB2, and both are significantly upregulated in IDD. This study revealed a novel mechanism through which circ_0040039 and circ_0004354 induce PAoptosis, a mixed inflammatory cell death, by competitively interacting with miR-345-3p and influencing the expression of its target genes FAF1 and TP73. In this study, the authors simultaneously studied two circRNAs, serving as a good demonstration for other researchers to study the effects and mechanisms of interactions between two or even multiple circRNAs in IDD pathogenesis.
Jia et al. (Jia et al., 2023) additionally studied the role of circ_0040039 (called circSNTB2) in regulating lactate dehydrogenase (LDH) development in vitro and in vivo. The authors verified that circSNTB2 is upregulated in LDH tissues through bioinformatic screening and qRT-PCR. The knockdown of circSNTB2 reverses the inhibitory effect of TNF-α treatment on the proliferation of NP cells and the promotion of their apoptosis in both the LDH cell and rat models. Finally, circSNTB2 regulates CUL4A by mediating miR-665, which inhibits NP cell proliferation and induces apoptosis. However, the study of circ_0040039 has been extended beyond this point. Huang et al. (Huang et al., 2022) showed via bioinformatic analysis that circ_0040039 may also act as a sponge for hsa-miR-424-5p and hsa-miR-15b-5p to regulate downstream genes involved in IDD-related developmental processes through the Wnt/β-catenin signalling pathway. However, this preliminary prediction needs to be supported by further experiments.
From the above results, it can be concluded that the miRNAs sponged by circ_0040039 (circSNTB2) are different from their target genes. This indicates that the same circRNA can bind to two or more miRNAs and act on different target genes by sponging miRNAs, playing different roles in regulating IDD progression. This suggests that we should pay attention to the crosstalk between circRNAs and the network of miRNAs when studying their mechanism of action and pathways to comprehensively analyse the influence of a certain circRNA on IDD.
9) N6-methyladenosine (m6A) modification is key in regulating gene expression and function and is currently a subject of extensive research. Among the various forms of methylation in eukaryotic cells, m6A modification is the most common and is closely associated with diseases such as tumours, obesity, autoimmune diseases, and neurological disorders (Zhang et al., 2020a). M6A modifications regulate the metabolism and function of circRNAs by affecting circRNA biosynthesis and degradation, cytoplasmic export, and translation; however, m6A modifications are also affected by circRNA interactions (Wang et al., 2021b; Lin et al., 2022). In studies on musculoskeletal disorders, m6A modifications have been found to play a crucial role in regulating the proliferation and migration of tumours, the differentiation of osteoblasts and osteoclasts, chondrocyte function, and the senescence and apoptosis of NP cells (Hu et al., 2022). However, studies on the effect of m6A modifications on circRNAs in IDD are limited.
Chen et al. (Chen et al., 2023) innovatively explored the mechanism and role of m6A methylation in IDD to regulate circGPATCH2L. They screened and verified that circGPATCH2L expression is upregulated in IDD, and m6A methylation is significantly reduced. They demonstrated that upregulated circGPATCH2L inhibits the ubiquitination and degradation of p53 by targeting TRIM28, which impairs DNA damage repair and apoptosis in NP cells. Through a series of experiments, including RNA pulldown, RNA immunoprecipitation, and co-immunoprecipitation, they found that circGPATCH2L regulates the function of TRIM28 proteins by binding to amino acids 402–452 of TRIM28 as a molecular bait, directly targeting TRIM28 proteins. A mouse model of IDD confirmed that the knockdown of circGPATCH2L improves the function of NP cells and ECM metabolism, inhibiting the progression of IDD. Finally, they successfully verified that m6A-methylated circGPATCH2L changes the circGPATCH2L expression levels by cleavage in vivo via the YTHDF2-RPL10-RNase P/MRP complex. The results confirmed, for the first time, that circRNAs can act as decoys to interact with target proteins in IDD, regulating the function of target proteins. They also found, for the first time, that m6A modifications alter the progression of IDD by regulating circRNA levels, providing a good model for researchers to study the roles and regulatory mechanisms of circRNAs.
6 Potential clinical applications of circRNAs
circRNAs have a great potential for use in various clinical diseases. CircRNAs are highly conserved sequences that lack a 5′ cap and 3′ end, which makes them resistant to degradation by ribonucleases such as ribonuclease R and gives them a longer half-life than linear ribonucleic acids (Harland and Misher, 1988). The technology for identifying and analysing disease-related circRNAs has greatly improved. We identified IDD-related circRNAs via microarray analysis of probes targeting the reverse splice site, which was further confirmed via RT-qPCR (Xu et al., 2021). Furthermore, circRNAs are stable in serum exosomes and human cell-free saliva and are readily detectable in the human body (Li et al., 2015a; Bahn et al., 2015). More importantly, circRNA expression is tissue- or cell-specific (Liu et al., 2016; Yang et al., 2017a).
CircRNAs are stable, easy to detect, and highly specific in the human body with significant potential for clinical diagnostic testing and treatment of IDD and are expected to become biomarkers for IDD and therapeutic targets. Kun et al. (Kun-Peng et al., 2018) demonstrated that serum circPVT1 is more reliable than the conventional alkaline phosphatase as a diagnostic tool for identifying patients with osteosarcoma. CircPVT1 expression is also significantly upregulated in several types of malignancies, including oesophageal carcinoma, colorectal cancer, gastric cancer, and ERα-positive breast cancer (Chen et al., 2017a; Zhong et al., 2019; Chen and Chen, 2021; RezaSoltani et al., 2023; Yi et al., 2023). This close correlation with a spectrum of clinical conditions makes circPVT1 a prospective diagnostic and prognostic biomarker of oncological diseases. CircRNAs are abnormally expressed in degenerating disc tissues and cells and are closely associated with the clinical staging of IDD. CircRNAs are promising biological indicators for the early diagnosis of disc degeneration, initial assessment of IDD severity, and determination of prognosis.
circRNAs bind to miRNAs and affect the expression of their target genes. Notably, a preliminary study by Chen et al. (Chen et al., 2023) found that circGPATCH2L directly binds to target proteins and influences the development of IDD, an area that deserves further research. Current research on circRNAs has revealed that they can accelerate disc degeneration and act as protective factors that slow the progression of IDD. Based on these findings, we can effectively treat degenerative disc disease by overexpressing the circRNAs that protect the disc or by silencing circRNAs that can damage and destroy the disc. This treatment strategy is very promising for the clinical management of IDD, as it is less invasive and can effectively reduce the complications and sequelae caused by surgical procedures.
With the increasing need to regulate miRNA activity, reduce therapeutic costs, and improve production efficiency for disease treatment, artificial circRNAs have been designed. Jost et al. (Jost et al., 2018) designed an artificial in vitro transcribed-and-ligated circRNA sponge to isolate miRNA-122 from the hepatitis C virus (HCV) in a manner similar to that of miravirsen. MiRNA-122 is closely related to HCV and promotes HCV propagation by binding to sites at the 5′ end of the virus genome. The authors designed an artificial circRNA that effectively inhibits viral protein production by sponging miRNA-122 into a full-length HCV cell culture system. MiR-21 is overexpressed in gastric cancer (GC) and is associated with the proliferation, invasion, and apoptosis of GC cells.
Liu et al. (Liu et al., 2018) synthesised an artificial circRNA containing five mir-21 bulged binding sites using an enzymatic binding method, which was able to effectively restore luciferase activity. The degradation rate of LRNA21 was 92% after 30 min in 4% FBS, whereas the synthetic circRNA was more stable and was only degraded at FBS concentrations of 7% or higher. Moreover, synthetic circRNAs inhibit the proliferation of GC cells by upregulating the expression of DAXX, a key tumour suppressor gene and a direct target of miR-21. Pfafenrot et al. (Pfafenrot et al., 2021) first combined the classical antisense-RNA approach with synthetic short circRNAs, which interfered with the expression of the SARS-COV-2 genome and viral proliferation by specifically targeting SARS-COV-2. Shu et al. (Shu et al., 2018) generated endogenous cRNA sponges via an atypical head-to-tail back-splicing mechanism. Moreover, a user-friendly system was developed to express circulating miRNA inhibitors, which inhibits the function of endogenous miRNAs more potently than its linear counterpart. These designed and synthesised artificial circRNAs are functionally efficient and inexpensive to produce, providing a new approach for inhibiting the in vitro functions of targeted miRNAs (Breuer and Rossbach, 2020). The development of synthetic circRNAs requires time for refinement. The mass production of artificial circRNAs in a more efficient manner, monitoring their intracellular stability, and controlling their purification and quality need to be further explored.
7 Conclusion and future prospects
This review categorises the circRNAs linked to IDD into two distinct groups: disc-protective and detrimental circRNAs. This classification was derived from a comprehensive examination of the pathogenesis of IDD and the fundamental functions of circRNAs. We scrutinised and dissected the most recent research exploring the correlation between circRNAs and IDD. Given that each study investigating the mechanisms of circRNAs in IDD displays both shared features and unique aspects, we conducted a comprehensive review on the functional mechanism of each circRNA involved in IDD to offer critical insights into each research piece.
Furthermore, we propose the potential of circRNAs as diagnostic indicators or therapeutic agents for the treatment in future clinical scenarios. There is a promising field in which engineered circRNAs could be incorporated into cells to modulate the expression of target genes. These advancements could significantly broaden the scope of genetically targeted therapeutics. Our analysis shows that circRNAs in IDD samples are either upregulated or downregulated, playing a crucial role in the development of IDD, primarily by sponging-related miRNAs to regulate the expression levels of target genes. Therefore, circRNAs affect the proliferation, apoptosis, and senescence of IVD cells and influence the balance between the synthesis and degradation of the ECM and the secretion of inflammatory factors. The conceptualisation and production of artificial circRNAs could help drive further clinical applications of circRNAs in treating related diseases.
Current research on circRNAs is an area of intense interest; however, this is still much to be explored and developed in this field. First, research on the mechanisms involving circRNA in IDD has been focused on sponging miRNAs; however, circRNAs have roles beyond this in interacting with proteins, even in encoding proteins, and influencing parental gene expression. However, there is insufficient research to confirm whether these other functions play profound roles in the development of IDD.
Second, many studies have used a single factor to treat disc cells to build an IDD model. The most commonly used method for inducing disc degeneration is by using single factors such as inflammatory factors (e.g., IL-1 and TNF-a), intermittent cyclic tension, or TBHP, applied to the NP or chondrocytes. However, disc degeneration results from a combination of factors such as cyclic tension, compressive stress, trauma, inflammatory factors, and oxidative stress. Most studies on the therapeutic effects of circRNA-mediated disc degeneration are limited to using the overexpression or silencing of circRNAs to observe their therapeutic effects in single-factor models of IDD. Therefore, even if the overexpression or silencing of circRNAs can reverse the influence of one factor on the IVD, further research is needed to determine whether it affects IDD in cells treated with another factor or a combination of factors.
Third, current research on the mechanisms involving circRNA in IDD focuses on the role played by a single circRNA in the IVD. Nonetheless, several aberrantly expressed circRNAs in degenerating disc tissues and cells have been identified, all of which suppress or accelerate disc degeneration. Li et al. (Li et al., 2022b) revealed the possibility of competition and interaction between two or more circRNAs. Therefore, research efforts need to focus not only on specific circRNAs but also on the interplay and primary and secondary effects of multiple circRNAs in IDD. Thus, we can better understand the underlying mechanism of the entire circRNA population in the IVD.
Fourth, there are many in vitro studies of IDD; however, there are few in vivo models and clinical studies. Owing to ethical, technical, cost considerations, most in vivo studies remain at the animal study stage, and there have been no studies on circRNAs in in vivo models. However, the exact roles and therapeutic effects of circRNAs in humans are not well understood. We need to conduct retrospective and prospective multicentre independent cohort studies in patients with IDD to determine the exact effects of circRNAs in humans. We should also identify a convenient, specific, and reliable method for detecting circRNAs in patients with IDD.
In summary, circRNAs have a great potential for application in degenerative disc diseases. However, our current understanding of the roles and mechanisms of circRNAs in IDD is limited. Thus, further research should be conducted to study the role and regulatory mechanism of circRNAs and build suitable models to verify their clinical value.
Author contributions
GX: Writing–original draft. TW: Writing–original draft. GJ: Conceptualization, Writing–original draft. HW: Conceptualization, Writing–original draft. YL: Investigation, Writing–original draft. BW: Supervision, Writing–review and editing. WH: Resources, Supervision, Writing–review and editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was funded by the Young Innovative Talents Project of Guangdong Higher Education Institutions (2021KQNCX023), Guangdong Medical Science Research Fund (B2023059), Zhanjiang Science and Technology Planning Project (2022A01178), Youth Training Fund project of Guangdong Medical University (GDMUQ2022001), and Discipline Construction Project of Guangdong Medical University (4SG22260G).
Acknowledgments
The authors thank all team members who participated in the cooperation.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
ADAMTS, A disintegrin and metalloproteinase with thrombospondin-like motifs; AF, annulus fibrosus; BHB, Bulge-helix-bulge; BMSC-exos, Bone marrow-derived mesenchymal stem cell exosomes; CEP, Cartilaginous vertebral endplates; ceRNA, Competitive endogenous; CiRNAs, Circular intronic RNAs; DISC, Death-inducing signalling complex; ECM, Extracellular matrix; EcRNAs, Exonic circular RNAs; EIcRNAs, exonic intronic circular RNAs; FBSS, Failed back surgery syndrome; GC, gastric cancer; HCV, Hepatitis C Virus; IDD, Intervertebral disc degeneration; circRNA:circular RNA; IL-1β, Interleukin-1β, IL-6, Interleukin-6; IVD, Intervertebral disc; LBP, Low back pain; MiRNAs, MicroRNAs; MMP, Matrix metalloproteinases; MREs, MiRNA response elements; NP, Nucleus pulposus; pre-mRNAs, precursor mRNAs; RNA, Δψm: mitochondrial membrane potential; SA-beta-gal, Senescence-associated beta-galactosidase; tgf-β3, Transforming growth factor 3; TNF, tumour necrosis factor; TSEN, tRNA splice endonuclease.
References
Abdelmohsen, K., Panda, A. C., Munk, R., Grammatikakis, I., Dudekula, D. B., De, S., et al. (2017). Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol. 14 (3), 361–369. doi:10.1080/15476286.2017.1279788
Adams, M. A., and Roughley, P. J. (2006). What is intervertebral disc degeneration, and what causes it? Spine (Phila Pa 1976) 31 (18), 2151–2161. doi:10.1097/01.brs.0000231761.73859.2c
Ashinsky, B. G., Gullbrand, S. E., Bonnevie, E. D., Mandalapu, S. A., Wang, C., Elliott, D. M., et al. (2019). Multiscale and multimodal structure-function analysis of intervertebral disc degeneration in a rabbit model. Osteoarthr. Cartil. 27 (12), 1860–1869. doi:10.1016/j.joca.2019.07.016
Ashrafi, G., and Schwarz, T. L. (2013). The pathways of mitophagy for quality control and clearance of mitochondria. Cell. Death Differ. 20 (1), 31–42. doi:10.1038/cdd.2012.81
Ashwal-Fluss, R., Meyer, M., Pamudurti, N. R., Ivanov, A., Bartok, O., Hanan, M., et al. (2014). circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell. 56 (1), 55–66. doi:10.1016/j.molcel.2014.08.019
Bahn, J. H., Zhang, Q., Li, F., Chan, T. M., Lin, X., Kim, Y., et al. (2015). The landscape of microRNA, Piwi-interacting RNA, and circular RNA in human saliva. Clin. Chem. 61 (1), 221–230. doi:10.1373/clinchem.2014.230433
Borchardt, E. K., Meganck, R. M., Vincent, H. A., Ball, C. B., Ramos, S. B. V., Moorman, N. J., et al. (2017). Inducing circular RNA formation using the CRISPR endoribonuclease Csy4. Rna 23 (5), 619–627. doi:10.1261/rna.056838.116
Breuer, J., and Rossbach, O. (2020). Production and purification of artificial circular RNA sponges for application in molecular Biology and medicine. Methods Protoc. 3 (2), 42. doi:10.3390/mps3020042
Chen, F., Liu, H., Wang, X., Li, Z., Zhang, J., Pei, Y., et al. (2020a). Melatonin activates autophagy via the NF-κB signaling pathway to prevent extracellular matrix degeneration in intervertebral disc. Osteoarthr. Cartil. 28 (8), 1121–1132. doi:10.1016/j.joca.2020.05.011
Chen, J., Li, Y., Zheng, Q., Bao, C., He, J., Chen, B., et al. (2017a). Circular RNA profile identifies circPVT1 as a proliferative factor and prognostic marker in gastric cancer. Cancer Lett. 388, 208–219. doi:10.1016/j.canlet.2016.12.006
Chen, K., Lv, X., Li, W., Yu, F., Lin, J., Ma, J., et al. (2017b). Autophagy is a protective response to the oxidative damage to endplate chondrocytes in intervertebral disc: implications for the treatment of degenerative lumbar disc. Oxid. Med. Cell. Longev. 2017, 4041768. doi:10.1155/2017/4041768
Chen, L., Wang, C., Sun, H., Wang, J., Liang, Y., Wang, Y., et al. (2021). The bioinformatics toolbox for circRNA discovery and analysis. Brief. Bioinform 22 (2), 1706–1728. doi:10.1093/bib/bbaa001
Chen, T., and Chen, F. (2021). The role of circular RNA plasmacytoma variant translocation 1 as a biomarker for prognostication of acute myeloid leukemia. Hematology 26 (1), 1018–1024. doi:10.1080/16078454.2021.1987649
Chen, Y. C., Lee, C. Y., and Chen, S. J. (2019). Narcotic addiction in failed back surgery syndrome. Cell. Transpl. 28 (3), 239–247. doi:10.1177/0963689718796072
Chen, Z., Song, J., Xie, L., Xu, G., Zheng, C., Xia, X., et al. (2023). N6-methyladenosine hypomethylation of circGPATCH2L regulates DNA damage and apoptosis through TRIM28 in intervertebral disc degeneration. Cell. Death Differ. 30 (8), 1957–1972. doi:10.1038/s41418-023-01190-5
Chen, Z., Zhang, W., Deng, M., Li, Y., and Zhou, Y. (2020b). CircGLCE alleviates intervertebral disc degeneration by regulating apoptosis and matrix degradation through the targeting of miR-587/STAP1. Aging (Albany NY) 12 (21), 21971–21991. doi:10.18632/aging.104035
Cheng, X., Zhang, L., Zhang, K., Zhang, G., Hu, Y., Sun, X., et al. (2018). Circular RNA VMA21 protects against intervertebral disc degeneration through targeting miR-200c and X linked inhibitor-of-apoptosis protein. Ann. Rheum. Dis. 77 (5), 770–779. doi:10.1136/annrheumdis-2017-212056
Choi, H., Merceron, C., Mangiavini, L., Seifert, E. L., Schipani, E., Shapiro, I. M., et al. (2016). Hypoxia promotes noncanonical autophagy in nucleus pulposus cells independent of MTOR and HIF1A signaling. Autophagy 12 (9), 1631–1646. doi:10.1080/15548627.2016.1192753
Chuang, T. J., Chen, Y. J., Chen, C. Y., Mai, T. L., Wang, Y. D., Yeh, C. S., et al. (2018). Integrative transcriptome sequencing reveals extensive alternative trans-splicing and cis-backsplicing in human cells. Nucleic Acids Res. 46 (7), 3671–3691. doi:10.1093/nar/gky032
Clouet, J., Vinatier, C., Merceron, C., Pot-Vaucel, M., Hamel, O., Weiss, P., et al. (2009). The intervertebral disc: from pathophysiology to tissue engineering. Jt. Bone Spine 76 (6), 614–618. doi:10.1016/j.jbspin.2009.07.002
Cocquerelle, C., Mascrez, B., Hétuin, D., and Bailleul, B. (1993). Mis-splicing yields circular RNA molecules. Faseb J. 7 (1), 155–160. doi:10.1096/fasebj.7.1.7678559
De Jong, O. G., Van Balkom, B. W., Schiffelers, R. M., Bouten, C. V., and Verhaar, M. C. (2014). Extracellular vesicles: potential roles in regenerative medicine. Front. Immunol. 5, 608. doi:10.3389/fimmu.2014.00608
Di Agostino, S., Riccioli, A., De Cesaris, P., Fontemaggi, G., Blandino, G., Filippini, A., et al. (2020). Circular RNAs in embryogenesis and cell differentiation with a focus on cancer development. Front. Cell. Dev. Biol. 8, 389. doi:10.3389/fcell.2020.00389
DiStefano, T. J., Vaso, K., Danias, G., Chionuma, H. N., Weiser, J. R., and Iatridis, J. C. (2022). Extracellular vesicles as an emerging treatment option for intervertebral disc degeneration: therapeutic potential, translational pathways, and regulatory considerations. Adv. Healthc. Mater 11 (5), e2100596. doi:10.1002/adhm.202100596
Du, X., Chen, S., Cui, H., Huang, Y., Wang, J., Liu, H., et al. (2022). Circular RNA hsa_circ_0083756 promotes intervertebral disc degeneration by sponging miR-558 and regulating TREM1 expression. Cell. Prolif. 55, e13205. doi:10.1111/cpr.13205
Enuka, Y., Lauriola, M., Feldman, M. E., Sas-Chen, A., Ulitsky, I., and Yarden, Y. (2016). Circular RNAs are long-lived and display only minimal early alterations in response to a growth factor. Nucleic Acids Res. 44 (3), 1370–1383. doi:10.1093/nar/gkv1367
Errichelli, L., Dini Modigliani, S., Laneve, P., Colantoni, A., Legnini, I., Capauto, D., et al. (2017). FUS affects circular RNA expression in murine embryonic stem cell-derived motor neurons. Nat. Commun. 8, 14741. doi:10.1038/ncomms14741
Farshbaf, M., Khosroushahi, A. Y., Mojarad-Jabali, S., Zarebkohan, A., Valizadeh, H., and Walker, P. R. (2020). Cell surface GRP78: an emerging imaging marker and therapeutic target for cancer. J. Control Release 328, 932–941. doi:10.1016/j.jconrel.2020.10.055
French, S. D., Downie, A. S., and Walker, B. F. (2018). Low back pain: a major global problem for which the chiropractic profession needs to take more care. Chiropr. Man. Ther. 26, 28. doi:10.1186/s12998-018-0199-6
Grunhagen, T., Wilde, G., Soukane, D. M., Shirazi-Adl, S. A., and Urban, J. P. (2006). Nutrient supply and intervertebral disc metabolism. J. Bone Jt. Surg. Am. 88 (Suppl. 2), 30–35. doi:10.2106/jbjs.E.01290
Guo, H., Ingolia, N. T., Weissman, J. S., and Bartel, D. P. (2010). Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466 (7308), 835–840. doi:10.1038/nature09267
Guo, H. Y., Guo, M. K., Wan, Z. Y., Song, F., and Wang, H. Q. (2020a). Emerging evidence on noncoding-RNA regulatory machinery in intervertebral disc degeneration: a narrative review. Arthritis Res. Ther. 22 (1), 270. doi:10.1186/s13075-020-02353-2
Guo, J., Shao, M., Lu, F., Jiang, J., and Xia, X. (2017). Role of Sirt1 plays in nucleus pulposus cells and intervertebral disc degeneration. Spine (Phila Pa 1976) 42 (13), E757-E766–e766. doi:10.1097/brs.0000000000001954
Guo, J. U., Agarwal, V., Guo, H., and Bartel, D. P. (2014). Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 15 (7), 409. doi:10.1186/s13059-014-0409-z
Guo, W., Mu, K., Zhang, B., Sun, C., Zhao, L., Dong, Z. Y., et al. (2020b). The circular RNA FAM169A functions as a competitive endogenous RNA and regulates intervertebral disc degeneration by targeting miR-583 and BTRC. Cell. Death Dis. 11 (5), 315. doi:10.1038/s41419-020-2543-8
Guo, W., Mu, K., Zhang, B., Sun, C., Zhao, L., Li, H. R., et al. (2020c). The circular RNA circ-GRB10 participates in the molecular circuitry inhibiting human intervertebral disc degeneration. Cell. Death Dis. 11 (8), 612. doi:10.1038/s41419-020-02882-3
Guo, W., Zhang, B., Mu, K., Feng, S. Q., Dong, Z. Y., Ning, G. Z., et al. (2018). Circular RNA GRB10 as a competitive endogenous RNA regulating nucleus pulposus cells death in degenerative intervertebral disk. Cell. Death Dis. 9 (3), 319. doi:10.1038/s41419-017-0232-z
Guo, W., Zhang, B., Sun, C., Duan, H. Q., Liu, W. X., Mu, K., et al. (2020d). Circular RNA derived from TIMP2 functions as a competitive endogenous RNA and regulates intervertebral disc degeneration by targeting miR-185-5p and matrix metalloproteinase 2. Int. J. Mol. Med. 46 (2), 621–632. doi:10.3892/ijmm.2020.4621
Hanifeh, M., and Ataei, F. (2022). XIAP as a multifaceted molecule in Cellular Signaling. Apoptosis 27 (7-8), 441–453. doi:10.1007/s10495-022-01734-z
Hansen, T. B., Jensen, T. I., Clausen, B. H., Bramsen, J. B., Finsen, B., Damgaard, C. K., et al. (2013). Natural RNA circles function as efficient microRNA sponges. Nature 495 (7441), 384–388. doi:10.1038/nature11993
Harland, R., and Misher, L. (1988). Stability of RNA in developing Xenopus embryos and identification of a destabilizing sequence in TFIIIA messenger RNA. Development 102 (4), 837–852. doi:10.1242/dev.102.4.837
Hiyama, A., Arai, F., Sakai, D., Yokoyama, K., and Mochida, J. (2012). The effects of oxygen tension and antiaging factor Klotho on Wnt signaling in nucleus pulposus cells. Arthritis Res. Ther. 14 (3), R105. doi:10.1186/ar3830
Holm, S., Holm, A. K., Ekström, L., Karladani, A., and Hansson, T. (2004). Experimental disc degeneration due to endplate injury. J. Spinal Disord. Tech. 17 (1), 64–71. doi:10.1097/00024720-200402000-00012
Horner, H. A., and Urban, J. P. (2001). 2001 Volvo Award Winner in Basic Science Studies: effect of nutrient supply on the viability of cells from the nucleus pulposus of the intervertebral disc. Spine (Phila Pa 1976) 26 (23), 2543–2549. doi:10.1097/00007632-200112010-00006
Hoy, D., Bain, C., Williams, G., March, L., Brooks, P., Blyth, F., et al. (2012). A systematic review of the global prevalence of low back pain. Arthritis Rheum. 64 (6), 2028–2037. doi:10.1002/art.34347
Hsiao, K. Y., Lin, Y. C., Gupta, S. K., Chang, N., Yen, L., Sun, H. S., et al. (2017). Noncoding effects of circular RNA CCDC66 promote colon cancer growth and metastasis. Cancer Res. 77 (9), 2339–2350. doi:10.1158/0008-5472.Can-16-1883
Hu, B., Xiao, L., Wang, C., Liu, C., Zhang, Y., Ding, B., et al. (2022). Circ_0022382 ameliorated intervertebral disc degeneration by regulating TGF-β3 expression through sponge adsorption of miR-4726-5p. Bone 154, 116185. doi:10.1016/j.bone.2021.116185
Huang, S., Zhong, J., Qi, Q., Liu, G., and Gong, M. (2022). CircRNA expression profile and potential role of hsa_circ_0040039 in intervertebral disc degeneration. Med. Baltim. 101 (32), e30035. doi:10.1097/md.0000000000030035
Huang, Y., Gao, J., Wang, J., Ye, H., Yao, T., Xu, Y., et al. (2021a). Inhibition of intervertebral disc disease progression via the circPKNOX1-miR-370-3p-KIAA0355 axis. Cell. Death Discov. 7 (1), 39. doi:10.1038/s41420-021-00420-4
Huang, Y., Zhang, Z., Wang, J., Shen, S., Yao, T., Xu, Y., et al. (2021b). circSPG21 protects against intervertebral disc disease by targeting miR-1197/ATP1B3. Exp. Mol. Med. 53 (10), 1547–1558. doi:10.1038/s12276-021-00674-z
Jeck, W. R., Sorrentino, J. A., Wang, K., Slevin, M. K., Burd, C. E., Liu, J., et al. (2013). Circular RNAs are abundant, conserved, and associated with ALU repeats. Rna 19 (2), 141–157. doi:10.1261/rna.035667.112
Jeong, S. W., Lee, J. S., and Kim, K. W. (2014). In vitro lifespan and senescence mechanisms of human nucleus pulposus chondrocytes. Spine J. 14 (3), 499–504. doi:10.1016/j.spinee.2013.06.099
Jia, Y., Huo, X., Wu, L., Zhang, H., Xu, W., and Leng, H. (2023). circSNTB2 and CUL4A induces dysfunction of nucleus pulposus cells by competitively binding miR-665. Biochem. Genet. doi:10.1007/s10528-023-10465-y
Johnson, W. E., Stephan, S., and Roberts, S. (2008). The influence of serum, glucose and oxygen on intervertebral disc cell growth in vitro: implications for degenerative disc disease. Arthritis Res. Ther. 10 (2), R46. doi:10.1186/ar2405
Jost, I., Shalamova, L. A., Gerresheim, G. K., Niepmann, M., Bindereif, A., and Rossbach, O. (2018). Functional sequestration of microRNA-122 from Hepatitis C Virus by circular RNA sponges. RNA Biol. 15 (8), 1032–1039. doi:10.1080/15476286.2018.1435248
Jost, P. J., and Vucic, D. (2020). Regulation of cell death and immunity by XIAP. Cold Spring Harb. Perspect. Biol. 12 (8), a036426. doi:10.1101/cshperspect.a036426
Kang, B. R., Yang, S. H., Chung, B. R., Kim, W., and Kim, Y. (2016). Cell surface GRP78 as a biomarker and target for suppressing glioma cells. Sci. Rep. 6, 34922. doi:10.1038/srep34922
Karki, R., Sharma, B. R., Tuladhar, S., Williams, E. P., Zalduondo, L., Samir, P., et al. (2021). Synergism of TNF-α and IFN-γ triggers inflammatory cell death, tissue damage, and mortality in SARS-CoV-2 infection and cytokine shock syndromes. Cell. 184 (1), 149–168.e17. doi:10.1016/j.cell.2020.11.025
Kim, K. W., Chung, H. N., Ha, K. Y., Lee, J. S., and Kim, Y. Y. (2009). Senescence mechanisms of nucleus pulposus chondrocytes in human intervertebral discs. Spine J. 9 (8), 658–666. doi:10.1016/j.spinee.2009.04.018
Knezevic, N. N., Mandalia, S., Raasch, J., Knezevic, I., and Candido, K. D. (2017). Treatment of chronic low back pain - new approaches on the horizon. J. Pain Res. 10, 1111–1123. doi:10.2147/jpr.S132769
Kong, D., Gu, R., Zhang, C., and Yin, R. (2020). Knockdown of hsa_circ_0059955 induces apoptosis and cell cycle arrest in nucleus pulposus cells via inhibiting itchy E3 ubiquitin protein ligase. Drug Des. Devel Ther. 14, 3951–3963. doi:10.2147/dddt.S253293
Krautbauer, S., Neumeier, M., Haberl, E. M., Pohl, R., Feder, S., Eisinger, K., et al. (2019). The utrophin-beta 2 syntrophin complex regulates adipocyte lipid droplet size independent of adipogenesis. Mol. Cell. Biochem. 452 (1-2), 29–39. doi:10.1007/s11010-018-3409-6
Kristensen, L. S., Hansen, T. B., Venø, M. T., and Kjems, J. (2018). Circular RNAs in cancer: opportunities and challenges in the field. Oncogene 37 (5), 555–565. doi:10.1038/onc.2017.361
Kun-Peng, Z., Xiao-Long, M., and Chun-Lin, Z. (2018). Overexpressed circPVT1, a potential new circular RNA biomarker, contributes to doxorubicin and cisplatin resistance of osteosarcoma cells by regulating ABCB1. Int. J. Biol. Sci. 14 (3), 321–330. doi:10.7150/ijbs.24360
Legnini, I., Di Timoteo, G., Rossi, F., Morlando, M., Briganti, F., Sthandier, O., et al. (2017). Circ-ZNF609 is a circular RNA that can Be translated and functions in myogenesis. Mol. Cell. 66 (1), 22–37. doi:10.1016/j.molcel.2017.02.017
Le Maitre, C. L., Freemont, A. J., and Hoyland, J. A. (2005). The role of interleukin-1 in the pathogenesis of human intervertebral disc degeneration. Arthritis Res. Ther. 7 (4), R732–R745. doi:10.1186/ar1732
Li, H., Tian, L., Li, J., Li, Y., Du, L., Huo, Z., et al. (2022a). The roles of circRNAs in intervertebral disc degeneration: inflammation, extracellular matrix metabolism, and apoptosis. Anal. Cell. Pathol. (Amst) 2022, 9550499. doi:10.1155/2022/9550499
Li, S., Hua, W., Wang, K., Gao, Y., Chen, S., Liu, W., et al. (2018). Autophagy attenuates compression-induced apoptosis of human nucleus pulposus cells via MEK/ERK/NRF1/Atg7 signaling pathways during intervertebral disc degeneration. Exp. Cell. Res. 370 (1), 87–97. doi:10.1016/j.yexcr.2018.06.012
Li, Y., Wu, X., Li, J., Du, L., Wang, X., Cao, J., et al. (2022b). Circ_0004354 might compete with circ_0040039 to induce NPCs death and inflammatory response by targeting miR-345-3p-FAF1/TP73 axis in intervertebral disc degeneration. Oxid. Med. Cell. Longev. 2022, 2776440. doi:10.1155/2022/2776440
Li, Y., Zheng, Q., Bao, C., Li, S., Guo, W., Zhao, J., et al. (2015a). Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell. Res. 25 (8), 981–984. doi:10.1038/cr.2015.82
Li, Y., Zhou, S., Peng, P., Wang, X., Du, L., Huo, Z., et al. (2020). Emerging role of circular RNA in intervertebral disc degeneration: knowns and unknowns (Review). Mol. Med. Rep. 22 (4), 3057–3065. doi:10.3892/mmr.2020.11437
Li, Z., Huang, C., Bao, C., Chen, L., Lin, M., Wang, X., et al. (2015b). Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 22 (3), 256–264. doi:10.1038/nsmb.2959
Liao, Z., Luo, R., Li, G., Song, Y., Zhan, S., Zhao, K., et al. (2019). Exosomes from mesenchymal stem cells modulate endoplasmic reticulum stress to protect against nucleus pulposus cell death and ameliorate intervertebral disc degeneration in vivo. Theranostics 9 (14), 4084–4100. doi:10.7150/thno.33638
Lin, H., Wang, Y., Wang, P., Long, F., and Wang, T. (2022). Mutual regulation between N6-methyladenosine (m6A) modification and circular RNAs in cancer: impacts on therapeutic resistance. Mol. Cancer 21 (1), 148. doi:10.1186/s12943-022-01620-x
Lin, Z., Lu, F., Ma, X., Xia, X., Zou, F., and Jiang, J. (2021). Roles of circular RNAs in the pathogenesis of intervertebral disc degeneration (Review). Exp. Ther. Med. 22 (5), 1221. doi:10.3892/etm.2021.10655
Liu, X., Abraham, J. M., Cheng, Y., Wang, Z., Wang, Z., Zhang, G., et al. (2018). Synthetic circular RNA functions as a miR-21 sponge to suppress gastric carcinoma cell proliferation. Mol. Ther. Nucleic Acids 13, 312–321. doi:10.1016/j.omtn.2018.09.010
Liu, Y., Xue, M., Du, S., Feng, W., Zhang, K., Zhang, L., et al. (2019). Competitive endogenous RNA is an intrinsic component of EMT regulatory circuits and modulates EMT. Nat. Commun. 10 (1), 1637. doi:10.1038/s41467-019-09649-1
Liu, Y. C., Li, J. R., Sun, C. H., Andrews, E., Chao, R. F., Lin, F. M., et al. (2016). CircNet: a database of circular RNAs derived from transcriptome sequencing data. Nucleic Acids Res. 44 (D1), D209–D215. doi:10.1093/nar/gkv940
Liuke, M., Solovieva, S., Lamminen, A., Luoma, K., Leino-Arjas, P., Luukkonen, R., et al. (2005). Disc degeneration of the lumbar spine in relation to overweight. Int. J. Obes. (Lond) 29 (8), 903–908. doi:10.1038/sj.ijo.0802974
Lu, T. X., and Rothenberg, M. E. (2018). MicroRNA. J. Allergy Clin. Immunol. 141 (4), 1202–1207. doi:10.1016/j.jaci.2017.08.034
Lyu, F. J., Cui, H., Pan, H., Mc Cheung, K., Cao, X., Iatridis, J. C., et al. (2021). Painful intervertebral disc degeneration and inflammation: from laboratory evidence to clinical interventions. Bone Res. 9 (1), 7. doi:10.1038/s41413-020-00125-x
Mahmmoud, Y. A. (2005). Curcumin modulation of Na,K-ATPase: phosphoenzyme accumulation, decreased K+ occlusion, and inhibition of hydrolytic activity. Br. J. Pharmacol. 145 (2), 236–245. doi:10.1038/sj.bjp.0706185
Molinos, M., Almeida, C. R., Caldeira, J., Cunha, C., Gonçalves, R. M., and Barbosa, M. A. (2015a). Inflammation in intervertebral disc degeneration and regeneration. J. R. Soc. Interface 12 (104), 20141191. doi:10.1098/rsif.2014.1191
Molinos, M., Almeida, C. R., Caldeira, J., Cunha, C., Gonçalves, R. M., and Barbosa, M. A. (2015b). Inflammation in intervertebral disc degeneration and regeneration. J. R. Soc. Interface 12 (108), 20150429. doi:10.1098/rsif.2015.0429
Mosley, G. E., Evashwick-Rogler, T. W., Lai, A., and Iatridis, J. C. (2017). Looking beyond the intervertebral disc: the need for behavioral assays in models of discogenic pain. Ann. N. Y. Acad. Sci. 1409 (1), 51–66. doi:10.1111/nyas.13429
Nicolet, B. P., Engels, S., Aglialoro, F., van den Akker, E., von Lindern, M., and Wolkers, M. C. (2018). Circular RNA expression in human hematopoietic cells is widespread and cell-type specific. Nucleic Acids Res. 46 (16), 8168–8180. doi:10.1093/nar/gky721
O'Conor, C. J., Case, N., and Guilak, F. (2013). Mechanical regulation of chondrogenesis. Stem Cell. Res. Ther. 4 (4), 61. doi:10.1186/scrt211
Ogawa, H., Cornelius, F., Hirata, A., and Toyoshima, C. (2015). Sequential substitution of K(+) bound to Na(+),K(+)-ATPase visualized by X-ray crystallography. Nat. Commun. 6, 8004. doi:10.1038/ncomms9004
Oichi, T., Taniguchi, Y., Oshima, Y., Tanaka, S., and Saito, T. (2020). Pathomechanism of intervertebral disc degeneration. JOR Spine 3 (1), e1076. doi:10.1002/jsp2.1076
Okholm, T. L. H., Sathe, S., Park, S. S., Kamstrup, A. B., Rasmussen, A. M., Shankar, A., et al. (2020). Transcriptome-wide profiles of circular RNA and RNA-binding protein interactions reveal effects on circular RNA biogenesis and cancer pathway expression. Genome Med. 12 (1), 112. doi:10.1186/s13073-020-00812-8
Ort, T., Voronov, S., Guo, J., Zawalich, K., Froehner, S. C., Zawalich, W., et al. (2001). Dephosphorylation of beta2-syntrophin and Ca2+/mu-calpain-mediated cleavage of ICA512 upon stimulation of insulin secretion. Embo J. 20 (15), 4013–4023. doi:10.1093/emboj/20.15.4013
Pagliarini, V., Jolly, A., Bielli, P., Di Rosa, V., De la Grange, P., and Sette, C. (2020). Sam68 binds Alu-rich introns in SMN and promotes pre-mRNA circularization. Nucleic Acids Res. 48 (2), 633–645. doi:10.1093/nar/gkz1117
Pamudurti, N. R., Bartok, O., Jens, M., Ashwal-Fluss, R., Stottmeister, C., Ruhe, L., et al. (2017). Translation of CircRNAs. Mol. Cell. 66 (1), 9–21. doi:10.1016/j.molcel.2017.02.021
Panagopoulos, A., Samant, S., Bakhos, J. J., Liu, M., Khan, B., Makadia, J., et al. (2022). Triggering receptor expressed on myeloid cells-1 (TREM-1) inhibition in atherosclerosis. Pharmacol. Ther. 238, 108182. doi:10.1016/j.pharmthera.2022.108182
Paushkin, S. V., Patel, M., Furia, B. S., Peltz, S. W., and Trotta, C. R. (2004). Identification of a human endonuclease complex reveals a link between tRNA splicing and pre-mRNA 3' end formation. Cell. 117 (3), 311–321. doi:10.1016/s0092-8674(04)00342-3
Pfafenrot, C., Schneider, T., Müller, C., Hung, L. H., Schreiner, S., Ziebuhr, J., et al. (2021). Inhibition of SARS-CoV-2 coronavirus proliferation by designer antisense-circRNAs. Nucleic Acids Res. 49 (21), 12502–12516. doi:10.1093/nar/gkab1096
Quero, L., Klawitter, M., Schmaus, A., Rothley, M., Sleeman, J., Tiaden, A. N., et al. (2013). Hyaluronic acid fragments enhance the inflammatory and catabolic response in human intervertebral disc cells through modulation of toll-like receptor 2 signalling pathways. Arthritis Res. Ther. 15 (4), R94. doi:10.1186/ar4274
Raj, P. P. (2008). Intervertebral disc: anatomy-physiology-pathophysiology-treatment. Pain Pract. 8 (1), 18–44. doi:10.1111/j.1533-2500.2007.00171.x
RezaSoltani, M., Forouzesh, F., Salehi, Z., Zabihi, M. R., Rejali, L., and Nazemalhosseini-Mojarad, E. (2023). Identification of LncPVT1 and CircPVT1 as prognostic biomarkers in human colorectal polyps. Sci. Rep. 13 (1), 13113. doi:10.1038/s41598-023-40288-1
Richardson, S. M., Doyle, P., Minogue, B. M., Gnanalingham, K., and Hoyland, J. A. (2009). Increased expression of matrix metalloproteinase-10, nerve growth factor and substance P in the painful degenerate intervertebral disc. Arthritis Res. Ther. 11 (4), R126. doi:10.1186/ar2793
Risbud, M. V., and Shapiro, I. M. (2014). Role of cytokines in intervertebral disc degeneration: pain and disc content. Nat. Rev. Rheumatol. 10 (1), 44–56. doi:10.1038/nrrheum.2013.160
Roberts, S., Evans, H., Trivedi, J., and Menage, J. (2006). Histology and pathology of the human intervertebral disc. J. Bone Jt. Surg. Am. 88 (Suppl. 2), 10–14. doi:10.2106/jbjs.F.00019
Roughley, P. J. (2004). Biology of intervertebral disc aging and degeneration: involvement of the extracellular matrix. Spine (Phila Pa 1976) 29 (23), 2691–2699. doi:10.1097/01.brs.0000146101.53784.b1
Rutges, J. P., Kummer, J. A., Oner, F. C., Verbout, A. J., Castelein, R. J., Roestenburg, H. J., et al. (2008). Increased MMP-2 activity during intervertebral disc degeneration is correlated to MMP-14 levels. J. Pathol. 214 (4), 523–530. doi:10.1002/path.2317
Rybak-Wolf, A., Stottmeister, C., Glažar, P., Jens, M., Pino, N., Giusti, S., et al. (2015). Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol. Cell. 58 (5), 870–885. doi:10.1016/j.molcel.2015.03.027
Salzman, J., Gawad, C., Wang, P. L., Lacayo, N., and Brown, P. O. (2012). Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One 7 (2), e30733. doi:10.1371/journal.pone.0030733
Schmidt, C. A., Giusto, J. D., Bao, A., Hopper, A. K., and Matera, A. G. (2019). Molecular determinants of metazoan tricRNA biogenesis. Nucleic Acids Res. 47 (12), 6452–6465. doi:10.1093/nar/gkz311
Séguin, C. A., Pilliar, R. M., Roughley, P. J., and Kandel, R. A. (2005). Tumor necrosis factor-alpha modulates matrix production and catabolism in nucleus pulposus tissue. Spine (Phila Pa 30 (17), 1940–1948. doi:10.1097/01.brs.0000176188.40263.f9
Serres, M. P., Shaughnessy, R., Escot, S., Hammich, H., Cuvelier, F., Salles, A., et al. (2023). MiniBAR/GARRE1 is a dual Rac and Rab effector required for ciliogenesis. Dev. Cell. 58, 2477–2494.e8. doi:10.1016/j.devcel.2023.09.010
Shu, Y., Wu, K., Zeng, Z., Huang, S., Ji, X., Yuan, C., et al. (2018). A simplified system to express circularized inhibitors of miRNA for stable and potent suppression of miRNA functions. Mol. Ther. Nucleic Acids 13, 556–567. doi:10.1016/j.omtn.2018.09.025
Song, J., Chen, Z. H., Zheng, C. J., Song, K. H., Xu, G. Y., Xu, S., et al. (2020). Exosome-transported circRNA_0000253 competitively adsorbs MicroRNA-141-5p and increases IDD. Mol. Ther. Nucleic Acids 21, 1087–1099. doi:10.1016/j.omtn.2020.07.039
Song, J., Wang, H. L., Song, K. H., Ding, Z. W., Wang, H. L., Ma, X. S., et al. (2018). CircularRNA_104670 plays a critical role in intervertebral disc degeneration by functioning as a ceRNA. Exp. Mol. Med. 50 (8), 94–12. doi:10.1038/s12276-018-0125-y
Spencer, E., Jiang, J., and Chen, Z. J. (1999). Signal-induced ubiquitination of IkappaBalpha by the F-box protein Slimb/beta-TrCP. Genes. Dev. 13 (3), 284–294. doi:10.1101/gad.13.3.284
Starke, S., Jost, I., Rossbach, O., Schneider, T., Schreiner, S., Hung, L. H., et al. (2015). Exon circularization requires canonical splice signals. Cell. Rep. 10 (1), 103–111. doi:10.1016/j.celrep.2014.12.002
Studer, R. K., Vo, N., Sowa, G., Ondeck, C., and Kang, J. (2011). Human nucleus pulposus cells react to IL-6: independent actions and amplification of response to IL-1 and TNF-α. Spine (Phila Pa 1976) 36 (8), 593–599. doi:10.1097/BRS.0b013e3181da38d5
Tay, Y., Rinn, J., and Pandolfi, P. P. (2014). The multilayered complexity of ceRNA crosstalk and competition. Nature 505 (7483), 344–352. doi:10.1038/nature12986
Thomson, S., and Jacques, L. (2009). Demographic characteristics of patients with severe neuropathic pain secondary to failed back surgery syndrome. Pain Pract. 9 (3), 206–215. doi:10.1111/j.1533-2500.2009.00276.x
Urban, J. P., and Roberts, S. (2003). Degeneration of the intervertebral disc. Arthritis Res. Ther. 5 (3), 120–130. doi:10.1186/ar629
Urban, J. P., Smith, S., and Fairbank, J. C. (2004). Nutrition of the intervertebral disc. Spine (Phila Pa 1976) 29 (23), 2700–2709. doi:10.1097/01.brs.0000146499.97948.52
Vasiliadis, E. S., Pneumaticos, S. G., Evangelopoulos, D. S., and Papavassiliou, A. G. (2014). Biologic treatment of mild and moderate intervertebral disc degeneration. Mol. Med. 20 (1), 400–409. doi:10.2119/molmed.2014.00145
Venkatesh, S., Li, H., Gogol, M. M., and Workman, J. L. (2016). Selective suppression of antisense transcription by Set2-mediated H3K36 methylation. Nat. Commun. 7, 13610. doi:10.1038/ncomms13610
Vlaeyen, J. W. S., Maher, C. G., Wiech, K., Van Zundert, J., Meloto, C. B., Diatchenko, L., et al. (2018). Low back pain. Nat. Rev. Dis. Prim. 4 (1), 52. doi:10.1038/s41572-018-0052-1
Vo, N. V., Hartman, R. A., Yurube, T., Jacobs, L. J., Sowa, G. A., and Kang, J. D. (2013). Expression and regulation of metalloproteinases and their inhibitors in intervertebral disc aging and degeneration. Spine J. 13 (3), 331–341. doi:10.1016/j.spinee.2012.02.027
Wang, F., Cai, F., Shi, R., Wang, X. H., and Wu, X. T. (2016). Aging and age related stresses: a senescence mechanism of intervertebral disc degeneration. Osteoarthr. Cartil. 24 (3), 398–408. doi:10.1016/j.joca.2015.09.019
Wang, H., He, P., Pan, H., Long, J., Wang, J., Li, Z., et al. (2018a). Circular RNA circ-4099 is induced by TNF-α and regulates ECM synthesis by blocking miR-616-5p inhibition of Sox9 in intervertebral disc degeneration. Exp. Mol. Med. 50 (4), 1–14. doi:10.1038/s12276-018-0056-7
Wang, H., Zhu, Y., Cao, L., Guo, Z., Sun, K., Qiu, W., et al. (2021a). circARL15 plays a critical role in intervertebral disc degeneration by modulating miR-431-5p/DISC1. Front. Genet. 12, 669598. doi:10.3389/fgene.2021.669598
Wang, J., Markova, D., Anderson, D. G., Zheng, Z., Shapiro, I. M., and Risbud, M. V. (2011). TNF-α and IL-1β promote a disintegrin-like and metalloprotease with thrombospondin type I motif-5-mediated aggrecan degradation through syndecan-4 in intervertebral disc. J. Biol. Chem. 286 (46), 39738–39749. doi:10.1074/jbc.M111.264549
Wang, L., Cho, Y. L., Tang, Y., Wang, J., Park, J. E., Wu, Y., et al. (2018b). PTEN-L is a novel protein phosphatase for ubiquitin dephosphorylation to inhibit PINK1-Parkin-mediated mitophagy. Cell. Res. 28 (8), 787–802. doi:10.1038/s41422-018-0056-0
Wang, M., Wey, S., Zhang, Y., Ye, R., and Lee, A. S. (2009). Role of the unfolded protein response regulator GRP78/BiP in development, cancer, and neurological disorders. Antioxid. Redox Signal 11 (9), 2307–2316. doi:10.1089/ars.2009.2485
Wang, S., Mao, C., and Liu, S. (2019a). Peptides encoded by noncoding genes: challenges and perspectives. Signal Transduct. Target Ther. 4, 57. doi:10.1038/s41392-019-0092-3
Wang, S., Sun, J., Yang, H., Zou, W., Zheng, B., Chen, Y., et al. (2019b). Profiling and bioinformatics analysis of differentially expressed circular RNAs in human intervertebral disc degeneration. Acta Biochim. Biophys. Sin. (Shanghai) 51 (6), 571–579. doi:10.1093/abbs/gmz036
Wang, W. J., Yu, X. H., Wang, C., Yang, W., He, W. S., Zhang, S. J., et al. (2015). MMPs and ADAMTSs in intervertebral disc degeneration. Clin. Chim. Acta 448, 238–246. doi:10.1016/j.cca.2015.06.023
Wang, X., Ma, R., Zhang, X., Cui, L., Ding, Y., Shi, W., et al. (2021b). Crosstalk between N6-methyladenosine modification and circular RNAs: current understanding and future directions. Mol. Cancer 20 (1), 121. doi:10.1186/s12943-021-01415-6
Wang, X., Wang, B., Zou, M., Li, J., Lü, G., Zhang, Q., et al. (2018c). CircSEMA4B targets miR-431 modulating IL-1β-induced degradative changes in nucleus pulposus cells in intervertebral disc degeneration via Wnt pathway. Biochim. Biophys. Acta Mol. Basis Dis. 1864 (11), 3754–3768. doi:10.1016/j.bbadis.2018.08.033
Wang, Y., Gao, W., Shi, X., Ding, J., Liu, W., He, H., et al. (2017). Chemotherapy drugs induce pyroptosis through caspase-3 cleavage of a gasdermin. Nature 547 (7661), 99–103. doi:10.1038/nature22393
Waxenbaum, J. A., Reddy, V., and Futterman, B. (2021). “Anatomy, back, intervertebral discs,” in StatPearls. (Treasure island (FL). (China: StatPearls Publishing).
Wei, L., Guo, J., Zhai, W., Xie, Y., and Jia, Y. (2023). CircRNA GRB10 is a novel biomarker for the accurate diagnosis of lumbar degenerative disc disease. Mol. Biotechnol. 65 (5), 816–821. doi:10.1007/s12033-022-00574-1
Willms, E., Cabañas, C., Mäger, I., Wood, M. J. A., and Vader, P. (2018). Extracellular vesicle heterogeneity: subpopulations, isolation techniques, and diverse functions in cancer progression. Front. Immunol. 9, 738. doi:10.3389/fimmu.2018.00738
Xia, X., Guo, J., Lu, F., and Jiang, J. (2015). SIRT1 plays a protective role in intervertebral disc degeneration in a puncture-induced rodent model. Spine (Phila Pa 1976) 40 (9), E515–E524. doi:10.1097/brs.0000000000000817
Xia, Y., Yang, R., Hou, Y., Wang, H., Li, Y., Zhu, J., et al. (2022). Application of mesenchymal stem cell-derived exosomes from different sources in intervertebral disc degeneration. Front. Bioeng. Biotechnol. 10, 1019437. doi:10.3389/fbioe.2022.1019437
Xiang, Q., Kang, L., Wang, J., Liao, Z., Song, Y., Zhao, K., et al. (2020). CircRNA-CIDN mitigated compression loading-induced damage in human nucleus pulposus cells via miR-34a-5p/SIRT1 axis. EBioMedicine 53, 102679. doi:10.1016/j.ebiom.2020.102679
Xiao, L., Ding, B., Xu, S., Gao, J., Yang, B., Wang, J., et al. (2020). circRNA_0058097 promotes tension-induced degeneration of endplate chondrocytes by regulating HDAC4 expression through sponge adsorption of miR-365a-5p. J. Cell. Biochem. 121 (1), 418–429. doi:10.1002/jcb.29202
Xie, L., Huang, W., Fang, Z., Ding, F., Zou, F., Ma, X., et al. (2019). CircERCC2 ameliorated intervertebral disc degeneration by regulating mitophagy and apoptosis through miR-182-5p/SIRT1 axis. Cell. Death Dis. 10 (10), 751. doi:10.1038/s41419-019-1978-2
Xu, D., Ma, X., Sun, C., Han, J., Zhou, C., Wong, S. H., et al. (2021). Circular RNAs in intervertebral disc degeneration: an updated review. Front. Mol. Biosci. 8, 781424. doi:10.3389/fmolb.2021.781424
Xu, G., Liu, C., Jiang, J., Liang, T., Yu, C., Qin, Z., et al. (2020). A novel mechanism of intervertebral disc degeneration: imbalance between autophagy and apoptosis. Epigenomics 12 (13), 1095–1108. doi:10.2217/epi-2020-0079
Yan, Z., Pan, Y., Wang, S., Cheng, M., Kong, H., Sun, C., et al. (2017). Static compression induces ECM remodeling and integrin α2β1 expression and signaling in a rat tail caudal intervertebral disc degeneration model. Spine (Phila Pa 1976) 42 (8), E448–e458. doi:10.1097/brs.0000000000001856
Yang, Q., Du, W. W., Wu, N., Yang, W., Awan, F. M., Fang, L., et al. (2017a). A circular RNA promotes tumorigenesis by inducing c-myc nuclear translocation. Cell. Death Differ. 24 (9), 1609–1620. doi:10.1038/cdd.2017.86
Yang, Y., Fan, X., Mao, M., Song, X., Wu, P., Zhang, Y., et al. (2017b). Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell. Res. 27 (5), 626–641. doi:10.1038/cr.2017.31
Yang, Z., Xie, L., Han, L., Qu, X., Yang, Y., Zhang, Y., et al. (2017c). Circular RNAs: regulators of cancer-related signaling pathways and potential diagnostic biomarkers for human cancers. Theranostics 7 (12), 3106–3117. doi:10.7150/thno.19016
Yi, J., Wang, L., Hu, G. S., Zhang, Y. Y., Du, J., Ding, J. C., et al. (2023). CircPVT1 promotes ER-positive breast tumorigenesis and drug resistance by targeting ESR1 and MAVS. Embo J. 42 (10), e112408. doi:10.15252/embj.2022112408
Zeng, Y., Du, W. W., Wu, Y., Yang, Z., Awan, F. M., Li, X., et al. (2017). A circular RNA binds to and activates AKT phosphorylation and nuclear localization reducing apoptosis and enhancing cardiac repair. Theranostics 7 (16), 3842–3855. doi:10.7150/thno.19764
Zhang, B., Zhang, Z., Li, L., Qin, Y. R., Liu, H., Jiang, C., et al. (2018). TSPAN15 interacts with BTRC to promote oesophageal squamous cell carcinoma metastasis via activating NF-κB signaling. Nat. Commun. 9 (1), 1423. doi:10.1038/s41467-018-03716-9
Zhang, F., Lin, F., Xu, Z., and Huang, Z. (2021a). Circular RNA ITCH promotes extracellular matrix degradation via activating Wnt/β-catenin signaling in intervertebral disc degeneration. Aging (Albany NY) 13 (10), 14185–14197. doi:10.18632/aging.203036
Zhang, G. Z., Liu, M. Q., Chen, H. W., Wu, Z. L., Gao, Y. C., Ma, Z. J., et al. (2021b). NF-κB signalling pathways in nucleus pulposus cell function and intervertebral disc degeneration. Cell. Prolif. 54 (7), e13057. doi:10.1111/cpr.13057
Zhang, J., Hu, S., Ding, R., Yuan, J., Jia, J., Wu, T., et al. (2021c). CircSNHG5 sponges mir-495-3p and modulates CITED2 to protect cartilage endplate from degradation. Front. Cell. Dev. Biol. 9, 668715. doi:10.3389/fcell.2021.668715
Zhang, J., Jiang, H., Li, M., and Ding, L. (2022). Knockdown of triggering receptor expressed on myeloid cells 1 (TREM1) inhibits endoplasmic reticulum stress and reduces extracellular matrix degradation and the apoptosis of human nucleus pulposus cells. Exp. Ther. Med. 24 (4), 607. doi:10.3892/etm.2022.11544
Zhang, L., Hou, C., Chen, C., Guo, Y., Yuan, W., Yin, D., et al. (2020a). The role of N(6)-methyladenosine (m(6)A) modification in the regulation of circRNAs. Mol. Cancer 19 (1), 105. doi:10.1186/s12943-020-01224-3
Zhang, T. W., Li, Z. F., Dong, J., and Jiang, L. B. (2020b). The circadian rhythm in intervertebral disc degeneration: an autophagy connection. Exp. Mol. Med. 52 (1), 31–40. doi:10.1038/s12276-019-0372-6
Zhang, X. O., Dong, R., Zhang, Y., Zhang, J. L., Luo, Z., Zhang, J., et al. (2016). Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res. 26 (9), 1277–1287. doi:10.1101/gr.202895.115
Zhang, Y., Sun, Z., Liu, J., and Guo, X. (2008). Advances in susceptibility genetics of intervertebral degenerative disc disease. Int. J. Biol. Sci. 4 (5), 283–290. doi:10.7150/ijbs.4.283
Zhang, Y., Zhang, X. O., Chen, T., Xiang, J. F., Yin, Q. F., Xing, Y. H., et al. (2013). Circular intronic long noncoding RNAs. Mol. Cell. 51 (6), 792–806. doi:10.1016/j.molcel.2013.08.017
Zhao, X., Yuan, J., Jia, J., Zhang, J., Liu, J., Chen, Q., et al. (2023). Role of non-coding RNAs in cartilage endplate (Review). Exp. Ther. Med. 26 (1), 312. doi:10.3892/etm.2023.12011
Zhong, R., Chen, Z., Mo, T., Li, Z., and Zhang, P. (2019). Potential Role of circPVT1 as a proliferative factor and treatment target in esophageal carcinoma. Cancer Cell. Int. 19, 267. doi:10.1186/s12935-019-0985-9
Keywords: lower back pain, intervertebral disc degeneration, nucleus pulposus cell, extracellular matrix, circular RNA (circRNA)
Citation: Xie G, Wu T, Ji G, Wu H, Lai Y, Wei B and Huang W (2023) Circular RNA and intervertebral disc degeneration: unravelling mechanisms and implications. Front. Mol. Biosci. 10:1302017. doi: 10.3389/fmolb.2023.1302017
Received: 25 September 2023; Accepted: 05 December 2023;
Published: 19 December 2023.
Edited by:
Alessio Colantoni, Sapienza University of Rome, ItalyReviewed by:
Nahid Arghiani, Stockholm University, SwedenArijita Sarkar, University of Southern California, United States
Copyright © 2023 Xie, Wu, Ji, Wu, Lai, Wei and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bo Wei, d2VpYm9AZ2RtdS5lZHUuY24=; Wenhua Huang, aHVhbmd3ZW5odWEyMDA5QDEzOS5jb20=
†These authors have contributed equally to this work
 Guohao Xie
Guohao Xie Tingrui Wu
Tingrui Wu Guangju Ji1
Guangju Ji1 Wenhua Huang
Wenhua Huang