- 1Department of Biomedical Engineering, Indian Institute of Technology Ropar, Punjab, Punjab, India
- 2Department of Molecular Biology and Biotechnology, Tezpur University, Napaam, Assam, India
An imbalance in microbial homeostasis, referred to as dysbiosis, is critically associated with the progression of obesity-induced metabolic disorders including type 2 diabetes (T2D). Alteration in gut microbial diversity and the abundance of pathogenic bacteria disrupt metabolic homeostasis and potentiate chronic inflammation, due to intestinal leakage or release of a diverse range of microbial metabolites. The obesity-associated shifts in gut microbial diversity worsen the triglyceride and cholesterol level that regulates adipogenesis, lipolysis, and fatty acid oxidation. Moreover, an intricate interaction of the gut-brain axis coupled with the altered microbiome profile and microbiome-derived metabolites disrupt bidirectional communication for instigating insulin resistance. Furthermore, a distinct microbial community within visceral adipose tissue is associated with its dysfunction in obese T2D individuals. The specific bacterial signature was found in the mesenteric adipose tissue of T2D patients. Recently, it has been shown that in Crohn’s disease, the gut-derived bacterium Clostridium innocuum translocated to the mesenteric adipose tissue and modulates its function by inducing M2 macrophage polarization, increasing adipogenesis, and promoting microbial surveillance. Considering these facts, modulation of microbiota in the gut and adipose tissue could serve as one of the contemporary approaches to manage T2D by using prebiotics, probiotics, or faecal microbial transplantation. Altogether, this review consolidates the current knowledge on gut and adipose tissue dysbiosis and its role in the development and progression of obesity-induced T2D. It emphasizes the significance of the gut microbiota and its metabolites as well as the alteration of adipose tissue microbiome profile for promoting adipose tissue dysfunction, and identifying novel therapeutic strategies, providing valuable insights and directions for future research and potential clinical interventions.
1 Introduction
Obesity is a severe human health problem that increasing globally at an alarming rate which significantly contributes to the development of several metabolic diseases including type 2 diabetes (T2D) (Pulgaron and Delamater, 2014). According to the World Health Organization report, more than 2 billion adults worldwide are overweight or obese, and the number of people with diabetes has risen from 151 million in 2000 to 537 million in 2021 (Ruze et al., 2023). Recent evidence suggests that diet and food habits strikingly influence the pathophysiological state of obesity potentiating the onset and progression of T2D (Guo et al., 2020).
The gut microbiota iscomprisedof about 10 trillion different bacteria that increase in density as these moves along the gastrointestinal (GI) tract (Cani et al., 2019). The host and gut microbiota represent a symbiotic connection in which the host provides a nutrient-rich habitat for the microbiome community while the microbes influence the host’s physiology, immunology, and metabolism (Thursby and Juge, 2017). The dynamic complexity of the microbial ecosystem and its composition is influenced by several factors, including diet and lifestyle, which play a vital role in the maintenance of host health by regulating the immune system, metabolizing nutrients, and producing essential vitamins and hormones (Hou et al., 2022). Moreover, the gut microbiome critically regulates the gut-brain axis wherein gut microbial metabolites such as short-chain fatty acids (SCFAs) and neurotransmitters can cross the blood-brain barrier and influence brain function (Silva et al., 2020).
Gut dysbiosis, a condition characterized by an imbalance in the composition and function of the gut microbiome, has been implicated in several chronic diseases, including inflammatory bowel disease, colorectal cancer, and T2D (Vijay and Valdes, 2022). Understanding the relationship between the gut microbiome and insulin resistance has been improved in the last few decades. Gut dysbiosis and the disrupted gut permeability (leaky gut) in obesity allow lipopolysaccharide (LPS) and other pro-inflammatory molecules to enter the bloodstream and trigger systemic inflammation, contributing to the progression of insulin resistance and T2D (Boulangé et al., 2016). Obesity has been attributed to several metabolic changes that are causally linked with glucose intolerance and insulin resistance associated with T2D (Martyn et al., 2008; Wondmkun, 2020). Few recent studies revealed the enrichment of microorganisms in mesenteric and omental adipose tissue during obesity, which acts as a key driver of adipose tissue inflammation that potentiates Crohn’s disease and insulin resistance (Ha et al., 2020; Massier et al., 2020). Thus, targeting the gut microbiome has emerged as a promising strategy for the prevention and treatment of obesity-induced T2D. Several therapeutic strategies, such as probiotics, prebiotics, and dietary modifications, have been proposed to modulate the gut microbiome to prevent or treat T2D (Huda et al., 2021).
This review aims to provide an overview of the current understanding of gut and adipose tissue dysbiosis in obesity-induced T2D and to identify future research directions and associated challenges in this field. We explored the mechanisms by which adipose tissue microbiome enriched in obesity and associated gut dysbiosis that correlated with altered gut microbiota-brain axis, and pancreas dysfunction through the emergence of microbial metabolites contributing to the development of T2D. We also examined the shreds of evidence supporting the use of various therapeutic strategies aimed at modulating the gut microbiome.
2 Gut dysbiosis linked with obesity and type 2 diabetes
The human gut accommodates a diverse community of enteric microflora, including Firmicutes and Bacteroidetes (up to 75% of total gut flora), which is also called a “virtual organ”, provides structural, metabolic, and protective benefits to intestinal epithelial cells (O’Hara and Shanahan, 2006; Foster and McVey Neufeld, 2013). The human gut environment can be viewed as a dynamic system formed by the host and its microbiota working together. The gut microbiota involves in carbohydrate and fat metabolism, vitamin and amino acid production, the proliferation of epithelial cells, defense against infections, and hormone regulation in the host body (Oliphant and Allen-Vercoe, 2019) and also helps in the digestion of plant polysaccharides, complex nutrients, and milk sugars (Xu and Gordon, 2003; Hersoug et al., 2018) those which were not properly digested by the host and thus contributing 10% of caloric value and aid in preserving intestinal health and anti-cancer properties (Wong et al., 2006; Blustein et al., 2013; Cox and Blaser, 2013).
2.1 Obesity induces alteration of gut microbiota profile
Obesity is a complex metabolic disorder resulting from an imbalance between energy intake and energy expenditure (Hill et al., 2012). It is associated with dysregulation of lipid and glucose metabolism leading to abnormal levels of blood lipids causally linked with hyperlipidemia, ectopic lipid deposition, glucose intolerance, insulin resistance, and T2D (Klop et al., 2013). Reportedly, lipid has bidirectional regulation with the gut microbiota. Dietary lipids break down into fatty acids that are majorly absorbed in the GI tract and are found to modulate bacterial diversity as several fatty acids exhibit antibiotic activity and reduce ATP production (Jackman et al., 2016; Schoeler and Caesar, 2019). In the distal colon, residual peptides and proteins, bile acids, and choline undergo fermentation that produces a more diverse range of products compared to the fermentation of carbohydrates in the proximal colon (Canfora et al., 2022). Some of the products generated in the distal colon include bacterial toxins such as LPS, hydrogen sulfide, bile acid derivatives like deoxycholate and lithocholate, branched-chain amino acids (BCAAs) and their metabolites, and branched-chain fatty acids (BCFAs) like isobutyrate, 2-methyl butyrate, and isovalerate (Zhai et al., 2021). Additionally, aromatic amino acids (AAAs) give rise to phenolic, indolic, skatolic, and p-cresolic compounds, ammonia, and polyamines. Choline, another substrate, produces dimethylamine (DMA) and trimethylamine (TMA) as end products in the distal colon (Zhai et al., 2021) (Table 1).
Specific gut bacteria, such as Akkermansia muciniphila and Bifidobacterium, have been shown to produce metabolites that directly affect glucose metabolism leading to increased insulin sensitivity and improved glucose tolerance. On the contrary, obesity-associated changes in gut bacteria have been shown to produce metabolites that can promote insulin resistance and glucose intolerance. Mice feeding with an HFD comprising 49.5% lipid content reduce Bifidobacterium spp., Eubacterium rectale, Clostridium coccoides as well as Bacteroides (Cani et al., 2007). Both high-fat and high-sugar diet feeding significantly altered the gut microbiota diversity, promoting the accumulation of Gram-negative bacteria (Cani et al., 2007). About 50% reduction in Bacteroidetes and a proportional increase in Firmicutes have been identified in genetically obese mice models (Ley et al., 2005).
Many bacterial genera are either positively or negatively correlated with obese T2D conditions. The overabundance of bacterial genera including Fusobacterium, Ruminococcus, and Blautia was found to be positively correlated with the pathophysiology of T2D (Table 2). In contrast, several bacterial genera including Faecalibacterium, Bifidobacterium, Akkermansia, Roseburia, and Bacteroides were negatively correlated with T2D and therefore beneficial to counteract the pathogenesis of T2D (Gurung et al., 2020) (Table 2). Certain gut bacterial populations were linked to metabolic alterations, body fat mass, and calorie consumption. The gut bacteria Faecalibacterium prausnitzii was reported to be linked with shifts in inflammatory status and insulin sensitivity (Furet et al., 2010). F. prausnitzii is relatively attuned to urinary metabolites, corpulence, and inflammation, and exhibits anti-inflammatory effects by targeting the NF-κB pathway. This bacteria species is recognized as a preserved and prominent species of healthy individuals’ faecal microbiota (Li et al., 2008; Sokol et al., 2008; Tap et al., 2009; Furet et al., 2010). Lactobacillus rhamnosus CNCM I-3690 as a probiotic candidate counteracts B. Wadsworth-mediated immunological dysfunction and also supports bolstering the intestinal barrier and lowering inflammation (Natividad et al., 2018). Blautia wexlerae administration reduces obesity and T2D through metabolic gut microbiota reconstruction (Hosomi et al., 2022). The microbial community regulates hormonal balance, bowel permeability, expression of genes regulating lipogenesis, insulin resistance, endotoxemia, interaction with bile acids, and changes in the distribution of brown adipose tissue (BAT) (Cornejo-Pareja et al., 2019). Moreover, an unbalanced gut flora in obese people might cause chronic inflammation followed by metabolic diseases including insulin resistance and T2D (Singh et al., 2017). People even trying to improve gut barrier function by using herbal medicine like Bofutsushosan causally associated with the blooms Akkermansia muciniphila and improves glucose metabolism in mice (Fujisaka et al., 2020). Nuciferine, a main bioactive component in the lotus leaf, when treated with HFD rats showed a significant decline in the ratio of Firmicutes/Bacteroidetes phyla which resulted in reduced blood endotoxemia and inflammation along with increased intestinal integrity and SCFA synthesis (Wang et al., 2020). Interestingly, blood glucose levels may also be adversely affected by the oral microbial compositions that cause both local and systemic inflammation (Omori et al., 2022). Alteration in the salivary microbiome has also been reported in T2D patients with a higher abundance of Streptococcus sp., Lactobacillus sp., Blautia wexlerae, Lactobacillus fermentum, Nocardia coeliaca, Selenomonas artemidis (Kampoo et al., 2014; Long et al., 2017) and reduction of phylum Actinobacteria, and Bifidobacterium (Kampoo et al., 2014; Long et al., 2017).
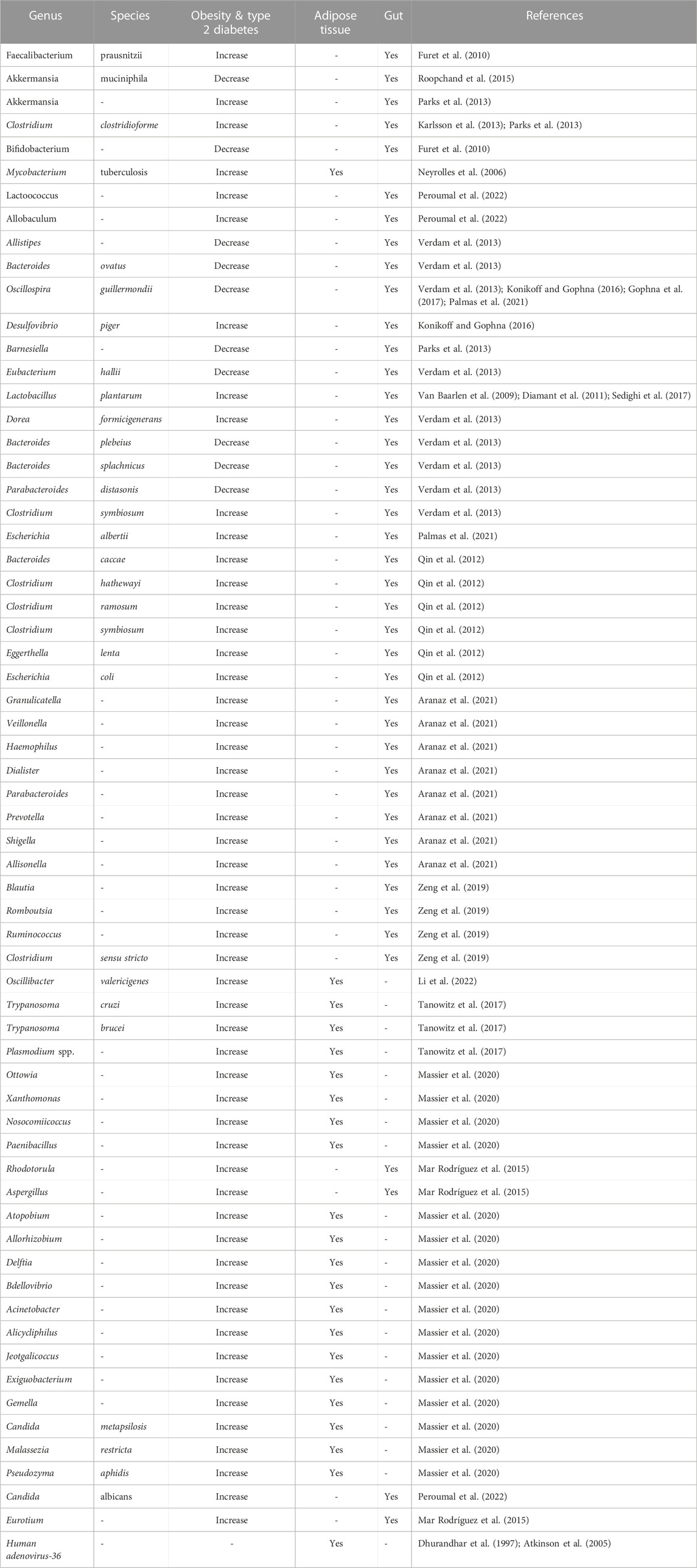
TABLE 2. Summary of microbial signature at different taxonomic level in the gut and adipose tissue of obesity and type 2 diabetes.
2.2 Gut microbial-derived metabolites modulate lipid metabolism in obesity
Microbiota-derived metabolites, such as bile acids, LPS, and SCFAs have been identified as important factors in the regulation of hyperlipidemia (Agus et al., 2021; Jia et al., 2021). SCFAs such as acetic acid, propionic acid, butyric acid, isobutyric acid, valeric acid, and isovaleric acid are produced from the fermentation of dietary fibre by gut microbes and are known to be associated with several health benefits (Rios-Covian et al., 2020). These fatty acids also influence adipogenesis, lipolysis, and fatty acid oxidation, all of which are related to lipid metabolism in non-obese and obese states (Breton et al., 2022). Propionate has been found to increase the expression of peroxisome proliferator-activated receptors (PPARs), which are key regulators of adipogenesis (Hong et al., 2005). Gut microbiota is known to inhibit adenosine monophosphate kinase (AMPK) activity, an enzyme that plays a crucial role in energy homeostasis, leading to the reduction of fatty acid oxidation coincided with higher cholesterol and triglycerides favouring lipogenesis. High-fat diet (HFD)-fed germ-free (GF) mice showed higher levels of phosphorylated AMPK and promoted fatty acid oxidation, compared to control animals (Bäckhed et al., 2007). Microbiota transfer from conventional (CV) mice to GF mice has been found to affect the fasting-induced adipose factor, angiopoietin-like 4 (ANGPTL4). ANGPTL4 inhibits lipoprotein lipase (LPL), which decreases the accumulation of triglycerides in adipocytes and relevantly regulates fat storage (Bäckhed et al., 2004).
Obesity-associated gut microbial imbalance leads to alterations in the production of these metabolites and signalling molecules causing T2D pathogenesis (Figure 1). It has been shown that a specific gut microbiota community belonging to the Prevotella group produced a small metabolite succinate that improved glucose homeostasis in mice by regulating intestinal gluconeogenesis (Ghanim et al., 2009). Similarly, it has been found that a membrane protein from A. muciniphila, a mucin-degrading gut bacterium, protected mice from obesity and associated complications (Buckley et al., 2014). Moreover, a novel microbially produced small molecule imidazole propionate is involved in the impairment of insulin signalling by targeting the mammalian target of rapamycin complex 1 (mTORC1) and thus promoting glucose intolerance and insulin resistance (Kindt et al., 2010). Furthermore, alteration of microbiota profile as well microbial leakage causes pancreatic infection and inflammation that impairs the production of insulin and promotes insulin resistance leading to glucose intolerance and T2D. Transplantation of faecal matter from Western diet-fed or genetically obese mice was capable of generating an obese phenotype in the standard diet-fed or non-obese mice, which resulted in a larger weight gain compared to treatment with wild-type microorganisms (De Groot et al., 2020). Furthermore, an obese pathophysiological state decreases the number of A. muciniphila by increasing NAD and riboflavin-biosynthesis. Together, these functional adjustments particularly allow glutathione to be recharged to its reduced state, enabling redox balance in microorganisms that may be exposed to the gut which is a potentially unfriendly, inflammatory, and oxidatively challenged environment (Yassour et al., 2016). It has also been observed that Bacteroidetes were decreased in number with a concomitant increase in Firmicutes in the gut of HFD-fed mice. This alteration in the bacterial balance may have a role in the emergence and development of obesity (Magne et al., 2020). Firmicutes bacteria are believed to have a greater capacity to obtain energy from food, which could increase caloric intake. On the contrary, Bacteroidetes are linked with weight management either by oxidation of SCFAs or the digestion of complex carbohydrates (Singh et al., 2017). Moreover, by transferring faecal microbiota from hyperlipidemic individuals to GF mice, researchers have been able to observe the pathogenic effects of the microbiota on lipid metabolism (Burz et al., 2021). Velagapudi et al.compared GF mice with CV mice and found differences in triglyceride (TG) levels in various tissues. The GF mice showed lower levels of TG in adipose tissue and the liver but higher levels of TG in the circulatory system compared to CV mice (Velagapudi et al., 2010). Interestingly, western diet-fed GF mice were found to have less hypercholesterolemia (high levels of cholesterol in the blood) and increased cholesterol excretion in the liver and faeces compared to CV mice (Velagapudi et al., 2010). This indicates that the absence of gut microbiota influences cholesterol metabolism and may lead to reduced cholesterol accumulation in the blood. These studies highlight the relationship between gut microbial compositions and metabolic disorders such as T2D in HFD feeding.
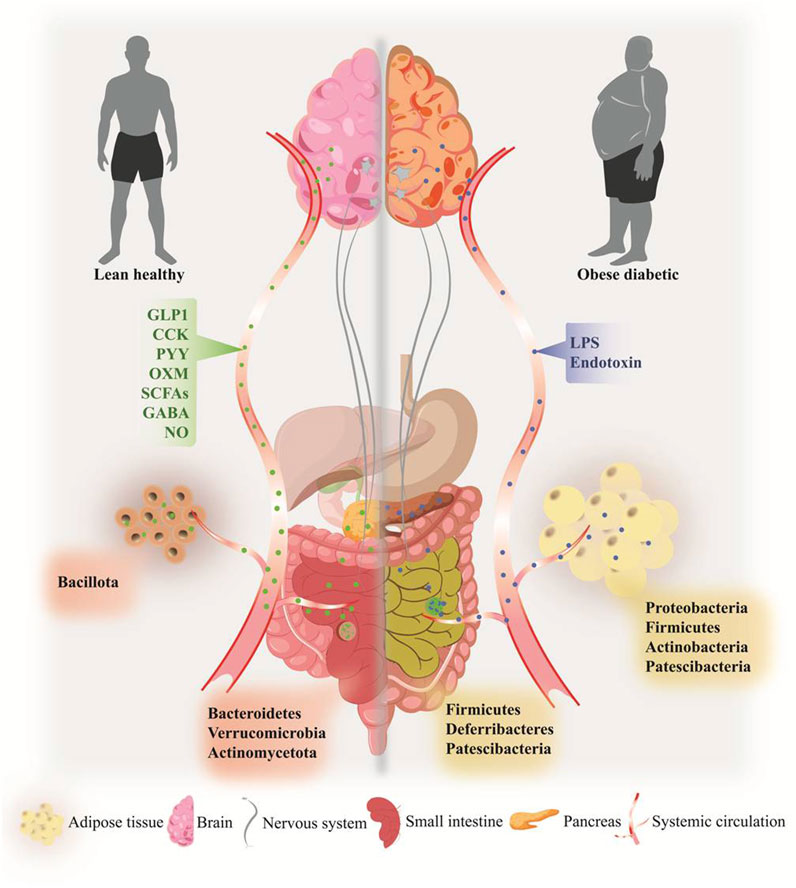
FIGURE 1. Overview of obesity-induced dysbiosis and associated pathogenesis in the gut, gut-brain axis, pancreas, and adipose tissue of type 2 diabetes. Specific microbial phyla signatures were observed in gut and adipose tissue in the onset of obesity-induced T2D (right side) compared to lean healthy individual (left side). A Plethora of gut microbiome-derived metabolites participates in organs function, including gut-brain homeostasis, pancreas function, and inflammatory homeostasis. Alteration of microbial metabolites including LPS, endotoxins, and MAMPs leads to the development of pathophysiological state in obesity and T2D, by rapid systemic inflammation, releasing pro-inflammatory molecules, gut-brain dysfunction, and pancreatic damages.
2.3 Gut microbiome components promote chronic inflammation in obesity
Obesity is associated with chronic low-grade systemic inflammation, often regulated by microbial signature in the host, which is associated with insulin resistance, a hallmark of T2D. Inflammatory molecules produced by gut bacteria can enter the bloodstream, promoting chronic inflammation and impairing insulin signalling. LPS, a common structural component of the outer membrane of Gram-negative bacteria, reportedly promoted the onset of obesity (Boulangé et al., 2016). LPS and other bacterial debris pass from the gut environment and enter into the circulatory system due to increased intestinal permeability triggering an immune reaction. As seen by enhanced adipose macrophage recruitment and hepatic NF-κB/IKKβ inflammatory-signaling pathways, this was linked to systemic inflammation, most likely via LPS or saturated fatty acids (Takeda et al., 2003; Kleinridders et al., 2009) Mice lacking toll-like receptor 4 (TLR4) and cluster of differentiation 14 (CD14), receptors for LPS, are resistant to HFD- or LPS-induced hyperinsulinemia and insulin resistance (Stavropoulou et al., 2020). Elevated LPS content in the gut decreases the tight junction proteinzona occludens-1 (ZO-1) and occludin expression and affects the permeability and integrity of intestinal epithelial cells (Jia et al., 2021). Overall, the release of inflammatory metabolites ultimately impairs lipid metabolism. In a recent study by Mishra et al., it has been shown that gut microbiota in obese mice and humans have reduced ability for ethanolamine metabolism that resulted in increased intestinal permeability (Mishra et al., 2023). Moreover, ethanolamine elevated the abundance of a specific microRNA, miR-101a-3p, and enhanced miR promoter binding ARID3a transcription factor, which leads to the loss of a critical tight junction protein ZO-1. As a result, the weakening of the intestinal barrier leads to increased gut permeability, inflammation, and abnormalities in glucose metabolism (Figure 2). Importantly, restoration of ethanolamine-metabolizing activity can be done by correcting ARID3a/miR-101a-3p/ZO-1 axis and improving the integrity of the intestinal barrier (Mishra et al., 2023).
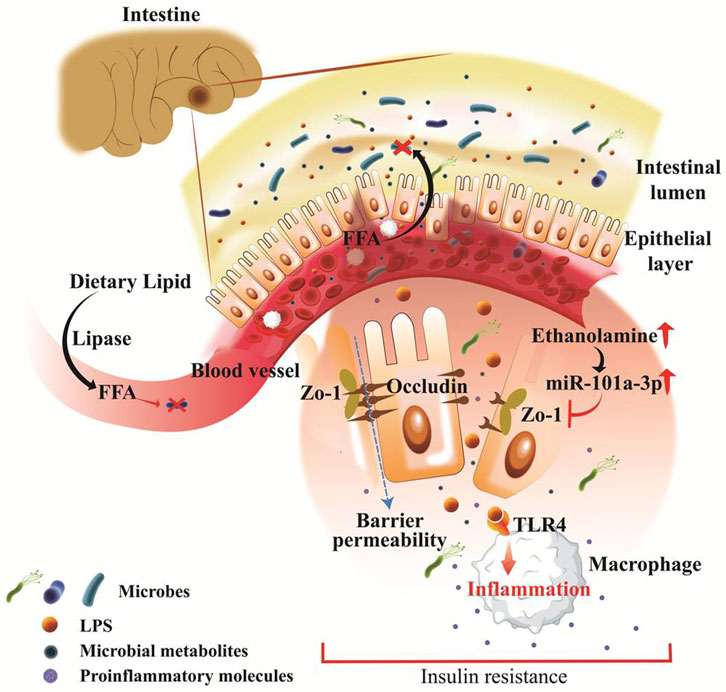
FIGURE 2. Pathophysiologically leaky gut associated with obesity and type 2 diabetes. Fat-enriched diet consumption is aligned with obesity and hyperlipidemia wherein dietary lipid breaks down into free fatty acids and absorbed in the intestine and embraces antibiotic response, and facilitates microbiome alteration in obesity. Intestinal epithelial layer barricades between gut microbiota and systemic circulation, that rapidly disrupted during gut pathogenesis. Pathogenic microbes and derived metabolites including LPS and ethanolamine are rapidly produced in obesity and directly found routes to systemic circulation. LPS endorses rapid inflammation through TLR4, CD14, and MD-2 which breaks down epithelial tight junctions. Ethanolamine activated microRNA species such as miR-101a-3p and inhibits the critical tight junction molecules zona occludens-1 (ZO-1). This result in increased barrier permeability and affiliates rapid inflammation, insulin resistance, and abnormal glucose metabolism.
2.4 Obesity-induced gut dysbiosis influences T2D-associated pathogenesis
Gut dysbiosis is a phenomenon that refers to a disruption in the composition and function of the microorganisms that inhabit the human body. Recent studies have demonstrated that gut dysbiosis is associated with obesity and T2D, two major health concerns that affect millions of people worldwide. Obesity and T2D are closely linked, with obesity being one of the major risk factors for developing T2D. Studies have indicated that alterations in gut dysbiosis may contribute to the development of insulin resistance, which are key driver of T2D. In a landmark study, researchers transplanted gut microbiota from lean or obese human donors into GF mice. Mice receiving microbiota from obese donors showed increased total body fat and insulin resistance compared to those receiving microbiota from lean donors. This study provided early evidence for a potential link between gut microbiota and obesity-related metabolic dysfunction (Turnbaugh et al., 2006). It has been reported that the gut microbiota of obese individuals is highly efficient at extracting energy from the diet, leading to increased energy storage, which promotes the hypertrophy of adipose tissue and weight gain. The gut microbiome is structured by diet composition (David et al., 2014; Von Schwartzenberg et al., 2021), and polyunsaturated fatty acids (PUFA)-enriched dietary lipids exhibited a positive effect on the gut microbiota, restoring obesity-induced gut microbial dysfunction in mice (Haneishi et al., 2023). Multiple population studies have consistently demonstrated associations between gut microbiota composition and obesity-related metabolic disorders, including T2D (Table 3). Qin et al. found significant differences in the gut microbial composition between obese and non-obese individuals, with lower bacterial diversity observed in the gut microbiota of obese individuals (Qin et al., 2012). Specific microbial taxa associated with obesity were identified, indicating a potential role of the gut microbiota in energy harvest and metabolic dysfunction. Le Chatelier et al. also identified compositional differences in the gut microbiota between lean and obese individuals, with certain bacterial groups associated with adiposity, insulin resistance, and metabolic parameters (Le Chatelier et al., 2013). Moreover, Karlsson et al. highlighted the importance of gut microbial diversity, as reduced bacterial richness was associated with a higher prevalence of insulin resistance and metabolic disorders (Karlsson et al., 2013). A large-scale population study was performed to examine the association between gut microbiota and metabolic health, specifically focusing on obesity and insulin resistance. It identified specific gut microbial signatures, or patterns, that were associated with obesity and insulin resistance, as well as observed distinct differences in the gut microbiota composition between individuals with normal and impaired glucose metabolism which altogether highlighted the potential importance of the microbiome in the development and progression of obesity-related diabetes (Forslund et al., 2015). Several reports on animal studies also provide compelling evidence that alterations in the gut microbiota can influence metabolic health, including obesity-related diabetes. They demonstrated that manipulating the gut microbiota composition or transferring microbiota from obese individuals can lead to metabolic abnormalities in healthy animals (Cani et al., 2008; Ridaura et al., 2013; Everard et al., 2014). These studies collectively support the notion that alterations in the gut microbiota composition are associated with obesity-related diabetes and provide insight into potential mechanisms underlying this relationship (Table 3).
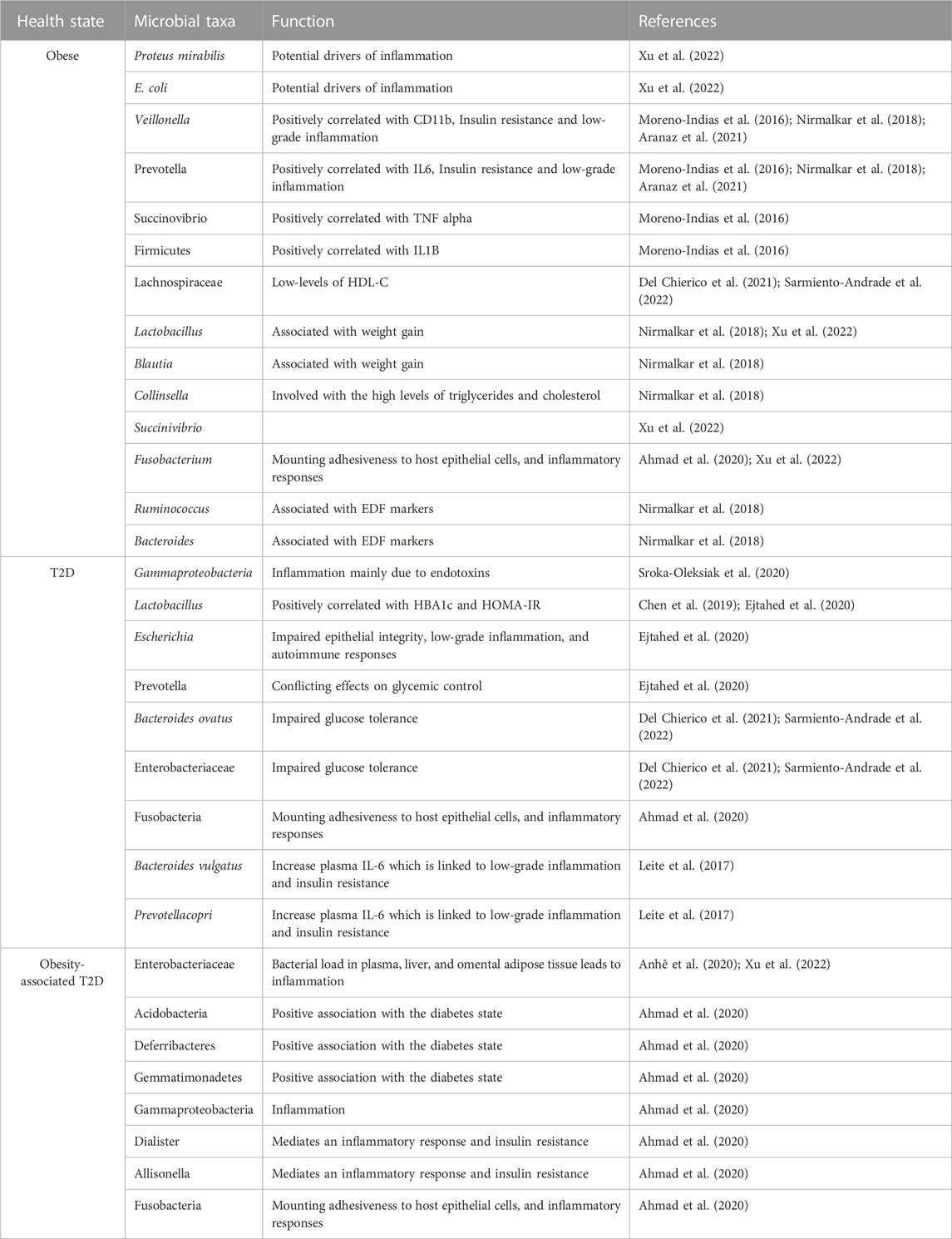
TABLE 3. Summary of predominant microbial taxa and their functions in different health conditions, such as obesity, T2D, and obesity-associated T2D.
It is also worth mentioning that the field of gut microbiota research is evolving, and new studies may provide additional insights into the specific mechanisms and relationships between microbial compositions and metabolic disorders. These findings highlight the important role of gut microbiota composition in metabolic disorders and provide valuable insights into specific bacterial genera that may be associated with either positive or negative outcomes in terms of metabolic health.
3 Adipose tissue dysbiosis promotes adipocyte dysfunction in obesity-induced type 2 diabetes
Adipose tissue is a metabolically active endocrine, immunological, energy storage organ critically regulating energy balance and glucose metabolism. The enriching complexity of adipose tissue is associated with its microbiome, which consists of a diverse community of microorganisms that reside within the tissue microenvironment and appear to play an important role in regulating adipose tissue inflammation and insulin sensitivity.
3.1 Microbiota-derived factors regulate adipose tissue function
Adipose tissue dysbiosis can also influence adipose tissue function by altering the production and release of various adipokines. Apart from the tissue microbiome, the intestinal microbial population was found to regulate adipocyte function. Genetically obese mice (ob/ob) inherit alteration in the microbiota with increasing Firmicutes population in comparison to lean heterozygous (ob/+) animals. Moreover, the microbial composition is very similar to the obese human samples (Ley et al., 2005). The fat-storing capacity is critically regulated by the microbiome and GF mice are protected from obesity even after being fed an HFD (Bäckhed et al., 2004). The absence of a microbiome leads to an overexpression of Fasting-Induced Adipose Factor (Fiaf), a protein involved in lipid metabolism by inhibiting the activity of lipoprotein lipase (LPL), an enzyme responsible for breaking down triglycerides. Additionally, it induces the expression of PGC-1α and activates AMPK, both of which impact cellular energy metabolism (Bäckhed et al., 2005). These combined effects enhance adipocyte lipid storage, potentially leading to increased fat accumulation. Few microbes become therapeutically significant as of their impact on leanness; such as the administration of Akkermansia muciniphila, Lactobacillus plantarum reportedly reduces the impact of an obesogenic diet on human and animals (Depommier et al., 2019; Heeney et al., 2019). Faecalmicrobiota transplantation (FMT) experiments involving the transfer of intestinal microbiota from obese patients into GF mice have provided evidence that microbiota transfer correlates with obesity. To minimize the inter-individual variability of FMT transplantation, a recent study was performed by FMT on the same morbidly obese patient before and after undergoing (RYGB) bariatric surgery in GF mice. The results showed that RYGB surgery led to improving metabolic health along with the changes in the microbiota composition in both patients and the mice receiving the FMTs from pre- and post-surgery stools compared to mice with pre-surgery microbiota. Moreover, visceral adipose tissue (VAT) and subcutaneous white adipose tissue (scWAT) function improved by reducing the expression of Tnf-a, Ccl2, Elane (neutrophil elastase), and increasing anti-inflammatory Sirtuin1 level in the post-RYGB FMT mice along with improvement of BAT function by enriching UCP1 positive areas, anti-inflammatory markers IL33, IL2Ra expression (Yadav et al., 2023). Another subset of microbiome-derived products including propionate, flavonoid, tryptophan-derived metabolites, and cell wall components actively regulates adipose tissue. Tryptophan serves as an essential amino acid, playing a vital role as a building block for various proteins. However, it is worth noting that tryptophan has additional significance beyond its role in protein synthesis. The microbiota, residing in the gut, possesses the ability to convert tryptophan into indole compounds. These indole compounds can accumulate in the gut lumen. A recent study investigated the impact of tryptophan-derived metabolites on a group of miRNAs known as the miR-181 family, which are known to be upregulated in obese white adipose tissue. Interestingly, deletion of the two most highly expressed miR-181 clusters in mice protected them from developing obesity caused by an HFD. A specific tryptophan-derived metabolite called indole-3-carboxylic acid was identified as being reduced in mice fed an HFD. It was found that this metabolite acts on adipocytes to inhibit the expression of miR-181. By regulating miR-181 expression, indole-3-carboxylic acid was shown to have an impact on energy expenditure and insulin sensitivity, suggesting its involvement in the regulation of metabolism (Virtue et al., 2019). Emerging evidence suggeststoll-like receptor (TLR) ligands and nucleotide-binding oligomerization domain-containing (NOD) proteins, specifically NOD1 and NOD2, play crucial roles in adipose tissue dysfunction (Chan et al., 2017). An abundance of microbiota-derived LPS causes activation of TLR signalling in adipocytes and other immune cells, which in turn results secretion of pro-inflammatory cytokines and chemokines, promoting a state of chronic low-grade inflammation; a characteristic feature of adipose tissue dysfunction observed in conditions such as obesity and associate with insulin resistance. Moreover, NOD1 and NOD2, members of the NOD-like receptor (NLR) family, are present in both adipocytes and immune cells within adipose tissue and play significant roles in adipose tissue dysfunction. Activation of NOD1 is associated with insulin resistance, whereas NOD2 null mice develop chronic inflammation and insulin resistance without change in adiposity upon being fed an HFD (Denou et al., 2015; Chan et al., 2017; Martínez-Montoro et al., 2022). Overall, these pattern recognition receptors contribute to adipose tissue inflammation, metabolic dysregulation, and impaired insulin sensitivity.
3.2 Microbial compartmentalization in the white adipose tissue depots during obesity
Similar to the intestinal microbiome, recent studies revealed independent adipose tissue microbiome exists and obese people exhibit an increase in Firmicutes and a reduction in Bacteroidetes. Among the different depots of adipose tissue, mesenteric white adipose tissue (mWAT) harbours the highest bacterial quantity and diversity. The mWAT is a continuous band of adipose tissue that wraps around the various segments of the intestines and acts as a gateway for the intestines to communicate with the rest of the body’s systems. Moreover, the adipose tissue microbiome’s composition and functionality are known to vary with obesity which regulates inflammation and metabolic dysfunction (Davis, 2016; Massier et al., 2020; Zheng et al., 2020). Burcelin et al. introduced the initial hypothesis of the “tissue microbiota” after finding bacterial DNA in different metabolic organs such as the liver, and adipose tissue of human beings (Burcelin et al., 2013). Moreover, the presence of bacterial DNA in distinct compartmentalization within the mesenteric adipose tissueand omental adipose tissue is associated with type 2 diabetes (T2D) (Anhê et al., 2020). Later, the bacterial diversity was compared in the white adipose tissue (WAT) of subcutaneous, visceral, and mesenteric depots and it was found bacterial population is predominant in mesenteric WAT (Massier et al., 2020). These findings suggest that WAT may operate as a critical institution to host bacterial populations and to metabolic dysfunction under obesity-associated T2D (Table 2).
Microbiota-mediated interruptions in OXPHOS/mitochondria lead to functional impairment of white adipose tissue, the primary governing factor of systemic glucose metabolism (Li et al., 2022). Consumption of a high-fat/high-sugar diet leads to the expansion of specific bacteria that produce TLR2 ligands, triggering the induction of Mmp12+ macrophages in WAT. Mmp12+ macrophages serve as a link between microbiota-dependent inflammation and OXPHOS damage in WAT. These macrophages exhibited a specific molecular signature that was associated with insulin resistance in obese patients and released MMP12, which acts as a bridge between inflammation and mitochondrial damage in WAT, ultimately leading to insulin resistance (Li et al., 2022).
Alteration in the microbiota may have a role in both chronic inflammation and insulin resistance (Boulangé et al., 2016; Foley et al., 2018; Kawai et al., 2021). Studies have shown that specific bacterial species prevalent in the adipose tissue of obese people can create inflammatory mediators like LPS, which can cause insulin resistance and other metabolic diseases (Massier et al., 2020). LPS plays a crucial role in macrophage polarization, specifically in the transition from an anti-inflammatory to a pro-inflammatory phenotype. The exposure of adipocytes to LPS, potentially influenced by their size, may play a role in adipocyte cell death with the formation of crown-like structures in inflamed adipose tissue. Furthermore, it has been observed that LPS present within adipocytes can activate caspase-4/5/11, which can induce a highly inflammatory form of programmed cell death known as pyroptosis; typically associated with intracellular pathogen infections (Hersoug et al., 2018). Changing microflora composition in obese adipose tissue exhibits similarities to the altered gut microbiome. In overweight individuals, studies have shown that adipose tissue contains higher amounts of bacterial strains such as Proteobacteria and Actinobacteria (Massier et al., 2020). These changes in the composition of adipose tissue microbiota may have implications for metabolic health. Further analysis using 16s rRNA sequencing revealed specific bacterial families in the VAT of obese and lean individuals. The presence of Streptococcaceae and Ruminococaceae families, along with other unnamed genera, was observed in obese VAT, and Marvinobryantia and Bacilliales were found in lean VAT (Shantaram et al., 2022). It is worth noting that the presence of Streptococcaceae and Ruminococaceae in the VAT has been associated with inflammatory milieu and the recruitment of VAT-specific neutrophils, suggesting potential implications for adipose tissue inflammation. Studies have also reported the presence of bacteria like Mycobacterium tuberculosis, parasites such as Trypanosoma cruzi, Trypanosoma brucei, Plasmodium berghei, and pathogens like Rickettsia prowazekii, Coxiella burnetii, as well as viruses like human immunodeficiency virus (HIV) and simian immunodeficiency virus (SIV) (Neyrolles et al., 2006; Tanowitz et al., 2017). These findings highlight the potential role of adipose tissue as a niche for various microorganisms (Table 2). Moreover, Table 3 highlights the predominant microbial taxa and their functions in different health conditions, such as obesity, T2D, and obesity-associated T2D. However, the significance of the dormant state observed in certain microorganisms, allowing them to evade host defence mechanisms and drugs, in the context of obesity and related health conditions is currently unclear. In a recent report, it has been shown that in Crohn’s disease, gut bacteria translocated to mesenteric adipose tissue (Ha et al., 2020). This process contributes to the formation of “creeping fat” and obstructs the systemic translocation of gut bacteria. The expanded mesenteric adipose tissue in the sites of the gut barrier enriched with C. innocuum remodels the macrophage population toward the M2 phenotype, which helps prevent potentially harmful bacterial antigens that have translocated across the barrier from the gut lumen (Ha et al., 2020; Smith and Bénézech, 2020). These findings provide insights into the specific microbial compositions associated with different inflammatory conditions and their relationship with adipose tissue (Figure 1).
Overall, these studies shed light on the diverse microbial communities present in adipose tissue and their potential implications for obesity-induced inflammation and related health conditions. Further research is needed to fully understand the complex interactions between adipose tissue and the gutmicrobiota for their role in metabolic health and disease.
4 Gut-brain axis linked with obesity-induced type 2 diabetes
The gut-brain axis, commonly referred to as the feeding system, is a sophisticated feedback mechanism between the gut and the brain. It has long been known that the gut-brain axis is crucial for maintaining energy balance. The relationship of the gut microbiota with the enteric nervous system (ENS) and central nervous system (CNS) is beginning to be shown by more recent research.
4.1 Gut-brain axis in metabolic homeostasis
The gut-brain axis is a bidirectional network system of hormonal and neurological signalling cascades that are involved in neurologic, endocrine, immune, and metabolic pathways linking the ENS and CNS systems, and connecting the effect of gut microbiota on physiological health (Appleton, 2018). Signals from the gut in response to an influx of nutrients during a meal are traditionally conveyed to the brain, notifying the CNS about meal quantity and composition (Lam et al., 2009; Thorens, 2014; Fournel et al., 2017). The brain, especially the hypothalamus, integrates this information as well as other gut-derived signals to regulate the balance of dietary intake, consumption of energy, and glucose homeostasis (Peterhoff et al., 2003; Gautam et al., 2006). This postprandial gut feedback is mediated by entero-endocrine cells (EECs), which are specialized neuroendocrine cells of the intestinal epithelium. The EECs are found throughout the gut epithelium and respond to nutrient and mechanical stimuli by secreting hormones and neurotransmitters such as glucagon-like peptide 1 (GLP-1), gastric inhibitory polypeptide (GIP), cholecystokinin (CCK) (Gribble and Reimann, 2016), Peptide YY (Batterham et al., 2006), oxyntomodulin (OXM) (Schepp et al., 1996), ghrelin (Date et al., 2000), nesfatin (Stengel et al., 2009), serotonin (5-hydroxytryptamine), and Insulin-like peptide 5 (Insl5) (Grosse et al., 2014) which informs CNS particularly hypothalamus to coordinate and maintain metabolic homeostasis (Schwartz et al., 2000; Lam et al., 2009). These molecules critically influence the secretion of insulin, gastric acid, and bile acids, as well as gut motility and food intake via vagal afferent neurons of ENS or through the circulatory route (Ley et al., 2005; Cani et al., 2007; Duparc et al., 2011). Indeed, metabolites generated from the gut microbiota including SCFAs, butyrate, propionate, lactate, or mimetics such as γ-aminobutyric acid (GABA), and melanocyte-stimulating hormone (MSH)-mimetic, ClpB, can influence the release of these hormones and neurotransmitters (Newgard et al., 2009; Qin et al., 2012; Molinaro et al., 2020). Nutrient-induced gut peptides can function in a paracrine fashion by activating vagal neurons that innervate tissue surrounding the intestinal epithelium and signal to the brain, or in an endocrine fashion by targeting the brain and other peripheral organs involved in metabolic regulation (Köhler et al., 2003). Under normal conditions, intestinal glucose sensors, like SGLT1, and TASR1/2 initiate a signal to the afferent neurons for generating nitric oxide (NO) in the hypothalamus to allow glucose entry into the tissue by stimulating the autonomic nervous system (ANS) (Fournel et al., 2017). The intervention of ANS in pancreatic islets’ parasympathetic and sympathetic nerves regulates the islet hormone secretion (Peterhoff et al., 2003; Gautam et al., 2006; Thorens, 2014).
4.2 Gut microbial metabolites modulate gut-brain axis in type 2 diabetes
Gut microbiota maintains glucose homeostasis by directly communicating with the brain via microbe-derived metabolites such as SCFAs. The SCFAs activate G-protein coupled receptors (GPRs), FFAR2 (free fatty acid receptor 2), and FFAR3 (free fatty acid receptor 3) localized in EECs (Kasubuchi et al., 2015), resulting in gut peptide release. In healthy intestinal microbiota, the majority (approximately 90%) of known phylogenetic categories consist of Bacteroidetes and Firmicutes, including genera such as Ruminococcus, Lactobacillus, Clostridium, and to a lesser extent Actinobacteria, Verrucomicrobia, and Fusobacteria (Eckburg et al., 2005). These bacteria play important roles in maintaining intestinal health and normal physiological functions. However, in individuals with T2D, there are notable changes in the composition of the intestinal microbiota that includes opportunistic pathogenic bacteria such as Bacteroides caccae, Clostridium hathewayi, Clostridium symbiosum, Eggerthella lenta, Clostridium ramosum, and Escherichia coli are found to be more abundant (Qin et al., 2012). Additionally, mucin-degrading bacteria like Akkermansia muciniphila, sulfate-reducing bacteria like Desulfovibrio Sp., and imidazole propionate-producing bacteria such as Clostridium baumannii, Clostridium parasymbiotics, and Ruminococcus gnavus have also been linked to T2D (Molinaro et al., 2020). On the other hand, butyrate-producing bacteria such as Clostridiales, Faecalibacterium prausnitzii, Roseburia intestinalis, E. rectale, and Roseburia inulinivorans, which are important for gut health and energy metabolism, are significantly decreased in individuals with T2D (Qin et al., 2012). In the case of db/db mice, a commonly used mouse model for studying T2D pathophysiology, it has been observed that these mice experience intestinal inflammation, which can disrupt glucose metabolism (Duparc et al., 2011). This inflammation may be induced by an increase in NO production which impacted the disturbances in glucose metabolism contributing to the progression and severity of T2D in these mice. In addition, prebiotic-induced improvements in glucose homeostasis are contingent on GLP-1R signaling implying that prebiotic-induced microbiome changes repair the gut-brain axis (Jin et al., 2013). Increasing peptide production in prebiotic therapy enhances gut barrier integrity in the context of high-fat eating and obesity, which ultimately lowers the circulatory levels of LPS (Thorens, 2011). Recent research also identified small intestinal microbiota as a key mediator of the gut-brain axis in glucose homeostasis. Direct small intestinal infusion of Lactobacillus gasseri rescues intestinal lipid sensing, which is consistent with changes in the small intestinal microbiota regulating nutrient-induced gut-brain transmission (Biessels and Reagan, 2015; Omori et al., 2022). Lactobacillus gasseri expresses bile salt hydrolase and improvements in lipid sensing were dependent on the decreased farnesoid-X receptor (FXR) signalling, emphasizing the role of bile acids in gut-brain signalling processes that regulate metabolic homeostasis (Omori et al., 2022). Changes in the proportions of Gram-positive compared to Gram-negative bacteria in the intestinal lumen have a major impact on LPS bioavailability (Qin et al., 2012). The HFD-induced leaky gut helps to form an interaction of the bacterium with the intestinal epithelial cells by three successive pathways: a) attachment and invasion, b) alteration in the epithelial barrier, and c) inducing inflammation (Köhler et al., 2003). This allows tissue microbe-associated molecular patterns (MAMPs) and LPS to translocate from the intestinal lumen to the circulatory system (Delzenne and Cani, 2011). A crucial defence against gut-derived bacterial products getting into the bloodstream is provided by intestinal inflammatory cells (Steenbergen et al., 2015). It was discovered that microbiota-driven LPS, through its interactions with myeloid differentiation factor 2 (MD-2), TLR4, and CD14 contributes to the low-grade tissue inflammation in metabolic diseases associated with endotoxemia (Delzenne and Cani, 2011; Steenbergen et al., 2015). Similarly, microbiota-emanated peptidoglycans upon binding with NOD2 can modulate the intestinal inflammatory milieu thus influencing glucose tolerance and insulin sensitivity (Cani et al., 2008).
Upon activation of the pro-inflammatory cytokines by intestinal innate and adaptive immune cells, the activities of intrinsic and extrinsic enteric sensory neurons are altered (Kindt et al., 2010; Buckley et al., 2014). Intestinal cells send an abnormal nerve message that does not increase hypothalamic NO release and leads to dysfunctionality in ENS neurons (Gonzalez-Correa et al., 2017). The elevated level of TNF-α negatively impacts gut motility by upregulating the expression of cyclooxygenase 2 (COX-2) (Rehn et al., 2005) and enteric neuron apoptosis (Chandrasekharan et al., 2013). Additionally, IL-1β accelerates the diarrhoea phase by depolarizing the membrane potential, and decreasing the membrane conductance in obese/diabetic patients (Acosta and Camilleri, 2014). It also reported that a high accumulation of inflammatory markers induces the expression of serotonin and GABA, inhibiting brain functionality (Wang et al., 2012; Jin et al., 2013). Dysfunctionality in the gut-brain axis is driven by the interruption of the brain-islet axis and leads to islet abnormalities (Thorens, 2011), which leads to the failure of the insulin-regulated cognition and neuronal plasticity in the CNS (Biessels and Reagan, 2015). Moreover, it has been discovered that E. coli generates ClpB which regulates food intake (Breton et al., 2016). GF mice that lack any microbiome exhibit lower adiposity and higher insulin sensitivity despite greater food intake (Chandrasekharan et al., 2013), and they are protected against diet-induced obesity (Acosta and Camilleri, 2014). In comparison to ordinary mice, GF mice have altered gut-brain metabolic communication with alterations in EEC quantity as well as differences in intestinal nutrient-sensing machinery and nutrient-induced gut peptide release (Wang et al., 2012). Thus, ENS signalling deficiencies in GF mice lead to changes in the gut-brain axis that modulates energy balance exhibiting diminished activation of brainstem neurons.
Overall, these findings highlight the importance of a balanced and diverse intestinal microbiota in maintaining metabolic health. Alterations in the composition of the microbiota, characterized by changes in specific bacterial genera, can contribute to the development and progression of T2D and associated metabolic dysfunctions.
5 Gut microbiome-derived metabolites promote pancreatic β-cell dysfunction in obesity
Obesity-associated metabolic disturbances can impair the function of the pancreas, particularly in insulin-secreting β-cells and it has been unequivocally proven in prior studies that β-cell mass reduces before the development of T2D (Weir et al., 2020). The molecular reasons for β-cell depletion were thoroughly discussed in several research articles and reviewed elsewhere (Kahn, 2001; Prentki and Nolan, 2006; Cerf, 2013; Swisa et al., 2017). Although none of the direct evidence showed an existing pancreatic microbiome, there are a plethora of reports that prove gut microbiome-derived metabolites are responsible for β-cell dysfunction and insulin resistance. Healthy gut microbiota supports pancreatic cell expansion and maintenance of cell growth, as such early zebrafish pancreatic cell growth required certain gut microbiota such as some Aeromonas strains that can release cell expansion factor A (BefA), which encouraged proliferation and thereby cell growth (Hill et al., 2016; Zhou et al., 2022). More crucially, the specific bacterial species in humans could also release proteins that functioned similarly to BefA-like proteins leading to the development of novel T2D therapy strategies (Hill et al., 2016). Obesity exerts a multifaceted impact on the pancreas, as well as the composition and function of the gut microbiome. These alterations contribute to metabolic disturbances and insulin resistance, playing a significant role in the development and progression of obesity-related metabolic disorders. The β-cell death in T2D has been linked to many variables, including hyperglycemia, amyloid build-up, oxidative or endoplasmic reticulum stress, inflammatory cytokines, dysfunctional autophagy, and lipotoxicity (Lv et al., 2022).
5.1 Metabolites of gut microbiome affect β-cell function and insulin sensitivity
BCAAs such as leucine, isoleucine, and valine serve as important signalling molecules in the body, exerting both direct and indirect effects on the insulin signalling pathway. While BCAAs have shown potential anti-obesity effects in rodent models, it is noteworthy that individuals with obesity often exhibit elevated levels of circulating BCAAs. One proposed mechanism linking elevated BCAA levels and T2D involves the activation of the mTORC1by leucine, one of the BCAAs. Activation of mTORC1 by leucine can lead to the uncoupling of insulin signalling at an early stage, potentially contributing to insulin resistance. On the contrary, the activation of mTORC1 in β-cells has been associated with a protective effect against the development of T2D. This activation is linked to compensatory increases in islet and β-cell mass, which help to prevent the onset of T2D. An alternative model, known as the BCAA dysmetabolism model, suggests that it is not the BCAAs themselves but the accumulation of metabolites derived from BCAA metabolism that contributes to β-cell mitochondrial dysfunction, stress signalling with mitotoxic effects, and apoptosis associated with T2D (Lynch and Adams, 2014).
L-tryptophan (Trp), an exogenous essential amino acid acts as a metabolic precursor, for melatonin (Leja-Szpak et al., 2004) and serotonin (Almaça et al., 2016). Melatonin rescues the pancreatic cells against acute pancreatic damage by oxidative stress (Leja-Szpak et al., 2004). Almaca et al. have revealed that serotonin acts as a paracrine signal released by pancreatic β-cells to regulate glucagon secretion. The study demonstrated that without serotonin signalling, α-cells fail to respond appropriately to glucose level alteration (Almaça et al., 2016). Tryptophan metabolism involves three main pathways: the kynurenine pathway through indoleamine 2,3-dioxygenase 1 (IDO1), the serotonin production pathway via Trp hydroxylase 1 (TpH1), and the direct transformation of Trp into various molecules by the gut microbiota, including ligands of the aryl hydrocarbon receptor (AhR). The kynurenine pathway is the predominant route of tryptophan breakdown in most mammalian cells. Within this pathway, kynurenines can inhibit proinsulin synthesis in pancreatic islets and form complexes with insulin, reducing its biological activity and promoting the development of insulin resistance (Song et al., 2017; Vangipurapu et al., 2020).
The gut microbiome produces various metabolites, such as SCFAs and bile acids which can influence pancreatic function and insulin secretion (Kaneto et al., 2022). Pancreatitis is associated with alteration in the composition of the gut microbiota, particularly at the phylum level, characterized by an increase in Proteobacteria and a decrease in strains that produce SCFAs. A study by Yu et al. revealed that Eubacterium hallii, a prominent bacterium responsible for producing butyrate, was significantly depleted in pancreatitis patients and the loss of butyrate-producing bacteria is attributed to increased oxidative stress in the pancreas (Yu et al., 2020). Moreover, butyrate has been shown to ameliorate pancreatitis by suppressing the activation of NF-κB and decreasing HMGB1 expression (Pan et al., 2019). The SCFAs such as propionate have been associated with insulin secretion and sensitivity. It has been shown that SCFAs stimulate the production of GLP-1 through the activation of FFAR2, also known as GPR43 (G-protein coupled receptor 43), regulate glucose-dependent insulin secretion from pancreatic β-cells, and inhibit glucagon secretion. This mechanism contributes to the maintenance of glucose homeostasis (Silva et al., 2020).
The gut-derived metabolite trimethylamine N-oxide (TMAO) has been linked to obesity, and elevated levels of TMAO have been found to positively correlate with the presence of T2D. Krueger et al. showed the effects of TMAO on insulin secretion by β cells were investigated, revealing that TMAO exposure did not contribute to the development of T2D but instead exhibited beneficial effects (Krueger et al., 2021). Interestingly, TMAO exposure demonstrated a protective effect against glucolipotoxicity (GLT)-induced damage to β-cells by reducing oxidative stress and preserving insulin granule formation, which suggests that TMAO promotes the preservation of functional β-cell mass, thus counteracting T2D-promoting mechanisms (Krueger et al., 2021).
5.2 Oxidative stress and inflammation in pancreatic β-cell associated with microbial components
Chronic hyperglycemia, a characteristic feature of T2D, is linked to the rapid formation of advanced glycation end products (AGEs) and glucose autoxidation (Robertson et al., 2003). These processes further lead to the production of reactive oxygen species (ROS) and oxidative stress. In β-cells, superoxide (O2-), hydroxyl (OH) radicals, hydrogen peroxide (H2O2), and other ROS molecules are among the many that are being intensively investigated for their harmful impact in exacerbating diabetes-related problems (Kulkarni et al., 2023). Reactive oxygen species (ROS) and reactive nitrogen species (RNS) overproduction or improper clearance lead to oxidative stress, which may have a variety of harmful consequences on cellular metabolism (Hasnain et al., 2016). ER stress during protein misfolding results in unfolded protein response (UPR) may impair insulin transcription and translation and trigger inflammation and death. The SAPKs, p38, and JNK are examples of stress-sensing pathways that are activated by oxidative stress and cause damage to cells (Houet al., 2008). Reports have indicated that malfunctioning pancreatic islets exhibit relatively low levels of antioxidant enzymes such as copper/zinc superoxide dismutase (Cu/Zn-SOD), manganese superoxide dismutase (Mn-SOD), catalase, and glutathione peroxidase (GPx) (Robertson et al., 2003; Stancill et al., 2019). As a result, they become more vulnerable to oxidative damage, eventually resulting in the death of β-cells and it is clearly showing that there is a clear connection between oxidative stress, mitochondrial dysfunction, and T2D (Rocha et al., 2020).
Obesity and T2D also feature higher release of free fatty acids (FFAs), which modulatesignalling pathways related to glucose metabolism, and β-cell function. Elevated FFAs inhibit glucose-stimulated insulin secretion and lead to β-cell dysfunction through cytotoxic mechanisms, including apoptosis. FFAs exposure is associated with ceramide synthesis, mitochondrial dysfunction, overexpression of apoptotic genes, and intracellular triglyceride accumulation in β-cells mediated by sterol regulatory element-binding proteins (SREBPs) (Martínez-Montoro et al., 2022). Visceral adipose tissue releases pro-inflammatory factors like IL-2, IL-6, IL-8, IL-12A, and MCP-1 in obese conditions, which can contribute to β-cell dysfunction. Macrophages present in adipose tissue are the early responders and play a crucial role in promoting the pro-inflammatory environment that negatively impacts pancreatic β-cells (Daniele et al., 2014). Vangipurapu et al. reported an increase in sphingolipids (e.g., myoinositol) and fatty acids from early analysis of pathway enrichment and plasma metabolite analysis of chronic pancreatitis patient samples (Vangipurapu et al., 2020). Gao et al. revealed that the accumulation of microbial DNAs contributes to inflammation and abnormalities in pancreatic islet β-cells through their observations of passing the extracellular vesicles containing microbial DNA from the gut to β-cells (Gao et al., 2022). Bacterial LPS trigger inflammatory responses and impact pancreatic β-cell function (Wu et al., 2023). These metabolites can directly affect pancreatic function and also influence systemic inflammation, gut barrier integrity, and other metabolic pathways. Individual variations in gut microbial composition and metabolism further contribute to the complexity of this relationship (Figure 3). Further research is needed to fully understand the mechanisms by which microbial metabolites contribute to pancreas dysfunctions in T2D and explore potential therapeutic targets.
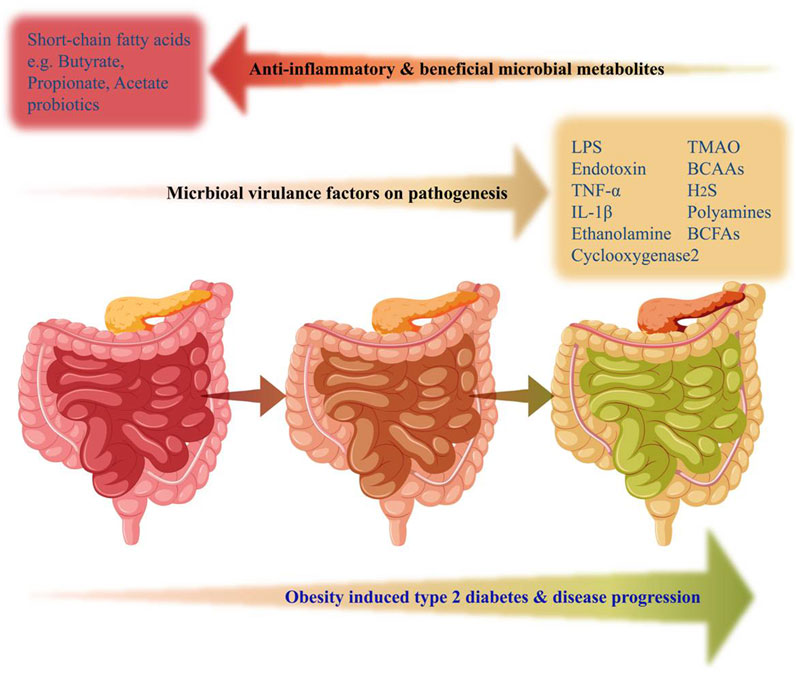
FIGURE 3. Gut microbial-derived factors alteration with the progression of obesity and type 2 diabetes. Increased production of several metabolites associated with gut dysbiosis such as LPS, TNFα, TMAO, H2S, BCAAs, BCFAs, and Polyamines were linked with the progression of obesity-induced T2D. On the contrary, beneficial microbiome-derived metabolites from the healthy gut, responsible for anti-inflammatory effects and physiological homeostasis, such as Propionate, Acetate, and Butyrate (short-chain fatty acids) were reduced in obesity-induced gut dysbiosis. The gut represented in pink color indicates a healthy state, whereas, brown color and green color indicate obesity-induced pathophysiological progression to T2D state.
6 Therapeutic approaches to manage type 2 diabetes by targeting gut microbiota
Therapeutics classes including probiotics, prebiotics, FMT, and microbial-derived molecules, offer potential strategies for controlling insulin secretion and insulin resistance (Zhang et al., 2019). Probiotics, such as Lactobacillus spp. and Bifidobacterium spp., have shown promise in improving glycemic control and insulin sensitivity (Salles et al., 2020). Prebiotics, as dietary fibers, can selectively promote the growth of beneficial gut bacteria and improve glucose metabolism (Holscher, 2017). Moreover, FMT is being explored for T2D management. Microbial-derived molecules like SCFAs have implications for metabolic processes. However, further research is needed to optimize these approaches and personalize interventions for effective microbiome-based therapies in T2D. Probiotics, live microorganisms that confer health benefits to the host, have been shown to improve insulin sensitivity and glucose tolerance in both animal and human studies. Several studies have reported that probiotics can lead to improvements in markers of inflammation, glycemic control, and blood pressure. Probiotic supplementation has been shown to reduce c-reactive protein (CRP) levels, indicating a potential anti-inflammatory effect, along with the improvement of HbA1c, fasting plasma glucose, and fasting insulin levels (Wang et al., 2021). Consumption of probiotics has been associated with a decrease in serum cholesterol levels and reduced cholesterol absorption in the intestines which inhibits the activity of the HMG-CoA reductase, an enzyme critically involved in endogenous cholesterol synthesis (Kumar et al., 2012).
The biological effects of Lactobacillus spp. and Bifidobacterium spp., on glucose intolerance and insulin resistance were majorly studied in animal models of T2D. Lactobacillus plantarum CCFM0236 has shown beneficial effects in improving insulin resistance, reducing systemic inflammation, and ameliorating pancreatic β-cell dysfunction in HFD-induced T2D animal models. This probiotic strain reduced insulin resistance and preserved pancreatic β-cell function, leading to better glycemic control (Li et al., 2016). Clinical trial studies revealed Lactobacillus reuteri DSM 17938, Lactobacillus case 431, Lactobacillus acidophilus, Bifidobacterium lactis, Bifidobacterium bifidum, Lactobacillus salivarius W24 improves insulin sensitivity index, reduces HbA1c and HOMA-IR. Moreover, strains of Lactobacillus, Lactococcus, Bifidobacterium, Propionibacterium, and Acetobacter genera supplementation significantly reduced pro-inflammatory cytokines including TNF-α, IL-1β and improved physiological glycemic control by lowering HOMA-IR and HbA1c (Zhai et al., 2021).
Combined supplementations of probiotics and prebiotics have also been shown to improve glucose metabolism and reduce insulin resistance. Prebiotics normalizes the GI tract’s pH value, reduce hyperlipidemia, and improve the absorption of cation ions, creating an environment favourable for the growth of beneficial bacteria (Megur et al., 2022). In addition, FMT has shown promise in improving glycemic control in patients with T2D. The recipients of FMT showed a significant improvement in insulin sensitivity, indicating a positive effect on glucose metabolism. This finding was further supported by a larger-scale follow-up study where the recipients exhibited a reduction in HbA1c levels at 6 weeks post-intervention (Kootte et al., 2017). However, the insulin sensitivity of the recipients and the composition of their gut microbiota returned to baseline after 18 weeks post-intervention (Kootte et al., 2017). This suggests that the effects of FMT on insulin sensitivity and gut microbiota composition may be transient and not sustained in the long term.
7 Summary
The altered gut microbial community leads to disruptions in metabolic homeostasis causally linked with the imbalance of lipid homeostasis and chronic inflammation which is associated with obesity and T2D. Remodelling of microbial profiles and microbiome-derived metabolites contributes to the development of insulin resistance by modulating the gut-brain axis, loss of pancreatic functionality, and white adipose tissue dysfunction. Additionally, specific microbial genera were found in the visceral adipose tissue that influences its normal function during obesity. Moreover, pathogenic microbes such as Escherichia and Shigella were abundantly present in obese diabetic patients. The applications of prebiotics, probiotics, or faecal microbial transplantation are considered contemporary approaches that can be offered to manage T2D. Here we consolidate the current knowledge in this field and underscore the urgent need for further research and clinical interventions to combat the rising epidemic of obesity-induced metabolic disorders.
8 Future perspective
In recent years, the study of gut and adipose tissue dysbiosis in obese individuals with T2D has attracted significant attention, and researchers are exploring several therapeutic approaches in this field. One potential way is the development of personalized microbiome-based therapies. Another important aspect is gaining a mechanistic understanding of how gut dysbiosis contributes to obesity and T2D for developing targeted therapeutics. It is needed to understand the potential implications and mechanisms underlying the relationship between microbial dormancy and obesity. This review raises a few fundamental questions to understand the basics of obesity-induced pathogenesis. Such as, how microbial compartmentalization occurs within the visceral adipose tissue (VAT) during obesity and how the obese adipose tissue microenvironment supports microbial survival. It also demands the ongoing search for the source and origin of tissue-specific microbes. Moreover, what are the factors that govern gut and adipose tissue dysbiosis in obesity and actively control microbial localization in insulin-responsive organs such as the liver and adipose tissue? Therefore, further research is needed to fully comprehend the gut-adipose tissue axis in obesity and its role in the progression of insulin resistance and T2D, as our understanding of this subject is still in its nascent stages.
Author contributions
DeP: Hypotheses proposal, Literature search, Writing, Review, Editing; DB: Literature search, Writing; PR: Literature search, Writing, and Table preparation; SR: Literature search, Writing, and Figure preparation; SD: Hypotheses proposal, Writing, Review, Editing, Supervision; DuP: Hypotheses proposal, Writing, Review, Editing, Supervision. All authors contributed to the article and approved the submitted version.
Acknowledgments
DeP, PR, and SR express their gratitude to the IIT Ropar and MoE, Govt. of India for the award of their research fellowships. SD and DuP gratefully acknowledge Tezpur University and IIT Ropar, respectively, for administrative support. We are also thankful to the Frontiers in Molecular Biosciences for the invitation to publish our manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Acosta, A., and Camilleri, M. (2014). Gastrointestinal morbidity in obesity. Ann. N. Y. Acad. Sci. 1311, 42–56. doi:10.1111/nyas.12385
Agus, A., Clément, K., and Sokol, H. (2021). Gut microbiota-derived metabolites as central regulators in metabolic disorders. Gut 70, 1174–1182. doi:10.1136/gutjnl-2020-323071
Ahmad, A., Yang, W., Chen, G., Shafiq, M., Javed, S., Ali Zaidi, S. S., et al. (2020). Analysis of gut microbiota of obese individuals with type 2 diabetes and healthy individuals. PLOS ONE 14, e0226372. doi:10.1371/journal.pone.0226372
Almaça, J., Molina, J., Menegaz, D., Pronin, A. N., Tamayo, A., Slepak, V., et al. (2016). Human beta cells produce and release serotonin to inhibit glucagon secretion from alpha cells. Cell Rep. 17, 3281–3291. doi:10.1016/j.celrep.2016.11.072
Anhê, F. F., Jensen, B. A. H., Varin, T. V., Servant, F., Van Blerk, S., Richard, D., et al. (2020). Type 2 diabetes influences bacterial tissue compartmentalisation in human obesity. Nat. Metab. 2, 233–242. doi:10.1038/s42255-020-0178-9
Appleton, J. (2018). The gut-brain Axis: influence of microbiota on mood and mental health. Integr. Med. (Encinitas) 17, 28–32.
Aranaz, P., Ramos-Lopez, O., Cuevas-Sierra, A., Martinez, J. A., Milagro, F. I., and Riezu-Boj, J. I. (2021). A predictive regression model of the obesity-related inflammatory status based on gut microbiota composition. Int. J. Obes. 45, 2261–2268. doi:10.1038/s41366-021-00904-4
Atkinson, R. L., Dhurandhar, N. V., Allison, D. B., Bowen, R. L., Israel, B. A., Albu, J. B., et al. (2005). Human adenovirus-36 is associated with increased body weight and paradoxical reduction of serum lipids. Int. J. Obes. 29, 281–286. doi:10.1038/sj.ijo.0802830
Bäckhed, F., Ding, H., Wang, T., Hooper, L. V., Koh, G. Y., Nagy, A., et al. (2004). The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl. Acad. Sci. U. S. A. 101, 15718–15723. doi:10.1073/pnas.0407076101
Bäckhed, F., Ley, R. E., Sonnenburg, J. L., Peterson, D. A., and Gordon, J. I. (2005). Host-bacterial mutualism in the human intestine. Science 307, 1915–1920. doi:10.1126/science.1104816
Bäckhed, F., Manchester, J. K., Semenkovich, C. F., and Gordon, J. I. (2007). Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc. Natl. Acad. Sci. U. S. A. 104, 979–984. doi:10.1073/pnas.0605374104
Batterham, R. L., Heffron, H., Kapoor, S., Chivers, J. E., Chandarana, K., Herzog, H., et al. (2006). Critical role for peptide YY in protein-mediated satiation and body-weight regulation. Cell Metab. 4, 223–233. doi:10.1016/j.cmet.2006.08.001
Biessels, G. J., and Reagan, L. P. (2015). Hippocampal insulin resistance and cognitive dysfunction. Nat. Rev. Neurosci. 16, 660–671. doi:10.1038/nrn4019
Blustein, J., Attina, T., Liu, M., Ryan, A. M., Cox, L. M., Blaser, M. J., et al. (2013). Association of caesarean delivery with child adiposity from age 6 weeks to 15 years. Int. J. Obes. 37, 900–906. doi:10.1038/ijo.2013.49
Boulangé, C. L., Neves, A. L., Chilloux, J., Nicholson, J. K., and Dumas, M.-E. (2016). Impact of the gut microbiota on inflammation, obesity, and metabolic disease. Genome Med. 8, 42. doi:10.1186/s13073-016-0303-2
Breton, J., Galmiche, M., and Déchelotte, P. (2022). Dysbiotic gut bacteria in obesity: an overview of the metabolic mechanisms and therapeutic perspectives of next-generation probiotics. Microorganisms 10, 452. doi:10.3390/microorganisms10020452
Breton, J., Tennoune, N., Lucas, N., Francois, M., Legrand, R., Jacquemot, J., et al. (2016). Gut commensal E. Coli proteins activate host satiety pathways following nutrient-induced bacterial growth. Cell Metab. 23, 324–334. doi:10.1016/j.cmet.2015.10.017
Buckley, M. M., O'halloran, K. D., Rae, M. G., Dinan, T. G., and O'malley, D. (2014). Modulation of enteric neurons by interleukin-6 and corticotropin-releasing factor contributes to visceral hypersensitivity and altered colonic motility in a rat model of irritable bowel syndrome. J. Physiol. 592, 5235–5250. doi:10.1113/jphysiol.2014.279968
Bui, T. I., Gill, A. L., Mooney, R. A., and Gill, S. R. (2022). Modulation of gut microbiota metabolism in obesity-related type 2 diabetes reduces osteomyelitis severity. Microbiol. Spectr. 10, 00170222–e100122. doi:10.1128/spectrum.00170-22
Burcelin, R., Serino, M., Chabo, C., Garidou, L., Pomié, C., Courtney, M., et al. (2013). Metagenome and metabolism: the tissue microbiota hypothesis. Diabetes Obes. Metab. 15, 61–70. doi:10.1111/dom.12157
Burz, S. D., Monnoye, M., Philippe, C., Farin, W., Ratziu, V., Strozzi, F., et al. (2021). Fecal microbiota transplant from human to mice gives insights into the role of the gut microbiota in non-alcoholic fatty liver disease (NAFLD). Microorganisms 9, 199. doi:10.3390/microorganisms9010199
Canfora, E. E., Hermes, G. D. A., Müller, M., Bastings, J., Vaughan, E. E., Van Den Berg, M. A., et al. (2022). Fiber mixture-specific effect on distal colonic fermentation and metabolic health in lean but not in prediabetic men. Gut Microbes 14, 2009297. doi:10.1080/19490976.2021.2009297
Canfora, E. E., Meex, R. C. R., Venema, K., and Blaak, E. E. (2019). Gut microbial metabolites in obesity, NAFLD and T2DM. Nat. Rev. Endocrinol. 15, 261–273. doi:10.1038/s41574-019-0156-z
Cani, P. D., Bibiloni, R., Knauf, C., Waget, A., Neyrinck, A. M., Delzenne, N. M., et al. (2008). Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes 57, 1470–1481. doi:10.2337/db07-1403
Cani, P. D., Amar, J., Iglesias, M. A., Poggi, M., Knauf, C., Bastelica, D., et al. (2007). Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes 56, 1761–1772. doi:10.2337/db06-1491
Cani, P. D., Van Hul, M., Lefort, C., Depommier, C., Rastelli, M., and Everard, A. (2019). Microbial regulation of organismal energy homeostasis. Nat. Metab. 1, 34–46. doi:10.1038/s42255-018-0017-4
Cerf, M. E. (2013). Beta cell dysfunction and insulin resistance. Front. Endocrinol. 4, 37. doi:10.3389/fendo.2013.00037
Chan, K. L., Tam, T. H., Boroumand, P., Prescott, D., Costford, S. R., Escalante, N. K., et al. (2017). Circulating NOD1 activators and hematopoietic NOD1 contribute to metabolic inflammation and insulin resistance. Cell Rep. 18, 2415–2426. doi:10.1016/j.celrep.2017.02.027
Chandrasekharan, B., Jeppsson, S., Pienkowski, S., Belsham, D. D., Sitaraman, S. V., Merlin, D., et al. (2013). Tumor necrosis factor-neuropeptide Y cross talk regulates inflammation, epithelial barrier functions, and colonic motility. Inflamm. Bowel Dis. 19, 2535–2546. doi:10.1097/01.MIB.0000437042.59208.9f
Chen, P.-C., Chien, Y.-W., and Yang, S.-C. (2019). The alteration of gut microbiota in newly diagnosed type 2 diabetic patients. Nutrition 63-64, 51–56. doi:10.1016/j.nut.2018.11.019
Cornejo-Pareja, I., Muñoz-Garach, A., Clemente-Postigo, M., and Tinahones, F. J. (2019). Importance of gut microbiota in obesity. Eur. J. Clin. Nutr. 72, 26–37. doi:10.1038/s41430-018-0306-8
Cox, L. M., and Blaser, M. J. (2013). Pathways in microbe-induced obesity. Cell Metab. 17, 883–894. doi:10.1016/j.cmet.2013.05.004
Daniele, G., Guardado Mendoza, R., Winnier, D., Fiorentino, T. V., Pengou, Z., Cornell, J., et al. (2014). The inflammatory status score including IL-6, TNF-α, osteopontin, fractalkine, MCP-1 and adiponectin underlies whole-body insulin resistance and hyperglycemia in type 2 diabetes mellitus. Acta Diabetol. 51, 123–131. doi:10.1007/s00592-013-0543-1
Date, Y., Kojima, M., Hosoda, H., Sawaguchi, A., Mondal, M. S., Suganuma, T., et al. (2000). Ghrelin, a novel growth hormone-releasing acylated peptide, is synthesized in a distinct endocrine cell type in the gastrointestinal tracts of rats and humans. Endocrinology 141, 4255–4261. doi:10.1210/endo.141.11.7757
David, L. A., Maurice, C. F., Carmody, R. N., Gootenberg, D. B., Button, J. E., Wolfe, B. E., et al. (2014). Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559–563. doi:10.1038/nature12820
Davis, C. D. (2016). The gut microbiome and its role in obesity. Nutr. Today 51, 167–174. doi:10.1097/NT.0000000000000167
De Groot, P., Scheithauer, T., Bakker, G. J., Prodan, A., Levin, E., Khan, M. T., et al. (2020). Donor metabolic characteristics drive effects of faecal microbiota transplantation on recipient insulin sensitivity, energy expenditure and intestinal transit time. Gut 69, 502–512. doi:10.1136/gutjnl-2019-318320
De La Cuesta-Zuluaga, J., Corrales-Agudelo, V., Velásquez-Mejía, E. P., Carmona, J. A., Abad, J. M., and Escobar, J. S. (2018). Gut microbiota is associated with obesity and cardiometabolic disease in a population in the midst of Westernization. Sci. Rep. 8, 11356. doi:10.1038/s41598-018-29687-x
Del Chierico, F., Manco, M., Gardini, S., Guarrasi, V., Russo, A., Bianchi, M., et al. (2021). Fecal microbiota signatures of insulin resistance, inflammation, and metabolic syndrome in youth with obesity: a pilot study. Acta Diabetol. 58, 1009–1022. doi:10.1007/s00592-020-01669-4
Delzenne, N. M., and Cani, P. D. (2011). Interaction between obesity and the gut microbiota: relevance in nutrition. Annu. Rev. Nutr. 31, 15–31. doi:10.1146/annurev-nutr-072610-145146
Denou, E., Lolmède, K., Garidou, L., Pomie, C., Chabo, C., Lau, T. C., et al. (2015). Defective NOD2 peptidoglycan sensing promotes diet-induced inflammation, dysbiosis, and insulin resistance. EMBO Mol. Med. 7, 259–274. doi:10.15252/emmm.201404169
Depommier, C., Everard, A., Druart, C., Plovier, H., Van Hul, M., Vieira-Silva, S., et al. (2019). Supplementation with Akkermansiamuciniphila in overweight and obese human volunteers: a proof-of-concept exploratory study. Nat. Med. 25, 1096–1103. doi:10.1038/s41591-019-0495-2
Dhurandhar, N. V., Kulkarni, P. R., Ajinkya, S. M., Sherikar, A. A., and Atkinson, R. L. (1997). Association of adenovirus infection with human obesity. Obes. Res. 5, 464–469. doi:10.1002/j.1550-8528.1997.tb00672.x
Diamant, M., Blaak, E. E., and De Vos, W. M. (2011). Do nutrient–gut–microbiota interactions play a role in human obesity, insulin resistance and type 2 diabetes? Obes. Rev. 12, 272–281. doi:10.1111/j.1467-789X.2010.00797.x
Duparc, T., Naslain, D., Colom, A., Muccioli, G. G., Massaly, N., Delzenne, N. M., et al. (2011). Jejunum inflammation in obese and diabetic mice impairs enteric glucose detection and modifies nitric oxide release in the hypothalamus. Antioxid. Redox Signal 14, 415–423. doi:10.1089/ars.2010.3330
Eckburg, P. B., Bik, E. M., Bernstein, C. N., Purdom, E., Dethlefsen, L., Sargent, M., et al. (2005). Diversity of the human intestinal microbial flora. Science 308, 1635–1638. doi:10.1126/science.1110591
Ejtahed, H.-S., Hoseini-Tavassol, Z., Khatami, S., Zangeneh, M., Behrouzi, A., Ahmadi Badi, S., et al. (2020). Main gut bacterial composition differs between patients with type 1 and type 2 diabetes and non-diabetic adults. J. Diabetes Metab. Disord. 19, 265–271. doi:10.1007/s40200-020-00502-7
Everard, A., Lazarevic, V., Gaïa, N., Johansson, M., Ståhlman, M., Backhed, F., et al. (2014). Microbiome of prebiotic-treated mice reveals novel targets involved in host response during obesity. ISME J. 8, 2116–2130. doi:10.1038/ismej.2014.45
Fournel, A., Drougard, A., Duparc, T., Marlin, A., Brierley, S. M., Castro, J., et al. (2017). Apelin targets gut contraction to control glucose metabolism via the brain. Gut 66, 258–269. doi:10.1136/gutjnl-2015-310230
Foley, K. P., Zlitni, S., Denou, E., Duggan, B. M., Chan, R. W., Stearns, J. C., et al. (2018). Long term but not short term exposure to obesity related microbiota promotes host insulin resistance. Nat. Commun. 9, 4681. doi:10.1038/s41467-018-07146-5
Forslund, K., Hildebrand, F., Nielsen, T., Falony, G., Le Chatelier, E., Sunagawa, S., et al. (2015). Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature 528, 262–266. doi:10.1038/nature15766
Foster, J. A., and McVey Neufeld, K.-A. (2013). Gut–brain axis: how the microbiome influences anxiety and depression. Trends Neurosci. 36, 305–312. doi:10.1016/j.tins.2013.01.005
Fujisaka, S., Usui, I., Nawaz, A., Igarashi, Y., Okabe, K., Furusawa, Y., et al. (2020). Bofutsushosan improves gut barrier function with a bloom of Akkermansia muciniphila and improves glucose metabolism in mice with diet-induced obesity. Sci. Rep. 10, 5544. doi:10.1038/s41598-020-62506-w
Furet, J. P., Kong, L. C., Tap, J., Poitou, C., Basdevant, A., Bouillot, J. L., et al. (2010). Differential adaptation of human gut microbiota to bariatric surgery-induced weight loss: links with metabolic and low-grade inflammation markers. Diabetes 59, 3049–3057. doi:10.2337/db10-0253
Gao, H., Luo, Z., Ji, Y., Tang, K., Jin, Z., Ly, C., et al. (2022). Accumulation of microbial DNAs promotes to islet inflammation and β cell abnormalities in obesity in mice. Nat. Commun. 13, 565. doi:10.1038/s41467-022-28239-2
Gautam, D., Han, S. J., Hamdan, F. F., Jeon, J., Li, B., Li, J. H., et al. (2006). A critical role for beta cell M3 muscarinic acetylcholine receptors in regulating insulin release and blood glucose homeostasis in vivo. Cell Metab. 3, 449–461. doi:10.1016/j.cmet.2006.04.009
Ghanim, H., Abuaysheh, S., Sia, C. L., Korzeniewski, K., Chaudhuri, A., Fernandez-Real, J. M., et al. (2009). Increase in plasma endotoxin concentrations and the expression of toll-like receptors and suppressor of cytokine signaling-3 in mononuclear cells after a high-fat, high-carbohydrate meal: implications for insulin resistance. Diabetes Care 32, 2281–2287. doi:10.2337/dc09-0979
Gonzalez-Correa, C. A., Mulett-Vásquez, E., Miranda, D. A., Gonzalez-Correa, C. H., and Gómez-Buitrago, P. A. (2017). The colon revisited or the key to wellness, health and disease. Med. Hypotheses 108, 133–143. doi:10.1016/j.mehy.2017.07.032
Gophna, U., Konikoff, T., and Nielsen, H. B. (2017). Oscillospira and related bacteria - From metagenomic species to metabolic features. Environ. Microbiol. 19, 835–841. doi:10.1111/1462-2920.13658
Gribble, F. M., and Reimann, F. (2016). Enteroendocrine cells: chemosensors in the intestinal epithelium. Annu. Rev. Physiol. 78, 277–299. doi:10.1146/annurev-physiol-021115-105439
Grosse, J., Heffron, H., Burling, K., Akhter Hossain, M., Habib, A. M., Rogers, G. J., et al. (2014). Insulin-like peptide 5 is an orexigenic gastrointestinal hormone. Proc. Natl. Acad. Sci. U. S. A. 111, 11133–11138. doi:10.1073/pnas.1411413111
Guo, Y., Huang, Z., Sang, D., Gao, Q., and Li, Q. (2020). The role of nutrition in the prevention and intervention of type 2 diabetes. Front. Bioeng. Biotechnol. 8, 575442. doi:10.3389/fbioe.2020.575442
Gurung, M., Li, Z., You, H., Rodrigues, R., Jump, D. B., Morgun, A., et al. (2020). Role of gut microbiota in type 2 diabetes pathophysiology. eBioMedicine 51, 102590. doi:10.1016/j.ebiom.2019.11.051
Ha, C. W. Y., Martin, A., Sepich-Poore, G. D., Shi, B., Wang, Y., Gouin, K., et al. (2020). Translocation of viable gut microbiota to mesenteric adipose drives formation of creeping fat in humans. Cell 183, 666–683. doi:10.1016/j.cell.2020.09.009
Haneishi, Y., Furuya, Y., Hasegawa, M., Takemae, H., Tanioka, Y., Mizutani, T., et al. (2023). Polyunsaturated fatty acids-rich dietary lipid prevents high fat diet-induced obesity in mice. Sci. Rep. 13, 5556. doi:10.1038/s41598-023-32851-7
Hasnain, S. Z., Prins, J. B., and Mcguckin, M. A. (2016). Oxidative and endoplasmic reticulum stress in β-cell dysfunction in diabetes. J. Mol. Endocrinol. 56, R33–R54. doi:10.1530/JME-15-0232
Heeney, D. D., Zhai, Z., Bendiks, Z., Barouei, J., Martinic, A., Slupsky, C., et al. (2019). Lactobacillus plantarum bacteriocin is associated with intestinal and systemic improvements in diet-induced obese mice and maintains epithelial barrier integrity in vitro. Gut Microbes 10, 382–397. doi:10.1080/19490976.2018.1534513
Hermes, G. D. A., Reijnders, D., Kootte, R. S., Goossens, G. H., Smidt, H., Nieuwdorp, M., et al. (2020). Individual and cohort-specific gut microbiota patterns associated with tissue-specific insulin sensitivity in overweight and obese males. Sci. Rep. 10, 7523. doi:10.1038/s41598-020-64574-4
Hersoug, L.-G., Møller, P., and Loft, S. (2018). Role of microbiota-derived lipopolysaccharide in adipose tissue inflammation, adipocyte size and pyroptosis during obesity. Nutr. Res. Rev. 31, 153–163. doi:10.1017/S0954422417000269
Hill, J. H., Franzosa, E. A., Huttenhower, C., and Guillemin, K. (2016). A conserved bacterial protein induces pancreatic beta cell expansion during zebrafish development. eLife 5, e20145. doi:10.7554/eLife.20145
Hill, J. O., Wyatt, H. R., and Peters, J. C. (2012). Energy balance and obesity. Circulation 126, 126–132. doi:10.1161/CIRCULATIONAHA.111.087213
Holscher, H. D. (2017). Dietary fiber and prebiotics and the gastrointestinal microbiota. Gut Microbes 8, 172–184. doi:10.1080/19490976.2017.1290756
Hong, Y. H., Nishimura, Y., Hishikawa, D., Tsuzuki, H., Miyahara, H., Gotoh, C., et al. (2005). Acetate and propionate short chain fatty acids stimulate adipogenesis via GPCR43. Endocrinology 146, 5092–5099. doi:10.1210/en.2005-0545
Hosomi, K., Saito, M., Park, J., Murakami, H., Shibata, N., Ando, M., et al. (2022). Oral administration of Blautiawexlerae ameliorates obesity and type 2 diabetes via metabolic remodeling of the gut microbiota. Nat. Commun. 13, 4477. doi:10.1038/s41467-022-32015-7
Hou, K., Wu, Z. X., Chen, X. Y., Wang, J. Q., Zhang, D., Xiao, C., et al. (2022). Microbiota in health and diseases. Signal Transduct. Target Ther. 7, 135. doi:10.1038/s41392-022-00974-4
Hou, N., Torii, S., Saito, N., Hosaka, M., and Takeuchi, T. (2008). Reactive oxygen species-mediated pancreatic β-cell death is regulated by interactions between Stress-Activated Protein Kinases, p38 and c-Jun N-Terminal Kinase, and Mitogen-Activated Protein Kinase Phosphatases. Endocrinology 149, 1654–1665. doi:10.1210/en.2007-0988
Huda, M. N., Kim, M., and Bennett, B. J. (2021). Modulating the microbiota as a therapeutic intervention for type 2 diabetes. Front. Endocrinol. 12, 632335. doi:10.3389/fendo.2021.632335
Jackman, J. A., Yoon, B. K., Li, D., and Cho, N. J. (2016). Nanotechnology formulations for antibacterial free fatty acids and monoglycerides. Molecules 21, 305. doi:10.3390/molecules21030305
Jia, X., Xu, W., Zhang, L., Li, X., Wang, R., and Wu, S. (2021). Impact of gut microbiota and microbiota-related metabolites on hyperlipidemia. Front. Cell Infect. Microbiol. 11, 634780. doi:10.3389/fcimb.2021.634780
Jin, Z., Mendu, S. K., and Birnir, B. (2013). GABA is an effective immunomodulatory molecule. Amino Acids 45, 87–94. doi:10.1007/s00726-011-1193-7
Kahn, S. E. (2001). Clinical review 135: the importance of beta-cell failure in the development and progression of type 2 diabetes. J. Clin. Endocrinol. Metab. 86, 4047–4058. doi:10.1210/jcem.86.9.7713
Kampoo, K., Teanpaisan, R., Ledder, R. G., and Mcbain, A. J. (2014). Oral bacterial communities in individuals with type 2 diabetes who live in southern Thailand. Appl. Environ. Microbiol. 80, 662–671. doi:10.1128/AEM.02821-13
Kaneto, H., Kimura, T., Shimoda, M., Obata, A., Sanada, J., Fushimi, Y., et al. (2022). Molecular mechanism of pancreatic β-cell failure in type 2 diabetes mellitus. Biomedicines 10, 818. doi:10.3390/biomedicines10040818
Karlsson, F. H., Tremaroli, V., Nookaew, I., Bergström, G., Behre, C. J., Fagerberg, B., et al. (2013). Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature 498, 99–103. doi:10.1038/nature12198
Kasubuchi, M., Hasegawa, S., Hiramatsu, T., Ichimura, A., and Kimura, I. (2015). Dietary gut microbial metabolites, short-chain fatty acids, and host metabolic regulation. Nutrients 7, 2839–2849. doi:10.3390/nu7042839
Kawai, T., Autieri, M. V., and Scalia, R. (2021). Adipose tissue inflammation and metabolic dysfunction in obesity. Am. J. Physiol. Cell Physiol. 320, C375–C391. doi:10.1152/ajpcell.00379.2020
Kim, M.-H., Yun, K. E., Kim, J., Park, E., Chang, Y., Ryu, S., et al. (2020). Gut microbiota and metabolic health among overweight and obese individuals. Sci. Rep. 10, 19417. doi:10.1038/s41598-020-76474-8
Kindt, S., Vanden Berghe, P., Boesmans, W., Roosen, L., and Tack, J. (2010). Prolonged IL-1L exposure alters neurotransmitter and electrically induced Ca2+ responses in the myenteric plexus. Neurogastroenterol. Motil. 22, 321–e85. doi:10.1111/j.1365-2982.2009.01414.x
Kleinridders, A., Schenten, D., Könner, A. C., Belgardt, B. F., Mauer, J., Okamura, T., et al. (2009). MyD88 signaling in the CNS is required for development of fatty acid-induced leptin resistance and diet-induced obesity. Cell Metab. 10, 249–259. doi:10.1016/j.cmet.2009.08.013
Klop, B., Elte, J. W., and Cabezas, M. C. (2013). Dyslipidemia in obesity: mechanisms and potential targets. Nutrients 5, 1218–1240. doi:10.3390/nu5041218
Köhler, H., Mccormick, B. A., and Walker, W. A. (2003). Bacterial-enterocyte crosstalk: cellular mechanisms in health and disease. J. Pediatr. Gastroenterol. Nutr. 36, 175–185. doi:10.1097/00005176-200302000-00005
Konikoff, T., and Gophna, U. (2016). Oscillospira: a central, enigmatic component of the human gut microbiota. Trends Microbiol. 24, 523–524. doi:10.1016/j.tim.2016.02.015
Kootte, R. S., Levin, E., Salojärvi, J., Smits, L. P., Hartstra, A. V., Udayappan, S. D., et al. (2017). Improvement of insulin sensitivity after lean donor feces in metabolic syndrome is driven by baseline intestinal microbiota composition. Cell Metab. 26, 611–619. doi:10.1016/j.cmet.2017.09.008
Krueger, E. S., Beales, J. L., Russon, K. B., Elison, W. S., Davis, J. R., Hansen, J. M., et al. (2021). Gut metabolite Trimethylamine N-Oxide protects INS-1-cell and rat islet function under diabetic glucolipotoxic conditions. Biomolecules 11, 1892. doi:10.3390/biom11121892
Kulkarni, A., Muralidharan, C., May, S. C., Tersey, S. A., and Mirmira, R. G. (2023). Inside the β cell: molecular stress response pathways in diabetes pathogenesis. Endocrinology 164, bqac184. doi:10.1210/endocr/bqac184
Kumar, M., Nagpal, R., Kumar, R., Hemalatha, R., Verma, V., Kumar, A., et al. (2012). Cholesterol-lowering probiotics as potential biotherapeutics for metabolic diseases. Exp. Diabetes Res. 2012, 902917. doi:10.1155/2012/902917
Lam, C. K., Chari, M., and Lam, T. K. (2009). CNS regulation of glucose homeostasis. Physiology 24, 159–170. doi:10.1152/physiol.00003.2009
Le Chatelier, E., Nielsen, T., Qin, J., Prifti, E., Hildebrand, F., Falony, G., et al. (2013). Richness of human gut microbiome correlates with metabolic markers. Nature 500, 541–546. doi:10.1038/nature12506
Lee, D.-S., Jang, J.-H., Ko, W., Kim, K.-S., Sohn, J. H., Kang, M.-S., et al. (2013). PTP1B inhibitory and anti-inflammatory effects of secondary metabolites isolated from the marine-derived fungus Penicillium sp. JF-55. Mar. Drugs 11, 1409–1426. doi:10.3390/md11041409
Leite, A. Z., Rodrigues, N. D. C., Gonzaga, M. I., Paiolo, J. C. C., De Souza, C. A., Stefanutto, N. a. V., et al. (2017). Detection of increased plasma Interleukin-6 levels and prevalence of Prevotella copri and Bacteroides vulgatus in the feces of type 2 diabetes patients. Front. Immunol. 8, 1107. doi:10.3389/fimmu.2017.01107
Leja-Szpak, A., Jaworek, J., Tomaszewska, R., Nawrot, K., Bonior, J., Kot, M., et al. (2004). Melatonin precursor; L-tryptophan protects the pancreas from development of acute pancreatitis through the central site of action. J. Physiol. Pharmacol. 55, 239–254.
Ley, R. E., Bäckhed, F., Turnbaugh, P., Lozupone, C. A., Knight, R. D., and Gordon, J. I. (2005). Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. U. S. A. 102, 11070–11075. doi:10.1073/pnas.0504978102
Li, M., Wang, B., Zhang, M., Rantalainen, M., Wang, S., Zhou, H., et al. (2008). Symbiotic gut microbes modulate human metabolic phenotypes. Proc. Natl. Acad. Sci. U. S. A. 105, 2117–2122. doi:10.1073/pnas.0712038105
Li, X., Wang, N., Yin, B., Fang, D., Jiang, T., Fang, S., et al. (2016). Effects of Lactobacillus plantarum CCFM0236 on hyperglycaemia and insulin resistance in high-fat and streptozotocin-induced type 2 diabetic mice. J. Appl. Microbiol. 121, 1727–1736. doi:10.1111/jam.13276
Li, Z., Gurung, M., Rodrigues, R. R., Padiadpu, J., Newman, N. K., Manes, N. P., et al. (2022). Microbiota and adipocyte mitochondrial damage in type 2 diabetes are linked by Mmp12+ macrophages. J. Exp. Med. 219, e20220017. doi:10.1084/jem.20220017
Lin, H. V., Frassetto, A., Kowalik, E. J., Nawrocki, A. R., Lu, M. M., Kosinski, J. R., et al. (2012). Butyrate and propionate protect against diet-induced obesity and regulate gut hormones via free fatty acid receptor 3-independent mechanisms. PLoS One 7, e35240. doi:10.1371/journal.pone.0035240
Long, J., Cai, Q., Steinwandel, M., Hargreaves, M. K., Bordenstein, S. R., Blot, W. J., et al. (2017). Association of oral microbiome with type 2 diabetes risk. J. Periodontal Res. 52, 636–643. doi:10.1111/jre.12432
Lv, C., Sun, Y., Zhang, Z. Y., Aboelela, Z., Qiu, X., and Meng, Z. X. (2022). β-cell dynamics in type 2 diabetes and in dietary and exercise interventions. J. Mol. Cell Biol. 14, mjac046. doi:10.1093/jmcb/mjac046
Lynch, C. J., and Adams, S. H. (2014). Branched-chain amino acids in metabolic signalling and insulin resistance. Nat. Rev. Endocrinol. 10, 723–736. doi:10.1038/nrendo.2014.171
Magne, F., Gotteland, M., Gauthier, L., Zazueta, A., Pesoa, S., Navarrete, P., et al. (2020). The firmicutes/bacteroidetes ratio: a relevant marker of gut dysbiosis in obese patients? Nutrients 12, 1474. doi:10.3390/nu12051474
Mar Rodríguez, M., Pérez, D., Javier Chaves, F., Esteve, E., Marin-Garcia, P., Xifra, G., et al. (2015). Obesity changes the human gut mycobiome. Sci. Rep. 5, 14600. doi:10.1038/srep14600
Martínez-Montoro, J. I., Damas-Fuentes, M., Fernández-García, J. C., and Tinahones, F. J. (2022). Role of the gut microbiome in beta cell and adipose tissue crosstalk: a review. Front. Endocrinol. 13, 869951. doi:10.3389/fendo.2022.869951
Martyn, J. A., Kaneki, M., and Yasuhara, S. (2008). Obesity-induced insulin resistance and hyperglycemia: etiologic factors and molecular mechanisms. Anesthesiology 109, 137–148. doi:10.1097/ALN.0b013e3181799d45
Massier, L., Chakaroun, R., Tabei, S., Crane, A., Didt, K. D., Fallmann, J., et al. (2020). Adipose tissue derived bacteria are associated with inflammation in obesity and type 2 diabetes. Gut 69, 1796–1806. doi:10.1136/gutjnl-2019-320118
Megur, A., Daliri, E. B., Baltriukienė, D., and Burokas, A. (2022). Prebiotics as a tool for the prevention and treatment of obesity and diabetes: classification and ability to modulate the gut microbiota. Int. J. Mol. Sci. 23, 6097. doi:10.3390/ijms23116097
Mishra, S. P., Wang, B., Jain, S., Ding, J., Rejeski, J., Furdui, C. M., et al. (2023). A mechanism by which gut microbiota elevates permeability and inflammation in obese/diabetic mice and human gut. Gut 72, 1848–1865. doi:10.1136/gutjnl-2022-327365
Molinaro, A., Bel Lassen, P., Henricsson, M., Wu, H., Adriouch, S., Belda, E., et al. (2020). Imidazole propionate is increased in diabetes and associated with dietary patterns and altered microbial ecology. Nat. Commun. 11, 5881. doi:10.1038/s41467-020-19589-w
Moreno-Indias, I., Sánchez-Alcoholado, L., García-Fuentes, E., Cardona, F., Queipo-Ortuño, M. I., and Tinahones, F. J. (2016). Insulin resistance is associated with specific gut microbiota in appendix samples from morbidly obese patients. Am. J. Transl. Res. 8, 5672–5684.
Natividad, J. M., Lamas, B., Pham, H. P., Michel, M.-L., Rainteau, D., Bridonneau, C., et al. (2018). Bilophilawadsworthia aggravates high fat diet induced metabolic dysfunctions in mice. Nat. Commun. 9, 2802. doi:10.1038/s41467-018-05249-7
Newgard, C. B., An, J., Bain, J. R., Muehlbauer, M. J., Stevens, R. D., Lien, L. F., et al. (2009). A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab. 9, 311–326. doi:10.1016/j.cmet.2009.02.002
Neyrolles, O., Hernández-Pando, R., Pietri-Rouxel, F., Fornès, P., Tailleux, L., Payán, J. a. B., et al. (2006). Is adipose tissue a place for Mycobacterium tuberculosis persistence? PLOS ONE 1, e43. doi:10.1371/journal.pone.0000043
Nirmalkar, K., Murugesan, S., Pizano-Zárate, M. L., Villalobos-Flores, L. E., García-González, C., Morales-Hernández, R. M., et al. (2018). Gut microbiota and endothelial dysfunction markers in obese Mexican children and adolescents. Nutrients 10, 2009. doi:10.3390/nu10122009
O’Hara, A. M., and Shanahan, F. (2006). The gut flora as a forgotten organ. EMBO Rep. 7, 688–693. doi:10.1038/sj.embor.7400731
Oliphant, K., and Allen-Vercoe, E. (2019). Macronutrient metabolism by the human gut microbiome: major fermentation by-products and their impact on host health. Microbiome 7, 91. doi:10.1186/s40168-019-0704-8
Omori, M., Kato-Kogoe, N., Sakaguchi, S., Kamiya, K., Fukui, N., Gu, Y. H., et al. (2022). Characterization of salivary microbiota in elderly patients with type 2 diabetes mellitus: a matched case-control study. Clin. Oral Investig. 26, 493–504. doi:10.1007/s00784-021-04027-y
Parks, B. W., Nam, E., Org, E., Kostem, E., Norheim, F., Hui, S. T., et al. (2013). Genetic control of obesity and gut microbiota composition in response to high-fat, high-sucrose diet in mice. Cell Metab. 17, 141–152. doi:10.1016/j.cmet.2012.12.007
Palmas, V., Pisanu, S., Madau, V., Casula, E., Deledda, A., Cusano, R., et al. (2021). Gut microbiota markers associated with obesity and overweight in Italian adults. Sci. Rep. 11, 5532. doi:10.1038/s41598-021-84928-w
Pan, X., Fang, X., Wang, F., Li, H., Niu, W., Liang, W., et al. (2019). Butyrate ameliorates caerulein-induced acute pancreatitis and associated intestinal injury by tissue-specific mechanisms. Br. J. Pharmacol. 176, 4446–4461. doi:10.1111/bph.14806
Peroumal, D., Sahu, S. R., Kumari, P., Utkalaja, B., and Acharya, N. (2022). Commensal fungus Candida albicans maintains a long-term mutualistic relationship with the host to modulate gut microbiota and metabolism. Microbiol. Spectr. 10, e0246222. doi:10.1128/spectrum.02462-22
Peterhoff, M., Sieg, A., Brede, M., Chao, C. M., Hein, L., and Ullrich, S. (2003). Inhibition of insulin secretion via distinct signaling pathways in alpha2-adrenoceptor knockout mice. Eur. J. Endocrinol. 149, 343–350. doi:10.1530/eje.0.1490343
Prentki, M., and Nolan, C. J. (2006). Islet beta cell failure in type 2 diabetes. J. Clin. Invest. 116, 1802–1812. doi:10.1172/JCI29103
Pulgaron, E. R., and Delamater, A. M. (2014). Obesity and type 2 diabetes in children: epidemiology and treatment. Curr. Diab Rep. 14, 508. doi:10.1007/s11892-014-0508-y
Qin, J., Li, Y., Cai, Z., Li, S., Zhu, J., Zhang, F., et al. (2012). A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 490, 55–60. doi:10.1038/nature11450
Reddy, P., Leong, J., and Jialal, I. (2018). Amino acid levels in nascent metabolic syndrome: a contributor to the pro-inflammatory burden. J. Diabetes Complicat 32, 465–469. doi:10.1016/j.jdiacomp.2018.02.005
Rehn, M., Hild, D., and Diener, M. (2005). Upregulation of cyclooxygenase-2 and thromboxane A2 production mediate the action of tumor necrosis factor-alpha in isolated rat myenteric ganglia. Am. J. Physiol. Gastrointest. Liver Physiol. 289, G586–G591. doi:10.1152/ajpgi.00020.2005
Ridaura, V. K., Faith, J. J., Rey, F. E., Cheng, J., Duncan, A. E., Kau, A. L., et al. (2013). Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science 341, 1241214. doi:10.1126/science.1241214
Rios-Covian, D., González, S., Nogacka, A. M., Arboleya, S., Salazar, N., Gueimonde, M., et al. (2020). An overview on fecal branched short-chain fatty acids along human life and as related with body mass index: associated dietary and anthropometric factors. Front. Microbiol. 11, 973. doi:10.3389/fmicb.2020.00973
Robertson, R. P., Harmon, J., Tran, P. O., Tanaka, Y., and Takahashi, H. (2003). Glucose toxicity in β-cells: type 2 diabetes, good radicals gone bad, and the glutathione connection. Diabetes 52, 581–587. doi:10.2337/diabetes.52.3.581
Rocha, M., Apostolova, N., Diaz-Rua, R., Muntane, J., and Victor, V. M. (2020). Mitochondria and T2D: role of autophagy, ER stress, and inflammasome. Trends Endocrinol. Metab. 31, 725–741. doi:10.1016/j.tem.2020.03.004
Roopchand, D. E., Carmody, R. N., Kuhn, P., Moskal, K., Rojas-Silva, P., Turnbaugh, P. J., et al. (2015). Dietary polyphenols promote growth of the gut bacterium Akkermansia muciniphila and attenuate high-fat diet–induced metabolic syndrome. Diabetes 64, 2847–2858. doi:10.2337/db14-1916
Ruze, R., Liu, T., Zou, X., Song, J., Chen, Y., Xu, R., et al. (2023). Obesity and type 2 diabetes mellitus: connections in epidemiology, pathogenesis, and treatments. Front. Endocrinol. 14, 1161521. doi:10.3389/fendo.2023.1161521
Salles, B. I. M., Cioffi, D., and Ferreira, S. R. G. (2020). Probiotics supplementation and insulin resistance: a systematic review. Diabetol. Metab. Syndr. 12, 98. doi:10.1186/s13098-020-00603-6
Sarmiento-Andrade, Y., Suárez, R., Quintero, B., Garrochamba, K., and Chapela, S. P. (2022). Gut microbiota and obesity: new insights. Front. Nutr. 9, 1018212. doi:10.3389/fnut.2022.1018212
Schepp, W., Dehne, K., Riedel, T., Schmidtler, J., Schaffer, K., and Classen, M. (1996). Oxyntomodulin: a cAMP-dependent stimulus of rat parietal cell function via the receptor for glucagon-like peptide-1 (7-36)NH2. Digestion 57, 398–405. doi:10.1159/000201367
Schoeler, M., and Caesar, R. (2019). Dietary lipids, gut microbiota and lipid metabolism. Rev. Endocr. Metab. Disord. 20, 461–472. doi:10.1007/s11154-019-09512-0
Schwartz, M. W., Woods, S. C., Porte, D., Seeley, R. J., and Baskin, D. G. (2000). Central nervous system control of food intake. Nature 404, 661–671. doi:10.1038/35007534
Sedighi, M., Razavi, S., Navab-Moghadam, F., Khamseh, M. E., Alaei-Shahmiri, F., Mehrtash, A., et al. (2017). Comparison of gut microbiota in adult patients with type 2 diabetes and healthy individuals. Microb. Pathog. 111, 362–369. doi:10.1016/j.micpath.2017.08.038
Segain, J.-P., Blétière, D. R. D. L., Bourreille, A., Leray, V., Gervois, N., Rosales, C., et al. (2000). Butyrate inhibits inflammatory responses through NFkappaB inhibition: implications for Crohn's disease. Gut 47, 397–403. doi:10.1136/gut.47.3.397
Shantaram, D., Blaszczak, A. M., Hoyd, R., Smith, A., Antwi, L., Liu, J. Z., et al. (2022). 1417-P: gut microbiome translocation to visceral adipose tissue promotes recruitment of adipose-specific neutrophils in human obesity. Diabetes 71, 1417. doi:10.2337/db22-1417-p
Silva, Y. P., Bernardi, A., and Frozza, R. L. (2020). The role of short-chain fatty acids from gut microbiota in gut-brain communication. Front. Endocrinol. 11, 25. doi:10.3389/fendo.2020.00025
Singh, R. K., Chang, H.-W., Yan, D., Lee, K. M., Ucmak, D., Wong, K., et al. (2017). Influence of diet on the gut microbiome and implications for human health. J. Transl. Med. 15, 73. doi:10.1186/s12967-017-1175-y
Smith, P., and Bénézech, C. (2020). Creeping fat in Crohn’s disease: innocuous or innocuum? Immunity 53, 905–907. doi:10.1016/j.immuni.2020.10.018
Sokol, H., Pigneur, B., Watterlot, L., Lakhdari, O., Bermúdez-Humarán, L. G., Gratadoux, J.-J., et al. (2008). Faecalibacteriumprausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl. Acad. Sci. U. S. A. 105, 16731–16736. doi:10.1073/pnas.0804812105
Song, P., Ramprasath, T., Wang, H., and Zou, M.-H. (2017). Abnormal kynurenine pathway of tryptophan catabolism in cardiovascular diseases. Cell Mol. Life Sci. 74, 2899–2916. doi:10.1007/s00018-017-2504-2
Sroka-Oleksiak, A., Młodzińska, A., Bulanda, M., Salamon, D., Major, P., Stanek, M., et al. (2020). Metagenomic analysis of duodenal microbiota reveals a potential biomarker of dysbiosis in the course of obesity and type 2 diabetes: a pilot study. J. Clin. Med. 9, 369. doi:10.3390/jcm9020369
Stancill, J. S., Broniowska, K. A., Oleson, B. J., Naatz, A., and Corbett, J. A. (2019). Pancreatic β-cells detoxify H2O2 through the peroxiredoxin/thioredoxin antioxidant system. J. Biol. Chem. 294, 4843–4853. doi:10.1074/jbc.RA118.006219
Stavropoulou, E., Kantartzi, K., Tsigalou, C., Konstantinidis, T., Romanidou, G., Voidarou, C., et al. (2020). Focus on the gut-kidney axis in health and disease. Front. Med. 7, 620102. doi:10.3389/fmed.2020.620102
Steenbergen, L., Sellaro, R., Van Hemert, S., Bosch, J. A., and Colzato, L. S. (2015). A randomized controlled trial to test the effect of multispecies probiotics on cognitive reactivity to sad mood. Brain Behav. Immun. 48, 258–264. doi:10.1016/j.bbi.2015.04.003
Stengel, A., Goebel, M., Yakubov, I., Wang, L., Witcher, D., Coskun, T., et al. (2009). Identification and characterization of nesfatin-1 immunoreactivity in endocrine cell types of the rat gastric oxyntic mucosa. Endocrinology 150, 232–238. doi:10.1210/en.2008-0747
Swisa, A., Glaser, B., and Dor, Y. (2017). Metabolic stress and compromised identity of pancreatic beta cells. Front. Genet. 8, 21. doi:10.3389/fgene.2017.00021
Takeda, K., Kaisho, T., and Akira, S. (2003). Toll-like receptors. Annu. Rev. Immunol. 21, 335–376. doi:10.1146/annurev.immunol.21.120601.141126
Tanowitz, H. B., Scherer, P. E., Mota, M. M., and Figueiredo, L. M. (2017). Adipose tissue: A safe haven for parasites? Trends Parasitol. 33, 276–284. doi:10.1016/j.pt.2016.11.008
Tap, J., Mondot, S., Levenez, F., Pelletier, E., Caron, C., Furet, J.-P., et al. (2009). Towards the human intestinal microbiota phylogenetic core. Environ. Microbiol. 11, 2574–2584. doi:10.1111/j.1462-2920.2009.01982.x
Thorens, B. (2011). Brain glucose sensing and neural regulation of insulin and glucagon secretion. Diabetes Obes. Metab. 13 (1), 82–88. doi:10.1111/j.1463-1326.2011.01453.x
Thorens, B. (2014). Neural regulation of pancreatic islet cell mass and function. Diabetes ObesMetab 16 (1), 87–95. doi:10.1111/dom.12346
Thursby, E., and Juge, N. (2017). Introduction to the human gut microbiota. Biochem. J. 474, 1823–1836. doi:10.1042/BCJ20160510
Turnbaugh, P. J., Ley, R. E., Mahowald, M. A., Magrini, V., Mardis, E. R., and Gordon, J. I. (2006). An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444, 1027–1031. doi:10.1038/nature05414
Vallianou, N. G., Stratigou, T., and Tsagarakis, S. (2018). Microbiome and diabetes: where are we now? Diabetes Res. Clin. Pract. 146, 111–118. doi:10.1016/j.diabres.2018.10.008
Van Baarlen, P., Troost, F. J., Van Hemert, S., Van Der Meer, C., De Vos, W. M., De Groot, P. J., et al. (2009). Differential NF-κB pathways induction by Lactobacillus plantarum in the duodenum of healthy humans correlating with immune tolerance. Proc. Natl. Acad. Sci. U. S. A. 106, 2371–2376. doi:10.1073/pnas.0809919106
Vangipurapu, J., Fernandes Silva, L., Kuulasmaa, T., Smith, U., and Laakso, M. (2020). Microbiota-related metabolites and the risk of type 2 diabetes. Diabetes Care 43, 1319–1325. doi:10.2337/dc19-2533
Velagapudi, V. R., Hezaveh, R., Reigstad, C. S., Gopalacharyulu, P., Yetukuri, L., Islam, S., et al. (2010). The gut microbiota modulates host energy and lipid metabolism in mice. J. Lipid Res. 51, 1101–1112. doi:10.1194/jlr.M002774
Verdam, F. J., Fuentes, S., De Jonge, C., Zoetendal, E. G., Erbil, R., Greve, J. W., et al. (2013). Human intestinal microbiota composition is associated with local and systemic inflammation in obesity. Obesity 21, E607–E615. doi:10.1002/oby.20466
Vijay, A., and Valdes, A. M. (2022). Role of the gut microbiome in chronic diseases: a narrative review. Eur. J. Clin. Nutr. 76, 489–501. doi:10.1038/s41430-021-00991-6
Virtue, A. T., Mccright, S. J., Wright, J. M., Jimenez, M. T., Mowel, W. K., Kotzin, J. J., et al. (2019). The gut microbiota regulates white adipose tissue inflammation and obesity via a family of microRNAs. Sci. Transl. Med. 11, eaav1892. doi:10.1126/scitranslmed.aav1892
Von Schwartzenberg, R. J., Bisanz, J. E., Lyalina, S., Spanogiannopoulos, P., Ang, Q. Y., Cai, J., et al. (2021). Caloric restriction disrupts the microbiota and colonization resistance. Nature 595, 272–277. doi:10.1038/s41586-021-03663-4
Wang, D. S., Zurek, A. A., Lecker, I., Yu, J., Abramian, A. M., Avramescu, S., et al. (2012). Memory deficits induced by inflammation are regulated by α5-subunit-containing GABAA receptors. Cell Rep. 2, 488–496. doi:10.1016/j.celrep.2012.08.022
Wang, X., Yang, J., Qiu, X., Wen, Q., Liu, M., Zhou, D., et al. (2021). Probiotics, pre-biotics and synbiotics in the treatment of pre-diabetes: A systematic review of randomized controlled trials. Front. Public Health 9, 645035. doi:10.3389/fpubh.2021.645035
Wang, Y., Yao, W., Li, B., Qian, S., Wei, B., Gong, S., et al. (2020). Nuciferine modulates the gut microbiota and prevents obesity in high-fat diet-fed rats. Exp. Mol. Med. 52, 1959–1975. doi:10.1038/s12276-020-00534-2
Weir, G. C., Gaglia, J., and Bonner-Weir, S. (2020). Inadequate β-cell mass is essential for the pathogenesis of type 2 diabetes. Lancet Diabetes Endocrinol. 8, 249–256. doi:10.1016/S2213-8587(20)30022-X
Wondmkun, Y. T. (2020). Obesity, insulin resistance, and type 2 diabetes: associations and therapeutic implications. Diabetes Metab. Syndr. Obes. 13, 3611–3616. doi:10.2147/DMSO.S275898
Wong, J. M. W., De Souza, R., Kendall, C. W. C., Emam, A., and Jenkins, D. J. A. (2006). Colonic health: fermentation and short chain fatty acids. J. Clin. Gastroenterol. 40, 235–243. doi:10.1097/00004836-200603000-00015
Wu, J., Yang, K., Fan, H., Wei, M., and Xiong, Q. (2023). Targeting the gut microbiota and its metabolites for type 2 diabetes mellitus. Front. Endocrinol. 14, 1114424. doi:10.3389/fendo.2023.1114424
Xu, J., and Gordon, J. I. (2003). Honor thy symbionts. Proc. Natl. Acad. Sci. U. S. A. 100, 10452–10459. doi:10.1073/pnas.1734063100
Xu, Z., Jiang, W., Huang, W., Lin, Y., Chan, F. K. L., and Ng, S. C. (2022). Gut microbiota in patients with obesity and metabolic disorders - a systematic review. Genes Nutr. 17, 2. doi:10.1186/s12263-021-00703-6
Yadav, J., Liang, T., Qin, T., Nathan, N., Schwenger, K. J. P., Pickel, L., et al. (2023). Gut microbiome modified by bariatric surgery improves insulin sensitivity and correlates with increased brown fat activity and energy expenditure. Cell Rep. Med. 4, 101051. doi:10.1016/j.xcrm.2023.101051
Yassour, M., Lim, M. Y., Yun, H. S., Tickle, T. L., Sung, J., Song, Y.-M., et al. (2016). Sub-clinical detection of gut microbial biomarkers of obesity and type 2 diabetes. Genome Med. 8, 17. doi:10.1186/s13073-016-0271-6
Yu, S., Xiong, Y., Xu, J., Liang, X., Fu, Y., Liu, D., et al. (2020). Identification of dysfunctional gut microbiota through rectal swab in patients with different severity of acute pancreatitis. Dig. Dis. Sci. 65, 3223–3237. doi:10.1007/s10620-020-06061-4
Zeng, Q., Li, D., He, Y., Li, Y., Yang, Z., Zhao, X., et al. (2019). Discrepant gut microbiota markers for the classification of obesity-related metabolic abnormalities. Sci. Rep. 9, 13424. doi:10.1038/s41598-019-49462-w
Zhai, L., Wu, J., Lam, Y. Y., Kwan, H. Y., Bian, Z. X., and Wong, H. L. X. (2021). Gut-microbial metabolites, probiotics and their roles in type 2 diabetes. Int. J. Mol. Sci. 22, 12846. doi:10.3390/ijms222312846
Zhang, Z., Mocanu, V., Cai, C., Dang, J., Slater, L., Deehan, E. C., et al. (2019). Impact of fecal microbiota transplantation on obesity and metabolic syndrome-A systematic review. Nutrients 11, 2291. doi:10.3390/nu11102291
Zheng, D., Liwinski, T., and Elinav, E. (2020). Interaction between microbiota and immunity in health and disease. Cell Res. 30, 492–506. doi:10.1038/s41422-020-0332-7
Keywords: adipose tissue, dysbiosis, gut microbiota, insulin resistance, obesity, type 2 diabetes
Citation: Patra D, Banerjee D, Ramprasad P, Roy S, Pal D and Dasgupta S (2023) Recent insights of obesity-induced gut and adipose tissue dysbiosis in type 2 diabetes. Front. Mol. Biosci. 10:1224982. doi: 10.3389/fmolb.2023.1224982
Received: 18 May 2023; Accepted: 14 September 2023;
Published: 28 September 2023.
Edited by:
Rungroch Sungthong, Chulalongkorn University, ThailandReviewed by:
Hong Jin Lee, Chung-Ang University, Republic of KoreaJing Li, Capital Medical University, China
Medha Priyadarshini, University of Illinois Chicago, United States
Asli Ekin Atici, Cedars Sinai Medical Center, United States
Yize Li, Washington University in St. Louis, United States
Copyright © 2023 Patra, Banerjee, Ramprasad, Roy, Pal and Dasgupta. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Suman Dasgupta, c3VtYW4uZHN1dEBnbWFpbC5jb20=; Durba Pal, ZHVyYmEucGFsQGlpdHJwci5hYy5pbg==
†These authors have contributed equally to this work
 Debarun Patra1†
Debarun Patra1† Durba Pal
Durba Pal Suman Dasgupta
Suman Dasgupta