
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
EDITORIAL article
Front. Mol. Biosci., 06 June 2023
Sec. Molecular Diagnostics and Therapeutics
Volume 10 - 2023 | https://doi.org/10.3389/fmolb.2023.1190116
This article is part of the Research TopicUsing Multi-Omics to Develop New Strategies to Improve Prognosis and Immunotherapy Outcomes in CancersView all 5 articles
Editorial on the Research Topic
Using multi-omics to develop new strategies to improve prognosis and immunotherapy outcomes in cancers
The identification and development of novel biomarkers to accurately predict the efficacy of immunotherapy is a critical challenge in the field of tumor immunotherapy. Next-generation sequencing has emerged as a powerful tool for detecting cancer-related molecular alterations on a large scale, refining the molecular classification of cancer, and facilitating research and development of new molecularly targeted drugs and individualized therapeutic strategies (Berger and Mardis, 2018). Further rapid advances in molecular analysis technologies have enabled us to decipher the molecular composition of tumors at a single-cell resolution (Lewis et al., 2021). Subcellular-level spatial genomics, transcriptomics, and proteomics describe cellular interactions between tumor cells and the tumor immune microenvironment (Marx, 2021; Zhao et al., 2022). Multi-omics analysis of biomarkers such as circulating tumor cells provides profound insight into the dynamics of tumor molecular structure during tumor progression and treatment (Corcoran and Chabner, 2018). In addition, functional analysis in vitro shed light on the exploration of drug sensitivity in tumor patients (Driehuis, Kretzschmar, and Clevers, 2020). Advancements in multi-omics technologies, such as genomics, transcriptomics, proteomics, metabolomics, and microbiomics, have led to the discovery of numerous biomarkers with high value in clinical practice. These biomarkers enable more accurate diagnoses, more effective treatments, and more precise judgment in disease prognosis for patients (Hasin, Seldin, and Lusis, 2017) (Figure 1). Multi-omics techniques possess tremendous potential in the treatment and prognosis of tumors such as ovarian cancer. They offer promising avenues for the treatment and prognosis of tumors, with potential applications in identifying new biomarkers and improving clinical assessment (Xiao et al., 2022).
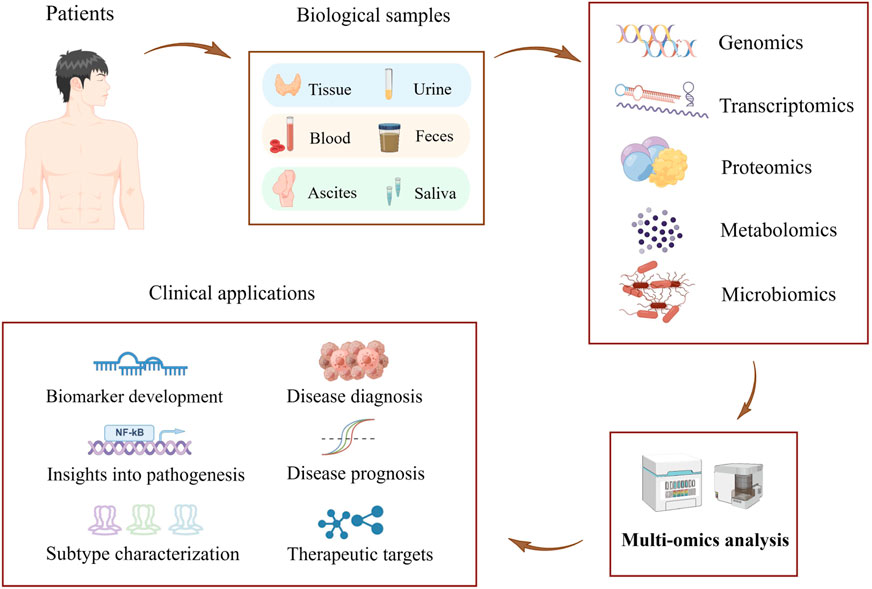
FIGURE 1. Schematic representation of a multi-omics approach to discovering biomarkers for the early diagnosis of tumors (The figure was created with Figdraw.com).
This Research Topic consisted of four articles authored by 29 experts that aimed to share novel strategies for developing multi-omics, improving immunotherapy efficacy, and enhancing patient prognosis. Two centers in these articles focused on the discovery and validation of biomarkers that could aid in predicting immunotherapy efficacy and patient prognosis, thus providing valuable insights for cancer patients. Among these biomarkers, albumin levels emerged as a strong prognostic indicator for cancer patients treated with immune checkpoint inhibitors (ICI) due to their association with both nutritional and inflammatory status. Guven et al. conducted a comprehensive analysis of 36 studies encompassing 8,406 advanced cancer patients from various databases and found that patients with lower albumin levels had a significantly higher risk of death compared to those with higher levels. Additionally, every 1 g/dL decrease in albumin levels led to a 10% increase in the risk of death, making albumin levels a critical sequential prognostic factor. Another promising biomarker was microbial status before ICI treatment initiation, which could predict patient outcomes. Shoji et al. analyzed the diversity of oral and gut microbiota in 28 non-small cell lung cancer (NSCLC) patients undergoing ICI treatment and revealed that gut microbiota composition significantly differed between responders and non-responders. Conversely, no statistically significant difference in oral microbiota composition was observed, thus further revealing a close relationship between ICI response and gut microbiota diversity in NSCLC patients. Extracellular vesicles (EVs) were broadly classified into three main categories based on their biosynthetic or secretory processes: exosomes, microvesicles/particles/extracellular bodies, and apoptotic vesicles (Yanez-Mo et al., 2015). With the arrival of the “omics era” and a deeper understanding of EVs, Lu et al. discussed a novel class of EV-centered immunotherapies for cancer, summarizing advances in the multi-omic analysis of EVs for the early diagnosis of precancer and hepatocellular carcinoma. Closely related to tumor progression, apoptosis has been recognized as one of the hot spots of research in recent years. IAGsPI and pAGsPI were potential biomarkers to predict the prognosis of patients with renal clear cell carcinoma (Lin et al., 2022). A model based on the pyroptosis-related genes (PRGs) was developed to predict the prognosis of colon adenocarcinoma with excellent performance in external cohorts (Luo et al., 2021). In addition, serving as one of the members of the gastrin family, GSDMD was differentially expressed in most cancers and could perform as a prognostic indicator for adrenocortical carcinoma (ACC) (Qiu, Hu, and Dong, 2021). Gao et al. utilized basic experimental validation and systematic bioinformatics analysis to model the prognosis of PRGs features in ACC patients and analyzed the correlation between immune infiltration and PRGs.
The Research Topic “Using Multi-Omics to Develop New Strategies for Improving Prognosis and Immunotherapy Outcomes in Cancer” serves as a platform for sharing innovative strategies developed through multi-omics to enhance the effectiveness of immunotherapies and improve patient outcomes. These studies integrate data from various layers, providing valuable insights into the molecular structure of tumors and expanding the scope of cancer biology. Although clinical applications of these techniques are still in the early stages, we believe that several of these new approaches will not only advance our understanding of tumor biology but also significantly shape the future of precision cancer therapies.
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
We would like to thank all authors for their contributions to this Research Topic.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Berger, M. F., and Mardis, E. R. (2018). 'The emerging clinical relevance of genomics in cancer medicine. Nat. Rev. Clin. Oncol. 15, 353–365. doi:10.1038/s41571-018-0002-6
Corcoran, R. B., and Chabner, B. A. (2018). 'Application of cell-free DNA analysis to cancer treatment. N. Engl. J. Med. 379, 1754–1765. doi:10.1056/NEJMra1706174
Driehuis, E., Kretzschmar, K., and Clevers, H. (2020). 'Establishment of patient-derived cancer organoids for drug-screening applications. Nat. Protoc. 15, 3380–3409. doi:10.1038/s41596-020-0379-4
Hasin, Y., Seldin, M., and Lusis, A. (2017). 'Multi-omics approaches to disease. Genome Biol. 18, 83. doi:10.1186/s13059-017-1215-1
Lewis, S. M., Asselin-Labat, M. L., Nguyen, Q., Berthelet, J., Tan, X., Wimmer, V. C., et al. (2021). 'Spatial omics and multiplexed imaging to explore cancer biology. Nat. Methods 18, 997–1012. doi:10.1038/s41592-021-01203-6
Lin, G., Feng, Q., Zhan, F., Yang, F., Niu, Y., and Li, G. (2022). 'Generation and analysis of pyroptosis-based and immune-based signatures for kidney renal clear cell carcinoma patients, and cell experiment. Front. Genet. 13, 809794. doi:10.3389/fgene.2022.809794
Luo, B., Lin, J., Cai, W., and Wang, M. (2021). 'Identification of the pyroptosis-related gene signature and risk score model for colon adenocarcinoma. Front. Genet. 12, 771847. doi:10.3389/fgene.2021.771847
Marx, V. (2021). 'Method of the year: Spatially resolved transcriptomics. Nat. Methods 18, 9–14. doi:10.1038/s41592-020-01033-y
Qiu, S., Hu, Y., and Dong, S. (2021). 'Pan-cancer analysis reveals the expression, genetic alteration and prognosis of pyroptosis key gene GSDMD. Int. Immunopharmacol. 101, 108270. doi:10.1016/j.intimp.2021.108270
Xiao, Y., Bi, M., Guo, H., and Li, M. (2022). 'Multi-omics approaches for biomarker discovery in early ovarian cancer diagnosis. EBioMedicine 79, 104001. doi:10.1016/j.ebiom.2022.104001
Yanez-Mo, M., Siljander, P. R., Andreu, Z., Zavec, A. B., Borras, F. E., Buzas, E. I., et al. (2015). 'Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles 4, 27066. doi:10.3402/jev.v4.27066
Keywords: multi-omics, immunotherapy, prognostic signature, molecular subtype, biomarker
Citation: Cao C, Chen S, Wang L, Liu Z and Han X (2023) Editorial: Using multi-omics to develop new strategies to improve prognosis and immunotherapy outcomes in cancers. Front. Mol. Biosci. 10:1190116. doi: 10.3389/fmolb.2023.1190116
Received: 20 March 2023; Accepted: 30 May 2023;
Published: 06 June 2023.
Edited by:
William C. Cho, QEH, Hong Kong SAR, ChinaReviewed by:
Yingcheng Wu, Fudan University, ChinaCopyright © 2023 Cao, Chen, Wang, Liu and Han. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zaoqu Liu, bGl1emFvcXVAMTYzLmNvbQ==; Xinwei Han, ZmNjaGFueHdAenp1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.