- Department of Molecular Biosciences, Kyoto Sangyo University, Kyoto, Japan
V/A-ATPase is a rotary molecular motor protein that produces ATP through the rotation of its central rotor. The soluble part of this protein, the V1 domain, rotates upon ATP hydrolysis. However, the mechanism by which ATP hydrolysis in the V1 domain couples with the mechanical rotation of the rotor is still unclear. Cryo-EM snapshot analysis of V/A-ATPase indicated that three independent and simultaneous catalytic events occurred at the three catalytic dimers (ABopen, ABsemi, and ABclosed), leading to a 120° rotation of the central rotor. Besides the closing motion caused by ATP bound to ABopen, the hydrolysis of ATP bound to ABsemi drives the 120° step. Our recent time-resolved cryo-EM snapshot analysis provides further evidence for this model. This review aimed to provide a comprehensive overview of the structure and function of V/A-ATPase from a thermophilic bacterium, one of the most well-studied rotary ATPases to date.
Introduction
ATP synthases are enzymes that play crucial roles in energy metabolism (Boyer, 1997; Yoshida et al., 2001; Kuhlbrandt, 2019; Frasch et al., 2022). They convert an electrochemical proton motive force across the membrane generated by respiration into chemical energy that is stored in the form of ATP. In the absence of a proton motive force (pmf), ATP synthases can hydrolyze ATP and use the energy released to transport protons across the membrane. Because of this ATP hydrolysis activity, ATP synthases are sometimes referred to as rotary ATPases. ATP synthases are classified into two major categories: F-type ATP synthases (FoF1-ATPase) and V-type ATP synthases (V/A-ATPase) (Figures 1A, B) (Yokoyama and Imamura, 2005; Forgac, 2007; Vasanthakumar and Rubinstein, 2020). FoF1-ATPases are found in the inner membrane of mitochondria, the thylakoid membrane of chloroplasts, and the plasma membrane of eubacteria (Frasch et al., 2022). V/A-ATPases are present in some eubacteria and archaea and are structurally similar to eukaryotic V-ATPases. The basic structure of both ATPases comprises a rotor complex comprising a hydrophobic c-ring and a stator apparatus surrounding the rotor (Figure 1C). The c-ring in the Fo/Vo domain rotates by the pmf, which drives ATP synthesis in the F1/V1 domain. Conversely, when ATP is hydrolyzed in the F1/V1 domain and the central rotary axis rotates, protons are transported across the membrane in the Fo/Vo domain.
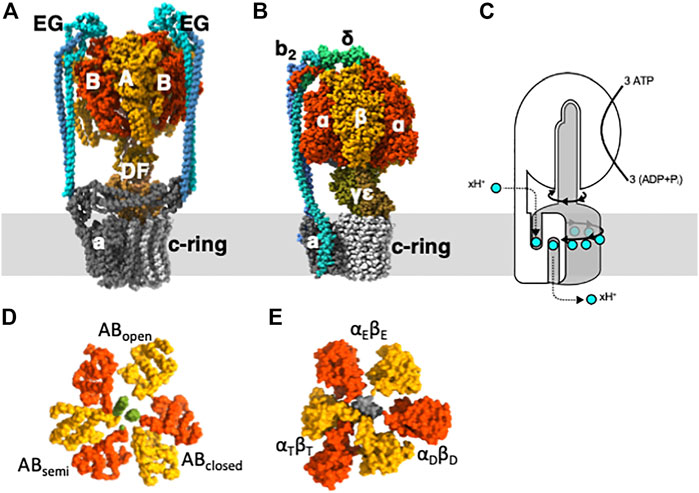
FIGURE 1. Subunit structure and function for V-type and F-type ATPases. (A) V/A-ATPase from Thermus. thermophilus. (B) bacterial FoF1 ATPase from Geobacillus stearothermophilus. (C) The schematic representation illustrates the rotary catalytic mechanism of rotary ATPases, where the rotation of the central rotor complex (grey) driven by ATP in the surrounding stator apparatus (white) rotates the membrane-embedded c-ring, leading to the translocation of protons across the membrane. Slice view of the C-terminal domain of V1 (D) and F1 (E).
The isolated hydrophilic F1/V1 domain exhibits ATP hydrolysis activity and is referred to as F1-ATPase and V1-ATPase, respectively (Yokoyama et al., 1989; Yokoyama et al., 1990; Weber and Senior, 1997; Suzuki et al., 2016). In the F1-ATPase, the catalytic site is located at the interface between the α and ß subunits, resulting in three catalytic sites. In V1-ATPase, the catalytic site is located at the interface between subunits A and B (Figure 1D) (Maher et al., 2009; Suzuki et al., 2016; Nakanishi et al., 2018). The hydrolysis of a single ATP molecule results in a 120° rotation of the rotor, and the hydrolysis of three ATP molecules results in a full 360° rotation (Imamura et al., 2003).
The theory of the binding change mechanism of FoF1-ATPase rotation was proposed by Boyer over 50 years ago (Boyer et al., 1973). This theory postulates that the three catalytic sites within the F1 domain of FoF1-ATPase sequentially adopt Loose, Tight, and Open forms, with differing nucleotide affinities, as the central γ subunit rotates (Figure 2A). Synchronized changes in the properties of these sites lead to ATP release. The asymmetric hexamer structure of the F1 domain predicted by Boyer was confirmed by the crystal structure of bovine heart mitochondrial F1-ATPase in 1994 (Abrahams et al., 1994).
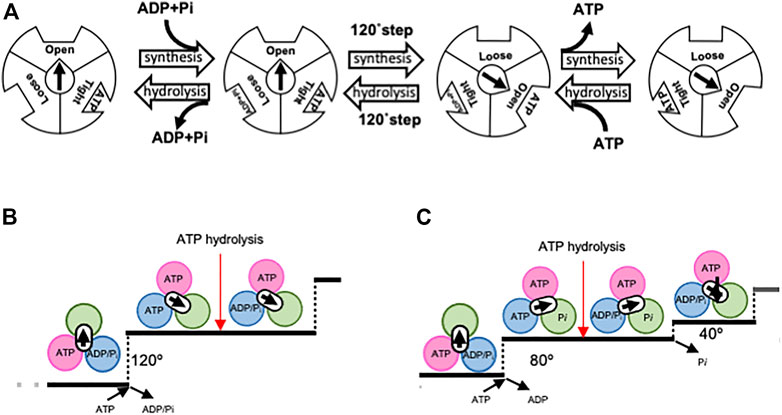
FIGURE 2. Proposed rotary mechanism of rotary ATPases. Boyer first proposed a model for the binding-change mechanism (A). Open, Loose, and Tight bonds correspond to αEβE, αDβD, and αTβT, respectively. The direction of synthesis is characterized by the binding of ADP and Pi to Loose, followed by a 120° step that results in Tight becoming Open and the release of ATP. Through the conversion of Loose to Tight, ADP is phosphorylated on Tight to form ATP. The reverse sequence of events occurred in the direction of ATP hydrolysis. Proposed chemo-mechanical cycles of V1-ATPase (B) and F1-ATPase (C). Each image was viewed from the V1/F1 side. In V1-ATPase, ATP binding results in a 120° rotation of the DF, followed by ATP hydrolysis. In contrast, F1-ATPase has been proposed to operate according to a model in which ATP binding initiates an 80° rotation, followed by ATP hydrolysis and Pi release, leading to a further 80° rotation.
The crystal structure of F1-ATPase revealed that the three catalytic dimers adopted distinct conformations (Figure 1E). One dimer exhibits an open catalytic interface without any nucleotides (αEβE), whereas the other dimer displays a closed structure with ADP bound (αDβD). The third dimer has a partially open structure that accommodates an analog of ATP (αTβT). Since then, numerous crystal structures of F1-ATPase from different species and under different conditions have been reported, but they are essentially identical to those of asymmetric hexamer (Groth and Pohl, 2001; Menz et al., 2001; Kagawa et al., 2004; Cingolani and Duncan, 2011; Rees et al., 2012; Ferguson et al., 2016).
The rotary catalytic mechanism of F1-ATPase was directly confirmed by single-molecule observation experiments (Noji et al., 1997). In these experiments, a probe was attached to the rotor γ subunit, whereas the stator α3β3 was fixed to the glass surface. The rotation of the probe was observed using an optical microscope, and upon ATP addition, the unidirectional rotation of the probe was observed, thereby establishing that F1-ATPase functions as a rotary molecular motor driven by ATP. Similarly, the rotary mechanism of the V1-ATPase was demonstrated 5 years later by our group (Imamura et al., 2003).
Since the demonstration of the rotation of F1-ATPase, numerous single-molecule observation experiments have furthered our understanding of the rotary mechanism of F1-ATPase (Yasuda et al., 1998; Yasuda et al., 2001; Shimabukuro et al., 2003; Itoh et al., 2004; Adachi et al., 2007; Furuike et al., 2008; Adachi et al., 2012; Saita et al., 2015). For instance, it has been demonstrated that the binding of one ATP molecule results in a 120° step, which comprises an 80° and 40° substep, with ATP hydrolysis occurring at the 80° position (Figures 2B, C) (Yasuda et al., 2001). In addition, high time-resolution analysis has also been performed on F1-ATPase from Escherichia coli using tiny gold rods as probes (Spetzler et al., 2006; Hornung et al., 2011). However, the precise mechanism by which the hydrolysis energy of ATP is converted into rotational force remains unclear.
This review aimed to describe the chemo-mechanical coupling mechanism between ATP hydrolysis and rotor rotation revealed by cryo-snapshot analysis of V/A-ATPase. The similarity between V/A-ATPase and FoF1 suggests that the F1 domain shares this mechanism.
Structure and rotation of V/A-ATPase from Thermus thermophilus
The V/A-ATPase from Thermus thermophilus is one of the best-studied ATP synthases. It comprises two domains: the V1 domain and the Vo. The V1 domain is a rotary motor that rotates its central rotor (DF) within A3B3, which contains three catalytic sites comprising AB dimers (Yokoyama and Imamura, 2005; Maher et al., 2009) (Figure 1D).
The Vo domain (E2G2d1a1c12) is composed of stator parts, including a subunit and two, EG peripheral stalks, and the d1c12 rotor complex (Kishikawa et al., 2020), which comprises a central rotor complex with the DF subunits of V1. During the rotation of the central rotor (DF d1c12), caused by the proton motive force, the DF induces changes in the three AB dimers, leading to the cooperative synthesis of ATP from ADP and Pi at the A3B3 hexamer. The reverse rotation of the central rotor caused by ATP hydrolysis by A3B3 drives the proton translocation in the Vo domain (Yokoyama et al., 1998; Toei et al., 2007).
A single-molecule rotation experiment showed that the V1 domain rotates in 120° steps, unlike the 80° steps observed for F1-ATPases (Figure 2B). This indicates that ATP binding and hydrolysis in the V1 domain occur at the 120° dwell position (Imamura et al., 2005; Furuike et al., 2011). These results were confirmed by a cryo-EM snapshot analysis.
Structural analysis of V/A- ATPase by cryo-electron microscopy
Although the crystal structures of both F1-ATPase and V1-ATPase have been determined, no crystal structures of intact V-ATPase or FoF1 have been reported, owing to the structural variability arising from the rotational state of the F1/V1 domain.
This situation has changed with the advent of the resolution revolution in cryo-electron microscopy (CryoEM) structural analysis. This has enabled us to obtain the overall structures of FoF1 and V-ATPases (Allegretti et al., 2015; Zhao et al., 2015; Zhou et al., 2015; Schep et al., 2016; Nakanishi et al., 2018), and further advances in analytical techniques have made it possible to produce high-resolution maps and build atomic models of rotary ATPases in a relatively short period (Hahn et al., 2018; Roh et al., 2018; Gu et al., 2019; Guo et al., 2019; Murphy et al., 2019; Sobti et al., 2019; Abbas et al., 2020).
The single-particle analysis allowed the separation of different structures present in a V/A-ATPase sample through classification into several classes (Cheng, 2018). In yeast V-ATPase, this technique has led to the isolation of three classes with different orientations of the central rotational axis (Zhou et al., 2015). The same approach was applied to V/A-ATPase, resulting in the isolation of three rotational state structures that showed the significant movement of the peripheral hydrophilic domains (EG and the hydrophilic domain of a subunit) in each state.
To capture the intermediate structure of V/A-ATPase during ATP hydrolysis, we used the TSSA mutant V/A-ATPase, which is less prone to the ADP-inhibited form and has a reduced affinity for ADP at catalytic sites (Nakano et al., 2008). This mutant V/A-ATPase, dialyzed with ethylenediamineetetraacetic acid (EDTA)-containing phosphate buffer to remove bound ADP, was reconstituted on nanodiscs and subjected to cryo-grid preparation. We prepared cryogrids under different reaction conditions, such as waiting for ATP binding with a low ATP concentration, waiting for a catalytic reaction with a saturated ATP concentration, and a saturated ATPγS concentration. These samples were then subjected to cryo-electron microscopy using a Titan Krios. The atomic-resolution structures of the V1 domain without nucleotides at the catalytic sites were obtained through single-particle analysis with focused refinement. The nucleotide-free V1 structure (Vnucfree) consisted of an open structure (ABopen), a slightly closed structure (ABsemi), and a closed structure (ABclosed) (Figure 3A). The Vnucfree structures were largely similar to the V1 structures occupied by nucleotides at three catalytic sites (V3nuc), implying that the asymmetry of the V1 domain arises from its intrinsic structure rather than conformational changes induced by nucleotide binding. This asymmetric structure of the V1 domain has been reported for other V-ATPases (Oot et al., 2016; Suzuki et al., 2016; Abbas et al., 2020; Tan et al., 2022), highlighting the robustness of the V1 domain structure.
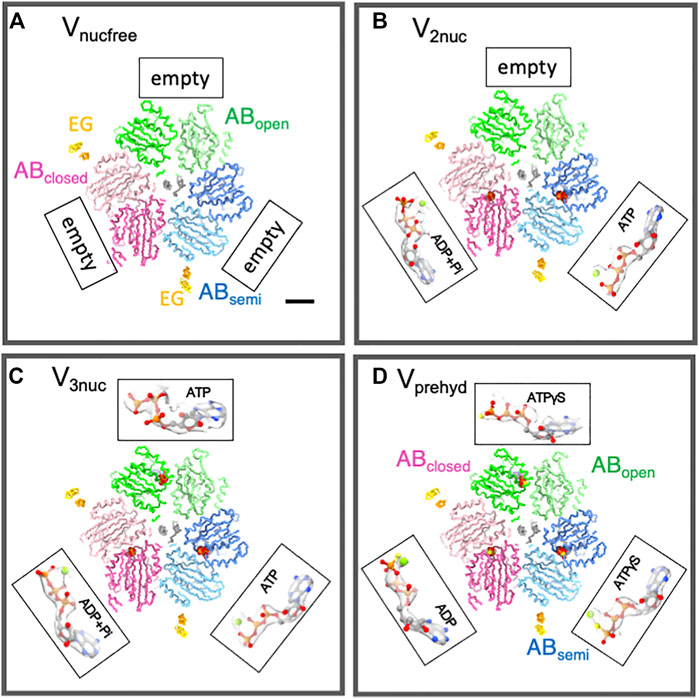
FIGURE 3. Structures of nucleotide binding sites in each condition. The slice of Vnucfree (A), Vprehyd (B), V3nuc (C), and V2nuc (D) viewed from the V1 (cytosolic) side were shown with bound nucleotides. The scale bar is 20 Å. Three catalytic dimers (ABopen, ABsemi, and ABclosed) in each structure are shown in green, blue, and pink, respectively. Two, EG peripheral stalks were shown in yellow. CryoEM maps were represented as semi-translucent. The magnified view of bound nucleotides and Mg ions was highlighted as a ball-and-stick and sphere representation.
Subsequently, we performed a single-particle analysis of V/A-ATPase in a reaction solution containing 50 μM or 6 mM ATP, and an ATP regeneration system. Given that ATP binding is the rate-limiting step in the ATP hydrolysis reaction containing 50 μM ATP (which is lower than its Km of 500 μM), most of the V/A-ATPase molecules in the reaction solution are in a waiting state for ATP binding. The V1 domain structure under these conditions was largely indistinguishable from that of Vnucfree, comprising ABopen, ABsemi, and ABclosed structures. Among the catalytic sites, ADP was bound to ABclosed and ATP to ABsemi, whereas ABopen had no bound nucleotide (Figure 3B), suggesting that the next ATP molecule should bind to ABopen. This structure was referred to as V2nuc as it contained two bound nucleotides.
In the presence of 6 mM ATP (>> Km), the dwell time for ATP binding to the enzyme was negligible, and most of the V/A-ATPase molecules in the reaction solution were in a waiting state for the catalytic reaction after ATP binding. The V1 domain under these conditions consisted of ABopen, ABsemi, and ABclosed, all of which were bound to nucleotides (V3nuc) (Figure 3C). ABclosed was bound to ATP or (ADP + Pi), while ATP binding was also confirmed at both ABopen and ABsemi. Therefore, it is clear that ATP will bind to the ABopen in the V2nuc, resulting in V3nuc formation. This indicates that the V3nuc represents the state just before the 120° rotation of the rotor takes place. In summary, the 120° rotation does not occur immediately after ATP binds to the V2nuc.
Finally, a single-particle analysis of V/A-ATPase was performed in a reaction solution containing 4 mM ATPγS, a slow-hydrolyzing ATP analog. Under these conditions, the reaction solution was substrate-saturated; therefore, most of the resulting structures were those waiting for ATP hydrolysis (Vprehyd). In Vprehyd, ATPγS and ADP were observed at all catalytic sites in the V1 domain of V/A-ATPase (Figure 3D). Notably, ADP was found at ABclosed, indicating that ATPγS-bound ABclosed had already been hydrolyzed. This also indicated that ATPγS bound to ABsemi was waiting for hydrolysis. In the catalytic site of ABsemi, the amino acid residues responsible for ATP hydrolysis were located further away from the ATP molecule (in this case, ATPγS), resulting in a slower ATPγS-bound ABsemi hydrolysis. The hydrolysis of ATPγS bound to ABsemi requires a conformational change from ABsemi to ABclosed, which involves 120° rotation in the DF rotor. Thus, Vprehyd represents a waiting state for the hydrolysis of ATPγS on ABsemi with 120° rotation.
The rotary mechanism of V/A-ATPase fueled by ATP
The structures of the Vnucfree and the nucleotide-containing forms (V2nuc and V3nuc) were nearly identical (Figure 3). The asymmetric structure of the V1 domain, composed of ABopen, ABsemi, and ABclosed, was maintained regardless of the nucleotide-binding state, suggesting that nucleotide binding to the V1 domain does not cause an asymmetric change in its structure. The conformational changes in the V1 domain coupled with ATP hydrolysis appear to be discrete, indicating that the power-stroke model, in which the axis rotation is driven by conformational changes in the catalytic subunit, may not be appropriate.
Previous single-molecule rotation experiments on both F1-ATPase and V1-ATPase indicated that the 120° step (80° step in F1) was concurrent with ATP binding, as the histogram analysis of the initiation dwell time indicated a first-order exponential relationship with ATP concentration (Yasuda et al., 2001; Furuike et al., 2011). The presence of the V3nuc structure, in which ATP is bound but the 120° step has not yet occurred, challenges the assumption that ATP binding to the V1 domain and the 120° step occur simultaneously.
Structural analysis of V/A-ATPase during rotation provides key insights into the coupling of ATP hydrolysis with the mechanical movement of the rotor. During the transition from V3nuc to V2nuc, the three AB dimers undergo the following conformational changes, as depicted in Figure 4: (a) from ABopen to ABsemi, (b) from ABsemi to ABclosed, and (c) from ABclosed to ABopen, which are accompanied by a 120° step in the DF rotor. (a) Is the process by which the AB dimer becomes more closed upon ATP binding. This has been confirmed experimentally, with the ß-subunit of F1 showing a closed conformation upon ATP binding (Akutsu, 2017). The difference in the nucleotide states bound to ABsemi (ATP) and ABclosed (ADP and Pi) generates a downhill chemical potential, which drives the 120° step in the process (b) as the AB dimer changes from ABsemi to ABclosed. The release of bound ADP and Pi from ABclosed and its conversion to ABopen during the process (c) is a result of the interaction between ADP or Pi and the amino acid residues at the catalytic site being cleaved, causing ABclosed to open (Kishikawa et al., 2022). This process is non-spontaneous and requires external work. If ADP is bound to ABclosed, it results in an ADP-inhibited state of the V1 domain (Yokoyama et al., 1998; Nakanishi et al., 2018), and the 120° step does not occur even if ATP is bound to ABopen.
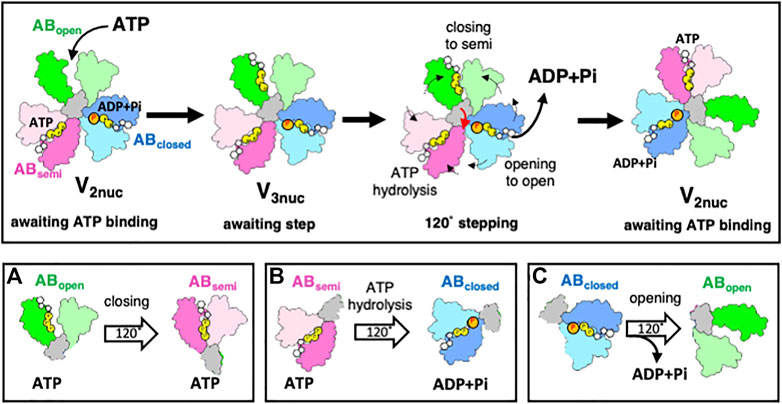
FIGURE 4. The rotary mechanism of the V1 domain powered by ATP hydrolysis. The figure shows schematic models of ABopen, ABsemi, and ABclosed in green, pink, and blue respectively. The grey region shows the coiled region of the D subunit in contact with A3B3. In the V2nuc state, waiting for ATP binding, binding of ATP onto ABopen does not result in a 120° step. In the V3nuc state, both the closing of ABopen and ATP hydrolysis in ABsemi result in the opening of ABclosed, accompanied by ADP and Pi release. The catalytic events in three AB dimers occur simultaneously with a 120° step of DF, leading to a structural transition from V3nuc to V2nuc. The lower part of the figure illustrates the simultaneous occurrence of catalytic events at the three catalytic dimers (ABopen, ABsemi, ABclosed) with a 120° step. The events involve changing ABopen to ABsemi by binding ATP (A). The ABsemi also changes ABclosed through the hydrolysis of bound ATP (B). ABclosed changes ABopen through a 120° step of DF, releasing bound ADP and Pi (C).
Our model represents an improvement over Boyer’s original model. Unlike Boyer’s bi-site model, which alternates between a state of one and two bound nucleotides and is characterized by a 120° step caused by bound ATP (Figure 2A), the model proposed here is a tri-site model that involves the cooperative interaction of chemical reactions occurring at three distinct sites. This study also sheds light on the role of ATP binding and hydrolysis in the different conformational states of the AB dimer that drives the 120°step of the DF rotor in V/A-ATPase. Both the closure of ABopen to ABsemi by binding ATP and the hydrolysis of ATP in ABsemi trigger the opening of ABclosed, leading to ADP and Pi release from the catalytic site. This process was synchronized with 120°rotation of the DF rotor. This model highlights the coupling between the downhill chemical potential generated by ATP hydrolysis and the mechanical 120° step of the rotor, as confirmed by the time-resolved structural analysis in the following chapter.
Time-resolved snapshot structural analysis of V/A-ATPase
The change in V/A-ATPase from a nucleotide-depleted state to a steady state with nucleotides was studied using the single-particle cryo-EM method. The experiments were performed as shown in Figure 5. V/A-ATPase reacted with a low concentration of ATP and 20 mM sulfate ions for 60 s (Figure 5A), followed by cryo-grid preparation. The resulting structure (Vsemi1ATP) showed ATP binding to ABsemi and a 120° rotation, as indicated by the absence of ATP binding to ABopen. The reaction solution containing V/A-ATPase was then reacted with high ATP concentrations for 5 and 30 s. The 5-s reaction solution yielded two structures, V1ATP and V2ATP, with ATP/ADP and sulfate bound to different sites (Figure 5B). V1ATP showed ATP binding to the catalytic site on ABopen and sulfate binding to both ABclosed and ABsemi. V2ATP contains two ATP molecules in the catalytic sites on ABopen and ABsemi and sulfate in the catalytic site on ABclosed. The 30-s reaction solution yielded two structures, V1ATP and V3ATP, with ATP/ADP bound to all three sites, which is similar to the V3nuc obtained under high ATP conditions in a previous study (Figure 5C).
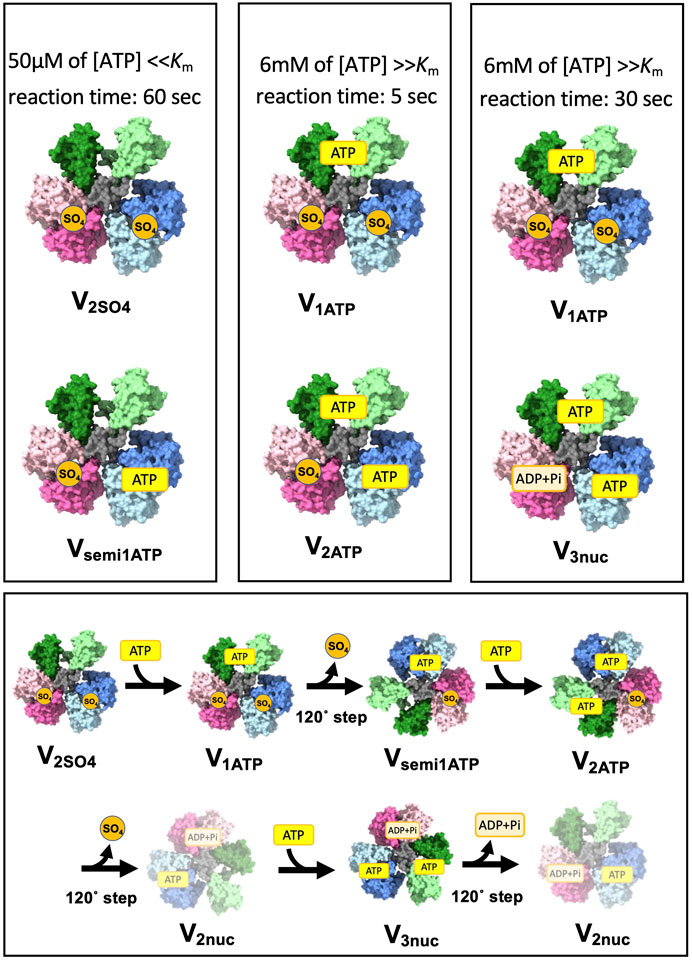
FIGURE 5. Short-lived initial intermediate V1 structure captured by a time-resolved cryo snapshot analysis. This figure shows the results of cryo-electron microscopy (CryoEM) analysis of the structural changes in a nucleotide-depleted V/A-ATPase enzyme upon reaction with different ATP concentrations. The upper columns represent the initial intermediate structures captured at different time points after the reaction with either 50 µM or 6 mM ATP. The lower column shows the structural transition from the initial intermediate structures to the final, steady-state structures (V2nuc and V3nuc).
The transition process from the initial state of V1 to the V3nuc is shown in Figure 5D. When ATP binds to V2SO4, it changes to V1ATP. Upon ATP binding to ABopen, resulting in a 120° step, one molecule of SO4 is released, resulting in the transition to Vsemi1ATP. ATP binds to Vsemi1ATP to produce V2ATP. This undergoes a 120° step and releases ADP and Pi, yielding V2nuc. ATP binding to the V2nuc produces V3nuc. The initially bound SO4 is released, and V1 eventually reaches a steady state in which V2nuc and V3nuc alternately appear as a result of ATP hydrolysis.
The presence of V1ATP in the 30-s dataset and the absence of V2ATP suggest that an immediate transition from V2ATP to V3ATP occurred. These results suggest that 120° rotation of the rotor does not occur immediately upon the closure of ABopen with ATP in the absence of ATP binding on ABsemi. This is consistent with our model, in which both the closure of ABopen with ATP and the hydrolysis of ATP in ABsemi drive the open motion of ABclosed, releasing ADP and Pi coupled with the simultaneous 120° rotation of the rotor.
Conclusion
The thermophilic F1-ATPase has been studied using single-molecule rotation observations, which have shown the presence of at least one intermediate state during the 120° step, in addition to the three rotational states. A recent cryo-electron microscopy (EM) structural study of F1-ATPase revealed six structures corresponding to these intermediate states (Sobti et al., 2021). The V1 domain comprises only the basic structures of ABopen, ABsemi, and ABclosed, while the βα dimer, the catalytic unit of F1-ATPase, adopts intermediate structures beyond these basic structures. This makes the rotational mechanism of the F1-ATPase more complex than that of the V/A-ATPase, but the underlying principle remains the same. Our recent structural snapshot analysis of FoF1 supports this, suggesting that FoF1 converts the downhill chemical potential generated by ATP hydrolysis into a mechanical step through a similar mechanism (Nakano et al., 2022). This highlights the importance of both ATP binding and hydrolysis in the generation of rotational forces by rotary ATPases.
Author contributions
The author confirms being the sole contributor of this work and has approved it for publication.
Funding
This work was supported by the Platform Project for Supporting Drug Discovery and Life Science Research (Basis for Supporting Innovative Drug Discovery and Life Science Research (BINDS)) from AMED under Grant Number JP17a.m.0101001 (support number 1312) and the Nanotechnology Platform of the Ministry of Education, Culture, Sports, Science and Technology (MEXT) (Project Number. JPMXP09A21OS0008).
Acknowledgments
We are grateful to Dr. Kishikawa, Dr. Nakanishi, and all the members of the Yokoyama Lab and Mitsuoka Lab for their continuous support and technical assistance. We acknowledge the support from the Japan Society for the Promotion of Science (KAKENHI; Grant No. 20H03231) and the Takeda Science Foundation.
Conflict of interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abbas, Y. M., Wu, D., Bueler, S. A., Robinson, C. V., and Rubinstein, J. L. (2020). Structure of V-ATPase from the mammalian brain. Science 367, 1240–1246. doi:10.1126/science.aaz2924
Abrahams, J. P., Leslie, A. G., Lutter, R., and Walker, J. E. (1994). Structure at 2.8 A resolution of F1-ATPase from bovine heart mitochondria. Nature 370, 621–628. doi:10.1038/370621a0
Adachi, K., Oiwa, K., Nishizaka, T., Furuike, S., Noji, H., Itoh, H., et al. (2007). Coupling of rotation and catalysis in F(1)-ATPase revealed by single-molecule imaging and manipulation. Cell 130, 309–321. doi:10.1016/j.cell.2007.05.020
Adachi, K., Oiwa, K., Yoshida, M., Nishizaka, T., and Kinosita, K. (2012). Controlled rotation of the F₁-ATPase reveals differential and continuous binding changes for ATP synthesis. Nat. Commun. 3, 1022. doi:10.1038/ncomms2026
Akutsu, H. (2017). Dynamic mechanisms driving conformational conversions of the beta and epsilon subunits involved in rotational catalysis of F1-ATPase. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 93, 630–647. doi:10.2183/pjab.93.040
Allegretti, M., Klusch, N., Mills, D. J., Vonck, J., Kühlbrandt, W., and Davies, K. M. (2015). Horizontal membrane-intrinsic α-helices in the stator a-subunit of an F-type ATP synthase. Nature 521, 237–240. doi:10.1038/nature14185
Boyer, P. D., Cross, R. L., and Momsen, W. (1973). A new concept for energy coupling in oxidative phosphorylation based on a molecular explanation of the oxygen exchange reactions. Proc. Natl. Acad. Sci. U. S. A. 70, 2837–2839. doi:10.1073/pnas.70.10.2837
Boyer, P. D. (1997). The ATP synthase-a splendid molecular machine. Annu. Rev. Biochem. 66, 717–749. doi:10.1146/annurev.biochem.66.1.717
Cheng, Y. (2018). Single-particle cryo-EM-How did it get here and where will it go. Science 361, 876–880. doi:10.1126/science.aat4346
Cingolani, G., and Duncan, T. M. (2011). Structure of the ATP synthase catalytic complex (F(1)) from Escherichia coli in an autoinhibited conformation. Nat. Struct. Mol. Biol. 18, 701–707. doi:10.1038/nsmb.2058
Ferguson, S. A., Cook, G. M., Montgomery, M. G., Leslie, A. G., and Walker, J. E. (2016). Regulation of the thermoalkaliphilic F1-ATPase from Caldalkalibacillus thermarum. Proc. Natl. Acad. Sci. U. S. A. 113, 10860–10865. doi:10.1073/pnas.1612035113
Forgac, M. (2007). Vacuolar ATPases: Rotary proton pumps in physiology and pathophysiology. Nat. Rev. Mol. Cell Biol. 8, 917–929. doi:10.1038/nrm2272
Frasch, W. D., Bukhari, Z. A., and Yanagisawa, S. (2022). F1FO ATP synthase molecular motor mechanisms. Front. Microbiol. 13, 965620. doi:10.3389/fmicb.2022.965620
Furuike, S., Hossain, M. D., Maki, Y., Adachi, K., Suzuki, T., Kohori, A., et al. (2008). Axle-less F1-ATPase rotates in the correct direction. Science 319, 955–958. doi:10.1126/science.1151343
Furuike, S., Nakano, M., Adachi, K., Noji, H., Kinosita, K., and Yokoyama, K. (2011). Resolving stepping rotation in Thermus thermophilus H(+)-ATPase/synthase with an essentially drag-free probe. Nat. Commun. 2, 233. doi:10.1038/ncomms1215
Groth, G., and Pohl, E. (2001). The structure of the chloroplast F1-ATPase at 3.2 A resolution. J. Biol. Chem. 276, 1345–1352. doi:10.1074/jbc.M008015200
Gu, J., Zhang, L., Zong, S., Guo, R., Liu, T., Yi, J., et al. (2019). Cryo-EM structure of the mammalian ATP synthase tetramer bound with inhibitory protein IF1. Science 364, 1068–1075. doi:10.1126/science.aaw4852
Guo, H., Suzuki, T., and Rubinstein, J. L. (2019). Structure of a bacterial ATP synthase. Elife 8, e43128. doi:10.7554/eLife.43128
Hahn, A., Vonck, J., Mills, D. J., Meier, T., and Kuhlbrandt, W. (2018). Structure, mechanism, and regulation of the chloroplast ATP synthase. Science 360, eaat4318. doi:10.1126/science.aat4318
Hornung, T., Martin, J., Spetzler, D., Ishmukhametov, R., and Frasch, W. D. (2011). Microsecond resolution of single-molecule rotation catalyzed by molecular motors. Methods Mol. Biol. 778, 273–289. doi:10.1007/978-1-61779-261-8_18
Imamura, H., Nakano, M., Noji, H., Muneyuki, E., Ohkuma, S., Yoshida, M., et al. (2003). Evidence for rotation of V1-ATPase. Proc. Natl. Acad. Sci. U. S. A. 100, 2312–2315. doi:10.1073/pnas.0436796100
Imamura, H., Takeda, M., Funamoto, S., Shimabukuro, K., Yoshida, M., and Yokoyama, K. (2005). Rotation scheme of V1-motor is different from that of F1-motor. Proc. Natl. Acad. Sci. U. S. A. 102, 17929–17933. doi:10.1073/pnas.0507764102
Itoh, H., Takahashi, A., Adachi, K., Noji, H., Yasuda, R., Yoshida, M., et al. (2004). Mechanically driven ATP synthesis by F1-ATPase. Nature 427, 465–468. doi:10.1038/nature02212
Kagawa, R., Montgomery, M. G., Braig, K., Leslie, A. G., and Walker, J. E. (2004). The structure of bovine F1-ATPase inhibited by ADP and beryllium fluoride. Embo J. 23, 2734–2744. doi:10.1038/sj.emboj.7600293
Kishikawa, J. I., Nakanishi, A., Furuta, A., Kato, T., Namba, K., Tamakoshi, M., et al. (2020). Mechanical inhibition of isolated V(o) from V/A-ATPase for proton conductance. Elife 9, e56862. doi:10.7554/eLife.56862
Kishikawa, J., Nakanishi, A., Nakano, A., Saeki, S., Furuta, A., Kato, T., et al. (2022). Structural snapshots of V/A-ATPase reveal the rotary catalytic mechanism of rotary ATPases. Nat. Commun. 13, 1213. doi:10.1038/s41467-022-28832-5
Kuhlbrandt, W. (2019). Structure and mechanisms of F-type ATP synthases. Annu. Rev. Biochem. 88, 515–549. doi:10.1146/annurev-biochem-013118-110903
Maher, M. J., Akimoto, S., Iwata, M., Nagata, K., Hori, Y., Yoshida, M., et al. (2009). Crystal structure of A3B3 complex of V-ATPase from Thermus thermophilus. EMBO J. 28, 3771–3779. doi:10.1038/emboj.2009.310
Menz, R. I., Walker, J. E., and Leslie, A. G. (2001). Structure of bovine mitochondrial F(1)-ATPase with nucleotide bound to all three catalytic sites: Implications for the mechanism of rotary catalysis. Cell 106, 331–341. doi:10.1016/s0092-8674(01)00452-4
Murphy, B. J., Klusch, N., Langer, J., Mills, D. J., Yildiz, Ö., and Kühlbrandt, W. (2019). Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F1-Fo coupling. Science 364, eaaw9128. doi:10.1126/science.aaw9128
Nakanishi, A., Kishikawa, J. I., Tamakoshi, M., Mitsuoka, K., and Yokoyama, K. (2018). Cryo EM structure of intact rotary H(+)-ATPase/synthase from Thermus thermophilus. Nat. Commun. 9, 89. doi:10.1038/s41467-017-02553-6
Nakano, A., Kishikawa, J., Mitsuoka, K., and Yokoyama, K. (2022). Mechanism of ATP hydrolysis dependent rotation of ATP synthases. bioRxiv. doi:10.1101/2022.12.23.521728
Nakano, M., Imamura, H., Toei, M., Tamakoshi, M., Yoshida, M., and Yokoyama, K. (2008). ATP hydrolysis and synthesis of a rotary motor V-ATPase from Thermus thermophilus. J. Biol. Chem. 283, 20789–20796. doi:10.1074/jbc.M801276200
Noji, H., Yasuda, R., Yoshida, M., and Kinosita, K. (1997). Direct observation of the rotation of F1-ATPase. Nature 386, 299–302. doi:10.1038/386299a0
Oot, R. A., Kane, P. M., Berry, E. A., and Wilkens, S. (2016). Crystal structure of yeast V1-ATPase in the autoinhibited state. Embo J. 35, 1694–1706. doi:10.15252/embj.201593447
Rees, D. M., Montgomery, M. G., Leslie, A. G., and Walker, J. E. (2012). Structural evidence of a new catalytic intermediate in the pathway of ATP hydrolysis by F1-ATPase from bovine heart mitochondria. Proc. Natl. Acad. Sci. U. S. A. 109, 11139–11143. doi:10.1073/pnas.1207587109
Roh, S. H., Stam, N. J., Hryc, C. F., Couoh-Cardel, S., Pintilie, G., Chiu, W., et al. (2018). The 3.5-A CryoEM structure of nanodisc-reconstituted yeast vacuolar ATPase Vo proton channel. Mol. Cell 69, 993–1004. doi:10.1016/j.molcel.2018.02.006
Saita, E., Suzuki, T., Kinosita, K., and Yoshida, M. (2015). Simple mechanism whereby the F1-ATPase motor rotates with near-perfect chemomechanical energy conversion. Proc. Natl. Acad. Sci. U. S. A. 112, 9626–9631. doi:10.1073/pnas.1422885112
Schep, D. G., Zhao, J., and Rubinstein, J. L. (2016). Models for the a subunits of the Thermus thermophilus V/A-ATPase and Saccharomyces cerevisiae V-ATPase enzymes by cryo-EM and evolutionary covariance. Proc. Natl. Acad. Sci. U. S. A. 113, 3245–3250. doi:10.1073/pnas.1521990113
Shimabukuro, K., Yasuda, R., Muneyuki, E., Hara, K. Y., Kinosita, K., and Yoshida, M. (2003). Catalysis and rotation of F1 motor: Cleavage of ATP at the catalytic site occurs in 1 ms before 40 degree substep rotation. Proc. Natl. Acad. Sci. U. S. A. 100, 14731–14736. doi:10.1073/pnas.2434983100
Sobti, M., Ishmukhametov, R., Bouwer, J. C., Ayer, A., Suarna, C., Smith, N. J., et al. (2019). Cryo-EM reveals distinct conformations of E. coli ATP synthase on exposure to ATP. Elife 8, e43864. doi:10.7554/eLife.43864
Sobti, M., Ueno, H., Noji, H., and Stewart, A. G. (2021). The six steps of the complete F(1)-ATPase rotary catalytic cycle. Nat. Commun. 12, 4690. doi:10.1038/s41467-021-25029-0
Spetzler, D., York, J., Daniel, D., Fromme, R., Lowry, D., and Frasch, W. (2006). Microsecond time scale rotation measurements of single F1-ATPase molecules. Biochemistry 45, 3117–3124. doi:10.1021/bi052363n
Suzuki, K., Mizutani, K., Maruyama, S., Shimono, K., Imai, F. L., Muneyuki, E., et al. (2016). Crystal structures of the ATP-binding and ADP-release dwells of the V(1) rotary motor. Nat. Commun. 7, 13235. doi:10.1038/ncomms13235
Tan, Y. Z., Keon, K. A., Abdelaziz, R., Imming, P., Schulze, W., Schumacher, K., et al. (2022). Structure of V-ATPase from citrus fruit. Structure 30, 1403–1410.e4. doi:10.1016/j.str.2022.07.006
Toei, M., Gerle, C., Nakano, M., Tani, K., Gyobu, N., Tamakoshi, M., et al. (2007). Dodecamer rotor ring defines H+/ATP ratio for ATP synthesis of prokaryotic V-ATPase from Thermus thermophilus. Proc. Natl. Acad. Sci. U. S. A. 104, 20256–20261. doi:10.1073/pnas.0706914105
Vasanthakumar, T., and Rubinstein, J. L. (2020). Structure and roles of V-type ATPases. Trends Biochem. Sci. 45, 295–307. doi:10.1016/j.tibs.2019.12.007
Weber, J., and Senior, A. E. (1997). Catalytic mechanism of F1-ATPase. Biochim. Biophys. Acta 1319, 19–58. doi:10.1016/s0005-2728(96)00121-1
Yasuda, R., Noji, H., Kinosita, K., and Yoshida, M. (1998). F1-ATPase is a highly efficient molecular motor that rotates with discrete 120 degree steps. Cell 93, 1117–1124. doi:10.1016/s0092-8674(00)81456-7
Yasuda, R., Noji, H., Yoshida, M., Kinosita, K., and Itoh, H. (2001). Resolution of distinct rotational substeps by submillisecond kinetic analysis of F1-ATPase. Nature 410, 898–904. doi:10.1038/35073513
Yokoyama, K., Hisabori, T., and Yoshida, M. (1989). The reconstituted alpha 3 beta 3 delta complex of the thermostable F1-ATPase. J. Biol. Chem. 264, 21837–21841. doi:10.1016/s0021-9258(20)88260-x
Yokoyama, K., and Imamura, H. (2005). Rotation, structure, and classification of prokaryotic V-ATPase. J. Bioenerg. Biomembr. 37, 405–410. doi:10.1007/s10863-005-9480-1
Yokoyama, K., Muneyuki, E., Amano, T., Mizutani, S., Yoshida, M., Ishida, M., et al. (1998). V-ATPase of Thermus thermophilus is inactivated during ATP hydrolysis but can synthesize ATP. J. Biol. Chem. 273, 20504–20510. doi:10.1074/jbc.273.32.20504
Yokoyama, K., Oshima, T., and Yoshida, M. (1990). Thermus thermophilus membrane-associated ATPase. Indication of a eubacterial V-type ATPase. J. Biol. Chem. 265, 21946–21950. doi:10.1016/s0021-9258(18)45830-9
Yoshida, M., Muneyuki, E., and Hisabori, T. (2001). ATP synthase - a marvellous rotary engine of the cell. Nat. Rev. Mol. Cell Biol. 2, 669–677. doi:10.1038/35089509
Zhao, J., Benlekbir, S., and Rubinstein, J. L. (2015). Electron cryomicroscopy observation of rotational states in a eukaryotic V-ATPase. Nature 521, 241–245. doi:10.1038/nature14365
Keywords: ATP synthase, V-ATPase, rotary motor protein, molecular motor, FOF1 ATP synthase
Citation: Yokoyama K (2023) Rotary mechanism of V/A-ATPases—how is ATP hydrolysis converted into a mechanical step rotation in rotary ATPases?. Front. Mol. Biosci. 10:1176114. doi: 10.3389/fmolb.2023.1176114
Received: 28 February 2023; Accepted: 13 April 2023;
Published: 24 April 2023.
Edited by:
Tibor Páli, ELKH, HungaryReviewed by:
Nicolas Vitale, Centre National de la Recherche Scientifique (CNRS), FranceKai Tittmann, University of Göttingen, Germany
Copyright © 2023 Yokoyama. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ken Yokoyama, eW9rb2tlbkBjYy5reW90by1zdS5hYy5qcA==
 Ken Yokoyama
Ken Yokoyama