
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mol. Biosci., 27 June 2023
Sec. Molecular Diagnostics and Therapeutics
Volume 10 - 2023 | https://doi.org/10.3389/fmolb.2023.1163089
This article is part of the Research TopicMethodological Advances of Mass Spectrometry-Based Techniques in Biomedical AnalysisView all 4 articles
 Weifang Zheng1*†
Weifang Zheng1*† Mingwei Wang2†
Mingwei Wang2† Xiaoyin Chai3
Xiaoyin Chai3 Fuzhen Pan1
Fuzhen Pan1 Meihui Xu1
Meihui Xu1 Yingchen Wang1
Yingchen Wang1 Liuhao Lan3
Liuhao Lan3 Feiran Hu1
Feiran Hu1 Zhe Zhang3
Zhe Zhang3 Zhu Chen1
Zhu Chen1The morbidity and mortality of colorectal cancer (CRC) have been increasing in recent years, and early detection of CRC can improve the survival rate of patients. RNA methylation plays crucial roles in many biological processes and has been implicated in the initiation of various diseases, including cancer. Serum contains a variety of biomolecules and is an important clinical sample for biomarker discovery. In this study, we developed a targeted metabolomics method for the quantitative analysis of nucleosides in human serum samples by using liquid chromatography with tandem mass spectrometry (LC-MS/MS). We successfully quantified the concentrations of nucleosides in serum samples from 51 healthy controls, 37 patients with colorectal adenomas, and 55 patients with CRC. The results showed that the concentrations of N6-methyladenosine (m6A), N1-methyladenosine (m1A), and 3-methyluridine (m3U) were increased in patients with CRC, whereas the concentrations of N2-methylguanosine (m2G), 2′-O-methyluridine (Um), and 2′-O-methylguanosine (Gm) were decreased in patients with CRC, compared with the healthy controls and patients with colorectal adenomas. Moreover, the levels of 2′-O-methyluridine and 2′-O-methylguanosine were lower in patients with colorectal adenomas than those in healthy controls. Interestingly, the levels of Um and Gm gradually decreased in the following order: healthy controls to colorectal adenoma patients to CRC patients. These results revealed that the aberrations of these nucleosides were tightly correlated to colorectal adenomas and CRC. In addition, the present work will stimulate future investigations about the regulatory roles of these nucleosides in the initiation and development of CRC.
The morbidity and mortality rate of colorectal cancer (CRC) have been increasing worldwide in recent years. CRC is the second leading cause of cancer-related mortality worldwide, posing a serious threat to human health and causing a huge social burden (Bray et al., 2018; Hyuna et al., 2021). By the early detection of colorectal adenomas and colorectal cancer through screening, the morbidity and mortality of CRC can be successfully reduced (Shaukat et al., 2021). Screening techniques for CRC and colorectal adenomas include the following four types: 1) fecal-based examination, 2) imaging examination, 3) endoscopy, and 4) liquid biopsy. Nevertheless, due to the invasive nature of the procedures and poor patient compliance, large-scale screening is difficult to perform (Jaaks et al., 2022; Zhou et al., 2022).
Nucleic acid modification is defined as the chemical modification of nucleobases and the ribose of DNA and RNA. Most often, these alterations do not alter DNA or RNA sequences, but they do affect gene expression and several physiological processes (Roundtree et al., 2017; Deng et al., 2022). To date, more than 170 RNA modifications have been identified, each of which plays a crucial role in numerous life processes. RNA methylation modification plays an essential regulatory role in numerous biological processes and is implicated in the incidence and progression of various diseases, including cancer (Ontiveros et al., 2019; Isaia and Tony, 2020). The most abundant internal modification in mRNA is N6-methyladenosine (m6A) (Liu et al., 2020), which was discovered in RNA as early as the 1970s (Dubin and Taylor, 1975; Perry et al., 1975). As a typical product of RNA methylation, with the discovery of m6A-associated methyltransferases (e.g., METTL3), demethyltransferases (e.g., FTO), and reader proteins (e.g., YTHDC1) (Liu et al., 2014; Zhang et al., 2015; Yang et al., 2020), there is growing evidence that the m6A level is dynamic in vivo and regulates RNA splicing, RNA stabilization, and translation (Shi et al., 2020; Sung and Yoon, 2020). Moreover, aberrant m6A levels are tightly associated with the onset and progression of various types of cancers (Wang et al., 2020; Jiang et al., 2021).
There are four types of nucleosides in RNA, namely, adenosine (A), guanosine (G), cytidine (C), and uridine (U). Methylated nucleosides can be produced when nitrogen or carbon atoms in the nucleobase are modified by methylation. Moreover, hydrogen in the 2′-hydroxyl group in a ribose substituted by the methyl group produced 2′-O-methylated nucleosides. According to a recent study, alterations of the levels of N1-methyladenosine (m1A) and m6A in human serum and urine are associated with increased risks of colorectal and gastric cancer (Guo et al., 2021; Hu et al., 2021). Additionally, decreased concentrations of N1-methylguanosine (m1G), 2′-O-methylcytidine (Cm), and 2′-O-methyluridine (Um) in serum may be associated with breast cancer (Fang et al., 2021). As a result, these methylated nucleosides may be indicators for the early detection of various types of cancers.
Most commonly, serum is used in biomarker discovery, is easy to obtain, and is less traumatic (Huang et al., 2018; Shen et al., 2021). The presence of methylated nucleosides in urine and serum has previously been linked to the development of cancer in a number of studies (Djukovic et al., 2010; Guo C et al., 2018; Fang et al., 2022; Zhang et al., 2022). Liquid chromatography with tandem mass spectrometry (LC-MS/MS) has the benefit of extremely high sensitivity to meet the efficient qualitative analysis and the accurate quantitative analysis of metabolites in complex biological samples (Guo et al., 2016; Guo M et al., 2018; Zheng et al., 2020). With the LC-MS/MS method established in this study, 11 nucleosides were quantified in 37 patients with CRC, 55 patients with colorectal adenomas, and 51 healthy controls. We demonstrated the differences of these nucleosides among these groups and evaluated the potential of these nucleosides as biomarkers of colorectal adenomas and CRC.
Acetonitrile of chromatographic grade was purchased from Merck KGaA (Darmstadt, Germany). A solution of formic acid (HCOOH) was purchased from Fluka (Muskegon, USA). Ammonium formate and malic acid were obtained from Sigma-Aldrich (St Louis, MO, United States). Adenosine, N6-methyladenosine, N1-methyladenosine, 2′-O-methylguanosine (Gm), N1-methylguanosine, N2-methylguanosine (m2G), 2′-O-methylcytidine, uridine, 3-methyluridine (m3U), 5-methyluridine (m5U), and 2′-O-methyluridine and isotopically labeled standards, including [13C5] A, [D3] m6A, [D3] m1A, [D3] Cm, [13CD3] m5C, [13C5N2] U, [D3] Um, [13C5] m5U, and [D6] m2,2G, were purchased from Toronto Research Chemicals (Toronto, Canada). In order to purify water, a Milli-Q water purification device was used (Millipore, Milford, MA, United States).
The ACQUITY UPLC system (Waters, Milford, MA, United States) was used to analyze nucleosides and methylated nucleosides. For the MS analysis, an AB SCIEX 4000 QTRAP mass spectrometer (Foster City, CA, United States) was used. A Waters BEH Amide column (2.1 × 100 mm; 1.7 µm) was used for chromatographic separation. Data were acquired using the multiple-reaction monitoring (MRM) mode in the electrospray ionization (ESI) positive-ion mode. Data collection and processing were conducted using Analyst 1.6.3 software.
In this study, colorectal cancer patients, colorectal adenoma patients, and healthy controls were recruited from the Lanxi Hospital of Traditional Chinese Medicine. This study complies with the Helsinki Declaration of the World Medical Association. A total of 37 colorectal cancer patients, 55 colorectal adenoma patients, and 51 healthy controls were studied. Information about these volunteers can be found in Supplementary Table S1. Serum was collected early in the morning and was stored at −80°C until analysis.
The serum sample of 100 μL was placed into a 1.5-mL centrifuge tube, thawed in ice, and 10 μL of the internal standard isotope (IS) was added. To thoroughly remove the protein, 330 μL of pre-cooled methanol/acetonitrile (2:1, v/v) was added, and the mixture was vortexed for 1 minute and placed at −20°C for 2 h. Extraction was performed by centrifuging the obtained mixture at 13,000 rpm at 4°C. A volume of 352 μL of the supernatant was taken and evaporated under vacuum. A volume of 80 μL of water/acetonitrile (9:1, v/v) was used to dissolve the residues. Finally, the solution was analyzed by LC-MS/MS.
The mobile phase A was a solution of 10 mM ammonium formate, 0.2% formic acid, and 0.05 mM malic acid in water. The mobile phase B was composed of a solution containing 2 mM ammonium formate, 0.2% formic acid, and 0.05 mM malic acid in acetonitrile. The 11 analytes were separated at a flow rate of 0.3 mL/min using gradient elution, shown as follows: 0–5.5 min, 5% A; 5.5–7 min, 5%–8% A; 7–9 min, 8%–15% A; 9–9.5 min, 15%–17% A; 9.5–11 min, 17%–20% A; 11–11.5 min, 20%–5% A; and 11.5–15 min, 5% A. Each sample was tested at an injection volume of 5 µL.
The voltage of 5.5 kV was used in the ion spray process. The ion source (TEM) was set at 550°C. Both ion source gases (GS1 and GS2) were set as 50 psi. The pressure of the curtain gas (CUR) was set at 40 psi. The declustering potential (DP), entry potential (EP), collision energy (CE), and collision cell exit potential (CXP) were optimized, as shown in Supplementary Table S2.
GraphPad Prism 8 was used to perform the statistical analysis. The levels of nucleosides in 51 healthy controls, 55 patients with colorectal adenomas, and 37 patients with colorectal cancer were compared using the Mann–Whitney test. The statistics were defined as significant when the p-value was less than 0.05.
The chemical structures of nucleosides are shown in Figure 1. In addition, a BEH Amide column (2.1 × 100 mm, 1.7 µm) was chosen. As depicted in Figure 2, the analysis was completed in 12 min. The target analytes were perfectly separated, demonstrating that the approach has high separation efficiency, fast analysis speed, and high throughput. Hence, it is suited for the analysis of a large number of clinical samples.
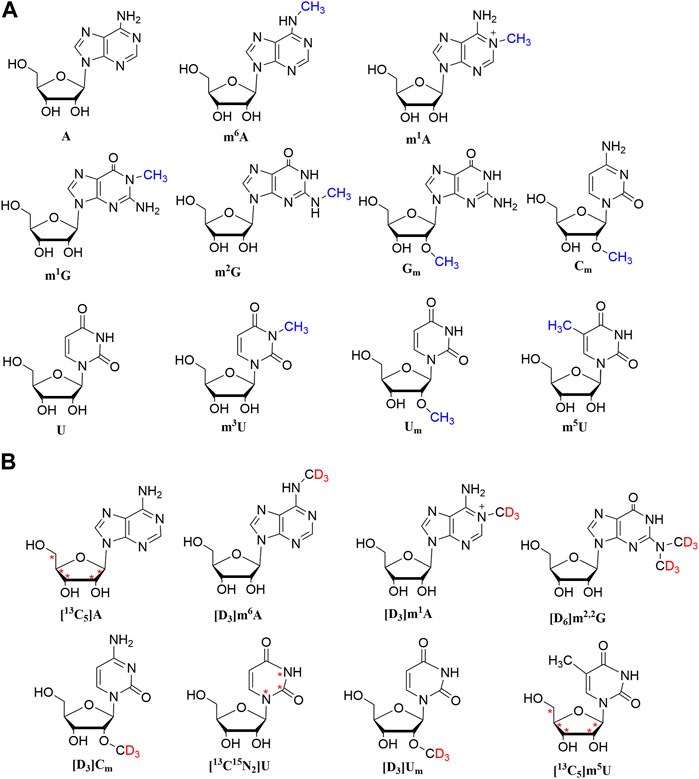
FIGURE 1. Chemical structures of A, m1A, m6A, U, m3U, m5U, Um, Cm, m2G, m1G, and Gm and their stable isotope-labeled internal standards. Asterisk (*) designates the of 13C labeling.
To optimize the MRM parameters, a direct infusion of the standard solution into the mass spectrometer by using a peristaltic pump was performed. In full-scan ESI-MS, [M + H]+ ions at m/z 268.1 were found for A. Abundant [M + H]+ and M+ ions at m/z 282.1 were detected in m6A and m1A, respectively. For m1G, m2G, and Gm, abundant [M + H]+ ions at m/z 298.1 were observed. For m3U, m5U, and Um, abundant [M + H]+ ions at m/z 259.1 were observed. In addition, [M + H]+ ions at m/z 245.1 for U and m/z 258.1 for Cm were observed. Subsequently, a collision-induced dissociation (CID) was carried out. For A, CID data showed that the most common fragment ion was at m/z 136.0. For m1A and m6A, the most abundant fragment ions were both at m/z 150.0, and for Gm, the most abundant fragment ion was at m/z 152.0. For m1G and m2G, the most abundant fragment ions were both at m/z 166.0. For m3U and m5U, the most abundant fragment ions were both at m/z 127.0. Furthermore, the most abundant fragment ions for U and Cm were found to be at m/z 112.0 and m/z 113.0, respectively. Then, the MRM parameters of these nucleosides and their isotope-labeled internal standards were optimized, as shown in Supplementary Table S2.
By establishing the LC-MS/MS method described previously, we detected these nucleosides in serum samples from 37 colorectal cancer patients, 55 colorectal adenoma patients, and 51 healthy volunteers. The findings indicated that in all human serum samples, m3U, Um, m6A, m5U, U, A, Cm, m1G, Gm, m2G, and m1A were detected. As shown in Figure 3, the retention times of m3U, Um, m6A, m5U, U, A, Cm, m1G, Gm, m2G, and m1A were 2.01, 2.25, 3.65, 4.14, 4.96, 5.62, 6.49, 7.80, 8.58, 10.11, and 11.68 min, respectively. Notably, the chromatographic retention times of Um, m6A, m5U, U, A, Cm, and m1A were consistent with their corresponding isotopically labeled internal standards. Moreover, m3U, m2G, Gm, and m1G were confirmed by comparing their retention time with that of their standards (Supplementary Figure S1). In summary, the presence of these nucleosides in human serum was confirmed.
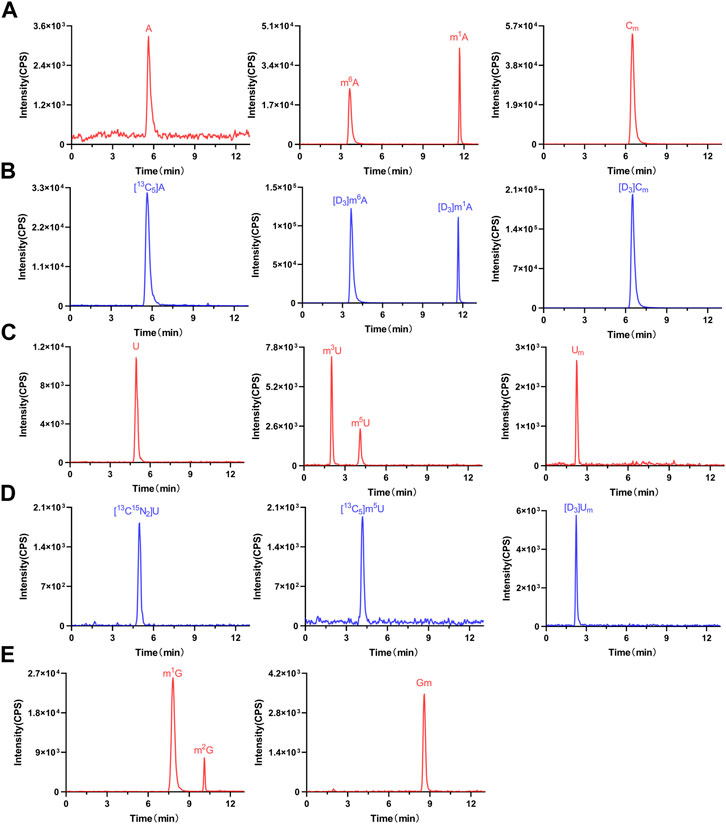
FIGURE 3. Representative MRM chromatograms of (A) A, m1A, m6A, Cm, (C) U, m3U, m5U, Um, (E) m1G, m2G, and Gm and (B) (D) spiked isotope-labeled internal standards in a serum sample.
To determine whether there were any differences in the levels of these nucleosides between colorectal adenoma patients, CRC patients, and healthy controls, we quantified the levels of these nucleosides. First, the calibration curves were established, and the results showed that good linearity (R2 > 0.999) was achieved (Table 1). The concentrations of nucleosides in serum were calculated according to the linear equations. For the quantification of m3U, [13C5] m5U was used as the internal standard. Also, for m2G, Gm, and m1G, [D6] m2,2G was used as the internal standard. A comparison of the mean concentrations of A, m1A, m6A, U, m3U, m5U, Um, Cm, m2G, m1G, and Gm in serum samples was shown in Supplementary Table S3. The levels of A, m1A, m6A, U, m3U, m5U, Um, Cm, m1G, m2G, and Gm in the serum samples from healthy controls were within the range of 0.25–1.93, 24.93–127.80, 6.97–37.71, 2,170.83–9,368.89,6.48–62.79, 141.31–356.98, 11.45–64.80, 8.08–54.77, 14.02–38.11, 4.33–23.96, and 7.85–25.54 nM, respectively. The mean concentrations were 0.66, 64.70, 16.62, 4,803.45, 29.23, 225.57, 31.17, 25.69, 25.87, 14.04, and 14.80 nM, respectively. In the serum from patients with colorectal adenomas, the levels of A, m1A, m6A, U, m3U, m5U, Um, Cm, m1G, m2G, and Gm were in the range of 0.18–1.18, 25.47–168.41, 6.36–51.35, 2039.52–7,855.11,4.36–76.20, 150.96–316.67, 6.76–55.78, 7.03–47.95, 12.49–43.36, 5.64–22.37, and 8.38–21.12 nM, respectively. The mean concentrations were 0.46, 77.32, 21.43, 4,634.80, 28.80, 210.41, 24.17, 26.89, 24.66, 13.85, and 13.18 nM, respectively. The concentrations of A, m1A, m6A, U, m3U, m5U, Um, Cm, m1G, m2G, and Gm in the serum from patients with CRC were in the range of 0.08–0.82, 32.10–200.98, 8.01–69.87, 2,389.30–7,951.33, 24.93–119.88, 106.71–305.06, 5.02–18.67, 3.70–49.25, 13.93–49.65, 4.09–18.86, and 7.93–14.39 nM, respectively. The mean concentrations were 0.39, 105.85, 40.09, 4,377.36, 51.31, 206.68, 10.72, 24.75, 25.06, 9.67, and 11.09 nM, respectively.
As shown in Figure 4, the levels of m1A, m6A, and m3U in the serum from patients with CRC were considerably higher than those of the healthy controls (p < 0.0001 for m1A, p < 0.0001 for m6A, and p < 0.0001 for m3U) and those of colorectal adenoma patients (p < 0.01 for m1A, p < 0.0001 for m6A, and p < 0.0001 for m3U). In contrast, the concentrations of Um, Gm, and m2G in CRC patients were considerably lower than those of the healthy controls (p < 0.05 for A, p < 0.0001 for Um, p < 0.001 for Gm, and p < 0.0001 for m2G) and those of colorectal adenoma patients (p < 0.0001 for Um, p < 0.01 for Gm, and p < 0.0001 for m2G). Compared with healthy controls, the level of A was also decreased in the serum from CRC patients, whereas there was no difference between colorectal adenoma patients and CRC patients. Moreover, the levels of Um and Gm were both decreased in colorectal adenoma patients, compared with the healthy controls. Interestingly, the concentrations of Um and Gm decreased gradually from the healthy control group to colorectal adenoma patients to CRC patients. However, there were no differences in the levels of m5U, U, m1G, and Cm between these groups (p > 0.05).
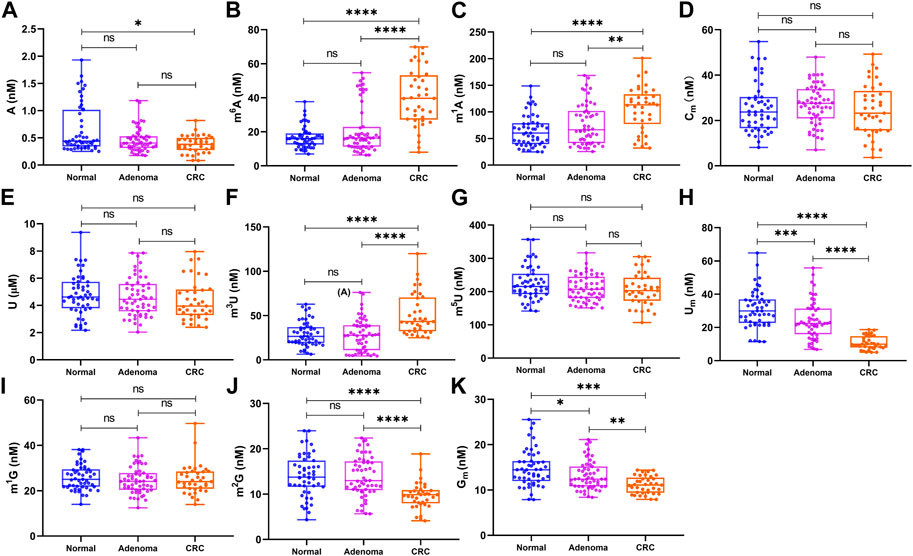
FIGURE 4. Concentration of (A) A, (B) m6A, (C) m1A, (D) Cm, (E) U, (F) m3U, (G) m5U, (H) Um, (I) m1G, (J) m2G, and (K) Gm in the serum samples and statistical analysis. ****, p < 0.0001; ***, p < 0.001; **, p < 0.01; *, p < 0.05; ns, p > 0.05.
In addition, receiver operating characteristic (ROC) curve analysis was carried out to evaluate the capacity of these nucleosides to differentiate patients with colorectal adenomas and patients with CRC from the healthy controls. As shown in Figure 5A, for colorectal adenoma patients and normal controls, the area under the curve (AUC) of Um and Gm were 0.69 and 0.63, respectively. For CRC patients and normal controls (Figure 5B), the AUC of m1A, m6A, A, m3U, Um, m2G, and Gm were 0.79, 0.90, 0.65, 0.81, 0.97, 0.79, and 0.74, respectively. For CRC patients and colorectal adenoma patients (Figure 5C), the AUC of m1A, m6A, m3U, Um, m2G, and Gm were 0.69, 0.80, 0.78, 0.89, 0.79, and 0.70, respectively. These results indicate that there is a close association between the levels of these nucleosides in the serum and the incidence of colorectal adenomas and CRC.
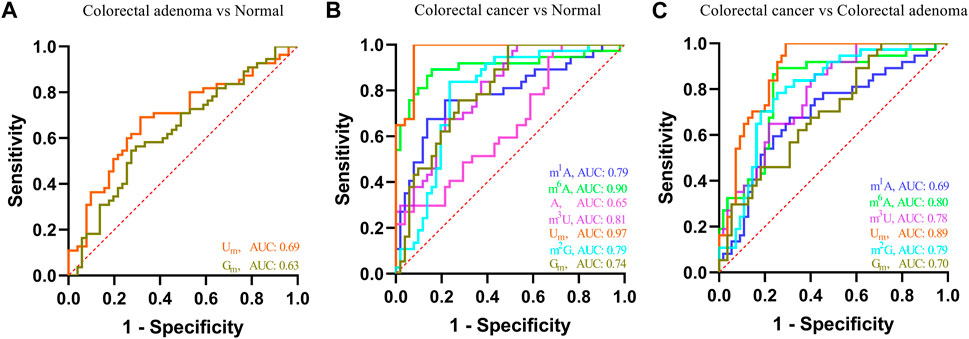
FIGURE 5. ROC analysis for these nucleosides in the serum. (A) Colorectal adenoma vs. normal control, (B) colorectal cancer vs. normal control, and (C) colorectal cancer vs. colorectal adenoma.
Both the morbidity and mortality rates of colorectal cancer are on the rise around the globe. Timely detection of specific biomarkers plays a vital role in early diagnosis, prevention, and monitoring during treatment. According to the findings of this study, an increase in m6A, m1A, and m3U in the serum and a decrease in A, Um, Gm, and m2G in the serum, may have great potential as non-invasive biomarkers for the early detection of CRC. Notably, decreased levels of Um and Gm may serve as non-invasive biomarkers for the early detection of colorectal adenomas.
In the present study, a targeted metabolomics method for the quantitative analysis of nucleosides in human serum samples by using LC-MS/MS was developed. We measured the concentration of A, m1A, m6A, U, m3U, m5U, Um, Cm, m1G, m2G, and Gm in the serum samples from 51 healthy volunteers, 37 CRC patients, and 55 colorectal adenoma patients. CRC patients had significantly higher levels of m6A, m1A, and m3U than the colorectal adenomas patients and healthy volunteers. The levels of Um, Gm, and m2G in CRC patients were significantly lower than those in the colorectal adenomas patients and healthy volunteers. Interestingly, there was a gradual decrease in Um and Gm concentrations from healthy controls to colorectal adenoma patients to CRC patients. All these results suggested that the levels of m6A, m1A, m3U, A, Um, Gm, and m2G in the serum were expected to be potential non-invasive biomarkers for the early detection of CRC, and the levels of Um and Gm might be non-invasive biomarkers for the early detection of colorectal adenomas. In addition, these results implied that these nucleosides might play an important role in the development and progression of colorectal adenomas and CRC.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
The studies involving human participants were reviewed and approved by the Institutional Review Board of Medical Research, the Lanxi Hospital of Traditional Chinese Medicine. The patients/participants provided their written informed consent to participate in this study. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
WZ designed the study; MW, XC, MX, YW, LL, and ZZ performed the experiments; FH and ZC collected the serum samples; MW, FP, and WZ analyzed and interpreted the data; MW and FP wrote the manuscript. All authors commented and approved the final manuscript.
This study was supported by the Zhejiang Provincial Medical Health Science and Technology Plan Project (2023XY205).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2023.1163089/full#supplementary-material
Bray, F., Ferlay, J., Soerjomataram, I., Siegel, R. L., Torre, L. A., and Jemal, A. (2018). Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68, 394–424. doi:10.3322/caac.21492
Deng, L. J., Deng, W. Q., Fan, S. R., Chen, M. F., Qi, M., Lyu, W. Y., et al. (2022). m6A modification: recent advances, anticancer targeted drug discovery and beyond. Mol. Cancer 21, 52. doi:10.1186/s12943-022-01510-2
Djukovic, D., Baniasadi, H. R., Kc, R., Hammoud, Z., and Raftery, D. (2010). Targeted serum metabolite profiling of nucleosides in esophageal adenocarcinoma. Rapid Commun. Mass Spectrom. 24, 3057–3062. doi:10.1002/rcm.4739
Dubin, D. T., and Taylor, R. H. (1975). The methylation state of poly A-containing messenger RNA from cultured hamster cells. Nucleic Acids Res. 2, 1653–1668. doi:10.1093/nar/2.10.1653
Fang, Z. H., Hu, Y. Q., Chen, J. N., Xu, K. L., Wang, K. L., Zheng, S., et al. (2021). Mass spectrometry-based targeted serum monomethylated ribonucleosides profiling for early detection of breast cancer. Front. Mol. Biosci. 8, 741603. doi:10.3389/fmolb.2021.741603
Fang, Z. H., Hu, Y. Q., Hong, X. J., Zhang, X. X., Pan, T., Pan, C., et al. (2022). Simultaneous determination of methylated nucleosides by HILIC-MS/MS revealed their alterations in urine from breast cancer patients. Metabolites 12, 973. doi:10.3390/metabo12100973
Guo, C., Hu, Y., Cao, X., and Wang, Y. (2021). HILIC-MS/MS for the determination of methylated adenine nucleosides in human urine. Anal. Chem. 93, 17060–17068. doi:10.1021/acs.analchem.1c03829
Guo, C., Li, X., Wang, R., Yu, J., Ye, M., Mao, L., et al. (2016). Association between oxidative DNA damage and risk of colorectal cancer: Sensitive determination of urinary 8-Hydroxy-2'-deoxyguanosine by UPLC-MS/MS analysis. Sci. Rep. 6, 32581. doi:10.1038/srep32581
Guo, C., Xie, C., Chen, Q., Cao, X., Guo, M., Zheng, S., et al. (2018). A novel malic acid-enhanced method for the analysis of 5-methyl-2'-deoxycytidine, 5-hydroxymethyl-2'-deoxycytidine, 5-methylcytidine and 5-hydroxymethylcytidine in human urine using hydrophilic interaction liquid chromatography-tandem mass spectrometry. Anal. Chim. Acta 1034, 110–118. doi:10.1016/j.aca.2018.06.081
Guo, M., Zhang, L., Du, Y., Du, W., Liu, D., Guo, C., et al. (2018). Enrichment and quantitative determination of 5-(Hydroxymethyl)-2'-deoxycytidine, 5-(Formyl)-2'-deoxycytidine, and 5-(Carboxyl)-2'-deoxycytidine in human urine of breast cancer patients by magnetic hyper-cross-linked microporous polymers based on polyionic liquid. Anal. Chem. 90, 3906–3913. doi:10.1021/acs.analchem.7b04755
Hu, Y., Fang, Z., Mu, J., Huang, Y., Zheng, S., Yuan, Y., et al. (2021). Quantitative analysis of methylated adenosine modifications revealed increased levels of N (6)-methyladenosine (m(6)A) and N (6),2'-O-dimethyladenosine (m(6)Am) in serum from colorectal cancer and gastric cancer patients. Front. Cell Dev. Biol. 9, 694673. doi:10.3389/fcell.2021.694673
Huang, J. Q., Weinstein, S. J., Moore, S. C., Derkach, A., Hua, X., Liao, L. M., et al. (2018). Serum metabolomic profiling of all-cause mortality: A prospective analysis in the alpha-tocopherol, beta-carotene cancer prevention (atbc) study cohort. Am. J. Epidemiol. 187, 1721–1732. doi:10.1093/aje/kwy017
Hyuna, S., Jacques, F., Rebecca, L. S., Mathieu, L., Isabelle, S., Ahmedin, J., et al. (2021). Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71, 209–249. doi:10.3322/caac.21660
Isaia, B., and Tony, K. (2020). Role of RNA modifications in cancer. Nat. Rev. Cancer 20, 303–322. doi:10.1038/s41568-020-0253-2
Jaaks, P., Coker, E. A., Vis, D. J., Edwards, O., Carpenter, E. F., Leto, S. M., et al. (2022). Effective drug combinations in breast, colon and pancreatic cancer cells. Nature 603, 166–173. doi:10.1038/s41586-022-04437-2
Jiang, X., Liu, B., Nie, Z., Duan, L., Xiong, Q., Jin, Z., et al. (2021). The role of m6A modification in the biological functions and diseases. Signal Transduct. Target Ther. 6, 74. doi:10.1038/s41392-020-00450-x
Liu, J. Z., Yue, Y. N., Han, D. L., Wang, X., Fu, Y., Zhang, L., et al. (2014). A METTL3-METTL14 complex mediates mammalian nuclear RNA N-6-adenosine methylation. Nat. Chem. Biol. 10, 93–95. doi:10.1038/nchembio.1432
Liu, J., Do, X. Y., Chen, C. Y., Chen, C., Liu, C., Xu, M. M., et al. (2020). N-6-methyladenosine of chromosome-associated regulatory RNA regulates chromatin state and transcription. Science 367, 580–586. doi:10.1126/science.aay6018
Ontiveros, R. J., Stoute, J., and Liu, K. F. (2019). The chemical diversity of RNA modifications. Biochem. J. 476, 1227–1245. doi:10.1042/BCJ20180445
Perry, R. P., Kelley, D. E., Friderici, K., and Rottman, F. (1975). The methylated constituents of L cell messenger RNA: Evidence for an unusual cluster at the 5' terminus. Cell 4, 387–394. doi:10.1016/0092-8674(75)90159-2
Roundtree, I. A., Evans, M. E., Pan, T., and He, C. (2017). Dynamic RNA modifications in gene expression regulation. Cell 169, 1187–1200. doi:10.1016/j.cell.2017.05.045
Shaukat, A., Kahi, C. J., Burke, C. A., Rabeneck, L., Sauer, B. G., and Rex, D. K. (2021). ACG clinical guidelines: Colorectal cancer screening 2021. Am. J. Gastroenterol. 116, 458–479. doi:10.14309/ajg.0000000000001122
Shen, F., Xiong, Y., Zhang, L., Li, H., Zhao, H., Liu, X., et al. (2021). Rapid sample preparation workflow for serum sample analysis with different mass spectrometry acquisition strategies. Anal. Chem. 93, 1578–1585. doi:10.1021/acs.analchem.0c03985
Shi, H. H., Chai, P. W., Jia, R. B., and Fan, X. Q. (2020). Novel insight into the regulatory roles of diverse RNA modifications: Re-Defining the bridge between transcription and translation. Mol. Cancer 19, 78. doi:10.1186/s12943-020-01194-6
Sung, H. B., and Yoon, K. K. (2020). The emerging role of RNA modifications in the regulation of mRNA stability. Exp. Mol. Med. 52, 400–408. doi:10.1038/s12276-020-0407-z
Wang, Q., Chen, C., Ding, Q. Q., Zhao, Y., Wang, Z. D., Chen, J. J., et al. (2020). METTL3-mediated m(6)A modification of HDGF mRNA promotes gastric cancer progression and has prognostic significance. Gut 69, 1193–1205. doi:10.1136/gutjnl-2019-319639
Yang, X., Zhang, S., He, C., Xue, P., Zhang, L., He, Z., et al. (2020). METTL14 suppresses proliferation and metastasis of colorectal cancer by down-regulating oncogenic long non-coding RNA XIST. Mol. Cancer 19, 46. doi:10.1186/s12943-020-1146-4
Zhang, G. Q., Huang, H., Liu, D., Cheng, Y., Liu, X. L., Zhang, W. X., et al. (2015). N-6-Methyladenine DNA modification in Drosophila. Cell 161, 893–906. doi:10.1016/j.cell.2015.04.018
Zhang, X. X., Hu, Y. Q., Hong, X. J., Wang, M. W., Fang, Z. H., Cao, X. J., et al. (2022). Determination of adenosine and its modifications in urine and plasma from breast cancer patients by hydrophilic interaction liquid chromatography-tandem mass spectrometry. J. Chromatogr. B-Analytical Technol. Biomed. Life Sci. 1209, 123428. doi:10.1016/j.jchromb.2022.123428
Zheng, F., Zhao, X., Zeng, Z., Wang, L., Lv, W., Wang, Q., et al. (2020). Development of a plasma pseudotargeted metabolomics method based on ultra-high-performance liquid chromatography-mass spectrometry. Nat. Protoc. 15, 2519–2537. doi:10.1038/s41596-020-0341-5
Keywords: colorectal cancer, colorectal adenomas, LC-MS/MS, nucleosides, serum, biomarker
Citation: Zheng W, Wang M, Chai X, Pan F, Xu M, Wang Y, Lan L, Hu F, Zhang Z and Chen Z (2023) Targeted metabolomics analysis of nucleosides and the identification of biomarkers for colorectal adenomas and colorectal cancer. Front. Mol. Biosci. 10:1163089. doi: 10.3389/fmolb.2023.1163089
Received: 10 February 2023; Accepted: 29 March 2023;
Published: 27 June 2023.
Edited by:
Kezhi Jiang, Hangzhou Normal University, ChinaReviewed by:
Yunfeng Chai, Tea Research Institute (CAAS), ChinaCopyright © 2023 Zheng, Wang, Chai, Pan, Xu, Wang, Lan, Hu, Zhang and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Weifang Zheng, emhlbmd3ZWlmYW5nMTk3MkAxNjMuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.