- 1Department of Microbiology, Ludwik Rydygier Collegium Medicum in Bydgoszcz, Nicolaus Copernicus University, Toruń, Poland
- 2Department of Food Hygiene and Consumer Health, Wrocław University of Environmental and Life Sciences, Wrocław, Poland
Listeria monocytogenes causes listeriosis, a disease characterized by a high mortality rate (up to 30%). Since the pathogen is highly tolerant to changing conditions (high and low temperature, wide pH range, low availability of nutrients), it is widespread in the environment, e.g., water, soil, or food. L. monocytogenes possess a number of genes that determine its high virulence potential, i.e., genes involved in the intracellular cycle (e.g., prfA, hly, plcA, plcB, inlA, inlB), response to stress conditions (e.g., sigB, gadA, caspD, clpB, lmo1138), biofilm formation (e.g., agr, luxS), or resistance to disinfectants (e.g., emrELm, bcrABC, mdrL). Some genes are organized into genomic and pathogenicity islands. The islands LIPI-1 and LIPI-3 contain genes related to the infectious life cycle and survival in the food processing environment, while LGI-1 and LGI-2 potentially ensure survival and durability in the production environment. Researchers constantly have been searching for new genes determining the virulence of L. monocytogenes. Understanding the virulence potential of L. monocytogenes is an important element of public health protection, as highly pathogenic strains may be associated with outbreaks and the severity of listeriosis. This review summarizes the selected aspects of L. monocytogenes genomic and pathogenicity islands, and the importance of whole genome sequencing for epidemiological purposes.
Introduction
General characteristics of Listeria monocytogenes
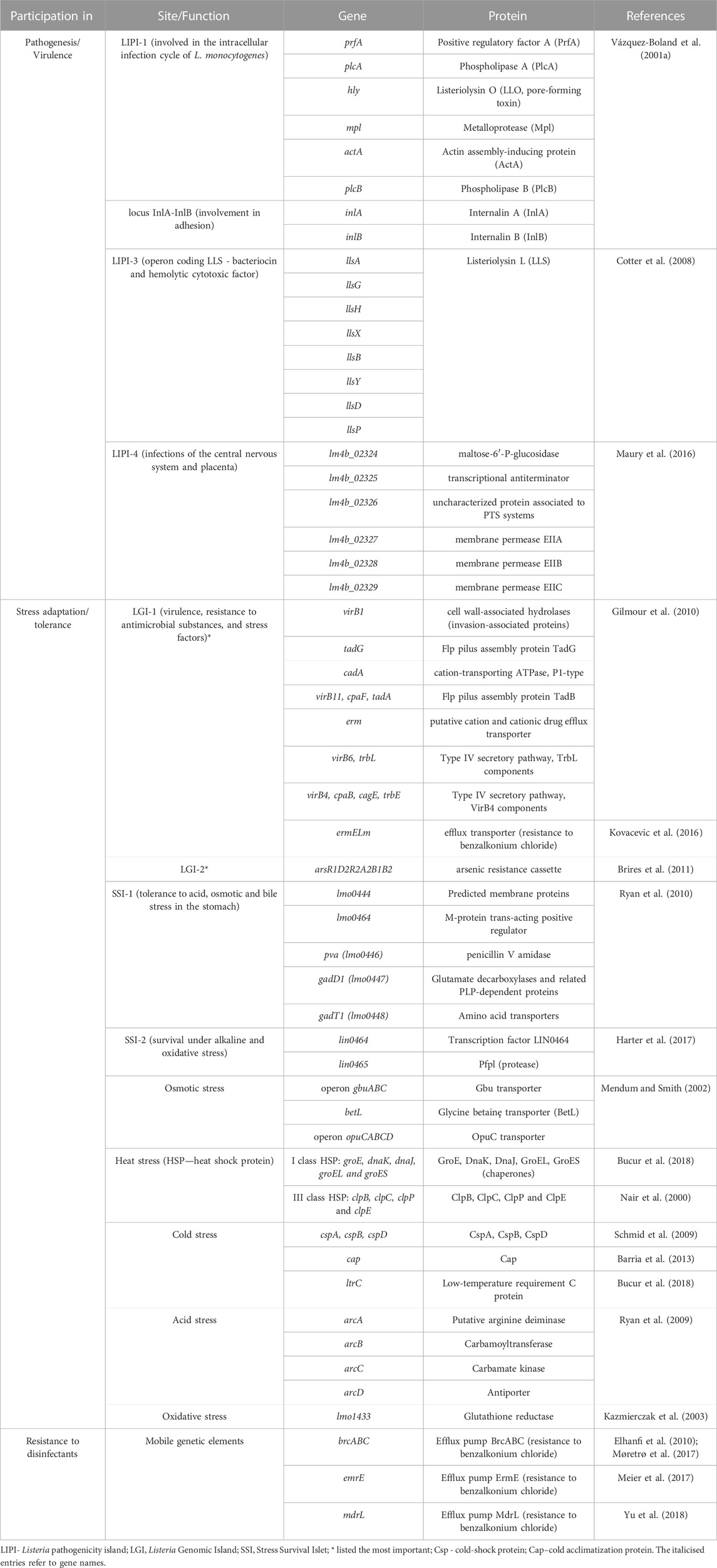
L. monocytogenes are Gram-positive, non-spore-forming, relatively anaerobic rods (Gandhi and Chikindas, 2007). L. monocytogenes easily adapt to environmental conditions (Muskalska and Szymczak, 2015), can grow in a wide range of temperatures (0°C–45°C), pH (4.3–9.6), tolerate high salt concentrations (up to 10.0% NaCl) and low water activity (Aw to 0.90) (Gandhi and Chikindas, 2007; Wiktorczyk-Kapischke et al., 2021). Adaptation to unfavorable environmental conditions is associated with the expression of many genes (Begley et al., 2005; Ryan et al., 2009; Kocaman and Sarimehmetoğlu, 2016; Wiktorczyk-Kapischke et al., 2021). Response to stressful conditions ensure genes localized on SSI-1 (stress survival islet-1): acid stress, osmotic stress, bile stress in the stomach (Ryan et al., 2010) and SSI-2 (stress survival islet-2): alkaline and oxidative stress (Harter et al., 2017). Table 1 presents a summary of the most important genes involved in pathogenesis and adaptation to stress.
L. monocytogenes were divided into three evolutionary lineages, 14 serotypes, grouped into four serogroups (1/2a-3a, 1/2b-3b-7, 1/2c-3c, and 4b-4d-4e) (Doumith et al., 2004) (Table 2). In 2019, Yin et al. (2019) have described serotype 4h, HSL-II hybrid sublineage. The results reported by Maury et al. (2016) distinguished three categories of the most common clones of L. monocytogenes, e.g., CC1, CC2, CC4 and CC6—associated with infection; CC9 and CC12—food related clones; intermediate clones. Additionally, L. monocytogenescan be divided into four epidemic clones - ECI, ECII, ECIII and ECIV (Kathariou, 2003; Chen et al., 2007).
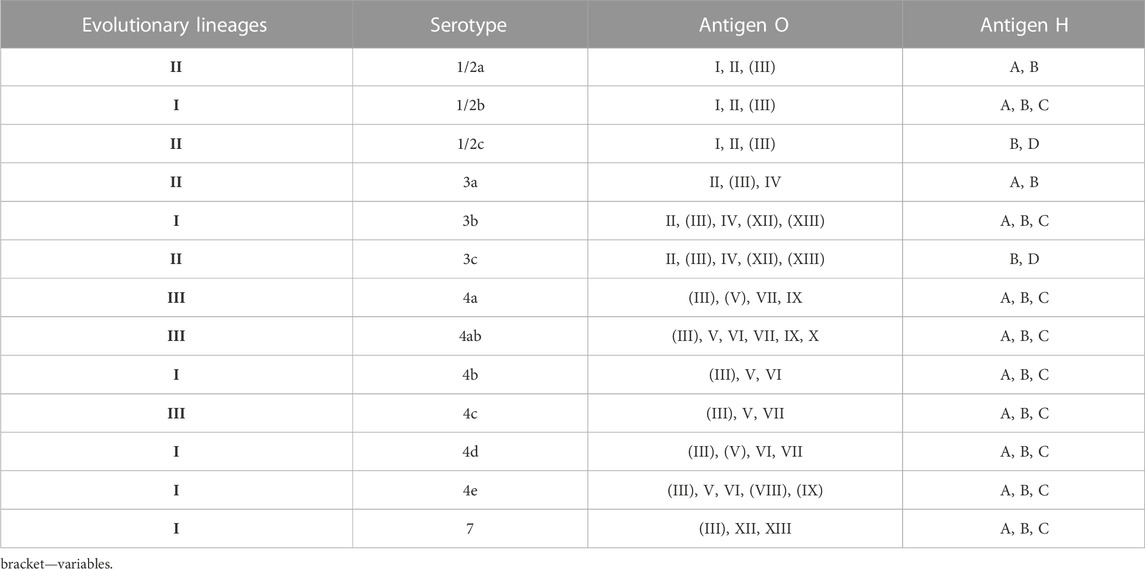
TABLE 2. Division into evolutionary lineages in L. monocytogenes serotypes (based on: Seeliger and Hőhne, 1979; Seeliger and Langer, 1989; Roberts et al., 2006; Ward et al., 2008; Orsi et al., 2011).
Occurrence of Listeria monocytogenes
The ability to survive unfavorable conditions determines the ubiquitous nature ofL. monocytogenes in the environment. The rods are isolated from, e.g., water, soil, sewage, rotting vegetation, and animal feed, as well as from various species of fish, birds, and mammals (Nightingale et al., 2005; Skowron et al., 2019; Liu et al., 2020; Al et al., 2022; Cavalcanti et al., 2022; Tahir et al., 2022). Food is the main source of L. monocytogenes for humans. This foodborne pathogen was isolated from variety of food, e.g., raw and smoked fish, meat products, unpasteurized milk products, as well as from ready-to-eat (RTE) products (European Food Safety Authority, 2018; European Centre for Disease Prevention and Control, 2019a; Skowron et al., 2019; Centers for Disease Control and Prevention, 2022a). Food of non-animal origin (FNAO) may also be a source of L. monocytogenes (EFSA Panel on Biological Hazards BIOHAZ Panel, 2013). FNAOs are products derived from plants and are an ingredient in almost every meal. FNAOs include fruits, vegetables, nuts and seeds, herbs, spices, or, mushrooms and algae (EFSA Panel on Biological Hazards BIOHAZ Panel, 2013). The first outbreak of listeriosis associated with FNAO was recorded in Boston (United States, 1979). The source of the rods was raw celery, tomatoes and lettuce (Ho, 1986). The feature of L. monocytogenes, which determines its presence in the food processing environment, is its resistance to disinfectants (Elhanafi et al., 2010; Ratani et al., 2012; Kovacevic et al., 2016; Meier et al., 2017; Møretrø et al., 2017; Yu et al., 2018) in L. monocytogenes (Table 1). Genes located on LGI-2 (Listeria Genomic Island 2) confer arsenic-cadmium resistance (Lee et al., 2013; Parsons et al., 2017; Haubert et al., 2019). High virulence and ubiquitous nature of L. monocytogenes may pose a relevant public health problem. Disinfectant resistance determines the presence of L. monocytogenes in the food processing environment, which can be a source of food contamination. Knowledge on the mechanisms conferring resistance and searching for new methods of L. monocytogeneseradication is indispensable to limit the spread of the rods and listeriosis outbreaks.
Pathogenicity
L. monocytogenes is the etiological factor of listeriosis, characterized by a high mortality rate (up to 30%) (World Health Organization, 2018a). L. monocytogenes serotypes: 4b, 1/2b, and 1/2c (98% of documented cases) are most often responsible for listeriosis (Wiedmann et al., 1997; Liu et al., 2006). The most vulnerable to infection are elderly (over 65 years old), pregnant women, newborns and people with reduced immunity (cancer, diabetes, transplant, HIV (human immunodeficiency virus-infected, alcoholics) (Gandhi and Chikindas, 2007; World Health Organization, 2018a). L. monocytogenes has the ability to colonize the intestine and cross the blood-brain and placenta barriers (Kammoun et al., 2022). Genes localized on LIPI-1 (Listeria Pathogenicity Island 1) and the InlA-InlB locus (Vázquez-Boland et al., 2001a) are implicated in the infectious cycle of L. monocytogenes. Transmission of the microorganism to newborns may occur in the womb or in the infected birth canal during childbirth (Allerberger and Wagner, 2010). In the case of the central nervous system infection, as much as 50.0% of disease cases are fatal (Godziszewska et al., 2015). Genes of LIPI-4 participate in neuroinfection and fetal infection (Maury et al., 2016). Cases of listeriosis are reported most often in late summer and early fall (Lamont et al., 2011). First-line drugs in the listeriosis treatment include ampicillin or gentamicin (Temple and Nahata, 2000; Hof, 2004). Also rifampicin, vancomycin, linezolid and carbapenems (Goulet and Marchetti, 1996; Benes et al., 2002; Hof, 2004) or trimethoprim (in patients allergic to beta-lactams) are recommended (Benes et al., 2002; Hof, 2004). In recent years there has been an increase in antibiotic resistance in L. monocytogenes (Morvan et al., 2010). This phenomenon may contribute to therapeutic difficulties in the following years, especially in the case of multi-antibiotic-resistant strains.
In recent years, many foodborne outbreaks of listeriosis have been registered worldwide, e.g., in Republic of South Africa (ready-to-eat processed meat products, 2017–2018) (Smith et al., 2019), Australia (rockmelons, 2018) (World Health Organization, 2018b), and several epidemics in the United States, e.g., mexican-style cheese, 1985 (Beckers et al., 1987) or deli meats, 2020 (Centers for Disease Control and Prevention, 2021). The last documented epidemics of listeriosis was associated with Big Olaf’s ice cream. This outbreak included 23 cases of listeriosis (22 hospitalized, one death) (Centers for Disease Control and Prevention, 2022b). A number of listeriosis outbreaks linked to FNAO have been reported in the United States, e.g., raw broccoli and cauliflower (Simpson, 1996), cantaloupe (Centers for Disease Control and Prevention, 2011), celery (Gaul et al., 2013), caramel apples, (Centers for Disease Control and Prevention, 2015a). Also, two independent listeriosis outbreaks associated with packaged salads have been reported. One in 2016 involved 9 states and 19 cases (1 death) (Centers for Disease Control and Prevention, 2016d). The second, in 2022, covered eight states, 10 confirmed hospitalizations and 1 death (Centers for Disease Control and Prevention, 2022c). These data indicate the need for continuous monitoring of FNAO for the presence of L. monocytogenes.
The epidemiology of listeriosis infections associated with FNAO in the European Union varies. Between 2007 and 2011, the EU have reported on pre-packed mixed salad vegetables (England) (Little et al., 2010) and mixed salads (Greece) (Gillespie et al., 2010) contaminated with L. monocytogenes. Prior to 2007, cases of listeriosis associated with FNAO included: rice salad (Italy) (Salamina et al., 1996), salted mushrooms (Finland) (Junttila and Brander, 1989), vegetable rennet England (Kerr et al., 1988). Lately L. monocytogenes was detected in vegan cheeses and one vegetable pâté produced in France between April and December 2022. Five people were sick, including four pregnant women who gave birth prematurely. Contaminated products were distributed in Austria, Belgium, Germany, Italy, the Netherlands, Singapore, Spain, Switzerland, and the United Kingdom (https://ask-bioexpert.com/news/in-france-recall-of-various-jay-and-joy-vegan-products-due-to-listeria-monocytogenes/; https://www.foodsafetynews.com/2023/01/five-sick-in-french-listeria-outbreak-linked-to-cheese-alternative/; https://webgate.ec.europa.eu/rasff-window/screen/notification/591930). The presented statistics indicated the need to evaluate the presence of L. monocytogenes in FNAO products. Wartha et al. (2023a) have identified L. monocytogenes among 1.72% of FNAO (Germany) product samples. L. monocytogenes strains isolated from FNAO were resistant to benzylpenicillin, fosfomycin, and moxifloxacin (Wartha et al., 2023b). Continuously reported cases of listeriosis confirm the pathogenic nature of L. monocytogenes and the presence of rods in food products. Phenotypic and genetic evaluation of L. monocytogenes strains responsible for epidemics would be a valuable element in understanding the ecology of these rods. The search for new genetic determinants underlying virulence, antibiotic and disinfectant resistance is crucial for better understanding these pathogenic rods.
Knowledge of the pathogenic and adaptive nature of L. monocytogenes, especially strains isolated from food and the food industry, is a relevant aspect allowing reduction the of listeriosis cases number (Disson et al., 2021). It is also essential to understand the genetic basis of L. monocytogenes virulence in combination with phenotypic features (Stratakos et al., 2020; Lakicevic et al., 2022). More, genes involved in the stress response and adaptation to changing conditions also influence the pathogenic nature of L. monocytogenes (Wiktorczyk-Kapischke et al., 2021).
This review aims to characterize selected aspects of the genomic and pathogenicity islands in L. monocytogenes. We emphasize the importance of the Whole Genome Sequencing (WGS) technique for the identification of new genetic elements in L. monocytogenes and its use in epidemiological research. Knowledge of the virulence of L. monocytogenes could help prevent future listeriosis outbreaks through the appropriate control strategies selection.
Pathogenicity and adaptation to stress conditions
The infectious cycle
L. monocytogenes is an intracellular pathogen (Tilney and Portnoy, 1989). The mechanisms by which the rod penetrates and then multiplies in the host cells are still being intensively studied. L. monocytogenes uses a wide variety of virulence factors to promote host cell invasion (InlA, InlB), phagosome escape (LLO, PlcA and PlcB), rapid cytoplasmic replication (Hpt), and spread from cell to cell (ActA, InlC) (Hamon et al., 2006; Krypotou et al., 2019) (Figure 1). The main virulence factors essential for invasion include internalins (InlA and InIB) and listeriolysin O (LLO, encoded by the hly gene) (Phelps et al., 2018). The first stage of infection is adhesion to the surface of cells and internalization of their cytoplasmic membrane (Camejo et al., 2011). The Ami protein is responsible for the cleavage of the amide bond in the peptidoglycan. The Lap protein is an adhesin and murein hydrolase involved in the invasion of non-phagocytic cells (Ireton, 2007; Camejo et al., 2011). The FbpA protein, which enables fibronectin binding and protects L. monocytogenes from identification by the human immune system, also plays an important role at this stage (Ireton, 2007; Camejo et al., 2011). In L. monocytogenes, 27 proteins belonging to the internalin family (surface proteins) have been identified. In order to bind to the membrane surface, the rods utilize anchoring domains, particularly the LPXTG motif mediated by sortase A (Sabet et al., 2005). Internalin A exhibits strict cell tropism during its invasion of host cells, limited only to cells of epithelial origin (Bonazzi et al., 2009). The InlA is covalently attached to the bacterial cell wall by the LPXTG motif (Sabet et al., 2005; Pizarro-Cerdá and Cossart, 2006). InlA binds to E-cadherin, which interact with catenines, leading to the reorganization of the host cell actin cytoskeleton and phagocytosis (Camejo et al., 2011). The InlA/E-cadherin interaction is species-specific (Lecuit et al., 1999; Lecuit, et al., 2001). Researchers have reported premature stop codons (PMSCs) in the inlA gene of L. monocytogenes strains (Jonquieres et al., 1998; Olier et al., 2003; Jacquet et al., 2004; Handa-Miya et al., 2007). These mutants had a reduced ability to invade host cells (Nightingale et al., 2008). Internalin B uses three receptors, i.e., c—Met (transmembrane hepatocyte growth factor receptor - HGF), heparin, heparin sulphate proteoglycan (HPSG) and the qC1q—R glycoprotein (Bonazzi et al., 2009; Camejo et al., 2011). Met is a ubiquitous tyrosine kinase receptor that controls cell migration and growth during embryogenesis, tumor cell invasion and metastasis (Trusolino and Comoglio, 2002). InlB enables the invasion of cells of various types and origins (the c-Met receptor is expressed in a wide range of cells) (Ireton, 2007). As a result, L. monocytogenes invades the cell through phagocytosis (bacteria closed in the vacuole). Next, rods escape from the vacuole. The most important virulence factors at this stage include listeriolysin O (LLO), phospholipases A and B (PlcA and PlcB), metalloprotease (Mpl). LLO is a pore-forming toxin, enabling the lysis of the vacuole membrane and the entry into the host cytoplasm (Kayal and Charbit, 2006). PlcA, PlcB and Mpl participate in vacuole lysis, supporting the action of LLO (Camejo et al., 2011). PlcA, PlcB and Mpl participate in vacuole lysis, supporting the action of LLO (Camejo et al., 2011). Next stage is intercellular multiplication (Hpt), and spread to neighboring cells (ActA, InlC, P60) (Wuenscher et al., 1993). L. monocytogenes enters a neighboring cell with the formation of a secondary vacuole. Once released into the cytoplasm, the cycle is initiated anew (Ireton, 2007; Camejo et al., 2011) (Figure 1).
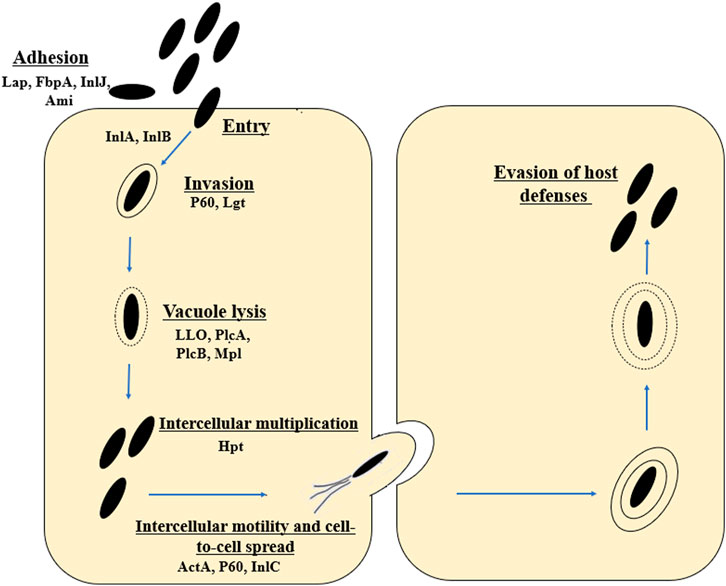
FIGURE 1. Stages of the life cycle of L. monocytogenes (virulence factors involved in each stage are included). The first stage includes adhesion (Lap, FbpA, inlJ, Ami, RecA), entry and internalization (InlA, InlB). Next stages are invasion (P60, Lgt), vacuole lysis (LLO, PlcA, PlcB, Mpl), intracellular proliferation (Hgt) and intracellular movement (ActA, P60, InlC). Lap, Listeria adhesion protein; FbpA, Fibronectin-binding protein; InlJ, interanlin J; Ami, autolysin amidase; InlA, internalin A; InlB, internalin B; Lgt, prolipoprotein diacylglyceryl transferase; LLO, Listeriolysin O; PlcA, secreted phosphatidylinositol-specific phospholipase C; PlcB, phospholipase C; Mpl, Metalloprotease; ActA, Actin assembly-inducing protein (according to: Southwick and Purich, 1996; Portnoy et al., 2002; Freitag et al., 2009; Ireton et al., 2021).
Other factors promoting the intracellular cycle are discussed later in this paper during LIPI-1description.
L. monocytogenes can survive and multiply in typical phagocytic cells but also attack and multiply in non-phagocytic cells (Lecuit, 2007; Seveau et al., 2007). The pathogen crosses the epithelial barrier by transcytosis invading the basal lamina propria (Nikitas et al., 2011). The bacterium reaches the mesenteric lymph nodes (MLN) through the lymphatic vessels and spreads to the liver and spleen via the lymph and blood. The bacteria can spread to secondary infection targets such as the central nervous system and the placenta. From the intestine, L. monocytogenes can also reach the liver via the hepatic portal vein (Melton-Witt et al., 2012). Next, L. monocytogenes can translocate into the gallbladder through the bile ducts. Due to extracellular replication in the bile ducts (Hardy et al., 2006; Eimerman, 2010) L. monocytogenes can be reintroduced into the gastrointestinal tract (Hardy et al., 2006).
In response to immune cells infection by L. monocytogenes, the host organism induces the production of many cytokines (Hansen et al., 2014). Induction of IFNα/ß and cytokines during L. monocytogenes infection results from the bacterial wall components recognition by Toll-like receptors (TLRs) and additional molecules of bacterial origin by intracellular TLR-independent mechanisms (Leber et al., 2008; Stockinger et al., 2009; Yang et al., 2010; Abdullah et al., 2012). The presence of L. monocytogenes DNA in the cytoplasm of host cells is a potent activator of the induction of IFN type I (Stetson and Medzhitov, 2006; Hansen et al., 2014). Moreover, listeriolysin O potentially stimulates IFN induction (Woodward et al., 2010; Burdette et al., 2011).
Adaptaion to stress factors
L. monocytogenes are present in many environments as the bacteria tolerate a wide range of variable conditions (Wiktorczyk-Kapischke et al., 2021), both during host invasion (Krypotou et al., 2019) and also in the food processing environment (O’Byrne and Karatzas, 2008). Some strains of L. monocytogenes are avirulent. On the other hand, some strains may increase virulence after exposure to environmental stress (Stratakos et al., 2020). Exposure to environmental stress influences cell morphology, antimicrobial resistance (AMR), pathogenicity and virulence of L. monocytogenes through the expression of a number of genes (Matereke and Okoh, 2020). More, sublethal stress may contribute to stress adaptation (resistance to higher levels of the same stress factor) or cross-resistance (resistance to other stress factors) (Faezi-Ghasemi and Kazemi, 2015). The pathogen has developed a number of mechanisms that allow adaptation to stressful conditions. The most frequently encountered adverse conditions by L. monocytogenes include: osmotic, heat, cold, acid, alkaline and nutrients stresses (Cotter et al., 2005; Giotis et al., 2010; Kocaman and Sarimehmetoğlu, 2016; Bucur et al., 2018; Manso et al., 2020). Stress response in L. monocytogenes involves many genes, followed by proteins that include general stress proteins and mechanisms related to adaptation to specific conditions (Wiktorczyk-Kapischke et al., 2021) (Table 1, Figure 2).
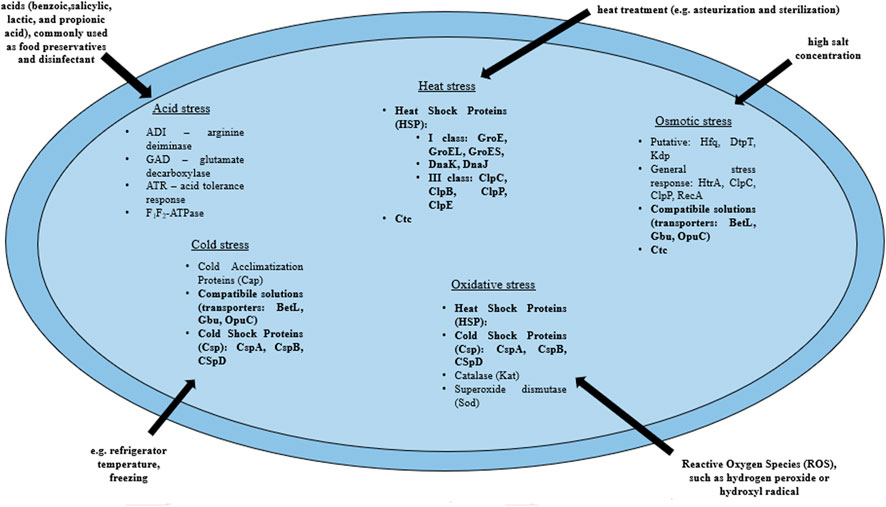
FIGURE 2. Mechanisms of the stress response in L. monocytogenes (bold print indicates the mechanisms involved in the response to several stressors). BetL, Glycine betaine transport system I; Gbu, Glycine betaine transport system II; OpuC, Carnitine transport system; ClpC, ATPase protein; ClpP, Serine protease; ClpB, Chaperone protein ClpB; ClpE, ATPase protein; GroES, GroE—Chaperone proteins which regulate HrcA posttranscriptionally; HtrA, Serine protease (according to: Wiktorczyk-Kapischke et al., 2021).
The response to osmotic stress relies on the compatible substances accumulation from the environment. This involves the respective transporters, i.e., BetL and Gbu (glycineabetaine transport) and OpuC (carnitine transport). In the absence of osmoprotectants in the environment, general stress proteins (Ryan et al., 2009) participate in the osmotic stress response. Heat shock protein (Hsp) synthesis begins in L. monocytogenes under high temperature conditions. Class I proteins act as chaperone intracellular proteins, and class III: ATP-dependent proteins with caseinolytic activity (Table 1). Also, GroEL and GroES proteins (regulation of basic cellular processes) and DnaK and DnaJ proteins (stabilization of unfolded proteins conformations) are involved in the heat stress response in L. monocytogenes (Nair et al., 2000; Bucur et al., 2018). In response to a drop in temperature, L. monocytogenes begins to synthesize cold shock proteins (Csp). Csp are molecular chaperone proteins that enable replication, transcription and translation at low temperatures (Schmid et al., 2009). The synthesis of cold acclimation proteins (Cap) also occurs during exposure to cold stress (Barria et al., 2013). Acid stress triggers mechanisms responsible for homeostasis maintaining, such as acid tolerance response (ATR), Glutamate decarbosylase activity (GAD), putative arginine deiminase (ADI) and F1F0-ATPase (Cotter et al., 2005) (Table 1; Figure 2). ATR protects the cell after short-term exposure to mild acids (Koutsoumanis et al., 2003). The GAD system enables survival in foods with low pH. GAD converts extracellular glutamate to gamma-aminobutyric acid (GABA), resulting in an increase in pHi (Werbrouck et al., 2009). Subsequently, GABA is exchanged for glutamate in the cell membrane via the GadT2 antiporter, contributing to the environment alkalization and pH homeostasis restoring (Ryan et al., 2009). In contrast, the ADI system is activated in response to extreme acid stress (low pH) (Soares and Knuckley, 2016). ADI converts, imported from the external environment, arginine to ornithine, CO2, ammonia and ATP. Ammonia, formed as a by-product, reacts with intracellular protons to produce NH4+, thereby increasing the pH of the cytoplasm and protecting the cell from an acidic environment (Soares and Knuckley, 2016). F1F0-ATPase generates a proton gradient, H+ efflux and restoration of homeostasis (Ryan et al., 2009) (Table 1; Figure 2).
The gastrointestinal stress-induced adaptive tolerance response to acid and osmotic stress can protect the pathogen from similar stresses in the gastrointestinal tract (GIT) and thus directly support its virulence potential. Moreover, in the GIT,L. monocytogenes switches from avirulent to a virulent state via reprogramming gene expression from stress survival-associated genes to virulence genes. The crosstalk between stress adaptation and pathogenicity is controlled by two overlapping and interrelated transcriptional networks regulated by sigma B factor and positive regulatory factor A (PrfA)) (Sibanda and Buys, 2022). The alternative sigma B factor - σB, controls more than 300 stress response and virulence genes (Kazmierczak et al., 2003; Lungu et al., 2009). Researchers have shown that σB is involved osmotic stress, cold and heat stress, or oxidative stress response (Becker et al., 1998; Ferreira et al., 2001; Moorhead and Dykes, 2004). σB also contributes to the transcriptional activation of the prfA gene, encoding PrfA, the central regulator of L. monocytogenes virulence gene expression (Nadon et al., 2002). Recently, scientists have discovered new genes conditioning adaptation to unfavorable conditions, e.g., SSI-1 (Ryan et al., 2010) and SSI-2 (Harter et al., 2017). Importantly, hitherto unidentified genes were detected among the L. monocytogenes strains responsible for the outbreaks, e.g., LG-1 in the L. monocytogenes 08–5578 strain (the Canadian deli meat listeriosis outbreak in 2008) (Gilmour et al., 2010) and bcrABC gene in L. monocytogenes strain H7550 (the multistate outbreak in 1998–1999) (Elhanafi et al., 2010).
Despite extensive knowledge about the L. monocytogenes genome, the role of many proteins remains unexplained. An important aspect is also expanding knowledge on genomic and pathogenicity islands. A complete understanding of the virulent potential of L. monocytogenes strains requires whole genome data analysis combined with phenotypic characterization (Stratakos et al., 2020). In our opinion, exposure to stressful conditions can affect changes in the phenotypic characteristics of L. monocytogenes, which affects their virulence.
Genomic and pathogenicity islands of Listeria monocytogenes
Genomic islands (GEIs) are gene clusters in the bacterial genome, most likely acquired through horizontal gene transfer (Gal-Mor and Finlay, 2006). Genomic islands contain genes that code for traits favorable under certain environmental conditions. GEIs are characterized by a large size (>10 kbp) and a different content of G + C (compared to the rest of the chromosome) (Dobrindt et al., 2004). GEIs can differ in the composition and sequence of genes even within one species (Hallstrom and McCormick, 2014). GEIs can evolve from mobile genetic elements, such as bacteriophages or plasmids, which can be transferred between unrelated microorganisms. A typical feature of many islands is the presence of a functional integrase gene, which allows for the insertion and removal of such elements (Dobrindt et al., 2004). This plasticity of GEIs enables pathogens adaptation to different environments (Dobrindt et al., 2004).
Pathogenicity islands (PAIs) are GEIs encoding virulence factors of pathogenic bacteria (Dobrindt et al., 2004; Hallstrom and McCormick, 2014). Non-pathogenic strains do not contain PAIs (Gal-Mor and Finlay, 2006). PAIs are a subclass of genomic islets obtained by horizontal transmission (Hallstrom and McCormick, 2014). PAIs can constitute large part of the chromosome (from 10 kbp to over 100 kbp) (Gal-Mor and Finlay, 2006). Expression of PAI genes, like other virulence genes, occurs in response to environmental cues (Gal-Mor and Finlay, 2006). Some strains also contain smaller pieces of DNA (1–10 kbp), termed “pathogenicity islets.” Expression of genes of PAIs is regulated by the transcription factors located on the island or externally (off the island) (Hallstrom and McCormick, 2014).
Infectious diseases, including listeriosis, remain a significant cause of mortality worldwide. The problem has been exacerbated recently by the increasing resistance of bacteria to antibiotics. Identifying the virulence factors used by these bacterial pathogens and understanding their evolution are relevant for both basic science and current medical challenges. The search and identification of various PAIs is essential from the medical point of view. Genes located on PAI can serve as markers in the molecular diagnostics of bacterial pathogens, assessment of their pathogenic potential, and even their antibiotic resistance pattern (Gal-Mor and Finlay, 2006).
The genome of L. monocytogenes is approximately 3 Mb, encoding approximately 2,910 core genes (den Bakker et al., 2010). L. monocytogenes virulence genes are organized within genomic and pathogenicity islands. Vázquez-Boland et al. (2001b) named the first Listeria spp. PAI LIPI-1, using a unified nomenclature to designate all large, genetically heterogeneous PAIs identified in Listeria spp. LIPI-1 plays a key role in the pathogenesis of L. monocytogenes due to the presence of genes required for the intracellular cycle (Portnoy et al., 1992; Vázquez-Boland et al., 2001b). Currently, scientists have identified in various strains of L. monocytogenes LIPI-1, LIPI-2 fragment (containing genes: smcL, i-inIF and i-inIE) (Yin et al., 2019), LIPI-3 (Cotter et al., 2008), LIPI-4 (Maury et al., 2016), and also genomic islands: LGI-1 (Gilmour et al., 2010), LGI-2 (Kuenne et al., 2013; Lee et al., 2013), LGI-3 (Palma et al., 2020) and SSI-1 (Ryan et al., 2009) and SSI-2 (Harter et al., 2017), which are discussed later in this paper.
Despite the broad knowledge of virulence and resistance determinants inL. monocytogenes, the functioning of many genes remains unexplained. More, scientists have been still reporting on new pathogenicity islands. We believe that understanding these genetic determinants is relevant for public health protection by limiting listeriosis outbreaks or reducing the rate of antibiotic resistance acquisition.
Patgogenicity islands
Listeria Pathogenicity Island 1 (LIPI-1)
Listeria Pathogenicity Island 1 (LIPI-1) contains virulence genes involved in the intracellular infection cycle of L. monocytogenes (Vázquez-Boland et al., 2001a; Vázquez-Boland et al., 2001b; Osman et al., 2020). This 9 kbp gene cluster is located between prs and orfX and consists of six genes: prfA, plcA, hly, mpl, actA, and plcB (Dussurget, 2008; Hadjilouka et al., 2018) (Figure 3A). Down-stream of the hly gene is the mpl-actA-plcB operon of 5.7 kbp transcribed in the same orientation (Vázquez-Boland et al., 1992). In turn, the genes upstream of the hly gene, organized in the plcA-prfA operon, are transcribed in the opposite orientation (bicistronic or monocistronic) (Menguad et al., 1991; Freitag et al., 1993) (Figure 3A). The most intensive expression occurs at the physiological temperature of mammals (37°C), while the temperature drop to 30°C contributes to gene silencing (Mandin et al., 2005). LIPI-1 is regulated by the transcription factor PrfA (Cossart, 2011).
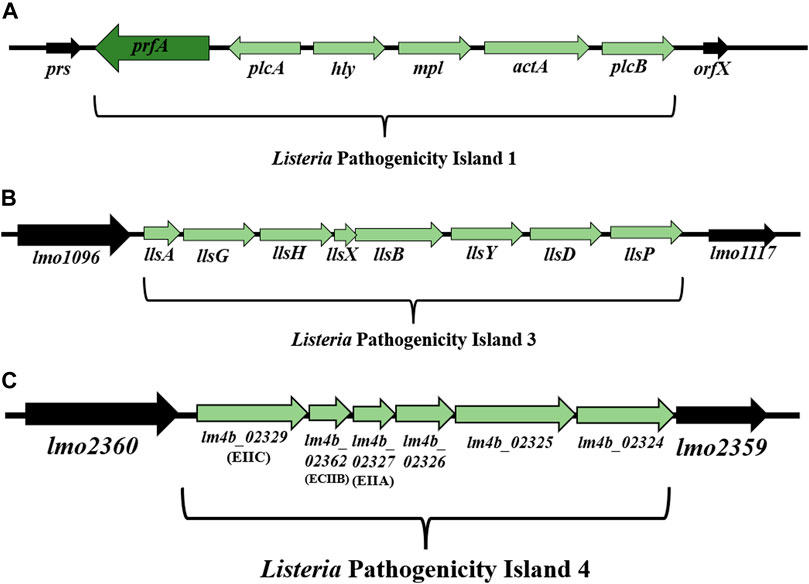
FIGURE 3. (A) Organization of Listeria Pathogenicity Island 1 (LIPI-1) (according to: Vázquez-Boland et al., 2001a; Vázquez-Boland et al., 2001b). (B) Organization of LIPI-3 among L. monocytogenes strains (according to: Cotter et al., 2008). (C) Organization of LIPI-4 among L. monocytogenes strains (according to: Maury et al., 2016). Direction of transcription is indicated by the respective arrows.
The hly gene (1,590 bp) encodes the pore-forming toxin listeriolysin O (LLO). LLO (58 kDa) was the first identified virulence factor of L. monocytogenes (Harvey and Faber, 1941; Gaillard et al., 1986; Geoffroy et al., 1987; Cossart et al., 1989). The main function of the LLO is to participate in the lysis of the phagocytic vacuole and the release of L. monocytogenes into the host cytoplasm (Arnett et al., 2014) (Figure 1). During spread to neighboring cells, L. monocytogenes is enclosed in a secondary vacuole (Figure 1) and then released by LLO to the host cytoplasm (Osborne and Brumell, 2017). Lam et al. (2018) have documented that LLO is critical for the human hepatocytes’ internalization (Figure 4). LLO participates in membrane binding (cholesterol-rich) and oligomerization (pre-pore complex). This complex passes into transmembrane pores that allow the influx of extracellular Ca2+. The increase of Ca2+ in the cytoplasm results in the translocation and activation of the conventional protein kinase C (cPKC). Activated cPKC signals induce Arp2/3-mediated remodeling of F-actin in the plasma membrane, leading to L. monocytogenes entry into the cell. The influx of extracellular Ca2+ also activates the membrane resealing pathway (Lam et al., 2018). LLO also forms small pores in the host cell membranes during other stages of the invasion cycle (Osborne and Brumell, 2017). LLO may play role in the cytoplasm of host cells. Pillich et al. (2012) have shown that the presence of LLO led to the induction of unfolded protein response (UPR). In contrast, damage to the endoplasmic reticulum (ER) (the site of intracellular calcium storage) caused by LLO is a source of calcium elevation during infection (Gekara et al., 2007).
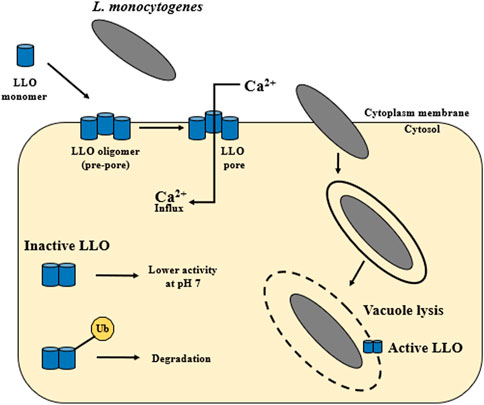
FIGURE 4. The activity of listeriolysin O. LLO is secreted as a water-soluble monomer that binds to cholesterol in host membranes. Then it oligomerizes into large complex—a toxin is formed, which can generate pores. The main function of LLO is the release of L. monocytogenes from the vacuole into the host cytoplasm. Additionally, the activity of LLO is pH dependent. Listeriolysin O has the so-called pH sensor. The highest activity of LLO is observed at the acidic pH of the vacuole (pH 5.5), while at the neutral pH of the host cytoplasm this activity decreases. Cytosolically synthesized LLO is first ubiquitylated and then LLO monomers are degraded by the proteasome. LLO participates in membrane internalization through membrane binding (cholesterol-rich) and oligomerization (pre-pore complex). This complex passes into transmembrane pores that allow the influx of extracellular Ca2+ (according to: Vadia et al., 2011; Arnett et al., 2014; Osborne and Brumell, 2017; Chen et al., 2018; Lam et al., 2018).
Listeriolysin O belongs to the cholesterol-dependent cytolysin (CDC) family (Heuck et al., 2010). The proteins of the CDCs family have four regions. One of them (C-terminal domain) is involved in targeting the action of the toxin on the cytoplasmic membrane, and the other three are responsible for the oligomerization of LLO (Pizarro-Cerdá and Cossart, 2006). Köster et al. (2014) revealed the LLO crystal structure in 2014. According to the crystal structure, the LLO consists of four distinct domains (D1—D4), which play a different role in the functioning of the LLO (Köster et al., 2014). The D1 domain possesses a structural motif in the C-terminal region, crucial for membrane binding and the cytotoxic activity of the LLO (Faezi-Ghasemi and Kazemi, 2015; Materake and Okoh, 2020). In turn, the D2 domain is the sequence that joins D1 to D4 via a glycine linker (Rosado et al., 2008; Köster et al., 2014). The D3 domain consists of a five-stranded anti-parallel β sheet surrounded by six helices (Köster et al., 2014). A significant feature of D3 is the presence of three residues (D208, E247 and D320), i.e., a pH sensor (Dubail et al., 2001; Schuerch et al., 2005).
The LLO protein is synthesized as a precursor with the SS signal sequence at its N-terminus. Upon cleavage of the SS sequence, the mature protein functions as a monomer (Kayal and Charbit, 2006). In the first step, LLO is secreted as a water-soluble monomer that binds to cholesterol in the host’s membranes. Next, LLO oligomerizes into large complex (from 30 to 50 subunits). This stage results in a transmembrane toxin formation, generating pores with a diameter of about 50 nm (Dunstone and Tweten, 2012; Arnett et al., 2014; Chen et al., 2018) (Figure 4). Additionally, LLO activity is pH-dependent. Listeriolysin O has a so-called pH sensor (a triad of acidic amino acid residues in D3). The highest activity of LLO is observed at the acidic pH of the vacuole (pH 5.5), while at the neutral pH of the host cytoplasm this activity decreases (Schuerch et al., 2005; Bavdek et al., 2012). It ensures a quick inactivation of the toxin after L. monocytogenes escape into the cytoplasm. LLO requires a 30–40 mol% Chol threshold in the lipid membrane for effective binding and pore formation (Bavdek et al., 2007). Decatur and Portnoy (2000) have identified a relevant region within listeriolysin O, the so-called PEST-like region (P: proline, E: glutamine, S: serine, T: teronine). The PEST sequence is not required for the hemolytic activity but is a key element during the phagosomal escape of L. monocytogenes (Decatur and Portnoy, 2000). Due to six prolines, the PEST sequence shows the so-called Type II polyproline (PPII) system. The sequence plays a regulatory role in the host cytoplasm, inhibiting or preventing LLO oligomerization and pore formation (Kӧster et al., 2014). The vacuole rupture and access to the host cytoplasm occurs 15–30 min after infection of epithelial cells and macrophages (Quereda et al., 2018). LLO can also induce cytolysis in infected host cells even at a low concentration of 5 ng/mL (Jacobs et al., 1998).
The mechanism of LLO-mediated apoptosis induction on activated T cells involves two processes: one through caspase-3 and caspase-6 activation (Carrero et al., 2008). Caspase activation depends on the expression of granzymes (Carrero et al., 2008). Conversely, the second mechanism is LLO-dependent but caspase-independent, inducing phosphatidylserine exposure and loss of plasma membrane potential (Carrero et al., 2008). Listeriolysin O stimulates the host’s immune system, influences the production of pro-inflammatory mediators (NO), cytokines, and activates the NF - κB pathway and the formation of antibodies. LLO can additionally induce apoptotic pathways, stimulate MAP kinases (mitogen-activated protein kinases) and increase the expression of adhesion molecules (Camejo et al., 2011).
L. monocytogenes mutants that do not synthesize listeriolysin O remain trapped inside the vacuole and are five orders of magnitude less virulent than wild-type rods (Lam et al., 2018). The incidence of non-hemolytic L. monocytogenes is approximately 0.1% (Maury et al., 2017). Non-hemolytic L. monocytogenes strains most commonly occur among isolates from the food processing environment, but some clinical isolates have been reported to exhibit reduced hemolysis (Maury et al., 2017; Kawacka et al., 2022). Henry et al. (2006) have demonstrated that LLO deletion mutants escaped from the vacuole before being internalized in non-phagocytic human cell lines such as HeLa, HepG2, Henle 407, HEp-2, HCT116, HEK-293, and dendritic cells, where PlcB plays a major role in vacuole fracture.
LLO is one of the main toxins of L. monocytogenes, which determines virulence. However, researchers have attempted to use the toxin as a vaccine. In experiments carried out in animal models, researchers assessed LLO as an adjuvant in protective vaccinations against allergies, cancer and pathogens. The LLO was also used in the experimental treatment of tumor models such as follicular lymphoma and head and neck cancers (Xiong et al., 1998; Alberti-Segui et al., 2007; Hernández-Flores and Vivanco-Cid, 2015). The above studies offer the possibility of using this pathogen. Therefore, research on L. monocytogenes should also include the potential use of the toxins produced.
L. monocytogenes synthesizes two phospholipases C, specific for phosphatidylinositol, phospholipase A (PlcA, encoded by plcA, 954 bp) and broad-spectrum phospholipase B (PlcB, encoded by plcB, 870 bp). Both plcA and plcB genes are regulated by the transcription activator PrfA. These phospholipases hydrolyze phospholipids and then damage the host’s cytoplasmic membrane (Vázquez-Boland et al., 2001a; Vázquez-Boland et al., 2001b).
PlcB is a zinc-dependent metalloenzyme secreted as an inactive 264 amino acid proenzyme (to prevent degradation of the phospholipids contained in the bacterial membrane). PlcB (29–30 kDa) is activated by proteolytic cleavage in the extracellular environment. There is also an Mpl-independent activation path. This pathway depends on the level of acidification of the vacuole environment (Vázquez-Boland et al., 2001a). PlcB has a wide range of optimum pH - from 5.5 to 8.0. PlcB hydrolyzes phosphatidylcholine, phosphatidylethanolamine, phosphatidylserine, and sphingomyelin. It shows weak phosphatidylinositol hydrolysis activity and weak calcium-independent hemolytic activity at 37°C (Vázquez-Boland et al., 2001a). PlcB plays a relevant role in vacuole escape and spread from cell to cell (Alberti-Segui et al., 2007; Quereda et al., 2018).
PlcA is a phospholipase that cleaves the signal between alanine (29) and tyrosine (30). This enzyme is specific for phosphatidylinositol (PI) but also slightly hydrolyzes eukaryotic PI glycosyl (GPI)—a membrane protein with an optimal pH range of 5.5–7.0 (Goldfine and Knob, 1992). PlcA helps in the escape from the primary phagosomes and the secondary binary vacuolar membrane (Alberti-Segui et al., 2007).
Surface protein ActA (Actin assembly-inducing protein) ensures intra- and intercellular movement ofL. monocytogenes (Py et al., 2007). The ActA protein is responsible for the polymerization of actin filaments in 1 cell pole, creating a structure resembling the so-called “Comet tail” (Camejo et al., 2011). The resulting force allows the rods to move within the host’s cytoplasm, form a secondary phagosome, and to enter the neighboring cell (Py et al., 2007; Camejo et al., 2011). ActA interacts with the Arp2/3 complex proteins, VASP (vasodilator-stimulated phospho-protein), profilin and cophilin (Pizarro-Cerdá and Cossart, 2006). The central ActA domain (proline-rich region) interacts with proteins of the Ena/VASP complex that modulate the speed and direction of rod movement. VASP recruits profilins (actin monomer-binding proteins), which enables actin polymerization. The protein reduces the frequency of branching of actin fibers, which promotes the formation of long and parallel filaments (Rafelska and Therio, 2006; Camejo et al., 2011). Also ERM family proteins (ezrin, radixin and moesin) contribute to efficient spread of L. monocytogenes to neighboring cell. ERM proteins connect the actin tail with the cytoplasmic membrane, forming and stabilizingsecondary sections in the membrane (Pust et al., 2005). The ActA protein may have other functions, e.g., binding heparan sulfate on the cell surface, which allows attachmemt and entering host cells cultured in vitro (Alvarez-Dominguez et al., 1997; Suárez et al., 2001). In addition, the ActA protein enables the spread of L. monocytogenes within the placenta (Bakardjiev et al., 2005; Le Monnier et al., 2007). There is also a model in which ActA is involved in crossing the blood-brain barrier, at least in part, through the so-called “Trojan horse” mechanism, which has not been formally confirmed yet (Drevets et al., 2001; Join-Lambert et al., 2005). ActA expression depends on the growth medium composition and temperature (Travier and Lecuit, 2014). Moreover, full activation of actA under varying environmental conditions, such as low temperature, requires both PrfA and σB (Tiensuu et al., 2013). Different versions of the actA gene are present in hypervirulent strains of L. monocytogenes. Hence, the effect of mutations in the actA gene on virulence and pathogenicity in humans is not fully understood (Lake et al., 2021).
Metalloprotease (Mpl) has the HEXXH motif characteristic of this family members. Metal-loprotease is synthesized in the form of a proenzyme (Bitar et al., 2008) and activates phospholipase B (PC - PLC) (Camejo et al., 2011). Mutants with a transposon insertion in mpl gene exhibited reduced virulence and lecithinase production (Menguad et al., 1991; Raveneau et al., 1992). Poyart et al. (1993) highlighted that a zinc-dependent metalloprotease is involved in the virulence of L. monocytogenes through its action on PC-PLC. A study by Alvarez and Agaisse (2016) showed that Mpl regulates ActA levels on the bacterial side in protrusions. The scientists have suggested that Mpl maintains ActA polymerization in protrusions which contributes to efficient actin polymerization (Alvarez and Agaisse, 2016). The mpl, plcB and actAgenes are organized in one operon (Hain et al., 2006).
The prfA gene can exist in two functional states, i.e., weakly or highly active. Its activity depends on temperature, presence of carbon, and easily metabolized sugars such as cellobiose (McGann et al., 2007). The PrfA protein (27 kDa) is a member of the CRP (cyclic AMP receptor protein)/FNR (fumarate and nitrate reduction regulator) family and is composed of 233 amino acids (Arnett et al., 2014). Polypeptide expression and activity regulation includes transcription, post-transcriptional and post-translational mechanisms (Port and Freitag, 2007). The prfA gene recognizes a 14 bp palindromic sequence termed “PrfA box” situated typically 40 nucleotides upstream of the target transcription start site. In the post-transcriptional mechanism, the level of PrfA activity depends on environmental factors (the presence of fermentable carbohydrates) and the physiological state of the rods (Ollinger et al., 2009). The fully active state of PrfA in the post-translational mechanism ensures the binding of a small cofactor molecule (Mueller and Freitag, 2005). The prfA gene has three promoters. PplcA positively directs prfA expression by binding to the so-called “PrfA box” (bicistronic expression). In contrast, PprfAP1 and PprfAP2 control monocistronic reactions. The PprfAP1 promoter has a 5′UTR untranslated region that acts as a temperature sensor (Figure 5A). Transcript translation is only effective at 37°C. At the temperature of 30°C, in the region containing the Shine-Dalgarno sequence, a stable spatial structure of the mRNA is formed, preventing the attachment of the ribosome and the polypeptide synthesis. At the physiological temperature of the host (37°C), destabilization of the structure occurs, enabling the PrfA synthesis (Figure 5B). The expression of the PplcA and PprfAP2 promoters is independent of temperature (Lemon et al., 2010). The PprfAP1 promoter is recognized by the factor σA, while PprfAP2 can be recognized by σA and σB factors (Lemon et al., 2010). Full PrfA activation requires cofactor, i.e., glutathione binding allosterically to the protein (Johannson and Freitag, 2019) (Figure 5C). According to Loh et al. (2009), expression of the prfA gene is almost 16-fold higher at 37°C compared to 30°C. PrfA regulates expression of LIPI-1 genes (Cossart, 2011).

FIGURE 5. Regulation of prfA expression and PrfA protein activity. (A) transcriptional control of prfA (three promoters of prfA gene). PplcA positively directs prfA expression by binding to the “PrfA box”. PprfAP1 and PprfAP2 control monocistronic reactions. The PprfAP1 promoter has a 5′UTR untranslated region that acts as a temperature sensor); (B) post-transcriptional control of prfA (at 30°C, a stable spatial structure, referred to as a “hairpin” is formed in the region containing the Shine-Dalgarno sequence. It unifies the attachment of the ribosome and the synthesis of the polypeptide. At 37°C, the “hairpin” structure is destabilized, allowing attachment of the ribosome and synthesis of the PrfA protein); (C) post-translational control of PrfA (attachment of a cofactor molecule (glutathione) to the PrfA protein) (according to: Loh et al., 2009; Xayarath and Freitag, 2012; Lemon et al., 2010; Johansson and Freitag, 2019).
A recent analysis by Prokop et al. (2017) has showed that the orfX gene (lmo0206) also plays a significant role in the virulence of L. monocytogenes. Researchers showed that OrfX is a small se-creted protein positively regulated by PrfA. The primary role of OrfX is to suppress the oxidative reaction of infected macrophages, which contributes to the intracellular survival of the bacteria. OrfX targets the nucleus and lowers the regulatory protein RybP levels (Prokop et al., 2017).
Genes located on LIPI-1, i.e., prfA and hly, are commonly used for L. monocytogenes detection, especially in food products (using PCR reaction) (Amagliani et al., 2004; Wang et al., 2004; Jofre et al., 2005; Germini et al., 2009). Ward et al. (2004) reported that phylograms from each of the genes present in LIPI-1 can differentiate the strains studied according to their origin. Also, Hadjilouka et al. (2018) have revealed that plcA, plcB, mpl, actA and intergenic regions plcA-prfA and plcA-hly are useful for serotypes differentiation. In turn, Poimenidou et al. (2018) have reported that LIPI-1 virulence genes follow different evolutionary paths. Evolutionary changes depend on the strain origin and serotype, as well as the epidemiological dominance of some subgroups. Additionally, research has shown that the most conserved genes are prfA and hly, and the actA gene is the most diverse (Poimenidou et al., 2018).
LIPI-1 plays essential role in the virulence of L. monocytogenes. Due to LIPI-1 genetic diversity it could be valuable to investigate the role of point mutations in the pathogenicity and stress adaptation of L. monocytogenes.
Listeria Pathogenicity Island 2 (LIPI-2)
The LIPI-2 region is specific for strains belonging to the species Listeria ivanovii. On LIPI-2 (22 kbp) the following genes are present: i-inlB2, i-inlL, i-inlK, i-inlB1, i-inlJ, i-inlI, i-inlH, i-inlG, smcL, i-inlF, i-inlE, surF3, mainly coding internalins (Sergeant et al., 1991; Guillet et al., 2010). LIPI-2 in L. ivanovii is located between lmo1240 and lmo1422 (Vázquez-Boland et al., 2001a). However, Yin et al. (2019) have documented the LIPI-2 locus (presence of LIPI-1 and absence of LIPI-3 and LIPI-4) in L. monocytogenes isolates (HSL-II, serovar 4h) responsible for the listeriosis outbreak in China. The identified LIPI-2 fragment contained the smcL, i-inIF and i-inIE, genes encoding sphingomyelinase and internalin, respectively. This fragment L. monocytogenes likely acquired through the exogenous DNA acquisition from L. ivanovii (Yin et al., 2019). The i-inlEF locus is characteristic of L. ivanovii. Both the i-inlE and i-inlF genes are under the control of PrfA. The i-inlF and i-inlE genes are arranged in tandem, which suggests they generation by gene duplication (Engelbrecht et al., 1998). In turn, the smcL gene (1,008 bp) is PrfA-independent, and sphingomyelinase C (335 amino acids) is responsible for the different hemolytic properties of L. ivanovii (bizonal hemolysis and CAMP-like reaction with Rhodococcus equi). González-Zorn et al. (2000) have shown that the 5′end of the smcL gene was contiguous with the i-inlFE locus. The role of the LIPI-2 locus among L. monocytogenes HSL-II has not been elucidated yet (Disson et al., 2021). According to Yin et al. (2019), the LIPI-2 fragment presence may result from the kinship and coexistence of L. ivanovii and L. monocytogenes in the same environment. Additionally, HSL-II strains possessed many other virulence factors associated with cases of listeriosis in humans (Yin et al., 2019). The accquisition of a new PAI can significantly affect the phenotype or lifestyle of L. monocytogenes. Therefore, there is a need for further studies on the identification and the role of LIPI-2 in L. monocytogenes.
Listeria Pathogenicity Island 3 (LIPI-3)
Cotter et al. (2008) have demonstrated the presence of Listeria Pathogenicity Island 3 (LIPI-3) among L. monocytogenes strains line I F2365 (SL1/CC1) and H7858 (SL6/CC6) (Cotter et al., 2008). LIPI-3 consists of eight genes: llsA, llsG, llsH, llsX, llsB, llsY, llsD, llsP (Figure 3B) (Cotter et al., 2008; Disson et al., 2021).
The operon LLS encodes listeriolysin S (LLS, a thiazole/oxazole–modified microcin (TOMM)), a post-translational modified peptide that exhibits properties of both bacteriocin and hemolytic cytotoxic factor (Cotter et al., 2008; Quereda et al., 2016; Meza-Torres et al., 2021). The LLS operon consists of a structural gene encoding a peptide (llsA), three genes that form the synthetase complex necessary for LLS maturation (llsB, llsY, llsD), an ABC transporter (llsG, llsH), a putative protease (llsP), and a gene of unknown function (llsX) (Clayton et al., 2011; Molloy et al., 2011; Quereda et al., 2016; Quereda et al., 2017; Lee, 2020). As a bacteriocin, LLS restricts the growth of other related Gram-positive bacteria such as Lactococcus lactis, Lactobacillus plantarum, Staphylococcus aureus and even L. monocytogenes line II (EGD and 10403S) that lack the LLS operon (Cotter et al., 2008; Mohammadzadeh et al., 2019). LLS causes only weak hemolysis of red blood cells in vitro, and is not cytotoxic to eukaryotic cells (Cotter et al., 2008). The LLS probably contains the Ala-Gly motif (amino acid 26), and the C-terminal core region with an extreme predominance of Cys, Ser and Thr residues allowing post-translational modifications resulting in a characteristic heterocyclic compound (Clayton et al., 2011). Meza-Torres et al. (2021) have shown that LLS remains bound to the bacterial cell membrane and cytoplasm and is not secreted into the extracellular space of the bacteria. LLS requires direct contact between LLS-producing bacteria and target bacteria in order to exhibit bactericidal activity and thus behaves like a contact-dependent bacteriocin. Contact exposure to LLS leads to permeabilization/depolarization of the target bacterial cell membrane and release of adenosine triphosphate (ATP) (Meza-Torres et al., 2021). Cotter et al. (2008) have noted that llsA promoter expression was negligible in vitro and only induced upon exposure to hydrogen peroxide. In turn, Quereda et al. (2016) have found slight and enhanced llsA expression under classical in vitro laboratory conditions and in infected mice, respectively. The discrepancy between the two studies requires further experimentation. The LLS cluster is present only in the subset of line I strains, responsible for the majority of human listeriosis outbreaks, and absent in line II and III strains of L. monocytogenes (Cotter et al., 2008).
The llsX gene encodes a potential membrane signal peptide of unknown function but very specific for L. monocytogenes. Researchers have already identified the llsX gene in strains with different origins and genetic profiles (Chen et al., 2018; Kim et al., 2018). Tavares et al. (2020) have shown that the L. monocytogenes strains belonging to line II did not possess LIPI-3. Researchers have also demonstrated llsX expression under acidic stress after 6-h incubation for one of the three tested strains of serogroup 4b (Tavares et al., 2020). In addition, Vilchis-Rangel et al. (2019) showed a strong relationship between llsX and the invasiveness of L. monocytogenes. Several studies (Clayton et al., 2011; Chen et al., 2019; Wang et al., 2019) have used the llsX gene as a marker of LIPI-3, provided that this gene is well conserved in various clonal complexes of L. monocytogenes, and even in atypical hemolytic Listeria innocua.
The llsB gene putatively plays an essential role in the systemic infection phase (Chen C. et al., 2018). The functions of the remaining genes still require analysis.
ThellsB gene likely plays an essential role in the systemic infection phase (Chen C. et al., 2018). The functions of the remaining genes still require analysis.
Since the discovery of LIPI-3, LIPI-3 has been documented among manyL. monocytogenes isolates worldwide (Kim et al., 2018; Camargo et al., 2019; Zhang et al., 2019; Chen Y. et al., 2020a; Wieczorek et al., 2020; Zhang et al., 2020), belonging to different lineages. Further research should focus on the role of all genes located on LIPI-3, especially of unknown function (llsX).
Listeria Pathogenicity Island 4 (LIPI-4)
The presence of Listeria Pathogenicity Island 4 (LIPI-4) has been demonstrated among clinical strains isolated from infections of the central nervous system and placenta (Maury et al., 2016), as well as among L. monocytogenes SL87 (CC87) strains widespread in Asia (Wang et al., 2019). LIPI-4 (6 kbp) is located between the genes lmo2360 and lmo2359 (Maury et al., 2016; Disson et al., 2021). LIPI-4 encodes the cellobiose family phosphotransferase system that determines the tropism of L. monocytogenes to the central nervous system (CNS) and placental cells (Maury et al., 2016). Genes located within LIPI-4 include: lm4b_02324 (maltose-6′-P-glucosidase), lm4b_02325 (transcriptional antiterminator), lm4b_02326 (uncharacterized protein associated to PTS systems), lm4b_02327 (membrane permease EIIA), lm4b_02328 (membrane permease EIIB), lm4b_02329 (membrane permease EIIC) (Figure 3C) (Maury et al., 2016). Lake et al. (2021) have found LIPI-4 among all CC4 (serogroup 4b-4d-4e) and CC87 strains (serogroup 1/2b-3b-7). In turn, Shen et al. (2022) have identified LIPI-4 in CC4 and CC388. The presence of LIPI-4 indicates the hypervirulent character of L. monocytogenes strains (Maury et al., 2016; Hurley et al., 2019; Raschle et al., 2021). Identification of LIPI-4 among subsequent clones of L. monocytogenes, both clinical and environmental strains (Raschle et al., 2021), confirms the need to continue research on the function of the remaining proteins and monitoring of L. monocytogenes in the environment. Due to the involvement of genes located on LIPI-4 in the nervous system and placenta infections, screening of strains from all evolutionary lineages is advisable.
Genomic islands
Listeria Genomic Island-1 (LGI-1)
Genomic islands may contain genes that potentially influence higher adaptation to unfavorable environmental conditions. It, in turn, may increase the pathogenic potential of bacteria. Gilmour et al. (2010) has demonstrated for the first time a horizontally acquired LGI-1 (Listeria Genomic Island 1, coordinates 1836435-1886209 of 08-5578; coding sequences LM5578_1850 to LM5578_1903) in strains of L. monocytogenes serotype 1/2a isolated from the 2008 listeriosis outbreak in Canada. It was the deadliest outbreak in Canada, and the source of the pathogen was deli meat. LGI-1 isolates studied so far belonged to serotype 1/2a except for the one isolate of serotype 3a (Knabel et al., 2012). The island of LGI-1 (50 kb) encodes genes responsible for virulence, resistance to antimicrobial substances, and stress factors. The canonical genes predicted in LGI-1 include virB4, virD4, and virB11, which encode ATPases recruiting substrates into the cell, and virB5 and virB6, or subunit genes that form the core of the membrane transfer complex. The presence of the cpa and tad genes indicates possible pilus-like outgrowth, whereas the dnaG gene presence suggests that the genetic island may be mobilized (Gilmour et al., 2010). Researchers have also identified the emrE gene encoding an efflux pump involved in the resistance to toxic cationic hydrophobic molecules such as quaternary ammonium compounds and tetracycline (Pornillos et al., 2005; Gilmour et al., 2010). A study by Kovacevic et al. (2016) has demonstrated that the minimal function of the LGI-1 island enhances L. monocytogenes tolerance to quaternary ammonium compounds (QAC) through ermELm. The data on LGI-1 presented so far indicate its great importance in the survival of L. monocytogenes within the food processing chain and during host invasion. To date, LGI-1 has not been identified among many strains in studies conducted around the world (Table 3).
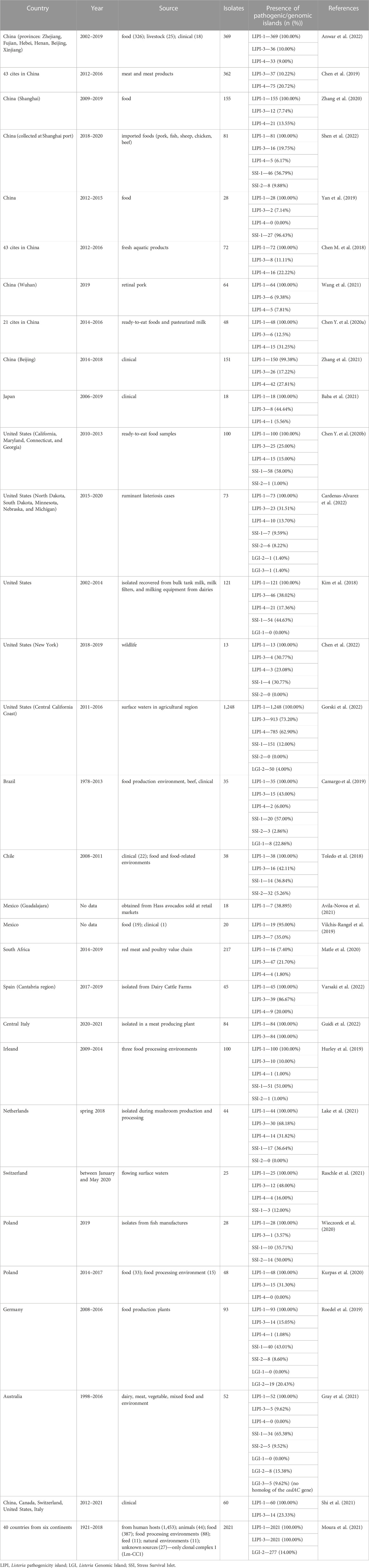
TABLE 3. Summary of selected papers (published in 2018–2022) describing the frequency of genomic/pathogenicity islands among L. monocytogenes strains isolated from various sources.
Listeria Genomic Island-2 (LGI-2)
The genome of L. monocytogenes Scott A possesses a 35 kb chromosomal region called Listeria Genomic Island 2 (LGI-2) (Kuenne et al., 2013; Lee et al., 2013; Lee et al., 2017). LGI-2 contains a cassette of arsenic resistance genes (arsR1D2R2A2B1B2), two additional preceding genes (arsD1A1), cadmium resistance gene (cadA4)), and genes putatively implicated in DNA integration, conjugation, and pathogenicity (Brires et al., 2011; Lee et al., 2013; Parsons et al., 2017). LGI-2 has been identified mainly among L. monocytogenes serotype 4b strains, including hypervirulent clones of serotype 4b CC1 and CC2 (Kathariou et al., 2017; Lee et al., 2017; Gelbicova et al., 2021), and several stable strains belonging to CC14 and CC204 line II (Fox et al., 2016; Lee et al., 2017; Pasquali et al., 2018). Lee et al. (2017) have observed content plasticity of LGI-2. Researchers have identified LGI-2 at multiple genomic locations, frequently disrupting open frame reading and shown sequence content variation (Lee et al., 2017).
Researchers have detected notably diverse variant of this island, designated LGI2-1, in some strains of L. monocytogenes CC1 (Lee et al., 2017). LGI2-1 contained a unique gene encoding a putative cystathionine gamma-synthase immediately upstream of arsenic resistance genes (Lee et al., 2017; Haubert et al., 2019). The ability of strains possessing LGI2-1 to tolerate higher levels of cadmium seems to be mediated by cadA5C5 (Lee et al., 2017). The authors have suggested that LGI-2 may serve as a model showing dynamic selection of arsenic-resistant subpopulations of L. monocytogenes after exposure to a strong environmental toxin such as arsenic (Lee et al., 2017).
There is limited information on the potential effect of heavy metal tolerance on the virulence and persistence of L. monocytogenes. However, understanding the ecology and evolution of LGI-2 among hypervirulent strains of L. monocytogenes capable of cadmium and arsenic detoxification would be significant.
Listeria Genomic Island-3 (LGI-3)
Palma et al. (2020) have revealed Listeria Genomic Island-3 (LGI3) (31.5 kbp) in L. monocytogenes strains (isolated in France) as a highly conserved and specific region for CC101 strains (persistent, RTE seafood processing plants). LGI-3, located downstream of the inIJ gene (homologue of strain EGD-e lmo1413) (Palma et al., 2020), contains 29 predicted coding sequences. It integrates the chromosomal cadmium resistance determinant cadA1C, flanked by recombinase and Tn3 transposase, and genes putatively implicated in DNA integration, conjugation, translocation, and recombination. Palma et al. (2020) have suggested the need for a more detailed characterization of LGI-3 in order to know the virulent potential of L. monocytogenes. Persistent strains of L. monocytogenes are defined as isolates repeatedly isolated from the same source or ecological niche over a period of time (Unrath et al., 2021). Scientists have recently confirmed the presence of persistent strains in the food processing environment (Ferreira et al., 2014; Leong et al., 2014; Wiktorczyk-Kapischke et al., 2022). Undoubtedly, surviving strains are a significant global problem. Understanding the genetic aspects of the environmental persistence of L. monocytogenes strains would be highly relevant.
Genomic island 7 (GI-7)
As already, Yin et al. (2019) have mentioned demonstrated the LIPI-2 fragment among the hybrid subline II strains of L. monocytogenes serovar 4h. The latest study by Jin et al. (2022) identified the LMxysn_1693 gene, a component of genomic island-7 (GI-7) (containing 20 ORFs) in the 4h serovar of L. monocytogenes XYSN strain (high-virulent). LMxysn_1693 gene (534 bp) synthetically interacts with genes involved in bile resistance and biofilm formation, thus contributing to resistance in the intestinal environment. More, LMxysn_1693 can upregulate the transcriptional expression of PrfA. Therefore, the interaction between PrfA and LMxysn_1693 affects the ability of L. monocytogenes to form a biofilm (Jin et al., 2022). The GI-7 island likely contains two ABC transporters involved in extracellular and surface proteins transport (Jin et al., 2022). ABC transporters are relevant for the functioning of bacteria, especially during invasion (Locher, 2016), but also participate in biofilm formation (Benda et al., 2021), resistance to environmental stress conditions (Kang et al., 2015; Grubaugh et al., 2018; Jiang et al., 2019) and metal utilization (Kang et al., 2015; Grubaugh et al., 2018; Jiang et al., 2019). Most GI-7 genes encode hypothetical proteins of unknown function. Therefore, further studies on the role of GI-7 are required.
Genomic islands determine virulence and resistance determinants in L. monocytogenes. However, the composition and sequence of genes within GEIs can vary, making their identification a bit challenging. We believe that whole genome sequencing can be a helpful tool to search for GEIs among L. monocytogenes.
Stress survival islets
Stress survival islet (SSI)-1
Stress survival islet (SSI-1) (8.7 kbp) is a region consisting of five genes: lmo0444, lmo0464, pva (lmo0446), gadD1 (lmo0447) and gadT1 (lmo0448) (Figure 6). These genes are associated with tolerance to acid, osmotic, and bile stress in the stomach (Begley et al., 2005; Cotter et al., 2005; Ryan et al., 2009; Begley et al., 2010) and are important for adaptation and survival in the food processing environment (Ryan et al., 2009). SSI-1 is located at the hypervariable region from lmo0442 to lmo0449 (Ryan et al., 2009). Lin0464 is a putative transcription regulator of the GntR family with a DNA helix-twist-helix binding domain. Lin0465 belongs to the DJ-1/PfpI protease superfamily with a type I glutamine amidotransferase-like domain characterized in Pyrococcus furiosus (Halio et al., 1998). Scientists have demonstrated the presence of SSI-1 among L. monocytogenes ST121 strains often isolated from the food processing environment (Hein et al., 2011; Schmitz-Esser et al., 2015; Rychli et al., 2017). Palacios-Gorba et al. (2021) have confirmed the presence of SSI-1 among L. monocytogenes strains (SL 1555) isolated from wild boar and deer tonsils. In turn, Liu et al. (Liu et al., 2022) have shown the presence of SSI-1 in L. monocytogenes ST5, ST121, and ST120, suggesting that these ST types may better tolerate the food processing environment than ST2. According to Hilliard et al. (2018), there is a relationship between the presence of SSI-1 and the persistence of L. monocytogenes. In addition, SSI-1 was strongly correlated with biofilm formation and truncated inlA gene (STOP codon) (Franciosa et al., 2009; Keeney et al., 2018; Lakicevic, Den Besten and De Biase, 2022). More recent findings confirmed the effect of SSI-1 and shortened inlA on increased biofilm production in L. monocytogenes (Lakicevic, Den Besten and De Biase, 2022). These data indicate the need for monitoring the environment and food processing area for virulent L. monocytogenes strains. Also, studies on the impact of SSI-1 on biofilm formation by L. monocytogenesunder environmental stress conditions would be valuable.
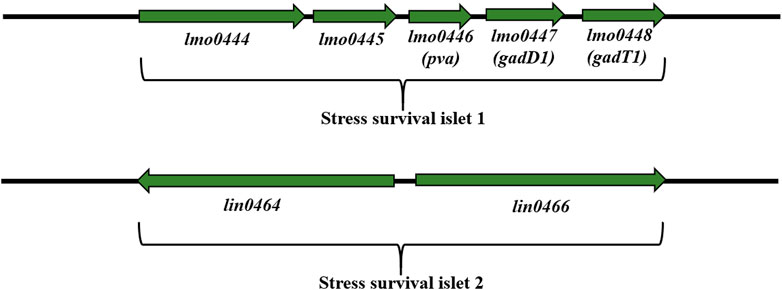
FIGURE 6. Organization of SSI-1 and SSI-2 among L. monocytogenes strains (according to: Ryan et al., 2009; Harter et al., 2017; Unrath et al., 2021). Direction of transcription is indicated by the respective arrows.
Stress survival islet (SSI-2)
Harter et al. (2017) have documemted the presence of stress survival islet (SSI-2) mainly among L. monocytogenes ST121 strains (persisting for months and even years in food processing environments). SSI-2 consists of two genes: lin0464 and lin0465 (Figure 6), coding respectively, the transcription factor LIN0464 and the Pfpl protease beneficial for survival under alkaline and oxidative stress. SSI-2 is located at the hypervariable genetic point from lmo0442 to lmo0449, which contains: SSI-1, SSI-2 and the homologue of the LMOf2365_0481 gene (Harter et al., 2017). SSI-2 is highly conserved and occurs mainly in L. monocytogenes ST121 strains, the most abundant group of isolates in the food and food processing environment (Harter et al., 2017). Additionally, Harter et al. (2017) have shown that SSI-2 was not under σB control suggesting σH or σL regulation.
Interestingly, Harter et al. (2017) have noted that the presence of the LMOf2365_0481 homologue is an alternative to SSI-2 and is common among clinical strains (function unknown). Gray et al. (2021) have found that the ST1 and ST2 isolates had the LMOf2365_0481 homologue.
The presence of SSI-2 may potentially support the adaptation and persistence of L. monocytogenes strains in the food processing environment. However, its role in the pathogens’ survival merits further investigation.
Occurrence of genomic and pathogenicity islands among strains of Listeria monocytogenes isolated from various sources
Table 3 summarizes several studies on the frequency of genomic/pathogenicity islands among L. monocytogenes strains isolated from different sources in different regions worldwide. The LIPI-1 occurs in most L. monocytogenes strains, proving their virulence and ability to cause infection. The presence of SSI-1 and SSI-2 is typical for strains isolated from food and the food processing environment. Both SSI-1 and SSI-2 determine adaptation to stress conditions encountered during food processing. Also, LIPI-3 and LIPI-4 were detected in both environmental and clinical strains, which may indicate the hypervirulent nature of these strains. Nonetheless, researchers have reported a much higher frequency of LIPI-4 in clinical strains. On the contrary, LGI-1 island rarely occurs among strains of L. monocytogenes. There are also data on the occurrence of the recently discovered LGI-3 island among L. monocytogenes (Table 3). Monitoring the presence of virulence determinants in L. monocytogenes is of great importance.
Transcription regulators of L. monocytogenes
L. monocytogenes possess four transcriptional regulators controlling virulence: PrfA, SigB, CodY, and VirR.
PrfA is the “major regulator of virulence” in L. monocytogenes because it directly activates all nine virulence genes (located on LIPI-1) and indirectly regulates more than 140 accessory genes (Milohanic et al., 2003; de las Heras et al., 2011). Other genes under the control of PrfA include internalins (involved in adhesion), the hpt gene (encodes a glucose-6-phosphate transport translocation involved in the proliferation of L. monocytogenes within the host cell cytoplasm) (Chico-Calero et al., 2002), and the lapB gene (encodes the surface protein LPXTG enabling adhesion into host cells) (Reis et al., 2010).
Most strains of L. monocytogenes have five sigma factors, including one main one - σA and four alternative ones - σB, σC, σH, and σL (O’Byrne and Karatzas, 2008). σB regulates the expression of virulence genes in L. monocytogenes in response to environmental stress. The alternative factor sigma Bdirects the expression of over 300 genes involved in the stress response (NicAogain and O’Byrne, 2016). For the first time, Becker et al. (1998) demonstrated the role of σB in response to stress (osmotic). Further studies confirmed its participation in response to a variety of environmental conditions, including osmotic (Utratna et al., 2012), pH (Wiedmann et al., 1998; Wemekamp-Kamphuis et al., 2004), temperature (Liu et al., 2002) and oxidative stress (Ferreira et al., 2001).
σB interacts with PrfA in gene regulation, which is necessary for L. monocytogenes to achieve full virulence (Ollinger et al., 2009). Cross-talk between σB and PrfA ensures adequate gene expression both inside and outside the host (Gaballa et al., 2019). Interactions between σB and PrfA occur at the transcriptional, post-transcriptional, and translational levels (Gaballa et al., 2019). A direct connection exists through the σB-dependent monocistronic P2prfA promoter. In addition, the PrfA-regulated inlAB locus is also controlled by σB (Kazmierczak et al., 2003; Kim et al., 2005).
CodY is a transcriptional regulator that activates the transcription of prfA and other genes in response to low concentrations of branched-chain amino acids (BCAAs) found in host cells (Lobel et al., 2012; Lobel et al., 2015). Intracellular growth conditions also require adaptation to carbon uptake and nitrogen fixation from available sources. The nutrient-responsive regulator CodY coordinates de novo synthesis of BCAAs and sugar catabolism (Bennett et al., 2007; Lobel et al., 2012; Lobel et al., 2015). CodY represses the biosynthesis of amino acids (mainly BCAAs and histidine), purines, riboflavin, and some carbon and nitrogen metabolism genes under nutrient-rich conditions (Lemos et al., 2008; Pohl et al., 2009). Lobel and Herskovits (2016) showed that CodY activates the critical tricarboxylic acid (TCA) cycle enzymes, including glutamate/glutamine derivatives and the arginine biosynthesis pathway. Under nutrient-rich conditions, CodY directs metabolic flux from pyruvate to the TCA cycle via pyruvate carboxylase (PycA). At the same time, CodY directly represses the pyruvate oxidase gene (poxB, and the ilv operon), inhibiting pyruvate flux to the BCAA biosynthesis pathway (Lobel and Herskovits, 2016). CodY activates prfA transcription, by direct binding, in its coding sequence 15 nucleotides downstream of the start codon and thus stimulates the expression of virulence genes in response to low BCAA availability (Cossart and Archambaud, 2009). In response to nutrient excess CodY represses expression of σB and other genes (including: sigB, arg, his, actA, glpF, gadG, gdhA, glnR and fla) associated with metabolism, motility and virulence (Lobel and Herskovits, 2016).
Another but least characterized transcription regulator is VirR. VirR is a response regulator in a two-component system (TCS) consisting of a sensor histidine kinase and a response regulator (Mandin et al., 2005; Williams and Whitworth, 2010). The two-component VirR/S system controls the expression of 17 genes, including its own operon (Cossart, 2011) and the dltABCD operon (whose products are responsible for the incorporation of D-alanine into lipoteichoic acid) (Perego et al., 1995; Abachin et al., 2002). Also, the MprF protein (lysinylates phospholipids in the cell membrane of L. monocytogenes) (Thedieck et al., 2006) and AnrAB (an ATP-binding cassette (ABC) transporter) (Mandin et al., 2005; Collins et al., 2010) are under VirR control. According to Mandin et al. (2005), regulation of dlt and mprF by VirR suggests importance VirR/VirS system as a regulatorof L. monocytogenes resistance to human defensins or cationic peptides. Additionally, VirR is implicated in resistance to food preservatives and antimicrobials, especially for food preservation (Cossart, 2011; Kang et al., 2015). L. monocytogenes ΔvirR mutants showed reduced virulence in a murine infection model (Cossart, 2011). The expression of VirR and many VirR-regulated genes arehighly induced during in vivo infection, suggesting that VirR participates in the detection of the host cell environment by L. monocytogenes (Cossart and Archambaud, 2009).
Whole genome sequencing (WGS) for epidemiological purposes
Whole genome sequencing (WGS) as a tool to search for new virulence determinants?
Whole genome sequencing (WGS) is a relevant tool in the virulence assessment ofL. monocytogenes. This technique allowed the identification of five genomic/pathogenicity islands in L. monocytogenes (Table 4). The WGS also enables the characterization of strains responsible for outbreaks in the past when such modern techniques were not available, e.g., analysis of L. monocytogenesScott A responsible for the listeriosis outbreak in Massachusetts (1983) (Briers et al., 2011).
Constant analysis of the variability of L. monocytogenes strains isolated from various envi-ronments, combined with clinical, environmental, and epidemiological data, will allow us to approximate the selection mechanisms that can stimulate the adaptation of L. monocytogenes to different conditions (Disson et al., 2021). Therefore, it is essential to combine phenotypic and genotypic data to improve the value of the information provided by WGS (Wagner et al., 2022). Examples of such studies are the works of Stratakos et al. (2020), Kuenne et al. (2013), and Yan et al. (2019).
The WGS method has also been used in the identification of persistent L. monocytogenes strains (Stasiewicz et al., 2015; Cherifi et al., 2018; Wieczorek et al., 2020), which undoubtedly constitute a serious problem in the food industry.
Whole genome sequencing (WGS) and epidemic
New features ofL. monocytogenes strains, determining their hypervirulent nature, are constantly being identified. Quick strain identification may limit the pathogens’ spread, which is especially relevant during the outbreak. So far, the PFGE (Pulsed-field gel electrophoresis) method has been used as the gold standard (Ruppitsch et al., 2015; Centers for Disease Control and Prevention, 2016a; Neoh et al., 2019). In 2014, the implementation of whole genome sequencing under an epidemic study confirmed that this technique is arelevant tool for detecting outbreaks and quickly solving problems (Garner and Kathariou, 2016; Jackson et al., 2016; Moura et al., 2016; Pietzka et al., 2019). The WGS development has led to more accurate detection and analysis of the strains responsible for epidemics (Table 5). WGS allows the identification of single nucleotide polymorphisms (SNPs) and, thus, the determination of relatedness between different isolates (Moran-Gilad, 2017; Datta and Burall, 2018). Global surveillance of the food production system is critical in limiting the spread of microbial threats, including L. monocytogenes. WGS helped, among others, link the strains responsible for the outbreak in Australia with cases in Singapore (Das, 2019) or the US outbreak with cases in Australia (Kwong et al., 2016). In Europe, the WGSallowed the association of sporadic cases with food products, source attribution, and identification of the transmission pathway and antimicrobial resistance (Centers for Disease Control and Prevention, 2015a) (Table 5).
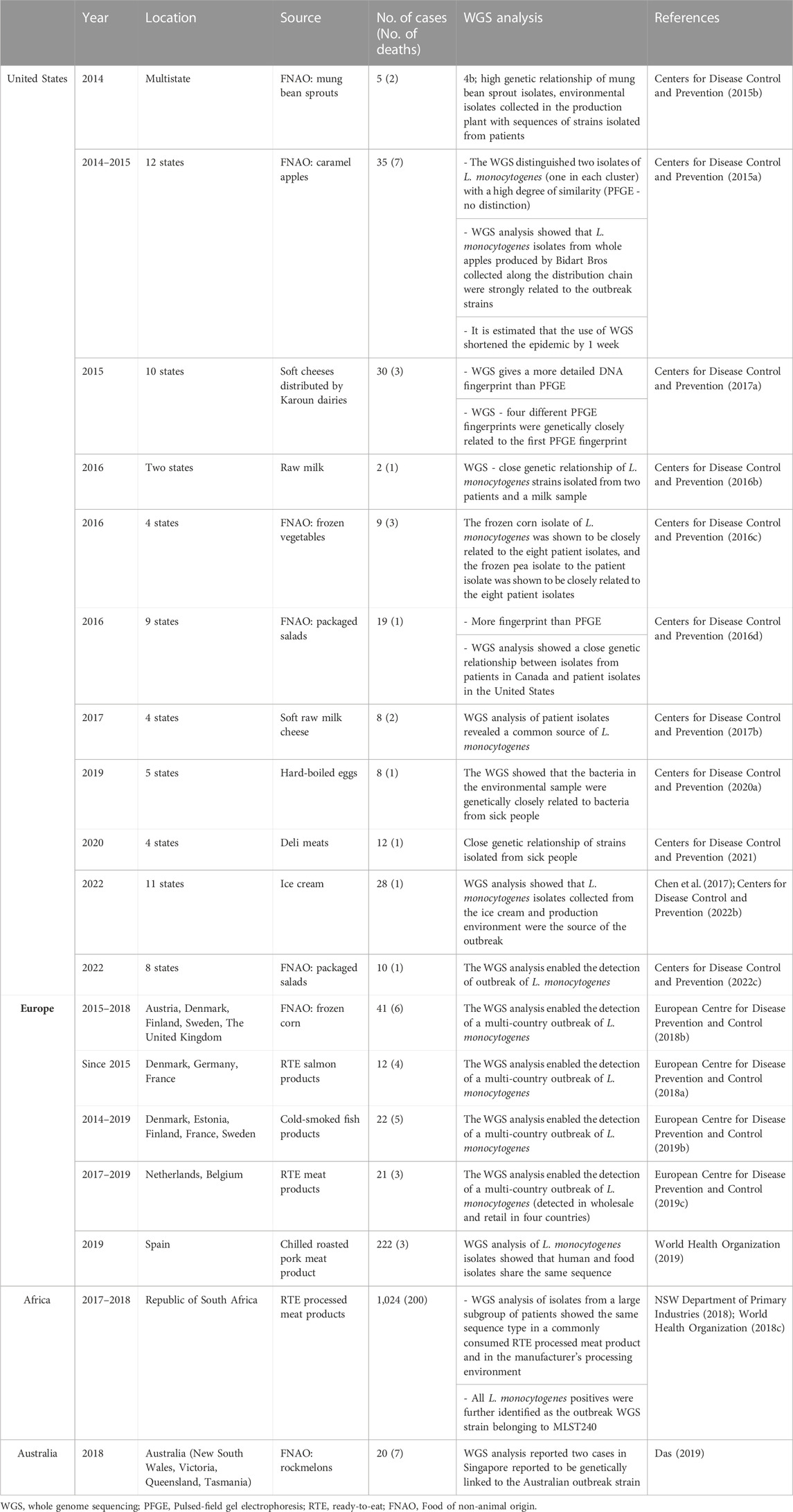
TABLE 5. The use of whole genome sequencing (WGS) for epidemiological purposes (chosen listeriosis outbreaks).
WGS is a helpful tool for strain genotype identification derived from different evolutionary lines and sources. When used for epidemiological purposes, WGS makes it possible to determine the source of L. monocytogenes.
Conclusion
The genomic and pathogenicity islands are essential for the survival and adaption of L. monocytogenes to various environmental conditions. There are still many unknowns about the function of individual proteins and their role in the pathogenesis and virulence of L. monocytogenes, as well as the compilation of this information in relation to strains isolated from different environments. Understanding the genetics of L. monocytogenes is indispensable to controlling the pathogen and reducing the risk of listeriosis. Future studies should include a comparison of pheno- and genotypic characteristics to get a full picture of the virulence and ecology of L. monocytogenes. Research on the effect of stress factors on changes in phenotypic traits and consideration of horizontal gene transfer between L. monocytogenes strains and also other species present in the environment and food processing (especially antibiotic resistance genes) is desirable.
Author contributions
Conceptualization, NW-K and KS; writing—original draft preparation, NW-K; writing—review and editing, KS and EW-Z; visualization, NW-K; supervision, KS and EW-Z; funding acquisition, NW-K. All authors contributed to the article and approved the submitted version.
Funding
The article is financed with funding of publication costs under the “Excellence Initiative—Research University” programme in Nicolaus Copernicus University in Toruń.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abachin, E., Poyart, C., Pellegrini, E., Milohanic, E., Fiedler, F., Berche, P., et al. (2002). Formation of d-alanyl-lipoteichoic acid is required for adhesion and virulence of Listeria monocytogenes. Mol. Microbiol. 43, 1–14. doi:10.1046/j.1365-2958.2002.02723.x
Abdullah, Z., Schlee, M., Roth, S., Mraheil, M. A., Barchet, W., Bottcher, J., et al. (2012). RIG-I detects infection with live Listeria by sensing secreted bacterial nucleic acids. EMBO J. 31, 4153–4164. doi:10.1038/emboj.2012.274
Al, S., Disli, H. B., Hizlisoy, H., Ertas Onmaz, N., Yildirim, Y., and Gonulalan, Z. (2022). Prevalence and molecular characterization of Listeria monocytogenes isolated from wastewater of cattle slaughterhouses in Turkey. J. Appl. Microbiol. 132 (2), 1518–1525. doi:10.1111/jam.15261
Alberti-Segui, C., Goeden, K. R., and Higgins, D. E. (2007). Differential function of Listeria monocytogenes listeriolysin O and phospholipases C in vacuolar dissolution following cell-to-cell spread. Cell Microbiol. 9 (1), 179–195. doi:10.1111/j.1462-5822.2006.00780.x
Allerberger, F., and Wagner, M. (2010). Listeriosis: A resurgent foodborne infection. Clin. Microbiol. Infect. 16, 16–23. doi:10.1111/j.1469-0691.2009.03109.x
Alvarez, D. E., and Agaisse, H. (2016). The metalloprotease mpl supports Listeria monocytogenes dissemination through resolution of membrane protrusions into vacuoles. Infect. Immun. 84 (6), 1806–1814. doi:10.1128/IAI.00130-16
Alvarez-Domínguez, C., Vázquez-Boland, J. A., Carrasco-Marín, E., López-Mato, P., and Leyva-Cobián, F. (1997). Host cell heparan sulfate proteoglycans mediate attachment and entry of Listeria monocytogenes, and the listerial surface protein ActA is involved in heparan sulfate receptor recognition. Infect. Immun. 65, 78–88. doi:10.1128/iai.65.1.78-88.1997
Amagliani, G., Brandi, G., Omiccioli, E., Casiere, A., Bruce, I. J., and Magnani, M. (2004). Direct detection of Listeria monocytogenes from milk by magnetic based DNA isolation and PCR. Food Microbiol. 21, 597–603. doi:10.1016/j.fm.2003.10.008
Anwar, T. M., Pan, H., Chai, W., -Dra, A., Fang, W., Li, Y., et al. (2022). Genetic diversity, virulence factors, and antimicrobial resistance of Listeria monocytogenes from food, livestock, and clinical samples between 2002 and 2019 in China. Int. J. Food Microbiol. 366, 109572. doi:10.1016/j.ijfoodmicro.2022.109572
Arnett, E., Vadia, S., Nackerman, C. C., Oghumu, S., Satoskar, A. R., McLeish, K. R., et al. (2014). The pore-forming toxin listeriolysin O is degraded by neutrophil metalloproteinase-8 and fails to mediate Listeria monocytogenes intracellular survival in neutrophils. J. Immunol. 192 (1), 234–244. doi:10.4049/jimmunol.1301302
Avila-Novoa, M. G., Navarrete-Sahagún, V., González-Gómez, J. P., Novoa-Valdovinos, C., Guerrero-Medina, P. J., García-Frutos, R., et al. (2021). Conditions of in vitro biofilm formation by serogroups of Listeria monocytogenes isolated from hass avocados sold at markets in Mexico. Foods 10, 2097. doi:10.3390/foods10092097
Baba, H., Kanamori, H., Kakuta, R., Sakurai, H., Oshima, K., Aoyagi, T., et al. (2021). Genomic characteristics of Listeria monocytogenes causing invasive listeriosis in Japan. Diagn Microbiol. Infect. Dis. 99 (3), 115233. doi:10.1016/j.diagmicrobio.2020.115233
Bakardjiev, A. I., Stacy, B. A., and Portnoy, D. A. (2005). Growth of Listeria monocytogenes in the Guinea pig placenta and role of cell-to-cell spread in fetal infection. J. Infect. Dis. 191, 1889–1897. doi:10.1086/430090
Barria, C., Malecki, M., and Arraiano, C. M. (2013). Bacterial adaptation to cold. Microbiology 159, 2437–2443. doi:10.1099/mic.0.052209-0
Bavdek, A., Gekara, N. O., Priselac, D., Gutiérrez Aguirre, I., Darji, A., Chakraborty, T., et al. (2007). Sterol and pH interdependence in the binding, oligomerization, and pore formation of listeriolysin O. Biochem. 46, 4425–4437. doi:10.1021/bi602497g
Bavdek, A., Kostanjšek, R., Antonini, V., Lakey, J. H., Dalla Serra, M., Gilbert, R. J., et al. (2012). pH dependence of listeriolysin O aggregation and pore-forming ability. FEBS J. 279, 126–141. doi:10.1111/j.1742-4658.2011.08405.x
Becker, L. A., Cetin, M. S., Hutkins, R. W., and Benson, A. K. (1998). Identification of the gene encoding the alternative sigma factor sigmaB from Listeria monocytogenes and its role in osmotolerance. J. Bacteriol. 80 (17), 4547–4554. doi:10.1128/JB.180.17.4547-4554.1998
Beckers, H. J., Soentoro, P. S. S., and Delfgou-van Asch, E. H. M. (1987). The occurrence of Listeria monocytogenes in soft cheeses and raw milk and its resistance to heat. Int. J. Food Microbiol. 4, 249–256. doi:10.1016/0168-1605(87)90041-9
Begley, M., Cotter, P. D., Hill, C., and Ross, R. P. (2010). Glutamate decarboxylase-mediated nisin resistance in Listeria monocytogenes. Appl. Environ. Microbiol. 76, 6541–6546. doi:10.1128/AEM.00203-10
Begley, M., Sleator, R. D., Gahan, C. G. M., and Hill, C. (2005). Contribution of three bile-associated loci, bsh, pva, and btlB, to gastrointestinal persistence and bile tolerance of Listeria monocytogenes. Infect. Immun. 73, 894–904. doi:10.1128/IAI.73.2.894-904.2005
Benda, M., Schulz, L. M., Stülke, J., and Rismondo, J. (2021). Influence of the ABC transporter YtrBCDEF of Bacillus subtilis on competence, biofilm formation and cell wall thickness. Front. Microbiol. 12, 587035. doi:10.3389/fmicb.2021.587035
Benes, J., Viechova, J., Kabelkova, M., and Horova, B. (2002). Listerial endocarditis in a penicillin-allergic woman successfully treated with a combination of 4 drugs. Scand. J. Infect. Dis. 34, 383–384. doi:10.1080/00365540110080430
Bennett, H. J., Pearce, D. M., Glenn, S., Taylor, C. M., Kuhn, M., Sonenshein, A. L., et al. (2007). Characterization of relA and codY mutants of Listeria monocytogenes: Identification of the CodY regulon and its role in virulence. Mol. Microbiol. 63, 1453–1467. doi:10.1111/j.1365-2958.2007.05597.x
Bitar, A. P., Cao, M., and Marquis, H. (2008). The metalloprotease of Listeria monocytogenes is activated by intramolecular autocatalysis. J. Bacteriol. 190 (1), 107–111. doi:10.1128/JB.00852-07
Bonazzi, M., Lecuit, M., and Cossart, P. (2009). Listeria monocytogenes internalin and E-cadherin: From bench to bedside. Cold Spring Harb. Perspect. Biol. 1 (4), a003087. doi:10.1101/cshperspect.a003087
Briers, Y., Klumpp, J., Schuppler, M., and Loessner, M. J. (2011). Genome sequence of Listeria monocytogenes Scott A, a clinical isolate from a food-borne listeriosis outbreak. J. Bacteriol. 193, 4284–4285. doi:10.1128/JB.05328-11
Bucur, F. I., Grigore-Gurgu, L., Crauwels, P., Riedel, C. U., and Nicolau, A. I. (2018). Resistance of Listeria monocytogenes to stress conditions encountered in food and food processing environments. Front. Microbiol. 9, 2700. doi:10.3389/fmicb.2018.02700
Burdette, D. L., Monroe, K. M., Sotelo-Troha, K., Iwig, J. S., Eckert, B., Hyodo, M., et al. (2011). STING is a direct innate immune sensor of cyclic di-GMP. Nature 478, 515–518. doi:10.1038/nature10429
Camargo, A. C., Moura, A., Avillan, J., Herman, N., McFarland, A. P., Sreevatsan, S., et al. (2019). Whole-genome sequencing reveals Listeria monocytogenes diversity and allows identification of long-term persistent strains in Brazil. Environ. Microbiol. 21 (12), 4478–4487. doi:10.1111/1462-2920.14726
Camejo, A., Carvalho, F., Reis, O., Leitão, E., Sousa, S., and Cabanes, D. (2011). The arsenal of virulence factors deployed by Listeria monocytogenes to promote its cell infection cycle. Virulence 2 (5), 379–394. doi:10.4161/viru.2.5.17703
Cardenas-Alvarez, M. X., Zeng, H., Webb, B. T., Mani, R., Muñoz, M., and Bergholz, T. M. (2022). Comparative genomics of Listeria monocytogenes isolates from ruminant listeriosis cases in the midwest United States. Microbiol. Spectr. 31, e0157922. doi:10.1128/spectrum.01579-22
Carrero, J. A., Calderon, B., and Unanue, E. R. (2008). Listeriolysin O from Listeria monocytogenes is a lymphocyte apoptogenic molecule. J. Immunol. 172 (8), 4866–4874. doi:10.4049/jimmunol.172.8.4866
Carrero, J. A., Vivanco-Cid, H., and Unanue, E. R. (2008). Granzymes drive a rapid listeriolysin O-induced T cell apoptosis. J. Immunol. 181 (2), 1365–1374. doi:10.4049/jimmunol.181.2.1365
Cavalcanti, A. A. C., Limeira, C. H., de Siqueira, I. N., de Lima, A. C., de Medeiros, F. J. C., de Souza, J. G., et al. (2022). The prevalence of Listeria monocytogenes in meat products in Brazil: A systematic literature review and meta-analysis. Res. Veterinary Sci. 145, 169–176. doi:10.1016/j.rvsc.2022.02.015
Centers for Disease Control and Prevention (CDC) (2015b). Fresh produce–associated listeriosis outbreaks, sources of concern, teachable moments, and insights. Sprouts and investigation of human listeriosis cases (final update). Available at: http://www.cdc.gov/listeria/outbreaks/bean-sprouts-11-14/index.html [Accessed December 10, 2022]
Centers for Disease Control and Prevention (CDC) (2022c). Listeria outbreak linked to packaged salads produced by fresh express. Available at: https://www.cdc.gov/listeria/outbreaks/packaged-salad-12-21-b/index.html [Accessed May 31, 2023]
Centers for Disease Control and Prevention (CDC) (2022a). Listeria outbreaks. Available at: https://www.cdc.gov/listeria/outbreaks/index.html [Accessed March 15, 2022]
Centers for Disease Control and Prevention (CDC) (2016c). Multistate outbreak of listeriosis linked to frozen vegetables (final update). Available at: https://www.cdc.gov/listeria/outbreaks/frozen-vegetables-05-16/index.html [Accessed December 10, 2022]
Centers for Disease Control and Prevention (CDC) (2016b). Multistate outbreak of listeriosis linked to raw milk produced by miller’s organic farm in Pennsylvania (final update). Available at: https://www.cdc.gov/listeria/outbreaks/raw-milk-03-16/index.html [Accessed December 10, 2022]
Centers for Disease Control and Prevention (CDC) (2017a). Multistate outbreak of listeriosis linked to soft cheeses distributed by karoun dairies, inc. (final update). Available at: https://www.cdc.gov/listeria/outbreaks/soft-cheeses-09-15/index.html [Accessed December 10, 2022]
Centers for Disease Control and Prevention (CDC) (2017b). Multistate outbreak of listeriosis linked to soft raw milk cheese made by vulto creamery (final update). Available at: https://www.cdc.gov/listeria/outbreaks/soft-cheese-03-17/index.html [Accessed December 10, 2022]
Centers for Disease Control and Prevention (CDC) (2020). Outbreak of Listeria infections linked to hard-boiled eggs. Available at: https://www.cdc.gov/listeria/outbreaks/eggs-12-19/index.html [Accessed December 10, 2022]
Centers for Disease Control and Prevention (CDC) (2016a). Pulsed-field gel electrophoresis (PFGE). Available at: https://www.cdc.gov/pulsenet/pathogens/pfge.html [Accessed December 14, 2022]
Centers of Disease Control and Prevention (CDC) (2022b). Listeria outbreak linked to ice cream. Available at: https://www.cdc.gov/listeria/outbreaks/monocytogenes-06-22/index.html [Accessed May 24, 2022]
Centers of Disease Control and Prevention (CDC) (2015a). Multistate outbreak of listeriosis linked to commercially produced, prepackaged caramel apples made from Bidart Bros. Apples (Final Update). Available at: https://www.cdc.gov/listeria/outbreaks/caramel-apples-12-14/index.html [Accessed March 26, 2022]
Centers of Disease Control and Prevention (CDC) (2016d). Multistate outbreak of listeriosis linked to packaged salads produced at springfield, Ohio dole processing facility (final update). Available at: https://www.cdc.gov/listeria/outbreaks/bagged-salads-01-16/index.html [Accessed March 30, 2022]
Centers of Disease Control and Prevention (CDC) (2021). Outbreak of Listeria infections linked to deli meats. Available at: https://www.cdc.gov/listeria/outbreaks/delimeat-10-20/index.html [Accessed April 14, 2022]
Centers of Disease Control and Prevention (CDC) (2011). Timeline of events: Multistate outbreak of listeriosis linked to whole cantaloupes from jensen farms, Colorado. Available at: https://www.cdc.gov/listeria/outbreaks/cantaloupes-jensen-farms/timeline.html [Accessed April 12, 2022]
Chen, C., Nguyen, B. N., Mitchell, G., Margolis, S. R., Ma, D., and Portnoy, D. A. (2018). The listeriolysin O PEST-like sequence Co-opts AP-2-mediated endocytosis to prevent plasma membrane damage during Listeria infection. Cell Host Microbe 23 (6), 786–795. doi:10.1016/j.chom.2018.05.006
Chen, M., Cheng, J., Wu, Q., Zhang, J., Chen, Y., Xue, L., et al. (2018). Occurrence, antibiotic resistance, and population diversity of Listeria monocytogenes isolated from fresh aquatic products in China. Front. Microbiol. 9, 2215. doi:10.3389/fmicb.2018.02215
Chen, M., Cheng, J., Zhang, J., Chen, Y., Zeng, H., Xue, L., et al. (2019). Isolation, potential virulence, and population diversity of Listeria monocytogenes from meat and meat products in China. Front. Microbiol. 10, 946. doi:10.3389/fmicb.2019.00946
Chen, T., Orsi, R. H., Chen, R., Gunderson, M., Roof, S., Wiedmann, M., et al. (2022). Characterization of Listeria monocytogenes isolated from wildlife in central New York. Vet. Med. Sci. 8 (3), 1319–1329. doi:10.1002/vms3.758
Chen, Y., Chen, M., Wang, J., Wu, Q., Cheng, J., Zhang, J., et al. (2020a). Heterogeneity, characteristics, and public health implications of Listeria monocytogenes in ready-to-eat foods and pasteurized milk in China. Front. Microbiol. 11, 642. doi:10.3389/fmicb.2020.00642
Chen, Y., Chen, Y., Pouillot, R., Dennis, S., Xian, Z., Luchansky, J. B., et al. (2020b). Genetic diversity and profiles of genes associated with virulence and stress resistance among isolates from the 2010-2013 interagency Listeria monocytogenes market basket survey. PLoS One 15 (4), e0231393. doi:10.1371/journal.pone.0231393
Chen, Y., Luo, Y., Curry, P., Timme, R., Melka, D., Doyle, M., et al. (2017). Assessing the genome level diversity of Listeria monocytogenes from contaminated ice cream and environmental samples linked to a listeriosis outbreak in the United States. PLoS One 12 (2), e0171389. doi:10.1371/journal.pone.0171389
Chen, Y., Zhang, W., and Knabel, S. J. (2007). Multi-virulence-locus sequence typing identifies single nucleotide polymorphisms which differentiate epidemic clones and outbreak strains of Listeria monocytogenes. J. Clin. Microbiol. 45, 835–846. doi:10.1128/jcm.01575-06
Cherifi, T., Carrillo, C., Lambert, D., Miniaï, I., Quessy, S., Larivière-Gauthier, G., et al. (2018). Genomic characterization of Listeria monocytogenes isolates reveals that their persistence in a pig slaughterhouse is linked to the presence of benzalkonium chloride resistance genes. BMC Microbiol. 18, 220. doi:10.1186/s12866-018-1363-9
Chico-Calero, I., Suarez, M., Gonzalez-Zorn, B., Scortti, M., Slaghuis, J., Goebel, W., et al. (2002). The European Listeria Genome Consortium. Hpt, a bacterial homolog of the microsomal glucose- 6-phosphate translocase, mediates rapid intracellular proliferation in Listeria. Proc. Natl. Acad. Sci. U. S. A. 99, 431–436. doi:10.1073/pnas.012363899
Clayton, E. M., Hill, C., Cotter, P. D., and Ross, R. P. (2011). Real-time PCR assay to differentiate Listeriolysin S-positive and negative strains of Listeria monocytogenes. Appl. Environ. Microbiol. 77, 163–171. doi:10.1128/AEM.01673-10
Collins, B., Curtis, N., Cotter, P. D., Hill, C., and Ross, R. P. (2010). The ABC transporter AnrAB contributes to the innate resistance of Listeria monocytogenes to nisin, bacitracin, and various beta-lactam antibiotics. Antimicrob. Agents Chemother. 54, 4416–4423. doi:10.1128/aac.00503-10
Cossart, P., and Archambaud, C. (2009). The bacterial pathogen Listeria monocytogenes: An emerging model in prokaryotic transcriptomics. J. Biol. 8, 107. doi:10.1186/jbiol202
Cossart, P. (2011). Illuminating the landscape of host-pathogen interactions with the bacterium Listeria monocytogenes. Proc. Natl. Acad. Sci. U. S. A. 108 (49), 19484–19491. doi:10.1073/pnas.1112371108
Cossart, P., Vicente, M. F., Mengaud, J., Baquero, F., Perez-Diaz, J. C., and Berche, P. (1989). Listeriolysin O is essential for virulence of Listeria monocytogenes: Direct evidence obtained by gene complementation. Infect. Immun. 57 (11), 3629–3636. doi:10.1128/iai.57.11.3629-3636.1989
Cotter, P. D., Draper, L. A., Lawton, E. M., Daly, K. M., Groeger, D. S., Casey, P. G., et al. (2008). Listeriolysin S, a novel peptide haemolysin associated with a subset of lineage I Listeria monocytogenes. PLoS Pathog. 4, e1000144. doi:10.1371/journal.ppat.1000144
Cotter, P. D., Ryan, S., Gahan, C. G., and Hill, C. (2005). Presence of GadD1 glutamate decarboxylase in selected Listeria monocytogenes strains is associated with an ability to grow at low pH. Appl. Environ. Microbiol. 71, 2832–2839. doi:10.1128/AEM.71.6.2832-2839.2005
Das, A., Sai Mala, G., Singh, R. S., Majumdar, A., Chatterjee, R., Chaudhuri, I., et al. (2019). Prelacteal feeding practice and maintenance of exclusive breast feeding in Bihar, India - identifying key demographic sections for childhood nutrition interventions: A cross-sectional study. Glob. Biosecurity 1, 1. doi:10.12688/gatesopenres.12862.3
Datta, A. R., and Burall, L. S. (2018). Serotype to genotype: The changing landscape of listeriosis outbreak investigations. Food Microbiol. 75, 18–27. doi:10.1016/j.fm.2017.06.013
de las Heras, A., Cain, R. J., Bielecka, M. K., and Vázquez-Boland, J. A. (2011). Regulation of Listeria virulence: PrfA master and commander. Curr. Opin. Microbiol. 14, 118–127. doi:10.1016/j.mib.2011.01.005
Decatur, A. L., and Portnoy, D. A. (2000). A PEST-like sequence in listeriolysin O essential for Listeria monocytogenes pathogenicity. Science 290 (5493), 992–995. doi:10.1126/science.290.5493.992
den Bakker, H. C., Cummings, C. A., Ferreira, V., Vatta, P., Orsi, R. H., Degoricija, L., et al. (2010). Comparative genomics of the bacterial genus Listeria: Genome evolution is characterized by limited gene acquisition and limited gene loss. BMC Genomics 11, 688. doi:10.1186/1471-2164-11-688
Disson, O., Moura, A., and Lecuit, M. (2021). Making sense of the biodiversity and virulence of Listeria monocytogenes. Trends Microbiol. 29 (9), 811–822. doi:10.1016/j.tim.2021.01.008
Dobrindt, U., Hochhut, B., Hentschel, U., and Hacker, J. (2004). Genomic islands in pathogenic and environmental microorganisms. Nat. Rev. Microbiol. 2, 414–424. doi:10.1038/nrmicro884
Doumith, M., Buchrieser, C., Glaser, P., Jacquet, C., and Martin, P. (2004). Differentiation of the major Listeria monocytogenes serovars by multiplex PCR. J. Clin. Microbiol. 42, 3819–3822. doi:10.1128/JCM.42.8.3819-3822.2004
Drevets, D. A., Jelinek, T. A., and Freitag, N. E. (2001). Listeria monocytogenes-infected phagocytes can initiate central nervous system infection in mice. Infect. Immun. 69, 1344–1350. doi:10.1128/iai.69.3.1344-1350.2001
Dubail, I., Autret, N., Beretti, J.-L., Kayal, S., Berche, P., and Charbit, A. (2001). Functional assembly of two membrane-binding domains in listeriolysin O, the cytolysin of Listeria monocytogenes. Microbiology 147 (10), 2679–2688. doi:10.1099/00221287-147-10-2679
Dunstone, M. A., and Tweten, R. K. (2012). Packing a punch: The mechanism of pore formation by cholesterol dependent cytolysins and membrane attack complex/perforin-like proteins. Curr. Opin. Struct. Biol. 22, 342–349. doi:10.1016/j.sbi.2012.04.008
Dussurget, O. (2008). New insights into determinants of Listeria monocytogenes virulence. Int. Rev. Cell Mol. Biol. 270, 1–38. doi:10.1016/S1937-6448(08)01401-9
EFSA Panel on Biological Hazards (BIOHAZ) Panel (2013). Scientific Opinion on the risk posed by pathogens in food of non-animal origin. Part 1 (outbreak data analysis and risk ranking of food/pathogen combinations). EFSA J. 11 (1), 3025. doi:10.2903/j.efsa.2013.3025
Eimerman, P. R. (2010). Characterization of Listeria monocytogenes growth and colonization of the murine gallbladder. Stanford, CA, USA: Stanford University.
Elhanafi, D., Dutta, V., and Kathariou, S. (2010). Genetic characterization of plasmid associated benzalkonium chloride resistance determinants in a Listeria monocytogenes strain from the 1998-1999 outbreak. Appl. Environ. Microbiol. 76, 8231–8238. doi:10.1128/AEM.02056-10
Engelbrecht, F., Domínguez-Bernal, G., Hess, J., Dickneite, C., Greiffenberg, L., Lampidis, R., et al. (1998). A novel PrfA-regulated chromosomal locus, which is specific for Listeria ivanovii, encodes two small, secreted internalins and contributes to virulence in mice. Mol. Microbiol. 30 (2), 405–417. doi:10.1046/j.1365-2958.1998.01076.x
European Centre for Disease Prevention and Control (ECDC) (2019a). ECDC strategic framework for the integration of molecular and genomic typing into European surveillance and multi-country outbreak investigations: 2019–2021. Stockholm: ECDC. doi:10.2900/805317
European Centre for Disease Prevention and Control (ECDC) (2018b). Epidemiological update: Multi-country outbreak of Listeria monocytogenes serogroup IVb, multi-locus sequence type 6 infections. Available at: https://www.ecdc.europa.eu/en/news-events/epidemiological-update-multi-country-outbreak-listeria-monocytogenes-serogroup-ivb [Accessed December 10, 2022]
European Centre for Disease Prevention and Control (ECDC) (2019b). Multicountry outbreak of Listeria monocytogenes clonal complex 8 infections linked to consumption of cold-smoked fish products. Available at: https://www.ecdc.europa.eu/en/publications-data/multi-country-outbreak-listeria-monocytogenes-fish-products [Accessed December 10, 2022]
European Centre for Disease Prevention and Control (ECDC) (2018a). Multicountry outbreak of Listeria monocytogenes infections linked to consumption of salmon products. Available at: https://www.ecdc.europa.eu/en/news-events/multi-country-outbreak-listeria-monocytogenes-infections-linked-consumption-salmon [Accessed December 10, 2022]
European Centre for Disease Prevention and Control (ECDC) (2019b). Rapid outbreak assessment: Multi-country outbreak of Listeria monocytogenes sequence type 6 infections linked to ready-to-eat meat products. Available at: https://www.ecdc.europa.eu/en/publications-data/rapid-outbreak-assessment-multi-country-outbreak-listeria-monocytogenes-sequence [Accessed December 10, 2022]
Faezi-Ghasemi, M., and Kazemi, S. (2015). Effect of sub-lethal environmental stresses on the cell survival and antibacterial susceptibility of Listeria monocytogenes PTCC1297. Zahedan J. Res. Med. Sci. 17, 1–6.
Ferreira, A., O'Byrne, C. P., and Boor, K. J. (2001). Role of sigma(B) in heat, ethanol, acid, and oxidative stress resistance and during carbon starvation in Listeria monocytogenes. Appl. Environ. Microbiol. 67 (10), 4454–4457. doi:10.1128/aem.67.10.4454-4457.2001
Ferreira, V., Wiedmann, M., Teixeira, P., and Stasiewicz, M. J. (2014). Listeria monocytogenes persistence in food-associated environments: Epidemiology, strain characteristics, and implications for public health. J. Food Prot. 77, 150–170. doi:10.4315/0362-028X.JFP-13-150
Fox, E. M., Allnutt, T., Bradbury, M. I., Fanning, S., and Chandry, P. S. (2016). Comparative genomics of the Listeria monocytogenes ST204 subgroup. Front. Microbiol. 7, 2057. doi:10.3389/fmicb.2016.02057
Franciosa, G., Maugliani, A., Scalfaro, C., Floridi, F., and Aureli, P. (2009). Expression of internalin A and biofilm formation among Listeria monocytogenes clinical isolates. Int. J. Immunopathol. Pharmacol. 22, 183–193. doi:10.1177/039463200902200121
Freitag, N. E., Rong, L., and Portnoy, D. A. (1993). Regulation of the prfA transcriptional activator of Listeria monocytogenes: Multiple promoter elements contribute to intracellular growth and cell-to-cell spread. Infect. Immun. 61, 2537–2544. doi:10.1128/iai.61.6.2537-2544.1993
Freitag, N., Port, G., and Miner, M. (2009). Listeria monocytogenes — From saprophyte to intracellular pathogen. Nat. Rev. Microbiol. 7, 623–628. doi:10.1038/nrmicro2171
Gaballa, A., Guariglia-Oropeza, V., Wiedmann, M., and Boor, K. J. (2019). Cross talk between SigB and PrfA in Listeria monocytogenes facilitates transitions between extra- and intracellular environments. Microbiol. Mol. Biol. Rev. 83, e00034. doi:10.1128/MMBR.00034-19
Gaillard, J. L., Berche, P., and Sansonetti, P. (1986). Transposon mutagenesis as a tool to study the role of hemolysin in the virulence of Listeria monocytogenes. Infect. Immun. 52, 50–55. doi:10.1128/iai.52.1.50-55.1986
Gal-Mor, O., and Finlay, B. B. (2006). Pathogenicity islands: A molecular toolbox for bacterial virulence. Cell Microbiol. 8 (11), 1707–1719. doi:10.1111/j.1462-5822.2006.00794.x
Gandhi, M., and Chikindas, L. M. (2007). Listeria: A foodborne pathogen that knows how to survive. Int. J. Food Microbiol. 113, 1–15. doi:10.1016/j.ijfoodmicro.2006.07.008
Garner, D., and Kathariou, S. (2016). Fresh produce-associated listeriosis outbreaks, sources of concern, teachable moments, and insights. J. Food Prot. 79 (2), 337–344. doi:10.4315/0362-028X.JFP-15-387
Gaul, L. K., Farag, N. H., Shim, T., Kingsley, M. A., Silk, B. J., and Hyytia-Trees, E. (2013). Hospital-acquired listeriosis outbreak caused by contaminated diced celery - Texas, 2010. Clin. Infect. Dis. 56, 20–26. doi:10.1093/cid/cis817
Gekara, N. O., Westphal, K., Ma, B., Rohde, M., Groebe, L., and Weiss, S. (2007). The multiple mechanisms of Ca2+ signalling by listeriolysin O, the cholesterol-dependent cytolysin of Listeria monocytogenes. Cell. Microbiol. 9, 2008–2021. doi:10.1111/j.1462-5822.2007.00932.x
Gelbicova, T., Florianova, M., Hluchanova, L., Kalova, A., Korena, K., Strakova, N., et al. (2021). Comparative analysis of genetic determinants encoding cadmium, arsenic, and benzalkonium chloride resistance in Listeria monocytogenes of human, food, and environmental origin. Front. Microbiol. 11, 599882. doi:10.3389/fmicb.2020.599882
Geoffroy, C., Gaillard, J. L., Alouf, J. E., and Berche, P. (1987). Purification, characterization, and toxicity of the sulfhydryl-activated hemolysin listeriolysin O from Listeria monocytogenes. Infect. Immun. 55, 1641–1646. doi:10.1128/iai.55.7.1641-1646.1987
Germini, A., Masola, A., Carnevali, P., and Marchelli, R. (2009). Simultaneous detection of Escherichia coli O157: H7, Salmonella spp. and Listeria monocytogenes by multiplex PCR. Food control. 20, 733–738. doi:10.1016/j.foodcont.2008.09.010
Gillespie, I. A., Mook, P., Little, C. L., Grant, K., and Adak, G. K. (2010). Listeria monocytogenes infection in the over-60s in England between 2005 and 2008: A retrospective case-control study utilizing market research panel data. Foodborne path. Dis. 7, 1373–1379. doi:10.1089/fpd.2010.0568
Gilmour, M. W., Graham, M., Van Domselaar, G., Tyler, S., Kent, H., Trout-Yakel, K. M., et al. (2010). High-throughput genome sequencing of two Listeria monocytogenes clinical isolates during a large foodborne outbreak. BMC Genomics 11, 120–215. doi:10.1186/1471-2164-11-120
Giotis, E. S., Muthaiyan, A., Natesan, S., Wilkinson, B. J., Blair, I. S., and McDowell, D. A. (2010). Transcriptome analysis of alkali shock and alkali adaptation in Listeria monocytogenes 10403S. Foodborne Pathog. Dis. 7, 1147–1157. doi:10.1089/fpd.2009.0501
Godziszewska, S., Musioł, E., and Duda, I. (2015). Listeriosis – A dangerous, contagious disease. Meningitis caused by Listeria monocytogenes – Case report. ANN.ACAD.MED. SILES (online). 69, 118–124. doi:10.18794/aams/33100
Goldfine, H., and Knob, C. (1992). Purification and characterization of Listeria monocytogenes phosphatidylinositol-specific phospholipase C. Infect. Immun. 60 (10), 4059–4067. doi:10.1128/iai.60.10.4059-4067.1992
González-Zorn, B., Domínguez-Bernal, G., Suárez, M., Ripio, M.-T., Vega, Y., Novella, S., et al. (2000). SmcL, a novel membrane-damaging virulence factor in Listeria. List. Int. J. Med. Microbiol. 290 (4-5), 369–374. doi:10.1016/S1438-4221(00)80044-2
Gorski, L., Cooley, M. B., Oryang, D., Carychao, D., Nguyen, K., Luo, Y., et al. (2022). Prevalence and clonal diversity of over 1,200 Listeria monocytogenes isolates collected from public access waters near produce production areas on the central California coast during 2011 to 2016. Appl. Environ. Microbiol. 88 (8), e0035722. doi:10.1128/aem.00357-22
Goulet, V., and Marchetti, P. (1996). Listeriosis in 225 non-pregnant patients in 1992: Clinical aspects and outcome in relation to predisposing conditions. Scand. J. Infect. Dis. 28, 367–374. doi:10.3109/00365549609037921
Gray, J. A., Scott Chandry, P., Kaur, M., Kocharunchitt, C., Bowman, J. P., and Fox, E. M. (2021). Characterisation of Listeria monocytogenes food-associated isolates to assess environmental fitness and virulence potential. Int. J. Food Microbiol. 350, 109247. doi:10.1016/j.ijfoodmicro.2021.109247
Grubaugh, D., Regeimbal, J. M., Ghosh, P., Zhou, Y., Lauer, P., Dubensky, T. W., et al. (2018). The VirAB ABC transporter is required for VirR regulation of Listeria monocytogenes virulence and resistance to nisin. Infect. Immun. 86, e00901–e00917. doi:10.1128/IAI.00901-17
Guidi, F., Lorenzetti, C., Centorotola, G., Torresi, M., Cammà, C., Chiaverini, A., et al. (2022). Atypical serogroup IVb-v1 of Listeria monocytogenes assigned to new ST2801, widely spread and persistent in the environment of a pork-meat producing plant of central Italy. Front. Microbiol. 13, 930895. doi:10.3389/fmicb.2022.930895
Guillet, C., Join-Lambert, O., Le Monnier, A., Leclercq, A., Mechaï, F., Mamzer-Bruneel, M. F., et al. (2010). Human listeriosis caused by Listeria ivanovii. Emerg. Infect. Dis. 16, 136–138. doi:10.3201/eid1601.091155
Hadjilouka, A., Paramithiotis, S., and Drosinos, E. H. (2018). Genetic analysis of the Listeria pathogenicity island 1 of Listeria monocytogenes 1/2a and 4b isolates. Curr. Microbiol. 75, 857–865. doi:10.1007/s00284-018-1458-4
Hain, T., Steinweg, C., and Chakraborty, T. (2006). Comparative and functional genomics of Listeria spp. J. Biotechnol. 126, 37–51. doi:10.1016/j.jbiotec.2006.03.047
Halio, S. B., Blumentals, I. I., Short, S. A., Merrill, B. M., and Kelly, R. M. (1998). Sequence, expression in Escherichia coli, and analysis of the gene encoding a novel intracellular protease (PfpI) from the hyperthermophilic archaeon Pyrococcus furiosus. J. Bacteriol. 178, 2605–2612. doi:10.1128/jb.178.9.2605-2612.1996
Hallstrom, K. N., and McCormick, B. A. (2014). Pathogenicity islands: Origins, structure, and roles in bacterial pathogenesis. Mol. Med. Microbiol. 2, 303–314. doi:10.1016/B978-0-12-397169-2.00016-0
Hamon, M., Bierne, H., and Cossart, P. (2006). Listeria monocytogenes: A multifaceted model. Nat. Rev. Microbiol. 4, 423–434. doi:10.1038/nrmicro1413
Handa-Miya, S., Kimura, B., Takahashi, H., Sato, M., Ishikawa, T., Igarashi, K., et al. (2007). Nonsense-mutated inlA and prfA not widely distributed in Listeria monocytogenes isolates from ready-to-eat seafood products in Japan. Int. J. Food Microbiol. 117, 312–318. doi:10.1016/j.ijfoodmicro.2007.05.003
Hansen, K., Prabakaran, T., Laustsen, A., Jørgensen, S. E., Rahbæk, S. H., Jensen, S. B., et al. (2014). Listeria monocytogenes induces IFNβ expression through an IFI16-cGAS- and STING-dependent pathway. EMBO J. 33 (15), 1654–1666. doi:10.15252/embj.201488029
Hardy, J., Margolis, J. J., and Contag, C. H. (2006). Induced biliary excretion of Listeria monocytogenes. Infect. Immun. 74, 1819–1827. doi:10.1128/IAI.74.3.1819-1827.2006
Harter, E., Wagner, E. M., Zaiser, A., Halecker, S., Wagner, M., and Rychli, K. (2017). Stress survival islet 2, predominantly present in Listeria monocytogenes strains of sequence type 121, is involved in the alkaline and oxidative stress responses. Appl. Environ. Microbiol. 83 (16), 008277–e917. doi:10.1128/AEM.00827-17
Harvey, P. C., and Faber, J. E. (1941). Studies on the listerella group: I. Biochemical and hemolytic reactions. J. Bacteriol. 42, 677–687. doi:10.1128/JB.42.5.677-687.1941
Haubert, L., Zehetmeyr, M. L., and da Silva, W. P. (2019). Resistance to benzalkonium chloride and cadmium chloride in Listeria monocytogenes isolates from food and food processing environments in southern Brazil. Can. J. Microbiol. 65, 429–435. doi:10.1139/cjm-2018-0618
Hein, I., Klinger, S., Dooms, M., Flekna, G., Stessl, B., Leclercq, A., et al. (2011). Stress survival islet 1 (SSI-1) survey in Listeria monocytogenes reveals an insert common to Listeria innocua in sequence type 121 L. monocytogenes strains. Appl. Environ. Microbiol. 77, 2169–2173. doi:10.1128/AEM.02159-10
Henry, R., Shaughnessy, L., Loessner, M. J., Alberti-Segui, C., Higgins, D. E., and Swanson, J. A. (2006). Cytolysin dependent delay of vacuole maturation in macrophages infected with Listeria monocytogenes. Cell Microbiol. 8 (1), 107–119. doi:10.1111/j.1462-5822.2005.00604.x
Hernández-Flores, K. G., and Vivanco-Cid, H. (2015). Biological effects of listeriolysin O: Implications for vaccination. Biomed. Res. Int. 360741, 360741. doi:10.1155/2015/360741
Heuck, A. P., Moe, P. C., and Johnson, B. B. (2010). The cholesterol-dependent cytolysin family of gram-positive bacterial toxins. Subcell. Biochem. 51, 551–577. doi:10.1007/978-90-481-8622-8_20
Hilliard, A., Leong, D., Callaghan, A. O., Culligan, E. P., Morgan, C. A., Delappe, N., et al. (2018). Genomic characterization of Listeria monocytogenes isolates associated with clinical listeriosis and the food production environment in Ireland. Genes 9, 171. doi:10.3390/genes9030171
Ho, J. L., Shands, K. N., Friedland, G., Eckind, P., and Fraser, D. W. (1986). An outbreak of type 4b Listeria monocytogenes infection involving patients from eight Boston hospitals. Arch. Intern Med. 146 (3), 520–524. doi:10.1001/archinte.146.3.520
Hof, H. (2004). An update on the medical management of listeriosis. Expert Opin. Pharmacother. 5, 1727–1735. doi:10.1517/14656566.5.8.1727
Hurley, D., Luque-Sastre, L., Parker, C. T., Huynh, S., Eshwar, A. K., Nguyen, S. V., et al. (2019). Whole-genome sequencing-based characterization of 100 Listeria monocytogenes isolates collected from food processing environments over a four-year period. mSphere 4 (4), 002522–e319. doi:10.1128/mSphere.00252-19
Ireton, K. (2007). Entry of the bacterial pathogen Listeria monocytogenes into mammalian cells. Cell. Microbiol. 9 (6), 1365–1375. doi:10.1111/j.1462-5822.2007.00933.x
Ireton, K., Mortuza, R., Gyanwali, G. C., Gianfelice, A., and Hussain, M. (2021). Role of internalin proteins in the pathogenesis of Listeria monocytogenes. Mol. Microbiol. 116 (6), 1407–1419. doi:10.1111/mmi.14836
Jackson, B. R., Tarr, C., Strain, E., Jackson, K. A., Conrad, A., Carleton, H., et al. (2016). Implementation of nationwide real-time whole-genome sequencing to enhance listeriosis outbreak detection and investigation. Clin. Infect. Dis. 63 (3), 380–386. doi:10.1093/cid/ciw242
Jacobs, T., Darji, A., Frahm, N., Rohde, M., Wehland, J., Chakraborty, T., et al. (1998). Listeriolysin O: Cholesterol inhibits cytolysis but not binding to cellular membranes. Mol. Microbiol. 28 (6), 1081–1089. doi:10.1046/j.1365-2958.1998.00858.x
Jacquet, C., Doumith, M., Gordon, J. I., Martin, P. M. V., Cossart, P., and Lecuit, M. (2004). A molecular marker for evaluating the pathogenic potential of foodborne Listeria monocytogenes. J. Infect. Dis. 189, 2094–2100. doi:10.1086/420853
Jiang, X., Geng, Y., Ren, S., Yu, T., Li, Y., Liu, G., et al. (2019). The VirAB-VirSR-AnrAB multicomponent system is involved in resistance of Listeria monocytogenes EGD-e to cephalosporins, bacitracin, nisin, benzalkonium chloride, and ethidium bromide. Appl. Environ. Microbiol. 85, 014700–e1519. doi:10.1128/AEM.01470-19
Jin, F., Feng, Y., Chen, C., Yao, H., Zhang, R., Zhang, Q., et al. (2022). Transmembrane protein LMxysn_1693 of serovar 4h Listeria monocytogenes is associated with bile salt resistance and intestinal colonization. Microorganisms 10, 1263. doi:10.3390/microorganisms10071263
Jofre, A., Martin, B., Garriga, M., Pla, M., Rodriguez-Lazaro, D., Aymerich, T., et al. (2005). Simultaneous detection of Listeria monocytogenes and Salmonella by multiplex PCR in cooked ham. Food Microbiol. 22, 109–115. doi:10.1016/j.fm.2004.04.009
Johansson, J., and Freitag, N. E. (2019). Regulation of Listeria monocytogenes virulence. Microbiol. Spectr. 7 (4). doi:10.1128/microbiolspec.GPP3-0064-2019
Join-Lambert, O. F., Ezine, S., Le Monnier, A., Jaubert, F., Okabe, M., Berche, P., et al. (2005). Listeria monocytogenes-infected bone marrow myeloid cells promote bacterial invasion of the central nervous system. Cell Microbiol. 7, 167–180. doi:10.1111/j.1462-5822.2004.00444.x
Jonquieres, R., Bierne, H., Mengaud, J., and Cossart, P. (1998). The inlA gene of Listeria monocytogenes L028 harbors a nonsense mutation resulting in release of internalin. Infect. Immun. 66, 3420–3422. doi:10.1128/IAI.66.7.3420-3422.1998
Junttila, J., and Brander, M. (1989). Listeria monocytogenes septicemia associated with consumption of salted mushrooms. Scand. J. Infect. Dis. 21, 339–342. doi:10.3109/00365548909035707
Kammoun, H., Kim, M., Hafner, L., Gaillard, J., Disson, O., and Lecuit, M. (2022). Listeriosis, a model infection to study host-pathogen interactions in vivo. Curr. Opin. Microbiol. 66, 11–20. doi:10.1016/j.mib.2021.11.015
Kang, J., Wiedmann, M., Boor, K. J., and Bergholz, T. M. (2015). VirR-mediated resistance of Listeria monocytogenes against food antimicrobials and cross-protection induced by exposure to organic acid salts. Appl. Environ. Microbiol. 81, 4553–4562. doi:10.1128/AEM.00648-15
Kathariou, S. (2003). “Foodborne outbreaks of listeriosis and epidemic-associated lineages of Listeria monocytogenes,” in Microbial food safety in animal agriculture. Editor M. E. Torrence, and R. E. Isaacson (Ames, IA: State University Press), 243–256.
Kathariou, S., Evans, P., and Dutta, V. (2017). “Strain-specific virulence differences in Listeria monocytogenes: Current perspectives in addressing an old and vexing issue,” in Foodborne pathogens 1st Editors J. B. Gurtler, M. P. Doyle, and J. L. Kornacki (Cham, Switzerland: Springer), 61–92.
Kawacka, I., Olejnik-Schmidt, A., and Schmidt, M. (2022). Nonhemolytic Listeria monocytogenes—prevalence rate, reasons underlying atypical phenotype, and methods for accurate hemolysis assessment. Microorganisms 10, 483. doi:10.3390/microorganisms10020483
Kayal, S., and Charbit, A. (2006). Listeriolysin O: A key protein of Listeria monocytogenes with multiple functions. FEMS Microbiol. Rev. 30(4), 514–529. doi:10.1111/j.1574-6976.2006.00021.x
Kazmierczak, M. J., Mithoe, S. C., Boor, K. J., and Wiedmann, M. (2003). Listeria monocytogenes sigma B regulates stress response and virulence functions. J. Bacteriol. 185 (19), 5722–5734. doi:10.1128/JB.185.19.5722-5734.2003
Keeney, K., Trmcic, A., Zhu, Z., Delaquis, P., and Wang, S. (2018). Stress survival islet 1 contributes to serotype-specific differences in biofilm formation in Listeria monocytogenes. Lett. Appl. Microbiol. 67, 530–536. doi:10.1111/lam.13072
Kerr, K., Dealler, S. F., and Lacey, R. W. (1988). Listeria in cook-chill food. Lancet 2, 37–38. doi:10.1016/s0140-6736(88)92960-1
Kim, H., Marquis, H., and Boor, K. J. (2005). SigmaB contributes to Listeria monocytogenes invasion by controlling expression of inlA and inlB. Microbiol. Read. 151, 3215–3222. doi:10.1099/mic.0.28070-0
Kim, S. W., Haendiges, J., Keller, E. N., Myers, R., Kim, A., Lombard, J. E., et al. (2018). Genetic diversity and virulence profiles of Listeria monocytogenes recovered from bulk tank milk, milk filters, and milking equipment from dairies in the United States (2002 to 2014. ). PLoS ONE 13 (5), e0197053. doi:10.1371/journal.pone.0197053
Knabel, S. J., Reimer, A., Verghese, B., Lok, M., Ziegler, J., Farber, J., et al. (2012). Sequence typing confirms that a predominant Listeria monocytogenes clone caused human listeriosis cases and outbreaks in Canada from 1988 to 2010. J. Clin. Microbiol. 50, 1748–1751. doi:10.1128/JCM.06185-11
Kocaman, N., and Sarimehmetoğlu, B. (2016). Stress responses of Listeria monocytogenes. Ank. Üniv Vet. Fak. Derg. 63, 421–427.
Köster, S., van Pee, K., Hudel, M., Leustik, M., Rhinow, D., Kühlbrandt, W., et al. (2014). Crystal structure of listeriolysin O reveals molecular details of oligomerization and pore formation. Nat. Commun. 5, 3690. doi:10.1038/ncomms4690
Koutsoumanis, K. P., Kendall, P. A., and Sofos, J. N. (2003). Effect of food processing-related stresses on acid tolerance of Listeria monocytogenes. Appl. Environ. Microbiol. 69, 7514–7516. doi:10.1128/AEM.69.12.7514-7516.2003
Kovacevic, J., Ziegler, J., Wałecka-Zacharska, E., Reimer, A., Kitts, D. D., and Gilmour, M. W. (2016). Tolerance of Listeria monocytogenes to quaternary ammonium sanitizers is mediated by a novel efflux pump encoded by emrE. Appl. Environ. Microbiol. 82 (3), 939–953. doi:10.1128/AEM.03741-15
Krypotou, E., Scortti, M., Grundström, C., Oelker, M., Luisi, B. F., Sauer-Eriksson, A. E., et al. (2019). Control of bacterial virulence through the peptide signature of the habitat. Cell Rep. 26 (7), 1815–1827. doi:10.1016/j.celrep.2019.01.073
Kuenne, C., Billion, A., Mraheil, M. A., Strittmatter, A., Daniel, R., Goesmann, A., et al. (2013). Reassessment of the Listeria monocytogenes pangenome reveals dynamic integration hotspots and mobile genetic elements as major components of the accessory genome. BMC Genomics 14, 47. doi:10.1186/1471-2164-14-47
Kurpas, M., Osek, J., Moura, A., Leclercq, A., Lecuit, M., and Wieczorek, K. (2020). Genomic characterization of Listeria monocytogenes isolated from ready-to-eat meat and meat processing environments in Poland. Front. Microbiol. 11, 1412. doi:10.3389/fmicb.2020.01412
Kwong, J. C., Mercoulia, K., Tomita, T., Easton, M., Li, H. Y., Bulach, D. M., et al. (2016). Prospective whole-genome sequencing enhances national surveillance of Listeria monocytogenes. List. Monocytogenes. J. Clin. Microbiol. 54, 333–342. doi:10.1128/JCM.02344-15
Lake, F. B., van Overbeek, L. S., Baars, J. J. P., Koomen, J., Abee, T., and den Besten, H. M. W. (2021). Genomic characteristics of Listeria monocytogenes isolated during mushroom (Agaricus bisporus) production and processing. Int. J. Food Microbiol. 360, 360109438. doi:10.1016/j.ijfoodmicro.2021.109438
Lakicevic, B. Z., Den Besten, H. M. W., and De Biase, D. (2022). Landscape of stress response and virulence genes among Listeria monocytogenes strains. Front. Microbiol. 12, 738470. doi:10.3389/fmicb.2021.738470
Lam, J. G. T., Vadia, S., Pathak-Sharma, S., McLaughlin, E., Zhang, X., Swanson, J., et al. (2018). Host cell perforation by listeriolysin O (LLO) activates a Ca2+-dependent cPKC/Rac1/Arp2/3 signaling pathway that promotes Listeria monocytogenes internalization independently of membrane resealing. Mol. Biol. Cell 29 (3), 270–284. doi:10.1091/mbc.E17-09-0561
Lamont, F. R., Sobel, J., Mazaki-Tovi, S., Kusanovic, J. P., Vaisbuch, E., Kim, S. K., et al. (2011). Listeriosis in human pregnancy: A systematic review. J. Perinat. Med. 39 (3), 227–236. doi:10.1515/jpm.2011.035
Le Monnier, A., Autret, N., Join-Lambert, O., Jaubert, F., Charbit, A., Berche, P., et al. (2007). ActA is required for crossing of the fetoplacental barrier by Listeria monocytogenes. Infect. Immun. 75, 950–957. doi:10.1128/IAI.01570-06
Leber, J. H., Crimmins, G. T., Raghavan, S., Meyer-Morse, N. P., Cox, J. S., and Portnoy, D. A. (2008). Distinct TLR- and NLR-mediated transcriptional responses to an intracellular pathogen. PLoS Pathog. 4, e6. doi:10.1371/journal.ppat.0040006
Lecuit, M., Dramsi, S., Gottardi, C., Fedor-Chaiken, M., Gumbiner, B., and Cossart, P. (1999). A single amino acid in E-cadherin responsible for host specificity towards the human pathogen Listeria monocytogenes. EMBO J. 18, 3956–3963. doi:10.1093/emboj/18.14.3956
Lecuit, M. (2007). Human listeriosis and animal models. Microbes Infect. 9, 1216–1225. doi:10.1016/j.micinf.2007.05.009
Lecuit, M., Vandormael-Pournin, S., Lefort, J., Huerre, M., Gounon, P., Dupuy, C., et al. (2001). A transgenic model for listeriosis: Role of internalin in crossing the intestinal barrier. Science 292, 1722–1725. doi:10.1126/science.1059852
Lee, S. (2020). Bacteriocins of Listeria monocytogenes and their potential as a virulence factor. Toxins (Basel) 12 (2), 103. doi:10.3390/toxins12020103
Lee, S., Rakic-Martinez, M., Graves, L. M., Ward, T. J., Siletzky, R. M., and Kathariou, S. (2013). Genetic determinants for cadmium and arsenic resistance among Listeria monocytogenes serotype 4b isolates from sporadic human listeriosis patients. Appl. Environ. Microbiol. 79 (7), 2471–2476. doi:10.1128/AEM.03551-12
Lee, S., Ward, T. J., Jima, D. D., Parsons, C., and Kathariou, S. (2017). The arsenic resistance-associated Listeria genomic island LGI2 exhibits sequence and integration site diversity and a propensity for three Listeria monocytogenes clones with enhanced virulence. Appl. Environ. Microbiol. 83 (21), 011899–e1217. doi:10.1128/AEM.01189-17
Lemon, P. K., Freitag, E. N., and Kolter, R. (2010). The virulence regulator PrfA promotes biofilm formation by Listeria monocytogenes. J. Bacteriol. 192 (15), 3969–3976. doi:10.1128/JB.00179-10
Lemos, J. A., Nascimento, M. M., Lin, V. K., Abranches, J., and Burne, R. A. (2008). Global regulation by (p)ppGpp and CodY in Streptococcus mutans. J. Bacter 190 (15), 5291–5299. doi:10.1128/JB.00288-08
Leong, D., Alvarez-Ordonez, A., and Jordan, K. (2014). Monitoring occurrence and persistence of Listeria monocytogenes in foods and food processing environments in the Republic of Ireland. Front. Microbiol. 5, 436. doi:10.3389/fmicb.2014.00436
Little, C. L., Pires, S. M., Gillespie, I. A., Grant, K., and Nichols, G. L. (2010). Attribution of human Listeria monocytogenes infections in England and wales to ready-to-eat food sources placed on the market: Adaptation of the hald Salmonella source attribution model. Foodborne path. Dis. 7, 749–756. doi:10.1089/fpd.2009.0439
Liu, D., Lawrence, M. L., Wiedmann, M., Gorski, L., Mandrell, R. E., Ainsworth, A. J., et al. (2006). Listeria monocytogenes subgroups IIIA, IIIB, and IIIC delineate genetically distinct populations with varied pathogenic potential. J. Clin. Microbiol. 44 (11), 4229–4233. doi:10.1128/JCM.01032-06
Liu, S., Graham, J. E., Bigelow, L., Morse, P. D., and Wilkinson, B. J. (2002). Identification of Listeria monocytogenes genes expressed in response to growth at low temperature. Appl. Environ. Microbiol. 68 (4), 1697–1705. doi:10.1128/AEM.68.4.1697-1705.2002
Liu, X., Chen, W., Fang, Z., Yu, Y., Bi, J., Wang, J., et al. (2022). Persistence of Listeria monocytogenes ST5 in ready-to-eat food processing environment. Foods 11, 2561. doi:10.3390/foods11172561
Liu, Y., Sun, W., Sun, T., Gorris, L. G. M., Wang, X., Liu, B., et al. (2020). The prevalence of Listeria monocytogenes in meat products in China: A systematic literature review and novel meta-analysis approach. Int. J. Food Microbiol. 312, 108358. doi:10.1016/j.ijfoodmicro.2019.108358
Lobel, L., and Herskovits, A. A. (2016). Systems level analyses reveal multiple regulatory activities of CodY controlling metabolism, motility and virulence in Listeria monocytogenes. PLoS Genet. 12, e1005870. doi:10.1371/journal.pgen.1005870
Lobel, L., Sigal, N., Borovok, I., Belitsky, B. R., Sonenshein, A. L., and Herskovits, A. A. (2015). The metabolic regulator CodY links Listeria monocytogenes metabolism to virulence by directly activating the virulence regulatory gene prfA. Mol. Microbiol. 95, 624–644. doi:10.1111/mmi.12890
Lobel, L., Sigal, N., Borovok, I., Ruppin, E., and Herskovits, A. A. (2012). Integrative genomic analysis identifies isoleucine and CodY as regulators of Listeria monocytogenes virulence. PLoS Genet. 8, e1002887. doi:10.1371/journal.pgen.1002887
Locher, K. P. (2016). Mechanistic diversity in ATP-binding cassette (ABC) transporters. Nat. Struct. Mol. Biol. 23, 487–493. doi:10.1038/nsmb.3216
Loh, E., Dussurget, O., Gripenland, J., Vaitkevicius, K., Tiensuu, T., Mandin, P., et al. (2009). A trans acting Riboswitch controls expression of the virulence regulator PrfA in Listeria monocytogenes. Cell 139 (4), 770–779. doi:10.1016/j.cell.2009.08.046
Lungu, B., Ricke, S. C., and Johnson, M. G. (2009). Growth, survival, proliferation and pathogenesis of Listeria monocytogenes under low oxygen or anaerobic conditions. Anaerobe 15, 7–17. doi:10.1016/j.anaerobe.2008.08.001
Mandin, P., Fsihi, H., Dussurget, O., Vergassola, M., Milohanic, E., Toledo-Arana, A., et al. (2005). VirR, a response regulator critical for Listeria monocytogenes virulence. Mol. Microbiol. 57 (5), 1367–1380. doi:10.1111/j.1365-2958.2005.04776.x
Manso, B., Melero, B., Stessl, B., Jaime, I., Wagner, M., Rovira, J., et al. (2020). The response to oxidative stress in Listeria monocytogenes is temperature dependent. Microorganisms 8, 521. doi:10.3390/microorganisms8040521
Matereke, L. T., and Okoh, A. I. (2020). Listeria monocytogenes virulence, antimicrobial resistance and environmental persistence: A review. Pathogens 9, 528. doi:10.3390/pathogens9070528
Matle, I., Mafuna, T., Madoroba, E., Mbatha, K. R., Magwedere, K., and Pierneef, R. (2020). Population structure of non-ST6 Listeria monocytogenes isolated in the red meat and poultry value chain in South Africa. Microorganisms 8 (8), 1152. doi:10.3390/microorganisms8081152
Maury, M. M., Chenal-Francisque, V., Bracq-Dieye, H., Han, L., Leclercq, A., Vales, G., et al. (2017). Spontaneous loss of virulence in natural populations of Listeria monocytogenes. Infect. Immun. 85, 005411–e617. doi:10.1128/IAI.00541-17
Maury, M. M., Tsai, Y. H., Charlier, C., Touchon, M., Chenal-Francisque, V., Leclercq, A., et al. (2016). Uncovering Listeria monocytogenes hypervirulence by harnessing its biodiversity. Nat. Genet. 48, 308–313. doi:10.1038/ng.3501
McGann, P., Wiedmann, M., and Boor, K. (2007). The alternative sigma factor σB and the virulence gene regulator PrfA both regulate transcription of Listeria monocytogenes internalin. Appl. Environ. Microbiol. 73 (9), 2919–2930. doi:10.1128/AEM.02664-06
Meier, A. B., Guldimann, C., Markkula, A., Pöntinen, A., Korkeala, H., and Tasara, T. (2017). Comparative phenotypic and genotypic analysis of Swiss and Finnish Listeria monocytogenes isolates with respect to benzalkonium chloride resistance. Front. Microbiol. 8, 397. doi:10.3389/fmicb.2017.00397
Melton-Witt, J. A., Rafelski, S. M., Portnoy, D. A., and Bakardjiev, A. I. (2012). Oral infection with signature-tagged Listeria monocytogenes reveals organ-specific growth and dissemination routes in Guinea pigs. Infect. Immun. 80 (2), 720–732. doi:10.1128/IAI.05958-11
Mendum, M. L., and Smith, L. T. (2002). Characterization of glycine betaine porter I from Listeria monocytogenes and its roles in salt and chill tolerance. Appl. Environ. Microbiol. 68, 813–819. doi:10.1128/AEM.68.2.813-819.2002
Mengaud, J., Dramsi, S., Gouin, E., Vazquez-Boland, J. A., Milon, G., and Cossart, P. (1991). Pleiotropic control of Listeria monocytogenes virulence factors by a gene that is autoregulated. Mol. Microbiol. 5, 2273–2283. doi:10.1111/j.1365-2958.1991.tb02158.x
Mengaud, J., Geoffroy, C., and Cossart, P. (1991). Identification of a new operon involved in Listeria monocytogenes virulence: Its first gene encodes a protein homologous to bacterial metalloproteases. Infect. Immun. 59, 1043–1049. doi:10.1128/iai.59.3.1043-1049.1991
Meza-Torres, J., Lelek, M., Quereda, J. J., Sachse, M., Manina, G., Ershov, D., et al. (2021). Listeriolysin S: A bacteriocin from Listeria monocytogenes that induces membrane permeabilization in a contact-dependent manner. Proc. Natl. Acad. Sci. 118 (40), e2108155118. doi:10.1073/pnas.2108155118
Milohanic, E., Glaser, P., Coppée, J.-Y., Frangeul, L., Vega, Y., Vázquez- Boland, J. A., et al. (2003). Transcriptome analysis of Listeria monocytogenes identifies three groups of genes differently regulated by PrfA. Mol. Microbiol. 47, 1613–1625. doi:10.1046/j.1365-2958.2003.03413.x
Mohammadzadeh, R., Azadegan, A., and Kalani, B. S. (2019). Listeriolysin S may inhibit the anti-listerial properties of Lactobacillus plantarum. Microb. Pathog. 137, 103744. doi:10.1016/j.micpath.2019.103744
Molloy, E. M., Cotter, P. D., Hill, C., Mitchell, D. A., and Ross, R. P. (2011). Streptolysin S-like virulence factors: The continuing sagA. Nat. Rev. Microbiol. 9, 670–681. doi:10.1038/nrmicro2624
Moorhead, S. M., and Dykes, G. A. (2004). Influence of the sigB gene on the cold stress survival and subsequent recovery of two Listeria monocytogenes serotypes. Int. J. Food Microbiol. 91 (1), 63–72. doi:10.1016/S0168-1605(03)00332-5
Moran-Gilad, J. (2017). Whole genome sequencing (WGS) for food-borne pathogen surveillance and control—taking the pulse. Eurosurveillance 22, 30547. doi:10.2807/1560-7917.ES.2017.22.23.30547
Møretrø, T., Schirmer, B. C. T., Heir, E., Fagerlund, A., Hjemli, P., and Langsrud, S. (2017). Tolerance to quaternary ammonium compound disinfectants may enhance growth of Listeria monocytogenes in the food industry. Int. J. Food Microbiol. 241, 215–224. doi:10.1016/j.ijfoodmicro.2016.10.025
Morvan, A., Moubareck, C., Leclercq, A., Hervé-Bazin, M., Bremont, S., Lecuit, M., et al. (2010). Antimicrobial resistance of Listeria monocytogenes strains isolated from humans in France. Antimicrob. Agents Chemother. 54 (6), 2728–2731. doi:10.1128/AAC.01557-09
Moura, A., Criscuolo, A., Pouseele, H., Maury, M. M., Leclercq, A., Tarr, C., et al. (2016). Whole genome-based population biology and epidemiological surveillance of Listeria monocytogenes. Nat. Microbiol. 2, 16185. doi:10.1038/nmicrobiol.2016.185
Moura, A., Lefrancq, N., Wirth, T., Leclercq, A., Borges, V., Gilpin, B., et al. (2021). Emergence and global spread of Listeria monocytogenes main clinical clonal complex. Sci. Adv. 7, eabj9805. doi:10.1126/sciadv.abj9805
Mueller, J. K., and Freitag, E. N. (2005). Pleiotropic enhancement of bacterial pathogenesis resulting from the constitutive activation of the Listeria monocytogenes regulatory factor PrfA. Infet Immun. 73 (4), 1917–1926. doi:10.1128/IAI.73.4.1917-1926.2005
Muskalska, B. K., and Szymczak, B. (2015). Postępy badań nad bakteriami rodzaju Listeria. Postępy Mikrobiol. 54 (2), 123–132.
Nadon, C. A., Bowen, B. M., Wiedmann, M., and Boor, K. J. (2002). Sigma B contributes to PrfA-mediated virulence in Listeria monocytogenes. Infect. Immun. 70 (7), 3948–3952. doi:10.1128/IAI.70.7.3948-3952.2002
Nair, S., Milohanic, E., and Berche, P. (2000). ClpC ATPase is required for cell adhesion and invasion of Listeria monocytogenes. Infect. Immun. 68, 7061–7068. doi:10.1128/IAI.68.12.7061-7068.2000
Neoh, H. M., Tan, X. E., Sapri, H. F., and Tan, T. L. (2019). Pulsed-field gel electrophoresis (PFGE): A review of the "gold standard" for bacteria typing and current alternatives. Infect. Genet. Evol. 74, 103935. doi:10.1016/j.meegid.2019.103935
NicAogain, K., and O’Byrne, C. P. (2016). The role of stress and stress adaptations in determining the fate of the bacterial pathogen Listeria monocytogenes in the food chain. Front. Microbiol. 7, 1865. doi:10.3389/fmicb.2016.01865
Nightingale, K. K., Ivy, R. A., Ho, A. J., Fortes, E. D., Njaa, B. L., Peters, R. M., et al. (2008). inlA premature stop codons are common among Listeria monocytogenes isolates from foods and yield virulence-attenuated strains that confer protection against fully virulent strains. Appl. Environ. Microbiol. 74 (21), 6570–6583. doi:10.1128/AEM.00997-08
Nightingale, K. K., Windham, K., and Wiedmann, M. (2005). Evolution and molecular phylogeny of Listeria monocytogenes isolated from human and animal listeriosis cases and foods. J. Bacteriol. 187 (16), 5537–5551. doi:10.1128/JB.187.16.5537-5551.2005
Nikitas, G., Deschamps, C., Disson, O., Niault, T., Cossart, P., and Lecuit, M. (2011). Transcytosis of Listeria monocytogenes across the intestinal barrier upon specific targeting of goblet cell accessible E-cadherin. J. Exp. Med. 208 (11), 2263–2277. doi:10.1084/jem.20110560
NSW Department of Primary Industries (2018). Listeria outbreak investigation - summary report for the melon industry. Available at: https://www.foodauthority.nsw.gov.au/sites/default/files/_Documents/foodsafetyandyou/listeria_outbreak_investigation.pdf [Accessed December 15, 2022]
O’Byrne, C. P., and Karatzas, K. A. (2008). The role of sigmaB (σB) in the stress adaptations of Listeria monocytogenes: Overlaps between stress adaptation and virulence. Adv. Appl. Microbiol. 65, 115–140. doi:10.1016/S0065-2164(08)00605-9
Olier, M., Pierre, F., Rousseaux, S., Lamaitre, J. P., Rousset, A., Piveteau, P., et al. (2003). Expression of truncated Internalin A is involved in impaired internalization of some Listeria monocytogenes isolates carried asymptomatically by humans. Infect. Immun. 71, 1217–1224. doi:10.1128/IAI.71.3.1217-1224.2003
Ollinger, J., Bowen, B., Wiedmann, M., Boor, K. J., and Bergholz, T. M. (2009). Listeria monocytogenes σB modulates PrfA mediated virulence factor expression. Infect. Immun. 75 (5), 2113–2124. doi:10.1128/IAI.01205-08
Orsi, R. H., den Bakker, H. C., and Wiedmann, M. (2011). Listeria monocytogenes lineages: Genomics, evolution, ecology, and phenotypic characteristics. Int. J. Med. Microbiol. 301, 79–96. doi:10.1016/j.ijmm.2010.05.002
Osborne, S. E., and Brumell, J. H. (2017). Listeriolysin O: From bazooka to Swiss army knife. Philos. Trans. R. Soc. Lond B Biol. Sci. 372 (1726), 20160222. doi:10.1098/rstb.2016.0222
Osman, K. M., Kappell, A. D., Fox, E. M., Orabi, A., and Samir, A. (2020). Prevalence, pathogenicity, virulence, antibiotic resistance, and phylogenetic analysis of biofilm producing Listeria monocytogenes isolated from different ecological niches in Egypt: Food, humans, animals, and environment. Pathogens 9, 5–19. doi:10.3390/pathogens9010005
Palacios-Gorba, C., Moura, A., Leclercq, A., Gómez-Martín, Á., Gomis, J., Jiménez-Trigos, E., et al. (2021). Listeria spp. isolated from tonsils of wild deer and boars: Genomic characterization. Appl. Environ. Microbiol. 87 (6), 026511–e2720. doi:10.1128/AEM.02651-20
Palma, F., Brauge, T., Radomski, N., Mallet, L., Felten, A., Mistou, M. Y., et al. (2020). Dynamics of mobile genetic elements of Listeria monocytogenes persisting in ready-to-eat seafood processing plants in France. BMC Genomics 21, 130. doi:10.1186/s12864-020-6544-x
Parsons, C., Lee, S., Jayeola, V., and Kathariou, S. (2017). Novel cadmium resistance determinant in Listeria monocytogenes. Appl. Environ. Microbiol. 83, 025800–e2616. doi:10.1128/AEM.02580-16
Pasquali, F., Palma, F., Guillier, L., Lucchi, A., De Cesare, A., and Manfreda, G. (2018). Listeria monocytogenes sequence types 121 and 14 repeatedly isolated within one year of sampling in a rabbit meat processing plant: Persistence and ecophysiology. Front. Microbiol. 9, 596. doi:10.3389/fmicb.2018.00596
Perego, M., Glaser, P., Minutello, A., Strauch, M. A., Leopold, K., and Fischer, W. (1995). Incorporation of d-alanine into lipoteichoic acid and wall teichoic acid in Bacillus subtilis. Identification of genes and regulation. J. Biol. Chem. 270, 15598–15606. doi:10.1074/jbc.270.26.15598
Phelps, C. C., Vadia, S., Arnett, E., Tan, Y., Zhang, X., Pathak-Sharma, S., et al. (2018). Relative roles of listeriolysin O, InlA, and InlB in Listeria monocytogenes uptake by host cells. Infect. Immun. 86 (10), 005555–e618. doi:10.1128/IAI.00555-18
Pietzka, A., Allerberger, F., Murer, A., Lennkh, A., Stöger, A., Cabal Rosel, A., et al. (2019). Whole genome sequencing based surveillance of L. monocytogenes for early detection and investigations of listeriosis outbreaks. Front. Public Health. 7, 139. doi:10.3389/fpubh.2019.00139
Pillich, H., Loose, M., Zimmer, K. P., and Chakraborty, T. (2012). Activation of the unfolded protein response by Listeria monocytogenes. Cell. Microbiol. 14, 949–964. doi:10.1111/j.1462-5822.2012.01769.x
Pizarro-Cerdá, J., and Cossart, P. (2006). Subversion of cellular functions by Listeria monocytogenes. J. Pathol. 208, 215–223. doi:10.1002/path.1888
Pohl, K., Francois, P., Stenz, L., Schlink, F., Geiger, T., Herbert, S., et al. (2009). CodY in Staphylococcus aureus: A regulatory link between metabolism and virulence gene expression. J. Bacteriol. 191 (9), 2953–2963. doi:10.1128/JB.01492-08
Poimenidou, S. V., Dalmasso, M., Papadimitriou, K., Fox, E. M., Skandamis, P. N., and Jordan, K. (2018). Virulence gene sequencing highlights similarities and differences in sequences in Listeria monocytogenes serotype 1/2a and 4b strains of clinical and food origin from 3 different geographic locations. Front. Microbiol. 9, 1103. doi:10.3389/fmicb.2018.01103
Pornillos, O., Chen, Y. J., Chen, A. P., and Chang, G. (2005). X-ray structure of the EmrE multidrug transporter in complex with a substrate. Science 310 (5756), 1950–1953. doi:10.1126/science.1119776
Port, C. G., and Freitag, E. N. (2007). Identification of novel Listeria monocytogenes secreted virulence factors following mutational activation of the central virulence regulator, PrfA. Infect. Immun. 75 (12), 5886–5897. doi:10.1128/IAI.00845-07
Portnoy, D. A., Auerbuch, V., and Glomski, I. J. (2002). The cell biology of Listeria monocytogenes infection: The intersection of bacterial pathogenesis and cell-mediated immunity. J. Cell Biol. 158 (3), 409–414. doi:10.1083/jcb.200205009
Portnoy, D. A., Chakraborty, T., Goebel, W., and Cossart, P. (1992). Molecular determinants of Listeria monocytogenes pathogenesis. Infect. Immun. 60 (4), 1263–1267. doi:10.1128/iai.60.4.1263-1267.1992
Poyart, C., Abachin, E., Razafimanantsoa, I., and Berche, P. (1993). The zinc metalloprotease of Listeria monocytogenes is required for maturation of phosphatidylcholine phospholipase C: Direct evidence obtained by gene complementation. Infect. Immun. 61 (4), 1576–1580. doi:10.1128/iai.61.4.1576-1580.1993
Prokop, A., Gouin, E., Villiers, V., Nahori, M. A., Vincentelli, R., Duval, M., et al. (2017). OrfX, a nucleomodulin required for Listeria monocytogenes virulence. mBio 8 (5), 015500–e1617. doi:10.1128/mBio.01550-17
Pust, S., Morrison, H., Wehland, J., Sechi, A. S., and Herrlich, P. (2005). Listeria monocytogenes exploits ERM protein functions to efficiently spread from cell to cell. EMBO J. 24, 1287–1300. doi:10.1038/sj.emboj.7600595
Py, F. B., Lipinski, M. M., and Yuan, J. (2007). Autophagy limits Listeria monocytogenes intracellular growth in the early phase of primary infection. Autophagy 3 (2), 117–125. doi:10.4161/auto.3618
Quereda, J. J., Cossart, P., and Pizarro-Cerdá, J. (2018). “Role of Listeria monocytogenes exotoxins in virulence,” in Microbial toxins. Toxinology Editors B. Stiles, A. Alape-Girón, J. Dubreuil, and M. Mandal (Dordrecht: Springer). doi:10.1007/978-94-007-6449-1_24
Quereda, J. J., Dussurget, O., Nahori, M. A., Ghozlane, A., Volant, S., Dillies, M. A., et al. (2016). Bacteriocin from epidemic Listeria strains alters the host intestinal microbiota to favor infection. Proc. Natl. Acad. Sci. U. S. A. 113, 5706–5711. doi:10.1073/pnas.1523899113
Quereda, J. J., Meza-Torres, J., Cossart, P., and Pizarro-Cerdá, J. (2017). Listeriolysin S: A bacteriocin from epidemic Listeria monocytogenes strains that targets the gut microbiota. Gut Microbes 8, 384–391. doi:10.1080/19490976.2017.1290759
Rafelski, M. S., and Therio, A. J. (2006). Mechanism of polarization of Listeria monocytogenes surface protein ActA. Mol. Microbiol. 59 (4), 1262–1279. doi:10.1111/j.1365-2958.2006.05025.x
Raschle, S., Stephan, R., Stevens, M. J. A., Cernela, N., Zurfluh, K., Muchaamba, F., et al. (2021). Environmental dissemination of pathogenic Listeria monocytogenes in flowing surface waters in Switzerland. Sci. Rep. 11, 9066. doi:10.1038/s41598-021-88514-y
Ratani, S. S., Siletzky, R. M., Dutta, V., Yildirim, S., Osborne, J. A., Lin, W., et al. (2012). Heavy metal and disinfectant resistance of Listeria monocytogenes from foods and food processing plants. Appl. Environ. Microbiol. 78, 6938–6945. doi:10.1128/AEM.01553-12
Raveneau, J., Geoffroy, C., Beretti, J.-L., Gaillard, J.-L., Alouf, J. E., and Berche, P. (1992). Reduced virulence of a Listeria monocytogenes phospholipase-deficient mutant obtained by transposon insertion into the zinc metalloprotease gene. Infect. Immun. 60, 916–921. doi:10.1128/iai.60.3.916-921.1992
Reis, O., Sousa, S., Camejo, A., Villiers, V., Gouin, E., Cossart, P., et al. (2010). LapB, a novel Listeria monocytogenes LPXTG surface adhesin, required for entry into eukaryotic cells and virulence. J. Infect. Dis. 202, 551–562. doi:10.1086/654880
European Food Safety Authority (EFSA) Ricci, A., Allende, A., Bolton, D., Chemaly, M., Davies, R., et al. (2018). Listeria monocytogenes contamination of ready-to-eat foods and the risk for human health in the EU. EFSA J. 16 (1), e05134. doi:10.2903/j.efsa.2018.5134
Roberts, A., Nightingale, K., Jeffers, G., Fortes, E., Kongo, J. M., and Wiedmann, M. (2006). Genetic and phenotypic characterization of Listeria monocytogenes lineage III. Microbiology 152, 685–693. doi:10.1099/mic.0.28503-0
Roedel, A., Dieckmann, R., Brendebach, H., Hammerl, J. A., Kleta, S., Noll, M., et al. (2019). Biocide-tolerant Listeria monocytogenes isolates from German food production plants do not show cross-resistance to clinically relevant antibiotics. Appl. Environ. Microbiol. 85 (20), 012533–e1319. doi:10.1128/AEM.01253-19
Rosado, C. J., Kondos, S., Bull, T. E., Kuiper, M. J., Law, R. H., Buckle, A. M., et al. (2008). The MACPF/CDC family of pore-forming toxins. Cell Microbiol. 10 (9), 1765–1774. doi:10.1111/j.1462-5822.2008.01191.x
Ruppitsch, W., Pietzka, A., Prior, K., Bletz, S., Fernandez, H. L., Allerberger, F., et al. (2015). Defining and evaluating a core genome multilocus sequence typing scheme for whole-genome sequence-based typing of Listeria monocytogenes. J. Clin. Microbiol. 53 (9), 2869–2876. doi:10.1128/JCM.01193-15
Ryan, S., Begley, M., Gahan, C. G. M., and Hill, C. (2009). Molecular characterization of the arginine deiminase system in Listeria monocytogenes: Regulation and role in acid tolerance. Environ. Microbiol. 11, 432–445. doi:10.1111/j.1462-2920.2008.01782.x
Ryan, S., Begley, M., Hill, C., and Gahan, C. G. M. (2010). A five-gene stress survival islet (SSI-1) that contributes to the growth of Listeria monocytogenes in suboptimal conditions. J. Appl. Microbiol. 109, 984–995. doi:10.1111/j.1365-2672.2010.04726.x
Rychli, K., Wagner, E. M., Ciolacu, L., Zaiser, A., Tasara, T., Wagner, M., et al. (2017). Comparative genomics of human and non-human Listeria monocytogenes sequence type 121 strains. PLoS One 12, e0176857. doi:10.1371/journal.pone.0176857
Sabet, C., Lecuit, M., Canames, D., Cossart, P., and Bierne, H. (2005). LPXTG protein InlJ, a newly identified internalin involved in Listeria monocytogenes virulence. Infect. Immun. 73 (10), 6912–6922. doi:10.1128/IAI.73.10.6912-6922.2005
Salamina, G., Dalle Donne, E., Niccolini, A., Poda, G., Cesaroni, D., Bucci, M., et al. (1996). A foodborne outbreak of gastroenteritis involving Listeria monocytogenes. Epidemiol. Infect. 117, 429–436. doi:10.1017/s0950268800059082
Schmid, B., Klumpp, J., Raimann, E., Loessner, M. J., Stephan, R., and Tasara, T. (2009). Role of cold shock proteins in growth of Listeria monocytogenes under cold and osmotic stress conditions. Appl. Environ. Microbiol. 75, 1621–1627. doi:10.1128/AEM.02154-08
Schmitz-Esser, S., Müller, A., Stessl, B., and Wagner, M. (2015). Genomes of sequence type 121 Listeria monocytogenes strains harbor highly conserved plasmids and prophages. Front. Microbiol. 6, 380. doi:10.3389/fmicb.2015.00380
Schuerch, D. W., Wilson-Kubalek, E. M., and Tweten, R. K. (2005). Molecular basis of listeriolysin O pH dependence. Proc. Natl. Acad. Sci. U. S. A. 102 (35), 12537–12542. doi:10.1073/pnas.0500558102
Seeliger, H. P., and Langer, B. (1989). Serological analysis of the genus Listeria. Its values and limitations. Int. J. Food Microbiol. 8, 245–248. doi:10.1016/0168-1605(89)90020-2
Seeliger, H. P. R., and Hőhne, K. (1979). Serotyping of Listeria monocytogenes and related species. Methods Microbiol. 13, 31–49.
Sergeant, E. S., Love, S. C., and McInnes, A. (1991). Abortions in sheep due to Listeria ivanovii. Aust. Vet. J. 68, 39. doi:10.1111/j.1751-0813.1991.tb09846.x
Seveau, S., Pizarro-Cerda, J., and Cossart, P. (2007). Molecular mechanisms exploited by Listeria monocytogenes during host cell invasion. Microbes Infect. 9, 1167–1175. doi:10.1016/j.micinf.2007.05.004
Shen, J., Zhang, G., Yang, J., Zhao, L., Jiang, Y., Guo, D., et al. (2022). Prevalence, antibiotic resistance, and molecular epidemiology of Listeria monocytogenes isolated from imported foods in China during 2018 to 2020. Int. J. Food Microbiol. 382, 109916. doi:10.1016/j.ijfoodmicro.2022.109916
Shi, D., Anwar, T. M., Pan, H., Chai, W., Xu, S., and Yue, M. (2021). Genomic determinants of pathogenicity and antimicrobial resistance for 60 global Listeria monocytogenes isolates responsible for invasive infections. Front. Cell. Infect. Microbiol. 11, 718840. doi:10.3389/fcimb.2021.718840
Sibanda, T., and Buys, E. M. (2022). Listeria monocytogenes pathogenesis: The role of stress adaptation. Microorganisms 10 (8), 1522. doi:10.3390/microorganisms10081522
Simpson, D. M. (1996). Microbiology and epidemiology in foodborne disease outbreaks: The whys and when nots. J. Food Prot. 59, 93–95. doi:10.4315/0362-028X-59.1.93
Skowron, K., Wiktorczyk, N., Grudlewska, K., Wałecka-Zacharska, E., Paluszak, Z., Kruszewski, S., et al. (2019). Phenotypic and genotypic evaluation of Listeria monocytogenes strains isolated from fish and fish processing plants. Ann. Microbiol. 69, 469–482. doi:10.1007/s13213-018-1432-1
Smith, A. M., Tau, N. P., Smouse, S. L., Allam, M., Ismail, A., Ramalwa, N. R., et al. (2019). Outbreak of Listeria monocytogenes in South Africa, 2017-2018: Laboratory activities and experiences associated with whole-genome sequencing analysis of isolates. Foodborne Pathog. Dis. 16, 524–530. doi:10.1089/fpd.2018.2586
Soares, C. A., and Knuckley, B. (2016). Mechanistic studies of the agmatine deiminase from Listeria monocytogenes. Biochem. J. 473, 1553–1561. doi:10.1042/BCJ20160221
Southwick, F. S., and Purich, D. L. (1996). Intracellular pathogenesis of listeriosis. N. Engl. J. Med. 21 (12), 770–776. doi:10.1056/NEJM199603213341206
Stasiewicz, M. J., Oliver, H. F., Wiedmann, M., and den Bakker, H. C. (2015). Whole-genome sequencing allows for improved identification of persistent Listeria monocytogenes in food-associated environments. Appl. Environ. Microbiol. 81 (17), 6024–6037. doi:10.1128/AEM.01049-15
Stetson, D. B., and Medzhitov, R. (2006). Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity 24, 93–103. doi:10.1016/j.immuni.2005.12.003
Stockinger, S., Kastner, R., Kernbauer, E., Pilz, A., Westermayer, S., Reutterer, B., et al. (2009). Characterization of the interferon-producing cell in mice infected with Listeria monocytogenes. PLoS Pathog. 5, e1000355. doi:10.1371/journal.ppat.1000355
Stratakos, A. Ch., Ijaz, U. Z., Ward, P., Linton, M., Kelly, C., Pinkerton, L., et al. (2020). In vitro and in vivo characterisation of Listeria monocytogenes outbreak isolates. Food control. 107, 106784. doi:10.1016/j.foodcont.2019.106784
Suárez, M., González-Zorn, B., Vega, Y., Chico-Calero, I., and Vázquez-Boland, J. A. (2001). A role for ActA in epithelial cell invasion by Listeria monocytogenes. Cell Microbiol. 3, 853–864. doi:10.1046/j.1462-5822.2001.00160.x
Tahir, R., Rabbani, M., Ahmad, A., Tipu, M. Y., Chaudhary, M. H., and Jayarao, B. M. (2022). Study on occurrence of Listeria monocytogenes in soil of Punjab Province and its associated risk factors. J. Anim. Plant Sci. 32 (1), 45–51. doi:10.36899/JAPS.2022.1.0400
Tavares, R. M., Silva, D. A. L. D., Camargo, A. C., Yamatogi, R. S., and Nero, L. A. (2020). Interference of the acid stress on the expression of llsX by Listeria monocytogenes pathogenic island 3 (LIPI-3) variants. Food Res. Int. 132, 109063. doi:10.1016/j.foodres.2020.109063
Temple, M. E., and Nahata, M. C. (2000). Treatment of listeriosis. Ann. Pharmacother. 34, 656–661. doi:10.1345/aph.19315
Thedieck, K., Hain, T., Mohamed, W., Tindall, B. J., Nimtz, M., Chakraborty, T., et al. (2006). The MprF protein is required for lysinylation of phospholipids in listerial membranes and confers resistance to cationic antimicrobial peptides (CAMPs) on Listeria monocytogenes. Mol. Microbiol. 62, 1325–1339. doi:10.1111/j.1365-2958.2006.05452.x
Tiensuu, T., Andersson, C., Rydẻn, P., and Johansson, J. (2013). Cycles of light and dark co-ordinate reversible colony differentiation in Listeria monocytogenes. Mol. Microbiol. 87, 909–924. doi:10.1111/mmi.12140
Tilney, L. G., and Portnoy, D. A. (1989). Actin filaments and the growth, movement, and spread of the intracellular bacterial parasite, Listeria monocytogenes. J. Cell Biol. 109, 1597–1608. doi:10.1083/jcb.109.4.1597
Toledo, V., Den Bakker, H. C., Hormazábal, J. C., González-Rocha, G., Bello-Toledo, H., Toro, M., et al. (2018). Genomic diversity of Listeria monocytogenes isolated from clinical and non-clinical samples in Chile. Genes 9, 396. doi:10.3390/genes9080396
Travier, L., and Lecuit, M. (2014). Listeria monocytogenes ActA: A new function for a 'classic' virulence factor. Curr. Opin. Microbiol. 17, 53–60. doi:10.1016/j.mib.2013.11.007
Trusolino, L., and Comoglio, P. M. (2002). Scatter-factor and semaphorin receptors: Cell signalling for invasive growth. Nat. Rev. Cancer 2, 289–300. doi:10.1038/nrc779
Unrath, N., McCabe, E., Macori, G., and Fanning, S. (2021). Application of whole genome sequencing to aid in deciphering the persistence potential of Listeria monocytogenes in food production environments. Microorganisms 9 (9), 1856. doi:10.3390/microorganisms9091856
Utratna, M., Cosgrave, E., Baustian, C., Ceredig, R., and O'Byrne, C. (2012). Development and optimization of an EGFP-based reporter for measuring the general stress response in Listeria monocytogenes. Bioeng. Bugs 3 (2), 93–103. doi:10.4161/bbug.19476
Vadia, S., Arnett, E., Haghighat, A. C., Wilson-Kubalek, E. M., Tweten, R. K., and Seveau, S. (2011). The pore-forming toxin listeriolysin O mediates a novel entry pathway of L. monocytogenes into human hepatocytes. PLoS Pathog. 7, e1002356. doi:10.1371/journal.ppat.1002356
Varsaki, A., Ortiz, S., Santorum, P., López, P., López-Alonso, V., Hernández, M., et al. (2022). Prevalence and population diversity of Listeria monocytogenes isolated from dairy cattle farms in the cantabria region of Spain. Anim. (Basel) 12 (18), 2477. doi:10.3390/ani12182477
Vázquez-Boland, J. A., Domínguez-Bernal, G., González-Zorn, B., Kreft, J., and Goebel, W. (2001b). Pathogenicity islands and virulence evolution in Listeria. Microbes Infect. 3, 571–584. doi:10.1016/s1286-4579(01)01413-7
Vázquez-Boland, J. A., Kocks, C., Dramsi, S., Ohayon, H., Geoffroy, C., Mengaud, J., et al. (1992). Nucleotide sequence of the lecithinase operon of Listeria monocytogenes and possible role of lecithinase in cell-to-cell spread. Infect. Immun. 60, 219–230. doi:10.1128/iai.60.1.219-230.1992
Vázquez-Boland, J. A., Kuhn, M., Berche, P., Chakraborty, T., Domínguez-Bernal, G., Goebel, W., et al. (2001a). Listeria pathogenesis and molecular virulence determinants. Clin. Microbiol. Rev. 14 (3), 584–640. doi:10.1128/CMR.14.3.584-640.2001
Vilchis-Rangel, R. E., Espinoza-Mellado, M. D. R., Salinas-Jaramillo, I. J., Martinez-Peña, M. D., and Rodas-Suárez, O. R. (2019). Association of Listeria monocytogenes LIPI-1 and LIPI-3 marker llsX with invasiveness. Curr. Microbiol. 76, 637–643. doi:10.1007/s00284-019-01671-2
Wagner, E., Fagerlund, A., Thalguter, S., Jensen, M. R., Heir, E., Møretrø, T., et al. (2022). Deciphering the virulence potential of Listeria monocytogenes in the Norwegian meat and salmon processing industry by combining whole genome sequencing and in vitro data. Int. J. Food Microbiol. 383, 109962. doi:10.1016/j.ijfoodmicro.2022.109962
Wang, X., Jothikumar, N., and Griffiths, M. W. (2004). Enrichment and DNA extraction protocols for the simultaneous detection of Salmonella and Listeria monocytogenes in raw sausage meat with multiplex real-time PCR. J. Food Prot. 67, 189–192. doi:10.4315/0362-028x-67.1.189
Wang, Y., Ji, Q., Li, S., and Liu, M. (2021). Prevalence and genetic diversity of Listeria monocytogenes isolated from retail pork in wuhan, China. Front. Microbiol. 12, 620482. doi:10.3389/fmicb.2021.620482
Wang, Y., Luo, L., Li, Q., Wang, H., Wang, Y., Sun, H., et al. (2019). Genomic dissection of the most prevalent Listeria monocytogenes clone, sequence type ST87, in China. BMC Genomics 20, 1014. doi:10.1186/s12864-019-6399-1
Ward, T. J., Ducey, T. F., Usgaard, T., Dunn, K. A., and Bielawski, J. P. (2008). Multilocus genotyping assays for single nucleotide polymorphism-based subtyping of Listeria monocytogenes isolates. Appl. Environ. Microbiol. 74, 7629–7642. doi:10.1128/AEM.01127-08
Ward, T. J., Gorski, L., Borucki, M. K., Mandrell, R. E., Hutchins, J., and Pupedis, K. (2004). Intraspecific phylogeny and lineage group identification based on the prfA virulence gene cluster of Listeria monocytogenes. J. Bacteriol. 186, 4994–5002. doi:10.1128/JB.186.15.4994-5002.2004
Wartha, S., Bretschneider, N., Dangel, A., Hobmaier, B., Hörmansdorfer, S., Huber, I., et al. (2023b). Genetic characterization of Listeria from food of non-animal origin products and from producing and processing companies in bavaria, Germany. Foods 12, 1120. doi:10.3390/foods12061120
Wartha, S., Huber, S., Kraemer, I., Alter, T., and Messelhäußer, U. (2023a). Presence of Listeria at primary production and processing of food of non-animal origin (FNAO) in Bavaria, Germany. J. Food Prot. 86 (1), 100015. doi:10.1016/j.jfp.2022.11.007
Wemekamp-Kamphuis, H. H., Wouters, J. A., de Leeuw, P. P., Hain, T., Chakraborty, T., and Abee, T. (2004). Identification of sigma factor sigma B-controlled genes and their impact on acid stress, high hydrostatic pressure, and freeze survival in Listeria monocytogenes EGD-e. Appl. Environ. Microbiol. 70 (6), 3457–3466. doi:10.1128/AEM.70.6.3457-3466.2004
Werbrouck, H., Vermeulen, A., Van Coillie, E., Messens, W., Herman, L., Devlieghere, F., et al. (2009). Influence of acid stress on survival, expression of virulence genes and invasion capacity into Caco-2 cells of Listeria monocytogenes strains of different origins. Int. J. Food Microbiol. 134, 140–146. doi:10.1016/j.ijfoodmicro.2009.03.022
Wieczorek, K., Bomba, A., and Osek, J. (2020). Whole-genome sequencing-based characterization of Listeria monocytogenes from fish and fish production environments in Poland. Int. J. Mol. Sci. 21, 9419. doi:10.3390/ijms21249419
Wiedmann, M., Arvik, T. J., Hurley, R. J., and Boor, K. J. (1998). General stress transcription factor sigmaB and its role in acid tolerance and virulence of Listeria monocytogenes. J. Bacteriol. 180 (14), 3650–3656. doi:10.1128/JB.180.14.3650-3656.1998
Wiedmann, M., Bruce, J. L., Keating, C., Johnson, A. E., McDonough, P. L., and Batt, C. A. (1997). Ribotypes and virulence gene polymorphisms suggest three distinct Listeria monocytogenes lineages with differences in pathogenic potential. Infect. Immun. 65, 2707–2716. doi:10.1128/iai.65.7.2707-2716.1997
Wiktorczyk-Kapischke, N., Skworon, K., Grudlewska-Buda, K., Wałecka-Zacharska, E., Korkus, J., and Gospodarek-Komkowska, E. (2021). Adaptive response of Listeria monocytogenes to the stress factors in the food processing environment. Front. Microbiol. 12, 710085. doi:10.3389/fmicb.2021.710085
Wiktorczyk-Kapischke, N., Wałecka-Zacharska, E., Skowron, K., Kijewska, A., Bernaciak, Z., Bauza-Kaszewska, J., et al. (2022). Comparison of selected phenotypic features of persistent and sporadic strains of Listeria monocytogenes sampled from fish processing plants. Foods 11 (10), 1492. doi:10.3390/foods11101492
Williams, R. H., and Whitworth, D. E. (2010). The genetic organisation of prokaryotic two-component system signalling pathways. BMC Genomics 11, 720. doi:10.1186/1471-2164-11-720
Woodward, J. J., Iavarone, A. T., and Portnoy, D. A. (2010). c-di-AMP secreted by intracellular Listeria monocytogenes activates a host type I interferon response. Science 328, 1703–1705. doi:10.1126/science.1189801
World Health Organization (WHO) (2018a). Listeriosis. Available at: https://www.who.int/news-room/fact-sheets/detail/listeriosis [Accessed May 25, 2022]
World Health Organization (WHO) (2018c). Listeriosis – South Africa. Available at: https://www.who.int/emergencies/disease-outbreak-news/item/28-march-2018-listeriosis-south-africa-en [Accessed January 21, 2023]
World Health Organization (WHO) (2019). Listeriosis– Spain. Available at: https://www.who.int/emergencies/disease-outbreak-news/item/2019-DON256 [Accessed April 17, 2022]
World Health Organization (WHO) (2018b). Listeriosis—Australia. Available at: https://www.who.int/emergencies/disease-outbreak-news/item/09-april-2018-listeriosis-australia-en [Accessed April 17, 2022]
Wuenscher, D. M., Kőhler, S., Bubert, A., Gerike, U., and Goebel, W. (1993). The iap gene of Listeria monocytogenes is essential for cell viability, and its gene product, p60, has bacteriolytic activity. J. Bacteriol. 175 (11), 3491–3501. doi:10.1128/jb.175.11.3491-3501.1993
Xayarath, B., and Freitag, N. E. (2012). Optimizing the balance between host and environmental survival skills: Lessons learned from Listeria monocytogenes. Future Microbiol. 7 (7), 839–852. doi:10.2217/fmb.12.57
Xiong, H., Tanabe, Y., Ohya, S., and Mitsuyama, M. (1998). Administration of killed bacteria together with listeriolysin O induces protective immunity against Listeria monocytogenes in mice. Immunology 94 (1), 14–21. doi:10.1046/j.1365-2567.1998.00477.x
Yan, S., Li, M., Luque-Sastre, L., Wang, W., Hu, Y., Peng, Z., et al. (2019). Susceptibility (re)-testing of a large collection of Listeria monocytogenes from foods in China from 2012 to 2015 and WGS characterization of resistant isolates. J. Antimicrob. Chemother. 74 (7), 1786–1794. doi:10.1093/jac/dkz126
Yang, P., An, H., Liu, X., Wen, M., Zheng, Y., Rui, Y., et al. (2010). The cytosolic nucleic acid sensor LRRFIP1 mediates the production of type I interferon via a beta-catenin-dependent pathway. Nat. Immunol. 11, 487–494. doi:10.1038/ni.1876
Yin, Y., Yao, H., Doijad, S., Kong, S., Shen, Y., Cai, X., et al. (2019). A hybrid sub-lineage of Listeria monocytogenes comprising hypervirulent isolates. Nat. Commun. 10, 4283. doi:10.1038/s41467-019-12072-1
Yu, T., Jiang, X., Zhang, Y., Ji, S., Gao, W., and Shi, L. (2018). Effect of benzalkonium chloride adaptation on sensitivity to antimicrobial agents and tolerance to environmental stresses in Listeria monocytogenes. Front. Microbiol. 9, 2906. doi:10.3389/fmicb.2018.02906
Zhang, H., Chen, W., Wang, J., Xu, B., Liu, H., Dong, Q., et al. (2020). 10-Year molecular surveillance of Listeria monocytogenes using whole-genome sequencing in shanghai, China, 2009–2019. Front. Microbiol. 11, 551020. doi:10.3389/fmicb.2020.551020
Zhang, X., Liu, Y., Zhang, P., Niu, Y., Chen, Q., and Ma, X. (2021). Genomic characterization of clinical Listeria monocytogenes isolates in beijing, China. Front. Microbiol. 12, 751003. doi:10.3389/fmicb.2021.751003
Keywords: Listeria monocytogenes, virulence, pathogenicity island (PAI), genomic island (GEI), genes, outbreaks, whole genome sequencing (WGS)
Citation: Wiktorczyk-Kapischke N, Skowron K and Wałecka-Zacharska E (2023) Genomic and pathogenicity islands of Listeria monocytogenes—overview of selected aspects. Front. Mol. Biosci. 10:1161486. doi: 10.3389/fmolb.2023.1161486
Received: 08 February 2023; Accepted: 01 June 2023;
Published: 14 June 2023.
Edited by:
Kurt Henry Piepenbrink, University of Nebraska-Lincoln, United StatesReviewed by:
Marta Laranjo, University of Evora, PortugalRajarshi P. Ghosh, Howard Hughes Medical Institute (HHMI), United States
Sujeet Bhoite, GlaxoSmithKline, United States
Copyright © 2023 Wiktorczyk-Kapischke, Skowron and Wałecka-Zacharska. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ewa Wałecka-Zacharska, ZXdhLndhbGVja2FAdXB3ci5lZHUucGw=
 Natalia Wiktorczyk-Kapischke
Natalia Wiktorczyk-Kapischke Krzysztof Skowron
Krzysztof Skowron Ewa Wałecka-Zacharska
Ewa Wałecka-Zacharska