- 1Department of Breast Surgery and Oncology (Key Laboratory of Cancer Prevention and Intervention, China National Ministry of Education, Key Laboratory of Molecular Biology in Medical Sciences, Zhejiang Province, China), Cancer Institute, The Second Affiliated Hospital, Zhejiang University School of Medicine, Zhejiang Provincial Clinical Research Center for Cancer, Cancer Center of Zhejiang University, Hangzhou, Zhejiang, China
- 2Department of Radiation Oncology, Shandong Cancer Hospital and Institute, Shandong First Medical University and Shandong Academy of Medical Sciences, Jinan, Shandong, China
Adenocarcinoma not otherwise specified (AC) and mucinous adenocarcinoma (MC) have different biological behaviors and clinical features. We utilized our previous proteomic data and public transcriptome, single-cell transcriptome, and spatial transcriptome databases to profile the molecular atlas of the tumor microenvironments of MC, AC, and normal colon tissues. By exploring the general and specific molecular features of AC and MC, we found that AC was immune-active but exposed to a hypoxic microenvironment. MC cells could protect against DNA damage, and the microenvironment was unfavorable to leukocyte transendothelial migration. We identified several potential molecular and cellular targets of AC and MC for future research. We also highlighted that the major difference between AC and MC was not the variety of cell types and functions but possibly cell interactions. Stromal and epithelial cell interactions play important roles in both MC and AC, but different regulatory pathways were involved.
Introduction
Colorectal cancer (CRC) is the second most common cancer and causes approximately 196,000 deaths each year in China alone (Cao et al., 2021). From 2000 to 2016, the incidence rate and mortality of colorectal cancer have shown an upward trend there. Most colorectal cancers are classified as adenocarcinoma not otherwise specified (AC). Mucinous adenocarcinoma (MC) is characterized by the formation of a tumor composed of >50% mucin and accounts for approximately 10%–15% of all primary CRCs (15% of colon cancer versus 9% of rectal cancer) (Hugen et al., 2015a). AC is arranged in solid masses or small strips with gland cavities or into tubular or gland-like structures. MC shows expansile growth with clusters, strips, or singly arranged tumor cells floating in mucin pools.
Apart from morphological differences, there are different biological behaviors and clinical characteristics between AC and MC (summarized in Supplementary Table S1). Patients with MC are more often correlated with advanced-stage tumors, lymph node metastases, a larger diameter, and localization in the proximal colon (Nozoe et al., 2000; Chiang et al., 2010; Melis et al., 2010). Compared with AC, MC is related to an earlier onset age, a more advanced clinical stage, a higher degree of difficulty in surgery, and higher rates of recurrence and metastasis (Ahnen et al., 2014; Luo et al., 2019). It is difficult to effectively prevent tumor recurrence and progression through various treatments (S. H. Kim et al., 2013). MC patients mostly die of malignant intestinal obstruction (Hugen et al., 2016). Most studies have shown that MC patients often have a lower progression-free survival rate and a shorter median overall survival time (Catalano et al., 2012; Numata et al., 2012; Ott et al., 2018). Unfortunately, MC is not sensitive to chemotherapy and radiotherapy (S. H. Kim et al., 2013; McCawley et al., 2016), and other available treatments—such as targeted molecular therapy, hyperthermic intraperitoneal chemotherapy, and immunotherapy—are also not satisfactory (Luo et al., 2019; Huang et al., 2021).
Regardless of advances in the molecular characterization of CRC, there are still few molecular targets or biomarkers that could be clinically applied to improve individual treatment. Clinical features, including histologic subtype, tumor grade, lymphatic and perineural invasion, tumor budding, and host immune responses, remain the main factors in CRC treatment. Molecular biomarkers (Biller and Schrag, 2021; Ciardiello et al., 2022), such as microsatellite instability (MSI), BRAF, KRAS, NRAS, RAS, HER2, and NTRK, have contributed greatly to CRC treatment. However, they are unable to clarify some malignant events in CRC or explain the different biological behaviors between AC and MC.
Thus, it is important to decipher the molecular signature of AC and MC to improve the individualized management of patients with CRC. It has been documented that MC is related to a higher frequency of KRAS and BRAF mutations, microsatellite instability, the CpG island methylator phenotype pathway, and a lower frequency of TP53 mutations (Luo et al., 2019), on which the current treatment is mainly based. The MUC (mucin protein) family is aberrantly expressed in colorectal MC, which might be related to tumor invasion, metastasis, and chemoradiotherapy resistance (Reynolds et al., 2019). Targeted drugs for mucins are expected to be sensitive to chemoradiotherapy or immunotherapy and to improve the prognosis of patients with MC. However, there has still been no promising research outcome. Identifying comprehensive molecular characteristics will hopefully provide new strategies.
In this study, we tried to profile the molecular atlas of AC and MC with our previous proteomic data and public transcriptome, single-cell transcriptome, and spatial transcriptome databases. We found that AC was immune-active and anaerobic. MC cells could protect against DNA damage, and the microenvironment was unfavorable to leukocyte transendothelial migration. Stromal and epithelial cell interactions play important roles in both AC and MC, and different regulatory pathways are involved. There were differences in the characteristics of the tumor environments of AC and MC. The major difference was not the variety of cell types and functions but may be cell interactions.
Materials and methods
Bioinformatic analysis of the proteomics dataset
The proteomic data were obtained from our previous study (Shao et al., 2022; Siegel et al., 2022). These data were from 16 normal colon tissues, 15 colorectal AC samples, and 15 MC colorectal cancer samples (Supplementary Table S2). Gene set enrichment analysis (GSEA) was performed using the OmicStudio tool (https://www.omicstudio.cn/tool) with hallmark gene sets from the Molecular Signatures Database (MSigDB, c2.cp.kegg.v7.4). Significantly enriched hallmarks were chosen according to the cutoff p-value of <0.05. Protein–protein interaction networks functional enrichment analysis by STRING (https://string-db.org/, v11.5) was employed to perform Gene Ontology (GO) analysis, including biological processes (BPs), cellular components (CCs), and molecular functions (MFs), and distribution of tissue expression. Network analysis was performed using Cytoscape v3.7.2 with network data downloaded from STRING. R v3.6.0 was used for statistical analysis.
Bioinformatic analysis of the single-cell RNA sequencing (scRNA-seq) dataset
Human colorectal cancer scRNA-seq data from a study by Hacohen et al. (Pelka et al., 2021) (GSE178341) were downloaded from GEO. Seurat 3.0 was used to perform data normalization, variable feature finding, data scale, and reduction; after cells were clustered, we used the metadata from the paper to annotate the single cell type. NicheNet analysis (Browaeys et al., 2020) was performed using tools at saeyslab/nichenetr: NicheNet: predict active ligand–target links between interacting cells (github.com).
Bioinformatic analysis of the spatial transcriptome dataset
Human colorectal cancer spatial transcriptome data from Hu et al. (Peng et al., 2022) were downloaded. Dimensionality reduction and clustering and integration of the spatial transcriptome data with the scRNA-seq data from GSE178341 were used to perform cell annotation.
Results
Common proteomic characteristics between two types of colorectal adenocarcinomas and normal colon
We analyzed proteomics data from our previous study to profile the protein characteristics of AC, MC, and normal colon samples. First, GSEA was performed for the AC vs. normal colon groups and MC vs. normal colon groups to determine whether any signatures were enriched in the specific groups. The results indicated that the signatures were similar when the MC and AC groups were compared to the normal colon group. The top three signatures enriched in the AC group (vs. the normal group) were related to peroxisomes, drug metabolism involving cytochrome P450, and starch and sucrose metabolism (Figure 1A). The top three signatures enriched in the MC group (vs. the normal group) were associated with the metabolism of xenobiotics by cytochrome P450, drug metabolism involving cytochrome P450, and retinol metabolism (Figure 1B), which were all downregulated.
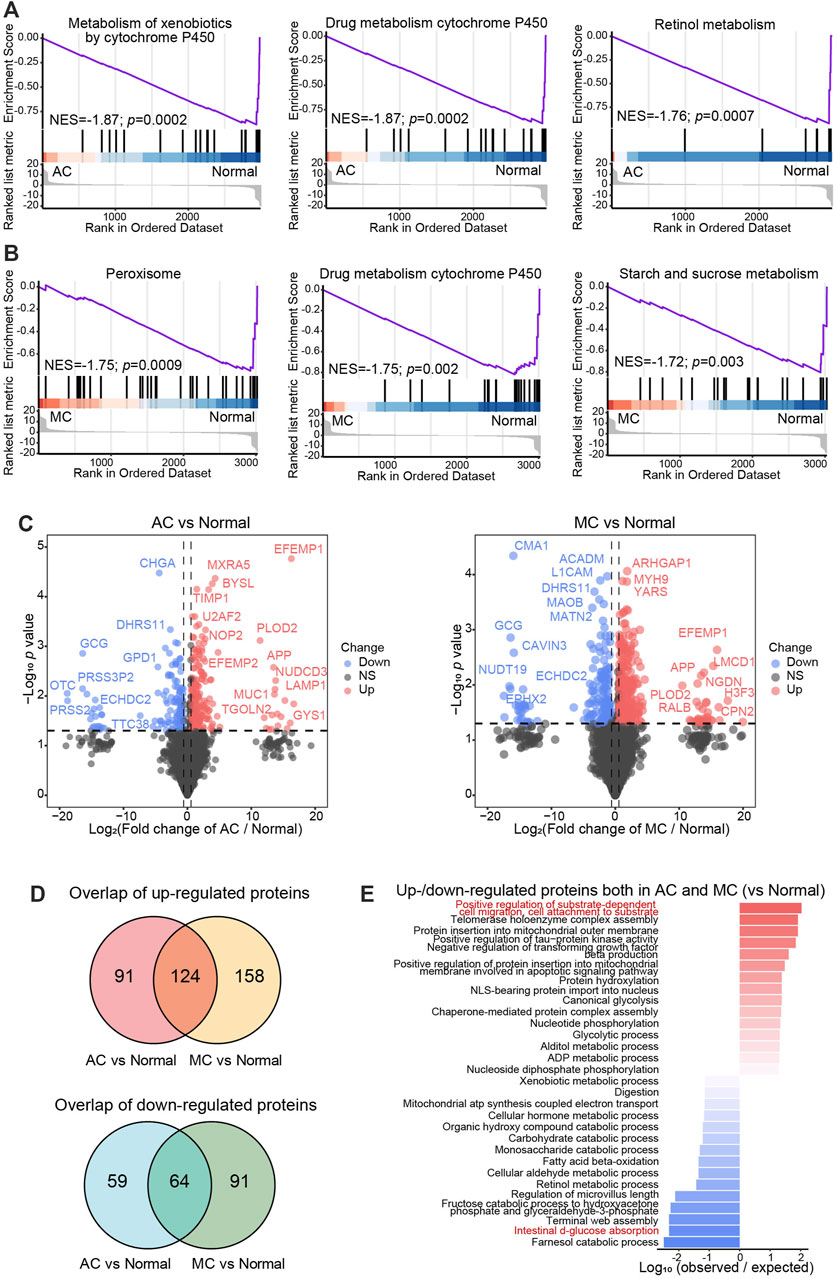
FIGURE 1. Common proteomics characteristics between two types of colorectal adenocarcinomas and normal colon. A–B: Gene set enrichment analysis (GSEA) showed the top three significantly enriched hallmarks between AC and normal tissue (A), and MC and normal tissue (B). NES: normal enrichment score. (C) Volcano plots showed the differentially expressed proteins between AC and normal tissue (left), and MC and normal tissue (right). Cutoff: fold change >1.5, p < 0.05. (D) Venn diagrams exhibited the overlap up- (upper) or down- (lower) regulated differentially expressed protein numbers between AC and normal tissue, and MC and normal tissue. (E) Bar plot showed the top -15 biological processes enriched with the overlap up- or downregulated differentially expressed proteins according to log10 (observed/expected proteins).
To further investigate the molecular characteristics of different pathologies with that of normal tissue, we compared the differentially expressed proteins between the AC group and the normal colon group and between the MC group and the normal colon group (fold change >1.5, p-value <0.05, Figure 1C, Supplementary Table S3). There were 215 proteins that were upregulated and 123 proteins that were downregulated in the AC group compared to the normal colon group, and 282 proteins that were upregulated and 155 proteins that were downregulated in the MC group compared to the normal colon group (Figure 1D). A total of 124 upregulated proteins and 64 downregulated proteins overlapped. The overlapping upregulated proteins were mostly related to the positive regulation of substrate-dependent cell migration and cell attachment to substrate (Figure 1E). The biological process of intestinal d-glucose absorption was enriched in the overlapping downregulated proteins. These findings indicate dysfunction of gut cells in CRC, which results in cachexia.
Collectively, we found that drug metabolism involving cytochrome P450 was inactive, and substrate-dependent cell migration and cell attachment to substrate processes were important for both the AC and MC groups.
Proteomic differences between two types of colorectal adenocarcinomas and normal colon tissue
To investigate differences in the proteomics between the two types of CRC and normal colon tissue, the biological processes of proteins up- or downregulated only in the AC or MC group compared with the normal colon group were enriched (Figure 2A), and proteins of some representative biological processes were assessed by network analysis (Figure 2B). The biological processes that positively regulate type I interferon production—leukocyte-mediated immunity and immune effector processes—were enriched in the proteins that were upregulated only in the AC group. Oxidative phosphorylation and other respiratory electron transport chain-related biological processes were enriched in the proteins that were downregulated only in the AC group. Among them, PECAM1 (platelet and endothelial cell adhesion molecule 1, also known as CD31) was one of the most significantly upregulated proteins found only in the AC group, and RETSAT (retinol saturase) was one of the most significantly downregulated proteins. Moreover, biological processes, including protein localization to the endoplasmic reticulum, DNA damage response, and detection of DNA damage, were enriched in the proteins upregulated only in the MC group. Extracellular matrix organization- and cellular respiration-related proteins were downregulated in the MC group. TRAM1 (translocation-associated membrane protein 1) and NDUFA7, NDUFA12, and NUBPL were some of the most significantly up/downregulated proteins found only in MC.
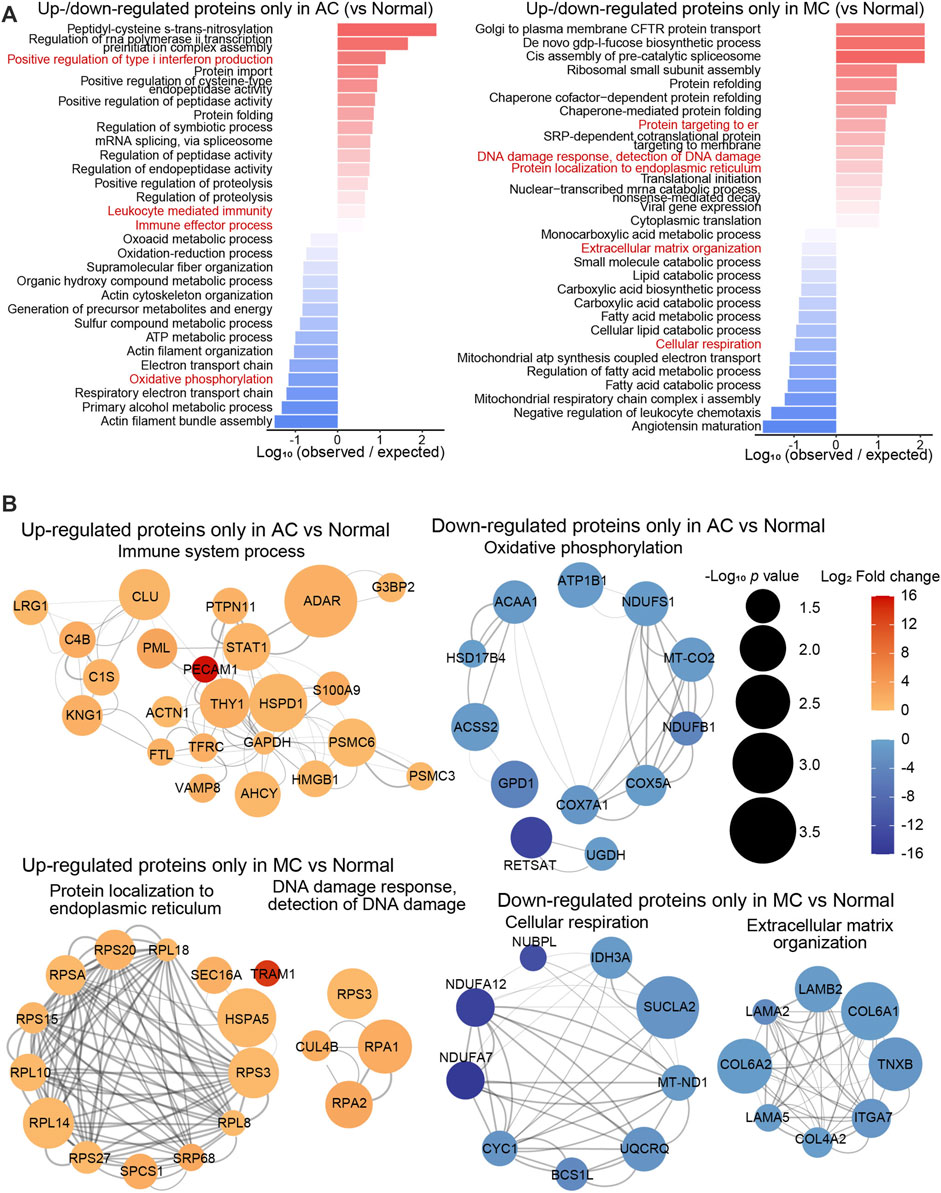
FIGURE 2. Proteomics difference between two types of colorectal adenocarcinomas and normal colon. (A) Bar plots showed the top 15 biological processes enriched with up- or downregulated differentially expressed proteins only in AC (left) or in MC (right). (B) Networks presented proteins of partial representative biological processes. Color bar: -log10 (protein expression p-value of AC/normal or MC/normal). Circle size: -log10 (protein expression fold change of AC/normal or MC/normal).
To gain further insight into the different protein features of AC and MC, we identified the differentially expressed proteins between the AC and MC groups (fold change >1.5, p-value <0.05, Figure 3A). The proteins upregulated in the AC and MC groups were mainly located in the digestive gland, whereas proteins upregulated in the MC group were also found to be present in the internal female genital organ. This was another organ system, which includes the ovary, that was at high risk of the occurrence of primary mucinous carcinoma and of colorectal mucinous carcinoma metastasis (Figures 3B, C). We inferred to a certain degree the homogeneity of MC in the digestive tract and internal female genital organs. The hypoxia-inducible factor-1 alpha signaling pathway, various metabolic processes, and leukocyte activation were significantly enriched in the proteins upregulated in the AC group compared to the MC group. For the cellular component, the proteins upregulated in the AC group were mainly located in the tertiary granule lumen. Moreover, proteins upregulated in the MC group were mostly secreted extracellularly with exosomes, which suggests active crosstalk in the tumor microenvironment. The leukocyte transendothelial migration pathway was the most significantly enriched pathway, as determined by GSEA between the AC and MC groups (Figure 3D). Compared with that in the AC group, the leukocyte transendothelial migration pathway was inhibited in the MC group; the involved factors include VAV1 (vav guanine nucleotide exchange factor 1), NCF4 (neutrophil cytosolic factor 4), MMP9 (matrix metalloproteinase-9), ITGB2 (integrin beta-2), ITGAM (integrin alpha-M), and PECAM1 (Figure 3E).
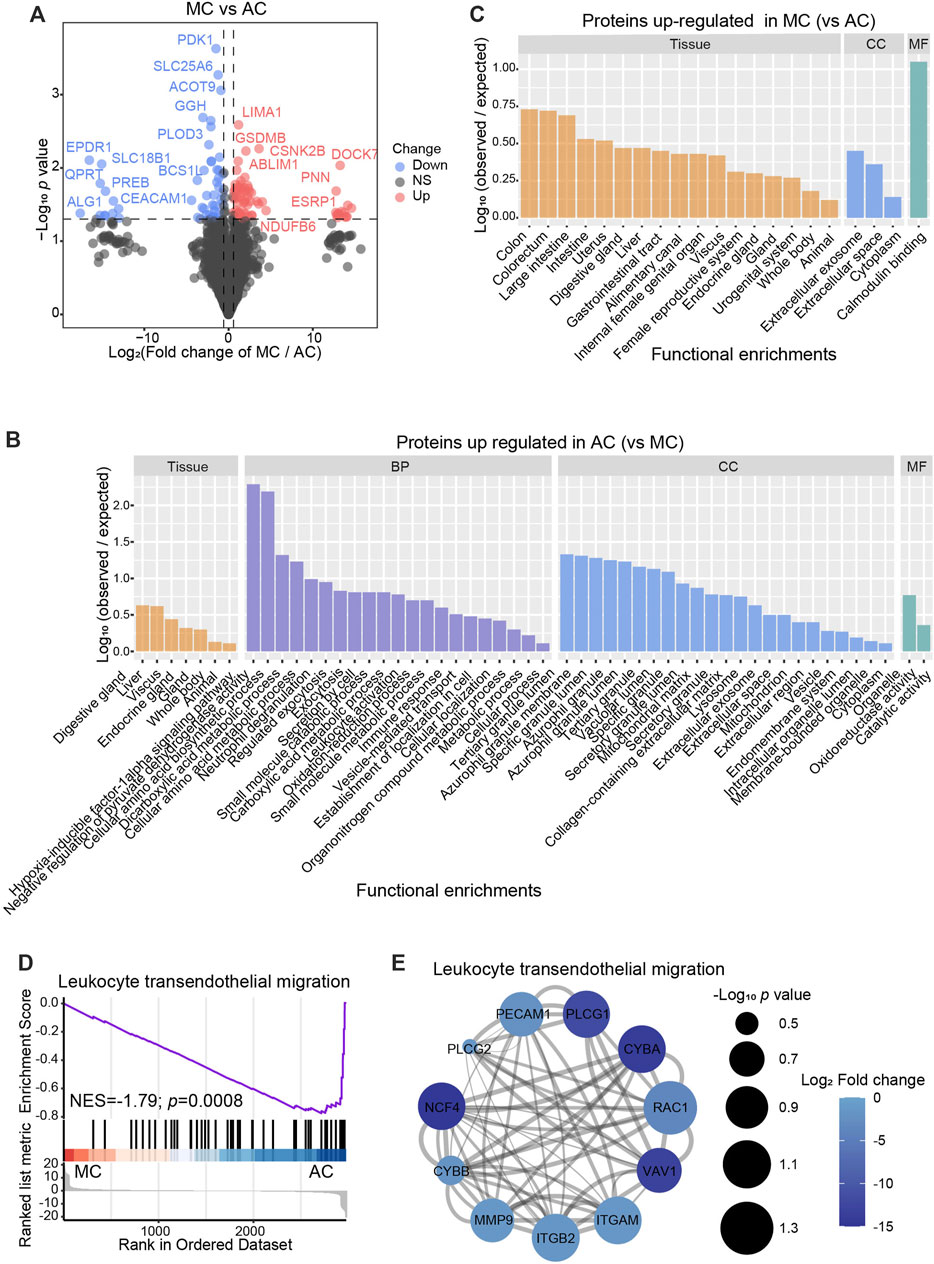
FIGURE 3. Proteomics difference between two types of colorectal adenocarcinomas. (A) Volcano plots showed the differentially expressed proteins between AC and MC. Cutoff: fold change >1.5, p < 0.05. B–C: Bar plots showed the distribution of tissue expression and gene ontology (GO) analysis, including biological process (BP), cellular component (CC), and molecular function (MF), which was enriched with proteins upregulated in AC (B) or MC (C). (D) Gene set enrichment analysis (GSEA) showed the top significantly enriched hallmark between AC and MC. (E) Networks presented proteins of the top GSEA between AC and MC. Color bar: -log10 (protein expression p-value of AC and MC). Circle size: -log10 (protein expression fold change of AC/MC).
In this section, we compared the differences in proteomics between two types of colorectal adenocarcinomas and normal colon tissues, with a direct comparison between the two types of CRC. AC was immune-active but was exposed to a hypoxic microenvironment. MC cells could protect from DNA damage, and the microenvironment was unfavorable to leukocyte transendothelial migration.
Single-cell transcriptome atlas of human colorectal adenocarcinoma not otherwise specified, mucinous adenocarcinoma, and normal colon tissue
We analyzed the GSE178341 scRNA-seq dataset, which includes 36 normal colon tissues, 55 AC, and 7 MC (Supplementary Table S2). We selected min.cell = 3 and min. feature = 200 when creating SeuratObject. Then, we used the metadata provided to perform the cell type annotation. After data normalization, we clustered cells based on significant principal components. There were seven major cell types, including B cells (n = 25,455) that expressed CD79A, CD79B, CD19, and MS4A1; epithelial cells (n = 163,146), expressing KRT8, KRT18, and EPCAM; mast cells (n = 3767), expressing TPSAB1, TPSB2, KIT, and CPA3; myeloid cells (n = 40,420), expressing LYZ and FCGR3A; plasma cells (n = 37,513), expressing JCHAIN; stromal cells (n = 15,041), expressing DCN and COL1A2; and T/NK cells (n = 74,438) that expressed CD3D and GZMB (Figure 4A). The defined marker genes are shown in Figure 4B. We then analyzed different cell proportions in MC, AC, and adjacent normal tissue (Figures 4C, D). Myeloid cells and T/NK cells were significantly increased in both AC and MC tissues. Plasma cells and stromal cells were significantly decreased in both AC and MC tissues. B cells in the AC and MC groups were also decreased but were only significantly decreased in the AC group. These cell proportion differences suggest a remodeling of the tumor microenvironment. However, we did not find a significant difference between the AC and MC groups, which might be due to the coarse cell annotation. Further cell subtype proportion analysis is needed to investigate the differences in cell type proportions between AC and MC.
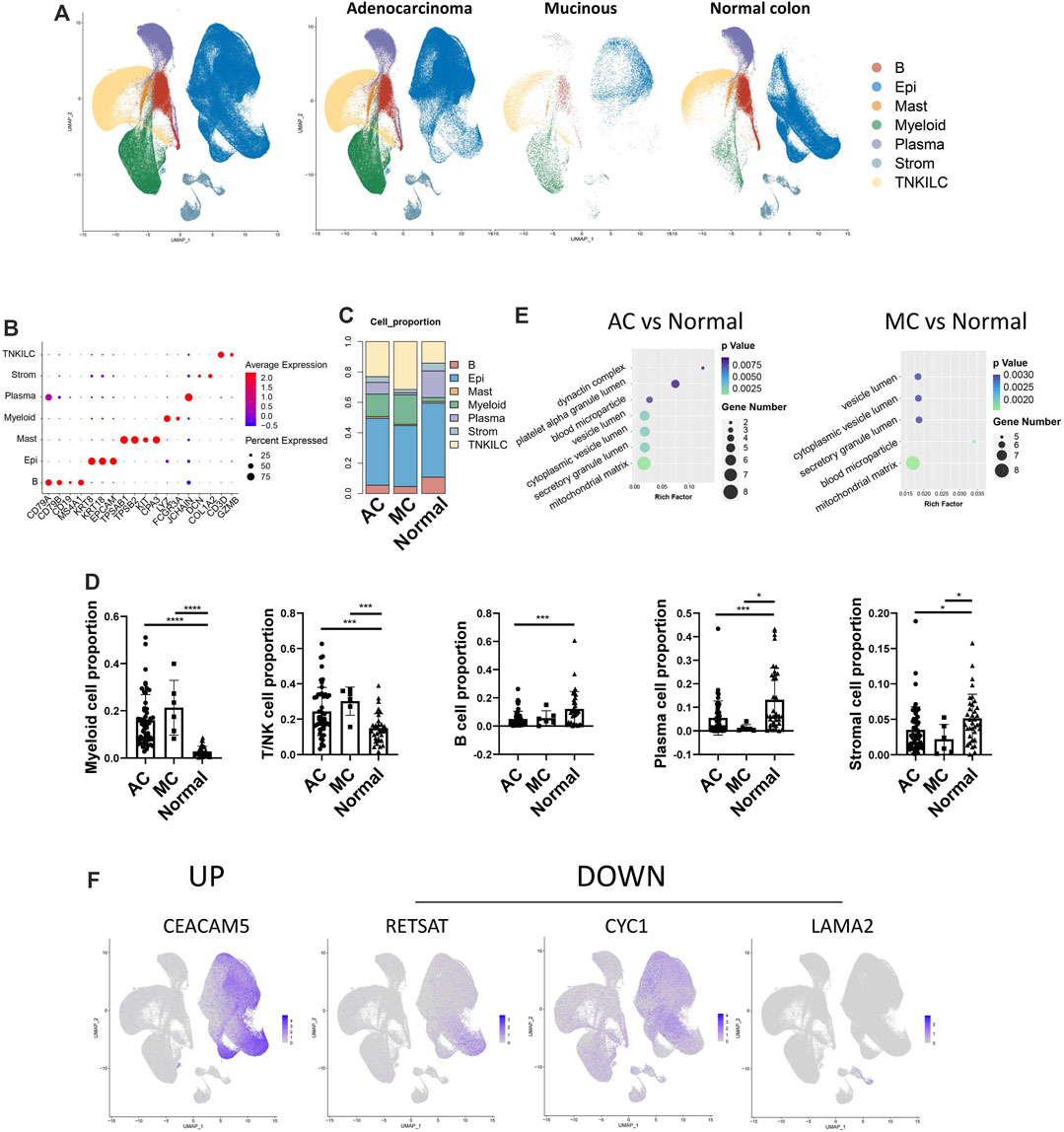
FIGURE 4. Single-cell transcriptome atlas of human colorectal adenocarcinoma not otherwise specified, mucinous adenocarcinoma, and normal colon. (A) Coarse cell annotation of scRNA-seq data of two types of colorectal adenocarcinomas and normal colon. (B) Marker genes of coarse cell type of scRNA-seq data on two types of colorectal adenocarcinomas and normal colon. (C) Coarse cell type proportion of two types of colorectal adenocarcinomas and normal colon. (D) Bar plot showing the significance of different cell-type proportions in two types of colorectal adenocarcinomas and normal colon. (E) BP pathways which were enriched in DEGs from AC and normal tissue, and MC and normal tissue. (F) Feature plot of mRNA in scRNA-seq data corresponding to differentially expressed proteins found in proteomics data.
Differentially expressed genes (DEGs) between MC and adjacent normal tissue and between AC and adjacent normal tissue were identified, and GO enrichment analysis was performed to analyze the involved pathways (Figure 4E). Most of the enriched pathways of the DEGs between MC and adjacent normal tissue and between AC and adjacent normal tissue overlapped. The dynactin complex pathway was only enriched in DEGs between AC and normal tissue. In MC tissue, we did not observe an enrichment of the dynactin pathway, which was consistent with our finding that AC tissue had a more favorable environment for cell migration than MC tissue. The vesicle lumen was the pathway that was most significantly enriched in DEGs between MC and normal colon tissue. This finding is consistent with the results from the proteomic analysis, which indicate that proteins upregulated in MC tissue would mostly be related to extracellular secretion.
We then investigated the expression of the differentially expressed proteins that we found in the proteomics data in the single-cell data to determine whether there was any overlap and in which cluster they were expressed. We selected the most variable differentially expressed proteins (fold change >1.5, p-value <0.05) among AC, MC, and normal colon tissues. These overlapping molecules might play essential roles in the special pathology of CRC and be potential targets for treatment. To investigate in which cell type these markers were expressed, we used feature plots to show the expression pattern of these markers (Figure 4F, Supplementary Figure S1A). Among the 30 most variable proteins, 18 corresponding mRNAs were detected in the scRNA-seq data. Of these, four mRNAs (CEACAM5, RETSAT, CYC1, and LAMA2) showed specific expression in one cell type. In our protein data, CEA cell adhesion molecule 5 (CEACAM5) was upregulated, while retinol saturase (RETSAT) was downregulated in the AC group compared with the normal colon group. Cytochrome (CYC1) was downregulated in the MC group compared with the normal colon group. These three corresponding mRNAs were mainly expressed in epithelial cells in the scRNA-seq data. Laminin subunit alpha 2 (LAMA2) was downregulated in the MC group compared with the normal colon group and was mainly expressed in stromal cells. CEACAM5 is the coding gene of CEA that is expressed in many epithelial malignancies, including colorectal cancer (DeLucia et al., 2021). CYC1 is a metabolism-related gene that is downregulated in colorectal cancer by several miRNAs after m6A modification, thereby reprogramming the mitochondrial metabolism in colorectal cancer (Yao et al., 2022). These findings suggest that the most variable proteins in AC and MC, compared to normal colon tissue, are mostly expressed in epithelial and stromal cells, indicating that epithelial and stromal cells might play a key role in CRC.
In this section, we analyzed the variation in cell types and DEGs among AC, MC, and normal colon tissues. The differences in cell type and function between AC and MC tissue were not significant, but we found that epithelial and stromal cells might play a key role in CRC. Further investigation is needed to explore the two cell types and their interactions, which we inferred contributed to the different biological behaviors and clinical outcomes of the two CRC subtypes.
Epithelial and stromal cell infiltration in colorectal adenocarcinoma not otherwise specified, mucinous adenocarcinoma, and normal colon tissue
CRCs, including ACs and MCs, mostly originate from epithelial cells (Joanito et al., 2022). To investigate the potential interaction between epithelial cells and stromal cells, we first analyzed the subcluster of stromal and epithelial cell proportions in AC, MC, and normal tissues (Figures 5A, D). The proliferating endothelial cell proportion was higher in AC and MC tissues than in normal tissue, which indicates angiogenesis in AC and MC tissues (Figure 5B). The proportions of most subtypes of fibroblasts were lower in AC and MC tissues than in normal tissue, whereas the pericyte proportion was higher in AC and MC tissues than in normal tissue. Pericyte–fibroblast transition has been reported to promote tumor growth and metastasis (Hosaka et al., 2016), indicating the importance of angiogenesis in CRC. CXCL14+ cancer-associated fibroblasts (CAFs) and CCL8 fibroblast-like cell proportions were also higher in AC and MC tissues than in normal tissue (Figure 5C). The stem-like epithelial cell proportion was higher and the enterocyte and goblet proportions were lower in AC and MC tissues than in normal tissue (Figure 5E). Moreover, the proportions of bestrophin 4-positive (BEST4+) cells and Turf cells were lower in AC and MC tissues than in normal tissue. These findings indicate active proliferation and angiogenesis in AC and MC tissues. CXCL14+ CAFs and CCL8 fibroblast-like cells might be involved in the immune response in AC and MC.
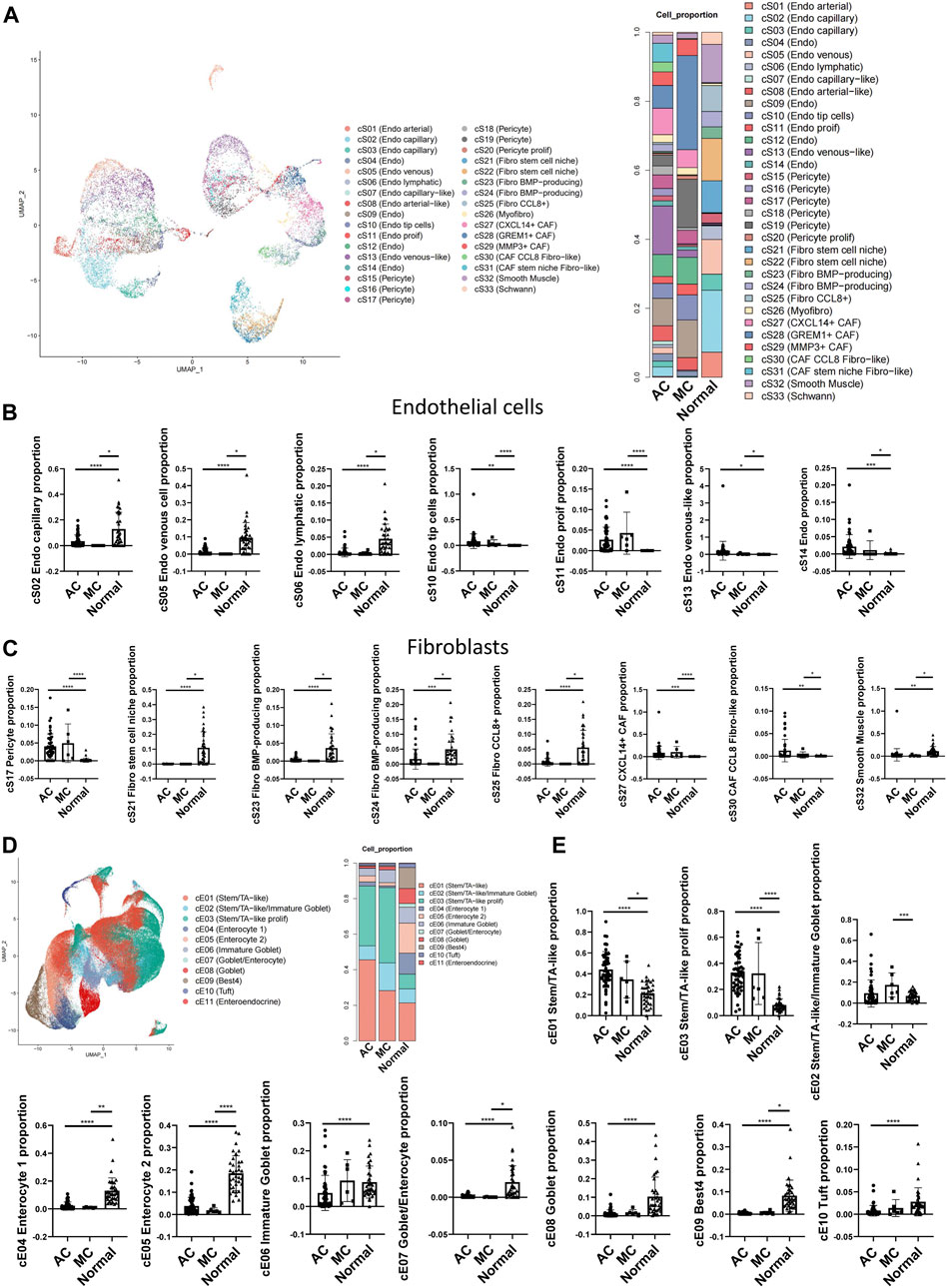
FIGURE 5. Stromal cell and epithelial cell proportions in human colorectal adenocarcinoma not otherwise specified, mucinous adenocarcinoma, and normal colon. (A) Stromal cell subcluster and proportion in human colorectal adenocarcinoma not otherwise specified, mucinous adenocarcinoma, and normal colon. (B) Bar plots showing the significance of different endothelial cell proportions in two types of colorectal adenocarcinomas and normal colon. (C) Bar plots showing the significance of different fibroblast proportions in two types of colorectal adenocarcinomas and normal colon. (D) Epithelial cell subcluster and proportion in human colorectal adenocarcinoma not otherwise specified, mucinous adenocarcinoma, and normal colon. (E) Bar plots showing the significance of different epithelial cell proportions in two types of colorectal adenocarcinomas and normal colon.
The changes in cell types between AC and MC tissues and normal tissue in our findings involved mostly immune reactivity-related cells, highlighting the importance of immune regulation in AC and MC. Thus, we then analyzed the immune cell subclusters and their proportion in AC and MC tissues (Supplementary Figures S1A, B). Many immune cell types that represent an active immune environment were increased in AC and MC tissues compared with normal tissue. Such cells include CXCL13+ CD4+ T cells, which have antitumor effects in colon cancer (Ukita et al., 2022), CXCL13+ CD8+ T cells, and CXCL13+ CD8+ proliferating T cells. Gamma delta (γδ)-like PDCD+ T cells were also increased in AC and MC tissues compared with normal tissue; these cells have been reported to be enriched in deficient mismatch repair (dMMR) colon cancer (Pelka et al., 2021). CD4+ Treg and CD4+ proliferating regulatory T (Treg) cells were also increased in AC and MC tissues compared with normal tissue. These findings are consistent with results from previous studies that demonstrated that the infiltration of Treg cells into the colon was significantly higher in CRC tissue than in healthy colon tissue and that they had been associated with CRC progression (Allinen et al., 2004; Loddenkemper et al., 2006) (Supplementary Figure S1C). We then analyzed whether there were immune cells whose proportions differed between AC and MC tissues. We found that plasmacytoid dendritic cells (pDCs), PLZF+ proliferating T cells, CD16A+ NK cells, GZMK+ NK cells, and XCL+ NK cells were all increased in MC tissue compared with AC tissue (Supplementary Figure S1D). These findings indicate that NK cells might be more important in MC and could be a potential target for immune therapy for MC.
Taken together, these findings suggest the reshaping of the tumor microenvironment during the tumorigenesis of AC and MC, and we emphasize the importance of epithelial and stromal cell infiltration. CXCL14+ cancer-associated fibroblasts and CCL8 fibroblast-like cell proportions were higher in both AC and MC tissues than in normal colon tissue. NK cells might be a potential target for immune therapy for MC.
Epithelial and stromal cell interactions in colorectal adenocarcinoma not otherwise specified, mucinous adenocarcinoma, and normal colon tissues
Next, we used NicheNet (Browaeys et al., 2020) analysis to investigate potential ligands in stromal cells and target genes in epithelial cells. First, we analyzed the top ligands and target genes in both AC and MC tissues (Figure 6A). We then performed GO enrichment of the top ligands, and the biological process pathways in which the DEGs were enriched included the regulation of transcription, signal transduction, positive regulation of cell proliferation, and cell differentiation. The cellular component pathways in which the DEGs were enriched included integral components of the membrane, extracellular space, and extracellular region. The molecular function pathways included ATP, nucleotide, and RNA binding (Figure 6B). Thence, we analyzed the ligands in stromal cells and target genes in epithelial cells in AC and MC tissues and compared them with those of normal tissue (Figures 6C, D). We then analyzed the GO terms (Supplementary Figure S2A). The biological process pathways in which the DEGs between AC and normal tissues were enriched included cell adhesion, positive regulation of cell proliferation, and positive regulation of cell migration; these pathways were also observed in the comparison between MC and normal tissues. Positive regulation of the ERK1 and ERK2 cascade pathways was only enriched in the DEGs between MC and normal tissues, a finding which is consistent with previous research indicating that MC is associated with mutations in genes involved in the Ras-Raf-MEK-ERK pathway (Hugen et al., 2015b).
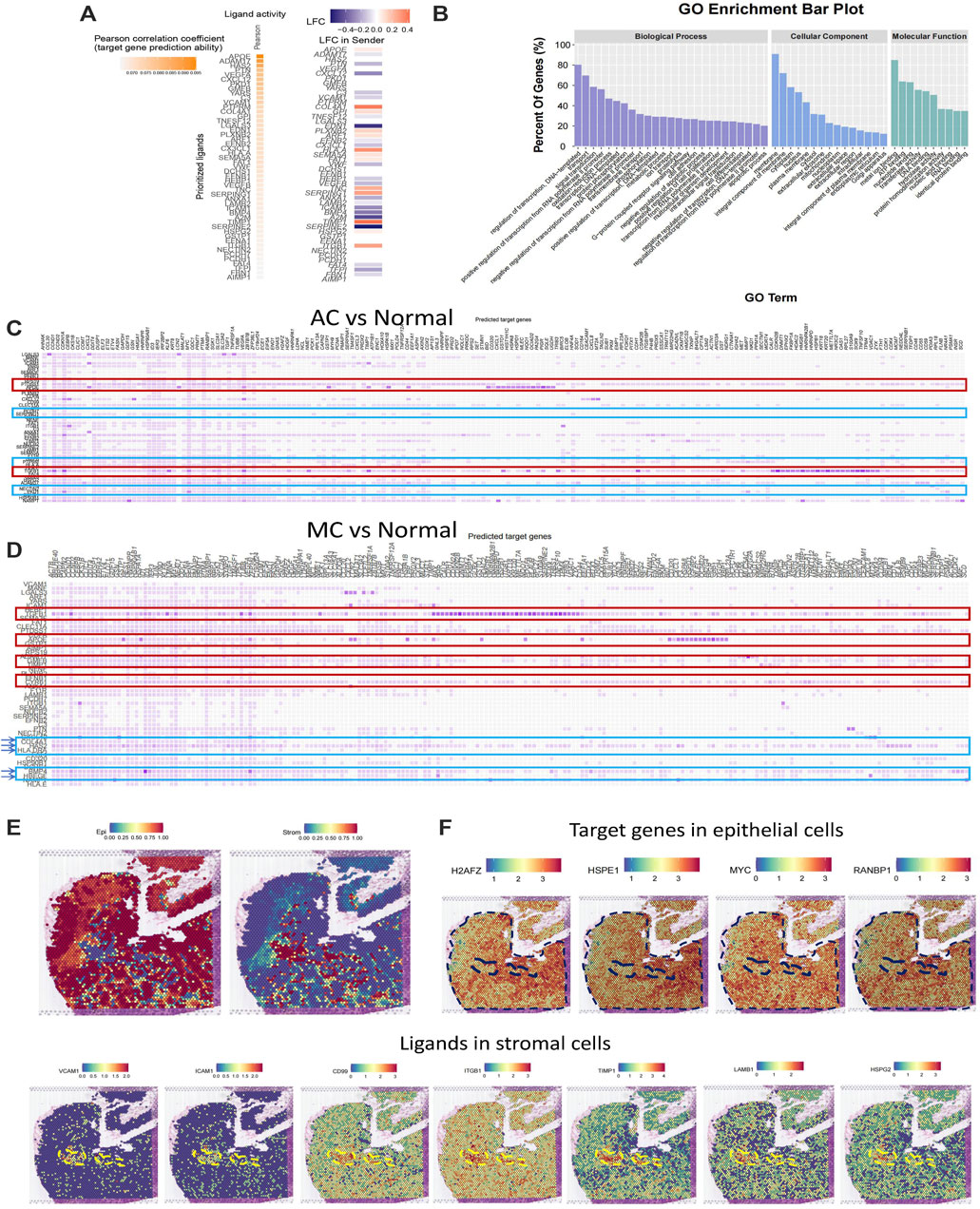
FIGURE 6. Stromal and epithelial cell interaction in human colorectal adenocarcinoma not otherwise specified, mucinous adenocarcinoma, and normal colon. (A) Top ligands in stromal cells regulating epithelial cells predicted by NicheNet from colorectal adenocarcinoma not otherwise specified and mucinous adenocarcinoma with normal colon as the reference. (B) GO enrichment of top ligands from colorectal adenocarcinoma not otherwise specified and mucinous adenocarcinoma with normal colon as the reference. (C) Top ligands in stromal cells predicted by NicheNet from colorectal adenocarcinoma not otherwise specified with normal colon as the reference and their target genes in epithelial cells. (D) Top ligands in stromal cells predicted by NicheNet from mucinous with normal colon as the reference and their target genes in epithelial cells. (E) Cell annotation on spatial transcriptome data on human colorectal adenocarcinoma not otherwise specified. (F) Spatial feature plot of target genes in epithelial cells and ligands in stromal cells predicted by NicheNet from colorectal adenocarcinoma not otherwise specified with normal colon as the reference.
To further investigate the ligands on stromal and epithelial cells in AC and MC tissues, we calculated the top ligands of AC and MC tissues and compared them with those of normal colon tissue to determine whether there were overlapping and specific ligands of AC and MC tissues. Such ligands might be key molecules connecting stromal cells and tumor epithelial cells. Among the top 50 ligands of AC tissues and the top 48 ligands of MC tissues, 43 were overlapping ligands. Additionally, seven ligands were found only in AC tissue, and five only in MC tissue (Supplementary Table S4). Among the 43 overlapping ligands, apolipoprotein-E (APOE) and transforming growth factor (TGF)-β1 showed high prediction scores with target genes from epithelial cells, indicating that the two ligands might play important roles in stromal and epithelial cell interactions. Among the seven ligands that were specifically found in AC tissues, heparan sulfate proteoglycan 2 (HSPG2) (Datta et al., 2006), glycosylphosphatidylinositol (GPI) (Nicolas et al., 1994), and CD99 (Manara et al., 2018) were involved in regulating cell adhesion and invasion. Among the five ligands that were found only in MC tissues, bone morphogenetic protein 4 (BMP4) (Lombardo et al., 2011), hyaluronic acid synthase 2 (HAS2) (Kim et al., 2019), and heparin-binding epidermal growth factor-like growth factor (HBEGF) (Yun et al., 2018) were reported to be involved in cell proliferation or cell migration in colorectal cancer.
To further validate the ligand and target gene interactions between stromal and epithelial cells in colorectal cancer, we used spatial transcriptome data from an AC sample from Hu et al. (Peng et al., 2022). After running a routine process for spatial transcriptome data, including dimensionality reduction and clustering, we integrated the spatial transcriptome data with the single-cell data used in this study and annotated the cell type on the spatial plot. We observed that the stromal and epithelial cells were spatially connected to each other (Figure 6E). We then generated a spatial feature plot of ligands and target genes predicted from the comparison between AC and normal tissues. We found that the ligands VCAM1, ICAM1, CD99, ITGB1, TIMP1, LAMB1, and HSPG2 are highly expressed in the epithelial region and that their target genes of H2AFZ, HSPE1, MYC, and RANBP1 are highly expressed in the stromal region (Figure 6F).
In this section, we have sought to analyze the interactions between stromal cells and epithelial cells in AC and MC tissues and aimed to provide a potential therapeutic target for the more specific treatment of AC and MC. The positive regulation of the ERK1 and ERK2 cascade pathways was enriched only in DEGs between MC and normal tissues, indicating its potential for target development.
Discussion
Regardless of improvements in therapeutic strategies for CRC patients, their overall prognosis is still not optimistic. With the development of biomarkers and drug targets, CRC treatment has become more individualized according to subtype classification with specific molecular and histological characteristics. With a higher proportion and more features of malignancy in young patients, the treatment and management of MC, as a unique subtype of CRC, have become a tough challenge in clinical practice. Although patients with MC account for a small proportion of CRC patients, they often suffer from poor prognoses, with the treatment of MC being a challenge for oncologists. Understanding molecular characteristics might impact clinical decisions for treatment strategies and promote the accurate treatment of CRC patients, especially with MC.
AC and MC are generally similar in tumor biological behavior. Cytochromes P450 (CYPs) are the major source of variability in drug response (Zanger and Schwab, 2013). We found that drug metabolism involving cytochrome P450 was inactive in AC and MC tissues. The dysfunction of CYP might be one of the reasons for poor drug response or even resistance in CRC.
Previous therapies have mainly depended on KRAS and BRAF mutations or MSI status, which are not pertinent to the MC subtype. This study aimed to identify more molecules and pathways that could be potential therapeutic targets for AC and MC.
It has been well-documented that hypoxia confers poor clinical outcomes via tumor propagation, malignant progression, and resistance to therapy (Hockel and Vaupel, 2001). Our proteomic analysis showed that cellular respiration-related biological processes were downregulated in both AC and MC tissues and that hypoxia-inducible factor-1 alpha signaling pathway-related proteins were upregulated in AC tissue compared to MC tissue. This finding reveals that, although hypoxia is a universal feature of tumor growth, it contributes more to the malignancy of AC but not MC. Therapy targeting hypoxia in the tumor environment would offer promising effects for AC but might not provide beneficial outcomes in AC treatment.
We found that the DNA damage response, which includes the CUL4B, RPS3, RPA1, and RPA2 proteins, was active in MC tissues, which might result in resistance to various therapies for MC. As an RNA-binding protein (RBP), RPS3 has been reported as a promising drug target for hepatocellular carcinoma (Zhao et al., 2019). The RPA (human replication protein A) family, including RPA1, RPA2, and RPA3, plays an essential role in eukaryotic DNA replication, homologous recombination, and excision repair; it has been recognized as an oncogene and therapeutic target for cancer (Chen et al., 2017; Wang et al., 2018).
The prevailing perspective is that the mucinous subtype elicits a worse response to immunotherapy than the non-mucinous subtype (Kim et al., 2013; Yan et al., 2022). Our analysis suggests that the positive regulation of type I interferon production and leukocyte-mediated immunity was enriched in the proteins upregulated only in AC tissue but not in MC tissue, while the leukocyte transendothelial migration pathway was inhibited in MC tissue. We infer that leukocyte transendothelial migration inhibition might be the major modality for insufficient leukocyte-mediated immunity and type I interferon production. Translocation-associated membrane protein 1 (TRAM1) was one of the most significantly upregulated proteins that were upregulated only in MC tissue. TRAM1 participates in human cytomegalovirus US2- and US11-mediated dislocation of an endoplasmic reticulum membrane glycoprotein and the subsequent degradation of major histocompatibility complex (MHC) class I heavy chains, thereby decreasing immune detection by cytotoxic T cells (Oresic et al., 2009). Together, both a lack of immune infiltration and MHC I binding led to immunotherapy resistance by MC and immunotherapy combined with targeting TRAM1 might improve the antitumor effect of immunotherapy for MC.
In addition, the proportion of bestrophin 4 (BEST4) + cell (Burclaff et al., 2022) was lower in AC and MC tissues than in normal tissue. A previous study reported that BMP3 was secreted by BEST4+ cells in the small intestine, with BMP3 being largely studied for its antitumor roles (Loh et al., 2008; Wen et al., 2019). Tuft cells are chemosensory cells that regulate type 2 immune reactions (Schneider et al., 2019) in the intestinal epithelial cell and could respond to pathogenic metabolites (Schneider et al., 2019). Therefore, BEST4+ and Tuft cells might act as potential targets for AC and MC immune therapy.
Apart from targeting cancer cells, anticancer therapies should target the stromal compartment to be effective and result in improved patient outcomes (Valkenburg et al., 2018). Previous studies have reported the pro-tumoral effects of CXCL14, based on evidence for the upregulation of CXCL14 in the tumor stroma and CAFs (Allinen et al., 2004; Augsten et al., 2009). In prostate cancer, CXCL14-producing fibroblasts have enhanced pro-tumoral effects (Augsten et al., 2009). In breast cancer, CCL8 is produced in stromal fibroblasts at the tumor margins and in tissues in which cancer cells tend to metastasize, such as the lungs and the brain (Farmaki et al., 2016). Therefore, CXCL14 fibroblasts and CCL8 fibroblast-like cells may have pro-tumorigenic functions in AC and MC. Interest has recently focused on NK cells for therapeutic interventions as these cells have antitumor properties (Chiossone et al., 2018). We found that NK cells might be important in MC and could be a potential target for immune therapy for MC.
Based on the different morphological characteristics, biological behaviors, and treatment responses (Johncilla and Yantiss, 2020), we explored the differences in cell types and functions between AC and MC. We found that there were few significantly different cell types and functions between AC and MC but noticed the important role of epithelial and stromal cells in CRC. We inferred that this difference contributed to the different biological behaviors and clinical outcomes of the two CRC subtypes. Moreover, we focused on ligands on stromal cells that might regulate the malignant behavior of tumor cells. Emerging evidence suggests that targeting the stromal compartment can shape the tumor environment and responsiveness to immunotherapy (Turley et al., 2015).
Apolipoprotein-E (APOE) and transforming growth factor (TGF)-β1 were identified as important ligands that interact between stromal cells and tumor epithelial cells and could be potential therapeutic targets for both AC and MC. APOE plays an important role in lipoprotein metabolism and is expressed at high levels by monocyte-derived macrophages (Mahley, 1988). It is highly expressed in colorectal liver metastases in colorectal cancer (Zhao et al., 2018). TGF-β1 is involved in almost every aspect of multipotent mesenchymal stromal cells (de Araújo Farias et al., 2018) and is shown to be a promoter of tumor progression and metastasis (Meulmeester and Ten Dijke, 2011). Furthermore, BMP4, which we identified as a specific ligand in MC, is a member of the TGF-β superfamily and is related to colorectal cancer progression (Li et al., 2022).
Collectively, we clarified the variations in the tumor environments among AC, MC, and normal colon tissues and tried to identify more molecular and cellular therapeutic targets for AC and MC.
However, our study does have certain limitations. First, the data source of proteomic, scRNAseq, and spatial transcriptome data belongs to different projects, which limited the analysis. It is difficult to warrant the sample information as comparable, such as sample gender, age, ethnicity, and race. Second, there has been until now no spatial transcriptome data on MC samples available to compare the spatial information between AC and MC. Third, the potential biomarkers or drug/therapy targets mentioned in this study are still hypothetical and need a lot of work to verify. We next aim to collect samples for proteomic, scRNAseq, and spatial transcriptome detection to further investigate this issue.
Conclusion
There were different characteristics of the tumor environment between colorectal adenocarcinoma not otherwise specified (AC) and mucinous adenocarcinoma (MC). The major difference between AC and MC was not the variety of cell types and functions but may be cell interactions. Stromal and epithelial cell interactions play important roles in both MC and AC, but different regulatory pathways were involved.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material; further inquiries can be directed to the corresponding authors.
Author contributions
KX performed data interpretation and analysis, and wrote the manuscript. SZ and BL offered resources and polished the article. YS provided founding, designed the study, and polished the article. XY designed the study, performed data interpretation and analysis, and polished the article. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (Grant No. 82203705) and the Program for Zhejiang Provincial Clinical Research Center for CANCER (Grant No. 2022E50008).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2023.1150362/full#supplementary-material
SUPPLEMENTARY FIGURE S1 | (A) Immune cell subcluster and proportion in human colorectal adenocarcinoma not otherwise specified, mucinous adenocarcinoma, and normal colon. (B) Bar plots showing the significance of different immune cell proportions that differed in two types of colorectal adenocarcinomas and normal colon. (C) Bar plots showing the significance of different immune cell proportions that differ in two types of colorectal adenocarcinoma.
SUPPLEMENTARY FIGURE S2 | (A) GO enrichment of top ligands from colorectal adenocarcinoma not otherwise specified with normal colon as the reference, and from mucinous adenocarcinoma with normal colon as the reference.
Abbreviations
AC, adenocarcinoma not otherwise specified; BP, biological process; CAFs, cancer-associated fibroblasts; CC, cellular component; CRC, colorectal cancer; DEGs, differentially expressed genes; GO, gene ontology; MC, mucinous adenocarcinoma; MF, molecular function; MSI, microsatellite instability; pDCs, plasmacytoid dendritic cells.
References
Ahnen, D. J., Wade, S. W., Jones, W. F., Sifri, R., Mendoza Silveiras, J., Greenamyer, J., et al. (2014). The increasing incidence of young-onset colorectal cancer: A call to action. Mayo Clin. Proc. 89 (2), 216–224. doi:10.1016/j.mayocp.2013.09.006
Allinen, M., Beroukhim, R., Cai, L., Brennan, C., Lahti-Domenici, J., Huang, H., et al. (2004). Molecular characterization of the tumor microenvironment in breast cancer. Cancer Cell 6 (1), 17–32. doi:10.1016/j.ccr.2004.06.010
Augsten, M., Hägglöf, C., Olsson, E., Stolz, C., Tsagozis, P., Levchenko, T., et al. (2009). CXCL14 is an autocrine growth factor for fibroblasts and acts as a multi-modal stimulator of prostate tumor growth. Proc. Natl. Acad. Sci. U. S. A. 106 (9), 3414–3419. doi:10.1073/pnas.0813144106
Biller, L. H., and Schrag, D. (2021). Diagnosis and treatment of metastatic colorectal cancer: A review. JAMA 325 (7), 669–685. doi:10.1001/jama.2021.0106
Browaeys, R., Saelens, W., and Saeys, Y. (2020). NicheNet: Modeling intercellular communication by linking ligands to target genes. Nat. Methods 17 (2), 159–162. doi:10.1038/s41592-019-0667-5
Burclaff, J., Bliton, R. J., Breau, K. A., Ok, M. T., Gomez-Martinez, I., Ranek, J. S., et al. (2022). A proximal-to-distal survey of healthy adult human small intestine and colon epithelium by single-cell transcriptomics. Cell Mol. Gastroenterol. Hepatol. 13 (5), 1554–1589. doi:10.1016/j.jcmgh.2022.02.007
Cao, W., Chen, H.-D., Yu, Y.-W., Li, N., and Chen, W.-Q. (2021). Changing profiles of cancer burden worldwide and in China: A secondary analysis of the global cancer statistics 2020. Chin. Med. J. 134 (7), 783–791. doi:10.1097/cm9.0000000000001474
Catalano, V., Loupakis, F., Graziano, F., Bisonni, R., Torresi, U., Vincenzi, B., et al. (2012). Prognosis of mucinous histology for patients with radically resected stage II and III colon cancer. Ann. Oncol. 23 (1), 135–141. doi:10.1093/annonc/mdr062
Chen, C. C., Juan, C. W., Chen, K. Y., Chang, Y. C., Lee, J. C., and Chang, M. C. (2017). Upregulation of RPA2 promotes NF-κB activation in breast cancer by relieving the antagonistic function of menin on NF-κB-regulated transcription. Carcinogenesis 38 (2), 196–206. doi:10.1093/carcin/bgw123
Chiang, J. M., Yeh, C. Y., Changchien, C. R., Chen, J. S., Tang, R., and Chen, J. R. (2010). Mucinous adenocarcinoma showing different clinicopathological and molecular characteristics in relation to different colorectal cancer subgroups. Int. J. Colorectal Dis. 25 (8), 941–947. doi:10.1007/s00384-010-0958-x
Chiossone, L., Dumas, P. Y., Vienne, M., and Vivier, E. (2018). Natural killer cells and other innate lymphoid cells in cancer. Nat. Rev. Immunol. 18 (11), 671–688. doi:10.1038/s41577-018-0061-z
Ciardiello, F., Ciardiello, D., Martini, G., Napolitano, S., Tabernero, J., and Cervantes, A. (2022). Clinical management of metastatic colorectal cancer in the era of precision medicine. CA Cancer J. Clin. 72 (4), 372–401. doi:10.3322/caac.21728
Datta, M. W., Hernandez, A. M., Schlicht, M. J., Kahler, A. J., DeGueme, A. M., Dhir, R., et al. (2006). Perlecan, a candidate gene for the CAPB locus, regulates prostate cancer cell growth via the Sonic Hedgehog pathway. Mol. Cancer 5, 9. doi:10.1186/1476-4598-5-9
de Araújo Farias, V., Carrillo-Gálvez, A. B., Martín, F., and Anderson, P. (2018). TGF-β and mesenchymal stromal cells in regenerative medicine, autoimmunity and cancer. Cytokine Growth Factor Rev. 43, 25–37. doi:10.1016/j.cytogfr.2018.06.002
DeLucia, D. C., Cardillo, T. M., Ang, L., Labrecque, M. P., Zhang, A., Hopkins, J. E., et al. (2021). Regulation of CEACAM5 and therapeutic efficacy of an anti-CEACAM5-SN38 antibody-drug conjugate in neuroendocrine prostate cancer. Clin. Cancer Res. 27 (3), 759–774. doi:10.1158/1078-0432.Ccr-20-3396
Farmaki, E., Chatzistamou, I., Kaza, V., and Kiaris, H. (2016). A CCL8 gradient drives breast cancer cell dissemination. Oncogene 35 (49), 6309–6318. doi:10.1038/onc.2016.161
Hockel, M., and Vaupel, P. (2001). Tumor hypoxia: Definitions and current clinical, biologic, and molecular aspects. J. Natl. Cancer Inst. 93 (4), 266–276. doi:10.1093/jnci/93.4.266
Hosaka, K., Yang, Y., Seki, T., Fischer, C., Dubey, O., Fredlund, E., et al. (2016). Pericyte-fibroblast transition promotes tumor growth and metastasis. Proc. Natl. Acad. Sci. U. S. A. 113 (38), E5618–E5627. doi:10.1073/pnas.1608384113
Huang, A., Yang, Y., Shi, J. Y., Li, Y. K., Xu, J. X., Cheng, Y., et al. (2021). Mucinous adenocarcinoma: A unique clinicopathological subtype in colorectal cancer. World J. Gastrointest. Surg. 13 (12), 1567–1583. doi:10.4240/wjgs.v13.i12.1567
Hugen, N., Brown, G., Glynne-Jones, R., de Wilt, J. H., and Nagtegaal, I. D. (2016). Advances in the care of patients with mucinous colorectal cancer. Nat. Rev. Clin. Oncol. 13 (6), 361–369. doi:10.1038/nrclinonc.2015.140
Hugen, N., Simons, M., Halilović, A., van der Post, R. S., Bogers, A. J., Marijnissen-van Zanten, M. A., et al. (2015a). The molecular background of mucinous carcinoma beyond MUC2. J. Pathol. Clin. Res. 1 (1), 3–17. doi:10.1002/cjp2.1
Hugen, N., Verhoeven, R. H., Lemmens, V. E., van Aart, C. J., Elferink, M. A., Radema, S. A., et al. (2015b). Colorectal signet-ring cell carcinoma: Benefit from adjuvant chemotherapy but a poor prognostic factor. Int. J. Cancer 136 (2), 333–339. doi:10.1002/ijc.28981
Joanito, I., Wirapati, P., Zhao, N., Nawaz, Z., Yeo, G., Lee, F., et al. (2022). Single-cell and bulk transcriptome sequencing identifies two epithelial tumor cell states and refines the consensus molecular classification of colorectal cancer. Nat. Genet. 54 (7), 963–975. doi:10.1038/s41588-022-01100-4
Johncilla, M., and Yantiss, R. K. (2020). Histology of colorectal carcinoma: Proven and purported prognostic factors. Surg. Pathol. Clin. 13 (3), 503–520. doi:10.1016/j.path.2020.05.008
Kim, S. H., Shin, S. J., Lee, K. Y., Kim, H., Kim, T. I., Kang, D. R., et al. (2013). Prognostic value of mucinous histology depends on microsatellite instability status in patients with stage III colon cancer treated with adjuvant FOLFOX chemotherapy: A retrospective cohort study. Ann. Surg. Oncol. 20 (11), 3407–3413. doi:10.1245/s10434-013-3169-1
Kim, Y. H., Lee, S. B., Shim, S., Kim, A., Park, J. H., Jang, W. S., et al. (2019). Hyaluronic acid synthase 2 promotes malignant phenotypes of colorectal cancer cells through transforming growth factor beta signaling. Cancer Sci. 110 (7), 2226–2236. doi:10.1111/cas.14070
Li, Y., Hu, J., Wang, M., Yuan, Y., Zhou, F., Zhao, H., et al. (2022). Exosomal circPABPC1 promotes colorectal cancer liver metastases by regulating HMGA2 in the nucleus and BMP4/ADAM19 in the cytoplasm. Cell Death Discov. 8 (1), 335. doi:10.1038/s41420-022-01124-z
Loddenkemper, C., Schernus, M., Noutsias, M., Stein, H., Thiel, E., and Nagorsen, D. (2006). In situ analysis of FOXP3+ regulatory T cells in human colorectal cancer. J. Transl. Med. 4, 52. doi:10.1186/1479-5876-4-52
Loh, K., Chia, J. A., Greco, S., Cozzi, S. J., Buttenshaw, R. L., Bond, C. E., et al. (2008). Bone morphogenic protein 3 inactivation is an early and frequent event in colorectal cancer development. Genes Chromosom. Cancer 47 (6), 449–460. doi:10.1002/gcc.20552
Lombardo, Y., Scopelliti, A., Cammareri, P., Todaro, M., Iovino, F., Ricci-Vitiani, L., et al. (2011). Bone morphogenetic protein 4 induces differentiation of colorectal cancer stem cells and increases their response to chemotherapy in mice. Gastroenterology 140 (1), 297–309. doi:10.1053/j.gastro.2010.10.005
Luo, C., Cen, S., Ding, G., and Wu, W. (2019). Mucinous colorectal adenocarcinoma: Clinical pathology and treatment options. Cancer Commun. 39 (1), 13. doi:10.1186/s40880-019-0361-0
Mahley, R. W. (1988). Apolipoprotein E: Cholesterol transport protein with expanding role in cell biology. Science 240 (4852), 622–630. doi:10.1126/science.3283935
Manara, M. C., Pasello, M., and Scotlandi, K. (2018). CD99: A cell surface protein with an oncojanus role in tumors. Genes (Basel) 9 (3), 159. doi:10.3390/genes9030159
McCawley, N., Clancy, C., O'Neill, B. D., Deasy, J., McNamara, D. A., and Burke, J. P. (2016). Mucinous rectal adenocarcinoma is associated with a poor response to neoadjuvant chemoradiotherapy: A systematic review and meta-analysis. Dis. Colon Rectum 59 (12), 1200–1208. doi:10.1097/DCR.0000000000000635
Melis, M., Hernandez, J., Siegel, E. M., McLoughlin, J. M., Ly, Q. P., Nair, R. M., et al. (2010). Gene expression profiling of colorectal mucinous adenocarcinomas. Dis. Colon Rectum 53 (6), 936–943. doi:10.1007/DCR.0b013e3181d320c4
Meulmeester, E., and Ten Dijke, P. (2011). The dynamic roles of TGF-β in cancer. J. Pathol. 223 (2), 205–218. doi:10.1002/path.2785
Nicolas, F., Tiveron, M. C., Davoust, J., and Reggio, H. (1994). GPI membrane anchor is determinant in intracellular accumulation of apical plasma membrane proteins in the non-polarized human colon cancer cell line HT-29 18. J. Cell Sci. 107 (10), 2679–2689. doi:10.1242/jcs.107.10.2679
Nozoe, T., Anai, H., Nasu, S., and Sugimachi, K. (2000). Clinicopathological characteristics of mucinous carcinoma of the colon and rectum. J. Surg. Oncol. 75 (2), 103–107. doi:10.1002/1096-9098(200010)75:2<103:Aid-jso6>3.0.Co;2-c
Numata, M., Shiozawa, M., Watanabe, T., Tamagawa, H., Yamamoto, N., Morinaga, S., et al. (2012). The clinicopathological features of colorectal mucinous adenocarcinoma and a therapeutic strategy for the disease. World J. Surg. Oncol. 10, 109. doi:10.1186/1477-7819-10-109
Oresic, K., Ng, C. L., and Tortorella, D. (2009). TRAM1 participates in human cytomegalovirus US2- and US11-mediated dislocation of an endoplasmic reticulum membrane glycoprotein. J. Biol. Chem. 284 (9), 5905–5914. doi:10.1074/jbc.M807568200
Ott, C., Gerken, M., Hirsch, D., Fest, P., Fichtner-Feigl, S., Munker, S., et al. (2018). Advanced mucinous colorectal cancer: Epidemiology, prognosis and efficacy of chemotherapeutic treatment. Digestion 98 (3), 143–152. doi:10.1159/000487710
Pelka, K., Hofree, M., Chen, J. H., Sarkizova, S., Pirl, J. D., Jorgji, V., et al. (2021). Spatially organized multicellular immune hubs in human colorectal cancer. Cell 184 (18), 4734–4752.e20. doi:10.1016/j.cell.2021.08.003
Peng, Z., Ye, M., Ding, H., Feng, Z., and Hu, K. (2022). Spatial transcriptomics atlas reveals the crosstalk between cancer-associated fibroblasts and tumor microenvironment components in colorectal cancer. J. Transl. Med. 20 (1), 302. doi:10.1186/s12967-022-03510-8
Reynolds, I. S., Fichtner, M., McNamara, D. A., Kay, E. W., Prehn, J. H. M., and Burke, J. P. (2019). Mucin glycoproteins block apoptosis; promote invasion, proliferation, and migration; and cause chemoresistance through diverse pathways in epithelial cancers. Cancer Metastasis Rev. 38 (1-2), 237–257. doi:10.1007/s10555-019-09781-w
Schneider, C., O'Leary, C. E., and Locksley, R. M. (2019). Regulation of immune responses by tuft cells. Nat. Rev. Immunol. 19 (9), 584–593. doi:10.1038/s41577-019-0176-x
Shao, Y., Xu, K., Zheng, X., Zhou, B., Zhang, X., Wang, L., et al. (2022). Proteomics profiling of colorectal cancer progression identifies PLOD2 as a potential therapeutic target. Cancer Commun. (Lond) 42 (2), 164–169. doi:10.1002/cac2.12240
Siegel, R. L., Miller, K. D., Fuchs, H. E., and Jemal, A. (2022). Cancer statistics, 2022. CA Cancer J. Clin. 72 (1), 7–33. doi:10.3322/caac.21708
Turley, S. J., Cremasco, V., and Astarita, J. L. (2015). Immunological hallmarks of stromal cells in the tumour microenvironment. Nat. Rev. Immunol. 15 (11), 669–682. doi:10.1038/nri3902
Ukita, M., Hamanishi, J., Yoshitomi, H., Yamanoi, K., Takamatsu, S., Ueda, A., et al. (2022). CXCL13-producing CD4+ T cells accumulate in the early phase of tertiary lymphoid structures in ovarian cancer. JCI Insight 7 (12), e157215. doi:10.1172/jci.insight.157215
Valkenburg, K. C., de Groot, A. E., and Pienta, K. J. (2018). Targeting the tumour stroma to improve cancer therapy. Nat. Rev. Clin. Oncol. 15 (6), 366–381. doi:10.1038/s41571-018-0007-1
Wang, J., Yang, T., Chen, H., Li, H., and Zheng, S. (2018). Oncogene RPA1 promotes proliferation of hepatocellular carcinoma via CDK4/Cyclin-D pathway. Biochem. Biophys. Res. Commun. 498 (3), 424–430. doi:10.1016/j.bbrc.2018.02.167
Wen, J., Liu, X., Qi, Y., Niu, F., Niu, Z., Geng, W., et al. (2019). BMP3 suppresses colon tumorigenesis via ActRIIB/SMAD2-dependent and TAK1/JNK signaling pathways. J. Exp. Clin. Cancer Res. 38 (1), 428. doi:10.1186/s13046-019-1435-1
Yan, X., Cheng, Y., Zhang, X., Hu, Y., Huang, H., Ren, J., et al. (2022). NICD3 regulates the expression of MUC5AC and MUC2 by recruiting SMARCA4 and is involved in the differentiation of mucinous colorectal adenocarcinoma. Mol. Oncol. 16 (19), 3509–3532. doi:10.1002/1878-0261.13296
Yao, L., Man, C.-F., He, R., He, L., Huang, J.-B., and Xiang, S.-Y. (2022). The interaction between N6-methyladenosine modification and non-coding RNAs in gastrointestinal tract cancers. Front. Oncol. 11. doi:10.3389/fonc.2021.784127
Yun, S., Kwak, Y., Nam, S. K., Seo, A. N., Oh, H. K., Kim, D. W., et al. (2018). Ligand-independent epidermal growth factor receptor overexpression correlates with poor prognosis in colorectal cancer. Cancer Res. Treat. 50 (4), 1351–1361. doi:10.4143/crt.2017.487
Zanger, U. M., and Schwab, M. (2013). Cytochrome P450 enzymes in drug metabolism: Regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol. Ther. 138 (1), 103–141. doi:10.1016/j.pharmthera.2012.12.007
Zhao, L., Cao, J., Hu, K., Wang, P., Li, G., He, X., et al. (2019). RNA-binding protein RPS3 contributes to hepatocarcinogenesis by post-transcriptionally up-regulating SIRT1. Nucleic Acids Res. 47 (4), 2011–2028. doi:10.1093/nar/gky1209
Keywords: colorectal cancer, mucinous adenocarcinoma, adenocarcinoma, not otherwise specified, tumor microenvironment, stromal cell, epithelial cell
Citation: Xu K, Zheng S, Li B, Shao Y and Yin X (2023) Molecular characterization of colorectal mucinous adenocarcinoma and adenocarcinoma, not otherwise specified, identified by multiomic data analysis. Front. Mol. Biosci. 10:1150362. doi: 10.3389/fmolb.2023.1150362
Received: 24 January 2023; Accepted: 27 February 2023;
Published: 05 April 2023.
Edited by:
Xiaoji Cao, Zhejiang University of Technology, ChinaReviewed by:
Hao Yang, Shanghai University of Medicine and Health Sciences, ChinaShaohua Qi, Houston Methodist Research Institute, United States
Copyright © 2023 Xu, Zheng, Li, Shao and Yin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yingkuan Shao, eWtzaGFvQHpqdS5lZHUuY24=; Xiaoyang Yin, eGlhb3lhbmd5MjAxOUAxNjMuY29t
 Kailun Xu
Kailun Xu Shu Zheng
Shu Zheng Baosheng Li
Baosheng Li Yingkuan Shao
Yingkuan Shao Xiaoyang Yin2*
Xiaoyang Yin2*