
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Mol. Biosci. , 24 February 2023
Sec. Molecular Diagnostics and Therapeutics
Volume 10 - 2023 | https://doi.org/10.3389/fmolb.2023.1137215
This article is part of the Research Topic Role of the SUMOylation in Cancer Regulation View all 6 articles
Post-translational modifications of proteins play very important roles in regulating RNA metabolism and affect many biological pathways. Here we mainly summarize the crucial functions of small ubiquitin-like modifier (SUMO) modification in RNA metabolism including transcription, splicing, tailing, stability and modification, as well as its impact on the biogenesis and function of microRNA (miRNA) in particular. This review also highlights the current knowledge about SUMOylation regulation in RNA metabolism involved in many cellular processes such as cell proliferation and apoptosis, which is closely related to tumorigenesis and cancer progression.
Small ubiquitin-like modifier (SUMO), a member of the ubiquitin-like post-translational modifiers are well conserved in eukaryotes (Wilkinson and Henley, 2010; Wang and Yu, 2021). Up to now, mass spectrometry and bioinformatics analysis provide proteomic evidence for SUMOylation of 3,617 proteins at 7,327 SUMOylation sites (Hendriks and Vertegaal, 2016). Although SUMO targets are distributed in various distinct sub-cellular structures, nucleoproteins still account for the majority. SUMOylation has an important regulatory role for most nuclear processes, including RNA transcription, primary RNA processing, nucleocytoplasmic transport, cell cycle progression, nuclear body formation and protein stabilization (Hendriks and Vertegaal, 2016). SUMOylation affects cancer progression by regulating tumorigenesis, proliferation, transformation, migration, invasion, senescence, inflammation, angiogenesis, etc. Therefore, SUMO-related enzymes that involve in tumorigenesis can be used as targets for drug development to treat human cancers.
SUMOylation is a reversible process of adding SUMO to substrate proteins at specific lysine residues as a post-translational modification (PTM) (Wang and Yu, 2021). In mammalian cells, SUMO proteins are conjugated to over thousands proteins. SUMO is conserved from yeast to mammalian cells. Thus far, there are five SUMO isoforms with different sequences in mammals: SUMO1, 2, 3, 4 (Han et al., 2018) and 5 (Liang et al., 2016). SUMO1, SUMO2 and SUMO3 are widely expressed in humans, while SUMO4 and SUMO5 are only found in specific tissues or organs (Guo et al., 2004). SUMO2 and SUMO3 share about 97% identity in humans and are often known as SUMO2/3. SUMO1 only have 53% identity with SUMO2/3. SUMO4 has an 86% amino acid homology with SUMO2, containing a unique proline-90 residue may prevent the maturation and disrupting conjugation to substrates (Owerbach et al., 2005). SUMO5 is reported as a novel SUMO that highly homologous to SUMO1, and poly-SUMO5 conjugation on promyelocytic leukaemia protein (PML) results in the recruitment of proteins to form PML nuclear bodies (PML NBs) (Liang et al., 2016). So far, the research on SUMO4 and SUMO5 is limited.
Similar to the biochemical process of ubiquitination, SUMO attaches to the Lys K) residues of target proteins via an isopeptide bond between its C-terminal glycine G) and the ε-NH2 group of a lysine residue in the target protein (Gill, 2004). SUMOylation is a multi-step enzymatic process, including activation, conjugation and ligation (Figure 1A). The first step of SUMO conjugation pathway is the maturation of SUMOs, whose COOH termini is cleaved by Sentrin-specific proteases (SENPs) to expose the di-glycine (GG) residues required for conjugation (Hay, 2007). Second, mature SUMOs are then activated through an ATP-dependent way by a heterodimeric SUMO E1-activating enzyme, SAE1/2, which is a heterodimer composed of Aos1/SAE1 and Uba2/SAE2 with a size of 110 kDa (Desterro et al., 1999). The SUMOs exclusively interact with the SAE2 subunit, and their C-terminal GG carboxyl group is activated by linking with cysteine C) residue of SAE2 through a thioester bond. Third, the activated SUMO is then transferred to a SUMO conjugating enzyme E2, Ubc9. Fourth, SUMO transferring of Ubc9-SUMO has two ways, by which Ubc9 catalyzes an isopeptide bond between the SUMO terminal GG and the K side chain of the substrate proteins by direct interacting with or without a E3 ligase enzyme. The final step, SUMOylation is reversed by the family of SUMO-specific proteases SENPs that break isopeptide bonds to remove SUMO from the substrate, recycling SUMO molecules (Yeh, 2009; Chang and Yeh, 2020).
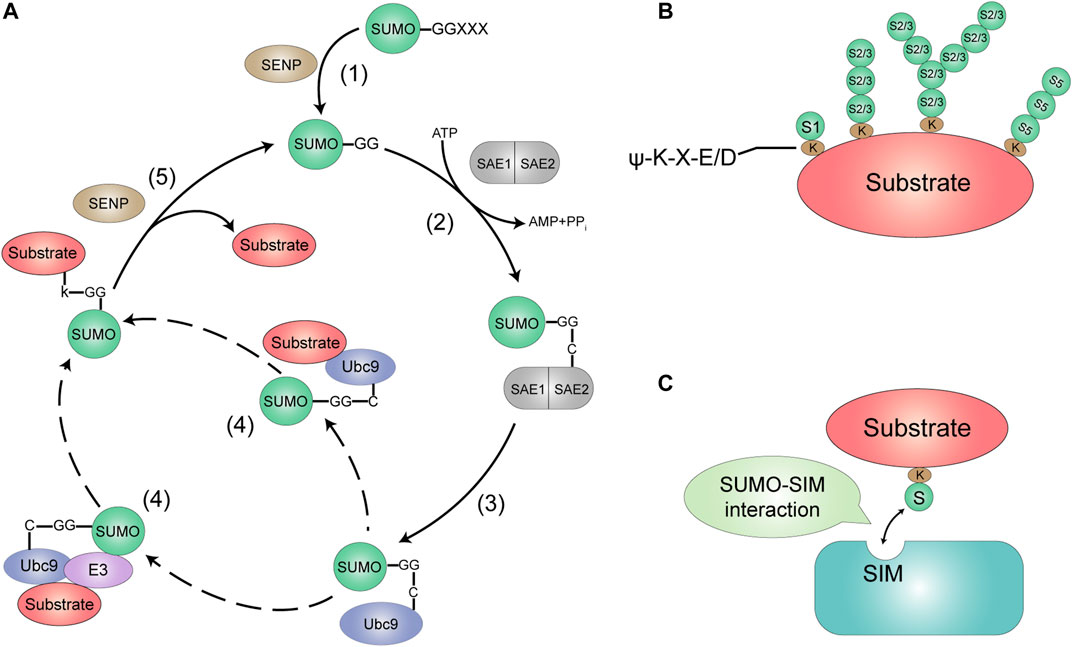
FIGURE 1. The SUMOylation system. (A) SUMO is encoded as an inactive precursor that is cleaved by members of the SENP family to expose the C-terminal diglycine motif (1). This mature form of SUMO is then activated by forming an ATP-dependent thioesler bond with the active site of the Et enzyme (a heterodimer of SAE1 and SAE2) (2). The activated SUMO is then delivered to the active site cysteine of the E2-conjugating enzyme Ubc9. Which then catalyzes the transfer of SUMO to the target protein, either alone or with the help of a SUMO E3 ligase (3, 4). SUMOylated sub-strates show phenotypic differences from the unmodified form. DeSUMOylation is mediated by the SENP protease family (5). This process releases unmodified target protein (not shown) and mature SUMO. which can then be used for further binding to the target protein. (B) SUMO modifications most commonly target proteins with the consensus motif w-K-X-FJD. to is a hydrophobic amino acid. K is the target Lys. X is any amino acid, and DIE is Asp or Glu. SUM01 modifies substrates into monomers. While SUMO2/315 modifies substrates in poly•SUMO chains. (C) Proteins contain-ing SIMs mediate non-covalent interactions with SUMO.
In general, the most common target proteins of SUMO modification are to have a common motif ψ-K-X-E/D (ψ is a hydrophobic amino acid, K is the target Lys, X is any amino acid, D/E is amino acid Asp/Glu), but there are also some proteins without classical motifs that have been more reported recently (Chang and Yeh, 2020). In usual, SUMO1 modifies the substrate proteins by one monomer, while SUMO2/3 can modify by poly-SUMO to form multiple SUMO chains. At present there is no evidence that SUMO4 is conjugated to K of substrate protein, but SUMO5 seems to be able to form a poly-SUMO chain (Figure 1B). Reader proteins can recognize and bind to SUMOylated proteins through their SUMO interaction motifs (SIMs). (Martin et al., 2007) (Figure 1C). Functionally, SUMO is covalently coupled to a large number of proteins to modulate their enzymatic activity, subcellular localization or their protein interactions. SUMOylation may also join with ubiquitin to degrade proteins through SUMO-targeting ubiquitin ligase (STUbL) (Hickey et al., 2012).
Importantly, SUMOylation plays an important role in RNA metabolism, including messenger RNA (mRNA) and non-coding RNA (ncRNA). Abnormal RNA metabolism caused by SUMOylation is often the main reason affecting the occurrence and development of tumors. On the one hand, mRNA metabolism regulated by SUMOylation involves in transcription, processing (e.g., capping, splicing and polyadenylation), mRNA translation and quality control. On the other hand, SUMOylation of regulation on ncRNA metabolism is mostly and broadly focused on microRNAs (miRNAs). In addition, researches on cellular processes compartmentalized by membrane-less organelles (MLOs) have become a hotspot. MLOs typically contain disordered proteins and RNAs by liquid-liquid phase separation (LLPS) (Alberti and Hyman, 2021). MLOs for molecular condensates in the nucleus include nucleoli (Tartakoff et al., 2022), paraspeckles (Yamazaki et al., 2021), nuclear speckles (NS) (Spector and Lamond, 2011), Cajal bodies (Nizami et al., 2010), PML NBs (Lallemand-Breitenbach and de The, 2018) and nuclear stress bodies (nSBs) (Biamonti and Vourc’h, 2010), and in the cytoplasm there are P-bodies and stress granules (SGs) (Ivanov et al., 2019). They function as RNA and protein quality control centers and are highly sensitive to cellular stress, including proteotoxic stress (Advani and Ivanov, 2019; Hirose et al., 2022). Growing evidence shows that SUMOylation is critically involved in regulating both the assembly and disassembly of MLOs, and thus therefore regulates RNA splicing, processing and RNA stability (Hofweber and Dormann, 2019).
This review provides insights into the role of SUMOylation in regulating RNA metabolism in the pathophysiology of diseases, especially cancers. We summarize the regulation of RNA metabolism by SUMOylation from the following aspects: 1) RNA transcription; 2) mRNA processing; 3) RNA stability; 4) RNA editing and RNA modification; 5) Non-coding RNA metabolism.
SUMOylation plays an important role in transcriptional regulation via SUMO conjugating to histones, histone-modifying enzymes, transcription factors, transcriptional co-regulators and transcriptional machinery-related proteins. The mechanisms underlying SUMO-mediated transcriptional regulation includes alterations in subcellular localization, DNA binding, protein-protein interaction, stability and enzymatic activity (Chymkowitch et al., 2015).
Histones undergo many modifications, including acetylation, methylation, phosphorylation, ubiquitination, glycosylation, ADP ribosylation and SUMOylation, some of which play key roles in the regulation of chromatin structure and functions. Most histones, including H1, H2A, H2B, H3, H4 and H2A.X are SUMOylated (Table 1). SUMOylation of H4 leads to chromatin compaction and transcriptional repression through the recruitment of histone deacetylase 1 (HDAC1) and heterochromatin protein 1 (HP1) (Shiio and Eisenman, 2003; Ryu and Hochstrasser, 2021) (Figure 2A; Figure 1). However, SUMOylation of H4K12 has also been reported to inhibit chromatin compaction (Dhall et al., 2014). Moreover, SUMOylation of linker histone H1 drives chromatin condensation and restriction of embryonic cell fate identity (Dhall et al., 2014). The transcriptional repression mechanisms of SUMOylation at other histones are not very clear.
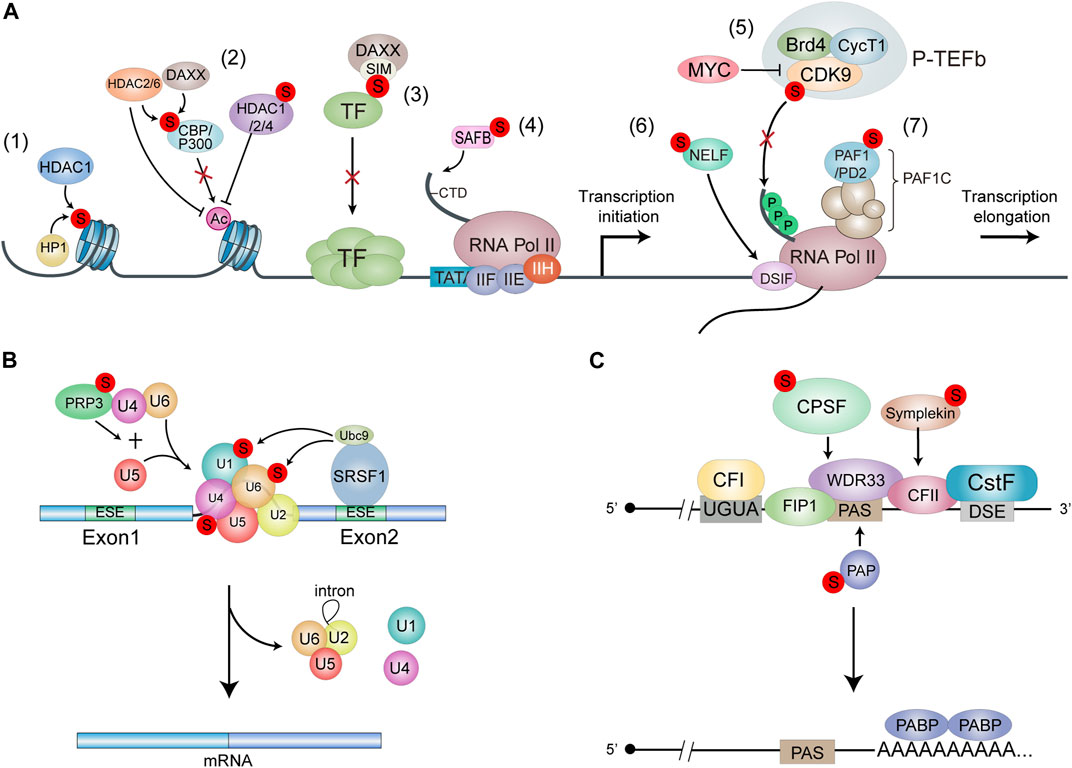
FIGURE 2. Models for the functions of SUMOylation in RNA transcription and processing. (A) SUMOylation leads to transcnptional repression. Histone SUMOylation mediates transcriptional repression through recruitment of HDAC1 and HP1 (1). SUMOylation of p300 and CBP recruits HDAC6 and DAXX/HDAC2 respechvehi, leading to SUMO-dependent transcnptional repression. SUMOylation of liADC1/2/4 promotes the deacetylase activity to repress transcription (2). The SIM domain of DAXX provides a molecular explanation for the interaction between DAXX and SUMO-mod-ified transcription factors, resulting In transcriptional repression (3) SUMOylation of SAFE enhances its binding with RNAPII, thereby promoting gene transoiption (4). SUMO and MYC antagonistically control global gene expression through regulating COK9 SUMOylation. P-TEFb formation and tran-scriptional elongation. SUMOylation of CDK9 blocks its interaction with Cyclin Ti. Thereby inhibiting the formation of active P-TEFb complex (5). NELF and OSIF bind to the potymerase in a manner that restricts Pol II mobility and impairs further RNA elongation. SUMOylation regulates the recruitment of NELF to promoters upon stress to drive transcriptional downregulabon (6). SUMOytabon of PAF1/PD2 facilitates its interaction with PML proteins (7). (B) SRSF1. Bound to the exonic splicing enhancer (ESE). Interacts with Ubc9 to regulate the SUMOylation of spliceosome protein components. Thuieby increasing splicing efficiency. PRP3 acts as a component of the 114/U6 cli-snRNP. and Its SUMOylatlon Promotes U41U6/1.15 tri-snRNP formation to effect splicing. (C) SUMOylation of CPSF. PAP and Sim-pleton affects the assembly and activity of pre-mRNA 3’complex.
As known, the acetylation level of histone is positively correlated with gene activation. Histone acetylation is catalyzed by histone acetyltransferases (HATs) such as CREB-binding protein (CBP) and p300 while it is removed by histone deacetylase (HDACs). CBP and p300 are co-regulators of many sequence-specific DNA binding proteins in response to a variety of signaling (Yu et al., 2004), and are regarded as key drivers of tumorigenesis (Hogg et al., 2021). Both p300 and CBP contain a cell cycle regulatory domain 1 (CRD1), which is a strong transcriptional repression domain. It has been reported that SUMOylation of p300 CDR1 domain mediates transcriptional repression by promoting the recruitment of HDAC6 (Girdwood et al., 2003) (Figure 2A–2). Similarly, SUMOylation of CBP negatively modulates transcriptional activity by recruiting Death domain-associated protein (DAXX) and then HDAC2 (Kuo et al., 2005; Park et al., 2017).
On the other hand, HDACs reverse chromatin acetylation by removing acetyl groups and alter the transcription of oncogenes and tumor suppressor genes governing cancer initiation and progression (Li and Seto, 2016). HDACs family members such as HDAC1, HDAC2 and HDAC4 can also be SUMOylated. SUMOylation of HDAC1/HDAC4 enhances their histone deacetylase activity, leading to the transcriptional repression (David et al., 2002; Kirsh et al., 2002). Moreover, SUMOylation of HADC4 can enhance its deacetylation of p53, thus blocking the recruitment of p53 into the promoter, which modulates the apoptotic response to genotoxic stress in cancer cells (Brandl et al., 2012). In addition to enhancing the deacetylase activity, SUMOylation of HDAC2 also increases its binding to the Elk-1 regulated promoters, which directly leads to a decrease of histone acetylation and hence transcriptional repression at Elk-1 target genes (Yang and Sharrocks, 2004) (Figure 2A). HDACs are abnormally high in a variety of cancers, which govern a wide array of biological processes including cancer initiation and progression, thus the development of HDACs-targeted imaging probes for cancer detection and targeting HDACs for cancer therapy have become a hotspot in epigenetic research (West and Johnstone, 2014; Tang et al., 2022). Since SUMOylation of HDACs plays an important role in the regulation of HDACs activity in cancer cells, it is worthwhile to design some small chemical molecules or other specific drugs to interfere with the SUMO modification of HDACs in cancers.
A wide range of transcription factors and co-transcriptional regulators have been reported as SUMOylated proteins. In most cases, SUMOylation is usually associated with transcriptional repression (Rouviere et al., 2013). Here, we list transcription regulators, especially those related to cancer, which can be modified by SUMOs to modulate transcriptional activation activity (Table 2). Remarkably, DAXX regulates gene expression as a transcriptional co-repressor or co-activator by interacting with diverse core histones, chromatin-associated proteins, transcription factors and epigenetic regulators (Mahmud and Liao, 2019). DAXX-regulated genes are important effectors in cell death, survival and tumorigenesis (Mahmud and Liao, 2019), so the expression level of DAXX is increased in several cancer types including prostate (Tsourlakis et al., 2013), ovarian (Pan et al., 2013) and gastric cancer (Xu et al., 2017). DAXX can be modified by SUMO1 increasing recruitment of DAXX to PML-NBs (Jang et al., 2002), which induces cancer cell apoptosis (Takahashi et al., 2004). Moreover, the conserved SIMs of DAXX provide a molecular explanation for the interactions of DAXX with SUMO-modified proteins, leading to transcriptional activation or repression (Figure 2A; Figure 3). For examples, DAXX can interact SUMO-modified androgen receptor (AR) and glucocorticoid receptor (GR) to repress their transcriptional activities by inhibiting their DNA binding (Lin et al., 2004; Lin et al., 2006). By binding with SUMOylated-SMAD4, DAXX inhibits the tumor suppressive effect of transforming growth factor-β (TGF-β) signaling mediated by SMAD4 (Chang et al., 2005), which affects the growth arrest and apoptosis of cancer cells (Zhao et al., 2018).
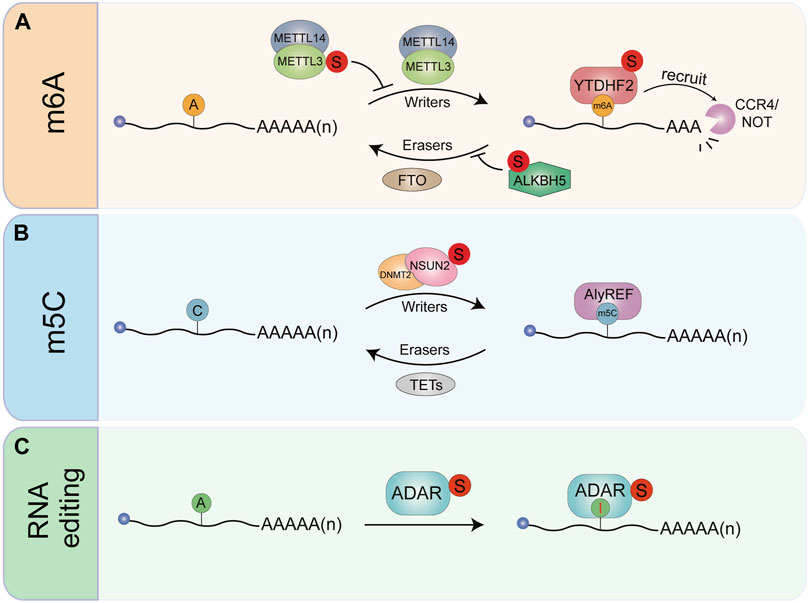
FIGURE 3. The effects of SUMOylation on RNA modification and RNA editing. (A) The N6-methylad-enosine writer METTL3. Reader YTHDF2 and eraser ALKBHS can be modified by SUMO. SUMOyIa-lion of METTL3 does not affect its stability. Subcellular localization. Or Interaction with MET11141 W-TAP. but significantly inhibits METTL3 m6A methyltransferase activity. YTHDF2 can be modified by SUMO1 under hypoxia. Enhancing its binding affinity to m6A-RNAs. Resulting in dysregulation of gene expression. ALKBHS SUMOylation loads to inhibition of ALKBHS m6A demethylase activity. Thereby increasing global mRNA m6A levels. (B) SUMOylation stabilizes m5C writer NSUN2 and mediates its transport into the nucleus. (C) The RNA editing enzyme ADAR1. Which converts ade-nosine to inosine, can be SUMOylated thereby inhibiting its RNA editing activity.
Although SUMOylation of most transcription factors and transcriptional co-regulators plays a negative role in transcription, it also activates several transcription factors, such as tumor suppressor p53 (Gostissa et al., 1999; Rodriguez et al., 1999), the G2/M transcription factor forkhead box protein M1 (FOXM1) (Schimmel et al., 2014), PAX6 (Yan et al., 2010), nuclear hormone receptor ESR1 (Sentis et al., 2005) and EGR1 (Yu et al., 2009) (Table 2). Our early data reveal that SUMO1 modification of EGR1 contributes to EGR1-mediated PTEN transcriptional activation (Yu et al., 2009).
There are also some evidences show that SUMOylation of transcriptional machinery-related proteins can promote transcriptional activation. SUMO1 marks the chromatin on the promoters of many housekeeping genes, which encode translation factors and ribosomal subunit proteins, to promote the transcription of these specific RNAs (Liu et al., 2012). The C-terminal domain (CTD) of the largest subunit of RNA polymerase II (RNAP II) is important in coupling RNA processing and transcription (Corden and Patturajan, 1997; Bentley, 2014). The chromatin scaffold protein SAFB (scaffold related factor B) interacts with the CTD of RNAPII and a subset of serine-/arginine-rich RNA processing factors (SR proteins) to form a ‘transcriptosomal’ complex that couple the transcription and RNA processing (Nayler et al., 1998). SUMO1-modified SAFB1 can promote the binding of RNAPII to the promoters of ribosomal protein genes and pre-mRNA splicing (Liu et al., 2015) (Figure 2A; Figure 4).
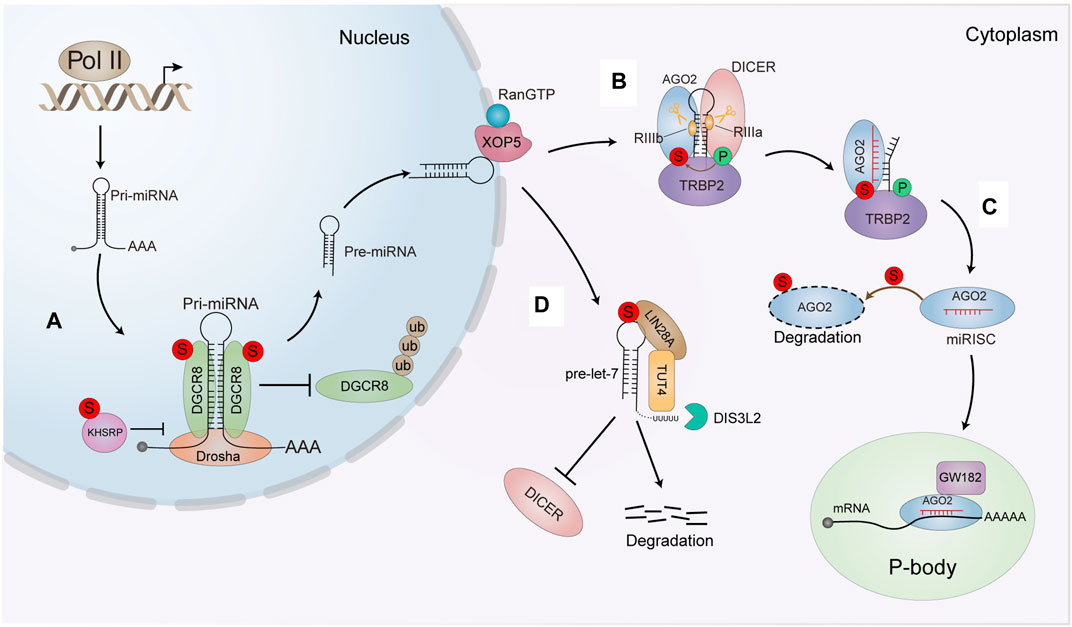
FIGURE 4. The effect of SUMO modification on the miRNA pathway. (A) OGCR8 SUMOyfation increases its protein stability by preventing the degradation via the ubiquitin proteasome pathway. The SUMOylation also enhances its affinity with pri-miRNA thus positively promoting the pri-miRNA direct recognition and repression of the targeted mRNA. SUMOylated KHSRP inhioits interaction with Dro-sha/DGCR8 and pri-miRNAs complex, sequentially downregulatlng a subset of miRNAS biogenesis. (B) SUMOylation of TARBP2 controls the efficiency of RNA-induced gene silencing by increasing its interaction with AGO2 and precursor miRNAs/siRNAs (C) SUMOytation enhances AGO2 turnover and antagonizes its stability. (D) SUMOylated of LIN28 A increases the binding affinity of LIN28 A and pre-let-7. Thereby leading to the degradation of pre•let-7 and reducing mature let-7 biogenesis. The intense interaction between LIN28 A and pre-let-7 can efficiently recruit TUT4 to urictylate pre-let-7 and block the processing of pre-let-7.
The transcription profiling reveals that SUMOylation represses the global transcription by inhibiting transcriptional elongation. SUMOs and MYC oppositely control global gene expression by regulating the dynamic SUMOylation and deSUMOylation of CDK9. CDK9 is the catalytic subunit of the P-TEFb kinase and is essential for productive transcriptional elongation. SUMOylation of CDK9 represses global transcription, while MYC amplifies global transcription by antagonizing CDK9 SUMOylation (Yu et al., 2018) (Figure 2A). The oncogene MYC is part of a superfamily of genes encoding the most commonly activated oncoproteins in human cancers (Schaub et al., 2018). MYC is highly associated with cell proliferation, differentiation, survival and death (Bernard and Eilers, 2006; Bretones et al., 2015; Stine et al., 2015; Baluapuri et al., 2020). Dysregulation of MYC expression or activation has been linked to many human cancers (Dang, 2012; Lourenco et al., 2021). Many therapeutic agents targeting MYC are under development, among which the inhibitors of MYC transcriptional activity may be an attractive therapeutic intervention (Dhanasekaran et al., 2022). Considering that MYC amplifies gene expression by antagonizing CDK9 SUMOylation, modulation of CDK9 SUMOylation may be a feasible therapeutic approach given that there are currently no approved direct inhibitors of MYC.
In response to stresses, the transcription of housekeeping genes is usually downregulated in cells. Stress-induced SUMOylation of negative elongation factor (NELF) is required for global transcriptional repression by promoting the binding of NELF to the promoters, which impairs RNAPII elongation. NELF forms nuclear condensations in a dephosphorylation-dependent and SUMOylation-dependent manner under stressful conditions. Biomolecular condensation facilitates enhanced recruitment of NELF to promoters upon stress to drive transcriptional downregulation (Rawat et al., 2021) (Figure 2A). RNA polymerase II-associated factor 1 (PAF1)/pancreatic differentiation 2 (PD2) is a core subunit of the human PAF1 complex (PAF1C) that regulates the RNA polymerase II function during transcriptional elongation. SUMOylation of PAF1/PD2 predominantly presents in the nucleus and promotes its interaction with PML protein in response to radiation. PML functions in multiple aspects of cellular function including transcription. PML-NBs are strongly associated with the SUMOylation process, since SUMOylation of PML induces recruitment of other SIM-containing factors to these bodies (Zhong et al., 2000). Furthermore, inhibition of SUMOylation or PML reduces the cell growth and proliferation of pancreatic ductal adenocarcinoma (PDAC) cells (Rauth et al., 2021) (Figure 2A).
Most mRNA precursors (pre-mRNAs) undergo three processing steps: the 5′end is capped by addition of 7-methylguanosine; introns are removed and exons ligated by splicing; and the 3′end is created by an endonucleolytic cleavage followed by addition of a 100–300 nt long poly(A) tail. The evidence linking SUMOylation with the process of RNA capping, splicing and tailing is majorly based upon proteomic analysis (Richard et al., 2017). In the past decade, the affinity purification strategy based on tagged-SUMO peptides has led to the development of mass spectrometry (MS) identifying large-scale SUMOylated proteins, many of which are involved in RNA processing events such as capping, splicing, polyadenylation and mRNA export in mammals (Li et al., 2004; Manza et al., 2004; Gocke et al., 2005; Guo et al., 2005; Rosas-Acosta et al., 2005; Vertegaal et al., 2006; Blomster et al., 2009; Golebiowski et al., 2009; Matafora et al., 2009; Matic et al., 2010; Tatham et al., 2011; Becker et al., 2013; Schimmel et al., 2014; Tammsalu et al., 2014; Yang and Paschen, 2015). In yeast, a significant proportion (17%) of the SUMO-modified proteins identified is found to be involved in RNA-related processes (Denison et al., 2005). It is reported that enzymes in SUMO pathway are co-located in some nucleosomes, which is believed to be closely related to RNA processing. Nuclear speckle NS, for an example, plays a major role in regulating the availability of splicing factors at the transcription site since more than 50% of NS proteins are involved in transcription or splicing regulation. NS-associated processers are well regulated by SUMOylation because SUMO E2 Ubc9 (Ihara et al., 2008) and E3 ligases PIAS family (Tan et al., 2002) localize to NS. However, how SUMOylation cooperatively modulates the function of individual NS proteins remains to be revealed (Galganski et al., 2017). In addition, SUMO1 and Ubc9 are also localized to Cajal bodies, which are implicated in RNA-related metabolic processes (Navascues et al., 2008).
Pre-mRNA splicing is one of most important RNA processing catalyzed by the spliceosome. Spliceosome assembly occurs by the ordered interaction of five small nuclear ribonucleoprotein particles, termed U1, U2, U4, U5, and U6 small nuclear ribonucleoproteins (snRNPs). Each snRNP consists of a small nuclear RNA (snRNA) and a large set of associated proteins. In the earliest cross-intron spliceosomal complex, U1 and U2 snRNP bound to the intron to form A complex. Then, the activated spliceosome B Complex generated by the recruitment of U4/U6/U5 tri-snRNP. After numerous RNA and protein rearrangements, including the dissociation of the U1 and U4 snRNPs, and splicing by the DEAH-box RNA helicase PRP2, the catalytic C complex is finally yielded. The C complex catalyzes intron excision and ligation of the exons followed by spliceosome disassembly (Wilkinson et al., 2020). Several spliceosomal proteins are SUMOylated in vitro and in cultured cells, including components of U1, U2, U4/U6, U5 snRNPs, non-snRNPs, heterogeneous nuclear ribonucleoproteins (hnRNPs) as well as a wide range of other spliceosome-associated factors, suggesting that SUMOylation may play a role in multiple steps of pre-mRNA splicing (Pozzi et al., 2017; Chanarat and Mishra, 2018).
Serine/arginine (SR) proteins, such as members of the hnRNP family and many splicing factors, are highly SUMOylated in response to various stresses (Golebiowski et al., 2009; Psakhye and Jentsch, 2012). The SR protein SRSF1 (known as SF2/ASF) acts as a regulator of the SUMOylation pathway by interacting with Ubc9 to modulate the activity of E3 ligase PIAS1, thus promoting SUMOylation of specific substrates. SRSF1 is upregulated in varies cancer type (Ghigna et al., 2005; Karni et al., 2007; de Miguel et al., 2014; Anczukow et al., 2015) and controls alternative splicing of many tumor-related genes, which affect cell apoptosis (Olsson and Zhivotovsky, 2011; Anczukow et al., 2012; Gautrey and Tyson-Capper, 2012), proliferation and migration (Ghigna et al., 2005) thereby promoting tumorigenesis and cancer development. Knockdown of SRSF1 suppresses SUMO modification of splicing factors as well as tumor progression (Pelisch et al., 2010). The splicing factor U4/U6 small nuclear ribonucleoprotein PRP3, a component of the U4/U6 di-snRNP, is another SUMOylation substrates during splicing and its SUMOylation is regulated by SRSF1 (Li N et al., 2020). SUMOylation of PRP3 is required for U4/U6/U5 tri-snRNP formation and recruitment to activate the spliceosome. The SUMOylation-deficient mutant PRP3/2 KR do not affect PRP3 binding to U4 or U6 snRNAs, but prevented its interaction with the associated splicing factors U2-SF3A1 and U5-Snu114, suggesting that SUMOylation of PRP3 promotes U4/U6/U5 tri-snRNP formation to affect splicing (Pozzi et al., 2017) (Figure 2B).
SAFB, which interacts with the CTD of RNAPII and RNA processing proteins such as SR proteins as mentioned before, is identified as an SUMO1 substrate binding to promoters of highly expressing genes. SUMOylation of SAFB also increases splicing rates of mRNAs encoding ribosomal proteins, suggesting a role for SUMOylated-SAFB in coupling transcription and RNA processing (Liu et al., 2015). SAFB is also a key player in tumorigenesis. Overexpression of SAFB1 leads to growth inhibition of breast cancer cells (Townson et al., 2000), and it involved in apoptosis and the immune system as a transcriptional repressor regulating genes in breast cancer cells (Hammerich-Hille et al., 2010). However, the role of SAFB SUMOylation in cancer development is unclear. Except for factors directly involved in splicing, AKT is also involved in the splicing process as a key sensor that translates extracellular signals into changes in splicing patterns. AKT is modified by SUMO at K276 and K301 within its kinase domain, and its mutation (Akt E17 K) increases its SUMOylation levels in several human cancer types, enhancing its capacity to regulate fibronectin and Bcl-x alternative splicing patterns (Risso et al., 2013). In general, SUMOylation modulates the activity of splicing-related proteins to promote spliceosome assembly and improve splicing efficiency, which regulates alternative splicing of many tumor-associated genes to affect cancer progression, but more studies are needed to clarify the detailed mechanisms of SUMOylation of splicing related proteins in the regulation of tumorigenesis, so as to promote the improvement of cancer treatment strategies.
SUMOylation is also involved in regulating another key mRNA processing step, polyadenylation, which processes the 3′-end of nascent transcripts and requires several complexes including CFI (cleavage factor I), CFII, CPSF (cleavage and polyadenylation specificity factor) and CstF (cleavage stimulation factor) and additional protein factors (Shi et al., 2009). The 3′ends of pre-mRNAs are formed in a two-step process, with an endonucleolytic cleavage generating a 3′OH end followed by the synthesis of a poly(A) tail (Colgan and Manley, 1997; Proudfoot and O'Sullivan, 2002). For canonical 3′-end processing in mammals, the cleavage site is located between an upstream polyadenylation signal (PAS), most frequently an AAUAAA hexamer, and a GU-rich downstream element (DSE) (Sun et al., 2020). The multi-subunit CPSF and CstF define the poly(A) site by binding cooperatively to PAS and DSE, respectively. Cleavage factors I and II help in the complex assembly and in the first step, then Poly(A) polymerase (PAP) catalyzes the addition of poly(A) (Takagaki et al., 1989; Raabe et al., 1991). It has found that CPSF, PAP and assembly factor Symplekin are modified by SUMO2/3 (Richard et al., 2017) (Figure 2C). Blocking SUMOylation of CPSF and Simplekin affects pre-mRNA 3′complex assembly and activity and is lethal to cell viability (Vethantham et al., 2007; 2008). CPSFs have multiple roles in different tumor types. CPSF1 (Zhang et al., 2017a) and CPSF4 (Tang et al., 2016; Yi et al., 2016; Zhang et al., 2017a) promote cell proliferation in ovarian cancer and lung adenocarcinoma (LUAD), respectively, while CPSF2 inhibits invasion and cancer stem cell growth (Sung et al., 2015). However, the mechanism of CPSF SUMOylation in cancer remains unclear. PAP is highly SUMOylated in its C-terminal regulatory domain by strong binding with Ubc9, which is required for its correct nuclear localization and stability. In vitro polyadenylation assays show that SUMOylated PAP has lower poly(A) synthesis activity, indicating that SUMOylation of PAP inhibits polyadenylation activity to profoundly affect gene expression (Vethantham et al., 2008). Dysfunction of PAP activity has been shown to associated with diverse diseases. The increasing activity of PAP is correlated with malignancy in breast cancer and leukemia (Papamichail et al., 1983). Conversely, the inhibition of PAP activity is associated with apoptosis (Scorilas et al., 1998). Thus, regulation of PAP mediated by SUMOylation may have important role in cancer development and more studies are needed to investigate its mechanism.
SUMOylation regulates some of the key factors controlling RNA stability under cellular stresses. Nucleolin is a multifunctional DNA and RNA binding protein, closely associates with regulation of cell proliferation and growth, which is abundant in growing and cancerous cells (Grinstein et al., 2007). SUMOylation of nucleolin is required for maintaining its nuclear localization and promoting its activity in mediating gadd45α mRNA stability, which in turn increases apoptosis under arsenic exposure (Zhang et al., 2015). Therefore, it is likely that the SUMOylation of nucleolin has an important regulatory role in specific stress-induced mRNA stabilization.
N6-methyladenosine (m6A) is one of the most common modifications in mRNA, rRNA, tRNA, microRNA and long non-coding RNA (Alarcon et al., 2015). Human YTH domain family 2 (YTHDF2) selectively recognized m6A-RNAs to regulate degradation by recruiting the CCR4-NOT complex through a direct interaction between the YTHDF2 N-terminal region and the SH domain of the CNOT1 subunit (Du et al., 2016) (Figure 3A). We recently found that YTHDF2 can be modified by SUMO1 under hypoxic conditions, which enhances its binding affinity to m6A RNAs, leading to accelerated degradation of mRNAs. Subsequently, SUMOylation of YTHDF2 changes the gene expression profile, thus promoting cancer progression (Hou et al., 2021).
In response to various environmental stresses, stress granules (SGs) rapidly form thus transiently storing mRNAs and RNA-binding proteins (RBPs) to limit protein synthesis. SGs are associated with various diseases including cancers, viral infections and neurodegenerative diseases (Ivanov et al., 2019). It has been demonstrated that SUMO-conjugating enzymes are recruited to SGs and SUMOylation regulates SG formation and disassembly (Keiten-Schmitz et al., 2020; Marmor-Kollet et al., 2020). As reported by several groups, the SUMOylation level of RBPs is regulated under stress conditions, however how SG regulates RNA metabolism remains unclear.
SUMOylation has been implicated in RNA modifications. m6A is the most prevalent internal RNA modification in eukaryotes. It is a dynamic and reversible modification that occurs primarily in 3′untranslated regions (3′UTRs) and near the stop codons (Meyer et al., 2012). The methyltransferase like 3 (METTL3) is a key component of the large N6-adenosine-methyltransferase complex in mammalian. Our laboratory found that METTL3 is modified by SUMO1, which does not affect its stability, subcellular localization, or interaction with methyltransferase complex METTL14/WTAP, but significantly inhibits its m6A methyltransferase activity. SUMOylation of METTL3 enhances tumor growth in human non-small cell lung carcinoma cell line H1299 by decreasing the m6A level in mRNAs and subsequently changing gene expression profile (Du et al., 2018). XU et al confirmed SUMOylation of METTL3 can regulate hepatocellular carcinoma (HCC) progression via controlling Snail mRNA homeostasis in a m6A methyltransferase activity-dependent manner (Xu et al., 2020). ROS induces a global increase in mRNA m6A via inhibition of the m6A demethylase ALKBH5. Mechanistically, ROS promotes ALKBH5 SUMOylation through activating ERK/JNK signaling, leading to inhibition of ALKBH5 m6A demethylase activity by blocking substrate accessibility. Increased global mRNA m6A levels lead to the rapid and efficient induction of thousands of genes involved in a variety of biological processes including DNA damage repair (Yu et al., 2021) (Figure 3A). Methylases and demethylases of m6A can affect the complexity of cancer progression (Zhang et al., 2017b; Weng et al., 2018; Huang et al., 2019; Li J et al., 2020), thus SUMOylation of remaining enzymes should be investigated for potential clinical application of cancer treatment.
5-methylcytosine (m5C) is another important posttranscriptional modification of RNA. The modification m5C in mRNAs is mainly catalyzed by the RNA methyltransferase NSUN2 and specifically recognized by the mRNA export adaptor ALYREF (Yang et al., 2017). NSUN2 is highly expressed in various tumors but extremely low in most normal tissues (Okamoto et al., 2012). NSUN2 is modified by SUMO2/3 stabilizing NSUN2 and mediating its nuclear transport, which regulates the m5C modification levels of mRNAs that are involved in GC progression (Hu et al., 2021) (Figure 3B).
ADAR1, an RNA-editing enzyme also undergo SUMOylation to regulate RNA metabolism. ADAR1 binds to double-stranded RNA and converts adenosine to inosine, resulting in changes in the amino acid coding that alter protein sequence/function (Baker and Slack, 2022). ADAR1 is commonly overexpressed in a growing number of cancer types, and its oncogenic role in cancers is attributed to multiple mechanisms. It has been found that ADAR1 can be modified by SUMO1. Although SUMOylation does not affect ADAR1 localization in the nucleolus, it appears to inhibit its RNA editing activity (Desterro et al., 2005) (Figure 3C).
Non-coding RNAs (ncRNAs) are a class of RNA molecules that do not encode proteins but can function except mRNA, tRNA and rRNA. miRNAs are small non-coding regulatory RNAs ranging from 20–25 nucleotides, which binding to partially complementary sequences within the 3′-untranslated regions of target mRNAs inducing translational repression or mRNA degradation (Treiber et al., 2019). miRNAs are extensively deregulated in human cancers, as miRNA expression is globally suppressed in tumor cells compared to normal cells (Lu et al., 2005). miRNA biogenesis is strictly controlled at several levels, such as transcription, processing, itself modification and decay and is closely related to tumors (Ha and Kim, 2014). Aberrant miRNA biogenesis in cancer occurs at different steps during miRNA maturation, and more importantly, key proteins of miRNA biogenesis are post-transnationally modified by SUMOs to regulate miRNA biogenesis and functions (Table 3).
Primary miRNAs (pri-miRNAs) are typically transcribed by RNA polymerase II in the nucleus (Krol et al., 2010), and then pri-miRNAs are cleaved into approximately 70 nucleotide stem-loop structures by the nuclear RNase III-type enzyme DROSHA, in a complex with its co-factor DGCR8 (DiGeorge syndrome critical region 8 homolog), which is essential for pri-miRNA processing by cleaving pri-miRNA into pre-miRNA in the nucleus. We first reported that DGCR8 can be modified by SUMO1 at K259 and K707. K707-SUMOylation of DGCR8 enhances protein stability by preventing degradation by the ubiquitin-proteasome pathway (Zhu et al., 2015), whereas K259-SUMOylation of DGCR8 is critical for its nuclear localization for its normal miRNA processing functions (Zhu et al., 2016). SUMOylation of DGCR8 at K707 does not alter its association with Drosha and miRNA biosynthesis. However, K707-SUMOylation affects its affinity for pri-miRNA to control the direct function of pri-miRNAs in recognizing and repressing target mRNAs (Zhu et al., 2015). SUMOylation of DGCR8 is apparently related to the function of DGCR8 in regulating tumorigenesis and cell migration in lung adenocarcinoma (Figure 4A). The hnRNP K homology (KH)-type splicing regulatory protein (KHSRP) is one major component of the DROSHA complex. KHSRP can promote the biogenesis of miRNAs, such as let-7 family, whose pri-miRNAs harboring short G-rich stretches in their terminal loops (TL). SUMO1 modification of KHSRP at K87 can be induced upon the microenvironmental hypoxia while reduced by the treatment with growth factors. SUMOylation of KHSRP probably increases its translocation from the nucleus to the cytoplasm, thus inhibiting its interaction with the pri-miRNA/DROSHA-DGCR8 complex, therefore SUMOylation of KHSRP results in the downregulation of TL-G-Rich miRNAs such as let-7 family, which is linked to tumorigenesis and cancer progression (Figure 4A) (Yuan et al., 2017).
In the cytoplasm, pre-miRNAs are cleaved into double-stranded RNAs (dsRNAs) by another RNase III-type enzyme, DICER with its co-factors TRBP (TAR RNA-binding protein 2, also known as TARBP2), resulting in an RNA duplex of ∼22 nucleotides. The short RNA duplex is bound by an AGO (Argonaute) protein, a component of a multi-subunit complex termed miRISC (miRNA-induced silencing complex) (Iwakawa and Tomari, 2022). Finally, miRISC mediates the recognition of targeted mRNAs by guiding miRNA to their specific mRNA targets through base pairing (Zealy et al., 2017). Interestingly, we found that SUMOylation of TARBP2 regulates the efficiency miRNA/short interfering RNA (siRNA) (Chen et al., 2015). TARBP2 is SUMOylated at K52, which can be enhanced by its phosphorylation. SUMOylation of TARBP2 significantly promotes its binding with pre-miRNAs to facilitating the efficiency miRNA-inducing gene silencing. SUMOylated TARBP2 also represses its ubiquitination and stabilizes AGO2 by increasing the binding with AGO2 via SIMs of AGO2. SUMOylation of TARBP2 are implicated in suppression of tumor growth and tumor cell migration (Figure 4B) (Chen et al., 2015). Moreover, AGO2 can be modified by SUMO1 and SUMO2/3 leading to reduce its stability (Figure 4C) (Sahin et al., 2014). However, our unpublished data suggested that SUMOylation of AGO2 does not affect its stability, but crosstalk with phosphorylation together modulates miRNA loading to AGO2, which affects the gene silencing activity.
In addition to key proteins in these classical miRNA biogenesis pathways, other proteins also undergo SUMO modification to affect miRNA metabolism. Let-7 family members (let-7 s) are known as important tumor suppressors, which are downregulated in multiple cancers, and associated with increased proliferation and invasion of cancer cells. LIN28 A is a conserved RNA-binding protein that inhibits processing of pre-let-7 miRNAs, thus promoting cancer progression. Our lab showed that LIN28 A is SUMOylated at K15, which is increased by hypoxia but reduced by chemotherapy drugs such as Cisplatin and Paclitaxel. SUMOylation of LIN28 A increases its binding affinity with the pre-let-7 and recruits of terminal uridylyltransferase TUT4 to block processing of pre-let-7. Subsequently, SUMOylation of LIN28 A exacerbates its inhibition of let-7 maturation, thereby reducing mature let-7 production. These effects promote cancer cell proliferation, migration, invasion and tumor growth in DU145 and T47D cell lines (Figure 4D) (Dou et al., 2020).
PIWI interacting RNAs (piRNAs) and long non-coding RNAs (lncRNAs) are also influenced by SUMOylation. Several studies in Drosophila have demonstrated the role of SUMOylation in primary piRNA biogenesis (Muerdter et al., 2013) and piRNA-guided transcriptional silencing and heterochromatin formation (Mugat et al., 2020; Ninova et al., 2020; Andreev et al., 2022; ElMaghraby et al., 2022). Moreover in C. elegans, PIE-1 is itself modified by SUMO and acts as a SUMO E3 together with the conserved SUMO-E3 GEI-17/PAIS1 to promote piRNA-dependent silencing. PIE-1 SUMOylation promotes SUMOylation of HDA-1 and the assembly of a MEP-1/Mi-2/HDA-1 chromatin remodeling complex in the adult germline to promote fertility and embryonic development (Kim et al., 2021). However, there is relatively limited study of SUMOylation on the function of piRNAs in cancers.
A growing body of evidence has shown that many lncRNAs contribute to regulate the SUMOylation of some proteins, leading to cancer development and progression. LncRNA myosin heavy chain associated RNA transcript (MHRT) can regulate the SUMOylation levels of NAD-dependent protein deacetylase sirtuin-1 (SIRT1), peroxisome proliferator-activated receptor γ coactivator-1 α (PGC-1α)/peroxisome proliferator-activated receptor-α (PPARα), specificity protein 1 (SP1)/HDAC4 in cardiac hypertrophy (Liu et al., 2022). LncRNA ELNAT1 promotes lymphangiogenesis and lymph node (LN) metastasis in bladder cancer (BCa) cell lines by inducing Ubc9 overexpression to catalyze SUMOylation of hnRNPA1 (Chen et al., 2021). LncRNA small nucleolar RNA host gene 1 (SNHG1) enhanced SUMOylation of Bhlhe40 protein by facilitating the binding of SUMO E3 ligase PIAS3 to Bhlhe40, resulting in increased nuclear translocation of Bhlhe40 (Li S et al., 2022). LncRNA SDCBP2-AS1 binds to hnRNP K to repress its SUMOylation, which facilitates the ubiquitination of hnRNP K and β-catenin, thereby promoting the degradation of β-catenin and suppressing tumorigenesis and metastasis in gastric cancer (Han et al., 2022). Oncogene glucose transporter 1 (GLUT1) associated lncRNA (GAL) interacts with GLUT1 protein, which increases GLUT1 SUMOylation and inhibits the ubiquitin proteasome system on the GLUT1 protein, thus promoting colorectal cancer cell migration and invasion (Li B et al., 2022). LncRNA rhabdomyosarcoma 2 associated transcript (RMST) enhances FUS SUMOylation, which contributes to the interaction between FUS and hnRNP D to suppress glioblastoma cell mitophagy (Liu C. et al., 2020). LncRNA RP11-214F16.8 decreases the protein level of tumor suppressor NISCH by replacing SUMOylation with ubiquitination, which drives breast cancer (Lv and Zhang, 2022). LncRNA p53-stabilizing and activating RNA (PSTAR) can bind to hnRNP K and enhance its SUMOylation, thereby strengthening the interaction between hnRNP K and p53, ultimately resulting in the accumulation and transactivation of p53 to suppress hepatocellular carcinoma (Qin et al., 2020). Interestingly, c-Myc promotes p53 polyubiquitination and turnover by reducing p53 SUMOylation through c-Myc-inducible LncRNA inactivating P53 (MILIP), driving cancer pathogenesis (Feng et al., 2020). In addition, lncRNA TINCR can encode an evolutionary conserved ubiquitin-like protein (UBL) named pTINCR. pTINCR increases CDC42 SUMOylation and promotes its activation, leading to epithelial differentiation and tumor suppression, which acts as a tumor suppressor in epithelial cancers (Boix et al., 2022). These findings suggest lncRNA regulating SUMOylation can be as a new potential direction for the cancer treatment strategies.
In the past decade, SUMOylation research has found a large set of SUMO target proteins, which regulate the different stages of RNA metabolism. In this article, we summarize the regulatory roles of SUMOylation in RNA metabolism in cancer cells. A tight control of mRNA metabolism is crucial to allow cells in response to physiological stresses. SUMO proteome with key proteins/enzymes of mRNA transcription, splicing or nuclear complex is subject to spatio-temporal regulation and cellular stress remodeling to control mRNA metabolism. Since most of SUMOylation substrate proteins are localized in the nucleus, the effect of SUMOylation is mainly to inhibit the global transcription activity. When SUMOylation sites are located within the inhibitory or repressive domains, mutations of these sites can dramatically enhance the transcriptional activity.
MiRNA biogenesis is closely regulated by SUMOylation, thus affecting tumorigenesis and cancer progression. SUMOylation has no uniform effect on miRNA biogenesis process and miRNA-induced RNA silencing efficiency. Considering that the activity and abundance of core proteins and their partners in different stages of miRNA biogenesis are not consistent in different cancers, there is no comprehensive understanding and definite conclusion on how SUMO specifically affects miRNA metabolism in specific types of cancer. Moreover, a lot of lncRNAs mediate SUMOylation of target proteins, which plays complex and precise regulatory roles in cancer progression. However, the role of SUMOylation in the transcription, processing, stability and functions of lncRNAs is limited and unclear. Finally, understanding the impact of SUMOylation on RNA metabolism may provide new therapeutic strategies for cancer treatment.
YC collected literatures and wrote the manuscript. CH designed and prepared the figures. XZ and JY designed the paper, critically revised the manuscript and approved the manuscript before submission. All authors listed have made a substantial, direct contribution to the work and approved the submitted version.
This work was supported by China’s National Key R&D Programmes (NKP) (No. 2019YFE0110600) and grants from the National Natural Science Foundation of China (82230100, 32271310, 82273143).
We thank all members of our group for constructive feedback and discussions.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Advani, V. M., and Ivanov, P. (2019). Translational control under stress: Reshaping the translatome. Bioessays 41 (5), e1900009. doi:10.1002/bies.201900009
Alarcon, C. R., Lee, H., Goodarzi, H., Halberg, N., and Tavazoie, S. F. (2015). N6-methyladenosine marks primary microRNAs for processing. Nature 519 (7544), 482–485. doi:10.1038/nature14281
Alberti, S., and Hyman, A. A. (2021). Biomolecular condensates at the nexus of cellular stress, protein aggregation disease and ageing. Nat. Rev. Mol. Cell Biol. 22 (3), 196–213. doi:10.1038/s41580-020-00326-6
Anczukow, O., Akerman, M., Clery, A., Wu, J., Shen, C., Shirole, N. H., et al. (2015). SRSF1-Regulated alternative splicing in breast cancer. Mol. Cell 60 (1), 105–117. doi:10.1016/j.molcel.2015.09.005
Anczukow, O., Rosenberg, A. Z., Akerman, M., Das, S., Zhan, L., Karni, R., et al. (2012). The splicing factor SRSF1 regulates apoptosis and proliferation to promote mammary epithelial cell transformation. Nat. Struct. Mol. Biol. 19 (2), 220–228. doi:10.1038/nsmb.2207
Andreev, V. I., Yu, C., Wang, J., Schnabl, J., Tirian, L., Gehre, M., et al. (2022). Panoramix SUMOylation on chromatin connects the piRNA pathway to the cellular heterochromatin machinery. Nat. Struct. Mol. Biol. 29 (2), 130–142. doi:10.1038/s41594-022-00721-x
Astrom, A., D'Amore, E. S., Sainati, L., Panarello, C., Morerio, C., Mark, J., et al. (2000). Evidence of involvement of the PLAG1 gene in lipoblastomas. Int. J. Oncol. 16 (6), 1107–1110. doi:10.3892/ijo.16.6.1107
Baker, A. R., and Slack, F. J. (2022). ADAR1 and its implications in cancer development and treatment. Trends Genet. 38 (8), 821–830. doi:10.1016/j.tig.2022.03.013
Baluapuri, A., Wolf, E., and Eilers, M. (2020). Target gene-independent functions of MYC oncoproteins. Nat. Rev. Mol. Cell Biol. 21 (5), 255–267. doi:10.1038/s41580-020-0215-2
Bawa-Khalfe, T., Cheng, J., Wang, Z., and Yeh, E. T. (2007). Induction of the SUMO-specific protease 1 transcription by the androgen receptor in prostate cancer cells. J. Biol. Chem. 282 (52), 37341–37349. doi:10.1074/jbc.M706978200
Becker, J., Barysch, S. V., Karaca, S., Dittner, C., Hsiao, H. H., Berriel Diaz, M., et al. (2013). Detecting endogenous SUMO targets in mammalian cells and tissues. Nat. Struct. Mol. Biol. 20 (4), 525–531. doi:10.1038/nsmb.2526
Bentley, D. L. (2014). Coupling mRNA processing with transcription in time and space. Nat. Rev. Genet. 15 (3), 163–175. doi:10.1038/nrg3662
Bernard, S., and Eilers, M. (2006). Control of cell proliferation and growth by Myc proteins. Results Probl. Cell Differ. 42, 329–342. doi:10.1007/400_004
Berta, M. A., Mazure, N., Hattab, M., Pouyssegur, J., and Brahimi-Horn, M. C. (2007). SUMOylation of hypoxia-inducible factor-1alpha reduces its transcriptional activity. Biochem. Biophys. Res. Commun. 360 (3), 646–652. doi:10.1016/j.bbrc.2007.06.103
Biamonti, G., and Vourc'h, C. (2010). Nuclear stress bodies. Cold Spring Harb. Perspect. Biol. 2 (6), a000695. doi:10.1101/cshperspect.a000695
Bies, J., Markus, J., and Wolff, L. (2002). Covalent attachment of the SUMO-1 protein to the negative regulatory domain of the c-Myb transcription factor modifies its stability and transactivation capacity. J. Biol. Chem. 277 (11), 8999–9009. doi:10.1074/jbc.M110453200
Blomster, H. A., Hietakangas, V., Wu, J., Kouvonen, P., Hautaniemi, S., and Sistonen, L. (2009). Novel proteomics strategy brings insight into the prevalence of SUMO-2 target sites. Mol. Cell Proteomics 8 (6), 1382–1390. doi:10.1074/mcp.M800551-MCP200
Boix, O., Martinez, M., Vidal, S., Gimenez-Alejandre, M., Palenzuela, L., Lorenzo-Sanz, L., et al. (2022). pTINCR microprotein promotes epithelial differentiation and suppresses tumor growth through CDC42 SUMOylation and activation. Nat. Commun. 13 (1), 6840. doi:10.1038/s41467-022-34529-6
Brandl, A., Wagner, T., Uhlig, K. M., Knauer, S. K., Stauber, R. H., Melchior, F., et al. (2012). Dynamically regulated sumoylation of HDAC2 controls p53 deacetylation and restricts apoptosis following genotoxic stress. J. Mol. Cell Biol. 4 (5), 284–293. doi:10.1093/jmcb/mjs013
Bretones, G., Delgado, M. D., and Leon, J. (2015). Myc and cell cycle control. Biochim. Biophys. Acta 1849 (5), 506–516. doi:10.1016/j.bbagrm.2014.03.013
Chanarat, S., and Mishra, S. K. (2018). Emerging roles of ubiquitin-like proteins in pre-mRNA splicing. Trends Biochem. Sci. 43 (11), 896–907. doi:10.1016/j.tibs.2018.09.001
Chang, C. C., Lin, D. Y., Fang, H. I., Chen, R. H., and Shih, H. M. (2005). Daxx mediates the small ubiquitin-like modifier-dependent transcriptional repression of Smad4. J. Biol. Chem. 280 (11), 10164–10173. doi:10.1074/jbc.M409161200
Chang, H. M., and Yeh, E. T. H. (2020). Sumo: From bench to bedside. Physiol. Rev. 100 (4), 1599–1619. doi:10.1152/physrev.00025.2019
Chauhan, K. M., Chen, Y., Chen, Y., Liu, A. T., Sun, X. X., and Dai, M. S. (2021). The SUMO-specific protease SENP1 deSUMOylates p53 and regulates its activity. J. Cell Biochem. 122 (2), 189–197. doi:10.1002/jcb.29838
Chen, C., Zheng, H., Luo, Y., Kong, Y., An, M., Li, Y., et al. (2021). SUMOylation promotes extracellular vesicle-mediated transmission of lncRNA ELNAT1 and lymph node metastasis in bladder cancer. J. Clin. Invest. 131 (8), e146431. doi:10.1172/JCI146431
Chen, C., Zhu, C., Huang, J., Zhao, X., Deng, R., Zhang, H., et al. (2015). SUMOylation of TARBP2 regulates miRNA/siRNA efficiency. Nat. Commun. 6, 8899. doi:10.1038/ncomms9899
Chen, W. T., Alpert, A., Leiter, C., Gong, F., Jackson, S. P., and Miller, K. M. (2013). Systematic identification of functional residues in mammalian histone H2AX. Mol. Cell Biol. 33 (1), 111–126. doi:10.1128/MCB.01024-12
Cheng, J., Wang, D., Wang, Z., and Yeh, E. T. (2004). SENP1 enhances androgen receptor-dependent transcription through desumoylation of histone deacetylase 1. Mol. Cell Biol. 24 (13), 6021–6028. doi:10.1128/MCB.24.13.6021-6028.2004
Chun, T. H., Itoh, H., Subramanian, L., Iniguez-Lluhi, J. A., and Nakao, K. (2003). Modification of GATA-2 transcriptional activity in endothelial cells by the SUMO E3 ligase PIASy. Circ. Res. 92 (11), 1201–1208. doi:10.1161/01.RES.0000076893.70898.36
Chymkowitch, P., Nguea, P. A., and Enserink, J. M. (2015). SUMO-Regulated transcription: Challenging the dogma. Bioessays 37 (10), 1095–1105. doi:10.1002/bies.201500065
Citro, S., Jaffray, E., Hay, R. T., Seiser, C., and Chiocca, S. (2013). A role for paralog-specific sumoylation in histone deacetylase 1 stability. J. Mol. Cell Biol. 5 (6), 416–427. doi:10.1093/jmcb/mjt032
Colgan, D. F., and Manley, J. L. (1997). Mechanism and regulation of mRNA polyadenylation. Genes Dev. 11 (21), 2755–2766. doi:10.1101/gad.11.21.2755
Cong, L., Pakala, S. B., Ohshiro, K., Li, D. Q., and Kumar, R. (2011). SUMOylation and SUMO-interacting motif (SIM) of metastasis tumor antigen 1 (MTA1) synergistically regulate its transcriptional repressor function. J. Biol. Chem. 286 (51), 43793–43808. doi:10.1074/jbc.M111.267237
Corden, J. L., and Patturajan, M. (1997). A CTD function linking transcription to splicing. Trends Biochem. Sci. 22 (11), 413–416. doi:10.1016/s0968-0004(97)01125-0
Daniel, A. R., Faivre, E. J., and Lange, C. A. (2007). Phosphorylation-dependent antagonism of sumoylation derepresses progesterone receptor action in breast cancer cells. Mol. Endocrinol. 21 (12), 2890–2906. doi:10.1210/me.2007-0248
David, G., Neptune, M. A., and DePinho, R. A. (2002). SUMO-1 modification of histone deacetylase 1 (HDAC1) modulates its biological activities. J. Biol. Chem. 277 (26), 23658–23663. doi:10.1074/jbc.M203690200
de Miguel, F. J., Sharma, R. D., Pajares, M. J., Montuenga, L. M., Rubio, A., and Pio, R. (2014). Identification of alternative splicing events regulated by the oncogenic factor SRSF1 in lung cancer. Cancer Res. 74 (4), 1105–1115. doi:10.1158/0008-5472.CAN-13-1481
Denison, C., Rudner, A. D., Gerber, S. A., Bakalarski, C. E., Moazed, D., and Gygi, S. P. (2005). A proteomic strategy for gaining insights into protein sumoylation in yeast. Mol. Cell Proteomics 4 (3), 246–254. doi:10.1074/mcp.M400154-MCP200
Desterro, J. M., Keegan, L. P., Jaffray, E., Hay, R. T., O'Connell, M. A., and Carmo-Fonseca, M. (2005). SUMO-1 modification alters ADAR1 editing activity. Mol. Biol. Cell 16 (11), 5115–5126. doi:10.1091/mbc.e05-06-0536
Desterro, J. M. P., Rodriguez, M. S., and Hay, R. T. (1998). SUMO-1 modification of IkappaBalpha inhibits NF-kappaB activation. Mol. Cell 2 (2), 233–239. doi:10.1016/s1097-2765(00)80133-1
Desterro, J. M., Rodriguez, M. S., Kemp, G. D., and Hay, R. T. (1999). Identification of the enzyme required for activation of the small ubiquitin-like protein SUMO-1. J. Biol. Chem. 274 (15), 10618–10624. doi:10.1074/jbc.274.15.10618
Dhall, A., Wei, S., Fierz, B., Woodcock, C. L., Lee, T. H., and Chatterjee, C. (2014). Sumoylated human histone H4 prevents chromatin compaction by inhibiting long-range internucleosomal interactions. J. Biol. Chem. 289 (49), 33827–33837. doi:10.1074/jbc.M114.591644
Dhall, A., Weller, C. E., Chu, A., Shelton, P. M. M., and Chatterjee, C. (2017). Chemically sumoylated histone H4 stimulates intranucleosomal demethylation by the LSD1-CoREST complex. ACS Chem. Biol. 12 (9), 2275–2280. doi:10.1021/acschembio.7b00716
Dhanasekaran, R., Deutzmann, A., Mahauad-Fernandez, W. D., Hansen, A. S., Gouw, A. M., and Felsher, D. W. (2022). The MYC oncogene - the grand orchestrator of cancer growth and immune evasion. Nat. Rev. Clin. Oncol. 19 (1), 23–36. doi:10.1038/s41571-021-00549-2
Dou, J., Zhang, H., Chen, R., Shu, Z., Yuan, H., Zhao, X., et al. (2020). SUMOylation modulates the LIN28A-let-7 signaling pathway in response to cellular stresses in cancer cells. Mol. Oncol. 14 (9), 2288–2312. doi:10.1002/1878-0261.12694
Du, H., Zhao, Y., He, J., Zhang, Y., Xi, H., Liu, M., et al. (2016). YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat. Commun. 7, 12626. doi:10.1038/ncomms12626
Du, L., Fakih, M. G., Rosen, S. T., and Chen, Y. (2020). SUMOylation of E2F1 regulates expression of EZH2. Cancer Res. 80 (19), 4212–4223. doi:10.1158/0008-5472.CAN-20-1259
Du, Y., Hou, G., Zhang, H., Dou, J., He, J., Guo, Y., et al. (2018). SUMOylation of the m6A-RNA methyltransferase METTL3 modulates its function. Nucleic Acids Res. 46 (10), 5195–5208. doi:10.1093/nar/gky156
ElMaghraby, M. F., Tirian, L., Senti, K. A., Meixner, K., and Brennecke, J. (2022). A genetic toolkit for studying transposon control in the Drosophila melanogaster ovary. Genetics 220 (1), iyab179. doi:10.1093/genetics/iyab179
Eloranta, J. J., and Hurst, H. C. (2002). Transcription factor AP-2 interacts with the SUMO-conjugating enzyme UBC9 and is sumolated in vivo. J. Biol. Chem. 277 (34), 30798–30804. doi:10.1074/jbc.M202780200
Feng, Y. C., Liu, X. Y., Teng, L., Ji, Q., Wu, Y., Li, J. M., et al. (2020). c-Myc inactivation of p53 through the pan-cancer lncRNA MILIP drives cancer pathogenesis. Nat. Commun. 11 (1), 4980. doi:10.1038/s41467-020-18735-8
Galganski, L., Urbanek, M. O., and Krzyzosiak, W. J. (2017). Nuclear speckles: Molecular organization, biological function and role in disease. Nucleic Acids Res. 45 (18), 10350–10368. doi:10.1093/nar/gkx759
Gautrey, H. L., and Tyson-Capper, A. J. (2012). Regulation of Mcl-1 by SRSF1 and SRSF5 in cancer cells. PLoS One 7 (12), e51497. doi:10.1371/journal.pone.0051497
Ghigna, C., Giordano, S., Shen, H., Benvenuto, F., Castiglioni, F., Comoglio, P. M., et al. (2005). Cell motility is controlled by SF2/ASF through alternative splicing of the Ron protooncogene. Mol. Cell 20 (6), 881–890. doi:10.1016/j.molcel.2005.10.026
Gill, G. (2004). SUMO and ubiquitin in the nucleus: Different functions, similar mechanisms? Genes Dev. 18 (17), 2046–2059. doi:10.1101/gad.1214604
Girdwood, D., Bumpass, D., Vaughan, O. A., Thain, A., Anderson, L. A., Snowden, A. W., et al. (2003). p300 transcriptional repression is mediated by SUMO modification. Mol. Cell 11 (4), 1043–1054. doi:10.1016/s1097-2765(03)00141-2
Gocke, C. B., Yu, H., and Kang, J. (2005). Systematic identification and analysis of mammalian small ubiquitin-like modifier substrates. J. Biol. Chem. 280 (6), 5004–5012. doi:10.1074/jbc.M411718200
Golebiowski, F., Matic, I., Tatham, M. H., Cole, C., Yin, Y., Nakamura, A., et al. (2009). System-wide changes to SUMO modifications in response to heat shock. Sci. Signal 2 (72), ra24. doi:10.1126/scisignal.2000282
Gong, L., Ji, W. K., Hu, X. H., Hu, W. F., Tang, X. C., Huang, Z. X., et al. (2014). Sumoylation differentially regulates Sp1 to control cell differentiation. Proc. Natl. Acad. Sci. U. S. A. 111 (15), 5574–5579. doi:10.1073/pnas.1315034111
Goodson, M. L., Hong, Y., Rogers, R., Matunis, M. J., Park-Sarge, O. K., and Sarge, K. D. (2001). Sumo-1 modification regulates the DNA binding activity of heat shock transcription factor 2, a promyelocytic leukemia nuclear body associated transcription factor. J. Biol. Chem. 276 (21), 18513–18518. doi:10.1074/jbc.M008066200
Gostissa, M., Hengstermann, A., Fogal, V., Sandy, P., Schwarz, S. E., Scheffner, M., et al. (1999). Activation of p53 by conjugation to the ubiquitin-like protein SUMO-1. EMBO J. 18 (22), 6462–6471. doi:10.1093/emboj/18.22.6462
Grinstein, E., Du, Y., Santourlidis, S., Christ, J., Uhrberg, M., and Wernet, P. (2007). Nucleolin regulates gene expression in CD34-positive hematopoietic cells. J. Biol. Chem. 282 (17), 12439–12449. doi:10.1074/jbc.M608068200
Guo, D., Han, J., Adam, B. L., Colburn, N. H., Wang, M. H., Dong, Z., et al. (2005). Proteomic analysis of SUMO4 substrates in HEK293 cells under serum starvation-induced stress. Biochem. Biophys. Res. Commun. 337 (4), 1308–1318. doi:10.1016/j.bbrc.2005.09.191
Guo, D., Li, M., Zhang, Y., Yang, P., Eckenrode, S., Hopkins, D., et al. (2004). A functional variant of SUMO4, a new I kappa B alpha modifier, is associated with type 1 diabetes. Nat. Genet. 36 (8), 837–841. doi:10.1038/ng1391
Ha, M., and Kim, V. N. (2014). Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 15 (8), 509–524. doi:10.1038/nrm3838
Hammerich-Hille, S., Kaipparettu, B. A., Tsimelzon, A., Creighton, C. J., Jiang, S., Polo, J. M., et al. (2010). SAFB1 mediates repression of immune regulators and apoptotic genes in breast cancer cells. J. Biol. Chem. 285 (6), 3608–3616. doi:10.1074/jbc.M109.066431
Han, J., Nie, M., Chen, C., Cheng, X., Guo, T., Huangfu, L., et al. (2022). SDCBP-AS1 destabilizes beta-catenin by regulating ubiquitination and SUMOylation of hnRNP K to suppress gastric tumorigenicity and metastasis. Cancer Commun. (Lond) 42 (11), 1141–1161. doi:10.1002/cac2.12367
Han, Z. J., Feng, Y. H., Gu, B. H., Li, Y. M., and Chen, H. (2018). The post-translational modification, SUMOylation, and cancer (Review). Int. J. Oncol. 52 (4), 1081–1094. doi:10.3892/ijo.2018.4280
Hay, R. T. (2007). SUMO-Specific proteases: A twist in the tail. Trends Cell Biol. 17 (8), 370–376. doi:10.1016/j.tcb.2007.08.002
Hendriks, I. A., and Vertegaal, A. C. (2016). A comprehensive compilation of SUMO proteomics. Nat. Rev. Mol. Cell Biol. 17 (9), 581–595. doi:10.1038/nrm.2016.81
Hickey, C. M., Wilson, N. R., and Hochstrasser, M. (2012). Function and regulation of SUMO proteases. Nat. Rev. Mol. Cell Biol. 13 (12), 755–766. doi:10.1038/nrm3478
Hirose, T., Ninomiya, K., Nakagawa, S., and Yamazaki, T. (2022). A guide to membraneless organelles and their various roles in gene regulation. Nat. Rev. Mol. Cell Biol. 23, s41580. doi:10.1038/s41580-022-00558-8
Hofweber, M., and Dormann, D. (2019). Friend or foe-Post-translational modifications as regulators of phase separation and RNP granule dynamics. J. Biol. Chem. 294 (18), 7137–7150. doi:10.1074/jbc.TM118.001189
Hogg, S. J., Motorna, O., Cluse, L. A., Johanson, T. M., Coughlan, H. D., Raviram, R., et al. (2021). Targeting histone acetylation dynamics and oncogenic transcription by catalytic P300/CBP inhibition. Mol. Cell 81 (10), 2183–2200.e13. doi:10.1016/j.molcel.2021.04.015
Hong, Y., Rogers, R., Matunis, M. J., Mayhew, C. N., Goodson, M. L., Park-Sarge, O. K., et al. (2001). Regulation of heat shock transcription factor 1 by stress-induced SUMO-1 modification. J. Biol. Chem. 276 (43), 40263–40267. doi:10.1074/jbc.M104714200
Hou, G., Zhao, X., Li, L., Yang, Q., Liu, X., Huang, C., et al. (2021). SUMOylation of YTHDF2 promotes mRNA degradation and cancer progression by increasing its binding affinity with m6A-modified mRNAs. Nucleic Acids Res. 49 (5), 2859–2877. doi:10.1093/nar/gkab065
Hu, Y., Chen, C., Tong, X., Chen, S., Hu, X., Pan, B., et al. (2021). NSUN2 modified by SUMO-2/3 promotes gastric cancer progression and regulates mRNA m5C methylation. Cell Death Dis. 12 (9), 842. doi:10.1038/s41419-021-04127-3
Huang, Y., Su, R., Sheng, Y., Dong, L., Dong, Z., Xu, H., et al. (2019). Small-molecule targeting of oncogenic FTO demethylase in acute myeloid leukemia. Cancer Cell 35 (4), 677–691.e10. doi:10.1016/j.ccell.2019.03.006
Ihara, M., Stein, P., and Schultz, R. M. (2008). UBE2I (UBC9), a SUMO-conjugating enzyme, localizes to nuclear speckles and stimulates transcription in mouse oocytes. Biol. Reprod. 79 (5), 906–913. doi:10.1095/biolreprod.108.070474
Imoto, S., Ohbayashi, N., Ikeda, O., Kamitani, S., Muromoto, R., Sekine, Y., et al. (2008). Sumoylation of Smad3 stimulates its nuclear export during PIASy-mediated suppression of TGF-beta signaling. Biochem. Biophys. Res. Commun. 370 (2), 359–365. doi:10.1016/j.bbrc.2008.03.116
Ivanov, P., Kedersha, N., and Anderson, P. (2019). Stress granules and processing bodies in translational control. Cold Spring Harb. Perspect. Biol. 11 (5), a032813. doi:10.1101/cshperspect.a032813
Iwakawa, H. O., and Tomari, Y. (2022). Life of RISC: Formation, action, and degradation of RNA-induced silencing complex. Mol. Cell 82 (1), 30–43. doi:10.1016/j.molcel.2021.11.026
Jacobs, A. M., Nicol, S. M., Hislop, R. G., Jaffray, E. G., Hay, R. T., and Fuller-Pace, F. V. (2007). SUMO modification of the DEAD box protein p68 modulates its transcriptional activity and promotes its interaction with HDAC1. Oncogene 26 (40), 5866–5876. doi:10.1038/sj.onc.1210387
Jang, M.-S., Ryu, S.-W., and Kim, E. (2002). Modification of Daxx by small ubiquitin-related modifier-1. Biochem. Biophysical Res. Commun. 295 (2), 495–500. doi:10.1016/s0006-291x(02)00699-x
Karni, R., de Stanchina, E., Lowe, S. W., Sinha, R., Mu, D., and Krainer, A. R. (2007). The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nat. Struct. Mol. Biol. 14 (3), 185–193. doi:10.1038/nsmb1209
Keiten-Schmitz, J., Wagner, K., Piller, T., Kaulich, M., Alberti, S., and Muller, S. (2020). The nuclear SUMO-targeted ubiquitin quality control network regulates the dynamics of cytoplasmic stress granules. Mol. Cell 79 (1), 54–67.e7. doi:10.1016/j.molcel.2020.05.017
Kim, H., Ding, Y. H., Lu, S., Zuo, M. Q., Tan, W., Conte, D., et al. (2021). PIE-1 SUMOylation promotes germline fates and piRNA-dependent silencing in C. elegans. Elife 10, e63300. doi:10.7554/eLife.63300
Kim, J., Cantwell, C. A., Johnson, P. F., Pfarr, C. M., and Williams, S. C. (2002). Transcriptional activity of CCAAT/enhancer-binding proteins is controlled by a conserved inhibitory domain that is a target for sumoylation. J. Biol. Chem. 277 (41), 38037–38044. doi:10.1074/jbc.M207235200
Kirsh, O., Seeler, J. S., Pichler, A., Gast, A., Muller, S., Miska, E., et al. (2002). The SUMO E3 ligase RanBP2 promotes modification of the HDAC4 deacetylase. EMBO J. 21 (11), 2682–2691. doi:10.1093/emboj/21.11.2682
Krol, J., Loedige, I., and Filipowicz, W. (2010). The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 11 (9), 597–610. doi:10.1038/nrg2843
Kuo, H.-Y., Chang, C.-C., Jeng, J.-C., Hu, H.-M., Lin, D.-Y., Maul, G. G., et al. (2005). SUMO modification negatively modulates the transcriptional activity of CREB-binding protein via the recruitment of Daxx. Proc. Natl. Acad. Sci. 102 (47), 16973–16978. doi:10.1073/pnas.0504460102
Lallemand-Breitenbach, V., and de The, H. (2018). PML nuclear bodies: From architecture to function. Curr. Opin. Cell Biol. 52, 154–161. doi:10.1016/j.ceb.2018.03.011
Lee, P. S., Chang, C., Liu, D., and Derynck, R. (2003). Sumoylation of Smad4, the common Smad mediator of transforming growth factor-beta family signaling. J. Biol. Chem. 278 (30), 27853–27863. doi:10.1074/jbc.M301755200
Li, B., Kang, H., Xiao, Y., Du, Y., Xiao, Y., Song, G., et al. (2022). LncRNA GAL promotes colorectal cancer liver metastasis through stabilizing GLUT1. Oncogene 41 (13), 1882–1894. doi:10.1038/s41388-022-02230-z
Li, J., Xie, H., Ying, Y., Chen, H., Yan, H., He, L., et al. (2020). YTHDF2 mediates the mRNA degradation of the tumor suppressors to induce AKT phosphorylation in N6-methyladenosine-dependent way in prostate cancer. Mol. Cancer 19 (1), 152. doi:10.1186/s12943-020-01267-6
Li, N., Zhang, S., Xiong, F., Eizirik, D. L., and Wang, C. Y. (2020). SUMOylation, a multifaceted regulatory mechanism in the pancreatic beta cells. Semin. Cell Dev. Biol. 103, 51–58. doi:10.1016/j.semcdb.2020.03.008
Li, S., Ni, Y., Li, C., Xiang, Q., Zhao, Y., Xu, H., et al. (2022). Long noncoding RNA SNHG1 alleviates high glucose-induced vascular smooth muscle cells calcification/senescence by post-transcriptionally regulating Bhlhe40 and autophagy via Atg10. J. Physiol. Biochem. 79, 83–105. doi:10.1007/s13105-022-00924-2
Li, T., Evdokimov, E., Shen, R. F., Chao, C. C., Tekle, E., Wang, T., et al. (2004). Sumoylation of heterogeneous nuclear ribonucleoproteins, zinc finger proteins, and nuclear pore complex proteins: A proteomic analysis. Proc. Natl. Acad. Sci. U. S. A. 101 (23), 8551–8556. doi:10.1073/pnas.0402889101
Li, Y., and Seto, E. (2016). HDACs and HDAC inhibitors in cancer development and therapy. Cold Spring Harb. Perspect. Med. 6 (10), a026831. doi:10.1101/cshperspect.a026831
Liang, Y. C., Lee, C. C., Yao, Y. L., Lai, C. C., Schmitz, M. L., and Yang, W. M. (2016). SUMO5, a novel poly-SUMO isoform, regulates PML nuclear bodies. Sci. Rep. 6, 26509. doi:10.1038/srep26509
Lin, D. Y., Fang, H. I., Ma, A. H., Huang, Y. S., Pu, Y. S., Jenster, G., et al. (2004). Negative modulation of androgen receptor transcriptional activity by Daxx. Mol. Cell Biol. 24 (24), 10529–10541. doi:10.1128/MCB.24.24.10529-10541.2004
Lin, D. Y., Huang, Y. S., Jeng, J. C., Kuo, H. Y., Chang, C. C., Chao, T. T., et al. (2006). Role of SUMO-interacting motif in Daxx SUMO modification, subnuclear localization, and repression of sumoylated transcription factors. Mol. Cell 24 (3), 341–354. doi:10.1016/j.molcel.2006.10.019
Lin, X., Liang, M., Liang, Y. Y., Brunicardi, F. C., Melchior, F., and Feng, X. H. (2003a). Activation of transforming growth factor-beta signaling by SUMO-1 modification of tumor suppressor Smad4/DPC4. J. Biol. Chem. 278 (21), 18714–18719. doi:10.1074/jbc.M302243200
Lin, X., Sun, B., Liang, M., Liang, Y.-Y., Gast, A., Hildebrand, J., et al. (2003b). Opposed regulation of corepressor CtBP by SUMOylation and PDZ binding. Mol. Cell 11 (5), 1389–1396. doi:10.1016/s1097-2765(03)00175-8
Liu, C., Peng, Z., Li, P., Fu, H., Feng, J., Zhang, Y., et al. (2020). lncRNA RMST suppressed GBM cell mitophagy through enhancing FUS SUMOylation. Mol. Ther. Nucleic Acids 19, 1198–1208. doi:10.1016/j.omtn.2020.01.008
Liu, H. W., Banerjee, T., Guan, X., Freitas, M. A., and Parvin, J. D. (2015). The chromatin scaffold protein SAFB1 localizes SUMO-1 to the promoters of ribosomal protein genes to facilitate transcription initiation and splicing. Nucleic Acids Res. 43 (7), 3605–3613. doi:10.1093/nar/gkv246
Liu, H. W., Zhang, J., Heine, G. F., Arora, M., Gulcin Ozer, H., Onti-Srinivasan, R., et al. (2012). Chromatin modification by SUMO-1 stimulates the promoters of translation machinery genes. Nucleic Acids Res. 40 (20), 10172–10186. doi:10.1093/nar/gks819
Liu, K., Wang, X., Li, D., Xu, D., Li, D., Lv, Z., et al. (2020). Ginkgolic acid, a SUMO-1 inhibitor, inhibits the progression of oral squamous cell carcinoma by alleviating SUMOylation of SMAD4. Mol. Ther. Oncolytics 16, 86–99. doi:10.1016/j.omto.2019.12.005
Liu, M. Y., Yue, L. J., Luo, Y. C., Lu, J., Wu, G. D., Sheng, S. Q., et al. (2022). SUMOylation of SIRT1 activating PGC-1α/PPARα pathway mediates the protective effect of LncRNA-MHRT in cardiac hypertrophy. Eur. J. Pharmacol. 930, 175155. doi:10.1016/j.ejphar.2022.175155
Long, J., Wang, G., He, D., and Liu, F. (2004). Repression of Smad4 transcriptional activity by SUMO modification. Biochem. J. 379, 23–29. doi:10.1042/BJ20031867
Lourenco, C., Resetca, D., Redel, C., Lin, P., MacDonald, A. S., Ciaccio, R., et al. (2021). MYC protein interactors in gene transcription and cancer. Nat. Rev. Cancer 21 (9), 579–591. doi:10.1038/s41568-021-00367-9
Lu, J., Getz, G., Miska, E. A., Alvarez-Saavedra, E., Lamb, J., Peck, D., et al. (2005). MicroRNA expression profiles classify human cancers. Nature 435 (7043), 834–838. doi:10.1038/nature03702
Lv, X., and Zhang, Q. (2022). LncRNA RP11-214F16.8 drives breast cancer tumorigenesis via a post-translational repression on NISCH expression. Cell Signal 92, 110271. doi:10.1016/j.cellsig.2022.110271
Mahmud, I., and Liao, D. (2019). DAXX in cancer: Phenomena, processes, mechanisms and regulation. Nucleic Acids Res. 47 (15), 7734–7752. doi:10.1093/nar/gkz634
Manza, L. L., Codreanu, S. G., Stamer, S. L., Smith, D. L., Wells, K. S., Roberts, R. L., et al. (2004). Global shifts in protein sumoylation in response to electrophile and oxidative stress. Chem. Res. Toxicol. 17 (12), 1706–1715. doi:10.1021/tx049767l
Marmor-Kollet, H., Siany, A., Kedersha, N., Knafo, N., Rivkin, N., Danino, Y. M., et al. (2020). Spatiotemporal proteomic analysis of stress granule disassembly using APEX reveals regulation by SUMOylation and links to ALS pathogenesis. Mol. Cell 80 (5), 876–891.e6. doi:10.1016/j.molcel.2020.10.032
Martin, S., Wilkinson, K. A., Nishimune, A., and Henley, J. M. (2007). Emerging extranuclear roles of protein SUMOylation in neuronal function and dysfunction. Nat. Rev. Neurosci. 8 (12), 948–959. doi:10.1038/nrn2276
Matafora, V., D'Amato, A., Mori, S., Blasi, F., and Bachi, A. (2009). Proteomics analysis of nucleolar SUMO-1 target proteins upon proteasome inhibition. Mol. Cell Proteomics 8 (10), 2243–2255. doi:10.1074/mcp.M900079-MCP200
Matic, I., Schimmel, J., Hendriks, I. A., van Santen, M. A., van de Rijke, F., van Dam, H., et al. (2010). Site-specific identification of SUMO-2 targets in cells reveals an inverted SUMOylation motif and a hydrophobic cluster SUMOylation motif. Mol. Cell 39 (4), 641–652. doi:10.1016/j.molcel.2010.07.026
Matsuzaki, K., Minami, T., Tojo, M., Honda, Y., Uchimura, Y., Saitoh, H., et al. (2003). Serum response factor is modulated by the SUMO-1 conjugation system. Biochem. Biophysical Res. Commun. 306 (1), 32–38. doi:10.1016/s0006-291x(03)00910-0
Meyer, K. D., Saletore, Y., Zumbo, P., Elemento, O., Mason, C. E., and Jaffrey, S. R. (2012). Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons. Cell 149 (7), 1635–1646. doi:10.1016/j.cell.2012.05.003
Mugat, B., Nicot, S., Varela-Chavez, C., Jourdan, C., Sato, K., Basyuk, E., et al. (2020). The Mi-2 nucleosome remodeler and the Rpd3 histone deacetylase are involved in piRNA-guided heterochromatin formation. Nat. Commun. 11 (1), 2818. doi:10.1038/s41467-020-16635-5
Nakagawa, K., and Yokosawa, H. (2002). PIAS3 induces SUMO-1 modification and transcriptional repression of IRF-1. FEBS Lett. 530 (1-3), 204–208. doi:10.1016/s0014-5793(02)03486-5
Navascues, J., Bengoechea, R., Tapia, O., Casafont, I., Berciano, M. T., and Lafarga, M. (2008). SUMO-1 transiently localizes to Cajal bodies in mammalian neurons. J. Struct. Biol. 163 (2), 137–146. doi:10.1016/j.jsb.2008.04.013
Nayler, O., Stratling, W., Bourquin, J. P., Stagljar, I., Lindemann, L., Jasper, H., et al. (1998). SAF-B protein couples transcription and pre-mRNA splicing to SAR/MAR elements. Nucleic Acids Res. 26 (15), 3542–3549. doi:10.1093/nar/26.15.3542
Ninova, M., Chen, Y. A., Godneeva, B., Rogers, A. K., Luo, Y., Fejes Toth, K., et al. (2020). Su(var)2-10 and the SUMO pathway link piRNA-guided target recognition to chromatin silencing. Mol. Cell 77 (3), 556–570.e6. doi:10.1016/j.molcel.2019.11.012
Nizami, Z., Deryusheva, S., and Gall, J. G. (2010). The Cajal body and histone locus body. Cold Spring Harb. Perspect. Biol. 2 (7), a000653. doi:10.1101/cshperspect.a000653
Okamoto, M., Hirata, S., Sato, S., Koga, S., Fujii, M., Qi, G., et al. (2012). Frequent increased gene copy number and high protein expression of tRNA (cytosine-5-)-methyltransferase (NSUN2) in human cancers. DNA Cell Biol. 31 (5), 660–671. doi:10.1089/dna.2011.1446
Olsson, M., and Zhivotovsky, B. (2011). Caspases and cancer. Cell Death Differ. 18 (9), 1441–1449. doi:10.1038/cdd.2011.30
Owerbach, D., McKay, E. M., Yeh, E. T., Gabbay, K. H., and Bohren, K. M. (2005). A proline-90 residue unique to SUMO-4 prevents maturation and sumoylation. Biochem. Biophys. Res. Commun. 337 (2), 517–520. doi:10.1016/j.bbrc.2005.09.090
Pan, W. W., Zhou, J. J., Liu, X. M., Xu, Y., Guo, L. J., Yu, C., et al. (2013). Death domain-associated protein DAXX promotes ovarian cancer development and chemoresistance. J. Biol. Chem. 288 (19), 13620–13630. doi:10.1074/jbc.M112.446369
Papamichail, M., Trangas, T., Courtis, N., Ioannides, C., Cosmidou, H., Pangalis, G. A., et al. (1983). Poly(A)-polymerase levels in leukemia. Haematol. Blood Transfus. 28, 28–29. doi:10.1007/978-3-642-68761-7_5
Park, S., Stanfield, R. L., Martinez-Yamout, M. A., Dyson, H. J., Wilson, I. A., and Wright, P. E. (2017). Role of the CBP catalytic core in intramolecular SUMOylation and control of histone H3 acetylation. Proc. Natl. Acad. Sci. U. S. A. 114 (27), E5335–E5342. doi:10.1073/pnas.1703105114
Pelisch, F., Gerez, J., Druker, J., Schor, I. E., Munoz, M. J., Risso, G., et al. (2010). The serine/arginine-rich protein SF2/ASF regulates protein sumoylation. Proc. Natl. Acad. Sci. U. S. A. 107 (37), 16119–16124. doi:10.1073/pnas.1004653107
Picard, N., Caron, V., Bilodeau, S., Sanchez, M., Mascle, X., Aubry, M., et al. (2012). Identification of estrogen receptor β as a SUMO-1 target reveals a novel phosphorylated sumoylation motif and regulation by glycogen synthase kinase 3β. Mol. Cell Biol. 32 (14), 2709–2721. doi:10.1128/MCB.06624-11
Poukka, H., Karvonen, U., Janne, O. A., and Palvimo, J. J. (2000). Covalent modification of the androgen receptor by small ubiquitin-like modifier 1 (SUMO-1). Proc. Natl. Acad. Sci. U. S. A. 97 (26), 14145–14150. doi:10.1073/pnas.97.26.14145
Pozzi, B., Bragado, L., Will, C. L., Mammi, P., Risso, G., Urlaub, H., et al. (2017). SUMO conjugation to spliceosomal proteins is required for efficient pre-mRNA splicing. Nucleic Acids Res. 45 (11), 6729–6745. doi:10.1093/nar/gkx213
Proudfoot, N., and O'Sullivan, J. (2002). Polyadenylation: A tail of two complexes. Curr. Biol. 12 (24), R855–R857. doi:10.1016/s0960-9822(02)01353-2
Psakhye, I., and Jentsch, S. (2012). Protein group modification and synergy in the SUMO pathway as exemplified in DNA repair. Cell 151 (4), 807–820. doi:10.1016/j.cell.2012.10.021
Qin, G., Tu, X., Li, H., Cao, P., Chen, X., Song, J., et al. (2020). Long noncoding RNA p53-stabilizing and activating RNA promotes p53 signaling by inhibiting heterogeneous nuclear ribonucleoprotein K deSUMOylation and suppresses hepatocellular carcinoma. Hepatology 71 (1), 112–129. doi:10.1002/hep.30793
Raabe, T., Bollum, F. J., and Manley, J. L. (1991). Primary structure and expression of bovine poly(A) polymerase. Nature 353 (6341), 229–234. doi:10.1038/353229a0
Rauth, S., Karmakar, S., Shah, A., Seshacharyulu, P., Nimmakayala, R. K., Ganguly, K., et al. (2021). SUMO modification of PAF1/PD2 enables PML interaction and promotes radiation resistance in pancreatic ductal adenocarcinoma. Mol. Cell Biol. 41 (12), e0013521. doi:10.1128/MCB.00135-21
Rawat, P., Boehning, M., Hummel, B., Aprile-Garcia, F., Pandit, A. S., Eisenhardt, N., et al. (2021). Stress-induced nuclear condensation of NELF drives transcriptional downregulation. Mol. Cell 81 (5), 1013–1026.e11. doi:10.1016/j.molcel.2021.01.016
Richard, P., Vethantham, V., and Manley, J. L. (2017). Roles of sumoylation in mRNA processing and metabolism. Adv. Exp. Med. Biol. 963, 15–33. doi:10.1007/978-3-319-50044-7_2
Risso, G., Pelisch, F., Pozzi, B., Mammi, P., Blaustein, M., Colman-Lerner, A., et al. (2013). Modification of Akt by SUMO conjugation regulates alternative splicing and cell cycle. Cell Cycle 12 (19), 3165–3174. doi:10.4161/cc.26183
Rodriguez, M. S., Desterro, J. M., Lain, S., Midgley, C. A., Lane, D. P., and Hay, R. T. (1999). SUMO-1 modification activates the transcriptional response of p53. EMBO J. 18 (22), 6455–6461. doi:10.1093/emboj/18.22.6455
Rosas-Acosta, G., Russell, W. K., Deyrieux, A., Russell, D. H., and Wilson, V. G. (2005). A universal strategy for proteomic studies of SUMO and other ubiquitin-like modifiers. Mol. Cell Proteomics 4 (1), 56–72. doi:10.1074/mcp.M400149-MCP200
Rouviere, J. O., Geoffroy, M. C., and Palancade, B. (2013). Multiple crosstalks between mRNA biogenesis and SUMO. Chromosoma 122 (5), 387–399. doi:10.1007/s00412-013-0408-y
Ryu, H. Y., and Hochstrasser, M. (2021). Histone sumoylation and chromatin dynamics. Nucleic Acids Res. 49 (11), 6043–6052. doi:10.1093/nar/gkab280
Ryu, H. Y., Zhao, D., Li, J., Su, D., and Hochstrasser, M. (2020). Histone sumoylation promotes Set3 histone-deacetylase complex-mediated transcriptional regulation. Nucleic Acids Res. 48 (21), 12151–12168. doi:10.1093/nar/gkaa1093
Sahin, U., Lapaquette, P., Andrieux, A., Faure, G., and Dejean, A. (2014). Sumoylation of human argonaute 2 at lysine-402 regulates its stability. PLoS One 9 (7), e102957. doi:10.1371/journal.pone.0102957
Sapetschnig, A., Rischitor, G., Braun, H., Doll, A., Schergaut, M., Melchior, F., et al. (2002). Transcription factor Sp3 is silenced through SUMO modification by PIAS1. EMBO J. 21 (19), 5206–5215. doi:10.1093/emboj/cdf510
Schaub, F. X., Dhankani, V., Berger, A. C., Trivedi, M., Richardson, A. B., Shaw, R., et al. (2018). Pan-cancer alterations of the MYC oncogene and its proximal network across the cancer genome atlas. Cell Syst. 6 (3), 282–300.e2. doi:10.1016/j.cels.2018.03.003
Schimmel, J., Eifler, K., Sigurethsson, J. O., Cuijpers, S. A., Hendriks, I. A., Verlaan-de Vries, M., et al. (2014). Uncovering SUMOylation dynamics during cell-cycle progression reveals FoxM1 as a key mitotic SUMO target protein. Mol. Cell 53 (6), 1053–1066. doi:10.1016/j.molcel.2014.02.001
Scorilas, A., Courtis, N., Yotis, J., Talieri, M., Michailakis, M., and Trangas, T. (1998). Poly(A)polymerase activity levels in breast tumour cytosols. J. Exp. Clin. Cancer Res. 17 (4), 511–518.
Sentis, S., Le Romancer, M., Bianchin, C., Rostan, M. C., and Corbo, L. (2005). Sumoylation of the estrogen receptor alpha hinge region regulates its transcriptional activity. Mol. Endocrinol. 19 (11), 2671–2684. doi:10.1210/me.2005-0042
Shi, Y., Di Giammartino, D. C., Taylor, D., Sarkeshik, A., Rice, W. J., Yates, J. R., et al. (2009). Molecular architecture of the human pre-mRNA 3' processing complex. Mol. Cell 33 (3), 365–376. doi:10.1016/j.molcel.2008.12.028
Shiio, Y., and Eisenman, R. N. (2003). Histone sumoylation is associated with transcriptional repression. Proc. Natl. Acad. Sci. U. S. A. 100 (23), 13225–13230. doi:10.1073/pnas.1735528100
Spector, D. L., and Lamond, A. I. (2011). Nuclear speckles. Cold Spring Harb. Perspect. Biol. 3 (2), a000646. doi:10.1101/cshperspect.a000646
Stine, Z. E., Walton, Z. E., Altman, B. J., Hsieh, A. L., and Dang, C. V. (2015). MYC, metabolism, and cancer. Cancer Discov. 5 (10), 1024–1039. doi:10.1158/2159-8290.CD-15-0507
Sun, Y., Hamilton, K., and Tong, L. (2020). Recent molecular insights into canonical pre-mRNA 3'-end processing. Transcription 11 (2), 83–96. doi:10.1080/21541264.2020.1777047
Sung, T. Y., Kim, M., Kim, T. Y., Kim, W. G., Park, Y., Song, D. E., et al. (2015). Negative expression of CPSF2 predicts a poorer clinical outcome in patients with papillary thyroid carcinoma. Thyroid 25 (9), 1020–1025. doi:10.1089/thy.2015.0079
Takagaki, Y., Ryner, L. C., and Manley, J. L. (1989). Four factors are required for 3'-end cleavage of pre-mRNAs. Genes Dev. 3 (11), 1711–1724. doi:10.1101/gad.3.11.1711
Takahashi, Y., Lallemand-Breitenbach, V., Zhu, J., and de The, H. (2004). PML nuclear bodies and apoptosis. Oncogene 23 (16), 2819–2824. doi:10.1038/sj.onc.1207533
Tammsalu, T., Matic, I., Jaffray, E. G., Ibrahim, A. F. M., Tatham, M. H., and Hay, R. T. (2014). Proteome-wide identification of SUMO2 modification sites. Sci. Signal 7 (323), rs2. doi:10.1126/scisignal.2005146
Tan, J. A., Hall, S. H., Hamil, K. G., Grossman, G., Petrusz, P., and French, F. S. (2002). Protein inhibitors of activated STAT resemble scaffold attachment factors and function as interacting nuclear receptor coregulators. J. Biol. Chem. 277 (19), 16993–17001. doi:10.1074/jbc.M109217200
Tang, C., Wang, X., Jin, Y., and Wang, F. (2022). Recent advances in HDAC-targeted imaging probes for cancer detection. Biochim. Biophys. Acta Rev. Cancer 1877 (5), 188788. doi:10.1016/j.bbcan.2022.188788
Tang, Z., Yu, W., Zhang, C., Zhao, S., Yu, Z., Xiao, X., et al. (2016). CREB-binding protein regulates lung cancer growth by targeting MAPK and CPSF4 signaling pathway. Mol. Oncol. 10 (2), 317–329. doi:10.1016/j.molonc.2015.10.015
Tartakoff, A., DiMario, P., Hurt, E., McStay, B., Panse, V. G., and Tollervey, D. (2022). The dual nature of the nucleolus. Genes Dev. 36 (13-14), 765–769. doi:10.1101/gad.349748.122
Tatham, M. H., Matic, I., Mann, M., and Hay, R. T. (2011). Comparative proteomic analysis identifies a role for SUMO in protein quality control. Sci. Signal 4 (178), rs4. doi:10.1126/scisignal.2001484
Townson, S. M., Sullivan, T., Zhang, Q. P., Clark, G. M., Osborne, C. K., Lee, A. V., et al. (2000). HET/SAF-B overexpression causes growth arrest and multinuclearity and is associated with aneuploidy in human breast cancer. Clin. Cancer Res. 6 (9), 3788–3796.
Treiber, T., Treiber, N., and Meister, G. (2019). Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat. Rev. Mol. Cell Biol. 20 (1), 5–20. doi:10.1038/s41580-018-0059-1
Tsourlakis, M. C., Schoop, M., Plass, C., Huland, H., Graefen, M., Steuber, T., et al. (2013). Overexpression of the chromatin remodeler death-domain-associated protein in prostate cancer is an independent predictor of early prostate-specific antigen recurrence. Hum. Pathol. 44 (9), 1789–1796. doi:10.1016/j.humpath.2013.01.022
Van Dyck, F., Delvaux, E. L., Van de Ven, W. J., and Chavez, M. V. (2004). Repression of the transactivating capacity of the oncoprotein PLAG1 by SUMOylation. J. Biol. Chem. 279 (34), 36121–36131. doi:10.1074/jbc.M401753200
Vertegaal, A. C., Andersen, J. S., Ogg, S. C., Hay, R. T., Mann, M., and Lamond, A. I. (2006). Distinct and overlapping sets of SUMO-1 and SUMO-2 target proteins revealed by quantitative proteomics. Mol. Cell Proteomics 5 (12), 2298–2310. doi:10.1074/mcp.M600212-MCP200
Vethantham, V., Rao, N., and Manley, J. L. (2007). Sumoylation modulates the assembly and activity of the pre-mRNA 3' processing complex. Mol. Cell Biol. 27 (24), 8848–8858. doi:10.1128/MCB.01186-07
Vethantham, V., Rao, N., and Manley, J. L. (2008). Sumoylation regulates multiple aspects of mammalian poly(A) polymerase function. Genes Dev. 22 (4), 499–511. doi:10.1101/gad.1628208
Wang, Y., and Yu, J. (2021). Dissecting multiple roles of SUMOylation in prostate cancer. Cancer Lett. 521, 88–97. doi:10.1016/j.canlet.2021.08.034
Weng, H., Huang, H., Wu, H., Qin, X., Zhao, B. S., Dong, L., et al. (2018). METTL14 inhibits hematopoietic stem/progenitor differentiation and promotes leukemogenesis via mRNA m(6)A modification. Cell Stem Cell 22 (2), 191–205.e9. doi:10.1016/j.stem.2017.11.016
West, A. C., and Johnstone, R. W. (2014). New and emerging HDAC inhibitors for cancer treatment. J. Clin. Invest. 124 (1), 30–39. doi:10.1172/JCI69738
Wilkinson, K. A., and Henley, J. M. (2010). Mechanisms, regulation and consequences of protein SUMOylation. Biochem. J. 428 (2), 133–145. doi:10.1042/BJ20100158
Wilkinson, M. E., Charenton, C., and Nagai, K. (2020). RNA splicing by the spliceosome. Annu. Rev. Biochem. 89, 359–388. doi:10.1146/annurev-biochem-091719-064225
Xu, H., Wang, H., Zhao, W., Fu, S., Li, Y., Ni, W., et al. (2020). SUMO1 modification of methyltransferase-like 3 promotes tumor progression via regulating Snail mRNA homeostasis in hepatocellular carcinoma. Theranostics 10 (13), 5671–5686. doi:10.7150/thno.42539
Xu, J. F., Zhao, Z. G., Ye, L. L., Zhuge, W., Han, Z., Zhang, T. M., et al. (2017). Prognostic significance of daxx NCR (Nuclear/Cytoplasmic ratio) in gastric cancer. Cancer Med. 6 (9), 2063–2075. doi:10.1002/cam4.1144
Yamazaki, T., Yamamoto, T., Yoshino, H., Souquere, S., Nakagawa, S., Pierron, G., et al. (2021). Paraspeckles are constructed as block copolymer micelles. EMBO J. 40 (12), e107270. doi:10.15252/embj.2020107270
Yan, Q., Gong, L., Deng, M., Zhang, L., Sun, S., Liu, J., et al. (2010). Sumoylation activates the transcriptional activity of Pax-6, an important transcription factor for eye and brain development. Proc. Natl. Acad. Sci. U. S. A. 107 (49), 21034–21039. doi:10.1073/pnas.1007866107
Yang, S.-H., Jaffray, E., Hay, R. T., and Sharrocks, A. D. (2003). Dynamic interplay of the SUMO and ERK pathways in regulating elk-1 transcriptional activity. Mol. Cell 12 (1), 63–74. doi:10.1016/s1097-2765(03)00265-x
Yang, S.-H., and Sharrocks, A. D. (2004). SUMO promotes HDAC-mediated transcriptional repression. Mol. Cell 13 (4), 611–617. doi:10.1016/s1097-2765(04)00060-7
Yang, W., and Paschen, W. (2015). SUMO proteomics to decipher the SUMO-modified proteome regulated by various diseases. Proteomics 15 (5-6), 1181–1191. doi:10.1002/pmic.201400298
Yang, X., Yang, Y., Sun, B. F., Chen, Y. S., Xu, J. W., Lai, W. Y., et al. (2017). 5-methylcytosine promotes mRNA export - NSUN2 as the methyltransferase and ALYREF as an m(5)C reader. Cell Res. 27 (5), 606–625. doi:10.1038/cr.2017.55
Yeh, E. T. (2009). SUMOylation and de-SUMOylation: Wrestling with life's processes. J. Biol. Chem. 284 (13), 8223–8227. doi:10.1074/jbc.R800050200
Yi, C., Wang, Y., Zhang, C., Xuan, Y., Zhao, S., Liu, T., et al. (2016). Cleavage and polyadenylation specific factor 4 targets NF-κB/cyclooxygenase-2 signaling to promote lung cancer growth and progression. Cancer Lett. 381 (1), 1–13. doi:10.1016/j.canlet.2016.07.016
Yu, F., Shi, G., Cheng, S., Chen, J., Wu, S. Y., Wang, Z., et al. (2018). SUMO suppresses and MYC amplifies transcription globally by regulating CDK9 sumoylation. Cell Res. 28 (6), 670–685. doi:10.1038/s41422-018-0023-9
Yu, F., Wei, J., Cui, X., Yu, C., Ni, W., Bungert, J., et al. (2021). Post-translational modification of RNA m6A demethylase ALKBH5 regulates ROS-induced DNA damage response. Nucleic Acids Res. 49 (10), 5779–5797. doi:10.1093/nar/gkab415
Yu, J., de Belle, I., Liang, H., and Adamson, E. D. (2004). Coactivating factors p300 and CBP are transcriptionally crossregulated by Egr1 in prostate cells, leading to divergent responses. Mol. Cell 15 (1), 83–94. doi:10.1016/j.molcel.2004.06.030
Yu, J., Zhang, S. S., Saito, K., Williams, S., Arimura, Y., Ma, Y., et al. (2009). PTEN regulation by Akt-EGR1-ARF-PTEN axis. EMBO J. 28 (1), 21–33. doi:10.1038/emboj.2008.238
Yuan, H., Deng, R., Zhao, X., Chen, R., Hou, G., Zhang, H., et al. (2017). SUMO1 modification of KHSRP regulates tumorigenesis by preventing the TL-G-Rich miRNA biogenesis. Mol. Cancer 16 (1), 157. doi:10.1186/s12943-017-0724-6
Zealy, R. W., Wrenn, S. P., Davila, S., Min, K. W., and Yoon, J. H. (2017). microRNA-binding proteins: specificity and function. Wiley Interdiscip. Rev. RNA 8 (5), e1414. doi:10.1002/wrna.1414
Zhang, D., Liang, Y., Xie, Q., Gao, G., Wei, J., Huang, H., et al. (2015). A novel post-translational modification of nucleolin, SUMOylation at Lys-294, mediates arsenite-induced cell death by regulating gadd45α mRNA stability. J. Biol. Chem. 290 (8), 4784–4800. doi:10.1074/jbc.M114.598219
Zhang, S., Liu, Y., Liu, D., and Yang, L. (2017a). Targeting cleavage and polyadenylation specific factor 1 via shRNA inhibits cell proliferation in human ovarian cancer. J. Biosci. 42 (3), 417–425. doi:10.1007/s12038-017-9701-x
Zhang, S., Zhao, B. S., Zhou, A., Lin, K., Zheng, S., Lu, Z., et al. (2017b). m(6)A demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell 31 (4), 591–606.e6. doi:10.1016/j.ccell.2017.02.013
Zhao, M., Mishra, L., and Deng, C. X. (2018). The role of TGF-β/SMAD4 signaling in cancer. Int. J. Biol. Sci. 14 (2), 111–123. doi:10.7150/ijbs.23230
Zheng, Z., Cai, C., Omwancha, J., Chen, S. Y., Baslan, T., and Shemshedini, L. (2006). SUMO-3 enhances androgen receptor transcriptional activity through a sumoylation-independent mechanism in prostate cancer cells. J. Biol. Chem. 281 (7), 4002–4012. doi:10.1074/jbc.M509260200
Zhong, S., Muller, S., Ronchetti, S., Freemont, P. S., Dejean, A., and Pandolfi, P. P. (2000). Role of SUMO-1-modified PML in nuclear body formation. Blood 95 (9), 2748–2752. doi:10.1182/blood.v95.9.2748.009k31a_2748_2752
Zhu, C., Chen, C., Chen, R., Deng, R., Zhao, X., Zhang, H., et al. (2016). K259-SUMOylation of DGCR8 promoted by p14ARF exerts a tumor-suppressive function. J. Mol. Cell Biol. 8 (5), 456–458. doi:10.1093/jmcb/mjw030
Keywords: sumoylation, mRNA transcription, mRNA processing, RNA editing, miRNA biogenesis, cancer
Citation: Cao Y, Huang C, Zhao X and Yu J (2023) Regulation of SUMOylation on RNA metabolism in cancers. Front. Mol. Biosci. 10:1137215. doi: 10.3389/fmolb.2023.1137215
Received: 04 January 2023; Accepted: 15 February 2023;
Published: 24 February 2023.
Edited by:
Yitao Qi, Shaanxi Normal University, ChinaReviewed by:
Hiroaki Kiyokawa, Northwestern University, United StatesCopyright © 2023 Cao, Huang, Zhao and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianxiu Yu, SmlhbnhpdS5ZdUBzaHNtdS5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.