- 1Department of Liver Disease, Shuguang Hospital Affiliated to Shanghai University of Traditional Chinese Medicine, Shanghai, China
- 2Institute of Infectious Disease, Shuguang Hospital Affiliated to Shanghai University of Traditional Chinese Medicine, Shanghai, China
Due to the lack of distinct early symptoms and specific biomarkers, most patients with hepatocellular carcinoma (HCC) are usually diagnosed at advanced stages, rendering the treatment ineffective and useless. Therefore, recognition of the malady at precancerous lesions and early stages is particularly important for improving patient outcomes. The interest in extracellular vesicles (EVs) has been growing in recent years with the accumulating knowledge of their multiple cargoes and related multipotent roles in the modulation of immune response and tumor progression. By virtue of the rapid advancement of high-throughput techniques, multiple omics, including genomics/transcriptomics, proteomics, and metabolomics/lipidomics, have been widely integrated to analyze the role of EVs. Comprehensive analysis of multi-omics data will provide useful insights for discovery of new biomarkers and identification of therapeutic targets. Here, we review the attainment of multi-omics analysis to the finding of the potential role of EVs in early diagnosis and the immunotherapy in HCC.
1 Introduction
Hepatocellular carcinoma (HCC) is the predominant type of primary liver cancer with comprising 75%–85% of cases, and the third leading cause of cancer-related death worldwide (Sung et al., 2021). The carcinogenesis mostly occurs in liver cirrhosis and is thought to be a complex multistep process (Sciarra et al., 2016). The precancerous stage is characterized with the appearance of dysplastic nodules in cirrhosis, which are divided into low-grade dysplastic nodules (LGDN) and high-grade dysplastic nodules (HGDN) according to the degree of cellular and structural atypia (International Consensus Group for Hepatocellular Neoplasia, 2009). HGDN was a potent predictor of hepatocarcinogenesis, with reported malignant transformation rates of 46.2%, 61.5%, and 80.8% at 1, 3, and 5 years (Borzio et al., 2003; Kobayashi et al., 2006; Sato et al., 2015). Patients with precancerous lesions and early-stage HCC can benefit from curative therapies including hepatic resection, liver transplantation, and radiofrequency ablation, leading to the attainment of 70%–80% of 5-year survival rate (Cho et al., 2011; Villanueva, 2019; Llovet et al., 2021). However, due to the lack of distinct early symptoms and specific biomarkers, most patients are usually diagnosed at advanced stages when the tumor is unresectable, with a median survival of only 6 months (Hollebecque et al., 2015; Hartke et al., 2017). Systemic therapy for advanced HCC has only been limited to anti-angiogenic tyrosine kinase inhibitors (TKIs) until 2017. In recent years, the deep research of immune checkpoint inhibitors (ICIs) favored the development of immune-based systemic therapy in clinical practice. However, immunotherapy has only shown strong antitumor response to some HCC patients, and main challenges are the identification of sensitive population and the optimization of therapeutic strategies (Sangro et al., 2021). Therefore, finding of early diagnostic specific markers and establishment of effective strategies for immunotherapy have attracted tremendous attention.
An interest in extracellular vesicles (EVs) sharply increased in recent years following the discovery that EVs act as carriers for the intercellular transport of proteins, nucleic acids, lipids, metabolites, etc. EVs are a heterogenous group of lipid bilayer-delimited particles that are naturally released by almost all types of cells and can be broadly sorted into three main classes according to their biosynthesis or secretion process: exosomes, microvesicles/microparticles/ectosomes, and apoptotic bodies (Yáñez-Mó et al., 2015). Exosomes originate from the endosomal system in which endosomes invaginate to form multivesicular bodies (MVBs), and further develop into intraluminal vesicles (ILVs). ILVs are released as “exosomes” upon fusion of MVBs with the plasma membrane. Ectosomes (microparticles/microvesocles) are generated by the outward budding and fission of the plasma membrane, and subsequently released into the extracellular space. Apoptotic bodies are blebs yielded and released by dying cells that undergo apoptosis (Colombo et al., 2014; van Niel et al., 2018). Since no specific markers have been found to identify EV subtypes, “extracellular vesicle” or analogous appellations are the currently preferred nomenclature for these particles (Théry et al., 2018). During the biogenesis, different types of EVs are selectively enriched with a variety of cellular bioactive molecules as cargos. Although some proteins may be shared among EV subtypes, the character of vesicle cargo is highly dependent on the type of donor cells, and their conditions in the physiological or pathological process (van Niel et al., 2018). In the tumor microenvironment, EVs can transfer molecular cargoes among tumor cells, stromal cells, and immune cells for intercellular communication (Becker et al., 2016). Studies have demonstrated the critical role of EVs in tumor progression and antitumor immune responses (Marar et al., 2021). As the properties of EVs as signaling vehicles continue to be revealed, their roles in cancer diagnosis and treatment are gradually emerging. Recent development and broad availability of multi-omics technologies, including genomics, transcriptomics, proteomics, metabolomics, and lipidomics, have promoted a more in-depth study of EVs, which provides profound prospect for the discovery of new candidate biomarkers and the establishment of effective strategies for cancer therapy (Figure 1). In this review, we summarize the progression of multiomics-integrated analysis of EVs for the early diagnosis of precancerous lesions and HCC and discuss a new class of EV-centered cancer immunotherapies.
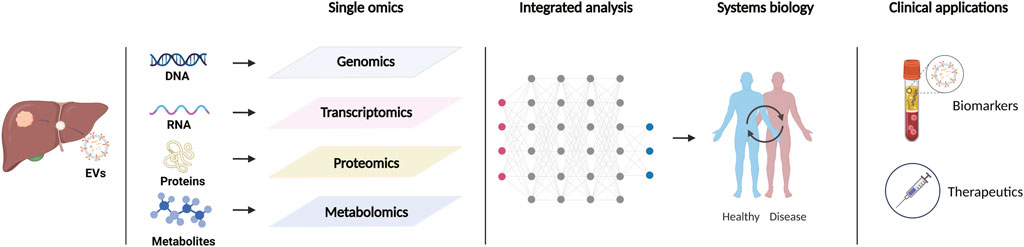
FIGURE 1. Schematic representation of multi-omics approaches to extracellular vesicles in the study of hepatic precancerous lesions and hepatocellular carcinoma. Created with https://BioRender.com.
2 Applications of EVs in early diagnosis
EVs are extensively found in different biological fluids, and carry various biomolecules from parental cells, including functional proteins, nucleic acids, and lipids metabolites. They initiate a diversity of physiological and pathological processes in the way of cell-cell communication via delivery of the cargo to recipient cells, and exert the prominent function in many diseases including cancers (Yáñez-Mó et al., 2015; Becker et al., 2016). EVs that are released from tumor cells into the blood can provide a survey of the entire tumor and represent as biomarkers that can be collected and analyzed from blood samples, offering great potential for early diagnosis of cancer. However, there is currently no single optimal separation method to obtain pure EVs, so the choice depends on the downstream application and the scientific question to be solved (Théry et al., 2018).
2.1 Genomics/transcriptomics
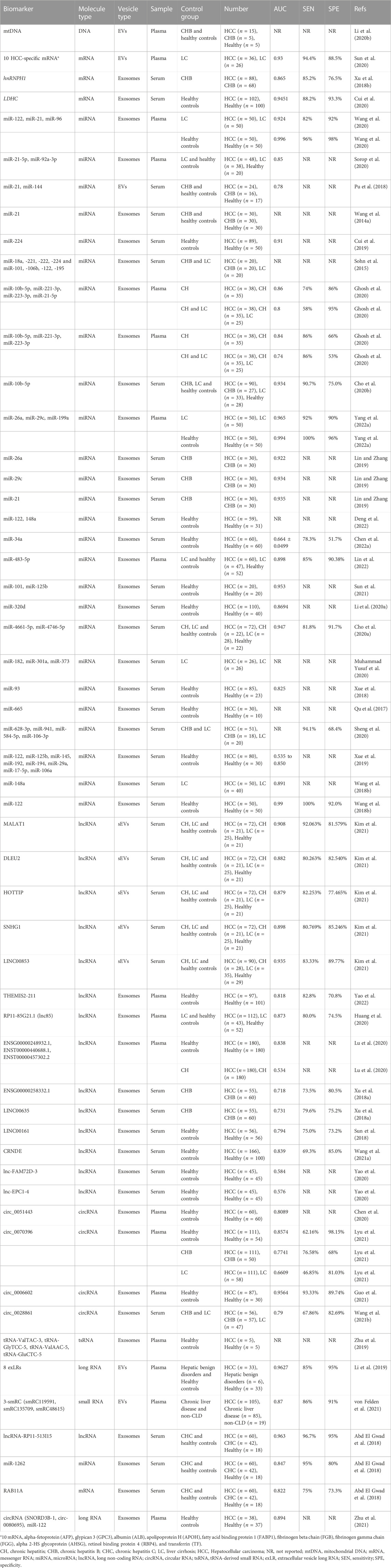
Recent studies have demonstrated the presence of complete genomic DNA (gDNA), mitochondrial DNA (mtDNA), and even viral DNA in EVs(Cai et al., 2013; Thakur et al., 2014; Sansone et al., 2017; Lázaro-Ibáñez et al., 2019; Saari et al., 2020; Liu H. et al., 2022). Bioactive DNA-carrying EVs play crucial roles in maintaining cellular homeostasis, regulating immune responses, and participating in tumor formation and progression (Takahashi et al., 2017; Torralba et al., 2018; Malkin and Bratman, 2020; Elzanowska et al., 2021; Hagey et al., 2021). It has been found that oxidized mtDNA-enriched EVs can synergize with acetaldehyde to activate oxidative stress and multiple oncogenic pathways in cancerous cells for the promotion of alcohol-related liver cancer development (Seo et al., 2019). Data analysis based on the examination of next-generation sequencing (NGS) technique showed that EV mtDNA covered the entire mitochondrial genome, in which mtDNA end sites, cleavage numbers, and copy numbers were significantly reduced in EVs from HCC patients compared with healthy controls, indicating the potential value of EV mtDNA signatures in the diagnosis of HCC(Li Y. et al., 2020). Although their biological significance has been recognized, EV DNAs are still less explored, and their role in precancerous lesions and HCC still needs to be further verified.
EVs are also rich in various types of RNAs, which exhibit great potential for early detection of HCC. A study containing 291 participants revealed that mRNA levels of serum exosomal heterogeneous ribonucleoprotein H1 (hnRNPH1) were significantly higher in HCC than in cirrhosis, chronic hepatitis B, and healthy controls (Xu et al., 2018b). The hnRPH1 gene encodes a splicing regulator that can stimulate pre-mRNA cleavage and polyadenylation, and is abnormally expressed in a number of human cancers such as esophageal cancer, pancreatic cancer, colon cancer, and prostate cancer (Honoré et al., 2004; Sun et al., 2016; Yang et al., 2016). Here exosomal hnRNPH1 mRNA was identified as an effective marker for HBV-related HCC evidenced by an AUC of 0.865 (95% CI = 0.808–0.922, p = 0.003), and its combination with AFP could further improve the diagnostic validity. Lactate dehydrogenase C (LDHC) is a germ cell-specific member of the lactate dehydrogenase family. LDHC activation in cancers may provide a metabolic rescue pathway in tumor cells by using lactate for ATP delivery. Studies have confirmed that LDHC mRNA is expressed in various tumor tissues such as lung cancer, melanoma, breast cancer, and kidney cancer (Koslowski et al., 2002; Wang et al., 2018a). Recently, Cui Z et al. found that the expression of LDHC mRNA was also significantly upregulated in serum EVs of HCC patients, which could clearly distinguish early-stage HCC patients from healthy controls (Cui et al., 2020). Compared to total circulating EVs, tumor-derived EVs have more potential to be used as disease-specific biomarkers. However, current conventional isolation techniques based on EVs’ physical properties fail to isolate tumor-specific EVs. To overcome this issue, Sun N et al. have developed a covalent chemistry-based HCC-specific EV purification system, EV Click Chips, which enables the isolation of plasma HCC-derived EVs in high efficiency and purity (Sun et al., 2020). Moreover, by coupling EV Click Chips with a downstream reverse-transcription droplet digital PCR assay, 10 well-validated HCC-specific mRNA transcripts, such as alpha-fetoprotein (AFP), glypican 3 (GPC3), albumin (ALB), apolipoprotein H (APOH), were selectively quantified for the calculation of a digital score. The resulting score in EVs exhibited great potential for non-invasive detection of early-staged HCC in high-risk cirrhosis patients, evidenced as an area under the ROC curve (AUC) of 0.93 (95% CI, 0.86 to 1.00; sensitivity = 94.4%, specificity = 88.5%).
In addition to mRNA, some non-coding RNAs (ncRNAs), including microRNAs (miRNAs), long non-coding RNAs (lncRNAs), circular RNAs (circRNAs), transfer RNA (tRNA), have also exhibited instrumental impact on HCC progression. MiRNAs are a class of endogenous small ncRNAs with the length of 19–25 nucleotides, which serve as critical regulators of post-transcriptional gene expression and play an important role in cell growth, differentiation, development, and apoptosis (Saliminejad et al., 2019). In recent years, a large number of researches have shown that many miRNAs can act as tumor suppressors or oncogenes to promote the occurrence and development of tumors (Di Leva et al., 2014). One of the representative paradigm is miR-21 that was early discovered to be widespread in human cells or tissues. Increasing studies have manifested that miR-21 functions as an oncogene to involve the post-transcriptional gene regulation, cell differentiation, proliferation and apoptosis, which are closely related to tumorigenesis (Singh et al., 2021). Furthermore, miR-21 was evidenced to be enriched in serum EVs of patients with liver diseases, and the level of EV miR-21 in HCC patients is significantly higher than in patients with chronic hepatitis B, liver cirrhosis or healthy donors, indicating the potential of miR21-containing EVs as a diagnostic marker for HCC(Wang H. et al., 2014; Pu et al., 2018; Mjelle et al., 2019; Sorop et al., 2020; Wang et al., 2020). In addition, other circulating miRNAs, such as miR-221/222, miR-224, miR-155, may also play an important role in the early stage of hepatocarcinogenesis (Wang et al., 2009; Zhang Y. et al., 2012; Cui et al., 2019). Analysis of miRNA expression in selected serum EVs showed that miR-18a, -221, -222, and -224 were specifically upregulated, while miR-101, -106b, - 122 and -195 were downregulated in HCC patients (Sohn et al., 2015). Among them, miR-221 was evidenced to promote hepatcarcinogenesis by dysregulating DNA damage-inducible transcript 4 (DDIT4), the modulator of mTOR pathway, whereas miR-101 that is downregulated in HCC may induce apoptosis and inhibit tumorigenicity via targeting Mcl-1 (Su et al., 2009; Pineau et al., 2010). Combining transcriptome techniques plus existing datasets from GEO and TCGA databases, Ghosh S et al. analyzed the miRNA profile of HBV- and HCV-infected HCC versus normal or adjacent tissues, and Panther/Gene Ontology enrichment/Cytoscape analysis indicated that the targets were mostly involved in carcinogenesis pathways (Ghosh et al., 2020). After validated to be in plasma-derived exosomes from HBV/HCV-infected non-HCC, such as chronic hepatitis and cirrhosis, and HCC samples, four miRNAs including miR-10b-5p, miR-221-3p, miR-223-3p and miR-21-5p were identified as novel potential biomarkers for early HCC diagnosis, and their combination showed better sensitivity in distinguishing HCC, especially with low AFP expression. Similarly, the study by Cho HJ et al. confirmed that miR-10b-5p in serum EVs is a promising diagnostic biomarker for early-stage HCC with the AUC equal to 0.934 (sensitivity = 90.7%, specificity = 75.0%) (Cho et al., 2020b). In another study, analysis of plasma miRNA microarray was performed to screen the miRNA with different expression in EVs derived from HCC, cirrhosis and healthy control. As a result, three miRNAs including miRNA-26a, miRNA-29c and miRNA-199a were identified to be significantly reduced in HCC-derived EVs. Further analysis indicated that these miRNAs may act as tumor suppressors in HCC, and the examination of EVs with the miRNAs-embedded panel showed higher accuracy in the diagnosis of HCC(Lin and Zhang, 2019; Yang J. et al., 2022). In addition, other numerous EVs miRNAs have been identified as potential early-diagnosed markers for HCC, including but not limited to miR-34a, miR-93, miR-101, miR-122, miR-125b, miR -148a, miR-182, miR-301a, miR-320d, miR-373, miR-483-5p, miR-665, miR-3129, miRNA-4661-5p, miR-4746-5p, and the combinations of multiple miRNAs would present higher diagnostic utility (Qu et al., 2017; Wang et al., 2018b; Xue et al., 2018; Xue et al., 2019; Cho et al., 2020a; Li W. et al., 2020; Muhammad Yusuf et al., 2020; Sheng et al., 2020; Sun et al., 2021; Yang et al., 2021; Chen S. et al., 2022; Deng et al., 2022; Lin et al., 2022). These markers are expected to construct new liquid biopsy modalities in the near future.
Recently, many lncRNAs and circRNAs derived from EVs have been proposed as potential candidate biomarkers for HCC diagnosis. LncRNAs are ncRNAs with a length of more than 200 nucleotides, which play an important role in regulating the transcriptome level. They were found to be differentially expressed in tumors and were directly associated with the transformation of normal cells into tumor cells (Huarte, 2015). By virtue of TCGA_LIHC and Catholic_LIHC projects (two human HCC whole transcriptome datasets), Kim SS et al. identified specific tumor-driving lncRNA candidates in serum EVs by comparing the expression of lncRNA in HCC and non-tumor tissues. In the end, the EV lncRNA-MALAT1, -DLEU2, -HOTTIP, -SNHG1, and -LINC00853 were identified as promising diagnostic markers for early-stage HCC, even in AFP-negative patients (Kim et al., 2020; Kim et al., 2021). MALAT1 is a representative onco-lncRNA that stimulates tumor growth and metastasis through multiple mechanisms in different tissues (Wang J. et al., 2014; Hou et al., 2017). DLEU2, HOTTIP, and SNHG1 have also been reported as onco-lncRNAs in HCC, while LINC00853 is currently poorly understood for the role in cancer and deserved for further study. RNA-seq technology coupled with multi-stage validation revealed many meaningful lncRNAs candidates with different expression, including lncRNA-THEMIS2-211, -RP11-85G21.1, -ENSG00000248932.1, -ENST00000440688.1, -ENST00000457302.2, -ENSG00000258332.1, and -LINC00635(Xu et al., 2018a; Huang et al., 2020; Lu et al., 2020; Yao et al., 2022). These lncRNAs are packed in EVs and play an instrumental role in inducing HCC formation and development. Their expression levels in EVs of HCC patients were significantly higher than those of chronic hepatitis/cirrhosis or healthy controls, presenting a potent diagnostic ability, and even better than AFP in diagnosing early-stage HCC. Other studies further added EV LINC00161, CRNDE, lnc-FAM72D-3, and lnc-EPC1-4 as potential biomarker candidates for HCC diagnosis (Sun et al., 2018; Yao et al., 2020; Wang T. et al., 2021). Moreover, lncRNA-HEIH showed specific diagnostic values in HCV-related HCC patients (Zhang et al., 2018). CircRNAs are a class of endogenous non-coding RNAs with a covalent closed loop structure, which are enriched and stable in EVs (Li et al., 2015; Kristensen et al., 2019). Hu K et al. found elevated levels of circCMTM3 in exosomes from HCC patients. Further experiments showed that exosomal circCMTM3 could promote angiogenesis through the miR-3619-5p/SOX9 axis, thereby promoting HCC tumorigenesis (Hu et al., 2021). In contrast, exosome-delivered circ_0051443 could inhibit the malignant biological behavior of HCC cells by inducing apoptosis and arresting cell cycle, and its expression in plasma EVs of HCC patients was significantly lower than that of healthy controls, evidenced by an AUC of 0.8089 (Chen et al., 2020). Another study showed that circ_0070396 was highly expressed in EVs of HCC and outperformed AFP in distinguishing HCC patients from chronic hepatitis B/cirrhosis or healthy donors (Lyu et al., 2021). Through transcriptome sequencing analysis and large sample clinical validation, circ_0006602 and circ_0028861 in EVs have been identified as candidate biomarkers for HCC, which can be used for early diagnosis and screening of HCC(Wang Y. et al., 2021; Guo et al., 2021). Functional experiments displayed that exo_circ_0006602 could significantly improve the proliferation, migration, and invasion of HCC cells, with the promoted expression of tumor proliferation-related protein. Exosomal circ_0028861 may affect HCC progression by regulating the miRNA targets and downstream tumor-related signaling pathways. Besides, other study reported the role of EVs tRNA-derived small RNA (tsRNA, a novel small non-coding RNA) in the diagnosis of HCC. It was found that tsRNAs are ubiquitous in exosomes, and the levels of four tsRNAs (tRNA-ValTAC-3, tRNA-GlyTCC-5, tRNA-ValAAC-5 and tRNA-GluCTC-5) were significantly elevated in plasma exosomes of HCC patients compared with healthy controls, revealing the diagnostic value of tsRNA in HCC (Zhu et al., 2019).
Comprehensive analysis of EV RNAs is a viable option in the diagnosis of patients with early-staged HCC (Wu Z. H. et al., 2022). Li Y et al. found that the sequencing profile of extracellular vesicle long RNA in blood (exLR, including mRNA, lncRNA, and circRNA) can distinguish cancer patients from healthy individuals, and in this way revealed that 8 exLRs had the potential as HCC diagnostic biomarkers in a multistage cohort (Li et al., 2019). The exLR-based HCC classifier has utility beyond AFP, not only for diagnosing early-stage or AFP-negative HCC, but also for distinguishing HCC patients from those with hepatitis, cirrhosis, or benign tumors. Another study evaluating a largely unexplored EVs-associated unannotated small RNA clusters (smRCs) demonstrated that the 3-smRC signature was significantly overexpressed in plasma EVs from HCC patients, which exhibited high sensitivity and specificity for the detection of early HCC in an independent validation cohort evidenced by the AUC of 0.87 (von Felden et al., 2021). Through a series of studies, Abd El Gwad A et al. demonstrated the role of three EV RNA-based biomarkers (lncRNA-RP11-513I15.6, miR-1262 and RAB11A) in inducing the formation of precancerous lesions in rats, and their excellent ability to differentiate HCC patients from chronic hepatitis and healthy volunteers (Abd El Gwad et al., 2018; Matboli et al., 2019; Hasanin et al., 2020). Comprehensive analysis of other extracellular RNAs also revealed the diagnostic value of different RNA detection panels in HCC, such as the combination of circRNA (SNORD3B-1, circ-0080695) with miR-122 (Zhu et al., 2021; Cabiati et al., 2022). Biomarkers identified by genomics/transcriptomics are summarized in Table 1. With the rapid development of liquid biopsy technology, circulating EV miRNAs, lncRNAs, and circRNAs may become more reliable biomarkers for HCC prediction in the future.
2.2 Proteomics
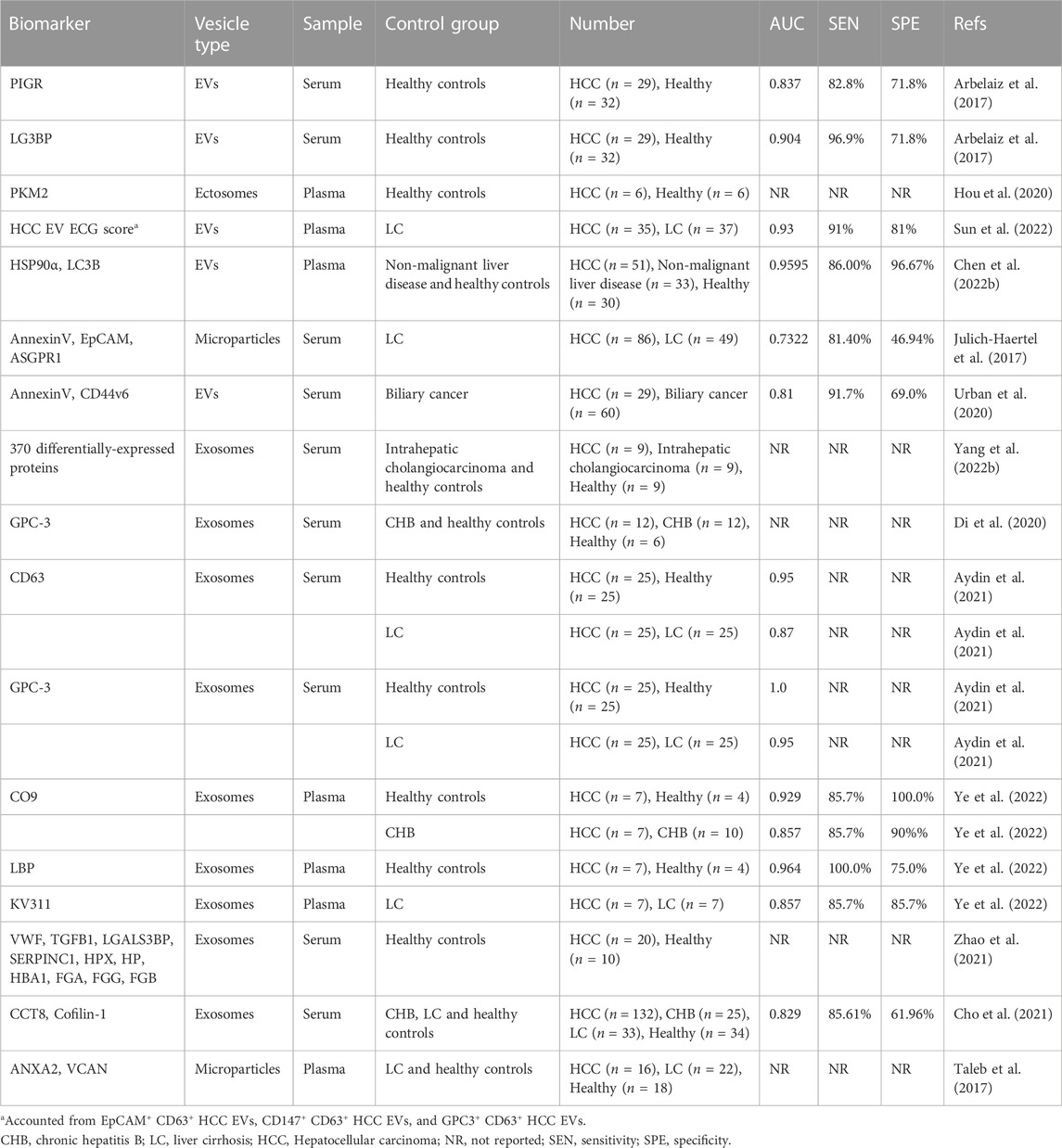
Besides nucleic acids, proteins are also critical cargoes in EVs. Proteins packed and transferred via EVs play a vital role in the progression of HCC(Wang et al., 2017; Fu et al., 2018; Jiang et al., 2020; Liu et al., 2021; Tey et al., 2022). Polymeric immunoglobulin receptor (pIgR) and galectin-3-binding protein (LG3BP) are both oncogenic proteins that promote cellular proliferation, transformation and invasion in HCC progression (Serizawa et al., 2015; Yue et al., 2017). EVs have been evidenced to be enriched with pIgR that can drive stemness-like conversion and tumorigenesis by activating PDK1/Akt/GSK3β/β-catenin signaling axis (Tey et al., 2022). In addition, pIgR and LG3BP were found to be significantly overexpressed in circulating EVs of HCC patients and exhibited higher diagnostic capacity than AFP(Arbelaiz et al., 2017). Pyruvate kinase M2 isoform (PKM2), an enzyme that plays a key role in the glucose metabolism of cancer cells, induces macrophage differentiation and remodels the tumor microenvironment through EV excretion, which level is positively correlated with hepatocarcinogenesis (Hou et al., 2020). The testing of clinical samples also validated a significant upregulation of plasma EV PKM2 levels from HCC patients. Sun N et al. developed and optimized an HCC-specific EV surface protein detection method (referred as HCC EV ECG score), based on three HCC EV subpopulations of EpCAM+CD63+, CD147+CD63+, and GPC3+ CD63+, by which EVs were isolated from peripheral blood, and HCC EV surface proteins were detected. This new established detection model was confirmed to have an accuracy of more than 93% (AUC >0.93) in distinguishing early HCC from cirrhosis with excellent detection performance (Sun et al., 2022). LC3B (MAP1LC3B) is a widely used marker of autophagy. Recent studies revealed that LC3B+ EVs carrying HSP90α are the master regulators of tumor progression, and the level of plasma HSP90α+ LC3B+ EVs in HCC patients were significantly higher than those in non-cancerous controls, suggesting the potential role as a diagnostic marker for HCC(Chen Y. Q. et al., 2022). Julich-Haertel H et al. isolated and detected various tumor-associated EV populations from patient serum and plasma via Fluorescence-activated cell scanning (FACS), and found that AnnexinV+ EpCAM+ ASGPR1+ EV and AnnV+ CD44v6+ EV were excellent in distinguishing liver malignancies from cirrhotic/control subjects (Julich-Haertel et al., 2017; Urban et al., 2020). These data suggest that the EV phenotype can serve as a novel HCC liquid biopsy biomarker.
To overcome the disadvantages of most traditional EV testing regarding huge samples, tedium, time consuming, low purity, and high cost, as well as the technical barriers of EV protein molecular analysis, some novel strategies for optimizing EV isolation, detection, and analysis have been developed (Di et al., 2020; Wu et al., 2020; Yang K. et al., 2022). Yang K et al. developed a new affinity method to isolate EVs by adhering highly abundant phosphatidylserine molecules on the surface of EVs to immobilized small peptide molecules (SiO2 microspheres), which can recover complete EVs in a short time with high yield and low cost (Yang K. et al., 2022). By comparing the biological information of differentially expressed proteins in serum EVs isolated by this method, early HCC patients and healthy individuals can be well differentiated. Another study also reported a nanozyme-assisted immunosorbent assay (NAISA), a platform based on the installation of 2 nm gold nanoparticles onto the phospholipid membrane of EVs, which enables rapid, sensitive, and specific analysis of EV proteins (Di et al., 2020). Protein expression in cell-derived EVs was measured by NAISA, and the results indicated that GPC-3 was specifically upregulated in HCC cell-derived EVs. GPC-3 is a member of the glypican family involved in the regulation of cell proliferation and apoptosis, which highly expresses as the indicator of malignant transformation of hepatocytes, and may mediate oncogenesis and oncogenic signaling pathways (Yao et al., 2016). Researchers further validated in clinical samples that both EV GPC-3 and CEA could effectively discriminate HCC patients from chronic hepatitis B patients and healthy controls, indicating the enormous potential of these EV proteins as effective biomarkers for early diagnosis of HCC. The study by Aydin Y et al. also confirmed the role of GPC3-positive circulating EVs in the detection of HCC in patients with liver cirrhosis (Aydin et al., 2021).
The nature of relatively high throughput in proteomic analysis shows great advantage in screening potential diagnostic biomarkers for HCC. Through differential proteomic analysis, multiple EV proteins were identified as potential candidate biomarkers and arrayed in testing panels, such as “LBP, KV311 and CO9,” “VWF, LGALS3BP, TGFB1, SERPINC1, HPX, HP, HBA1, FGA, FGG and FGB” and “Cofilin-1 and CCT8” (Cho et al., 2021; Zhao et al., 2021; Ye et al., 2022). These proteins play important roles in regulating cell growth, proliferation, differentiation, and innate and adaptive immune responses. Another study also showed that annexin A2 (ANXA2) and versican core protein (VCAN) were elevated in plasma EVs in HCV-related HCC patients, which can be used to characterize HCV-induced HCC(Taleb et al., 2017). ANXA2 belongs to the calcium-dependent phospholipid-binding protein family and plays an important role in the malignant transformation of HCC(Mohammad et al., 2008). VCAN, an extracellular matrix component, has been shown to also be involved in fibrotic deposition in the liver, which may limit its applicability as a diagnostic marker for HCC in the setting of cirrhosis. Biomarkers identified by proteomics are summarized in Table 2.
2.3 Metabolomics/lipidomics
Like nucleic acids and proteins, metabolites are also an important type of molecules carried by EVs. The metabolome of EVs mainly includes lipids, amino acids, organic acids, carnitine, steroids, vitamins, sugars and their complexes, as well as nucleotides and nucleosides (Zebrowska et al., 2019; Luo et al., 2020). In cancers, these molecular metabolites could be transferred to parenchymal cells and their surrounding stromal cells through the EV pathway, and influence the metabolism of recipient cells (Puhka et al., 2017). Metabolic reprogramming has been shown to be a hallmark of cancer, and researchers have recently increasingly focused on characterizing signatures of altered metabolism in cancer. The emerging role of EV-mediated metabolic reprogramming in tumor microenvironment remodeling and cancer progression has gradually emerged (Yang et al., 2020). Liu et al. (2021) observed reduced levels of triosephosphate isomerase 1 (TPI1), a glycolytic enzyme, in EVs released from HCC cells enhanced aerobic glycolysis-driven hepatocarcinogenesis. Metabolites represent downstream products of genes, transcripts, and proteins in multiple biological reactions, providing a more comprehensive molecular perspective and thus more likely to reveal dynamic changes in biological state (Luo et al., 2018; Tian et al., 2018). However, studies involving the intact metabolites present in EVs are scarce, and the reported metabolic analyses of EVs have mainly focused on lipids so far. Lipids are not only involved in building the structure of EV membranes, but also play an important role in the formation and release of EVs into the extracellular environment (Skotland et al., 2019). With the development of lipidomic analysis methods, the identification of novel lipid-based biomarkers has become available in practice. Sanchez JI et al. analyzed the difference of lipids in plasma EVs from 31 HCC patients and 41 cirrhotic patients using ultra-high resolution mass spectrometry (for untargeted lipidomic analysis), and discovered that ten classes of lipids were significantly enriched in EVs from HCC patients compared with non-HCC ones, while four classes were depleted (Sanchez et al., 2021). Logistic regression analysis revealed the close association of HCC with the enriched lipids, including sphingosine (SPH), disolysin (DLCL), lysophosphatidylserine (LysoPS), (O-acyl)-1-hydroxy fatty acids (OAHFA), and the depleted ones, such as gangliosides (GD1a) and fatty acids (FA). The difference in the composition and abundance of lipids could facilitate to distinguish HCC from non-HCC patients (p < 0.001), suggesting that the lipidomic signature of EVs has the potential to be a candidate biomarker for early detection of HCC in the future.
However, due to the paucity of current knowledge about lipid metabolism and the defects of current corresponding technologies, the research of metabolomics/lipidomics is far from as sufficient as that of proteomics and transcriptomics, and its clinical application is still limited. Therefore, the EVs metabolome remains to be digged more deeply and extensively.
2.4 Integromics and systems biology
2.4.1 Multi-omics combined
Single omics analyses can provide information on biological processes that differs in different life configurations, or on disease conditions compared to normal ones. However, these analyses can only provide limited insights, not suggesting comprehensive information on interactions among DNA, RNA, proteins, and metabolites. The most important question is whether the markers will eventually have the power to test efficacy in clinical practice. A single marker seems difficult to meet this requirement. Therefore, the combination of multi-omics is another promising direction to discover early cancer diagnostic markers. The integrated analysis of data obtained by different omics approaches is expected to provide new insights into complex biological systems and reveal the interaction network between all molecular-level processes. The multi-omics analysis of EVs is to use a variety of high-throughput technologies, including genomics, transcriptomics, proteomics, metabolomics, etc., to comprehensively analyze various cargo molecules in EVs from multiple aspects, in order to understand their biological function and clinical application prospect. The advantage of EV multi-omics analysis is that by analyzing the composition of various active molecules in EV, the origin, biological function and mechanism of action of EV can be more accurately determined, thereby revealing its role in physiology and disease, and providing a theoretical basis for clinical diagnosis and treatment. Meanwhile, EV multi-omics analysis is characterized by high throughput, high sensitivity, high specificity, and non-invasiveness, allowing simultaneous analysis of multiple types of molecules, thus improving research efficiency and accuracy. Integrative omics analysis of EVs in the context of hepatic precancerous lesions and hepatocellular carcinoma has been seldom reported, but multi-omics studies with applications to other cancers are continuously emerging, especially in the area of biomarker discovery. Using a combination of proteomics, metabolomics, and lipidomics, Eylem et al. (2020) characterized colorectal cancer (CRC)-derived exosomes and identified signature molecules that could differentiate CRC patients from healthy individuals. Luo et al. (2021) dissected the transcriptome and proteome profiles of exosomes released from human lung adenocarcinoma stem-like cells (LSLC) and found that several LSLC markers were highly expressed and associated with poor survival, which may serve as promising liquid biopsy biomarkers for the diagnosis of lung adenocarcinoma. Given the biodiversity and heterogeneity of EVs, such integrative multi-omics data analysis can be used to reveal the multi-level biological data of EVs to explore the functions of EVs more deeply and accurately.
However, multi-omics analysis of EVs also faces some challenges. First, the purification and quantification of EVs are still problematic, thus, it remains a difficult task to extract pure EVs from complex body fluids accurately and efficiently. Secondly, the molecular composition of EVs is heterogeneous and complex, and how identifying and classifying different types of EVs and their molecular compositions is a difficult problem. In addition, EV multi-omics analysis requires the analysis and integration of a large amount of data, so how to effectively integrate data and perform bioinformatics analysis to systematically and comprehensively annotate complex biological network regulation is also a challenge.
2.4.2 Systems biology
Systems biology, as a rookie in the post-genomic era, is becoming a more sophisticated approach to integromices. This approach, characterized by the integration of systems theory and experimental and computational methods, combines experimental data at multiple molecular levels with computational models and treats the system as a whole, providing a new perspective for revealing the role of EVs in human health and disease (Gézsi et al., 2019; Çelebier and Ercan, 2021). The integrated analysis of the interactions of different molecules in the genome, transcriptome, proteome, metabolome, etc., helps to identify the best biomarker candidates. With the development of multi-omics diagnostic tools, and of new computational methods for data integration, the combination of EVs with multi-omics for the diagnosis of early HCC will illuminate a profound understanding and progress in the future.
3 Applications of EVs to immunotherapy
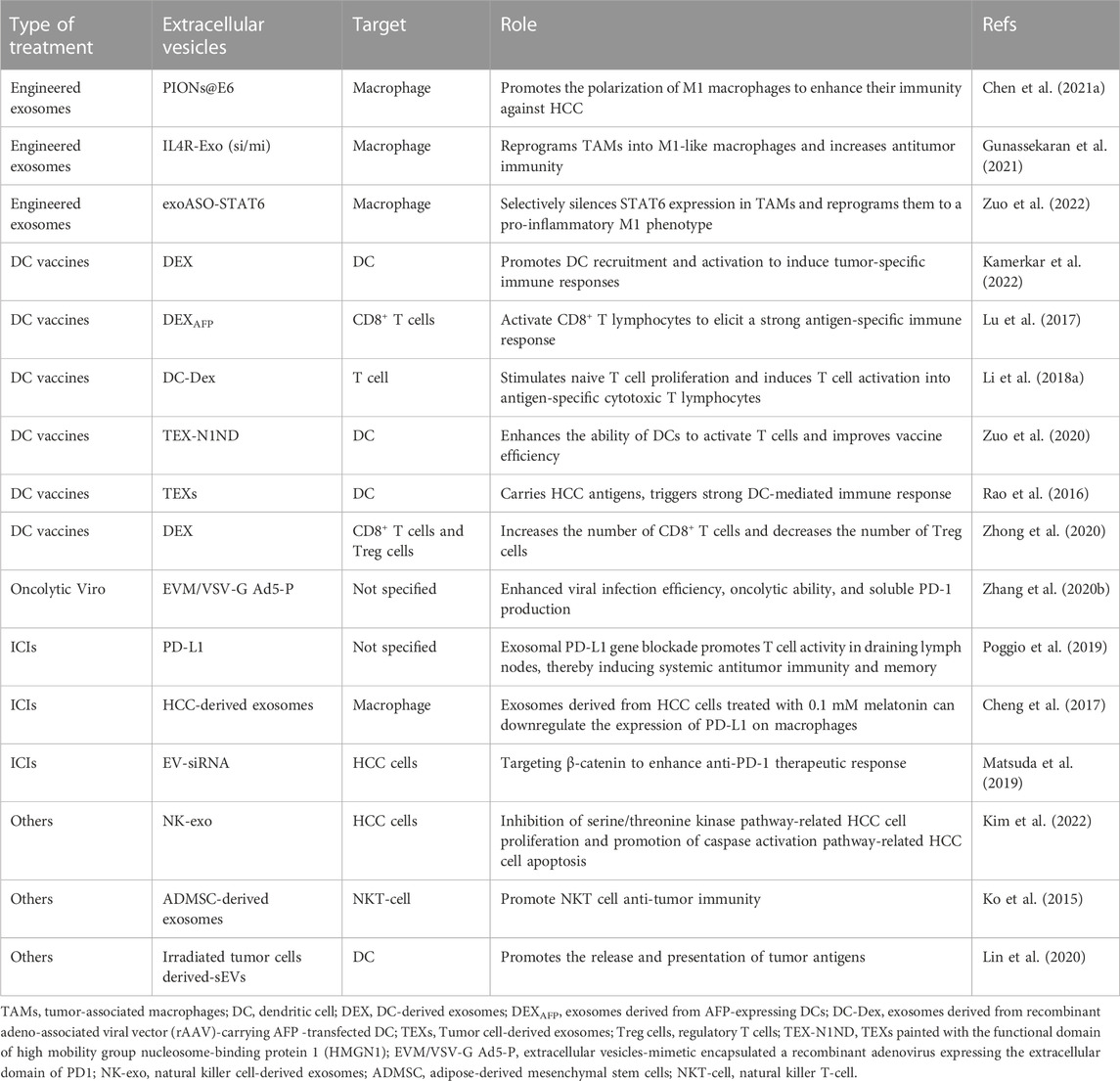
Since the immunoregulatory potential of EVs was first discovered in 1998, novel EVs-centered cancer immunotherapy has been extensively explored (Zitvogel et al., 1998). Recently, EVs have been proposed as a cell-free therapeutic strategy that has attracted considerable attention in the field of cancer immunotherapy (Liu C. et al., 2022). The involvement of EVs in the crosstalk between tumor cells and immune cells in the microenvironment provides many opportunities for cancer immunotherapy. Macrophages are one of the prominent immune cells in the tumor microenvironment and can be polarized to assume different phenotypes under different microenvironment (Lewis and Pollard, 2006). There are two main subgroups designated M1 (classically activated macrophages) and M2 (alternatively activated macrophages) (Mosser and Edwards, 2008). Tumor-associated macrophages (TAMs) are mainly of the M2 phenotype, which play an important role in tumor progression and immunosuppression (Galdiero et al., 2013). Studies have shown that tumor cell-derived EVs can target neighboring macrophages and modulate their conversion to M2 (Li X. et al., 2018); in turn, M2 macrophage-derived EVs can facilitate tumor progression and CD8+ T cell exhaustion in HCC mice (Chen J. et al., 2021; Pu et al., 2021). Several engineered EV therapeutic candidates targeting macrophages have been demonstrated to reprogram TAMs to a pro-inflammatory M1 phenotype and enhance immunity against HCC, resulting in marked inhibition of tumor growth (Chen H. et al., 2021; Gunassekaran et al., 2021; Kamerkar et al., 2022).
Immune cell-derived EVs also display the potential for HCC immunotherapy. It was reported that dendritic cell (DC)-derived EVs (DEX) could elicit strong antigen-specific antitumor immune responses and significant tumor regression in HCC mice after intravenous administration (Zuo et al., 2022). Some previous studies also showed that EVs from tumor antigen-expressing DCs can reshape the tumor immune microenvironment, resulting in significant tumor growth retardation and survival prolongation in HCC mice (Lu et al., 2017; Li J. et al., 2018). In addition, pulsed stimulation with tumor cell-derived EVs (TEX) carrying multiple HCC antigens can potentiate DCs immunogenicity and improve vaccines efficiency (Rao et al., 2016; Zuo et al., 2020). Another study also suggested that DEX could induce an equivalent antitumor immune-enhancing effect to DC when combined with microwave ablation (Zhong et al., 2020). These results indicate that DEX has a great application prospect as a cell-free tumor vaccine or a natural antitumor adjuvant, opening up a new avenue for HCC immunotherapy. Moreover, EVs derived from other immune cells such as natural killer cells (NK) have also shown specific tumor-killing effects on HCC cells (Kim et al., 2022); Wu J. et al. (2022) have developed a motion bioreactor that can produce NK EVs on a large scale, which provides technical support for promoting the clinical translation of EVs. The cytotoxic effect of immune cell-derived EVs on tumors greatly enriches the role of EVs in cell-free immunotherapy. With the understanding of the mechanisms of these phenomena, more advances will be elucidated.
Another potential usage of EVs in the HCC immunotherapy is to enhance the efficacy of immune checkpoint inhibitors (ICIs). Systemic therapy based on ICIs has gradually been the mainstay of HCC immunotherapy, but only about 20% of patients show effective response to the treatment. Recent studies have demonstrated that specific types of TEX could promote the immune escape of HCC by inducing the expansion of regulatory T cells and regulatory B cells (Ye et al., 2018; Huang et al., 2022). Wei et al. (2021)found that the HMGB1-RICTOR crosstalk networks epigenetically impede the response to anti-PD-L1 immunotherapy by upregulating the activity of PD-L1+ exosomes in HCC. In addition to T cells and B cells, TEX can also inhibit various innate immune cells, such as NK cells and macrophages. Studies have indicated that specific types of TEX could inhibit the function of NK cells by inhibiting the secretion of IFN-γ and TNF-α in NK cells, and reduce the proportion of NK cells and the tumor infiltration, thus leading to the resistance to anti-PD1 therapy in HCC (Zhang P. F. et al., 2020). Other studies have also found that HCC-derived EVs promote immunosuppression and resistance to anti-PD1 treatment through the elevation of CD39 expression, while inhibition of the ATP-adenosine pathway by targeting CD39 on macrophages can rescue anti-PD1 therapy in HCC (Lu et al., 2021). These findings provide potential therapeutic strategies for HCC patients. Besides, a recent study demonstrated that PD-L1 secreted by TEXs can inhibit T cell activation in draining lymph nodes and promote tumor growth in an immune-dependent manner; whereas blocking EV PD-L1 suppresses tumor growth and overcome resistance to anti-PD-L1 antibodies (Poggio et al., 2019). In addition, altering the function of TEXs is able to downregulate the expression of PD-L1 on macrophages and attenuate the immunosuppressive state, providing a way to improve the efficacy of immunotherapy in HCC patients (Cheng et al., 2017). These studies suggested that immune checkpoint blockade based on EVs is a promising strategy for cancer immunotherapy.
Apart from as carriers to transport various molecules, EVs could act as the barriers to protect the content in a relatively stable milieu by isolating the inside from the outside. Oncolytic virus is a class of natural viruses, which has been genetically reconstructed to function as the tumor vaccine via specifically infecting and killing tumor cells (Zhang K. J. et al., 2012). However, owing to possessing strong immunogenicity, oncolytic virus usually triggers vehement self-attacking immune response to eliminate themselves. To avoid this situation, Zhang Y. et al. (2020) proposed a new strategy for using EVs to pack the oncolytic adenoviruses, and the results showed that EVs-mimetic encapsulation technology can increase virus infection and shield them from neutralizing antibodies in the serum, thus significantly improving the efficacy of oncolytic virotherapy. In another study, EVs was reformed to deliver small interfering RNA specific for β-catenin with the protection from the degradation, which could effectively strengthen the therapeutic response of HCC to anti-PD-1antibodies (Matsuda et al., 2019). In summary, EVs can exert an immunotherapeutic effect through one or more strategies, which provides a new idea for HCC immunotherapy.
Although EVs-based immunotherapy has shown great potential in preclinical studies, there are no reports of its application in the immunotherapy of HCC patients so far. Therefore, there is still a long way to go in implementing it into routine clinical practice. Current tentative applications of extracellular vesicles in clinical HCC immunotherapies were summarized in Table 3.
4 Conclusion
A growing body of research has provided valuable knowledge and understanding of the role of EVs in HCC. Although EVs hold great promise as potential early diagnostic biomarkers as well as novel cancer immunotherapeutic targets, their use in clinical settings first requires methodological standardization. It has been recognized that EV-based studies are particularly challenging because of their complex biological origins and enormous heterogeneity in size, composition, and source (van Niel et al., 2018). The advancement of high-throughput omics technology provides a good perspective for deciphering the molecular cargo of EVs and enables the collection and integration of data and information at different molecular levels. The information obtained through multi-omics techniques will provide new opportunities for the diagnosis and treatment of HCC and will improve the prognosis of HCC patients. Although further experimental confirmation and clinical validations are required for findings made through emerging omics approaches, we believe these approaches will transform medical practice in the near future. In the future, the implementation of various omics-based technologies and the integration of systems biology methods for the development of EV-based HCC diagnostics will become a promising trend. On the other hand, although significant progress has been made in the application of EVs in the immunotherapy of HCC, it is still in a secondary position in clinical practice at present. Therefore, future work should simultaneously focus on the rational design of engineered EVs in immune regulation and expand their application in cancer immunotherapy.
In addition, it is a little regrettable that although hepatic precancerous lesion was identified clinically more than 10 years ago, the underlying molecular mechanisms involved in its progression and transformation remain unclear, and little is known about the role that EVs play in it. With the advent of the “omics era” and the deepening understanding of EVs, we expect more research on hepatic precancerous lesions and tumorigenesis to provide new strategies for early diagnosis and intervention of disease.
Author contributions
ZY conceived the idea for the manuscript. XL wrote the first draft of the manuscript. XZ and JS conducted literature collection. YYL and YL draw the figure and tables. YG and HF edited the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by National Natural Science Foundation of China (Nos. 82222074, 81874436, 82074154, 81774240), Three-year Action Plan for the Development of Chinese Medicine in Shanghai [Nos. ZY (2018-2020)-CCCX-2003-01], Shanghai Key Clinical Specialty Construction Project (No. shslczdzk01201), The Siming Scholar from Shanghai Shuguang Hospital (SGXZ-201904), Youth Tip-top Talent program in Shanghai, Xinglin Youth Scholar from Shanghai University of Traditional Chinese Medicine.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abd El Gwad, A., Matboli, M., El-Tawdi, A., Habib, E. K., Shehata, H., Ibrahim, D., et al. (2018). Role of exosomal competing endogenous RNA in patients with hepatocellular carcinoma. J. Cell Biochem 119 (10), 8600–8610. doi:10.1002/jcb.27109
Arbelaiz, A., Azkargorta, M., Krawczyk, M., Santos-Laso, A., Lapitz, A., Perugorria, M. J., et al. (2017). Serum extracellular vesicles contain protein biomarkers for primary sclerosing cholangitis and cholangiocarcinoma. Hepatology 66 (4), 1125–1143. doi:10.1002/hep.29291
Aydin, Y., Koksal, A. R., Thevenot, P., Chava, S., Heidari, Z., Lin, D., et al. (2021). Experimental validation of novel glypican 3 exosomes for the detection of hepatocellular carcinoma in liver cirrhosis. J. Hepatocell. Carcinoma 8, 1579–1596. doi:10.2147/jhc.S327339
Becker, A., Thakur, B. K., Weiss, J. M., Kim, H. S., Peinado, H., and Lyden, D. (2016). Extracellular vesicles in cancer: Cell-to-Cell mediators of metastasis. Cancer Cell 30 (6), 836–848. doi:10.1016/j.ccell.2016.10.009
Borzio, M., Fargion, S., Borzio, F., Fracanzani, A. L., Croce, A. M., Stroffolini, T., et al. (2003). Impact of large regenerative, low grade and high grade dysplastic nodules in hepatocellular carcinoma development. J. Hepatol. 39 (2), 208–214. doi:10.1016/s0168-8278(03)00190-9
Cabiati, M., Salvadori, C., Basta, G., Del Turco, S., Aretini, P., Cecchettini, A., et al. (2022). miRNA and long non-coding RNA transcriptional expression in hepatocellular carcinoma cell line-secreted extracellular vesicles. Clin. Exp. Med. 22 (2), 245–255. doi:10.1007/s10238-021-00744-6
Cai, J., Han, Y., Ren, H., Chen, C., He, D., Zhou, L., et al. (2013). Extracellular vesicle-mediated transfer of donor genomic DNA to recipient cells is a novel mechanism for genetic influence between cells. J. Mol. Cell Biol. 5 (4), 227–238. doi:10.1093/jmcb/mjt011
Çelebier, M., and Ercan, A. (2021). Recent approaches to integrate multiomics data on system biology. Curr. Anal. Chem. 17 (9), 1243–1251. doi:10.2174/1573411016999200525003730
Chen, H., Jiang, S., Zhang, P., Ren, Z., and Wen, J. (2021a). Exosomes synergized with PIONs@E6 enhance their immunity against hepatocellular carcinoma via promoting M1 macrophages polarization. Int. Immunopharmacol. 99, 107960. doi:10.1016/j.intimp.2021.107960
Chen, J., Lin, Z., Liu, L., Zhang, R., Geng, Y., Fan, M., et al. (2021b). GOLM1 exacerbates CD8(+) T cell suppression in hepatocellular carcinoma by promoting exosomal PD-L1 transport into tumor-associated macrophages. Signal Transduct. Target Ther. 6 (1), 397. doi:10.1038/s41392-021-00784-0
Chen, S., Mao, Y., Chen, W., Liu, C., Wu, H., Zhang, J., et al. (2022a). Serum exosomal miR-34a as a potential biomarker for the diagnosis and prognostic of hepatocellular carcinoma. J. Cancer 13 (5), 1410–1417. doi:10.7150/jca.57205
Chen, W., Quan, Y., Fan, S., Wang, H., Liang, J., Huang, L., et al. (2020). Exosome-transmitted circular RNA hsa_circ_0051443 suppresses hepatocellular carcinoma progression. Cancer Lett. 475, 119–128. doi:10.1016/j.canlet.2020.01.022
Chen, Y. Q., Zheng, L., Zhou, J., Wang, P., Wang, L., Zhang, Y., et al. (2022b). Evaluation of plasma LC3B(+)extracellular vesicles as a potential novel diagnostic marker for hepatocellular carcinoma. Int. Immunopharmacol. 108, 108760. doi:10.1016/j.intimp.2022.108760
Cheng, L., Liu, J., Liu, Q., Liu, Y., Fan, L., Wang, F., et al. (2017). Exosomes from melatonin treated hepatocellularcarcinoma cells alter the immunosupression status through STAT3 pathway in macrophages. Int. J. Biol. Sci. 13 (6), 723–734. doi:10.7150/ijbs.19642
Cho, H. J., Baek, G. O., Seo, C. W., Ahn, H. R., Sung, S., Son, J. A., et al. (2020a). Exosomal microRNA-4661-5p-based serum panel as a potential diagnostic biomarker for early-stage hepatocellular carcinoma. Cancer Med. 9 (15), 5459–5472. doi:10.1002/cam4.3230
Cho, H. J., Baek, G. O., Yoon, M. G., Ahn, H. R., Son, J. A., Kim, S. S., et al. (2021). Overexpressed proteins in HCC cell-derived exosomes, CCT8, and cofilin-1 are potential biomarkers for patients with HCC. Diagn. (Basel) 11 (7), 1221. doi:10.3390/diagnostics11071221
Cho, H. J., Eun, J. W., Baek, G. O., Seo, C. W., Ahn, H. R., Kim, S. S., et al. (2020b). Serum exosomal MicroRNA, miR-10b-5p, as a potential diagnostic biomarker for early-stage hepatocellular carcinoma. J. Clin. Med. 9 (1), 281. doi:10.3390/jcm9010281
Cho, Y. K., Wook Chung, J., Kim, Y., Je Cho, H., and Hyun Yang, S. (2011). Radiofrequency ablation of high-grade dysplastic nodules. Hepatology 54 (6), 2005–2011. doi:10.1002/hep.24589
Colombo, M., Raposo, G., and Théry, C. (2014). Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev. Cell Dev. Biol. 30, 255–289. doi:10.1146/annurev-cellbio-101512-122326
Cui, Y., Xu, H. F., Liu, M. Y., Xu, Y. J., He, J. C., Zhou, Y., et al. (2019). Mechanism of exosomal microRNA-224 in development of hepatocellular carcinoma and its diagnostic and prognostic value. World J. Gastroenterol. 25 (15), 1890–1898. doi:10.3748/wjg.v25.i15.1890
Cui, Z., Li, Y., Gao, Y., Kong, L., Lin, Y., and Chen, Y. (2020). Cancer-testis antigen lactate dehydrogenase C4 in hepatocellular carcinoma: A promising biomarker for early diagnosis, efficacy evaluation and prognosis prediction. Aging (Albany NY) 12 (19), 19455–19467. doi:10.18632/aging.103879
Deng, P., Li, M., and Wu, Y. (2022). The predictive efficacy of serum exosomal microRNA-122 and microRNA-148a for hepatocellular carcinoma based on smart healthcare. J. Healthc. Eng. 2022, 5914541. doi:10.1155/2022/5914541
Di, H., Mi, Z., Sun, Y., Liu, X., Liu, X., Li, A., et al. (2020). Nanozyme-assisted sensitive profiling of exosomal proteins for rapid cancer diagnosis. Theranostics 10 (20), 9303–9314. doi:10.7150/thno.46568
Di Leva, G., Garofalo, M., and Croce, C. M. (2014). MicroRNAs in cancer. Annu Rev. Pathol. 9, 287–314. doi:10.1146/annurev-pathol-012513-104715
Elzanowska, J., Semira, C., and Costa-Silva, B. (2021). DNA in extracellular vesicles: Biological and clinical aspects. Mol. Oncol. 15 (6), 1701–1714. doi:10.1002/1878-0261.12777
Eylem, C. C., Yilmaz, M., Derkus, B., Nemutlu, E., Camci, C. B., Yilmaz, E., et al. (2020). Untargeted multi-omic analysis of colorectal cancer-specific exosomes reveals joint pathways of colorectal cancer in both clinical samples and cell culture. Cancer Lett. 469, 186–194. doi:10.1016/j.canlet.2019.10.038
Fu, Q., Zhang, Q., Lou, Y., Yang, J., Nie, G., Chen, Q., et al. (2018). Primary tumor-derived exosomes facilitate metastasis by regulating adhesion of circulating tumor cells via SMAD3 in liver cancer. Oncogene 37 (47), 6105–6118. doi:10.1038/s41388-018-0391-0
Galdiero, M. R., Garlanda, C., Jaillon, S., Marone, G., and Mantovani, A. (2013). Tumor associated macrophages and neutrophils in tumor progression. J. Cell Physiol. 228 (7), 1404–1412. doi:10.1002/jcp.24260
Gézsi, A., Kovács, Á., Visnovitz, T., and Buzás, E. I. (2019). Systems biology approaches to investigating the roles of extracellular vesicles in human diseases. Exp. Mol. Med. 51 (3), 1–11. doi:10.1038/s12276-019-0226-2
Ghosh, S., Bhowmik, S., Majumdar, S., Goswami, A., Chakraborty, J., Gupta, S., et al. (2020). The exosome encapsulated microRNAs as circulating diagnostic marker for hepatocellular carcinoma with low alpha-fetoprotein. Int. J. Cancer 147 (10), 2934–2947. doi:10.1002/ijc.33111
Gunassekaran, G. R., Poongkavithai Vadevoo, S. M., Baek, M. C., and Lee, B. (2021). M1 macrophage exosomes engineered to foster M1 polarization and target the IL-4 receptor inhibit tumor growth by reprogramming tumor-associated macrophages into M1-like macrophages. Biomaterials 278, 121137. doi:10.1016/j.biomaterials.2021.121137
Guo, S., Hu, C., Zhai, X., and Sun, D. (2021). Circular RNA 0006602 in plasma exosomes: A new potential diagnostic biomarker for hepatocellular carcinoma. Am. J. Transl. Res. 13 (6), 6001–6015.
Hagey, D. W., Kordes, M., Görgens, A., Mowoe, M. O., Nordin, J. Z., Moro, C. F., et al. (2021). Extracellular vesicles are the primary source of blood-borne tumour-derived mutant KRAS DNA early in pancreatic cancer. J. Extracell. Vesicles 10 (12), e12142. doi:10.1002/jev2.12142
Hartke, J., Johnson, M., and Ghabril, M. (2017). The diagnosis and treatment of hepatocellular carcinoma. Semin. Diagn Pathol. 34 (2), 153–159. doi:10.1053/j.semdp.2016.12.011
Hasanin, A. H., Matboli, M., and Seleem, H. S. (2020). Hesperidin suppressed hepatic precancerous lesions via modulation of exophagy in rats. J. Cell Biochem 121 (2), 1295–1306. doi:10.1002/jcb.29363
Hollebecque, A., Malka, D., Ferté, C., Ducreux, M., and Boige, V. (2015). Systemic treatment of advanced hepatocellular carcinoma: From disillusions to new horizons. Eur. J. Cancer 51 (3), 327–339. doi:10.1016/j.ejca.2014.12.005
Honoré, B., Baandrup, U., and Vorum, H. (2004). Heterogeneous nuclear ribonucleoproteins F and H/H' show differential expression in normal and selected cancer tissues. Exp. Cell Res. 294 (1), 199–209. doi:10.1016/j.yexcr.2003.11.011
Hou, P. P., Luo, L. J., Chen, H. Z., Chen, Q. T., Bian, X. L., Wu, S. F., et al. (2020). Ectosomal PKM2 promotes HCC by inducing macrophage differentiation and remodeling the tumor microenvironment. Mol. Cell 78 (6), 1192–1206. doi:10.1016/j.molcel.2020.05.004
Hou, Z., Xu, X., Fu, X., Tao, S., Zhou, J., Liu, S., et al. (2017). HBx-related long non-coding RNA MALAT1 promotes cell metastasis via up-regulating LTBP3 in hepatocellular carcinoma. Am. J. Cancer Res. 7 (4), 845–856.
Hu, K., Li, N. F., Li, J. R., Chen, Z. G., Wang, J. H., and Sheng, L. Q. (2021). Exosome circCMTM3 promotes angiogenesis and tumorigenesis of hepatocellular carcinoma through miR-3619-5p/SOX9. Hepatol. Res. 51 (11), 1139–1152. doi:10.1111/hepr.13692
Huang, M., Huang, X., and Huang, N. (2022). Exosomal circGSE1 promotes immune escape of hepatocellular carcinoma by inducing the expansion of regulatory T cells. Cancer Sci. 113 (6), 1968–1983. doi:10.1111/cas.15365
Huang, X., Sun, L., Wen, S., Deng, D., Wan, F., He, X., et al. (2020). RNA sequencing of plasma exosomes revealed novel functional long noncoding RNAs in hepatocellular carcinoma. Cancer Sci. 111 (9), 3338–3349. doi:10.1111/cas.14516
Huarte, M. (2015). The emerging role of lncRNAs in cancer. Nat. Med. 21 (11), 1253–1261. doi:10.1038/nm.3981
International Consensus Group for Hepatocellular Neoplasia (2009). Pathologic diagnosis of early hepatocellular carcinoma: A report of the international consensus group for hepatocellular neoplasia. Hepatology 49 (2), 658–664. doi:10.1002/hep.22709
Jiang, K., Dong, C., Yin, Z., Li, R., Mao, J., Wang, C., et al. (2020). Exosome-derived ENO1 regulates integrin α6β4 expression and promotes hepatocellular carcinoma growth and metastasis. Cell Death Dis. 11 (11), 972. doi:10.1038/s41419-020-03179-1
Julich-Haertel, H., Urban, S. K., Krawczyk, M., Willms, A., Jankowski, K., Patkowski, W., et al. (2017). Cancer-associated circulating large extracellular vesicles in cholangiocarcinoma and hepatocellular carcinoma. J. Hepatol. 67 (2), 282–292. doi:10.1016/j.jhep.2017.02.024
Kamerkar, S., Leng, C., Burenkova, O., Jang, S. C., McCoy, C., Zhang, K., et al. (2022). Exosome-mediated genetic reprogramming of tumor-associated macrophages by exoASO-STAT6 leads to potent monotherapy antitumor activity. Sci. Adv. 8 (7), eabj7002. doi:10.1126/sciadv.abj7002
Kim, H. Y., Min, H. K., Song, H. W., Yoo, A., Lee, S., Kim, K. P., et al. (2022). Delivery of human natural killer cell-derived exosomes for liver cancer therapy: An in vivo study in subcutaneous and orthotopic animal models. Drug Deliv. 29 (1), 2897–2911. doi:10.1080/10717544.2022.2118898
Kim, S. S., Baek, G. O., Ahn, H. R., Sung, S., Seo, C. W., Cho, H. J., et al. (2020). Serum small extracellular vesicle-derived LINC00853 as a novel diagnostic marker for early hepatocellular carcinoma. Mol. Oncol. 14 (10), 2646–2659. doi:10.1002/1878-0261.12745
Kim, S. S., Baek, G. O., Son, J. A., Ahn, H. R., Yoon, M. K., Cho, H. J., et al. (2021). Early detection of hepatocellular carcinoma via liquid biopsy: Panel of small extracellular vesicle-derived long noncoding RNAs identified as markers. Mol. Oncol. 15 (10), 2715–2731. doi:10.1002/1878-0261.13049
Ko, S. F., Yip, H. K., Zhen, Y. Y., Lee, C. C., Lee, C. C., Huang, C. C., et al. (2015). Adipose-derived mesenchymal stem cell exosomes suppress hepatocellular carcinoma growth in a rat model: Apparent diffusion coefficient, natural killer T-cell responses, and histopathological features. Stem Cells Int. 2015, 853506. doi:10.1155/2015/853506
Kobayashi, M., Ikeda, K., Hosaka, T., Sezaki, H., Someya, T., Akuta, N., et al. (2006). Dysplastic nodules frequently develop into hepatocellular carcinoma in patients with chronic viral hepatitis and cirrhosis. Cancer 106 (3), 636–647. doi:10.1002/cncr.21607
Koslowski, M., Türeci, O., Bell, C., Krause, P., Lehr, H. A., Brunner, J., et al. (2002). Multiple splice variants of lactate dehydrogenase C selectively expressed in human cancer. Cancer Res. 62 (22), 6750–6755.
Kristensen, L. S., Andersen, M. S., Stagsted, L. V. W., Ebbesen, K. K., Hansen, T. B., and Kjems, J. (2019). The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 20 (11), 675–691. doi:10.1038/s41576-019-0158-7
Lázaro-Ibáñez, E., Lässer, C., Shelke, G. V., Crescitelli, R., Jang, S. C., Cvjetkovic, A., et al. (2019). DNA analysis of low- and high-density fractions defines heterogeneous subpopulations of small extracellular vesicles based on their DNA cargo and topology. J. Extracell. Vesicles 8 (1), 1656993. doi:10.1080/20013078.2019.1656993
Lewis, C. E., and Pollard, J. W. (2006). Distinct role of macrophages in different tumor microenvironments. Cancer Res. 66 (2), 605–612. doi:10.1158/0008-5472.Can-05-4005
Li, J., Huang, S., Zhou, Z., Lin, W., Chen, S., Chen, M., et al. (2018a). Exosomes derived from rAAV/AFP-transfected dendritic cells elicit specific T cell-mediated immune responses against hepatocellular carcinoma. Cancer Manag. Res. 10, 4945–4957. doi:10.2147/cmar.S178326
Li, W., Ding, X., Wang, S., Xu, L., Yin, T., Han, S., et al. (2020a). Downregulation of serum exosomal miR-320d predicts poor prognosis in hepatocellular carcinoma. J. Clin. Lab. Anal. 34 (6), e23239. doi:10.1002/jcla.23239
Li, X., Lei, Y., Wu, M., and Li, N. (2018b). Regulation of macrophage activation and polarization by HCC-derived exosomal lncRNA TUC339. Int. J. Mol. Sci. 19 (10), 2958. doi:10.3390/ijms19102958
Li, Y., Guo, X., Guo, S., Wang, Y., Chen, L., Liu, Y., et al. (2020b). Next generation sequencing-based analysis of mitochondrial DNA characteristics in plasma extracellular vesicles of patients with hepatocellular carcinoma. Oncol. Lett. 20 (3), 2820–2828. doi:10.3892/ol.2020.11831
Li, Y., Zhao, J., Yu, S., Wang, Z., He, X., Su, Y., et al. (2019). Extracellular vesicles long RNA sequencing reveals abundant mRNA, circRNA, and lncRNA in human blood as potential biomarkers for cancer diagnosis. Clin. Chem. 65 (6), 798–808. doi:10.1373/clinchem.2018.301291
Li, Y., Zheng, Q., Bao, C., Li, S., Guo, W., Zhao, J., et al. (2015). Circular RNA is enriched and stable in exosomes: A promising biomarker for cancer diagnosis. Cell Res. 25 (8), 981–984. doi:10.1038/cr.2015.82
Lin, H., and Zhang, Z. (2019). Diagnostic value of a microRNA signature panel in exosomes for patients with hepatocellular carcinoma. Int. J. Clin. Exp. Pathol. 12 (4), 1478–1487.
Lin, J., Lin, W., Bai, Y., Liao, Y., Lin, Q., Chen, L., et al. (2022). Identification of exosomal hsa-miR-483-5p as a potential biomarker for hepatocellular carcinoma via microRNA expression profiling of tumor-derived exosomes. Exp. Cell Res. 417 (2), 113232. doi:10.1016/j.yexcr.2022.113232
Lin, W., Xu, Y., Chen, X., Liu, J., Weng, Y., Zhuang, Q., et al. (2020). Radiation-induced small extracellular vesicles as "carriages" promote tumor antigen release and trigger antitumor immunity. Theranostics 10 (11), 4871–4884. doi:10.7150/thno.43539
Liu, B. H. M., Tey, S. K., Mao, X., Ma, A. P. Y., Yeung, C. L. S., Wong, S. W. K., et al. (2021). TPI1-reduced extracellular vesicles mediated by Rab20 downregulation promotes aerobic glycolysis to drive hepatocarcinogenesis. J. Extracell. Vesicles 10 (10), e12135. doi:10.1002/jev2.12135
Liu, C., Wang, Y., Li, L., He, D., Chi, J., Li, Q., et al. (2022a). Engineered extracellular vesicles and their mimetics for cancer immunotherapy. J. Control Release 349, 679–698. doi:10.1016/j.jconrel.2022.05.062
Liu, H., Tian, Y., Xue, C., Niu, Q., Chen, C., and Yan, X. (2022b). Analysis of extracellular vesicle DNA at the single-vesicle level by nano-flow cytometry. J. Extracell. Vesicles 11 (4), e12206. doi:10.1002/jev2.12206
Llovet, J. M., Kelley, R. K., Villanueva, A., Singal, A. G., Pikarsky, E., Roayaie, S., et al. (2021). Hepatocellular carcinoma. Nat. Rev. Dis. Prim. 7 (1), 6. doi:10.1038/s41572-020-00240-3
Lu, J. C., Zhang, P. F., Huang, X. Y., Guo, X. J., Gao, C., Zeng, H. Y., et al. (2021). Amplification of spatially isolated adenosine pathway by tumor-macrophage interaction induces anti-PD1 resistance in hepatocellular carcinoma. J. Hematol. Oncol. 14 (1), 200. doi:10.1186/s13045-021-01207-x
Lu, Y., Duan, Y., Xu, Q., Zhang, L., Chen, W., Qu, Z., et al. (2020). Circulating exosome-derived bona fide long non-coding RNAs predicting the occurrence and metastasis of hepatocellular carcinoma. J. Cell Mol. Med. 24 (2), 1311–1318. doi:10.1111/jcmm.14783
Lu, Z., Zuo, B., Jing, R., Gao, X., Rao, Q., Liu, Z., et al. (2017). Dendritic cell-derived exosomes elicit tumor regression in autochthonous hepatocellular carcinoma mouse models. J. Hepatol. 67 (4), 739–748. doi:10.1016/j.jhep.2017.05.019
Luo, H. T., Zheng, Y. Y., Tang, J., Shao, L. J., Mao, Y. H., Yang, W., et al. (2021). Dissecting the multi-omics atlas of the exosomes released by human lung adenocarcinoma stem-like cells. NPJ Genom Med. 6 (1), 48. doi:10.1038/s41525-021-00217-5
Luo, P., Mao, K., Xu, J., Wu, F., Wang, X., Wang, S., et al. (2020). Metabolic characteristics of large and small extracellular vesicles from pleural effusion reveal biomarker candidates for the diagnosis of tuberculosis and malignancy. J. Extracell. Vesicles 9 (1), 1790158. doi:10.1080/20013078.2020.1790158
Luo, P., Yin, P., Hua, R., Tan, Y., Li, Z., Qiu, G., et al. (2018). A Large-scale, multicenter serum metabolite biomarker identification study for the early detection of hepatocellular carcinoma. Hepatology 67 (2), 662–675. doi:10.1002/hep.29561
Lyu, L., Yang, W., Yao, J., Wang, H., Zhu, J., Jin, A., et al. (2021). The diagnostic value of plasma exosomal hsa_circ_0070396 for hepatocellular carcinoma. Biomark. Med. 15 (5), 359–371. doi:10.2217/bmm-2020-0476
Malkin, E. Z., and Bratman, S. V. (2020). Bioactive DNA from extracellular vesicles and particles. Cell Death Dis. 11 (7), 584. doi:10.1038/s41419-020-02803-4
Marar, C., Starich, B., and Wirtz, D. (2021). Extracellular vesicles in immunomodulation and tumor progression. Nat. Immunol. 22 (5), 560–570. doi:10.1038/s41590-021-00899-0
Matboli, M., ElGwad, A. A., Hasanin, A. H., El-Tawdi, A., Habib, E. K., Elmansy, R. A., et al. (2019). Pantoprazole attenuates tumorigenesis via inhibition of exosomal secretion in a rat model of hepatic precancerous lesion induced by diethylnitrosamine and 2-acetamidofluorene. J. Cell Biochem 120 (9), 14946–14959. doi:10.1002/jcb.28757
Matsuda, A., Ishiguro, K., Yan, I. K., and Patel, T. (2019). Extracellular vesicle-based therapeutic targeting of β-catenin to modulate anticancer immune responses in hepatocellular cancer. Hepatol. Commun. 3 (4), 525–541. doi:10.1002/hep4.1311
Mjelle, R., Dima, S. O., Bacalbasa, N., Chawla, K., Sorop, A., Cucu, D., et al. (2019). Comprehensive transcriptomic analyses of tissue, serum, and serum exosomes from hepatocellular carcinoma patients. BMC Cancer 19 (1), 1007. doi:10.1186/s12885-019-6249-1
Mohammad, H. S., Kurokohchi, K., Yoneyama, H., Tokuda, M., Morishita, A., Jian, G., et al. (2008). Annexin A2 expression and phosphorylation are up-regulated in hepatocellular carcinoma. Int. J. Oncol. 33 (6), 1157–1163.
Mosser, D. M., and Edwards, J. P. (2008). Exploring the full spectrum of macrophage activation. Nat. Rev. Immunol. 8 (12), 958–969. doi:10.1038/nri2448
Muhammad Yusuf, A. N., Raja Ali, R. A., Muhammad Nawawi, K. N., and Mokhtar, N. M. (2020). Potential biomarkers in NASH-induced liver cirrhosis with hepatocellular carcinoma: A preliminary work on roles of exosomal miR-182, miR-301a, and miR-373. Malays J. Pathol. 42 (3), 377–384.
Pineau, P., Volinia, S., McJunkin, K., Marchio, A., Battiston, C., Terris, B., et al. (2010). miR-221 overexpression contributes to liver tumorigenesis. Proc. Natl. Acad. Sci. U S A 107 (1), 264–269. doi:10.1073/pnas.0907904107
Poggio, M., Hu, T., Pai, C. C., Chu, B., Belair, C. D., Chang, A., et al. (2019). Suppression of exosomal PD-L1 induces systemic anti-tumor immunity and memory. Cell 177 (2), 414–427. doi:10.1016/j.cell.2019.02.016
Pu, C., Huang, H., Wang, Z., Zou, W., Lv, Y., Zhou, Z., et al. (2018). Extracellular vesicle-associated mir-21 and mir-144 are markedly elevated in serum of patients with hepatocellular carcinoma. Front. Physiol. 9, 930. doi:10.3389/fphys.2018.00930
Pu, J., Xu, Z., Nian, J., Fang, Q., Yang, M., Huang, Y., et al. (2021). M2 macrophage-derived extracellular vesicles facilitate CD8+T cell exhaustion in hepatocellular carcinoma via the miR-21-5p/YOD1/YAP/β-catenin pathway. Cell Death Discov. 7 (1), 182. doi:10.1038/s41420-021-00556-3
Puhka, M., Takatalo, M., Nordberg, M. E., Valkonen, S., Nandania, J., Aatonen, M., et al. (2017). Metabolomic profiling of extracellular vesicles and alternative normalization methods reveal enriched metabolites and strategies to study prostate cancer-related changes. Theranostics 7 (16), 3824–3841. doi:10.7150/thno.19890
Qu, Z., Wu, J., Wu, J., Ji, A., Qiang, G., Jiang, Y., et al. (2017). Exosomal miR-665 as a novel minimally invasive biomarker for hepatocellular carcinoma diagnosis and prognosis. Oncotarget 8 (46), 80666–80678. doi:10.18632/oncotarget.20881
Rao, Q., Zuo, B., Lu, Z., Gao, X., You, A., Wu, C., et al. (2016). Tumor-derived exosomes elicit tumor suppression in murine hepatocellular carcinoma models and humans in vitro. Hepatology 64 (2), 456–472. doi:10.1002/hep.28549
Saari, H., Turunen, T., Lõhmus, A., Turunen, M., Jalasvuori, M., Butcher, S. J., et al. (2020). Extracellular vesicles provide a capsid-free vector for oncolytic adenoviral DNA delivery. J. Extracell. Vesicles 9 (1), 1747206. doi:10.1080/20013078.2020.1747206
Saliminejad, K., Khorram Khorshid, H. R., Soleymani Fard, S., and Ghaffari, S. H. (2019). An overview of microRNAs: Biology, functions, therapeutics, and analysis methods. J. Cell Physiol. 234 (5), 5451–5465. doi:10.1002/jcp.27486
Sanchez, J. I., Jiao, J., Kwan, S. Y., Veillon, L., Warmoes, M. O., Tan, L., et al. (2021). Lipidomic profiles of plasma exosomes identify candidate biomarkers for early detection of hepatocellular carcinoma in patients with cirrhosis. Cancer Prev. Res. (Phila) 14 (10), 955–962. doi:10.1158/1940-6207.Capr-20-0612
Sangro, B., Sarobe, P., Hervás-Stubbs, S., and Melero, I. (2021). Advances in immunotherapy for hepatocellular carcinoma. Nat. Rev. Gastroenterol. Hepatol. 18 (8), 525–543. doi:10.1038/s41575-021-00438-0
Sansone, P., Savini, C., Kurelac, I., Chang, Q., Amato, L. B., Strillacci, A., et al. (2017). Packaging and transfer of mitochondrial DNA via exosomes regulate escape from dormancy in hormonal therapy-resistant breast cancer. Proc. Natl. Acad. Sci. U S A 114 (43), E9066–E9075. doi:10.1073/pnas.1704862114
Sato, T., Kondo, F., Ebara, M., Sugiura, N., Okabe, S., Sunaga, M., et al. (2015). Natural history of large regenerative nodules and dysplastic nodules in liver cirrhosis: 28-year follow-up study. Hepatol. Int. 9 (2), 330–336. doi:10.1007/s12072-015-9620-6
Sciarra, A., Di Tommaso, L., Nakano, M., Destro, A., Torzilli, G., Donadon, M., et al. (2016). Morphophenotypic changes in human multistep hepatocarcinogenesis with translational implications. J. Hepatol. 64 (1), 87–93. doi:10.1016/j.jhep.2015.08.031
Seo, W., Gao, Y., He, Y., Sun, J., Xu, H., Feng, D., et al. (2019). ALDH2 deficiency promotes alcohol-associated liver cancer by activating oncogenic pathways via oxidized DNA-enriched extracellular vesicles. J. Hepatol. 71 (5), 1000–1011. doi:10.1016/j.jhep.2019.06.018
Serizawa, N., Tian, J., Fukada, H., Baghy, K., Scott, F., Chen, X., et al. (2015). Galectin 3 regulates HCC cell invasion by RhoA and MLCK activation. Lab. Invest 95 (10), 1145–1156. doi:10.1038/labinvest.2015.77
Sheng, L. Q., Li, J. R., Qin, H., Liu, L., Zhang, D. D., Zhang, Q., et al. (2020). Blood exosomal micro ribonucleic acid profiling reveals the complexity of hepatocellular carcinoma and identifies potential biomarkers for differential diagnosis. World J. Gastrointest. Oncol. 12 (10), 1195–1208. doi:10.4251/wjgo.v12.i10.1195
Singh, A., Singh, A. K., Giri, R., Kumar, D., Sharma, R., Valis, M., et al. (2021). The role of microRNA-21 in the onset and progression of cancer. Future Med. Chem. 13 (21), 1885–1906. doi:10.4155/fmc-2021-0096
Skotland, T., Hessvik, N. P., Sandvig, K., and Llorente, A. (2019). Exosomal lipid composition and the role of ether lipids and phosphoinositides in exosome biology. J. Lipid Res. 60 (1), 9–18. doi:10.1194/jlr.R084343
Sohn, W., Kim, J., Kang, S. H., Yang, S. R., Cho, J. Y., Cho, H. C., et al. (2015). Serum exosomal microRNAs as novel biomarkers for hepatocellular carcinoma. Exp. Mol. Med. 47 (9), e184. doi:10.1038/emm.2015.68
Sorop, A., Iacob, R., Iacob, S., Constantinescu, D., Chitoiu, L., Fertig, T. E., et al. (2020). Plasma small extracellular vesicles derived miR-21-5p and miR-92a-3p as potential biomarkers for hepatocellular carcinoma screening. Front. Genet. 11, 712. doi:10.3389/fgene.2020.00712
Su, H., Yang, J. R., Xu, T., Huang, J., Xu, L., Yuan, Y., et al. (2009). MicroRNA-101, down-regulated in hepatocellular carcinoma, promotes apoptosis and suppresses tumorigenicity. Cancer Res. 69 (3), 1135–1142. doi:10.1158/0008-5472.Can-08-2886
Sun, L., Su, Y., Liu, X., Xu, M., Chen, X., Zhu, Y., et al. (2018). Serum and exosome long non coding RNAs as potential biomarkers for hepatocellular carcinoma. J. Cancer 9 (15), 2631–2639. doi:10.7150/jca.24978
Sun, L., Xu, M., Zhang, G., Dong, L., Wu, J., Wei, C., et al. (2021). Identification of circulating exosomal miR-101 and miR-125b panel act as a potential biomarker for hepatocellular carcinoma. Int. J. Genomics 2021, 1326463. doi:10.1155/2021/1326463
Sun, N., Lee, Y. T., Zhang, R. Y., Kao, R., Teng, P. C., Yang, Y., et al. (2020). Purification of HCC-specific extracellular vesicles on nanosubstrates for early HCC detection by digital scoring. Nat. Commun. 11 (1), 4489. doi:10.1038/s41467-020-18311-0
Sun, N., Zhang, C., Lee, Y. T., Tran, B. V., Wang, J., Kim, H., et al. (2022). HCC EV ECG score: An extracellular vesicle-based protein assay for detection of early-stage hepatocellular carcinoma. Hepatology 77, 774–788. doi:10.1002/hep.32692
Sun, Y. L., Liu, F., Liu, F., and Zhao, X. H. (2016). Protein and gene expression characteristics of heterogeneous nuclear ribonucleoprotein H1 in esophageal squamous cell carcinoma. World J. Gastroenterol. 22 (32), 7322–7331. doi:10.3748/wjg.v22.i32.7322
Sung, H., Ferlay, J., Siegel, R. L., Laversanne, M., Soerjomataram, I., Jemal, A., et al. (2021). Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71 (3), 209–249. doi:10.3322/caac.21660
Takahashi, A., Okada, R., Nagao, K., Kawamata, Y., Hanyu, A., Yoshimoto, S., et al. (2017). Exosomes maintain cellular homeostasis by excreting harmful DNA from cells. Nat. Commun. 8, 15287. doi:10.1038/ncomms15287
Taleb, R. S. Z., Moez, P., Younan, D., Eisenacher, M., Tenbusch, M., Sitek, B., et al. (2017). Quantitative proteome analysis of plasma microparticles for the characterization of HCV-induced hepatic cirrhosis and hepatocellular carcinoma. Proteomics Clin. Appl. 11, 11–12. doi:10.1002/prca.201700014
Tey, S. K., Wong, S. W. K., Chan, J. Y. T., Mao, X., Ng, T. H., Yeung, C. L. S., et al. (2022). Patient pIgR-enriched extracellular vesicles drive cancer stemness, tumorigenesis and metastasis in hepatocellular carcinoma. J. Hepatol. 76 (4), 883–895. doi:10.1016/j.jhep.2021.12.005
Thakur, B. K., Zhang, H., Becker, A., Matei, I., Huang, Y., Costa-Silva, B., et al. (2014). Double-stranded DNA in exosomes: A novel biomarker in cancer detection. Cell Res. 24 (6), 766–769. doi:10.1038/cr.2014.44
Théry, C., Witwer, K. W., Aikawa, E., Alcaraz, M. J., Anderson, J. D., Andriantsitohaina, R., et al. (2018). Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the international society for extracellular vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 7 (1), 1535750. doi:10.1080/20013078.2018.1535750
Tian, Y., Wang, Z., Liu, X., Duan, J., Feng, G., Yin, Y., et al. (2018). Prediction of chemotherapeutic efficacy in non-small cell lung cancer by serum metabolomic profiling. Clin. Cancer Res. 24 (9), 2100–2109. doi:10.1158/1078-0432.Ccr-17-2855
Torralba, D., Baixauli, F., Villarroya-Beltri, C., Fernández-Delgado, I., Latorre-Pellicer, A., Acín-Pérez, R., et al. (2018). Priming of dendritic cells by DNA-containing extracellular vesicles from activated T cells through antigen-driven contacts. Nat. Commun. 9 (1), 2658. doi:10.1038/s41467-018-05077-9
Urban, S. K., Sänger, H., Krawczyk, M., Julich-Haertel, H., Willms, A., Ligocka, J., et al. (2020). Synergistic effects of extracellular vesicle phenotyping and AFP in hepatobiliary cancer differentiation. Liver Int. 40 (12), 3103–3116. doi:10.1111/liv.14585
van Niel, G., D'Angelo, G., and Raposo, G. (2018). Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 19 (4), 213–228. doi:10.1038/nrm.2017.125
Villanueva, A. (2019). Hepatocellular carcinoma. N. Engl. J. Med. 380 (15), 1450–1462. doi:10.1056/NEJMra1713263
von Felden, J., Garcia-Lezana, T., Dogra, N., Gonzalez-Kozlova, E., Ahsen, M. E., Craig, A., et al. (2021). Unannotated small RNA clusters associated with circulating extracellular vesicles detect early stage liver cancer. Gut 71, 2069–2080. doi:10.1136/gutjnl-2021-325036
Wang, B., Majumder, S., Nuovo, G., Kutay, H., Volinia, S., Patel, T., et al. (2009). Role of microRNA-155 at early stages of hepatocarcinogenesis induced by choline-deficient and amino acid-defined diet in C57BL/6 mice. Hepatology 50 (4), 1152–1161. doi:10.1002/hep.23100
Wang, H., Hou, L., Li, A., Duan, Y., Gao, H., and Song, X. (2014a). Expression of serum exosomal microRNA-21 in human hepatocellular carcinoma. Biomed. Res. Int. 2014, 864894. doi:10.1155/2014/864894
Wang, J., Su, L., Chen, X., Li, P., Cai, Q., Yu, B., et al. (2014b). MALAT1 promotes cell proliferation in gastric cancer by recruiting SF2/ASF. Biomed. Pharmacother. 68 (5), 557–564. doi:10.1016/j.biopha.2014.04.007
Wang, S., Chen, G., Lin, X., Xing, X., Cai, Z., Liu, X., et al. (2017). Role of exosomes in hepatocellular carcinoma cell mobility alteration. Oncol. Lett. 14 (6), 8122–8131. doi:10.3892/ol.2017.7257
Wang, S., Yang, Y., Sun, L., Qiao, G., Song, Y., and Liu, B. (2020). Exosomal MicroRNAs as liquid biopsy biomarkers in hepatocellular carcinoma. Onco Targets Ther. 13, 2021–2030. doi:10.2147/ott.S232453
Wang, T., Zhu, H., Xiao, M., and Zhou, S. (2021a). Serum exosomal long noncoding RNA CRNDE as a prognostic biomarker for hepatocellular carcinoma. J. Clin. Lab. Anal. 35 (11), e23959. doi:10.1002/jcla.23959
Wang, Y., Li, G., Wan, F., Dai, B., and Ye, D. (2018a). Prognostic value of D-lactate dehydrogenase in patients with clear cell renal cell carcinoma. Oncol. Lett. 16 (1), 866–874. doi:10.3892/ol.2018.8782
Wang, Y., Pei, L., Yue, Z., Jia, M., Wang, H., and Cao, L. L. (2021b). The potential of serum exosomal hsa_circ_0028861 as the novel diagnostic biomarker of HBV-derived hepatocellular cancer. Front. Genet. 12, 703205. doi:10.3389/fgene.2021.703205
Wang, Y., Zhang, C., Zhang, P., Guo, G., Jiang, T., Zhao, X., et al. (2018b). Serum exosomal microRNAs combined with alpha-fetoprotein as diagnostic markers of hepatocellular carcinoma. Cancer Med. 7 (5), 1670–1679. doi:10.1002/cam4.1390
Wei, Y., Tang, X., Ren, Y., Yang, Y., Song, F., Fu, J., et al. (2021). An RNA-RNA crosstalk network involving HMGB1 and RICTOR facilitates hepatocellular carcinoma tumorigenesis by promoting glutamine metabolism and impedes immunotherapy by PD-L1+ exosomes activity. Signal Transduct. Target Ther. 6 (1), 421. doi:10.1038/s41392-021-00801-2
Wu, D., Yu, Y., Jin, D., Xiao, M. M., Zhang, Z. Y., and Zhang, G. J. (2020). Dual-aptamer modified graphene field-effect transistor nanosensor for label-free and specific detection of hepatocellular carcinoma-derived microvesicles. Anal. Chem. 92 (5), 4006–4015. doi:10.1021/acs.analchem.9b05531
Wu, J., Wu, D., Wu, G., Bei, H. P., Li, Z., Xu, H., et al. (2022a). Scale-out production of extracellular vesicles derived from natural killer cells via mechanical stimulation in a seesaw-motion bioreactor for cancer therapy. Biofabrication 14 (4), 045004. doi:10.1088/1758-5090/ac7eeb
Wu, Z. H., Li, C., Zhang, Y. J., and Lin, R. (2022b). Bioinformatics study revealed significance of exosome transcriptome in hepatocellular carcinoma diagnosis. Front. Cell Dev. Biol. 10, 813701. doi:10.3389/fcell.2022.813701
Xu, H., Chen, Y., Dong, X., and Wang, X. (2018a). Serum exosomal long noncoding RNAs ENSG00000258332.1 and LINC00635 for the diagnosis and prognosis of hepatocellular carcinoma. Cancer Epidemiol. Biomarkers Prev. 27 (6), 710–716. doi:10.1158/1055-9965.Epi-17-0770
Xu, H., Dong, X., Chen, Y., and Wang, X. (2018b). Serum exosomal hnRNPH1 mRNA as a novel marker for hepatocellular carcinoma. Clin. Chem. Lab. Med. 56 (3), 479–484. doi:10.1515/cclm-2017-0327
Xue, X., Wang, X., Zhao, Y., Hu, R., and Qin, L. (2018). Exosomal miR-93 promotes proliferation and invasion in hepatocellular carcinoma by directly inhibiting TIMP2/TP53INP1/CDKN1A. Biochem Biophys. Res. Commun. 502 (4), 515–521. doi:10.1016/j.bbrc.2018.05.208
Xue, X., Zhao, Y., Wang, X., Qin, L., and Hu, R. (2019). Development and validation of serum exosomal microRNAs as diagnostic and prognostic biomarkers for hepatocellular carcinoma. J. Cell Biochem 120 (1), 135–142. doi:10.1002/jcb.27165
Yáñez-Mó, M., Siljander, P. R., Andreu, Z., Zavec, A. B., Borràs, F. E., Buzas, E. I., et al. (2015). Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles 4, 27066. doi:10.3402/jev.v4.27066
Yang, E., Wang, X., Gong, Z., Yu, M., Wu, H., and Zhang, D. (2020). Exosome-mediated metabolic reprogramming: The emerging role in tumor microenvironment remodeling and its influence on cancer progression. Signal Transduct. Target Ther. 5 (1), 242. doi:10.1038/s41392-020-00359-5
Yang, J., Dong, W., Zhang, H., Zhao, H., Zeng, Z., Zhang, F., et al. (2022a). Exosomal microRNA panel as a diagnostic biomarker in patients with hepatocellular carcinoma. Front. Cell Dev. Biol. 10, 927251. doi:10.3389/fcell.2022.927251
Yang, K., Jia, M., Cheddah, S., Zhang, Z., Wang, W., Li, X., et al. (2022b). Peptide ligand-SiO(2) microspheres with specific affinity for phosphatidylserine as a new strategy to isolate exosomes and application in proteomics to differentiate hepatic cancer. Bioact. Mater 15, 343–354. doi:10.1016/j.bioactmat.2021.12.017
Yang, Y., Jia, D., Kim, H., Abd Elmageed, Z. Y., Datta, A., Davis, R., et al. (2016). Dysregulation of miR-212 promotes castration resistance through hnRNPH1-mediated regulation of AR and AR-V7: Implications for racial disparity of prostate cancer. Clin. Cancer Res. 22 (7), 1744–1756. doi:10.1158/1078-0432.Ccr-15-1606
Yang, Y., Mao, F., Guo, L., Shi, J., Wu, M., Cheng, S., et al. (2021). Tumor cells derived-extracellular vesicles transfer miR-3129 to promote hepatocellular carcinoma metastasis by targeting TXNIP. Dig. Liver Dis. 53 (4), 474–485. doi:10.1016/j.dld.2021.01.003
Yao, J., Hua, X., Shi, J., Hu, X., Lui, K., He, K., et al. (2022). LncRNA THEMIS2-211, a tumor-originated circulating exosomal biomarker, promotes the growth and metastasis of hepatocellular carcinoma by functioning as a competing endogenous RNA. Faseb J. 36 (4), e22238. doi:10.1096/fj.202101564R
Yao, M., Wang, L., Fang, M., Zheng, W., Dong, Z., and Yao, D. (2016). Advances in the study of oncofetal antigen glypican-3 expression in HBV-related hepatocellular carcinoma. Biosci. Trends 10 (5), 337–343. doi:10.5582/bst.2016.01176
Yao, Z., Jia, C., Tai, Y., Liang, H., Zhong, Z., Xiong, Z., et al. (2020). Serum exosomal long noncoding RNAs lnc-FAM72D-3 and lnc-EPC1-4 as diagnostic biomarkers for hepatocellular carcinoma. Aging (Albany NY) 12 (12), 11843–11863. doi:10.18632/aging.103355
Ye, B., Shen, Y., Chen, H., Lin, S., Mao, W., Dong, Y., et al. (2022). Differential proteomic analysis of plasma-derived exosomes as diagnostic biomarkers for chronic HBV-related liver disease. Sci. Rep. 12 (1), 14428. doi:10.1038/s41598-022-13272-4
Ye, L., Zhang, Q., Cheng, Y., Chen, X., Wang, G., Shi, M., et al. (2018). Tumor-derived exosomal HMGB1 fosters hepatocellular carcinoma immune evasion by promoting TIM-1(+) regulatory B cell expansion. J. Immunother. Cancer 6 (1), 145. doi:10.1186/s40425-018-0451-6
Yue, X., Ai, J., Xu, Y., Chen, Y., Huang, M., Yang, X., et al. (2017). Polymeric immunoglobulin receptor promotes tumor growth in hepatocellular carcinoma. Hepatology 65 (6), 1948–1962. doi:10.1002/hep.29036
Zebrowska, A., Skowronek, A., Wojakowska, A., Widlak, P., and Pietrowska, M. (2019). Metabolome of exosomes: Focus on vesicles released by cancer cells and present in human body fluids. Int. J. Mol. Sci. 20 (14), 3461. doi:10.3390/ijms20143461
Zhang, C., Yang, X., Qi, Q., Gao, Y., Wei, Q., and Han, S. (2018). lncRNA-HEIH in serum and exosomes as a potential biomarker in the HCV-related hepatocellular carcinoma. Cancer Biomark. 21 (3), 651–659. doi:10.3233/cbm-170727
Zhang, K. J., Qian, J., Wang, S. B., and Yang, Y. (2012a). Targeting Gene-Viro-Therapy with AFP driving Apoptin gene shows potent antitumor effect in hepatocarcinoma. J. Biomed. Sci. 19 (1), 20. doi:10.1186/1423-0127-19-20
Zhang, P. F., Gao, C., Huang, X. Y., Lu, J. C., Guo, X. J., Shi, G. M., et al. (2020a). Cancer cell-derived exosomal circUHRF1 induces natural killer cell exhaustion and may cause resistance to anti-PD1 therapy in hepatocellular carcinoma. Mol. Cancer 19 (1), 110. doi:10.1186/s12943-020-01222-5
Zhang, Y., Wei, W., Cheng, N., Wang, K., Li, B., Jiang, X., et al. (2012b). Hepatitis C virus-induced up-regulation of microRNA-155 promotes hepatocarcinogenesis by activating Wnt signaling. Hepatology 56 (5), 1631–1640. doi:10.1002/hep.25849
Zhang, Y., Wu, J., Zhang, H., Wei, J., and Wu, J. (2020b). Extracellular vesicles-mimetic encapsulation improves oncolytic viro-immunotherapy in tumors with low coxsackie and adenovirus receptor. Front. Bioeng. Biotechnol. 8, 574007. doi:10.3389/fbioe.2020.574007
Zhao, L., Shi, J., Chang, L., Wang, Y., Liu, S., Li, Y., et al. (2021). Serum-derived exosomal proteins as potential candidate biomarkers for hepatocellular carcinoma. ACS Omega 6 (1), 827–835. doi:10.1021/acsomega.0c05408
Zhong, X., Zhou, Y., Cao, Y., Ding, J., Wang, P., Luo, Y., et al. (2020). Enhanced antitumor efficacy through microwave ablation combined with a dendritic cell-derived exosome vaccine in hepatocellular carcinoma. Int. J. Hyperth. 37 (1), 1210–1218. doi:10.1080/02656736.2020.1836406
Zhu, L., Li, J., Gong, Y., Wu, Q., Tan, S., Sun, D., et al. (2019). Exosomal tRNA-derived small RNA as a promising biomarker for cancer diagnosis. Mol. Cancer 18 (1), 74. doi:10.1186/s12943-019-1000-8
Zhu, Y., Wang, S., Xi, X., Zhang, M., Liu, X., Tang, W., et al. (2021). Integrative analysis of long extracellular RNAs reveals a detection panel of noncoding RNAs for liver cancer. Theranostics 11 (1), 181–193. doi:10.7150/thno.48206
Zitvogel, L., Regnault, A., Lozier, A., Wolfers, J., Flament, C., Tenza, D., et al. (1998). Eradication of established murine tumors using a novel cell-free vaccine: Dendritic cell-derived exosomes. Nat. Med. 4 (5), 594–600. doi:10.1038/nm0598-594
Zuo, B., Qi, H., Lu, Z., Chen, L., Sun, B., Yang, R., et al. (2020). Alarmin-painted exosomes elicit persistent antitumor immunity in large established tumors in mice. Nat. Commun. 11 (1), 1790. doi:10.1038/s41467-020-15569-2
Keywords: multi-omics, extracellular vesicle, hepatic precancerous lesions, hepatocellular carcinoma, early diagnosis, immunotherapy
Citation: Lu X, Li Y, Li Y, Zhang X, Shi J, Feng H, Gao Y and Yu Z (2023) Advances of multi-omics applications in hepatic precancerous lesions and hepatocellular carcinoma: The role of extracellular vesicles. Front. Mol. Biosci. 10:1114594. doi: 10.3389/fmolb.2023.1114594
Received: 02 December 2022; Accepted: 06 March 2023;
Published: 16 March 2023.
Edited by:
Long Liu, First Affiliated Hospital of Zhengzhou University, ChinaReviewed by:
Danfeng Guo, First Affiliated Hospital of Zhengzhou University, ChinaZhijin Fan, Sun Yat-sen University, China
Copyright © 2023 Lu, Li, Li, Zhang, Shi, Feng, Gao and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hai Feng, Zm9nc2VhQDE2My5jb20=; Yueqiu Gao, Z2FveXVlcWl1QHNodXRjbS5lZHUuY24=; Zhuo Yu, emh1b3l1QHNodXRjbS5lZHUuY24=
 Xiaona Lu
Xiaona Lu Yuyao Li1
Yuyao Li1 Hai Feng
Hai Feng Yueqiu Gao
Yueqiu Gao Zhuo Yu
Zhuo Yu