
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Mol. Biosci. , 21 April 2023
Sec. Molecular Diagnostics and Therapeutics
Volume 10 - 2023 | https://doi.org/10.3389/fmolb.2023.1113249
This article is part of the Research Topic Insights in Molecular Diagnostics and Therapeutics: 2022 View all 9 articles
Autophagy is a contentious issue in leishmaniasis and is emerging as a promising therapeutic regimen. Published research on the impact of autophagic regulation on Leishmania survival is inconclusive, despite numerous pieces of evidence that Leishmania spp. triggers autophagy in a variety of cell types. The mechanistic approach is poorly understood in the Leishmania parasite as autophagy is significant in both Leishmania and the host. Herein, this review discusses the autophagy proteins that are being investigated as potential therapeutic targets, the connection between autophagy and lipid metabolism, and microRNAs that regulate autophagy and lipid metabolism. It also highlights the use of systems biology to develop novel autophagy-dependent therapeutics for leishmaniasis by utilizing artificial intelligence (AI), machine learning (ML), mathematical modeling, network analysis, and other computational methods. Additionally, we have shown many databases for autophagy and metabolism in Leishmania parasites that suggest potential therapeutic targets for intricate signaling in the autophagy system. In a nutshell, the detailed understanding of the dynamics of autophagy in conjunction with lipids and miRNAs unfolds larger dimensions for future research.
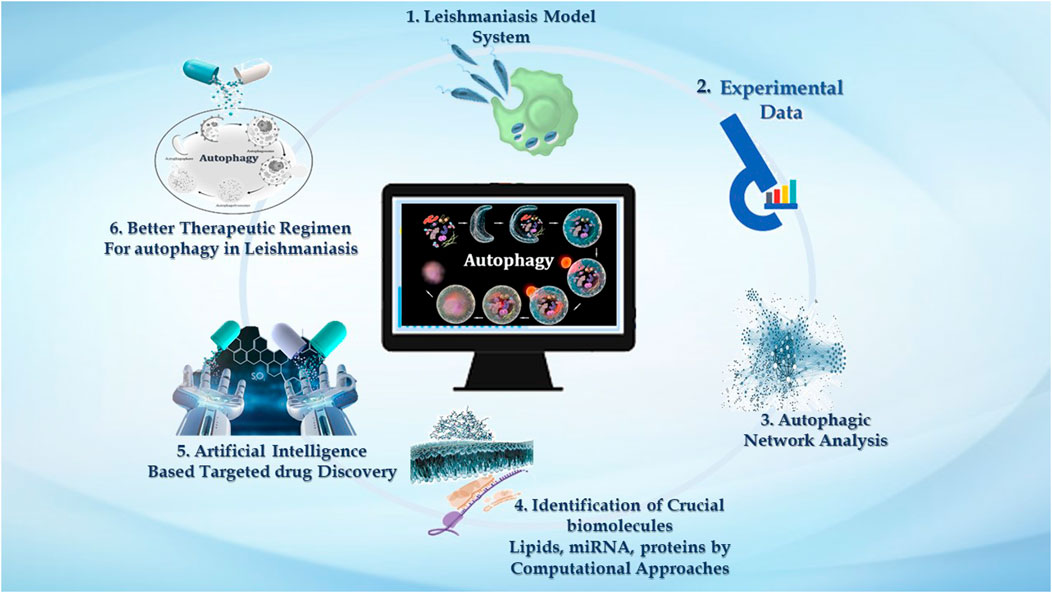
GRAPHICAL ABSTRACT | We demonstrate the significance of computational approaches in studying autophagy in leishmaniasis. 1. Macrophages are the major phagocytic cells where these Leishmania parasites reside, and via invading immune signaling, it establishes successful infection. 2. Very large data reported in the literature accentuating experimental evidence related to Leishmaniasis embrace host autophagy and parasite autophagy, and the impact of host autophagy on parasite survival. 3. Experimental knowledge can be used to analyze the intricate autophagic network. Numerous online databases contribute to providing insight into the autophagic process in disease. 4. Systems approaches such as mathematical model reconstruction and network analysis help emphasize the autophagy process—interconnected with three pivotal biomolecules, i.e., lipids, proteins, and miRNAs. 5. Computational network analysis is decisive for target identification, which can further be utilized by AI-based tools to screen new drugs for leishmaniasis that impacts autophagy. 6. Drug targets identified by AI-based techniques emphasized targeting autophagy as an improved treatment regimen to combat leishmaniasis.
The obligate intracellular parasitic protozoa of genus Leishmania are an etiological agent of a complex vector-borne zoonotic disease named leishmaniasis. It gets transmitted by more than 20 species of sand fly vectors of the genera Phlebotomus and Lutzomyia (Djune Yemeli et al., 2021). According to the 2010 WHO expert committee report, different clinical manifestations that occurred in the Old World are classified into three primary forms: 1) visceral leishmaniasis (VL), also known as kala-azar, caused by L. donovani and L. infantum; 2) cutaneous leishmaniasis (CL), most frequently caused by L. tropica, L. major, and L. aethiopica; and 3) mucocutaneous leishmaniasis (MCL) (can be caused by any species). 4) diffused cutaneous leishmaniasis (caused by L. aethiopica) and 5) post kala-azar dermal leishmaniasis (present in all areas with L. donovani). While CL is the most common form of the disease, VL is the most serious and is almost always fatal if untreated (Colmenares et al., 2002).
This group of neglected diseases occurs in 98 countries with 12 million cases at risk and 20,000–40,000 deaths per year, according to the World Health Organization (WHO) report (https://www. who. int/news-room/fact-sheets/detail/leishmaniasis). The annual incidence of new cases varies from 0.2 to 0.4 million and 0.7–1.2 million cases per year for visceral and cutaneous leishmaniasis, respectively (Sabzevari et al., 2021). Leishmaniasis is prevalent in Africa, Latin America, Asia, the Mediterranean basin, and the Middle East. Although the cutaneous form (CL) of the disease accounts for more than 50% of new cases of leishmaniasis, 90% of CL cases are found in South America, the Middle East, and Afghanistan. Five nations that account for the majority of VL cases are viz., India, Bangladesh, Ethiopia, Sudan, and Brazil (Modabber, 2010). Although humans are the only known hosts for L. donovani, the disease is mostly zoonotic in origin, with canine species serving as the primary animal reservoir. Within a few years after surviving VL, between 20% and 60% of L. donovani-infected patients would acquire post kala-azar dermal leishmaniasis (PKDL). Patients with PKDL are believed to be a major source of parasites for new infections because of the large number of parasites identified in their skin (Kedzierski & Evans, 2014).
The life cycle of Leishmania is digenetic, i.e., it alternates between two hosts, i.e., mammalian and insect (Figure 1). In the case of leishmaniasis, the insect vector is the sandfly belonging to the Phlebotomus genus in Old World Leishmania species and Lutzomyia genus for New World Leishmania species. After consuming blood from an infected host, the sandfly becomes infected for the first time. The parasite differentiates once it is inside the sandfly, developing into procyclic promastigotes. Promastigotes are flagellated and motile forms of the parasite (Killick-Kendrick, 1974). Their elongated, ovoid body is 1.5–3.5 mm wide and 15–20 mm long. The parasite binds to the sandfly’s gut because the length of its flagella varies from 15 to 28 μm. The procyclic forms split in the midgut of the sandfly, where they produce nectomonad promastigotes that do not divide. These nectomonad promastigotes migrate from the abdominal midgut to the anterior midgut and then transform into leptomonad promastigotes. In due course, the leptomonad promastigotes finally differentiate into metacyclic promastigotes and travel to the sandfly’s proboscis, where they are prepared for transmission to a mammalian host (Harizanov and Kaftandjiev Iskren, 2014). Metacyclic promastigotes are introduced into the host through bite, and upon adhering to the plasma membrane, these promastigotes initiate a phagocytic process. In this manner, the promastigotes enter the macrophage and infect the parasitophorous vacuole. The promastigotes develop into ovoid amastigotes, which have a diameter of 2–4 μm. Inside the parasitophorous vacuole, the amastigotes grow and multiply to the point where the macrophage ruptures, releasing all of the mature amastigotes. The cycle continues ultimately, leading to one of the clinical manifestations discussed previously (Pulvertaft and Hoyle, 1960).
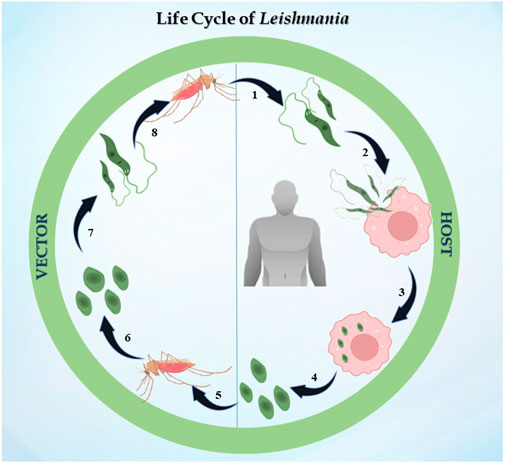
FIGURE 1. Life cycle of Leishmania: the figure represents the digenetic life cycle of 1. promastigotes injected in the host by sandfly, 2. promastigotes engulfed by macrophages of the host, 3. promastigotes transformed into amastigotes inside macrophages, 4. amastigotes transmitted to other healthy macrophages, 5. an infected sandfly taking its next blood meal, 6. amastigotes transforming into the promastigote stage in the midgut, 7. dividing into the midgut and migrating to the proboscis, and 8. the life cycle continues.
Pentavalent antimony was once thought to be the first-line pharmacological treatment for leishmaniasis; however, it is now known to cause cardiotoxicity, cirrhosis, pancreatic toxicity, and resistance risks (Frézard et al., 2009). Paromomycin was found to be helpful for Indian patients with VL, but less effective for Sudanese patients (Sundar and Olliaro, 2007). Amphotericin B (and lipid formulation) emerged as the second-line therapy as a result of this drug’s developing resistance (Pradhan et al., 2022). Since then, miltefosine has been used in VL and CL. Its advantages include being an oral medicine with good efficacy and a brief course, but its drawbacks include teratogenicity and drug resistance. Miltefosine is still a significant alternative in VL chemotherapy, even though its efficacy as a monotherapy has declined in recent years, especially when combined with other medications (Sundar et al., 2007). Repurposing of available drugs for leishmaniasis includes amphotericin B, miltefosine, paromomycin, and pentamidine (Andrade-neto et al., 1996). Leishmaniasis has also been investigated using azole antifungals, and itraconazole was found to be more effective than fluconazole and ketoconazole at preventing the growth of most Leishmania strains (Alrajhi et al., 2002).
Combination chemotherapy has been developed to prevent drug resistance, enhance compliance, decrease the length of treatment, and hence lower the cost of therapy. The other combinations are miltefosine plus paromomycin, miltefosine plus liposomal amphotericin B, and sodium stibogluconate/meglumine antimoniate plus paromomycin (Sundar & Olliaro, 2007), (Omollo et al., 2011).
Local therapies have been proven to be safe alternatives to systemic drug administration for CL-specific conditions. Thermotherapy, cryotherapy, and photodynamic therapy (PDT) have all been tested in CL. PDT uses topically administered aminolevulinic acid (ALA) or methyl-aminolevulinate, followed by laser or intense pulsed light irradiation. A few mechanistic investigations have focused on the ideas underlying the use of PDT for treatment of CL (Akilov et al., 2007). Eradication of host cells is the mechanism through which ALA-PDT exerts its anti-leishmanial effects for CL. Topical ALA-PDT is not suggested in clinical practice because of insufficient data (Tsai et al., 2002). At temperatures below 0, cryotherapy destroys infected cells, kills amastigotes, and modifies cell membranes to produce intracellular and extracellular ice crystals. Cryonecrosis results in the release of antigenic compounds that stimulate immune responses and repair further lesions. Since chemotherapy has several drawbacks, cryotherapy may be a useful alternative for treating CL. It has shown excellent response in patients with skin lesions ranging in size from 10 to 30 mm, those with fewer lesions, and those in whom the development was under 3 months. Cryotherapy and intralesional sodium stibogluconate were highly efficient, resulting in 100% healing of CL lesions. CO2 laser administration and thermotherapy based on the principle of directly destroying the Leishmania parasites is a simple way to deliver external heat on infected tissues, causing damage to specific areas with parasitism (Valencia et al., 2013). Heat sensitivity prevents Leishmania species from growing or surviving in environments hotter than 39°C. Thus, thermotherapy has been considered a possible treatment for CL lesions. Patients with CL have tried radiofrequency (RF) therapy, a type of thermotherapy. The cure rate for thermotherapy administered once every 3 weeks was 73%, whereas the cure rate for thermotherapy administered once every week was 81% (Sadeghian et al., 2007).
Immunotherapy is a well-established treatment for leishmaniasis, and immunomodulators may be developed to achieve this goal under the hypothesis that a non-protective anti-leishmanial immune response could be transformed into a protective phenotype. The most promising method, when immunomodulators are considered, is therapeutic vaccination, which is based on cytokines that encourage macrophages to eradicate Leishmania parasites (Nicholls et al., 2010). IFN-γ immunotherapy for VL patients increased parasitological control and improved the therapeutic effectiveness of traditional pentavalent antimony (Sbv) therapy, enabling a more than 80% cure rate (Robledo et al., 1999).
The field of drug discovery and design has undergone a revolution primarily because of nanotechnology, which has additionally proven to be a potential tool for parasitic diseases. It has managed to invent nanoparticles, as carriers, for drug delivery; liposomal formulation of amphotericin B reduces its toxicity profile (Date et al., 2007). Nanotechnology has significantly enhanced the conventional leishmanization procedure by inducing a Th1-type immune response in BALB/c during L. major infection using a liposome–protamine–DNA nanoparticle with immunostimulatory CpG (Alavizadeh et al., 2012), (Fakhraee et al., 2016). Nano-liposomal formulation of 1,2-dioleoyl-3-trimethylammonium-propane (DOTAP) and soluble leishmanial antigen (SLA) causes protection against L. major infection and enhances Th1-type immune response. Modified DOTAP, a second-generation vaccine candidate that has been reported as a conjugated nanoparticle with amastigote class I nuclease, makes for a promising candidate for the development of a CL vaccine (Fakhraee et al., 2016). By increasing its therapeutic efficacy, nanotechnology has thereby pioneered the door for the development of vaccines and medications (Sundar and Singh, 2018).
In the past few years, phytotherapy has grown in popularity as researchers look for inexpensive anti-leishmanial treatments. The use of Kalanchoe pinnata, with components including triterpenes, sterols, and flavonoids, indicate substantial antileishmanial activity, but significant research in the field remains to be undertaken (Da Silva et al., 1995). Naphthoquinone is another class of secondary metabolite from plants with potential anti-leishmanial activity. Plumbagin, a naphthoquinone, has been reported to inhibit trypanothione reductase from L. donovani and induces mitochondria-mediated cell death (Awasthi et al., 2016).
Recent research has shown that important proteins and macromolecules like lipids are great pharmaceutical targets by analyzing several metabolic as well as immunological signaling networks. While targeting metabolic pathways, identification of homology between the host and the parasite proteins is still the primary therapeutic target issue. Blocking energy metabolism remains a choice of target (Verlinde et al., 2001). Since Kreb’s cycle, glycolysis, and oxidative phosphorylation occur in the glycosome and mitochondria, these structures serve as the primary energy production houses. Interruption between any steps of glycolysis or Krebs’s cycle arrest the energy flux, leading to parasite killing. The potential drug target chalcone was discovered to be an anti-leishmanial prospective pharmacological target that targets the ultrastructure and functions of mitochondria, and later its ability to block fumarate reductase (Zhai et al., 1999), (Zhai et al., 1995).
Endochin-like quinolones (ELQs) have been demonstrated to be harmful to L. donovani and L. mexicana amastigotes, and hydroxynaphthoquinone buparvaquone is a more effective inhibitor of electron transport, ATP synthesis, and parasite multiplication, raising concerns about targeting cytochrome bc1 as a potential therapeutic strategy (Ortiz et al., 2016).
Polyamine metabolism is still an important prospective pathway for medication development. The first enzyme in the polyamine biosynthesis pathway is arginase (E. R. Da Silva et al., 2015), whereas ornithine decarboxylase (Boitz et al., 2009), S-adenosylmethionine decarboxylase (Brun et al., 1996), polyamine oxidase (Baumann et al., 1990), trypanothione synthetase, trypanothione reductase, tryparedoxin peroxidase, and deoxyhypusine synthase act as potential drug targets (Khan, 2007), (Wyllie et al., 2004).
It has long been understood that the generation of sterols is indispensable for cellular health and preservation of cell structure. Ergosterol and 24-methyl sterol are the most important sterols in trypanosomatids for growth and viability. As a result, the sterol and fatty acid metabolic pathways are appealing therapeutic targets. The mitochondria are the primary sites for fatty acid metabolism, with 3-hydroxy-3-methylglutaryl-CoA (HMG-CoA) reductase from L. donovani reported as a potential therapeutic target (Dinesh et al., 2014). Fatty acyl-CoA ligase affects cellular lipid homeostasis and has been found to be differently regulated in antimony-resistant L. donovani, confirming its potential as a therapeutic target (Kaur et al., 2011). Other than this, enzymes involved in sterol biosynthesis including farnesyl diphosphate synthase (Docampo & Moreno, 2008), sterol methyl transferase (Lorente et al., 2004), sterol 14 alpha-demethylase (McCall et al., 2015), and squalene synthase (Rodrigues et al., 2008) are reported as drug targets involved in chemical inhibition of the biosynthesis pathway. Immune signaling, energy metabolism, polyamine pathways, and fatty acid metabolism are examined by researchers to highlight potential molecules in the signaling cascade. Despite phenomenal research being conducted to create therapeutic targets for leishmaniasis, chemotherapy still serves as the cornerstone of effective treatment (Figure 2 depicting the treatment regimen of leishmaniasis). The details of therapeutics in leishmaniasis are listed in Table 1.
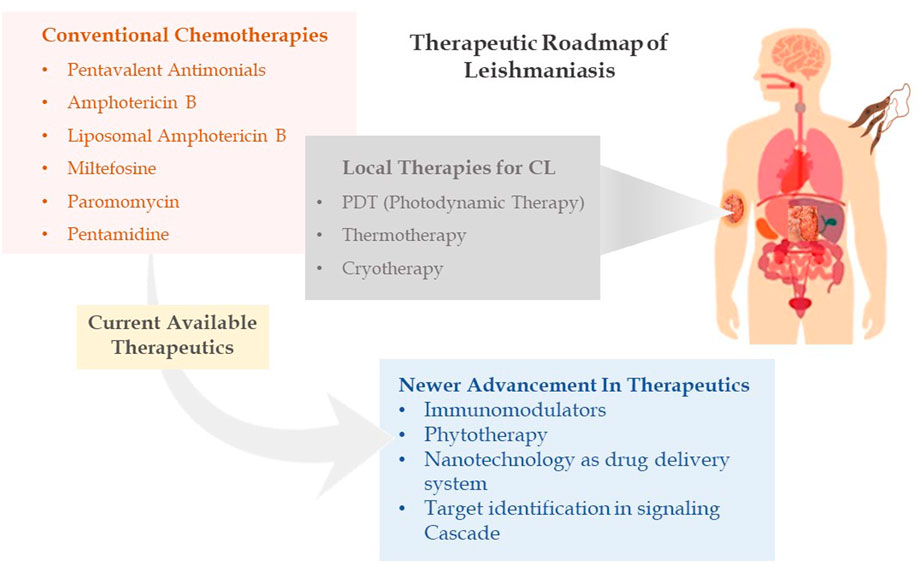
FIGURE 2. Therapeutic roadmap of leishmaniasis illustrating conventional chemotherapies, local therapies, and newer advancement in therapeutics.
Researchers and international health organizations are compelled to develop novel strategies to combat and control this serious, untreated disease leishmaniasis due to the emergence of drug-resistant strains (Sundar, 2001), high toxicity, co-infections like HIV/Leishmania spp., limited chemotherapeutics available for disease treatment, and low investment for the discovery/development of new drugs for a particular disease. Thus, to investigate novel therapeutic options to effectively eradicate parasites, an understanding of host–parasite interactions is a prerequisite. Autophagy is found to be one of the crucial fundamental processes involved in leishmaniasis. Here, in this review, we elucidate the role of autophagy in leishmaniasis.
Autophagy is a climacteric cellular catabolic process to maintain cellular homeostasis, and it is conserved in a hierarchy from yeasts to mammals. The term autophagy was coined by Christian de Duve in 1963, but it remained a biological enigma until the early 1990s (De_Duve, 1974- Lysotropic. pdf). Furthermore, in the early 1990s, Yoshinori Ohsumi, an Assistant Professor at Tokyo University, studied autophagy using the budding yeast Saccharomyces cerevisiae as a model system and revealed some basic phenomena of the autophagy. He also discovered gene autophagy 1 (APG1), which is responsible for autophagy in yeast, and reported 15 more APG genes responsible for autophagy in the eukaryotic cell yeast. Autophagic genes were further identified in yeast and other species, and this specific group of proteins is abbreviated as ATG (Tsukada and Ohsumi, 1993), (Matsuura et al., 1997). All cells engage in low-level basal autophagy to carry out homeostatic functions like protein and organelle turnover. When cells produce intracellular nutrients and energy, such as during deprivation, growth factor depletion, or high bioenergetic demands, it is quickly activated. Furthermore, autophagy is activated when cells are ready to undergo structural remodeling, such as during developmental transitions, or when they need to get rid of potentially harmful cytoplasmic elements, such as under oxidative stress, infection, or the buildup of protein aggregates. Regulation of autophagy is significantly influenced by nutritional status, hormonal factors, and other factors like temperature, oxygen concentrations, and cell density (Klionsky, 2007), (Maiuri et al., 2007).
To date, collectively 41 types of proteins are known to play an important role in autophagy in different groups on the basis of their functions. These proteins are classified into six functional groups: ATG1-kinase/ULK1 complex, phosphatidylinositol (PI) 3-kinase complex (PI3K), membrane protein ATG9, ATG2–ATG18 complex, ATG16L conjugation system, and ATG8 conjugation system (Mizushima et al., 2011). These proteins and their homologs are well-studied in yeast as well as in mammals (C. He & Klionsky, 2009).
Initiation of autophagosome formation in mammals is a complicated process, orchestrated by three major proteins ULK1 (unc-51-like kinase 1) ATG13, FIP200 (focal adhesion kinase family interacting protein of 200 kDa), and ATG101 combined to form the ULK1 complex. Succeeding autophagy induction, the ULK1 complex translocates to the autophagophore membrane or autophagy initiation site and further regulates PI3K complex recruitment over the autophagophore. This PI3K complex includes the VPS (vacuolar protein sorting) 34 (VPS34) complex, class III phosphatidylinositol 3-kinase VPS34, Beclin-1, VPS15, and ATG14L (ATG14-like), which govern the production of the phospholipid phosphatidylinositol 3-phosphate (PI3P) at the autophagophore. WIPI (WD repeat domain phosphoinositide interacting protein 2) and DFCP1 (double FYVE-containing protein 1) are recruited at the PI3P binding site and contribute to autophagophore expansion (Zachari & Ganley, 2017), (Bento et al., 2016). PI3P acts as a crucial component where numerous proteins are recruited and helps in autophagosome formation. ATG2 interacts with ATG18, which acts as a PI3P-binding partner, and forms a complex that further translocates to autophagophores and contributes to autophagosome biogenesis (Kotani et al., 2018).
In participation with the PI3K complex, ATG9 is playing a crucial role in lipid accumulation on the autophagophore and its expansion; ATG9 is a multi-spanning membrane protein (Geng et al., 2008), (Webber & Tooze, 2010), (Kotani et al., 2018). The exact molecular role of ATG2, as well as ATG9, is still mysterious, but directly or indirectly, they contribute to the PI3K complex for autophagosome initiation and expansion. Along with these complexes, two major autophagy-specific ubiquitin-like protein conjugation systems play a crucial role in autophagosome formation and maturation. It begins with the ATG16L conjugation system that consists of the ATG12–ATG5 conjugate covalently linked by ubiquitin-like enzymes ATG7 and ATG10 that act as E1 and E2-like enzymes, respectively. In addition, for localization of this conjugate on the autophagosome membrane, ATG16L is required. The ATG12–ATG5 conjugate and ATG16L that interact non-covalently are recruited on the membrane, whereas ATG5 mediates the interaction leading to autophagosome expansion (Romanov et al., 2012), (Noda et al., 2013). The ATG12–ATG5 conjugate has been shown to mediate LC3II–PE complex formation, while the ATG16L complex is observed to stabilize this complex on the membrane. In addition to acting as a marker for autophagy, the LC3II–PE conjugate is an essential step in the production of autophagosomes. Formation of the LC3II–PE conjugate is mediated by ATG4, which acts as a cysteine protease and cleaves ATG8 from the C-terminus at arginine and exposes the glycine residue and allows it to interact with PE (phosphatidyl ethanolamine) (Maruyama and Noda, 2018). This glycine-exposed LC3 and PE interaction is facilitated by ATG7, an E1-like enzyme that transfers LC3I to an E3-like enzyme and subsequently transfers LC3I to the membrane lipid PE, and LC3II complex translocates on the autophagosome. Autophagosome maturation takes place by various ATG protein complexes, as shown in Figure 3.
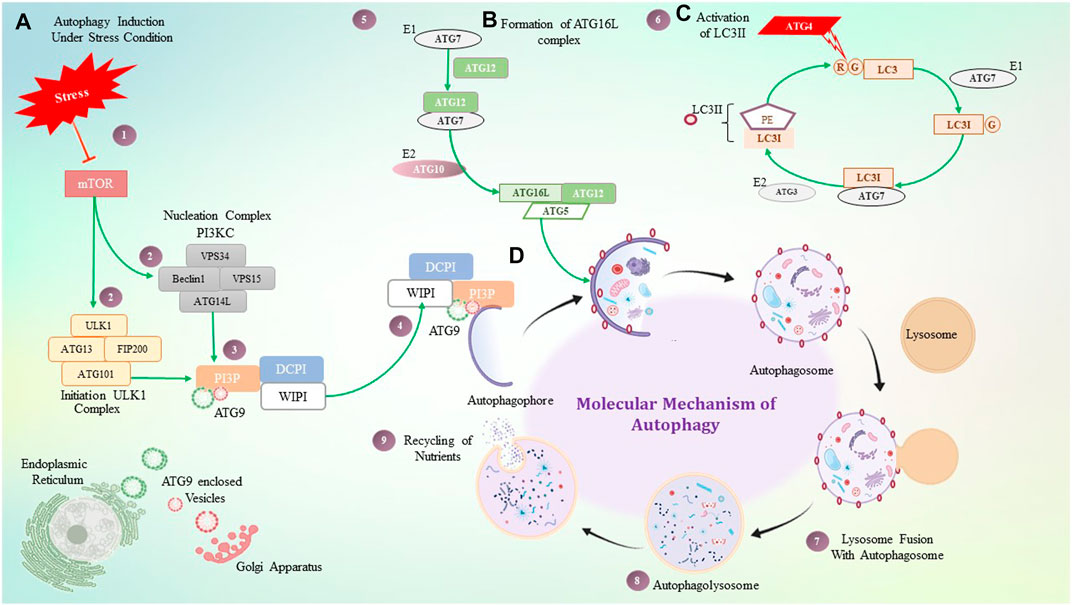
FIGURE 3. Molecular mechanism of autophagy. (A) Under stressful conditions, mTOR induces autophagy by activating the ULK1 and PI3KC3 complex, which then activates PI3P. PI3P is interacting with ATG9, DCPI, and WIPI proteins, which help with autophagosome elongation. (B) ATG16 forms a complex non-covalently with the ATG12 and ATG5 conjugate, where ATG7 plays a role as an E1-like enzyme. (C) ATG4 is a cysteine protease that cleaves LC3 protein at the glycine residue, and with the help of ATG7- and ATG3-like enzymes, it binds to PE which is termed LC3II. LC3II gets stabilized on the autophagosome with the help of the ATG16L1 complex. (D) Step-by-step diagrammatic description of autophagy shows the initial formation of the autophagosome membrane, followed by engulfment of damaged cellular components, their fusion with the lysosome, and the formation of the autophagolysosome.
Autophagy processes are involved in strategies to eliminate invasive pathogens through the production of reactive oxygen species, adjustment of vital nutrients and cations, destruction by proteolytic enzymes (Galán and Cossart, 2005), and by playing a crucial role in host–pathogen interaction, especially in intracellular pathogenesis. Research conducted over the last few years suggests that autophagy is one of the most astonishing intracellular host cell defense machinery tools that pathogens must spread during cell invasion. During infection, autophagosomes engulf intracellular pathogens; this process is known as xenophagy since it results in the eradication of foreign material. Pathogens are engulfed during autophagy and bind to the autophagosome membrane protein LC3 (light-chain 3)/Atg8 (autography-related protein 8).
To establish a persistent infection, most intracellular pathogens manipulate the autophagy pathway at the molecular level (Campoy and Colombo, 2009). Toll-like receptors (TLRs) that identify pathogen-associated molecular patterns induce autophagy during infection and allow autophagy to kill the pathogen. TLR signaling cascade and autophagy are two innate defense mechanisms that allow intracellular parasite clearance (Delgado et al., 2008). TLRs recognize conserved microbial components and induce a variety of antimicrobial activities, such as xenophagy via Myd88 and TRIF interacting with Beclin 1 (Campos-Salinas et al., 2013), (Shi and Kehrl, 2008). Autophagy also gets induced by cell-to-cell signaling. In human cells, IFN-γ stimulates autophagy through IRGM1, whereas CD40 ligation causes autophagy through PI3K and Rab7, enabling cells to fight intracellular infections like Toxoplasma gondii (Andrade et al., 2006).
Several molecules, including Nod-1 and Nod-2, recognize microbial antigen peptidoglycans and target bacteria for xenophagy (Cooney et al., 2010), (Travassos et al., 2010). However, many pathogens escape the phagosome after phagocytosis and proliferate in the cytosol. Microbes can also promote xenophagy via a variety of cell stress mechanisms, whereas T. gondii increases intracellular calcium levels to promote autophagy (Singh et al., 2009). Studies of Group A streptococci (GAS) also show evidence of autophagy. Getting rid of GAS (escapes from endosomes by encoding the hemolytic toxin streptolysin O) is a great illustration of autophagy, where ATG5 associates with ATG8/LC3 to form autophagic vacuoles for clearance of GAS (Nakagawa et al., 2004).
In the extensively studied pathogen Mycobacterium tuberculosis (Mtb), the removal of bacteria by autophagy, IFN (interferon)-induced IRGs (immunity-related p47 GTPases), and the overexpression of LRG-47 (one of the 23 IRGs in mice) promote autophagosome formation and autophagy-dependent clearance of phagosomes containing Mtb in mice (Gutierrez et al., 2004). Both mouse IRGs and autophagy can target the phagosomes that T. gondii uses for replication during toxoplasmosis. In contrast to the macroautophagy upregulating effect found for IRGs during Mtb infection, the mouse IRGs, IIGP1 and IGTP, appear to disrupt the T. gondii-contained PV (parasitophorous vacuole) membrane and even the parasite’s membrane, exposing it for destruction via macroautophagy (Tang et al., 2006), (Romanov et al., 2012). An effective illustration of evading autophagosome engulfment is T. cruzi (the causative agent of Chagas disease) infection to mammalian cells and activation of a signal transduction cascade that results in the establishment of a parasitophorous vacuole. Lysosomes have a significant impact on how T. cruzi infections progress, according to studies. The T. cruzi vacuole is decked with the host cell autophagic protein LC3. Fasting or pharmacologically induced autophagy increased the number of infected cells dramatically before infection, while inhibitors of this process significantly decreased the invasion, showing that mammalian autophagy is an important process that favors colonization of T. cruzi in the host cell (Alvarez et al., 2008). Skin lesions from BALB/c mice having CL also exhibit autophagy. In cultured macrophages, autophagy inhibitor 3-methyladenine (3 MA) decreased the infection index, while autophagy-inducing agents like rapamycin or fasting had no effects. This finding suggests that one function of autophagy is to provide nutritional support during infection (Cyrino et al., 2012).
In a comparative study of L. major and L. amazonensis, both species of infected macrophages demonstrated an increase in the LC3-II/Act (autophagic marker) ratio after 24 h, although treatment with autophagic inhibitors had no discernible effect on the parasite burden or infection rate. It is noteworthy to observe that pharmacological autophagy inducers increase parasite survival, whereas pharmacological autophagy inhibitors have no effect on it (Dias et al., 2018).
Researchers have become more interested in autophagy in leishmaniasis despite contradictory findings concerning the importance of autophagy in leishmaniasis. The reason is autophagy has been evidenced to be a therapeutic target in several intracellular pathogeneses, inflammatory illnesses, metabolic disorders, diabetes, cancer, osteoporosis, and other conditions.
Literature on leishmaniasis focuses on the interaction between lipid metabolism and autophagy. Both autophagy and lipid metabolism are necessary for creation of new perspectives on the investigation of leishmanial therapy. We are aware of very few articles that discuss the significance of lipid metabolism and autophagy. In this review, we try to highlight these crucial processes concerning therapeutics.
According to earlier studies, Leishmania infection alters the cell’s lipid metabolic pathways, where lipid droplets play a critical role (Bouazizi-Ben Messaoud et al., 2017). Lipid droplets (lipid bodies, LDs) are dynamic organelles that have beneficial roles in modulating lipid metabolism, energy homeostasis, signaling, membrane trafficking, and inflammation. Lipid droplets are roughly spherical structures consisting mostly of triacylglycerols and sterol esters which lack a traditional bilayer membrane but are surrounded by a monolayer of phospholipids and cholesterol as well as a few related proteins (Vallochi et al., 2018).
LDs are organelles that are closely associated to the ER. According to the first and most widely used theory of how LDs formed, these organelles originate from the accumulation of recently synthesized lipids within the double layer of the ER membrane, which then bud off into the cytoplasm once they reach a crucial size (S. Martin and Parton, 2006). Due to their varied protein and lipid components and involvement in essential cellular activities, LDs have become an important specialized and inducible cytoplasmic organelle. Beyond managing lipid metabolism, LDs additionally play a role in cell signaling, immunological activation, membrane trafficking, and formation and secretion of inflammatory mediators.
The LDs in infectious disorders also point to roles that go beyond simple interactions with numerous viral proteins or bacteria; they can take part in significant immune system cellular processes. Recently, researchers have been concentrating on the downstream pathways connected to the production of LD once an infection occurs (Figure 4). It has been determined how signaling pathways regulate the expression of genes linked to lipid influx/efflux and de novo synthesis during infectious processes. In accordance with a recent report, bacterial components may alter the expression and functioning of PPARγ. The lipid-activated nuclear receptor family includes PPARγ, a member of which directly controls a few genes involved in fatty acid intake, lipid storage, and inflammatory response. PPARγ has emerged as a crucial regulator of lipid metabolism and inflammatory genes in macrophages and dendritic cells (Odegaard et al., 2007). LD generation during protozoan parasite infection is mainly mediated by TLR2. TLR2-dependent LD formation was triggered in macrophages by T. cruzi infections. Macrophages’ absorption of apoptotic cells enhanced LD formation and PGE2 generation through TGFβ-signaling (D’Avila et al., 2011).
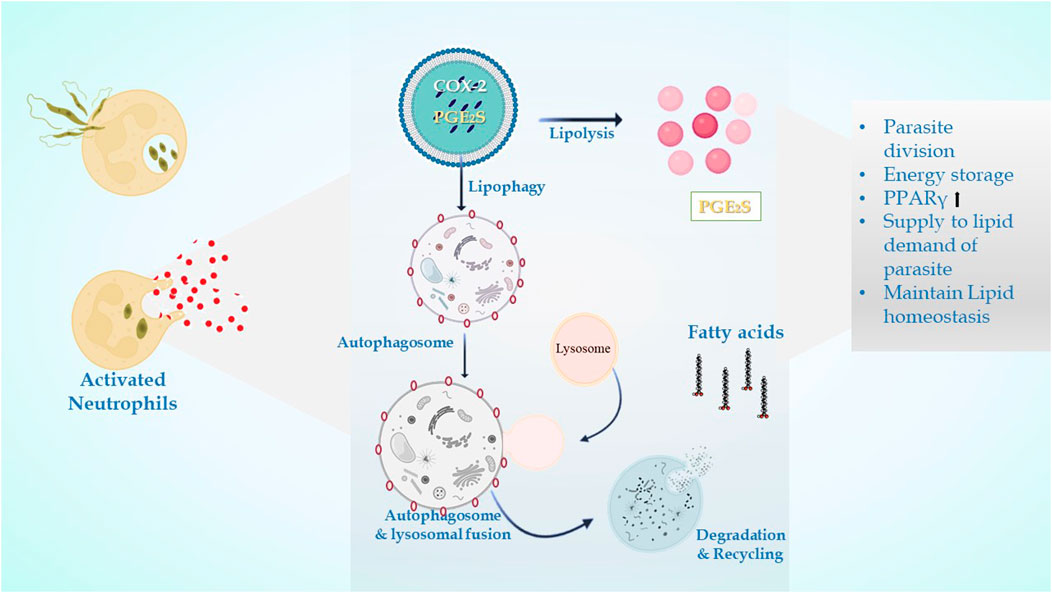
FIGURE 4. In leishmanial infection, lipid droplets are induced in the host during infection in response to provide lipids for parasite survival and for their differentiation. Within the LDs of stimulated cells acquired under infectious circumstances, the key eicosanoid-forming enzymes, cyclooxygenase, 5-lipoxygenase (5-LO), 15-LO, and PGE2 synthase were discovered. The synthesis of prostaglandin E2 aids in parasite survival. The production of signals that cause LD formation, fatty acid intake, lipid storage, and inflammatory response has been identified to involve PPARγ.
There is a huge involvement of lipids for the rapid multiplication of parasites and establishment of infection. Leishmania infection is efficient in modulating cholesterol metabolism in the host tissue in both acute and chronic patients (Johndrow et al., 2014). Cholesterol or ergosterols are essential components of plasma membranes found in lipid rafts and membrane microdomains due to their physiochemical properties. The available chemotherapeutic drug for leishmaniasis, amphotericin B, preferentially binds to ergosterol, causing the membrane’s osmotic integrity in target cells to be compromised (Chattopadhyay and Jafurulla, 2011), (Xu et al., 2014). Recently, quantified sterols of L. major through gas chromatography–mass spectrometry allowed (Yao and Wilson, 2016) comparing both qualitative and quantitative characteristics of sterols in infectious and non-infectious parasite forms.
The Leishmania parasite lacks a de novo mechanism for cholesterol production and thus must scavenge this lipid from the host environment (Biagiotti et al., 2017). According to reports, sphingomyelin degradation in the host acts as an enticing driver of Leishmania parasite infection (Martínez and Ruiz, 2019). Sphingolipids (SLs) are a group of cardinal membrane compounds that are produced abundantly in mammalian cells, in the form of sphingomyelin and glycosylsphingolipids, which do not exist in parasite cells (Ghosh et al., 2012).
Promastigotes synthesize most of their lipids, including glycerophospholipids, sterols, and sphingolipids, by de novo synthesis. Amastigotes, on the other hand, are able to carry out de novo synthesis but obtain most of their lipids from the host. The change in Leishmania from rapidly replicating promastigotes to slowly growing, metabolically dormant amastigotes is reflected in the transition from de novo synthesis to salvage (Zhang, 2021). Phosphatidylcholine (PC), phosphatidylethanolamine (PE), and phosphatidylinositol (PI) are the three phospholipids most abundant in Leishmania promastigotes, with other phospholipids found in smaller amounts. In comparison to promastigotes, amastigotes have higher levels of phosphatidylserine and sphingomyelin and lower levels of PE and PI (Martínez and Ruiz, 2019). Additionally, the parasite’s non-infectious form is distinguished by having more unsaturated fatty acids than saturated ones and a significant generation of n-6 polyunsaturated fatty acids in comparison to n-3 forms (which is lowered in amastigotes) (Biagiotti et al., 2017).
It has been reported that the percentage of cholesterol almost doubled in the amastigotes, whereas the concentration of ergosterol decreases significantly (80%) (Bouazizi-Ben Messaoud et al., 2017). Cholesterol is scavenged by intracellular parasites from the host. As a source of energy storage, to maintain metabolic balance, and to escape the host reaction, intracellular parasites have their LDs. As de novo synthesis is absent in parasites, lipids needed by many intracellular parasites are obtained from host LDs. Intracellular parasites have acquired methods to entice host LDs to their phagosome compartment and engulf them whole (Melo and Dvorak, 2012). It is unclear how lipids are incorporated by Leishmania parasites at the cellular and molecular levels.
Murine macrophages infected with Leishmania exhibit increased cholesterol absorption and triacylglycerol production, resulting in the development of lipid droplets near or inside parasitophorous vacuoles (PVs) (Rodríguez et al., 2017). Amastigote forms replicate inside the host macrophages within PVs, specialized membrane-bound organelles of the endocytic pathway with late endosomal/lysosomal characteristics. PV morphology differs depending on the species of Leishmania. Large communal PVs characterize infection with both L. mexicana and L. amazonensis and are formed by the fusion of small individual vacuoles with compartments of the endocytic pathway. According to several studies, host cell macromolecules are transported to the PV lumen and endocytosed by the parasite. PVs get cytosolic macromolecules from L. mexicana-infected macrophages by a process resembling host cell autophagy (Pinheiro et al., 2009). The interrelationship between autophagy and lipid metabolism is termed macrolipophagy (Singh et al., 2009), as autophagy regulates lipid content. Singh et al. observed that inhibition of autophagy increases TGs and LDs in vitro and in vivo, whereas loss of autophagy decreases TG breakdown and LC3 associated with LDs. There is also a reverse relationship that suggests an abnormal increase in intracellular lipids impairs autophagic clearance, based on decreased LD/LAMP1 co-localization, absence of autophagic upregulation in hepatocytes cultured with lipids, and diminished association of autophagic vacuoles with LDs in response to starvation in HFD-fed mice. This interrelationship shows that decreased autophagy promotes lipid accumulation, which then further suppresses autophagic function, thereby additionally increasing lipid retention (Figure 5).
Studies have demonstrated that depending on the type of parasite and the host interaction, autophagy can either support infection or facilitate parasite expulsion. To date, studies have examined the involvement of autophagy in Leishmania infection. Although it has been repeatedly demonstrated that the Leishmania parasite induces autophagy in different cells, but how autophagic modulation affects Leishmania survival is still conflicting.
Earlier studies have shown that L. mexicana has developed access to endosomes, phagosomes, and autophagosomes as sources of host cell material; such a strategy might offer the parasite two different adaptive benefits. In the beginning, it offers an additional supply of nutrients. Furthermore, PVs are part of the host’s route for processing and presenting antigens (Schaible et al., 1999). For the first time, Mitroulis, Ioannis, et al. discovered induction of the autophagic machinery (i.e., LC3B conversion) during acute natural bone marrow infection by L. donovani under real-time circumstances, although the role of autophagy in parasites was discovered earlier. For obtaining essential nutrients, Leishmania species trigger autophagy. The expression of the autophagy genes Atg7 and LC3, as well as the LAP-like accumulation of LC3, around the parasite vacuoles, appears to be related to the severity of the L. infantum infection (Esch et al., 2015). Nutrient deprivation is a potential inducer of autophagy, L. amazonensis was administered to BALB/c macrophages, and durations of 1–24 h of amino acid and serum fasting were used to induce autophagy. The result demonstrated the elevation of positively stained vacuoles for MDC (monodansylcadaverine), a marker of autophagy. The intracellular burden of L. amazonensis in BALB/c macrophages increased in response to inducers rapamycin and glucagon. In response to starvation, the intracellular load of L. amazonensis increases up to 2–12 h without altering macrophage viability, but it unexpectedly decreases at 24 h by loss of macrophage viability. Altogether, our findings suggested that, following the onset of autophagy, intrinsic host cell factors regulate the course of infection (Pinheiro et al., 2009). Also, it was highlighted that one function of autophagy may provide the parasite with nutritional support, as 3-methyladenine (3 MA), the autophagy inhibitor, decreased the infection index, while autophagy inducers rapamycin or fasting had no effect (Cyrino et al., 2012).
Autophagy stimulation was observed by IFN-γ or fasting-boosted L. amazonensis infection in BALB/c mice, but not by L. major infection. As L. amazonensis amastigotes do not co-localize with IFN- γ-induced, double-membrane vacuoles, starvation or autophagy-inducing agents may make it favorable for the parasites to be consumed and proliferate (Pinheiro et al., 2009). Not only in macrophages, Leishmania can also induce autophagy in neutrophils. This study uncovered that canonical autophagy was prevalent in Leishmania infection after 3 h and that L. donovani infection encouraged a time-dependent enhanced autophagy, responsive to blocking by 3-methyladenine but sensitive to ULK1/2 inhibition only after 3 h (Pitale et al., 2019). Thomas et al. (2018) reported that in THP-1 cells, knockdown of Atg5 and Atg9 reduced intracellular L. donovani survival. On the other hand, L. major parasite load was increased by Atg5 knockdown in BALB/c and C57BL/6 macrophages (Franco et al., 2017). Leishmania fine-tunes the time and inhibits mTOR-regulated autophagy while concurrently enhancing parasite survival (Thomas et al., 2018).
Pattern-recognition receptor (PRR), TLR, is also responsible for induction of autophagy in infected macrophages as macrophages deficient in TLR3, 7, and 9; UNC93B1; or MyD88 failed to undergo L. major-induced autophagy (Franco et al., 2017). Matte et al., 2016 focused on LC3-associated phagocytosis, the non-canonical aspect of autophagy in leishmaniasis. During L. major infection, LC3 retention in the autophagosome is diminished, which occurs as a result of VAMP8, and NOX2 NADPH oxidase—required for the formation of ROS—is inhibited in a way that is dependent on the parasite surface metalloprotease GP63. Overall, the results showed that VAMP8 is important for LC3-associated phagocytosis, while GP63 is necessary for L. major survival. Beatriz R. S. Dias, in 2018, reported an increase in the LC3-II/Act ratio upon infection with L. major and L. amazonensis.
Although LC3 requirement on parasitophorous vacuoles was enhanced after 24 h, no impact on parasite load or infection rate was imposed by autophagy inducers or inhibitors in infected macrophages. However, by reducing NO production, the inducer helps L. major to become more viable than L. amazonensis, whereas the inhibitor had no effect (Dias et al., 2018). In glomerulonephritis caused by L. infantum, elevated levels of the glomerular nucleotide-binding domain leucine-rich repeat-containing-like receptor family, pyrin domain-containing 3, and autophagosome-associated LC3 within glomeruli and tubules were observed to be indicative of the induction or response to glomerular deposition of immune complexes and antigen, according to immune-histopathology and transcriptional analysis (Esch et al., 2015). Apoptotic-like Leishmania uses the autophagic machinery of the host cells to inhibit CD4+ T-cell proliferation, which limits the parasite’s ability to survive intracellularly (Crauwels et al., 2015).
Several studies have examined how autophagic activation impacts the pathophysiology of Leishmania infection, but the interactions between the parasite and the host have produced contradictory and varying results. It is evident that more mechanistic research and explanation are needed for the precise model of interaction between Leishmania species and the host autophagy mechanism. Mechanistic insights into autophagy in disease, which differentiate the autophagic machinery in the host and in the parasite, are proven to be critical for a clearer understanding.
Research on trypanosomatids has indicated the significance of autophagy in the differentiation and remodeling of the parasite under starvation, and it has been proposed that preventing autophagy might be a newer approach to battling parasite illness (Vanrell et al., 2017), (Besteiro et al., 2006), (Besteiro et al., 2007). As autophagy is an essential lysosomal network in response to nutrient deprivation, similar illustrations have been observed in parasite L. major. In Leishmania, autophagy is critical for maintaining cellular homeostasis, recycling damaged organelles, turnover of glycosomes via the Atg5 protein (Cull et al., 2014), and protein turnover (Besteiro et al., 2007). Moreover, metacyclogenesis, the process by which epimastigotes differentiate into metacyclic trypomastigotes, and polyamine metabolism both depend on autophagy (Vanrell et al., 2017).
The function of MVBs (multivesicular bodies) in L. major is examined by characterizing the leishmanial Vps4 homolog. A similar function of VPS4 was observed in L. major when the dominant negative mutant VPS4 (VPS4E235Q) accumulated the mutated protein around vesicular structures of the endocytic system and showed a defect in transport to the MVT lysosome, signifying conservation of the role of this protein in MVB architecture from the early branching kinetoplastid flagellate lineage to mammals. VPS4E235Q overexpressing L. major was impaired in differentiation, and their resistance to starvation proved the VPS4 and MVB compartment role in this process (Besteiro et al., 2006). The role of ATG4.2 in metacyclogenesis (Besteiro et al., 2006) as well as in virulence of the parasite (Williams et al., 2013) was reported. Other than ATG4, around 25 ATG8-like family proteins were discovered and designated as ATG8, ATG8A, ATG8B, and ATG8C. ATG8 has been revealed to form putative autophagosomes during differentiation and starvation of L. major; ATG8A has a role in starvation-induced autophagy, whereas ATG8B and ATG8C both have a distinctive subcellular location near the flagellar pocket, but the incidence of the GFP-labeled puncta recommends that they do not have a role in autophagy. In addition to this role, cysteine peptidase also plays roles in L. major similar to those of yeast and mammals, where ATG4.1 cleaves sessile glycine in ATG8, ATG8B, and ATG8C, while ATG4.2 is able to cleave ATG8A (Williams et al., 2009), (Williams et al., 2013). The parasite homolog of ATG12 has been found on parts of ATG8 containing puncta, indicating toward the ATG5–ATG12 conjugate in the parasite (Williams et al., 2009). The importance of ATG8 in parasite survival and infectivity was reported by Williams et al. (2012), and Giri and Shaha (2019) also reported the role of ATG5 in ATG8-dependent autophagy and mitochondrial homeostasis in L. major, which ultimately showed their significance in parasite virulence.
Leishmania parasite comprises various homologs of ATG proteins, even though the exact autophagic mechanism is not yet deciphered. Research points to a crucial function for autophagy and the protein it produces in the differentiation and infectivity of parasites. Thus, anti-leishmanial research may direct toward autophagy proteins in the parasite. Moreover, strategies developed by parasites to circumvent the host defense mechanism such as autophagy bring autophagy into the spotlight and give weightage to the enticing role of autophagy as an emerging target for therapeutics in leishmaniasis.
The effect of autophagy on parasite survival and its modulation made by the parasite for its own survival establishing the host autophagy pathway serves as a possible therapeutic target in the treatment of leishmaniasis. Available information highlighted a few of the protein molecules which can serve as a target for future therapeutic purposes against leishmaniasis.
Leishmania-infected human monocyte-derived macrophages (hMDMs) lead to stimulation of autophagy LC3-associated phagocytosis (LAP), which has an even sharper effect on decreasing proliferation by inhibiting CD4+ T-cell growth and proliferation of CD4+ T-cells, leading to a reduced intracellular parasite survival. This further implies that host cell autophagy modulates the T-cell response, enhances parasite persistence, and yet has no effect on the viability of the parasite. Apparently, Leishmania utilizes the autophagy mechanism of the host cells to inhibit T-cell proliferation and highlights the autophagy mechanism (Crauwels et al., 2015). CD4+ Th1 cell-mediated immunity is required for protective immunity against leishmaniasis (Okwor et al., 2014), (Peters et al., 2014) because the progression of the parasite is due to the exhaustion of T cells; however, the mechanism involved is not clearly understood (Esch et al., 2013).
Blocking PDL-1 signaling in vivo leads to reinstatement of protective type 1 response by both CD4+ and CD8+ T cells, and as a result, the parasite burden was significantly reduced. Inhibition of PDL-1 enhanced the production of TNF, IL-12, reactive oxygen species (ROS), and nitric oxide (NO) and significantly reduced parasite survival (Roy et al., 2019). Mechanistically, PDL-1 inhibition blocked the autophagy process (Robainas et al., 2017) hijacked by Leishmania to acquire host cell nutrients for their survival (Crauwels et al., 2015). The restoration of effector arms of protective immunity against leishmaniasis and subsequent parasite clearance can be achieved with anti-PDL-1 antibody therapy (Robainas et al., 2017).
The anti-pathogen properties of mammalian target of rapamycin (mTOR) have been proven, as well as its ability to prevent autophagy (Rangwala et al., 2014). Leishmania promotes the PI3K pathway in the host, and PI3K activation inhibits autophagy via enhancing mTOR (Ruhland et al., 2007). The ability of different mTOR inhibitors such as rapamycin, GSK-2126458, and KU-0063794 was evaluated by administering to infected BALB/c mice. In contrast to KU-0063794, rapamycin or GSK-2126458 significantly reduced the parasite load (Khadir et al., 2018) in the draining lymph node as well as the swelling of the footpad. Other studies demonstrated that pre-treatment of host cells with rapamycin to induce autophagy prior to infection was inhibitory to survival of parasites in a concentration-dependent manner, and once infection is established, the stimulation of autophagy by Leishmania confers the advantage of parasite survival.
Thomas et al. (2018) suggested that Leishmania infection delayed induction of host cell autophagy that is essential for the best possible intracellular survival. L. donovani boosted AKT activation to control the GSK-3/-catenin/FOXO-1 axis, preventing host cell apoptosis and immune response necessary for the survival of the parasite in macrophages (Gupta et al., 2016). Taken together, mTOR inhibitors or the PI3K/Akt pathway are used in therapeutic settings for managing leishmaniasis (Madeira Da Silva and Beverley, 2010), (Khadir et al., 2018), (Thomas et al., 2018).
Leishmania infection significantly induces the expression of SIRT1, which inactivated FOXO-1 through deacetylation, constitutively active FOXO-1 led to increased cell death, thereby suggesting that nuclear FOXO-1 might be inactivated (Gutierrez et al., 2004). SIRT1 knockdown led to increased apoptosis and significantly decreased parasite survival along with increased production of TNF-α, ROS, and NO. SIRT1 inhibitor sirtinol in infected mice shows a synergistic effect found with the PD-1 inhibitor. Although they are interconnected, the SIRT1/FOXO-1 axis is used by Leishmania to differentially regulate PD-1 signaling, and both pathways alone support intracellular parasite persistence (Roy et al., 2019).
The parasite uses the PERK/ATF4 pathway, which is crucial for maintaining protein homeostasis and establishing infection (Abhishek et al., 2018). Additionally, PERK signaling reduces translation and promotes ATF4 expression, which either promotes cell survival or causes cell death. Cytoprotective autophagy genes, ATG5 and ATG7, are known to be upregulated as a result of PERK/ATF4 pathway activation (Zheng et al., 2019), which favors cellular survival and parasite infection (Aoki et al., 2019). GSK2606414 previously used as an anti-cancerous drug was discovered through chemical inhibition tests to assess prospective drugs since it targets the kinase domain of PERK (Rangwala et al., 2014), and its therapeutic implications and role in leishmanial infection may substantiate it as a repurposed drug for treatment options for leishmaniasis.
PGE2 (prostaglandin E2) is responsible for the increment in parasite load in macrophages (Ribeiro-Gomes et al., 2004), and the physiological stimulation of autophagy promotes the intracellular survival of L. amazonensis by a process involving increases in the synthesis of lipid bodies and PGE2, as well as lower NO levels (Vallochi et al., 2018). Also, Martínez and Ruiz (2019) stated that lysosomes destroy LDs after autophagosomes sequester it during lipophagy. Growing evidence points to the involvement of LDs in the control of the autophagic process and their eradication by autophagy via lipophagy. Thus, blocking autophagy-mediated process via lipid droplets may serve as a newer area to combat leishmaniasis.
Recent research has shown that the host microRNA, MIR30A-3p, may regulate host autophagy following L. donovani infection. After L. donovani infection, MIR30A-3p expression is significantly increased in a time-dependent manner. MIR30A-3p mimic decreased and with antagomir increased the expression of BECN1 protein in L. donovani-infected macrophages, demonstrating the role of MIR30A-3p in autophagy in leishmaniasis (Singh et al., 2016). Autophagy and miRNAs are interconnected (Frank et al., 2015). BMDM showed intracellular amastigote clearance by autophagy in BMDM through mTOR phosphorylation-dependent counteracting mechanism after in vitro infection with L. major. Additionally, the study demonstrates a relationship between the infection-specific overexpression of (BCL2/adenovirus E1B 19 kDa protein-interacting protein) BNIP3 and (cathepsin-E) CTSE and autophagy-related protein 5 (ATG5). There is evidence for showing the involvement of miR-101c, miR-129, and miR-210 in RNA-level autophagy regulation during L. major infection and miRNAs have been linked to lipid metabolism, not in leishmaniasis but in other disorders (Latorre et al., 2017).
Altogether in L. major-infected host macrophages, autophagy is a tightly controlled cellular process at the RNA and protein levels (Frank et al., 2015), suggesting a role of RNAs in the modulation of autophagy in leishmaniasis.
A new era is emerging where people are trying to combat fatal situations with the use of microRNAs. So, in this review, we elaborate on the collaborative functioning of microRNAs with lipid-mediated autophagy, and the respective probability of incorporation of miRNA and lipids concerning autophagy in therapeutic approaches. The capacity of miRNAs to disrupt various signaling pathways and consequently alter the cellular response and the outcome of diseases is a current hotspot in medical research science (Yang and Wang, 2016), (Butterworth, 2018), (Barbu et al., 2020), (Gorabi et al., 2020), (Gholamrezaei et al., 2020). Gholamrezaei et al. (2020) have studied that the miR-155 inhibitor and mimic effect in L. major-infected macrophages can induce apoptosis and reduce parasite burden and hence highlighting it as an essential biomolecule that may be utilized as a therapeutic target against the disease. However, Taganov et al. (2006) have suggested that induction of miR-146a/b is triggered by those TLRs that mainly sense viral nucleic acids and localize in the intracellular area such as TLR2, TLR4, and TLR5 and proposed a role of a miR-146 in control of TLR and cytokine signaling through a negative feedback regulation loop involving downregulation of IRAK1 and TRAF6 protein levels. Pauley et al. (2010) have shown that the production of 5 cytokines/chemokines (IL-6, IP-10, IL-8, MCP-1, and IL-1b) was reduced after transfection of miR-146a mimic. Thus, it is demonstrated that miR-146a controls acute inflammation caused by LPS stimulation by limiting the production of cytokines after the initial innate response. O’Connell et al. (2007) Findings imply that the JNK pathway, which activates the AP-1 complex, is involved in miR-155 induction by both TLRs and TNF-α, presumably through transcriptional activation of the miR155 encoding BIC gene. Members of the let-7 family of miRNAs are well-represented in the data set in the study by Geraci et al. (2015). Most noticeable are the let-7a and let-7b dichotomous expression patterns, where only DCs and MPs infected with L. donovani display upregulation of those miRNAs, and cells infected with L. major were observed to downregulate the same miRNA (Geraci et al., 2015). For VL (Singh et al., 2016), results confirm that BECN1 is a potential target of MIR30A-3p in both THP-1 cells and HsMDMs infected with L. donovani. Di Loria et al. (2020) have studied that Leishmania organisms can disrupt the production of miR122, which will disrupt the metabolism of cholesterol and ensure its proliferation in the infected host, emphasizing its capacity to hamper lipid metabolism of the host.
The review clarifies that the invading pathogen’s main targets for modulating and maintaining infection in the host are the post-transcriptional and post-translational stages, miRNAs are significant contenders for considering their role in lipid-mediated autophagy.
While it is important to control autophagy brought on by infection to manage parasite survival, it is also important to pay attention to proteins that may serve as targets for autophagy in parasites. Numerous research studies attempt to uncover proteins or chemicals that might impair parasite diversity, infectiousness, or survival.
Unsatisfactory leishmaniasis treatment encourages researchers to investigate more modern options, which now target the parasite’s functioning proteins or molecules. The heat shock protein 90 (Hsp90) is believed to be a commendable drug target against parasitic diseases (Debnath et al., 2014), (Stofberg et al., 2021), (O. Angel et al., 2013), (Silva et al., 2013), (De Andrade et al., 1992). 17-N-allylamino-17-demethoxygeldanamycin (17-AAG), a Hsp90 inhibitor, caused a leishmanicidal effect in vitro (Petersen et al., 2012) and in vivo (Santos et al., 2014) in leishmaniasis. In promastigotes, 17-AAG induces an autophagy pathway, leading to parasite death. Following treatment with 17-AAG, ∆atg5 parasites are less prone to cell death, which shows that autophagy is involved in inhibitor-induced parasite death, where ATG5 is playing an important role (Petersen et al., 2021).
Transporter proteins of the ABC subfamily, LABCG2, are essential for Leishmania virulence (Campos-Salinas et al., 2013). It is necessary for the externalization of phosphatidylserine, a macrophage invasion adaptation strategy. Additionally, it was shown that LABCG1 was a crucial component, cooperating with LABCG2 to control metacyclogenesis, infectivity, oxidative stress, and autophagy (Manzano et al., 2017). Studies on this transporter’s mutations showed that the parasite’s pathogenesis and virulence were both reduced. As a result, these transporters may be exploited as therapeutic targets, and in combination with them, it may be possible to overcome drug resistance (Kabra et al., 2020).
Natural indoloquinoline alkaloid cryptolepine (CLP) induces cellular dysfunction in L. donovani promastigotes, which leads to the contribution of initial autophagic response by showing monodansylcadaverine (MDC)-labeled autophagic vacuoles. However, cells later exhibit characteristics of cell death which may not be because of autophagy but due to prolonged intracellular stress due to CLP (Sengupta et al., 2011). The toxicity of quinoline derivative salts (QDS) was evaluated in mammalian cells, and the mechanism of action proved it as a promising compound (Calixto et al., 2018), while the in vivo effect of the qunoline derivative (AMQ-j) in BALB/c mice was evaluated (Antinarelli et al., 2018). QDS highlighted the anti-leishmanial impact of this substance by inhibiting the formation of autophagic vacuoles, which may have contributed to parasite death by impeding autophagic mechanisms in the clearance of damaged organelles. Additionally, an increase in ROS levels was observed, reinforcing that the induction of oxidative stress triggered the death of intracellular parasites (Calixto et al., 2018). On the other hand, the quinoline derivative (AMQ-j) exhibited several morphological and biochemical changes caused by AMQ-j, including intense mitochondrial swelling, collapse of the mitochondrial membrane potential, abnormal chromatin condensation, externalization of phosphatidylserine, accumulation of lipid bodies, disruption of the cell cycle, formation of autophagic vacuoles, and increase in acidic compartments (Antinarelli et al., 2018). These changes could be related to both QDS and AMQ-j and show a strong effect of autophagy-related and apoptosis-like processes against L. amazonensis and its anti-leishmanial activity (Calixto et al., 2018), (Antinarelli et al., 2018). Thiosemicarbazone molecule (BZTS) in synergy with the available drug miltefosine shows a significant increase in reactive oxygen and nitrogen species as a result of mitochondrial malfunction, leading to severe cell damage, an extensive autophagic process, and the ensuing apoptotic cell death in parasites by least damaging mammalian cells.
Thus, these findings support the synergetic impact on several metabolic pathways in leishmaniasis and encourage the notion of therapeutics toward metabolic pathways and further target identification (Scariot et al., 2017). Anti-leishmanial activity of apigenin in vitro and in vivo was described. Apigenin induced reactive oxygen species (ROS) generation, and the number of autophagosomes after the infection is suggestive of the involvement of host autophagy in macrophages infected with L. amazonensis (Fonseca-Silva et al., 2016). The role of ATG8 in L. donovani for its survival mechanism and infectivity showed it as a drug target. Molecular docking and molecular dynamic simulation studies show the enticing role of thiabendazole derivatives that impede the survival mechanisms by affecting ATG8 (Guhe et al., 2022), while virtual screening of natural ligand library had been performed against ATG8 of L. donovani, showing that urolithin-A which is formed by combinations of coumarin and isocoumarin stably blocked ATG8 (Rahman et al., 2022).
The attention of phenomenal research in leishmaniasis toward therapeutics considering limitations of current treatments is shifting toward autophagy. Targets identified in the Leishmania parasite and respective drugs are enlisted in Table 2. However, there is also a need for a novel approach due to the intricacy of the autophagy network, its connections to other metabolic pathways in the host, and homological conflicts between the presence of autophagy in the host and parasite. Given the complexity of the Leishmania–host relationship, further studies should concentrate on the genetic investigation of autophagic pathway alteration.
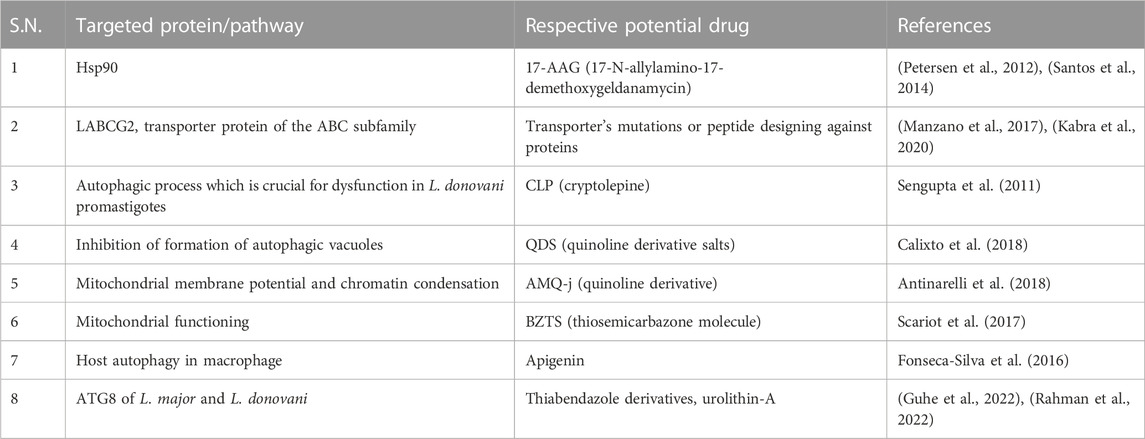
TABLE 2. Drugs showing anti-leishmanial effect by modulating the autophagy process and their respective targets.
The enormous growth in genomic data for pathogens, particularly the available genome sequence, has created a capacity to discover potential vaccines or chemotherapeutic targets. Strategies to accomplish clinical objectives have been made clear by the advent of functional genomics techniques, such as microarray and more recently deep sequencing technologies, as well as proteomics (Meissner et al., 2022). For instance, the proteomic technique is utilized for identifying the binding proteins for a given small molecule. Due to its arduousness and length, this strategy has not been very effective in identifying targets (Ivens et al., 2005). Additionally, in the post-genomic timeframe, these advancements enabled bioinformatics to arise, significantly accelerating the research process. A variety of confirmatory assays, computational algorithms, and high-throughput screening techniques all considerably aid in the identification and characterization of emerging, powerful drug targets (Tao et al., 2004), (Tang et al., 2006). In addition, several computational (in silico) technologies have also been designed.
In diseases caused by external pathogens like bacteria, viruses, and parasites, a comparative functional genomics from humans to the respective pathogen is found to be crucial for identification of novel targets (García-Lara et al., 2005). Genome availability is a key regulator when genetic techniques and bioinformatics approaches are taken into consideration for the understanding of the system. In the post-genomic era, genome information of various pathogens was deciphered, including Leishmania (Ivens et al., 2005), disclosing the haploid genome of L. major (Friedlin strain), where genes involved in host–pathogen interactions were highlighted. Based on the annotation released by the Wellcome Trust Sanger Institute and the genomic sequence of L. major (v5.2), the LeishCyc database was originally developed. LeishCyc is a platform for the analysis, interpretation, and visualization of Leishmania omics data (transcriptomics, proteomics, and metabolomics) in the context of metabolic pathways and offers a systematic approach to structuring the increasing information regarding Leishmania biochemical networks (Doyle et al., 2009). After the post-genomic era, various studies switched to computational approaches for discovering new drug targets as genomic data are available for Leishmania as well. Target-based drug development methods that block a single target molecule hardly ever produce the desired results, but bioinformatics approaches provided a ray of hope for this hypothesis (Schreiber et al., 2016).
A potential field in systems biology, the use of artificial intelligence (AI) is receiving much attention as a result of the present data explosion. Over the decade, AI has revolutionized the process of drug discovery as a computational aspect. Autophagy has been studied using AI-based methods, as it has many other biological processes. He et al. (2020) used machine learning (ML) methods to categorize renal cell cancer (RCC) subtypes using autophagy proteins. Recently, various software that incorporate a wide range of ML algorithms have been created. For instance, Serrano et al. (2018) studied the impact of mRNA variations of a few autophagic genes, one pro-apoptotic gene and one anti-apoptotic gene in HIV-infected patients who were successfully treated with combined antiretroviral therapy using scikit-learn. Numerous applications of AI in drug discovery, including virtual screening and drug design, have been used (Walters and Barzilay, 2021).
Due to the growing accessibility of enormous pharmacological and chemical datasets, leishmaniasis and other NTDs (neglected tropical diseases) are being thoroughly investigated using ML models that use a non-clinical approach. Utilizing ML and virtual screening technologies, an organic compound dataset with activity against L. amazonensis was used to identify novel potential therapeutic options such as leishmanicides (Mollalo et al., 2018), (Tuon et al., 2022). Ornithine decarboxylase (ODC), a key enzyme in Leishmania proliferation, is inhibited by deguelin. Deguelin derivatives showed greater affinity and specificity against ODC than the original molecule using an in silico-generated combinatorial library and docking techniques. Thus, deguelin has been revealed as a promising approach for treating the infection by computer-aided drug design (CADD) (Vicente-Barrueco et al., 2022). The computational technique uses ML to build prediction models for molecular classification to find new leishmaniasis treatment components (Jamal and Scaria, 2013). Computational approaches also manifest crucial derivatives as a repurposing of the available drug to target specific processes. Since ATG8 of Leishmania plays a crucial role in infectivity and virulence (Giri and Shaha, 2019), it can be a potential target by using computational approaches for drug screening. Thiabendazole and urolithin A were shown to be possible drug molecules to combat against autophagy in leishmaniasis (Guhe et al., 2022), (Rahman et al, 2022.)
On the other hand, chemical agents that disrupt this pathway at the right molecule can be chosen if a crucial pathway is a target. Evidently, research on complex biological systems should acknowledge the constraints of simple systems in order to devise effective and safe therapeutics to treat disease. A systems biology approach, or a combination of experimental and computational study, is necessary to comprehend complex biological systems.
Cells can respond by changing their transcriptional activity, metabolism, or other regulatory processes in response to changes in their environment (internal stimuli and external stimuli) through signaling cascades. The role of these cells is crucial since it aids in cell survival and differentiation in various circumstances. It also controls the plasticity of cells in multicellular organisms. To understand the complex behavior of signaling networks, computational system modeling techniques were utilized to recreate conceptual models that highlight some fundamental characteristics of signaling routes and interpret prototypic signaling networks. Computational biology offers a solid platform to approach pertinent scientific issues through practical modeling and theoretical analysis.
Signaling pathways permit cells to perceive changes in their environment, external and internal stimuli, and respond by altering their transcriptional activity, metabolism, or other regulatory processes. Because it helps cell survival and differentiates in varied situations, the function of these cells is essential. Additionally, it controls the plasticity of cells in multicellular organisms. Computational systems modeling techniques were used to interpret prototypic signaling networks by reconstructing conceptual models that underline some key attributes of signaling pathways in order to comprehend the intricate behavior of signaling networks. Through pragmatic modeling and theoretical examination, computational biology provides a robust framework to address relevant scientific problems.
Network analysis in systems biology can be divided into two groups. Enrichment analysis falls under the first group. The objective of the second category, which is based on algorithm-related activities, is to identify possible targets or significant nodes in a network. It leads to centrality analysis, which has been used to identify the key nodes in the network. These techniques include radiality, clustering coefficient, betweenness centrality, and degree centrality. For the network-based study, the most widely used visualization and analysis software are Cytoscape (Shannon et al., 1971), Gephi (Bastian et al., 2009), Tulip (Auber et al., 2004), and Pajek (Mrvar and Batagelj, 2016). According to a bioinformatics study, NRF2 was interconnected to several signaling pathways while analyzing the complex autophagic network, including the AGE–RAGE pathway, MAPK pathway, NF-kappa B signaling, PI3K–Akt signaling, and VEGF signaling. This pathway enrichment was performed using “edgeR” R, and KEGG enrichment analysis was carried out with the DAVID system. Based on data analysis and experimental validation, NRF2 may be considered a tumor suppressor in tumorigenesis but promotes prostate cancer. Further confirmations offer more evidence that NRF2 is a crucial regulator and that blocking NRF2 and autophagy may be effective CRPC (castration-resistant prostate cancer) therapy (Zhang et al., 2022).
When it comes to network analysis, systems biology studies mainly rely on biological databases. They provide access to a wide range of biologically significant data, such as the PPI (protein–protein interaction) network, disease–protein association data, microarray, next-generation sequencing, protein localization, post-translational modification, structural information about a protein or compound, and pathways related to proteins. However, there are no many databases that provide data about autophagy and about leishmaniasis. Databases available to the best of our knowledge based on host–pathogen interaction are enlisted in Table 3:
A mathematical model serves as a reflector of the complexity of biological processes. A biological system’s complexity arises from a variety of causes. One of these is the system’s fundamental hierarchy, which extends from the level of cells to that of organisms. The hierarchical levels are all dynamic. Acute significant and stochastic alterations may occur at any time, even for minute changes in the cellular environment, even though they mimic regular and predictable activity. These uncertainties provide complications that are challenging for experimental research to fully understand. Even if they exist, it is difficult to understand the actual pathways and patterns of complex evolution. Because most autophagy biochemical events are non-linear, a small alteration in any one of its steps may not have a noticeable impact on the system as a whole.
To pinpoint the autophagy steps that oversee a specific system behavior, mathematical modeling supports reduced abstractions and approximations. Additionally, the system’s ongoing behavioral change introduces randomness into the autophagy process. In order to study the dynamics of the system in response to any changes in the environment brought on by numerous external or internal perturbations or signals, mathematical modeling of autophagy keeps track of these elements.
Experimental research and mathematical models have been used to demonstrate the intricate dynamics of autophagy. Studying the impact of starvation-induced autophagy on cell (yeast) population dynamics is a mathematical modeling application (Jin and Lei, 2014). Models investigate the dynamics of the proteins that control and influence various phases of macroautophagy. The fundamental set of proteins or pathways in various cellular conditions and diseases, such as cancer and Alzheimer’s disease, have been mathematically explored to capture the process of disease progression and to extract the significant parameter to restore cell homeostasis under disease situations (Han et al., 2012; Martin et al., 2013; Hao and Friedman, 2016; Enderling et al., 2007; Louzoun et al., 2014).
The computational model generated novel hypotheses that can be tested experimentally regarding the effects of ATG9 depletion and variation in LC3 copy number in mammalian cells (Martin et al., 2013). In conclusion, a mathematical model focuses on unraveling the intricate web of related autophagic processes in order to highlight the crucial ATG proteins or other molecules within the cascade. In processes like the destruction of pancreatic beta cells that cause diabetes or treatment of cancer, reactive oxygen species (ROS) dynamics are crucial. However, proposed data still fall short of a comprehensive explanation of existing data of cell metabolic and ROS dynamics. A computationally assisted mathematical model emphasized on the relationship between the metabolic processes in the cytoplasm, such as glycolysis, lactate and ATP production and consumption, and in the mitochondria, such as pyruvate reduction, NAD(P)H production, the function of the electron transport chain complexes, and others that control ROS dynamics (Fridlyand and Philipson, 2017).
By developing an ODE model, Kapuy et al. investigated Beclin1-mediated autophagy and caspase-mediated apoptosis. The B-cell lymphoma 2 (BCL2)–Beclin1–caspase minimal network was the focus of the model’s construction. The observation led to the hypothesis that the transition from autophagy to apoptosis is controlled by a bistable switch and that the sequential activation of cellular response can be started by a combination of BCL2-dependent regulation and feedback loops between Beclin1 and caspases depending on the intensity and duration of stress levels (Kapuy et al., 2013). Another study also suggested that TLR2/6 activates proinflammatory cytokines, whereas TNF-α activates PI3P to activate autophagy while causing autophagy inhibition by ATG9, showing the ATG9–PI3P axis acts as a negative feedback loop in autophagic machinery in the infection model of leishmaniasis (Guhe et al., 2022).
Although computational approaches have been studied in relation to other diseases, there are very few papers that highlight the autophagic behavior in leishmaniasis utilizing a systems biology approach, notably through mathematical modeling and network analysis. In order to help the researchers, the complexity of the autophagy network relevant to leishmaniasis is lessened, and this review recommends them to concentrate more of their efforts on computationally assisted techniques.
A fascinating field called synthetic biology emerged at the beginning of this century. It builds genetic programming systems for desired cellular behavior using molecular biology tools and advanced engineering ideas (Cameron et al., 2014). The goal is to replicate emergent biological processes using synthetic chemicals to develop artificial life. The other goals involve assembling systems with abnormal behavior using replaceable components from natural biology.
The term “synthetic biology” was introduced by Eric Kool and other speakers at the American Chemical Society’s annual meeting in San Francisco in 2000 (Benner and Sismour, 2005). Synthetic biology aims to expand the range of potential biological functions for therapeutic and scientific applications by making biology more efficient, dependable, and predictable to construct. By altering, recombining, and synthesizing wholly new genetic components, synthetic biologists rewire biological systems. This strategy is now feasible because there are more genetic building pieces available and we have a better grasp of how to assemble bimolecular modules, such as protein interaction networks and DNA regulatory sequences. The use of synthetic biology also benefits from developments in mathematical modeling and from engineering-related principles (Endy, 2005). Very few recent pieces of evidence are published that included autophagy modulation via genetic or synthetic circuits (Dey and Rangarajan, 2021), (Gupta et al., 2020). Literature also envisions the importance of the synthetic circuit in developing a leishmaniasis treatment plan (Mol et al., 2014), (Soni and Singh, 2022). There is still opportunity for research and the creation of new strategies for treating the disease by targeting the autophagic processes because, as far as we are aware, there are no studies that describe an autophagy regulation-based synthetic circuit in leishmaniasis.
A less explored and cardinal area in leishmanial infection is lipid metabolism, and it has become the focus nowadays showing its relationship with the autophagy process. The process of autophagy has been a key topic of research due to the intricate mechanism in many biological processes and diseases, because it is still a mystery with many unsolved issues, just like in leishmaniasis. The best methods to find these components might be systems biology-based ones. These methods can be used to gain a thorough understanding of the relationship and operation between lipids, miRNA, and autophagy. This study aims to highlight the interactions between essential biomolecules including RNA, proteins, and lipids that help maintain homeostasis and proper functioning of the cellular process of autophagy and also to identify novel strategies for the treatment of critical elements associated with these three key proteins for autophagy. Recent research on the function of autophagy in leishmaniasis and how it affects parasite survival has been widely disseminated, highlighting the growing significance of this process. The role of several biomolecules, including lipids, proteins, and mRNA, particularly miRNA, in modifying the autophagy process is another research focus. Recently, different ATG proteins such as LC3B, ATG9, ATG5, and ATG7 and other signaling molecules such as mTOR, FOXO, PI3K, and PDL have been shown to play a role in autophagy regulation in infection conditions. Study of autophagy in leishmaniasis is giving more attention to the PI3K/AKT pathway, PERK/ATF4 signaling, and FOXO1/SIRT1 axis. Moreover, LABCG2, an ABC family transporter; HSP90; ATG5; and an ATG8 homolog in Leishmania are a few more targets that have been found important for understanding leishmaniasis in experimental studies (Table 4).
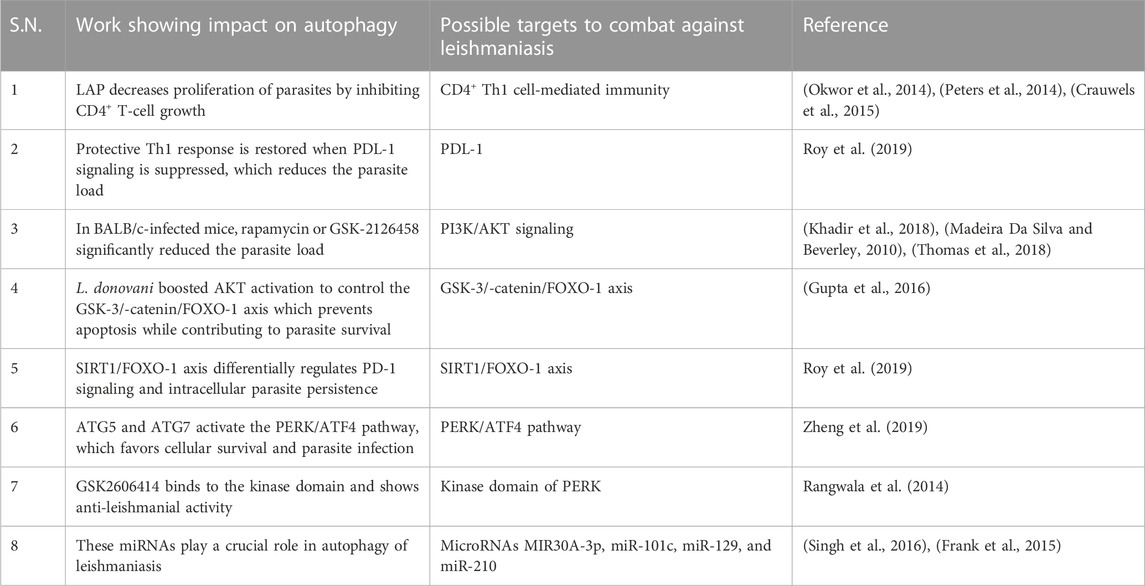
TABLE 4. Probable targets in the host for regulating the autophagy process in leishmaniasis and its impact on the fate of parasite.
However, there is very little computational research compared to experimental studies. The review encourages filling these gaps to gain a better understanding of the dynamics of the autophagy pathways and their regulation in leishmaniasis. The literature has described the use of a variety of computational techniques in systems biology, including models based on mathematical modeling, AI-based approaches, synthetic biology-based techniques, genetic techniques, and image-based modeling, among many others. However, these techniques have not been sufficiently studied concerning leishmaniasis.
From the perspective of systems biology analysis, a handful of literature reported on autophagy under Leishmania infection. Such a study will help decipher the change in behavior of autophagy and its impact on parasites and help identify potential drug targets against disease. A structure-based study can be performed to determine the possible binding sites of the targets. The identification of binding sites of a protein can help in the rational designing of the therapeutic agents, which could be used to target processes by synthetic biology approaches via genetic circuits. This review also facilitates mathematical modeling to check the robustness of the network and for the identification of important nodes. A pipeline-based study is required in leishmaniasis for targeting the autophagic process and will surely aid to deliver fruitful insights into the role of autophagy in leishmaniasis and its impact on the proliferation and progression of the disease.
The need for new therapeutics against leishmaniasis is driven by the toxicity and drug resistance limitations of currently used chemotherapeutics. In leishmaniasis, it is unclear whether autophagy plays a role in disease progression or regression. Autophagy is a fundamental biological process that eliminates unwanted cellular components and pathogens in order to maintain cellular homeostasis. Due to its significant influence on cellular fate, autophagy is still dominating many cellular processes and signaling networks. Monitoring of genes involved in autophagy and signals that control autophagy have been used to identify and manage the autophagic pathways, and any disruption to any of them may result in several diseases. Given the complexity of autophagy and immunological signaling in disease systems and the growing attention to autophagy research in leishmaniasis, synthetic techniques may prove to be a successful therapeutic approach to cure leishmaniasis. Following many years of research, the experimental evidence on autophagy has grown, creating a thin-line intersection between lipids, proteins, and miRNA for the regulation of autophagy that needs to be investigated. Application of computational biology tools to these datasets may help in understanding information on the crucial proteins that regulate the autophagic processes and their underlying dynamics, which may significantly advance ongoing leishmaniasis research on its therapeutics. In conclusion, our study contributed to the advancement of autophagy research and showed how important biomolecules including lipids, proteins, and miRNA regulate this process. We advocate that in leishmaniasis, computational system biology tools may lead to a promising therapeutic approach for finding newer treatment regimens in the autophagic processes mediated by lipid, protein, and miRNAs.
VG, PI, and AT contributed in the original writing of the manuscript. VG and PI prepared the figures of the manuscript. SS laid the conceptual framework and editing the manuscript.
We would like to thank extra mural funding from the ICMR (Indian Council of Medical Research), New Delhi, Project No. ICMR 58/39/20-PHA/BMS.
VG would like to acknowledge her Senior Research Fellowship from UGC. PI would like to acknowledge her ICMR-Senior Research Fellowship. AT would like to acknowledge his Junior Research Fellowship from the ICMR.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abhishek, K., Das, S., Kumar, A., Kumar, A., Kumar, V., Saini, S., et al. (2018). Leishmania donovani induced Unfolded Protein Response delays host cell apoptosis in PERK dependent manner. PLoS Neglected Trop. Dis. 12 (7), 00066466–e6722. doi:10.1371/journal.pntd.0006646
Akilov, O. E., Kosaka, S., O’riordan, K., and Hasan, T. (2007). Parasiticidal effect of δ-aminolevulinic acid-based photodynamic therapy for cutaneous leishmaniasis is indirect and mediated through the killing of the host cells. Exp. Dermatol. 16 (8), 651–660. doi:10.1111/j.1600-0625.2007.00578.x
Alavizadeh, S. H., Badiee, A., Khamesipour, A., Jalali, S. A., Firouzmand, H., Abbasi, A., et al. (2012). The role of liposome-protamine-DNA nanoparticles containing CpG oligodeoxynucleotides in the course of infection induced by Leishmania major in BALB/c mice. Exp. Parasitol. 132 (3), 313–319. doi:10.1016/j.exppara.2012.06.014
Alrajhi, A. A., Ibrahim, E. A., De Vol, E. B., Khairat, M., Faris, R. M., and Maguire, J. H. (2002). Fluconazole for the treatment of cutaneous leishmaniasis caused by Leishmania major. Leishmania major N. Engl. J. Med. 346 (12), 891–895. doi:10.1056/nejmoa011882
Alvarez, V. E., Kosec, G., Sant’Anna, C., Turk, V., Cazzulo, J. J., and Turk, B. (2008). Autophagy is involved in nutritional stress response and differentiation in Trypanosoma cruzi. J. Biol. Chem. 283 (6), 3454–3464. doi:10.1074/jbc.M708474200
Andrade, R. M., Wessendarp, M., Gubbels, M. J., Striepen, B., and Subauste, C. S. (2006). CD40 induces macrophage anti-Toxoplasma gondii activity by triggering autophagy-dependent fusion of pathogen-containing vacuoles and lysosomes. J. Clin. Investigation 116 (9), 2366–2377. doi:10.1172/JCI28796
Andrade-neto, V. V., Cunha-junior, E. F., Faioes, S., Pereira, T., Silva, R. L., Leon, L. L., et al. (1996). Drug repurposing for leishmaniases, 967–996.
Angel, S. O., Matrajt, M., and Echeverria, P. C. (2013). A review of recent patents on the Protozoan parasite HSP90 as a drug target. Recent Pat. Biotechnol. 7 (1), 2–8. doi:10.2174/1872208311307010002
Antinarelli, L. M. R., de Oliveira Souza, I., Zabala Capriles, P. V., Gameiro, J., Britta, E. A., Nakamura, C. V., et al. (2018). Antileishmanial activity of a 4-hydrazinoquinoline derivative: Induction of autophagy and apoptosis-related processes and effectiveness in experimental cutaneous leishmaniasis. Exp. Parasitol. 195, 78–86. doi:10.1016/j.exppara.2018.10.007
Aoki, J. I., Laranjeira-Silva, M. F., Muxel, S. M., and Floeter-Winter, L. M. (2019). The impact of arginase activity on virulence factors of Leishmania amazonensis. Curr. Opin. Microbiol. 52, 110–115. doi:10.1016/j.mib.2019.06.003
Auber, D., Farin, G., Hege, H.-C., Hoffman, D., Johnson, C. R., and Polthier, K. (2004). “Tulip - a huge graph visualization software,” in Graph drawing software, 105–126. Available at: http://dept-info.labri.fr/∼auber/documents/publi/auberChapterTulipGDSBook.pdf.
Awasthi, B. P., Kathuria, M., Pant, G., Kumari, N., and Mitra, K. (2016). Plumbagin, a plant-derived naphthoquinone metabolite induces mitochondria mediated apoptosis-like cell death in Leishmania donovani: An ultrastructural and physiological study. Apoptosis 21 (8), 941–953. doi:10.1007/s10495-016-1259-9
Barbu, M. G., Condrat, C. E., Thompson, D. C., Bugnar, O. L., Cretoiu, D., Toader, O. D., et al. (2020). MicroRNA involvement in signaling pathways during viral infection. Front. Cell Dev. Biol. 8, 143–222. doi:10.3389/fcell.2020.00143
Bastian, M., Heymann, S., and Jacomy, M. (2009). “Gephi: An open source software for exploring and manipulating networks visualization and exploration of large graphs,” in Proceedings of the international AAAI conference on web and social media, 361–362. www.aaai.org.
Baumann, R. J., Hanson, W. L., McCann, P. P., Sjoerdsma, A., and Bitonti, A. J. (1990). Suppression of both antimony-susceptible and antimony-resistant Leishmania donovani by a bis(benzyl)polyamine analog. Antimicrob. Agents Chemother. 34 (5), 722–727. doi:10.1128/AAC.34.5.722
Benner, S. A., and Sismour, A. M. (2005). Synthetic biology. Nat. Rev. Genet. 6 (7), 533–543. doi:10.1038/nrg1637
Bento, C. F., Renna, M., Ghislat, G., Puri, C., Ashkenazi, A., Vicinanza, M., et al. (2016). Mammalian autophagy: How does it work? Annu. Rev. Biochem. 85, 685–713. doi:10.1146/annurev-biochem-060815-014556
Besteiro, S., Williams, R. A. M., Coombs, G. H., and Mottram, J. C. (2007). Protein turnover and differentiation in Leishmania. Int. J. Parasitol. 37 (10), 1063–1075. doi:10.1016/j.ijpara.2007.03.008
Besteiro, S., Williams, R. A. M., Morrison, L. S., Coombs, G. H., and Mottram, J. C. (2006). Endosome sorting and autophagy are essential for differentiation and virulence of Leishmania major. J. Biol. Chem. 281 (16), 11384–11396. doi:10.1074/jbc.M512307200
Biagiotti, M., Dominguez, S., Yamout, N., and Zufferey, R. (2017). Lipidomics and anti-trypanosomatid chemotherapy. Clin. Transl. Med. 6 (1), 27. doi:10.1186/s40169-017-0160-7
Boitz, J. M., Yates, P. A., Kline, C., Gaur, U., Wilson, M. E., Ullman, B., et al. (2009). Leishmania donovani ornithine decarboxylase is indispensable for parasite survival in the mammalian host. Infect. Immun. 77 (2), 756–763. doi:10.1128/IAI.01236-08
Bouazizi-Ben Messaoud, H., Guichard, M., Lawton, P., Delton, I., and Azzouz-Maache, S. (2017). Changes in lipid and fatty acid composition during intramacrophagic transformation of Leishmania donovani complex promastigotes into amastigotes. Lipids 52 (5), 433–441. doi:10.1007/s11745-017-4233-6
Brun, R., Bühler, Y., Sandmeier, U., Kaminsky, R., Bacchi, C. J., Rattendi, D., et al. (1996). In vitro trypanocidal activities of new S-adenosylmethionine decarboxylase inhibitors. Antimicrob. Agents Chemother. 40 (6), 1442–1447. doi:10.1128/aac.40.6.1442
Butterworth, M. B. (2018). Role of microRNAs in aldosterone signaling. Curr. Opin. Nephrol. Hypertens. 27 (5), 390–394. doi:10.1097/MNH.0000000000000440
Calixto, S. L., Glanzmann, N., Xavier Silveira, M. M., da Trindade Granato, J., Gorza Scopel, K. K., Torres de Aguiar, T., et al. (2018). Novel organic salts based on quinoline derivatives: The in vitro activity trigger apoptosis inhibiting autophagy in Leishmania spp. Chemico-Biological Interact. 293, 141–151. doi:10.1016/j.cbi.2018.08.003
Cameron, D. E., Bashor, C. J., and Collins, J. J. (2014). A brief history of synthetic biology. Nat. Rev. Microbiol. 12 (5), 381–390. doi:10.1038/nrmicro3239
Campos-Salinas, J., León-Guerrero, D., González-Rey, E., Delgado, M., Castanys, S., Pérez-Victoria, J. M., et al. (2013). LABCG2, a new ABC transporter implicated in phosphatidylserine exposure, is involved in the infectivity and pathogenicity of leishmania. PLoS Neglected Trop. Dis. 7 (4), e2179. doi:10.1371/journal.pntd.0002179
Campoy, E., and Colombo, M. I. (2009). Autophagy in intracellular bacterial infection. Biochimica Biophysica Acta - Mol. Cell Res. 1793 (9), 1465–1477. doi:10.1016/j.bbamcr.2009.03.003
Chattopadhyay, A., and Jafurulla, M. (2011). A novel mechanism for an old drug: Amphotericin B in the treatment of visceral leishmaniasis. Biochem. Biophysical Res. Commun. 416 (1–2), 7–12. doi:10.1016/j.bbrc.2011.11.023
Colmenares, M., Kar, S., Goldsmith-Pestana, K., and McMahon-Pratt, D. (2002). Mechanisms of pathogenesis: Differences amongst Leishmania species. Trans. R. Soc. Trop. Med. Hyg. 96 (1), S3–S7. doi:10.1016/s0035-9203(02)90044-1
Cooney, R., Baker, J., Brain, O., Danis, B., Pichulik, T., Allan, P., et al. (2010). NOD2 stimulation induces autophagy in dendritic cells influencing bacterial handling and antigen presentation. Nat. Med. 16 (1), 90–97. doi:10.1038/nm.2069
Crauwels, P., Bohn, R., Thomas, M., Gottwalt, S., Jäckel, F., Krämer, S., et al. (2015). Apoptotic-like Leishmania exploit the host´s autophagy machinery to reduce T-cell-mediated parasite elimination. Autophagy 11 (2), 285–297. doi:10.1080/15548627.2014.998904
Croft, S. L., Sundar, S., and Fairlamb, A. H. (2006). Drug resistance in leishmaniasis. Clin. Microbiol. Rev. 19 (1), 111–126. doi:10.1128/CMR.19.1.111-126.2006
Cull, B., Godinho, J. L. P., Rodrigues, J. C. F., Frank, B., Schurigt, U., Williams, R. A. M., et al. (2014). Glycosome turnover in Leishmania major is mediated by autophagy. Autophagy 10 (12), 2143–2157. doi:10.4161/auto.36438
Cyrino, L. T., Araújo, A. P., Joazeiro, P. P., Vicente, C. P., and Giorgio, S. (2012). In vivo and in vitro Leishmania amazonensis infection induces autophagy in macrophages. Tissue Cell 44 (6), 401–408. doi:10.1016/j.tice.2012.08.003
Da Silva, E. R., Boechat, N., Pinheiro, L. C. S., Bastos, M. M., Costa, C. C. P., Bartholomeu, J. C., et al. (2015). Novel selective inhibitor of leishmania (leishmania) amazonensis arginase. Chem. Biol. Drug Des. 86 (5), 969–978. doi:10.1111/cbdd.12566
Da Silva, S. A. G., Costa, S. S., Mendonça, S. C. F., Silva, E. M., Moraes, V. L. G., and Rossi-Bergmann, B. (1995). Therapeutic effect of oral Kalanchoe pinnata leaf extract in murine leishmaniasis. Acta Trop. 60 (3), 201–210. doi:10.1016/0001-706X(95)00128-2
Date, A. A., Joshi, M. D., and Patravale, V. B. (2007). Parasitic diseases: Liposomes and polymeric nanoparticles versus lipid nanoparticles. Adv. Drug Deliv. Rev. 59 (6), 505–521. doi:10.1016/j.addr.2007.04.009
D’Avila, H., Freire-de-Lima, C. G., Roque, N. R., Teixeira, L., Barja-Fidalgo, C., Silva, A. R., et al. (2011). Host cell lipid bodies triggered by Trypanosoma cruzi infection and enhanced by the uptake of apoptotic cells are associated with prostaglandin E₂ generation and increased parasite growth. J. Infect. Dis. 204 (6), 951–961. doi:10.1093/infdis/jir432
De Andrade, C. R., Kirchhoff, L. V., Donelson, J. E., and Otsu, K. (1992). Recombinant Leishmania Hsp90 and Hsp70 are recognized by sera from visceral leishmaniasis patients but not Chagas’ disease patients. J. Clin. Microbiol. 30 (2), 330–335. doi:10.1128/jcm.30.2.330-335.1992
Debnath, A., Shahinas, D., Bryant, C., Hirata, K., Miyamoto, Y., Hwang, G., et al. (2014). Hsp90 inhibitors as new leads to target parasitic diarrheal diseases. Antimicrob. Agents Chemother. 58 (7), 4138–4144. doi:10.1128/AAC.02576-14
Delgado, M. A., Elmaoued, R. A., Davis, A. S., Kyei, G., and Deretic, V. (2008). Toll-like receptors control autophagy. EMBO J. 27 (7), 1110–1121. doi:10.1038/emboj.2008.31
Deng, Y., Zhu, L., Cai, H., Wang, G., and Liu, B. (2018). Autophagic compound database: A resource connecting autophagy-modulating compounds, their potential targets and relevant diseases. Cell Prolif. 51 (3), e12403–e12406. doi:10.1111/cpr.12403
Dey, T., and Rangarajan, P. N. (2021). Carbon starvation-induced synthesis of GDH2 and PEPCK is essential for the survival of Pichia pastoris. Biochem. Biophysical Res. Commun. 581, 25–30. doi:10.1016/j.bbrc.2021.10.015
Di Loria, A., Dattilo, V., Santoro, D., Guccione, J., De Luca, A., Ciaramella, P., et al. (2020). Expression of serum exosomal miRNA 122 and lipoprotein levels in dogs naturally infected by Leishmania infantum: A preliminary study. Animals 10 (1), 1–10. doi:10.3390/ani10010100
Dias, B. R. S., de Souza, C. S., Almeida, N. de J., Lima, J. G. B., Fukutani, K. F., dos Santos, T. B. S., et al. (2018). Autophagic induction Greatly Enhances Leishmania major intracellular survival compared to Leishmania amazonensis in CBA/j-infected macrophages. Front. Microbiol. 9, 1890–1915. doi:10.3389/fmicb.2018.01890
Dinesh, N., Pallerla, D. S. R., Kaur, P. K., Kishore Babu, N., and Singh, S. (2014). Exploring Leishmania donovani 3-hydroxy-3-methylglutaryl coenzyme A reductase (HMGR) as a potential drug target by biochemical, biophysical and inhibition studies. Microb. Pathog. 66, 14–23. doi:10.1016/j.micpath.2013.11.001
Djune Yemeli, L., Domche, A., Nana Djeunga, H. C., Lenou Nanga, C. G., Njih Tabah, E., Nko’Ayissi, G. B., et al. (2021). Leishmaniasis in Cameroon: What is known and is done so far? A protocol for systematic review. BMJ Open 11 (4), e047530–e047534. doi:10.1136/bmjopen-2020-047530
Docampo, R., and Moreno, S. (2008). The acidocalcisome as a target for chemotherapeutic agents in Protozoan parasites. Curr. Pharm. Des. 14 (9), 882–888. doi:10.2174/138161208784041079
Doyle, M. A., MacRae, J. I., De Souza, D. P., Saunders, E. C., McConville, M. J., and Likić, V. A. (2009). LeishCyc: A biochemical pathways database for Leishmania major. BMC Syst. Biol. 3, 57–12. doi:10.1186/1752-0509-3-57
Enderling, H., Chaplain, M. A. J., Anderson, A. R. A., and Vaidya, J. S. (2007). A mathematical model of breast cancer development, local treatment and recurrence. J. Theor. Biol. 246, 245–259. doi:10.1016/j.jtbi.2006.12.010
Endy, D. (2005). Foundations for engineering biology. Nature 438 (7067), 449–453. doi:10.1038/nature04342
Esch, K. J., Juelsgaard, R., Martinez, P. A., Jones, D. E., and Petersen, C. A. (2013). Programmed death 1–mediated T cell exhaustion during visceral leishmaniasis impairs phagocyte function. J. Immunol. 191 (11), 5542–5550. doi:10.4049/jimmunol.1301810
Esch, K. J., Schaut, R. G., Lamb, I. M., Clay, G., Morais Lima, Á. L., Do Nascimento, P. R. P., et al. (2015). Activation of autophagy and nucleotide-binding domain leucine-rich repeat-containing-like receptor family, pyrin domain-containing 3 inflammasome during Leishmania infantum-associated glomerulonephritis. Am. J. Pathology 185 (8), 2105–2117. doi:10.1016/j.ajpath.2015.04.017
Fakhraee, F., Badiee, A., Alavizadeh, S. H., Jalali, S. A., Chavoshian, O., Khamesipour, A., et al. (2016). Coadminstration of L. major amastigote class I nuclease (rLmaCIN) with LPD nanoparticles delays the progression of skin lesion and the L. major dissemination to the spleen in BALB/c mice-based experimental setting. Acta Trop. 159, 211–218. doi:10.1016/j.actatropica.2016.04.004
Fonseca-Silva, F., Inacio, J. D. F., Canto-Cavalheiro, M. M., Menna-Barreto, R. F. S., and Almeida-Amaral, E. E. (2016). Oral efficacy of apigenin against cutaneous leishmaniasis: Involvement of reactive oxygen species and autophagy as a mechanism of action. PLoS Neglected Trop. Dis. 10 (2), 00044422–e4516. doi:10.1371/journal.pntd.0004442
Franco, L. H., Fleuri, A. K. A., Pellison, N. C., Quirino, G. F. S., Horta, C. V., De Carvalho, R. V. H., et al. (2017). Autophagy downstream of endosomal Toll-like receptor signaling in macrophages is a key mechanism for resistance to Leishmania major infection. J. Biol. Chem. 292 (32), 13087–13096. doi:10.1074/jbc.M117.780981
Frank, B., Marcu, A., De Oliveira Almeida Petersen, A. L., Weber, H., Stigloher, C., Mottram, J. C., et al. (2015). Autophagic digestion of Leishmania major by host macrophages is associated with differential expression of BNIP3, CTSE, and the miRNAs miR-101c, miR-129, and miR-210. Parasites Vectors 8, 1–31. doi:10.1186/s13071-015-0974-3
Frézard, F., Demicheli, C., and Ribeiro, R. R. (2009). Pentavalent antimonials: New perspectives for old drugs. Molecules 14 (7), 2317–2336. doi:10.3390/molecules14072317
Fridlyand, L., and Philipson, L. H. (2017). Computational model of cellular energy homeostasis and ROS dynamics for nonmuscle and cancer cells. Biophysical J. 112 (3), 283a. doi:10.1016/j.bpj.2016.11.1533
Galán, J. E., and Cossart, P. (2005). Host-pathogen interactions: A diversity of themes, a variety of molecular machines. Curr. Opin. Microbiol. 8 (1), 1–3. doi:10.1016/j.mib.2004.12.015
García-lara, J., Masalha, M., Foster, S. J., García-lara, J., and Foster, S. J. (2005). Staphylococcus aureus: The search for novel targets. Drug Discov. Today 10 (9), 643–651. doi:10.1016/S1359-6446(05)03432-X
Geng, J., Baba, M., Nair, U., and Klionsky, D. J. (2008). Quantitative analysis of autophagy-related protein stoichiometry by fluorescence microscopy. J. Cell Biol. 182 (1), 129–140. doi:10.1083/jcb.200711112
Geraci, N. S., Tan, J. C., and Mcdowell, M. A. (2015). Characterization of microRNA expression profiles in Leishmania-infected human phagocytes. Parasite Immunol. 37 (1), 43–51. doi:10.1111/pim.12156
Gholamrezaei, M., Rouhani, S., Mohebali, M., Mohammadi-Yeganeh, S., Haji Molla Hoseini, M., Haghighi, A., et al. (2020). MicroRNAs expression induces apoptosis of macrophages in response to Leishmania major (MRHO/IR/75/ER): An in-vitro and in-vivo study. Iran. J. Parasitol. 15 (4), 475–487. doi:10.18502/ijpa.v15i4.4851
Ghosh, J., Das, S., Guha, R., Ghosh, D., Naskar, K., Das, A., et al. (2012). Hyperlipidemia offers protection against Leishmania donovani infection: Role of membrane cholesterol. J. Lipid Res. 53 (12), 2560–2572. doi:10.1194/jlr.M026914
Giri, S., and Shaha, C. (2019). Leishmania donovani parasite requires Atg8 protein for infectivity and survival under stress. Cell Death Dis. 10 (11), 808. doi:10.1038/s41419-019-2038-7
Gorabi, A. M., Kiaie, N., Sathyapalan, T., Al-Rasadi, K., Jamialahmadi, T., and Sahebkar, A. (2020). The role of MicroRNAs in regulating cytokines and growth factors in coronary artery disease: The ins and outs. J. Immunol. Res. 2020, 5193036. doi:10.1155/2020/5193036
Guhe, V., Anjum, F., Shafie, A., Hassan, M. I., Pasupuleti, V. R., and Singh, S. (2022). Infection dynamics of ATG8 in leishmania: Balancing autophagy for therapeutics. Molecules 27 (10), 3142–3221. doi:10.3390/molecules27103142
Gupta, P., Srivastav, S., Saha, S., Das, P. K., and Ukil, A. (2016). Leishmania donovani inhibits macrophage apoptosis and pro-inflammatory response through AKT-mediated regulation of β-catenin and FOXO-1. Cell Death Differ. 23 (11), 1815–1826. doi:10.1038/cdd.2016.101
Gupta, S., Silveira, D. A., and Mombach, J. C. M. (2020). Towards DNA-damage induced autophagy: A boolean model of p53-induced cell fate mechanisms. DNA Repair 96, 102971. doi:10.1016/j.dnarep.2020.102971
Gutierrez, M. G., Master, S. S., Singh, S. B., Taylor, G. A., Colombo, M. I., and Deretic, V. (2004). Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell 119 (6), 753–766. doi:10.1016/j.cell.2004.11.038
Han, K., Kwon, H. W., Kang, H., Kim, J., Lee, M. S., and Choi, M. Y. (2012). Dynamics of macroautophagy: Modeling and oscillatory behavior. Phys. A Stat. Mech. Its Appl. 391 (3), 686–692. doi:10.1016/j.physa.2011.08.046
Hao, W., and Friedman, A. (2016). Mathematical model on Alzheimer’s disease. BMC Syst. Biol. 10, 1–18. doi:10.1186/s12918-016-0348-2
Harizanov, R. N., and Kaftandjiev Iskren, T. (2014). Interactions between parasite and host in human visceral leishmaniasis. Cytol. Tissue Biol. 1 (1), 1–8. doi:10.24966/ctb-9107/100001
He, C., and Klionsky, D. J. (2009). Regulation mechanisms and signaling pathways of autophagy. Annu. Rev. Genet. 43, 67–93. doi:10.1146/annurev-genet-102808-114910
He, Z., Liu, H., Moch, H., and Simon, H. U. (2020). Machine learning with autophagy-related proteins for discriminating renal cell carcinoma subtypes. Sci. Rep. 10 (1), 720–727. doi:10.1038/s41598-020-57670-y
Homma, K., Suzuki, K., and Sugawara, H. (2011). The autophagy database: An all-inclusive information resource on autophagy that provides nourishment for research. Nucleic Acids Res. 39 (1), 986–990. doi:10.1093/nar/gkq995
Ivens, A. C., Peacock, C. S., Worthey, E. A., Murphy, L., Aggarwal, G., Berriman, M., et al. (2005). Ivens_Tritryp_Science_Leish_2005, 1–7. July sftp://cerca@192.168.2.5/home/cerca/Desktop/data/laptop_files/info/biologia/parasitas/leishmania/genoma_leish/ivens_tritryp_science_leish_2005.pdf%5Cnpapers2://publication/uuid/ECC15CE2-1DC8-4578-9B8D-561DCB8CCC5B.
Jamal, S., and Scaria, V. (2013). Cheminformatic models based on machine learning for pyruvate kinase inhibitors of Leishmania mexicana. Leishmania Mex. BMC Bioinforma. 14 (1), 329. doi:10.1186/1471-2105-14-329
Jin, H., and Lei, J. (2014). A mathematical model of cell population dynamics with autophagy response to starvation. Math. Biosci. 258, 1–10. doi:10.1016/j.mbs.2014.08.014
Johndrow, C., Nelson, R., Tanowitz, H., Weiss, L. M., and Nagajyothi, F. (2014). Trypanosoma cruzi infection results in an increase in intracellular cholesterol. Microbes Infect. 16 (4), 337–344. doi:10.1016/j.micinf.2014.01.001
Kabra, R., Ingale, P., and Singh, S. (2020). Computationally designed synthetic peptides for transporter proteins imparts allostericity in Miltefosine resistant L. major. Biochem. J. 447 (10), 2007–2026. doi:10.1042/BCJ20200176
Kapuy, O., Vinod, P. K., Mandl, J., and Bánhegyi, G. (2013). A cellular stress-directed bistable switch controls the crosstalk between autophagy and apoptosis. Mol. Biosyst. 9 (2), 296–306. doi:10.1039/c2mb25261a
Kaur, J., Tiwari, R., Kumar, A., and Singh, N. (2011). Bioinformatic analysis of leishmania donovani long-chain fatty acid-CoA ligase as a novel drug target. Mol. Biol. Int. 2011, 278051–278114. doi:10.4061/2011/278051
Kedzierski, L., and Evans, K. J. (2014). Immune responses during cutaneous and visceral leishmaniasis. Parasitology 141 (12), 1544–1562. doi:10.1017/S003118201400095X
Khadir, F., Shaler, C. R., Oryan, A., Rudak, P. T., Mazzuca, D. M., Taheri, T., et al. (2018). Therapeutic control of leishmaniasis by inhibitors of the mammalian target of rapamycin. PLoS Neglected Trop. Dis. 12 (8), e0006701–e0006719. doi:10.1371/journal.pntd.0006701
Khan, M. O. F. (2007). Trypanothione reductase: A viable chemotherapeutic target for antitrypanosomal and antileishmanial drug design. Drug Target Insights 2 (1999), 117739280700200. doi:10.1177/117739280700200007
Klionsky, D. J. (2007). Autophagy: From phenomenology to molecular understanding in less than a decade. Nat. Rev. Mol. Cell Biol. 8 (11), 931–937. doi:10.1038/nrm2245
Kotani, T., Kirisako, H., Koizumi, M., Ohsumi, Y., and Nakatogawa, H. (2018). The Atg2-Atg18 complex tethers pre-autophagosomal membranes to the endoplasmic reticulum for autophagosome formation. Proc. Natl. Acad. Sci. U. S. A. 115 (41), 10363–10368. doi:10.1073/pnas.1806727115
Kumar Saha, A., Mukherjee, T., and Bhaduri, A. (1986). Mechanism of action of amphotericin B on Leishmania donovani promastigotes. Mol. Biochem. Parasitol. 19 (3), 195–200. doi:10.1016/0166-6851(86)90001-0
Lai A Fat, E. J. S. K., Vrede, M. A., Soetosenojo, R. M., and Lai A Fat, R. F. M. (2002). Pentamidine, the drug of choice for the treatment of cutaneous leishmaniasis in Surinam. Int. J. Dermatology 41 (11), 796–800. doi:10.1046/j.1365-4362.2002.01633.x
Latorre, J., Moreno-Navarrete, J. M., Mercader, J. M., Sabater, M., RoviraGironès, J., Ricart, W., et al. (2017). Decreased lipid metabolism but increased FA biosynthesis are coupled with changes in liver microRNAs in obese subjects with NAFLD. Int. J. Obes. 41 (4), 620–630. doi:10.1038/ijo.2017.21
Leibovici, V., and Aram, H. (1986). Cryotherapy in acute cutaneous leishmaniasis. Int. J. Dermatol. 25, 473–475. doi:10.1111/j.1365-4362.1986.tb03461.x
Lorente, S. O., Rodrigues, J. C. F., Jiménez, C. J., Joyce-Menekse, M., Rodrigues, C., Croft, S. L., et al. (2004). Novel azasterols as potential agents for treatment of leishmaniasis and trypanosomiasis. Antimicrob. Agents Chemother. 48 (8), 2937–2950. doi:10.1128/AAC.48.8.2937-2950.2004
Louzoun, Y., Xue, C., Lesinski, G. B., and Friedman, A. (2014). A mathematical model for pancreatic cancer growth and treatments. J. Theor. Biol. 351, 74–82. doi:10.1016/j.jtbi.2014.02.028
Madeira Da Silva, L., and Beverley, S. M. (2010). Expansion of the target of rapamycin (TOR) kinase family and function in Leishmania shows that TOR3 is required for acidocalcisome biogenesis and animal infectivity. Proc. Natl. Acad. Sci. U. S. A. 107 (26), 11965–11970. doi:10.1073/pnas.1004599107
Maiuri, M. C., Zalckvar, E., Kimchi, A., and Kroemer, G. (2007). Self-eating and self-killing: Crosstalk between autophagy and apoptosis. Nat. Rev. Mol. Cell Biol. 8 (9), 741–752. doi:10.1038/nrm2239
Manzano, J. I., Perea, A., León-Guerrero, D., Campos-Salinas, J., Piacenza, L., Castanys, S., et al. (2017). Leishmania LABCG1 and LABCG2 transporters are involved in virulence and oxidative stress: Functional linkage with autophagy. Parasites Vectors 10 (1), 267–312. doi:10.1186/s13071-017-2198-1
Martin, K. R., Barua, D., Kauffman, A. L., Westrate, L. M., Posner, R. G., Hlavacek, W. S., et al. (2013). Computational model for autophagic vesicle dynamics in single cells. Autophagy 9 (1), 74–92. doi:10.4161/auto.22532
Martin, S., and Parton, R. G. (2006). Lipid droplets: A unified view of a dynamic organelle. Mol. Cell 7, 373–378. doi:10.1038/nrm1912
Martínez, C. R., and Ruiz, C. J. (2019). Alterations in host lipid metabolism produced during visceral leishmaniasis infections. Curr. Trop. Med. Rep. 6 (4), 250–255. doi:10.1007/s40475-019-00187-w
Maruyama, T., and Noda, N. N. (2018). Autophagy-regulating protease Atg4: Structure, function, regulation and inhibition. J. Antibiotics 71 (1), 72–78. doi:10.1038/ja.2017.104
Matsuura, A., Tsukada, M., Wada, Y., and Ohsumi, Y. (1997). Apg1p, a novel protein kinase required for the autophagic process in Saccharomyces cerevisiae. Gene 192 (2), 245–250. doi:10.1016/S0378-1119(97)00084-X
Matte, C., Casgrain, P. A., Séguin, O., Moradin, N., Hong, W. J., and Descoteaux, A. (2016). Leishmania major promastigotes evade LC3-associated phagocytosis through the action of GP63. PLoS Pathog. 12 (6), 10056900–e1005717. doi:10.1371/journal.ppat.1005690
McCall, L. I., El Aroussi, A., Choi, J. Y., Vieira, D. F., De Muylder, G., Johnston, J. B., et al. (2015). Targeting ergosterol biosynthesis in Leishmania donovani: Essentiality of sterol 14alpha-demethylase. PLoS Neglected Trop. Dis. 9 (3), 00035888–e3617. doi:10.1371/journal.pntd.0003588
Meissner, F., Geddes-McAlister, J., Mann, M., and Bantscheff, M. (2022). The emerging role of mass spectrometry-based proteomics in drug discovery. Nat. Rev. Drug Discov. 21 (9), 637–654. doi:10.1038/s41573-022-00409-3
Melo, R. C. N., and Dvorak, A. M. (2012). Lipid body-phagosome interaction in macrophages during infectious diseases: Host defense or pathogen survival strategy? PLoS Pathog. 8 (7), e1002729. doi:10.1371/journal.ppat.1002729
Mizushima, N., Yoshimori, T., and Ohsumi, Y. (2011). The role of atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 27, 107–132. doi:10.1146/annurev-cellbio-092910-154005
Modabber, F. (2010). Leishmaniasis vaccines: Past, present and future. Int. J. Antimicrob. Agents 36 (1), S58–S61. doi:10.1016/j.ijantimicag.2010.06.024
Mol, M., Patole, M. S., and Singh, S. (2014). Immune signal transduction in leishmaniasis from natural to artificial systems: Role of feedback loop insertion. Biochimica Biophysica Acta - General Subj. 1840 (1), 71–79. doi:10.1016/j.bbagen.2013.08.018
Mollalo, A., Sadeghian, A., Israel, G. D., Rashidi, P., Sofizadeh, A., and Glass, G. E. (2018). Machine learning approaches in GIS-based ecological modeling of the sand fly Phlebotomus papatasi, a vector of zoonotic cutaneous leishmaniasis in Golestan province, Iran. Acta Trop. 188, 187–194. doi:10.1016/j.actatropica.2018.09.004
Mrvar, A., and Batagelj, V. (2016). Analysis and visualization of large networks with program package Pajek. Complex Adapt. Syst. Model. 4 (1), 6. doi:10.1186/s40294-016-0017-8
Nakagawa, I., Amano, A., Mizushima, N., Yamamoto, A., Yamaguchi, H., Kamimoto, T., et al. (2004). Autophagy defends cells against invading group A Streptococcus. Science 306 (5698), 1037–1040. doi:10.1126/science.1103966
Nanduri, R., Kalra, R., Bhagyaraj, E., Chacko, A. P., Ahuja, N., Tiwari, D., et al. (2019). AutophagySMDB: A curated database of small molecules that modulate protein targets regulating autophagy. Autophagy 15 (7), 1280–1295. doi:10.1080/15548627.2019.1571717
Nicholls, E. F., Madera, L., and Hancock, R. E. W. (2010). Immunomodulators as adjuvants for vaccines and antimicrobial therapy. Ann. N. Y. Acad. Sci. 1213 (1), 46–61. doi:10.1111/j.1749-6632.2010.05787.x
Noda, N. N., Fujioka, Y., Hanada, T., Ohsumi, Y., and Inagaki, F. (2013). Structure of the Atg12-Atg5 conjugate reveals a platform for stimulating Atg8-PE conjugation. EMBO Rep. 14 (2), 206–211. doi:10.1038/embor.2012.208
O’Connell, R. M., Taganov, K. D., Boldin, M. P., Cheng, G., and Baltimore, D. (2007). MicroRNA-155 is induced during the macrophage inflammatory response. Proc. Natl. Acad. Sci. U. S. A. 104 (5), 1604–1609. doi:10.1073/pnas.0610731104
Odegaard, J. I., Ricardo-Gonzalez, R. R., Goforth, M. H., Morel, C. R., Subramanian, V., Mukundan, L., et al. (2007). Macrophage-specific PPARgamma controls alternative activation and improves insulin resistance. Nature 447 (7148), 1116–1120. doi:10.1038/nature05894
Okwor, I. B., Jia, P., Mou, Z., Onyilagha, C., and Uzonna, J. E. (2014). CD8+ T cells are preferentially activated during primary low dose Leishmania major infection but are completely dispensable during secondary anti-leishmania immunity. PLoS Neglected Trop. Dis. 8 (11), e3300–e3311. doi:10.1371/journal.pntd.0003300
Omollo, R., Alexander, N., Edwards, T., Khalil, E. A. G., Younis, B. M., Abuzaid, A. A., et al. (2011). Safety and Efficacy of miltefosine alone and in combination with sodium stibogluconate and liposomal amphotericin B for the treatment of primary visceral leishmaniasis in East Africa: Study protocol for a randomized controlled trial. Trials 12, 166. doi:10.1186/1745-6215-12-166
Ortiz, D., Forquer, I., Boitz, J., Soysa, R., Elya, C., Fulwiler, A., et al. (2016). Targeting the cytochrome bc1 complex of Leishmania parasites for discovery of novel drugs. Antimicrob. Agents Chemother. 60 (8), 4972–4982. doi:10.1128/AAC.00850-16
Patel, P., Mandlik, V., and Singh, S. (2016). LmSmdB: An integrated database for metabolic and gene regulatory network in Leishmania major and Schistosoma mansoni. Genomics Data 7, 115–118. doi:10.1016/j.gdata.2015.12.012
Pauley, K. M., Satoh, M., Pauley, B. A., Dominguez-Gutierrez, P. R., Wallet, S. M., Holliday, L. S., et al. (2010). Formation of GW/P bodies as marker for microRNA-mediated regulation of innate immune signaling in THP-1 cells. Immunol. Cell Biol. 88 (2), 205–212. doi:10.1038/icb.2009.84
Peters, N. C., Pagán, A. J., Lawyer, P. G., Hand, T. W., Henrique Roma, E., Stamper, L. W., et al. (2014). Chronic parasitic infection maintains high frequencies of short-lived Ly6C+CD4+ effector T cells that are required for protection against Re-infection. PLoS Pathog. 10 (12), e1004538. doi:10.1371/journal.ppat.1004538
Petersen, A. L. D O. A., Cull, B., Dias, B. R. S., Palma, L. C., Luz, Y. da S., de Menezes, J. P. B., et al. (2021). 17-AAG-induced activation of the autophagic pathway in leishmania is associated with parasite death. Microorganisms 9 (5), 1089–1115. doi:10.3390/microorganisms9051089
Petersen, A. L. de O. A., Guedes, C. E. S., Versoza, C. L., Lima, J. G. B., de Freitas, L. A. R., Borges, V. M., et al. (2012). 17-AAG kills intracellular Leishmania amazonensis while reducing inflammatory responses in infected macrophages. PLoS ONE 7 (11), 494966–e49512. doi:10.1371/journal.pone.0049496
Pinheiro, R. O., Nunes, M. P., Pinheiro, C. S., D’Avila, H., Bozza, P. T., Takiya, C. M., et al. (2009). Induction of autophagy correlates with increased parasite load of Leishmania amazonensis in BALB/c but not C57BL/6 macrophages. Microbes Infect. 11 (2), 181–190. doi:10.1016/j.micinf.2008.11.006
Pitale, D. M., Gendalur, N. S., Descoteaux, A., and Shaha, C. (2019). Leishmania donovani induces autophagy in human blood–derived neutrophils. J. Immunol. 202 (4), 1163–1175. doi:10.4049/jimmunol.1801053
Pradhan, S., Schwartz, R. A., Patil, A., Grabbe, S., and Goldust, M. (2022). Treatment options for leishmaniasis. Clin. Exp. Dermatology 47 (3), 516–521. doi:10.1111/ced.14919
Pulvertaft, R. J. V., and Hoyle, G. F. (1960). Stages in the life-cycle of leishmania donovani. Trans. R. Soc. Trop. Med. Hyg. 54 (2). cultures of normal hamster macrophages derived from adult spleen explants are described . Preparation of Mammalian Cells.
Rahman, F., Ali, R., Tabrez, S., Mobeen, A., Akand, S. K., Arish, M., et al. (2022). Exploration of potential inhibitors for autophagy-related protein 8 as antileishmanial agents. Chem. Biol. Drug Des. 99 (6), 816–827. doi:10.1111/cbdd.14029
Rangwala, R., Chang, Y. C., Hu, J., Algazy, K. M., Evans, T. L., Fecher, L. A., et al. (2014). Combined MTOR and autophagy inhibition: Phase I trial of hydroxychloroquine and temsirolimus in patients with advanced solid tumors and melanoma. Autophagy 10 (8), 1391–1402. doi:10.4161/auto.29119
Ribeiro-Gomes, F. L., Otero, A. C., Gomes, N. A., Moniz-de-Souza, M. C. A., Cysne-Finkelstein, L., Arnholdt, A. C., et al. (2004). Macrophage interactions with neutrophils regulate Leishmania major infection. J. Immunol. 172 (7), 4454–4462. doi:10.4049/jimmunol.172.7.4454
Robainas, M., Otano, R., Bueno, S., and Ait-Oudhia, S. (2017). Understanding the role of PD-L1/PD1 pathway blockade and autophagy in cancer therapy. OncoTargets Ther. 10, 1803–1807. doi:10.2147/OTT.S132508
Robledo, S. M., Valencia, A. Z., and Saravia, N. G. (1999). Sensitivity to Glucantime® of Leishmania viannia isolated from patients prior to treatment. J. Parasitol. 85 (2), 360–366. doi:10.2307/3285647
Rodrigues, J. C. F., Concepcion, J. L., Rodrigues, C., Caldera, A., Urbina, J. A., and De Souza, W. (2008). In vitro activities of ER-119884 and E5700, two potent squalene synthase inhibitors, against Leishmania amazonensis: Antiproliferative, biochemical, and ultrastructural effects. Antimicrob. Agents Chemother. 52 (11), 4098–4114. doi:10.1128/AAC.01616-07
Rodríguez, N. E., Lockard, R. D., Turcotte, E. A., Araújo-Santos, T., Bozza, P. T., Borges, V. M., et al. (2017). Lipid bodies accumulation in Leishmania infantum-infected C57BL/6 macrophages. Parasite Immunol. 39 (8), e12443–e12445. doi:10.1111/pim.12443
Romanov, J., Walczak, M., Ibiricu, I., Schüchner, S., Ogris, E., Kraft, C., et al. (2012). Mechanism and functions of membrane binding by the Atg5-Atg12/Atg16 complex during autophagosome formation. EMBO J. 31 (22), 4304–4317. doi:10.1038/emboj.2012.278
Roy, S., Saha, S., Gupta, P., Ukil, A., and Das, P. K. (2019). Crosstalk of PD-1 signaling with the SIRT1/FOXO-1 axis during the progression of visceral leishmaniasis. J. Cell Sci. 132 (9), jcs226274. doi:10.1242/jcs.226274
Ruhland, A., Leal, N., and Kima, P. E. (2007). Leishmania promastigotes activate PI3K/Akt signalling to confer host cell resistance to apoptosis. Cell. Microbiol. 9 (1), 84–96. doi:10.1111/j.1462-5822.2006.00769.x
Sabzevari, S., Teshnizi, S. H., Shokri, A., Bahrami, F., and Kouhestani, F. (2021). Cutaneous leishmaniasis in Iran: A systematic review and meta-analysis. Microb. Pathog. 152, 104721. doi:10.1016/j.micpath.2020.104721
Sadeghian, G., Nilfroushzadeh, M. A., and Iraji, F. (2007). Efficacy of local heat therapy by radiofrequency in the treatment of cutaneous leishmaniasis, compared with intralesional injection of meglumine antimoniate. Clin. Exp. Dermatology 32 (4), 371–374. doi:10.1111/j.1365-2230.2007.02405.x
Santos, D. M., Petersen, A. L. O. A., Celes, F. S., Borges, V. M., Veras, P. S. T., and de Oliveira, C. I. (2014). Chemotherapeutic potential of 17-AAG against cutaneous leishmaniasis caused by leishmania (viannia) braziliensis. PLoS Neglected Trop. Dis. 8 (10), e3275. doi:10.1371/journal.pntd.0003275
Scariot, D. B., Britta, E. A., Moreira, A. L., Falzirolli, H., Silva, C. C., Ueda-Nakamura, T., et al. (2017). Induction of early autophagic process on Leishmania amazonensis by synergistic effect of miltefosine and innovative semi-synthetic thiosemicarbazone. Front. Microbiol. 8, 1–16. doi:10.3389/fmicb.2017.00255
Schaible, U. E., Schlesinger, P. H., Steinberg, T. H., Mangel, W. F., Kobayashi, T., and Russell, D. G. (1999). Parasitophorous vacuoles of Leishmania mexicana acquire macromolecules from the host cell cytosol via two independent routes. J. Cell Sci. 112 (5), 681–693. doi:10.1242/jcs.112.5.681
Schreiber, S. L., Ciba, M., and Schreiber, S. L. (2016). Linked references are available on JSTOR for this article: Target-oriented and diversity-oriented organic synthesis in drug discovery. 287 (5460), 1964–1969.
Sengupta, S., Chowdhury, S., BoseDasgupta, S., Wright, C. W., and Majumder, H. K. (2011). Cryptolepine-induced cell death of Leishmania donovani promastigotes is augmented by inhibition of autophagy. Mol. Biol. Int., 2011, 187850–187912. doi:10.4061/2011/187850
Serrano, A., Haddad, S. E., Moal, F., Prazuck, T., Legac, E., Robin, C., et al. (2018). Dysregulation of apoptosis and autophagy gene expression in peripheral blood mononuclear cells of efficiently treated HIV-infected patients. Aids 32 (12), 1579–1587. doi:10.1097/QAD.0000000000001851
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (1971). Cytoscape: A software environment for integrated models. Genome. Res. 13, 426. doi:10.1101/gr.1239303.metabolite
Shi, C. S., and Kehrl, J. H. (2008). MyD88 and Trif target Beclin 1 to trigger autophagy in macrophages. J. Biol. Chem. 283 (48), 33175–33182. doi:10.1074/jbc.M804478200
Silva, K. P., Seraphim, T. V., and Borges, J. C. (2013). Structural and functional studies of Leishmania braziliensis Hsp90. Biochimica Biophysica Acta - Proteins Proteomics 1834 (1), 351–361. doi:10.1016/j.bbapap.2012.08.004
Singh, A. K., Pandey, R. K., Shaha, C., and Madhubala, R. (2016). MicroRNA expression profiling of Leishmania donovani-infected host cells uncovers the regulatory role of MIR30A-3p in host autophagy. Autophagy 12 (10), 1817–1831. doi:10.1080/15548627.2016.1203500
Singh, R., Kaushik, S., Wang, Y., Xiang, Y., Novak, I., Komatsu, M., et al. (2009). Autophagy regulates lipid metabolism. Nature 458 (7242), 1131–1135. doi:10.1038/nature07976
Soni, B., and Singh, S. (2022). Synthetic perturbations in il6 biological circuit induces dynamical cellular response. Molecules 27 (1), 124. doi:10.3390/molecules27010124
Stofberg, M. L., Caillet, C., de Villiers, M., and Zininga, T. (2021). Inhibitors of the plasmodium falciparum hsp90 towards selective antimalarial drug design: The past, present and future. Cells 10 (11), 2849. doi:10.3390/cells10112849
Sundar, S. (2001). Drug resistance in Indian visceral leishmaniasis. Trop. Med. Int. Health 6 (11), 849–854. doi:10.1046/j.1365-3156.2001.00778.x
Sundar, S., Jha, T. K., Thakur, C. P., Sinha, P. K., and Bhattacharya, S. K. (2007). Injectable paromomycin for Visceral leishmaniasis in India. N. Engl. J. 356, 2571–2581. doi:10.1056/NEJMoa066536
Sundar, S., and Olliaro, P. L. (2007). Miltefosine in the treatment of leishmaniasis: Clinical evidence for informed clinical risk management. Ther. Clin. Risk Manag. 3 (5), 733–740.
Sundar, S., and Chakravarty, J. (2008). Paromomycin in the treatment of leishmaniasis. Expert Opin. Investig. Drugs 17, 787–794. doi:10.1517/13543784.17.5.787
Sundar, S., and Singh, B. (2018). Emerging therapeutic targets for treatment of leishmaniasis. Expert Opin. Ther. Targets 22 (6), 467–486. doi:10.1080/14728222.2018.1472241
Taganov, K. D., Boldin, M. P., Chang, K. J., and Baltimore, D. (2006). NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. U. S. A. 103 (33), 12481–12486. doi:10.1073/pnas.0605298103
Tang, Y., Zhu, W., Chen, K., and Jiang, H. (2006). New technologies in computer-aided drug design: Toward target identification and new chemical entity discovery. Drug Discov. Today Technol. 3 (3), 307–313. doi:10.1016/j.ddtec.2006.09.004
Tao, Y., Liu, Y., Friedman, C., and Lussier, Y. A. (2004). Information visualization techniques in bioinformatics during the postgenomic era. Drug Discov. Today BIOSILICO 2 (6), 237–245. doi:10.1016/S1741-8364(04)02423-0
Thomas, S. A., Nandan, D., Kass, J., and Reiner, N. E. (2018). Countervailing, time-dependent effects on host autophagy promotes intracellular survival of Leishmania. J. Biol. Chem. 293 (7), 2617–2630. doi:10.1074/jbc.M117.808675
Travassos, L. H., Carneiro, L. A. M., Ramjeet, M., Hussey, S., Kim, Y. G., Magalhes, J. G., et al. (2010). Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat. Immunol. 11 (1), 55–62. doi:10.1038/ni.1823
Tsai, J. C., Chen, I. H., Wong, T. W., and Lo, Y. L. (2002). In vitro/in vivo correlations between transdermal delivery of 5-aminolaevulinic acid and cutaneous protoporphyrin IX accumulation and effect of formulation. Br. J. Dermatology 146 (5), 853–862. doi:10.1046/j.1365-2133.2002.04715.x
Tsukada, M., and Ohsumi, Y. (1993). Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 333 (1–2), 169–174. doi:10.1016/0014-5793(93)80398-E
Tuon, F. F., Amato, V. S., Zequinao, T., and Cruz, J. A. W. (2022). Emerging computational technologies in human leishmaniasis: Where are we? Trans. R. Soc. Trop. Med. Hyg. 116 (11), 981–985. doi:10.1093/trstmh/trac047
Türei, D., Földvári-Nagy, L., Fazekas, D., Módos, D., Kubisch, J., Kadlecsik, T., et al. (2015). Autophagy regulatory network - a systemslevel bioinformatics resource for studying the mechanism and regulation of autophagy. Autophagy 11 (1), 155–165. doi:10.4161/15548627.2014.994346
Valencia, B. M., Miller, D., Witzig, R. S., Boggild, A. K., and Llanos-Cuentas, A. (2013). Novel low-cost thermotherapy for cutaneous leishmaniasis in Peru. PLoS Neglected Trop. Dis. 7 (5), e2196–e2199. doi:10.1371/journal.pntd.0002196
Vallochi, A. L., Teixeira, L., Oliveira, K. da S., Maya-Monteiro, C. M., and Bozza, P. T. (2018). Lipid droplet, a key player in host-parasite interactions. Front. Immunol. 9, 1022. doi:10.3389/fimmu.2018.01022
Vanrell, M. C., Losinno, A. D., Cueto, J. A., Balcazar, D., Fraccaroli, L. V., Carrillo, C., et al. (2017). The regulation of autophagy differentially affects Trypanosoma cruzi metacyclogenesis. PLoS Neglected Trop. Dis. 11 (11), 00060499–e6123. doi:10.1371/journal.pntd.0006049
Verlinde, C. L. M. J., Hannaert, V., Blonski, C., Willson, M., Périé, J. J., Fothergill-Gilmore, L. A., et al. (2001). Glycolysis as a target for the design of new anti-trypanosome drugs. Drug Resist. Updat. 4 (1), 50–65. doi:10.1054/drup.2000.0177
Vicente-Barrueco, A., Román, Á. C., Ruiz-Téllez, T., and Centeno, F. (2022). In silico research of new therapeutics rotenoids derivatives against Leishmania amazonensis infection. Biology 11 (1), 133–214. doi:10.3390/biology11010133
Walters, W. P., and Barzilay, R. (2021). Critical assessment of AI in drug discovery. Expert Opin. Drug Discov. 16 (9), 937–947. doi:10.1080/17460441.2021.1915982
Wang, N. N., Dong, J., Zhang, L., Ouyang, D., Cheng, Y., Chen, A. F., et al. (2018). HAMdb: A database of human autophagy modulators with specific pathway and disease information. J. Cheminformatics 10 (1), 34–38. doi:10.1186/s13321-018-0289-4
Webber, J. L., and Tooze, S. A. (2010). New insights into the function of Atg9. FEBS Lett. 584 (7), 1319–1326. doi:10.1016/j.febslet.2010.01.020
Williams, R. A. M., Mottram, J. C., and Coombs, G. H. (2013). Distinct roles in autophagy and importance in infectivity of the two ATG4 cysteine peptidases of Leishmania major. J. Biol. Chem. 288 (5), 3678–3690. doi:10.1074/jbc.M112.415372
Williams, R. A. M., Smith, T. K., Cull, B., Mottram, J. C., and Coombs, G. H. (2012). ATG5 is essential for ATG8-dependent autophagy and mitochondrial homeostasis in Leishmania major. PLoS Pathog. 8 (5), e1002695. doi:10.1371/journal.ppat.1002695
Williams, R. A. M., Woods, K. L., Juliano, L., Mottram, J. C., and Coombs, G. H. (2009). Characterization of unusual families of ATG8-like proteins and ATG12 in the protozoan parasite Leishmania major. Autophagy 5 (2), 159–172. doi:10.4161/auto.5.2.7328
Wyllie, S., Cunningham, M. L., and Fairlamb, A. H. (2004). Dual action of antimonial drugs on thiol redox metabolism in the human pathogen Leishmania donovani. J. Biol. Chem. 279 (38), 39925–39932. doi:10.1074/jbc.M405635200
Xu, W., Hsu, F. F., Baykal, E., Huang, J., and Zhang, K. (2014). Sterol biosynthesis is required for heat resistance but not extracellular survival in leishmania. PLoS Pathog. 10 (10), e1004427. doi:10.1371/journal.ppat.1004427
Yang, Y., and Wang, J. K. (2016). The functional analysis of MicroRNAs involved in NF-κB signaling. Eur. Rev. Med. Pharmacol. Sci. 20 (9), 1764–1774.
Yao, C., and Wilson, M. E. (2016). Dynamics of sterol synthesis during development of Leishmania spp. parasites to their virulent form. Parasites Vectors 9 (1), 200–212. doi:10.1186/s13071-016-1470-0
Zachari, M., and Ganley, I. G. (2017). The mammalian ULK1 complex and autophagy initiation. Essays Biochem. 61 (6), 585–596. doi:10.1042/EBC20170021
Zhai, L., Blom, J., Chen, M., Christensen, S. B., and Kharazmi, A. (1995). The antileishmanial agent licochalcone A interferes with the function of parasite mitochondria. Antimicrob. Agents Chemother. 39 (12), 2742–2748. doi:10.1128/AAC.39.12.2742
Zhai, L., Chen, M., Blom, J., Theander, T. G., Christensen, S. B., and Kharazmi, A. (1999). The antileishmanial activity of novel oxygenated chalcones and their mechanism of action. J. Antimicrob. Chemother. 43 (6), 793–803. doi:10.1093/jac/43.6.793
Zhang, K. (2021). Balancing de novo synthesis and salvage of lipids by Leishmania amastigotes. Curr. Opin. Microbiol. 63, 98–103. doi:10.1016/j.mib.2021.07.004
Zhang, Y., Xin, Z., Dong, B., and Xue, W. (2022). Combination of the NRF2 inhibitor and autophagy inhibitor significantly inhibited tumorigenicity of castration-resistant prostate cancer. Comput. Math. Methods Med. 2022, 4182401. doi:10.1155/2022/4182401
Keywords: autophagy, leishmaniasis, lipid droplets, miRNA, systems biology, artificial intelligence, anti-leishmanial drugs, computational biology
Citation: Guhe V, Ingale P, Tambekar A and Singh S (2023) Systems biology of autophagy in leishmanial infection and its diverse role in precision medicine. Front. Mol. Biosci. 10:1113249. doi: 10.3389/fmolb.2023.1113249
Received: 01 December 2022; Accepted: 05 April 2023;
Published: 21 April 2023.
Edited by:
William C. Cho, QEH, Hong Kong SAR, ChinaReviewed by:
Sudharsana Ande, University of Manitoba, CanadaCopyright © 2023 Guhe, Ingale, Tambekar and Singh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shailza Singh, c2luZ2hzQG5jY3MucmVzLmlu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.