
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Mol. Biosci. , 15 February 2023
Sec. Metabolomics
Volume 10 - 2023 | https://doi.org/10.3389/fmolb.2023.1111482
This article is part of the Research Topic Reviews in Metabolomics: Personalized and Predictive Medicine of Infectious Diseases View all 6 articles
COVID-19 currently represents one of the major health challenges worldwide. Albeit its infectious character, with onset affectation mainly at the respiratory track, it is clear that the pathophysiology of COVID-19 has a systemic character, ultimately affecting many organs. This feature enables the possibility of investigating SARS-CoV-2 infection using multi-omic techniques, including metabolomic studies by chromatography coupled to mass spectrometry or by nuclear magnetic resonance (NMR) spectroscopy. Here we review the extensive literature on metabolomics in COVID-19, that unraveled many aspects of the disease including: a characteristic metabotipic signature associated to COVID-19, discrimination of patients according to severity, effect of drugs and vaccination treatments and the characterization of the natural history of the metabolic evolution associated to the disease, from the infection onset to full recovery or long-term and long sequelae of COVID.
Undoubtedly, COVID-19 outbreak emerged as one of the biggest medical challenges worldwide. The virus responsible for COVID-19 is the Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), named after SARS-CoV for its genetic similarity (Velavan and Meyer, 2020). Shortly after its first detection in Wuhan (China) in December 2019 the virus quickly spread out, and the World Health Organization (WHO) declared a state of pandemic in March 2020 (Cucinotta and Vanelli, 2020). Now a days the virus has caused more than 624 million infections and 6.5 million deaths1. SARS-CoV-2 is very easily transmitted from person to person through droplets inhalation together with aerosol emission and consequent contact of the virus with nose, mouth or eyes mucous (Ciotti et al., 2020; Esakandari et al., 2020), providing an explanation for the high virulence associated with this pathogen.
At first, COVID-19 was characterized primarily as a lung disease since the main symptoms targeted the respiratory track. Indeed, infection of SARS-CoV-2 starts by binding the virus’ spike protein to the angiotensin-converting-enzyme 2 (ACE2) receptor in the host cells surface (Li et al., 2003). ACE2 is principally expressed into the nasal epithelium cells and in the bronchial epithelia (Sungnak et al., 2020). Yet, ACE2 receptor is also present in other organs, providing a mechanistic explanation for the onset of secondary issues in other organs like renal insufficiency, heart or nervous system (Esakandari et al., 2020; Ashraf et al., 2021).
As any infectious disease, COVID-19 immediately triggers the immunological response, which is highly variable among patients and it depends on personal characteristics and different involved risk factors (Jackson et al., 2022). For a subset of patients, an excessive immune response (cytokine storm) is associated with a severe disease phenotype (Coperchini et al., 2020; Rothan and Byrareddy, 2020). Associated to it, the related acute respiratory distress syndrome (ARDS) consists in an increased release of immune system cells that has been associated with organ failure. Therefore, the combination of the cytokine storm and ARDS are considered the main causes of death among patients (Montazersaheb et al., 2022). Any case, the interplay between SARS-CoV-2 infection and the host immunological system largely regulates the disease progression and is ultimately responsible for the severity of the disease (Gallo Marin et al., 2021).
Altogether, considering the highly variable phenotypic response and the systemic character of the disease, it becomes clear the need of investigating COVID-19 using alternative techniques such as the ones used in precision medicine. The aim of precision medicine is to provide an individualised solution to health problems that a subject can present (Bissonnette and Bergeron, 2012; König et al., 2017). To account for it, the “omics” technologies can help building a suitable description of the individual’s specific characteristics to find personalized treatment for the cure of specific diseases (Zhang, 2015). Genomics, transcriptomic, proteomics and metabolomics investigated on non-invasive fluid samples (i.e., serum, plasma or urine) can provide useful information to understand the disparate response to a given disorder from different subjects (Ward et al., 2021). In this regard, metabolomics is specially suitable because it is more sensitive to any phenotypic alteration (Nicholson, 2021). Thus, metabolomics has been extensively applied to COVID-19 to provide mechanistic information of the disease, to find robust diagnostic and prognostic biomarkers and to investigate the natural history course of the infection. The present review aims to discuss all the advances in the knowledge of COVID-19 disease as provided by the metabolomic analysis of patient samples.
As expected, COVID-19 has been extensively investigated using metabolomics approaches. We scrutinized the databases (Web of Science, Scopus and Pubmed, 2020–2022), using general keywords for a maximum coverage (Table 1). The outcome shows a very large number of contributions, in proportion with the emergency produced by the pandemic. After manually curating the results (Table 1), we found 90 search items that correspond to original contributions where metabolomics has been applied to COVID-19 patient cohorts. Only studies involving humans have been considered. All these studies involved either mass spectrometry coupled to gas chromatography (GC-MS) or liquid chromatography (LC-MS) (70%) and/or nuclear magnetic resonance (NMR) experiments, except for one (Robertson et al., 2022) that used Raman spectroscopy to investigate urine samples. LC-MS and GC-MS show an exquisite sensitivity that enables monitoring the entire metabolism with little signal overlapping but at the cost of limited reproducibility (Pan and Raftery, 2007). Additionally, they may require sample derivatization of the results and the quantification requires the use of standards (Bingol, 2018). In turn, NMR spectroscopy is highly complementary since it is fully quantitative, requires no derivatization and it is very reproducible, but with low sensitivity, which largely limits the accessible metabolome that can be investigated by this technique (Bruzzone et al., 2021). Remarkably, some contributions correspond to the development of new methodology for the application of these techniques to COVID-19 (Holmes et al., 2021b; Lodge et al., 2021c; Nitschke et al., 2022a; 2022b) or ring studies between laboratories (Masuda et al., 2021).
In terms of the nature of the specimen, serum and plasma followed by urine (Kurano et al., 2022b; Kida et al., 2022) are the most studied biofluids, while other samples also scrutinized include saliva (Spick et al., 2022b; Frampas et al., 2022; Saheb Sharif-Askari et al., 2022), faeces (He et al., 2021; Lv et al., 2021; Ren et al., 2021), platelets (Schuurman et al., 2022), exhaled breath (Barberis et al., 2021a; Grassin-Delyle et al., 2021; Bennet et al., 2022; Remy et al., 2022), sebum from skin (Spick et al., 2021) and breastmilk (Zhao et al., 2020). Due to the high volume of information (Table 1), we here complement other recent reviews on COVID-19 metabolomics (Hasan et al., 2021; Lin et al., 2021; Costanzo et al., 2022) by selecting a set of contributions to be discussed in the present review, based on our evaluation of novelty, impact and originality.
In addition to the consubstantial symptomatology of COVID-19, very early studies corroborated the fact that SARS-CoV-2 infection results in a much-altered metabolism, as determined from serum samples of hospitalized patients (Shen et al., 2020; Thomas et al., 2020). This metabotype is characteristic and differs from the one observed in flu-induced ARDS patients (Lorente et al., 2021). Significant alterations in the kynerurate/tryptophan pathway and abnormal glucose levels were amongst the first proposed metabolic markers associated to COVID-19 (Shen et al., 2020). These observations were further confirmed by other studies (Blasco et al., 2020; Bruzzone et al., 2020; Kimhofer et al., 2020; Song et al., 2020) that also overcame the technical limitations from the early cohorts (i.e., reduced size of the cohorts, sample inactivation and non-fasting conditions). Elevated kynurenic acid is gender-specific (Cai et al., 2021) and, in conjunction with other gender sensitive metabolites (Escarcega et al., 2022), it provides a rationale for the poorer clinical outcome in males than in females. Tryptophan is an essential amino acid and a neurotransmitter, and with phenylalanine also involved in the modulation of the immune response (Fernstrom and Wurtman, 1971) and inflammatory processes in lung and kidney diseases, or other infections like HIV or sepsis (Darcy et al., 2011; Kimhofer et al., 2020). SARS-CoV-2 infection also alters many other metabolic pathways (Albóniga et al., 2022), with changes in the serum amino acid signature (for instance, elevated glutamine/glutamate and Fischer’s ratios) (Doğan et al., 2021; Páez-Franco et al., 2022). Glutamate alteration in COVID-19 is in part mediated by the α-glutathione S-transferase, associated with certain processes such as liver failure, skeletal muscle metabolism, cancer or immunodeficiency (Kinscherf et al., 1996; Cruzat et al., 2018). Other found alterations include the circulating exosome (Alzahrani et al., 2021; Lam et al., 2021), the serum fatty acids (Chen et al., 2022) and other serum lipids such as carnitines (Castañé et al., 2022), ceramides (Dei Cas et al., 2021; Khodadoust, 2021) and phospholipids (Barberis et al., 2020; Shen et al., 2020; Janneh et al., 2021; Masoodi et al., 2022). The serum lipoprotein composition is also largely dysregulated (Bruzzone et al., 2020; Kimhofer et al., 2020; Lodge et al., 2021c; 2021a; Gray et al., 2021; Masuda et al., 2021), showing a pathogenic redistribution of the lipoprotein particle size and composition to increase the atherosclerotic risk.
Several studies used metabolic models to discriminate with great success (AUROC >0.95) between COVID-19 patients and healthy individuals (Bruzzone et al., 2020; Ruffieux et al., 2022). A simplified COVID-19 metabolic signature of the serum/plasma metabolomic dysregulation in acute patients can be obtained from the integrative analysis of the abovementioned studies, and it is shown in Figure 1. Undoubtedly, the strongest metabolic signal associated to SARS-CoV-2 is tryptophan metabolism (pink in Figure 1), whose catabolysis is upregulated with accumulation of kynurenine and other intermediates and depletion of trigonelline in COVID-19 patients. Related to this, upregulation of purine metabolites and some components of the urea cycle (blue in Figure 1) are also consubstantial to COVID-19. The increase in lactate and the dysregulation of essential metabolites in the central metabolism suggests a mitochondrial impairment. Finally, dyslipidemia is observed at the lipid level, with some lipids consistently showing upregulation (ceramides, PE, cholesterol) or downregulation (sphingomyelin and LPC), but also in the serum lipoproteins composition (yellow in Figure 1). All these studies are consistent with a model in which SARS-CoV-2 infection induces damage in the liver, the kidney, and other organs during the acute phase, also associated with dyslipidemia and oxidative stress.
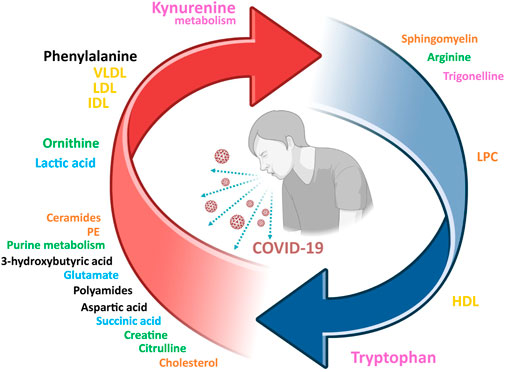
FIGURE 1. A consensus view of the metabolic signature of COVID-19 patients. Arrow colors indicate if metabolites are upregulated (red) or downregulated (blue). The letter size is proportional to the supporting experimental evidence. Color legend for the metabolic pathways: tryptophan metabolism (pink), central metabolism (blue), purine metabolism and urea cycle (green), lipids (orange), lipoproteins (yellow), other (black). VLDL, very low-density lipoproteins; LDL, low density lipoproteins; HDL, high density lipoproteins; IDL, intermediate density lipoproteins; PE, phosphatidylethanolamine.
This metabolic signature seems to be rather homogeneous worldwide, with equivalent results obtained when considering very distant geographical and cultural regions such as Mexico (López-Hernández et al., 2021), Italy (Saccon et al., 2021; Ghini et al., 2022b; Ciccarelli et al., 2022), and Africa (Li X. et al., 2022). Instead, characteristic but distinct metabotypes can be associated to pregnant women (Id et al., 2022), newborn (Kontou et al., 2021) and infantile (Wang et al., 2021) COVID-19 population. Finally, the COVID-19 metabotype changes concertedly with other subcellular elements (i.e., proteins, gene expression), that can be integrated to obtain the trans-omic landscape of COVID-19 (Wu et al., 2021).
Finally, COVID-19 is associated with an exacerbated inflammatory response and many studies have investigated the associated metabolic changes in conjunction with inflammatory markers (Lodge et al., 2021a; Yang et al., 2022). Specifically, IL-18, IL-6, IFN-γ, IP-10 and RANTES exhibited strong positive correlations with the pro-atherogenic LDL sub-particles and negative correlation with HDL particles and sub-particles (Lodge et al., 2021a), while M-CSF and IL-12p40 correlate with plasma levels of glycylproline and long-chain acylcarnitines in COVID-19 patients (Yang et al., 2022). That said, the COVID-19 interplay with the inflammatory system is not unique (Liu et al., 2022) and more than 20 metabolites were associated equally strongly to other unrelated severe pneumonia events (Julkunen et al., 2021). In line with this observation, the vast majority of tuberculosis (TB) patients experienced a severe SARS-CoV-2 post-TB infection (Diboun et al., 2022).
From the very beginning it became clear that metabolic pathways detectable in plasma, but not other signaling pathways, could be used to stratify COVID-19 patients (Wu et al., 2020) and metabolomic analysis of several biofluids has been extensively used to discriminate patients according to severity of SARS-CoV-2 infection (Table 2). One problem is the heterogeneity in the severity criteria, but most studies abided the guidelines for diagnosis of SARS-CoV-2 issued by the WHO or by the National Health Commission of the People’s Republic of China. In Table 2, cohorts from the different studies have been unified to a single classification: asymptomatic (AS), mild (M), moderate (MO), severe (S) or critical/deceased (C) according to severity, with special classes for COVID-19 non-infected subjects (controls, CO) and people with other pathologies (other, O). Hospitalized patients are mainly S, C and O classes.
In an early study, Danlos et al. (2021) investigated plasma samples from a well-stratified cohort of COVID-19 patients to identify up to 77 metabolites (amino acids, polyamines, sugars and their derivatives among others) that differ between critical and mild patients. These results were further confirmed and extended by subsequent studies in plasma as well (Albóniga et al., 2022; Dillard et al., 2022), using other matrices (Buyukozkan et al., 2022; Dewulf et al., 2022) or as a part of a multi-omic study (Su et al., 2020; Wu et al., 2021). All these contributions highlight the critical role that kynerurate pathway has in the metabotype associated to the disease severity (D’Amora et al., 2021; Marín-Corral et al., 2021; Roberts et al., 2022). Dysregulated amino acids (Ibarra-gonza et al., 2022), alterations in intermediates of amino acid catabolism (Páez-Franco et al., 2021) and elevated porphyrins (San Juan et al., 2020; Oliveira et al., 2022) are also linked to severe phenotypes of the disease.
Not only metabolites but also lipids such as carnitines and phosphatidylcholine (Caterino et al., 2021b; D’alessandro et al., 2021; Wu et al., 2022) and an NMR-determined pro-atherogenic lipoprotein profile have been associated to COVID-19 severity (Schmelter et al., 2021; Rendeiro et al., 2022). In addition, the metabolic changes associated with severity are also correlated with immune response markers: between plasma oxylipins (Karu et al., 2022) or acetylcholine (Pérez et al., 2022) with chemokines/neutrophiles or between lysophosphatidyl choline (LPC) with IL-6 (Sindelar et al., 2021).
This information has predictive power and a machine learning based model can predict COVID-19 prognosis employing only 22 plasma metabolites, most of them LPCs (Sindelar et al., 2021). Other equivalent models can be obtained when combining the information from 12 urine metabolites with proteomic data (Li T. et al., 2021), with the concerted analysis of an extended panel of lipids and metabolites with cytokines (Byeon et al., 2022) or by the combination of 21 lipids with four protein markers (Li Y. et al., 2021) (Figure 2).
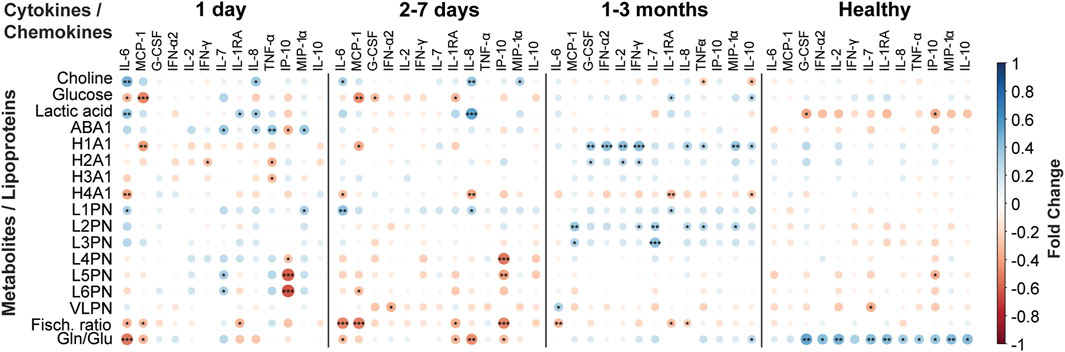
FIGURE 2. Immunometabolic correlation plots between a panel of cytokines/chemokines related with COVID-19 disease versus a set of metabolites and lipoproteins quantified in blood serum samples from COVID-19 patients. The plots represent the correlations from disease onset to up 3 months after infection. Negative correlations are coloured in red, while positive correlations are coloured in blue, as indicated in the figure legend. The calculations were performed in R.
Metabonomics aims to measure the overall metabolic response of living systems to biological stimuli or to genetic manipulation (Nicholson and Lindon, 2008). In the context of COVID-19, metabonomic analyses of non-invasive samples have been widely used to investigate the effect of drugs. Spick and co-workers used serum metabonomics in combination with other omic techniques to investigate the mechanism of glucocorticoids in the palliation of ARDS in COVID-19 patients, which was also used as a surrogate marker for severity (Spick et al., 2022a). Consistently, the effect of corticoids or liver protective drugs (i.e., arbitol) could be discriminated using untargeted metabonomics, but not the combined effect of two antiviral agents (lopinavir and ritonavir), that showed similar metabotype as for untreated patients (Shi et al., 2021). Furthermore, Meoni et al. (2021) employed NMR-based metabolomics and lipidomics in plasma samples to demonstrate that treatment with tocilizumab partially reverts the metabolic alterations due to SARS-CoV-2 infection.
An impressive scientific and economic effort allowed the early appearance of prophylactic measures for SAR-CoV-2 infection and several studies have investigated the putative metabolic alterations induced by mRNA vaccines. Using a combination of LC-MS and NMR spectroscopy, Dagla et al. (2022) analyzed plasma samples from patients up to 3 months after the first dose and observed distinct plasma metabotypes in relation to the level of immune response, highlighting the role of amino acid metabolism and the lipid profile as predictive markers of response to vaccination (Dagla et al., 2022). Equivalent variations in the lipoproteins (but not in the metabolome) were observed by serum NMR profiling of vaccinated individuals (Ghini et al., 2022a). In turn, He et al. (2022) have investigated the serum metabolic profiles associated with a proper response of the host immune antibodies and cytokines. Finally, the effect of nutrition as a protective factor for COVID-19 prognosis has also been investigated by lipidomic analysis (Barberis et al., 2021b).
Metabolomics has been extensively used to investigate the natural history of COVID-19 and patient’s recovery at the metabolic level. Several studies have investigated the metabolic recovery of COVID-19 patients by analyzing blood (serum or plasma) of patients from onset to discharge from hospital and the subsequent follow up check-ups, spanning up to more than 1 year from the disease onset (Table 3). Characterization of metabolic phenoreversion, a concept introduced by Jeremy Nicholson in the context of COVID-19 to describe its metabolic evolution (Lodge et al., 2021b), reveals that partial reversion of the metabolic phenotype can be associated to severity of the disease (Holmes et al., 2021a), and it correlates with an abnormal pulmonary function after three (Xu et al., 2021) or even six (Li H. et al., 2022) months from the disease onset. Other studies also suggest a slower metabolic phenoreversion as compared to the patient’s discharge time (Zhang et al., 2021), with dysregulated lipoprotein profile after hospital discharge (Bizkarguenaga et al., 2022). Persistent alterations of the metabolism concentrate in the amino acids, organic acids, purine, fatty acids (Valdés et al., 2022) and lipid metabolism (Kurano et al., 2022a), while the kynurenate pathway returned to normal levels (Li F. et al., 2022).
Prospective observational studies have proven useful to investigate the natural history of metabolic phenoreversion (Liptak et al., 2022). These studies evidence the intimate relationship between metabolic phenoreversion and the normalization of the exacerbated immune response (Jing et al., 2022; Zhang et al., 2022). In a large prospective study (Table 3), acute patients showed a metabolic and lipidomic dysregulation that accompanies the exacerbated immunological response, resulting in a slow metabolic recovery time with a maximum probability around 62 days (unpublished data). As an example, Figure 3 shows the correlation between a panel of COVID-19 associated inflammatory markers as compared to disease-altered metabolites and lipoproteins, as a function of the recovery time. The slow metabolic normalization in acute patients is lineage dependent (Lewis et al., 2022) and it maintains for months a lipoprotein profile compatible with enhanced atherosclerotic risk (unpublished data), providing an explanation for the elevated number of cardiovascular episodes found in postCOVID-19 cohorts (Xie et al., 2022). In line with this idea, survivors from non-severe COVID-19 from Wuhan still show metabolic abnormalities after 6 months (Li F. et al., 2022). This is consistent with previous studies on other related viruses such as MERS and SARS-CoV-1.

FIGURE 3. Metabolites that are associated with severity prognosis of COVID-19, sorted according to the disease severity (arrow axis).
Unfortunately, COVID-19 does not always evolve towards a full restoration of the metabolism and post-acute sequelae of COVID-19 represent an emerging global crisis (Su et al., 2022) and the need to find biomarkers for long COVID is pressing. In this context, deconvolution of NMR spectra from COVID-19 sera identified three diagnostic subregions of the supramolecular phospholipid composite signal envelope that provide insight about the increased cardiovascular risk in COVID-19 patients and the risk persistence in post-acute COVID-19 syndrome (Masuda et al., 2022). Notably, lipoproteins emerge as an important prognostic biomarker for the prediction of long COVID effects (Bai et al., 2021).
SARS-CoV-2 infection produces a profound metabolic dysregulation that can be adequately characterized by metabolomics and lipidomics. That said, the different techniques used, and the inherent variability impede a proper comparison between studies, particularly when considering the quantification of the changes associated to COVID-19. Many altered metabolites and lipoproteins can be used to evaluate disease severity and to monitor drug intervention, with a subset of biomarkers that also show prognostic value to evaluate long-term sequelae of the disease. The long-standing dysregulation of lipoprotein metabolism in COVID-19 patients provide an explanation for the elevated risk of cardiovascular episodes detected in post-infection individuals.
CB, RC, and NE performed initial literature review. CB and OM wrote the manuscript. JM and NE provided revisions. JM and OM approved the final version of the manuscript.
This research was funded by the SPRI I + D COVID-19 fund (Basque Government, bG-COVID-19), BIOEF EITB Maratoia (BIO21/COV/037) and PID 2019-107956RA-I00.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Albóniga, O. E., Jiménez, D., Sánchez-Conde, M., Vizcarra, P., Ron, R., Herrera, S., et al. (2022). Metabolic snapshot of plasma samples reveals new pathways implicated in SARS-CoV-2 pathogenesis. J. Proteome Res. 21, 623–634. doi:10.1021/acs.jproteome.1c00786
Alzahrani, F. A., Mohammed, M. R. S., Alkarim, S., Azhar, E. I., El-Magd, M. A., Hawsawi, Y., et al. (2021). Untargeted metabolic profiling of extracellular vesicles of sars-cov-2-infected patients shows presence of potent anti-inflammatory metabolites. Int. J. Mol. Sci. 22, 10467. doi:10.3390/ijms221910467
Ashraf, U. M., Abokor, A. A., Edwards, J. M., Waigi, E. W., Royfman, R. S., Hasan, S. A. M., et al. (2021). Sars-cov-2, ace2 expression, and systemic organ invasion. Physiol. Genomics 53, 51–60. doi:10.1152/physiolgenomics.00087.2020
Bai, Y., Huang, W., Li, Y., Lai, C., Huang, S., Wang, G., et al. (2021). Lipidomic alteration of plasma in cured COVID-19 patients using ultra high-performance liquid chromatography with high-resolution mass spectrometry. Biosci. Rep. 41, BSR20204305–12. doi:10.1042/BSR20204305
Barberis, E., Amede, E., Khoso, S., Castello, L., Sainaghi, P. P., Bellan, M., et al. (2021a). Metabolomics diagnosis of Covid-19 from exhaled breath condensate. Metabolites 11, 847. doi:10.3390/metabo11120847
Barberis, E., Amede, E., Tavecchia, M., Marengo, E., Cittone, M. G., Rizzi, E., et al. (2021b). Understanding protection from SARS-CoV-2 using metabolomics. Sci. Rep. 11, 13796–13810. doi:10.1038/s41598-021-93260-2
Barberis, E., Timo, S., Amede, E., Vanella, V. V., Puricelli, C., Cappellano, G., et al. (2020). Large-scale plasma analysis revealed new mechanisms and molecules associated with the host response to sars-cov-2. Int. J. Mol. Sci. 21, 8623–8625. doi:10.3390/ijms21228623
Bennet, S., Kaufmann, M., Takami, K., Sjaarda, C., Douchant, K., Moslinger, E., et al. (2022). Small-molecule metabolome identifies potential therapeutic targets against COVID-19. Sci. Rep. 12, 10029–10111. doi:10.1038/s41598-022-14050-y
Bi, X., Liu, W., Ding, X., Liang, S., Zheng, Y., Zhu, X., et al. (2022). Proteomic and metabolomic profiling of urine uncovers immune responses in patients with COVID-19. Cell Rep. 38, 110271. doi:10.1016/j.celrep.2021.110271
Bingol, K. (2018). Recent advances in targeted and untargeted metabolomics by NMR and MS/NMR methods. High. Throughput 7, 9. doi:10.3390/ht7020009
Bissonnette, L., and Bergeron, M. G. (2012). Infectious disease management through point-of-care personalized medicine molecular diagnostic technologies. J. Pers. Med. 2, 50–70. doi:10.3390/jpm2020050
Bizkarguenaga, M., Bruzzone, C., Gil-Redondo, R., SanJuan, I., Martin-Ruiz, I., Barriales, D., et al. (2022). Uneven metabolic and lipidomic profiles in recovered COVID-19 patients as investigated by plasma NMR metabolomics. NMR Biomed. 35, 46377–e4710. doi:10.1002/nbm.4637
Blasco, H., Bessy, C., Plantier, L., Lefevre, A., Piver, E., Bernard, L., et al. (2020). The specific metabolome profiling of patients infected by SARS-COV-2 supports the key role of tryptophan-nicotinamide pathway and cytosine metabolism. Sci. Rep. 10, 16824–16912. doi:10.1038/s41598-020-73966-5
Bruzzone, C., Bizkarguenaga, M., Gil-Redondo, R., Diercks, T., Arana, E., García de Vicuña, A., et al. (2020). SARS-CoV-2 infection dysregulates the metabolomic and lipidomic profiles of serum. iScience 23, 101645. doi:10.1016/j.isci.2020.101645
Bruzzone, C., Gil-Redondo, R., Seco, M., Barragán, R., de la Cruz, L., Cannet, C., et al. (2021). A molecular signature for the metabolic syndrome by urine metabolomics. Cardiovasc Diabetol. 20, 155. doi:10.1186/s12933-021-01349-9
Buyukozkan, M., Alvarez-Mulett, S., Racanelli, A. C., Schmidt, F., Batra, R., Hoffman, K. L., et al. (2022). Integrative metabolomic and proteomic signatures define clinical outcomes in severe COVID-19. iScience 25, 104612. doi:10.1016/j.isci.2022.104612
Byeon, S. K., Madugundu, A. K., Garapati, K., Ramarajan, M. G., Saraswat, M., Kumar-M, , P., et al. (2022). Development of a multiomics model for identification of predictive biomarkers for COVID-19 severity: A retrospective cohort study. Lancet Digit. Health 4, e632–e645. doi:10.1016/S2589-7500(22)00112-1
Cai, Y., Kim, D. J., Takahashi, T., Broadhurst, D. I., Yan, H., Ma, S., et al. (2021). Kynurenic acid may underlie sex-specific immune responses to COVID-19. Sci. Signal 14, eabf8483–11. doi:10.1126/scisignal.abf8483
Castañé, H., Iftimie, S., Baiges-Gaya, G., Rodríguez-Tomàs, E., Jiménez-Franco, A., López-Azcona, A. F., et al. (2022). Machine learning and semi-targeted lipidomics identify distinct serum lipid signatures in hospitalized COVID-19-positive and COVID-19-negative patients. Metabolism 131, 155197. doi:10.1016/j.metabol.2022.155197
Caterino, M., Costanzo, M., Fedele, R., Cevenini, A., Gelzo, M., Di Minno, A., et al. (2021a). The serum metabolome of moderate and severe COVID-19 patients reflects possible liver alterations involving carbon and nitrogen metabolism. Int. J. Mol. Sci. 22, 9548. doi:10.3390/IJMS22179548
Caterino, M., Gelzo, M., Sol, S., Fedele, R., Annunziata, A., Calabrese, C., et al. (2021b). Dysregulation of lipid metabolism and pathological inflammation in patients with COVID-19. Sci. Rep. 11, 2941. doi:10.1038/s41598-021-82426-7
Ceballos, F. C., Virseda-Berdices, A., Resino, S., Ryan, P., Martínez-González, O., Peréz-García, F., et al. (2022). Metabolic profiling at COVID-19 onset shows disease severity and sex-specific dysregulation. Front. Immunol. 13, 925558–925616. doi:10.3389/fimmu.2022.925558
Chen, W., Yao, M., Chen, M., Ou, Z., Yang, Q., He, Y., et al. (2022). Using an untargeted metabolomics approach to analyze serum metabolites in COVID-19 patients with nucleic acid turning negative. Front. Pharmacol. 13, 964037–964112. doi:10.3389/fphar.2022.964037
Ciccarelli, M., Merciai, F., Carrizzo, A., Sommella, E., Di Pietro, P., Caponigro, V., et al. (2022). Untargeted lipidomics reveals specific lipid profiles in COVID-19 patients with different severity from Campania region (Italy). J. Pharm. Biomed. Anal. 217, 114827. doi:10.1016/j.jpba.2022.114827
Ciotti, M., Ciccozzi, M., Terrinoni, A., Jiang, W. C., Wang, C. B., and Bernardini, S. (2020). The COVID-19 pandemic. Crit. Rev. Clin. Lab. Sci. 57, 365–388. doi:10.1080/10408363.2020.1783198
Coperchini, F., Chiovato, L., Croce, L., Magri, F., and Rotondi, M. (2020). The cytokine storm in COVID-19: An overview of the involvement of the chemokine/chemokine-receptor system. Cytokine Growth Factor Rev. 53, 25–32. doi:10.1016/j.cytogfr.2020.05.003
Correia, B. S. B., Ferreira, V. G., Piagge, P. M. F. D., Almeida, M. B., Assunção, N. A., Raimundo, J. R. S., et al. (2022). 1H qNMR-based metabolomics discrimination of covid-19 severity. J. Proteome Res. 21, 1640–1653. doi:10.1021/acs.jproteome.1c00977
Costanzo, M., Caterino, M., Fedele, R., Cevenini, A., Pontillo, M., Barra, L., et al. (2022). COVIDomics: The proteomic and metabolomic signatures of COVID-19. Int. J. Mol. Sci. 23, 2414. doi:10.3390/ijms23052414
Cruzat, V., Rogero, M. M., Keane, K. N., Curi, R., and Newsholme, P. (2018). Glutamine: Metabolism and immune function, supplementation and clinical translation. Nutrients 10, 1564. doi:10.3390/NU10111564
Cucinotta, D., and Vanelli, M. (2020). WHO declares COVID-19 a pandemic. Acta Biomed. 91, 157–160. doi:10.23750/abm.v91i1.9397
Dagla, I., Iliou, A., Benaki, D., Gikas, E., Mikros, E., Bagratuni, T., et al. (2022). Plasma metabolomic alterations induced by COVID-19 vaccination reveal putative biomarkers reflecting the immune response. Cells 11, 1241. doi:10.3390/cells11071241
D’alessandro, A., Thomas, T., Akpan, I. J., Reisz, J. A., Cendali, F. I., Gamboni, F., et al. (2021). Biological and clinical factors contributing to the metabolic heterogeneity of hospitalized patients with and without Covid-19. Cells 10, 2293–2323. doi:10.3390/cells10092293
D’Amora, P., Silva, I. D. C. G., Budib, M. A., Ayache, R., Silva, R. M. S., Silva, F. C., et al. (2021). Towards risk stratification and prediction of disease severity and mortality in COVID-19: Next generation metabolomics for the measurement of host response to COVID-19 infection. PLoS One 16, e0259909–e0259916. doi:10.1371/journal.pone.0259909
Danlos, F. X., Grajeda-Iglesias, C., Durand, S., Sauvat, A., Roumier, M., Cantin, D., et al. (2021). Metabolomic analyses of COVID-19 patients unravel stage-dependent and prognostic biomarkers. Cell Death Dis. 12, 258. doi:10.1038/s41419-021-03540-y
Darcy, C. J., Davis, J. S., Woodberry, T., McNeil, Y. R., Stephens, D. P., Yeo, T. W., et al. (2011). An observational cohort study of the kynurenine to tryptophan ratio in sepsis: Association with impaired immune and microvascular function. PLoS One 6, e21185. doi:10.1371/journal.pone.0021185
Dei Cas, M., Ottolenghi, S., Morano, C., Rinaldo, R., Roda, G., Chiumello, D., et al. (2021). Link between serum lipid signature and prognostic factors in COVID-19 patients. Sci. Rep. 11, 21633–21712. doi:10.1038/s41598-021-00755-z
Dewulf, J. P., Martin, M., Marie, S., Oguz, F., Belkhir, L., De Greef, J., et al. (2022). Urine metabolomics links dysregulation of the tryptophan-kynurenine pathway to inflammation and severity of COVID-19. Sci. Rep. 12, 9959–9968. doi:10.1038/s41598-022-14292-w
Diboun, I., Cyprian, F. S., Anwardeen, N. R., Yassine, H. M., Elrayess, M. A., Rahmoon, S. M., et al. (2022). Identification of prognostic metabolomic biomarkers at the interface of mortality and morbidity in pre-existing TB cases infected with SARS-CoV-2. Front. Cell Infect. Microbiol. 12, 929689–929713. doi:10.3389/fcimb.2022.929689
Dillard, L. R., Wase, N., Ramakrishnan, G., Park, J. J., Sherman, N. E., Carpenter, R., et al. (2022). Leveraging metabolic modeling to identify functional metabolic alterations associated with COVID-19 disease severity. Metabolomics 18, 51–14. doi:10.1007/s11306-022-01904-9
Doğan, H. O., Şenol, O., Bolat, S., Yıldız, Ş. N., Büyüktuna, S. A., Sarıismailoğlu, R., et al. (2021). Understanding the pathophysiological changes via untargeted metabolomics in COVID-19 patients. J. Med. Virol. 93, 2340–2349. doi:10.1002/jmv.26716
Esakandari, H., Nabi-Afjadi, M., Fakkari-Afjadi, J., Farahmandian, N., Miresmaeili, S. M., and Bahreini, E. (2020). A comprehensive review of COVID-19 characteristics. Biol. Proced. Online 22, 19–10. doi:10.1186/s12575-020-00128-2
Escarcega, R. D., Honarpisheh, P., Colpo, G. D., Ahnstedt, H. W., Couture, L., Juneja, S., et al. (2022). Sex differences in global metabolomic profiles of COVID-19 patients. Cell Death Dis. 13, 461. doi:10.1038/s41419-022-04861-2
Fernstrom, J. D., and Wurtman, R. J. (1971). Brain serotonin content: Physiological dependence on plasma tryptophan levels. Science 173, 149–152. doi:10.1126/science.173.3992.149
Frampas, C. F., Longman, K., Spick, M., Lewis, H. M., Costa, C. D. S., Stewart, A., et al. (2022). Untargeted saliva metabolomics by liquid chromatography-Mass spectrometry reveals markers of COVID-19 severity. PLoS One 17. e0274967. doi:10.1371/journal.pone.0274967
Gallo Marin, B., Aghagoli, G., Lavine, K., Yang, L., Siff, E. J., Chiang, S. S., et al. (2021). Predictors of COVID-19 severity: A literature review. Rev. Med. Virol. 31, 1–10. doi:10.1002/rmv.2146
Ghini, V., Maggi, L., Mazzoni, A., Spinicci, M., Zammarchi, L., Bartoloni, A., et al. (2022a). Serum NMR profiling reveals differential alterations in the lipoproteome induced by pfizer-BioNTech vaccine in COVID-19 recovered subjects and naïve subjects. Front. Mol. Biosci. 9, 839809–839818. doi:10.3389/fmolb.2022.839809
Ghini, V., Meoni, G., Pelagatti, L., Celli, T., Veneziani, F., Petrucci, F., et al. (2022b). Profiling metabolites and lipoproteins in COMETA, an Italian cohort of COVID-19 patients. PLoS Pathog. 18, 10104433–e1010516. doi:10.1371/journal.ppat.1010443
Grassin-Delyle, S., Roquencourt, C., Moine, P., Saffroy, G., Carn, S., Heming, N., et al. (2021). Metabolomics of exhaled breath in critically ill COVID-19 patients: A pilot study. EBioMedicine 63, 103154. doi:10.1016/j.ebiom.2020.103154
Gray, N., Lawler, N., Zeng, A., Ryan, M., Bong, S., Boughton, B., et al. (2021). Diagnostic potential of the plasma lipidome in infectious disease: Application to acute SARS-CoV-2 infection. Metabolites 11, 467. doi:10.3390/metabo11070467
Gu, M., Pan, H., Yuan, Y., Zhou, X., Chen, L., Wang, X., et al. (2022). Sera metabolomics characterization of patients at different stages in wuhan identifies critical biomarkers of COVID-19. Front. Cell Infect. Microbiol. 12, 882661–882715. doi:10.3389/fcimb.2022.882661
Hao, Y., Zhang, Z., Feng, G., Chen, M., Wan, Q., Lin, J., et al. (2021). Distinct lipid metabolic dysregulation in asymptomatic COVID-19. iScience 24. 102974. doi:10.1016/j.isci.2021.102974
Hasan, M. R., Suleiman, M., and Pérez-López, A. (2021). Metabolomics in the diagnosis and prognosis of COVID-19. Front. Genet. 12, 721556. doi:10.3389/fgene.2021.721556
He, F., Zhang, T., Xue, K., Fang, Z., Jiang, G., Huang, S., et al. (2021). Fecal multi-omics analysis reveals diverse molecular alterations of gut ecosystem in COVID-19 patients. Anal. Chim. Acta 1180, 338881. doi:10.1016/j.aca.2021.338881
He, M., Huang, Y., Wang, Y., Liu, J., Han, M., Xiao, Y., et al. (2022). Metabolomics-based investigation of SARS-CoV-2 vaccination (Sinovac) reveals an immune-dependent metabolite biomarker. Front. Immunol. 13, 954801–954814. doi:10.3389/fimmu.2022.954801
Holmes, E., Wist, J., Masuda, R., Lodge, S., Nitschke, P., Kimhofer, T., et al. (2021b). Incomplete systemic recovery and metabolic phenoreversion in post-acute-phase nonhospitalized COVID-19 patients: Implications for assessment of post-acute COVID-19 syndrome. J. Proteome Res. 20, 3315–3329. doi:10.1021/acs.jproteome.1c00224
Holmes, E., Nicholson, J. K., Lodge, S., Nitschke, P., Kimhofer, T., Wist, J., et al. (2021a). Diffusion and relaxation edited proton NMR spectroscopy of plasma reveals a high-fidelity supramolecular biomarker signature of SARS-CoV-2 infection. Anal. Chem. 93, 3976–3986. doi:10.1021/acs.analchem.0c04952
Ibarra-gonza, I., Mart, L. E., Tusie, T., Moreno-mac, H., Jimenez-gutierrez, G. E., Va, P., et al. (2022). Metabolic reprogramming in SARS-CoV-2 infection impacts the outcome of COVID-19 patients. Front. Immunol. 13, 936106–936112. doi:10.3389/fimmu.2022.936106
Id, M. A., Mok, T., Id, C. J., Yeo, S., Quach, A., and Id, Y. A. (2022). Severe COVID-19 in pregnancy has a distinct serum profile, including greater complement activation and dysregulation of serum lipids. 1–12. doi: doi:10.1371/journal.pone.0276766
Jackson, C. B., Farzan, M., Chen, B., and Choe, H. (2022). Mechanisms of SARS-CoV-2 entry into cells. Nat. Rev. Mol. Cell Biol. 23, 3–20. doi:10.1038/s41580-021-00418-x
Janneh, A. H., Kassir, M. F., Dwyer, C. J., Chakraborty, P., Pierce, J. S., Flume, P. A., et al. (2021). Alterations of lipid metabolism provide serologic biomarkers for the detection of asymptomatic versus symptomatic COVID-19 patients. Sci. Rep. 11, 14232. doi:10.1038/s41598-021-93857-7
Jing, Y., Wang, J., Zhang, H., Yang, K., Li, J., Zhao, T., et al. (2022). Alterations of urinary microbial metabolites and immune indexes linked with COVID-19 infection and prognosis. Front. Immunol. 13, 841739. doi:10.3389/fimmu.2022.841739
Julkunen, H., Cichońska, A., Slagboom, P. E., and Würtz, P. (2021). Metabolic biomarker profiling for identification of susceptibility to severe pneumonia and COVID-19 in the general population. Elife 10, 630333–e63120. doi:10.7554/eLife.63033
Karu, N., Kindt, A., Lamont, L., van Gammeren, A. J., Ermens, A. A. M., Harms, A. C., et al. (2022). Plasma oxylipins and their precursors are strongly associated with COVID-19 severity and with immune response markers. Metabolites 12, 619–710. doi:10.3390/metabo12070619
Khodadoust, M. M. (2021). Inferring a causal relationship between ceramide levels and COVID-19 respiratory distress. Sci. Rep. 11, 20866–20913. doi:10.1038/s41598-021-00286-7
Kida, M., Nakamura, T., Kobayashi, K., Shimosawa, T., and Murata, T. (2022). Urinary lipid profile of patients with coronavirus diseases 2019. Front. Med. (Lausanne) 9, 941563–941567. doi:10.3389/fmed.2022.941563
Kimhofer, T., Lodge, S., Whiley, L., Gray, N., Loo, R. L., Lawler, N. G., et al. (2020). Integrative modeling of quantitative plasma lipoprotein, metabolic, and amino acid data reveals a multiorgan pathological signature of SARS-CoV-2 infection. J. Proteome Res. 19, 4442–4454. doi:10.1021/acs.jproteome.0c00519
Kinscherf, R., Hack, V., Fischbach, T., Friedmann, B., Weiss, C., Edler, L., et al. (1996). Low plasma glutamine in combination with high glutamate levels indicate risk for loss of body cell mass in healthy individuals: The effect of N- acetyl-cysteine. J. Mol. Med. 74, 393–400. doi:10.1007/BF00210633
König, I. R., Fuchs, O., Hansen, G., von Mutius, E., and Kopp, M. V. (2017). What is precision medicine? Eur. Respir. J. 50, 1700391–1700412. doi:10.1183/13993003.00391-2017
Kontou, A., Virgiliou, C., Mouskeftara, T., Begou, O., Meikopoulos, T., Thomaidou, A., et al. (2021). Plasma lipidomic and metabolomic profiling after birth in neonates born to SARS-CoV-19 infected and non-infected mothers at delivery: Preliminary results. Metabolites 11, 830–914. doi:10.3390/METABO11120830
Kurano, M., Aoki, J., and Moriya, K. (2022a). Dynamic modulations of sphingolipids and glycerophospholipids in COVID-19. doi: doi:10.1002/ctm2.1069
Kurano, M., Jubishi, D., Okamoto, K., Hashimoto, H., Sakai, E., Morita, Y., et al. (2022b). Dynamic modulations of urinary sphingolipid and glycerophospholipid levels in COVID-19 and correlations with COVID-19-associated kidney injuries. J. Biomed. Sci. 29, 94. doi:10.1186/s12929-022-00880-5
Lam, S. M., Zhang, C., Wang, Z., Ni, Z., Zhang, S., Yang, S., et al. (2021). A multi-omics investigation of the composition and function of extracellular vesicles along the temporal trajectory of COVID-19. Nat. Metab. 3, 909–922. doi:10.1038/s42255-021-00425-4
Lewis, H. M., Liu, Y., Frampas, C. F., Longman, K., Spick, M., Stewart, A., et al. (2022). Metabolomics markers of COVID-19 are dependent on collection wave. Metabolites 12, 713. doi:10.3390/metabo12080713
Li, F., Fu, L., Liu, X., Liu, X. A., Liang, Y., Lv, Y., et al. (2022a). Serum metabolomic abnormalities in survivors of non-severe COVID-19. Heliyon 8, e10473. doi:10.1016/j.heliyon.2022.e10473
Li, H., Li, X., Wu, Q., Wang, X., Qin, Z., Wang, Y., et al. (2022b). Plasma proteomic and metabolomic characterization of COVID-19 survivors 6 months after discharge. Cell Death Dis. 13, 235. doi:10.1038/s41419-022-04674-3
Li, T., Ning, N., Li, B., Luo, D., Qin, E., Yu, W., et al. (2021a). Longitudinal metabolomics reveals ornithine cycle dysregulation correlates with inflammation and coagulation in COVID-19 severe patients. Front. Microbiol. 12, 723818–723912. doi:10.3389/fmicb.2021.723818
Li, W., Moore, M. J., Vasilieva, N., Sui, J., Wong, S. K., Berne, M. A., et al. (2003). Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426, 450–454. doi:10.1038/nature02145
Li, X., Tu, B., Zhang, X., Xu, W., Chen, J., Xu, B., et al. (2022c). Dysregulation of glutamine/glutamate metabolism in COVID-19 patients: A metabolism study in african population and mini meta-analysis. J. Med. Virol. 0, e28150–e28152. doi:10.1002/jmv.28150
Li, Y., Hou, G., Zhou, H., Wang, Y., Tun, H. M., Zhu, A., et al. (2021b). Multi-platform omics analysis reveals molecular signature for COVID-19 pathogenesis, prognosis and drug target discovery. Signal Transduct. Target Ther. 6, 155. doi:10.1038/s41392-021-00508-4
Lin, B., Liu, J., Liu, Y., and Qin, X. (2021). Progress in understanding COVID-19: Insights from the omics approach. Crit. Rev. Clin. Lab. Sci. 58, 242–252. doi:10.1080/10408363.2020.1851167
Liptak, P., Baranovicova, E., Rosolanka, R., Simekova, K., Bobcakova, A., Vysehradsky, R., et al. (2022). Persistence of metabolomic changes in patients during post-COVID phase: A prospective, observational study. Metabolites 12, 641. doi:10.3390/metabo12070641
Liu, J., Li, Z. B., Lu, Q. Q., Yu, Y., Zhang, S. Q., Ke, P. F., et al. (2022). Metabolite profile of COVID-19 revealed by UPLC-MS/MS-based widely targeted metabolomics. Front. Immunol. 13, 894170–894213. doi:10.3389/fimmu.2022.894170
Lodge, S., Nitschke, P., Kimhofer, T., Coudert, J. D., Begum, S., Bong, S. H., et al. (2021a). NMR spectroscopic windows on the systemic effects of SARS-CoV-2 infection on plasma lipoproteins and metabolites in relation to circulating cytokines. J. Proteome Res. 20, 1382–1396. doi:10.1021/acs.jproteome.0c00876
Lodge, S., Nitschke, P., Kimhofer, T., Wist, J., Bong, S.-H., Loo, R. L., et al. (2021b). Diffusion and relaxation edited proton NMR spectroscopy of plasma reveals a high-fidelity supramolecular biomarker signature of SARS-CoV-2 infection. Anal. Chem. 93, 3976–3986. doi:10.1021/acs.analchem.0c04952
Lodge, S., Nitschke, P., Loo, R. L., Kimhofer, T., Bong, S.-H., Richards, T., et al. (2021c). Low volume in vitro diagnostic proton NMR spectroscopy of human blood plasma for lipoprotein and metabolite analysis: Application to SARS-CoV-2 biomarkers. J. Proteome Res. 20, 1415–1423. doi:10.1021/acs.jproteome.0c00815
López-Hernández, Y., Monárrez-Espino, J., Oostdam, A. S. H., Delgado, J. E. C., Zhang, L., Zheng, J., et al. (2021). Targeted metabolomics identifies high performing diagnostic and prognostic biomarkers for COVID-19. Sci. Rep. 11, 14732–14813. doi:10.1038/s41598-021-94171-y
Lorente, J. A., Nin, N., Villa, P., Vasco, D., Miguel-Coello, A. B., Rodriguez, I., et al. (2021). Metabolomic diferences between COVID-19 and H1N1 influenza induced ARDS. Crit. Care 25, 390–411. doi:10.1186/s13054-021-03810-3
Lv, L., Jiang, H., Chen, Y., Gu, S., Xia, J., Zhang, H., et al. (2021). The faecal metabolome in COVID-19 patients is altered and associated with clinical features and gut microbes. Anal. Chim. Acta 1152, 338267. doi:10.1016/j.aca.2021.338267
Marín-Corral, J., Rodríguez-Morató, J., Gomez-Gomez, A., Pascual-Guardia, S., Muñoz-Bermúdez, R., Salazar-Degracia, A., et al. (2021). Metabolic signatures associated with severity in hospitalized Covid-19 patients. Int. J. Mol. Sci. 22, 4794. doi:10.3390/ijms22094794
Masoodi, M., Peschka, M., Schmiedel, S., Haddad, M., Frye, M., Maas, C., et al. (2022). Disturbed lipid and amino acid metabolisms in COVID-19 patients. J. Mol. Med. 100, 555–568. doi:10.1007/s00109-022-02177-4
Masuda, R., Lodge, S., Nitschke, P., Spraul, M., Schaefer, H., Bong, S.-H., et al. (2021). Integrative modeling of plasma metabolic and lipoprotein biomarkers of SARS-CoV-2 infection in Spanish and Australian COVID-19 patient cohorts. J. Proteome Res. 20, 4139–4152. doi:10.1021/acs.jproteome.1c00458
Masuda, R., Lodge, S., Whiley, L., Gray, N., Lawler, N., Nitschke, P., et al. (2022). Exploration of human serum lipoprotein supramolecular phospholipids using statistical heterospectroscopy in n-dimensions (SHY- n): Identification of potential cardiovascular risk biomarkers related to SARS-CoV-2 infection. Anal. Chem. 94, 4426–4436. doi:10.1021/acs.analchem.1c05389
Meoni, G., Ghini, V., Maggi, L., Vignoli, A., Mazzoni, A., Salvati, L., et al. (2021). Metabolomic/lipidomic profiling of COVID-19 and individual response to tocilizumab. PLoS Pathog. 17, 10092433–e1009314. doi:10.1371/JOURNAL.PPAT.1009243
Montazersaheb, S., Hosseiniyan Khatibi, S. M., Hejazi, M. S., Tarhriz, V., Farjami, A., Ghasemian Sorbeni, F., et al. (2022). COVID-19 infection: An overview on cytokine storm and related interventions. Virol. J. 19, 92–15. doi:10.1186/s12985-022-01814-1
Nicholson, J. K. (2021). Molecular phenomic approaches to deconvolving the systemic effects of SARS-CoV-2 infection and post-acute COVID-19 syndrome. Phenomics 1, 143–150. doi:10.1007/S43657-021-00020-3
Nicholson, J. K., and Lindon, J. C. (2008). Systems biology: Metabonomics. Metabonomics 455, 1054–1056. doi:10.1038/4551054a
Nitschke, P., Lodge, S., Hall, D., Schaefer, H., Spraul, M., Embade, N., et al. (2022a). Direct low field J-edited diffusional proton NMR spectroscopic measurement of COVID-19 inflammatory biomarkers in human serum. Analyst 147, 4213–4221. doi:10.1039/d2an01097f
Nitschke, P., Lodge, S., Kimhofer, T., Masuda, R., Bong, S. H., Hall, D., et al. (2022b). J-edited DIffusional proton nuclear magnetic resonance spectroscopic measurement of glycoprotein and supramolecular phospholipid biomarkers of inflammation in human serum. Anal. Chem. 94, 1333–1341. doi:10.1021/acs.analchem.1c04576
Oliveira, L. B., Mwangi, V. I., Sartim, M. A., Delafiori, J., Sales, G. M., de Oliveira, A. N., et al. (2022). Metabolomic profiling of plasma reveals differential disease severity markers in COVID-19 patients. Front. Microbiol. 13, 844283–844315. doi:10.3389/fmicb.2022.844283
Páez-Franco, J. C., Maravillas-Montero, J. L., Mejía-Domínguez, N. R., Torres-Ruiz, J., Tamez-Torres, K. M., Pérez-Fragoso, A., et al. (2022). Metabolomics analysis identifies glutamic acid and cystine imbalances in COVID-19 patients without comorbid conditions. Implications on redox homeostasis and COVID-19 pathophysiology. PLoS One 17, e0274910. doi:10.1371/journal.pone.0274910
Páez-Franco, J. C., Torres-Ruiz, J., Sosa-Hernández, V. A., Cervantes-Díaz, R., Romero-Ramírez, S., Pérez-Fragoso, A., et al. (2021). Metabolomics analysis reveals a modified amino acid metabolism that correlates with altered oxygen homeostasis in COVID-19 patients. Sci. Rep. 11, 6350–6412. doi:10.1038/s41598-021-85788-0
Pan, Z., and Raftery, D. (2007). Comparing and combining NMR spectroscopy and mass spectrometry in metabolomics. Anal. Bioanal. Chem. 387, 525–527. doi:10.1007/s00216-006-0687-8
Pérez, M. M., Pimentel, V. E., Fuzo, C. A., da Silva-Neto, P. V., Toro, D. M., Fraga-Silva, T. F. C., et al. (2022). Acetylcholine, fatty acids, and lipid mediators are linked to COVID-19 severity. J. Immunol. 209, 250–261. doi:10.4049/jimmunol.2200079
Remy, R., Kemnitz, N., Trefz, P., Fuchs, P., Bartels, J., Klemenz, A.-C., et al. (2022). Profiling of exhaled volatile organics in the screening scenario of a COVID-19 test center. iScience 25, 105195. doi:10.1016/j.isci.2022.105195
Ren, Z., Wang, H., Cui, G., Lu, H., Wang, L., Luo, H., et al. (2021). Alterations in the human oral and gut microbiomes and lipidomics in COVID-19. Gut 70, 1253–1265. doi:10.1136/gutjnl-2020-323826
Rendeiro, A. F., Vorkas, C. K., Krumsiek, J., Singh, H. K., Kapadia, S. N., Cappelli, L. V., et al. (2022). Metabolic and immune markers for precise monitoring of COVID-19 severity and treatment. Front. Immunol. 12, 809937–810013. doi:10.3389/fimmu.2021.809937
Roberts, I., Wright Muelas, M., Taylor, J. M., Davison, A. S., Xu, Y., Grixti, J. M., et al. (2022). Untargeted metabolomics of COVID-19 patient serum reveals potential prognostic markers of both severity and outcome. Metabolomics 18, 6–19. doi:10.1007/s11306-021-01859-3
Robertson, J. L., Senger, R. S., Talty, J., Du, P., Sayed-Issa, A., Avellar, M. L., et al. (2022). Alterations in the molecular composition of COVID-19 patient urine, detected using Raman spectroscopic/computational analysis. PLoS One 17, e0270914–e0270916. doi:10.1371/journal.pone.0270914
Rothan, H. A., and Byrareddy, S. N. (2020). Epidemiology and pathogenesis of coronavirus disease (COVID-19). Nov. Res. Microbiol. J. 4, 675–687. doi:10.21608/nrmj.2020.84016
Ruffieux, H., Hanson, A. L., Lodge, S., Lawler, N. G., Whiley, L., Gray, N., et al. (2022). A patient-centric modeling framework captures recovery from SARS-CoV-2 infection. doi: doi:10.1038/s41590-022-01380-2
Saccon, E., Bandera, A., Sciumè, M., Mikaeloff, F., Lashari, A. A., Aliberti, S., et al. (2021). Distinct metabolic profile associated with a fatal outcome in COVID-19 patients during the early epidemic in Italy. Microbiol. Spectr. 9, e0054921–e0054927. doi:10.1128/spectrum.00549-21
Saheb Sharif-Askari, N., Soares, N. C., Mohamed, H. A., Saheb Sharif-Askari, F., Alsayed, H. A. H., Al-Hroub, H., et al. (2022). Saliva metabolomic profile of COVID-19 patients associates with disease severity. Metabolomics 18, 81. doi:10.1007/s11306-022-01936-1
San Juan, I., Bruzzone, C., Bizkarguenaga, M., Bernardo-Seisdedos, G., Laín, A., Gil-Redondo, R., et al. (2020). Abnormal concentration of porphyrins in serum from COVID-19 patients. Br. J. Haematol. 190, e265–e267. doi:10.1111/bjh.17060
Schmelter, F., Föh, B., Mallagaray, A., Rahmöller, J., Ehlers, M., Lehrian, S., et al. (2021). Metabolic and lipidomic markers differentiate COVID-19 from non-hospitalized and other intensive care patients. Front. Mol. Biosci. 8, 737039. doi:10.3389/FMOLB.2021.737039
Schuurman, A. R., Léopold, V., Pereverzeva, L., Chouchane, O., Reijnders, T. D. Y., Brabander, J. de, et al. (2022). The platelet lipidome is altered in patients with COVID-19 and correlates with platelet reactivity. Thromb. Haemost. 122, 1683–1692. doi:10.1055/s-0042-1749438
Shen, B., Yi, X., Sun, Y., Bi, X., Du, J., Zhang, C., et al. (2020). Proteomic and metabolomic characterization of COVID-19 patient sera. Cell 182, 59–72.e15. doi:10.1016/j.cell.2020.05.032
Shi, D., Yan, R., Lv, L., Jiang, H., Lu, Y., Sheng, J., et al. (2021). The serum metabolome of COVID-19 patients is distinctive and predictive. Metabolism 118, 154739. doi:10.1016/j.metabol.2021.154739
Sindelar, M., Stancliffe, E., Schwaiger-Haber, M., Anbukumar, D. S., Adkins-Travis, K., Goss, C. W., et al. (2021). Longitudinal metabolomics of human plasma reveals prognostic markers of COVID-19 disease severity. Cell Rep. Med. 2, 100369. doi:10.1016/j.xcrm.2021.100369
Song, J. W., Lam, S. M., Fan, X., Cao, W. J., Wang, S. Y., Tian, H., et al. (2020). Omics-driven systems interrogation of metabolic dysregulation in COVID-19 pathogenesis. Cell Metab. 32, 188–202.e5. doi:10.1016/j.cmet.2020.06.016
Spick, M., Campbell, A., Baricevic-Jones, I., von Gerichten, J., Lewis, H. M., Frampas, C. F., et al. (2022a). Multi-omics reveals mechanisms of partial modulation of COVID-19 dysregulation by glucocorticoid treatment. Int. J. Mol. Sci. 23, 12079. doi:10.3390/ijms232012079
Spick, M., Lewis, H. M., Frampas, C. F., Longman, K., Costa, C., Stewart, A., et al. (2022b). An integrated analysis and comparison of serum, saliva and sebum for COVID-19 metabolomics. Sci. Rep. 12, 11867–11912. doi:10.1038/s41598-022-16123-4
Spick, M., Longman, K., Frampas, C., Lewis, H., Costa, C., Walters, D. D., et al. (2021). Changes to the sebum lipidome upon COVID-19 infection observed via rapid sampling from the skin. EClinicalMedicine 33, 100786. doi:10.1016/j.eclinm.2021.100786
Su, Y., Chen, D., Yuan, D., Lausted, C., Choi, J., Dai, C. L., et al. (2020). Multi-omics resolves a sharp disease-state shift between mild and moderate COVID-19. Cell 183, 1479–1495.e20. doi:10.1016/J.CELL.2020.10.037
Su, Y., Yuan, D., Chen, D. G., Ng, R. H., Wang, K., Choi, J., et al. (2022). Multiple early factors anticipate post-acute COVID-19 sequelae. Cell 185, 881–895.e20. doi:10.1016/j.cell.2022.01.014
Sungnak, W., Huang, N., Bécavin, C., Berg, M., Queen, R., Litvinukova, M., et al. (2020). SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat. Med. 26, 681–687. doi:10.1038/s41591-020-0868-6
Thomas, T., Stefanoni, D., Reisz, J. A., Nemkov, T., Bertolone, L., Francis, R. O., et al. (2020). COVID-19 infection alters kynurenine and fatty acid metabolism, correlating with IL-6 levels and renal status. JCI Insight 5, e140327. doi:10.1172/jci.insight.140327
Torretta, E., Garziano, M., Poliseno, M., Capitanio, D., Biasin, M., Santantonio, T. A., et al. (2021). Severity of covid-19 patients predicted by serum sphingolipids signature. Int. J. Mol. Sci. 22, 10198. doi:10.3390/ijms221910198
Valdés, A., Moreno, L. O., Rello, S. R., Orduña, A., Bernardo, D., and Cifuentes, A. (2022). Metabolomics study of COVID-19 patients in four different clinical stages. Sci. Rep. 12, 1650. doi:10.1038/S41598-022-05667-0
Velavan, T. P., and Meyer, C. G. (2020). The COVID-19 epidemic. Trop. Med. Int. Health 25, 278–280. doi:10.1111/tmi.13383
Wang, C., Li, X., Ning, W., Gong, S., Yang, F., Fang, C., et al. (2021). Multi-omic profiling of plasma reveals molecular alterations in children with COVID-19. Theranostics 11, 8008–8026. –8026. doi:10.7150/THNO.61832
Ward, R. A., Aghaeepour, N., Bhattacharyya, R. P., Clish, C. B., Gaudillière, B., Hacohen, N., et al. (2021). Harnessing the potential of multiomics studies for precision medicine in infectious disease. Open Forum Infect. Dis. 8, ofab483–12. doi:10.1093/ofid/ofab483
Wu, D., Shu, T., Yang, X., Song, J. X., Zhang, M., Yao, C., et al. (2020). Plasma metabolomic and lipidomic alterations associated with COVID-19. Natl. Sci. Rev. 7, 1157–1168. doi:10.1093/NSR/NWAA086
Wu, J., Cyr, A., Gruen, D. S., Lovelace, T. C., Benos, P. V., Das, J., et al. (2022). Lipidomic signatures align with inflammatory patterns and outcomes in critical illness. Nat. Commun. 13, 6789. doi:10.1038/s41467-022-34420-4
Wu, P., Chen, D., Ding, W., Wu, P., Hou, H., Bai, Y., et al. (2021). The trans-omics landscape of COVID-19. Nat. Commun. 12, 4543–4616. doi:10.1038/s41467-021-24482-1
Xie, Y., Xu, E., Bowe, B., and Al-Aly, Z. (2022). Long-term cardiovascular outcomes of COVID-19. Nat. Med. 28 (3 28), 583–590. doi:10.1038/s41591-022-01689-3
Xu, J., Zhou, M., Luo, P., Yin, Z., Wang, S., Liao, T., et al. (2021). Plasma metabolomic profiling of patients recovered from coronavirus disease 2019 (COVID-19) with pulmonary sequelae 3 Months after discharge. Clin. Infect. Dis. 73, 2228–2239. doi:10.1093/CID/CIAB147
Xue, M., Zhang, T., Cheng, Z. J., Guo, B., Zeng, Y., Lin, R., et al. (2022). Effect of a functional phospholipid metabolome-protein association pathway on the mechanism of COVID-19 disease progression. Int. J. Biol. Sci. 18, 4618–4628. doi:10.7150/ijbs.72450
Yang, Z., Wu, D., Lu, S., Qiu, Y., Hua, Z., Tan, F., et al. (2022). Plasma metabolome and cytokine profile reveal glycylproline modulating antibody fading in convalescent COVID-19 patients. Proc. Natl. Acad. Sci. U. S. A. 119, 21170891199–e2117089211. doi:10.1073/pnas.2117089119
Zhang, C. S., Zhang, B., Li, M., Wei, X., Gong, K., Li, Z., et al. (2022). Identification of serum metabolites enhancing inflammatory responses in COVID-19. Sci. China Life Sci. 65, 1971–1984. doi:10.1007/s11427-021-2099-7
Zhang, S., Luo, P., Xu, J., Yang, L., Ma, P., Tan, X., et al. (2021). Plasma metabolomic profiles in recovered Covid-19 patients without previous underlying diseases 3 months after discharge. J. Inflamm. Res. 14, 4485–4501. doi:10.2147/JIR.S325853
Zhang, X. D. (2015). Precision medicine, personalized medicine, omics and big data: Concepts and relationships. J. Pharmacogenomics Pharmacoproteomics 06, 1–2. doi:10.4172/2153-0645.1000e144
Keywords: COVID-19, SARS-CoV-2 infection, metabolomics, lipidomics, NMR, LC/GC-MS, phenoreversion, long covid
Citation: Bruzzone C, Conde R, Embade N, Mato JM and Millet O (2023) Metabolomics as a powerful tool for diagnostic, pronostic and drug intervention analysis in COVID-19. Front. Mol. Biosci. 10:1111482. doi: 10.3389/fmolb.2023.1111482
Received: 29 November 2022; Accepted: 06 February 2023;
Published: 15 February 2023.
Edited by:
Nguyen Phuoc Long, Inje University College of Medicine, Republic of KoreaReviewed by:
Aihua Zhang, Heilongjiang University of Chinese Medicine, ChinaCopyright © 2023 Bruzzone, Conde, Embade, Mato and Millet. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Oscar Millet, b21pbGxldEBjaWNiaW9ndW5lLmVz
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.