- 1Institute of Translational Medicine, The Affiliated Hospital of Qingdao University, Qingdao University, Qingdao, China
- 2School of Basic Medicine, Qingdao University, Qingdao, China
- 3National and Kapodistrian University of Athens, Athens, Greece
- 4Basic Medical Department, Graduate School, Chinese PLA General Hospital, Beijing, China
Gastric cancer (GC) is a malignant cancer that reduces life expectancy worldwide. Although treatment strategies have improved, patients with GC still have poor prognoses. Hence, it is necessary to understand the molecular mechanisms of GC and to find new therapeutic targets. Mitochondrial dynamics and mitochondrial dysfunction are associated with cancer cell growth and progression. Numerous studies have reported that non-coding RNAs (ncRNAs) can participate in the occurrence and development of GC by regulating mitochondrial dynamics. Elucidating the crosstalk between ncRNAs and mitochondria would be helpful in preventing and treating GC. Herein, we review and summarize the functions of oncogenes and tumor suppressors in suppressing ncRNAs and regulating mitochondrial dynamics in GC tumor growth, proliferation, invasion and metastasis. This review provides new insights into the pathogenesis of and intervention for GC.
1 Introduction
Gastric cancer (GC) is a malignant cancer with the fifth highest incidence and fourth highest mortality in the world (Sung et al., 2021). The pathogenesis of GC is related to several factors, including dietary, infectious, genetic, and epigenetic alterations. Chronic Helicobacter pylori infection is the leading cause of non-cardia GC (Plummer et al., 2015), but most patients with early GC have no obvious symptoms; diagnosed patients are usually in the intermediate and advanced stages of GC, and have a 5-year survival rate below 30% (Wei et al., 2022). The proliferation and metastasis of tumor cells require large amounts of material and energy to support tumorigenesis. Mitochondria serve as metabolic centers, generating energy through oxidative phosphorylation in eukaryotic cells (Nunnari and Suomalainen, 2012). Mitochondrial dynamics refer to how mitochondria change their shape, distribution, size, and function through the dynamic balance between continuous fission and fusion to meet the energy and product requirements of host cells and to respond to environmental changes. Mitochondrial fusion allows gene products to be transferred between mitochondria for optimal functioning. In contrast, fission is crucial for mitochondrial division and quality control. In addition, mitochondrial dynamics are linked to cellular activity, such as cell cycle, apoptosis, and Ca2+ signaling (Giacomello et al., 2020; Peng et al., 2020; Schell et al., 2020), and studies have demonstrated that mitochondrial dysfunction is related to the occurrence and development of GC (Ng et al., 2021; Poole and Macleod, 2021). Therefore, reviewing and understanding the dynamic regulation of and functional changes in mitochondria is important for the clinical treatment of GC.
RNAs are regarded as transmitters of genetic information and with the development of molecular technology, the understanding of RNAs has improved (Dutta et al., 2021) About 80% of the human genome is transcribed into RNAs, but non-coding RNAs (ncRNAs) account for the majority (Zhang et al., 2019; Zhou et al., 2022). According to length of RNAs, regulatory ncRNAs can be further divided into two categories: small ncRNAs with lengths less than 200 nucleotides, including microRNAs (miRNAs) and PIWI-interacting RNAs (piRNAs), and long non-coding RNAs (lncRNAs) with lengths over 200 nucleotides. In addition, another class of ncRNA with a covalently closed circular structure [circular RNAs (circRNAs)] have emerged as critical regulator of gene expression. Even the ncRNAs are not involved in coding proteins, play important roles in many cellular activities. Many studies have shown that ncRNAs are expressed abnormally in many diseases, such as heart disease and cancer (Zhou et al., 2018; Gowda et al., 2022; Ren et al., 2022). NcRNAs can regulate the expression of other endogenous competing RNAs, regulating transcription gene translation and protein localization.
Some research has shown that ncRNAs contribute to the synchronization of a series of important cellular and mitochondrial biological processes (Song et al., 2019; Zhang et al., 2021). Some ncRNAs involved in the signal pathways of GC have numerous crosslinks with those involved in mitochondria. The targeted regulation of ncRNAs in genes is closely related to mitochondrial function. NcRNAs play a vital biological role by directly influencing mitochondrial dynamics in cardiovascular diseases (Aung et al., 2021). Therefore, it is important to understand the crosstalk between ncRNAs and mitochondrial functions for GC prevention and treatment. In this study, we reviewed the role of ncRNAs in regulating mitochondrial dynamics in the pathological process of GC. This work may provide new insights for further investigation into the invasion and proliferation mechanisms of GC and provide some promising molecularly targeted therapies.
2 NcRNA biogenesis and functions
NcRNAs are synthesized in the nuclei and play a biological role in the cytoplasm. Because they cannot encode proteins, ncRNAs have long been regarded as “transcriptional waste products” of transcriptase II. Studies discovered that these “waste products” participate in nearly all life processes, such as cell growth and death (Slack and Chinnaiyan, 2019; Zhao X. et al., 2020; Sati and Parhar, 2021) (Figure 1).
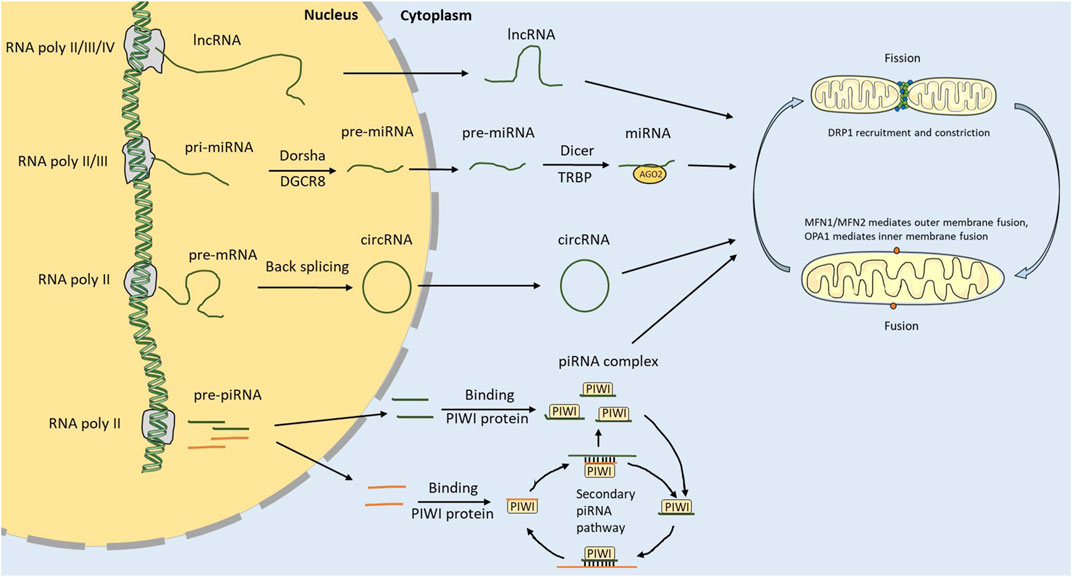
FIGURE 1. ncRNA biosynthesis and regulation of mitochondrial dynamics. ncRNA is transcribed by RNA polymerase in the nucleus. lncRNAs and circRNAs can be transported out of the nucleus through nuclear pores. pri-miRNAs are cleaved into pre-miRNAs by DGCR8 and Dorsha in the nucleus, then transported out of the nucleus and cleaved into miRNAs by Dicer and TRBP. The strand miRNAs attach to AGO2 to form the RISC. The piRNAs are transcribed from the piRNA cluster. Mature piRNAs can be synthesized through the primary and secondary piRNA pathways. ncRNA can regulate the mitochondrial dynamics by regulating core genes related to fusion and fission.
miRNAs of 22 nucleotides in length are the earliest most widely studied ncRNAs (Shukla et al., 2011). Mature miRNAs covalently bind mRNAs to induce degradation or inhibit their expression by forming silencing complexes with specific protein compositions (Ha and Kim, 2014). miRNAs are synthesized through RNA polymerase II/III, transcribing genes containing miRNA sites to form primary miRNAs with two stem-loops (Mousavi et al., 2022). The DiGeorge syndrome critical region 8 (DGCR8) and Drosha subsequently cut the primary miRNAs, resulting in a single stem-loop structural precursor with approximately 70 nucleotides. The precursor miRNAs cross the nuclear envelopes into the cytoplasm through Ran-GTP-Exportin-5 transporters. Precursor miRNAs are recognized and processed by the transactivation response element RNA-binding protein (TRBP) and dicer in the cytoplasm. The modified precursor miRNAs separate into form two strands: one strand that rapidly degrades, and another that is bound by argonaute 2 (AGO2) protein to form RNA-induced silencing complex (RISC), which can bind to the 3′UTR of mRNAs and affect their expression.
piRNAs are 24–31 nucleotides long and mainly derive from transposons, coding regions and specific intergenic sites in the genome (Iwasaki et al., 2015). The function of piRNAs is to protect the genome from transposons, especially germ cells. Studies have shown that although their expression is low in somatic cells, they have additional functions therein (Ortogero et al., 2014; Ozata et al., 2019). They play a functional role in posttranscriptional gene silencing by forming functional complexes with PIWI proteins (Iwasaki et al., 2015). Generally, two piRNA-producing pathways are conserved in species, from sponges to higher mammals (Yamashiro and Siomi, 2018; Ozata et al., 2019; Chen S. et al., 2021). The primary processing pathway occurs in somatic cells and consists of single-stranded piRNA clusters, double-stranded piRNA clusters, gene-derived piRNAs, and transposon-derived generation pathways. Single-stranded piRNA precursors are processed into piRNA intermediates that bind to PIWI proteins and undergo nuclear or cytoplasmic silencing. The secondary processing pathway, also called the ping-pang pathway, is related to the posttranscriptional silencing functions of piRISC. In this pathway, the main piRISC proteins are AUB and AGO3. AUB binds to a piRNA to form a complex that recognizes and silences the mRNA. Secondary piRNAs are generated through transcript, cleavage, and then shorter cleaved product is loaded into the AGO3 protein, forming an AGO3-piRNA complex precursor. The mature AGO3-piRNA complex recognizes and cleaves the piRNA precursor transcript, and the cleaved product can initiate a new cycle to continuously produce substrates.
LncRNAs are remarkably similar to mRNAs (Kanduri, 2016; Quinn and Chang, 2016). Their transcriptional genomic sites are similar to mRNA transcriptional genomic sites in chromatin status, and they are usually capped, spliced, and polyadenylated at the 5’ ends. In most cases, except for untranslated open reading frames, there is no biochemical difference between lncRNAs and mRNAs, but there are also many differences. For instance, lncRNAs are shorter in length than mRNAs with fewer exons. Moreover, lncRNAs have relatively low expression levels and poor first-order sequence conservation. Several studies have revealed that lncRNAs can function as competing endogenous RNAs to bind with miRNAs and circRNAs (Guo C. J. et al., 2020; Statello et al., 2021). LncRNA can also bind to mRNAs and specific protein sites. In addition, lncRNAs can function as precursors for some small molecules that target RNA.
circRNAs were first discovered in plants and suspected to be a pathogenic viroid (Sanger et al., 1976). Later studies found that circular RNA molecules were also distributed in prokaryotes, eukaryotes, and mammals (Capel et al., 1993; Ford and Ares, 1994). With the help of high-throughput sequencing and bioanalysis techniques, cirRNAs have gradually been identified and found to play an important role in biological processes. circRNAs are not easily degraded by exonuclease due to their ring structure, which makes them more stable than linear RNAs (Zhu et al., 2022). A large number of circRNAs exist in the cytoplasm of eukaryotic cells, mainly derived from exons, and small numbers of intron-derived circRNAs exist in the nuclei. circRNAs are generated from the back-splicing of precursor mRNAs and are divided into three types according to the positions of their transcription loops: intronic RNAs, exonic circRNAs, and exon-intron circRNAs. Exonic circRNAs account for the majority (Tang et al., 2021). circRNA rings are formed in two main ways. In the first, lariat RNA is generated by the intron during splicing, which is formed by a 2′,5′-phosphodiester bond between the 5′splice site and the branch site of the intron, and a loop is formed by the exon of the gene when the downstream 5′splice site forms a 3′,5′phosphodiester bond with the upstream 3′splice site. The second type of ring formation mainly depends on the complementary pairing of repeated sequences of introns or the interactions between specific RNA-binding proteins. circRNAs have many functions, including binding to related binding proteins as transcriptional regulators, acting as miRNA sponges, and generating pseudogenes (Yang et al., 2021; Liu and Chen, 2022). Most circRNAs are non-coding, but a few contain internal ribosome entry site sequences (IRESs) that enable them to translate (Legnini et al., 2017; Biswas et al., 2021).
3 Altered mitochondrial dynamics associated with tumorigenesis
The earliest research found that cancer absorbs and ferments glucose to form lactic acid under aerobic conditions, so mitochondrial respiratory defects are the basis of cancer (Warburg, 1956). However, later studies found that not all cancers have this glycolytic feature; most still retain mitochondrial function (Vasan et al., 2020). Some cancer cells are robustly involved in the glycolysis and TCA cycle metabolism processes in mitochondria (DeBerardinis and Chandel, 2016). The increased rate of the metabolic process can activate various oncogenic controllers related to many signaling pathways. Tumor cells lost their mitochondrial ubiquinone regeneration ability, and the mitochondrial complex III subunit is a crucial part of tumor cell proliferation (Wang et al., 2020). Therefore, functional electron transport chain is required to oxidize ubiquinol for tumor growth (Meyers et al., 2017). Metabolites produced by the TCA cycle synthesize nucleotides, lipids, amino acids, and heme. In addition, the cycle produces oncogenic metabolites in some tumor contexts (Birsoy et al., 2015). Although some tumors rely only on glycolysis to meet their biological needs, they still need products from the mitochondrial oxidation process to promote cell proliferation (Sullivan et al., 2015). Synthetic requirements and the production of oncogenic metabolites promote tumorigenesis (Cluntun et al., 2017). Moreover, mitochondria are the signal transmission centers in cells, participating in cell apoptosis, autophagy, calcium transport, and other processes (Averbeck and Rodriguez-Lafrasse, 2021). Mitochondrial dynamics help maintain the mitochondrial pool and optimal mitochondrial oxidative phosphorylation activity in cells, allowing efficient transport and distribution of mitochondrial content.
3.1 Mitochondrial fission
The maintenance of normal mitochondrial function depends on a balance between mitochondrial fission and fusion, but this dynamism is dysregulated in most cancers. Mitochondrial fission is the division of a mitochondrion into two mitochondria. This process is important for maintaining mitochondrial numbers, distribution, and cell apoptosis activation (Tait and Green, 2010). Mitochondrial fission is related to the activation of dynamin-related protein 1 (DRP1) and mitochondrial fission one protein (FIS1) (Westermann, 2010; Wai and Langer, 2016). DRP1 is an evolutionarily conserved GTPase in approximately 97% of cytoplasm, and it lyses mitochondria by localizing the adaptor protein FIS1 in the mitochondrial membrane. Other proteins, such as dynamin 2, human mitochondrial dynamics protein 49 (MID49), and MID51 are also related to the fission process (Chan, 2012). Abnormal DRP1 expression has been identified in various human cancers, and its upregulation promotes the growth and metastasis of pancreatic cancer cells through increased mitochondrial fission and aerobic glycolysis (Liang et al., 2020). High glucose levels can increase the expression of DRP1, resulting in cell dysfunction and promoting the migration and invasion of endometrial cancer cells (Guo J. et al., 2020). The high expression of DRP1 was correlated with poor survival of head and neck cancer patients (Huang T. L. et al., 2022). DRP1 deletion can affect aerobic glycolysis and suppress tumor growth and metastasis (Huang T. L. et al., 2022). FIS1 is overexpressed in GC, and its expression is correlated with cancer metastasis (Karimi et al., 2022). Fan et al. (Fan et al., 2015) found that FIS1 knockdown could reduce mitochondrial fission and cisplatin sensitivity.
3.2 Mitochondrial fusion
Mitochondrial fusion is the process by which two mitochondria join to become one. Previous studies have found that different regulatory molecules are required for the fusion of the inner and outer membranes of mitochondria (Mattie et al., 2018; Adebayo et al., 2021). The major proteins regulating outer membrane fusion are mitofusin 1 (MFN1) and mitofusin 2 (MFN2) (Hales and Fuller, 1997). Optic atrophy 1 (OPA1) is located in and participates in inner mitochondrial membrane fusion. Mitochondrial fusion is necessary for exchanging genetic material and maintaining normal function. In addition, the fusion process can be triggered by some treatments, such as autophagy and starvation (Gomes et al., 2011; Rambold et al., 2011). Mitochondrial fusion is overactivated in liver cancer and cholangiocarcinoma. OPA1 inhibition can effectively reduce the proliferation and migration of breast cancer cells (Zamberlan et al., 2022). MFN2 has previously been reported to regulate cell proliferation, apoptosis and differentiation (Papanicolaou et al., 2012; Gong et al., 2015). Xu et al. (Xu et al., 2017) found that MFN2 knockout promoted cell viability, colony formation, and invasion of breast cancer cells. GC patients with high levels of MFN2 have a worse overall survival rate, and these high levels could be prognostic markers for GC (Fang et al., 2017).
4 ncRNAs control mitochondrial dynamics in GC
Mitochondria are important organelles in cells and hubs of many important biological processes. As intracellular gene regulators, ncRNAs can regulate mitochondrial functions and participate in GC through the direct or indirect targeting of the mitochondria-related genes or pathways (Figure 2; Table 1).
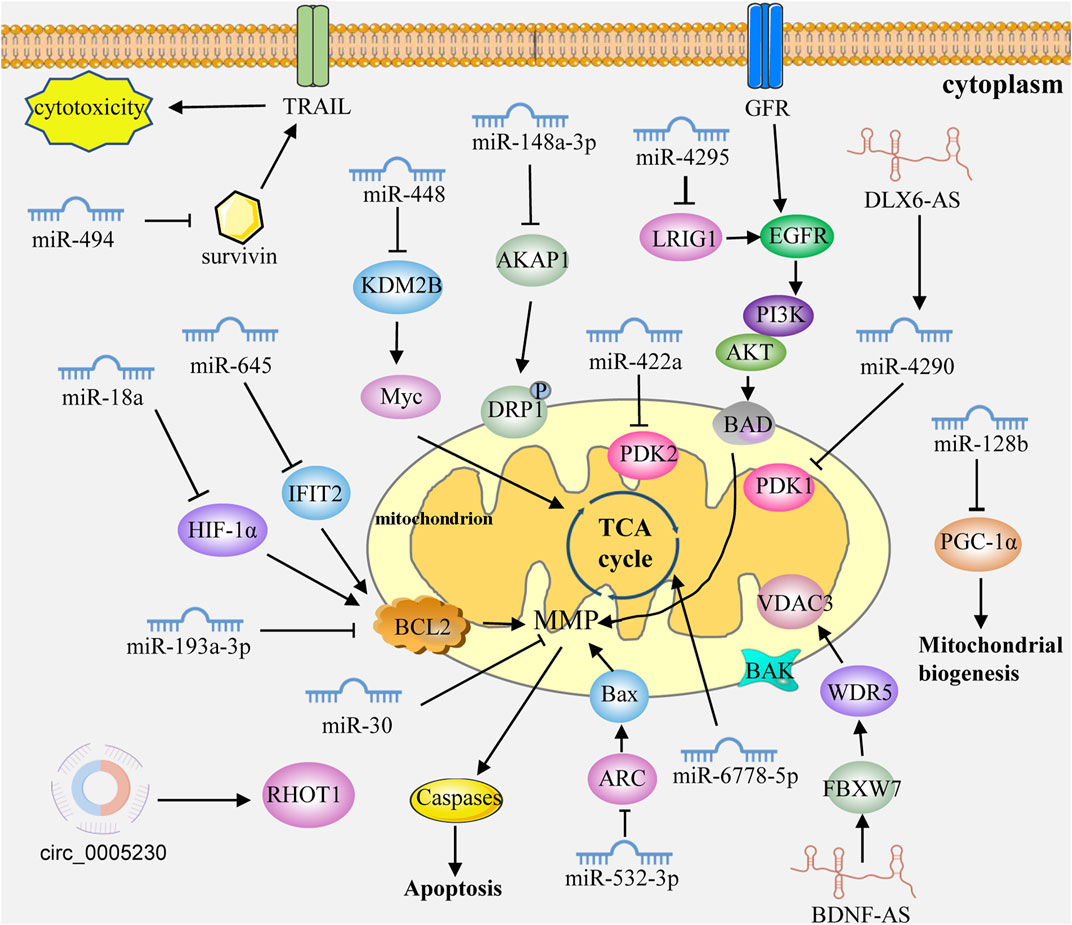
FIGURE 2. ncRNAs control mitochondrial dynamics in GC cells. ncRNAs can regulate mitochondrial functions and participate in GC through the direct or indirect targeting of the mitochondria-related genes or pathways. Caspases: caspase family protein (caspase3, 9 and 7). MMP: mitochondrial membrane permeabilization. GFR: growth factor receptor.
4.1 miRNA control of GC through mitochondrial pathways
miRNA are the most studied ncRNAs and play oncogenic and tumor-suppressive roles in GC progression. miR-645 is upregulated in GC tissues and cell lines. Interferon-induced protein with tetratricopeptide repeats 2 (IFIT2) was shown to be a mitochondrial apoptosis protein predicted and verified as the target of miR-645 (Feng et al., 2014). Knockdown of miR-645 increases drug sensitivity and promotes the apoptosis of cancer cells by regulating IFIT2 expression (Feng et al., 2014). The overexpression of miR-18 promotes apoptosis and inhibits proliferation and invasion by targeting hypoxia-inducible factor-1α (HIF-1α) (Wu et al., 2015). The overexpression of miR-18 in tumor cells also activates the expression of mitochondria-mediated apoptosis genes, such as Bax, caspase 3 and caspase 9 (Wu et al., 2015). Overexpression of miR-448 can promote cell proliferation and increase the glycolytic response by inhibiting lysine (K)-specific demethylase 2B (KDM2B) expression. Moreover, Myc proved to be a target of KDM28, which controls the metabolism switch, and miR-488/KDM2B/Myc was a key axis controlling the occurrence and development of GC (Hong et al., 2016). Li et al. (2017) found that miR-148a-3p enhanced the resistance of GC cells to cisplatin by promoting mitochondrial fission and reducing the expression levels of A-Kinase anchoring protein 1 (AKAP1) and DRP1 dephosphorylation. miR-30 was significantly overexpressed in GC tissues and cell lines, and miR-30 inhibition decreased mitochondrial oxygen consumption and activated mitochondria-mediated apoptosis (Wang et al., 2017).
He et al., 2018 found that miR-422a was significantly downregulated in GC, and overexpression of miR-422a inhibited the expression of pyruvate dehydrogenase kinase 2 and affected the cell cycle, ultimately inhibiting cancer cell proliferation. Overexpression of miR-494 sensitized GC cells to TNF-related apoptosis-inducing ligand (TRAIL)-induced cytotoxicity. Overexpression of miR-494 promoted TRAIL-induced mitochondrial collapse and apoptosis pathways by targeting survivin (Xu et al., 2018). Yan et al., 2018 found that miR-4295 could target leucine-rich repeats and immunoglobulin-like domain 1 to activate the EGFR/PI3K/Akt signaling pathway and promote GC cell proliferation. Lee et al., 2019 found that miR-193a-3p could regulate the resistance of GC through the mitochondrial apoptosis pathway, and the peroxisome proliferator-activated receptor γ coactivator 1α (PGC-1α) mediated mitochondrial biogenesis. miR-128 could affect the expression of PGC-1α, and the knockdown of PGC-1α inhibited cancer cell metabolic activity and increased apoptosis (Wang et al., 2019). miR-6778-5p could regulate its host gene SHMT1 by targeting YWHAE to affect the mitochondrial carbon metabolic pathway (Zhao M. et al., 2020). miR-532-3p can inhibit GC cell proliferation in vitro and in vitro by activating the ARC/Bax/mitochondria-mediated apoptosis pathway (Chen Z. et al., 2021).
4.2 Other ncRNAs regulate GC through mitochondrial pathways
Studies have shown that lncRNAs are aberrantly expressed in GC tissues, suggesting that lncRNAs could serve as the target candidates for tumor diagnosis and therapy. LncRNA DLX6-AS1 is overexpressed in GC and DLX6-AS1 knockdown inhibits tumor growth and aerobic glycolysis by targeting miR-4290 and 3-phosphoinositide-dependent protein kinase 1 (PDK1) (Qian et al., 2021). Huang G. et al. (2022) found that lncRNA BDNF-AS aggregates WDR5 to mediate the expression of FBXW7, resulting in changes in the transcription of FBXW7, which in turn regulates the expression level of voltage-dependent anion channel 3 in the mitochondrial outer membrane through ubiquitination. The abnormal expression of circRNAs was found to be related to tumorigenesis and the prognosis of GC. Peng et al., 2022 found that circ_0005230 was upregulated in GC tissues, and circ_0005230 knockdown could inhibit GC cell proliferation and mitophagy by regulating Ras homolog family member T1 (RHOT1) expression via sponging miR-1299.
5 Conclusion
GC remains a serious public health problem associated with high mortality and poor prognosis. Gastroscopy is the usual method of screening for and diagnosing of GC, but it is unsuitable for all cases (Xie et al., 2020). Traditional screening biomarkers for diagnosing GC are often inadequate due to a lack of specificity. It is urgently necessary to find a diagnostic method for GC. Accumulating evidence has demonstrated that ncRNAs participate in tumorigenesis by regulating invasion, proliferation, and migration (Fonseca Cabral et al., 2020; Tan et al., 2020; Lu et al., 2021; Yang et al., 2022). As mentioned previously, ncRNAs are abnormally expressed in GC and their expression is significantly related to patients’ prognoses. Therefore, miRNAs, lncRNAs, circRNAs, and piRNAs may be biomarkers for the diagnosis and/or prognosis of GC. Compared with protein coding biomarkers, ncRNAs have certain advantages as biomarkers for the diagnosis and/or prognosis of GC. First, compared with protein coding mRNAs, ncRNAs are tissue specific. Second, ncRNAs are more stable in various clinical specimens (such as serum, plasma, urine, and gastric juice), which may allow non-invasive GC detection.
ncRNAs can be used not only as potential biological diagnostic indicators but also as therapeutic targets or combined with other therapies to improve their anti-tumor effects. Some studies have shown that RNA-based therapies have good clinical potential because RNA can directly target specific genes and their products (Lin et al., 2020; Wu et al., 2022). At present, some RNA-based therapies approved worldwide target specific genes in liver or muscle cells and change gene expression by injecting siRNA or oligonucleotide chains (Shaw et al., 2022). Since the important role of ncRNAs in GC progression has been fully demonstrated, they are becoming a new class of targets for RNA-based drug discovery. As a therapeutic intervention, ncRNAs have some advantages over mRNAs because they are more diverse. The pleiotropic nature of ncRNAs makes them particularly attractive drug targets for multifactorial diseases. However, there is no ncRNA-based drug for GC. In addition, ncRNA-based therapy poses many problems, including the stability and toxicity of drugs and the need to avoid immune reactions caused by foreign nucleic acids. The means of administration and the efficiency of drugs after entering the body should also be thoroughly considered.
Mitochondrial dynamics are closely related to the activity of neuronal cells, but some studies have shown that phenotypic changes in cancer cells are also affected by mitochondrial dynamic defects (Sehrawat et al., 2016; Shen and Zhan, 2022). The substances required for tumor metabolism include amino acids and are closely related to mitochondrial metabolism. As mediators of energy metabolism and the signaling pathway, dynamic changes in mitochondria can regulate the signaling pathway. Moreover, changes in the mitochondrial respiratory chain can affect electron transport, energy supply, redox state, metabolism, and apoptosis (Xia et al., 2022). At present, there have been studies targeting mitochondria for treatment of cancer. For example, Lin et al., 2013 found that SOCS6 could promote cell apoptosis and increase the accumulation of Bax in mitochondria; it could also control mitochondrial fission by targeting DRP1. Chuang et al., 2020 reported that imiquimod could significantly increase the apoptosis of skin cancer cells by regulating mitochondrial dynamic. However, the FDA has approved few drugs for mitochondrial therapy. The specific molecular mechanisms of mitochondrial therapy still need to be identified. As an independent organelle, the role of mitochondrial dynamics in cancer development is complex; hence, modes of administration that target mitochondria remain to be studied.
In this review, we focused on the functions of ncRNAs in tumorigenesis and tumor suppression via the regulation of mitochondria. Although many ncRNAs have been identified, this is only the beginning. The sensitivity and specificity of ncRNA as diagnostic indexes need to be improved. As a therapeutic target, the drug metabolism, pharmacodynamics, biological distribution, and cellular uptake mechanisms of ncRNAs require further research. This study revealed crosstalk between ncRNAs and mitochondria in the progression of GC. Understanding these networks will provide a theoretical basis for discovering new targets for diagnosing and treating GC and will provide a new approach to clinical GC research.
Author contributions
PL, YL, and XC contributed to conception and design of the study. KS supervised the manuscript. XC, CW and LH wrote sections of the manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
Funding
This work was supported by the Major Research Program of the National Natural Science Foundation of China, grant number 91849209.
Acknowledgments
The graphical abstract was modified from Servier Medical Art (https://smart.servier.com/), licensed under a Creative Common Attribution 3.0 Generic License (https://creativecommons.org/licenses/by/3.0/).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Adebayo, M., Singh, S., Singh, A. P., and Dasgupta, S. (2021). Mitochondrial fusion and fission: The fine-tune balance for cellular homeostasis. Faseb J. 35 (6), e21620. doi:10.1096/fj.202100067R
Aung, L. H. H., Chen, X., Cueva Jumbo, J. C., Li, Z., Wang, S. Y., Zhao, C., et al. (2021). Cardiomyocyte mitochondrial dynamic-related lncRNA 1 (CMDL-1) may serve as a potential therapeutic target in doxorubicin cardiotoxicity. Mol. Ther. Nucleic Acids 25, 638–651. doi:10.1016/j.omtn.2021.08.006
Averbeck, D., and Rodriguez-Lafrasse, C. (2021). Role of mitochondria in radiation responses: Epigenetic, metabolic, and signaling impacts. Int. J. Mol. Sci. 22 (20), 11047. doi:10.3390/ijms222011047
Birsoy, K., Wang, T., Chen, W. W., Freinkman, E., Abu-Remaileh, M., and Sabatini, D. M. (2015). An essential role of the mitochondrial electron transport chain in cell proliferation is to enable aspartate synthesis. Cell 162 (3), 540–551. doi:10.1016/j.cell.2015.07.016
Biswas, A., Chowdhury, N., and Bagchi, A. (2021). Structural characterization of the hidden peptide SHPRH-146aa encoded by non-coding circ-SHPRH to act as tumor suppressor. Appl. Biochem. Biotechnol. 193 (7), 2076–2086. doi:10.1007/s12010-021-03520-0
Capel, B., Swain, A., Nicolis, S., Hacker, A., Walter, M., Koopman, P., et al. (1993). Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell 73 (5), 1019–1030. doi:10.1016/0092-8674(93)90279-y
Chan, D. C. (2012). Fusion and fission: Interlinked processes critical for mitochondrial health. Annu. Rev. Genet. 46, 265–287. doi:10.1146/annurev-genet-110410-132529
Chen, S., Ben, S., Xin, J., Li, S., Zheng, R., Wang, H., et al. (2021). The biogenesis and biological function of PIWI-interacting RNA in cancer. J. Hematol. Oncol. 14 (1), 93. doi:10.1186/s13045-021-01104-3
Chen, Z., Liang, Y., Feng, X., Liang, Y., Shen, G., Huang, H., et al. (2021). Vitamin-B12-conjugated PLGA-PEG nanoparticles incorporating miR-532-3p induce mitochondrial damage by targeting apoptosis repressor with caspase recruitment domain (ARC) on CD320-overexpressed gastric cancer. Mater Sci. Eng. C Mater Biol. Appl. 120, 111722. doi:10.1016/j.msec.2020.111722
Chuang, K. C., Chang, C. R., Chang, S. H., Huang, S. W., Chuang, S. M., Li, Z. Y., et al. (2020). Imiquimod-induced ROS production disrupts the balance of mitochondrial dynamics and increases mitophagy in skin cancer cells. J. Dermatol Sci. 98 (3), 152–162. doi:10.1016/j.jdermsci.2020.03.009
Cluntun, A. A., Lukey, M. J., Cerione, R. A., and Locasale, J. W. (2017). Glutamine metabolism in cancer: Understanding the heterogeneity. Trends Cancer 3 (3), 169–180. doi:10.1016/j.trecan.2017.01.005
DeBerardinis, R. J., and Chandel, N. S. (2016). Fundamentals of cancer metabolism. Sci. Adv. 2 (5), e1600200. doi:10.1126/sciadv.1600200
Dutta, K., Das, R., Medeiros, J., Kanjilal, P., and Thayumanavan, S. (2021). Charge-conversion strategies for nucleic acid delivery. Adv. Funct. Mater 31 (24), 2011103. doi:10.1002/adfm.202011103
Fan, S., Chen, W. X., Lv, X. B., Tang, Q. L., Sun, L. J., Liu, B. D., et al. (2015). miR-483-5p determines mitochondrial fission and cisplatin sensitivity in tongue squamous cell carcinoma by targeting FIS1. Cancer Lett. 362 (2), 183–191. doi:10.1016/j.canlet.2015.03.045
Fang, C. L., Sun, D. P., Chen, H. K., Lin, C. C., Hung, S. T., Uen, Y. H., et al. (2017). Overexpression of mitochondrial GTPase MFN2 represents a negative prognostic marker in human gastric cancer and its inhibition exerts anti-cancer effects. J. Cancer 8 (7), 1153–1161. doi:10.7150/jca.17986
Feng, X., Wang, Y., Ma, Z., Yang, R., Liang, S., Zhang, M., et al. (2014). MicroRNA-645, up-regulated in human adencarcinoma of gastric esophageal junction, inhibits apoptosis by targeting tumor suppressor IFIT2. BMC Cancer 14, 633. doi:10.1186/1471-2407-14-633
Fonseca Cabral, G., Azevedo Dos Santos Pinheiro, J., Vidal, A. F., Santos, S., and Ribeiro-Dos-Santos, Â. (2020). piRNAs in gastric cancer: A new approach towards translational research. Int. J. Mol. Sci. 21 (6), 2126. doi:10.3390/ijms21062126
Ford, E., and Ares, M. (1994). Synthesis of circular RNA in bacteria and yeast using RNA cyclase ribozymes derived from a group I intron of phage T4. Proc. Natl. Acad. Sci. U. S. A. 91 (8), 3117–3121. doi:10.1073/pnas.91.8.3117
Giacomello, M., Pyakurel, A., Glytsou, C., and Scorrano, L. (2020). The cell biology of mitochondrial membrane dynamics. Nat. Rev. Mol. Cell Biol. 21 (4), 204–224. doi:10.1038/s41580-020-0210-7
Gomes, L. C., Di Benedetto, G., and Scorrano, L. (2011). During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nat. Cell Biol. 13 (5), 589–598. doi:10.1038/ncb2220
Gong, G., Song, M., Csordas, G., Kelly, D. P., Matkovich, S. J., Dorn, G. W., et al. (2015). Parkin-mediated mitophagy directs perinatal cardiac metabolic maturation in mice. Science 350 (6265), aad2459. doi:10.1126/science.aad2459
Gowda, P., Reddy, P. H., and Kumar, S. (2022). Deregulated mitochondrial microRNAs in Alzheimer's disease: Focus on synapse and mitochondria. Ageing Res. Rev. 73, 101529. doi:10.1016/j.arr.2021.101529
Guo, C. J., Xu, G., and Chen, L. L. (2020). Mechanisms of long noncoding RNA nuclear retention. Trends Biochem. Sci. 45 (11), 947–960. doi:10.1016/j.tibs.2020.07.001
Guo, J., Ye, F., Jiang, X., Guo, H., Xie, W., Zhang, Y., et al. (2020). Drp1 mediates high glucose-induced mitochondrial dysfunction and epithelial-mesenchymal transition in endometrial cancer cells. Exp. Cell Res. 389 (1), 111880. doi:10.1016/j.yexcr.2020.111880
Ha, M., and Kim, V. N. (2014). Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 15 (8), 509–524. doi:10.1038/nrm3838
Hales, K. G., and Fuller, M. T. (1997). Developmentally regulated mitochondrial fusion mediated by a conserved, novel, predicted GTPase. Cell 90 (1), 121–129. doi:10.1016/s0092-8674(00)80319-0
He, Z., Li, Z., Zhang, X., Yin, K., Wang, W., Xu, Z., et al. (2018). MiR-422a regulates cellular metabolism and malignancy by targeting pyruvate dehydrogenase kinase 2 in gastric cancer. Cell Death Dis. 9 (5), 505. doi:10.1038/s41419-018-0564-3
Hong, X., Xu, Y., Qiu, X., Zhu, Y., Feng, X., Ding, Z., et al. (2016). MiR-448 promotes glycolytic metabolism of gastric cancer by downregulating KDM2B. Oncotarget 7 (16), 22092–22102. doi:10.18632/oncotarget.8020
Huang, G., Xiang, Z., Wu, H., He, Q., Dou, R., Lin, Z., et al. (2022). The lncRNA BDNF-AS/WDR5/FBXW7 axis mediates ferroptosis in gastric cancer peritoneal metastasis by regulating VDAC3 ubiquitination. Int. J. Biol. Sci. 18 (4), 1415–1433. doi:10.7150/ijbs.69454
Huang, T. L., Chang, C. R., Chien, C. Y., Huang, G. K., Chen, Y. F., Su, L. J., et al. (2022). DRP1 contributes to head and neck cancer progression and induces glycolysis through modulated FOXM1/MMP12 axis. Mol. Oncol. 16 (13), 2585–2606. doi:10.1002/1878-0261.13212
Iwasaki, Y. W., Siomi, M. C., and Siomi, H. (2015). PIWI-interacting RNA: Its biogenesis and functions. Annu. Rev. Biochem. 84, 405–433. doi:10.1146/annurev-biochem-060614-034258
Kanduri, C. (2016). Long noncoding RNAs: Lessons from genomic imprinting. Biochim. Biophys. Acta 1859 (1), 102–111. doi:10.1016/j.bbagrm.2015.05.006
Karimi, D., Pedram, N., Kakaei, F., Asadi, M., Poursaei, E., and Kermani, T. A. (2022). FIS1 overexpression is correlated with tumor metastasis in gastric adenocarcinoma. J. Gastrointest. Cancer 53 (2), 466–471. doi:10.1007/s12029-021-00639-5
Lee, S. D., Yu, D., Lee, D. Y., Shin, H. S., Jo, J. H., and Lee, Y. C. (2019). Upregulated microRNA-193a-3p is responsible for cisplatin resistance in CD44(+) gastric cancer cells. Cancer Sci. 110 (2), 662–673. doi:10.1111/cas.13894
Legnini, I., Di Timoteo, G., Rossi, F., Morlando, M., Briganti, F., Sthandier, O., et al. (2017). Circ-ZNF609 is a circular RNA that can Be translated and functions in myogenesis. Mol. Cell 66 (1), 22–37. doi:10.1016/j.molcel.2017.02.017
Li, B., Wang, W., Li, Z., Chen, Z., Zhi, X., Xu, J., et al. (2017). MicroRNA-148a-3p enhances cisplatin cytotoxicity in gastric cancer through mitochondrial fission induction and cyto-protective autophagy suppression. Cancer Lett. 410, 212–227. doi:10.1016/j.canlet.2017.09.035
Liang, J., Yang, Y., Bai, L., Li, F., and Li, E. (2020). DRP1 upregulation promotes pancreatic cancer growth and metastasis through increased aerobic glycolysis. J. Gastroenterol. Hepatol. 35 (5), 885–895. doi:10.1111/jgh.14912
Lin, H. Y., Lai, R. H., Lin, S. T., Lin, R. C., Wang, M. J., Lin, C. C., et al. (2013). Suppressor of cytokine signaling 6 (SOCS6) promotes mitochondrial fission via regulating DRP1 translocation. Cell Death Differ. 20 (1), 139–153. doi:10.1038/cdd.2012.106
Lin, Y. X., Wang, Y., Blake, S., Yu, M., Mei, L., Wang, H., et al. (2020). RNA nanotechnology-mediated cancer immunotherapy. Theranostics 10 (1), 281–299. doi:10.7150/thno.35568
Liu, C. X., and Chen, L. L. (2022). Circular RNAs: Characterization, cellular roles, and applications. Cell 185 (12), 2016–2034. doi:10.1016/j.cell.2022.04.021
Lu, Y., Li, K., Gao, Y., Liang, W., Wang, X., and Chen, L. (2021). CircRNAs in gastric cancer: Current research and potential clinical implications. FEBS Lett. 595 (21), 2644–2654. doi:10.1002/1873-3468.14196
Mattie, S., Riemer, J., Wideman, J. G., and McBride, H. M. (2018). A new mitofusin topology places the redox-regulated C terminus in the mitochondrial intermembrane space. J. Cell Biol. 217 (2), 507–515. doi:10.1083/jcb.201611194
Meyers, R. M., Bryan, J. G., McFarland, J. M., Weir, B. A., Sizemore, A. E., Xu, H., et al. (2017). Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nat. Genet. 49 (12), 1779–1784. doi:10.1038/ng.3984
Mousavi, S. M., Derakhshan, M., Baharloii, F., Dashti, F., Mirazimi, S. M. A., Mahjoubin-Tehran, M., et al. (2022). Non-coding RNAs and glioblastoma: Insight into their roles in metastasis. Mol. Ther. Oncolytics 24, 262–287. doi:10.1016/j.omto.2021.12.015
Ng, M. Y. W., Wai, T., and Simonsen, A. (2021). Quality control of the mitochondrion. Dev. Cell 56 (7), 881–905. doi:10.1016/j.devcel.2021.02.009
Nunnari, J., and Suomalainen, A. (2012). Mitochondria: In sickness and in health. Cell 148 (6), 1145–1159. doi:10.1016/j.cell.2012.02.035
Ortogero, N., Schuster, A. S., Oliver, D. K., Riordan, C. R., Hong, A. S., Hennig, G. W., et al. (2014). A novel class of somatic small RNAs similar to germ cell pachytene PIWI-interacting small RNAs. J. Biol. Chem. 289 (47), 32824–32834. doi:10.1074/jbc.M114.613232
Ozata, D. M., Gainetdinov, I., Zoch, A., O'Carroll, D., and Zamore, P. D. (2019). PIWI-Interacting RNAs: Small RNAs with big functions. Nat. Rev. Genet. 20 (2), 89–108. doi:10.1038/s41576-018-0073-3
Papanicolaou, K. N., Kikuchi, R., Ngoh, G. A., Coughlan, K. A., Dominguez, I., Stanley, W. C., et al. (2012). Mitofusins 1 and 2 are essential for postnatal metabolic remodeling in heart. Circ. Res. 111 (8), 1012–1026. doi:10.1161/circresaha.112.274142
Peng, W., Wong, Y. C., and Krainc, D. (2020). Mitochondria-lysosome contacts regulate mitochondrial Ca(2+) dynamics via lysosomal TRPML1. Proc. Natl. Acad. Sci. U. S. A. 117 (32), 19266–19275. doi:10.1073/pnas.2003236117
Peng, Y. Y., Sun, D., and Xin, Y. (2022). Hsa_circ_0005230 is up-regulated and promotes gastric cancer cell invasion and migration via regulating the miR-1299/RHOT1 axis. Bioengineered 13 (3), 5046–5063. doi:10.1080/21655979.2022.2036514
Plummer, M., Franceschi, S., Vignat, J., Forman, D., and de Martel, C. (2015). Global burden of gastric cancer attributable to Helicobacter pylori. Int. J. Cancer 136 (2), 487–490. doi:10.1002/ijc.28999
Poole, L. P., and Macleod, K. F. (2021). Mitophagy in tumorigenesis and metastasis. Cell Mol. Life Sci. 78 (8), 3817–3851. doi:10.1007/s00018-021-03774-1
Qian, Y., Song, W., Wu, X., Hou, G., Wang, H., Hang, X., et al. (2021). DLX6 antisense RNA 1 modulates glucose metabolism and cell growth in gastric cancer by targeting microRNA-4290. Dig. Dis. Sci. 66 (2), 460–473. doi:10.1007/s10620-020-06223-4
Quinn, J. J., and Chang, H. Y. (2016). Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 17 (1), 47–62. doi:10.1038/nrg.2015.10
Rambold, A. S., Kostelecky, B., Elia, N., and Lippincott-Schwartz, J. (2011). Tubular network formation protects mitochondria from autophagosomal degradation during nutrient starvation. Proc. Natl. Acad. Sci. U. S. A. 108 (25), 10190–10195. doi:10.1073/pnas.1107402108
Ren, B., Guan, M. X., Zhou, T., Cai, X., and Shan, G. (2022). Emerging functions of mitochondria-encoded noncoding RNAs. Trends Genet. doi:10.1016/j.tig.2022.08.004
Sanger, H. L., Klotz, G., Riesner, D., Gross, H. J., and Kleinschmidt, A. K. (1976). Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl. Acad. Sci. U. S. A. 73 (11), 3852–3856. doi:10.1073/pnas.73.11.3852
Sati, I., and Parhar, I. (2021). MicroRNAs regulate cell cycle and cell death pathways in glioblastoma. Int. J. Mol. Sci. 22 (24), 13550. doi:10.3390/ijms222413550
Schell, M., Chudoba, C., Leboucher, A., Alfine, E., Flore, T., Ritter, K., et al. (2020). Interplay of dietary fatty acids and cholesterol impacts brain mitochondria and insulin action. Nutrients 12 (5), 1518. doi:10.3390/nu12051518
Sehrawat, A., Croix, C. S., Baty, C. J., Watkins, S., Tailor, D., Singh, R. P., et al. (2016). Inhibition of mitochondrial fusion is an early and critical event in breast cancer cell apoptosis by dietary chemopreventative benzyl isothiocyanate. Mitochondrion 30, 67–77. doi:10.1016/j.mito.2016.06.006
Shaw, T. I., Zhao, B., Li, Y., Wang, H., Wang, L., Manley, B., et al. (2022). Multi-omics approach to identifying isoform variants as therapeutic targets in cancer patients. Front. Oncol. 12, 1051487. doi:10.3389/fonc.2022.1051487
Shen, L., and Zhan, X. (2022). Mitochondrial dysfunction pathway alterations offer potential biomarkers and therapeutic targets for ovarian cancer. Oxid. Med. Cell Longev. 2022, 5634724. doi:10.1155/2022/5634724
Shukla, G. C., Singh, J., and Barik, S. (2011). MicroRNAs: Processing, maturation, target recognition and regulatory functions. Mol. Cell Pharmacol. 3 (3), 83–92.
Slack, F. J., and Chinnaiyan, A. M. (2019). The role of non-coding RNAs in oncology. Cell 179 (5), 1033–1055. doi:10.1016/j.cell.2019.10.017
Song, R., Hu, X. Q., and Zhang, L. (2019). Mitochondrial MiRNA in cardiovascular function and disease. Cells 8 (12), 1475. doi:10.3390/cells8121475
Statello, L., Guo, C. J., Chen, L. L., and Huarte, M. (2021). Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 22 (2), 96–118. doi:10.1038/s41580-020-00315-9
Sullivan, L. B., Gui, D. Y., Hosios, A. M., Bush, L. N., Freinkman, E., and Vander Heiden, M. G. (2015). Supporting aspartate biosynthesis is an essential function of respiration in proliferating cells. Cell 162 (3), 552–563. doi:10.1016/j.cell.2015.07.017
Sung, H., Ferlay, J., Siegel, R. L., Laversanne, M., Soerjomataram, I., Jemal, A., et al. (2021). Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71 (3), 209–249. doi:10.3322/caac.21660
Tait, S. W., and Green, D. R. (2010). Mitochondria and cell death: Outer membrane permeabilization and beyond. Nat. Rev. Mol. Cell Biol. 11 (9), 621–632. doi:10.1038/nrm2952
Tan, H., Zhang, S., Zhang, J., Zhu, L., Chen, Y., Yang, H., et al. (2020). Long non-coding RNAs in gastric cancer: New emerging biological functions and therapeutic implications. Theranostics 10 (19), 8880–8902. doi:10.7150/thno.47548
Tang, X., Ren, H., Guo, M., Qian, J., Yang, Y., and Gu, C. (2021). Review on circular RNAs and new insights into their roles in cancer. Comput. Struct. Biotechnol. J. 19, 910–928. doi:10.1016/j.csbj.2021.01.018
Vasan, K., Werner, M., and Chandel, N. S. (2020). Mitochondrial metabolism as a target for cancer therapy. Cell Metab. 32 (3), 341–352. doi:10.1016/j.cmet.2020.06.019
Wai, T., and Langer, T. (2016). Mitochondrial dynamics and metabolic regulation. Trends Endocrinol. Metab. 27 (2), 105–117. doi:10.1016/j.tem.2015.12.001
Wang, J., Jiao, Y., Cui, L., and Jiang, L. (2017). miR-30 functions as an oncomiR in gastric cancer cells through regulation of P53-mediated mitochondrial apoptotic pathway. Biosci. Biotechnol. Biochem. 81 (1), 119–126. doi:10.1080/09168451.2016.1238294
Wang, P., Guo, X., Zong, W., Li, Y., Liu, G., Lv, Y., et al. (2019). PGC-1α/SNAI1 axis regulates tumor growth and metastasis by targeting miR-128b in gastric cancer. J. Cell Physiol. 234 (10), 17232–17241. doi:10.1002/jcp.28193
Wang, W., Liparulo, I., Rizzardi, N., Bolignano, P., Calonghi, N., Bergamini, C., et al. (2020). Coenzyme Q depletion reshapes MCF-7 cells metabolism. Int. J. Mol. Sci. 22 (1), 198. doi:10.3390/ijms22010198
Warburg, O. (1956). On the origin of cancer cells. Science 123 (3191), 309–314. doi:10.1126/science.123.3191.309
Wei, C., Li, M., Lin, S., and Xiao, J. (2022). Characterization of tumor mutation burden-based gene signature and molecular subtypes to assist precision treatment in gastric cancer. Biomed. Res. Int. 2022, 4006507. doi:10.1155/2022/4006507
Westermann, B. (2010). Mitochondrial fusion and fission in cell life and death. Nat. Rev. Mol. Cell Biol. 11 (12), 872–884. doi:10.1038/nrm3013
Wu, F., Huang, W., and Wang, X. (2015). microRNA-18a regulates gastric carcinoma cell apoptosis and invasion by suppressing hypoxia-inducible factor-1α expression. Exp. Ther. Med. 10 (2), 717–722. doi:10.3892/etm.2015.2546
Wu, Q., Feng, L., Wang, Y., Mao, Y., Di, X., Zhang, K., et al. (2022). Multi-omics analysis reveals RNA splicing alterations and their biological and clinical implications in lung adenocarcinoma. Signal Transduct. Target Ther. 7 (1), 270. doi:10.1038/s41392-022-01098-5
Xia, L., Zhang, C., Lv, N., Liang, Z., Ma, T., Cheng, H., et al. (2022). AdMSC-derived exosomes alleviate acute lung injury via transferring mitochondrial component to improve homeostasis of alveolar macrophages. Theranostics 12 (6), 2928–2947. doi:10.7150/thno.69533
Xie, S., Chang, Y., Jin, H., Yang, F., Xu, Y., Yan, X., et al. (2020). Non-coding RNAs in gastric cancer. Cancer Lett. 493, 55–70. doi:10.1016/j.canlet.2020.06.022
Xu, K., Chen, G., Li, X., Wu, X., Chang, Z., Xu, J., et al. (2017). MFN2 suppresses cancer progression through inhibition of mTORC2/Akt signaling. Sci. Rep. 7, 41718. doi:10.1038/srep41718
Xu, S., Li, D., Li, T., Qiao, L., Li, K., Guo, L., et al. (2018). miR-494 sensitizes gastric cancer cells to TRAIL treatment through downregulation of survivin. Cell Physiol. Biochem. 51 (5), 2212–2223. doi:10.1159/000495867
Yamashiro, H., and Siomi, M. C. (2018). PIWI-interacting RNA in Drosophila: Biogenesis, transposon regulation, and beyond. Chem. Rev. 118 (8), 4404–4421. doi:10.1021/acs.chemrev.7b00393
Yan, R., Li, K., Yuan, D. W., Wang, H. N., Zhang, Y., Dang, C. X., et al. (2018). Downregulation of microRNA-4295 enhances cisplatin-induced gastric cancer cell apoptosis through the EGFR/PI3K/Akt signaling pathway by targeting LRIG1. Int. J. Oncol. 53 (6), 2566–2578. doi:10.3892/ijo.2018.4595
Yang, Q., Li, F., He, A. T., and Yang, B. B. (2021). Circular RNAs: Expression, localization, and therapeutic potentials. Mol. Ther. 29 (5), 1683–1702. doi:10.1016/j.ymthe.2021.01.018
Yang, Y., Huang, Y., Lin, W., Liu, J., Chen, X., Chen, C., et al. (2022). Host miRNAs-microbiota interactions in gastric cancer. J. Transl. Med. 20 (1), 52. doi:10.1186/s12967-022-03264-3
Zamberlan, M., Boeckx, A., Muller, F., Vinelli, F., Ek, O., Vianello, C., et al. (2022). Inhibition of the mitochondrial protein Opa1 curtails breast cancer growth. J. Exp. Clin. Cancer Res. 41 (1), 95. doi:10.1186/s13046-022-02304-6
Zhang, G. Q., Wang, S. Q., Chen, Y., Fu, L. Y., Xu, Y. N., Li, L., et al. (2021). MicroRNAs regulating mitochondrial function in cardiac diseases. Front. Pharmacol. 12, 663322. doi:10.3389/fphar.2021.663322
Zhang, Z., Liu, J., Zeng, Z., Fan, J., Huang, S., Zhang, L., et al. (2019). lncRNA Rmst acts as an important mediator of BMP9-induced osteogenic differentiation of mesenchymal stem cells (MSCs) by antagonizing Notch-targeting microRNAs. Aging (Albany NY) 11 (24), 12476–12496. doi:10.18632/aging.102583
Zhao, M., Hou, Y., Du, Y. E., Yang, L., Qin, Y., Peng, M., et al. (2020). Drosha-independent miR-6778-5p strengthens gastric cancer stem cell stemness via regulation of cytosolic one-carbon folate metabolism. Cancer Lett. 478, 8–21. doi:10.1016/j.canlet.2020.02.040
Zhao, X., Su, L., He, X., Zhao, B., and Miao, J. (2020). Long noncoding RNA CA7-4 promotes autophagy and apoptosis via sponging MIR877-3P and MIR5680 in high glucose-induced vascular endothelial cells. Autophagy 16 (1), 70–85. doi:10.1080/15548627.2019.1598750
Zhou, J., Duan, X., Wang, J., Feng, Y., and Yuan, J. (2022). Valsartan regulates PI3K/AKT pathways through lncRNA GASL1 to improve isoproterenol-induced heart failure. Dis. Markers 2022, 1447399. doi:10.1155/2022/1447399
Zhou, Z., Lin, Z., He, Y., Pang, X., Wang, Y., Ponnusamy, M., et al. (2018). The long noncoding RNA D63785 regulates chemotherapy sensitivity in human gastric cancer by targeting miR-422a. Mol. Ther. Nucleic Acids 12, 405–419. doi:10.1016/j.omtn.2018.05.024
Keywords: non-coding RNA, mitochondrial dynamic, gastric cancer, therapeutic strategy, cancer progression
Citation: Chen X, Wei C, Huang L, Syrigos K, Li Y and Li P (2023) Non-coding RNAs regulate mitochondrial dynamics in the development of gastric cancer. Front. Mol. Biosci. 10:1107651. doi: 10.3389/fmolb.2023.1107651
Received: 25 November 2022; Accepted: 03 January 2023;
Published: 12 January 2023.
Edited by:
Tao Yuan, Jiangxi Normal University, ChinaCopyright © 2023 Chen, Wei, Huang, Syrigos, Li and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuzhen Li, emhxZDE5NjNAMTI2LmNvbQ==; Peifeng Li, cGVpZmxpQHFkdS5lZHUuY24=
†These authors have contributed equally to this work
 Xiatian Chen
Xiatian Chen Chuang Wei1,2†
Chuang Wei1,2† Peifeng Li
Peifeng Li