- 1Department of Cellular and Molecular Medicine, University of California, San Diego, San Diego, CA, United States
- 2Stem Cell Program, University of California, San Diego, San Diego, CA, United States
- 3Institute for Genomic Medicine, University of California, San Diego, San Diego, CA, United States
- 4Neurosciences Graduate Program, University of California, San Diego, San Diego, CA, United States
Disordered RNA-binding proteins and repetitive RNA sequences are the main genetic causes of several neurodegenerative diseases, including amyotrophic lateral sclerosis and Huntington’s disease. Importantly, these components also seed the formation of cytoplasmic liquid-like granules, like stress granules and P bodies. Emerging evidence demonstrates that healthy granules formed via liquid-liquid phase separation can mature into solid- or gel-like inclusions that persist within the cell. These solidified inclusions are a precursor to the aggregates identified in patients, demonstrating that dysregulation of RNA granule biology is an important component of neurodegeneration. Here, we review recent literature highlighting how RNA molecules seed proteinaceous granules, the mechanisms of healthy turnover of RNA granules in cells, which biophysical properties underly a transition to solid- or gel-like material states, and why persistent granules disrupt the cellular homeostasis of neurons. We also identify various methods that will illuminate the contributions of disordered proteins and RNAs to neurodegeneration in ongoing research efforts.
Introduction
The compartmentalization of cellular contents into membrane-bound organelles is a long-established paradigm in biology. However, recent evidence demonstrates that lipid membranes are not strictly required to segregate macromolecules into distinct structures. Instead, discrete liquid phases—like oil in water—may form when proteins containing intrinsically disordered regions (IDRs) bind to RNA, forming multimeric complexes. These multimeric complexes eventually support a phase transition, in which a dense proteinaceous granule forms by liquid-liquid phase separation (LLPS). A variety of cytoplasmic liquid-like granules form via LLPS, including stress granules (SGs), P bodies, G bodies, and others (Banani et al., 2017). Importantly, many of these granules are seeded in response to stress, and they are disassembled once the stress event ends (Buchan et al., 2008; Wheeler et al., 2016; Jin et al., 2017). However, a granule may persist beyond the stress event if it undergoes a material transition into a solid- or gel-like status (Zhang et al., 2019; Lu et al., 2021). When granules mature into gels, they have reduced interactions with the surrounding cellular milieu and can further transition into fibrils or other aggregated structures (Patel et al., 2015). Solid- or gel-like granules are disastrous for neurons, most of which are not replenished throughout an organism’s lifetime (Lim and Yue, 2015). Dysregulation of the material state of cellular granules is linked with a variety of neurodegenerative diseases, including amyotrophic lateral sclerosis (ALS), Huntington’s, Alzheimer’s, and others (Patel et al., 2015; Li et al., 2016; Wegmann et al., 2018).
RNA is a critical component of phase-separated granules. Many proteins that are enriched within granules contain not only IDRs but also canonical RNA-binding domains (Markmiller et al., 2018). These RNA-binding proteins (RBPs) can therefore multimerize with themselves via IDRs and RNA via their RNA-binding domains (Rhine et al., 2020b). Biochemical studies demonstrate that RNA can reduce the amount of protein needed to form proteinaceous droplets in vitro (Kato et al., 2012; Molliex et al., 2015; Niaki et al., 2020; Yang et al., 2020), and elegant cellular experiments are consistent with the idea that cytoplasmic RNA promotes LLPS (Fuller et al., 2020; Bauer et al., 2022). Conversely, an overabundance of RNA may buffer phase separation by diluting multimeric interactions among many RNA and RBP molecules, though this phenomenon is more pronounced in the nucleus (Maharana et al., 2018). The cytoplasmic concentration of RNA can be tuned by stress events, especially translational arrest at polysomes that cause an acute increase in available RNA for RBP binding (Bounedjah et al., 2014; Iserman et al., 2020). In general, longer RNAs are more effectively recruited into granules, but biases toward certain sequence or structure motifs heavily depend on the RBP recognizing the RNA molecule and other polymers in the cell (Khong et al., 2017; Hallegger et al., 2021; Rhine et al., 2022a). Repetitive RNAs may also contribute to granule formation independently of proteins by undergoing self-associations like those found in G-quadruplexes (Boeynaems et al., 2019).
Aside from promoting granule formation, RNA also alters the viscoelastic properties of granules (Roden and Gladfelter, 2021; Laghmach et al., 2022). As mentioned above, RNA promotes RBP multimerization by acting as a scaffold to which proteins may bind (Schwartz et al., 2013; Rhine et al., 2020a). Scaffolds naturally stabilize the resulting condensate (Decker et al., 2022; Sanchez-Burgos et al., 2022), which can accelerate coarsening into solid- or gel-like material states (Bose et al., 2022). In addition, the physical interaction between RNA and its cognate proteins may lead to conformational rearrangements in the protein that shield certain domains, inhibiting efficient dissolution by chaperones that resolve granules (Yoshizawa et al., 2018). Finally, mutations in RBPs can further promote solid-like transitions (Zhu et al., 2014; Niaki et al., 2020). Indeed, many mutations identified in ALS affect RBPs containing IDRs, including FUS, TDP-43, hnRNPA1, and others (Sreedharan et al., 2008; Vance et al., 2009; Kim et al., 2013). RNA promotes condensation of all these proteins, indirectly supporting an aberrant material state transition consistent with inclusions found in neurodegeneration patients.
Therefore, RNA has a critical role in establishing the biophysical properties of condensates. Given the vast number of biological processes that require RNA, sequestration of RNA into disease-associated granules is an inherently perturbative outcome that destabilizes cellular homeostasis and eventually promotes cell death (Pushpalatha et al., 2022). In the following sections, we review the various regulatory and biophysical processes that contribute to RNA granule aging, especially in the context of neurodegeneration as persistent RNA granules disrupt cellular homeostasis. We also present various techniques that may be used to further our understanding of how RNA impacts granule maturation.
Turnover of stress granules in health and disease
The normal life cycle of a stress granule involves rapid formation and quick dissolution regulated by several mechanisms. Upon different stress conditions, multiple pathways including inhibition of mTOR, phosphorylation of eIF2a and disruption of eIF4F complex induce SG formation (Harding et al., 2000; Gilks et al., 2004; Thedieck et al., 2013). All three pathways converge to stall and disassemble polysomes, which is arguably the common trigger of SG formation (Buchan and Parker, 2009). When SGs form, RBPs and mRNA molecules play a major role. Two RBPs, in particular, have been thoroughly studied for the assembly of cytoplasmic SG: T-cell intracellular antigen 1 (TIA-1) and Ras-GTPase-activating protein SH3-domain-binding protein 1 (G3BP1) (Tourriere et al., 2003; Gilks et al., 2004). G3BP1 and its homolog G3BP2 are the core components of stress granules (Figure 1), which undergo RNA-dependent LLPS by sequestering free RNA to generate protein-RNA condensates (Guillen-Boixet et al., 2020; Yang et al., 2020). G3BP1 can also interact with many IDR-containing RBPs, including Caprin-1 and TIA-1, and both promote G3BP1/2-mediated LLPS (Guillen-Boixet et al., 2020; Yang et al., 2020). The size and liquid properties of RNA-protein condensates in cells are affected by RNA recruitment (Garcia-Jove Navarro et al., 2019; Roden and Gladfelter, 2021). Furthermore, the availability of RNA–RNA assemblies is important for granule assembly in vitro (Van Treeck et al., 2018). Trcek et al. demonstrated in Drosophila germ cells that different mRNA features govern mRNA localization to granules and self-assembly within granules; they noted that localization is encoded by specific RNA regions, whereas self-assembly is RNA sequence-independent (Trcek et al., 2020).
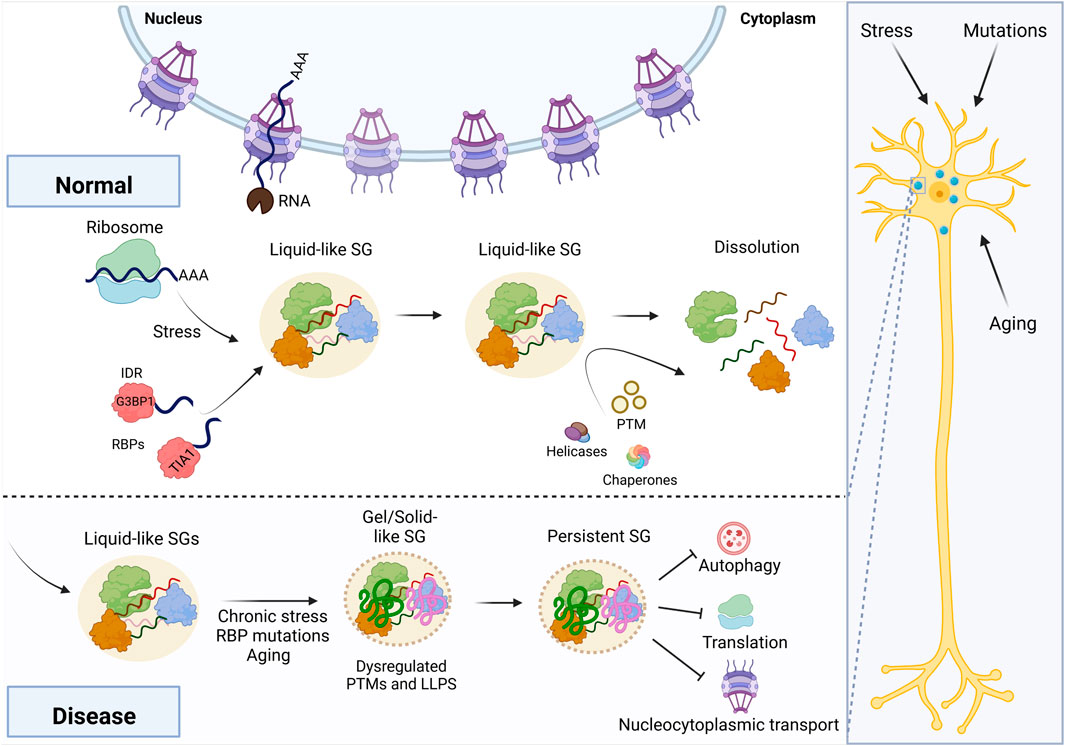
FIGURE 1. Maturation of RNA granules into disease-associated states. Stress, aging, and mutations can all stall neuronal translation, initiating stress granule assembly via G3BP1. Chronic stress exposure or other mechanisms may lead to a disease-associated outcome, in which the granule transitions to a gel- or solid-like state. In turn, this persistent granule disrupts cellular homeostasis, including autophagy, translation, and nucleocytoplasmic transport. This figure was created using BioRender.
Canonical RNA turnover in SGs occurs in a stepwise manner (Wheeler et al., 2016). First, translation is slowed during stress, and polysomes release their respective messenger ribonucleoproteins (mRNPs). The free mRNP particles oligomerize via protein-protein, protein–RNA, or RNA–RNA interactions. When more RNAs enter the non-translating pool, oligomers form stable core assemblies (Decker et al., 2022). As a result of fusion and mRNP recruitment, a mature SG with a distinct core-shell substructure is generated. Even mature SGs (especially the “shell”) are still in dynamic equilibrium, exchanging materials with polysomes and P bodies (Wheeler et al., 2016). It is worth noting that a recent super-resolution imaging study showcased comparable translation efficiency of their reporter in SGs over outside of SGs, suggesting that the SG environment is not inhibitory to translation and that global translation inhibition in response to stress is more likely to be upstream of rather than the consequence of SG formation (Mateju et al., 2020). SGs begin to break down via shell loss followed by core dispersal, and mRNPs re-enter translation. Translationally-stalled mRNAs are titrated out of SGs, producing structural instability in the protein complexes and eventually gradual deconstruction of the visible SGs (Wheeler et al., 2016). Several lines of evidence have shown that RNA can mitigate excessive protein-protein interactions, which lead to pathological aggregates seen in a variety of neurodegenerative disorders (Maharana et al., 2018; Mann et al., 2019; Zacco et al., 2019), emphasizing the critical balance between protein-protein, RNA–protein, and RNA–RNA interactions required for proper SG assembly and disassembly. Post-translational modifications also play a critical role in SG disassembly, Maxwell et al. revealed an important function of heat-induced polyubiquitylation, which is instrumental in preparing cells for the restart of cellular activities upon stress release (Maxwell et al., 2021). Furthermore, the SG scaffold protein G3BP1 is a key substrate for polyubiquitin-dependent disassembly of heat-induced SGs (Gwon et al., 2021). One crucial mechanism in this recovery phase is the restart of translation, which is accompanied by the ubiquitin-dependent disassembly of SGs (Maxwell et al., 2021). Lastly, increased recruitment of small ubiquitin-like modifier ligases into the SGs is observed upon stress exposure leading to SUMOylation of proteins necessary for SG disassembly (Marmor-Kollet et al., 2020).
RNA helicases also have a major role in RNA granule dynamics and could contribute to the interconnection between mRNA storage, translation, and decay (Hondele et al., 2019; Weis and Hondele, 2022). Recent work shows that assembly and disassembly of RNA granules are monitored by DEAD-Box helicase 6 (DDX6), an essential P body component, in neuronal maturation both in vitro and in vivo (Bauer et al., 2022). ATP, which is consumed by RNA helicases, is required for the fast construction, remodeling, and disassembly of stress granule components (Wolozin and Ivanov, 2019). Furthermore, DDX6 granule condensation requires Staufen-2-dependent RNA interaction during synaptic inhibition. This is most likely owing to Stau2’s participation in mRNA transport and redistribution (Bauer et al., 2022). In addition, recent work has shown that sex chromosome-encoded RNA helicases such as DDX3X and DDX3Y influence the formation of RNA granules in an ATP-independent manner but have different LLPS propensities (Shen H. et al., 2022).
Nuclear import of granule-associated proteins is another important layer of SG regulation (Guo et al., 2018), and defects in nucleocytoplasmic transport are a significant pathogenic factor in ALS (Zhang et al., 2015; Gleixner et al., 2022). Cytoplasmic protein clumps, which are frequently observed in many neurodegenerative illnesses, impair nucleocytoplasmic transport, implying that abnormalities in nucleocytoplasmic transport might constitute a general cause of neurodegeneration (Hutten and Dormann, 2020; Gonzalez et al., 2021; Odeh et al., 2022). Critical nucleocytoplasmic transport components such as karyopherins (importins and exportins), Ran GTPase, and nucleoporins translocate to stress granules in response to cellular stress, resulting in inefficient nucleocytoplasmic transport (Zhang et al., 2018). Importantly, in C9ORF72 ALS models, blocking stress granule construction decreases these abnormalities as well as neurodegeneration (Zhang et al., 2018). These discoveries linked two pathophysiological processes, stress granule construction and nucleocytoplasmic transport disruption, into a single pathway that leads to pathogenesis. Thus, an intriguing issue of the present work is analyzing how changes to essential stress granule components or remodeling machinery affect the various phases of this assembly and disassembly process.
RNA promotes the maturation of granules to solid-like material states
Following granule formation, the cell has a variety of mechanisms to promote dissolution of granules, including posttranslational modifications that signal for turnover of critical RBPs, expression of chaperones that recognize granule proteins, and helicase activity that ejects RBPs from RNA (Guo et al., 2018; Hondele et al., 2019; Maxwell et al., 2021). However, complete resolution of RNA granules may not be possible if the granule has coarsened into a solid- or gel-like state (Figure 1). Many in vitro studies demonstrate the effect of aging, but there are two separate biophysical processes through which a granule may achieve a solid- or gel-like state. A recent study by Jawerth et al. proposed that condensates do not become gels per se and instead behave as a Maxwell fluid with strongly increasing viscosity as a function of condensate age (Jawerth et al., 2020). Other studies indicate that RBPs bound to RNA may undergo dynamical arrest via percolation, halting exchange with the surrounding dilute phase (Harmon et al., 2017; Rhine et al., 2020a; Choi et al., 2020; Bose et al., 2022; Linsenmeier et al., 2022; Mittag and Pappu, 2022). Granules may also partially transition to gels, as was observed with the RBP FUS (Shen Y. et al., 2022). In principle, these mechanisms likely arrive at the same outcome: an RNA granule that is resistant to dissolution because of a change in its material properties.
Many neurodegenerative diseases are linked to the formation of fibrils or solid-like inclusions (Kim et al., 2013; Nomura et al., 2014; Bowden and Dormann, 2016; French et al., 2019), which have similar material properties to aged RNA-containing SGs (Zhu et al., 2014; Patel et al., 2015; Murray et al., 2017; Fonda et al., 2021). Proteins associated with ALS (FUS, TDP-43, hnRNPA1, etc.), Alzheimer’s (Tau), and Parkinson’s (α-synuclein) undergo both LLPS and aggregation into fibers (Gotz et al., 2001; Roberson et al., 2007; Sreedharan et al., 2008; Vance et al., 2009; Kim et al., 2013; Wegmann et al., 2018; Ray et al., 2020). Although fibers are thought to be the causative agent of disease, certain ALS-associated RBPs incorporate into liquid-like stress granules (Bentmann et al., 2012; Markmiller et al., 2018; Reber et al., 2021; An et al., 2022). ALS-linked mutations in these RBPs accelerate the granule aging process or inhibit recognition by protein chaperones (Guo et al., 2018; Hofweber et al., 2018; Niaki et al., 2020). Aged RNA granules can be deleterious on their own by stalling translation via RNA sequestration and by preventing cellular recovery from stress (Reineke and Neilson, 2019; Pushpalatha et al., 2022). Disease-linked mutations cause some proteins like TDP-43 to bypass the liquid phase altogether, forming aggregates or fibrils at physiological concentrations (Patel et al., 2015; Cao et al., 2019; Mathieu et al., 2020). In these cases, LLPS may act as a protective agent to prevent or decelerate deleterious solid- or gel-like transitions.
Recent evidence highlights the role of RNA in promoting the transition of granules to solid-like material states. The most common genetic cause of ALS—a repeat expansion of the C9ORF72 locus—leads to the expression of the highly toxic (GGGGCC)n RNA motif and translation of repetitive dipeptide chains, especially poly-RG and poly-PR (DeJesus-Hernandez et al., 2011). Importantly, this RNA repeat engages in multivalent base pairing, promoting granules that transition into a gel-like state (Jain and Vale, 2017). Other disease-associated RNA repeats also impact granule aging in vitro (Ma et al., 2022), and the self-association of G-rich RNA forms solid-like fibers (Boeynaems et al., 2019). Normal mRNA sequences may also contribute to RNA self-assembly or gelation, as has been observed in worms and flies (Lee et al., 2020; Trcek et al., 2020). It is possible that P-body-associated RNA helicases are required for proper resolution of multivalent RNA tangles (Hondele et al., 2019; Majerciak et al., 2021; Linsenmeier et al., 2022), which can age condensates into gels. Together, these studies demonstrate that RNA can promote solid- or gel-like transitions in RNA granules, and sequencing-based technologies like CLIP-seq and others may help identify changes in RNA biology during disease (Van Nostrand et al., 2016; Hallegger et al., 2021; Wollny et al., 2022) (Table 1). Although we do not yet know how exactly aged RNA granules and fibers lead to cell death, the next section will discuss how the persistence of aged granules and fibers may disrupt cellular homeostasis.
Persistent granules disrupt cellular homeostasis and could potentially serve as therapeutic targets
The persistence of RNA granules may disrupt cellular homeostasis through multiple mechanisms, including posing as a cytotoxic stimulus, sequestering proteins important for maintaining cellular homeostasis, and altering SG assembly and disassembly (Figure 1). In Alzheimer’s disease, amyloid-beta aggregates stimulate persistent SGs (Ghosh and Geahlen, 2015), and SG assembly promotes the phosphorylation of tau (Vanderweyde et al., 2012), potentially accelerating the onset of neurodegeneration. In the case of multiple polyglutamine (polyQ) diseases like Huntington’s, the polyQ aggregates could trigger various types of stresses (Labbadia and Morimoto, 2013; Matos et al., 2019), including misfolded protein stress. The polyQ aggregates also sequester SG components (Uchihara et al., 2001; Furukawa et al., 2009; Sleigh et al., 2020), leading to aberrant SG composition. These SGs contain misfolded proteins, further sequestering components of autophagy (Mateju et al., 2017). As one of the SG clearance pathways (Ryu et al., 2014), impaired autophagy could in turn disrupt the disassembly of SGs. In ALS, SGs colocalize with TDP-43 and poly-GR aggregates (Liu-Yesucevitz et al., 2010; Chew et al., 2019), and chronic SGs could directly contribute to ALS onset (Daigle et al., 2016; Zhang et al., 2019). The poly-PR aggregates could directly interact with ribosomes and impair protein translation (Zhang et al., 2018). Global splicing and RNA localization may also be altered due to the sequestration of important regulators in these cellular processes (Charizanis et al., 2012; Markmiller et al., 2021).
The unique formation of persistent granules in these neurodegenerative diseases enables the development of new strategies for therapeutic treatment. antisense oligonucleotides and RNA-targeting CRISPR have been demonstrated to eliminate toxic RNA granules (Lee et al., 2012; Batra et al., 2017) when particular RNAs are known to form these granules. Small molecule drugs could potentially be developed to target specific proteins responsible for the assembly or preventing disassembly of persistent granules. More efforts are needed to better depict the components of these granules under different disease conditions (Table 1). Meanwhile, explorative drug screens could be performed to repurpose existing drugs for resolving persistent granules in different disease contexts (Fang et al., 2019).
Technologies for investigating RNA granules
Pioneering research has laid the groundwork for our present understanding of RNA granules in neurodegenerative disease, expanding our understanding of the fundamental mechanisms of granule assembly and disassembly, as well as their composition and structural organization. However, there remains many gaps in the field. To conclude, we review new methods to aid our understanding of RNA granules (Table 1).
Over the past decades, there have been a variety of different approaches and methods to characterize and identify different stress granule components and functions (Table 1). A starting point of analysis for any granule-associated client protein is to identify its components with imaging technology. Initially many components of stress granules were identified using specific antibodies via immunofluorescence or immunohistochemistry. These imaging-based experiments detected several different types of stress granule components, including RBPs and translation machinery components. Recent advances in microscopy enable robust live-cell imaging of diffusing granule proteins, providing biophysical data of granule material properties. Fluorescence in situ hybridization and the MS2 tagging system can be used for fixed and live tracking of RNAs (Le et al., 2022).
Alternatively, omics-based techniques identify components of the proteome and transcriptome of RNA granules. Previously published work from our lab used ascorbate peroxidase (APEX2) proximity labeling to identify components of SGs in human cells (Markmiller et al., 2018). APEX can also be repurposed to pull down RNAs instead of proteins (Padron et al., 2019). Other techniques such as BioID provide similarly robust datasets of the granule proteome (Youn et al., 2018). We have also pioneered enhanced CLIP technologies, which identify bound RNAs in granules (Van Nostrand et al., 2016; Corley et al., 2020; Blue et al., 2022). Coupling these technologies with different biological conditions can identify stress-responsive changes in granule components, including shifts that are precursors to neurodegeneration (Markmiller et al., 2021).
Biochemical techniques allow the most robust characterization of granules properties and even enable the purification of whole granules from cells. Matheny et al. compared two granule isolation methods, and they demonstrated that more SG enrichment comes from differential centrifugation and immunopurification (Matheny et al., 2019). The RNAs from purified granules were similar to the RNAs found in the cellular granules confirmed with FISH. Biophysical techniques such as FRAP and microrheology also identify the material properties of in vitro and in vivo granules, which may help pinpoint the transition from a normal granule to a disease-associated granule. Overall, the approaches discussed here give a wealth of information for understanding stress granule production and disassembly, function, and regulation, which we anticipate will be important for identifying how and why RNA granules transition into neurodegeneration-linked fibers.
Conclusion
Sequestering critical RBPs and RNA molecules into granules is an inherently risky maneuver for cells. If a cell does not pause translation, splicing, and other energetically expensive processes, it may not survive the stress event. However, if the stress response persists for too long, the cell may permanently entangle essential machinery for translation, splicing, and other processes into granules. As we discussed above, persistent granules may undergo a phase transition into nondynamic, solid- or gel-like structure, which is a hallmark of neurodegenerative diseases. Therefore, it is necessary to better understand how and why RNA granules mature into nonresolvable aggregates so that we can develop appropriate remedies to counteract deleterious phase transitions.
Author contributions
KR, NA-A, and TY contributed equally to the writing, editing, and figure preparation for this review. GY edited the writing and figures and supervised the preparation of the review.
Funding
RNA and neurodegeneration research in the Yeo laboratory is supported by the National Institutes of Health via I132122, HG011864, NS103172, EY029166, HG004659 and HG009889. GY is also supported by an Allen Distinguished Investigator Award, a Paul G. Allen Frontiers Group advised grant of the Paul G. Allen Family Foundation.
Acknowledgments
We thank the members of the Yeo laboratory for helpful discussions while writing this review.
Conflict of interest
GY is an SAB member of Jumpcode Genomics and a co-founder, member of the Board of Directors, on the SAB, equity holder, and paid consultant for Locanabio and Eclipse BioInnovations. GY is a visiting professor at the National University of Singapore. GY’s interests have been reviewed and approved by the University of California, San Diego in accordance with its conflict-of-interest policies. All other authors declare no competing.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
An, H., Litscher, G., Watanabe, N., Wei, W., Hashimoto, T., Iwatsubo, T., et al. (2022). ALS-linked cytoplasmic FUS assemblies are compositionally different from physiological stress granules and sequester hnRNPA3, a novel modifier of FUS toxicity. Neurobiol. Dis. 162, 105585. doi:10.1016/j.nbd.2021.105585 | |
Banani, S. F., Lee, H. O., Hyman, A. A., and Rosen, M. K. (2017). Biomolecular condensates: Organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 18, 285–298. doi:10.1038/nrm.2017.7 | |
Batra, R., Nelles, D. A., Pirie, E., Blue, S. M., Marina, R. J., Wang, H., et al. (2017). Elimination of toxic microsatellite repeat expansion RNA by RNA-targeting Cas9. Cell 170, 899–912. doi:10.1016/j.cell.2017.07.010 | |
Bauer, K. E., Bargenda, N., Schieweck, R., Illig, C., Segura, I., Harner, M., et al. (2022). RNA supply drives physiological granule assembly in neurons. Nat. Commun. 13, 2781. doi:10.1038/s41467-022-30067-3 | |
Benhalevy, D., Anastasakis, D. G., and Hafner, M. (2018). Proximity-CLIP provides a snapshot of protein-occupied RNA elements in subcellular compartments. Nat. Methods 15, 1074–1082. doi:10.1038/s41592-018-0220-y | |
Bentmann, E., Neumann, M., Tahirovic, S., Rodde, R., Dormann, D., and Haass, C. (2012). Requirements for stress granule recruitment of fused in sarcoma (FUS) and TAR DNA-binding protein of 43 kDa (TDP-43). J. Biol. Chem. 287, 23079–23094. doi:10.1074/jbc.M111.328757 | |
Blue, S. M., Yee, B. A., Pratt, G. A., Mueller, J. R., Park, S. S., Shishkin, A. A., et al. (2022). Transcriptome-wide identification of RNA-binding protein binding sites using seCLIP-seq. Nat. Protoc. 17, 1223–1265. doi:10.1038/s41596-022-00680-z | |
Boeynaems, S., Holehouse, A. S., Weinhardt, V., Kovacs, D., Van Lindt, J., Larabell, C., et al. (2019). Spontaneous driving forces give rise to protein-RNA condensates with coexisting phases and complex material properties. Proc. Natl. Acad. Sci. U. S. A. 116, 7889–7898. doi:10.1073/pnas.1821038116 | |
Bose, M., Lampe, M., Mahamid, J., and Ephrussi, A. (2022). Liquid-to-solid phase transition of oskar ribonucleoprotein granules is essential for their function in Drosophila embryonic development. Cell 185, 1308–1324.e23. doi:10.1016/j.cell.2022.02.022 | |
Bounedjah, O., Desforges, B., Wu, T. D., Pioche-Durieu, C., Marco, S., Hamon, L., et al. (2014). Free mRNA in excess upon polysome dissociation is a scaffold for protein multimerization to form stress granules. Nucleic Acids Res. 42, 8678–8691. doi:10.1093/nar/gku582 | |
Bowden, H. A., and Dormann, D. (2016). Altered mRNP granule dynamics in FTLD pathogenesis. J. Neurochem. 138 (1), 112–133. doi:10.1111/jnc.13601 | |
Buchan, J. R., Muhlrad, D., and Parker, R. (2008). P bodies promote stress granule assembly in Saccharomyces cerevisiae. J. Cell Biol. 183, 441–455. doi:10.1083/jcb.200807043 | |
Buchan, J. R., and Parker, R. (2009). Eukaryotic stress granules: The ins and outs of translation. Mol. Cell 36, 932–941. doi:10.1016/j.molcel.2009.11.020 | |
Cao, Q., Boyer, D. R., Sawaya, M. R., Ge, P., and Eisenberg, D. S. (2019). Cryo-EM structures of four polymorphic TDP-43 amyloid cores. Nat. Struct. Mol. Biol. 26, 619–627. doi:10.1038/s41594-019-0248-4 | |
Charizanis, K., Lee, K. Y., Batra, R., Goodwin, M., Zhang, C., Yuan, Y., et al. (2012). Muscleblind-like 2-mediated alternative splicing in the developing brain and dysregulation in myotonic dystrophy. Neuron 75, 437–450. doi:10.1016/j.neuron.2012.05.029 | |
Chew, J., Cook, C., Gendron, T. F., Jansen-West, K., Del Rosso, G., Daughrity, L. M., et al. (2019). Aberrant deposition of stress granule-resident proteins linked to C9orf72-associated TDP-43 proteinopathy. Mol. Neurodegener. 14, 9. doi:10.1186/s13024-019-0310-z | |
Choi, J. M., Hyman, A. A., and Pappu, R. V. (2020). Generalized models for bond percolation transitions of associative polymers. Phys. Rev. E 102, 042403. doi:10.1103/PhysRevE.102.042403 | |
Corley, M., Flynn, R. A., Lee, B., Blue, S. M., Chang, H. Y., and Yeo, G. W. (2020). Footprinting SHAPE-eCLIP reveals transcriptome-wide hydrogen bonds at RNA-protein interfaces. Mol. Cell 80, 903–914. doi:10.1016/j.molcel.2020.11.014 | |
Daigle, J. G., Krishnamurthy, K., Ramesh, N., Casci, I., Monaghan, J., Mcavoy, K., et al. (2016). Pur-alpha regulates cytoplasmic stress granule dynamics and ameliorates FUS toxicity. Acta Neuropathol. 131, 605–620. doi:10.1007/s00401-015-1530-0 | |
Decker, C. J., Burke, J. M., Mulvaney, P. K., and Parker, R. (2022). RNA is required for the integrity of multiple nuclear and cytoplasmic membrane-less RNP granules. EMBO J. 41, e110137. doi:10.15252/embj.2021110137 | |
Dejesus-Hernandez, M., Mackenzie, I. R., Boeve, B. F., Boxer, A. L., Baker, M., Rutherford, N. J., et al. (2011). Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 72, 245–256. doi:10.1016/j.neuron.2011.09.011 | |
Elbaum-Garfinkle, S., Kim, Y., Szczepaniak, K., Chen, C. C., Eckmann, C. R., Myong, S., et al. (2015). The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proc. Natl. Acad. Sci. U. S. A. 112, 7189–7194. doi:10.1073/pnas.1504822112 | |
Elmsaouri, S., Markmiller, S., and Yeo, G. W. (2022). APEX proximity labeling of stress granule proteins. Methods Mol. Biol. 2428, 381–399. doi:10.1007/978-1-0716-1975-9_23 | |
Fang, M. Y., Markmiller, S., Vu, A. Q., Javaherian, A., Dowdle, W. E., Jolivet, P., et al. (2019). Small-molecule modulation of TDP-43 recruitment to stress granules prevents persistent TDP-43 accumulation in ALS/FTD. Neuron 103, 802–819. doi:10.1016/j.neuron.2019.05.048 | |
Fonda, B. D., Jami, K. M., Boulos, N. R., and Murray, D. T. (2021). Identification of the rigid core for aged liquid droplets of an RNA-binding protein low complexity domain. J. Am. Chem. Soc. 143, 6657–6668. doi:10.1021/jacs.1c02424 | |
French, R. L., Grese, Z. R., Aligireddy, H., Dhavale, D. D., Reeb, A. N., Kedia, N., et al. (2019). Detection of TAR DNA-binding protein 43 (TDP-43) oligomers as initial intermediate species during aggregate formation. J. Biol. Chem. 294, 6696–6709. doi:10.1074/jbc.RA118.005889 | |
Fuller, G. G., Han, T., Freeberg, M. A., Moresco, J. J., Ghanbari Niaki, A., Roach, N. P., et al. (2020). RNA promotes phase separation of glycolysis enzymes into yeast G bodies in hypoxia. Elife 9, e48480. doi:10.7554/eLife.48480 | |
Furukawa, Y., Kaneko, K., Matsumoto, G., Kurosawa, M., and Nukina, N. (2009). Cross-seeding fibrillation of Q/N-rich proteins offers new pathomechanism of polyglutamine diseases. J. Neurosci. 29, 5153–5162. doi:10.1523/JNEUROSCI.0783-09.2009 | |
Ganser, L. R., and Myong, S. (2020). Methods to study phase-separated condensates and the underlying molecular interactions. Trends biochem. Sci. 45, 1004–1005. doi:10.1016/j.tibs.2020.05.011 | |
Garcia-Jove Navarro, M., Kashida, S., Chouaib, R., Souquere, S., Pierron, G., Weil, D., et al. (2019). RNA is a critical element for the sizing and the composition of phase-separated RNA–protein condensates. Nat. Commun. 10, 3230. doi:10.1038/s41467-019-11241-6 | |
Ghosh, S., and Geahlen, R. L. (2015). Stress granules modulate SYK to cause microglial cell dysfunction in alzheimer's disease. EBioMedicine 2, 1785–1798. doi:10.1016/j.ebiom.2015.09.053 | |
Gilks, N., Kedersha, N., Ayodele, M., Shen, L., Stoecklin, G., Dember, L. M., et al. (2004). Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol. Biol. Cell 15, 5383–5398. doi:10.1091/mbc.e04-08-0715 | |
Gleixner, A. M., Verdone, B. M., Otte, C. G., Anderson, E. N., Ramesh, N., Shapiro, O. R., et al. (2022). NUP62 localizes to ALS/FTLD pathological assemblies and contributes to TDP-43 insolubility. Nat. Commun. 13, 3380. doi:10.1038/s41467-022-31098-6 | |
Gonzalez, A., Mannen, T., Çağatay, T., Fujiwara, A., Matsumura, H., Niesman, A. B., et al. (2021). Mechanism of karyopherin-β2 binding and nuclear import of ALS variants FUS(P525L) and FUS(R495X). Sci. Rep. 11, 3754. doi:10.1038/s41598-021-83196-y | |
Gotz, J., Chen, F., Van Dorpe, J., and Nitsch, R. M. (2001). Formation of neurofibrillary tangles in P301l tau transgenic mice induced by Abeta 42 fibrils. Science 293, 1491–1495. doi:10.1126/science.1062097 | |
Guillen-Boixet, J., Kopach, A., Holehouse, A. S., Wittmann, S., Jahnel, M., Schlussler, R., et al. (2020). RNA-induced conformational switching and clustering of G3BP drive stress granule assembly by condensation. Cell 181, 346–361. doi:10.1016/j.cell.2020.03.049 | |
Guo, L., Kim, H. J., Wang, H., Monaghan, J., Freyermuth, F., Sung, J. C., et al. (2018). Nuclear-import receptors reverse aberrant phase transitions of RNA-binding proteins with prion-like domains. Cell 173, 677–692. doi:10.1016/j.cell.2018.03.002 | |
Gwon, Y., Maxwell, B. A., Kolaitis, R. M., Zhang, P., Kim, H. J., and Taylor, J. P. (2021). Ubiquitination of G3BP1 mediates stress granule disassembly in a context-specific manner. Science 372, eabf6548. doi:10.1126/science.abf6548 | |
Hallegger, M., Chakrabarti, A. M., Lee, F. C. Y., Lee, B. L., Amalietti, A. G., Odeh, H. M., et al. (2021). TDP-43 condensation properties specify its RNA-binding and regulatory repertoire. Cell 184, 4680–4696.e22. doi:10.1016/j.cell.2021.07.018 | |
Harding, H. P., Novoa, I., Zhang, Y., Zeng, H., Wek, R., Schapira, M., et al. (2000). Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell 6, 1099–1108. doi:10.1016/s1097-2765(00)00108-8 | |
Harmon, T. S., Holehouse, A. S., Rosen, M. K., and Pappu, R. V. (2017). Intrinsically disordered linkers determine the interplay between phase separation and gelation in multivalent proteins. Elife 6, e30294. doi:10.7554/eLife.30294 | |
Hofweber, M., Hutten, S., Bourgeois, B., Spreitzer, E., Niedner-Boblenz, A., Schifferer, M., et al. (2018). Phase separation of FUS is suppressed by its nuclear import receptor and arginine methylation. Cell 173, 706–719. doi:10.1016/j.cell.2018.03.004 | |
Hondele, M., Sachdev, R., Heinrich, S., Wang, J., Vallotton, P., Fontoura, B. M. A., et al. (2019). DEAD-box ATPases are global regulators of phase-separated organelles. Nature 573, 144–148. doi:10.1038/s41586-019-1502-y | |
Horvathova, I., Voigt, F., Kotrys, A. V., Zhan, Y., Artus-Revel, C. G., Eglinger, J., et al. (2017). The dynamics of mRNA turnover revealed by single-molecule imaging in single cells. Mol. Cell 68, 615–625. doi:10.1016/j.molcel.2017.09.030 | |
Hutten, S., and Dormann, D. (2020). Nucleocytoplasmic transport defects in neurodegeneration - cause or consequence? Semin. Cell Dev. Biol. 99, 151–162. doi:10.1016/j.semcdb.2019.05.020 | |
Irgen-Gioro, S., Walling, V., and Chong, S. (2022). Fixation can change the appearance of phase separation in living cells, bioRxiv 2005. 490956.
Iserman, C., Desroches Altamirano, C., Jegers, C., Friedrich, U., Zarin, T., Fritsch, A. W., et al. (2020). Condensation of Ded1p promotes a translational switch from housekeeping to stress protein production. Cell 181, 818–831. doi:10.1016/j.cell.2020.04.009 | |
Ivanov, P., Kedersha, N., and Anderson, P. (2011). Stress puts TIA on TOP. Genes Dev. 25, 2119–2124. doi:10.1101/gad.17838411 | |
Jain, A., and Vale, R. D. (2017). RNA phase transitions in repeat expansion disorders. Nature 546, 243–247. doi:10.1038/nature22386 | |
Jawerth, L., Fischer-Friedrich, E., Saha, S., Wang, J., Franzmann, T., Zhang, X., et al. (2020). Protein condensates as aging Maxwell fluids. Science 370, 1317–1323. doi:10.1126/science.aaw4951 | |
Jin, M., Fuller, G. G., Han, T., Yao, Y., Alessi, A. F., Freeberg, M. A., et al. (2017). Glycolytic enzymes coalesce in G bodies under hypoxic stress. Cell Rep. 20, 895–908. doi:10.1016/j.celrep.2017.06.082 | |
Kato, M., Han, T. W., Xie, S., Shi, K., Du, X., Wu, L. C., et al. (2012). Cell-free formation of RNA granules: Low complexity sequence domains form dynamic fibers within hydrogels. Cell 149, 753–767. doi:10.1016/j.cell.2012.04.017 | |
Khong, A., Matheny, T., Jain, S., Mitchell, S. F., Wheeler, J. R., and Parker, R. (2017). The stress granule transcriptome reveals principles of mRNA accumulation in stress granules. Mol. Cell 68, 808–820. e805. doi:10.1016/j.molcel.2017.10.015 | |
Kim, D. I., Jensen, S. C., Noble, K. A., Kc, B., Roux, K. H., Motamedchaboki, K., et al. (2016). An improved smaller biotin ligase for BioID proximity labeling. Mol. Biol. Cell 27, 1188–1196. doi:10.1091/mbc.E15-12-0844 | |
Kim, H. J., Kim, N. C., Wang, Y.-D., Scarborough, E. A., Moore, J., Diaz, Z., et al. (2013). Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature 495, 467–473. doi:10.1038/nature11922 | |
Labbadia, J., and Morimoto, R. I. (2013). Huntington's disease: Underlying molecular mechanisms and emerging concepts. Trends biochem. Sci. 38, 378–385. doi:10.1016/j.tibs.2013.05.003 | |
Laghmach, R., Alshareedah, I., Pham, M., Raju, M., Banerjee, P. R., and Potoyan, D. A. (2022). RNA chain length and stoichiometry govern surface tension and stability of protein-RNA condensates. iScience 25, 104105. doi:10.1016/j.isci.2022.104105 | |
Le, P., Ahmed, N., and Yeo, G. W. (2022). Illuminating RNA biology through imaging. Nat. Cell Biol. 24, 815–824. doi:10.1038/s41556-022-00933-9 | |
Lee, C. S., Putnam, A., Lu, T., He, S., Ouyang, J. P. T., and Seydoux, G. (2020). Recruitment of mRNAs to P granules by condensation with intrinsically-disordered proteins. Elife 9, e52896. doi:10.7554/eLife.52896 | |
Lee, J. E., Bennett, C. F., and Cooper, T. A. (2012). RNase H-mediated degradation of toxic RNA in myotonic dystrophy type 1. Proc. Natl. Acad. Sci. U. S. A. 109, 4221–4226. doi:10.1073/pnas.1117019109 | |
Li, L., Liu, H., Dong, P., Li, D., Legant, W. R., Grimm, J. B., et al. (2016). Real-time imaging of Huntingtin aggregates diverting target search and gene transcription. Elife 5, e17056. doi:10.7554/eLife.17056 | |
Lim, J., and Yue, Z. (2015). Neuronal aggregates: Formation, clearance, and spreading. Dev. Cell 32, 491–501. doi:10.1016/j.devcel.2015.02.002 | |
Linsenmeier, M., Hondele, M., Grigolato, F., Secchi, E., Weis, K., and Arosio, P. (2022). Dynamic arrest and aging of biomolecular condensates are modulated by low-complexity domains, RNA and biochemical activity. Nat. Commun. 13, 3030. doi:10.1038/s41467-022-30521-2 | |
Liu-Yesucevitz, L., Bilgutay, A., Zhang, Y. J., Vanderweyde, T., Citro, A., Mehta, T., et al. (2010). Tar DNA binding protein-43 (TDP-43) associates with stress granules: Analysis of cultured cells and pathological brain tissue. PLoS One 5, e13250. doi:10.1371/journal.pone.0013250 | |
Lu, S., Hu, J., Aladesuyi, B., Goginashvili, A., Vazquez-Sanchez, S., Diedrich, J., et al. (2021). Heat shock chaperone HSPB1 regulates cytoplasmic TDP-43 phase separation and liquid-to-gel transition. bioRxiv 2010, 464447.
Ma, Y., Li, H., Gong, Z., Yang, S., Wang, P., and Tang, C. (2022). Nucleobase clustering contributes to the formation and hollowing of repeat-expansion RNA condensate. J. Am. Chem. Soc. 144, 4716–4720. doi:10.1021/jacs.1c12085 | |
Maharana, S., Wang, J., Papadopoulos, D. K., Richter, D., Pozniakovsky, A., Poser, I., et al. (2018). RNA buffers the phase separation behavior of prion-like RNA binding proteins. Science 360, 918–921. doi:10.1126/science.aar7366 | |
Majerciak, V., Zhou, T., and Zheng, Z.-M. (2021). RNA helicase DDX6 in P-bodies is essential for the assembly of stress granules. bioRxiv 2009, 461736.
Mann, J. R., Gleixner, A. M., Mauna, J. C., Gomes, E., Dechellis-Marks, M. R., Needham, P. G., et al. (2019). RNA binding antagonizes neurotoxic phase transitions of TDP-43. Neuron 102, 321–338. doi:10.1016/j.neuron.2019.01.048 | |
Markmiller, S., Sathe, S., Server, K. L., Nguyen, T. B., Fulzele, A., Cody, N., et al. (2021). Persistent mRNA localization defects and cell death in ALS neurons caused by transient cellular stress. Cell Rep. 36, 109685. doi:10.1016/j.celrep.2021.109685 | |
Markmiller, S., Soltanieh, S., Server, K. L., Mak, R., Jin, W., Fang, M. Y., et al. (2018). Context-dependent and disease-specific diversity in protein interactions within stress granules. Cell 172, 590–604. doi:10.1016/j.cell.2017.12.032 | |
Marmor-Kollet, H., Siany, A., Kedersha, N., Knafo, N., Rivkin, N., Danino, Y. M., et al. (2020). Spatiotemporal proteomic analysis of stress granule disassembly using APEX reveals regulation by SUMOylation and links to ALS pathogenesis. Mol. Cell 80, 876–891. doi:10.1016/j.molcel.2020.10.032 | |
Mateju, D., Eichenberger, B., Voigt, F., Eglinger, J., Roth, G., and Chao, J. A. (2020). Single-molecule imaging reveals translation of mRNAs localized to stress granules. Cell 183, 1801–1812. doi:10.1016/j.cell.2020.11.010 | |
Mateju, D., Franzmann, T. M., Patel, A., Kopach, A., Boczek, E. E., Maharana, S., et al. (2017). An aberrant phase transition of stress granules triggered by misfolded protein and prevented by chaperone function. EMBO J. 36, 1669–1687. doi:10.15252/embj.201695957 | |
Matheny, T., Rao, B. S., and Parker, R. (2019). Transcriptome-Wide comparison of stress granules and P-bodies reveals that translation plays a major role in RNA partitioning. Mol. Cell. Biol. 39, e00313-19. doi:10.1128/MCB.00313-19 | |
Mathieu, C., Pappu, R. V., and Taylor, J. P. (2020). Beyond aggregation: Pathological phase transitions in neurodegenerative disease. Science 370, 56–60. doi:10.1126/science.abb8032 | |
Matos, C. A., De Almeida, L. P., and Nobrega, C. (2019). Machado-joseph disease/spinocerebellar ataxia type 3: Lessons from disease pathogenesis and clues into therapy. J. Neurochem. 148, 8–28. doi:10.1111/jnc.14541 | |
Maxwell, B. A., Gwon, Y., Mishra, A., Peng, J., Nakamura, H., Zhang, K., et al. (2021). Ubiquitination is essential for recovery of cellular activities after heat shock. Science 372, eabc3593. doi:10.1126/science.abc3593 | |
Mittag, T., and Pappu, R. V. (2022). A conceptual framework for understanding phase separation and addressing open questions and challenges. Mol. Cell 82, 2201–2214. doi:10.1016/j.molcel.2022.05.018 | |
Molliex, A., Temirov, J., Lee, J., Coughlin, M., Kanagaraj, A. P., Kim, H. J., et al. (2015). Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 163, 123–133. doi:10.1016/j.cell.2015.09.015 | |
Moon, S. L., Morisaki, T., Stasevich, T. J., and Parker, R. (2020). Coupling of translation quality control and mRNA targeting to stress granules. J. Cell Biol. 219, e202004120. doi:10.1083/jcb.202004120 | |
Murray, D. T., Kato, M., Lin, Y., Thurber, K. R., Hung, I., Mcknight, S. L., et al. (2017). Structure of FUS protein fibrils and its relevance to self-assembly and phase separation of low-complexity domains. Cell 171, 615–627. doi:10.1016/j.cell.2017.08.048 | |
Niaki, A. G., Sarkar, J., Cai, X., Rhine, K., Vidaurre, V., Guy, B., et al. (2020). Loss of dynamic RNA interaction and aberrant phase separation induced by two distinct types of ALS/FTD-Linked FUS mutations. Mol. Cell 77, 82–94. doi:10.1016/j.molcel.2019.09.022 | |
Niranjanakumari, S., Lasda, E., Brazas, R., and Garcia-Blanco, M. A. (2002). Reversible cross-linking combined with immunoprecipitation to study RNA-protein interactions in vivo. Methods 26, 182–190. doi:10.1016/S1046-2023(02)00021-X | |
Nomura, T., Watanabe, S., Kaneko, K., Yamanaka, K., Nukina, N., and Furukawa, Y. (2014). Intranuclear aggregation of mutant FUS/TLS as a molecular pathomechanism of amyotrophic lateral sclerosis. J. Biol. Chem. 289, 1192–1202. doi:10.1074/jbc.M113.516492 | |
Odeh, H. M., Fare, C. M., and Shorter, J. (2022). Nuclear-import receptors counter deleterious phase transitions in neurodegenerative disease. J. Mol. Biol. 434, 167220. doi:10.1016/j.jmb.2021.167220 | |
Padron, A., Iwasaki, S., and Ingolia, N. T. (2019). Proximity RNA labeling by APEX-seq reveals the organization of translation initiation complexes and repressive RNA granules. Mol. Cell 75, 875–887. doi:10.1016/j.molcel.2019.07.030 | |
Patel, A., Lee, H. O., Jawerth, L., Maharana, S., Jahnel, M., Hein, M. Y., et al. (2015). A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell 162, 1066–1077. doi:10.1016/j.cell.2015.07.047 | |
Pushpalatha, K. V., Solyga, M., Nakamura, A., and Besse, F. (2022). RNP components condense into repressive RNP granules in the aging brain. Nat. Commun. 13, 2782. doi:10.1038/s41467-022-30066-4 | |
Ray, S., Singh, N., Kumar, R., Patel, K., Pandey, S., Datta, D., et al. (2020). α-Synuclein aggregation nucleates through liquid–liquid phase separation. Nat. Chem. 12, 705–716. doi:10.1038/s41557-020-0465-9 | |
Reber, S., Jutzi, D., Lindsay, H., Devoy, A., Mechtersheimer, J., Levone, B. R., et al. (2021). The phase separation-dependent FUS interactome reveals nuclear and cytoplasmic function of liquid-liquid phase separation. Nucleic Acids Res. 49, 7713–7731. doi:10.1093/nar/gkab582 | |
Reineke, L. C., and Neilson, J. R. (2019). Differences between acute and chronic stress granules, and how these differences may impact function in human disease. Biochem. Pharmacol. 162, 123–131. doi:10.1016/j.bcp.2018.10.009 | |
Rhine, K., Dasovich, M., Yoniles, J., Badiee, M., Skanchy, S., Ganser, L. R., et al. (2022a). Poly(ADP-ribose) drives condensation of FUS via a transient interaction. Mol. Cell 82, 969–985.e11. e911. doi:10.1016/j.molcel.2022.01.018 | |
Rhine, K., Makurath, M. A., Liu, J., Skanchy, S., Lopez, C., Catalan, K. F., et al. (2020a). ALS/FTLD-Linked mutations in FUS Glycine residues cause accelerated gelation and reduced interactions with wild-type FUS. Mol. Cell 80, 666–681. doi:10.1016/j.molcel.2020.10.014 | |
Rhine, K., Skanchy, S., and Myong, S. (2022b). Single-molecule and ensemble methods to probe RNP nucleation and condensate properties. Methods 197, 74–81. doi:10.1016/j.ymeth.2021.02.012 | |
Rhine, K., Vidaurre, V., and Myong, S. (2020b). RNA droplets. Annu. Rev. Biophys. 49, 247–265. doi:10.1146/annurev-biophys-052118-115508 | |
Roberson, E. D., Scearce-Levie, K., Palop, J. J., Yan, F., Cheng, I. H., Wu, T., et al. (2007). Reducing endogenous tau ameliorates amyloid beta-induced deficits in an Alzheimer's disease mouse model. Science 316, 750–754. doi:10.1126/science.1141736 | |
Roden, C., and Gladfelter, A. S. (2021). RNA contributions to the form and function of biomolecular condensates. Nat. Rev. Mol. Cell Biol. 22, 183–195. doi:10.1038/s41580-020-0264-6 | |
Roux, K. J., Kim, D. I., Raida, M., and Burke, B. (2012). A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol. 196, 801–810. doi:10.1083/jcb.201112098 | |
Ryu, H. H., Jun, M. H., Min, K. J., Jang, D. J., Lee, Y. S., Kim, H. K., et al. (2014). Autophagy regulates amyotrophic lateral sclerosis-linked fused in sarcoma-positive stress granules in neurons. Neurobiol. Aging 35, 2822–2831. doi:10.1016/j.neurobiolaging.2014.07.026 | |
Sanchez-Burgos, I., Espinosa, J. R., Joseph, J. A., and Collepardo-Guevara, R. (2022). RNA length has a non-trivial effect in the stability of biomolecular condensates formed by RNA-binding proteins. PLoS Comput. Biol. 18, e1009810. doi:10.1371/journal.pcbi.1009810 | |
Schwartz, J. C., Wang, X., Podell, E. R., and Cech, T. R. (2013). RNA seeds higher-order assembly of FUS protein. Cell Rep. 5, 918–925. doi:10.1016/j.celrep.2013.11.017 | |
Shen, H., Yanas, A., Owens, M. C., Zhang, C., Fritsch, C., Fare, C. M., et al. (2022a). Sexually dimorphic RNA helicases DDX3X and DDX3Y differentially regulate RNA metabolism through phase separation. Mol. Cell 82, 2588–2603.e9. doi:10.1016/j.molcel.2022.04.022 | |
Shen, Y., Chen, A., Wang, W., Shen, Y., Ruggeri, F. S., Aime, S., et al. (2022b). Solid/liquid coexistence during aging of FUS condensates. bioRxiv 2008, 503964. doi:10.1101/2022.08.15.503964 |
Sleigh, J. N., Tosolini, A. P., Gordon, D., Devoy, A., Fratta, P., Fisher, E. M. C., et al. (2020). Mice carrying ALS mutant TDP-43, but not mutant FUS, display in vivo defects in axonal transport of signaling endosomes. Cell Rep. 30, 3655–3662. doi:10.1016/j.celrep.2020.02.078 | |
Sreedharan, J., Blair, I. P., Tripathi, V. B., Hu, X., Vance, C., Rogelj, B., et al. (2008). TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science 319, 1668–1672. doi:10.1126/science.1154584 | |
Thedieck, K., Holzwarth, B., Prentzell, M. T., Boehlke, C., Klasener, K., Ruf, S., et al. (2013). Inhibition of mTORC1 by astrin and stress granules prevents apoptosis in cancer cells. Cell 154, 859–874. doi:10.1016/j.cell.2013.07.031 | |
Tourriere, H., Chebli, K., Zekri, L., Courselaud, B., Blanchard, J. M., Bertrand, E., et al. (2003). The RasGAP-associated endoribonuclease G3BP assembles stress granules. J. Cell Biol. 160, 823–831. doi:10.1083/jcb.200212128 | |
Trcek, T., Douglas, T. E., Grosch, M., Yin, Y., Eagle, W. V. I., Gavis, E. R., et al. (2020). Sequence-independent self-assembly of germ granule mRNAs into homotypic clusters. Mol. Cell 78, 941–950. doi:10.1016/j.molcel.2020.05.008 | |
Uchihara, T., Fujigasaki, H., Koyano, S., Nakamura, A., Yagishita, S., and Iwabuchi, K. (2001). Non-expanded polyglutamine proteins in intranuclear inclusions of hereditary ataxias – triple-labeling immunofluorescence study. Acta Neuropathol. 102, 149–152. doi:10.1007/s004010100364 | |
Van Nostrand, E. L., Pratt, G. A., Shishkin, A. A., Gelboin-Burkhart, C., Fang, M. Y., Sundararaman, B., et al. (2016). Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP). Nat. Methods 13, 508–514. doi:10.1038/nmeth.3810 | |
Van Treeck, B., Protter, D. S. W., Matheny, T., Khong, A., Link, C. D., and Parker, R. (2018). RNA self-assembly contributes to stress granule formation and defining the stress granule transcriptome. Proc. Natl. Acad. Sci. U. S. A. 115, 2734–2739. doi:10.1073/pnas.1800038115 | |
Vance, C., Rogelj, B., Hortobagyi, T., De Vos, K. J., Nishimura, A. L., Sreedharan, J., et al. (2009). Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science 323, 1208–1211. doi:10.1126/science.1165942 | |
Vanderweyde, T., Yu, H., Varnum, M., Liu-Yesucevitz, L., Citro, A., Ikezu, T., et al. (2012). Contrasting pathology of the stress granule proteins TIA-1 and G3BP in tauopathies. J. Neurosci. 32, 8270–8283. doi:10.1523/JNEUROSCI.1592-12.2012 | |
Wegmann, S., Eftekharzadeh, B., Tepper, K., Zoltowska, K. M., Bennett, R. E., Dujardin, S., et al. (2018). Tau protein liquid-liquid phase separation can initiate tau aggregation. EMBO J. 37, e98049. doi:10.15252/embj.201798049 | |
Weis, K., and Hondele, M. (2022). The role of DEAD-box ATPases in gene expression and the regulation of RNA-protein condensates. Annu. Rev. Biochem. 91, 197–219. doi:10.1146/annurev-biochem-032620-105429 | |
Wheeler, J. R., Matheny, T., Jain, S., Abrisch, R., and Parker, R. (2016). Distinct stages in stress granule assembly and disassembly. Elife 5, e18413. doi:10.7554/eLife.18413 | |
Wollny, D., Vernot, B., Wang, J., Hondele, M., Safrastyan, A., Aron, F., et al. (2022). Characterization of RNA content in individual phase-separated coacervate microdroplets. Nat. Commun. 13, 2626. doi:10.1038/s41467-022-30158-1 | |
Wolozin, B., and Ivanov, P. (2019). Stress granules and neurodegeneration. Nat. Rev. Neurosci. 20, 649–666. doi:10.1038/s41583-019-0222-5 | |
Yang, P., Mathieu, C., Kolaitis, R. M., Zhang, P., Messing, J., Yurtsever, U., et al. (2020). G3BP1 is a tunable switch that triggers phase separation to assemble stress granules. Cell 181, 325–345. doi:10.1016/j.cell.2020.03.046 | |
Yoshizawa, T., Ali, R., Jiou, J., Fung, H. Y. J., Burke, K. A., Kim, S. J., et al. (2018). Nuclear import receptor inhibits phase separation of FUS through binding to multiple sites. Cell 173, 693–705. doi:10.1016/j.cell.2018.03.003 | |
Youn, J. Y., Dunham, W. H., Hong, S. J., Knight, J. D. R., Bashkurov, M., Chen, G. I., et al. (2018). High-density proximity mapping reveals the subcellular organization of mRNA-associated granules and bodies. Mol. Cell 69, 517–532. e511. doi:10.1016/j.molcel.2017.12.020 | |
Zacco, E., Grana-Montes, R., Martin, S. R., De Groot, N. S., Alfano, C., Tartaglia, G. G., et al. (2019). RNA as a key factor in driving or preventing self-assembly of the TAR DNA-binding protein 43. J. Mol. Biol. 431, 1671–1688. doi:10.1016/j.jmb.2019.01.028 | |
Zhang, K., Daigle, J. G., Cunningham, K. M., Coyne, A. N., Ruan, K., Grima, J. C., et al. (2018). Stress granule assembly disrupts nucleocytoplasmic transport. Cell 173, 958–971. doi:10.1016/j.cell.2018.03.025 | |
Zhang, K., Donnelly, C. J., Haeusler, A. R., Grima, J. C., Machamer, J. B., Steinwald, P., et al. (2015). The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature 525, 56–61. doi:10.1038/nature14973 | |
Zhang, P., Fan, B., Yang, P., Temirov, J., Messing, J., Kim, H. J., et al. (2019). Chronic optogenetic induction of stress granules is cytotoxic and reveals the evolution of ALS-FTD pathology. Elife 8, e39578. doi:10.7554/eLife.39578 | |
Zhu, L., Xu, M., Yang, M., Yang, Y., Li, Y., Deng, J., et al. (2014). An ALS-mutant TDP-43 neurotoxic peptide adopts an anti-parallel beta-structure and induces TDP-43 redistribution. Hum. Mol. Genet. 23, 6863–6877. doi:10.1093/hmg/ddu409 | |
Keywords: neurodegeneration, RNA granules, stress granules, RNA, liquid-liquid phase separation
Citation: Rhine K, Al-Azzam N, Yu T and Yeo GW (2022) Aging RNA granule dynamics in neurodegeneration. Front. Mol. Biosci. 9:991641. doi: 10.3389/fmolb.2022.991641
Received: 11 July 2022; Accepted: 22 August 2022;
Published: 16 September 2022.
Edited by:
Natalie Gilks Farny, Worcester Polytechnic Institute, United StatesReviewed by:
Pavel Ivanov, Brigham and Women’s Hospital, Harvard Medical School, United StatesEric D. Ross, Colorado State University, United States
Copyright © 2022 Rhine, Al-Azzam, Yu and Yeo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gene W. Yeo, Z2VuZXllb0B1Y3NkLmVkdQ==
 Kevin Rhine
Kevin Rhine Norah Al-Azzam1,2,3,4
Norah Al-Azzam1,2,3,4 Tao Yu
Tao Yu