
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mol. Biosci., 06 September 2022
Sec. Protein Biochemistry for Basic and Applied Sciences
Volume 9 - 2022 | https://doi.org/10.3389/fmolb.2022.989851
This article is part of the Research TopicADAM, ADAMTS and Astacin Proteases: Challenges and Breakthroughs in the -Omics Era - Volume IIView all 8 articles
 Lauren K. Wareham1†
Lauren K. Wareham1† Amy E. Whitener1†
Amy E. Whitener1† Hang-Jing Wu1†
Hang-Jing Wu1† Shu-Yu Wu2
Shu-Yu Wu2 Hassane S. Mchaourab2
Hassane S. Mchaourab2 Douglas P. Mortlock2
Douglas P. Mortlock2 Rachel W. Kuchtey1,2
Rachel W. Kuchtey1,2 John Kuchtey1*
John Kuchtey1*Although mutations in ADAMTS10 have long been known to cause autosomal recessive Weill-Marchesani Syndrome which is characterized by short stature and ocular abnormalities, more recent work has shown that certain mutations in ADAMTS10 cause glaucoma in dogs. In humans, glaucoma is the leading cause of irreversible vision loss that affects tens of millions of people world-wide. Vision loss in glaucoma is a result of neurodegeneration of retinal ganglion cells that form the inner-most layer of the retina and whose axons form the optic nerve which relays visual information to the brain. ADAMTS10 contributes to the formation of microfibrils which sequester latent transforming growth factor β (TGFβ). Among its many biological functions, TGFβ promotes the development of retinal ganglion cells and is also known to play other roles in glaucoma pathogenesis. The aim of this study was to test the hypothesis that ADAMTS10 plays a role in retinal ganglion cell development through regulation of TGFβ signaling. To this end, Adamts10 expression was targeted for reduction in zebrafish embryos carrying either a fluorescent reporter that labels retinal ganglion cells, or a fluorescent reporter of pSmad3-mediated TGFβ family signaling. Loss of adamts10 function in zebrafish embryos reduced retinal ganglion cell reporter fluorescence and prevented formation of an ordered retinal ganglion cell layer. Targeting adamts10 expression also drastically reduced constitutive TGFβ signaling in the eye. Direct inhibition of the TGFβ receptor reduced retinal ganglion cell reporter fluorescence similar to the effect of targeting adamts10 expression. These findings unveil a previously unknown role for Adamts10 in retinal ganglion cell development and suggest that the developmental role of Adamts10 is mediated by active TGFβ family signaling. In addition, our results show for the first time that Adamts10 is necessary for pSmad3-mediated constitutive TGFβ family signaling.
Members of the ADAMTS family of secreted metalloproteases are highly conserved across vertebrate species and contribute to a variety of biological processes, including remodeling of extracellular matrix and development (Apte, 2009; Brunet et al., 2015; Kelwick et al., 2015). Previously, we identified a G661R mutation in ADAMTS10 as disease-causative for a colony of Beagle dogs with autosomal recessive inheritance of glaucoma (Kuchtey et al., 2011). Protein structure modeling of the G661R mutation predicts disruption of normal ADAMTS10 folding (Kuchtey et al., 2011), though this does not prevent secretion of ADAMTS10 (unpublished data). Involvement of ADAMTS10 with glaucoma has been verified in another dog breed and extended to include ADAMTS17 which is structurally and functionally highly similar to ADAMTS10 (Kuchtey et al., 2013; Ahonen et al., 2014; Forman et al., 2015; Oliver et al., 2015; Karoulias et al., 2020b). In humans, glaucoma is the leading cause of irreversible vision loss affecting tens of millions of people world-wide. Vision loss in glaucoma is a result of neurodegeneration of retinal ganglion cells (RGCs) that form the inner-most layer of the retina and whose axons form the optic nerve which relays visual information to the brain (Calkins, 2012; Jonas et al., 2017).
In addition to glaucoma, autosomal recessive mutations in ADAMTS10 cause Weill-Marchesani Syndrome (WMS), a rare connective tissue disorder characterized by short stature and ocular abnormalities including glaucoma (Faivre et al., 2003a; Dagoneau et al., 2004; Kutz et al., 2008). A clinically indistinguishable autosomal dominant form of WMS is caused by mutations in the fibrillin-1 gene (FBN1) (Faivre et al., 2003a; Faivre et al., 2003b; Sengle et al., 2012), suggesting overlapping function of ADAMTS10 and FBN1 proteins (Apte, 2009; Karoulias et al., 2020b). FBN1 assembles to form microfibrils, which are widely expressed components of the extracellular matrix that contribute to tissue biomechanics and are essential for the formation of elastic fibers (Ramirez and Sakai, 2010; Kielty, 2017). Microfibrils also act as extracellular storage sites for latent TGFβ, thereby controlling its activation and bioavailability (Charbonneau et al., 2004; Ramirez and Rifkin, 2009). Mutations in the genes encoding LTBP2 and ADAMTS17, both of which bind fibrillin-1, also cause WMS (Haji-Seyed-Javadi et al., 2012; Karoulias et al., 2020a). ADAMTS10 has been shown to bind FBN1 with high affinity and to colocalize with microfibrils and promote their formation (Kutz et al., 2011; Hubmacher and Apte, 2015; Cain et al., 2016; Matsuzaki et al., 2020). Recent studies with Adamts10 mutant mice have shown that ADAMTS10 plays a role in a developmental switch of microfibrils from an immature form with predominant fibrillin-2 immunoreactivity, to a mature form with predominant fibrillin-1 immunoreactivity (Mularczyk et al., 2018; Wang et al., 2019; Wu et al., 2021). Mutations in FBN1 cause Marfan Syndrome, for which defective microfibril formation is a key component of pathogenesis, along with dysregulation of TGFβ signaling (Sakai et al., 2016). Therefore, disruption of microfibrils can lead to dysregulation of TGFβ signaling, which may have detrimental consequences that cause disease. TGFβ plays a role in neuronal development, including in the retina, where it participates in axonal growth and protects against apoptosis (Dunker et al., 2001; Braunger et al., 2013; Carrella et al., 2015; Tachibana et al., 2016).
ADAMTS10 is abundantly expressed in the developing mouse eye and up-regulation of adamts10 mRNA has been shown in the developing zebrafish embryo (Brunet et al., 2015; Wang et al., 2019). In the present study, we found that targeting adamts10 expression in zebrafish embryos using a morpholino oligonucleotide (MO) resulted in abnormal RGC development accompanied by a large reduction in endogenous pSmad3-mediated TGFβ family signaling. Similar defective RGC development was induced by inhibition of TGFβ receptors. Our results suggest that Adamts10 plays a role in RGC development that is mediated by TGFβ family signaling.
Adult zebrafish (Danio rerio) were maintained in aquatic housing units on a monitored recirculating system at 26°C with standard light/dark (14/10 h) cycle. Experiments were performed using embryos from outcrosses of wildtype AB strain to a transgenic reporter line that expresses GFP under control of a pou4f1 (brn3a) enhancer element, Tg (pou4f1-hsp70l:GFP)rw0110bTg formerly Tg (brn3a-hsp70l:GFP)rw0110bTg provided by Dr. Takanori Hamaoka and Dr. Hitoshi Okamoto of the Riken Brain Institute through the European Zebrafish Resource Center (Aizawa et al., 2005) or to a transgenic reporter line that expresses GFP in response to activation of a Smad3 binding element, Tg(12xSBE:EGFP)ia16Tg, generated by Dr. Francesco Argenton, Universita di Padova, obtained through the European Zebrafish Resource Center (Casari et al., 2014). Experiments using zebrafish embryos were conducted in accordance with the Association for Research in Vision and Ophthalmology statement for the Use of Animals in Ophthalmic and Vision Research and the Institutional Animal are and Use Committee of Vanderbilt University.
Zebrafish embryos or adult eyes were embedded in 20% sucrose/OCT and 7 μm-thick cryosections were made as described below for immunohistochemistry. RNA In Situ Hybridization was carried out on cryosections by the RNAscope technique (Wang et al., 2012) using a custom designed probe set for zebrafish adamts10 mRNA and proprietary amplification reagents (mRNA in situ RNAscope 2.5 Chromogenic Assay, Advanced Cell Diagnostics, Hayward, CA) following the manufacturer’s protocol. The adamts10 probe set was checked to avoid cross detection of the adamts6 which has high homology with adamts10. A probe set for DapB, a bacterial gene, was used as negative control. Briefly, embryo sections were post-fixed in 4% PFA/PBS for 15 min at 4°C, dehydrated through gradient ethanol then treated with hydrogen peroxide for 10 min at room temperature. Target retrieval was conducted by submerging slides in retrieval solution for 5 min at 100°C. Following 30 min protease plus treatment at 40°C in a hybridization oven (HybEZ II Hybridization System, Advanced Cell Diagnostics), samples were hybridized with probes, subjected to signal amplification steps and then reacted with Fast Red. Bright field images were captured using a Nikon Eclipse microscope equipped with a ×40 objective.
Injections of 1–2 nl were made into the yoke adjacent to the embryo at the one-cell stage at approximately 12:00 p.m. To visualize injections, 0.025% phenol red was included in the injection solution. After injection, embryos were incubated at 29°C in egg water (Instant Ocean, Blacksburg, VA). At approximately 10:00 a.m. on post-injection days 1, 2 or 3 (referred to from here on as 24, 48, and 72 h post-fertilization (hpf), embryos were euthanized in 300 mg/L tricaine methanesulfonate, then fixed in 4% PFA/PBS. For fluorescence microscopy, 0.2 mM phenylthiourea (PTU) was added post-injection to prevent pigmentation.
MO obtained from Gene Tools (Philomath, OR), were reconstituted to 1 mM with RNase/DNase-free water and stored at room temperature as suggested by the manufacturer. Translation-blocking MO (5′-CCAAACTCCTCCACACCGTTTCCAT-3′) targeting the translation initiation complex of adamts10 mRNA (referred to from here on as adamts10 MO) was injected at 4–5 ng/embryo. Human ADAMTS10 mRNA was transcribed from expression plasmids described previously (Kuchtey et al., 2011) using the in vitro T7 ultra mMessage mMachine™ kit with poly-A tail addition (Thermo Fisher Scientific). For MO rescue experiments, embryos were co-injected with 4 ng adamts10 MO with 400 ng of either normal or G661R mutated human ADAMTS10 mRNA.
To block TGFβ signaling, 100 µM SB431542 (Sigma), a concentration previously shown to drastically reduce reporter fluorescence of Tg(12xSBE:EGFP) embryos within 8 h after treatment (Casari et al., 2014), or vehicle control (DMSO) was added at 11 hpf to wells of 12 well-plates containing 10–12 GFP-positive Tg (pou4f1-hsp70l:GFP) or Tg(12xSBE:EGFP) embryos. This treatment protocol did not induce gross disruption of eye morphology, though treated embryos were slightly smaller than untreated (Supplementary Figure S1). At 48 hpf, embryos were euthanized in 300 mg/L tricaine methanesulfonate, fixed overnight in 4% PFA and stored in PBS at 4°C until examination by fluorescence microscopy.
GFP-positive embryos were dechorionated, fixed in 4% PFA/PBS for 2 h at room temperature or at 4°C overnight, then stored in PBS at 4°C. For imaging, embryos were placed ventral side up in 3% (w/v) methyl cellulose and fluorescence and brightfield images taken with a ×4 objective with a Nikon AZ100M fluorescence microscope.
Embryos were embedded in 20% sucrose in optimal cutting temperature compound (OCT) as described by Barthel and Raymond. (1990) with modifications as follows: euthanized embryos were dechorionated then fixed in 4% PFA/5% sucrose in PBS for 1 h then incubated in increasing concentrations of sucrose (10%, 12.5%, and 15%) in PBS for 1 h and finally transferred to 20% sucrose/PBS and incubated overnight at 4°C. To facilitate visualization of embryos during orientation and sectioning, the 20% sucrose/PBS contained 0.002% toluidine blue. Embryos were then embedded and frozen in a 2:1 solution of 20% sucrose in OCT. Transverse cryosections 7 µm thick were cut with a CryoStar NX50 cryostat (Thermo Fisher Scientific), placed on glass slides and stored at -80°C until use.
For immunostaining, cryosections from the center of the eye were rehydrated with PBS before addition of blocking buffer (5% normal donkey serum/0.05% Triton X-100/PBS) for 1 h. Blocking buffer was removed and primary antibody solution in incubation buffer (3% normal donkey serum/0.05% triton X-100/PBS) was added. The antibodies used were 1:1,000 rabbit anti-GFP (Torrey Pines Biolabs) and 1:25 mouse anti-Isl1 (clone 40.2D6, Development Studies Hybridoma Bank, University of Iowa). After overnight incubation at 4°C in a humidified chamber, sections were washed 3 times in PBS for 10 min and incubated in 1:1,000 Alexa Fluor-donkey-anti-mouse-546 and 1:1,000 Alexa Fluor-donkey-anti-rabbit-488 (Invitrogen) in secondary incubation buffer (1% normal donkey serum/0.05% Triton X-100/PBS) for 1 h. Sections were then washed 3 times with PBS and mounted with Prolong Gold mounting fluid with DAPI (Invitrogen).
Integrated fluorescence density of fluorescent embryo images, defined as the product of the area of the region of interest (ROI) and the mean gray value, was measured by drawing an ROI around the retina using the ImageJ polygon tool. 3D surface plots were generated from single-channel GFP images using the NIH ImageJ interactive 3D surface plot plug-in (http://rsb.info.nih.gov/ij/plugins/surface-plot-3d.html). Statistical analyses were performed in Graphpad Prism. Statistical tests and sample sizes are indicated in the figure legends.
Expression of adamts10 mRNA in the developing zebrafish embryos and in the adult zebrafish eye has been shown in a previous RT-PCR study (Brunet et al., 2015). By RNAscope in situ hybridization (Advanced Cell Diagnostics), we found adamts10 mRNA expression in the eye of zebrafish embryos at 24, 48, and 72 hpf (Figure 1A) as well as in adult zebrafish 2 years of age (Figure 1B). Expression of adamts10 mRNA was found along the retinal pigment epithelium and dispersed within the whole retina at 24, 48, and 72 hpf. In the eyes of 72 hpf embryos, robust signal extending from the inner retina and traversing the retinal pigment epithelium was suggestive of expression of adamts10 in the optic nerves. In 2-year-old adult eyes, adamts10 mRNA was found in the outer nuclear layer, inner nuclear layer and ganglion cell layer. Given the retinal expression of adamts10, we chose to target its expression for reduction to investigate its role in RGC development.
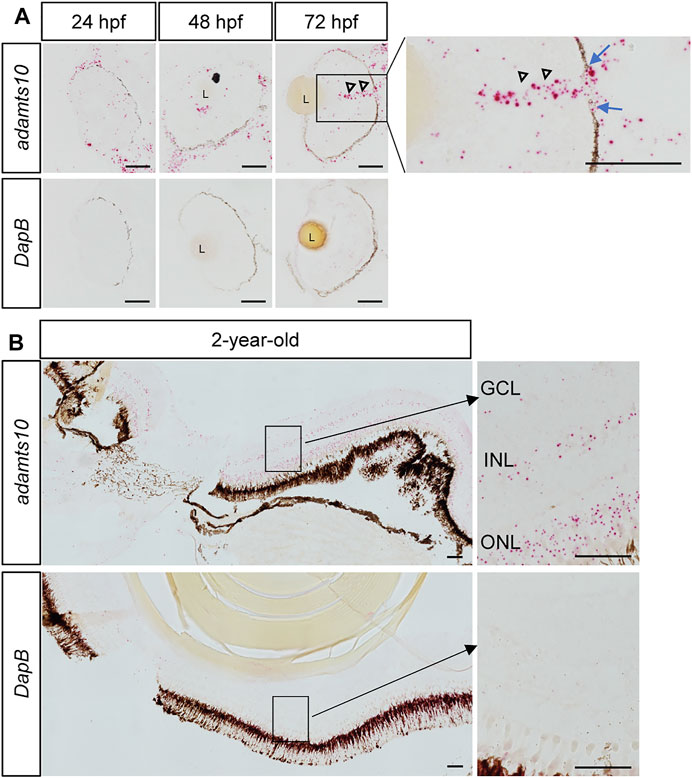
FIGURE 1. Ocular expression of adamts10 mRNA in zebrafish. (A) Expression of adamts10 mRNA (red dots, top row) was observed along the retinal pigment epithelium (dark brown/black structure) and dispersed throughout the retina of embryos at 24, 48, and 72 hpf. In 72 hpf eyes, robust signal (open arrowheads) extending from the inner retina and traversing a gap in the retinal pigmented epithelium (blue arrows, insert) is suggestive of optic nerve expression of adamts10 mRNA. (B) In 2-year-old adult eyes, expression of adamts10 mRNA was found in the outer nuclear layer (ONL), inner nuclear layer (INL), and ganglion cell layer (GCL). Hybridization probe against DapB, a bacterial gene, served as negative control on adjacent sections (bottom rows, A and B). Scale bar: 50 µm. L, Lens.
Within the retina, Pou4f1 (also known as brn3a) is specifically expressed in ∼80% of developing and differentiated RGCs (Nguyen-Ba-Charvet and Rebsam, 2020). We used the Tg(pou4f1-hsp70l:GFP) line of zebrafish, which expresses GFP driven by a pou4f1 enhancer element (Aizawa et al., 2005; Sato et al., 2015) to test the hypothesis that Adamts10 plays a role in RGC development. Embryos were injected at the one cell stage with adamts10 MO or left uninjected. At 48 or 72 hpf, embryos were imaged to evaluate GFP fluorescence (Figure 2). In uninjected embryos at 48 hpf, GFP strongly labeled the retina and the optic nerve, which is composed of RGC axons (Figure 2A). As shown in 3D surface plots, GFP fluorescence in uninjected embryos appeared uniform in intensity along the retina, with faint fluorescence in the optic nerves (Figure 2B). However, in embryos injected with adamts10 MO, GFP fluorescence was reduced and uneven, exhibiting patches of high and low intensity with barely visible optic nerves (Figures 2C,D). At 48 hpf, retinal fluorescence was reduced 50.4% in adamts10 MO-injected embryos as compared to uninjected controls (p < 0.0001, Figure 2E). Decreased fluorescence was still evident at 72 hpf, although not as reduced as at 48 hpf, possibly due to dilution of MO with cell divisions, with a 26% reduction compared to uninjected control (p < 0.0001, Figure 2F). The reduction in GFP expression driven by a pou4f1 enhancer suggests that injecting embryos with adamts10 MO resulted in fewer RGCs and/or altered RGC development.
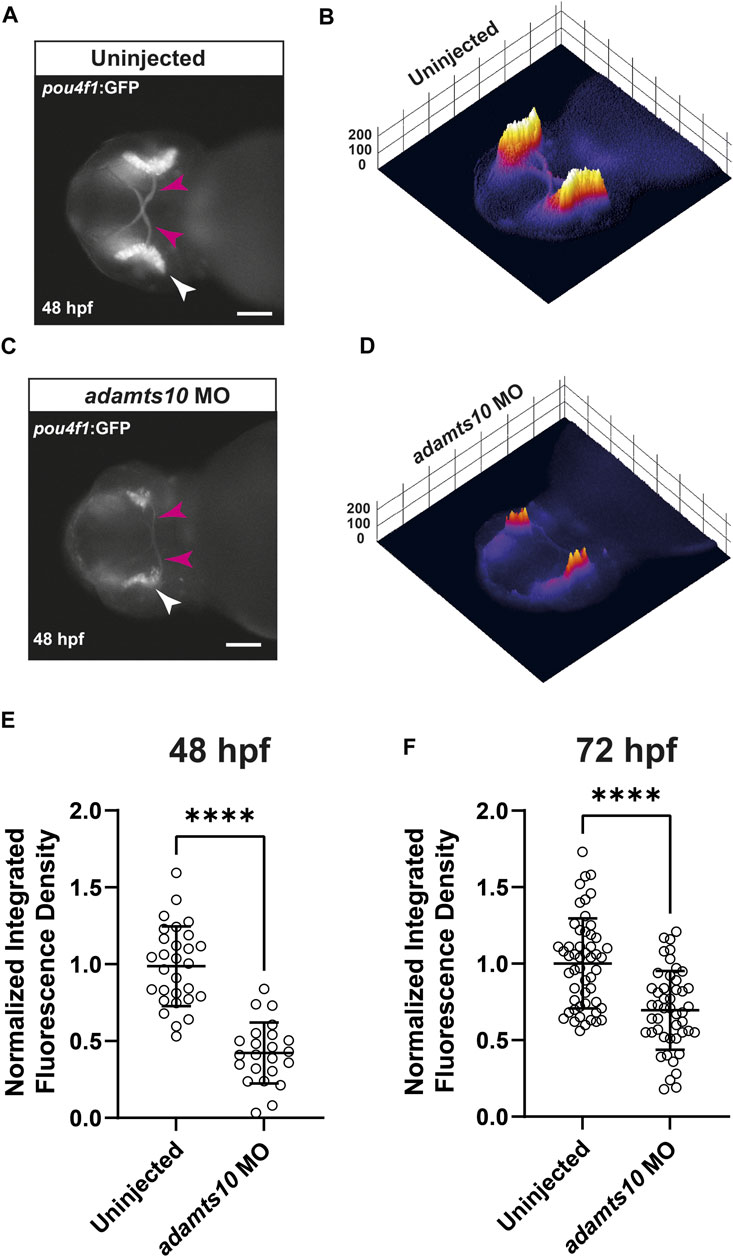
FIGURE 2. Adamts10 MO reduces pou4f1 enhancer-driven GFP fluorescence in zebrafish retinae. Representative fluorescent images of an uninjected (A) and an adamts10 MO-injected (C) Tg(pou4f1-hsp70l:GFP) embryo at 48 hpf showing pou4f1 enhancer-driven GFP fluorescence in the retina (white arrows) and optic nerves (magenta arrows, scale = 100 µm), with corresponding 3D surface plots of fluorescence intensity (B,D). (E) Normalized integrated fluorescence density of uninjected and adamts10 MO-injected embryos at 48 hpf (n = 29 and 23, respectively, ****p < 0.0001, Welch’s t-test). (F) Normalized integrated fluorescence density of uninjected and adamts10 MO-injected embryos at 72 hpf (n = 52 and n = 47, respectively, ****p < 0.0001; Welch’s t-test).
To confirm that MO-induced phenotypes were specific to adamts10, rescue experiments were performed in which Tg(pou4f1-hsp70l:GFP) embryos were co-injected with adamts10 MO and human ADAMTS10 mRNA (which is not targeted by the zebrafish adamts10 MO). GFP fluorescence was compared at 48 hpf to embryos that were uninjected or injected with adamts10 MO only. In uninjected embryos, the retina displayed strong fluorescence with staining of the optic nerve consistent with labeling of RGCs (Figure 3A). Fluorescence was uniformly distributed along the retina, as shown in the 3D surface plot (Figure 3E). Injection of adamts10 MO alone resulted in a 51% decrease in GFP fluorescence in the retina compared with uninjected controls (p < 0.0001, Figure 3I), with a patchy distribution of low intensity GFP fluorescence in the retina (Figure 3F), similar to the MO experiment shown in Figure 2. Co-injection of adamts10 MO with human ADAMTS10 mRNA resulted in a milder 24% reduction of GFP intensity compared to uninjected embryos (p = 0.03), with a more uniform distribution of fluorescence intensity along the retina (Figures 3C,G,I), similar to uninjected embryos (Figures 3A,E). This partial rescue of the MO-induced retinal phenotype suggests that human ADAMTS10 mRNA compliments a deficit of endogenous adamts10 in MO-injected embryos. These results support the specificity of the MO effects for targeting of adamts10 expression.
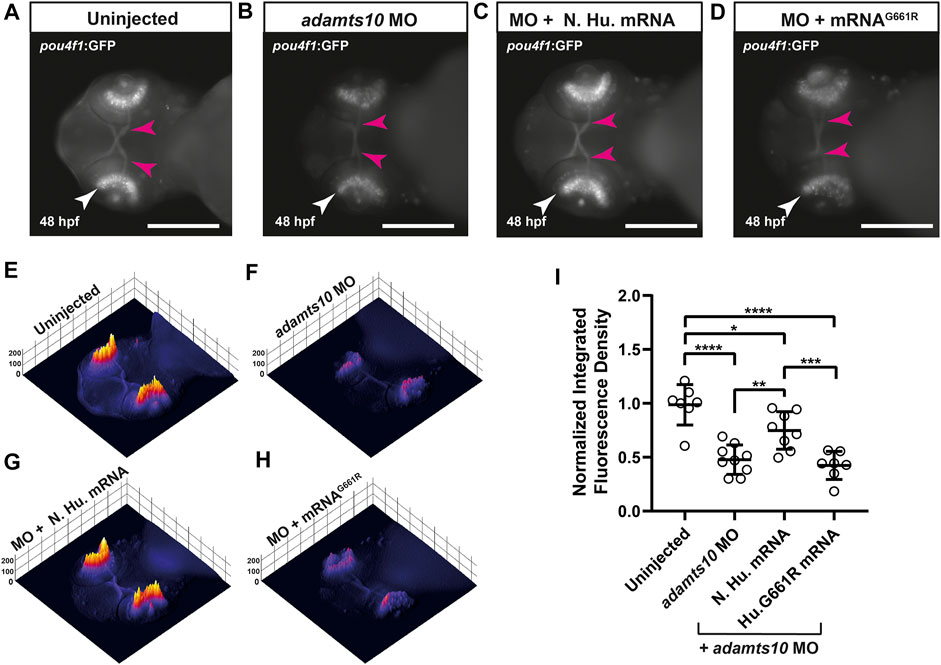
FIGURE 3. Adamts10 MO-induced reduction in pou4f1 enhancer-driven GFP expression is decreased with normal human ADAMTS10 mRNA but not ADAMTS10G661R mutant mRNA. Representative fluorescent images of a Tg(pou4f1-hsp70l:GFP) embryo uninjected (A) injected with adamts10 MO (B), co-injected with ADAMTS10 MO and normal human mRNA (C) and co-injected with adamts10 MO and ADAMTS10G661R mRNA (D), showing pou4f1 enhancer-driven fluorescence in the retina (white arrows) and optic nerve (magenta arrows); scale = 250 µm. Corresponding 3D surface plots showing fluorescence intensity for each condition (E–H). Normalized integrated fluorescence density (I) of each condition (uninjected n = 7, adamts10 MO-injected n = 9, normal human mRNA-injected n = 8 and ADAMTS10G661R mRNA n = 7, *p = 0.03, **p = 0.008,***p = 0.003, ****p < 0.0001; one-way ANOVA Tukey’s multiple comparisons test).
To determine if the glaucoma-causative G661R mutation affects the developmental function of ADAMTS10, rescue experiments were performed with G661R ADAMTS10 mRNA. Co-injection of G661R mutant mRNA did not rescue the decreased GFP fluorescence which displayed retinal fluorescence intensity and pattern of expression similar to adamts10 MO only injection, with a 56.3% reduction (p < 0.0001) in integrated fluorescence density (Figures 3D,H,I). The failure of G661R ADAMTS10 mRNA to complement targeting of endogenous adamts10 suggests that the glaucoma-causing mutation is deleterious for the developmental function of Adamts10.
To further investigate the effect of adamts10 MO on retinal development, immunohistochemistry was performed on cryosections of 72 hpf Tg(pou4f1-hsp70l:GFP) embryos using an antibody against GFP and an antibody against Isl1, which is expressed in postmitotic RGCs and is required for RGC development (Pan et al., 2008). In uninjected controls, GFP-positive cell bodies with a range of fluorescence intensities defined the ganglion cell layer with radially oriented neurite processes (Figure 4A). The inner plexiform layer was clearly delineated by GFP-positive RGC dendritic processes. The ganglion cell and inner plexiform layers presented as well-defined layers of uniform thickness and semicircular shape adjacent to the lens. Isl1 immunostaining labeled cell bodies in the ganglion cell layer that overlapped with GFP positive cells (Figures 4B,D) confirming their identity as RGCs, though it appeared that more cells were labeled for Isl1 compared to GFP. These findings confirm that pou4f1 enhancer-driven GFP specifically labels RGCs in the retina.
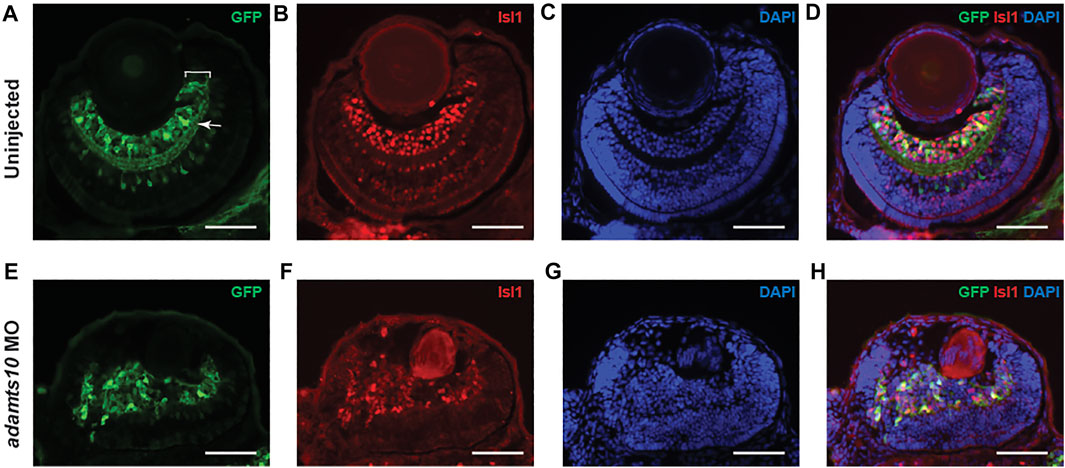
FIGURE 4. Adamts10 MO prevents proper formation of the ganglion cell layer. Representative images of stained sections from uninjected (A–D) and MO-injected (E–H) Tg(pou4f1-hsp70l:GFP) embryos at 72 hpf showing GFP staining (A,E), Isl1 staining (B,F), DAPI staining (C,G) and merged images (D,H). In A, white bracket = ganglion cell layer, white arrow = inner plexiform layer. Scale = 40 µm).
In adamts10 MO-injected embryos, rather than forming a well-defined layer, GFP fluorescence labeled disordered groups of cells near the lens that often extended towards the outer retina with randomly oriented neurite processes and no defined inner plexiform layer (Figure 4E), demonstrating defective retinal lamination. Isl1 immunostaining overlapped with GFP-positive cells (Figures 4F,H), confirming their identity as differentiated RGCs, though the Isl1 fluorescence intensity appeared to be lower than that of eyes from uninjected embryos. Co-expression of Isl1 with Pou4f1 suggests that RGC development proceeds normally to some extent. However, the lamination defect suggests an inability of RGCs to migrate to their proper location.
ADAMTS10 plays a role in microfibril structure and function (Kutz et al., 2011; Sengle et al., 2012; Mularczyk et al., 2018; Wang et al., 2019). Since microfibrils regulate localization and activation of TGFβ signaling (Charbonneau et al., 2004; Ramirez and Rifkin, 2009), we tested the hypothesis that targeting Adamts10 expression would disrupt TGFβ family signaling. For these experiments, we used the Tg(12xSBE:EGFP) line of zebrafish which expresses GFP in response to activation of a pSmad3 binding element, thereby acting as a reporter of active TGFβ superfamily signaling (Casari et al., 2014). In uninjected embryos at 48 hpf, GFP fluorescence was found throughout the retina, as well as in the brain and lens of Tg(12xSBE:EGFP) embryos (Figure 5A), indicating constitutive TGFβ superfamily signaling in those tissues. Injection of adamts10 MO resulted in a 66.6% reduction in fluorescence as compared to uninjected (Figures 5B,C, p < 0.0001). These results indicate that targeting Adamts10 expression drastically reduces constitutive pSmad3-mediated TGFβ superfamily signaling in the retina.
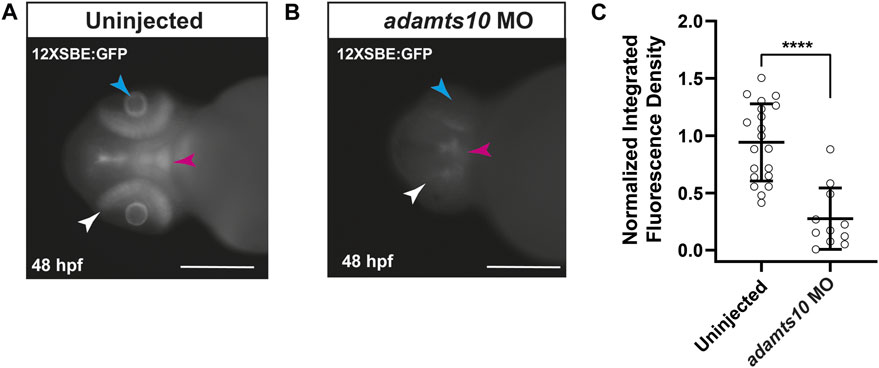
FIGURE 5. Adamts10 MO reduces pSmad3-Mediated TGFβ family signaling in zebrafish retinae. Representative fluorescent images of an uninjected (A) and adamts10 MO-injected (B) Tg(12xSBE:EGFP) embryo at 48 hpf showing fluorescence reporting of active pSmad3-mediated signaling in the retina (white arrows), brain (magenta arrows) and lens (cyan arrows, scale = 250 µm). Normalized integrated fluorescence density of uninjected, and adamts10 MO-injected Tg(12xSBE:EGFP) embryos at 48 hpf [(C), n = 21 and n = 11, respectively, ****p < 0.0001; Welch’s t-test].
The strong reduction in retinal pSmad3-mediated signaling by adamts10 MO suggested that Adamts10 may exert its effect on RGC development through suppression of TGFβ signaling. To test this hypothesis, embryos were treated with an inhibitor of the TGFβ receptor, SB431542, at 11 hpf, a time at which the optic primordium has just formed (Kimmel et al., 1995), and fluorescence examined at 48 hpf. Embryos of the Tg(12xSBE:EGFP) line treated with SB431542 showed nearly complete reduction of pSmad3-driven GFP fluorescence, verifying effective inhibition of TGFβ signaling, while vehicle control (DMSO) had no effect (Figures 6A,C). To investigate the effect of TGFβ receptor inhibition on RGC development, Tg(pou4f1-hsp70l:GFP) embryos were treated with SB431542. Treatment with SB431542, but not vehicle control, strongly reduced pou4f1 enhancer-driven GFP expression (76% reduction, p < 0.0001, Figures 6B,C), The reduction of RGC reporter fluorescence was similar to the reductions resulting from targeting adamts10 expression with MO (Figures 2, 3). These results support the hypothesis that effect of adamts10 on RGC development is mediated by TGFβ.
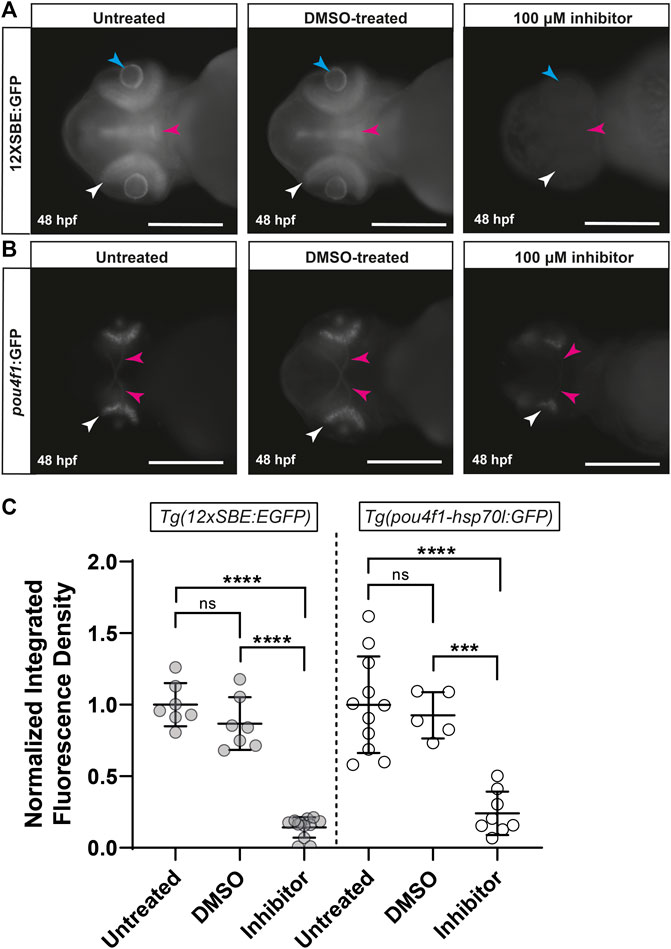
FIGURE 6. Inhibition of TGFβ signaling with SB431542 reduces pou4f1 enhancer-driven GFP expression in the retina. Representative images of untreated, DMSO-treated and SB431542-treated Tg(12xSBE:EGFP) embryos at 48 hpf (A) showing GFP fluorescence reporting of pSmad3-mediated signaling in the retina (white arrows), brain (magenta arrows) and lens (blue arrows); scale = 250 µm. Representative images of untreated, DMSO-treated and SB431542-treated Tg(pou4f1-hsp70l:GFP) embryos at 48 hpf (B) showing pou4f1 enhancer-driven GFP fluorescence in the retina (white arrows) and optic nerves (magenta arrows); scale = 250 µm. Normalized integrated fluorescence density (C) of each condition (Tg(12xSBE:EGFP); untreated: n = 7, DMSO-treated: n = 7, SB431542-treated: n = 12: ****p < 0.0001, ***p = 0.0004, ns = non-significant; Tg(pou4f1-hsp70l:GFP); untreated: n = 11, DMSO-treated: n = 5 and SB431542-treated: n = 8, ****p < 0.0001, ***p = 0.0004, ns = non-significant; one-way ANOVA Tukey’s multiple comparisons test.
A developmental role for ADAMTS10 is suggested by its association with WMS, a dysmorphic connective tissue disorder characterized by short stature and ocular phenotypes, including glaucoma (Faivre et al., 2003a). Recent studies suggest a role for ADAMTS10 in eye development by showing that Adamts10 null mice retain their hyaloid vasculature into adulthood and are defective in a developmental fibrillin isotype switch (Mularczyk et al., 2018; Wang et al., 2019; Wu et al., 2021). Our study in zebrafish revealed for the first time that ADAMTS10 plays an important role in RGC development.
Mutations in ADAMTS10 and in ADAMTS17, which is genetically and functionally highly similar to ADAMTS10 (Karoulias et al., 2020b) cause glaucoma in dogs (Kuchtey et al., 2011; Ahonen et al., 2014; Forman et al., 2015; Oliver et al., 2015). In humans, ADAMTS8 is associated with key glaucoma endophenotypes (Wain et al., 2011; Springelkamp et al., 2014; Springelkamp et al., 2017), suggesting a role for ADAMTS genes in human glaucoma. Delineation of the biological functions of ADAMTS genes may lead to identification of novel targets for treatment of glaucoma patients. Homeobox genes such as SIX6/SIX1, LMX1b and MEIS (Carnes et al., 2014; Shiga et al., 2018), which play prominent roles in development, have been shown to be associated with glaucoma. FOXC1, a gene involved in ocular development, is associated with adult onset glaucoma in addition to congenital glaucoma (Souzeau et al., 2017). Also, ATOH7, a transcription factor expressed in post-mitotic RGC precursors and necessary for their development is strongly associated with optic disc cupping, an important risk factor for developing glaucomatous neurodegeneration (Ramdas et al., 2010). Our study suggests that ADAMTS10 is another example of an ocular developmental gene that plays a role in glaucoma pathogenesis and suggests that the glaucoma-causative G661R mutation interferes with this developmental role.
Since glaucoma is defined as an optic neuropathy due to degeneration of RGC axons (Calkins, 2012), we focused on RGC development by looking at transcriptional activation of pou4f1 (also known as brn3a) which is expressed early in post-mitotic RGC precursors and plays an important role in their development (Badea et al., 2009; Nguyen-Ba-Charvet and Rebsam, 2020). RGCs are the first cell type to differentiate in the developing retina, with RGC progenitors becoming postmitotic between 27 and 28 hpf in zebrafish (Hu and Easter, 1999; Avanesov and Malicki, 2010). Using the Tg(pou4f1-hsp70l:GFP) line of zebrafish that express GFP driven by a pou4f1 enhancer element (Aizawa et al., 2005; Sato et al., 2015), we found that injecting embryos with a MO designed to block translation of adamts10 mRNA resulted in reduced GFP expression in the retina at 48 and 72 hpf (Figures 2, 3), suggesting abnormal or delayed RGC development and/or a reduction in the number of RGC progenitor cells.
Since MOs can have non-specific effects, we performed rescue experiments and found that normal human ADAMTS10 mRNA complimented the effect of co-injected adamts10 MO (Figure 3). Interestingly, ADAMTS10 mRNA carrying the glaucoma-causing G661R mutation was unable to compliment the MO effect, suggesting that this mutation interferes with the developmental function of adamts10, adding further support to the idea that defects in ocular development contribute to glaucoma pathogenesis. These results support specificity of the MO effect for targeting expression of adamts10, and support our conclusion that Adamts10 has a previously unknown role in retinal development.
In zebrafish, the RGC layer becomes discernable by 34–36 hpf and by 60 hpf, over 90% of the cells in the retina are postmitotic and the neuronal layers are established (Hu and Easter, 1999; Avanesov and Malicki, 2010). We performed immunohistochemistry on transverse sections from uninjected and adamts10 MO-injected Tg(pou4f1-hsp70l:GFP) embryos using antibodies against GFP and Isl1 which is expressed in post-mitotic RGCs and plays key roles in RGC development (Wu et al., 2015). Expression of Isl1 in MO-injected embryos suggests that RGCs were able to undergo differentiation from retinal neural progenitors. However, Isl1 and GFP immunohistochemistry showed that RGCs from retinas of adamts10 MO-injected embryos did not migrate to their proper position to form an ordered ganglion cell layer but instead formed disordered groups of cells (Figure 4). In addition, instead of forming an inner plexiform layer, RGC neuronal processes appeared randomly oriented. The disordered assembly of RGCs and lack of lamination suggests that targeting adamts10 expression may deleteriously affect apical to basal migration of RGCs preventing formation of a normal ganglion cell layer (Icha et al., 2016), resulting in a retinal lamination defect.
Hyperactivated TGFβ signaling is well established in mouse models of microfibril deficiencies due to mutations in FBN1 (Ramirez and Rifkin, 2009). Given that ADAMTS10 promotes microfibril formation (Kutz et al., 2011; Sengle et al., 2012), we hypothesized that reduction of Adamts10 expression would alter TGFβ signaling. TGFβ signal transduction is initiated by TGFβ binding to the receptors ALK5 (TβR1) and TβRII resulting in phosphorylation of Smad2 and Smad3 which form complexes with Smad4 that are translocated into the nucleus where they activate transcription (Massague, 2012). To test our hypothesis, we used the Tg(12xSBE:EGFP) line of zebrafish that expresses GFP in response to activation of a pSmad3 binding element. We found that uninjected Tg(12xSBE:EGFP) embryos expressed abundant GFP throughout the retina at 48 hpf (Figure 5), indicating constitutive pSmad3-mediated signaling. Injection of adamts10 MO caused a large decrease in pSmad3-driven fluorescence, supporting our hypothesis that reduction of Adamts10 expression alters TGFβ signaling. It is interesting to note that the large decrease in pSmad3-mediated signaling resulting from disruption of Adamts10 expression is the opposite of the hyperactivation of TGFβ signaling found in fibrillin-1 deficiencies (Neptune et al., 2003; Ng et al., 2004; Habashi et al., 2006). Our result shows for the first time a role for Adamts10 in regulating pSmad3-mediated TGFβ family signaling and suggest a requirement for normal expression of adamts10 to support constitutive TGFβ family signaling in the developing zebrafish retina.
A role for ADAMTS10 in TGFβ family signaling contrasts with the results of Mularczyk et al., who found that a premature termination mutation of Adamts10 in mice did not affect pSMAD2/3-mediated TGFβ signaling but instead reduced pSMAD1/5/8-mediated bone morphogenic protein (BMP) signaling in mouse embryo fibroblasts (Mularczyk et al., 2018). Although we did not address BMP signaling since our focus was on TGFβ due to its importance in glaucoma, the effects of ADAMTS10 deficiency could vary between tissues, developmental stages and species investigated. Another signaling role for ADAMTS10 described by Cain et al. that we did not investigate is its role in formation of focal adhesions and epithelial cell-cell junctions (Cain et al., 2016).
TGFβ is involved in neuronal development, including in the retina, in programmed cell death and in axon specification (Braunger et al., 2013; Carrella et al., 2015; Kaiser et al., 2020). This led us to form the hypothesis that the RGC developmental defect resulting from targeting Adamts10 expression is mediated by TGFβ. Consistent with our hypothesis, inhibiting the TGFβ receptor with SB431542 greatly reduced Pou4f1 enhancer-driven expression, similar to the effect of adamts10 MO (Figure 6). However, it is important to note that in addition to the Alk5 TGFβ receptor, SB431542 inhibits the activin and nodal receptors, Alk4 and Alk7 (Inman et al., 2002). Although a TGFβ-mediated effect is consistent with a role for ADAMTS10 in microfibril formation, another possibility is that the pSmad3 signaling was initiated through activation of Alk4/Alk7 by Activin/Nodal. Smad3 signaling is activated only by the TGFβ family members TGFβ, activin or nodal (Pauklin and Vallier, 2015). Our results suggest that Adamts10 mediates its effects on retinal development via a pSmad3-mediated TGFβ family pathway.
Isl1 and Pou4f1 are expressed in early post-mitotic RGCs and are known to play important roles in their differentiation (Nguyen-Ba-Charvet and Rebsam, 2020). Expression of Isl1 and Pou4f1 reporter in retinas of adamts10 MO-injected embryos (Figure 4) suggest that production of post-mitotic RGCs was not prevented by targeting adamts10 expression. This indicates that the primary defect of RGC development in adamts10 MO treated embryos is likely a failure to migrate to the appropriate position resulting in a retinal lamination defect.
In summary, we have discovered a previously unknown role for Adamts10 in RGC development, possibly contributing to their apical to basal translocation and formation of an ordered ganglion cell layer. Our results suggest that the developmental role of Adamts10 is mediated by active TGFβ family signaling. In addition, our results show for the first time that Adamts10 is necessary for pSmad3-mediated constitutive TGFβ family signaling in the developing retina.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
The animal study was reviewed and approved by Vanderbilt University Institutional Animal Care and Use Committee.
LW performed experiments, analyzed data, and drafted the manuscript. AW performed most of the experiments, designed experiments and analyzed data. H-JW performed and designed experiments, analyzed data and edited the manuscript. S-YW designed initial studies, performed experiments, and analyzed data. HM designed initial studies and analyzed data. DM designed experiments and analyzed data. RK conceived the project, analyzed results, and edited the manuscript. JK conceived the project, designed experiments, analyzed data, and wrote the manuscript.
This work was Supported by the Vanderbilt Eye Institute and by NIH grants R01EY027746 (JK), R01EY12018 (HSM), Training Grant in Vision Research EY007135 and Vanderbilt Vision Research Center (P30 EY08126). Also supported by a Departmental Unrestricted Award from Research to Prevent Blindness, Inc. Imaging was performed through use of the Vanderbilt Cell Imaging Shared Resource, which is supported by NIH grants CA68485, DK20593, DK58404, DK59637 and EY08126.
We thank Abudi Nashabi for zebrafish husbandry and histology.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2022.989851/full#supplementary-material
hpf, hours post-fertilization; RGC, retinal ganglion cell.
Ahonen, S. J., Kaukonen, M., Nussdorfer, F. D., Harman, C. D., Komaromy, A. M., and Lohi, H. (2014). A novel missense mutation in adamts10 in norwegian elkhound primary glaucoma. PLoS One 9, e111941. doi:10.1371/journal.pone.0111941
Aizawa, H., Bianco, I. H., Hamaoka, T., Miyashita, T., Uemura, O., Concha, M. L., et al. (2005). Laterotopic representation of left-right information onto the dorso-ventral axis of a zebrafish midbrain target nucleus. Curr. Biol. 15, 238–243. doi:10.1016/j.cub.2005.01.014
Apte, S. S. (2009). A disintegrin-like and metalloprotease (reprolysin-type) with thrombospondin type 1 motif (adamts) superfamily: Functions and mechanisms. J. Biol. Chem. 284, 31493–31497. doi:10.1074/jbc.R109.052340
Avanesov, A., and Malicki, J. (2010). Analysis of the retina in the zebrafish model. Methods Cell Biol. 100, 153–204. doi:10.1016/B978-0-12-384892-5.00006-2
Badea, T. C., Cahill, H., Ecker, J., Hattar, S., and Nathans, J. (2009). Distinct roles of transcription factors brn3a and brn3b in controlling the development, morphology, and function of retinal ganglion cells. Neuron 61, 852–864. doi:10.1016/j.neuron.2009.01.020
Barthel, L. K., and Raymond, P. A. (1990). Improved method for obtaining 3-microns cryosections for immunocytochemistry. J. Histochem. Cytochem. 38, 1383–1388. doi:10.1177/38.9.2201738
Braunger, B. M., Pielmeier, S., Demmer, C., Landstorfer, V., Kawall, D., Abramov, N., et al. (2013). Tgf-beta signaling protects retinal neurons from programmed cell death during the development of the mammalian eye. J. Neurosci. 33, 14246–14258. doi:10.1523/JNEUROSCI.0991-13.2013
Brunet, F. G., Fraser, F. W., Binder, M. J., Smith, A. D., Kintakas, C., Dancevic, C. M., et al. (2015). The evolutionary conservation of the a disintegrin-like and metalloproteinase domain with thrombospondin-1 motif metzincins across vertebrate species and their expression in teleost zebrafish. BMC Evol. Biol. 15, 22. doi:10.1186/s12862-015-0281-9
Cain, S. A., Mularczyk, E. J., Singh, M., Massam-Wu, T., and Kielty, C. M. (2016). Adamts-10 and -6 differentially regulate cell-cell junctions and focal adhesions. Sci. Rep. 6, 35956. doi:10.1038/srep35956
Calkins, D. J. (2012). Critical pathogenic events underlying progression of neurodegeneration in glaucoma. Prog. Retin. Eye Res. 31, 702–719. doi:10.1016/j.preteyeres.2012.07.001
Carnes, M. U., Liu, Y. P., Allingham, R. R., Whigham, B. T., Havens, S., Garrett, M. E., et al. (2014). Discovery and functional annotation of six6 variants in primary open-angle glaucoma. PLoS Genet. 10, e1004372. doi:10.1371/journal.pgen.1004372
Carrella, S., Barbato, S., D'agostino, Y., Salierno, F. G., Manfredi, A., Banfi, S., et al. (2015). Tgf-beta controls mir-181/erk regulatory network during retinal axon specification and growth. PLoS One 10, e0144129. doi:10.1371/journal.pone.0144129
Casari, A., Schiavone, M., Facchinello, N., Vettori, A., Meyer, D., Tiso, N., et al. (2014). A smad3 transgenic reporter reveals tgf-beta control of zebrafish spinal cord development. Dev. Biol. 396, 81–93. doi:10.1016/j.ydbio.2014.09.025
Charbonneau, N. L., Ono, R. N., Corson, G. M., Keene, D. R., and Sakai, L. Y. (2004). Fine tuning of growth factor signals depends on fibrillin microfibril networks. Birth Defects Res. C Embryo Today 72, 37–50. doi:10.1002/bdrc.20000
Dagoneau, N., Benoist-Lasselin, C., Huber, C., Faivre, L., Megarbane, A., Alswaid, A., et al. (2004). Adamts10 mutations in autosomal recessive weill-marchesani syndrome. Am. J. Hum. Genet. 75, 801–806. doi:10.1086/425231
Dunker, N., Schuster, N., and Krieglstein, K. (2001). Tgf-beta modulates programmed cell death in the retina of the developing chick embryo. Development 128, 1933–1942. doi:10.1242/dev.128.11.1933
Faivre, L., Dollfus, H., Lyonnet, S., Alembik, Y., Megarbane, A., Samples, J., et al. (2003a). Clinical homogeneity and genetic heterogeneity in weill-marchesani syndrome. Am. J. Med. Genet. A 123A, 204–207. doi:10.1002/ajmg.a.20289
Faivre, L., Gorlin, R. J., Wirtz, M. K., Godfrey, M., Dagoneau, N., Samples, J. R., et al. (2003b). In frame fibrillin-1 gene deletion in autosomal dominant weill-marchesani syndrome. J. Med. Genet. 40, 34–36. doi:10.1136/jmg.40.1.34
Forman, O. P., Pettitt, L., Komaromy, A. M., Bedford, P., and Mellersh, C. (2015). A novel genome-wide association study approach using genotyping by exome sequencing leads to the identification of a primary open angle glaucoma associated inversion disrupting adamts17. PLoS One 10, e0143546. doi:10.1371/journal.pone.0143546
Habashi, J. P., Judge, D. P., Holm, T. M., Cohn, R. D., Loeys, B. L., Cooper, T. K., et al. (2006). Losartan, an at1 antagonist, prevents aortic aneurysm in a mouse model of marfan syndrome. Science 312, 117–121. doi:10.1126/science.1124287
Haji-Seyed-Javadi, R., Jelodari-Mamaghani, S., Paylakhi, S. H., Yazdani, S., Nilforushan, N., Fan, J. B., et al. (2012). Ltbp2 mutations cause weill-marchesani and weill-marchesani-like syndrome and affect disruptions in the extracellular matrix. Hum. Mutat. 33, 1182–1187. doi:10.1002/humu.22105
Hu, M., and Easter, S. S. (1999). Retinal neurogenesis: the formation of the initial central patch of postmitotic cells. Dev. Biol. 207, 309–321. doi:10.1006/dbio.1998.9031
Hubmacher, D., and Apte, S. S. (2015). Adamts proteins as modulators of microfibril formation and function. Matrix Biol. 47, 34–43. doi:10.1016/j.matbio.2015.05.004
Icha, J., Kunath, C., Rocha-Martins, M., and Norden, C. (2016). Independent modes of ganglion cell translocation ensure correct lamination of the zebrafish retina. J. Cell Biol. 215, 259–275. doi:10.1083/jcb.201604095
Inman, G. J., Nicolas, F. J., Callahan, J. F., Harling, J. D., Gaster, L. M., Reith, A. D., et al. (2002). Sb-431542 is a potent and specific inhibitor of transforming growth factor-beta superfamily type i activin receptor-like kinase (alk) receptors alk4, alk5, and alk7. Mol. Pharmacol. 62, 65–74. doi:10.1124/mol.62.1.65
Jonas, J. B., Aung, T., Bourne, R. R., Bron, A. M., Ritch, R., and Panda-Jonas, S. (2017). Glaucoma. Lancet 390, 2183–2193. doi:10.1016/S0140-6736(17)31469-1
Kaiser, J., Maibach, M., Piovesana, E., Salpeter, I., Escher, N., Ormen, Y., et al. (2020). TGFβ1 induces axonal outgrowth via ALK5/PKA/SMURF1-Mediated degradation of RhoA and stabilization of PAR6. eNeuro 7, ENEURO.0104-20.2020. doi:10.1523/ENEURO.0104-20.2020
Karoulias, S. Z., Beyens, A., Balic, Z., Symoens, S., Vandersteen, A., Rideout, A. L., et al. (2020a). A novel adamts17 variant that causes weill-marchesani syndrome 4 alters fibrillin-1 and collagen type i deposition in the extracellular matrix. Matrix Biol. 88, 1–18. doi:10.1016/j.matbio.2019.11.001
Karoulias, S. Z., Taye, N., Stanley, S., and Hubmacher, D. (2020b). The adamts/fibrillin connection: Insights into the biological functions of adamts10 and adamts17 and their respective sister proteases. Biomolecules 10, E596. doi:10.3390/biom10040596
Kelwick, R., Desanlis, I., Wheeler, G. N., and Edwards, D. R. (2015). The adamts (a disintegrin and metalloproteinase with thrombospondin motifs) family. Genome Biol. 16, 113. doi:10.1186/s13059-015-0676-3
Kielty, C. M. (2017). Fell-muir lecture: Fibrillin microfibrils: structural tensometers of elastic tissues? Int. J. Exp. Pathol. 98, 172–190. doi:10.1111/iep.12239
Kimmel, C. B., Ballard, W. W., Kimmel, S. R., Ullmann, B., and Schilling, T. F. (1995). Stages of embryonic development of the zebrafish. Dev. Dyn. 203, 253–310. doi:10.1002/aja.1002030302
Kuchtey, J., Olson, L. M., Rinkoski, T., Mackay, E. O., Iverson, T. M., Gelatt, K. N., et al. (2011). Mapping of the disease locus and identification of adamts10 as a candidate gene in a canine model of primary open angle glaucoma. PLoS Genet. 7, e1001306. doi:10.1371/journal.pgen.1001306
Kuchtey, J., Kunkel, J., Esson, D., Sapienza, J. S., Ward, D. A., Plummer, C. E., et al. (2013). Screening adamts10 in dog populations supports gly661arg as the glaucoma-causing variant in beagles. Invest. Ophthalmol. Vis. Sci. 54, 1881–1886. doi:10.1167/iovs.12-10796
Kutz, W. E., Wang, L. W., Dagoneau, N., Odrcic, K. J., Cormier-Daire, V., Traboulsi, E. I., et al. (2008). Functional analysis of an adamts10 signal peptide mutation in weill-marchesani syndrome demonstrates a long-range effect on secretion of the full-length enzyme. Hum. Mutat. 29, 1425–1434. doi:10.1002/humu.20797
Kutz, W. E., Wang, L. W., Bader, H. L., Majors, A. K., Iwata, K., Traboulsi, E. I., et al. (2011). Adamts10 protein interacts with fibrillin-1 and promotes its deposition in extracellular matrix of cultured fibroblasts. J. Biol. Chem. 286, 17156–17167. doi:10.1074/jbc.M111.231571
Massague, J. (2012). TGFβ signalling in context. Nat. Rev. Mol. Cell Biol. 13, 616–630. doi:10.1038/nrm3434
Matsuzaki, T., Keene, D. R., Nishimoto, E., and Noda, M. (2020). Reversion-inducing cysteine-rich protein with kazal motifs and mt1-mmp promote the formation of robust fibrillin fibers. J. Cell. Physiol. 236, 1980–1995. doi:10.1002/jcp.29982
Mularczyk, E. J., Singh, M., Godwin, A. R. F., Galli, F., Humphreys, N., Adamson, A. D., et al. (2018). Adamts10-mediated tissue disruption in weill-marchesani syndrome. Hum. Mol. Genet. 27, 3675–3687. doi:10.1093/hmg/ddy276
Neptune, E. R., Frischmeyer, P. A., Arking, D. E., Myers, L., Bunton, T. E., Gayraud, B., et al. (2003). Dysregulation of tgf-beta activation contributes to pathogenesis in marfan syndrome. Nat. Genet. 33, 407–411. doi:10.1038/ng1116
Ng, C. M., Cheng, A., Myers, L. A., Martinez-Murillo, F., Jie, C., Bedja, D., et al. (2004). Tgf-beta-dependent pathogenesis of mitral valve prolapse in a mouse model of marfan syndrome. J. Clin. Invest. 114, 1586–1592. doi:10.1172/JCI22715
Nguyen-Ba-Charvet, K. T., and Rebsam, A. (2020). Neurogenesis and specification of retinal ganglion cells. Int. J. Mol. Sci. 21, E451. doi:10.3390/ijms21020451
Oliver, J. A., Forman, O. P., Pettitt, L., and Mellersh, C. S. (2015). Two independent mutations in adamts17 are associated with primary open angle glaucoma in the basset hound and basset fauve de bretagne breeds of dog. PLoS One 10, e0140436. doi:10.1371/journal.pone.0140436
Pan, L., Deng, M., Xie, X., and Gan, L. (2008). Isl1 and brn3b co-regulate the differentiation of murine retinal ganglion cells. Development 135, 1981–1990. doi:10.1242/dev.010751
Pauklin, S., and Vallier, L. (2015). Activin/nodal signalling in stem cells. Development 142, 607–619. doi:10.1242/dev.091769
Ramdas, W. D., Van Koolwijk, L. M., Ikram, M. K., Jansonius, N. M., De Jong, P. T., Bergen, A. A., et al. (2010). A genome-wide association study of optic disc parameters. PLoS Genet. 6, e1000978. doi:10.1371/journal.pgen.1000978
Ramirez, F., and Rifkin, D. B. (2009). Extracellular microfibrils: Contextual platforms for tgfbeta and bmp signaling. Curr. Opin. Cell Biol. 21, 616–622. doi:10.1016/j.ceb.2009.05.005
Ramirez, F., and Sakai, L. Y. (2010). Biogenesis and function of fibrillin assemblies. Cell Tissue Res. 339, 71–82. doi:10.1007/s00441-009-0822-x
Sakai, L. Y., Keene, D. R., Renard, M., and De Backer, J. (2016). Fbn1: The disease-causing gene for marfan syndrome and other genetic disorders. Gene 591, 279–291. doi:10.1016/j.gene.2016.07.033
Sato, T., Sato, F., Kamezaki, A., Sakaguchi, K., Tanigome, R., Kawakami, K., et al. (2015). Neuregulin 1 type ii-erbb signaling promotes cell divisions generating neurons from neural progenitor cells in the developing zebrafish brain. PLoS One 10, e0127360. doi:10.1371/journal.pone.0127360
Sengle, G., Tsutsui, K., Keene, D. R., Tufa, S. F., Carlson, E. J., Charbonneau, N. L., et al. (2012). Microenvironmental regulation by fibrillin-1. PLoS Genet. 8, e1002425. doi:10.1371/journal.pgen.1002425
Shiga, Y., Akiyama, M., Nishiguchi, K. M., Sato, K., Shimozawa, N., Takahashi, A., et al. (2018). Genome-wide association study identifies seven novel susceptibility loci for primary open-angle glaucoma. Hum. Mol. Genet. 27, 1486–1496. doi:10.1093/hmg/ddy053
Souzeau, E., Siggs, O. M., Zhou, T., Galanopoulos, A., Hodson, T., Taranath, D., et al. (2017). Glaucoma spectrum and age-related prevalence of individuals with foxc1 and pitx2 variants. Eur. J. Hum. Genet. 25, 839–847. doi:10.1038/ejhg.2017.59
Springelkamp, H., Hohn, R., Mishra, A., Hysi, P. G., Khor, C. C., Loomis, S. J., et al. (2014). Meta-analysis of genome-wide association studies identifies novel loci that influence cupping and the glaucomatous process. Nat. Commun. 5, 4883. doi:10.1038/ncomms5883
Springelkamp, H., Iglesias, A. I., Mishra, A., Hohn, R., Wojciechowski, R., Khawaja, A. P., et al. (2017). New insights into the genetics of primary open-angle glaucoma based on meta-analyses of intraocular pressure and optic disc characteristics. Hum. Mol. Genet. 26, 438–453. doi:10.1093/hmg/ddw399
Tachibana, N., Cantrup, R., Dixit, R., Touahri, Y., Kaushik, G., Zinyk, D., et al. (2016). Pten regulates retinal amacrine cell number by modulating akt, Tgfβ, and erk signaling. J. Neurosci. 36, 9454–9471. doi:10.1523/JNEUROSCI.0936-16.2016
Wain, L. V., Verwoert, G. C., O'reilly, P. F., Shi, G., Johnson, T., Johnson, A. D., et al. (2011). Genome-wide association study identifies six new loci influencing pulse pressure and mean arterial pressure. Nat. Genet. 43, 1005–1011. doi:10.1038/ng.922
Wang, F., Flanagan, J., Su, N., Wang, L. C., Bui, S., Nielson, A., et al. (2012). Rnascope: a novel in situ rna analysis platform for formalin-fixed, paraffin-embedded tissues. J. Mol. Diagn. 14, 22–29. doi:10.1016/j.jmoldx.2011.08.002
Wang, L. W., Kutz, W. E., Mead, T. J., Beene, L. C., Singh, S., Jenkins, M. W., et al. (2019). Adamts10 inactivation in mice leads to persistence of ocular microfibrils subsequent to reduced fibrillin-2 cleavage. Matrix Biol. 77, 117–128. doi:10.1016/j.matbio.2018.09.004
Wu, F., Kaczynski, T. J., Sethuramanujam, S., Li, R., Jain, V., Slaughter, M., et al. (2015). Two transcription factors, pou4f2 and isl1, are sufficient to specify the retinal ganglion cell fate. Proc. Natl. Acad. Sci. U. S. A. 112, E1559–E1568. doi:10.1073/pnas.1421535112
Keywords: ADAMTS10, transforming growth factor beta (TGFβ), retinal development, zebrafish, morpholino (MO), retina, retinal ganglion cell (RGC)
Citation: Wareham LK, Whitener AE, Wu H-J, Wu S-Y, Mchaourab HS, Mortlock DP, Kuchtey RW and Kuchtey J (2022) Adamts10 controls transforming growth factor β family signaling that contributes to retinal ganglion cell development. Front. Mol. Biosci. 9:989851. doi: 10.3389/fmolb.2022.989851
Received: 08 July 2022; Accepted: 05 August 2022;
Published: 06 September 2022.
Edited by:
Salvatore Santamaria, Imperial College London, United KingdomReviewed by:
Dirk Hubmacher, Icahn School of Medicine at Mount Sinai, United StatesCopyright © 2022 Wareham, Whitener, Wu, Wu, Mchaourab, Mortlock, Kuchtey and Kuchtey. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: John Kuchtey, am9obi5rdWNodGV5QHZ1bWMub3Jn
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.