- 1Biomedical Research and Drug Discovery Research Group, Faculty of Health Sciences, Higher Colleges of Technology, Sharjah, United Arab Emirates
- 2Department of Bioinformatics, Hans Raj Mahila Maha Vidyalaya, Jalandhar, India
- 3Vice Chancellery, The University of Notre Dame Australia, Fremantle, WA, Australia
- 4The Institute of Immunology and Infectious Diseases, Murdoch University, Perth, WA, Australia
- 5PathWest Nedlands, QEII Medical Centre, Nedlands, WA, Australia
- 6Biomedical Biotechnology Research Unit, Department of Biochemistry and Microbiology, Rhodes University, Grahamstown, South Africa
The heat shock protein 40 (Hsp40) family, also called J domain proteins (JDPs), regulate their Hsp70 partners by ensuring that they are engaging the right substrate at the right time and in the right location within the cell. A number of JDPs can serve as co-chaperone for a particular Hsp70, and so one generally finds many more JDPs than Hsp70s in the cell. In humans there are 13 Hsp70s and 49 JDPs. The human malaria parasite, Plasmodium falciparum, has dedicated an unusually large proportion of its genome to molecular chaperones, with a disproportionately high number of JDPs (PfJDPs) of 49 members. Interestingly, just under half of the PfJDPs are exported into the host cell during the asexual stage of the life cycle, when the malaria parasite invades mature red blood cells. Recent evidence suggests that these PfJDPs may be functionalizing both host and parasite Hsp70s within the infected red blood cell, and thereby driving the renovation of the host cell towards pathological ends. PfJDPs have been found to localize to the host cytosol, mobile structures within the host cytosol (so called “J Dots”), the host plasma membrane, and specialized structures associated with malaria pathology such as the knobs. A number of these exported PfJDPs are essential, and there is growing experimental evidence that they are important for the survival and pathogenesis of the malaria parasite. This review critiques our understanding of the important role these exported PfJDPs play at the host-parasite interface.
Introduction
The malaria parasite, Plasmodium falciparum, invades the cells of its human host, enabling it to evade the immune system and ultimately harness the cellular machinery to propagate itself and cause severe pathology. Surrounded by a self-created parasitophorous vacuole (PV) within human erythrocytes, the malaria parasite renovates the host cell by exporting over 400 parasite proteins, including a number of heat shock proteins, which oversee protein folding as molecular chaperones and co-chaperones (Hiller et al., 2004; Marti et al., 2004; Jonsdottir et al., 2021). An important co-chaperone family, the P. falciparum J domain proteins (PfJDPs; also called heat shock protein 40 s, Hsp40s), are localized to almost every compartment of the infected erythrocyte, with arguably the greatest number of exported members of any protein family (Sargeant et al., 2006; Botha et al., 2007; Dutta et al., 2021a; Figure 1). Furthermore, many of the exported PfJDPs are expressed in the early stages of the asexual phase of the malaria parasite life cycle (Bozdech et al., 2003), consistent with their proposed critical role in the export of other malaria proteins and survival during febrile episodes (Dutta et al., 2021a). Indeed, there is growing evidence that these exported PfJDPs are key players in the survival and pathogenesis of the malaria parasite. The following sections of this review provide a critique of our understanding of the important role that these exported PfJDPs play at the host-parasite interface of malaria pathology.

FIGURE 1. Schematic representation of the exported PfJDPs. The PlasmoDB accession numbers/common name, essential nature, type, domain organization (not to scale), and number of amino acids are shown. The domains of the PfJDPs shown are the signature J domain (blue) which stimulates the ATPase activity of Hsp70, the glycine/phenylalanine/serine (GFS)-rich region (green) involved in regulation of protein substrate binding by Hsp70, the C-terminal domain (grey) involved in substrate binding and dimerization, the PEXEL motif required for export through the PTEX, and the PHIST domain (yellow) and MEC motif (red) involved in binding to the erythrocyte cytoskeleton. Certain PfJDPs (type IVs) have a J domain where the highly conserved histidine-proline-aspartic acid (HPD) motif is altered (blue with dashed border).
The diversity, structure and function of PfJDPs
There are at least 49 PfJDPs encoded on the P. falciparum genome, far more than any other Plasmodium species (Njunge et al., 2013). All JDPs by definition contain a signature J domain (Kampinga et al., 2019), which contains a highly conserved histidine-proline-aspartic acid (HPD) motif, and is essential for regulation of the chaperone activity of partner heat shock protein 70 s (Hsp70s) (Hennessy et al., 2005). There are a number of other domains which have been used to categorize JDPs into four types (types I-IV; Botha et al., 2007). Interestingly, the HPD motif was thought to be invariant, until the discovery of the type IV JDPs containing a non-conserved HPD motif (Botha et al., 2007).
The Plasmodium export element (PEXEL; Hiller et al., 2004; Marti et al., 2004) has been shown to tag many P. falciparum proteins for export through the Plasmodium translocon of exported proteins (PTEX; de Koning-Ward et al., 2009; Beck et al., 2014; Elsworth et al., 2014; Elsworth et al., 2016). While the PEXEL was initially identified on 19 PfJDPs (Botha et al., 2007; Njunge et al., 2013; Pesce and Blatch 2014), recent revisions have indicated 18 PEXEL-containing PfJDPs (Dutta et al., 2021a). Therefore, at least 18 PfJDPs are proposed to be exported into the infected erythrocyte based on the presence of a PEXEL; three type II, four type III and eleven type IV PfJDPs (Figure 1).
The exported PfJDPs appear to be critical for survival and pathogenesis of the malaria parasite, through their essential nature (4/18; Zhang et al., 2018; Figure 1), involvement in protein folding of exported virulence proteins (e.g., PFE0055c and PFA0660w associated with “J Dots”; Külzer et al., 2010; Külzer et al., 2012; Behl et al., 2019), requirement for growth or survival under febrile conditions (e.g., PFA0110w, the ring-infected erythrocyte surface antigen protein, RESA; Silva et al., 2005; Diez-Silva et al., 2012) and involvement in pathogenesis (e.g., PF10_0381; knockout causes loss of knobs; Maier et al., 2008). Of the 6 PfHsp70s expressed by the malaria parasite (Shonhai et al., 2007; Przyborski et al., 2015; Shonhai et al., 2021), PfHsp70-x appears to be the only member exported into the cytosol of the infected host erythrocyte (Külzer et al., 2010; Külzer et al., 2012; Grover et al., 2013), even though it lacks a PEXEL motif (Rhiel et al., 2016). Interestingly, the host cytosol appears to contain human chaperones and co-chaperones, including JDPs (which are likely to be non-functional remnants; Pasini et al., 2006; van Gestel et al., 2010) and significant levels of functional human Hsp70. (e.g., HSPA1A, also called Hsp72; referred to here as hHsp70), occurring free or in complex with Hsp90 and the Hsp70/Hsp90 organising protein, Hop (Banumathy et al., 2002). Furthermore, evidence is emerging that certain exported PfJDPs are capable of functionally interacting with PfHsp70-x, or hHsp70, or both Hsp70s (Dutta et al., 2021a; Diehl et al., 2021).
The structures of the ATPase (Day et al., 2019) and substrate binding (Schmidt and Vakonakis, 2020) domains of PfHsp70-x have been elucidated, as has the structure of the J domain of the exported PfJDP, PFA0660w (Day et al., 2019). In addition, molecular modelling revealed that helix II of the PFA0660w J domain makes a primary interface with the ATPase domain of PfHsp70-x (Day et al., 2019). Comparative molecular modelling suggested that the PFA0660w J domain-PfHsp70-x complex was less stable than that of another exported PfJDP (PFE0055c J domain-PfHsp70-x complex) (Dutta et al., 2021b). The functional differences between PFA0660w and PFE0055c were attributed to the J domain helix II of PFA0660w being less positively charged than the more typical J domain of PFE0055c. Interestingly, a multiple sequence alignment and surface electrostatic potential analysis of the J domains of all the exported PfJDPs revealed that the positive nature of the J domain helix II appears to decrease going from type II to type IV, with the type IVs exhibiting significant negative charge or hydrophobicity (Figure 2; Supplementary Figure S1). These J domain surface differences together with the considerable variations in the HPD motifs for the type IVs, suggests that the structural and functional nature of the association, if any, between the different types of exported PfJDPs and Hsp70 (PfHsp70-x or hHsp70) are likely to be considerably different.
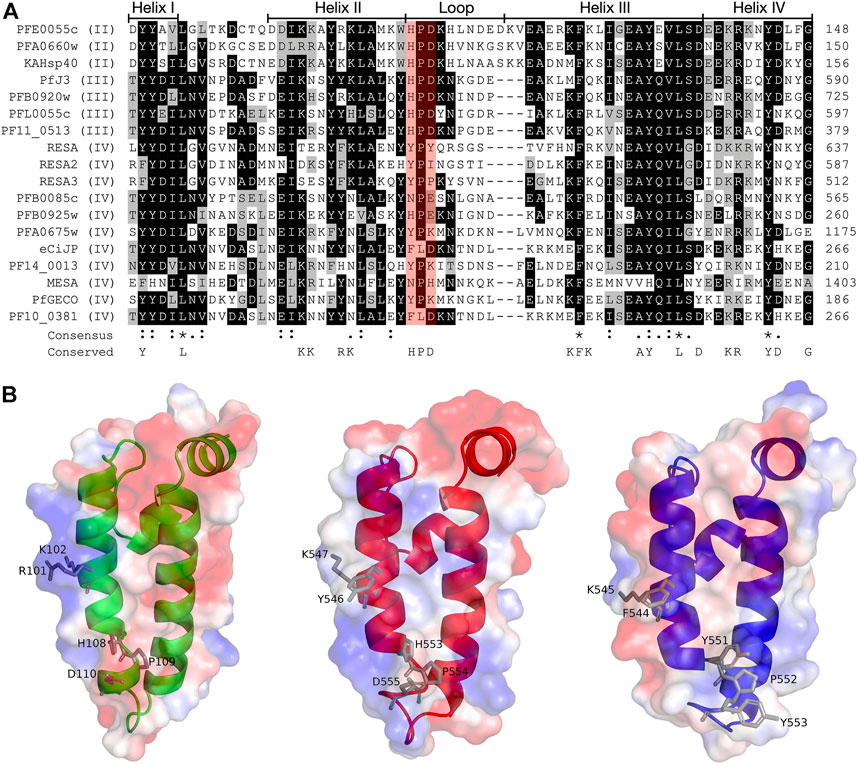
FIGURE 2. Multiple sequence alignment and molecular modelling of the J domains of exported PfJDPs. (A) Multiple sequence alignment of the J domains of the 18 exported PfJDPs proteins. The proteins are defined by either their PlasmoDB accession number or common name in the first column, and the roman numerals in brackets refer to the type of JDP. Colored in black are identical amino acids (in at least 50% of the aligned sequences), colored in light grey are similar amino acids (in at least 50% of the aligned sequences), and colored in white are the amino acids with no identity or similarity. The default categories for similar amino acids were applied to the multiple sequence alignment (ILV, FWY, KRH, DE, GAS, P, C and TNQM). The row titled “Consensus” are the common consensus symbols of the multiple sequence alignment: an * (asterisk) indicates positions which have a single, fully conserved residue; a: (colon) indicates conservation between groups of strongly similar properties; and a. (period) indicates conservation between groups of weakly similar properties. The row titled “Conserved” refers to residues previously found to be highly conserved across J domains of different origins, with the residues defined at the bottom of the alignment (Hennessy et al., 2000; Hennessy et al., 2005). The protein helices and loop region are defined by bidirectional lines on top of the alignment. Highlighted in red shading is the HPD motif. The alignment was created using Clustal Omega (Sievers and Higgins, 2018) and rendered with box shading using Multiple Align Show (Stothard, 2000). (B) Three dimensional models of the J domains of PFE0055c type II (green), PfJ3 type III (red), and RESA type IV (blue) to illustrate the conserved HPD and RK motif (grey sticks). The positive charge is shown in blue colored surface, the negative charge is shown in red colored surface and the neutral potential are shown in white colored surface. The surface electrostatic potential was calculated by APBS. The models were prepared using SWISS-MODEL (Waterhouse et al., 2018; the template structures are listed in Supplementary Table S1) and graphically rendered using PyMol 2.5.2 (PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC).
J Dots in the host cytosol contain PfJDPs and PfHsp70-x
J Dots are highly mobile lipid-containing protein complexes found within the malaria parasite-infected erythrocyte cytosol (Külzer et al., 2010; Külzer et al., 2012; Petersen et al., 2016). Localization and immunoprecipitation studies have convincingly detected the exported type II PfJDPs, PFA0660w and PFE0055c, and PfHsp70-x in J Dots, which suggested that they formed a functional partnership in the host cytosol. However, these may not be the only J Dot chaperone partnerships, as an exported type IV PfJDP, called eCiJP (PF11_0034; a paralogue of PF10_0381), has recently been reported to localize to J-Dots (Sahu et al., 2022). There is also evidence that J Dots associate with P. falciparum erythrocyte membrane protein 1 (PfEMP1), leading to the proposal that they may be involved in the trafficking of this major virulence factor (Külzer et al., 2012). These findings have been corroborated by biochemical studies on PFA0660w, PFE0055c and PfHsp70-x. PFA0660w was found to be highly effective at protein aggregation suppression, and both PFA0660w and PFE0055c were shown to be capable of specifically stimulating the basal ATPase activity of PfHsp70-x and not hHsp70 (Daniyan et al., 2016; Daniyan and Blatch 2017; Dutta et al., 2021b). These steady-state ATPase assays were conducted under saturating ATP concentrations which favour chaperone-co-chaperone over chaperone-substrate interactions, and in the case of PFE0055c could be inhibited by a J domain-specific inhibitor (chalcone C86), suggesting that PFA0660w and PFE0055c form specific co-chaperones partnerships with PfHsp70-x. PFE0055c stimulated the ATPase activity to a much greater extent than PFA0660w, and molecular modelling suggested that PFE0055c formed a more stable complex with PfHsp70-x than did PFA0660w (Dutta et al., 2021b). Furthermore, biochemical studies have shown that PfHsp70-x was capable of simultaneously associating with cholesterol-bound PFA0660w and PfEMP1 to form a stable complex (Behl et al., 2019). In contrast, biochemical studies using just the J domain of PFA0660w and PFE0055c (Day et al., 2019) or a PFA0660w J domain-substrate fusion construct (Diehl et al., 2021) have provided evidence that they may be capable of functional interaction with both PfHsp70-x and hHsp70. The inconsistency between the biochemical studies on full-length versus J domain proteins with respect to the hHsp70 findings, could be due to differences in the nature of the assays. However, it is also likely that the full-length PfJDPs contain regions beyond the J domain that determine co-chaperone-chaperone specificity. This interpretation is consistent with structural analyses of the interaction between PFA0660w and PfHsp70-x, which indicated that they associated in a bipartite manner requiring both the J domain and the G/F region of PFA0660w (Behl and Mishra 2019).
Exported PfJDPs play a key role in malaria pathology
The third exported type II PfJDP (PFB0090c, also called knob-associated Hsp40, KAHsp40; Acharya et al., 2012), has not been reported to associate with PfHsp70-x or with J Dots, despite significant structural similarity to the other type II PfJDPs. In contrast, PFB0090c has been shown to associate with PTEX and knobs, and is proposed to be involved in the trafficking, folding, and assembly of knob protein complexes (Acharya et al., 2012). Knob complexes appear to contain human chaperones and co-chaperones in a highly complexed state (Hsp70, Hsp90 and Hop), and there is evidence that hHsp70 plays a role in assembly of knobs (Banumathy et al., 2002; Alampalli et al., 2018). It is tempting to speculate that PFB0090c is the co-chaperone recruiting hHsp70 to knobs. However, knock-out studies and associated genetic and biochemical analyses suggested that PFA0660w was also involved in knob formation and cytoadherence in collaboration with hHsp70 (Diehl et al., 2021). Interestingly, all of the exported type II PfJDPs (PFB0090c, PFA0660w, and PFE0055c) have been shown to functionally interact with hHsp70 using a heterologous yeast two-hybrid complementation assay (Jha et al., 2017). However, this assay did not test for functional association between PfJDPs and PfHsp70-x, and the biochemical and biophysical nature of the interactions were not investigated.
There are four exported type III PfJDPs (PFB0920w, PFL0055c, PF10_0378/Pfj3 and PF11_0513), three of which contain the Plasmodium helical interspersed sub-telomeric (PHIST) domain (Sargeant et al., 2006; Oakley et al., 2007; Oakley et al., 2011; Figure 1). Four exported type IV PfJDPs (PF11_0509/RESA3, PF11_0512/RESA2, PFA0110w/RESA and PFB0085c; Figure 1) also contain the PHIST domain (Figure 1). All of these PHIST-containing PfJDPs also contain the MEC (MESA erythrocyte cytoskeleton-binding) motif found in five other exported type IV PfJDPs (PFE0040c/mature parasite-infected erythrocyte surface antigen [MESA], PFA0675w, PFB0925w, PF10_0381, PF11_0034/eCiJP; Bennett et al., 1997; Kilili and LaCount 2011; Njunge et al., 2013; Figure 1; Supplementary Figure S2). PHIST proteins are proposed to play an important role in trafficking of PfEMP1 proteins and the modulation of membrane rigidity and cytoadherence in parasite-infected erythrocytes (Kumar et al., 2019). Therefore, the presence of the MEC motif and the PHIST domain in these PfJDPs, is consistent with the proposed role of many of these proteins in remodelling at the cytoskeleton-membrane interface of the infected erythrocyte. RESA is arguably the most extensively studied exported PfJDP, and has been shown to play a key role in modulation of the rigidity of the infected erythrocyte membrane (Silva et al., 2005; Maier et al., 2008). RESA associates with spectrin and stabilizes its tertiary structure (Pei et al., 2007), and its effect on the cytoskeleton decreases the deformability of the infected erythrocyte, and protects the membrane during febrile episodes (Silva et al., 2005; Mills et al., 2007; Diez-Silva et al., 2012). The MEC motif enables MESA to bind to erythrocyte protein 4.1, thereby tethering it to the cytoskeleton (Coppel 1992; Bennett et al., 1997; Waller et al., 2003; Black et al., 2008); and MESA has been implicated in erythrocyte membrane modification (Coppel et al., 1988). However, the absence of MESA does not seem to have an influence on cytoadhesion (Petersen et al., 1989; Cooke et al., 2002). PF10_0381 has been implicated in knob formation (Maier et al., 2008), while its paralogue, eCiJP, has been shown to associate with the erythrocyte cytoskeleton, and potentially recruit hHsp70 to this location (Sahu et al., 2022; Figure 1). Apart from this report for eCiJP, there are virtually no reports on the interaction of type IV PfJDPs with Hsp70s (PfHsp70-x or hHsp70). Whether these proteins function as co-chaperones of Hsp70s remains to be determined.
Exported PfJDPs as drug targets
Very few small molecule inhibitors have been identified that bind specifically to JDPs (e.g., phenoxy-N-arylacetamides; Cassel et al., 2012). In addition, the identification of inhibitors that can directly disrupt the interaction between JDPs and Hsp70s is challenging, as the interaction is transient and the binding sites are located on the surface of the protein as shallow exposed clefts which are recalcitrant to small molecule association (Pesce et al., 2016). Nevertheless, recent advances in our understanding of PfJDP-PfHsp70 interactions and their inhibition, suggests that they are a promising target for anti-malarial drug development (Daniyan and Blatch, 2017; Dutta et al., 2021a). Pyrimidinones have shown potential as protein-protein interaction inhibitors when tested on the parasite-resident PfJDP-PfHsp70 system (Botha et al., 2011). In contrast, naphthoquinones and prenylated alkaloids were identified that functionally disrupted the exported PfHsp70-x and its interaction with JDPs (Cockburn et al., 2014). The most promising inhibitor was the prenylated alkaloid, malonganenone A (MalA), where side-by-side inhibition assays conducted on JDP-PfHsp70-x and JDP-hHsp70 showed that MalA had no effect on the basal ATPase activity of both Hsp70s, and yet significantly inhibited the JDP-stimulated ATPase activity of PfHsp70-x but not that of hHsp70 (Cockburn et al., 2014). MalA has significant anti-malarial activity, but low cytotoxicity to human cell lines (Cockburn et al., 2011), suggesting that it is a promising small-molecule candidate for the development of specific inhibitors of the exported PfJDP-PfHsp70-x system. Chalcones represent another class of small molecules found to have anti-malarial activity (Sinha et al., 2019; Qin et al., 2020), and to inhibit JDP-hHsp70 interaction (Moses et al., 2018). Interestingly, chalcone C86 (3-nitro-2′,4′,6′-trimethoxychalcone) was found to bind to the J domain of JDP families A-C, indicating that it was a pan-inhibitor of JDPs (Moses et al., 2018). Recently, it was found that C86 also inhibited PfJDP-PfHsp70-x interaction (Dutta et al., 2021b). While C86 had no effect on the basal ATPase activity of PfHsp70-x, it significantly inhibited the PFE0055c-stimulated ATPase activity of PfHsp70-x (Dutta et al., 2021b). Notably, significant inhibition was only observed when C86 was pre-incubated with PFE0055c prior to the addition of PfHsp70-x. This finding is consistent with C86 binding specifically to the region of the J domain of PFE0055c that makes contact with PfHsp70-x (Dutta et al., 2021a). Molecular modelling of a predicted complex suggests that C86 makes major contacts with previously identified residues of helix II of the PFE0055c J domain involved in binding to the PfHsp70-x ATPase domain (Figure 3). This model provides a rationale for the development of C86 chemotypes, with the aim of identifying high affinity derivatives with specificity for PfJDPs. Given that the exported PfJDP, PFE0055c, is essential (Zhang et al., 2018), the inhibition of its functional interaction with PfHsp70-x represents an important interface for anti-malarial drug development.
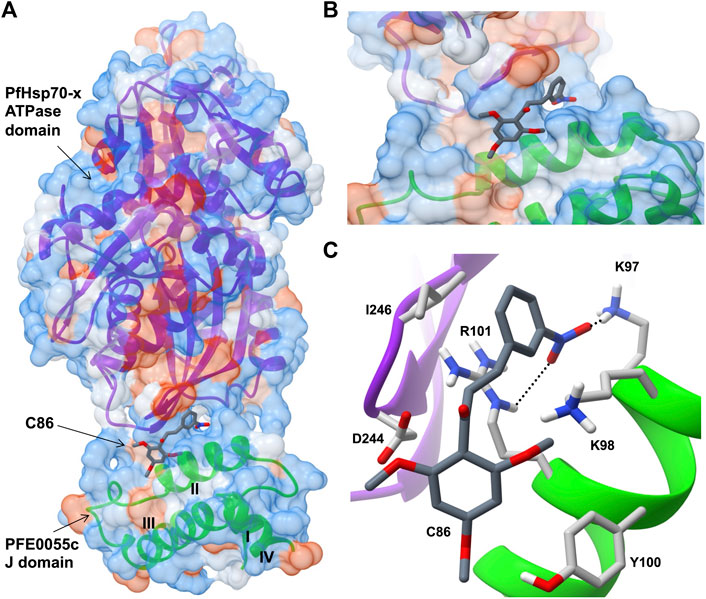
FIGURE 3. Predicted model of a complex of the small molecule inhibitor C86, PFE0055c J domain and PfHsp70-x ATPase domain. (A) Overall binding pose of C86 (middle) to the complex formed between the PfHsp70-x ATPase domain (top) and the PFE0055c J domain (bottom). (B) Focused view of c86 binding to the complex. (C) Detailed view of residues interacting with C86. The surface (set at 60% transparency) is colored according to amino acid hydrophobicity using the Kyte-Doolittle scale (with dodger blue representing more hydrophilic, to white, to orange red representing more hydrophobic residues). The cartoon representation underneath the surface depicts the secondary structure elements in the PfHsp70-x ATPase domain (purple) and PFE0055c J domain (green). The roman numerals (I to IV) mark the four helices of the PFE0055c J domain. C86 as well the interacting residues of the complex have been shown using the element color scheme (using red, blue and grey for oxygen, nitrogen and carbon, respectively). The carbons in C86 are depicted using dark grey while its interacting residues are colored in light grey. The residues are shown as sticks, and the numbering refers to their position in the full length protein. The predicted hydrogen bonds were identified using LigPlot + version 2.2.5 (https://www.ebi.ac.uk/thornton-srv/software/LigPlus/; Laskowski and Swindells, 2011), and have been rendered using FindHBond option in Chimera in relaxed mode and depicted using dotted lines (black). The lengths of the larger and the smaller hydrogen bonds are 2.74 Å and 2.32 Å, respectively. The PfHsp70-x ATPase domain-PFE0055c J domain complex was obtained using HADDOCK2.2 (Dutta et al., 2020a) while AutoDock Vina (https://vina.scripps.edu/; Trott and Olson, 2010) was used to dock C86 into this complex. Images for the 3D structures were rendered using UCSF Chimera 1.10.1 (https://www.cgl.ucsf.edu/chimera/), using one of the nine best binding conformations of C86.
Conclusion
There is solid evidence that certain exported type II PfJDPs (PFA0660w and PFE0055c) functionally associate with PfHsp70-x (Daniyan et al., 2016; Dutta et al., 2021b), and are core components of J Dots potentially involved in the trafficking and folding of virulence factors (Külzer et al., 2010; Külzer et al., 2012; Petersen et al., 2016; Behl et al., 2019). However, there is also emerging evidence that these PfJDPs play a role in knob formation (PFB0090c, Acharya et al., 2012), potentially in association with hHsp70 (PFA0660w, Diehl et al., 2021). Furthermore, a type IV PfJDP (eCiJP) was recently reported to associate with both J Dots and the erythrocyte cytoskeleton, with the latter potentially involving interaction with hHsp70 (Sahu et al., 2022). Based on the presence of the PHIST domain and MEC motif, it would not be surprising if many of the exported type III and type IV PfJDPs are found to associate at the cytoskeleton-membrane interface, and to be involved in recruitment of hHsp70 for remodeling purposes. Detailed mechanistic studies will no doubt shed light on this fascinating host-parasite interface, and elucidate the novel manner in which these exported PfJDPs interact with their partner Hsp70s (PfHsp70-x or hHsp70) to trigger the assembly of erythrocyte cytoskeleton and knob-associated proteins. While there is a need to continue exploring the role of exported PfJDPs in P. falciparum-infected erythrocytes, there is a pressing need to investigate their role in other stages of the parasite life cycle. For example, apart from studies on the type IV PfJDP, P. falciparum gametocyte erythrocyte cytosolic protein (PfGECO/PFL2550w; Morahan et al., 2011; Figure 1), there have been limited studies on PfJDPs expressed in the gametocyte stage. Given the role of these exported PfJDPs in the pathogenesis of malaria, and the evidence indicating that their function can be inhibited, the elucidation of their protein networks across the entire parasite life cycle is needed to fully understand their biological role, and to reveal further novel anti-malarial drug targets.
Author contributions
Conceptualization, GB; Figure 1, GB; bioinformatics analyses and Figure 2, SA; bioinformatics analyses and Figure 3, HS; writing-original draft preparation, GB; writing—review and editing, SA, TD, HS, and GB. All authors have read and agreed to the published version of the manuscript.
Funding
GB acknowledges the financial support of Higher Colleges of Technology, UAE (Interdisciplinary Research Grant, IRG; grant number 213471), and Rhodes University, South Africa (Rated Researcher Grant, RRG; project number IFRR100006). HS acknowledges institutional funding from the Department of Science and Technology, Government of India, for the development of computational resources (“Fund for Improvement of Science and Technology Infrastructure (FIST)” grant D. O. No. SR/FST/PG College/2009).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2022.978663/full#supplementary-material
References
Acharya, P., Chaubey, S., Grover, M., and Tatu, U. (2012). An exported heat shock protein 40 associates with pathogenesis-related knobs in Plasmodium falciparum infected erythrocytes. PloS ONE 7, e44605. doi:10.1371/journal.pone.0044605
Alampalli, S. V., Grover, M., Chandran, S., Tatu, U., and Acharya, P. (2018). Proteome and structural organization of the knob complex on the surface of the Plasmodium infected red blood cell. Proteomics. Clin. Appl. 12, e1600177. doi:10.1002/prca.201600177
Banumathy, G., Singh, V., and Tatu, U. (2002). Host chaperones are recruited in membrane-bound complexes by Plasmodium falciparum. J. Biol. Chem. 277, 3902–3912. doi:10.1074/jbc.M110513200
Beck, J. R., Muralidharan, V., Oksman, A., and Goldberg, D. E. (2014). PTEX component HSP101 mediates export of diverse malaria effectors into host erythrocytes. Nature 511, 592–595. doi:10.1038/nature13574
Behl, A., Kumar, V., Bisht, A., Panda, J. J., Hora, R., and Mishra, P. C. (2019). Cholesterol bound Plasmodium falciparum co-chaperone ‘PFA0660w’ complexes with major virulence factor ‘PfEMP1’ via chaperone ‘PfHsp70-x. Sci. Rep. 9, 2664–2667. doi:10.1038/s41598-019-39217-y
Behl, A., and Mishra, P. C. (2019). Structural insights into the binding mechanism of Plasmodium falciparum exported Hsp40-Hsp70 chaperone pair. Comput. Biol. Chem. 83, 107099. doi:10.1016/j.compbiolchem.2019.107099
Bennett, B. J., Mohandas, N., and Coppel, R. L. (1997). Defining the minimal domain of the Plasmodium falciparum protein MESA involved in the interaction with the red cell membrane skeletal protein 4.1. J. Biol. Chem. 272, 15299–15306. doi:10.1074/jbc.272.24.15299
Black, C. G., Proellocks, N. I., Kats, L. M., Cooke, B. M., Mohandas, N., Coppel, R. L., et al. (2008). In vivo studies support the role of trafficking and cytoskeletal-binding motifs in the interaction of MESA with the membrane skeleton of Plasmodium falciparum-infected red blood cells. Mol. Biochem. Parasitol. 160, 143–147. doi:10.1016/j.molbiopara.2008.04.001
Botha, M., Chiang, A. N., Needham, P. G., Stephens, L. L., Hoppe, H. C., Külzer, S., et al. (2011). Plasmodium falciparum encodes a single cytosolic type I Hsp40 that functionally interacts with Hsp70 and is upregulated by heat shock. Cell Stress Chaperones 16, 389–401. doi:10.1007/s12192-010-0250-6
Botha, M., Pesce, E.-R., and Blatch, G. L. (2007). The Hsp40 proteins of Plasmodium falciparum and other apicomplexa: Regulating chaperone power in the parasite and the host. Int. J. Biochem. Cell Biol. 39, 1781–1803. doi:10.1016/j.biocel.2007.02.011
Bozdech, Z., Llinás, M., Pulliam, B. L., Wong, E. D., Zhu, J., and DeRisi, J. L. (2003). The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum. PLoS Biol. 1, E5. doi:10.1371/journal.pbio.0000005
Cassel, J. A., Ilyin, S., McDonnell, M. E., and Reitz, A. B. (2012). Novel inhibitors of heat shock protein Hsp70-mediated luciferase refolding that bind to DnaJ. Bioorg. Med. Chem. 20, 3609–3614. doi:10.1016/j.bmc.2012.03.067
Cockburn, I. L., Boshoff, A., Pesce, E. R., and Blatch, G. L. (2014). Selective modulation of plasmodial Hsp70s by small molecules with antimalarial activity. Biol. Chem. 395, 1353–1362. doi:10.1515/hsz-2014-0138
Cockburn, I. L., Pesce, E. R., Pryzborski, J. M., Davies-Coleman, M. T., Clark, P. G., Keyzers, R. A., et al. (2011). Screening for small molecule modulators of Hsp70 chaperone activity using protein aggregation suppression assays: Inhibition of the plasmodial chaperone PfHsp70-1. Biol. Chem. 392, 431–438. doi:10.1515/BC.2011.040
Cooke, B. M., Glenister, F. K., Mohandas, N., and Coppel, R. L. (2002). Assignment of functional roles to parasite proteins in malaria-infected red blood cells by competitive flow-based adhesion assay. Br. J. Haematol. 117, 203–211. doi:10.1046/j.1365-2141.2002.03404.x
Coppel, R. L., Lustigman, S., Murray, L., and Anders, R. F. (1988). MESA is a Plasmodium falciparum phosphoprotein associated with the erythrocyte membrane skeleton. Mol. Biochem. Parasitol. 31, 223–231. doi:10.1016/0166-6851(88)90152-1
Coppel, R. L. (1992). Repeat structures in a Plasmodium falciparum protein (MESA) that binds human erythrocyte protein 4.1. Mol. Biochem. Parasitol. 50, 335–347. doi:10.1016/0166-6851(92)90231-8
Daniyan, M. O., and Blatch, G. L. (2017). Plasmodial Hsp40s: New avenues for antimalarial drug discovery. Curr. Pharm. Des. 23, 4555–4570. doi:10.2174/1381612823666170124142439
Daniyan, M. O., Boshoff, A., Prinsloo, E., Pesce, E. R., and Blatch, G. L. (2016). The malarial exported PFA0660w is an Hsp40 co-chaperone of PfHsp70-x. PLoS ONE 11, e0148517. doi:10.1371/journal.pone.0148517
Day, J., Passecker, A., Beck, H. P., and Vakonakis, I. (2019). The Plasmodium falciparum Hsp70-x chaperone assists the heat stress response of the malaria parasite. FASEB J. 33, 14611–14624. doi:10.1096/fj.201901741R
de Koning-Ward, T. F., Gilson, P. R., Boddey, J. A., Rug, M., Smith, B. J., Papenfuss, A. T., et al. (2009). A newly discovered protein export machine in malaria parasites. Nature 459, 945–949. doi:10.1038/nature08104
Diehl, M., Roling, L., Rohland, L., Weber, S., Cyrklaff, M., Sanchez, C. P., et al. (2021). Co-chaperone involvement in knob biogenesis implicates host-derived chaperones in malaria virulence. PLoS Pathog. 17, e1009969. doi:10.1371/journal.ppat.1009969
Diez-Silva, M., Park, Y., Huang, S., Bow, H., Mercereau-Puijalon, O., Deplaine, G., et al. (2012). Pf155/RESA protein influences the dynamic microcirculatory behavior of ring-stage Plasmodium falciparum infected red blood cells. Sci. Rep. 2, 614. doi:10.1038/srep00614
Dutta, T., Pesce, E. R., Maier, A. G., and Blatch, G. L. (2021a). Role of the J domain protein family in the survival and pathogenesis of Plasmodium falciparum. Adv. Exp. Med. Biol. 1340, 97–123. doi:10.1007/978-3-030-78397-6_4
Dutta, T., Singh, H., Gestwicki, J. E., and Blatch, G. L. (2021b). Exported plasmodial J domain protein, PFE0055c, and PfHsp70-x form a specific co-chaperone-chaperone partnership. Cell Stress Chaperones 26, 355–366. doi:10.1007/s12192-020-01181-2
Elsworth, B., Matthews, K., Nie, C. Q., Kalanon, M., Charnaud, S. C., Sanders, P. R., et al. (2014). PTEX is an essential nexus for protein export in malaria parasites. Nature 511, 587–591. doi:10.1038/nature13555
Elsworth, B., Sanders, P. R., Nebl, T., Batinovic, S., Kalanon, M., Nie, C. Q., et al. (2016). Proteomic analysis reveals novel proteins associated with the Plasmodium protein exporter PTEX and a loss of complex stability upon truncation of the core PTEX component. PTEX150. Cell. Microbiol. 18, 1551–1569. doi:10.1111/cmi.12596
Grover, M., Chaubey, S., Ranade, S., and Tatu, U. (2013). Identification of an exported heat shock protein 70 in Plasmodium falciparum. Parasite 20, 2. doi:10.1051/parasite/2012002
Hennessy, F., Cheetham, M. E., Dirr, H. W., and Blatch, G. L. (2000). Analysis of the levels of conservation of the J domain among the various types of DnaJ-like proteins. Cell Stress Chaperones 5 (4), 347–358.
Hennessy, F., Nicoll, W. S., Zimmermann, R., Cheetham, M. E., and Blatch, G. L. (2005). Not all J domains are created equal: Implications for the specificity of hsp40-hsp70 interactions. Protein Sci. 14, 1697–1709. doi:10.1110/ps.051406805
Hiller, N. L., Bhattacharjee, S., van Ooij, C., Liolios, K., Harrison, T., Lopez-Estraño, C., et al. (2004). A host-targeting signal in virulence proteins reveals a secretome in malarial infection. Science 306, 1934–1937. doi:10.1126/science.1102737
Jha, P., Laskar, S., Dubey, S., Bhattacharyya, M. K., and Bhattacharyya, S. (2017). Plasmodium Hsp40 and human Hsp70: A potential cochaperone-chaperone complex. Mol. Biochem. Parasitol. 214, 10–13. doi:10.1016/j.molbiopara.2017.03.003
Jonsdottir, T. K., Gabriela, M., and Gilson, P. R. (2021). The role of malaria parasite heat shock proteins in protein trafficking and remodelling of red blood cells. Adv. Exp. Med. Biol. 1340, 141–167. doi:10.1007/978-3-030-78397-6_6
Kampinga, H. H., Andreasson, C., Barducci, A., Cheetham, M. E., Cyr, D., Emanuelsson, C., et al. (2019). Function, evolution, and structure of J-domain proteins. Cell Stress Chaperones 24, 7–15. doi:10.1007/s12192-018-0948-4
Kilili, G. K., and LaCount, D. J. (2011). An erythrocyte cytoskeleton-binding motif in exported Plasmodium falciparum proteins. Eukaryot. Cell 10, 1439–1447. doi:10.1128/EC.05180-11
Külzer, S., Charnaud, S., Dagan, T., Riedel, J., Mandal, P., Pesce, E. R., et al. (2012). Plasmodium falciparum-encoded exported hsp70/hsp40 chaperone/co-chaperone complexes within the host erythrocyte. Cell. Microbiol. 14, 1784–1795. doi:10.1111/j.1462-5822.2012.01840.x
Külzer, S., Rug, M., Brinkmann, K., Cannon, P., Cowman, A., Lingelbach, K., et al. (2010). Parasite-encoded Hsp40 proteins define novel mobile structures in the cytosol of the P. falciparum-infected erythrocyte. Cell. Microbiol. 12, 1398–1420. doi:10.1111/j.1462-5822.2010.01477.x
Kumar, V., Behl, A., Sharma, R., Sharma, A., and Hora, R. (2019). Plasmodium helical interspersed subtelomeric family-an enigmatic piece of the Plasmodium biology puzzle. Parasitol. Res. 118, 2753–2766. doi:10.1007/s00436-019-06420-9
Laskowski, R. A., and Swindells, M. B. (2011). LigPlot+: Multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 51, 2778–2786. doi:10.1021/ci200227u
Maier, A. G., Rug, M., O'Neill, M. T., Brown, M., Chakravorty, S., Szestak, T., et al. (2008). Exported proteins required for virulence and rigidity of Plasmodium falciparum-infected human erythrocytes. Cell 134, 48–61. doi:10.1016/j.cell.2008.04.051
Marti, M., Good, R. T., Rug, M., Knuepfer, E., and Cowman, A. F. (2004). Targeting malaria virulence and remodeling proteins to the host erythrocyte. Science 306, 1930–1933. doi:10.1126/science.1102452
Mills, J. P., Diez-Silva, M., Quinn, D. J., Dao, M., Lang, M. J., Tan, K. S., et al. (2007). Effect of plasmodial RESA protein on deformability of human red blood cells harboring Plasmodium falciparum. Proc. Natl. Acad. Sci. U. S. A. 104, 9213–9217. doi:10.1073/pnas.0703433104
Morahan, B. J., Strobel, C., Hasan, U., Czesny, B., Mantel, P. Y., Marti, M., et al. (2011). Functional analysis of the exported type IV HSP40 protein PfGECO in Plasmodium falciparum gametocytes. Eukaryot. Cell 10, 1492–1503. doi:10.1128/EC.05155-11
Moses, M. A., Kim, Y. S., Rivera-Marquez, G. M., Oshima, N., Watson, M. J., Beebe, K. E., et al. (2018). Targeting the Hsp40/Hsp70 chaperone axis as a novel strategy to treat castration-resistant prostate cancer. Cancer Res. 78, 4022–4035. doi:10.1158/0008-5472.CAN-17-3728
Njunge, J. M., Ludewig, M. H., Boshoff, A., Pesce, E. R., and Blatch, G. L. (2013). Hsp70s and J proteins of Plasmodium parasites infecting rodents and primates: Structure, function, clinical relevance, and drug targets. Curr. Pharm. Des. 19, 387–403. doi:10.2174/138161213804143734
Oakley, M. S., Gerald, N., McCutchan, T. F., Aravind, L., and Kumar, S. (2011). Clinical and molecular aspects of malaria fever. Trends Parasitol. 27, 442–449. doi:10.1016/j.pt.2011.06.004
Oakley, M. S., Kumar, S., Anantharaman, V., Zheng, H., Mahajan, B., Haynes, J. D., et al. (2007). Molecular factors and biochemical pathways induced by febrile temperature in intraerythrocytic Plasmodium falciparum parasites. Infect. Immun. 75, 2012–2025. doi:10.1128/IAI.01236-06
Pasini, E. M., Kirkegaard, M., Mortensen, P., Lutz, H. U., Thomas, A. W., and Mann, M. (2006). In-depth analysis of the membrane and cytosolic proteome of red blood cells. Blood 108, 791–801. doi:10.1182/blood-2005-11-007799
Pei, X., Guo, X., Coppel, R., Bhattacharjee, S., Haldar, K., Gratzer, W., et al. (2007). The ring-infected erythrocyte surface antigen (RESA) of Plasmodium falciparum stabilizes spectrin tetramers and suppresses further invasion. Blood 110, 1036–1042. doi:10.1182/blood-2007-02-076919
Pesce, E.-R., Blatch, G. L., and Edkins, A. L. (2016). Hsp40 co-chaperones as drug targets: Towards the development of specific inhibitors. Top. Med. Chem. 19, 163–196. doi:10.1007/7355_2015_92
Pesce, E. R., and Blatch, G. L. (2014). Plasmodial Hsp40 and Hsp70 chaperones: Current and future perspectives. Parasitology 141, 1167–1176. doi:10.1017/S003118201300228X
Petersen, C., Nelson, R., Magowan, C., Wollish, W., Jensen, J., and Leech, J. (1989). The mature erythrocyte surface antigen of Plasmodium falciparum is not required for knobs or cytoadherence. Mol. Biochem. Parasitol. 36, 61–65. doi:10.1016/0166-6851(89)90200-4
Petersen, W., Külzer, S., Engels, S., Zhang, Q., Ingmundson, A., Rug, M., et al. (2016). J-dot targeting of an exported HSP40 in Plasmodium falciparum-infected erythrocytes. Int. J. Parasitol. 46, 519–525. doi:10.1016/j.ijpara.2016.03.005
Przyborski, J. M., Diehl, M., and Blatch, G. L. (2015). Plasmodial HSP70s are functionally adapted to the malaria parasite life cycle. Front. Mol. Biosci. 2, 34. doi:10.3389/fmolb.2015.00034
Qin, H. L., Zhang, Z. W., Lekkala, R., Alsulami, H., and Rakesh, K. P. (2020). Chalcone hybrids as privileged scaffolds in antimalarial drug discovery: A key review. Eur. J. Med. Chem. 193, 112215. doi:10.1016/j.ejmech.2020.112215
Rhiel, M., Bittl, V., Tribensky, A., Charnaud, S. C., Strecker, M., Müller, S., et al. (2016). Trafficking of the exported P. falciparum chaperone PfHsp70x. Sci. Rep. 6, 36174. doi:10.1038/srep36174
Sahu, W., Bai, T., Panda, P. K., Mazumder, A., Das, A., Ojha, D. K., et al. (2022). Plasmodium falciparum HSP40 protein eCiJp traffics to the erythrocyte cytoskeleton and interacts with the human HSP70 chaperone HSPA1. FEBS Lett. 596, 95–111. doi:10.1002/1873-3468.14255
Sargeant, T. J., Marti, M., Caler, E., Carlton, J. M., Simpson, K., Speed, T. P., et al. (2006). Lineage-specific expansion of proteins exported to erythrocytes in malaria parasites. Genome Biol. 7, R12. doi:10.1186/gb-2006-7-2-r12
Schmidt, J., and Vakonakis, I. (2020). Structure of the substrate-binding domain of Plasmodium falciparum heat-shock protein 70-x. Acta Crystallogr. F. Struct. Biol. Commun. 76, 495–500. doi:10.1107/S2053230X2001208X
Shonhai, A., and Blatch, G. L. (2021). Heat shock proteins of malaria: Highlights and future prospects. Adv. Exp. Med. Biol. 1340, 237–246. doi:10.1007/978-3-030-78397-6_10
Shonhai, A., Boshoff, A., and Blatch, G. L. (2007). The structural and functional diversity of Hsp70 proteins from Plasmodium falciparum. Protein Sci. 16, 1803–1818. doi:10.1110/ps.072918107
Sievers, F., and Higgins, D. G. (2018). Clustal Omega for making accurate alignments of many protein sequences. Protein Sci. 27, 135–145. doi:10.1002/pro.3290
Silva, M. D., Cooke, B. M., Guillotte, M., Buckingham, D. W., Sauzet, J. P., Le Scanf, C., et al. (2005). A role for the Plasmodium falciparum RESA protein in resistance against heat shock demonstrated using gene disruption. Mol. Microbiol. 56, 990–1003. doi:10.1111/j.1365-2958.2005.04603.x
Sinha, S., Batovska, D. I., Medhi, B., Radotra, B. D., Bhalla, A., Markova, N., et al. (2019). In vitro anti-malarial efficacy of chalcones: Cytotoxicity profile, mechanism of action and their effect on erythrocytes. Malar. J. 18, 421. doi:10.1186/s12936-019-3060-z
Stothard, P. (2000). The sequence manipulation suite: JavaScript programs for analyzing and formatting protein and DNA sequences. BioTechniques 28, 1102–1104. doi:10.2144/00286ir01
Trott, O., and Olson, A. J. (2010). AutoDock vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J. Comput. Chem. 31, 455–461. doi:10.1002/jcc.21334
van Gestel, R. A., van Solinge, W. W., van der Toorn, H. W., Rijksen, G., Hec, k. A. J., van Wijk, R., et al. (2010). Quantitative erythrocyte membrane proteome analysis with Blue-native/SDS PAGE. J. Proteomics 2010 73, 456–465. doi:10.1016/j.jprot.2009.08.010
Waller, K. L., Nunomura, W., An, X., Cooke, B. M., Mohandas, N., and Coppel, R. L. (2003). Mature parasite-infected erythrocyte surface antigen (MESA) of Plasmodium falciparum binds to the 30-kDa domain of protein 4.1 in malaria-infected red blood cells. Blood 102, 1911–1914. –1914. doi:10.1182/blood-2002-11-3513
Waterhouse, A., Bertoni, M., Bienert, S., Studer, G., Tauriello, G., Gumienny, R., et al. (2018). SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303. doi:10.1093/nar/gky427
Keywords: heat shock proteins, J domain proteins, molecular chaperones and co-chaperones, Plasmodium falciparum, protein export, protein folding
Citation: Almaazmi SY, Singh H, Dutta T and Blatch GL (2022) Exported J domain proteins of the human malaria parasite. Front. Mol. Biosci. 9:978663. doi: 10.3389/fmolb.2022.978663
Received: 26 June 2022; Accepted: 02 August 2022;
Published: 31 August 2022.
Edited by:
Stanley Makumire, University of Cape Town, South AfricaReviewed by:
Jude Przyborski, Justus-Liebig-University Gießen, GermanyDhanasekaran Shanmugam, National Chemical Laboratory (CSIR), India
Copyright © 2022 Almaazmi, Singh, Dutta and Blatch. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gregory L. Blatch, Zy5ibGF0Y2hAcnUuYWMuemE=
†These authors have contributed equally to this work
‡ORCID: Shaikha Y. Almaazmi, orcid.org/0000-0002-8145-7998Harpreet Singh, orcid.org/0000-0001-7202-3912Tanima Dutta, orcid.org/0000-0002-0882-2602Gregory L. Blatch, orcid.org/0000-0003-0778-8577
 Shaikha Y. Almaazmi
Shaikha Y. Almaazmi Harpreet Singh
Harpreet Singh Tanima Dutta3,4,5†‡
Tanima Dutta3,4,5†‡ Gregory L. Blatch
Gregory L. Blatch