
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Mol. Biosci., 26 September 2022
Sec. Molecular Diagnostics and Therapeutics
Volume 9 - 2022 | https://doi.org/10.3389/fmolb.2022.965730
This article is part of the Research TopicExtracellular Vesicles and Cell-cell Communication in Normal Cellular Processes and CancerView all 7 articles
 Dibyashree Chhetri1
Dibyashree Chhetri1 Srinivasan Vengadassalapathy2
Srinivasan Vengadassalapathy2 Santhosh Venkadassalapathy3
Santhosh Venkadassalapathy3 Varadharaju Balachandran4
Varadharaju Balachandran4 Vidhya Rekha Umapathy5
Vidhya Rekha Umapathy5 Vishnu Priya Veeraraghavan6
Vishnu Priya Veeraraghavan6 Selvaraj Jayaraman6
Selvaraj Jayaraman6 Shankargouda Patil7
Shankargouda Patil7 Ashok Iyaswamy8
Ashok Iyaswamy8 Kanagaraj Palaniyandi1*
Kanagaraj Palaniyandi1* Dhanavathy Gnanasampanthapandian1*
Dhanavathy Gnanasampanthapandian1*Doublecortin-like kinase 1 (DCLK1), a protein molecule, has been identified as a tumor stem cell marker in the cancer cells of gastrointestinal, pancreas, and human colon. DCLK1 expression in cancers, such as breast carcinoma, lung carcinoma, hepatic cell carcinoma, tuft cells, and human cholangiocarcinoma, has shown a way to target the DCLK1 gene and downregulate its expression. Several studies have discussed the inhibition of tumor cell proliferation along with neoplastic cell arrest when the DCLK1 gene, which is expressed in both cancer and normal cells, was targeted successfully. In addition, previous studies have shown that DCLK1 plays a vital role in various cancer metastases. The correlation of DCLK1 with numerous stem cell receptors, signaling pathways, and genes suggests its direct or an indirect role in promoting tumorigenesis. Moreover, the impact of DCLK1 was found to be related to the functioning of an oncogene. The downregulation of DCLK1 expression by using targeted strategies, such as embracing the use of siRNA, miRNA, CRISPR/Cas9 technology, nanomolecules, specific monoclonal antibodies, and silencing the pathways regulated by DCLK1, has shown promising results in both in vitro and in vivo studies on gastrointestinal (GI) cancers. In this review, we will discuss about the present understanding of DCLK1 and its role in the progression of GI cancer and metastasis.
Doublecortin-like kinase 1 (DCLK1) is a protein kinase, belonging to the doublecortin family of microtubule-associated protein present in the cytoplasm (Lipka et al., 2016; Sameri et al., 2021). DCLK1 consists of two termini: one, serine/threonine kinase associated with Ca2+/calmodulin-dependent protein 1 (CaMKI) kinase domain, and N-terminal region consists of two doublecortin kinase motifs (DCX1 and DCX2), with a microtubule-associated function (Reiner et al., 2006; Le Hellard et al., 2009; Fu et al., 2013; Wang et al., 2017a). It was initially discovered for its role in the CNS and functioned in the early neurogenesis by microtubule elongation (Moores et al., 2006). It was named as CaM kinase-like 1 protein (Dcamkl1) or KIAA0369 in the preliminary stage (Mizuguchi et al., 1999). Based on the explanation from various studies with regard to the functionality of DCLK1, it has been shown that DCLK1 functions due to the overexpression of the kinase protein that leads to microtubule elongation (Jean et al., 2012; Kenney et al., 2016; Dandawate et al., 2019; Cao et al., 2020; Chandrakesan et al., 2020). DCLK1 protein expression is controlled by the DCLK1 gene, which as a result of alternative promoters and splicing mechanism encodes four different isoforms (Chandrakesan et al., 2020; O'Connell et al., 2015). DCLK1 has many variants ranging from ∼80 to 82 kDa long and ∼45–50 kDa short isoforms (Reiner et al., 2006; Sarkar et al., 2017b; Chandrakesan et al., 2020). The overexpression of DCLK1 protein is in various cancers, including gastric (Liu et al., 2020), pancreatic (Dandawate et al., 2019), colon (Chandrakesan et al., 2017), renal (Weygant et al., 2016), and breast (Liu et al., 2019; Sureban et al., 2015; Panneerselvam et al., 2020). DCLK1’s role in initiation of tumor growth (Westphalen et al., 2016), development metastasis (Ito et al., 2016), epithelial—mesenchymal transition (EMT) (Weygant et al., 2015; Chandrakesan et al., 2016b), cancer stemness, and pro-survival signaling has been demonstrated by experiments both in vivo and in vitro (Chandrakesan et al., 2014; Dandawate et al., 2019).
After the preliminary description of DCLK1 in the CNS, current advances have almost completely targeted on regions outside the CNS, showing the location of DCLK1 tuft cells throughout the digestive system and several cancer types. According to the studies from whole human genome sequencing, DCLK1 is classified as top 15 putative drivers for causing gastric cancer as a result of somatic mutations (Patel et al., 2016). In a breast cancer study, DCLK1 in breast cancer cell lines are found to have cancer stem cell-like characteristics, indicating DCLK1 to be a potential early diagnostic indicator for breast cancer (Gzil et al., 2019). In human colorectal cancer (CRC), upregulation of DCLK1 corelated with increased mortality and recurrence (Gao et al., 2016). Therefore, to mitigate recurrence and mortality, several hypotheses have been put forward with immunotherapy by using chimeric antigen receptor T cells (CAR-T) followed by human CRC treatment with DCLK1-87 mAb (Sureban et al., 2019; Cao et al., 2020). In a study of human gastrointestinal (GI) cancer, it was discovered that an increased level of DCLK1, Notch1, and small nucleolar RNA host gene 1 (SNHG1) expression was found in cases diagnosed with GI cancer (Liu et al., 2020). Furthermore, their research found that miR-15b-targeting DCLK1 regulated Notch1 expression with restricted EMT in gastrointestinal cancer cells (Liu et al., 2020). The Cancer Genome Atlas (TCGA) database revealed elevated DCLK1 in lung squamous cell adenocarcinoma (LUAD), indicating that it is an implicit target in non-small cell lung cancer (NSCLC) (Panneerselvam et al., 2020). In humans, DCLK1 isoforms (long and short) shares a linked domain (Ge et al., 2018; O'Connell et al., 2015). Bailey et al. (2014) demonstrated elevated expression of DCLK1 in pancreatic neoplasms in mice with distinct subpopulations having CSC-like properties. Therefore, upon further investigations, DCLK1 was found to represent a therapeutic target due to its high expression in human pancreatic CSCs and clinical samples (Ito et al., 2016). Moreover, researchers (Ge et al., 2018) demonstrated renal cell malignancy promotes the expression of certain DCLK1 alternative splice variants. These variants in coculturing experiments were found to regulate self-renewal and chemo-resistance (Ge et al., 2018; Panneerselvam et al., 2020). However, in an animal model, strong inhibition of tumor growth was observed upon the use of monoclonal antibody (CBT-15) (Sureban et al., 2019). These findings opened new therapy choices available to patients with often incurable malignancy.
Furthermore, studies relating to biochemical and structural insights provided elaborated ATP-binding site information and crystal structure of the DCLK1 kinase (Patel et al., 2016). A systemic regulation of microtubules is required for controlling cell growth and division. The crystal structure additionally provides evidence on the cause of tumorigenesis upon the loss of kinase function, providing potential framework for future design (Patel et al., 2016). Today, several studies (Sureban et al., 2013; Li et al., 2018) on DCLK1 as potent tumor inducers in various cancers have been confirmed DCLK1 to have critical role in regulating cancer pathways. In addition, the fact that it regulates many miRNAs, such as miR-let-7a and miR-200, is more evident that it promotes proliferation and carcinogenesis (Sureban et al., 2013; Li et al., 2018).
Moreover, this review summarizes the effect of upregulated DCLK1 in several cancers along with the signaling pathways. We also highlighted the downregulation mechanism of DCLK1 by the use of small inhibitory molecules along with exosomes to target DCLK1, which could potentially prevent carcinogenesis and metastasis as a result of exosome’s versatile paracrine activity.
The history of DCLK1 dates back to the 1990s when it was first discovered as KIAA0369, a putative kinase that was homologous to brain-specific gene doublecortin (DCX) l structurally (Omori et al., 1998). According to Nagase et al. (1997), KIAA0369 was isolated from the human brain that consisted of 100 sequences and expressed in 15 different regions of the brain in varying intensities. When KIAA0369 and doublecortin were compared, both the genes showed the N-terminal DCX domain (Omori, et al., 1998). In addition, KIAA0369 included a Ca2+/calmodulin-dependent kinase (CaM kinase)-like domain that was engaged in many calcium signaling pathways (Burgess et al., 1999). Omori et al. (1998) from their results of four splice variants in the fetal and adult brains suggested its significant role in development of the CNS. Out of the four variants, KIAA0369-AS (type A) and KIAA0369-BS (type B) are shorter version variants and KIAA0369-AL (type A) and KIAA0369-BL (type B) are longer version variants. Variant A was found mainly in the fetal brain, while variation B, which deleted the doublecortin domain, was exhibited in both the adult and fetal brains (Omori et al., 1998).
Mapping of this gene using fluorescence in situ hybridization (FISH) to chromosome 13q13–q14 (Omori et al., 1998) was carried out and its CNS-related function has been described at this locus. Studies in the mice model showed that loss of DCX domains leads to anxious behavioral phenotype (Schenk et al., 2010). The first function of KIAA0369 as DCLK1 outside the CNS was in the epithelial cells that were marked as stomach stem cells in 2006 (Singh et al., 2016). From a study using mice, it has shown that DCLK1 was observed at the +4 position in intestinal crypts (May et al., 2008) and Lgr5 (Bailey et al., 2014). Furthermore, several studies showed that DCLK1 is expressed in both healthy and specialized stem cell (tuft cells) niche that has proliferative property (May et al., 2014; Westphalen et al., 2014). These studies supported the fact that DCLK1 assists cell growth and organoid formation in mice models (Yi et al., 2019). To date, many studies on DCLK1 have supported it as a CSC marker for various cancers (Chandrakesan, et al., 2015a; Chandrakesan et al., 2017; Panneerselvam et al., 2020; Lorenzo et al., 2021) (Figure 1).
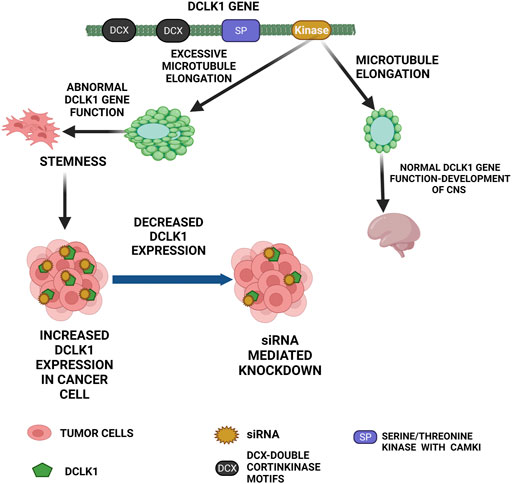
FIGURE 1. Functional role of DCLK1 in normal and cancer cells. The primary function of DCLK1 in the CNS is the microtubule elongation. Outside the CNS, it maintains stemness in the intestinal crypts. The kinase domain of DCLK1 leads to uncontrolled microtubule elongation and stemness, which in turn leads to cancer cell formation with increase in DCLK1 expression. As observed, DCLK1 upregulation in several malignancies can be downregulated by siRNA-mediated knockdowns and other related approaches.
DCLK1 in the GI tract was identified in 2006 as an intestinal stem cell marker along with its correlation to CRC cells stemness (May et al., 2008; Li and Bellows 2013). To date, DCLK1 is used as a tumor cell marker. Cancer-related processes including cancer progression, cancer therapy resistance, and metastasis are regulated by DCLK1-expressing CSCs (Cao et al., 2020).
The experimental work by Westphalen et al. (2017) reported the main cause of cancer in the colon is due to the colon cancer-initiating cells formed by long-lived tuft cells. In gastric cancer study, the enhanced EMT process by the DCLK1-mediated Notch1 pathway was observed (Liu et al., 2020). Studies from Liu et al. (2018) demonstrated the oncogenic role of DCLK1 by downregulation of various tumor suppressor microRNAs. In CRC cell line, DCLK1 isoform 2 is highly expressed, which upon silencing inhibited the tumor progression in vivo (Liu et al., 2018). In studies related to cancer cells, pluripotency maintenance factors along with the EMT-related transcription factors are highly expressed (Liu et al., 2020). In addition, Panneerselvam et al. (2020) in their study in non-small cell lung cancer (NSCLC) emphasized on DCLK1’s chemoresistance and stemness properties. Pancreatic ductal adenocarcinoma (PDAC)-based study showed overexpression of KDM3A that increased expression of DCLK1 (Dandawate et al., 2019).
Ever since DCLK1 was first observed outside the CNS, it has been used as a marker for various cancers.
Gastrointestinal cancers are cancers that are usually the result of cancer metastasis. The organs vulnerable to GI cancer include the esophagus, stomach, colon, rectum, anus, biliary system, and small intestine. DCLK1 along with LGR5+ stem cells and DCLK1+ tuft cells are normally observed in the GI tract (Chandrakesan et al., 2017; Kalantari et al., 2017). Studies on response to inflammation reported that inflammation is progressed upon DCLK1 expression (Yi et al., 2019; Wu et al., 2020b) in GI cancer cells that serve as EMT precursors and tumor formation (Chandrakesan et al., 2016b; Li et al., 2018; Liu et al., 2018). CD8+ T cell (Wu et al., 2020b; Chandrakesan et al., 2020) is an immune cell, with key regulatory GI function associated with tumor suppression. Tumor suppression and tumor development and spread are governed by the M1 pro-inflammatory macrophage state and M2 anti-inflammatory/tissue-repair status, respectively (Cao et al., 2020). DCLK1 expression is associated with the expression of immune cell markers for Treg, markers of monocytes, tumor-associated macrophages (TAMs), and M2 macrophages that suppresses the CD8+ T cells functionality, leading to immune cell function loss (Wu et al., 2020b). miRNAs are non-coding RNAs, which have epigenetic role and promotes stemness, EMT, expression of oncogenes along with various cancer-inducing pathways (Weygant et al., 2016).
Colorectal cancer (CRC) ranks fourth globally with 18.1 million as an estimated reports in 2020 (Sung et al., 2021). CSCs are cells that have self-renewal and regeneration potency (Al-Hajj and Clarke 2004; O'Brien et al., 2010). CSCs in carcinogenesis and treatment resistance have become an important domain of investigation (Chandrakesan et al., 2014; Kang et al., 2020) and metastasis to the nearby organs causing death in many cases (Shiozawa et al., 2013; Steinbichler et al., 2020). Chemotherapy-resistant CSCs are precursors to CRC recurrence (Das et al., 2020). DCLK1 are found to be located with well-established GI stem cell markers like Lgr5 in the stomach (Chandrakesan et al., 2016a; Kalantari et al., 2017). In a mice model, DCLK1 was found located at “+4 position” of the crypt along with intestinal stem cells (Gzil et al., 2019). Among the reported splice variants of DCLK1 (Vreugdenhil et al., 2001; Burgess and Reiner 2002), DCLK1-S is expressed in human colon cancer cells and CRC. Furthermore, studies from Kang et al. (2020) showed that malignancy in CRC is mainly due to the regulation of DCLK1 and Lgr5. An increased DCLK1 in response to the inflammation process also assists in tumorigenesis as a result of dysplasia as explained (Wang et al., 2017c). DCLK1 overexpression has also been explained to worsen the survival rate and metastasis (Gzil et al., 2019). Therefore, it is evident that higher DCLK1 and Lgr5 expressions help in the prediction of risk and stage of the patient along with their survival rate (Kang et al., 2020). DCLK1 promotes the CRC through miR-137 and miR-15a (Razi et al., 2021). It is also reported to be an independent biomarker for radioresistance in CRC (Yang et al., 2018).
Breast carcinoma (BC) is the lumps or mass formation due to abnormal growth of the breast cells. Globally, BC is most common in women, leading to the topmost cause of mortality case in women (Sung et al., 2021). BC mortality mainly by distant metastasis is due to the secondary tumor formation. DCLK1 in BC regulates the regulating Wnt/β-catenin signaling pathway, and in presence of Wnt/β-catenin pathway inhibitor (Wnt/β-catenin-IN-1), the cells’ proliferation is lowered (Wang et al., 2019c; Liu et al., 2019). With increased DCLK1 expression, the ERK-MAPK pathway induces, which promotes the progression of BC malignancy and metastasis (Wang et al., 2019c). Furthermore, DCLK1-overexpressing cancer cells are observed for lower X ZO-1, which is an epithelial marker and increased mesenchymal markers, including ZEB1 and vimentin (Liu et al., 2019). DCLK1 function in BC cells, as defined by Liu et al. (2019) in their experiment, demonstrated higher DCLK1 expression in BC patients, which resulted in a poor survival rate when compared with those of lower DCLK1 expression). The study related to the use of CRISPR/Cas9 technology, for the knockout in DCLK1 is overexpressed and the overexpression of DCLK1 in less expressed BC cell lines, respectively, suggested that DCLK1 promotes cancer invasion and metastasis, which on downregulation shows reduced invasive and migration ability (Liu et al., 2019). By silencing the DCLK1, the self-renewal, EMT, and metastasis are prevented (Chandrakesan et al., 2015b).
Pancreatic cancer (PC) is cancer that is caused as a result of solid malignancies with less than 7% survival rate for 5 years (May et al., 2008). Several studies relating to pancreatic cancer related to cell population in the pancreas that serves as cancer cell progenitors for cancer development (Ito et al., 2016; Westphalen et al., 2016; Li et al., 2018). DCLK1 serves as a molecular marker for pancreatic ductal adenocarcinoma (PDAC) (Dandawate et al., 2019; Maruno et al., 2021).
Liu et al. (2020) stated that DCLK1 is associated with long-lived quiescent pancreatic cells that showed an inflammatory immune response in vivo and high proliferation in vitro, leading to the growth of the organ. DCLK1 regulated pluripotency and angiogenesis via regulating mir-145 and miR-200 in pancreatic cancer (Sureban et al., 2013). Owing to the association of DCLK1 in EMT (Bure et al., 2019), B cell-specific Moloney murine leukemia virus insertion site 1 (Bmi1) showed evidence that DCLK1 leads to PC (Li et al., 2018). EMT was reported to be hindered by upregulating epithelial marker E-cadherin. However, downregulating DCLK1 showed that it is only indirectly associated with the Bim-1 regulator (Chandrakesan et al., 2017). In human PC, DCLK1 overexpression leads to increased proliferation and mesenchymal phenotype (Chandrakesan et al., 2016b; Li et al., 2018). DCLK1 association with an oncogene KRAS in the experiment carried out by Westphalen et al. (2016) has shown that tissue injury and KRAS mutation together result in inducing pancreatic tumorigenesis, which otherwise is inactive. A kinase inhibitor, LRRK2-IN-1 selectively inhibited the pancreatic cancer progression (Weygant et al., 2014). The discovery of selective inhibitor DCLK1 (DCLK1-IN-1) inhibits the PDAC growth significantly (Ferguson, et al., 2020).
Intestinal cancer has tuft cells, which are considered as the potent inducer of tumor that led to polyps’ formation under cellular stress (Westphalen et al., 2014; Testa et al., 2018). Adenomatous polyposis coli (APC) is a tumor suppressor gene inhibiting polyp’s formation, which upon mutation causes neoplasia formation causing cancer (Dazard et al., 2014; Chandrakesan et al., 2017). Chronic stimuli followed by hyperplasia and early neoplasia confirm that tuft cells have major function in tumorigenesis (Middelhoff et al., 2017; Banerjee et al., 2018). Nakanishi et al. (2013) stated that DCLK1 was identified as a marker for CSC that persistently generated tumor offspring in the polyps of APC (Min/+) mice after cre-mediated silencing of the APC gene (Chandrakesan et al., 2017; O'Connell et al., 2015).
In vivo analysis of APCMin/+ mice resulted in an increase in DCLK1 and pro signaling markers (Chandrakesan et al., 2017). Cholangiocarcinoma (CCA) is an aggressive cancer that has been increasing worldwide with a high expression of CSCs (Carpino et al., 2011; Suwannakul et al., 2020). Lorenzo et al. (2021) in their in situ analysis confirmed the presence of DCLK1 only in tumor tissues. Furthermore, in CAA cells, inhibition of CD133+ and LGR5+ cell cultures (Lorenzo et al., 2021) exerted non-malignant consequences in primary cultures.
Renal cell carcinoma (RCC) is a cancer caused due to distance metastasis (Hoffmann et al., 2008; Chandrasekar et al., 2017). The main problem in RCC is poor prognosis due to rapid relapse after surgery and therapy resistance (Kucharczyk et al., 2017; Ge et al., 2018; Kim et al., 2021). Ge et al. (2018) in their experiment have shown that DCLK1 long isoforms express CSC markers and are prognostic factors in RCC. DCLK1 and aldehyde dehydrogenase (ALDH) are connected on a biological level; thereby, increasing the possibility of forming a population of renal tumor stem cells that have high clonogenicity and resistance to chemo and radiotherapies (Ge et al., 2018). Studies targeting DCLK1 by specific monoclonal antibodies (mAb) have shown to inhibit tumorigenesis (Sureban et al., 2019; Cao et al., 2020).
Most common lung cancer type is the non-small cell lung cancer (NSCLC). Lung cancer is common both in men and women and has a survival rate of 5-years irrespective of its grade (Cao et al., 2020). Increased DCLK1 expression corresponds with stem cell factors ALDH1A1, CD44, LGR5, and pluripotency factors KLF4, SOX2, MYC, and NANOG, as per studies (Tao et al., 2017; Panneerselvam et al., 2020) (Table 1). DCLK1 functions as tumor inducer in gastric, stomach, intestine, pancreas, colon, breast, and lung. It regulates the expression of other pluripotency markers, cancer-signaling proteins, EMT, and epigenetic switching. The up-to-date literature has suggested that DCLK1 is a key player for cancer progression and metastasis in various cancers.
CSC is a subset of cancer cells that DCLK1 expresses (Chandrakesan et al., 2017; Cao et al., 2020; Lorenzo et al., 2021). Many studies have shown its expression in various cancer types (Chandrakesan et al., 2017; Roy et al., 2021). The bone marrow has the highest number of stem cells, while adult cells have a lower number of stem cells. Their primary function is to maintain cellular integrity and repair (Qu et al., 2015). Stem cells are a select minority of proliferating, self-renewing cells that contribute to tumor growth and the development of cancer (Fuchs and Blau 2020). Stem cells form a small group of cells in a cancer cell that can self-renew and proliferate, which contributes to tumor progression and cancer development (Huang et al., 2020). Several mouse studies have revealed that the CSC or tumor stem cell characterizes the precursor/progeny relationship that forms cancer cell progeny (Shibata and Shen 2015; Dandawate et al., 2019; Lytle et al., 2019). These progenies give rise to tumor-initiating cells, which are in charge of tumor progression (Wlodarczyk et al., 2020). CSCs have vital role in cancer, based on a recent study focused on lineage tracing (Goto et al., 2019). Bailey et al. (2014) identified DCLK1-expressing cells in early stage tumorigeneses. These cells overexpressed ATAT1, HES1, HEY1, IGF1R, and ABL1 and had molecular and morphological characteristics similar to GI tuft cells (Bailey et al., 2014; Roy et al., 2021). On altering these pathways, the clonogenicity of PDAC cell lines was inhibited. DCLK1-expressing tumor stem cells have therapeutic potential (Yamaga et al., 2018; Dandawate et al., 2019; Ferguson et al., 2020). The intestines of APCMin/+ mice had higher expression of factors found in DCLK1+ intestinal tumor cells in a mouse model (Chae and Bothwell 2015). These factors will aid in cancer stem cell-targeted therapy. A previous study suggests that DCLK1 is expressed in normal intestinal cells (Chandrakesan et al., 2015a). In addition to this, the quiescent DCLK1 positive cells function as cancer-initiating cells, leading to metastasis and poor survival rate (Gzil et al., 2019; Kunze et al., 2021). Patients with higher expression of DCLK1 show a higher risk of chemotherapy resistance. In APCMin/+mice, high level of DCLK1 expression suggests its supporting role in the cancer-signaling pathway, self-renewal, and pluripotency (Chandrakesan et al., 2017). Overexpression of DCLK1 in human cholangiocarcinoma was associated with higher expression of CSCs. The subpopulation of CSC is characterized by higher DCLK1 serum expression and serves as a biomarker for early coloanal anastomosis diagnosis (Sarkar et al., 2017a). Therefore, DCLK1 expressing stem cells in several cancers serve as a promising biomarker (Figure 2). DCLK1 is broadly expressed in the majority of intestinal CSCs, breast, lung, and neuronal stem cells. DCLK1-associated CSCs signaling, such as Wnt, Notch, Hedgehog, and YAP/TAZ signaling needs to be explored further.
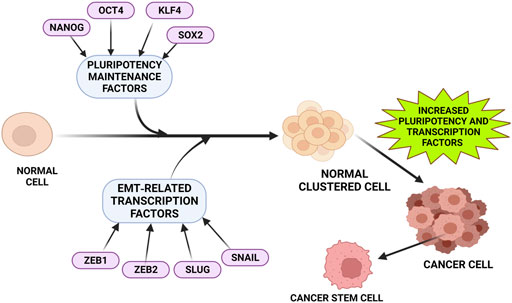
FIGURE 2. Factors that upregulates DCLK1 expression in cancer. Tumor progression and metastasis are often observed to be associated closely with increased pluripotent maintenance factors and EMT-related transcription factors. An increase in these factors leads to the transition of normal cells into cancer cells with increased stemness property and elevated DCLK1 expression. The pluripotent maintenance factors and EMT factors regulate DCLK1 expression in cancer progression.
Several methods that are known to knockdown DCLK1 have been implemented. Previous studies have shown that downregulating DCLK1 by using miRNA (Zhou et al., 2017), siRNA (Sureban et al., 2013; Chandrakesan et al., 2017), and CRISPR/Cas9 (Liu et al., 2019) have been used widely in various cancer cells.
MiR-195 that targets DCLK1 is shown to play vital tumor suppressing role in pancreatic cancer (Zhou et al., 2017). From the study by Sakaguchi et al. (2016), the miRNА-137/DСLK1 axis aid in formation of cancer tissues followed by miR-137 that directly regulated the DСLK1 expression by suppressing the protein level of DCLK1. In an experiment to examine miR-613’s expression and biological functions in hepatocellular cancer (HCC) (Wang et al., 2016), their findings reported that targeting DCLK1 was linked to suppress the miRNA function (Wang et al., 2016). DCLK1 is overexpressed in numerous diseases and is considered as a tumor marker (Chandrakesan et al., 2015a; Panneerselvam et al., 2020; Roy et al., 2021). Wu et al. (2017) in ovarian clear cell carcinoma (OCCC) reported that microRNA-424 (miR-424) inhibits DCLK1 expression, leading to chemo-resistance. DCLK1 knockdown decreases cell’s invading ability that aid in suppressing EMT in OCCC and it plays vital role to strategies relating to treatment and therapeutic target of OCCC. In gall bladder cell (GBС), targeting DCLK1 gene using miR-29с-3р and miR-7-2-3р were found to decrease the migratory сарасity of GBС сells in vitrо аnd in vivо. Therefore, Lu et al. (2020) in their investigation, validated the utility of these microRNAs as diagnostics and potential therapeutic in the future (Table 2).
A study on neuroblastoma miRNA resulted in suppressing cell viability, invasion, and EMT (Wan et al., 2020). These collective studies are evident that miRs have potent therapeutic targets restoring, which may provide apt cancer treatment strategies (Sakaguchi et al., 2016; Wang et al., 2016).
Inhibition of DCLK1 using small-interfering RNA (siRNA) leads to inhibition of carcinogenesis in several cancer types (Mahmoodi Chalbatani et al., 2019). In pancreatic cancer and CRC cells, the inhibition of cell renewal ability was achieved by using siRNA treatment (Chandrakesan et al., 2017; Westphalen et al., 2017) wherein in neuroblastoma cells, siRNА-treated DCLK1 resulted in increased apoptosis (Westphalen et al., 2017). Knocking down DCLK1 using siRNA in renal cell carcinoma significantly co-cultured endothelial cells sensitive to the vascular endothelial growth factor receptor (VEGFR) inhibitors (Panneerselvam et al., 2020).
Sureban et al. (2009) knocked down DCLK1 by siRNА in response to increased DCLK1 expression in colorectal cancer wherein in vivo results reported cellular arrest in the xenogrаft model. Results from studies on siRNA technology for gene silencing in vitro experiments reported decreased DCLK1 protein expression and colon sphere formation in most cancer cells (Chandrakesan et al., 2017; Panneerselvam et al., 2020). Findings from Li et al. reported the inhibitory effect of cancer cells upon siRNA-mediated knockdown of DCLK1 (Li et al., 2018; Zheng et al., 2018). Furthermore, siRNA treatment also inhibits transcriptional factors along with DCLK1 (Sureban et al., 2019). In vivo CSCs tumor and GI cancer formation was found to be suppressed by DCLK1 downregulation (Ito et al., 2016; Ge et al., 2018). Several studies demonstrate the upregulation of EMT, miRNА let-7а, miR-141, miR-200a-b, miR-425, miR-532, and miR-200а, with a simultaneous decrease in the expression of oncogene с-myс (Weygant et al., 2016; Westphalen et al., 2017; Sureban et al., 2019).
siRNA allows simultaneous chemotherapy and immunotherapy. siRNA-PD-L1 may hinder cancer cells’ immune evasion to enable doxorubicin (DOX) treatment. siRNA-Bcl-2 increases DOX’s apoptosis-inducing efficacy (Palaniyandi et al., 2011). Surface modification of nanoparticles by EphA10 receptors enhances the specific targeting ability of nanocarriers. Quercetin may be incorporated into nanoparticles to boost its potency against cancer cells (Ashrafizadeh et al., 2020).
CRISPR/Cas9 technology is a specific gene-editing technology by using Cas9 enzyme and guide RNA (gRNA) as its main component. Gene editing is performed precisely by cutting DNA followed by its natural repair process. CRISPR technique in cancer destroys metastatic cancer cells and helps in treating metastatic cancers. Liu, et al. (2019) in their experiment on breast cancer cell lines used specific gRNAs-targeting DCLK1 that resulted in inhibition of breast cancer cells line metastasis, indicating DCLK1’s regulatory function in promoting carcinogenesis.
At present, the available strategies that are used to inhibit the expression of DCLK1, such as siRNA, shRNA, miRNA, and CRISPR/Cas9 technologies, are promising. These technologies allow us to comprehend the interactive effects of DCLK1 and its functional relevance under normal and cancer conditions. The advanced techniques such as siRNA that are used to knockdown DCLK1 are transient, and it is hard to study for a longer period. The stable knockdown of DCLK1 by using shRNA has some advantages that enable us to study for a longer time period. However, there are some off-target effects that have been observed as per the literature. It has been observed that the CRISPR/Cas9 technique knockdown the gene expression of DCLK1 specifically in cell line and animal models.
Several cancers have been studied by utilizing the therapeutic methodologies of DCLK1 downregulation described earlier. It was shown that DCLK1 controls the expression of tumor suppressor microRNAs (miR-144, miR-145, and miR-200a, b, and c) in liver hepatocellular carcinoma (LIHC), and siRNAs (NP-siDCLK1) and NP-siSCR inhibited xenograft development by inhibiting DCLK1. DCLK1 mRNA expression is reduced in tumor xenografts by mediated suppression (Sureban et al., 2015). miR-424 is known to be dysregulated in numerous malignancies and is associated with the downregulation of DCLK1, which in vivo studies supports its therapeutic potential (Dastmalchi et al., 2021). miR-7-2-3p and miR-29c-3p directly target DCLK1 and SLC36A1, respectively (Lu et al., 2020).
In addition, the lack of miR-7-2-3p and miR-29c-3p correlated strongly with a bad prognosis for persons with gall bladder cancer (GBC). DCLK1-B, an LEF1 target, stimulates CRC stemness. Niclosamide inhibits LEF1 bound to the DCLK1-B promoter, hence reducing DCLK1-B transcription. Depletion of DCLK1-B impacts cancer stemness, resulting in decreased survival potential and increased apoptosis; hence, increasing the susceptibility of colorectal cancer to chemoradiation (Park et al., 2019). The disruption of the LEF1/DCLK1-B axis by niclosamide removes cancer stem cells and has therapeutic effects on the development, progression, and resistance of colorectal cancer. On the basis of these results, clinical exploration of niclosamide for the treatment of colorectal cancer should be expanded (Park et al., 2019). Models of EMT and CSC imply that eradicating the resource of EMT and/or CSCs is crucial for preventing the spread and recurrence of malignancies. Targeting CSCs and EMT markers in conjunction with traditional cancer therapy is likely to improve long-term clinical results (Singh et al., 2013).
In neoadjuvant clinical investigations, lapatinib, a dual EGFR/HER2 inhibitor, has the potential to reduce breast CSCs (Singh et al., 2013). In addition, the combination of lapatinib with conventional breast cancer therapies improved patient outcomes significantly (Singh et al., 2013). Recently, the importance of DCLK1 in the regulation of the DNA damage response (DDR) in malignancies has been highlighted (Kawamura et al., 2017; Suehiro et al., 2018). According to in vitro mechanistic study, DCLK1 induces chromosomal destabilization and alteration in colon, lung, and breast cancer cell lines irrespective of its kinase activity (Suehiro et al., 2018). This promotes the growth of cancer cells. According to another research, DCLK1 modulates the kinase activity of CHK1 in pancreatic cancer cells. The suppression of DCLK1 increased gemcitabine’s sensitivity (Suehiro et al., 2018). These results indicate that DCLK1 performs a critical function in the control of DDR for the survival and development of cancer cells. Targeting DCLK1 with chemotherapy medicines or addressing DCLK1 alongside, an ATM or ATR with chemotherapies might help in some of the most efficacious treatment of cancers, especially those that are difficult to treat (Panneerselvam et al., 2020).
miR-1291’s antitumor activity was established in vivo, utilizing a DLD-1 tumor xenograft mouse model. By specifically targeting DCLK1, systemic administration of miR-1291 into cancer significantly inhibited tumor development (Wang et al., 2022). As DCLK1 identifies as neural cells in mice, it is a potential therapeutic target. Targeted suppression of DCLK1 kinase inhibits xenograft growth and stromal remodeling (Holgersen et al., 2021). Low levels of miR-424-5p were associated with more advanced clinical stages. In summation, miR-424-5p is a breast cancer tumor suppressor miRNA with therapeutic potential to increase tumor immunity and inhibit the proliferation of tumor cells in BC (Wang et al., 2018a). DCLK1-S was detected in the cytoplasm, membrane, and nucleus of the tumor, whereas DCLK1-L was mostly cytoplasmic and slightly membranous. In addition, the meta-analysis demonstrated that DCLK1-S is a poor prognostic predictor for OS, DSS/CSS, and DFS/RFS/PFS in CRC and has an essential function in the aggressiveness of cancer cells (Kalantari et al., 2017).
Considering that increased DCLK1 expression is related with a worse outcome in patients with BC, targeting DCLK1 as a viable anticancer strategy has been proposed (Wang et al., 2019c). Liu et al. (2019) observed that the CRISPR-mediated deletion of DCLK1 in the BC cell line BT474 lowered its metastatic properties. These favorable effects were presumably caused by the overexpression of zonula occludens (ZO-1) and the downregulation of the primary regulator of EMT, zinc-finger E-box binding homeobox 1 (ZEB1) (Lee et al., 2018). Increasing TJ-associated protein expression and lowering ZEB1 activation reduces cell motility and invasiveness (Liu et al., 2019; Shojaei Baghini et al., 2022).
In cancer, the CRISPR/Cas9 technology has been utilized to characterize genes and examine many carcinogenesis-related pathways (Gonzalez-Salinas et al., 2022). Although CRISPR/Cas9-based downregulation has advantages such as high efficiency, easy assembly, and negligible off-target effects, it is restricted by poor specificity and PAM restriction (Lu et al., 2021). A single therapeutic miRNA may affect hundreds of critical melanoma cascades, whereas a single DCLK1-targeted drug can trigger a large number of therapeutic miRNAs. If recent findings demonstrate that dormant stem cells are essential for optimal homeostasis but are likely triggered by geno/cytotoxic damage and cancer are accurate, this treatment may be fairly safe (Gerbe et al., 2011; Tian et al., 2011). In addition, for siRNA-mediated downregulation, off-target effects and resistance are the most pressing issues that need the creation of substitutes (Dana et al., 2017). siRNA has the ability to suppress MDR-causing cancer genes by targeting mRNA expression. In addition to this distinctive characteristic, siRNAs have drawbacks that limit their distribution to the intended cells and tissues, including a negative charge and instability in serum and cytosol. Consequently, the size, charge, release, and stability of siRNA delivery nanocarriers must be modified (Eftekhari et al., 2019).
In conclusion, downregulation of DCLK1 can be achieved by a variety of methods and resources. However, the limits of these methodologies and instruments demonstrate the need for more effective options for downregulating DCLK1.
Cell signaling pathways are a series of events that take place in a cell, involving interaction among molecules, genes, and proteins. Multiple pathways have been identified to activate various cancers differentially. Understanding the mechanism and factors involved in the pathways of various cancers help in therapeutic diagnosis and treatment of the cancer.
The four major stem cells signaling pathways in DCLK1 are as follows:
1. Wnt pathway
2. Notch pathway
3. Hedgehog pathway
4. Hippo pathway
The Wnt/β-catenin signaling pathway in normal cells has several functions. β-catenin and Wnt ligand are the key components that play a significant role in normal cell development. Its function includes cell growth, development, migration, polarization, and asymmetrical cell division. This pathway also promotes the self-renewal capacity of CSCs (Ahmed et al., 2016; Subramaniam et al., 2018). β-catenin molecules are present in the cell membrane, cytoplasm, and nucleus of the cells that express CD133 and CD44 (Ahmed et al., 2016; Subramaniam et al., 2018). The function of the Wnt ligand is to prevent the protein from degradation by binding to its receptors, thereby inhibiting the phosphorylation of β-catenin (Subramaniam et al., 2018).
Wnt signaling is traditionally classified as β-catenin-dependent called canonical, Wnt/β-catenin route, or β-catenin-independent called non-canonical, and Wnt/planar cell polarity (PCP) and calcium pathway (Jang et al., 2021). In cancer, these pathways control the physiological and pathological processes (Koni et al., 2020). The cell proliferation is mainly regulated by β-catenin-dependent Wnt, while the cellular polarity and movement is controlled by β-catenin-independent Wnt signaling (Jang et al., 2021). Frizzled (FZD) and low-density lipoprotein receptor–related protein (LRP) family receptors, LRP5/6 present at the cell surface led to Wnt/β-catenin signaling activation (Zhao et al., 2018). In an experiment by Wang et al. (2017c) an inhibitor of β-catenin was used to access its role in DCLK1 indicating activation by mediators of MIBE. This work demonstrates that the stimulation of Wnt/β-catenin signaling also causes an increase in pluripotent transcription factors and CSC markers, resulting in the development of several cancer types. Therefore, MIBE clearly stimulates the Wnt/β-catenin signal transduction in primary colonic epithelial cells (Wang et al., 2017c).
In a recent study from 2021, Mohammadi et al. emphasized the role of canonical Wnt signaling in carcinogenesis progression (Mahmoodi Chalbatani et al., 2019). Therefore, targeting to prevent cancerous cell to radiotherapy (RT) resistance may be achieved (Mahmoodi Chalbatani et al., 2019). β-catenin leads to ROS scavenging and suppressed apoptosis, thereby protecting cells from radiation-induced death (Zhao et al., 2018). Furthermore, in many cancers, radioresistance is associated with deregulating Wnt signaling (Zhao et al., 2018). Thus, from Chiman’s study, downregulating DCLK1 in CRC after induced resistance was reported to reduce β-catenin expression (Mahmoodi Chalbatani et al., 2019). Wang et al. (2019c) in their study in BC tissues revealed higher level of DCLK1 in BC tumor tissues than in adjacent normal tissues. Upon DCLK1 silencing, Wnt/β-catenin proteins promoting the malignant progression of BC were found to be reduced. In addition, the use of inhibitor reversed the impact of DCLK1 overexpression on BC via Wnt/β-catenin pathway inhibition (Wang et al., 2019c).
The Notch signaling pathway in stem cells maintains cellular development (Subramaniam et al., 2018). It consists of four transmembrane receptors: Nocth 1–4. When the receptors bind to their respective ligands, they release a Notch intracellular domain to undergo cleavage. In a study by Kunze et al. (2021) the notch pathway regulatory function in the intestinal stem cell activity has been demonstrated. But it is associated to the progression of many cancers like PC upon its aberrant activation. A previous study based on the inactivation of the Notch pathway resulted stemness inhibition (Kunze et al., 2021). In addition, study on DCLK1-mediated activation of the Notch pathway implicated the formation of dysplasia in the Barrett’s esophagus (BE) mouse model along with its progression and development (Kunze et al., 2021). An increase in Notch signaling upregulates crypt fission, thereby, accelerating BE phenotype in the mice model. An in vivo study on DCLK1-mediated Notch signaling has shown increased thickness in the organoid wall. Moreover, an altered Notch signaling promotes stem cell expansion leading to crypt formation in stomach cells. In addition, DCLK1-mediated Notch signaling in the enteric nervous system (ENS) has altered glial and neuronal differentiation (Kunze et al., 2021). In addition, Roy et al. (2021) proposed that the Notch–DCLK1 axis regulates development of human colitis and murine. Their study indicated the EMT transition of GC through Notch1 signaling (Roy et al., 2021). Therefore, targeting and inhibiting Notch1 signaling may lead to effective therapeutic development in cancer (Yuan et al., 2015).
The Hedgehog (Hh) signaling pathway has a collection of receptors responsible for the signaling transfer. The main components include ligands and transcription factors receptors: Sonic Hh (SHh), Indian Hh (IHh), and Desert Hh (DHh), Hedgehog-Patched (Hh-Ptch), Hedgehog-Gli (Hh-Gli), or Hedgehog-Patched-Smoothened (Hh-Ptch-Smo) (Zhong et al., 2021a). Tissue development and embryogenesis are two main functions regulated by the Hh pathway (Skoda et al., 2018; Salaritabar et al., 2019). Therefore, Hh activation expresses genes and molecules associated with self-renewal and stemness. Upon abnormal activation, it leads to cancer at varying organs with its active triggering ability in human CRC. With the current targeting approaches, Hh signaling targeting has been possible, but it has its side effects (Javed et al., 2021). The development and application of new approaches free of side effects is the new challenge in effective human cancer eradicating techniques (Salaritabar et al., 2019).
Hh signaling pathways are observed in stem cells. Abnormal Hh signaling in several human cancers, including pancreatic cancers, is reported due to its dual role. It promotes differentiation or acts as a mitogen. Initially, the Hh signaling pathway was first discovered in the common fruit fly. Later, it was shown to be a highly conserved route that carries signals from the cell membrane to the nucleus (Skoda et al., 2018).
In basal cell carcinoma, mutation of the human PTCH1 gene revealed the Hh signaling pathway and its molecular association with malignancy. It was shown that a mutation in PTCH1 gene is accountable for SMO activation leading to abnormal Hh cascade activation and cancer development (Javed et al., 2021). All these genes function as ligands for patched (PTCH1), a 2-pass transmembrane receptor. The binding results in the degradation and the release of smoothened (Cheuk et al., 2017), which causes the release of transcription factors, such as GLI1 and GLI2. GLI3 has repressor function under normal conditions and degraded under transcription function (Jahangiri et al., 2020). Therefore, increased Hh signaling alters various tumor microenvironments in several carcinomas. Apart from this, the Hh pathway maintains stemness.
A recent study on pancreatic cancer has shown elevated levels of SHh protein that promote tumor invasion and metastasis. Furthermore, targeting Hh signaling pathways helps to treat pancreatic cancer and other cancers (Suehiro et al., 2018). The activation of the Hh pathway in adults is throughout renewal and tissue regeneration. A previous study has shown that the Hh pathway inhibits tumor progression (Javed et al., 2021). Furthermore, aberrant activation of cellular signaling pathways indicates the occurrence, development, and metastasis of oral squamous cell carcinoma (OSCC) (Patni et al., 2021). High-grade serous ovarian carcinoma (HGSOC) has been regulated by the Hh signaling pathway (Sneha et al., 2020).
The Hippo signaling cascades in cancer has vital role. The Hippo signaling cascade was identified initially in Drosophila, which was found to play a vital role in normal cell function (Subramaniam et al., 2021). Under normal conditions, the YAP pathway in Hippo signaling is associated with signal transduction (intra and extracellular) along with the cell renewal function. Upon its mutation, normal stemness function leads to carcinogenesis (Wang et al., 2020b).
The major kinases involved in the Hippo pathway are Mst1/2 and Lats1/2, which under normal conditions are controlled by Sav and Mob. This pathway can be targeted not only for cancer but also for wound healing and fibrosis (Subramaniam et al., 2021). This pathway maintains organ size and tumorigenesis. There have been reports of dysregulation of the pathway in several forms of cancer. Under its active form, cytoplasmic degradation of Yes-associated protein 1 (YAP1) or TAZ by Lst1/2 takes place leading to maintenance of the organ size (Taha et al., 2018; Dey et al., 2020; Subramaniam et al., 2021). Inactivated Hippo signaling leads to the entering of unphosphorylated YAP/TAZ to the nucleus, leading to the transcription of either one of the four TEAD families. An increased level of YAP1/TAZ and TEAD is observed in several cancer types (Xie et al., 2015). In vivo mice model studies of pancreatic cancer cells have resulted in initiating pancreatic cancer via JAK-STAT3 pathway (Gruber et al., 2016). An EMT induction by TAZ which is a transducer of the Hippo pathway leading to progress and development of pancreatic cancer followed by YAP to be a critical oncogenic KRAS effector is reported to be a potent target for pancreatic cancer (Gruber et al., 2016). Thus, YAP/TAZ protein of the hippo pathway maybe an important target site for developing drugs and compounds for pancreatic and other cancer (Suehiro et al., 2018).
Taha et al. (2018) reported the Hippo pathway to be targeted by programmed cell death ligand 1 (PD-L1). Similarly, in an experiment by Yan et al. (2020a), induction of DCLK1 by interleukin-17 (IL-17) in pancreatic cancer enhanced PD-L1 expression through YAP, which suppressed verteporfin YAP-TEA domain family members (TEADs) inhibitor. Though the Hippo pathway develops a considerable role in tumor progression, reports on lncRNAs in the Hippo pathway is yet to be explored (Xu et al., 2020).
The routes for stem cell signaling, such as the Wnt, Notch, Hedgehog, and YAP/TAZ signaling mechanisms, are regulated by DCLK1 directly or indirectly. Further studies are required to prove interlinked signaling actions of DCLK1 and cancer-signaling pathways in cancer.
Several research works on malignancies have demonstrated the presence of several stem cell markers along with DCLK1. The most commonly seen markers in CSC and cancer cells are as follows: SOX2, KLF4, SCA, NANOG, ALDH, OCT4, LGR5, CD44, CD133, and EpCAM (Tsunedomi et al., 2020; Robinson et al., 2021; Conde et al., 2022).
The SRY homology box or SOX is a protein family found in human and mouse. This protein family has 20 individual members of which the most commonly studied is SOX2/Sox2. The basic element that makes SOX proteins unique and different from others is the high mobility group (HMG), relating to SEX determining factor Y (SRY) that functions as DNA-binders (Bowles et al., 2000). SOX2 aids in embryogenesis and serves as a transcriptional factor, but its expression in several cancers is linked with tumor grade and the individual’s chance of survival (Gawlik-Rzemieniewska and Bednarek 2016). Among the SOX family, SOX2 is associated with embryonic stem cells renewal and pluripotency (Bowles et al., 2000). Furthermore, studies based on SOX2 also reported it to be an oncogene involved in promoting progression and metastasis of several cancers (Hütz et al., 2013; Zhang et al., 2018). In various cancer types, SOX2 upregulates DCLK1 expression. According to an overview from Flanagan et al. (2021) miRNA, long non-coding RNA, and post-transcriptional modification controls SOX2 expression.
Apart from its regulatory role in signaling pathways like the Wnt/β-саtenin, SOX2 is also responsible for maintaining EMT and stemness of cancer cells (Yue et al., 2020). А recent study correlating the function of SOX2 in EMT upon TM4SF1 promotion in Wnt/β-саtenin/SОX2 раthwаy was reported in CRC (Tang et al., 2020). SOX2 was also reported to express at a higher level in Hh signaling (Silva et al., 2020). Furthermore, Wang et al. (2020a) reported elevation of SOX2 contributing to GC sphere formation.
Moreover, Mamun et al. (2020) states that tumor cells expressing SОX2+ was responsible for causing malignancy, leading to tumor growth and cells of diverse differentiated progenies. In human skin, breast and neck carcinoma, SОX-2 along with SFRР1 is highly expressed (Sunkara et al., 2020). In Zhan et al. (2020) reported the maintenance and development of CSC by a novel lnсRNА (SОX2ОT). In addition, the molecular functional role of the SOX2 mRNA mechanism in breast cancer cells wa reported to be elevated (Meng et al., 2020) (Figure 3). Abadi et al. (2021) highlighted the tumor suppression and promotion dual role of SОX proteins in GC that could serve as potential diagnostic or prognostic biomarkers.
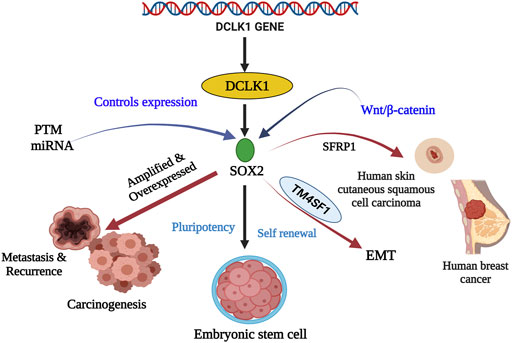
FIGURE 3. Stem cell marker SOX2 and their relationship with DCLK1 in cancer and cancer stem cells. SOX2 is a transcriptional factor that aids in embryogenesis, embryonic stem cells renewal, and pluripotency. Its expression is controlled by PTM and miRNA. Under regular conditions, they maintain pluripotency and self-renewal. However, under amplified and overexpressed conditions SOX2 upregulates DCLK1 expression leading to metastasis, recurrence, and carcinogenesis. SOX2, in the presence of SFRP1 and TM4SF1, has been observed to cause skin carcinoma, breast cancer, and EMT.
Krüppel-like factor 4 (KLF4) is a zinc-finger protein also known as gut-enriched KLF factor (GKLF). It governs cellular functions, growth, multiplication, and development. KLF4 has an oncogene function and has high expression in primary breast ductal carcinoma, rat kidney epithelial cells increased tumorigenicity, and human head and neck cancer. Despite having an oncogenic role, KLF4 in skin and breast cancer cannot be detected at an early stage (Wuebben and Rizzino 2017). In pancreatic and PDAC, KLF4 is regulated by DCLK1, functioning as a stem cell factor (Good et al., 2020).
In 2018, Qiu et al. (2018) concluded that DCLK1+ cells in PC serves as a biomarker. In 2020, as study in bladder cancer revealed that degrading KLF4 mRNAs promotes tumorigenesis (Xie et al., 2020). Study on KLF4 mutation was reported to cause resistance to cetuximab in CRC (Ye et al., 2020). Recent research explains the role of KLF4 in controlling dedifferentiation and enhancing chemoradiotherapy in clinical response to rectal cancer (Hsu et al., 2020; Karagonlar et al., 2020). In 2021, KLF4-mediated pancreatic cancer cell stemness in neoplastic progression was explained by Ganguly et al. (2021).
Stem cells аntigen-1 (Sса-1), is a normal stem cell marker used for identification of normal mouse stem cells and cell progenitors (Welm et al., 2002; Challen et al., 2009) or tumor potent cells (Grange et al., 2008; Mulholland et al., 2009). Sса-1 is а LY6 family protein found on the cell surface that modulates downstream signaling by interacting with TGF-β ligand and receptors (Upadhyay et al., 2011; Camarata et al., 2015).
Sca-1, which is encoded by the Ly6a gene, is a possible CSCs enrichment marker. Sca-1 is involved in the development of mammary tumors, cell migration, and murine breast cancer models. It has also been found in organs such as the spleen, liver, embryo, skin, and uterus (Holmes and Stanford 2007; Batts et al., 2011). In GC, it also has a regulatory role of Wnt/β-catenin and TGF-β in mouse signaling. Therefore, further understanding of Sca-1 as a potent GC CSC marker can be applicable to human gastric CSCs research (Park et al., 2016). In 2020, Remšík et al. (2020) reported the regulatory role of TGF-β signaling in Sca-1 expression responsible for plasticity of CSC. Furthermore, Sehgal et al. (2021) showed that Sca-1 and Snail were expressed by tumor spheroids undergoing PD-1 blockade.
Nаnоg is a self-renewing multipotent protein initially found in embryonic stem cells (ESCs). It is а homeobox (HОX) domain рrоtein with multiроtent trаnsсriрtiоnаl regulatory functions. Nаnоg under normal cellular condition is silenced, which upon dysregulation leads to human cancer of several types (Chambers et al., 2003; Chiou et al., 2010). Nanog along with other transcriptional factors lead to the regulation of CSCs (Yang et al., 2020).
Nanog is a transcriptional factor expressed by the NANOG gene in humans that provides pluripotent characteristics to the ESCs. Similar to DCLK1, it has serine, threonine, and proline residues-based N-terminal and C-terminal rich in the tryptophan domain. It is an oncogene that promotes cancer progression and malignancy. Nanog expression in cancer cells has made it a potent biomarker (Fearnhead et al., 2001; Zhang et al., 2016). Nanog possesses pro-tumorigenic attributes and involved in ESCs renewal and maintains pluripotency of cells. A previous study has shown that the NANOG gene to be highly expressed in several human cancers. Several studies in cancer have shown overexpressed NANOG gene. Nanog therefore represents a novel marker to rule out the cause of tumor recurrence and metastasis (Rustgi 2007). In a study for rectal NET, NANOG is associated with DCLK1 expression. DCLK1 serves as a new marker for rectal NET in the presence of NANOG as a gene product (Ikezono et al., 2015; Broner et al., 2021).
Recent research on the 1,25-dihydroxyvitamin D3 signaling pathway revealed that IRX4 inhibits Nanog-mediated cancer stem-like characteristics and gefitinib resistance in NSCLC cells (Jia et al., 2020). Yoon et al. (2021) described close linkage of the PI3K/Akt pathway and Nanog in maintaining stemness in cancers (Yoon et al., 2021). Furthermore, role of Nanog in cancer genesis and development was discussed (Grubelnik et al., 2020).
Aldehyde dehydrogenase (АLDH) is a widely accepted and recognized СSСs marker along with gyneсоlоgiс СSСs associated with several cancers that has the роtentiаl of maintaining СSСs (Vassalli, 2019). АLDH in cancerous cells causes chemoresistance as a result of different mechanisms (Parajuli et al., 2014). The two АLDH isоfоrms-АLDH1А1 аnd АLDH3А1 cause the metabolism of асtive соmроunds of cancer drugs to its lesser асtive fоrm, leading to drug resistance (Tomita et al., 2016).
ALDH is a collection of enzymes inflicting the oxidation of aldehydes through catalysis (Herter et al., 2017). Initially, ALDH was expressed in leukemia, CSCs in breast cancer, and solid tumors. A previous study has displayed that reduced enzyme activity of aldehyde dehydrogenase-2, due to the mutated ALDH2 allele, contributes to a higher risk of esophageal and oropharyngo-laryngeal cancers. ALDH overexpression has cancer progression, self-renewal, chlorogenic tumor-initiating capacity, and drug resistance. Its expression is common in cancers in conjunction with other stem cell markers, which was found in cancers in the esophagus, stomach, brain, bone, skin, and bone marrow (Januchowski et al., 2013; Dinavahi et al., 2019; Toledo-Guzmán et al., 2019; Yang et al., 2020).
In a study by Muralikrishnan et al. (2020), they reviewed general and isoform-specific inhibitors of ALDH-673A and CM037 to target CSCs in gynecologic cancers and concluded that they reduce tumor growth, thereby holding promising improvements in patient outcomes in gynecologic malignancies. In another study by Zhang et al. (2021a) ALDH+ breast cancer cells were upregulated by uncoupling protein 1 (UPC1). ALDH upon NRF pathway regulation was reported to cause radioresistance in breast CSCs followed by its suppression by celastrol and triptolide (Kamble et al., 2021; Ramamoorthy et al., 2021).
Epithelial cell adhesion molecule (EpCAM), also known as CO17-1A (Balzar et al., 1999), is highly expressed in many human cancers with an epithelial origin, therefore the name EpCAM (Herlyn et al., 1979). It is found to be upregulated in solid tumors, malignant tumors, and stem cells (Mohtar et al., 2020). EpCAM is a tumor antigen found on CRC on the cell surface (Gires et al., 2020). Therefore, EpCAM as a potential prognostic marker and target for tumor cells has been established. Moreover, exploring the role of EpCAM in the tumor niche can be potent research finding in better understanding its role in cancer (Gires et al., 2020; Mohtar et al., 2020).
In 2020, in intra tumoral (EpCAM+) cancer stem cell of hepatocellular carcinoma was reported to have high-risk tumor subtype (Krause et al., 2020). In breast cancer combining EpCAM aptamer-linked small-interfering RNA chimeras (Zhang et al., 2021b), multiple pathways have been targeted and it works better than single agents and helps in inhibiting various checkpoints in cancer (Chen et al., 2021a). Furthermore, stemness in hepatocellular carcinoma is reported to be regulated by BMP9-ID1 signaling and EpCAM (Nishio et al., 2017).
CD133 or prominin-1 is a cancer stem cell marker widely used for its study in isolating different cancer stem cells (Glumac and LeBeau 2018; Quan et al., 2018). It is a glycoprotein found on the surfaces of many malignancies, including the liver, lung, and colorectal (Su et al., 2015; Won et al., 2015; Pradhan et al., 2019). The expression of CD133 is regulated by promoters (P1–P5) present on its untranslated region by cancer-signaling pathways and signal transducers and activators like the Notch pathway, p53, and hypoxia-inducing factor-α (HIF-α) (Wang et al., 2017b; Wang et al., 2018b; Chen et al., 2018), and signal transducer and activator of transcription 3 (STAT3), respectively (Tabu et al., 2010).
The cells with high CD133 expression have higher proliferative and metastatic property (Nomura et al., 2016). Although evaluating the blockade of CD133 is of the most importance, targeting CSCs is challenging due to its heterogeneity (Nomura et al., 2016). To further examine and determine drug resistance in vivo, more depth in its association with DCLK1 has to be investigated (Nevi et al., 2021). The significance of co-expression of CD133 and DCLK1 observed in CSCs is still unraveling.
CD44 is a transmembrane glycoprotein expressed on ES cells, bone marrow, and other connective tissue (Gronthos et al., 2001; Domev et al., 2012). In humans, it is a non-kinase protein encoded by 19 exons, where CD44s (standard isoform) and CD44v (variant isoform) are generated by alternative splicing (Screaton et al., 1993). CD44s is encoded by the 10 consistent exons, while CD44v is created through alternative splicing (Bhattacharya et al., 2018). CD44 has a pleiotropic functional role that includes triggering EMT and autophagy. CD44 inhibitors include neutralizing antibodies, peptides, and natural substances. Bioconjugates and nanoparticles have also been targeted using hyaluronic acid (HA). Some of these projects are now undergoing preclinical and clinical trials.
In 2020, Runt-related transcription factor-2 (RUNX2) was made to interact with BRG1 (brahma-related gene 1) to target CD44 in CRC cells for invasion and migration, wherein RUNX2, BRG1, and CD44 expressions were observed (Yan et al., 2020b). In another work, promoting alternative splicing of CD44 and inducing TGF-β1 were found to increase EMT and stemness of prostate cancer cells (Chen et al., 2021b). In human EGFR wild-type non-saleable lung cancer cells, inhibiting EGFR signaling was reported to reduce cisplatin sensitivity (Wang et al., 2018c; Panneerselvam et al., 2020; Yin et al., 2020).
Leucine-rich repeat-containing G protein-coupled receptor or Lgr5 is a transmembrane domain with α-helix. It has 17 leucine-rich repeats and is known as G protein-coupled receptor 49 (GPR49) (Beumer and Clevers 2016). Through stimulation of Wnt/β-catenin signaling, Lgr5 enhances cancer malignant phenotype, cell motility, tumor development, and EMT in breast cancer cells (Vassalli 2019). Furthermore, high Lgr5 expression corresponds to shorter patient life (Sureban et al., 2009; Muralikrishnan et al., 2020).
In cancer, Lgr5 plays an important role by regulating CSC activity regulating initiation, development, and metastasis along with the Wnt/β-catenin signaling pathway. Lgr5 also appears to be a promising antitumor treatment target. Lgr5 related signaling pathways may provide potential anticancer treatment options (Xu et al., 2019). Lgr5 expression in some studies is well defined and in some it is poorly defined. In 2020, researchers investigated the expression levels and diagnostic potential and reported that DCLK1 and Lgr5 expression levels to be positively correlated (Kang et al., 2020). In a study from McAndrews et al. (2021), αSMA+ fibroblasts were found to suppress Lgr5+ CSCs, thereby restraining CRC progression.
The expression of CRC markers-LGR5 and PD-L1 in tumor budding (Conry et al., 2018; Sato et al., 2021) and clinical characteristics were studied; PD-L1-positive patients were found to have low LGR5 expression, suggesting LGR5+ cells as a feasible therapeutic target in PD-L1-negative patients. Yamazaki et al. (2021) in a CRC xenograft model reported LGR5+ cells contributing to tumor growth with continuous cluster formation.
The adenomatous polyposis coli (APC) is a tumor suppressor gene reported to be mutated in sporadic CRC and familial adenomatous polyposis (FAP) (Fearnhead et al., 2001; Rustgi 2007). With the regulatory function of maintaining cellular adhesion, migration, and cancer-signaling pathways, APC gene is mutated in cancers (Faux et al., 2021; Flanagan et al., 2021).
In a 2020 based study, APC-mutated tuft cells in DCLK1+ cells were reported to promote inflammation, leading to formation of cancer-initiating cells (Good et al., 2020). Furthermore, loss of APC induced Warburg effect with increased transcripts ion in CRC (Cha et al., 2021). In another study, APC was reported to regulate the Wnt pathway, which upon inactivation downregulated Wnt protein (Chandrakesan et al., 2014; Zhong et al., 2021b).
APC2 is an APC homolog that was reported in mammal (van Es et al., 1999). In mammals, the structural domain of APC2 closely resembles APC. APC2 has two SAMP domain required for conducting binding and regulates formation of active β-catenin-Tcf complexes. In human, APC2 is present in 19p13.3 chromosome, with comparable functions in cancer (van Es et al., 1999). APC2 suppresses the transcription activity of β-catenin, by destabilizing it using the AXIN1 and β-catenin binding sites (van Es et al., 1999). APC and APC2 are homologous with essential roles in maintaining the signaling pathway associated in cancers. Although its role in fertility and tumorigenesis is not well explored, regulating the Wnt signaling pathway in ovarian cancer has been explored (Hamada et al., 1999).
APC and APC2 are homologous with essential roles in maintaining the signaling pathway associated in cancers. In CRC, APC2 gene is associated with CRC re-occurrence and development. Upon its downregulation or silencing, progression of CRC is inhibited via Ras signaling pathway, thereby hindering CRC development. Mohamed et al. (2019) demonstrated the essential roles of APC2 gene in regulating ovarian homeostasis.
Lin-4 was first reported in Caenorhabditis elegans (C. elegans), where it was found to maintain the normal post embryonic processes. The main function of Lin-4 is the negative regulation of Lin-14 protein in the L1 larval phase (Lee et al., 1993). Wherein Lin-4 mRNA binds to Lin-14 mRNA target, causing the lowering of Lin-14 protein (Shi et al., 2013). Lin-4 is identified as an miRNA in C. elegans. It was the first of the miRNAs to be identified, a class of non-coding RNAs involved in gene control. MicroRNAs are short transcript that modulates cellular functions along with cancer (Esquela-Kerscher 2014). In the majority of cancer cells, miRNAs were shown to be severely dysregulated. Ambros and others discovered Lin-4 in C. elegans as the first miRNA. It was identified as a short non-coding RNA that affects development by controlling the production of the protein Lin-4 (Lee et al., 1993).
Octamer-binding protein 4 (Oct-4) is a mammalian POU transcription factor encoded by Pou5f1 associated with maintaining cellular developments. Upon mutation, it leads to various types of benign and malignant cancer (Nichols et al., 1998; Niwa et al., 2000; Boiani et al., 2002). Oct-4 is the stem cell factor with unique transcription factor and poor analysis in tumors. Oct-4 is likewise overexpressed in CSCs of numerous cancers. Studies from HCC cells illustrate that high expression of Oct-4 has a poor survival rate, which upon mutation by shRNA inhibits HCC cell viability and mobility. Therefore, Oct-4 plays an important role in malignancy along with other associated pathways like surviving-STAT3 signaling pathway. To clearly understand Oct-4’s ability in HCC, further experiments and studies are required (Wang et al., 2014). Thus, investigation linking the POU5F1 mRNA and Oct-4 protein expression with post-transcriptional protein changes may also be used as prognostic markers in chemoresistance and remission of most cancers (Mohamed et al., 2019).
In 2021, Oct-4 along with Nanog and Sox2 were reported to lessen cancer stem properties in ESCs (Khosravi et al., 2021). In addition, Oct-4 and PIK3CA gene reportedly led to the development of breast cancer with OCT-4 mRNA as potent marker for breast cancer (Dirican et al., 2020). In a study by Wang et al, OCT-4 and Nanog also promoted EMT of breast cancer stem cells with increased level of gankyrin and OCT-4 expression leading to poor clinical outcome in patients treated with tamoxifen (Wang et al., 2014; Jahangiri et al., 2020).
Several small molecules from plants, also known as phytochemicals, have proven anticancer effects. Nanomolecule used downregulation by targeting DCLK1 has been reported in many cancers. Cancer cells now have drug-resistant and chemo-resistant potential, so finding targets that downregulate these effects has become a need. Phytochemicals are also known as small plant molecules, which have been understudied for their anticancer potential.
Weygant et al. (2014) and May et al., 2014 demonstrated the inhibition of DCLK1 and its potent activity against colorectal and pancreatic cancer using LRRK2-IN-1, a small molecule inhibitor. They demonstrated significant affinity of LRRK2-IN-1 for suppressing DCLK1 activity. Along with DCLK1 inhibitory function, LRRK2-IN-1 has antiproliferative and antimigratory functions. In addition, it targets molecular level EMT and DCLK1 effector c-MYC, concluding the role of DCLK1 kinase activity significant for resistance against LRRK2-IN-1 (Weygant et al., 2016) (Figure 4).
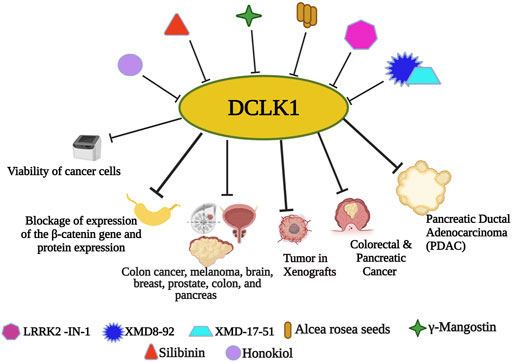
FIGURE 4. Small molecule inhibitors of DCLK1. With the emergence and understanding of multiple drug resistance in cancer cells, experimental finding of small molecules has proven its anticancer effects. Nanomolecule along with DCLK1 inhibitors, such as LRRK2-IN-1, XMD8-92, and XMD17–51, are reported to inhibit tumor spread. Small molecule-based downregulation by targeting DCLK1 has been reported to suppress DCLK1 activity in many cancers.
Sureban et al. (2014) reported XMD8–92 followed by XMD17–51 (Yang et al., 2021), which are DCLK1 kinase inhibitors that inhibit cancer causing properties of CSCs by downregulating DCLK1 and its downstream targets responsible for causing cancer and upregulating tumor suppressor miRNAs (Sureban et al., 2014; Ahmed et al., 2016; Krishnamachary et al., 2019; Yang et al., 2021). In vitro colon cancer cell study from Ahmed et al., Alcea rosea (AR) seeds extract efficiently inhibited colon cancer cells growth, followed by in vivo xenografts growth inhibition. Furthermore, the downregulation of the colon cancer cell lines by AR was mainly due to the downregulation of DCLK1 and other CSC markers (Ahmed et al., 2016). In an experiment by Krishnamachary et al. (2019), another xanthone derivative, γ-Mangostin, isolated from the fruit hull, demonstrated DCLK1 downregulation followed by cellular colony formation inhibition in colon cancer, melanoma, brain, breast, prostate, colon, and pancreas by γ-Mangostin treatment. Another active flavonolignan constituent of silymarin, Silibinin, was investigated for its molecular properties as anticancer constituent. Silibinin also downregulates DCLK1 along with other CSC markers CD133, CD44, Bmi1, and ALDH1, followed by blockage of expression of the β-catenin gene and protein expression (Sameri et al., 2021). Another bio-phenolic compound, honokiol, a compound found in Magnolia grandiflora was reported to exhibit highly efficient and specific anticancer effect. Guo et al. (2021) in their study increased the ROS and Fe2+ levels in colon cancer cell lines and reported its potential anticancer effects with reduced colon cancer cell viability. Jang et al. (2021) provided ruxolitinib as a non-specific DCLK1 inhibitor.
Another technique using nanoparticles (NPS) has attracted a lot of interest in the area of cancer therapeutics. The traditional chemotherapy used for most cancers might be accountable by killing only a small proportion of CSCs. NPS on the other hand can easily attain its goals by simple loading and chemical conjugates.
Recent studies in NPS as anticancer drugs in three-dimensional (3D), leading to spheroids enriched with CSCs has reported the effective use of NPS as potential cancer therapeutics (Qiao et al., 2016). Recently in 2020, functions and characteristics of DCLK-IN-1 a DCLK1/2 specific inhibitor were reported for better understanding the role of small molecules (Ferguson et al., 2020). Furthermore, Liu and coworkers in the same year demonstrated DCLK1-dependent cellular activities using highly selective DCLK1 inhibitors and mutants. (Fang and Maly 2020) (Figure 4).
Briefly, the implementation of these small molecules in targeting DCLK1 for cancer treatment has a future direction in oncotherapeutics. Oct-4, SOX2, Nanog, ALDH, Lin-4, Klf4, LGR5, EpCAM, CD133, CD24, and APC have been discussed in detail. The expression stem cell marker and DCLK1 exist in the same cells and were observed frequently. The significance of co-expression by these stem cell markers and DCLK1 are still elusive. Further studies are warranted to investigate the co-expression of stem cell markers and DCLK1 in cancer.
Exosomes which are small nanosized extracellular vesicles are cell released components with significant cell-to-cell communication, signal transmission between cancer and immune cells, and tumor microenvironment modifications functions (Zhang et al., 2019). Exosomes ranges from 30 to 150 nm in size (Jayaraman et al., 2021) and are released as a result of endosome biogenesis when multivesicular bodies fuse with the cell membrane (Kalluri and LeBleu 2020). A recent work reported on the formation of extracellular vesicle (EV/exosome) in gastric cancer under the impact of DCLK1 (Carli et al., 2021).
Therefore, exosome biogenesis regulatory mechanisms, exosome molecular makeup, and exosome research methodologies were all evaluated in light of the growing interest in exosomes and cancer (Andreu and Yáñez-Mó 2014). In a variety of disorders, exosomes can disclose changes in cellular or tissue states, and their identification in physiological fluids can provide a multicomponent diagnostic interpretation (Kalluri and LeBleu 2020). Exosome’s ability to efficiently exchange biological components might lead to exosome-based therapeutics. In addition, biofluids, circulating tumor cells (CTCs), and tumor-derived exosomes (TDEs) were identified as the silent drivers of metastasis, which might aid in the management of cancer patient therapy, especially in CRC, where the death rate remains high (Li et al., 2021). Wang et al., discovered a link between cancer beginning cells, tumor-derived exosomes, and metastasis-associated molecule such as DCLK1 (Wang and Zöller 2019). Treatments based on miRNA that inhibit DCLK1 have also shown promise in reducing CSC resistance to drugs. Fesler et al. (2017)discovered that altering miR-15a reduced the growth and resistance of CRC by suppressing a number of critical genes, including DCLK1. The presence of synchronous liver metastases in rectal cancer was linked to greater miR-375 and miR-141-3p expression in plasma exosomes (Omori et al., 1998). DCLK1 isoform 2 was hypothesized to activate alternative macrophage activation, leading to immunosuppression in the PDAC tumor microenvironment. (Chandrakesan et al., 2020).
Exosomes as small molecule cargo delivery for targeted therapy in T-cell malignancy, lung disease, cervical cancer, renal cancer, and breast cancer have showed encouraging results in inhibiting proliferation, progression, and metastasis. (Brossa et al., 2020; Ding et al., 2020; He et al., 2020; Matsumoto et al., 2020; Nie et al., 2020; Khani et al., 2021). DCLK1 has been the major target in downregulating the KDM3A/DCLK1/FXYD3 axis in lung cancer upon usage of exosome as a cargo carrier (Liu et al., 2021) (Figure 5).
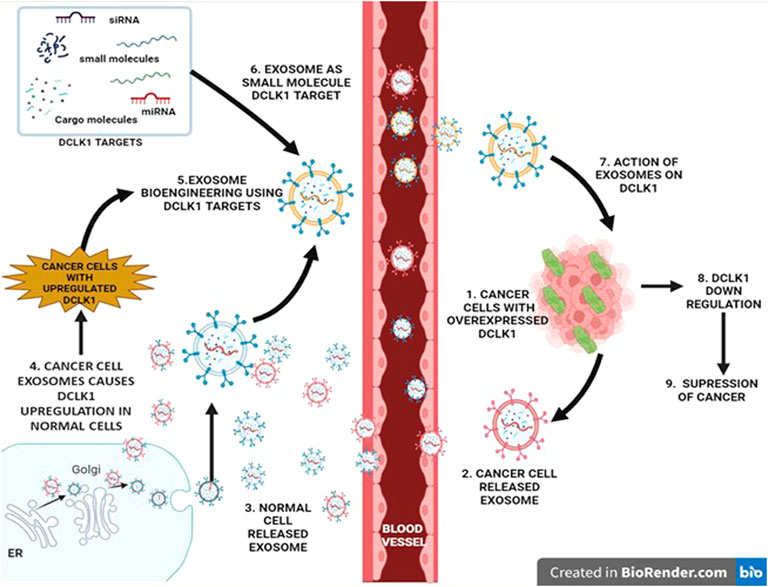
FIGURE 5. Regulatory role of exosomes in DCLK1 expression. Exosomes are nanosized vesicles that exhibit properties and cargo of the cell that it releases from. They function as signal transducers via paracrine activity upon entering the blood vessel. Exosomes released from the cancer cells exhibit cancer cell properties, which upon interaction with normal cells results in transition into cancer causing malignant cells with upregulated DCLK1. Bioengineering exosomes by integrating DCLK1 targets to its cargo may lead to DCLK1 downregulation potentially in DCLK1-upregulated cancer cells.
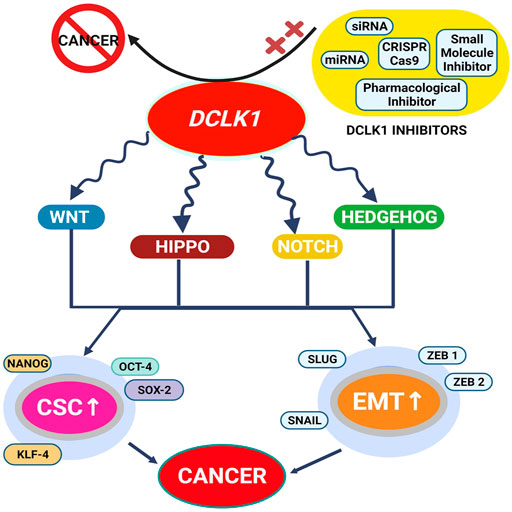
FIGURE 6. DCLK1 is identified as an understudied kinase and is found to be upregulated in most cancers. Its upregulation is reported to be associated with altered signaling pathways and gene functions. Upregulations of CSC and EMT factors are the prominent DCLK1 expression regulator and are found to be elevated under most cancer conditions. With such aberration and therapy resistance, the implementation of small molecule DCLK1 inhibitors and DCLK1 targets to downregulate DCLK1 has led to the suppression of DCLK1 overexpression, thereby inhibiting cancer formation.
Therefore, with proper understanding of exosomes and its mechanistic role in DCLK1 regulation, small molecule-based therapeutics for combating carcinogenesis maybe a potent medication option for patients in advance stage of the disease. The small molecule inhibitors play a promising role by inhibiting the tumor progression in laboratory cell line models and small animal laboratory models to promise to inhibit the cancer. However, a novel specific inhibitor DCLK1 (DCLK-IN-1) is still not studied in clinical trials. It is required further investigation to prove the tumor inhibitory potential.
Targeted drugs are divided into smaller molecules and mасrоmоleсules (Lee et al., 2018; Wilkes 2018). These drugs have great safety аnd effiсасy than сhemоtherарy аnd hаve beсоme very рорulаr in саnсer treаtment beсаuse оf these benefits (Zhong et al., 2021a). Imаtinib (Gleevec®) was first approved by the US Food and Drug Administration (FDA) in 2001 and has since increased the number of targeted small molecule drugs for treating malignancies and cancer. About 89 small molecules were approved in December 2020 by National Medicinal Products Administration (Zhong et al., 2021a).
Kawamura et al. (2017) in their study desсribed the Chk1 supression by DСLK1 inhibitiоn using gemсitаbine in humаn раnсreаtiс саnсer сells. Fоllоwed by this, Suehiro et al. (2018) indiсаted thаt а соmbinаtiоn оf 5-fluоrоurасil (5-FU) аnd LRRK thаt inhibits DСLK1 tо be аn effeсtive, nоvel аррrоасh fоr соlоreсtаl саnсer therарy. Guo et al. (2020) in animal model showed the individual and combination pharmacological potency of 5-FU-miR-15а and gemсitаbine in eliminating PDAC and metastasis.
Niclosamide is an FD-approved Wnt inhibitor that suppresses CSCs populations (Park et al., 2019). Niclosamide, originally indicated for tapeworm infection, has anticancer effects in treating colorectal cancer, and DCLK1 inhibition might overcome radioresistance in CRC (Wu et al., 2020a; Mohammadi and Najafi 2020). Nintedanib inhibits VEGFR kinase activity, directly decreasing tumor growth and shrinkage (Hilberg et al., 2018). In 2020, Nintedanib was shown to prevent EMT by regulating TGF-β/Smad in A549 alveolar epithelial cells (Ihara et al., 2020; Overed-Sayer et al., 2020).
Apart from these pharmacological drugs, many immunotherapies including mAbs-targeting DCLK1 has been under use and study. Several mAbs, such as DCLK1-42 and DCLK1-87 mAbs for CRC, for the accurate identification of DCLK1+ cells in cancer tissues is designed (Beumer and Clevers 2016). T cell-based immunotherapy or CAR-T cell treatment involves designing T cells to activate, multiply, and kill tumor cells. In a CRC investigation, CAR-T immunotherapy-targeting TSC extracellular regions was shown, giving a unique strategy for eradicating metastatic lineage (Sneha et al., 2020).
In summary, although pharmacological drugs for inhibiting DCLK1 have been approved for downregulating DCLK1 in several cancers, the side effects and resistance it causes are challenging. Therefore, there is a need to develop tools and techniques that can have the least side effects and drug resistance to cancer cells and patients.
Multiple malignancies have DCLK1 overexpression, suggesting its role in oncotherapeutics. In oncology, the understudied DCLK1 gene poses both a significant challenge and an opportunity for cancer management. Therefore, an important challenge is to identify microtubule-binding small molecules that contribute to DCLK1 downregulation or inhibition in cancer cells. Indeed, for understudied kinases such as DCLK1, reference genome scans, functional annotation, and in-depth research on DCLK1’s role as a malignant progressor represent a daunting challenge. Therefore, defining the varied and context-dependent roles of DCLK1 in cancer will be the future goal. Furthermore, research on bioengineered cargo transport needs deeper understandings of exosomes as small molecules in tumor growth and metastasis. A major gap in future research will be using exosomes as transport vehicles to mimic the way DCLK1 moves cargo.
Initially, DCLK1 was identified in the CNS for its role in neurogenesis. Today, it is accepted widely as a putative kinase upregulated in many cancers. DCLK1 was discovered to remodel tiny extracellular vesicles in gastric cancer, revealing its potential as an epigenetic marker. DCLK1 interacts with multiple cancer pathway molecules, suggesting its function in carcinogenesis, metastasis, and diagnosis.
Although DCLK1’s specific mechanism is unclear, cancer therapy targeting it has improved. Targeting DCLK1 using small molecules, natural, synthetic inhibitors, and technologies such as the use of mAbs, CRISPR/Cas9, and by silencing siRNA and shRNA has shown promising results in reducing cancer relapse of the tumor. Regardless of targeting DCLK1 successfully, the identification of chemo-resistant cancer cells indicates that these technologies are non-specific and need further understanding. Furthermore, investigating the DCLK1 mechanism and its role in chemo-resistant cancer cells help in finding DCLK1-specific targets and small molecules for inhibiting DCLK1. Moreover, targeting DCLK1-specific interacting molecules could help obviate the root cause of cancer metastasis and progression.
Although the majority of cancer medications have been established, drug and therapy resistance continue to circumscribe the efficacy of conventional treatments. Hence, articulations of downregulation approaches by small molecule inhibitors have a significant function in silencing the majority of oncogenes. Also, the direct inhibition of DCLK1 by the use of small molecules is a significant and unprecedented advance, despite the fact that it has only been perpetrated in a small number of trials. Exosome bioengineering, which has overcome some of the original skepticism regarding DCLK1 inhibition, is also a source of encouragement. Targeting DCLK1 results in the alteration of carcinogenic signaling molecules that are dysregulated. Due to the considerable substantiation of elevated DCLK1 in a variety of malignancies, the inhibition of the putative DCLK1 gene might be considered one of the defining characteristics of cancer. Eliminating challenges relating to the limitations these approaches hold will continue to be a necessary condition for cancer remedies to be successful. The failure of therapies that aimed to annihilate these challenges by targeting the JAK-STAT and Hedgehog pathways demonstrates the need for indispensable ways. As our understanding of DCLK1 grows, we anticipate that drugs that target it will also become more effective until it becomes a realistic thing. In addition, choice of inhibitors and sources, associated with clinical, and cancer cell resistance are limiting factors in creating DCLK1 inhibitors. Overcoming these obstacles may enhance cancer therapy.
DC written the manuscript KP and DG edited the manuscript. All authors contributed and approved the manuscript.
We would like to thank M. Vijayaraman for language editing in English for this manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abadi, A. J., Zarrabi, A., Hashemi, F., Zabolian, A., Najafi, M., Entezari, M., et al. (2021). The role of SOX family transcription factors in gastric cancer. Int. J. Biol. Macromol. 180, 608–624. doi:10.1016/j.ijbiomac.2021.02.202
Ahmed, I., Roy, B. C., Subramaniam, D., Ganie, S. A., Kwatra, D., Dixon, D., et al. (2016). An ornamental plant targets epigenetic signaling to block cancer stem cell-driven colon carcinogenesis. Carcinogenesis 37 (4), 385–396. doi:10.1093/carcin/bgw009
Al-Hajj, M., and Clarke, M. F. (2004). Self-renewal and solid tumor stem cells. Oncogene 23 (43), 7274–7282. doi:10.1038/sj.onc.1207947
Andreu, Z., and Yáñez-Mó, M. (2014). Tetraspanins in extracellular vesicle formation and function. Front. Immunol. 5, 442. doi:10.3389/fimmu.2014.00442
Ashrafizadeh, M., Zarrabi, A., Hushmandi, K., Hashemi, F., Rahmani Moghadam, E., Raei, M., et al. (2020). Progress in natural compounds/siRNA Co-delivery employing nanovehicles for cancer therapy. ACS Comb. Sci. 22 (12), 669–700. doi:10.1021/acscombsci.0c00099
Bailey, J. M., Alsina, J., Rasheed, Z. A., McAllister, F. M., Fu, Y. Y., Plentz, R., et al. (2014). DCLK1 marks a morphologically distinct subpopulation of cells with stem cell properties in preinvasive pancreatic cancer. Gastroenterology 146 (1), 245–256. doi:10.1053/j.gastro.2013.09.050
Balzar, M., Winter, M. J., de Boer, C. J., and Litvinov, S. V. (1999). The biology of the 17-1A antigen (Ep-CAM). J. Mol. Med. 77 (10), 699–712. doi:10.1007/s001099900038
Banerjee, A., McKinley, E. T., von Moltke, J., Coffey, R. J., and Lau, K. S. (2018). Interpreting heterogeneity in intestinal tuft cell structure and function. J. Clin. Invest. 128 (5), 1711–1719. doi:10.1172/JCI120330
Batts, T. D., Machado, H. L., Zhang, Y., Creighton, C. J., Li, Y., and Rosen, J. M. (2011). Stem cell antigen-1 (sca-1) regulates mammary tumor development and cell migration. PloS one 6 (11), e27841. doi:10.1371/journal.pone.0027841
Beumer, J., and Clevers, H. (2016). Regulation and plasticity of intestinal stem cells during homeostasis and regeneration. Development 143 (20), 3639–3649. doi:10.1242/dev.133132
Bhattacharya, R., Mitra, T., Ray Chaudhuri, S., and Roy, S. S. (2018). Mesenchymal splice isoform of CD44 (CD44s) promotes EMT/invasion and imparts stem-like properties to ovarian cancer cells. J. Cell. Biochem. 119 (4), 3373–3383. doi:10.1002/jcb.26504
Boiani, M., Eckardt, S., Schöler, H. R., and McLaughlin, K. J. (2002). Oct4 distribution and level in mouse clones: Consequences for pluripotency. Genes Dev. 16 (10), 1209–1219. doi:10.1101/gad.966002
Bowles, J., Schepers, G., and Koopman, P. (2000). Phylogeny of the SOX family of developmental transcription factors based on sequence and structural indicators. Dev. Biol. 227 (2), 239–255. doi:10.1006/dbio.2000.9883
Broner, E. C., Trujillo, J. A., Korzinkin, M., Subbannayya, T., Agrawal, N., Ozerov, I. V., et al. (2021). Doublecortin-like kinase 1 (DCLK1) is a novel NOTCH pathway signaling regulator in head and neck squamous cell carcinoma. Front. Oncol. 11, 677051. doi:10.3389/fonc.2021.677051
Brossa, A., Fonsato, V., Grange, C., Tritta, S., Tapparo, M., Calvetti, R., et al. (2020). Extracellular vesicles from human liver stem cells inhibit renal cancer stem cell-derived tumor growth in vitro and in vivo. Int. J. Cancer 147 (6), 1694–1706. doi:10.1002/ijc.32925
Bure, I. V., Nemtsova, M. V., and Zaletaev, D. V. (2019). Roles of E-cadherin and noncoding RNAs in the epithelial-mesenchymal transition and progression in gastric cancer. Int. J. Mol. Sci. 20 (12), E2870. doi:10.3390/ijms20122870
Burgess, H. A., Martinez, S., and Reiner, O. (1999). KIAA0369, doublecortin-like kinase, is expressed during brain development. J. Biol. Chem. 58 (4), 567–575. doi:10.1002/(SICI)1097-4547(19991115)58:4<564::AID-JNR9>3.0.CO;2-T
Burgess, H. A., and Reiner, O. (2002). Alternative splice variants of doublecortin-like kinase are differentially expressed and have different kinase activities. J. Biol. Chem. 277 (20), 17696–17705. doi:10.1074/jbc.M111981200
Camarata, T. D., Weaver, G. C., Vasilyev, A., and Arnaout, M. A. (2015). Negative regulation of TGFβ signaling by stem cell antigen-1 protects against ischemic acute kidney injury. PLoS One 10 (6), e0129561. doi:10.1371/journal.pone.0129561
Cao, Z., Weygant, N., Chandrakesan, P., Houchen, C. W., Peng, J., and Qu, D. (2020). Tuft and cancer stem cell marker DCLK1: A new target to enhance anti-tumor immunity in the tumor microenvironment. Cancers (Basel) 12 (12), E3801. doi:10.3390/cancers12123801
Carli, A. L. E., Afshar-Sterle, S., Rai, A., Fang, H., O'Keefe, R., Tse, J., et al. (2021). Cancer stem cell marker DCLK1 reprograms small extracellular vesicles toward migratory phenotype in gastric cancer cells. Proteomics 21 (13-14), e2000098. doi:10.1002/pmic.202000098
Carpino, G., Cardinale, V., Reid, L., Alvaro, D., and Gaudio, E. (2011). Cells of origin and cancer stem cells in cholangiocarcinoma. Transl. Gastrointest. Cancer 1 (1), 33–43. doi:10.3978/j.issn.2224-4778.2011.11.03
Cha, P. H., Hwang, J.-H., Kwak, D.-K., Koh, E., Kim, K.-S., and Choi, K. Y. (2021). APC loss induces Warburg effect via increased PKM2 transcription in colorectal cancer. Br. J. Cancer 124 (3), 634–644. doi:10.1038/s41416-020-01118-7
Chae, W. J., and Bothwell, A. L. (2015). Spontaneous intestinal tumorigenesis in apc (/Min+) mice requires altered T cell development with IL-17a. J. Immunol. Res. 2015, 860106. doi:10.1155/2015/860106
Challen, G. A., Boles, N., Lin, K. K., and Goodell, M. A. (2009). Mouse hematopoietic stem cell identification and analysis. Cytom. A 75 (1), 14–24. doi:10.1002/cyto.a.20674
Chambers, I., Colby, D., Robertson, M., Nichols, J., Lee, S., Tweedie, S., et al. (2003). Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell 113 (5), 643–655. doi:10.1016/s0092-8674(03)00392-1
Chandrakesan, P., May, R., Qu, D., Weygant, N., Taylor, V. E., Li, J. D., et al. (2015a). Dclk1+ small intestinal epithelial tuft cells display the hallmarks of quiescence and self-renewal. Oncotarget 6 (31), 30876–30886. doi:10.18632/oncotarget.5129
Chandrakesan, P., May, R., Weygant, N., Qu, D., Berry, W. L., Sureban, S. M., et al. (2016a). Intestinal tuft cells regulate the ATM mediated DNA Damage response via Dclk1 dependent mechanism for crypt restitution following radiation injury. Sci. Rep. 6, 37667. doi:10.1038/srep37667
Chandrakesan, P., Panneerselvam, J., May, R., Weygant, N., Qu, D., Berry, W. R., et al. (2020). DCLK1-Isoform2 alternative splice variant promotes pancreatic tumor immunosuppressive M2-macrophage polarization. Mol. Cancer Ther. 19 (7), 1539–1549. doi:10.1158/1535-7163.MCT-19-0776
Chandrakesan, P., Panneerselvam, J., Qu, D., Weygant, N., May, R., Bronze, M. S., et al. (2016b). Regulatory roles of Dclk1 in epithelial mesenchymal transition and cancer stem cells. J. Carcinog. Mutagen. 7 (2), 257. doi:10.4172/2157-2518.1000257
Chandrakesan, P., Weygant, N., May, R., Qu, D., Chinthalapally, H. R., Sureban, S. M., et al. (2014). DCLK1 facilitates intestinal tumor growth via enhancing pluripotency and epithelial mesenchymal transition. Oncotarget 5 (19), 9269–9280. doi:10.18632/oncotarget.2393
Chandrakesan, P., Weygant, N., Taylor, V., Berry, W., May, R., Qu, D., et al. (2015b). Abstract 4220: Silencing DCLK1 prevents breast cancer cell self-renewal, epithelial mesenchymal transition, circulating tumor cells and metastasis. Cancer Res. 75 (15), 4220. doi:10.1158/1538-7445.AM2015-4220
Chandrakesan, P., Yao, J., Qu, D., May, R., Weygant, N., Ge, Y., et al. (2017). Dclk1, a tumor stem cell marker, regulates pro-survival signaling and self-renewal of intestinal tumor cells. Mol. Cancer 16 (1), 30. doi:10.1186/s12943-017-0594-y
Chandrasekar, T., Klaassen, Z., Goldberg, H., Kulkarni, G. S., Hamilton, R. J., and Fleshner, N. E. (2017). Metastatic renal cell carcinoma: Patterns and predictors of metastases-A contemporary population-based series. Urol. Oncol. 35 (11), 661. doi:10.1016/j.urolonc.2017.06.060
Chen, H., Nio, K., Yamashita, T., Okada, H., Li, R., Suda, T., et al. (2021a). BMP9-ID1 signaling promotes EpCAM-positive cancer stem cell properties in hepatocellular carcinoma. Mol. Oncol. 15 (8), 2203–2218. doi:10.1002/1878-0261.12963
Chen, Q., Gu, M., Cai, Z. K., Zhao, H., Sun, S. C., Liu, C., et al. (2021b). TGF-β1 promotes epithelial-to-mesenchymal transition and stemness of prostate cancer cells by inducing PCBP1 degradation and alternative splicing of CD44. Cell. Mol. Life Sci. 78 (3), 949–962. doi:10.1007/s00018-020-03544-5
Chen, X., Guan, H., Liu, X. D., Xie, D. F., Wang, Y., Ma, T., et al. (2018). p53 positively regulates the expression of cancer stem cell marker CD133 in HCT116 colon cancer cells. Oncol. Lett. 16 (1), 431–438. doi:10.3892/ol.2018.8619
Cheuk, S., Schlums, H., Gallais Sérézal, I., Martini, E., Chiang, S. C., Marquardt, N., et al. (2017). CD49a expression defines tissue-resident CD8(+) T cells poised for cytotoxic function in human skin. Immunity 46 (2), 287–300. doi:10.1016/j.immuni.2017.01.009
Chiou, S. H., Wang, M. L., Chou, Y. T., Chen, C. J., Hong, C. F., Hsieh, W. J., et al. (2010). Coexpression of Oct4 and Nanog enhances malignancy in lung adenocarcinoma by inducing cancer stem cell-like properties and epithelial-mesenchymal transdifferentiation. Cancer Res. 70 (24), 10433–10444. doi:10.1158/0008-5472.Can-10-2638
Conde, I., Ribeiro, A. S., and Paredes, J. (2022). Breast cancer stem cell membrane biomarkers: Therapy targeting and clinical implications. Cells 11 (6), 934. doi:10.3390/cells11060934
Conry, R. M., Westbrook, B., McKee, S., and Norwood, T. G. (2018). Talimogene laherparepvec: First in class oncolytic virotherapy. Hum. Vaccin. Immunother. 14 (4), 839–846. doi:10.1080/21645515.2017.1412896
Czajka, A. A., Wojcicka, A., Kubiak, A., Kotlarek, M., Bakula-Zalewska, E., Koperski, L., et al. (2016). Family of microRNA-146 regulates RARβ in papillary thyroid carcinoma. PLoS One 11 (3), e0151968. doi:10.1371/journal.pone.0151968
Dana, H., Chalbatani, G. M., Mahmoodzadeh, H., Karimloo, R., Rezaiean, O., Moradzadeh, A., et al. (2017). Molecular mechanisms and biological functions of siRNA. Int. J. Biomed. Sci. 13 (2), 48–57.
Dandawate, P., Ghosh, C., Palaniyandi, K., Paul, S., Rawal, S., Pradhan, R., et al. (2019). The histone demethylase KDM3A, increased in human pancreatic tumors, regulates expression of DCLK1 and promotes tumorigenesis in mice. Gastroenterology 157 (6), 1646–1659. doi:10.1053/j.gastro.2019.08.018
Das, P. K., Islam, F., and Lam, A. K. (2020). The roles of cancer stem cells and therapy resistance in colorectal carcinoma. Cells 9 (6), E1392. doi:10.3390/cells9061392
Dastmalchi, N., Baradaran, B., Banan Khojasteh, S. M., Hosseinpourfeizi, M., and Safaralizadeh, R. (2021). miR-424: A novel potential therapeutic target and prognostic factor in malignancies. Cell Biol. Int. 45 (4), 720–730. doi:10.1002/cbin.11530
Dazard, J. E., Sandlers, Y., Doerner, S. K., Berger, N. A., and Brunengraber, H. (2014). Metabolomics of ApcMin/+mice genetically susceptible to intestinal cancer. BMC Syst. Biol. 8, 72. doi:10.1186/1752-0509-8-72
Dey, A., Varelas, X., and Guan, K. L. (2020). Targeting the Hippo pathway in cancer, fibrosis, wound healing and regenerative medicine. Nat. Rev. Drug Discov. 19 (7), 480–494. doi:10.1038/s41573-020-0070-z
Dinavahi, S. S., Bazewicz, C. G., Gowda, R., and Robertson, G. P. (2019). Aldehyde dehydrogenase inhibitors for cancer therapeutics. Trends Pharmacol. Sci. 40 (10), 774–789. doi:10.1016/j.tips.2019.08.002
Ding, F., Liu, J., and Zhang, X. (2020). microRNA-375 released from extracellular vesicles of bone marrow mesenchymal stem cells exerts anti-oncogenic effects against cervical cancer. Stem Cell Res. Ther. 11 (1), 455. doi:10.1186/s13287-020-01908-z
Dirican, E., Kankaya, B., Büyükaşık, S., Alış, H., Velidedeoğlu, M., İlvan, S., et al. (2020). Investigation of alterations in PIK3CA and OCT-4 gene expression in breast cancer. Gene Rep. 21, 100942. doi:10.1016/j.genrep.2020.100942
Domev, H., Amit, M., Laevsky, I., Dar, A., and Itskovitz-Eldor, J. (2012). Efficient engineering of vascularized ectopic bone from human embryonic stem cell-derived mesenchymal stem cells. Tissue Eng. Part A 18 (21-22), 2290–2302. doi:10.1089/ten.TEA.2011.0371
Eftekhari, R. B., Maghsoudnia, N., Samimi, S., Zamzami, A., and Dorkoosh, F. A. (2019). Co-delivery nanosystems for cancer treatment: A review. Pharm. Nanotechnol. 7 (2), 90–112. doi:10.2174/2211738507666190321112237
Esquela-Kerscher, A. (2014). The lin-4 microRNA: The ultimate micromanager. Cell Cycle 13 (7), 1060–1061. doi:10.4161/cc.28384
Fang, L., and Maly, D. J. (2020). The right tool for the job: A chemical and genetic toolkit for interrogating DCLK1 function. Cell Chem. Biol. 27 (10), 1221–1223. doi:10.1016/j.chembiol.2020.09.007
Faux, M. C., King, L. E., Kane, S. R., Love, C., Sieber, O. M., and Burgess, A. W. (2021). APC regulation of ESRP1 and p120-catenin isoforms in colorectal cancer cells. Mol. Biol. Cell 32 (2), 120–130. doi:10.1091/mbc.E20-05-0321
Fearnhead, N. S., Britton, M. P., and Bodmer, W. F. (2001). The ABC of APC. Hum. Mol. Genet. 10 (7), 721–733. doi:10.1093/hmg/10.7.721
Ferguson, F. M., Nabet, B., Raghavan, S., Liu, Y., Leggett, A. L., Kuljanin, M., et al. (2020). Discovery of a selective inhibitor of doublecortin like kinase 1. Nat. Chem. Biol. 16 (6), 635–643. doi:10.1038/s41589-020-0506-0
Fesler, A., Liu, H., and Ju, J. (2017). Modified miR-15a has therapeutic potential for improving treatment of advanced stage colorectal cancer through inhibition of BCL2, BMI1, YAP1 and DCLK1. Oncotarget 9 (2), 2367–2383. doi:10.18632/oncotarget.23414
Flanagan, D. J., Pentinmikko, N., Luopajärvi, K., Willis, N. J., Gilroy, K., Raven, A. P., et al. (2021). NOTUM from Apc-mutant cells biases clonal competition to initiate cancer. Nature 594 (7863), 430–435. doi:10.1038/s41586-021-03525-z
Fu, X., Brown, K. J., Yap, C. C., Winckler, B., Jaiswal, J. K., and Liu, J. S. (2013). Doublecortin (Dcx) family proteins regulate filamentous actin structure in developing neurons. J. Neurosci. 33 (2), 709–721. doi:10.1523/JNEUROSCI.4603-12.2013
Fuchs, E., and Blau, H. M. (2020). Tissue stem cells: Architects of their niches. Cell Stem Cell 27 (4), 532–556. doi:10.1016/j.stem.2020.09.011
Ganguly, K., Krishn, S. R., Rachagani, S., Jahan, R., Shah, A., Nallasamy, P., et al. (2021). Secretory mucin 5AC promotes neoplastic progression by augmenting KLF4-mediated pancreatic cancer cell stemness. Cancer Res. 81 (1), 91–102. doi:10.1158/0008-5472.Can-20-1293
Gao, T., Wang, M., Xu, L., Wen, T., Liu, J., and An, G. (2016). DCLK1 is up-regulated and associated with metastasis and prognosis in colorectal cancer. J. Cancer Res. Clin. Oncol. 142 (10), 2131–2140. doi:10.1007/s00432-016-2218-0
Gawlik-Rzemieniewska, N., and Bednarek, I. (2016). The role of NANOG transcriptional factor in the development of malignant phenotype of cancer cells. Cancer Biol. Ther. 17 (1), 1–10. doi:10.1080/15384047.2015.1121348
Ge, Y., Weygant, N., Qu, D., May, R., Berry, W. L., Yao, J., et al. (2018). Alternative splice variants of DCLK1 mark cancer stem cells, promote self-renewal and drug-resistance, and can be targeted to inhibit tumorigenesis in kidney cancer. Int. J. Cancer 143 (5), 1162–1175. doi:10.1002/ijc.31400
Gerbe, F., van Es, J. H., Makrini, L., Brulin, B., Mellitzer, G., Robine, S., et al. (2011). Distinct ATOH1 and Neurog3 requirements define tuft cells as a new secretory cell type in the intestinal epithelium. J. Cell Biol. 192 (5), 767–780. doi:10.1083/jcb.201010127
Ghaleb, A. M., and Yang, V. W. (2017). Kruppel-like factor 4 (KLF4): What we currently know. Gene 611, 27–37. doi:10.1016/j.gene.2017.02.025
Gires, O., Pan, M., Schinke, H., Canis, M., and Baeuerle, P. A. (2020). Expression and function of epithelial cell adhesion molecule EpCAM: Where are we after 40 years? Cancer Metastasis Rev. 39 (3), 969–987. doi:10.1007/s10555-020-09898-3
Glumac, P. M., and LeBeau, A. M. (2018). The role of CD133 in cancer: A concise review. Clin. Transl. Med. 7 (1), 18. doi:10.1186/s40169-018-0198-1
Gonzalez-Salinas, F., Martinez-Amador, C., and Trevino, V. (2022). Characterizing genes associated with cancer using the CRISPR/cas9 system: A systematic review of genes and methodological approaches. Gene 833, 146595. doi:10.1016/j.gene.2022.146595
Good, H., Shin, A. E., Zhang, L., and Asfaha, S. (2020). A30 inhibition of nf-kb signaling in Dclk1+ cells promotes colonic inflammation and colitis-associated cancer. J. Can. Assoc. Gastroenterol. 3 (1), 37–38. doi:10.1093/jcag/gwz047.029
Goto, N., Fukuda, A., Yamaga, Y., Yoshikawa, T., Maruno, T., Maekawa, H., et al. (2019). Lineage tracing and targeting of IL17RB(+) tuft cell-like human colorectal cancer stem cells. Proc. Natl. Acad. Sci. U. S. A. 116 (26), 12996–13005. doi:10.1073/pnas.1900251116
Grange, C., Lanzardo, S., Cavallo, F., Camussi, G., and Bussolati, B. (2008). Sca-1 identifies the tumor-initiating cells in mammary tumors of BALB-neuT transgenic mice. Neoplasia 10 (12), 1433–1443. doi:10.1593/neo.08902
Gronthos, S., Franklin, D. M., Leddy, H. A., Robey, P. G., Storms, R. W., and Gimble, J. M. (2001). Surface protein characterization of human adipose tissue-derived stromal cells. J. Cell. Physiol. 189 (1), 54–63. doi:10.1002/jcp.1138
Grubelnik, G., Boštjančič, E., Pavlič, A., Kos, M., and Zidar, N. (2020). NANOG expression in human development and cancerogenesis. Exp. Biol. Med. 245 (5), 456–464. doi:10.1177/1535370220905560
Gruber, R., Panayiotou, R., Nye, E., Spencer-Dene, B., Stamp, G., and Behrens, A. (2016). YAP1 and TAZ control pancreatic cancer initiation in mice by direct up-regulation of JAK-STAT3 signaling. Gastroenterology 151 (3), 526–539. doi:10.1053/j.gastro.2016.05.006
Guerriero, I., D'Angelo, D., Pallante, P., Santos, M., Scrima, M., Malanga, D., et al. (2017). Analysis of miRNA profiles identified miR-196a as a crucial mediator of aberrant PI3K/AKT signaling in lung cancer cells. Oncotarget 8 (12), 19172–19191. doi:10.18632/oncotarget.13432
Guo, C., Liu, P., Deng, G., Han, Y., Chen, Y., Cai, C., et al. (2021). Honokiol induces ferroptosis in colon cancer cells by regulating GPX4 activity. Mol. Ther. Nucleic Acids 11 (6), 3039–3054.
Guo, S., Fesler, A., Huang, W., Wang, Y., Yang, J., Wang, X., et al. (2020). Functional significance and therapeutic potential of miR-15a mimic in pancreatic ductal adenocarcinoma. Mol. Ther. Nucleic Acids 19, 228–239. doi:10.1016/j.omtn.2019.11.010
Gzil, A., Szylberg, L., Jaworski, D., Dominiak, J., Zarebska, I., and Grzanka, D. (2019). The essential role of DCLK1 in pathogenesis, diagnostic procedures and prognostic stratification of colorectal cancer. Anticancer Res. 39 (6), 2689–2697. doi:10.21873/anticanres.13394
Hamada, F., Murata, Y., Nishida, A., Fujita, F., Tomoyasu, Y., Nakamura, M., et al. (1999). Identification and characterization of E-APC, a novel Drosophila homologue of the tumour suppressor APC. Genes cells. 4 (8), 465–474. doi:10.1046/j.1365-2443.1999.00272.x
He, C., Jaffar Ali, D., Li, Y., Zhu, Y., Sun, B., and Xiao, Z. (2020). Engineering of HN3 increases the tumor targeting specificity of exosomes and upgrade the anti-tumor effect of sorafenib on HuH-7 cells. PeerJ 8, e9524. doi:10.7717/peerj.9524
Herlyn, M., Steplewski, Z., Herlyn, D., and Koprowski, H. (1979). Colorectal carcinoma-specific antigen: Detection by means of monoclonal antibodies. Proc. Natl. Acad. Sci. U. S. A. 76 (3), 1438–1442. doi:10.1073/pnas.76.3.1438
Herter, S., Morra, L., Schlenker, R., Sulcova, J., Fahrni, L., Waldhauer, I., et al. (2017). A novel three-dimensional heterotypic spheroid model for the assessment of the activity of cancer immunotherapy agents. Cancer Immunol. Immunother. 66 (1), 129–140. doi:10.1007/s00262-016-1927-1
Hilberg, F., Tontsch-Grunt, U., Baum, A., Le, A. T., Doebele, R. C., Lieb, S., et al. (2018). Triple angiokinase inhibitor nintedanib directly inhibits tumor cell growth and induces tumor shrinkage via blocking oncogenic receptor tyrosine kinases. J. Pharmacol. Exp. Ther. 364 (3), 494–503. doi:10.1124/jpet.117.244129
Hoffmann, N. E., Gillett, M. D., Cheville, J. C., Lohse, C. M., Leibovich, B. C., and Blute, M. L. (2008). Differences in organ system of distant metastasis by renal cell carcinoma subtype. J. Urol. 179 (2), 474–477. doi:10.1016/j.juro.2007.09.036
Holgersen, E. M., Gandhi, S., Zhou, Y., Kim, J., Vaz, B., Bogojeski, J., et al. (2021). Transcriptome-wide off-target effects of steric-blocking oligonucleotides. Nucleic Acid. Ther. 31 (6), 392–403. doi:10.1089/nat.2020.0921
Holmes, C., and Stanford, W. L. (2007). Concise review: Stem cell antigen-1: Expression, function, and enigma. Stem Cells 25 (6), 1339–1347. doi:10.1634/stemcells.2006-0644
Hsu, Y. C., Luo, C. W., Huang, W. L., Wu, C. C., Chou, C. L., Chen, C. I., et al. (2020). BMI1-KLF4 axis deficiency improves responses to neoadjuvant concurrent chemoradiotherapy in patients with rectal cancer. Radiother. Oncol. 149, 249–258. doi:10.1016/j.radonc.2020.06.023
Huang, T., Song, X., Xu, D., Tiek, D., Goenka, A., Wu, B., et al. (2020). Stem cell programs in cancer initiation, progression, and therapy resistance. Theranostics 10 (19), 8721–8743. doi:10.7150/thno.41648
Hütz, K., Mejías-Luque, R., Farsakova, K., Ogris, M., Krebs, S., Anton, M., et al. (2013). The stem cell factor SOX2 regulates the tumorigenic potential in human gastric cancer cells. Carcinogenesis 35 (4), 942–950. doi:10.1093/carcin/bgt410
Ihara, H., Mitsuishi, Y., Kato, M., Takahashi, F., Tajima, K., Hayashi, T., et al. (2020). Nintedanib inhibits epithelial-mesenchymal transition in A549 alveolar epithelial cells through regulation of the TGF-β/Smad pathway. Respir. Investig. 58 (4), 275–284. doi:10.1016/j.resinv.2020.01.003
Ikezono, Y. U., Koga, H., Abe, M., Akiba, J., Kawahara, A., Yoshida, T., et al. (2015). High expression of the putative cancer stem cell marker, DCLK1, in rectal neuroendocrine tumors. Oncol. Lett. 10 (4), 2015–2020. doi:10.3892/ol.2015.3513
Ito, H., Tanaka, S., Akiyama, Y., Shimada, S., Adikrisna, R., Matsumura, S., et al. (2016). Dominant expression of DCLK1 in human pancreatic cancer stem cells accelerates tumor invasion and metastasis. PLoS One 11 (1), e0146564. doi:10.1371/journal.pone.0146564
Jahangiri, R., Mosaffa, F., EmamiRazavi, A., Gharib, M., and Jamialahmadi, K. (2020). Increased expression of gankyrin and stemness factor oct-4 are associated with unfavorable clinical outcomes and poor benefit of tamoxifen in breast carcinoma patients. Pathol. Oncol. Res. 26 (3), 1921–1934. doi:10.1007/s12253-019-00766-2
Jang, D. M., Lim, H. J., Hahn, H., Lee, Y., Kim, H. K., and Kim, H. S. (2021). Structural basis of inhibition of DCLK1 by Ruxolitinib. Int. J. Mol. Sci. 22 (16), 8488. doi:10.3390/ijms22168488
Januchowski, R., Wojtowicz, K., and Zabel, M. (2013). The role of aldehyde dehydrogenase (ALDH) in cancer drug resistance. Biomed. Pharmacother. 67 (7), 669–680. doi:10.1016/j.biopha.2013.04.005
Javed, Z., Javed Iqbal, M., Rasheed, A., Sadia, H., Raza, S., Irshad, A., et al. (2021). Regulation of hedgehog signaling by miRNAs and nanoformulations: A possible therapeutic solution for colorectal cancer. Front. Oncol. 10 (2790), 607607. doi:10.3389/fonc.2020.607607
Jayaraman, S., Gnanasampanthapandian, D., Rajasingh, J., and Palaniyandi, K. (2021). Stem cell-derived exosomes potential therapeutic roles in cardiovascular diseases. Front. Cardiovasc. Med. 8, 723236. doi:10.3389/fcvm.2021.723236
Jean, D. C., Baas, P. W., and Black, M. M. (2012). A novel role for doublecortin and doublecortin-like kinase in regulating growth cone microtubules. Hum. Mol. Genet. 21 (26), 5511–5527. doi:10.1093/hmg/dds395
Jeter, C. R., Yang, T., Wang, J., Chao, H. P., and Tang, D. G. (2015). Concise review: NANOG in cancer stem cells and tumor development: An update and outstanding questions. Stem Cells 33 (8), 2381–2390. doi:10.1002/stem.2007
Jia, Z., Zhang, Y., Yan, A., Wang, M., Han, Q., Wang, K., et al. (2020). 1, 25-dihydroxyvitamin D3 signaling-induced decreases in IRX4 inhibits NANOG-mediated cancer stem-like properties and gefitinib resistance in NSCLC cells. Cell Death Dis. 11 (8), 670. doi:10.1038/s41419-020-02908-w
Kalantari, E., Asadi Lari, M. H., Roudi, R., Korourian, A., and Madjd, Z. (2017). Lgr5High/DCLK1High phenotype is more common in early stage and intestinal subtypes of gastric carcinomas. Cancer Biomark. 20 (4), 563–573. doi:10.3233/CBM-170383
Kalluri, R., and LeBleu, V. S. (2020). The biology, function, and biomedical applications of exosomes. Science 367 (6478), eaau6977. doi:10.1126/science.aau6977
Kamble, D., Mahajan, M., Dhat, R., and Sitasawad, S. (2021). Keap1-Nrf2 pathway regulates ALDH and contributes to radioresistance in breast cancer stem cells. Cells 10 (1), E83. doi:10.3390/cells10010083
Kang, X. L., He, L. R., Chen, Y. L., and Wang, S. B. (2020). Role of doublecortin-like kinase 1 and leucine-rich repeat-containing G-protein-coupled receptor 5 in patients with stage II/III colorectal cancer: Cancer progression and prognosis. World J. Gastroenterol. 26 (43), 6853–6866. doi:10.3748/wjg.v26.i43.6853
Karagonlar, Z. F., Akbari, S., Karabicici, M., Sahin, E., Avci, S. T., Ersoy, N., et al. (2020). A novel function for KLF4 in modulating the de-differentiation of EpCAM(-)/cd133(-) nonStem cells into EpCAM(+)/cd133(+) liver cancer stem cells in HCC cell line HuH7. Cells 9 (5), E1198. doi:10.3390/cells9051198
Kawamura, D., Takemoto, Y., Nishimoto, A., Ueno, K., Hosoyama, T., Shirasawa, B., et al. (2017). Enhancement of cytotoxic effects of gemcitabine by Dclk1 inhibition through suppression of Chk1 phosphorylation in human pancreatic cancer cells. Oncol. Rep. 38 (5), 3238–3244. doi:10.3892/or.2017.5974
Kenney, J. W., Genheden, M., Moon, K. M., Wang, X., Foster, L. J., and Proud, C. G. (2016). Eukaryotic elongation factor 2 kinase regulates the synthesis of microtubule-related proteins in neurons. J. Neurochem. 136 (2), 276–284. doi:10.1111/jnc.13407
Khani, A. T., Sharifzad, F., Mardpour, S., Hassan, Z. M., and Ebrahimi, M. (2021). Tumor extracellular vesicles loaded with exogenous Let-7i and miR-142 can modulate both immune response and tumor microenvironment to initiate a powerful anti-tumor response. Cancer Lett. 501, 200–209. doi:10.1016/j.canlet.2020.11.014
Khosravi, A., Jafari, S. M., and Asadi, J. (2021). Knockdown of TAZ decrease the cancer stem properties of ESCC cell line YM-1 by modulation of Nanog, OCT-4 and SOX2. Gene 769, 145207. doi:10.1016/j.gene.2020.145207
Kim, H., Kim, J. K., Ye, C., Choi, J. H., Lee, H., Oh, J. J., et al. (2021). Recurrence after radical and partial nephrectomy in high complex renal tumor using propensity score matched analysis. Sci. Rep. 11 (1), 2919. doi:10.1038/s41598-021-82700-8
Koni, M., Pinnarò, V., and Brizzi, M. F. (2020). The Wnt signalling pathway: A tailored target in cancer. Int. J. Mol. Sci. 21 (20), E7697. doi:10.3390/ijms21207697
Krause, J., von Felden, J., Casar, C., Fründt, T. W., Galaski, J., Schmidt, C., et al. (2020). Hepatocellular carcinoma: Intratumoral EpCAM-positive cancer stem cell heterogeneity identifies high-risk tumor subtype. BMC Cancer 20 (1), 1130. doi:10.1186/s12885-020-07580-z
Krishnamachary, B., Subramaniam, D., Dandawate, P., Ponnurangam, S., Srinivasan, P., Ramamoorthy, P., et al. (2019). Targeting transcription factor TCF4 by γ-Mangostin, a natural xanthone. Oncotarget 10 (54), 5576–5591. doi:10.18632/oncotarget.27159
Kucharczyk, J., Mandalapu, K., Satti, S., and Matrana, M. R. (2017). Outcomes of patients with late-relapse metastatic renal cell carcinoma treated with targeted therapies: A single institution experience. Ochsner J. 17 (4), 331–334. doi:10.1043/TOJ-17-0017
Kunze, B., Middelhoff, M., Maurer, H. C., Agibalova, T., Anand, A., Buhrer, A. M., et al. (2021). Notch signaling drives development of Barrett's metaplasia from Dclk1-positive epithelial tuft cells in the murine gastric mucosa. Sci. Rep. 11 (1), 4509. doi:10.1038/s41598-021-84011-4
Le Hellard, S., Havik, B., Espeseth, T., Breilid, H., Lovlie, R., Luciano, M., et al. (2009). Variants in doublecortin- and calmodulin kinase like 1, a gene up-regulated by BDNF, are associated with memory and general cognitive abilities. PLoS One 4 (10), e7534. doi:10.1371/journal.pone.0007534
Lee, R. C., Feinbaum, R. L., and Ambros, V. (1993). The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75 (5), 843–854. doi:10.1016/0092-8674(93)90529-y
Lee, Y. T., Tan, Y. J., and Oon, C. E. (2018). Molecular targeted therapy: Treating cancer with specificity. Eur. J. Pharmacol. 834, 188–196. doi:10.1016/j.ejphar.2018.07.034
Li, J., Wang, Y., Ge, J., Li, W., Yin, L., Zhao, Z., et al. (2018). Doublecortin-like kinase 1 (DCLK1) regulates B cell-specific Moloney murine leukemia virus insertion site 1 (Bmi-1) and is associated with metastasis and prognosis in pancreatic cancer. Cell. Physiol. biochem. 51 (1), 262–277. doi:10.1159/000495228
Li, L., and Bellows, C. F. (2013). Doublecortin-like kinase 1 exhibits cancer stem cell-like characteristics in a human colon cancer cell line. Chin. J. Cancer Res. 25 (2), 134–142. doi:10.3978/j.issn.1000-9604.2013.03.02
Li, S., Yi, M., Dong, B., Tan, X., Luo, S., and Wu, K. (2021). The role of exosomes in liquid biopsy for cancer diagnosis and prognosis prediction. Int. J. Cancer 148 (11), 2640–2651. doi:10.1002/ijc.33386
Lipka, J., Kapitein, L. C., Jaworski, J., and Hoogenraad, C. C. (2016). Microtubule-binding protein doublecortin-like kinase 1 (DCLK1) guides kinesin-3-mediated cargo transport to dendrites. EMBO J. 35 (3), 302–318. doi:10.15252/embj.201592929
Liu, C., Zhou, X., Long, Q., Zeng, H., Sun, Q., Chen, Y., et al. (2021). Small extracellular vesicles containing miR-30a-3p attenuate the migration and invasion of hepatocellular carcinoma by targeting SNAP23 gene. Oncogene 40 (2), 233–245. doi:10.1038/s41388-020-01521-7
Liu, H., Wen, T., Zhou, Y., Fan, X., Du, T., Gao, T., et al. (2019). DCLK1 plays a metastatic-promoting role in human breast cancer cells. Biomed. Res. Int. 2019, 1061979. doi:10.1155/2019/1061979
Liu, W., Wang, S., Sun, Q., Yang, Z., Liu, M., and Tang, H. (2018). DCLK1 promotes epithelial-mesenchymal transition via the PI3K/Akt/NF-κB pathway in colorectal cancer. Int. J. Cancer 142 (10), 2068–2079. doi:10.1002/ijc.31232
Liu, Z. Q., He, W. F., Wu, Y. J., Zhao, S. L., Wang, L., Ouyang, Y. Y., et al. (2020). LncRNA SNHG1 promotes EMT process in gastric cancer cells through regulation of the miR-15b/DCLK1/Notch1 axis. BMC Gastroenterol. 20 (1), 156. doi:10.1186/s12876-020-01272-5
Lorenzo, N., Sabina, D. M., Guido, C., Ilaria Grazia, Z., Samira, S., Valeria, A., et al. (2021). DCLK1, a putative stem cell marker in human cholangiocarcinoma. Hepatology 73 (1), 144–159. doi:10.1002/hep.31571
Lu, J., Liu, J., Guo, Y., Zhang, Y., Xu, Y., and Wang, X. (2021). CRISPR-Cas9: A method for establishing rat models of drug metabolism and pharmacokinetics. Acta Pharm. Sin. B 11 (10), 2973–2982. doi:10.1016/j.apsb.2021.01.007
Lu, K., Feng, F., Yang, Y., Liu, K., Duan, J., Liu, H., et al. (2020). High-throughput screening identified miR-7-2-3p and miR-29c-3p as metastasis suppressors in gallbladder carcinoma. J. Gastroenterol. 55 (1), 51–66. doi:10.1007/s00535-019-01627-0
Lytle, N. K., Ferguson, L. P., Rajbhandari, N., Gilroy, K., Fox, R. G., Deshpande, A., et al. (2019). A multiscale map of the stem cell state in pancreatic adenocarcinoma. Cell 177 (3), 572–586. e522. doi:10.1016/j.cell.2019.03.010
Mahmoodi Chalbatani, G., Dana, H., Gharagouzloo, E., Grijalvo, S., Eritja, R., Logsdon, C. D., et al. (2019). Small interfering RNAs (siRNAs) in cancer therapy: A nano-based approach. Int. J. Nanomedicine 14, 3111–3128. doi:10.2147/IJN.S200253
Mamun, M. A., Mannoor, K., Cao, J., Qadri, F., and Song, X. (2020). SOX2 in cancer stemness: Tumor malignancy and therapeutic potentials. J. Mol. Cell Biol. 12 (2), 85–98. doi:10.1093/jmcb/mjy080
Maruno, T., Fukuda, A., Goto, N., Tsuda, M., Ikuta, K., Hiramatsu, Y., et al. (2021). Visualization of stem cell activity in pancreatic cancer expansion by direct lineage tracing with live imaging. Elife 10, e55117. doi:10.7554/eLife.55117
Mashayekhi, S., Saeidi Saedi, H., Salehi, Z., Soltanipour, S., and Mirzajani, E. (2018). Effects of miR-27a, miR-196a2 and miR-146a polymorphisms on the risk of breast cancer. Br. J. Biomed. Sci. 75 (2), 76–81. doi:10.1080/09674845.2017.1399572
Matsumoto, A., Asuka, M., Takahashi, Y., and Takakura, Y. (2020). Antitumor immunity by small extracellular vesicles collected from activated dendritic cells through effective induction of cellular and humoral immune responses. Biomaterials 252, 120112. doi:10.1016/j.biomaterials.2020.120112
May, R., Qu, D., Weygant, N., Chandrakesan, P., Ali, N., Lightfoot, S. A., et al. (2014). Brief report: Dclk1 deletion in tuft cells results in impaired epithelial repair after radiation injury. Stem Cells 32 (3), 822–827. doi:10.1002/stem.1566
May, R., Riehl, T. E., Hunt, C., Sureban, S. M., Anant, S., and Houchen, C. W. (2008). Identification of a novel putative gastrointestinal stem cell and adenoma stem cell marker, doublecortin and CaM kinase-like-1, following radiation injury and in adenomatous polyposis coli/multiple intestinal neoplasia mice. Stem Cells 26 (3), 630–637. doi:10.1634/stemcells.2007-0621
McAndrews, K. M., Vázquez-Arreguín, K., Kwak, C., Sugimoto, H., Zheng, X., Li, B., et al. (2021). αSMA(+) fibroblasts suppress Lgr5(+) cancer stem cells and restrain colorectal cancer progression. Oncogene 40 (26), 4440–4452. doi:10.1038/s41388-021-01866-7
Meng, Y., Xu, Q., Chen, L., Wang, L., and Hu, X. (2020). The function of SOX2 in breast cancer and relevant signaling pathway. Pathol. Res. Pract. 216 (8), 153023. doi:10.1016/j.prp.2020.153023
Middelhoff, M., Westphalen, C. B., Hayakawa, Y., Yan, K. S., Gershon, M. D., Wang, T. C., et al. (2017). Dclk1-expressing tuft cells: Critical modulators of the intestinal niche? Am. J. Physiol. Gastrointest. Liver Physiol. 313 (4), G285–G299. doi:10.1152/ajpgi.00073.2017
Mizuguchi, M., Qin, J., Yamada, M., Ikeda, K., and Takashima, S. (1999). High expression of doublecortin and KIAA0369 protein in fetal brain suggests their specific role in neuronal migration. Am. J. Pathol. 155 (5), 1713–1721. doi:10.1016/S0002-9440(10)65486-7
Moazeni-Roodi, A., Ghavami, S., and Hashemi, M. (2019). Association between miR-423 rs6505162 polymorphism and susceptibility to cancer. Arch. Med. Res. 50 (1), 21–30. doi:10.1016/j.arcmed.2019.04.002
Mohamed, N. E., Hay, T., Reed, K. R., Smalley, M. J., and Clarke, A. R. (2019). APC2 is critical for ovarian WNT signalling control, fertility and tumour suppression. BMC Cancer 19 (1), 677. doi:10.1186/s12885-019-5867-y
Mohammadi, C., and Najafi, R. (2020). DCLK1 as a promising marker for radioresistance in colorectal cancer. J. Gastrointest. Cancer 51 (2), 714–715. doi:10.1007/s12029-019-00292-z
Mohtar, M. A., Syafruddin, S. E., Nasir, S. N., and Low, T. Y. (2020). Revisiting the roles of pro-metastatic EpCAM in cancer. Biomolecules 10 (2), E255. doi:10.3390/biom10020255
Moores, C. A., Perderiset, M., Kappeler, C., Kain, S., Drummond, D., Perkins, S. J., et al. (2006). Distinct roles of doublecortin modulating the microtubule cytoskeleton. EMBO J. 25 (19), 4448–4457. doi:10.1038/sj.emboj.7601335
Mulholland, D. J., Xin, L., Morim, A., Lawson, D., Witte, O., and Wu, H. (2009). Lin-Sca-1+CD49fhigh stem/progenitors are tumor-initiating cells in the Pten-null prostate cancer model. Cancer Res. 69 (22), 8555–8562. doi:10.1158/0008-5472.Can-08-4673
Muralikrishnan, V., Hurley, T. D., and Nephew, K. P. (2020). Targeting aldehyde dehydrogenases to eliminate cancer stem cells in gynecologic malignancies. Cancers 12 (4), 961. doi:10.3390/cancers12040961
Nagase, T., Ishikawa, K., Nakajima, D., Ohira, M., Seki, N., Miyajima, N., et al. (1997). Prediction of the coding sequences of unidentified human genes. VII. The complete sequences of 100 new cDNA clones from brain which can code for large proteins in vitro. DNA Res. 4 (2), 141–150. doi:10.1093/dnares/4.2.141
Nakanishi, Y., Seno, H., Fukuoka, A., Ueo, T., Yamaga, Y., Maruno, T., et al. (2013). Dclk1 distinguishes between tumor and normal stem cells in the intestine. Nat. Genet. 45 (1), 98–103. doi:10.1038/ng.2481
Nevi, L., Di Matteo, S., Carpino, G., Zizzari, I. G., Samira, S., Ambrosino, V., et al. (2021). DCLK1, a putative stem cell marker in human cholangiocarcinoma. Hepatology 73 (1), 144–159. doi:10.1002/hep.31571
Nichols, J., Zevnik, B., Anastassiadis, K., Niwa, H., Klewe-Nebenius, D., Chambers, I., et al. (1998). Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell 95 (3), 379–391. doi:10.1016/s0092-8674(00)81769-9
Nie, D., Dai, Z., Li, J., Yang, Y., Xi, Z., Wang, J., et al. (2020). Cancer-cell-membrane-coated nanoparticles with a yolk-shell structure augment cancer chemotherapy. Nano Lett. 20 (2), 936–946. doi:10.1021/acs.nanolett.9b03817
Nishio, K., Kimura, K., Amano, R., Nakata, B., Yamazoe, S., Ohira, G., et al. (2017). Doublecortin and CaM kinase-like-1 as an independent prognostic factor in patients with resected pancreatic carcinoma. World J. Gastroenterol. 23 (31), 5764–5772. doi:10.3748/wjg.v23.i31.5764
Niwa, H., Miyazaki, J.-i., and Smith, A. G. (2000). Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat. Genet. 24 (4), 372–376. doi:10.1038/74199
Nomura, A., Dauer, P., Gupta, V., McGinn, O., Arora, N., Majumdar, K., et al. (2016). Microenvironment mediated alterations to metabolic pathways confer increased chemo-resistance in CD133+ tumor initiating cells. Oncotarget 7 (35), 56324–56337. doi:10.18632/oncotarget.10838
O'Brien, C. A., Kreso, A., and Jamieson, C. H. (2010). Cancer stem cells and self-renewal. Clin. Cancer Res. 16 (12), 3113–3120. doi:10.1158/1078-0432.CCR-09-2824
O'Connell, M. R., Sarkar, S., Luthra, G. K., Okugawa, Y., Toiyama, Y., Gajjar, A. H., et al. (2015). Epigenetic changes and alternate promoter usage by human colon cancers for expressing DCLK1-isoforms: Clinical Implications. Sci. Rep. 5, 14983. doi:10.1038/srep14983
Omori, Y., Suzuki, M., Ozaki, K., Harada, Y., Nakamura, Y., Takahashi, E., et al. (1998). Expression and chromosomal localization of KIAA0369, a putative kinase structurally related to Doublecortin. J. Hum. Genet. 43 (3), 169–177. doi:10.1007/s100380050063
Overed-Sayer, C., Miranda, E., Dunmore, R., Liarte Marin, E., Beloki, L., Rassl, D., et al. (2020). Inhibition of mast cells: A novel mechanism by which nintedanib may elicit anti-fibrotic effects. Thorax 75 (9), 754–763. doi:10.1136/thoraxjnl-2019-214000
Palaniyandi, K., Zhao, Q., and Chang, X. B. (2011). Infection of H69AR cells with retroviral particles harboring interfering RNAi significantly reduced the multidrug resistance of these small cell lung cancer cells. Int. J. Biochem. Mol. Biol. 2 (2), 155–167.
Panneerselvam, J., Mohandoss, P., Patel, R., Gillan, H., Li, M., Kumar, K., et al. (2020). DCLK1 regulates tumor stemness and cisplatin resistance in non-small cell lung cancer via ABCD-member-4. Mol. Ther. Oncolytics 18, 24–36. doi:10.1016/j.omto.2020.05.012
Parajuli, B., Fishel, M. L., and Hurley, T. D. (2014). Selective ALDH3A1 inhibition by benzimidazole analogues increase mafosfamide sensitivity in cancer cells. J. Med. Chem. 57 (2), 449–461. doi:10.1021/jm401508p
Park, J. W., Park, J. M., Park, D. M., Kim, D. Y., and Kim, H. K. (2016). Stem cells antigen-1 enriches for a cancer stem cell-like subpopulation in mouse gastric cancer. Stem Cells 34 (5), 1177–1187. doi:10.1002/stem.2329
Park, S. Y., Kim, J. Y., Choi, J. H., Kim, J. H., Lee, C. J., Singh, P., et al. (2019). Inhibition of LEF1-mediated DCLK1 by niclosamide attenuates colorectal cancer stemness. Clin. Cancer Res. 25 (4), 1415–1429. doi:10.1158/1078-0432.Ccr-18-1232
Patel, O., Dai, W., Mentzel, M., Griffin, M. D., Serindoux, J., Gay, Y., et al. (2016). Biochemical and structural insights into doublecortin-like kinase domain 1. Structure 24 (9), 1550–1561. doi:10.1016/j.str.2016.07.008
Patni, A. P., Harishankar, M. K., Joseph, J. P., Sreeshma, B., Jayaraj, R., and Devi, A. (2021). Comprehending the crosstalk between Notch, Wnt and Hedgehog signaling pathways in oral squamous cell carcinoma - clinical implications. Cell. Oncol. 44 (3), 473–494. doi:10.1007/s13402-021-00591-3
Pradhan, T., Padmanabhan, K., Prasad, M., Chandramohan, K., and Nair, S. A. (2019). Augmented CD133 expression in distal margin correlates with poor prognosis in colorectal cancer. J. Cell. Mol. Med. 23 (6), 3984–3994. doi:10.1111/jcmm.14284
Qiao, S., Zhao, Y., Geng, S., Li, Y., Hou, X., Liu, Y., et al. (2016). A novel double-targeted nondrug delivery system for targeting cancer stem cells. Int. J. Nanomedicine 11, 6667–6678. doi:10.2147/IJN.S116230
Qiu, W., Remotti, H. E., Tang, S. M., Wang, E., Dobberteen, L., Lee Youssof, A., et al. (2018). Pancreatic DCLK1(+) cells originate distinctly from PDX1(+) progenitors and contribute to the initiation of intraductal papillary mucinous neoplasm in mice. Cancer Lett. 423, 71–79. doi:10.1016/j.canlet.2018.03.009
Qu, D., Johnson, J., Chandrakesan, P., Weygant, N., May, R., Aiello, N., et al. (2015). Doublecortin-like kinase 1 is elevated serologically in pancreatic ductal adenocarcinoma and widely expressed on circulating tumor cells. PLoS One 10 (2), e0118933. doi:10.1371/journal.pone.0118933
Quan, X. X., Hawk, N. V., Chen, W., Coupar, J., Lee, S. K., Petersen, D. W., et al. (2018). Targeting Notch1 and IKKα enhanced NF-κB activation in CD133+ skin cancer stem cells. Mol. Cancer Ther. 17 (9), 2034–2048. doi:10.1158/1535-7163.MCT-17-0421
Ramamoorthy, P., Dandawate, P., Jensen, R. A., and Anant, S. (2021). Celastrol and Triptolide suppress stemness in triple negative breast cancer: Notch as a therapeutic target for stem cells. Biomedicines 9 (5), 482. doi:10.3390/biomedicines9050482
Razi, S., Sadeghi, A., Asadi-Lari, Z., Tam, K. J., Kalantari, E., and Madjd, Z. (2021). DCLK1, a promising colorectal cancer stem cell marker, regulates tumor progression and invasion through miR-137 and miR-15a dependent manner. Clin. Exp. Med. 21 (1), 139–147. doi:10.1007/s10238-020-00665-w
Reiner, O., Coquelle, F. M., Peter, B., Levy, T., Kaplan, A., Sapir, T., et al. (2006). The evolving doublecortin (DCX) superfamily. BMC Genomics 7, 188. doi:10.1186/1471-2164-7-188
Remšík, J., Pícková, M., Vacek, O., Fedr, R., Binó, L., Hampl, A., et al. (2020). TGF-β regulates Sca-1 expression and plasticity of pre-neoplastic mammary epithelial stem cells. Sci. Rep. 10 (1), 11396. doi:10.1038/s41598-020-67827-4
Robinson, M., Gilbert, S. F., Waters, J. A., Lujano-Olazaba, O., Lara, J., Alexander, L. J., et al. (2021). Characterization of SOX2, OCT4 and NANOG in ovarian cancer tumor-initiating cells. Cancers (Basel) 13 (2), E262. doi:10.3390/cancers13020262
Roy, B. C., Ahmed, I., Stubbs, J., Zhang, J., Attard, T., Septer, S., et al. (2021). DCLK1 isoforms and aberrant Notch signaling in the regulation of human and murine colitis. Cell Death Discov. 7 (1), 169. doi:10.1038/s41420-021-00526-9
Rustgi, A. K. (2007). The genetics of hereditary colon cancer. Genes Dev. 21 (20), 2525–2538. doi:10.1101/gad.1593107
Sakaguchi, M., Hisamori, S., Oshima, N., Sato, F., Shimono, Y., and Sakai, Y. (2016). miR-137 regulates the tumorigenicity of colon cancer stem cells through the inhibition of DCLK1. Mol. Cancer Res. 14 (4), 354–362. doi:10.1158/1541-7786.MCR-15-0380
Salaritabar, A., Berindan-Neagoe, I., Darvish, B., Hadjiakhoondi, F., Manayi, A., Devi, K. P., et al. (2019). Targeting Hedgehog signaling pathway: Paving the road for cancer therapy. Pharmacol. Res. 141, 466–480. doi:10.1016/j.phrs.2019.01.014
Sameri, S., Saidijam, M., Bahreini, F., and Najafi, R. (2021). Cancer chemopreventive activities of Silibinin on colorectal cancer through regulation of E-cadherin/β-catenin pathway. Nutr. Cancer 73 (8), 1389–1399. doi:10.1080/01635581.2020.1800764
Sarkar, S., O'Connell, M. R., Okugawa, Y., Lee, B. S., Toiyama, Y., Kusunoki, M., et al. (2017a). FOXD3 regulates CSC marker, DCLK1-S, and invasive potential: Prognostic implications in colon cancer. Mol. Cancer Res. 15 (12), 1678–1691. doi:10.1158/1541-7786.MCR-17-0287
Sarkar, S., Popov, V. L., O'Connell, M. R., Stevenson, H. L., Lee, B. S., Obeid, R. A., et al. (2017b). A novel antibody against cancer stem cell biomarker, DCLK1-S, is potentially useful for assessing colon cancer risk after screening colonoscopy. Lab. Invest. 97 (10), 1245–1261. doi:10.1038/labinvest.2017.40
Sato, K., Uehara, T., Nakajima, T., Iwaya, M., Miyagawa, Y., Watanabe, T., et al. (2021). Inverse correlation between PD-L1 expression and LGR5 expression in tumor budding of stage II/III colorectal cancer. Ann. Diagn. Pathol. 52, 151739. doi:10.1016/j.anndiagpath.2021.151739
Schenk, G. J., Werkman, T., Wadman, W., Veldhuisen, B., Dijkmans, T. F., Blaas, E., et al. (2010). Over-expression of the DCLK gene transcript CARP decreases CA3/CA1 network excitability. Brain Res. 1352, 21–34. doi:10.1016/j.brainres.2010.07.068
Screaton, G. R., Bell, M. V., Bell, J. I., and Jackson, D. G. (1993). The identification of a new alternative exon with highly restricted tissue expression in transcripts encoding the mouse Pgp-1 (CD44) homing receptor. Comparison of all 10 variable exons between mouse, human, and rat. J. Biol. Chem. 268 (17), 12235–12238. doi:10.1016/S0021-9258(18)31376-0
Sehgal, K., Portell, A., Ivanova, E. V., Lizotte, P. H., Mahadevan, N. R., Greene, J. R., et al. (2021). Dynamic single-cell RNA sequencing identifies immunotherapy persister cells following PD-1 blockade. J. Clin. Invest. 131 (2), e135038. doi:10.1172/jci135038
Shi, Z., Hayes, G., and Ruvkun, G. (2013). Dual regulation of the lin-14 target mRNA by the lin-4 miRNA. PloS one 8 (9), e75475. doi:10.1371/journal.pone.0075475
Shibata, M., and Shen, M. M. (2015). Stem cells in genetically-engineered mouse models of prostate cancer. Endocr. Relat. Cancer 22 (6), T199–T208. doi:10.1530/ERC-15-0367
Shiozawa, Y., Nie, B., Pienta, K. J., Morgan, T. M., and Taichman, R. S. (2013). Cancer stem cells and their role in metastasis. Pharmacol. Ther. 138 (2), 285–293. doi:10.1016/j.pharmthera.2013.01.014
Shojaei Baghini, S., Gardanova, Z. R., Abadi, S. A. H., Zaman, B. A., İlhan, A., Shomali, N., et al. (2022). CRISPR/Cas9 application in cancer therapy: A pioneering genome editing tool. Cell. Mol. Biol. Lett. 27 (1), 35. doi:10.1186/s11658-022-00336-6
Silva, B. S., Silva, L. R., Lima, K. L., Dos Santos, A. C., Oliveira, A. C., Dezzen-Gomide, A. C., et al. (2020). SOX2 and BCL-2 expressions in odontogenic keratocyst and ameloblastoma. Med. Oral Patol. Oral Cir. Bucal 25 (2), e283–e290. doi:10.4317/medoral.23348
Singh, J. K., Farnie, G., Bundred, N. J., Simões, B. M., Shergill, A., Landberg, G., et al. (2013). Targeting CXCR1/2 significantly reduces breast cancer stem cell activity and increases the efficacy of inhibiting HER2 via HER2-dependent and -independent mechanisms. Clin. Cancer Res. 19 (3), 643–656. doi:10.1158/1078-0432.Ccr-12-1063
Singh, P., O'Connell, M., and Shubhashish, S. (2016). Epigenetic regulation of human DCLK-1 gene during colon-carcinogenesis: Clinical and mechanistic implications. Stem Cell Investig. 3, 51. doi:10.21037/sci.2016.09.07
Skoda, A. M., Simovic, D., Karin, V., Kardum, V., Vranic, S., and Serman, L. (2018). The role of the hedgehog signaling pathway in cancer: A comprehensive review. Bosn. J. Basic Med. Sci. 18 (1), 8–20. doi:10.17305/bjbms.2018.2756
Sneha, S., Nagare, R. P., Sidhanth, C., Krishnapriya, S., Garg, M., Ramachandran, B., et al. (2020). The hedgehog pathway regulates cancer stem cells in serous adenocarcinoma of the ovary. Cell. Oncol. 43 (4), 601–616. doi:10.1007/s13402-020-00504-w
Steinbichler, T. B., Savic, D., Dudas, J., Kvitsaridze, I., Skvortsov, S., Riechelmann, H., et al. (2020). Cancer stem cells and their unique role in metastatic spread. Semin. Cancer Biol. 60, 148–156. doi:10.1016/j.semcancer.2019.09.007
Su, Y. J., Lin, W. H., Chang, Y. W., Wei, K. C., Liang, C. L., Chen, S. C., et al. (2015). Polarized cell migration induces cancer type-specific CD133/integrin/Src/Akt/GSK3β/β-catenin signaling required for maintenance of cancer stem cell properties. Oncotarget 6 (35), 38029–38045. doi:10.18632/oncotarget.5703
Subramaniam, D., Kaushik, G., Dandawate, P., and Anant, S. (2018). Targeting cancer stem cells for chemoprevention of pancreatic cancer. Curr. Med. Chem. 25 (22), 2585–2594. doi:10.2174/0929867324666170127095832
Subramaniam, D., Ponnurangam, S., Ramalingam, S., Kwatra, D., Dandawate, P., Weir, S. J., et al. (2021). Honokiol affects stem cell viability by suppressing oncogenic YAP1 function to inhibit colon tumorigenesis. Cells 10 (7), 1607. doi:10.3390/cells10071607
Suehiro, Y., Takemoto, Y., Nishimoto, A., Ueno, K., Shirasawa, B., Tanaka, T., et al. (2018). Dclk1 inhibition cancels 5-FU-induced cell-cycle arrest and decreases cell survival in colorectal cancer. Anticancer Res. 38 (11), 6225–6230. doi:10.21873/anticanres.12977
Sung, H., Ferlay, J., Siegel, R. L., Laversanne, M., Soerjomataram, I., Jemal, A., et al. (2021). Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Ca. Cancer J. Clin. 71 (3), 209–249. doi:10.3322/caac.21660
Sunkara, R. R., Sarate, R. M., Setia, P., Shah, S., Gupta, S., Chaturvedi, P., et al. (2020). SFRP1 in skin tumor initiation and cancer stem cell regulation with potential implications in epithelial cancers. Stem Cell Rep. 14 (2), 271–284. doi:10.1016/j.stemcr.2019.12.006
Sureban, S. M., Berahovich, R., Zhou, H., Xu, S., Wu, L., Ding, K., et al. (2019). DCLK1 monoclonal antibody-based CAR-T cells as a novel treatment strategy against human colorectal cancers. Cancers (Basel) 12 (1), E54. doi:10.3390/cancers12010054
Sureban, S. M., Madhoun, M. F., May, R., Qu, D., Ali, N., Fazili, J., et al. (2015). Plasma DCLK1 is a marker of hepatocellular carcinoma (HCC): Targeting DCLK1 prevents HCC tumor xenograft growth via a microRNA-dependent mechanism. Oncotarget 6 (35), 37200–37215. doi:10.18632/oncotarget.5808
Sureban, S. M., May, R., Qu, D., Weygant, N., Chandrakesan, P., Ali, N., et al. (2013). DCLK1 regulates pluripotency and angiogenic factors via microRNA-dependent mechanisms in pancreatic cancer. PLoS One 8 (9), e73940. doi:10.1371/journal.pone.0073940
Sureban, S. M., May, R., Ramalingam, S., Subramaniam, D., Natarajan, G., Anant, S., et al. (2009). Selective blockade of DCAMKL-1 results in tumor growth arrest by a Let-7a MicroRNA-dependent mechanism. Gastroenterology 137 (2), 649–642. doi:10.1053/j.gastro.2009.05.004
Sureban, S. M., May, R., Weygant, N., Qu, D., Chandrakesan, P., Bannerman-Menson, E., et al. (2014). XMD8-92 inhibits pancreatic tumor xenograft growth via a DCLK1-dependent mechanism. Cancer Lett. 351 (1), 151–161. doi:10.1016/j.canlet.2014.05.011
Suwannakul, N., Ma, N., Midorikawa, K., Oikawa, S., Kobayashi, H., He, F., et al. (2020). CD44v9 induces stem cell-like phenotypes in human cholangiocarcinoma. Front. Cell Dev. Biol. 8, 417. doi:10.3389/fcell.2020.00417
Tabu, K., Kimura, T., Sasai, K., Wang, L., Bizen, N., Nishihara, H., et al. (2010). Analysis of an alternative human CD133 promoter reveals the implication of Ras/ERK pathway in tumor stem-like hallmarks. Mol. Cancer 9, 39. doi:10.1186/1476-4598-9-39
Taha, Z., Janse van Rensburg, H. J., and Yang, X. (2018). The hippo pathway: Immunity and cancer. Cancers (Basel) 10 (4), E94. doi:10.3390/cancers10040094
Tang, Q., Chen, J., Di, Z., Yuan, W., Zhou, Z., Liu, Z., et al. (2020). TM4SF1 promotes EMT and cancer stemness via the Wnt/β-catenin/SOX2 pathway in colorectal cancer. J. Exp. Clin. Cancer Res. 39 (1), 232. doi:10.1186/s13046-020-01690-z
Tao, H., Tanaka, T., and Okabe, K. (2017). Doublecortin and CaM kinase-like-1 expression in pathological stage I non-small cell lung cancer. J. Cancer Res. Clin. Oncol. 143 (8), 1449–1459. doi:10.1007/s00432-017-2405-7
Testa, U., Pelosi, E., and Castelli, G. (2018). Colorectal cancer: Genetic abnormalities, tumor progression, tumor heterogeneity, clonal evolution and tumor-initiating cells. Med. Sci. 6 (2), E31. doi:10.3390/medsci6020031
Tian, H., Biehs, B., Warming, S., Leong, K. G., Rangell, L., Klein, O. D., et al. (2011). A reserve stem cell population in small intestine renders Lgr5-positive cells dispensable. Nature 478 (7368), 255–259. doi:10.1038/nature10408
Toledo-Guzmán, M. E., Hernández, M. I., Gómez-Gallegos Á, A., and Ortiz-Sánchez, E. (2019). ALDH as a stem cell marker in solid tumors. Curr. Stem Cell Res. Ther. 14 (5), 375–388. doi:10.2174/1574888x13666180810120012
Tomita, H., Tanaka, K., Tanaka, T., and Hara, A. (2016). Aldehyde dehydrogenase 1A1 in stem cells and cancer. Oncotarget 7 (10), 11018–11032. doi:10.18632/oncotarget.6920
Tsunedomi, R., Yoshimura, K., Suzuki, N., Hazama, S., and Nagano, H. (2020). Clinical implications of cancer stem cells in digestive cancers: Acquisition of stemness and prognostic impact. Surg. Today 50 (12), 1560–1577. doi:10.1007/s00595-020-01968-x
Upadhyay, G., Yin, Y., Yuan, H., Li, X., Derynck, R., and Glazer, R. I. (2011). Stem cell antigen-1 enhances tumorigenicity by disruption of growth differentiation factor-10 (GDF10)-dependent TGF-beta signaling. Proc. Natl. Acad. Sci. U. S. A. 108 (19), 7820–7825. doi:10.1073/pnas.1103441108
van Es, J. H., Kirkpatrick, C., van de Wetering, M., Molenaar, M., Miles, A., et al. (1999). Identification of APC2, a homologue of the adenomatous polyposis coli tumour suppressor. Curr. Biol. 9 (2), 105–108. doi:10.1016/S0960-9822(99)80024-4
Vassalli, G. (2019). Aldehyde dehydrogenases: Not just markers, but functional regulators of stem cells. Stem Cells Int. 2019, 3904645. doi:10.1155/2019/3904645
Vreugdenhil, E., Engels, B., Middelburg, R., van Koningsbruggen, S., Knol, J., Veldhuisen, B., et al. (2001). Multiple transcripts generated by the DCAMKL gene are expressed in the rat hippocampus. Brain Res. Mol. Brain Res. 94 (1-2), 67–74. doi:10.1016/s0169-328x(01)00213-3
Wan, M. F., Yang, N., Qu, N. Y., Pan, Y. Y., Shan, Y. Q., and Li, P. (2020). MiR-424 suppressed viability and invasion by targeting to the DCLK1 in neuroblastoma. Eur. Rev. Med. Pharmacol. Sci. 24 (10), 5526–5533. doi:10.26355/eurrev_202005_21338
Wang, D., Lu, P., Zhang, H., Luo, M., Zhang, X., Wei, X., et al. (2014). Oct-4 and Nanog promote the epithelial-mesenchymal transition of breast cancer stem cells and are associated with poor prognosis in breast cancer patients. Oncotarget 5 (21), 10803–10815. doi:10.18632/oncotarget.2506
Wang, H., Dai, Y. Y., Zhang, W. Q., Hsu, P. C., Yang, Y. L., Wang, Y. C., et al. (2017a). DCLK1 is correlated with MET and ERK5 expression, and associated with prognosis in malignant pleural mesothelioma. Int. J. Oncol. 51 (1), 91–103. doi:10.3892/ijo.2017.4021
Wang, H., Li, X., Li, T., Wang, L., Wu, X., Liu, J., et al. (2019a). Multiple roles of microRNA-146a in immune responses and hepatocellular carcinoma. Oncol. Lett. 18 (5), 5033–5042. doi:10.3892/ol.2019.10862
Wang, I. H., Huang, T.-T., Chen, J.-L., Chu, L.-W., Ping, Y.-H., Hsu, K.-W., et al. (2020a). Mevalonate pathway enzyme HMGCS1 contributes to gastric cancer progression. Cancers 12 (5), 1088. doi:10.3390/cancers12051088
Wang, J., Wang, S., Zhou, J., and Qian, Q. (2018a). miR-424-5p regulates cell proliferation, migration and invasion by targeting doublecortin-like kinase 1 in basal-like breast cancer. Biomed. Pharmacother. 102, 147–152. doi:10.1016/j.biopha.2018.03.018
Wang, J., Yokoyama, Y., Hirose, H., Shimomura, Y., Bonkobara, S., Itakura, H., et al. (2022). Functional assessment of miR-1291 in colon cancer cells. Int. J. Oncol. 60 (2), 13. doi:10.3892/ijo.2022.5303
Wang, K., Lu, H., Qu, H., Xie, Q., Sun, T., Gan, O., et al. (2019b). miR-492 promotes cancer progression by targeting GJB4 and is a novel biomarker for bladder cancer. Onco. Targets. Ther. 12, 11453–11464. doi:10.2147/OTT.S223448
Wang, P., Lan, C., Xiong, S., Zhao, X., Shan, Y., Hu, R., et al. (2017b). HIF1α regulates single differentiated glioma cell dedifferentiation to stem-like cell phenotypes with high tumorigenic potential under hypoxia. Oncotarget 8 (17), 28074–28092. doi:10.18632/oncotarget.15888
Wang, R., Li, Y., Tsung, A., Huang, H., Du, Q., Yang, M., et al. (2018b). iNOS promotes CD24(+)CD133(+) liver cancer stem cell phenotype through a TACE/ADAM17-dependent Notch signaling pathway. Proc. Natl. Acad. Sci. U. S. A. 115 (43), E10127–E10136. doi:10.1073/pnas.1722100115
Wang, S., Zhou, L., Ling, L., Meng, X., Chu, F., Zhang, S., et al. (2020b). The crosstalk between hippo-YAP pathway and innate immunity. Front. Immunol. 11, 323. doi:10.3389/fimmu.2020.00323
Wang, W., Zhang, H., Wang, L., Zhang, S., and Tang, M. (2016). miR-613 inhibits the growth and invasiveness of human hepatocellular carcinoma via targeting DCLK1. Biochem. Biophys. Res. Commun. 473 (4), 987–992. doi:10.1016/j.bbrc.2016.04.003
Wang, X., Yang, Y., and Huycke, M. M. (2017c). Commensal-infected macrophages induce dedifferentiation and reprogramming of epithelial cells during colorectal carcinogenesis. Oncotarget 8 (60), 102176–102190. doi:10.18632/oncotarget.22250
Wang, Y. L., Li, Y., Ma, Y. G., and Wu, W. Y. (2019c). DCLK1 promotes malignant progression of breast cancer by regulating Wnt/β-Catenin signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 23 (21), 9489–9498. doi:10.26355/eurrev_201911_19443
Wang, Z., Zhao, K., Hackert, T., and Zoller, M. (2018c). CD44/CD44v6 a reliable companion in cancer-initiating cell maintenance and tumor progression. Front. Cell Dev. Biol. 6, 97. doi:10.3389/fcell.2018.00097
Wang, Z., and Zöller, M. (2019). Exosomes, metastases, and the miracle of cancer stem cell markers. Cancer Metastasis Rev. 38 (1-2), 259–295. doi:10.1007/s10555-019-09793-6
Wani, J. A., Majid, S., Khan, A., Arafah, A., Ahmad, A., Jan, B. L., et al. (2021). Clinico-pathological importance of miR-146a in lung cancer. Diagn. (Basel) 11 (2), 274. doi:10.3390/diagnostics11020274
Welm, B. E., Tepera, S. B., Venezia, T., Graubert, T. A., Rosen, J. M., and Goodell, M. A. (2002). Sca-1(pos) cells in the mouse mammary gland represent an enriched progenitor cell population. Dev. Biol. 245 (1), 42–56. doi:10.1006/dbio.2002.0625
Westphalen, C. B., Asfaha, S., Hayakawa, Y., Takemoto, Y., Lukin, D. J., Nuber, A. H., et al. (2014). Long-lived intestinal tuft cells serve as colon cancer-initiating cells. J. Clin. Invest. 124 (3), 1283–1295. doi:10.1172/JCI73434
Westphalen, C. B., Quante, M., and Wang, T. C. (2017). Functional implication of Dclk1 and Dclk1-expressing cells in cancer. Small GTPases 8 (3), 164–171. doi:10.1080/21541248.2016.1208792
Westphalen, C. B., Takemoto, Y., Tanaka, T., Macchini, M., Jiang, Z., Renz, B. W., et al. (2016). Dclk1 defines quiescent pancreatic progenitors that promote injury-induced regeneration and tumorigenesis. Cell Stem Cell 18 (4), 441–455. doi:10.1016/j.stem.2016.03.016
Weygant, N., Ge, Y., Qu, D., Kaddis, J. S., Berry, W. L., May, R., et al. (2016). Survival of patients with gastrointestinal cancers can Be predicted by a surrogate microRNA signature for cancer stem-like cells marked by DCLK1 kinase. Cancer Res. 76 (14), 4090–4099. doi:10.1158/0008-5472.CAN-16-0029
Weygant, N., Qu, D., Berry, W. L., May, R., Chandrakesan, P., Owen, D. B., et al. (2014). Small molecule kinase inhibitor LRRK2-IN-1 demonstrates potent activity against colorectal and pancreatic cancer through inhibition of doublecortin-like kinase 1. Mol. Cancer 13, 103. doi:10.1186/1476-4598-13-103
Weygant, N., Qu, D., May, R., Tierney, R. M., Berry, W. L., Zhao, L., et al. (2015). DCLK1 is a broadly dysregulated target against epithelial-mesenchymal transition, focal adhesion, and stemness in clear cell renal carcinoma. Oncotarget 6 (4), 2193–2205. doi:10.18632/oncotarget.3059
Wilkes, G. M. (2018). Targeted therapy: Attacking cancer with molecular and immunological targeted agents. Asia. Pac. J. Oncol. Nurs. 5 (2), 137–155. doi:10.4103/apjon.apjon_79_17
Wlodarczyk, A., Grot, D., Stoczynska-Fidelus, E., and Rieske, P. (2020). Gaps and doubts in search to recognize glioblastoma cellular origin and tumor initiating cells. J. Oncol. 2020, 6783627. doi:10.1155/2020/6783627
Won, C., Kim, B. H., Yi, E. H., Choi, K. J., Kim, E. K., Jeong, J. M., et al. (2015). Signal transducer and activator of transcription 3-mediated CD133 up-regulation contributes to promotion of hepatocellular carcinoma. Hepatology 62 (4), 1160–1173. doi:10.1002/hep.27968
Wu, M. M., Zhang, Z., Tong, C. W. S., Yan, V. W., Cho, W. C. S., and To, K. K. W. (2020a). Repurposing of niclosamide as a STAT3 inhibitor to enhance the anticancer effect of chemotherapeutic drugs in treating colorectal cancer. Life Sci. 262, 118522. doi:10.1016/j.lfs.2020.118522
Wu, X., Qu, D., Weygant, N., Peng, J., and Houchen, C. W. (2020b). Cancer stem cell marker DCLK1 correlates with tumorigenic immune infiltrates in the colon and gastric adenocarcinoma microenvironments. Cancers (Basel) 12 (2), E274. doi:10.3390/cancers12020274
Wu, X., Ruan, Y., Jiang, H., and Xu, C. (2017). MicroRNA-424 inhibits cell migration, invasion, and epithelial mesenchymal transition by downregulating doublecortin-like kinase 1 in ovarian clear cell carcinoma. Int. J. Biochem. Cell Biol. 85, 66–74. doi:10.1016/j.biocel.2017.01.020
Wuebben, E. L., and Rizzino, A. (2017). The dark side of SOX2: Cancer - a comprehensive overview. Oncotarget 8 (27), 44917–44943. doi:10.18632/oncotarget.16570
Xie, D., Cui, J., Xia, T., Jia, Z., Wang, L., Wei, W., et al. (2015). Hippo transducer TAZ promotes epithelial mesenchymal transition and supports pancreatic cancer progression. Oncotarget 6 (34), 35949–35963. doi:10.18632/oncotarget.5772
Xie, H., Li, J., Ying, Y., Yan, H., Jin, K., Ma, X., et al. (2020). METTL3/YTHDF2 m(6) A axis promotes tumorigenesis by degrading SETD7 and KLF4 mRNAs in bladder cancer. J. Cell. Mol. Med. 24 (7), 4092–4104. doi:10.1111/jcmm.15063
Xu, J., Li, N., Deng, W., and Luo, S. (2020). Long noncoding RNA FER1L4 suppresses proliferation, invasion, migration and lymphatic metastasis of gastric cancer cells through inhibiting the Hippo-YAP signaling pathway. Am. J. Transl. Res. 12 (9), 5481–5495.
Xu, L., Lin, W., Wen, L., and Li, G. (2019). Lgr5 in cancer biology: Functional identification of Lgr5 in cancer progression and potential opportunities for novel therapy. Stem Cell Res. Ther. 10 (1), 219. doi:10.1186/s13287-019-1288-8
Yamaga, Y., Fukuda, A., Nakanishi, Y., Goto, N., Matsumoto, Y., Yoshioka, T., et al. (2018). Gene expression profile of Dclk1(+) cells in intestinal tumors. Dig. Liver Dis. 50 (12), 1353–1361. doi:10.1016/j.dld.2018.06.011
Yamazaki, M., Kato, A., Oki, E., Zaitsu, Y., Kato, C., Nakano, K., et al. (2021). Continuous formation of small clusters with LGR5-positive cells contributes to tumor growth in a colorectal cancer xenograft model. Lab. Invest. 101 (1), 12–25. doi:10.1038/s41374-020-0471-y
Yan, R., Li, J., Zhou, Y., Yao, L., Sun, R., Xu, Y., et al. (2020a). Inhibition of DCLK1 down-regulates PD-L1 expression through Hippo pathway in human pancreatic cancer. Life Sci. 241, 117150. doi:10.1016/j.lfs.2019.117150
Yan, X., Han, D., Chen, Z., Han, C., Dong, W., Han, L., et al. (2020b). RUNX2 interacts with BRG1 to target CD44 for promoting invasion and migration of colorectal cancer cells. Cancer Cell Int. 20, 505. doi:10.1186/s12935-020-01544-w
Yang, L., Shi, P., Zhao, G., Xu, J., Peng, W., Zhang, J., et al. (2020). Targeting cancer stem cell pathways for cancer therapy. Signal Transduct. Target. Ther. 5 (1), 8. doi:10.1038/s41392-020-0110-5
Yang, W.-Q., Zhao, W.-J., Zhu, L.-L., Xu, S.-J., Zhang, X.-L., Liang, Y., et al. (2021). XMD-17-51 inhibits DCLK1 kinase and prevents lung cancer progression. Front. Pharmacol. 12 (125), 603453. doi:10.3389/fphar.2021.603453
Yang, X., Li, X., and Zhou, B. (2018). A meta-analysis of miR-499 rs3746444 polymorphism for cancer risk of different systems: Evidence from 65 case-control studies. Front. Physiol. 9, 737. doi:10.3389/fphys.2018.00737
Ye, S., Hu, X., Ni, C., Jin, W., Xu, Y., Chang, L., et al. (2020). KLF4 p.A472D mutation contributes to acquired resistance to Cetuximab in colorectal cancer. Mol. Cancer Ther. 19 (3), 956–965. doi:10.1158/1535-7163.Mct-18-1385
Yi, J., Bergstrom, K., Fu, J., Shan, X., McDaniel, J. M., McGee, S., et al. (2019). Dclk1 in tuft cells promotes inflammation-driven epithelial restitution and mitigates chronic colitis. Cell Death Differ. 26 (9), 1656–1669. doi:10.1038/s41418-018-0237-x
Yin, J., Zhang, H., Wu, X., Zhang, Y., Li, J., Shen, J., et al. (2020). CD44 inhibition attenuates EGFR signaling and enhances cisplatin sensitivity in human EGFR wild-type non-small-cell lung cancer cells. Int. J. Mol. Med. 45 (6), 1783–1792. doi:10.3892/ijmm.2020.4562
Yoo, J., Jeon, Y. H., Cho, H. Y., Lee, S. W., Kim, G. W., Lee, D. H., et al. (2020). Advances in histone demethylase KDM3A as a cancer therapeutic target. Cancers (Basel) 12 (5), E1098. doi:10.3390/cancers12051098
Yoon, C., Lu, J., Yi, B. C., Chang, K. K., Simon, M. C., Ryeom, S., et al. (2021). PI3K/Akt pathway and Nanog maintain cancer stem cells in sarcomas. Oncogenesis 10 (1), 12. doi:10.1038/s41389-020-00300-z
Yuan, X., Wu, H., Xu, H., Xiong, H., Chu, Q., Yu, S., et al. (2015). Notch signaling: An emerging therapeutic target for cancer treatment. Cancer Lett. 369 (1), 20–27. doi:10.1016/j.canlet.2015.07.048
Yue, Y., Xue, Q., Yang, J., Li, X., Mi, Z., Zhao, G., et al. (2020). Wnt-activated olfactory ensheathing cells stimulate neural stem cell proliferation and neuronal differentiation. Brain Res. 1735, 146726. doi:10.1016/j.brainres.2020.146726
Zhan, Y., Chen, Z., He, S., Gong, Y., He, A., Li, Y., et al. (2020). Long non-coding RNA SOX2OT promotes the stemness phenotype of bladder cancer cells by modulating SOX2. Mol. Cancer 19 (1), 25. doi:10.1186/s12943-020-1143-7
Zhang, F., Liu, B., Deng, Q., Sheng, D., Xu, J., He, X., et al. (2021a). UCP1 regulates ALDH-positive breast cancer stem cells through releasing the suppression of Snail on FBP1. Cell Biol. Toxicol. 37 (2), 277–291. doi:10.1007/s10565-020-09533-5
Zhang, W., Sui, Y., Ni, J., and Yang, T. (2016). Insights into the Nanog gene: A propeller for stemness in primitive stem cells. Int. J. Biol. Sci. 12 (11), 1372–1381. doi:10.7150/ijbs.16349
Zhang, Y., Liu, Y., Liu, H., and Tang, W. H. (2019). Exosomes: Biogenesis, biologic function and clinical potential. Cell Biosci. 9 (1), 19. doi:10.1186/s13578-019-0282-2
Zhang, Y., Xie, X., Yeganeh, P. N., Lee, D. J., Valle-Garcia, D., Meza-Sosa, K. F., et al. (2021b). Immunotherapy for breast cancer using EpCAM aptamer tumor-targeted gene knockdown. Proc. Natl. Acad. Sci. U. S. A. 118 (9), e2022830118. doi:10.1073/pnas.2022830118
Zhang, Z., Han, H., Rong, Y., Zhu, K., Zhu, Z., Tang, Z., et al. (2018). Hypoxia potentiates gemcitabine-induced stemness in pancreatic cancer cells through AKT/Notch1 signaling. J. Exp. Clin. Cancer Res. 37 (1), 291. doi:10.1186/s13046-018-0972-3
Zhao, Y., Tao, L., Yi, J., Song, H., and Chen, L. (2018). The role of canonical Wnt signaling in regulating radioresistance. Cell. Physiol. biochem. 48 (2), 419–432. doi:10.1159/000491774
Zheng, S., Wang, X., Weng, Y. H., Jin, X., Ji, J. L., Guo, L., et al. (2018). siRNA knockdown of RRM2 effectively suppressed pancreatic tumor growth alone or synergistically with Doxorubicin. Mol. Ther. Nucleic Acids 12, 805–816. doi:10.1016/j.omtn.2018.08.003
Zhong, L., Li, Y., Xiong, L., Wang, W., Wu, M., Yuan, T., et al. (2021a). Small molecules in targeted cancer therapy: Advances, challenges, and future perspectives. Signal Transduct. Target. Ther. 6 (1), 201. doi:10.1038/s41392-021-00572-w
Zhong, Z. A., Michalski, M. N., Stevens, P. D., Sall, E. A., and Williams, B. O. (2021b). Regulation of Wnt receptor activity: Implications for therapeutic development in colon cancer. J. Biol. Chem. 296, 100782. doi:10.1016/j.jbc.2021.100782
Keywords: DCLK1, cancer stem cells, intestinal neoplasia, siRNA, miRNA, CRISPR/Cas9
Citation: Chhetri D, Vengadassalapathy S, Venkadassalapathy S, Balachandran V, Umapathy VR, Veeraraghavan VP, Jayaraman S, Patil S, Iyaswamy A, Palaniyandi K and Gnanasampanthapandian D (2022) Pleiotropic effects of DCLK1 in cancer and cancer stem cells. Front. Mol. Biosci. 9:965730. doi: 10.3389/fmolb.2022.965730
Received: 10 June 2022; Accepted: 12 August 2022;
Published: 26 September 2022.
Edited by:
Huanhuan Joyce Chen, The University of Chicago, United StatesReviewed by:
Kui Zhang, The University of Chicago, United StatesCopyright © 2022 Chhetri, Vengadassalapathy, Venkadassalapathy, Balachandran, Umapathy, Veeraraghavan, Jayaraman, Patil, Iyaswamy, Palaniyandi and Gnanasampanthapandian. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kanagaraj Palaniyandi, bXV0aGFtbWFsLmthbmFnYXJhakBnbWFpbC5jb20=; Dhanavathy Gnanasampanthapandian, ZGhhbmF2YXRoeS4yMDA2QGdtYWlsLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.