- Research Center for Micro-Nano Technology, Hosei University, Tokyo, Japan
A non-canonical DNA/RNA structure, G-quadruplex (G4), is a unique structure formed by two or more guanine quartets, which associate through Hoogsteen hydrogen bonding leading to form a square planar arrangement. A set of RNA-binding proteins specifically recognize G4 structures and play certain unique physiological roles. These G4-binding proteins form ribonucleoprotein (RNP) through a physicochemical phenomenon called liquid-liquid phase separation (LLPS). G4-containing RNP granules are identified in both prokaryotes and eukaryotes, but extensive studies have been performed in eukaryotes. We have been involved in analyses of the roles of G4-containing RNAs recognized by two G4-RNA-binding proteins, TDP-43 and FUS, which both are the amyotrophic lateral sclerosis (ALS) causative gene products. These RNA-binding proteins play the essential roles in both G4 recognition and LLPS, but they also carry the risk of agglutination. The biological significance of G4-binding proteins is controlled through unique 3D structure of G4, of which the risk of conformational stability is influenced by environmental conditions such as monovalent metals and guanine oxidation.
Introduction
A non-canonical DNA/RNA structure, G-quadruplex (G4) is formed by two or more guanine quartets, which associate through Hoogsteen base pairs leading to form a square planar arrangement stabilized by a central cation (Rodes and Lipps, 2015). The G4 structure first discovered in vitro was found to be telomere-formed structures, and later abundant G4-foming sequences have been identified in the genome (Sen and Gilbert, 1988, Sundquist and Klug, 1989; Rodes and Lipps, 2015). Such G4-forming sequences are present in both prokaryotes and eukaryotes. For instance, a total of 52 G4 motifs are present in the E. coli genome while more than 700,000 G4 motifs are present in the human genome (Métifiot et al., 2014; Doluca et al., 2016; Kaplan et al., 2016; Marsico et al., 2019; Ruggiero and Richter, 2020; Tu et al., 2021). Some of these G4 motifs are involved in DNA replication, recombination, telomere regulation and transcriptional regulation (Figure 1) (Métifiot et al., 2014; Yang, 2019; Prorok et al., 2019; Cammas and Millevoi, 2017). Some G4 motifs are copied into RNAs, thereby leading to the regulation of RNA functions such as processing, stability, translation and transport (Figure 1) (Bugaut and Balasubramanian, 2012; Cammas and Millevoi, 2017; Yang, 2019; Shao et al., 2020; Dumas et al., 2021; Georgakopoulos-Soares et al., 2022). G4-RNAs are often assembled into unique ribonucleoprotein (RNP) granules without boundary membranes, which were identified as precipitats in the brain tissue and cultured cell extracts using biotinylated isoxazole (Subramanian et al., 2011; Han et al., 2012). The membraneless RNP organelles formed through liquid-liquid phase separation (LLPS) are classified into stress granule, germ granule, P-bodie, neuronal granule, nuclear puncta, RNA-transport granule, RNA foci, and paraspeckle (Rhine et al., 2020). It has been suggested that the G4 structure supports the formation of condensates (Rhine et al., 2020; Roden and Gladfelter, 2021), which is the opposite to the inhibition of condensation by promiscuous RNA assembly (Maharana et al., 2018). Eventually, in 2021, four laboratories experimentally demonstrated that G4 promotes the condensation of DNA/RNA-protein complexes through LLPS (DNA: Liu et al., 2021; Gao et al., 2021; DNA: Mimura et al., 2021; RNA: Ishiguro et al., 2021).
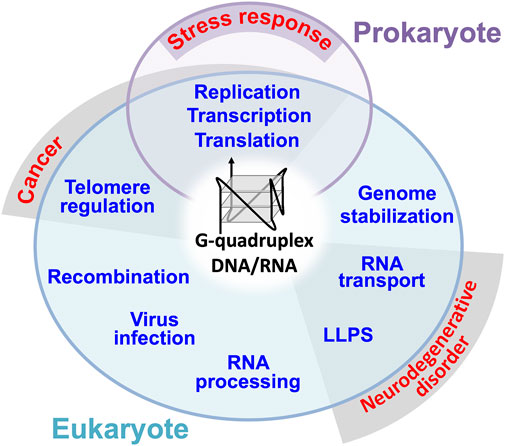
FIGURE 1. General roles of DNA/RNA G4s in prokaryote and eukaryote. G4 has also been suggested to be involved in prokaryotic stress responses through replication, transcription, and translation. Dysregulation of telomere regulation, transcription, translation and replication by G4 are suggested to be involved in the pathogenic mechanism of human cancer. Mutations in G4 containing RNAs and G4 binding proteins involved in RNA transport and LLPS have been found in patients with neurodegenerative diseases such as ALS.
G4s and G4-binding proteins in eukaryotes are presumed to exhibit the stress response and pathologically related functions including cancer and neurodegenerative disorder (see Figure 1). Therefore, G4s and G4-binding proteins could be either the potential protection targets or pathogenic drivers in the pathogenic cascade of various diseases including neurological disorders (Simone et al., 2015; Wang et al., 2021). The amyotrophic lateral sclerosis (ALS), a neurodegenerative disease, is a multifunctional disorder involving both genetic and non-genetic risk factors (Taylor et al., 2016; Deng et al., 2017; Scarlino et al., 2020). Genes encoding many RNA-binding proteins have been identified for genetic risks, and recent studies suggest their involvement in G4 and RNP granules as a common function. In this review article, we summarize the roles and risks of G4-contaning RNAs and two G4-RNA binding proteins, TDP-43 (43 kDa TAR DNA-binding protein) and FUS (fused in sarcoma).
Dedicated Methods of G4-RNA
Research on molecular mechanisms of G4s and G4 binding proteins is still in its infancy stage. In fact, some RNA binding proteins have recently been re-identified one after another as G4-RNA binding proteins (Byrd et al., 2016; Ishiguro et al., 2016; Ramesh et al., 2020; Simko et al., 2020; Zheng et al., 2020; He et al., 2021; Ishiguro et al., 2021). Attempts to identify bound proteins with known G4s have been performed by mass spectrometry and proteome microarray (Mori et al., 2013; Zhang et al., 2021). On the other hand, however, attempts to identify binding target RNAs from the protein side are often difficult because of technical problems. Thus, the correct choice of analytical methods for identification of G4-RNAs could be the driving force for discovering unidentified G4-RNA regulatory systems.
Identification of non-canonical G4 motifs is different from that of canonical RNA sequences (Guo and Bartel, 2016; Yang et al., 2020). For example, cross-linking and immunoprecipitation (CLIP), one of the most used methods for identifying RNAs as protein binding targets from whole cellular RNA, often result in selective loss of most non-canonical G4 motifs. Unlike Watson-Click pairing, the high stability of G4 structure due to extensive hydrogen-bonding and base-stacking interactions interfere with reverse transcription and PCR in the process (Guo and Bartel, 2016; Stevens et al., 2017). Therefore, several methods have been developed such as rG4-seq (Guo and Bartel, 2016; Kwok et al., 2016; Yang et al., 220) and ADO (allelic drop) (Stevens et al., 2017). Due to incomplete identification of in the high-throughput screening, it is necessary to confirm the prediction by biochemical analysis of direct interactions between the proteins and the target RNA candidates using such as the gel shift assay. However, the sharply bent DNA/RNA-protein complexes do not migrate normally in native acrylamide gel electrophoresis and G4-protein complex often retained in the top of gel, if the complex is correctly formed (Carey et al., 2010; Al-Furoukh et al., 2013; Ariyo et al., 2015; Huang et al., 2017; Mou et al., 2022). Furthermore, it is difficult to increase the concentration of monocations in the gel during electrophoretic fractionation, which is necessary for the stabilization of G4 tetrads. thus often leading to incorrect identification (Yakhnin et al., 2012; Kim et al., 2015). To avoid these problems, use in combination with other methods such as the filter-binding assay, thermophoresis assay or surface plasmon resonance analysis is effective to determine the specific interaction between G4-RNA and G4-RNA binding protein (von Hacht et al., 2014; Ishiguro et al., 2016; Mou et al., 2022).
G4-RNAs and G4-DNAs in Prokaryote
G4-forming sequence in bacteria genome has often been identified in the promoter region (Dey et al., 2021; Yadav et al., 2021), suggesting the involvement of G4 in transcription regulation (see Figure 1). In fact, the expression of some stress response genes such as radioprotection-related genes are under the control of G4-containing promoters (Beaume et al., 2013; Kumari and Raghavan, 2021). Roles for G4s have also been posited in the antigenic variation systems of bacteria (Harris and Merrick, 2015). Along this line, G4 could be used for the development of antimicrobial target (Yadav et al., 2021). G4 is also suggested in the maintenance of bacterial genome. For instance, E. coli Rep helicase and RecA recombinase unwind the G4 DNA, thus stabilizing toleration of toxicity induced by G4-stabilizing ligands (Paul et al., 2020; Xue et al., 2020). As in the case of eukaryote, E. coli RNA polymerase forms clusters, together with transcription anti-terminator NusA, LLPS-mediated biomolecular condensates, (Ladouceur et al., 2020). However, the biological relevance of G4 DNA/RNA in prokaryotes has only begun to emerge even though their very important and conserved biological functions.
G4-RNAs and G4-Binding RNA Proteins as the Genetic Causes of Amyotrophic Lateral Sclerosis
As to the G4-RNAs and G4-RNA binding proteins in eukaryote, the well-analyzed systems are those involved in ALS, the neurodegenerative disorder characterized by the progressive death of upper and lower motor neurons (Taylor et al., 2016) (see Figure 1). Approximately 90% cases of ALS are called sporadic, and a history of ALS has been identified for only about 10% of familial cases (Renton et al., 2014; Scarlino et al., 2020). The peak age of ALS onset is 58–63 years for sporadic, and 47–52 years for familial (Kiernan et al., 2011). More than 50 ALS-associated genes have been identified, of which gain or loss of the gene function led to the risk (Mejzini et al., 2019). These gene products are involved in various cellular processes such as redox regulation, signal transduction, and axonal transport of RNAs. The most abundant gene species encode RNAs and RNA-binding proteins, such as intronic expansion of the human C9orf72 (chromosome 9 open reading frame 72) gene, TDP-43, FUS, hnRNPA1 (heterogeneous nuclear ribonucleoprotein A1), hnRNP A2/B1, hnRNPA3, EWSR1 (Ewing’s sarcoma RNA binding protein 1), and TIA1 (T cell-restricted intracellular antigen-1) (Taylor et al., 2016; Ishiguro et al., 2021). Surprisingly, these RNA binding-proteins recognize and bind to mRNA containing G4 motifs (Ishiguro et al., 2021). For instance, ALS-linked intronic expansion of the human C9orf72 gene and potentially causative two long noncoding RNAs, NEAT1 (nuclear enriched abundant transcript 1) and MALAT1 (metastasis associated in lung adenocarcinoma transcript 1/NEAT2) contain abundant G4 motifs (Grigg et al., 2014; Haeusler et al., 2014; Simko et al., 2020; Mou et al., 2022). More interestingly, these ALS-related proteins/RNAs constitute RNP granules or regulate the formation of RNP granules (Tourrière et al., 2003; Fujii et al., 2005; Guil et al., 2006; Bosco et al., 2010; Molliex et al., 2015; Mensch et al., 2018; Ishiguro et al., 2021). RNP granules assembled by liquid-liquid phase separation (LLPS) are in turn involved in the regulation of the functions of many RNAs (Freibaum et al., 2021). RNP granules can also control the long-distance transport of mRNA, local translation, and mRNA silencing under stress conditions (Freibaum et al., 2021). Dysregulation of the system might contribute to the development and progression of ALS (Rotem et al., 2017; Gamarra et al., 2021; Milicevic et al., 2022).
A Structural Feature Common to G4 Recognition and Liquid-Liquid Phase Separation
The G4-binding proteins often carry one or more intrinsically disordered regions (IDRs) (Lin et al., 2015; Molliex et al., 2015; Wheeler et al., 2016; Uversky, 2017; Martin and Holehouse, 2020). In generally, the interfaces of DNA/RNA-binding proteins are enriched in positively charged and aromatic residues, lacking negatively charged and Pro residues (Zhang et al., 2019; Bartas et al., 2021). These G4 RNA-binding proteins all have low complexity IDRs (Bartas et al., 2021; Dettori et al., 2021), which contain two to five different amino acids making up at least 50%, critically enriched in Glu, Ser, Lys, Pro, Gly, Ala, and Arg, but lacking aromatic residues (Dettori et al., 2021).
Intrinsically disordered proteins drive protein clustering and subsequent LLPS using their IDRs (Darling et al., 2018; Jo et al., 2021). These G4-RNA binding proteins form condensates mediated through LLPS. The LLPS formation can potentially be mediated by disorder-to-order transitions of dynamic IDR-RNA complexes. In addition, cation-π and π-π interactions have been suggested to regulate the phase behavior of constituent proteins in driving LLPS (Vernon et al., 2018; Paloni et al., 2020). G4 composed of guanine tetrads stacked by π-π interactions may provide more π-interactions for condensate formation (Raguseo et al., 2020; Ishiguro et al., 2021).
Diversity of G4-RNA Binding Proteins: TDP-43 and FUS
The best characterized ALS-linked G4-RNA binding proteins, TDP-43 and FUS, share structural and functional similarities, including participation in ALS pathogenesis (Lagier-Tourenne and Cleveland, 2009; da Cruz and Cleveland, 2011; Strong and Volkening, 2011; Guerrero et al., 2016; Ratti and Buratti, 2016; Ishiguro et al., 2021). RNP granules formed by TDP-43 and FUS play roles in axonal transport (Fujii et al., 2005; Subramanian et al., 2011; Alami et al., 2014; Ishiguro et al., 2016; Fay et al., 2017). Although both proteins recognize G4 structures, their binding specificity is different, thereby leading to different mode of action (Ishiguro et al., 2021). Three typical G4 topologies are known: parallel, anti-parallel and hybrid, of which most G4-RNAs carry the adopt parallel formation. TDP-43 binds only to the parallel-stranded G4 DNA/RNA (Figure 2A). The selective G4 recognition of TDP-43 to parallel stranded G4 is probably due to the dimerization of TDP-43, which has two RNA recognition motifs (RRMs) and a C-terminal Gly-rich IDR. The RRM fragment, and the full-length TDP-43 that does not form a homo dimer, exhibit binding to wide range of UG-rich sequences (Buratti and Baralle, 2001; Lukavsky et al., 2013; Rengifo-Gonzalez et al., 2021). Under physiological conditions, full-length TDP-43 forms a homodimer, which loses the binding priority to the UG-rich sequence, suggesting the presence of a complex binding mechanisms using multiple RNA-binding modules (Ishiguro et al., 2016; Ishiguro et al., 2021). In contrast, FUS shows binding to all three types of G4 and shows binding to G-rich hairpin structure (Loughlin et al., 2019; Ishiguro et al., 2021) (Figure 2A). We also found that TDP-43 induces stable G4 conformation of the target RNA, whereas FUS destabilizes G4 conformation into an open-structure with close association between 3′ and 5′ termini (Ishiguro et al., 2021) (Figure 2B).
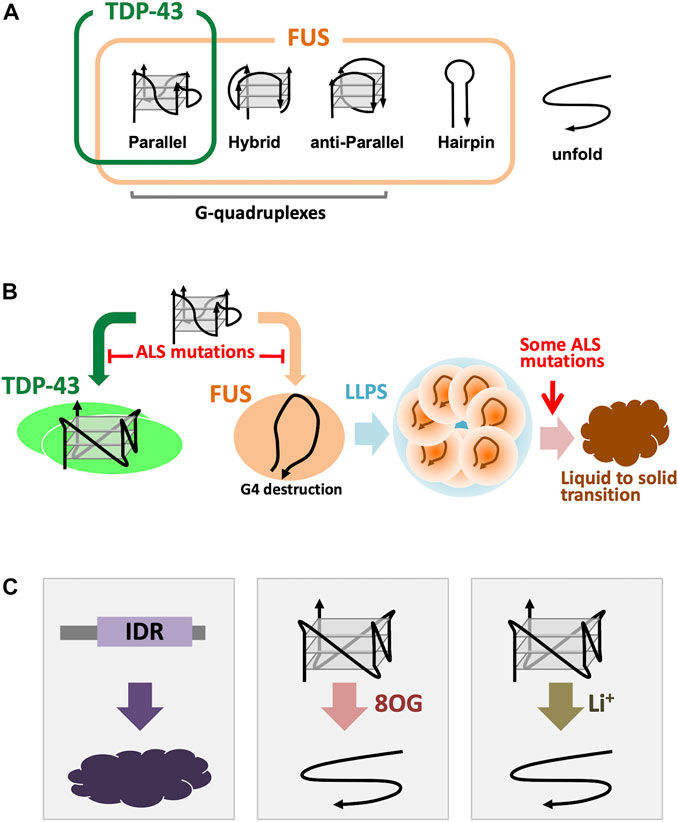
FIGURE 2. ALS-linked G4 binding proteins. (A), Binding specificity of TDP-43 and FUS. TDP-43 can bind only for the parallel-stranded G4-DNA/RNAs, however, FUS binds to parallel, hybrid, and even G-rich hairpin forms (Ishiguro et al., 2021). (B), Different G4 binding modes for TDP-43 and FUS. TDP-43 protects and stabilizes the G4 structure, while FUS destabilizes the G4 conformation and recreates it into a “terminal associated” configuration. In both cases, the G4-protein interaction was inhibited by introducing amino acid substitutions from ALS patients (Ishiguro et al., 2021). In some FUS mutations have been found that promotion of the phase transition of liquid to solid phase of the droplet formed by LLPS (Ishiguro et al., 2021). (C), Concerned risks for G4s and G4 binding proteins. Low-complexity IDRs have considered to be tended to be aggregated (van der Lee et al., 2014; Taylor et al., 2016). 8OG produced by oxidation of guanine interferes with the normal formation of G4. Li+ is the same monocation as K+ and Na+, but it is known that it may not contribute to the structural stabilization of G4.
The effects of amino acid substitution mutations from ALS patients are common to these two proteins, and TDP-43 and FUS mutant proteins show reduced interaction with G4. (Ishiguro et al., 2021; Ishiguro et al., 2021) (Figure 2B). ALS-linked FUS mutations induced altered formation of the G4-dependent droplets (Ishiguro et al., 2021) (Figure 2B). In addition, the phase transition from droplets to aggregates is also altered by FUS mutations (Figure 2B). These mutations are considered to result in the loss of function, but the possibility of a gain of function is yet not excluded.
The Essential Risks for G4-RNAs and G4-RNA Binding Proteins
The assembly of RNP granules by G4-RNA and G4-RNA binding proteins control many biological processes, including mRNA transport from soma to distal neurites for local translation. However, there are several risks associated with the molecular properties of G4-RNA and G4-RNA binding proteins that lead to perturbation of the interplay between G4-RNA and G4-RNA binding protein including the pathogenesis of ALS. Several factors are shown below (Figure 2C):
Protein Aggregation
ALS is characterized by the accumulation of protein aggregates in motor neurons (Blokhuis et al., 2013; Malik and Wiedau, 2020). However, it remains unsolved whether the aggregated proteins themselves are toxic due to gain of function or accompanied by loss of normal function. The presence of protein aggregation is, however, not a sufficient condition for the development and progression of ALS (Baloh, 2011; Blokhuis et al., 2013). G4 recognition and phase separation is mediated by low-complexity IDRs that are present in ALS-linked RNA binding proteins. The contribution of low-complexity IDR to those processes, however, carries the risk because IDRs intend to aggregate (van der Lee et al., 2014; Taylor et al., 2016).
Guanine Oxidation
Oxidative stress is a risk factor of neurodegeneration and also a therapeutic target (Barber et al., 2006). Mutations of copper zinc superoxide dismutase 1 (SOD1) gene were found in 10%–20% of familial ALS cases (Rosen et al., 1993). Guanine has the lowest oxidation potential than other nucleobases and is readily oxidized to 8-oxoguanine (8OG), with elevated level of 8OG confirmed in the cervical cord of ALS patients (Chang et al., 2008). Cellular RNA is more susceptible target for guanine oxidation than DNA, and the oxidative damage to RNA could be a major focus in the investigation of neurodegenerative diseases (Hofer et al., 2005; Castellani et al., 2008). Since G4 is composed of many guanine bases, it should be sensitive to guanine oxidation. In fact, the insertion of 8OG into G4 motif causes structural destruction (Cheong et al., 2015).
Lithium-Ion
G-quartets are stabilized in the presence of cations, but different cations result in different structural properties (Wang et al., 2014). Li+ is known as a factor that interferes with the uptake of Na+ and K+, which stabilizes the G-quadruplex structure in vitro. Although Na+ and K+ are the physiologically relevant monovalent ions in the context of G4 structures, however, Li+ with smaller ionic radius than Na+ and K+ causes destabilization of G4 conformation (Balaratnam and Basu, 2015; Bhattacharyya et al., 2016). Although induction of ALS due to extensive uptake of Li+ has not been observed, administration of lithium to ALS patients resulted in almost 70% of adverse events, including death (Chiò et al., 2010).
Conclusion and Outlook
G4-associated regulations are involved in varieties of biological systems, including replication, recombination, telomere regulation, transcriptional regulation, RNA processing, RNA stability, translation, RNA transport, and stress responses. Although many of them have long been overlooked without experimental confirmation, but the advent of a new age arrived to reveal molecular details underlying the interaction of G4-RNAs and G4-binding proteins. The dysregulation of G4-RNAs and G4-binding proteins has been identified as neurodegenerative diseases in human. Risk factors in G4 and G4 binding proteins are involved in the pathogenic mechanism of ALS (for instance, Ishiguro et al., 2016; Ishiguro et al., 2020; Ishiguro et al., 2021). This review gives an overview on the interaction of G4-RNA with binding proteins and discussed dysregulation due to their structural properties, focusing the ALS-linked proteins and their interplay with G4-conteining RNAs. Unknown features affecting the risks along the interplay between G4-containing RNAs/DNAs and G4-binding proteins will be more clearly embodied in further investigation in this area.
As an extension of this line research, we have started to identify the role of G4 in the most well-characterized prokaryote E. coli. It may provide a platform for analyzing the formation of complex structures in G4.
Author Contributions
AI (1st author) and AI (2nd author) wrote the manuscript, elaborated the figures and approved the review for publication.
Funding
This work was supported by Japan Society for the Promotion of Science (JSPS) Grant-in-Aid for Scientific Research (17K07291 and 22K07032 to AI (1st author).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher
References
Al-Furoukh, N., Goffart, S., Szibor, M., Wanrooij, S., and Braun, T. (2013). Binding to G-Quadruplex RNA Activates the Mitochondrial GTPase NOA1. Biochimica Biophysica Acta (BBA) - Mol. Cell. Res. 1833 (12), 2933–2942. doi:10.1016/j.bbamcr.2013.07.022
Alami, N. H., Smith, R. B., Carrasco, M. A., Williams, L. A., Winborn, C. S., Han, S. S. W., et al. (2014). Axonal Transport of TDP-43 mRNA Granules Is Impaired by ALS-Causing Mutations. Neuron 81 (3), 536–543. doi:10.1016/j.neuron.2013.12.018
Ariyo, E. O., Booy, E. P., Patel, T. R., Dzananovic, E., McRae, E. K., Meier, M., et al. (2015). Biophysical Characterization of G-Quadruplex Recognition in the PITX1 mRNA by the Specificity Domain of the Helicase RHAU. PloS one 10 (12), e0144510. doi:10.1371/journal.pone.0144510
Balaratnam, S., and Basu, S. (2015). Divalent Cation-Aided Identification of Physico-Chemical Properties of Metal Ions that Stabilize RNA G-Quadruplexes. Biopolymers 103 (7), 376–386. doi:10.1002/bip.22628
Baloh, R. H. (2011). TDP-43: the Relationship between Protein Aggregation and Neurodegeneration in Amyotrophic Lateral Sclerosis and Frontotemporal Lobar Degeneration. FEBS J. 278 (19), 3539–3549. doi:10.1111/j.1742-4658.2011.08256.x
Barber, S. C., Mead, R. J., and Shaw, P. J. (2006). Oxidative Stress in ALS: a Mechanism of Neurodegeneration and a Therapeutic Target. Biochimica Biophysica Acta (BBA) - Mol. Basis Dis. 1762 (11-12), 1051–1067. doi:10.1016/j.bbadis.2006.03.008
Bartas, M., Červeň, J., Guziurová, S., Slychko, K., and Pečinka, P. (2021). Amino Acid Composition in Various Types of Nucleic Acid-Binding Proteins. Ijms 22 (2), 922. doi:10.3390/ijms22020922
Beaume, N., Pathak, R., Yadav, V. K., Kota, S., Misra, H. S., Gautam, H. K., et al. (2013). Genome-wide Study Predicts Promoter-G4 DNA Motifs Regulate Selective Functions in Bacteria: Radioresistance of D. Radiodurans Involves G4 DNA-Mediated Regulation. Nucleic Acids Res. 41, 76–89. doi:10.1093/nar/gks1071
Bhattacharyya, D., Mirihana Arachchilage, G., and Basu, S. (2016). Metal Cations in G-Quadruplex Folding and Stability. Front. Chem. 4, 38. doi:10.3389/fchem.2016.00038
Blokhuis, A. M., Groen, E. J. N., Koppers, M., van den Berg, L. H., and Pasterkamp, R. J. (2013). Protein Aggregation in Amyotrophic Lateral Sclerosis. Acta Neuropathol. 125 (6), 777–794. doi:10.1007/s00401-013-1125-6
Bosco, D. A., Lemay, N., Ko, H. K., Zhou, H., Burke, C., Kwiatkowski, T. J., et al. (2010). Mutant FUS Proteins that Cause Amyotrophic Lateral Sclerosis Incorporate into Stress Granules. Hum. Mol. Gene. 19 (21), 4160–4175. doi:10.1093/hmg/ddq335
Bugaut, A., and Balasubramanian, S. (2012). 5'-UTR RNA G-Quadruplexes: Translation Regulation and Targeting. Nucleic Acids Res. 40 (11), 4727–4741. doi:10.1093/nar/gks068
Buratti, E., and Baralle, F. E. (2001). Characterization and Functional Implications of the RNA Binding Properties of Nuclear Factor TDP-43, a Novel Splicing Regulator ofCFTR Exon 9. J. Biol. Chem. 276 (39), 36337–36343. doi:10.1074/jbc.M104236200
Byrd, A. K., Zybailov, B. L., Maddukuri, L., Gao, J., Marecki, J. C., Jaiswal, M., et al. (2016). Evidence that G-Quadruplex DNA Accumulates in the Cytoplasm and Participates in Stress Granule Assembly in Response to Oxidative Stress. J. Biol. Chem. 291 (34), 18041–18057. doi:10.1074/jbc.M116.718478
Cammas, A., and Millevoi, S. (2017). RNA G-Quadruplexes: Emerging Mechanisms in Disease. Nucleic Acids Res. 45 (4), gkw1280–1595. doi:10.1093/nar/gkw1280
Carey, M. F., Peterson, C. L., and Smale, S. T. (2010). Magnesium-agarose Electrophoretic Mobility Shift Assay (EMSA) of Transcription Factor IID Binding to DNA. Cold Spring Harb. Protoc. 2010, pdb.prot5514. doi:10.1101/pdb.prot5514
Castellani, R., Nunomura, A., Rolston, R., Moreira, P., Takeda, A., Perry, G., et al. (2008). Sublethal RNA Oxidation as a Mechanism for Neurodegenerative Disease. Ijms 9 (5), 789–806. doi:10.3390/ijms9050789
Chang, Y., Kong, Q., Shan, X., Tian, G., Ilieva, H., Cleveland, D. W., et al. (2008). Messenger RNA Oxidation Occurs Early in Disease Pathogenesis and Promotes Motor Neuron Degeneration in ALS. PloS One 3 (8), e2849. doi:10.1371/journal.pone.0002849
Cheong, V. V., Heddi, B., Lech, C. J., and Phan, A. T. (2015). Xanthine and 8-oxoguanine in G-Quadruplexes: Formation of a G·G·X·O Tetrad. Nucleic Acids Res. 43 (21), gkv826–10514. doi:10.1093/nar/gkv826
Chiò, A., Borghero, G., Calvo, A., Capasso, M., Caponnetto, C., Corbo, M., et al. (2010). Lithium Carbonate in Amyotrophic Lateral Sclerosis: Lack of Efficacy in a Dose-Finding Trial. Neurology 75 (7), 619–625. doi:10.1212/WNL.0b013e3181ed9e7c
da Cruz, S., and Cleveland, D. W. (2011). Understanding the Role of TDP-43 and FUS/TLS in ALS and beyond. Curr. Opin. Neurobiol. 21 (6), 904–919. doi:10.1016/j.conb.2011.05.029
Darling, A. L., Liu, Y., Oldfield, C. J., and Uversky, V. N. (2018). Intrinsically Disordered Proteome of Human Membrane‐Less Organelles. Proteomics 18 (5-6), 1700193. doi:10.1002/pmic.201700193
Deng, Z., Sheehan, P., Chen, S., and Yue, Z. (2017). Is Amyotrophic Lateral Sclerosis/frontotemporal Dementia an Autophagy Disease? Mol. Neurodegener. 12 (1), 90. doi:10.1186/s13024-017-0232-6
Dettori, L. G., Torrejon, D., Chakraborty, A., Dutta, A., Mohamed, M., Papp, C., et al. (2021). A Tale of Loops and Tails: The Role of Intrinsically Disordered Protein Regions in R-Loop Recognition and Phase Separation. Front. Mol. Biosci. 8, 691694. doi:10.3389/fmolb.2021.691694
Dey, U., Sarkar, S., Teronpi, V., Yella, V. R., and Kumar, A. (2021). G-Quadruplex Motifs Are Functionally Conserved in Cis-Regulatory Regions of Pathogenic Bacteria: An In-Silico Evaluation. Biochimie 184, 40–51. doi:10.1016/j.biochi.2021.01.017
Doluca, O., Kaplan, O. I., Berber, B., and Hekim, N. (2016). G-Quadruplex Prediction in E.Coli Genome Reveals a Conserved Putative G-Quadruplex-Hairpin-Duplex Switch. bioRxiv, 032615. doi:10.1101/032615
Dumas, L., Herviou, P., Dassi, E., Cammas, A., and Millevoi, S. (2021). G-Quadruplexes in RNA Biology: Recent Advances and Future Directions. Trends Biochem. Sci. 46 (4), 270–283. doi:10.1016/j.tibs.2020.11.001
Fay, M. M., Lyons, S. M., and Ivanov, P. (2017). RNA G-Quadruplexes in Biology: Principles and Molecular Mechanisms. J. Mol. Biol. 429 (14), 2127–2147. doi:10.1016/j.jmb.2017.05.017
Freibaum, B. D., Messing, J., Yang, P., Kim, H. J., and Taylor, J. P. (2021). High-fidelity Reconstitution of Stress Granules and Nucleoli in Mammalian Cellular Lysate. J. Cell. Biol. 220 (3), e202009079. doi:10.1083/jcb.202009079
Fujii, R., Okabe, S., Urushido, T., Inoue, K., Yoshimura, A., Tachibana, T., et al. (2005). The RNA Binding Protein TLS Is Translocated to Dendritic Spines by mGluR5 Activation and Regulates Spine Morphology. Curr. Biol. 15 (6), 587–593. doi:10.1016/j.cub.2005.01.058
Gamarra, M., de la Cruz, A., Blanco-Urrejola, M., and Baleriola, J. (2021). Local Translation in Nervous System Pathologies. Front. Integr. Neurosci. 15, 689208. doi:10.3389/fnint.2021.689208
Gao, J., Gao, Z., Putnam, A. A., Byrd, A. K., Venus, S. L., Marecki, J. C., et al. (2021). G-Quadruplex DNA Inhibits Unwinding Activity but Promotes Liquid-Liquid Phase Separation by the DEAD-Box Helicase Ded1p. Chem. Commun. 57 (60), 7445–7448. doi:10.1039/d1cc01479j
Georgakopoulos-Soares, I., Parada, G. E., Wong, H. Y., Miska, E. A., Kwok, C. K., and Hemberg, M. (2022). Alternative Splicing Modulation by G-Quadruplexes. Nat. Commun. 13 (1), 2404. bioRxiv 700575. doi:10.1101/700575
Grigg, J. C., Shumayrikh, N., and Sen, D. (2014). G-Quadruplex Structures Formed by Expanded Hexanucleotide Repeat RNA and DNA from the Neurodegenerative Disease-Linked C9orf72 Gene Efficiently Sequester and Activate Heme. PloS one 9 (9), e106449. doi:10.1371/journal.pone.0106449
Guerrero, E. N., Wang, H., Mitra, J., Hegde, P. M., Stowell, S. E., Liachko, N. F., et al. (2016). TDP-43/FUS in Motor Neuron Disease: Complexity and Challenges. Prog. Neurobiol. 145-146, 78–97. doi:10.1016/j.pneurobio.2016.09.004
Guil, S., Long, J. C., and Cáceres, J. F. (2006). hnRNP A1 Relocalization to the Stress Granules Reflects a Role in the Stress Response. Mol. Cell. Biol. 26 (15), 5744–5758. doi:10.1128/MCB.00224-06
Guo, J. U., and Bartel, D. P. (2016). RNA G-Quadruplexes Are Globally Unfolded in Eukaryotic Cells and Depleted in Bacteria. Science 353 (6306), aaf5371. doi:10.1126/science.aaf5371
Haeusler, A. R., Donnelly, C. J., Periz, G., Simko, E. A. J., Shaw, P. G., Kim, M.-S., et al. (2014). C9orf72 Nucleotide Repeat Structures Initiate Molecular Cascades of Disease. Nature 507, 195–200. doi:10.1038/nature13124
Han, T. W., Kato, M., Xie, S., Wu, L. C., Mirzaei, H., Pei, J., et al. (2012). Cell-free Formation of RNA Granules: Bound RNAs Identify Features and Components of Cellular Assemblies. Cell. 149 (4), 768–779. doi:10.1016/j.cell.2012.04.016
Harris, L. M., and Merrick, C. J. (2015). G-Quadruplexes in Pathogens: a Common Route to Virulence Control? PLoS Pathog. 11, e1004562. doi:10.1371/JOURNAL.PPAT.1004562
He, X., Yuan, J., and Wang, Y. (2021). G3BP1 Binds to Guanine Quadruplexes in mRNAs to Modulate Their Stabilities. Nucleic Acids Res. 49 (19), 11323–11336. doi:10.1093/nar/gkab873
Hofer, T., Badouard, C., Bajak, E., Ravanat, J.-L., Mattsson, Å., and Cotgreave, I. A. (2005). Hydrogen Peroxide Causes Greater Oxidation in Cellular RNA Than in DNA. Biol. Chem. 386 (4), 333–337. doi:10.1515/BC.2005.040
Huang, H., Zhang, J., Harvey, S. E., Hu, X., and Cheng, C. (2017). RNA G-Quadruplex Secondary Structure Promotes Alternative Splicing via the RNA-Binding Protein hnRNPF. Genes. Dev. 31 (22), 2296–2309. doi:10.1101/gad.305862.117
Ishiguro, A., Kimura, N., Noma, T., Shimo‐Kon, R., Ishihama, A., and Kon, T. (2020). Molecular Dissection of ALS‐linked TDP‐43 - Involvement of the Gly‐rich Domain in Interaction with G‐quadruplex mRNA. FEBS Lett. 594, 2254–2265. doi:10.1002/1873-3468.13800
Ishiguro, A., Kimura, N., Watanabe, Y., Watanabe, S., and Ishihama, A. (2016). TDP-43 Binds and Transports G-Quadruplex-Containing mRNAs into Neurites for Local Translation. Genes. cells. 21, 466–481. doi:10.1111/gtc.12352
Ishiguro, A., Lu, J., Ozawa, D., Nagai, Y., and Ishihama, A. (2021). ALS-Linked FUS Mutations Dysregulate G-quadruplex-dependent Liquid-Liquid Phase Separation and Liquid-To-Solid Transition. J. Biol. Chem. 297 (5), 101284. ISSN 0021-9258. doi:10.1016/j.jbc.2021.101284
Jo, Y., Jang, J., Song, D., Park, H., and Jung, Y. (2021). Determinants for Intrinsically Disordered Protein Recruitment into Phase-Separated Protein Condensates. Chem. Sci. 13 (2), 522–530. doi:10.1039/d1sc05672g
Kaplan, O. I., Berber, B., Hekim, N., and Doluca, O. (2016). G-Quadruplex Prediction in E. coli Genome Reveals a Conserved Putative G-Quadruplex-Hairpin-Duplex switchE. coli Genome Reveals a Conserved Putative G-Quadruplex-Hairpin-Duplex Switch. Nucleic Acids Res. 44 (19), 9083–9095. doi:10.1093/nar/gkw769
Kiernan, M. C., Vucic, S., Cheah, B. C., Turner, M. R., Eisen, A., Hardiman, O., et al. (2011). Amyotrophic Lateral Sclerosis. Lancet 377 (9769), 942–955. doi:10.1016/S0140-6736(10)61156-7
Kim, B. G., Evans, H. M., Dubins, D. N., and Chalikian, T. V. (2015). Effects of Salt on the Stability of a G-Quadruplex from the Human C-MYC Promoter. Biochemistry 54 (22), 3420–3430. doi:10.1021/acs.biochem.5b00097
Kumari, N., and Raghavan, S. C. (2021). G-Quadruplex DNA Structures and Their Relevance in Radioprotection. Biochimica Biophysica Acta (BBA) - General Subj. 1865 (5), 129857. doi:10.1016/j.bbagen.2021.129857
Kwok, C. K., Marsico, G., Sahakyan, A. B., Chambers, V. S., and Balasubramanian, S. (2016). rG4-seq Reveals Widespread Formation of G-Quadruplex Structures in the Human Transcriptome. Nat. Methods 13, 841–844. doi:10.1038/nmeth.3965
Ladouceur, A.-M., Parmar, B. S., Biedzinski, S., Wall, J., Tope, S. G., Cohn, D., et al. (2020). Clusters of Bacterial RNA Polymerase Are Biomolecular Condensates that Assemble through Liquid-Liquid Phase Separation. Proc. Natl. Acad. Sci. U.S.A. 117 (31), 18540–18549. doi:10.1073/pnas.2005019117
Lagier-Tourenne, C., and Cleveland, D. W. (2009). Rethinking ALS: the FUS about TDP-43. Cell. 136 (6), 1001–1004. doi:10.1016/j.cell.2009.03.006
Lin, Y., Protter, D. S. W., Rosen, M. K., and Parker, R. (2015). Formation and Maturation of Phase-Separated Liquid Droplets by RNA-Binding Proteins. Mol. Cell. 60 (2), 208–219. doi:10.1016/j.molcel.2015.08.018
Liu, X., Xiong, Y., Zhang, C., Lai, R., Liu, H., Peng, R., et al. (2021). G-Quadruplex-Induced Liquid-Liquid Phase Separation in Biomimetic Protocells. J. Am. Chem. Soc. 143 (29), 11036–11043. doi:10.1021/jacs.1c03627
Loughlin, F. E., Lukavsky, P. J., Kazeeva, T., Reber, S., Hock, E.-M., Colombo, M., et al. (2019). The Solution Structure of FUS Bound to RNA Reveals a Bipartite Mode of RNA Recognition with Both Sequence and Shape Specificity. Mol. Cell. 73 (3), 490–504. doi:10.1016/j.molcel.2018.11.012
Lukavsky, P. J., Daujotyte, D., Tollervey, J. R., Ule, J., Stuani, C., Buratti, E., et al. (2013). Molecular Basis of UG-Rich RNA Recognition by the Human Splicing Factor TDP-43. Nat. Struct. Mol. Biol. 20 (12), 1443–1449. doi:10.1038/nsmb.2698
Maharana, S., Wang, J., Papadopoulos, D. K., Richter, D., Pozniakovsky, A., Poser, I., et al. (2018). RNA Buffers the Phase Separation Behavior of Prion-like RNA Binding Proteins. Science 360 (6391), 918–921. doi:10.1126/science.aar7366
Malik, R., and Wiedau, M. (2020). Therapeutic Approaches Targeting Protein Aggregation in Amyotrophic Lateral Sclerosis. Front. Mol. Neurosci. 13, 98. doi:10.3389/fnmol.2020.00098
Marsico, G., Chambers, V. S., Sahakyan, A. B., McCauley, P., Boutell, J. M., Antonio, M. D., et al. (2019). Whole Genome Experimental Maps of DNA G-Quadruplexes in Multiple Species. Nucleic Acids Res. 47 (8), 3862–3874. doi:10.1093/nar/gkz179
Martin, E. W., and Holehouse, A. S. (2020). Intrinsically Disordered Protein Regions and Phase Separation: Sequence Determinants of Assembly or Lack Thereof. Emerg. Top. Life Sci. 4 (3), 307–329. doi:10.1042/ETLS20190164
Mejzini, R., Flynn, L. L., Pitout, I. L., Fletcher, S., Wilton, S. D., and Akkari, P. A. (2019). ALS Genetics, Mechanisms, and Therapeutics: Where Are We Now? Front. Neurosci. 13, 1310. doi:10.3389/fnins.2019.01310
Mensch, A., Meinhardt, B., Bley, N., Hüttelmaier, S., Schneider, I., Stoltenburg-Didinger, G., et al. (2018). The p.S85C-Mutation in MATR3 Impairs Stress Granule Formation in Matrin-3 Myopathy. Exp. Neurol. 306, 222–231. doi:10.1016/j.expneurol.2018.05.012
Métifiot, M., Amrane, S., Litvak, S., and Andreola, M.-L. (2014). G-Quadruplexes in Viruses: Function and Potential Therapeutic Applications. Nucleic Acids Res. 42 (20), 12352–12366. doi:10.1093/nar/gku999
Milicevic, K., Rankovic, B., Andjus, P. R., Bataveljic, D., and Milovanovic, D. (2022). Emerging Roles for Phase Separation of RNA-Binding Proteins in Cellular Pathology of ALS. Front. Cell. Dev. Biol. 10, 840256. doi:10.3389/fcell.2022.840256
Mimura, M., Tomita, S., Shinkai, Y., Hosokai, T., Kumeta, H., Saio, T., et al. (2021). Quadruplex Folding Promotes the Condensation of Linker Histones and DNAs via Liquid-Liquid Phase Separation. J. Am. Chem. Soc. 143 (26), 9849–9857. doi:10.1021/jacs.1c03447
Molliex, A., Temirov, J., Lee, J., Coughlin, M., Kanagaraj, A. P., Kim, H. J., et al. (2015). Phase Separation by Low Complexity Domains Promotes Stress Granule Assembly and Drives Pathological Fibrillization. Cell. 163 (1), 123–133. doi:10.1016/j.cell.2015.09.015
Mori, K., Lammich, S., Mackenzie, I. R., Forné, I., Zilow, S., Kretzschmar, H., et al. (2013). hnRNP A3 Binds to GGGGCC Repeats and Is a Constituent of P62-positive/TDP43-Negative Inclusions in the hippocampus of Patients with C9orf72 Mutations. Acta Neuropathol. 125 (3), 413–423. doi:10.1007/s00401-013-1088-7
Mou, X., Liew, S. W., and Kwok, C. K. (2022). Identification and Targeting of G-Quadruplex Structures in MALAT1 Long Non-coding RNA. Nucleic Acids Res. 50 (1), 397–410. doi:10.1093/nar/gkab1208
Paloni, M., Bailly, R., Ciandrini, L., and Barducci, A. (2020). Unraveling Molecular Interactions in Liquid-Liquid Phase Separation of Disordered Proteins by Atomistic Simulations. J. Phys. Chem. B 124 (41), 9009–9016. doi:10.1021/acs.jpcb.0c06288
Paul, T., Voter, A. F., Cueny, R. R., Gavrilov, M., Ha, T., Keck, J. L., et al. (2020). E. coli Rep Helicase and RecA Recombinase Unwind G4 DNA and Are Important for Resistance to G4-Stabilizing Ligands. Nucleic Acids Res. 48 (12), 6640–6653. doi:10.1093/nar/gkaa442
Prorok, P., Artufel, M., Aze, A., Coulombe, P., Peiffer, I., Lacroix, L., et al. (2019). Involvement of G-Quadruplex Regions in Mammalian Replication Origin Activity. Nat. Commun. 10 (1), 3274. doi:10.1038/s41467-019-11104-0
Raguseo, F., Chowdhury, S., Minard, A., and Di Antonio, M. (2020). Chemical-biology Approaches to Probe DNA and RNA G-Quadruplex Structures in the Genome. Chem. Commun. (Camb) 56 (9), 1317–1324. doi:10.1039/c9cc09107f
Ramesh, N., Kour, S., Anderson, E. N., Rajasundaram, D., and Pandey, U. B. (2020). RNA-Recognition Motif in Matrin-3 Mediates Neurodegeneration through Interaction with hnRNPM. Acta Neuropathol. Commun. 8 (1), 138. doi:10.1186/s40478-020-01021-5
Ratti, A., and Buratti, E. (2016). Physiological Functions and Pathobiology of TDP-43 and FUS/TLS Proteins. J. Neurochem. 138 Suppl 1 (1), 95–111. doi:10.1111/jnc.13625
Rengifo-Gonzalez, J. C., El Hage, K., Clément, M. J., Steiner, E., Joshi, V., Craveur, P., et al. (2021). The Cooperative Binding of TDP-43 to GU-rich RNA Repeats Antagonizes TDP-43 Aggregation. eLife 10, e67605. doi:10.7554/eLife.67605
Renton, A. E., Chiò, A., and Traynor, B. J. (2014). State of Play in Amyotrophic Lateral Sclerosis Genetics. Nat. Neurosci. 17 (1), 17–23. doi:10.1038/nn.3584
Rhine, K., Vidaurre, V., and Myong, S. (2020). RNA Droplets. Annu. Rev. Biophys. 49, 247–265. doi:10.1146/annurev-biophys-052118-115508
Rhodes, D., and Lipps, H. J. (2015). G-Quadruplexes and Their Regulatory Roles in Biology. Nucleic Acids Res. 43 (18), 8627–8637. doi:10.1093/nar/gkv862
Roden, C., and Gladfelter, A. S. (2021). RNA Contributions to the Form and Function of Biomolecular Condensates. Nat. Rev. Mol. Cell. Biol. 22 (3), 183–195. doi:10.1038/s41580-020-0264-6
Rosen, D. R., Siddique, T., Patterson, D., Figlewicz, D. A., Sapp, P., Hentati, A., et al. (1993). Mutations in Cu/Zn Superoxide Dismutase Gene Are Associated with Familial Amyotrophic Lateral Sclerosis. Nature 362 (6415), 59–62. doi:10.1038/362059a0
Rotem, N., Magen, I., Ionescu, A., Gershoni-Emek, N., Altman, T., Costa, C. J., et al. (2017). ALS along the Axons - Expression of Coding and Noncoding RNA Differs in Axons of ALS Models. Sci. Rep. 7, 44500. doi:10.1038/srep44500
Ruggiero, E., and Richter, S. N. (2020). Viral G-Quadruplexes: New Frontiers in Virus Pathogenesis and Antiviral Therapy. Annu. Rep. Med. Chem. 54, 101–131. doi:10.1016/bs.armc.2020.04.001
Scarlino, S., Domi, T., Pozzi, L., Romano, A., Pipitone, G. B., Falzone, Y. M., et al. (2020). Burden of Rare Variants in ALS and Axonal Hereditary Neuropathy Genes Influence Survival in ALS: Insights from a Next Generation Sequencing Study of an Italian ALS Cohort. Int. J. Mol. Sci. 21 (9), 3346. doi:10.3390/ijms21093346
Sen, D., and Gilbert, W. (1988). Formation of Parallel Four-Stranded Complexes by Guanine-Rich Motifs in DNA and its Implications for Meiosis. Nature 334, 364–366. doi:10.1038/334364a0
Shao, X., Zhang, W., Umar, M. I., Wong, H. Y., Seng, Z., Xie, Y., et al. (2020). RNA G-Quadruplex Structures Mediate Gene Regulation in Bacteria. mBio 11 (1), e02926–19. doi:10.1128/mBio.02926-19
Simko, E. A. J., Liu, H., Zhang, T., Velasquez, A., Teli, S., Haeusler, A. R., et al. (2020). G-Quadruplexes Offer a Conserved Structural Motif for NONO Recruitment to NEAT1 Architectural lncRNA. Nucleic Acids Res. 48 (13), 7421–7438. doi:10.1093/nar/gkaa475
Simone, R., Fratta, P., Neidle, S., Parkinson, G. N., and Isaacs, A. M. (2015). G-Quadruplexes: Emerging Roles in Neurodegenerative Diseases and the Non-coding Transcriptome. FEBS Lett. 589 (14), 1653–1668. doi:10.1016/j.febslet.2015.05.003
Stevens, A. J., Taylor, M. G., Pearce, F. G., and Kennedy, M. A. (2017). Allelic Dropout During Polymerase Chain Reaction Due to G-Quadruplex Structures and DNA Methylation Is Widespread at Imprinted Human Loci. G3 (Bethesda) 7 (3), 1019–1025. doi:10.1534/g3.116.038687
Strong, M. J., and Volkening, K. (2011). TDP-43 and FUS/TLS: Sending a Complex Message about Messenger RNA in Amyotrophic Lateral Sclerosis? FEBS J. 278 (19), 3569–3577. doi:10.1111/j.1742-4658.2011.08277.x
Subramanian, M., Rage, F., Tabet, R., Flatter, E., Mandel, J. L., and Moine, H. (2011). G-Quadruplex RNA Structure as a Signal for Neurite mRNA Targeting. EMBO Rep. 12 (7), 697–704. doi:10.1038/embor.2011.76
Sundquist, W. I., and Klug, A. (1989). Telomeric DNA Dimerizes by Formation of Guanine Tetrads between Hairpin Loops. Nature 342, 825–829. doi:10.1038/342825a0
Taylor, J. P., Brown, R. H., and Cleveland, D. W. (2016). Decoding ALS: from Genes to Mechanism. Nature 539 (7628), 197–206. doi:10.1038/nature20413
Tourrière, H., Chebli, K., Zekri, L., Courselaud, B., Blanchard, J. M., Bertrand, E., et al. (2003). The RasGAP-Associated Endoribonuclease G3BP Assembles Stress Granules. J. Cell. Biol. 160 (6), 823–831. doi:10.1083/jcb.200212128
Tu, J., Duan, M., Liu, W., Lu, N., Zhou, Y., Sun, X., et al. (2021). Direct Genome-wide Identification of G-Quadruplex Structures by Whole-Genome Resequencing. Nat. Comm. 12 (1), 6014. doi:10.1038/s41467-021-26312-w
Uversky, V. N. (2017). Intrinsically Disordered Proteins in Overcrowded Milieu: Membrane-Less Organelles, Phase Separation, and Intrinsic Disorder. Curr. Opin. Struct. Biol. 44, 18–30. doi:10.1016/j.sbi.2016.10.015
van der Lee, R., Buljan, M., Lang, B., Weatheritt, R. J., Daughdrill, G. W., Dunker, A. K., et al. (2014). Classification of Intrinsically Disordered Regions and Proteins. Chem. Rev. 114 (13), 6589–6631. doi:10.1021/cr400525m
Vernon, R. M., Chong, P. A., Tsang, B., Kim, T. H., Bah, A., Farber, P., et al. (2018). Pi-Pi Contacts Are an Overlooked Protein Feature Relevant to Phase Separation. Elife 7, e31486. doi:10.7554/eLife.31486
von Hacht, A., Seifert, O., Menger, M., Schütze, T., Arora, A., Konthur, Z., et al. (2014). Identification and Characterization of RNA Guanine-Quadruplex Binding Proteins. Nucleic Acids Res. 42 (10), 6630–6644. doi:10.1093/nar/gku290
Wang, E., Thombre, R., Shah, Y., Latanich, R., and Wang, J. (2021). G-Quadruplexes as Pathogenic Drivers in Neurodegenerative Disorders. Nucleic Acids Res. 49 (9), 4816–4830. doi:10.1093/nar/gkab164
Wang, Z. F., Li, M. H., Hsu, S. T., and Chang, T. C. (2014). Structural Basis of Sodium-Potassium Exchange of a Human Telomeric DNA Quadruplex without Topological Conversion. Nucleic Acids Res. 42 (7), 4723–4733. doi:10.1093/nar/gku083
Wheeler, J. R., Matheny, T., Jain, S., Abrisch, R., and Parker, R. (2016). Distinct Stages in Stress Granule Assembly and Disassembly. Elife 5, e18413. doi:10.7554/eLife.18413
Xue, Z. Y., Wu, W. Q., Zhao, X. C., KumarKumar, A., Ran, X., Zhang, X. H., et al. (2020). Single-molecule Probing the Duplex and G4 Unwinding Patterns of a RecD Family Helicase. Int. J. Biol. Macromol. 164, 902–910. doi:10.1016/j.ijbiomac.2020.07.158
Yadav, P., Kim, N., Kumari, M., Verma, S., Sharma, T. K., Yadav, V., et al. (2021). G-Quadruplex Structures in Bacteria: Biological Relevance and Potential as an Antimicrobial Target. J. Bacteriol. 203, e0057720. doi:10.1128/JB.00577-20
Yakhnin, A. V., Yakhnin, H., and Babitzke, P. (2012). Gel Mobility Shift Assays to Detect Protein-RNA Interactions. Methods Mol. Biol. 905, 201–211. doi:10.1007/978-1-61779-949-5_12
Yang, D. (2019). G-Quadruplex DNA and RNA. Methods Mol. Biol. Clift. N.J.), 2035, 1–24. doi:10.1007/978-1-4939-9666-7_1
Yang, X., Cheema, J., Zhang, Y., Deng, H., Duncan, S., Umar, M. I., et al. (2020). RNA G-Quadruplex Structures Exist and Function In Vivo in Plants. Genome Biol. 21 (1), 226. doi:10.1186/s13059-020-02142-9
Zhang, J., Ma, Z., and Kurgan, L. (2019). Comprehensive Review and Empirical Analysis of Hallmarks of DNA-, RNA- and Protein-Binding Residues in Protein Chains. Brief. Bioinform 20 (4), 1250–1268. doi:10.1093/bib/bbx168
Zhang, L., Zhou, J., Xu, M., and Yuan, G. (2021). Exploration of the Hsa-miR-1587-Protein Interaction and the Inhibition to CASK. Int. J. Mol. Sci. 22 (19), 10716. doi:10.3390/ijms221910716
Keywords: G-quadruplex (G4), amyotrophic lateral sclerosis (ALS), liquid-liquid phase separation (LLPS), RNP granule, TDP-43 (43 kDa TAR DNA-binding protein), FUS (fused in sarcoma)
Citation: Ishiguro A and Ishihama A (2022) Essential Roles and Risks of G-Quadruplex Regulation: Recognition Targets of ALS-Linked TDP-43 and FUS. Front. Mol. Biosci. 9:957502. doi: 10.3389/fmolb.2022.957502
Received: 31 May 2022; Accepted: 21 June 2022;
Published: 11 July 2022.
Edited by:
Olga N. Ozoline, Institute of Cell Biophysics (RAS), RussiaReviewed by:
Jiang Zhou, Peking University, ChinaCopyright © 2022 Ishiguro and Ishihama. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Akira Ishiguro, YWtpcmEuaXNoaWd1cm8uaXVAaG9zZWkuYWMuanA=
 Akira Ishiguro
Akira Ishiguro Akira Ishihama
Akira Ishihama